Abstract
Background
Immunocompromised individuals are highly susceptible to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Whether vaccine-induced immunity in these individuals involves oral cavity, a primary site of infection, is presently unknown.
Methods
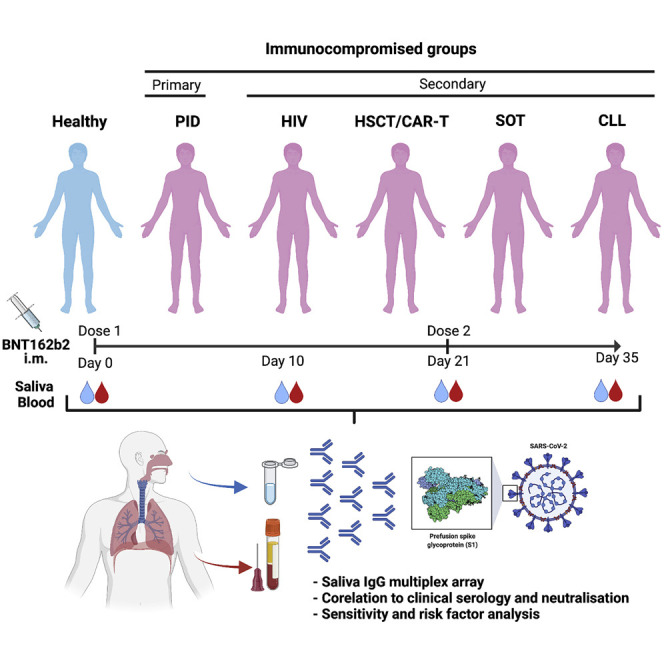
Immunocompromised patients (n = 404) and healthy controls (n = 82) participated in a prospective clinical trial (NCT04780659) encompassing two doses of the mRNA BNT162b2 vaccine. Primary immunodeficiency (PID), secondary immunodeficiencies caused by human immunodeficiency virus (HIV) infection, allogeneic hematopoietic stem cell transplantation (HSCT)/chimeric antigen receptor T cell therapy (CAR-T), solid organ transplantation (SOT), and chronic lymphocytic leukemia (CLL) patients were included. Salivary and serum immunoglobulin G (IgG) reactivities to SARS-CoV-2 spike were measured by multiplex bead-based assays and Elecsys anti-SARS-CoV-2 S assay.
Findings
IgG responses to SARS-CoV-2 spike antigens in saliva in HIV and HSCT/CAR-T groups were comparable to those of healthy controls after vaccination. The PID, SOT, and CLL patients had weaker responses, influenced mainly by disease parameters or immunosuppressants. Salivary responses correlated remarkably well with specific IgG titers and the neutralizing capacity in serum. Receiver operating characteristic curve analysis for the predictive power of salivary IgG yielded area under the curve (AUC) = 0.95 and positive predictive value (PPV) = 90.7% for the entire cohort after vaccination.
Conclusions
Saliva conveys vaccine responses induced by mRNA BNT162b2. The predictive power of salivary spike IgG makes it highly suitable for screening vulnerable groups for revaccination.
Funding
Knut and Alice Wallenberg Foundation, Erling Perssons family foundation, Region Stockholm, Swedish Research Council, Karolinska Institutet, Swedish Blood Cancer Foundation, PID patient organization of Sweden, Nordstjernan AB, Center for Medical Innovation (CIMED), Swedish Medical Research Council, and Stockholm County Council (ALF).
Keywords: COVID-19, vaccination, immunodeficiency, HIV, cancer, transplantation, saliva, serum, antibody
Graphical abstract
Context and significance
People with a weakened immune system may respond less well to vaccination and are more vulnerable to infections. This work investigates the predictive value of saliva antibodies in immunocompromised patients. We report that IgG to SARS-CoV-2 spike in saliva correlated remarkably well to that detected in blood after Pfizer mRNA vaccination. Among the immunosuppressive conditions studied, low spike-IgG responses were mainly associated with genetic immune disorders, organ transplantation, chronic lymphatic leukemia, and immunosuppressants, while people living with human immunodeficiency virus or stem cell transplant responded comparably to healthy participants. The clear correlation between anti-spike-IgG in saliva and blood extends to the neutralizing capacity in serum. In conclusion, saliva is suitable and efficient for monitoring vaccine immunity and revaccination.
Healy et al. report a clear correlation between salivary and blood IgG to SARS-CoV-2 spike in immunocompromised patients after mRNA vaccination. The specific IgG also correlates to the serum-neutralizing capacity. Their findings indicate that saliva is highly suitable for monitoring vaccine immunity in these extremely vulnerable patients.
Introduction
Vaccine development has been a success story of the coronavirus disease 2019 (COVID-19) pandemic. Among approved vaccines, the BNT162b2 vaccine (Comirnaty, Pfizer-BioNTech) relies on novel mRNA technology, where mRNA is packaged into lipid nanoparticles to deliver genetic instructions for human cells to produce the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein.1 Accumulating data from the general population in Israel and early studies in US healthcare workers confirmed that vaccination with a two-dose regimen confers 94.6% and 95% protection against symptomatic infection and severe disease, respectively, 1 to 2 weeks after the second dose.2, 3, 4 In a more recent UK study, two doses were shown to be approximately 85%–90% effective in adults aged 70 years and older.5 In contrast, data from studies in older adults receiving a single dose of BNT162b2 have yielded mixed results.6, 7, 8
Adult patients with primary immunodeficiency (PID) or secondary immunodeficiency (SID) generally display higher morbidity and mortality rates from COVID-19 than immunocompetent individuals.9, 10, 11 The overall infection fatality rates (IFR) for PID and SID have been reported to be as high as 20% (PID) and 33% (SID), compared with less than 1% in the general population.9 Around six million people worldwide are estimated to live with a PID,12 , 13 while SID disorders are frequent consequences of underlying medical conditions, e.g., human immunodeficiency virus (HIV) infection, malignant diseases, or clinical interventions with immunosuppressive drugs.14 Patients receiving immunosuppression after undergoing hematopoietic stem cell transplantation (HSCT) or specific cellular therapies (e.g., chimeric antigen receptor T cell [CAR-T] cell therapy) or having hematological malignancies often show prolonged virus shedding and transmission dynamics in which shedding of infectious SARS-CoV-2 could be prolonged up to 2 months or more due to weakened immunity.15 , 16 Notably, people with compromised immunity have been mostly excluded from large clinical trials addressing mRNA vaccine effectiveness.2 , 17 Recent published reports have, however, indicated weak or absent immune responses in several groups of immunocompromised persons.18, 19, 20, 21, 22
Mucosal immunity in the aerodigestive tract is considered a front-line defense against SARS-CoV-2 infection. The oral cavity is an important site for SARS-CoV-2 infection, and saliva is considered a potential route of virus transmission.23 Transmission can occur by activities involving the oral cavity, such as breathing, coughing, sneezing, speaking, or singing.24, 25, 26 Oral manifestations, such as taste loss, dry mouth, and oral lesions, are present in about half of confirmed COVID-19 cases.27 Viral entry factors, such as ACE-2 and TMPRSS2, TMPRSS4, and TMPRSS11D are expressed in the oral cavity (buccal mucosa, ventral tongue, and the dorsal tongue) and the oropharynx (soft palate and tonsils), including salivary glands and epithelial cells in saliva.23 It was recently shown that saliva antibodies correlate with seroconversion in mRNA-vaccinated healthcare workers,28 and that the new Omicron virus variant is detected easily in saliva of infected individuals.29 However, whether mRNA vaccines, such as the BNT162b2 vaccine, can induce mucosal immunity at distal sites, such as the oral cavity, following intramuscular injection in immunocompromised patients is presently unknown. Immunocompromised patients in this context represent a large and highly important risk group in need of continuous monitoring of vaccination efficacy.
To fill the knowledge gap in respect to COVID-19 vaccine efficacy, we recently conducted a prospective open-label clinical trial (COVAXID, EudraCT, no. 2021-000175-37) investigating the immunogenicity of the BNT162b2 vaccine in immunocompromised patients and healthy controls.30 The aim of the present study was to investigate vaccine-induced humoral immunity in the oral cavity in the same cohort.
Results
Study design and patient demographics
From the COVAXID clinical trial (539 participants; 449 patients and 90 controls), 486 participants, 404 immunocompromised and 82 healthy participants, were eligible for inclusion in the present study. Patient parameters are shown in Table S1. As presented in the accompanying flowchart (Figure S1), eligible participants had to be SARS-CoV-2 seronegative at baseline and not meet exclusion criteria, such as PCR positivity at any point of the study, missing baseline serum antibody data, or fewer than two vaccine doses. Saliva and serum samples were collected at four time points: days 0 (D0); 10 (D10); 21 (D21); and 35 (D35) from first vaccine dose. The second vaccine dose was administered at D21. A total of 1,870 saliva samples were obtained with 1,829 paired serum samples across all time points and were subjected to analysis of spike-specific antibodies.31, 32, 33 The saliva flow rate in most participants was above normal (>0.1 mL/min) at each time point measured, albeit a lower mean flow rate was seen in the PID (p = 0.0392) and chronic lymphocytic leukemia (CLL) groups (p < 0.0001; Table S1; Figure S2).
