Abstract
Deleted in Breast Cancer 1 (DBC1) is an important metabolic sensor. Previous studies have implicated DBC1 in various cellular functions, notably cell proliferation, apoptosis, histone modification, and adipogenesis. However, current reports about the role of DBC1 in tumorigenesis are controversial and designate DBC1 alternatively as a tumor suppressor or a tumor promoter. In the present study, we report that polyoma small T antigen (PyST) associates with DBC1 in mammalian cells, and this interaction leads to the posttranslational downregulation of DBC1 protein levels. When coexpressed, DBC1 overcomes PyST-induced mitotic arrest and promotes the exit of cells from mitosis. Using both transient and stable modes of PyST expression, we also show that cellular DBC1 is subjected to degradation by LKB1, a tumor suppressor and cellular energy sensor kinase, in an AMP kinase-independent manner. Moreover, LKB1 negatively regulates the phosphorylation as well as activity of the prosurvival kinase AKT1 through DBC1 and its downstream pseudokinase substrate, Tribbles 3 (TRB3). Using both transient transfection and stable cell line approaches as well as soft agar assay, we demonstrate that DBC1 has oncogenic potential. In conclusion, our study provides insight into a novel signaling axis that connects LKB1, DBC1, TRB3, and AKT1. We propose that the LKB1–DBC1–AKT1 signaling paradigm may have an important role in the regulation of cell cycle and apoptosis and consequently tumorigenesis.
Keywords: DBC1, CCAR2, KIAA 1967, small T antigen, AKT, LKB1, mitosis, apoptosis, tumor cell biology, tumor virus
Abbreviations: CCAR2, cell cycle and apoptosis regulator 2; DBC1, deleted in breast cancer 1; HSP, heat shock protein; OA, okadaic acid; PARylation, poly ADP ribosylation; pH3, phospho-histone 3; PP2A, protein phosphatase 2A; PyST, polyoma small T antigen; TAP, tandem affinity purification
In this study, we have used polyoma virus small T antigen (PyST) to elucidate the role of deleted in breast cancer 1 (DBC1) in cell cycle regulation, cellular signaling, and tumorigenesis. PyST is a ∼22 kDa protein encoded by the early set of polyoma virus genome. It consists of an N-terminal J domain that binds heat shock proteins (HSPs) and a C-terminal domain that binds protein phosphatase 2A (PP2A) (Fig. 1A) (1). Expression of small T antigen in mammalian cells induces striking mitotic arrest as indicated by cell rounding and activation of spindle assembly checkpoint, followed by extensive apoptosis within 2 to 3 days, leading to the almost total annihilation of the host cells (2, 3). For inducing mitotic arrest and apoptosis, interaction of PyST with PP2A is essential. We have recently shown that small T induced apoptosis in host mammalian cells also involves the activation of dependence receptor UNC5B, in a PP2A dependent manner (4). However, the molecular details of PyST-induced mitotic arrest are not fully understood yet.
Figure 1.
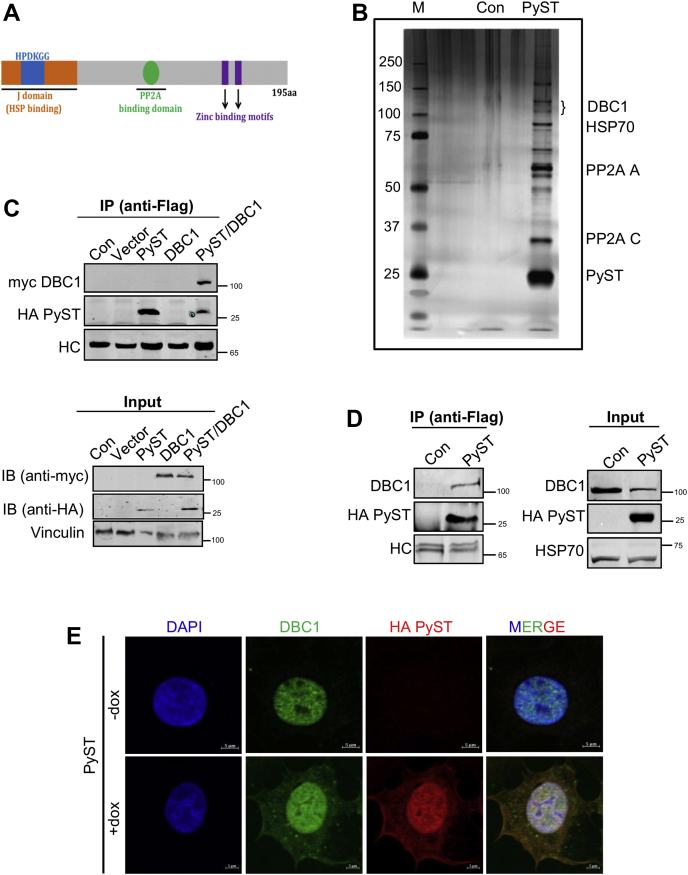
Polyoma small T (PyST) interacts with DBC1.A, schematic representation of polyoma virus small T antigen and its structural features, showing an N-terminal J domain and a C-terminal PP2A-binding domain. B, PyST-associated complexes isolated from pTREX-PyST-HA-FLAG cell lines by tandem affinity purification (TAP) using consecutive anti-FLAG and anti-HA affinity matrices were run on an SDS-PAGE and visualized by silver staining. C, 293T cells were cotransfected with pcDNA3-PyST-HA-Flag (1.5 μg), myc-DBC1 (1.5 μg), and empty vector (pCDNA3.1, 1.5 μg). PyST-HA-Flag was immunoprecipitated with anti-Flag antibody and subjected to SDS-PAGE and immunoblotting (IB) with anti-myc and anti-HA antibodies, as indicated. D, immunoprecipitation of endogenous DBC1 from 293T cells. These cells were transfected with pcDNA3-PyST-HA-Flag (1.5 μg). PyST was immunoprecipitated using anti-Flag antibody. Endogenous DBC1 was detected using anti-DBC1 antibody. In C and D immunoprecipitate panels, HC designates heavy chain of IgG. E, colocalization results of PyST (red) and DBC1 (green) using immunofluorescence microscopy obtained at about 24 h of doxycycline addition. Nuclei were stained with DAPI (63× magnification; the scale bar represents 5 μm). The results are representative of three independent experiments. DBC1, deleted in breast cancer 1; HSP, heat shock protein; PP2A, protein phosphatase 2A.
DBC1, also known as cell cycle and apoptosis regulator 2 (CCAR2) or KIAA1967, is a critical regulator of multiple cellular functions including transcription, mRNA splicing, glucose/fat metabolism, apoptosis, circadian rhythms, and epigenetic regulation (5, 6, 7, 8, 9, 10). Identification of DBC1-binding partners using proteomic approaches has shown its association predominantly with the proteins involved in cell cycle regulation, gene expression/splicing, chromatin remodeling, and circadian cycles (5, 11). The diverse role of DBC1 in numerous cellular processes can be attributed to its structural features. It consists of an N-terminal S1 RNA-binding motif, a nuclear localization signal, a leucine zipper, a nudix domain, EF hand, and a coiled coil domain (9). The N-terminal part of DBC1 is known to bind and hence regulate the functions of SIRT1, HDAC3, and SUV39H1 (12, 13, 14). Of these, SIRT1 is the most well-known binding partner for DBC1. By binding to SIRT1, DBC1 sequesters and hence negatively regulates the deacetylase activity of SIRT1. DBC1–SIRT1 interaction thus has an important role in fat and energy metabolism, chromatin remodeling, as well as epigenetic regulations.
Although DBC1 has been reported to interact with many proteins involved in cell division (11), its specific role in cell cycle has not been studied in detail, and its role in tumorigenesis is still controversial. A few studies have reported it to be overexpressed in some cancers (15, 16, 17), thus proposing it to have a tumor-promoting role. It has also been shown to bind BRCA1, a well-known tumor suppressor, through its BRCT domain, resulting in the suppression of transcriptional activity of BRCA1 (18). Furthermore, DBC1 positively regulates AKT1 activity via the transcriptional repression of TRB3 which is known to sequester AKT1 and prevent its activation (19). In contrast, other reports support the role of DBC1 as a tumor suppressor. For example, a study using DBC1 knockout mice has shown to stabilize p53 through the displacement of MDM2, which usually sequesters and degrades p53 (20). These mice were more susceptible to tumor formation than the WT mice. Recently, it was shown that DBC1 negatively regulates the functions of MDM2-CREB binding protein ubiquitinating complex, thus preventing the polyubiquitination and subsequent proteasomal degradation of p53 (21). Similarly, by inhibiting SIRT1, DBC1 leads to the hyperacetylation and stabilization/activation of p53 and FOXO3, thus promoting their proapoptotic activity (20). SIRT1 is known to be over expressed in many types of cancers such as colorectal, prostate, and breast cancer (22, 23, 24). Further, by also negatively regulating SIRT1 activity, DBC1 could function as a tumor suppressor.
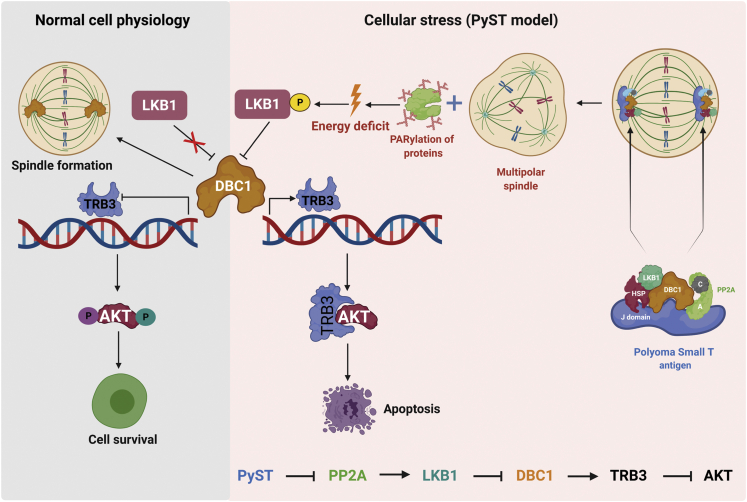
How DBC1 regulates cell cycle and tumorigenesis is mostly unclear, though several studies have demonstrated its role in the regulation of p53 (20), c-MYC (25), BRCA1 (18), MCC, and β-catenin (26, 27). In this study, using PyST as a tool, we show that DBC1 has an important role in cell cycle regulation by promoting the progression of cells through the mitotic phase, particularly when cells are exposed to agents like small T, that lead to mitotic aberrations. Interestingly, we also identified DBC1 as a potential downstream target of LKB1 signaling. LKB1 (also called STK11), a ubiquitously expressed serine/threonine kinase, is known to phosphorylate the family of 14 kinases referred to as AMPK-related kinases and is therefore termed a “master kinase” (28). Germline mutations in human LKB1 are the main cause of Peutz–Jeghers Syndrome (29) and such patients are at a high risk of developing hamartomatous polyps, especially in the gastrointestinal tract, that can develop into tumors (30). We also found that LKB1 negatively regulates AKT1 activity through DBC1 and Tribbles 3 (TRB3). We therefore propose that down-regulation of DBC1 leading to the inactivation of AKT1 may be one of the mechanisms by which LKB1 exerts its tumor suppressor functions.
Results
Identification of DBC1 as a novel PyST interacting protein
To understand the detailed mechanism of mitotic arrest and apoptosis that is induced by PyST, we set out to identify its potential cellular interacting partners. For that purpose, we made use of an inducible stable U2OS cell line expressing PyST having HA-FLAG tags at the C-terminus. This cell line, designated as pTREX-PyST-HA-Flag, has been previously described (2). Addition of doxycycline causes the expression of PyST in these cells and leads to the appearance of a remarkable rounding up phenotype within 16 to 20 h, a feature usually associated with cells undergoing mitosis (31, 32). Prolonged induction of small T expression (>24 h) leads to the initiation of apoptosis, which kills most of the cells within 48 h (4).
We harvested these cells, most of which were arrested in the mitotic phase, as was apparent by their predominantly rounded up phenotype, before they would initiate apoptosis. Tandem affinity purification (TAP) on the cell lysates was performed at about 20 h of doxycycline addition, using anti-HA and anti-Flag antibodies sequentially. A small aliquot of the immunoprecipitates was run on SDS-PAGE and analyzed by silver staining. In this gel, bands for PyST and some of its well-known binding partners like A and C subunits of PP2A and HSP70 were clearly seen, as indicated by their molecular weight positions (Fig. 1B). The rest of the immunoprecipitates were subjected to LC-MS-MS analysis. DBC1 (CCAR2) was found to be one of the notable and novel interacting partners of PyST and showed significant coverage of peptides in these mass spectrometry results (Tables S1 and S2). The raw mass spectrometry data files are available at ftp://massive.ucsd.edu/MSV000087239/.
