Abstract
Cell density shows very little variation within a given cell type. For example, in humans variability in cell density among cells of a given cell type is 100 times smaller than variation in cell mass. This tight control indicates that maintenance of a cell type-specific cell density is important for cell function. Indeed, pathological conditions such as cellular senescence are accompanied by changes in cell density. Despite the apparent importance of cell-type specific density we know very little about how cell density affects cell function, how it is controlled and how it sometimes changes as part of a developmental process or in response to changes in the environment. The recent development of new technologies to accurately measure cell density of single cells in suspension and in tissues, is likely to provide answers to these important questions.
Keywords: Cell Density, Molecular Crowding, Cell Growth, Cell Volume
Introduction
The density (ρ) of a cell is defined by its relative water (ρ=1g/mL) content and composition of dry mass. Cellular water content ranges from 60–80% depending on cell type and culture conditions [1–4]. The major constituents of cellular dry mass are proteins (ρ=1.3–1.43g/mL,), nucleic acids (ρ= 2g/mL,), carbohydrates (ρ=1.55–1.62g/mL) and lipids (ρ= 0.91–1.01g/mL) [5–8]. The relative composition of dry mass varies between different cell types and growth conditions (Table 1). Proteins are the largest contributor to a cell’s dry mass (40–60%), while the relative contribution of the other dry mass constituents varies substantially between E. coli, S. cerevisiae and cultured mammalian cells (Table 1). Dry mass composition depends on the presence and size of subcellular structures (cell wall, vacuole and endomembrane systems). Changing cellular water content or the composition of dry mass affects a cell’s density, which in turn is likely to have wide-ranging effects on cell physiology. Exploring the interplay between cell density and function as well as describing how cell density is regulated will be the focus of this review.
Table 1:
Composition of different cell types (exponential growth conditions)
| E. coli a | S. cerevisiae b | Mammalian Cell | |
|---|---|---|---|
| Water mass (% total mass) | 70 (70–74) | 68 (60–80) | 70 |
| Dry Mass (% total mass) | 30 (26–30) | 32 (20–40) | 30 |
| DNA (% dry mass) | 3 (1.6–3.4) | 0.3 (0.3–0.6) | 1 |
| RNA (% dry mass) | 21 (10–30) | 11 (6–12) | 4 |
| Protein (% dry mass) | 55 (42–76) | 51 (40–60) | 60 |
| Lipids (% dry mass) | 9 (9–10) | 7 (3–8) | 16 |
| Carbohydrates (% dry mass) | 8 (6.5–8) | 27 (17–50) | 7 |
| Inorganic Ions & Metabolites (% dry mass) | 4 | 4 (4–5) | 13 |
| References | [1,3,32,109,114–117] | [3,30,117–121] | [4,122] |
Values for E. coli grown at 37 °C in aerobic glucose containing minimal medium at a doubling time of 40 minutes [114]. Numbers in brackets indicate the range of reported values for various growth conditions.
Values for S. cerevisiae grown at 30 °C in aerobic 0.5% glucose containing minimal medium at a doubling time of 160 minutes [119]. Numbers in brackets indicate the range of reported values for various growth conditions.
Measuring cell density
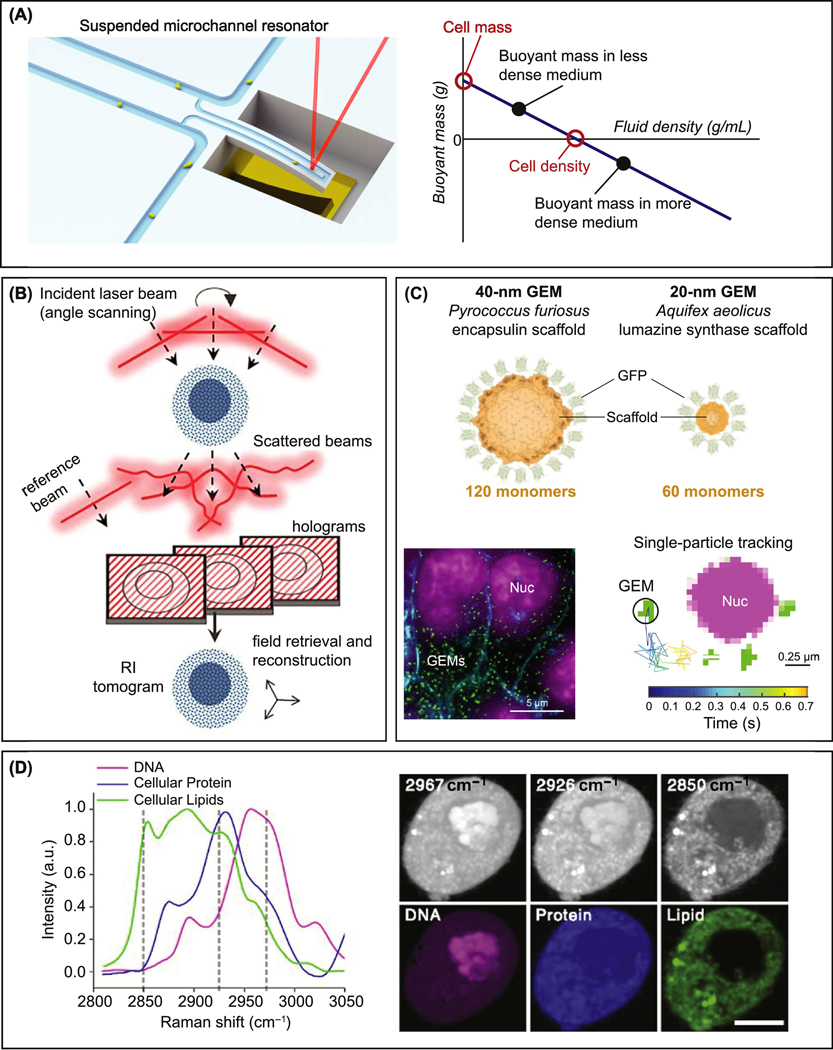
To understand cell density we must be able to accurately assess its value. A number of methods have been developed that do just that (reviewed in [9]; Table 2). Density gradient centrifugation has been used for many decades to detect and isolate cells of a particular density [10–13]. More recently, suspended microchannel resonators (SMRs) have been developed to precisely measure the buoyant mass of single cells in suspension [14]. SMR devices consist of a vibrating cantilever with an embedded microfluidic channel. The resonance frequency of the cantilever is proportional to the mass of the cantilever and changes as a cell passes through the microchannel, allowing for a precise measurement of the buoyant (floating) mass of the passing cell (Figure 1A) [14]. Newer versions of this technology enable measurement of buoyant mass of the same cell in two media with different densities. Using Archimede’s principle, this then allows calculation of the cell mass, cell volume and cell density (Figure 1A) [15,16].
Table 2:
Methods to measure cell density
| Method | Measured Parameter | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|
| Density Gradient Centrifugation | Buoyant density | Can be used to isolate cells, Easily accessible technology | Batch analysis, Suspended cells only | [10,40, 52] |
| Suspended microchannel resonator (SMR ) | Buoyant density | High accuracy, Single cell resolution | Suspended cells only | [15,16] |
| Particle tracking | Diffusion (large particles) | Spatial resolution, Takes particle size into account | Indirect density measurement, Sensitive to pH and specific interactions | [20,22–24,26] |
| Fluorescence correlation spectroscopy (FCS) | Diffusion (small particles) | Additional Information: concentration and complex stoichiometry | Indirect density measurement, Sensitive to pH and specific interactions | [27] |
| Fluorescence | Diffusion | Distinguishes | Indirect density | [28,29] |
| recovery after photobleaching (FRAP) | (small particles) | between mobile and immobile fraction | measurement, Sensitive to pH and specific interactions | |
| Refractive index tomography | Refractive index (proportional to density) | Spatial resolution, tag free, suitable for live imaging | [18,19] | |
| Cryo-electron tomography | Number and density of large structures | High spatial resolution | Fixed specimens only | [20,21] |
| Stimulated raman scattering microscopy (SRS) | Biomass composition and concentration | Distinguishes different classes of macromolecules, can be applied to tissues | Sensitive to autofluorescence | [35–37,39,123] |
Figure 1: Methods to measure cell density.
