Abstract
DNA polymerases catalyze nucleotidyl transfer, the central reaction in synthesis of DNA polynucleotide chains. They function not only in DNA replication, but also in diverse aspects of DNA repair and recombination. Some DNA polymerases can perform translesion DNA synthesis, facilitating damage tolerance and leading to mutagenesis. In addition to these functions, many DNA polymerases conduct biochemically distinct reactions. This review presents examples of DNA polymerases that carry out nuclease (3ʹ—5′ exonuclease, 5′ nuclease, or end-trimming nuclease) or lyase (5′ dRP lyase) extracurricular activities. The discussion underscores how DNA polymerases have a remarkable ability to manipulate DNA strands, sometimes involving relatively large intramolecular movement.
Keywords: DNA polymerases, nucleotidyl transfer, DNA repair, nuclease activity, lyase activity, proofreading
Introduction
DNA polymerases have been intensively studied for decades because of their fundamental importance in DNA replication. Organisms throughout nature possess an array of polymerases encoded by their genomes, specialized for functions in DNA repair, recombination, and DNA damage tolerance. The canonical DNA polymerase reaction is the addition of a nucleotide, usually a deoxynucleoside triphosphate, to the 3′ end of a growing DNA chain, liberating pyrophosphate (Figure 1). The reverse reaction, pyrophosphorolysis, catalyzed by some DNA polymerases, is driven backwards by an excess of pyrophosphate. The purpose of this review is to highlight the existence of additional activities associated with DNA polymerases beyond canonical nucleotidyl transfer (Figure 2). We briefly summarize each activity, emphasizing recent results and unsolved issues.
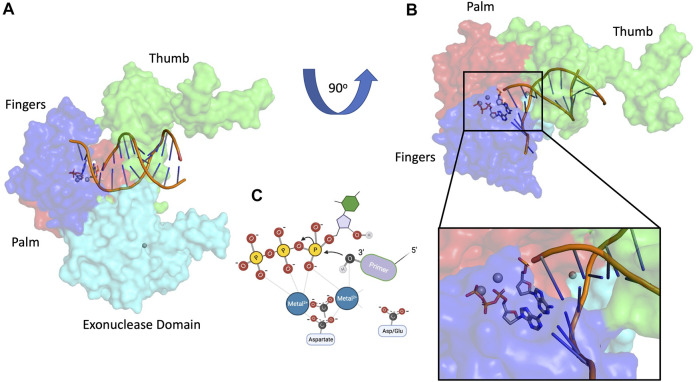
FIGURE 1.
Canonical DNA polymerase nucleotidyl transfer activity. (A) T7 DNA polymerase structure 1SKR (Li et al., 2004), (B) T7 DNA polymerase rotated 90o with magnified inset, (C) two-metal DNA polymerase mechanism. Subdomain color scheme: fingers (blue), palm (red), thumb (green), 3′-5′ exonuclease (cyan).
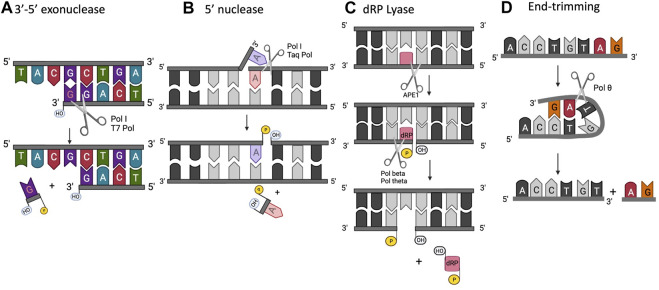
FIGURE 2.
Extracurricular functions of DNA polymerases. (A) 3′-5′ exonuclease, (B) 5′ structure-specific nuclease, (C) dRP lyase, (D) end-trimming nuclease.
3′–5′ Exonuclease Activity
All DNA polymerases share a common polymerase fold, which has been compared to a human right hand, composed of three subdomains: fingers, palm, and thumb (Steitz, 1999). In addition, the DNA polymerase toolbox can include a 3′–5′ exonuclease domain whose main role is to proofread new DNA synthesis to remove nucleotides that have been incorrectly incorporated. This reduces the error rate of DNA polymerases by one or more orders of magnitude during DNA replication (Joyce, 2013). In certain contexts, this 3′–5′ exonuclease activity is essential. Loss of editing activity results in increased incidence of cancer and pronounced genome instability (Shevelev and Hübscher, 2002; Shanbhag et al., 2018).
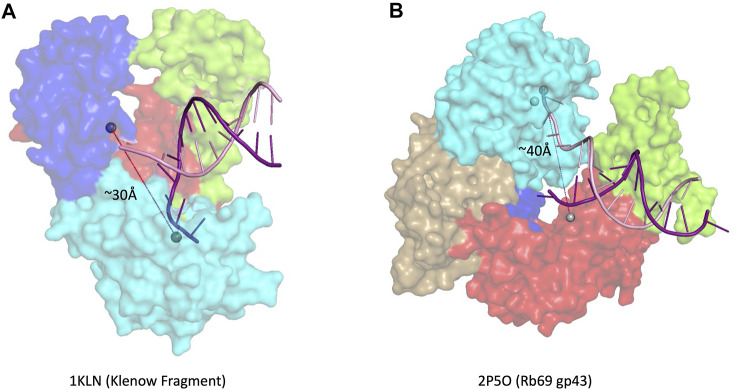
Multiple families of DNA polymerases harbor a 3′–5′ exonuclease activity: B-, C-, and D-family replicative polymerases (δ and ε in eukaryotes, Pol III in E. coli, PolD in euryarchaeota) and A-family repair polymerases, such as E. coli Pol I (Filée et al., 2002). Many A-family and some B-family polymerases have a conserved 3′–5′ exonuclease domain located in the N-terminal region of the larger polymerase domain (Khare and Eckert, 2002). In other B-family polymerases, the location of the exonuclease domain is found in a structurally distant region. The exonuclease active site in E. coli DNA polymerase I, an A-family polymerase, is ∼30 Å away from the polymerase active site (Figure 3A), whereas the distance between the two active sites is closer to ∼40 Å in B-family polymerases (Figure 3B) (Franklin et al., 2001; Hogg et al., 2004). Most B-family DNA polymerases have an extended β-hairpin loop that facilitates primer movement by holding the template strand in place as the primer switches into the exonuclease active site, a feature not found in A-family polymerases with 3′–5′ exonuclease activity (Hogg et al., 2007).
FIGURE 3.
3′–5′ exonuclease. (A) Family- A polymerase Klenow fragment with DNA in the editing site (1KLN) (Beese et al., 1993), (B) Family-B polymerase RB69 gp43 with DNA in the editing site (2P5O) (Hogg et al., 2004). Subdomain color scheme same as Figure 1, with RB69 gp43 N-terminal domain in gold.
Although homology among DNA polymerases varies significantly, the 3′–5′ exonuclease active site itself is very conserved within A-family polymerases. There are three highly conserved amino acid regions containing critical residues that coordinate two divalent metal ions, ssDNA and deoxyribonucleoside monophosphates (dNMPs) (Bernad et al., 1989; Pinz and Bogenhagen, 2000). A key catalytic step required for 3′–5′ exonuclease activity is the chelation of two metal ions by active-site aspartates and glutamates (carboxylates) (Park et al., 1997). The Exo I, Exo II, and Exo III regions are essential for the exonuclease activity, as they contain the carboxylates that directly bind the metal ions required for catalysis (Bernad et al., 1989). Recent studies have identified other residues that help facilitate catalysis in the exonuclease active site. In bacteriophage ϕ29 DNA polymerase, tyrosine, threonine, and glutamine residues help coordinate the binding of DNA to the exonuclease active site. Specifically, Tyr 101 and Thr 189 assist in melting dsDNA to form ssDNA that can bind the active site (Rodriguez et al., 2019). Although Thermus aquaticus DNA polymerase (Taq) comprises an exonuclease domain, the enzyme lacks the three aspartates and one glutamate essential for catalysis and therefore has no proofreading activity (Kim et al., 1995); nevertheless exonuclease activity can be conferred to some extent by introducing the four carboxylates into the Taq sequence (Park et al., 1997).
