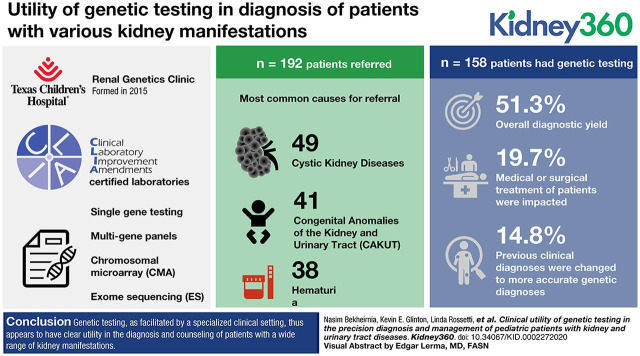
Visual Abstract
Keywords: genetics, data collection, genetic testing, kidney disease, Renal Genetics Clinic, urologic diseases
Abstract
Background
As genetic testing increasingly integrates into the practice of nephrology, our understanding of the basis of many kidney disorders has exponentially increased. Given this, we recently initiated a Renal Genetics Clinic (RGC) at our large, urban children’s hospital for patients with kidney disorders.
Methods
Genetic testing was performed in Clinical Laboratory Improvement Amendments–certified laboratories using single gene testing, multigene panels, chromosomal microarray, or exome sequencing.
Results
A total of 192 patients were evaluated in this clinic, with cystic kidney disease (49/192) being the most common reason for referral, followed by congenital anomalies of the kidney and urinary tract (41/192) and hematuria (38/192). Genetic testing was performed for 158 patients, with an overall diagnostic yield of 81 out of 158 (51%). In the 16 out of 81 (20%) of patients who reached a genetic diagnosis, medical or surgical treatment of the patients were affected, and previous clinical diagnoses were changed to more accurate genetic diagnoses in 12 of 81 (15%) patients.
Conclusions
Our genetic testing provided an accurate diagnosis for children and, in some cases, led to further diagnoses in seemingly asymptomatic family members and changes to overall medical management. Genetic testing, as facilitated by such a specialized clinical setting, thus appears to have clear utility in the diagnosis and counseling of patients with a wide range of kidney manifestations.
Introduction
Genetic testing has increasingly integrated into the practice of different specialties in medicine and surgery. Within the field of nephrology, in particular, the availability of such testing led to the rapid growth and expansion of our knowledge of the clinical spectrum of monogenic kidney diseases. The genetic etiology of kidney diseases, such as polycystic kidney disease, Alport syndrome, several forms of monogenic steroid-resistant nephrotic syndrome (SRNS), and nephronophthisis has grown and can now be identified in a significant portion of affected individuals. In patients with SRNS, 30% of those diagnosed before age 25 will have a pathogenic variant in one of 30 known SRNS genes (1). Even in a condition not commonly associated with genetic causes, such as nephrolithiasis, around 15% of individuals have a specific underlying genetic etiology (2). Given the growing number of recognized disease-causing gene defects, multigene panels are now available and, in some cases, can provide adequate diagnostic coverage (3). Similarly, exome sequencing (ES) has immense utility in the diagnosis of adults and children with a variety of disorders (4,5).
With the expanding number of candidate genes and the increasing complexity of genetic testing available, the need for more comprehensive diagnostic evaluations for such patients has also increased. To address this need, a Renal Genetics Clinic (RGC) at Texas Children’s Hospital (TCH) was formed in February 2015. Patients are referred from a variety of care settings, including the Pediatric Nephrology Clinic and various inpatient/outpatient services at TCH. Through this clinic, patients undergo a thorough genetic evaluation with a focus on kidney-specific malformations, complications, or diseases. Furthermore, given the nature of the clinic, family members of affected individuals can be evaluated, allowing us to provide guidance, if needed, for family planning. Extensive research shows the key roles genetic defects play in pediatric kidney disorders, and a growing number of studies are evaluating the utility of clinical genetics evaluation and genetic testing in the clinical practice (6,7). However, there is still a need to expand the knowledge in the intersection of clinical nephrology and clinical genetics. The specific objective of this study is to assess the role of clinical genetics in precision diagnosis and management of early onset pediatric kidney diseases. We hypothesized that genetic evaluation improves patient care in pediatric nephrology. Diagnostic yield and effect on medical management is reported for the first 4 years of this clinic’s operations.
Materials and Methods
Study Participants
Patients were all evaluated within the RGC at TCH. The clinic was initially held on only one half day per month, but this was increased to a full clinical day monthly after approximately 18 months. Patients were referred by pediatric nephrologists on the basis of their expert opinions. Patients were interviewed and examined by a clinical geneticist, and appropriate genetic testing was recommended on the basis of their clinical history, presentation, and family history. Pretest counseling was provided. Patients consented for ES on the basis of the performing Clinical Laboratory Improvement Amendment (CLIA) laboratories’ consent forms that include secondary findings (medically actionable and carrier status). The genetic variants reported in this study were classified only by CLIA laboratories. Reported variants by CLIA laboratories were evaluated by a clinical geneticist during clinical visits, aiming to provide a clinical diagnosis and to discuss pertinent management. Clinical information on subjects was collected retrospectively for the period of February 2015 to June 2019. All patients seen during this timeframe were eligible to participate. Institutional-review-board approval was obtained to perform a retrospective cross-sectional study to investigate the yield and effect of genetics evaluation. The outcome of the study was defined as the “impact” of genetic evaluation on diagnosis and management of patients, and this impact was classified into five categories: (1) effect on medical and/or surgical treatment (L1), (2) change of medical diagnosis (L2), (3) providing diagnostic certainty (L3), (4) subsequent evaluation of other body-system involvement (L4), and (5) cascade family member testing (L5). This definition, with the associated categories, has not been previously published; however, it is proposed as a future methodology for other RGCs, because it provides an overarching scoring system to combine impact stratification between clinical genetics and pediatric nephrology. The scoring system was created by a clinical geneticist and a pediatric nephrologist. To minimize the bias regarding scoring of the outcome, a pediatric nephrologist, blinded to the identifiers, reviewed the allocated scores.
Definition of Proteinuria and Nephrotic Syndrome
Patients with proteinuria and hematuria were referred from pediatric nephrology service at TCH. Proteinuria was defined as urinary protein excretion >100 mg/m2 per day or 4 mg/m2 per hour. Nephrotic-range proteinuria was defined as ≥1000 mg/m2 per day or 40 mg/m2 per hour. Microscopic hematuria was defined as the presence of more than five red blood cells per high-power field (40× magnification). CKD was defined on the basis of fulfilling one of the following clinical criteria (8):
GFR of <60 ml/min per 1.73 m2 for >3 months with implications for health, regardless of whether other CKD markers are present.
GFR >60 ml/min per 1.73 m2 that is accompanied by evidence of structural damage or other markers of functional kidney abnormalities, including proteinuria, albuminuria, renal tubular disorders, or pathologic abnormalities detected by histology or inferred by imaging. ESKD was defined as a GFR <15 ml/min per 1.73 m2.
Genetic Testing
Testing performed by CLIA laboratories include disease-specific panels (Supplemental Table 1), chromosomal microarray (CMA), expanded next-generation sequencing panels (Total BluePrint), and ES (trio or proband only [when both parents were not available]). When appropriate, combinations of these tests were also performed to optimize diagnostic yield in cases with atypical or unclear phenotypes. Overall, for patients with isolated hematuria or proteinuria, specific panels were recommended first. For cystic kidney with suspicion of autosomal dominant polycystic kidney disease (ADPKD), a PKD panel was recommended. For patients with congenital anomalies of the kidney and urinary tract (CAKUT), ESKD of unknown etiology, and suspected nephronophthisis, broad genetic testing was recommended. Specific panels were also recommended for specific rare kidney diseases (e.g., Gitelman syndrome and renal tubular acidosis). In general, for CAKUT, we tried to order CMA first and use ES when CMA was not diagnostic. For proteinuria, when we did not have an identifiable genetic variant by the panel, ES was recommended (9). Comparison analysis of detection rate between different testing modalities was not performed because the choice of genetic testing was not randomized, and, therefore, is biased to compare the detection rate of different genetic testing modalities. We expect that broad genetic testing (e.g., CMA/ES) has a higher yield; however, this requires further investigation and depends on other factors, such as patient population and reasons for referral. Genomic DNA was isolated from peripheral leukocytes obtained via venipuncture or, less commonly, from saliva.
