Abstract
During secondary growth, meristematic cells in the cambium can either proliferate to maintain the stem cell population or differentiate into xylem or phloem. The balance between these two developmental trajectories is tightly regulated by many environmental and endogenous cues. Strigolactones (SLs), a class of plant hormones, were previously reported to regulate secondary growth by promoting cambium activity. However, the underlying molecular mechanisms of SL action in plant secondary growth are not well understood. We performed histological, genetic, and biochemical analyses using genetic materials in Arabidopsis (Arabidopsis thaliana) with altered activity of the transcription factors BRI1-EMS-SUPPRESSOR1 (BES1) or WUSCHEL-related HOMEOBOX4 (WOX4) or lacking MORE AXILLARY SHOOT2 (MAX2), a key positive component in the SL signaling pathway. We found that BES1, a downstream regulator in the SL signaling pathway that promotes shoot branching and xylem differentiation, also inhibits WOX4 expression, a key regulator of cambium cell division in the intercellular TRACHEARY ELEMENT DIFFERENTIATION INHIBITORY FACTOR (TDIF)–TDIF RECEPTOR (TDR) signaling pathway. The antagonistic roles of BES1 and WOX4 in the regulation of cambium activity may integrate intercellular TDIF signals to efficiently and bidirectionally modulate cambium cell proliferation and differentiation. As both BES1 and WOX4 are widely involved in various endogenous signals and responses to environmental stimuli, these findings may provide insight into the dynamic regulation of cambium development.
Transcription factors BES1 and WOX4 play antagonistic roles downstream of strigolactone and intercellular TDIF signaling to regulate secondary growth in Arabidopsis.
Introduction
The plant vascular system provides routes to transport water, various mineral and organic nutrients, and signaling molecules. Secondary growth of the plant vascular system, which manifests as the thickening of the stems and roots to strengthen the plant body, contributes to plant adaptation to various changing environments throughout development. Plant secondary growth takes place in the vascular cambium (VC) cells, as they divide anticlinally to add new cambium initial cells, and periclinally to produce xylem or phloem mother cells (Spicer and Groover, 2010). The alternation of cambium cell division and differentiation is tightly regulated by internal and environmental cues, such as light, cold, drought, various nutrients, and many phytohormones, including auxin, cytokinin, ethylene, brassinosteroids (BRs), gibberellins (GAs), and strigolactones (SLs) (Jouannet et al., 2015; Nieminen et al., 2015; Bhalerao and Fischer, 2017).
SLs have been reported to play an important role in secondary growth by promoting cambium activity (Agusti et al., 2011; Li et al., 2014). Both SL biosynthetic and signaling mutants show reduced cambium cell layers, and localized treatment with GR24, an SL analog, enhances the meristematic activity of the VC (Agusti et al., 2011). Furthermore, a loss of function mutation in MORE AXILLARY SHOOT2 (MAX2) with reduced secondary growth can be fully rescued by the expression of MAX2 driven by a cambium-specific promoter (Agusti et al., 2011). In Arabidopsis (Arabidopsis thaliana), the downstream components involved in the SL-regulated branching, including the DWARF53 (D53)-like SUPPRESSOR OF MAX2-1 LIKE (SMXL) family and BRI1-EMS-SUPPRESSOR1 (BES1), have been identified, and they are ubiquitinated and degraded by the SCFMAX2 complex in an SL-dependent manner (Jiang et al., 2013; Wang et al., 2013, 2015; Zhou et al., 2013; Soundappan et al., 2015; Liang et al., 2016; Wallner et al., 2017; Hu et al., 2019, 2020). However, what role, if any, they play in the SL-regulated vascular development is unclear. Indeed, both overexpression and loss-of-function mutants of SMXL6, SMXL7, and SMXL8 are characterized by a reduced stem diameter, illustrating the complex role played by SMXL proteins in secondary growth (Liang et al., 2016). Likewise, BES1 has been reported to promote xylem differentiation downstream of the TRACHEARY ELEMENT DIFFERENTIATION INHIBITORY FACTOR (TDIF)–TDIF RECEPTOR (TDR)–GLYCOGEN SYNTHASE KINASE3s (GSK3s) pathway (Kondo et al., 2014; Saito et al., 2018). However, whether BES1 functions downstream of the SL signaling to regulate cambium cell proliferation is unknown.
The transcription factor WUSCHEL-related HOMEOBOX4 (WOX4) and its homolog, WOX14, act downstream of TDIF, encoded by CLAVATA3/EMBRYO SURROUNDING REGION-related 41 (CLE41) and CLE44 in Arabidopsis, a small peptide secreted from the phloem, and its procambial-localized receptor TDR. The recognition of TDIF by TDR induces WOX4 expression, which initiates the self-division of cambium cells. In addition, although TDIF-TDR signaling inhibits the BES1-mediated transcriptional regulation by activating a GSK3-like kinase, BRASSINOSTEROID-INSENSITIVE2 (BIN2), thus repressing xylem differentiation independently of the TDIF–TDR–WOX4 pathway (Ito et al., 2006; Fisher and Turner, 2007; Hirakawa et al., 2008; Whitford et al., 2008; Kondo et al., 2014), how TDIF induces WOX4 expression to promote cell proliferation and simultaneously inhibit cell differentiation in the cambium is unknown.
In this study, we cytologically examined stem cross sections undergoing secondary growth from the mutated lines with compromised function in MAX2, BES1, and WOX4 in Arabidopsis. We demonstrate that BES1 plays an important role in inhibiting cambium cell proliferation by repressing WOX4, which also accounts for a reduced cambium activity in the max2-1 mutant. Furthermore, we show that BES1 directly binds to the WOX4 promoter to inhibit its expression. Based on these observations, we propose that BES1 and WOX4 exert antagonistic roles that enable intercellular TDIF signals to efficiently maintain the appropriate balance between cambium proliferation and differentiation in response to various signals.
Results
BES1 negatively regulates cambium activity in the SL signaling pathway
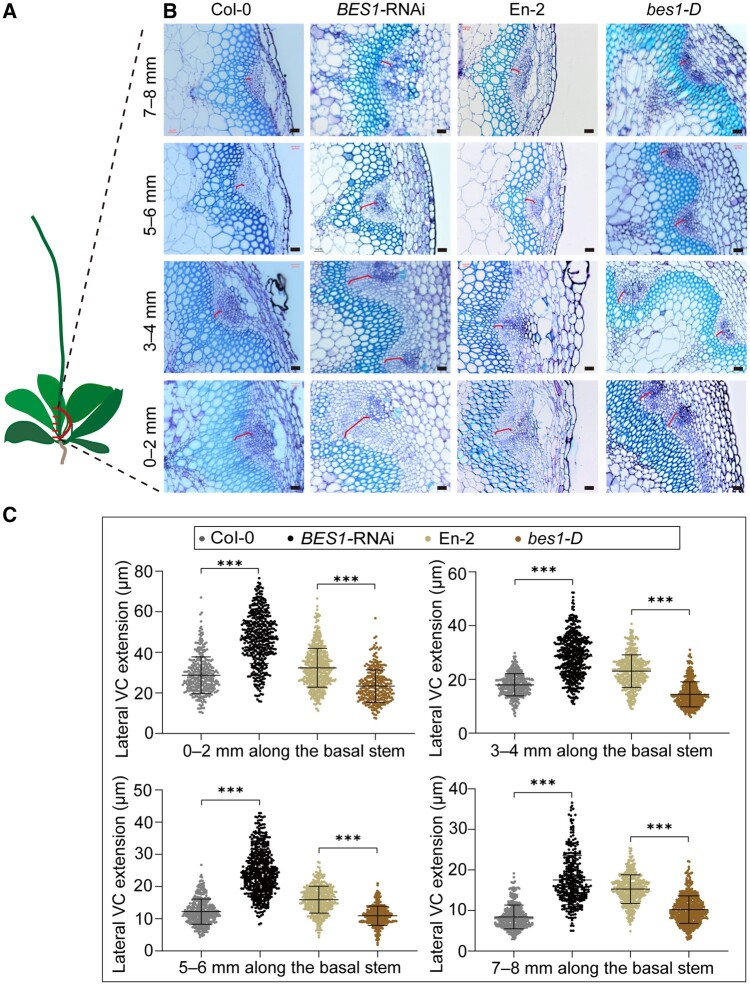
SL signaling promotes cambium cell proliferation during plant secondary growth by an unknown mechanism (Agusti et al., 2011). Recently, several independent studies demonstrated that BES1 and its homologs conservatively act downstream of the D14-MAX2 signaling module in shoot branching (Wang et al., 2013; Fang et al., 2019; Hu et al., 2019, 2020). In addition, because BES1 has been reported to function in vascular development by promoting xylem and phloem differentiation (Kondo et al., 2014; Saito et al., 2018), we hypothesized that BES1 might play a role in the SL-regulated VC cell proliferation during secondary growth. To test this hypothesis, we first characterized the VC in cross sections from the basal stems of the gain-of-function mutant bes1-D and a BES1 RNA interference (BES1-RNAi) line, immediately above the joint of the hypocotyl and shoot. We recorded cambium activity by scoring the width of the lateral VC expanded between the phloem and xylem of the basal stem (Supplemental Figure S1). We divided the 8-mm stem segments into four equal fragments, which we labeled as 0–2, 3–4, 5–6, and 7–8 mm (Figure 1, A). The cambium region in the BES1-RNAi line was much wider than that in wild-type Col-0 (Figure 1, B and C, and Supplemental Figure S2). By contrast, the corresponding VC region was reduced in the bes1-D mutant compared with wild-type En-2 (Figure 1, B and C, and Supplemental Figure S2), which was similar in their hypocotyl diameter (Supplemental Figure S3). Thus, it suggests that BES1 plays a negative role in regulating cambium activity. We next analyzed BES1 expression pattern in the vasculature with the β-GLUCURONIDASE (GUS) reporter line harboring the pBES1(L)pro:GUS and pBES1(S)pro:GUS reporters (BES1(L), an Arabidopsis-specific long isoform of BES1 that contains an additional N-terminal bipartite nuclear localization signal and has stronger activity in promoting BR signaling than the canonical and widely used short BES1(S) [Jiang et al., 2015]). GUS staining was strong for both reporter lines in the basal stem and the cambium (Supplemental Figure S4). These results indicate that BES1 plays an important role in inhibiting cambium activity.
