Abstract
Immune responses are triggered when pattern recognition receptors recognize microbial molecular patterns. The Arabidopsis (Arabidopsis thaliana) receptor-like cytoplasmic kinase BOTRYTIS-INDUCED KINASE1 (BIK1) acts as a signaling hub of plant immunity. BIK1 homeostasis is maintained by a regulatory module in which CALCIUM-DEPENDENT PROTEIN KINASE28 (CPK28) regulates BIK1 turnover via the activities of two E3 ligases. Immune-induced alternative splicing of CPK28 attenuates CPK28 function. However, it remained unknown whether CPK28 is under proteasomal control. Here, we demonstrate that CPK28 undergoes ubiquitination and 26S proteasome-mediated degradation, which is enhanced by flagellin treatment. Two closely related ubiquitin ligases, ARABIDOPSIS TÓXICOS EN LEVADURA31 (ATL31) and ATL6, specifically interact with CPK28 at the plasma membrane; this association is enhanced by flagellin elicitation. ATL31/6 directly ubiquitinate CPK28, resulting in its proteasomal degradation. Furthermore, ATL31/6 promotes the stability of BIK1 by mediating CPK28 degradation. Consequently, ATL31/6 positively regulate BIK1-mediated immunity. Our findings reveal another mechanism for attenuating CPK28 function to maintain BIK1 homeostasis and enhance immune responses.
The negative immune regulator CPK28 is subjected to ubiquitination and proteasomal degradation to positively regulate immune signaling.
Introduction
Plants have a sophisticated innate immune system to defend against pathogen invasion. Microbe-associated molecular patterns or pathogen-associated molecular patterns (PAMPs) trigger immune responses in plants, a process known as pattern-triggered immunity (PTI). PTI confers the host defense against a wide range of phytopathogens (Jones and Dangl, 2006). In Arabidopsis (Arabidopsis thaliana), the bacterial flagellin or its derived peptide flg22 is recognized by FLAGELLIN-SENSING2 (FLS2), a well-studied plasma membrane-localized leucine-rich repeat receptor kinase (LRR-RK; Gomez-Gomez and Boller, 2000). BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1) serves as a co-receptor for FLS2 and other pattern recognition receptors (PRRs), such as EF-TU RECEPTOR (EFR), which perceives the bacterial elongation factor EF-Tu and the derived epitope elf18. Upon ligand binding, FLS2 and EFR rapidly interact with BAK1 and the related SOMATIC-EMBRYOGENESIS RECEPTOR-LIKE KINASE proteins to activate downstream immune signaling (Zipfel et al., 2006; Chinchilla et al., 2007; Heese et al., 2007; Roux et al., 2011). Additionally, PRRs sense endogenous damage-associated molecular patterns that are released upon wounding or pathogen perception (Zipfel, 2014). Arabidopsis PLANT ELICITOR PEPTIDEs (Peps) are perceived by two LRR-RKs, PEP RECEPTOR1 (PEPR1) and PEPR2, triggering immune responses (Yamaguchi et al., 2006, 2010; Krol et al., 2010).
BOTRYTIS-INDUCED KINASE 1 (BIK1) is a receptor-like cytoplasmic kinase (RLCK) that associates with multiple PRRs and functions as a signaling hub immediately downstream of these PRRs. BIK1 undergoes hyperphosphorylation upon PAMP perception in a BAK1-depdendent manner (Lu et al., 2010; Zhang et al., 2010). To date, various substrates have been identified for BIK1. BIK1 directly phosphorylates a plasma membrane-resident NADPH oxidase, RESPIRATORY BURST OXIDASE HOMOLOG D (RbohD), resulting in reactive oxygen species (ROS) burst and stomatal immunity (Kadota et al., 2014; Li et al., 2014). In addition, BIK1 phosphorylates the Ca2+-permeable channel OSCA1.3 within minutes of flg22 treatment and enhances its channel activity, which is critical for PAMP-induced stomatal closure during immunity (Thor et al., 2020). Upon pathogen attack, BIK1 also phosphorylates and activates two CYCLIC NUCLEOTIDE-GATED CHANNEL (CNGC) proteins, CNGC2 and CNGC4, which triggers calcium entry into the cytosol (Tian et al., 2019).
Immune signaling is finely tuned to ensure its appropriate magnitude and duration. BAK1-INTERACTING RECEPTOR-LIKE KINASE2 (BIR2) and BIR3 block the association of FLS2 with BAK1 in the resting state, whereas they are released from BAK1 after flg22 perception, facilitating the interaction between BAK1 and FLS2 (Halter et al., 2014; Imkampe et al., 2017). Protein Ser/Thr phosphatase 2A (PP2A) negatively regulates immunity by dephosphorylating BAK1 (Segonzac et al., 2014). Some endogenous peptides, such as RAPID ALKALINIZATION FACTOR23 (RALF23) and RALF33, are rapidly released and bind to their receptor FERONIA (FER) to inhibit immunity in Arabidopsis (Stegmann et al., 2017). Another Arabidopsis RLCK, AvrPphB SUSCEPTIBLE1-LIKE13 (PBL13), phosphorylates the C terminus of RbohD to affect its activity and stability, thereby negatively regulating immunity (Lin et al., 2015; Lee et al., 2020). The transcription factors TARGET OF EAT1 (TOE1) and TOE2 directly bind to the FLS2 promoter and inhibit its activity to suppress plant immunity during the early stages of seedling development (Zou et al., 2018, 2020).
Protein ubiquitination is a widespread type of post-translational modification that is involved in the regulation of signal transduction. In the presence of ATP, ubiquitin is activated and transferred to a specific target protein in a stepwise manner by three types of enzymes: ubiquitin activating enzyme (UBA, E1), ubiquitin conjugating enzyme (UBC, E2), and ubiquitin ligase (E3). During ubiquitination, E3 ubiquitin ligases play a critical role in determining the substrate specificity. A major consequence of ubiquitination is the targeting of the substrate proteins to the 26S proteasome for degradation (Callis, 2014). Ubiquitination plays an important role in regulating immune signaling. FLS2 is ubiquitinated by two ubiquitin ligases PLANT U-BOX12 (PUB12) and PUB13, which leads to the attenuation of immune signaling (Lu et al., 2011). The ubiquitination of RbohD by PBL13 INTERACTING RING DOMAIN E3 LIGASE is further enhanced upon RbohD phosphorylation catalyzed by PBL13, which results in the degradation of RbohD and the negative regulation of immune signaling (Lee et al., 2020). PUB22 ubiquitinates EXOCYST SUBUNIT 70 PROTEIN B2 (Exo70B2), a subunit of the exocyst complex required for activation of immune responses, leading to the degradation of Exo70B2 and the attenuation of immune signaling (Stegmann et al., 2012; Furlan et al., 2017).
The immune signaling hub BIK1 is subjected to tight regulation. BIK1 homeostasis is maintained by a regulatory module that consists of CALCIUM-DEPENDENT PROTEIN KINASE28 (CPK28), two closely related ubiquitin ligases (PUB25 and PUB26), and heterotrimeric G proteins. In this module, PUB25/26 ubiquitinate BIK1, leading to its degradation. The ligase activity of PUB25 is enhanced when it is phosphorylated at Thr95 by CPK28, which promotes the polyubiquitination and degradation of BIK1 (Monaghan et al., 2014; Wang et al., 2018).
It was recently reported that a retained intron (RI) variant of CPK28, known as CPK28-RI, is induced after treatment with flg22 (Bazin et al., 2020) or AtPeps (Dressano et al., 2020). CPK28-RI encodes a truncated CPK28 isoform that exhibits impaired kinase activity (Dressano et al., 2020). The immune-induced alternative splicing of the CPK28 transcripts plays a regulatory role in dynamically amplifying immune signaling (Bazin et al., 2020; Dressano et al., 2020). Furthermore, a more recent proteomics study showed that CPK28 is ubiquitinated at multiple Lys residues (Grubb et al., 2021). However, the underlying mechanisms of CPK28 ubiquitination and whether CPK28 undergoes proteasomal degradation to maintain immune homeostasis remained unknown.
Here, we demonstrate that CPK28 undergoes ubiquitination and 26S proteasome-mediated degradation that are enhanced by flg22 stimulation. Two closely related ubiquitin ligases, ARABIDOPSIS TÓXICOS EN LEVADURA 31 (ATL31) and ATL6, interact with CPK28 in an flg22-enhanced manner. Furthermore, ATL31 and ATL6 directly ubiquitinate CPK28, resulting in the proteasomal degradation of CPK28. As a result, ATL31 and ATL6 positively regulate BIK1 stability and immune responses. Our work reveals how CPK28 is targeted by ATL31/6 for proteasome-mediated degradation to maintain BIK1 homeostasis and dynamically regulate immune signaling.
Results
CPK28 undergoes ubiquitination and 26S proteasome-mediated degradation
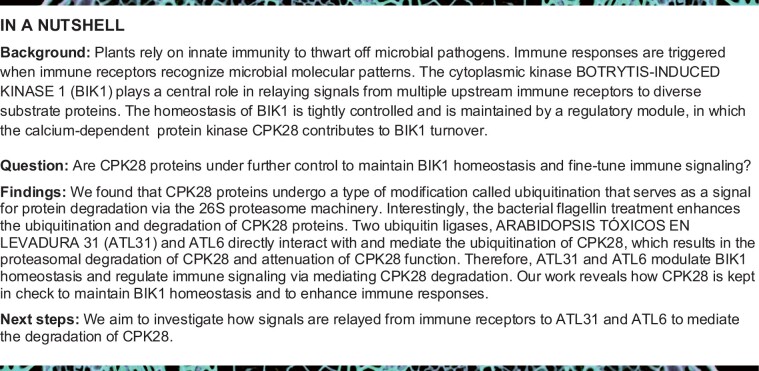
CPK28 is a key component of the recently characterized immune regulatory module that modulates BIK1 stability (Monaghan et al., 2014; Wang et al., 2018). To examine whether CPK28 is under proteolytic control to keep this regulatory module in check, we examined its ubiquitination and proteasomal degradation. We performed in vivo ubiquitination assays by transiently expressing ubiquitin tagged with the FLAG epitope (FLAG-Ub) and CPK28 tagged with hemagglutinin (CPK28-HA) in Arabidopsis protoplasts; a similar approach was previously used to examine the ubiquitination of BIK1 (Ma et al., 2020) and FLS2 (Lu et al., 2011) in vivo. Following immunoprecipitation (IP) with anti-FLAG antibodies, the ubiquitinated CPK28 proteins were detected by immunoblotting with anti-HA antibodies. CPK28 exhibited ladder-like smears, which are indicative of protein ubiquitination (Figure 1A). Notably, a recent proteomics study identified multiple ubiquitination sites on CPK28 proteins (Grubb et al., 2021), suggesting that CPK28 indeed undergoes ubiquitination. Furthermore, we found that the ubiquitination of CPK28 was enhanced by flg22 treatment (Figure 1A).
Figure 1.
CPK28 undergoes ubiquitination and 26S proteasome-mediated degradation. A, flg22 treatment enhances CPK28 ubiquitination in vivo. FLAG-Ub and CPK28-HA were co-expressed in Arabidopsis protoplasts. In the presence of 20-μM MG132, the protoplasts were treated with 2-μM flg22 for 30 min before harvesting. Total ubiquitinated proteins were immunoprecipitated with anti-FLAG antibodies, and ubiquitinated CPK28 proteins were detected by immunoblotting with anti-HA antibodies. B, CPK28 undergoes proteasomal degradation. Seven-day-old Col-0 seedlings (10 seedlings per sample) were treated with 100-μM CHX for the indicated times in the presence or absence of 50-μM MG132. Total proteins were isolated from whole seedlings. CPK28 proteins were detected with anti-CPK28 antibodies. ACTIN was used as a loading control. C, flg22 treatment enhances the proteasomal degradation of CPK28. Seven-day-old Col-0 seedlings (10 seedlings per sample) were treated with 100-μM CHX in the presence or absence of 50-μM MG132, followed 1 h later by treatment with 2-μM flg22 for the indicated times. Total proteins were isolated from whole seedlings. CPK28 proteins were detected with anti-CPK28 antibodies. ACTIN was used as a loading control.
