Abstract
Tau protein neurofibrillary tangles are closely linked to neuronal/synaptic loss and cognitive decline in Alzheimer’s disease and related dementias. Our knowledge of the pattern of neurofibrillary tangle progression in the human brain, critical to the development of imaging biomarkers and interpretation of in vivo imaging studies in Alzheimer’s disease, is based on conventional two-dimensional histology studies that only sample the brain sparsely.
To address this limitation, ex vivo MRI and dense serial histological imaging in 18 human medial temporal lobe specimens (age 75.3 ± 11.4 years, range 45 to 93) were used to construct three-dimensional quantitative maps of neurofibrillary tangle burden in the medial temporal lobe at individual and group levels. Group-level maps were obtained in the space of an in vivo brain template, and neurofibrillary tangles were measured in specific anatomical regions defined in this template.
Three-dimensional maps of neurofibrillary tangle burden revealed significant variation along the anterior-posterior axis. While early neurofibrillary tangle pathology is thought to be confined to the transentorhinal region, we found similar levels of burden in this region and other medial temporal lobe subregions, including amygdala, temporopolar cortex, and subiculum/cornu ammonis 1 hippocampal subfields.
Overall, the three-dimensional maps of neurofibrillary tangle burden presented here provide more complete information about the distribution of this neurodegenerative pathology in the region of the cortex where it first emerges in Alzheimer’s disease, and may help inform the field about the patterns of pathology spread, as well as support development and validation of neuroimaging biomarkers.
Keywords: neurofibrillary tangles, Alzheimer’s disease, neurodegeneration, medial temporal lobe
See Khan et al. (doi:10.1093/brain/awab314) for a scientific commentary on this article.
Yushkevich et al. use MRI and dense serial histology to construct the first three-dimensional maps of tau neurofibrillary tangle pathology at individual and group levels. The maps show a more extensive involvement of medial temporal lobe regions in tau deposition than previously thought.
See Khan et al. (doi:10.1093/brain/awab314) for a scientific commentary on this article.
Introduction
Tau neurofibrillary tangle (NFT) pathology is closely linked to neurodegeneration and cognitive decline in Alzheimer’s disease.1 The topographic characterization of the spread of NFT pathology through the human brain by Braak and others2-10 has had tremendous impact in many areas of Alzheimer’s disease research, including the diagnosis of Alzheimer’s disease.10,11 Yet this characterization is based on histological examination of post-mortem tissue that is inherently two-dimensional (2D) and samples the brain at a sparse set of locations, limiting our knowledge of the spread of NFT pathology. In particular, since histological sectioning is almost always done in the coronal plane, little is known about the distribution of NFT pathology along the anterior-posterior axis.
Recent advances in PET imaging enabled in vivo detection and mapping of tau pathology in three dimensions (3D). Tau PET imaging suggests a more diffuse pattern of early spread of tau pathology12 than what is suggested by the Braak and Braak staging system,5,7,10 according to which early NFT pathology is largely confined to the transentorhinal cortex, a small region located on the medial portion of Brodmann area 35 (BA35) in the anterior medial temporal lobe (MTL). However, tau PET is not a direct measure of NFT burden in the brain, since it has limited spatial resolution and has variable binding to multiple types of tau pathology. Structural MRI studies examining patterns of neurodegeneration in the MTL also suggest that, while BA35 is clearly impacted early in the disease, other MTL structures undergo similar rates of atrophy.13,14 The analysis of in vivo PET and MRI is significantly hampered by the lack of a comprehensive ‘gold standard’ post-mortem reference that would characterize the distribution and spread of NFT pathology in 3D and would be compatible with tools used to analyse in vivo PET and MRI data.
Quantitative 3D mapping of NFT and other neurodegenerative proteinopathies may also hold the key to solving the problem of mixed pathology in Alzheimer’s disease. Most patients diagnosed with Alzheimer’s at autopsy also harbour one or more concomitant neurodegenerative pathologies, such as TDP-43 and α-synuclein proteinopathies, non-Alzheimer’s tauopathies, as well as vascular disease.10,15–21 Unlike amyloid-β and tau, these pathologies cannot be detected reliably in vivo using current technology, although efforts are underway to develop PET tracers that would allow their detection. Histological staging studies and antemortem–pathology correlation studies suggest that these concomitant pathologies follow a pattern of spread that is different from NFT pathology, leading to distinct patterns of brain tissue loss and atrophy.21–27 Precise 3D characterization of the spread of NFT pathology, contrasted to 3D characterization of the spread of concomitant pathologies may help define ‘hotspots’ in the brain where atrophy is more likely to be linked to one pathology than another, potentially allowing in vivo detection. Furthermore, such hotspots could help clinical trials measure treatment efficacy more reliably by quantifying the brain’s response to treatment in relevant hotspots, rather than less specific regions such as the hippocampus. Additionally, tools that allow 3D mapping of TDP-43 and α-synuclein proteinopathies could prove useful for validation of novel PET tracers.
In this study, we develop a framework that can generate quantitative 3D maps of neurodegenerative proteinopathies in the human brain, which can then serve as a 3D reference for in vivo MRI analysis. We restrict our attention to the MTL, an essential component of the human memory system and the site of early neurodegeneration in Alzheimer’s disease. Tau, TDP-43, α-synuclein and vascular pathologies all affect the MTL in their early stages, making the MTL a hotbed of early neurodegenerative activity.10,21,28 We also restrict our attention to generating 3D maps of tau NFT burden, although the underlying approach is amendable to similar mapping of other pathologies. Our framework combines ex vivo MRI, image-guided tissue processing, serial histology imaging, and advanced machine learning algorithms. We leverage it to generate 3D maps of NFT burden for 18 brain donors, culminating in group-level maps that reveal significant early involvement of MTL structures beyond the transentorhinal cortex, as well as a marked anterior to posterior gradient of NFT deposition. These 3D maps of NFT deposition, defined in the space of an in vivo brain MRI template, are provided in digital form to facilitate their use in in vivo MRI and PET analysis.
Materials and methods
Donor cohort
Brain hemisphere specimens from 18 donors 45–93 years of age were obtained from the archive cases from the Human Neuroanatomy Laboratory at the University of Castilla La Mancha (UCLM, n = 12) and the Center for Neurodegenerative Disease Research at the University of Pennsylvania (UPenn, n = 6). In each donation, the donor’s next of kin provided consent to autopsy. Donors from UCLM were from the general population served by the brain bank, and included mostly older adults with no known neurological disease. Donors from UPenn were participants in in vivo ageing and dementia research, and included patients from the Penn Frontotemporal Degeneration Center and the Penn Alzheimer’s Disease Core Center. Table 1 provides summary demographic and diagnostic data for the brain donor cohort, with additional details in Supplementary Table 2.
Table 1.
Demographic composition of the brain donor cohort, primary and secondary post-mortem diagnoses, and global neuropathological staging using the Hyman et al.10 protocol
| Brain donor cohort | ||||
|---|---|---|---|---|
| n | 18 | |||
| Age | 75.2 ± 11.4 | 45–93 | ||
| Sex, female:male | 7:11 | |||
|
| ||||
| Diagnosis (from contralateral sampling) | Primary | Secondary | ||
|
| ||||
| Unremarkable brain | 1 | – | ||
| Primary age-related pathology (PART) | 4 | – | ||
| Low ADNC | 6 | 1 | ||
| Intermediate ADNC | 1 | – | ||
| Corticobasal degeneration (CBD) | 2 | 1 | ||
| Lewy body disease (LBD) | 1 | 2 | ||
| Argyrophilic grain disease (AGD) | 1 | – | ||
| Frontotemporal lobar degeneration (TDP-43 type) | 1 | 1 | ||
| Cerebrovascular disease (CVD) | 1 | 1 | ||
| Progressive supranuclear palsy (PSP) | – | 1 | ||
|
| ||||
| Neuropathological staging (from contralateral sampling) | Stage | |||
| 0 | 1 | 2 | 3 | |
|
| ||||
| Amyloid (A) | 7 | 10 | 0 | 1 |
| Braak (B)a | 3 | 10 | 4 | 0 |
| CERAD (C) | 13 | 4 | 0 | 1 |
| α-Synuclein | 15 | 2 | 1 | 0 |
| TDP-43 | 16 | 0 | 0 | 2 |
Staging and diagnoses were derived from the opposite hemisphere from the one scanned with MRI. ADNC = Alzheimer’s disease neuropathological change.
For the brain donor with the diagnosis of argyrophilic grain disease, the Braak (B) score was undeterminable.
