Abstract
Ralstonia solanacearum is one of the most destructive pathogens worldwide. In the last 30 years, the molecular mechanisms at the origin of R. solanacearum pathogenicity have been studied in depth. However, the nutrition status of the pathogen once inside the plant has been poorly investigated. Yet, the pathogen needs substrates to sustain a fast‐enough growth, maintain its virulence and subvert the host immunity. This study aimed to explore in‐depth the xylem environment where the pathogen is abundant, and its trophic preferences. First, we determined the composition of tomato xylem sap, where fast multiplication of the pathogen occurs. Then, kinetic growth on single and mixtures of carbon sources in relation to this environment was performed to fully quantify growth. Finally, we calculated the concentration of available metabolites in the xylem sap flux to assess how much it can support bacterial growth in planta. Overall, the study underlines the adaptation of R. solanacearum to the xylem environment and the fact that the pathogen assimilates several substrates at the same time in media composed of several carbon sources. It also provides metrics on key physiological parameters governing the growth of this major pathogen, which will be instrumental in the future to better understand its metabolic behavior during infection.
Keywords: diauxic growth, metabolism, plant pathogen, putrescine, substrate yield, tomato
The lifecycle of the plant pathogen Ralstonia solanacearum, the causative agent of bacterial wilt, includes a central and critical phase of intense proliferation in plant xylem vessels. The nutritive sources sustaining such an intense and fast growth were not clearly identified so far. Through a combination of xylem sap quantitative metabolomics and the detailed monitoring of the pathogen growth kinetics in numerous minimal media complemented with single and mixtures of carbon substrates, we identified glutamine and asparagine as potential nutritional substrates that allow the rapid growth of the pathogen in vascular tissues.

1. INTRODUCTION
Ralstonia solanacearum is a plant pathogen responsible for the lethal vascular wilt disease of more than 200 plant species from 50 different families (Hayward, 1991; Safni et al., 2014), including economically important plants such as tomato, potato, eggplant, banana, ginger, custard apple, peanut, and eucalyptus (Genin, 2010). This group of phytopathogenic bacteria causes important economic losses in most countries affected by this pathogen. In addition, the host range expansion of this pathogen is constantly reported, highlighting its great adaptation potential. R. solanacearum is a soil‐borne bacterium, able to survive in the soil for years as a saprophytic bacterium (Van Elsas et al., 2000). It generally enters plant hosts through the roots from wounds, root tips, and secondary root emerging points. It then progressively invades the xylem vessels and disperses rapidly to the aerial part of the plant (stems and leaves) through the vascular system (Peeters et al., 2013; Plener et al., 2012). Wilting symptoms probably result from the extensive bacterial colonization of the xylem and the production of a large amount of exopolysaccharides (EPS) leading to the obstruction of the vessels and plant water transport (Ingel et al., 2021).
Many cellular functions associated with or directly contributing to R. solanacearum pathogenicity have been identified. Among them are the Type III Secretion System (T3SS; Peeters et al., 2013) and a quorum‐sensing regulatory system controlling the expression of multiple virulence factors (Ujita et al., 2019). There is increasing evidence that metabolism and virulence are closely linked in R. solanacearum, as in many other pathogens. First, cases of metabolic adaptation to the conditions encountered in the plant by the pathogen are documented (Lowe‐Power et al., 2018a; Peyraud et al., 2018; Plener et al., 2012). Second, there is evidence that effectors translocated into plant cells through T3SS can enable the pathogen to exploit host resources (Lowe‐Power et al., 2018b; Peyraud et al., 2016; Wu et al., 2019; Xian et al., 2020), such as modifying the plant metabolism to start or increase production of preferred pathogen's substrates. In addition, the metabolic cost of virulence was recently demonstrated by Peyraud et al. (2016), in which different mutants with a progressive loss of virulence (mutant eps, mutant xpsR, and mutant phcA) had corresponding increased growth rate. Once inside the plant, the pathogen needs nutrients to proliferate inside the host, but also to mobilize matter and energy for the production of essential virulence factors to subvert host immunity (Peyraud et al., 2016). The purpose of this study was to explore and understand the trophic preferences of R. solanacearum, the environment it experiences in the plant, and the impact of this environment on pathogen growth. We also attempted to address if xylem sap is rich enough to sustain the fast growth of the pathogen in the host.
The trophic preferences of R. solanacearum are a central issue that has been investigated using targeted genetics (Dalsing & Allena, 2014; Hamilton et al., 2021; Jacobs et al., 2012; Lowe‐Power et al., 2018a; 2018b; Plener et al., 2012; Xian et al., 2020), but few studies have addressed this issue from the perspective of bacterial physiology. Metabolic activity phenotyping was performed by Peyraud et al., (2016) and Yabuuchi et al., (1992). They found that 36 carbon sources were used by R. solanacearum. However, no kinetic experiments were performed (except for glutamate in Peyraud et al. (2016)). Velocity and biomass yield in relation to available substrates are also key parameters to explain the exponential growth usually observed in planta. In this study, we investigated the trophic behavior of the R. pseudosolanacearum strain GMI1000, a reference strain of the R. solanacearum species complex. We monitored strain GMI1000 growth in liquid cultures on several major sugars and amino acids. Kinetics and growth yield in relation to single substrates and a mixture of substrates were determined. This provided a broad picture of R. solanacearum trophic preferences and a prediction of the nutrient regime once in the plant. Analysis was focused on the plant xylem tissue, which is the main environment in which fast multiplication of R. solanacearum occurs. Only carbon assimilation was considered since carbon was assumed as the limiting substrate inside the xylem. Indeed, inorganic nitrogen sources (NH4, NO2, and NO3) are present in high quantities in the xylem or are directly supplied in non–limiting conditions by the carbon source (amino acids; Bialczyk et al., 2004; Dalsing et al., 2015; Zuluaga et al., 2013).
2. MATERIALS AND METHODS
2.1. Xylem sap extraction method
Xylem fluid was extracted from 4 weeks old (3 weeks after germination), non‐inoculated, healthy tomato plants (Solanum lycopersicum cv Marmande), grown in 30 × 30 cm trays filled with compost, and cultivated in a greenhouse. Twenty‐four hours before extraction, tomato plants were introduced in a climate‐controlled growth chamber (75% humidity, 12h/12h cycle of night/light, 28°C, drenched when needed).
The plants were drenched 30 minutes before the extraction. The stems were cut using a sharp scalpel, approximately 1.5 cm above cotyledons. The cut surfaces were rinsed using autoclaved distilled water to eliminate vegetal and cellular debris and avoid phloem contaminations. Samples were then collected in an Eppendorf tube on ice for approximately 30 minutes using a pipette. The samples from each tray composed of 16 plants were pooled into one sample which was then stored at −80°C, until NMR analysis. The experiment was performed on 10 tomato trays at two different periods.
2.2. NMR analysis of xylem sap
Organic molecules in the xylem sap were quantified by NMR. Samples were analyzed by 1D 1H NMR on the MetaToul analytics platform (UMR5504, UMR792, CNRS, INRAE, INSA 135 Avenue de Rangueil 31077 Toulouse Cedex 04, France), using the Bruker Avance 800 MHz. Each xylem sap sample was centrifuged to remove the residues, then placed in 3 mm NMR tubes. TSP‐d4 standard (Sodium 3‐(trimethylsilyl)(1‐13C,2H4)propanoate) was used as a reference. The samples were kept at a temperature of 280 K (6.85°C) all along the analysis. Resonances of metabolites were manually integrated using Bruker TopSpin software and the concentrations were calculated based on the number of equivalent protons for each integrated signal and on the TSP final concentration.
2.3. Bacterial strains and inoculum
The R. pseudosolanacearum strain used for this study is the wild‐type strain GMI1000 (Salanoubat et al., 2002). The bacteria were plated on Phi medium (with g L−1: Bactopeptone: 10; casamino acid: 1; yeast extract: 1), supplied with glucose 20% (5 g L−1) and TTC (0.05 g L−1) for 48 h at 28°C. Then, a colony of bacteria was grown in minimal medium (with g L−1: FeSO4, 7H2O: 1.25 × 10–4, (NH4)2, SO4: 0.5 MgSO4, 7H2O: 0.05; KH2PO4: 3.4) supplemented with glutamate (100 mM C). pH was adjusted to 6.5 using KOH. 250 mL shake flasks containing 50 mL of culture were used. Inocula were shaken at 180 rpm on an orbital shaker placed in an incubator at 28°C.
2.4. Microplate growth experiment
GMI1000 was grown in a 96‐well‐plate (GREINER 96 F‐BOTTOM) in minimal medium, each supplemented with different carbon sources. Growth for each carbon source was monitored in three different wells. Volume culture per well was 200 µL. OD measurement was performed at 600 nm every five minutes for 72 hours using a FLUOstar OMEGA reader. A triplicate blank was also monitored in each microplate experiment, to ensure no contamination of the medium as well as to monitor evaporation. Only growth kinetics, maximal OD, and the maximum specific growth rate of the bacteria were concluded from these experimentations.
