Abstract
We recently described X-linked acrogigantism (X-LAG), a condition of early childhood-onset pituitary gigantism associated with microduplications of the GPR101 receptor. The expression of GPR101 in hyperplastic pituitary regions and tumors in X-LAG patients, and GPR101’s normally transient pituitary expression during fetal development, suggest a role in the regulation of growth. Nevertheless, little is still known about GPR101’s physiological functions, especially during development. By using zebrafish models, we investigated the role of gpr101 during embryonic development and somatic growth. Transient ectopic gpr101 expression perturbed the embryonic body plan but did not affect growth. Loss of gpr101 led to a significant reduction in body size that was even more pronounced in the absence of maternal transcripts, as well as subfertility. These changes were accompanied by gastrulation and hypothalamic defects. In conclusion, both gpr101 loss- and gain-of-function affect, in different ways, fertility, embryonic patterning, growth and brain development.
Keywords: X-linked acrogigantism, GPR101, zebrafish, embryonic development, growth
Introduction
Somatic growth in vertebrates is under the influence of diverse cellular, hormonal, and environmental factors, but it is mainly controlled by the hypothalamo–pituitary–IGF-1 (also called hypothalamo–pituitary–somatotropic [HPS]) axis (Murray et al., 2015). In 2014 we reported a rare syndrome of human pituitary gigantism characterized by extreme physical size and stature (Trivellin et al., 2014). This condition, which we named X-linked acrogigantism (X-LAG), is caused by large and commonly mixed growth hormone (GH)- and prolactin-secreting pituitary lesions (tumors and/or hyperplasia) that lead to a dramatic phenotype of early childhood-onset gigantism (Beckers et al., 2015). Thirty-six X-LAG patients have been identified so far (Trivellin et al., 2020), accounting for an estimated 10% of all cases of pituitary gigantism (Rostomyan et al., 2015,Iacovazzo et al., 2016).
Genetically, X-LAG is caused by microduplications on chromosome Xq26.3 involving the orphan G protein-coupled receptor (GPCR)-encoding gene GPR101 (Trivellin et al., 2014,Iacovazzo et al., 2016). GPR101 duplications in X-LAG cause exceptionally high expression of the receptor in the pituitary lesions, which in turn constitutively activates the cyclic AMP (cAMP) and IP3 signaling pathways (Trivellin et al., 2014,Abboud et al., 2020). Nevertheless, how this ultimately translates to elevated circulating GH and IGF-1 levels and abnormal somatic growth has not been fully determined, although considerable progress has been recently made regarding GPR101’s role in mouse pituitary (Abboud et al., 2020). In normal tissues, GPR101 is highly expressed in the fetal pituitary (but not in the normal adult pituitary) and in specific regions of the brain, in particular the nucleus accumbens and the hypothalamus, while very low or absent expression is observed in almost all other organs (Trivellin et al., 2016,Bates et al., 2006,Nilaweera et al., 2007,Regard et al., 2008). The involvement of GPR101 in X-LAG, its restricted tissue-specific expression pattern, and its function in terminally differentiated GH-secreting cells suggest that GPR101 is important for the development of the HPS axis.
Recently, we characterized gpr101 expression during wild-type (WT) zebrafish (Danio rerio) development and demonstrated an evolutionary conservation in brain expression (Trivellin et al., 2016). Zebrafish is a suitable animal model to study early vertebrate development (Bruce, 2016), neural and endocrine organs (Gupta et al., 2018,Lohr and Hammerschmidt, 2011), and determinate growth, as seen in mammals (Biga and Goetz, 2006,Hu et al., 2019). We, therefore, decided to use zebrafish to study these three aspects in the context of perturbed gpr101/GPR101 expression. We show here that loss of gpr101 in zebrafish led to a significant reduction in body size that was even more pronounced in the absence of maternal transcripts. These changes in size were accompanied by hypothalamic defects. Loss- and gain-of-function experiments also showed that gpr101 is important for normal gastrulation and formation of the embryonic body plan.
Results
Transient gpr101/GPR101 upregulation affects embryonic development but not juvenile growth
We have previously shown that although the human GPR101 and zebrafish gpr101 orthologues share just 47.5% identity at the protein level (https://www.ncbi.nlm.nih.gov/homologene/), both genes are strongly expressed in the central nervous system (CNS) (Trivellin et al., 2016) and synteny is conserved in the genomic region surrounding gpr101 (Trivellin et al., 2018). Using a cAMP response element (CRE) luciferase reporter, we now show that gpr101 over-expression in mammalian cells constitutively activates the cAMP signaling pathway, although with lower potency compared to GPR101 (Supplementary Fig. 1). Consistent with our findings, it was recently reported that mouse Gpr101 can also increase cAMP levels (Abboud et al., 2020). This suggests that vertebrate GPR101 orthologues can convey intracellular signals in a similar way and can do so in the absence of a ligand.
To mimic what happens in X-LAG patients and to study the pathological effects exerted by GPR101 upregulation on vertebrate growth, we developed transgenic zebrafish that ubiquitously express gpr101 using the Gal4-UAS system. We crossed SAGFF(LF)73A;UAS:GFP fish (Asakawa and Kawakami, 2009,Asakawa et al., 2008) with UAS:gpr101 fish and obtained transgenic gpr101 fish that we confirmed expressed the GFP reporter in the whole body until adulthood (Supplementary Fig. 2a). Despite robust GFP expression, when we measured gpr101 mRNA expression we could not detect ubiquitous and strong gpr101 expression (Supplementary Fig. 2b). We excluded that the UAS-GFP transgene reporter can lead to lower gpr101 expression levels by depleting the amount of Gal4 protein available to activate the UAS-gpr101 transgene. (Supplementary Fig. 2c). When we measured body size at different time points throughout juvenile development – a stage defined by active hunting and rapid body growth – we did not observe significant differences between transgenic gpr101 and control siblings (Supplementary Fig. 2d). To try to achieve a more restricted expression pattern, we then generated animals with pituitary-specific (Xa207:Gal4 driver line, Supplementary Fig. 3) and pan-neuronal (including the hypothalamus) gpr101 expression (HuC:Gal4 driver line, Supplementary Fig. 4). We tracked fish growth in both transgenic lines but again we did not see any effect on juvenile body growth compared to controls (Supplementary Fig. 5).
Failure to elicit strong gpr101 expression analogous to that observed in X-LAG, made us to consider an alternative approach that consisted in transiently over-expressing both zebrafish and human GPR101 orthologues in embryos. Synthetic mRNAs encoding gpr101 and GPR101 were in vitro synthesized and microinjected into WT embryos. By injecting 75 pg into 1-cell stage embryos, we obtained high expression levels (on average, 1,467.5-fold increase of gpr101 vs. uninjected controls as assessed by RT-qPCR at 90% epiboly, Supplementary Fig. 6), comparable to those observed in X-LAG lesions (Trivellin et al., 2014). Two major embryonic phenotypes resulting from the transient over-expression were observed at 1 dpf (day post fertilization, pharyngula stage, Fig. 1a–b). One phenotype, dorsalization, was related to anamniotic dorsoventral axis patterning, with anterior structures being expanded at the expense of more posterior ones (Kishimoto et al., 1997); the other phenotype, cyclopia, was related to eye and brain development (Varga et al., 1999). Over-expression of both orthologues resulted in these two phenotypes with similar proportions, indicating sufficient evolutionary conservation of structure for both orthologues of this GPCR to activate the same targets in zebrafish.
Figure 1.
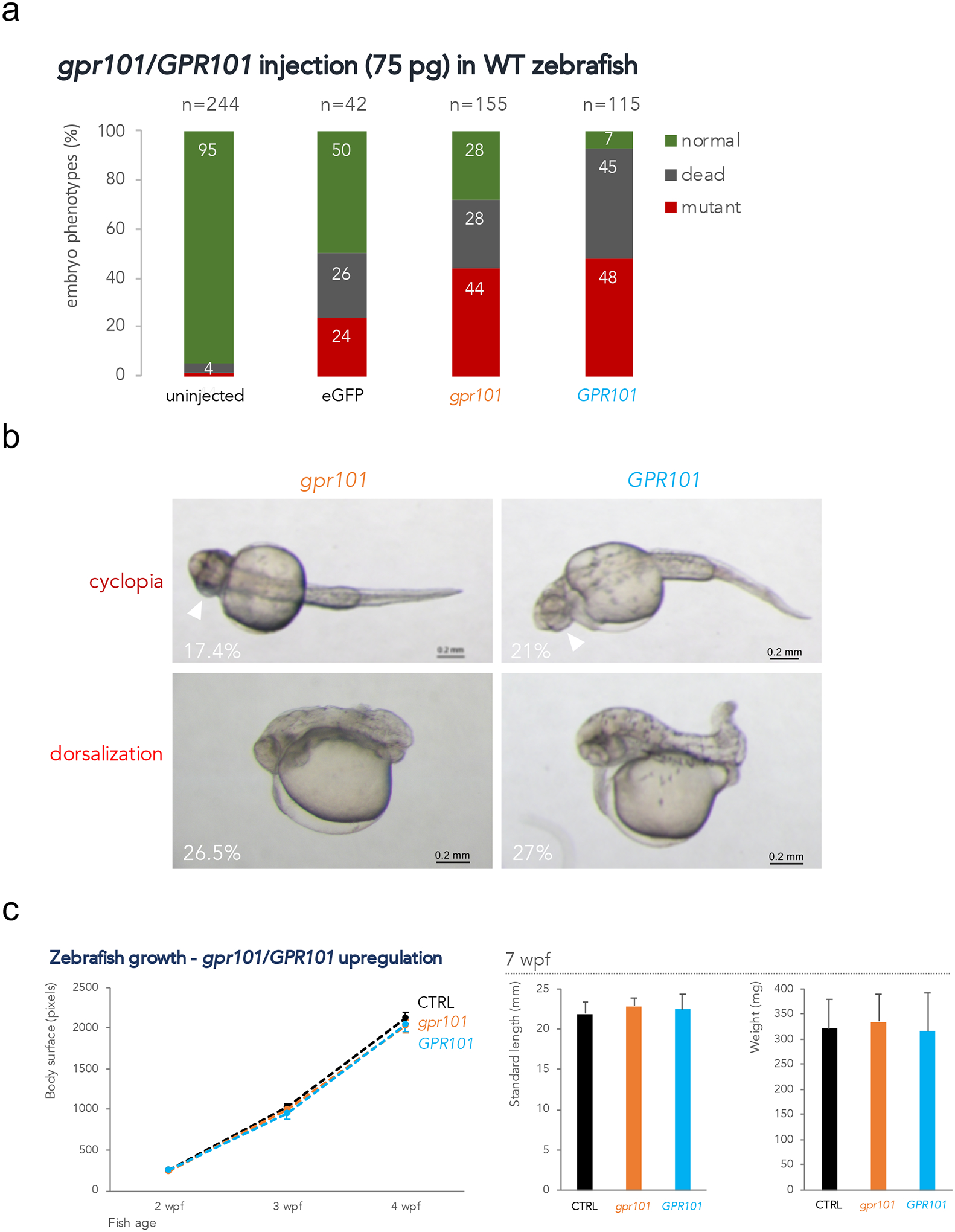
gpr101/GPR101 exogenous expression experiments. a) WT embryos were injected with 75 pg of mRNA and scored at 1 dpf. About 40% of embryos showed a mutant phenotype after either gpr101 and GPR101 injection. Injection with eGFP mRNA resulted in 24% of embryos with non-specific phenotypes. The total number of analyzed embryos is shown at the top of each column. Data shown are from one representative experiment. For each experiment, two expert operators performed injections. b) The two main phenotypes observed in gpr101/GPR101 injected embryos – cyclopia and dorsalization – are shown, together with their frequency. The white arrowheads indicate the fused eyes. Upper panels: ventral view; lower panels: lateral view. Anterior is left. Scale bar: 200 μm. c) Growth trajectories in eGFP (controls, black), gpr101 (orange) and GPR101 (light blue) injected embryos. Body surface was measured in larvae at 2 (eGFP, n = 17; gpr101, n = 21; GPR101, n = 20), 3 (eGFP, n = 17; gpr101, n = 21; GPR101, n = 19), and 4 (eGFP, n = 16; gpr101, n = 19; GPR101, n = 19) wpf (left panel), while standard length and body weight were measured in 7 wpf juvenile fish (eGFP, n = 16; gpr101, n = 18; GPR101, n = 19; right panel). No significant differences in growth among the three groups were observed at any of the analyzed time points.
We then investigated whether such potent but transient insults had long-lasting effects on fish growth throughout juvenile development in the subgroup of WT-looking embryos. As shown in Fig. 1c, neither gpr101 nor GPR101 over-expression had a significant effect on body size from 2 to 7 wpf (weeks post fertilization) compared to controls (eGFP-injected embryos).
gpr101 knockout (KO) decreases juvenile growth
Since we did not observe any obvious effect on fish growth when we over-expressed GPR101, we decided to perform loss-of-function (LOF) experiments. We generated two LOF gpr101 alleles by CRISPR-Cas9 resulting in out-of-frame deletions. Both deletions were located at the N-terminus of gpr101, on codon 23: a deletion of 5 (c.69_73del, hereafter referred to as gpr101y629) and a deletion of 13 (c.68_80del, gpr101y630) nucleotides (Supplementary Fig. 7). Both frameshifts resulted in nonsense mutations and predicted protein truncation. After confirming by Sanger sequencing that both mutations were present at the mRNA level, we selectively propagated the frame-shifting alleles from F0 founders.
We then crossed heterozygous F1 parents (one gpr101y629/+ and one gpr101y630/+) and tracked growth in the F2 progeny. Genotype frequencies in the progeny of all crosses followed Mendelian distribution at all time points. Interestingly, we observed a significant decrease in standard length and body weight in KOs at 3, 6, and 9 wpf (main effect of genotype, p < 0.0001 for all variables, Fig. 2a and 2b, and Supplementary Fig. 8). Variation in the expression of hormones, growth factors and their downstream signaling pathways may explain the observed differences in growth rate. Therefore, we measured the expression of pituitary and hypothalamic hormones encoding genes (gh1, prl, pomca, pomcb, trh), of a transcription factor involved in pituitary development (pou1f1), and of a GH-dependent growth regulator (igf1a) at the juvenile stage (6 wpf). However, the smaller body size was not accompanied by significant differences in the expression of these genes (Fig. 2c).