In addition to CPK28, other negative immune regulators, such as BIR2 and BIR3, also undergo ubiquitination (Supplemental Figure S1, A and B); consistently, ubiquitination sites were also identified on both BIR2 and BIR3 by the recent proteomics study (Grubb et al., 2021). However, ubiquitination of PBL13 and RCN1 (PP2A subunit A1) was not observed under the conditions we used (Supplemental Figure S1, C and D), demonstrating the specificity of this in vivo ubiquitination system.
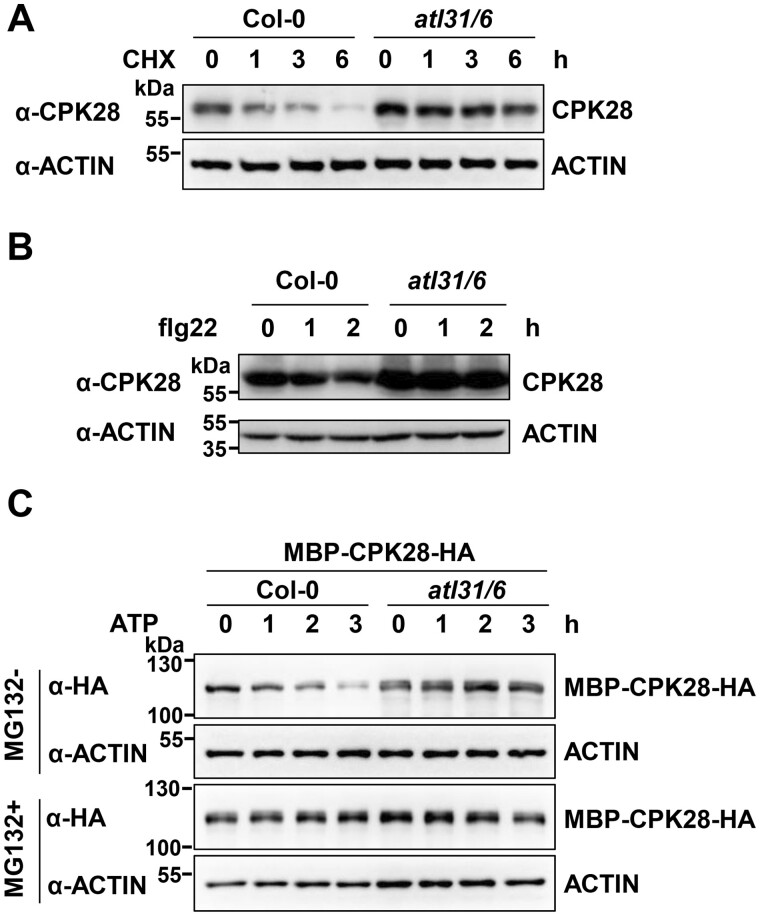
We then examined the turnover of CPK28 protein. We raised antibodies that specifically detected CPK28 in Col-0 but not in cpk28 mutants (Monaghan et al., 2014; Supplemental Figure S2, A–E). The anti-CPK28 antibodies specifically recognized recombinant CPK28 proteins, but not other tested CPKs, such as CPK8, CPK6, CPK1, CPK3, and CPK16 that is closely related to CPK28 (Boudsocq and Sheen, 2013; Supplemental Figure S2F). Importantly, the anti-CPK28 antibodies did not recognize recombinant CPK28-RI proteins (Supplemental Figure S2G). CPK28 protein accumulation increased in Col-0 seedlings treated with the 26S proteasome inhibitor MG132, as measured by immunoblotting with anti-CPK28 antibodies (Supplemental Figure S3A). Moreover, when Col-0 seedlings were treated with cycloheximide (CHX) to block protein synthesis, CPK28 protein accumulation declined over time. However, the decrease in CPK28 protein level was largely blocked by MG132 treatment (Figure 1B), suggesting that CPK28 proteins are under proteasomal control. Notably, in the presence of CHX, we observed a significant reduction in CPK28 protein levels within 2 h after flg22 treatment. This degradation was inhibited by MG132 treatment (Figure 1C), indicating that flg22 treatment enhances the proteasomal degradation of CPK28. Moreover, like CPK28, both BIR2 and BIR3 also undergo proteasomal degradation, as assayed using proteins transiently expressed in Arabidopsis protoplasts (Supplemental Figure S3, B and C).
Identification of ubiquitin ligases that mediate CPK28 degradation
We then sought to identify E3 ubiquitin ligases that mediate the ubiquitination of CPK28. Considering that the interaction between ubiquitin ligase and its target may result in degradation of the target protein, instead of directly screening for CPK28 interactors, we used an alternative approach to identify the cognate ubiquitin ligases of CPK28. We performed RNA sequencing (RNA-seq) to identify ubiquitin ligase genes that were differentially expressed upon flg22 treatment using RNA isolated from Arabidopsis seedlings treated with flg22 or water (Supplemental Figure S4A). Forty-nine ubiquitin ligase genes were upregulated upon flg22 treatment, and 22 were downregulated (Supplemental Figure S4B and Supplemental Table S1), and their expression was confirmed by reverse transcription- quantitative polymerase chain reaction (RT-qPCR; Supplemental Figure S4C).
To examine which ubiquitin ligases could mediate the degradation of CPK28, we performed a leaf protoplast cell-based screen by co-expressing CPK28 together with a selected ubiquitin ligase gene. To validate the feasibility of this screening strategy, we co-expressed PUB25-HA together with BIK1-FLAG in protoplasts, using a green fluorescent protein (GFP)-FLAG fusion as a transfection control. Consistent with a previous report (Wang et al., 2018), PUB25 overexpression led to the reduced accumulation of BIK1 in this transient system (Supplemental Figure S5). As flg22 enhanced the ubiquitination and proteasomal degradation of CPK28, we screened the flg22-induced ubiquitin ligase genes. We screened 16 of the ubiquitin ligase genes described above and found that the expression of ATL31-HA significantly reduced CPK28-MYC protein abundance (Supplemental Figure S6).
ATL31 is a RING-H2 ubiquitin ligase with a well-characterized role in plant carbon/nitrogen (C/N)-nutrient responses (Sato et al., 2009, 2011; Yasuda et al., 2017). ATL31 contains a transmembrane domain at its N terminus. ATL31-GFP proteins were mainly localized to the cell periphery when introduced into onion (Allium cepa) epidermal cells (Sato et al., 2009), and ATL31-GFP co-localized with the integral plasma membrane protein BRASSINOSTEROID INSNESITIVE1 (BRI1) fused with red fluorescent protein (RFP; Wang et al., 2001) at the plasma membrane, when these two proteins were co-expressed in Arabidopsis protoplasts (Supplemental Figure S7), suggesting that ATL31 is localized to the plasma membrane in this expression system.
We also analyzed the expression patterns of ATL31 and CPK28 in response to flg22 treatment. CPK28 has a canonically spliced transcript and the CPK28-RI splice variant (Dressano et al., 2020; Bazin et al., 2020). The proportion of CPK28-RI transcript levels but not those of CPK28 significantly increased after AtPep1 treatment (Dressano et al., 2020). We found that when the seedlings were treated with flg22, CPK28 transcript levels started to increase after 15 min of treatment, peaked at 30 min (∼16-fold), and returned to the initial level after 120 min (Supplemental Figure S8, A and D). In contrast, CPK28-RI transcript levels increased to a peak after 15 min of flg22 treatment (∼49-fold), and then decreased to the initial level after 60 min (Supplemental Figure S8, B and D). These results demonstrate that induction of CPK28 by flg22 was weaker and slower than that of CPK28-RI. Moreover, the expression pattern of ATL31 after flg22 treatment was more similar to that of CPK28 than to that of CPK28-RI (Supplemental Figure S8, A–C).
ATL31/6 interact with CPK28 at the plasma membrane
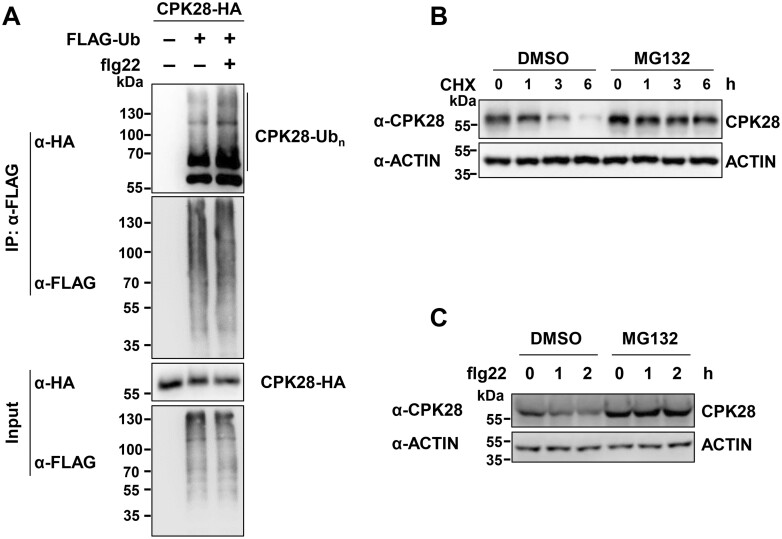
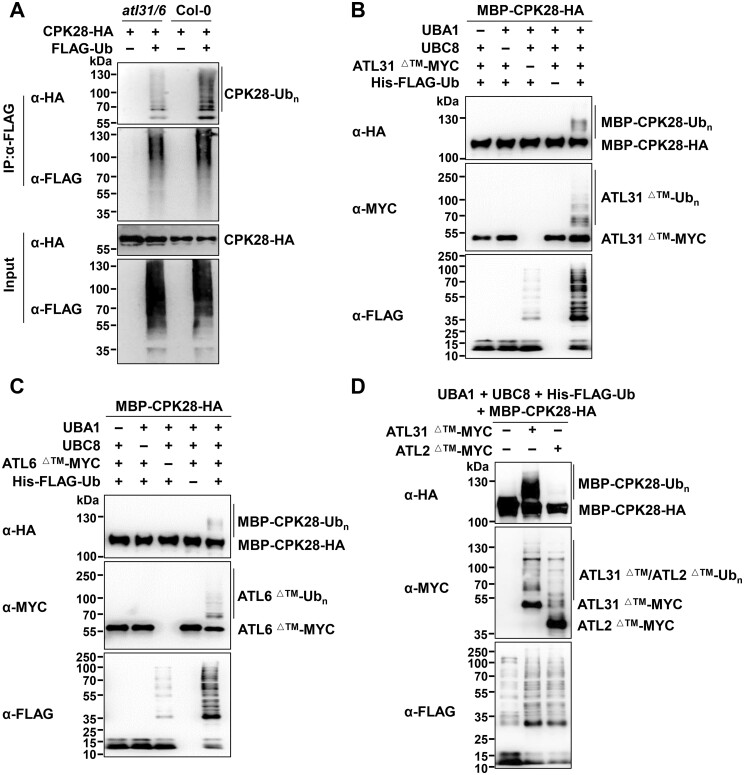
To examine whether ATL31 interacts with CPK28, we performed co-IP assays in which we co-expressed CPK28-HA and ATL31C143H145A-FLAG in Arabidopsis protoplasts. ATL31C143H145A is an ATL31 variant in which both Cys-143 (Sato et al., 2009) and His-145 in the RING domain were replaced by Ala to impair its activity and decrease the potential target degradation. CPK28-HA associated with ATL31C143H145A-FLAG in this assay (Figure 2A). Similarly, ATL31C143H145A-HA was also present in the CPK28-FLAG immunoprecipitates (Figure 2B). Importantly, we observed that flg22 treatment enhanced the association of ATL31C143H145A and CPK28 (Figure 2, A and B). Moreover, the association of CPK28 with ATL31C143H145A was much stronger than its association with wild-type (WT) ATL31 (Figure 2C), perhaps mainly due to the degradation of CPK28 mediated by ATL31. To examine whether the association of ATL31 and CPK28 is dependent on the FLS2-BAK1 receptor complex, we performed co-IP using protoplasts isolated from fls2 and bak1. ATL31 still associated with CPK28 in both fls2 and bak1 plants. However, the flg22-enhanced association between ATL31 and CPK28 was no longer observed in the fls2 or bak1 mutant (Figure 2, D and E). These results indicate that the association of ATL31 and CPK28 occurs before and after flg22 treatment but is further enhanced upon the recognition of flg22 by the FLS2–BAK1 complex.
Figure 2.