Imaging procedures
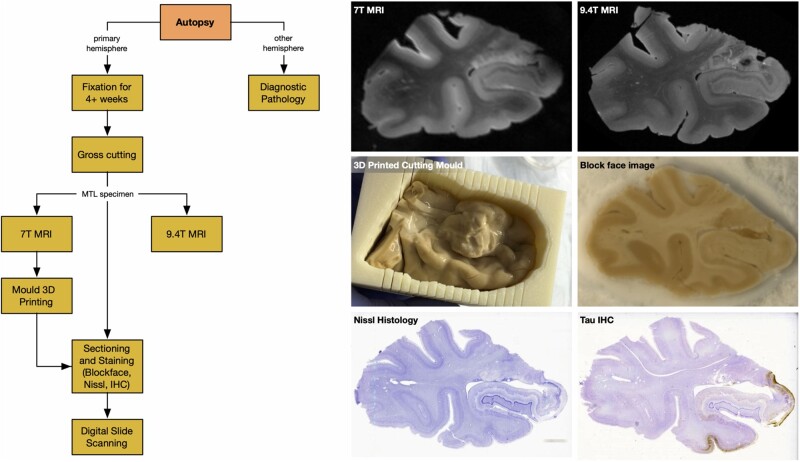
For each brain donation, a series of imaging procedures summarized in Fig. 1 were performed. Tissue from one brain hemisphere was used for imaging (MRI and histology) and the opposite hemisphere was sampled for diagnostic pathology following the NIA-AA protocol.10 After fixation, a specimen containing the intact MTL was dissected from the hemisphere. Each intact MTL specimen was scanned overnight on a 9.4 T animal MRI scanner with 0.2 × 0.2 × 0.2 mm3 native resolution using a standard T2-weighted sequence. A separate T2-weighted MRI scan was obtained on a 7 T human MRI scanner with 0.4 × 0.4 × 0.4 mm3 resolution. Registration between the 9.4 T and 7 T scans was performed to correct the 9.4 T scan for geometric distortions due to the non-linearity of the magnetic gradient field that increases towards the ends of the sample. A custom mould that tightly fits the MTL specimen was 3D printed using the 7 T scan and used to guide tissue sectioning. The mould (Fig. 1) orients sectioning orthogonal to the main axis of the hippocampus and reduces the complexity of subsequent registration between MRI and histology, as the plane of histological sectioning relative to the 7 T MRI scan is known. Using the custom mould, specimens were cut into 20-mm thick blocks, with most specimens yielding four blocks. Cryoprotected blocks were frozen using dry ice and sectioned using a sliding microtome coupled to a freezing unit into 50-µm sections, with no gaps between sections. Before cutting each section, a digital photograph of the block was taken using a mounted overhead camera (called ‘block-face images’). Every 10th section (i.e. sections 10, 20, 30, …) was stained for the Nissl series using the thionin stain. Every 20th section (i.e. sections 19, 39, 59, …) was stained using AT8, a human phosphorylated tau antibody immunohistochemistry (IHC) stain, and were counterstained for Nissl. Thus, Nissl-stained sections were at 0.5 mm intervals (∼40 per block) and anti-tau sections were adjacent to the Nissl sections and at 1-mm intervals (∼20 per block). Sections were mounted on 75 mm × 50 mm glass slides, digitally scanned at 20× resolution, and uploaded to an in-house created cloud-based digital histology archive that supports web-based visualization, anatomical labelling, and machine learning classifier training.
Figure 1.
The workflow of specimen preparation and imaging, along with examples of different imaging modalities and a sample 3D printed mould. The six images show a coronal section from the 0.4 × 0.4 × 0.4 mm3 7 T MRI, which is used to generate 3D printed moulds and to correct 9.4 T scans for distortion; a coronal section from the 0.2 × 0.2 × 0.2 mm3 9.4 T MRI; intact MTL specimen placed in the cutting mould 3D printed from the 7 T MRI; a block-face photograph taken during cryosectioning; a Nissl-stained histology slide; and an anti-tau IHC slide counterstained for Nissl.
Additional details on tissue preparation, imaging, and 3D printing procedures are provided in the Supplementary material, section 1.2.
Image processing
Deriving 2D maps of neurofibrillary tangle burden using weakly supervised learning
To quantify the burden of NFT pathology on individual anti-tau IHC sections, we adapted the weakly supervised learning (WSL) approach from Computer Vision. WSL performs segmentation at the level of pixels (e.g. outlines animals in photos) using training data that contains only image-level labels (e.g. ‘image 1 contains a cat’). WSL occupies a middle ground between simply classifying image regions into tangle/non-tangle classes (which fails to discriminate between regions with dense tangles and sparse tangles) and full-blown segmentation of individual tangles (which requires costly pixel-level training data, and is unnecessary when the objective is to derive NFT burden maps at the resolution of the ex vivo MRI).
Training data for WSL were generated by 12 raters using a custom web-based slide annotation system over the course of two day-long ‘Tanglethon’ events. Raters had access to full-resolution whole-slide IHC slides and were asked to use boxes to mark examples of tau pathologies, normal-appearing tissue, and background. Over 11 000 512 × 512 pixel patches (examples in Supplementary Fig. 2) were extracted from 176 slides in six MTL specimens and assigned into tangle-like (NFTs and pre-tangles) and non-tangle (tau neuropil threads, astroglial tau, tau coils in the white matter, normal tissue, slide background, artefacts, tissue folds) classes. The WSL algorithm WildCat29 was trained to classify patches into tangle versus non-tangle classes. In addition to assigning each input patch a class, WildCat generates a spatial heat map indicating the location and intensity of the features that drive its classification decision (Supplementary Fig. 2). We hypothesized that these heat maps capture the regional burden of NFT pathology.
We evaluated WSL as follows. First, we measured the accuracy of WildCat tangle/non-tangle classification in six leave-one-out cross-validation experiments, using slides from five specimens for training/validation and using the remaining specimen for testing. Then a single WildCat model was then trained on all six Tanglethon specimens and applied to all anti-tau whole-slide IHC images in the study, yielding whole-slide heat maps. We sampled 48 boxes of 2048 × 2048 pixels from the grey matter in three specimens not used for WildCat training, and compared the WildCat heat map integrated over each box to two conventional histopathology measures: total tangle count and ordinal semi-quantitative rating of NFT pathology, with categories ranging from 0 (none) to 3 (severe).
Additional details on training and evaluation of WildCat and application to whole-slide images are in the Supplementary material, section 1.3.
3D reconstruction of histology and matching to 9.4 T MRI for individual specimens
For each specimen, Nissl and anti-tau IHC histology images were reconstructed in the space of the 9.4 T MRI scan using a multi-stage pipeline that incorporates multiple image registration steps and takes advantage of the known orientation of the histology slicing plane with respect to the MRI. The pipeline, detailed in the Supplementary material, section 1.4, is largely automated, with a small number of manual initialization steps that together require under 2 h per specimen. A common feature in the pipeline is that when dealing with multi-modality registration problems (between MRI and block-face images or between MRI and Nissl histology), we transform the intensity of the block-face or histology images to have an ‘MRI-like’ appearance. This transformation allows us to perform image deformable registration using the normalized cross-correlation image similarity metric,30 which we found to result in better matching than the mutual information metric, which is formulated for multimodality image registration. The pipeline consists of the following stages:
MRI processing. Deformable registration is performed between the 9.4 T MRI and 7 T MRI to correct for the geometric distortions in the 9.4 T MRI scans (Supplementary material, section 1.4.2).
Alignment of MRI to 3D reconstructed block-face volume. The block-face photographs from each block are stacked in 3D and aligned to the distortion-corrected 9.4 T MRI scan using affine registration (Supplementary material, section 1.4.3 and Supplementary Fig. 5).
Nissl to MRI intensity remapping. To facilitate image registration between these two very distinct modalities, we generate an ‘MRI-like’ appearance from whole-slide Nissl histology images. Full-resolution patches in the Nissl slide are transformed to feature vectors using a deep learning-based image clustering algorithm,31 and robust linear regression is used to fit these feature vectors to MRI intensity. This yields Nissl-derived images with resolution and appearance characteristics similar to the 9.4 T MRI (Supplementary material, section 1.4.4 and Supplementary Figs 6 and 7).
Nissl to MRI registration. Each Nissl slide is aligned and deformed to the corresponding cross-section of the distortion-corrected 9.4 T MRI using the initial block-face-to-MRI alignment as initialization. An iterative registration pipeline that interleaves between all slides in a block is used (Supplementary material, section 1.4.5 and Supplementary Fig. 8).
IHC to Nissl registration. Each IHC slide is aligned and deformed to the corresponding Nissl slide. To account for tearing that is frequent during histological processing, the foreground region of the Nissl slide is divided into regions using a graph partition algorithm, and registration is applied in a piece-wise fashion to the partitions (Supplementary material, section 1.4.6 and Supplementary Fig. 9).
Final reconstruction. The transformations computed at the different stages of the pipeline are composed and applied to the IHC-derived NFT burden maps to transform them into the space of the distortion-corrected 9.4 T MRI scan (Supplementary material, section 1.4.7).
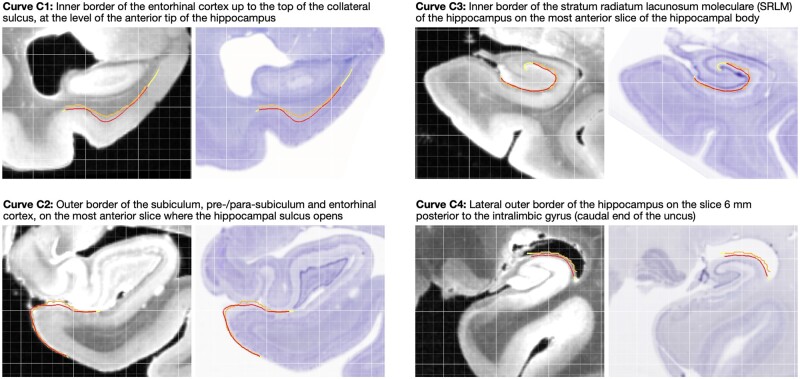
Evaluation. To evaluate the accuracy of MRI to histology reconstruction, four anatomical boundaries in the hippocampus and the entorhinal cortex (defined in Supplementary Table 3, examples in Fig. 4 and Supplementary Fig. 11) were traced by the same rater (S.A.L.) independently in Nissl and MRI space. Each boundary curve was traced on a single Nissl slice and on multiple consecutive MRI slices (typically nine) to account for out-of-plane mismatch (Supplementary Fig. 12). Symmetric root mean squared distance (RMSD) was measured between the corresponding boundaries at different stages of 3D reconstruction.