Anti-spike IgG responses in saliva are related to immunodeficiency status
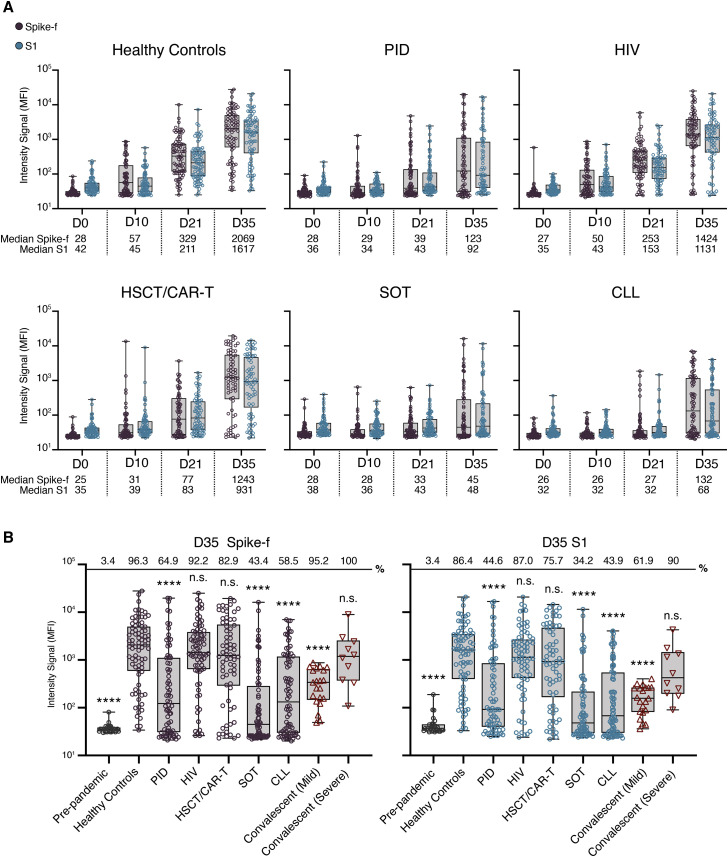
All groups showed a steady induction of anti-spike immunoglobulin G (IgG) reactivities in saliva after the first vaccine dose, where people living with HIV, hereafter referred to as “HIV,” and healthy controls exhibited the earliest and largest increase relative to baseline (D0; Figure 1A). From baseline to D21 (before second dose), the IgG reactivities to Spike-f (full-length spike, trimeric form stabilized in prefusion-conformation) increased by 12- and 9-fold in healthy controls and the HIV group, respectively, thereafter to 74- and 53-fold after the second dose (D35). In these groups, most participants (>90%) developed anti-spike IgG (both Spike-f and S1) in saliva at D35. In the HSCT/CAR-T group, a moderate 3-fold increase in salivary Spike-f IgG reactivity was observed at D21. After the second dose, it rose to 50-fold relative to baseline by D35, indicating a potent response after full vaccination. In contrast, weak responses were seen in the PID, SOT, and CLL groups in which a discrete increase (1- to 2-fold) in D21 samples was found relative to baseline. Not until D35 did a sizeable fraction of PID, SOT, and CLL patients demonstrate a detectable anti-Spike-f IgG reactivity in saliva with median values of 4- to 5-fold over baseline in PID and CLL groups and less in SOT group, albeit many patients in these three groups remained negative. It was noted that Spike-S1-specific IgG reactivities were similar as seen for the Spike-f antigen.
Figure 1.
SARS-CoV-2 spike-specific IgG responses in saliva in immunocompetent or immunocompromised individuals
(A) Levels of salivary Spike-f and S1 IgG responses on days 0, 10, 21, and 35 after first vaccine dose. Healthy controls: D0; D10; D21; and D35 (n = 82; n = 77; n = 81; and n = 81), PID: D0; D10; D21; and D35 (n = 78; n = 77; n = 78; and n = 74), HIV: D0; D10; D21; and D35 (n = 79; n = 78; n = 77; and n = 77), HSCT/CAR-T: D0; D10; D21; and D35 (n = 73; n = 74; n = 73; and n = 70), SOT: D0; D10; D21; and D35 (n = 78; n = 80; n = 80; and n = 76), and CLL: D0; D10; D21; and D35 (n = 85; n = 78; n = 82; and n = 82) are shown. MFI, median fluorescence intensity. Error bars indicate minumum and maximum for whiskers.
(B) Comparison of D35 Spike-f and S1 IgG responses in saliva of vaccinated healthy controls relative to indicated patient group or control non-vaccinated groups (pre-pandemic [n = 29] and respective mild or severe [n = 21 or n = 10] convalescent individuals). Lines, boxes, and whiskers represent the median, interquartile range (IQR), and min-max range, respectively. Percentage positive samples over technical cutoff are indicated on top. The Mann-Whitney U test was used for group comparisons against healthy controls in (B). ∗∗∗∗p < 0.0001. ns, not significant. Error bars indicate minumum and maximum for whiskers.
Saliva data collected at D35, i.e., 14 days after the second vaccine dose, from all groups were compared, using COVID-19 convalescence saliva and pre-pandemic saliva samples also as references (Figure 1B). Among immunocompromised patient groups, the strongest magnitude of anti-Spike-f and anti-S1 responses in saliva was observed in the HIV and HSCT/CAR-T groups at D35, which did not differ from the healthy controls. In contrast, the PID, SOT, and CLL patient groups all had lower SARS-CoV-2-specific responses in saliva on D35 relative to healthy controls (p < 0.001). In addition, salivary IgG to both spike antigens in the healthy controls, HIV, and HSCT/CAR-T groups was higher than convalescence saliva collected from mild COVID-19 patients (p < 0.001) and was of similar magnitude as the severe COVID-19 convalescent saliva.
SARS-CoV-2-specific IgG responses in saliva and serum strongly correlate
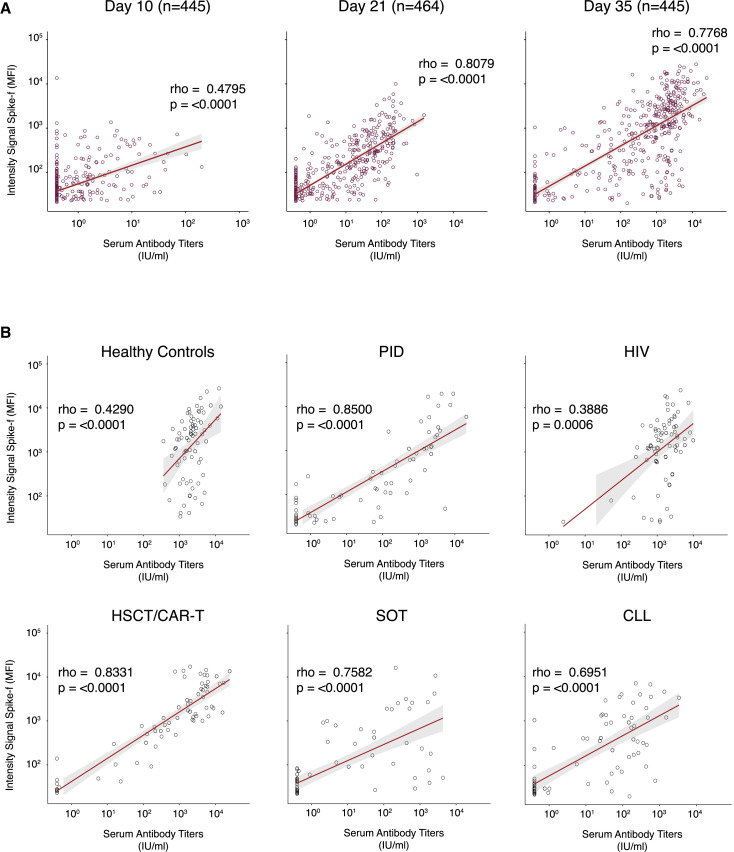
To evaluate whether SARS-CoV-2-specific IgG responses in saliva corresponded to those in serum, paired analyses were performed across all time points. Serum anti-S1 antibody data were generated using the quantitative test Elecsys Anti-SARS-CoV-2 S34 that has been validated on serum samples from patients and against the World Health Organization (WHO) reference standard (WHO/BS/2020.2402). To assess saliva as a diagnostic indicator of serum responses, Spearman correlation analysis in paired samples was performed for the entire cohort at D10 (n = 445), D21 (n = 464), and D35 (n = 445). As shown in Figure 2A, a moderate correlation was observed by D10 (rho = 0.4795; p < 0.0001), followed by strong correlations at D21 (rho = 0.8079; p < 0.0001) and D35 (rho = 0.7768; p < 0.0001). The slightly lower rho value noted for day 35 could be attributed by a minor subset of samples with very high antibody levels in serum but very low in saliva or some highly concentrated serum samples hitting the maximum detection level in some groups. Similar temporal correlations were also found between anti-S1 IgG in saliva and paired serum spike receptor binding domain (RBD) IgG levels (Figure S3). Correlating the D35 salivary Spike-f IgG reactivities on a group level to serum anti-S1 antibody titers demonstrated moderate correlations in the healthy controls (rho = 0.4290; p < 0.0001) and HIV (rho = 0.3886; p = 0.0006) groups and strong correlations in the PID (rho = 0.8500; p < 0.0001), HSCT/CAR-T (rho = 0.8331; p < 0.0001), SOT (rho = 0.7582; p < 0.0001), and CLL (rho = 0.6951; p < 0.0001) groups (Table S2; Figure 2B). Taken together, these data confirmed there is a strong agreement between the salivary spike IgG (irrespective of Spike-f or S1) and serum spike IgG, with the latter measured by an independent clinical laboratory. This concordance was seen at the cohort level as well as patient group level, particularly after D21 from first vaccine dose.
