Abstract
Plants are sensitive to a variety of stresses that cause various diseases throughout their life cycle. However, they have the ability to cope with these stresses using different defense mechanisms. The endoplasmic reticulum (ER) is an important subcellular organelle, primarily recognized as a checkpoint for protein folding. It plays an essential role in ensuring the proper folding and maturation of newly secreted and transmembrane proteins. Different processes are activated when around one-third of newly synthesized proteins enter the ER in the eukaryote cells, such as glycosylation, folding, and/or the assembling of these proteins into protein complexes. However, protein folding in the ER is an error-prone process whereby various stresses easily interfere, leading to the accumulation of unfolded/misfolded proteins and causing ER stress. The unfolded protein response (UPR) is a process that involves sensing ER stress. Many strategies have been developed to reduce ER stress, such as UPR, ER-associated degradation (ERAD), and autophagy. Here, we discuss the ER, ER stress, UPR signaling and various strategies for reducing ER stress in plants. In addition, the UPR signaling in plant development and different stresses have been discussed.
Keywords: plants, ER, ER stress, UPR, IRE1, bZIP17, bZIP28, bZIP60
1. Introduction
Plants are becoming exposed to numerous environmental changes during their lifecycle and use complex integrated mechanisms to sense and adapt to these conditions for their growth and development [1,2]. Plants have evolved a number of strategies to respond to various types of stresses at diverse levels, from gene expression alterations to changes in morphology [3,4,5]. The sensing and transduction of environmental signals have been extensively studied in stressed plants, revealing potential strategies for improving agricultural productivity and plant tolerance against different stresses [6]. The environmental sensors ultimately induce changes in metabolic pathways, protein synthesis, and gene expression to enhance plant tolerance against various stresses [6]. This review paper discusses the ER and its functions, the ER stress, and different strategies that play a crucial role in reducing ER stress in plants. Moreover, the role of the unfolded protein response (UPR) signaling in plant development and in various stresses has been discussed.
2. Endoplasmic Reticulum (ER)
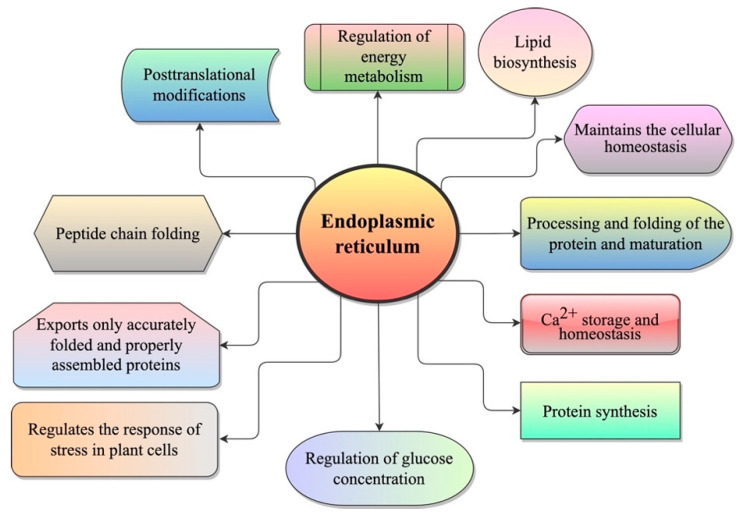
ER is a large, complex, and highly dynamic cytoplasmic membrane system of eukaryotic cells and is considered to be a central network of interconnected tubules and flattened cisternae that extend across the cytoplasm [7,8]. The ER network occupies a significant portion of the cytoplasm with its membrane, accounting for ~50% of total cellular membranes [9]. ER plays a crucial role in protein synthesis, peptide chain folding and processing, post-translational modifications, lipid biosynthesis, Ca2+ storage and homeostasis, and the regulation of glucose concentration [10,11] (Figure 1). This organelle provides an oxidative environment to facilitate the formation of a disulfide bond and is loaded with molecular chaperones [12]. In addition, the ER is involved in regulating the stress responses in animal and plant cells [13].
Figure 1.
Different functions of the ER in plants.
3. ER Stress
ER plays a vital role in maintaining cellular homeostasis in different cellular processes through its functions, such as initial modification and folding of transmembrane and secretory proteins. Endogenous factors and environmental conditions can increase the demand for protein folding machinery. Many factors induce ER stress in plants, such as pathogens, environmental stresses, salinity, and drought, resulting in a higher load on secretory proteins in the ER [14,15]. These stresses accumulate misfolded or unfolded proteins that induce ER homeostasis imbalance, which is called ER stress [16,17,18]. Protein synthesis and modification lead to errors in almost one-third of the nascent proteins in the ER [19,20,21]. ER stress often leads to growth retardation in Arabidopsis (Arabidopsis thaliana) [22]. To overcome ER stress, the stress sensors localized in the ER activate distinct downstream organelle-nucleus signaling pathways to invoke a cytoprotective response, which is known as UPR [23]. UPR has recently been recognized as an important intracellular signaling pathway for linking ER proteostasis with gene regulation in the nucleus to reduce ER stress [6]. Although the molecular mechanism of ER stress in plants is not as well understood as it is in animals [24]. The expansion and abundance of genes related to ER stress, revealed by the genome sequencing of various plant species, suggest that plants use more ER stress responses than animals in order to adapt to the environment [24].
4. Chemical Inducers for the Accumulation of the Unfolded Protein
4.1. Tunicamycin (TM) Stress
TM and Dithiothreitol (DTT) have been found to induce UPR by interfering with protein folding in the ER [25,26]. TM triggers ER-mediated stress in various eukaryotic species, such as plants, yeast, and humans [27,28,29,30]. TM inhibits the N-linked glycosylation (N-glycans formation) by interfering with the GlcNac phosphotransferase enzyme, which is responsible for the initial glycosylation steps [31]. The N-glycans are essential for the proper folding and stability of proteins, as well as their transportation to the Golgi apparatus for the packaging of vesicles and secretion. Besides, N-glycans are involved in the post-translational alteration of microbe-associated molecular patterns (MAMPs) receptors and immune response in plants [30,32,33].
4.2. Dithiothreitol (DTT) Stress
DTT is a powerful reducing agent that induces acute ER stress by disrupting the redox conditions required to form disulfide bridges in proteins [34,35]. DTT is a robust reducing agent commonly used to promote reductive pressure. It can cross membranes and inhibit the formation of a disulfide bond. DTT treatment induces reductive stress, leading to the accumulation of misfolded proteins in the ER [36,37]. However, as a strong inducer of ER stress, DTT has not been considered as an ideal UPR triggering agent for in vivo studies because it can inhibit disulfide bond formation in the ER and cytosol during protein formation, making it unspecific to ER stress [38]. Besides, TM and DTT have differential effects on ER stress kinetics and can influence the expression of UPR target genes [30,39]. Arabidopsis basic leucine zipper 28 (bZIP28), an ER membrane-associated transcription factor (TF) (ER membrane-associated basic leucine zipper), was triggered by an ER stress response induced by exposure to DTT and TM or adverse environmental conditions [37,40]. In addition, TM or DTT may chemically induce basic leucine zipper TF 60 (bZIP60) splicing [41,42].
5. UPR Signaling in Plant Development
UPR has been broadly studied in the context of ER stress, although recently more attention has been diverted to the role of UPR in plant development and defense. UPR has been found to play a crucial role in both reproductive and vegetative development [14]. Additionally, UPR plays an unexpected role in hormone biology, which may explain the effect of UPR on vegetative growth and development [14]. Normal plant growth and development require UPR. Normal growth and development require the mobilization of basic leucine zipper 17 (bZIP17) into the nucleus [43]. The triple mutant inositol-requiring enzyme 1 (IRE1a IRE1b) bZIP17 grew abnormally under normal growth conditions and was also defective in the stress signaling pathways. The functions of bZIP17 in A. thaliana development were observed in another study using genomic and genetic approaches. In contrast to bZIP28 and bZIP60, the bZIP17 is not a primary UPR activator, but works in conjunction with bZIP28 to regulate development-related genes, particularly stress maintenance and root elongation [44]. In A. thaliana, ER stress induces the expression of NAC103. NAC103 overexpression has pleiotropic effects on plant growth, plays a vital role in inducing the expression of some UPR downstream genes under normal growth conditions [45]. BLISTER (BLI) protein is localized to the Golgi, which negatively regulates IRE1a/IRE1b activity under normal growth conditions. A BLI loss-of-function mutation results in prolonged up-regulation of non-canonical UPR downstream genes and canonical UPR genes, resulting in growth retardation and cell death [46]. A. thaliana aquaporins; SIP1;1, SIP1;2, and SIP2;1 are localized in the ER. The aquaporin SIP2;1 plays an important role in alleviating ER stress. A reduction in the elongation of the pollen tube and pollen germination was observed in the absence of SIP2;1 [47]. The basal mRNA level of binding protein 3 (BiP3) is an essential ER stress-induced gene in pollen, suggesting that pollen has experienced ER stress under normal growth conditions [47]. Plants lacking SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 6 (SPL6) showed hyperactivation of IRE1, leading to cell death in rice panicles, which indicates that the SPL6 is an important survival factor in suppressing persistent or extreme ER stress conditions [48]. DERLIN-like protein (OsDER1) is a homolog of yeast and mammal DER1 localized in the ER and has been observed to be accumulated considerably in rice under ER stress. Overexpression or suppression of OsDER1 leads to the activation of UPR and hypersensitivity to the ER stress, and suppression results in shrunken and floury seeds [49].
