Abstract
African swine fever virus (ASFV) is the causative agent of African swine fever (ASF) which reaches up to 100% case fatality in domestic pigs and wild boar and causes significant economic losses in the swine industry. Lack of knowledge of the function of ASFV genes is a serious impediment to the development of the safe and effective vaccine. Herein, I267L was identified as a relative conserved gene and an early expressed gene. A recombinant virus (SY18ΔI267L) with I267L gene deletion was produced by replacing I267L of the virulent ASFV SY18 with enhanced green fluorescent protein (EGFP) cassette. The replication kinetics of SY18ΔI267L is similar to that of the parental isolate in vitro. Moreover, the doses of 102.0 TCID50 (n = 5) and 105.0 TCID50 (n = 5) SY18ΔI267L caused virulent phenotype, severe clinical signs, viremia, high viral load, and mortality in domestic pigs inoculated intramuscularly as the virulent parental virus strain. Therefore, the deletion of I267L does not affect the replication or the virulence of ASFV. Utilizing the fluorescent-tagged virulence deletant can be easy to gain a visual result in related research such as the inactivation effect of some drugs, disinfectants, extracts, etc. on ASFV.
Keywords: African swine fever virus (ASFV), I267L, deletion, virulence, replication
1. Introduction
African swine fever (ASF) is a highly contagious and severe viral disease that infects domestic and wild pigs, which has caused devastating economic losses to the swine industry worldwide. The causative agent, African swine fever virus (ASFV), has a large double stranded DNA genome ranging from 170 to 190 kilobase pairs [1]. At present, 24 genotypes have been identified based on variation of C-terminal sequence of B646L genes (coding p72 protein) among different isolates [2,3]. The continuous epidemic of ASF will lead to constant genomic changes of ASFV. Differential virulence strains including high- [4,5,6], low- [7,8,9,10], and non-virulence strains [11] have been isolated from naturally occurring and cell lines-adapted strains.
ASF occurred for the first time in Kenya in 1909 [12] and was first identified and reported by Montgomery in 1921 [13]. The genotype I of ASFV mainly emerged in African and part of European countries from 1921 to the mid-1990s. European countries of ASFV outbreaks, except Sardinia, eradicated this disease until the late 1990s. The genotype II of ASFV emerged in Georgia in 2007, and it soon spread to Russia and more European countries. In August 2018, ASF was first reported in China. Since then, the spread of ASF in Asia has begun. At present, 15 countries, including Mongolia, Vietnam, Cambodia, etc. have successively reported the ASF epidemic. At present, ASF outbreaks are in the Africa, Europe, Asia, North America (Dominican Republic and Haiti), and Oceania (Papua New Guinea) region currently. There are more than 60 countries and territories that have reported the disease [14,15,16]. The continued extension of ASF has caused a serious threat to the countries without ASF.
In most cases, vaccines are the first choice for disease prevention. However, ASF vaccine researches based on inactivated virus [17], naturally attenuated virus [18], DNA plasmids, proteins DNAs/antigens [19,20,21,22], live viral vector [23,24,25,26,27,28,29,30], and deletion of virulence gene [31,32,33,34,35,36,37,38,39,40] are all still in the experimental stage. The inactivated viruses and naturally-occurring attenuated viruses had been verified either ineffective or causing strong adverse reactions [41]. DNA- and protein-based subunit vaccines showed a partial protection [19,20,21]. Currently, ASF vaccine research concentrates on genetically modified live attenuated ASFV and virus-vectored subunit vaccines. Researchers have reported gene deletions, such as 9GL [32,42], 9GL/UK [43], MGF505/MGF360/CD2v [37], DP148R [33], I177L [38], L7-L11L [39], A137R [40], and I226R [44], which can attenuate ASFV virulence and protect inoculated pigs against the challenge of homologous or virulent parental isolate. On the other side, a subunit vaccine, comprised of the ASFV genes and vectored by replication-deficient human adenovirus 5 (rAd) and modified vaccinia Ankara (MVA), led to 100% protection against a fatal ASFV [23]. Noteworthily, deleting the same gene from different virulent isolates may appear different attenuation of virulence, such as 9GL in Georgia and Malawi Lil-20/1. Additionally, doses and routes of immunization may induce differential protection effect [45].
The I267L gene locates at the right end of ASFV genome and consists of 267 amino acids which encodes a 30.9-Kilodalton protein. No significant similarity has been found among the known genes in the genetic database with I267L. Here, we reported that the sequence of I267L gene is conserved among ASFV isolates and the transcription of I267L gene occurs in early stage of the infection. We studied the function of I267L gene by constructing a recombinant ASFV (SY18∆I267L) deleting I267L gene and evaluated the virulence of SY18∆I267L on domestic pigs inoculated intramuscularly with 102.0TCID50 and 105.0TCID50. All animals developed similar fever, clinical presentation, and viremia with the same dose of the virulent parental ASFV SY18. The result demonstrated that the deletion of I267L gene from ASFV SY18 isolate does not influence the replication in vitro or the virulence of the parental virus in vivo.
2. Materials and Methods
2.1. Viruses and Cells
The virulent ASFV SY18 was isolated from the spleen of a domestic pig infected with ASFV in July 2018 in China. Bone Marrow-Derived Macrophages (BMDMs) were prepared from 2 to 3-month-old Landrace piglets. Briefly, the piglet was euthanized using pentobarbital. The ribs and leg bones were used to separate BMDMs and the red blood cells were lysed with Red Blood Cell Lysis Buffer (BOSTER, Pleasanton, CA, USA). The BMDMs were cultured in RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum (Gibco, Waltham, MA, USA) and 10 ng·mL−1 granulocyte-macrophage colony-stimulating factor (GM-CSF) (E. coli-derived porcine GM-CSF protein prepared by our lab) for 7–10 days to stimulate the differentiation of BMDMs and make them infectious.
