Abstract
Sequence detection by the 5′ nuclease TaqMan assay uses online detection of internal fluorogenic probes in closed PCR tubes. Primers and probe were chosen from a part of the omlA gene common to all serotypes of Actinobacillus pleuropneumoniae, which gave an amplicon of 92 bp. The test was evaluated with 73 lung isolates and 120 tonsil isolates of A. pleuropneumoniae as well as with a collection of reference strains. By using a Ct value (cycle number in which the fluorescence exceeds the threshold defined by the software) of 30 as the cutoff limit, the 5′ nuclease assay represents a test with 100% sensitivity and 100% specificity. A high degree of reproducibility of the test was demonstrated. If samples with Ct values of ≤30 are considered positive, the detection limit of the assay was 1 CFU/reaction tube, corresponding to a 10-fold higher number of DNA templates. After cycle 30, nonspecific reactions appeared when testing dilutions of DNA templates or pure cultures of A. pleuropneumoniae, as well as when testing tonsil scrapings from specific-pathogen-free herds. The diagnostic sensitivity, as evaluated with 586 tonsil scrapings from animals infected with A. pleuropneumoniae, is the same level as that of a PCR test based on the omlA gene described previously. The 5′ nuclease assay represents a fast method for species-specific detection and identification of A. pleuropneumoniae in pure and mixed cultures. The evaluation shows, however, that a Ct value cutoff limit of ≤30 must be chosen in order to obtain reliable results. The investigation emphasizes that a thorough evaluation of the criteria used to define a positive test result is necessary.
Actinobacillus pleuropneumoniae is the causative agent of porcine pleuropneumonia, which is one of the most important diseases in swine production and which has a big impact on animal welfare and production economy. Fourteen serotypes have been described (5, 19, 20), all of which are potentially pathogenic, although considerable differences in the virulences of the various serotypes have been reported (6). Attempts to control the disease by vaccination, treatment with antibiotics, and establishment of herds free of the infection, i.e., herds that are specific pathogen free (SPF), have been made.
Although serological surveillance has been efficient in controlling porcine pleuropneumonia (2, 21), this method has some limitations. Animals carrying the bacterium may be serologically negative (16, 23), or animals might be seropositive without showing clinical signs of disease. In these cases isolation of the bacterium is important for confirming the presence of the infection in the animals or herds. Isolation of A. pleuropneumoniae from acutely diseased animals normally poses no problem. However, in latently infected animals the bacterium can reside in the upper respiratory tract in low numbers (14, 17), where the commensal bacterial flora makes the isolation of the bacterium difficult. Identification of such latently infected animals is of great importance for avoiding introduction of the bacterium into herds free of A. pleuropneumoniae or in eradication programs. To overcome these difficulties a special medium for isolation of A. pleuropneumoniae has been composed (12). Furthermore, PCR tests for detection of A. pleuropneumoniae without isolation of the organism have been developed, with some of these being species specific (8), while others are used for serotype-specific detection (15).
Although the PCR assays might offer high degrees of specificity and sensitivity, they are presently not easily adapted to the processing of large numbers of samples. The use of gel electrophoresis for visualization of the PCR products can lead to problems with cross-contamination in the laboratory. In addition, the presence of weak bands can make the results difficult to interpret. Furthermore, identification of the PCR amplification product necessitates additional time-consuming procedures.
We consequently decided to use the 5′ nuclease assay (11) which exploits the 5′ nuclease activity of the Taq polymerase to cleave an internal oligonucleotide probe labeled with two fluorescent dyes (9) referred to as “reporter” and “quencher” (TaqMan probe). Due to the proximity of the dyes, energy absorbed by the reporter is transferred to the quencher. During elongation of the PCR primers the probe is cleaved and the reporter and quencher molecules are released to the solution, resulting in an increased level of emission from the reporter. The amount of reporter released is proportional to the amount of DNA being amplified by PCR and increases for each cycle. The assay uses the ABI 7700 Sequence Detection System, which makes it possible to monitor the PCR amplification from cycle to cycle and which also allows quantification of the starting DNA, provided a series of standards are included (9). The results are displayed as a value termed ΔRn, the ratio between the emission intensities of the reporter and the quencher minus the ratio measured before PCR amplification. In addition, the software calculates Ct, the cycle number when the ΔRn exceeds a threshold defined as 10 standard deviations (SDs) above the mean baseline observed in the no-template controls (NTCs) between cycles 3 and 15, which is the software's default value (9). As all reactions take place in closed tubes, the risk for cross-contamination in the laboratory is greatly reduced. At the same time the degradation of the probe represents a way to directly identify the amplification product. In addition, the laser detection device might make the reproducible detection of small amounts of DNA template feasible.
