Abstract
Background
Hypertension and proteinuria are common bevacizumab-induced toxicities. No validated biomarkers are available for identifying patients at risk of these toxicities.
Methods
A genome-wide association study (GWAS) meta-analysis was performed in 1039 bevacizumab-treated patients of European ancestry in four clinical trials (CALGB 40502, 40503, 80303, 90401). Grade ≥2 hypertension and proteinuria were recorded (CTCAE v.3.0). Single-nucleotide polymorphism (SNP)-toxicity associations were determined using a cause-specific Cox model adjusting for age and sex.
Results
The most significant SNP associated with hypertension with concordant effect in three out of the four studies (p-value <0.05 for each study) was rs6770663 (A > G) in KCNAB1, with the G allele increasing the risk of hypertension (p-value = 4.16 × 10−6). The effect of the G allele was replicated in ECOG-ACRIN E5103 in 582 patients (p-value = 0.005). The meta-analysis of all five studies for rs6770663 led to p-value = 7.73 × 10−8, close to genome-wide significance. The most significant SNP associated with proteinuria was rs339947 (C > A, between DNAH5 and TRIO), with the A allele increasing the risk of proteinuria (p-value = 1.58 × 10−7).
Conclusions
The results from the largest study of bevacizumab toxicity provide new markers of drug safety for further evaluations. SNP in KCNAB1 validated in an independent dataset provides evidence toward its clinical applicability to predict bevacizumab-induced hypertension.
ClinicalTrials.gov Identifier: NCT00785291 (CALGB 40502); NCT00601900 (CALGB 40503); NCT00088894 (CALGB 80303) and NCT00110214 (CALGB 90401).
Subject terms: Cancer genetics, Predictive markers
Background
Bevacizumab is a recombinant humanised monoclonal antibody that targets the vascular endothelial growth factor (VEGF), inhibiting binding to its receptors VEGFR1 and VEGFR2 [1]. It is approved by the U.S. Food and Drug Administration (FDA) for the treatment of metastatic colorectal cancer, advanced nonsquamous lung cancer, metastatic renal cell carcinoma, recurrent glioblastoma, advanced cervical cancer and ovarian cancer and hepatocellular carcinoma [2]. The antitumour efficacy of bevacizumab relies on inhibition of the VEGF-signaling pathway, which is involved in endothelial survival, vascular permeability and therefore tumour angiogenesis [3].
Despite the demonstrated efficacy of bevacizumab in combination regimens, patients frequently experience toxicity that limits both the duration of therapy with bevacizumab and the efficacy of the regimen. The most frequent bevacizumab toxicities are hypertension and proteinuria, with a prevalence that varies in different studies (proteinuria 21–41%, hypertension 3–43%) [4–6]. Hypertension and proteinuria can be occasionally life threatening [5].
The underlying mechanisms of bevacizumab-induced hypertension and proteinuria are not well understood, but are postulated to involve nitric oxide (NO) inhibition and increased peripheral vascular resistance, renal dysfunction and glomerular damage by inhibition of VEGF produced by podocytes [4–6]. It is not clear whether the occurrence of proteinuria shares the same mechanism responsible for hypertension, or whether the kidney damage is a secondary effect.
Currently, there are no validated biomarkers to predict bevacizumab-induced hypertension or proteinuria. Previous genetic studies of bevacizumab-induced hypertension focused on a few single-nucleotide polymorphisms (SNPs) in genes of the VEGF pathway [7, 8], and only one genome-wide association study (GWAS) has been performed to date [9]. Despite the reported associations, none of the studies generated evidence that is robust enough to allow for utilisation of these markers in the clinic, mostly due to lack of replication of the signal across multiple studies.
We performed a GWAS meta-analysis of four randomised phase III clinical trials from the Cancer and Leukemia Group B (CALGB, now part of the Alliance for Clinical Trials in Oncology, Alliance) in cancer patients treated with bevacizumab. The primary aim of this study was to identify novel genes and genetic variants that showed a consistent association with either hypertension or proteinuria across the different studies. We also aimed to replicate SNPs associated with hypertension in another external study of patients treated with bevacizumab.
Materials and methods
Patients and phase III clinical trials
All four studies were randomised, either placebo-controlled versus bevacizumab (CALGB 80303, 40503 and 90401) or bevacizumab combined with different chemotherapies (CALGB 40502). Cancer patients included in each study and their respective treatment regimens are shown in Table 1. Two studies included only female patients (breast cancer, CALGB 40503 and 40502), one study included only male patients (prostate cancer, CALGB 90401), and one study included both female and male patients (pancreatic cancer, CALGB 80303). More details on patient eligibility, characteristics, stratifications and treatments can be found in the Supplementary File and in the individual publication of each trial [10–13].
Table 1.
Patients of genetically determined European ancestry treated with bevacizumab and analyzed in the GWAS.
| CALGB trial | 80303 n = 154 | 40503 n = 105 | 90401 n = 314 | 40502 n = 466 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Age | Mean (SD) | 64.4 (10.5) | 56.9 (11.7) | 68.4 (8.3) | 57.2 (10.7) | |||||
| Sex | Male | 90 | 0 | 314 | 0 | |||||
| Female | 64 | 105 | 0 | 466 | ||||||
| Cancer type | Advanced pancreatic cancer | Hormone receptor-positive, advanced-stage breast cancer | Metastatic castration-resistant prostate cancer | Recurrent or metastatic breast cancer | ||||||
| Treatment | Gemcitabine 1000 mg/m2 on days 1, 8 and 15 plus either placebo or bevacizumab 10 mg/kg on days 1 and 15 | Letrozole 2.5 mg orally/day plus either placebo or bevacizumab 15 mg/kg every 21 days | Docetaxel 75 mg/m2 in combination with prednisone 5 mg orally on day 1 plus either placebo or bevacizumab 15 mg/kg every 21 days | Paclitaxel 90 mg/m2 or nab-paclitaxel 150 mg/m2 or ixabepilone 16 mg/m2 on days 1, 8 and 15 plus bevacizumab 10 mg/kg on days 1 and 15 | ||||||
| Genotype platform | Illumina HumanHap550-Quad | Illumina Human OmniExpressExome-8 | Illumina HumanHap610-Quad | Illumina Human OmniExpressExome-8 | ||||||
| Grade ≥ 2 | Grade 3 | Grade ≥ 2 | Grade 3 | Grade ≥ 2 | Grade 3 | Grade ≥ 2 | Grade 3 | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Hypertension | (n, %) | 26 (16.9) | 19 (12.3) | 53 (50.5) | 29 (27.6) | 47 (15.0) | 22 (7.0) | 143 (30.7) | 50 (10.7) | |
| Proteinuria | (n, %) | 26 (16.9) | 9 (5.8) | 32 (30.5) | 10 (9.5) | 20 (6.4) | 5 (1.6) | – | ||
| Composite toxicity | (n, %) | 43 (27.9) | 26 (16.9) | 65 (61.9) | 34 (32.4) | 61 (19.4) | 26 (8.3) | – | ||
No grade 4 toxicity was reported. “–“ not available. Grade≥ 2: Recurrent or persistent (≥24 h) or symptomatic increase by >20 mm Hg (diastolic) or to >150/100 if previously within normal limits; monotherapy may be indicated. Grade 3: Requiring more than one drug or more intensive therapy than previously.
