Abstract
In bacteria, the DsbA oxidoreductase is a crucial factor responsible for the introduction of disulfide bonds to extracytoplasmic proteins, which include important virulence factors. A lack of proper disulfide bonds frequently leads to instability and/or loss of protein function; therefore, improper disulfide bonding may lead to avirulent phenotypes. The importance of the DsbA function in phytopathogens has not been extensively studied yet. Dickeya solani is a bacterium from the Soft Rot Pectobacteriaceae family which is responsible for very high economic losses mainly in potato. In this work, we constructed a D. solani dsbA mutant and demonstrated that a lack of DsbA caused a loss of virulence. The mutant bacteria showed lower activities of secreted virulence determinants and were unable to develop disease symptoms in a potato plant. The SWATH-MS-based proteomic analysis revealed that the dsbA mutation led to multifaceted effects in the D. solani cells, including not only lower levels of secreted virulence factors, but also the induction of stress responses. Finally, the outer membrane barrier seemed to be disturbed by the mutation. Our results clearly demonstrate that the function played by the DsbA oxidoreductase is crucial for D. solani virulence, and a lack of DsbA significantly disturbs cellular physiology.
Keywords: plant pathogen, Dickeya solani, virulence factors, plant infection, oxidoreductase DsbA, disulfide bonds, proteomic analysis, secretome
1. Introduction
The success of plant host infection by phytopathogenic bacteria depends on the coordinated action of numerous virulence factors and virulence determinants. In most cases, these factors are extracellular secreted proteins or proteins associated with the surface of a pathogen’s cell [1]. The ability to withstand unfavorable environmental conditions (both in and outside of the host) is an equally important factor. As an outer part of the bacterium, the bacterial cellular envelope is particularly exposed to fluctuations of the environment. Therefore, the ability of bacteria to survive various external stress factors is strictly linked with the correct functioning of the envelope as the cell/environment barrier. Since secreted or envelope proteins are particularly exposed to harsh environmental conditions, which may result in their rapid denaturation, their structure is frequently stabilized by disulfide (S-S) bonds formed between cysteine residues. Conversely, a lack of proper disulfide bonds in these proteins frequently results in unfolding, loss of function and subsequent protein degradation. In bacteria, oxidoreductases of the Dsb (Disulfide bond) protein family are responsible for introducing proper S-S bonds between cysteine residues [2,3,4], reviewed in [5].
In the model Gram-negative bacterium Escherichia coli, the cysteine residues of exported proteins are oxidized by the DsbA/DsbB system in the periplasm [6]. Proteins that contain more than two cysteine residues frequently need rearrangement of the S-S bonds, and the DsbC/DsbD isomerase system catalyzes this process. Additionally, DsbE and DsbG proteins act as specialized reducing enzymes [7].
The substrates of DsbA comprise a large variety of proteins whose functions are necessary to maintain cellular homeostasis as well as proteins associated with bacterial pathogenesis. The latter include diverse bacterial virulence factors, for example, toxins, secretion systems, adhesins and flagellar motility machinery. As a result, bacterial cells deprived of the functional dsbA gene exhibit a pleiotropic phenotype, frequently show reduced fitness in animal infection models and/or display attenuated virulence, reviewed in [5,8]. The vast majority of previous studies concerning Dsb proteins were performed in E. coli. Likewise, only a few animal/human bacterial pathogens (e.g., Burkholderia pseudomallei, uropathogenic E. coli or Salmonella enterica Typhimurium) were used to assess the role of Dsb proteins in the interaction between these bacteria and their hosts [8]. In contrast, knowledge about the involvement of these enzymes in the interaction between phytopathogenic bacteria and their hosts is still very limited.
According to the published genome sequence [9] and Uniprot [10] analysis, Dickeya solani generally encodes all components of the archetypal Dsb system from E. coli K12: DsbA/DsbB (oxidative pathway), DsbC/DsbD (isomerization pathway) as well as DsbG as a specialized reducing enzyme (a gene coding for DsbE seems to be absent in D. solani). The DsbA protein from D. solani (thiol:disulfide interchange protein of D. solani RNS 08.23.3.1.A; UniProtID: A0A2K8W006) shares 66.8% amino acid sequence identity and 82.2% similarity with E. coli strain K12 DsbA (UniProtID: P0AEG4) (EMBOSS Needle sequence alignment tool). The sizes of both DsbA homologs are similar as well (208 and 213 amino acids in D. solani and E. coli, respectively), and the composition of their catalytic sites is identical (CPHC). Active site cysteine residues are localized at positions 49 and 52 (E. coli) and 55 and 58 (D. solani).
This work aimed to assess, in detail and at the molecular level, the role of the protein DsbA in the environmental fitness and virulence of a member of the Soft Rot Pectobacteriaceae (SRP) group, D. solani, during its interaction with its natural host plant, potato. The SRP bacteria cause significant losses in various crops and ornamentals worldwide and therefore were included among the top ten most important plant pathogens [11]. In Europe, the SRPs cause economic losses mainly in potato, estimated to be approximately 46 M Euro annually [12]. Among the many SRP species, D. solani has recently been recognized as one of the most virulent plant pathogens. D. solani causes the rotting of tubers in the field and under storage conditions and the blackleg disease of stems of field-growing potato plants. Compared to other Dickeya species, D. solani causes more severe symptoms in potato plants, has a wider optimal temperature range for growth and disease development and can develop blackleg disease from lower inoculum levels [13]. Furthermore, over the past 20 years, D. solani became the most abundant Dickeya species isolated from infected potato plants in Europe [14,15,16,17,18,19,20].
The SRP group of plant pathogenic bacteria, including D. solani, adopted an invasion strategy based on the massive production of extracellular virulence factors whose role is to degrade components of the plant cell wall (plant-cell-wall-degrading enzymes; PCWDEs). The essential PCWDEs of SRPs are pectin-degrading enzymes (pectinases), cellulases and proteases, which are directly responsible for decomposing plant tissues. Pectinases are a group of enzymes indispensable for bacterial virulence, comprising pectate lyases, endopolygalacturonases, pectin lyases and pectin esterases. Among them, pectate lyases are considered the most important, and they cause the rapid degradation of pectins and polygalacturonic acid, reviewed in [21]. Some literature reports suggest the necessity of DsbA for the activity of PCWDEs. For example, in Dickeya dadantii 3937 (formerly Erwinia chrysanthemi 3937), DsbA is required for the stability and secretion of several pectate lyases and of the cellulase CelZ [22]. Studies performed on Pectobacterium atrosepticum (formerly Erwinia carotovora subsp. atroseptica) demonstrated that DsbA deficiency impairs the production of almost all secreted virulence factors. As a result, the secretome of the P. atrosepticum dsbA mutant lacked the major secreted proteins. Likewise, the total amount of secreted proteins was lower than in the wild-type (wt) strain [23].
The plant host infection caused by the SRPs can be divided into three main steps: (I) the adhesion of bacterial cells to the plant surface and their entry into plant tissues, (II) colonization of the apoplast and finally (III) degradation of the plant cell wall components, resulting in the development of visible disease symptoms. Therefore, to successfully colonize and cause disease symptoms in the host, bacteria need to coordinate the expression and activity of many virulence factors. In the first stage of interaction (adhesion to host cell and entry into plant tissue), the motility and chemotaxis of the pathogen play a crucial role. These determinants are essential for bacteria when searching for favorable sites to adhere to and enter the plant through. For this reason, non-motile D. dadantii mutants encounter difficulties in penetrating the plant tissue and in establishing a systemic infection [24]. There are two major types of bacterial movement, swimming and swarming; both types depend on functional flagella and the sensing of chemical signals (attractants or repellents). Swimming cells perceive chemical signals via methyl-accepting chemotaxis proteins and trigger appropriate chemotactic responses to the signals [25]. Swarming is a movement of tight groups of bound bacterial cells on semisolid surfaces [26] and is associated with a differentiation process of bacterial cells [27,28]. During the colonization of the apoplast, bacteria must adjust to rather harsh environmental conditions, including plant cell defense responses and the low availability of iron ions, which are crucial cofactors of many enzymes. For this purpose, bacteria produce additional virulence determinants, including systems related to iron acquisition (siderophores and iron storage proteins) and the reactive oxygen species scavenger indigoidine [29,30].
In this work, we examined the importance of the oxidoreductase DsbA for the growth, virulence and activity of essential virulence determinants of the plant pathogenic D. solani type strain, IPO2222. We also undertook a global investigation into the role of DsbA in maintaining cellular proteostasis and protein secretion. We found that the dsbA mutants showed no growth defects under stressful conditions but were unable to infect plants and to produce disease symptoms. Proteomic analysis revealed a global impact of the dsbA inactivation on the D. solani proteome, involving the envelope and both cytoplasmic and secreted proteins. In addition, the dsbA mutant was deprived of most secreted virulence factors, including PCWDEs, siderophores and flagellar proteins. To our knowledge, this is the first such extensive work focused on the function of DsbA in a bacterial phytopathogen of the genus Dickeya.
2. Results
2.1. Inactivation of the dsbA Gene Does Not Cause an Increased Sensitivity to Stressful Conditions
To investigate the potential contribution of the DsbA protein in D. solani fitness and virulence, the dsbA gene of the D. solani IPO2222 chromosome was inactivated by marker exchange mutagenesis to produce the D. solani dsbA::cmR mutant (TP02) (Figure S1). To ensure that the potential differences between the mutant and parental strains were solely due to the presence/absence of the DsbA function, we constructed a complemented strain (TP03), where the wt dsbA gene was returned into its native location in the D. solani chromosome.
To evaluate the phenotypic effects of the dsbA mutation, the growth curves of the D. solani dsbA mutant, complemented and parental D. solani IPO2222 strains were determined. The dsbA mutant did not show any growth defects compared to the control strains when grown under physiological conditions (Figure S2A). Next, the phenotype of D. solani dsbA was evaluated under stressful conditions: heat shock, osmotic/saline, oxidative and acid stresses. We found that the inactivation of DsbA did not enhance the negative effects of either stressor (Figures S2B, S3–S6).
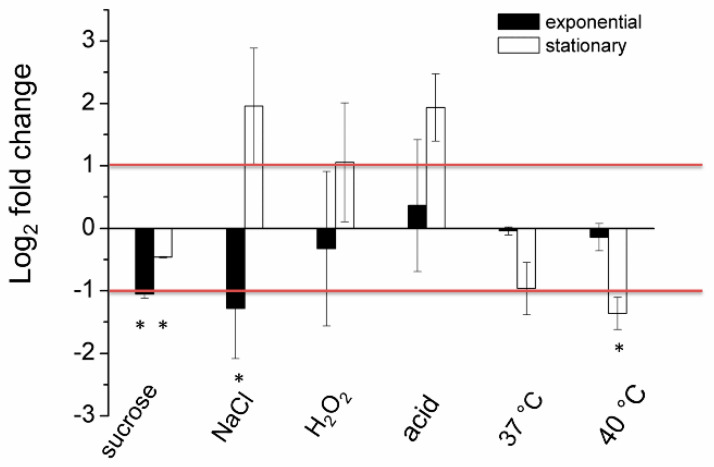
To confirm that DsbA does not play a significant role in the protection of the D. solani cells against stress consequences, we examined the expression of the dsbA gene under selected stressful conditions. As shown in Figure 1, treatment with stressors generally did not lead to a significant induction of the dsbA transcription. However, we observed dsbA repression under several growth conditions, particularly in the exponentially growing cells in the presence of osmotica (both ionic and non-ionic) or in stationary-phase growing cells at 40 °C. Only three culture conditions, treatment of stationary-phase cells with sodium chloride (NaCl), hydrogen peroxide (H2O2) or acid pH, seemed to increase the dsbA mRNA content, but the differences were not statistically significant. Hence, DsbA does not seem to be a significant factor in the D. solani stress response.
Figure 1.
The relative log2 fold change of expression levels of the dsbA gene in the D. solani cells under stressful conditions analyzed via qPCR. The data correspond to the means ± standard deviation (SD) of three biological replicates with three technical repeats. The red horizontal lines denote a relative two-fold increase/decrease in the dsbA expression level. * indicates statistically significant (95% Confidence Interval) fold change in expression level according to the REST 2009 software.
2.2. The D. solani dsbA Mutant Is Impaired in Motility
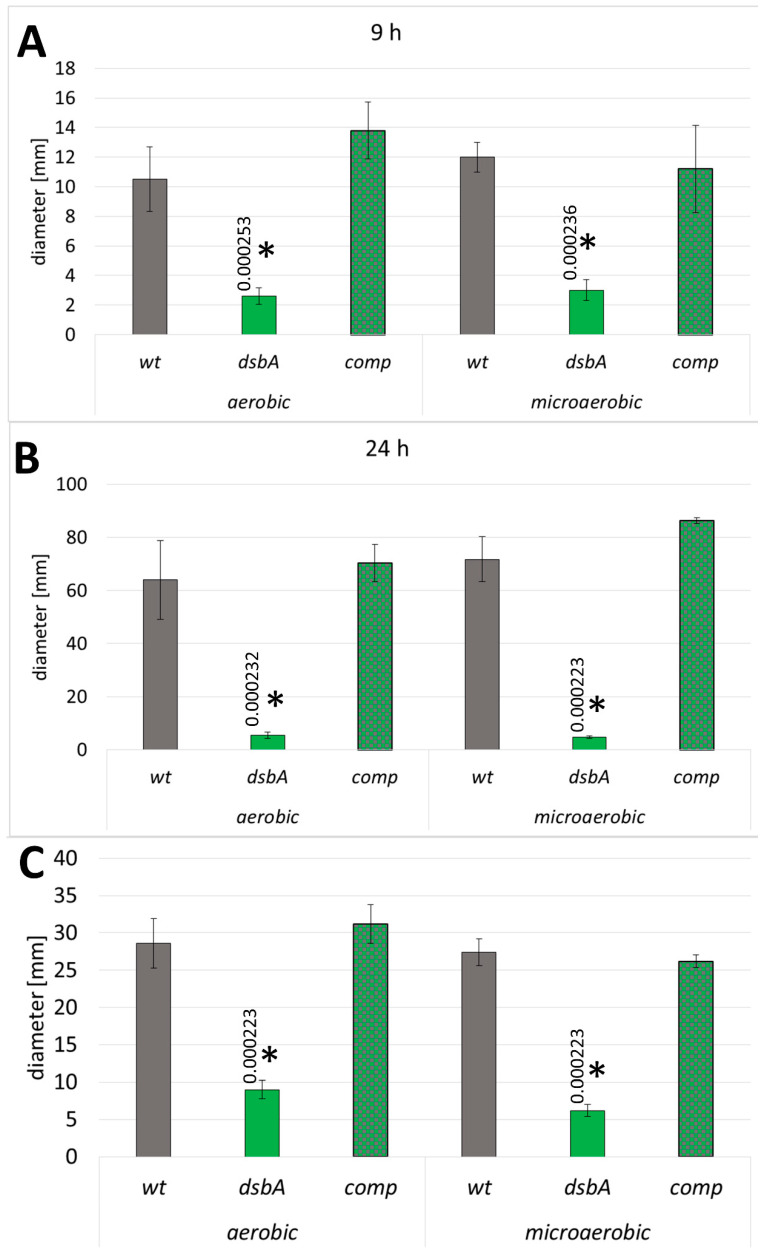
We found that the dsbA mutant exhibited a dramatic reduction in swarming and swimming motility both under microaerobic and aerobic conditions. On swarming plates (0.5% agar), the migration of the mutant cells was hardly detectable when the wild-type bacteria migrated efficiently and formed large swarming growth zones after 24 h (Figure 2A,B and Figure S7A). Non-swarming dsbA cells grew as a confined colony in the center of the plate. Under prolonged incubation, the colony diameter of non-swarming cells may increase owing to the contribution of sliding motility [31], as observed on plates incubated for 24 h. The dsbA mutant failed to exhibit swimming motility in 0.3% agar (Figure 2C and Figure S7B). The expression of DsbA in the complemented strain restored both types of motilities (Figure 2 and Figure S7).
Figure 2.
Effect of the dsbA knockout on the motility of D. solani. Mean values of swarming zones after 9 and 24 h of incubation (A,B) and swimming zones (C) are shown; wt—wild-type strain; dsbA—mutant strain TP02; comp—complemented strain TP03. The data represent values for five biological replicates. The statistical analysis of differences was determined by two-way analysis of variance (ANOVA) test followed by a post hoc Tukey test. The asterisks indicate statistical significance (p < 0.001); the numbers next to asterisks show the actual p-values.