Both ATL31 and ATL6 associate with CPK28. A, flg22 treatment enhances the association of the ATL31C143H145A-FLAG variant and CPK28-HA. CPK28-HA and ATL31C143H145A-FLAG were co-expressed in Arabidopsis protoplasts, and the protoplasts were treated with 2-μM flg22 for 30 min before harvesting. IPs were performed using anti-FLAG antibodies. The immunoprecipitated ATL31C143H145A-FLAG proteins were detected by immunoblotting with anti-FLAG antibodies, and the associated proteins were detected by immunoblotting with anti-HA antibodies (top two parts). The levels of ATL31C143H145A-FLAG and CPK28-HA are shown in the bottom two parts. B, flg22 treatment enhances the association of the ATL31C143H145A-HA variant and CPK28-FLAG. CPK28-FLAG and ATL31C143H145A-HA were co-expressed in Arabidopsis protoplasts, and the protoplasts were treated with 2-μM flg22 for 30 min before harvesting. C, The association of CPK28 with the ATL31C143H145A variant was much stronger than that with WT ATL31. WT ATL31-FLAG or the ATL31C143H145A-FLAG variant was co-expressed with CPK28-HA in Arabidopsis protoplasts. IPs were performed with anti-FLAG antibodies. In the top part, equal amounts of immunoprecipitates were loaded (1:1:1); in the second part, the ATL31C143H145A-FLAG immunoprecipitate and control sample were diluted 10-fold (0.1:1:0.1). The associated CPK28-HA proteins were detected by immunoblotting with anti-HA antibodies. D, E, The association of ATL31 and CPK28 in the fls2 and bak1 mutants. ATL31-FLAG was co-expressed with CPK28-HA in protoplasts isolated from Col-0, fls2, or bak1-4 plants. F, flg22 treatment enhances the association of CPK28 and ATL6. ATL6-FLAG was co-expressed with CPK28-HA in protoplasts, and the protoplasts were treated with 2-μM flg22 for 30 min before harvesting. G, CPK28 does not associate with ATL2. ATL2-FLAG was co-expressed with CPK28-HA in protoplasts. D–G, IPs were performed using anti-FLAG antibodies, and the associated proteins were detected by immunoblotting with anti-HA antibodies.
ATL6 has the highest level of amino acid sequence similarity with ATL31 (Sato et al., 2009). CPK28 also associated with ATL6, but not with ATL2 (Figure 2, F and G), another ATL family member that was previously shown to be involved in plant defense responses (Serrano and Guzman, 2004). Moreover, flg22 treatment enhanced the association of CPK28 and ATL6 (Figure 2F). Finally, ATL31 was not associated with CPK8, another CPK family member (Supplemental Figure S9). These results demonstrate the specificity of the association between ATL31/6 and CPK28.
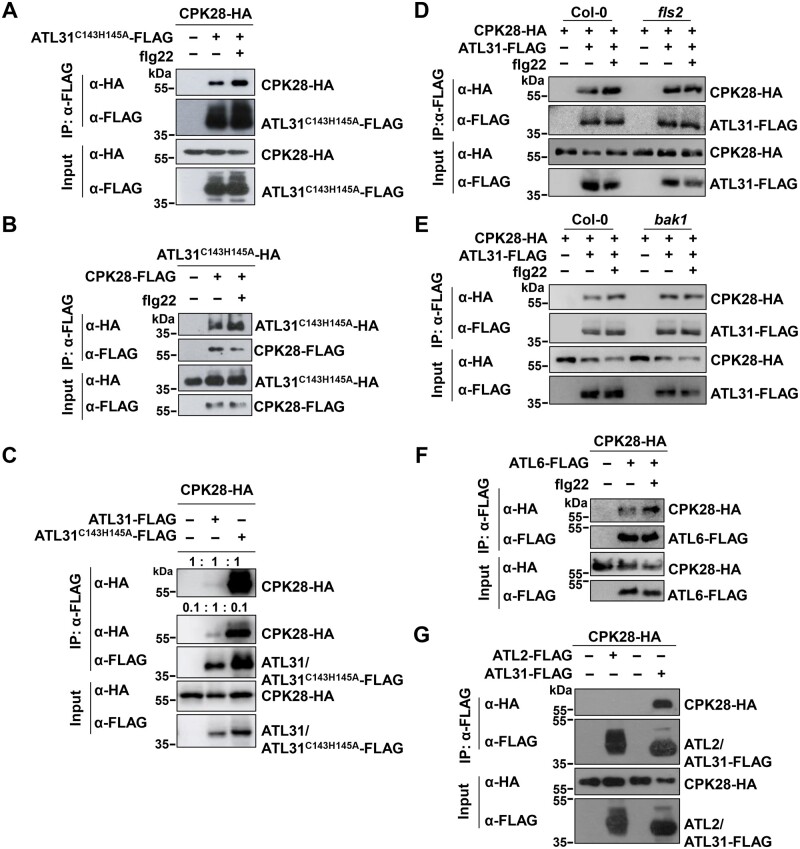
CPK28 associates with the plasma membrane via its myristoylation (Monaghan et al., 2014). Therefore, the CPK28G2A variant containing a mutation in its myristoylation site loses its plasma membrane localization when transiently expressed in Nicotiana benthamiana (Monaghan et al., 2014). Consistently, the loss of plasma membrane localization of CPK28G2A was also observed in Arabidopsis protoplasts (Supplemental Figure S10). Accordingly, a co-IP assay showed that ATL31 was no longer associated with CPK28G2A (Figure 3A). We also performed bimolecular fluorescence complementation (BiFC) assays in Arabidopsis protoplasts using ATL31C143H145A tagged with the N-terminal half of yellow fluorescent protein (nYFP) tag and CPK28 tagged with the C-terminal half of YFP (cYFP) tag. ATL31C143H145A associated with CPK28 at the plasma membrane (Figure 3B). In line with a previous report (Huang et al., 2019), the plasma membrane-associated protein PCRK1 (pattern-triggered immunity compromised RLCK1; Kong et al., 2016) associated with the plasma membrane-resident protein BRI1 (Wang et al., 2001; Figure 3B). However, ATL31C143H145A did not associate with PCRK1, and CPK28 did not associate with BRI1. Consistently, the CPK28 mutant CPK28G2A did not associate with ATL31C143H145A in the BiFC assays (Figure 3B). These results further confirm the specificity of the association between ATL31 and CPK28 at the plasma membrane.
Figure 3.
ATL31 interacts with CPK28 at the plasma membrane. A, ATL31 does not associate with CPK28G2A. CPK28G2A-HA and ATL31-FLAG were co-expressed in protoplasts. IPs were performed using anti-FLAG antibodies, and the associated proteins were detected by immunoblotting with anti-HA antibodies. B, ATL31C143H145A associates with CPK28 at the plasma membrane. The indicated BiFC constructs were transfected into Arabidopsis protoplasts, and fluorescence was visualized by confocal microscopy. Scale bars, 10 μm. C, ATL31/6 directly interact with CPK28 in vitro. Recombinant MBP-HA, MBP-ATL31△TM-HA, or MBP-ATL6△TM -HA proteins immobilized on amylose resin were incubated with His-FLAG-CPK28 proteins, and MBP PD assays were performed. △TM, lacking the transmembrane domain. Washed resin was subjected to immunoblotting with anti-HA and anti-FLAG antibodies to detect the immobilized proteins and the pulled-down proteins, respectively (top two parts). Input proteins were detected by immunoblotting with the indicated antibodies (bottom two parts).
Furthermore, CPK28 was pulled down by the intracellular region of ATL31 (ATL31△TM, amino acids 67–368, lacking the transmembrane domain) and by ATL6 (ATL6△TM, amino acids 71–368) fused to a maltose-binding protein (MBP) tag in MBP pull-down (PD) assays (Figure 3C). These results suggest that ATL31/6 directly interact with CPK28.
ATL31/6 ubiquitinate CPK28
ATL31/6 were shown to have ubiquitin ligase activity (Sato et al., 2009; Maekawa et al., 2012). To investigate their roles in CPK28 ubiquitination, we isolated the ATL31 and ATL6 null mutants, atl31 and atl6 (Sato et al., 2009, 2011), and generated the homozygous atl31 atl6 double mutant (Sato et al., 2009, 2011; Supplemental Figure S11, A–F). atl31 atl6 was slightly smaller than Col-0, but was morphologically very similar to Col-0 during vegetative growth and after the transition from the vegetative to the reproductive stage (Supplemental Figure S11G). CPK28-HA and FLAG-Ub were co-expressed in protoplasts isolated from atl31 atl6 or Col-0 plants, and the ubiquitination of CPK28 was weaker in atl31 atl6 than in Col-0 plants (Figure 4A). Notably, CPK28 still underwent ubiquitination even in atl31 atl6 plants, suggesting that in addition to ATL31/6, there are likely other unidentified ubiquitin ligases involved in CPK28 ubiquitination (Figure 4A).
Figure 4.
ATL31/6 ubiquitinate CPK28. A, CPK28 ubiquitination is weaker in atl31 atl6 than in Col-0 plants. CPK28-HA and FLAG-Ub were co-expressed in protoplasts isolated from atl31 atl6 or Col-0 plants. Following IP with anti-FLAG antibodies, the ubiquitination of CPK28 was detected by immunoblotting with anti-HA antibodies. B–D, ATL31/6 but not ATL2 directly ubiquitinate CPK28. The combination of ATL31△TM/ATL6△TM/ATL2△TM-MYC, AtUBA1 (E1), AtUBC8 (E2), His-FLAG-Ub, and MBP-CPK28-HA, or similar combinations lacking one component, were co-expressed in E. coli. The bacterial lysates were subjected to immunoblotting analysis with anti-HA antibodies to detect CPK28 ubiquitination, with anti-MYC antibodies to detect ATL31△TM/ATL6△TM/ATL2△TM autoubiquitination, or with anti-FLAG antibodies to detect ubiquitin conjugates and free ubiquitin chains.
To further confirm the ubiquitination of CPK28 by ATL31/6, we transfected Col-0 protoplasts with FLAG-Ub, CPK28-HA, and ATL31-GFP or ATL31C143H145A-GFP and then performed IP with anti-FLAG antibodies. CPK28 was ubiquitinated, and ATL31 overexpression enhanced its ubiquitination (Supplemental Figure S12). Additionally, WT ATL31 underwent substantial autoubiquitination, whereas the ATL31C143H145A variant exhibited much weaker autoubiquitination, suggesting that the ubiquitin ligase activity of ATL31C143H145A is markedly weakened. Accordingly, ATL31C143H145A was less effective than WT ATL31 at promoting CPK28 ubiquitination (Supplemental Figure S12).
To determine whether ATL31/6 directly ubiquitinate CPK28, we used a bacterial ubiquitination system that was previously developed in our laboratory (Han et al., 2017). ATL31/6-MYC, AtUBA1 (E1), AtUBC8 (E2), His- and FLAG-tagged ubiquitin (His-FLAG-Ub), and MBP-CPK28-HA were co-expressed in Escherichia coli BL21 strain. ATL31 and ATL6 exhibited autoubiquitination activity in this system, as detected with anti-MYC or anti-FLAG antibodies (Figure 4, B and C), which is consistent with previous reports (Sato et al., 2009; Maekawa et al., 2012). Both ATL31 and ATL6 directly ubiquitinated CPK28 in this bacterial system when UBC8 was used as the E2. The lack of E1, E2, ATL31/6, or ubiquitin resulted in the loss of CPK28 ubiquitination (Figure 4, B and C). Moreover, the mutation of C143 and H145 in ATL31 largely impaired its own ubiquitin ligase activity and resulted in decreased ubiquitination of CPK28 (Supplemental Figure S13A).
CPK28 interacts with BIK1 and promotes its turnover (Monaghan et al., 2014). However, ATL31 did not ubiquitinate either BIK1 or the cytosolic domain of BAK1 (BAK1CD; Supplemental Figure S13, B and C). Moreover, ATL2 did not ubiquitinate CPK28 (Figure 4D). These data further support the specificity of CPK28 ubiquitination by ATL31/6. Notably, we frequently observed some ubiquitination bands in the presence of E1, E2, and ubiquitin but not E3 during the detection of ubiquitin conjugates by immunoblotting with anti-FLAG antibodies (Figure 4, B–D;Supplemental Figure S13A). These ubiquitin conjugates were likely free ubiquitin chains generated by UBC8, as E2s such as UBC8, UBC7, UBC13, and UBC35 (with ubiquitin conjugating enzyme variant D together) could all generate free ubiquitin chains in the absence of E3, as previously shown (Zhao et al., 2013; Han et al., 2017; Turek et al., 2018).