Figure 4.
Examples of curve-based evaluation of registration between 9.4 T MRI and reconstructed Nissl slides. Four anatomical curves (C1–C4) were traced independently in Nissl space (orange) and MRI space (red). To account for mismatch in the direction orthogonal to the Nissl slice plane, curves in MRI were traced on consecutive MRI slices (typically nine), and the red curve in the figure is the intersection of the resulting surface with the Nissl slice plane. After registration, the symmetric RMSD between the registered curves was measured, with regions where curves overhang (yellow portions) excluded. The four examples shown here correspond to median registration performance for each curve.
Group-level analysis of neurofibrillary tangle burden
To allow group-level analysis of three-dimensional NFT burden maps, the distortion-corrected 9.4 T MRI scans from all 18 specimens were jointly co-registered and brought into the reference space of an in vivo MRI brain template (Supplementary material, section 1.5 and Supplementary Fig. 13). Maps of average NFT burden were computed and visualized in template space (Supplementary material, section 3.4).
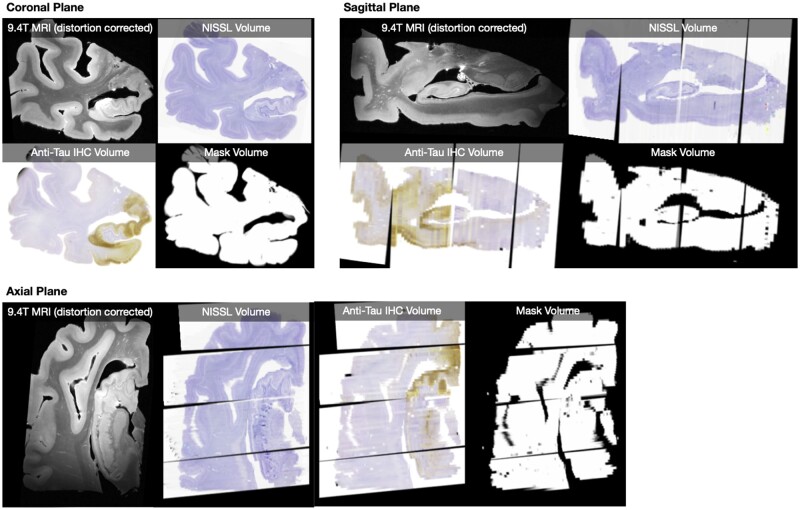
Two sets of anatomical labels available in the in vivo MRI template were used to compare summary measures of NFT burden between anatomical regions of interest: annotations of hippocampal subfields and MTL subregions with the Berron et al.32 protocol; and whole-brain annotations from the Glasser et al.33 multimodal atlas. A single summary measure of NFT burden for each region of interest in each specimen was obtained by taking the 90th percentile of the NFT burden map across all voxels in that region of interest for which IHC-based measures were available (i.e. the masked region in Fig. 3).
Figure 3.
Histological reconstructions. Example reconstruction, for a single specimen, of serial Nissl histology (one slide every 1 mm) and serial anti-tau IHC histology (one slide every 2 mm) in the space of the distortion-corrected 9.4 T MRI. Reconstructions for all 18 specimens are included in the digital archive. The mask volume specifies where in the MRI image space histology measures are available.
Data availability
Raw data were generated at the University of Pennsylvania and University of Castilla La Mancha. Derived data supporting the findings of this study are publicly available in the OpenNeuro repository (https://openneuro.org/datasets/ds003417). Processing scripts for this study are available on GitHub (https://github.com/pyushkevich/tau_maps_brain_2021). The source code for the web-based platform used for histology annotation is available at (https://github.com/pyushkevich/histoannot).
Results
WildCat tau neurofibrillary tangle burden measures agree with conventional histopathology
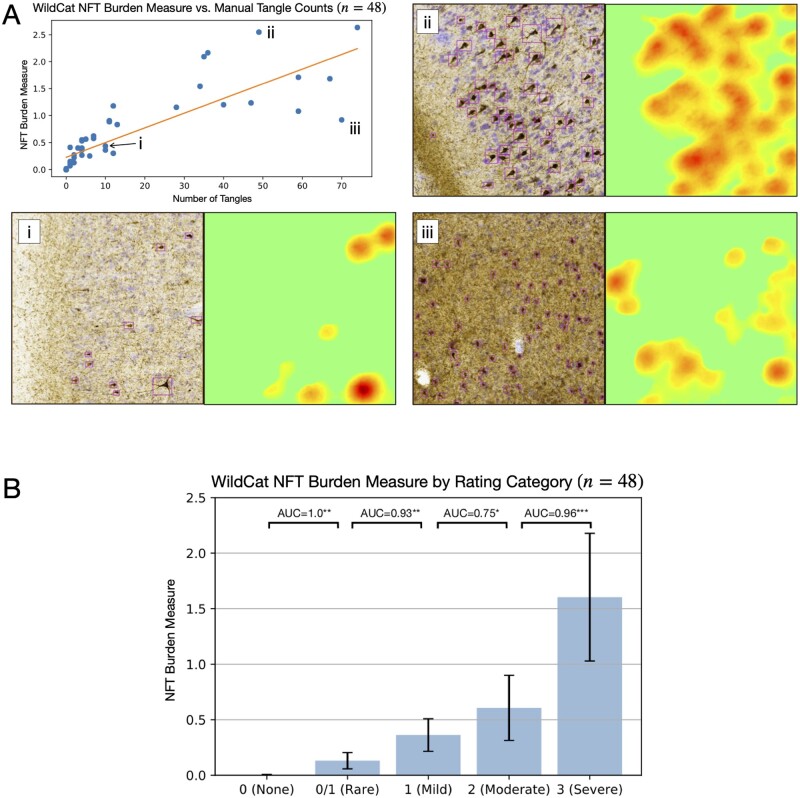
Across the six cross-validation experiments described in the ‘Deriving 2D maps of neurofibrillary tangle burden using weakly supervised learning’ section, WildCat assigned testing patches to the correct class (tangle or non-tangle) with high accuracy (95.9 ± 2.0%; range 93.1–98.4%). The agreement between WildCat heat maps integrated over 48 sample boxes and conventional histopathology measures in those boxes is reported in Fig. 2. The WildCat measure showed strong correlation with manual tangle counts (Kendall’s τ = 0.81. Spearman’s ρ = 0.94). Examining the outliers in this correlation suggests that the WildCat measure is greater than expected for a given number of tangles per box when the tangles are large and prominent [Fig. 2A(ii)], and less than expected when pre-tangles are present [Fig. 2A(iii)]. This suggests that the WildCat NFT burden measure captures both the number and prominence of tangle-like pathological inclusions. Furthermore, the WildCat NFT burden measure was significantly different between adjacent ordinal semi-quantitative rating categories, with Mann-Whitney area under the receiver operating characteristic curves (AUCs) exceeding 0.9, except AUC = 0.75 between ‘mild’ and ‘moderate’ categories (Fig. 2B). The discrimination between categories ‘none’ and ‘rare’ is particularly strong, indicating that this burden measure is sensitive to early NFT pathology. Overall, the strong discrimination between clinical categories and the strong concordance of NFT burden with manual counts suggest that our automated surrogate measure of NFT load may be used as a stand-in for these manual measures, especially for the purpose of differentiating between regions of low and high NFT density.
Figure 2.
NFT burden measure derived using the WSL algorithm WildCat compared to manual counting of NFTs and semi-quantitative ordinal ratings. (A) Regression between the WildCat burden measure and manual NFT count in 48 boxes from specimens left out from WildCat training is plotted in the top left panel. For three boxes, including two outliers, histology (with counted NFTs outlined in purple) and WildCat-derived burden maps are shown. (B) Plot of the average WildCat burden measure across five NFT severity categories into which the 48 boxes were assigned by an expert (D.J.I.) based on visual rating. Mann-Whitney AUC [U / (n1n2)] is shown between adjacent categories (*P < 0.05, **P < 0.01. ***P < 0.001, uncorrected).
Evaluation of the 3D reconstruction of serial histology guided by MRI and 3D printing
The reconstruction of Nissl and anti-tau IHC slides in the space of the distortion-corrected 9.4 T MRI is illustrated for one specimen in Fig. 3. At the final reconstruction stage, the mean RMSD between the anatomical curves traced independently in MRI and Nissl images was 0.37 ± 0.14 mm for curve C1, 0.29 ± 0.24 mm for curve C2, 0.21 ± 0.15 mm for curve C3, and 0.37 ± 0.269 mm for curve C4. Examples of median registration performance for each curve are shown in Fig. 4. Overall, the registration accuracy is high, with average RMSD below two MRI voxel widths for all four curves, and close to one voxel width for curves C2 and C3. Additional results describing curve mismatch at different stages of the registration pipeline are reported in the Supplementary material, section 2.1.