2.3. The dsbA Mutant Shows Impaired Activity of Secreted Virulence Factors
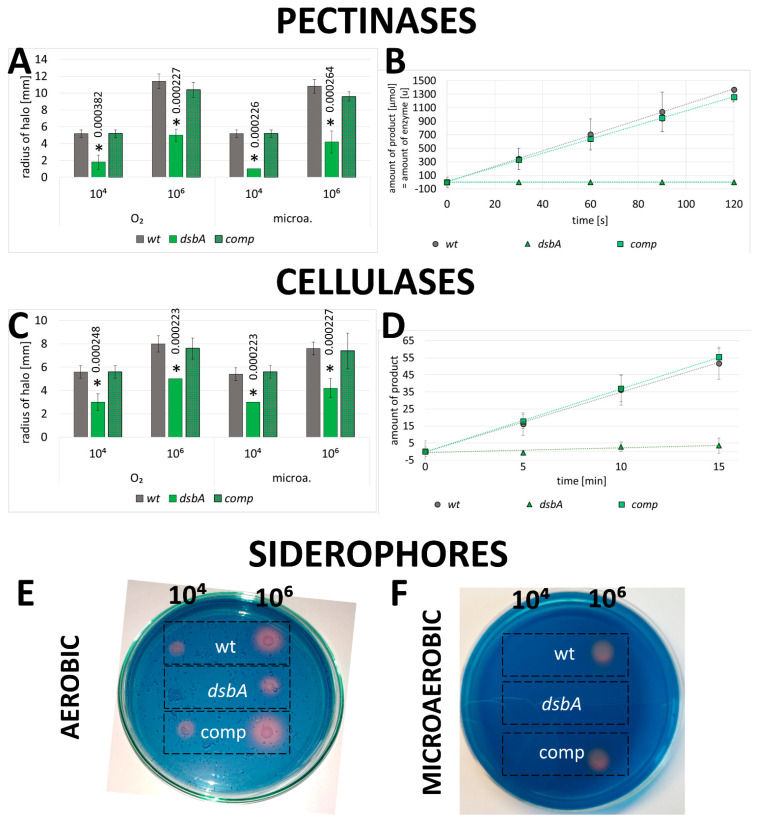
As a part of the phenotypic characterization of the D. solani dsbA mutants, we measured the activity of secreted PCWDEs (pectinases, cellulases and proteases) using plate assays. We found that bacteria lacking the functional dsbA gene were severely defective in the secretion of active pectinases into the growth medium. As demonstrated in Figure 3A and Figure S8A, the pectinolytic activity of the dsbA mutant was very low in plate assays. In contrast, the wt and complemented strains secreted comparable amounts of active enzymes. To further quantify the effect of DsbA on the secreted pectate lyase production, we used a quantitative spectrophotometric assay to measure pectinolytic activity in culture supernatants. While no pectate lyase activity was detected in the D. solani dsbA secretome, polygalacturonate was efficiently degraded in the presence of the control bacteria supernatants (Figure 3B).
Figure 3.
Impact of the dsbA knockout on the secreted virulence factors of D. solani: (A,B) pectinolytic activity, (C,D) cellulolytic activity and (E,F) siderophore secretion. Panels A and C show the mean values of the halo radii determined by plate assays. The statistical analysis of differences was determined by two-way analysis of variance (ANOVA) test followed by a post hoc Tukey test. The asterisks indicate statistical significance (p < 0.001); the numbers next to asterisks show the actual p-values. B and D—spectrophotometric determination of the enzymes’ activity in culture supernatants. Error bars represent SD from five experimental repetitions. Representative examples of Chromo-Azurol stained plates used to detect siderophore production are shown in E, F. In every assay, five independent replicates of each strain were analyzed. Abbreviations: wt—wild-type strain; dsbA—mutant strain TP02; comp—complemented strain TP03.
Similarly, the dsbA knockout significantly affected the activity of secreted cellulases. The hydrolysis of the cellulase substrate was markedly less efficient in the dsbA mutant secretome than in the control bacteria secretomes (Figure 3C,D and Figure S8B).
In contrast, the proteolytic activity was not impaired by the dsbA knockout, as judged from the casein degradation zone diameters observed in aerobic or anaerobic growth conditions (Figure S9).
D. solani produces water-soluble siderophores that strongly bind ferric iron under iron-limiting conditions. We observed that siderophore secretion was significantly reduced in the dsbA mutant when compared to the control bacteria (Figure 3E,F).
2.4. DsbA Is Required to Establish the Infection in Potato Plants and to Cause Disease Symptoms
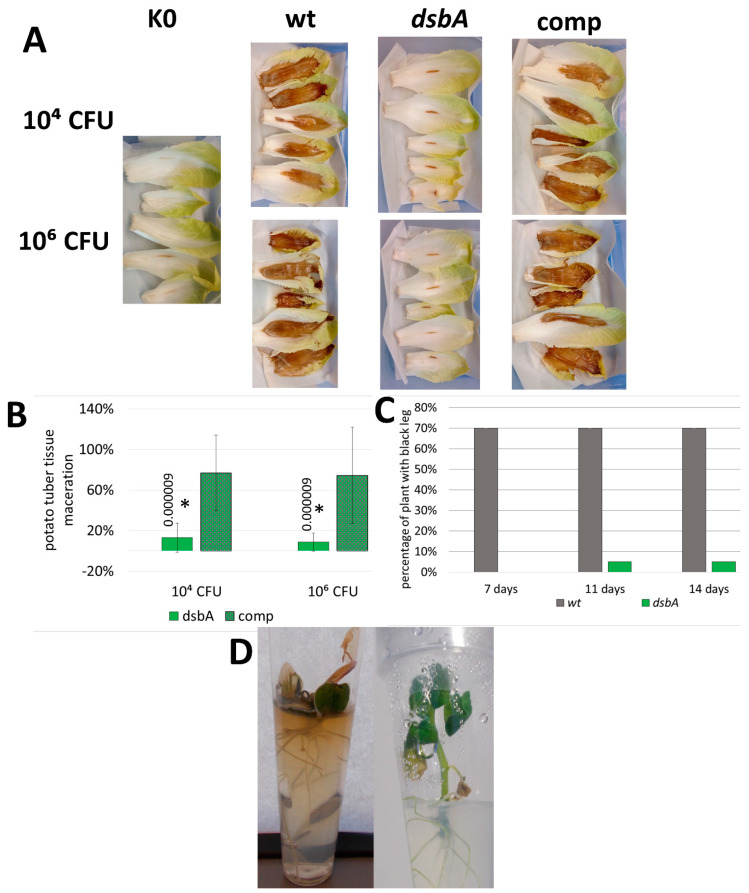
To determine the effect of the dsbA mutation on D. solani virulence, we conducted various infectivity tests using potato (Solanum tuberosum) and chicory (Cichorium intybus) as host plants. In each test, a decreased ability of the dsbA mutant to infect plant tissues and develop disease symptoms was observed. First, the D. solani dsbA mutant did not cause the maceration of chicory leaves, contrary to the control strains. The efficiencies of tissue maceration caused by the wt and complemented strains were very similar, confirming that the avirulent phenotype of the dsbA mutant was indeed a result of a lack of the DsbA function (Figure 4A). Secondly, the dsbA mutant was inefficient in the maceration of whole potato tubers (Figure 4B and Figure S10). Using a potato tuber slice assay, the maceration level was dependent on the bacteria density used for inoculation. Tuber tissue rotting occurred when at least 104 CFU of bacteria were used for inoculation. With that number of bacteria, the efficiency of rotting was clearly higher in the case of the control bacteria, while at 106 CFU, a moderate maceration of tuber tissues was also visible in the case of the dsbA mutant (Figure S11). Good oxygen availability in the tuber slice assay can at least partially suppress the effects of the dsbA knockout. In the whole tuber assay, the oxygen access is much more limited. Whole tubers infected with the dsbA mutant showed minor rotting symptoms, and the weight of macerated tissue was approximately five-fold lower than that produced by the wt D. solani strain. Disease symptoms caused by the complemented strain were similar to those produced by the wt D. solani strain (Figure 4B). These data prove that the phenotype of the D. solani dsbA mutant directly results from the dsbA inactivation. Under microaerobic conditions, the presence of a functional dsbA gene is necessary for potato tuber tissue maceration.
Figure 4.
Influence of the dsbA mutation on the D. solani virulence. For the virulence tests, maceration of plant tissues was evaluated on isolated plant parts (A,B) and in planta infection assays were performed on whole plants (C,D). (A) Representative examples of chicory leaf infections. Five independent biological replicates of each strain and bacterial inoculum were analyzed. (B) Relative values of potato tuber maceration caused by the mutant TP02 (dsbA) and the complemented strain TP03 (comp) expressed as percentages of maceration caused by the wt D. solani cells. The weight of tissue macerated by wild-type bacteria was set at 100%. Per set of strains, nine individual tubers were inoculated in two independent experiments (n = 18). Statistical analysis of differences was determined by two-way analysis of variance (ANOVA) test followed by a post hoc Dunnett test. The result was considered significant at p < 0.05; the numbers next to asterisks show the actual p-values. (C) Percentage of the potato plants showing black leg symptoms in the pot assay. Five independently grown cultures of each bacterial strain were used in the assay. Each culture was used to infect 4 potato plants, giving 20 plants per strain (n = 20). Inoculated plants were visually inspected 7, 11 and 14 days post infection for wilting, typical blackleg symptoms and plant death. (D) Representative examples of the potato plantlets infected with D. solani wt (left) and D. solani dsbA (right).
Then, the ability of the D. solani dsbA mutant to infect the whole potato plants and develop disease symptoms was examined. In planta experiments were performed using two models: potted potato plants (cv. Vineta) and in vitro grown potato plantlets (cv. Kondor). The latter approach allowed for extended studies under strictly controlled and sterile conditions. It turned out that the D. solani dsbA mutant was unable to efficiently develop disease symptoms. After inoculation with the dsbA mutant, only one pot-grown potato plant (out of twenty) showed black leg symptoms (Figure 4C), while none of the in vitro plantlets were symptomatic (Figure 4D). In contrast, the majority of potato plants infected with the wt D. solani quickly developed disease symptoms (Figure 4C,D). In plants inoculated with sterile Ringer buffer, no disease symptoms were observed during the entire course of the experiments.
The use of culture tubes-grown plants allowed us to check in detail whether the mutants could penetrate the host, disseminate and colonize the plant tissues. We observed that the mutant cells were present in the upper parts of the plantlets (1 cm above the surface of the culture medium) (Figure S12). Two days post inoculation (dpi), the numbers of dsbA or wt cells in the plantlet stems were comparable. At the later stages of infection (9th or 13th dpi), the CFU counts were higher for the control strain than for the dsbA mutant (although the changes were not statistically significant).
2.5. The Lack of a Functional dsbA Gene Causes Pronounced Changes in the Overall D. solani Cell Proteome
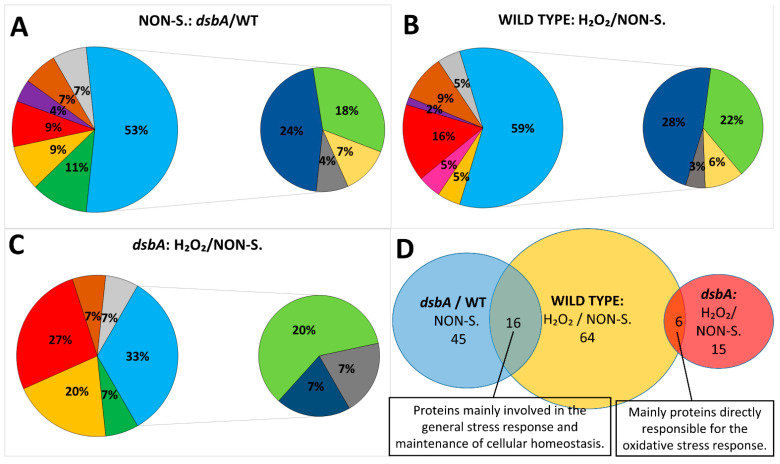
To gain more insights into the characteristics of the D. solani dsbA cells, the proteomes of the mutant and wt bacteria were compared by the means of Sequential Window Acquisition of All Theoretical Mass Spectra (SWATH-MS) analysis. This analysis identified a total of 607 proteins for which at least two peptides per protein were quantified (Table S1). Inactivation of the dsbA gene caused strong changes in the D. solani proteome. For further analysis, we narrowed the number of differentially expressed proteins by applying the following cut-off criteria: p < 0.05, as well as fold changes below 0.5 or above 2. As a result, we selected 45 proteins with significantly altered abundance. These proteins were grouped based on their functions: motility and chemotaxis, transport, stress response and protein quality control, virulence and attachment, primary metabolism, transcription and translation, membrane and cell wall biogenesis and finally, other or unknown functions (Table 1, Figure 5A).
Table 1.
Summary of proteins whose level depends on DsbA activity.
| UniProt ID | Name | p-Value | Fold Change | Description | Number of Cys | Predicted Localization |
|---|---|---|---|---|---|---|
| MOTILITY AND CHEMOTAXIS | ||||||
| A0A2K8VVH9 A0A6N0AT44 |
CheA | 0.0023 | 0.4227 | Signal transduction histidine kinase CheA | 3 | C |
| A0A6N0AAK8 | CheV | 0.0062 | 2.1167 | Chemotaxis protein | 1 | C |
| A0A6N0AH46 | CheW | 0.0269 | 0.4451 | Chemotaxis protein | 0 | C |
| A0A6N0B2V1 | FlgE | 0.0085 | 0.3043 | Flagellar hook protein | 0 | EX |
| A0A6N0AG83 | FliC | <0.0001 | 0.0080 | Flagellin 1 | 0 | EX |
| TRANSPORT | ||||||
| A0A6N0AQD6 | GltI | 0.0010 | 0.2706 | Glutamate/aspartate periplasmic-binding protein | 2 | EX |
| A0A6N0B4D0 | OppA | 0.0234 | 0.3684 | Periplasmic oligopeptide-binding protein | 0 | EX |
| A0A6N0AX80 | PstS | 0.0214 | 2.6510 | Phosphate-binding protein | 0 | EX |
| A0A2K8W558 A0A6N0B1L8 |
SitA | 0.0025 | 2.6286 | Manganese ABC transporter, periplasmic-binding protein | 0 | EX |
| STRESS RESPONSE AND PROTEIN QUALITY CONTROL | ||||||
| A0A6N0AF95 | DegP | 0.0162 | 2.9982 | Periplasmic serine endoprotease | 2 | EX |
| A0A6N0AY12 | GroES | 0.0004 | 0.2959 | 10 kDa chaperonin | 0 | C |
| A0A6N0BH32 | GshB | 0.0043 | 2.7279 | Glutathione synthetase | 4 | C |
| A0A6N0ARS9 | RecA | <0.0001 | 2.5880 | Recombinase A | 4 | C |
| A0A6N0ABQ5 | RcsB | 0.0274 | 2.1452 | Transcriptional regulatory protein | 0 | C |
| VIRULENCE AND ATTACHMENT | ||||||
| A0A6N0ADK4 | OmpW | 0.0465 | 0.4662 | Outer membrane protein W | 0 | EX |
| A0A2K8W5L9 A0A6N0BBS5 |
OmpX | 0.0001 | 2.0530 | Attachment invasion locus protein precursor | 0 | EX |
| PRIMARY METABOLISM | ||||||
| A0A6N0AP75 | MaeA | 0.0202 | 3.0996 | NAD-dependent malic enzyme | 8 | C |
| A0A2K8VWM7 | SuhB | 0.0004 | 5.5655 | Inositol-1-monophosphatase | 3 | C |
| A0A6N0AXG4 | AccA | 0.0005 | 2.2560 | Acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha | 4 | C |
| A0A2K8W5K9 A0A6N0B1Z5 |
0.0059 | 0.3826 | acidPPc domain-containing protein | 3 | EX | |
| A0A6N0AKV1 | 0.0452 | 2.8777 | Cytochrome bo(3) ubiquinol oxidase subunit 1 | 4 | EX | |
| A0A2K8VUJ2 A0A6N0AFZ5 |
0.0009 | 3.6718 | 2-dehydropantoate 2-reductase | 5 | C | |
| A0A6N0AZD1 | 0.0260 | 0.1796 | L-asparaginase | 0 | C | |
| A0A2K8W0U8 A0A6N0AIW6 |
0.0472 | 0.3564 | NADH dehydrogenase | 6 | C | |
| A0A6N0AL19 | 0.0108 | 2.1397 | NAD(P)H-flavin reductase | 2 | C | |
| A0A6N0AAE7 | 0.0089 | 6.8300 | Phosphoglucomutase | 5 | C | |
| A0A6N0B8R8 | 0.0279 | 10.8132 | Ribulose-phosphate 3-epimerase | 1 | C | |
| → protein synthesis | ||||||
| A0A6N0B0C3 | HisD | 0.0057 | 2.5386 | Histidinol dehydrogenase, histidine biosynthesis |
7 | C |
| A0A6N0AIR2 | LeuA | 0.0357 | 0.4026 | 2-isopropylmalate synthase | 6 | C |
| A0A6N0AQ52 | L32 (RpmF) | 0.0294 | 0.4699 | 50S ribosomal protein L32 | 0 | C |
| A0A6N0AJL8 | TrpS | 0.0024 | 2.1145 | Tryptophan-tRNA ligase | 4 | C |
| A0A6N0AWV6 | TypA (BipA) | 0.0012 | 0.4929 | GTP-binding protein TypA/BipA (ribosome biogenesis) | 5 | C |
| A0A2K8VZB6 A0A6N0AI45 |
YxeP | 0.0013 | 3.3600 | N-acetyl-L,L-diaminopimelate deacetylase-like protein (Leu biosynthesis) | 2 | C |
| A0A6N0BB83 | 0.0012 | 3.9494 | Elongation factor Tu | 3 | C | |
| A0A6N0AJ97 | 0.0051 | 3.9942 | Elongation factor Tu | 3 | C | |
| → nucleic acids AND cofactors synthesis and homeostasis | ||||||
| A0A6N0BGD6 | PurF | 0.0098 | 0.4078 | Amidophosphoribosyltransferase | 6 | C |
| A0A2K8VZA5 A0A6N0AL00 |
0.0002 | 2.1056 | Adenylosuccinate lyase (de novo purine synthesis) |
2 | C | |
| → polyamine biosynthesis | ||||||
| A0A6N0ASX1 | SpeA | 0.0010 | 3.1524 | Biosynthetic arginine decarboxylase | 8 | EX |
| A0A6N0AWG0 | SpeD | 0.0253 | 2.0829 | S-adenosylmethionine decarboxylase proenzyme | 6 | C |
| LIPOPROTEINS, MEMBRANE AND CELL WALL BIOGENESIS | ||||||
| A0A6N0BB31 | BamA | 0.0019 | 2.5034 | Outer membrane protein assembly factor | 2 | EX |
| A0A2K8VZQ3 A0A6N0AK79 |
DcrB | 0.0126 | 2.3492 | Periplasmic bacteriophage sensitivity protein DcrB | 1 | EX |
| A0A6N0BHR4 | MurC | 0.0060 | 2.9827 | UDP-N-acetylmuramate-L-alanine ligase | 2 | C |
| OTHERS, UNIDENTIFIED | ||||||
| A0A2K8W438 A0A6N0APE9 |
VirK | 0.0480 | 0.1451 | VirK protein (D. dadantii YbjX homologue) | 2 | EX |
| A0A2K8VTQ1 A0A6N0AFA2 |
0.0001 | 0.3205 | CaMKII-AD, RNS: SgcJ/EcaC family oxidoreductase | 2 | EX | |
| A0A6N0AEH1 | 0.0233 | 0.4976 | Uncharacterized protein | 0 | N | |
UniProtID in gray are the archival numbers. The fold change values of downregulated proteins are in italics; number of cysteine residues in a protein (Number of. Cys); predicted protein localization: the cytoplasm (C), the extracytoplasmic space (EX), not known (N).
