Abstract
Determining mechanisms that underlie reproductive isolation (RI) is key to understanding how species boundaries are maintained in nature. Transposable elements (TEs) are ubiquitous across eukaryotic genomes. However, the role of TEs in modulating the strength of RI between species is poorly understood. Several species of Drosophila have been found to harbor P-elements (PEs), yet only D. simulans is known to be currently polymorphic for their presence in wild populations. PEs can cause RI between PE-containing (P) and PE-lacking (M) lineages of the same species. However, it is unclear whether they also contribute to the magnitude of RI between species. Here, we use the simulans species complex to assess whether differences in PE status between D. simulans and its sister species, which do not harbor PEs, contribute to multiple barriers to gene flow between species. We show that crosses involving a P D. simulans father and an M mother from a sister species exhibit lower F1 female fecundity than crosses involving an M D. simulans father and an M sister-species mother. We also find that another TE, I-element, might play a minor role in determining the frequency of dysgenesis between species. Our results suggest that the presence of PEs in a species can strengthen isolation from its sister species, providing evidence that TEs can play a role in RI.
Keywords: Drosophila, P-elements, Hybrids
Impact Summary
Transposable elements (TEs) are repetitive genetic units found across the tree of life. They play a fundamental role in the evolution of each species’ genome. TEs have been implicated in diversification, extinction, and the origin of novelty. However, their potential role in contributing to the maintenance of species boundaries remains largely understudied. Using whole genome sequences, we compared the relative content of TEs across the three species of the Drosophila simulans complex. We find that the presence of one TE, P-element (PE), in D. simulans, and its absence in the sister taxa, differentiates the three species. PEs cause a suite of fitness defects in Drosophila pure-species individuals if their father has PEs but their mother does not, a phenomenon known as hybrid dysgenesis (HD). We thus studied the possibility that PEs enhance isolation between recently diverged species. In particular, we studied whether the progeny from interspecific crosses was more prone to suffer from HD than pure species. We found that the presence of paternal PEs reduces hybrid female fecundity, similar to observations of HD described within species. The effect of PEs is stronger in the interspecific hybrids than in pure species. A second TE, I-element, has a weaker, but nonnegligible, effect on the magnitude of female fecundity. Our results suggest that PEs can strengthen reproductive isolation in well-formed sister species that still hybridize in nature and pose the question of whether other TEs are involved in the formation of species or in their persistence over time.
Barriers to gene flow can be categorized depending on when in the reproductive cycle they occur (Dobzhansky 1937; Ramsey et al. 2003; Sobel et al. 2010). Prezygotic isolation includes all reproductiveisolation (RI) traits that preclude the formation of a hybrid zygote, such as ecological, behavioral, and gametic incompatibilities (Dobzhansky 1937). Postzygotic barriers occur after a hybrid zygote is formed and include phenotypes as extreme as hybrid sterility and inviability (Orr and Presgraves 2000; Orr 2005), but also more nuanced traits, such as hybrid behavioral defects (Turissini et al. 2017; McQuillan et al. 2018) and delays in development (Matute and Coyne 2010; Ishikawa et al. 2011). Although both types of barriers can lead to speciation, prezygotic barriers tend to be completed faster than postzygotic ones (Sasa et al. 1998; Moyle et al. 2004; Turissini et al. 2018). Nonetheless, postzygotic barriers seem to be crucial to the persistence of species in the face of gene flow once lineages come into secondary contact (Rosenblum et al. 2012; Coughlan and Matute 2020).
The genetic basis of postzygotic isolation usually stems from deleterious interactions between parental genomes in hybrids, which can occur at the allelic level (reviewed in Maheshwari and Barbash, 2011; Nosil and Schluter, 2011) or the genomic level (i.e., “genome clashes”; Nolte et al. 2009; Dion-Côté et al. 2014; Dion-Côté and Barbash 2017). Transposable elements (TEs) are a candidate to cause the latter (Ginzburg et al. 1984; Castillo and Moyle 2012; Watson and Demuth 2013; Serrato-Capuchina and Matute 2018). TEs are repetitive genetic units found ubiquitously across taxa and have been linked to molecular and phenotypic novelty (McClintock 1953; Werren 2011). TEs can facilitate rapid genetic change by causing genic interruptions, duplications, or gene expression changes through their transposition (Trizzino et al. 2017). In pure species, TEs are repressed by the interaction between PIWI genes and noncoding RNAs called PIWI-interacting RNAs (piRNAs). piRNAs are small (24−35 nucleotides long) and suppress TE expression across multiple taxa (reviewed in Michalak 2009). Derepression of TEs through a mismatch of regulatory systems might activate dormant transposons, which in turn can have deleterious fitness implications in hybrids (Kelleher 2016; Kelleher et al. 2020). Comparative gene expression surveys suggest TE misexpression is common in hybrids but not in pure species (Pélissier et al. 1995, 1996; Metcalfe et al. 2007; Dion-Côté et al. 2014), which might be an indication of the role of TE derepression in hybrid breakdown.
P-elements (PEs) in Drosophila melanogaster are one of the best-studied TEs in terms of molecular mechanisms and phenotypic consequences (reviewed in Kelleher 2016). The invasion of PEs into all populations of D. melanogaster occurred rapidly (Kidwell et al. 1977). PEs in D. melanogaster cause a variety of phenotypes collectively named hybrid dysgenesis (HD), a syndrome of deleterious effects in F1s from crosses between a mother who lacks PEs (♀M) and a father who carries them (♂P). F1s from these crosses show a suite of defects that include sterility, chromosomal breaks, and increased mutation rates (Kidwell and Kidwell 1975; Kidwell et al. 1977; Schaefer et al. 1979; Engels and Preston 1980; Kidwell 1983a). Conversely, in crosses where the mother carries PEs (♀P), F1s do not show HD, regardless of the genotype of the father, because piRNAs and associated PIWI genes inherited from the mother confer protection from deleterious effects of paternally inherited PEs. Additionally, environmental conditions also affect the severity of HD, which tends to be stronger at higher temperatures (Hill et al. 2016; Kidwell 1983b). This multiway interaction between paternally inherited PEs, maternally inherited piRNAs, and temperature ultimately determines whether an F1 suffers from HD due to this TE (Slotkin and Martienssen 2007; Siomi et al. 2011; Saito 2013).
Although it is clear that PEs can cause HD within D. melanogaster and D. simulans (Kidwell et al. 1977; Bingham et al. 1982; Hill et al. 2016), the role of PEs in contributing to RI between well-formed species remains largely untested. If two potentially hybridizing species differ in their TE content (e.g., PEs), hybrid progeny might suffer from HD, likely due to TE and suppression system mismatch (Serrato-Capuchina and Matute 2018). Examining the potential effect of the presence versus absence of PEs on RI between species requires the study of a group of species in which: (i) some species harbor PEs while others do not, (ii) species hybridize, and (iii) interspecific crosses yield fertile progeny. Drosophila melanogaster, the classical model for the study of HD, can intercross with several other species, but all viable hybrids are sterile and consequently do not fulfill these criteria (but see Davis et al. 1996; Barbash and Ashburner 2003; Brideau et al. 2006). Species from the willistoni and saltans species groups all seem to have PEs (Mizrokhi and Mazo 1990; Clark et al. 1995; Brookfield and Badge 1997; Silva and Kidwell 2000; De Setta et al. 2007) and thus do not satisfy the criteria either.
The simulans species group, which contains D. simulans, D. sechellia, and D. mauritiana, is an ideal system to assess the potential effects of TEs in RI for at least three reasons. First, PEs recently invaded D. simulans (Kofler et al. 2015). Longitudinal sampling from 1988 to 2014 and genome-wide screening revealed PEs invaded D. simulans in the early 2000s and to date remain polymorphic (i.e., are only present in some lines). Although most D. simulans individuals collected after 2015 harbor functional PEs, to date neither D. sechellia nor D. mauritiana have shown any evidence for their presence, past or present (Kofler et al. 2015). Second, all pairwise crosses of species in this group produce fertile female F1 hybrids (Turissini et al. 2018). Third, there is evidence of hybridization and admixture in nature among these three species. Drosophila simulans and D. sechellia hybridize in the central islands of the Seychelles archipelago (Matute and Ayroles 2014) and show signatures of gene flow (Garrigan et al. 2012; Schrider et al. 2018). Despite the absence of a contemporary hybrid zone between D. mauritiana and D. simulans, these two species also show evidence of introgression (Garrigan et al. 2012; Brand et al. 2013; Meiklejohn et al. 2018). The simulans species group is thus a powerful system to assess whether TEs modulate the strength of RI in nature because the natural variation in D. simulans lines allows for the test of whether populations with PEs show stronger RI toward the sister species than populations without PEs do.
Here, we leverage the recent PE invasion of D. simulans to test whether the presence of PEs and environmental temperature affects the magnitude of postzygotic RI between species that differ in their PE content. We first compared relative content of TEs across species to assess the extent to which PE uniquely explains TE variance across species. Next, we found that PEs do affect F1 female fecundity, particularly at high temperature, mirroring observations of HD described within species (i.e., HD only occurs in ♀M/♂P F1s). This effect is only observable in F1 females, as F1 males from interspecific crosses between species of the simulans species complex are invariably sterile. As observed in pure species, progeny from fathers with higher PE copy numbers showed stronger HD, but this effect was stronger in the interspecific hybrids. Our results serve as a formal test for the putative role of PEs in strengthening RI between sister species.
Materials and Methods
TE DETECTION: SEQUENCING AND READ MAPPING
Genome sequencing, short reads
To assess the number of TE copies in each of the three species in the simulans complex, we used short reads of 115 lines (42 D. sechellia, 15 D. mauritiana, and 58 D. simulans). Table S1 describes which lines were previously sequenced and lists the accession numbers of all lines. To sequence new lines (two D. sechellia and two D. mauritiana), we obtained genomic DNA using the Qiagen®DNeasy® 96 Tissue Kit following the protocol and recommendations from the manufacturer. Next, we outsourced library construction and sequencing to the High Throughput Sequencing Facility at the University of North Carolina, Chapel Hill. The libraries were built using the Nextera protocol as specified by the manufacturer. We obtained read quality information using HiSeq Control Software 2.0.5 and RTA 1.17.20.0 (real-time analysis). CASAVA-1.8.2 generated and reported run statistics of each of the final FASTQ files. Resulting reads ranged from 100 to 150 bp, and the target average coverage for each line was 20×. The mean coverage and accession numbers for these sequences is listed in Table S1.
Genome sequencing, single-molecule sequencing
We sequenced two D. sechellia (Anro71 and Denis72) and two D. mauritiana lines (R42 and R50) with Oxford Nanopore Technology (ONT) to assess if any of these lines harbored complete PE copies. For each of these four lines, we extracted DNA from 20 whole male flies of each line using a modified phenol:chloroform protocol (Sambrook and Russell 2006). We prepared libraries using the Ligation Sequencing Kit 1D with native barcoding (SQK-LSK109 and EXP-NBD104, Oxford Nanopore) according to the manufacturer’s protocol. Libraries were sequenced on six R9.4 flow cells with an average of 1526 available pores and run for 48 h, or until no pores were available, on a GridION running MinKNOW version 3.1.8. Reads were basecalled using Guppy (ONT) with the dna_r9.4.1_450bps_flipflop model. For each line, we mapped reads in a pairwise fashion using Minimap2 version 2.15-r905 (Li 2018) and assembled them using Miniasm version 0.3-r179 (Li 2016). We then corrected and generated consensus assemblies with four iterations of Racon version 1.3.2 (Vaser et al. 2017), followed by Medaka version 0.6.2 (https://github.com/nanoporetech/medaka). The mean coverage and accession numbers for these sequences are listed in Table S2.