(A) Left: Cartoon of a suspended microchannel resonator. The vibration frequency of the cantilever is measured with a laser (red line) and changes as a cell passes through the microchannel. Right: Graph illustrating how buoyant mass measurement in two fluids of different density allows for the determination of absolute cell mass and density. (Image credit: Francisco Feijo Delgado & Scott Manalis, Koch Institute Image Gallery, Collection: Image Awards 2011. Graph adapted with permission from [15])
(B) Schematic overview of refractive index tomography: A specimen is illuminated from different angles. The phase information is retrieved from the acquired holograms and used to construct a 3D tomograph of the refractive index inside the specimen (Figure adapted with permission from [18]).
(C) GFP tagged encapsulin and lumazine synthase scaffold proteins self-assemble into spherical particles of a stereotypical size. Measuring particle displacement over time in live cells yields information about cytoplasm or nuclear density. GEM: Genetically encoded multimeric nanoparticle. Nuc: Nucleus (Figure adapted with permission from [20])
(D) Left: Raman spectra of DNA, protein and lipids extracted from HeLa cells. Right: Images acquired using stimulated Raman scattering microscopy at the wabelengths indicated in the Raman spectra before (top) and after (bottom) spectral unmixing (Figure adapted with permission from [39]).
Optical methods have also been developed to determine cell density. When light passes through a translucent object like a cell, it is slowed down resulting in a phase shift. The extent of this phase shift (optical depth) is proportional to the sample thickness and to its average refractive index, which correlates with mass density. Quantitative phase microscopy retrieves the phase information for each pixel using interferometric microscopy [17]. A 3D-tomographic image of the refractive index can be computationally reconstructed from a series of 2D quantitative phase images acquired at different illumination angles (Figure 1B) [18,19]. Cryo-electron tomography uses a similar principle to convert 2D electron density images into 3D electron density maps. Because the resolution of electron microscopy images is much higher than light microscopy based methods, this technique can be used to identify and locate large, dense particles, such as ribosomes, within a cell [20,21]. The advantage of these methods over measuring buoyant mass is that they provide spatial resolution within cells and thus may provide information about potential causes of alterations in cell density.
The dense crowding of the cytoplasm restricts diffusion of macromolecules. This property can also be exploited to indirectly detect changes in cell density. Diffusion of large fluorescently tagged macromolecular assemblies and organelles can be measured using particle tracking. Fluorescent proteins that bind to engineered DNA or RNA sequences can be used to track individual genetic loci or mRNPs, respectively [22–24]. Diffusion of endogenous particles can be affected by specific interactions. To overcome this limitation, exogenous particles can be employed for this tracking method: In mammalian cells, synthetic quantum dots are taken up through endocytosis or can be injected [25,26]. Alternatively, expression of GFP fusion proteins that self-assemble into globular structures, such as viral proteins or encapsulin, can be used to express genetically encoded multimeric nanoparticles (GEMs) of different sizes (Figure 1C) [20,22]. Diffusion of smaller particles can be measured using fluorescence correlation spectroscopy (FCS), where fluctuations in fluorescence intensity in a small volume are monitored [27]. Similarly, measuring, fluorescence recovery after photobleaching (FRAP) in a specific region yields information about the local mobility of fluorescent particles [28,29]. While changes in cell density do affect diffusion of these molecules, it is important to keep in mind that other parameters also affect intracellular diffusion. In particular, a reduction in intracellular pH slows diffusion in the cytoplasm [23,24]. Thus, one needs to interpret such indirect measurements of cell density with caution.
A drawback of all the above-described density measurement techniques is that they do not distinguish between different classes of macromolecules. Quantification of the relative contribution of protein, nucleic acids and lipids typically requires separate quantification of cell volume, total biomass and quantity of the different types of macromolecules [30–32]. Because measuring density in this manner is indirect and relies on more than one measured parameter, the measurement error is inherently high. On the other hand, this type of analysis can be combined with RNA sequencing and mass spectrometry thereby allowing for determination of the concentration of specific transcripts and proteins, respectively [31,33,34]. Direct, spatially resolved measurement of the concentration of different types of macromolecules within a cell has recently become possible through stimulated Raman scattering (SRS) microscopy [35,36]. In Raman scattering, a photon transfers a part of its energy to the vibrational energy of a molecule and emerges red-shifted (Stokes Raman shift). Because each chemical bond has characteristic vibrational modes, the size of Raman shift is characteristic for each substance. Thus, different molecules display characteristic Raman spectra (Figure 1D). SRS enables fast, label-free, high sensitivity and high resolution imaging of Raman spectra of biological samples. Measuring Raman scattering at different wavelengths, followed by spectral unmixing and normalization allows absolute quantification of protein, lipid, nucleic acids and water within a cell (Figure 1D) [37–39]. The two major advantages of this technique are that it can be used to measure cellular density in the context of tissues and that it allows absolute density quantification of different classes of macromolecules.
Which method should one use to measure cell density? The SMR technology is the most accurate and sensitive method to assess cell density, but can only be applied to cells in suspension. Stimulated Raman microscopy can be applied to cells cultured on a slide and on tissues and also reveals spatially resolved information about dry mass composition.
Physiological cell density changes
Cell density is narrowly defined for a given cell type [15], yet can change in response to environmental and developmental cues (Table 3). This finding indicates that mechanisms are in place that set a specific cell density. As mentioned above, we know very little about how cell density is set. However, we do know that density is not an unalterable parameter.
Table 3 –
Examples of naturally occurring changes in cell density
| Occasion | Density change | Effect on cell function | Reference |
|---|---|---|---|
| Cell cycle | |||
| Budding yeast | G1: ↑ G2/M: ↓ |
Not known | [11,16,40] |
| Mammalian cells | M: ↓ | Not known | [41–43] |
| Metabolic status | |||
| Lymphocyte inactivation | ↑ | Correlates with decreased metabolic activity | [3,45] |
| Carbon starvation in yeast | ↑ | Potentially cytoprotective | [23,24] |
| Amino acid starvation in yeast | ↓ | Not known | [20] |
| Carbon starvation in bacteria | ↑ | Not known | [3,22] |
| Spore formation (fungi and bacteria) | ↑ | Contributes to stress resistance of spores in bacteria | [47,48,51] |
| Differentiation | |||
| Hematopoiesis | ↑ | Not known | [52] |
| Chondrocyte differentiation | ↓ | Contributes to bone growth | [53] |
| Pathological conditions | |||
| Cell senescence yeast | ↓ | Not known | [31] |
| Cell senescence human cells | ↓ | Not known | [31] |
| Apoptosis | ↑ | Required for caspase activation | [55–57] |
| Sickle cell disease | ↑ | Promotes polymerization of Sickle β-globin, altering cell shape and stiffness. | [15,60] |
Changes in cell density have been described to occur under a number of physiological conditions. Fluctuations in cell density during the cell cycle have been described in budding yeast and human cells [16,40–43]. In budding yeast density is highest in late G1 and early S phase and lowest in early G1 [16,40]. In human cells, density is remarkably constant throughout the cell cycle, except during mitosis, when cells experience a rapid cell volume increase and a corresponding decrease in cell density [41–43]. With roughly 0.5% change in cell density, these fluctuations are relatively small and correspond to the density change caused by a 10% increase in cell volume by water influx [16,40–42]. A potential explanation for the density fluctuations during the budding yeast cell cycle is that synthesis of different classes of macromolecules is to some degree cell cycle regulated: expression of ribosome biogenesis genes is highest during G1 (when cell density increases), while lipogenic proteins are more highly expressed later in the cell cycle (when cell density decreases) [44]. In human cells, mitotic swelling depends on Na-H ion exchange pumps [41]. Whether and how these fluctuations in cell density affect cell division and cell function has not been explored.
