Abstract
The clinical outcome of Helicobacter pylori infection may be associated with the cagA bacterial genotype. To investigate the cagA status of H. pylori-infected patients and the relationship between cagA and peptic ulcer disease, gastric biopsy specimens from 103 Caucasian patients in Brazil were analyzed by PCR. Since allelic variation in cagA exists and distinct H. pylori subgenotypes may circulate in different regions, PCR using primers for a variable 3′ region of the cagA gene according to a Japanese methodology and for a consensus cagA 3′ region used in Western methods was used for cagA detection. cagA was present in 53 (71%) of 75 H. pylori-positive cases when analyzed by the consensus region method and was associated with duodenal ulcer disease (P = 0.02), but not with gastric ulcer (P = 0.26), when compared to patients with duodenitis or gastritis. The variable region PCR method was able to detect 43 (57%) cagA-positive cases within the same group of H. pylori-positive patients and showed three subtypes of cagA (A, B/D, and C) that were not associated with clinical outcome. However, in 8 (18%) of the cases, more than one subtype was present, and an association between patients with multiple subtypes and disease outcome was observed when compared to patients with isolated subtypes (P = 0.048). cagA was a marker of H. pylori strains for duodenal ulcer disease in our population, and in spite of the differences in the 3′ region of the cagA gene, the Japanese methodology was able to detect the cagA status in most cases. The presence of multiple subgenotypes of cagA was associated with gastric ulcer.
Helicobacter pylori infection affects more than half of the world population, but symptomatic disease appears in only a minority of infected subjects (4, 16, 24, 32). The clinical spectrum of this infection ranges from asymptomatic gastritis to peptic ulcer and gastric cancer (19, 29). It is unclear why only a minority of the subjects infected by H. pylori develop peptic ulcer and gastric cancer while most people harboring this bacterium remain asymptomatic or have only chronic gastritis.
The causes of the different outcomes of H. pylori infection may include host and environmental factors and differences in prevalence or expression of bacterial virulence factors (2, 17, 45). Several genes have been identified that may play a role in the pathogenicity of the bacterium (8, 28, 40). The cagA, vacA, and iceA genes have been used as molecular markers of H. pylori virulence (44). The cagA gene (cytotoxin-associated gene) is considered a marker for the presence of one pathogenicity island of about 40 kbp (5). The structure of the gene reveals a 5′ highly conserved region and a variable 3′ region, in which the presence of a variable number of repeat sequences results in a protein with a molecular mass of 120 to 140 kDa (7, 40, 48). CagA-producing H. pylori strains induce inflammatory cytokines, especially interleukin-8 (9, 10). The presence of cagA is associated with duodenal ulcer, gastric mucosal atrophy, and gastric cancer (12, 38, 44), and the majority of H. pylori strains can be classified into one of two groups (cag positive or cag negative), based on the possession of the cagA gene and the associated genes in the cag pathogenicity island (37). The gene vacA encodes the vacuolating cytotoxin that is involved in epithelial cell injury (8). The production of the cytotoxin is associated with pathogenicity and depends on the bacterium genotype (14). The gene is present in all H. pylori strains and comprises two variable parts. The s region (encoding the signal peptide) exists as an s1 or s2 allele, and s1 can be further subdivided into subtype s1a, s1b, or s1c. The m region (middle) occurs as an m1 or m2 allele (43). vacA s1m1 strains produce a large amount of toxin, s1m2 strains produce moderate amounts, and s2m2 strains produce very little or no toxin (1). The presence of cytotoxic activity has been suggested as a marker for strains with enhanced virulence acting either directly via cytotoxicity or indirectly via an increased inflammatory and immune response (1). Since the vast majority of vacA s1 strains are also cagA positive, the two markers are closely related (1, 35, 42). The iceA gene (induced by contact with epithelium) has two main allelic variants, designated iceA1 and iceA2. The expression of iceA1 is up-regulated on contact between H. pylori and human epithelial cells and may be associated with peptic ulcer disease (28). As suggested by van Doorn et al. (44), strains typed as vacA s1 cagA+ iceA1 can be considered the most pathogenic and are found predominantly in patients with ulcer disease. In contrast, strains typed as vacA s2m2 iceA2 (cagA negative) appear to be the least pathogenic and do not occur in peptic ulcer patients (44).
The proportion of H. pylori isolates which are cagA positive varies from one geographic region to another (13). Studies from Japan (21, 37), Korea (26), and China (25) have shown that more than 90% of H. pylori strains are cagA positive, while in the United States (27), Canada (30), and Europe (18, 35, 43) these percentages are lower. Therefore, cagA cannot be used as a marker for the presence of peptic ulcer disease in those regions where the prevalence of cagA-positive H. pylori strains is uniformly high. Since allelic variation in cagA exists and distinct H. pylori subgenotypes may circulate in different regions (25), differences in cagA subgenotype may provide a marker for differences in virulence among cagA-positive H. pylori strains (48). Some studies have demonstrated that PCR using primers designed from Western H. pylori isolates were ineffective in the detection of Eastern Asia H. pylori strains, and vice versa (25, 41, 47). The aim of this study was to verify the cagA genotype of H. pylori isolates, using primers designed according to the 3′ variable region of Japanese isolates (48) and primers designed according to a consensus region (3), and to determine the prevalence of the cagA genotype and the relationship between cagA subgenotypes and peptic ulcer disease in Caucasian patients from southern Brazil.
MATERIALS AND METHODS
Patients and endoscopy method.