TE relative frequency
We aligned Illumina short-read data from the lines sequenced to a list of canonical Drosophila TE sequences (version 10.1; https://github.com/bergmanlab/transposons/blob/master/current/transposon_sequence_set.fa) using minimap2 (Li 2018) with the short-read option (“-cx sr”). We then tabulated the number of base pairs from each sample aligning to each TE and calculated the effective copy number per TE per sample by first dividing that quantity by the canonical length of the TE. This, in turn, was normalized for the depth of sequencing by dividing by the ratio of total base pairs sequenced to genome size. For the latter, we used a fixed estimate of 125 Mb. We generated consensus sequences per TE per line, defining each base as the major allele mapping to that site and leaving as undefined any site for which no allele had more than five supporting reads. This approach differs from previous approaches (Serrato-Capuchina et al. 2020), in that we used the total number of mapped base pairs to calculate the TE copy number and not the number of mapped reads. The results were similar with the two approaches (see below). To assess whether our inference was affected by the way we scored copy number, we tried two different cutoff schemes and counted a copy as complete if it was missing (i.e., the alignment to the reference contained ambiguous bases for) no more than 0% and 25% of the sequence. This approach potentially reveals the number of copies in each isofemale at the time of sequencing. We verified the results from this approach with the long-read sequencing.
TE DETECTION: STATISTICAL ANALYSES
To determine whether the three species of the simulans species complex differed in TE content, we used a linear discriminant analysis (LDA) for the species group. We used the function “lda” (library MASS; Venables et al. 2003) to find the best two linear discriminant (LD) functions, LD1 and LD2. For these analyses, we only retained TEs for which we had inferred at least one full copy in more than one sequenced line. We plotted the scores of these two functions using the function plot (library graphics; R Core Team 2016). We extracted the relative contribution of each PE from the object scaling. To determine which TEs differed between species, we fit a one-way ANOVAs for each TE using lm (library “stats”; R Core Team 2016) where estimated copy number was the response, and species was the fixed factor. We then performed pairwise comparisons using a Tukey HSD test using the function glht (library “multcomp”; Hothorn et al. 2008).
FLY STOCKS
All experiments reported in this manuscript used isofemale lines; all the lines have been previously used and reported elsewhere. We used at least seven lines for each species of the simulans species group. The details of each line are listed in Table S3 and are described by species as follows.
Drosophila simulans
PEs have been present in D. simulans for the last 15 years and have rapidly increased in frequency during that time (Kofler et al. 2015). For crosses, we used seven lines that showed evidence of harboring PEs (denoted as P): 18MPALA11, cascade1, 18MU05, 18KARI10, 18MPALA13, 18MPALA02C, and BiokoLB1. We also used seven lines with no evidence for the presence of PEs (denoted as M): Md06, Md105, Md106, Md199, NS05, NS40, and NS113. Donors and collections details of the lines are listed in Table S3. These lines were collected in Africa between 2000 and 2019 (Rogers et al. 2014; Comeault et al. 2017) and those that contain PEs vary in their copy number (Serrato-Capuchina et al. 2020). All of these lines have been sequenced; Tables S1 and S2 lists the SRA number of each line.
Drosophila sechellia
Unlike D. simulans, D. sechellia lacks functional PEs (Daniels et al. 1990; Kofler et al. 2015; Hill et al. 2016; see Results). For crosses (see below), we used seven lines collected on the islands of La Digué, Denis, and Mahé (Anse Royale Beach) by D. R. Matute and J. F. Ayroles in 2012: LD11, LD16, Denis72, DenisNF13, DenisNF100, Denis124, and Anro71. Instead of banana-yeast traps, females were collected using bottles seeded with ripe Morinda fruits, due to resource preference. Additional details for the lines have been published elsewhere (Matute and Ayroles 2014). Six lines have been previously sequenced (Turissini et al. 2017; Schrider et al. 2018). We obtained short-read and long-read sequencing data for Anro71 and Denis72 (see above). Tables S1 and S2 list the collection location, collectors, and SRA number of each line.
Drosophila mauritiana
Similar to D. sechellia, D. mauritiana shows no evidence of PE presence (Brookfield et al. 1984; Daniels et al. 1990; Kofler et al. 2015; see Results). For crosses, we used seven lines collected in the islands of Mauritius and Rodrigues: R8, R23, R31, R32, R61, R42, and R50. Donors and details of the lines are listed in Table S3. The first five lines have been previously sequenced (Garrigan et al. 2012; Brand et al. 2013; Meiklejohn et al. 2018; Matute et al. 2020). We obtained sequencing data (short- and long-read) for R42 and R50 (see above).
F1 PRODUCTION
Our goal was to study the effect of TEs in interspecific F1 fecundity (see below) at two different temperatures. In all instances, we present all crosses ordered as female × male. For ♀M × ♂P and ♀M × ♂M crosses, we crossed females from seven lines of each of M D. simulans, seven D. mauritiana, and seven D. sechellia lines to males from seven M and seven P D. simulans lines for a total of 294 types of crosses (21 × 14). For ♀P × ♂M crosses, we crossed females from seven lines of each of M and P D. simulans to seven lines of each of M D. simulans, D. sechellia, and D. mauritiana, for a total of 294 types of crosses (14 × 21). For both within- and between-species crosses, we placed 4- to 9-day-old virgins of each species in 30-mL bottles with yeast and cornmeal medium. For conspecific crosses, we used a 1:1 ratio of females to males in each cross. For heterospecific crosses, we used a 1:2 ratio of females to males in each cross to account for greater female choosiness (Lachaise et al. 1986; Turissini et al. 2018), with no more than 50 total flies per vial. Right after mixing, we maintained the vials at 24°C for 24 h. After this time, vials were placed in a 25 or 29°C incubator (Percival DR-36VL; Percival Scientific Inc., Boone, NC). Every 6 days, we transferred the adults to a new vial and inserted a pupation substrate (KimWipes; Kimberly-Clark, Roswell, NM) treated with 0.5% propionic acid to the old vial. Vials were inspected daily for the production of progeny. F1s were collected for up to 25 days after parentals were removed. Our goal was to study at least 20 individuals per cross. We dissected 20 females for each cross with the exception of the crosses listed in Table S4, for which we dissected 40 females per cross.
OVARY NUMBER, COUNTS
One of the most pronounced phenotypes of the HD syndrome is reduced female fecundity, as evidenced by the presence of atrophied gonads (ovaries). We scored the number of functional ovaries in pure species and F1 hybrids in the simulans species complex at two temperatures. The procedure for conspecific and heterospecific crosses was identical. We produced F1 females as described above (see F1 PRODUCTION). To score female fecundity, we counted the number of functional gonads in each F1 female (i.e., 0, 1, or 2 ovaries). We collected females aged 4–9 days posteclosion, anesthetized them with CO2, and extracted their reproductive tracts using forceps (Wong and Schedl 2006). The ovaries from each individual were then fixed on a precleaned glass slide with chilled Drosophila Ringer’s solution (Cold Spring Harbor Protocols; Serrato-Capuchina et al. 2020). We counted the number of functional and atrophied gonads for each individual. An ovary was considered functional if it contained at least one ovariole (egg chamber). We used a Leica S6E stereoscopic microscope (Leica Microsystems, Brønshøj, Denmark) to score all dissections. We scored 20 females for each of the conspecific and heterospecific combinations (Table S4 lists the exceptions).
OVARY NUMBER, STATISTICAL ANALYSIS
PE effect
A distinct trait of HD is a reduced number of ovaries in females from ♀M × ♂P crosses (Kidwell et al. 1977; Engels and Preston 1980; Rubin et al. 1982; Kidwell 1985; Hill et al. 2016). Our goal was to assess whether the presence of PEs led to a more pronounced HD phenotype in heterospecifics than in conspecifics. First, we compared whether different crosses (e.g., ♀M × ♂P vs. ♀M × ♂M) showed differences in the proportion of dysgenic females they produced. We pooled the observations for all crosses that involved the same maternal line and the same type of male (i.e., same species, either ♂M or ♂P), and calculated the binomial confidence interval of each proportion using the function “binom.cloglog” (library binom; Dorai-Raj 2015) and plotted the means and confidence intervals using the function “plotCI” (library gplots; Warnes; Warnes et al. 2016). We compared whether proportions of dysgenic progeny differed between ♀ sim M × ♂ mau and ♀ sim P × ♂ mau, between ♀ sim M × ♂ sech and ♀ sim P × ♂ sech, between ♀ mau × ♂ sim M and ♀ mau × ♂ sim P, and between ♀ sech × ♂ sim M and ♀ sech × ♂ sim P using two-sample tests for equality of proportions with continuity correction (2sEP) as implemented in the function “prop.test” (library stats; R Core Team 2016).
I-elements and hobo effect
PEs are not the only type of TE that can be involved in HD. We studied whether the presence of any of two other TEs known to cause dysgenesis—hobo (Blackman et al. 1987; Ladevèze et al. 1998) and I-element (IE; Picard et al. 1978; Orsi et al. 2010)—varied among species and was involved in HD in F1 hybrid females. Ideally, we would be able to compare the prevalence of HD in a combination of crosses in which both females and males have all the possible combinations of TEs. This was not the case for our sample. The experimental crosses were originally designed to study the effect of PEs on HD. A small sample of our ♀sim × ♂sim crosses involved females who were IE−, whereas the majority involved females who were IE+. We used the same approach described for PEs, using 2sEPs, to assess whether females from ♀IE− × ♂IE+ crosses were more likely to show HD than females from ♀IE− × ♂IE− crosses. We compared the power of these comparisons and the ones involving PEs (described immediately above) with the function “pwr.2p2n.test” (library pwr; Champely et al. 2018). A similar approach was not possible for hobo because all the assayed D. simulans lines were hobo+ (i.e., our experiment did not include hobo− females). We restricted our analyses to the two crosses that showed dysgenic progeny: ♀mau × ♂sim, and ♀sech × ♂sim.
Copy number effect
We studied the effect of PE, hobo, and IE copy number in the paternal genome in crosses with D. simulans M, D. mauritiana, and D. sechellia females. For each of two types of interspecific crosses for which we observed evidence of HD (i.e., ♀sech × ♂sim and ♀mau × ♂sim) and for each temperature, we fitted a Generalized Linear Mixed Model using Markov chain Monte Carlo where the response of the model was whether a female was dysgenic or not, the type of cross (either conspecific or heterospecific) was a fixed effect, and the number of TE copies in the paternal genome (of each of PE, hobo, and IE with a 0% missing sequence threshold) was a continuous variable. We included three interactions between the type of cross and TE type. We also included two random effects, one for the mother line identity and one for the father line identity. Each of these datasets included interspecific data and intraspecific data (within D. simulans). We fitted a regression using the function “MCMCglmm” (library MCMCglmm; Hadfield 2010). Each of the four fully factorial models followed the form:
To determine if the MCMC chains had converged, we used the “gelman.diag” function (library MCMCglmm; Hadfield 2010) for two chains ran in parallel. To quantify the significance of each effect, we used the function summary and determined whether the regression coefficient differed from zero by examining the confidence interval of each interaction and their associated pMCMC value.