2.5. Shake‐flasks growth experiments
GMI1000 was grown in 1 L shake flasks shaken at 180 rpm on an orbital shaker placed in an incubator at 28°C. The starting volume culture was 250 mL. The culture medium was the same minimal medium as the inocula supplemented with a different carbon source. Dynamic sampling was performed for monitoring biomass growth (optical density (OD) measurement at 600 nm using a spectrometer), substrate consumption (NMR analysis), and products excretion (NMR analysis, Bradford essay).
Sampling time was chosen according to the kinetic evolution with intensive sampling during the exponential growth phase. After OD measurement, the collected volume for each point was filtered with a 0.22 µm micro‐filter and conserved at −20°C for further analysis.
2.6. NMR analysis of culture filtrate
Small‐size organic metabolites from the liquid culture in the shake flask were quantified by NMR. Samples were analyzed by 1D 1H NMR on the MetaToul analytics platform (UMR5504, UMR792, CNRS, INRAE, INSA 135 Avenue de Rangueil 31077 Toulouse Cedex 04, France), using the Bruker Avance 500 MHz. The samples were kept all along the analysis at a temperature of 298°K or 280°K for culture supplemented with glucose. Note, TSP‐d4 standard (Sodium 3‐(trimethylsilyl)(1‐13C,2H4)propanoate) was used as a reference. Resonances of metabolites were manually integrated using Bruker TopSpin software and the concentrations were calculated based on the number of equivalent protons for each integrated signal and on the TSP final concentration.
2.7. Bradford assay
Proteins quantification of the culture filtrate was performed using the Bradford assay. 100 µL of Bradford 1X Dye reagent (Sigma‐Aldrich) was added to 100 µL sample and absorption was measured at 595 nm after 30 minutes of incubation in a FLUOstar OMEGA. Bovine serum albumin was used as a standard for the quantification.
2.8. µmax measurement
The maximum specific growth rate (µmax) was computed using linear regression of ln(OD600NM) data plotted against time using the following equation:
2.9. Yield measurements
Biomass yields were computed using a linear regression between biomass dry weight (DW) and substrate:
Conversion of OD at 600 nm to biomass in dry weight was obtained using calibration data from Peyraud et al., (2016).
Putrescine yields were estimated using a linear regression between putrescine and substrate data:
Proteins yields were computed similarly. For the mixture of a substrate, the linear regression was made against the sum of substrates (in mg).
2.10. Substrate, proteins, and putrescine assimilation/excretion fluxes measurements
To compute fluxes of assimilation or excretion, the following formula was used:
2.11. Estimation of xylem substrate fluxes
Xylem surface compared to stem surface (Sxylem/Sstem) was estimated to be 1.063% using Figure 7 of Planas‐Marquès et al. (2020) and ImageJ software. Tomato plant density was estimated to be 0.8 g mL−1. Plant fresh weight was estimated to be 10.3 ± 1.55 g from Gerlin et al. (2021a). Xylem volume was thus estimated to be 0.136 mL for a 4‐week old tomato plant using the following formulae:
Xylem flux of metabolites for a 4‐week old plant can thus be estimated, for each metabolite, using the following formulae:
With Qtranspi the transpiration rate (38.44 ± 19.78 mL day−1) and [Metabolite]xylem the concentration of a given metabolite in the xylem (see Table 1).
TABLE 1.
Xylem organic composition of 4 week‐old tomato plants cv Marmande
| Compound | Rep 1 (mM C) | Rep 2 (mM C) | Growth of GMI1000 |
|---|---|---|---|
| Glutamine | 4.25 ± 0.53 | 3.59 ± 1.27 | +++ |
| Proline | 0.81 ± 0.41 | 0.53 ± 0.09 | + |
| Asparagine | 0.52 ± 0.03 | 0.68 ± 0.2 | +++ |
| Valine | 0.74 ± 0.13 | 0.28 ± 0.07 | ‐ |
| Leucine | 0.72 ± 0.08 | 0.22 ± 0.07 | + |
| Isoleucine | 0.7 ± 0.13 | 0.24 ± 0.06 | ‐ |
| Arginine | 0.61 ± 0.23 | 0.16 ± 0.05 | ‐ |
| Threonine | 0.46 ± 0.12 | 0.17 ± 0.05 | ‐ |
| Tyrosine | 0.43 ± 0.06 | 0.12 ± 0.03 | ‐ |
| Phenylalanine | 0.32 ± 0.06 | 0.12 ± 0.02 | + |
| Lysine | 0.28 ± 0.08 | 0.14 ± 0.03 | + |
| Aspartate | 0.15 ± 0.1 | 0.08 ± 0.03 | +++ |
| Glucose | 0.32 ± 0.19 | 0.46 ± 0.17 | +++ |
| Sucrose | 0.54 ± 0.23 | 0.44 ± 0.04 | + |
| Ethanol | 0.33 ± 0.22 | 0.51 ± 0.44 | + |
| Total mM C | 11.18 ± 2.60 | 7.72 ± 2.60 | |
| Total mM N | 3.24 ± 0.62 | 2.25 ± 0.70 | |
| C/N ratio | 3.45 | 3.43 | |
| %AA | 89% | 82% | |
| Growth support mM C | 5.46 ± 0.89 | 4.78 ± 0.89 | |
| %Growth support | 49% | 62% | |
| C/N ratio support | 2.8 | 2.7 |
Abbreviations: ‐, cannot support growth as a carbon substrate+, can support a slow growth as a carbon substrate; +++, can support growth as a carbon substrate.
2.12. Conversion of CFU/gFW into OD600nm and mg/L of biomass
From Table S4 from Peyraud et al. (2016), we have access to conversion between CFU/mL and OD, and OD to mg/L of biomass, using the following formulae:
In addition, thanks to the estimation of the volume of the xylem, we can obtain conversion formulae between CFU/FW and CFU/mLxyl:
Finally, we can straightforwardly estimate a conversion between CFU/FW and OD or biomass concentration in mg/Lxyl using the formulae:
So, 109 CFU/gFW thus represents an OD of 17.9, which is equivalent to a biomass concentration of 7.41 g/L. 1010 CFU/gFW thus represents an OD of 179, which is equivalent to a biomass concentration of 74.1 g/L.
3. RESULTS
3.1. The trophic environment of strain GMI1000 in plant xylem
To be in the same condition as standard pathogenicity assays, we analyzed xylem sap of 4 week‐old tomato plants (Solanum lycopersicum cv Marmande) infected by strain GMI1000. The metabolic composition of xylem obtained from two independent experiments and the different concentrations for each compound were measured using NMR analysis (Table S1, for detailed experimental values, see Supplementary Data). Fourteen compounds were identified, with the presence of a vast majority of amino acids (11/14), whose concentration ranges between 0.08 and 4.25 mM of carbon (mM C). The other three compounds are sugar (sucrose and glucose), and ethanol. Concentrations of the substrate between the different samples differ quite a lot as illustrated by the high standard deviation values found, which represents on average 31% of the nominal value (Table 1). The dominant compound was always glutamine, which represents 38%–47% of total carbon content. Amino acids represent 82%–89% of the xylem content, and the average C/N ratio is around 3.4.
3.2. The growth rate in presence of single carbon sources
To understand the link between metabolites available in the environment and R. solanacearum growth in planta, we performed microplate growth cultures of GMI1000 in standard minimal medium supplemented with substrates potentially present in the xylem as carbon sources. The list of the compounds to test was chosen after an investigation of tomato xylem sap in literature (Bialczyk et al., 2004; Hiery et al., 2013; Lowe‐Power et al., 2018b; Senden et al., 1992; White et al., 1981; Zuluaga et al., 2013) and the previous results. Tested molecules include sugars, amino acids, and organic acids from the Krebs cycle.
Strain GMI1000 had fast growth on aspartate, glutamine, glutamate, malate, glucose, asparagine, and pyruvate as illustrated by their specific growth rate µmax above 0.1 h−1 (Table 2). GMI10000 had a slower growth on GABA, histidine, alanine, alpha keto‐glutarate, and galacturonate. Growth was very slow on the other sugars and amino acids such as galactose, sucrose, proline, phenylalanine with a µmax lower than 0.04 h−1. For all the other compounds, no growth was observed.
TABLE 2.