A total of 103 Caucasian patients (48 male and 55 female; mean age = 51.9 ± 15.4 years; range, 15 to 81 years) with dyspeptic symptoms underwent esophagogastroduodenoscopy in a private clinic in Porto Alegre, Rio Grande do Sul, Brazil. Endoscopies were performed by one of the investigators (J. C. P.-L.), and the diagnosis was based on visual examination of the stomach and the duodenum. Twenty-six patients presented with duodenal ulcer, 31 with gastric ulcer, 21 with duodenitis, and 21 with erosive gastritis, and 4 were considered to have a normal endoscopic examination. The endoscopic diagnosis was grouped into three main categories: (i) nonulcer patients, for those patients presenting only gastritis or duodenitis (n = 42); (ii) gastric ulcer patients (n = 31); and (iii) duodenal ulcer patients (n = 26). The four patients who were considered to have a normal endoscopy were excluded from analysis. During endoscopic examination, four biopsy specimens were obtained from the antrum. The specimens were examined for the presence of H. pylori by histopathology (two specimens), urease test, and PCR. Exclusion criteria were the use of nonsteroidal anti-inflammatory drugs (NSAIDs) or recent (within the previous month) intake of antibiotics. The patients were asked about documented history of peptic ulcer disease and previous anti-Helicobacter therapy. Informed consent was obtained from all participants. The Ethics Committee of our institutions granted ethical approval.
After each endoscopic examination, endoscopes were cleansed with combined manual and mechanic cleaning procedures for washing and disinfection. A new autoclaved biopsy forceps was used for each patient. PCR for detection of H. pylori was randomly performed in the last cleaning-water wash as a contamination control. All wash samples were negative for H. pylori by PCR, even when the analysis was performed after H. pylori-positive patients (data not shown).
Histological analysis.
The biopsy specimens for each patient were fixed in Formol solution, dehydrated, and embedded in paraffin wax. Sections were cut and stained with hematoxylin and eosin to grade the severity of gastritis and with Giemsa stain to detect H. pylori. The classification and grading of gastritis were made in accordance with the Sydney system (33).
Urease test.
The biopsy specimens were introduced with a sterile needle into solid urea agar and incubated at room temperature. Results were recorded up to 24 h after inoculation. If urease enzyme of H. pylori is present in the specimen, the resulting degradation of urea causes the pH to rise and the color of the gel turns from yellow to pink.
Preparation of DNA for PCR amplification.
The biopsy specimens for DNA analysis were placed in 0.85% NaCl, and the DNA was isolated directly from the biopsy specimens using the QIAamp tissue kit (Qiagen Inc., Santa Clarita, Calif.) according to the manufacturer's instructions. The DNA was eluted in 200 μl of elution buffer, and 10 μl was used for the amplification reaction.
H. pylori detection by PCR.
PCR primer sets specific for the H. pylori 16S rRNA (15) and ureA (6) genes were used. The primer pair HPU18 and HPU54 was designed by Clayton et al. (6) and optimized by Furuta et al. (15). The primers HPU18N (5′-CCCATTTGACTCAATGCGATG-3′) and HPU54N (5′-TGGGATTAGCGAGTATGTCGG-3′) amplify a 132-bp product from the 16S rRNA gene. To confirm H. pylori identification, another PCR test, with primers annealing to the H. pylori urease structural gene subunit A (ureA) (6), was performed. The primers employed, UREA1 (5′-GCCAATGGTAAATTAGTT-3′) and UREA2 (5′-CTCCTTAATTGTTTTTAC-3′) amplify a 394-bp product from the ureA gene. PCRs were performed in a volume of 50 μl containing 20 nmol of Tris-HCl (pH 8.4) per liter, 50 mmol of KCl per liter, 2.5 (for 16S rRNA) and 1.5 (for ureA) mmol of MgCl2 per liter, 200 μmol of deoxynucleoside triphosphate per liter, 2.5 U of Taq DNA polymerase (GIBCO BRL, Grand Island, N.Y.), and 25 pmol of both forward and reverse primers. PCR was performed in a PTC-1196 DNA thermocycler (MJ Research Inc., Watertown, Mass.) under the following conditions: 5 min of preincubation at 94°C followed by 35 cycles of 1 min at 94°C, 1 min at 55°C (for 16S rRNA) or 45°C (for ureA), and 1 min at 72°C. Final extension was performed for 7 min at 72°C. The 16S rRNA and ureA amplimers (20-μl aliquots) were examined by electrophoresis on 3% and 2% agarose gels according to standard procedures (36). The results were considered positive when the product that was equivalent to the fragment described was found. Negative and positive H. pylori controls, as well as water as a negative contamination PCR control, were included in each assay. PCR amplification was performed in duplicate for each DNA sample.
cagA detection by PCR.
The primers 5′-ACCCTAGTCGGTAATGGGTTA-3′ (CAG1) and 5′-GTAATTGTCTAGTTTCGC-3′ (CAG2) (48) were used to amplify the 3′ region of the cagA gene. These primers were designed to amplify the variable 3′ region of the cagA gene in a Japanese population. Depending on the type and number of repeats present in the sequence of the gene, the products were classified into fragment A (sizes ranging from 600 to 650 bp), fragment C (around 800-bp products), and fragments B/D (around 750 bp). Another primer set for a constant region near the 3′ end of cagA described by Bukanov and Berg (3) was also used to detect the cagA gene. These forward and reverse primers, CagA/ConF (5′-GTGCCTGCTAGTTTGTCAGCG-3′) and CagA/Con-R (5′-TTGGAAACCACCTTTTGTATTAGC-3′), amplify a 402-bp fragment. PCRs were performed in a volume of 50 μl containing 20 nmol of Tris-HCl (pH 8.4) per liter, 50 mmol of KCl per liter, 2.5 (for CagA/Con) and 1.5 (for CAG1/2) mmol of MgCl2 per liter, 200 μmol of deoxynucleoside triphosphate per liter, 2.5 U of Taq DNA polymerase (GIBCO BRL), and 25 pmol of both forward and reverse primers. PCR was performed in a PTC-1196 DNA thermocycler (MJ Research Inc.) under the following conditions for the CAG1/2 amplification: 35 cycles of 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C. For the CagA/Con amplification, each reaction mixture was amplified for 35 cycles as follows: 30 s at 94°C, 30 s at 50°C, and 30 s at 72°C. A preincubation of 3 min at 94°C and a final extension for 7 min at 72°C were performed in both cagA detection procedures. PCR amplification was performed in duplicate for each DNA sample, and positive and negative controls were included in each assay. The products of amplification were examined by electrophoresis on 2% agarose gels according to standard procedures (36).