Interpretation of Genetic Findings
Sequencing or copy number data were generated in CLIA-certified laboratories, and data were reviewed and interpreted by clinical molecular geneticists at the performing laboratories. The reported results were correlated with clinical and family history of the patients, and a final diagnosis was confirmed by a clinical geneticist at RGC. American College of Medical Genetics and Genomics guidelines were followed throughout this process for consistent interpretation (10).
Venn-Diagram Generation
The scoring system was processed by the library in Pandas. Because this is a five-class comparison, circular Venn diagrams are challenging. Therefore, oval-shaped Venn diagrams were chosen. Modified application-programming-interface calls were used in the Python environment to create the Venn diagram in this manuscript.
Results
A total of 192 patients were evaluated in this clinic from February 2015 to June 2019 (Table 1). Patients ranged in age from 1 day of life to 25 years of age, with a mean age of 8.7 years (SD, 6.0 years). Patients were from diverse ethnic backgrounds. The most common reason for referral was cystic kidney disease in 49 patients (26%), followed by CAKUT in 41 patients (21%), 38 patients with hematuria (20%), and 21 patients with proteinuria (11%). A further 43 patients (23%) were seen for “other” clinical diagnoses, including nephronophthisis, nephrocalcinosis, developmental delay combined with kidney disease, or overlapping phenotypes (Supplemental Table 2). Of the 192 patients, three were asymptomatic with a positive family history of ADPKD. Considering the ethics of genetic screening in asymptomatic children, genetic testing was only recommended for their affected parent. In addition, parents of two patients were not interested in genetic testing at the initial visit. Genetic testing was performed for 158 of 187 patients (85%). We were not able to perform genetic testing for 29 individuals (12 because of insurance denial, 16 families were not interested in pursuing genetic testing, and one was not available at the time of testing). Information regarding detection rates of different tests among variable indications for referral can be found in Supplemental Table 3. Among 158 patients, 81 (51%) had positive diagnostic results (Table 2). The type of genetic testing (e.g., panel, CMA, ES, and Total BluePrint) and post-test recommendations are summarized in Table 2. In an additional five patients, the patients’ phenotypes were partially explained by genetic workup (Supplemental Table 4).
Table 1.
Demographics and indications for referrals among 192 patients evaluated at Renal Genetics Clinic between 2/2015 and 6/2019
| Characteristic | Value |
|---|---|
| Sex, n (%) | |
| Male | 102 (53) |
| Female | 90 (47) |
| Total patients evaluated, n | 192 |
| Ethnicity, n (%) | |
| White | 67 (35) |
| Latino | 67 (35) |
| Black | 29 (15) |
| Mixed | 25 (13) |
| Other | 3 (2) |
| Asian | 1 (0.1) |
| Age at evaluation | |
| Mean (SD) in yr | 8.7 (6.0) |
| Range | 1 d–25 yr |
| Initial indications for referral, n (%) | |
| Cystic kidney disease | 49 (26) |
| Congenital anomalies of the kidney and urinary tract | 41 (21) |
| Other | 43 (23) |
| Hematuria | 38 (20) |
| Proteinuria | 21 (11) |
Table 2.
Demographics, genetic information, and effect on management for patients with diagnostic genetic results
| Patient Number | Sex | Age(yr) | L1 | L2 | L3 | L4 | L5 | Type of Genetic Testing (1, 2, 3, 4, 5)a | Gene/Locus | Genetic Finding (SNV/Indel/CNV) | Phenotype (Indication for Referral) | Comment |
| RGC-0001 | F | 10 | − | − | + | + | − | 1 | PKD1 | NM_001009944.2: c.7987C>T (p.Q2663*) (het) | Bilateral renal cysts | |
| RGC-0003 | M | 0.8 | − | − | + | + | + | 4 | PKD1 | Partial PKD1 gene deletion (at least exons 27–38) (het) (novel) | Bilateral renal cysts | Subsequently mother was found have cysts in her kidneys |
| RGC-0004 | M | 13 | − | − | + | + | + | 2 | HNF1B | arr[GRCh37]2q36.3 (227999132_228097605)x1 | Chromosomal abnormality | Deletion was found to be maternally inherited |
| RGC-0009 | M | 10 | − | − | + | + | + | 1 | PKD1 | NM_001009944.2: c.7483T>C (p.C2495R) (het) | Bilateral renal cysts and duplicated collecting system | Symptomatic sibling tested positive for KFM |
| RGC-0010 | M | 16 | − | − | − | + | + | 1 | COL4A5 | NM_000495.4: c.152G>T (p.G51V) (hem) | Hematuria and proteinuria | Symptomatic sibling tested positive for KFM |
| RGC-0013 | M | 3 | − | − | − | + | − | 1 | COL4A5 | NM_000495.4: c.3197G>C (p.G1066A) (hem) | Alport syndrome | |
| RGC-0014 | F | 10 | − | − | + | + | − | 2 | 1q21 del | arr[GRCh37]1q21.1q21.2 (146618988–147825855)x1 | Learning disability, VUR, cataracts, microcephaly | Patient also has 16p11.2 0.521 Mb duplication |
| RGC-0018 | M | 1.5 | − | − | + | + | − | 2 | HNF1B | arr[GRCh37] 17q12 (34842059–36214026)x1 | Unilateral multicystic dysplastic kidney, VUR, hypercalcemia, developmental delay, hypotonia | |
| RGC-0019 | F | 16 | − | − | + | − | − | 2, 3 | WDR19 | NM_025132: c.3703G>A (p.E1235K) (het) arr[GRCh37]4p14 (39215680–39219295)x1 | ESKD, dysautonomia, migraines, choledochal and pancreas cyst | |
| RGC-0021 | F | 2.7 | − | − | + | + | + | 2, 4 | PKD1 | NM_001009944.2: c.1259A>G (p.Y420C) (het) | Cystic kidney and Chiari malformation | PKD1 variant is de novo |
| RGC-0026 | F | 4 | − | − | + | − | + | 2, 4 | EYA1 | arr[GRCh37]8q13.2q13.3 (69901440–72586292)x1 | Branchio-oto-renal syndrome | EYA1 variant is de novo |
| RGC-0029 | M | 2.9 | − | − | + | + | − | 1 | PKD1 | NM_001009944.2: c.2659delT (p.W887Gfs*11) (het) | Bilateral renal cysts | |
| RGC-0030 | F | 1.5 | + | − | + | − | + | 1 | NPHS2 | NM_014625: c.790G>C (p.E264Q) (het), and c.779T>A (p.V260E) (het) | Infantile nephrotic syndrome | Phase of the variants were determined (opposite chromosomes), subsequently symptomatic siblings tested positive for these variants |
| RGC-0032 | M | 12 | − | − | + | − | + | 2, 4 | DYRK1A | NM_001396: c. 501delA (p.G168fs) (het) | Intellectual disability and hypospadias | DYRK1A variant is de novo |
| RGC-0034 | F | 2.6 | + | − | + | + | + | 1, 2, 4 | WT1 | NM_024426.4: c.1390G>A (p.D464N) (het) | Atypical HUS | WT1 variant is de novo |