To ensure replication fidelity the exonuclease activity operates in a delicate balance with the polymerase activity, with the exonuclease favored when a mismatch is inserted (Doublié and Zahn, 2014). The enzyme contributes to this regulation by monitoring the newly incorporated bases. The polymerase senses a mismatch by using strategically positioned protein residues that detect the minor groove N3 and exocyclic O2 positions of incorrect base pairs after incorporation, such as Arg 429 and Gln 615 in T7 DNA polymerase, and Tyr 567 and Lys 706 in RB69 gp43, respectively (Doublié et al., 1998; Franklin et al., 2001; Hogg et al., 2004). In Family B DNA polymerase ϵ, conserved Lys 967 and Arg 988 were found essential in regulating the switching of the DNA primer between the polymerase active site and exonuclease active site. Both residues lie between the palm and thumb domains and directly interact with the minor groove (Ganai et al., 2015). When a mismatch is present, the melting temperature of the bases at the 3′ terminus is lower favoring the formation of ssDNA ends. In fact, the binding of at least 3-4 bp ssDNA in the exonuclease active site is required to favor the cleavage reaction over the polymerase reaction, (Derbyshire et al., 1988; Beese et al., 1993). The functional separation of the two active sites also helps in the regulation of the exonuclease activity, to ensure it is used sparingly and only when mismatches are introduced. To achieve proper binding in the exonuclease active site, the enzyme must translocate backwards, leading to sufficient DNA unwinding to allow the primer terminus to leave the polymerase active site and bind the exonuclease active site (Beese et al., 1993). More recent FRET experiments mapped the trajectories recorded by a DNA primer translocating from the polymerase active site to the exonuclease active site of the Klenow fragment of DNA pol I. These data revealed that the enzyme can shuttle the primer between the two sites without dissociating from DNA (intramolecular transfer) (Lamichhane et al., 2013).
In some B-family polymerases, the two domains may be more structurally distant, complicating this active site switching step. To overcome this obstacle, some polymerases can bend the DNA in the active site to bring the exonuclease domain closer to the polymerase domain, facilitating the cleavage reaction. In the bacteriophage ϕ29 DNA pol, the primer terminus moves in one dimension from the polymerization site to the exonuclease site by a 40° rotation in the helix of the DNA (Khare and Eckert, 2002; Del Prado et al., 2018). In addition to the conserved catalytic aspartates in the exonuclease domain discussed above, a non-catalytic conserved aspartate with a regulatory role has recently been identified (Del Prado et al., 2018). In ϕ29 DNA pol, this residue, Asp 121, is located between the Exo II and Exo III motifs. This Asp facilitates the binding of ssDNA substrates at the 3′–5′ exonuclease active site, but, interestingly, it does not come into contact with the metal ions or the ssDNA. Asp 121 facilitates the melting of dsDNA to form the ssDNA primer required to catalyze the cleavage reaction. Similar invariant aspartates have also been identified in other enzymes, such as Asp 150 in E. coli RNase T (Zuo and Deutscher, 2002). In bacteriophage T7 DNA pol, mutating the predicted invariant Asp90 in the active site of the exonuclease domain completely ablated the exonuclease activity of the enzyme (Del Prado et al., 2018). The proximity of Asp 90 to multiple residues required for the exonuclease reaction suggested to the authors that this residue functions in a “Romanesque vault” structure, facilitating bending of the DNA to bring the primer into the correct orientation in the active site for cleavage (Del Prado et al., 2018).
Even further redundancies and regulators have been found that facilitate the switch between polymerase activity and exonuclease activity, which is essential for replication fidelity. The switch between polymerase active site and exonuclease active site is also controlled by the concentration of dNTP present in the active site (Beechem et al., 1998). Decreased occupancy of the polymerase active site by incoming dNTP shifts the balance towards exonuclease catalysis. More recently, it was shown that phosphorylation of yeast Pol ε during replication fork stalling can also shift the equilibrium. Checkpoint kinases mediate the phosphorylation of Ser 430 in S. cerevisiae pol2, preventing primer DNA switching to exonuclease sites. This post-translational modification regulates the DNA switching between the polymerase active site and the exonuclease site, limiting fork resection and subsequent collapse (Pellicanò et al., 2021).
Beyond proofreading, polymerase 3′–5′ exonuclease activity is important in promoting virus genetic recombination, as seen in Poxviruses. Vaccinia DNA Polymerase mutants lacking 3′–5′ exonucleolytic activity showed a dramatic reduction in recombination (Gammon and Evans, 2009). The authors propose a model in which the polymerase may use 3′–5′ exonuclease activity to rescue a double-strand break (DSB) by exposing sufficient homology in ssDNA to allow annealing at the break, repair by DNA polymerase and ligase, and subsequent resumption of DNA replication (Gammon and Evans, 2009).
5′-Nuclease Activity
Some DNA polymerases, such as Taq, also possess a 5′- nuclease activity located in a separate domain (Figure 4) (Longley et al., 1990; Lyamichev et al., 1999). The 5′-nuclease activity was originally referred to as a 5′–3′ exonuclease activity, but it has since been established as a structure specific-cleavage of a 5′-ssDNA end joined to duplex DNA, a structure that is formed during lagging strand DNA synthesis (Joyce, 2013). Conserved residues in the 5’ nuclease domain (Gutman and Minton, 1993) share homology with the flap endonuclease (FEN1/XPG) family of proteins (Harrington and Lieber, 1994; Robins et al., 1994; Patel et al., 2001; Tsutakawa et al., 2011). In E. coli Pol I, the 5′-nuclease domain is tethered to the polymerase domain by an unstructured 16 amino acid peptide that is susceptible to proteolytic cleavage, allowing isolation of a Pol I “Klenow fragment” that lacks the 5′-nuclease domain (Klenow and Overgaard-Hansen, 1970). As such, the Pol I 5′-nuclease primarily operates during lagging strand synthesis to remove 5′ flaps formed when Pol I encounters Okazaki fragments during synthesis. The 5′-nuclease activity recognizes the 5′ flap and cuts between the first two bases, creating a nick on the DNA that is subsequently filled by DNA ligase (Ceska and Sayers, 1998; Xu et al., 2000).
FIGURE 4.
5′ structure specific nuclease. (A): Taq polymerase with 5′ nuclease domain shown in purple, (B): Taq polymerase rotated 180o to show 5′ nuclease residue conservation across 150 homologs (1TAQ) (Kim et al., 1995). Subdomain color scheme same as Figure 1.
DNA travels between the polymerase and 5′- nuclease domains through several conformational transitions (Joyce et al., 1992). The 5′-nuclease domain is flexible and adopts different positions within the Pol I-DNA complex to remove 5′ flaps (Ceska and Sayers, 1998). The polymerase and 5′ nuclease active sites compete for access to DNA ends, with dramatic shifts between the two active sites occurring during ongoing lagging strand synthesis. The 5’ nuclease activity also requires 2 divalent metal ions for cleavage. The selection of activity is likely intricately regulated by molecular “gates” in the active site, although details of this regulation are unknown (Joyce, 2013). However, the intricate gating mechanisms in FEN family proteins that help discern between different substrates can be used to glean insights into how 5′- nuclease is regulated in Pol I (Grasby et al., 2012; Pauszek et al., 2021).