Figure 2.
Correlation between salivary Spike-f IgG and serum spike IgG levels in paired samples
(A) Salivary Spike-f IgG MFI signal intensity (y axis) was measured by a multiplex bead-based assay, and serum SARS-CoV-2 spike IgG levels expressed as international units (x axis) were measured by the quantitative test Elecsys Anti-SARS-CoV-2 S. Correlation plots of the entire COVAXID cohort at D10, D21, and D35 post-vaccination are shown.
(B) Correlation plots of each respective study group. IU, international units; MFI, median fluorescence intensity. Spearman correlation analysis was used to determine rho and p values.
Sex- and age-based influences on SARS-CoV-2-specific responses in saliva and blood
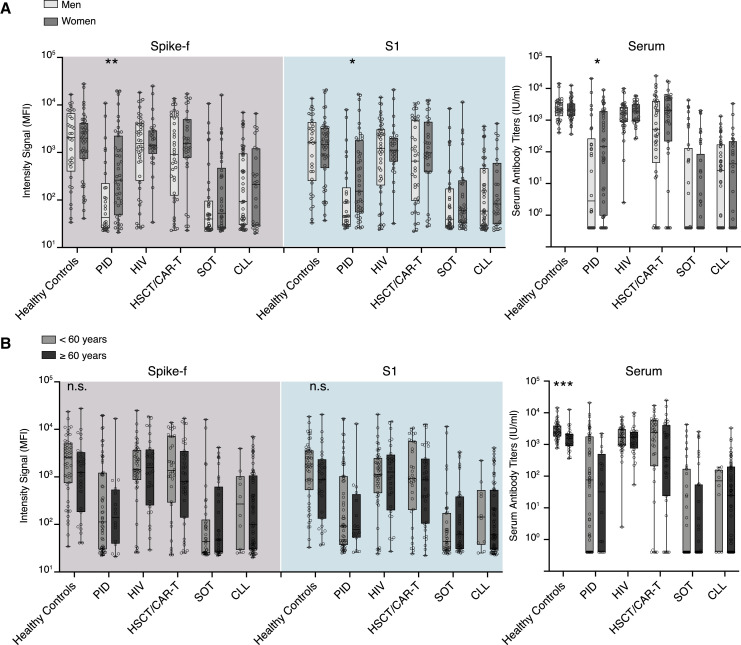
To evaluate whether sex and age impacted SARS-CoV-2 vaccine responses in the study cohort, these parameters were analyzed on a group level in paired saliva and serum samples collected 2 weeks after the second dose (D35). A significant sex-based difference was observed in both serum and saliva in the PID group, with women demonstrating significantly stronger responses, while none of the other groups showed any significant sex-based influence (Figure 3A). Subgrouping the patient cohort by age (<60 years/≥60 years) did not reveal any significant differences in saliva. However, a stronger serum SARS-CoV-2-specific IgG magnitude was found in the younger subgroup (<60 years) of healthy controls (Figure 3B). Taken together, except for the PID group, sex and age appeared to have little impact on the saliva results.
Figure 3.
Sex and age have minimal impact on antibody responses detected in saliva and serum
(A) Sex- and (B) age-based comparisons of salivary IgG to Spike-f and S1 MFI and serum IgG to spike of paired D35 saliva and serum samples from fully vaccinated individuals. Lines, boxes, and whiskers represent the median, IQR, and min-max range, respectively. The Mann-Whitney U test was used to test. Significance: ∗∗∗∗p < 0.0001; ∗∗∗p < 0.0002; ∗∗p < 0.0021; ∗p < 0.0332.
Influences of disease and treatment status observed in both serum and saliva
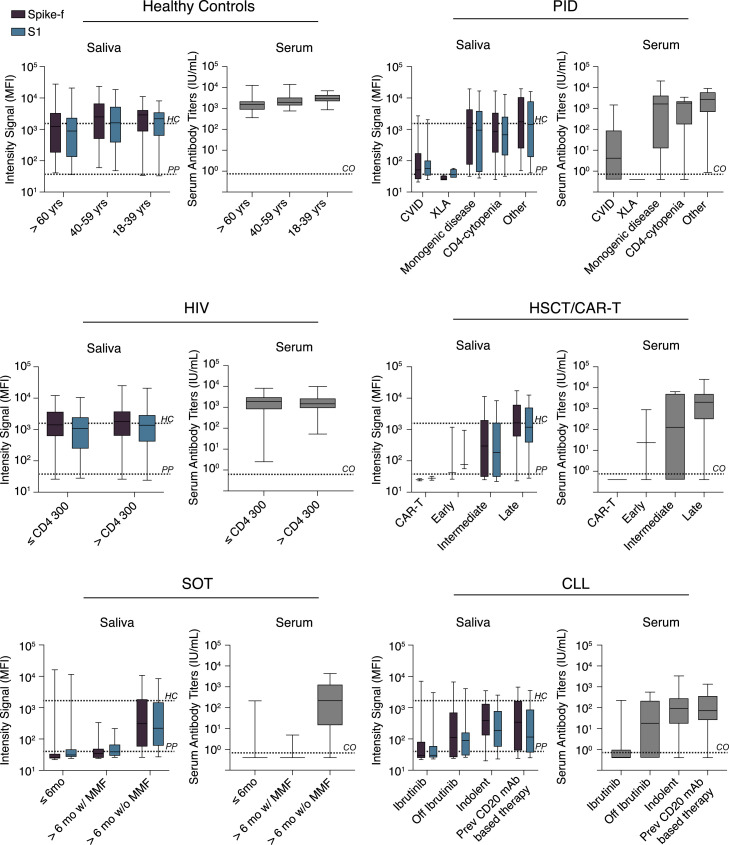
The influence of disease status or treatment regimens on vaccine-induced SARS-CoV-2-specific responses were further examined on a patient subgroup level in paired saliva and serum samples on D35. As shown in Figure 4 , further age stratification revealed no difference among healthy participants. Among PID patients, the common variable immunodeficiency (CVID) and X-linked agammaglobulinemia (XLA) subgroups (n = 39 and n = 4, respectively) showed the lowest median antibody response in both saliva and serum, while subgroups with monogenic PID disease (n = 9), CD4 cytopenia (n = 11), or other PID disorders (n = 10) generated responses close to healthy control levels. On the other hand, participants living with HIV with either low (n = 22) or high (n = 52) CD4 T cell counts had a similar range of median antibody levels in both saliva and serum compared with healthy controls. In the HSCT/CAR-T group, the lowest responses were seen in those patients receiving CAR-T treatment (n = 2) and in those being in an early or intermediate phase (<6 months) post-HSCT transplantation (n = 3 and n = 11, respectively). The responses were, however, close to healthy levels in the subgroup in the late-phase post-HSCT transplantation (n = 53). SOT patients had the lowest overall antibody response in both serum and saliva, with a particularly poor response in patients receiving mycophenolate mofetil (MMF) as a part of their immunosuppression regimen (n = 46), while patients without MMF (n = 30, all vaccinated >6 months after transplantation) had a moderate vaccine response. In the CLL subgroups, lowest antibody responses were observed in those receiving ibrutinib treatment (n = 26) followed by those being off ibrutinib treatment (n = 8), a BTK inhibitor that suppresses B cell signaling. Although the responses varied among the CLL subgroups, a significant proportion of indolent or previously chemoimmunotherapy-treated (including CD20 monoclonal antibody [mAb] therapy) CLL patients produced antibody responses in both serum and saliva. Based on these observations, the striking similarities in the SARS-CoV-2-specific IgG profile in saliva and serum observed even at the subgroup levels strengthen the usefulness for saliva as an indicator for seroconversion, which was measured by the quantitative clinical serology assay.
Figure 4.
Patient subgroup analysis
Disease or treatment status subgrouping of immunocompromised individuals included in the COVAXID study on D35 saliva or serum samples of fully vaccinated individuals. Lines, boxes, and error bars represent the median, IQR, and min-max range. The black and gray dashed line indicate the spike-specific IgG MFI for healthy controls at D35 and pre-pandemic samples, respectively. HC, healthy controls; PP, pre-pandemic.
