Abstract
Emergent approaches in regenerative medicine look towards the use of extracellular vesicles (EVs) as a next generation treatment strategy for intervertebral disc (IVD) degeneration (IVDD) because of their ability to attenuate chronic inflammation, reduce apoptosis, and stimulate proliferation in a number of tissue systems. Yet, there are no FDA-approved EV therapeutics on the market with an indication for IVDD which motivates this article to review the current state of the field and provide an IVD-specific framework to assess its efficacy. In this systematic review, we identify 29 preclinical studies that investigate EVs in relation to the IVD, and additionally review the regulatory approval process in an effort to accelerate emerging EV-based therapeutics towards FDA submission and timeline-to-market. The majority of studies focus on nucleus pulposus responses to EV treatment, where main findings show that stem cell-derived EVs can decelerate the progression of IVDD on the molecular, cellular, and organ level. Findings also highlight the importance of the EV parent cell’s pathophysiological and differentiation state, which affects downstream treatment responses and therapeutic outcomes. This systematic review substantiates the use of EVs as a promising cell-free strategy to treat IVDD and enhance endogenous repair.
Keywords: Intervertebral Disc, Regenerative Medicine, Extracellular Vesicles, Exosomes, Tissue Engineering
Table of Contents
Extracellular vesicles are an emerging cell-free treatment strategy for intervertebral disc degeneration with demonstrated therapeutic potential to promote endogenous repair. This systematic review identifies all preclinical studies to-date that investigate the use of stem cell- or primary cell-derived extracellular vesicles to treat models of intervertebral disc degeneration, aggregates their outcomes, and highlights future directions to advance this next-generation treatment strategy.
1. Introduction
The intervertebral disc (IVD) is the largest avascular organ in the body, and by consequence, has a poor intrinsic ability to heal itself upon injury and degeneration.[1–4] Accumulation of irreparable tissue damage can result in painful IVD degeneration (IVDD) involving loss of function, chronic pain, and disability from spinal pathologies such as IVD herniation which can warrant surgical intervention.[5,6] Although surgical treatment options are effective in relieving neuropathic and radicular pain, they do not restore the IVD’s native structure or biomechanical function and may accelerate IVDD.[7,8] Next generation treatment strategies call upon regenerative medicine to develop therapies that prevent back and leg pain by retarding degenerative processes and enhancing repair.[9] These strategies are broadly categorized as cell-based or cell-free therapies, where cellular therapies have gained much attention as a biologically active treatment option for IVDD.[10–13] Although cell-based approaches demonstrate some functional improvement compared to untreated controls, results are variable in preclinical IVDD models primarily due to the IVD’s harsh microenvironment and biomechanical loading patterns.[14–17] Given these challenging biological and mechanical conditions, it’s unsurprising that cell-based strategies have mixed outcomes and can be ineffective or lead to unfavorable outcomes that undermine the translational success of such approaches; notable adverse outcomes include injectate leakage, poor cell viability, and ectopic osteophyte formation following an intradiscal injection of an exogenous supply of cells.[18–21] Moreover, the regulatory pathway and bioethical use of human-sourced cell products pose additional obstacles for the translation of cell therapies for IVDD.[22]
While cell delivery strategies still hold promise to treat IVDD, cell-free alternatives may offer similar or greater therapeutic benefits with fewer translational obstacles, bioethical superiority, and a more straightforward pathway to regulatory approval.[10] As growing evidence suggests that cell therapies principally impart their therapeutic effects through paracrine signaling factors, scientists are aiming to identify and apply the soluble and vesicular fractions in the secretome as cell-free alternatives for therapy.[23–26] Extracellular vesicles (EVs) are a heterogeneous population of nanoparticles produced by nearly all cell types and are key mediators of intercellular communication that can efficiently transfer its molecular cargo from source cell to target cell.[27,28] The molecular contents encapsulated within EVs are directly representative of its source cell, rendering EVs as highly suitable agents for biomarkers as well as natural drug delivery systems.[29] EVs effector molecules such as microRNAs (miRs) emanate from the EV source cell and modulate target cell function through post-transcriptional regulation by binding to mRNA.[30] Therapeutic use of EVs initially emerged as a cell-free treatment strategy to repair cardiovascular, neurological, pulmonary, hepatic, renal, and dermal tissue systems, thus offering promise for musculoskeletal repair applications.[31–41] The majority of preclinical studies derive EVs from mesenchymal stem cells (MSCs), which elicit pro-regenerative and immunomodulatory responses by attenuating inflammation, reducing fibrotic remodeling, decreasing oxidative stress, increasing cell proliferation, and stimulating resident cell migration.[31–33,42–50]
EVs use as a therapeutic agent for musculoskeletal repair and regeneration is still in its infancy, with most studies published in the last 3–5 years.[32,51] Given their therapeutic benefits in other organ systems, it is clear that EVs also have the potential for the treatment of degenerative joint diseases such as osteoarthritis (OA) and IVDD in an effort to slow or prevent their progression in addition to relieving painful symptoms.[26,32,51–55] In OA-specific applications, MSC-derived EV treatment led to a downregulation of proinflammatory cytokines (IL-1β, IL-6, IL-8, etc.), inhibition of hyperalgesia, increase in ECM synthesis, as well as preservation of condylar cartilage and subchondral bone.[56] Although there are distinct differences in the etiology and pathophysiology of OA and IVDD, the phenotypic hallmarks between the two degenerative joint diseases are quite similar suggesting that MSC-derived EVs also could be a new treatment paradigm for IVDD (Figure 1).[57] When evaluating the effectiveness of EV treatment in the context of IVDD, it is imperative to consider tissue-specific evaluation criteria that are prescribed by the complex anatomy and physiology of the IVD. Ultimately, these assessments are related to the biomechanical function of the IVD as well as the biology of resident cells within the IVD that retains that function. However, there is currently no guiding framework to scientifically assess the efficacy of EV therapies in an IVD-specific manner.
Figure 1:
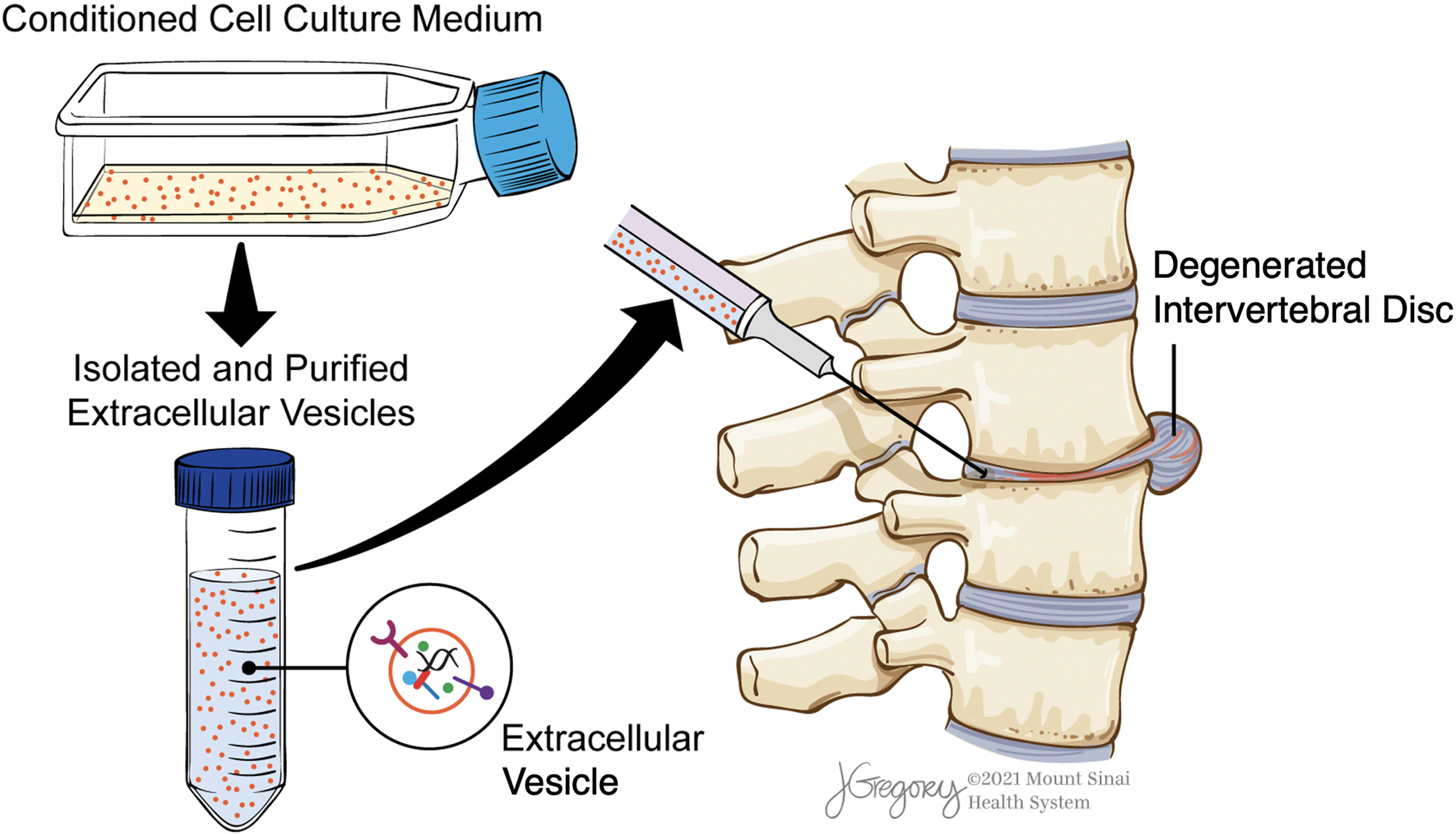
Workflow of EV therapy for IVDD. Cell culture platforms are first used to generate conditioned medium, which is then collected and processed for EV isolation and purification. Resuspended EVs are then delivered to the degenerated IVD via intradiscal injection for treatment.
In order to postulate a framework that evaluates EVs as a biologic for IVDD, it is first necessary to define the mechanism of action for EVs as a drug delivery system and compile outcomes from previous studies that use EVs to treat IVDD in preclinical models. To that end, we provide a systematic review that addresses these gaps in the literature by answering four primary questions: (1) What is the cellular mechanism of EV biogenesis and how do they transfer effector molecules from cell to cell?; (2) What post-isolation characterization methods do investigators use to examine biophysical and biochemical properties of EVs and ensure quality control?; (3) What are the known outcomes from previous in vitro and in vivo studies that use EVs as a treatment strategy for IVDD?; and (4) On the molecular, cellular, and tissue levels, what are key functional assessments needed to demonstrate effectiveness of EVs in ameliorating hallmarks of painful IVDD? We then identify critical avenues of future investigation and provide an overview of the regulatory approval pathway in order to advance the translation of EVs as a next generation cell-free alternative for IVDD therapy.
2. Literature Review Methods
PubMed, Scopus, and MEDLINE were the three literature databases used in this systematic review, where a total of 44 citations were identified in February 2021 from the primary search using the following search terms: (1) “Extracellular Vesicles” and “Intervertebral Disc”, (2) “Extracellular Vesicles” and “Annulus Fibrosus”, (3) “Extracellular Vesicles” and “Nucleus Pulposus”, (4) “Exosomes” and “Intervertebral Disc”, (5) “Exosomes” and “Annulus Fibrosus”, and (6) “Exosomes” and “Nucleus Pulposus” (Figure 2). After carefully examining each citation according to the inclusion and exclusion criteria, 29 non-duplicate original research articles were retrieved and used to identify EV characterization methods (Section 4), compile preclinical outcomes (Section 5), and propose an IVDD-specific conceptual framework for therapeutic evaluation (Section 6). Data regarding EV source cell type, source cell species, culture conditions, IVDD model system, target cell type, target cell species, EV dosage, and EV treatment outcomes were collected for all studies. Across the 29 articles, large heterogeneity was observed in EV source cell type and treatment outcomes, thus motivating a review of EV biogenesis and their mechanisms of action (Section 3). Given the translational promise of EV-based therapies for IVDD demonstrated in the 29 articles, regulatory and manufacturing considerations are included in this review and informed by FDA documentation (Section 7).
Figure 2:
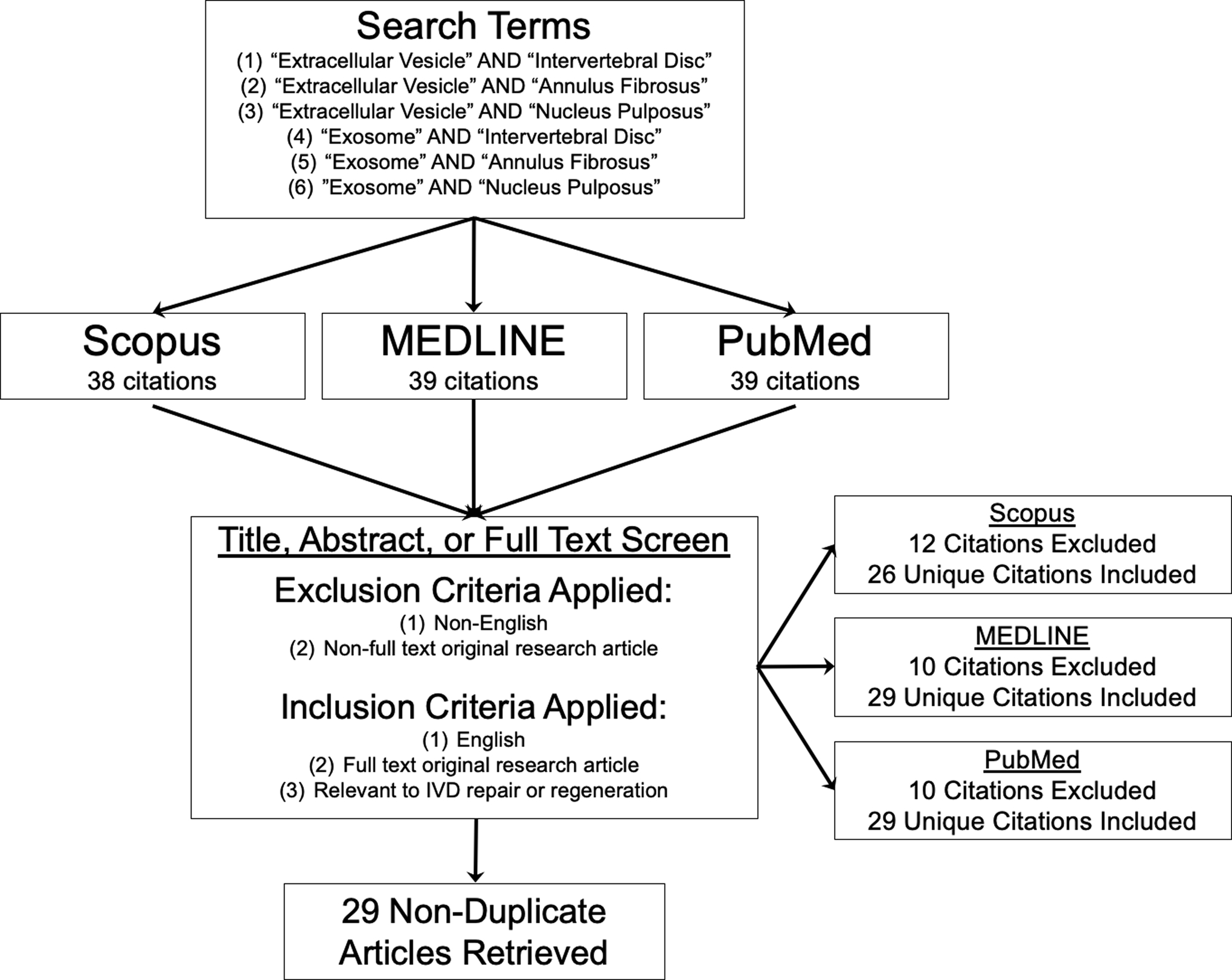
Literature review methods for this systematic review article. Six independent literature searches with the defined search terms were performed using three literature databases, and all identified articles were screened for exclusion or inclusion. Twenty-nine non-duplicate original research articles were included in this systematic review after identification and screening.