MALE HYBRID STERILITY
F1 hybrid males between all interspecific crosses in the simulans species subgroup are sterile (Turissini et al. 2018; Lachaise et al. 1986). We evaluated whether PE infection status affected this barrier. We made hybrid males as described above at two different temperatures: 25 and 29°C. Four- to 8-day-old males from conspecific and heterospecific crosses were then lightly anesthetized with CO2. Their testes were then extracted with Dumont #5 forceps and mounted in chilled Drosophila Ringer’s solution (Cold Spring Harbor Protocols; Serrato-Capuchina et al. 2020). An average of five to 10 pairs of testes were mounted per slide. Using a microscope, we scored whether each pair of testes had motile sperm within 5 min of having mounted the samples. For each interspecific genotype combination, we scored sperm motility for 100 males per cross (398 combinations: 39,800 total males).
Results
TE SURVEY IN THE simulans SPECIES COMPLEX
First, we explored whether the genomes of the three species from the simulans complex differed in their TE content. Figure 1A shows an LDA of the TEs that contribute to differences among genotypes (i.e., species and lines). We show the results when we allowed for up to 25% of a sequence to be missing in a TE to count it as present in that line. The first discriminant function, LD1, separates the three species (86% of the trace) more effectively than LD2 (14% of the trace). Figure 1B shows the loadings in LD1 for 27 TEs. Of the three TEs that have been implicated in HD (PE, hobo, and IE), PE has the largest contribution to the discriminant function (Fig. 1B). Figure S1 shows the loadings for LD2, but this function is at least five times less effective at separating species than LD1. These results suggest that the differences in overall TE content in the simulans species group are multifaceted, and cannot be explained by a single type of TE. Figures S2 and S3 show LDAs for a different, and more stringent, cutoff to estimate TE copy number (0% of the consensus sequence missing to count it as present in that line); regardless of the cutoff, TE content differs between species in the simulans complex, and PE copy number is a large contributor to that total difference.
Figure 1.
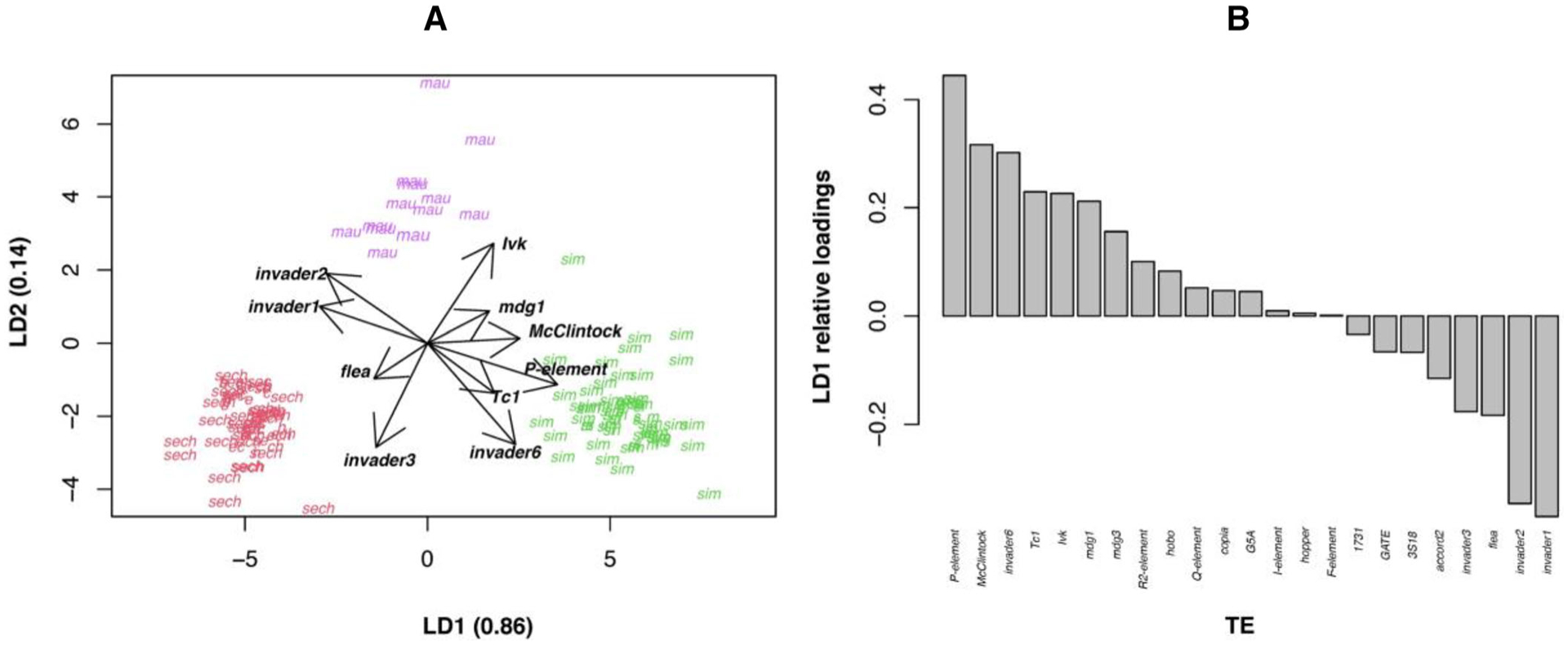
Linear discriminant analysis (LDA) of TEs present in the three species of the simulans species complex. (A) Discriminant function scores of individuals from the three species of the simulans species complex. D. sechellia: red; D. mauritiana: purple; D. simulans: green. The axes show the proportion of trace (i.e., separation achieved) by each of the two discriminant functions. Arrows show the 10 TEs with the largest loadings. (B) LD1 relative loadings by each of the 23 included TEs. Figure S1 shows the LD2 relative loadings.
Next, we studied whether any TE was present in one of the species of the simulans clade but not in the other ones. Figure 2 shows the estimated number of copies for the 12 TEs that explain most of the variance in LD1 (listed in Fig. 1B). Figures S4 and S5 show the other 38 TEs. The majority of TEs were either present in all the three species—but in different copy numbers (Fig. 2)—or absent in the three species (Fig. S4). This similarity reflects the close genealogical relationship between the triad of species. Different cutoffs had no effect on our results. Figure S5 shows boxplots for a more stringent cutoff to estimate TE copy number. Unlike D. simulans, which carry complete and incomplete copies of PEs (Kofler et al. 2015; Hill et al. 2016; Serrato-Capuchina et al. 2020), the latter two species have no traces of the PE sequence (i.e., no read coverage). The use of different cutoffs of sequence completeness to infer TE copy number had no effect on the inference of PE copy number (Fig. 2A vs. S5). Long-reads confirmed that a subset of D. mauritiana and D. sechellia do not have complete—or incomplete—copies of PE (Table S5).
Figure 2.
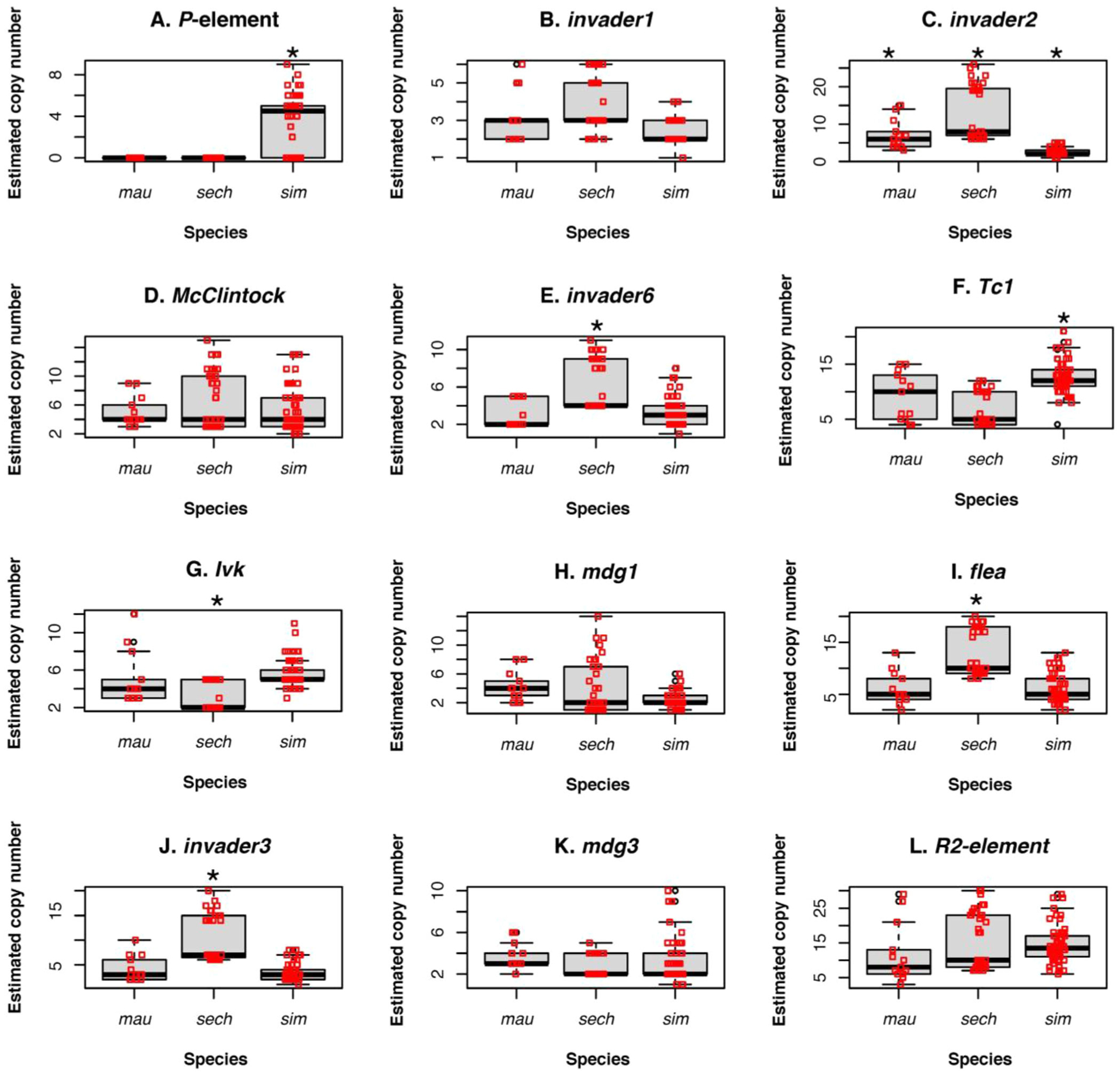
The abundance of TEs varies in the three species of the simulans complex. We filtered out copies in which at least 25% of the sequence of the TE was missing. We show the 12 TEs with the largest loadings in LD1; other 11 TEs are shown in Figure S4. Results from a different filtering scheme are shown in Figure S5. Please note that the y-axes differ across panels. The only TE present in one species but completely absent in the others is P-element (panel A). Asterisks show instances where one species differed from the other two at a given TE copy number (Tukey post hoc test).