Evolution of anti-spike IgG in saliva and serum is harmonized and strongly correlated after vaccination
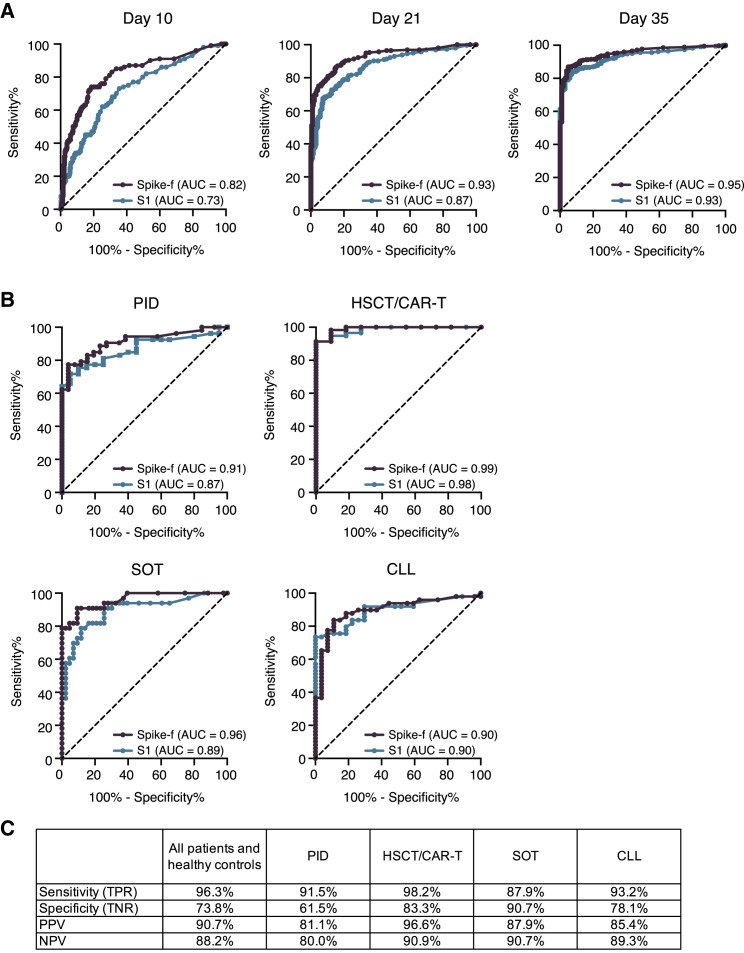
Next, a receiver operating characteristic (ROC) curve analysis was performed to determine the performance of the salivary anti-spike IgG conversion classified by the present assay relative to the clinical serology result. As shown in Figure 5A, area under the curve (AUC) scores raised from 0.82 (D10) to 0.93 (D21) and 0.95 (D35) for anti-Spike-f and 0.73 (D10) to 0.87 (D21) and 0.87 (D35) for the anti-S1 responses. This was also assessed at the respective patient group level (Figure 5B). Here, we found that AUC scores in PID, HSCT/CAR-T, SOT, and CLL reached 0.92, 0.99, 0.96, and 0.90, respectively, for anti-Spike-f and 0.87, 0.99, 0.89, and 0.90 for the anti-S1 antibodies, respectively. Due to the very high rates of seroconversion in the healthy control and HIV groups, they were excluded from the analysis. Because the Spike-f antigen appears superior in detecting seroconverted participants, it was chosen for further evaluations against serology data. Based on the serology data and the adjusted cutoff at >50 median fluorescence intensity (MFI) for Spike-f (Figure S4), the endpoint (D35) saliva antibody result yielded 96.3.8% in sensitivity and 73.8% in specificity, relative to the paired serology data when the entire cohort, i.e., all patients and healthy controls, were considered. The D35 data also yielded a positive predictive value (PPV) of 90.7% and negative predictive value (NPV) of 88.2% (Figure 5C). Similarly high levels of performance were seen when PID, HSCT, SOT, and CLL groups’ anti-Spike-f data were analyzed separately (Figure 5C). Altogether, this result confirms the consistently strong serum-to-saliva correlations seen in Figures 3 and 4. These data indicate that saliva is functional and accurate in predicting seroconversion as measured in blood.
Figure 5.
Determination of seroconversion in blood using saliva samples
(A) Graphical representation of the ROC AUC curves for salivary Spike-f and S1 IgG reactivities for the entire study cohort on indicated day after initial vaccination, using the clinical IgG serology result determined by Elecsys Anti-SARS-CoV-2 S test as reference.
(B) ROC AUC curve analysis of indicated patient groups on D35 saliva samples.
(C) Performance summary on D35 salivary anti-Spike-f IgG to detect seroconversion in paired serum classified by the Elecsys Anti-SARS-CoV-2 S test. AUC, area under the curve; PPV, positive predictive value; NPV, negative predictive value; ROC, receiver operating characteristic.
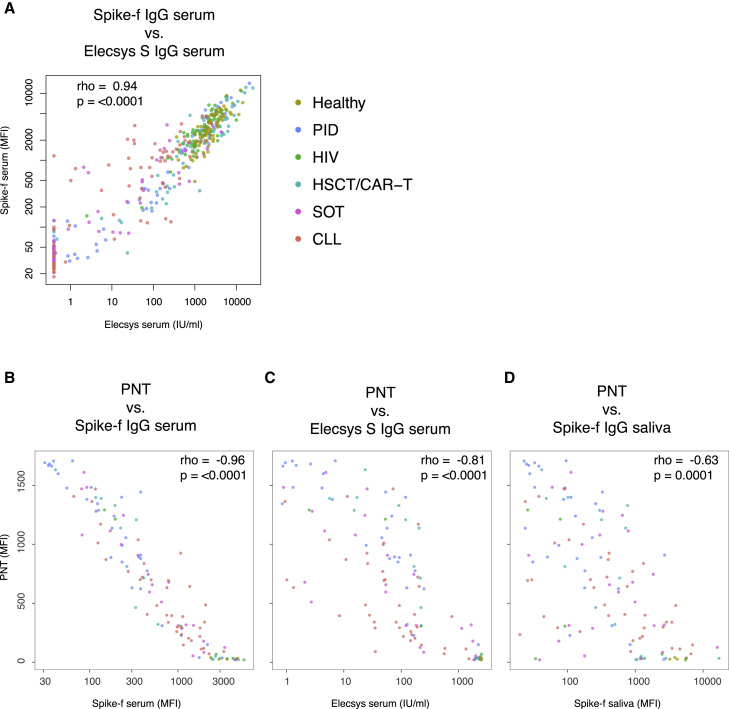
Because serum antibody neutralization is reported as a key protective correlate,35 we next measured the serum neutralizing capacity33 among D35 samples for a correlation analysis to the anti-spike IgG reactivities of the two biological sites. We first performed a technical validation of the multiplex serology platform in serum to the Elecsys assay, using only samples for which paired data were available (n = 435), and saw a striking correlation (Figure 6A; rho = 0.94; p < 0.0001). We next performed the pseudoneutralization (PNT) assay on selected samples (n = 120), where we also saw a clear and significant correlation between neutralization capacity and Spike-f serum bead serology data (Figure 6B; rho = −0.96; p < 0.0001), Elecsys serum IgG (Figure 6C; rho = −0.81; p < 0.0001), and saliva Spike-f (Figure 6D; rho = −0.63; p = 0.0001). Conclusively, the proximate relationships found here between the salivary and serum spike IgG support the assumption that these specific antibodies detected in saliva in naive RNA-vaccinated individuals are mainly derived from the circulating blood after vaccination.
Figure 6.
Antibody neutralization in relation to the serum and salivary spike IgG
All included samples are collected at day 35 from the beginning of vaccination.
(A) Correlation between anti-Spike serum antibodies detected by the quantitative test Elecsys Anti-SARS-CoV-2 S and the multiplex bead-based assay (Spike-f; n = 435).
(B–D) 120 samples were tested by PNT assay (B–D). For PNT, a low signal intensity indicates the blocking of Spike-ACE2 binding (y axis) and the data are used to correlate with the serum or salivary Spike-specific IgG levels (x axis). All correlations were tested by Spearman’s test, and rho coefficient and p value are reported in the plots. Each dot represents one sample, and the color code refers to indicated group. PNT, pseudoneutralization.
Assessment of negative predictors for salivary IgG response after vaccination
Next, we evaluated the risk factors associated with failure of salivary antibody conversion after vaccination, where the anti-Spike-f positivity in the endpoint (D35) saliva samples were considered (Table 1 ). As shown in the univariable analysis, age and sex had little impact on the salivary IgG response. However, the strongest risk for a failure of salivary anti-Spike-f IgG conversion was found in the SOT patients (odds ratio [OR], 32.14; p < 0.001), followed by CLL (OR, 17.94; p < 0.001), PID (OR, 13.65; p < 0.001), and lastly HSCT patients (OR, 5.19; p < 0.01). The exact same rank order for these groups was found for serum regarding the risk for failure of seroconversion.30 Within the patient groups, the attributable risk factors among disease- or treatment-specific parameters were also assessed. Notably, being in an early-phase post-HSCT transplantation was a strong negative predictor (OR, 19.2; p < 0.02). The MMF or ibrutinib drug usage, which are critical medications for the SOT and CLL patients, respectively, also negatively impacted the salivary response substantially (OR, 16.44; p < 0.001 and OR, 24.44; p < 0.001). These results were confirmed by the multivariate analysis as summarized in Table 1.
Table 1.