Figure 5.
Graphic representations of proteomic data. (A–C) Functional categories and percentages of proteins whose levels were changed in the D. solani proteome due to (A) dsbA mutation, (B) oxidative stress or (C) in the D. solani dsbA mutant under oxidative stress. Areas in big circles: green: motility and chemotaxis; yellow: transport; red: stress response and protein quality control; pink: chaperones; purple: virulence and attachment; brown: lipoproteins, membrane and cell wall biogenesis; gray: others, unidentified; blue: metabolism. Areas in small circles: dark blue: general metabolism; green: protein synthesis; yellow: nucleotide and cofactors synthesis and homeostasis; gray: polyamine biosynthesis. (D) Diagram of unique and shared differentially expressed proteins identified in the proteome samples. Blue: proteins differentially expressed in the dsbA mutant compared to the parental wt D. solani cells, both grown under non-stressful conditions (NON-S.); yellow: proteins differentially expressed in the wt cells in the presence of the oxidant compared to the bacteria grown under non-stressful conditions; red: proteins differentially expressed in the dsbA mutant cells in the presence of the oxidant compared to cells grown under non-stressful conditions.
First, we focused on proteins that could be related to the D. solani pathogenicity. Almost all of the mass spectra (MS) identified proteins linked to motility and chemotaxis, except CheV, showed decreased levels in the D. solani dsbA proteome compared to that of the wild-type bacteria. The protein showing the most reduced content (app. 100-fold) in the dsbA mutant was the flagellin FliC. The second protein involved in flagella structure, the flagellar hook protein FlgE, was also less abundant. Of the proteins involved in chemotaxis, two proteins showed significantly reduced levels, including CheA, a signal transduction kinase which participates in the transmission of signals from the chemoreceptors to the flagellar motors [32].
A lack of DsbA function led to the induction of proteins related to protein quality control (PQC) and to general stress responses. Among the periplasmic PQC components, only the DegP protease was markedly upregulated. The proteomic analysis suggested an increased abundance of some other periplasmic chaperones/folding helpers (SurA, FkpA); albeit their fold changes were lower than 2. Several cytoplasmic stress-related proteins were also upregulated, such as the glutathione synthetase GshB, the transcriptional regulator RcsB, the protein RecA required for the SOS response and probably an aldo/keto reductase (A0A2K8VWF4) (the fold changes were not statistically significant in the latter case).
The level of some proteins specifically involved in transport was either increased or decreased in the dsbA mutant. SitA, a metal transport protein, and PstS, a phosphate-binding protein, were present in larger quantities, while the concentrations of GltI, a glutamate/aspartate binding protein, and OppA, a periplasmic oligopeptide binding protein, were reduced in comparison to the control cells.
Enzymes related to primary metabolism constituted a large group of proteins with altered levels in the dsbA mutant. The ribulose-phosphate 3-epimerase RPE, an enzyme of the pentose phosphate pathway, was highly induced (app. 10-fold). Other abundant proteins included components of sugar metabolic pathways (SuhB and phosphoglucomutase), polyamine biosynthesis pathways (SpeA and SpeD), membrane and cell wall biogenesis (BamA, MurC and DcrB), amino-acid- and protein-synthesis-related pathways (HisD, TrpS, YxeP and elongation factors EF-Tu) and electron transfer (Cytochrome bo(3) ubiquinol oxidase, NAD(P)H-flavin reductase, NADH dehydrogenase and NAD-dependent malic enzyme). Some metabolism-related proteins were less abundant in the dsbA mutant cells, such as L-asparaginase (catalyzing the transformation of L-asparagine into L-aspartic acid and ammonia), a putative periplasmic phosphatase (A0A2K8W5K9), 2-isopropylmalate synthase, TypA (50S ribosomal subunit assembly factor) and BipA (a member of the ribosome-binding GTPase superfamily).
2.6. Exposure to Oxidative Stress Causes More Significant Changes in the Proteome of the wt D. solani Cells Than in That of the dsbA Mutant
As the dsbA mutation is related to disturbances in cellular redox balance, we also compared proteomes of bacteria treated with oxidative stress. Treatment with the oxidant H2O2 caused pronounced effects on the cellular proteome of D. solani IPO2222. The levels of 64 proteins were significantly altered: 51 proteins were upregulated, while 13 were down-regulated in comparison to protein levels found in bacterial cells grown under non-stressful conditions (Table 2, Figure 5B,C).
Table 2.
The influence of oxidative stress on the proteomes of the D. solani wild-type strain and the derived dsbA mutant.
| UniProt ID | Name | p-Value | Fold Change | p-Value | Fold Change | Description | No. of Cys | Pred. Local. |
|---|---|---|---|---|---|---|---|---|
| WILD TYPE | dsbA MUTANT | |||||||
| MOTILITY AND CHEMOTAXIS | ||||||||
| A0A2K8W3X3 A0A6N0ABX5 |
0.0272 | 0.3321 | Methyl-accepting chemotaxis protein I, serine chemoreceptor protein |
3 | EX | |||
| TRANSPORT | ||||||||
| A0A6N0B6V8 | OmpF | <0.0001 | 0.2333 | Outer membrane protein F | 0 | EX | ||
| A0A6N0AX80 | PstS | 0.0013 | 4.4512 | Phosphate-binding protein | 0 | EX | ||
| A0A6N0A6N3 | SecB | 0.0139 | 2.2809 | Protein-export protein SecB | 4 | C | ||
| A0A2K8VZD5 A0A6N0B8Y3 |
0.0453 | 0.4820 | ABC-type polar amino acid transport system, ATPase component | 1 | EX | |||
| A0A6N0AZI4 | 0.0145 | 0.4237 | Putative Fe(2+)-trafficking protein | 1 | EX | |||
| A0A2K8W4X3 A0A6N0AAN4 |
0.0138 | 2.0647 | Outer membrane receptor protein, involved in siderophore uptake | 0 | EX | |||
| CHAPERONES | ||||||||
| A0A6N0B884 | IbpB | <0.0001 | 0.3876 | Small heat shock protein | 0 | C | ||
| A0A6N0BK24 | SlyD | 0.0053 | 0.4416 | FKBP-type peptidyl-prolyl cis-trans isomerase | 5 | C | ||
| A0A2K8VY47 | SurA | 0.0098 | 2.0860 | Chaperone | 0 | EX | ||
| STRESS RESPONSE AND PROTEIN QUALITY CONTROL | ||||||||
| A0A6N0AZJ6 | ClpP | 0.0003 | 2.0636 | ATP-dependent Clp protease proteolytic subunit | 2 | C | ||
| A0A6N0AN16 | Dps | 0.0010 | 6.5526 | <0.0001 | 5.6274 | DNA protection during starvation protein | 0 | C |
| A0A6N0AYR4 | KatG | 0.0017 | 2.0880 | 0.0014 | 2.3612 | Catalase-peroxidase | 1 | C |
| A0A6N0AGM0 | OhrB | 0.0039 | 6.8570 | Organic hydroperoxide resistance protein | 2 | EX | ||
| A0A6N0AY64 | Spy | 0.0309 | 2.9841 | Spheroplast protein Y | 0 | EX | ||
| A0A6N0ADU5 | UspE | 0.0320 | 0.4192 | Universal stress protein E | 4 | C | ||
| A0A6N0A9Y2 | <0.0001 | 4.7646 | <0.0001 | 6.3572 | Alkyl hydroperoxide reductase subunit C | 2 | C | |
| A0A6N0BH32 | 0.0012 | 3.0958 | Glutathione synthetase | 4 | C | |||
| A0A6N0A6E0 | <0.0001 | 3.0858 | 0.0034 | 2.0317 | Superoxide dismutase | 1 | C | |
| A0A6N0BD39 | EC-YbbN | 0.0067 | 2.1508 | Thioredoxin | 2 | C | ||
| VIRULENCE AND ATTACHMENT | ||||||||
| A0A2K8VV27 A0A6N0ARC4 |
0.0076 | 2.4047 | Endo-1,4-beta-xylanase A, cellulolythic enzyme | 2 | EX | |||
| METABOLISM | ||||||||
| A0A6N0AXG4 | AccA | 0.0011 | 2.1010 | Acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha | 4 | C | ||
| A0A6N0AKX6 | CoaBC | 0.0380 | 2.6397 | Coenzyme A biosynthesis bifunctional protein | 6 | C | ||
| A0A6N0AEA5 | FabA | 0.0124 | 2.9558 | 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase | 2 | C | ||
| A0A6N0AP75 | MaeA | 0.0379 | 2.4525 | NAD-dependent malic enzyme | 8 | C | ||
| A0A6N0B091 | MenB | 0.0002 | 2.0532 | 1,4-Dihydroxy-2-naphthoyl-CoA synthase | 8 | C | ||
| A0A6N0BMC3 | MetK | 0.0005 | 2.2850 | S-adenosyl-methionine synthase | 4 | C | ||
| A0A6N0B5I8 | PanB | 0.0029 | 0.4842 | 3-methyl-2-oxobutanoate hydroxymethyl-transferase | 5 | C | ||
| A0A6N0AI34 | Ppa | 0.0001 | 0.2987 | Inorganic pyrophosphatase | 2 | C | ||
| A0A6N0B8R8 | Rpe | <0.0001 | 9.3293 | Ribulose-phosphate 3-epimerase | 1 | C | ||
| A0A2K8VWM7 | SuhB | 0.0425 | 2.0263 | Inositol-1-monophosphatase | 3 | C | ||
| A0A2K8VW53 A0A6N0ARP4 |
YfdZ | 0.0291 | 2.4133 | Putative PLP-dependent aminotransferase YfdZ | 4 | C | ||
| A0A2K8VWF4 A0A6N0B632 |
YhdN_3 | 0.0010 | 3.6295 | Oxidoreductase | 0 | C | ||
| A0A2K8VUJ2 A0A6N0AFZ5 |
<0.0001 | 3.3844 | 2-dehydropantoate 2-reductase | 5 | C | |||
| A0A6N0BJD2 | 0.0027 | 0.1940 | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase | 5 | C | |||
| A0A2K8VYG6 A0A6N0AT13 |
<0.0001 | 7.0231 | 0.0042 | 7.5702 | Exported lipase | 1 | EX | |
| A0A2K8VZB6 A0A6N0AI45 |
0.0010 | 3.3037 | N-acetyl-L,L-diaminopimelate deacetylase-like protein | 2 | C | |||
| A0A6N0B1V3 | 0.0312 | 0.4177 | NADH:flavin oxidoreductase | 1 | C | |||
| A0A6N0AAE7 | 0.0275 | 2.5083 | Phosphoglucomutase | 5 | C | |||
| → Protein synthesis | ||||||||
| A0A6N0BHG2 | HisB | 0.0023 | 2.5244 | Histidine biosynthesis bifunctional protein | 6 | C | ||
| A0A6N0B0C3 | HisD | 0.0025 | 3.1096 | Histidinol dehydrogenase, histidine biosynthesis | 7 | C | ||
| A0A6N0BKQ0 | LeuC | 0.0003 | 0.4741 | 3-isopropylmalate dehydratase large subunit 1, L-leucine biosynthesis | 7 | C | ||
| A0A6N0B5Q4 | LeuD | 0.0013 | 0.4868 | 3-isopropylmalate dehydratase small subunit 1, L-leucine biosynthesis |
2 | C | ||
| A0A6N0AIZ0 | RplL | <0.0001 | 2.7155 | 50S ribosomal protein L7/L12 | 0 | C | ||
| A0A6N0BJY9 | RplX | <0.0001 | 0.4967 | 50S ribosomal protein L24 | 0 | C | ||
| A0A2K8VYU2 A0A6N0B5N4 |
RpsJ | 0.0005 | 0.4470 | 30S ribosomal protein S10 | 0 | C | ||
| A0A6N0BIP4 | RpsT | 0.0048 | 0.4073 | 30S ribosomal protein S20 | 0 | C | ||
| A0A2K8VTM5 A0A6N0BHV7 |
TrpD | 0.0020 | 3.6390 | Anthranilate phosphoribosyl-transferase | 2 | C | ||
| A0A6N0AJL8 | TrpS | 0.0003 | 2.2887 | Tryptophan-tRNA ligase | 4 | C | ||
| A0A6N0BHM5 | YfiA | <0.0001 | 2.0523 | Ribosome-associated inhibitor A | 1 | C | ||
| A0A6N0B1J2 | YgfZ | 0.0299 | 0.4667 | tRNA-modifying protein | 3 | C | ||
| A0A6N0AMQ7 | 0.0114 | 2.0908 | DNA-binding protein H-NS | 0 | C | |||
| A0A6N0BB83 | 0.0224 | 3.2046 | Elongation factor Tu | 3 | C | |||
| A0A6N0AJ97 | 0.0307 | 2.7311 | Elongation factor Tu | 3 | C | |||
| A0A6N0ALX1 | 0.0057 | 3.0829 | Peptide chain release factor 3 | 6 | C | |||
| A0A6N0B620 | 0.0377 | 2.0594 | Sulfite reductase (NADPH) flavoprotein alpha-component, cysteine biosynthesis | 1 | C | |||
| → Nucleic acids and cofactor synthesis and homeostasis | ||||||||
| A0A6N0AB00 | Apt | 0.0208 | 0.4420 | Adenine phosphoribosyl-transferase | 1 | C | ||
| A0A6N0BBV4 | CspE | 0.0051 | 0.4385 | Cold shock-like protein | 0 | C | ||
| A0A6N0AQX1 | IhfA | 0.0172 | 2.0893 | Integration host factor subunit alpha | 0 | C | ||
| A0A6N0BHM3 | 0.0232 | 2.0453 | Exoribonuclease 2 | 7 | C | |||
| → Poliamine biosynthesis | ||||||||
| A0A6N0ASX1 | SpeA | 0.0102 | 2.5712 | Biosynthetic arginine decarboxylase | 8 | EX | ||
| A0A2K8VWZ1 A0A6N0BJQ5 |
SpeE | 0.0170 | 3.2343 | 0.0056 | 3.0386 | Polyamine aminopropyltransferase | 8 | C |
| LIPOPROTEINS, MEMBRANE AND CELL WALL BIOGENESIS | ||||||||
| A0A6N0BJT9 | ArnA | 0.0129 | 2.4335 | Bifunctional polymyxin resistance protein ArnA | 7 | C | ||
| A0A6N0AHH6 | BamD | 0.0005 | 2.6820 | Outer membrane protein assembly factor | 1 | EX | ||
| A0A6N0AZB8 | GlmU | 0.0086 | 2.7221 | Bifunctional protein, cell wall biogenesis |
4 | C | ||
| A0A6N0BB99 | YbiS | 0.0120 | 2.5079 | L,D-transpeptidase | 1 | EX | ||
| A0A6N0ARZ3 | 0.0371 | 2.9322 | Osmotically inducible lipoprotein E | 4 | EX | |||
| A0A6N0BHR4 | 0.0042 | 2.6863 | UDP-N-acetylmuramate-L-alanine ligase | 2 | C | |||
| A0A6N0AV98 | 0.0025 | 3.6632 | UDP-N-acetylmuramoyl-tripeptide-D-alanyl-D-alanine ligase | 4 | C | |||
| OTHERS, UNIDENTIFIED | ||||||||
| A0A2K8VZQ3 A0A6N0AK79 |
DcrB | 0.0020 | 3.0076 | Periplasmic bacteriophage sensitivity protein DcrB | 1 | EX | ||
| A0A6N0AWQ4 | ZapB | 0.0094 | 3.1295 | Cell division protein ZapB | 0 | C | ||
| A0A2K8VTW3 | 0.0009 | 5.9074 | Uncharacterized protein | 3 | N | |||
| A0A6N0AH49 | 0.0032 | 2.6561 | Uncharacterized protein | 2 | N | |||
UniProtID in gray are the archival numbers. The fold change values of downregulated proteins are in italics; number of cysteine residues in a protein (Number of. Cys); predicted protein localization: the cytoplasm (C), the extracytoplasmic space (EX), not known (N).
