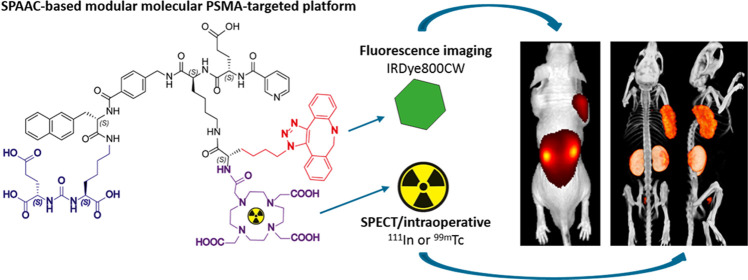
Abstract
Strain-promoted azide–alkyne cycloaddition (SPAAC) is a straightforward and multipurpose conjugation strategy. The use of SPAAC to link different functional elements to prostate-specific membrane antigen (PSMA) ligands would facilitate the development of a modular platform for PSMA-targeted imaging and therapy of prostate cancer (PCa). As a first proof of concept for the SPAAC chemistry platform, we synthesized and characterized four dual-labeled PSMA ligands for intraoperative radiodetection and fluorescence imaging of PCa. Ligands were synthesized using solid-phase chemistry and contained a chelator for 111In or 99mTc labeling. The fluorophore IRDye800CW was conjugated using SPAAC chemistry or conventional N-hydroxysuccinimide (NHS)–ester coupling. Log D values were measured and PSMA specificity of these ligands was determined in LS174T-PSMA cells. Tumor targeting was evaluated in BALB/c nude mice with subcutaneous LS174T-PSMA and LS174T wild-type tumors using μSPECT/CT imaging, fluorescence imaging, and biodistribution studies. SPAAC chemistry increased the lipophilicity of the ligands (log D range: −2.4 to −4.4). In vivo, SPAAC chemistry ligands showed high and specific accumulation in s.c. LS174T-PSMA tumors up to 24 h after injection, enabling clear visualization using μSPECT/CT and fluorescence imaging. Overall, no significant differences between the SPAAC chemistry ligands and their NHS-based counterparts were found (2 h p.i., p > 0.05), while 111In-labeled ligands outperformed the 99mTc ligands. Here, we demonstrate that our newly developed SPAAC-based PSMA ligands show high PSMA-specific tumor targeting. The use of click chemistry in PSMA ligand development opens up the opportunity for fast, efficient, and versatile conjugations of multiple imaging moieties and/or drugs.
Introduction
Prostate cancer (PCa) is the second most common cancer in men worldwide, leading to substantial morbidity and mortality.1 About 90% of PCa patients have a localized tumor at initial screening and are candidates for surgery.2 The prostate is located between critical structures, and as a consequence, the surgeon has to perform very narrow tumor resections.3,4 In approximately 15–65% of PCa patients, dependent on the disease stage, tumor resection is incomplete, caused by positive surgical tumor margins upon removal of the primary tumor and/or incomplete removal of tumor positive lymph nodes in the pelvis.5,6 Innovative approaches to improve intraoperative tumor detection can increase the chance of complete surgical resection of all tumor tissue.
To specifically detect prostate cancer cells during surgery, prostate-specific membrane antigen (PSMA) targeting ligands conjugated to one or multiple imaging moieties can be used.7,8 One of these imaging moieties is a fluorophore. Fluorescence imaging allows direct visualization of tumor tissue during surgery.9 This enables a more precise removal of the primary tumor with less positive surgical margins as a result. Another approach is radioguided surgery using a γ-emitting radionuclide, which allows for an intraoperative detection of deeper-seated tumor lesions and metastatic lymph nodes.10 Moreover, dual-labeling of PSMA-targeting ligands can provide a powerful combination of the two complementary modalities mentioned above.4 Therefore, we focused on fluorescence imaging combined with the radionuclide detection of PCa using dual-labeled PSMA-targeting ligands.
To develop a versatile platform that enables easy synthesis of PSMA ligands, strain-promoted azide–alkyne cycloaddition (SPAAC, N3-DBCO) can be used, a well-known form of click chemistry.11−13 Click chemistry reactions proceed with ease under mild nontoxic conditions (i.e., at room temperature in water) tolerating the presence of a wide range of functional groups. Hence, strain-promoted cycloadditions provide fast and highly efficient chemistry to link PSMA ligands to different functional elements.11,14 Nonetheless, there is still uncertainty about the influence of the hydrophobic functional group dibenzocyclooctene that is introduced with SPAAC on the pharmacokinetics and nonspecific uptake of the ligands.11,15,16 Therefore, the aim of the current study is to develop dual-labeled small-molecule PSMA ligands that can be conjugated to different functional components using SPAAC chemistry (N3-DBCO) and to evaluate the effects of the hydrophobic dibenzocyclooctene group on the binding affinity, pharmacokinetics, and biodistribution of the ligand.
As a first proof of concept for the SPAAC chemistry-based PSMA-targeting platform, we synthesized PSMA ligands that were conjugated with the near-infrared (NIR) fluorophore IRDye800CW using SPAAC chemistry. Moreover, a chelator for either technetium-99m (99mTc) or indium-111 (111In) radiolabeling was added. We compared the SPAAC-based PSMA-targeting ligands with similar ligands where conventional N-hydroxysuccinimide (NHS)–ester coupling of IRDye800CW was used. With these in hand, we could determine the effect of conjugation strategy (SPAAC vs NHS) on the PSMA-binding affinity, biodistribution, and pharmacokinetics of these novel dual-labeled ligands. As a secondary aim, we evaluated the differences in affinity and biodistribution of the 111In-labeled ligands and their 99mTc-labeled equivalents.
Results
Design and Synthesis
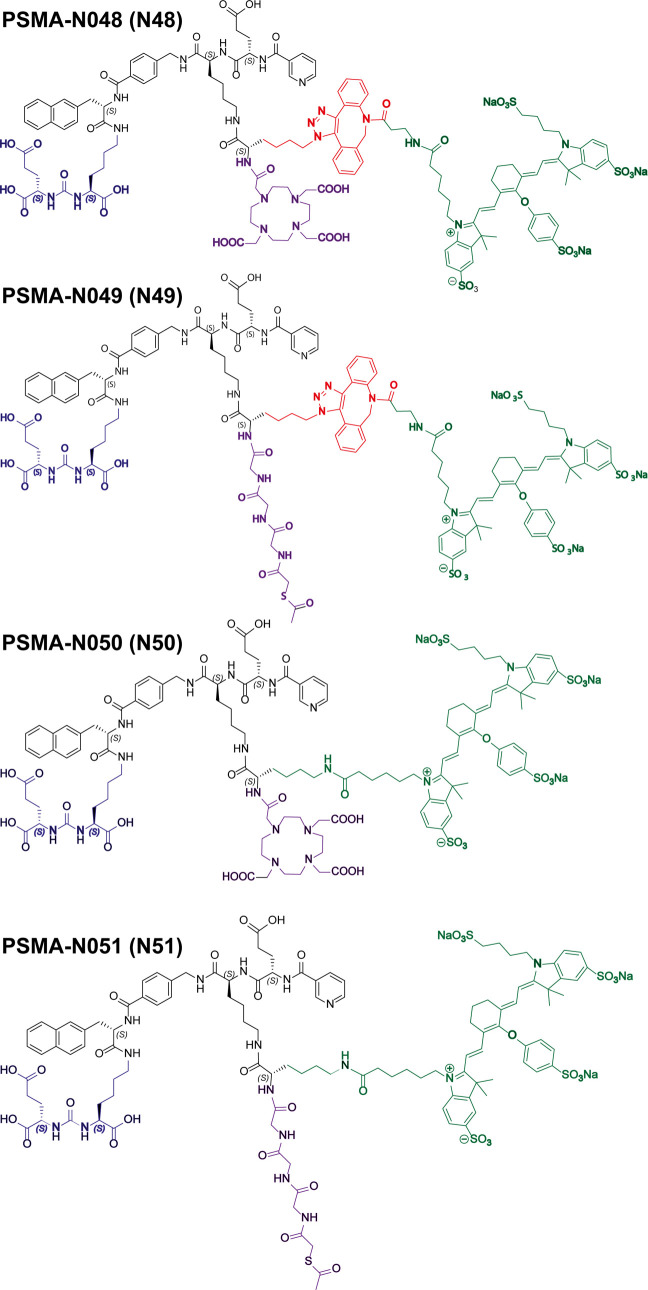
PSMA-1007 was previously reported to perfectly fit the active site as well as the entrance funnel of PSMA (Figure S4).17−19 Therefore, the design of our ligands is based on this high-affinity ligand, meaning that it consists of a naphthylalanine, aminomethyl benzoic acid, a glutamic acid, and a nicotinic acid (nonfluorinated). Importantly, the backbone of PSMA-1007 also contains two glutamic acid residues, of which the most C-terminal glutamic acid is oriented toward the exterior of PSMA (Figure S4, red circle). To synthesize dual-labeled ligands, we replaced the most C-terminal glutamic acid residue in the backbone of PSMA-1007 with a lysine. This lysine is then oriented toward the exterior of PSMA in the same manner as the side chain of the original glutamic acid is, providing space for conjugation of multiple functional moieties. To the side chain of this lysine, we added another lysine or azidolysine to introduce two groups that could be further functionalized with a metal chelator as well as a fluorophore. Most of the synthesis was performed on the solid phase, including the incorporation of a 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA) or mercaptoacetylglycylglycylglycine (MAG3) chelator. After cleavage from the resin, we conjugated all ligands in solution with IRDye800CW via either SPAAC (N3-DBCO) or NHS-ester-based chemistry. The following ligands were synthesized: PSMA-N048 (KuE-linker-DOTA-SPAAC-IRDye800CW), PSMA-N049 (KuE-linker-MAG3-SPAAC-IRDye800CW), PSMA-N050 (KuE-linker-DOTA-NHS-IRDye800CW), and PSMA-N051 (KuE-linker-MAG3-NHS-IRDye800CW) (Figure 1). The ligands are further referred to as N48, N49, N50, and N51.