Statistical analysis.
Data were analyzed by using SPSS software, version 6.1.3 (SPSS Inc., Chicago, Ill.). Pearson's chi-square and Fisher exact tests were used to assess relationships between categorical variables. Analysis of variance with post hoc tests (Tukey's) was used for statistical comparisons of continuous variables among the diagnostic groups. Relative risk and 95% confidence intervals (CI) were calculated by using EPI-INFO's version 6.04 Statcalc. Significance was defined as P values of <0.05.
RESULTS
Patients.
H. pylori was analyzed in biopsy specimens of 103 Caucasian Brazilian patients who underwent endoscopy. Fifty-seven (55%) patients had peptic ulcer disease: 26 (25%) with duodenal ulcer and 31 (30%) with gastric ulcer. Twenty-one (20%) of the patients had gastritis only and 21 (20%) had duodenitis; these patients were classified as nonulcer cases (n = 42; 41%). Four patients (4%) were normal and since they were H. pylori negative, they were excluded from the statistical analysis. Patients with gastric ulcer were significantly older than the nonulcer cases (P = 0.02) (Table 1).
TABLE 1.
Characteristics of the study groups and prevalence of H. pylori infection evaluated by different methods in patients with peptic ulcer and in nonulcer patients
| Characteristic | Clinical statusa (n)
|
P value | |||
|---|---|---|---|---|---|
| Nonulcer
|
Peptic ulcer
|
||||
| G (21) | D (21) | GU (31) | DU (26) | ||
| Mean age, yr (±SD) | 48.6 (±16.9) | 46.1 (±12.3) | 59.2 (±13.6) | 50.8 (±15.3) | 0.02 |
| Sex ratio (M/F)b | 7/14 | 14/7 | 13/18 | 14/12 | 0.06 |
| No. (%) positive for H. pylori infection as determined by: | |||||
| Histology | 9 (43) | 14 (67) | 27 (87) | 15 (63) | |
| Urease test | 8 (38) | 13 (62) | 21 (68) | 16 (67) | |
| ureA PCR | 9 (43) | 10 (48) | 20 (65) | 19 (73) | |
| 16S rRNA PCR | 10 (48) | 13 (62) | 28 (90) | 24 (92) | |
Abbreviations: G, gastritis; D, duodenitis; GU, gastric ulcer; DU, duodenal ulcer.
M, male; F, female.
Prevalence of H. pylori infection.
PCR with primers for the 16S rRNA gene was performed on DNA extracted directly from biopsy specimens of all 103 patients. A total of 75 (75%) samples were positive, including 24 (92%) of the 26 patients with duodenal ulcer, 28 (90%) of the 31 patients with gastric ulcer, and 23 (55%) of the 42 patients in the nonulcer group. PCR using primers for the ureA gene showed that all of the 28 samples negative for the 16S rRNA primers were also negative for ureA, but of the 75 16S rRNA-positive samples only 58 were ureA positive. The analysis of the 17 16S rRNA-positive and ureA-negative samples showed that 11 were also cagA positive. On histological examination, 66 patients were positive for H. pylori, 34 were negative, and in 3 patients the examination was not possible. Using the urease test, 58 patients were positive, 41 were negative, and the test was not performed in 4 patients. H. pylori status was defined according to the agreement of 16S rRNA positivity and two or more of the diagnostic tests used. However, three patients were positive only by 16S rRNA PCR, and they were considered H. pylori positive because of the cagA positivity, as determined by two different assays. With the PCR using 16S rRNA as the “gold standard,” the sensitivity of the ureA PCR was 77.3% (95% CI, 65.0 to 85.9), the urease test sensitivity was 78.1% (95% CI, 68.6 to 87.6), and the histology sensitivity was 83.3% (95% CI, 74.7 to 91.9). The specificity of the ureA PCR was 100% (95% CI, 85.0 to 100.0), the urease test specificity was 96.2% (95% CI, 88.9 to 100.0), and the histology sensitivity was 78.6% (95% CI, 63.9 to 93.6).
The prevalence of H. pylori in patients with duodenal ulcer (24 of 26; 92%) and gastric ulcer (28 of 31; 90%) was found to be significantly higher than in the nonulcer group (23 of 42; 55%; χ2 for trend = 14.0; P < 0.001). The relative risk for duodenal ulcer in patients positive for H. pylori was 5.36 (95% CI, 1.39 to 20.63; P = 0.003) and the relative risk for gastric ulcer in H. pylori-positive patients was 4.03 (95% CI, 1.37 to 11.86; P = 0.003), when compared to H. pylori-negative patients.
Detection of cagA.