First of all, we observed a significant increase in the abundance of proteins directly engaged in cellular defense against reactive oxygen species (ROS) or in general stress response. These include ROS-neutralizing proteins (catalase KatG, superoxide dismutase, organic hydroperoxide resistance protein OhrB and alkyl hydroperoxide reductase), proteins providing reducing power (glutathione reductase and thioredoxin), proteins protecting DNA (Dps and universal stress protein E—UspE), protein quality control protease ClpP and envelope stress-induced periplasmic chaperone Spy. Additionally, an elevated level of enzymes participating in polyamine biosynthesis (SpeA, SpeE) can be considered as part of the stress response. Furthermore, a group of enzymes involved in membrane and cell wall biogenesis was upregulated. These include the outer membrane protein assembly factor BamD, the osmotically inducible lipoprotein E, L,D-transpeptidase, the UDP-N-acetylmuramate-L-alanine ligase and the UDP-N-acetylmuramoyl-tripeptide-D-alanyl-D-alanine ligase. The AccA protein, which provides the malonyl-CoA substrate for the biosynthesis of fatty acids [33], was also more abundant in H2O2-stressed cells.
The oxidative stress also caused changes in the cellular content of many primary metabolic enzymes. The ribulose-phosphate 3-epimerase RPE was the most strongly upregulated protein. Since this enzyme is particularly sensitive to oxidative damage [34], an increased synthesis is most probably necessary to maintain proper function of the pentose phosphate pathway.
We also observed that several pathways which are related to protein synthesis were upregulated. In particular, we detected increased levels of some proteins engaged in the translation process (TrpS, YfiA, EF-Tu and peptide chain release factor 3) and enzymes involved in the de novo amino acid biosynthesis (HisB, HisD, TrpD and sulfite reductase (NADPH) flavoprotein) and metabolism (S-adenosylmethionine synthase MetK). In contrast, the abundance of certain ribosomal proteins (L24, S10 and S20) decreased. Moreover, the levels of two proteins related to the transcription process were altered. The content of CspE, whose activity is important for antitermination and the decrease in transcription pausing in E. coli [35], was reduced, while the Exoribonuclease 2, involved in mRNA degradation [36], was upregulated, suggesting a downregulation of the transcription process in H2O2-stressed cells.
Interestingly, the dsbA mutant cells treated with H2O2 showed significantly less changes in their proteome compared to the wt cells. In fact, only 15 proteins showed altered abundance, and the changes were mainly related to proteins typically induced under oxidative stress conditions (KatG, superoxide dismutase, alkyl hydroperoxide reductase and Dps). Other elevated proteins, which were also found in the stressed wt cells, were SpeE and lipase (A0A2K8VYG6). Interestingly, we detected some proteins whose content was increased in the dsbA mutant but not in the parental wt D. solani. These include the protein export chaperone SecB, an outer membrane receptor involved in siderophore uptake (A0A2K8W4X3), RplL, the bifunctional polymyxin resistance protein ArnA and the cell division protein ZapB.
2.7. The Lack of the Functional dsbA Gene Affects the Secretion of Virulence Factors
As the function of the DsbA oxidase is particularly important to stabilize the structure of secreted proteins, we also compared the protein content of the extracellular compartment (secretome) of the wt and mutant strains. Our MS analysis identified 573 proteins which were present in the growth medium of the D. solani culture (Table S2). The secretome profile of the dsbA mutant was very different from that of the wt D. solani strain. First of all, we observed that the content of secreted virulence factors was drastically reduced (Table 3). This effect was particularly pronounced in the case of the PCWDEs: pectate lyases (PelA, PelC_1, PelC_2, PelE, PelL_1 and PelL_2), pectin lyase (Pnl), pectin esterase A (PemA), glucuronoxylanase (XynC), the protease PrtA and a neutral metalloproteinase. Another group of underrepresented proteins in the dsbA mutant secretome includes flagellum components: the flagellin FliC, the filament cap protein FliD, the hook-filament junction (FlgL and FlgK), the hook FlgE, the distal rod FlgG and the proximal rod (FlgF, FliE, FlgB, and FlgC).
Table 3.
Extracellular proteins whose level is downregulated due to the lack of a functional DsbA protein.
| Peak Name | Name | p-Value | Fold Change | Description | Number of Cys |
|---|---|---|---|---|---|
| MOTILITY AND CHEMOTAXIS | |||||
| A0A6N0AH46 | CheW | 0.0168 | 0.4267 | Chemotaxis protein CheW | 0 |
| A0A6N0AF74 | CheY | 0.0012 | 0.0683 | Chemotaxis protein CheY | 0 |
| A0A6N0ADL9 | FlgB | 0.0006 | 0.0973 | Flagellar basal body rod protein FlgB | 0 |
| A0A6N0B762 | FlgC | <0.0001 | 0.2802 | Flagellar basal body rod protein FlgC | 0 |
| A0A6N0ADS2 | FlgD | 0.0011 | 0.1424 | Basal body rod modification protein FlgD | 0 |
| A0A6N0B2V1 | FlgE | <0.0001 | 0.1296 | Flagellar hook protein FlgE | 0 |
| A0A6N0BFR1 | FlgF | <0.0001 | 0.1241 | Flagellar basal body rod protein FlgF | 0 |
| A0A6N0B547 | FlgG_1 | 0.0006 | 0.0664 | Flagellar basal body rod protein FlgG | 0 |
| A0A6N0AH33 | FlgK | <0.0001 | 0.0324 | Flagellar hook-associated protein 1 | 0 |
| A0A6N0ADK6 | FlgL | <0.0001 | 0.0458 | Flagellar hook-associated protein 3 | 0 |
| A0A6N0AGC2 | FlgM | <0.0001 | 0.0238 | Negative regulator of flagellin synthesis | 0 |
| A0A6N0AG83 | FliC_1 | <0.0001 | 0.0291 | Flagellin 1 | 0 |
| A0A6N0ASL9 | FliD | <0.0001 | 0.0201 | Flagellar hook-associated protein 2 | 0 |
| A0A6N0B5D6 | FliE | <0.0001 | 0.0420 | Flagellar hook–basal body complex protein FliE | 0 |
| A0A6N0B4U8 | FliK | 0.0013 | 0.0256 | Flagellar hook-length control protein | 0 |
| VIRULENCE, ATTACHMENT | |||||
| A0A2K8VTS7 | <0.0001 | 0.0402 | Putative cellulase | 2 | |
| A0A2K8VVJ2 A0A6N0BEF2 |
<0.0001 | 0.0255 | Endo-1,4-beta-xylanase A | 4 | |
| A0A6N0AEA4 | PelA | <0.0001 | 0.0118 | Pectate lyase A | 10 |
| A0A6N0ATI6 | PelC_1 | <0.0001 | 0.0471 | Pectate lyase C | 4 |
| A0A6N0BJ90 | PelC_2 | <0.0001 | 0.0227 | Pectate lyase C | 4 |
| A0A6N0ARF7 | PelE | <0.0001 | 0.0181 | Pectate lyase E | 2 |
| A0A6N0ANC2 | PelL_1 | <0.0001 | 0.0255 | Pectate lyase L | 3 |
| A0A6N0AG85 | PelL_2 | 0.0017 | 0.0200 | Pectate lyase L | 4 |
| A0A2K8VZ10 A0A6N0AYE1 |
<0.0001 | 0.0700 | Pectate lyase | 1 | |
| A0A6N0B698 | PemA | <0.0001 | 0.1544 | Pectinesterase A | 2 |
| A0A6N0BBW4 | Pnl | <0.0001 | 0.0408 | Pectin lyase | 3 |
| A0A6N0B2L1 | PrtA | 0.0364 | 0.3663 | Serralysin A | 0 |
| A0A2K8VWN4 A0A6N0BGU2 |
0.0114 | 0.2795 | Neutral metalloproteinase | 2 | |
| A0A6N0AC94 | XynC | 0.0003 | 0.1446 | Glucuronoxylanase XynC | 3 |
UniProtID in gray are the archival numbers. The fold change values of downregulated proteins are in italics.
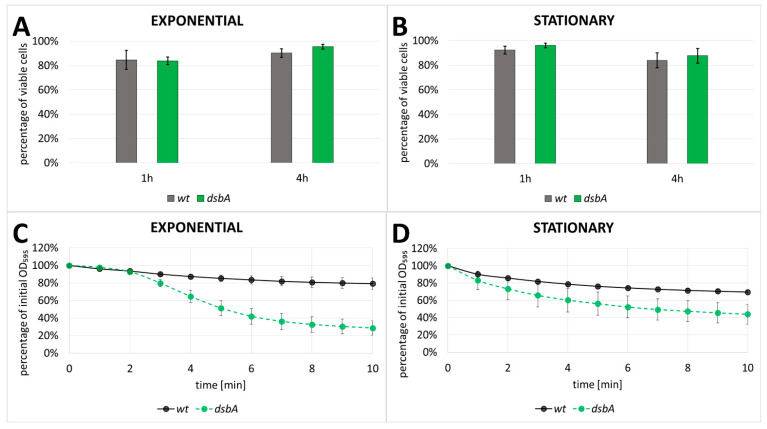
Since the levels of many cellular proteins were increased in the secretome fraction of the dsbA mutant, we deduced that the membrane integrity might have been weakened in the mutant cells, rendering them more prone to lysis. To verify this assumption, we tested the permeability of the membranes against compounds that have restricted access to intact cells. These include propidium iodide (PI) and sodium dodecyl sulfate (SDS). PI is a membrane-impermeable DNA-intercalating fluorescent dye which is regarded as the inner membrane permeability indicator [37]. SDS is an ionic detergent which disrupts the phospholipid bilayer, but due to the presence of lipopolysaccharide (LPS), Gram-negative bacteria are resistant to low concentrations of SDS [38]; therefore, SDS may serve as an indicator for outer membrane disturbance. We found that PI stained comparable numbers of the wt and mutant D. solani cells (Figure 6A,B), suggesting that the inner membrane integrity was not affected by the dsbA mutation. However, we observed a pronounced negative effect of SDS on the dsbA mutant. The dsbA cells were sensitive to the presence of 1% SDS, which induces frequent cell lysis (Figure 6C,D). In this condition, the wt cell lysis was only observed at a low frequency. Taken together, these results indicate that a lack of DsbA led to outer membrane alterations, while the inner membrane seemed to be unaffected.
Figure 6.
Analysis of membrane permeability using LIVE/DEAD fluorescence microscope assay (A,B) and sensitivity to SDS (C,D). The assays were conducted in exponential (A,C) and stationary (B,D) growth phases (wt—wild-type strain; dsbA—mutant strain, TP02). In LIVE/DEAD assay, data obtained from three independent biological replicates with at least three technical replicates per sample were analyzed. In the SDS sensitivity assay, the error bars represent SD from five independent biological replicates in two independent experiments (n = 10).
3. Discussion
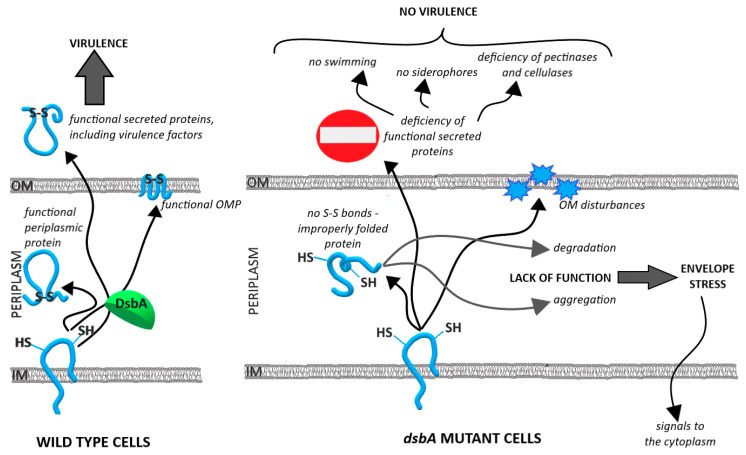
The DsbA homologs are oxidoreductases present in most bacterial species where they are responsible for introducing disulfide bonds in extracytoplasmic proteins. The majority of the dsbA mutants exhibit pleiotropic phenotypes due to the incorrect folding of numerous periplasmic and secreted proteins. As a result, the mutants show reduced fitness and attenuated virulence in the animal infection models due to improper folding/stability of several crucial virulence factors [8].
In this study, we explored the role of DsbA in the fitness and pathogenicity of a potato pathogen, D. solani. First of all, we found that the inactivation of dsbA in D. solani IPO2222 strongly reduced the bacterial virulence. Using both a natural host plant, potato, and a model host plant, chicory, we demonstrated that the dsbA mutant exhibited a markedly lower ability to macerate plant tissues than the wild-type strain IPO2222. Likewise, the dsbA mutant could not develop disease symptoms in whole potato plants grown in potting compost under phytochamber conditions. The observed lack of virulence can be due to several reasons: (i) the inability of the bacterial cells to penetrate the host, (ii) problems with enduring stressful conditions during host colonization, (iii) defects in growth and multiplication inside the host and finally, (iv) disturbances in the production and secretion of virulence factors during infection.
At the initial stages of infection, the capability to actively move towards a plant host, driven by the perception of appropriate chemical signals and the subsequent penetration of bacteria into the apoplast, are of particular importance [29]. Therefore, both motility and chemotaxis are major virulence determinants of SRP bacteria, including D. solani. As reported for the dsbA mutants of other bacterial pathogens (E. coli, P. syringae pv. Tomato, P. fluorescens and P. atrosepticum) [23,39,40,41], the D. solani dsbA mutant exhibited reduced swimming and swarming motility. Both types of movement require the presence of functional flagella, and as shown in E. coli, DsbA is required to introduce proper disulfide bonds in the P-ring protein (FlgI) of the flagella motor [39]. Dickeya sp. also contains an FlgI homolog with very well conserved pairs of cysteines (according to UniProtKB), also expected to require DsbA for stabilizing disulfide bonding. Therefore, the non-motile phenotype exhibited by the D. solani dsbA mutant could be due to the non-functional flagella motor component FlgI. Although our proteomic MS analysis did not detect the protein FlgI, the content of the other flagella components was strongly reduced in the D. solani dsbA mutant proteome and secretome. For efficient motility, bacteria need to perceive appropriate chemotactic signals. Thus, aberrances in chemotactic response negatively affect cell motility [42,43,44,45]. According to our proteome analysis, several proteins involved in chemotaxis were less abundant in the dsbA mutant. Hence, the substantial reduction in dsbA mutant cell motility resulted from both the deficiency of the flagella motor and decreased chemotactic response. Similar observations were reported in the case of P. atrosepticum, where 9 proteins related to flagella synthesis were present at a reduced level in the dsbA mutant secretome, and 25 genes encoding components of flagella and chemotactic response were downregulated in the dsbA mutant in comparison with the wild-type strain [23]. As flagellar gene expression occurs sequentially, according to the order of flagella assembly, blocking at the flagellar basal body level leads to the repression of late genes (e.g., fliC encoding flagellin). It was noticed that the P. atrosepticum dsbA mutation also causes partial repression of the early and middle flagella synthesis genes [23]. Our proteomic data indicate that the levels of all detected flagella proteins decreased in the D. solani dsbA mutant. One reason for this may be the envelope stress generated by the dsbA inactivation. In S. enterica serovar Typhimurium, the general envelope stress turns off the production of flagella [46]. Therefore, in the D. solani dsbA mutant, the synthesis of flagellar proteins may also be repressed by the envelope stress caused by the DsbA lack of function-dependent redox imbalance. This assumption agrees with our finding that the D. solani dsbA mutant contained an elevated level of the protein RcsB (app. 2-fold increase). RcsB is a transcriptional regulator that responds to envelope stress, and it is known to negatively regulate motility by directly repressing the operon flhDC encoding the master regulator of flagellar gene transcription [47,48,49].
Once inside the plant, the D. solani cells are challenged with several hostile factors, such as osmotic, oxidative and acidic stresses. We observed that the inactivation of dsbA in D. solani did not impair bacterial growth under neither physiological nor stressful conditions. This result was in agreement with a lack of induction of the dsbA expression in most stress-exposed D. solani cultures. Hence, it seems that the stress response in D. solani does not require elevated levels of the protein DsbA. The lack of growth defects due to the dsbA mutation in D. solani is not exceptional, as the dsbA mutants of several other bacterial species grow similarly to their parental strains under many tested conditions [50,51]. This phenomenon can be explained by a functional redundancy or overlapping activity among the extracytoplasmic folding factors [52,53]. The effects of the dsbA mutation become visible in the presence of additional mutations affecting protein quality control. For example, double inactivations of dsbA and degP (htrA) [54,55] or dsbA and surA [56] result in an increased sensitivity to several stresses. The genes degP and surA encode essential components of the protein quality-control system. DegP is a periplasmic protease/chaperone from the HtrA serine proteases family, responsible for removing aberrant proteins from the cellular envelope [57]. SurA serves as a general periplasmic chaperone for outer membrane proteins. Indeed, we found that the proteomes of the D. solani dsbA mutant contained increased concentrations of the HtrA homologs DegP (app. 3-fold increase ) and DegQ (app. 1.5-fold increase). A deficiency of DsbA also led to increased levels of various periplasmic chaperones and folding helpers: FkpA (1.6-fold increase), outer membrane protein assembly factor BamA (2.5-fold increase) and BamD (1.9-fold increase) (Table 1 and Table S1). The p-values of all of the above-mentioned proteins were significant.