Figure 1.
BES1 inhibits VC cell proliferation. A, Schematic showing the regions from the basal stem used for sections in (B). B, Representative cross sections show the extension of lateral VC in the basal stems from Col-0, BES1-RNAi, En-2, and bes1-D plants. A segment of 8 mm collected immediately above the shoot–hypocotyl junction was cut into four equal parts, marked as 0–2, 3–4, 5–6, and 7–8 mm according to their position along the stem. Cross sections of each fragment were observed and counted randomly. The red bracket indicates the measured areas. Bars represent 20 μm. C, Quantification of lateral VC extension in (B) by ImageJ. Data are means ± se. n > 300 for each sample were analyzed using ANOVA with post hoc Tukey’s test. ***P < 0.001.
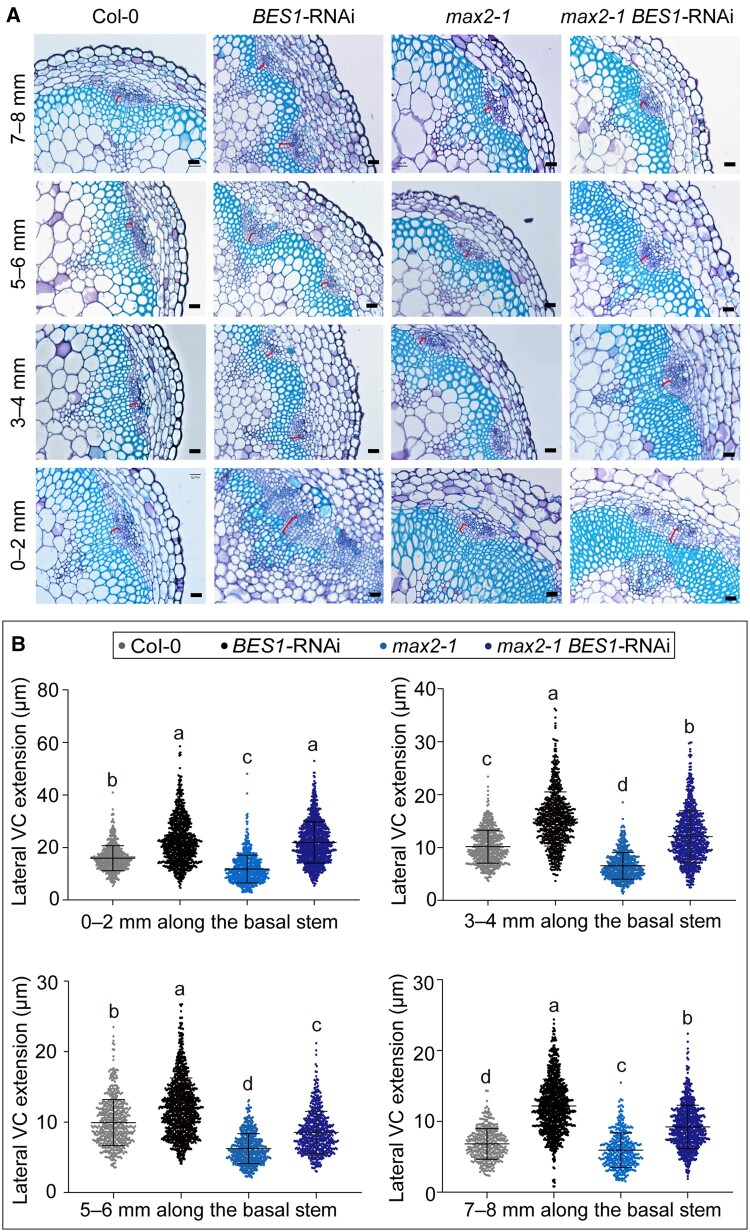
To test whether the BES1-regulated VC activity accounts for the SL-mediated secondary growth, we then generated a line in which the max2-1 mutant also carries the BES1-RNAi construct and quantified the cambium activity of the max2-1 BES1-RNAi line as above (Supplemental Figure S5). A stem segment of 8 mm was equally divided into four fragments: 0–2, 2–4, 5–6, and 7–8 mm. Knockdown of BES1 and its homologs significantly rescued VC activity of secondary stem and hypocotyls in the max2-1 BES1-RNAi line, which resembled that in the BES1-RNAi line but not in the max2-1 mutant (Figure 2, A and B, and Supplemental Figures S2, S3), suggesting that BES1 acts downstream of SL signaling to negatively regulate cambium activity during the secondary growth.
Figure 2.
BES1 acts downstream of SL signaling to regulate cambium activity. A, Representative cross sections show the extension of lateral VC in the basal stems of Col-0, max2-1, BES1-RNAi, and max2-1 BES1-RNAi plants. A segment of 8 mm collected immediately above the shoot–hypocotyl junction was cut into four equal parts, marked as 0–2, 3–4, 5–6, and 7–8 mm. The red bracket indicates the measured areas. Bars represent 20 μm. B, Quantification of lateral VC extension in (A) by ImageJ. Data are means ± se. n > 300 for each sample were analyzed using ANOVA with post hoc Tukey’s test. Means with a P-value ≤ 0.05 were considered statistically different among multiple comparisons. Distinct letters (a–c) indicate statistically significant difference among all samples.