Maximum specific growth rates (µmax) in microplate experiments of GMI1000 for different carbon substrates
| Substrate | µmax (h−1) | Max OD (600nm) | Productivity in 24h (OD) |
|---|---|---|---|
| Aspartate a | 0.301 ± 0.003 | 0.64 ± 0.01 | 4.61 |
| Glutamine a | 0.184 ± 0.008 | 0.92 ± 0.05 | 4.06 |
| Glutamate | 0.156 ± 0.005 | 0.78 ± 0.03 | 2.92 |
| Malate a | 0.149 ± 0.001 | 0.40 ± 0.05 | 1.43 |
| Glucose a | 0.145 ± 0.001 | 1.45 ± 0.01 | 5.05 |
| Asparagine a | 0.104 ± 0.001 | 0.51 ± 0.01 | 1.27 |
| Pyruvate a | 0.100 ± 0.004 | 0.69 ± 0.04 | 1.66 |
| Gamma‐aminobutyrate | 0.074 ± 0.004 | 0.77 ± 0.04 | 1.37 |
| Histidine | 0.074 ± 0.002 | 0.60 ± 0.01 | 1.07 |
| Alanine a | 0.051 ± 0.002 | 0.27 ± 0.01 | 0.33 |
| Alpha‐ketoglutarate | 0.049 ± 0.001 | 0.33 ± 0.02 | 0.39 |
| Galacturonate | 0.049 ± 0.001 | ||
| Galactose | 0.034 ± 0.001 | ||
| Sucrose | 0.026 ± 0.001 | ||
| Gluconate | 0.026 ± 0.004 | ||
| Trehalose | 0.025 ± 0.002 | ||
| Saccharose | 0.024 ± 0.001 | ||
| Threonine | 0.023 ± 0.001 | ||
| Proline | 0.021 ± 0.002 | ||
| Fructose | 0.021 ± 0.001 | ||
| Mannitol | 0.020 ± 0.001 | ||
| Ethanol | 0.019 ± 0.001 | ||
| Phenylalanine | 0.017 ± 0.001 | ||
| Lysine | 0.011 ± 0.001 | ||
| Leucine | 0.011 ± 0.003 | ||
| Arginine | ‐ | ||
| Isoleucine | ‐ | ||
| Valine | ‐ | ||
| Serine | ‐ | ||
| Tyrosine | ‐ | ||
| Methionine | ‐ | ||
| Tryptophane | ‐ | ||
| Cysteine | ‐ | ||
| Glycine | ‐ | ||
| Fumarate | ‐ | ||
| Citrate | ‐ | ||
| Maleate | ‐ | ||
| Succinate | ‐ | ||
| Oxalate | ‐ | ||
| Xylose | ‐ | ||
| Arabinose | ‐ | ||
| Rhamnose | ‐ |
Max OD was determined for growth finished in <60 hours only. All experiments were performed with an initial carbon concentration of 100 mM C in a minimal medium.
Abbreviation: ‐, absence of growth.
Carbon sources used for shake‐flasks experiments.
All substrates used for the experiments have a normalized initial carbon concentration (100 mM C). This allowed comparing the final bacterial density (OD600) since the same quantity of carbon is available for each substrate. Among substrates for which a plateau phase was reached during growth (growth in less than 60 hours), glucose led to the highest biomass yield. This is probably because oxygen might be limited in such a growing system and glucose, when catabolized along Entner‐Doudoroff (Jyoti et al., 2020), generates ATP and NADH before reaching the TCA cycle, contrary to other substrates that need O2 to generate ATP using ATP synthetase. In terms of biomass productivity, glucose, even if it leads to a slower growth rate compared to other substrates, had better productivity since it has a better biomass yield. Glutamine led also to both a good biomass yield and a good growth rate. Glucose and glutamine thus seem to be preferential substrates for the growth of GMI1000. Aspartate, even if it leads to a good growth rate, results in a lower biomass yield, and thus appears less favorable for GM1000 growth.
On a growth/no‐growth basis, these results are in good agreement with BIOLOG results performed by Peyraud et al. (2016). Disagreement is for proline, phenylalanine, lysine, and leucine, for which we observed growth and there is none in BIOLOG microplates. Except for proline, growth is slow, which could explain the difference between the two experiments. There is also a discrepancy for serine, fumarate, and citrate, for which we observed no growth in our microplates experiment whereas growth is observed in BIOLOG microplates.
Asparagine growth is much slower than glutamine growth and has two exponential phases (second growth rate: and 0.011 ± 0.001 h−1). The presence of these two growth phases is dependent on the initial concentration of asparagine since it was observed for the initial concentration of 50 mM C or 100 mM C but not for 10 mM C (Figure 1).
FIGURE 1.
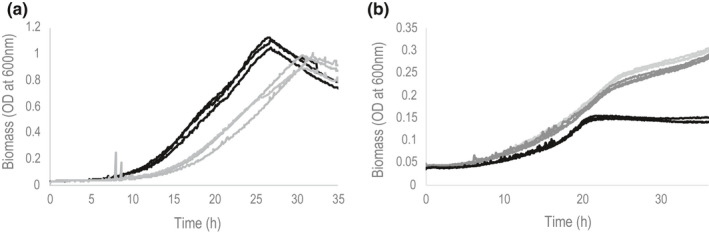
Growth curve of GMI1000 on glutamine (panel A, light grey curve), a mixture of glutamine and asparagine (left panel, black curve) and asparagine (panel B, 100 mM C light grey curve, 50 mM C dark grey curve, 10 mM C black curve)
3.3. Monitoring growth in carbon source mixtures
Diauxic growth is a phenomenon generally observed when microorganisms, in presence of two carbon sources (most of the time two sugars), exhibit a bi‐phasic exponential growth with a lag phase between each phase, which reflects the sequential use of each substrate (Monod, 1942). In a diauxic growth, only one substrate at a time is consumed in each phase. Hence, the maximum growth rate of each phase corresponds to the maximum growth rate of each substrate when the bacteria are grown alone. Diauxic growth in bacteria can thus reveal mechanisms of genetic repression of uptake of a secondary nutrient in the presence of another one that is preferentially metabolized. We tested mixtures of substrates on the growth of strain GMI1000 to know if such diauxic growth occurs (Table 3). When there was only one exponential growth phase observed (13 cases out of 32), there is no diauxic growth. When there is a second exponential phase, in 13 cases out of 32 cases, the growth rate of the first exponential phase is higher than the growth rate on each substrate alone. Thus, for these 13 mixtures, there is no diauxic growth. The second exponential phases are present because of the exhaustion of one substrate before the other. For the mixtures asparagine + aspartate, glucose + aspartate, and malate + aspartate, the growth rate is not as fast as on aspartate alone but is faster than with asparagine alone, malate alone, or glucose alone. So there is probably no diauxic growth for these three cases. Only in the cases of glutamate + asparagine, glutamate + galactose, and glutamine + galactose, there are two growth phases and a growth rate equal or lower than for glutamate (resp. glutamine) alone. In these three cases, catabolic repression might take place.
TABLE 3.
Maximum specific growth rates (µmax) in microplate experiments of GMI1000 for different mixtures of carbon substrates
| Substrates | µmax 1 (h−1) | µmax 2 (h−1) |
|---|---|---|
| Glutamine + Aspartate | 0.262 ± 0.007 | ‐ |
| Glutamine + Malate | 0.242 ± 0.005 | 0.055 ± 0.009 |
| Glutamine + Succinate | 0.230 ± 0.027 | ‐ |
| Glutamine + Asparagine a | 0.220 ± 0.008 | 0.094 ± 0.009 |
| Glutamine + Glucose a | 0.211 ± 0.010 | 0.062 ± 0.003 |
| Glutamine + Gamma‐aminobutyrate | 0.203 ± 0.008 | 0.036 ± 0.004 |
| Glutamine + Alanine | 0.199 ± 0.005 | 0.088 ± 0.001 |
| Glutamine + Histidine | 0.190 ± 0.031 | 0.039 ± 0.008 |
| Glutamine + Proline | 0.167 ± 0.005 | ‐ |
| Glutamine + Galactose | 0.161 ± 0.002 | 0.045 ± 0.003 |
| Glutamine + Alpha‐ketoglutarate | 0.090 ± 0.010 | ‐ |
| Asparagine + Alanine | 0.221 ± 0.022 | ‐ |
| Asparagine + Glucose a | 0.218 ± 0.005 | 0.092 ± 0.008 |
| Asparagine + Succinate | 0.213 ± 0.018 | 0.070 ± 0.005 |
| Asparagine + Malate | 0.210 ± 0.002 | 0.170 ± 0.004 |
| Asparagine + Alpha‐ketoglutarate | 0.184 ± 0.025 | 0.163 ± 0.020 |
| Asparagine + Aspartate | 0.179 ± 0.005 | 0.032 ± 0.002 |
| Asparagine + Proline | 0.148 ± 0.002 | 0.068 ± 0.003 |
| Asparagine + Galactose | 0.127 ± 0.002 | ‐ |
| Asparagine + Histidine | 0.108 ± 0.001 | 0.044 ± 0.001 |
| Aspartate + Glucose a | 0.207 ± 0.006 | 0.096 ± 0.013 |
| Aspartate + Malate | 0.249 ± 0.030 | 0.064 ± 0.004 |
| Casamino acid | 0.219 ± 0.006 | ‐ |
| Glutamate + Aspartate | 0.215 ± 0.013 | ‐ |
| Glutamate + Succinate | 0.175 ± 0.006 | ‐ |
| Glutamate + Alpha‐ketoglutarate | 0.163 ± 0.005 | ‐ |
| Glutamate + Glucose | 0.160 ± 0.008 | ‐ |
| Glutamate + Gamma‐aminobutyrate | 0.158 ± 0.006 | 0.036 ± 0.007 |
| Glutamate + Galactose | 0.154 ± 0.007 | 0.037 ± 0.002 |
| Glutamate + Proline | 0.151 ± 0.013 | ‐ |
| Glutamate + Asparagine | 0.148 ± 0.005 | 0.096 ± 0.002 |
| Glutamate + Alanine | 0.148 ± 0.007 | ‐ |
| Glutamate + Histidine | 0.139 ± 0.012 | ‐ |
All experiments were performed with a total initial carbon concentration of 100 mM C, 50 mM C for each substrate in the mixture.