ATL31/6 mediate the proteasomal degradation of CPK28
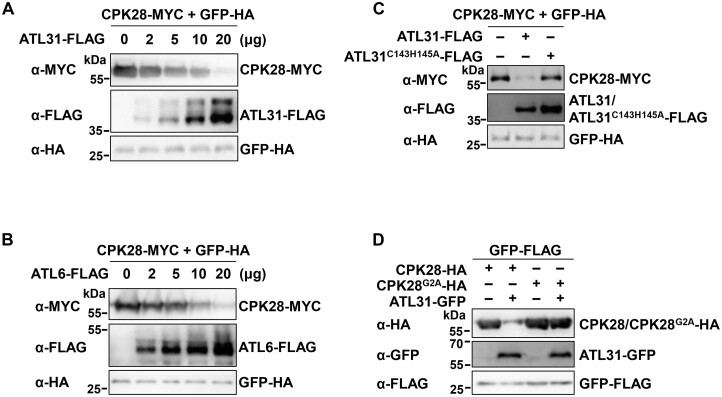
We have shown that overexpressing ATL31 in protoplasts significantly reduced CPK28-MYC protein abundance, but overexpressing ATL41, ATL45, and ATL80 did not (Supplemental Figure S6). Like ATL31, its closest homolog ATL6 also mediated CPK28 degradation, but ATL2 did not, when they were co-expressed with CPK28-HA in Arabidopsis protoplasts (Supplemental Figure S14, A–C). PUB25 is known to interact with and serve as a substrate of CPK28 (Wang et al., 2018). Nonetheless, PUB25 did not affect CPK28 protein accumulation (Supplemental Figure S14D). Furthermore, ATL31/6 promoted CPK28 degradation in a dose-dependent manner (Figure 5, A and B). In contrast to WT ATL31, the ATL31C143H145A variant was much less effective at promoting CPK28 protein degradation (Figure 5C). Although ATL31 mediated the degradation of WT CPK28, it barely contributed to the turnover of the CPK28G2A variant (Figure 5D).
Figure 5.
ATL31/6 promote the degradation of CPK28 in Arabidopsis protoplasts. A, B, ATL31/6 promote CPK28 degradation in a dose-dependent manner. Different amounts of ATL31- (A) or ATL6-FLAG (B) were co-expressed with CPK28-MYC in protoplasts. GFP-HA was used as an internal transfection control. CPK28-MYC/ATL31-FLAG/ATL6-FLAG/GFP-HA proteins were detected by immunoblotting with the indicated antibodies. C, ATL31C143H145A is much less effective than ATL31 at promoting CPK28 protein degradation. ATL31- or ATL31C143H145A-FLAG was co-expressed with CPK28-MYC in protoplasts. D, Overexpressing ATL31 hardly reduces CPK28G2A protein accumulation. ATL31-GFP was co-expressed with CPK28G2A-HA or CPK28-HA in protoplasts. GFP-HA or GFP-FLAG was used as an internal transfection control (C and D).
To further verify the role of ATL31/6 in mediating CPK28 degradation in plants, we monitored CPK28 protein accumulation in the atl31 atl6 double mutant plants using anti-CPK28 antibodies. CPK28 protein levels were lower in Col-0 than in atl31 atl6 (Supplemental Figure S15), while CPK28 transcript levels in Col-0 were comparable to those in atl31 atl6 plants (Supplemental Figure S16). Furthermore, in the presence of CHX, CPK28 degraded much more rapidly in Col-0 than in atl31 atl6 plants, as measured with anti-CPK28 antibodies (Figure 6A). These results strongly support the notion that ATL31/6 mediate CPK28 degradation. Furthermore, in the presence of CHX, CPK28 protein levels decreased in Col-0 within 2 h of flg22 treatment (Figure 6B). However, this phenomenon did not occur in atl31 atl6 plants (Figure 6B). These results suggest that ATL31/6 mediate flg22-enhanced CPK28 degradation. CPK28 directly phosphorylates BIK1 (Monaghan et al., 2014), and BIK1 was recently shown to transphosphorylate CPK28 (Bredow et al., 2021; Bredow and Monaghan, 2021). To test whether BIK1 affects CPK28 degradation, we measured CPK28 protein accumulation in bik1 plants. CPK28 protein levels in bik1 were comparable to those in Col-0 plants (Supplemental Figure S15).
Figure 6.
The degradation of CPK28 in atl31 atl6 and Col-0 plants. A, CPK28 protein turnover in atl31 atl6 and Col-0 plants. Seven-day-old Col-0 and atl31 atl6 seedlings (10 seedlings per sample) were treated with 100-μM CHX for different times. B, flg22-induced CPK28 degradation is largely blocked by loss of ATL31/6 function. Seven-day-old Col-0 and atl31 atl6 seedlings (10 seedlings per sample) were pretreated with 100-μM CHX, followed 1 h later by treatment with 2-μM flg22 for the indicated times. A, B, Total proteins were isolated from whole seedlings. CPK28 proteins were detected by immunoblotting with anti-CPK28 antibodies. ACTIN was used as a loading control. C, ATL31/6 mediate the proteasomal degradation of CPK28, as demonstrated by cell-free degradation assays. Purified recombinant MBP-CPK28-HA proteins were incubated with total protein extracts isolated from 14-day-old Col-0 or atl31 atl6 seedlings in the presence of 1-mM ATP for the indicated times, with or without 50-μM MG132. MBP-CPK28-HA was detected with anti-HA antibodies. ACTIN was detected as a loading control.
We also performed cell-free degradation assays to confirm that ATL31/6 mediate the degradation of CPK28. In these assays, recombinant MBP-CPK28-HA was incubated with total protein extracts from Col-0 or atl31 atl6 plants in the presence of ATP. The degradation rate of MBP-CPK28-HA was much slower in extracts from atl31 atl6 than from Col-0. Moreover, when MG132 treatment was applied, the degradation of MBP-CPK28 was largely inhibited (Figure 6C). These results support the notion that ATL31/6 mediate CPK28 degradation in a 26S proteasome-dependent manner.
ATL31/6 modulate BIK1 homeostasis through CPK28
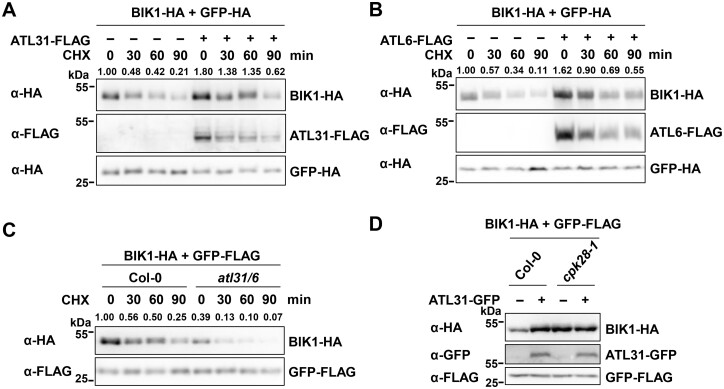
CPK28 is known to promote the degradation of BIK1 through PUB25/26 (Monaghan et al., 2014; Wang et al., 2018). We found that CPK28 accumulation was further regulated by ATL31/6. Therefore, we sought to determine whether ATL31/6 affect BIK1 accumulation. To this end, we transiently expressed BIK1-HA together with ATL31/6-FLAG in protoplasts. Overall, the accumulation of BIK1 was enhanced by ATL31/6 overexpression, which slowed BIK1 turnover (Figure 7, A and B;Supplemental Figure S17A), likely due to the degradation of CPK28 mediated by ATL31/6 (Figure 6). In contrast, overexpressing ATL31C143H145A or ATL2 in protoplasts had little effect on BIK1 accumulation (Supplemental Figure S17, A and B), and overexpressing ATL31 barely affected the accumulation of FLS2 and BAK1 (Supplemental Figure S17, C and D). Furthermore, when BIK1-HA was transiently expressed in protoplasts isolated from alt31 atl6 and Col-0, overall BIK1-HA protein levels were lower in alt31 atl6 than in Col-0, and BIK1 turnover was accelerated in alt31 atl6 compared to that in Col-0 (Figure 7C). These results suggest that ATL31/6 promote BIK1 protein accumulation.
Figure 7.
ATL31/6 modulate BIK1 homeostasis. A, B, ATL31/6 promote BIK1 accumulation. BIK1-HA was co-expressed with ATL31/6-FLAG in protoplasts. The protoplasts were treated with 50-μM CHX for the indicated times before harvesting. BIK1-HA proteins were detected by immunoblotting with anti-HA antibodies. GFP-HA was used as an internal transfection control. The relative protein levels of BIK1-HA were normalized to those of GFP-HA. The relative protein level of BIK1-HA at the beginning of the CHX treatment (in the absence of ATL31-FLAG) was set to 1. The density of the BIK1-HA and GFP-HA protein bands was quantified using EvolutionCapt v18.10 software. C, BIK1-HA protein levels are lower in atl31 atl6 than in Col-0 protoplasts. BIK1-HA was expressed in protoplasts isolated from atl31 atl6 or Col-0 plants. The protoplasts were treated with 50-μM CHX for the indicated times. GFP-FLAG was used as an internal transfection control. The relative protein levels of BIK1-HA were normalized to those of GFP-FLAG. The relative protein level of BIK1-HA in Col-0 at the beginning of the CHX treatment was set to 1. D, ATL31 modulates BIK1 homeostasis via CPK28. BIK1-HA was co-expressed together with or without ATL31-GFP in protoplasts isolated from Col-0 or cpk28-1 plants. GFP-FLAG was used as an internal transfection control. BIK1-HA proteins were detected by immunoblotting with anti-HA antibodies.
To confirm that ATL31/6 modulate the stability of BIK1 via CPK28, we examined the effect of ATL31 overexpression on BIK1 accumulation in the cpk28 mutant (Monaghan et al., 2014). BIK1-HA protein levels were higher in cpk28 protoplasts than in Col-0, which is consistent with previous reports (Monaghan et al., 2014; Wang et al., 2018). When BIK1 was co-expressed with ATL31, BIK1 protein accumulation in Col-0 protoplasts was enhanced to a level comparable to that in cpk28 protoplasts. However, ATL31 had little effect on promoting BIK1 protein accumulation in cpk28 protoplasts (Figure 7D). These results demonstrate that ATL31 modulates BIK1 homeostasis through CPK28.
ATL31/6 positively regulate BIK1-mediated immunity
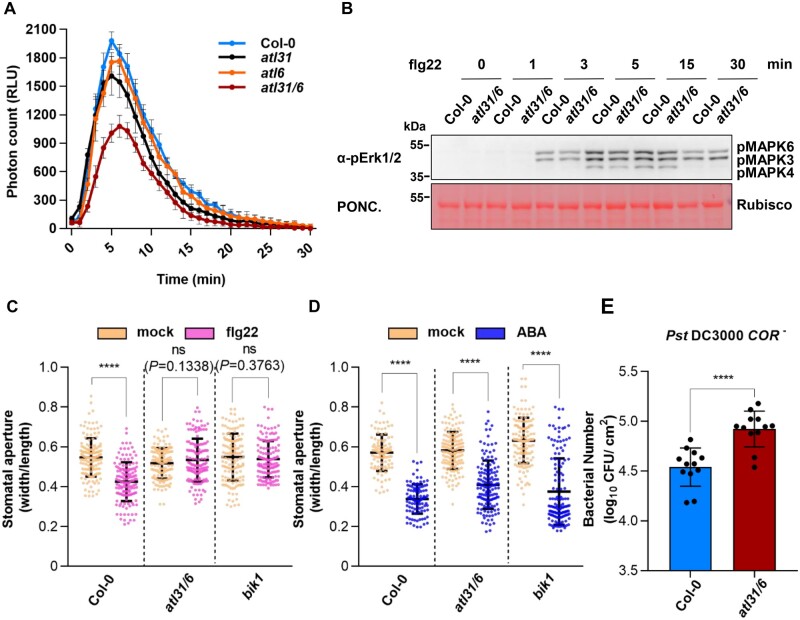
BIK1 plays a critical role in flg22-triggered production of ROS (Kadota et al., 2014; Li et al., 2014), and cpk28 mutant plants produce more ROS than Col-0 plants upon flg22 treatment (Monaghan et al., 2014). We therefore tested whether ATL31/6 regulate the flg22-triggered ROS burst. After flg22 treatment, the atl31 and atl6 single mutants exhibited less ROS production than Col-0 plants, and the atl31 atl6 double mutant plants produced even less ROS than either single mutant (Figure 8A), suggesting that ATL31 and ATL6 play redundant roles in positively regulating PTI signaling.