CALGB Cancer and Leukemia Group B, SD standard deviation.
Toxicity
Hypertension and proteinuria were recorded by the CALGB (Alliance) Statistics and Data Center per protocol [10–13] according to Common Toxicity Criteria for Adverse Events (CTCAE) version 3.0 (Table S1). These criteria are standardised and widely used in clinical trials. Composite toxicity is defined as the first occurrence of either hypertension or proteinuria or both. For CALGB 40502, data on proteinuria (and hence composite toxicity) were not available. In all trials, blood pressure was measured prior to bevacizumab treatment on day 1 of each cycle. Urine protein levels were measured prior to bevacizumab treatment on day 1 of the 1st cycle and every 4 weeks after starting treatment in CALGB 80303; prior to bevacizumab treatment on day 1 of the 1st cycle and up to 5 days before or prior to bevacizumab treatment on the day 1 of subsequent cycles in CALGB 40503; and within 48 h prior to every treatment cycle in CALGB 90401.
Only toxicities with an attribution of “possibly related” or higher were included, according to standardised criteria that are uniform across all study protocols. According to the National Cancer Institute (NCI) Guidelines [14], possibly related toxicities are those adverse events related to the investigational drug. Treatment-terminating events included death, disease progression, other toxicities defined per protocol, withdrawal of consent for treatment, loss to follow-up or unknown recorded events. The time to event was reconstructed manually for each patient in each study, as we did previously [15]. The time to event was calculated as the time from the first administration of bevacizumab to the first date of experiencing the toxicity of interest, or other treatment-terminating events, whichever occurred first.
According to the CTCAE version 3.0, grade 2 hypertension is defined as recurrent or persistent (≥24 h) or symptomatic increase by >20 mm Hg (diastolic) or >150/100, whereas grade 3 hypertension is defined as the same blood pressure levels as grade 2 but requiring more than one drug or more intensive therapy than previously administered (Table S1). Because grade 3 hypertension classification depends on the physician assessment and the patient response to anti-hypertensive therapy, we employed a cut-off of grade ≥2 for our analysis.
Genotyping and quality control
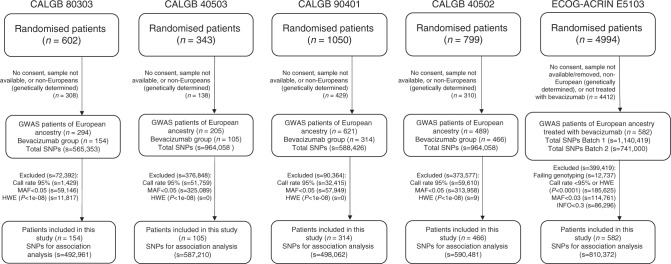
Germline DNA was obtained from peripheral blood. The genotyping platforms used in each study are described in Table 1, and the number of SNPs used for analysis after quality control (QC) are described in Fig. 1. Additional information on the QC procedures can be found in the individual publications of the GWAS data of CALGB 80303, 90401 and 40502 [16–18]. The genotyping and QC for CALGB 40503 has not been previously reported and it is described in the Supplementary File. Following QC, the total number of SNPs found in common among the four CALGB studies was 261,099. Each SNP that passed QC and has genotype data for at least one study was tested for association.
Fig. 1. CONSORT and quality control flowchart for CALGB 80303, 40503, 90401 and 40502, and ECOG-ACRIN E5103.
MAF minor allele frequency, HWE Hardy-Weinberg Equilibrium.
For additional GWAS study level QC, quantile–quantile (Q–Q) plots were generated (Fig. S1). In addition, scatter plots of minor allele frequencies (MAF) for the top fifty SNPs in each CALGB study and in Europeans (EUR) subjects from the 1000 Genomes project were generated (Figs. S2, S3, and S4).
Statistical analysis
The primary objective of the study was to test the association between SNPs and grade ≥2 toxicities in patients of genetically determined European ancestry treated with bevacizumab in each trial. A GWAS meta-analysis was performed including the results of the four trials, which have never been reported before.
For the principal component analysis, the genetic ancestral origin of patients was estimated based on clustering by Eigenstrat [19]. Following linkage disequilibrium (LD) pruning using an LD threshold of 0.2, pruned genotype data were compared to genotype data for the set of pruned variants from the 1000 Genomes Project Phase 3 samples, including individuals from ASW, CEU, CHB, MEX and YRI populations. Patients within each dataset that grouped with 1000 Genomes Project CEU samples based upon the first and second principal components were categorised as genetically determined European ancestry.
A cause-specific Cox model, where the outcome is defined as the pair of time to event and the censoring indicator, was fitted to obtain the estimate of the SNP effect (and the corresponding standard error estimate) on toxicity in each individual study. The analyses assumed an additive effect of each risk allele (additive genetic model) and were adjusted for age and sex. The inverse variance formula was used to combine the SNP effect in each study to obtain the meta-analysis estimate (β) of the SNP-toxicity association and its standard error. The heterogeneity across studies was examined by Cochran’s Q test and the reported SNPs are those with Cochran’s Q p-value > 0.20. The top fifty most statistically significant SNPs in the meta-analysis in LD with R2 > 0.10 based on 1000 Genomes Project Phase 3 CEU population and within 500 kb of distance were pruned. The adjusted (for age and sex) and also unadjusted results for the top fifty SNPs after pruning and their respective Cochran’s Q p-values are shown in Tables S2, S3, and S4. The reported p-values are not corrected for multiple comparisons and the cut-off for genome-wide significance is p-value < 5.00 × 10−8.
The meta-analysis p-values and β values have been used as the starting point to examine the performance in the sub-studies and conduct further analysis, including replication and bioinformatics. First, the SNPs were ordered according to the most statistically significant p-values of the meta-analysis and then the concordance of the effect was evaluated across the studies used in the meta-analysis. The SNPs that had the same direction of effect in at least three out of the four studies for hypertension (and two out of the three studies for proteinuria and composite toxicity) were investigated further for bioinformatic analysis. Codes used for analyses will be provided upon request.
Functional annotation of SNPs
Functional annotation of SNPs and genes was performed to describe our findings using the SCAN database [20]. SNPs were analyzed by LDlink for analyses of LD [21]. The UCSC Genome Browser (http://genome.ucsc.edu/) [22], RegulomeDB [23] and Haploreg v4 [24] were used for functional inference. The Genotype-Tissue Expression project (GTEx v7) [25] and NephQTL [26] were used for analyses of expression quantitative trait loci (eQTL).
Replication of associations with hypertension in ECOG-ACRIN E5103
Data from a GWAS of bevacizumab-induced hypertension from the Eastern Cooperative Oncology Group and the American College of Radiology Imaging Network (ECOG-ACRIN) 5103 trial was used as an independent replication set. The most statistically significant SNP in the meta-analysis with a concordant direction of effect and a p-value <0.05 in the three out of four studies for hypertension was tested for replication in ECOG-ACRIN E5103.