5′-dRP Lyase Activity
Steps in base excision repair (BER) occur in a coordinated manner and enzymes are sequentially displaced to facilitate the next step in the pathway. During BER, a damaged base (for example, one modified by reactive oxygen species) is removed by a lesion-specific DNA glycosylase. This process leaves an apurinic/apyrimidinic (AP) site or abasic site that is recognized and cleaved by an AP endonuclease. This cleavage leaves a 5′-terminal deoxyribophosphate (dRP) residue that must be removed in concert with replacement of the base. In mammalian cells, Pol β is the main DNA polymerase involved in BER, and it is able to carry out both of these enzymatic steps. Pol β removes 5′- dRP and fills the gap (Matsumoto and Kim, 1995; Sobol et al., 1996; Allinson et al., 2001).
The 5′-dRP lyase activity is contained within an 8 kDa N-terminal domain of Pol β (Prasad et al., 1994; Prasad et al., 1998) (Figure 5A). A metal ion is required for release of the 5′dRP from double-stranded DNA allowing the gap in the DNA to be filled and subsequently ligated (Matsumoto and Kim, 1995).
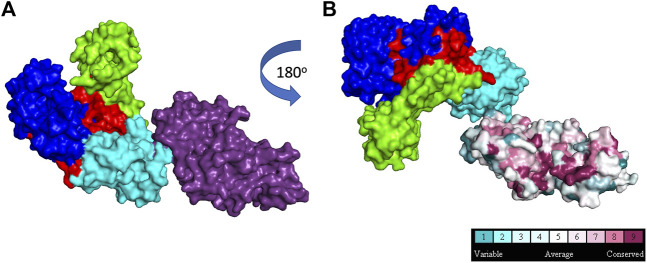
FIGURE 5.
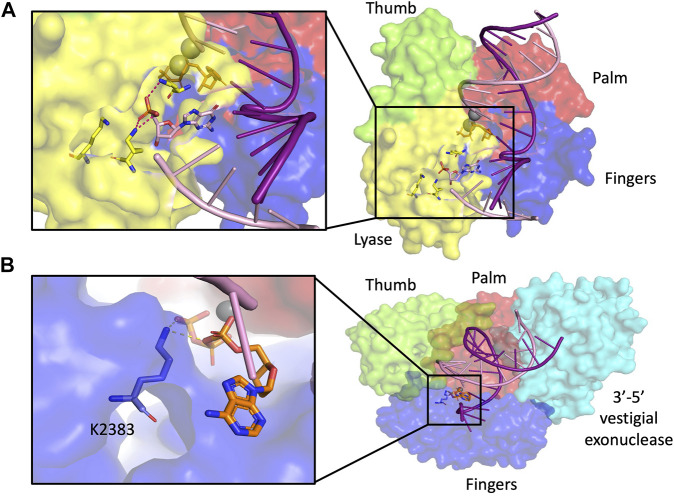
dRP lyase. (A) Pol β structure with dRP lyase residues highlighted in yellow in magnified inset (2FMS) (Batra et al., 2006), (B) Pol θ structure with dRP lyase residue highlighted in blue in magnified inset (4×0P) (Zahn et al., 2015). Subdomain color scheme same as Figure 1.
A nick adjacent to a 5′-dRP residue is a small target, and Pol β must bind non-specifically along the DNA as it searches for the right substrate to cleave. PARP1 has a high affinity for this substrate and may help recruit pol β (Khodyreva et al., 2010; Prasad et al., 2014). Recently, it was found that three lysines in the lyase active site of Pol β facilitate the search for the 5′- flap substrate on the dsDNA and destabilize non-specific DNA binding. Mutating these three lysine residues to alanines in the lyase active site of Pol β (Lys 35, Lys 68, and Lys 72), increased the binding affinity of Pol β for nonspecific DNA, indicating a role of the 5′dRP lyase in the specificity of lesion recognition (Howard et al., 2020). A tumor-associated variant in the N-terminal 8 kDa domain of Pol β retains polymerase activity but is dRP lyase deficient (Dalal et al., 2008). DNA methylating agents such as methyl methane sulfonate (MMS) create base lesions that are removed by BER. Fibroblasts of Pol β null mice are hypersensitive to MMS. This sensitivity can be rescued by complementation with the 5′dRP lyase activity of Pol β, without requiring the DNA polymerase activity (Sobol et al., 2000). In this case, another DNA polymerase such as pol δ accomplishes gap filling (Klungland and Lindahl, 1997; Fortini et al., 1998). Furthermore, Pol β 5′dRP lyase activity is suggested to be important in preventing trinucleotide repeat instability. Pol β defective in 5′-dRP lyase activity can be tightly bound to a repair site, forcing slippage that can result in deletions of trinucleotides (Lai et al., 2018).
Other polymerases also carry out 5′-dRP lyase activity. Y-Family pol iota (Pol ι) possesses 5′-dRP lyase activity and can alleviate BER deficiency in vitro (Bebenek et al., 2001; Prasad et al., 2003). Loss of Pol ι sensitizes the cells to treatment with oxidative damaging agents (Petta et al., 2008). Pol ι accumulates in sites of oxidative DNA damage and associates with BER modulator XRCC1 to facilitate repair. The recruitment of Pol ι to sites of DNA damage in human cells requires the 5-dRP lyase domain of the enzyme. It is not clear whether the dRPase of Pol ι is functionally relevant to its operation in a pathway to counteract DNA replication stress (Wang et al., 2020), involving p53-dependent reactivation of DNA replication forks (Hampp et al., 2016; Biber et al., 2020).
The A-Family polymerase Pol θ also possesses 5′-dRP lyase activity within its polymerase domain. In vitro, Pol θ can participate in BER reactions (Prasad et al., 2009). Lys 2383, a residue critical for AP lyase, is also important for the DNA polymerase activity of Pol θ (Figure 5B) (Laverty et al., 2018). The 5′-dRP lyase activity of Pol θ can also operate in structurally clustered lesions (Laverty and Greenberg, 2018). In these clustered regions, Pol β is 15–20 fold slower at excising the 5′-dRP lyase compared to Pol θ, which suggests that the two polymerases may have evolved to take care of a larger variety of substrates, with Pol β being favored for excising lesions with opposite polarity than those excised by Pol θ (Laverty and Greenberg, 2018). BER is also important for repair of oxidative lesions in mitochondrial DNA. Mitochondrial DNA polymerase y (Pol γ) also possesses 5′-dRP lyase activity, although with a much slower release of the dRP group from the enzyme compared to Pol β. In fact, the 5′-dRP lyase activity in A-family polymerases such as Pol γ, Pol θ, and Pol I is significantly slower than in Pol β, which suggests that this function may not be sufficient to complete repair of abundant AP sites (Pinz and Bogenhagen, 2000).