BES1 inhibits cambium cell proliferation through WOX4
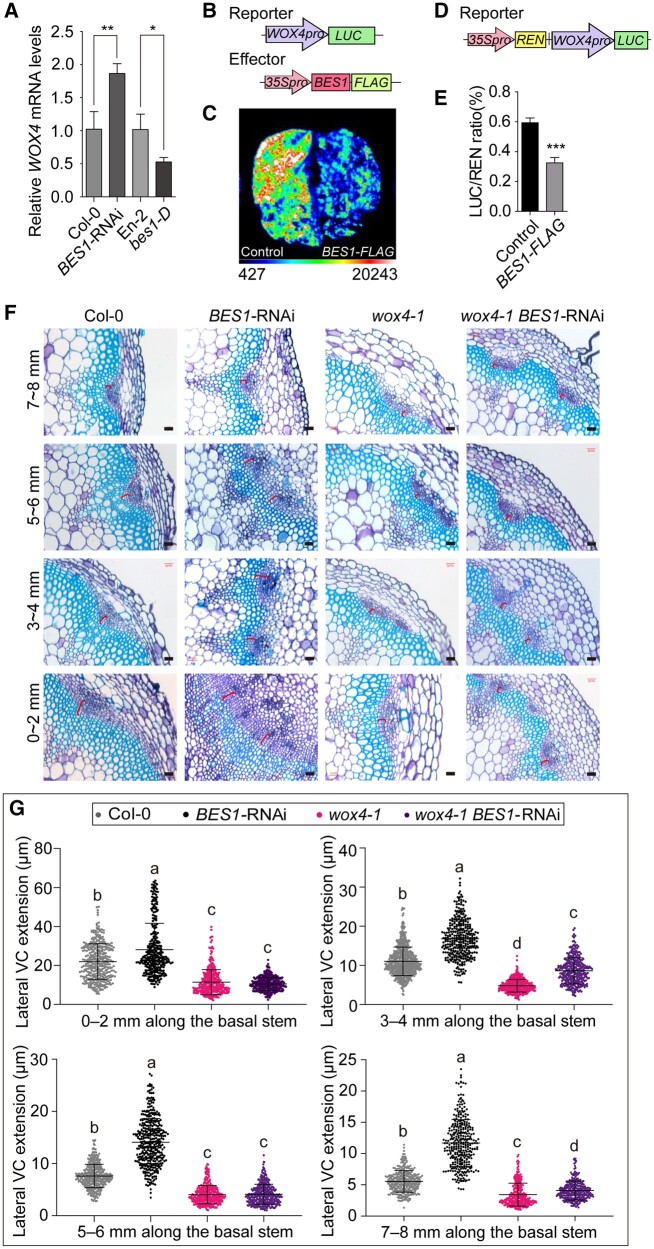
Cell division in the cambium has been proposed to be regulated by the intercellular TDIF–TDR–WOX4 signaling pathway in Arabidopsis (Etchells et al., 2013; Campbell and Turner, 2017). In addition, BES1 activity is repressed by GSK3s in a TDIF-dependent manner to regulate xylem differentiation (Kondo et al., 2014). Therefore, we asked whether BES1 might regulate cambium activity through WOX4. We first tested WOX4 transcript levels in the seedlings of the BES1-RNAi line, the bes1-D mutant, and their corresponding wild types. The WOX4 transcript levels were significantly higher in the BES1-RNAi seedlings and lower in the bes1-D mutant when compared with the respective wild types (Figure 3, A), indicating that BES1 possibly inhibits WOX4 transcription to regulate the cambium activity.
Figure 3.
BES1 inhibits cambial cell proliferation by repressing WOX4 transcription. A, Relative expression levels of WOX4 in Col-0, BES1-RNAi, En-2, and bes1-D. Data are means ± sd (n = 3). P values were determined by Student’s t test; **P < 0.01; *P < 0.05. B, Diagrams representing the luciferase reporter and effector constructs used in the luciferase assays (C). The WOX4 promoter drives the expression of the firefly LUC reporter. The BES1-FLAG effector construct is driven by the 35S promoter. We used 35Spro:GFP as a negative control. C, Transient luciferase assays in N. benthamiana leaves demonstrate the repression of WOX4pro:LUC activity by co-expression with BES1-FLAG. D, Diagram of the reporter used for the dual bioluminescence assay (E). The reporter consists of the WOX4pro:LUC cassette placed downstream of 35Spro:REN. E, Dual-luciferase assays in N. benthamiana leaves confirm the repression of luciferase activity from the WOX4pro:LUC reporter by the BES1-FLAG effector. Data are means ± sd (n = 3). P values were determined by Student’s t test. ***P < 0.001. F, Representative cross sections illustrating the extension of lateral VC in the basal stems of Col-0, BES1-RNAi, wox4-1, and wox4-1 BES1-RNAi plants. A segment of 8 mm was collected at the base of the stem immediately above the shoot–hypocotyl junction and cut into four equal parts, marked as 0–2, 3–4, 5–6, and 7–8 mm. The red bracket indicates the measured areas. Bars represent 20 μm. G, Quantification of lateral VC extension in (F) by ImageJ. Data are means ± se. n > 300 for each sample were analyzed using ANOVA with post hoc Tukey’s test. Means with a P value ≤ 0.05 were considered statistically different among multiple comparisons. Distinct letters (a–c) indicate statistically significant difference among all samples.
We then used a luciferase (LUC) reporter system in Nicotiana benthamiana leaves to test whether BES1 can directly regulate the WOX4 transcription. To this end, we cloned the WOX4 promoter driving the LUC gene to generate a WOX4pro:LUC reporter, 35Spro:BES1-FLAG as the effector and 35Spro:GFP as a negative control (Figure 3, B). We found that luciferase activity emitted from N. benthamiana leaves infiltrated with the WOX4pro:LUC reporter was much lower when the reporter was co-infiltrated with the 35Spro:BES1-FLAG compared with the 35Spro:GFP (Figure 3, C and Supplemental Figure S6). Using a dual bioluminescence assay in N. benthamiana leaves with a construct harboring both the WOX4pro:LUC reporter and the 35Spro:REN (Renilla) as an internal control (Figure 3, D), we observed that luciferase activity from the WOX4pro:LUC decreased ∼50% when the 35Spro:BES1-FLAG effector was co-infiltrated (Figure 3, E). Thus, we conclude that BES1 inhibits WOX4 transcription.
To test whether the altered expression of WOX4 in the BES1-RNAi plants may account for the enhanced cambium activity in the BES1-RNAi plants, we crossed BES1-RNAi with the wox4-1 mutant to generate the wox4-1 BES1-RNAi line. We then measured cambium activity in Col-0, the wox4-1, the BES1-RNAi, and the wox4-1 BES1-RNAi plants. The wox4-1 BES1-RNAi line had reduced VC region than that in BES1-RNAi plants, but comparable to that in the wox4-1 mutant, indicating that the loss of WOX4 function largely suppresses the enhanced VC length (Figure 3, F and G, and Supplemental Figure S2) and the hypocotyl diameter (Supplemental Figure S3) in the BES1-RNAi line. We conclude that BES1 inhibits the proliferation of VC cells by repressing WOX4 expression.
BES1 directly binds to the WOX4 promoter
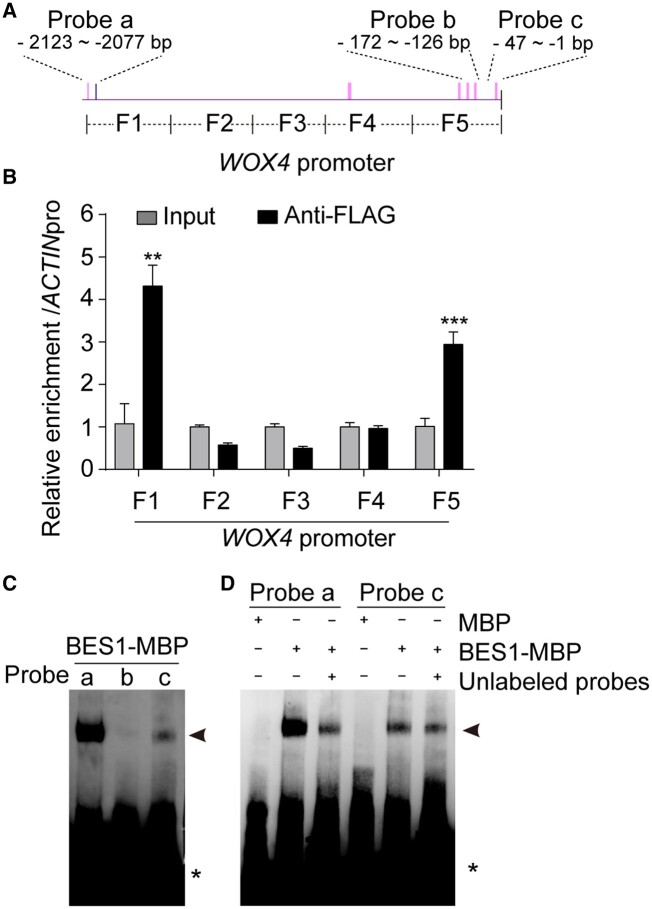
To test whether BES1 directly inhibits WOX4 expression, we performed chromatin immunoprecipitation-quantitative polymerase chain reaction (ChIP-qPCR) assays using the anti-FLAG beads and chromatin purified from the 35Spro:BES1-FLAG and Col-0 seedlings. We determined the extent of binding by qPCR using five fragments (F1–F5) spanning the WOX4 promoter (Figure 4, A). The results showed that fragments F1 and F5, which contain the cis-E-box (CANNTG) or the BRRE (BR response element) box (CGTGT/CG), were highly enriched by immunoprecipitation with the BES1-FLAG in vivo (Figure 4, B).