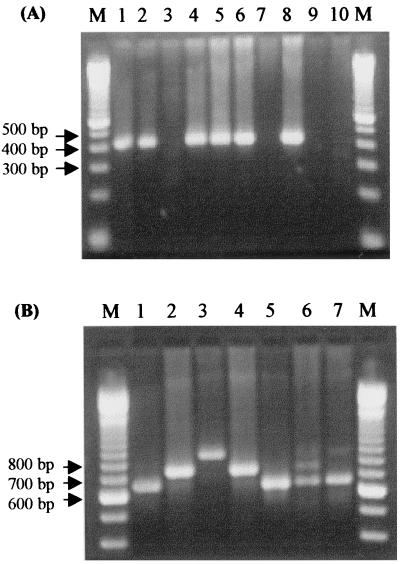
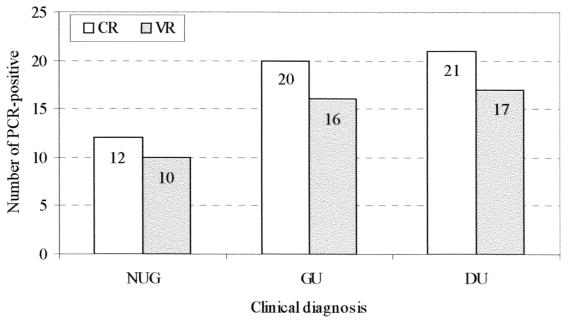
The cagA gene was detected in 43 (57%) samples among 75 H. pylori samples positive for 16S rRNA by PCR when the 3′ variable sequence primers were used, while 53 (71%) samples were cagA positive using the consensus sequence primers. According to the presence of repeat sequences in the 3′ variable region, as determined by the analysis of the molecular size of the PCR products (17), the following amplification products were observed: 28 (65%) patients were subtype A (products ranging from 600 to 650 bp), 5 (12%) patients were subtypes B or D (around 750 bp), and 2 (5%) patients showed subtype C (around 850 bp). Since the subtypes B and D have the same PCR product length and can be distinguished only by sequencing, they were grouped together. Eight (18%) patients presented two or three subgenotypes: three (7%) patients had A, B/D, and C genotypes; three (7%) patients had A and B/D; one (2%) patient had C and B/D; and another (2%) patient had A and C genotypes. PCR products of the type A subgenotype were always more intense on gel electrophoresis than B/D or C in cases with mixed strain infections. The amplification products of the PCR methods using the consensus region primers (400 bp) or the variable region primers (ranging from 600 to 850 bp) are shown in Fig. 1. The consensus region primers could detect 10 patients that were cagA negative by the technique using the variable region primers. Four of the patients belonged to the duodenal ulcer group, four belonged to the gastric ulcer group, and two belonged to the nonulcer group (Fig. 2).
FIG. 1.
Analysis of the cagA gene by PCR using primers for the consensus region (A) and for the variable 3′ region (B). (A) PCR products (400 bp) from positive cagA isolates (lanes 1, 2, 4, 5, and 6) and negative isolates (absence of PCR products) (lanes 3 and 7). Lane 8, positive control; and lanes 9 and 10, negative controls. (B) PCR products ranging from 650 to 850 bp from cagA-positive isolates. Fragments of 650 bp (type A) (lanes 1 and 5), fragments of 750 bp (types B/D) (lanes 2 and 4), and fragments of 850 bp (type C) (lane 3) were obtained. Lane 6, from a patient with types A, B/D, and C; lane 7, from a patient with types A and C; lane M, molecular size marker ladder, 100 bp.
FIG. 2.
PCR for cagA gene detection using primers for the consensus region (CR) or variable region (VR). NUG, nonulcer group; GU, gastric ulcer; DU, duodenal ulcer.
Relationship between cagA genotype and pathogenicity.
The cagA status of H. pylori isolates correlated with duodenal ulcer disease when the consensus sequence primers were used to detect cagA strains. The cagA gene was detected using the consensus primers in 21 of the 24 (87%) H. pylori-positive duodenal ulcer patients, in 20 of the 28 (72%) H. pylori-positive gastric ulcer patients, and in 12 of the 23 (52%) isolates from H. pylori-positive patients with gastritis only or duodenitis (P = 0.009) (Table 2). In contrast, using the variable sequence primers, 17 (71%) duodenal ulcer patients, 16 (57%) gastric ulcer patients, and 10 (43%) nonulcer patients showed the cagA gene (P = 0.16). The presence of cagA did show a significant association with duodenal ulcer disease, as opposed to gastritis only or duodenitis (nonulcer group), when the cagA gene was determined by the consensus PCR primer set. cagA-positive strains were significantly more prevalent among patients with duodenal ulcer (P = 0.02) but not among gastric ulcer patients (P = 0.26). In patients without ulcer, the prevalence of cagA-positive and cagA-negative strains was approximately equal. Table 2 shows the statistical analysis for the consensus primer amplification assay. The 3′ variable region subgenotypes (A, B or D, and C) did not show a significant association with specific gastroduodenal disease (P = 0.84). However, when patients having more than one subtype (multiple subtypes) were compared with those having a single subtype, there was a significant association between peptic ulcer and multiple cagA subgenotypes (P = 0.048). The most frequent type of cagA 3′ region was type A (81%), followed by types B/D (28%) and C (16%). In 18% of the cases, more than one subtype was observed, and in 75% of the cases with multiple subtypes, this was related to gastric ulcer. Table 3 summarizes these results.
TABLE 2.
Association between cagA status of H. pylori isolates and duodenal ulcer, gastric ulcer, and nonulcer patients
| Clinical status | n | No. with H. pylori cagAa | RRc | 95% CI | P valueb |
|---|---|---|---|---|---|
| Nonulcer | 23 | 12 (52) | |||
| Gastric ulcer | 28 | 20 (72) | 1.48 | 0.82–2.68 | 0.26 |
| Duodenal ulcer | 24 | 21 (87) | 2.97 | 1.05–8.37 | 0.02 |
cagA was analyzed by PCR using the consensus region primers.
χ2 for trend = 6.77; P = 0.009.
RR, relative risk.
TABLE 3.
Association between cagA subtypes of H. pylori isolates and duodenal ulcer, gastric ulcer, and nonulcer patients
| Clinical statusa (n) | No. (%) with cagA subtypebc
|
||||||
|---|---|---|---|---|---|---|---|
| Single subtype
|
Multiple subtypes
|
||||||
| A | B/D | C | A + B/D + C | A + B/D | B/D + C | A + C | |
| NU (10) | 7 (70) | 2 (20) | 1 (10) | ||||
| GU (16) | 8 (50) | 1 (6) | 1 (6) | 3 (19) | 2 (13) | 1 (6) | |
| DU (17) | 13 (76) | 2 (12) | 1 (6) | 1 (6) | |||
Abbreviations: NU, nonulcer; GU, gastric ulcer; DU, duodenal ulcer.
cagA was analyzed by PCR with variable region primers.
χ2 for isolated versus multiple subtypes, P = 0.048.