6. UPR Signaling in Different Stresses
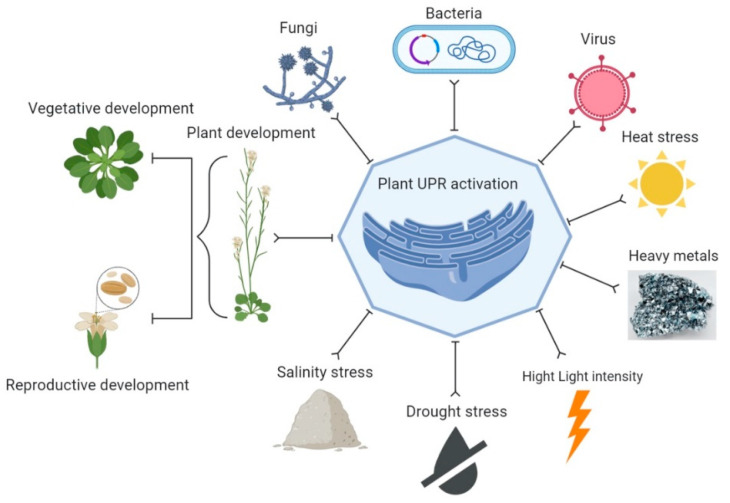
The UPR can be activated by various stresses in plants that induce the accumulation of unfolded proteins in the ER lumen [50,51] (Figure 2). UPR has been involved in the immunity and development of plants and provides defense against different stresses [14,42,44], such as heat [52], drought [53], salinity [54,55], osmotic pressure, high light intensity and heavy metals. These stresses disturb protein folding [15,56]. In addition, UPR is activated under protein synthesis overload conditions when the need for protein folding simply does not meet demands [57,58]. However, cells commit to programmed cell death (PCD) during the failure of UPR in chronic or unresolved ER stress conditions [23,59,60,61]. Biotic agents have more complex effects on the UPR. Various biotic agents have been reported to induce the UPR or require the UPR to infect plants. A study observed that when Nicotiana benthamiana was inoculated with the host-pathogen and non-host pathogens, Pseudomonas syringae and Pseudomonas cichorii, respectively, the host pathogen P. syringae did not induce the expression of bZIP60. At the same time, the expression of bZIP60P was induced by the non-host pathogen, P. cichorii. However, the plants became more susceptible to P. syringae when virus-induced gene silencing (VIGS) was used to silence the bZIP60 in N. benthamiana [62]. UPR is induced and required by various plant viruses for successful infection. In another study, a knockout in bZIP60 was observed to suppress the viral symptoms and the transgenic expression of an activated form of bZIP60 that could suppress the symptoms in the bZIP60 knockout [63]. Various genes are associated with the ER stress response by multiple stresses and plant development in A. thaliana and many other major crops/plants (Table 1).
Figure 2.
Activation of the UPR. A variety of stresses (biotic and abiotic) and plant development processes (vegetative and reproductive) trigger the UPR by excessive accumulation of unfolded proteins in the ER or cause an imbalance in the supply of amino acids, which leads to the activation of one or more UPR arms. This figure was created by using BioRender software.
Table 1.
Involvement of different genes in ER and other stresses.
| Gene | Function | Stress | Plant/Crop | Reference |
|---|---|---|---|---|
| HRD3A | Defects in HRD3A cause alteration in the UPR, increased plant sensitivity to salt, and retention of ERAD substrates in plant cells. | Salt and ER stresses | Arabidopsis (A. thaliana) | [66] |
| UBC32 | UBC32 affects the stability of barley powdery mildew O (MLO) mutant MLO-12, a known ERAD substrate. | ER stress | Arabidopsis (A. thaliana) | [67] |
| AtOS9 | AtOS9 is an ER-localized glycoprotein and co-expresses with various predicted/known ER chaperones. | ER stress | Arabidopsis (A. thaliana) | [68] |
| NAC103 | ER stress induces the expression of NAC103. Overexpression of NAC103 has pleiotropic effects on plant growth. It plays a crucial role in inducing the expression of some UPR downstream genes under normal growth conditions. | ER stress | Arabidopsis (A. thaliana) | [45] |
| EBS7 | Arabidopsis ethyl methane sulfonate-mutagenized brassinosteroid insensitive 1 suppressor 7 (EBS7) gene observed to be accumulated under ER stress, and its mutations lead to hypersensitivity to salt and ER stresses. | ER and salt stresses | Arabidopsis (A. thaliana) | [69] |
| WRKY75 | WRKY75 is an ER-stress cellular response regulator. Plants expressing WRKY75 show tolerance to salt stress, which connects the ER and abiotic stress responses. | ER and salt stresses | Arabidopsis (A. thaliana) | [71] |
| AtNRP1, AtNRP2 and AtNRPs; (ANAC036 and gVPE) | Loss-of-function of AtNRP1 and AtNRP2 attenuates the cell death caused by ER stress. Osmotic and ER stresses have been shown to induce AtNRPs; (gVPE and ANAC036). | ER stress | Arabidopsis (A. thaliana) | [88] |
| AtHSPR | AtHSPR (A. thaliana Heat Shock Protein Related) is involved in ER stress signaling and cell death caused by salt stress. | ER stress | Arabidopsis (A. thaliana) | [89] |
| SAL1 | SAL1 is a negative regulator of stress signaling and is linked to plant stress responses. Loss-of-function of SAL1 resulted in a significant reduction in ER stress and a significant increase in Cd tolerance. | ER and cadmium (Cd) stresses | Arabidopsis (A. thaliana) | [77] |
| HOP | HSP70-HSP90 organizing protein (HOP) is a member of the cytosolic cochaperones family. HOP3 interacts in vivo with cytosolic HSP70 and HSP90, and with binding immunoglobulin protein (BiP), an HSP70 protein is localized in the ER. | ER stress | Arabidopsis (A. thaliana) | [90] |
| CER9 and HRD1A/1B | Arabidopsis ERAD genes, HRD1A/1B and CER9 might regulate the heat stress response. HRD1A/1B and CER9 collaboratively regulate plant thermos tolerance. | ER and heat stresses | Arabidopsis (A. thaliana) | [70] |
| AtNTL7 | AtNTL7 is a membrane-tethered NAC TF that leads to resistance to ER stress. Overexpression of AtNTL7 exhibits strong resistance to ER stress. | ER stress | Arabidopsis (A. thaliana) | [91] |
| HY5 | Mutation of a main light signaling component, ELONGATED HYPOCOTYL 5 (HY5), leads to ER stress tolerance. HY5 negatively regulates the UPR by competing with bZIP28 for binding to the G-box-like element present in the ER stress response element. | ER stress | Arabidopsis (A. thaliana) | [56] |
| OsDER1 | Suppression or overexpression of OsDER1 results in the activation of UPR and hypersensitivity to ER stress and suppression leads to shrunken and floury seeds. | ER stress | Rice (O. sativa L.) |
[49] |
| SPL6 | Mutation of SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE 6 (SPL6) up-regulates the expression of IRE1 and persistent UPR, which causes cell death and the abortion of rice apical panicles. | ER stress | Rice (O. sativa L.) |
[48] |
| EMR | ERAD-mediating RING finger protein (EMR) plays an essential role in the plant ERAD system, affecting the BR signaling under ER stress conditions. | ER stress | Arabidopsis (A. thaliana) | [92] |
| GAAP1 | GAAP1 (Arabidopsis Golgi anti-apoptotic protein 1) regulates the PCD and UPR. GAAP1 prevents cell death induced by ER stress and encourages the recovery of plant growth by attenuating the UPR process mediated by IRE1 after ER stress relief. | ER stress | Arabidopsis (A. thaliana) | [78,79] |
| BLI | BLISTER (BLI) protein loss-of-function mutation up-regulates the canonical UPR of non-canonical UPR downstream genes, inducing growth retardation and cell death. | ER stress | Arabidopsis (A. thaliana) | [46] |
| hyl1 | HYPONASTIC LEAVES1 (hyl1) mutant plants are more susceptible to TM, which causes ER stress. | ER stress | Arabidopsis (A. thaliana) | [93] |
| FAD2 | The 7 fatty acid desaturases (FADs) desaturate each glycerolipid class differently in plastids and ER. FAD2 mutants have resulted in a hypersensitive response to TM through systematic screening of FAD mutants. | ER stress | Arabidopsis (A. thaliana) | [94] |
| NF-YC14 | NF-YC14 involves in regulating the ER stress response. NF-YC14 overexpression improves plant tolerance to ER stress and increases the expression of downstream genes for ER stress response. | ER stress | Arabidopsis (A. thaliana) | [95] |
| GAAP1, GAAP3, and MAPR3 | Arabidopsis Golgi anti-apoptotic proteins 1 and 3 (GAAP1, 3) resist PCD against ER stress and negatively modulate the IRE1-bZIP60 pathway. Mutations in GAAP1/GAAP3 or/and Membrane-associated progesterone receptor 3 (MAPR3) increase the vulnerability of seedlings to ER stress. | ER stress | Arabidopsis (A. thaliana) | [96] |
| MfSTMIR | MfSTMIR plays a crucial role in salt and ER stress response. The expression of MfSTMIR was observed to be induced by TM and salt. | ER and salt stresses | Sickle medic (Medicago falcata) |
[75] |
| SIP1;1, SIP1;2 | A. thaliana aquaporins; SIP1;1, SIP1;2 and SIP2;1 are localized in the ER. The aquaporin SIP2;1 involves alleviating the ER stress. The absence of SIP2;1 reduces pollen tube elongation and pollen germination. | ER stress | Arabidopsis (A. thaliana) | [47] |
| BiP3 | The basal mRNA level of BiP3 is an important gene induced by ER stress in pollen. | ER stress | Arabidopsis (A. thaliana) | [47] |
| PAWH1 and PAWH2 | PAWH1 and PAWH2 are localized in the ER membrane and associated with Hrd1 through EMS-mutagenized Bri1 Suppressor 7 (EBS7). Removal of two PAWHs constitutively triggers the UPR and compromises the resistance to stress. | ER stress | Arabidopsis (A. thaliana) | [97] |
| AtOTU1 | AtOTU1 selectively hydrolyzes various forms of ubiquitin chains. AtOTU1 is required to process plant ERAD substrates. | ER stress | Arabidopsis (A. thaliana) | [98] |
| AtSec62 | Arabidopsis Sec62 (AtSec62) is required for plant development and may function as an ER-phagy receptor in plants. | ER stress | Arabidopsis (A. thaliana) | [99] |