Changes in cellular density also occur in response to alterations in metabolic state. Depletion of pro-inflammatory cytokines inactivates lymphocytes and causes their differentiation into memory cells. This developmental process is accompanied by a reduction in metabolic activity and an increase in cell density [15,45]. Reduced metabolic activity also correlates with an increase in cell density in unicellular organisms. Carbon starvation and energy depletion increase cell density in bacteria and yeast [22–24]. Furthermore, during sustained periods of starvation, many unicellular organisms form durable, stress resistant spores that are also extremely dense [46–48]. It is worth noting that increased density in carbon starved cells and spores correlates with a decrease in cytosolic pH [23,24,49,50]. This drop in pH contributes to increased cell density and is important for survival under energy depleted conditions [24]. Whether decreased cytosolic pH exerts its cytoprotective effect through increasing cell density is not clear. In bacteria, increased density in spores correlates with increased tolerance to heat stress [48,51]. This observation suggests that increased cell density indeed contributes to stress resistance and prolonged survival of dormant, metabolically inactive cells.
Changes in cell density have also been documented during differentiation in mammalian cells. Cell density increases during myeloid maturation, with neutrophils and erythrocytes being the most dense cell types [15,52]. Changes in cell density also accompany cell differentiation during bone development: Bone expansion is driven by a dramatic increase in chondrocyte cell volume. The roughly 10-fold increase in cell volume is largely driven by water influx and coincides with a 50% decrease in density [53]. In summary, in mammals, differentiation-induced changes in cell function correlate with changes in cellular density, raising the interesting possibility that cell type specific cell density contributes to specify cell function.
The most compelling evidence that a cell-type specific cell density is important for cell function stems from the analysis of circumstances where deviations in cell density correlate with a loss of cell function. For example, cell density is decreased in senescent budding yeast and human cells [31], raising the interesting possibility that the pleiotropic defects that characterize senescent cells are caused by reduced cytoplasm density. Apoptosis on the other hand is accompanied by a decrease in cell volume and increase in cell density [54,55]. Inhibition of this cell volume reduction prevents caspase activation and DNA fragmentation [55–57]. It has been proposed that increased macromolecule concentration in the cytoplasm may promote caspase oligomerization and thereby induce autocatalytic caspase activation [58,59].
Pathological conditions associated with mis-regulated red blood cell volume, most famously sickle cell anemia, also lead to increased cell density. The increase in intracellular protein concentration associated with this increased density promotes polymerization of a mutant version of β-globin (hemoglobin S) into filaments. The filaments change the mechanical properties of erythrocytes, giving them their characteristic sickle shape, which can lead to clogging of capillaries and cell lysis [60]. In summary, deviation from the cell type specific cell density can have detrimental consequences. Understanding the molecular basis of cell density control is thus also medically important.
How cell density could affect cell function
Cell density is influenced by a number of parameters. Changing the volume to surface ratio or differential scaling of intracellular organelles, such as disproportionately increasing vacuolar volume, will affect overall cell density. Of particular interest are changes in cytoplasm density. This has long been recognized to be critically important by scientists working with cell free extracts. Dilution of extracts leads to a rapid loss of extract function [61].
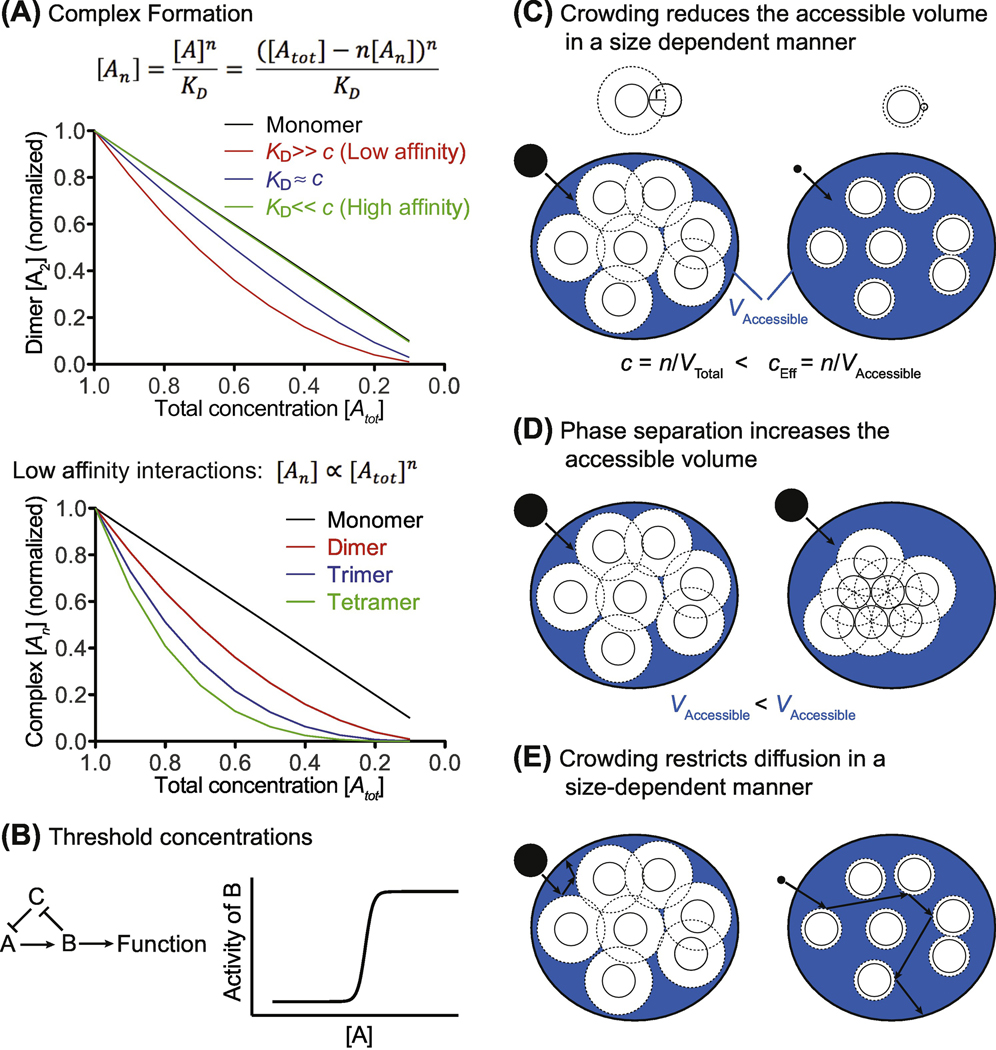
The most direct consequence of altering cytoplasm density is that overall protein concentration changes. Because the probability of reaction partners to collide is concentration dependent, all protein-catalyzed reactions in the cell will be affected by altering total protein concentration. Changing protein concentration will also affect the assembly of protein complexes, with low affinity interactions and multimeric complexes being most sensitive to changes in protein concentration (Figure 2A) [62]. It follows that assembly of dynamic polymers is strongly concentration dependent. This is well documented for the dynamic assembly and disassembly of actin filaments and microtubules [63–65]. Decreased tubulin concentration resulting from mitotic swelling of mammalian cells [41,42] could therefore contribute to the decreased stability of microtubules observed during mitosis [66].