During the later stages of plant infection, bacteria proliferate to reach a high density favorable to start the production of secreted virulence factors, such as PCWDEs, to rapidly destroy plant tissues and acquire nutrients reviewed in [29]. Several virulence factors of pathogenic bacteria require S-S bonds for their activity and stability. We found that activities of the crucial extracellular PCWDEs, pectinases and cellulases were strongly reduced in the D. solani dsbA mutant compared to the wt strain. According to this finding, the secretomes of the dsbA mutant contained negligible amounts of several pectate and pectin lyases and a low amount of pectinesterase A. Each of these enzymes contain at least two cysteine residues which are engaged in S-S bonding [58,59,60,61]. The lack of DsbA function can also affect the secretion system involved in export of PCWDEs. Some components of the D. dadantii Type 2 Secretion System (T2SS) have disulfide bonds which are essential for their correct folding and functioning [62]. Furthermore, genes encoding pectate lyases are under the negative control of the RcsB transcriptional regulator [63]. Therefore, an extremely low amount of secreted pectate lyases may also result from a decreased expression of their genes due to the elevated RcsB level observed in the dsbA mutant. The cellulase CelZ, which is responsible for the major endoglucanase activity in D. dadantii [64] and presumably also in D. solani, and a putative cellulase (A0A2K8VTS7) (Uniprot) were also strongly reduced in abundance. CelZ is known to contain S-S bonds [65], and the other enzyme also contains two cysteine residues. Interestingly, the secreted proteolytic activity was similar in the dsbA mutant and the parental D. solani strain. In the P. atrosepticum dsbA mutant, the enzymatic activities of all secreted enzymes were dramatically reduced: pectinase and cellulase activities were negligible and the protease activity was reduced to one-third that of the wild-type strain [23]. In the D. solani secretome, two proteases, PrtA and a neutral metalloproteinase, were approximately 2–3-fold less abundant (Table 1). Nevertheless, it is possible that the decreased amount of these two proteases was masked by the redundancy of proteases in the secretome. Although only a few proteases are known to participate in D. solani virulence, most of them can degrade milk proteins. As a result, the global proteolytic activity in the secretomes of both strains, dsbA and wt, was similar in the plate assays.
An interesting phenotype of the D. solani dsbA mutant, not previously observed in other bacteria, was a deficiency of secreted siderophores. Dickeya species secrete two types of siderophores: chrysobactin and achromobactin, which are necessary to acquire iron from an iron-poor environment in a plant host. Mutations that disable any of the siderophore-mediated iron transport prevent bacteria from spreading throughout the plant [66]. Hence, the secretion of siderophores is an important virulence determinant. Unfortunately, at the moment, we cannot explain the mechanism that leads to the impairment of the iron-chelating ability of the dsbA mutant, as none of the proteins directly involved in the siderophore synthesis or transport were detected in our proteome analysis. Nevertheless, we found that one of the proteins involved in iron uptake (A0A6N0B1L8, NCBI protein AYQ47458.1; Table 1), the periplasmic-binding protein SitA, was induced approximately by a two-fold factor in the D. solani dsbA mutant. SitA is homologous to YfeA of Yersinia or SitA of Rhizobium meliloti (according to the Paperblast database) [67,68]. YfeA, a component of the siderophore-independent, Yfe ABC permease, can bind manganese and iron ions. The yfe mutants show growth defects only under conditions of limited free iron availability, suggesting that iron acquisition is the main function of the Yfe system [67]. Our MS analysis indicates that in the D. solani dsbA mutant cells, the Yfe iron acquisition system is induced. The increased content of the Yfe system may compensate, at least partially, for a siderophore deficiency, and it may allow the mutant cells to colonize the host plant (albeit without developing disease symptoms).
The detailed analysis of the proteomes and secretomes of D. solani dsbA mutant and the parental strain enabled us to highlight the physiological effects of the DsbA deficiency. First of all, we noticed that in the dsbA mutant, several stress-response-related proteins were upregulated. This phenomenon concerned both envelope and cytoplasmic proteins. The first group can be attributed to the extracytoplasmic stress induced by the presence of misfolded proteins lacking proper disulfide bonds. Defects in the envelope protein folding and membrane alterations lead to the induction of the two major and partially overlapping envelope stress response systems: the Cpx two-component signal transduction pathway and the extracytoplasmic sigma factor σE (rpoE) dependent pathway. The induction of these systems leads to the stimulation of the synthesis of several extracytoplasmic protein quality control components, including chaperones and folding catalysts (e.g., Skp, spy, PpiA and PpiD), and the protease DegP [69,70,71]. Furthermore, the synthesis of components of the machineries involved in the transport and assembly of LPS and outer membrane proteins is also upregulated in an σE -dependent manner [72]. Indeed, our proteomic analysis revealed that the dsbA mutant contained elevated levels of Skp, spy, PpiA, SurA and BamA. Moreover, an increased abundance of the protein CpxP was detected in the dsbA mutant secretome. CpxP is a negative regulator associated with the sensor protein CpxA, but it also serves as a chaperone whose function is to present the unfolded proteins to the DegP protease. A high level of CpxP is a hallmark of Cpx pathway induction [73]. As mentioned previously, we observed an increased content of the transcriptional regulatory protein RcsB, a component of the Rcs system which senses damage or defects within the envelope and regulates the transcriptome to counteract stress. In particular, this system detects outer membrane damage, LPS synthesis defects, peptidoglycan perturbation and the mislocalization of lipoproteins [74]. The induction of the Rcs system has been reported in the S. enterica dsbA mutant [75]. In the D. solani dsbA mutant, we also found an increased content of some members of pathways maintaining envelope homeostasis. They include enzymes involved in: (I) the generation of energy and reducing power (components of the central carbon metabolism), (II) the synthesis of precursors needed for cell wall and membrane biogenesis (peptidoglycan and LPS synthesis pathways, the synthesis of CoA and fatty acid biosynthesis), and (III) general protein synthesis (amino acid biosynthesis pathways, ribosomal proteins and translation factors—EF-Tu and peptide chain release factor RF3). The central carbon metabolism plays essential roles in a cell by providing energy and precursors for many biosynthetic pathways. Under stress conditions, the central carbon metabolism and other metabolic pathways become altered to provide the metabolites necessary to overcome the stress effects reviewed in [76]. For example, we observed increased amounts of the pentose phosphate pathway steps generating the reductant NADPH and nucleotide precursors for DNA repair [77,78]. Ribulose-phosphate 3-epimerase, which participates in the nonoxidative phase of the pentose phosphate pathway, was the protein that showed the most increased level (>10-fold). The involvement of this enzyme in the stress response has not yet been reported in bacteria. However, in the yeast Saccharomyces cerevisiae, a lack of ribulose 5-phosphate epimerase resulted in an increased sensitivity to H2O2 [77]. Upregulation of the cell wall biogenesis enzymes may indicate issues with maintaining the integrity of the envelope (proper structure/function of the peptidoglycan and cell membranes). Increased expression of 25 genes involved in protein synthesis was also observed in the P. atrosepticum dsbA mutant, and this phenomenon has also been attributed to the stress response [23].
When analyzing the D. solani secretomes, we observed many proteins whose location were assigned to the periplasm or even to the cytoplasm. The pattern of most cellular proteins was generally comparable in both strains, indicating that the presence of these proteins in the growth medium was a result of cell lysis during the bacterial culture and possibly at the centrifugation step. However, many cytoplasmic and periplasmic proteins were enriched in the dsbA mutant secretome. This observation and the increased level of proteins involved in the cell wall and membrane biogenesis in the D. solani dsbA mutant proteome suggest that the cellular barriers do not work correctly in the mutant. This assumption was confirmed by the increased sensitivity of the dsbA mutant to low concentrations of SDS. A properly structured outer membrane provides protection against anionic detergents (e.g., SDS) [79], and permeabilization of the outer membrane can be manifested as increased susceptibility to the bacteriolytic action of detergents [80]. On the other hand, the dsbA mutant and wild-type strain of D. solani did not differ in PI entry; we thus concluded that the cytoplasmic membrane permeability was not affected by the lack of DsbA. It seems possible that the presence of not properly folded proteins due to the absence of DsbA leads to disturbances in the outer membrane and possibly to the peptidoglycan layer, making a cell more susceptible to lysis. There is also another explanation for the presence of cellular proteins in the growth medium. The removal of damaged proteins from the periplasm is one of the ways to deal with envelope protein folding stress. For example, protein leakage across the outer membrane was observed in the absence of the key protein quality control protease DegP (HtrA) [81]. In Gram-negative bacteria, degP/htrA mutants are characterized by an increased production of outer membrane vesicles, a phenomenon explained by the need to remove damaged proteins [82,83]. Therefore, the increased abundance of some periplasmic and outer membrane proteins in the dsbA mutant secretome may result from both protein leakage across damaged outer membrane and the elevated production of outer membrane vesicles.
The extracytoplasmic protein folding stress undoubtedly affects the protein homeostasis of the whole cell. The misfolding or deficiency of some membrane proteins can lead to disturbances in cell membrane functionality, for example, nutrient uptake. Indeed, the dsbA mutant proteome contained a lower content of some transport-related proteins, such as GltI (glutamate/aspartate periplasmic-binding protein), OppA (periplasmic oligopeptide-binding protein), inositol transport system sugar-binding protein (secretome) and hemin-binding protein (secretome). Moreover, as described above, the siderophore-dependent iron uptake system seems to be severely disturbed in the D. solani dsbA mutant. Dysfunctions of the envelope systems must have consequences for cytoplasmic homeostasis. Therefore, it was not surprising that the stress-related response was also induced in the cytoplasm. Several cytoplasmic proteins involved in stress response were significantly upregulated in the D. solani dsbA mutant, such as an organic hydroperoxide resistance protein of the Ohr/OsmC family, a glutathione synthetase and an aldo/keto reductase previously assigned as general stress protein 69. Moreover, we detected an increased content of the biosynthetic arginine decarboxylase SpeA and the S-adenosylmethionine decarboxylase SpeD, both involved in the biosynthesis of polyamines. Polyamines, e.g., putrescine and spermidine, play an essential role in stabilizing nucleic acids, increasing the efficiency of translation and providing resistance to environmental stress, including temperature changes, reactive oxygen species, osmotic pressure or the presence of toxic compounds [84].
Interestingly, the proteins mentioned above became induced in the wild-type D. solani strain by treatment with the oxidant H2O2. Several other proteins which were upregulated in the dsbA mutant were also induced in the wild-type strain under oxidative stress (Table 2, Figure 5D). Therefore, the impacts of the dsbA mutation in the cytoplasm seem to be similar to those of oxidative stress. This observation explains why oxidative stress caused slight changes in the dsbA mutant proteome. In fact, in the D. solani dsbA mutant treated with H2O2, mainly the proteins specific to oxidative stress response were upregulated (Table 2, Figure 5D). They included superoxide dismutase, DNA protection during starvation protein Dps, spermidine synthase, alkyl hydroperoxide reductase and catalase. The high level of an exported lipase observed in both strains after H2O2 treatment may reflect a necessity to modulate fatty acid composition in the oxidatively damaged membranes.
It is worth noting that the levels of the general cytoplasmic chaperones DnaK, ClpB, DnaJ and GroEL, were not significantly modified in the dsbA mutant, as well as in the wild-type strain under oxidative stress. Most probably, neither mutation nor the oxidant affected the structure of the cytoplasmic proteins significantly. This observation is in agreement with our previous finding, demonstrating that the dnaK, dnaJ and groEL genes were not induced by oxidative stress in D. solani [85].
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
The wild-type Dickeya strains used in this study were D. dadantii 3937 [86] and D. solani IPO2222 [20]. The φ-EC2 generalized transducing phage was used for transduction of the Dickeya strains [87]. Bacterial strains and plasmids used in this study are listed in Table 4.
Table 4.
Bacterial strains and plasmids.
| Strains | Relevant Genotype | Reference or Source |
|---|---|---|
| Dickeya dadantii 3937 | WT | [88] |
| D. dadantii 3937 dsbA::cm (TP01) | dsbA gene inactivated by CmR insertion | This work |
| D. dadantii 3937 dsbA-kn | Complementation strain with a functional dsbA gene and a KnR cassette | This work |
| D. solani IPO2222 | WT | [89] |
| D. solani IPO2222 dsbA::cm (TP02) | dsbA gene inactivated by CmR | This work |
| D. solani IPO2222 dsbA-kn (TP03) | Complementation strain with functional dsbA gene and KnR | This work |
| Plasmids | Feature | Reference or Source |
| pGEM-7 | Cloning vector | Promega |
| pCYC-184-cm | Donor of CmR cassette | [90], New England Biolabs |
| pDOC-K | Donor of KnR cassette | [91] |
| pDF-dsbA::cm | pGEM-7 with D. dadantii dsbA gene inactivated by CmR cassette | This work |
| pDF-dsbA-kn | pGEM-7 with D. dadantii functional dsbA gene and KnR cassette | This work |
Bacteria were cultivated at 30 or 37 °C in minimal medium M63Y (0.1 M KH2PO4, 15 mM (NH4)2SO4, 9 μM FeSO4, 1 mM MgSO4, 1 mg L−1 vitamin B1 and 3 g L−1 glycerol, pH = 7.0) [92] in aerobic or microaerobic conditions, as described in [85]. When required, chloramphenicol (Cm) was added at a concentration of 12.5 µg mL−1. The oxidative stress was induced by 0.1 or 0.25 mM hydrogen peroxide (H2O2). For combined H2O2 and acid stress, the medium was acidified to pH = 5.0 by malic acid [93]. The osmotic stress was induced and analyzed as described in [85]. Briefly, ionic stress was induced by an increase in sodium chloride (NaCl) content to a concentration of 0.8 M in liquid medium or 0.1–0.3 M in solid medium. To induce osmotic non-ionic stress, sucrose was added to a final concentration of 0.32 M (liquid medium) or 0.3–0.6 M (solid medium). The thermal stress was induced by transferring the liquid bacterial cultures from 30 to 40 °C and subsequent 5 h incubation at elevated temperature. The assessments of cell cultivability and viability were performed as described in [85].
4.2. Construction of the D. solani dsbA Knockout Strain
The D. solani dsbA mutant was generated in two steps, according to [94]. In the first stage, the dsbA gene of the D. dadantii 3937 chromosome was disrupted by the insertion of a Cm resistance cassette. Briefly, the marker exchange plasmid containing the D. dadantii dsbA gene interrupted by the Cm cassette was constructed after amplification of the dsbA gene by PCR using the primers Dd-dsbA-L and Dd-dsbA-R (Table 5). The PCR product was cloned into the pGEM-7 vector (PROMEGA, Madison, WI, USA) into the SmaI restriction site, generating the plasmid pDF-dsbA. The Cm cassette used for the gene inactivation was amplified from the pCYC-184-cm plasmid using primers Cm-Sawitzke-L and Cm-Sawitzke-R modified from [90]. Then, the dsbA gene sequence was interrupted by introduction of the Cm cassette into the AfeI restriction site at one-third of the gene length to generate the marker-exchange plasmid pDF-dsbA::cm. The pDF-dsbA::cm plasmid was introduced to D. dadantii cells via electroporation. Marker exchange recombination was obtained after growth in a low-phosphate medium, as described by [94]. The presence of the Cm cassette within the dsbA gene in the D. dadantii chromosome was verified via PCR using the primers Cm-Sawitzke-L and Cm-Sawitzke-R.
Table 5.
Primer sequences.
| Name | Sequence |
|---|---|
| Dd-dsbA-L | GGACAATGACGACCAACTGGAG |
| Dd-dsbA-R | GCTCGCTCATTACGCTTTTTGC |
| Cm-Sawitzke-L | ACCAGCAATAGACATAAGCG |
| Cm-Sawitzke-R | TGTGACGGAAGATCACTTC |
| Ds-dsbA-L | GGCACCGATGTACTGACACTTA |
| Ds-dsbA-R | GCAGTAAGCAGAAATGTCCAAAC |
| Kan-L | GACCGGTCAATTGGCTGGAG |
| Kan-R | GAATATCCTCCTTAGTTCC |
Subsequently, the inactivated dsbA (dsbA::cmR) gene was transferred from D. dadantii dsbA::cmR (TP01 strain) into the D. solani IPO2222 chromosome by φ-EC2 phage transduction. First, a phage stock was prepared on the D. dadantii dsbA::cm. Then, the phage stock was used to infect D. solani IPO2222, and the mutants were selected on LB + Cm plates. Generally, five different CmR colonies were recovered from each transduction. The presence of the mutation in the D. solani genome was confirmed via PCR using the primers Ds-dsbA-L and Ds-dsbA-R.
4.3. Construction of the D. solani dsbA Complementation Strain
The dsbA gene from D. dadantii and a kanamycin (Kn) resistance cassette were cloned into the pGEM-7 vector generating the pDF-dsbA-kn plasmid. The dsbA gene was cloned into the SmaI restriction site. The Kn cassette was amplified from the pDOC-K plasmid [91] using the primers Kan-L and Kan-R and cloned into the PmlI restriction site, which is located 48 nucleotides downstream from the dsbA gene. Subsequently, the pDF-dsbA-kn plasmid was introduced via electroporation into the D. dadantii cells. The next steps were similar to the construction of the D. solani dsbA::cm mutant, except that selection was performed on LB + Kn plates. The reintroduction of the wild-type dsbA gene to its native locus in the D. solani dsbA::cmR genome was confirmed via PCR using the primers Dd-dsbA-L and DddsbA-R.