| RGC-0039 | F | 7 | − | − | + | + | + | 2, 4 | COL4A3 | NM_000091: c.1407delA (p.G470fs) (het) and c.40_63del (p.L14_L21del) (het) | Hereditary nephritis | Each variant is inherited from one parent |
| RGC-0041 | M | 15 | − | − | + | − | + | 2 | 22q11 triplication | arr[GRCh37]22q11.1q11.21 (17289827–18640328)x3 | Facial asymmetry, imperforate anus, neurogenic bladder | This triplication is de novo |
| RGC-0043 | M | 11 | − | − | + | − | + | 2, 4 | KAT6B | NM_012330: c.3280delG (p.E1094fs) (het) | Bilateral undescended testes, a mild hypospadias, and Ohdo syndrome | KAT6B variant is de novo |
| RGC-0046 | M | 3 | + | − | + | − | − | 1 | NPHS2 | NM_014625: c.790G>C (p.E264Q) (het), and c.779T>A (p.V260E) (het) | Positive family history of infantile nephrotic syndrome | Avoid immune suppression |
| RGC-0047 | F | 2 | + | − | + | − | − | 1 | NPHS2 | NM_014625: c.790G>C (p.E264Q) (het) and c.779T>A (p.V260E) (het) | Infantile nephrotic syndrome | Avoid immune suppression |
| RGC-0050 | M | 18 | − | − | + | − | + | 2, 4 | TMEM67 | NM_153704: c. 515G>T (p.R172L) (het) and c.1021G>A (p.G341R) (het) (novel) | Joubert syndrome | Each variant is inherited from one parent |
| RGC-0052 | M | 0.8 | − | − | + | + | + | 2, 4 | NSD1 | NM_022455.4: c.3423_3424insCC (p.N1142PfsX11) (het) (novel) | Macrosomia and nephromegaly | NSD1 variant is de novo |
| RGC-0054 | M | 1.9 | + | − | + | − | + | 2, 4 | PLCE1 | NM_016341.3: c.4675_4678delITTAG (p.L1559fs) (hom) (novel) | Nephrotic-range proteinuria | Each variant is inherited from one parent |
| RGC-0055 | M | 10 | − | − | + | + | − | 1 | PKD1 | NM_001009944.2: c.7483T>C (p.C2495R) (het) | Family history of ADPKD, bilateral cystic kidney disease, and duplicated collecting system | |
| RGC-0058 | M | 3 | − | − | − | + | − | 1 | ATP6V0A4 | NM_020632.2: c.1231G>T (p.D411Y) (hom) | Distal renal tubular acidosis | Hearing evaluation was normal |
| RGC-0063 | F | 3 | − | − | − | + | − | 1 | PKD1 | NM_001009944.2: c.7111del (p.V2371Cfs*11) (het) | Bilateral renal cysts | Echocardiogram |
| RGC-0066 | F | 19 | − | + | − | + | + | 2, 4 | USP9X | NM_001039590.2: c.5606_5607dupTC (p.V1870SfsX37) (het) (novel) | Hypertension and Townes–Brocks syndrome | USP9X variant is de novo |
| RGC-0067 | M | 4.9 | − | − | + | + | + | 2, 4 | COL4A5 | NM_000495: c.5034T>A (p.C1678X) (hem) (novel) | Hematuria and thin basement membrane nephropathy | COL4A5 variant is maternally inherited |
| RGC-0068 | M | 14 | − | + | − | + | + | 2, 4 | OCRL | NM_000276.3: c.2531_2539delGAGAACTC TinsAAG (p.R844_L847delinsQV) (hem) (novel) | Cataracts and proteinuria | OCRL variant is maternally inherited, subsequently sibling tested positive for KFM |
| RGC-0070 | F | 13 | − | + | − | + | − | 2, 3 | NPHP4 | NM_015102.3: c.3611C>T (p.P1204L) (hom) | CKD | |
| RGC-0072 | M | 11 | − | − | + | + | − | 2, 3 | PKD1 | NM_001009944: c.9859_9861del (p.L3287del) (het) | Bilateral renal cysts | |
| RGC-0075 | F | 14 | − | − | + | + | − | 2, 3 | DCDC2 | NM_016356: c.383C>G (p.S128X) (hom) | ESKD and liver fibrosis | |
| RGC-0076 | M | 4 | − | − | − | + | + | 1 | COL4A5 | NM_000495.4: c.1948G>A (p.G650S) (hem) | Alport syndrome | Siblings tested negative for KFM |
| RGC-0077 | F | 6 | − | − | + | + | + | 1 | PKD1 | NM_001009944.2: likely pathogenic c.8948+1G>T (het), VUS c.955GG>C (p.Vl3184L) (het) | Bilateral renal cysts | Each variant in PKD1 is inherited from one parent |
| RGC-0078 | F | 1.9 | − | − | + | + | + | 1 | PKD1 | Likely pathogenic c.9829C>T (p.R3277C) (het), VUS c.3494A>G (p. D1165G) (het) | Bilateral renal cysts | Each variant in PKD1 is inherited from one parent |
| RGC-0080 | M | 12 | + | − | + | − | + | 1 | PKHD1 | NM_138694.3: likely pathogenic (c.3761_3762delinsG) (p.A1254Gfs*49) (het), VUS c.4292G>A (p.C1431Y) (het) | Bilateral renal cysts | Pseudodominant ARPKD, each variant is inherited form one parent; both father and paternal aunt are clinically diagnosed with ARPKD |
| RGC-0081 | M | 13 | − | − | − | + | + | 2, 4 | COL4A5 | arr[GRCh37]Xq22.3 (107802035–107802303)x0 (Novel) | Alport syndrome, developmental delay, autism, ADHD | This deletion is maternally inherited |
| RGC-0083 | M | 16 | + | + | − | + | + | 2, 4 | COL4A5 | NM_000495: c.3059dupT (p.G1021fs) (hem) (novel) | FSGS | Both patient and his affected mother’s diagnosis has been changed and avoid immune suppression |
| RGC-0084 | F | 4.9 | + | − | + | − | + | 2, 4 | RMND1 | NM_017909: c.713A>G (p.N238S) (het) and c.533C>T (p.T178M) (het) | CKD, congenital hearing loss, and developmental delay | Each variant is inherited from one parent |
| RGC-0085 | M | 0.5 | − | − | + | − | + | 2, 4 | CASK | NM_003688: c.1721dupA (p.S575fs) (hem) (novel) | Microcephaly, dysmorphic features, right club feet, neurologic dysfunction, hypotonia, pontocerebellar hypoplasia, and right cryptorchidism | Variant in CASK is de novo |
| RGC-0086 | M | 0.2 | − | − | + | + | − | 2 | 1q23.2q25.1 deletion | arr[GRCh37]1q23.2q25.1 (160369890–175796325)x1 | Multiple congenital anomalies including dysplastic ears, dysplastic kidney, bilateral undescended testes, dysmorphic features, and abnormality of the shape of hands | |
| RGC-0087 | M | 9 | − | − | + | + | + | 1 | PKD1 | NM_001009944.2: c.11017–10C>A (IVS37–10C>A) (het) | Bilateral renal cysts | PKD1 variant is inherited from father; subsequently, father and PGF were diagnosed with ADPKD |
| RGC-0088 | F | 6 | − | − | − | + | − | 1 | PKD1 | NM_001009944.2: c.6806C>G (p.S2269*) (het) | Bilateral renal cysts | |
| RGC-0090 | F | 18 | − | − | − | + | − | 1 | COL4A5 | NM_000495.4: c.4602del (p.Y1535Ifs*13) (het) (novel) | Alport syndrome | |
| RGC-0091 | M | 8 | − | − | + | + | − | 2, 3 | PKD1 | NM_001009944.2: c.8043_8046delCTCG (p.S2682Afs*2) (het) (novel) | Bilateral renal cysts | |
| RGC-0092 | F | 2 | − | − | − | + | + | 2, 4 | PKHD1 | NM_138694.3: pathogenic variant c.3761_3762delCCinsG (het), VUS c.10666C>T (p.R3558C) (het) | Bilateral renal cysts | One variant is inherited from one parent and the other one is de novo |
| RGC-0097 | F | 17 | − | − | + | + | + | 4 | COL4A5 | NM_033380.1: c.3631G>A (p.G1211R) (het) | Hereditary nephritis | COL4A5 variant is de novo |