DBC1/CCAR2 attracted our attention because of its known role in apoptosis (7, 33, 34), which correlated well with the apoptotic phenotype induced by PyST expression. To confirm that PyST and DBC1 interact with each other in the cells, we transiently coexpressed PyST-HA-Flag and myc-DBC1 in 293T cells and subjected the lysates to immunoprecipitation using anti-Flag antibodies. Western blotting results using anti-myc tag antibody confirmed that DBC1 interacts with PyST (Fig. 1C). To confirm if PyST interacts with the endogenous DBC1 also, 293T cells were transfected with PyST-HA-Flag construct. After immunoprecipitation of PyST using anti-Flag antibodies, endogenous DBC1 was detected using anti-DBC1 antibody (Fig. 1D). Altogether, immunoprecipitation results showed that PyST interacts with exogenous and endogenous DBC1 protein. Next, we wanted to find out whether the two proteins colocalize within the cells. DBC1 is known to be nuclear in its localization (7). Our results also showed that it is almost exclusively localized to the nucleus. In presence of doxycycline (+dox), PyST was expressed and was also found predominantly in the nucleus (Fig. 1E), though a lesser proportion was also seen in the cytoplasm, which is consistent with the published reports (2). Consistent with PyST’s localization pattern, DBC1 was also seen to localize in both these compartments in the presence of PyST expression (+dox), thus further supporting the argument that DBC1 and PyST may interact with each other.
Polyoma small T antigen causes posttranslational down-regulation of DBC1
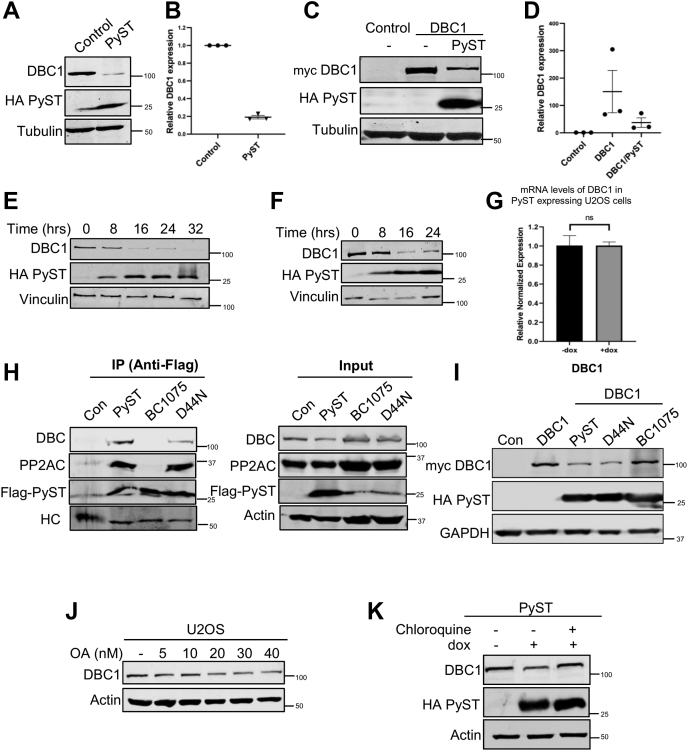
To study whether PyST has any impact on DBC1 expression and/or functions, both these proteins were cotransfected in 293T cells. The results showed that PyST coexpression causes profound down-regulation of both endogenous and exogenous protein levels of DBC1, respectively (Fig. 2, A–D). These results were consistent with the lower DBC1 protein levels in the presence of PyST, as was also evident in Figure 1, C and D. Next, we wanted to find out whether the down-regulation of DBC1 is also valid in inducible PyST-expressing U2OS cell line (pTREX-PyST-HA-Flag) mentioned above. Upon doxycycline addition, we observed a time-dependent decrease in the endogenous and exogenously expressing protein levels of DBC1 (Fig. 2, E and F). RT-qPCR results showed that PyST expression has no effect on the mRNA levels of DBC1, implicating that down-regulation of DBC1 occurs at translational or post-translational levels but not at transcriptional level (Fig. 2G). To find out the particular domain of PyST that is required for down-regulation of DBC1, we cotransfected WT PyST, or its mutants PyST-BC1075 (PyST that does not bind PP2A) and PyST-D44N (PyST J domain mutant) (35) along with DBC1, followed by immunoprecipitation. The interaction between PyST and the endogenous DBC1 was seen with WT PyST and PyST-D44N, but not with the PyST-BC1075 mutant (Fig. 2H). To further support these results, we cotransfected myc-DBC1 with WT PyST or its mutants. Western blotting results showed down-regulation of DBC1 protein levels in presence of the WT PyST and D44N mutant, but not PyST-BC1075 (Fig. 2I). To independently confirm the involvement of PP2A in PyST-mediated degradation of DBC1, U2OS cells were treated for about 16 h with different concentrations of okadaic acid (OA), a known inhibitor of PP2A (36). As shown in Figure 2J, OA also down-regulated the protein levels of DBC1 in a dose-dependent manner, particularly at higher concentrations (i.e., 20–40 nM). All these results thus suggested the role of PP2A in regulating DBC1 protein levels and/or activity inside the cells. Next, we treated the PyST expressing cells with various inhibitors of protein degradation like MG132, bortezomib (proteasomal inhibitors), or chloroquine (lysosomal inhibitor). The results showed that DBC1 degradation was prevented by the treatment of cells with chloroquine (Fig. 2K), but not MG132 or bortezomib (data not shown), thus suggesting that DBC1 degradation by PyST occurs through the lysosomal pathway.
Figure 2.
Polyoma small T expression leads to thedown-regulationof DBC1 protein levels in aPP2A-dependentmanner.A, 293T cells were transfected with pCDNA3-PyST-HA-Flag (1.5 μg). After 48 h, the cell extracts were blotted for endogenous DBC1 using anti-DBC1 antibody and PyST with anti-HA antibody. B, graphical representation of relative DBC1 expression (endogenous) in PyST-transfected cells compared with control cells. Protein expression in control cells was arbitrarily taken as one in all of the Western blotting quantifications. C, 293T cells were cotransfected with pCDNA3 PyST-HA-Flag (1.5 μg) and myc-DBC1 (1.5 μg) constructs. Cell lysates were blotted for exogenously expressed myc-DBC1 using anti-myc tag antibody and PyST using anti-HA antibody. D, graph representing relative myc-DBC1 expression in the cells transfected with empty vector (control), myc-DBC1, and myc-DBC1/PyST. The values indicate mean ± SEM; n = 3 biological replicates. E, Western blotting of pTREX PyST-HA-Flag expressing cell extracts at different time periods of doxycycline addition using anti-DBC1 for detecting DBC1 (endogenous) and anti-HA antibodies for PyST. F, effect of PyST overexpression on exogenously expressing myc-DBC1 protein levels in pTREX PyST-HA-Flag cell line at different time points as seen by Western blotting. G, effects of overexpression of PyST on DBC1 mRNA at about 24 h of doxycycline addition. After cDNA synthesis, RT-qPCR was carried out using primers for DBC1 and Actin. Fold change was calculated by ΔΔCq method, and p-value was calculated using student’s t test; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. The error bars represent mean ± SEM. H, 293T cells were transfected with PyST (1.0 μg), BC1075 (1.5 μg), or D44N (1.5 μg). HA-Flag-tagged PyST (WT and mutants) was immunoprecipitated by anti-Flag antibody, and the lysates were immunoblotted with indicated antibodies. HC:heavy chain of IgG. I, 293T cells were cotransfected with myc-DBC1 (2 μg) and different PyST-HA-Flag constructs like WT PyST (1.5 μg), BC1075 (1.5 μg) (PyST: PP2A-), or D44N (1.5 μg) (PyST: HSPs-) constructs. Western blot for DBC1 and PyST was carried out using indicated antibodies. J, U2OS cells were treated with different concentrations of okadaic acid (OA) (for 16 h) to inhibit PP2A. Endogenous DBC1 was detected with DBC1 antibody. K, PyST expressing cells were treated with chloroquine (50 μM). Twenty hours posttreatment, the cell lysates were obtained and used for Western blotting. Endogenous levels of DBC1 were detected using anti-DBC1 antibody. DBC1, deleted in breast cancer 1; HSPs, heat shock proteins; PP2A, protein phosphatase 2A.
DBC1 plays a role in the regulation of mitosis
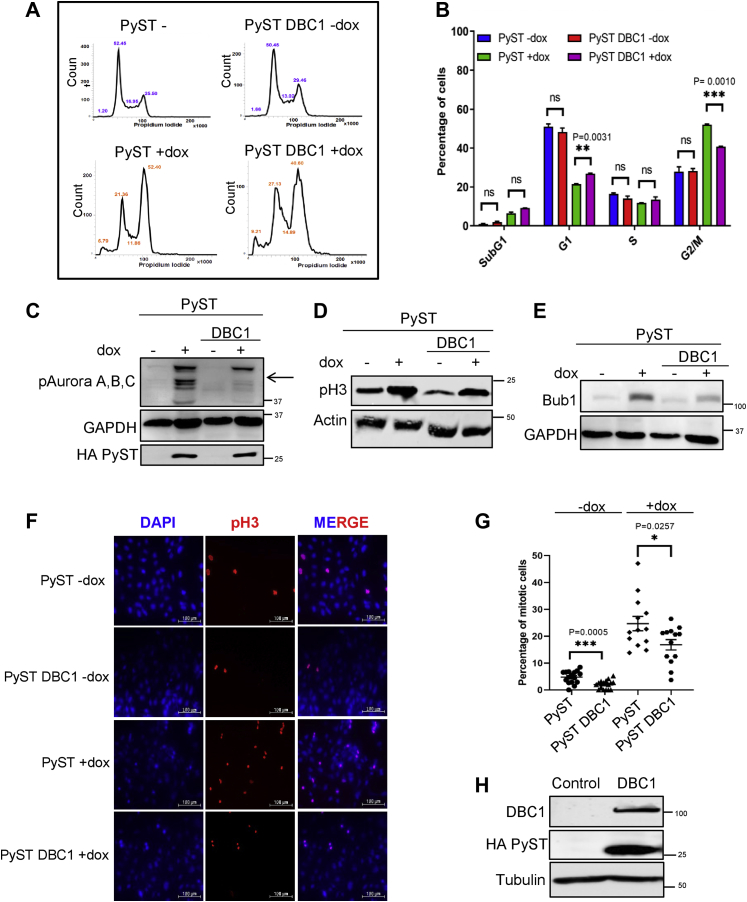
Because PyST induces a striking mitotic arrest in the host cells and also leads to the degradation of DBC1, therefore it was intriguing to find out whether DBC1 has any role in the regulation of mitosis. We used pTREX-PyST-DBC1 (double stable) cell line for cell cycle analysis using flow cytometry. Consistent with our previously published reports (2), the expression of PyST in U2OS cell lines induced a very robust mitotic arrest at about 24 h of PyST induction. In contrast, pTREX-PyST-DBC1 (+dox) cells exhibited a decreased G2/M peak vis-à-vis an increase in G0/G1 peak (Fig. 3, A and B). This was also supported by Western blotting results, using several known mitotic markers. Phosphorylation levels of Aurora A, B, C, and phospho-Histone 3 (pH3), as well as total amounts of Bub1 were clearly reduced in presence of DBC1 overexpression (Fig. 3, C–E). Immunofluorescence results, using pH3 (ser10) staining, also showed a clear decrease in the mitotic index in DBC1 overexpressing cells (Fig. 3, F and G). Altogether, these results thus indicated that DBC1 enables PyST expressing cells to overcome the mitotic arrest and hence exit mitosis. The expression of DBC1 and PyST in pTREX-PyST-DBC1 cell line was confirmed by Western blotting (Fig. 3H).
Figure 3.
DBC1 promotes mitotic progression in mammalian cells.A, FACS analysis of cells expressing PyST and DBC1 at 24 h of doxycycline addition. B, graphical representation of the FACS data, as shown in A. The values indicate mean ± SEM; n = 3 replicates (two-tailed unpaired student’s t test). p < 0.05 was taken as significant. The error bars represent mean ± SEM. C, DBC1 coexpression reduced phosphorylation levels of Aurora A, B, C levels in PyST expressing cells, after doxycycline addition for about 24 h. D and E, DBC1 expression reduced the phospho-Histone H3 and Bub1 signals, which are otherwise enhanced by PyST expression. F and G, immunofluorescence of phosphorylated histone 3 (pH3) in U2OS cells expressing PyST and DBC1. PyST cells overexpressing DBC1 showed decreased percentage of mitotic cells, as observed by pH3 staining (10× magnification; the scale bar represents 100 μm). The data represent the mean ± SEM; n = 3 replicates; N = 950 cells for PyST-dox, 799 cells for PyST DBC1-dox, 730 cells for PyST+dox, and 1008 cells for PyST DBC1+dox. H, Western blot showing the expression of DBC1 and PyST in U2OS double stable cell line, using anti DBC1 and anti HA antibodies respectively. Tubulin was used as a loading control. DBC1, deleted in breast cancer 1; FACS, fluorescence-activated cell sorting; pH3, phospho-histone 3; PyST, polyoma small T antigen.