Figure 4.
BES1 represses WOX4 transcription by directly binding to its promoter. A, Schematic representation of the WOX4 promoter fragment and position of probes used for ChIP and EMSAs. Pink vertical bars indicate E-box (CANNTG) elements. Blue vertical bars indicate the position of BR response elements (BRREs). B, BES1-FLAG immunoprecipitates the F1 and F5 fragments of the WOX4 promoter in ChIP-qPCR assays. The relative enrichment of the WOX4 promoter relative to the ACTIN promoter is normalized to the input. Data are means ± sd (n = 3). P values were determined by Student’s t test. ***P < 0.001, **P < 0.01. C, BES1-MBP binds to probes a and c. D, BES1-MBP specifically binds to the WOX4 promoter. Black arrowheads indicate the position of the shifted BES1-MBP-WOX4 promoter complexes. Asterisks (*) indicate free probe.
We further validated the binding of BES1 to these promoter fragments by performing electrophoretic mobility shift assays (EMSAs). Accordingly, we synthesized the biotin-labeled probes (probes a, b, and c) covering the potential binding sites in fragments F1 and F5 (Figure 4, A). We incubated the purified recombinant BES1-maltose binding protein (MBP) with each probe and observed the BES1-MBP binding to probes a and c (Figure 4, C). The binding was specific, as successfully competed binding was observed between the recombinant protein and the labeled probes by unlabeled probes (Figure 4, D). These results indicate that BES1 binds to the WOX4 promoter to directly inhibit WOX4 expression and repress VC cell proliferation in Arabidopsis.
SL signaling regulates secondary growth through WOX4
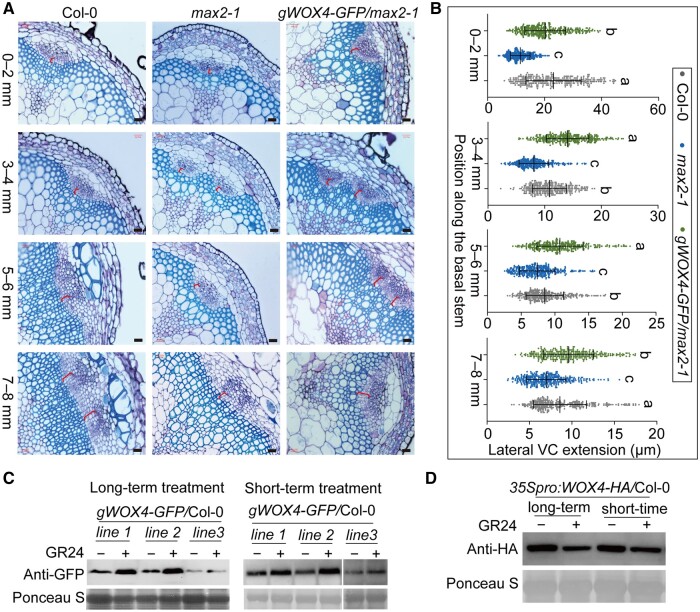
WOX4 not only acts downstream of the TDIF–TDR signaling pathway to control vascular cell proliferation, but also acts as a signal integrator that links TDIF signaling and phytohormone signaling, such as auxin, GAs, and ethylene (Hirakawa et al., 2010; Suer et al., 2011; Etchells et al., 2012; Denis et al., 2017; Kucukoglu et al., 2017). Thus, we asked whether WOX4 expression is also regulated by the SL signaling. We first measured the WOX4 transcript levels in the SL biosynthesis mutant max3-1 and the signaling mutants max2-1 and Atd14-1. The WOX4 expression was lower in these SL mutants (Supplemental Figure S7), which is consistent with a previous study that showed reduced WOX4 transcription in the SL biosynthesis mutant max1-1 (Agusti et al., 2011). Next, to detect whether WOX4 genetically acts downstream of SL signaling pathway in regulating secondary growth, we introduced the genomic WOX4-GFP driven by the WOX4 promoter into the max2-1 background to generate the gWOX4-GFP/max2-1 plants (Supplemental Figure S8). We noticed a marked improvement of VC development in the basal stem of the gWOX4-GFP/max2-1 plants relative to max2-1 (Figure 5, A and B, and Supplemental Figure S2). In agreement with this result, the hypocotyl of the gWOX4-GFP/max2-1 plants was thicker than those of the max2-1 mutant (Supplemental Figures S3, S8, S9). In fact, the increase in hypocotyl diameter was positively associated with the WOX4 expression levels across various gWOX4-GFP/max2-1 transgenic lines (Supplemental Figure S9).
Figure 5.
The SL signaling pathway regulates cambium activity by inducing WOX4 accumulation at the transcriptional level. A, Representative cross sections showing the extension of lateral VC in the basal stems of Col-0, max2-1, and gWOX4-GFP/max2-1 plants. A segment of 8 mm at the base of the main stem was collected immediately above the shoot–hypocotyl junction and cut into four equal parts, marked as 0–2, 3–4, 5–6, and 7–8 mm. The red brackets indicate the measured areas. Bars represent 20 μm. B, Quantification of lateral VC extension in (A) by ImageJ. Data are means ± se. n > 300 for each sample were analyzed using ANOVA with post hoc Tukey’s test. Means with a P value ≤ 0.05 were considered statistically different among multiple comparisons. Distinct letters (a–c) indicate statistically significant difference among all samples. C, WOX4-GFP protein accumulation is induced by GR24 treatment. The gWOX4-GFP/Col-0 seedlings were grown for one week on 1/2 MS medium under mock treatment (–) or with 5 μM GR24 (+) as the long-term treatment. The short-term treatment was performed with either mock (–) or 5 μM GR24 treated for 1 h using the one-week-old gWOX4-GFP/Col-0 seedlings. D, WOX4-HA protein accumulation decreases in 35Spro:WOX4-HA/Col-0 seedlings after GR24 treatment. 35Spro:WOX4-HA/Col-0 seedlings were grown for 1 week on half-strength MS medium with mock (–) or with 5 μM GR24 (+) (long-term), or they were treated with either mock (–) or 5 μM GR24 for 1 h after growth on half-strength MS medium for 1 week.
Because protein stability of the downstream components in the SL signaling pathway is a major regulatory mechanism for SL signal transduction (Jiang et al., 2013; Wang et al., 2013, 2015; Zhou et al., 2013; Soundappan et al., 2015), we next tested whether SL signals also regulate the WOX4 protein stability. We introduced the gWOX4-GFP, mentioned above, into Col-0 and grew the seedlings on half-strength Murashige and Skoog (MS) medium with or without 5 μM GR24 for 1 week. When we measured the WOX4-GFP protein levels in the transgenic lines carrying gWOX4-GFP, we found that the WOX4-GFP protein levels were enhanced in response to a long-term treatment with GR24 (Figure 5, C). Even a 1-h treatment with 5 μM GR24 was sufficient to raise the WOX4-GFP protein levels over those of the mock control (Figure 5, C). By contrast, the 35S:WOX4-HA transgenic plants subjected to the same treatment of 5 μM GR24 showed a reduction in the WOX4-HA protein levels after both short-term (1 h) and long-term (1 week) GR24 treatments (Figure 5, D). We further detected that the increased protein level of WOX4 was caused by a higher WOX4 transcript level in Col-0 seedlings. Indeed, the WOX4 transcription was increased in the GR24-treated seedlings when compared with the mock-treated seedlings (Figure 6, A). Therefore, the SL signaling may not directly regulate the WOX4 protein stability, but rather the WOX4 expression levels through BES1. The altered cambium activity caused by WOX4 in the BES1-RNAi line, bes1-D, and SL-deficient mutants may thus depend on the regulation of WOX4 transcription through BES1. We conclude that SL signaling regulates cambium activity during secondary growth by regulating BES1 stability, which in turn alters WOX4 transcription to regulate cambium cell proliferation.