Figure 2: Theoretical impact of alterations in cytoplasm density on biological processes.
(A) Simulation of multi subunit protein complex concentration [An] as a function of changing total protein concentration [Atot] as described by the law of mass action. For simplicity we are considering homomultimers, but the conclusions also apply to heteromeric complexes. KD dissociation constant. Top: Dimers [A2] that bind with low affinity are more sensitive to changes in total protein concentration than dimers that bind with high affinity. Bottom: For low affinity interactions, the concentration of total protein is much larger than the concentration of the complex [Atot] >> [An]. The concentration of monomeric protein is therefore nearly the same as the total protein concentration [A] ≈ [Atot]. The concentration of multimers with weakly interacting subunits is therefore proportional to the product of the concentrations of all subunits [An] α [Atot]n.
(B) Feedback regulation can generate sharp transitions between different cellular states. The components A, B and C represent a feedback loop. Activity of A is inhibited by C. If A surpasses a threshold concentration, it initiates the feedback loop by activating B, which in turn inactivates C, allowing for full activation of A and B. Cytoplasm density can influence whether or not key regulatory proteins reach threshold concentrations that trigger such transitions.
(C) Large macromolecules, such as ribosomes (white circles), occupy up to 20% of the cytoplasm volume, reducing the accessible volume (blue, VAccessible) for other macromolecules (black filled circles). As a result, the effective concentration of a molecule (cEff) in the cytoplasm is higher than the total concentration (c) inside the cell, and biochemical reactions are accelerated by the excluded volume effect. The center of macromolecules (black filled circles) can only approach other macromolecules (white circles) up to a distance of half its diameter (dashed line). Large molecules (left cartoon) are therefore excluded from a larger volume than small molecules (right cartoon). One consequence of this phenomenon is that compact protein conformations are entropically favored in a crowded environment. Crowding thus facilitates protein folding. n=number of molecules, V=volume.
(D) Molecular crowding promotes the generation of phase-separated condensates (right cartoon) because this reduces the excluded cell volume and increases the accessible volume for other molecules. This state is therefore entropically favored.
(E) Molecular crowding restricts lateral diffusion. Large molecules (left cartoon) are more affected by this than small molecules (right cartoon), because their center cannot move as closely to other particles for steric reasons.
Cellular decisions often depend on threshold concentrations: Feedback mechanisms generate sharp transitions in cellular states that are initiated by small concentration changes of critical regulators (Figure 2B). For example, in budding yeast the balance between the accumulation of proteins that promote cell cycle progression and dilution of factors that inhibit cell division have been suggested to determine when cells activate a positive feedback loop that defines the transition from G1 into S-phase [67–69]. Changing overall cellular protein concentration can affect whether and how quickly such threshold concentrations are reached.
The dense packing of macromolecules inside the cytoplasm also creates a number of emergent properties that are often summarized under the concept of molecular crowding (reviewed in [70]). Many effects of macromolecular crowding can be attributed to volume exclusion, meaning that the volume occupied by one specific type of macromolecule is not available for all other macromolecules. Volume exclusion increases the effective concentration of molecules, accelerates biochemical reactions and promotes protein folding and stability (Figure 2C) [70]. Furthermore, molecular crowding entropically favors the formation of large molecular assemblies and phase separated compartments both, in vitro and in vivo (Figure 2D) [20,71–73]. Such phase separated compartments have recently been proposed to play a role in diverse cellular processes such as DNA damage repair, transcription initiation, mRNA translation and cell surface receptor signaling [74–80]. Indeed, addition of synthetic crowding agents like dextran, polyethylene glycol (PEG) or ficoll are often necessary to recapitulate biochemical processes in vitro. In vitro DNA replication, transcription and assembly of viral capsid proteins occur more efficiently in crowded environments [71,81,82], emphasizing the importance of molecular crowding for different aspects of cell physiology.
While a high concentration of macromolecules in the cytoplasm is required for many processes, overcrowding has deleterious consequences. Complete loss of intracellular water due to desiccation is detrimental for most organisms. Membrane damage and protein denaturation are thought to be the most deleterious consequences of desiccation and subsequent rehydration [83,84]. In desiccation resistant organisms, such as yeasts, certain nematodes and tardigrades, the non-reducing disaccharide trehalose and small disordered charged proteins act as chaperones and reduce membrane rupture and protein denaturation due to desiccation and rehydration [85–88].
Water loss induced by hypertonic stress or mechanical compression represents a less severe challenge for cells than desiccation, but the resulting increase in cytoplasm density also has far reaching consequences for cell physiology. Large macromolecular assemblies, such as chromatin or ribosomes [20,89], are mostly responsible for nuclear and cytoplasmic crowding, respectively. In budding yeast, ribosomes occupy up to 20% of the total cytoplasm volume [20]. The dense packing of ribosomes severely restricts diffusion of large structures, such as ribosomes themselves or organelles, but affects diffusion of smaller particles to a lesser extent (Figure 2E). A reduction of cell volume by as little as 20% in budding yeast and 30% in human cells is sufficient to induce a complete loss of mobility of large particles in the cytoplasm, while smaller particles can still move relatively unrestricted [20,22–24,90–94].
It is remarkable that the concentration of macromolecules inside the cytoplasm is kept so close to this critical point. Maintaining a high macromolecule concentration likely maximizes the beneficial effects of molecular crowding outlined above. Under conditions where cells go beyond this maximally beneficial crowded state, such as occurs during carbon starvation in budding yeast and bacteria, the cytoplasm transitions into a glass-like state [22–24,95–97]. As mentioned above, whether this increase in cell density is critical for preserving cell function during periods of starvation remains to be determined. It is however tempting to speculate that transition to a cytoplasmic state characterized by complete immobilization of large macromolecular structures may represent an effective way to stop metabolic activity and stabilize the proteome during stress conditions. In summary, changing the density of the cytoplasm is predicted to affect many, perhaps all cellular functions through multiple mechanisms.
Regulation of cell density
A number of instances have been reported where a biological process is associated with a change in cell density. One such situation - osmotic challenge – has been studied in detail. When cells are exposed to a hypertonic environment, cell volume decreases rapidly because of water efflux. As a result, cytoplasm density increases. Cells respond to this water efflux by increasing the concentration of osmotically active molecules, which then reverse the water loss and return the intracellular concentration of macromolecules to values that are similar to isotonic conditions.