3′-End-Trimming and Single-Strand Extension
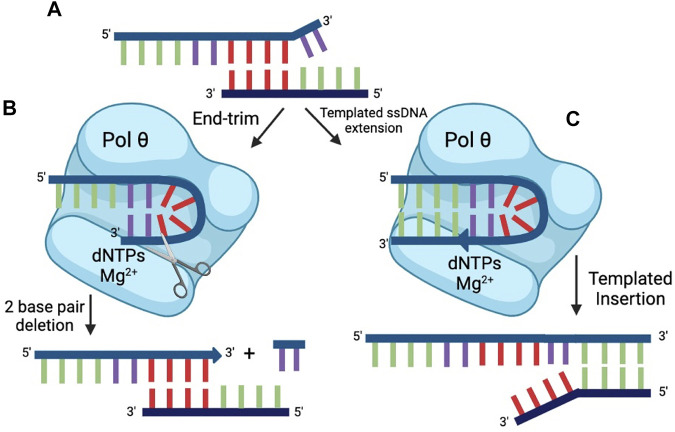
A DNA end-trimming activity, distinct from the nuclease activities discussed above, was identified in human Pol θ (Zahn et al., 2021). The rapid end-trimming activity acts on single-stranded DNA when placed in a transient and appropriate self-pairing conformation within pol θ (Figure 6). This end-trimming activity is different from 3′ to 5′ exonuclease activity. Although Pol θ contains a proofreading domain, it lacks two of the conserved amino acids necessary for 3′–5′ exonuclease activity. Further, a short loop in the exonuclease-like domain of pol θ would impede access of DNA to the exonuclease active site (Zahn et al., 2015).
FIGURE 6.
End-trimming. (A) The top strand of DNA cannot serve as a primer for successful repair because the microhomology (red) is internal. Two terminal 3′ nt (violet) are unpaired. Pol θ can process ssDNA ends by two modes: (B) Transient self-pairing of a 3′ end within pol θ (violet nt) can lead to end-trimming catalysis, resulting in removal of 2 nt (in this case). This allows the internal microhomology (red) to engage in productive priming. (C) Transient self-pairing of a 3′ end within pol θ (violet nt) can alternatively result in limited synthesis. In this case, 4 nt (green) that are complementary to an internal microhomology on the bottom strand of DNA are synthesized. The result of this process is a short, templated insertion.
This end-trimming activity shares many of the dependencies and catalytic residues that are required for the DNA polymerase reaction. For example, end-trimming is dependent on the presence of divalent cation (Mg2+ or Mn2+), on catalytic residues including invariant aspartate Asp 2330 that coordinates Mg2+ in the active site, and on the addition of dNTPs (Zahn et al., 2021). Catalysis of both extension and nuclease activity from the same active site is plausible in principle because the two-metal ion active sites of nucleotidyl transfer enzymes share close similarities with the active sites of many nucleases (Yang et al., 2006).
Experimentally, dNTP-dependent end-trimming activity is observed with some, but not all single-stranded oligonucleotides. For those substrates that are end-trimmed, a unifying feature is that the 3′ terminus is capable of potential self-pairing with a short sequence within the oligonucleotide, suggesting that a hairpin-type configuration is transiently formed within the active site of pol θ (Figure 6B). On the most-studied substrate, 2 nt are cleaved from the 3’ terminus in an endonucleolytic reaction; on some substrates 1 nt is removed; on others, possibly more than 2 nt may be end-trimmed. The end-trimming configuration appears to be unimolecular, because all substrate is quickly consumed. Further, the proposed end-trimming configuration explains another activity of pol θ, the ability to extend single-stranded DNA substrate.
Pol θ DNA polymerase can extend some single-stranded DNA molecules, an activity that is not shared by other known A-family polymerases (Hogg et al., 2012). In the presence of Mg2+, the products observed are both primed and largely templated by the same oligonucleotide (Figure 6C) (Hogg et al., 2012; Yousefzadeh et al., 2014). It was suggested that the single-stranded extension activity of Pol θ was the result of non-templated terminal transferase activity (Kent et al., 2016). However, Pol θ is not a terminal transferase. Experiments in the study suggesting terminal transferase activity were conducted using Mn2+. This metal leads to promiscuous synthesis and a high rate of mismatch incorporation, and it is known that Pol θ can extend readily from a 3′-terminal mismatch (Seki and Wood, 2008; He and Yang, 2018). Like other A family polymerases, Pol θ can add an additional dATP to a blunt DNA end, but otherwise shows predictable template requirements in a first round of DNA synthesis (He and Yang, 2018).
DNA polymerase θ is the defining enzyme in a double strand break repair pathway, termed theta-mediated end-joining (TMEJ). During TMEJ, Pol θ engages single stranded 3′ ends by microhomology. It thus makes sense that Pol θ recognizes and processes ssDNA in several ways. Pol θ apparently manipulates the 3′ end of the ssDNA in the active site, forming a transitory hairpin that serves as the intermediate for either cleavage or extension of the 3′-end (Zahn et al., 2021). The factors that dictate the choice between these two activities are being investigated, as well as how processed single-strands are then advanced to a repair stage that uses pairing at microhomologies and primed extension on double-stranded DNA. Statistically, microhomologies will rarely be found at the ends of a double-strand break but will more often occur internal to the break (Figure 6A). The two steps of processing of DNA ends and templated extension appear to largely explain the mutational “signature” of TMEJ, involving short sequence insertions and deletions at the site of repair (Schimmel et al., 2019; Carvajal-Garcia et al., 2020; Hwang et al., 2020).
Endonucleolytic cleavage of a polynucleotide chain is also a feature of RNA polymerases, which edit transcripts by cleaving off 1 or 2 nucleotides from the 3′ end (Izban and Luse, 1992; Reines, 1992; Borukhov et al., 1993). This editing reactions are conducted in a Mg2+ - dependent manner from the RNA polymerase active site. The regulation of extension vs. cleavage is enhanced by protein cofactors that participate directly in reorganizing the amino acids in the catalytic site, referred to as “active center tuning” (Sosunova et al., 2013). For example, the endonuclease activity of E. coli RNA polymerase is stimulated by Gre proteins, and eukaryotic RNA pol II by TFIIS. Stimulated by Gre, RNA polymerase clips off the two terminal nt of a synthetic RNA fragment (Sosunov et al., 2003; Sosunova et al., 2013). In contrast to the pol θ end-trimming reaction, RNA polymerase editing does not seem to be directly dNTP- or NTP-dependent, although it can be modulated by nucleotide concentration. Other nucleotidyl transferases may also engage in a form of end-trimming or editing. For example, telomerase, including highly purified human telomerase, can shorten products as well as extend them (Collins and Greider, 1993; Rubtsova et al., 2012; Fouquerel et al., 2016).
Activity on RNA
Some DNA polymerases perform nucleotidyl transferase reactions that do not involve the usual DNA substrates or primers, and are briefly mentioned here to further illustrate the enzymatic versatility of DNA polymerases.
Reverse Transcriptase
Reverse transcriptases are RNA-dependent DNA polymerases, which use an RNA template to synthesize DNA. Some DNA-dependent DNA polymerases have been reported to have reverse transcriptase activity. Two bacterial A-family polymerases, Bst DNA polymerase and the Klenow fragment of E. coli Pol I, were successfully used to reverse transcribe RNA in vitro (Shi et al., 2015). Taq DNA polymerase can also engage in this activity, after mutating a single aspartate into asparagine (Barnes et al., 2021). Human pol η can use either a DNA duplex or and RNA/DNA mixed duplex during synthesis (Franklin et al., 2004; Su et al., 2017), and was shown to function as a reverse transcriptase in a cellular environment (Su et al., 2019). Pol θ was recently reported to also have a reverse transcriptase activity, although additional studies are warranted to establish the physiological relevance of this finding (Chandramouly et al., 2021).