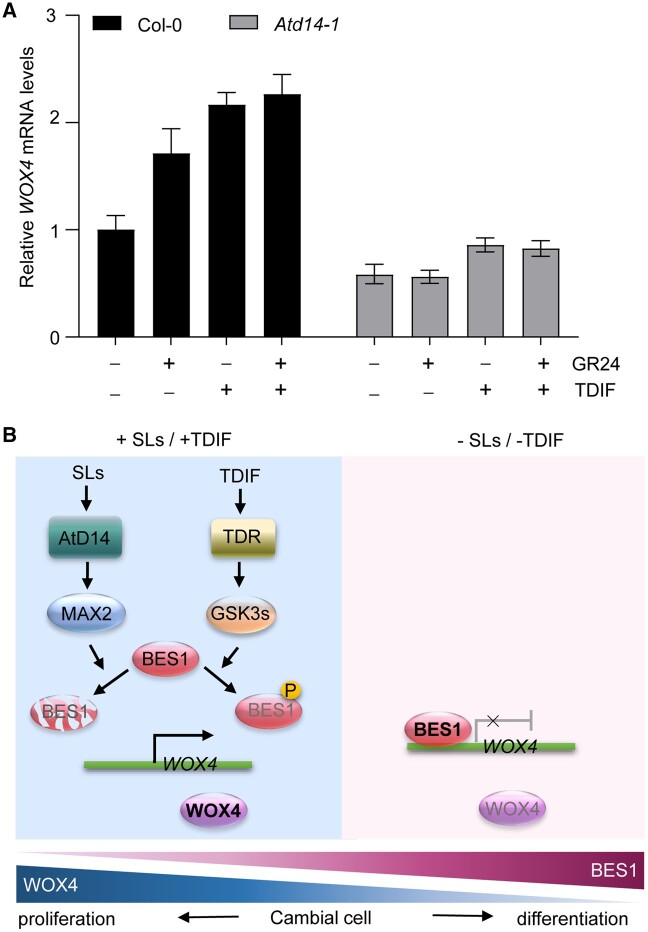
Figure 6.
A proposed model illustrating that BES1 and WOX4 mediate SL and TDIF signaling to regulate cambial cell proliferation and differentiation in Arabidopsis. A, Relative WOX4 mRNA levels in Col-0 and Atd14-1 seedlings grown for 1 week on 1/2 MS medium with 5 μM GR24 (+), 1 μM TDIF, or both, or a mock treatment (–). Mean ± sd, n = 3. B, The model of SL and TDIF signaling in secondary growth. During secondary growth, BES1 and WOX4 antagonistically regulate bidirectional activity of cambial cell, during which WOX4 promotes cell proliferation while BES1 represses WOX4 transcription to promote xylem and phloem differentiation. SL signaling promotes cambium activity by inducing BES1 degradation, which leads to higher WOX4 expression and cambial proliferation. Similarly, the TDIF-induced signaling inhibits BES1 transcriptional activity through the TDR-GSK3s pathway, which also promotes WOX4 expression and cambium proliferation. In contrast, when the SL/TDIF signaling is reduced, BES1 stability or transcriptional activity will increase, which then suppresses WOX4 expression and cambium proliferation, but enhances cambial cell differentiation (TDIF, Tracheary element differentiation inhibitory factor).
Discussion
In this study, we provide several lines of evidence to demonstrate that BES1 acts downstream of SL perception to inhibit cambium cell proliferation by directly repressing WOX4 transcription. First, we showed the altered cambium width in the basal stems of the BES1-RNAi line and the bes1-D mutant (Figure 1), as well as strong GUS staining from the pBES1(L)pro:GUS and pBES1(S)pro:GUS reporter constructs in the secondary VC (Supplemental Figure S4), which supports a role for BES1 as a negative regulator in cambium proliferation during secondary growth. Second, the analysis of the BES1-RNAi line and the max2-1 mutant provides genetic evidence that BES1 acts downstream of SL signaling in regulating cambium activity (Figure 2 and Supplemental Figure S5), in addition to its known role as an essential component of the SL–AtD14–MAX2 pathway for shoot branching (Wang et al., 2013; Fang et al., 2019; Hu et al., 2019, 2020). Third, BES1 inhibits cambium proliferation by directly repressing WOX4 transcription (Figures 3, 4 and Supplemental Figure S6). The characterization of wox4-1 BES1-RNAi and gWOX4-GFP/max2-1 plants demonstrated that the altered WOX4 expression fully explains the altered cambial phenotypes observed in the max2-1 and BES1-RNAi plants (Figures 3, F and G, 5, A and B, and Supplemental Figures S3, S8, S9). Therefore, we have uncovered the molecular mechanism by which SL signaling regulates secondary growth in Arabidopsis.
BES1 was previously reported to promote xylem differentiation, but our study expands the roles of BES1 in the promotion of cambium cell proliferation during plant secondary growth. Upon activation of intercellular TDIF signals, GSK3 family kinases phosphorylate and repress BES1 protein, leading to an inhibition of xylem differentiation (Kondo et al., 2014, 2016). Here, analysis of paraffin sections of the cambium region in the stem of bes1-D and BES1-RNAi plants revealed that BES1 can also positively regulate cambium cell proliferation.
Furthermore, our study revealed that TDIF signaling induces WOX4 expression by inhibiting the transcriptional activity of BES1 to efficiently and simultaneously regulate cambium cell division and differentiation. TDIF signals are thought to be transduced by two independent signaling branches: TDIF–TDR–WOX4 and TDIF–TDR–BIN2–BES1. The TDIF–TDR–WOX4 signaling module promotes the self-proliferation of cambium cells by inducing WOX4 expression (Hirakawa et al., 2010; Etchells et al., 2013). In agreement, the wox4 mutant displays reduced cambium cell proliferation, but normal xylem differentiation in secondary growth (Suer et al., 2011). The second signaling module, TDIF–TDR–BIN2–BES1, was initially thought to repress xylem differentiation, based on the observation that wox4 mutant plants treated with bikinin showed additive effects on the ratio of adjacent xylem and phloem in root vasculature (Kondo et al., 2014). However, how procambium cell numbers might change in the above pathways was not determined, leaving open the question of whether BIN2-BES1 affects cambium cell proliferation. We propose here that the activation of TDIF signaling induces WOX4 transcription likely by relieving the direct binding of BES1 to the WOX4 promoter.
It is likely that BES1 and WOX4 form a signaling hub to regulate the dynamic coordination of cambium cell proliferation and differentiation in response to changing environmental and phytohormonal cues. Secondary growth is regulated by many climatic factors, including light, photoperiod, and temperature, as well as edaphic factors, such as drought, salt, and other stresses, and various phytohormones (Arend and Fromm, 2007; Escalante-Perez et al., 2009; Berta et al., 2010; Tanino et al., 2010; Balducci et al., 2013; Begum et al., 2013; Maurya and Bhalerao, 2017). WOX4 is involved in several phytohormone-regulated mechanisms of cambium cell proliferation (Agusti et al., 2011; Etchells et al., 2012; Denis et al., 2017; Kucukoglu et al., 2017; Brackmann et al., 2018). For example, auxin induces WOX4 transcription, and the wox4 mutant fails to enhance cambium activity upon auxin treatment (Suer et al., 2011). Similarly, the transcription factor AUXIN RESPONSE FACTOR5 (ARF5) attenuated cambium proliferation by directly repressing WOX4 expression (Brackmann et al., 2018). Ethylene has also been reported to positively regulate vascular cell division by interacting with the TDR–WOX4 pathway (Etchells et al., 2012). BES1 and its homologs function in several signaling cascades and modulate various aspects of plant growth in response to environmental factors, such as light, temperature, and drought stress (Sun et al., 2015; Nolan et al., 2017; Yang et al., 2017; Yang and Wang, 2017; Li et al., 2018). As the long-distance transport system of plants, the vasculature must be dynamically regulated to ensure appropriate xylem and phloem development to meet the changing demands for water and various nutrients. For instance, light promotes enhanced cambium cell proliferation, which is accompanied by reduced xylem differentiation, which might be attributed to the repression of BES1 transcriptional activity (Li et al., 2018; Wang and Lu, 2018; Wu et al., 2019) and the resulting induction of WOX4 transcription to promote cambium cell divisions. As expected, TDIF and GR24 treatment on the Atd14 mutant and the wild type Col-0 demonstrated that both SL and TDIF can positively regulate WOX4 expression, and only TDIF treatment can induce WOX4 expression in the Atd14 mutant (Figure 6, A). Taken together, it was demonstrated that BES1-WOX4 may act as a downstream hub for multiple signaling pathways in secondary growth.