2.2. Homology Analysis of I267L Gene among ASFV Isolates
The amino acid sequences of I267L gene from different genotype isolates (GenBank accession numbers: NC_044957.1, NC_044943.1, NC_044941.1, NC_044958.1, NC_044956.1, U18466.2, LS478113.1, MH766894.2, MK333180.1, NC_044959.2, AY261365.1, AY261364.1, AY261361.1, MH025916.1, AY261360.1, and AY261363.1) were downloaded from NCBI (https://pubmed.ncbi.nlm.nih.gov/, accessed on 27 December 2021). The multiple alignment of the amino acid sequences was performed using MAFFT online website [46] (https://www.ebi.ac.uk/Tools/msa/mafft/, accessed on 27 December 2021) and Jalview 2.11.1.5 software (http://www.jalview.org/, accessed on 27 December 2021) to value the conservation of I267L.
2.3. Characteristic of the I267L Gene Expression
The BMDMs prepared in 12 well-plates were infected with ASFV SY18 at 3 MOI and the mock infected BMDMs were used as a control. The cultures were collected at 2, 4, 6, 8, 10, 12, 15, 18, 21, 24 hpi and all-time point has three repeats. The total RNA was extracted from the BMDMs using the RNAsimple Total RNA Kit (Qiagen, Dusseldorf, Germany). The elimination of ASFV genomic DNA and the transcription of RNA in vitro were completed following the instructions of PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, Tokyo, Japan). The cDNA was amplified by the primes of B646L, CP204L, I267L, and GAPDH genes based on SYBR Green I-based quantitative PCR. The results were analyzed by the GraphPad Prism 8.0.2 software (https://www.graphpad.com/, accessed on 27 December 2021). The primers were shown in Table 1.
Table 1.
Primers were used to assay gene expression by real-time quantitative PCR.
| Gene | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|
| B646L | CGAACTTGTGCCAATCTC | ACAATAACCACCACGATGA |
| CP204L | TTCTTCTTGAGCCTGATGTT | TAGCGGTAGAATTGTTACGA |
| I267L | GCCAATGCTTGAAGAGATG | ACCGTCCAGAACTTGAAC |
| GAPDH | CCTTCATTGACCTCCACTACA | GATGGCCTTTCCATTGATGAC |
2.4. Construction of the SY18ΔI267L
To investigate the role of I267L in virulent ASFV SY18, a recombinant virus with deletion of the I267L gene was constructed by homologous recombination. The recombinant plasmid contains the two flanking sequences of the I267L and EGFP gene. The left arm of the I267L gene was located at 168,410–169,522 bp and the right arm was located at 170,483–171,645 bp in the ASFV genome. The expression of the EGFP gene was controlled by the p72 promoter (Pp72) [47] cloned from ASFV genome and SV40 polyA, which was located between the left and right arms of I267L gene. The SY18∆I267L was constructed according to the recombination between SY18 genome and the recombinant plasmid. Briefly, the recombinant plasmid was transfected into the BMDMs using jetPEI®-Macrophage DNA Transfection (Polyplus, Strasbourg, France). After 4 h, the BMDMs were further infected with ASFV SY18 at 3 MOI. The fluorescent cells were presented about 12 h post infection (hpi), which were screened under the Fluorescence microscope (Olmpus-IX73, Tokyo, Japan) and further purified in BMDMs through limiting dilution. The purity of SY18ΔI267L was detected by PCR. The forward primer (5′-CGTATATCTTGTGATAATGG-3′) and the reverse primer (5′-GGACTACATCTCTTCAAGCA-3′) were designed in the I267L gene and the right flanking region of I267L gene by Primer Premier 6 software, respectively. A 364 bp fragment could be amplified if the parental SY18 exists.
2.5. Growth Characteristic of SY18ΔI267L In Vitro
To evaluate the effect on replication while deleting the I267L gene from ASFV SY18, BMDMs were infected with SY18ΔI267L and ASFV SY18 at 0.01 MOI, respectively. Then the cultures were collected at 2, 12, 24, 36, 48, 72, and 96 hpi and repeatedly frozen–thawed between liquid nitrogen and water. In order to reduce the error, all times points were repeated thrice. The virus titer at every time point was tested according to a tenfold serial dilution and add 100 μL/well for each dilution to 8 wells of 96-well plate. The number of fluorescence wells for each dilution were calculated according to the Reed-Muench method for Tissue Culture Infectious Dose 50 (TCID50). After 72 hpi, the BMDMs infected with SY18ΔI267L and ASFV SY18 were stained by FITC-labeled p30 monoclonal antibody (diluted 1:500 in PBS) prepared by our lab. The number of the well staining fluorescence were calculated by Reed-Muench method. The cells infected with SY18∆I267L can also be directly observed under the fluorescence microscope.
2.6. Animal Experiments
To evaluate the effect of deleting the I267L gene on ASFV SY18 virulence, fifteen Landrace Pigs weighting about 20 kg were divided randomly into three groups. The pigs in the first two groups were intramuscularly (I.M.) inoculated with 102.0 TCID50 or 105.0 TCID50 of SY18ΔI267L, respectively. The pigs in the last group were inoculated with 102.0 TCID50 of ASFV SY18 as a control. After 28 days, all pigs survived the inoculation will be challenged by intramuscular injection with 102.0 TCID50 SY18. Clinical symptoms such as high fever, inappetence, depression, diarrhea, waddling, reluctance to stand, skin cyanosis, and arthrocele were observed and recorded daily throughout the experiment. The whole peripheral blood was collected in EDTA-containing tubes and the serum was isolated from normal blood every two days post inoculation. The pigs showing severe clinical signs were euthanized in extremis using pentobarbital and the tissues, including submandibular lymph node, tonsil, heart, lung, thymus, marrow, liver, spleen, kidney, stomach, colon, jejunum, bladder, inguinal lymph nodes, joint fluid, and muscle, were assessed the development of viral load. All samples were detected in CFX96TM Real-Time System (Thermo, Waltham, MA, USA).