Figure 2.
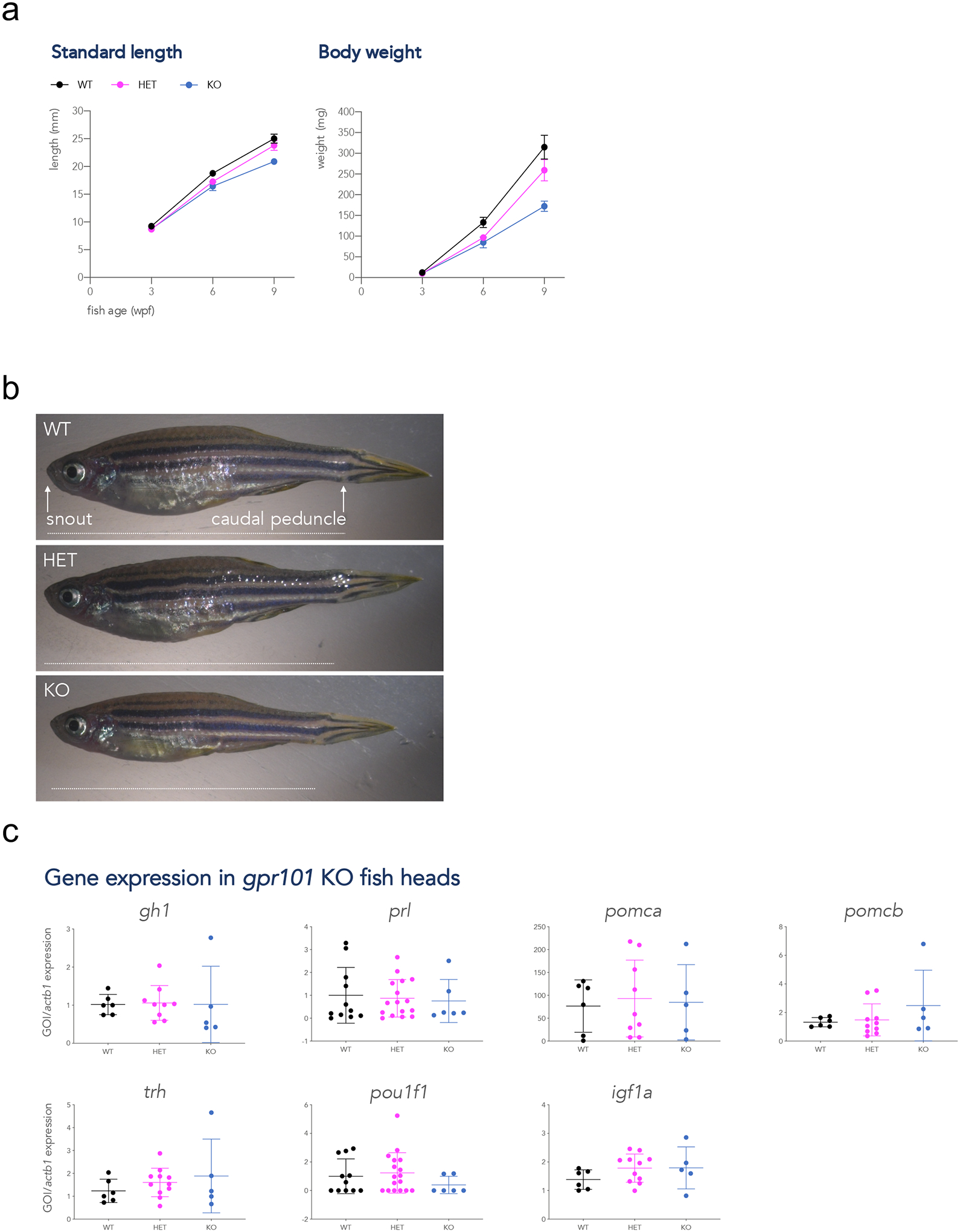
Growth trajectories and gene expression in gpr101 KO zebrafish. A) Standard length and body weight were measured at 3, 6, and 9 wpf in WT (black), heterozygous (HET, purple), and full gpr101 KO (blue) fish. The experiment was performed in two independent sets of fish and the data presented together. The number of fish for each genotype at all time points is given in Supplementary Figure 7a. Data were analyzed by a two-way ANOVA; main effect of genotype, p < 0.0001 for all variables. b) Representative images of 9 wpf female fish belonging to the three genotypes. The way standard length (distance snout-caudal peduncle) was measured is shown as a horizontal white dashed line in the upper panel. The vertical black dashed line in the upper panel shows where deeply anesthetized fish were decapitated for gene expression experiments. c) The mRNA expression of a transcription factor involved in pituitary development (pou1f1), a GH-dependent growth regulator (igf1a), and pituitary and hypothalamic hormones (the rest), was measured by RT-qPCR at a juvenile stage (6 wpf) in WT (n=11 for prl and pou1f1, n=6 for the rest), HET (n=17 for prl and pou1f1, n=11 for the rest), and KO (n=11 for prl and pou1f1, n=6 for the rest), HET (n=6 for prl and pou1f1, n=5 for the rest). actb1 was used as endogenous control. None of these comparisons showed a significant difference between genotypes. GOI: gene of interest.
Surviving maternal-zygotic (MZ) gpr101 mutants manifest a stronger impairment of somatic growth and an enlarged hypothalamus
We previously showed that gpr101 is a maternal factor during embryogenesis, being found in blastomeres during the first stages of the cleavage period (Trivellin et al., 2016). To investigate the contribution of both maternally and zygotically supplied gpr101 on zebrafish growth, we also generated MZgpr101 embryos by in-crossing homozygous mutant F3 parents. We noticed a dearth of developing MZgpr101 embryos at 2–3 hpf (65% unfertilized eggs), indicating very low fertility. Out of the embryos surviving to 1 dpf, 58% appeared as WT-looking at morphological inspection (Fig. 3a). We then tracked the growth of the WT-appearing group of embryos throughout the larval and juvenile stages (6 dpf – 44 dpf). MZgpr101 mutants were consistently smaller than WT controls at all time points (main effect of genotype, p < 0.0001 for all variables, Fig. 3b and Supplementary Fig. 9a). The effect in maternal zygotic mutants was about 2-fold greater in magnitude compared to the reduction in size also observed in zygotic KOs (respectively, 28.8% vs. 12.4% difference in standard length [p = 0.003] and 63.0% vs. 36.2% difference in weight [p = 0.0413] relative to WT controls at 6 wpf). Moreover, a difference in standard length was already appreciable in MZgpr101 mutants at 6 dpf, before feeding started (Supplementary Fig. 9a). Individually-housed mutants fed ad libitum showed a reduction in body size relative to WT that was not significantly different from those housed in groups (means of mutants in group vs. individually-housed mutants where compared, p = 0.84 for standard length and p = 0.27 for weight at 6 wpf, Supplementary Fig. 9b). These data a) suggest that lack of maternal and zygotic gpr101 impairs zebrafish growth at a very early stage during embryonic/larval development, and b) excludes the possibility that the mutants are smaller because they are less able to compete for food.
Figure 3.
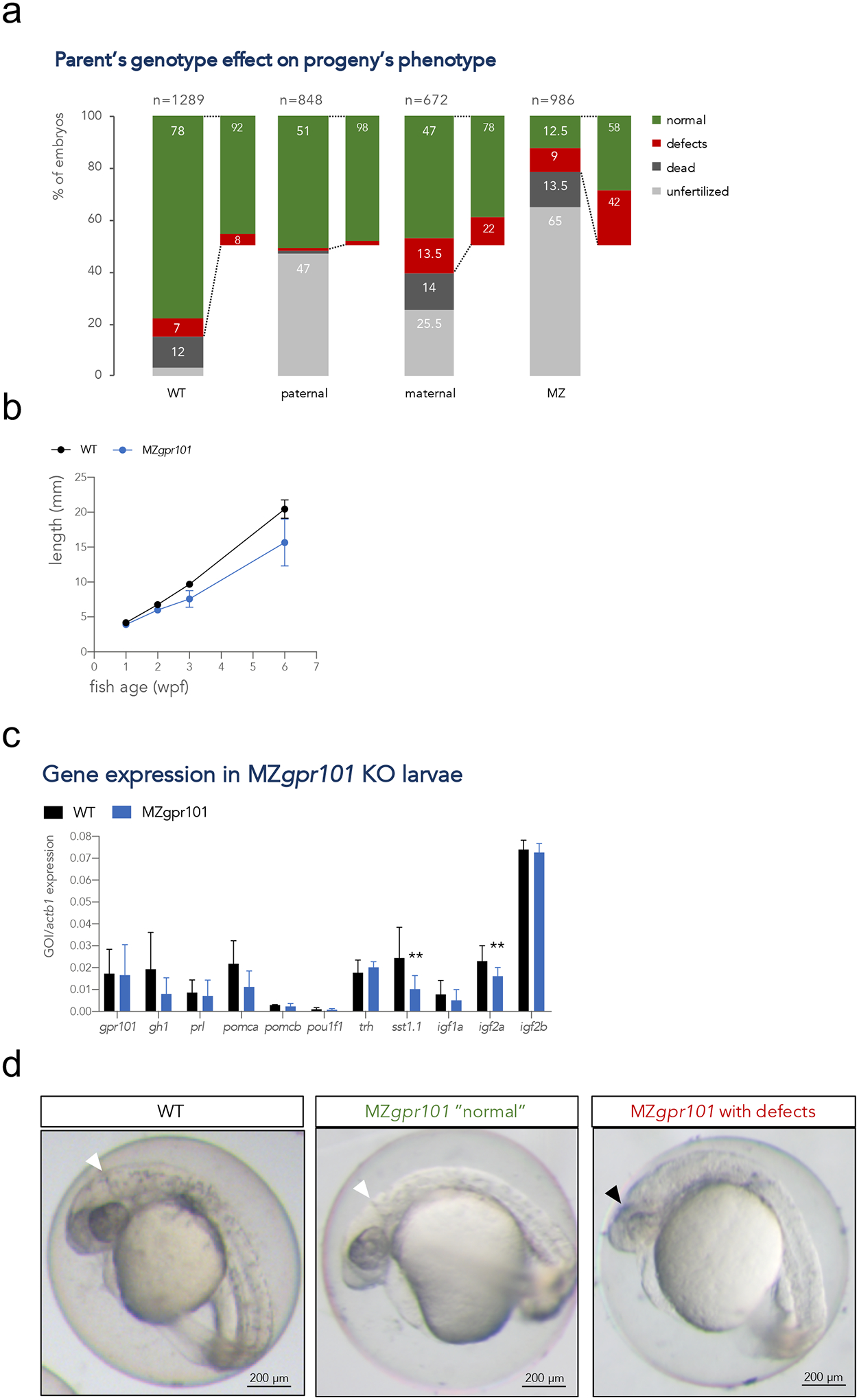
a) Embryos from parents of four different genotypic combinations were generated to assess the parent-of-origin’s contribution to phenotypes: WT (gpr101+/+ embryos from 4 crosses), paternal mutants (gpr101+/− embryos from 4 crosses, which have the maternal and zygotic gpr101, but lack potential paternal gpr101), maternal mutants (gpr101+/− embryos from 1 harem cross consisting of two females and two males, which lack maternal, but contain zygotic and potential paternal gpr101), maternal-zygotic (MZ, gpr101−/− embryos from five crosses, which lack maternal, zygotic and potential paternal gpr101). Each column indicates the percentage of phenotypes observed at morphological inspection at 1 dpf. The total number of analyzed embryos is shown at the top of each column. For the WT controls crosses, 485 fertilized embryos out of 1289 eggs were assessed; for the maternal cross, 128 fertilized embryos out of 672 eggs were assessed. b) Growth measurements for single-housed WT (black) and MZgpr101 mutants (blue). Standard length was measured at 1, 2, 3, and 6 wpf. The number of fish for each genotype at all time points is given in Supplementary Figure 8a. Data were analyzed by a two-way ANOVA; p < 0.001. c) The mRNA expression of gpr101, pituitary and hypothalamic genes, and growth regulators was measured by RT-qPCR in 1 wpf larvae. actb1 was used as endogenous control. GOI: gene of interest. *, p < 0.05; **, p < 0.01. d) Representative MZgpr101 embryos showing a WT-looking (normal) embryo (left panel) and an embryo lacking the midbrain-hindbrain boundary and showing brain necrosis (right panel). Seven out of 61 fertilized embryos (11.5%) presented with midbrain-hindbrain boundary defects and necrotic areas in the brain. The white arrowhead indicates the midbrain-hindbrain boundary, while the black arrowhead indicates a necrotic area. Anterior is right. Scale bar: 200 μm.
We then measured the expression of gpr101, pituitary and hypothalamic hormones encoding genes, and growth regulators in larvae at 1 wpf. gpr101 expression between MZgpr101 mutants and WT controls was similar, which is important because reduced expression due to nonsense-mediated mRNA decay in some cases leads to genetic compensation (El-Brolosy et al., 2019). We observed significant decreases in igf2a and somatostatin 1 (sst1.1) expression levels (p = 0.0096 and p = 0.0015, respectively) and a tendency for gh1 (p = 0.0669, Fig. 3c). While these results seem contradictory (lower sst1.1 expression is expected to cause higher gh1 and igf2a levels), they nonetheless suggest that the HPS axis is perturbed in MZgpr101 mutants. To investigate whether this was the case, we conducted an unbiased morphometric analysis of the larval zebrafish brain using a voxel-based approach; this technique identifies quantitative changes in the size and/or shape of individual brain regions (Gupta et al., 2018). This analysis revealed that WT-looking MZgpr101 mutants had changes in the volumes of specific brain regions. Specifically, we observed that subdivisions of the hypothalamic-pituitary unit, and the sensory ganglia were larger, while the dorsal diencephalon was smaller (p < 0.01 for hypothalamus, p < 0.01 for dorsal diencephalon, p < 0.001 for sensory ganglia, Fig. 4).
Figure 4.