This report describes a 5′ nuclease assay based on the omlA gene, which was previously shown to be a promising basis for species-specific detection of A. pleuropneumoniae (8). The specificity and the detection limit of the assay were evaluated, and the diagnostic sensitivity of the assay was compared with those of bacterial cultivation and the previously described PCR test (8) with scrapings from pig tonsils.
MATERIALS AND METHODS
Bacterial strains.
A total of 203 Danish field isolates of A. pleuropneumoniae, 73 strains isolated from pigs with pleuropneumonia and 120 strains isolated from tonsils, representing serotypes 2 (n = 18), 5 (n = 16), 6 (n = 45), 8 (n = 1), 7 (n = 3), 10 (n = 41), 12 (n = 16), K2:O7 (strains with capsular polysaccharides and lipopolysaccharide similar to serotypes 2 and 7, respectively [20]) (n = 25), and nontypeable strains (n = 18), were included in the study, in addition to reference strains representing serotypes 1 to 14 (see Table 1). All strains were identified as A. pleuropneumoniae by phenotypic criteria (13) and by a species-specific PCR test (8). Reference strains for genetically related species, as well as other species normally found in the respiratory tracts of pigs, were used to test the specificity of the 5′ nuclease assay (Table 1). Sixteen strains of Actinobacillus lignieresii were included due to its phylogenetic proximity to A. pleuropneumoniae. All V factor-dependent strains were grown on PPLO agar (Difco, Detroit, Mich.), whereas all other strains tested were grown on Columbia agar supplemented with 5% bovine blood. All cultures were incubated at 37°C in atmospheric air for 16 h. The cultures were stored at −80°C in brain heart infusion broth (Difco) with 10% glycerol.
TABLE 1.
Reference strains used for evaluation of the species specificity of the 5′ nuclease assay
| Species | Strain designationa | Ct valueb |
|---|---|---|
| Actinobacillus pleuropneumoniae | ||
| Serotype 1 | Shope 4074T | 16.6 |
| Serotype 2 | S 1536 | 16.8 |
| Serotype 2 (Danish isolate) | 4226 | 15.6 |
| Serotype 3 | S 1421 | 17.8 |
| Serotype 4 | M62 | 18.7 |
| Serotype 5A | K17 | 17.7 |
| Serotype 5B | L20 | 19.1 |
| Serotype 6 | Femø | 17.1 |
| Serotype 7 | WF83 | 17.8 |
| Serotype 8 | 405 | 18.1 |
| Serotype 9 | CVJ 13261 | 15.8 |
| Serotype 10 | D 13039 | 17.5 |
| Serotype 11 | 56153 | 19.3 |
| Serotype 12 | 8329 | 17.6 |
| Serotype 13 (biovar 2) | N-273 | 16.0 |
| Serotype 14 (biovar 2) | 3906 | 15.3 |
| Actinobacillus lignieresii | ATCC 49236T | 39.1 |
| CCUG 215 | 38.7 | |
| CCUG 12401 | 40.0 | |
| CCUG 18727 | 40.0 | |
| CCUG 18728 | 40.0 | |
| CCUG 18729 | 38.9 | |
| CCUG 22227 | 40.0 | |
| CCUG 22228 | 40.0 | |
| CCUG 22229 | 40.0 | |
| CCUG 23132 | 40.0 | |
| CCUG 23133 | 40.0 | |
| CCUG 27160 | 40.0 | |
| CCUG 27361 | 40.0 | |
| CCUG 37052 | 36.3 | |
| CCUG 38958 | 40.0 | |
| NCTC 4985 | 40.0 | |
| Actinobacillus ureae | NCTC 10219T | 39.0 |
| P1144 | 36.3c | |
| Actinobacillus capsulatus | NCTC 11408T | 36.4d |
| P1364 | 39.0 | |
| Actinobacillus hominis | NCTC 11529T | 37.0c |
| P1336 | 38.4 | |
| Actinobacillus equuli | NCTC 8529T | 39.5 |
| P1284 | 39.3 | |
| Actinobacillus rossii | ATCC 27072T | 37.7d |
| Actinobacillus suis | CAPM 5586T | 39.6 |
| P1143 | 39.3 | |
| Actinobacillus minor | NM 305T | 40.0 |
| Actinobacillus porcinus | NM 319T | 40.0 |
| 18765-5(T1) | 39.2 | |
| 18870-2(T5) | 39.0 | |
| 18870-2(T7) | 35.8c | |
| 595 | 39.3 | |
| Actinobacillus indolicus | 46KC2T | 40.0 |
| 39Cl | 39.4 | |
| Haemophilus parasuis | NCTC 4557T | 38.0d |
| Mannheimia haemolytica | NCTC 9380T | 38.9d |
| Mannheimia varigena | CAPM 5786 | 40.0 |
| CCUG 16500 | 39.9 | |
| Pasteurella multocida | NCTC 10320 | 38.1d |
| HIM 746–6 | 38.5d | |
| Pasteurella avium biovar 2 | Strain 5 | 39.3 |
| Pasteurella canis biovar 2 | Strain 25 | 39.4 |
| Pasteurella aerogenes | ATCC 27883T | 38.5 |
| Streptococcus suis | 735 | 38.0 |
| 8074 | 39.9 | |
| 24636/74 | 37.7d | |
| Bordetella bronchiseptica | 1317 | 38.2 |
ATCC, American Type Culture Collection, Manassas, Va.; CAPM, Collection of Animal Pathogenic Microorganisms, Brno. Czech Republic; CCUG, Culture Collection University of Gothenburg, Gothenburg, Sweden; NCTC, National Collection of Type Cultures, Colindale, United Kingdom.