As a post-hoc, exploratory analysis performed during the revision process, the ten most statistically significant SNPs from the meta-analysis (Table S2) have been also tested for replication in ECOG-ACRIN E5103.
ECOG-ACRIN E5103 was an adjuvant study in breast cancer patients treated with doxorubicin and cyclophosphamide for four cycles, followed by four cycles of weekly paclitaxel. Concurrently to chemotherapy, patients were randomised to receive either placebo (arm A) or bevacizumab 10–15 mg/kg (arms B and C). Patients in arm C continued bevacizumab monotherapy (15 mg/kg every 3 weeks) for an additional 10 cycles. Hypertension was recorded according to either the Joint National Committee (JNC-7) or the CTCAE version 3.0 (Table S1). Bevacizumab-induced hypertension was defined as either the first occurrence of a systolic blood pressure (SBP) > 160 mm Hg (stage II of JNC-7) or as the first occurrence of hypertension grade ≥3 (CTCAE) after the start of treatment in arms B and C. Patients in arms B and C were used as controls (no hypertension) as defined upon meeting the following criteria: SBP < 140 mm Hg at baseline without use of an anti-hypertensive; maintained a median SBP < 140 mm Hg throughout therapy; did not meet any of the definitions of cases as outlined above; and received all prescribed doses of bevacizumab with follow-up for at least 3 months after the last dose. The treatment regimen and genotyping platforms are described in Table S5 and additional information on the QC procedures can be found in the GWAS publication [9].
The association between SNPs and hypertension in ECOG-ACRIN E5103 was tested in patients of genetically determined European ancestry (based upon Eigenstrat) and the number of SNPs used for analysis after QC are described in Fig. 1. A binary model with a standard case-control association was performed to identify SNPs associated with the presence or absence of hypertension (either SBP > 160 mm Hg or grade ≥3), where patients with SBP > 160 mm Hg (cases) and/or grade ≥3 hypertension were tested versus controls. Age and obesity (body mass index (BMI) > 30) were included as covariates. Because we do not have access to information on BMI for the CALGB studies, we have also performed an association analysis in ECOG-ACRIN E5103 using only age as a covariate.
For the SNP replicated in ECOG-ACRIN E5103, the inverse variance formula was used to combine the SNP effect in each of the four CALGB studies (for grade ≥2) and ECOG-ACRIN E5103 (for SBP > 160 mm Hg) to obtain the meta-analysis p-value, estimate (β) and the odds ratio (OR) of the association in the five studies overall.
Sex-stratified analysis
Because of the higher prevalence of bevacizumab-induced hypertension in female than male patients in CALGB studies (Table 1 and Table S6), we have also performed meta-analyses for the association between the SNP replicated in ECOG-ACRIN E5103 and grade ≥2 hypertension only in either female or male patients in the CALGB studies.
Results
A total of 1039 cancer patients of genetically determined European ancestry treated with bevacizumab were included in this study (Table 1). Grade ≥2 hypertension (15.0–50.5%) was more prevalent than grade ≥2 proteinuria (6.4–30.5%). The prevalence of grade ≥2 composite toxicity (first occurrence of either hypertension, proteinuria or both) was 19.4–61.9%. No grade 4 toxicity was reported. The prevalence of these toxicities was higher in females (breast cancer patients, CALGB 40502 and 40503) than in males (prostate cancer patients, CALGB 90401), and the same trend was also observed in the pancreatic cancer study (CALGB 80303) (Table S6).
Manhattan and Q–Q plots for the meta-analysis of SNPs association with toxicity are shown in Fig. S5.
SNPs associated with bevacizumab-induced hypertension
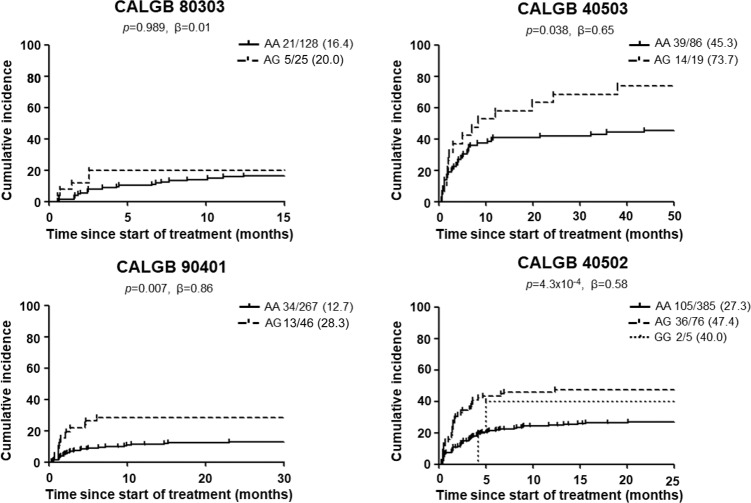
The ten most statistically significant SNPs that had the same direction of effect in at least three out of the four studies are shown in Table 2. None of the SNPs reached the genome-wide significance cut-off (p-value < 5.00 × 10−8). Variant rs13135230 (G > A, MAF 0.25–0.28), located 3.8 kb 3’ from LGI2, was the most statistically significant (p-value = 1.36 × 10−6, β = 0.47), with the A allele increasing the risk of hypertension. Two of these ten SNPs had a p-value < 0.05 and the same direction of effect in three out of the four studies (Table 2) and, out of these two SNPs, rs6770663 in KCNAB1 (Fig. S6) was the most statistically significant (p-value = 4.16 × 10−6, β = 0.58). The G allele of rs6770663 (A > G, MAF 0.08–0.09) was associated with an increased risk of hypertension (Fig. 2 and Fig. S7).
Table 2.
Top ten SNPs ranked by adjusted (for age and sex) p-value for association with grade ≥ 2 hypertension, proteinuria and composite toxicity with the same direction of effect (either reduced or increased risk) in at least two out of three trials for proteinuria and composite toxicity, and three out of four trials for hypertension.