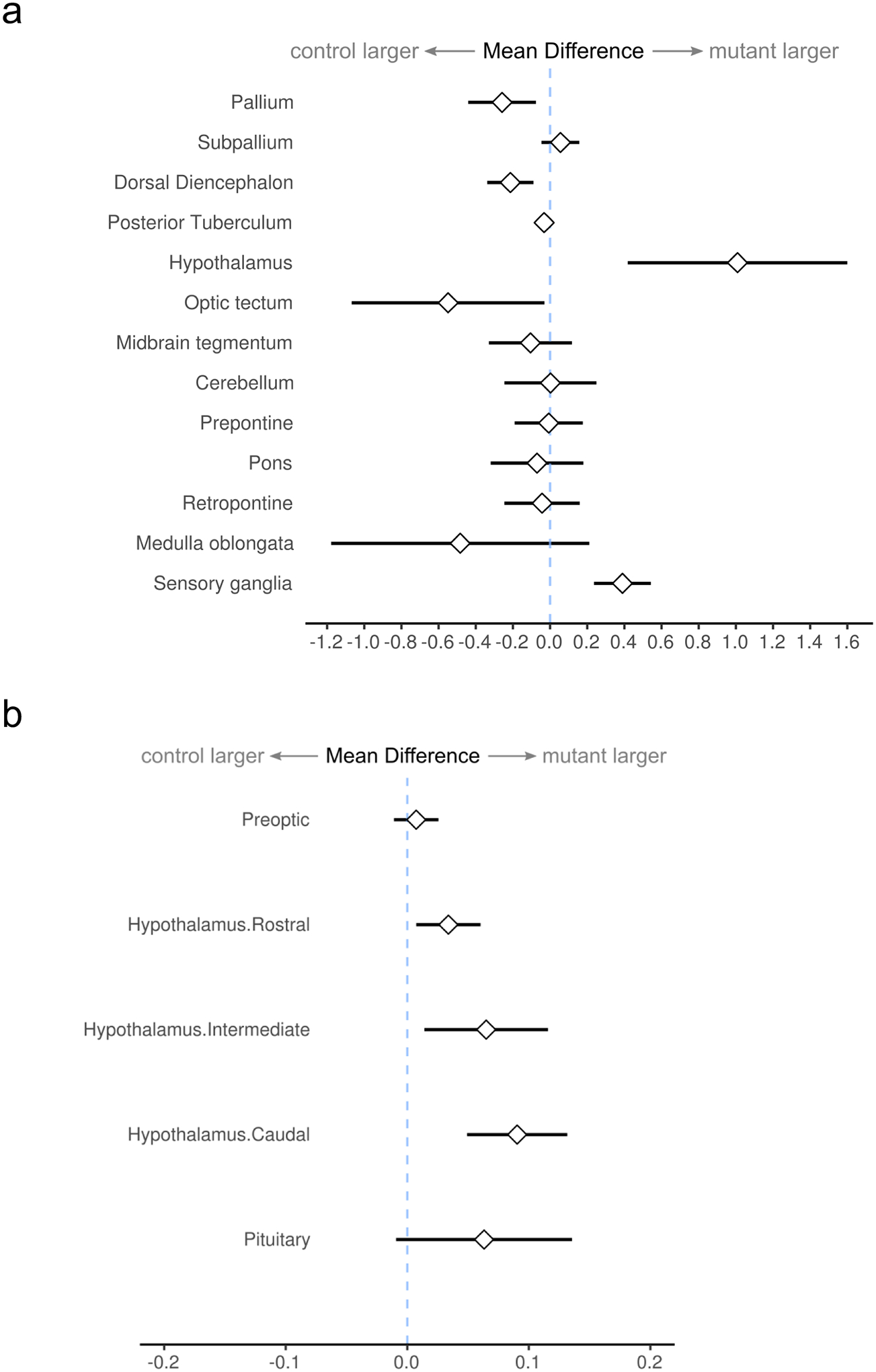
a) Morphometric analysis of MZgpr101 mutants, displaying changes in the relative volume of major brain divisions at 6 dpf (n=21, 22 control, mutant). Graph shows the mean difference (and 95% confidence interval) between the percentage of the brain occupied by each division in mutants compared to controls. b) Subdivisions of the hypothalamic-pituitary unit are expanded in mutants.
To identify the signaling pathways underlying these morphological alterations, we performed RNA sequencing (RNA-Seq) in WT-looking 1 wpf larvae reared at the optimal growth temperature of 28.5 °C. The Wnt signaling pathway emerged as the pathway harboring the highest number of differentially expressed (DE) genes, with the majority (11 out of 13) being upregulated (Supplementary Fig. 10a and 10b). Nervous system was also classified as the most significant enrichment cluster by Metascape (Supplementary Fig. 9c). Indeed, an overactive canonical Wnt pathway during embryonic development has been shown to lead to an enlarged hypothalamus in zebrafish (Wang et al., 2012) and mice (Newman et al., 2018). We then analyzed more closely the more severely affected MZgpr101 mutants that presented with morphological defects when assessed at an earlier developmental stage (1 dpf). As shown in Fig. 3d, the most consistent phenotypes were the lack of the midbrain-hindbrain boundary, a region with active neurogenesis, and necrotic areas in the brain. All these data support an involvement of gpr101 in developmental neurogenesis.
A parental effect on fertilization competence
In light of the critical role for maternal Gpr101 in the context of zygotic null (gpr101−/−) embryonic phenotypes, we next asked how a lack of maternal or paternal sources of Gpr101 might affect heterozygous embryos (gpr101+/−). To test for a paternal effect, we crossed WT females with gpr101−/− males to obtain gpr101+/− embryos that had functional RNA and possibly Gpr101 protein deposited in the egg cytoplasm, but no possible contribution from sperm. To test for a maternal effect, we crossed WT males with gpr101−/− females to obtain gpr101+/− embryos that theoretically might have received Gpr101 protein or RNA from the sperm of their WT fathers, but with no possible egg deposition from their gpr101−/− mothers. Heterozygous progeny of null males were frequently unfertilized (47%), but for those that were successfully fertilized, development was normal. Heterozygous progeny of null females were also unfertilized at elevated rates, but to a lesser degree (23.5%). An elevated incidence of developmental defects was also seen among maternal mutants (22% of those surviving beyond 1 dpf vs. 8% in WT controls), but no consistent phenotype was noted, in contrast to the characteristic head defects seen among the 42% defective MZgpr101 embryos surviving beyond 1 dpf (Fig. 3a). These results indicate that a) the consistent phenotypes seen in MZgpr101 embryos represent developmental requirements that can be relieved from either maternal or zygotic sources; b) there may be a low-penetrance maternal-only developmental requirement for Gpr101 with variable phenotypic consequences; and c) there is an additional requirement for Gpr101 protein or RNA in gametes or gametogenesis, especially for sperm or spermatogenesis, for fertilization competence. This biparental effect on fertilization competence is of sufficient magnitude to account for the low fertilization rates (35%) among MZgpr101 progeny, which were generated from gpr101−/− × gpr101−/− crosses.
Several maternal factors are deregulated in eggs devoid of gpr101
Having established that the defects in somatic growth observed in MZgpr101 mutants are caused by early developmental deficits, we decided to investigate whether loss of maternal gpr101 affects the levels of other deposited maternal mRNAs. Maternal transcripts, deposited in the yolk during oogenesis, drive the first stages of embryo development and are involved in the control of zygotic gene activation, starting at the midblastula transition (Zhang et al., 2017). We collected mature, unfertilized eggs from gpr101 KO females and performed RNA-Seq analysis. Pathway analysis showed an enrichment for gene sets involved in collagen formation (Fig. 5a); most of the genes in this pathway were downregulated (Supplementary Fig. 11a and Supplementary Table 5). These findings are interesting in light of a recent study reporting a role for maternal collagen type I transcripts during the earliest stages of zebrafish development (Gistelinck et al., 2016). We then examined individual genes whose expression was completely lost in the mutant eggs (Fig. 5b). Among them, we validated casp23, dachb, dachc, and adgrl3.1 by RT-PCR/RT-qPCR (Supplementary Fig. 11b). casp23 is a zebrafish-specific inflammatory caspase that is only maternally expressed (Spead et al., 2018), while the two dachshund paralogues dachb and dachc, which are critically involved in eye formation, and the adhesion GPCR adgrl3.1, are expressed in the brain of the developing embryo (Hammond et al., 2002,Lange et al., 2012). Based on the exclusively maternal expression pattern of casp23, we decided to test whether Casp23 re-expression in MZgpr101 embryos could rescue the mutant phenotype. Therefore, we injected MZgpr101 embryos at the 1-cell stage with 5 pg of casp23 mRNA. Casp23 expression did not rescue the phenotype (31% vs. 57% mutant-looking embryos in uninjected vs. casp23-injected MZgpr101 embryos, respectively, Supplementary Fig. 12a and 12b), but at a molecular level it was able to increase by about 3-fold the expression of adgrl3.1 at bud stage (Supplementary Fig. 12c). This suggests that the impaired expression of adgrl3.1 in MZgpr101 mutants might be a consequence of casp23 loss. No effects were seen on dachb and dachc expression (Supplementary Fig. 12c).
Figure 5.
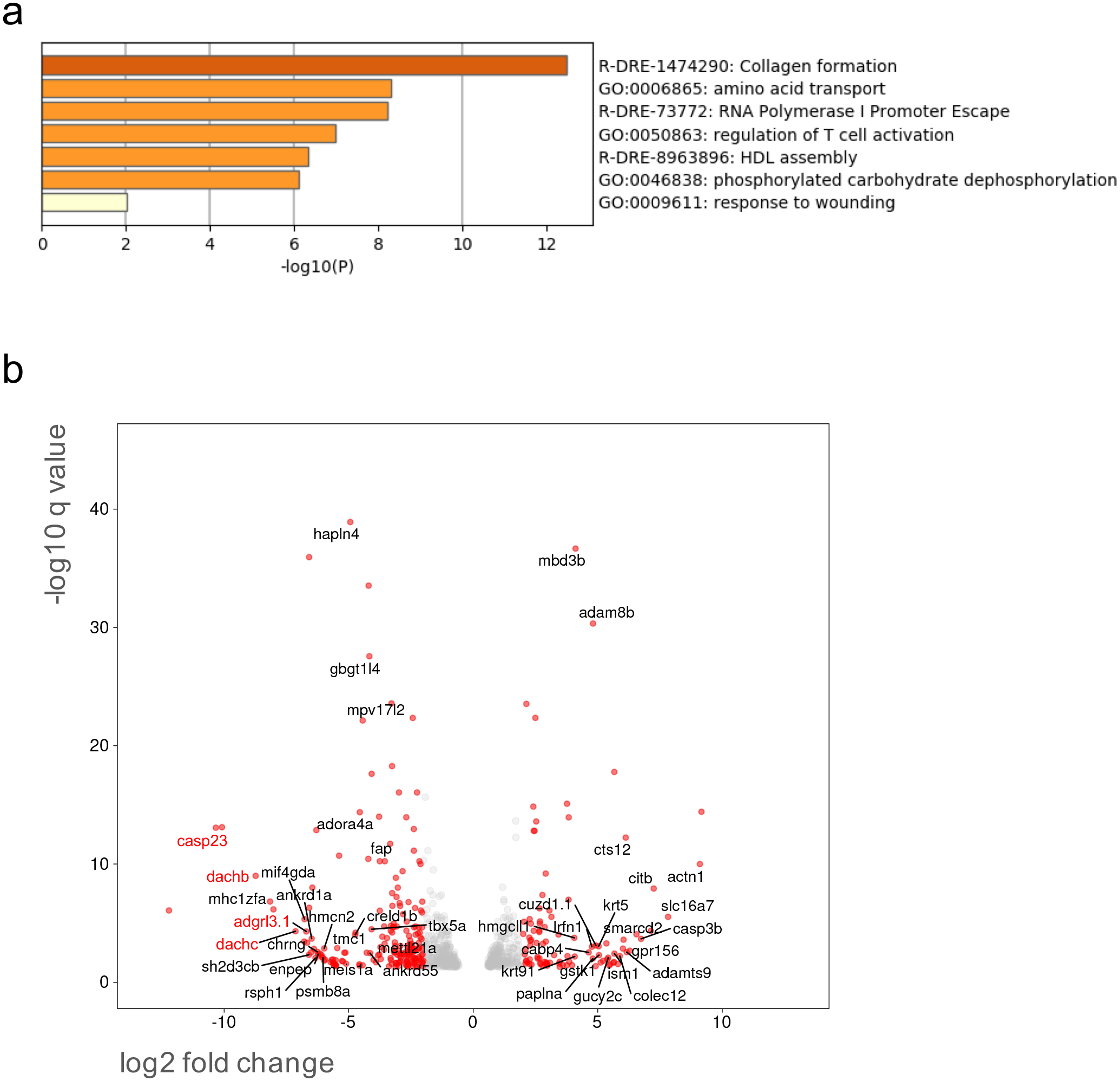
a) Pathway enrichment analysis in mature, unfertilized eggs collected from WT (n=4) and gpr101 KO (n=3) females was performed with Metascape by selecting the custom analysis option and uploading a background list of genes appropriate for the developmental stage under study (1-cell stage). The list was extracted from White et al. (White et al., 2017). Out of the 613 differentially expressed (DE) genes (±1 log2 fold change threshold), 557 genes were recognized by the software when applying these parameters. The top non-redundant enrichment cluster was collagen formation. b) Volcano plot showing significant DE genes (q < 0.05). Genes with a log2 fold change < −1 or > 1 are shown as red dots, the rest as grey dots. The names of genes (only those with an assigned name) with a log2 fold change < −4 or > 4 and q < 0.01 are shown. Selected genes showing the highest negative fold change and significance values were validated by RT-qPCR/RT-PCR and their names are reported in red.