Abbreviation: ‐, absence of second growth phase
Carbon sources used for shake‐flasks experiments.
Most mixtures of two substrates could sustain a rather fast growth, the highest growth rate being observed for glutamine and aspartate. This was not surprising given the fact that GMI1000 has fast growth on both substrates independently. Interestingly, growth in presence of casamino acids, a hydroxychloric acid hydrolysate of casein composed of many kinds of amino acids (Bonnemain, 1966), resulted in a growth rate of 0.22h−1. Yet this growth rate is lower than several mixtures of substrate tested such as glutamine + aspartate, glutamine + malate, glutamine + succinate, or aspartate + malate, showing that casamino‐acids are not the optimal substrate for growth.
Most of the time, mixtures of substrates sustained a faster growth than the best growth rate obtained with the two substrates alone, showing a synergistic effect of the presence of the two substrates (19/32). For example, this is the case for glutamine + asparagine (Figure 1). For other mixtures of substrates (13/32), the growth rate was lower than the best growth rate obtained on each substrate alone. For most of them (10/13), the growth rate was better than the mean growth rate of the two substrates alone. For example, this is the case for glutamine + proline. All mixtures containing aspartate also behave like this. Since aspartate sustains a very high growth rate of R. solanacearum, this result could be explained by the presence of two distinct populations, one specialized on each substrate, some internal regulation taking place, or that a resource allocation limit is reached, such as a limited space of transporters in the cell membrane. Finally, it is interesting to emphasize that we did not detect any significant growth of GMI1000 on succinate alone under the experimental conditions used, whereas a mutant deficient for the global virulence regulator phcA can (Peyraud et al., 2016), and mixtures of succinate with an amino acid yielded a higher growth rate compared to the amino acid alone. Succinate can therefore act as a complementary nutrition source, and the lack of growth cannot be explained by a lack of transporter.
3.4. Determining substrate assimilation rates, product excretion, and biomass yields
Microplate experiments gave first insights into trophic preferences of GMI1000. Yet, to have a more complete picture, in particular substrate consumption rates, we performed kinetic experiments in shaken flasks. Shake‐flasks experiments allowed having better oxygenation of the culture media compared to microplates. In addition, shake‐flasks experiments enabled to sample media at the different time point and analyze it using NMR technology, as well as to fully characterize substrate consumption and metabolite excretion. This allowed verifying that no catabolic repression occurs, with a fine tracking on substrate concentration over time. Eleven shake‐flask growth experiments were performed using single substrates and mixtures of substrates selected for their ability to sustain fairly rapid growth as carbon sources. From these experiments, maximal growth rates, putrescine yield, protein yield, and substrate yield were calculated (Table 4).
TABLE 4.
Maximum specific growth rates (µmax) in shake‐flasks experiments of GMI1000 for different carbon substrates
| Substrate(s) | µmax (h−1) | Substrate Yield (mgB/mgS) | Putrescine Yield (mg/mgS) | Protein yield (mg/mgS) | Substrate flux (mg/mgB/h) | Putrescine flux (mg/mgB/h) | Protein flux (mg/mgB/h) |
|---|---|---|---|---|---|---|---|
| Aspartate | 0.43 ± 0.005 | 0.32 ± 0.03 | 0.010 ± 0.0006 | 0.0027 ± 0.0002 | 1.33 ± 0.09 | 0.013 ± 0.0002 | 0.0036 ± 0.0004 |
| Malate | 0.30 ± 0.009 | 0.30 ± 0.04 | 0.015 ± 0.0024 | 0.0032 ± 0.0014 | 0.99 ± 0.11 | 0.015 ± 0.0006 | 0.0030 ± 0.0012 |
| Glutamine | 0.29 ± 0.002 | 0.53 ± 0.01 | 0.022 ± 0.0019 | 0.0053 ± 0.0002 | 0.53 ± 0.01 | 0.012 ± 0.0009 | 0.0029 ± 0.0002 |
| Glucose | 0.23 ± 0.001 | 0.45 ± 0.01 | 0.036 ± 0.0004 | 0.0080 ± 0.0007 | 0.50 ± 0.01 | 0.018 ± 0.0003 | 0.0040 ± 0.0003 |
| Pyruvate | 0.18 ± 0.002 | 0.38 ± 0.02 | 0.029 ± 0.0006 | 0.0026 ± 0.0022 | 0.48 ± 0.02 | 0.014 ± 0.0004 | 0.0013 ± 0.0011 |
| Asparagine |
0.13 ± 0.004, then 0.02 ± 0.001 |
0.23 ± 0.03 | 0.022 ± 0.0031 | 0.0014 ± 0.0003 | 0.54 ± 0.05 | 0.013 ± 0.0002 | 0.0008 ± 0.0001 |
| Alanine | 0.12 ± 0.004 | 0.32 ± 0.01 | 0.030 ± 0.0002 | 0.0048 ± 0.0001 | 0.34 ± 0.01 | 0.011 ± 0.0001 | 0.0017 ± 0.0001 |
| Aspartate + Glucose | 0.41 ± 0.012 then 0.20 ± 0.002 | 0.36 ± 0.01 | 0.017 ± 0.0006 | 0.0045 ± 0.0003 | 1.09 ± 0.06 | 0.018 ± 0.0014 | 0.0049 ± 0.0004 |
| Glutamine + Glucose | 0.40 ± 0.036 then 0.17 ± 0.002 | 0.43 ± 0.02 | 0.026 ± 0.0009 | 0.0080 ± 0.002 | 0.90 ± 0.1 | 0.023 ± 0.0026 | 0.0066 ± 0.0014 |
| Asparagine + Glucose | 0.39 ± 0.007 then 0.13 ± 0.008 | 0.41 ± 0.01 | 0.013 ± 0.0003 | 0.0041 ± 0.0002 | 0.94 ± 0.02 | 0.012 ± 0.0002 | 0.0038 ± 0.0002 |
| Glutamine + Asparagine | 0.30 ± 0.016 | 0.42 ± 0.03 | 0.015 ± 0.0004 | 0.0036 ± 0.0007 | 0.70 ± 0.03 | 0.011 ± 0.0006 | 0.0025 ± 0.0005 |
All experiments were performed with an initial carbon concentration of 100 mM C. Yield and fluxes were determined only for the first exponential phase if two exponential phases were present. For mixtures of a substrate, substrate yield and substrate fluxes were computed on the mass sum of both substrates.
GMI1000 growth kinetic observed was in accordance with classical bacterial growth curves (Figure 2, Figure 3), comprising a lag phase, an exponential phase, and a death (decline) phase. However, the stationary phase was not observed (or only rarely). For growth with asparagine, again we observed two exponential phases with an inflection point between them, as for microplate experiments (Figure 2B). All the growth rates were significantly higher in shake‐flask experiments than in microplate experiments, confirming that growth conditions are better in shake‐flasks, probably due to better oxygenation. Aspartate led to the best growth rate (0.43h−1), followed by glutamine and malate (0.29–0.30h−1), in accordance with microplate experiments. For pyruvate and glucose, we observed a good growth rate (0.18–0.22h−1), whereas growth rates were significantly lower for asparagine and alanine (0.12–0.13h−1). For mixed substrates, growth rates were as high as the one of aspartate alone. Thus the rate of 0.41–0.43 h−1 therefore appears as the maximum growth rate of strain GMI1000, probably due to the resource allocation trade‐off between ribosomes, transporters, and enzymes in the cytoplasm (Goelzer et al., 2015).
FIGURE 2.
