Abstract
Since Saccharomyces cerevisiae appears to be an emerging pathogen, there is a need for a valuable molecular marker able to distinguish among strains. In this work, we investigated the potential value of microsatellite length polymorphism with a panel of 91 isolates, including 41 clinical isolates, 14 laboratory strains, and 28 strains with industrial relevance. Testing seven polymorphic regions (five trinucleotide repeats and two dinucleotide repeats) in a subgroup of 58 unrelated strains identified a total of 69 alleles (6 to 13 per locus) giving 52 different patterns with a discriminatory power of 99.03%. We found a cluster of clinical isolates sharing their genotype with a bakery strain, suggesting a digestive colonization following ingestion of this strain with diet. With the exception of this cluster of isolates and isolates collected from the same patient or from patients treated with Saccharomyces boulardii, all clinical isolates gave different and unique patterns. The genotypes are stable, and the method is reproducible. The possibility to make the method portable is of great interest for further studies using this technique. This work shows the possibility to readily identify S. boulardii (a strain increasingly isolated from invasive infections) using a unique and specific microsatellite allele.
The hemiascomycetous yeast Saccharomyces cerevisiae has been used for centuries for the production of fermented food such as cider, wine, beer, and bread. More recently, it has also been considered as a nutritional supplement specially used by sportsmen. Saccharomyces boulardii, now considered a strain of S. cerevisiae (17), is prescribed as a biotherapeutic agent for the treatment or the prevention of diarrhea, notably that related to Clostridium difficile or associated with enteral and parenteral nutrition (20).
It is not known if S. cerevisiae is a true commensal of the human intestinal flora or only a transient agent of gastrointestinal colonization, possibly related to its ingestion with food. Yet over the past several years S. cerevisiae has been identified as an occasional human pathogen. Virulence traits have been documented both in vitro (pseudohypha formation, growth at 42°C) and in animal models (18). In women, S. cerevisiae is mainly responsible for vaginitis, clinically comparable to recurrent candidal vaginitis, which can be effectively treated by the extensive use of azole derivatives, against which S. cerevisiae is usually resistant (27, 30). S. cerevisiae may also be responsible for severe disease in immunocompromised patients (1, 7) and is now considered an emerging opportunist pathogen (12). Over the past decade, a growing number of reports on systemic and disseminated infections due to Saccharomyces have been published, and a recent epidemiological study showed that S. cerevisiae was responsible for 3.6% of all fungemias (24). It has been recently demonstrated that some of these infections are related to S. boulardii therapy (13, 24), but the epidemiology of human Saccharomyces infections remains poorly understood.
Thus, there is a need for molecular markers able to distinguish strains. Despite the availability of several molecular methods, data on S. cerevisiae typing are still limited. Moreover, some of these methods, such as karyotyping or mitochondrial DNA polymorphism, are incompletely evaluated because of the low number of strains tested or because of the lack of studies on stability and reproducibility (28). In addition, the discriminatory power of some methods appears insufficient when these methods are tested alone. In a comparative study of several techniques including randomly amplified polymorphic DNA, PCR fingerprinting, and enzymatic restriction of amplified DNA, Baleiras Couto et al. concluded that there was no single PCR-mediated typing technique able to discriminate at the strain level (2). More-laborious pulsed-field gel electrophoresis of NotI-digested DNA appeared to be a valuable typing method since it could distinguish 62 distinct patterns from 76 clinical isolates (30). Restriction fragment length polymorphism (RFLP) generated by EcoRI digestion of total DNA allowed the differentiation of 41 subtypes among 60 isolates (49 clinical and 11 nonclinical) (6). Moreover, the patterns could be divided into two genetic groups that were distinguishable by the virulence of the strains when tested in a murine model (5).
Microsatellites are short (usually less than 10-bp) sequence repeats which have been shown to exhibit a substantial level of polymorphism (∼10−2 to 10−5) in a number of eukaryotic genomes (26). In humans, they have been used in paternity tests, forensic medicine, and population structure studies. They have been successfully applied for typing fungi such as Candida albicans (16) and Aspergillus fumigatus (3). The complete sequence of the S. cerevisiae genome allows the identification of these regions and thus their use for the development of this novel molecular tool for typing. Here, we report the use of microsatellite polymorphism as a new tool for the identification of S. cerevisiae strains. It is shown that the patterns are stable and that the method is discriminant, making it a powerful tool for epidemiological purposes.
MATERIALS AND METHODS
Strains.
Ninety-one strains were tested including 14 laboratory strains among which was the S288c strain, which has been completely sequenced (11), 28 strains with industrial relevance collected in eight countries, i.e., France, Spain, Russia, China, Taiwan, Vietnam, Japan, and the Czech Republic, seven reference strains of unknown origin, and 41 isolates collected from 36 patients (Table 1). These patients were either outpatients or hospitalized in eight different institutions from three European countries (France, Spain, and The Netherlands). Isolates were collected from routine cultures of vaginal discharges, stools, sputum, bronchoalveolar lavages, and blood cultures. Nineteen patients (21 isolates) were receiving S. boulardii therapy when their isolates were collected. Additionally, three isolates of S. boulardii (one reference strain and two isolates collected from commercial preparations) were tested. Strains from closely related species, i.e., Saccharomyces pastorianus (CLIB 176, CLIB 180, CLIB 277, CLIB 1260, and CLIB 1486), Saccharomyces paradoxus (CLIB 97, CLIB 228, CLIB 2980, and CLIB 7400), and Saccharomyces bayanus (CLIB 251 and CLIB 106) were also tested. Strains were stored at −80°C in 10% glycerol until tested.
TABLE 1.