Figure 8.
ATL31/6 positively regulate BIK1-mediated immunity. A, atl31, atl6, and atl31 atl6 produce less ROS than Col-0 after flg22 treatment. Leaf discs from 30-day-old plants of the indicated genotypes were treated with 100-nM flg22, and RLU (representing the relative amounts of H2O2) were immediately measured. Values are means ± sd (n = 8, n means leaf disc number). B, flg22-induced MPK3/6 activation in atl31 atl6 and Col-0 plants. Seven-day-old seedlings (10 seedlings per sample) were treated with 100-nM flg22 for the indicated times. Total proteins were isolated from whole seedlings. MAPK activation was detected by immunoblotting with anti-pErk1/2 antibodies. Ponceau (PONC.) staining of the membrane was used as the loading control. C, flg22-triggered stomatal closure in atl31 atl6 and Col-0 plants. Leaves from 5-week-old Col-0, atl31 atl6, and bik1 plants were treated with 10-μM flg22 for 1 h. The stomatal aperture was measured using ImageJ software. Each data point represents a single stoma; values are means ± sd for n = 144 stomata from three biological repeats using independent plant samples grown and treated with flg22 under the same conditions. D, ABA-triggered stomatal closure in atl31 atl6 and Col-0 plants. Leaves from 5-week-old Col-0, atl31 atl6, and bik1 plants were treated with 10-μM ABA for 1 h. The stomatal aperture was measured using ImageJ software. Each data point represents a single stoma; values are means ± sd for n > 100 stomata from three biological repeats using independent plant samples grown and treated with ABA under the same conditions. C, D, Statistical significance compared to mock-treated plants was determined by Student’s t tests: ****P < 0.0001; ns, no significant difference. Number of stomata counted and statistical analysis are described in detail in the “Materials and methods” and Supplemental Table S3. E, Growth of Pst DC3000 COR– in Col-0 and atl31 atl6 plants. Four-week-old Col-0 and atl31 atl6 plants were dipping-inoculated with the bacteria. Bacterial growth was evaluated as colony-forming units per cm2 of leaf area (cfu/cm2) and was determined 3 days post inoculation. Individual data points are shown with means ± sd (n = 12 leaves from three biological replicates using plant samples grown and inoculated under the same conditions). Statistical significance compared to Col-0 plants was determined by Student’s t tests: ****P < 0.0001 (Supplemental Table S3).
flg22 also triggers MITOGEN-ACTIVATED PROTEIN KINASE (MAPK) activation (Segonzac et al., 2014, Xu et al., 2014). flg22-induced MPK3/6 activation was not affected by the loss of ATL31 and ATL6 (Figure 8B), which is in line with a previous observation that overexpression of CPK28 had no impact on MPK3/6 activation (Monaghan et al., 2014).
As BIK1 plays an important role in stomatal immunity (Li et al., 2014; Thor et al., 2020), we also measured flg22-induced stomatal closure in Col-0 and atl31 atl6 plants. As observed in the immune-deficient mutant bik1 (Li et al., 2014) and in transgenic plants overexpressing CPK28 (Monaghan et al., 2014), flg22-induced stomatal closure was almost abolished in atl31 atl6 mutant plants compared to that in Col-0 (Figure 8C). However, abscisic acid (ABA)-triggered stomatal closure was not affected in atl31 atl6 (Figure 8D), suggesting that ATL31/6 specifically function in stomatal closure during innate immunity. Moreover, atl31 atl6 plants were more susceptible than Col-0 to the hypovirulent bacterial pathogen Pseudomonas syringae pv. tomato (Pst) DC3000 COR–, a strain deficient in the production of the phytotoxin coronatine (COR), which stimulates stomatal opening (Ma et al., 1991; Melotto et al., 2006; Figure 8E). We also inoculated atl31 atl6, cpk28-1, and Col-0 plants with the nonvirulent type III secretion mutant strain Pst DC3000 hrcC–. atl31 atl6 also supported significantly higher bacterial growth than Col-0 plants, whereas the cpk28 mutant exhibited stronger resistance to bacteria than Col-0 (Supplemental Figure S18), which is consistent with a previous report (Monaghan et al., 2014). These results demonstrate that ATL31/6 positively regulate BIK1-mediated immunity.
Discussion
Immune responses must be tightly regulated to ensure their proper timing, appropriate amplitude, and optimal duration. BIK1 functions as a signaling hub of plant immunity and is subjected to different modes of modification, including phosphorylation and poly- and monoubiquitination (Lu et al., 2010; Zhang et al., 2010; Monaghan et al., 2014; Wang et al., 2018; Ma et al., 2020). Importantly, an immune signaling module forms to modulate BIK1 stability and immune homeostasis in which CPK28 phosphorylates PUB25/26 to enhance their activity, thereby promoting BIK1 degradation (Monaghan et al., 2014; Wang et al., 2018). Here, we demonstrated that CPK28 is further subjected to negative regulation by ATL31/6. Our findings indicate that the PRR–BIK1 immune complex is subjected to multi-layered regulation, and CPK28 is kept in check to maintain BIK1 homeostasis. Furthermore, we demonstrate how ATL31/6-mediated CPK28 degradation is dynamically controlled to regulate immune singling. In the resting state, ATL31/6 regulate CPK28 turnover to maintain immune homeostasis. Following flg22 treatment, the degradation of CPK28 was enhanced, which resulted in the relief of the CPK28-mediated negative regulation of the immune receptor complex to further enhance immune responses (Figure 9). It was recently reported that CPK28 transcripts are alternatively spliced after flg22 (Bazin et al., 2020) and AtPep1 treatment (Dressano et al., 2020), resulting in the accumulation of the truncated CPK28 variants, which has been proposed as a major immune-induced CPK28 attenuation mechanism. Therefore, flg22-induced CPK28 degradation has emerged as another CPK28 attenuation mechanism. Notably, the canonically spliced CPK28 transcript was also induced upon flg22 treatment; however, its induction was weaker and slower than that of CPK28-RI (Supplemental Figure S8). Therefore, it is likely that the induction of WT CPK28 by flg22 acts as a negative feedback regulation to maintain immune homeostasis. On the other hand, this result indicates that the negative immune regulator genes can also be induced by PAMP treatment, like PUB22, PUB23, and PUB24 (Trujillo et al., 2008).
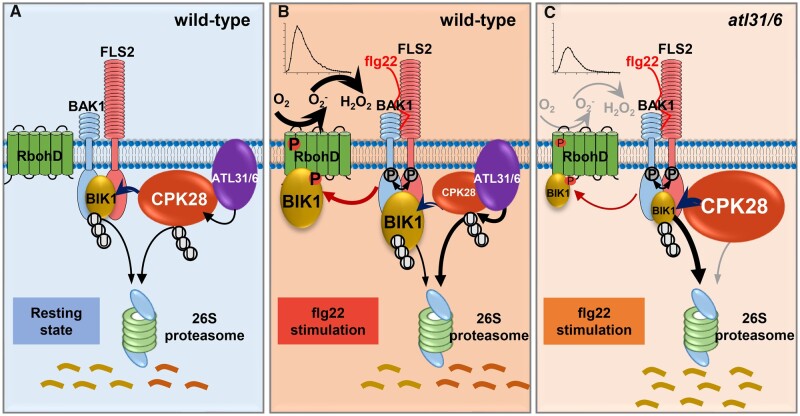
Figure 9.
CPK28 is targeted by ATL31 and ATL6 for proteasome-mediated degradation to fine-tune immune signaling. BIK1 acts as a signaling hub of plant immunity, which relays the immune signal from the FLS2-BAK1 complex to RbohD to promote ROS production. BIK1 homeostasis is maintained by a regulatory module in which CPK28 contributes to BIK1 turnover. Based on a previous conceptual model (Monaghan et al., 2014), we proposed a model for maintenance of BIK1 homeostasis by ATL31/6 via CPK28. Our work demonstrates that CPK28 is under proteasomal control to fine-tune immune signaling. The ubiquitin ligases ATL31 and ATL6 interact with CPK28 at the plasma membrane. ATL31/6 directly ubiquitinate CPK28, which results in the degradation of CPK28 via the 26S proteasome. A, In the resting state, ATL31/6 mediate CPK28 turnover to maintain BIK1 homeostasis. B, Following flg22 treatment, the association of CPK28 and ATL31/6 is enhanced, which promotes the degradation of CPK28 and leads to the relief of the CPK28-mediated negative regulation of the immune receptor complex to enhance immune responses. C, CPK28 protein accumulation is higher in atl31 atl6 than in Col-0. Consequently, BIK1 protein levels are lower in atl31 atl6 than in Col-0. Therefore, the immune responses in atl31 atl6 are reduced compared to those of Col-0.
The ubiquitination and proteasomal degradation of positive regulators of plant immunity has often been shown to contribute to their degradation and the attenuation of immune signaling, except for the monoubiquitination of BIK1 by RHA3A and RHA3B (Stegmann et al., 2012; Monaghan et al., 2014; Lee et al., 2020; Wang et al., 2018; Ma et al., 2020). In contrast, less is known about the ubiquitination and proteasomal degradation of negative immune regulators, which may contribute to activation of immune signaling. Here, we showed that negative immune regulators such as CPK28, BIR2, and BIR3 also undergo ubiquitination and proteasomal degradation (Figure 1B;Supplemental Figure S1, A and B). In line with this, a recent proteomics study identified ubiquitination sites on CPK28, BIR2, and BIR3 proteins (Grubb et al., 2021). We showed that CPK28 was ubiquitinated by ATL31/6, leading to its degradation. However, which E3 ligases mediate the ubiquitination and degradation of BIR2 or BIR3 should be investigated in the near future.
ATL is a RING-type ubiquitin ligase gene family unique to plants that consists of 91 members in Arabidopsis (Aguilar-Hernández et al., 2011; Guzmán, 2014). Several ATL family members are involved in defense response. Ectopic expression of ATL2 leads to the induction of many defense-related genes (Serrano and Guzman, 2004). Here, we demonstrated that ATL2 does not associate with and ubiquitinate CPK28 (Figure 2, G and D); consequently, it does not mediate CPK28 degradation (Supplemental Figure S14C). Moreover, ATL9, another ATL family member that is localized to the endoplasmic reticulum, positively regulates plant resistance to the biotrophic fungal pathogen Golovinomyces cichoracearum (Berrocal-Lobo et al., 2010). Although ATL31 and ATL6 positively regulate plant resistance to Pst DC3000 infection (Maekawa et al., 2012), the underlying mechanism remains elusive. Here, by screening for ubiquitin ligases that mediate CPK28 degradation, we identified ATL31 and ATL6 as the cognate E3s that target CPK28 for ubiquitination and proteasomal degradation. However, the substrates of ATL2 and ATL9 have yet to be identified and warrant further investigation.
The E3 ligases ATL44 and ATL45, also known as RHA3A and RHA3B, respectively, were recently shown to play positive roles in activating immune signaling by mediating the monoubiquitination of BIK1, resulting in the release of BIK1 from the FLS2 PRR complex (Ma et al., 2020). Therefore, ATL-mediated protein ubiquitination not only has proteolytic functions, but also plays a nonproteolytic role in regulating immune responses.
Ub chains with different linkage types encode different signals that dictate the fates of their modified substrates. The Lys48 Ub chain usually encodes a signal for 26S proteasomal degradation, while Lys63 Ub chains often play nonproteolytic roles (Komander and Rape 2012). E2s play a critical role in ubiquitin chain assembly (Komander and Rape 2012; Stewart et al., 2016). The Arabidopsis genome encodes 37 E2 enzymes. UBC8 is one of the Group VI E2s that consist almost exclusively of the UBC domain that binds to E3s (Kraft et al., 2005; Turek et al., 2018). We previously showed that UBC8 had broad lysine specificity, and likely mediated the formation of Ub chains at both the Lys48 and Lys63 sites or at other Lys sites of Ub moieties (Han et al., 2017). UBC8, as well as other group VI E2 members, functions with a wide range of E3 ubiquitin ligases (Kraft et al., 2005). Therefore, UBC8 is widely used for in vitro ubiquitination assays to examine the ubiquitination of various substrates by cognate E3s (Lu et al., 2011; Stegmann et al., 2012; Wang et al., 2018; Ma et al., 2020). A set of E2s that paired with PUB22 in vivo were recently identified, including various members of group VI, such as UBC8 and UBC30 (Turek et al., 2018). Here, we also used UBC8 as the E2 and demonstrated the ubiquitination of CPK28 by ATL31/6 (Figure 4, B–D). However, to uncover the type of CPK28 ubiquitination mediated by ATL31/6 and to fully understand the distinct cellular roles played by ATL31/6, an endogenous E2 for ATL31/6 needs to be identified.