| RGC-0100 | M | 15 | − | − | + | + | − | 1 | HNF1B | NM_000458.2: c.513G>A (p.W171X) (het) | Bilateral renal cysts | |
| RGC-0101 | M | 16 | − | − | − | + | − | 1 | COL4A5 | arr[GRCh37]Xq22.3 (107868501–107869156)x0 (novel) | Alport syndrome | |
| RGC-0105 | F | 8 | − | + | − | + | − | 1 | COL4A4 | NM_000092.4: c.1334G>C (p.G445A) (het) and c.2570C>T (p.P857L) (het) | Steroid-sensitive nephrotic syndrome | |
| RGC-0108 | M | 15 | + | + | − | − | + | 1, 4 | OCRL | NM_000276.3: c.239delG (p.S80MfsX26) (hem) (novel) | Proteinuria | OCRL variant was maternally inherited |
| RGC-0110 | M | 5 | − | − | + | + | + | 5 | WDR19 | NM_025132: pathogenic c.1122_1123insT (p.P375fs) (het) (novel) and VUS c.817A>G (p.N273D) (het) | ESKD | Each variant in WDR19 is inherited from one parent |
| RGC-0112 | M | 14 | − | − | + | + | − | 2 | NPHP1 | arr[GRCh37]2q13 (110862477–110970270)x1 | CKD | |
| RGC-0113 | F | 6 | − | + | − | + | + | 2, 4 | PKD2 | NM_000297.3: c. 965G>A (p.R322Q) (het) | VUR, duplicated collecting system, and bilateral cystic kidney | PKD2 variant is paternally inherited |
| RGC-0115 | F | 7 | − | − | + | + | − | 1 | PKD2 | NM_000297.3: c.2614C>T (p.R872*) (het) | Unilateral renal cysts | |
| RGC-0116 | F | 1 | − | − | + | + | + | 1, 2, 5 | RPS19 | NM_001022: c.185G>A (p.R62Q) (het) | CKD and Diamond–Blackfan anemia | RPS19 variant is de novo |
| RGC-0117 | M | 0.16 | − | − | + | + | + | 2, 4 | BBS12 | NM_152618.2: pathogenic c.1115_1116delTT (p.F372*) (het), and VUS c.1277G>A (p.C426Y) (het) | Polydactyly and bilateral renal cysts | Each variant in BBS12 is inherited from one parent |
| RGC-0118 | M | 9 | + | + | − | + | + | 2, 4 | KCNJ1 | NM_000220.3: c.924C>A (p.C308*) (hom) | Renal dysplasia | Both parents are heterozygous for variant in KCNJ1 |
| RGC-0120 | M | 17 | − | − | − | + | + | 2, 4 | INVS | NM_014425.3: c.2695C>T (p.R899*) (hom) | Nephronophthisis | Both parents are heterozygous for variant in INVS |
| RGC-0124 | F | 17 | − | − | − | + | − | 1 | COL4A4 | NM_000092.4: c.1580del, (p.G527Vfs*126) (het) | Microscopic hematuria | |
| RGC-0128 | M | 2 | − | − | − | + | − | 1 | PKD1 | NM_01009944.2: c.8016+2T>C (IVS21+2T>C) (het) | Bilateral renal cysts | |
| RGC-0129 | M | 14 | − | − | − | − | + | 2, 4 | SLC7A9 | NM_014270.4: c.419T>C (p.F140S) (het) and c.164T>A (p.V55E) (het) (novel) | Cystine stones and dysplastic kidney | Each variant in SLC7A9 is inherited from one parent |
| RGC-0132 | F | 17 | − | − | + | + | − | 1 | PKD1 | NM_001009944.2: c.11712+1G>A (het) | Bilateral renal cysts | |
| RGC-0143 | M | 2 | + | − | + | − | + | 1 | NPHS1 | NM_004646.3; c.1747G>A (p.S910P) (het) (likely pathogenic), and c.1747G>A (p.E583K) (het) (VUS) | Nephrotic syndrome | Avoid immune suppression |
| RGC-0145 | M | 16 | − | + | − | − | + | 1 | NEK8 | NM_178170.2: c.1523T>A (p.Met508Lys) (het), and c.673G>C (p.Asp225His) (het) | Cystic kidney disease | Clinical diagnosis of ARPKD was changed to nephronophthisis |
| RGC-0147 | M | 14 | − | − | − | + | + | 2, 4 | KCNJ1 | NM_000220.3: c.924C>A, (p.C308*) (hom) | Bartter syndrome | Both parents are heterozygous for variant in KCNJ1 |
| RGC-0152 | F | 10 | − | − | + | + | − | 1 | COL4A5 | NM_000495.4: c.994_998delinsTCCC (p.Q332Sfs*14) (het) (novel) | Alport syndrome | |
| RGC-0156 | M | 14 | − | − | + | + | + | 1 | COL4A5 | NM_000495.4: c.4688+1G>T (hem) | Alport syndrome | COL4A5 variant is maternally inherited and sibling was tested negative for KFM |
| RGC-0157 | M | 12 | − | − | + | − | + | 1 | COL4A4 | NM_000092.4; c.1325G>C (p.G442A) (het) | Microscopic hematuria | |
| RGC-0159 | M | 2 | − | − | + | − | − | 1 | COL4A4 | NM_000092.4: c.1697–1G>A (het) | Microscopic hematuria | |
| RGC-0160 | M | 5 | − | − | + | − | + | 1 | AVPR2 | NM_000054.4: c.337C>T (p.R113W) (hem) | Diabetes insipidus | Subsequently sibling was tested positive for KFM |
| RGC-0162 | F | 8 | − | − | + | − | − | 1 | COL4A5 | NM_000495.4: c.2678G>A (p.G893D) (het) | Microscopic hematuria | |
| RGC-0164b | M | 1.3 | − | + | − | + | + | 4 | HNF1B | Arr[GRCh37]17q12 (34856055–36248918)x1dn | Bilateral renal cysts | This deletion is de novo and secondary finding of BRCA2 is maternally inherited |
| RGC-0171 | M | 2 | + | − | + | + | + | 2, 4 | WT1 | NM_024426.4: c.1432+4C>T (het) | Proteinuria, recurrent UTI, and hypospadias | WT1 variant was de novo |
| RGC-0182 | F | 11 | − | + | − | + | − | 1 | COL4A5 | NM_000495: c.557G>A (p.Gly186Asp) (het) (novel) | Microscopic hematuria | Familial diagnosis of FSGS changed to Alport syndrome |
| RGC-0183 | F | 16 | − | − | + | + | − | 1 | SLC12A3 | NM_000339; c.1001G>A (p.R334Q) (hom) | Gitelman syndrome | |
| RGC-0185 | F | 0.1 | + | − | + | − | − | 2, 5 | TPRM6 | NM_017662.4: c.5488–1G>C (hom) (novel) | Hypomagnesemia | Hypocalcemia and hypomagnesemia are due to defect in intestinal absorption of magnesium |
| RGC-0186 | M | 0.9 | + | − | − | + | − | 1 | WT1 | NM_0024426.3: c.1288C>T (p.R430*) (het) (novel) | Bilateral Wilms tumor | Impacted nephrectomy |
| RGC-0190 | F | 10 | + | − | + | + | + | 2, 4 | WT1 | NM_024426.4: c.1432+5 G>A (het) | ESKD and nephrotic range proteinuria | CMA revealed patient is XY female, and WT1 variant is de novo |
| RGC-0191 | F | 11 | + | − | + | − | − | 1 | CACNA1S | NM_000069.2: c.3715C>G (p.R1239G) (het) | Hypokalemia | Treatment with acetazolamide |
| RGC-0192 | M | 18 | − | + | − | + | + | 1, 2, 4 | COL4A5 | NM_000495.4: c.4298–20T>A (hem) (novel) | Hematuria and proteinuria | COL4A5 variant is maternally inherited |
L1, effect on medical and/or surgical treatment; L2, change of medical diagnosis; L3, providing diagnostic certainty; L4, subsequent evaluation of other body-system involvement; L5, cascade family member testing; SNV, single nucleotide variant; CNV, copy number variant; F, female; M, male; het, heterozygous; KFM, known familial mutation; hem, hemizygous; del, deletion; VUR, vesicoureteral reflux; HUS, hemolytic uremic syndrome; ins, insertion; hom, homozygous; ADPKD, autosomal dominant polycystic kidney disease; VUS, variant of uncertain significance; ARPKD, autosomal recessive polycystic kidney disease; ADHD, attention deficit hyperactivity disorder; PGF, paternal grandfather; UTI, urinary tract infection; CMA, chromosomal microarray.