Strains tested for microsatellite polymorphism in this study
| No. | Reference no. | Origin | City and/or Country of origin, hospitalg |
|---|---|---|---|
| S. boulardii isolates | |||
| 1 | UL G84F88I90 | Lichee (reference strain Biocodex) | Vietnam |
| 90 | Lot 1127 | Isolate collected from UltraLevure packet | Paris, France |
| 91 | Lot 98 D 15 | Isolate collected from Pérentérol capsule | Brussels, Belgium |
| Unknown strainsh | |||
| 14 | CLIB 156 | Russia | |
| 17 | CLIB 155 | Russia | |
| 18 | CLIB 158 | Russia | |
| 21 | CLIB 268 | Czech Republic | |
| 22 | CLIB 269 | Czech Republic | |
| 23 | CLIB 273 | Czech Republic | |
| Laboratory strains | |||
| 10 | S288c | ||
| 11 | CLIB 193 (ATCC 42296) | ||
| 12 | ATCC 44067 | ||
| 13 | bACC+ | ||
| 16 | CLIB 154 | ||
| 24 | CLIB 112 (YNN 295) | ||
| 25 | DBY 746 | ||
| 26 | DBY 745 | ||
| 27 | 101-4B | ||
| 28 | p7ax170 | ||
| 29 | D6 | ||
| 30 | D224-1B | ||
| 31 | 777-3A | ||
| 32 | HC9-7 | ||
| Bioindustrial strains | |||
| 2 | CLIB 319 | Bakery | France |
| 15 | CLIB 157 | Wine | Spain |
| 19 | CLIB 219 (CBS 5287) | Grape | Russia |
| 20 | CLIB 227 (ATCC 18824) | Type strain; brewer's top yeast | The Netherlands |
| 33 | C1 | Bakerya | China |
| 34 | C2 | Bakery | China |
| 35 | CLIB 408 | Bakery | Taiwan |
| 36 | CLIB 409 (ATCC 4126) | Sugar cane | Vietnam |
| 37 | CLIB 410 | Sake | Japan |
| 38 | CLIB 411 (CBS 435) | Sake | Japan |
| 39 | CLIB 412 | Sake | Japan |
| 40 | CLIB 413 | Fermented rice | China |
| 41 | CLIB 414 | High-sugar food | Japan |
| 42 | CLIB 415 | Fermented rice | China |
| 43 | CLIB 2011 | Wine | Champagne, France |
| 44 | CLIB 2013 | Wine | Burgundy, France |
| 45 | CLIB 2018 | Wine | Cognac, France |
| 46 | CLIB 2019 | Wine | Cognac, France |
| 47 | CLIB 2020 | Wine | Cognac, France |
| 48 | C2A | Wine | Narbonne, France |
| 49 | CLIB 2022 | Wine | Cognac, France |
| 50 | MtBzn 2 | Wine | Narbonne, France |
| 51 | SRC 120 | Cider | France |
| 52 | SRC 147 | Cider | Britanny, France |
| 53 | SRC 213 | Cider | Britanny, France |
| 54 | TL 299 | Cheeseb | France |
| 55 | TL 230 | Cheeseb | France |
| 57 | FD31 | Wine | France |
| Clinical isolates | |||
| 3 | 3285 | Bronchoalveolar lavage | Paris, France, H3 |
| 4 | MB1 | Vagina | Paris, France, H4 |
| 5 | MB2 | Feces | Paris, France, H4 |
| 6 | MB4 | Feces | Paris, France, H4 |
| 7 | MB5 | Feces | Paris, France, H4 |
| 8 | 98-3312 | Feces | Amiens, France |
| 9 | 98-2601 | Feces | Amiens, France |
| 56 | FD29 | Vagina | France |
| 58 | FD32 | Feces | France |
| 59 | FD33 | Fecesc | France |
| 60 | P11 | Mouth | Spain |
| 61 | P12 | Mouth | Spain |
| 62 | P13 | Mouth | Spain |
| 63 | P14 | Feces | Spain |
| 64 | PB11 | Feces | The Netherlands |
| 65 | PB12 | Feces | The Netherlands |
| 66 | PB13 | Sputum | The Netherlands |
| 67 | PB14 | Sputumd | The Netherlands |
| 68 | PB15 | Feces | The Netherlands |
| Therapy isolatesi | |||
| 69 | 459 | Blood | Paris, France, H1 |
| 70 | 96-150017 | Feces | Paris, France, H2 |
| 71 | 98-125391 | Blood | Paris, France, H3 |
| 72 | 97-139287 | Blood | Paris, France, H2 |
| 73 | 138633 | Fecese | Paris, France, H2 |
| 74 | 97-141076 | Feces | Paris, France, H2 |
| 75 | 96-150865 | Feces | Paris, France, H2 |
| 76 | MB3 | Feces | Paris, France, H4 |
| 77 | 98-125391 | Mouth | Amiens, France |
| 78 | 98-3161 | Feces | Amiens, France |
| 79 | 98-2574 | Feces | Amiens, France |
| 80 | 98-2831 | Feces | Paris, France, H5 |
| 81 | M2402A | Feces | Paris, France, H2 |
| 82 | 97/00129 | Blood | Paris, France, H2 |
| 83 | 97/106043 | Feces | Paris, France, H2 |
| 84 | STA6436 | Blood | Paris, France, H6 |
| 85 | STA6441 | Bloodf | Paris, France, H6 |
| 86 | STA6442 | Catheterf | Paris, France, H6 |
| 87 | STA6452 | Blood | Paris, France, H6 |
| 88 | STA6553 | Blood | Paris, France, H6 |
| 89 | STA6766 | Blood | Paris, France, H6 |
While used in Asia, this strain may well be imported from Europe (Austria).
Collected from the same cheese.
Collected from the same patient as isolate 58.
Collected from the same patient as isolate 66.
Collected from the same patient as isolate 72.
Collected from the same patient as isolate 84.
H1, Boucicault Hospital; H2, Necker-Enfants Malades Hospital; H3, Laënnec Hospital; H4, Broussais Hospital; H5, Cochin Hospital; H6, Saint Antoine Hospital.
Unknown strains, strains of unknown origin or utilization.