2.7. Detection of ASFV Genome in Blood and Tissues
A probe-based real-time quantitative PCR (qPCR) targeting the ASFV B646L gene was performed to quantify the ASFV genomic DNA copy in the blood and tissue samples. The primer synthesis and the reaction condition were recommended by the World Organization for Animal Health (OIE) [48]. According to the standard plasmid, the standard curve is y = −3.34x + 40.1. The value of y represents the value of Ct and the value of x represents the value of Log10 copies/μL. Additionally, the sensitivity is about 50 copies via detecting the standard plasmid using the OIE recommended method. To establish a standard curve for absolute quantification by qPCR, the B646L gene was cloned into the pMD18-T vector and used as a standard plasmid. The standard curve was synthesized according to detecting the 10-fold gradient p72 plasmid. The blood and tissue samples were processed by the following methods. The tissues added PBS were ground in an automatic sample grinding machine. The blood and the supernatant of tissues were lysed by a lysis buffer (prepared by our lab) in volume ratio of 1:1. The mixture was oscillated for 5 s, boiled for 5 min, and centrifuged for 1 min. The supernatant was used as the template and was detected in CFX96TM Real-Time System.
2.8. Detection of Anti-p54 Antibodies
The level of antibodies against ASFV-specific protein p54 antibodies in serum was measured using an indirect ELISA (developed by our lab). The detailed process was described in the previous article [39]. Briefly, the ELISA plates (Corning, New York, NY, USA) were coated with purified p54 protein (1 μg·mL−1) which was expressed in prokaryotic expression system and was blocked with 5% skimmed milk. The serum sample (S) and positive control (P) were added to the ELISA plates and incubated for 1 h at room temperature (RT). The horseradish peroxidase (HRP)-labeled sheep anti-pig IgG (CWBIO, Haimen, China) used for second antibody was incubated for 1 h at RT. The chromogenic reaction began with the addition of 3,3′,5,5′-Tetramethylbenzidine (TMB) substrate (SeraCare, Delaware, USA) and ended with 2 M sulfuric acid (BEIJING SHIJI, Beijing, China). The optical density (OD) values at 450 nm were read by iMarkTM Microplate Reader (BIO-RAD, Hercules, CA, USA). The ratio of S/P above 0.25 is recognized as a positive sample.
2.9. Statistical Analysis
Statistical significance was determined using the Holm–Sidak test. A p-value < 0.05 was considered statistically significant and a p-value ≥ 0.05 was considered statistically non-significant. Similar results were obtained from three independent experiments. Statistically significant differences between groups were analyzed using GraphPad Prism 8.0.2 software (https://www.graphpad.com/, accessed on 27 December 2021).
3. Results
3.1. A Relative Conserved I267L Gene
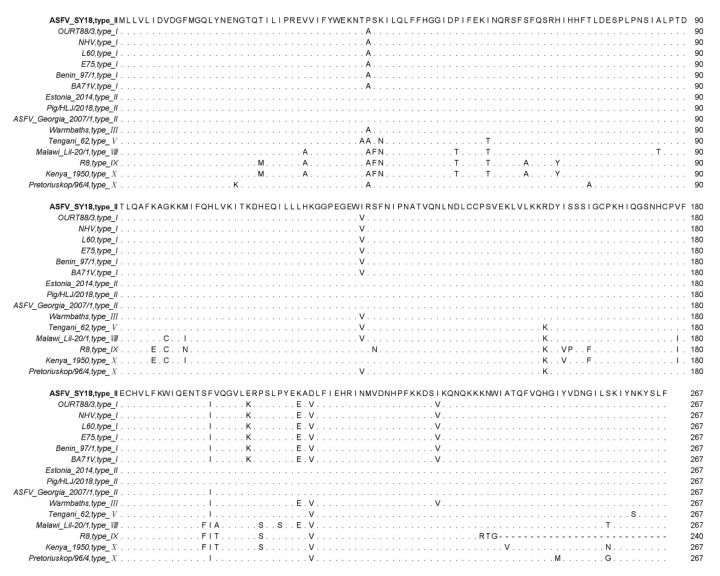
The amino acid sequences of different isolates were analyzed using a MAFFT online website and Jalview software. The results showed that the I267L gene encoded 267 amino acids in most of the ASFV strains. There are several ASFV isolates—R7, R8, R25, R35, and N10—encoding 240 amino acids due to a single-base mutation, which caused an early termination of the ORF (some sequences are not shown except R8) in Figure 1. The truncation of the 27 amino acids in 3′ end sequence did not affect virus virulence [49]. The I267L ORFs between genotype I and II are relatively conserved. Mutations mostly occur in virus isolates among type VIII, IX, and X. The homology of the pI267L amino acids is 88.4–100%, which is relatively conserved.
Figure 1.
Multiple sequence alignment of ASFV pI267L amino acids residues. In multiple sequence alignment, the same amino acids are displayed by ‘.’, the absence of amino acids are displayed by ‘-’, and the differential amino acids are displayed by abbreviated letters of amino acids.