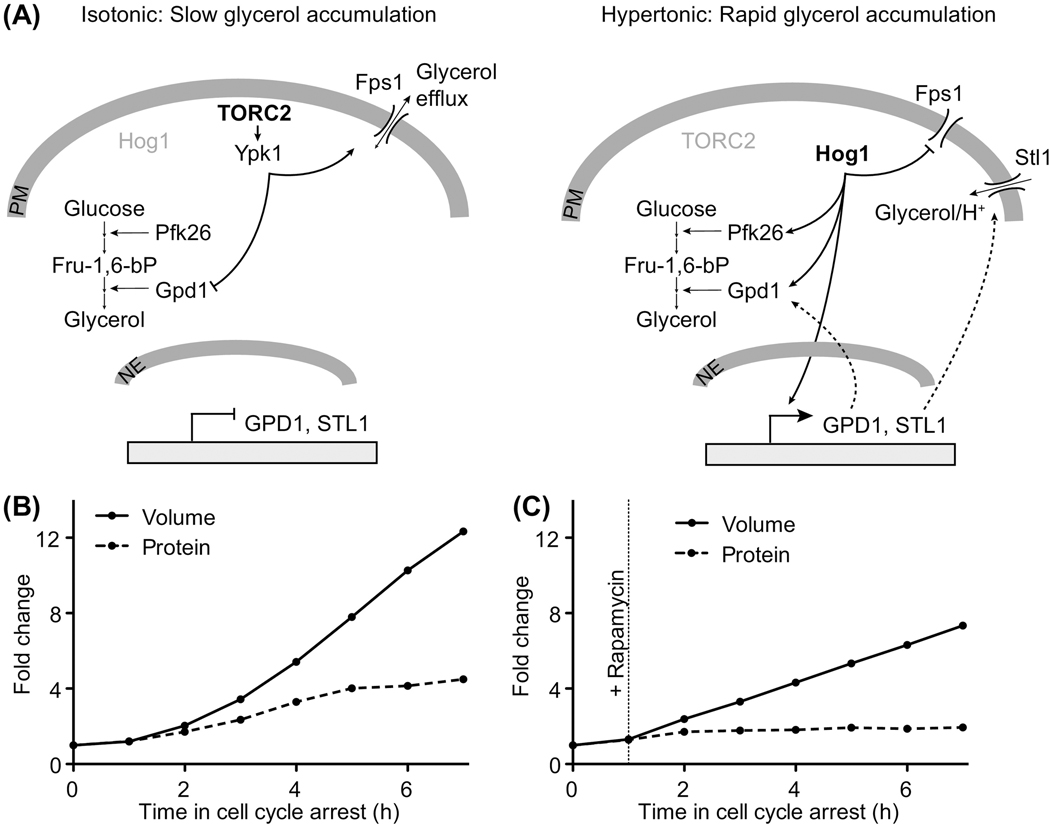
The most important osmolyte that budding yeast cells produce in response to hypertonic stress is glycerol (reviewed in [98]). Two mechanisms have been shown to contribute to glycerol accumulation after hypertonic stress. A MAP kinase pathway known as the high osmolarity glycerol response (Hog1) induces expression of genes required for glycerol synthesis, including the rate limiting enzyme Gpd1 and the glycerol import channel Stl1 (Figure 3A) [98,99]. Activation of this signaling pathway also inhibits the aquaglyceroporin Fps1 [100] thereby preventing glycerol release from cells. Concomitant with activation of the high osmolarity MAP kinase pathway, hypertonic stress inactivates the TORC2 complex (target of rapamycin complex 2) [79,101]. Under isotonic conditions, when TORC2 is active, the TORC2 effector protein kinases Ypk1 and Ypk2 phosphorylate Gpd1 thereby reducing its activity [102]. Ypk1 also phosphorylates the aquaglyceroporin Fps1, which keeps Fps1 in an open conformation and allows efflux of glycerol from the cell (Figure 3A) [103]. TORC2 inactivation after hypertonic stress therefore leads to increased Gpd1 activity and reduced glycerol efflux through Fps1 thereby contributing to glycerol accumulation. Glycerol accumulation and the resulting influx of water re-establishes turgor pressure and reduces the intracellular protein concentration to levels that are similar to the pre-stress conditions. Last but not least, hypertonic stress activates a generic stress response, known as the environmental stress response (ESR), which promotes the downregulation of genes required for ribosome biogenesis [104]. Reduced protein synthesis rates during recovery from hypertonic stress further aid in the rapid return to lower intracellular protein concentrations.
Figure 3. Regulation of cell density.
(A) Regulation of glycerol metabolism in budding yeast in isotonic (left) and hypertonic (right) growth conditions. TORC2 reduces glycerol accumulation under isotonic conditions by inactivating the rate limiting enzyme Gpd1 and by maintaining the aquaglyceroporin Fps1 in an open conformation. Under hypotonic conditions, TORC2 is inactivated and the stress activated MAP kinase Hog1 becomes active. Hog1 promotes glycerol accumulation by stimulating glycolytic flux through activation of phosphofructokinase (Pfk26), through direct activation of Gpd1 and by inducing the expression of GPD1 and of the gene encoding the glycerol importer Stl1. PM: plasma membrane. NE: nuclear envelope.
(B) Budding yeast cells arrest in G1 phase upon inactivation of a conditional allele of the gene encoding the cyclin dependent kinase Cdc28 (cdc28–13). Arrested cells continue to synthesize RNA and protein (quantified using a protein specific dye and RNA seq) and grow very large (volume determined by coulter counter measurement). Initially, protein content increases in accordance with cell size, but when cells grow too large, protein synthesis and cell volume increase become uncoupled, leading to a dilution of the cytoplasm. Protein synthesis and volume increase are therefore to some extent regulated independently.
(C) As in (B), but cells were treated with the TORC1 inhibitor rapamycin. Rapamycin treatment prevents further protein accumulation while cell volume continues to increase, but at a reduced rate. This shows that protein accumulation contributes to cell volume increase, but other processes must also contribute to enlargements of cells. Data from (B, C) are reprinted with permission from [31].
In mammalian cells water loss in response to hypertonic stress is rapidly counteracted by the influx of potassium, sodium and chloride ions (reviewed in [105]). Because the resulting high ionic strength interferes with cytoplasm function, these ions are subsequently replaced by non-charged organic molecules such as polyols and neutral amino acids, which are better tolerated than ions at high concentrations [106]. Like in yeast, MAP kinases play a key role in the adaptation to hypertonic stress in mammalian cells. The MAP kinases ERK1/ERK2, and the stress induced MAP kinases p38 and JNK are activated in response to hypertonic stress and mediate both, rapid ion influx and expression of genes required for the synthesis of compatible osmolytes [105].
How cells regulate their density in the absence of osmotic challenge is less clear. A cell type specific constant density requires the coordination of RNA and protein synthesis with cell volume increase. RNA and protein synthesis affect osmolarity indirectly through their negative charge at physiological pH. Accumulation of negatively charged molecules in the cell will cause influx of cations, which promotes water influx and cell volume increase [9]. Reducing protein synthesis rates by blocking the target of rapamycin complex 1 (TORC1), a master regulator of ribosome biogenesis and protein synthesis [107,108], reduces the rate of cell volume increase (Figure 3B, C) [31]. Conversely, high expression of a “useless” protein in E. coli increases cell volume in otherwise small cells [109]. RNA and protein accumulation inside the cell are therefore a driving force for cell volume increase and a determinant of cell density. Recent evidence in budding yeast however suggests that cell volume increase is to some extent regulated independently of macromolecule accumulation: When eukaryotic cells arrest in the cell cycle, macromolecule biosynthesis continues and cells increase in cell size [110]. In budding yeast, protein and nucleic acid synthesis is initially coupled with cell volume increase, but once cells have reached a certain large size, RNA and protein synthesis rates decline because the DNA becomes limiting [31,34]. But – for reasons unknown – cell volume continues to increase (Figure 3B) [31]. As a result, cytoplasm density declines. Similarly, direct inhibition of protein synthesis by blocking TORC1 does not completely block a further increase in cell volume (Figure 3C) [31] and therefore also results in a reduced cytoplasm density [20]. Blocking cell volume increase on the other hand, by either preventing secretion of new cell surface or by treating cells with repeated osmotic shocks does not stop protein production and cells become more dense [10,111–113]. These observations demonstrate that macromolecule synthesis and cell volume increase are only loosely coupled. The fact that two such fundamental processes are regulated independently is surprising, especially considering that changes in cytoplasm density have far-reaching functional consequences. It is tempting to speculate that the independent regulation of cell volume and protein content is required for cells to modulate cytoplasm density as a regulatory mechanism to simultaneously control multiple cellular processes.