It is worth noting that two other TEs have been implicated in HD in Drosophila: hobo (Blackman et al. 1987; Ladevèze et al. 1998) and IE (Picard et al. 1978; Orsi et al. 2010). When we used short-reads and a 25% missing sequence cutoff, IE and hobo were inferred to be polymorphic in the three species of the simulans complex (Fig. S4). Long-read data (with a 60% and 70% accuracy threshold) indicated that functional copies of IE and hobo might be present in all species of the simulans species complex depending on the accuracy threshold (Table S5). When we used short-reads and a more stringent cutoff (0% missing sequence), IE was inferred to be absent in D. sechellia and D. mauritiana; hobo was inferred to be absent in D. sechellia and polymorphic in D. mauritiana. These results emphasize the importance of reporting the relevant thresholds used for the detection in TEs.
The species difference in the presence of PEs and D. simulans’ polymorphism for PE copy number make the simulans species complex ideal to study the effects of different types of TEs in RI in well-formed species.
EFFECTS OF PE ON FEMALE FECUNDITY
PEs have been associated with reduced fecundity and higher rates of female sterility in crosses within D. melanogaster and D. simulans (Bingham et al. 1982; Kidwell 1983b; Hill et al. 2016). PEs also differ in presence–absence between D. simulans and the other two species of the simulans complex (see above). We studied whether PEs had an effect on female hybrid fecundity. Specifically, we studied whether F1 hybrid females from ♀mau × ♂sim and ♀sech × ♂sim crosses were more likely to show atrophied ovaries, a proxy of HD if the fathers carried PEs. Figure 3 shows examples of progeny with atrophied ovaries from conspecific and heterospecific crosses. Three results are notable from these counts. First, females from ♀M × ♂M crosses showed few females with atrophied gonads in both conspecific and heterospecific crosses (~0.1%; Table 1). Second, ♀M/♂P females from heterospecific crosses showed a higher rate of gonad atrophy than females from conspecific crosses at both temperatures (Fig. 4; Table 1). ♀M/♂P F1 females from interspecific crosses were more likely to have atrophied ovaries than ♀M/♂P females from crosses within D. simulans (Table 1). Third, we found dysgenic females in all ♀M × ♂P crosses at both temperatures but, as expected (Kidwell and Novy 1979), the frequency of gonad atrophy in ♀M × ♂P crosses is higher at 29°C than at 25°C in conspecific crosses (2sEP: χ2 = 159.37, df = 1, P < 1 × 10−10) and heterospecific crosses (2sEPmau × sim: χ2 = 122.76, df = 1, P < 1 × 10−10; 2sEP sech × sim: χ2 = 18.593, df = 1, P = 1.618 × 10−5; Fig. 4A vs. 4C and Fig. 4B vs. 4D). Previous studies have shown that temperature increases the severity of HD in conspecific crosses (Kidwell 1983b; Serrato-Capuchina et al. 2020). These results suggest that PE-induced HD—at least in the form of ovary atrophy—is more pronounced in heterospecific than in conspecific crosses.
Figure 3.
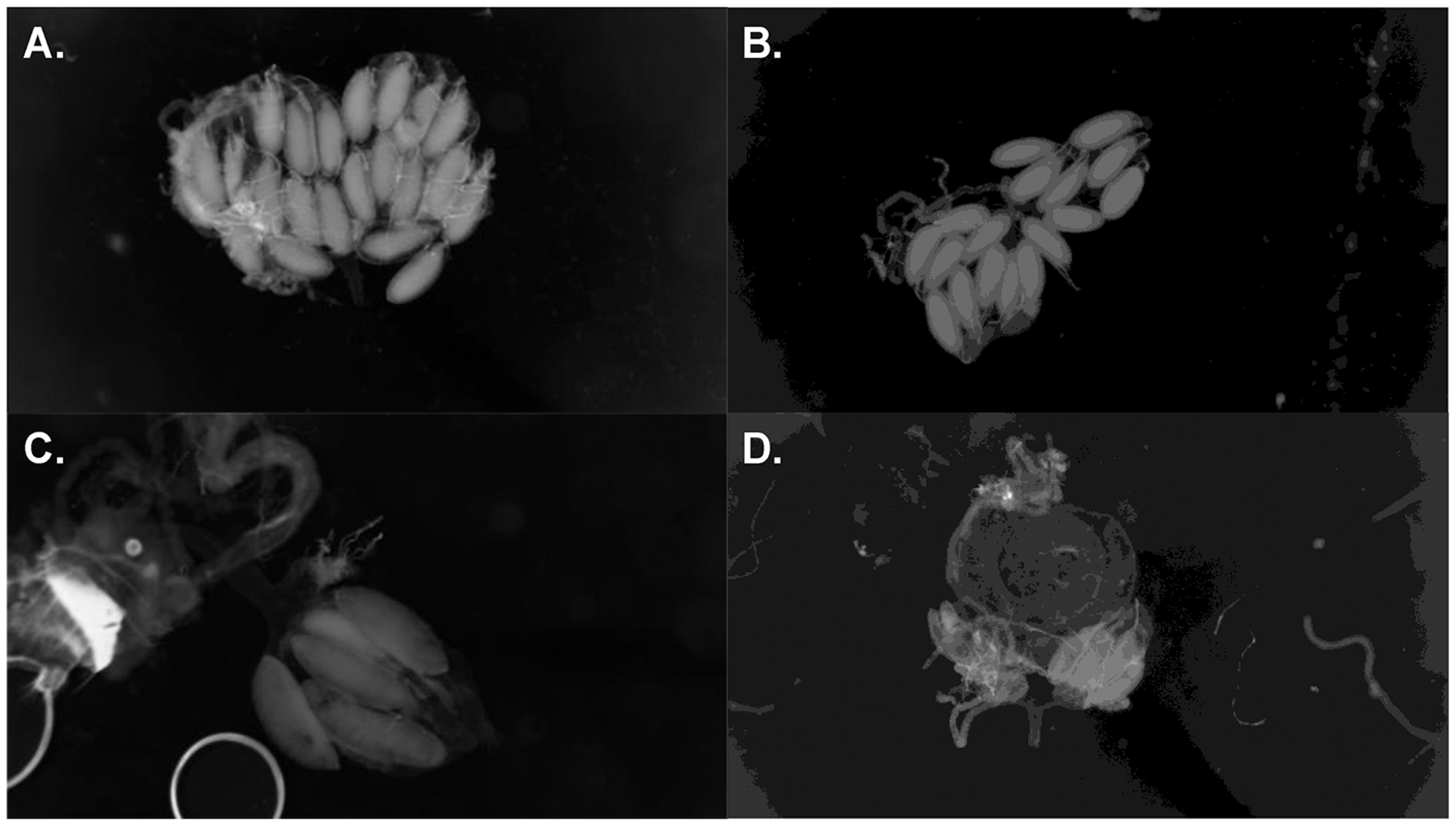
Examples of nondysgenic and dysgenic female reproductive tracts from conspecific and heterospecific F1s. (A) Nondysgenic ♀M/♂M D. simulans/D. simulans F1 female. (B) Nondysgenic ♀M/♂M F1 D. mauritiana female. (C) Dysgenic ♀M/♂P D. simulans/D. simulans F1 female. (D) Dysgenic ♀M/♂P D. mauritiana/D. simulans F1 female. Both ♀M/♂P females (C and D) have only one functional ovary.
Table 1.
♀M/♂P females from heterospecific crosses are more likely to be dysgenic than ♀M/♂P females from conspecific crosses.
| Temp (°C) | ♀M/♂P | ♀M/♂M | Proportion test | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | Mean | CI | Total | Mean | CI | χ 2 | df | P-value | ||
| ♀ sim × ♂ sim | 25 | 2100 | 0.982 | [0.975, 0.987] | 1960 | 0.999 | [0.996, 1.000] | 28.574 | 1 | <0.0001 |
| ♀ mau × ♂ sim | 25 | 980 | 0.851 | [0.827, 0.872] | 980 | 1.000 | [0.996, 1.000] | 44.175 | 1 | <0.0001 |
| ♀ sech × ♂ sim | 25 | 980 | 0.821 | [0.796, 0.844] | 980 | 0.986 | [0.976, 0.992] | 18.593 | 1 | <0.0001 |
| ♀ sim × ♂ sim | 29 | 2100 | 0.829 | [0.812, 0.844] | 1960 | 0.988 | [0.982, 0.992] | 299.22 | 1 | <0.0001 |
| ♀ mau × ♂ sim | 29 | 980 | 0.728 | [0.699, 0.754] | 980 | 1.000 | [0.996, 1.000] | 44.175 | 1 | <0.0001 |
| ♀ sech × ♂ sim | 29 | 980 | 0.740 | [0.711, 0.766] | 980 | 1.000 | [0.996, 1.000] | 18.593 | 1 | <0.0001 |
Females with atrophied ovaries are more common in heterospecific than in conspecific crosses in this direction of the cross. Each row shows a type of cross. Temp (°C): Temperature (°C); Mean: number of nondysgenic females; CI: Binomial confidence interval. We used two-sample tests for equality of proportions (2sEP) for all comparisons and adjusted the P-values with a Bonferroni correction.
Figure 4.