3.2. Transcription of I267L Occurs at the Early Stage of Infection
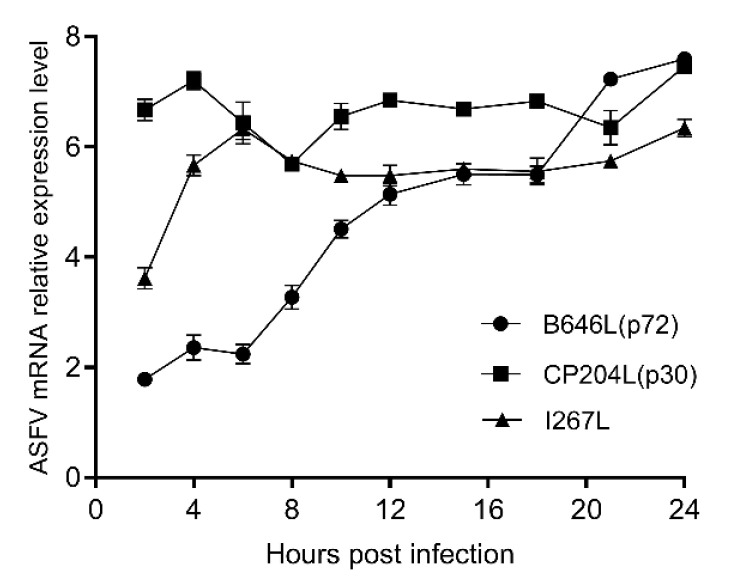
Total RNA of BMDMs infected with ASFV SY18 or mock infection were detected by the late viral gene B646L (p72), the early viral gene CP204L (p30), I267L and GAPDH (housekeeping gene). The results showed that the CP204L gene maintained high expression from 2 to 24 h post infection, and the expression of the B646L gene kept rising from 2 to 24 h during infection in Figure 2. The I267L gene presented a rapid expression at the first 6 hpi and then maintained high expression. The expressing trend of I267L gene was similar to the CP204L gene. Therefore, we speculate that the I267L gene is an early transcription gene, and this result is consistent with the results of Cackett’s ASFV transcriptome data [50].
Figure 2.
The relative expression levels of mRNA of I267L CP204L and B646L. The relative expression level of mRNA of I267L, CP204L, and B646L genes were quantified between BMDMs infected with SY18 and mock-infected BMDMs. The values of Y axis were expressed by the base-10 logarithm (log10) of the relative expression level.
3.3. Generation of SY18 ∆I267L
The illustrations of the recombinant plasmid can be seen in Figure 3a. The diagram illuminated the design of ASFV SY18ΔI267L and the position of the I267L gene in ASFV SY18 in Figure 3b. The purified SY18∆I267L expressed the green fluorescence in Figure 3c. SY18ΔI267L displayed an identical growth kinetic comparing to that of the parental virus in Figure 3d. There was no significant difference (p > 0.5) in replication in vitro between SY18∆I267L and ASFV SY18 from the beginning to the end of the infection. Therefore, the I267L is not a replication-related gene for ASFV.
Figure 3.
Construction of SY18∆I267L. (a) The recombinant plasmid, pUC-∆I267L-EGFP, was constructed. (b) Schematic representation of SY18ΔI226R construction. The location of the I267L gene was replaced with the EGFP cassette via homologous recombination between pUC-ΔI267L-EGFP and ASFV SY18 genomic DNA in vitro. The red dotted frame represents the position of the homology arms and the green square represents the position of EGFP. (c) BMDMs were infected with purified SY18ΔI267L and expressed green fluorescence (Left). The mock-infected BMDMs were non-fluorescence (Right). Bar 50 μm. (d) The viral titers of the two viruses were measured at 0, 12, 24, 36, 48, 72, and 96 hpi and exhibited using log10 TCID50/mL. They were non-significant differences (ns) at specific times (“ns” p ≥ 0.05).
3.4. Animal Experiments
There were no pigs that survived the inoculation of 102.0 TCID50/mL SY18ΔI267L, 105.0 TCID50/mL SY18ΔI267L, and 102.0 TCID50/mL ASFV SY18 (Table 2). All pigs inoculated with ASFV SY18ΔI267L presented ASF-associated clinical signs.
Table 2.
Survival and fever responses of pigs inoculated with SY18ΔI267L and ASFV SY18.
| Virus | No. of Survivors (n = 5) |
Fever | Days of Viremia Onset (±SD) |
Clinical Sympotoms | Mean Days to Death (±SD) | ||
|---|---|---|---|---|---|---|---|
| Days of Onset (±SD) |
Days of Duration (±SD) | Maximum Daily Temp.°C (±SD) | |||||
| ASFV SY18 (102.0TCID50) |
0/5 | 4.0 (±0) | 5.4 (±1.14) | 41.7 (±0.08) | 6 (±0) | 1. Fever (5/5) 2. Inappetence (4/5) 3. Diarrhea (4/5) 4. Arthrocele (3/5) |
10.2 (±0.45) |
| SY18ΔI267L (102.0TCID50) |
0/5 | 6.0 (±0.71) | 5.4 (±0.55) | 41.7 (±0.18) | 8 (±0) | 1. Fever (5/5) 2. Inappetence (5/5) 3. Diarrhea (3/5) 4. Arthrocele (3/5) |
11.4 (±0.89) |
| SY18ΔI267L (105.0TCID50) |
0/5 | 2.8 (±0.84) | 6.4 (±1.14) | 41.8 (±0.12) | 5.2 (±1.10) | 1. Fever (5/5) 2. Inappetence (4/5) 3. Diarrhea (3/5) 4. Arthrocele (2/5) |
9.4 (±0.55) |
Note: 1. Numero sign is abbreviated as No.. 2. Standard deviation is abbreviated as SD.
The animals inoculated with 105.0 TCID50 SY18ΔI267L showed early fever, viremia, clinical signs, and death comparing with the low dose of SY18ΔI267L and ASFV SY18 (Figure 4a–c). The animals inoculated with 102.0 TCID50 SY18ΔI267L presented delay fever, viremia, clinical signs, and death comparing with the same dose of ASFV SY18.
Figure 4.
The results of survival rate, temperature, viremia, antibodies, and viral load of tissues. (a) The survival rate of the animal post inoculation with 102.0 TCID50 and 105.0 TCID50 SY18ΔI267L and 102.0 TCID50 ASFV SY18. (b) The temperature of the animal post inoculation. (c) The viremia of the animal post inoculation. (d) The value of anti-p54 antibody of the animal post inoculation. The value of P (OD450)/N (OD450) greater than 0.25 is considered positive. The red dotted line represents P (OD450)/N (OD450) equal to 0.25. (e) The viral load in the tissues of the animal euthanized in extremis. The black dots represent the viral load of the individual in the tissue.