3. Exosome Biogenesis and Mechanisms of Action
Since the majority of studies included in this systematic review (22 of 29 articles) investigate the exosome subpopulation of EVs, this section is exclusively focused on exosome biogenesis and their mechanisms of action, given that exosome-related mechanisms are distinctly different than other EV subpopulations (i.e. microvesicles and apoptotic bodies). Exosomes constitute a vesicular fraction of the secretome and are a heterogeneous population of lipid-bound nanoparticles by composition and size.[58] Nearly all cell types produce exosomes, which carry a variety of biologically active effector molecules and range from 30 nm to 150 nm in diameter.[59,60] While their precise contents depend on the originating cell type and culture conditions, they are known to carry lipids, nucleic acids, amino acids, metabolites, an assortment of proteins (e.g. tetraspanins, ALIX, Flotillin, TSG101, heat shock proteins, Rab family proteins, enzymes, etc.), mRNAs, short non-coding RNAs (e.g. miRNAs, lncRNAs, tRNAs, etc.), and DNA.[61] These contents are encapsulated by plasma membranes that resemble lipid rafts in composition, containing high proportions of sphingomyelin, phosphatidylcholine, cholesterol, ceramide, and diacylglycerol.[62]
Exosome biogenesis starts with the formation of an early sorting endosome after endocytosis, which initially contains extracellular content and is subsequently loaded with molecular cargo from mitochondria, endoplasmic reticulum, and the trans-Golgi complex.[63,64] The early sorting endosome matures into the late sorting endosome and continues to exchange cargo in and out of the endosome via the trans-Golgi network.[65] Late sorting endosomes then undergo inward budding to form intraluminal vesicles (ILVs, also known as pre-exosomes) within newly formed multivesicular bodies (MVB). These MVBs fuse with the cell membrane through docking proteins and exocytose their ILVs into the extracellular space, which are then called exosomes.[51]
Exosomes serve as key mediators of intercellular communication involving several mechanisms of biogenesis and cellular uptake (Figure 3). Internalization of exosomes occurs through six possible pathways: (1) soluble signaling, (2) juxtacrine signaling, (3) fusion, (4) receptor-/raft-mediated endocytosis, (5) macropinocytosis, and (6) phagocytosis.[66,67] Through soluble and juxtacrine signaling, proteins on the surface of exosomes can bind to one or more receptors on target cells, inducing a cellular response through a downstream signaling cascade.[68] Through fusion, exosomes merge directly with the plasma membrane of the recipient cell, releasing their contents into the cytoplasm where they influence cellular expression and function.[69] In receptor-/raft-mediated endocytosis, exosomes can undergo clathrin-mediated endocytosis, caveolae-mediated endocytosis, as well as RhoA-, CDC42-, and ARF6-regulated endocytosis, leading to a cellular response or clearance.[70] In micropinocytosis, cellular protrusions on the recipient cell invaginate extracellular fluid containing particles, which can then lead to lysosomal degradation or transfer of molecular cargo that induces a cellular response.[67] Following cellular uptake, exosomes can either transfer their material to induce a cellular response, undergo lysosomal degradation, or take part in endosomal recycling.[67] Lastly, exosomes can undergo phagocytosis, in which these particles first bind with complement receptors or Fc receptors and are subsequently processed for lysosomal degradation.[71] Small exosomes are likely internalized via non-phagocytic processes, while larger exosomes are likely internalized via phagocytosis.[72]
Figure 3:
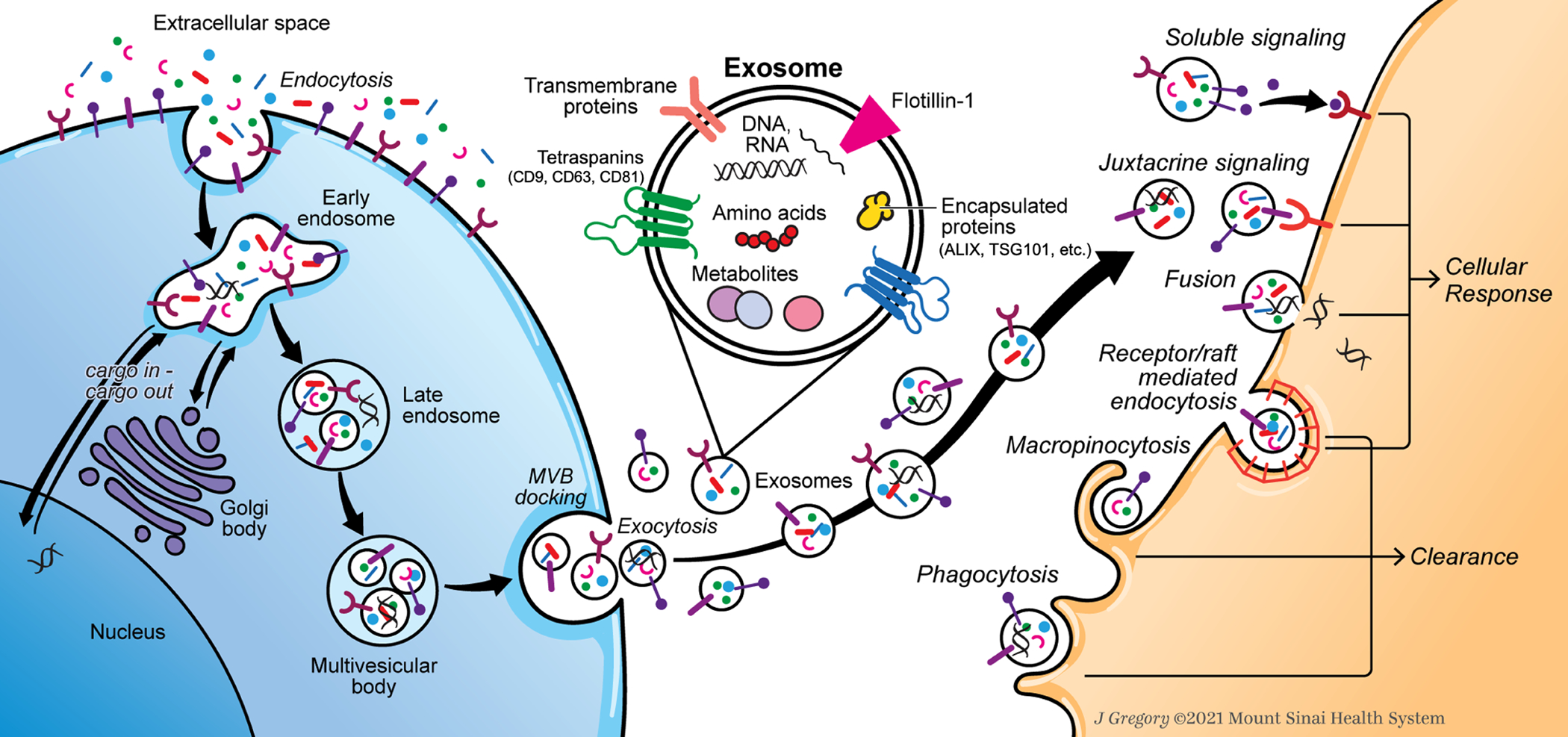
Exosome biogenesis and internalization mechanisms. Extracellular contents are first internalized and processed by the source cell, ultimately forming intraluminal vesicles contained within multivesicular bodies. Exosomes are then released into the extracellular environment and internalized by recipient cells through six known mechanisms, which can either induce a cellular response or lead to cellular clearance.
Exosome internalization can lead to the uptake and processing of its molecular cargo in recipient cells, where exosomal proteins and genetic material can induce a cellular response. miRs are one of the main types of exosomal effector molecules that modulate target cell expression and function.[59] There are four known methods in which miRs are sorted into naïve exosomes: (1) the neural sphingomyelinase 2-dependent pathway, (2) the miRNA motif and sumoylated heterogeneous nuclear ribonucleoproteins-dependent pathway, (3) the 3’-end of the miRNA sequence-dependent pathway, and (4) the miRNA induced silencing complex-related pathway.[73] By composition, miRs are short (~22 base pair) non-coding RNA strands that bind to complementary mRNA sequences and functionally promote target mRNA degradation or translational repression.[74] Also, miRs can serve as physiological ligands to specific Toll-like receptors (TLR) and yield an immune response through the TLR signaling cascade.[75] Exosomes characteristically contain a higher proportion of miRs than their parent cells, underscoring their key role in mediating miR transfer and post-transcriptionally regulating target cells.[76] A given miR transcript can have multiple downstream targets, thereby affecting a large number of gene networks and featuring high regulatory diversity in recipient cells.[59] This pleiotropic phenomena enables miRs to produce a wide variety of functional effects that can mediate tissue homeostasis, disease pathophysiology, and therapy.
4. EV Characterization Methods
EV characterization is important with respect to quality control since EVs are classified into three different groups based on size, molecular composition, and biogenesis mechanism. Exosomes are the smallest class of EVs, with sizes ranging from 30 nm to 150 nm.[51] Microvesicles are generally larger, ranging in size from 50 nm – 1000 nm, while apoptotic bodies are the largest, ranging in size from 50 nm – 5 μm.[77] Given the overlaps in size across each group, it is necessary to measure specific biochemical and biophysical properties unique to the EV subtype in order to confirm the class and quantity of EVs.
Among the 29 research articles in this review, there were a variety of EV isolation methods as well as biochemical and biophysical characterization techniques used to confirm the presence, type, and quantity of EVs in biological samples. (Table 1) We retained the nomenclature of the isolation product used in the original article for consistency with the literature and also provide the isolation methods described. We note that isolation methods are not always fully detailed in the text and can influence whether the EV products are pure exosomes. Nearly every study used a combination of at least one biochemical technique and at least one biophysical technique to characterize EV samples. The most common technique for biochemical characterization was western blot, where CD9, CD63, TSG101, and ALIX were the most frequent positive protein markers for blotting. This analysis was most often used in tandem with transmission electron microscopy (TEM) to physically characterize EV morphology and size. Nanoparticle tracking analysis (NTA) was the second most common biophysical characterization technique to determine EV size distribution. While dynamic light scattering (DLS), flow cytometry (FC), and scanning electron microscopy (SEM) are other common techniques, they were less frequently used for EV physical characterization. Associated disadvantages of these techniques may explain why DLS, FC, and SEM are less commonly used for biophysical characterization of EVs. Although DLS methods acquire data on the EV size distribution for a given sample, it fails to report the concentration of EVs at a given hydrodynamic diameter unlike NTA methods and generally requires more concentrated samples.[78] Conventional FC using immune-captured EVs or nano-FC may require instruments with a high degree of sensitivity and fluorophores with a large fluorescent intensity since EVs are limited by the number of antigen molecules due to their small size.[79–81] SEM methods can result in a ‘coffee ring phenomena’ when imaging EVs, which creates bias in size and quantity measurements of EVs in a given field of view.[82,83] In addition to biophysical characterization methods, a number of biochemical techniques were used across the 29 studies to characterize and quantify EV protein or RNA content. Methods such as the Qubit Protein Assay Kit, μBCA Protein Assay Kit, Fourier-transform infrared spectroscopy, and liquid chromatography-mass spectrometry were used in 15 studies for the quantification and/or identification of proteins in EV sample preparations. With respect to RNA quantification, quantitative polymerase chain reaction was used in 4 studies to determine levels of expression for specific RNA transcripts in EV samples.
Table 1:
EV Isolation and Characterization Methods.
| Isolation Product (Method) | Biophysical Characterization Techniques | Biochemical Characterization Techniques | Reference | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TEM | SEM | DLS | NTA | FC | Protein Quantification | PCR | WB | Positive EV Marker/Label | ||
| Exosomes (Differential UC with 30% sucrose cushion) | X | X | X | CD9, CD63, TSG101 | Xia et al, 2019[99] | |||||
| Exosomes (Differential UC) | X | CD63, TSG101 | Lu et al., 2017[100] | |||||||
| Extracellular vesicles (Differential UC & SEC) | X | PKH67 | Bach et al., 2017[101] | |||||||
| Exosomes (Differential UC) | X | X | CD63, CD81, TSG101 | Lan et al., 2019[102] | ||||||
| Exosomes (Differential UC) | X | X | Qi et al., 2019[103] | |||||||
| Exosomes (Differential UC) | X | TSG101, ALIX, CD9, CD63 | Cheng et al., 2018[104] | |||||||
| Apoptotic bodies (Sequential centrifugation) | X | X | Yuan et al., 2019[105] | |||||||
| Exosome-like vesicles (Differential UC) | X | X | X | X | CD9, TSG101, ALIX | Moen et al., 2017[106] | ||||
| Extracellular vesicles (Differential UC) | X | Bach et al., 2016[107] | ||||||||
| Lyo-secretome extracellular vesicles (Ultrafiltration) | X | X | X | Bari et al., 2018[108] | ||||||
| Exosomes (Low-speed centrifugation with Total Exosome Isolation Reagent) | X | X | X | CD63, TSG101, ALIX | Liao et al., 2019[109] | |||||
| Exosomes (Differential UC) | X | X | CD63, TSG101 | Chen et al., 2020[110] | ||||||
| Small extracellular vesicles (sEVs)/Exosomes (Differential UC) | X | X | X | X | X | CD9, CD63, CD81, FLOT1 | Hingert et al., 2020[111] | |||
| Exosomes (Differential UC) | X | X | X | TSG101, ALIX | Hu et al., 2020[112] | |||||
| Exosomes (Differential UC) | X | X | X | X | CD63, TSG101 | Li et al., 2020[113] | ||||
| Exosomes (Ultrafiltration with Total Exosome Isolation Reagent) | X | X | CD9, CD63 | Li et al., 2020[114] | ||||||
| Exosomes (Differential UC) | X | X | X | CD9, CD63, CD81, TSG101, ALIX | Luo et al., 2021[115] | |||||
| Exosomes (Total Exosome Isolation ExoQuick PLUS Exosome Purification Kit) | X | X | X | X | CD9, CD63, CD81 | Song et al., 2020[116] | ||||
| Exosomes (Differential UC) | X | X | X | X | CD63, TSG101, PKH67 | Sun et al., 2020[117] | ||||
| Exosomes (Differential UC) | X | X | X | CD63, TSG101, ALIX | Sun et al., 2021[118] | |||||
| Extracellular vesicles (Differential UC) | X | Tang et al., 2021[119] | ||||||||
| Extracellular vesicles (Differential UC) | X | X | X | CD68, CD81, TSG101 | Wen et al., 2021[120] | |||||
| Exosomes (Differential UC) | X | X | X | CD63, TSG101 | Xiang et al., 2020[121] | |||||
| Exosomes (Differential UC) | X | X | X | CD63, TSG101 | Xie et al., 2020[122] | |||||
| Exosomes (Differential UC) | X | X | X | X | CD9, CD63 | Yuan et al., 2020[123] | ||||
| Exosomes (Differential UC with 30% sucrose cushion) | X | X | X | CD9, CD63, CD81, TSG101 | Zhang et al., 2020[124] | |||||
| Exosomes (Differential UC) | X | X | X | CD9, CD63, TSG101, ALIX | Zhang et al., 2020[125] | |||||
| Exosomes (Differential UC) | X | X | X | CD63, TSG101 | Zhu et al., 2020[126] | |||||
| Exosomes (Chromatography) | X | X | X | CD9, CD63, CD81 | Zhu et al., 2020[127] | |||||
Abbreviations: UC = Ultracentrifugation; SEC = Size Exclusion Chromatography; TEM = Transmission Electron Microscopy; SEM = Scanning Electron Microscopy; DLS= Dynamic Light Scattering; NTA = Nanoparticle Tracking Analysis; FC = Flow Cytometry; PCR = Polymerase Chain Reaction; WB = Western Blot; CD = Cluster of Differentiation; TSG101 = Tumor Susceptibility Gene 101; ALIX = ALG-2 Interacting Protein X; ARF6 = ADP-Ribosylation Factor 6; FLOT1 = Flotillin-1.
Biochemical characterization techniques qualitatively and quantitatively describe the molecular composition and identity of EVs, which commonly includes western blotting to detect canonical EV proteins in reconstituted samples. It is recommended that three categories of markers be analyzed in bulk EV preparations to confirm the presence of EVs.[84] The first category includes the presence of transmembrane proteins, with some of the most commonly used non-cell specific markers being CD47, CD55, CD59, CD63, CD81, CD82, and FLOT1/2, as well as commonly used MSC-specific markers being CD9 and CD90.[84] The other two categories of EV markers include the presence of cytosolic proteins (TSG101, ALIX, etc.), as well as the presence of protein contaminants (e.g. apolioproteins A1/2 and B, albumin, and uromodulin) that are often co-isolated with EVs isolated from biofluids.[84] There exists a number of other transmembrane and cytosolic proteins that are less-commonly used as markers. This includes transmembrane proteins that are non-cell specific markers, as well as markers that are specific to cells and tissues other than MSCs. Additional techniques used to characterize biochemical properties are immunosorbent assays, which are techniques derived from enzyme-linked immunosorbent protein assays.[85] These techniques generally involve capturing EVs on a supporting surface coated with an antibody targeting EV-associated transmembrane proteins, such as CD9, CD63 and CD81. Once the antibodies are labeled with an enzyme to induce conversion of a fluorescent substrate, a spectrophotometer is commonly used to quantify the conversion. Additionally, captured EVs could be identified using fluorophore-linked immunosorbent assay or time-resolved-fluorescence immunoassay.[85] For the quantification of RNA transcripts in EVs, next generation sequencing methods can be used for a comprehensive transcriptomic analysis of RNA content or quantitative polymerase chain reaction can be used to determine levels of expression for specific RNA transcripts.