Therapy isolates, clinical isolates collected during S. boulardii therapy.
Microsatellite polymorphism. (i) DNA extraction.
DNA was extracted according to the method previously described (15). Briefly, yeast cells were grown overnight at 30°C in 3 ml of YPGlu medium (1% [wt/vol] yeast extract [Difco Laboratories, Detroit, Mich.], 1% [wt/vol] Bacto peptone [Difco Laboratories], 2% [wt/vol] glucose). Yeast cells were pelleted and resuspended in 200 μl of lysis buffer (50 mM Tris-HCl [pH 8], 25 mM EDTA, 1% [vol/vol] β-mercaptoethanol, 100,000 U of Zymolyase [ICN Pharmaceuticals, Costa Mesa, Calif.]). After 1 h at 37°C, 200 μl of a solution containing 200 mM diethanolamine, pH 9 (Sigma), 80 mM EDTA, pH 9, and 1% (wt/vol) sodium dodecyl sulfate was added and the mixture was incubated at 65°C for 30 min followed by 5 min on ice. A volume of 100 μl of potassium acetate, 5 M, was added at 4°C for 45 min. After centrifugation for 5 min at 10,000 × g, the supernatant was precipitated by adding ammonium acetate at a final concentration of 2.5 M plus 3 volumes of 100% ethanol. DNA was pelleted by centrifugation for 10 min at 10,000 × g, rinsed with 70% ethanol, dried, and resuspended in 50 μl of Tris-EDTA buffer.
(ii) Microsatellite amplification.
Seven microsatellites (five trinucleotide and two dinucleotide simple repeats) were selected (Table 2). Oligonucleotide primers were designed from the sequences of the flanking regions to obtain PCR products ranging in size between 110 and 170 bp. Amplification was performed with a mixture containing [α-32P]dATP (500 μCi), 500 mM KCl, 100 mM Tris-HCl (pH 8.3), 15 mM MgCl2, 0.1% gelatin, deoxyribonucleotide triphosphate (0.2 mM), forward and reverse primers (20 pmol each), Taq polymerase (1.5 U; Eurogentec, Seraing, Belgium), and 0.5 to 1 μg of DNA. Amplification was achieved through 30 cycles (94°C for 15 s, 55°C for 1 min, 72°C for 1 min 30 s) and a final extension step at 72°C for 10 min.
TABLE 2.
Characteristics of the seven polymorphic microsatellite loci tested in this study
| Locus no. | Gene | Function | Repeata
|
Sequencec of:
|
Fragment size expected (bp) | ||
|---|---|---|---|---|---|---|---|
| Sequence | No. | Upper primer | Lower primer | ||||
| Trinucleotide repeats in ORFd | |||||||
| 1 | YKL172w | Similar to human nucleolar protein P40 | GAA | 10 | CAGGACGCTACCGAAGCTCAAAAG | ATCTTGAGAAATTGGCCAAAAGT | 126 |
| 2 | YKR072c | Cell cycle-specific gene expression | GAC | 13 | AGATACAGAAGATAAGAACGAAAA | GGTATAATAGATAAGCATCAATAA | 154 |
| 3 | YKL139w | Kinase, carboxy-terminal domain, alpha subunit | AAT | 8 | AAGCGTCCTAACATACTATCCACC | TAAGGATATATAGCCAATTGAAAT | 117 |
| 4 | YLR177w | Similar to suppressor protein PSP5 | CAG | 10 | CTTAAACAACAGCTCCCAAA | ATTTCTGATGCGCTGATTCAT | 133 |
| 5 | YDR289c | Protein of unknown function | ATC | 11 | AATTGCTGTCATTGGATCTAT | CACTTCATACGTAGGAATAAT | 121 |
| Dinucleotide repeats | |||||||
| 6 | YMR057c | Unknown | AT | 15 | ATGCACTCAAACAGTCGATCCTT | TTGTTCTATAAGCGATGGATAGG | 142 |
| 7 | ylrb | TC | 31 | CTGGAATGAAATTAAACAAAAGC | GAGAAGATAGTAGAAAAGGAAGA | 170 | |
Data valid for strain S288c (11).
Intergene between YLR346c and YLR347c
From 5′ end.
ORFs, open reading frames.
(iii) Length polymorphism analysis.
Radiolabeled PCR fragments were electrophoresed in 7.5 M urea–12.5% (1:30) polyacrylamide vertical gels at 90 mA for 3 h. Length polymorphisms were evaluated by comparison with the migration of the S288c reference strain tested in triplicate in each run.
Evaluation of the method.
To test genotype stability, DNA from six independent subclones of S288c strain were extracted and tested simultaneously for the seven loci. In addition, an S288c strain was grown in a fermentor for 35 weeks (about 690 generations). Twice a week, aliquots were collected and DNA was extracted and tested by PCR for three loci (loci 1 to 3). Genotypes of strains 25 to 32 were determined and compared to results of the same analysis performed 3 years ago. The reproducibility was assessed by testing the S288c strain 12 times in four separate experiments for each microsatellite. Also, S. boulardii strain UL G84F88I90 was included in each of these experiments.
Excluding pairs of strains collected from the same patient, strains related to S. boulardii, i.e., collected from patients taking S. boulardii therapy and from commercial preparations, and the laboratory strains which derived from a small panel of ancestral strains (22), we consider that 58 strains are epidemiologically unrelated. Discriminatory power was calculated for this group using the Simpson index of diversity
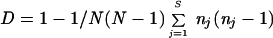 |
where N is the number of strains tested, S is the number of different types, and nj is the number of strains exhibiting the j type (14).
Phylogenetic analyses.
We considered that the number of repeats at each locus was equiprobable for each isolate. Thus, all alleles were considered as phylogenetic markers with two character states, i.e., present (1) and absent (0). The matrix was analyzed using the maximum-parsimony approach (unweighted heuristic search; 100 random stepwise addition sequences) with PAUP 4 software (29). A strict consensus tree was drawn using the midpoint rooting option. Statistical robustness of nodes was evaluated using bootstrap resamplings (100 iterations). Because of computation time the “fast stepwise addition” option was used for each replicate. The consensus tree is given, with branch lengths obtained using the Minf optimization.