There were several pigs that produced anti-p54 antibody in the three groups in Figure 4d. The positive seroconversion rates were 4/5, 2/5, and 3/5 in the groups of 102.0 TCID50 SY18ΔI267L, 105.0 TCID50 SY18ΔI267L, and 102.0 TCID50 ASFV SY18, respectively, which occurred on the 8th day post inoculation. The time of positive seroconversion is similar with our previous study on a gene-deleted attenuated SY18ΔMGF/ΔCD2v [51] and SY18ΔI226R [44].
The viral load was detected in multiple tissues of the pigs. Almost all tissues had a high viral load (up to 108.0 copies/g) in Figure 4e. The tissues, including spleen, joint fluid, bladder, jejunum, and colon, had a high detection rate (15/15) in the three groups.
4. Discussion
ASFV is spreading in more and more countries. Strict measures, including disinfection of pigpen, surveillance of pathogen, quarantine, and cull of infected and in-contact animals have been applied to prevent and control of ASF, which cost a lot of manpower, material, and financial resources. People urgently need a safe and effective vaccine. ASFV encodes more than 150 ORFs. However, the function of about half the genes is still unknown. A comprehensive understanding of the ASFV gene will help the development of vaccines.
I267L is a gene of unknown function and there are no known genes or proteins to match up with it in gene and protein databases. The homology of I267L amino acid residues is 88.4–100%, which is relatively conserved among ASFV isolates of the different serotypes. Mazloum et al. identified I267L as possible genetic markers to discriminate between the closely related genotype II viruses based on the identification of SNPs or Indels within ASFV genomes [52]. In several ASFV isolates, the translation of I267L was terminated prematurely due to nucleotide mutations, leaving 27 amino acids untranslated, but the mutation does not influence the virulence of these isolates. The result indicates that the 27 amino acids have nothing to do with ASFV virulence. Currently, there is no research on the role of I267L in the process of ASFV infection. The transcription of I267L gene starts at an early stage and maintains a high level during the infection process.
In vitro, the deletion of I267L of virulent SY18 did not lead to the replication deficiency. In vivo, the animal inoculated with the same dose of SY18ΔI267L and ASFV SY18 developed the similar clinical result. The results demonstrate that I267L is not replication- or virulence-related gene for ASFV. There are parts of the animal that developed positive seroconversion at 8th day post infection in the three groups, which is similar to our previous research on gene-deletion attenuated SY18∆MGF/∆CD2v and SY18∆I226R. Although neither ASFV-resistant swine serum nor attenuated strain immunized pig serum can neutralize ASFV, the positive outcome of the serum is positively correlated with whether it can survive the challenge of virulent ASFV (the data were not presented). We detected the high level ASFV genome in almost all the tissues. The viral load of the bladder and colon has the high copy and detection rate. The results demonstrate that the animal mainly excretes the virus from urine and feces. This is one of the main reasons that ASFV spread rapidly and is hard to control.
The new deletant has similar characteristic in vivo and vitro with virulent ASFV SY18, which has been considered as a marker ASFV. Borac et al. developed a fluorescent ASFV strain that replaced the genes of MGF360-13L and MGF360-14L and retained the ability to cause disease in swine. Besides, the deletants, such as deletion of X69R [53] and C962R [36], also have the potential to be utilized in related research. The modified ASFV is a suitable tool to research on pathogenesis, virus-macrophage interaction, and viral antigen-based assays [54].
In summary, I267L is expressed at an early stage during the infection and encodes a relative conserved protein. The absence of I267L does not affect the replication of ASFV on primary swine macrophage in vitro. The domestic pigs could not survive the I.M. challenge with high or low doses of SY18∆I267L.
Acknowledgments
We thank the National Key R&D Program of China (approval no. 2021YFD1801400) and National Natural Science Foundation of China for financial support (approval no. U19A2039). We thank the teachers and staffs working in Animal Management Centre of Changchun Veterinary Research Institute for their excellent work.
Author Contributions
Conceptualization, R.H. and Y.L.; methodology, R.H., Y.Z. and J.Z.; validation, J.Z., T.C., Q.L. and J.K.; formal analysis, R.H. and Y.Z.; investigation, J.K., X.Z., Y.Q., R.Z., X.H., N.L. and L.W.; resources, S.Z. and L.M.; data curation, J.K. and Y.Z.; writing—original draft preparation, Y.Z.; writing—review and editing, R.H., Y.L., Q.L., T.C. and S.W.; visualization, J.K., H.Y., J.Y. (Jinmei Yang) and J.Y. (Jinjin Yang); supervision, F.M. and S.Z.; project administration, T.C. and S.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Key R&D Program of China (approval no. 2021YFD1801400) and National Natural Science Foundation (approval no. U19A2039) of China for financial support.
Institutional Review Board Statement
All experimental procedures involving ASFV and animal experiment were performed in Changchun Veterinary Research Institute. The animal experiment was conducted according to the standard procedures approved by both the Animal Welfare and Ethics Committee of the Changchun Veterinary Research Institute (review ID: IACUC of AMMS-11-2020-018).