| TIN1 | Transcriptional induction of Tunicamycin induced 1 (TIN1) by ER stress was partially regulated by AtbZIP60. The accumulation of TIN1 protein was observed in response to TM treatment. | ER stress | Arabidopsis (A. thaliana) | [28] |
Many studies in rice (Oryza sativa L.) (OsbZIP50), maize (Zea mays L.) [41] and A. thaliana [64,65] have reported IRE1 splicing of the heat-induced bZIP60. Furthermore, HRD3A is an influential part of plant ERAD and plays a crucial role in plant UPR. In A. thaliana, a defect in HRD3A results in UPR alteration, increased sensitivity of the plant to salt, and the retention of ERAD substrates in plant cells [66]. The stress-induced and ER membrane-localized functional ubiquitin conjugation enzyme (E2) UBC32 connects the process of ERAD and brassinosteroid (BR)-mediated growth promotion and salt stress tolerance in A. thaliana. UBC32 affects the stability of barley powdery mildew O (MLO) mutant MLO-12, a known ERAD substrate [67]. Arabidopsis homolog (AtOS9) of an ER luminal lectin Yos9 plays a vital role in recognizing a unique asparagine-linked glycan on misfolded proteins. AtOS9 is a glycoprotein localized to the ER and co-expressed with various predicted/known ER chaperones [68]. Arabidopsis ethyl methane sulfonate-mutagenized brassinosteroid insensitive 1 suppressor 7 (EBS7) gene encodes an ERAD component localized in the ER membrane. The accumulation of EBS7 has been observed under ER stress, and its mutations cause hypersensitivity to salt and ER stresses [69]. Arabidopsis ERAD genes, HRD1A/1B, and CER9 might regulate the heat stress response. HRD1A/1B and CER9 collaboratively regulate plant thermos tolerance and the expression of both UPR and Cytosolic Protein Response (CPR) genes, no matter under heat stress or normal conditions [70]. WRKY75 is an ER-stress cellular response regulator as its expression directly responds to ER stress-inducing chemicals, such as TM and DTT. Plants that express WRKY75 show tolerance to salt stress, connecting ER and abiotic stress responses [71].
The overexpression of BhbZIP60, an AtbZIP60 homologous from the Boea hygrometrica plant, has resulted in increased resistance to mannitol and drought stresses [72]. Expression profile analyses of soybean plants treated with an osmotic stress inducer (polyethylene glycol) or ER stress inducers (TM/azidothymidine) indicate a correlation between the osmotic stress pathway and ER stress [73]. BiP expression is up-regulated in wheat (Triticum aestivum L.) during osmotic stress-related cell death [74]. The MfSTMIR, which encodes a highly conserved ER-membrane-localized RING E3 ligase in leguminous plants, plays an essential role in combatting salt and ER stress in Medicago. In another study, the expression of MfSTMIR was found to be induced by TM and salt. The mtstmir loss-of-function mutants showed impaired induction of BiP1/2 and BiP3 ER stress-responsive genes under TM treatment and sensitivity to salt stress [75].
In A. thaliana, the overexpression of bZIP60 improved tolerance against salt stress [76]. Xi et al. showed that SAL1 loss-of-function caused improved Cd tolerance and reduced ER stress in A. thaliana [77]. Arabidopsis Golgi anti-apoptotic proteins 1 and 3 (GAAP1, 3) were observed to resist PCD against ER stress and negatively modulate the IRE1-bZIP60 pathway. Mutations in GAAP1/GAAP3 or/and Membrane-associated progesterone receptor 3 (MAPR3) increase the vulnerability of seedlings to ER stress [78]. Moreover, GAAP1 and GAAP3 are involved in regulating cell death and UPR. GAAP1 to GAAP3 were observed to play redundant roles in delaying the UPR activation induced by ER stress and inhibiting cell death [79].
Many phytohormones are involved in UPR signaling. Increasing evidence supports newly emerging roles for plant hormones, such as jasmonic acid (JA) [80], salicylic acid (SA) [81], auxin, and Ethylene (ETH) [80,82], secondary messengers (e.g., Ca2+) [83], as well as other signaling molecules such as Reactive oxygen species (ROS) and sugars, as essential regulators of the UPR in plants [84]. JA signaling pathway is involved in defense against necrotrophic pathogens. A study showed that transcriptional levels of chaperone protein genes, such as BiP, calreticulin (CRT), calnexin 1-like (CNX 1-like), and protein disulfide isomerase (PDI), and genes involved in the IRE1-bZIP60 pathway, were all significantly induced in Nicotiana attenuata leaves after the inoculation of A. alternata. The silencing of bZIP60 or IRE1 gene increased the susceptibility of N. attenuata plants to A. alternata. IRE1-bZIP60 pathway is needed for the resistance of N. attenuata to A. alternata, and JA signaling pathway plays an essential role in eliciting the IRE1-bZIP60 pathway and chaperone protein genes [85]. SA has been observed to play a crucial role in ER stress signaling and UPR regulation under stress conditions [24], although its mode of action is unknown. In a study, the relationship between ER stress and SA-mediated defense responses was postulated, and spatiotemporal change was described [86]. In another study, Wang et al. [87] discovered that SA-induced master regulator protein NPR1 (nonexpressor of pathogenesis-related (PR) genes 1) regulates numerous ER stress and UPR components induced by SA during systemic acquired resistance (SAR) development in A. thaliana.
7. Strategies to Reduce ER Stress
Many strategies have been used to reduce ER stress, including the UPR, ER-associated degradation (ERAD), and autophagy.
7.1. Unfolded Protein Response (UPR)
UPR is an intracellular signaling mechanism activated by ER stress and has been designed to restore the ER function and to ignite the PCD processes when ER stress remains unresolved [100]. ER stress leads to the accumulation and aggregation of the unfolded proteins in the ER lumen. Moreover, UPR originates at the ER, where it overcomes the ER stress, restores the ER homeostasis, and leads to the ER chaperones and foldases synthesis to attenuate the ER stress [95,101]. These ER chaperones and foldases are BiP, protein disulfide isomerase (PDI), glucose-regulated protein (GRP94), peptidyl-prolyl isomerases (PPI) or immunophilins, calnexin and calreticulin [102]. Both signaling pathways eventually result in the upregulation of genes to either correctly fold or degrade misfolded proteins and regulate translation and transcription for restoring the ER homeostasis [28,103]. UPR may relieve the transient ER stress, whereas persistent ER stress can result in PCD [104,105].
7.2. Mechanism of UPR Signaling Pathway in Plants
7.2.1. Regulated IRE-1 Dependent Splicing (RIDS)
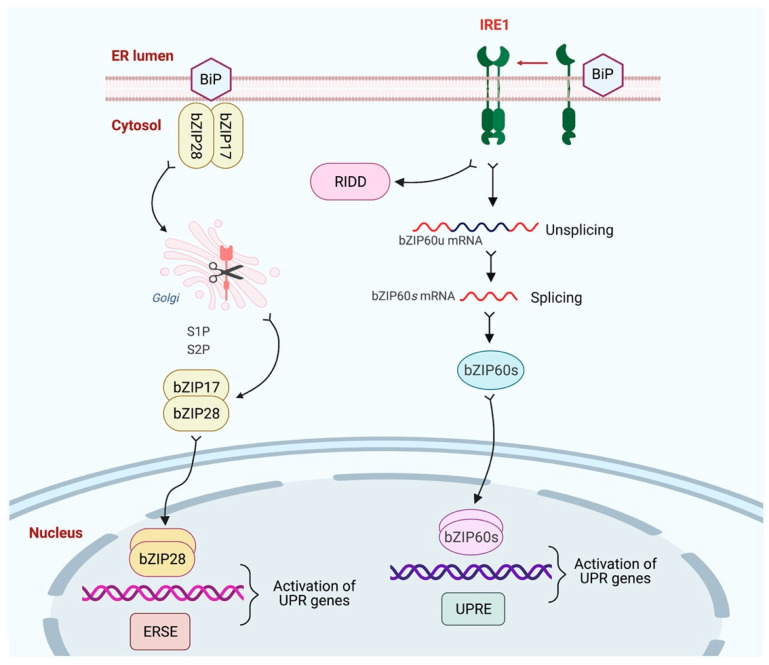
A wide range of stresses affect protein folding, causing ER stress that is communicated to the nucleus via the UPR, a cellular homeostatic response to ER stress [106,107]. As a result, genes involved in the folding, import, export, and quality control of proteins are up-regulated. In plants, signal transducers mediate the signaling that forms two arms of the UPR signaling pathway [14,38,43]. One arm includes membrane-associated TFs, for instance, bZIP17 and bZIP28, and the other arm includes IRE1, which is an RNA splicing factor. These two arms shape the stress transcriptome, the upregulation and the downregulation of the expression of genes to combat the stress effects [103]. On the other hand, cells undergo PCD if the adaptation mechanisms are inadequate to manage the unfolded protein load. In plants, the PCD regulatory mechanism and the key factors that regulate various outputs of ER stress receptors remain unclear. The bZIP17/28 are retained in the ER under normal conditions, associating with UPR regulator BiP. However, when unfolded proteins accumulate under stress conditions, BiP is sequestered and released from bZIP17/28 [108,109]. In response to ER stress, bZIP17 and/or bZIP28 are mobilized and transported to the Golgi, where they are proteolytically cleaved by two proteases: Site 1 Protease (S1P), a processing site and Site 2 Protease (S2P), a recognition site. The S1P cleaves them in the Golgi’s C-terminal region and the S2P in its cytosolic end. These two proteases release their TF domains [bZIP17(p) and/or bZIP28(p)] into the cytoplasm for further importation into the nucleus, where they upregulate the expression of stress response genes and restore the ER homeostasis [108,110] (Figure 3). The regulated intramembrane proteolysis (RIP)-mediated activation of the bZIP28 is the first-hand response for mitigating the ER stress in plants. The molecular structure of a type II membrane protein bZIP28 reveals that it comprises a cytoplasmic DNA-binding bZIP domain at its N-terminus, a single transmembrane domain, and a luminal domain at its C-terminus [15].