MZgpr101 mutants are temperature sensitive and exhibit epiboly defects
During our characterization of MZgpr101 embryos, we noticed that when they were not immediately incubated at the optimal temperature of 28.5 °C, a significant number were found dead at 1 dpf. This observation suggested that MZgpr101 embryos are temperature sensitive. To confirm this hypothesis, embryos were reared at suboptimal temperatures ranging from 22 °C to 24.5 °C. The vast majority of fertilized MZgpr101 embryos died by 1 dpf when reared at 24.5 °C, and none were fertilized at 22 °C vs. 34% of WT (of which only 5.5% survived and had defects, Fig. 6a). Thus, comprised fertilization competence and lethality during the first 24 hpf in the absence of Gpr101 are exacerbated under hypothermic conditions. Having established the critical temperature impacting the embryonic development of MZgpr101 embryos, and their fertilization, we then precisely monitored their development from fertilization to 1 dpf by time-lapse microscopy. All embryos underwent asymmetrical cell divisions during the cleavage stage and a cell mound appeared on top of an enlarged cytoplasm. Then, the vast majority died at either the blastula (59.6%) or gastrula (27.7%) stages (Fig. 6b). From an epiboly standpoint, 49% of embryos died at early epiboly stages (epiboly initiation) and 28% at mid-late epiboly stages (epiboly progression). Some cell death was visible in all embryos during epiboly, and then the most common outcome was an abrupt burst of the yolk cell (Supplementary Fig. 13 and Supplementary Videos 1–4).
Figure 6.
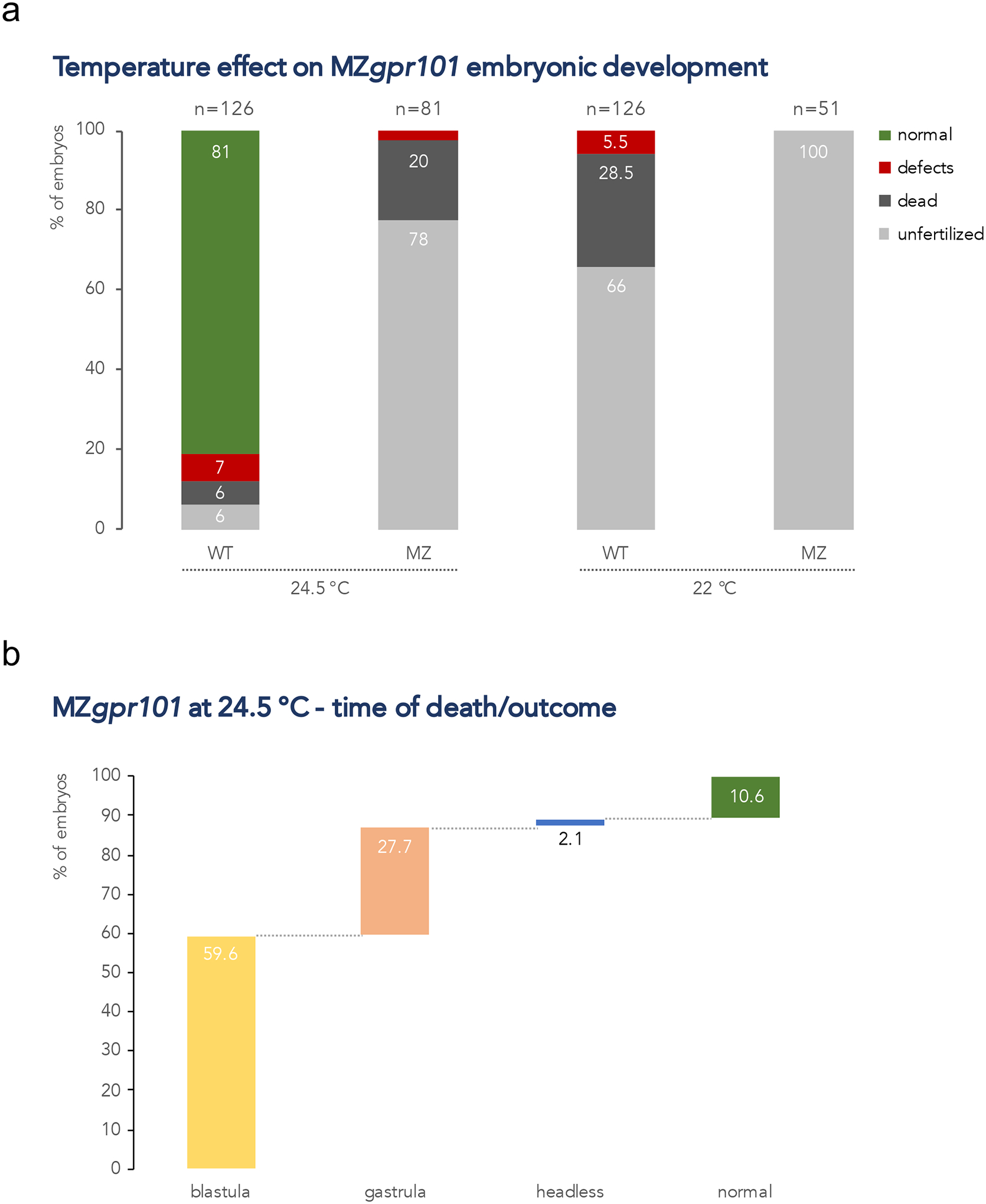
MZgpr101 mutants are temperature sensitive. a) Lower incubation temperatures (22 and 24.5 °C) adversely affect MZgpr101 embryonic development. Each column indicates the percentage of phenotypes observed at morphological inspection at 1 dpf. The total number of analyzed embryos is shown at the top of each column. b) MZgpr101 (n=48) and WT (n=17) fertilized embryos were reared at 24.5 °C and analyzed by time-lapse microscopy from 1-cell stage to 24 hpf. All WT embryos developed normally. The graph shows the percentage of MZgpr101 embryos that died at either the blastula or gastrula stages, and the phenotypes and percentage of the surviving embryos.
gpr101 misexpression affects developmental growth by perturbing multiple biological pathways
The temperature sensitivity exhibited by MZgpr101 mutants pointed to stages of early embryonic development when gpr101 function is critical. To identify the cellular pathways perturbed by gpr101 misexpression (loss- or gain-of-function, i.e. gpr101 upregulation and MZgpr101), we performed RNA-Seq in mutant embryos collected at 50% epiboly, the end of early epiboly and the time of the blastula to gastrula transition. Pathway analysis in MZgpr101 embryos showed an enrichment of gene ontology (GO) terms related to cell morphogenesis, cell adhesion, and drug/ion transport (Fig. 7). The enrichment for gene sets involved in cell morphogenesis and cell adhesion is consistent with the perturbed cell movements observed in the mutants. Among the genes coding for membrane transporters, many were members of the solute carrier (Slc) gene superfamily, most of which were upregulated (Supplementary Table 6). A statistical over-representation test for protein class showed an enrichment for zinc finger (ZnF) transcription factors; the majority of ZnF genes were downregulated (Supplementary Tables 6 and 7). In WT embryos, the expression of ZnF genes has been shown to start to rise sharply and coordinately from the onset of zygotic transcription – occurring at the midblastula stage – through 75% epiboly (White et al., 2017). The downregulation observed in the mutants suggests that activation of the zygotic genome may be impaired. Supporting this hypothesis, genes known to activate transcription during the maternal-to-zygotic transition, such as nanog, pou5f3, and sox3 (a member of the Soxb1 family) (Lee et al., 2013) were also downregulated (Supplementary Fig. 14b). At the same time, her3 expression was upregulated (Supplementary Fig. 14b), as observed in MZnanog (Veil et al., 2018).
Figure 7.
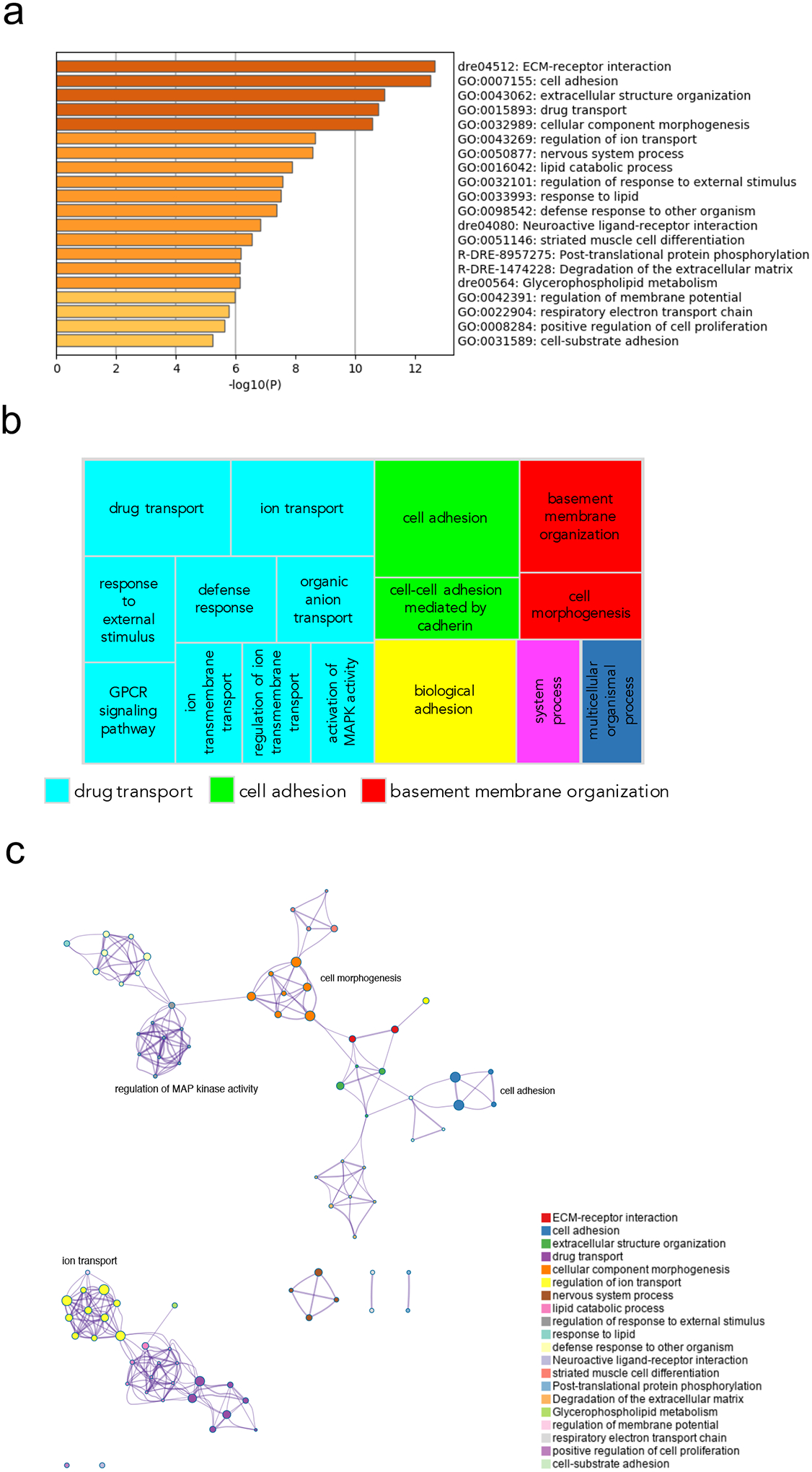
Pathway and network analysis at the onset of gastrulation (50% epiboly) in MZgpr101 mutants. a) Pathway enrichment analysis was performed with Metascape by selecting the custom analysis option and uploading a background list of genes appropriate for the developmental stage under study. The list was extracted from White et al. (White et al., 2017). Out of the 1008 DE genes (±2 log2 fold change threshold), 785 genes were recognized by the software when applying these parameters. The top non-redundant enrichment clusters are shown. b) Gene ontology (GO) treemap based on the enrichment of the most differentially expressed transcripts. GOrilla was used to determine enrichment of transcripts based on the minimum hypergeometric scoring method. Redundant GO terms were filtered with the GO Trimming tool and then visualized with REViGO. Each box is a single cluster representative. Box sizes correlate to the −log10 p value of the GO-term enrichment. Boxes with the same color are grouped by semantic similarity (SimRel, similarity = 0.7) into superclusters of loosely related terms. Drug transport and cell adhesion were confirmed among the most enriched superclusters. c) Network analysis was performed with Metascape using the settings outlined above for pathway analysis.
Pathway analysis in gain-of-function embryos in which gpr101 was exogenously expressed revealed an enrichment of GO terms related to early patterning, morphogenetic processes, and cell fate commitment (Fig. 8). Several genes showing the strongest upregulation are involved in the formation of the neural tube during neurulation (e.g., olig3, zic4, mdka), or are expressed in the brain and/or pituitary (e.g., scg3, tac3b, pax5) (Supplementary Fig. 6, 14a, and 14c). These transcriptomic results are consistent with the defective midline (cyclopia) and dorsoventral patterning (dorsalized embryos) we observed (Fig. 1b).
Figure 8.