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wang N., Zhao D., Wang J., Zhang Y., Wang M., Gao Y., Li F., Wang J., Bu Z., Rao Z., et al. Architecture of African swine fever virus and implications for viral assembly. Science. 2019;366:640–644. doi: 10.1126/science.aaz1439. [DOI] [PubMed] [Google Scholar]
- 2.Achenbach J., Gallardo C., Nieto-Pelegrín E., Rivera-Arroyo B., Degefa-Negi T., Arias M., Jenberie S., Mulisa D.D., Gizaw D., Gelaye E., et al. Identification of a New Genotype of African Swine Fever Virus in Domestic Pigs from Ethiopia. Transbound. Emerg. Dis. 2017;64:1393–1404. doi: 10.1111/tbed.12511. [DOI] [PubMed] [Google Scholar]
- 3.Simulundu E., Lubaba C., van Heerden J., Kajihara M., Mataa L., Chambaro H.M., Sinkala Y., Munjita S.M., Munang’Andu H.M., Nalubamba K.S., et al. The Epidemiology of African Swine Fever in “Nonendemic” Regions of Zambia (1989–2015): Implications for Disease Prevention and Control. Viruses. 2017;9:236. doi: 10.3390/v9090236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rowlands R., Michaud V., Heath L., Hutchings G., Oura C., Vosloo W., Dwarka R., Onashvili T., Albina E., Dixon L.K. African swine fever virus isolate, Georgia, 2007. Emerg. Infect. Dis. 2008;14:1870–1874. doi: 10.3201/eid1412.080591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou X., Li N., Luo Y., Miao F., Chen T., Zhang S., Cao P., Li X., Tian K., Qiu H.J., et al. Emergence of African Swine Fever in China, 2018. Transbound. Emerg. Dis. 2018;65:1482–1484. doi: 10.1111/tbed.12989. [DOI] [PubMed] [Google Scholar]
- 6.Neilan J., Zsak L., Lu Z., Kutish G.F., Afonso C.L., Rock D.L. Novel swine virulence determinant in the left variable region of the African swine fever virus genome. J. Virol. 2002;76:3095–3104. doi: 10.1128/JVI.76.7.3095-3104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun E., Huang L., Zhang X., Zhang J., Shen D., Zhang Z., Wang Z., Huo H., Wang W., Huangfu H., et al. Genotype I African swine fever viruses emerged in domestic pigs in China and caused chronic infection. Emerg. Microbes Infect. 2021;10:2183–2193. doi: 10.1080/22221751.2021.1999779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallardo C., Soler A., Rodze I., Nieto R., Cano-Gómez C., Fernandez-Pinero J., Arias M. Attenuated and non-haemadsorbing (non-HAD) genotype II African swine fever virus (ASFV) isolated in Europe, Latvia 2017. Transbound. Emerg. Dis. 2019;66:1399–1404. doi: 10.1111/tbed.13132. [DOI] [PubMed] [Google Scholar]
- 9.Leitão A., Cartaxeiro C., Coelho R., Cruz B., Parkhouse R.M.E., Portugal F.C., Vigário J.D., Martins C.L.V. The non-haemadsorbing African swine fever virus isolate ASFV/NH/P68 provides a model for defining the protective anti-virus immune response. J. Gen. Virol. 2001;82:513–523. doi: 10.1099/0022-1317-82-3-513. [DOI] [PubMed] [Google Scholar]
- 10.Boinas F., Hutchings G., Dixon L., Wilkinson P.J. Characterization of pathogenic and non-pathogenic African swine fever virus isolates from Ornithodoros erraticus inhabiting pig premises in Portugal. J. Gen. Virol. 2004;85:2177–2187. doi: 10.1099/vir.0.80058-0. [DOI] [PubMed] [Google Scholar]
- 11.Rodríguez J., Moreno L., Alejo A., Lacasta A., Rodríguez F., Salas M.L. Genome Sequence of African Swine Fever Virus BA71, the Virulent Parental Strain of the Nonpathogenic and Tissue-Culture Adapted BA71V. PLoS ONE. 2015;10:e0142889. doi: 10.1371/journal.pone.0142889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Penrith M., Vosloo W., Jori F., Bastos A. African swine fever virus eradication in Africa. Virus Res. 2013;173:228–246. doi: 10.1016/j.virusres.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 13.Montgomery E.R. On A Form of Swine Fever Occurring in British East Africa (Kenya Colony) J. Comp. Pathol. Ther. 1921;34:159–191. doi: 10.1016/S0368-1742(21)80031-4. [DOI] [Google Scholar]
- 14.Dixon L., Stahl K., Jori F., Vial L., Pfeiffer D.U. African Swine Fever Epidemiology and Control. Annu. Rev. Anim. Biosci. 2020;8:221–246. doi: 10.1146/annurev-animal-021419-083741. [DOI] [PubMed] [Google Scholar]
- 15.Ministry of Agriculture and Rural Affairs of the People’s Republic of China, One Week’s International Animal Epidemic Situation. [(accessed on 27 September 2021)]; Available online: http://www.moa.gov.cn/
- 16.OIE Global Situation of African Swine Fever. [(accessed on 27 September 2021)]. Available online: https://www.oie.int/app/uploads/2021/03/report-47-global-situation-asf.pdf.