Statistical analysis.
To test the hypothesis of a nonrandom distribution of the allelic sizes, we calculated for each locus the chi-square value by comparison of the observed distributions to theoretically normal distributions characterized by the same geometric mean and standard deviation. A P value below 0.05 was considered significant.
RESULTS
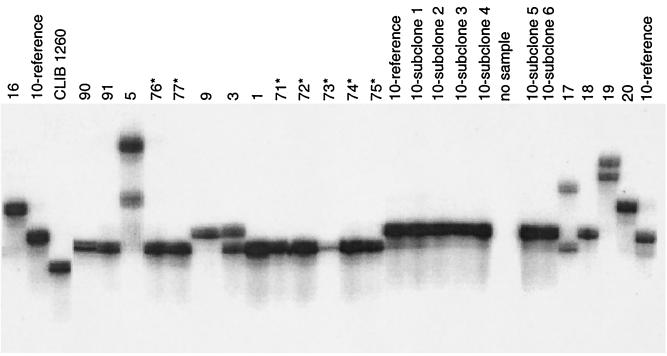
All the S. cerevisiae strains, including S. boulardii, gave 100% positive PCR results with all microsatellites tested. In contrast, typeability, defined as the proportion of strains that are assigned to a type, was 0 for S. bayanus and ranged between 0.25 and 0.5 for S. paradoxus and between 0 and 1 for S. pastorianus, depending on the locus tested. Each locus appeared polymorphic with 12, 13, 6, 11, 8, 6, and 13 alleles for microsatellites 1 to 7, respectively (Table 3). For each locus considered and each strain, the size of the microsatellite on gel was estimated. Figure 1 gives an example of the stability of the patterns and the discriminatory power of the method. In some of the strains analyzed, we found two bands at a given locus. We concluded that these strains were diploid and heterozygous for the allele considered (Table 3). In contrast, when an isolate exhibited only one band for all loci, no conclusion on the ploidy of the isolate could be drawn. For different loci, allele sizes were distributed somewhat differently (data not shown). In some cases, one allele was predominant and smaller or larger alleles were found in the analyzed population, demonstrating a progressive continuum for increasing or decreasing numbers of repeats (microsatellites 2, 5, 6, and 7). In other cases, no predominant allele was observed but the sizes were distributed among six or more categories (microsatellites 1, 3, and 4). However, none of the loci studied demonstrated what could be considered a normal (Gaussian) distribution (P < 0.05) (data not shown).
TABLE 3.
Repeat numbers for strains and microsatellites tested in this work
| No. | Reference no. | No. of repeatsa at microsatellite locus (from Table 2):
|
||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
| S. boulardii isolates | ||||||||
| 1 | UL G84F88I90 | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| 90 | UltraLevure | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| 91 | Perenterol | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| Unknown strainsc | ||||||||
| 14 | CLIB 156 | 9 | 10 | 16 | 8 | 12 | 9 | 17 |
| 17 | CLIB 155 | 9/13 | 10/11 | 14/16 | 8 | 10 | 9/10 | 17 |
| 18 | CLIB 158 | 10 | 10 | 8 | 11 | 11 | 14 | 31 |
| 21 | CLIB 268 | 10 | 10 | 13 | 6 | 10 | 12 | 17 |
| 22 | CLIB 269 | 10 | 10 | 8 | 10 | 10 | 14 | 31 |
| 23 | CLIB 273 | 15 | 21 | 13/16 | 7 | 10 | 9 | 17/26 |
| Laboratory strains | ||||||||
| 10 | S288c | 10 | 13 | 8 | 10 | 11 | 14 | 31 |
| 11 | CLIB 193 | 10 | 13 | 8 | 10 | 14 | 14 | 31 |
| 12 | ATCC 44067 | 10 | 10 | 9 | 10 | 14 | 14 | 17 |
| 13 | bACC+ | 12/16 | 10 | 13/14 | 2 | 7 | 10 | 21/26 |
| 16 | CLIB 154 | 12 | 10 | 13 | 7 | 10 | 8 | 24 |
| 24 | CLIB 295 | 10 | 13 | 8 | 10 | 11 | 14 | 31 |
| 25 | DBY 746 | 9 | 13 | 16 | 10 | 11 | 14 | 31 |
| 26 | DBY 745 | 10 | 10 | 8 | 10 | 11 | 9 | 26 |
| 27 | 101-4B | 10 | 10 | 16 | 10 | 11 | 14 | 17 |
| 28 | p7ax170 | 10 | 10 | 8 | 10 | 11 | 14 | 17 |
| 29 | D6 | 10 | 9 | 8 | 7 | 10 | 9 | 31 |
| 30 | D224-1B | 6 | 10 | 8 | 10 | 11 | 9 | 17 |
| 31 | 777-3A | 6 | 10 | 8 | 10 | 11 | 9 | 17 |
| 32 | HC9-7 | 6/10 | 10 | 8 | 10 | 11 | 9 | 17 |
| Bioindustrial strains | ||||||||
| 2 | CLIB 319 | 12/16 | 9/10 | NDb | 2 | 10/12 | 8/9 | 17 |
| 15 | CLIB 157 | 9 | 10 | 16 | 8 | 13 | 9/12 | 17 |
| 19 | CLIB 219 | 14/15 | 9 | 13 | 7 | 11 | 12 | 26 |
| 20 | CLIB 227 | 12 | 5/10 | 13 | 6 | 10/11 | 8 | 24 |
| 33 | C1 | ND | 10/14 | 13 | 8 | ND | 9 | 17/26 |
| 34 | C2 | ND | 11 | 13 | 6/13 | 9 | 8 | 28 |
| 35 | CLIB 408 | 9 | 10 | 13 | 2 | 12 | 9/11 | 24 |
| 36 | CLIB 409 | 11 | 11 | 14 | 5 | 9 | 8 | 23 |
| 37 | CLIB 410 | 11 | 11 | 13 | 7 | 9 | 8 | 19 |
| 38 | CLIB 411 | 11 | 13 | 13/15 | 7 | 9 | ND | ND |
| 39 | CLIB 412 | 11 | 11 | 13 | 7 | 9 | ND | ND |