Figure 1.
Chemical structures of PSMA-N048 (N48), PSMA-N049 (N49), PSMA-N050 (N50), and PSMA-N051 (N51). Ligands consisting of KuE (blue), linker (black), SPAAC (red), IRDye800CW (green), and MAG3 or DOTA chelator (purple).
Radiolabeling of the Ligands
RCY of 111In-labeled ligands (N48 and N50) exceeded 93% in all experiments (molar activity 30–33 MBq/nmol). RCY of 99mTc-labeled ligands ranged between 15 and 69% (molar activity 22–100 MBq/nmol). All ligands were purified, resulting in a radiochemical purity >95% as determined by instant thin-layer chromatography (ITLC) and high-performance liquid chromatography (HPLC). The stability of the radiolabeled ligands in human serum was determined by RP-HPLC. After incubation for 2 h at 37 °C, there were no changes in the pattern of the HPLC peaks, indicating that the radiolabel was stably coupled and there was no decomposition of the ligand (Figure S5).
In Vitro Characterization of the Ligands
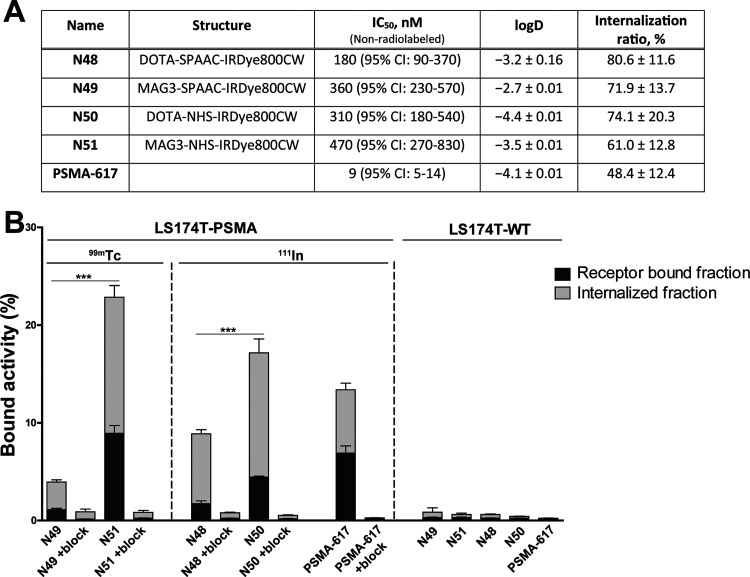
To characterize the four ligands, we first measured the distribution coefficient (log D at pH 7.4), to determine the effect of the dibenzocyclooctene group on the lipophilicity of the PSMA ligand (Figure 2A). The log D of 99mTc-N49 was −2.4 compared with −3.5 for its NHS-based equivalent 99mTc-N51 (p < 0.01). The log D of 111In-N48 (SPAAC) was −3.2 compared with −4.4 for 111In-PSMA-N50 (NHS) (p < 0.001). Hence, as expected the ligands with the dibenzocyclooctene group for SPAAC chemistry were more lipophilic. The use of MAG3 compared with DOTA as a chelator increased lipophilicity as well (p < 0.01). In addition to the lipophilicity, we determined the IC50 values of the nonradiolabeled ligands, which ranged between 184 and 475 nM (p = 0.11) (Figures 2A and S6).
Figure 2.
In vitro characterization of N48, N49, N50, and N51. (A) IC50 values of ligands as determined in competitive binding assays using LS174T-PSMA cells. IC50 values were determined using a nonradiolabeled ligand (N48, N49, N50, and N51) in competition with 111In-labeled PSMA-617. Lipophilicity of ligands expressed in log D values. Internalization ratio as determined in LS174T-PSMA cells. (B) Membrane binding and internalization kinetics of N48, N49, N50, and N51 in LS174T-PSMA-positive and -negative cells. Nonspecific binding was determined by blocking with an excess of 2-PMPA (50 μg). PSMA-617 was added as a positive control.
Next, we verified the PSMA-binding potential of our ligands in an in vitro binding and internalization assay using PSMA-positive and -negative cells, in which all four ligands showed PSMA-specific binding (Figure 2B). The use of SPAAC chemistry led to a significantly lower total binding of the ligand; 3.9 ± 0.5% for 99mTc-N49 and 8.9 ± 0.6% for 111In-N48, compared with NHS-conjugated variants; 22.9 ± 1.6% for 99mTc-N51 and 17.2 ± 3.5% for 111In-N50 (Figure 2B, p < 0.001). As a reference, we determined the total binding of 111In-PSMA-617 in our assay, which was 13.4 ± 3.5%. For all ligands, the internalized fraction ranged between 61.0 and 80.6% of the total cell-associated activity and did not differ significantly (Figure 2A). Minimal binding and internalization were observed in PSMA- negative cells or PSMA-positive cells incubated with an excess of 2-PMPA, demonstrating that, despite their higher lipophilicity, the SPAAC chemistry-based ligands do not suffer from nonspecific binding and uptake (Figure 2B).
Pharmacokinetics of SPAAC-Based Ligands
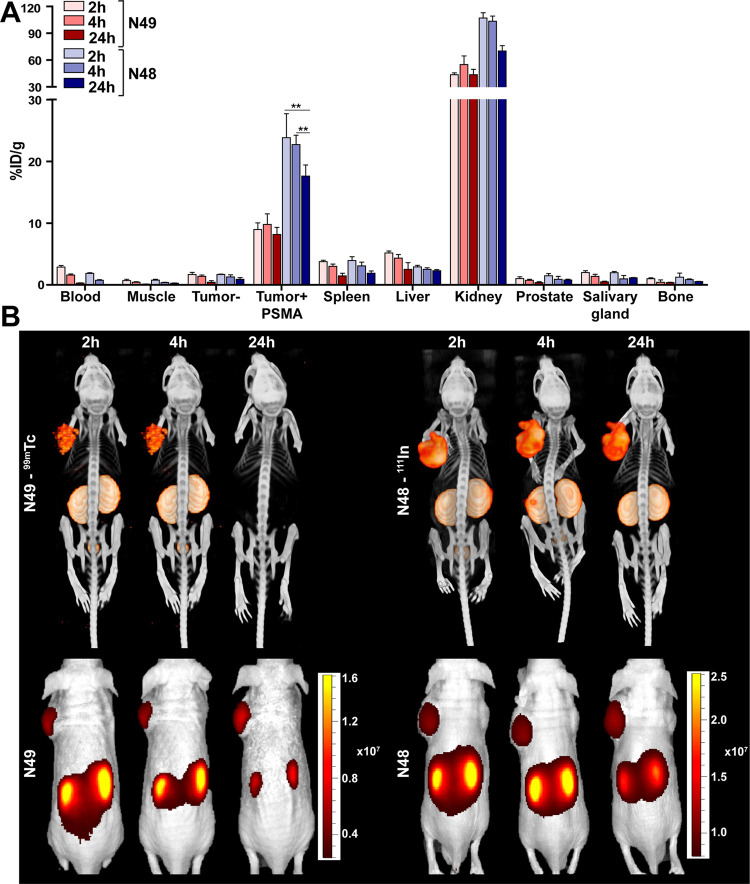
To establish the pharmacokinetics of the SPAAC-based PSMA ligands, the ex vivo biodistribution of 99mTc-N49 and 111In-N48 was determined 2, 4, and 24 h p.i. in mice bearing LS174T-PSMA-positive and -negative tumors. 99mTc-N49 showed PSMA-specific tumor uptake >8%ID/g, which was stable up to 24 h after injection (Figure 3A and Supplementary Table 1). Tumor uptake of 111In-N48 was 23.8 ± 3.9%ID/g and 22.7 ± 1.5%ID/g 2 and 4 h p.i., respectively. Yet, after 24 h p.i. tumor uptake of 111In-N48 significantly decreased to 17.6 ± 1.8%ID/g (p < 0.05). Tumor uptake of the 111In-labeled ligand N48 was significantly higher compared with the 99mTc-labeled ligand N49 at all three timepoints measured (p < 0.05 for all timepoints).