Biophysical characterization techniques qualitatively and quantitatively describe EV physical properties and can be used to identity the subtype of EVs in a given sample preparation. Biophysical characterization includes determination of average particle diameter, size distribution/polydispersity, and morphology. Electron microscopy techniques enable investigators to observe particle morphology and size by obtaining high resolution images on the nanoscale. TEM is the most common technique and is used to confirm whether a sample contains EVs and to visually examine sample purity for downstream applications.[86] TEM yields a 2D image of EV particles typically stained with uranyl acetate, where the characteristic morphology of an EV is a lipid-bound cup-shaped structure.[87] Additionally, cryo-TEM combined with immunogold labeling is used to differentiate between the three EV groups, analyze EV proteins, and track EV uptake by recipient cells.[83] SEM and atomic force microscopy (AFM) are used to determine the surface topography of EVs, with a round or saucer-shaped morphology characteristic of SEM and a cup or spherical morphology characteristic of AFM.[83,88]
Light scattering methodologies are an integral component of biophysical characterization techniques to measure particle size distribution. DLS involves the fluctuations of scattered light as a function of time due to the Brownian motion of suspended particles.[89] DLS measures hydrodynamic particle diameter ranging from 1 nm - 6 μm as well as the particle size distribution, where monodisperse suspensions yield the most accurate light scattering measurements.[90,91] NTA is another light scattering technique based on Brownian motion of particles in suspension, and allows for the determination of average particle size, modal value, and size distribution.[92] NTA allows for minimal sample preparation, particle size measurements as low as 30 nm, as well as the recovery of samples after analysis.[91] Like DLS, NTA is best suited for monodisperse samples, and although fluorescent labeling is used for the detection of antigens on EVs, it is limited to very bright fluorescent signals. Bead-free and bead-based FC can quantitatively characterize biophysical and biochemical EV properties by measuring scattered light at different angles to determine particle size as well as the presence of specific markers.[93,94] Forward scattered light in FC provides data regarding EV particle size and side scattered light in FC provides data on the granularity of internal structures.[88] Additionally, EVs can be labeled with fluorescent dyes or antibodies to detect the presence of specific proteins, lipids, or nucleic acids.[93] However, bead-free FC is limited by its ability to accurately size particles 500 nm in diameter and greater, since EVs below this size scatter laser light in the range of electronic noise, making it difficult to characterize smaller EVs.[91] There exists a number of other techniques to characterize EV physical properties, including tunable resistive pulse sensing and small-angle X-ray scattering, however these techniques are less commonly used due to the technical challenges of such techniques or other associated disadvantages.[91,95,96] Tunable resistive pulse sensing methods can be technically challenging for heterogeneous EV fractions, where large EVs and EV aggregates can frequently clog the nanopore during data acquisition.[97] Determining the size distribution of polydisperse EV fractions via small-angle X-ray scattering is difficult since large differences in EV diameter will result in large differences in the scattering signal.[98] Moreover, the low electron density contrast between EVs and aqueous buffers requires intense monochromatic X-rays, in which instruments with such specialized capabilities are located at specific synchrotron radiation facilities.[98]
EV characterization techniques provide important measures to qualitatively and quantitatively analyze EV samples, where western blot was most commonly used with NTA and TEM to determine EV protein expression, size distribution, concentration, and morphology. As EVs advance beyond preclinical development towards regulatory review, comprehensive proteomic and transcriptomic analyses will enable investigators to establish quality control criteria for EV manufacturing and determine therapeutic mechanisms of action. When assessing EV preparation methods, only 1 of the 29 studies incorporated a cryoprotectant to preserve the integrity and stability of EV fractions upon freeze-thaw cycles, where Bari et al. used 0.5% (w/v) Mannitol to treat lyophilized EVs.[108] Moreover, only 1 study by Cheng et al. examined miR stability by incorporating 0.4mg/mL RNase A and 0.1mg/mL Proteinase K in EV fractions and Xie et al. added these reagents to EVs for qRT-PCR analysis.[104,122] With respect to protein integrity for EV characterization, Zheng et al. added a 1X protease inhibitor cocktail to lysed EVs for western blot procedures.[125] Notably, 25 of the 29 studies do not report the addition of RNase and/or protease inhibitors in EV fractions to enhance product stability or report the use of RNases and/or proteases for biochemical characterization methods.
5. Overview of Preclinical Studies
Experimental parameters (Table 2) and associated outcomes (Table 3) were extracted from the 29 original research articles included in this systematic review and were broadly categorized across in vitro and in vivo studies that: (1) investigate the use of EVs derived from stem cells to treat terminally differentiated cells in vitro or the IVD in vivo, and (2) investigate the use of EVs derived from terminally differentiated cells in the IVD to treat another cell population.
Table 2:
Experimental Parameters and Configuration of Preclinical Studies.
| Reference | Cell Source of EV | Species of Cell Source | Cell Source Culture Conditions | Test Method of Application | Species of Target Cell or Tissue | Dosage of EVs Used* | Target Cell Type or Tissue | Biochemical Challenge? | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AF | NP | non-CEPCs | CEPCs | MSC | Intradiscal Injection (Whole IVD) | Dorsal Nerve Root | Other | ||||||||
| Xia et al., 2019[99] | BM-MSCs | Mouse | Normoxic | in vitro | Rat | 100 μg/mL | X | H2O2 (500μM) | |||||||
| in vivo | X (L4/5) | ||||||||||||||
| Lu et al., 2017[100] | BM-MSCs | Human | Normoxic | in vitro | Human | 50μg/mL | X | No | |||||||
| NPCs | Human | Normoxic | in vitro | Human | 1, 10, 100 μg/mL | X | |||||||||
| Bach et al., 2017[101] | NCs | Porcine | Hypoxic (5% O2) | in vitro | Canine | 1:1, 1:2, 1:4, 1:8, 1:16 | X | No | |||||||
| Human | X | ||||||||||||||
| Lan et al., 2019[102] | NPCs | Rat | Normoxic | in vitro | Rat | 0, 25, 50, 75, 100 μg/mL | X | No | |||||||
| Qi et al., 2019[103] | UC-MSCs | Human | Normoxic | in vitro | Human | N/A | X | High Glucose (35mM) | |||||||
| Cheng et al., 2018[104] | BM-MSCs | Human | Normoxic | in vitro | Rat | 1μg/mL | X | TNFα (5ng/mL) | |||||||
| in vivo | 1.5×106 particles/2 μL | X (Co6/7, Co8/9, Co 10/11) | No | ||||||||||||
| Yuan et al., 2019[105] | CEPCs | Rat | Not Specified | in vitro | Rat | 1 μg/mL | X | H2O2 (1–2mM) | |||||||
| Moen et al., 2017[106] | NPCs | Rat | Normoxic | in vivo | Rat | N/A | X | No | |||||||
| Bach et al., 2016[107] | NCs | Porcine | Hypoxic (5% O2) | in vitro | Bovine | N/A | X | No | |||||||
| Canine | Canine | 1X, 10X | X | No | |||||||||||
| Bari et al., 2018[108] | ASCs | Human | Normoxic | in vitro | Human | 5, 12.5, 25, 50, 75, 100, 150, 200 mg/mL | X | X | Fibroblasts | H2O2 (1mM) | |||||
| Liao et al., 2019[109] | BM-MSCs | Human | Not Specified | in vitro | Human | 10, 50, 100 μg/mL | X | AGEs (200μg/mL) | |||||||
| in vivo | Rat | 100 μg/mL | X (Co7/8, Co8/9, Co 9/10) | ||||||||||||
| Chen et al., 2020[110] | NPCs | Rat | Normoxic | in vitro | Rat | Not Specified | X | IL-1β (10ng/mL) | |||||||
| Hingert et al., 2020[111] | BM-MSCs | Human | Normoxic | in vitro | Human | 5×1010 particles/mL | X | X | No | ||||||
| Hu et al., 2020[112] | NPCs | Rat | Normoxic | in vitro | N/A | N/A | Rapamycin (100nM) | ||||||||
| Li et al., 2020[113] | BM-MSCs | Human | Normoxic | in vitro | Human | 1, 5, 10, 15, 20, 25, 30 μg/mL | X | HCl pH(Medium) = 6.5–6.7 & 5.9–6.1 | |||||||
| Li et al., 2020[114] | BM-MSCs | Human | Normoxic | in vitro | Human | Not Specified | X | IL-1β (10ng/mL) | |||||||
| Luo et al., 2021[115] | CESCs | Rat | Normoxic | in vitro | Rat | 40μg/mL | X | TBHP (100μmol/mL) | |||||||
| in vivo | X (Levels not specified) | LY294002 (20μmol/mL) | |||||||||||||
| Song et al., 2020[116] | NPCs | Human | Normoxic | in vitro | Human | Not Specified | X | No | |||||||
| in vivo | Rat | X (Co5/6, Co6/7, Co7/8) | |||||||||||||
| Sun et al., 2020[117] | NCs | Rat | Normoxic | in vitro | Human | 0, 50, 100, 150 μg | HUVEC | No | |||||||
| in vivo | Mouse | Not Specified | X (C4/5) | ||||||||||||
| Sun et al., 2021[118] | AFCs | Human | Normoxic | in vitro | Human | 100μg/mL | HUVEC | No | |||||||
| Tang et al., 2021[119] | NPCs | Human | Normoxic | in vitro | Human | 1×109 particles/mL | X | No | |||||||
| PMEFs | Mouse | in vivo | Mouse | 2.6×108 particles/2μL | X (L4/5, L5/6, L6/S1) | ||||||||||
| Wen et al., 2021[120] | BM-MSCs | Rat | Normoxic | in vitro | Mouse | 20μM | X | No | |||||||
| in vivo | 100μg/mL | X (L2/3, L3/4, L4/5) | |||||||||||||
| Xiang et al., 2020[121] | USCs | Human | Normoxic | in vitro | Human | 10, 50, 100 μg/mL | X | 1.0MPa – 90% N2, 5% CO2, 5% O2 | |||||||
| in vivo | Rat | 100μg/mL | X (Co4/5) | No | |||||||||||
| Xie et al., 2020[122] | MSCs | Rat | Normoxic | in vitro | Rat | Not Specified | X | TBHP (20–60μM) | |||||||
| in vivo | X (Co7/8) | No | |||||||||||||
| Yuan et al., 2020[123] | PLMSCs | Human | Normoxic | in vitro | Human | 1×1010 particles/mL | X | TNFα (10ng/mL) | |||||||
| in vivo | Mouse | X (L5/6, L6/S1) | No | ||||||||||||
| Zhang et al., 2020[124] | MSCs | Human | Normoxic | in vitro | Mouse | 20μg/mL | X | LPS (5mmol/L) | |||||||
| in vivo | X (Levels not specified) | No | |||||||||||||
| Zhang et al., 2020[125] | NPCs | Rat | Normoxic | in vitro | Rat | Not Specified | X | IL-1β (Concentration not specified) | |||||||
| Zhu et al., 2020[126] | BM-MSCs | Mouse | Normoxic | in vitro | Mouse | 50μg/mL | X | IL-1β (10ng/mL) | |||||||
| Zhu et al., 2020[127] | BM-MSCs | Rat | Normoxic | in vitro | Rat | Not Specified | X | TNFα (20ng/mL) | |||||||
MSC = Mesenchymal Stem Cell; BM-MSC = Bone Marrow-derived Mesenchymal Stem Cell; NPC = Nucleus Pulposus Cell; NC = Notochordal Cell; AFC = Annulus Fibrosus Cell; UC-MSC = Umbilical Cord-derived Mesenchymal Stem Cell; ASC = Adipose-derived Mesenchymal Stromal Cell; CEPC = Cartilage Endplate Chondrocyte; CESC = Cartilage Endplate Stem Cell; PMEF = Primary Mouse Embryonic Fibroblast; USC = Urine-derived Stem Cell; PLMSC = Placental Mesenchymal Stem Cell; HUVEC = Human Umbilical Vein Endothelial Cell;
Bold = EV concentration that elicited the strongest effect sizes in downstream cellular and/or tissue responses
Table 3:
Regenerative Outcomes in Preclinical EV Studies.
| Reference | Cell Source of EV | Test Method of Application | Species of Target Cell or Tissue | Outcome |
|---|---|---|---|---|
| Xia et al., 2019[99] | BM-MSCs | in vitro | Rat | 1. EV treatment attenuated NPC apoptosis after H2O2 exposure |
| 2. EV treatment restored iNOS, IL6, MMP3, MMP13, COL2A1, CASP1, IL1b, TXNIP, NLRP3, and SOX9 to untreated control levels | ||||
| 3. EV treatment increased the number of mitochondria and reduced mitochondrial dysfunction | ||||
| in vivo | 1. Intradiscal injection of EVs at 1μg/μL slowed the decrease in disc height index through 8 weeks compared to injury group | |||
| 2. Intradiscal delivery of EVs slowed the progression of IVDD through 8 weeks assessed by histological scoring | ||||
| 3. Intradiscal delivery of EVs restored MMP13 and COL2A1 equivalent to uninjured control through 8 weeks | ||||
| Lu et al., 2017[100] | BM-MSC | in vitro | Human | 1. EV treatment increased proliferation rate over 12-day period |
| 2. EV treatment increased ACAN, COL2A1, SOX9, and TIMP1 over 21 days in culture | ||||
| 3. EV treatment decreased MMP1 and MMP3 over 21 days in culture | ||||
| NPC | in vitro | 1. Migration activity increased with an increase in NPC-EV concentration | ||
| 2. NPC-EV treatment increased MSC ACAN, SOX9, COL2A1, HIF1a, CA12, and KRT19 expression | ||||
| 3. Changes in MSC expression were greater after EV treatment than indirect co-culture model with NPCs | ||||
| Bach et al., 2017[101] | NCs | in vitro | Canine | 1. EV treatment increased GAG and GAG/DNA in chondrocyte-like cell aggregates |
| 2. EV treatment increased GAG and collagen content in culture medium | ||||
| 3. Increase in EV treatment concentration increased DNA content, GAG content, and GAG/DNA ratio in a 7-day culture period | ||||
| 4. Significant positive correlation between total number of EVs used to treat chondrocyte-like cell aggregates and GAG content and GAG/DNA ratio | ||||
| Human | 1. EV treatment increased DNA, GAG and GAG/DNA in chondrocyte-like cell aggregates | |||
| 2. EV treatment increased GAG and collagen content in culture medium | ||||
| Lan et al., 2019[102] | NPCs | in vitro | Rat | 1. EV treatment increased ACAN, SOX9, COL2A1 expression in hBM-MSCs |
| 2. Knock down of Notch1 in MSCs resulted in higher upregulation of ACAN, SOX9, COL2A1 after EV treatment than controls | ||||
| Qi et al., 2019[103] | UC-MSCs | in vitro | Human | 1. EV treatment protected NPMSCs from high glucose induced injury |
| Cheng et al., 2018[104] | BM-MSC | in vitro | Rat | 1. Lower apoptosis rate for NPCs in EV treatment group when compared to untreated controls after application of TNFα |
| 2. miR-21 delivery via EVs inhibited TNFα-induced NPC apoptosis by targeting PTEN in the PI3K-Akt pathway | ||||
| in vivo | 1. Intradiscal injection of EVs alleviated TNFα induced NPC apoptosis in vivo | |||
| 2. No difference in Pfirmann grade between uninjured control and EVs treated IVDs | ||||
| 3. EV-treated IVDs appeared histologically similar to uninjured control IVDs through H&E staining | ||||
| Yuan et al., 2019[105] | CEPCs | in vitro | Rat | 1. Treatment with apoptotic bodies (Abs) promoted mineralization and upregulation of ALP, RUNX2, OCN, and COL1A1 in endplate chondrocytes |
| 2. Abs treatment promoted PPi metabolism modifications in endplate chondrocytes with an increase in Pi and decrease in PPi | ||||
| 3. Abs treatment decreased levels of ENPP1 and ANK expression, but increased TNAP expression | ||||
| 4. Treatment with H2O2 significantly increased the generation of Abs due to oxidative stress | ||||
| Moen et al., 2017[106] | NPCs | in vivo | Rat | 1. Application of miR-223-3p onto dorsal nerve roots decreased C-fiber responses (indirect application of NPC-EVs) |
| 2. miR-223 upregulated in NPC-EVs when the NP tissue is exposed to dorsal nerve roots | ||||
| Bach et al., 2016[107] | NCs | in vitro | Bovine | 1. The effects of porcine NCCM-P factors were negligible on bovine CLCs |
| Canine | 1. Canine NCCM pelletable factors increased the canine CLC GAG, GAG/DNA and COL2 content compared with controls | |||
| 2. Canine NCCM pelletable factors decreased VEGF and increased KRT19 expression | ||||
| 3. At least 4 d of freezing at −70 °C did not influence the biological activity of canine Canine NCCM pelletable factors on canine CLC micro-aggregates compared to non-frozen controls | ||||
| 4. Protein aggregates and EVs exerted a moderate concentration-dependent anabolic effect, but only on canine CLCs | ||||
| Bari et al., 2018[108] | ASCs | in vitro | Human | 1. Exosomes were less abundant than microvesicles in lyo-secretome |
| 2. Lyo-secretome was not hematolytic at any of the tested concentrations | ||||
| 3. Cell metabolic activity remained at least ≥60% when treated with lyo-secretome | ||||
| 4. Lyo-secretome became cytotoxic to NPCs at a concentration of over 50 mg/mL | ||||
| 5. Lyo-secretome (5–50 mg/mL) protected NPCs from the oxidative stress damages induced by H2O2 | ||||
| Liao et al., 2019[109] | BM-MSCs | in vitro | Human | 1. EVs led to protective effect by reducing ER stress-induced apoptosis |
| 2. EVs regulated UPR activation in response to AGEs-induced ER stress in human NPCs | ||||
| 3. EVs protected against ER stress-related apoptosis partly through the AKT and ERK activation in human NPCs | ||||
| in vivo | Rat | 1. EVs inhibited the activation of AGEs-induced ER stress-related cell apoptosis and slowed the progression of IVDD | ||
| Chen et al., 2020[110] | NPCs | in vitro | Rat | 1. Senescent NPC EVs showed an increase in the relative expression of P21 and P53 |
| 2. Senescent NPC-EV treatment led to a lower growth rate, fewer colony forming units, and higher SA-β-gal positivity in healthy NPCs | ||||
| 3. Senescent NPC-EV treatment led to more G1 phase cells and fewer S phase cells compared to the control group | ||||
| 4. siRNA transfection of EV treated NPCs led to a decrease in P21 and P53 expression, higher growth rate, and lower SA-β-gal positivity | ||||
| Hingert et al., 2020[111] | BM-MSCs | in vitro | Human | 1. EV treatment increased cell proliferation and decreased cellular apoptosis in degenerated disc cells |
| 2. EV-treated disc cell pellets demonstrated 3X greater ECM production compared to control disc cell pellets | ||||
| 3. EV treatment suppressed secretion of MMP-1 in disc cells | ||||
| Hu et al., 2020[112] | NPCs | N/A | N/A | 1. Rapamycin and bafilomycin A1 led to induction of NPC autophagy and EV secretion in an autophagy-dependent manner |