| 40 | CLIB 413 | 9/11 | 9 | 13 | 6/13 | 9 | 9 | ND |
| 41 | CLIB 414 | 13 | 9 | 13 | 7 | 9 | ND | 31 |
| 42 | CLIB 415 | 9 | 11 | 13 | 13 | 9 | 8 | 24 |
| 43 | CLIB 2011 | ND | 10/13 | 16 | 8 | ND | 9/10 | 17 |
| 44 | CLIB 2013 | 9 | 10 | 16 | 8 | 10 | 9 | 17 |
| 45 | CLIB 2018 | 9 | 10 | 16 | 8 | 10 | 9 | 17 |
| 46 | CLIB 2019 | 9 | 10 | 16 | 7 | 10 | 9 | 17 |
| 47 | CLIB 2020 | 9 | 10 | 16 | 8 | 10 | 9 | 17 |
| 48 | C2A | 9 | 10 | 16 | 7 | 10 | 9 | 17 |
| 49 | CLIB 2022 | 9 | 12 | 16 | 7 | 10 | 9 | 17 |
| 50 | MtBzn 2 | 9 | 10 | 16 | 7 | 10 | 9 | 17 |
| 51 | SRC 120 | 9 | 11 | 14 | 8 | 10 | 9 | 17 |
| 52 | SRC 147 | 9 | 12 | 16 | 7 | 10 | 9 | 17 |
| 53 | SRC 213 | 9 | 12 | 16 | 7 | 10 | 9 | 17 |
| 54 | TL 229 | ND | 8 | 13/14 | 6 | 14/16 | 9/11 | 17 |
| 55 | TL 230 | ND | 8 | 13/14 | 6 | 14/16 | 9/11 | 17 |
| 57 | FD31 | 8 | 13/14 | 16 | 6 | 10 | 10 | 17 |
| Clinical isolates | ||||||||
| 3 | 3285 | 9/10 | 10/13 | 8/13 | 8 | 10 | 8 | 17 |
| 4 | MB1 | 18 | 22 | 13 | 13 | 9 | 8 | 28 |
| 5 | MB2 | 12/16 | 9/10 | ND | 2 | 10/13 | 8/9 | 21/26 |
| 6 | MB4 | 8/9 | 8/13 | 14/16 | 5/7 | ND | 9 | 17 |
| 7 | MB5 | 12/16 | 9/10 | ND | 2 | 10/13 | 8/9 | 21/26 |
| 8 | 98-3312 | 12/16 | 9/10 | ND | 2 | 10/13 | 8/9 | 21/26 |
| 9 | 98-2601 | 10 | 10 | 13/13 | 10/13 | 10 | 8 | 26/30 |
| 56 | FD29 | 12/16 | 9/10 | 13/16 | 2 | 10/13 | 9/9 | 21/26 |
| 58 | FD32 | 10 | 10/13 | 14 | 8 | 10 | 9 | 18 |
| 59 | FD33 | 10 | 10/13 | 14 | 8 | 10 | 9 | 18 |
| 60 | P11 | 12 | 17 | 14 | 10/11 | 9 | 8/12 | 27 |
| 61 | P12 | 10 | 21 | 14 | 12 | 9 | 9 | 29 |
| 62 | P13 | 9/14 | 18 | 14 | 8 | 10/12 | 9 | 28 |
| 63 | P14 | 13 | 20 | 15 | 8 | 10 | 9/12 | 24 |
| 64 | PB11 | 10/11 | 18 | 15 | 14 | 10 | 8/9 | 22 |
| 65 | PB12 | 10 | 17 | 14 | 10/11 | 9 | 8/9 | 31 |
| 66 | PB13 | 7/14 | 18 | 14/16 | 12 | 10/13 | 9/12 | 24 |
| 67 | PB14 | 7/14 | 18 | 14/16 | 12 | 10/13 | 9/12 | 24 |
| 68 | PB15 | 10/11 | ND | 14/15 | 10/11 | 10 | 8/9 | 22 |
| Therapy isolatesd | ||||||||
| 69 | 459 | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| 70 | 96-150017 | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| 71 | 98-125391 | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| 72 | 97-139287 | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| 73 | 138633 | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| 74 | 97-141076 | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| 75 | 96-150865 | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| 76 | MB3 | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| 77 | 98-125391 | 9 | 10 | 16 | 9 | 10 | ND | 17 |
| 78 | 98-3161 | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| 79 | 98-2574 | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| 80 | 98-2831 | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| 81 | M2402A | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| 82 | 97/00129 | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| 83 | 97/106043 | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| 84 | ST6436 | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| 85 | STA6441 | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| 86 | STA6442 | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| 87 | STA6452 | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| 88 | STA6553 | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
| 89 | ST6766 | 9 | 10 | 16 | 9 | 10 | 9 | 17 |
Repeat numbers were calculated by comparison to the length (in nucleotides) of the PCR fragments using the S288c sequence as a reference. Diploid strains were heterozygous for some loci, displaying two different alleles for the locus considered.
ND, not determined.
Unknown strains, strains of unknown origin or utilization.
Therapy isolates, clinical isolates collected during S. boulardii therapy.
FIG. 1.
Denaturing polyacrylamide gel electrophoresis of products of PCR amplification of the YKL172w microsatellite (microsatellite 1). S. cerevisiae strains are described in Table 1. Note that CLIB 1260 (lane 3) corresponds to an S. pastorianus strain. *, clinical isolates that were collected from patients taking S. boulardii therapy.