DBC1 localizes to centrosome, spindle poles, and mid-body in mitotic cells
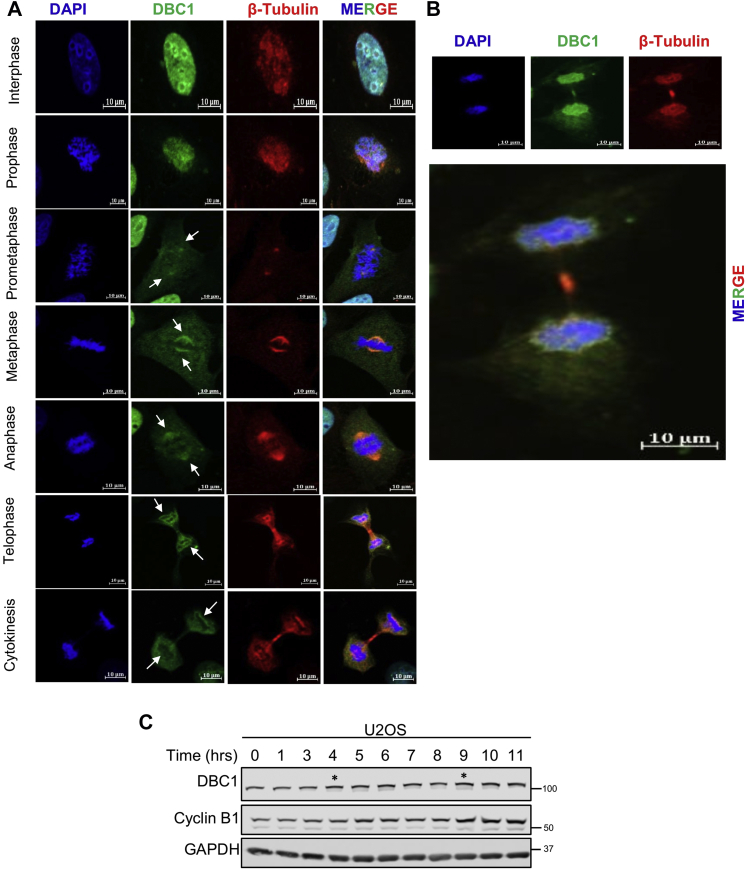
The above results further prompted us to study the localization of DBC1 through the cell cycle progression, using immunofluorescence microscopy. During interphase, DBC1 was seen to be exclusively nuclear (Fig. 4A), as was also seen in Figure 1E. At prophase, it showed similar staining as that of the condensing chromatin and also leaked into the cytoplasm, probably in response to the nuclear envelope breakdown. At prometaphase, DBC1 was spread all over the cell including the cytoplasm due to nuclear envelope breakdown, but was also prominently found at the spindle poles. During metaphase and anaphase, DBC1 was clearly seen to be located at the spindle poles. At telophase and cytokinesis, DBC1 staining was seen prominently concentrated around the chromosomal regions, forming a ring-like structure. Also, it was more visible at the central spindle and mid-body regions in anaphase and telophase/cytokinesis, respectively (Fig. 4A). During cytokinesis, DBC1 was seen distinctly at the mid-body (Fig. 4B).
Figure 4.
DBC1 localization and expression during cell cycle progression.A, immunofluorescence microscopic analysis of DBC1 in different stages of cell cycle in U2OS cells, as indicated. DAPI staining (blue) was used to stain the DNA and ascertain the cell cycle stage. Green staining shows DBC1 and red staining indicates β-Tubulin. The images are representative of three independent experiments (63× magnification; the scale bar represents 10 μm). The data are representative of at least three independent experiments. B, localization of DBC1 (green) and tubulin (red) at the mid-body during cytokinesis. A magnified size of the merged image of the same experiment is also shown. C, Western blot showing increased DBC1 protein amounts in U2OS cells, after release from thymidine block. Mitotic stage was apparent at about 9 to 11 h after thymidine release, as was confirmed by enhanced cyclin B1 signals. The asterisks (∗) at 4 h and 9 h indicate that the DBC1 levels clearly increase during the S and G2/M phases, respectively. DBC1, deleted in breast cancer 1.
To study the expression levels of DBC1 during cell cycle progression, U2OS cells were subjected to synchronization by double thymidine block, released and harvested at different time points. Although DBC1 protein levels were seen to increase as the cells proceeded to G2/M phase (starting from S phase at ∼4 h post release and later), the most significant increase was seen during the mitotic phase, which was concomitant with high levels of cyclin B1 amounts at ∼9 to 11 h post release of cells from the thymidine block (Fig. 4C).
PyST expression promotes high ADP ribosylation on the spindle and decreases interaction between DBC1 and its cellular interacting partners
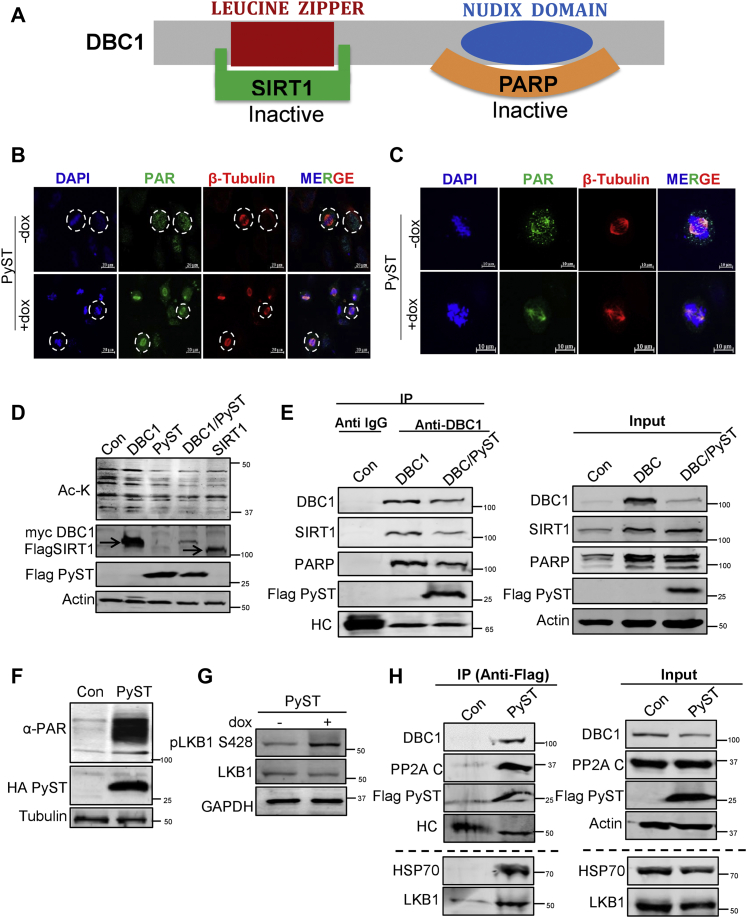
DBC1 has been shown to interact with SIRT1 through its N-terminal leucine zipper region (12) and with PARP-1 through its C-terminal Nudix region (37), thereby inhibiting their corresponding enzymatic activities (Fig. 5A)
Figure 5.
PySTdown-regulatesDBC1viaactivation of LKB1.A, schematic representation of different domains of DBC1 that interact with SIRT1 and PARP-1. B, detection of PARylation in PyST expressing cell lines using immunofluorescence. The cells with circles represent the ones in mitosis in control (−dox) and PyST expressing cells (+dox), (20× magnification; the scale bar represents 20 μm). C, one cell was chosen from the control (−dox) and PyST expressing cells (+dox) from the experiments, as shown in (B) and magnified to highlight the expression of PAR in mitosis. The magnified cells as shown in panel C are different images but from the same set of experiments (63× magnification; the scale bar represents 10 μm). D, 293T cells were transfected with myc DBC1 (1.0 μg), PyST-HA-Flag (1.0 μg), and Flag SIRT1 (1.0 μg), and Western blotting was performed with anti-acetyl lysine (Ac-K) antibody. E, 293T cells were transfected with PyST-HA-Flag (1.5 μg) and myc-DBC1 (1 μg) constructs. Immunoprecipitation was carried out with anti-DBC1 and anti-IgG (control) antibody, followed by Western blotting to confirm the interaction between PyST, DBC1, SIRT1, and PARP-1, using the indicated antibodies. F, Western blotting of α-PAR levels in the cell lysates obtained from PyST-U2OS cell line. G, U2OS cells stably over-expressing PyST were induced for 24 h. The cell lysates were blotted for pLKB1 (S428) and total LKB1. H, Western blot showing immunoprecipitation (IP) of PyST-HA-FLAG using anti-FLAG antibody and detection of DBC1, PP2A-C, HSP70, and LKB1 using the respective antibodies, as indicated. Please note that this experiment is the same as was carried out for Figure 2H. Although DBC1, PP2A-C, and FLAG-PyST were shown in Figure 2H, these bands (only for the control (Con) and PyST lanes) are reproduced here as they are part of the H also. This is indicated by the dotted line. The bands of LKB1 and HSP-70 of this experiment are shown here, below the dotted line. Similarly, for the “input” part of this experiment (right panel), the bands of DBC1, PP2A-C, Flag-PyST, and Actin are reproduced from Figure 2H (“Input”). These two parts (in Figs. 2H and 5H) are assembled again here for the sake of understanding and supporting the model in Figure 9. DBC1, deleted in breast cancer 1; HC, heavy chain; HSP, heat shock protein; PARylation, poly ADP ribosylation; PP2A, protein phosphatase 2A; PyST, polyoma small T antigen.
Because DBC1 binds and inhibits PARP-1 in response to low energy/NAD+ levels (37), we wanted to confirm if the degradation of DBC1 by PyST would lead to enhanced PARP-1 activity, which would be reflected as higher poly ADP ribosylation (PARylation) of the cellular substrates. It has been previously reported that ADP ribosylation by PARP-1 has a very important role in the spindle formation and its stabilization in normally dividing cells (38). While cells are in interphase, PARylation is seen mostly in the cytoplasm; however, during mitosis, PARylation levels are substantially increased and it occurs on the spindle also, including poles, kinetochores, and microtubules. We noticed a similar staining pattern of spindle apparatus in control mitotic cells (−dox), although it was prominent in the cytoplasm also. On the other hand, in PyST expressing cells (+dox) that had predominantly abnormal spindles, PARylation staining was much more concentrated on the spindle as compared to the cytoplasm (Fig. 5, B and C).
To further explore the cellular consequences of PyST-mediated down-regulation of DBC1, we focused on examining the interaction between DBC1 and its binding partners, SIRT1 and PARP-1 and the corresponding effect on their respective activities. We first tested whether PyST expression affected the deacetylase activity of SIRT1. The cell lysates from control and PyST expressing cells were analyzed by Western blotting, using anti-acetyl lysine antibody. The results showed that expression of PyST led to a substantial decrease in the overall cellular substrate acetylation levels, consistent with higher SIRT1 deacetylase activity. This was similar to the deacetylation caused by the coexpression of SIRT1, which was used as a positive control for deacetylation (Fig. 5D). Next, we cotransfected DBC1 and PyST in 293T cells followed by immunoprecipitation of DBC1 using anti-DBC1 antibody. The results showed that DBC1 protein levels were clearly down-regulated in presence of PyST expression, thus resulting in a reduced interaction of DBC1 with SIRT1 and PARP-1 (Fig. 5E), consequently enhancing the respective activities of SIRT1 and PARP-1.
PyST induces energy deficit and activates LKB1
We have also previously reported very high levels of ADP ribosylation of various cellular proteins in PyST expressing cells, which occurs because of extensive cellular stress after DNA/chromosomal damages and very high mitotic index, following PyST expression (3). Consistent with those findings and our immunofluorescence results (Fig. 5, B and C), we observed much higher ADP ribosylation in the PyST expressing cells (Fig. 5F). ADP ribosylation is a high-energy consuming process and occurs during stressful cellular conditions like mitotic arrest. It induces extreme energy deficit by depleting its precursor cellular NAD+ pools and is thus expected to activate the energy sensing machinery. Because LKB1 is the major cellular energy sensor, we therefore expected LKB1 to be activated in PyST expressing cells. Indeed, we observed significantly elevated levels of phosphorylated LKB1 (pS428) in these cells, indicating its enhanced activation (39) (Fig. 5G). In light of our results that PyST interacts with PP2A- and PyST-mediated down-regulation of DBC1 is PP2A dependent (Fig. 2, H and I), we hypothesized that inhibition of PP2A by PyST might be one of the reasons for the activation of LKB1 in PyST expressing cells and that PyST may function as a scaffolding protein, capable of stringing together PP2A, LKB1, HSP40, and DBC1. To test our hypothesis, we transfected 293T cells with PyST-HA-Flag and subjected it to coimmunoprecipitation (Fig. 5H). We found that endogenous LKB1 and HSP70 coimmunoprecipitated with PyST besides DBC1, indicating that all these proteins form a complex in the cells which ultimately translates to the down-regulation of DBC1.