| SNP (base change) | Ch | Gene | Feature | Flanking genes | MAF per study | Effect size (β) per study | p-value per study | Effect size (β) | SE | p-value adjusted | p-value unadjusted |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Hypertension | |||||||||||
| rs13135230 (G > A) | 4 | NA | NA | CCDC149/ LGI2 | 0.27/0.28/0.25/0.28 | 0.57/0.47/0.22/0.54 | 0.076/0.026/0.311/6.7 × 10−5 | 0.47 | 0.10 | 1.36 × 10−6 | 1.26 × 10−6 |
| rs6770663 (A > G) | 3 | KCNAB1 | Intron | LOC751837/SSR3 | 0.09/0.09/0.08/0.09 | 0.01/0.65/0.86/0.58 | 0.989/0.038/0.007/4.3 × 10−4 | 0.58 | 0.13 | 4.16 × 10−6 | 4.79 × 10−6 |
| rs4611262 (G > A) | 12 | NUP37 | Intron | CCDC53/C12orf48 | 0.20/0.25/0.23/0.23 | 0.47/0.21/0.23/0.51 | 0.220/0.369/0.302/3.1 × 10−5 | 0.40 | 0.09 | 1.59 × 10−5 | 3.74 × 10−5 |
| rs10519829 (G > A) | 5 | NA | NA | LOC644659/LOC100130551 | 0.44/0.46/0.47/0.47 | 0.16/0.62/0.36/0.33 | 0.562/0.003/0.083/0.004 | 0.37 | 0.09 | 1.84 × 10−5 | 2.28 × 10−5 |
| rs2000611 (A > G) | 14 | KCNK10 | Intron | KCNK10/SPATA7 | 0.44/0.43/0.50/0.48 | 0.08/−0.49/−0.59/−0.36 | 0.760/0.021/0.005/0.003 | −0.38 | 0.09 | 2.25 × 10−5 | 3.49 × 10−5 |
| rs1530837 (A > G) | 15 | PLA2G4E | Intron | EHD4/PLA2G4D | 0.23/0.19/0.18/0.17 | 0.55/0.19/0.62/0.42 | 0.107/0.444/0.004/0.004 | 0.44 | 0.10 | 2.28 × 10−5 | 2.59 × 10−5 |
| rs11133274 (G > A) | 4 | FIP1L1 | Intron | LOC100129913/CHURC1 | 0.21/0.20/0.21/0.20 | 0.41/0.28/0.52/0.41 | 0.239/0.199/0.021/0.002 | 0.41 | 0.10 | 2.46 × 10−5 | 7.50 ×10−5 |
| rs10842185 (A > G) | 12 | SOX5 | Intron | LOC100131418/LOC100129937 | 0.10/0.13/0.10/0.10 | 0.30/0.75/0.03/0.59 | 0.505/0.013/0.935/1.9 × 10−4 | 0.52 | 0.12 | 2.92 × 10−5 | 4.97 × 10−5 |
| rs1268663 (G > A) | 14 | SPTB | Intron | LOC100130982/LNX1 | 0.20/0.22/0.20/0.21 | 0.49/0.35/0.77/0.30 | 0.155/0.121/0.001/0.037 | 0.42 | 0.10 | 5.26 × 10−5 | 7.34 × 10−5 |
| rs17787479 (C > A) | 8 | NA | NA | COL22A1/KCNK9 | 0.21/0.26/0.20/0.20 | 0.63/0.33/0.56/0.34 | 0.114/0.125/0.011/0.015 | 0.40 | 0.10 | 5.88 × 10−5 | 4.68 × 10−5 |
| Proteinuria | |||||||||||
| rs339947 (C > A) | 5 | NA | NA | DNAH5/TRIO | 0.12/_/0.14/. | 1.09/_/1.42/. | 0.001/_/2.9 × 10−5/. | 1.26 | 0.24 | 1.58 × 10−7 | 7.66 × 10−8 |
| rs7959783 (A > G) | 12 | NA | NA | MDM/LOC729376 | 0.31/0.26/0.33/. | 0.66/0.66/0.94/. | 0.009/0.014/0.004/. | 0.73 | 0.16 | 6.56 × 10−6 | 7.01 × 10−6 |
| rs12612153 (A > C) | 2 | C2orf34 | Intron | PREPL/LOC100130502 | 0.11/_/0.11/. | 1.01/_/1.24/. | 0.011/_/1.8 × 10−4/. | 1.15 | 0.26 | 6.93 × 10−6 | 3.89 × 10−6 |
| rs4916009 (G > A) | 1 | JAK1 | Intron | RAVER2/LOC100130270 | 0.08/_/0.09/. | 0.99/_/1.37/. | 0.042/_/6.3 × 10−5/. | 1.24 | 0.28 | 8.86 × 10−6 | 2.10 × 10−5 |
| rs1509247 (A > G) | 4 | LOC643751 | Intron | GBA3/PPARGC1A | 0.43/0.43/0.52/. | 1.03/0.58/0.64/. | 0.001/0.014/0.075/. | 0.72 | 0.16 | 1.09 × 10−5 | 1.83 × 10−5 |
| rs10486445 (G > A) | 7 | OSBPL3 | Intron | DFNA5/CYCS | 0.42/0.35/0.39/. | 0.53/1.14/0.49/. | 0.114/2.6 × 10−5/0.134/. | 0.78 | 0.18 | 1.11 × 10−5 | 1.17 × 10−5 |
| rs17345945 (G > A) | 8 | CPA6 | Intron | LOC100132812/PREX2 | 0.09/_/0.08/. | 0.84/_/1.50/. | 0.034/_/6.4 × 10−5/. | 1.19 | 0.27 | 1.30 × 10−5 | 1.36 × 10−5 |
| rs627761 (A > G) | 11 | LOC338667 | Intron | DDX25/CDON | 0.10/_/0.09/. | 1.38/_/0.87/. | 8.0 × 10−5/_/0.039/. | 1.18 | 0.27 | 1.34 × 10−5 | 7.49 × 10−6 |
| rs12345440 (G > A) | 9 | RP11-35N6.1 | Intron | LOC392374/LOC347275 | 0.09/_/0.10/. | 0.89/_/1.50/. | 0.071/_/4.79 × 10−5/. | 1.28 | 0.30 | 1.45 × 10−5 | 1.89 × 10−5 |
| rs17624773 (G > A) | 12 | NA | NA | ASSP14/SYT10 | 0.10/0.06/0.14/. | 1.44/0.68/0.82/. | 4.8 × 10−4/0.138/0.013/. | 0.97 | 0.22 | 1.59 × 10−5 | 1.79 × 10−5 |
| Composite toxicity | |||||||||||
| rs16945809 (A > G) | 17 | YWHAE | Intron | TUSC5/CRK | 0.08/0.08/0.06/. | 1.26/0.82/0.88/. | 0.003/0.007/0.009/. | 0.94 | 0.20 | 2.16 × 10−6 | 2.00 × 10−5 |
| rs7038808 (G > A) | 9 | NA | NA | LOC286370/OR7E31P | 0.13/_/0.15/. | 1.19/_/0.77/. | 0.006/_/1.6 × 10−4/. | 0.85 | 0.18 | 4.55 × 10−6 | 9.17 × 10−5 |
| rs1564470 (C > A) | 8 | NA | NA | ZFAT/LOC286094 | 0.30/_/0.32/. | 0.72/_/0.76/. | 0.018/_/9.8 × 10−5/. | 0.75 | 0.16 | 5.09 × 10−6 | 6.02 × 10−5 |
| rs7204266 (A > G) | 16 | GRIN2A | Intron | LOC653737/LOC727844 | 0.28/_/0.28/. | 0.69/_/0.69/. | 0.027/_/8.9 × 10−5/. | 0.69 | 0.15 | 6.84 × 10−6 | 1.44 × 10−5 |
| rs222151 (G > A) | 21 | NA | NA | APP/CYYR1 | 0.25/0.23/0.22/. | −0.73/−0.77/−0.69/. | 0.063/0.002/0.010/. | −0.73 | 0.17 | 9.10 × 10−6 | 9.49 × 10−5 |
| rs1000032 (C > A) | 2 | NA | NA | KIAA1715/EVX2 | 0.13/0.18/0.19/. | 0.46/0.60/0.70/. | 0.235/0.005/0.001/. | 0.63 | 0.14 | 9.26 × 10−6 | 3.21 × 10−6 |
| rs7821773 (A > G) | 8 | NA | NA | TNKS/LINC00599 | 0.08/0.05/0.08/. | 0.19/1.08/0.73/. | 0.703/2.8 × 10−4/0.006/. | 0.80 | 0.19 | 1.75 × 10−5 | 5.98 × 10−5 |
| rs461409 (G > A) | 5 | NA | NA | LIX/RGMB-AS1 | 0.31/0.30/0.32/. | 0.49/0.36/0.71/. | 0.085/0.077/2.4 × 10−4/. | 0.54 | 0.13 | 2.16 × 10−5 | 2.32 × 10−4 |
| rs693996 (G > A) | 11 | OR8U8 | Intron | OR5M8/OR5M11 | 0.34/_/0.38/. | 0.88/_/0.65/. | 0.007/_/0.001/. | 0.71 | 0.17 | 2.16 × 10−5 | 7.58 × 10−5 |
| rs8006648 (G > A) | 14 | NA | NA | LOC730121/GALC | 0.27/0.28/0.26/. | 0.84/0.56/0.43/. | 0.013/0.004/0.41/. | 0.55 | 0.13 | 2.87 × 10−5 | 2.33 × 10−6 |
SNPs in LD with R2 > 0.10 within 500 kb distance were pruned, Rows in bold are for SNPs with p-value < 0.05 in at least two out of three trials for proteinuria or composite toxicity, or three out of four trials for hypertension. “_” SNP not present in the genotype platform of the trial, “.” data on proteinuria and composite toxicity were not available. MAF, Effect size (β) and p-value per study: in CALGB 80303, 40503, 90401 and 40502, respectively.