Figure 3.
In vivo pharmacokinetics of 99mTc-N49 and 111In-N48. (A) Biodistribution as determined after dissection of 99mTc-N49 (3 MBq/mouse) and 111In-N48 (10 MBq/mouse) 2, 4, and 24 h p.i. (0.3 nmol, n = 5/group). Biodistribution was determined in mice bearing subcutaneous LS174T-PSMA and LS174T wild-type xenografts. Data are expressed as %ID/g ± SD; ** indicates p < 0.01. (B) Representative μSPECT/CT scans and fluorescence images of mice with s.c. LS174T-PSMA (left) and wild-type LS174T (right) tumors after i.v. injection of 99mTc-N49 (3 MBq/mouse) and 111In-N48 (10 MBq/mouse) 2, 4, and 24 h p.i.
For both ligands, the amount of tracer present in the blood over time decreased from 2–3%ID/g (2 h p.i.) to <0.3%ID/g (24 h p.i.). Furthermore, accumulation of the two ligands in other organs including the PSMA-negative tumor, spleen, liver, prostate, and salivary glands was low (<5%ID/g) at the 2 h p.i. timepoint and further decreased over the course of 24 h, leading to high tumor-to-background ratios at all timepoints measured (Supplementary Table 1). μSPECT/CT and fluorescence imaging of the mice revealed that both ligands clearly visualized the subcutaneous LS174T-PSMA tumors (left flank) up to 4 h p.i. (Figure 3B).
Biodistribution Comparison of SPAAC- and NHS-Based Ligands
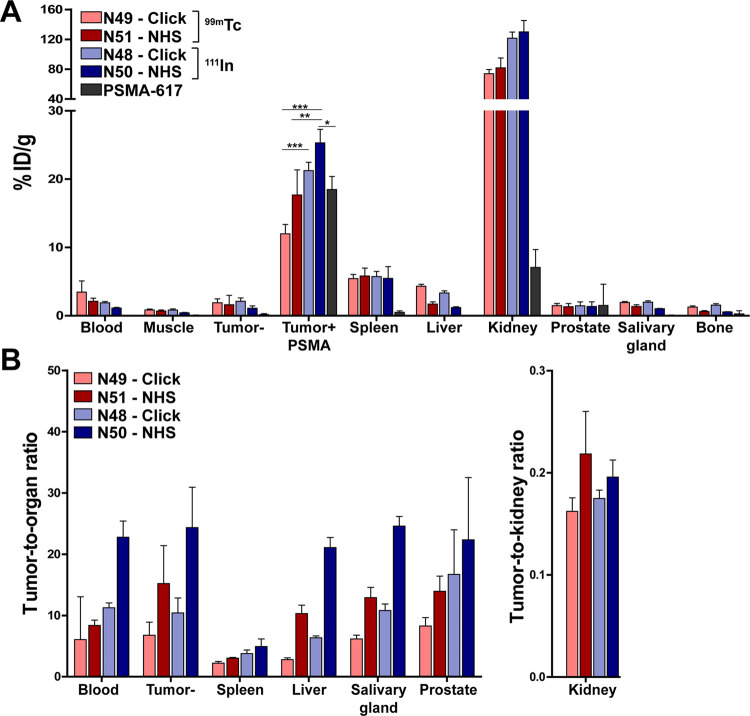
To determine the effect of conjugation chemistry (SPAAC vs NHS) on the in vivo performance of the PSMA ligands, their tumor and normal tissue uptake was determined in mice bearing both a PSMA-positive and -negative LS174T tumor 2 h after injection (Figure 4A and Supplementary Table 2). Despite the high tumor uptake of all four ligands tested, the NHS-based ligands showed higher tumor accumulation compared to their SPAAC-based counterparts (not significant, p > 0.05). Tumor uptake of N48 (SPAAC) and N50 (NHS) labeled with 111In were 21.2 ± 1.2%ID/g and 25.3 ± 2.0%ID/g, respectively. The lowest uptake in the PSMA-expressing tumors was measured for the 99mTc-labeled ligands; 12.0 ± 1.4%ID/g and 17.7 ±3.7%ID/g for N49 (SPAAC) and N51 (NHS), respectively. In our LS174T s.c. tumor model, uptake of 111In-N50 was significantly higher than both 99mTc-labeled ligands (p < 0.01) and control ligand 111In-PSMA-617 (18.5 ± 1.9%ID/g, p < 0.05). All four ligands showed a significantly higher uptake in the PSMA-positive tumor compared to the PSMA-negative tumor (p < 0.001) (Figure 4A and Supplementary Table 2).
Figure 4.
In vivo comparison of N48, N49, N50, and N51. (A) Biodistribution as determined after dissection and (B) resulting tumor-to-organ ratios of four 111In- (10 MBq/mouse) or 99mTc-labeled (15 MBq/mouse) ligands and positive control PSMA-617 (0.3 nmol, 2 h p.i., n = 5/group). Biodistribution was determined in mice bearing subcutaneous LS174T-PSMA and LS174T wild-type xenografts. Data are expressed as %ID/g ± SD; * indicates p < 0.05, ** indicates p < 0.01, and *** indicates p < 0.001.
All ligands showed rapid blood clearance and minimal uptake in muscle, bone, salivary glands, and prostate at 2 h after injection, leading to high tumor-to-organ ratios (Figure 4B and Supplementary Table 2). The SPAAC chemistry-based ligands showed a higher liver uptake compared to their NHS-based equivalents; 4.3 ± 0.3%ID/g (SPAAC) vs 1.7 ± 0.3%ID/g (NHS) for 99mTc and 3.4 ± 0.3%ID/g (SPAAC) vs 1.2 ± 0.1%ID/g (NHS) for 111In (p < 0.05). Ligand accumulation in the excretory organ, the kidneys, was similar for both conjugation strategies but lower for the 99mTc-labeled ligands (range 73.9–81.6%ID/g), compared with the 111In-labeled ligands (>120%ID/g, p < 0.001).