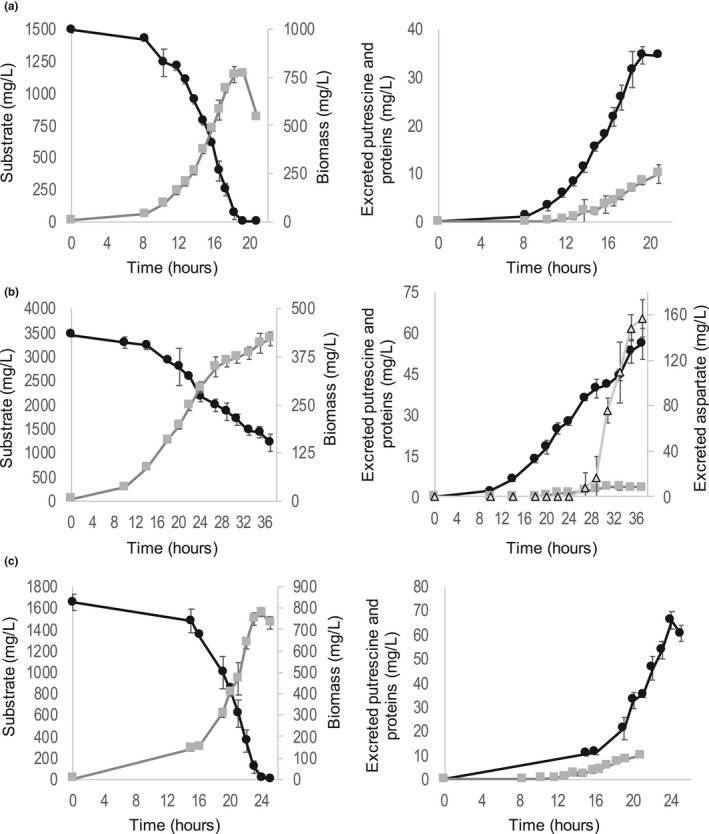
Kinetics of substrate consumption, biomass production, and excreted products for glutamine only (a), asparagine only (b), and glucose only (c). Left graphs: substrate are black dots, biomass are grey squares. Right graphs: putrescine are black dots, proteins are grey squares, aspartate (for b only) are light grey triangles. Three triplicates were performed for each different set of substrates
FIGURE 3.
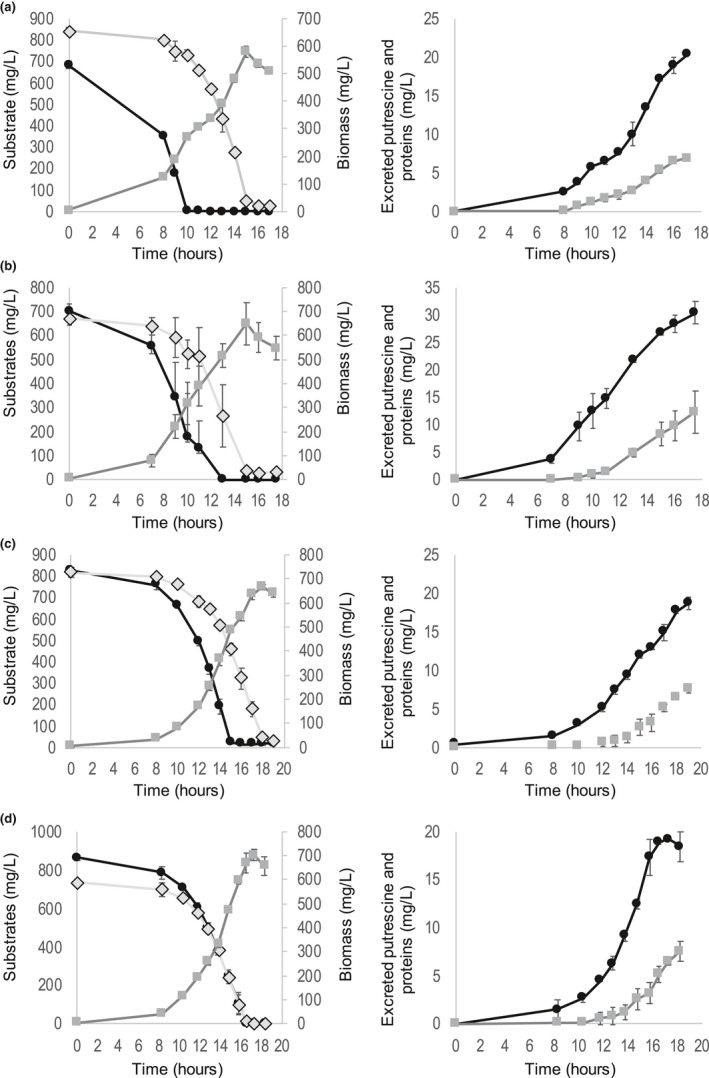
Kinetics of substrates consumption, biomass production, and excreted products for aspartate and glucose (a), glutamine and glucose (b), asparagine and glucose (c), and glutamine and asparagine (d). Three triplicates were performed for each different set of substrates. Right graphs: putrescine are black dots and proteins are grey squares. Left graphs: substrates are black dots or grey diamonds, biomass is grey squares. a: dots: aspartate, diamonds: glucose. b: dots: glutamine, diamonds: glucose. c. dots: asparagine, diamonds: glucose. d: dots: glutamine, diamonds: asparagine
Monitoring of substrate consumption showed a total depletion of all carbon sources used for the different cultures, except for DL‐Malate, confirming that carbon is the limiting element for growth. For DL‐Malate, this can be explained by the fact that the substrate was composed of a mixture of D and L‐Malate and that GMI1000 can only grow with the natural enantiomer L‐Malate and not with the D‐ form, as reported in previous studies (Peyraud et al., 2016; Tunchai et al., 2017). Results also showed a co‐consumption of substrates in the case of a mixture of carbon sources, confirming the absence of diauxic behaviors (Figure 3). Bacterial density was also monitored during growth and an increase over time was observed (Figure 2, Figure 3), confirming that all the tested substrates were sustaining bacterial growth.
Regarding yields, glutamine is the substrate that allowed achieving the best biomass yield: 0.53 mg of biomass per mg of the substrate. Glucose also yielded a high quantity of biomass (0.45 mg of biomass per mg of the substrate) whereas aspartate, pyruvate, and alanine yielded good biomass quantity (0.32–0.38 mg of biomass per mg of the substrate). Asparagine yielded only 0.23 mg of biomass per mg of the substrate.
The NMR analysis of media also revealed, in accordance with Lowe‐Power et al. (2018b); Peyraud et al. (2016), that GMI1000 excretes a non‐negligible quantity of putrescine during growth (Table 4, Figure 2). A look at yields showed that putrescine is excreted in large quantities for glucose, alanine, and pyruvate (0.29–0.36 mg putrescine per mg of the substrate), in medium quantity for asparagine and glutamine (0.15–0.22 mg putrescine per mg of the substrate), and lower quantity for malate and aspartate (0.10–0.15 mg putrescine per mg of the substrate). When looking at the putrescine excretion flux, the flux is rather constant whatever the substrate, around 0.137 mg of putrescine per mg of biomass per hour, except for glutamine and glucose. Note, GMI1000 is also known to excrete a large number of proteins, including many virulence factors (Lonjon et al., 2016) (Figure 2, Table 4). Protein production yields also differed among substrates, with glucose yielding the highest quantity of proteins. Of note, aspartate was detected as excreted and then re‐assimilated during the second exponential growth phase in presence of asparagine, emphasizing the different metabolic behavior that occurs in this second growth phase (Figure 2B).
3.5. Estimation of the xylem sap flow rate reveals that this environment is not nutrient‐limiting for R. solanacearum fast growth
It is often stated that xylem sap is a relatively nutrient‐poor environment (Lowe‐Power et al., 2018a) since only 9.45 mM C (Table 1) are present in total, and not all metabolites can support the growth of the pathogen. The fast growth of R. solanacearum, which can reach 109–1010 CFU/gFW in 3 days after stem inoculation at 104 CFU for the whole plant, thus appears paradoxical (Guidot et al., 2014; McGarvey et al., 1999). Yet, xylem sap is often viewed as a static medium containing a certain quantity of carbon substrates, probably because the only one‐time measurement is performed. Xylem sap should rather be seen as a continuous flow of nutrients. Based on data from this study, from Gerlin et al. (2021a) and Planas‐Marquès et al. (2020), we estimated the continuous flow of nutrients to which R. solanacearum has access. Briefly (see details in Material & Methods), we estimated the volume of the xylem tissue from the plant density and the proportional area represented by the xylem in the stem (Planas‐Marquès et al., 2020). Then, based on the plant transpiration rate (Gerlin et al., 2021a) and the concentration of metabolites that compose the xylem sap (Table 1), we deduced the quantity of nutrients carried in the xylem flow.