C and N in plants function not only as energy sources but also as signaling molecules. C and N are reciprocally regulated and tightly integrated to maintain their proper balance. The ratio of C and N (C/N) is sensed in plant cells, and C/N-nutrient responses are initiated to regulate plant growth and development (Vidal and Gutierrez, 2008). ATL31/6 negatively regulate plant C/N responses via the ubiquitination and subsequent degradation of 14-3-3 proteins (Sato et al., 2009, 2011). In this study, we showed that ATL31/6 function as positive regulators of plant immune responses by mediating CPK28 ubiquitination. C and N metabolism is thought to be related to plant defense responses (Gómez-Ariza et al., 2007; Ros et al., 2008; Maekawa et al., 2012). Therefore, it is likely that ATL31/6 function in the crosstalk between plant immune signaling and C/N responses, but this requires further investigation. Additionally, whether CPK28 is involved in regulating plant C/N responses remains to be determined.
Our results demonstrate that ATL31/6 target CPK28 for ubiquitination and degradation. A previous study revealed that eight 14-3-3 isoforms are associated with ATL31. Of these 14-3-3s, 14-3-3χ and 14-3-3λ directly interact with ATL31 (Sato et al., 2011). Furthermore, 14-3-3χ serves as a substrate of ATL31 and thus is ubiquitinated and targeted for 26S proteasome-mediated degradation in response to cellular C/N changes (Sato et al., 2011). 14-3-3 proteins bind to the specific phosphorylated motifs in a wide array of target proteins and play important roles in various cellular processes, including plant defense responses (Roberts et al., 2002; Chevalier et al., 2009). 14-3-3 proteins play positive roles in regulating early PTI responses in Arabidopsis, such as PAMP-triggered ROS burst and stomatal immunity, as chemical disruption of the interaction between 14-3-3s and their client proteins greatly suppressed these immune responses (Lozano-Durán et al., 2014). In addition, 14-3-3λ binds to RPW8.2 (RESISTANCE TO POWDERY MILDEW8.2) and positively regulates RPW8.2-mediated resistance to the fungal pathogens Golovinomyces spp. in Arabidopsis via the salicylic acid signaling pathway (Yang et al., 2009). Notably, CPK28 and other CPKs phosphorylated 14-3-3χ and 14-3-3ε in vitro kinase assays. Furthermore, CPK28 harbors a canonical 14-3-3 binding motif R-S/T-X-S43-X-P; and Ser43 in this motif is an autophosphorylation site of CPK28 (Swatek et al., 2014). However, whether these 14-3-3 isoforms are involved in ATL31-mediated CPK28 degradation and in regulating CPK28-mediated immunity remain to be determined.
CPK28 plays dual roles in regulating immune signaling and stem elongation (Matschi et al., 2013, 2015). After the induction of bolting, cpk28 mutant plants displayed reduced stem elongation compared to the WT plants (Matschi et al., 2013, 2015). Like CPK28 overexpression lines (Monaghan et al., 2014), atl31 atl6 plants did not show a growth defect after the transition to the reproductive stage (Supplemental Figure S11G). However, whether ATL31/6 regulate stem elongation and whether they mediate CPK28 degradation in a stage-specific manner remain to be investigated.
The phosphorylation status of a kinase affects its ubiquitination by cognate E3 ligases. BIK1 phosphorylation is a prerequisite for its monoubiquitination at the plasma membrane (Ma et al., 2020). In contrast, flg22-induced phosphorylation at Ser236/Thr237 of BIK1 inhibits its polyubiquitination by PUB25/26 (Wang et al., 2018). CPK28 undergoes autophosphorylation at multiple sites (Hegeman et al., 2006; Matschi et al., 2013; Swatek et al., 2014; Bender et al., 2017). The kinase activity of CPK28 is required for its roles in stem elongation and immune homeostasis (Matschi et al., 2013; Monaghan et al., 2014). flg22 treatment can promote CPK28 kinase activity (Wang et al., 2018). However, the sites undergoing flg22-induced phosphorylation have not been identified for CPK28. CPK28 was recently shown to undergo intermolecular autophosphorylation at Ser318, which can also be transphosphorylated by BIK1 (Bredow et al., 2021). Moreover, phosphorylation of Ser318 is required for CPK28 activation at physiological [Ca2+] and CPK28-mediated immune homeostasis, but not required for stem elongation (Bredow et al., 2021). Additionally, Thr76 of CPK28, a conserved BIK1 phosphorylation site but not an autophosphorylation site, is also required for CPK28-mediated immune regulation (Bredow and Monaghan, 2021). It would be interesting to examine whether the autophosphorylation of CPK28 and the phosphorylation of CPK28 at Ser318/Thr76 are required for its ubiquitination by ATL31/6 and subsequent proteosomal degradation. We observed that CPK28 protein levels in bik1 plants were comparable to those of Col-0 plants (Supplemental Figure S15). It is likely that phosphorylation of CPK28 by BIK1 does not affect CPK28 protein accumulation. However, BIK1 and its closest homolog PBL1 function additively in PTI signaling (Zhang et al., 2010). To investigate the role of BIK1 in CPK28 accumulation, it is necessary to use the bik1 pbl1 double mutant, as a recent epistasis analysis demonstrates CPK28-mediated immune signaling is dependent on both BIK1 and PBL1 (Bredow et al., 2021). Intriguingly, CPK28-RI has been shown to have impaired kinase activity and compromised autophosphorylation (Dressano et al., 2020), but whether CPK28-RI is ubiquitinated by ATL31/6 and undergoes degradation await more investigation.
Materials and methods
Plant materials and growth conditions
For protoplast isolation, the stomatal aperture assay, and the pathogen infection assay, A. thaliana plants were grown in soil (TS 1, Lithuania) in a growth room at 22°C with a 60% relative humidity and under 70 μE m−2 s−1 light (white fluorescent bulbs) with a 12-h photoperiod for 30 days. For assays using Arabidopsis seedlings, seeds were surface sterilized, germinated and grown on 1/2 Murashige and Skoog plates with 0.5% sucrose and 0.8% agar. The plates were kept in the dark at 4°C for 2 days, and then the plants were grown under the same conditions described above. The fls2, bak1-4, and bik1 mutants were described previously (Shan et al., 2008; Lu et al., 2010). The T-DNA insertion mutants atl31 (GK_746D08, Sato et al., 2009), atl6 (Salk_149614, Sato et al., 2009), cpk28-1 (WiscDslox246D03, Monaghan et al., 2014), and cpk28-3 (GABI_523B08, Monaghan et al., 2014) were obtained from the Arabidopsis Biological Resource Center (ABRC), genotyped by PCR-based methods, and confirmed by RT-PCR analysis. The atl31 atl6 double mutant was generated through a genetic cross between atl31 and atl6 (Sato et al., 2009, 2011) and confirmed by molecular genotyping and RT-PCR analysis. The primers used for genotyping are listed in Supplemental Table S2.
Elicitors and antibodies
The flg22 peptide (QRLSTGSRINSAKDDAAGLQIA) was synthesized by the Shanghai Biotech Company (Shanghai, China).
Anti-CPK28 antibodies were generated in mouse by the Animal Facility, Institute of Genetics and Developmental Biology (Beijing), using full-length recombinant CPK28 proteins as antigens. The rabbit polyclonal anti-Phospho-p44/42 MAPK (Erk1/2) antibody (Cat# 9101) was purchased from Cell Signaling Technology (USA). The anti-HA antibody (Cat# H3663), mouse monoclonal anti-FLAG antibody (Cat# F1804), mouse monoclonal anti-HA-peroxidase (Cat# H6533), mouse monoclonal anti-FLAG-peroxidase (Cat# A8592), and mouse monoclonal anti-C-MYC-peroxidase (Cat# A5598) were purchased from Sigma (USA). The goat polyclonal anti-GFP-peroxidase (Cat# ab6663) was purchased from Abcam (USA). The mouse monoclonal anti-plant beta-ACTIN antibody (Cat# AT0004) was purchased from CMCTAG (USA) and used at 1:10,000 dilution. The goat anti-rabbit & mouse IgG-HRP (Cat#M21003-S) was purchased from Abmart (China) and used at 1:10,000 dilution.
Plasmid construction
The full-length coding sequences of Arabidopsis CPK28, CPK8, CPK6, CPK1, CPK3, CPK16, CPK28-RI, BIR2, BIR3, ATL31, ATL6, ATL2, PUB25, PBL13, RCN1 and a set of E3 ubiquitin ligase genes were amplified by PCR from Col-0 cDNA and cloned into the plant expression vector pHBT, where they were fused to either an HA, FLAG, MYC, cYFP, nYFP, GFP, or RFP tag.
Arabidopsis FLS2, BAK1, BIK1, UBQ10, BRI1, PCRK1, and GFP constructs were described previously (Huang et al., 2019; Lu et al., 2010, 2011). CPK28, CPK8, CPK6, CPK1, CPK3, CPK16, CPK28-RI, ATL31△TM (amino acids 67–368), and ATL6△TM (amino acids 71–398) were subcloned into the modified His-tagged or MBP-tagged fusion protein expression vector pET28a (Novagen) or pMAL-C2 (New England Biolabs). The primers used are listed in Supplemental Table S2.
RT-qPCR
Total RNA was extracted from 7-day-old seedlings using TRIzol (Invitrogen) following the manufacturer’s instructions. The first-strand cDNA was synthesized in 20-μL reactions from 1 μg DNase I-treated total RNA using a reverse transcription kit (Promega, USA). qPCR was then performed on a Bio-Rad CFX-96 Real-Time PCR system using a SYBR Green qPCR kit (Promega, USA). Gene expression levels were normalized to that of GAPC, a stably expressed reference gene (Czechowski et al., 2005).
Transcriptome sequencing
Seven-day-old Arabidopsis seedlings were treated with 4-μM flg22 or water for 30 min. Total RNA was isolated from the seedlings using TRIzol reagent (Invitrogen). Library construction and sequencing were performed by Novogene (Tianjing, China). In-house Perl scripts were used to process the raw reads, and the clean data were obtained by calculating Q20, Q30, and GC content. The clean reads were aligned to the reference Arabidopsis genome using TopHat v2.0.12. The number of fragments per kilobase of transcript sequence per million base pairs sequenced was counted using HTSeq v0.6.1 (Trapnell et al., 2009). Differential expression analysis of seedlings treated with flg22 or water for two biological replicates (independent seedling samples grown and treated with flg22 or water under the same conditions) was performed using the DESeq R package (1.18.0). A model based on the negative binomial distribution was used to calculate P-values for differential expression, which were further adjusted using the Benjamini and Hochberg approach. Differentially expressed genes were defined as those with P < 0.05.
Transient gene expression in protoplasts
Arabidopsis leaf protoplasts were isolated and used for transient gene expression as described previously (Huang et al., 2019). Protein expression in protoplasts was then detected by immunoblotting with anti-HA, anti-FLAG, anti-MYC, or anti-GFP antibodies.
Co-IP assays
Co-IP assays were performed as described by Huang et al. (2019). Protoplasts were transfected and incubated for 10 h. Before being harvested, the protoplasts were treated with 2-μM flg22 or water for 30 min. Total proteins were extracted from the protoplasts with protein extraction buffer containing 1-mM EDTA, 0.5% Triton X-100 and a protease inhibitor cocktail tablet (Roche). After centrifugation (12,470g) at 4°C for 10 min, the supernatant was incubated with anti-FLAG antibodies (Sigma) at 4°C for 2 h and then protein-G-agarose beads (Roche) were added and the incubation was continued for another 2 h. The agarose beads were collected by centrifugation (100g) at room temperature for 2 min, washed three times with washing buffer containing 1-mM EDTA and 0.1% Triton X-100, and washed once with 50-mM Tris–HCl at pH 7.5. The immunoprecipitated proteins were detected by immunoblotting.