Figure 3.
Overview of the pathway for UPR signaling. UPR signaling pathways in plants have two arms/branches. One branch contains the dual protein kinase and ribonuclease, IRE1, responsible for splicing bZIP60 mRNA when activated. The other branch is mediated by bZIP17 and bZIP28, two ER membrane-anchored TFs. A frameshift is introduced by splicing bZIP60(u) mRNA so that the resulting spliced type bZIP60(s) mRNA is translated into a nucleus-targeted TF. In response to ER stress, the bZIP17 and/or bZIP28 are mobilized and transported to the Golgi, where resident site-1 and site-2 proteases process them and release their cytosolic TF domains [bZIP17(p) and/or bZIP28(p)] into the cytoplasm for further importation into the nucleus. The bZIP17(p) and bZIP28(p) can homodimerize or heterodimerize in the nucleus, where they bind to the promoters and regulate the expression of genes that respond to stress. This figure was created using BioRender software.
Additionally, IRE1, a bifunctional protein kinase/ribonuclease, is an essential plant UPR regulator that mediates the cytoplasmic splicing of RNA encoding the TF bZIP60. This triggers the signaling pathway of UPR and regulates the canonical UPR genes. However, it is largely unknown how IRE1’s protein activity is controlled during the growth and development of plants [46]. Two isoforms, IRE1-IRE1a and IRE1b are found in the A. thaliana [111]. Both IRE1a and IRE1b isoforms are classified as type I single-pass transmembrane proteins. These comprise multifunctional domains, for instance, a protein kinase domain, an N-terminal signal peptide, a cytosol-facing C-terminal ribonuclease domain, and an ER-stress sensing domain that faces the ER lumen [15]. Both IRE1a and IRE1b specifically activate the bZIP60 mRNA’s unconventional splicing in response to the biotic and abiotic stresses [42,64]. The plant IRE1a and IRE1b can form homo/heterodimers to activate the IRE1-dependent UPR signaling pathway, similar to how mammals and yeast activate the IRE pathway [63,112].
In the IRE1 pathway, luminal BiPs interact with the ER-membrane protein IRE1 in the ER lumen. After accumulating unfolded proteins, BiPs bind them and release IRE1 proteins that form dimers that, unusually, splice bZIP60 mRNAs in the cytosol. The spliced mRNA translates into a functional TF, which moves to the nucleus and upregulates the genes containing ER stress elements (ERSEs) and UPR responsive elements (UPREs) in their regulatory regions [45,105,113]. The IRE1 pathway promotes the cytosplicing of bZIP60. Moreover, it promotes ER-localized mRNA degradation, which is known as regulated IRE1-dependent decay (RIDD), and autophagy, in order to reduce protein load in ER or timely eliminate the damaged ER [96,114]. The degradation creates a frameshift to encode a type of bZIP60(s) that is transportable to the nucleus [64]. For the activation of stress response genes in the nucleus, the bZIP17(p), bZIP28(p), and bZIP60(s) can homodimerize or heterodimerize [43,110]. RIDD typically contributes to pro-life processes by reducing the abundance of ER client mRNA in ER stress environments [107,115]. Nonetheless, RIDD activity against targeted non-ER client mRNAs has widened the species-specific mannered downstream effects of IRE1 activation [100].
7.2.2. ER-Associated Degradation (ERAD)
ERAD is part of the ER protein quality-control system (ERQC), which is considered essential for the conformation fidelity of most of the membrane and secretory proteins in eukaryotes. The ERAD process is associated with the ubiquitin/proteasome system (UPS), which relieves ER stress. The UPS function involves the use of a ubiquitin-activating enzyme (E1), the ubiquitin-conjugating enzyme (E2), the ubiquitin ligase (E3), and the 26S proteasome [116]. ERAD is accomplished through multistep reactions involving the sequential recruitment of E1, E2, and E3 enzymes. E2 and E3 enzymes are responsible for the specificity of the substrate [92,117]. Since the plants are sessile species, they respond to environmental changes by regulating the signaling pathways from seed germination to the mature organism. Consequently, plant cells express more E3 ligase family members than mammals and yeast cells [92,118]. ERAD is believed to work in plants with core machineries that are highly conserved to those found in yeast and humans, but the ERAD system in plants is poorly understood [98]. The protein folding process is complex and can be easily disrupted [38,66,67,110]. Therefore, conserved ERQC strictly monitors protein folding and identified misfolded proteins to be eliminated by ERAD [66].
7.2.3. Autophagy
Autophagy is a self-eating cellular process that has been conserved throughout evolution. Autophagy functions as a degradation process in recycling cellular cytoplasmic contents and removing damaged proteins or organelles under adverse growth conditions [101]. There are three main types of autophagy on the basis of their mechanism; chaperone-mediated autophagy [119], macroautophagy [120], and microautophagy [121]. Chaperone-mediated autophagy is highly selective, while macroautophagy and microautophagy may be selective or non-selective [122,123]. In plants, the following two types of autophagy are known: macroautophagy and microautophagy [124]. During macroautophagy, the majority of cytosolic constituents are sequestered into a double-membrane structure, known as an autophagosome. The autophagosome is a specific organelle that mediates macroautophagy. For degradation, autophagy delivers cytoplasmic materials or organelles to the vacuole/lysosome by forming autophagosome [99,125]. The autophagosome’s outer membrane fuse with the vacuolar membrane upon delivery to the vacuole, delivering the inner membrane structure and its cargo, i.e., the autophagic body into the vacuolar lumen for degradation by vacuolar hydrolases [126,127]. During microautophagy, the invagination of the vacuolar membrane (autophagic bodies) delivers a part of the cytoplasm to the vacuolar lumen and then the resident vacuolar hydrolases digest them [128]. A number of eukaryotic-conserved autophagy-related (ATG) proteins play a significant role in this process [129].
A selective autophagic pathway for resolving ER stress and restoring the ER homeostasis, ER-phagy, has been defined to remove misfolded or unfolded proteins accumulating in the ER [130]. Similarly, ER-phagy, the autophagy receptors that act as a bridge between autophagic cargoes selection and autophagosome formation, are also needed [131]. Under these ER stress conditions, ER-phagy delivers the unfolded or misfolded ER proteins and fragments for degradation into the vacuole [101], which indicates that ER-phagy receptors are possibly involved in plants [99]. Autophagy can also be significantly induced in plants by ER stress agents, including DTT and TM which prevent proper protein folding in the ER [101]. It was shown by the electron and confocal microscopy that ER portions are engulfed by autophagosomes and delivered to the vacuole, most likely for degradation. Furthermore, one of the ER stress sensors, IRE1b is needed for inducing autophagy by ER stress [101]. In response to ER stress, autophagy upregulation requires IRE1 RIDD activity by degrading the mRNA of proteins involved in autophagy in A. thaliana, for instance, b-glucosidase 21 and PR protein 14 [132]. It is still unknown if RIDD is essential for plant survival [100].
8. Concluding Remarks and Future Perspectives
Plants have evolved sophisticated signal transduction mechanisms and sensitive detection systems for coping with various stress conditions. Due to climate change, changes in the agricultural environment cause significant reductions in crop yields. To assist crops in coping with newly emerging stresses, it is critical to understand resistance mechanisms of plants. ER is a large, architecturally variable, and functionally complex organelle in eukaryotes. Different intracellular and extracellular stresses may increase the number of misfolded proteins, resulting in an ER homeostasis imbalance, which is referred to as ER stress. Adverse conditions interfere with several sensitive cellular processes in plants that accumulate the misfolded proteins. In plants, UPR mediates the response to many biotic and abiotic stresses. UPR is necessary for the homeostasis of proteins in the ER when adverse environmental conditions challenge plants. In A. thaliana, IRE1 is one of the major sensors which activates the bZIP60. The bZIP28 is another sensor that triggers another ER arm, and bZIP17 induces downstream genes. Significant progress has been made in elucidating the UPR signaling in plants and various techniques have been developed to study ER stress in A. thaliana and other major crops/plants. The molecular mechanism of ER stress in plants is not as well understood as it is in animals. However, further work is needed to expand the understanding of the mechanism of ER stress and the UPR signaling pathway against various stresses in major crops/plants.