Ch chromosome, NA Intergenic SNP, MAF minor allele frequencies, SE standard error.
Fig. 2. Cumulative incidence of grade ≥ 2 hypertension for rs6770663 in KCNAB1.
The numbers next to each genotype correspond to the number of patients with hypertension grade ≥2 / total patients (%). One patient in CALGB 80303 and one patient in CALGB 90401 have the GG genotype of rs6770663 and did not report bevacizumab-induced grade ≥ 2 hypertension.
SNPs associated with bevacizumab-induced proteinuria
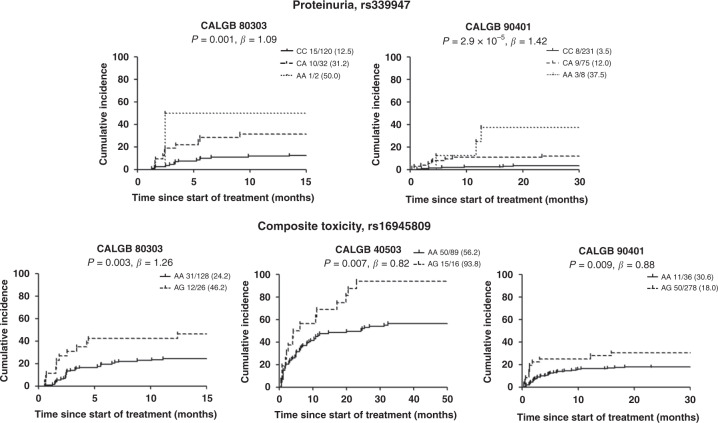
The ten most statistically significant SNPs that had the same direction of effect in at least two out of the three studies are shown in Table 2. None of the SNPs reached the genome-wide significance cut-off (p-value < 5.00 × 10−8). Variant rs339947 (C > A, MAF 0.12–0.14), intergenic (73 kb 5’ from DNAH5 and 130 kb 5’ from TRIO, Fig. S6), was the most statistically significant (p-value = 1.58 × 10−7, β = 1.26), with the A allele increasing the risk of proteinuria (Fig. 3 and Fig. S7). Eight of these ten variants had a p-value < 0.05 and the same direction of effect in at least two studies (Table 2).
Fig. 3. Cumulative incidence of grade ≥ 2 proteinuria for rs339947 between DNAH5 and TRIO and grade ≥ 2 composite toxicity for rs16945809 in YWHAE.
rs339947 was not present in the genotype platform of CALGB 40503. The numbers next to each genotype correspond to the number of patients with hypertension grade ≥2 / total patients (%).
SNPs associated with bevacizumab-induced composite toxicity
The ten most statistically significant SNPs that had the same direction of effect in at least two out of the three studies are shown in Table 2. None of the SNPs reached the genome-wide significance cut-off (p-value < 5.00 × 10−8). Variant rs16945809 in YWHAE (A > G, MAF 0.06–0.08, Fig. S6) was the most statistically significant (p-value = 2.16 × 10−6, β = 0.94), with the A allele increasing the risk of composite toxicity (Fig. 2 and Fig. S7). Nine of these ten variants had a p-value < 0.05 and the same direction of effect in at least two studies (Table 2).
Replication of rs6770663 in KCNAB1 for hypertension in ECOG-ACRIN E5103
One SNP associated with grade ≥2 hypertension in our study, rs6770663 in KCNAB1, was evaluated for replication in patients of genetically determined European ancestry from ECOG-ACRIN E5103. This variant was evaluated because it was among the top ten most statistically significant SNPs and also was the most statistically significant SNP that had a concordant effect in three out of the four studies (p-value < 0.05 for each study) (Table 2). The G allele of rs6770663 (A > G, MAF 0.10) was associated with a higher risk of SBP > 160 mm Hg (OR = 1.76, p-value = 0.005) in ECOG-ACRIN E5103 (Table S7), similar to the increased risk of grade ≥2 hypertension in the CALGB studies (p-value = 4.16 × 10−6, β = 0.58) (Table 2). For grade ≥3 hypertension in ECOG-ACRIN E5103, the increased risk (OR = 1.32) conferred by the G allele of rs6770663 did not reach statistical significance (p-value = 0.213).
In the association analysis in ECOG-ACRIN E5103 using only age as a covariate, the G allele of rs6770663 (A > G) was associated with a higher risk of SBP > 160 mm Hg with OR = 1.83 and p-value =0.007. For grade ≥3 hypertension, the association did not reach statistical significance (OR = 1.35, p-value = 0.227).
In the meta-analysis of rs6770663 in CALGB studies and ECOG-ACRIN E5103, the G allele of rs6770663 (A > G) was associated with a higher risk of bevacizumab-induced hypertension with β = 0.59, OR = 1.81, and p-value = 7.73 × 10−8 adjusted for age and sex (CALGB studies) and age and BMI (ECOG-ACRIN E5103), and β = 0.57, OR = 1.77, and p-value = 9.07 × 10−8 in an unadjusted analysis.