DISCUSSION
H. pylori infection leads to the development of chronic gastritis and is associated with peptic ulcer disease, gastric adenocarcinoma, and gastric mucosa-associated lymphoid tissue-type lymphoma (19). However, it has not been determined to what extent differences in infecting strains, characteristics of the host, interactions with other ulcerogenic factors, and environmental factors explain why most infected subjects will never be affected by peptic ulcer disease or gastric cancer (2, 17, 45). Specific H. pylori genotypes, e.g., vacA s1 cagA+ iceA1, are related to more severe morbidity, whereas others, e.g., vacA s2m2 iceA2 (cagA negative), appear to be less pathogenic. Genotyping of iceA and cagA offers the most effective combination for the identification of patients with ulcers. However, because cagA and vacA s-region genotypes are so strongly associated, the combination of iceA and vacA s type is almost as effective as the combination of iceA and cagA (44). Although the association between H. pylori-infected, cagA-positive patients and peptic ulcer disease is still controversial, most studies to date show that indeed such an association exists. Therefore, the aim of this study was to investigate the association of cagA genotype and subgenotype and clinical outcome, as well as to compare the detection of cagA positivity between an Eastern Asia-based assay and a Western sample-based assay in a subset of H. pylori-infected Caucasian Brazilian patients.
In this study, the prevalence of cagA strains detected by PCR using primers designed according to the sequence that includes the repeat sequences in the 3′ region from Japanese strains was 57%. Yamaoka et al. (48) examined the possibility that the different structural subtypes of the cagA gene (A to D) may be preferentially associated with specific H. pylori-related gastric disease, and they found that 86% of type C variants were obtained from patients with gastric cancer. In this study from Japan, the most frequent type of cagA 3′ region was type A (93%). In our study, type A was also the most frequent type encountered (81%). However, in contrast to what was found in the Japanese study, we observed a much higher frequency of the subtypes B or D (28% in our study versus 2% in the Japanese study) and C (16% versus 4%). Since the cagA subtypes were not sequenced but were determined by the analysis of the molecular weight of the PCR products, subtypes B and D, which have the same molecular weight, were grouped together.
A considerable proportion of our cases (18%) contained multiple genotypes, confirming that patients may have more than one H. pylori strain (39, 42). The proportion of specimens that have multiple strains may be underestimated if all of them have identical cagA genotypes. As described in other studies, the use of nonsterile endoscopes may be an alternative explanation for the high percentage of multiple strains (34), but it seems unlikely that this occurred in our study because we followed several steps for contamination control. Another explanation for the patients presenting with different cagA status could be the existence of subclones within the secluded strain. Enroth et al. (11) found that in subclones of H. pylori the entire cag pathogenicity island or part of it is deleted, which may affect binding capacity and virulence. Finally, the observation that on PCR amplification there seems to be a predominance of a specific cagA subgenotype (A) suggests that template competition might be occurring during the PCR amplification process and/or in mixed infections per se, and, therefore, we might have underestimated the number of mixed infections in our patients.
The subgenotypes did not show a significant correlation with specific gastroduodenal disease in our study (P = 0.84). However, there was a trend for the association of subtype C as well as subtype B/D, when analyzed as multiple subtypes, with gastric ulcer. When patients having more than one subtype (multiple subtypes) were compared with those having a single subtype, there was a significant association between peptic ulcer and multiple cagA subgenotypes (P = 0.048). However, if there was an underestimation of mixed-strain infections within our patient group, this conclusion must be reconsidered. Yamaoka et al. (48) reported that both types B and C are associated with severe atrophic gastritis, the precursor lesion of gastric cancer. In our study, six of the eight cases with multiple cagA subgenotypes were encountered in patients with gastric ulcer. In another study by Yamaoka et al. (46), they reported that strains with more than three repeat regions were associated with significantly higher scores for gastric mucosal atrophy and metaplasia than those strains with fewer repeat regions. H. pylori strains with three repeat regions were also significantly more susceptible to pH 3 than isolates with fewer repeat regions. The variation in the pattern of repeats in strains isolated from western countries and from East Asian strains may reflect different conditions within the stomach in different geographic regions and eventually the outcome disease (46). Type A may be the most prevalent subgenotype around the world, as shown in our study as well as in the study from Japan, i.e., from two widely separated geographic locations (48). However, we observed a higher frequency of the type C subgenotype (16%) than that reported previously in the analysis by Yamaoka et al. in a Japanese population (4%) (48) or in a Korean population where no type C H. pylori strains were found, although the sequences of cagA repeat regions from Korean strains were very homologous to those from Japanese strains (46). Furthermore, depending on the geographic area analyzed, there may be a variation in the subtypes that are more frequently related to symptomatic H. pylori-associated disease.
It has been suggested that the presence of repeat sequences in the 3′ region of the cagA gene may result in proteins with different immunogenic properties (7). The structural cagA variants result in larger-sized CagA proteins. In a study by Yamaoka et al. (48), the molecular weight of CagA proteins was significantly greater in strains with larger-size PCR products than in strains with smaller-size PCR fragments, and the enzyme-linked immunosorbent assay titer of serum anti-CagA antibody in patients infected with type C strains was significantly higher than that in patients infected with type A strains (48). These data suggest that cagA variants may provide new markers for other factors involved in gastric carcinogenesis, or they may be associated with higher levels of immune response, possibly influencing the outcome of H. pylori infection (48). In the near future, we will need more cagA-positive cases so that we can evaluate the prevalence of the different subtypes and determine whether there is a relationship between cagA subgenotype and H. pylori infection outcome. For cancer cases it will also be necessary to verify the cagA subgenotype and its association with neoplasia in our population.
Because the sequence of the cagA gene differs markedly from one geographic region to another (Western versus East Asia isolates) (40, 46, 47), we tested our H. pylori-positive samples using a primer set designed according to a consensus sequence of the cagA gene (3). Ten patients that were cagA negative after amplification with primers for the variable sequence were positive using primers for the consensus region, showing that some H. pylori strains from Brazil were not detectable by PCR using primers that included the repeat sequences from the Japanese isolates (48). The prevalence of cagA positivity by the former method was 71%, showing an association between cagA and disease outcome (P = 0.009). The presence of cagA showed a stronger correlation with duodenal ulcer disease (87%) than with gastritis only or duodenitis (52%; P = 0.020). In the gastric ulcer group, the prevalence of cagA positivity was 72%, and there was probably not enough statistical power to demonstrate the association of gastric ulcer and cagA (P = 0.26). The fact that the prevalence of cagA-positive and cagA-negative strains was approximately equal in patients without ulcer implies that individuals without ulcer can be infected with high-risk H. pylori genotypes. It has been speculated that patients infected with the more pathogenic strains but who do not have peptic ulcer disease may be younger than ulcer patients with these strains and they may have not yet developed the disease (44). Indeed, in our study, patients without ulcer were significantly younger than the patients with gastric ulcer (P = 0.021).