Multimodal Imaging of Subcutaneous PSMA-Positive Tumors
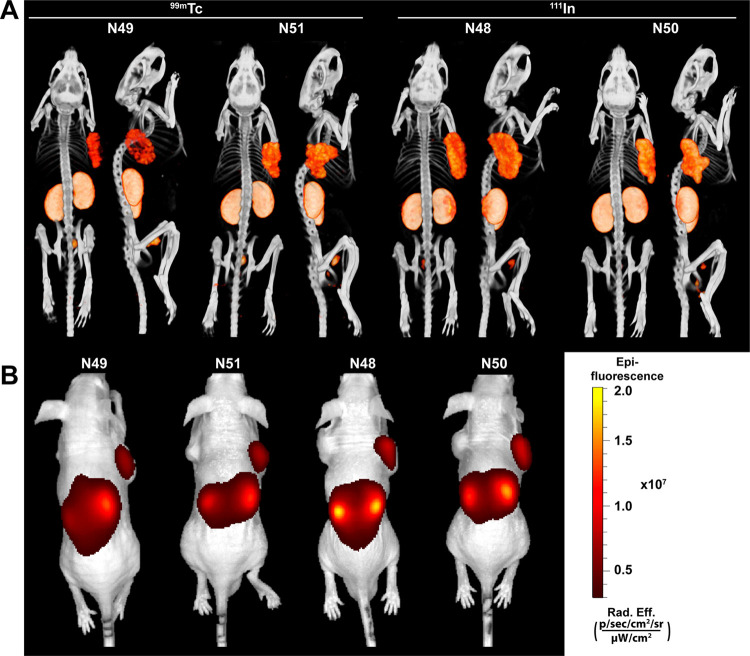
To compare the multimodal imaging potential of our SPAAC chemistry-based ligands to that of their NHS-based equivalents, we scanned mice using both a μSPECT/CT and a fluorescence scanner. Figure 5 shows representative back- and side-view μSPECT/CT images (Figure 5A) and the corresponding fluorescence images (Figure 5B) of all four ligands 2 h after injection. No large differences in imaging potential were observed between the SPAAC and NHS-based ligands. All ligands could clearly visualize the subcutaneous LS174T-PSMA tumors (right flank), without showing clearly visible uptake in the PSMA-negative LS174T tumors (left flank). In addition, multimodal images visualized high renal tracer accumulation in all mice.
Figure 5.
All ligands clearly visualize PSMA-positive tumors using both μSPECT/CT and fluorescence imaging. Representative μSPECT/CT scans (A) and fluorescence images (B) of mice with s.c. LS174T-PSMA (right) and wild-type LS174T (left) tumors after i.v. injection of 111In- (10 MBq/mouse) or 99mTc-labeled (15 MBq/mouse) ligands (0.3 nmol, 2 h p.i.).
Multimodal Imaging of Intraperitoneal Tumors
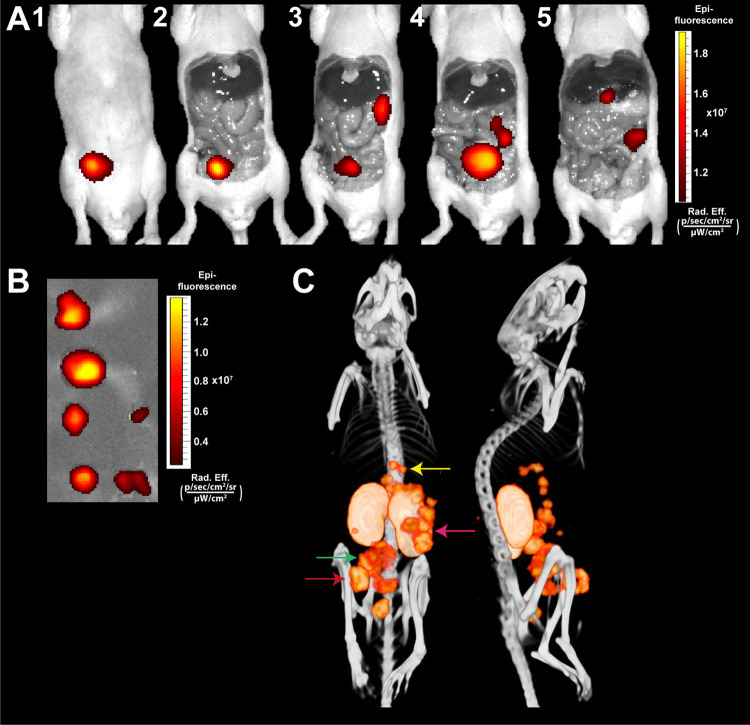
To evaluate the applicability of the dual-labeled PSMA ligands for image-guided resections in a more clinically relevant setting, we injected six mice with intraperitoneal LS174T-PSMA-positive tumors with our ligand 111In-N50. Two hours after injection of the ligand, a preoperative μSPECT/CT scan was acquired, on which multiple i.p. tumors could be visualized (Figure 6C). Next, we carried out image-guided resection of the i.p. tumors, of which Figure 6 shows one illustrative example. NIR fluorescence imaging immediately revealed a tumor at the injection site, also visible on μSPECT/CT (Figure 6A1,2,6C, red arrow). After removal of the tumor at the injection site, fluorescence imaging revealed a large tumor in the abdomen (Figure 6A3,4) and multiple tumors close to the spleen and pancreas (Figure 6A3–5), all visible on μSPECT/CT as well (Figure 6C green and pink arrows). Finally, small nodules beneath the liver were found (Figure 6A5,6C, yellow arrow). All resected tumors showed fluorescent uptake (Figure 6B). The kidneys were highly fluorescent as well, which hampered the detection of tumor lesions in close proximity to these execratory organs in mice.
Figure 6.
Multimodal fluorescence and μSPECT/CT imaging of intraperitoneal PSMA-positive tumors using 111In-N50. Mouse with several intraperitoneal LS174T-PSMA tumors located at different depths in the peritoneal cavity. (A) Same-scale NIRF images of mouse with several intraperitoneal tumors after i.v. injection of 111In-labeled N50 (0.3 nmol, 10 MBq/mouse, 2 h p.i.). (B) NIRF image of removed tumors. (C) Corresponding μSPECT/CT images in supine and left lateral positions.
Discussion
In this study, we used strain-promoted azide–alkyne cycloaddition (SPAAC) chemistry to synthesize dual-labeled PSMA-targeting ligands that could aid in the intraoperative detection and resection of PCa. The literature has indicated that the dibenzocyclooctene group, present in the ligand after SPAAC chemistry, is rather hydrophobic and could negatively impact the affinity and nonspecific binding of small PSMA-targeting ligands. Therefore, we compared the performance of our SPAAC-based ligands with conventional NHS-ester-based PSMA ligands. Withal, the SPAAC-based ligands showed similar high affinity toward PSMA in vitro, and specific uptake and retention in PSMA-positive tumors in vivo, compared to NHS-based ligands. This indicates the feasibility of SPAAC chemistry as a versatile conjugation strategy in high-affinity PSMA ligand design.