- 17.Blome S., Gabriel C., Beer M. Modern adjuvants do not enhance the efficacy of an inactivated African swine fever virus vaccine preparation. Vaccine. 2014;32:3879–3882. doi: 10.1016/j.vaccine.2014.05.051. [DOI] [PubMed] [Google Scholar]
- 18.Krug P., Holinka L., O’Donnell V., Reese B., Sanford B., Fernandez-Sainz I., Gladue D., Arzt J., Rodriguez L., Risatti G.R., et al. The progressive adaptation of a georgian isolate of African swine fever virus to vero cells leads to a gradual attenuation of virulence in swine corresponding to major modifications of the viral genome. J. Virol. 2015;89:2324–2332. doi: 10.1128/JVI.03250-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sunwoo S., Pérez-Núñez D., Morozov I., Sánchez E.G., Gaudreault N.N., Trujillo J.D., Mur L., Nogal M., Madden D., Urbaniak K., et al. DNA-Protein Vaccination Strategy Does Not Protect from Challenge with African Swine Fever Virus Armenia 2007 Strain. Vaccines. 2019;7:12. doi: 10.3390/vaccines7010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pérez-Núñez D., Sunwoo S., Sánchez E., Haley N., García-Belmonte R., Nogal M., Morozov I., Madden D., Gaudreault N.N., Mur L., et al. Evaluation of a viral DNA-protein immunization strategy against African swine fever in domestic pigs. Vet. Immunol. Immunopathol. 2019;208:34–43. doi: 10.1016/j.vetimm.2018.11.018. [DOI] [PubMed] [Google Scholar]
- 21.Argilaguet J., Pérez-Martín E., Nofrarías M., Gallardo C., Accensi F., Lacasta A., Mora M., Ballester M., Galindo-Cardiel I., López-Soria S., et al. DNA vaccination partially protects against African swine fever virus lethal challenge in the absence of antibodies. PLoS ONE. 2012;7:e40942. doi: 10.1371/journal.pone.0040942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lacasta A., Ballester M., Monteagudo P., Rodriguez J.M., Salas M.L., Accensi F., Pina-Pedrero S., Bensaid A., Argilaguet J., Lopez-Soria S., et al. Expression library immunization can confer protection against lethal challenge with African swine fever virus. J. Virol. 2014;88:13322–13332. doi: 10.1128/JVI.01893-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goatley L., Reis A., Portugal R., Goldswain H., Shimmon G.L., Hargreaves Z., Ho C.-S., Montoya M., Sánchez-Cordón P.J., Taylor G., et al. A Pool of Eight Virally Vectored African Swine Fever Antigens Protect Pigs Against Fatal Disease. Vaccines. 2020;8:234. doi: 10.3390/vaccines8020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Netherton C., Goatley L., Reis A., Portugal R., Nash R.H., Morgan S.B., Gault L., Nieto R., Norlin V., Gallardo C., et al. Identification and Immunogenicity of African Swine Fever Virus Antigens. Front. Immunol. 2019;10:1318. doi: 10.3389/fimmu.2019.01318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cadenas-Fernández E., Sánchez-Vizcaíno J., Kosowska A., Rivera B., Mayoral-Alegre F., Rodríguez-Bertos A., Yao J., Bray J., Lokhandwala S., Mwangi W., et al. Adenovirus-vectored African Swine Fever Virus Antigens Cocktail Is Not Protective against Virulent Arm07 Isolate in Eurasian Wild Boar. Pathogens. 2020;9:171. doi: 10.3390/pathogens9030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lokhandwala S., Waghela S., Bray J., Sangewar N., Charendoff C., Martin C.L., Hassan W.S., Koynarski T., Gabbert L., Burrage T.G., et al. Adenovirus-vectored novel African Swine Fever Virus antigens elicit robust immune responses in swine. PLoS ONE. 2017;12:e0177007. doi: 10.1371/journal.pone.0177007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lokhandwala S., Waghela S., Bray J., Martin C.L., Sangewar N., Charendoff C., Shetti R., Ashley C., Chen C.-H., Berghman L.R., et al. Induction of Robust Immune Responses in Swine by Using a Cocktail of Adenovirus-Vectored African Swine Fever Virus Antigens. Clin. Vaccine Immunol. CVI. 2016;23:888–900. doi: 10.1128/CVI.00395-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopera-Madrid J., Osorio J., He Y., Xiang Z., Adams L.G., Laughlin R.C., Mwangi W., Subramanya S., Neilan J., Brake D., et al. Safety and immunogenicity of mammalian cell derived and Modified Vaccinia Ankara vectored African swine fever subunit antigens in swine. Vet. Immunol. Immunopathol. 2017;185:20–33. doi: 10.1016/j.vetimm.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neilan J., Zsak L., Lu Z., Burrage T., Kutish G., Rock D. Neutralizing antibodies to African swine fever virus proteins p30, p54, and p72 are not sufficient for antibody-mediated protection. Virology. 2004;319:337–342. doi: 10.1016/j.virol.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 30.Lokhandwala S., Petrovan V., Popescu L., Sangewar N., Elijah C., Stoian A., Olcha M., Ennen L., Bray J., Bishop R.P., et al. Adenovirus-vectored African Swine Fever Virus antigen cocktails are immunogenic but not protective against intranasal challenge with Georgia 2007/1 isolate. Vet. Microbiol. 2019;235:10–20. doi: 10.1016/j.vetmic.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 31.Zsak L., Caler E., Lu Z., Kutish G.F., Neilan J.G., Rock D.L. A nonessential African swine fever virus gene UK is a significant virulence determinant in domestic swine. J. Virol. 1998;72:1028–1035. doi: 10.1128/JVI.72.2.1028-1035.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Donnell V., Holinka L., Krug P., Gladue D., Carlson J., Sanford B.J., Alfano M., Kramer E.J., Lu Z., Arzt J., et al. African Swine Fever Virus Georgia 2007 with a Deletion of Virulence-Associated Gene 9GL (B119L), when Administered at Low Doses, Leads to Virus Attenuation in Swine and Induces an Effective Protection against Homologous Challenge. J. Virol. 2015;89:8556–8566. doi: 10.1128/JVI.00969-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reis A., Goatley L., Jabbar T., Sanchez-Cordon P.J., Netherton C.L., Chapman D.A.G., Dixon L.K. Deletion of the African Swine Fever Virus Gene DP148R Does Not Reduce Virus Replication in Culture but Reduces Virus Virulence in Pigs and Induces High Levels of Protection against Challenge. J. Virol. 2017;91:e01428-17. doi: 10.1128/JVI.01428-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Donnell V., Holinka L., Sanford B., Krug P.W., Carlsona J., Pacheco J.M., Reese B., Risatti G.R., Gladue D.P., Borcaa M.V. African swine fever virus Georgia isolate harboring deletions of 9GL and MGF360/505 genes is highly attenuated in swine but does not confer protection against parental virus challenge. Virus Res. 2016;221:8–14. doi: 10.1016/j.virusres.