Overall, there is 2671 mM C.day−1 and 776 mM N.day−1 (total compounds), including 1446 mM C.day−1 that can promote growth. For three days, the substrate concentration to which R. solanacearum can have access is therefore 8013 mM C and 2328 mM N, including 3438 mM C which can support growth. Final density, around 109–1010 CFU/gFW (Guidot et al., 2014; McGarvey et al., 1999) is equivalent to an OD of 17.8–178 and biomass concentration in the xylem of 7.41–74.1 g.L−1. In comparison, 50 mM C of glutamine in shake‐flasks experiments in this study yields 0.775 g of biomass. Using substrates yields measured on glutamine, glucose, asparagine, and aspartate from Table 4 and xylem concentrations from Table 1, the xylem flux of metabolites could yield 62.0 g L−1 of R. solanacearum biomass if these four substrates are entirely consumed. This value is in the same order of magnitude as what can be measured 3 days after a stem inoculation. This estimate of the flux of available nutrients in tomato xylem sap suggests that there are sufficient metabolic resources in the vascular tissue for R. solanacearum to achieve the high level of growth observed during infection.
4. DISCUSSION
4.1. The variability of the xylem sap environment
In our study, we identified 14 compounds with a concentration above 1 μM (detection limit) in xylem sap of tomato cv Marmande, with a vast majority of amino acids and glutamine as the main compound. We confirmed that the presence of ethanol was not due to the chemical contamination of the sample. Its presence might result from the activity of cells from the root and the center of the stem that switch to a fermentation metabolism due to micro‐oxygenic conditions. Ethanol was already detected in sap from other plants species such as pea (Jackson et al., 1982), olive trees (Anguita‐Maeso et al., 2021), or tomatoes (Gerlin et al., 2021a). Sugar presence might be due to phloem contamination since tomato was demonstrated to have ascending phloem ((Bonnemain, 1966)). However, previous studies have shown that mutants defective for the assimilation of sugars have only a slightly slower infection rate compared to the wild‐type, showing that sugars are present in xylem sap (Gerlin et al., 2021a; Hamilton et al., 2021; Jacobs et al., 2012).
The total organic carbon concentration we measured in the xylem sap of tomato Marmande is about 10 mM and the nitrogen concentration is around 3 mM. The overall C/N ratio measured in our samples is about 3.5. Our results are consistent with those reported in the literature (Bialczyk et al., 2004; Gerlin et al., 2021a; Hiery et al., 2013; Lowe‐Power et al., 2018b; Senden et al., 1992; White et al., 1981; Zuluaga et al., 2013). Indeed, according to these different studies, the tomato xylem is mainly composed of ions, amino acids, sugars, and acids from the Krebs cycle (citrate, malate, fumarate). Prevalent molecules are glutamine, the most concentrated amino acid, as well as asparagine, aspartate, alanine, arginine, leucine, isoleucine, valine, lysine, tyrosine, and phenylalanine, in lower concentrations. In these studies, total organic carbon concentration varies significantly between 0.67 and 22 mM C, total organic nitrogen varies between 0.24 and 5 mM and the carbon over nitrogen ratio (C/N) has a wide range of 3.28‐17.38. The disparity of molecules found and the wide range of concentrations is probably due to different experimental conditions between studies. The large variability in the C/N ratio depends on how much sugar and acids from the Krebs cycle are present in the xylem, and whether or not they were measured (due to different analytical techniques used). Age of the plants and culture growth conditions can also explain some of these differences and variations in xylem sap composition between two tomato cultivars have already been highlighted (Georgoulisa et al., 2020). The methods of extraction of xylem fluids can differ, some of which may lead to phloem fluid contamination. Finally, analytical techniques also differ. In this study, we used NMR technology to describe and quantify the xylem composition of tomato (cv Marmande) aged 4 weeks after sewing. The advantage of NMR technology is that no a priori knowledge on the detectable molecules is necessary.
In a previous study, we determined the xylem sap content of another tomato cultivar, M82 (Gerlin et al., 2021a) whose composition is relatively similar to the results obtained on cultivar Marmande for the nature of substrates, but the M82 xylem sap was found to be four times more concentrated, with a higher proportion of glutamine. The plants had the same age, and the xylem extraction technique and analysis of the xylem content were similar. The difference observed might be due to the cultivar (Georgoulisa et al., 2020), but also to the fertilizer (Bialczyk et al., 2004), since in the M82 experiment, tomato plants were grown in soil (SB2, Proveen, The Netherlands) supplemented with Osmocote® (Sierra Chemical Company), whereas in this study tomato plants were grown in the same soil without any supplementation by fertilizer.
4.2. Adaptation of the R. solanacearum metabolism to the tomato xylem
On the 14 compounds detected in xylem sap, strain GMI1000 can assimilate all of them either as carbon or nitrogen source (Peyraud et al., 2016). However, from the results of the growth kinetics from microplates and shaken flask experiments, only part of these compounds can sustain sufficient growth (i.e. a rapid growth with a good substrate yield): glutamine, asparagine, aspartate, and glucose. Their proportion represents 50%–60% of the xylem organic content. The other compounds can either sustain a slow growth or can be used as complementary sources of nutrition, boosting growth. The observed increase of growth is probably due to the non‐necessity of synthesizing de novo the additional molecules present in the media.
The composition of xylem sap is quite conserved among plants (Andersen & Brodbeck, 1989; Gerlin et al., 2021a; Hocking, 1980; Anguita Maeso et al., 2021), with glutamine being the most abundant amino acid in most cases. As common xylem sap molecules can be efficiently metabolized by GMI1000, thus confirming it is overall well‐adapted to the xylem sap environment, in which it can thrive easily (Gerlin et al., 2021a; Lowe‐Power et al., 2018a). This could certainly contribute to the wide host range of this pathogen. Interestingly, GMI1000 grows also well on malate and moderately on pyruvate. Both of these organic acids are found in the soil (Liebeke et al., 2009) and strain GMI1000 was found to be chemically attracted by malate (Hida et al., 2015; Tunchai et al., 2017).
The average xylem C/N ratio measured in our xylem sap samples is 3.4, and the C/N ratio of substrates sustaining growth is lower, situated around 2.8. Given the fact that GMI1000 uses a C/N of at least 7.25 for growth in presence of glutamate (Peyraud et al., 2016) (4 for biomass, the rest for generating energy through the TCA cycle, with CO2 as waste), organic carbon is thus the limiting element for GMI1000 growth in xylem sap. The C/N ratio unbalance is mainly due to the dominance of amino acids (around 85%). This unbalance will likely have consequences on GMI1000 growth, such as the necessity to get rid of the nitrogen excess, under the form of organic and inorganic compounds (Fyson et al., 2017), such as ammonium. Putrescine could be considered a nitrogenous waste product, but putrescine excretion only accounts for 25% of excess nitrogen on glutamate, and protein excretion 0.8%. In addition, putrescine excretion is observed on carbon‐only substrates such as glucose, thus emphasizing a different role for putrescine, such as, possibly, virulence (Gerlin et al., 2021a, 2021b; Lowe‐Power et al., 2018b).
4.3. Xylem sap is sufficient to support R. solanacearum growth to high densities
From the xylem data we obtained and from other previous studies (Gerlin et al., 2021a; Planas‐Marquès et al., 2020) we inferred the concentration of metabolites in the xylem stream to which R. solanacearum has access for growth. At the beginning of the infection process, bacteria do not grow fast enough to consume all the incoming flux of substrate, and plant defenses such as ROS probably provoke some cell death. On the other hand, other amino acids and sucrose in the xylem sap can boost growth and increase yields on these four major metabolized substrates (glutamine, glucose, asparagine, aspartate). It is therefore difficult to conclude from our data that the continuous flow rate of metabolites in the xylem is enough to support all the growth observed in planta but there is potential to reach high population levels (up to at least 109 CFU/gFW). It can also be concluded that xylem sap is not nutrient‐poor, contrary to what is often stated in the literature.
5. CONCLUSIONS
By determining the content of the environment where Ralstonia solanacearum multiplies in tomato plants and by revealing the trophic preferences of the bacterium using kinetic experiments in microplates and shake flasks, we showed that glutamine and asparagine are the preferential substrates likely to be assimilated by strain GMI1000 when in plant xylem. The other amino acids, as well as sugars, likely act as complementary sources of nutrition, to boost growth. Finally, through a calculation relying on plant physiology and bacteria substrate yields, the xylem sap flux was shown to be rich enough to sustain GMI1000 growth to high densities in the plant.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
Caroline Baroukh: Conceptualization (lead); Data curation (lead); Formal analysis (lead); Funding acquisition (lead); Investigation (equal); Methodology (lead); Project administration (lead); Resources (equal); Supervision (lead); Writing – original draft (lead); Writing – review and editing (equal). Meriem Zemouri: Data curation (supporting); Formal analysis (supporting); Investigation (equal); Writing – original draft (supporting); Writing – review and editing (supporting). Stéphane Genin: Conceptualization (supporting); Methodology (supporting); Resources (equal); Writing – review and editing (equal).
ETHICS STATEMENT
None required.