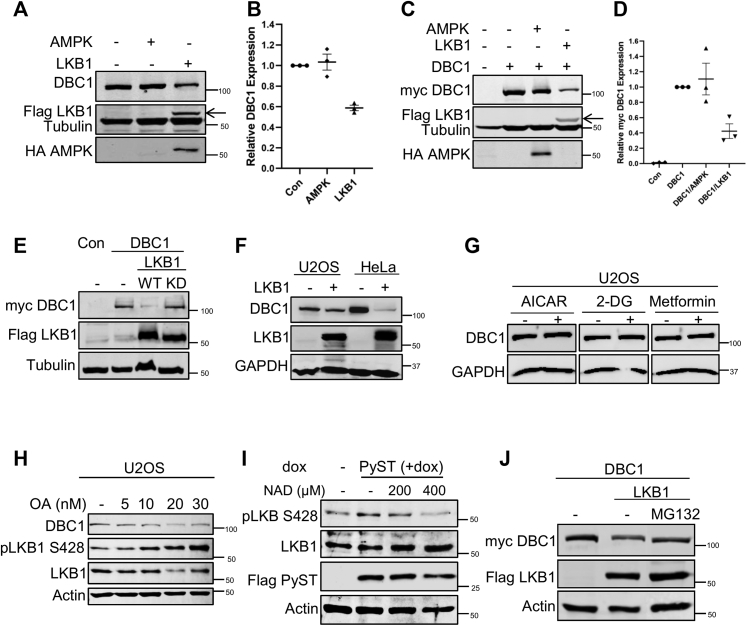
Next, we wanted to test whether LKB1 also has any impact on DBC1 protein levels. Because LKB1 is known to predominantly work through AMPK1 by phosphorylating its catalytic subunit (AMPKα at T172) as its downstream target, we also included AMPKα1 in these assays to check whether the effect of LKB1, if at all, occurs via AMP Kinase. Myc-DBC1 was coexpressed with Flag-LKB1 or HA-AMPK constructs in 293T cell lines, followed by Western blotting. The results showed that LKB1 caused an appreciable down-regulation of endogenous DBC1 (Fig. 6, A and B) and exogenously expressed myc-DBC1 protein levels (Fig. 6, C and D). Interestingly, neither AMPK (Fig. 6, A and C) nor LKB1 kinase dead mutant (Fig. 6E) had any such effect on the protein levels of DBC1.
Figure 6.
LKB1 degrades DBC1 independently of AMPK.A, 293T cells were transiently transfected with HA-AMPK (1.5 μg) and Flag-LKB1 (1.5 μg) constructs, and the cell lysates were blotted using indicated antibodies. C, 293T cells were transfected with HA-AMPK or Flag-LKB1 constructs as in A. In addition, they were also cotransfected with myc DBC1 (2 μg). At 48 h posttransfection, the cells were lysed and protein extracts were analyzed by Western blotting, using indicated antibodies (Flag-LKB1 band is indicated by black arrow). B and D, graphs representing relative DBC1 expression in the indicated control, AMPK, and LKB1 transfected 293T cells. The values indicate mean ± SEM; n = 3 biological replicates. Protein expression in control cells was arbitrarily taken as 1 in all of the Western blotting quantifications. E, DBC1 levels were not decreased by kinase dead (KD) mutant of LKB1. 293T cell lines were transfected with myc-DBC1, Flag-LKB1 (WT), and mutant of Flag-LKB1 (KD) constructs (2 μg each). Western blot of the lysates was performed using anti-myc for DBC1 and anti-Flag antibody for LKB1. F, effect of LKB1 over-expression on DBC1 amounts in stable cell lines. Lysates from U2OS and HeLa cells stably overexpressing LKB1 were blotted for endogenous DBC1 using anti-DBC1 antibody. G, LKB1 mediated down-regulation of DBC1 is independent of AMPK. U2OS cells stably expressing DBC1 were treated with different AMPK activators (AICAR (1 mM), 2-deoxy glucose (25 mM) and metformin (25 mM)) for 12 h. The lysates were blotted for DBC1 using anti-DBC1 antibody. H, U2OS cells were treated with different concentrations of okadaic acid (OA) (for 16 h) to inhibit PP2A. Western blot was carried out with DBC1, pLKB1, and LKB1 antibodies. I, U2OS cells stably expressing PyST (U2OS pTREX-PyST-HA-FLAG) were treated with different concentrations of NAD for 24 h. pLKB1 and LKB1 amounts were determined by Western blotting. J, 293T Cells were transiently transfected with myc-DBC1 (1 μg) and FLAG-LKB1 (1 μg) constructs. The cells were treated with 10 μM of MG132 for 5 h followed by Western blotting of the cell lysates. Proteasome inhibitor MG132 prevented LKB1-induced degradation of DBC1. DBC1, deleted in breast cancer 1; PyST, polyoma small T antigen.
Similar results of LKB1 on DBC1 levels were also obtained in U2OS and HeLa stable cell lines overexpressing Flag-LKB1 (pBABE-puro-Flag-LKB1). We particularly chose HeLa cells as these are known to be genetically LKB1 null cells (−/−), and hence provided a good model for comparing the DBC1 levels with U2OS cells that have WT LKB1 (40). Because HeLa cells do not have LKB1 expression, hence they presumably have relatively higher levels of DBC1 than in the U2OS cells. However, in both U2OS and HeLa cells stably overexpressing Flag-LKB1, DBC1 levels were clearly down-regulated (Fig. 6F), which is consistent with what we had observed in transient transfections (Fig. 6, A and C).
To further confirm that LKB1 degrades DBC1 independently of AMPK, we stimulated cells with various agonists that activate AMPK but not LKB1. The stimulation of cells with AICAR, 2-deoxyglucose, and metformin also did not affect the protein levels of DBC1 (Fig. 6G). These results clearly showed that the regulation of DBC1 by LKB1 is independent of AMPK. Because PyST does most of the functions through the regulation of PP2A and most of the serine-threonine kinases are also regulated by PP2A, we wanted to further investigate the role of PP2A in PyST- and LKB1-mediated down-regulation of DBC1. We treated U2OS cells with different concentrations of OA for 16 h. As shown in Figure 6H (also in Fig. 2J), with increasing concentration of OA, there was a significant decrease in the protein levels of DBC1. This was concomitant with a corresponding increase in activating phosphorylation (S428) of LKB1. To gain further support for the role of energy deficit and LKB1 in PyST-mediated signaling, we treated PyST cell lines with different concentrations of NAD. As expected, by increasing the concentration of NAD in PyST expressing cells, there was a corresponding decrease in the activation of LKB1 (Fig. 6I). All these results thus demonstrated that PyST-mediated inhibition of PP2A results in the constitutive activation of LKB1, which ultimately leads to the down-regulation of DBC1.
We also wanted to find out if the degradation of DBC1 is mediated by the proteasomes, because numerous proteins with a role in cell cycle regulation and signal transduction are degraded by proteasomes. Western blotting results reproducibly showed the protection of DBC1 in the presence of proteasomal inhibitor MG132, even when LKB1 was coexpressed (Fig. 6J).
DBC1 negatively regulates Tribbles3 in PyST expressing cells
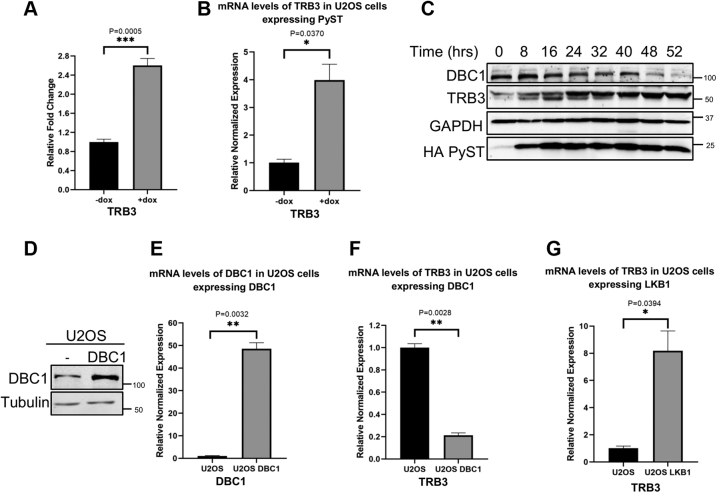
DBC1 is known to positively regulate AKT1 S473 phosphorylation via Tribbles 3 (TRB3) (19). It does so by the transcriptional repression of TRB3 expression. In the absence of growth signals, TRB3 binds and sequesters AKT1 and prevents its phosphorylation and activation (41). Because the expression of PyST causes down-regulation of DBC1, it meant that PyST expressing cells would have higher amounts of TRB3 mRNA and hence reduced the phosphorylation of AKT1 at S473. We had previously performed whole genome microarray analysis on mRNAs obtained from the inducible PyST expressing cells (pTREX-PyST-HA-Flag), using Human Genome U133 Plus 2 Array (4). Microarray data revealed that PyST expressing cells (+dox) had about 2.8 fold increase in TRB3 mRNA levels (Fig. 7A) (NCBI Microarray GEO accession number: GSE149525 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE149525)). RT-qPCR results also confirmed that PyST expressing cells (+dox) had about five-fold increase in TRB3 mRNA (Fig. 7B). Western blotting results in PyST expressing cells also showed an inverse relationship between DBC1 and TRB3 protein levels (Fig. 7C). Consistent with these results, in DBC1 overexpressing stable cell lines (Fig. 7, D and E), the mRNA levels of TRB3 were decreased by about five-fold (Fig. 7F). In further support of our data, RT-qPCR results in U2OS LKB1 cell lines showed that over-expression of LKB1 resulted in about eight-fold increase in the mRNA levels of TRB3 (Fig. 7G).
Figure 7.
LKB1 regulates Tribbles 3 expression through DBC1.A, graphical representation of microarray data from control (−dox) and PyST expressing U2OS cells (+dox) for Tribbles 3 (TRB3). These experiments were carried out as biological triplicates. B, fold change for TRB3 mRNA in PyST expressing cells (+dox, 24 h) as compared to control cells (−dox), as shown by RT-qPCR. The data were normalized by using actin as control. C, PyST increases protein levels of TRB3. pTREX-PyST cells were induced for expression of PyST for different time periods as indicated (0 to 52 h), and cell lysates were then analyzed by Western blotting for DBC1, TRB3, and PyST using antibodies against respective proteins. D and E, confirmation of DBC1 expression in U2OS stable cells at protein and mRNA levels by Western blotting (using anti DBC1 antibody) and RT-qPCR, respectively. F and G, effect of the overexpression of DBC1 and LKB1 respectively, on TRB3 mRNA levels using RT-qPCR. For all these experiments, total cellular RNA was extracted from respective cell lines. After cDNA synthesis, RT-qPCR was carried out using appropriate primers and normalization was carried out using actin as control. The experiments were carried out in duplicates and the gene expression normalized to actin expression and fold change was calculated using ΔΔCq method. p-values were calculated using student’s t test. Shown are ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. The error bars represent mean ± SEM. DBC1, deleted in breast cancer 1; PyST, polyoma small T antigen.