Numbers represent the following testing types: 1, panel; 2, CMA; 3, proband exome sequencing; 4, trio exome sequencing; 5, Total BluePrint.
Patient had a secondray finding of pathogenic variant in BRCA2.
Among 158 patients, 115 variants of uncertain significance were detected in 42 patients. These variants were all reviewed by a clinical geneticist, their significance was reevaluated on the basis of the patient’s history, and further recommendations were provided to clarify their significance. The challenges of interpretation of these variants of uncertain significance are summarized in Supplemental Table 5.
Given the breadth of diagnoses encountered, no single test was universally applicable to every patient. Different tests, or a combination of tests, were recommended and completed for patients depending on the specificity of their clinical phenotype or reported history through different CLIA laboratories. For patients with CAKUT (41 patients; tests completed for 33), for instance, CMA or a combination of CMA with ES ultimately led to a diagnostic yield of 42% (14/33; including the partially diagnosed cases). However, in patients who presented with cystic kidney, the use of a multigene panel was the most successful approach to provide a genetic diagnosis in 79% (15/19) patients. Multigene panel testing also had a high detection rate for patients with proteinuria (seven out of ten patients; 70%) and hematuria (ten out of 15 patients; 67%).
Our testing approach led to the identification of pathogenic or likely pathogenic single nucleotide variants (SNVs) in 34 genes (Table 2). Similarly, 11 different pathogenic or likely pathogenic copy number variants (CNVs) were also identified, ranging from single exon deletions to large megabase-sized deletions of multiple genes. Pathogenic SNVs or CNVs were found most commonly in PKD1 (15), followed by COL4A5 (14), HNF1B (4), COL4A4 (4), WT1 (4), and PKHD1 (4). Secondary findings of BRCA2 pathogenic variants were identified in two families; they were provided with appropriate genetic counseling.
Pathogenic or likely pathogenic variants in PKD1, their strength, and age of diagnoses are summarized in Table 3. Out of 15 variants in PKD1, seven are truncating, four are missense, one was a partial gene deletion, one was an in-frame indel, and two were splice-site variants that likely do not cause truncation, but cause exon skipping. Missense variants all have a Combined Annotation Dependent Depletion score of >20, which put them in the top 1% of deleterious variants in the human genome. Therefore, these variants are likely to put the patients at high risk of progression. However, truncating variants pose a higher chance of reaching ESKD at a younger age (11).
Table 3.
Genetic information and the strength of the genetic variants for patients diagnosed with pathogenic PKD1 variant
| Family Identifier | Variant | Type of Variant | Indication for testing | Age of Diagnosis | CADD Score |
| RGC-001 | c.7987C>T (p.Q2663*) (het) | Stop gain | Family history of cystic kidney disease but not definitive for ADPKD and symptomatic | 2 yr | Truncating |
| RGC-003 | Partial PKD1 gene deletion (at least exons 27–38) (het) | Partial gene deletion | No family history, but symptomatic | 3 mo | NA |
| RGC-009 | c.7483T>C (p.C2495R) (het) | Missense | Family history of ADPKD and symptomatic | 9 yr | 24.2 |
| RGC-0021 | c.1259A>G (p.Y420C) (het) | Missense | No family history but symptomatic | 18 mo | 23.6 |
| RGC-0029 | c.2659delT (p.W887Gfs*11) (het) | Frameshift | No family history but symptomatic | 2 yr | Truncating |
| RGC-0055 | c.7483T>C (p.C2495R) (het) | Missense | Family history of ADPKD, bilateral cystic kidney disease, and duplicated collecting system | 10 yr | 24.2 |
| RGC-0063 | c.7111del (p.V2371Cfs*11) (het) | Frameshift | Positive family history of ADPKD and symptomatic | 1 yr | Truncating |
| RGC-0072 | c.9859_9861del (p.L3287del) (het) | In-frame deletion | Family history of kidney disease and symptomatic | 10 yr | NA |
| RGC-0077 | Likely pathogenic c.8948+1G>T (het) (novel), VUS c.9550G>C (p.V3184L) (het) | Splice site, Missense | Family history of cystic kidney disease but not definitive for ADPKD and symptomatic | Prenatal | 33 |
| 23.9 | |||||
| RGC-0078 | Likely pathogenic c.9829C>T (p.R3277C) (het), c.3494A>G (p. D1165G) (het) | Missense | Family history of cystic kidney disease but not definitive for ADPKD and symptomatic | Prenatal | 23.9 |
| 24.6 | |||||
| RGC-0087 | c.11017–10C>A (IVS37–10C>A) (het) | Splice site | No family history but symptomatic | 5 yr | Predicted to skip exon 38 likely to be nontruncating (12) |
| RGC-0088 | c.6806C>G (p.S2269*) (het) | Stop gain | Family history of ADPKD and symptomatic | 6 yr | Truncating |
| RGC-0091 | c.8043_8046delCTCG (p.S2682Afs*2) (het) | Frameshift | Family history of kidney disease and symptomatic | 6 yr | Truncating |
| RGC-0128 | c.8016+2T>C (IVS21+2T>C) (het) (novel) | Splice site | Family history of cystic kidney disease but not definitive for ADPKD and symptomatic | 2 yr | Truncating |
| RGC-0132 | c.11712+1G>A (het) | Splice site | Family history of cystic kidney disease but not definitive for ADPKD and symptomatic | 16 yr | Truncating (13) |
CADD, Combined Annotation Dependent Depletion; het, heterozygous; ADPKD, autosomal dominant polycystic kidney disease; NA, not applicable; VUS, variant of uncertain significance.
Effect on Precision Diagnosis and Management
To assess the effect of genetic testing and evaluation on patients’ management, each patient with a positive result was scored according to a five-level scoring system as defined in the Methods. Out of 81 positive diagnostic results, 16 (20%) affected immediate medical or L1, and 12 (15%) prior L2 Details regarding L1 and L2 effects on management are summarized in Table 4. The most common indication of referral among these patients with L1 impact was nephrotic syndrome or proteinuria, a condition where medication adjustment, by avoiding immunosuppression, became possible. Other immediate benefits of genetic evaluation included surgical decision making regarding the need for prophylactic (patient number RGC-0034) or therapeutic nephrectomy (RGC-0186) in patients with pathogenic variants in WT1. In three patients (RGC-0118, RGC-0185, and RGC-0191), targeted treatment recommendations with directed pharmacotherapy (indomethacin, magnesium, and acetazolamide) became possible after identification of underlying genetic diagnosis (KCNJ1, TPRM6, and CACNA1S).
Table 4.