Several important aspects can influence the occurrence of peptic ulcer in patients with or without H. pylori infection (23, 31). Not all patients in whom peptic ulcer disease was diagnosed were infected with the virulent genotype of H. pylori. Conversely, the virulent genotype was found in patients without peptic ulcer disease. Since the use of NSAIDs can be directly associated with ulcer disease (22), we excluded from our study any patient with current NSAID use. Even so, we were unable to control for past use of these drugs, with H. pylori presenting in these patients as an innocent bystander. This could explain why 29% of the H. pylori-positive gastric ulcer patients were cagA negative. Conversely, patients using acid-suppressing drugs may present without peptic ulcer at endoscopy. Host factors, such as smoking, acid output, and human leukocyte antigen status, may also influence the clinical outcome of H. pylori infection and may interact with bacterial virulence factors (31). Additional bacterial virulence factors may also play an important role, such as the bacterial Leb-binding phenotype, which has been associated with the presence of cagA among clinical isolates of H. pylori (20). Regarding our sampling methods, a single biopsy specimen from the antrum was analyzed by PCR, and so sampling errors cannot be excluded. In addition, the primer sets used might be unable to detect specific Brazilian sequences. A characterization of our population may be necessary to investigate differences in the primary structure of the gene and to define subgenotype prevalence.
Considering that gastritis caused by H. pylori infection could progress to ulcer disease (38), patients with only gastritis at the time of endoscopy may still develop ulcer disease later in life. The quantity and virulence of H. pylori strains in the duodenum may play a critical role in the development of duodenal ulcer. Evidence of a high density of cagA-positive strains in the duodenum of patients with severe duodenitis is an important determinant of duodenal ulcer disease (18), and it is speculated that this is a relevant factor in the reason why duodenal ulcers develop in only a minority of H. pylori-infected subjects.
The unequal distribution of genotypes may contribute to the different prevalence rates of H. pylori-associated disease in different parts of the world (44). A recent study reported that cagA genotyping is useful for molecular epidemiological studies, showing that strains can be completely separated (East Asian and non-Asian strains) by differences in the cagA 3′ region. Yamaoka et al. (47) reported that PCR using specific Japanese cagA primers was not able to detect non-Asian strains and could be used to identify the population of origin of H. pylori isolates. In our study, we used a primer set that, although not identical to that of Yamaoka et al., was also originally designed for a Japanese population and which nevertheless allowed the identification of several positive strains with similar subgenotype patterns isolated from non-Oriental Brazilian patients. This finding suggests that it might be necessary to sequence a representative number of H. pylori isolates from individual populations in order to affirm that one specific method can be used to identify a particular population.
In summary, our results showed a cagA prevalence of 71% and a specific association with duodenal ulcer disease when cagA gene status was determined by the consensus PCR primer set, suggesting that cagA is an important marker for this disease in our country, as already observed by other investigators (13). To completely elucidate whether specific cagA subgenotypes have different prognostic implications in H. pylori-associated disease, it will be necessary to further study the sensitivity of PCR methods designed to detect different subgenotypes, and this could be achieved by sequencing a representative number of cagA-positive H. pylori isolates in our population. The identification of the most virulent H. pylori strain seems to be an important prognostic factor and may guide prophylactic anti-Helicobacter treatment to prevent peptic ulcer disease later in life or to prevent high-risk patients from developing gastric cancer.
ACKNOWLEDGMENTS
This work was supported by the Laboratory of Molecular and Cellular Biology of Netlab—Laboratório Bioclínico.
We thank Patrícia Ashton-Prolla and João Carlos Prolla for critical review of the manuscript and useful discussions. We thank Luciano P. Krug, Hermides Pinto Junior, Gefrem Nunes da Silva, and Elmo Cardoso for valuable contributions and Lúcia Pellanda Zimmer for help with statistical analyses.