A total of 69 different alleles were observed, generating 52 distinct patterns. Pattern identity was observed for French strains used in the agroalimentary industry: strains 54 and 55; strains 44, 45, and 47; strains 46, 48, and 50; and strains 49, 52, and 53. Similarly, among laboratory strains, strains 30 and 31 and 10 and 11 were indistinguishable. The last two were indistinguishable from an apparently non-epidemiologically related strain (strain 24).
The patterns of the 21 isolates collected from the patients receiving S. boulardii therapy were indistinguishable. These patterns were also indistinguishable from those of the two isolates of S. boulardii collected from commercial preparations and the reference strain from the manufacturer. S. boulardii-related isolates exhibited an allele for locus 4 that was not shared with any other strain. Among the 19 isolates collected from 17 patients not taking S. boulardii therapy, we found 14 unique patterns. Four French clinical isolates gave a pattern indistinguishable from that of a French bakery strain (strain 2). There were two pairs of clinical isolates that could not be differentiated. In both cases, they were isolated from the same patient (isolates 66 and 67 and 58 and 59). None of the patients not treated with S. boulardii had an isolate with a pattern characteristic of S. boulardii.
Performance of the method.
Genomic stability of assayed microsatellites was confirmed by analysis of six independent subclones of the S288c strain. In addition, no change of genotype was observed over the 690 generations collected during 35 weeks of growth in a fermentor (see Materials and Methods). Moreover, stability was also confirmed by comparison between experiments performed 3 years ago and the present study. Independent PCRs run using the same DNA (strain S288c and UL G84F88I90) and the same primers gave the same pattern, demonstrating the total reproducibility of the technique. The discriminatory power calculated for a sample of 58 strains considered unrelated was 99.03%.
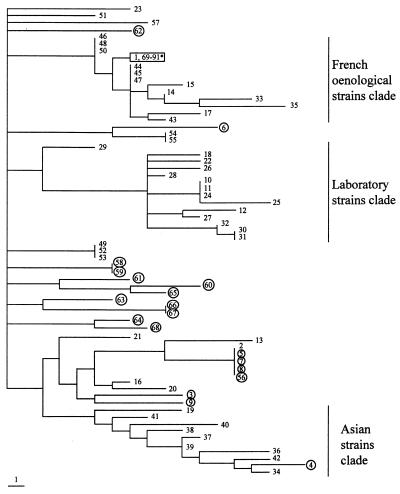
Phylogenetic analysis.
The phylogenetic analysis provided 1,479 equiparsimonious trees of 208 steps, with a consistency index of 0.332 and a retention index of 0.7 (Fig. 2). Four clades exhibited bootstrap proportions over 70%: the clades comprising strains 2, 5, 7, 8, and 56 (95%); strains 58 and 59 (85%); strains 66 and 67 (99%); and strains 54 and 55 (95%). Also, it was noted that 12 of the 14 laboratory strains form a clade. Similarly, 8 (strains 34, 36, 37, 38, 39, 40, 41, and 42) out of 10 Asian isolates are members of the same clade. Finally, 7 (strains 43 to 48 and 50) out of 11 French wine strains showed a strong identity. However, it was not possible to distinguish a clade comprising clinical isolates. Also, there was no difference between strains isolated in true pathogenic conditions (isolated from blood and cases of vaginitis) and those isolated as probably commensal organisms (feces).
FIG. 2.
Strict consensus of the 1,479 most-parsimonious phylogenetic trees. The phylogram is constructed using the midpoint rooting method. Tree length, 208; consistency index, 0.332; retention index, 0.7. Strain numbering refers to Table 1. Clinical isolates from patients not taking S. boulardii are circled. ∗ (boxed), S. boulardii reference strain (1), isolates from S. boulardii commercial preparations (90 and 91), and isolates from patients treated with S. boulardii (69 to 89).
DISCUSSION
The increasing importance of fungi among nosocomial infections creates a need for the development of molecular markers to investigate the epidemiology of such infections. To be useful, these methods have to be carefully evaluated to ascertain their performance. Using the guidelines proposed by the European Study Group on Epidemiological Markers (28), we find that the application of microsatellite polymorphism to S. cerevisiae is a powerful method. The development of such molecular markers is important for a better understanding of Saccharomyces infections. For example, in case of relapse, the method could be used to trace isolates, to test whether the infection is due to the persistence of the microorganism or reinfection with a new strain. Also, since the risk of transmission of S. cerevisiae from patient to patient has been suggested (30), it is important to identify isolates in order to demonstrate the origin of the infection and further apply appropriate prevention measures. This is particularly true for S. boulardii, which is increasingly reported as responsible for bloodstream infection and for which contamination of catheters related to hand carriage has been hypothesized (13). In the present work, reproducibility of the method and stability of the genomic regions were confirmed. To our knowledge, the level of discrimination between strains obtained with this method has never been previously reached with other methods applied to S. cerevisiae typing, confirming the high degree of polymorphism of these regions among strains of S. cerevisiae. All clinical strains collected from patients not treated with S. boulardii exhibited a distinct pattern, with the exception of isolates collected from the same patients and a cluster of four isolates collected from French patients that shared their pattern with that of a commercial baker yeast. In this case it may be postulated that the colonization of the patients results from the use of this strain in the diet, as has already been shown in another study (23).
Another important advantage of the method is its portability. Since all results can be expressed as a number of repeats, computer translation is easy, offering the opportunity to compare results from different groups via the Internet. This is an important issue for further comparisons of larger sets of strains isolated in very different settings, e.g., human medicine, environmental studies, and wine production. A similar approach involving the multilocus sequence typing method applied to bacteria has been undertaken (8, 9). Also it should be underlined that the technique can be used with a nonradioactive method, as has already been done for A. fumigatus (3).