3D maps of neurofibrillary tangle density in 18 medial temporal lobe specimens
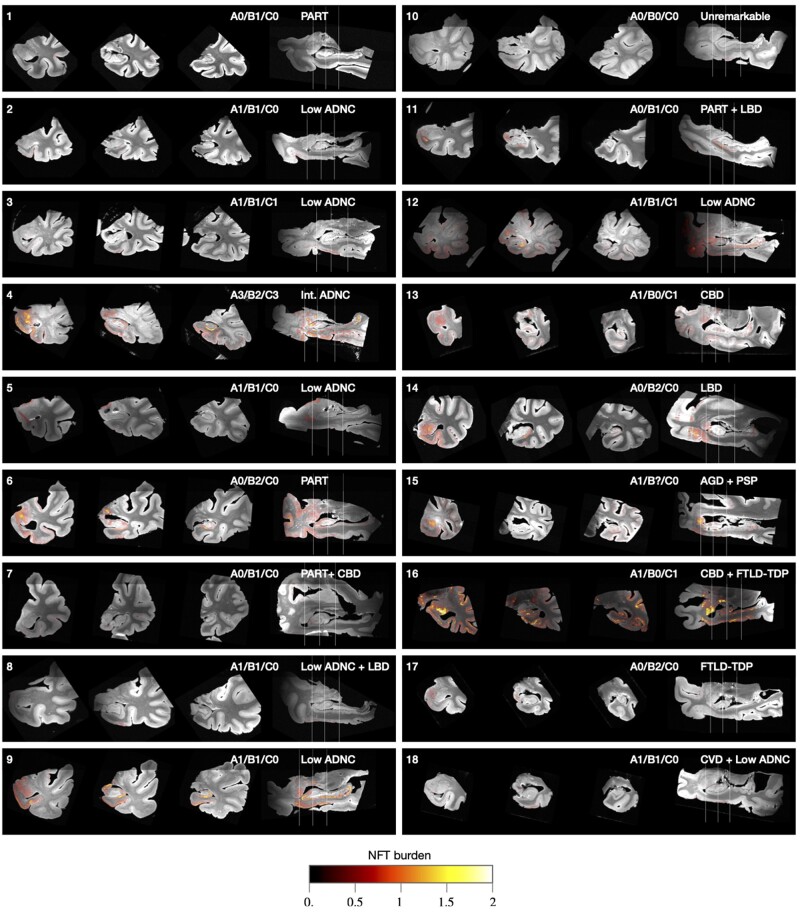
Following 3D reconstruction, whole-slide maps of NFT burden generated by WildCat were transformed into the space of the distortion-corrected 9.4 T MRI for all 18 specimens. Coronal and sagittal sections through these reconstructed 3D NFT burden maps are shown in Fig. 5, and the full 3D maps are distributed in the digital archive. While some small errors due to registration can be observed in Fig. 5, the overall reconstruction demonstrates excellent ability to project NFT burden measures into anatomical space, allowing inferences to be drawn about the 3D distribution of NFT pathology in the MTL.
Figure 5.
Coronal and sagittal cross-sections of 3D NFT burden maps derived from serial histology and overlaid on the 9.4 T MRI scans of medial temporal lobe specimens from 18 brain donors. Each specimen is annotated with the brain donor identifier (cross-referencing Supplementary Table 2), NIA-AA neuropathological A/B/C staging,10 and primary/secondary pathological diagnoses. The A/B/C staging is from the contralateral hemisphere and encompasses Thal amyloid plaque staging (A0–A3), Braak staging (B0–B3) and CERAD neuritic plaque staging (C0–C3). The coronal slices are through the amygdala, anterior hippocampus and hippocampal body. Additional analysis comparing quantitative NFT burden measures to clinical Braak staging is reported in the Supplementary material, section 2.2.
In most specimens, the intensity of the NFT burden map agrees with the ordinal Braak (B) tau pathology score obtained in the contralateral hemisphere with the NIA-AA Alzheimer’s disease staging protocol.10 One exception is Case 16, which had a neuropathological diagnosis of corticobasal degeneration with widespread tau pathology and Braak stage B0. Corticobasal degeneration is a 4R tauopathy and a subtype of frontotemporal lobar degeneration-Tau (FTLD-Tau).34 Alzheimer’s disease Braak staging in FTLD-Tau is challenging due to the difficulty in distinguishing Alzheimer’s disease neurofibrillary tau pathology from FTLD-associated tauopathy in neurons and glia. Use of the Alzheimer’s disease-specific tau antibody GT3835–37 in this case revealed minimal Alzheimer’s NFTs consistent with a Braak B0 stage, while our machine learning approach detected more extensive FTLD related tau. An example IHC slide from Case 16 is shown in Supplementary Fig. 14, highlighting the visually similar appearance of these two types of tau inclusions. Future work will examine the ability of our machine learning pipeline to distinguish Alzheimer’s disease-type NFTs from glial and neuronal tau in FTLD-Tau. In the current paper, we excluded primary FTLD-Tau cases (i.e. Cases 13, 15, and 16) from subsequent group-level analysis, since our burden maps in these cases may be capturing non-Alzheimer’s forms of tau. Among the remaining 15 cases, there is general agreement between clinical Braak (B) rating and NFT burden, as reported in the Supplementary material, section 2.2 and Supplementary Fig. 16.
Group-level 3D analysis of neurofibrillary tangle burden
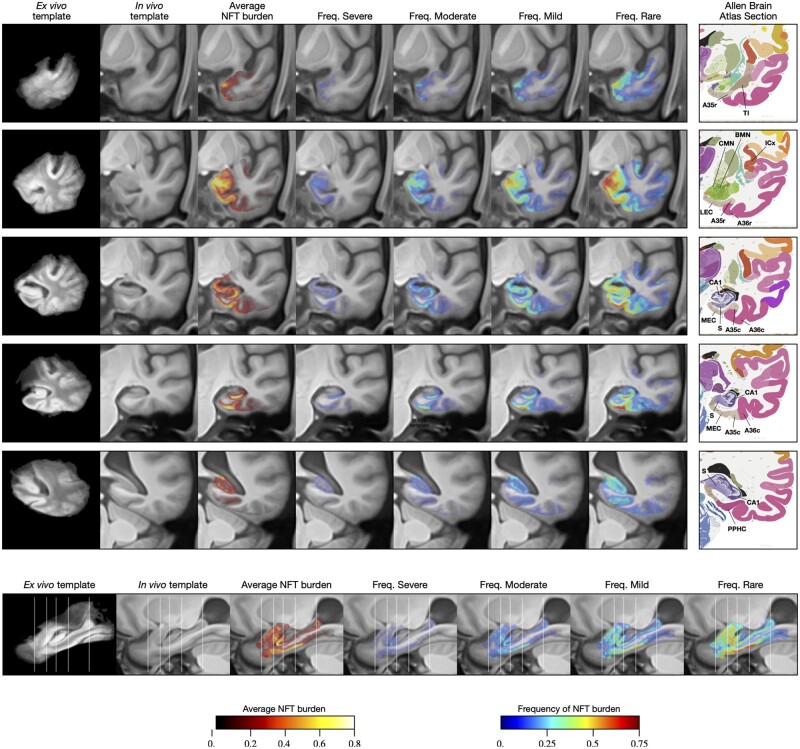
The population 9.4 T MRI template and population-level NFT burden maps are shown in Fig. 6. The average NFT burden map for the 15 non-FTLD-Tau specimens is plotted in the third column of Fig. 6, revealing a clear topographic distribution of NFT burden, with regions of high average burden following anatomical features such as the cornu ammonis and subiculum layers of the hippocampus, the entorhinal cortex, BA35, and amygdala. This conspicuous anatomical pattern indicates the success of the various data fusion and registration stages, as consistent failures in any of the stages (IHC to Nissl, Nissl to 9.4 T MRI, 9.4 T MRI to template) would have resulted in a blurry average map. However, this average map may offer limited information about the stereotypical progression of NFT burden because of the variability in this sample of brain donors. Figure 6 also plots four frequency maps (columns 4–7), each indicating how frequently among these 15 cases the NFT burden exceeds a certain threshold, with thresholds corresponding to severe, moderate, mild and rare levels of NFT burden, as inferred from Fig. 2A. Anatomical regions with higher frequency are likely to be involved earlier in the disease.
Figure 6.
Average and summary maps of tau NFT burden in the space of an anatomical template. These maps use data from 15 of 18 cases in Fig. 5, excluding three cases with primary diagnosis of FTLD-Tau diagnosis. The top five rows in the figure show coronal cross-sections through the medial temporal lobe at the levels of the temporal pole, amygdala, hippocampal head, hippocampal body and hippocampal tail, respectively. The bottom row shows a sagittal section through the hippocampus. Each row includes a visualization of the average NFT burden map, as well as four frequency maps. The frequency maps at each voxel describe the fraction of cases for which the NFT burden at that voxel was above a given threshold. Thresholds were chosen based on the analysis in Fig. 2B and correspond to different levels of pathological burden (>1.0 for ‘severe’; >0.5 for ‘moderate’; >0.25 for ‘mild’; >0.1 for ‘rare’). Coronal sections are also accompanied by roughly corresponding slices from the Allen Brain Atlas.38 Anatomical abbreviations (following Allen Brain Atlas nomenclature): A35r/A35c = rostral/caudal BA35; A36r/A36c = rostral/caudal BA36; BMN = mediobasal nucleus of the amygdala; CA1 = cornu ammonis 1 region of the hippocampus; CMN = corticomedial nuclear group of the amygdala; ICx = insular neocortex; LEC/MEC = lateral/medial entorhinal cortex; PPHC = posterior parahippocampal cortex; S = subiculum; TI = temporal agranular insular cortex (area TI).