Based on this and previous studies, we propose a model in which BES1 generally represses WOX4 transcription to promote xylem and phloem differentiation (Figure 6, B). When the TDIF signaling peptide is upregulated by other environmental or phytohormonal factors, the transcriptional activity of BES1 is inhibited by TDR-GSK3s, which in turn promotes WOX4 transcription and results in cambium cell proliferation. Similarly, in SL signaling, SLs induce BES1 degradation, which will promote WOX4 expression to enhance cambium cell proliferation. Therefore, under various endogenous and environmental cues, the regulation of BES1 activity and WOX4 expression provides a dynamic balance between cell differentiation and proliferation of cambium cells. How phytohormones and environmental signals modulate cambium development and intersect with the BES1-WOX4 axis is a fascinating direction for future investigations.
Materials and methods
Plant materials and growth conditions
Arabidopsis (A. thaliana) accessions Columbia-0 (Col-0) and Enkheim-2 (En-2) were used as wild-type controls in these experiments. The Arabidopsis mutant alleles and transgenic lines used in this study were wox4-1 (SALK_210239), max2-1 (SALK_092836), BES1-RNAi, and bes1-D. We introduced the BES1-RNAi transgene into single mutants by genetic crossing to generate max2-1 BES1-RNAi and wox4-1 BES1-RNAi. The BES1-RNAi line and the bes1-D mutant were obtained from Yin et al. (2002). Transgenic plants were generated in the Col-0 background: 35Spro:BES1-FLAG, 35Spro:WOX4-HA, and gWOX4-GFP (genomic WOX4 driven by ∼2 kb of WOX4 upstream regulatory sequences, primers WOX4pro-F and WOX4-CDs-R are listed in Supplemental Table S1). The gWOX4-GFP transgene was also introduced into the max2-1 mutant. Transgenic lines homozygous for the BES1(S)pro:GUS and BES1(L)pro:GUS reporters were previously described (Jiang et al., 2015). Surfaced-sterilized seeds were sown on 0.8% agar plates containing half-strength MS medium (pH 5.8). Plates were kept in the dark for 2 d at 4°C, before release at 22°C under a long-day photoperiod (16-h light/8-h dark) for 7–10 d. Seedlings were then transferred to soil and grown in the same conditions as above.
For long-term GR24 treatment, surfaced-sterilized seeds were sown on 0.8% agar plates containing half-strength MS medium (pH 5.8) with 5 μM GR24, 1 μM TDIF, both or mock (with acetone at the same volume as that used for GR24), and grow under a long-day photoperiod (16-h light/8-h dark) for 7 d after being kept in the dark for 2 d at 4°C. For the short-term GR24 treatment, seedlings grown under the above conditions on 0.8% agar plates containing half-strength MS medium (pH 5.8) for 7 d were collected in half-strength MS medium (pH 5.8) with 5 μM GR24 or mock (acetone) and 0.01% (v/v) Silwet L-77 for 1 h.
Construction of plasmids and generation of transgenic lines
The backbone for all plasmid constructs used to generate transgenic lines in this study was pCAMBIA1300 with different tags. All the recombinant vectors were generated using Seamless Cloning Kit (Vazyme). To generate the transgenic plants 35Spro:WOX4-HA in Col-0, we cloned the full-length coding sequences of WOX4 into pCAMBIA1300, with a HA tag to place them under the control of the Cauliflower Mosaic Virus (CaMV) 35S promoter for GR24 treatment assays. The short-form BES1 coding sequence with a FLAG tag placed under the control of the CaMV 35S promoter was linked into pCAMBIA1300 to obtain the 35Spro:BES1-FLAG effector for LUC assays.
Likewise, we cloned the WOX4 full-length genomic sequence (including the single intron) into pCAMBIA1302 containing the GREEN FLUORESCENT PROTEIN (GFP) coding sequence to generate gWOX4-GFP in Col-0 and max2-1. Primers WOX4pro-F and WOX4-CDs-R used to amplify the genomic DNA and primers WOX4-CDs-F and WOX4-CDs-R used to amplify coding sequences of WOX4 are listed in Supplemental Table S1. All the plasmids and restriction sites used to generate transgenic lines are listed in Supplemental Table S2.
Histological analysis
We grew plants on soil for about 2 months before harvesting 8-mm-long sections of the inflorescence basal stem, immediately above the junction between the main shoot and the hypocotyl. We then divided these stem segments into four equal sections according to their positions along the basal stem, marked as 0–2, 3–4, 5–6, and 7–8 mm (Supplemental Figure S1). The diameter of hypocotyl (2 mm immediately below the shoot-hypocotyl junction) of 4-week-old plants (or about 15 cm high) grown on soil was immediately measured after being pulling out from soil. All tissues were fixed in formalin/acetic acid/alcohol (FAA; 1:1:18) with vacuum infiltration at room temperature and incubated for at least 24 h. Fixed tissue samples were then transferred to a graded ethanol series (50%, 70%, 80%, 85%, 95%, and 100% [v/v]; in this order) for 1 h each. Tissues were then subjected to a graded chloroform series (20%, 40%, 60%, 80%, and 100% chloroform [v/v], in this order) for 1 h each. All samples were then embedded in 100% wax at 56°C after immersion successively in 50%, 70%, and 100% wax (w/v) for 2 h each. The embedded samples were cut into 6–10-µm-thick sections with a LEICA rotary microtome (model RM2165). Sliced samples were stained with 0.1% toluidine blue for 1 min. The radial width of the VC (defined as the lateral VC extension) was scored randomly across multiple sections from at least five independent plants per line and measured by ImageJ. Cross sections from five plants were observed and randomly collected at least 300 numbers per genotype. The VC lengths were measured as shown in Supplemental Figure S1.
GUS staining assay
Histochemical analysis of GUS expression was performed as described (Jiang et al., 2015). Transgenic plants harboring the BES1(S)pro:GUS or BES1(L)pro:GUS reporter construct were grown on soil for 3 weeks before harvesting. We collected the base of the main stem, about 2 cm from the shoot–root junction, before soaking in pre-chilled acetone for 30 min for fixation before washing and staining in X-Gluc solution (100 mM NaH2PO4, pH 7.2, 2 mM K-ferrocyanide, 2 mM K-ferricyanide, 2 mM X-Gluc, and 0.1% Triton X-100) at 37°C in the dark for 12 h (overnight). Samples were then rinsed several times with 70% (v/v) ethanol to remove chlorophyll. The samples were then embedded in paraffin and sectioned as above.
LUC reporter assay and dual bioluminescence assay
For the luciferase (LUC) reporter assays, we cloned the WOX4 promoter (using primers WOX4pro-F and WOX4pro-R) into pCAMBIA1300 harboring the firefly LUC gene as the reporter. For the effector construct BES1-FLAG, the full-length short-form BES1 coding sequence was introduced into pCAMBIA1306 and placed under the control of the 35S promoter, using pCAMBIA1300 carrying a 35Spro:GFP cassette as control, the 35S:HsfA1d:GFP was used as an unrelated transcription factor control (Supplemental Figure S6).