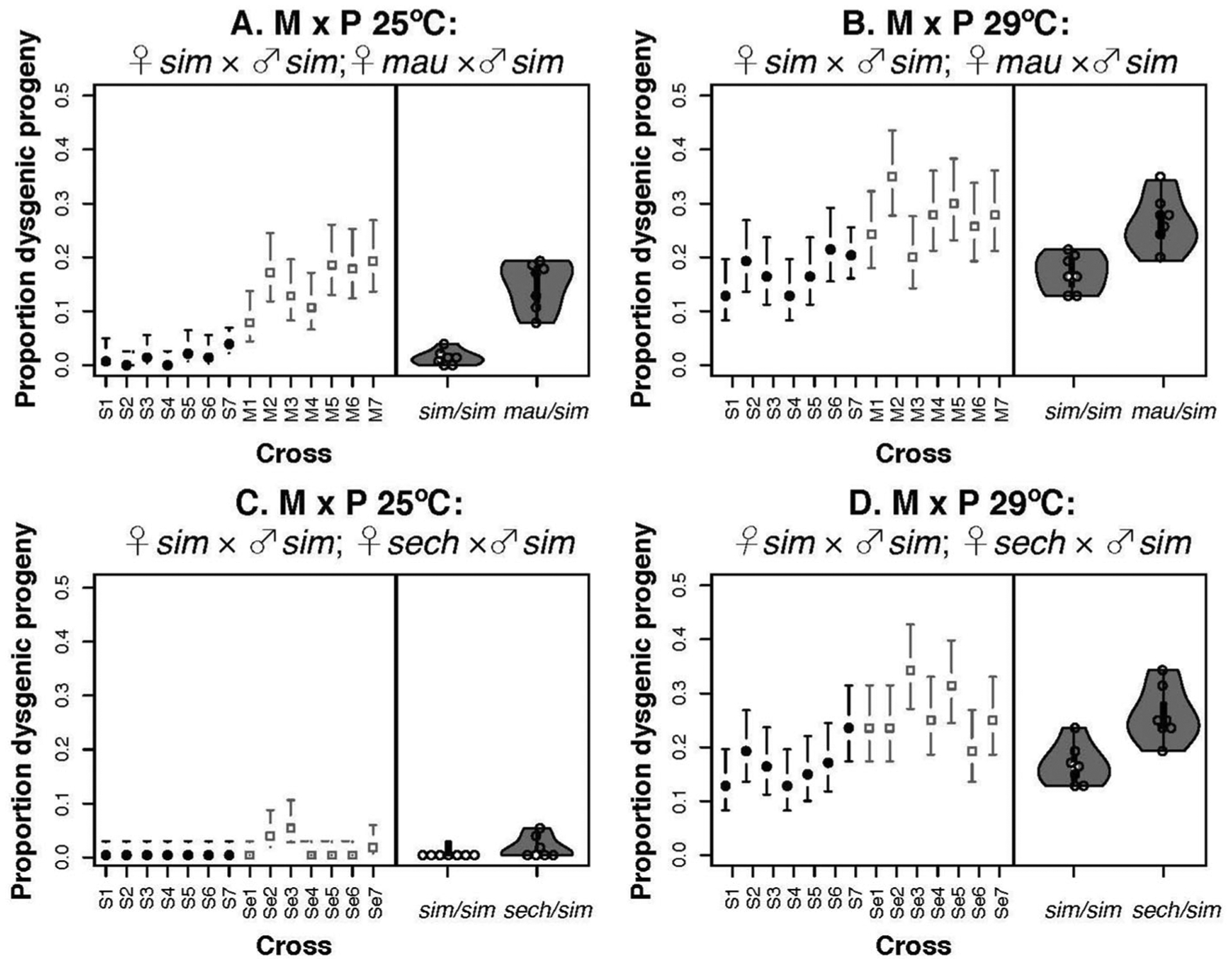
The proportion of dysgenic progeny is higher in heterospecific than in conspecific ♀M/♂P crosses. Left panels: Solid black circles represent conspecific crosses within D. simulans (♀ D. simulans × ♂ D. simulans); open squares represent interspecific crosses (either ♀ D. mauritiana × ♂ D. simulans or ♀ D. sechellia × ♂ D. simulans). Bars show the Bayesian confidence interval of the proportion. Right panels: Violin plots showing the distribution of the means for each maternal line (shown in left panels) for each type of cross (conspecific vs. heterospecific). (A) ♀ sim M × ♂ sim P and ♀ mau M × ♂ sim P at 25°C. (B) ♀ sim M × ♂ sim P and ♀ mau M × ♂ sim P at 29°C. (C) ♀ sim M × ♂ sim P and ♀ sech M × ♂ sim P at 25°C. (D) ♀ sim M × ♂ sim P and ♀ sech M × ♂ sim P at 29°C. S1: Md06, S2: Md105, S3: Md106, S4: Md199, S5: NS05, S6: NS113, and S7: NS40; D. mauritiana lines: M1: R23, M2: R31, M3: R32, M4: R42, M5: R50, M6: R61, and M7: R8; D. sechellia lines: Se1: Anro71, Se2: Denis124, Se3: Denis72, Se4: DenisNF100, Se5: DenisNF13, Se6: LD11, and Se7: LD16.
An additional prediction of PE-induced HD is that F1 females from ♀P × ♂M crosses (♀P/♂M) should show similar ovary counts to F1 ♀M/♂M females in conspecific and heterospecific crosses. Regardless of the cross and the temperature, the vast majority of conspecific and hybrid females with a D. simulans mother had two ovaries (Table S6). As predicted, ♀P/♂M F1s from either conspecific or heterospecific crosses are nondysgenic.
POTENTIAL EFFECT OF OTHER TES IN HD
Two other TEs, IE and hobo, have been implicated in HD in D. melanogaster (IE: Bucheton et al. 1984, Chaboissier et al. 1995, Orsi et al. 2010; hobo: Blackman et al. 1987, Yannopoulos et al. 1987). Our experiment was designed to assess the effect of PEs in interspecific hybrids but a subset of the data in which the mother had no complete copies of IE (as determined by the 0% missing filter) allowed us to inquire whether IE is also associated with the prevalence of HD. In spite of being a subset of the whole dataset, the power of the 2sEP involving IE was similar to the ones involving PE (powerIE = 0.865, powerPE = 0.821). Please note that for consistency, we included two D. mauritiana (R42 and R50) and D. sechellia (Anro71 and Denis72) lines as IE− because the short-read calculations allowing for a 0% of missing sequence threshold showed no functional copies, even though the long-read estimates and the short-read calculations allowing for a 25% of missing sequence show they might harbor IEs. The expectation was that if IEs cause HD, then the presence of IE should increase the prevalence of dysgenic females in crosses between females without IE (IE−) and males with IE (IE+). First, we assessed whether IEs cause HD in either pure D. simulans crosses or any of the two interspecific crosses at any of the two studied temperatures. The results for these pairwise comparisons are shown in Table 3. We find no differences in the proportion of dysgenic females between any type of cross at any of the two studied temperatures. This result ran contrary to the expectation that IE would cause HD in the simulans species group. A similar analysis was not possible for hobo because all surveyed D. simulans had evidence of complete hobo elements, even with the most astringent filter (number of copies estimated with 0% sequence missing). Similarly, we did not conduct this analysis for IE allowing for a 25% of missing sequence because all assayed lines showed the potential of harboring IEs at this more permissive threshold.
Table 3.
The effect of paternal copy number of PE, IE, and hobo in crosses with M females depends on temperature and the species of the mother.
| Cross | Temp (°C) | Effect | CI | pMCMC |
|---|---|---|---|---|
| ♀ mau × ♂ sim | 25 | Cross | [−0.012, 0.135] | 0.090 |
| ♀ mau × ♂ sim | 25 | PE copy number | [−0.025, −0.013] | <0.001 |
| ♀ mau × ♂ sim | 25 | hobo copy number | [0.000, 0.004] | 0.064 |
| ♀ mau × ♂ sim | 25 | IE copy number | [−0.002, 0.001] | 0.650 |
| ♀ mau × ♂ sim | 25 | Cross × PE copy number | [0.012, 0.024] | <0.001 |
| ♀ mau × ♂ sim | 25 | Cross × hobo copy number | [−0.005, 0.000] | 0.122 |
| ♀ mau × ♂ sim | 25 | Cross × IE copy number | [−0.001, 0.002] | 0.654 |
| ♀ mau × ♂ sim | 29 | Cross | [0.069, 0.150] | 0.480 |
| ♀ mau × ♂ sim | 29 | PE copy number | [−0.051, −0.032] | <0.001 |
| ♀ mau × ♂ sim | 29 | hobo copy number | [−0.001, 0.008] | 0.128 |
| ♀ mau × ♂ sim | 29 | IE copy number | [−0.003, 0.002] | 0.558 |
| ♀ mau × ♂ sim | 29 | Cross × PE copy number | [0.003, 0.021] | 0.020 |
| ♀ mau × ♂ sim | 29 | Cross × hobo copy number | [−0.006, 0.003] | 0.670 |
| ♀ mau × ♂ sim | 29 | Cross × IE copy number | [− 0.003, 0.003] | 0.736 |
| ♀ sech × ♂ sim | 25 | Cross | [−0.059, 0.085] | 0.722 |
| ♀ sech × ♂ sim | 25 | PE copy number | [−0.032, −0.023] | <0.001 |
| ♀ sech × ♂ sim | 25 | hobo copy number | [0.001, 0.006] | 0.010 |
| ♀ sech × ♂ sim | 25 | IE copy number | [−0.008, −0.002] | 0.004 |
| ♀ sech × ♂ sim | 25 | Cross × PE copy number | [0.018, 0.032] | <0.001 |
| ♀ sech × ♂ sim | 25 | Cross × hobo copy number | [−0.007, 0.001] | 0.084 |
| ♀ sech × ♂ sim | 25 | Cross × IE copy number | [0.000, 0.010] | 0.026 |
| ♀ sech × ♂ sim | 29 | Cross | [0.075, 0.292] | 0.002 |
| ♀ sech × ♂ sim | 29 | PE copy number | [−0.044, −0.015] | <0.001 |
| ♀ sech × ♂ sim | 29 | hobo copy number | [0.000, 0.013] | 0.060 |
| ♀ sech × ♂ sim | 29 | IE copy number | [−0.006, 0.003] | 0.464 |
| ♀ sech × ♂ sim | 29 | Cross × PE copy number | [−0.012, 0.008] | 0.614 |
| ♀ sech × ♂ sim | 29 | Cross × hobo copy number | [−0.010, −0.001] | 0.010 |
| ♀ sech × ♂ sim | 29 | Cross × IE copy number | [−0.002, 0.004] | 0.672 |
The significance of each variable is determined by whether the confidence interval of the coefficient (CI) overlaps zero (i.e., no effect on the likelihood of HD). Positive values in the interaction coefficients indicate a more pronounced increased in the frequency of HD with the increment of copy number in interspecific crosses. Gelman diagnostics for all factors and interactions equaled 1.00.
TE COPY NUMBER EFFECT ON FEMALE FECUNDITY
Paternal lines with more PEs cause stronger HD in conspecific crosses with M females (Bingham et al. 1982; Hill et al. 2016; Srivastav and Kelleher 2017; Serrato-Capuchina et al. 2020 but see Bergman et al. 2017; Srivastav et al. 2019). We next assessed, using MCMC linear models, whether the paternal copy number of PE, hobo, and IE each had an effect on the likelihood of HD in the progeny produced when crossed to M lines. We used a 0% missing sequence threshold for these analyses. Table 2 shows all the results for the linear models. The comparisons between F1(♀ sim M × ♂ sim) and F1(♀ mau × ♂ sim) crosses yielded qualitatively similar results at the two temperatures. PE copy number was the only significant continuous variable at both temperatures. The only interaction that was significant was between cross and number of PE copies. These results suggest that a higher PE copy number increases the likelihood of dysgenesis more pronouncedly in hybrid F1(♀ mau × ♂ sim) females than in (♀ sim M × ♂ sim) crosses. More generally, in F1(♀ mau × ♂ sim) female hybrids the copy number of PE, and not of other TEs, is a crucial determinant of the frequency of HD.
Table 2.