4.4. Quantitative Real-Time PCR (qPCR)
Bacterial cultures for gene expression analyses, subsequent RNA isolation, reverse transcription and quantitative real-time PCR were performed as described in [95]. The characteristics of the primers are shown in Table 6.
Table 6.
Primers used for qPCR.
| FWD Primer Sequence (5′-3′) |
REV Primer Sequence (5′-3′) |
Amplicon Length | PCR Efficiency | R2 | Concentration [µM] | |
|---|---|---|---|---|---|---|
| dsbA | AACAGCATCAAACATCAGCGGG | TCTGGGAGAATTGGGTAAGGAGC | 97 | 1.99 | 0.99 | 0.225 |
| 16S rRNA | GCTCGTGTTGTGAAATGTTGGGTT | GCAGTCTCCCTTGAGTTCCCAC | 94 | 1.96 | 1.0 | 0.225 |
4.5. Virulence Determinants Assays
The motility of bacteria was evaluated under aerobic or microaerobic conditions. A swimming test was performed on the 0.3% agar MMA minimal medium (K2HPO4 7g L−1, KH2PO4 3 g L−1, MgSO4 × 7 H2O 0.1 g L−1, (NH4)2SO4 1 g L−1 and 1 mM galactose as an attractant). Swarming was tested on the 0.5% agar TSA (Tryptic Soy Agar). In both tests, individual colonies of the wt or mutant strains were stab inoculated into the center of the motility plate and incubated right-side-up at 30 °C. The radius of outward expansion from the inoculation point was measured at 72 (swimming) or 9 and 24 h (swarming) post inoculation.
Activities of the secreted PCWDEs were determined using plate and spectrophotometric assays. The experiments on plates were performed essentially as described in [96]. In brief, 5 μL aliquots of bacterial suspensions of stationary grown cultures containing 104 or 106 CFU were spotted on appropriate indicatory plates and incubated for 72 h at 30 °C. Proteases were detected on a medium containing skim milk (12.5 g L−1) [97]. Pectinolytic activity was tested on M63Y medium with polygalacturonic acid (PGA), as described in [98]. The ability to secrete active cellulases was analyzed on M63Y agar plates supplemented with 2 g L−1 glycerol and 10 g L−1 carboxymethylcellulose (CMC), as described by [99].
The spectrophotometric assay of pectate lyase activity was based on [100,101]. The rates of increase in absorbance at λ = 235 nm (A235) due to the formation of unsaturated products from polygalacturonate [102] were measured every 30 s for 2 min. The enzyme activity (units) (micromoles of 4,5-unsaturated product released in one minute) was calculated using the formula:
The cellulase activity spectrophotometric assay was conducted based on [101,103]. The cellulolytic activity was expressed as the amount of digested product of ostazin brilliant red cellulose using the following formula:
The value represents a difference between absorbance values of the test and control samples; OD595 is the optical density of cultures. The control sample was fresh growth medium with the ostazin brilliant red cellulose solution. Both spectrophotometric tests were performed using the U-5100 spectrophotometer (Hitachi, Tokyo, Japan).
To detect secreted siderophores, the Chrome-Azurol-S agar was used according to [104]. After incubation, the diameter of the orange halo around bacterial colonies was measured.
4.6. Infection Tests
The virulence of the D. solani dsbA mutant and the control strains (the parental wt strain and the complemented strain) was assessed on a model plant, chicory (Cichorium intybus), and the natural host, potato (Solanum tuberosum).
The chicory leaf virulence test was performed as previously described [105]. Briefly, each leaf was infected with 20 μL aliquots of bacterial solutions containing approximately 104 or 106 CFU per inoculation site. After incubation in a dew chamber for two days at 30 °C, the sizes of rotten tissue were evaluated and compared. At least five chicory leaves were used for each strain and each bacterial suspension.
The potato tubers (cv. Vineta) obtained locally in Gdansk, Poland were prepared and infected basically as described in [105]. For each tuber, the mass of tissue rotten caused by the wt D. solani strain was set at 100%, and a degree of maceration caused by the other strains was calculated as the percentage of the wt value. This approach enabled us to minimize the maceration differences due to the different properties of each tuber. The experiments were performed twice. Nine tubers were used for each biological replicate.
For the potato tuber slice test, the tubers of potatoes cv. Vineta were sterilized as described in [13] and sterile cut into approximately 1 cm thick slices. In the middle of each slice, a hole was made and 10 µL aliquots of suspensions of each strain containing 104 or 106 CFU were inoculated. As a negative control, 10 µL of sterile Ringer solution was injected. For disease development, the slices were incubated at 30 °C for 72 h in a humid box, then the diameters of rotting tissue around the wells were compared.
Statistical analysis of differences was determined by a two-way analysis of variance (ANOVA) test followed by a post hoc Dunnett test. A result was considered significant at p < 0.05.
The pot-grown potato plants were obtained from potato tubers cv. Vineta and subsequently cultivated as described in [95]. The plants were infected after root injury by soaking potting compost in each pot with 30 mL of bacterial suspensions containing 108 CFU mL−1. Five independent repeats were used for the infection of twenty plants with each strain (D. solani wt or dsbA mutant). As a negative control, twenty plants were inoculated with sterile Ringer’s solution. On the 7th, 11th and 14th day post infection (dpi), symptoms were visually assessed for wilting, blackleg, stem desiccation and plant death, and the percentage of diseased plants was calculated.
The in vitro grown potato plantlets of cultivar Kondor were propagated on Murashige and Skoog (MS) medium [106] with 30 g L−1 sucrose and 7 g L−1 agar in culture tubes as previously described [107]. The plantlets were grown at 20–22 °C for 20 days prior to inoculation with bacteria. For inoculation, 10 μL aliquots of bacterial suspensions in water containing 105 CFU mL−1 of D. solani dsbA::cm (three independently obtained isolates) or the parental D. solani IPO2222 (positive control) or sterile Ringer solution (negative control) were deposited at the interspace between the stem base and MS medium of each individual plant. Thirty-six individual plantlets were used per bacterial strain or negative control. The inoculated plantlets were grown at 20–22 °C under white fluorescent light with a 16 h photoperiod (white cool fluorescent light, Philips, TLD 58 W/84o, 30–35 μmol m−2 s−1). On the 2nd, 6th, 9th and 13th dpi, the plantlets were visually assessed for disease symptoms (wilting, typical blackleg, turbidity around the roots, stem desiccation and plant death), and the percentage of blackleg-diseased plants was calculated per time point. At the same time, 9 plants per strain were aseptically removed from the tubes and 1 cm long stem fragments were cut out from each plant approx. 1 cm above the stem base and separately suspended in 1 mL of 1/4-strength Ringer’s buffer (Merck, Darmstadt, Germany) supplemented with 0.02% diethyldithiocarbamic acid (Acros Organics, Geel, Belgium). To release bacteria from plant tissues, samples were grinded using sterile metal pestles, and serial dilutions were made in Ringer’s buffer and spotted onto LB agar plates to estimate the bacterial density (CFU per 1 g of plant tissue).
4.7. Cell Viability and Membrane Permeability Assays
The viability of cells was monitored by means of a LIVE/DEAD BacLight Bacterial Viability Kit (Thermo Fisher Scientific, Eugene, OR, USA), as described in [108]. Depending on the growth phase and culture density, 100–400 µL aliquots of bacterial cultures were centrifuged, washed and suspended in 100 µL of Ringer’s buffer. A freshly prepared mixture of fluorescent dyes (SYTO 9 and PI) was added to 10 µL portions of bacterial suspensions, according to the manufacturer’s instructions. Next, bacteria were pipetted onto glass slides, and pictures were taken after 8 min of incubation using a fluorescence microscope (AXIO Scop, Carl Zeiss, Jena, Germany). Data obtained from at least three experiments were analyzed with IMAGEJ software (http://imagej.nih.gov/ij/ accessed date: the 18 October 2017; Research Services Branch, National Institute of Mental Health, Bethesda, MD, USA.).
The permeability of the outer membrane was assessed by a degree of cell lysis in the presence of sodium dodecyl sulphate (SDS), determined by a decrease in the optical density (OD) of the cell culture. The method was partially based on [79,109]. The bacterial cultures were centrifuged, washed twice, and suspended in 1 mL of buffer PBS to OD in the range of 0.642–1.096 or 0.417–0.610 in the case of the exponential or stationary growth phase, respectively. Then, 900 µL aliquots of bacterial suspensions were mixed with 100 µL of 10% SDS (final concentration: 1%), and the optical density was measured at one-minute intervals for 10 min.
4.8. Mass Spectrometry
4.8.1. Sample Preparation for Mass Spectrometry
Five independently inoculated cultures of each D. solani strain (wt or dsbA) were grown overnight in the minimal medium M63Y at 30 °C with aeration. Then, the OD of the cultures was normalized to 0.7, and the bacterial suspensions were inoculated 1:50 into fresh medium. Bacteria were further grown with shaking (180 rpm) for 16 h under microaerobic conditions. To induce oxidative stress, H2O2 was added to a final concentration of 0.25 mM, and the cultures were incubated for 30 min.
For the whole-cell proteome analysis, 14 mL of each culture was centrifuged (5000× g, 10 min), the cells were suspended in the lysis solution (4% SDS, 100 mM Tris/HCl pH 7.6, 0.1 M DTT) and incubated 9 min at 96–98 °C. Suspensions were cooled on ice for 10 min and stored at −80 °C.
To isolate the secreted protein fraction (secretome), 45 mL of each bacterial culture was centrifuged for 10 min at 5000× g. The supernatant, containing secreted proteins, was collected and filtered using syringe filters (PVDF membrane, 0.22 µm, Bionovo, Legnica, Poland) to remove the remaining bacterial cells. EDTA was added to a final concentration of 1.0 mM and incubated for 5 min on ice. Then, the supernatant volumes were reduced to 2 mL via centrifugation (6000× g, 4 °C) using VIVASPIN 20 concentrators (Membrane 3000 MWCO, Sartorius, Stonehouse, UK). The secretome samples were supplemented with protease inhibitors (Roche, Basel, Switzerland) and were further concentrated to a volume of 1 mL. The concentrated protein solutions (approx. 1 mL) were mixed with 9 mL of 99% cold ethanol and centrifuged (30,882× g, 4 °C, 30 min). The precipitates were washed in 2 mL of ethanol, centrifuged (30,882× g, 4 °C, 10 min) and dried at −20 °C overnight. Next, the secretome precipitates were dissolved in the lysis solution and stored at −80 °C.
4.8.2. Protein Digestion
Proteins were digested by trypsin in a standard FASP (Filter-Aided Sample Preparation) procedure [110]. Briefly, 100 µg of protein was placed in a Microcon with a 10 kDa cutoff membrane (Merck-Millipore, Darmstadt, Germany) and washed multiple times with urea-containing buffer (8 M urea, 100 mM Tris-HCl, pH 8.5) via centrifugation at 10,000× g. Next, the cysteine residues were alkylated using the 55 mM iodoacetamide solution in the urea buffer with a 20 min incubation period in darkness at room temperature. The excess iodoacetamide was washed away with subsequent centrifugations with the urea buffer followed by the digestion buffer (50 mM Tris-HCl, pH 8.5). Filters were placed in new tubes, and samples were incubated with trypsin (1:50 enzyme-to-substrate weight ratio) overnight at 37 °C. After incubation, proteolytic peptides were eluted via centrifugation at 10,000× g. Samples were then desalted using a STAGE Tips procedure [111] on in-house-prepared tips containing Empore C18 resin (3M). Tips were prepared via subsequent centrifugations with methanol, 60% acetonitrile and 1% acetic acid in water and 1% acetic acid in water. Then, 10 µg of peptide was placed in each tip, and tips were washed with 1% acetic acid in water before the final elution with 60% acetonitrile and 1% acetic acid in water to new tubes. Samples were concentrated to 40 µL in a SpeedVac before mass spectrometry measurements.
4.8.3. Liquid Chromatography and Mass Spectrometry in Data-Dependent Acquisition Mode
LC-MS/MS measurements were conducted in a positive ion mode on a Triple TOF 5600+ mass spectrometer (SCIEX, Framingham, MA, USA) coupled with an Ekspert MicroLC 200 Plus System (Eksigent, Redwood City, CA, USA). The instrument was operated by Analyst TF 1.7.1 software (SCIEX, Framingham, MA, USA). LC separations were carried out on the ChromXP C18CL column (3 µm, 120 Å, 150 × 0.3 mm, Eksigent, Framingham, MA, USA) in 60 min gradients of 3.5–20% buffer B or 7–35% buffer B for proteome and secretome analysis, respectively (buffer A: 0.1% formic acid in water, buffer B: 0.1% formic acid in acetonitrile). MS runs of whole-proteome samples in data-dependent analysis (DDA) mode consisted of a TOF scan in the m/z range of 400–1200 Da in 50 ms and a Product ion scan in the m/z range of 100–1800 m/z in 40 ms, resulting in a cycle time of 0.9 s. DDA analysis of secretome samples consisted of a TOF scan in the m/z range of 400–1200 Da in 100 ms and a Product ion scan in the m/z range of 100–1800 m/z in 50 ms, resulting in a cycle time of 1.15 s. Precursor ions were fragmented using collision-induced dissociation (CID). All samples were analyzed once for the purpose of spectral library generation.
4.8.4. SWATH Mass Spectrometry Experiments
MS analysis in data-independent acquisition (DIA) mode was conducted with the SWATH-MS method [112] focused on high sensitivity. MS methods were adjusted separately for the whole proteome and secretome samples. In both cases, the m/z range of 400–1200 Da was covered by 25 overlapping windows of variable width constructed in the SWATHTuner software [113] with a focus on equalized ion frequency. All SWATH-MS measurements comprised a TOF survey scan in the m/z range of 400–1200 Da in 50 ms and Product Ion scans in the m/z range of 100–1800 Da in 40 ms, resulting in a cycle time of 1.1 s. All samples were measured in triplicate.
4.8.5. Data Analysis
All data analysis steps were performed separately for the whole proteome and secretome samples. A database search was conducted in ProteinPilot 5.0.2 software (SCIEX, Framingham, MA, USA) against the UniprotKB D. solani D s0432-1 database (version from 19.10.2020) with an automated false discovery rate analysis. Only proteins identified at 1% FDR were considered valid identifications. SWATH-MS data were analyzed in a similar manner as described before [114]. Database search results were imported to the PeakView 2.2 software (SCIEX, Framingham, MA, USA) to construct spectral libraries with the exclusion of shared peptides. SWATH-MS measurement files were uploaded into software and analyzed using relevant libraries. The results were exported to MarkerView software (SCIEX, Framingham, MA, USA) and normalized using the Total Area Sums (TAS) approach. T-tests between studied groups were performed, and the protein concentration changes were considered to be statistically significant if the p-value was lower than 0.05 and the fold change was lower than 0.5 or greater than 2.
The mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium via the PRIDE [115] partner repository with the dataset identifier PXD028047.
5. Conclusions
The lack of a functional dsbA gene resulted in the loss of virulence of D. solani IPO2222. Consequently, the mutant was unable to develop disease symptoms in the natural host, the potato plant.
The dsbA mutant was non-motile and deficient in the secretion of most extracellular virulence factors (pectinases, cellulases and siderophores).
The lack of the DsbA protein led to multifaceted effects in the D. solani cells, with pronounced changes observed in the cellular proteome and secretome.
The proteomic analysis suggested the induction of the extracytoplasmic and cytoplasmic stress responses in the dsbA mutant cells, that could lead to repression of biosynthesis pathways involved in virulence (flagella and PCWDE). A graphical summary of the conclusions is shown in Figure 7.
Figure 7.
Model depicting the functioning of the D. solani cell in the presence and absence of the oxidoreductase DsbA. The properly functioning DsbA enzyme maintains the homeostasis of the cell envelope and ensures production of active virulence factors (left panel). The lack of DsbA disrupts the cell envelope homeostasis and causes an avirulent phenotype (right panel).
To our knowledge, our work provides the first comprehensive study regarding the importance of the DsbA function in the processes related to D. solani pathogenicity, which are based on detailed proteomic analyses and phenotypic and virulence assays.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms23020697/s1.