Details of effect on management (L1 and L2) among patients with diagnostic results
| Patient Identifier | L1/L2 | Initial Diagnosis | Changed Diagnosis | Variant Found | Effect on Management |
| RGC-0030 | L1 | Infantile nephrotic syndrome | NPHS2 | Avoidance of immune suppression | |
| RGC-0034 | L1 | Atypical HUS | WT1 | Bilateral nephrectomy, pelvic MRI, tapering eculizumab | |
| RGC-0046 | L1 | Positive family history of infantile nephrotic syndrome | NPHS2 | Avoidance of immune suppression | |
| RGC-0047 | L1 | Infantile nephrotic syndrome | NPHS2 | Avoidance of immune suppression | |
| RGC-0054 | L1 | Nephrotic-range proteinuria | PLCE1 | Avoidance of immune suppression | |
| RGC-0066 | L2 | Townes–Brocks syndrome | USP9X-related disorder | USP9X | |
| RGC-0068 | L2 | FSGS | Lowe syndrome | OCRL | |
| RGC-0070 | L2 | Developmental delay and kidney problem | Nephronophthisis | NPHP4 | |
| RGC-0080 | L1 | ARPKD/ADPKD | PKHD1 | Pseudodominant ARPKD | |
| RGC-0083 | L1, L2 | FSGS | Alport syndrome | COL4A5 | Avoidance of immune suppression |
| RGC-0084 | L1 | Mitochondrial disease | RMND1 | Kidney transplantation is indicated for patients with RMND1 variants if needed | |
| RGC-0105 | L2 | Nephrotic syndrome | Alport syndrome | COL4A4 | |
| RGC-0108 | L1, L2 | Proteinuria/Alport syndrome | Dent syndrome | OCRL | Avoidance of immune suppression, management related to Dent disease |
| RGC-0113 | L2 | CAKUT | ADPKD | PKD2 | |
| RGC-0118 | L1, L2 | CAKUT | Bartter syndrome | KCNJ1 | Indomethacin treatment recommended and DEXA bone scan showed low bone density |
| RGC-0143 | L1 | Nephrotic syndrome | NPHS1 | Avoidance of immune suppression | |
| RGC-0145 | L2 | Polycystic kidney disease | Nephronophthisis | NEK8 | Clinical diagnosis of ARPKD was changed to nephronophthisis |
| RGC-0164a | L2 | Cystic kidney disease | 17q12 deletion syndrome | HNF1B and BRCA2 | Secondary finding of BRCA2 |
| RGC-0171 | L1 | Proteinuria | WT1-associated disease | WT1 | Followed by cancer prevention clinic |
| RGC-0182 | L2 | FSGS | Alport syndrome | COL4A5 | |
| RGC-0185 | L1 | Hypomagnesemia | TRPM6 | Hypocalcemia and hypomagnesemia are due to defect in intestinal absorption of magnesium | |
| RGC-0186 | L1 | Wilms tumor | WT1-associated syndrome | Affected surgical nephrectomy of patient | |
| RGC-0190 | L1 | Renal failure, proteinuria | WT1-associated syndrome | WT1 | CMA revealed patient is XY female. Risk of gonad blastoma in an XY female patient was discussed |
| RGC-0191 | L1 | Periodic hypokalemic paralysis | CACNA15 | Treatment with Acetazolamide | |
| RGC-0192 | L2 | CKD | Alport syndrome | COL4A5 |
L1, effect on medical and/or surgical treatment; L2, change of medical diagnosis; HUS, hemolytic uremic syndrome; MRI, magnetic resonance imaging; ARPKD, autosomal recessive polycystic kidney disease; ADPKD, autosomal dominant polycystic kidney disease; CAKUT, congenital anomalies of the kidney and urinary tract; DEXA, dual-energy x-ray absorptiometry; CMA, chromosomal microarray.
Patient had a secondray finding of pathogenic variant in BRCA2.
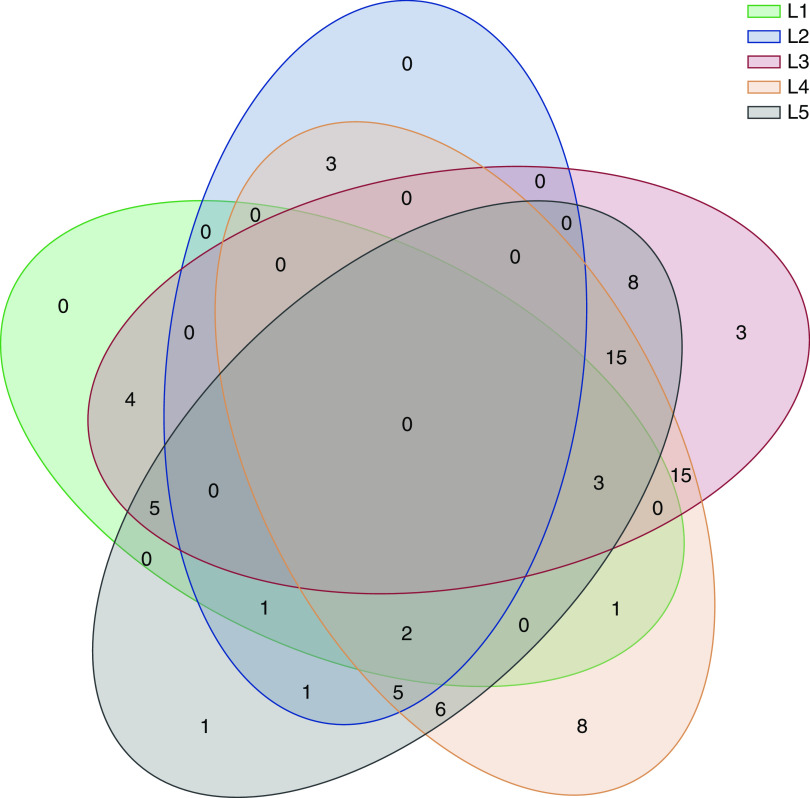
Among 12 patients with L2 impact, four diagnoses were changed from FSGS to Alport syndrome. Additionally, in 53 patients, diagnostic certainty became possible only with genetic testing (L3). Other effects on management included evaluation of other body organ systems (L4) and cascade family testing (L5) in 58 and 47 patients, respectively. Although cascade testing should have been done for every patient with kidney disease attributable to an autosomal dominant genetic variant, this was not possible in some families due to health-insurance coverage of parents or other family members. In three families, reproductive genetic counseling immediately affected the family’s decision making for their family planning. Figure 1 summarizes the overlaps and relationships between five levels of the proposed scoring system. Other important effects on management included screening of potential living related kidney donors, planning for solid organ transplantation, and accurate genetic counseling. The discovery of inherited pathogenic variants in autosomal dominant disease genes led, for instance, to the discovery of previously unrecognized clinical abnormalities in parents (e.g., patients RGC-0003 and RGC-0087) and the illumination of unusual inheritance patterns (e.g., pseudodominance in patient RGC-0080).
Figure 1.
Overlaps between L1, L2, L3, L4, and L5 are demonstrated as a Venn diagram. In 3 patients both L1 and L2 were noted. L1, effect on medical and/or surgical treatment; L2, change of medical diagnosis; L3, providing diagnostic certainty; L4, subsequent evaluation of other body-system involvement; L5, cascade family member testing.
Discussion
In this study, the detection rate (81/158, 51%) and the clinical utility of genetic evaluation/testing was demonstrated for pediatric kidney disorders in an RGC setting. In 31% (25/81) of the patients with positive results, immediate medical/surgical treatment was affected, or the prior diagnoses (achieved by either biopsy or clinical evaluation) were changed.
This clinic is staffed by several pediatric nephrologists with an interest in inherited kidney diseases, a clinical geneticist, and a genetic counselor, and is supported by a strong clinical and human genetics program at Baylor College of Medicine. The detection rate of 51% is within the range of other centers around the world and in the United States. We believe the detection rate can vary on the basis of the reasons for referral and the number of patients assessed. The clinical impact scoring system proposed in this study can potentially be applicable to other centers.
RGCs are the optimal mechanism for integrating a comprehensive genetic evaluation with appropriate molecular testing on a clinical basis (14). This kind of clinic also allows for a family-centered approach, where unaffected relatives may also be evaluated and counseled on their risks for kidney disease. Examples of these clinics in Australia, the United Kingdom, and China showed diagnostic yields of 46% (15), 42% (16), and 42% (7), respectively. In the United States, there are several kidney genetics clinics. In a recent publication, a detection rate of 60% was identified among 41 patients who are mostly in the adult age range (17).Our diagnosis yield is at the same scale, and the variations in detection rate could be explained by following factors. First, the indications of referral among these different clinic models are not the same. Second, broad genetic testing options were available in our center. Lastly, patients were referred with rigorous initial evaluation by pediatric nephrologists. A less-stringent referral criterion may lead to a larger number of patients being seen with a higher number of total positive diagnoses, but with an overall lower diagnosis rate.
Our testing approach used various combinations of targeted panels, CMA, and ES (by CLIA laboratories). This resulted in the identification of pathogenic SNVs in 34 different genes and 11 unique pathogenic CNVs. Of these changes, 21 are novel and have not previously been reported in published databases (Table 2). These novel variants, although not previously reported, are classified as pathogenic or likely pathogenic on the basis of American College of Medical Genetics and Genomics criteria by board-certified clinical molecular geneticists at CLIA-certified laboratories. In terms of testing performance, our diagnostic yield is higher than the reported yield of ES for adult patients with kidney disease in one study (18), although a higher detection rate was reported in another study with more selective criteria for testing (17).Overall, these findings may highlight the increased contribution of genetic abnormalities in the pediatric population. The diagnostic rate of CAKUT in this cohort is higher than that expected from the literature (19). This is likely due to stringent referral criteria that select patients who are syndromic.