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 35.Abrams C., Goatley L., Fishbourne E., Chapman D., Cooke L., Oura C.A., Netherton C.L., Takamatsu H.-H., Dixon L.K. Deletion of virulence associated genes from attenuated African swine fever virus isolate OUR T88/3 decreases its ability to protect against challenge with virulent virus. Virology. 2013;443:99–105. doi: 10.1016/j.virol.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramirez-Medina E., Vuono E., Rai A., Pruitt S., Silva E., Velazquez-Salinas L., Zhu J., Borca M.V., Gladue D.P. The C962R ORF of African Swine Fever Strain Georgia Is Non-Essential and Not Required for Virulence in Swine. Viruses. 2020;12:676. doi: 10.3390/v12060676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen W., Zhao D., He X., Liu R., Wang Z., Zhang X., Li F., Shan D., Chen H., Zhang J., et al. A seven-gene-deleted African swine fever virus is safe and effective as a live attenuated vaccine in pigs. Science China. Life Sci. 2020;63:623–634. doi: 10.1007/s11427-020-1657-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borca M., Ramirez-Medina E., Silva E., Vuono E., Rai A., Pruitt S., Holinka L.G., Velazquez-Salinas L., Zhu J., Gladue D.P. Development of a Highly Effective African Swine Fever Virus Vaccine by Deletion of the I177L Gene Results in Sterile Immunity against the Current Epidemic Eurasia Strain. J. Virol. 2020;94:e02017-19. doi: 10.1128/JVI.02017-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J., Zhang Y., Chen T., Yang J., Yue H., Wang L., Zhou X., Qi Y., Han X., Ke J., et al. Deletion of the L7L-L11L Genes Attenuates ASFV and Induces Protection against Homologous Challenge. Viruses. 2021;13:255. doi: 10.3390/v13020255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gladue D., Ramirez-Medina E., Vuono E., Silva E., Rai A., Pruitt S., Espinoza N., Velazquez-Salinas L., Borca M.V. Deletion of A137R gene from the pandemic strain of African swine fever virus is attenuated and offers protection against virulent pandemic virus. J. Virol. 2021;95:JVI0113921. doi: 10.1128/JVI.01139-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.King K., Chapman D., Argilaguet J., Fishbourne E., Hutet E., Cariolet R., Hutchings G., Oura C.A., Netherton C.L., Moffat K., et al. Protection of European domestic pigs from virulent African isolates of African swine fever virus by experimental immunisation. Vaccine. 2011;29:4593–4600. doi: 10.1016/j.vaccine.2011.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis T., Zsak L., Burrage T., Lu Z., Kutish G.F., Neilan J.G., Rock D.L. An African swine fever virus ERV1-ALR homologue, 9GL, affects virion maturation and viral growth in macrophages and viral virulence in swine. J. Virol. 2000;74:1275–1285. doi: 10.1128/JVI.74.3.1275-1285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Donnell V., Risatti G., Holinka L., Krug P.W., Carlson J., Velazquez-Salinas L., Azzinaro P.A., Gladue D.P., Borca M.V. Simultaneous Deletion of the 9GL and UK Genes from the African Swine Fever Virus Georgia 2007 Isolate Offers Increased Safety and Protection against Homologous Challenge. J. Virol. 2017;91:e01760-16. doi: 10.1128/JVI.01760-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y., Ke J., Zhang J., Yang J., Yue H., Zhou X., Qi Y., Zhu R., Miao F., Li Q., et al. ASFV bearing an I226R gene-deletion elicits a robust immunity in pigs to African swine fever. J. Virol. 2021;95:e01199-21. doi: 10.1128/JVI.01199-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sánchez-Cordón P., Jabbar T., Berrezaie M., Chapman D., Reis A., Sastre P., Rueda P., Goatley L., Dixon L.K. Evaluation of protection induced by immunisation of domestic pigs with deletion mutant African swine fever virus BeninΔMGF by different doses and routes. Vaccine. 2018;36:707–715. doi: 10.1016/j.vaccine.2017.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MAFFT Online Website. [(accessed on 27 December 2021)]. Available online: https://www.ebi.ac.uk/Tools/msa/mafft/
- 47.Portugal R., Bauer A., Keil G. Selection of differently temporally regulated African swine fever virus promoters with variable expression activities and their application for transient and recombinant virus mediated gene expression. Virology. 2017;508:70–80. doi: 10.1016/j.virol.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 48.King D., Reid S., Hutchings G., Grierson S.S., Wilkinson P.J., Dixon L.K., Bastos A.D., Drew T.W. Development of a TaqMan PCR assay with internal amplification control for the detection of African swine fever virus. J. Virol. Methods. 2003;107:53–61. doi: 10.1016/S0166-0934(02)00189-1. [DOI] [PubMed] [Google Scholar]
- 49.Masembe C., Sreenu V., Da Silva Filipe A., Wilkie G.S., Ogweng P., Mayega F.J., Muwanika V.B., Biek R., Palmarini M., Davison A.J. Genome Sequences of Five African Swine Fever Virus Genotype IX Isolates from Domestic Pigs in Uganda. Microbiol. Resour. Announc. 2018;7:e01018-18. doi: 10.1128/MRA.01018-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cackett G., Matelska D., Sýkora M., Portugal R., Malecki M., Bähler J., Dixon L., Werner F. The African Swine Fever Virus Transcriptome. J. Virol. 2020;94:e00119-20. doi: 10.1128/JVI.00119-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y., Chen T., Zhang J., Qi Y., Miao F., Bo Z., Wang L. Construction and immunoprotective characterization of gene deleted African swine fever virus vaccine and candidates. Chin. J. Vet. Sci. 2019;39:1421–1427. [Google Scholar]
- 52.Mazloum A., van Schalkwyk A., Shotin A., Igolkin A., Shevchenko I., Gruzdev K., Vlasova N. Comparative Analysis of Full Genome Sequences of African Swine Fever Virus Isolates Taken from Wild Boars in Russia in 2019. Pathogens. 2021;10:521. doi: 10.3390/pathogens10050521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borca M., O’Donnell V., Holinka L., Sanford B., Azzinaro P.A., Risatti G.R., Gladue D.P. Development of a fluorescent ASFV strain that retains the ability to cause disease in swine. Sci. Rep. 2017;7:46747. doi: 10.1038/srep46747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramirez-Medina E., Vuono E., Pruitt S., Rai A., Silva E., Zhu J., Velazquez-Salinas L., Gladue D.P., Borca M.V. X69R Is a Non-Essential Gene That, When Deleted from African Swine Fever, Does Not Affect Virulence in Swine. Viruses. 2020;12:918. doi: 10.3390/v12090918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.