Acknowledgments
Because of space limitations, we apologize to the authors whose works are not cited.
Abbreviations
| AtHSPR | A. thaliana Heat Shock Protein Related |
| AtSec62 | Arabidopsis Sec62 |
| BiP | Binding protein/Binding immunoglobulin protein |
| BiP3 | Binding protein 3 |
| BLI | Golgi-localized protein BLISTER |
| bZIP17 | BASIC LEUCINE ZIPPER 17 |
| bZIP60 | Leucine zipper transcription factor 60 |
| CNX 1-like | Calnexin 1-like |
| CPR | Cytosolic Protein Response |
| CRT | Calreticulin |
| DTT | Dithiothreitol |
| EMR | ERAD-mediating RING finger protein |
| EMS | Ethyl methanesulphonate |
| ER | Endoplasmic reticulum |
| ERAD | ER-associated degradation |
| ERQC | ER protein quality-control system |
| ERSE | ER stress response element |
| ETH | Ethylene |
| FAD2 | FATTY ACID DESATURASE 2 |
| GRP94 | Glucose-regulated protein 94 |
| HOP | HSP70-HSP90 organizing protein |
| HY5 | ELONGATED HYPOCOTYL 5 |
| hyl1 | HYPONASTIC LEAVES1 |
| IRE1 | Inositol requiring enzyme1 |
| MAPR3 | Membrane-associated progesterone receptor 3 |
| MLO | Barley powdery mildew O |
| NPR1 | Nonexpressor of pathogenesis-related (PR) genes 1 |
| PCD | Programmed cell death |
| PDI | Protein disulfide isomerase |
| PPI | Peptidyl-prolyl isomerases |
| PR | Pathogenesis-related |
| RIDD | IRE1-dependent decay of mRNAs |
| RIP | Regulated intramembrane proteolysis |
| ROS | Reactive oxygen species |
| S1P | Site 1 Protease |
| S2P | Site 2 Protease |
| SA | Salicylic acid |
| SAR | Systemic acquired resistance |
| SPL6 | SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE 6 |
| TF/TFs | Transcription factor/Transcription factors |
| TM | Tunicamycin |
| UPR | Unfolded protein response |
| UPS | Ubiquitin/proteasome system |
| VIGS | Virus-induced gene silencing |
Author Contributions
The authors confirm their contributions to this work: H.M. in conceptualization, original draft preparation, and writing; and J.L. in revising, and review. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from National Natural Science Foundation of China (No. NSFC31730019 to Jianming Li) and the China Postdoctoral Science Foundation (No. 2020M682721 to Hakim Manghwar).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Millar A.J. The intracellular dynamics of circadian clocks reach for the light of ecology and evolution. Ann. Rev. Plant Biol. 2016;67:595–618. doi: 10.1146/annurev-arplant-043014-115619. [DOI] [PubMed] [Google Scholar]
- 2.de Souza A., Wang J.-Z., Dehesh K. Retrograde signals: Integrators of interorganellar communication and orchestrators of plant development. Ann. Rev. Plant Biol. 2017;68:85–108. doi: 10.1146/annurev-arplant-042916-041007. [DOI] [PubMed] [Google Scholar]
- 3.Nakashima K., Ito Y., Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009;149:88–95. doi: 10.1104/pp.108.129791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su Z., Ma X., Guo H., Sukiran N.L., Guo B., Assmann S.M., Ma H. Flower development under drought stress: Morphological and transcriptomic analyses reveal acute responses and long-term acclimation in Arabidopsis. Plant Cell. 2013;25:3785–3807. doi: 10.1105/tpc.113.115428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hakim, Ullah A., Hussain A., Shaban M., Khan A.H., Alariqi M., Gul S., Jun Z., Lin S., Li J., et al. Osmotin: A plant defense tool against biotic and abiotic stresses. Plant Physiol. Biochem. 2018;123:149–159. doi: 10.1016/j.plaphy.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 6.Liu L., Li J. Communications Between the Endoplasmic Reticulum and Other Organelles During Abiotic Stress Response in Plants. Front. Plant Sci. 2019;10:749. doi: 10.3389/fpls.2019.00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuldiner M., Schwappach B. From rags to riches—The history of the endoplasmic reticulum. Biochim. Biophys. Acta. Mol. Cell Res. 2013;1833:2389–2391. doi: 10.1016/j.bbamcr.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Westrate L., Lee J., Prinz W., Voeltz G. Form follows function: The importance of endoplasmic reticulum shape. Ann. Rev. Biochem. 2015;84:791–811. doi: 10.1146/annurev-biochem-072711-163501. [DOI] [PubMed] [Google Scholar]
- 9.Stefano G., Brandizzi F. Advances in plant ER architecture and dynamics. Plant Physiol. 2018;176:178–186. doi: 10.1104/pp.17.01261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z., Lv Y., Zhao N., Guan G., Wang J. Protein kinase R-like ER kinase and its role in endoplasmic reticulum stress-decided cell fate. Cell Death Dis. 2015;6:e1822. doi: 10.1038/cddis.2015.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwarz D.S., Blower M.D. The endoplasmic reticulum: Structure, function and response to cellular signaling. Cell Mol. Life Sci. 2016;73:79–94. doi: 10.1007/s00018-015-2052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benham A.M. Endoplasmic Reticulum redox pathways: In sickness and in health. FEBS J. 2019;286:311–321. doi: 10.1111/febs.14618. [DOI] [PubMed] [Google Scholar]
- 13.Schröder M., Kaufman R.J. The mammalian unfolded protein response. Ann. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 14.Bao Y., Howell S.H. The unfolded protein response supports plant development and defense as well as responses to abiotic stress. Front. Plant Sci. 2017;8:344. doi: 10.3389/fpls.2017.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nawkar G.M., Lee E.S., Shelake R.M., Park J.H., Ryu S.W., Kang C.H., Lee S.Y. Activation of the transducers of unfolded protein response in plants. Front. Plant Sci. 2018;9:214. doi: 10.3389/fpls.2018.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merksamer P.I., Trusina A., Papa F.R. Real-time redox measurements during endoplasmic reticulum stress reveal interlinked protein folding functions. Cell. 2008;135:933–947. doi: 10.1016/j.cell.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merksamer P.I., Papa F.R. The UPR and cell fate at a glance. J. Cell Sci. 2010;123:1003–1006. doi: 10.1242/jcs.035832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kabir M.F., Kim H.-R., Chae H.-J. Endoplasmic Reticulum. IntechOpen; London, UK: 2018. Endoplasmic Reticulum Stress and Autophagy. [Google Scholar]
- 19.Chen Q., Yu F., Xie Q. Insights into endoplasmic reticulum-associated degradation in plants. New Phytol. 2020;226:345–350. doi: 10.1111/nph.16369. [DOI] [PubMed] [Google Scholar]
- 20.Schubert U., Anton L.C., Gibbs J., Norbury C.C., Yewdell J.W., Bennink J.R. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 21.Hetz C., Chevet E., Oakes S.A. Proteostasis control by the unfolded protein response. Nat. Cell Biol. 2015;17:829–838. doi: 10.1038/ncb3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S., Choi Y., Kwon C., Yun H.S. Endoplasmic reticulum stress-induced accumulation of VAMP721/722 requires CALRETICULIN 1 and CALRETICULIN 2 in Arabidopsis. J. Integr. Plant Biol. 2019;61:974–980. doi: 10.1111/jipb.12728. [DOI] [PubMed] [Google Scholar]
- 23.Ruberti C., Kim S.-J., Stefano G., Brandizzi F. Unfolded protein response in plants: One master, many questions. Curr. Opin. Plant Biol. 2015;27:59–66. doi: 10.1016/j.pbi.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park C.-J., Park J.M. Endoplasmic reticulum plays a critical role in integrating signals generated by both biotic and abiotic stress in plants. Front. Plant Sci. 2019;10:399. doi: 10.3389/fpls.2019.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martínez I.M., Chrispeels M.J. Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell. 2003;15:561–576. doi: 10.1105/tpc.007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carvalho H.H., Brustolini O.J., Pimenta M.R., Mendes G.C., Gouveia B.C., Silva P.A., Silva J.C.F., Mota C.S., Soares-Ramos J.R., Fontes E.P. The molecular chaperone binding protein BiP prevents leaf dehydration-induced cellular homeostasis disruption. PLoS ONE. 2014;9:e86661. doi: 10.1371/journal.pone.0086661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hauptmann P., Riel C., Kunz-Schughart L.A., Fröhlich K.U., Madeo F., Lehle L. Defects in N-glycosylation induce apoptosis in yeast. Mol. Microbiol. 2006;59:765–778. doi: 10.1111/j.1365-2958.2005.04981.x. [DOI] [PubMed] [Google Scholar]
- 28.Iwata Y., Nishino T., Takayama S., Koizumi N. Characterization of a plant-specific gene induced by endoplasmic reticulum stress in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2010;74:2087–2091. doi: 10.1271/bbb.100487. [DOI] [PubMed] [Google Scholar]