Primase-Polymerase
Some DNA polymerases, for example eukaryotic DNA pol α, have a separate primase subunit that allows RNA priming for DNA replication. PrimPol is the only known eukaryotic DNA polymerase with intrinsic priming activity. Both primase and DNA synthesis are mediated by the same active site. Recently it was found that a priming activity of DNA polymerase PrimPol is used for replication restart after DNA damage (Garcia-Gomez et al., 2013; Mouron et al., 2013). PrimPol is primarily a TLS polymerase which is found in both the nucleus and the mitochondria, and is capable of bypassing 7,8-dihydro-8-oxoguanine (oxoG) and cyclobutane pyrimidine dimers (CPD) (Mouron et al., 2013; Rechkoblit et al., 2021). PrimPol can synthesize short DNA primers which other polymerases, like mitochondrial polymerase γ, can use to restart replication promoting overall mitochondrial genome maintenance. Recent studies have found that PrimPol repriming activity is used by stalled replication forks to skip DNA lesions and restart replication, leaving behind ssDNA gaps that can be filled by other repair polymerases (Quinet et al., 2020; Tirman et al., 2021).
Conclusion
DNA polymerases catalyze the nucleotidyl transfer reaction to generate a DNA polymer. In practice, however, this class of enzymes is able to catalyze a host of other DNA processing reactions that make DNA polymerases a multi-tool of genomic integrity. Redundancy in the activities of DNA polymerases is critical for replication and maintenance of the cell. If a DNA polymerase is unable to perform its function, other DNA polymerases are poised to intervene and assume the role of replication and repair typically filled by the compromised enzyme. These redundancies also facilitate the sheer amount of DNA replication and repair that must continuously occur in the cell throughout its life. It must be noted that the existence of redundant polymerase roles makes targeting DNA polymerases in cancer a challenge due to the ability of the cell to compensate for the loss by employing a different polymerase.
By using a DNA polymerase to execute more than one aspect of DNA processing, the cell can benefit energetically. Requiring a separate enzyme for each step would necessitate a greater energetic commitment from the cell to synthesize and recruit these enzymes than employing an existing polymerase. Furthermore, repurposing the active site of a DNA polymerase can facilitate spatial organization of enzymatic activities. By utilizing a common active site for multiple reactions, the polymerase becomes a hub where each activity may be performed sequentially. This arrangement is advantageous to the cell as a polymerase can process DNA in multiple ways before diffusing from the DNA, further minimizing the amount of energy required to attain appropriately processed DNA.
While DNA polymerases have been investigated for close to 60 years, we continue to discover noncanonical polymerase activities. From the addition of an enzymatic domain to exploiting an existing active site, DNA polymerases act like Swiss army knives of DNA processing. We can look forward to discovering what other functions these DNA multi-tools utilize to tackle DNA processing errors. Conveniently these multiple functions present additional opportunities to design small molecule inhibitors against these multitasking enzymes.
Acknowledgments
We acknowledge Prof. Tom Steitz, whose pioneering structural insights into nucleic acid polymerases made the work presented here possible.
Author Contributions
All authors contributed to planning of the article, writing, designing figures, and editing.
Funding
Work on DNA polymerases in our laboratories is funded by National Institutes of Health grant T32 AI055402 (LD), P01 CA247773 (SD and RW), and the J. Ralph Meadows Chair in Carcinogenesis Research (RW).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Allinson S. L., Dianova I. I., Dianov G. L. (2001). DNA Polymerase Beta Is the Major dRP Lyase Involved in Repair of Oxidative Base Lesions in DNA by Mammalian Cell Extracts. EMBO J. 20 (23), 6919–6926. 10.1093/emboj/20.23.6919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes W. M., Zhang Z., Kermekchiev M. B. (2021). A Single Amino Acid Change to Taq DNA Polymerase Enables Faster PCR, Reverse Transcription and Strand-Displacement. Front. Bioeng. Biotechnol. 8. 10.3389/fbioe.2020.553474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra V. K., Beard W. A., Shock D. D., Krahn J. M., Pedersen L. C., Wilson S. H. (2006). Magnesium-induced Assembly of a Complete DNA Polymerase Catalytic Complex. Structure 14 (4), 757–766. 10.1016/j.str.2006.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebenek K., Tissier A., Frank E. G., McDonald J. P., Prasad R., Wilson S. H., et al. (2001). 5'-Deoxyribose Phosphate Lyase Activity of Human DNA Polymerase Ɩ In Vitro . Science 291(5511), 2156–2159. 10.1126/science.1058386 [DOI] [PubMed] [Google Scholar]
- Beechem J. M., Otto M. R., Bloom L. B., Eritja R., Reha-Krantz L. J., Goodman M. F. (1998). Exonuclease−Polymerase Active Site Partitioning of Primer−Template DNA Strands and Equilibrium Mg2+ Binding Properties of Bacteriophage T4 DNA Polymerase. Biochemistry 37 (28), 10144–10155. 10.1021/bi980074b [DOI] [PubMed] [Google Scholar]
- Beese L. S., Derbyshire V., Steitz T. A. (1993). Structure of DNA Polymerase I Klenow Fragment Bound to Duplex DNA. Science 260 (5106), 352–355. 10.1126/science.8469987 [DOI] [PubMed] [Google Scholar]
- Bernad A., Blanco L., Lázaro J., Martín G., Salas M. (1989). A Conserved 3′→5′ Exonuclease Active Site in Prokaryotic and Eukaryotic DNA Polymerases. Cell 59 (1), 219–228. 10.1016/0092-8674(89)90883-0 [DOI] [PubMed] [Google Scholar]
- Biber S., Pospiech H., Gottifredi V., Wiesmüller L. (2020). Multiple Biochemical Properties of the P53 Molecule Contribute to Activation of Polymerase iota-dependent DNA Damage Tolerance. Nucleic Acids Res. 48 (21), 12188–12203. 10.1093/nar/gkaa974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borukhov S., Sagitov V., Goldfarb A. (1993). Transcript Cleavage Factors from E. coli. Cell 72 (3), 459–466. 10.1016/0092-8674(93)90121-6 [DOI] [PubMed] [Google Scholar]
- Carvajal-Garcia J., Cho J.-E., Carvajal-Garcia P., Feng W., Wood R. D., Sekelsky J., et al. (2020). Mechanistic Basis for Microhomology Identification and Genome Scarring by Polymerase Theta. Proc. Natl. Acad. Sci. USA 117 (15), 8476–8485. 10.1073/pnas.1921791117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceska T., Sayers J. R. (1998). Structure-specific DNA Cleavage by 5′ Nucleases. Trends Biochem. Sci. 23 (9), 331–336. 10.1016/s0968-0004(98)01259-6 [DOI] [PubMed] [Google Scholar]