Logistic regression and univariable and multivariable analysis, assessing variables for failure of salivary antibody conversion to Spike-f after two doses of BNT162b2 in D35 saliva samples
| Univariatea |
Multivariatea |
|||
|---|---|---|---|---|
| All | p Value | OR (CI) | p Value | OR (CI) |
| Age | 0.04 | 1.01 (1-1.03) | – | – |
| Sex (M/F) | 0.03 | 1.61 (1.06–2.47) | 0.02 | 1.73 (1.08–2.82) |
| Patient groups | ||||
| PID | <0.001 | 13.65 (4.49–59.49) | <0.001 | 14.12 (4.62–61.76) |
| HIV | 0.28 | 2.18 (0.55–10.63) | 0.34 | 1.99 (0.5–9.77) |
| HSCT | 0.01 | 5.19 (1.56–23.58) | 0.02 | 4.94 (1.48–22.5) |
| SOT | <0.001 | 32.14 (10.74–139.33) | <0.001 | 32.16 (10.7–139.76) |
| CLL | <0.001 | 17.94 (5.97–77.75) | <0.001 | 16.26 (5.38–70.69) |
| Healthy | – | reference group | – | reference group |
| PID | p Value | OR (CI) | p Value | OR (CI) |
|---|---|---|---|---|
| Age | 0.74 | – | – | – |
| Sex (M/F) | 0.03 | 2.91 (1.09–8.03) | – | – |
| Subgroups | ||||
| CD4 cytop | 0.04 | 0.11 (0.01–0.63) | – | 0.11 (0.01–0.63) |
| Monogenic disease | 0.07 | – | – | – |
| Other | 0.05 | 0.12 (0.01–0.71) | – | 0.12 (0.01–0.71) |
| XLA | 0.99 | – | – | – |
| CVID | – | reference group | – | reference group |
| HSCT | p Value | OR (CI) | p Value | OR (CI) |
|---|---|---|---|---|
| Age | 0.22 | – | 0.16 | – |
| Sex (M/F) | 0.4 | – | – | – |
| Subgroups | ||||
| Early | 0.02 | 19.2 (1.58–460) | 0.03 | 17.16 (1.39–415) |
| Intermediate | 0.12 | 3.6 (0.64–17.95) | 0.06 | 5.68 (0.9–36.6) |
| Late | – | reference group | – | – |
| SOT | p Value | OR (CI) | p Value | OR (CI) |
|---|---|---|---|---|
| Age | 0.97 | – | – | – |
| Sex | 0.25 | – | – | – |
| MMF (yes/no)b | <0.001 | 16.44 (5.51–56.53) | <0.001 | 16.41 (5.32–59.36) |
| CLL | p Value | OR (CI) | p Value | OR (CI) |
|---|---|---|---|---|
| Age | 0.56 | – | – | – |
| Sex (M/F) | 0.64 | – | – | – |
| Subgroups | ||||
| Ibrutinib | <0.001 | 24.44 (6.07–132.85) | <0.001 | 17.13 (4–96.37) |
| Off ibrutinib | 0.03 | 7.33 (1.21–52.21) | 0.14 | – |
| Prev CD20 mAb therapy | 0.17 | – | 0.28 | – |
| Indolent | – | reference group | – | – |
CD20 mAb, cluster of differentiation monoclonal antibody; CI, 95% confidence interval; CVID, common variable immunodeficiency; indolent, indolent and not treated; MMF, mycophenolate mofetil; off ibrutinib, off ibrutinib treatment for >2 months; OR, odds ratio; XLA, X-linked agammaglobulinemia.
aVariables with p ≤ 0.35 in univariable analysis were submitted to multivariable analysis. In the multivariable analysis, the variables retained in the final model after stepwise selection procedure are shown (odds ratios and CI are shown only for the significant variables).
bFor variables with categories of yes (Y) or no (N), no was set as reference group.
Discussion
A central clinical question in the global COVID-19 vaccination effort is the effectiveness of the new vaccines in immunocompromised individuals, including how effective a parenterally administered new vaccine may provide mucosal immunity in these vulnerable individuals. While most studies focus on the immune markers in blood, this study focused on salivary immunity markers in these individuals. We report here that BNT162b2-vaccine-induced IgG levels in saliva in immunocompromised vaccinees may vary but could also reach levels normally acquired from natural SARS-CoV-2 infection, as recently shown in healthcare workers vaccinated with mRNA vaccine.28 Moreover, we identified risk factors for poor antibody induction in saliva, which pointed out several significant negative predictors, such as SOT, CLL, PID, and disease-related treatment regimens, i.e., MMF and ibrutinib, among the conditions studied here. These risk factors interestingly mirror the observations we made recently on a study using serum samples from this cohort.30 Therefore, our results not only demonstrate that the presence of salivary antibodies to the viral spike is strongly connected to the circulating antibodies, also it shows a strong performance in assessing seroconversion in blood that possibly could serve for a diagnostic purpose. To our knowledge, this is the largest SARS-CoV-2 vaccine study using saliva as a biofluid for tracking seroconversion, encompassing four different time points of sample collection on 445 age- and sex-matched (baseline seronegative) study participants donating over 1,800 paired saliva and serum samples.
The oral compartment is covered by mucosal surfaces that are susceptible to SARS-CoV-2 infection.23 Oral mucosa is endowed with various salivary immunochemical mechanisms to repel foreign intruders and is well explored by mucosal infection and vaccine experts.23 , 36 Primary mechanisms supporting oral mucosa permeability for systemic biomarkers include (1) passive diffusion, (2) carrier-mediated transport, and (3) endocytosis and exocytosis where material is actively taken up and excreted by cells via the endocytic pathway.37 Saliva is therefore a functional biofluid and has the potential to mirror systemic antibody responses. Both clinical and experimental data have shown that induction of mucosal immune responses after vaccination might significantly contribute to protection against mucosal infections of respiratory or enteric pathogens.23 Our data further confirm that the mechanism through which serum to saliva transudation occurs appears to operate well in some immunocompromised patients, particularly those living with HIV infection or who have undergone HSCT. In the HIV group, we did not find differences in humoral responses when stratified by low and high CD4 T cell count (at 300 cells/μL). In HSCT patients at least 6 months after immunocyte reconstitution (the late group), two doses were required to reach the antibody level of healthy controls. Concerning the subset of D35 samples showing very high specific IgG in serum but low in saliva, we could not find a clear association to any known demographic variables. Whether this may relate to individual variations in the oral mucosa integrity or other factors, including FcR expression or other factors of importance for IgG transport to the mucosa,38 , 39 remain to be determined. Apart from that, sex and age appeared to have little influence on the salivary antibody responses, which is in line with serum data reported in earlier cohort studies of HIV and hematological malignancy patients.23 The present data are, to the best of our knowledge, the first to report that swift mucosal antibody responses are in place after COVID-19 mRNA vaccination in certain immunocompromised risk groups.
In patients with PID, SOT, or CLL, salivary antibody responses were highly variable. As expected, the XLA and CVID patients were poor antibody responders due to absent (XLA) or impaired (CVID) immunoglobulin production. An Israeli study of 26 PID patients reported seroconversion in the majority of CVID patients, but anti-spike antibody levels remained low relative to healthy controls,20 which is consistent with the data presented here. Importantly, no detectable salivary spike IgG was observed in the XLA patients, which supports that the saliva analysis employed in our study is highly specific. We further report that other vaccinated PID subgroups— monogenic diseases and CD4 T cell cytopenia—had detectable saliva antibodies, with some patients responding normally in both saliva and serum. However, the median levels were 10- to 20-fold lower relative to healthy controls and two vaccine doses appeared insufficient for induction of robust immunity.
Next, we observed that SOT patients with a post-transplantation time of more than 6 months and no MMF treatment were more likely to develop salivary responses after two doses of BNT162b2. This not only mirrors the systemic responses shown in paired serum but also is in line with a blood serology study by Boyarsky et al.,40 who showed that the use of antimetabolites, including MMF, was persistently associated with poor humoral response in SOT patients after two doses of the BNT162b2 vaccine. Adding a third BNT162b2 vaccine dose could be an option to increase the level of protection in SOT patients, especially in those with an initially weak serological response,41 and a non-invasive antibody screening strategy could be helpful to identify this subgroup. Our data further confirm that neither sex nor age in any marked way impacted salivary or serum antibody responses in the SOT patients, which is consistent with the latter two studies.41 Furthermore, drugs used in CLL treatment may impair humoral immunity during COVID-1942 and often diminish vaccine responses, resulting in very low salivary IgG levels. This was found mainly in active or past ibrutinib (a BTK inhibitor) subgroups, although indolent CLL and previous CD20 mAb-based therapy were also impacted to some extent. Whether the variation observed could relate to drug compliance is unknown, as no biomarkers exist for validation.
Mucosal antibodies specific to SARS-CoV-2 are considered important in reducing transmission potential in vaccinated individuals.43 The magnitude of anti-spike IgG responses in the saliva of vaccinated individuals, which exceeded those seen in mild convalescent individuals, is encouraging, as it indicates that vaccination might confer a sterilizing immune response in the oral cavity and thereby lower virus transmission. The observation that vaccine-induced IgG efficiently translocates into saliva, with high predictive values of BNT162b2-induced seroconversion, is beneficial for immune surveys. Many risk groups are vulnerable to SARS-CoV-2 infection and need regular monitoring. Therefore, saliva and home sampling represent a safe and convenient alternative to even allow sensitive detection of emerging virus variants including the Omicron.29 Our finding that the potential sensitivity of saliva antibody detection is lower than serum antibody detection may not be a major concern in real life, as if saliva underreports the true degree of seroconversion, it may be more acceptable than overreporting, especially in ongoing vaccine campaigns to ensure protective immunity is in place.