In the post-hoc, exploratory analysis (see Methods section), none of the ten most statistically significant SNPs from the meta-analysis were associated with SBP > 160 mm Hg in ECOG-ACRIN E5103 (p-value > 0.05, Table S8). In a similar post-hoc, exploratory analysis performed during the revision process, rs2000611 in KCNK10 was also tested, due to similarity in gene function between KCNAB1 (harboring rs6770663) and KCNK10. Variant rs2000611 in KCNK10 was not associated with SBP > 160 mm Hg in ECOG-ACRIN E5103 (OR = 0.85, 95% CI 0.66–1.08, p-value = 0.382).
Sex-stratified analysis for rs6770663 in KCNAB1
The meta-analyses for the association between rs6770663 in KCNAB1 and grade ≥2 hypertension only in either female (n = 661) or male (n = 378) patients in the CALGB studies showed that the G allele of KCNAB1 (A > G) was associated with an increased risk of grade ≥2 hypertension in both female (p-value = 2.0 × 10−4, β = 0.53) and male (p-value = 0.0025, β = 0.87) patients.
Discussion
To the best of our knowledge, this is the largest GWAS meta-analysis of bevacizumab-induced hypertension and proteinuria, which included 1039 cancer patients from four randomised phase III clinical trials conducted in a single cancer network. The relatively large sample size, the randomised design, and the standardised collection of the phenotypic and genotypic data improve the validity of the results from testing genetic associations of drug response in patients. The use of genomic data from different studies allows the evaluation of the concordance of the effect, increasing the reproducibility of these associations. Further confirmation of a variant in KCNAB1 for bevacizumab-induced hypertension from an independent, external dataset provides even stronger evidence toward clinical actionability.
We have established evidence of replication of the effect of rs6770663 (A > G), intronic in KCNAB1, as a variant for the risk of hypertension (Fig. 2). Similar to the CALGB studies, where it was associated with an increased risk of grade ≥2 hypertension (p-value = 4.16 × 10−6), rs6770663 was also associated with a higher risk of SBP > 160 mm Hg in ECOG-ACRIN E5103 (p-value = 0.005). Moreover, the association between rs6770663 and bevacizumab-induced hypertension was close to the genome-wide significance cut-off (p-value = 7.73 × 10−8) in the meta-analysis of the CALGB studies and ECOG-ACRIN E5103. In ECOG-ACRIN E5103, rs6770663 showed the same direction of effect, increasing the risk of grade ≥3 hypertension, but without reaching statistical significance. Grade 3 hypertension is based upon a physician’s assessment of the need for therapeutic intervention as well as patient response to the anti-hypertensive treatment. This might explain the lack of statistical association with grade 3 hypertension in ECOG-ACRIN E5103, as the classification of grade 3 hypertension is not as reliable as the classification for grade 2 [9].
The biology of KCNAB1 is consistent with the phenotype of bevacizumab-induced hypertension. KCNAB1 encodes the K+ voltage activated channel subfamily 1 subunit β1 (Kvβ1.1–1.3) and is highly expressed in arteries (aorta, coronary, tibial) [25] and endothelial cells [27]. This subunit affects the channel function and/or its localisation [28, 29]. The conductance of K+ is the main regulator of membrane potential in vascular smooth muscle and endothelial cells, and the deactivation or closure of the channel leads to vasoconstriction [30]. The function of K+ channels is altered in hypertension [30], and is significantly reduced in artery cells in hypertensive rats [31]. KCNAB1 has been shown to be important for the normal function of the heart, and a deletion in KCNAB1 was found in a patient who suffered sudden cardiac death [32]. When KCNAB1 was knocked out in female mice, elevated blood pressure and cardiac hypertrophy were observed [33]. Variant rs6770663 (A > G) is located in a region of H3K4me1 enhancers in the aorta (Table S9), showing a possible regulatory role. The serum-response factor (SRF) is more likely to bind the A allele than the G allele of rs6770663 [24, 34]. SRF is a cardiac-enriched transcription factor, which activates a variety of genes expressed in the heart and vascular smooth muscle cells [35], including KCNAB1 [36]. With this body of biological evidence, we postulate that patients who present a genetic predisposition for lower expression of KCNAB1 (mediated by reduced KCNAB1 activation by SRF) will have an impaired activation of K+ channels and increased vasoconstriction. When patients with the G allele of rs6770663 are treated with bevacizumab, rs6770663 might act as a genetic modifier that increases the risk of hypertension. Variant rs6770663 has a frequency of about 10% in Europeans and a global frequency of 30% [37], and it might impact a significant proportion of patients treated with bevacizumab.
This study also proposes DNAH5 and TRIO as novel candidate genes based on their association with the intergenic variant rs339947, the most statistically significant SNP associated with an increased risk of proteinuria (p-value = 1.58 × 10−7). DNAH5 (dynein axonemal heavy chain 5) is expressed in kidney and endothelial cells [25, 27] and encodes a dynein protein that is important for the normal function of ciliated cells in many tissues, including the kidney tubule. Mutations in DNAH5 cause a genetic disorder associated with polycystic kidney disease [38]. TRIO (trio rho guanine nucleotide exchange factor) is also expressed in kidney and endothelial cells [25, 27] and encodes a GDP to GTP exchange factor promoting the reorganisation of the actin cytoskeleton, thereby playing a role in cell migration and growth. TRIO is highly expressed in podocytes and regulates the attachment of podocytes to glomerular basement membrane [39]. TRIO induces Rac1 activity, which contributes to podocyte injury and proteinuria [39]. Because of these biological functions, DNAH5, TRIO, or both could be involved in the mechanism of bevacizumab-induced proteinuria. Interestingly, our analysis showed that rs429023, in LD R2 = 0.51 with rs339947, is an eQTL increasing TRIO expression in the glomerulus (Table S9), potentially pointing toward TRIO as the putative gene of proteinuria.
We observed a higher prevalence of bevacizumab-induced toxicities in female versus male patients (Table 1 and Table S6), as previously reported [40]. The bevacizumab label also indicates a lower drug clearance in females compared to males [2]. Whether female sex is a risk factor for bevacizumab-induced toxicities requires further evaluation. We have replicated the effect of rs6770663 (A > G) in KCNAB1 for bevacizumab-induced hypertension in an independent female cohort and it should also be validated in additional male cohorts, although our results already provide evidence of the effect of rs6770663 in both female and male patients.
This study has some limitations. We performed a replication study only for hypertension, and we were not able to find existing, available GWAS datasets for proteinuria and more effort is required to replicate the findings for proteinuria. Only rs339947 (proteinuria, p-value = 1.58 × 10−7) and rs6770663 (hypertension p-value = 7.73 × 10−8 in the meta-analysis of all five studies) were close to the cut-off for genome-wide significance. However, signals not reaching the genome-wide cut-off for statistical significance could still be replicated across studies [41–43]. This study included four clinical trials that differ in tumour types, bevacizumab dosing, patient demographics and clinical characteristics. The selection of SNPs based on a Cochran’s Q p-value > 0.20 for heterogeneity minimises the contribution of confounding effects originating from these differences across studies. We consider the concordance of the genetic effect across different studies as a strength for associations that are likely to be more directly related to the pharmacological effects of bevacizumab and potentially refractory to the influences of other factors. We do not have access to data of anti-hypertensive therapies before treatment with bevacizumab, and we were not able to evaluate if patients with pre-existing (albeit controlled) essential hypertension have a higher risk of developing bevacizumab-induced hypertension. Lastly, we did not perform imputation to our data. This approach will be further applied to future studies aimed at discovering additional variants.