REFERENCES
- 1.Atherton J C, Cao P, Peek R M., Jr Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 2.Blaser M J, Parsonnet J. Parasitism by the “slow” bacterium Helicobacter pylori leads to altered gastric homeostasis and neoplasia. J Clin Investig. 1994;94:4–8. doi: 10.1172/JCI117336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bukanov N O, Berg D E. Ordered cosmid library and high-resolution physical genetic map of H. pylori strain NCTC 11638. Mol Microbiol. 1994;11:509–523. doi: 10.1111/j.1365-2958.1994.tb00332.x. [DOI] [PubMed] [Google Scholar]
- 4.Cave D R. Transmission and epidemiology of Helicobacter pylori. Am J Med. 1996;100:12S–18S. doi: 10.1016/s0002-9343(96)80224-5. [DOI] [PubMed] [Google Scholar]
- 5.Censini S, Lange C, Xiang Z, Crabtree J E, Ghiara P, Borodovsky M, Rappuoli R, Lovacci A. cagA pathogenicity island of H. pylori encodes type 1-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clayton C L, Kleanthous H, Coates P J, Morgan D D, Tabaqchali S. Sensitive detection of H. pylori by using polymerase chain reaction. J Clin Microbiol. 1992;30:192–200. doi: 10.1128/jcm.30.1.192-200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N, Rappuoli R. Molecular characterization H. pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993;90:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cover T L. The vacuolating cytotoxin of H. pylori. Mol Microbiol. 1996;20:241–246. doi: 10.1111/j.1365-2958.1996.tb02612.x. [DOI] [PubMed] [Google Scholar]
- 9.Crabtree J E, Farmery S M, Lindley I J D, Figura N, Peichi P, Tompkins D S. CagA/cytotoxic strains of Helicobacter pylori and interleukin-8 in gastric epithelial cell lines. J Can Pathol. 1994;47:945–950. doi: 10.1136/jcp.47.10.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crabtree J E, Covacci A, Farmery S M, Xiang Z, Tompkins D S, Perry S, Lindley I J D, Rappuoli R. Helicobacter pylori induced interleukin-8 expression in gastric epithelial cells is associated with CagA positive phenotype. J Clin Pathol. 1995;48:41–45. doi: 10.1136/jcp.48.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enroth H, Nyrén O, Engstrand L. One stomach—one strain. Does Helicobacter pylori strain variation influence disease outcome? Dig Dis Sci. 1999;44:102–107. doi: 10.1023/a:1026658301825. [DOI] [PubMed] [Google Scholar]
- 12.The Eurogast Study Group. An international association between Helicobacter pylori infection and gastric cancer. Lancet. 1993;34:1359–1362. [PubMed] [Google Scholar]
- 13.Evans D G, Queiroz D M M, Mendes E N, Evans D J., Jr Helicobacter pylori cagA status s and m alleles of vacA in isolates from individuals with a variety of H. pylori-associated gastric diseases. Am J Gastroenterol. 1999;94:1525–1531. doi: 10.1128/jcm.36.11.3435-3437.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Figueiredo Soares T, Queiroz D M M, Mendes E N, Rocha G A, Oliveira A M R, Cabral M M D A, Oliveira C A. The interrelationship between Helicobacter pylori vacuolating cytotoxin and gastric carcinoma. Am J Gastroenterol. 1998;93:1841–1847. doi: 10.1111/j.1572-0241.1998.533_d.x. [DOI] [PubMed] [Google Scholar]
- 15.Furuta T, Kaneko E, Suzuki M, Arai H, Futami H. Quantitative study of Helicobacter pylori in gastric mucus by competitive PCR using synthetic DNA fragments. J Clin Microbiol. 1996;34:2421–2425. doi: 10.1128/jcm.34.10.2421-2425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham D Y, Malaty H M, Evans D G, Evans D J, Klein P D, Adma E. Epidemiology of Helicobacter pylori in an asymptomatic population in the United States. Gastroenterology. 1991;100:1495–1501. doi: 10.1016/0016-5085(91)90644-z. [DOI] [PubMed] [Google Scholar]
- 17.Gunn M C, Stephens J C, Stewart J A D, Rathbone B J, West K P. The significance of cagA and vacA subtypes of Helicobacter pylori in the pathogenesis of inflammation and peptic ulceration. J Clin Pathol. 1998;51:761–764. doi: 10.1136/jcp.51.10.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamlet A, Thoreson A C, Nilsson O, Sevennerholm A M, Olbe L. Duodenal Helicobacter pylori infection differs in cagA genotype between asymptomatic subjects and patients with duodenal ulcers. Gastroenterology. 1999;116:259–268. doi: 10.1016/s0016-5085(99)70121-6. [DOI] [PubMed] [Google Scholar]
- 19.Howden C W. Clinical expressions of Helicobacter pylori infection. Am J Med. 1996;100:27S–34S. doi: 10.1016/s0002-9343(96)80226-9. [DOI] [PubMed] [Google Scholar]
- 20.Ilver D, Arnqvist A, Ögren J, Frick I M, Kersulyte D, Incecik E T, Berg D E, Covacci A, Engstrand L, Borén T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 21.Ito Y, Azuma T, Ito S, Miyaji H, Hirai M, Yamazaki Y, Sto F, Kato T, Kohli Y, Kuriyama M. Analysis and typing of the vacA gene from cagA-positive strains of Helicobacter pylori isolated in Japan. J Clin Microbiol. 1997;35:1710–1714. doi: 10.1128/jcm.35.7.1710-1714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laine A L. Helicobacter pylori and complicated ulcer disease. Am J Med. 1996;100:52S–59S. doi: 10.1016/s0002-9343(96)80229-4. [DOI] [PubMed] [Google Scholar]
- 23.Mapstone N P, Lynch D A F, Lewis F A, Axon A T R, Tompkins D S, Dixon M F, Quirke P. Identification of Helicobacter pylori DNA in the mouths and stomachs of patients with gastritis using PCR. J Clin Pathol. 1993;46:540–543. doi: 10.1136/jcp.46.6.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mobley H L T. Defining Helicobacter pylori as a pathogen: strain heterogeneity and virulence. Am J Med. 1996;100:2S–12S. doi: 10.1016/s0002-9343(96)80223-3. [DOI] [PubMed] [Google Scholar]
- 25.Pan Z J, van der Hulst R W M, Feller M, Xiao S D, Tytgat G N J, Dankert J, van der Ende A. Equally high prevalences of infection with cagA-positive Helicobacter pylori in Chinese patients with peptic ulcer disease and those with chronic gastritis-associated dyspepsia. J Clin Microbiol. 1997;35:1344–1347. doi: 10.1128/jcm.35.6.1344-1347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park S M, Park J, Kim J G, Cho H D, Cho J H, Lee D H, Cha Y J. Infection with Helicobacter pylori expressing the cagA gene is not associated with an increased risk of developing peptic ulcer diseases in Korean patients. Scand J Gastroenterol. 1998;33:923–927. doi: 10.1080/003655298750026921. [DOI] [PubMed] [Google Scholar]