| 2. siRNA against ATG5 induced accumulation of ILVs and decrease in isolated EVs | ||||
| 3. Knockdown of RhoC and ROCK2 with siRNA inhibited secretion of EVs | ||||
| Li et al., 2020[113] | BM-MSCs | in vitro | Human | 1. Proliferation activity, collagen II, and aggrecan expression decreased in NPCs cultured at pH 5.9 – 6.7 |
| 2. Caspase-3 and MMP-13 expression increased in NPCs cultured at pH 5.9 – 6.7 | ||||
| 3. EV treatment led to an upregulation of collagen II and aggrecan, and a downregulation of matrix-degrading enzymes | ||||
| Li et al., 2020[114] | BM-MSCs | in vitro | Human | 1. EVs suppressed IL1β-induced inflammation and apoptosis of AF cells by suppressing autophagy |
| 2. EVs supported AF cell viability after IL1β treatment | ||||
| 3. EVs inhibited AF cell autophagy by activating the PI3K/AKT/mTOR signaling pathway | ||||
| Luo et al., 2021[115] | CESCs | in vitro | Rat | 1. Treatment with healthy CESC-EVs inhibited apoptosis compared to degenerated CEP stem cell-derived EVs |
| 2. Healthy CESC-EVs inhibited apoptosis of NPCs by activating the PI3K/AKT pathways | ||||
| in vivo | 1. Healthy CESC-EVs alleviated IVDD via activation of PI3K/AKT pathways | |||
| Song et al., 2020[116] | NPCs | in vitro | Human | 1. circRNA_0000253 was highly upregulated in degenerative NPC-EVs |
| 2. circRNA_0000253 promoted an IVDD phenotype by adsorbing miRNA-141-5p and downregulating SIRT1 in vitro | ||||
| in vivo | Rat | 1. circRNA_0000253 accelerated IVDD by adsorbing miRNA-141-5p and downregulating SIRT1 in vivo | ||
| Sun et al., 2020[117] | NCs | in vitro | Human | 1. 0.5MPa-conditioned EVs inhibit endothelial cell angiogenesis through miR-140-5p and regulate Wnt/β-catenin signaling |
| 2. NP EV-derived miR-140-5p is negatively associated with angiogenesis in clinical samples | ||||
| in vivo | Mouse | 1. 0.5MPa-conditioned EV treatment reduced vascularization in degenerated IVDs | ||
| Sun et al., 2021[118] | AFCs | in vitro | Human | 1. HUVECs phagocytose AFC-EVs |
| 2. Degenerated AFC-EVs promoted cell migration and upregulation of IL-6, TNF-α, MMP-3, MMP-13, and VEGF, while non-degenerated AF cell-derived EVs demonstrated inverse effects | ||||
| Tang et al., 2021[119] | NPCs | in vitro | Human | 1. Bulk electroporation of cells with FOXF1 led to FOXF1 plasmids in designer EVs and demonstrated efficient cell uptake |
| PMEFs | in vivo | Mouse | 1. Injection of FOXF1-loaded EVs into IVDs showed significant upregulation of FOXF1 and Brachyury compared to controls | |
| Wen et al., 2021[120] | BM-MSCs | in vitro | Mouse | 1. EV treatment led to an increase in COL2 and ACAN staining intensity and decrease in SA-β and TUNEL positive NPCs |
| 2. A reduction in EV-derived miR-199a led to an impaired protective effect of EVs on NPCs | ||||
| 3. EV-derived miR-199a promotes repair by targeting GREM1 and downregulating TGFβ pathway | ||||
| in vivo | 1. EV treatment led to increased levels of miR-199a and decreased levels of MMP3-, MMP6-, TIMP1-, and TUNEL-positive cells | |||
| Xiang et al., 2020[121] | USCs | in vitro | Human | 1. EV treatment led to a decrease in GRP78, GRP94, Caspase 3, and Caspase 12 expression under stress-induced conditions |
| 2. EVs inhibit excessive activation of unfolded protein response under stress-induced conditions | ||||
| 3. EVs regulate stress by activating AKT and ERK signaling pathways in NPCs under stress-induced conditions | ||||
| in vivo | Rat | 1. EVs inhibited ER stress-associated cell apoptosis and decelerated IVDD progression in vivo | ||
| Xie et al., 2020[122] | MSCs | in vitro | Rat | 1. EVs inhibited apoptosis and TBHP-induced CEP calcification |
| 2. Downregulation of miR-31-5p impaired EV protective effects | ||||
| 3. miR-31-5p negatively regulated ATF6-related ER stress and inhibited CEP apoptosis and calcification | ||||
| in vivo | 1. Sub-endplate injection of EVs ameliorate IVDD hallmarks | |||
| 2. Downregulation of EV-derived miR-31-5p inhibited EV protective effects in vivo | ||||
| Yuan et al., 2020[123] | PLMSCs | in vitro | Human | 1. EV-derived AntagomiR-4450 ameliorates NPC damage by promoting proliferation and migration |
| 2. EV-derived AntagomiR-4450 decreased MMP13, IL6, IL1β, CASP3 expression, and increased COL2 and ACAN expression | ||||
| in vivo | Mouse | 1. EV-derived AntagomiR-4450 attenuated IVDD damage by repressing miR-4450 and increasing ZNF121 expression | ||
| 2. EV-derived AntagomiR-4450 ameliorated gait abnormality | ||||
| Zhang et al., 2020[124] | MSCs | in vitro | Mouse | 1. EV treatment inhibited pyroptosis by suppressing the NLRP3 pathway |
| 2. EV treatment inhibited LPS-induced pyroptosis in NPCs | ||||
| 3. EV-derived miR-410 suppressed LPS-induced pyroptosis in NPCs | ||||
| in vivo | 1. EV treatment and miR-410 treatment alleviated IVDD severity | |||
| Zhang et al., 2020[125] | NPCs | in vitro | Rat | 1. Rapamycin treatment led to an increase in miR-27a in NPCs and their EVs |
| 2. EV-derived miR-27a alleviated IL1β-induced ECM degradation by downregulating MMP13 in NPCs | ||||
| Zhu et al., 2020[126] | BM-MSCs | in vitro | Mouse | 1. EV treatment attenuated NPC apoptosis by reducing inflammatory cytokine secretion and activating MAPK pathway |
| 2. EV-derived miR-142-3p targets mixed MLK3 and inhibits NPC apoptosis and promotes MAPK signaling | ||||
| 3. MLK3 overexpression abolished EV effects on inflammation, NPC apoptosis, and MAPK signaling activation | ||||
| Zhu et al., 2020[127] | BM-MSCs | in vitro | Rat | 1. EV treatment led to inhibition of apoptosis, ECM catabolism, and fibrosis in TNFα-treated NPCs |
| 2. miR-532-5p was abundant in TNFα-treated MSC-derived EVs and was less abundant in apoptotic NPCs | ||||
| 3. RASSF5 is an empirically validated target of miR-532-5p |
MSC = Mesenchymal Stem Cell; BM-MSC = Bone Marrow-derived Mesenchymal Stem Cell; NPC = Nucleus Pulposus Cell; NC = Notochordal Cell; AFC = Annulus Fibrosus Cell; UC-MSC = Umbilical Cord-derived Mesenchymal Stem Cell; ASC = Adipose-derived Mesenchymal Stromal Cell; CEPC = Cartilage Endplate Chondrocyte; CESC = Cartilage Endplate Stem Cell; PMEF = Primary Mouse Embryonic Fibroblast; USC = Urine-derived Stem Cell; PLMSC = Placental Mesenchymal Stem Cell; HUVEC = Human Umbilical Vein Endothelial Cell.
5.1. Evaluation of EVs using in vitro model systems
In vitro systems enable the investigation of EVs in highly controlled environments and allow investigators to determine therapeutic mechanisms of action, where 28 of the 29 studies in this literature review evaluated EVs using in vitro experimental configurations.[99–105,107–127]
5.1.1. Stem cell-derived EVs rescue biochemically challenged IVD cells
Stem cell-derived EVs were applied in to in vitro challenge experiments to rescue hallmarks of degeneration in 14 of the 29 studies (Table 2).[99,103,104,108,109,113–115,121–124,126,127] The degenerative microenvironment was emulated in vitro through cellular challenges including exposure to advanced glycation end products (AGEs), hydrogen peroxide (H2O2), tumor necrosis factor alpha (TNFα), tert-butyl hydroperoxide (TBHP), lipopolysaccharide (LPS), interleukin-1β (IL1β), high concentrations of glucose, acidic pH, and high pneumatic pressure. Each biochemical challenge has the ability to induce a cellular stress response that is implicated in progressive degeneration. Despite some differences in their intracellular signaling pathways, these biochemical challenges at their respective working concentrations can all serve as damage-associated molecular patterns and produce common cellular responses such as NLRP3 inflammasome activation.[99,128–131] Upon NLRP3 activation, there is an increase in caspase 1 activity and upregulation of IL1β and IL18 cytokines, resulting in a proinflammatory state that emulates the degenerative environment.[132,133] Investigators used these in vitro systems of simulated degeneration to screen the therapeutic efficacy of EVs in attenuating damage-associated molecular pattern-induced apoptosis, catabolism, and inflammation.
EVs in these 15 rescue studies were derived from various cell sources, including MSCs from bone marrow (7 studies), adipose (1 study), umbilical cord (1 study), placental (1 study), and unspecified (2 studies) tissues, urine-derived stem cells (1 study), and cartilage endplate (CEP)-derived stem cells (1 study). Regardless of cell source, EVs surprisingly demonstrated protective effects when primary cells were exposed to biochemical challenges, such as a reduction in apoptosis, attenuated pro-inflammatory cytokine production, decreased catabolic activity, alleviated oxidative stress, and ameliorated endoplasmic reticulum stress. EV treatment also demonstrated partial restoration of gene expression levels to that of control nucleus pulposus cells (NPCs) (e.g. COL2A1, ACAN, SOX9, etc.), supported annulus fibrosus cell viability, and inhibited calcification of CEP chondrocytes.[99,103,104,108,109,113–115,121–127] Despite the variety of MSC tissue sources across studies, MSC-derived EVs may impart similar protective effects upon treatment given that 60% of EV protein content was conserved between EVs from differing MSC sources.[43,134] Although MSC-EV miR signatures are more sensitive to MSC tissue source than protein content, similar regenerative outcomes were observed for other tissues when systematically comparing treatment effects of MSC-EVs derived from bone marrow, umbilical cord, and adipose tissues.[135–137]
The mechanisms of action by which EVs alleviated cellular stress in these rescue experiments corresponded to differences in the respective mechanisms of action for each biochemical challenge, despite common therapeutic effects such as a reduction of programmed cell death observed across studies (Table 3). Two studies showed that exosomal miR-21 and miR-532-5p prevented TNFα-induced apoptosis in NPCs by targeting the PI3K/AKT pathway and RASSF5, respectively, after EV treatment.[104,127] A study that induced NPC apoptosis through IL1β treatment found that exosomal miR-142-3p reduced apoptosis by targeting mixed MLK3 in MAPK signaling.[126] Another study demonstrated that exosomal miR-410 targeted the NLRP3 3’UTR and reduced NPC pyroptosis after LPS treatment.[124] Although treatment with EVs containing these miRs led to a reduction in programmed cell death through multiple pathways, a number of small RNAs and proteins in MSC-derived EVs are responsible for other protective effects, highlighting additional avenues of research to comprehensively determine mechanisms of action for this type of biologic therapy.
5.1.2. Effects of stem cell-derived EVs on IVD cells without a biochemical challenge
Three studies evaluated MSC-EV treatment effects on NPCs without the use of biochemical agents that instigate damage-associated molecular pattern responses in culture.[100,111,120] These 3 studies show that MSC-EVs promoted NPC proliferation and inhibited apoptosis, generally supporting cell growth and survival. In particular, NPC proliferation rate increased with time in culture, suggesting that sustained exposure to MSC-EVs leads to the greatest increase in NPC proliferation.[100,111] Evaluation of ECM markers demonstrated that treatment with MSC-EVs led to increases in ACAN, SOX9, COL2, and TIMP1 expression, as well as decreases in MMP1 and MMP3 expression. Together, these outcomes indicate that MSC-EVs can stimulate pro-regenerative activities in terminally differentiated cells derived from degenerative IVD tissue by attenuating catabolic activity, promoting ECM elaboration, and supporting cell proliferation.[100,111,120]
5.1.3. Effects of primary IVD cell-derived EVs on primary cells and stem cells
Another subset of in vitro studies evaluate the regenerative capacity of EVs derived from terminally differentiated cells in the nucleus pulposus (NP).[100–102,107,125] Bach et al. parsed out the effects of soluble factors (i.e. peptides and proteins) and pelletable factors (i.e. EVs) from notochordal cell conditioned media (NCCM) on bovine and canine chondrocyte-like cell (CLC) proliferation and ECM anabolism.[101,107] The investigators reported that pelletable factors from porcine NCCM had negligible effects on bovine CLCs but showed that pelletable factors from canine NCCM significantly enhanced GAG and collagen type II production in canine CLCs.[107] A follow up study by Bach et al. further investigated the effects of EVs from porcine NCCM on human and canine CLCs in 3D culture, and reported outcomes that did not necessarily corroborate with their previous work.[101] In this subsequent study, Bach et al. reported that EVs derived from porcine NCCM increased GAG deposition in both human and canine CLC aggregates as well as showed an increase in DNA content in human CLC aggregates. These outcomes suggest that the species of the EV source may play a role in the regenerative potential of EV therapeutics and that congruence between species of EV source and target tissue/cell may be necessary to elicit proliferative and anabolic effects. Since EVs principally act by transcriptional regulation to modulate target cell function, discrepancy of species between the EV source and target tissue/cell may lead to null outcomes after treatment if the target genes of interest are not conserved across species. To ensure that EV treatment elicits a response in a target cell, it is imperative that small RNAs within the EV feature exact antisense sequences in the regions of interest for the mature mRNAs that it aims to regulate.
The regenerative effects of NPC-EVs engineered with FOXF1 plasmids on degenerate NPCs in 3D agarose gels was investigated by Tang and colleagues. EVs were successfully engineered by electroporation to encapsulate FOXF1 plasmids and NPCs efficiently internalized their cargo. Delivery of engineered NPC-EVs significantly modified degenerate NPC phenotype by upregulating FOXF1 and KRT19, downregulating IL1β, IL6, and MMP13, and increasing in GAG production, demonstrating that FOXF1 could upregulate healthy NP markers while attenuating effects of inflammation and catabolism. This study suggests that engineered EVs may have greater therapeutic potential than naïve EVs if the molecular contents are modified to promote a healthy phenotype.
NPC-EVs were investigated for their ability to induce differentiation of MSCs towards NP-like cells in two studies.[100,102] Lu et al. showed that NPC-EVs promoted an NP-like phenotype in hBM-MSCs over a 14-day culture period, demonstrated by a monotonic increase in ACAN, SOX9, COL2, HIF1α, CA12, and KRT19 expression. Lan et al. reproduced these findings and also showed that NPC-EVs were more effective in inducing MSC differentiation when compared to an indirect co-culture system with NPCs. Both studies postulate different mechanisms in which this differentiation response is elicited. Lu et al. suggests that this response is due to a high abundance of TGFβ in the EV samples, where TGFβ is a necessary factor for chondrogenic differentiation. Lan et al. attempted to investigate the Notch1 pathway through application of DAPT, a gamma-secretase inhibitor, to their MSC culture to knockdown Notch1. They showed that inhibition of the Notch1 pathway led to enhanced MSC differentiation towards an NP-like phenotype after EV treatment, where COL2A1, ACAN, and SOX9 were significantly upregulated compared to controls. Moreover, they applied SJAG1, a Notch ligand, to their MSC culture to enhance Notch1 signaling, and showed the opposite trends found in their DAPT-treated cultures, suggesting that inhibition of the Notch1 pathway facilitates NPC-EV induced differentiation of MSCs.
EVs from NP and AF cells can also affect cellular and pathological processes such as autophagy, angiogenesis, and vascularization implicated in the progression of IVDD. Hu et al. investigated the relationship between autophagy and EV secretion in NPCs, where rapamycin-activated autophagy increased the number of NPC-EVs while the inhibition of autophagy through bafilomycin A1 demonstrated the opposite effect.[112] The use of siRNA to silence the expression of ATG5, a gene implicated in autophagy, resulted in a decrease in the number of NPC-EVs and validated their original finding. Additional siRNA knockdown experiments identified that the RhoC/ROCK2 pathway modulates autophagy-regulated EV secretion, which may serve as a target for the synthesis of EVs as a therapeutic for IVDD. Sun et al. investigated the effect of mechanical loading on notochordal cell (NC)-EVs and their ability to inhibit angiogenesis, which is a pathological signature of IVDD.[117] NC-EVs collected under a 0.5MPa compressive load demonstrated an ability to inhibit angiogenesis by transferring miR-140-5p to endothelial cells and regulating the Wnt11/β-catenin signaling pathway. Notably, NC-EVs collected under 0MPa and 1MPa compressive loads did not differentially express miR-140-5p, highlighting the important role of the cellular culture environment in determining the EV molecular signature and regulatory capabilities. Sun et al. went on to investigate AF cell-EVs and determine if EVs originating from AF cells also possess a regulatory role in IVD vascularization.[118] EVs originating from degenerated AF cells promoted endothelial cell migration and expression of IL6, TNFα, MMP3, MMP13, and VEGF, whereas EVs obtained from healthy AF cells showed inverse effects. These findings indicate that degeneration grade influences the regulatory landscape of EVs derived from terminally differentiated cells in the IVD, where degenerated AF cell-EVs promote sustained inflammation and vascularization.