Mean Ct values from three repeats (A. pleuropneumoniae strains) or six repeats (other species).
Strains with ΔRn values significantly different from those for the NTCs (P < 0.01).
Strains with ΔRn values significantly different from those for NTCs (P < 0.05).
Tonsil samples.
Five hundred eighty-six samples of tonsil scrapings from pigs were also included in the study. The pigs came from a herd known to be infected with A. pleuropneumoniae. These scrapings were obtained from sows and piglets in 15 different litters and were taken at regular intervals from 4 weeks of age until slaughter at 22 to 24 weeks of age. The tonsil scrapings were transported in a bouillon containing calf blood, NAD, neomycin, and lincomycin described earlier (12) and were generally subcultured on the same day. When this was not possible the bouillon was stored at 4°C overnight and the scrapings were subcultured on the next day. After vortexing of the sample, approximately 20 μl was transferred to two selective blood agar plates (12) and the plates were incubated at 37°C for 16 h. One of the plates was used for isolation of A. pleuropneumoniae, and the other plate was used for PCR detection (8) and later on for the 5′ nuclease assay. The 120 tonsil isolates of A. pleuropneumoniae included in the study all originated from these 15 litters.
In addition, 40 tonsil scrapings originating from one SPF breeding herd and one SPF multiplying herd were included in the study and were analyzed as described above. Clinical signs of pleuropneumonia had never been observed in these herds. These herds tested negative every month for the presence of antibodies against serotypes 2 and 6, tested negative every third month for the presence of antibodies against serotype 12, and tested negative once a year for the presence of antibodies against serotypes 1, 5, 7, and 10. The serological testing was performed by a blocking enzyme-linked immunosorbent assay for serotype 2 (21) and by a complement fixation test for the other serotypes (18).
Preparation of samples for the 5′ nuclease assay and PCR detection.
Bacterial growth from the tissue scrapings was harvested by washing the agar plates with 2 ml of sterile water, and 200 μl was lysed by boiling for 10 min. The lysates were spun down at 13,000 × g for 2 min, and 150 μl was kept for analysis. For preparation of lysates of the field isolates and the reference strains, 10 μl of a 16-h-old pure culture were taken from the surface of an agar plate with a loop, mixed with 200 μl of sterile water, and lysed by boiling for 10 min. Lysates of pure cultures were diluted 1:100 before use. Plate washes were stored at −80°C and lysates were stored at −20°C until use.
Primers and probe.
The sequences of the omlA genes of A. pleuropneumoniae reference strains of serotypes 1 to 12 have been published previously (8) and are available through GenBank. The sequences were aligned by using DNASIS (version 2.5; Hitachi Software), and primers and probes from the conserved 3′ end of the gene were chosen by using Primer Express (Perkin-Elmer, Foster City, Calif.). The forward primer (FP) was 5′-AGT GCT TAC CGC ATG TAG TGG C-3′ (nucleotides 202 to 223 in the sequence with GenBank accession number L06318), and the reverse primer (RP) was 5′-TTG GTG CGG ACA TAT CAA CCT TA-3′ (nucleotides 271 to 293 in the sequence with GenBank accession number L06318). This should give an amplicon of 92 bp. The probe, located on the nonsense strand immediately upstream of the forward primer, was 5′-FAM-CGA TGA ACC CGA TGA GCC GCC-3′-TAMRA (nucleotides 224 to 244 in the sequence with GenBank accession number L06318), with a 3′ phosphate block used to prevent elongation of the probe (produced by Scandinavian Gene Synthesis AB, Köping, Sweden). FAM is the reporter dye 6-carboxyfluorescein, and TAMRA is the quencher dye 6-carboxytetramethylrhodamine.