- 29.Oslowski C.M., Urano F. Methods in Enzymology. Volume 490. Elsevier; Amsterdam, The Netherlands: 2011. Measuring ER stress and the unfolded protein response using mammalian tissue culture system; pp. 71–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCormack M.E., Liu X., Jordan M.R., Pajerowska-Mukhtar K.M. An improved high-throughput screening assay for tunicamycin sensitivity in Arabidopsis seedlings. Front. Plant Sci. 2015;6:663. doi: 10.3389/fpls.2015.00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koizumi N., Ujino T., Sano H., Chrispeels M.J. Overexpression of a gene that encodes the first enzyme in the biosynthesis of asparagine-linked glycans makes plants resistant to tunicamycin and obviates the tunicamycin-induced unfolded protein response. Plant Physiol. 1999;121:353–362. doi: 10.1104/pp.121.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saijo Y., Tintor N., Lu X., Rauf P., Pajerowska-Mukhtar K., Häweker H., Dong X., Robatzek S., Schulze-Lefert P. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 2009;28:3439–3449. doi: 10.1038/emboj.2009.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Häweker H., Rips S., Koiwa H., Salomon S., Saijo Y., Chinchilla D., Robatzek S., von Schaewen A. Pattern recognition receptors require N-glycosylation to mediate plant immunity. J. Biol. Chem. 2010;285:4629–4636. doi: 10.1074/jbc.M109.063073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng Y., Srivastava R., Howell S.H. Endoplasmic reticulum (ER) stress response and its physiological roles in plants. Int. J. Mol. Sci. 2013;14:8188–8212. doi: 10.3390/ijms14048188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu X., Wang T., Zhu M., Zhang L., Zhang F., Jing E., Ren Y., Wang Z., Xin Z., Lin T. Transcriptome and physiological analyses for revealing genes involved in wheat response to endoplasmic reticulum stress. BMC Plant Biol. 2019;19:193. doi: 10.1186/s12870-019-1798-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rand J.D., Grant C.M. The thioredoxin system protects ribosomes against stress-induced aggregation. Mol. Biol. Cell. 2006;17:387–401. doi: 10.1091/mbc.e05-06-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altuntaş C., Terzi R. Dithiothreitol and PEG Induced Combined Stress May Affect the Expressions of ABA Aldehyde Oxidase, Sucrose Synthase and Proline Metabolic Genes in Maize Seedlings. Phyton. 2020;89:487. doi: 10.32604/phyton.2020.08919. [DOI] [Google Scholar]
- 38.Howell S.H. Endoplasmic reticulum stress responses in plants. Ann. Rev. Plant Biol. 2013;64:477–499. doi: 10.1146/annurev-arplant-050312-120053. [DOI] [PubMed] [Google Scholar]
- 39.Li B., Yi P., Zhang B., Xu C.J., Liu Q.Y., Pi Z.J., Xu X.L., Chevet E., Liu J.F. Differences in endoplasmic reticulum stress signalling kinetics determine cell survival outcome through activation of MKP-1. Cell Signal. 2011;23:35–45. doi: 10.1016/j.cellsig.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 40.Srivastava R., Chen Y., Deng Y., Brandizzi F., Howell S.H. Elements proximal to and within the transmembrane domain mediate the organelle-to-organelle movement of bZIP28 under ER stress conditions. Plant J. 2012;70:1033–1042. doi: 10.1111/j.1365-313X.2012.04943.x. [DOI] [PubMed] [Google Scholar]
- 41.Li Y., Humbert S., Howell S.H. ZmbZIP60 mRNA is spliced in maize in response to ER stress. BMC Res. Notes. 2012;5:144. doi: 10.1186/1756-0500-5-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreno A.A., Mukhtar M.S., Blanco F., Boatwright J.L., Moreno I., Jordan M.R., Chen Y., Brandizzi F., Dong X., Orellana A. IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS ONE. 2012;7:e31944. doi: 10.1371/journal.pone.0031944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bao Y., Bassham D.C., Howell S.H. A functional unfolded protein response is required for normal vegetative development. Plant Physiol. 2019;179:1834–1843. doi: 10.1104/pp.18.01261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J.-S., Yamaguchi-Shinozaki K., Shinozaki K. ER-anchored transcription factors bZIP17 and bZIP28 regulate root elongation. Plant Physiol. 2018;176:2221–2230. doi: 10.1104/pp.17.01414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun L., Yang Z.T., Song Z.T., Wang M.J., Sun L., Lu S.J., Liu J.X. The plant-specific transcription factor gene NAC 103 is induced by b ZIP 60 through a new cis-regulatory element to modulate the unfolded protein response in A rabidopsis. Plant J. 2013;76:274–286. doi: 10.1111/tpj.12287. [DOI] [PubMed] [Google Scholar]
- 46.Hong Z.-H., Qing T., Schubert D., Kleinmanns J.A., Liu J.-X. BLISTER-regulated vegetative growth is dependent on the protein kinase domain of ER stress modulator IRE1A in Arabidopsis thaliana. PLoS Genet. 2019;15:e1008563. doi: 10.1371/journal.pgen.1008563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato R., Maeshima M. The ER-localized aquaporin SIP2; 1 is involved in pollen germination and pollen tube elongation in Arabidopsis thaliana. Plant Mol. Biol. 2019;100:335–349. doi: 10.1007/s11103-019-00865-3. [DOI] [PubMed] [Google Scholar]
- 48.Wang Q.-L., Sun A.-Z., Chen S.-T., Chen L.-S., Guo F.-Q. SPL6 represses signalling outputs of ER stress in control of panicle cell death in rice. Nat. Plants. 2018;4:280–288. doi: 10.1038/s41477-018-0131-z. [DOI] [PubMed] [Google Scholar]
- 49.Qian D., Chen G., Tian L. OsDER1 is an ER-associated protein degradation factor that responds to ER stress. Plant Physiol. 2018;178:402–412. doi: 10.1104/pp.18.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu J.-X., Howell S.H. Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell. 2010;22:2930–2942. doi: 10.1105/tpc.110.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Urade R. The endoplasmic reticulum stress signaling pathways in plants. Biofactors. 2009;35:326–331. doi: 10.1002/biof.45. [DOI] [PubMed] [Google Scholar]
- 52.Neill E.M., Byrd M.C., Billman T., Brandizzi F., Stapleton A.E. Plant growth regulators interact with elevated temperature to alter heat stress signaling via the Unfolded Protein Response in maize. Sci. Rep. 2019;9:10392. doi: 10.1038/s41598-019-46839-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carvalho H.H., Silva P.A., Mendes G.C., Brustolini O.J., Pimenta M.R., Gouveia B.C., Valente M.A.S., Ramos H.J., Soares-Ramos J.R., Fontes E.P. The endoplasmic reticulum binding protein BiP displays dual function in modulating cell death events. Plant Physiol. 2014;164:654–670. doi: 10.1104/pp.113.231928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guan P., Wang J., Li H., Xie C., Zhang S., Wu C., Yang G., Yan K., Huang J., Zheng C. Sensitive to SALT1, an endoplasmic reticulum-localized chaperone, positively regulates salt resistance. Plant Physiol. 2018;178:1390–1405. doi: 10.1104/pp.18.00840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henriquez-Valencia C., Moreno A.A., Sandoval-Ibañez O., Mitina I., Blanco-Herrera F., Cifuentes-Esquivel N., Orellana A. bZIP17 and bZIP60 regulate the expression of BiP3 and other salt stress responsive genes in an UPR-independent manner in Arabidopsis thaliana. J. Cell. Biochem. 2015;116:1638–1645. doi: 10.1002/jcb.25121. [DOI] [PubMed] [Google Scholar]
- 56.Nawkar G.M., Kang C.H., Maibam P., Park J.H., Jung Y.J., Chae H.B., Chi Y.H., Jung I.J., Kim W.Y., Yun D.-J. HY5, a positive regulator of light signaling, negatively controls the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2017;114:2084–2089. doi: 10.1073/pnas.1609844114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kørner C.J., Du X., Vollmer M.E., Pajerowska-Mukhtar K.M. Endoplasmic reticulum stress signaling in plant immunity—at the crossroad of life and death. Int. J. Mol. Sci. 2015;16:26582–26598. doi: 10.3390/ijms161125964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu J.X., Howell S.H. Managing the protein folding demands in the endoplasmic reticulum of plants. New Phytol. 2016;211:418–428. doi: 10.1111/nph.13915. [DOI] [PubMed] [Google Scholar]
- 59.Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 60.Angelos E., Ruberti C., Kim S.J., Brandizzi F. Maintaining the factory: The roles of the unfolded protein response in cellular homeostasis in plants. Plant J. 2017;90:671–682. doi: 10.1111/tpj.13449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Angelos E., Brandizzi F. NADPH oxidase activity is required for ER stress survival in plants. Plant J. 2018;96:1106–1120. doi: 10.1111/tpj.14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tateda C., Ozaki R., Onodera Y., Takahashi Y., Yamaguchi K., Berberich T., Koizumi N., Kusano T. NtbZIP60, an endoplasmic reticulum-localized transcription factor, plays a role in the defense response against bacterial pathogens in Nicotiana tabacum. J. Plant Res. 2008;121:603–611. doi: 10.1007/s10265-008-0185-5. [DOI] [PubMed] [Google Scholar]