Saliva sampling is entirely non-invasive, easy, and can be repeated multiple times. It is therefore ideal for real-time monitoring of frail patient groups that are sensitive to infections. It will be a safe and efficient approach for tracking vaccine immunity to support informed decisions and agreement on protective strategies for these patients, especially towards a re-opening of society. In this context, saliva is highly suitable for vaccine follow-up studies and can be used for monitoring seroconversion and antibody memory after vaccination. Furthermore, saliva-specific antibody studies also depict the local immunity at a crucial site for the SARS-CoV-2 infection. Swift comparisons of vaccine responses in immunocompromised individuals will improve vaccination strategies and identify those likely to remain at risk for COVID-19 for a revaccination. Our data merit a call to accentuate the diagnostic significance of salivary testing.
Limitations of the study
Our study is not without limitations. These include the lack of antibody isotype analysis and virus-neutralizing capacity of the salivary antibodies. Mutant spike antigens were not available for current study. The most effective vaccines known today, including the BNT 162b2, are capable of inducing neutralization antibodies of such levels considered protective in the majority of the general population, and such data are highly valuable in prediction analysis on protective antibody levels in vaccination.35 Such prediction is not possible with our present data, as neutralization data were not our primary endpoint and our vaccine-monitoring survey is yet ongoing. However, our preliminary observations do indicate that the few breakthrough cases with symptomatic COVID-19 noted so far appear to have weak spike IgG reactivities in their D35 saliva and serum, i.e., within of the lower 25th percentile of healthy controls (only preliminary unpublished observation), which remains to be confirmed when our surveillance study is completed. Moreover, the antibody durability in saliva and exploration of local memory B and T cell immunity also remain to be investigated. Vaccines other than BNT162b2 also need to be compared in similar cohorts. Although our data in saliva are consistent with Sahin et al.44 that, after full vaccination, healthy volunteers showed a 10-fold increase in producing spike-binding IgG, the present data suggest that the vast majority of HIV and HSCT patients also displayed similar levels as healthy controls. We did not measure IgA, as it had already been shown that the BNT162b2 vaccine elicits mainly IgG and less IgA in saliva in healthcare workers.28 Apart from sex, gender, and age, we did not compute ethnicity or socioeconomic status in our analysis.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| F(ab')2-Goat anti-Human IgG Fc Secondary Antibody, PE | Thermo Fisher Scientific | Cat# H10104; RRID:AB_2536546 |
| Biological samples | ||
| Serum | Human donors | Covaxid clinical trial |
| Saliva | Human donors | Covaxid clinical trial |
| Chemicals, peptides, and recombinant proteins | ||
| MES Hydrate | Sigma-Aldrich | Cat# M2933-100G |
| Sodium Phosphate Monobasic | Sigma-Aldrich | Cat# S3139-250G |
| NHS | Thermo Scientific | Cat#24510 |
| EDC 65mg | Thermo Scientific | Cat# 77149 |
| Blocking Reagent | Roche | Cat# 11112589001 |
| Proclin | Sigma-Aldrich | Cat# 48912-U |
| Software and algorithms | ||
| Luminex xPONENT software | Luminex Corp | |
| Prism software | GraphPad | Version 9 |
| Other | ||
| Spike S1 antigen | KTH – Royal Institute of technology, Stockholm, Sweden. Prof. Sophia Hober | N/A |
| Spike-foldon antigen | KTH – Royal Institute of technology, Stockholm, Sweden. Prof. Sophia Hober | N/A |
| MagPlex® Microspheres | Luminex Corp | Depending on selected region |
| FLEXMAP 3D System | Luminex Corp | FLEXMAP-3D |
Resource availability
Lead contact
Additional information and requests for resources and reagents should be directed to the lead contact, Professor Margaret Sällberg Chen (margaret.chen@ki.se).
Materials availability
This study did not generate new unique reagents.
Experimental models and subject details
Human subjects
We conducted a prospective, open-label clinical trial of BNT162b2 (Comirnaty®, Pfizer/BioNTech) with two doses given to immunocompromised patients and healthy controls at Karolinska University Hospital, Sweden. Evaluated in the study was safety and efficacy.30 The two doses of vaccine were given 21 days apart. Immunocompromised patients (n = 449) had either PID (n = 90), or SID due to infection with HIV infection (n = 90), allogeneic hematopoietic stem cell transplantation (HSCT)/chimeric antigen receptor T (CAR-T) cell therapy (n = 90), solid organ transplantation (SOT) (n = 89), or chronic lymphocytic leukemia (CLL) (n = 90). Healthy controls were age and sex matched (n = 90). The number of available saliva samples in each patient group was 79 in PID group, 80 in HIV group, 74 in HSCT/CART-T group, 83 in SOT group and 88 in CLL group. Eligible were men and women ≥ 18 years of age, with no known history of SARS-CoV-2 infection. The study was approved by the Swedish Medical Product Agency (ID 5.1-2021-5881) and the Swedish Ethical Review Authority (ID 2021-00451 and 2020-06381). All participants provided written informed consent.
Trial registration
ClinicalTrials.gov Identifier: NCT04780659.
A total of 486 patients’ saliva were included from the clinical vaccine study, with 445 paired serum samples for the D35 endpoint analysis. As positive controls, samples donated by COVID-19 convalescent patients were used. These patients were SARS-CoV-2 infected during February to March 2020 with mild (n=21) or severe (n=10) COVID-19. They were recruited from a post-COVID-19 follow-up study at Karolinska University Hospital and sampled 3-9 months after infection (mean: 7.03 months). Negative controls were pre- pandemic saliva samples (n=41) collected during 2016-2018.
Method details
Sample collection and SARS-CoV-2 antibody detection in saliva
All saliva samples were processed by a standardized protocol in the same laboratory. Briefly, unstimulated whole saliva was self-collected by fasted study participants as described earlier using standardized picture instructions.32 Participants were instructed to passively drool into a clean cup for five minutes after which the saliva was aliquoted in tubes using a transfer pipette. Samples were either submitted at the study site or mailed in by overnight post. All samples were immediately placed at 4°C upon arrival thereafter stored at -80°C on the same day. Prior to antibody analysis, saliva samples were thawed at 4°C and centrifuged at 400 xg for one min at 4°C to separate any debris. The supernatant was transferred to 96-well PCR plates (100 μL/well) and sealed using qPCR foil seals. Inactivation was then performed at 56°C for 30 min in plate format using a thermal cycler and cooled immediately to 4°C before transferring to −20°C for antibody analysis.
Antibodies binding to the full-length spike glycoprotein in trimeric form (Spike-f) and the S1 subunit were measured by means of a multiplex bead-based assay in the 384-well plate format,31 , 32 as previously described. Briefly, the antigens were immobilized on the surface of uniquely color-coded bead identities (IDs) (MagPlex-C, Luminex corp.), and the IDs pooled to generate the bead-array. Saliva samples were diluted 1:5 in assay buffer and incubated with the array. After cross-linking of the antibody-antigen complexes, a R-phycoerythrine-conjugated anti-human IgG antibody (H10104, Invitrogen) was applied for detection of IgG bound to spike. The assay readout was performed using a FlexMap3D instrument and the Luminex xPONENT software (Luminex Corp.). Each assay run included the same set of 12 negative and 4 positive saliva controls. The negative controls were selected among pre- pandemic saliva samples as representative of the background distribution and therefore used to calculate the antigen and assay specific cutoff, allowing to account for inter-assay variability. The positive controls were selected among convalescent samples with mild disease showing clear reactivity to spike. The inter-assay variability, evaluated as the % CV of the 16 control samples included in each assay run, was 10.8% for Spike-f and 12% for Spike S1 on average.
Sample collection and SARS-CoV-2 antibody detection in serum
Serum samples were analyzed for detection of antibodies to SARS-CoV-2 spike protein receptor binding domain (RBD), using the quantitative Elecsys® Anti-SARS-CoV-2 S test (Roche Diagnostics)34 on the Cobas 8000 e801pro. The measuring range is between 0.40 to 250 U/mL, and the cut-off value for positive results is ≥ 0.80 U/mL Positive samples with antibody titers of >250 U/mL were re-tested following 1/10 dilution, and in some cases 1/100 dilution with the upper level of measuring range 25,000 U/mL. Serum samples at day 35 were also tested to detect antibodies binding the full-length spike glycoprotein in trimeric form (Spike-f) and measure their neutralizing capability. Spike-f binding antibodies were measured by means of the same multiplex bead-based assay used for the detection of salivary antibodies (38, 39). Briefly, serum samples were diluted 1:5000 in assay buffer (38) and incubated with the array, followed by anti-Spike-f antibodies detection with an R-phycoerythrine-conjugated anti-human IgG antibody (H10104, Invitrogen) and read-out in FlexMap3D instrument and the Luminex xPONENT software (Luminex Corp.). The neutralizing capability was measured on serum diluted 1:50 and by means of a pseudoneutralization (PNT) assay as previously described.33 In brief, samples were pre-incubated with biotin-conjugated Spike-f followed by incubation with magnetic beads (Luminex Corp.) coupled to ACE-2. Only non-inhibited Spike-f binds to the ACE-2 receptor. Subsequently, a streptavidin-conjugated phycoerythrine is added the assay readout performed using a FlexMap3D instrument and the Luminex xPONENT software (Luminex Corp.). The signal intensity is reported in MFI and inversely proportional to the neutralization.