The sagittal cross-sections of the average burden and frequency maps (Fig. 6, bottom row) reveal a clear anterior-posterior gradient, with anterior portion of the parahippocampal gyrus having greater frequency and average burden values than the posterior portion, and similarly greater anterior involvement in the hippocampus. The coronal sections of the maps reveal high frequency of pathology and high average burden levels in BA35 (occupying the medial portion of the perirhinal cortex), entorhinal cortex, amygdala (particularly the medial superior portion, corresponding to the accessory basal and mediobasal nuclei), subiculum and the cornu ammonis 1 (CA1) subfield of the hippocampus, and the more medial aspect of the temporal polar cortex (possibly corresponding to the very anterior portion of BA35 or to area TI). Cortical regions outside of the entorhinal cortex and BA35 exhibit varying degrees of burden, most prominently BA36 (lateral portion of perirhinal cortex), insular gyri and the inferior temporal gyrus.
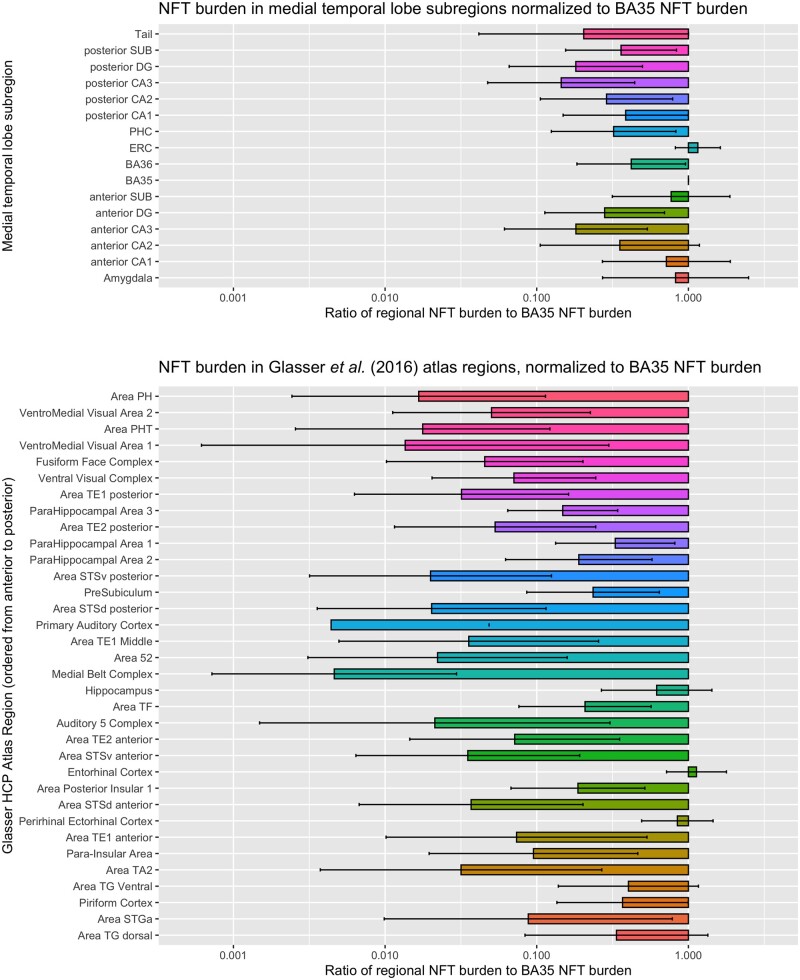
Region of interest analysis
Figure 7 plots NFT burden in the anatomical regions of interest defined in MRI template space using the Berron et al.32 and Glasser et al.33 atlases. For each region, the summary NFT burden measure is normalized to the summary NFT burden measure for the BA35 region of interest, and the mean and standard deviation across 15 specimens are plotted. BA35 was chosen as the reference region because it includes the transentorhinal region, described by Braak and Braak2 as the first cortical site of tau pathology in Alzheimer’s disease. The regions of interest with greatest average NFT burden relative to BA35 are the entorhinal cortex, amygdala, hippocampal subfields anterior subiculum and anterior CA1. The second tier of relatively impacted regions of interest includes other areas of the parahippocampal gyrus (BA36, parahippocampal cortex), temporopolar regions (ventral and dorsal area TG), piriform cortex (although because of its proximity to the amygdala, it is likely that this region in the Glasser et al.33 atlas is picking up NFT burden from the amygdala), and to a lesser extent, insular cortex. Additionally, the region of interest analysis highlights the presence of an anterior-posterior gradient in NFT burden, with greater sparing of posterior regions. A more detailed plot of summary region of interest NFT burden measures in Supplementary Fig. 15 shows that in individual specimens, including specimens with relatively mild BA35 NFT burden, it is not uncommon for other anatomical regions to exhibit NFT burden similar or greater to that of BA35. Nonetheless, the group analysis quantitatively supports the notion that BA35 and entorhinal cortex are the regions with greatest NFT burden.
Figure 7.
Summary measures of tau NFT burden in two sets of anatomical regions defined in template space. The top plot includes hippocampal subfields and subregions of the parahippocampal gyrus. The bottom plot includes 35 regions from the Human Connectome Project Multi-Modality Parcellation (MMP) atlas33 that cover the temporal lobe. For each region, the horizontal bars plot the ratio of NFT burden in that region to the NFT burden in BA35, which is thought to encompass the earliest site of cortical NFT formation in Alzheimer’s disease. For each region in each specimen, the summary measure of NFT burden is obtained by taking the 90th percentile of the regional NFT burden measure. Larger values in the plot indicate greater vulnerability to NFT pathology relative to BA35, while smaller values indicate that regions are spared from NFT pathology relative to BA35. Each set of regions is roughly arranged in the order from more posterior (top of plot) to more anterior (bottom of plot), indicating a presence of a posterior-anterior gradient in relative NFT burden, with posterior structures more spared relative to the anterior structures. These plots use data from 15 of 18 cases in Fig. 5, excluding three cases with primary FTLD-Tau diagnosis.
Discussion
3D maps of tau neurofibrillary tangle pathology generated for 18 brain donors
To our knowledge, this work is the first to perform 3D mapping of tau NFT pathology in a sizable collection of human brain specimens (n = 18). In a recent preprint, Alegro et al.39 describe a pipeline for generating whole-brain 3D tau density maps and present results for two specimens. They use deep learning to extract and quantify tau inclusions from IHC slides and use registration to ex vivo MRI to reconstruct 3D maps. Alegro et al.39 perform ex vivo MRI and CT in situ, an advantage over the current work since this approach eliminates MRI artefacts due to air bubbles and makes co-registration with in vivo MRI simpler. However, in situ scanning is not always feasible to integrate into existing autopsy protocols, including at the brain banks used in our study. While the whole-brain mapping and in situ imaging are relative advantages of the Alegro et al.39 pipeline, the amount of manual interaction in their pipeline is greater, requiring manual initialization for spline-based deformable registration for each histology slide. Tward et al.40 describe a new deformable registration approach for aligning serial anti-tau IHC slides to ex vivo MRI without a need for intensity transfer, and implement it for several tissue blocks from one brain donor. They also use deep learning to generate tangle heat maps; however, with a simpler patch classification strategy (assigning each patch a tangle/non-tangle label, as opposed to WSL used in this paper, although once the maps are brought to the resolution of the MRI it is unclear that this distinction is significant) and without evaluating their heat maps against manual tangle counts or semi-quantitative scores. Whereas previous tau 3D mapping efforts39,40 were applied to one to two specimens, the current work is the first to analyse histologically-derived 3D maps of tau pathology in a group template space and derive average and summary maps of NFT burden (Fig. 6). The fact that we were successful in applying our approach to 18 specimens also speaks to the generalizability and scalability of our approach to other individuals with other neurological conditions.
3D pattern of tau neurofibrillary tangle burden recapitulates earlier neuropathological studies
The patterns of tau NFT distribution in individual specimens (Fig. 5) and in the group template space (Fig. 6) are consistent with earlier histological literature that characterized NFT spread in the human brain. For instance, Arnold et al.4 quantified NFT density in 39 cortical regions in patients with clinically intractable dementia confirmed pathologically as Alzheimer’s disease. Greatest average NFT density was found in BA28 (ventral entorhinal cortex), subiculum/CA1, BA38 (temporopolar cortex), BA35, posterior parahippocampal areas TF and TH, amygdala, and BA51 (piriform cortex). Essentially the same areas come out at the top of our analysis. However, whereas in Arnold et al.,4 the NFT density in many regions was only slightly lower than in the most affected regions (e.g. 2.8 in the anterior insula versus 3.9 in BA28), in our analysis the distribution is less uniform, with most regions outside of the medial temporal lobe and temporopolar cortex having NFT burden reduced threefold or more relative to BA35. These similarities and differences are consistent with the brain donors in our study being in earlier stages of disease progression.