The 5′ nuclease assay.
The test was carried out with the ABI PRISM Sequence Detection System with the reagents in the TaqMan PCR core reagent kit and 20% glycerol. The 5′ nuclease assay was performed with 5 μl of lysate by using 0.05 U of AmpliTaq Gold polymerase (Perkin-Elmer) in a total volume of 50 μl containing 1× TaqMan Buffer A, 8% glycerol, dATP, dCTP, and dGTP at concentration of 200 μM each, 400 μM dUTP, 5 mM MgCl2, and 0.01 U of AmpErase UNG. Probe and primer concentrations were determined by optimizing the test according to the guidelines described by the manufacturer (TaqMan quantitation test development quick-start guide; Perkin-Elmer). The optimal test conditions were obtained with a probe concentration of 175 nM and primer concentrations of 300 nM (FP) and 50 nM (RP). Amplification was performed in a 96-well plate with optical caps using the following settings: 2 min at 50°C (activation of uracil N-glycosylase [UNG]) and 10 min at 95°C (deactivation of UNG and activation of AmpliTaq Gold), followed by 40 cycles of 15 s at 95°C and 1 min of annealing and extension at 62°C.
PCR amplification.
All lysates of bacterial strains and plate washings were also tested for the presence of A. pleuropneumoniae by using the earlier published PCR test based on the omlA gene (8). This PCR test produces an amplicon of approximately 900 bp. The FP used in this test is, except for being 3 bp shorter, the reverse complement of the RP used in the 5′ nuclease assay (nucleotides 225 to 242 in the sequence with GenBank accession number L06318). The PCR test with the tonsil scrapings had been performed before the 5′ nuclease assay was developed and was initially based on 1 μl of lysate for amplification and 30 PCR cycles. When a discrepancy between the results of the PCR test and those of the 5′ nuclease assay was observed, both tests were repeated with 5 μl of lysate and 40 PCR cycles to obtain comparable conditions for the two tests.
Test of detection limit for 5′ nuclease assay.
For determination of the detection limit of A. pleuropneumoniae in the 5′ nuclease assay, overnight cultures of A. pleuropneumoniae strains 1536 and 4226 (serotype 2) were suspended in 0.9% NaCl, and the suspension was adjusted to a density corresponding to that of a McFarland 0.5 standard. This solution was diluted 1:100, resulting in a solution containing approximately 106 CFU/ml; thereafter, 10-fold dilutions were prepared. Plate counts from each dilution were performed in triplicate. A total of 200 μl from each dilution was lysed as described above, and 5 μl was used for the 5′ nuclease assay. Each test was repeated three times.
A total of 1,279 bp was amplified from the omlA gene of strain K17 with primers LPF1 and LPR1 as described previously (8). The products from five PCR were purified with a QIAquick PCR purification kit (Qiagen, Hilden, Germany). The DNA concentration was determined with a spectrophotometer and was diluted to 104 templates/5 μl. Tenfold dilutions were made, and 5 μl from each dilution was used for both the PCR test and the 5′ nuclease assay. Each test was repeated three times.
Repeatability.
All 5′ nuclease tests were repeated at least three times for the reference strains listed in Table 1 and twice for the Danish field strains of A. pleuropneumoniae. All tests with the tonsil scrapings were performed in duplicate. A lysate of strain 1536 was used as a positive control in all experiments. Descriptive statistics were obtained by importing data on Ct and ΔRn into Excel 97 (Microsoft Corp.). To determine whether the ΔRn values for a sample were significantly different from the NTCs, a one-sided t test was performed by assuming unequal variances. Regression analysis and t tests were performed with SAS software (version 6.12; SAS Institute, Inc.). To estimate whether the slope of the regression line was significantly different from the theoretical value obtained with a PCR amplification with an efficiency equal to 1, a two-sided t test was performed.
RESULTS
Specificity of the 5′ nuclease test.
The serotype reference strains of A. pleuropneumoniae were positive between cycles 15 and 19 and had on average a Ct value of 17.3 (SD, 1.2) (Table 1). Seventy-three strains isolated from Danish pigs suffering from pleuropneumonia were positive between cycles 15 and 22, with an average Ct value of 17.8 (SD, 1.7). One hundred twenty A. pleuropneumoniae strains isolated from tonsils were positive between cycles 17 and 26, with an average Ct value of 19.7 (SD, 2.3).