Despite the clinical use of bevacizumab for many years, hypertension and proteinuria still represent obstacles to full delivery of effective therapy and pose a threat to patient health and quality of life. This study has identified and validated rs6770663 in KCNAB1 as a novel marker of bevacizumab-induced hypertension that can be used to guide decisions on the risk assessment of female patients of European descent treated with bevacizumab. This marker should be further evaluated in patients of different ancestries than European. Additionally, we have identified TRIO and DNAH5 as novel candidate genes associated with proteinuria. Because hypertension and proteinuria are also shared by many other anti-angiogenesis drugs, the availability of these results in the public domain will expedite their translation into clinical application for other drugs with a similar mechanism of action.
Supplementary information
Supplementary Tables, Figures, Methods and References
Acknowledgements
We acknowledge the PI of ECOG-ACRIN E5103, Kathy Miller, MD.
Author contributions
JCFQ, ASE and FI wrote the manuscript; FI designed the research; JCFQ, JW, ABS, CJ, ASE, FS, FM, JNP, DLH, ECD, HLM, MB, HR, HLK, WKK, MJR, DLK, KO, BS, DL and F.I. performed the research; JCFQ, JW, ABS, CJ, ASE, GJ, KO and FI analyzed the data; JW, ABS, CJ, ASE and DL contributed new reagents/analytical tools.
Funding
This work was supported by the National Cancer Institute of the National Institutes of Health under Award Numbers U10CA180821 and U10CA180882 (to the Alliance for Clinical Trials in Oncology), U24CA196171, UG1CA233253, UG1CA233327 and UG1CA233373. JCFQ was supported by the São Paulo Research Foundation-FAPESP (2018/04491-2). Also supported in part by funds from Abraxis BioScience, Bristol Meyers Squibb, Celgene and Genentech. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. https://acknowledgments.alliancefound.org
Data availability
The datasets generated during and/or analysed during the current study are available in the NHGRI-EBI GWAS Catalog under study accession numbers GCST90026609, GCST90026610 and GCST90026611.
Ethics approval and consent to participate
All participants provided written informed consent for sample collection and pharmacogenetic analysis, and all trials were conducted in accordance with recognised ethical guidelines. The study was performed in accordance with the Declaration of Helsinki and was approved by the local IRB.
Consent for publication
Not applicable.
Competing interests
JCFQ, JW, DL, KO and FI are coinventors of a patent application, serial number 16/932,002. FI is an advisor for Emerald Lake Safety. These relationships have been disclosed to and are under management by UNC-Chapel Hill.
Footnotes
The original online version of this article was revised: Due to an error in an affiliation.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
12/1/2021
A Correction to this paper has been published: 10.1038/s41416-021-01617-1
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-021-01557-w.
References
- 1.Ferrara N, Adamis AP. Ten years of anti-vascular endothelial growth factor therapy. Nat Rev Drug Disov. 2016;15:385–403. doi: 10.1038/nrd.2015.17. [DOI] [PubMed] [Google Scholar]
- 2.FDA AVASTIN® Prescribing Information. 2020. https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/125085s332lbl.pdf. Accessed on 05 Oct 2020.
- 3.Ellis LM, Kirkpatrick P. Bevacizumab. Nat Rev Drug Discov. 2005;3:995–996. doi: 10.1038/nrd1601. [DOI] [PubMed] [Google Scholar]
- 4.Zhu X, Wu S, Dahut WL, Parikh CR. Risks of proteinuria and hypertension with bevacizumab, an antibody against vascular endothelial growth factor: systematic review and meta-analysis. Am J Kidney Dis. 2007;49:186–93. doi: 10.1053/j.ajkd.2006.11.039. [DOI] [PubMed] [Google Scholar]
- 5.Izzedine H, Ederhy S, Goldwasser F, Soria JC, Milano G, Cohen A, et al. Management of hypertension in angiogenesis inhibitor-treated patients. Ann Oncol. 2009;20:807–15. doi: 10.1093/annonc/mdn713. [DOI] [PubMed] [Google Scholar]
- 6.Mir O, Coriat R, Cabanes L, Ropert S, Billemont B, Alexandre J, et al. An observational study of bevacizumab-induced hypertension as a clinical biomarker of antitumor activity. Oncologist. 2011;16:1325–32. doi: 10.1634/theoncologist.2010-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morita S, Uehara K, Nakayama G, Shibata T, Oguri T, Inada-Inoue M, et al. Association between bevacizumab-related hypertension and vascular endothelial growth factor (VEGF) gene polymorphisms in Japanese patients with metastatic colorectal cancer. Cancer Chemother Pharmacol. 2013;71:405–11. doi: 10.1007/s00280-012-2028-2. [DOI] [PubMed] [Google Scholar]
- 8.Schneider BP, Wang MM, Radovich M, Sledge GW, Badve S, Thor A, et al. Association of vascular endothelial growth factor and vascular endothelial growth factor receptor-2 genetic polymorphisms with outcome in a trial of paclitaxel compared with paclitaxel plus bevacizumab in advanced breast cancer: ECOG 2100. J Clin Oncol. 2008;28:4672–4678. doi: 10.1200/JCO.2008.16.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneider BP, Li L, Shen F, Miller KD, Radovich M, O’Neill A, et al. Genetic variant predicts bevacizumab-induced hypertension in ECOG-5103 and ECOG-2100. Br J Cancer. 2014;111:1241–1248. doi: 10.1038/bjc.2014.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kindler HL, Niedzwiecki D, Hollis D, Sutherland S, Schrag D, Hurwitz H, et al. Gemcitabine plus bevacizumab compared with gemcitabine plus placebo in patients with advanced pancreatic cancer: Phase III trial of the Cancer and Leukemia Group B (CALGB 80303) J Clin Oncol. 2010;28:3617–22. doi: 10.1200/JCO.2010.28.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickler MN, Barry WT, Cirrincione CT, Ellis MJ, Moynahan ME, Innocenti F, et al. Phase III trial evaluating letrozole as first-line endocrine therapy with or without bevacizumab for the treatment of postmenopausal women with hormone receptor-positive advanced-stage breast cancer: CALGB 40503 (Alliance) J Clin Oncol. 2016;34:2602–2609. doi: 10.1200/JCO.2015.66.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly WK, Halabi S, Carducci M, George D, Mahoney JF, Stadler WM, et al. Randomized, double-blind, placebo-controlled phase III trial comparing docetaxel and prednisone with or without bevacizumab in men with metastatic castration-resistant prostate cancer: CALGB 90401. J Clin Oncol. 2012;30:1534–40. doi: 10.1200/JCO.2011.39.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rugo HS, Barry WT, Moreno-Aspitia A, Lyss AP, Cirrincione C, Leung E, et al. Randomized phase III trial of paclitaxel once per week compared with nanoparticle albumin-bound nab-paclitaxel once per week or ixabepilone with bevacizumab as first-line chemotherapy for locally recurrent or metastatic breast cancer: CALGB 40502/NCCTG N0. J Clin Oncol. 2015;33:2361–2369. doi: 10.1200/JCO.2014.59.5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Institute of Cancer (NCI) Guidelines for Investigators. 2021. https://ctep.cancer.gov/protocoldevelopment/electronic_applications/docs/aeguidelines.pdf. Accessed on 08 Jan 2021.