LKB1 negatively regulates AKT1 through DBC1 and Tribbles3
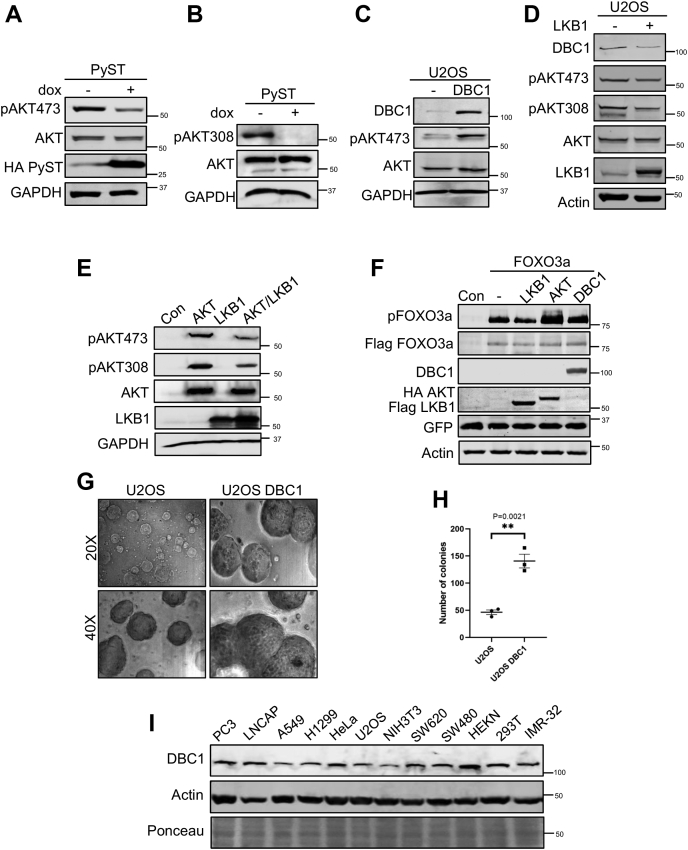
In light of the above results of the impact of PyST and LKB1 on DBC1 and the previous report that DBC1 activates AKT1 by repressing TRB3 (19), we wanted to check if PyST expression has any effect on AKT1 phosphorylation. The expression of PyST (+dox) led to decreased AKT1 phosphorylation at S473 and T308 (Fig. 8, A and B). These results are consistent with our previous report that PyST expression causes decreased AKT1 phosphorylation at S473 residue, but the mechanism was not studied (3). If LKB1, DBC1, and TRB3 were truly connected, then LKB1 and DBC1 would have contrasting impacts on AKT1 phosphorylation. Consistent with this prediction, it was observed that phosphorylation of AKT1 on S473 was substantially higher in DBC1 over-expressing cell line (Fig. 8C). As was expected, LKB1 over-expression caused down-regulation of DBC1, as was already seen in Figure 6, A–F. This was accompanied by a concomitant decrease in AKT1 S473 and T308 phosphorylations in a U2OS cell line stably expressing LKB1 (Fig. 8D). To directly test whether LKB1 affects the phosphorylation status of AKT1 as observed in stable cell lines, we transiently coexpressed AKT1 and LKB1 in 293T cells. The overexpression of LKB1 considerably decreased phosphorylation levels of AKT1 at S473 and T308 residues, thus further confirming the inhibition of AKT1 by LKB1 (Fig. 8E). We then examined whether LKB1 truly affects the activity of AKT1 by over-expressing LKB1 in 293T cells and blotting for forkhead protein (FOXO3a), using AKT1 and DBC1 as positive controls. FOXO1/3 proteins are transcription factors that are well-known to be the direct substrates of AKT1. Our findings showed that the over-expression of LKB1 caused significant decrease in pFOXO3a signals as well (Fig. 8F), in response to the inhibition of AKT1 by LKB1.
Figure 8.
LKB1 regulates AKT1 via DBC1 and TRB3.A and B, PyST inhibits phosphorylation of AKT1. Lysates from PyST-U2OS cell lines were blotted with antibodies specific for phosphorylation of S473 and T308 residues on AKT1. The membrane was stripped and blotted for total AKT protein levels. C, effect of DBC1 overexpression on AKT1 phosphorylation. Protein lysates from U2OS cells stably expressing DBC1 were blotted for AKT1 S473 phosphorylation. D, effect of LKB1 overexpression on AKT S473 and T308 phosphorylation levels in U2OS cell lines stably expressing LKB1. Proteins were detected with the indicated antibodies. E, 293T cells were transiently transfected with AKT1, LKB1, and AKT1/LKB1 constructs (1.5 μg each). pAKT 473, pAKT 308, total AKT, and LKB1 were detected in Western blotting using respective antibodies. F, 293T cells were transiently transfected with FLAG-FOXO3a (2 μg) alone or together with AKT1, LKB1, or DBC1 constructs (1.5 μg each). Cell lysates were blotted for pFOXO3a and total FOXO3a using indicated antibodies. Other transfected proteins like myc-DBC1, HA-AKT1, and Flag-LKB1 were also detected using indicated antibodies. GFP was used as transfection control and was detected with anti-GFP antibody. G, soft agar assay for U2OS cells stably over-expressing DBC1 was done in triplicates. Pictures of colonies as seen in the figure were taken after 3 weeks of growth. H, quantitative representation of the soft-agar assay. After 4 weeks of growth, the colonies were stained with iodonitrotetrazolium for overnight. After staining, colonies of 2 mm size or bigger were counted, and the data was analyzed by GraphPad Prism 8 (∗∗p < 0.01). I, Western blot showing the expression of DBC1 in different cancer cell lines as indicated. DBC1, deleted in breast cancer 1; PyST, polyoma small T antigen.
DBC1 enhances tumorigenesis via positive regulation of AKT1
AKT1 is a major survival kinase and a known proto-oncogene in cells. Based on our results on AKT1 activation, DBC1 is expected to be a positive regulator of cellular proliferation. To validate the role of DBC1 in tumorigenesis, we checked the ability of DBC1 over-expressing cells to form colonies in soft agar, a gold standard for testing the transforming property of cells. U2OS being a cancerous cell line was expected to form colonies in soft agar. Expectedly, our results showed that U2OS-DBC1 cells formed colonies that were larger in both size and number as compared to the U2OS cells (Fig. 8, G and H). Because there is usually a good correlation between in vitro transformation and in vivo carcinogenesis, these results implicated that DBC1 promotes tumorigenesis. If DBC1 has oncogenic potential, then its expression is expected to be higher in at least some cancer cell lines. As shown in Figure 8I, the expression level of DBC1 was seen to be higher in several cancer cell lines, particularly in prostrate (LnCAP and PC3), HeLa, and colorectal cancer cell lines (SW620 and SW480). Further support for the role of DBC1 in promoting tumorigenesis came from the patient tumor samples. In a pilot study, we observed that three out of four colorectal tumor samples had several-fold increase in the DBC1 protein levels as compared to the control (nontumor) samples from the adjacent tissues of the same patients (data not shown).
Discussion
Using immunoprecipitation and mass spectrometry approaches on the cell lysates of PyST expressing cells, we identified DBC1/CCAR2 as a novel binding partner of PyST. Our results also showed that PyST binds and leads to DBC1 degradation in a PP2A-dependent manner (Fig. 2, H and I), though there may be some contribution by a few amino acid residues from the HSP-binding J domain region as well, as is the case with SV40 small T antigen (42, 43). This is because, as for PyST, DBC1 is also known to bind to both PP2A (A and B subunits) and to HSP70/HSA8 (44, 45, 46). Using immunoprecipitation, we have also shown that PyST binds both DBC1 and PP2A (A and C subunits) (Fig. 2H). The interactions of DBC1 with various cellular proteins are also curated on various protein–protein interaction portals like Biogrid (https://thebiogrid.org), String (https://string-db.org), and HuRI (http://www.interactome-atlas.org) etc.
Our results also showed that DBC1 promotes the progression of cells through the M-phase during genotoxic conditions like mitotic arrest as induced by PyST expression. Thus, it is facilitating mitotic progression under such adverse conditions. The overexpression of DBC1 in PyST expressing cells decreased the percentage of cells in mitosis and prevented or at least substantially delayed mitotic catastrophe (Fig. 3). So, what is the role of DBC1 in mitosis? As the cells progress into mitosis (prophase and prometaphase), it localizes to the spindle poles, probably at the centrosomes. In anaphase and telophase, it is seen at the spindle poles as well as the central spindle region, followed by prominent localization at the mid-body during cytokinesis (Fig. 4). These findings are not entirely surprising because Numa1, a protein that is predominantly found at the spindle poles and has an important role in spindle organization and maintenance (47, 48), was also detected in the PyST–DBC1 interactome (Table S1). Using immunofluorescence microscopy, we have also seen that both DBC1 and Numa1 are present at the spindle poles (data not shown). Also, in several high throughput interactome analyses, DBC1 has been already reported to interact with CEP170 (44, 45, 49), a microtubule-binding protein present in the centrioles that also has a role in mitotic spindle formation (50). Further studies, however, need to be undertaken to confirm these interactions and study their detailed mechanistic role in the regulation of spindle dynamics.
Another role of DBC1 is to regulate the activity of PARP-1. Interestingly, the latter also plays a role in mitosis by PARylating numerous substrates including the spindle apparatus, leading to its stabilization (38). We showed that the extent of PARylation was much higher in PyST expressing cells, which is required to fix the extensive damage during mitotic arrest after PyST expression. So, DBC1 must function as a rheostat to regulate the extent of PARP-1 activation. Using immunoprecipitation experiments, we showed that PyST reduces the interaction of DBC1 with SIRT1 and with PARP-1 (Fig. 5E). This occurs because expression of PyST leads to the degradation of DBC1.
We also showed that the expression of small T causes severe energy deficit in the host mammalian cells, which activates LKB1, as was seen by enhanced LKB1 phosphorylation at S428 residue (Fig. 5G). So, what is the mechanism of LKB1 activation in PyST expressing cells? We have previously shown that PyST expression promotes abnormally high levels of ADP ribosylation (3) (Fig. 5F). This would lead to severe energy depletion, leading to LKB1 activation, and consequently promoting degradation of DBC1, directly or indirectly. The enhanced level of phosphorylation and activation of LKB1 can also be explained by the interaction of PyST and PP2A. PyST is known to bind to the A and C subunits of PP2A, displacing the regulatory B subunit, and hence forming the PP2A-A-C-PyST complex. This PP2A complex has altered phosphatase activity toward numerous substrates including several serine-threonine kinases like p38 and pJNK (51, 52). We have also reported higher phosphorylated amounts of these kinases in PyST expressing cells (3). Using immunoprecipitation experiments, we also showed that besides binding DBC1, PyST binds HSP70 and LKB1 as well (Fig. 5H). This is not entirely unexpected as both PyST (Fig. 1B and Table S1) and LKB1 are known to bind HSPs (HSP70/90) (45, 53). It is therefore quite possible that PyST may bind LKB1, and at the same time prevent its dephosphorylation by precluding the binding of an appropriate B subunit to the PP2A A/C complex, and hence the assembly of a functional PP2A holoenzyme that may be needed for LKB1 dephosphorylation. This may lead to a constitutive state of LKB1 phosphorylation and hence activation. This is illustrated in the proposed model (Fig. 9).
Figure 9.
This proposed model summarizes how PyST affects the mitotic progression and cell survival of host cells. PyST probably works like a scaffold that brings together many proteins and leads to their altered functions within the cell. As a result of PyST expression, LKB1 activation, and hence DBC1 degradation, spindle may not be properly formed. DBC1 degradation also leads to the transcriptional activation of TRB3. TRB3 in turn binds and sequesters AKT, preventing its phosphorylation and activation, thus inhibiting the survival signal, leading to apoptosis. DBC1, deleted in breast cancer 1; TRB3, Tribbles 3; PyST, polyoma small T antigen.
Our data identifies LKB1 as a negative regulator of DBC1, as the latter is degraded in the presence of LKB1 expression. Although most of the effects of LKB1 are known to occur via AMPK, our study reports that LKB1 regulates DBC1 independently of AMP kinase (Fig. 6, A–D and G). It has been previously reported that LKB1 plays a role in mitotic regulation in hematopoietic cells independently of AMP kinase. These cells from LKB1−/− mice showed various abnormalities like supernumerary centrosomes, abnormal spindles, and aneuploidy (54, 55). Our findings are thus consistent with these reports. We believe that LKB1 may either phosphorylate DBC1 directly or indirectly through some other kinase, amongt its down-stream set of kinase targets, other than AMPK, which needs to be investigated in the future studies.
In an interesting study, it has been recently reported that LKB1 phosphorylates SIRT1, which leads to the disruption of interaction between DBC1 and SIRT1, thus activating SIRT1 activity (56). In this study, we report that LKB1 degrades DBC1, probably by phosphorylation at some ser/thr site(s). It is quite possible that LKB1 may phosphorylate both these proteins, DBC1 and SIRT1. By phosphorylating DBC1, LKB1 leads to its degradation, whereas by phosphorylating SIRT1, it leads to its (SIRT1) activation. The resultant of both these phosphorylations on the two substrates (DBC1 and SIRT1) ultimately is the cumulative activation of SIRT1.
The role of DBC1 in human cancer has been largely paradoxical. It has been previously reported that DBC1 has a tumor suppressor activity by stabilizing p53 through the displacement of MDM2 (20) and also through the inhibition of SIRT1, a deacetylase. Using immunoprecipitation (Fig. 5E) and immunofluorescence (data not shown), we also confirmed that DBC1 interacts with SIRT1 in mammalian cells. By inhibiting SIRT1 and hence preventing the deacetylation of p53 and FOXO transcriptional factors, DBC1 expression leads to their stabilization and activation, thereby promoting their tumor suppressor activity. A recent study has shown that DBC1 negatively regulates the activity of CBP–MDM2 complex, which prevents the ubiquitination and degradation of p53 (21). In contrast, some other studies have reported that DBC1 has a tumor promoting role (15, 17, 18, 26, 34, 57, 58, 59, 60, 61, 62). We also believe that DBC1 must have an oncogenic potential based on the following reasons: (1) DBC1 helps cells in the progression of cells through mitosis, particularly under adverse conditions like mitotic arrest, probably through the correction of abnormalities. (2) DBC1 has a role in the activation of AKT1, as has been previously reported (19), and was also shown in the current study. AKT1 has a well-known role in cell survival and tumorigenesis. (3) DBC1 was seen to further promote colony formation in soft agar in U2OS cells, a property of transformed cells. (4) DBC1 protein levels are higher in several cancer cell lines (Fig. 8H) and in tumor samples (data not shown). It is also possible that it may have both these apparently contradictory roles, which may be context or cell-line specific property of this gene. Further studies need to be carried out to get a deeper insight into these functions of DBC1.