Supporting information
Supplementary Material
ACKNOWLEDGEMENTS
ZM was funded by a grant from the Centre National de la Recherche Scientifique, PEPs Modélisation de Processus Infectieux (2017‐2019). The study was funded by the Centre National de la Recherche Scientifique, PEPs Modélisation de Processus Infectieux (2017‐2019). Work in our laboratory is also supported by the French Laboratory of Excellence project “TULIP” (grant number ANR‐10‐LABX‐41; ANR‐11‐IDEX‐0002‐02). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript. We acknowledge the MetaToul platform (Metabolomics & Fluxomics Facilities, Toulouse, France, www.metatoul.fr), which is part of the MetaboHUB‐ANR‐11‐INBS‐0010 national infrastructure (www.metabohub.fr) and its staff: Cécilia Berges, Edern Cahoreau, and Lindsay Peyriga for access to NMR facilities.
Baroukh, C. , Zemouri, M. , & Genin, S. (2022). Trophic preferences of the pathogen Ralstonia solanacearum and consequences on its growth in xylem sap. MicrobiologyOpen, 11, e1240. 10.1002/mbo3.1240
DATA AVAILABILITY STATEMENT
All data supporting the findings of this study are available within the paper and the Supplementary Data file (Detailed content of xylem sap samples for tomato cv Marmande and tomato cv M82 plants).
REFERENCES
- Andersen, P. C. , & Brodbeck, B. V. (1989). Diurnal and temporal changes in the chemical profile of xylem exudate from Vitis rotundifolia. Physiologia Plantarum, 75, 63–70. 10.1111/j.1399-3054.1989.tb02064.x [DOI] [Google Scholar]
- Anguita‐Maeso, M. , Haro, C. , Montes‐Borrego, M. , De La Fuente, L. , Navas‐Cortés, J. A. , Landa, & B. B. (2021). Metabolomic, Ionomic and Microbial Characterization of Olive Xylem Sap Reveals Differences According to Plant Age and Genotype. Agronomy, 11(6), 1179. 10.3390/agronomy11061179 [DOI] [Google Scholar]
- Bialczyk, J. , Lechowski, Z. , & Dziga, D. (2004). Composition of the xylem sap of tomato seedlings cultivated on media with HCO3− and nitrogen source as NO3− or NH4+. Plant and Soil, 263, 265–272. 10.1023/B:PLSO.0000047739.11698.ca [DOI] [Google Scholar]
- Bonnemain, J. (1966). Sur les modalités de la distribution des assimilats chez la tomate et sur ses mécanismes. Comptes Rendus Hebdomadaires Des Seances De L Academie Des Sciences, 262, 1106–1109. [Google Scholar]
- Dalsing, B. L. , Trunchon, A. N. , Gonzalez‐Orta, E. T. , Milling, A. S. , & Allen, C. (2015). Ralstonia solanacearum uses inorganic nitrogen metabolism for virulence, ATP production, and detoxification in the oxygen‐limited. Mbio, 6(2), e02471‐14. 10.1128/mBio.02471-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalsing, B. L. , & Allena, C. (2014). Nitrate assimilation contributes to Ralstonia solanacearum root attachment, stem colonization, and virulence. Journal of Bacteriology, 196(5), 949–960. 10.1128/JB.01378-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyson, N. , King, J. , Belcher, T. , Preston, A. , & Colijn, C. (2017). A curated genome‐scale metabolic model of Bordetella pertussis metabolism. PLOS Computational Biology, 13(7), e1005639. 10.1371/journal.pcbi.1005639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin, S. (2010). Molecular traits controlling host range and adaptation to plants in Ralstonia solanacearum. New Phytologist, 187(2), 920–928. 10.1111/j.1469-8137.2010.03397.x [DOI] [PubMed] [Google Scholar]
- Georgoulisa, S. , Shalvarjian, K. E. , Helmann, T. C. , Hamilton, C. D. , Carlson, H. , Deutschbauer, A. M. , & Lowe‐Power, T. M. (2020). Genome‐wide identification of tomato xylem sap fitness factors for Ralstonia pseudosolanacearum and Ralstonia syzygii. bioRxiv. 10.1101/2020.08.31.276741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlin, L. , Baroukh, C. , & Genin, S. (2021a). Polyamines: double agents in disease and plant immunity. Trends in Plant Science, 26(10), 1061–1071. 10.1016/j.tplants.2021.05.007 [DOI] [PubMed] [Google Scholar]
- Gerlin, L. , Escourrou, A. , Cassan, C. , Maviane Macia, F. , Peeters, N. , Genin, S. , & Baroukh, C. (2021b). Unravelling physiological signatures of tomato bacterial wilt and xylem metabolites exploited by Ralstonia solanacearum. Environmental Microbiology. 10.1111/1462-2920.15535 [DOI] [PubMed] [Google Scholar]
- Goelzer, A. , Muntel, J. , Chubukov, V. , Jules, M. , Prestel, E. , Nölker, R. , Mariadassou, M. , Aymerich, S. , Hecker, M. , Noirot, P. , & Becher, D. (2015). Quantitative prediction of genome‐wide resource allocation in bacteria. Metabolic Engineering, 32, 232–243. 10.1016/j.ymben.2015.10.003 [DOI] [PubMed] [Google Scholar]
- Guidot, A. , Jiang, W. , Ferdy, J. B. , Thébaud, C. , Barberis, P. , Gouzy, J. , & Genin, S. (2014). Multihost experimental evolution of the pathogen Ralstonia solanacearum unveils genes involved in adaptation to plants. Molecular Biology and Evolution, 31(11), 2913–2928. 10.1093/molbev/msu229 [DOI] [PubMed] [Google Scholar]
- Hamilton, C. D. , Steidl, O. R. , MacIntyre, A. M. , Hendrich, C. G. , & Allen, C. (2021). Ralstonia solanacearum Depends on Catabolism of Myo‐Inositol, Sucrose, and Trehalose for Virulence in an Infection Stage–Dependent Manner. Molecular Plant‐Microbe Interactions®, 34(6), 669–679. 10.1094/mpmi-10-20-0298-r [DOI] [PubMed] [Google Scholar]
- Hayward, C. A. (1991). Biology and epidemiology of bacterial wilt caused by Pseudomonas Solanacearum. Annual Review of Phytopathology, 29, 65–87. 10.1146/annurev.py.29.090191.000433 [DOI] [PubMed] [Google Scholar]
- Hida, A. , Oku, S. , Kawasaki, T. , Nakashimada, Y. , Tajima, T. , & Kato, J. (2015). Identification of the mcpA and mcpM genes, encoding methyl‐accepting proteins involved in amino acid and L‐malate chemotaxis, and involvement of McpM‐mediated chemotaxis in plant infection by Ralstonia pseudosolanacearum Formerly Ralstonia solanacearum phylotype I and III). Applied and Environment Microbiology, 81(21), 7420–7430. 10.1128/AEM.01870-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiery, E. , Adam, S. , Reid, S. , Hofmann, J. , Sonnewald, S. , & Burkovski, A. (2013). Genome‐wide transcriptome analysis of Clavibacter michiganensis subsp. michiganensis grown in xylem mimicking medium. Journal of Biotechnology, 168(4), 348–354. 10.1016/j.jbiotec.2013.09.006 [DOI] [PubMed] [Google Scholar]
- Hocking, P. J. (1980). The composition of phloem exudate and xylem sap from tree tobacco (Nicotiana glauca Grah.). Annals of Botany, 45(6), 633–643. 10.1093/oxfordjournals.aob.a085871 [DOI] [Google Scholar]
- Ingel, B. , Caldwel, D. , Duong, F. , Parkinson, D. Y. , McCulloh, K. A. , Iyer‐Pascuzzi, A. S. , McElrone, A. J. , & Lowe‐Power, T. M. (2021). Revisiting the source of wilt symptoms: X‐ray microcomputed tomography provides direct evidence that Ralstonia biomass clogs xylem vessels. PhytoFrontiers. 10.1094/phytofr-06-21-0041-r [DOI] [Google Scholar]
- Jackson, M. B. , Herman, B. , & Goodenough, A. (1982). An examination of the importance of ethanol in causing injury to flooded plants. Plant, Cell and Environment, 5(2), 163–172. 10.1111/1365-3040.ep11571590 [DOI] [Google Scholar]
- Jacobs, J. M. , Babujee, L. , Meng, F. , Milling, A. , & Allen, C. (2012). The in planta transcriptome of Ralstonia solanacearum: conserved physiological and virulence strategies during bacterial wilt of tomato. Mbio, 3, e00114‐12(4), 10.1128/mBio.00114-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jyoti, P. , Shree, M. , Joshi, C. , Prakash, T. , Ray, S. K. , Satapathy, S. S. , & Masakapalli, S. K. (2020). The entner‐doudoroff and nonoxidative pentose phosphate pathways bypass glycolysis and the oxidative pentose phosphate pathway in Ralstonia solanacearum. mSystems, 5(2), e00091‐20. 10.1128/msystems.00091-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebeke, M. , Brözel, V. S. , Hecker, M. , & Lalk, M. (2009). Chemical characterization of soil extract as growth media for the ecophysiological study of bacteria. Applied Microbiology and Biotechnology, 83, 161–173. 10.1007/s00253-009-1965-0 [DOI] [PubMed] [Google Scholar]