- Chandramouly G., Zhao J., McDevitt S., Rusanov T., Hoang T., Borisonnik N., et al. (2021). Pol θ Reverse Transcribes RNA and Promotes RNA-Templated DNA Repair. Sci. Adv. 7, eabf1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K., Greider C. W. (1993). Tetrahymena Telomerase Catalyzes Nucleolytic Cleavage and Nonprocessive Elongation. Genes Develop. 7 (7b), 1364–1376. 10.1101/gad.7.7b.1364 [DOI] [PubMed] [Google Scholar]
- Dalal S., Chikova A., Jaeger J., Sweasy J. B. (2008). The Leu22Pro Tumor-Associated Variant of DNA Polymerase Beta Is dRP Lyase Deficient. Nucleic Acids Res. 36 (2), 411–422. 10.1093/nar/gkm1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Prado A., Franco-Echevarría E., González B., Blanco L., Salas M., de Vega M. (2018). Noncatalytic Aspartate at the Exonuclease Domain of Proofreading DNA Polymerases Regulates Both Degradative and Synthetic Activities. Proc. Natl. Acad. Sci. USA 115 (13), E2921–E2929. 10.1073/pnas.1718787115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire V., Freemont P. S., Sanderson M. R., Beese L., Friedman J. M., Joyce C. M., et al. (1988). Genetic and Crystallographic Studies of the 3′,5′-Exonucleolytic Site of DNA Polymerase I. Science 240 (4849), 199–201. 10.1126/science.2832946 [DOI] [PubMed] [Google Scholar]
- Doublié S., Tabor S., Long A. M., Richardson C. C., Ellenberger T. (1998). Crystal Structure of a Bacteriophage T7 DNA Replication Complex at 2.2 Å Resolution. Nature 391 (6664), 251–258. 10.1038/34593 [DOI] [PubMed] [Google Scholar]
- Doublié S., Zahn K. E. (2014). Structural Insights into Eukaryotic DNA Replication. Front. Microbiol. 5, 444. 10.3389/fmicb.2014.00444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filée J., Forterre P., Sen-Lin T., Laurent J. (2002). Evolution of DNA Polymerase Families: Evidences for Multiple Gene Exchange between Cellular and Viral Proteins. J. Mol. Evol. 54 (6), 763–773. 10.1007/s00239-001-0078-x [DOI] [PubMed] [Google Scholar]
- Fortini P., Pascucci B., Parlanti E., Sobol R. W., Wilson S. H., Dogliotti E. (1998). Different DNA Polymerases Are Involved in the Short- and Long-Patch Base Excision Repair in Mammalian Cells. Biochemistry 37 (11), 3575–3580. 10.1021/bi972999h [DOI] [PubMed] [Google Scholar]
- Fouquerel E., Lormand J., Bose A., Lee H.-T., Kim G. S., Li J., et al. (2016). Oxidative Guanine Base Damage Regulates Human Telomerase Activity. Nat. Struct. Mol. Biol. 23 (12), 1092–1100. 10.1038/nsmb.3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin A., Milburn P. J., Blanden R. V., Steele E. J. (2004). Human DNA Polymerase-η, an A-T Mutator in Somatic Hypermutation of Rearranged Immunoglobulin Genes, is a Reverse Transcriptase. Immunol. Cell Biol. 82, 219–225. [DOI] [PubMed] [Google Scholar]
- Franklin M. C., Wang J., Steitz T. A. (2001). Structure of the Replicating Complex of a Pol α Family DNA Polymerase. Cell 105 (5), 657–667. 10.1016/s0092-8674(01)00367-1 [DOI] [PubMed] [Google Scholar]
- Gammon D. B., Evans D. H. (2009). The 3′-to-5′ Exonuclease Activity of Vaccinia Virus DNA Polymerase Is Essential and Plays a Role in Promoting Virus Genetic Recombination. J. Virol. 83 (9), 4236–4250. 10.1128/jvi.02255-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganai R. A., Bylund G. O., Johansson E. (2015). Switching between Polymerase and Exonuclease Sites in DNA Polymerase ε. Nucleic Acids Res. 43 (2), 932–942. 10.1093/nar/gku1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gomez S., Reyes A., Martinez-Jimenez M. I., Chocron E. S., Mouron S., Terrados G., et al. (2013). PrimPol, an Archaic Primase/Polymerase Operating in Human Cells. Mol. Cell 52, 541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasby J. A., Finger L. D., Tsutakawa S. E., Atack J. M., Tainer J. A. (2012). Unpairing and Gating: Sequence-independent Substrate Recognition by FEN Superfamily Nucleases. Trends Biochem. Sci. 37 (2), 74–84. 10.1016/j.tibs.2011.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman P. D., Minton K. W. (1993). Conserved Sites in the 5'-3' Exonuclease Domain of Escherichia coli DNA Polymerase. Nucl. Acids Res. 21 (18), 4406–4407. 10.1093/nar/21.18.4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampp S., Kiessling T., Buechle K., Mansilla S. F., Thomale J., Rall M., et al. (2016). DNA Damage Tolerance Pathway Involving DNA Polymerase ι and the Tumor Suppressor P53 Regulates DNA Replication fork Progression. Proc. Natl. Acad. Sci. USA 113 (13), E4311–E4319. 10.1073/pnas.1605828113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington J. J., Lieber M. R. (1994). The Characterization of a Mammalian DNA Structure-specific Endonuclease. EMBO J. 13 (5), 1235–1246. 10.1002/j.1460-2075.1994.tb06373.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P., Yang W. (2018). Template and Primer Requirements for DNA Pol θ-mediated End Joining. Proc. Natl. Acad. Sci. USA 115 (30), 7747–7752. 10.1073/pnas.1807329115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg M., Aller P., Konigsberg W., Wallace S. S., Doublié S. (2007). Structural and Biochemical Investigation of the Role in Proofreading of a β Hairpin Loop Found in the Exonuclease Domain of a Replicative DNA Polymerase of the B Family. J. Biol. Chem. 282 (2), 1432–1444. 10.1074/jbc.m605675200 [DOI] [PubMed] [Google Scholar]
- Hogg M., Sauer-Eriksson A. E., Johansson E. (2012). Promiscuous DNA Synthesis by Human DNA Polymerase θ. Nucleic Acids Res. 40(6), 2611–2622. 10.1093/nar/gkr1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg M., Wallace S. S., Doublié S. (2004). Crystallographic Snapshots of a Replicative DNA Polymerase Encountering an Abasic Site. EMBO J. 23 (7), 1483–1493. 10.1038/sj.emboj.7600150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard M. J., Horton J. K., Zhao M.-L., Wilson S. H. (2020). Lysines in the Lyase Active Site of DNA Polymerase β Destabilize Nonspecific DNA Binding, Facilitating Searching and DNA gap Recognition. J. Biol. Chem. 295 (34), 12181–12187. 10.1074/jbc.ra120.013547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang T., Reh S., Dunbayev Y., Zhong Y., Takata Y., Shen J., et al. (2020). Defining the Mutation Signatures of DNA Polymerase θ in Cancer Genomes. NAR Cancer 2 (3), zcaa017. 10.1093/narcan/zcaa017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izban M. G., Luse D. S. (1992). The RNA Polymerase II Ternary Complex Cleaves the Nascent Transcript in a 3'----5' Direction in the Presence of Elongation Factor SII. Genes Dev. 6 (7), 1342–1356. 10.1101/gad.6.7.1342 [DOI] [PubMed] [Google Scholar]
- Joyce C. M. (2013). “DNA Polymerase I, Bacterial,” in Encyclopedia of Biological Chemistry. Editors Lennarz W.J., Lane M.D.. Second Edition (Waltham: Academic Press; ), 87–91. 10.1016/b978-0-12-378630-2.00306-6 [DOI] [Google Scholar]