Click chemistry has been used in many research fields because of its beneficial characteristics, including high yield, high specificity, and simplicity.20 More specifically, it enables reasonably fast kinetics under aqueous conditions in the presence of a wide range of functional groups and no high temperatures are required.21 However, the dibenzocyclooctene group used in SPAAC chemistry is rather lipophilic and could alter the affinity, internalization, and nonspecific binding of small PSMA-targeting ligands in vitro.22,23 On the contrary, Wirtz et al. found that higher lipophilicity of their PSMA-I&T-based ligands is beneficial in terms of affinity and internalization, possibly because of ligand interaction with a lipophilic binding pocket of PSMA.24 log D determination in our study showed that SPAAC chemistry conjugation of IRDye800CW indeed leads to more lipophilic ligands. We determined the IC50 values of all four ligands to be in the same order of magnitude (184–475 nM). Importantly, IC50 values were determined using the nonradiolabeled ligand and without a nonradioactive metal (Re/In) present in the chelator. Complexation of a metal in the chelator could alter ligand properties (e.g., charge, hydrophilicity, etc.) and with it ligand affinity.25 Overall, the affinity of the ligands was lower than that of the high-affinity PSMA-617 ligand (IC50: 8.5 nM). Yet, other IRDye800CW conjugated PSMA ligands reported in the literature also showed higher affinities (25 nM–1.7 μM) compared to the nonfluorescent ones like PSMA-617.25,26
Despite their lower affinity, ligand uptake in the PSMA-positive tumor of our four ligands was comparable with PSMA-617. Possible explanations for this discrepancy might be the increased internalization of the dual-labeled ligands compared with PSMA-617, which could lead to increased ligand retention in the tumor. Moreover, the dual-labeled ligands show a longer blood circulatory half-life (possibly due to larger size/charge/lipophilicity) compared to PSMA-617, which could also result in increased tumor uptake (Figure 4A).27 In line with this, the study of Baranski et al. showed that the addition of a fluorophore (including IRDye800CW) also decreased PSMA ligand affinity, while it increased ligand uptake in PSMA-positive LNCaP tumors.25
Next to the addition of the dye, higher ligand lipophilicity (due to the DBCO group) was reported to increase, bus also decrease tumor uptake of various click-chemistry-based tracers. Moreover, a lipophilic character of the tracer can increase nonspecific uptake in other organs.20,22−24,28 In the case of PSMA ligands, higher ligand lipophilicity of PSMA-I&T-based ligands led to an increase in tumor uptake in LNCaP xenografts.24 In addition, Böhmer et al. developed and characterized a copper(I)-catalyzed azide–alkyne cycloaddition (CuAAC) based PSMA ligand for PET imaging called [18F]PSMA-MIC01.29 This CuAAC-based ligand showed specific tumor uptake in LNCaP xenografts (11.7 ± 4.2%ID/g, 1 h p.i.) with only minor nonspecific uptake in other organs. In our study, the SPAAC chemistry PSMA ligands similarly showed high PSMA-specific tumor uptake with low nonspecific accumulation in other organs (e.g., PSMA-negative tumor, prostate, salivary glands). Yet, as might be expected, a more than 2-fold higher liver uptake was measured for the SPAAC variants. These data are in line with the statement of Notni et al. that a lipophilic character of the ligand, induced by large hydrophobic groups (i.e., aromatics such as in dibenzocyclooctene), is prone to increase the fraction of slow hepatobiliary clearance.23
Tumor-specific uptake values of clinically used ligands such as PSMA-617, PSMA-I&T, and PSMA-1007 reported in the literature range from 5 to 13%ID/g (LNCaP, 1/2 h p.i).17,30−33 Tumor uptake of N49 (9%ID/g) was in a similar range and uptake of N48 (21%ID/g) was perhaps even higher (LS174T-PSMA, 2 h p.i). Nonetheless, no direct comparison could be made due to the use of the LS174T-PSMA xenografts in this study compared with LNCaP xenografts used in the literature. However, a direct comparison of the LNCaP and LS174T-PSMA xenograft models did not show major differences in PSMA-I&T tracer uptake between these models,32 strongly indicating that the performance of our SPAAC ligands was in a similar range to those of the clinically available ligands.33−37
For radioguided surgery, 99mTc and 111In are used because they emit γ-photons detectable with a handheld γ probe. In the present study, as a secondary aim, we evaluated differences between DOTA-based ligands suitable for 111In-labeling (N48, N50) and MAG3-based ligands for labeling with 99mTc (N49, N51). Chelation of 111In in DOTA leads to a neutral charge, whereas chelation of 99mTc in MAG3 leads to a net charge of -1, which might be advantageous since the introduction of negative charges to increase PSMA affinity and ligand uptake in PSMA-positive tumors was reported in multiple studies.27,38−40 However, the use of MAG3 compared with DOTA as a chelator increased lipophilicity, which could also influence tumor uptake and affinity of the PSMA ligand. Our results show that the 111In-DOTA ligands have a significantly higher uptake in the s.c. LS174T-PSMA-positive tumors compared with the 99mTc-MAG3 ligands. Besides favorable in vitro and in vivo properties of the 111In-labeled ligands, labeling of 99mTc in the MAG3 chelator resulted in low RCY (15–69%), which was highly variable between each labeling. In comparison, labeling with 111In always resulted in high RCY (≥93%). Furthermore, the use of MAG3 as a chelator led to more difficulties with the synthesis and overall stability of the N51 ligand (data not shown). Consequently, DOTA-based ligands might be preferred over MAG3-based variants.
As shown in the i.p. model presented in this study, tumors located deeper inside the surgical cavity may not be visualized with NIR fluorescence imaging alone due to the absorption of the emitted fluorescent light in overlying tissues (2–3 mm).41 This further emphasizes the importance of dual-labeled NIR fluorescence and radionuclide targeting agents that, in addition to preoperative tumor localization, allow intraoperative mapping of more deeply situated tumor lesions with a γ probe. The findings of the current study encourage clinical studies with PSMA-targeted dual-labeled ligands to enable image-guided complete resection of all PCa lesions during radical prostatectomy, preventing cancer recurrences and improving the chances for curative PCa surgeries.
The SPAAC-based conjugation strategy presented in this study provides a versatile platform in which PSMA ligands can easily be coupled to different chelators, fluorophores or anticancer drugs. In the future, it offers the opportunity to click various imaging moieties (e.g., fluorophores, radionuclides or MRI contrast agents) to the ligand for preclinical microscopy, as well as clinical diagnostic, pre- and intraoperative imaging of PCa. In addition, theranostic tracers could be synthesized that include therapeutic elements such as α- or β–-emitting radionuclides for radioligand therapy, photosensitizers for PSMA-targeted photodynamic therapy, or anticancer drugs including immunomodulatory agents and chemotherapeutics. Finally, moieties that improve the pharmacokinetics of the ligands could be easily incorporated. For example, albumin binders such as Evans blue could be added. In the study of Wang et al. addition of Evans blue to PSMA-617 already led to major improvements in the pharmacokinetics of the ligand (e.g., significantly higher tumor accumulation and highly radiotherapeutic efficacy).42 In addition, Kuo et al. recently showed that albumin binder optimization and use of these optimal albumin binders could lead to enhanced tumor uptake and tumor-to-kidney absorbed dose ratios in 177Lu-labeled PSMA ligands.43
Conclusions
We developed four dual-labeled ligands which all showed high PSMA affinity and excellent PSMA-specific tumor uptake. We compared an NHS and SPAAC chemistry-based approach to attach the fluorophore IRDye800CW. Overall, no significant differences between the SPAAC chemistry ligands and their NHS-based counterparts were found, while 111In-labeled ligands outperformed the 99mTc ligands. These results inspire the use of click chemistry conjugations in PSMA ligand development to enable fast, efficient, and easy coupling of various chelators, dyes, or even anticancer drugs.
Experimental Procedures
Synthesis of Ligands
We synthesized KuE-based PSMA ligands with the chelator mercaptoacetylglycylglycylglycine (MAG3) for 99mTc labeling, or the chelator 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA) for 111In labeling using standard Fmoc solid-phase chemistry. After cleavage from the resin and intermediate preparative HPLC purification, fluorophore IRDye800CW was conjugated to the ligands via either SPAAC or NHS-ester coupling in solution. Full synthetic procedures can be found in the supplementals (Page 1–6, Figures S1–S3). The ligands are further referred to as N48 (KuE-linker-DOTA-SPAAC-IRDye800CW), N49 (KuE-linker-MAG3-SPAAC-IRDye800CW), N50 (KuE-linker-DOTA-NHS-IRDye800CW), and N51 (KuE-linker-MAG3-NHS-IRDye800CW) (Figures 1 and 2A).
Cell Culture
The LS174T colon carcinoma cell line was purchased from the American Type Culture Collection. LS174T-PSMA cells were created by stable transfection with human PSMA using the plasmid pcDNA3.1-hPSMA.5 Wild-type LS174T colon carcinoma cells were used as a control. All cells were cultured in RPMI 1640 medium supplemented with 10% FCS and 2 mM glutamine (5% CO2, 37 °C). Additionally, LS174T-PSMA cells were cultured in the presence of 0.3 mg/mL G418 geneticin.
Radiolabeling
Indium-111: Ligands (1–20 μg) were radiolabeled under metal-free conditions with 111InCl3 (Curium) in 0.5 M 2-(N-morpholino)ethanesulfonic acid (MES) buffer (pH 5.5, twice volume of 111InCl3). Labeling was performed at 90 °C for 30 min.44 Ethylenediamine-tetraacetic acid (EDTA, 50 mM) was added to a final concentration of 5 mM after the incubation. Molar activity after labeling ranged from 30 to 33 MBq/nmol. Ligands were purified by a Sep-Pak C18 light cartridge (Waters) and eluted from the cartridge with 50% ethanol in water.
Technetium-99m: Ligands (1–20 μg,) were radiolabeled in 45 μL of ammonium acetate (NH4Ac, 0.25 M, pH 8) and 15 μL of freshly prepared disodium tartrate buffer (50 mg/mL in 0.25 M NH4Ac), under metal-free conditions. Ascorbic acid buffer was prepared just before labeling (3 mg/mL in 10 mM HCl). Next, 5 μL of freshly prepared stannous chloride dihydrate (SnCl2) buffer (4 μg/mL in ascorbic acid buffer) was added simultaneously with 99mTcO4– in saline, followed by incubation for 30 min at 90 °C. Molar activity after labeling ranged from 16 to 100 MBq/nmol. Ligands were purified by a Sep-Pak C18 light cartridge (Waters) and eluted from the cartridge with 50% ethanol in water.