The method also appears to be specific to the S. cerevisiae species since isolates belonging to the closely related species S. pastorianus, S. paradoxus, and S. bayanus failed to be reproducibly amplified. This must be compared to results obtained for C. albicans (10), where Candida krusei and Candida parapsilosis failed to be amplified. This could be due to the frequent mutations observed in the DNA sequences flanking repeats, leading to negative PCR results (25). The positive PCR results obtained with S. boulardii isolates strongly suggest that S. boulardii is a strain of S. cerevisiae rather than a different species. Until now, it has not been possible to identify S. boulardii in a routine manner. Phenotypic traits such as the lack of galactose assimilation and the lack of alpha-glucosidase activity are characteristics of S. boulardii (19). However none of these traits allows a definitive identification, and confirmation requires multiple enzymatic restrictions of mitochondrial DNA (M. Maillé, P. V. Nguyen, S. Bertout, and J. Bastide, Program abstr. Int. Soc. Hum. Anim. Mycol., abstr. P511, 2000). Interestingly, even the highly discriminatory randomly amplified polymorphis DNA method failed to differentiate between isolates of S. cerevisiae and S. boulardii (21). In contrast, we found that sequence (CAG)9 at locus 4 is specific for this strain and thus constitutes a rapid alternative for an accurate identification of S. boulardii. These data allow us to confirm that Saccharomyces fungemias diagnosed in our patients treated with S. boulardii therapy (eight cases) are indeed due to this particular strain. The lack of polymorphism among S. boulardii isolates is in accordance with results obtained with EcoRI-generated RFLP, while virulence studies with an animal model demonstrated significant differences among isolates (17).
Considering the small number of clinical isolates different from S. boulardii, it was not possible to test the hypothesis of a nonrandom distribution depending on the site of isolation. The fact that clinical isolates did not cluster argues against the hypothesis of common virulent traits characteristic for these isolates. This may suggest that any thermotolerant (viability at 37°C) Saccharomyces strain is able to transiently colonize the human gastrointestinal tract. However, this idea conflicts with previous reports demonstrating that virulence as tested in a murine model is more frequent in clinical isolates and is associated with particular patterns generated with an RFLP typing method (5, 6). The gathering of laboratory strains supports the opinion of Mortimer, i.e., that yeast molecular biology studies were mostly carried out on a very limited panel of strains, possibly derived from Lindegren's original strain (22). This factor must be taken into consideration in further medical studies.
The close relationship between most French wine strains may be due either to the use of identical starter strains to initiate the fermentation process or to the clonal reproduction of natural strains originating from the same geographic area. The latter possibility is supported by the clustering of Asian isolates. Similarly, strains of the yeast Cryptococcus neoformans var. gatti, which have an environmental ecological niche, show geographical clustering, suggesting clonal reproduction (4).
In conclusion, the analysis of microsatellite polymorphism is a reliable method for S. cerevisiae strain identification, including S. boulardii, which is an increasing cause of fungemia in hospitalized patients. The phylogenetic data obtained with this method will be useful in further studies in the field of population genetics.
ACKNOWLEDGMENTS
We are indebted to F. Dromer, V. Lavarde, J. Meiss, A. Paugam, J. L. Poirot, J. Ponton, and the Bureau National Interprofessional du Cognac for providing isolates and R. Longin for his help in the continuous fermentor cultures. T. Ancelle is thanked for advice in statistical analysis. We thank C. Raccurt for support and interest. We thank the members of the Unité de Génétique Moléculaire des Levures for their fruitful comments.
REFERENCES
- 1.Aucott J N, Fayen J, Grossnicklas H, Morrissey A, Lederman M M, Salata R A. Invasive infection with Saccharomyces cerevisiae: report of three cases and review. Rev Infect Dis. 1990;12:406–411. doi: 10.1093/clinids/12.3.406. [DOI] [PubMed] [Google Scholar]
- 2.Baleiras Couto M M, Eijsma B, Hofstra H, Huis in't Veld J H, van der Vossen J M. Evaluation of molecular typing techniques to assign genetic diversity among Saccharomyces cerevisiae strains. Appl Environ Microbiol. 1996;62:41–46. doi: 10.1128/aem.62.1.41-46.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bart-Delabesse E, Humbert J F, Delabesse E, Bretagne S. Microsatellite markers for typing Aspergillus fumigatus isolates. J Clin Microbiol. 1998;36:2413–2418. doi: 10.1128/jcm.36.9.2413-2418.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boekhout T, van Belkum A, Leenders A C, Verbrugh H A, Mukamurangwa P, Swinne D, Scheffers W A. Molecular typing of Cryptococcus neoformans: taxonomic and epidemiological aspects. Int J Syst Bacteriol. 1997;47:432–442. doi: 10.1099/00207713-47-2-432. [DOI] [PubMed] [Google Scholar]
- 5.Clemons K V, McCusker J H, Davis R W, Stevens D A. Comparative pathogenesis of clinical and nonclinical isolates of Saccharomyces cerevisiae. J Infect Dis. 1994;169:859–867. doi: 10.1093/infdis/169.4.859. [DOI] [PubMed] [Google Scholar]