For dual bioluminescence assays, the same WOX4 promoter was cloned into pGreenII 0800-LUC, which also carries a 35Spro:REN reporter as control. The effector BES1-FLAG was as above. Primers are listed in Supplemental Table S1. These vectors were introduced into Agrobacterium (Agrobacterium tumefaciens) strain GV3101 and then infiltrated into young leaves of N. benthamiana. Plants were incubated in the dark for 1 d and transferred to a long-day photoperiod (16-h light/8-h dark) for 2 d. For LUC reporter assays, luminescence signals from pavement cells were detected after applying 2 mM luciferin by a charge-coupled device (CCD) system (LUMAZONE PYLON2048B). For the dual-bioluminescence assay, the luminescence from LUC and REN was detected using the Dual-Luciferase Reporter assay kit on a Mithras LB940 microplate reader.
Chromatin immunoprecipitation
The method described by Fiil et al. (2008) was followed. Briefly, 2-week-old Col-0 and BES1-FLAG seedlings were harvested in Fix Buffer (0.4 M sucrose, 10 mM Tris–HCl, pH 8.0, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1% formaldehyde). Seedlings were infiltrated under vacuum for 30 min for cross-linking. Beads linked to anti-FLAG (40 µL) were used to immunoprecipitate BES1-DNA complexes by incubation at 4°C overnight using a rotating mixer wheel. Finally, DNA was isolated by phenol:chloroform and resuspended in 50 μL of MilliQ water. Primers used to detect the enrichment for various WOX4 promoter fragments and for the control ACTIN promoter are listed in Supplemental Table S1.
Electrophoretic mobility shift assay
The amplified BES1 coding sequence was cloned in frame with the MBP gene and introduced into Escherichia coli. The production of the recombinant proteins BES1-MBP and MBP was induced by 0.4 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 24 h at 16°C. Purified recombinant proteins were incubated with biotin-labeled probes or with corresponding unlabeled probes for 30 min in EMSA-binding buffer (Thermo Fisher). Reaction mixtures were separated on non-denaturing polyacrylamide gels, and DNA signals were detected by chemiluminescence. Probes used in this assay are listed in Supplemental Table S1.
Reverse transcription quantitative PCR
For reverse transcription quantitative PCR (RT-qPCR) analysis, seedlings were frozen in liquid nitrogen immediately following harvesting. The frozen samples were ground to a powder in a pre-chilled mortar and pestle. Total RNA was isolated using a Plant Total RNA Extraction Kit (Tiangen), according to the manufacturer’s manual. First-strand cDNA was synthesized from 1 μg total RNA using oligo(dT) primers and a cDNA Synthesis Kit (Takara). RT-qPCR was performed using gene-specific primers (Supplemental Table S1) on a CFX 96 or CFX384 real-time PCR detection system (Bio-Rad) in a total volume of 10 μL containing 2 μL diluted cDNA, 0.25 mM gene-specific primers, and 5 μL SYBR Green Supermix (Bio-Rad). The Arabidopsis U-BOX (At5g15400) gene was used as the internal control. Primers used for RT-qPCR are listed in Supplemental Table S1.
Quantification and statistical analysis
The significance of related comparisons in Figures 1–3, 5 was analyzed using ANOVA with post hoc Tukey’s test. Means with a significant value lower than 0.05 (P-value ≤ 0.05) were considered statistically different among multiple comparisons. Distinct letters (a, b, c) indicate statistically significant difference among all samples in Figures 1–3, 5. Statistical parameters, including the number of replicates (n), the error bars (standard deviation [SD] or standard error [SE]), and statistical significance, are presented in the figure legends. Here, n means the number of measured data for phenotypic analysis or the numbers of technical replicates for RT-qPCR. Cross sections from five plants were observed and randomly collected at least 300 numbers per genotype for phenotypic analysis.
In Figures 3, A and E, 4, B, data were subjected to one-tailed or two-tailed Student’s t tests according to F test results. Asterisks denote statistical significance tests (***P < 0.001; **P < 0.01; *P < 0.05; NS, P > 0.05.) compared with the corresponding controls, unless otherwise specified by lines connecting the sets to be compared.
Accession numbers
Accession numbers of genes involved in this paper are listed in Supplemental Table S3.
Supplemental data
Supplemental Figure S1. Measured regions of lateral VC.
Supplemental Figure S2. Representative images of complete stem sections of different plants.
Supplemental Figure S3. Hypocotyl phenotype of different plants.
Supplemental Figure S4. Expression pattern of BES1(L) and BES1(S) in the secondary stem and vasculature.
Supplemental Figure S5. Identification and phenotypic analysis of the max2-1 BES1-RNAi line.
Supplemental Figure S6. Transient WOX4pro:LUC luciferase assay in N. benthamiana leaves.
Supplemental Figure S7. WOX4 transcript levels in the SL biosynthetic and signaling mutants max3-1, max2-1, and Atd14-1.
Supplemental Figure S8. Phenotype of the stem and hypocotyl in gWOX4-GFP/max2-1 whole plant.
Supplemental Figure S9. Hypocotyl phenotype of gWOX4-GFP/max2-1.
Supplemental Table S1. Primers for genotyping, ChIP, EMSA, qPCR, and recombinant vectors.
Supplemental Table S2. Plasmids and restriction sites used for recombinant vectors.
Supplemental Table S3. Accession numbers of genes.
Supplementary Material
Acknowledgments
Thanks for Yanhai Yin (Development and Cell Biology, Iowa State University) for providing the BES1-RNAi line and bes1-D mutant.
Funding
This work was supported by grant 2015CB910200 of the National Key Basic Research Foundation of China (to X.W.), grants 31870257, 91535104, and 31430046 of the National Natural Science Foundation of China (to X.W.), and grant CX3050A092004 of the Outstanding Talents Fund of Henan University of China (to X.W.).
Conflict of interest statement. The authors declare no potential conflict of interest.
X.W. and J.H. designed the experiments and wrote the manuscript. J.H., X.H., and Y.Y. did most of the experiments and analyzed the data. C.H. and J.H. helped with some experiments.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is Xuelu Wang (xueluw@henu.edu.cn).
One-sentence summary: Transcription factors BES1 and WOX4 play antagonistic roles downstream of strigolactone and intercellular TDIF signaling to regulate secondary growth in Arabidopsis.