- Joyce C. M., Sun X. C., Grindley N. D. (1992). Reactions at the Polymerase Active Site that Contribute to the Fidelity of Escherichia coli DNA Polymerase I (Klenow Fragment). J. Biol. Chem. 267 (34), 24485–24500. 10.1016/s0021-9258(18)35792-2 [DOI] [PubMed] [Google Scholar]
- Kent T., Mateos-Gomez P. A., Sfeir A., Pomerantz R. T. (2016). Polymerase θ Is a Robust Terminal Transferase that Oscillates between Three Different Mechanisms during End-Joining. eLife 5, e13740. 10.7554/eLife.13740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare V., Eckert K. A. (2002). The Proofreading 3′→5′ Exonuclease Activity of DNA Polymerases: a Kinetic Barrier to Translesion DNA Synthesis. Mutat. Research/Fundamental Mol. Mech. Mutagenesis 510 (1–2), 45–54. 10.1016/s0027-5107(02)00251-8 [DOI] [PubMed] [Google Scholar]
- Khodyreva S. N., Prasad R., Ilina E. S., Sukhanova M. V., Kutuzov M. M., Liu Y., et al. (2010). Apurinic/apyrimidinic (AP) Site Recognition by the 5'-dRP/AP Lyase in poly(ADP-Ribose) Polymerase-1 (PARP-1). Proc. Natl. Acad. Sci. 107 (51), 22090–22095. 10.1073/pnas.1009182107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Eom S. H., Wang J., Lee D.-S., Suh S. W., Steitz T. A. (1995). Crystal Structure of Thermus Aquaticus DNA Polymerase. Nature 376(6541), 612–616. 10.1038/376612a0 [DOI] [PubMed] [Google Scholar]
- Klenow H., Overgaard-Hansen K. (1970). Proteolytic Cleavage of DNA Polymerase from Escherichia Coli B into an Exonuclease Unit and a Polymerase Unit. FEBS Lett. 6 (1), 25–27. 10.1016/0014-5793(70)80032-1 [DOI] [PubMed] [Google Scholar]
- Klungland A., Lindahl T. (1997). Second Pathway for Completion of Human DNA Base Excision-Repair: Reconstitution with Purified Proteins and Requirement for DNase IV (FEN1). EMBO J. 16 (11), 3341–3348. 10.1093/emboj/16.11.3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Weizmann Y., Liu Y. (2018). The Deoxyribose Phosphate Lyase of DNA Polymerase β Suppresses a Processive DNA Synthesis to Prevent Trinucleotide Repeat Instability. Nucleic Acids Res. 46 (17), 8940–8952. 10.1093/nar/gky700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane R., Berezhna S. Y., Gill J. P., Van der Schans E., Millar D. P. (2013). Dynamics of Site Switching in DNA Polymerase. J. Am. Chem. Soc. 135 (12), 4735–4742. 10.1021/ja311641b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverty D. J., Greenberg M. M. (2018). Expanded Substrate Scope of DNA Polymerase θ and DNA Polymerase β: Lyase Activity on 5′-Overhangs and Clustered Lesions. Biochemistry 57 (42), 6119–6127. 10.1021/acs.biochem.8b00911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverty D. J., Mortimer I. P., Greenberg M. M. (2018). Mechanistic Insight through Irreversible Inhibition: DNA Polymerase θ Uses a Common Active Site for Polymerase and Lyase Activities. J. Am. Chem. Soc. 140 (29), 9034–9037. 10.1021/jacs.8b04158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Dutta S., Doublié S., Bdour H. M. d., Taylor J.-S., Ellenberger T. (2004). Nucleotide Insertion Opposite a Cis-Syn Thymine Dimer by a Replicative DNA Polymerase from Bacteriophage T7. Nat. Struct. Mol. Biol. 11 (8), 784–790. 10.1038/nsmb792 [DOI] [PubMed] [Google Scholar]
- Longley M. J., Bennett S. E., Mosbaugh D. W. (1990). Characterization of the 5′ to 3′ Exonuclease Associated with Thermus aquaticus DNA Polymerase. Nucl. Acids Res. 18 (24), 7317–7322. 10.1093/nar/18.24.7317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamichev V., Brow M. A. D., Varvel V. E., Dahlberg J. E. (1999). Comparison of the 5' Nuclease Activities of Taq DNA Polymerase and its Isolated Nuclease Domain. Proc. Natl. Acad. Sci. 96 (11), 6143–6148. 10.1073/pnas.96.11.6143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y., Kim K. (1995). Excision of Deoxyribose Phosphate Residues by DNA Polymerase β during DNA Repair. Science 269 (24), 699–702. 10.1126/science.7624801 [DOI] [PubMed] [Google Scholar]
- Mouron S., Rodriguez-Acebes S., Martinez-Jimenez M. I., Garcia-Gomez S., Chocron S., Blanco L., et al. (2013). Repriming of DNA Synthesis at Stalled Replication Forks by Human PrimPol. Nat. Struct. Mol. Biol. 20, 1383–1389. [DOI] [PubMed] [Google Scholar]
- Park Y., Choi H., Lee D. S., Kim Y. (1997). Improvement of the 3'-5' Exonuclease Activity of Taq DNA Polymerase by Protein Engineering in the Active Site. Mol. Cells 7 (3), 419–424. [PubMed] [Google Scholar]
- Patel P. H., Suzuki M., Adman E., Shinkai A., Loeb L. A. (2001). Prokaryotic DNA Polymerase I: Evolution, Structure, and "base Flipping" Mechanism for Nucleotide Selection. J. Mol. Biol. 308 (5), 823–837. 10.1006/jmbi.2001.4619 [DOI] [PubMed] [Google Scholar]
- Pauszek R. F., III, Lamichhane R., Rajkarnikar Singh A., Millar D. P. (2021). Single-molecule View of Coordination in a Multi-Functional DNA Polymerase. eLife 10, e62046. 10.7554/elife.62046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicanò G., Al Mamun M., Jurado-Santiago D., Villa-Hernández S., Yin X., Giannattasio M., et al. (2021). Checkpoint-mediated DNA Polymerase ε Exonuclease Activity Curbing Counteracts Resection-Driven fork Collapse. Mol. Cell 81 (13), 2778–e4. 10.1016/j.molcel.2021.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petta T. B., Nakajima S., Zlatanou A., Despras E., Couve-Privat S., Ishchenko A., et al. (2008). Human DNA Polymerase iota Protects Cells against Oxidative Stress. EMBO J. 27 (21), 2883–2895. 10.1038/emboj.2008.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinz K. G., Bogenhagen D. F. (2000). Characterization of a Catalytically Slow AP Lyase Activity in DNA Polymerase γ and Other Family A DNA Polymerases. J. Biol. Chem. 275 (17), 12509–12514. 10.1074/jbc.275.17.12509 [DOI] [PubMed] [Google Scholar]
- Prasad R., Beard W. A., Chyan J. Y., Maciejewski M. W., Mullen G. P., Wilson S. H. (1998). Functional Analysis of the Amino-Terminal 8-kDa Domain of DNA Polymerase β as Revealed by Site-Directed Mutagenesis. J. Biol. Chem. 273(18), 11121–11126. 10.1074/jbc.273.18.11121 [DOI] [PubMed] [Google Scholar]
- Prasad R., Beard W. A., Wilson S. H. (1994). Studies of Gapped DNA Substrate Binding by Mammalian DNA Polymerase Beta. Dependence on 5'-phosphate Group. J. Biol. Chem. 269 (27), 18096–18101. 10.1016/s0021-9258(17)32422-5 [DOI] [PubMed] [Google Scholar]
- Prasad R., Bebenek K., Hou E., Shock D. D., Beard W. A., Woodgate R., et al. (2003). Localization of the Deoxyribose Phosphate Lyase Active Site in Human DNA Polymerase ι by Controlled Proteolysis. J. Biol. Chem. 278 (32), 29649–29654. 10.1074/jbc.m305399200 [DOI] [PubMed] [Google Scholar]