- 63.Zhang L., Chen H., Brandizzi F., Verchot J., Wang A. The UPR branch IRE1-bZIP60 in plants plays an essential role in viral infection and is complementary to the only UPR pathway in yeast. PLoS Genet. 2015;11:e1005164. doi: 10.1371/journal.pgen.1005164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deng Y., Humbert S., Liu J.-X., Srivastava R., Rothstein S.J., Howell S.H. Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2011;108:7247–7252. doi: 10.1073/pnas.1102117108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deng Y., Srivastava R., Quilichini T.D., Dong H., Bao Y., Horner H.T., Howell S.H. IRE 1, a component of the unfolded protein response signaling pathway, protects pollen development in Arabidopsis from heat stress. Plant J. 2016;88:193–204. doi: 10.1111/tpj.13239. [DOI] [PubMed] [Google Scholar]
- 66.Liu L., Cui F., Li Q., Yin B., Zhang H., Lin B., Wu Y., Xia R., Tang S., Xie Q. The endoplasmic reticulum-associated degradation is necessary for plant salt tolerance. Cell Res. 2011;21:957–969. doi: 10.1038/cr.2010.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cui F., Liu L., Zhao Q., Zhang Z., Li Q., Lin B., Wu Y., Tang S., Xie Q. Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell. 2012;24:233–244. doi: 10.1105/tpc.111.093062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Su W., Liu Y., Xia Y., Hong Z., Li J. The Arabidopsis homolog of the mammalian OS-9 protein plays a key role in the endoplasmic reticulum-associated degradation of misfolded receptor-like kinases. Mol. Plant. 2012;5:929–940. doi: 10.1093/mp/sss042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu Y., Zhang C., Wang D., Su W., Liu L., Wang M., Li J. EBS7 is a plant-specific component of a highly conserved endoplasmic reticulum-associated degradation system in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2015;112:12205–12210. doi: 10.1073/pnas.1511724112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li L.-M., Lü S.-Y., Li R.-J. The Arabidopsis endoplasmic reticulum associated degradation pathways are involved in the regulation of heat stress response. Biochem. Biophys. Res. Commun. 2017;487:362–367. doi: 10.1016/j.bbrc.2017.04.066. [DOI] [PubMed] [Google Scholar]
- 71.Hossain M., Henríquez-Valencia C., Gómez-Páez M., Medina J., Orellana A., Vicente-Carbajosa J., Zouhar J. Identification of novel components of the unfolded protein response in Arabidopsis. Front. Plant Sci. 2016;7:650. doi: 10.3389/fpls.2016.00650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang B., Du H., Zhang Z., Xu W., Deng X. BhbZIP60 from resurrection plant Boea hygrometrica is an mRNA splicing-activated endoplasmic reticulum stress regulator involved in drought tolerance. Front. Plant Sci. 2017;8:245. doi: 10.3389/fpls.2017.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Irsigler A.S., Costa M.D., Zhang P., Reis P.A., Dewey R.E., Boston R.S., Fontes E.P. Expression profiling on soybean leaves reveals integration of ER-and osmotic-stress pathways. BMC Genom. 2007;8:431. doi: 10.1186/1471-2164-8-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang L., Xin Z., Yu X., Ma C., Liang W., Zhu M., Cheng Q., Li Z., Niu Y., Ren Y. Osmotic stress induced cell death in wheat is alleviated by tauroursodeoxycholic acid and involves endoplasmic reticulum stress–related gene expression. Front. Plant Sci. 2017;8:667. doi: 10.3389/fpls.2017.00667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang R., Chen H., Duan M., Zhu F., Wen J., Dong J., Wang T. Medicago falcata MfSTMIR, an E3 ligase of endoplasmic reticulum-associated degradation, is involved in salt stress response. Plant J. 2019;98:680–696. doi: 10.1111/tpj.14265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fujita M., Mizukado S., Fujita Y., Ichikawa T., Nakazawa M., Seki M., Matsui M., Yamaguchi-Shinozaki K., Shinozaki K. Identification of stress-tolerance-related transcription-factor genes via mini-scale Full-length cDNA Over-eXpressor (FOX) gene hunting system. Biochem. Biophys. Res. Commun. 2007;364:250–257. doi: 10.1016/j.bbrc.2007.09.124. [DOI] [PubMed] [Google Scholar]
- 77.Xi H., Xu H., Xu W., He Z., Xu W., Ma M. A SAL1 loss-of-function Arabidopsis mutant exhibits enhanced cadmium tolerance in association with alleviation of endoplasmic reticulum stress. Plant Cell Physiol. 2016;57:1210–1219. doi: 10.1093/pcp/pcw069. [DOI] [PubMed] [Google Scholar]
- 78.Guo K., Wang W., Fan W., Wang Z., Zhu M., Tang X., Wu W., Yang X., Shao X., Sun Y. Arabidopsis GAAP1 and GAAP3 modulate the unfolded protein response and the onset of cell death in response to ER stress. Front. Plant Sci. 2018;9:348. doi: 10.3389/fpls.2018.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang W., Li X., Zhu M., Tang X., Wang Z., Guo K., Zhou Y., Sun Y., Zhang W., Li X. Arabidopsis GAAP1 to GAAP3 play redundant role in cell death inhibition by suppressing the upregulation of salicylic acid pathway under endoplasmic reticulum stress. Front. Plant Sci. 2019;10:1032. doi: 10.3389/fpls.2019.01032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang X., Auwerx J. Systems phytohormone responses to mitochondrial proteotoxic stress. Mol. Cell. 2017;68:540–551.e545. doi: 10.1016/j.molcel.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 81.Poór P., Czékus Z., Tari I., Ördög A. The multifaceted roles of plant hormone salicylic acid in endoplasmic reticulum stress and unfolded protein response. Int. J. Mol. Sci. 2019;20:5842. doi: 10.3390/ijms20235842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen Y., Aung K., Rolčík J., Walicki K., Friml J., Brandizzi F. Inter-regulation of the unfolded protein response and auxin signaling. Plant J. 2014;77:97–107. doi: 10.1111/tpj.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wilkins K.A., Matthus E., Swarbreck S.M., Davies J.M. Calcium-mediated abiotic stress signaling in roots. Front. Plant Sci. 2016;7:1296. doi: 10.3389/fpls.2016.01296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Depaepe T., Hendrix S., van Rensburg H.C.J., Van den Ende W., Cuypers A., Van Der Straeten D. At the Crossroads of Survival and Death: The Reactive Oxygen Species–Ethylene–Sugar Triad and the Unfolded Protein Response. Trends Plant Sci. 2021;26:338–351. doi: 10.1016/j.tplants.2020.12.007. [DOI] [PubMed] [Google Scholar]
- 85.Xu Z., Song N., Ma L., Wu J. IRE1-bZIP60 pathway is required for Nicotiana attenuata resistance to fungal pathogen Alternaria alternata. Front. Plant Sci. 2019;10:263. doi: 10.3389/fpls.2019.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vlot A.C., Dempsey D.M.A., Klessig D.F. Salicylic acid, a multifaceted hormone to combat disease. Ann. Rev. Phytopathol. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- 87.Wang D., Weaver N.D., Kesarwani M., Dong X. Induction of protein secretory pathway is required for systemic acquired resistance. Science. 2005;308:1036–1040. doi: 10.1126/science.1108791. [DOI] [PubMed] [Google Scholar]
- 88.Reis P.A., Carpinetti P.A., Freitas P.P., Santos E.G., Camargos L.F., Oliveira I.H., Silva J.C.F., Carvalho H.H., Dal-Bianco M., Soares-Ramos J.R. Functional and regulatory conservation of the soybean ER stress-induced DCD/NRP-mediated cell death signaling in plants. BMC Plant Biol. 2016;16:156. doi: 10.1186/s12870-016-0843-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang T., Zhang P., Wang C. AtHSPR may function in salt-induced cell death and ER stress in Arabidopsis. Plant Signal. Behav. 2016;11:e1197462. doi: 10.1080/15592324.2016.1197462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fernández-Bautista N., Fernández-Calvino L., Muñoz A., Castellano M.M. HOP3, a member of the HOP family in Arabidopsis, interacts with BiP and plays a major role in the ER stress response. Plant Cell Environ. 2017;40:1341–1355. doi: 10.1111/pce.12927. [DOI] [PubMed] [Google Scholar]
- 91.Chi Y.H., Melencion S.M.B., Alinapon C.V., Kim M.J., Lee E.S., Paeng S.K., Park J.H., Nawkar G.M., Jung Y.J., Chae H.B. The membrane-tethered NAC transcription factor, AtNTL7, contributes to ER-stress resistance in Arabidopsis. Biochem. Biophys. Res. Commun. 2017;488:641–647. doi: 10.1016/j.bbrc.2017.01.047. [DOI] [PubMed] [Google Scholar]
- 92.Park J.H., Kang C.H., Nawkar G.M., Lee E.S., Paeng S.K., Chae H.B., Chi Y.H., Kim W.Y., Yun D.J., Lee S.Y. EMR, a cytosolic-abundant ring finger E3 ligase, mediates ER-associated protein degradation in Arabidopsis. N. Phytol. 2018;220:163–177. doi: 10.1111/nph.15279. [DOI] [PubMed] [Google Scholar]