Our patients were placed into one of five categories on the basis of their clinical presentation and presumed diagnosis. Each category varied in terms of which genetic testing was felt to be the most appropriate both initially and upon follow-up. For instance, panel testing (known to be cost-effective and specific) was very useful in cases of both cystic kidney disease and hematuria. For patients with cystic kidneys in particular, a panel appeared to be a good initial diagnostic choice because of the high prevalence of PKD1 pathogenic variants. If this test result was negative, or if patients had other concerning physical or clinical abnormalities, expanded testing could be pursued with ES or CMA. This allowed us to identify diagnostic variants in genes not previously considered. For instance, a patient initially referred for cystic kidney disease was later found to have a pathogenic variant in HNF1B, more commonly associated with CAKUT (patient RGC-0164); whereas another patient was diagnosed with biallelic variants in BBS12, indicative of Bardet–Biedl syndrome (patient RGC-0117).
Although ADPKD can be diagnosed by imaging studies, genetic diagnoses add certainty and might be the only option for an accurate diagnosis in young children. In this study, only children with cystic kidneys who had a positive family history or clinical suspicion of ADPKD underwent genetic testing. As shown in recent literature (20,21), genotype information in patients with ADPKD can provide prognostic value and can also be used to manage patients differently on the basis of newly developed therapies. Certainly, this is true when the patients reach the age of 18 when therapy can be provided, if indicated.
Most importantly, genetic evaluation resulted in recommendations for immediate medical or surgical treatment in 20% (16/81) of patients. In addition, the original diagnosis in 15% (12/81) of patients was changed. The benefits of L1 impact on management included targeted therapies and preventing the use of inappropriate treatments (i.e., corticosteroids where there was no expectation of benefit). We compared diagnosis pre- and postgenetic evaluation and concluded that genetic testing improved diagnostic accuracy given that the diagnosis might be different from what was previously achieved by clinical or pathologic evaluations. The change of diagnosis from FSGS to Alport syndrome, reported in this study, was also published by other investigators (22). Additional benefits included reducing the use of invasive diagnostic procedures, such as kidney biopsy. Reduction of genetic testing costs will ultimately result in the precise diagnosis of patients for whom an initial syndromic diagnosis was not clinically suspected. In addition to a confirmatory diagnosis, a genetic diagnosis may also provide prognostic information, establish a targeted surveillance of other organs, and facilitate kidney transplant and reproductive planning (6).
However, this study has the following limitations. First, the design of this study is retrospective and there is still a need for larger, prospective studies similar to the recent research published from an Australian group (23). Second, we did not study the patients’ viewpoints of genetic or genomic testing. Third, although our study included a range of diagnoses, the relatively small overall number/type of patients evaluated in this clinic may affect generalization of our data. Fourth, only a pediatric population was studied. Finally, although we have investigated the health effects of genetic testing, the economic effect of this testing in kidney disease was not studied.
Strengths of this study include the following: (1) the ability to perform advanced clinical genetic testing for a large proportion of our patients; (2) the diversity of the cohort, specifically their ethnicity, kidney phenotypes, and clinical diagnoses; (3) access to world-class pediatric nephrology and clinical genetics groups; and (4) affiliation with one of the largest children’s hospitals in the United States.
In conclusion, results of RGC in a single center is summarized to define the effect of genetic testing and evaluation on management of patients in a pediatric nephrology clinical setting. An overall detection rate of 51% is in line with other reports across the world and in the United States. A new classification for the effect of clinical genetic evaluation on management of patients is provided. In 20% of the patients, medical or surgical management was modified, and clinical diagnosis was changed to a more accurate genetic diagnosis in 15% of the patients.
Disclosures
W. Chen reports receiving other from PreventionGenetics LLC, during the conduct of the study, and other from PreventionGenetics LLC, outside the submitted work. D.J. Lamb reports receiving other from Celmatix, and other from Fellow, outside the submitted work. All remaining authors have nothing to disclose.
Funding
This study was supported, in part, by National Institute of Diabetes and Digestive and Kidney Diseases grants K12DK0083014 (under the Multidisciplinary K12 Urologic Research Career Development Program; M. R. Bekheirnia was a K12 scholar) and R01DK078121 to D.J. Lamb. D.J. Lamb is also supported, in part, by the New York Community Trust Frederick J. and Theresa Dow Wallace Fund. This study was also supported by start-up funds from the Renal Section, Department of Pediatrics at TCH, Baylor College of Medicine, and Woods Family Foundation.
Acknowledgments
The authors would like to acknowledge the many patients and their family members. The authors would like to thank Mr. Yash R. Choksi (B.S.) for his kind contribution to create a Venn diagram demonstrating the overlap of different levels of the clinical impact scoring system. The authors would additionally like to thank all referring physicians from either within, or outside of, the TCH medical system. The authors would also like to thank the following diagnostic laboratories for their superb genetic/genomic testing: Baylor Genetics, GeneDx, and PreventionGenetics.
Author Contributions
M.R. Bekheirnia was responsible for funding acquisition and validation; M.R. Bekheirnia and N. Bekheirnia were responsible for formal analysis and resources; M.R. Bekheirnia, N. Bekheirnia, and M.C. Braun were responsible for investigation; M.R. Bekheirnia, N. Bekheirnia, M.C. Braun, and K. E. Glinton were responsible for methodology; M.R. Bekheirnia, N. Bekheirnia, M.C. Braun, K.E. Glinton, and D.J. Lamb provided supervision; M.R. Bekheirnia, N. Bekheirnia, W. Chen, K.E. Glinton, J. Manor, and L. Rossetti were responsible for data curation; M.R. Bekheirnia, N. Bekheirnia, and K.E. Glinton conceptualized the study and were responsible for project administration and visualization; and all authors wrote the original draft and reviewed and edited the manuscript.
Supplemental Material
This article contains supplemental material online at http://kidney360.asnjournals.org/lookup/suppl/doi:10.34067/KID.0002272020/-/DCSupplemental.