Forty-four reference strains within the family Pasteurellaceae as well as other bacteria normally found in pigs were also tested (Table 1). Seven of these strains passed the threshold between cycles 35.8 and 38.1 and achieved ΔRn values that were significantly different at a 5% level from that for the NTCs after 40 cycles. Only two strains achieved ΔRn values that were significant at the 1% level. When testing the type strain of A. lignieresii without dilution, a Ct value of 33 was obtained. However, after a dilution of 1:100, neither the Ct nor the ΔRn value was significantly different from the values for the NTCs. A similar amount of A. pleuropneumoniae strain 1536 had to be diluted 1:108 before it achieved Ct and ΔRn values similar to those for the NTCs.
Detection limit.
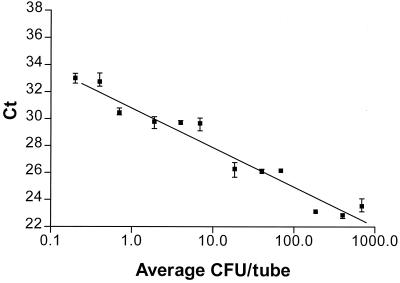
In the dilution experiments with suspensions of bacteria grown on agar plates, a linear relationship was observed between Ct values and log CFU count between cycles 23 and 32 (Fig. 1). For each 10-fold dilution the Ct value was raised on average by 2.93, close to but significantly different (P < 0.01) from the value (3.32 = ln 10/ln 2) expected for a PCR with an efficiency equal to 1 (implying that the amount of DNA is doubled for each cycle). A Ct value of approximately 30 corresponded to the presence of 1 CFU in the tube. There was some divergence between the different CFU counts associated with a given Ct value in the three experiments (Fig. 1).
FIG. 1.
Plot showing the relationship between Ct value in the 5′ nuclease assay and the number of CFU of A. pleuropneumoniae per reaction tube. The log-linear regression line and SDs are indicated.
A similar log-linear relationship was observed when a purified DNA template was used (Fig. 2; the slope of −3.41 was not significantly different from the theoretical value [P = 0.40]). However, here a Ct value of 30 corresponded to the presence of 10 templates in the tube. When the samples were diluted beyond this point, the correlation with the Ct value disappeared and the Ct value fluctuated between 30 and 35 in a random manner (data shown are only for one DNA template per tube in Fig. 2). The PCR test (which contained 5 μl of sample and which was run for 40 cycles) was positive for all samples with more than 10 templates per tube. With one DNA template per tube, one of three tests was positive. At higher dilutions all tests were negative.
FIG. 2.
Plot showing the relationship between Ct value in the 5′ nuclease assay and the number of omlA gene templates per reaction tube. The log-linear regression line and SDs are indicated.
Test of tonsil scrapings.
The 40 samples from the SPF herds passed the detection limit on average at cycle 34.4 (SD, 2.0; range, 31.6 to 38.7). The PCR tests (run for 30 cycles) were negative for all samples. However, two of the samples were weakly positive when the PCR test was run for 40 cycles.
A total of 586 tonsil samples from pigs originating from a conventional herd were investigated in duplicate and were distributed across 18 different runs. The average differences between the two measurements of the Ct and ΔRn values among the samples were 0.76 (SD, 1.42) and 0.10 (SD, 0.15), respectively. On average, the positive controls passed the threshold at a Ct value of 17.1 (SD, 2.0) and had a final ΔRn value of 1.74 (SD, 0.84) after 40 cycles. The NTCs passed the threshold on average at a Ct value of 39.7 (SD, 0.75), with a final ΔRn value of 0.07 (SD, 0.14) after 40 cycles.
The number of positive samples in the 5′ nuclease test depended on how the detection limit was determined. If the criterion was that the ΔRn value of the samples after 40 cycles should be significantly different (one-sided t test) from the NTCs, then 329 of the samples were positive (56%). This corresponds to the use of a Ct value of 38 as the detection limit. Alternatively, a sample might be regarded as positive if the Ct value was ≤33 (corresponding to the presence of one DNA template according to the regression line in Fig. 2), meaning that 149 of the samples (25%) were positive. By using a Ct value of ≤30 as the detection limit (considering the test values obtained for the SPF herds and the signal noise observed beyond this point), 112 of the samples (19%) should be regarded as positive.
A. pleuropneumoniae could be isolated from 92 (16%) of the tonsils, and 26 of these were negative by both the PCR test and the 5′ nuclease test when a Ct value of ≤30 was used as the detection limit. The 5′ nuclease test and the PCR test were positive for 112 (19%) and 134 (23%) of the samples, respectively. Forty samples were positive by the PCR test and negative by the 5′ nuclease test. When the tests for which a discrepancy between the two PCR-based tests were repeated, all tests negative by the 5′ nuclease test were negative by the PCR test as well.