- Lonjon, F. , Turner, M. , Henry, C. , Rengel, D. , Lohou, D. , van de Kerkhove, Q. , Cazalé, A. C. , Peeters, N. , Genin, S. , & Vailleau, F. (2016). Comparative secretome analysis of ralstonia solanacearum type 3 secretion‐associated mutants reveals a fine control of effector delivery, essential for bacterial pathogenicity. Molecular & Cellular Proteomics, 15(2), 598–613. 10.1074/mcp.M115.051078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe‐Power, T. M. , Khokhani, D. , & Allen, C. (2018a). How Ralstonia solanacearum exploits and thrives in the flowing plant xylem environment. Trends in Microbiology, 26(11), 929–942. 10.1016/j.tim.2018.06.002 [DOI] [PubMed] [Google Scholar]
- Lowe‐Power, T. M. , Hendrich, C. G. , von Roepenack‐Lahaye, E. , Li, B. , Wu, D. , Mitra, R. , Dalsing, B. L. , Ricca, P. , Naidoo, J. , Cook, D. , Jancewicz, A. , Masson, P. , Thomma, B. , Lahaye, T. , Michael, A. J. , & Allen, C. (2018b). Metabolomics of tomato xylem sap during bacterial wilt reveals Ralstonia solanacearum produces abundant putrescine, a metabolite that accelerates wilt disease. Environmental Microbiology, 20(4), 1330–1349. 10.1111/1462-2920.14020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey, J. A. , Denny, T. P. & Schekk, M. A. (1999). Spatial‐temporal and quantitative analysis of growth and EPS I production by Ralstonia solanacearum in resistant and susceptible tomato cultivars. Phytopathology, 89(12), 1233–1239. 10.1094/PHYTO.1999.89.12.1233 [DOI] [PubMed] [Google Scholar]
- Monod, J. (1942). Recherches sur la croissance des cultures bacteriennes. Annales de l'Institut Pasteur, 69, 179. [Google Scholar]
- Peeters, N. , Carrère, S. , Anisimova, M. , Plener, L. , Cazalé, A.‐C. , & Genin, S. (2013). Repertoire, unified nomenclature and evolution of the Type III effector gene set in the Ralstonia solanacearum species complex. BMC Genomics, 14, 859. 10.1186/1471-2164-14-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyraud, R. , Cottret, L. , Marmiesse, L. , Gouzy, J. , & Genin, S. (2016). A Resource Allocation Trade‐Off between Virulence and Proliferation Drives Metabolic Versatility in the Plant Pathogen Ralstonia solanacearum. PLOS Pathogens, 12(10), e1005939. 10.1371/journal.ppat.1005939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyraud, R. , Cottret, L. , Marmiesse, L. , & Genin, S. (2018). Control of primary metabolism by a virulence regulatory network promotes robustness in a plant pathogen. Nature Communications, 9(418). 10.1038/s41467-017-02660-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas‐Marquès, M. , Kressin, J. P. , Kashyap, A. , Panthee, D. R. , Louws, F. J. , Coll, N. S. , & Valls, M. (2020). Four bottlenecks restrict colonization and invasion by the pathogen Ralstonia solanacearum in resistant tomato. Journal of Experimental Botany, 71(6), 2157–2171. 10.1093/jxb/erz562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plener, L. , Boistard, P. , González, A. , Boucher, C. , & Genin, S. (2012) Metabolic adaptation of Ralstonia solanacearum during plant infection: a methionine biosynthesis case study. PLoS ONE, 7(5), e36877. 10.1371/journal.pone.0036877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safni, I. , Cleenwerck, I. , De Vos, P. , Fegan, M. , Sly, L. , & Kappler, U. (2014). Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii subsp. syzygii subsp. nov., R. solanacearum phylotype IV strains as Ralstonia syzygii subsp. indonesiensis subsp. nov., banana blood disease bacterium strains as Ralstonia syzygii subsp. celebesensis subsp. nov. and R. solanacearum phylotype I and III strains as Ralstonia pseudosolanacearum sp. nov. International Journal of Systematic and Evolutionary Microbiology, 64, 3087–3103. 10.1099/ijs.0.066712-0 [DOI] [PubMed] [Google Scholar]
- Salanoubat, M. , Genin, S. , Artiguenave, F. , Gouzy, J. , Mangenot, S. , Arlat, M. , Billault, A. , Brottier, P. , Camus, J. C , Cattolico, L. , Chandler, M. , Choisne, N. , Claudel‐Renard, C. , Cunnac, S. , Demange, N. , Gaspin, C. , Lavie, M. , Moisan, A. , Robert, C. , … Boucher, C. A (2002). Genome sequence of the plant pathogen Ralstonia solanacearum. Nature, 415, 497–502. 10.1038/415497a [DOI] [PubMed] [Google Scholar]
- Senden, M. H. M. N. , Van Der Meer, A. J G M , Limborgh, J. , & Wolterbeek, H. TH (1992). Analysis of major tomato xylem organic acids and PITC‐derivatives of amino acids by RP‐HPLC and UV detection. Plant and Soil, 142, 81–89. 10.1007/BF00010177 [DOI] [Google Scholar]
- Tunchai, M. , Hida, A. , Oku, S. , Nakashimada, Y. , Tajima, T. , & Kato, J. (2017). Identification and characterization of chemosensors for D‐malate, unnatural enantiomer of malate, in Ralstonia pseudosolanacearum. Microbiology, 163(2), 233–242. 10.1099/mic.0.000408 [DOI] [PubMed] [Google Scholar]
- Ujita, Y. , Sakata, M. , Yoshihara, A. , Hikichi, Y. , & Kai, K. (2019). Signal production and response specificity in the phc quorum sensing systems of Ralstonia solanacearum species complex. ACS Chemical Biology, 14(10), 2243–2251. 10.1021/acschembio.9b00553 [DOI] [PubMed] [Google Scholar]
- Van Elsas, J. D. , Kastelein, P. , Van Bekkum, P. , Van der Wolf, J. M. , de Vries, P. M. , & Van Overbeek, L. S. (2000). Survival of Ralstonia solanacearum biovar 2, the causative agent of potato brown rot, in field and microcosm soils in temperate climates. Phytopathology, 90(12), 1358–1366. 10.1094/PHYTO.2000.90.12.1358 [DOI] [PubMed] [Google Scholar]
- White, M. C. , Chaney, R. L. , & Decker, A. M. (1981). Metal complexation in xylem fluid. Plant Physiology, 67, 311–315. 10.1104/pp.67.2.311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, D. , von Roepenack‐Lahaye, E. , Buntru, M. , de Lange, O. , Schandry, N. , Pérez‐Quintero, A. L. , Weinberg, Z. , Lowe‐Power, T. M. , Szurek, B. , Michael, A. J. , Allen, C. , Schillberg, S. , & Lahaye, T. (2019). A plant pathogen type III effector protein subverts translational regulation to boost host polyamine levels. Cell Host & Microbe, 26(5), 638–649. 10.1016/j.chom.2019.09.014 [DOI] [PubMed] [Google Scholar]
- Xian, L. , Yu, G. , Wei, Y. , Rufian, J. S. , Li, Y. , Zhuang, H. , Xue, H. , Morcillo, R. J. L. , & Macho, A. P. (2020). A bacterial effector protein hijacks plant metabolism to support pathogen nutrition. Cell Host & Microbe, 28, 1–10. 10.1016/j.chom.2020.07.003 [DOI] [PubMed] [Google Scholar]
- Yabuuchi, E. , Kosako, Y. , Oyaizu, H. , Yano, I. , Hotta, H. , Hashimoto, Y. , Ezaki, T. , & Arakawa, M. et al (1992). Proposal of Burkholderia gen. nov. and transfer of seven of the genus Pseudomona homology group II to the new genus, with type species Burkholderia cepecia (Palleroni and Holmes 1981) comb. nov. Microbiology and Immunology, 40(12), 1251–1275. [DOI] [PubMed] [Google Scholar]
- Zuluaga, A. P. , Puigvert, M. , & Valls, M. (2013). Novel plant inputs influencing Ralstonia solanacearum during infection. Frontiers in Microbiology, 4, 1–7. 10.3389/fmicb.2013.00349 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
All data supporting the findings of this study are available within the paper and the Supplementary Data file (Detailed content of xylem sap samples for tomato cv Marmande and tomato cv M82 plants).