5.1.4. Effects of EV source cell pathophysiological state on downstream responses
EVs represent the pathophysiological state of their parent cell and a subset of 3 in vitro studies examined the effects of EVs produced by terminally differentiated cells from degenerated IVD tissues and assessed their role in IVDD pathogenesis. Song et al. isolated EVs from NPCs of varying degeneration grades and identified circRNA_0000253 and miR-141-5p as the most upregulated RNAs in EVs derived from degenerative tissue.[116] It was determined that circRNA_0000253 could competitively adsorb miR-141-5p and in turn downregulate SIRT1 to promote the expression of inflammatory and catabolic markers and decrease collagen II and aggrecan production, suggesting that circRNA_0000253 could serve as a potential therapeutic target to treat IVDD. Chen et al. investigated senescent NPC-EVs and their role in NPC senescence, which is a known hallmark of IVDD.[110] Senescent NPC-EVs were used to treat healthy NPCs, resulting in increased expression of senescence-related protein markers P53 and P21, as well as reduced proliferation and colony formation. siRNA transfection of the P53/P21 pathway resulted in significantly decreased expressions of P53 and P21, as well as increased NPC proliferation, indicating that senescent NPC-EVs can induce a degenerative phenotype that can be reversed through inhibition of the P53/P21 pathway.
The role of apoptotic bodies, a type of EV produced by cells undergoing apoptosis, in IVDD pathogenesis via CEP calcification was investigated by Yuan et al.[105] H2O2 was demonstrated to induce CEP chondrocyte oxidative stress in a dose-dependent manner and led to an increase in mineralization and production of apoptotic bodies. CEP chondrocyte-derived apoptotic bodies were then isolated and used to treat other CEP chondrocytes in culture, which led to an increase in mineralization and decrease of extracellular inorganic pyrophosphate (PPi) content. The investigators showed that apoptotic bodies produced under conditions of oxidative stress modified chondrocyte metabolism through increased TNAP expression, resulting in the conversion of PPi to Pi and consequentially promoting mineralization. CEP calcification as a result of chondrocyte mineralization accelerated the degenerative cascade by preventing the transport of nutrients into the disc space and waste products out of the IVD in order to maintain homeostasis.[138,139] These 3 studies demonstrate that the EV’s identity and molecular composition is a direct representation of the health or pathophysiological state of their originating cell and can contribute to IVDD pathophysiology if derived from a cell with an aberrant degenerative phenotype.
5.1.5. Future directions for in vitro EV studies
Taken together, 28 of the 29 studies utilized in vitro systems to demonstrate that EVs hold significant promise for IVDD therapy or as a potential biomarker for IVDD with several critically important unanswered questions remaining for future research. The majority of in vitro studies determine the effect of EVs on NPCs with a much smaller number of studies reporting the effects of EVs on AF cells (2 studies) and CEP chondrocytes (2 studies). As EVs undergo future investigation for therapeutic application and regulatory review, it is important to comprehensively understand the protective and regenerative effects of this treatment strategy on all terminally differentiated cell types within the IVD. For studies that investigate EVs from primary cells in the human IVD to induce MSC differentiation, it is not yet understood how the IVD degeneration grade corresponding to the EV’s parent cell affects MSC differentiation to IVD-like phenotypes. Given the outcomes from Song et al. and Chen et al., NPC-EVs from a degenerative IVD might yield a comparatively weaker NP-like phenotype as compared to NPC-EVs from a healthy IVD, however, this has yet to be tested.[110,116,140] Moreover, there is currently no study that systematically compares the extent and efficiency of MSC directed differentiation between NPC-EV treatment and traditional chondrogenic differentiation protocols with TGFβ supplementation. Studies published by Lu et al. and Lan et al. demonstrate that NPC-EV treatment could be a highly effective factor, where future work could engineer EVs to contain overexpressed small RNA transcripts that knockdown the Notch1 pathway in order to enhance MSC directed differentiation. Section 6 presents additional avenues of investigation using in vitro systems that evaluate EVs on the molecular and cellular level.
5.2. Evaluation of EVs with in vivo models of degeneration
In vivo systems facilitate the investigation of EVs in a physiologically relevant environment, however these studies are more complex when compared to in vitro systems and only 13 of the 29 studies in this literature review evaluated the effects of EVs using preclinical animal models of IVDD.[99,104,106,109,115–117,119–124]
5.2.1. Stem cell-derived EVs decelerate IVDD in vivo
Stem cell-derived EVs were investigated for their capacity to decelerate IVDD using in vivo IVD puncture models with a biochemical challenge or small molecule inhibitor in 2 (of the 13 in vivo studies). Liao and colleagues used an in vivo rat model, where coccygeal IVDs were punctured and injected with AGEs and MSC-EVs concomitantly.[109] At 4- and 8- weeks, intradiscal injection of EVs led to an improvement in disc height index, MRI grade, and histological score compared to AGE-challenged IVDs without EV supplementation. Moreover, EVs were able to attenuate AGE-induced apoptosis as demonstrated by a reduction in the number of TUNEL positive cells. In the same animal model, Luo and colleagues compared treatment effects between normal and degenerated CEP stem cell-derived EVs and demonstrated that normal CEP stem cell-derived EVs could inhibit NPC apoptosis and alleviate IVDD via activation of the PI3K/AKT/autophagy pathway.[115] AKT activation plays an important role in EV rescue since delivering LY294002, an AKT inhibitor, led to less prominent therapeutic effects.
Stem cell-derived EVs for therapeutic use in IVD needle puncture models without a biochemical challenge or small molecule inhibitor was investigated in 8 additional in vivo studies. Xie and colleagues used an in vivo rat model to evaluate the therapeutic effect of weekly sub-endplate injection of MSC-EVs for 9 weeks, where EV treatment led to a lower MRI score, better preservation of CEP and NP tissues, inhibition of apoptosis, and reduction in CEP calcification.[122] Other studies provide additional evidence that substantiate the therapeutic potential of stem cell-derived EVs in mouse and rabbit models, where EV treatment led to a reduction in NPC necrosis and apoptosis, decreased catabolic enzyme and proinflammatory cytokine production, increased GAG content, and improvements in gait pattern, disc height index, MRI signal intensity, and histological score.[99,104,109,120,121,123,124] Some studies attribute these therapeutic effects to specific miRs found in EVs, where Cheng et al. highlighted the role of miR-21 in preventing apoptosis, Zhang et al. elucidated the role of miR-410 in reducing pyroptosis, and Xie et al. specified the protective role of miR-31-5p in the ATF6/Endoplasmic Reticulum (ER) stress pathway.[104,122,124] Conversely, other studies attribute therapeutic effects to a downregulation of specific miRs, where Yuan et al. used MSC-EVs to deliver AntagomiR-4450 and block miR-4450, which in turn alleviated IVDD.[123]
5.2.2. Primary cell-derived EVs alleviate hallmarks of IVDD in vivo
Primary cell-derived EVs were evaluated using in vivo IVDD models in 4 studies, and includes EVs derived from mature NCs, NPCs, and primary mouse embryonic fibroblasts. IVD treatment with NC-EVs from 0.5MPa compressive load culture led to improved disc height index and decreased CD43 expression, indicating that EVs could elicit anti-angiogenesis effects in an in vivo mouse model.[117] Primary mouse embryonic fibroblast-derived EVs were engineered to efficiently deliver FOXF1 mRNA in vivo as a therapeutic strategy to promote a healthy NP phenotype and enhance FOXF1 and Brachyury expression.[119] Furthermore, Moen et al. found that miR-223 in NPC-EVs is associated with a reduced likelihood of persistent pain following herniation.[106] Through in vivo electrophysiological measurements in a rat model, Moen and colleagues showed that prolonged exposure of miR-223 onto dorsal nerve roots decreases C-fiber response, indicating that this specific miR has an anti-nociceptive effect. The investigators also found that miR-223 was one of three miRs that were significantly increased more than 5-fold from NP tissue. These findings suggest that the EV’s heterogenous cargo regulates a variety of downstream targets with distinct biological functions and produce a number of therapeutic outcomes. However, Song and colleagues showed that NPC-EVs could exacerbate the degenerative cascade if EVs were derived from degenerate NPCs.[116] Rat IVDs injected with degenerative NPC-EVs resulted in a higher degeneration grade, lower disc height index, increased expression of caspase-3, MMP3, MMP13, and ADAMTS4, as well as decreased expression of collagen II and aggrecan. Song et al. mechanistically determined that degenerative NPC-EVs transport circular RNAs such as circRNA_0000253 to regulate IVD degeneration by competitively adsorbing miR-141-5p. Overall findings from these 4 studies suggest that EVs derived from healthy primary cells demonstrate therapeutic potential comparable to that of EVs derived from stem cells, whereas EVs derived from primary cells with a degenerative phenotype can deleteriously affect cells downstream.
5.2.3. Future directions for EVs applied to in vivo models of IVDD
The in vivo studies in this literature review collectively demonstrate that EVs are promising to decelerate IVDD, yet there many open questions remain for future investigation regarding the in vivo administration of EVs to treat IVDD. These studies primarily provide insight regarding the effects of EVs on ECM remodeling and apoptosis, but they do not provide data on immune system responses and pain behaviors. Given that Yuan et al. was the only study to assess effects of EV treatment on biomechanical properties, more data is necessary to understand the effects of EVs on potential changes in functional IVD biomechanical behaviors. A key advantage of in vivo models is the ability to holistically evaluate the treatment response from multiple interconnected systems. However, there is a lack of understanding with regards to how these different systems work in concert to promote endogenous repair and attenuate pain following EV treatment. With respect to clinical translatability, two important factors of consideration are the dosage and frequency of EV administration. Although injection of EVs derived from healthy cells led to protective effects in all studies, there is no consensus with respect to the dose of EVs in a given injection and the frequency of administration. Section 6 touches upon avenues of investigation in which preclinical in vivo models of IVDD would be helpful in understanding the effect of EVs on the cellular and organ levels.
6. Evaluating Functional Metrics of EV Efficacy and Regenerative Potential
The pathophysiology of IVDD occurs between interrelated factors on the molecular, cellular, and tissue scales, thus warranting the development of an tissue-specific conceptual framework that factors functional assessments within these scales of biological organization and complexity.[141,142] Evaluation criteria on the molecular scale are first proposed and then related assessments on the cellular and tissue levels are proposed to comprehensively determine the therapeutic efficacy of a naïve or engineered EV of interest (Figure 4). This framework was constructed according to the evaluation methods and output measures previously reported in preclinical studies for IVDD therapy (Section 5) in addition to targeting the known mechanisms and/or factors that contribute to IVDD pathophysiology.[141–145] The primary goal of this conceptual framework is to highlight key avenues of preclinical investigation in order to determine if a naïve or engineered EV can slow or reverse progressive IVDD across all scales of action.
Figure 4:
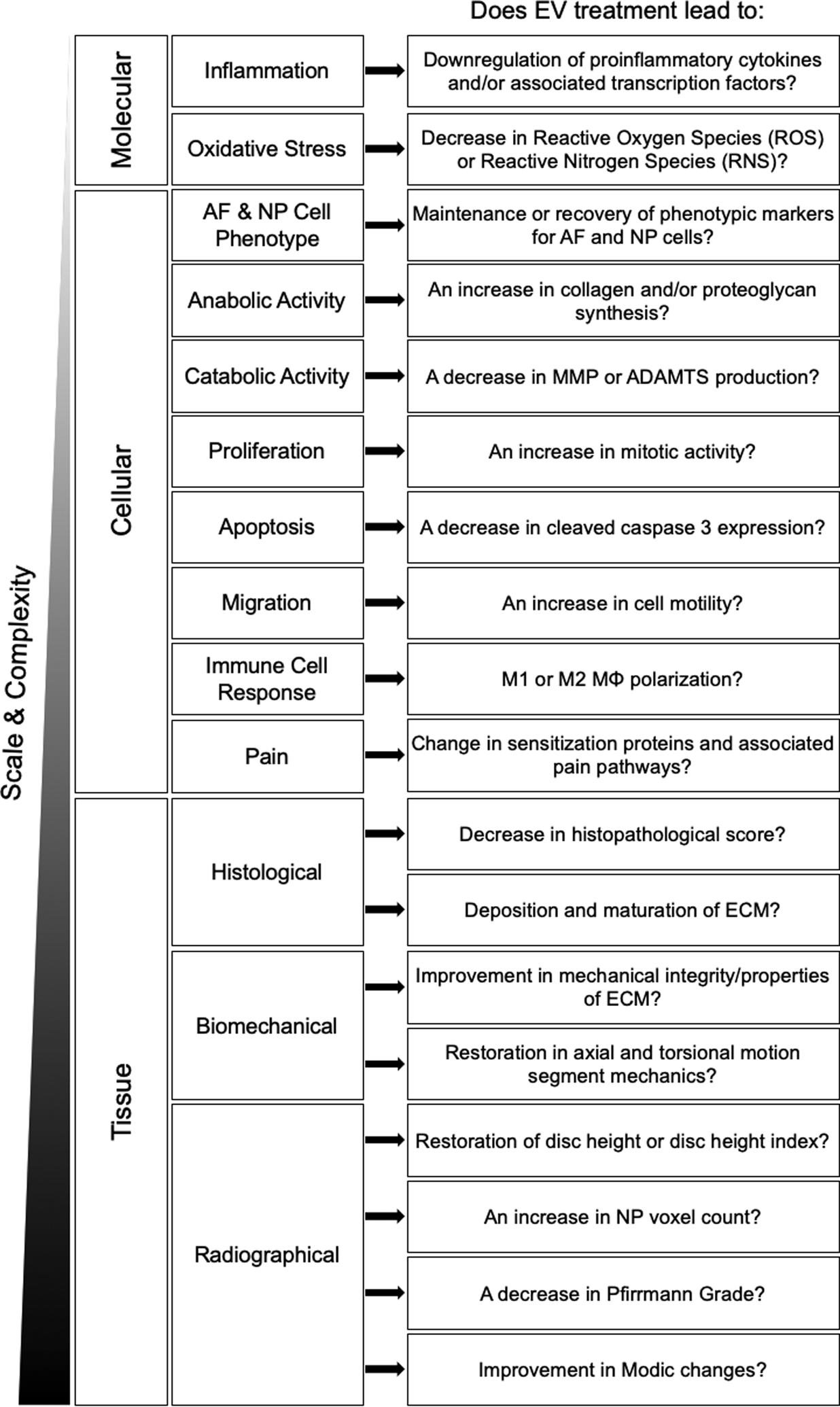
Conceptual framework to evaluate EV-based therapeutics for IVDD across biological levels of scale and complexity. Questions regarding therapeutic outcomes are grouped by functional response type and biological scale (i.e. molecular-, cellular-, and tissue-level).
6.1. Evaluation of therapeutic efficacy on the molecular level
The literature review identified that EVs have strong potential to regulate molecular factors implicated in the progression of IVDD.[146,147] Nutrient transport bears major importance for the survival and proper metabolic function of resident cells in the IVD.[17] In IVDD, there is limited nutrient transport and sub-physiological glucose concentrations result in a reduction in cell viability under acidic conditions.[148] Additionally, decreased transport leads to dysregulated metabolism and lactic acid accumulation, which not only drives degenerative processes, but is associated with the buildup of metabolites that induce oxidative stress, including reactive oxygen species and reactive nitrogen species.[149–151] Under oxidative stress, the proteomic profile of the AF and NP secretome changes and alterations occur in growth factor and cytokine production, particularly those involved in proinflammatory and catabolic processes.[147,152,153] Reagents such as H2O2 and TBHP, as used in 5 preclinical studies in this literature review, can experimentally contrive oxidative stress conditions to determine if EVs can alleviate metabolic and oxidative stress phenotypes associated with IVDD and elucidate EV mechanisms of action. Molecular targets for therapy that enhance solute diffusivity or glucose uptake into the cell (i.e. GLUT4) upon EV treatment may provide protective effects and aid in slowing the progression of IVDD by promoting cell viability. Given the bidirectional flow of mass transport within the IVD, lactic acid would be able to diffuse out of the disc space and prevent sub-physiological decreases in pH or aberrant reactive oxygen species/reactive nitrogen species production. The attenuation of reactive oxygen species/reactive nitrogen species would in turn prevent proinflammatory cytokine production and damage to resident cells and ECM, contributing to the maintenance of cell viability and ECM integrity following EV treatment. Since EVs contain a heterogeneous group of effector molecules with multiple downstream targets, they may have the ability to regulate signaling pathways and proteomic profiles associated with oxidative and metabolic stress by means of their molecular cargo, offering numerous protective effects in the context of IVDD treatment.
6.2. Evaluation of therapeutic efficacy on the cellular level
Cellular internalization of EVs can therapeutically target molecular pathways associated with IVDD and in turn modulate changes in cellular function under aberrant physiological conditions. AF and NP cells demonstrate shifts in their canonical markers from healthy to degenerative conditions, thus leading to changes in cellular behavior and homeostasis.[154,155] In the healthy IVD, cells maintain a balance of anabolic and catabolic activity resulting in normal ECM turnover. However, in the degenerative IVD, cells exhibit disproportionately high levels of catabolic activity, leading to significant breakdown of soft tissue that ultimately manifests in mechanical failure.[141,156] Preclinical outcomes in this literature review demonstrate that EVs affect ECM remodeling processes and treating degenerative IVD cells with EVs from healthy cells may be able to promote ECM elaboration in the AF, NP, and CEP and inhibit ECM breakdown. To remain consistent with previously reported outcomes, a naïve or engineered EV for therapeutic application is recommended to elicit similar responses in expression for anabolic and catabolic proteins. Restoration of anabolic activity includes the synthesis of collagen and proteoglycan content that aligns with the distinct species found in the AF, NP, and CEP. Repression of catabolic activity includes a reduction in proteases significantly associated with IVDD, such as enzymes belonging to the MMP and/or ADAMTS families.[157]
Preclinical outcomes in this systematic review indicate that EVs can influence cellular senescence, the senescence-associated secretory phenotype, and apoptosis depending on the pathophysiological state of the EV source cell.[99,104,109–111,114,115,120,122,126,127] Biophysical and biochemical changes in the ECM lead to aberrant cues that promote cellular senescence, where IVD cells residing in a degenerative microenvironment exhibit lower levels of mitotic activity compared to those in the healthy IVD.[158] Not only do resident cells slow their proliferation rate in IVDD, but they also undergo a marked increase in apoptosis, as evidenced by an upregulation of cleaved caspase 3 and corresponding loss in the number of viable cells.[159–161] Caspase 3 silencing and enhanced proliferation prevented IVDD in a rabbit model and EV treatment may achieve this therapeutic outcome by regulating these biological processes in that fashion.[162,163] Given the ability of EVs to target pathways involved in apoptosis and proliferation, investigations determining naïve and/or engineered EV treatment responses related to cellular senescence and proliferation are of interest for IVDD therapy.