DISCUSSION
The present investigation confirms that the omlA gene is a good choice for use in an A. pleuropneumoniae-specific test (8). Most strains of A. pleuropneumoniae investigated were positive before cycle 20. A few nontypeable strains had Ct values as high as 26, indicating that variation also exists in the omlA gene in those regions that were found to be conserved among the serotype reference strains. Weak and late reactions were observed among strains representing other species (Table 1), indicating that homologous DNA regions might exist in other species as well. At comparable concentrations, there were at least 10 cycle differences between strains of A. pleuropneumoniae and all other species. If the 5′ nuclease assay is read after 30 cycles, the test represents a species-specific test with 100% sensitivity and 100% specificity with pure cultures.
From the dilution experiments a given Ct value seems to correspond to a 10-fold higher number of DNA templates than the number of CFU. As the omlA gene is present at only one copy per genome (7), this can be interpreted only as detection of nonviable bacteria present in the solution. From Fig. 2 a log-linear relationship between the number of DNA templates and the Ct value is seen over a range from 1 to 104 templates per tube. For each 10-fold dilution the Ct value is increased by 3, close to the theoretical value for a PCR with an efficiency of 1. When the DNA concentration goes down to one template per tube, the SD of the experiments increases. This reflects the fact that the test should be expected to be negative for some of these repeated experiments. One of three PCR tests was positive at this concentration, which corresponds well to the expected proportion (63%), assuming a Poisson distribution and assuming that the DNA concentration was determined exactly by spectrophotometry. However, when the samples were diluted beyond 10 templates/tube, apparently randomly distributed Ct values in the range of 30 to 35 were observed by the 5′ nuclease assay. On the basis of these results, we chose a Ct value of 30 as the cutoff value, corresponding to a detection limit of 1 CFU per tube for the 5′ nuclease assay.
We were a bit surprised to find that, despite considerable efforts and repetitions, we were not able to dilute the samples to a level where the Ct values were as low as those for the NTCs (which were close to 40). The cause could not be nonspecific degradation of the probe, as the presence of an amplicon of approximately 92 bp could be confirmed by gel electrophoresis. Consequently, this might be the result of cross-contamination in the laboratory, possibly due to aerosol formation. This further indicates that under normal laboratory conditions a reliable detection limit of one template is difficult to obtain.
Different detection limits have been reported when the 5′ nuclease assay is applied with pure cultures of other bacterial species. Everett et al. (4) could detect as few as 1.7 cells of Clamydia spp. per reaction (amplicon of 100 to 150 bp), whereas Chen et al. (3) reported a detection limit of 2 CFU of Salmonella per tube (amplicon of 287 bp). Witham et al. (24) could detect down to 10 ± 5 CFU of Escherichia coli per PCR (amplicons in the range 161 to 164 bp were investigated). Oberst et al. (22) detected 4 CFU of E. coli O157 per reaction with an amplicon of 631 bp. Other tests have been less sensitive; e.g., Woo et al. (25) reported a detection limit of 400 cells of Leptospira per reaction (amplicon 428 bp), and Higgins et al. (10) could detect at best 333 genomes of Yersinia pestis per reaction (amplicon of 344 bp). Finally, Bassler et al. (1) reported a detection limit of 50 CFU of Listeria monocytogenes per PCR (amplicon of 858 bp). The detection limit seems to some extent to reflect the size of the amplicon produced. The present assay produces an amplicon of only 92 bp, with the probe being directly continuous with one of the primers. From the different dilution experiments, we have shown that the assay achieves a performance that is near the theoretical optimum for a PCR over a wide dilution range and has a detection limit of 1 CFU per tube. Our findings that the 5′ nuclease assay seems to be able to detect more DNA templates than the number of CFU present in a solution has not been reported by any of the other investigations cited above. This might reflect the fact that A. pleuropneumoniae might be more likely than other species to become nonviable in an agar colony.
The SPF herds tested must be assumed to be free of all serotypes of A. pleuropneumoniae. Considering the high Ct values found when we tested other species that are normally found in swine tonsils (Table 1), it was a bit surprising that we observed Ct values down to 31.6 among the samples from these herds. We must consequently regard these test results as nonspecific reactions. This supports our earlier decision of using a Ct value of ≤30 as a cut off limit for the test. This was also supported by the finding that two of the samples were PCR positive when the test was run for 40 cycles, whereas all were negative after 30 cycles.