IE+ fathers are not less likely than IE− fathers to produce dysgenic female progeny when crossed to IE− females.
| Temp (°C) | ♀ IE−/♂IE+ | ♀ IE−/♂IE+ | Proportion test | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | Mean | CI | Total | Mean | CI | χ 2 | df | P-value | ||
| ♀ sim × ♂ sim | 25 | 200 | 1 | [0.982,1] | 1340 | 0.986 | [0.979,0.992] | 1.680 | 1 | 1.000 |
| ♀ mau × ♂ sim | 25 | 280 | 1 | [0.987, 1] | 1680 | 0.913 | [0.899, 0.926] | 25.046 | 1 | 3.358 × 10−6 |
| ♀ sech × ♂ sim | 25 | 280 | 0.975 | [0.948, 0.988] | 1680 | 0.891 | [0.892, 0.876] | 18.184 | 1 | 1.204 × 10−4 |
| ♀ sim × ♂ sim | 29 | 200 | 0.995 | [0.965, 0.999] | 1340 | 0.884 | [0.866, 0.900] | 32.169 | 1 | 8.478 × 10−8 |
| ♀ mau × ♂ sim | 29 | 280 | 1 | [0.987, 1] | 1680 | 0.841 | [0.823, 0.858] | 50.176 | 1 | <1 × 10−10 |
| ♀ sech × ♂ sim | 29 | 280 | 0.989 | [0.967,0.997] | 1680 | 0.836 | [0.812, 0.853] | 44.894 | 1 | 1.248 × 10−10 |
Females with atrophied ovaries are rare in both, heterospecific and conspecific crosses. Each row shows a type of cross. Temp (°C): Temperature (°C); Mean: number of nondysgenic females; CI: binomial confidence interval. We used two-sample tests for equality of proportions (2sEP) for all comparisons and adjusted the P-values with a Bonferroni correction.
The comparisons between F1(♀ sim × ♂ sim) and F1(♀ sech × ♂ sim) crosses differed between the two temperatures, and from the results in F1(♀ mau × ♂ sim) crosses. At 25°C, copy number of the three TEs significantly affected the likelihood of dysgenesis but not in the same way. Although increasing the PE and IE copy number increased the likelihood of HD, increasing the hobo copy number decreased it. PE copy number showed a significant interaction with the type of cross. Unlike for F1(♀ mau × ♂ sim) female hybrids, IE copy number also showed a significant interaction with cross type. For both PE and IE, the propensity of HD to increase with copy number was higher in F1(♀ sech × ♂ sim) hybrids than in (♀ sim M × ♂ sim) females. At 29°C, only PE copy number increased the likelihood of dysgenesis; PE copy number had no strong interaction with cross indicating no difference in the effect of copy number between within-species and interspecific crosses. We found a significant interaction between cross and hobo copy number, which suggested that the effect of hobo in the prevalence of dysgenesis was more severe in (♀ sim M × ♂ sim) than in F1(♀ sech × ♂ sim) hybrids. The results from the comparisons between within-species D. simulans and F1(♀ sech × ♂ sim) hybrid females suggest that the nature of HD in interspecific hybrids is multifaceted, and several TEs play a role in determining its relative occurrence. Collectively, the study of HD in F1(♀ mau × ♂ sim), F1(♀ sech × ♂ sim), and pure D. simulans females suggests that the factors that determine the occurrence of HD in hybrids vary across species and that not a single TE is responsible for the totality of the phenotype, at least in the different interspecific hybridizations of the simulans species complex.
EFFECTS OF PE ON MALE FECUNDITY
We scored sperm motility in hybrid males involving D. sechellia, or D. mauritiana, mothers and P D. simulans fathers. All F1 males, from all interspecific crosses (N = 100 per cross), were completely sterile and produced no sperm, regardless of the identity of the cross. The PE status of D. simulans did not affect hybrid male fertility.
Discussion
PEs in Drosophila cause HD and remain one of the best-understood cases of how TEs interact with their host genome (reviewed in Kelleher 2016). In this report, we studied the role of PEs in RI between three closely related species of the simulans species complex by crossing D. simulans lines that varied in whether they carry PEs to two sister species that do not carry PEs. We find that PE status of the father (and the number of PE copies) affects F1 female fertility in interspecific crosses. HD in F1 ♀M/♂P females is stronger in hybrid females than in within-species females, suggesting that hybrids are more susceptible to the effects of HD than pure-species individuals. We find some evidence that IE also might affect the likelihood of HD in F1(♀sech × ♂sim) hybrid females but only at 25°C. Although it is certainly possible that other TEs different from PEs might modulate the magnitude of RI in hybrids from the simulans clade species, PE is the most clear example of this phenomenon in our survey.
Our study has caveats worth mentioning. First, TE count varies slightly on sequencing technology (Serrato-Capuchina et al. 2020). Because the magnitude and direction of the inferred patterns of HD are similar between short- and long-read data, this is not a major concern. Second, although we find that PEs are associated with the strength of HD, it remains possible that polymorphic genetic incompatibilities may account for some of the reduced fertility in M × P crosses. Differences in the genetic background among isofemale lines might contribute to the strength of isolation in interspecific crosses. If alleles involved in polymorphic hybrid incompatibilities were to explain the results we observed here, then they would have to be more frequent in individuals with higher PE copy number and their action should be temperature dependent. Both of these requirements seem unlikely.
The importance of TEs in the speciation process remains largely unexplored (Watson and Demuth 2013; Serrato-Capuchina and Matute 2018). In the case reported here, PEs cause HD in both pure species and hybrid F1 females. F1 ♀M/♂P females should suffer the deleterious effects of PEs because they do not have the maternally deposited piRNAs that confer protection against TE misregulation (Michalak 2009; Majumdar and Rio 2015; Kelleher 2016). This should be true of ♀M/♂P F1s from either pure-species or interspecific crosses. Our results show that even though PEs induce female HD in conspecific and interspecific ♀M/♂P F1s, the magnitude of HD is moderately stronger in interspecific hybrids than in within-species ♀M/♂P F1s. There must, therefore, be genetic interactions between the two parental genomes and PEs that affect the magnitude of HD in F1s. There is precedent supporting this hypothesis. In interspecific crosses involving a D. lummei mother and a D. virilis father, interspecific hybrids show exacerbated gonadal dystrophy (a proxy of HD) when the father has the TE Penelope. Genetic analyses suggest that Penelope interacts with other autosomal and sex-linked elements by modifying the penetrance of hybrid incompatibilities (Castillo and Moyle, 2019).
TE expression is regularly higher in hybrids than in pure species. In hybrid rice (Shan et al. 2005), hybrid sunflowers (Ungerer et al. 2006), and hybrid fish (Dion-Côté et al. 2014; Dennenmoser et al. 2019; Carleton et al. 2020), there is extensive TE mobilization and proliferation, indicating that these defects might be common among hybrids. In Drosophila, TEs are often over-expressed in Drosophila hybrids (Kelleher et al. 2012; Lopez-Maestre et al. 2017; Romero-Soriano et al. 2017). Misregulation of TEs in hybrids might be related to the divergence of PIWI and piRNA pathways across species, either by natural selection, genetic conflict, or systems drift (i.e., divergence in the molecular underpinnings of homologous traits not due directly to selection; reviewed in True and Haag 2001). Genes from the PIWI pathway show signatures of positive selection since the divergence between D. simulans and D. melanogaster (Vermaak et al. 2005; Heger and Ponting 2007; Obbard et al. 2009; Kolaczkowski et al. 2011; Lee and Langley 2012; Simkin et al. 2013; Wang et al. 2020 reviewed in Blumenstiel et al. 2016). At least five piRNA-interacting genes—aubergine (Kelleher et al. 2012; Wang et al. 2020), rhino, deadlock (Parhad et al. 2017), Armitage, and Spindle-E (Wang et al. 2020)—have functionally diverged between D. simulans and D. melanogaster. Functional divergence in these proteins leads to off-target effects on host transcripts in D. melanogaster transgenics carrying the D. simulans allele. This divergence might also play a role in reduced hybrid fitness. A decrease in genomic autoimmunity in hybrids caused by adaptive divergence of genes in the PIWI pathway might be a cause of PE-induced HD in interspecific hybrids and conspecific F1s.
The relatively recent invasions of PEs in both D. simulans and D. melanogaster seem to be explained by horizontal gene transfer through an unknown vehicle, which poses the question of how often TEs move between species and establish successfully in the host genome. A similarly intriguing question is why PEs have not been found in D. mauritiana and D. sechellia. In both of these island endemics, there is evidence for interspecific introgression with D. simulans (Garrigan et al. 2012; Brand et al. 2013; Meiklejohn et al. 2018), and even though alleles have crossed those species boundaries, PEs have not been transferred from D. simulans to D. mauritiana. Hybridization can be an effective mechanism of TE transfer across species boundaries (Vela et al. 2014). One possibility is that introgression is ancient and precedes the invasion of PEs in D. simulans. The case of D. sechellia is even more puzzling. Not only do D. sechellia and D. simulans show signatures of reciprocal introgression (Schrider et al. 2018), these two species form a contemporary hybrid zone in the central islands of the Seychelles archipelago (Matute and Ayroles 2014). This indicates that, at present, there is ample opportunity for gene exchange and for interspecific transfer of PEs. Alternatively, the D. sechellia and D. mauritiana genomes could be more refractory to the invasion of PEs than D. simulans. The evolutionary dynamics of TE copy number in admixed lineages can differ strongly even among evolutionarily closely related hybrids (Hénault et al. 2020, Heil et al. 2021). Experimental infections with PEs showed the potential of D. simulans to host PEs (Daniels et al. 1989). The same type of experiments could reveal if PEs are less successful at invading some genomes than others, and this, in turn, could explain the absence of PEs in the D. sechellia and D. mauritiana genomes.
The relative importance of different mechanisms of successful genome invasion by TEs (horizontal gene transfer vs. introgression) remains largely unexplored. Multiple epistatic and gene by environment interactions affect the magnitude of effects caused by TE transposition, both between and within species, and the extent to which these patterns affect the evolution of species remains underexplored. It is likely that multiple mechanisms play a role in the transmission of PEs, and only a systematic assessment of the relative frequency of PEs in different species of Drosophila and their putative vectors will reveal the relative importance of each mechanism in the spread of PEs within and between species. Our results show that the dynamics of PEs, and possibly of TEs in general, should not only be addressed exclusively with a lens on the fitness effects that PEs have on within-species crosses, but also on the effects that they might have on interspecific hybrids.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank A. A. Comeault, B. Cooper, and the members of the Matute lab for helpful scientific discussions and comments. We would like to thank the Bioko Biodiversity Protection Program (Equatorial Guinea), the Ministry of Environment (Republic of São Tomé and Príncipe), and the Zambia Wildlife Agency (Zambia) for permission to collect and export specimens for study. This work was supported by National Institutes of Health award R01GM121750. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
DATA ARCHIVING
All data and analytical code can be found at https://doi.org/10.5061/dryad.c2fqz6188
Supporting Information
Additional supporting information may be found online in the Supporting Information section at the end of the article.