A critically important process necessary for endogenous tissue repair is cellular migration.[164] The literature points to stem/progenitor cells residing in the perichondrium area outside the epiphyseal plate increasing migratory behavior and infiltrating the IVD upon degeneration, which is stimulated by intercellular communication via signaling factors.[165–170] NPCs release chemokines including HIF1α, VEGF, SDF1, and CCL5 into the degenerative environment and consequentially promote chemotaxis of non-resident MSCs into the IVD, suggesting that cell recruitment is an innate mechanism that drives cell-based repair.[165,171,172] However, due to the avascular nature of the IVD and harsh microenvironment, cell motility and tissue infiltration is limited, contributing to the IVD’s poor capacity to heal itself via endogenous repair processes.[173–176] Preclinical outcomes in this literature review suggest that EVs may promote cellular migration by establishing chemotactic gradients, particularly if the EVs were derived from NPCs.[100] When evaluating the cell homing capabilities of naïve or engineered EVs, investigators can use transwell in vitro systems to measure migratory behavior or use in vivo preclinical models following intradiscal injection. Since non-resident stem cells must migrate into a nutrient-deprived and acidic environment for endogenous IVD repair, experimental systems determining if cells can exhibit high levels of motility in the presence of aberrant biochemical cues following EV treatment are of importance.
Intradiscal injection of EV therapeutics not only would affect resident AF and NP cells, but also affect resident macrophages since the IVD contains a heterogeneous cell population including cells from the immune system. It is not yet well understood how EV treatment modulates resident macrophage responses, which play important roles in inflammation and tissue repair. Macrophages are shown to be directly involved with the pathophysiology of IVDD, where their polarization state is significantly correlated with degeneration grade and can either enhance the progression of IVDD or attenuate the progress.[177,178] Macrophage dysfunction and sustained activation towards the M1-type results in an overproduction of inflammatory factors and leads to ECM degradation, which in turn catalyzes the degenerative cascade.[179] In contrast, activation towards the pro-regenerative M2-type results in the production of anti-inflammatory cytokines and promotes tissue repair and remodeling, ultimately preventing degeneration.[177] Given the plasticity of macrophage activation state, cellular assessments may include the temporal characterization of macrophage polarization post-EV treatment and determine if EVs can transcriptionally regulate activation state preferentially towards the M2-type instead of the M1-type.[180,181]
Neuronal responses to EV treatment in dorsal root ganglion (DRG) are also of considerable interest due to the proximity of the nerve roots to the delivery site and neoinnervation in IVDD, where EVs could potentially modulate molecular markers of pain and corresponding sensitization pathways.[182–184] In IVDD, biomechanical and biochemical insult to the nerve roots leads to hyperalgesia and allodynia with an associated upregulation of TAC1 and CALCA, which are the encoding genes for substance P and calcitonin gene-related peptide, respectively.[185,186] Evaluation of an EV therapeutic offers potential to attenuate the expression of some of these factors associated with neoinnervation, which are implicated in pain sensitization and hyperalgesia.[187] Moreover, pain behavioral assays in preclinical animal models could support more strongly a functional reduction of pain upon EV treatment, as demonstrated in a rat model of temporomandibular joint osteoarthritis.[56,188]
6.3. Evaluation of therapeutic efficacy on the organ level
Degenerative changes on the molecular and cellular scales manifest in functional changes on the organ level, and are detected by histological, biomechanical, and radiographic methods. In the degenerative state, there are observable differences in matrix abundance and quality, thus leading to significant changes in histopathological scores.[189] These deleterious changes in IVD matrix architecture and composition affect the micromechanical properties of the ECM and may result in organ-level changes in motion segment axial and torsional biomechanics.[190,191] Additionally, significant alterations in matrix composition lead to distinguishable features through MRI, where degenerative IVDs exhibit significant decreases in NP voxel count (NP hydration state), Modic changes (vertebral endplate quality), and higher Pfirrmann grades.[192] Along with detectable changes via MRI, X-Ray imaging can detect degeneration-associated changes in disc height and disc height index, which can lead to neuropathic or inflammation-related pain if there are large enough decreases in either parameter.[193,194] An efficacious EV treatment would principally aim to either reverse or slow the progression of these histological, biomechanical, and radiographical degeneration-related changes. By modulating target cell expression, EVs may collectively influence cell behavior that manifests in organ-level changes and functional responses. Ex vivo and/or in vivo animal systems serve as preclinical IVD models to assess histological, biomechanical, and radiographical outcomes and determine whether a naïve or engineered EV can effectively treat these hallmarks of IVDD.
7. Regulatory and Manufacturing Considerations
As EVs emerge as a prominent candidate for cell-free therapy, it is critical that investigators understand the pertinent regulatory pathway for approval in order to advance these technologies towards the clinic. First, the FDA regulatory review process is described, which comprehensively evaluates EV characterization metrics, methods of administration, safety and efficacy profiles, and production processes. Following an overview of the FDA review process, important manufacturing considerations are highlighted for scale up. EV characterization methods (Section 4), experimental configurations and model systems (Section 5), and evaluation criteria (Section 6) inform the contents of an FDA application with the goal for favorable regulatory review.
7.1. FDA Regulatory Review Process for EV Therapeutics
EV-based therapeutics as novel and therapeutically active substances for intended use in clinical studies are considered an investigational new drug (IND). In order to advance an IND into clinical trials, the submission of an IND application following preclinical development is required.[195] These applications must provide information regarding previously conducted animal studies, manufacturing information, clinical protocols, and investigator information.[196] Additionally, EVs as, “a medicine that contains one or more active substances made by or derived from a biological cell,” are classified as biological medicinal products and the regulation of such products in the United States is under the purview of the Center for Biologics Evaluation and Research (CBER), a subsidiary of the Food and Drug Administration (FDA).[197] CBER reviews IND applications and ultimately decides if new therapeutics can progress into early phase clinical trials.
In an IND application, investigators are also required to report technical specifications that influence the clinical feasibility and outcomes of EV therapeutics, including the optimal EV source (e.g., donors, cells, tissues, fluids) as well as strategies for EV production, isolation, purification, characterization, and storage. Investigators must establish quality control requirements in which biological, molecular, and physical characteristics are consistent amongst batches of EVs to ensure consistent effects upon administration. In addition to technical specifications, investigators have to provide extensive in vitro and in vivo data that demonstrate adequate pharmacological and toxicological profiles for therapeutic safety and efficacy.[197] Alongside preclinical research and development, investigators are required to establish clinical protocols for treatment, including the route and frequency of administration. Ideal administration of EV-based therapeutics targeting IVDD would involve a localized injection, given the avascular composition of the IVD, as well as a single dose so as to prevent numerous needle injections that would instigate progressive degeneration.[198]
Since most EV-based therapeutics are in preclinical development, it is critically important to understand the necessary data required to file an IND application for favorable review. Notable sections of the IND application include 21CFR312.23(a)(7) ‘Chemistry, Manufacturing, and Control Information’ as well as 21CFR312.23(a)(8) ‘Pharmacology and Toxicology Information’.[199] To meet the requirements of 21CFR312.23(a)(7), investigators need to supply quantitative data regarding the identity, quality, purity, stability, and composition of a naïve or engineered EV therapeutic. Moreover, 21CFR312.23(a)(8) requires investigators to provide pharmacological and toxicological data on in vitro and in vivo effects, absorption, distribution, metabolism, excretion, toxicity, and, if possible, the corresponding mechanisms of action. A key challenge for EV therapeutics is defining clear mechanisms of action, given that EVs contain a wide variety of effector molecules and each effector molecule can have multiple downstream targets that induce pleiotropic effects.[195,200] Furthermore, data pertaining to these two key sections of the IND application should align with FDA-implemented safety requirements prescribed by the by The International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH), given that the production of EVs requires the use of biological matter (i.e. cells).[201] Ultimately, preclinical studies must provide compelling evidence regarding preliminary quality, safety, and efficacy of a naïve or engineered EV to treat a target disease.[202]
Following the completion of clinical testing and IND review phase, investigators then prepare a Biologics License Application (BLA) in order for a biological product to be legally marketed within the United States and enter interstate commerce.[203] A complete BLA submission includes a full description of manufacturing methods, representative samples of the product, summaries of preclinical and clinical test results, as well as proposed labels, enclosures, and containers for the product.[203] A BLA ultimately serves to demonstrate that a product meets pre-determined requirements in safety, quality, purity, and potency, demonstrated through data obtained in both preclinical and clinical studies.[204]
Although there are currently no FDA approved EV therapeutics on the market for any indication, results from early clinical trial testing are promising. Ongoing studies involving the treatment of melanoma, non-small cell lung cancer, colon cancer, and chronic kidney disease with EVs have all demonstrated high levels of safety and efficacy in human subjects.[205–208] Of particular note, chronic kidney disease patients treated with MSC-EVs display improved kidney function (increased levels of s-creatinine, blood urea, and eGFR) and decreased inflammation (increased levels of IL-10, decreased levels of TNFα, increased levels of TGF-β1).[208] These positive clinical outcomes to treat non-musculoskeletal tissues substantiate the promise of this biologic as a next generation treatment strategy for cell-free therapy of the musculoskeletal system. However, there are currently no clinical trials to evaluate EVs to treat degenerative musculoskeletal diseases, such as OA or IVDD.
7.2. Manufacturing Considerations for EV Therapeutics
Large-scale EV production is necessary to meet the requirements for distribution and manufacturing processes must be compliant with the appropriate regulatory standards to ensure quality control. The development of a manufacturing process to produce EV-based therapeutics requires an adequate reactor platform, associated control system, as well as an established quality management system to comply with good manufacturing practice (cGMP) standards.[209] One of the most significant barriers in producing EV-based therapeutics to scale is that there is a lack of established upstream and downstream manufacturing processes.[210] Recent efforts focus on developing bioreactors for large scale production of EVs, which include the use of hollow fiber membrane bioreactors that can culture significantly more cells than traditional cell culture flasks.[211] However, it is unclear how the operating conditions of such large-scale reactors impact the molecular composition and function of EVs, a concern which must be addressed when developing EV production processes. Other factors of consideration in addition to reactor operating conditions include cell culture parameters such as biological donor, cell type, seeding density, and population doubling level, which are known to impact the homogeneity and composition of EVs and therefore must be considered in the design of large-scale manufacturing processes.[210]
8. Conclusions
This systematic review provides a comprehensive overview of extracellular vesicles as an emerging therapeutic platform for cell-free treatment of IVDD. We identified 29 original research articles that investigate EVs for IVD applications and report EV characterization techniques employed in each study as well as associated outcomes in preclinical models of IVDD. Most studies characterize EVs using at least one biophysical method and one biochemical method to determine EV size, concentration, and protein levels. EVs derived from terminally differentiated NPCs had the ability to promote MSC migration and MSC differentiation towards an NP-like phenotype while MSC-EVs demonstrated remarkable therapeutic potential in vitro to alleviate hallmarks of degeneration, including a reduction in NPC apoptosis, inflammation, and ECM catabolism. Studies employing in vivo models further demonstrate MSC-derived and healthy primary cell-derived EVs yield partial functional restoration including an increase in disc height index when compared to untreated IVDD controls. Most in vitro and in vivo studies examine NPC or organ-level responses to treatment with markedly fewer studies examining EV treatment effects on AF or CEP cell types, highlighting important avenues of future investigation for tissue-specific outcomes. Other important directions of future research include immune cell and DRG neuronal responses to examine immunomodulation and pain sensitization, respectively, following EV treatment. Additional important areas of future investigation include the optimization of stem cell source, culture conditions, and molecular cargo of EVs to maximize their therapeutic potential. This review offers a guiding conceptual framework to advance EVs beyond the discovery phase and presents a holistic and systematic evaluation of the therapeutic efficacy of EVs spanning the molecular, cellular, and organ levels, and describes the regulatory approval pathway with the goal to accelerate naïve or engineered EVs towards the clinic as a novel treatment strategy for IVDD.
Acknowledgements
This work was supported by NIH/NIAMS Grant # R01 AR057397 and NIH/NIGMS Grant # T32 GM062754.
Author Biographies

Tyler DiStefano, PhD is a recent graduate from the Department of Orthopaedics at the Icahn School of Medicine at Mount Sinai. He received his B.Eng. degree in Mechanical Engineering from The Cooper Union. His research interests are in the areas of tissue engineering and regenerative medicine, polymeric biomaterials, biomechanics, drug delivery, and nanomedicine.

Jennifer R. Weiser, PhD is an Assistant Professor of Chemical Engineering at The Cooper Union for the Advancement of Science and Art in New York City where her research focuses on drug delivery, wound healing, and the development of polymeric biomaterials. Dr. Weiser earned her BS in Chemical Engineering from Rensselaer Polytechnic Institute and her MS and PhD from Cornell University in Biomedical Engineering, and was a postdoctoral fellow at Yale University in Biomedical Engineering. She is a member of the ORS, AIChE, and ASEE.

James C. Iatridis, PhD is Professor and Vice Chair for Research in the Department of Orthopaedics at the Icahn School of Medicine at Mount Sinai in New York City. Dr. Iatridis’ research program is focused on the prevention and treatment of painful intervertebral disc degeneration where his lab applies novel biomaterial strategies to repair intervertebral discsand deliver therapeutics to reduce disabling back pain. Dr. Iatridis is a Fellow of the Orthopaedic Research Society, International Combined Orthopaedic Research Societies, and the American Institute for Medical and Biological Engineering.
Footnotes
Declaration of competing interests
None.