Arguably, the most influential work on characterizing the spread of NFT pathology is by Braak and Braak,5,7 whose six-stage staging system is used universally for neuropathological diagnosis of Alzheimer’s disease.10 Braak and Braak5 used the term ‘transentorhinal’ to describe the early stages I and II, and argued that the earliest manifestations of NFT pathology are largely confined to the transentorhinal region (medial BA35), with some involvement of the entorhinal cortex as well. While many of the brain donors in our cohort were classified as B1 (corresponding to the ‘transentorhinal’ stages I and II), our 3D analysis suggests a more widespread distribution of NFT burden. In several cases with mild transentorhinal involvement, the amygdala, anterior CA1 and subiculum, and anterior temporal pole regions exhibit NFT burden on par or even greater than in BA35 (Figs 5, 6 and Supplementary Fig. 15). This discrepancy could be attributed to asymmetry (Braak staging was performed using the contralateral hemisphere) or limited sampling of tissue for Braak staging. Overall, however, our results suggest that in studies of early Alzheimer’s disease, a broader focus beyond the transentorhinal region, and inclusive of hippocampal subregions, amygdala, and temporopolar cortex may be warranted.
The vulnerability of temporopolar cortex and amygdala to NFT pathology has been described in the neuropathology literature.3,6,41 Kromer-Vogt et al.3 identified the basomedial and cortical nuclei of the amygdala as having highest NFT density. Interestingly these regions are strongly connected to regions typically thought to harbour early NFT pathology, such as the entorhinal cortex and perirhinal cortex.42 Our group-level 3D map appears to reveal elevated frequency of supra-threshold NFT burden in the cortical and basomedial nuclei relative to the rest of the amygdala (Fig. 6), although the precise distribution of tau burden in the amygdala is difficult to infer in the groupwise template, and will require a focused follow-up study with cytoarchitecture-based labelling of amygdala subnuclei. In the temporopolar cortex, Ding et al.41 reported greatest density of anti-tau-labelled neurons in the anterior BA35, followed by areas TG and TI, and to a lesser extent anterior BA36. The most anterior slice in Fig. 6 reveals two peaks in the NFT frequency maps whose locations are consistent with anterior BA35 and area TG as defined by Ding et al.41 This finding suggests that the early tau pathology of BA35, as well as the lateral entorhinal area, which also overlapped high levels of NFT in the frequency maps, extend much more anterior in these regions than usually considered in segmentation protocols measuring these areas on in vivo studies of Alzheimer’s disease.43
3D pattern of tau neurofibrillary tangle burden exhibits similarities with in vivo neuroimaging
Examining the NFT burden maps in Figs 5 and 6 along the sagittal axis reveals an anatomical pattern that cannot be visualized with conventional 2D histological analysis. Indeed, the regional analysis of prior histological studies describes NFT involvement in monolithic terms whereas the current 3D mapping suggests gradients of involvement even within regions. Namely, we observed a directional gradient in the NFT burden, with greater involvement of anterior areas of the parahippocampal gyrus (anterior hippocampus, entorhinal cortex, BA35, amygdala). Ranganath and Ritchey44 hypothesized that memory function is subserved by two MTL networks, anterior temporal and posterior medial networks. The anterior temporal network, which involves the perirhinal cortex (BA35), anterior hippocampus, lateral entorhinal cortex, amygdala, ventral temporopolar cortex, and orbitofrontal cortex, is linked to familiarity-based recognition memory, social cognition, and semantic memory; while the posterior medial network involves medial entorhinal cortex, posterior hippocampus, parahippocampal cortex, and retrosplenial cortex, and is linked to episodic, or recollection-based memory, spatial navigation, and situational models or schemas.44 It was proposed in this model, that semantic dementia best maps on to the anterior temporal system while Alzheimer’s disease most prominently involves the posterior medial network, which overlaps with the default mode network, but not completely to the exclusion of the anterior temporal. However, the 3D distribution of NFT burden in our results, with strong involvement of anterior structures, aligns closely with the anterior temporal network, with BA35, amygdala, anterior CA1/subiculum, and temporopolar regions, particularly the anterior extension of BA35, having most pronounced NFT burden. In fact, a number of studies have suggested alterations in cognition in very early Alzheimer’s disease more linked to the anterior temporal network, including object short and long-term memory, familiarity, and semantic memory.45–49 One explanation for the relative sparing of the posterior medial system in our 3D NFT data may be that non-tangle forms of tau pathology, including neuropil threads and tau in neuritic plaques, contribute to the deterioration of the posterior medial network in Alzheimer’s disease. Another explanation may be that in the presence of extensive amyloid-β pathology, the distribution of NFTs is more posterior than in the majority low amyloid-β cases that compose our cohort, although 2D histology studies have found primary age-related tauopathy to have a similar distribution to that of Alzheimer’s disease.50 Future analysis including more cases with more pronounced amyloid-β pathology and examining additional tau species may shed light on anterior temporal and posterior medial network involvement in Alzheimer’s disease and primary age-related tauopathy.
That said, in vivo studies of the anterior temporal and posterior medial systems using tau and amyloid PET are largely consistent with our 3D map of NFT burden. Schwartz et al.51 identify patterns of 18F-AV-1451 tau tracer uptake associated with progressive Braak stages, and report the pattern in the ‘transentorhinal’ stages I and II impacting the transentorhinal region (which, given the resolution of PET encompasses entorhinal cortex and BA35), hippocampus, and amygdala. Lowe et al.52 compared 18F-AV-1451 uptake across brain regions in cognitively impaired and unimpaired individuals with and without cognitive decline, and report amygdala and temporal pole as the two regions with greatest tau uptake in cognitively unimpaired, amyloid-negative group, consistent with the high NFT burden in these regions in our analysis. They also report findings of elevated tau uptake in regions outside of the MTL in unimpaired, amyloid-negative individuals, which is consistent with our findings of elevated NFT burden in a number of cortical regions even when NFT burden in BA35 was low. Maass et al.48 examined 18F-FTP-tau PET uptake in anterior temporal and posterior medial network regions in normal ageing and Alzheimer’s disease, and found elevated tau PET uptake in anterior temporal regions relative to posterior medial regions in both ageing and Alzheimer’s disease, whereas amyloid PET uptake was greater in the posterior medial network regions and concluded that ‘posterior-medial regions are affected by tau later in the course of the disease and do not “catch up” while anterior-temporal tau accumulation accelerates further’. This finding of increased anterior temporal versus posterior medial tau is consistent with the anterior-posterior gradient of NFT burden in our ex vivo analysis. It is worth noting that given the greater amyloid burden in the posterior medial network, it is possible that the tau in these regions are disproportionately neuritic while the anterior temporal network may reflect NFTs, as measured here. Given these general similarities between patterns of tau distribution in our NFT burden maps and in vivo tau PET uptake patterns, it would be promising in future research to carry out analyses that directly contrast these data on a voxel-wise basis.
Limitations and future work
Our approach incorporates a complex image analysis pipeline that, in contrast to our earlier work on MRI/histology co-registration,53 is largely automated, with only a few manual initialization steps (i.e. initial alignment) that involve whole specimens or whole blocks, as opposed to individual 2D slides. As a result, the human and computational effort to create a new reconstruction for a single specimen is significantly smaller (∼1 day alternating between manual steps and automated processing) than the time to perform imaging and histology procedures. The accuracy of the registration is on the order of one to two ex vivo MRI voxels on average in the four anatomical regions examined, and visually, it appears that histology and MRI are well aligned through most of the temporal lobe. While this error is significant on the scale of individual tangles and even cortical layers, it is quite small relative to the resolution of in vivo MRI or PET and to errors encountered for in vivo MRI registration, e.g. between individual MRI scans and population templates. Hence, for applications where 3D patterns of NFT burden are needed to inform in vivo MRI analysis (e.g. to derive hotspots in which to measure atrophy linked to tau pathology), the histology-to-MRI registration errors can be considered almost negligible.
Even though the WildCat classifier achieves >95% accuracy for individual tangle/non-tangle patches, at the whole-slide level, WildCat classification is not free of error. In particular, our training set includes a very small number (<2%) of astrocytic tau inclusions, which can be similar in visual appearance to tangles. Indeed some areas of age-related tau astrogliopathy (ARTAG)54 are incorrectly identified as areas of high NFT burden by the algorithm. Overcoming this limitation requires extending the training set with a large number of ARTAG examples and ensuring that the high accuracy of WildCat is maintained. ARTAG misclassified as NFT may contribute to elevated measures of NFT burden in some cortical regions and amygdala, where ARTAG is more common.54 Our algorithm also does not distinguish tangles from pre-tangles, which may lead to confounding of different stages of NFT pathology. However, visual assessment of the data suggests that ARTAG and pre-tangles are only present in a small fraction of cases and location is not consistent from case to case, making it likely that their impact on NFT burden maps reported here is relatively minor. Lastly, the WildCat algorithm was not trained to differentiate NFTs from similarly appearing 4R-tau inclusions in FTLD-Tau (Supplementary Fig. 14), necessitating the exclusion of FTLD-Tau cases from group analysis.
The groupwise registration used a conventional population-based template approach,55,56 whereas in earlier work we showed that a method initialized with shape-based registration of semi-automatic segmentations of anatomical regions of interest (hippocampus, MTL cortex) results in better groupwise registration of these regions.53,57 However, given the large anatomical extent considered in this study (entire temporal lobe, not just MTL) and variation in extent between individuals, this shape-based approach would be difficult to implement. Some improvements to registration can likely be realized by incorporating more specimens and considering more advanced groupwise registration strategies.58,59 A related limitation is that the anatomical labels for the analysis were derived from in vivo atlases, and may not match individual anatomical structures well in group template space. We are currently generating cytoarchitecture-guided segmentations of MTL subregions in the specimens in this study (a process requiring weeks of manual segmentation for each specimen), and intend to conduct more granular analyses, including analyses linking NFT burden to regional alterations in MTL cortical thickness in future work. Thus, the present work serves as a foundation upon which future work can build to characterize the relationships between tau and other proteinopathies in a variety of forms (e.g. ARTAG versus NFTs) and structural changes in the MTL. Such future studies will enhance our understanding of the local neurodegenerative consequences of these pathologies.