LITERATURE CITED
- Barbash DA, and Ashburner M. 2003. A novel system of fertility rescue in Drosophila hybrids reveals a link between hybrid lethality and female sterility. Genetics 163:217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, and Walker S. 2013. lme4: linear mixed-effects models using Eigen and S4. R package version 1.1–7 [Google Scholar]
- Bergman CM, Han S, Nelson MG, Bondarenko V, and Kozeretska I. 2017. Genomic analysis of P elements in natural populations of Drosophila melanogaster. PeerJ 5:e3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham PM, Kidwell MG, and Rubin GM. 1982. The molecular basis of P-M hybrid dysgenesis: the role of the P element, a P-strain-specific transposon family. Cell 29:995–1004. [DOI] [PubMed] [Google Scholar]
- Blackman RK, Grimaila R, Macy M, Koehler D, and Gelbart WM. 1987. Mobilization of hobo elements residing within the decapentaplegic gene complex: suggestion of a new hybrid dysgenesis system in Drosophila melanogaster. Cell 49:497–505. [DOI] [PubMed] [Google Scholar]
- Blumenstiel JP, Erwin AA, and Hemmer LW. 2016. What drives positive selection in the Drosophila piRNA machinery? The genomic autoimmunity hypothesis. Yale J. Biol. Med 89:499–512. [PMC free article] [PubMed] [Google Scholar]
- Brand CL, Kingan SB, Wu L, and Garrigan D. 2013. A selective sweep across species boundaries in Drosophila. Mol. Biol. Evol 30:2177–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau NJ, Flores HA, Wang J, Maheshwari S, Wang X, and Barbash DA. 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314:1292–1295. [DOI] [PubMed] [Google Scholar]
- Brookfield JF, and Badge RM. 1997. Population genetics models of transposable elements. Genetica 100:281–294. [PubMed] [Google Scholar]
- Brookfield JFY, Montgomery E, and Langley CH. 1984. Apparent absence of transposable elements related to the P elements of D. melanogaster in other species of Drosophila. Nature 310:330–332. [DOI] [PubMed] [Google Scholar]
- Bucheton A, Paro R, Sang HM, Pelisson A, and Finnegan DJ, 1984. The molecular basis of IR hybrid dysgenesis in Drosophila melanogaster: identification, cloning, and properties of the I factor. Cell 38:153–163. [DOI] [PubMed] [Google Scholar]
- Carleton KL, Conte MA, Malinsky M, Nandamuri SP, Sandkam BA, Meier JI, Mwaiko S, Seehausen O, and Kocher TD, 2020. Movement of transposable elements contributes to cichlid diversity. Mol. Ecol 29, pp.4956–4969. [DOI] [PubMed] [Google Scholar]
- Castillo DM, and Moyle LC. 2012. Evolutionary implications of mechanistic models of TE-mediated hybrid incompatibility. Int. J. Evol. Biol 2012:698198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo DM and Moyle LC 2019. Hybrid incompatibility is stronger in the presence of transposable elements in the Drosophila virilis clade. bioRxiv 753814; 10.1101/753814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaboissier MC, Bornecque C, Busseau I, and Bucheton A, 1995. A genetically tagged, defective I element can be complemented by actively transposing I factors in the germaine of IR dysgenic females in Drosophila melanogaster. Mol. Gen. Genet 248:434–438. [DOI] [PubMed] [Google Scholar]
- Champely S, Ekstrom C, Dalgaard P, Gill J, Weibelzahl S, Anandkumar A, Ford C, Volcic R, De Rosario H, and De Rosario MH, 2018. Package ‘pwr’. R package version, 1(2). [Google Scholar]
- Clark JB, Altheide TK, Schlosser MJ, and Kidwell MG. 1995. Molecular evolution of P transposable elements in the genus Drosophila. I. The saltans and willistoni species groups. Mol. Biol. Evol 12:902–913. [DOI] [PubMed] [Google Scholar]
- Comeault AA, Serrato Capuchina A, Turissini DA, McLaughlin PJ, David JR, and Matute DR. 2017. A nonrandom subset of olfactory genes is associated with host preference in the fruit fly Drosophila orena. Evol. Lett 1:73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlan JM and Matute DR, 2020. The importance of intrinsic postzygotic barriers throughout the speciation process. Philos. Trans. R. Soc. Lond., B, Biol. Sci 375:20190533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels SB, Chovnick A, and Kidwell MG. 1989. Hybrid dysgenesis in Drosophila simulans lines transformed with autonomous P elements. Genetics 121:281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels SB, Peterson KR, Strausbaugh LD, Kidwell MG, and Chovnik A. 1990. Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics 124:339–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AW, Roote J, Morley T, and Sawamura K. 1996. Rescue of hybrid sterility in crosses between D. melanogaster and D. simulans. Nature 380:157–159. [DOI] [PubMed] [Google Scholar]
- De Setta N, Costa APP, Lopes FR, Van Sluys MA, and Carareto CMA. 2007. Transposon display supports transpositional activity of P elements in species of the saltans group of Drosophila. J. Genet 86:37–43. [DOI] [PubMed] [Google Scholar]
- Dennenmoser S, Sedlazeck FJ, Schatz MC, Altmüller J, Zytnicki M, and Nolte AW, 2019. Genome-wide patterns of transposon proliferation in an evolutionary young hybrid fish. Mol. Ecol 28:1491–1505. [DOI] [PubMed] [Google Scholar]
- Dion-Côté A-M, Renaut S, Normandeau E, and Bernatchez L. 2014. RNA-seq reveals transcriptomic shock involving transposable elements reactivation in hybrids of young lake whitefish species. Mol. Biol. Evol 31:1188–1199. [DOI] [PubMed] [Google Scholar]
- Dion-Côté AM, and Barbash DA. 2017. Beyond speciation genes: an overview of genome stability in evolution and speciation. Curr. Opin. Genet. Dev 47:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T 1937. Genetics and the origin of species. Columbia Univ. Press. New York. [Google Scholar]
- Dorai-Raj S 2015. Package “binom”: binomial confidence intervals for several parameterizations CRAN R Project. [Google Scholar]
- Engels WR, and Preston CR. 1980. Components of hybrid dysgenesis in a wild population of Drosophila melanogaster. Genetics 95:111–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett DH, Lister CK, Kellett E, and Finnegan DJ, 1986. Transposable elements controlling IR hybrid dysgenesis in D. melanogaster are similar to mammalian LINEs. Cell 47:1007–1015. [DOI] [PubMed] [Google Scholar]
- Fox J, and Sanford W. 2011. car: an R companion to applied regression R package. [Google Scholar]
- Fox J, and Weisberg S. 2019. An R companion to applied regression SAGE, Los Angeles, CA. [Google Scholar]
- Garrigan D, Kingan SB, Geneva AJ, Andolfatto P, Clark AG, Thornton KR, and Presgraves DC. 2012. Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res 22:1499–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginzburg LR, Bingham PM, and Yoo S. 1984. On the theory of speciation induced by transposable elements. Genetics 107:331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield JD, 2010. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J. Stat. Softw 33:1–22.20808728 [Google Scholar]
- Heger A, and Ponting CP. 2007. Evolutionary rate analyses of orthologs and paralogs from 12 Drosophila genomes. Genome Res 17:1837–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil CS, Patterson K, Hickey AS-M, Alcantara E, and Dunham MJ. 2021. Transposable element mobilization in interspecific yeast hybrids. Genome Biol. Evol 13:evab033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hénault M, Marsit S, Charron G and Landry CR, 2020. The effect of hybridization on transposable element accumulation in an undomesticated fungal species. Elife, 9, e60474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T, Schlotterer C, and Betancourt AJ. 2016. Hybrid dysgenesis in Drosophila simulans associated with a rapid invasion of the P-element. PLOS Genet 12:e1005920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn AT, Zeileis A, Millo G, and Mitchell D. 2011. Package ‘lmtest.’ R News
- Hothorn T, Bretz F, and Westfall P. 2008. Simultaneous inference in general parametric models. Biometrical J 50:346–363. [DOI] [PubMed] [Google Scholar]
- Ishikawa R, Ohnishi T, Kinoshita Y, Eiguchi M, Kurata N, and Kinoshita T. 2011. Rice interspecies hybrids show precocious or delayed developmental transitions in the endosperm without change to the rate of syncytial nuclear division. Plant J 65:798–806. [DOI] [PubMed] [Google Scholar]
- Kelleher ES 2016. Reexamining the P-element invasion of Drosophila melanogaster through the lens of piRNA silencing. Genetics 203:1513–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher ES, Edelman NB, and Barbash DA. 2012. Drosophila interspecific hybrids phenocopy piRNA-pathway mutants. PLoS Biol 10:e1001428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher ES, Barbash DA, and Blumenstiel JP, 2020. Taming the turmoil within: new insights on the containment of transposable elements. Trends Genet 36:474–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG 1983a. Evolution of hybrid dysgenesis determinants in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 80:1655–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1983b. Hybrid dysgenesis in Drosophila melanogaster: factors affecting chromosomal contamination in the P-M system. Genetics 104:317–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1985. Hybrid dysgenesis in Drosophila melanogaster: nature and inheritance of P element regulation. Genetics 111:337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG, and Kidwell JF. 1975. Cytoplasm-chromosome interactions in Drosophila melanogaster. Nature 253:755–756. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, and Novy JB. 1979. Hybrid dysgenesis in Drosophila melanogaster: sterility resulting from gonadal dysgenesis in the P-M system. Genetics 92:1127–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG, Kidwell JF, and Sved JA. 1977. Hybrid dysgenesis in Drosophila melanogaster: a syndrome of aberrant traits including mutation, sterility and male recombination. Genetics 86:813–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Hill T, Nolte V, Betancourt AJ, and Schlötterer C. 2015. The recent invasion of natural Drosophila simulans populations by the P-element. Proc. Natl. Acad. Sci. USA 112:6659–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowski B, Hupalo DN, and Kern AD. 2011. Recurrent adaptation in RNA interference genes across the Drosophila phylogeny. Mol. Biol. Evol 28:1033–1042. [DOI] [PubMed] [Google Scholar]
- Lachaise D, David JR, Lemeunier F, Tsacas L, and Ashburner M. 1986. The reproductive relationships of Drosophila sechellia with D. mauritiana, D. simulans, and D. melanogaster from the Afrotropical region. Evolution 40:262–271. [DOI] [PubMed] [Google Scholar]
- Ladevèze V, Aulard S, Chaminade N, Périquet G, and Lemeunier F. 1998. Hobo transposons causing chromosomal breakpoints. Proc. Biol. Sci 265:1157–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YCG, and Langley CH. 2012. Long-term and short-term evolutionary impacts of transposable elements on Drosophila. Genetics 192:1411–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H 2016. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32:2103–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Maestre H, Carnelossi EAG, Lacroix V, Burlet N, Mugat B, Chambeyron S, Carareto CMA, and Vieira C. 2017. Identification of misexpressed genetic elements in hybrids between Drosophila-related species. Sci. Rep 7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari S and Barbash DA, 2011. The genetics of hybrid incompatibilities. Annu. Rev. Genet 45:331–355. [DOI] [PubMed] [Google Scholar]