Concluding Remarks
Intracellular macromolecule concentration is narrowly defined within a specific cell type. Moreover, changes in cytoplasm density correlate with altered cell function under both, physiological and pathological conditions. These observations together with theoretical considerations argue that a cell-type specific cellular density is important for cell function. Which cellular processes are particularly sensitive to changes in cytoplasm density and why is still poorly understood (see Outstanding Questions). Newly developed methods have enabled us to better detect changes in cell density. These tools will allow the field to gain a deeper understanding of how cells regulate intracellular density during cell division, differentiation, starvation, senescence, apoptosis and disease and will facilitate studies to understand how cell density affects cell function.
Outstanding Questions.
How are protein synthesis and cytoplasm volume coordinated with each other to ensure that intracellular protein and RNA concentration remains constant?
How can protein synthesis and cell volume increase become uncoupled from each other?
Are changes in cytoplasm density beneficial for cellular fitness under certain conditions?
Which cellular functions are affected by cytoplasm density and how does cytoplasm density affect these processes?
How does cytoplasm dilution contribute to loss of cell function during cellular senescence?
Highlights.
Recently developed methods allow precise measurement of cell density and composition.
Cell density is tightly regulated and varies very little within a given cell type.
Changes in cytoplasm density occur during cell division, cell differentiation, apoptosis and cellular senescence and correlate with altered metabolic activity and cell function.
Changing macromolecule concentration in the cytoplasm affects both individual biochemical reactions as well as basic biophysical properties of this compartment.
In eukaryotes, protein synthesis and cell volume increase can become uncoupled from each other and are therefore to some degree regulated independently.
Acknowledgements
We thank Xiaoxue Zhou, Pei-Hsin Hsu and Seungeun Oh for fruitful discussions and critical reading of the manuscript. Work from the Amon lab described here was supported by an NIH grant (HD085866) to A.A., who is an investigator of the Howard Hughes Medical Institute and the Paul F. Glenn Center for Biology of Aging Research at MIT. GEN was funded by SNSF (P2SKP3_148494, P300PA_160996) and EMBO (ALTF 33–2013).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Albe KR et al. (1990) Cellular concentrations of enzymes and their substrates. Journal of Theoretical Biology 143, 163–195 [DOI] [PubMed] [Google Scholar]
- 2.Illmer P. et al. (1999) A practicable and accurate method to differentiate between intra- and extracellular water of microbial cells. FEMS Microbiol. Lett. 178, 135–139 [DOI] [PubMed] [Google Scholar]
- 3.Feijó Delgado F. et al. (2013) Intracellular water exchange for measuring the dry mass, water mass and changes in chemical composition of living cells. PLoS ONE 8, e67590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Savitz D. et al. (1964) Osmotic properties of human red cells. J. Gen. Physiol. 48, 79–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson NG et al. (1966) Separation of subcellular components and viruses by combined rate- and isopycnic-zonal centrifugation. Natl Cancer Inst Monogr 21, 253–283 [PubMed] [Google Scholar]
- 6.Fischer H. et al. (2004) Average protein density is a molecular-weight-dependent function. Protein Sci. 13, 2825–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson SM and Buttress N. (1973) The osmotic insensitivity of sonicated liposomes and the density of phospholipid-cholesterol mixtures. Biochim. Biophys. Acta 307, 20–26 [DOI] [PubMed] [Google Scholar]
- 8.Martin AD et al. (1994) Adipose tissue density, estimated adipose lipid fraction and whole body adiposity in male cadavers. Int. J. Obes. Relat. Metab. Disord. 18, 79–83 [PubMed] [Google Scholar]
- 9.Model MA and Petruccelli JC (2018) Intracellular Macromolecules in Cell Volume Control and Methods of Their Quantification. Curr Top Membr 81, 237–289 [DOI] [PubMed] [Google Scholar]
- 10.Novick P. et al. (1980) Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell 21, 205–215 [DOI] [PubMed] [Google Scholar]
- 11.Hartwell LH (1970) Periodic density fluctuation during the yeast cell cycle and the selection of synchronous cultures. J. Bacteriol. 104, 1280–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makinoshima H. et al. (2002) Fractionation of Escherichia coli cell populations at different stages during growth transition to stationary phase. Mol. Microbiol. 43, 269–279 [DOI] [PubMed] [Google Scholar]
- 13.Okumura N. et al. (2015) Density-gradient centrifugation enables the purification of cultured corneal endothelial cells for cell therapy by eliminating senescent cells. Sci Rep 5, 15005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burg TP et al. (2007) Weighing of biomolecules, single cells and single nanoparticles in fluid. Nature 446, 1066–1069 [DOI] [PubMed] [Google Scholar]
- 15.Grover WH et al. (2011) Measuring single-cell density. Proceedings of the National Academy of Sciences 108, 10992–10996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryan AK et al. (2010) Measurement of mass, density, and volume during the cell cycle of yeast. Proceedings of the National Academy of Sciences 107, 999–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park YK et al. (2018) Quantitative phase imaging in biomedicine. Nature Photonics 12, 578–589 [Google Scholar]
- 18.Kim YS et al. (2018) Combining Three-Dimensional Quantitative Phase Imaging and Fluorescence Microscopy for the Study of Cell Pathophysiology. Yale J Biol Med 91, 267–277 [PMC free article] [PubMed] [Google Scholar]
- 19.Choi W. et al. (2007) Tomographic phase microscopy. Nat Meth 4, 717–719 [DOI] [PubMed] [Google Scholar]
- 20.Delarue M. et al. (2018) mTORC1 Controls Phase Separation and the Biophysical Properties of the Cytoplasm by Tuning Crowding. Cell 174, 338–349.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asano S. et al. (2016) In Situ Cryo-Electron Tomography: A Post-Reductionist Approach to Structural Biology. Journal of Molecular Biology 428, 332–343 [DOI] [PubMed] [Google Scholar]
- 22.Parry BR et al. (2014) The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell 156, 183–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joyner RP et al. (2016) A glucose-starvation response regulates the diffusion of macromolecules. eLife 5, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munder MC et al. (2016) A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. eLife 5, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaiswal JK and Simon SM (2015) Imaging Live Cells Using Quantum Dots. Cold Spring Harb Protoc 2015, 619–625 [DOI] [PubMed] [Google Scholar]
- 26.Jaiswal JK et al. (2004) Use of quantum dots for live cell imaging. Nat Meth 1, 73–78 [DOI] [PubMed] [Google Scholar]
- 27.Elson EL (2011) Fluorescence correlation spectroscopy: Past, present, future. Biophysical Journal 101, 2855–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lippincott-Schwartz J. et al. (2001) Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2, 444–456 [DOI] [PubMed] [Google Scholar]
- 29.Lorén N. et al. (2015) Fluorescence recovery after photobleaching in material and life sciences: putting theory into practice. Q. Rev. Biophys. 48, 323–387 [DOI] [PubMed] [Google Scholar]
- 30.Fonseca GG et al. (2007) Physiology of the yeast Kluyveromyces marxianusduring batch and chemostat cultures with glucose as the sole carbon source. FEMS Yeast Res 7, 422–435 [DOI] [PubMed] [Google Scholar]
- 31.Neurohr GE et al. (2019) Excessive Cell Growth Causes Cytoplasm Dilution And Contributes to Senescence. Cell 176, 1083–1097.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beck AE et al. (2018) Measuring cellular biomass composition for computational biology applications. Processes 6, [Google Scholar]