In conclusion, we propose a mechanism by which PyST may serve as a scaffold that brings together HSPs, DBC1, LKB1, and PP2A (A/C) complex. By activating LKB1’s phosphorylation and hence kinase activity due to PARylation-induced energy deficit, coupled with constitutive phosphorylation of LKB1 due to the simultaneous inhibition of PP2A, PyST promotes the degradation of DBC1. This would release SIRT1 and PARP-1 from DBC1 interaction and hence stimulate their enzymatic activities, thus resulting in higher PARylation and energy deficit, as well as enhanced deacetylation of substrates by SIRT1. Because the mitotic arrest by PyST occurs for a prolonged time period, this would lead to the depletion of energy levels below a threshold level and hence initiation of apoptosis in these cells. The proposed model is explained in Figure 9.
Nevertheless, some issues remain to be addressed. We showed that LKB1 down-regulates DBC1 but the actual mechanism is not well understood. Do LKB1 and DBC1 interact directly with each other or is there any other protein involved? Another question that requires further investigation is whether LKB1–DBC1 signaling pathway is mandatory for PyST mediated apoptosis or is it just one of the many pathways that are activated by PyST. Although the data we present implies that DBC1 has an important role in mitosis, particularly at the spindle, but the exact role of DBC1 in mitotic regulation needs further investigation.
Experimental procedures
Materials
The primers for cloning and RT qPCR were purchased from Integrated DNA Technologies. Kits for cDNA synthesis and RT qPCR were purchased from BioRad (iScriptcDNA Synthesis Kit and Sso Advanced Universal SYBR Green Supermix). Cell culture reagents like Dulbecco’s modified Eagle’s medium (DMEM), FCS, and antibiotics were purchased from Invitrogen/Thermofisher Scientific.
Antibody details
Primary antibodies used were anti-DBC1 (5693 and 5857; CST; 1:1000 dilution), anti-FLAG (F3165; Sigma: 1:5000 dilution), anti-FLAG (14793; CST; 1:5000 dilution), anti-HA (3724; CST; 1:5000 dilution), anti-LKB1 (CST; 3047; 1:1000 dilution), anti-LKB1 (AHO1392; Invitrogen; 1:1000 dilution), anti-AKT (2920; CST; 1:1000 dilution), anti-LKB1 S428 (3482; CST; 1:1000 dilution), anti-AKT 473 (4060; CST; 1:1000 dilution), anti-AKT 308 (13038; CST; 1:1000 dilution), anti-myc (CST; 2276; 1:10,000 dilution), anti-Vinculin (CAB839Hu22; Cloudclone; 1:1000 dilution), anti-Actin (3700; CST; 1:10,000 dilution), anti-Tubulin (236-10501; Invitrogen; 1:5000 dilution), anti-HSP70 (4872; CST; 1:1000 dilution), anti-GAPDH (CST; 2118; 1:10,000 dilution), anti-PP2A C (2038; CST; 1:1000 dilution), anti-pAurora A, B, C (2914; CST; 1:1000 dilution), anti-pH3 (53348; CST; 1:1000 dilution), anti-Bub1 (MAB-3610; Chemicon; 1:1000 dilution), anti-Cyclin B1 (12231; CST; 1:1000 dilution), anti-SIRT1 (2493; CST; 1:1000 dilution), anti-PARP (46D11; CST; 1:1000 dilution), anti-PAR (4335-MC-100; Trevigen; 1:1000 dilution), anti-TRB3 (T8076; Sigma; 1:1000 dilution), anti-pFOXO3a (2599; CST; 1:1000 dilution), and pAMPK (2535; CST; 1:1000 dilution).
Cloning
myc-tagged DBC1 (myc-DBC1) cloned in pcDNA3 was obtained from Addgene. For making stable cell lines, DBC1 was cloned in retroviral vector pWZL-Blast using EcoR I and Xho I sites, which were compatible between the two vectors.
Transient transfections
For transient transfections, PEI method was routinely used. Briefly, 293T cells were grown at a confluence of 60 to 70% in 60 mm plates. A total of not more than 5 μg of the DNA was mixed with 400 μl of DMEM and then 15 μl of PEI (stock concentration of 1 mg/ml) was added dropwise. The mixture was incubated for 15 to 20 min and was then added slowly to the plates and incubated overnight. Sixteen hours post transfection, the plates were washed with PBS, and then supplemented with fresh DMEM containing 10% fetal calf serum and 1% penicillin-streptomycin. Forty eight hours post transfection, the cells were harvested, and the extracts were used for further experiments.
Retroviral/lentiviral transductions
Phoenix cells stably expressing packaging proteins were transiently transfected with pWZL-Blast-DBC1 using PEI as a transfecting reagent. Briefly, 3 μg pWZl-DBC1, 1 μg gag-pol, and 1 μg of VSVG were mixed in 400 μl of DMEM, and 15 μl of PEI was added dropwise. The transfection mixtures were then added to cells grown in 60-mm plates at a confluence of about 60 to 80%. Next day, the cells were washed with (PBS), and fresh DMEM was added. After 48 h, the cells that were split a day before were infected with the viral supernatants obtained from the above transfected cells and supplemented with 8 μg of polybrene/ml (Sigma) for at least 6 h. This process was repeated the next day also. For selection of DBC1-expressing cells, the cells were grown in the presence of blasticidin (10 μg/ml). For making double stable cell lines for PyST and DBC1, U2OS cells stably expressing pTREX-PyST-HA-Flag were infected with viral titer generated by using pWZL-blast-DBC1. Cells were then selected using puromycin (5 μg/ml) for PyST and blasticidin for DBC1 (10 μg/ml). Similarly, for making stable LKB1 overexpressing cell lines, viral titer was generated in Phoenix cells using pBABE-puro-Flag-LKB1. U2OS and HeLa cells were infected with the viral supernatant and the cells were selected using puromycin (5 μg/ml for U2OS and 2 μg/ml for HeLa).
Western blotting
Cells were washed with ice cold PBS, centrifuged, and suspended in lysis buffer [Tris-Cl 50 mM, NaCl 150 mM, Glycerol 10%, SDS 0.1%, and NP-40 1%, EDTA 2 mM, supplemented with protease and phosphatase inhibitors (Complete, Roche)] for about 30 min on ice. Equal amounts of protein were loaded, as estimated by Bradford assay (Bio-Rad Laboratories). The blots were then detected using 481 LICOR Odyssey (for Infra-red detection) or Chemidoc MP (Bio-Rad laboratories, for chemiluminescence). Quantifications for all Western blots were carried out by densitometric analysis using Image J software. Normalization was performed to an internal loading control or to a coexpressing protein as per the requirements of the experiment. The data was analyzed using GraphPad Prism software. All the Western blots were reproduced three times as biological replicates.
Real time quantitative PCR (RT-qPCR)
For RT-qPCR, the cells were grown in 60 mm dishes. RNA was isolated by using Trizol method (Invitrogen). cDNA synthesis was carried out using Bio-Rad iScript cDNA Synthesis Kit. Bio-Rad Sso Advanced Universal SYBR Green Supermix was used for RT-qPCR. The following primer pairs were used for RT-qPCR: DBC1 5′-AAGGTGCAAACGCTCTCCAACCAG -3′ and 3′-GGATGTTTGGAAGAGACTCAGAG -5′, Actin 5′- TTAGTTGCGTTACACCCTTTC-3′ and 3′- ACC TTC ACC GTT CCA GTT T -5′, and TRB3 5′-TGCCCTACAGGCACTGAGTA-3′ and 3′-GTCCGAGTGAAAAAGGCGTA-5′, LKB1 5′-ACTGAGGAGGTTACGGCACA-3′ and 3′-CCTGGCACACTGGGAAAC-3′.
RT qPCR data processing
Base line correction and threshold setting were performed using automatic calculation of Bio-Rad CFX Manager Software 3.1. Relative quantities of all the samples were assessed by taking their validated efficiency into account and by assuming 100% overall efficiency. Fold change was calculated using ΔΔCq method. p-values were calculated using student’s t test, graphs were plotted in GraphPad Prism 7.0, and the error bars represent mean ± SEM.
Cell cycle analysis by fluorescence-activated cell sorting
U2OS cells stably expressing DBC1 and polyoma small T were grown to a confluence of 60 to 70% in 60 mm plates. The cells were washed with PBS/EDTA (0.1%) and then incubated at 37 °C for 5 min in PBS/EDTA, scraped from the plate, and collected in 15 ml falcon tubes. The cells were pelleted at 1000g for 5 min and washed once with PBS/serum (1%). Finally, the cells were fixed by adding 10 ml of cold ethanol while vortexing to prevent clumping. The fixed cells were stored at 4 °C at least overnight in the dark. Before acquisition on Flow Cytometer, the cells were centrifuged at 1000g for 5 min and then washed once with PBS/serum (1%). Pellets were resuspended in 500 μl of propidium iodide-RNase solution (50 μg/ml propidium iodide, 10 mM Tris, pH 7.5, 5 mM MgCl2, and 20 μg/ml RNase A) and incubated at 37 °C for 30 min. The samples were then acquired by using BD FACSverse and analyzed by BD FACSverse software. Statistical significance was calculated by using GraphPad Prism 8.0 software.
Immunoprecipitation
293T cells were grown in 60 mm plates, washed once with cold PBS, harvested, and lysed in 100 μl of lysis buffer in presence of protease and phosphatase inhibitors for 30 min on ice. The extracts were spun down at 10,000 rpm for 10 min. 20 μl of IgG sepharose beads were premixed with 5 μl of antibodies for 30 min to 1 h. About 20 μl of the lysate was aliquoted for the whole cell lysate blot. Equal amounts of protein as determined by Bradford assay were then added to the beads-antibody mixture and were kept on rotator overnight at 4 °C. The mixtures were then washed twice with cold PBS followed by a final wash with cold water. The beads were mixed with 2× sample buffer, boiled, and the proteins were analyzed by Western blotting.
Microarray
Total cellular RNA was isolated from pTREX-PyST cells (in triplicates) at about 20 h of doxycycline addition using Trizol method (Invitrogen). RNA was purified using RNAeasy kit (Qiagen) as per manufacturer’s specifications and used for microarray analysis, using Affymetrix Human Genome U133 Plus 2 Array chips. Microarray experiments were performed at DFCI, Harvard Medical School microarray core facility. Gene expression changes with >log 2-fold changes were considered significant (4).
Mass spectrometry analysis of TAP samples
Purified protein complexes were analyzed by mass spectrometry (LC/MS-MS), as described (63) with minor modifications. Proteins from the TAP samples were directly processed in solution. Cysteine residues were reduced with 10 mM DTT for 30 min at 56 °C in presence of 0.1% RapiGest SF (Waters). The proteins were digested overnight at 37 °C using 5 μg of trypsin after adjusting the pH to 8.0 with Tris buffer. Cys-HA peptide was removed from the TAP samples by incubating the digests with 20 μl of a 50% slurry of Activated Thiol Sepharose 4B (GE healthcare) for 30 min at room temperature. The HA peptide-depleted solutions were acidified by adding TFA to a final concentration of 1%, desalted by batch-mode reverse phase (RP, Poros 10R2, Applied Biosystems) solid phase extraction, and concentrated in a vacuum concentrator. The peptides were solubilized in 0.1% formic acid containing 25% Acetonitrile and further purified by Strong Cation Exchange (SCX Poros 10HS, Applied Biosystems). The peptides were sequentially eluted with 0.1% Formic Acid containing 25% Acetonitrile and the following concentrations of KCl: 10 mM, 30 mM, 50 mM, 100 mM, 150 mM, 250 mM, and 300 mM. Fractionated peptides were concentrated in a vacuum concentrator and reconstituted with 20 μl of 0.1% TFA. Peptides from each fraction were loaded onto a precolumn (4 cm POROS 10R2, Applied Biosystems) and eluted with an HPLC gradient (NanoAcquity UPLC system, Waters; 2%–35% B in 45 min; A = 0.2 M acetic acid in water, B = 0.2 M acetic acid in acetonitrile). The peptides were resolved on a self-packed analytical column (12 cm Monitor C18, Column Engineering) and introduced in the mass spectrometer (LTQ-Orbitrap-XL mass spectrometer, Thermo) equipped with a Digital PicoView electrospray source platform (64) (New Objective). The spectrometer was operated in data-dependent mode where the top eight most abundant ions in each MS scan were subjected to CAD (35% normalized collision energy) and subjected to MS2 scans (isolation width = 2.8 Da, threshold = 20,000). Dynamic exclusion was enabled with a repeat count of one and exclusion duration of 30 s. ESI voltage was set to 2.2 kV.