- Majumdar S, and Rio DC. 2015. P Transposable elements in Drosophila and other Eukaryotic organisms. Chapter 33. pp. 727–752 in Mobile DNA III, Edited by: Chandler M; Gellert M; Lambowitz AM; Rice PA; Sandmeyer SB Washington, DC: ASM Press. [Google Scholar]
- Matute DR, and Ayroles JF. 2014. Hybridization occurs between Drosophila simulans and D. sechellia in the Seychelles archipelago. J. Evol. Biol 27:1057–1068. [DOI] [PubMed] [Google Scholar]
- Matute DR, and Coyne JA. 2010. Intrinsic reproductive isolation between two sister species of Drosophila. Evolution 64:903–920. [DOI] [PubMed] [Google Scholar]
- Matute DR, Comeault AA, Earley E, Serrato-Capuchina A, Peede D, Monroy-Eklund A, Huang W, Jones CD, Mackay TFC, and Coyne JA. 2020. Rapid and predictable evolution of admixed populations between two Drosophila species pairs. Genetics 214:211–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B 1953. Induction of Instability at selected loci in maize. Genetics 38:579–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuillan MA, Roth TC, Huynh AV, and Rice AM. 2018. Hybrid chickadees are deficient in learning and memory. Evolution 72:1155–1164. [DOI] [PubMed] [Google Scholar]
- Meiklejohn CD, Landeen EL, Gordon KE, Rzatkiewicz T, Kingan SB, Geneva AJ, Vedanayagam JP, Muirhead CA, Garrigan D, Stern DL, et al. 2018. Gene flow mediates the role of sex chromosome meiotic drive during complex speciation. Elife 7:e35468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe CJ, Bulazel KV, Ferreri GC, Schroeder-Reiter E, Wanner G, Rens W, Obergfell C, Eldridge MDB, and O’Neill RJ. 2007. Genomic instability within centromeres of interspecific marsupial hybrids. Genetics 177:2507–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak P 2009. Epigenetic, transposon and small RNA determinants of hybrid dysfunctions. Heredity 102:45–50. [DOI] [PubMed] [Google Scholar]
- Mizrokhi LJ, and Mazo AM. 1990. Evidence for horizontal transmission of the mobile element jockey between distant Drosophila species. Proc. Natl. Acad. Sci. USA 87:9216–9220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle LC, Olson MS, and Tiffin P. 2004. Patterns of reproductive isolation in three angiosperm genera. Evolution 58:1195–1208. [DOI] [PubMed] [Google Scholar]
- Nolte AW, Renaut S, and Bernatchez L. 2009. Divergence in gene regulation at young life history stages of whitefish (Coregonus sp.) and the emergence of genomic isolation. BMC Evol. Biol 9:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P and Schluter D, 2011. The genes underlying the process of speciation. Trends Ecol. Evol 26:160–167. [DOI] [PubMed] [Google Scholar]
- Obbard DJ, Welch JJ, Kim KW, and Jiggins FM. 2009. Quantifying adaptive evolution in the Drosophila immune system. PLoS Genet 5:e1000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA 2005. The genetic basis of reproductive isolation: insights from Drosophila. Proc. Natl. Acad. Sci. USA 102:6522–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA, and Presgraves DC. 2000. Speciation by postzygotic isolation: forces, genes and molecules. BioEssays 22:1085–1094. [DOI] [PubMed] [Google Scholar]
- Orsi GA, Joyce EF, Couble P, McKim KS, and Loppin B. 2010. Drosophila I-R hybrid dysgenesis is associated with catastrophic meiosis and abnormal zygote formation. J. Cell Sci 123:3515–3524. [DOI] [PubMed] [Google Scholar]
- Parhad SS, Tu S, Weng Z, and Theurkauf WE, 2017. Adaptive evolution leads to cross-species incompatibility in the piRNA transposon silencing machinery. Dev. Cell 43:60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pélissier T, Tutois S, Deragon JM, Tourmente S, Genestier S, and Picard G. 1995. Athila, a new retroelement from Arabidopsis thaliana. Plant Mol. Biol 29:441–452. [DOI] [PubMed] [Google Scholar]
- Pélissier T, Tutois S, Tourmente S, Deragon JM, and Picard G. 1996. DNA regions flanking the major Arabidopsis thaliana satellite are principally enriched in Athila retroelement sequences. Genetica 97:141–151. [DOI] [PubMed] [Google Scholar]
- Picard G, Bregliano JC, Bucheton A, Lavige JM, Pelisson A, and Kidwell MG. 1978. Non-mendelian female sterility and hybrid dysgenesis in Drosophila melanogaster. Genet. Res 32:275–287. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2016. R: a language and environment for statistical computing R Foundation for Statistical Computing, Vienna. Available via https://www.R-project.org/. [Google Scholar]
- Ramsey J, Bradshaw HD, and Schemske DW. 2003. Components of reproductive isolation between the monkeyflowers Mimulus lewisii and M. cardinalis (Phrymaceae). Evolution 57:1520–1534. [DOI] [PubMed] [Google Scholar]
- Rogers RL, Cridland JM, Shao L, Hu TT, Andolfatto P, and Thornton KR. 2014. Landscape of standing variation for tandem duplications in Drosophila yakuba and Drosophila simulans. Mol. Biol. Evol 31:1750–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Soriano V, Modolo L, Lopez-Maestre H, Mugat B, Pessia E, Chambeyron S, Vieira C, and Garcia Guerreiro MP. 2017. Transposable element misregulation is linked to the divergence between parental piRNA pathways in Drosophila hybrids. Genome Biol. Evol 9:1450–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum EB, Sarver BAJ, Brown JW, Des Roches S, Hardwick KM, Hether TD, Eastman JM, Pennell MW, and Harmon LJ. 2012. Goldilocks meets Santa Rosalia: an ephemeral speciation model explains patterns of diversification across time scales. Evol. Biol 39:255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Kidwell MG, and Bingham PM. 1982. The molecular basis of P-M hybrid dysgenesis: the nature of induced mutations. Cell 29:987–994. [DOI] [PubMed] [Google Scholar]
- Saito K 2013. The epigenetic regulation of transposable elements by PIWI-interacting RNAs in Drosophila. Genes Genet. Syst 88:9–17. [DOI] [PubMed] [Google Scholar]
- Sambrook J, and Russell DW. 2006. Purification of RNA from cells and tissues by acid phenol-guanidinium thiocyanate-chloroform extraction. Cold Spring Harb. Protoc 10.1101/pdb.prot4045. [DOI] [PubMed] [Google Scholar]
- Sasa MM, Chippindale PT, and Johnson NA. 1998. Patterns of postzygotic isolation in frogs. Evolution 52:1811–1820. [DOI] [PubMed] [Google Scholar]
- Schaefer RE, Kidwell MG, and Fausto-Sterling A. 1979. Hybrid dysgenesis in Drosophila melanogaster: morphological and cytological studies of ovarian dysgenesis. Genetics 92:1141–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Ayroles J, Matute DR, and Kern AD. 2018. Supervised machine learning reveals introgressed loci in the genomes of Drosophila simulans and D. sechellia. PLOS Genet 14:e1007341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrato-Capuchina A, and Matute D. 2018. The role of transposable elements in speciation. Genes 9:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrato-Capuchina A, Wang J, Earley E, Peede D, Isbell K, and Matute DR. 2020. Paternally inherited P-element copy number affects the magnitude of hybrid dysgenesis in Drosophila simulans and D. melanogaster. Genome Biol. Evol 12:808–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X, Liu Z, Dong Z, Wang Y, Chen Y, Lin X, Long L, Han F, Dong Y, and Liu B. 2005. Mobilization of the active MITE transposons mPing and Pong in rice by introgression from wild rice (Zizania latifolia Griseb.). Mol. Biol. Evol 22:976–990. [DOI] [PubMed] [Google Scholar]
- Silva JC, and Kidwell MG. 2000. Horizontal transfer and selection in the evolution of P elements. Mol. Biol. Evol 17:1542–1557. [DOI] [PubMed] [Google Scholar]
- Simkin A, Wong A, Poh YP, Theurkauf WE, and Jensen JD. 2013. Recurrent and recent selective sweeps in the piRNA pathway. Evolution 67:1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, and Aravin AA. 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol 12:246–258. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, and Martienssen R. 2007. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet 8:272–285. [DOI] [PubMed] [Google Scholar]
- Sobel JM, Chen GF, Watt LR, and Schemske DW. 2010. The biology of speciation. Evolution 64:295–315. [DOI] [PubMed] [Google Scholar]
- Srivastav SP, and Kelleher ES. 2017. Paternal induction of hybrid dysgenesis in Drosophila melanogaster is weakly correlated with both P-element and hobo element dosage. G3 7:1487–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastav SP, Rahman R, Ma Q, and Lau NC. 2019. Har-P, a short P-element variant, weaponizes P-transposase to severely impact Drosophila gonad development. eLife 8:e49948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trizzino M, Park YS, Holsbach-Beltrame M, Aracena K, Mika K, Caliskan M, Perry GH, Lynch VJ, and Brown CD. 2017. Transposable elements are the primary source of novelty in primate gene regulation. Genome Res 27:1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True JR, and Haag ES. 2001. Developmental system drift and flexibility in evolutionary trajectories. Evol. Dev 3:109–119. [DOI] [PubMed] [Google Scholar]
- Turissini DA, Comeault AA, Liu G, Lee YCG, and Matute DR. 2017. The ability of Drosophila hybrids to locate food declines with parental divergence. Evolution 71:960–973. [DOI] [PubMed] [Google Scholar]
- Turissini DA, McGirr JA, Patel SS, David JR, and Matute DR. 2018. The rate of evolution of postmating-prezygotic reproductive isolation in Drosophila. Mol. Biol. Evol 35:312–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer MC, Strakosh SC, and Zhen Y. 2006. Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. Curr. Biol 16:R872–R873. [DOI] [PubMed] [Google Scholar]
- Vaser R, Sović I, Nagarajan N, and Šikić M. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 27:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vela D, Fontdevila A, Vieira C, and Pilar García Guerreiro M. 2014. A genome-wide survey of genetic instability by transposition in Drosophila hybrids. PLoS ONE 9:e88992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables WN, Ripley BD, and Venables WN. 2003. Modern applied statistics with S. Technometrics 45:111–111. [Google Scholar]
- Vermaak D, Henikoff S, and Malik HS. 2005. Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila. PLoS Genet 1:96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Barbash DA, and Kelleher ES. 2020. Adaptive evolution among cytoplasmic piRNA proteins leads to decreased genomic autoimmunity. PLoS Genet 16: p. e1008861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes GR, Bolker B, Bonebakker L, Gentleman R, Liaw WHA, Lumley T, Maechler M, Magnusson A, Moeller S, Schwartz M, et al. 2016. Package “gplots”: various R programming tools for plotting data. R Package version 2.17.0 [Google Scholar]
- Watson ET, and Demuth JP. 2013. The role of transposable elements in speciation. Pp. 227–250 in Michalak P, ed. Speciation: natural processes, genetics and biodiversity Nova Science Publishers, Inc, Hauppauge, NY. [Google Scholar]
- Werren JH 2011. Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc. Natl. Acad. Sci. USA 108:10863–10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LC, and Schedl P. 2006. Dissection of Drosophila ovaries. J. Vis. Exp 8:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannopoulos G, Stamatis N, Monastirioti M, Hatzopoulos P, and Louis C, 1987. hobo is responsible for the induction of hybrid dysgenesis by strains of Drosophila melanogaster bearing the male recombination factor 23.5 MRF. Cell 49, pp.487–495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