- 6.Clemons K V, Park P, McCusker J H, McCullough M J, Davis R W, Stevens D A. Application of DNA typing methods and genetic analysis to epidemiology and taxonomy of Saccharomyces isolates. J Clin Microbiol. 1997;35:1822–1828. doi: 10.1128/jcm.35.7.1822-1828.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eng R H, Drehmel R, Smith S M, Goldstein E J. Saccharomyces cerevisiae infections in man. Sabouraudia. 1984;22:403–407. [PubMed] [Google Scholar]
- 8.Enright M C, Day N P, Davies C E, Peacock S J, Spratt B G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright M C, Spratt B G. Multilocus sequence typing. Trends Microbiol. 1999;7:482–487. doi: 10.1016/s0966-842x(99)01609-1. [DOI] [PubMed] [Google Scholar]
- 10.Field D, Eggert L, Metzgar D, Rose R, Wills C. Use of polymorphic short and clustered coding-region microsatellites to distinguish strains of Candida albicans. FEMS Immunol Med Microbiol. 1996;15:73–79. doi: 10.1111/j.1574-695X.1996.tb00056.x. [DOI] [PubMed] [Google Scholar]
- 11.Goffeau A, Aert R, Agostini-Carbone M L, Ahmed A, Aigle M, Alberghina L, Albermann K, Albers M, Aldea M, Alexandraki D, Aljinovic G, Allen E, Alt-Morbe J, Andre B, Andrews S, Ansorge W, Antoine G, Anwar R, Aparicio A, Araujo R, Arino J, Arnold F, Arroyo J, Aviles E, Backes U. The yeast genome directory. Nature. 1997;387(Suppl.):1–105. [Google Scholar]
- 12.Hazen K. New and emerging yeast pathogens. Clin Microbiol Rev. 1995;8:462–478. doi: 10.1128/cmr.8.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hennequin C, Kauffmann-Lacroix C, Jobert A, Viard J P, Ricour C, Jacquemin J L, Berche P. Saccharomyces boulardii fungemia: the possible role of catheter. Eur J Clin Microbiol Infect Dis. 2000;19:16–20. doi: 10.1007/s100960050003. [DOI] [PubMed] [Google Scholar]
- 14.Hunter P. Reproducibility and indices of discriminatory power of microbial typing method. J Clin Microbiol. 1990;28:1903–1905. doi: 10.1128/jcm.28.9.1903-1905.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnston J R, Contopoulou C R, Mortimer R K. Karyotyping of yeast strains of several genera by field inversion gel electrophoresis. Yeast. 1988;4:191–198. doi: 10.1002/yea.320040304. [DOI] [PubMed] [Google Scholar]
- 16.Lunel F V, Licciardello L, Stefani S, Verbrugh H A, Melchers W J, Meis J F, Scherer S, van Belkum A. Lack of consistent short sequence repeat polymorphisms in genetically homologous colonizing and invasive Candida albicans strains. J Bacteriol. 1998;180:3771–3778. doi: 10.1128/jb.180.15.3771-3778.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCullough M J, Clemons K V, McCusker J H, Stevens D A. Species identification and virulence attributes of Saccharomyces boulardii (nom. inval.) J Clin Microbiol. 1998;36:2613–2617. doi: 10.1128/jcm.36.9.2613-2617.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCusker J H, Clemons K V, Stevens D A, Davis R W. Saccharomyces cerevisiae virulence phenotype as determined with CD-1 mice is associated with the ability to grow at 42°C and form pseudohyphae. Infect Immun. 1994;62:5447–5455. doi: 10.1128/iai.62.12.5447-5455.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McFarland L V. Saccharomyces boulardii is not Saccharomyces cerevisiae. Clin Infect Dis. 1996;22:200–201. doi: 10.1093/clinids/22.1.200. [DOI] [PubMed] [Google Scholar]
- 20.McFarland L V, Bernasconi P. Saccharomyces boulardii: a review of an innovative biotherapeutic agent. Microb Ecol Health Dis. 1993;343:171–172. [Google Scholar]
- 21.Molnar O, Messner R, Prillinger H, Stahl U, Slavikova E. Genotyping identification of Saccharomyces species using random amplified polymorphic DNA analysis. Syst Appl Microbiol. 1995;18:136–145. [Google Scholar]
- 22.Mortimer R. Evolution and variation of the yeast (Saccharomyces) genome. Genome Res. 2000;10:403–409. doi: 10.1101/gr.10.4.403. [DOI] [PubMed] [Google Scholar]
- 23.Nyirjesy P, Vazquez J A, Ufberg D D, Sobel J D, Boikov D A, Buckley H R. Saccharomyces cerevisiae vaginitis: transmission from yeast used in baking. Obstet Gynecol. 1995;86:326–329. doi: 10.1016/0029-7844(95)00174-P. [DOI] [PubMed] [Google Scholar]
- 24.Piarroux R, Millon L, Bardonnet K, Vagner O, Koenig H. Are live Saccharomyces yeasts harmful to patients? Lancet. 1999;353:1851–1852. doi: 10.1016/S0140-6736(99)02001-2. [DOI] [PubMed] [Google Scholar]
- 25.Richard G F, Dujon B. Distribution and variability of trinucleotide repeats in the genome of the yeast Saccharomyces cerevisiae. Gene. 1996;174:165–174. doi: 10.1016/0378-1119(96)00514-8. [DOI] [PubMed] [Google Scholar]
- 26.Richard G F, Hennequin C, Thierry A, Dujon B. Trinucleotide repeats and other microsatellites in yeasts. Res Microbiol. 1999;150:589–602. doi: 10.1016/s0923-2508(99)00131-x. [DOI] [PubMed] [Google Scholar]
- 27.Sobel J D, Vazquez J, Lynch M, Meriwether C, Zervos M J. Vaginitis due to Saccharomyces cerevisiae: epidemiology, clinical aspects, and therapy. Clin Infect Dis. 1993;16:93–99. doi: 10.1093/clinids/16.1.93. [DOI] [PubMed] [Google Scholar]
- 28.Struelens M the Members of the European Study Group on Epidemiological Markers. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin Microbiol Infect. 1996;2:2–11. doi: 10.1111/j.1469-0691.1996.tb00193.x. [DOI] [PubMed] [Google Scholar]
- 29.Swofford D I. PAUP: phylogenetic analysis using parsimony, version 4. Champaign, Ill: Illinois Natural History Survey; 1999. [Google Scholar]
- 30.Zerva L, Hollis R J, Pfaller M A. In vitro susceptibility testing and DNA typing of Saccharomyces cerevisiae clinical isolates. J Clin Microbiol. 1996;34:3031–3034. doi: 10.1128/jcm.34.12.3031-3034.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]