MS spectra were recalibrated using the background ion (Si(CH3)2O)6 at m/z 445.12 ± 0.03 and converted into a Mascot generic file format (.mgf) using multiplierz scripts (64, 65). The spectra were searched using Mascot (version 2.6) against two databases consisting of human protein sequences (downloaded from Uniprot on 09/27/2017 containing 21,082 protein entries) and common lab contaminants, with automatic Mascot Decoy search enabled. Precursor tolerance was set to 15 ppm and product ion tolerance to 0.6 Da. The search parameters included trypsin specificity, up to two missed cleavages, fixed carbamido-methylation (C, +57 Da), and variable oxidation (M, +16 Da). The spectra matching to peptides from the reverse database were used to calculate a global false discovery rate and were discarded. The data were further processed to remove peptide spectral matches to the forward database with an FDR greater than 1.0%. A fast peptide matching algorithm was used to map peptide sequences to all possible human genes in the search database (65). Peptides shared by two or more genes were excluded from consideration when constructing the final protein list. Any protein identified in more than 1% of 108 negative TAP controls (66) were removed from the sets of interactors. Proteins identified in the control TAP experiments were also removed.
Immunofluorescence
U2OS and HeLa cells were grown on coverslips up to 50% confluence. The cells were washed with chilled PBS and fixed with 4% paraformaldehyde in PBS. The cells were permeabilized with 0.15 v/v% Triton-X 100 for 3 min. Nonspecific sites were blocked with 2% bovine serum albumin (Sigma-Aldrich) in PBS for 45 min. Fixed cells were stained with the following antibodies (for 3 h at 37 °C): β-Tubulin (1:100, Sigma-Aldrich), DBC1 (1:100, Cell signaling), and HA (1:100, Invitrogen). Primary antibodies were visualized by secondary antibodies conjugated to Alexa fluor 488 and Alexa fluor 594 (1:5000, Jackson ImmunoResearch) at 37 °C for 1 h. The cells were stained with DAPI (BioRad) for 10 min. Pictures were taken with an inverted fluorescence microscope (Axio Observer, Colibri7, Carl Zeiss MicroImaging, Inc) using filters for DAPI, GFP and mRF12, X20, X63, and DIC Apotome.2 oil objectives. The pictures were generated with ZEN 2.3 pro (Zeiss).
Statistical analysis
All data are representative of atleast three independent experiments unless otherwise mentioned in the figure legends. Statistical difference between the control and the experimental groups was analyzed by student’s t test in GraphPad Prism 7.0. The error bars indicate mean ± SEM.
Densitometric analysis
Densitometric analysis of Western blots were performed, as described previously using ImageJ© software developed by the National Institutes of Health.
Software used for making model figure
We want to mention that the proposed model Figure 8 was generated with the help of Biorender software (https://biorender.com).
Data availability
Most of the data that was generated in this study is already incorporated in the article. The mass spectrometry data is currently available on ftp://massive.ucsd.edu/MSV000087239/. The microarray data is already available at NCBI Microarray GEO accession number: GSE149525 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE149525). If any further details are needed, please feel free to contact Dr Shaida Andrabi, University of Kashmir through email at shaida.andrabi@uok.edu.in (corresponding author).
Supporting information
This article contains supporting information.
Conflict of interest
J. A. M. serves on the SAB of 908 Devices and receives sponsored research support from Astra Zeneca, Vertex, and Taiho Pharmaceuticals.
Acknowledgments
We acknowledge the support of DST-FIST grants to the Department of Biochemistry, University of Kashmir. We thank Prof. Shakil Wani, Dr Zahid Kashoo, and their colleagues for allowing us to carry out the FACS analysis at SKUAST-K campus, Shuhama, Srinagar. We also want to thank Prof. Brian Schaffhausen (Tufts University, USA), Prof. Thomas Roberts (DFCI, USA), and Dr Ole Gjoerup (DFCI, USA) for reviewing this article and providing valuable criticisms.
Author contributions
Z. S., N. N., S. A. B., S. Q. G., I. R., M. U. N., G. A., J. A. M., and S. A. methodology; Z. S., G. A., J. A. M., and S. A. data curation; Z. S., S. A. B., and S. A. writing–original draft; Z. S., N. N., S. A. B., S. Q. G., I. R., M. U. N, G. A., J. A. M., and S. A. investigation; Z. S., N. N., S. A. B., S. Q. G., I. R., M. U. N, G. A., J. A. M., and S. A. formal analysis; Z. S., S. A. B., and S. A. software; Z. S., N. N., S. A. B., S. Q. G., I. R., M. U. N., G. A., J. A. M., and S. A. validation; Z. S. and S. Q. G. visualization; I. R. and S. A. supervision; J. A. M. and S. A. writing–review and editing; S. A. conceptualization; S. A. resources; S. A. funding acquisition; S. A. project administration.
Funding and additional information
S. A. was supported by the Department of Biotechnology (DBT), India through the following grants: Ramalingaswami Fellowship, BT/PR5743/BRB/10/1100/2012, BT/PR13605/MED/30/1525/2015, and BT/PR2283/AGR/36/692/2011. The Department of Biochemistry was also supported through the FIST grant (SR/FST/LS-I/2017-3(C) by the Department of Science and Technology (DST)-India. We also thank DST, India for providing INSPIRE fellowships to Z. S., S. A. B., S. Q. G. and also the CSIR-UGC for providing fellowship to I. R., N. N., and M. U. N.
Edited by Alex Toker
Supporting information
References
- 1.Andrabi S., Hwang J.H., Choe J.K., Roberts T.M., Schaffhausen B.S. Comparisons between murine polyomavirus and Simian virus 40 show significant differences in small T antigen function. J. Virol. 2011;85:10649–10658. doi: 10.1128/JVI.05034-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pores Fernando A.T., Andrabi S., Cizmecioglu O., Zhu C., Livingston D.M., Higgins J.M.G., Schaffhausen B.S., Roberts T.M. Polyoma small T antigen triggers cell death via mitotic catastrophe. Oncogene. 2015;34:2483–2492. doi: 10.1038/onc.2014.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrabi S., Gjoerup O.V., Kean J.A., Roberts T.M., Schaffhausen B. Protein phosphatase 2A regulates life and death decisions via Akt in a context-dependent manner. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19011–19016. doi: 10.1073/pnas.0706696104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhat S.A., Sarwar Z., Gillani Q., Un Nisa M., Reshi I., Nabi N., Xie S., Fazili K.M., Roberts T.M., Andrabi S. Polyomavirus small T antigen induces apoptosis in mammalian cells through the UNC5B pathway in a PP2A-dependent manner. J. Virol. 2020;94 doi: 10.1128/JVI.02187-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joshi P., Quach O.L., Giguere S.S.B., Cristea I.M. A functional proteomics perspective of DBC1 as a regulator of transcription. J. Proteomics Bioinform. 2013;Suppl 2:002. [PMC free article] [PubMed] [Google Scholar]
- 6.Close P., East P., Dirac-Svejstrup A.B., Hartmann H., Heron M., Maslen S., Chariot A., Soding J., Skehel M., Svejstrup J.Q. DBIRD complex integrates alternative mRNA splicing with RNA polymerase II transcript elongation. Nature. 2012;484:386–389. doi: 10.1038/nature10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sundararajan R., Chen G., Mukherjee C., White E. Caspase-dependent processing activates the proapoptotic activity of deleted in breast cancer-1 during tumor necrosis factor-alpha-mediated death signaling. Oncogene. 2005;24:4908–4920. doi: 10.1038/sj.onc.1208681. [DOI] [PubMed] [Google Scholar]
- 8.Kim W., Kim J.-E. Deleted in breast cancer 1 (DBC1) deficiency results in apoptosis of breast cancer cells through impaired responses to UV-induced DNA damage. Cancer Lett. 2013;333:180–186. doi: 10.1016/j.canlet.2013.01.026. [DOI] [PubMed] [Google Scholar]
- 9.Chini E.N., Chini C.C.S., Nin V., Escande C. Deleted in breast cancer-1 (DBC-1) in the interface between metabolism, aging and cancer. Biosci. Rep. 2013;33 doi: 10.1042/BSR20130062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basu S., Barad M., Yadav D., Nandy A., Mukherjee B., Sarkar J., Chakrabarti P., Mukhopadhyay S., Biswas D. DBC1, p300, HDAC3, and Siah1 coordinately regulate ELL stability and function for expression of its target genes. Proc. Natl. Acad. Sci. U. S. A. 2020;117:6509–6520. doi: 10.1073/pnas.1912375117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giguere S.S.B., Guise A.J., Jean Beltran P.M., Joshi P.M., Greco T.M., Quach O.L., Kong J., Cristea I.M. The proteomic profile of deleted in breast cancer 1 (DBC1) interactions points to a multifaceted regulation of gene expression. Mol. Cell. Proteomics. 2016;15:791–809. doi: 10.1074/mcp.M115.054619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J.-E., Chen J., Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 13.Chini C.C.S., Escande C., Nin V., Chini E.N. HDAC3 is negatively regulated by the nuclear protein DBC1. J. Biol. Chem. 2010;285:40830–40837. doi: 10.1074/jbc.M110.153270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z., Chen L., Kabra N., Wang C., Fang J., Chen J. Inhibition of SUV39H1 methyltransferase activity by DBC1. J. Biol. Chem. 2009;284:10361–10366. doi: 10.1074/jbc.M900956200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J.-E., Chen J., Lou Z. p30 DBC is a potential regulator of tumorigenesis. Cell Cycle. 2009;8:2932–2935. doi: 10.4161/cc.8.18.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y., Gu Y., Sha S., Kong X., Zhu H., Xu B., Li Y., Wu K. DBC1 is over-expressed and associated with poor prognosis in colorectal cancer. Int. J. Clin. Oncol. 2014;19:106–112. doi: 10.1007/s10147-012-0506-5. [DOI] [PubMed] [Google Scholar]
- 17.Kim S.-H., Kim J.H., Yu E.J., Lee K.-W., Park C.K. The overexpression of DBC1 in esophageal squamous cell carcinoma correlates with poor prognosis. Histol. Histopathol. 2012;27:49–58. doi: 10.14670/HH-27.49. [DOI] [PubMed] [Google Scholar]
- 18.Hiraike H., Wada-Hiraike O., Nakagawa S., Koyama S., Miyamoto Y., Sone K., Tanikawa M., Tsuruga T., Nagasaka K., Matsumoto Y., Oda K., Shoji K., Fukuhara H., Saji S., Nakagawa K., et al. Identification of DBC1 as a transcriptional repressor for BRCA1. Br. J. Cancer. 2010;102:1061–1067. doi: 10.1038/sj.bjc.6605577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Restelli M., Magni M., Ruscica V., Pinciroli P., De Cecco L., Buscemi G., Delia D., Zannini L. A novel crosstalk between CCAR2 and AKT pathway in the regulation of cancer cell proliferation. Cell Death Dis. 2016;7 doi: 10.1038/cddis.2016.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin B., Minter-Dykhouse K., Yu J., Zhang J., Liu T., Zhang H., Lee S., Kim J., Wang L., Lou Z. DBC1 functions as a tumor suppressor by regulating p53 stability. Cell Rep. 2015;10:1324–1334. doi: 10.1016/j.celrep.2015.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akande O.E., Damle P.K., Pop M., Sherman N.E., Szomju B.B., Litovchick L.V., Grossman S.R. DBC1 regulates p53 stability via inhibition of CBP-dependent p53 polyubiquitination. Cell Rep. 2019;26:3323–3335.e4. doi: 10.1016/j.celrep.2019.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X., Sun K., Jiao S., Cai N., Zhao X., Zou H., Xie Y., Wang Z., Zhong M., Wei L. High levels of SIRT1 expression enhance tumorigenesis and associate with a poor prognosis of colorectal carcinoma patients. Sci. Rep. 2014;4:7481. doi: 10.1038/srep07481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elangovan S., Ramachandran S., Venkatesan N., Ananth S., Gnana-Prakasam J.P., Martin P.M., Browning D.D., Schoenlein P.V., Prasad P.D., Ganapathy V., Thangaraju M. SIRT1 is essential for oncogenic signaling by estrogen/estrogen receptor alpha in breast cancer. Cancer Res. 2011;71:6654–6664. doi: 10.1158/0008-5472.CAN-11-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huffman D.M., Grizzle W.E., Bamman M.M., Kim J., Eltoum I.A., Elgavish A., Nagy T.R. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67:6612–6618. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- 25.Menssen A., Hydbring P., Kapelle K., Vervoorts J., Diebold J., Luscher B., Larsson L.-G., Hermeking H. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E187–E196. doi: 10.1073/pnas.1105304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pangon L., Mladenova D., Watkins L., Van Kralingen C., Currey N., Al-Sohaily S., Lecine P., Borg J.-P., Kohonen-Corish M.R.J. MCC inhibits beta-catenin transcriptional activity by sequestering DBC1 in the cytoplasm. Int. J. Cancer. 2015;136:55–64. doi: 10.1002/ijc.28967. [DOI] [PubMed] [Google Scholar]