ITLC/HPLC: Radiochemical yield (RCY) was determined by instant thin-layer chromatography (ITLC) using silica gel-coated paper (Agilent Technologies) and 0.1 M ammonium acetate containing 0.1 M EDTA pH 5.5 (111In) or 0.1 M Sodium Citrate pH 6.0 (99mTc), as the mobile phase. In addition, RCY was measured using reverse-phase high-performance liquid chromatography (RP-HPLC) on an Agilent 1200 system (Agilent Technologies) with an in-line radiodetector (Elysia-Raytest). A C18 column (5 μm, 4.6 × 250 mm2; HiChrom) was used at a flow rate of 1 mL/min. We used the following buffer system: buffer A, triethylammonium acetate (TEAA, 10 mM, pH 7); buffer B, 100% methanol; and a gradient of 97–0% buffer A (35 min).
Binding, Internalization, and IC50 Assay
Internalization assay: Binding and internalization characteristics of all ligands were compared using LS174T-PSMA and wild-type LS174T cells, cultured to confluency in six-well plates. The cells were incubated with 50 000 counts per minute (cpm) 111In- or 99mTc-labeled PSMA ligand (0.1–0.25 pmol/well) in 1 mL of binding buffer (RPMI/0.5% BSA) for 2 h at 37 °C. Nonspecific binding was determined by co-incubation with 2-(phosphonomethyl)pentane-1,5-dioic acid (2-PMPA, 21.57 μM). The cells were washed with PBS and incubated with acid buffer (0.1 M acetic acid, 154 mM NaCl, pH 2.6) for 10 min at 0 °C to retrieve the membrane-bound fraction. After this, the membrane-bound fraction was collected, the cells were washed and lysed with 1.5 mL of 0.1 M NaOH, and cell lysis (intracellular activity) was collected. Membrane-bound activity and intracellular activity fractions were measured in a γ-counter (2480 WIZARD2 Automatic γ Counter, PerkinElmer).5,45 IC50: The 50% inhibitory concentration (IC50) of the ligands was determined using LS174T-PSMA cells in a competitive binding assay. The LS174T-PSMA cells were cultured to confluency in six-well plates, followed by incubation on ice for 2 h in 1 mL of binding buffer (RPMI/0.5% BSA) with 50 000 cpm of 111In-labeled PSMA-617 and a series of increasing concentrations (0.01–300 nM) of unlabeled PSMA ligands. After incubation, the cells were washed with 2 mL of PBS and lysed with 1.5 mL of 0.1 M NaOH. Cell lysis was collected from the plate and the cell-associated activity was measured in a γ-counter and IC50 values were calculated using GraphPad Prism software version 5.03.
Lipophilicity
Log D values of all radiolabeled ligands were determined by adding 300 000 cpm (0.6–1.5 pmol) to a mixture of 3 mL of PBS (pH 7.4) and 3 mL of n-octanol. Tubes were vortexed vigorously for 1 min and centrifuged for 5 min at 201g. The concentration of radioactivity in a defined volume of each layer was measured in a well-type γ-counter.
Subcutaneous Tumor Model
Animal experiments were performed in 8- to 10-week-old male BALB/c nude mice (Janvier). The animals were housed under nonsterile conditions in individually ventilated cages (Blue line IVC, 4–5 mice per cage) with cage enrichment present and free access to water and chlorophyll-free animal chow (Sniff GmbH). The mice were subcutaneously inoculated with 3.0 × 106 LS174T-PSMA cells in the right flank and 1.5 × 106 LS174T wild-type cells in the left flank, diluted in 200 μL of complete RPMI 1640 medium. When xenografts were approximately 0.1 cm3 (10–14 days after tumor inoculation), tracers were injected intravenously in the tail vein. The biotechnicians performing the s.c. and i.v. injections were blinded for the experimental groups and tumor-bearing mice were block-randomized into groups based on tumor size. All experiments were conducted in accordance with the guidelines of the Revised Dutch Act on Animal Experimentation and approved by the institutional Animal Welfare Committee of the Radboud university medical center.
Biodistribution, Fluorescence Imaging, and μSPECT/CT Imaging
N48, N49, N50, and N51 were radiolabeled with 111In or 99mTc. Radio-HPLC chromatograms before and after Sep-Pak C18 purification show the radiochemical purity of the product before injection in mice (Figure S7). The mice were injected intravenously with 10 MBq 111In-labeled N48, N50, or PSMA-617 as control (0.3 nmol, molar activity 33.3 MBq/nmol) or 15 MBq 99mTc-labeled N49 or N50 (0.3 nmol, molar activity 50 MBq/nmol) in PBS/0.5% BSA. Two hours post injection (p.i.), the mice were euthanized by CO2/O2 asphyxiation, and images were acquired with the IVIS fluorescence imaging system (Xenogen VivoVision IVIS Lumina II, PerkinElmer), using an acquisition time of 30 s. Subsequently, μSPECT/CT images were acquired (U-SPECT II, MILabs) with a 1.0 mm diameter pinhole mouse collimator tube.46 The mice were scanned for 30 min followed by a CT scan (spatial resolution 160 μm, 65 kV, 615 μA) for anatomical reference. μSPECT/CT scans were reconstructed with MILabs reconstruction software, using an ordered-subset expectation maximization algorithm, energy windows 154–188 keV and 220–270 keV for 111In, and 126–154 keV for 99mTc, 1 iteration, 16 subsets, voxel size of 0.4 mm. μSPECT/CT scans were analyzed and maximum intensity projections (MIPs) were created using the Inveon Research Workplace software version 4.1 (Siemens Preclinical Solutions). NIRF images were analyzed using Living Image software version 4.2 (Caliper Life Sciences). Tumors, blood, and relevant organs and tissues were dissected, weighed, and radioactivity in each sample was quantified using a well-type γ-counter. The results were expressed as the percentage of injected dose per gram of tissue (%ID/g).
Pharmacokinetics
To determine the pharmacokinetics of the ligands, nine groups of five mice received an intravenous injection of 0.3 nmol 111In-labeled N48 or N50 (10 MBq/mouse, molar activity 33.3 MBq/nmol) or 99mTc-labeled N49 (5 MBq/mouse, molar activity 16.7 MBq/nmol) in PBS/0.5% BSA. At 2, 4, and 24 h p.i., the mice were euthanized followed by dissection. Tissues of interest were dissected, weighed, and measured for radioactivity in a γ-counter as described above. For each ligand, two mice from the 24 h groups underwent repeated μSPECT/CT and NIRF imaging (2, 4 and 24 h p.i.). During imaging, the mice were anesthetized with 2.5% isoflurane inhalation anesthesia and kept warm with a heating pad. Images were acquired and analyzed as described above.
Intraperitoneal Tumor Model
LS174T-PSMA cells (1.0 × 106) in 200 μL of complete RPMI 1640 medium were injected intraperitoneally and grew for 28 days after inoculation. Six male BALB/c nude mice with intraperitoneally growing LS174T-PSMA tumors were intravenously injected with 111In-labeled N50 (10 MBq and 0.3 nmol/mouse). Two hours p.i., μSPECT/CT imaging was performed preoperatively (30 min), followed by NIRF imaging of the mice in the supine position after surgical removal of skin, abdominal muscle layers, and peritoneum. After in vivo image acquisition, the visualized tumors were resected, followed by NIRF imaging to control whether residual tumor tissue was in situ.
Statistics
Statistical analyses were performed with GraphPad Prism, version 5.03. Results are presented as mean ± standard deviation (SD). Differences in in vitro affinity and tumor/organ uptake in vivo were tested for significance using a one-way ANOVA with a Tukey multiple comparison post-test. Differences were considered significant at p < 0.05, two-sided.
Acknowledgments
The authors thank Bianca Lemmers-van de Weem, Kitty Lemmens-Hermans, and Karin de Haas-Cremers for technical assistance with the animal experiments.
Glossary
Abbreviations
- 111In
indium-111
- 99mTc
technetium-99m
- DBCO
dibenzylcyclooctyne
- NHS
N-hydroxysuccinimide
- NIR
near infrared
- PCa
prostate cancer
- PSMA
prostate-specific membrane antigen
- SPAAC
strain-promoted azide–alkyne cycloaddition
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.bioconjchem.1c00537.
Supplemental methods on the synthesis of the ligands, HPLC analysis, and conjugation of the IRDye800CW; supplemental results on the crystal structure of PSMA-1007, serum stability, and IC50 of the ligands; radio-HPLCs before and after C18 purification; biodistribution and pharmacokinetics of the ligands; and the MALDI-ToF/HPLC spectra of all ligands (PDF)
This work was funded by Else Kröner-Fresenius-Stiftung (2016-A64) and the Dutch Cancer Society (NKB-KWF 10443/2016-1).