- 93.Hirata R., Mishiba K.-i., Koizumi N., Iwata Y. Deficiency in the double-stranded RNA binding protein HYPONASTIC LEAVES1 increases sensitivity to the endoplasmic reticulum stress inducer tunicamycin in Arabidopsis. BMC Res. Notes. 2019;12:580. doi: 10.1186/s13104-019-4623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nguyen V.C., Nakamura Y., Kanehara K. Membrane lipid polyunsaturation mediated by FATTY ACID DESATURASE 2 (FAD 2) is involved in endoplasmic reticulum stress tolerance in Arabidopsis thaliana. Plant J. 2019;99:478–493. doi: 10.1111/tpj.14338. [DOI] [PubMed] [Google Scholar]
- 95.Wang L., Mei X.P., Nan J., Liu C.X., Zhou L., Cai Y.L. Overexpression of ZmNF-YC14 confers plant ER stress tolerance and ABA sensitivity in Arabidopsis. Acta Physiol. Plant. 2019;41:138. doi: 10.1007/s11738-019-2922-x. [DOI] [Google Scholar]
- 96.Zhu M., Tang X., Wang Z., Xu W., Zhou Y., Wang W., Li X., Li R., Guo K., Sun Y. Arabidopsis GAAPs interacting with MAPR3 modulate the IRE1-dependent pathway upon endoplasmic reticulum stress. J. Exp. Bot. 2019;70:6113–6125. doi: 10.1093/jxb/erz402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lin L., Zhang C., Chen Y., Wang Y., Wang D., Liu X., Wang M., Mao J., Zhang J., Xing W. PAWH1 and PAWH2 are plant-specific components of an Arabidopsis endoplasmic reticulum-associated degradation complex. Nat. Commun. 2019;10:3492. doi: 10.1038/s41467-019-11480-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zang Y., Gong Y., Wang Q., Guo H., Xiao W. Arabidopsis OTU 1, a linkage-specific deubiquitinase, is required for endoplasmic reticulum-associated protein degradation. Plant J. 2020;101:141–155. doi: 10.1111/tpj.14524. [DOI] [PubMed] [Google Scholar]
- 99.Hu S., Ye H., Cui Y., Jiang L. AtSec62 is critical for plant development and is involved in ER-phagy in Arabidopsis thaliana. J. Integr. Plant Biol. 2020;62:181–200. doi: 10.1111/jipb.12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pastor-Cantizano N., Ko D.K., Angelos E., Pu Y., Brandizzi F. Functional Diversification of ER Stress Responses in Arabidopsis. Trends Biochem. Sci. 2020;45:123–136. doi: 10.1016/j.tibs.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu Y., Burgos J.S., Deng Y., Srivastava R., Howell S.H., Bassham D.C. Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell. 2012;24:4635–4651. doi: 10.1105/tpc.112.101535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gupta D., Tuteja N. Chaperones and foldases in endoplasmic reticulum stress signaling in plants. Plant Signal. Behav. 2011;6:232–236. doi: 10.4161/psb.6.2.15490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Srivastava R., Li Z., Russo G., Tang J., Bi R., Muppirala U., Chudalayandi S., Severin A., He M., Vaitkevicius S.I. Response to persistent ER stress in plants: A multiphasic process that transitions cells from Prosurvival activities to cell death. Plant Cell. 2018;30:1220–1242. doi: 10.1105/tpc.18.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Moreno A.A., Orellana A. The physiological role of the unfolded protein response in plants. Biol. Res. 2011;44:75–80. doi: 10.4067/S0716-97602011000100010. [DOI] [PubMed] [Google Scholar]
- 105.Alcântara A., Seitner D., Navarrete F., Djamei A. A high-throughput screening method to identify proteins involved in unfolded protein response of the endoplasmic reticulum in plants. Plant Methods. 2020;16:4. doi: 10.1186/s13007-020-0552-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ellgaard L., Helenius A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 107.Walter P., Ron D. The unfolded protein response: From stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 108.Gao H., Brandizzi F., Benning C., Larkin R.M. A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2008;105:16398–16403. doi: 10.1073/pnas.0808463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Srivastava R., Deng Y., Howell S.H. Stress sensing in plants by an ER stress sensor/transducer, bZIP28. Front. Plant Sci. 2014;5:59. doi: 10.3389/fpls.2014.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu J.-X., Howell S.H. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell. 2010;22:782–796. doi: 10.1105/tpc.109.072173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Koizumi N., Martinez I.M., Kimata Y., Kohno K., Sano H., Chrispeels M.J. Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol. 2001;127:949–962. doi: 10.1104/pp.010636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhou J., Liu C.Y., Back S.H., Clark R.L., Peisach D., Xu Z., Kaufman R.J. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc. Natl. Acad. Sci. USA. 2006;103:14343–14348. doi: 10.1073/pnas.0606480103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hayashi S., Takahashi H., Wakasa Y., Kawakatsu T., Takaiwa F. Identification of a cis-element that mediates multiple pathways of the endoplasmic reticulum stress response in rice. Plant J. 2013;74:248–257. doi: 10.1111/tpj.12117. [DOI] [PubMed] [Google Scholar]
- 114.Mishiba K.-i., Nagashima Y., Suzuki E., Hayashi N., Ogata Y., Shimada Y., Koizumi N. Defects in IRE1 enhance cell death and fail to degrade mRNAs encoding secretory pathway proteins in the Arabidopsis unfolded protein response. Proc. Natl. Acad. Sci. USA. 2013;110:5713–5718. doi: 10.1073/pnas.1219047110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Maurel M., Chevet E., Tavernier J., Gerlo S. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem. Sci. 2014;39:245–254. doi: 10.1016/j.tibs.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 116.Römisch K. Endoplasmic reticulum–associated degradation. Annu. Rev. Cell Dev. Biol. 2005;21:435–456. doi: 10.1146/annurev.cellbio.21.012704.133250. [DOI] [PubMed] [Google Scholar]
- 117.Hoseki J., Ushioda R., Nagata K. Mechanism and components of endoplasmic reticulum-associated degradation. J. Biochem. 2010;147:19–25. doi: 10.1093/jb/mvp194. [DOI] [PubMed] [Google Scholar]
- 118.Mazzucotelli E., Belloni S., Marone D., De Leonardis A., Guerra D., Di Fonzo N., Cattivelli L., Mastrangelo A. The E3 ubiquitin ligase gene family in plants: Regulation by degradation. Curr. Genom. 2006;7:509–522. doi: 10.2174/138920206779315728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Orenstein S.J., Cuervo A.M. Chaperone-mediated autophagy: Molecular mechanisms and physiological relevance. Semin. Cell Dev. Biol. 2010;21:719–726. doi: 10.1016/j.semcdb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang Z., Klionsky D.J. Autophagy in Infection and Immunity. Springer; Berlin/Heidelberg, Germany: 2009. An overview of the molecular mechanism of autophagy; pp. 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mijaljica D., Prescott M., Devenish R.J. Microautophagy in mammalian cells: Revisiting a 40-year-old conundrum. Autophagy. 2011;7:673–682. doi: 10.4161/auto.7.7.14733. [DOI] [PubMed] [Google Scholar]
- 122.Li W.-w., Li J., Bao J.-k. Microautophagy: Lesser-known self-eating. Cell. Mol. Life Sci. 2012;69:1125–1136. doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Floyd B.E., Morriss S.C., MacIntosh G.C., Bassham D.C. What to Eat: Evidence for Selective Autophagy in Plants F. J. Integr. Plant Biol. 2012;54:907–920. doi: 10.1111/j.1744-7909.2012.01178.x. [DOI] [PubMed] [Google Scholar]
- 124.Bassham D.C., Laporte M., Marty F., Moriyasu Y., Ohsumi Y., Olsen L.J., Yoshimoto K. Autophagy in development and stress responses of plants. Autophagy. 2006;2:2–11. doi: 10.4161/auto.2092. [DOI] [PubMed] [Google Scholar]
- 125.Zhang H., Zhang H. Autophagy: A self-eating mechanism for maintaining cellular homeostasis. Chin. Sci. Bull. 2016;61:3903–3906. doi: 10.1360/N972016-01167. [DOI] [Google Scholar]
- 126.Liu Y., Bassham D.C. Autophagy: Pathways for self-eating in plant cells. Ann. Rev. Plant Biol. 2012;63:215–237. doi: 10.1146/annurev-arplant-042811-105441. [DOI] [PubMed] [Google Scholar]
- 127.Pu Y., Bassham D.C. Links between ER stress and autophagy in plants. Plant Signal. Behav. 2013;8:e24297. doi: 10.4161/psb.24297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Islam M.S., Proshad R., Kormoker T., Tusher T.R. Autophagy-mediated Nutrient Recycling and Regulation in Plants: A Molecular View. J. Plant Biol. 2019;62:307–319. doi: 10.1007/s12374-019-0213-0. [DOI] [Google Scholar]
- 129.Mizushima N., Yoshimori T., Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 130.Grumati P., Dikic I., Stolz A. ER-phagy at a glance. J. Cell Sci. 2018;131:jcs217364. doi: 10.1242/jcs.217364. [DOI] [PubMed] [Google Scholar]
- 131.Rogov V., Dötsch V., Johansen T., Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 132.Bao Y., Pu Y., Yu X., Gregory B.D., Srivastava R., Howell S.H., Bassham D.C. IRE1B degrades RNAs encoding proteins that interfere with the induction of autophagy by ER stress in Arabidopsis thaliana. Autophagy. 2018;14:1562–1573. doi: 10.1080/15548627.2018.1462426. [DOI] [PMC free article] [PubMed] [Google Scholar]