- 27.Kim H.J., Moon S.J., Kim S.-H., Heo K., Kim J.H. DBC1 regulates Wnt/beta-catenin-mediated expression of MACC1, a key regulator of cancer progression, in colon cancer. Cell Death Dis. 2018;9:831. doi: 10.1038/s41419-018-0899-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardie D.G., Alessi D.R. LKB1 and AMPK and the cancer-metabolism link - ten years after. BMC Biol. 2013;11:36. doi: 10.1186/1741-7007-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hemminki A., Markie D., Tomlinson I., Avizienyte E., Roth S., Loukola A., Bignell G., Warren W., Aminoff M., Hoglund P., Jarvinen H., Kristo P., Pelin K., Ridanpaa M., Salovaara R., et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 30.Entius M.M., Keller J.J., Westerman A.M., van Rees B.P., van Velthuysen M.L., de Goeij A.F., Wilson J.H., Giardiello F.M., Offerhaus G.J. Molecular genetic alterations in hamartomatous polyps and carcinomas of patients with Peutz-Jeghers syndrome. J. Clin. Pathol. 2001;54:126–131. doi: 10.1136/jcp.54.2.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoijman E., Rubbini D., Colombelli J., Alsina B. Mitotic cell rounding and epithelial thinning regulate lumen growth and shape. Nat. Commun. 2015;6:7355. doi: 10.1038/ncomms8355. [DOI] [PubMed] [Google Scholar]
- 32.Stewart M.P., Helenius J., Toyoda Y., Ramanathan S.P., Muller D.J., Hyman A.A. Publisher correction: Hydrostatic pressure and the actomyosin cortex drive mitotic cell rounding. Nature. 2019;571 doi: 10.1038/s41586-019-1327-8. [DOI] [PubMed] [Google Scholar]
- 33.Zannini L., Buscemi G., Kim J.-E., Fontanella E., Delia D. DBC1 phosphorylation by ATM/ATR inhibits SIRT1 deacetylase in response to DNA damage. J. Mol. Cell. Biol. 2012;4:294–303. doi: 10.1093/jmcb/mjs035. [DOI] [PubMed] [Google Scholar]
- 34.Park S.H., Riley P., 4th, Frisch S.M. Regulation of anoikis by deleted in breast cancer-1 (DBC1) through NF-κB. Apoptosis. 2013;18:949–962. doi: 10.1007/s10495-013-0847-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martens I., Nilsson S.A., Linder S., Magnusson G. Mutational analysis of polyomavirus small-T-antigen functions in productive infection and in transformation. J. Virol. 1989;63:2126–2133. doi: 10.1128/jvi.63.5.2126-2133.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho W.S., Wang H., Maggio D., Kovach J.S., Zhang Q., Song Q., Marincola F.M., Heiss J.D., Gilbert M.R., Lu R., Zhuang Z. Pharmacologic inhibition of protein phosphatase-2A achieves durable immune-mediated antitumor activity when combined with PD-1 blockade. Nat. Commun. 2018;9:2126. doi: 10.1038/s41467-018-04425-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J., Bonkowski M.S., Moniot S., Zhang D., Hubbard B.P., Ling A.J.Y., Rajman L.A., Qin B., Lou Z., Gorbunova V., Aravind L., Steegborn C., Sinclair D.A. A conserved NAD(+) binding pocket that regulates protein-protein interactions during aging. Science. 2017;355:1312–1317. doi: 10.1126/science.aad8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang P., Jacobson M.K., Mitchison T.J. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- 39.Xie Z., Dong Y., Scholz R., Neumann D., Zou M.-H. Phosphorylation of LKB1 at serine 428 by protein kinase C-zeta is required for metformin-enhanced activation of the AMP-activated protein kinase in endothelial cells. Circulation. 2008;117:952–962. doi: 10.1161/CIRCULATIONAHA.107.744490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tiainen M., Ylikorkala A., Makela T.P. Growth suppression by Lkb1 is mediated by a G(1) cell cycle arrest. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9248–9251. doi: 10.1073/pnas.96.16.9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du K., Herzig S., Kulkarni R.N., Montminy M. TRB3: A tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science. 2003;300:1574–1577. doi: 10.1126/science.1079817. [DOI] [PubMed] [Google Scholar]
- 42.Cho U.S., Morrone S., Sablina A.A., Arroyo J.D., Hahn W.C., Xu W. Structural basis of PP2A inhibition by small t antigen. PLoS Biol. 2007;5 doi: 10.1371/journal.pbio.0050202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y., Xu Y., Bao Q., Xing Y., Li Z., Lin Z., Stock J.B., Jeffrey P.D., Shi Y. Structural and biochemical insights into the regulation of protein phosphatase 2A by small T antigen of SV40. Nat. Struct. Mol. Biol. 2007;14:527–534. doi: 10.1038/nsmb1254. [DOI] [PubMed] [Google Scholar]
- 44.Huttlin E.L., Ting L., Bruckner R.J., Gebreab F., Gygi M.P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K., Dong R., Guarani V., Vaites L.P., Ordureau A., Rad R., et al. The BioPlex network: A systematic exploration of the human interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huttlin E.L., Bruckner R.J., Paulo J.A., Cannon J.R., Ting L., Baltier K., Colby G., Gebreab F., Gygi M.P., Parzen H., Szpyt J., Tam S., Zarraga G., Pontano-Vaites L., Swarup S., et al. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017;545:505–509. doi: 10.1038/nature22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yadav L., Tamene F., Göös H., van Drogen A., Katainen R., Aebersold R., Gstaiger M., Varjosalo M. Systematic analysis of human protein phosphatase interactions and dynamics. Cell Syst. 2017;4:430–444.e5. doi: 10.1016/j.cels.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 47.Silk A.D., Holland A.J., Cleveland D.W. Requirements for NuMA in maintenance and establishment of mammalian spindle poles. J. Cell Biol. 2009;184:677–690. doi: 10.1083/jcb.200810091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ban K.H., Torres J.Z., Miller J.J., Mikhailov A., Nachury M.V., Tung J.J., Rieder C.L., Jackson P.K. The END network couples spindle pole assembly to inhibition of the anaphase-promoting complex/cyclosome in early mitosis. Dev. Cell. 2007;13:29–42. doi: 10.1016/j.devcel.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 49.Boldt K., van Reeuwijk J., Lu Q., Koutroumpas K., Nguyen T.-M.T., Texier Y., van Beersum S.E.C., Horn N., Willer J.R., Mans D.A., Dougherty G., Lamers I.J.C., Coene K.L.M., Arts H.H., Betts M.J., et al. An organelle-specific protein landscape identifies novel diseases and molecular mechanisms. Nat. Commun. 2016;7:11491. doi: 10.1038/ncomms11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welburn J.P.I., Cheeseman I.M. The microtubule-binding protein Cep170 promotes the targeting of the kinesin-13 depolymerase Kif2b to the mitotic spindle. Mol. Biol. Cell. 2012;23:4786–4795. doi: 10.1091/mbc.E12-03-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee T., Kim S.J., Sumpio B.E. Role of PP2A in the regulation of p38 MAPK activation in bovine aortic endothelial cells exposed to cyclic strain. J. Cell. Physiol. 2003;194:349–355. doi: 10.1002/jcp.10211. [DOI] [PubMed] [Google Scholar]
- 52.Avdi N.J., Malcolm K.C., Nick J.A., Worthen G.S. A role for protein phosphatase-2A in p38 mitogen-activated protein kinase-mediated regulation of the c-Jun NH(2)-terminal kinase pathway in human neutrophils. J. Biol. Chem. 2002;277:40687–40696. doi: 10.1074/jbc.M204455200. [DOI] [PubMed] [Google Scholar]
- 53.Choi Y.Y., Kang H.-K., Choi J.E., Jang J.S., Kim E.J., Cha S.I., Lee W.K., Kam S., Kim C.H., Han S.B., Jung T.H., Park J.Y. Comprehensive assessment of P21 polymorphisms and lung cancer risk. J. Hum. Genet. 2008;53:87–95. doi: 10.1007/s10038-007-0222-6. [DOI] [PubMed] [Google Scholar]
- 54.Gan B., Hu J., Jiang S., Liu Y., Sahin E., Zhuang L., Fletcher-Sananikone E., Colla S., Wang Y.A., Chin L., Depinho R.A. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 2010;468:701–704. doi: 10.1038/nature09595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gurumurthy S., Xie S.Z., Alagesan B., Kim J., Yusuf R.Z., Saez B., Tzatsos A., Ozsolak F., Milos P., Ferrari F., Park P.J., Shirihai O.S., Scadden D.T., Bardeesy N. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468:659–663. doi: 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang Y., Lu J., Zhan L., Wang M., Shi R., Yuan X., Gao X., Liu X., Zang J., Liu W., Yao X. Resveratrol-induced Sirt1 phosphorylation by LKB1 mediates mitochondrial metabolism. J. Biol. Chem. 2021;297:100929. doi: 10.1016/j.jbc.2021.100929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim W., Cheon M.G., Kim J.-E. Mitochondrial CCAR2/DBC1 is required for cell survival against rotenone-induced mitochondrial stress. Biochem. Biophys. Res. Commun. 2017;485:782–789. doi: 10.1016/j.bbrc.2017.02.131. [DOI] [PubMed] [Google Scholar]
- 58.Ha S.Y., Kim J.H., Yang J.W., Bae H., Cho H.Y., Park C.-K. Expression of DBC1 is associated with poor prognosis in hepatitis virus-related hepatocellular carcinoma. Pathol. Res. Pract. 2016;212:616–621. doi: 10.1016/j.prp.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 59.Wagle S., Park S.-H., Kim K.M., Moon Y.J., Bae J.S., Kwon K.S., Park H.S., Lee H., Moon W.S., Kim J.R., Jang K.Y. DBC1/CCAR2 is involved in the stabilization of androgen receptor and the progression of osteosarcoma. Sci. Rep. 2015;5:13144. doi: 10.1038/srep13144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Won K.Y., Cho H., Kim G.Y., Lim S.-J., Bae G.E., Lim J.U., Sung J.-Y., Park Y.-K., Kim Y.W., Lee J. High DBC1 (CCAR2) expression in gallbladder carcinoma is associated with favorable clinicopathological factors. Int. J. Clin. Exp. Pathol. 2015;8:11440–11445. [PMC free article] [PubMed] [Google Scholar]
- 61.Yu E.J., Kim S.-H., Kim H.J., Heo K., Ou C.-Y., Stallcup M.R., Kim J.H. Positive regulation of β-catenin-PROX1 signaling axis by DBC1 in colon cancer progression. Oncogene. 2016;35:3410–3418. doi: 10.1038/onc.2015.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cho D., Park H., Park S.-H., Kim K., Chung M., Moon W., Kang M., Jang K. The expression of DBC1/CCAR2 is associated with poor prognosis of ovarian carcinoma. J. Ovarian Res. 2015;8:2. doi: 10.1186/s13048-015-0129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adelmant G., Calkins A.S., Garg B.K., Card J.D., Askenazi M., Miron A., Sobhian B., Zhang Y., Nakatani Y., Silver P.A., Iglehart J.D., Marto J.A., Lazaro J.-B. DNA ends alter the molecular composition and localization of Ku multicomponent complexes. Mol. Cell. Proteomics. 2012;11:411–421. doi: 10.1074/mcp.M111.013581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ficarro S.B., Zhang Y., Lu Y., Moghimi A.R., Askenazi M., Hyatt E., Smith E.D., Boyer L., Schlaeger T.M., Luckey C.J., Marto J.A. Improved electrospray ionization efficiency compensates for diminished chromatographic resolution and enables proteomics analysis of tyrosine signaling in embryonic stem cells. Anal. Chem. 2009;81:3440–3447. doi: 10.1021/ac802720e. [DOI] [PubMed] [Google Scholar]
- 65.Askenazi M., Marto J.A., Linial M. The complete peptide dictionary--a meta-proteomics resource. Proteomics. 2010;10:4306–4310. doi: 10.1002/pmic.201000270. [DOI] [PubMed] [Google Scholar]
- 66.Rozenblatt-Rosen O., Deo R.C., Padi M., Adelmant G., Calderwood M.A., Rolland T., Grace M., Dricot A., Askenazi M., Tavares M., Pevzner S.J., Abderazzaq F., Byrdsong D., Carvunis A.-R., Chen A.A., et al. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature. 2012;487:491–495. doi: 10.1038/nature11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Most of the data that was generated in this study is already incorporated in the article. The mass spectrometry data is currently available on ftp://massive.ucsd.edu/MSV000087239/. The microarray data is already available at NCBI Microarray GEO accession number: GSE149525 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE149525). If any further details are needed, please feel free to contact Dr Shaida Andrabi, University of Kashmir through email at shaida.andrabi@uok.edu.in (corresponding author).