BiFC assay in Arabidopsis protoplasts and confocal laser-scanning microscopy
Arabidopsis protoplasts were transfected with the BiFC vectors ATL31C143H145A-nYFP, CPK28-cYFP, CPK28G2A-cYFP, BRI1-nYFP, and PCRK1-cYFP. Fluorescent signals were detected by confocal laser-scanning microscopy (Leica, Germany). YFP was excited at 514 nm using an argon laser, and emission was collected from 525 to 570 nm. GFP was excited at 488 nm using an argon laser, and emission was collected at 500–540 nm. RFP was excited at 552 nm using an argon laser, and emission was collected between 589 and 625 nm. The chlorophyll autofluorescence was collected at wavelengths between 630 and 700 nm.
Recombinant protein expression, purification, and MBP PD assay
The recombinant His–FLAG–CPK28 proteins were purified from E. coli using Ni-NTA agarose. MBP-ATL31ΔTM/ATL6ΔTM-HA or MBP-HA proteins immobilized on amylose resin (New England BioLabs) were incubated with purified His-tagged recombinant proteins. The incubation was carried out at 4°C for 2 h with gentle shaking. The incubation buffer contained 20-mM Tris–HCl (pH 7.5), 150-mM NaCl, 3-mM EDTA, and 0.5% Triton X-100. The amylose resin was collected and washed five times with buffer containing 20-mM Tris–HCl (pH 7.5), 3-mM EDTA, 150-mM NaCl, and 0.1% Triton X-100. The bound His-tagged proteins were detected by immunoblotting using anti-FLAG antibodies (Sigma).
In vitro ubiquitination assay
The ubiquitination assay using a bacterial reconstituted system was performed as previously described (Han et al., 2017). The pCDFDuet-MBP-CPK28/BIK1/BAK1CD-HA-UBA1-S, pACYCDuet-ATL31ΔTM/ATL6ΔTM/ATL2ΔTM/PUB25-MYC- UBC8-S, and pET28a-FLAG-Ub plasmids were co-transformed into E. coli BL21 (DE3) competent cells. The bacteria were cultured at 37°C in 500-μL liquid Luria-Bertani (LB) medium. When the absorbance at 600 nm reached 0.5, 0.5-mM isopropyl β-d-1-thiogalactopyranoside was applied to induce the expression of recombinant proteins. The bacteria were grown at 28°C for 10 h. Protein ubiquitination was analyzed by immunoblotting with anti-HA, anti-FLAG, and anti-MYC antibodies.
Cell-free protein degradation assay
Total cell-free extracts were obtained from 14-day-old Col-0 or atl31 alt6 mutant seedlings using protein extraction buffer (50-mM Tris–MES, pH 8.0, 0.5-M sucrose, 1-mM MgCl2, 10-mM EDTA, pH 8.0, 5-mM DTT). Purified recombinant MBP-CPK28-HA proteins were incubated with the above cell-free extracts in the presence of 1-mM ATP, with or without 50-mM MG132 (at 25°C). The degradation of MBP-CPK28-HA proteins was detected by immunoblotting with anti-HA antibodies.
MAPK activation assay
Seven-day-old seedlings were treated with 100-nM flg22 or water. MPK3/6 activation was detected by immunoblotting with anti-pErk1/2 antibodies (Cell Signaling Technology). ACTIN was used as the loading control, which was detected by immunoblotting with anti-beta-ACTIN antibodies (Cell Signal Pathway Research Tools Supplier).
ROS burst assay
ROS production was measured as described previously (Li et al., 2014). Leaf discs (4 mm in diameter) were collected from the leaves of 30-day-old plants. The leaf discs were soaked overnight in sterile water in a 96-well plate. The water was then replaced with ROS measurement solution containing 10-μg·mL−1 horseradish peroxidase (Sigma), 50-μM luminol (Sigma), and 100-nM flg22. Luminescence was captured and ROS production was measured immediately using a GLOMAX 96 microplate luminometer (Promega). The values for ROS production are represented by Relative Light Units (RLU), which were determined based on the data obtained from eight leaf discs per treatment.
Stomatal aperture measurement
Stomatal aperture was measured as described by Li et al. (2014) with some modifications. Before flg22 or ABA treatment, 4-week-old plants were kept under the light for 2 h so that most of the stomata were opened. The leaves were collected and soaked in buffer containing 10-mM MES (pH 6.15), 10-mM KCl, and 10-mM CaCl2. The leaves were then treated with 10-μM flg22 or 10-μM ABA for 1 h and the stomata were observed under a light microscope (Leica). Image J software (NIH, Bethesda, MD, USA) was used to measure the stomatal aperture. At least eight peels from eight different plants were examined for each treatment. Number of stomata counted in Figure 8C are: Col-0 mock: n = 144, Col-0 flg22: n = 144, atl31 atl6 mock: n = 144, atl31 atl6 flg22: n = 144, bik1 mock: n = 144, bik1 flg22: n = 144. Number of stomata counted in Figure 8D are: Col-0 mock: n = 100, Col-0 flg22: n = 100, atl31 atl6 mock: n = 144, atl31 atl6 flg22: n = 144, bik1 mock: n = 144, bik1 flg22: n = 144.
Pathogen infection assay
Pseudomonas syringae pv. tomato (Pst) strain DC3000 COR– or DC3000 hrcC– was grown overnight at 28°C in liquid King’s B (KB) medium supplemented with 50-μg·mL−1 rifampicin. Bacteria were collected by centrifugation (376g) at room temperature for 5 min, and washed with H2O. The bacteria were diluted with H2O to the desired density (optical density [OD]600 = 0.1). Thirty-day-old plants were dipped into 5 × 108 cfu·mL−1Pst DC3000 COR– solution containing 0.15% gelatin (Amresco) for 15 s (Jiang et al., 2013), or infiltrated with 5 × 108 cfu·mL−1Pst DC3000 hrcC– solution using a needleless syringe. Three-day post inoculation, leaf discs were ground in 100-μL H2O, and 10-fold serial dilutions of the bacteria solution were prepared to measure bacterial growth. Bacteria were grown on KB plates with 50-μg·mL−1 rifampicin at 28°C for 3 days, and bacterial colony forming units were counted.
Statistical analysis
Statistical significance was determined by one-way analysis of variance (ANOVA) and Student’s t tests; P-values were generated using GraphPad Prism 7.04.
Accession numbers
Sequence data from this article can be found in GenBank (https://www.ncbi.nlm.nih.gov/gene/) and the Arabidopsis Information Resource (http://www.arabidopsis.org/) under the following accession numbers: FLS2: NP_199445.1, AT5G46330; BAK1: NP_567920.1, AT4G33430; BIK1: NP_181496.1, AT2G39660; ATL31: NP_198094.1, AT5G27420; ATL6: NP_566249.1, AT3G05200; CPK8: NP_197446.1, AT5G19450; ATL2: NP_188294.1, AT3G16720; BRI1: NP_195650.1, AT4G39400; PCRK1: NP_187594.1, AT3G09830; PUB25: NP_566632.1, AT3G19380; BIR2: NP_189486.1, AT3G28450; BIR3: NP_174039.1, AT1G27190; PBL13: NP_198408.2, AT1G27190; RCN1: NP_174039.1, AT1G25490; BSK1: NP_567980.1, AT4G35230; CPK6: NP_565411.2, AT2G17290; CPK1: NP_196107.1, AT5G04870; CPK3: NP_194096.1, AT4G23650; CPK16: NP_179379.1, AT2G17890. Mutants used in this article can be obtained from ABRC under the following accession numbers: atl31 (GK_746D08), atl6 (SALK_149614), cpk28-1 (WiscDslox246D03), cpk28-3 (GABI_523B08), fls2 (SALK_141277), bak1-4 (SALK_116202), bik1(Salk_005291). The RNA-seq data were deposited into National Center for Biotechnology Information and are accessible via BioProject accession number PRJNA742510.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. BIR2 and BIR3 undergo ubiquitination.
Supplemental Figure S2. Identification of cpk28 mutants and verification of the specificity of anti-CPK28 antibodies.
Supplemental Figure S3. Various negative immune regulators undergo proteasomal degradation.
Supplemental Figure S4. Screening of differentially expressed ubiquitin ligase genes upon flg22 treatment.
Supplemental Figure S5. Validation of the feasibility of the protoplast cell-based screening system.
Supplemental Figure S6. Screening of ubiquitin ligases that can potentially mediate CPK28 degradation.
Supplemental Figure S7. ATL31 is localized to the plasma membrane in Arabidopsis protoplasts.
Supplemental Figure S8. Measurement of transcript levels of ATL31, CPK28, and the CPK28-RI splice variant upon flg22 treatment.
Supplemental Figure S9. ATL31 does not associate with CPK8.
Supplemental Figure S10. The CPK28G2A variant loses its plasma membrane localization in protoplasts.
Supplemental Figure S11. Identification of the homozygous T-DNA insertion mutants atl31 and atl6 and generation of the atl31 atl6 double mutant.
Supplemental Figure S12. ATL31 promotes the ubiquitination of CPK28.
Supplemental Figure S13. The specificity of CPK28 ubiquitination by ATL31.
Supplemental Figure S14 . Overexpressing ATL31/6 reduces CPK28 protein accumulation in Arabidopsis protoplasts.
Supplemental Figure S15. CPK28 protein accumulation in Col-0, atl31 atl6, and bik1 plants.
Supplemental Figure S16. CPK28 transcript levels in atl31 atl6 and Col-0.
Supplemental Figure S17. ATL31 specifically promotes BIK1 protein accumulation in protoplasts.
Supplemental Figure S18. Growth of Pst DC3000 hrcC– in atl31 atl6, cpk28-1, and Col-0 plants.
Supplemental Table S1. The differentially expressed ubiquitin ligase genes upon flg22 treatment.
Supplemental Table S2. Primers used for genotyping, cloning, and RT-qPCR.
Supplemental Table S3. Student’ s t test tables.
Supplemental Table S4. ANOVA tables.
Supplementary Material
Acknowledgments
We thank Drs Ping He and Libo Shan (Texas A&M University) for the bik1, fls2, and bak1-4 seeds.
Funding
The work was supported by grants from the National Natural Science Foundation of China (32070555, 31800222), the Natural Science Foundation of Hebei Province (C2019503069), and the State Key Laboratory of Plant Genomics.
Conflict of interest statement. The authors declare no conflict of interests.
X.Liu., Y.Z., M.D., and D.L. designed the study. X.Liu., Y.Z., M.D., X.L., F.F., G.H., and J.B. performed the experiments. X.Liu., Y.Z., M.D., Y.Zou., and D.L. analyzed the data. D.L. and X.Liu. wrote the paper.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Dongping Lu (dplu@sjziam.ac.cn).