Quantification and statistical analysis
The salivary antibody data were acquired as median fluorescence intensities (MFI) for each sample and antigen. The antigen and assay specific cutoff for positivity was calculated as the mean plus 6x standard deviation (SD) of the intensity signals of the 12 selected negative controls. The inter-assay variability was estimated for Spike-f and S1 as the average percent CV of the 16 control samples included in all 6 assay runs required to test the samples included in the current study. Statistical analysis was performed using R and R studio45 for correlation analyses and logistic regression analyses and Prism software v.9 (Graphpad) for all other comparisons. Datasets initially underwent a data normality distribution test. Differences between groups of samples were analyzed by Mann-Whitney U test for univariate analysis. Correlations were determined using Spearman rank correlation. Logistic regression, univariable or multivariable, was used to analyze possible negative predictive factors. P values <0.05 were considered statistically significant. Two-sided p values <0.05 were considered significant.
Additional resources
The sponsor of the study was Karolinska University Hospital. This trial was registered at EudraCT (no. 2021-000175-37), and clinicaltrials.gov (no. 2021-000175-37). The full clinical study protocol is available via the SciLifeLab Data Repository (English version: https://doi.org/10.17044/scilifelab.15059364; Swedish version https://doi.org/10.17044/scilifelab.15059355).
Acknowledgments
We thank all the participants enrolled in this study and all the research and clinical staff at Karolinska University Hospital, especially research/clinical nurses Sonja Sönnert Husa, Kirsti Niemalä, Begüm Eker, Eva Martell, Helena Pettersson, Charlotta Hausmann, Maria Abramsson, Ruza Milosavljevic, Karin Fransson, Karin Linderståhl, Linn Wursé, Cecilia Lång, Anna Löwhagen Welander, Susanne Hansen, Douglas Carrick, Katarina Stigsäter, Susanne Cederberg, Annika Olsson, Ingrid Andrén, and Margareta Gustafsson. For assistance with biobanking, we would like to thank Agne Kvedaraite, Nazila Samimi, Rosita Mario, and Daniela Sofia Ambrosio. We thank Prof. Jan Albert from Clinical Microbiology, Karolinska University Hospital for fruitful discussions. Funding for this study was provided from Knut and Alice Wallenberg Foundation, Erling Perssons family foundation, Region Stockholm, Swedish Research Council, Karolinska Institutet, The Swedish Blood Cancer Foundation, the organization for PID patient group in Sweden, Nordstjernan AB, Center for Medical Innovation (CIMED), Swedish Medical Research Council, and Stockholm County Council (ALF).
Author contributions
S.A., E.P., H.-G.L., P. Nilsson, and M.S.C. conceived the project. K.H., P.C., G.G., K.A.-M., H.A., M.J.S., M.A., A.C., and M.S.C. established the protocols for handling clinical material. G.S., P. Nowak, S. Mielke, L.H., P.B., C.I.E.S., P.L., X.X., O.B., and A.Ö. collected and compiled clinical data. E.P., C.H., S. Mravinacova, G.B., J.Y., and S. Muschiol performed measurements and analyzed the laboratory data. K.H., P.C., and D.V. performed statistics. S.A., E.P., S.H., P. Nilsson, H.-G.L., and M.S.C. provided resources and supervised the project. K.H., E.P., S.A., and M.S.C. wrote the first draft of the manuscript, reviewed it, and edited it. All authors agreed to submit the manuscript, read, and approved the final draft and take full responsibility of its content, including the accuracy of the data and the fidelity of the trial to the registered protocol and its statistical analysis. M.S.C. and S.A. have unrestricted access to all data.
Declaration of interests
S. Mielke received honoraria via his institution from Celgene/BMS, Novartis, Gilead/Kite, and DNA Prime for lectures and educational events and as a member and/or head of data safety monitoring boards from Miltenyi Biotec and Immunicum outside the submitted work. K.L. reports grants from Knut and Alice Wallenberg Foundation VC-2021-0018. H.-G.L. reports grants from Knut and Alice Wallenberg Foundation and Nordstjernan AB for studies on COVID-19. P.L. reports grants from Pfizer, grants from MSD, grants and personal fees from Takeda, personal fees from AiCuris, and personal fees from OctaPharma, Enanta Pharmaceuticals, and BMS outside the submitted work. S.A. has received honoraria for lectures and educational events, not related to this work, from Gilead, AbbVie, MSD, Biogen, and Netdoktor and reports grants from Knut and Alice Wallenberg Foundation for this study. M.S.C. reports grants from CIMED for this study and is co-founder of SVF AB.
Published: February 12, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.medj.2022.01.001.
Supplemental information
Data and code availability
Data
De-identified patient data presented in this manuscript will be made available upon request from the lead author, in a format compliant with local regulatory requirements with respect to the handling of patient data, and in adherence with the policies of the Karolinska University Hospital and Karolinska Institutet.
Code
This paper does not report original code.
Any additional information required to analyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., et al. Safety and immunogenicity of two RNA-based covid-19 vaccine candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dagan N., Barda N., Kepten E., Miron O., Perchik S., Katz M.A., Hernán M.A., Lipsitch M., Reis B., Balicer R.D. BNT162b2 mRNA covid-19 vaccine in a nationwide mass vaccination setting. N. Engl. J. Med. 2021;384:1412–1423. doi: 10.1056/NEJMoa2101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haas E.J., Angulo F.J., McLaughlin J.M., Anis E., Singer S.R., Khan F., Brooks N., Smaja M., Mircus G., Pan K., et al. Impact and effectiveness of mRNA BNT162b2 vaccine against SARS-CoV-2 infections and COVID-19 cases, hospitalisations, and deaths following a nationwide vaccination campaign in Israel: an observational study using national surveillance data. Lancet. 2021;397:1819–1829. doi: 10.1016/s0140-6736(21)00947-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernal J.L., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., Stowe J., Tessier E., Groves N., Dabrera G., et al. Effectiveness of COVID-19 vaccines against the B.1.617.2 variant. medRxiv. 2021 doi: 10.1101/2021.05.22.21257658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Müller L., Andrée M., Moskorz W., Drexler I., Walotka L., Grothmann R., Ptok J., Hillebrandt J., Ritchie A., Rabl D., et al. Age-dependent immune response to the Biontech/Pfizer BNT162b2 COVID-19 vaccination. Clin. Infect Dis. 2021 doi: 10.1093/cid/ciab381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collier D.A., De Marco A., Ferreira I., Meng B., Datir R.P., Walls A.C., Kemp S.A., Bassi J., Pinto D., Silacci-Fregni C., et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PubMed] [Google Scholar]
- 8.Subbarao S., Warrener L.A., Hoschler K., Perry K.R., Shute J., Whitaker H., O'Brien M., Baawuah F., Moss P., Parry H., et al. Robust antibody responses in 70-80-year-olds 3 weeks after the first or second doses of Pfizer/BioNTech COVID-19 vaccine, United Kingdom, January to February 2021. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.Es.2021.26.12.2100329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shields A.M., Burns S.O., Savic S., Richter A.G. COVID-19 in patients with primary and secondary immunodeficiency: the United Kingdom experience. J. Allergy Clin. Immunol. 2021;147:870–875.e871. doi: 10.1016/j.jaci.2020.12.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asano T., Boisson B., Onodi F., Matuozzo D., Moncada-Velez M., Maglorius Renkilaraj M.R.L., Zhang P., Meertens L., Bolze A., Materna M., et al. X-linked recessive TLR7 deficiency in ∼1% of men under 60 years old with life-threatening COVID-19. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abl4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bastard P., Gervais A., Le Voyer T., Rosain J., Philippot Q., Manry J., Michailidis E., Hoffmann H.H., Eto S., Garcia-Prat M., et al. Autoantibodies neutralizing type I IFNs are present in ∼4% of uninfected individuals over 70 years old and account for ∼20% of COVID-19 deaths. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abl4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCusker C., Warrington R. Primary immunodeficiency. Allergy Asthma Clin. Immunol. 2011;7:S11. doi: 10.1186/1710-1492-7-s1-s11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brittish Society for Immunology Immunodeficiency. 2017 https://www.immunology.org/policy-and-public-affairs/briefings-and-position-statements/immunodeficiency [Google Scholar]