When applying the 5′ nuclease assay for detection of A. pleuropneumoniae in clinical samples, the choice of detection threshold is of great importance, as we can see from the results for the 586 tonsil samples investigated. In some reports a threshold based on a 99% confidence interval from the mean for the NTCs has been used (1, 22, 24), implicitly assuming that the variances of the ΔRn values for the samples and the NTCs are equal, an assumption that can be questioned. By using this criterion 61% of the tonsils should be regarded as A. pleuropneumoniae positive, as most of the NTCs had ΔRn values close to 0.00 after 40 cycles. Without this assumption and by using a one-sided t test, 56% of the tonsils should still be regarded as positive. If the threshold was calculated in a similar manner when the 5′ nuclease assay was run for only 35 cycles, 39% of the tonsils tested positive. However, on the basis of the results of our dilution experiments and with the samples from the SPF herds, we found that the detection limit should be set to a level at which only 19% of the tonsils should be regarded as positive.
By use of this detection level, the diagnostic sensitivity of the 5′ nuclease assay was apparently lower than that of the PCR test (23%). However, when the tests for which a discrepancy between the two methods was observed were repeated, all samples negative by the 5′ nuclease assay were also negative by the PCR test. As all lysates had been stored at −20°C for up to 6 months before the 5′ nuclease assay was run, a certain amount of template degradation might be expected. The diagnostic sensitivity of the assay might have been higher if it had been applied to freshly prepared templates. A. pleuropneumoniae was isolated from 26 tonsil samples for which both PCR-based tests were negative. This can be explained by the fact that different agar plates, were used for bacterial isolation and preparation of the lysates for PCR. Consequently, the presence of very small numbers of A. pleuropneumoniae in the solution might lead to a stochastic distribution of the bacteria on the two agar plates.
The expenses for the consumables were approximately twice as high for the 5′ nuclease test as for the PCR test. On the other hand, the 5′ nuclease assay is quicker and less time-consuming to perform and can reduce contamination problems in the laboratory, but it is dependent on the acquisition of expensive technical equipment. The choice of whether to use the 5′ nuclease test in a diagnostic setting will depend on the relative importance of these factors.
Conclusions.
We have shown that the 5′ nuclease assay represents a fast method for species specific detection and identification of A. pleuropneumoniae in pure or mixed cultures. The evaluation shows, however, that a Ct value of ≤30 should be chosen as the cutoff limit in order to obtain reliable results. By using this criterion, the diagnostic sensitivity of the 5′ nuclease assay seems to be the same as that of the PCR test described previously. The investigation emphasizes that a thorough evaluation of the criteria used to define a positive test result is necessary.
ACKNOWLEDGMENTS
We thank Lars Melin for assistance with the Primer Express software and Henrik Stryhn for help with the statistical tests.
The tonsil scrapings were collected in connection with a project financed by The Research Center for the Management of Animal Production and Health (CEPROS).
REFERENCES
- 1.Bassler H A, Flood S J A, Livak K J, Marmaro J, Knorr R, Batt C A. Use of a fluorogenic probe in a PCR-based assay for the detection of Listeria monocytogenes. Appl Environ Microbiol. 1995;61:3724–3728. doi: 10.1128/aem.61.10.3724-3728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bossé J T, Johnson R P, Rosendahl S. Serodiagnosis of pleuropneumonia using enzyme-linked immunosorbent assay with capsular polysaccharide of serotype 1, 2, 5 and 7. Can J Vet Res. 1990;54:427–431. [PMC free article] [PubMed] [Google Scholar]
- 3.Chen S, Yee A, Griffiths M, Larkin C, Yamashiro C T, Behari R, Paszko-Kolva C, Rahn K, De Grandis S A. The evaluation of a fluorogenic polymerase chain reaction assay for the detection of Salmonella species in food commodities. Int J Food Microbiol. 1997;35:239–250. doi: 10.1016/s0168-1605(97)01241-5. [DOI] [PubMed] [Google Scholar]
- 4.Everett K D E, Hornung L J, Andersen A A. Rapid detection of the Chlamydiaceae and other families in the order Chlamydiales: three PCR tests. J Clin Microbiol. 1999;37:575–580. doi: 10.1128/jcm.37.3.575-580.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fodor L, Varga J, Molnar E, Hajtos I. Biochemical and serological properties of Actinobacillus pleuropneumoniae biotype 2 strains isolated from swine. Vet Microbiol. 1989;20:173–180. doi: 10.1016/0378-1135(89)90040-0. [DOI] [PubMed] [Google Scholar]