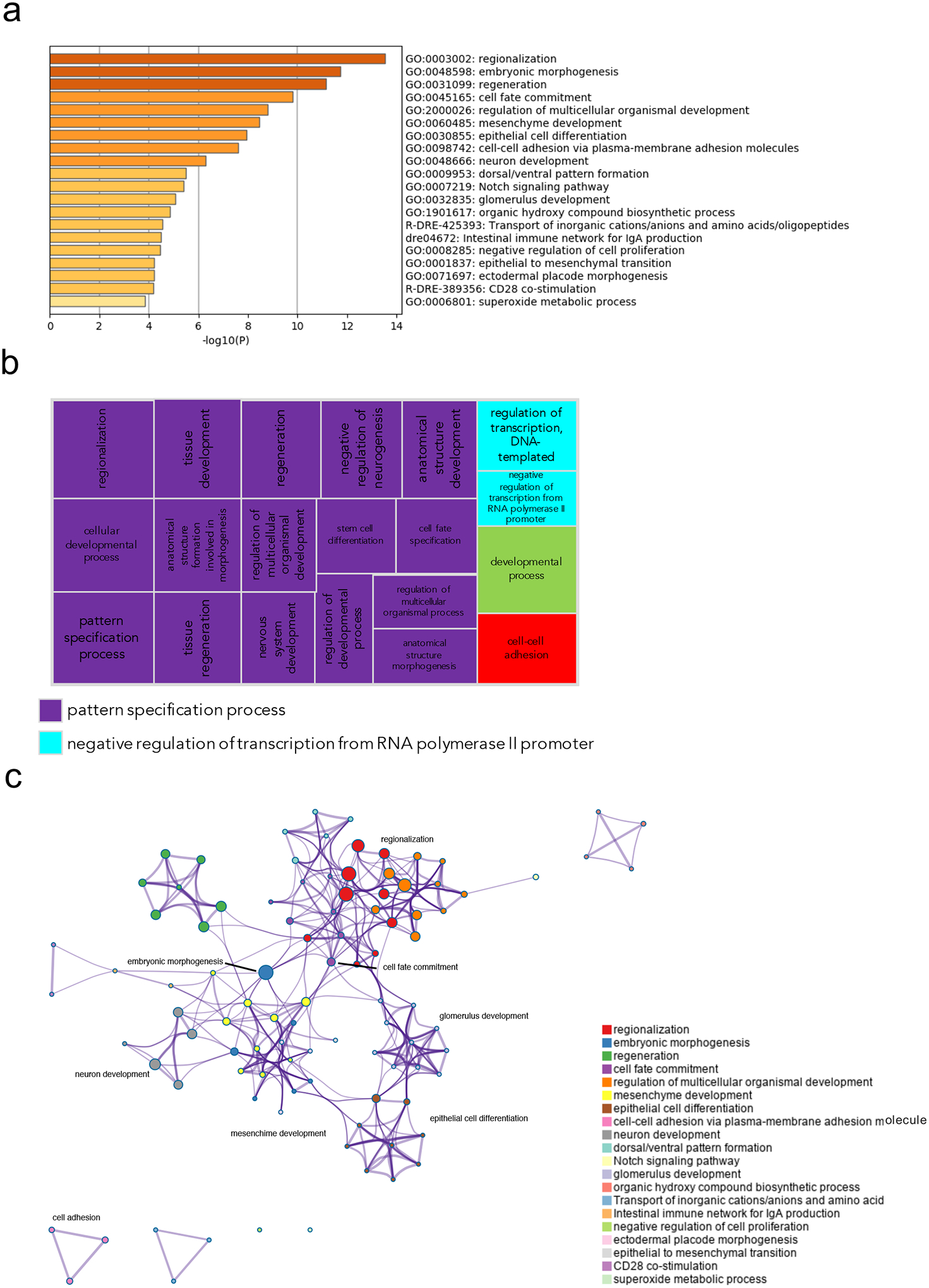
Pathway and network analysis at the onset of gastrulation (50% epiboly) in embryos with exogenous gpr101 expression. a) Pathway enrichment analysis was performed with Metascape by selecting the custom analysis option and uploading a background list of genes appropriate for the developmental stage under study. The list was extracted from White et al. (White et al., 2017). Out of the 313 DE genes (no log2 fold change threshold chosen), 300 genes were recognized by the software when applying these parameters. The top non-redundant enrichment clusters are shown. b) Gene ontology (GO) treemap based on the enrichment of the most differentially expressed transcripts. GOrilla was used to determine enrichment of transcripts based on the minimum hypergeometric scoring method. Redundant GO terms were filtered with the GO Trimming tool and then visualized with REViGO. Each box is a single cluster representative. Box sizes correlate to the −log10 p value of the GO-term enrichment. Boxes with the same color are grouped by semantic similarity (SimRel, similarity = 0.7) into superclusters of loosely related terms. Pattern specification process was the most predominant supercluster. c) Network analysis was performed with Metascape using the settings outlined above for pathway analysis.
Taken together, these results show that gpr101 affects developmental growth in different ways: loss of gpr101 disrupts morphogenesis, leading to cell death, while upregulation of gpr101 alters early patterning and differentiation (Supplementary Fig. 15).
Discussion
GPR101 exerts important but yet to be completely deciphered roles in the regulation of human growth, as illustrated by its de-regulation in X-LAG (Trivellin et al., 2014), a disease of extreme tall stature, and in human embryonic development, as we demonstrated by its strong expression in the fetal pituitary gland (Trivellin et al., 2016). By a combination of loss- and gain-of-function experiments performed in zebrafish, in this study we provide the first in vivo evidence in a zebrafish model that gpr101 regulates developmental growth. We also uncovered essential roles for this gene for fertilization competence and, at non-permissive temperatures, for viable morphogenesis during blastula/gastrula stages of early embryonic development. At permissive temperatures, gpr101 deficiency resulted in brain anomalies.
We previously showed that GPR101 expression is significantly upregulated (up to 1,000-fold) in the pituitary GH-secreting tumors of X-LAG patients (Trivellin et al., 2014). To study in vivo the effects of such a strong expression, we initially employed transgenic fish expressing gpr101 ubiquitously or in specific tissues relevant for X-LAG. However, that approach failed to induce potent gpr101 expression, thus preventing us to faithfully reproduce what happens in the human disease. Therefore, we resorted to injecting zebrafish embryos with gpr101/GPR101 mRNA. This potent, but transient, upregulation caused early axis patterning defects that manifested as cyclopean and dorsalizing phenotypes. Both orthologues produced the same phenotypes in similar numbers. These results support an evolutionary conservation of structure between the human GPR101 gene and its zebrafish orthologue, suggesting zebrafish is a suitable genetic model for uncovering functions of this gene that are also relevant to human biology.
Moreover, these findings suggest that the ectopic expression of gpr101 perturbs dorsoventral axis formation and midline development. Accordingly, a transcriptomic analysis conducted at the onset of gastrulation revealed an enrichment for gene sets responsible for patterning the embryonic axes and coordinating morphogenetic movements. The genes showing the strongest differential expression are known to participate in the formation of the neural tube or to be later expressed in the CNS and the pituitary. Interestingly, the two endogenous genes with the highest expression were secretogranin III (scg3) and neurokinin B (tac3b). Both genes were still strongly upregulated at the end of gastrulation. scg3 belongs to the granin family, a group of proteins involved in neuroendocrine cell secretion, and is expressed in the zebrafish pituitary gland, telencephalon, and midbrain (Toro et al., 2009). tac3b encodes for neurokinin B, a neurohormone expressed in the forebrain and ovary in sexually mature zebrafish (Biran et al., 2012), that, by controlling GnRH secretion, is involved in the endocrine regulation of reproduction in vertebrates. Neurokinin B was also recently reported to regulate the expression of growth-related genes in another fish species (Zhang et al., 2019). Given what we know about GPR101 and its roles in regulating hormone secretion, growth, and possibly reproduction (Trivellin et al., 2020,Abboud et al., 2020), these genes represent important targets that warrant further future studies.
It is also interesting to note that holoprosencephaly (HPE), the human equivalent of cyclopia, is frequently associated with abnormalities of anterior midline structures such as the hypothalamus and pituitary (Kelberman et al., 2009). Point mutations and copy number variations in several genes, mostly transcription factors, have been identified in patients, but the majority of cases still do not have a genetic diagnosis (Tekendo-Ngongang et al., 2000 [Updated 2020]). We believe our findings should prompt the investigation of GPR101’s role and possibly defects in patients with HPE.
We then examined whether transient gpr101 upregulation had a long-lasting effect on zebrafish growth trajectories. While any injected synthetic mRNA rarely lasts more than 3 days, the modified expression of some of the genes it regulates during embryonic development can have long-lasting consequences, for example in growth (Leinonen et al., 2019). However, in our model, we did not observe a significant increase in size in juvenile fish compared to controls.
The failure to drive zebrafish growth increase via transient expression of mRNA or with three distinct Gal4 drivers of our transgene initially suggested three possibilities to us. 1) That GPR101 mis-expression is only part of the human phenotype and is not sufficient for driving gigantism in model organisms. 2) That some peculiarity of zebrafish biology prevents misexpression of Gpr101 from driving additional growth. 3) That each of our strategies failed to deliver sufficient levels of Gpr101 at the right time or place to drive additional growth. Recently, Abboud et al. reported excessive growth and biochemical evidence of GH excess in a pituitary-specific transgenic mouse model for Gpr101 (GhrhrGpr101) (Abboud et al., 2020). They reported an approximate 15-fold increase in mouse Gpr101 expression in terminally differentiated GH-secreting cells. This finding narrowed our list of possibilities to two: a peculiarity of zebrafish biology or a failure to deliver enough Gpr101 to its target. We cannot yet distinguish between these two possibilities, but given how basic molecular pathways are so often conserved between vertebrate animals, and recognizing several limitations of our misexpression studies, we lean toward the failed delivery hypothesis. Some specific caveats in our misexpression studies were as follows. For the mRNA injection, we had to remove all dorsalized and cyclopean embryos, leaving us with a pool of embryos that by definition had received less gpr101 mRNA, and because injected mRNA degrades within a few days in developing embryos, even this low dose could not be expected to persist over time. For those transgenic experiments in which we measured gpr101 levels (Supplementary Fig. 2b), we saw only a 2-fold increase in the head, far below the up to 1,000-fold increase seen in X-LAG patients or the 15-fold increase seen in the mouse model. To resolve the question of whether zebrafish are capable of modeling Gpr101-driven excessive growth, alternative strategies might be employed in future studies including, but not limited, to the following. (1) Use of human, rather than zebrafish gpr101, which was more potent in our cAMP induction study. (2) Use of a more direct transgenic system in which GPR101 is linked to a strong promoter. (3) Identifying founders with multiple integrations, which might drive higher expression levels. (4) Using a version of the Gal4 response element with 14 UAS sites rather than the 4 UAS version we used.
A more feasible approach to gaining functional insights was the creation of a LOF mutant. This enabled us to investigate whether loss of gpr101 has an effect on body growth that is reciprocal to what is seen in the human and mouse pathology and to identify the perturbed developmental processes. The gpr101 null mutants that we generated by CRISPR/Cas9 were indeed found to be significantly thinner and shorter than their WT siblings, while the heterozygous fish showed an intermediate phenotype. However, we were not able to find significant differences in the expression of pituitary/hypothalamic hormones or downstream growth effectors such as IGFs. Further studies are thus necessary to find out what factors are causing the impaired juvenile growth.
In zebrafish, distinct from mammals, zygotic gene expression begins later in embryonic development, around the 1,000-cell stage (midblastula transition) (Harvey et al., 2013). Thus, initial embryogenesis is regulated by maternal factors stored in the blastomeres and yolk (Abrams and Mullins, 2009). Moreover, maternally-supplied nutrients (e.g., mRNAs, proteins, and lipids) deposited in the protruding yolk sac sustain the metabolic functions and growth of the embryo until the onset of exogenous feeding. Likewise, during the first trimester of human embryonic development, the yolk carries out similar functions until the onset of placental-fetal exchange (Sant and Timme-Laragy, 2018). Zebrafish, therefore, is an ideal model organism to investigate the contribution of maternal factors.
Since we know that gpr101 is expressed both maternally and zygotically (Trivellin et al., 2016), we decided to investigate whether the presence of maternal gpr101 products attenuated the phenotype of the zygotic mutants by fully or partially compensating for the loss of gene function. To this end, we generated maternal zygotic gpr101 mutants (MZgpr101). We observed that MZgpr101 mutants presented a stronger impairment of growth than zygotic KOs and we excluded that this phenotype was caused by the mutants being outcompeted for food by WT controls. Moreover, the difference in body size between MZgpr101 mutants and controls was already appreciable at the larval stage, implying that a complete lack of gpr101 affects early growth. Gene expression studies conducted at the larval stage showed a perturbation of the HPS axis. Our findings were corroborated by a morphometric analysis of the larval brain showing an enlarged hypothalamus. We observed that several members of the Wnt signaling pathway were upregulated in the mutants, in agreement with previous studies reporting that an overactive canonical Wnt pathway leads to an enlarged hypothalamus during embryonic development (Wang et al., 2012,Newman et al., 2018). These results, therefore, suggest that gpr101 might be a negative regulator of the Wnt pathway and provide the first evidence that impaired expression of gpr101 affects the development of the hypothalamic-pituitary unit. How these findings can be reconciled with the pathological proliferation of pituitary cells seen in XLAG giants is at present difficult to explain. We can speculate that an enlarged hypothalamic-pituitary unit either represents a compensatory mechanism trying to counteract an impaired growth-enhancing pathway caused by loss of gpr101 or that it is the consequence of a perturbed developmental process that affects different cell types than those sensitive to GPR101 duplication. In this respect, the observed differences in expression for Wnt-related genes may reflect tissue patterning defects in the mutants.
We then investigated whether loss of maternal gpr101 affects the deposition of maternal mRNAs in the yolk. The accumulation of maternal mRNAs is a non-random process related to specific biological pathways relevant for early embryogenesis (Aanes et al., 2011,Rauwerda et al., 2016,Mehjabin et al., 2019). Not surprisingly, the overall composition of the maternal transcriptome was found to be tightly regulated (Cheung et al., 2019). Transcriptomic studies conducted in mature, unfertilized eggs deriving from gpr101 KO females revealed that maternally-deposited gpr101 transcripts were required for the expression of several genes, such as casp23, dachb, dachc, and adgrl3.1. casp23 is an inflammatory caspase expressed only prior to gastrulation, which suggests an important role during the initial stages of embryonic development (Spead et al., 2018). Since multiprotein complexes termed inflammasomes are required for the activation of inflammatory caspases and seem to have a function in reproduction, it was speculated that inflammatory caspases may link some aspects of innate immunity to reproductive biology (i.e. gamete maturation and the early stages of embryonic development). Supporting this hypothesis, mice deficient in different components of the inflammasome displayed defects at the early stages of embryonic development (Martinon and Tschopp, 2007). To test this hypothesis, we verified whether re-expression of casp23 rescued the phenotype of MZgpr101 embryos. We were not able to rescue the phenotype in the mutants although partial recovery, in the form of increased adgrl3.1 expression, was observed.
In MZgpr101 embryos, the expression of adgrl3.1, dachb, and dachc remained downregulated at later stages. adgrl3.1 is an adhesion GPCR broadly expressed in the zebrafish brain, where it plays an essential role in establishing accurate dopaminergic signaling during development. Reduced adgrl3.1 has previously been shown to lead to misplacement of dopamine-secreting neurons in the ventral diencephalon (Lange et al., 2012). This finding requires further investigation given the well-known role of dopamine in negatively regulating prolactin secretion in the pituitary. The loss of expression of the two dachshund paralogues – genes fundamental for compound eye development (Chen et al., 1997) – is also intriguing based on the cyclopean phenotype we observed when over-expressing gpr101.
Our characterization of MZgpr101 embryos also revealed remarkable subfertility, with the main contribution being paternal, and an incompletely penetrant phenotype consisting in the lack of the midbrain-hindbrain boundary and brain necrosis. These results further support a role for gpr101 in the regulation of developmental neurogenesis.