- 14.Na I.K., Buckland M., Agostini C., Edgar J.D.M., Friman V., Michallet M., Sánchez-Ramón S., Scheibenbogen C., Quinti I. Current clinical practice and challenges in the management of secondary immunodeficiency in hematological malignancies. Eur. J. Haematol. 2019;102:447–456. doi: 10.1111/ejh.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aydillo T., Gonzalez-Reiche A.S., Aslam S., van de Guchte A., Khan Z., Obla A., Dutta J., van Bakel H., Aberg J., García-Sastre A., et al. Shedding of viable SARS-CoV-2 after immunosuppressive therapy for cancer. N. Engl. J. Med. 2020;383:2586–2588. doi: 10.1056/NEJMc2031670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goubet A.G., Dubuisson A., Geraud A., Danlos F.X., Terrisse S., Silva C.A.C., Drubay D., Touri L., Picard M., Mazzenga M., et al. Prolonged SARS-CoV-2 RNA virus shedding and lymphopenia are hallmarks of COVID-19 in cancer patients with poor prognosis. Cell Death Differ. 2021:1–19. doi: 10.1038/s41418-021-00817-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diefenbach C., Caro J., Koide A., Grossbard M., Goldberg J.D., Raphael B., Hymes K., Moskovits T., Kreditor M., Kaminetzky D., et al. Impaired humoral immunity to SARS-CoV-2 vaccination in non-hodgkin lymphoma and CLL patients. medRxiv. 2021 doi: 10.1101/2021.06.02.21257804. [DOI] [Google Scholar]
- 19.Firket L., Descy J., Seidel L., Bonvoisin C., Bouquegneau A., Grosch S., Jouret F., Weekers L. Serological response to mRNA SARS-CoV-2 BNT162b2 vaccine in kidney transplant recipients depends on prior exposure to SARS-CoV-2. Am. J. Transpl. 2021 doi: 10.1111/ajt.16726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagin D., Freund T., Navon M., Halperin T., Adir D., Marom R., Levi I., Benor S., Alcalay Y., Freund N.T. Immunogenicity of Pfizer-BioNTech COVID-19 vaccine in patients with inborn errors of immunity. J. Allergy Clin. Immunol. 2021 doi: 10.1016/j.jaci.2021.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herzog Tzarfati K., Gutwein O., Apel A., Rahimi-Levene N., Sadovnik M., Harel L., Benveniste-Levkovitz P., Bar Chaim A., Koren-Michowitz M. BNT162b2 COVID-19 vaccine is significantly less effective in patients with hematologic malignancies. Am. J. Hematol. 2021 doi: 10.1002/ajh.26284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rabinowich L., Grupper A., Baruch R., Ben-Yehoyada M., Halperin T., Turner D., Katchman E., Levi S., Houri I., Lubezky N., et al. Low immunogenicity to SARS-CoV-2 vaccination among liver transplant recipients. J. Hepatol. 2021;75:435–438. doi: 10.1016/j.jhep.2021.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang N., Pérez P., Kato T., Mikami Y., Okuda K., Gilmore R.C., Conde C.D., Gasmi B., Stein S., Beach M., et al. SARS-CoV-2 infection of the oral cavity and saliva. Nat. Med. 2021;27:892–903. doi: 10.1038/s41591-021-01296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghinai I., McPherson T.D., Hunter J.C., Kirking H.L., Christiansen D., Joshi K., Rubin R., Morales-Estrada S., Black S.R., Pacilli M., et al. First known person-to-person transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the USA. Lancet. 2020;395:1137–1144. doi: 10.1016/s0140-6736(20)30607-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pung R., Chiew C.J., Young B.E., Chin S., Chen M.I., Clapham H.E., Cook A.R., Maurer-Stroh S., Toh M., Poh C., et al. Investigation of three clusters of COVID-19 in Singapore: implications for surveillance and response measures. Lancet. 2020;395:1039–1046. doi: 10.1016/s0140-6736(20)30528-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamner L., Dubbel P., Capron I., Ross A., Jordan A., Lee J., Lynn J., Ball A., Narwal S., Russell S., et al. High SARS-CoV-2 attack rate following exposure at a choir practice - Skagit County, Washington, March 2020. MMWR Morb Mortal Wkly Rep. 2020;69:606–610. doi: 10.15585/mmwr.mm6919e6. [DOI] [PubMed] [Google Scholar]
- 27.Amorim Dos Santos J., Normando A.G.C., Carvalho da Silva R.L., Acevedo A.C., De Luca Canto G., Sugaya N., Santos-Silva A.R., Guerra E.N.S. Oral manifestations in patients with COVID-19: a living systematic review. J. Dent Res. 2021;100:141–154. doi: 10.1177/0022034520957289. [DOI] [PubMed] [Google Scholar]
- 28.Becker M., Dulovic A., Junker D., Ruetalo N., Kaiser P.D., Pinilla Y.T., Heinzel C., Haering J., Traenkle B., Wagner T.R., et al. Immune response to SARS-CoV-2 variants of concern in vaccinated individuals. Nat. Commun. 2021;12:3109. doi: 10.1038/s41467-021-23473-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marais G., Hsiao N.-Y., Iranzadeh A., Doolabh D., Enoch A., Chu C.-Y., et al. 2021. [DOI]
- 30.Bergman P., Blennow O., Hansson L., Mielke S., Nowak P., Chen P. Safety and efficacy of the mRNA BNT162b2 vaccine against SARS-CoV-2 in five groups of immunocompromised patients and healthy controls in a prospective open-label clinical trial. BioMedicine. 2021;74:103705. doi: 10.1016/j.ebiom.2021.103705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hober S., Hellström C., Olofsson J., Andersson E., Bergström S., Jernbom Falk A., Bayati S., Mravinacova S., Sjöberg R., Yousef J., et al. Systematic evaluation of SARS-CoV-2 antigens enables a highly specific and sensitive multiplex serological COVID-19 assay. Clin. Transl Immunol. 2021;10:e1312. doi: 10.1002/cti2.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alkharaan H., Bayati S., Hellström C., Aleman S., Olsson A., Lindahl K., Bogdanovic G., Healy K., Tsilingaridis G., De Palma P., et al. Persisting salivary IgG against SARS-CoV-2 at 9 months after mild COVID-19: a complementary approach to population surveys. J. Infect Dis. 2021;224:407–414. doi: 10.1093/infdis/jiab256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mravinacova S., Jönsson M., Christ W., Klingström J., Yousef J., Hellström C., Hedhammar M., Havervall S., Thålin C., Pin E., et al. A cell-free high throughput assay for assessment of SARS-CoV-2 neutralizing antibodies. N. Biotechnol. 2021;66:46–52. doi: 10.1016/j.nbt.2021.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Higgins V., Fabros A., Kulasingam V. Quantitative measurement of anti-SARS-CoV-2 antibodies: analytical and clinical evaluation. J. Clin. Microbiol. 2021;59 doi: 10.1128/jcm.03149-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 36.Isho B., Abe K.T., Zuo M., Jamal A.J., Rathod B., Wang J.H., Li Z., Chao G., Rojas O.L., Bang Y.M., et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in patients with COVID-19. Sci. Immunol. 2020;5:eabe5511. doi: 10.1126/sciimmunol.abe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hearnden V., Sankar V., Hull K., Juras D.V., Greenberg M., Kerr A.R., Lockhart P.B., Patton L.L., Porter S., Thornhill M.H. New developments and opportunities in oral mucosal drug delivery for local and systemic disease. Adv. Drug Deliv. Rev. 2012;64:16–28. doi: 10.1016/j.addr.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 38.Dickinson B.L., Badizadegan K., Wu Z., Ahouse J.C., Zhu X., Simister N.E., Blumberg R.S., Lencer W.I. Bidirectional FcRn-dependent IgG transport in a polarized human intestinal epithelial cell line. J. Clin. Invest. 1999;104:903–911. doi: 10.1172/jci6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu Y., Sitaraman S., Gewirtz A.T. Intestinal epithelial cell regulation of mucosal inflammation. Immunol. Res. 2004;29:55–68. doi: 10.1385/ir:29:1-3:055. [DOI] [PubMed] [Google Scholar]
- 40.Boyarsky B.J., Werbel W.A., Avery R.K., Tobian A.A.R., Massie A.B., Segev D.L., Garonzik-Wang J.M. Antibody Response to 2-Dose SARS-CoV-2 mRNA vaccine series in solid organ transplant recipients. Jama. 2021;325:2204–2206. doi: 10.1001/jama.2021.7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benotmane I., Gautier G., Perrin P., Olagne J., Cognard N., Fafi-Kremer S., Caillard S. Antibody response after a third dose of the mRNA-1273 SARS-CoV-2 vaccine in kidney transplant recipients with minimal serologic response to 2 doses. Jama. 2021 doi: 10.1001/jama.2021.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blixt L., Bogdanovic G., Buggert M., Gao Y., Hober S., Healy K., Johansson H., Kjellander C., Mravinacova S., Muschiol S., et al. Covid-19 in patients with chronic lymphocytic leukemia: clinical outcome and B- and T-cell immunity during 13 months in consecutive patients. Leukemia. 2021:1–6. doi: 10.1038/s41375-021-01424-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sterlin D., Mathian A., Miyara M., Mohr A., Anna F., Claër L., Quentric P., Fadlallah J., Devilliers H., Ghillani P., et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Translational Med. 2021;13:eabd2223. doi: 10.1126/scitranslmed.abd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sahin U., Muik A., Vogler I., Derhovanessian E., Kranz L.M., Vormehr M., Quandt J., Bidmon N., Ulges A., Baum A., et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature. 2021;595:572–577. doi: 10.1038/s41586-021-03653-6. [DOI] [PubMed] [Google Scholar]
- 45.Team R.C. R Foundation for Statistical Computing; 2021. R: A Language and Environment for Statistical Computing. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data
De-identified patient data presented in this manuscript will be made available upon request from the lead author, in a format compliant with local regulatory requirements with respect to the handling of patient data, and in adherence with the policies of the Karolinska University Hospital and Karolinska Institutet.
Code
This paper does not report original code.
Any additional information required to analyze the data reported in this paper is available from the lead contact upon request.