References
- Aguilar-Hernández V, Aguilar-Henonin L, Guzmán P (2011) Diversity in the architecture of ATLs, a family of plant ubiquitin-ligases, leads to recognition and targeting of substrates in different cellular environments. PLoS ONE 6:e23934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin J, Mariappan K, Jiang Y, Blein T, Voelz R, Crespi M, Hirt H (2020) Role of MPK4 in pathogen-associated molecular pattern-triggered alternative splicing in Arabidopsis. PLoS Pathog 16:e1008401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender KW, Blackburn RK, Monaghan J, Derbyshire P, Menke FL, Zipfel C, Goshe MB, Zielinski RE, Huber SC (2017) Autophosphorylation-based calcium (Ca2+) sensitivity priming and Ca2+/calmodulin inhibition of Arabidopsis thaliana Ca2+-dependent Protein Kinase 28 (CPK28). J Biol Chem 292:3988–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Stone S, Yang X, Antico J, Callis J, Ramonell KM, Somerville S (2010) ATL9, a RING zinc finger protein with E3 ubiquitin ligase activity implicated in chitin- and NADPH oxidase-mediated defense responses. PLoS ONE 5:e14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M, Sheen J (2013) CDPKs in immune and stress signaling. Trends Plant Sci 18:30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredow M, Bender KW, Johnson Dingee A, Holmes DR, Thomson A, Ciren D, Tanney CAS, Dunning KE, Trujillo M, Huber SC, et al. (2021). Phosphorylation-dependent subfunctionalization of the calcium-dependent protein kinase CPK28. Proc Natl Acad Sci USA 118:e2024272118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredow M, Monaghan J (2021) Differential regulation of the calcium-dependent protein kinase CPK28 by site-specific modification. Plant Physiol 186:1358–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis J (2014) The ubiquitination machinery of the ubiquitin system. Arab B 12:e0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier D, Morris ER, Walker JC (2009) 14-3-3 and FHA domains mediate phosphoprotein interactions. Annu Rev Plant Biol 60:67–91 [DOI] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nurnberger T, Jones JD, Felix G, Boller T (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448:497–500 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressano K, Weckwerth PR, Poretsky E, Takahashi Y, Villarreal C, Shen Z, Schroeder JI, Briggs SP, Huffaker A (2020) Dynamic regulation of Pep-induced immunity through post-translational control of defence transcript splicing. Nat Plants 6:1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlan G, Nakagami H, Eschen-Lippold L, Jiang X, Majovsky P, Kowarschik K, Hoehenwarter W, Lee J, Trujillo M (2017) Changes in PUB22 ubiquitination modes triggered by MITOGEN-ACTIVATED PROTEIN KINASE3 dampen the immune response. Plant Cell 29:726–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Ariza J, Campo S, Rufat M, Estopà M, Messeguer J, San Segundo B, Coca M (2007) Sucrose-mediated priming of plant defense responses and broad-spectrum disease resistance by overexpression of the maize pathogenesis-related PRms protein in rice plants. Mol Plant Microbe Interact 20:832–842 [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5:1003–1011 [DOI] [PubMed] [Google Scholar]
- Grubb LE, Derbyshire P, Dunning KE, Zipfel C, Menke FLH, Monaghan J (2021) Large-scale identification of ubiquitination sites on membrane-associated proteins in Arabidopsis thaliana seedlings. Plant Physiol 185:1483–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán P (2014) ATLs and BTLs, plant-specific and general eukaryotic structurally-related E3 ubiquitin ligases. Plant Sci 215–216:69–75 [DOI] [PubMed] [Google Scholar]
- Halter T, Imkampe J, Mazzotta S, Wierzba M, Postel S, Bücherl C, Kiefer C, Stahl M, Chinchilla D, Wang X, et al. (2014). The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr Biol 24:134–143 [DOI] [PubMed] [Google Scholar]
- Han Y, Sun J, Yang J, Tan Z, Luo J, Lu D (2017) Reconstitution of the plant ubiquitination cascade in bacteria using a synthetic biology approach. Plant J 91:766–776 [DOI] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104:12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegeman AD, Rodriguez M, Han BW, Uno Y, Phillips GN Jr, Hrabak EM, Cushman JC, Harper JF, Harmon AC, Sussman MR (2006) A phyloproteomic characterization of in vitro autophosphorylation in calcium-dependent protein kinases. Proteomics 6:3649–3664 [DOI] [PubMed] [Google Scholar]
- Huang G, Sun J, Bai J, Han Y, Fan F, Wang S, Zhang Y, Zou Y, Han Z, Lu D (2019) Identification of critical cysteine sites in brassinosteroid-insensitive 1 and novel signaling regulators using a transient expression system. New Phytol 222:1405–1419 [DOI] [PubMed] [Google Scholar]
- Imkampe J, Halter T, Huang S, Schulze S, Mazzotta S, Schmidt N, Manstretta R, Postel S, Wierzba M, Yang Y, et al. (2017) The Arabidopsis leucine-rich repeat receptor kinase BIR3 negatively regulates BAK1 receptor complex formation and stabilizes BAK1. Plant Cell 29:2285–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Yao J, Ma KW, Zhou H, Song J, He SY, Ma W (2013) Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog 9:e1003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444:323–329 [DOI] [PubMed] [Google Scholar]
- Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A, et al. (2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell 54:43–55 [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81:203–229 [DOI] [PubMed] [Google Scholar]
- Kong Q, Sun T, Qu N, Ma J, Li M, Cheng YT, Zhang Q, Wu D, Zhang Z, Zhang Y (2016) Two redundant receptor-like cytoplasmic kinases function downstream of pattern recognition receptors to regulate activation of SA biosynthesis. Plant Physiol 171:1344–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau OS, Deng XW, Callis J (2005) Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol 139:1597–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol E, Mentzel T, Chinchilla D, Boller T, Felix G, Kemmerling B, Postel S, Arents M, Jeworutzki E, Al-Rasheid KA, et al. (2010) Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J Biol Chem 285:13471–13479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Lal NK, Lin ZD, Ma S, Liu J, Castro B, Toruno T, Dinesh-Kumar SP, Coaker G (2020) Regulation of reactive oxygen species during plant immunity through phosphorylation and ubiquitination of RBOHD. Nat Commun 11:1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, Cai G, Gao L, Zhang X, Wang Y, et al. (2014) The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15:329–338 [DOI] [PubMed] [Google Scholar]
- Lin ZJ, Liebrand TW, Yadeta KA, Coaker G (2015) PBL13 is a serine/threonine protein kinase that negatively regulates Arabidopsis immune responses. Plant Physiol 169:2950–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R, Bourdais G, He SY, Robatzek S (2014) The bacterial effector HopM1 suppresses PAMP-triggered oxidative burst and stomatal immunity. New Phytol 202:259–269 [DOI] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332:1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, He P (2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA 107:496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SW, Morris VL, Cuppels DA. ( 1991. ) Characterization of a DNA region required for production of the phytotoxin coronatine by Pseudomonas syringae pv. tomato. Mol Plant Microbe Interact 4:69–77 [Google Scholar]
- Ma X, Claus LAN, Leslie ME, Tao K, Wu Z, Liu J, Yu X, Li B, Zhou J, Savatin DV, et al. (2020). Ligand-induced monoubiquitination of BIK1 regulates plant immunity. Nature 581:199–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S, Sato T, Asada Y, Yasuda S, Yoshida M, Chiba Y, Yamaguchi J (2012) The Arabidopsis ubiquitin ligases ATL31 and ATL6 control the defense response as well as the carbon/nitrogen response. Plant Mol Biol 79:217–227 [DOI] [PubMed] [Google Scholar]
- Matschi S, Hake K, Herde M, Hause B, Romeis T (2015) The calcium-dependent protein kinase CPK28 regulates development by inducing growth phase-specific, spatially restricted alterations in jasmonic acid levels independent of defense responses in Arabidopsis. Plant Cell 27:591–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschi S, Werner S, Schulze WX, Legen J, Hilger HH, Romeis T (2013) Function of calcium-dependent protein kinase CPK28 of Arabidopsis thaliana in plant stem elongation and vascular development. Plant J 73:883–896 [DOI] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126:969–980 [DOI] [PubMed] [Google Scholar]
- Monaghan J, Matschi S, Shorinola O, Rovenich H, Matei A, Segonzac C, Malinovsky FG, Rathjen JP, MacLean D, Romeis T, et al. (2014). The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 16:605–615 [DOI] [PubMed] [Google Scholar]
- Roberts MR, Salinas J, Collinge DB (2002) 14-3-3 proteins and the response to abiotic and biotic stress. Plant Mol Biol 50:1031–1039 [DOI] [PubMed] [Google Scholar]
- Ros B, Mohler V, Wenzel G, Thummler F (2008) Phytophthora infestans-triggered response of growth- and defense-related genes in potato cultivars with different levels of resistance under the influence of nitrogen availability. Physiol Plant 133:386–396 [DOI] [PubMed] [Google Scholar]
- Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tor M, de Vries S, Zipfel C (2011) The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23:2440–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Maekawa S, Yasuda S, Domeki Y, Sueyoshi K, Fujiwara M, Fukao Y, Goto DB, Yamaguchi J (2011) Identification of 14-3-3 proteins as a target of ATL31 ubiquitin ligase, a regulator of the C/N response in Arabidopsis. Plant J 68:137–146 [DOI] [PubMed] [Google Scholar]
- Sato T, Maekawa S, Yasuda S, Sonoda Y, Katoh E, Ichikawa T, Nakazawa M, Seki M, Shinozaki K, Matsui M, et al. (2009). CNI1/ATL31, a RING-type ubiquitin ligase that functions in the carbon/nitrogen response for growth phase transition in Arabidopsis seedlings. Plant J 60:852–864 [DOI] [PubMed] [Google Scholar]
- Segonzac C, Macho AP, Sanmartin M, Ntoukakis V, Sanchez-Serrano JJ, Zipfel C (2014) Negative control of BAK1 by protein phosphatase 2A during plant innate immunity. EMBO J 33:2069–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Guzman P (2004) Isolation and gene expression analysis of Arabidopsis thaliana mutants with constitutive expression of ATL2, an early elicitor-response RING-H2 zinc-finger gene. Genetics 167:919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, He P, Li J, Heese A, Peck SC, Nurnberger T, Martin GB, Sheen J (2008) Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4:17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M, Anderson RG, Ichimura K, Pecenkova T, Reuter P, Žársky V, McDowell JM, Shirasu K, Trujillo M (2012) The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 24:4703–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C (2017) The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355:287–289 [DOI] [PubMed] [Google Scholar]
- Stewart MD, Ritterhoff T, Klevit RE, Brzovic PS (2016) E2 enzymes: more than just middle men. Cell Res 26:423–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swatek KN, Wilson RS, Ahsan N, Tritz RL, Thelen JJ (2014) Multisite phosphorylation of 14-3-3 proteins by calcium-dependent protein kinases. Biochem J 459:15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thor K, Jiang S, Michard E, George J, Scherzer S, Huang S, Dindas J, Derbyshire P, Leitão N, DeFalco TA, et al. (2020). The calcium-permeable channel OSCA1.3 regulates plant stomatal immunity. Nature 585:569–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Hou C, Ren Z, Wang C, Zhao F, Dahlbeck D, Hu S, Zhang L, Niu Q, Li L, et al. (2019) A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 572:131–135 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo M, Ichimura K, Casais C, Shirasu K (2008) Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr Biol 18:1396–1401 [DOI] [PubMed] [Google Scholar]
- Turek I, Tischer N, Lassig R, Trujillo M (2018) Multi-tiered pairing selectivity between E2 ubiquitin-conjugating enzymes and E3 ligases. J Biol Chem 293:16324–16336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Gutierrez RA (2008) A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr Opin Plant Biol 11:521–529 [DOI] [PubMed] [Google Scholar]
- Wang J, Grubb LE, Wang J, Liang X, Li L, Gao C, Ma M, Feng F, Li M, Li L, et al. (2018) A regulatory module controlling homeostasis of a plant immune kinase. Mol Cell 69:493–504 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 411:219. [DOI] [PubMed] [Google Scholar]
- Xu J, Xie J, Yan C, Zou X, Ren D, Zhang S (2014) A chemical genetic approach demonstrates that MPK3/MPK6 activation and NADPH oxidase-mediated oxidative burst are two independent signaling events in plant immunity. Plant J 77:222–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA (2010) PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22:508–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Pearce G, Ryan CA (2006) The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc Natl Acad Sci USA 103:10104–10109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wang W, Coleman M, Orgil U, Feng J, Ma X, Ferl R, Turner JG, Xiao S (2009) Arabidopsis 14-3-3 lambda is a positive regulator of RPW8-mediated disease resistance. Plant J 60:539–550 [DOI] [PubMed] [Google Scholar]
- Yasuda S, Aoyama S, Hasegawa Y, Sato T, Yamaguchi J (2017) Arabidopsis CBL-interacting protein kinases regulate carbon/nitrogen-nutrient response by phosphorylating ubiquitin ligase ATL31. Mol Plant 10:605–618 [DOI] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. (2010) Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7:290–301 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Tian M, Li Q, Cui F, Liu L, Yin B, Xie Q (2013) A plant-specific in vitro ubiquitination analysis system. Plant J 74:524–533 [DOI] [PubMed] [Google Scholar]
- Zipfel C (2014) Plant pattern-recognition receptors. Trends Immunol 35:345–351 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125:749–760 [DOI] [PubMed] [Google Scholar]
- Zou Y, Wang S, Lu D (2020) MiR172b-TOE1/2 module regulates plant innate immunity in an age-dependent manner. Biochem Biophys Res Commun 531:503–507 [DOI] [PubMed] [Google Scholar]
- Zou Y, Wang S, Zhou Y, Bai J, Huang G, Liu X, Zhang Y, Tang D, Lu D (2018) Transcriptional regulation of the immune receptor FLS2 controls the ontogeny of plant innate immunity. Plant Cell 30:2779–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.