- 6.Frey J. Virulence in Actinobacillus pleuropneumoniae and RTX toxins. Trends Microbiol. 1995;3:257–261. doi: 10.1016/s0966-842x(00)88939-8. [DOI] [PubMed] [Google Scholar]
- 7.Gerlach G F, Anderson C, Klashinsky S, Rossi-Campos A, Potter A A, Willson P J. Molecular characterization of a protective outer membrane lipoprotein (OmlA) from Actinobacillus pleuropneumoniae serotype 1. Infect Immun. 1993;61:565–572. doi: 10.1128/iai.61.2.565-572.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gram T, Ahrens P. Improved diagnostic PCR assay for Actinobacillus pleuropneumoniae based on the nucleotide sequence of an outer membrane protein. J Clin Microbiol. 1998;36:443–448. doi: 10.1128/jcm.36.2.443-448.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 10.Higgins J A, Ezzell J, Hinnebusch J, Shipley M, Henchal E A, Ibrahim M S. 5′ nuclease PCR assay to detect Yersinia pestis. J Clin Microbiol. 1998;36:2284–2288. doi: 10.1128/jcm.36.8.2284-2288.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product utilizing the 5′-3′ exonuclease activity of Thermus aquaticus. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobsen M J, Nielsen J P. Development and evaluation of a selective and indicative medium for isolation of Actinobacillus pleuropneumoniae from tonsils. Vet Microbiol. 1995;47:191–197. doi: 10.1016/0378-1135(95)00062-f. [DOI] [PubMed] [Google Scholar]
- 13.Kilian M, Nicolet J, Biberstein E L. Biochemical and serological characterization of Haemophilus pleuropneumoniae (Matthews and Pattison 1961) Shope 1964 and proposal of a neotype strain. Int J Syst Bacteriol. 1978;28:20–26. [Google Scholar]
- 14.Kume K, Nakai T, Sawata A. Isolation of Haemophilus pleuropneumoniae from the nasal cavities of healthy pigs. Jpn J Vet Sci. 1984;46:641–647. doi: 10.1292/jvms1939.46.641. [DOI] [PubMed] [Google Scholar]
- 15.Lo T M, Ward C K, Inzana T J. Detection and identification of Actinobacillus pleuropneumoniae serotype 5 by multiplex PCR. J Clin Microbiol. 1998;36:1704–1710. doi: 10.1128/jcm.36.6.1704-1710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Møller K, Andersen L V, Christensen G, Kilian M. Optimalization of the detection of NAD dependent Pasteurellaceae from the respiratory tract of slaughterhouse pigs. Vet Microbiol. 1993;36:261–271. doi: 10.1016/0378-1135(93)90093-m. [DOI] [PubMed] [Google Scholar]
- 17.Møller K, Kilian M. V factor-dependent members of the family Pasteurellaceae in the porcine upper respiratory tract. J Clin Microbiol. 1990;28:2711–2716. doi: 10.1128/jcm.28.12.2711-2716.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen R. Haemophilus pleuropneumoniae infection in pigs. Thesis. Copenhagen, Denmark: Danish Veterinary Laboratory; 1982. [Google Scholar]
- 19.Nielsen R. New diagnostic techniques: a review of the HAP group of bacteria. Can J Vet Res. 1990;54(Suppl.):S68–S72. [PubMed] [Google Scholar]
- 20.Nielsen R, Andresen L O, Plambeck T. Serological characterization of Actinobacillus pleuropneumoniae biotype 1 strains antigenetically related to both serotypes 2 and 7. Acta Vet Scand. 1996;37:327–336. doi: 10.1186/BF03548098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen R, Plambeck T, Foged N T. Blocking enzyme-linked immunosorbent assay for detection of antibodies to Actinobacillus pleuropneumoniae serotype 2. J Clin Microbiol. 1991;29:794–797. doi: 10.1128/jcm.29.4.794-797.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oberst R D, Hays M P, Bohra L K, Phebus R K, Yamashiro C T, Paszko-Kolva C, Flood S J A, Sargeant J M, Gillespie J R. PCR-based DNA amplification and presumptive detection of Escherichia coli O157:H7 with an internal fluorogenic probe and the 5′ nuclease (TaqMan) assay. Appl Environ Microbiol. 1998;64:3389–3396. doi: 10.1128/aem.64.9.3389-3396.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sibidé M, Messier S, Larivière S, Gottschalk M, Mittal K T. Detection of Actinobacillus pleuropneumoniae in the upper respiratory tract as a complement to serological tests. Can J Vet Res. 1993;57:204–208. [PMC free article] [PubMed] [Google Scholar]
- 24.Witham P K, Yamashiro C T, Livak K J, Batt C A. A PCR-based assay for the detection of Escherichia coli Shiga-like toxin genes in ground beef. Appl Environ Microbiol. 1996;62:1347–1353. doi: 10.1128/aem.62.4.1347-1353.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woo T H S, Patel B K C, Smythe L D, Norris M A, Symond M L, Dohnt M F. Identification of pathogenic Leptospira by TaqMan probe in a LightCycler. Anal Biochem. 1998;256:132–134. doi: 10.1006/abio.1997.2503. [DOI] [PubMed] [Google Scholar]