The authors declare the following competing financial interest(s): Y. Derks, D. Lwik, S. Heskamp, P. Laverman and M. Rijpkema are applicants of patent: EP21155853 - PSMA-targeting ligands for multimodal applications. No other potential conflicts of interest relevant to this article exist.
Supplementary Material
References
- Sung H.; Ferlay J.; Siegel R. L.; Laversanne M.; Soerjomataram I.; Jemal A.; Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca-Cancer J. Clin. 2021, 71, 209. 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- Wang X.; Huang S. S.; Heston W. D.; Guo H.; Wang B. C.; Basilion J. P. Development of targeted near-infrared imaging agents for prostate cancer. Mol. Cancer Ther. 2014, 13, 2595–2606. 10.1158/1535-7163.Mct-14-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütje S.; Heskamp S.; Franssen G. M.; Frielink C.; Kip A.; Hekman M.; Fracasso G.; Colombatti M.; Herrmann K.; Boerman O. C.; et al. Development and characterization of a theranostic multimodal anti-PSMA targeting agent for imaging, surgical guidance, and targeted photodynamic therapy of PSMA-expressing tumors. Theranostics 2019, 9, 2924–2938. 10.7150/thno.35274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derks Y. H. W.; Löwik D. W. P. M.; Sedelaar J. P. M.; Gotthardt M.; Boerman O. C.; Rijpkema M.; Lütje S.; Heskamp S. PSMA-targeting agents for radio- and fluorescence-guided prostate cancer surgery. Theranostics 2019, 9, 6824–6839. 10.7150/thno.36739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütje S.; Rijpkema M.; Franssen G. M.; Fracasso G.; Helfrich W.; Eek A.; Oyen W. J.; Colombatti M.; Boerman O. C. Dual-Modality Image-Guided Surgery of Prostate Cancer with a Radiolabeled Fluorescent Anti-PSMA Monoclonal Antibody. J. Nucl. Med. 2014, 55, 995–1001. 10.2967/jnumed.114.138180. [DOI] [PubMed] [Google Scholar]
- Yossepowitch O.; Briganti A.; Eastham J. A.; Epstein J.; Graefen M.; Montironi R.; Touijer K. Positive surgical margins after radical prostatectomy: a systematic review and contemporary update. Eur. Urol. 2014, 65, 303–313. 10.1016/j.eururo.2013.07.039. [DOI] [PubMed] [Google Scholar]
- Hensbergen A. W.; van Willigen D. M.; van Beurden F.; van Leeuwen P. J.; Buckle T.; Schottelius M.; Maurer T.; Wester H. J.; van Leeuwen F. W. B. Image-Guided Surgery: Are We Getting the Most Out of Small-Molecule Prostate-Specific-Membrane-Antigen-Targeted Tracers?. Bioconjugate Chem. 2020, 31, 375–395. 10.1021/acs.bioconjchem.9b00758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüstemann T.; Haberkorn U.; Babich J.; Mier W. Targeting prostate cancer: Prostate-specific membrane antigen based diagnosis and therapy. Med. Res. Rev. 2019, 39, 40–69. 10.1002/med.21508. [DOI] [PubMed] [Google Scholar]
- Lütje S.; Rijpkema M.; Helfrich W.; Oyen W. J.; Boerman O. C. Targeted radionuclide and fluorescence dual-modality imaging of cancer: preclinical advances and clinical translation. Mol. Imaging Biol. 2014, 16, 747–755. 10.1007/s11307-014-0747-y. [DOI] [PubMed] [Google Scholar]
- Rauscher I.; Eiber M.; Jilg C. A.; Gschwend J. E.; Maurer T. [PSMA-radioguided surgery in localized recurrent prostate cancer: Current and future aspects]. Urologe 2017, 56, 18–23. 10.1007/s00120-016-0275-2. [DOI] [PubMed] [Google Scholar]
- Mushtaq S.; Yun S. J.; Jeon J. Recent Advances in Bioorthogonal Click Chemistry for Efficient Synthesis of Radiotracers and Radiopharmaceuticals. Molecules 2019, 24, 3567 10.3390/molecules24193567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwe K.; Brechbiel M. W. Growing applications of “click chemistry” for bioconjugation in contemporary biomedical research. Cancer Biother. Radiopharm. 2009, 24, 289–302. 10.1089/cbr.2008.0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J.; Saxena M.; Rishi N. An Overview of Recent Advances in Biomedical Applications of Click Chemistry. Bioconjugate Chem. 2021, 32, 1455–1471. 10.1021/acs.bioconjchem.1c00247. [DOI] [PubMed] [Google Scholar]
- Rossin R.; Robillard M. S. Pretargeted imaging using bioorthogonal chemistry in mice. Curr. Opin. Chem. Biol. 2014, 21, 161–169. 10.1016/j.cbpa.2014.07.023. [DOI] [PubMed] [Google Scholar]
- Kim E.; Koo H. Biomedical applications of copper-free click chemistry: in vitro, in vivo, and ex vivo. Chem. Sci. 2019, 10, 7835–7851. 10.1039/c9sc03368h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensbergen A. W.; van Willigen D. M.; Welling M. M.; van der Wijk F. A.; de Korne C. M.; van Oosterom M. N.; Schottelius M.; Wester H. J.; Buckle T.; van Leeuwen F. W. B. Click Chemistry in the Design and Production of Hybrid Tracers. ACS Omega 2019, 4, 12438–12448. 10.1021/acsomega.9b01484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale J.; Schäfer M.; Benešová M.; Bauder-Wüst U.; Leotta K.; Eder M.; Neels O. C.; Haberkorn U.; Giesel F. L.; Kopka K. Preclinical Evaluation of (18)F-PSMA-1007, a New Prostate-Specific Membrane Antigen Ligand for Prostate Cancer Imaging. J. Nucl. Med. 2017, 58, 425–431. 10.2967/jnumed.116.181768. [DOI] [PubMed] [Google Scholar]
- Barinka C.; Byun Y.; Dusich C. L.; Banerjee S. R.; Chen Y.; Castanares M.; Kozikowski A. P.; Mease R. C.; Pomper M. G.; Lubkowski J. Interactions between human glutamate carboxypeptidase II and urea-based inhibitors: structural characterization. J. Med. Chem. 2008, 51, 7737–7743. 10.1021/jm800765e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barinka C. N. Z.; Motlova L.. X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a urea based inhibitor PSMA 1007. Protein Data Bank 2018, PDB ID: 505T.