Author Contributions
Conceptualization: J.S.-G., T.P.; funding acquisition: J.S.-G.; investigation: T.P., D.F., A.B.; methodology: T.P., D.F., A.B., J.F.-K.; formal analysis: T.P., A.B.; supervision: J.S.-G.; validation: T.P.; D.F., J.S.-G.; visualization: T.P., D.F.; writing—original draft: T.P., A.B., J.S.-G.; writing—review and editing: T.P., D.F., A.B., R.C., J.F.-K., N.H.-C.-P., J.S.-G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Science Centre, Poland, NCN OPUS-7UMO-2014/13/B/NZ9/02021.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium via the PRIDE [115] partner repository with the dataset identifier PXD028047.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Charkowski A., Blanco C., Condemine G., Expert D., Franza T., Hayes C., Hugouvieux-Cotte-Pattat N., Solanilla E.L., Low D., Moleleki L., et al. The Role of Secretion Systems and Small Molecules in Soft-Rot Enterobacteriaceae Pathogenicity. Annu. Rev. Phytopathol. 2012;50:425–449. doi: 10.1146/annurev-phyto-081211-173013. [DOI] [PubMed] [Google Scholar]
- 2.Yu J., Kroll J.S. DsbA: A Protein-Folding Catalyst Contributing to Bacterial Virulence. Microbes Infect. 1999;1:1221–1228. doi: 10.1016/S1286-4579(99)00239-7. [DOI] [PubMed] [Google Scholar]
- 3.Missiakas D., Raina S. Protein Folding in the Bacterial Periplasm. J. Bacteriol. 1997;179:2465–2471. doi: 10.1128/jb.179.8.2465-2471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ha U.-H., Wang Y., Jin S. DsbA of Pseudomonas Aeruginosa Is Essential for Multiple Virulence Factors. Infect. Immun. 2003;71:1590–1595. doi: 10.1128/IAI.71.3.1590-1595.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heras B., Shouldice S.R., Totsika M., Scanlon M.J., Schembri M.A., Martin J.L. DSB Proteins and Bacterial Pathogenicity. Nat. Rev. Microbiol. 2009;7:215–225. doi: 10.1038/nrmicro2087. [DOI] [PubMed] [Google Scholar]
- 6.Bardwell J.C.A., McGovern K., Beckwith J. Identification of a Protein Required for Disulfide Bond Formation in Vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 7.Depuydt M., Leonard S.E., Vertommen D., Denoncin K., Morsomme P., Wahni K., Messens J., Carroll K.S., Collet J.-F. A Periplasmic Reducing System Protects Single Cysteine Residues from Oxidation. Science. 2009;326:1109–1111. doi: 10.1126/science.1179557. [DOI] [PubMed] [Google Scholar]
- 8.Smith R.P., Paxman J.J., Scanlon M.J., Heras B. Targeting Bacterial Dsb Proteins for the Development of Anti-Virulence Agents. Molecules. 2016;21:811. doi: 10.3390/molecules21070811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golanowska M., Galardini M., Bazzicalupo M., Hugouvieux-Cotte-Pattat N., Mengoni A., Potrykus M., Slawiak M., Lojkowska E. Draft Genome Sequence of a Highly Virulent Strain of the Plant Pathogen Dickeya Solani, IFB0099. Genome Announc. 2015;3:e00109–e00115. doi: 10.1128/genomeA.00109-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khayi S., Mondy S., Beury-Cirou A., Moumni M., Hélias V., Faure D. Genome Sequence of the Emerging Plant Pathogen Dickeya Solani Strain RNS 08.23.3.1A. Genome Announc. 2014;2 doi: 10.1128/genomeA.01270-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mansfield J., Genin S., Magori S., Citovsky V., Sriariyanum M., Ronald P., Dow M., Verdier V., Beer S.V., Machado M.A., et al. Top 10 Plant Pathogenic Bacteria in Molecular Plant Pathology. Mol. Plant Pathol. 2012;13:614–629. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupuis B., Nkuriyingoma P., Van Gijsegem F. Economic Impact of Pectobacterium and Dickeya Species on Potato Crops: A Review and Case Study. In: Van Gijsegem F., van der Wolf J.M., Toth I.K., editors. Plant Diseases Caused by Dickeya and Pectobacterium Species. Springer International Publishing; Berlin/Heidelberg, Germany: 2021. pp. 263–282. [Google Scholar]
- 13.Czajkowski R., de Boer W.J., Velvis H., van der Wolf J.M. Systemic Colonization of Potato Plants by a Soilborne, Green Fluorescent Protein-Tagged Strain of Dickeya Sp. Biovar 3. Phytopathology. 2010;100:134–142. doi: 10.1094/PHYTO-100-2-0134. [DOI] [PubMed] [Google Scholar]
- 14.Degefu Y., Potrykus M., Golanowska M., Virtanen E., Lojkowska E. A New Clade of Dickeya Spp. Plays a Major Role in Potato Blackleg Outbreaks in North Finland. Ann. Appl. Biol. 2013;162:231–241. doi: 10.1111/aab.12020. [DOI] [Google Scholar]
- 15.Laurila J., Ahola V., Lehtinen A., Joutsjoki T., Hannukkala A., Rahkonen A., Pirhonen M. Characterization of Dickeya Strains Isolated from Potato and River Water Samples in Finland. Eur. J. Plant Pathol. 2008;122:213–225. doi: 10.1007/s10658-008-9274-5. [DOI] [Google Scholar]
- 16.Potrykus M., Golanowska M., Sledz W., Zoledowska S., Motyka A., Kolodziejska A., Butrymowicz J., Lojkowska E. Biodiversity of Dickeya Spp. Isolated from Potato Plants and Water Sources in Temperate Climate. Plant Dis. 2016;100:408–417. doi: 10.1094/PDIS-04-15-0439-RE. [DOI] [PubMed] [Google Scholar]
- 17.Sławiak M., Łojkowska E., van der Wolf J.M. First Report of Bacterial Soft Rot on Potato Caused by Dickeya Sp. (Syn. Erwinia Chrysanthemi) in Poland. Plant Pathol. 2009;58 doi: 10.1111/j.1365-3059.2009.02028.x. [DOI] [Google Scholar]
- 18.Sławiak M., van Beckhoven J.R.C.M., Speksnijder A.G.C.L., Czajkowski R., Grabe G., van der Wolf J.M. Biochemical and Genetical Analysis Reveal a New Clade of Biovar 3 Dickeya Spp. Strains Isolated from Potato in Europe. Eur. J. Plant Pathol. 2009;125:245–261. doi: 10.1007/s10658-009-9479-2. [DOI] [Google Scholar]
- 19.Toth I.K., van der Wolf J.M., Saddler G., Lojkowska E., Hélias V., Pirhonen M., Tsror L., Elphinstone J.G. Dickeya Species: An Emerging Problem for Potato Production in Europe. Plant Pathol. 2011;60:385–399. doi: 10.1111/j.1365-3059.2011.02427.x. [DOI] [Google Scholar]
- 20.van der Wolf J.M., Nijhuis E.H., Kowalewska M.J., Saddler G., Parkinson N., Elphinstone J.G., Pritchard L., Toth I.K., Lojkowska E., Potrykus M., et al. Dickeya Solani Sp. Nov., a Pectinolytic Plant-Pathogenic Bacterium Isolated from Potato (Solanum Tuberosum) Int. J. Syst. Evol. Microbiol. 2014;64:768–774. doi: 10.1099/ijs.0.052944-0. [DOI] [PubMed] [Google Scholar]
- 21.Hugouvieux-Cotte-Pattat N., Condemine G., Shevchik V.E. Bacterial Pectate Lyases, Structural and Functional Diversity. Environ. Microbiol. Rep. 2014;6:427–440. doi: 10.1111/1758-2229.12166. [DOI] [PubMed] [Google Scholar]
- 22.Shevchik V.E., Bortoli-Gernnan I., Robert-Baudouy J., Robinet S., Barras F., Condemine G. Differential Effect of DsbA and DsbC Mutations on Extracellular Enzyme Secretion in Erwinia Chrysanthemi. Mol. Microbiol. 1995;16:745–753. doi: 10.1111/j.1365-2958.1995.tb02435.x. [DOI] [PubMed] [Google Scholar]
- 23.Coulthurst S.J., Lilley K.S., Hedley P.E., Liu H., Toth I.K., Salmond G.P.C. DsbA Plays a Critical and Multifaceted Role in the Production of Secreted Virulence Factors by the Phytopathogen Erwinia Carotovora Subsp. Atroseptica. J. Biol. Chem. 2008;283:23739–23753. doi: 10.1074/jbc.M801829200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antúnez-Lamas M., Cabrera-Ordóñez E., López-Solanilla E., Raposo R., Trelles-Salazar O., Rodríguez-Moreno A., Rodríguez-Palenzuela P. 2009 Role of Motility and Chemotaxis in the Pathogenesis of Dickeya Dadantii 3937 (Ex Erwinia Chrysanthemi 3937) Microbiology. 2009;155:434–442. doi: 10.1099/mic.0.022244-0. [DOI] [PubMed] [Google Scholar]
- 25.Yi T.-M., Huang Y., Simon M.I., Doyle J. Robust Perfect Adaptation in Bacterial Chemotaxis through Integral Feedback Control. Proc. Natl. Acad. Sci. USA. 2000;97:4649–4653. doi: 10.1073/pnas.97.9.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harshey R.M. Bacterial Motility on a Surface: Many Ways to a Common Goal. Annu. Rev. Microbiol. 2003;57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 27.Allison C., Hughes C. Bacterial Swarming: An Example of Prokaryotic Differentiation and Multicellular Behaviour. Sci. Prog. 1991;75:403–422. [PubMed] [Google Scholar]
- 28.Fraser G.M., Hughes C. Swarming Motility. Curr. Opin. Microbiol. 1999;2:630–635. doi: 10.1016/S1369-5274(99)00033-8. [DOI] [PubMed] [Google Scholar]
- 29.Reverchon S., Nasser W. Dickeya Ecology, Environment Sensing and Regulation of Virulence Programme. Environ. Microbiol. Rep. 2013;5:622–636. doi: 10.1111/1758-2229.12073. [DOI] [PubMed] [Google Scholar]
- 30.Reverchon S., Muskhelisvili G., Nasser W. Chapter Three-Virulence Program of a Bacterial Plant Pathogen: The Dickeya Model. Prog. Mol. Biol. Transl. Sci. 2016;142:51–92. doi: 10.1016/bs.pmbts.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 31.Hölscher T., Kovács Á.T. Sliding on the Surface: Bacterial Spreading without an Active Motor. Environ. Microbiol. 2017;19:2537–2545. doi: 10.1111/1462-2920.13741. [DOI] [PubMed] [Google Scholar]
- 32.Wolfe A.J., Conley M.P., Kramer T.J., Berg H.C. Reconstitution of Signaling in Bacterial Chemotaxis. J. Bacteriol. 1987;169:1878–1885. doi: 10.1128/jb.169.5.1878-1885.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alberts A.W., Vagelos P.R. 2 Acyl-CoA Carboxylases. Enzymes. 1972;6:37–82. doi: 10.1016/S1874-6047(08)60037-2. [DOI] [Google Scholar]
- 34.Sobota J.M., Imlay J.A. Iron Enzyme Ribulose-5-Phosphate 3-Epimerase in Escherichia Coli Is Rapidly Damaged by Hydrogen Peroxide but Can Be Protected by Manganese. Proc. Natl. Acad. Sci. USA. 2011;108:5402–5407. doi: 10.1073/pnas.1100410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bae W., Xia B., Inouye M., Severinov K. Escherichia Coli CspA-Family RNA Chaperones Are Transcription Antiterminators. Proc. Natl. Acad. Sci. USA. 2000;97:7784–7789. doi: 10.1073/pnas.97.14.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nossal N.G., Singer M.F. The Processive Degradation of Individual Polyribonucleotide Chains: I. Escherichia Coli Ribonuclease II. J. Biol. Chem. 1968;243:913–922. doi: 10.1016/S0021-9258(18)93603-3. [DOI] [PubMed] [Google Scholar]
- 37.Lopez-Amoros R., Comas J., Vives-Rego J. Flow Cytometric Assessment of Escherichia Coli and Salmonella Typhimurium Starvation-Survival in Seawater Using Rhodamine 123, Propidium Iodide, and Oxonol. Appl. Environ. Microbiol. 1995;61:2521–2526. doi: 10.1128/aem.61.7.2521-2526.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikaido H. Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dailey F.E., Berg H.C. Mutants in Disulfide Bond Formation That Disrupt Flagellar Assembly in Escherichia Coli. Proc. Natl. Acad. Sci. USA. 1993;90:1043–1047. doi: 10.1073/pnas.90.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kloek A.P., Brooks D.M., Kunkel B.N. A DsbA Mutant of Pseudomonas Syringae Exhibits Reduced Virulence and Partial Impairment of Type III Secretion. Mol. Plant Pathol. 2000;1:139–150. doi: 10.1046/j.1364-3703.2000.00016.x. [DOI] [PubMed] [Google Scholar]
- 41.Mavrodi O.V., Mavrodi D.V., Park A.A., Weller D.M., Thomashow L.S.Y. 2006 The Role of DsbA in Colonization of the Wheat Rhizosphere by Pseudomonas Fluorescens Q8r1-96. Microbiology. 2006;152:863–872. doi: 10.1099/mic.0.28545-0. [DOI] [PubMed] [Google Scholar]
- 42.Mariconda S., Wang Q., Harshey R.M. A Mechanical Role for the Chemotaxis System in Swarming Motility. Mol. Microbiol. 2006;60:1590–1602. doi: 10.1111/j.1365-2958.2006.05208.x. [DOI] [PubMed] [Google Scholar]
- 43.Novak E.A., Sekar P., Xu H., Moon K.H., Manne A., Wooten R.M., Motaleb M.A. The Borrelia Burgdorferi CheY3 Response Regulator Is Essential for Chemotaxis and Completion of Its Natural Infection Cycle. Cell. Microbiol. 2016;18:1782–1799. doi: 10.1111/cmi.12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng C., Dong Z., Han X., Wang H., Jiang L., Sun J., Yang Y., Ma T., Shao C., Wang X., et al. Thioredoxin A Is Essential for Motility and Contributes to Host Infection of Listeria Monocytogenes via Redox Interactions. Front. Cell. Infect. Microbiol. 2017;7 doi: 10.3389/fcimb.2017.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corral J., Pérez-Varela M., Barbé J., Aranda J. Direct Interaction between RecA and a CheW-like Protein Is Required for Surface-Associated Motility, Chemotaxis and the Full Virulence of Acinetobacter Baumannii Strain ATCC 17978. Virulence. 2020;11:315–326. doi: 10.1080/21505594.2020.1748923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spöring I., Felgner S., Preuße M., Eckweiler D., Rohde M., Häussler S., Weiss S., Erhardt M. Regulation of Flagellum Biosynthesis in Response to Cell Envelope Stress in Salmonella Enterica Serovar Typhimurium. mBio. 2018;9:e00736-17. doi: 10.1128/mBio.00736-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bontemps-Gallo S., Madec E., Dondeyne J., Delrue B., Robbe-Masselot C., Vidal O., Prouvost A.-F., Boussemart G., Bohin J.-P., Lacroix J.-M. Concentration of Osmoregulated Periplasmic Glucans (OPGs) Modulates the Activation Level of the RcsCD RcsB Phosphorelay in the Phytopathogen Bacteria Dickeya Dadantii. Environ. Microbiol. 2013;15:881–894. doi: 10.1111/1462-2920.12054. [DOI] [PubMed] [Google Scholar]
- 48.Madec E., Bontemps-Gallo S., Lacroix J.-M. 2014 Increased Phosphorylation of the RcsB Regulator of the RcsCDB Phosphorelay in Strains of Dickeya Dadantii Devoid of Osmoregulated Periplasmic Glucans Revealed by Phos-Tag Gel Analysis. Microbiology. 2014;160:2763–2770. doi: 10.1099/mic.0.081273-0. [DOI] [PubMed] [Google Scholar]
- 49.Wall E., Majdalani N., Gottesman S. The Complex Rcs Regulatory Cascade. Annu. Rev. Microbiol. 2018;72:111–139. doi: 10.1146/annurev-micro-090817-062640. [DOI] [PubMed] [Google Scholar]
- 50.Lee Y., Kim Y., Yeom S., Kim S., Park S., Jeon C.O., Park W. The Role of Disulfide Bond Isomerase A (DsbA) of Escherichia Coli O157:H7 in Biofilm Formation and Virulence. FEMS Microbiol. Lett. 2008;278:213–222. doi: 10.1111/j.1574-6968.2007.00993.x. [DOI] [PubMed] [Google Scholar]
- 51.Rosadini C.V., Wong S.M.S., Akerley B.J. The Periplasmic Disulfide Oxidoreductase DsbA Contributes to Haemophilus Influenzae Pathogenesis. Infect. Immun. 2008;76:1498–1508. doi: 10.1128/IAI.01378-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rizzitello A.E., Harper J.R., Silhavy T.J. Genetic Evidence for Parallel Pathways of Chaperone Activity in the Periplasm of Escherichia Coli. J. Bacteriol. 2001;183:6794–6800. doi: 10.1128/JB.183.23.6794-6800.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Missiakas D., Raina S. The Extracytoplasmic Function Sigma Factors: Role and Regulation. Mol. Microbiol. 1998;28:1059–1066. doi: 10.1046/j.1365-2958.1998.00865.x. [DOI] [PubMed] [Google Scholar]
- 54.Önder Ö., Turkarslan S., Sun D., Daldal F. Overproduction or Absence of the Periplasmic Protease DegP Severely Compromises Bacterial Growth in the Absence of the Dithiol: Disulfide Oxidoreductase DsbA. Mol. Cell. Proteom. 2008;7:875–890. doi: 10.1074/mcp.M700433-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skorko-Glonek J., Sobiecka-Szkatula A., Narkiewicz J., Lipinska B. 2008 The Proteolytic Activity of the HtrA (DegP) Protein from Escherichia Coli at Low Temperatures. Microbiology. 2008;154:3649–3658. doi: 10.1099/mic.0.2008/020487-0. [DOI] [PubMed] [Google Scholar]