- 27.Peek R M, Jr, Miller G G, Tham K T, Pérez-Pérez G I, Cover T L, Atherton J C, Dunn G D, Blaser M J. Detection of Helicobacter pylori gene expression in human gastric mucosa. J Clin Microbiol. 1995;33:28–32. doi: 10.1128/jcm.33.1.28-32.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peek R M, Jr, Thompson S A, Donahue J P, Tham K T, Atherton J C, Blaser M J. Adherence to gastric epithelial cells of Helicobacter pylori, iceA, that is associated with clinical outcome. Proc Assoc Am Physicians. 1998;110:531–544. [PubMed] [Google Scholar]
- 29.Pereira-Lima J C, Scholl J, Pinheiro J B, Pereira-Lima L, Riemann J F. Helicobacter pylori-associated gastritis: does it play a role in functional dyspepsia? Z Gastroenterol. 1995;33:421–425. [PubMed] [Google Scholar]
- 30.Perez-Perez G J, Bhat N, Gaensbauer J, Fraser A, Taylor D N, Kuipers E J, Zhang L, You W C, Blaser M J. Country-specific constancy by age in cagA+ proportion of Helicobacter pylori infections. Int J Cancer. 1997;72:453–456. doi: 10.1002/(sici)1097-0215(19970729)72:3<453::aid-ijc13>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 31.Peura D A. Helicobacter pylori and ulcerogenesis. Am J Med. 1996;100:19S–26S. doi: 10.1016/s0002-9343(96)80225-7. [DOI] [PubMed] [Google Scholar]
- 32.Pounder R E. The prevalence of Helicobacter pylori infection in different countries. Aliment Pharmacol Ther. 1995;9:33–39. [PubMed] [Google Scholar]
- 33.Price A B. The Sydney system: histological division. J Gastroenterol Hepatol. 1991;6:209–222. doi: 10.1111/j.1440-1746.1991.tb01468.x. [DOI] [PubMed] [Google Scholar]
- 34.Roosendaal R, Kuipers E, van den Brule A J C, Peña A S, Uyterlinde A M, Walboomers J M M, Meuwissen S G M, de Graaff J. Importance of the fiberoptic endoscope cleaning procedure for detection of Helicobacter pylori in gastric biopsy specimens by PCR. J Clin Microbiol. 1994;32:1123–1126. doi: 10.1128/jcm.32.4.1123-1126.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudi J, Rudi A, Maiwald M, Kuck D, Sieg A, Stremmel W. Direct determination of Helicobacter pylori vacA genotypes and cagA gene in gastric biopsies and relationship to gastrointestinal disease. J Clin Microbiol. 1998;36:944–948. doi: 10.1111/j.1572-0241.1999.1138_a.x. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Shimoyama T, Fukuda S, Tanaka M, Mikami T, Saito Y, Munakata A. High prevalence of the cagA-positive Helicobater pylori strains in Japanese asymptomatic patients and gastric cancer patients. Scand J Gastroenterol. 1997;32:465–468. doi: 10.3109/00365529709025082. [DOI] [PubMed] [Google Scholar]
- 38.Sipponen P. Gastric cancer—a long-term consequence of Helicobacter pylori infection? Scand J Gastroenterol. 1994;29:24–27. [PubMed] [Google Scholar]
- 39.Taylor N S, Fox J G, Akopyants N S, Berg D E, Thompson N, Shames B, Yan L, Fontham E, Janney F, Hunter F M, Correa P. Long-term colonization with single and multiple strains of Helicobacter pylori assessed by DNA fingerprinting. J Clin Microbiol. 1995;33:918–923. doi: 10.1128/jcm.33.4.918-923.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tumuru M K R, Cover T L, Blaser M J. Cloning and expression of a high-molecular-mass major antigen of H. pylori: evidence of linkage of cytotoxin production. Infect Immun. 1993;61:1799–1809. doi: 10.1128/iai.61.5.1799-1809.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Ende A, Pan Z-J, Bart A, van der Hulst R W, Feller M, Xiao S-D, Tytgat G, Dankert J. cagA-positive Helicobacter pylori populations in China and The Netherlands are distinct. Infect Immun. 1998;66:1822–1826. doi: 10.1128/iai.66.5.1822-1826.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Doorn L J, Figueiredo C, Mégraud F, Pena S, Midolo P, Queiroz D M M, Carneiro F, Vanderborght B, Pegado M G F, Sanna R, De Boer W, Schneeberger P M, Correa P, Ng E K W, Atherton J, Blaser M J, Quint W G V. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology. 1999;116:823–830. doi: 10.1016/s0016-5085(99)70065-x. [DOI] [PubMed] [Google Scholar]
- 43.van Doorn L J, Figueiredo C, Sanna R, Pena S, Midolo P, Ng E K W, Atherton J C, Blaser M J, Quint W G V. Expanding allelic diversity of Helicobacter pylori vacA. J Clin Microbiol. 1998;36:2597–2603. doi: 10.1128/jcm.36.9.2597-2603.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Doorn L J, Figueiredo C, Sanna R, Plaisier A, Schneeberger P, De Boer W, Quint W. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology. 1998;115:58–66. doi: 10.1016/s0016-5085(98)70365-8. [DOI] [PubMed] [Google Scholar]
- 45.Warburton V J, Everett S, Mapstone N P, Axon A T R, Hawkey P, Dixon M F. Clinical and histological associations of cagA and vacA genotypes in Helicobacter pylori gastritis. J Clin Pathol. 1998;51:55–61. doi: 10.1136/jcp.51.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamaoka Y, El-Zimaity H M T, Gutierrez O, Figura N, Kim J G, Kodama T, Kashima K, Graham D Y. Relationship between the cagA 3′ repeat region of Helicobacter pylori, gastric histology, and susceptibility to low pH. Gastroenterology. 1999;117:342–349. doi: 10.1053/gast.1999.0029900342. [DOI] [PubMed] [Google Scholar]
- 47.Yamaoka Y, Osato M S, Sepulveda A R, Gutierrez O, Figura N, Kim J G, Kodama T, Kashima K, Graham D Y. Molecular epidemiology of Helicobacter pylori: separation of H. pylori from East Asian and non-Asian countries. Epidemiol Infect. 2000;124:91–96. doi: 10.1017/s0950268899003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamaoka Y, Kodama T, Kashima K, Graham D Y, Sepulveda A R. Variants of the 3′ region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases. J Clin Microbiol. 1998;36:2258–2263. doi: 10.1128/jcm.36.8.2258-2263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]