- Prasad R., Horton J. K., Chastain P. D., Gassman N. R., Freudenthal B. D., Hou E. W., et al. (2014). Suicidal Cross-Linking of PARP-1 to AP Site Intermediates in Cells Undergoing Base Excision Repair. Nucleic Acids Res. 42 (10), 6337–6351. 10.1093/nar/gku288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R., Longley M. J., Sharief F. S., Hou E. W., Copeland W. C., Wilson S. H. (2009). Human DNA Polymerase Possesses 5'-dRP Lyase Activity and Functions in Single-Nucleotide Base Excision Repair In Vitro . Nucleic Acids Res. 37 (6), 1868–1877. 10.1093/nar/gkp035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinet A., Tirman S., Jackson J., Šviković S., Lemacon D., Carvajal-Maldonado D., et al. (2020). PRIMPOL-Mediated Adaptive Response Suppresses Replication Fork Reversal in BRCA-Deficient Cells. Mol. Cell 77, 461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechkoblit O., Johnson R. E., Gupta Y. K., Prakash L., Prakash S., Aggarwal A. K. (2021). Structural Basis of DNA Synthesis Opposite 8-Oxoguanine by Human PrimPol Primase-Polymerase. Nat. Commun. 12, 4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reines D. (1992). Elongation Factor-dependent Transcript Shortening by Template-Engaged RNA Polymerase II. J. Biol. Chem. 267 (6), 3795–3800. 10.1016/s0021-9258(19)50596-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins P., Pappin D. J., Wood R. D., Lindahl T. (1994). Structural and Functional Homology between Mammalian DNase IV and the 5'-nuclease Domain of Escherichia coli DNA Polymerase I. J. Biol. Chem. 269 (46) 28535–28538. 10.1016/s0021-9258(19)61935-6 [DOI] [PubMed] [Google Scholar]
- Rubtsova M. P., Vasilkova D. P., Malyavko A. N., Naraikina Y. V., Zvereva M. I., Dontsova O. A. (2012). Telomere Lengthening and Other Functions of Telomerase. Acta Naturae 4 (2), 44–61. 10.32607/actanaturae.10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel J., van Schendel R., den Dunnen J. T., Tijsterman M. (2019). Templated Insertions: A Smoking Gun for Polymerase Theta-Mediated End Joining. Trends Genet. 35 (9), 632–644. 10.1016/j.tig.2019.06.001 [DOI] [PubMed] [Google Scholar]
- Seki M., Wood R. D. (2008). DNA Polymerase θ (POLQ) Can Extend from Mismatches and from Bases Opposite a (6-4) Photoproduct. DNA Repair 7 (1), 119–127. 10.1016/j.dnarep.2007.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag V., Sachdev S., Flores J., Modak M., Singh K. (2018). Family A and B DNA Polymerases in Cancer: Opportunities for Therapeutic Interventions. Biology 7 (1), 5. 10.3390/biology7010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevelev I. V., Hübscher U. (2002). The 3′-5′ Exonucleases. Nat. Rev. Mol. Cel Biol 3 (5), 364–376. 10.1038/nrm804 [DOI] [PubMed] [Google Scholar]
- Shi C., Shen X., Niu S., Ma C. (2015). Innate Reverse Transcriptase Activity of DNA Polymerase for Isothermal RNA Direct Detection. J. Am. Chem. Soc. 137, 13804–13806. [DOI] [PubMed] [Google Scholar]
- Sobol R. W., Horton J. K., Kühn R., Gu H., Singhal R. K., Prasad R., et al. (1996). Requirement of Mammalian DNA Polymerase-β in Base-Excision Repair. Nature 379 (6561), 183–186. 10.1038/379183a0 [DOI] [PubMed] [Google Scholar]
- Sobol R. W., Prasad R., Evenski A., Baker A., Yang X.-P., Horton J. K., et al. (2000). The Lyase Activity of the DNA Repair Protein β-polymerase Protects from DNA-Damage-Induced Cytotoxicity. Nature 405 (6788), 807–810. 10.1038/35015598 [DOI] [PubMed] [Google Scholar]
- Sosunov V., Sosunova E., Mustaev A., Bass I., Nikiforov V., Goldfarb A. (2003). Unified Two-Metal Mechanism of RNA Synthesis and Degradation by RNA Polymerase. EMBO J. 22 (9), 2234–2244. 10.1093/emboj/cdg193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosunova E., Sosunov V., Epshtein V., Nikiforov V., Mustaev A. (2013). Control of Transcriptional Fidelity by Active Center Tuning as Derived from RNA Polymerase Endonuclease Reaction. J. Biol. Chem. 288 (9), 6688–6703. 10.1074/jbc.m112.424002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A. (1999). DNA Polymerases: Structural Diversity and Common Mechanisms. J. Biol. Chem. 274 (25), 17395–17398. 10.1074/jbc.274.25.17395 [DOI] [PubMed] [Google Scholar]
- Su Y., Egli M., Guengerich F. P. (2017). Human DNA Polymerase eta Accommodates RNA for Strand Extension. J. Biol. Chem. 292, 18044–18051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Ghodke P. P., Egli M., Li L., Wang Y., Guengerich F. P. (2019). Human DNA Polymerase η has Reverse Transcriptase Activity in Cellular Environments. J. Biol. Chem. 294, 6073–6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirman S., Quinet A., Wood M., Meroni A., Cybulla E., Jackson J., et al. (2021). Temporally Distinct Post-Replicative Repair Mechanisms Fill PRIMPOL-Dependent ssDNA Gaps in Human Cells. Mol. Cell 81, 4026–4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa S. E., Classen S., Chapados B. R., Arvai A. S., Finger L. D., Guenther G., et al. (2011). Human Flap Endonuclease Structures, DNA Double-Base Flipping, and a Unified Understanding of the FEN1 Superfamily. Cell 145 (2), 198–211. 10.1016/j.cell.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Lenoir W. F., Wang C., Su D., McLaughlin M., Hu Q., et al. (2020). DNA Polymerase ι Compensates for Fanconi Anemia Pathway Deficiency by Countering DNA Replication Stress. Proc. Natl. Acad. Sci. USA 117 (52), 33436–33445. 10.1073/pnas.2008821117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Grindley N. D. F., Joyce C. M. (2000). Coordination between the Polymerase and 5′-Nuclease Components of DNA Polymerase I of Escherichia coli. J. Biol. Chem. 275 (27), 20949–20955. 10.1074/jbc.m909135199 [DOI] [PubMed] [Google Scholar]
- Yang W., Lee J. Y., Nowotny M. (2006). Making and Breaking Nucleic Acids: Two-Mg2+-Ion Catalysis and Substrate Specificity. Mol. Cel 22 (1), 5–13. 10.1016/j.molcel.2006.03.013 [DOI] [PubMed] [Google Scholar]
- Yousefzadeh M. J., Wyatt D. W., Takata K.-I., Mu Y., Hensley S. C., Tomida J., et al. (2014). Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ. Plos Genet. 10 (10), e1004654. 10.1371/journal.pgen.1004654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn K. E., Averill A. M., Aller P., Wood R. D., Doublié S. (2015). Human DNA Polymerase θ Grasps the Primer Terminus to Mediate DNA Repair. Nat. Struct. Mol. Biol. 22 (4), 304–311. 10.1038/nsmb.2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn K. E., Jensen R. B., Wood R. D., Doublié S. (2021). Human DNA Polymerase θ Harbors DNA End-Trimming Activity Critical for DNA Repair. Mol. Cel 81 (7), 1534–1547. 10.1016/j.molcel.2021.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zuo Y., Deutscher M. P. (2002). The Physiological Role of RNase T Can Be Explained by its Unusual Substrate Specificity. J. Biol. Chem. 277 (33), 29654–29661. 10.1074/jbc.m204252200 [DOI] [PubMed] [Google Scholar]