References
- [1].Smith JW, Walmsley R, J. Bone Joint Surg. Br 1951, 33–B, 612. [DOI] [PubMed] [Google Scholar]
- [2].Hampton D, Laros G, McCarron R, Franks D, Spine 1989, 14, 398. [DOI] [PubMed] [Google Scholar]
- [3].Smith LJ, Nerurkar NL, Choi K-S, Harfe BD, Elliott DM, Dis. Model. Mech 2011, 4, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Torre OM, Das R, Berenblum RE, Huang AH, Iatridis JC, FASEB J 2018, 32, 4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Buckwalter JA, Spine 1995, 20, 1307. [DOI] [PubMed] [Google Scholar]
- [6].Weinstein JN, Lurie JD, Tosteson TD, Skinner JS, Hanscom B, Tosteson ANA, Herkowitz H, Fischgrund J, Cammisa FP, Albert T, Deyo RA, JAMA 2006, 296, 2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mariconda M, Galasso O, Attingenti P, Federico G, Milano C, Eur. Spine J 2010, 19, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ammerman J, Watters WC, Inzana JA, Carragee G, Groff MW, Cureus 2019, 11, e4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].van Uden S, Silva-Correia J, Oliveira JM, Reis RL, Biomater. Res 2017, 21, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Burdick JA, Mauck RL, Gorman JH, Gorman RC, Sci. Transl. Med 2013, 5, 176ps4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bowles RD, Setton LA, Biomaterials 2017, 129, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Panebianco CJ, Meyers JH, Gansau J, Hom WW, Iatridis JC, eCM 2020, 40, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sakai D, Schol J, J. Orthop. Translat 2017, 9, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sakai D, Andersson GBJ, Nat. Rev. Rheumatol 2015, 11, 243. [DOI] [PubMed] [Google Scholar]
- [15].Tong W, Lu Z, Qin L, Mauck RL, Smith HE, Smith LJ, Malhotra NR, Heyworth MF, Caldera F, Enomoto-Iwamoto M, Zhang Y, Transl Res 2017, 181, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bendtsen M, Bunger C, Colombier P, Le Visage C, Roberts S, Sakai D, Urban JPG, Acta Orthop 2016, 87, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Huang Y-C, Urban JPG, Luk KDK, Nat. Rev. Rheumatol 2014, 10, 561. [DOI] [PubMed] [Google Scholar]
- [18].Vadalà G, Sowa G, Hubert M, Gilbertson LG, Denaro V, Kang JD, J Tissue Eng Regen Med 2012, 6, 348. [DOI] [PubMed] [Google Scholar]
- [19].Varden LJ, Nguyen DT, Michalek AJ, JOR Spine 2019, 2, e1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Loibl M, Wuertz-Kozak K, Vadala G, Lang S, Fairbank J, Urban JP, JOR Spine 2019, 2, e1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Buckley CT, Hoyland JA, Fujii K, Pandit A, Iatridis JC, Grad S, JOR Spine 2018, 1, e1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].King NM, Perrin J, Stem Cell Res Ther 2014, 5, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gnecchi M, Zhang Z, Ni A, Dzau VJ, Circ. Res 2008, 103, 1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Burdon TJ, Paul A, Noiseux N, Prakash S, Shum-Tim D, Bone Marrow Res 2011, 2011, 207326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Liang X, Ding Y, Zhang Y, Tse H-F, Lian Q, Cell Transplant 2014, 23, 1045. [DOI] [PubMed] [Google Scholar]
- [26].Mianehsaz E, Mirzaei HR, Mahjoubin-Tehran M, Rezaee A, Sahebnasagh R, Pourhanifeh MH, Mirzaei H, Hamblin MR, Stem Cell Res Ther 2019, 10, 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yoon YJ, Kim OY, Gho YS, BMB Rep 2014, 47, 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Elsharkasy OM, Nordin JZ, Hagey DW, de Jong OG, Schiffelers RM, Andaloussi SE, Vader P, Adv. Drug Deliv. Rev 2020, 159, 332. [DOI] [PubMed] [Google Scholar]
- [29].Bernardi S, Balbi C, Biology (Basel) 2020, 9, 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].O’Brien K, Breyne K, Ughetto S, Laurent LC, Breakefield XO, Nat. Rev. Mol. Cell Biol 2020, 21, 585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tsiapalis D, O’Driscoll L, Cells 2020, 9, 991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Malda J, Boere J, van de Lest CHA, van Weeren PR, Wauben MHM, Nat. Rev. Rheumatol 2016, 12, 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rani S, Ryan AE, Griffin MD, Ritter T, Mol. Ther 2015, 23, 812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Liu B, Lee BW, Nakanishi K, Villasante A, Williamson R, Metz J, Kim J, Kanai M, Bi L, Brown K, Di Paolo G, Homma S, Sims PA, Topkara VK, Vunjak-Novakovic G, Nat. Biomed. Eng 2018, 2, 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ruppert KA, Nguyen TT, Prabhakara KS, Toledano Furman NE, Srivastava AK, Harting MT, Cox CS, Olson SD, Sci. Rep 2018, 8, 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Huang J-H, Yin X-M, Xu Y, Xu C-C, Lin X, Ye F-B, Cao Y, Lin F-Y, J. Neurotrauma 2017, 34, 3388. [DOI] [PubMed] [Google Scholar]
- [37].Shentu TP, Wong S, Espinoza C, Cernelc‐Kohan M, Hagood J, FASEB J 2016. [Google Scholar]
- [38].Morrison TJ, Jackson MV, Cunningham EK, Kissenpfennig A, McAuley DF, O’Kane CM, Krasnodembskaya AD, Am. J. Respir. Crit. Care Med 2017, 196, 1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Nojima H, Freeman CM, Schuster RM, Japtok L, Kleuser B, Edwards MJ, Gulbins E, Lentsch AB, J. Hepatol 2016, 64, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bruno S, Tapparo M, Collino F, Chiabotto G, Deregibus MC, Soares Lindoso R, Neri F, Kholia S, Giunti S, Wen S, Quesenberry P, Camussi G, Tissue Eng. Part A 2017, 23, 1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhao B, Zhang Y, Han S, Zhang W, Zhou Q, Guan H, Liu J, Shi J, Su L, Hu D, J Mol Histol 2017, 48, 121. [DOI] [PubMed] [Google Scholar]
- [42].Harrell CR, Jovicic N, Djonov V, Arsenijevic N, Volarevic V, Cells 2019, 8, 1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Cai J, Wu J, Wang J, Li Y, Hu X, Luo S, Xiang D, Cell Biosci 2020, 10, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gomzikova MO, James V, Rizvanov AA, Front. Immunol 2019, 10, 2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Shiue S-J, Rau R-H, Shiue H-S, Hung Y-W, Li Z-X, Yang KD, Cheng J-K, Pain 2019, 160, 210. [DOI] [PubMed] [Google Scholar]
- [46].Chen F, Li X, Zhao J, Geng J, Xie J, Xu B, In Vitro Cell Dev Biol Anim 2020, 56, 567. [DOI] [PubMed] [Google Scholar]
- [47].Arslan F, Lai RC, Smeets MB, Akeroyd L, Choo A, Aguor ENE, Timmers L, van Rijen HV, Doevendans PA, Pasterkamp G, Lim SK, de Kleijn DP, Stem Cell Res 2013, 10, 301. [DOI] [PubMed] [Google Scholar]
- [48].de Godoy MA, Saraiva LM, de Carvalho LRP, Vasconcelos-Dos-Santos A, Beiral HJV, Ramos AB, de P. Silva LR, Leal RB, Monteiro VHS, Braga CV, de Araujo-Silva CA, Sinis LC, Bodart-Santos V, Kasai-Brunswick TH, de L. Alcantara C, Lima APCA, da Cunha-E Silva NL, Galina A, Vieyra A, De Felice FG, Mendez-Otero R, Ferreira ST, J. Biol. Chem 2018, 293, 1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zhang B, Shen L, Shi H, Pan Z, Wu L, Yan Y, Zhang X, Mao F, Qian H, Xu W, Stem Cells Int 2016, 2016, 1929536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Shabbir A, Cox A, Rodriguez-Menocal L, Salgado M, Van Badiavas E, Stem Cells Dev 2015, 24, 1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Li JJ, Hosseini-Beheshti E, Grau GE, Zreiqat H, Little CB, Nanomaterials (Basel) 2019, 9, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Boere J, Malda J, van de Lest CHA, van Weeren PR, Wauben MHM, Front. Immunol 2018, 9, 2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Piazza N, Dehghani M, Gaborski TR, Wuertz-Kozak K, Front. Bioeng. Biotechnol 2020, 8, 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Alcaraz MJ, Compañ A, Guillén MI, Cells 2019, 9, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Tofiño-Vian M, Guillén MI, Alcaraz MJ, Biochem. Pharmacol 2018, 153, 134. [DOI] [PubMed] [Google Scholar]
- [56].Zhang S, Teo KYW, Chuah SJ, Lai RC, Lim SK, Toh WS, Biomaterials 2019, 200, 35. [DOI] [PubMed] [Google Scholar]
- [57].Rustenburg CME, Emanuel KS, Peeters M, Lems WF, Vergroesen P-PA, Smit TH, JOR Spine 2018, 1, e1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Willms E, Cabañas C, Mäger I, Wood MJA, Vader P, Front. Immunol 2018, 9, 738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zhang ZG, Buller B, Chopp M, Nat. Rev. Neurol 2019, 15, 193. [DOI] [PubMed] [Google Scholar]
- [60].EL Andaloussi S, Mäger I, Breakefield XO, Wood MJA, Nat. Rev. Drug Discov 2013, 12, 347. [DOI] [PubMed] [Google Scholar]
- [61].De Jong OG, Van Balkom BWM, Schiffelers RM, Bouten CVC, Verhaar MC, Front. Immunol 2014, 5, 608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Skotland T, Sandvig K, Llorente A, Prog. Lipid Res 2017, 66, 30. [DOI] [PubMed] [Google Scholar]
- [63].Jovic M, Sharma M, Rahajeng J, Caplan S, Histol Histopathol 2010, 25, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Villarroya-Beltri C, Baixauli F, Gutiérrez-Vázquez C, Sánchez-Madrid F, Mittelbrunn M, Semin. Cancer Biol 2014, 28, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kalluri R, LeBleu VS, Science 2020, 367, eaau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zhang Y, Liu Y, Liu H, Tang WH, Cell Biosci 2019, 9, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].McKelvey KJ, Powell KL, Ashton AW, Morris JM, McCracken SA, J. circ. biomark 2015, 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Buzás EI, Tóth EÁ, Sódar BW, Szabó-Taylor KÉ, Semin Immunopathol 2018, 40, 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Prada I, Meldolesi J, Int. J. Mol. Sci 2016, 17, 1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Gonda A, Kabagwira J, Senthil GN, Wall NR, Mol. Cancer Res 2019, 17, 337. [DOI] [PubMed] [Google Scholar]
- [71].Mulcahy LA, Pink RC, Carter DRF, J Extracell Vesicles 2014, 3, 24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Colombo M, Raposo G, Théry C, Annu. Rev. Cell Dev. Biol 2014, 30, 255. [DOI] [PubMed] [Google Scholar]
- [73].Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S, Genomics Proteomics Bioinformatics 2015, 13, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Ambros V, Nature 2004, 431, 350. [DOI] [PubMed] [Google Scholar]
- [75].Bayraktar R, Bertilaccio MTS, Calin GA, Front. Immunol 2019, 10, 1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Nolte-’t Hoen ENM, Buermans HPJ, Waasdorp M, Stoorvogel W, Wauben MHM, ‘t Hoen PAC, Nucleic Acids Res 2012, 40, 9272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Vader P, Mol EA, Pasterkamp G, Schiffelers RM, Adv. Drug Deliv. Rev 2016, 106, 148. [DOI] [PubMed] [Google Scholar]
- [78].Malloy A, Materials Today 2011, 14, 170. [Google Scholar]
- [79].Nolan JP, Duggan E, Methods Mol. Biol 2018, 1678, 79. [DOI] [PubMed] [Google Scholar]
- [80].Morales-Kastresana A, Jones JC, Methods Mol. Biol 2017, 1545, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Choi D, Montermini L, Jeong H, Sharma S, Meehan B, Rak J, ACS Nano 2019, 13, 10499. [DOI] [PubMed] [Google Scholar]
- [82].Chernyshev VS, Rachamadugu R, Tseng YH, Belnap DM, Jia Y, Branch KJ, Butterfield AE, Pease LF, Bernard PS, Skliar M, Anal. Bioanal. Chem 2015, 407, 3285. [DOI] [PubMed] [Google Scholar]
- [83].Chuo ST-Y, Chien JC-Y, Lai CP-K, J Biomed Sci 2018, 25, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach J-M, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina Zavec A, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, Borràs FE, Bosch S, Boulanger CM, Breakefield X, Breglio AM, Brennan MÁ, Brigstock DR, Brisson A, Broekman ML, Bromberg JF, Bryl-Górecka P, Buch S, Buck AH, Burger D, Busatto S, Buschmann D, Bussolati B, Buzás EI, Byrd JB, Camussi G, Carter DR, Caruso S, Chamley LW, Chang Y-T, Chen C, Chen S, Cheng L, Chin AR, Clayton A, Clerici SP, Cocks A, Cocucci E, Coffey RJ, Cordeiro-da-Silva A, Couch Y, Coumans FA, Coyle B, Crescitelli R, Criado MF, D’Souza-Schorey C, Das S, Datta Chaudhuri A, de Candia P, De Santana EF, De Wever O, Del Portillo HA, Demaret T, Deville S, Devitt A, Dhondt B, Di Vizio D, Dieterich LC, Dolo V, Dominguez Rubio AP, Dominici M, Dourado MR, Driedonks TA, Duarte FV, Duncan HM, Eichenberger RM, Ekström K, El Andaloussi S, Elie-Caille C, Erdbrügger U, Falcón-Pérez JM, Fatima F, Fish JE, Flores-Bellver M, Försönits A, Frelet-Barrand A, Fricke F, Fuhrmann G, Gabrielsson S, Gámez-Valero A, Gardiner C, Gärtner K, Gaudin R, Gho YS, Giebel B, Gilbert C, Gimona M, Giusti I, Goberdhan DC, Görgens A, Gorski SM, Greening DW, Gross JC, Gualerzi A, Gupta GN, Gustafson D, Handberg A, Haraszti RA, Harrison P, Hegyesi H, Hendrix A, Hill AF, Hochberg FH, Hoffmann KF, Holder B, Holthofer H, Hosseinkhani B, Hu G, Huang Y, Huber V, Hunt S, Ibrahim AG-E, Ikezu T, Inal JM, Isin M, Ivanova A, Jackson HK, Jacobsen S, Jay SM, Jayachandran M, Jenster G, Jiang L, Johnson SM, Jones JC, Jong A, Jovanovic-Talisman T, Jung S, Kalluri R, Kano S-I, Kaur S, Kawamura Y, Keller ET, Khamari D, Khomyakova E, Khvorova A, Kierulf P, Kim KP, Kislinger T, Klingeborn M, Klinke DJ, Kornek M, Kosanović MM, Kovács ÁF, Krämer-Albers E-M, Krasemann S, Krause M, Kurochkin IV, Kusuma GD, Kuypers S, Laitinen S, Langevin SM, Languino LR, Lannigan J, Lässer C, Laurent LC, Lavieu G, Lázaro-Ibáñez E, Le Lay S, Lee M-S, Lee YXF, Lemos DS, Lenassi M, Leszczynska A, Li IT, Liao K, Libregts SF, Ligeti E, Lim R, Lim SK, Linē A, Linnemannstöns K, Llorente A, Lombard CA, Lorenowicz MJ, Lörincz ÁM, Lötvall J, et al. , J Extracell Vesicles 2018, 7, 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Hartjes TA, Mytnyk S, Jenster GW, van Steijn V, van Royen ME, Bioengineering (Basel) 2019, 6, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Rikkert LG, Nieuwland R, Terstappen LWMM, Coumans FAW, J Extracell Vesicles 2019, 8, 1555419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Jung MK, Mun JY, J. Vis. Exp 2018, 131, 56482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Chiang C-Y, Chen C, J Biomed Sci 2019, 26, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Palmieri V, Lucchetti D, Gatto I, Maiorana A, Marcantoni M, Maulucci G, Papi M, Pola R, De Spirito M, Sgambato A, J. Nanopart. Res 2014, 16, 2583. [Google Scholar]
- [90].van der Pol E, Hoekstra AG, Sturk A, Otto C, van Leeuwen TG, Nieuwland R, J. Thromb. Haemost 2010, 8, 2596. [DOI] [PubMed] [Google Scholar]
- [91].Szatanek R, Baj-Krzyworzeka M, Zimoch J, Lekka M, Siedlar M, Baran J, Int. J. Mol. Sci 2017, 18, 1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Dragovic RA, Gardiner C, Brooks AS, Tannetta DS, Ferguson DJP, Hole P, Carr B, Redman CWG, Harris AL, Dobson PJ, Harrison P, Sargent IL, Nanomedicine 2011, 7, 780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Pospichalova V, Svoboda J, Dave Z, Kotrbova A, Kaiser K, Klemova D, Ilkovics L, Hampl A, Crha I, Jandakova E, Minar L, Weinberger V, Bryja V, J Extracell Vesicles 2015, 4, 25530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Suárez H, Gámez-Valero A, Reyes R, López-Martín S, Rodríguez MJ, Carrascosa JL, Cabañas C, Borràs FE, Yáñez-Mó M, Sci. Rep 2017, 7, 11271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Anderson W, Kozak D, Coleman VA, Jämting ÅK, Trau M, J. Colloid Interface Sci 2013, 405, 322. [DOI] [PubMed] [Google Scholar]
- [96].Tatischeff I, Larquet E, Falcón-Pérez JM, Turpin P-Y, Kruglik SG, J Extracell Vesicles 2012, 1, 19179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Maas SLN, Broekman MLD, de Vrij J, Methods Mol. Biol 2017, 1545, 21. [DOI] [PubMed] [Google Scholar]
- [98].van der Pol E, Coumans F, Varga Z, Krumrey M, Nieuwland R, J. Thromb. Haemost 2013, 11 Suppl 1, 36. [DOI] [PubMed] [Google Scholar]
- [99].Xia C, Zeng Z, Fang B, Tao M, Gu C, Zheng L, Wang Y, Shi Y, Fang C, Mei S, Chen Q, Zhao J, Lin X, Fan S, Jin Y, Chen P, Free Radic. Biol. Med 2019, 143, 1. [DOI] [PubMed] [Google Scholar]
- [100].Lu K, Li H-Y, Yang K, Wu J-L, Cai X-W, Zhou Y, Li C-Q, Stem Cell Res Ther 2017, 8, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Bach F, Libregts S, Creemers L, Meij B, Ito K, Wauben M, Tryfonidou M, Oncotarget 2017, 8, 88845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Lan W-R, Pan S, Li H-Y, Sun C, Chang X, Lu K, Jiang C-Q, Zuo R, Zhou Y, Li C-Q, Stem Cells Int 2019, 2019, 8404168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Qi L, Wang R, Shi Q, Yuan M, Jin M, Li D, J Bone Miner Metab 2019, 37, 455. [DOI] [PubMed] [Google Scholar]