In this study, we also showed that MZgpr101 embryos are temperature-sensitive mutants that manifest a highly penetrant phenotype when raised at the restrictive temperature of 24.5 °C. Temperature sensitive mutations are rare in higher organisms but in multiple instances in zebrafish (Tian et al., 2003,Zeng et al., 2015,Wang et al., 2019) have provided fundamental insight into our understanding of gene function. In our experiments, the restrictive temperature impaired cell organization at the cleavage stage, resulting in epiboly failure and finally, most commonly, lethality. In teleost fish, gastrulation, the process that generates the primary germ layers and the embryonic axis, is coordinated with epiboly (Bruce, 2016). Transcriptomic analysis in MZgpr101 embryos collected at the onset of gastrulation (50% epiboly), showed an enrichment for pathways underlying cell morphogenesis and cell adhesion, reflecting the perturbed cell movements observed in the mutants. Drug/ion transport was another strongly enriched pathway. The transport and gradient of different factors is important for embryo’s growth during development. Moreover, several non-endogenous compounds can use these transporters and lead to death or malformation, with the early developmental stages being particularly sensitive to metal intoxication (Jezierska et al., 2009). This finding raises the possibility that MZgpr101 embryos are more sensitive to the toxic action of exogenous drugs and heavy metals.
Finally, the observed downregulation of ZnF and other transcription factors with important roles during the maternal-to-zygotic transition, suggests an impairment in the activation of the zygotic genome. All together, these complementary LOF experiments led us to those embryonic stages and processes most affected by gpr101 loss.
In conclusion, the findings presented here show that both the loss and ectopic activation of gpr101 affect, in different ways, fertility, embryonic events surrounding gastrulation, the subsequent morphogenetic program of brain development, and, ultimately, somatic growth. Defective somatic growth is likely the consequence of the perturbed early developmental processes. We also identified several targets and pathways, opening up new research avenues for this orphan GPCR in both animal models and patients.
Methods
Zebrafish husbandry and embryo care
The WT Ekkwill (EK) zebrafish strain was used for all experiments. For breeding, males and females were placed in tanks overnight with a transparent separating divider that was removed in the morning, allowing them to spawn naturally and enabling synchronization of developmental stages. Unless otherwise stated, embryos were raised in egg water (Westerfield, 2007) at 28.5 °C. Embryos were observed in their chorions using a dissecting microscope and scored at 1 dpf (pharyngula period, when most organ systems have formed) for morphological defects and survival. Fertilization rates were also documented for all crosses.
Mature, unfertilized eggs were collected from seven females (four WT and three gpr101 KO) by the squeezing procedure described in http://zfin.org/zf_info/zfbook/chapt2/2.8.html#4 and (Cheung et al., 2019). Briefly, females and males were placed in tanks overnight with a transparent separating divider. In the morning the females were anesthetized briefly in egg water with 0.16 mg/ml tricaine (A5040, Sigma-Aldrich, St Louis, MO, USA) and gently squeezed to extrude their eggs. Eggs were then incubated in egg water for 5 minutes in order to confirm their viability, as healthy eggs undergo rapid stereotypical changes known as “activation” during this time. Activated eggs were then snap-frozen in dry ice and stored at −80°C.
All zebrafish lines were maintained in the aquatic animal facility at 28.5°C under a 14 h light:10 h dark photoperiodic regime in a filtered freshwater recirculation system according to our Animal Use Protocol (AUP), approved by the Institutional Animal Care and Use Committee (IACUC) of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, MD, USA.
Growth measurements
For experiments tracking zebrafish body growth during larval and juvenile stages, the number of fish per tank was kept similar between groups. For these experiments, the growth of individual fish was not tracked over time but rather each fish was measured only once at a single time point. Depending on the experiment, samples were collected at different time points, including 6 dpf, 12 dpf, 3, 4, 6, 7, and 9 wpf. Before measuring body growth, fish were anesthetized with 0.16 mg/ml tricaine. Each fish was then gently moved to a tissue to quickly wick away water on the body surface, photographed next to a ruler to measure standard length (distance from the snout to the caudal peduncle, or in pre-flexion larvae that do not have a caudal peduncle, distance from the snout to the posterior tip of the notochord, as defined in (Parichy et al., 2009)), and weighed on an analytical balance. After morphometric measurement, fish were culled for tissue collection. Total brain tissue (including braincase) was collected from 6 wpf fish by dissection. Standard length measurements were obtained using ImageJ software (NIH, Bethesda, MD, USA).
Female zebrafish initiate puberty around 45 dpf (Chen and Ge, 2013). To determine whether sex influenced body growth among groups, three individuals blind to the genotype independently judged the sex of 9 wpf specimens. A consensus was reached and sex was matched to genotype.
For longitudinal growth studies in which individual fish were kept separate from each other, larvae were first kept in 6-well plates from 5 to 12 dpf, then transferred to a modified DC-96 tank (R&D Aquatics), in which 24 zebrafish can be reared in isolated chambers.
Temperature stress exposure
MZgpr101 embryos obtained from matings of gpr101 homozygous KO fish were split into three groups at the 1-cell stage, and raised at 22, 24.5, and 28.5 °C until the end of the experiment.
Generation of a transgenic gpr101 line
gpr101 (ENSDARG00000039218) was cloned into a Tol2 construct downstream of 4xUAS between essential cis-sequences of Tol2. This construct harbored a crystallin promoter driving CFP expression in lens that allows for monitoring transgene insertion. Zebrafish carrying the UAS:gpr101 transgene were generated by injecting WT EK embryos at the 1-cell stage with 50 pf of plasmid and 50 pg of Tol2 transposase mRNA. Transgene-carrying F0 founders were identified by looking for CFP expression in the eyes. By performing inverse PCR (Potter and Luo, 2010) in twenty positive fish we identified one male founder with a single transgene insertion. Primer sequences for inverse PCR are reported in Supplementary Table 1. Database analysis identified that the insertion was located within an intron of the antisense transcript to irx6a on chromosome 7. Zebrafish carrying the SAGFF(LF)73A;UAS:GFP transgenes were obtained by Dr. Koichi Kawakami at the National Institute of Genetics, Japan (Asakawa et al., 2008). These transgenic fish express Gal4FF in the whole body and can be identified by looking for the expression of a GFP reporter. Zebrafish carrying the Xa207:Gal4;UAS:dsred and the HuC:Gal4;UAS:nuclear-mCardinal transgenes were available in our Institute’s zebrafish facility (Dr. Harold A. Burgess lab). Primer sequences for genotyping are reported in Supplementary Table 1. Female Gal4FF drivers were then crossed with the male UAS:gpr101 F0 founder. F1 and F2 zebrafish were used to track body growth and for quantitative RT-PCR experiments.
Generation of gpr101 KO lines
Zebrafish carrying gpr101 mutations were generated by CRISPR/Cas9-mediated gene targeting as follows. We used the UCSC Genome Browser’s NHGRI “GG” track on Zv9 to identify a target on the non-coding strand of gpr101’s exon 1: GGCAGCCCTGTCGCCCTCTTTGG (PAM underlined) and a full-length gRNA was synthesized in vitro using the methods of Varshney et al. (Varshney et al., 2015). Briefly, a single-stranded oligonucleotide (ss-oligo) hybrid of gene-derived PAM-proximal sequence and sequence complementary to a “universal” ss-oligo that is used for all such reactions was annealed to the universal ss-oligo. Non-complementary single-stranded portions of the duplex were then end-filled by a single round of PCR. The double-stranded product was then used as template for in vitro synthesis of a full-length gRNA containing both Cas9-binding and gene-targeting sequence. This gRNA was injected into freshly fertilized WT zebrafish embryos together with mRNA encoding Cas9 that had been synthesized in vitro using the plasmid pT3TS-nCas9n as a template (Jao et al., 2013). Most surviving injected embryos were raised as potential F0 founders, but a representative sample was reserved for total DNA extraction at 2–5 dpf. The gRNA cutting efficiency was determined by fluorescent PCR of DNA from these individual F0 embryos’ genomes, using the methods of Carrington et al. (Carrington et al., 2015), and quantified as a function of the relative abundance of WT sized amplicons versus aberrantly large (indicating insertions) or small (indicating deletions) amplicons. We observed an average (n=6) rate of 19% somatic allele disruption, which we find to be sufficient for reliable germline transmission. Sibling injected F0 embryos were accordingly raised to sexual maturity (2–4 months), mated with WT zebrafish, and the F1 progeny were analyzed as above by fluorescent PCR, revealing a variety of gpr101 heterozygous F1 embryos and thus identifying several F0 fish harboring this variety of altered gpr101 alleles in their germlines. Two frame-shifting gpr101 alleles – a deletion of 13 nucleotides and a deletion of 5 nucleotides – were selectively propagated from these F0 founders for genotype-phenotype analyses. MZgpr101 indicates maternal zygotic mutants obtained from the offspring of homozygous (gpr101y629/y629 or gpr101y630/y630) in-crosses. To generate embryos lacking maternal but not zygotic gpr101 activity, we crossed gpr101Δ5/Δ5 females to WT males. To generate embryos lacking paternal but not zygotic gpr101 activity, we crossed WT females to gpr101y629/y629 or gpr101y630/y630 males.
Possible off-target effects generated by CRISPR/Cas9 were bioinformatically predicted by CRISPOR (Haeussler et al., 2016) and Sanger sequencing found no deleterious coding SNVs in the closest theoretical hits in coding genes: ep400 and hivep3a (primer sequences reported in Supplementary Table 1).
Genotyping
To genotype gpr101 KO and transgenic mutants, genomic DNA was extracted from either whole embryos/larvae or the caudal fin of adults with DNA extraction buffer (10 mM Tris pH 8.2, 10 mM EDTA, 200 mM NaCl, 0.5% SDS and 200 μg/ml proteinase K). Genomic DNA was then used as template for genotyping PCR. Genotypes were then determined by either Sanger sequencing or fluorescent PCR as reported above (primer sequences for both methods are reported in Supplementary Table 1).
Expression constructs
A 1,438 bp genomic sequence corresponding to the entire zebrafish gpr101 gene (ENSDARG00000039218) (Trivellin et al., 2016), and a 1,527 bp genomic sequence corresponding to the entire human GPR101 coding sequence (NM_054021.1), were cloned into the pCS2 vector. The zebrafish casp23 genomic sequence (ENSDARG00000014657), originally cloned into a PCRII vector(Spead et al., 2018), was subcloned into the pCS2 vector using the BamHI and XhoI restriction sites. The pCS2+ vector containing the eGFP coding sequence was a generous gift of Dr. Igor Dawid (Gothilf et al., 2002). The nucleotide sequence of each construct was verified by Sanger sequencing.
mRNA synthesis and microinjection
The gpr101, GPR101, and eGFP plasmids were linearized with NotI, while the casp23 plasmid was linearized with KpnI. Quality and quantity of RNA were assessed using both a Bioanalyzer (Agilent, Inc., Santa Clara, CA, USA) and NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA). Sense strand capped mRNA was synthesized with SP6 RNA polymerase using the mMessage mMachine system (Thermo Fisher Scientific) following the manufacturer’s protocol. In vitro synthesized RNA was microinjected into the yolk of embryos at the 1-cell stage using a World Precision Instruments pneumatic PicoPump PV 830. For gpr101/GPR101 exogenous expression experiments, WT embryos were injected with 75 pg of mRNA. The specificity of phenotypic effects was assessed by using an equal amount of eGFP mRNA as control. Optimization experiments (not shown) were performed using higher amounts of mRNA (200 and 600 pg). We decided to use 75 pg as this was the lowest dose able to induce a phenotype but causing reduced mortality.
For rescue experiments performed in MZgpr101 embryos, 5 pg of casp23 mRNA were injected and embryos subsequently incubated at 24.5 °C.
Luciferase assays
The Human Embryonic Kidney (HEK)-293 AD cell line was purchased from ATCC (CRL-1573). Cells were tested for mycoplasma contamination and tested negative. Short Tandem Repeat (STR) profiling was performed by WiCell Cytogenetics (Madison, WI, USA) and the identity of the cells was confirmed. Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, high glucose, pyruvate, no glutamine; 10313, Gibco) supplemented with 10% foetal bovine serum (100–106, Gemini Bio Products) and 1% antibiotic–antimycotic (15240–062, Gibco) in a humidified atmosphere at 37°C with 5% CO2.
HEK-293 AD cells were seeded in 12-well plates at a density of 2.5 × 105 cells per well. After 24 h, cells were transfected with TurboFect (R0531, Thermo Fisher Scientific) according to the manufacturer’s protocol, using 1 μg of GPR101/gpr101 expression vectors, 800 ng of the pGL4.29[luc2P/CRE/Hygro] vector containing a cAMP response element (CRE) that drives the transcription of the Firefly luciferase reporter gene (Promega, Madison, WI, USA), and 40 ng of the Renilla vector (pRL-SV40, Promega). The empty pCS2 vector was used as negative control. 24 h after transfection, cells were lysed and Firefly and Renilla luciferase activities were measured consecutively in the same sample using the Dual-Luciferase Reporter Assay System (Promega) following the manufacturer’s protocol. Ratios of Firefly vs. Renilla luminescence signals, serving as a measure for reporter activity normalized for transfection efficiency, were determined using a FLUOstar Omega microplate reader (BMG Labtech, Ortenberg, Germany).
RNA extraction and sequencing
Embryos, larvae, and juvenile fish were lysed and mechanically homogenized in TRIzol (Thermo Fisher Scientific) using a Next Advance Bullet Blender Homogenizer (Scientific Instrument Services, Ringoes, NJ, USA). Chloroform was then added and phase separation was obtained following vortexing and centrifugation. The top aqueous phase containing total RNA was isolated and purified using an RNA Clean and Concentrator kit (Zymo Research, Irvine, CA, USA). An on-column DNase I treatment to remove genomic DNA contamination was also performed. Quality and quantity of RNA were assessed using both a Bioanalyzer (Agilent, Inc.) and NanoDrop (Thermo Fisher Scientific).