- 33.Marguerat S. et al. (2012) Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 151, 671–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhurinsky J. et al. (2010) A Coordinated Global Control over Cellular Transcription. Current Biology 20, 2010–2015 [DOI] [PubMed] [Google Scholar]
- 35.Cheng J-X and Xie XS (2015) Vibrational spectroscopic imaging of living systems: An emerging platform for biology and medicine. Science 350, aaa8870–aaa8870 [DOI] [PubMed] [Google Scholar]
- 36.Freudiger CW et al. (2008) Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. 322, 1857–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh S. et al. (2019) In situ measurement of absolute concentrations by Normalized Raman Imaging. bioRxiv 295, 629543 [Google Scholar]
- 38.Fu D. et al. (2013) Hyperspectral imaging with stimulated Raman scattering by chirped femtosecond lasers. J Phys Chem B 117, 4634–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu F-K et al. (2015) Label-free DNA imaging in vivo with stimulated Raman scattering microscopy. Proceedings of the National Academy of Sciences 112, 11624–11629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baldwin WW and Kubitschek HE (1984) Buoyant density variation during the cell cycle of Saccharomyces cerevisiae. J. Bacteriol. 158, 701–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Son S. et al. (2015) Resonant microchannel volume and mass measurements show that suspended cells swell during mitosis. The Journal of Cell Biology 211, 757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zlotek-Zlotkiewicz E. et al. (2015) Optical volume and mass measurements show that mammalian cells swell during mitosis. The Journal of Cell Biology 211, 765–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolff DA and Pertoft H. (1972) Separation of HeLa cells by colloidal silica density gradient centrifugation. I. Separation and partial synchrony of mitotic cells. The Journal of Cell Biology 55, 579–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blank HM et al. (2017) Translational control of lipogenic enzymes in the cell cycle of synchronous, growing yeast cells. The EMBO Journal 36, 487–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hecht VC et al. (2016) Biophysical changes reduce energetic demand in growth factor-deprived lymphocytes. The Journal of Cell Biology 212, 439–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang H. et al. (2010) Quantitative 3D imaging of whole, unstained cells by using X-ray diffraction microscopy. Proceedings of the National Academy of Sciences 107, 11234–11239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chatterjee A. et al. (1982) Isolation of yeast ascospores free of vegetative cell contamination. J. Gen. Microbiol. 128, 2725–2728 [Google Scholar]
- 48.Beaman TC et al. (1982) Bacterial spore heat resistance correlated with water content, wet density, and protoplast/sporoplast volume ratio. J. Bacteriol. 150, 870–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aon JC and Cortassa S. (1997) Fluorescent measurement of the intracellular pH during sporulation of Saccharomyces cerevisiae. FEMS Microbiol. Lett. 153, 17–23 [DOI] [PubMed] [Google Scholar]
- 50.Barton JK et al. (1980) Measurement of the internal pH of yeast spores by 31P nuclear magnetic resonance. Proc. Natl. Acad. Sci. U.S.A. 77, 2470–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beaman TC et al. (1984) Dehydration partitioned within core protoplast accounts for heat resistance of bacterial spores. FEMS Microbiol. Lett. 24, 47–51 [Google Scholar]
- 52.Zipursky A. et al. (1976) Leukocyte density and volume in normal subjects and in patients with acute lymphoblastic leukemia. Blood 48, 361–371 [PubMed] [Google Scholar]
- 53.Cooper KL et al. (2013) Multiple phases of chondrocyte enlargement underlie differences in skeletal proportions. Nature 495, 375–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wyllie AH and Morris RG (1982) Hormone-induced cell death. Purification ad properties of thymocytes undergoing apoptosis after glucocorticoid treatment. The American Journal of Pathology 109, 78–87 [PMC free article] [PubMed] [Google Scholar]
- 55.Maeno E. et al. (2000) Normotonic cell shrinkage because of disordered volume regulation is an early prerequisite to apoptosis. Proc. Natl. Acad. Sci. U.S.A. 97, 9487–9492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ernest NJ et al. (2008) Cytoplasmic condensation is both necessary and sufficient to induce apoptotic cell death. Journal of Cell Science 121, 290–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bortner CD and Cidlowski JA (2011) Life and death of lymphocytes: a volume regulation affair. Cell. Physiol. Biochem. 28, 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang X. et al. (1998) Autoproteolytic activation of pro-caspases by oligomerization. Molecular Cell 1, 319–325 [DOI] [PubMed] [Google Scholar]
- 59.Chang DW et al. (2003) Oligomerization is a general mechanism for the activation of apoptosis initiator and inflammatory procaspases. Journal of Biological Chemistry 278, 16466–16469 [DOI] [PubMed] [Google Scholar]
- 60.Gallagher PG (2017) Disorders of erythrocyte hydration. Blood 130, 2699–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lohka MJ and Maller JL (1985) Induction of nuclear envelope breakdown, chromosome condensation, and spindle formation in cell-free extracts. The Journal of Cell Biology 101, 518–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Veitia RA (2019) DNA Content, Cell Size, and Cell Senescence. Trends in Biochemical Sciences 44, 645–647 [DOI] [PubMed] [Google Scholar]
- 63.Mitchison T. and Kirschner M. (1984) Microtubule assembly nucleated by isolated centrosomes. Nature 312, 232–237 [DOI] [PubMed] [Google Scholar]
- 64.Mitchison T. and Kirschner M. (1984) Dynamic instability of microtubule growth. Nature 312, 237–242 [DOI] [PubMed] [Google Scholar]
- 65.Korn ED et al. (1987) Actin polymerization and ATP hydrolysis. 238, 638–644 [DOI] [PubMed] [Google Scholar]
- 66.Saxton WM et al. (1984) Tubulin dynamics in cultured mammalian cells. The Journal of Cell Biology 99, 2175–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmoller KM et al. (2015) Dilution of the cell cycle inhibitor Whi5 controls budding-yeast cell size. Nature 526, 268–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Topacio BR et al. (2019) Cyclin D-Cdk4,6 Drives Cell-Cycle Progression via the Retinoblastoma Protein’s C-Terminal Helix. Molecular Cell 74, 758–770.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heldt FS et al. (2018) Dilution and titration of cell-cycle regulators may control cell size in budding yeast. PLoS Comput. Biol. 14, e1006548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rivas G. and Minton AP (2016) Macromolecular Crowding In Vitro, In Vivo, and In Between. Trends in Biochemical Sciences 41, 970–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.del Alamo M. et al. (2005) Effect of macromolecular crowding agents on human immunodeficiency virus type 1 capsid protein assembly in vitro. J. Virol. 79, 14271–14281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Munishkina LA et al. (2004) The effect of macromolecular crowding on protein aggregation and amyloid fibril formation. J. Mol. Recognit. 17, 456–464 [DOI] [PubMed] [Google Scholar]
- 73.Woodruff JB et al. (2017) The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell 169, 1066–1077.e10 [DOI] [PubMed] [Google Scholar]
- 74.Boija A. et al. (2018) Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 175, 1842–1855.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cho W-K et al. (2018) Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. 361, 412–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kilic S. et al. (2019) Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. The EMBO Journal 38, e101379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pessina F. et al. (2019) Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nature Cell Biology 21, 1286–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Case LB et al. (2019) Regulation of Transmembrane Signaling by Phase Separation. Annu Rev Biophys 48, 465–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Riggi M. et al. (2018) Decrease in plasma membrane tension triggers PtdIns(4,5)P2 phase separation to inactivate TORC2. Nature Cell Biology 20, 1043–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berchowitz LE et al. (2015) Regulated Formation of an Amyloid-like Translational Repressor Governs Gametogenesis. Cell 163, 406–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fuller RS et al. (1981) Enzymatic replication of the origin of the Escherichia coli chromosome. Proc. Natl. Acad. Sci. U.S.A. 78, 7370–7374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sokolova E. et al. (2013) Enhanced transcription rates in membrane-free protocells formed by coacervation of cell lysate. Proceedings of the National Academy of Sciences 110, 11692–11697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Crowe JH et al. (1992) Anhydrobiosis. Annu. Rev. Physiol. 54, 579–599 [DOI] [PubMed] [Google Scholar]