- 56.Weski J., Ehrmann M. Genetic Analysis of 15 Protein Folding Factors and Proteases of the Escherichia Coli Cell Envelope. J. Bacteriol. 2012;194:3225–3233. doi: 10.1128/JB.00221-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meltzer M., Hasenbein S., Mamant N., Merdanovic M., Poepsel S., Hauske P., Kaiser M., Huber R., Krojer T., Clausen T., et al. Structure, Function and Regulation of the Conserved Serine Proteases DegP and DegS of Escherichia Coli. Res. Microbiol. 2009;160:660–666. doi: 10.1016/j.resmic.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 58.Creze C., Castang S., Derivery E., Haser R., Hugouvieux-Cotte-Pattat N., Shevchik V.E., Gouet P. The Crystal Structure of Pectate Lyase PelI from Soft Rot Pathogen Erwinia Chrysanthemi in Complex with Its Substrate. J. Biol. Chem. 2008;283:18260–18268. doi: 10.1074/jbc.M709931200. [DOI] [PubMed] [Google Scholar]
- 59.Thomas L.M., Doan C.N., Oliver R.L., Yoder M.D. Structure of Pectate Lyase A: Comparison to Other Isoforms. Acta Cryst. D Biol. Cryst. 2002;58:1008–1015. doi: 10.1107/S0907444902005851. [DOI] [PubMed] [Google Scholar]
- 60.Lietzke S.E., Scavetta R.D., Yoder M.D., Jurnak F. The Refined Three-Dimensional Structure of Pectate Lyase E from Erwinia Chrysanthemi at 2.2 A Resolution. Plant Physiol. 1996;111:73–92. doi: 10.1104/pp.111.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jenkins J., Mayans O., Smith D., Worboys K., Pickersgill R.W. Three-Dimensional Structure of Erwinia Chrysanthemi Pectin Methylesterase Reveals a Novel Esterase Active Site1 1Edited by J. Thornton. J. Mol. Biol. 2001;305:951–960. doi: 10.1006/jmbi.2000.4324. [DOI] [PubMed] [Google Scholar]
- 62.Korotkov K.V., Sandkvist M., Hol W.G.J. The Type II Secretion System: Biogenesis, Molecular Architecture and Mechanism. Nat. Rev. Microbiol. 2012;10:336–351. doi: 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu X., Zeng Q., Koestler B.J., Waters C.M., Sundin G.W., Hutchins W., Yang C.-H. Deciphering the Components That Coordinately Regulate Virulence Factors of the Soft Rot Pathogen Dickeya Dadantii. Mol. Plant Microbe Interact. 2014;27:1119–1131. doi: 10.1094/MPMI-01-14-0026-R. [DOI] [PubMed] [Google Scholar]
- 64.Boyer M.-H., Cami B., Kotoujansky A., Chambost J.-P., Frixon C., Cattanéo J. Isolation of the Gene Encoding the Major Endoglucanase of Erwinia Chrysanthemi. Homology between Cel Genes of Two Strains of Erwinia Chrysanthemi. FEMS Microbiol. Lett. 1987;41:351–356. doi: 10.1111/j.1574-6968.1987.tb02225.x. [DOI] [Google Scholar]
- 65.Bortoli-German I., Brun E., Py B., Chippaux M., Barras F. Periplasmic Disulphide Bond Formation Is Essential for Cellulase Secretion by the Plant Pathogen Erwinia Chrysanthemi. Mol. Microbiol. 1994;11:545–553. doi: 10.1111/j.1365-2958.1994.tb00335.x. [DOI] [PubMed] [Google Scholar]
- 66.Franza T., Expert D. Role of Iron Homeostasis in the Virulence of Phytopathogenic Bacteria: An ‘à La Carte’ Menu. Mol. Plant Pathol. 2013;14:429–438. doi: 10.1111/mpp.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perry R.D., Bobrov A.G., Kirillina O., Rhodes E.R., Actis L.A., Fetherston J.D. Yersinia Pestis Transition Metal Divalent Cation Transporters. Adv. Exp. Med. Biol. 2012;954:267–279. doi: 10.1007/978-1-4614-3561-7_34. [DOI] [PubMed] [Google Scholar]
- 68.Capela D., Barloy-Hubler F., Gouzy J., Bothe G., Ampe F., Batut J., Boistard P., Becker A., Boutry M., Cadieu E., et al. Analysis of the Chromosome Sequence of the Legume Symbiont Sinorhizobium Meliloti Strain 1021. Proc. Natl. Acad. Sci. USA. 2001;98:9877–9882. doi: 10.1073/pnas.161294398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hews C.L., Cho T., Rowley G., Raivio T.L. Maintaining Integrity Under Stress: Envelope Stress Response Regulation of Pathogenesis in Gram-Negative Bacteria. Front. Cell. Infect. Microbiol. 2019;9 doi: 10.3389/fcimb.2019.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Price N.L., Raivio T.L. Characterization of the Cpx Regulon in Escherichia Coli Strain MC4100. J. Bacteriol. 2009;191:1798–1815. doi: 10.1128/JB.00798-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bontemps-Gallo S., Madec E., Lacroix J.-M. The Two-Component System CpxAR Is Essential for Virulence in the Phytopathogen Bacteria Dickeya Dadantii EC3937. Environ. Microbiol. 2015;17:4415–4428. doi: 10.1111/1462-2920.12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Skovierova H., Rowley G., Rezuchova B., Homerova D., Lewis C., Roberts M., Kormanec J. 2006 Identification of the ΣE Regulon of Salmonella Enterica Serovar Typhimurium. Microbiology. 2006;152:1347–1359. doi: 10.1099/mic.0.28744-0. [DOI] [PubMed] [Google Scholar]
- 73.Danese P.N., Silhavy T.J. CpxP, a Stress-Combative Member of the Cpx Regulon. J. Bacteriol. 1998;180:831–839. doi: 10.1128/JB.180.4.831-839.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meng J., Young G., Chen J. The Rcs System in Enterobacteriaceae: Envelope Stress Responses and Virulence Regulation. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.627104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin D., Rao C.V., Slauch J.M. The Salmonella SPI1 Type Three Secretion System Responds to Periplasmic Disulfide Bond Status via the Flagellar Apparatus and the RcsCDB System. J. Bacteriol. 2008;190:87–97. doi: 10.1128/JB.01323-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Minic Z. Proteomic Studies of the Effects of Different Stress Conditions on Central Carbon Metabolism in Microorganisms. J. Proteom. Bioinform. 2015;8 doi: 10.4172/jpb.1000355. [DOI] [Google Scholar]
- 77.Juhnke H., Krems B., Kötter P., Entian K.-D. Mutants That Show Increased Sensitivity to Hydrogen Peroxide Reveal an Important Role for the Pentose Phosphate Pathway in Protection of Yeast against Oxidative Stress. Mol. Gen. Genet. 1996;252:456–464. doi: 10.1007/BF02173011. [DOI] [PubMed] [Google Scholar]
- 78.Calderini E., Celebioglu H.U., Villarroel J., Jacobsen S., Svensson B., Pessione E. Comparative Proteomics of Oxidative Stress Response of Lactobacillus Acidophilus NCFM Reveals Effects on DNA Repair and Cysteine de Novo Synthesis. Proteomics. 2017;17:1600178. doi: 10.1002/pmic.201600178. [DOI] [PubMed] [Google Scholar]
- 79.Samsonov V.V., Samsonov V.V., Sineoky S.P. DcrA and DcrB Escherichia Coli Genes Can Control DNA Injection by Phages Specific for BtuB and FhuA Receptors. Res. Microbiol. 2002;153:639–646. doi: 10.1016/S0923-2508(02)01375-X. [DOI] [PubMed] [Google Scholar]
- 80.Hancock R.E.W. Alterations in Outer Membrane Permeability. Annu. Rev. Microbiol. 1984;38:237–264. doi: 10.1146/annurev.mi.38.100184.001321. [DOI] [PubMed] [Google Scholar]
- 81.Strauch K.L., Johnson K., Beckwith J. Characterization of DegP, a Gene Required for Proteolysis in the Cell Envelope and Essential for Growth of Escherichia Coli at High Temperature. J. Bacteriol. 1989;171:2689–2696. doi: 10.1128/jb.171.5.2689-2696.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McBroom A.J., Kuehn M.J. Release of Outer Membrane Vesicles by Gram-Negative Bacteria Is a Novel Envelope Stress Response. Mol. Microbiol. 2007;63:545–558. doi: 10.1111/j.1365-2958.2006.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schwechheimer C., Sullivan C.J., Kuehn M.J. Envelope Control of Outer Membrane Vesicle Production in Gram-Negative Bacteria. Biochemistry. 2013;52:3031–3040. doi: 10.1021/bi400164t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gevrekci A.Ö. The Roles of Polyamines in Microorganisms. World J. Microbiol. Biotechnol. 2017;33:204. doi: 10.1007/s11274-017-2370-y. [DOI] [PubMed] [Google Scholar]
- 85.Przepiora T., Figaj D., Radzinska M., Apanowicz M., Sieradzka M., Ambroziak P., Hugouvieux-Cotte-Pattat N., Lojkowska E., Skorko-Glonek J. Effects of Stressful Physico-Chemical Factors on the Fitness of the Plant Pathogenic Bacterium Dickeya Solani. Eur. J. Plant Pathol. 2020;156:519–535. doi: 10.1007/s10658-019-01902-z. [DOI] [Google Scholar]
- 86.Kotoujansky A., Diolez A., Boccara M., Bertheau Y., Andro T., Coleno A. Molecular Cloning of Erwinia Chrysanthemi Pectinase and Cellulase Structural Genes. EMBO J. 1985;4:781–785. doi: 10.1002/j.1460-2075.1985.tb03697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Resibois A., Colet M., Faelen M., Schoonejans E., Toussaint A. ΦEC2, a New Generalized Transducing Phage of Erwinia Chrysanthemi. Virology. 1984;137:102–112. doi: 10.1016/0042-6822(84)90013-8. [DOI] [PubMed] [Google Scholar]
- 88.Glasner J.D., Yang C.-H., Reverchon S., Hugouvieux-Cotte-Pattat N., Condemine G., Bohin J.-P., Van Gijsegem F., Yang S., Franza T., Expert D., et al. Genome Sequence of the Plant-Pathogenic Bacterium Dickeya Dadantii 3937. J. Bacteriol. 2011;193:2076–2077. doi: 10.1128/JB.01513-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Khayi S., Blin P., Chong T.M., Chan K.-G., Faure D. Complete Genome Anatomy of the Emerging Potato Pathogen Dickeya Solani Type Strain IPO 2222T. Stand. Genom. Sci. 2016;11:87. doi: 10.1186/s40793-016-0208-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sawitzke J.A., Thomason L.C., Bubunenko M., Li X., Costantino N., Court D.L. Chapter Seven-Recombineering: Using Drug Cassettes to Knock out Genes in Vivo. Methods Enzymol. 2013;533:79–102. doi: 10.1016/B978-0-12-420067-8.00007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee D.J., Bingle L.E., Heurlier K., Pallen M.J., Penn C.W., Busby S.J., Hobman J.L. Gene Doctoring: A Method for Recombineering in Laboratory and Pathogenic Escherichia Colistrains. BMC Microbiol. 2009;9:252. doi: 10.1186/1471-2180-9-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Miller J.H. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia Coli and Related Bacteria. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY, USA: 1922. [Google Scholar]
- 93.Lohaus G., Pennewiss K., Sattelmacher B., Hussmann M., Muehling K.H. Is the Infiltration-Centrifugation Technique Appropriate for the Isolation of Apoplastic Fluid? A Critical Evaluation with Different Plant Species. Physiol. Plant. 2001;111:457–465. doi: 10.1034/j.1399-3054.2001.1110405.x. [DOI] [PubMed] [Google Scholar]
- 94.Roeder D.L., Collmer A. Marker-Exchange Mutagenesis of a Pectate Lyase Isozyme Gene in Erwinia Chrysanthemi. J. Bacteriol. 1985;164:51–56. doi: 10.1128/jb.164.1.51-56.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Figaj D., Czaplewska P., Przepióra T., Ambroziak P., Potrykus M., Skorko-Glonek J. Lon Protease Is Important for Growth Under Stressful Conditions and Pathogenicity of the Phytopathogen, Bacterium Dickeya Solani. Int. J. Mol. Sci. 2020;21:3687. doi: 10.3390/ijms21103687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Golanowska M., Kielar J., Lojkowska E. The Effect of Temperature on the Phenotypic Features and the Maceration Ability of Dickeya Solani Strains Isolated in Finland, Israel and Poland. Eur. J. Plant Pathol. 2017;147:803–817. doi: 10.1007/s10658-016-1044-1. [DOI] [Google Scholar]
- 97.Ji J., Hugouvieux-Cotte-Pattat N., Robert-Baudouy J. 1987 Use of Mu-Lac Insertions to Study the Secretion of Pectate Lyases by Erwinia Chrysanthemi. Microbiology. 1987;133:793–802. doi: 10.1099/00221287-133-3-793. [DOI] [Google Scholar]
- 98.Reverchon S., Van Gijsegem F., Rouve M., Kotoujansky A., Robert-Baudouy J. Organization of a Pectate Lyase Gene Family in Erwinia Chrysanthemi. Gene. 1986;49:215–224. doi: 10.1016/0378-1119(86)90282-9. [DOI] [PubMed] [Google Scholar]
- 99.Wood P.J. The Use of Dye—Polysaccharide Interactions in β-d-Glucanase Assay. Carbohydr. Res. 1981;94:C19–C23. doi: 10.1016/S0008-6215(00)80727-2. [DOI] [Google Scholar]
- 100.Collmer A., Ried J.L., Mount M.S. Assay Methods for Pectic Enzymes. Methods Enzymol. 1988;161:329–335. doi: 10.1016/0076-6879(88)61037-8. [DOI] [Google Scholar]
- 101.Coulthurst S.J., Lilley K.S., Salmond G.P.C. Genetic and Proteomic Analysis of the Role of LuxS in the Enteric Phytopathogen, Erwinia Carotovora. Mol. Plant Pathol. 2006;7:31–45. doi: 10.1111/j.1364-3703.2005.00314.x. [DOI] [PubMed] [Google Scholar]
- 102.Tardy F., Nasser W., Robert-Baudouy J., Hugouvieux-Cotte-Pattat N. Comparative Analysis of the Five Major Erwinia Chrysanthemi Pectate Lyases: Enzyme Characteristics and Potential Inhibitors. J. Bacteriol. 1997;179:2503–2511. doi: 10.1128/jb.179.8.2503-2511.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Biely P., Mislovičová D., Toman R. Soluble Chromogenic Substrates for the Assay of Endo-1,4-β-Xylanases and Endo-1,4-β-Glucanases. Anal. Biochem. 1985;144:142–146. doi: 10.1016/0003-2697(85)90095-8. [DOI] [PubMed] [Google Scholar]
- 104.Schwyn B., Neilands J.B. Universal Chemical Assay for the Detection and Determination of Siderophores. Anal. Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 105.Hugouvieux-Cotte-Pattat N. The RhaS Activator Controls the Erwinia Chrysanthemi 3937 Genes RhiN, RhiT and RhiE Involved in Rhamnogalacturonan Catabolism. Mol. Microbiol. 2004;51:1361–1374. doi: 10.1046/j.1365-2958.2003.03908.x. [DOI] [PubMed] [Google Scholar]
- 106.Murashige T., Skoog F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 107.Czajkowski R., van der Wolf J.M., Krolicka A., Ozymko Z., Narajczyk M., Kaczynska N., Lojkowska E. Salicylic Acid Can Reduce Infection Symptoms Caused by Dickeya Solani in Tissue Culture Grown Potato (Solanum Tuberosum L.) Plants. Eur. J. Plant Pathol. 2015;141:545–558. doi: 10.1007/s10658-014-0561-z. [DOI] [Google Scholar]
- 108.Figaj D., Gieldon A., Bartczak M., Koper T., Zarzecka U., Lesner A., Lipinska B., Skorko-Glonek J. The LD Loop as an Important Structural Element Required for Transmission of the Allosteric Signal in the HtrA (DegP) Protease from Escherichia Coli. FEBS J. 2016;283:3471–3487. doi: 10.1111/febs.13822. [DOI] [PubMed] [Google Scholar]
- 109.Taylor A.J., Zakai S.A.I., Kelly D.J. The Periplasmic Chaperone Network of Campylobacter Jejuni: Evidence That SalC (Cj1289) and PpiD (Cj0694) Are Involved in Maintaining Outer Membrane Integrity. Front. Microbiol. 2017;8 doi: 10.3389/fmicb.2017.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wiśniewski J.R. Quantitative Evaluation of Filter Aided Sample Preparation (FASP) and Multienzyme Digestion FASP Protocols. Anal. Chem. 2016;88:5438–5443. doi: 10.1021/acs.analchem.6b00859. [DOI] [PubMed] [Google Scholar]
- 111.Rappsilber J., Mann M., Ishihama Y. Protocol for Micro-Purification, Enrichment, Pre-Fractionation and Storage of Peptides for Proteomics Using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 112.Gillet L.C., Navarro P., Tate S., Röst H., Selevsek N., Reiter L., Bonner R., Aebersold R. Targeted Data Extraction of the MS/MS Spectra Generated by Data-Independent Acquisition: A New Concept for Consistent and Accurate Proteome Analysis. Mol. Cell. Proteom. 2012;11:O111.016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang Y., Bilbao A., Bruderer T., Luban J., Strambio-De-Castillia C., Lisacek F., Hopfgartner G., Varesio E. The Use of Variable Q1 Isolation Windows Improves Selectivity in LC–SWATH–MS Acquisition. J. Proteome Res. 2015;14:4359–4371. doi: 10.1021/acs.jproteome.5b00543. [DOI] [PubMed] [Google Scholar]
- 114.Lewandowska A.E., Macur K., Czaplewska P., Liss J., Łukaszuk K., Ołdziej S. Human Follicular Fluid Proteomic and Peptidomic Composition Quantitative Studies by SWATH-MS Methodology. Applicability of High PH RP-HPLC Fractionation. J. Proteom. 2019;191:131–142. doi: 10.1016/j.jprot.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 115.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium via the PRIDE [115] partner repository with the dataset identifier PXD028047.