Several embryos/larvae were pooled together for extracting RNA samples that then underwent RNA sequencing (Supplementary Table 2). For these experiments, embryos were reared at 28.5 °C. The library construction and sequencing were performed by Novogene (Novogene Corporation, Beijing, P.R. China). Briefly, after the quality control steps, mRNA was enriched using oligo(dT) beads. Then, the mRNA was fragmented randomly to short fragments of 200–700 nucleotides in length by adding fragmentation buffer consisting of divalent cations. These short fragments were used as templates to synthesize the first-strand cDNA using random hexamer-priming. The second-strand cDNA was synthesized using second-strand synthesis buffer (Illumina) including dNTPs, RNaseH, and DNA polymerase I. After a series of terminal repair, A ligation, and sequencing adaptor ligation, the double-stranded cDNA library was completed through size selection and PCR enrichment. Finally, the qualified paired-end cDNA library was sequenced with an Illumina Hiseq2000 sequencer.
Differential gene expression and Pathway analyses
FastQC (2019) was used for quality control, trimmomatic) Bolger et al., 2014(for conditional reads trimming, and Star (Dobin et al., 2013) for alignment to the GRCz11 reference genome. HTSeq (Anders et al., 2015) was used for reads quantification and DESeq2 (Love et al., 2014) for data normalization and downstream differential expression analysis. The final analysis was performed on R Studio (Version 1.2.5001, R Studio Inc, 2009–2019). The DE gene lists are available in Supplementary Table 3. Ensembl gene IDs were converted to gene names with g:Profiler (https://biit.cs.ut.ee/gprofiler/convert) (Reimand et al., 2016). Volcano plots were generated in R using the ggplot2 package. The differentially expressed genes obtained from the RNA-Seq analysis were then subjected to over-representation and functional classification analyses using three different softwares: Metascape, Protein ANalysis THrough Evolutionary Relationships (PANTHER), and GOrilla. For these analyses, we used for each gene list different log2 fold change thresholds to limit the number of analyzed genes to less than 1,000. Pathway enrichment and network analysis was performed with Metascape by selecting the custom analysis option and uploading a background list of genes appropriate for the developmental stages under study. The lists were extracted from White et al. (White et al., 2017). DE gene lists were also inputted into the PANTHER gene ontology database (http://geneontology.org/), where genes are classified according to family and subfamily, molecular function, biological process or pathway (Mi et al., 2019). For PANTHER statistical over-representation analyses, only statistically significant enrichment results (p < 0.05 after Bonferroni correction for multiple tests) are shown. GOrilla was used to determine enrichment of transcripts based on the minimum hypergeometric scoring method (Eden et al., 2009). Redundant gene ontology (GO) terms were filtered with the GO Trimming tool and then visualized as treemap with REViGO (Supek et al., 2011). Box sizes correlate to the −log10 p value of the GO-term enrichment. Boxes with the same color are grouped by semantic similarity (SimRel, similarity = 0.7).
Reverse transcription PCR (RT-PCR) and quantitative RT-PCR (RT-qPCR)
Equal amounts of RNA were reverse transcribed to cDNA using the SuperScript III First-Strand Synthesis SuperMix for qRT-PCR Kit (Thermo Fisher Scientific) according to the manufacturer’s protocols. The expression levels of the different zebrafish genes analyzed in this study were measured by RT-PCR or RT-qPCR using the primers listed in Supplementary Table 1 and the TaqMan assays reported in Supplementary Table 4, respectively. Zebrafish beta-actin (actb1) was used as endogenous control in all reactions (McCurley and Callard, 2008). Band intensities from RT-PCR reactions were manually quantified by drawing boxes of the same area using the Image Lab software (Bio-Rad Laboratories, Hercules, CA, USA). RT-qPCR reactions were performed according to the manufacturer’s protocol using ddPCR SuperMix for Probes (no dUTP, Bio-Rad Laboratories 1863024) and a QX200 Droplet Digital PCR System (Bio-Rad Laboratories). Results were analyzed with the Quanta Soft software 1.7.4.0917 (Bio-Rad Laboratories) and are presented as absolute quantification values (normalized to actb1).
Imaging of live embryos
All embryos were staged according to Kimmel et al. (Kimmel et al., 1995). Live zebrafish larvae or adults were immobilized by adding 0.16 mg/ml of tricaine to the egg or system water prior to imaging. After larvae or adults were immobilized, they were mounted on a layer on a glass depression slide with 3% methylcellulose solution with or without fresh 0.16 mg/ml tricaine as needed. Images of live zebrafish were obtained using a Leica MZ16F stereomicroscope (Leica Microsystems, Wetzlar, Germany) with an AxioCam HRc CCD camera (Carl Zeiss, Jena, Germany).
For time-lapse imaging, embryos at the 1-cell stage were laid out in a 96-well plate and imaged by time-lapse microscopy until 24 hpf. Cell movements were recorded under an upright microscope (Keyence BZ-X710) using bright field light microscopy. The incubation temperature was set to 25 °C. The embryos were imaged every 6 min with a 4x objective, using 20% aperture. Image sequences play at 3 frames per second. Time-lapse movies were generated using the BZ-X Viewer v01.03.01.01 software (Keyence).
Brain morphometry
MZgpr101 and control larvae were raised in E3 medium (Westerfield, 2007) supplemented with 1.5 mM HEPES (pH 7.3) and 300 μM N-Phenylthiourea (PTU, Sigma P7629) starting at 8–22 hpf to inhibit melanogenesis. At 5 dpf, larvae were immersed in E3 with 300 μM PTU, 1% DMSO and 1 μM LysoTracker Deep Red (ThermoFisher L12492) and incubated for 16–24 hours at 28 °C. At 6 dpf, the larvae were transferred directly to E3 containing 0.24 mg/mL tricaine methanesulfonate (MS-222) to anesthetize them, mounted in 2.5% low melting point agarose, and imaged using a Leica TCS SP5 II inverted laser-scanning confocal microscope (Marquart et al., 2017). Images were acquired at isotropic 2 μm per voxel using a 633 nm laser with spectral window set to 638–795 nm. Image volumes were stitched using Fiji (Preibisch et al., 2009,Schindelin et al., 2012), then registered to a lysotracker reference brain using ANTs (Avants et al., 2011). For each brain, we applied the inverse of the transformation matrix derived during registration to a zebrafish reference brain atlas to measure the size of individual brain regions in the original control and mutant brain scans (Gupta et al., 2018). After combining small regions into 13 major brain divisions (pallium, subpallium, etc.), we then calculated the confidence interval for the mean difference between mutant and WT brain divisions using R. We performed this experiment using two independent sets of larvae (n=11 per genotype, and n=10 per genotype, respectively). As the changes for the 13 brain subdivisions were highly correlated (Pearson’s r=0.94 between experiments) the results were combined in Fig. 4.
Statistical analysis
All graphs were plotted as mean ± standard deviation (SD), except Fig. 4 which shows means and 95% confidence intervals. All data distributions were assessed for approximate normality. Genotype distribution of the gpr101 KO crosses was analyzed by chi square. Other differences between experimental groups were analyzed by two-tailed Student’s t-test, 1-way ANOVA with Tukey’s post hoc test, and an ordinary (not repeated measures) 2-way ANOVA with Bonferroni’s post hoc test, or corresponding non-parametric tests, as appropriate. Data were analyzed using GraphPad Prism (GraphPad, San Diego, CA, USA) and R Studio. p values < 0.05 were considered statistically significant.
Supplementary Material
Acknowledgements
The authors would like to thank the aquatics staff members of NICHD and Charles River Laboratories for daily zebrafish husbandry in the zebrafish facility at NICHD. We thank Dr. Fabienne E. Poulain (University of South Carolina, SC, USA) and Dr. Igor Dawid for kindly providing the casp23-pCRII and the pCS2+-eGFP vectors, respectively. We also thank Dr. Alberto Rissone, Dr. Erica Bresciani, Dr. Martina Absinta, Dr. Craig Abbott, and all the members of Dr. Stratakis laboratory (all at the NIH, Bethesda, MD, USA), for productive discussions and technical help.
Funding
This work was supported by the Intramural Research Program, Eunice Kennedy Shriver National Institute of Child Health & Human Development (NICHD), National Institutes of Health (NIH). It also received research funding support by Pfizer Inc. for investigations on growth hormone-producing pituitary adenomas (2016 US ASPIRE ENDOCRINE grant number WI215907).
Disclosure Statement
Dr. Stratakis holds patents on technologies involving PRKAR1A, PDE11A, GPR101 and related genes causing adrenal, pituitary and other tumors. In addition, his laboratory has received research funding support by Pfizer Inc. for investigations on growth-hormone producing pituitary adenomas. Dr. Faucz and Dr. Trivellin hold a patent on the GPR101 gene and its function (US Patent No. 10,350,273, Treatment of Hormonal Disorders of Growth). The authors declare that they have no conflicts of interest with the contents of this article.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Murray PG, Higham CE and Clayton PE, 2015. 60 YEARS OF NEUROENDOCRINOLOGY: The hypothalamo-GH axis: the past 60 years, J Endocrinol. 226, T123–40. [DOI] [PubMed] [Google Scholar]
- [2].Trivellin G, Daly AF, Faucz FR, Yuan B, Rostomyan L, Larco DO, Schernthaner-Reiter MH, Szarek E, Leal LF, Caberg JH, Castermans E, Villa C, Dimopoulos A, Chittiboina P, Xekouki P, Shah N, Metzger D, Lysy PA, Ferrante E, Strebkova N, Mazerkina N, Zatelli MC, Lodish M, Horvath A, de Alexandre RB, Manning AD, Levy I, Keil MF, Sierra Mde L, Palmeira L, Coppieters W, Georges M, Naves LA, Jamar M, Bours V, Wu TJ, Choong CS, Bertherat J, Chanson P, Kamenicky P, Farrell WE, Barlier A, Quezado M, Bjelobaba I, Stojilkovic SS, Wess J, Costanzi S, Liu P, Lupski JR, Beckers A and Stratakis CA, 2014. Gigantism and acromegaly due to Xq26 microduplications and GPR101 mutation, N Engl J Med. 371, 2363–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Beckers A, Lodish MB, Trivellin G, Rostomyan L, Lee M, Faucz FR, Yuan B, Choong CS, Caberg JH, Verrua E, Naves LA, Cheetham TD, Young J, Lysy PA, Petrossians P, Cotterill A, Shah NS, Metzger D, Castermans E, Ambrosio MR, Villa C, Strebkova N, Mazerkina N, Gaillard S, Barra GB, Casulari LA, Neggers SJ, Salvatori R, Jaffrain-Rea ML, Zacharin M, Santamaria BL, Zacharieva S, Lim EM, Mantovani G, Zatelli MC, Collins MT, Bonneville JF, Quezado M, Chittiboina P, Oldfield EH, Bours V, Liu P, W W.d.H., Pellegata N, Lupski JR, Daly AF and Stratakis CA, 2015. X-linked acrogigantism syndrome: clinical profile and therapeutic responses, Endocr Relat Cancer. 22, 353–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Trivellin G, Faucz FR, Daly AF, Beckers A and Stratakis CA, 2020. GPR101, an orphan GPCR with roles in growth and pituitary tumorigenesis, Endocr Relat Cancer. [DOI] [PubMed] [Google Scholar]
- [5].Rostomyan L, Daly AF, Petrossians P, Nachev E, Lila AR, Lecoq AL, Lecumberri B, Trivellin G, Salvatori R, Moraitis AG, Holdaway I, Kranenburg-van Klaveren DJ, Chiara Zatelli M, Palacios N, Nozieres C, Zacharin M, Ebeling T, Ojaniemi M, Rozhinskaya L, Verrua E, Jaffrain-Rea ML, Filipponi S, Gusakova D, Pronin V, Bertherat J, Belaya Z, Ilovayskaya I, Sahnoun-Fathallah M, Sievers C, Stalla GK, Castermans E, Caberg JH, Sorkina E, Auriemma RS, Mittal S, Kareva M, Lysy PA, Emy P, De Menis E, Choong CS, Mantovani G, Bours V, De Herder W, Brue T, Barlier A, Neggers SJ, Zacharieva S, Chanson P, Shah NS, Stratakis CA, Naves LA and Beckers A, 2015. Clinical and genetic characterization of pituitary gigantism: an international collaborative study in 208 patients, Endocr Relat Cancer. 22, 745–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Iacovazzo D, Caswell R, Bunce B, Jose S, Yuan B, Hernandez-Ramirez LC, Kapur S, Caimari F, Evanson J, Ferrau F, Dang MN, Gabrovska P, Larkin SJ, Ansorge O, Rodd C, Vance ML, Ramirez-Renteria C, Mercado M, Goldstone AP, Buchfelder M, Burren CP, Gurlek A, Dutta P, Choong CS, Cheetham T, Trivellin G, Stratakis CA, Lopes MB, Grossman AB, Trouillas J, Lupski JR, Ellard S, Sampson JR, Roncaroli F and Korbonits M, 2016. Germline or somatic GPR101 duplication leads to X-linked acrogigantism: a clinico-pathological and genetic study, Acta Neuropathol Commun. 4, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Abboud D, Daly AF, Dupuis N, Bahri MA, Inoue A, Chevigné A, Ectors F, Plenevaux A, Pirotte B, Beckers A and Hanson J, 2020. GPR101 drives growth hormone hypersecretion and gigantism in mice via constitutive activation of Gs and Gq/11, Nature Communications. 11, 4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Trivellin G, Bjelobaba I, Daly AF, Larco DO, Palmeira L, Faucz FR, Thiry A, Leal LF, Rostomyan L, Quezado M, Schernthaner-Reiter MH, Janjic MM, Villa C, Wu TJ, Stojilkovic SS, Beckers A, Feldman B and Stratakis CA, 2016. Characterization of GPR101 transcript structure and expression patterns, J Mol Endocrinol. 57, 97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bates B, Zhang L, Nawoschik S, Kodangattil S, Tseng E, Kopsco D, Kramer A, Shan Q, Taylor N, Johnson J, Sun Y, Chen HM, Blatcher M, Paulsen JE and Pausch MH, 2006. Characterization of Gpr101 expression and G-protein coupling selectivity, Brain Res. 1087, 1–14. [DOI] [PubMed] [Google Scholar]
- [10].Nilaweera KN, Ozanne D, Wilson D, Mercer JG, Morgan PJ and Barrett P, 2007. G protein-coupled receptor 101 mRNA expression in the mouse brain: altered expression in the posterior hypothalamus and amygdala by energetic challenges, J Neuroendocrinol. 19, 34–45. [DOI] [PubMed] [Google Scholar]
- [11].Regard JB, Sato IT and Coughlin SR, 2008. Anatomical profiling of G protein-coupled receptor expression, Cell. 135, 561–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bruce AE, 2016. Zebrafish epiboly: Spreading thin over the yolk, Dev Dyn. 245, 244–58. [DOI] [PubMed] [Google Scholar]