- Takayama Y.; Kusamori K.; Nishikawa M. Click Chemistry as a Tool for Cell Engineering and Drug Delivery. Molecules 2019, 24, 172 10.3390/molecules24010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J.; Amor-Coarasa A.; Nikolopoulou A.; Kim D.; Williams C. Jr.; Ponnala S.; Babich J. W. Synthesis and pre-clinical evaluation of a new class of high-affinity (18)F-labeled PSMA ligands for detection of prostate cancer by PET imaging. Eur. J. Nucl. Med. Mol. Imaging 2017, 44, 647–661. 10.1007/s00259-016-3556-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoye N. C.; Baumeister J. E.; Khosroshahi F. N.; Hennkens H. M.; Jurisson S. S. Chelators and metal complex stability for radiopharmaceutical applications. Radiochim. Acta 2019, 107, 1087–1120. 10.1515/ract-2018-3090. [DOI] [Google Scholar]
- Notni J.; Wester H. J. A Practical Guide on the Synthesis of Metal Chelates for Molecular Imaging and Therapy by Means of Click Chemistry. Chem. - Eur. J. 2016, 22, 11500–11508. 10.1002/chem.201600928. [DOI] [PubMed] [Google Scholar]
- Wirtz M.; Schmidt A.; Schottelius M.; Robu S.; Günther T.; Schwaiger M.; Wester H. J. Synthesis and in vitro and in vivo evaluation of urea-based PSMA inhibitors with increased lipophilicity. EJNMMI Res. 2018, 8, 84 10.1186/s13550-018-0440-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranski A. C.; Schäfer M.; Bauder-Wüst U.; Roscher M.; Schmidt J.; Stenau E.; Simpfendörfer T.; Teber D.; Maier-Hein L.; Hadaschik B.; et al. PSMA-11-Derived Dual-Labeled PSMA Inhibitors for Preoperative PET Imaging and Precise Fluorescence-Guided Surgery of Prostate Cancer. J. Nucl. Med. 2018, 59, 639–645. 10.2967/jnumed.117.201293. [DOI] [PubMed] [Google Scholar]
- Kovar J. L.; Cheung L. L.; Simpson M. A.; Olive D. M. Pharmacokinetic and Biodistribution Assessment of a Near Infrared-Labeled PSMA-Specific Small Molecule in Tumor-Bearing Mice. Prostate Cancer 2014, 2014, 104248 10.1155/2014/104248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derks Y. H. W.; Rijpkema M.; Amatdjais-Groenen H. I. V.; Kip A.; Franssen G. M.; Sedelaar J. P. M.; Somford D. M.; Simons M.; Laverman P.; Gotthardt M.; et al. Photosensitizer-based multimodal PSMA-targeting ligands for intraoperative detection of prostate cancer. Theranostics 2021, 11, 1527–1541. 10.7150/thno.52166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Wu Y.; Zeng Q.; Xie T.; Yao S.; Zhang J.; Cui M. Synthesis, Preclinical Evaluation, and First-in-Human PET Study of Quinoline-Containing PSMA Tracers with Decreased Renal Excretion. J. Med. Chem. 2021, 64, 4179–4195. 10.1021/acs.jmedchem.1c00117. [DOI] [PubMed] [Google Scholar]
- Böhmer V. I.; Szymanski W.; van den Berg K. O.; Mulder C.; Kobauri P.; Helbert H.; van der Born D.; Reeβing F.; Huizing A.; Klopstra M.; et al. Modular Medical Imaging Agents Based on Azide-Alkyne Huisgen Cycloadditions: Synthesis and Pre-Clinical Evaluation of (18) F-Labeled PSMA-Tracers for Prostate Cancer Imaging. Chem. - Eur. J. 2020, 26, 10871–10881. 10.1002/chem.202001795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S. R.; Pullambhatla M.; Byun Y.; Nimmagadda S.; Foss C. A.; Green G.; Fox J. J.; Lupold S. E.; Mease R. C.; Pomper M. G. Sequential SPECT and optical imaging of experimental models of prostate cancer with a dual modality inhibitor of the prostate-specific membrane antigen. Angew. Chem., Int. Ed. 2011, 50, 9167–9170. 10.1002/anie.201102872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensbergen A. W.; Buckle T.; van Willigen D. M.; Schottelius M.; Welling M. M.; van der Wijk F. A.; Maurer T.; van der Poel H. G.; van der Pluijm G.; van Weerden W. M.; et al. Hybrid Tracers Based on Cyanine Backbones Targeting Prostate-Specific Membrane Antigen: Tuning Pharmacokinetic Properties and Exploring Dye-Protein Interaction. J. Nucl. Med. 2020, 61, 234–241. 10.2967/jnumed.119.233064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatalic K. L.; Heskamp S.; Konijnenberg M.; Molkenboer-Kuenen J. D.; Franssen G. M.; Clahsen-van Groningen M. C.; Schottelius M.; Wester H. J.; van Weerden W. M.; Boerman O. C.; et al. Towards Personalized Treatment of Prostate Cancer: PSMA I&T, a Promising Prostate-Specific Membrane Antigen-Targeted Theranostic Agent. Theranostics 2016, 6, 849–861. 10.7150/thno.14744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benešová M.; Schäfer M.; Bauder-Wüst U.; Afshar-Oromieh A.; Kratochwil C.; Mier W.; Haberkorn U.; Kopka K.; Eder M. Preclinical Evaluation of a Tailor-Made DOTA-Conjugated PSMA Inhibitor with Optimized Linker Moiety for Imaging and Endoradiotherapy of Prostate Cancer. J. Nucl. Med. 2015, 56, 914–920. 10.2967/jnumed.114.147413. [DOI] [PubMed] [Google Scholar]
- Gourni E.; Canovas C.; Goncalves V.; Denat F.; Meyer P. T.; Maecke H. R. (R)-NODAGA-PSMA: A Versatile Precursor for Radiometal Labeling and Nuclear Imaging of PSMA-Positive Tumors. PLoS One 2015, 10, e0145755 10.1371/journal.pone.0145755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourni E.; Henriksen G. Metal-Based PSMA Radioligands. Molecules 2017, 22, 523 10.3390/molecules22040523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schottelius M.; Wirtz M.; Eiber M.; Maurer T.; Wester H. J. [(111)In]PSMA-I&T: expanding the spectrum of PSMA-I&T applications towards SPECT and radioguided surgery. EJNMMI Res. 2015, 5, 68 10.1186/s13550-015-0147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weineisen M.; Schottelius M.; Simecek J.; Baum R. P.; Yildiz A.; Beykan S.; Kulkarni H. R.; Lassmann M.; Klette I.; Eiber M.; et al. 68Ga- and 177Lu-Labeled PSMA I&T: Optimization of a PSMA-Targeted Theranostic Concept and First Proof-of-Concept Human Studies. J. Nucl. Med. 2015, 56, 1169–1176. 10.2967/jnumed.115.158550. [DOI] [PubMed] [Google Scholar]
- Huang S. S.; Wang X.; Zhang Y.; Doke A.; DiFilippo F. P.; Heston W. D. Improving the biodistribution of PSMA-targeting tracers with a highly negatively charged linker. Prostate 2014, 74, 702–713. 10.1002/pros.22789. [DOI] [PubMed] [Google Scholar]
- Banerjee S. R.; Foss C. A.; Castanares M.; Mease R. C.; Byun Y.; Fox J. J.; Hilton J.; Lupold S. E.; Kozikowski A. P.; Pomper M. G. Synthesis and evaluation of technetium-99m- and rhenium-labeled inhibitors of the prostate-specific membrane antigen (PSMA). J. Med. Chem. 2008, 51, 4504–4517. 10.1021/jm800111u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uspenskaya A. A.; Nimenko E. A.; Machulkin A. E.; Beloglazkina E. K.; Majouga A. G. The importance of linkers in the structure of PSMA ligands. Curr. Med. Chem. 2021, 10.2174/0929867328666210804092200. [DOI] [PubMed] [Google Scholar]
- Kovar J. L.; Simpson M. A.; Schutz-Geschwender A.; Olive D. M. A systematic approach to the development of fluorescent contrast agents for optical imaging of mouse cancer models. Anal. Biochem. 2007, 367, 1–12. 10.1016/j.ab.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Tian R.; Niu G.; Ma Y.; Lang L.; Szajek L. P.; Kiesewetter D. O.; Jacobson O.; Chen X. Single Low-Dose Injection of Evans Blue Modified PSMA-617 Radioligand Therapy Eliminates Prostate-Specific Membrane Antigen Positive Tumors. Bioconjugate Chem. 2018, 29, 3213–3221. 10.1021/acs.bioconjchem.8b00556. [DOI] [PubMed] [Google Scholar]
- Kuo H. T.; Lin K. S.; Zhang Z.; Uribe C. F.; Merkens H.; Zhang C.; Bénard F. (177)Lu-Labeled Albumin-Binder-Conjugated PSMA-Targeting Agents with Extremely High Tumor Uptake and Enhanced Tumor-to-Kidney Absorbed Dose Ratio. J. Nucl. Med. 2021, 62, 521–527. 10.2967/jnumed.120.250738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brom M.; Joosten L.; Oyen W. J.; Gotthardt M.; Boerman O. C. Improved labelling of DTPA- and DOTA-conjugated peptides and antibodies with 111In in HEPES and MES buffer. EJNMMI Res. 2012, 2, 4. 10.1186/2191-219x-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütje S.; van Rij C. M.; Franssen G. M.; Fracasso G.; Helfrich W.; Eek A.; Oyen W. J.; Colombatti M.; Boerman O. C. Targeting human prostate cancer with 111In-labeled D2B IgG, F(ab′)2 and Fab fragments in nude mice with PSMA-expressing xenografts. Contrast Media Mol. Imaging 2015, 10, 28–36. 10.1002/cmmi.1596. [DOI] [PubMed] [Google Scholar]
- van der Have F.; Vastenhouw B.; Ramakers R. M.; Branderhorst W.; Krah J. O.; Ji C.; Staelens S. G.; Beekman F. J. U-SPECT-II: An Ultra-High-Resolution Device for Molecular Small-Animal Imaging. J. Nucl. Med. 2009, 50, 599–605. 10.2967/jnumed.108.056606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.