- 15.Innocenti F, Jiang C, Sibley AB, Denning S, Etheridge AS, Watson D, et al. An initial genetic analysis of gemcitabine-induced high-grade neutropenia in pancreatic cancer patients in CALGB 80303 (Alliance) Pharmacogenet Genomics. 2019;29:123–31. doi: 10.1097/FPC.0000000000000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Innocenti F, Owzar K, Cox NL, Evans P, Kubo M, Zembutsu H, et al. A genome-wide association study of overall survival in pancreatic cancer patients treated with gemcitabine in CALGB 80303. Clin Cancer Res. 2012;18:577–84. doi: 10.1158/1078-0432.CCR-11-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rashkin SR, Chua KC, Ho C, Mulkey F, Jiang C, Mushiroda T, et al. A pharmacogenetic prediction model of progression-free survival in breast cancer using genome-wide genotyping data from CALGB 40502 (Alliance) Clin Pharmacol Ther. 2019;105:738–45. doi: 10.1002/cpt.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hertz DL, Owzar K, Lessans S, Wing C, Jiang C, Kelly WK, et al. Pharmacogenetic discovery in CALGB (alliance) 90401 and mechanistic validation of a VAC14 polymorphism that increases risk of docetaxel-induced neuropathy. Clin Cancer Res. 2016;22:4890–4900. doi: 10.1158/1078-0432.CCR-15-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 20.Zhang W, et al. SCAN database: Facilitating integrative analyses of cytosine modification and expression QTL. Database. 2015;27:bav025. doi: 10.1093/database/bav025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Machiela MJ, Chanock SJ. LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.James Kent W, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward LD, Kellis M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gillies CE, Putler R, Menon R, Otto E, Yasutake K, Nair V, et al. An eQTL landscape of kidney tissue in human nephrotic syndrome. Am J Hum Genet. 2018;103:232–444. doi: 10.1016/j.ajhg.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 28.González C, Baez-Nieto D, Valencia I, Oyarzún I, Rojas P, Naranjo D, et al. K+ channels: Function-structural overview. Compr Physiol. 2012;2:2087–2149. doi: 10.1002/cphy.c110047. [DOI] [PubMed] [Google Scholar]
- 29.Tipparaju SM, Liu SQ, Barski OA, Bhatnagar A. NADPH binding to β-subunit regulates inactivation of voltage-gated K+ channels. Biochem Biophys Res Commun. 2007;359:269–276. doi: 10.1016/j.bbrc.2007.05.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sobey CG. Potassium channel function in vascular disease. Arterioscler Thromb Vasc Biol. 2001;21:28–38. doi: 10.1161/01.ATV.21.1.28. [DOI] [PubMed] [Google Scholar]
- 31.Martens JR, Gelband CH. Alterations in rat interlobar artery membrane potential and K+ channels in genetic and nongenetic hypertension. Circ Res. 1996;79:295–301. doi: 10.1161/01.RES.79.2.295. [DOI] [PubMed] [Google Scholar]
- 32.Banerjee B, Peiris DN, Koo SH, Chui P, Lee EJD, Hande MP. Genomic imbalances in key ion channel genes and telomere shortening in sudden cardiac death victims. Cytogenet Genome Res. 2009;122:350–355. doi: 10.1159/000167822. [DOI] [PubMed] [Google Scholar]
- 33.Tur J, Chapalamadugu KC, Padawer T, Badole SL, Kilfoil PJ, Bhatnagar A, et al. Deletion of Kvβ1.1 subunit leads to electrical and haemodynamic changes causing cardiac hypertrophy in female murine hearts. Exp Physiol. 2016;101:494–508. doi: 10.1113/EP085405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kulakovskiy IV, Vorontsov IE, Yevshin IS, Sharipov RN, Fedorova AD, Rumynskiy EI, et al. HOCOMOCO: Towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 2018;46:D252–D259. doi: 10.1093/nar/gkx1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niu Z, Li A, Zhang SX, Schwartz RJ. Serum response factor micromanaging cardiogenesis. Curr Opin Cell Biol. 2007;19:618–627. doi: 10.1016/j.ceb.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG, et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford). 2016;2016:baw100. [DOI] [PMC free article] [PubMed]
- 37.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leigh MW, Pittman JE, Carson JL, Ferkol TW, Dell SD, Davis SD, et al. Clinical and genetic aspects of primary ciliary dyskinesia/kartagener syndrome. Genet Med. 2009;11:473–487. doi: 10.1097/GIM.0b013e3181a53562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maier M, Baldwin C, Aoudjit L, Takano T. The role of trio, a rho guanine nucleotide exchange factor, in glomerular podocytes. Int J Mol Sci. 2018;19:E479. doi: 10.3390/ijms19020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brahmer JR, Dahlberg SE, Gray RJ, Schiller JH, Perry MC, Sandler A, et al. Sex differences in outcome with bevacizumab therapy: analysis of patients with advanced-stage non-small cell lung cancer treated with or without bevacizumab in combination with paclitaxel and carboplatin in the eastern cooperative oncology group trial 4599. J Thorac Oncol. 2011;6:1031–1038. doi: 10.1097/JTO.0b013e3181fa8efd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider BP, Li L, Radovich M, Shen F, Miller KD, Flockhart DA, et al. Genome-wide association studies for taxane-induced peripheral neuropathy in ECOG-5103 and ECOG-1199. Clin Cancer Res. 2015;21:5082–5091. doi: 10.1158/1078-0432.CCR-15-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baldwin RM, Owzar K, Zembutsu H, Chhibber A, Kubo M, Jiang C, et al. A genome-wide association study identifies novel loci for paclitaxel-induced sensory peripheral neuropathy in CALGB 40101. Clin Cancer Res. 2012;18:5099–5109. doi: 10.1158/1078-0432.CCR-12-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schirmer MA, Lüske CM, Roppel S, Schaudinn A, Zimmer C, Pflüger R, et al. Relevance of Sp binding site polymorphism in WWOX for treatment outcome in pancreatic cancer. J Natl Cancer Inst. 2016;108:djv387. doi: 10.1093/jnci/djv387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables, Figures, Methods and References
Data Availability Statement
The datasets generated during and/or analysed during the current study are available in the NHGRI-EBI GWAS Catalog under study accession numbers GCST90026609, GCST90026610 and GCST90026611.