- [13].Gupta T, Marquart GD, Horstick EJ, Tabor KM, Pajevic S and Burgess HA, 2018. Morphometric analysis and neuroanatomical mapping of the zebrafish brain, Methods. 150, 49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lohr H and Hammerschmidt M, 2011. Zebrafish in endocrine systems: recent advances and implications for human disease, Annu Rev Physiol. 73, 183–211. [DOI] [PubMed] [Google Scholar]
- [15].Biga PR and Goetz FW, 2006. Zebrafish and giant danio as models for muscle growth: determinate vs. indeterminate growth as determined by morphometric analysis, Am J Physiol Regul Integr Comp Physiol. 291, R1327–37. [DOI] [PubMed] [Google Scholar]
- [16].Hu Z, Ai N, Chen W, Wong QW and Ge W, 2019. Loss of Growth Hormone Gene (gh1) in Zebrafish Arrests Folliculogenesis in Females and Delays Spermatogenesis in Males, Endocrinology. 160, 568–586. [DOI] [PubMed] [Google Scholar]
- [17].Trivellin G, Hernandez-Ramirez LC, Swan J and Stratakis CA, 2018. An orphan G-protein-coupled receptor causes human gigantism and/or acromegaly: Molecular biology and clinical correlations, Best Pract Res Clin Endocrinol Metab. 32, 125–140. [DOI] [PubMed] [Google Scholar]
- [18].Asakawa K and Kawakami K, 2009. The Tol2-mediated Gal4-UAS method for gene and enhancer trapping in zebrafish, Methods. 49, 275–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Asakawa K, Suster ML, Mizusawa K, Nagayoshi S, Kotani T, Urasaki A, Kishimoto Y, Hibi M and Kawakami K, 2008. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish, Proc Natl Acad Sci U S A. 105, 1255–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kishimoto Y, Lee KH, Zon L, Hammerschmidt M and Schulte-Merker S, 1997. The molecular nature of zebrafish swirl: BMP2 function is essential during early dorsoventral patterning, Development. 124, 4457–66. [DOI] [PubMed] [Google Scholar]
- [21].Varga ZM, Wegner J and Westerfield M, 1999. Anterior movement of ventral diencephalic precursors separates the primordial eye field in the neural plate and requires cyclops, Development. 126, 5533–46. [DOI] [PubMed] [Google Scholar]
- [22].El-Brolosy MA, Kontarakis Z, Rossi A, Kuenne C, Gunther S, Fukuda N, Kikhi K, Boezio GLM, Takacs CM, Lai SL, Fukuda R, Gerri C, Giraldez AJ and Stainier DYR, 2019. Genetic compensation triggered by mutant mRNA degradation, Nature. 568, 193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang X, Kopinke D, Lin J, McPherson AD, Duncan RN, Otsuna H, Moro E, Hoshijima K, Grunwald DJ, Argenton F, Chien CB, Murtaugh LC and Dorsky RI, 2012. Wnt signaling regulates postembryonic hypothalamic progenitor differentiation, Dev Cell. 23, 624–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Newman EA, Wu D, Taketo MM, Zhang J and Blackshaw S, 2018. Canonical Wnt signaling regulates patterning, differentiation and nucleogenesis in mouse hypothalamus and prethalamus, Dev Biol. 442, 236–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhang M, Skirkanich J, Lampson MA and Klein PS, 2017. Cell Cycle Remodeling and Zygotic Gene Activation at the Midblastula Transition, Adv Exp Med Biol. 953, 441–487. [DOI] [PubMed] [Google Scholar]
- [26].Gistelinck C, Gioia R, Gagliardi A, Tonelli F, Marchese L, Bianchi L, Landi C, Bini L, Huysseune A, Witten PE, Staes A, Gevaert K, De Rocker N, Menten B, Malfait F, Leikin S, Carra S, Tenni R, Rossi A, De Paepe A, Coucke P, Willaert A and Forlino A, 2016. Zebrafish Collagen Type I: Molecular and Biochemical Characterization of the Major Structural Protein in Bone and Skin, Sci Rep. 6, 21540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Spead O, Verreet T, Donelson CJ and Poulain FE, 2018. Characterization of the caspase family in zebrafish, PLoS One. 13, e0197966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hammond KL, Hill RE, Whitfield TT and Currie PD, 2002. Isolation of three zebrafish dachshund homologues and their expression in sensory organs, the central nervous system and pectoral fin buds, Mech Dev. 112, 183–9. [DOI] [PubMed] [Google Scholar]
- [29].Lange M, Norton W, Coolen M, Chaminade M, Merker S, Proft F, Schmitt A, Vernier P, Lesch KP and Bally-Cuif L, 2012. The ADHD-susceptibility gene lphn3.1 modulates dopaminergic neuron formation and locomotor activity during zebrafish development, Mol Psychiatry. 17, 946–54. [DOI] [PubMed] [Google Scholar]
- [30].White RJ, Collins JE, Sealy IM, Wali N, Dooley CM, Digby Z, Stemple DL, Murphy DN, Billis K, Hourlier T, Fullgrabe A, Davis MP, Enright AJ and Busch-Nentwich EM, 2017. A high-resolution mRNA expression time course of embryonic development in zebrafish, Elife. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES and Giraldez AJ, 2013. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition, Nature. 503, 360–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Veil M, Schaechtle MA, Gao M, Kirner V, Buryanova L, Grethen R and Onichtchouk D, 2018. Maternal Nanog is required for zebrafish embryo architecture and for cell viability during gastrulation, Development. 145. [DOI] [PubMed] [Google Scholar]
- [33].Toro S, Wegner J, Muller M, Westerfield M and Varga ZM, 2009. Identification of differentially expressed genes in the zebrafish hypothalamic-pituitary axis, Gene Expr Patterns. 9, 200–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Biran J, Palevitch O, Ben-Dor S and Levavi-Sivan B, 2012. Neurokinin Bs and neurokinin B receptors in zebrafish-potential role in controlling fish reproduction, Proc Natl Acad Sci U S A. 109, 10269–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhang Z, Wen H, Li Y, Li Q, Li W, Zhou Y, Wang L, Liu Y, Lyu L and Qi X, 2019. TAC3 Gene Products Regulate Brain and Digestive System Gene Expression in the Spotted Sea Bass (Lateolabrax maculatus), Front Endocrinol (Lausanne). 10, 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kelberman D, Rizzoti K, Lovell-Badge R, Robinson IC and Dattani MT, 2009. Genetic regulation of pituitary gland development in human and mouse, Endocr Rev. 30, 790–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Tekendo-Ngongang C, Muenke M and Kruszka P, 2000. [Updated 2020]. Holoprosencephaly Overview, in: Adam MP et al. (Eds.), GeneReviews((R)). Seattle (WA). [Google Scholar]
- [38].Leinonen JT, Chen YC, Tukiainen T, Panula P and Widen E, 2019. Transient modification of lin28b expression - Permanent effects on zebrafish growth, Mol Cell Endocrinol. 479, 61–70. [DOI] [PubMed] [Google Scholar]
- [39].Harvey SA, Sealy I, Kettleborough R, Fenyes F, White R, Stemple D and Smith JC, 2013. Identification of the zebrafish maternal and paternal transcriptomes, Development. 140, 2703–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Abrams EW and Mullins MC, 2009. Early zebrafish development: it’s in the maternal genes, Curr Opin Genet Dev. 19, 396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sant KE and Timme-Laragy AR, 2018. Zebrafish as a Model for Toxicological Perturbation of Yolk and Nutrition in the Early Embryo, Curr Environ Health Rep. 5, 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Aanes H, Winata CL, Lin CH, Chen JP, Srinivasan KG, Lee SG, Lim AY, Hajan HS, Collas P, Bourque G, Gong Z, Korzh V, Alestrom P and Mathavan S, 2011. Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition, Genome Res. 21, 1328–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rauwerda H, Wackers P, Pagano JF, de Jong M, Ensink W, Dekker R, Nehrdich U, Spaink HP, Jonker M and Breit TM, 2016. Mother-Specific Signature in the Maternal Transcriptome Composition of Mature, Unfertilized Zebrafish Eggs, PLoS One. 11, e0147151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mehjabin R, Xiong L, Huang R, Yang C, Chen G, He L, Liao L, Zhu Z and Wang Y, 2019. Full-Length Transcriptome Sequencing and the Discovery of New Transcripts in the Unfertilized Eggs of Zebrafish (Danio rerio), G3 (Bethesda). 9, 1831–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cheung CT, Nguyen TV, Le Cam A, Patinote A, Journot L, Reynes C and Bobe J, 2019. What makes a bad egg? Egg transcriptome reveals dysregulation of translational machinery and novel fertility genes important for fertilization, BMC Genomics. 20, 584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Martinon F and Tschopp J, 2007. Inflammatory caspases and inflammasomes: master switches of inflammation, Cell Death Differ. 14, 10–22. [DOI] [PubMed] [Google Scholar]
- [47].Chen R, Amoui M, Zhang Z and Mardon G, 1997. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila, Cell. 91, 893–903. [DOI] [PubMed] [Google Scholar]
- [48].Tian J, Yam C, Balasundaram G, Wang H, Gore A and Sampath K, 2003. A temperature-sensitive mutation in the nodal-related gene cyclops reveals that the floor plate is induced during gastrulation in zebrafish, Development. 130, 3331–42. [DOI] [PubMed] [Google Scholar]
- [49].Zeng Z, Johnson SL, Lister JA and Patton EE, 2015. Temperature-sensitive splicing of mitfa by an intron mutation in zebrafish, Pigment Cell Melanoma Res. 28, 229–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wang T, Yan B, Lou L, Lin X, Yu T, Wu S, Lu Q, Liu W, Huang Z, Zhang M, Zhang W and Wen Z, 2019. Nlrc3-like is required for microglia maintenance in zebrafish, J Genet Genomics. 46, 291–299. [DOI] [PubMed] [Google Scholar]
- [51].Jezierska B, Lugowska K and Witeska M, 2009. The effects of heavy metals on embryonic development of fish (a review), Fish Physiol Biochem. 35, 625–40. [DOI] [PubMed] [Google Scholar]
- [52].Westerfield M, 2007. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio), 5th ed. University of Oregon Press, Eugene. [Google Scholar]
- [53].Parichy DM, Elizondo MR, Mills MG, Gordon TN and Engeszer RE, 2009. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish, Dev Dyn. 238, 2975–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Chen W and Ge W, 2013. Gonad differentiation and puberty onset in the zebrafish: evidence for the dependence of puberty onset on body growth but not age in females, Mol Reprod Dev. 80, 384–92. [DOI] [PubMed] [Google Scholar]
- [55].Potter CJ and Luo L, 2010. Splinkerette PCR for mapping transposable elements in Drosophila, PLoS One. 5, e10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Varshney GK, Pei W, LaFave MC, Idol J, Xu L, Gallardo V, Carrington B, Bishop K, Jones M, Li M, Harper U, Huang SC, Prakash A, Chen W, Sood R, Ledin J and Burgess SM, 2015. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9, Genome Res. 25, 1030–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Jao LE, Wente SR and Chen W, 2013. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system, Proc Natl Acad Sci U S A. 110, 13904–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Carrington B, Varshney GK, Burgess SM and Sood R, 2015. CRISPR-STAT: an easy and reliable PCR-based method to evaluate target-specific sgRNA activity, Nucleic Acids Res. 43, e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Haeussler M, Schonig K, Eckert H, Eschstruth A, Mianne J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, Joly JS and Concordet JP, 2016. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR, Genome Biol. 17, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Gothilf Y, Toyama R, Coon SL, Du SJ, Dawid IB and Klein DC, 2002. Pineal-specific expression of green fluorescent protein under the control of the serotonin-N-acetyltransferase gene regulatory regions in transgenic zebrafish, Dev Dyn. 225, 241–9. [DOI] [PubMed] [Google Scholar]
- [61].Babraham Bioinformatics - FastQC A Quality Control tool for High Throughput Sequence Data. (2019). [Google Scholar]
- [62].Bolger AM, Lohse M and Usadel B, 2014. Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics. 30, 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR, 2013. STAR: ultrafast universal RNA-seq aligner, Bioinformatics. 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Anders S, Pyl PT and Huber W, 2015. HTSeq--a Python framework to work with high-throughput sequencing data, Bioinformatics. 31, 166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Love MI, Huber W and Anders S, 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H and Vilo J, 2016. g:Profiler-a web server for functional interpretation of gene lists (2016 update), Nucleic Acids Res. 44, W83–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Mi H, Muruganujan A, Huang X, Ebert D, Mills C, Guo X and Thomas PD, 2019. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0), Nat Protoc. 14, 703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Eden E, Navon R, Steinfeld I, Lipson D and Yakhini Z, 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists, BMC Bioinformatics. 10, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Supek F, Bosnjak M, Skunca N and Smuc T, 2011. REVIGO summarizes and visualizes long lists of gene ontology terms, PLoS One. 6, e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].McCurley AT and Callard GV, 2008. Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue type, developmental stage and chemical treatment, BMC Mol Biol. 9, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Kimmel CB, Ballard WW, Kimmel SR, Ullmann B and Schilling TF, 1995. Stages of embryonic development of the zebrafish, Dev Dyn. 203, 253–310. [DOI] [PubMed] [Google Scholar]
- [72].Marquart GD, Tabor KM, Horstick EJ, Brown M, Geoca AK, Polys NF, Nogare DD and Burgess HA, 2017. High-precision registration between zebrafish brain atlases using symmetric diffeomorphic normalization, Gigascience. 6, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Preibisch S, Saalfeld S and Tomancak P, 2009. Globally optimal stitching of tiled 3D microscopic image acquisitions, Bioinformatics. 25, 1463–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P and Cardona A, 2012. Fiji: an open-source platform for biological-image analysis, Nat Methods. 9, 676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Avants BB, Tustison NJ, Song G, Cook PA, Klein A and Gee JC, 2011. A reproducible evaluation of ANTs similarity metric performance in brain image registration, Neuroimage. 54, 2033–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