References
- Agusti J, Herold S, Schwarz M, Sanchez P, Ljung K, Dun EA, Brewer PB, Beveridge CA, Sieberer T, Sehr EM, et al. (2011) Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proc Natl Acad Sci USA 108: 20242–20247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arend M, Fromm J (2007) Seasonal change in the drought response of wood cell development in poplar. Tree Physiol 27: 985–992 [DOI] [PubMed] [Google Scholar]
- Balducci L, Deslauriers A, Giovannelli A, Rossi S, Rathgeber CB (2013) Effects of temperature and water deficit on cambial activity and woody ring features in Picea mariana saplings. Tree Physiol 33: 1006–1017 [DOI] [PubMed] [Google Scholar]
- Begum S, Nakaba S, Yamagishi Y, Oribe Y, Funada R (2013) Regulation of cambial activity in relation to environmental conditions: understanding the role of temperature in wood formation of trees. Physiol Plant 147: 46–54 [DOI] [PubMed] [Google Scholar]
- Berta M, Giovannelli A, Sebastiani F, Camussi A, Racchi ML (2010) Transcriptome changes in the cambial region of poplar (Populus alba L.) in response to water deficit. Plant Biol (Stuttg) 12: 341–354 [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Fischer U (2017) Environmental and hormonal control of cambial stem cell dynamics. J Exp Bot 68: 79–87. [DOI] [PubMed] [Google Scholar]
- Brackmann K, Qi J, Gebert M, Jouannet V, Schlamp T, Grunwald K, Wallner ES, Novikova DD, Levitsky VG, Agusti J, et al. (2018) Spatial specificity of auxin responses coordinates wood formation. Nat Commun 9: 875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell L, Turner S (2017) Regulation of vascular cell division. J Exp Bot 68: 27–43 [DOI] [PubMed] [Google Scholar]
- Denis E, Kbiri N, Mary V, Claisse G, Conde ESN, Kreis M, Deveaux Y (2017) WOX14 promotes bioactive gibberellin synthesis and vascular cell differentiation in Arabidopsis. Plant J 90: 560–572 [DOI] [PubMed] [Google Scholar]
- Escalante-Perez M, Lautner S, Nehls U, Selle A, Teuber M, Schnitzler JP, Teichmann T, Fayyaz P, Hartung W, Polle A, et al. (2009) Salt stress affects xylem differentiation of grey poplar (Populus × canescens). Planta 229: 299–309 [DOI] [PubMed] [Google Scholar]
- Etchells JP, Provost CM, Turner SR (2012) Plant vascular cell division is maintained by an interaction between PXY and ethylene signalling. PLoS Genet 8: e1002997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchells JP, Provost CM, Mishra L, Turner SR (2013) WOX4 and WOX14 act downstream of the PXY receptor kinase to regulate plant vascular proliferation independently of any role in vascular organisation. Development 140: 2224–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z, Ji Y, Hu J, Guo R, Sun S, Wang X (2019) Strigolactones and brassinosteroids antagonistically regulate the stability of D53-OsBZR1 complex to determine FC1 expression in rice tillering. Mol Plant 13: 586–597 [DOI] [PubMed] [Google Scholar]
- Fiil BK, Qiu JL, Petersen K, Petersen M, Mundy J (2008) Coimmunoprecipitation (co-IP) of nuclear proteins and chromatin immunoprecipitation (ChIP) from Arabidopsis. CSH Protoc 2008: pdb.prot5049. [DOI] [PubMed] [Google Scholar]
- Fisher K, Turner S (2007) PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr Biol 17: 1061–1066 [DOI] [PubMed] [Google Scholar]
- Hirakawa Y, Kondo Y, Fukuda H (2010) TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 22: 2618–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y, Shinohara H, Kondo Y, Inoue A, Nakanomyo I, Ogawa M, Sawa S, Ohashi-Ito K, Matsubayashi Y, Fukuda H (2008) Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc Natl Acad Sci USA 105: 15208–15213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Sun S, Wang X (2020) Regulation of shoot branching by strigolactones and brassinosteroids: conserved and specific functions of Arabidopsis BES1 and Rice BZR1. Mol Plant 13: 808–810 [DOI] [PubMed] [Google Scholar]
- Hu J, Ji Y, Hu X, Sun S, Wang X (2019) BES1 functions as co-regulator of D53-like SMXLs to inhibit BRC1 expression in strigolactone-regulated shoot branching in Arabidopsis. Plant Commun 1: 100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H (2006) Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313: 842–845 [DOI] [PubMed] [Google Scholar]
- Jiang J, Zhang C, Wang X (2015) A recently evolved isoform of the transcription factor BES1 promotes brassinosteroid signaling and development in Arabidopsis thaliana. Plant Cell 27: 361–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Liu X, Xiong G, Liu H, Chen F, Wang L, Meng X, Liu G, Yu H, Yuan Y, et al. (2013) DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504: 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouannet V, Brackmann K, Greb T (2015) (Pro)cambium formation and proliferation: two sides of the same coin? Curr Opin Plant Biol 23: 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Ito T, Nakagami H, Hirakawa Y, Saito M, Tamaki T, Shirasu K, Fukuda H (2014) Plant GSK3 proteins regulate xylem cell differentiation downstream of TDIF-TDR signalling. Nat Commun 5: 3504. [DOI] [PubMed] [Google Scholar]
- Kondo Y, Nurani AM, Saito C, Ichihashi Y, Saito M, Yamazaki K, Mitsuda N, Ohme-Takagi M, Fukuda H (2016) Vascular cell induction culture system using Arabidopsis leaves (VISUAL) reveals the sequential differentiation of sieve element-like cells. Plant Cell 28: 1250–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukoglu M, Nilsson J, Zheng B, Chaabouni S, Nilsson O (2017) WUSCHEL-RELATED HOMEOBOX4 (WOX4)-like genes regulate cambial cell division activity and secondary growth in Populus trees. New Phytol 215: 642–657 [DOI] [PubMed] [Google Scholar]
- Li QF, Lu J, Yu JW, Zhang CQ, He JX, Liu QQ (2018) The brassinosteroid-regulated transcription factors BZR1/BES1 function as a coordinator in multisignal-regulated plant growth. Biochim Biophys Acta Gene Regul Mech 1861: 561–571 [DOI] [PubMed] [Google Scholar]
- Li X, Sun S, Li C, Qiao S, Wang T, Leng L, Shen H, Wang X (2014) The strigolactone-related mutants have enhanced lamina joint inclination phenotype at the seedling stage. J Genet Genomics 41: 605–608 [DOI] [PubMed] [Google Scholar]
- Liang Y, Ward S, Li P, Bennett T, Leyser O (2016) SMAX1-LIKE7 signals from the nucleus to regulate shoot development in Arabidopsis via partially EAR motif-independent mechanisms. Plant Cell 28: 1581–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurya JP, Bhalerao RP (2017) Photoperiod- and temperature-mediated control of growth cessation and dormancy in trees: a molecular perspective. Ann Bot 120: 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieminen K, Blomster T, Helariutta Y, Mahonen AP (2015) Vascular cambium development. Arabidopsis Book 13: e0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan T, Chen J, Yin Y (2017) Cross-talk of brassinosteroid signaling in controlling growth and stress responses. Biochem J 474: 2641–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Kondo Y, Fukuda H (2018) BES1 and BZR1 redundantly promote phloem and xylem differentiation. Plant Cell Physiol 59: 590–600 [DOI] [PubMed] [Google Scholar]
- Soundappan I, Bennett T, Morffy N, Liang Y, Stanga JP, Abbas A, Leyser O, Nelson DC (2015) SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27: 3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer R, Groover A (2010) Evolution of development of vascular cambia and secondary growth. New Phytol 186: 577–592 [DOI] [PubMed] [Google Scholar]
- Suer S, Agusti J, Sanchez P, Schwarz M, Greb T (2011) WOX4 imparts auxin responsiveness to cambium cells in Arabidopsis. Plant Cell 23: 3247–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Chen D, Li X, Qiao S, Shi C, Li C, Shen H, Wang X (2015) Brassinosteroid signaling regulates leaf erectness in Oryza sativa via the control of a specific U-type cyclin and cell proliferation. Dev Cell 34: 220–228 [DOI] [PubMed] [Google Scholar]
- Tanino KK, Kalcsits L, Silim S, Kendall E, Gray GR (2010) Temperature-driven plasticity in growth cessation and dormancy development in deciduous woody plants: a working hypothesis suggesting how molecular and cellular function is affected by temperature during dormancy induction. Plant Mol Biol 73: 49–65 [DOI] [PubMed] [Google Scholar]
- Wallner ES, Lopez-Salmeron V, Belevich I, Poschet G, Jung I, Grunwald K, Sevilem I, Jokitalo E, Hell R, Helariutta Y, et al. (2017) Strigolactone- and karrikin-independent SMXL proteins are central regulators of phloem formation. Curr Biol 27: 1241–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang B, Jiang L, Liu X, Li X, Lu Z, Meng X, Wang Y, Smith SM, Li J (2015) Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27: 3128–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lu X (2018) Photoexcited CRYPTOCHROME1 interacts with dephosphorylated BES1 to regulate brassinosteroid signaling and photomorphogenesis in Arabidopsis. Plant Cell 30: 1989–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sun S, Zhu W, Jia K, Yang H, Wang X (2013) Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Dev Cell 27: 681–688 [DOI] [PubMed] [Google Scholar]
- Whitford R, Fernandez A, De Groodt R, Ortega E, Hilson P (2008) Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc Natl Acad Sci USA 105: 18625–18630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Wang W, Xu P, Pan J, Zhang T, Li Y, Li G, Yang H, Lian H (2019) phyB interacts with BES1 to regulate brassinosteroid signaling in Arabidopsis. Plant Cell Physiol 60: 353–366 [DOI] [PubMed] [Google Scholar]
- Yang M, Wang X (2017) Multiple ways of BES1/BZR1 degradation to decode distinct developmental and environmental cues in plants. Mol Plant 10: 915–917 [DOI] [PubMed] [Google Scholar]
- Yang M, Li C, Cai Z, Hu Y, Nolan T, Yu F, Yin Y, Xie Q, Tang G, Wang X (2017) SINAT E3 ligases control the light-mediated stability of the brassinosteroid-activated transcription factor BES1 in Arabidopsis. Dev Cell 41: 47–58.e44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Moragarcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N, Wu F, Mao H, Dong W, Gan L, et al. (2013) D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504: 406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.