Supplementary Material
Acknowledgements
We gratefully acknowledge the generosity and altruism of the brain donors and their families. We thank the members of the brain banks at the University of Pennsylvania Center for Neurodegenerative Disease Research and at the University of Castilla La Mancha at Albacete for their contributions to this research.
Funding
This work was supported bythe US National Institutes of Health grants P30 AG010124, R01 AG056014, and R01 EB017255.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
Glossary
- ARTAG
age-related tau astrogliopathy
- BA
Brodmann area
- CA1
cornu ammonis field 1
- FTLD
frontotemporal lobar degeneration
- IHC
immunohistochemistry
- MTL
medial temporal lobe
- NFT
neurofibrillary tangle
- TDP-43
transactive response DNA-binding protein 43
- WSL
weakly supervised learning
Contributor Information
Paul A Yushkevich, Department of Radiology, University of Pennsylvania, Philadelphia, USA.
Mónica Muñoz López, Human Neuroanatomy Laboratory, Neuromax CSIC Associated Unit, University of Castilla-La Mancha, Albacete, Spain.
María Mercedes Iñiguez de Onzoño Martin, Human Neuroanatomy Laboratory, Neuromax CSIC Associated Unit, University of Castilla-La Mancha, Albacete, Spain.
Ranjit Ittyerah, Department of Radiology, University of Pennsylvania, Philadelphia, USA.
Sydney Lim, Department of Radiology, University of Pennsylvania, Philadelphia, USA.
Sadhana Ravikumar, Department of Radiology, University of Pennsylvania, Philadelphia, USA.
Madigan L Bedard, Department of Radiology, University of Pennsylvania, Philadelphia, USA.
Stephen Pickup, Department of Radiology, University of Pennsylvania, Philadelphia, USA.
Weixia Liu, Department of Radiology, University of Pennsylvania, Philadelphia, USA.
Jiancong Wang, Department of Radiology, University of Pennsylvania, Philadelphia, USA.
Ling Yu Hung, Department of Radiology, University of Pennsylvania, Philadelphia, USA.
Jade Lasserve, Department of Radiology, University of Pennsylvania, Philadelphia, USA.
Nicolas Vergnet, Department of Radiology, University of Pennsylvania, Philadelphia, USA.
Long Xie, Department of Radiology, University of Pennsylvania, Philadelphia, USA.
Mengjin Dong, Department of Radiology, University of Pennsylvania, Philadelphia, USA.
Salena Cui, Department of Neurology, University of Pennsylvania, Philadelphia, USA.
Lauren McCollum, Department of Neurology, University of Pennsylvania, Philadelphia, USA.
John L Robinson, Department of Pathology, University of Pennsylvania, Philadelphia, USA.
Theresa Schuck, Department of Pathology, University of Pennsylvania, Philadelphia, USA.
Robin de Flores, Institut National de la Santé et de la Recherche Médicale (INSERM), Caen, France.
Murray Grossman, Department of Neurology, University of Pennsylvania, Philadelphia, USA.
M Dylan Tisdall, Department of Radiology, University of Pennsylvania, Philadelphia, USA.
Karthik Prabhakaran, Department of Neurology, University of Pennsylvania, Philadelphia, USA.
Gabor Mizsei, Department of Radiology, University of Pennsylvania, Philadelphia, USA.
Sandhitsu R Das, Department of Neurology, University of Pennsylvania, Philadelphia, USA.
Emilio Artacho-Pérula, Human Neuroanatomy Laboratory, Neuromax CSIC Associated Unit, University of Castilla-La Mancha, Albacete, Spain.
Marı’a del Mar Arroyo Jiménez, Human Neuroanatomy Laboratory, Neuromax CSIC Associated Unit, University of Castilla-La Mancha, Albacete, Spain.
Marı’a Pilar Marcos Raba, Human Neuroanatomy Laboratory, Neuromax CSIC Associated Unit, University of Castilla-La Mancha, Albacete, Spain.
Francisco Javier Molina Romero, Human Neuroanatomy Laboratory, Neuromax CSIC Associated Unit, University of Castilla-La Mancha, Albacete, Spain.
Sandra Cebada Sánchez, Human Neuroanatomy Laboratory, Neuromax CSIC Associated Unit, University of Castilla-La Mancha, Albacete, Spain.
José Carlos Delgado González, Human Neuroanatomy Laboratory, Neuromax CSIC Associated Unit, University of Castilla-La Mancha, Albacete, Spain.
Carlos de la Rosa-Prieto, Human Neuroanatomy Laboratory, Neuromax CSIC Associated Unit, University of Castilla-La Mancha, Albacete, Spain.
Marta Córcoles Parada, Human Neuroanatomy Laboratory, Neuromax CSIC Associated Unit, University of Castilla-La Mancha, Albacete, Spain.
Edward B Lee, Department of Pathology, University of Pennsylvania, Philadelphia, USA.
John Q Trojanowski, Department of Pathology, University of Pennsylvania, Philadelphia, USA.
Daniel T Ohm, Department of Neurology, University of Pennsylvania, Philadelphia, USA.
Laura E M Wisse, Department of Diagnostic Radiology, University of Lund, Lund, Sweden.
David A Wolk, Department of Neurology, University of Pennsylvania, Philadelphia, USA.
David J Irwin, Department of Neurology, University of Pennsylvania, Philadelphia, USA.
Ricardo Insausti, Human Neuroanatomy Laboratory, Neuromax CSIC Associated Unit, University of Castilla-La Mancha, Albacete, Spain.
References
- 1. Bobinski M, Wegiel J, Tarnawski M, et al. Relationships between regional neuronal loss and neurofibrillary changes in the hippocampal formation and duration and severity of Alzheimer disease. J Neuropathol Exp Neurol. 1997;56(4):414–420. [DOI] [PubMed] [Google Scholar]
- 2. Braak H, Braak E. On areas of transition between entorhinal allocortex and temporal isocortex in the human brain. Normal morphology and lamina-specific pathology in Alzheimer’s disease. Acta Neuropathol. 1985;68(4):325–332. [DOI] [PubMed] [Google Scholar]
- 3. Kromer Vogt LJ, Hyman BT, Van Hoesen GW, Damasio AR. Pathological alterations in the amygdala in Alzheimer’s disease. Neuroscience. 1990;37(2):377–385. [DOI] [PubMed] [Google Scholar]
- 4. Arnold SE, Hyman BT, Flory J, Damasio AR, Van Hoesen GW. The topographical and neuroanatomical distribution of neurofibrillary tangles and neuritic plaques in the cerebral cortex of patients with Alzheimer’s disease. Cereb Cortex. 1991;1(1):103–116. [DOI] [PubMed] [Google Scholar]
- 5. Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–259. [DOI] [PubMed] [Google Scholar]
- 6. Arnold SE, Hyman BT, Van Hoesen GW. Neuropathologic changes of the temporal pole in Alzheimer’s disease and Pick’s disease. Arch Neurol. 1994;51(2):145–150. [DOI] [PubMed] [Google Scholar]
- 7. Braak H, Braak E. Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiol Aging. 1995;16(3):271–278. discussion 278–84. [DOI] [PubMed] [Google Scholar]
- 8. Braak H, Braak E. Frequency of stages of Alzheimer-related lesions in different age categories. Neurobiol Aging. 1997;18(4):351–357. [DOI] [PubMed] [Google Scholar]
- 9. Braak H, Thal DR, Ghebremedhin E, Del Tredici K. Stages of the pathologic process in Alzheimer disease: Age categories from 1 to 100 years. J Neuropathol Exp Neurol. 2011;70(11):960–969. [DOI] [PubMed] [Google Scholar]
- 10. Hyman BT, Phelps CH, Beach TG, et al. National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease. Alzheimers Dement. 2012;8(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jack CR Jr, Bennett DA, Blennow K, et al. ; Contributors. NIA-AA research framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018;14(4):535–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jagust W. Imaging the evolution and pathophysiology of Alzheimer disease. Nat Rev Neurosci. 2018;19(11):687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wolk DA, Das SR, Mueller SG, Weiner MW, Yushkevich PA, Alzheimer’s Disease Neuroimaging Initiative. Medial temporal lobe subregional morphometry using high resolution MRI in Alzheimer’s disease. Neurobiol Aging. 2017;49:204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xie L, Wisse LEM, Sandhitsu D, et al. ; for the Alzheimer's Disease Neuroimaging Initiative. Longitudinal atrophy in early Braak regions in preclinical Alzheimer’s disease. Hum Brain Mapp. 2020;41(16):4704–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schneider JA, Boyle PA, Arvanitakis Z, Bienias JL, Bennett DA. Subcortical infarcts, Alzheimer’s disease pathology, and memory function in older persons. Ann Neurol. 2007;62(1):59–66. [DOI] [PubMed] [Google Scholar]
- 16. Toledo JB, Arnold SE, Raible K, et al. Contribution of cerebrovascular disease in autopsy confirmed neurodegenerative disease cases in the National Alzheimer’s Coordinating Centre. Brain. 2013;136(Pt 9):2697–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Attems J, Jellinger KA. The overlap between vascular disease and Alzheimer’s disease–lessons from pathology. BMC Med. 2014;12:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robinson JL, Lee EB, Xie SX, et al. Neurodegenerative disease concomitant proteinopathies are prevalent, age-related and APOE4-associated. Brain. 2018;141(7):2181–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Walker L, McAleese KE, Thomas AJ, et al. Neuropathologically mixed Alzheimer’s and Lewy body disease: Burden of pathological protein aggregates differs between clinical phenotypes. Acta Neuropathol. 2015;129(5):729–748. [DOI] [PubMed] [Google Scholar]