- 84.Koshland D. and Tapia H. (2019) Desiccation tolerance: an unusual window into stress biology. Molecular Biology of the Cell 30, 737–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tapia H. and Koshland DE (2014) Trehalose is a versatile and long-lived chaperone for desiccation tolerance. Curr. Biol. 24, 2758–2766 [DOI] [PubMed] [Google Scholar]
- 86.Kim SX et al. (2018) Synergy between the small intrinsically disordered protein Hsp12 and trehalose sustain viability after severe desiccation. eLife 7, 959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boothby TC et al. (2017) Tardigrades Use Intrinsically Disordered Proteins to Survive Desiccation. Molecular Cell 65, 975–984.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tapia H. et al. (2015) Increasing intracellular trehalose is sufficient to confer desiccation tolerance to Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences 112, 6122–6127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gray WT et al. (2019) Nucleoid Size Scaling and Intracellular Organization of Translation across Bacteria. Cell 177, 1632–1648.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Miermont A. et al. (2013) Severe osmotic compression triggers a slowdown of intracellular signaling, which can be explained by molecular crowding. Proceedings of the National Academy of Sciences 110, 5725–5730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Babazadeh R. et al. (2013) Osmostress-induced cell volume loss delays yeast Hog1 signaling by limiting diffusion processes and by Hog1-specific effects. PLoS ONE 8, e80901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yu S. et al. (2018) Subdiffusion of loci and cytoplasmic particles are different in compressed Escherichia coli cells. Commun Biol 1, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mika JT et al. (2010) Molecular sieving properties of the cytoplasm of Escherichia coli and consequences of osmotic stress. Mol. Microbiol. 77, 200–207 [DOI] [PubMed] [Google Scholar]
- 94.Zhou EH et al. (2009) Universal behavior of the osmotically compressed cell and its analogy to the colloidal glass transition. Proceedings of the National Academy of Sciences 106, 10632–10637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ablett S. et al. (1999) Glass formation and dormancy in bacterial spores. International Journal of Food Science and Technology 34, 59–69 [Google Scholar]
- 96.Cowan AE et al. (2003) A soluble protein is immobile in dormant spores of Bacillus subtilis but is mobile in germinated spores: implications for spore dormancy. Proc. Natl. Acad. Sci. U.S.A. 100, 4209–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dijksterhuis J. et al. (2007) High Viscosity and Anisotropy Characterize the Cytoplasm of Fungal Dormant Stress-Resistant Spores. Eukaryotic Cell 6, 157–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hohmann S. (2015) An integrated view on a eukaryotic osmoregulation system. Curr. Genet. DOI: 10.1007/s00294-015-0475-0 [DOI] [PubMed] [Google Scholar]
- 99.O’Rourke SM and Herskowitz I. (2004) Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Molecular Biology of the Cell 15, 532–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mollapour M. and Piper PW (2007) Hog1 mitogen-activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Molecular and Cellular Biology 27, 6446–6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Berchtold D. et al. (2012) Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nature Cell Biology 14, 542–547 [DOI] [PubMed] [Google Scholar]
- 102.Lee YJ et al. (2012) Reciprocal Phosphorylation of Yeast Glycerol-3-Phosphate Dehydrogenases in Adaptation to Distinct Types of Stress. Molecular and Cellular Biology 32, 4705–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Muir A. et al. (2015) Down-regulation of TORC2-Ypk1 signaling promotes MAPK-independent survival under hyperosmotic stress. eLife 4, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gasch AP et al. (2000) Genomic expression programs in the response of yeast cells to environmental changes. Molecular Biology of the Cell 11, 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhou X. et al. (2016) Mitogen-activated protein kinases as key players in osmotic stress signaling. Biochim. Biophys. Acta 1860, 2037–2052 [DOI] [PubMed] [Google Scholar]
- 106.Yancey PH et al. (1982) Living with water stress: evolution of osmolyte systems. Science 217, 1214–1222 [DOI] [PubMed] [Google Scholar]
- 107.Saxton RA and Sabatini DM (2017) mTOR Signaling in Growth, Metabolism, and Disease. Cell 168, 960–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.González A. and Hall MN (2017) Nutrient sensing and TOR signaling in yeast and mammals. The EMBO Journal 36, 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Basan M. et al. (2015) Inflating bacterial cells by increased protein synthesis. 11, 836–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Johnston GC et al. (1977) Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Experimental Cell Research 105, 79–98 [DOI] [PubMed] [Google Scholar]
- 111.Henry SA et al. (1977) Growth and metabolism of inositol-starved Saccharomyces cerevisiae. J. Bacteriol. 130, 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Novick P. and Schekman R. (1979) Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 76, 1858–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Knapp BD et al. (2019) Decoupling of Rates of Protein Synthesis from Cell Expansion Leads to Supergrowth. Cell Syst 9, 434–445.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Neidhardt FC et al. (1990) Physiology of the Bacterial Cell, Sinauer Associates Incorporated. [Google Scholar]
- 115.Bratbak G. and Dundas I. (1984) Bacterial dry matter content and biomass estimations. Appl. Environ. Microbiol. 48, 755–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bremer H. and Dennis PP (2008) Modulation of Chemical Composition and Other Parameters of the Cell at Different Exponential Growth Rates. EcoSal Plus 3, [DOI] [PubMed] [Google Scholar]
- 117.Kamihira M. et al. (1987) Sterilization of microorganisms with supercritical carbon dioxide. Agricultural and Biological Chemistry 51, 407–412 [Google Scholar]
- 118.Nissen TL et al. (1997) Flux distributions in anaerobic, glucose-limited continuous cultures of Saccharomyces cerevisiae. Microbiology (Reading, Engl.) 143, 203–218 [DOI] [PubMed] [Google Scholar]
- 119.Gombert AK et al. (2001) Network Identification and Flux Quantification in the Central Metabolism of Saccharomyces cerevisiae under Different Conditions of Glucose Repression. J. Bacteriol. 183, 1441–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Furukawa K. et al. (1983) Influence of oxygen on the growth of Saccharomyces cerevisiae in continuous culture. Biotechnol. Bioeng. 25, 2293–2317 [DOI] [PubMed] [Google Scholar]
- 121.Illmer P. et al. (1999) A practicable and accurate method to differentiate between intra- and extracellular water of microbial cells. FEMS Microbiol. Lett. 178, 135–139 [DOI] [PubMed] [Google Scholar]
- 122.Alberts B. et al. (2002) The Chemical Components of a Cell. In Molecular Biology of the Cell (4 edn) [Google Scholar]
- 123.Fu D. et al. (2012) Quantitative chemical imaging with multiplex stimulated Raman scattering microscopy. J. Am. Chem. Soc. 134, 3623–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]