- [104].Cheng X, Zhang G, Zhang L, Hu Y, Zhang K, Sun X, Zhao C, Li H, Li YM, Zhao J, Cell Mol J. Med 2018, 22, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Yuan F-L, Xu R-S, Ye J-X, Zhao M-D, Ren L-J, Li X, Cell Mol J. Med 2019, 23, 3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Moen A, Jacobsen D, Phuyal S, Legfeldt A, Haugen F, Røe C, Gjerstad J, J. Transl. Med 2017, 15, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Bach FC, de Vries SA, Riemers FM, Boere J, van Heel FW, van Doeselaar M, Goerdaya SS, Nikkels PG, Benz K, Creemers LB, Maarten Altelaar AF, Meij BP, Ito K, Tryfonidou MA, Eur Cell Mater 2016, 32, 163. [DOI] [PubMed] [Google Scholar]
- [108].Bari E, Perteghella S, Di Silvestre D, Sorlini M, Catenacci L, Sorrenti M, Marrubini G, Rossi R, Tripodo G, Mauri P, Marazzi M, Torre ML, Cells 2018, 7, DOI 10.3390/cells7110190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Liao Z, Luo R, Li G, Song Y, Zhan S, Zhao K, Hua W, Zhang Y, Wu X, Yang C, Theranostics 2019, 9, 4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Chen C-C, Chen J, Wang W-L, Xie L, Shao C-Q, Zhang Y-X, Orthop. Surg 2020, 13, 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Hingert D, Ekström K, Aldridge J, Crescitelli R, Brisby H, Stem Cell Res Ther 2020, 11, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Hu S-Q, Zhang Q-C, Meng Q-B, Hu A-N, Zou J-P, Li X-L, Exp. Cell Res 2020, 395, 112239. [DOI] [PubMed] [Google Scholar]
- [113].Li M, Li R, Yang S, Yang D, Gao X, Sun J, Ding W, Ma L, Med. Sci. Monit 2020, 26, e922928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Li Z-Q, Kong L, Liu C, Xu H-G, Am. J. Med. Sci 2020, 360, 693. [DOI] [PubMed] [Google Scholar]
- [115].Luo L, Jian X, Sun H, Qin J, Wang Y, Zhang J, Shen Z, Yang D, Li C, Zhao P, Liu M, Tian Z, Zhou Y, Stem Cells 2021, 39, 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Song J, Chen Z-H, Zheng C-J, Song K-H, Xu G-Y, Xu S, Zou F, Ma X-S, Wang H-L, Jiang J-Y, Mol. Ther. Nucleic Acids 2020, 21, 1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Sun Z, Liu B, Liu Z-H, Song W, Wang D, Chen B-Y, Fan J, Xu Z, Geng D, Luo Z-J, Mol. Ther. Nucleic Acids 2020, 22, 1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Sun Z, Zhao H, Liu B, Gao Y, Tang W-H, Liu Z-H, Luo Z-J, Life Sci 2021, 265, 118778. [DOI] [PubMed] [Google Scholar]
- [119].Tang S, Salazar-Puerta A, Richards J, Khan S, Hoyland JA, Gallego-Perez D, Walter B, Higuita-Castro N, Purmessur D, Eur Cell Mater 2021, 41, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Wen T, Wang H, Li Y, Lin Y, Zhao S, Liu J, Chen B, Cell Cycle 2021, 20, 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Xiang H, Su W, Wu X, Chen W, Cong W, Yang S, Liu C, Qiu C, Yang S-Y, Wang Y, Zhang G, Guo Z, Xing D, Chen B, Oxid. Med. Cell. Longev 2020, 2020, 6697577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Xie L, Chen Z, Liu M, Huang W, Zou F, Ma X, Tao J, Guo J, Xia X, Lyu F, Wang H, Zheng C, Jiang J, Mol. Ther. Nucleic Acids 2020, 22, 601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Yuan Q, Wang X, Liu L, Cai Y, Zhao X, Ma H, Zhang Y, Stem Cells Dev 2020, 29, 1038. [DOI] [PubMed] [Google Scholar]
- [124].Zhang J, Zhang J, Zhang Y, Liu W, Ni W, Huang X, Yuan J, Zhao B, Xiao H, Xue F, J. Cell Mol. Med 2020, 24, 11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Zhang Q-C, Hu S-Q, Hu A-N, Zhang T-W, Jiang L-B, Li X-L, J. Orthop. Res 2020, 1. [Google Scholar]
- [126].Zhu L, Shi Y, Liu L, Wang H, Shen P, Yang H, Cell Cycle 2020, 19, 1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Zhu G, Yang X, Peng C, Yu L, Hao Y, Exp. Cell Res 2020, 393, 112109. [DOI] [PubMed] [Google Scholar]
- [128].Song Y, Wang Y, Zhang Y, Geng W, Liu W, Gao Y, Li S, Wang K, Wu X, Kang L, Yang C, J. Cell Mol. Med 2017, 21, 1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Wang J, Shen X, Liu J, Chen W, Wu F, Wu W, Meng Z, Zhu M, Miao C, Cell Death Dis 2020, 11, 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].McGeough MD, Wree A, Inzaugarat ME, Haimovich A, Johnson CD, Peña CA, Goldbach-Mansky R, Broderick L, Feldstein AE, Hoffman HM, J. Clin. Invest 2017, 127, 4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Wan Z, Fan Y, Liu X, Xue J, Han Z, Zhu C, Wang X, Diabetes. Metab. Syndr. Obes 2019, 12, 1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Molinos M, Almeida CR, Caldeira J, Cunha C, Gonçalves RM, Barbosa MA, J. R. Soc. Interface 2015, 12, 20141191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Johnson ZI, Schoepflin ZR, Choi H, Shapiro IM, Risbud MV, Eur Cell Mater 2015, 30, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Angulski ABB, Capriglione LG, Batista M, Marcon BH, Senegaglia AC, Stimamiglio MA, Correa A, Stem Cell Rev and Rep 2017, 13, 244. [DOI] [PubMed] [Google Scholar]
- [135].Álvarez-Viejo M, World J. Stem Cells 2020, 12, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Qiu G, Zheng G, Ge M, Wang J, Huang R, Shu Q, Xu J, Stem Cell Res Ther 2018, 9, 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Tang Y, Zhou Y, Li H-J, Stem Cell Res Ther 2021, 12, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Roberts S, Urban JP, Evans H, Eisenstein SM, Spine 1996, 21, 415. [DOI] [PubMed] [Google Scholar]
- [139].Peng B, Hou S, Shi Q, Jia L, Chin. Med. J 2001, 114, 308. [PubMed] [Google Scholar]
- [140].Maas SLN, Breakefield XO, Weaver AM, Trends Cell Biol 2017, 27, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Vergroesen PPA, Kingma I, Emanuel KS, Hoogendoorn RJW, Welting TJ, van Royen BJ, van Dieën JH, Smit TH, Osteoarthr. Cartil 2015, 23, 1057. [DOI] [PubMed] [Google Scholar]
- [142].Hadjipavlou AG, Tzermiadianos MN, Bogduk N, Zindrick MR, J. Bone Joint Surg. Br 2008, 90, 1261. [DOI] [PubMed] [Google Scholar]
- [143].Choi Y-S, Asian Spine J 2009, 3, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Richardson SM, Freemont AJ, Hoyland JA, in The Intervertebral Disc (Eds.: Shapiro IM, Risbud MV), Springer Vienna, Vienna, 2014, pp. 177–200. [Google Scholar]
- [145].Oichi T, Taniguchi Y, Oshima Y, Tanaka S, Saito T, JOR Spine 2020, 3, e1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Kepler CK, Ponnappan RK, Tannoury CA, Risbud MV, Anderson DG, Spine J 2013, 13, 318. [DOI] [PubMed] [Google Scholar]
- [147].Rider SM, Mizuno S, Kang JD, Spine Surg. Relat. Res 2019, 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Bibby SRS, Urban JPG, Eur. Spine J 2004, 13, 695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Feng C, Yang M, Lan M, Liu C, Zhang Y, Huang B, Liu H, Zhou Y, Oxid. Med. Cell. Longev 2017, 2017, 5601593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Shi J, Zhou X, Wang Z, Kurra S, Niu J, Yang H, BMC Musculoskelet. Disord 2019, 20, 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Kohyama K, Saura R, Doita M, Mizuno K, Kobe J Med Sci 2000, 46, 283. [PubMed] [Google Scholar]
- [152].Sarath Babu N, Krishnan S, Brahmendra Swamy CV, Venkata Subbaiah GP, Gurava Reddy AV, Idris MM, Spine J 2016, 16, 989. [DOI] [PubMed] [Google Scholar]
- [153].Risbud MV, Shapiro IM, Nat. Rev. Rheumatol 2014, 10, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Tang X, Jing L, Richardson WJ, Isaacs RE, Fitch RD, Brown CR, Erickson MM, Setton LA, Chen J, J. Orthop. Res 2016, 34, 1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Torre OM, Mroz V, Bartelstein MK, Huang AH, Iatridis JC, Ann. N. Y. Acad. Sci 2019, 1442, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Adams MA, Dolan P, J. Anat 2012, 221, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Vo NV, Hartman RA, Yurube T, Jacobs LJ, Sowa GA, Kang JD, Spine J 2013, 13, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Patil P, Niedernhofer LJ, Robbins PD, Lee J, Sowa G, Vo N, Curr. Mol. Biol. Rep 2018, 4, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Ding F, Shao Z, Xiong L, Apoptosis 2013, 18, 777. [DOI] [PubMed] [Google Scholar]
- [160].Zhao C-Q, Jiang L-S, Dai L-Y, Apoptosis 2006, 11, 2079. [DOI] [PubMed] [Google Scholar]
- [161].Bonnevie ED, Gullbrand SE, Ashinsky BG, Tsinman TK, Elliott DM, Chao P-HG, Smith HE, Mauck RL, Nat. Biomed. Eng 2019, 3, 998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Sudo H, Minami A, Arthritis Rheum 2011, 63, 1648. [DOI] [PubMed] [Google Scholar]
- [163].Masuda K, An HS, Eur. Spine J 2006, 15 Suppl 3, S422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Qu F, Guilak F, Mauck RL, Nat. Rev. Rheumatol 2019, 15, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Ma K, Chen S, Li Z, Deng X, Huang D, Xiong L, Shao Z, Osteoarthr. Cartil 2019, 27, 41. [DOI] [PubMed] [Google Scholar]
- [166].Henriksson H, Thornemo M, Karlsson C, Hägg O, Junevik K, Lindahl A, Brisby H, Spine 2009, 34, 2278. [DOI] [PubMed] [Google Scholar]
- [167].Henriksson HB, Svala E, Skioldebrand E, Lindahl A, Brisby H, Spine 2012, 37, 722. [DOI] [PubMed] [Google Scholar]
- [168].Barreto Henriksson H, Lindahl A, Skioldebrand E, Junevik K, Tängemo C, Mattsson J, Brisby H, Stem Cell Res Ther 2013, 4, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Sakai D, Nishimura K, Tanaka M, Nakajima D, Grad S, Alini M, Kawada H, Ando K, Mochida J, Spine J 2015, 15, 1356. [DOI] [PubMed] [Google Scholar]
- [170].Ying J, Han Z, Pei S, Su L, Ruan D, Stem Cells Int 2019, 2019, 9147835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Xu W, Xu R, Li Z, Wang Y, Hu R, J. Cell Mol. Med 2019, 23, 1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Pattappa G, Peroglio M, Sakai D, Mochida J, Benneker LM, Alini M, Grad S, Eur Cell Mater 2014, 27, 124. [DOI] [PubMed] [Google Scholar]
- [173].Han B, Wang H, Li H, Tao Y, Liang C, Li F, Chen G, Chen Q, Cells Tissues Organs (Print) 2014, 199, 342. [DOI] [PubMed] [Google Scholar]
- [174].Li H, Tao Y, Liang C, Han B, Li F, Chen G, Chen Q, Cells Tissues Organs (Print) 2013, 198, 266. [DOI] [PubMed] [Google Scholar]
- [175].Liu J, Tao H, Wang H, Dong F, Zhang R, Li J, Ge P, Song P, Zhang H, Xu P, Liu X, Shen C, Stem Cells Dev 2017, 26, 901. [DOI] [PubMed] [Google Scholar]
- [176].Vadalà G, Ambrosio L, Russo F, Papalia R, Denaro V, Stem Cells Int 2019, 2019, 2376172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Hou Y, Shi G, Guo Y, Shi J, Aging (Albany, NY) 2020, 12, 6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Nakazawa KR, Walter BA, Laudier DM, Krishnamoorthy D, Mosley GE, Spiller KL, Iatridis JC, Spine J 2018, 18, 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Yang C, Cao P, Gao Y, Wu M, Lin Y, Tian Y, Yuan W, Sci. Rep 2016, 6, 22182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Wynn TA, Vannella KM, Immunity 2016, 44, 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Das A, Sinha M, Datta S, Abas M, Chaffee S, Sen CK, Roy S, Am. J. Pathol 2015, 185, 2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Ito K, Creemers L, Global Spine J 2013, 3, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].D’Agnelli S, Gerra MC, Bignami E, Arendt-Nielsen L, Mol. Pain 2020, 16, 1744806920957800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Ren J, Liu N, Sun N, Zhang K, Yu L, Curr. Stem Cell Res. Ther 2019, 14, 644. [DOI] [PubMed] [Google Scholar]
- [185].Ohtori S, Takahashi K, Chiba T, Yamagata M, Sameda H, Moriya H, Ann. Anat 2002, 184, 235. [DOI] [PubMed] [Google Scholar]
- [186].Coppes MH, Marani E, Thomeer RT, Groen GJ, Spine 1997, 22, 2342. [DOI] [PubMed] [Google Scholar]
- [187].Simeoli R, Montague K, Jones HR, Castaldi L, Chambers D, Kelleher JH, Vacca V, Pitcher T, Grist J, Al-Ahdal H, Wong L-F, Perretti M, Lai J, Mouritzen P, Heppenstall P, Malcangio M, Nat. Commun 2017, 8, 1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Mosley GE, Evashwick-Rogler TW, Lai A, Iatridis JC, Ann. N. Y. Acad. Sci 2017, 1409, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Rutges JPHJ, Duit RA, Kummer JA, Bekkers JEJ, Oner FC, Castelein RM, Dhert WJA, Creemers LB, Osteoarthr. Cartil 2013, 21, 2039. [DOI] [PubMed] [Google Scholar]
- [190].Stokes IAF, Iatridis JC, Spine 2004, 29, 2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Iatridis JC, Nicoll SB, Michalek AJ, Walter BA, Gupta MS, Spine J 2013, 13, 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Kushchayev SV, Glushko T, Jarraya M, Schuleri KH, Preul MC, Brooks ML, Teytelboym OM, Insights Imaging 2018, 9, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Apfel CC, Cakmakkaya OS, Martin W, Richmond C, Macario A, George E, Schaefer M, Pergolizzi JV, BMC Musculoskelet. Disord 2010, 11, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Akeda K, Yamada T, Inoue N, Nishimura A, Sudo A, BMC Musculoskelet. Disord 2015, 16, 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Rohde E, Pachler K, Gimona M, Cytotherapy 2019, 21, 581. [DOI] [PubMed] [Google Scholar]
- [196].Ferkany JW, Williams M, Curr. Protoc. Pharmacol 2008, Chapter 9, Unit 9.10. [DOI] [PubMed] [Google Scholar]
- [197].Lener T, Gimona M, Aigner L, Börger V, Buzas E, Camussi G, Chaput N, Chatterjee D, Court FA, Del Portillo HA, O’Driscoll L, Fais S, Falcon-Perez JM, Felderhoff-Mueser U, Fraile L, Gho YS, Görgens A, Gupta RC, Hendrix A, Hermann DM, Hill AF, Hochberg F, Horn PA, de Kleijn D, Kordelas L, Kramer BW, Krämer-Albers E-M, Laner-Plamberger S, Laitinen S, Leonardi T, Lorenowicz MJ, Lim SK, Lötvall J, Maguire CA, Marcilla A, Nazarenko I, Ochiya T, Patel T, Pedersen S, Pocsfalvi G, Pluchino S, Quesenberry P, Reischl IG, Rivera FJ, Sanzenbacher R, Schallmoser K, Slaper-Cortenbach I, Strunk D, Tonn T, Vader P, van Balkom BWM, Wauben M, Andaloussi SE, Théry C, Rohde E, Giebel B, J Extracell Vesicles 2015, 4, 30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Blanquer SBG, Grijpma DW, Poot AA, Adv. Drug Deliv. Rev 2015, 84, 172. [DOI] [PubMed] [Google Scholar]
- [199].Food and Drug Administration, “CFR - Code of Federal Regulations Title 21 § 312.23 IND content and format,” can be found under https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/cfrsearch.cfm?fr=312.23, 2020.
- [200].Armstrong JPK, Stevens MM, Adv. Drug Deliv. Rev 2018, 130, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH), “ICH Safety Guidelines,” can be found under https://www.ich.org/page/safety-guidelines, n.d.
- [202].Gimona M, Pachler K, Laner-Plamberger S, Schallmoser K, Rohde E, Int. J. Mol. Sci 2017, 18, 1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Food and Drug Administration (FDA), “Biologics License Applications (BLA) Process (CBER),” can be found under https://www.fda.gov/vaccines-blood-biologics/development-approval-process-cber/biologics-license-applications-bla-process-cber, 2020.
- [204].Food and Drug Administration, “Applications for biologics licenses; procedures for filing. 21 C.F.R. § 601.2, ” can be found under https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/CFRSearch.cfm?fr=601.2, 2020.
- [205].Escudier B, Dorval T, Chaput N, André F, Caby M-P, Novault S, Flament C, Leboulaire C, Borg C, Amigorena S, Boccaccio C, Bonnerot C, Dhellin O, Movassagh M, Piperno S, Robert C, Serra V, Valente N, Le Pecq J-B, Spatz A, Lantz O, Tursz T, Angevin E, Zitvogel L, J. Transl. Med 2005, 3, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Morse MA, Garst J, Osada T, Khan S, Hobeika A, Clay TM, Valente N, Shreeniwas R, Sutton MA, Delcayre A, Hsu D-H, Le Pecq J-B, Lyerly HK, J. Transl. Med 2005, 3, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Dai S, Wei D, Wu Z, Zhou X, Wei X, Huang H, Li G, Mol. Ther 2008, 16, 782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Nassar W, El-Ansary M, Sabry D, Mostafa MA, Fayad T, Kotb E, Temraz M, Saad A-N, Essa W, Adel H, Biomater. Res 2016, 20, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Soekmadji C, Li B, Huang Y, Wang H, An T, Liu C, Pan W, Chen J, Cheung L, Falcon-Perez JM, Gho YS, Holthofer HB, Le MTN, Marcilla A, O’Driscoll L, Shekari F, Shen TL, Torrecilhas AC, Yan X, Yang F, Yin H, Xiao Y, Zhao Z, Zou X, Wang Q, Zheng L, J Extracell Vesicles 2020, 9, 1809766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Patel DB, Santoro M, Born LJ, Fisher JP, Jay SM, Biotechnol. Adv 2018, 36, 2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Watson DC, Bayik D, Srivatsan A, Bergamaschi C, Valentin A, Niu G, Bear J, Monninger M, Sun M, Morales-Kastresana A, Jones JC, Felber BK, Chen X, Gursel I, Pavlakis GN, Biomaterials 2016, 105, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]