- 20. James BD, Wilson RS, Boyle PA, Trojanowski JQ, Bennett DA, Schneider JA. TDP-43 stage, mixed pathologies, and clinical Alzheimer’s-type dementia. Brain. 2016;139(11):2983–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matej R, Tesar A, Rusina R. Alzheimer’s disease and other neurodegenerative dementias in comorbidity: A clinical and neuropathological overview. Clin Biochem. 2019;73:26–31. [DOI] [PubMed] [Google Scholar]
- 22. Dickson DW. Neuropathology of non-Alzheimer degenerative disorders. Int J Clin Exp Pathol. 2009;3(1):1–23. [PMC free article] [PubMed] [Google Scholar]
- 23. Colom-Cadena M, Gelpi E, Charif S, et al. Confluence of -synuclein, tau, and -amyloid pathologies in dementia with Lewy bodies. J Neuropathol Exp Neurol. 2013;72(12):1203–1212. [DOI] [PubMed] [Google Scholar]
- 24. Brettschneider J, Del Tredici K, Lee VM-Y, Trojanowski JQ. Spreading of pathology in neurodegenerative diseases: A focus on human studies. Nat Rev Neurosci. 2015;16(2):109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Josephs KA, Murray ME, Whitwell JL, et al. Updated TDP-43 in Alzheimer’s disease staging scheme. Acta Neuropathol. 2016;131(4):571–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bejanin A, Murray ME, Martin P, et al. Antemortem volume loss mirrors TDP-43 staging in older adults with non-frontotemporal lobar degeneration. Brain. 2019;142(11):3621–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. R de F, Wisse LEM, Das SR, et al. Contribution of mixed pathology to medial temporal lobe atrophy in Alzheimer’s disease. Alzheimers Dement. 2020;16(6):843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Small SA, Schobel SA, Buxton RB, Witter MP, Barnes CA. A pathophysiological framework of hippocampal dysfunction in ageing and disease. Nat Rev Neurosci. 2011;12(10):585–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Durand T, Mordan T, Thome N, Cord M. Wildcat: Weakly supervised learning of deep convnets for image classification, pointwise localization and segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2017:642–651. [Google Scholar]
- 30. Avants B, Epstein C, Grossman M, Gee J. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12(1):26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Caron M, Bojanowski P, Joulin A, Douze M. Deep clustering for unsupervised learning of visual features. In: Proceedings of the European Conference on Computer Vision (ECCV); 2018:132–149. [Google Scholar]
- 32. Berron D, Vieweg P, Hochkeppler A, et al. A protocol for manual segmentation of medial temporal lobe subregions in 7 tesla MRI. Neuroimage Clin. 2017;15:466–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Glasser MF, Coalson TS, Robinson EC, et al. A multi-modal parcellation of human cerebral cortex. Nature. 2016;536(7615):171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mackenzie IRA, Baker M, Pickering-Brown S, et al. The neuropathology of frontotemporal lobar degeneration caused by mutations in the progranulin gene. Brain. 2006;129(Pt 11):3081–3090. [DOI] [PubMed] [Google Scholar]
- 35. Gibbons GS, Banks RA, Kim B, et al. Detection of Alzheimer disease (AD)-specific tau pathology in AD and nonAD tauopathies by immunohistochemistry with novel conformation-selective tau antibodies. J Neuropathol Exp Neurol. 2018;77(3):216–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gibbons GS, Kim S-J, Robinson JL, et al. Detection of Alzheimer’s disease (AD) specific tau pathology with conformation-selective anti-tau monoclonal antibody in co-morbid frontotemporal lobar degeneration-tau (FTLD-tau). Acta Neuropathol Commun. 2019;7(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Robinson JL, Yan N, Caswell C, et al. Primary tau pathology, not copathology, correlates with clinical symptoms in PSP and CBD. J Neuropathol Exp Neurol. 2020;79(3):296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sunkin SM, Ng L, Lau C, et al. Allen brain atlas: An integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res. 2013;41(Database issue):D996–D1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Alegro M, Chen Y, Ovando D, et al. Deep learning for Alzheimer’s disease: Mapping large-scale histological tau protein for neuroimaging biomarker validation. bioRxiv. [Preprint] doi:10.1101/698902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tward D, Brown T, Kageyama Y, et al. Diffeomorphic registration with intensity transformation and missing data: Application to 3D digital pathology of Alzheimer’s disease. Front Neurosci. 2020;14:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ding S-L, Van Hoesen GW, Cassell MD, Poremba A. Parcellation of human temporal polar cortex: A combined analysis of multiple cytoarchitectonic, chemoarchitectonic, and pathological markers. J Comp Neurol. 2009;514(6):595–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Price JL. Comparative aspects of amygdala connectivity. Ann N Y Acad Sci. 2003;985:50–58. [DOI] [PubMed] [Google Scholar]
- 43. Yushkevich PA, Pluta JB, Wang H, et al. Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment. Hum Brain Mapp. 2015;36(1):258–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ranganath C, Ritchey M. Two cortical systems for memory-guided behaviour. Nat Rev Neurosci. 2012;13(10):713–726. [DOI] [PubMed] [Google Scholar]
- 45. Kivisaari SL, Tyler LK, Monsch AU, Taylor KI. Medial perirhinal cortex disambiguates confusable objects. Brain. 2012;135(Pt 12):3757–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wolk DA, Mancuso L, Kliot D, Arnold SE, Dickerson BC. Familiarity-based memory as an early cognitive marker of preclinical and prodromal AD. Neuropsychologia. 2013;51(6):1094–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Das SR, Mancuso L, Olson IR, Arnold SE, Wolk DA. Short-term memory depends on dissociable medial temporal lobe regions in amnestic mild cognitive impairment. Cereb Cortex. 2016;26(5):2006–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Maass A, Berron D, Harrison TM, et al. Alzheimer’s pathology targets distinct memory networks in the ageing brain. Brain. 2019;142(8):2492–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Besson G, Simon J, Salmon E, Bastin C. Familiarity for entities as a sensitive marker of antero-lateral entorhinal atrophy in amnestic mild cognitive impairment. Cortex. 2020;128:61–72. [DOI] [PubMed] [Google Scholar]
- 50. Crary JF, Trojanowski JQ, Schneider JA, et al. Primary age-related tauopathy (PART): A common pathology associated with human aging. Acta Neuropathol. 2014;128(6):755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schwarz AJ, Yu P, Miller BB, et al. Regional profiles of the candidate tau PET ligand 18F-AV-1451 recapitulate key features of Braak histopathological stages. Brain. 2016;139(Pt 5):1539–1550. [DOI] [PubMed] [Google Scholar]
- 52. Lowe VJ, Wiste HJ, Senjem ML, et al. Widespread brain tau and its association with ageing, Braak stage and Alzheimer’s dementia. Brain. 2018;141(1):271–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Adler DH, Wisse LEM, Ittyerah R, et al. Characterizing the human hippocampus in aging and Alzheimer’s disease using a computational atlas derived from ex vivo MRI and histology. Proc Natl Acad Sci U S A. 2018;115(16):4252–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kovacs GG, Xie SX, Robinson JL, et al. Sequential stages and distribution patterns of aging-related tau astrogliopathy (ARTAG) in the human brain. Acta Neuropathol Commun. 2018;6(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Joshi S, Davis B, Jomier M, Gerig G. Unbiased diffeomorphic atlas construction for computational anatomy. Neuroimage. 2004;23 (Suppl 1):S151–S160. [DOI] [PubMed] [Google Scholar]
- 56. Avants BB, Gee JC. Shape averaging with diffeomorphic flows for atlas creation. In: Proceedings of the IEEE International Symposium on Biomedical Imaging; 2004:595–598. [Google Scholar]
- 57. Ravikumar S, Wisse LEM, Ittyerah R, et al. Building an ex vivo atlas of the earliest brain regions affected by Alzheimer’s disease pathology. In: 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI); 2020:113–117. [Google Scholar]
- 58. Yushkevich PA, Wang H, Pluta J, Avants BB. From label fusion to correspondence fusion: A new approach to unbiased groupwise registration. In: IEEE International Conference on Computer Vision and Pattern Recognition; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ahmad S, Fan J, Dong P, Cao X, Yap P-T, Shen D. Deep learning deformation initialization for rapid groupwise registration of inhomogeneous image populations. Front Neuroinform. 2019;13:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data were generated at the University of Pennsylvania and University of Castilla La Mancha. Derived data supporting the findings of this study are publicly available in the OpenNeuro repository (https://openneuro.org/datasets/ds003417). Processing scripts for this study are available on GitHub (https://github.com/pyushkevich/tau_maps_brain_2021). The source code for the web-based platform used for histology annotation is available at (https://github.com/pyushkevich/histoannot).