Panels (utilized in this study) and the genes included in each of them. Download Supplemental Table 1, PDF file, 263 KB (263KB, pdf)
"Other" indications for referral. Download Supplemental Table 2, PDF file, 263 KB (263KB, pdf)
Detection rates of different tests among indications for referral. Download Supplemental Table 3, PDF file, 263 KB (263KB, pdf)
Impact on management in patients with partial diagnosis. Download Supplemental Table 4, PDF file, 263 KB (263KB, pdf)
Demographics, phenotype, genetic variants’ information, clinician’s comments and recommendations for patients without diagnostic result who were found to have variants of uncertain significance (VUS). Download Supplemental Table 5, PDF file, 263 KB (263KB, pdf)
References
- 1.Sadowski CE, Lovric S, Ashraf S, Pabst WL, Gee HY, Kohl S, Engelmann S, Vega-Warner V, Fang H, Halbritter J, Somers MJ, Tan W, Shril S, Fessi I, Lifton RP, Bockenhauer D, El-Desoky S, Kari JA, Zenker M, Kemper MJ, Mueller D, Fathy HM, Soliman NA, Hildebrandt F; SRNS Study Group: A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol 26: 1279–1289, 2015. 10.1681/ASN.2014050489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halbritter J, Baum M, Hynes AM, Rice SJ, Thwaites DT, Gucev ZS, Fisher B, Spaneas L, Porath JD, Braun DA, Wassner AJ, Nelson CP, Tasic V, Sayer JA, Hildebrandt F: Fourteen monogenic genes account for 15% of nephrolithiasis/nephrocalcinosis. J Am Soc Nephrol 26: 543–551, 2015. 10.1681/ASN.2014040388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bullich G, Domingo-Gallego A, Vargas I, Ruiz P, Lorente-Grandoso L, Furlano M, Fraga G, Madrid Á, Ariceta G, Borregán M, Piñero-Fernández JA, Rodríguez-Peña L, Ballesta-Martínez MJ, Llano-Rivas I, Meñica MA, Ballarín J, Torrents D, Torra R, Ars E: A kidney-disease gene panel allows a comprehensive genetic diagnosis of cystic and glomerular inherited kidney diseases. Kidney Int 94: 363–371, 2018. 10.1016/j.kint.2018.02.027 [DOI] [PubMed] [Google Scholar]
- 4.Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, Ward P, Braxton A, Wang M, Buhay C, Veeraraghavan N, Hawes A, Chiang T, Leduc M, Beuten J, Zhang J, He W, Scull J, Willis A, Landsverk M, Craigen WJ, Bekheirnia MR, Stray-Pedersen A, Liu P, Wen S, Alcaraz W, Cui H, Walkiewicz M, Reid J, Bainbridge M, Patel A, Boerwinkle E, Beaudet AL, Lupski JR, Plon SE, Gibbs RA, Eng CM: Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 312: 1870–1879, 2014 10.1001/jama.2014.14601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Posey JE, Rosenfeld JA, James RA, Bainbridge M, Niu Z, Wang X, Dhar S, Wiszniewski W, Akdemir ZH, Gambin T, Xia F, Person RE, Walkiewicz M, Shaw CA, Sutton VR, Beaudet AL, Muzny D, Eng CM, Yang Y, Gibbs RA, Lupski JR, Boerwinkle E, Plon SE: Molecular diagnostic experience of whole-exome sequencing in adult patients. Genet Med 18: 678–685, 2016. 10.1038/gim.2015.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jayasinghe K, Stark Z, Patel C, Mallawaarachchi A, McCarthy H, Faull R, Chakera A, Sundaram M, Jose M, Kerr P, Wu Y, Wardrop L, Goranitis I, Best S, Martyn M, Quinlan C, Mallett AJ: Comprehensive evaluation of a prospective Australian patient cohort with suspected genetic kidney disease undergoing clinical genomic testing: A study protocol. BMJ Open 9: e029541, 2019. 10.1136/bmjopen-2019-029541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao J, Liu X, Mao J, Tang X, Shen Q, Li G, Sun L, Bi Y, Wang X, Qian Y, Wu B, Wang H, Zhou W, Ma D, Zheng B, Shen Y, Chen Z, Luan J, Wang X, Wang M, Dang X, Wang Y, Wu Y, Hou L, Sun S, Li Q, Liu X, Bai H, Yang Y, Shao X, Li Y, Zheng S, Han M, Liu C, Cao G, Zhao L, Qiu S, Dong Y, Zhu Y, Wang F, Zhang D, Li Y, Zhao L, Yang C, Luo X, Chen L, Jiang X, Zhang A, Xu H; for “Internet Plus” Nephrology Alliance of National Center for Children’s Care: Genetic spectrum of renal disease for 1001 Chinese children based on a multicenter registration system. Clin Genet 96: 402–410, 2019. 10.1111/cge.13606 [DOI] [PubMed] [Google Scholar]
- 8.Kirsztajn GM, Filho NS, Draibe SA, de Pádua Netto MV, Thomé FS, Souza E, Bastos MG: Fast reading of the KDIGO 2012: guidelines for evaluation and management of chronic kidney disease in clinical practice. J Bras Nefrol 36: 63–73, 2014. 10.5935/0101-2800.20140012 [DOI] [PubMed] [Google Scholar]
- 9.Warejko JK, Tan W, Daga A, Schapiro D, Lawson JA, Shril S, Lovric S, Ashraf S, Rao J, Hermle T, Jobst-Schwan T, Widmeier E, Majmundar AJ, Schneider R, Gee HY, Schmidt JM, Vivante A, van der Ven AT, Ityel H, Chen J, Sadowski CE, Kohl S, Pabst WL, Nakayama M, Somers MJG, Rodig NM, Daouk G, Baum M, Stein DR, Ferguson MA, Traum AZ, Soliman NA, Kari JA, El Desoky S, Fathy H, Zenker M, Bakkaloglu SA, Müller D, Noyan A, Ozaltin F, Cadnapaphornchai MA, Hashmi S, Hopcian J, Kopp JB, Benador N, Bockenhauer D, Bogdanovic R, Stajić N, Chernin G, Ettenger R, Fehrenbach H, Kemper M, Munarriz RL, Podracka L, Büscher R, Serdaroglu E, Tasic V, Mane S, Lifton RP, Braun DA, Hildebrandt F: Whole exome sequencing of patients with steroid- resistant nephrotic syndrome. Clin J Am Soc Nephrol 13: 53–62, 2018. 10.2215/CJN.04120417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL; ACMG Laboratory Quality Assurance Committee: Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med 17: 405–424, 2015. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang YH, Conklin J, Chan W, Roslin NM, Liu J, He N, Wang K, Sundsbak JL, Heyer CM, Haider M, Paterson AD, Harris PC, Pei Y: Refining genotype-phenotype correlation in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 27: 1861–1868, 2016. 10.1681/ASN.2015060648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossetti Sandro, Consugar Mark B, Chapman Arlene B, Torres Vicente E, Guay-Woodford Lisa M, Grantham Jared J, Bennett William M, Meyers Catherine M, Walker Denise L, Bae Kyongtae, Zhang Qin Jean, Thompson Paul A, Miller J Philip, Harris Peter C; CRISP Consortium : Comprehensive molecular diagnostics in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 18[7]: 2143–2160, 2007. 10.1681/ASN.2006121387 [DOI] [PubMed] [Google Scholar]
- 13.Kinoshita Moritoshi, Higashihara Eiji, Kawano Haruna, Higashiyama Ryo, Koga Daisuke, Fukui Takafumi, Gondo Nobuhisa, Oka Takehiko, Kawahara Kozo, Rigo Krisztina, Hague Tim, Katsuragi Kiyonori, Sudo Kimiyoshi, Takeshi Masahiko, Horie Shigeo, Nutahara Kikuo: Technical Evaluation: Identification of Pathogenic Mutations in PKD1 and PKD2 in Patients with Autosomal Dominant Polycystic Kidney Disease by Next-Generation Sequencing and Use of a Comprehensive New Classification System. PLoS One 11[11]: e0166288, 2016. 10.1371/journal.pone.0166288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mallett A, Corney C, McCarthy H, Alexander SI, Healy H: Genomics in the renal clinic - translating nephrogenetics for clinical practice. Hum Genomics 9: 13, 2015. 10.1186/s40246-015-0035-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mallett AJ, McCarthy HJ, Ho G, Holman K, Farnsworth E, Patel C, Fletcher JT, Mallawaarachchi A, Quinlan C, Bennetts B, Alexander SI: Massively parallel sequencing and targeted exomes in familial kidney disease can diagnose underlying genetic disorders. Kidney Int 92: 1493–1506, 2017. 10.1016/j.kint.2017.06.013 [DOI] [PubMed] [Google Scholar]
- 16.Alkanderi S, Yates LM, Johnson SA, Sayer JA: Lessons learned from a multidisciplinary renal genetics clinic. QJM 110: 453–457, 2017. 10.1093/qjmed/hcx030 [DOI] [PubMed] [Google Scholar]
- 17.Thomas CP, Freese ME, Ounda A, Jetton JG, Holida M, Noureddine L, Smith RJ: Initial experience from a renal genetics clinic demonstrates a distinct role in patient management [published correction appears in Genet Med, 2020 10.1038/s41436-020-01000-0]. Genet Med 22: 1025–1035, 2020. 10.1038/s41436-020-0772-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groopman EE, Marasa M, Cameron-Christie S, Petrovski S, Aggarwal VS, Milo-Rasouly H, Li Y, Zhang J, Nestor J, Krithivasan P, Lam WY, Mitrotti A, Piva S, Kil BH, Chatterjee D, Reingold R, Bradbury D, DiVecchia M, Snyder H, Mu X, Mehl K, Balderes O, Fasel DA, Weng C, Radhakrishnan J, Canetta P, Appel GB, Bomback AS, Ahn W, Uy NS, Alam S, Cohen DJ, Crew RJ, Dube GK, Rao MK, Kamalakaran S, Copeland B, Ren Z, Bridgers J, Malone CD, Mebane CM, Dagaonkar N, Fellström BC, Haefliger C, Mohan S, Sanna-Cherchi S, Kiryluk K, Fleckner J, March R, Platt A, Goldstein DB, Gharavi AG: Diagnostic Utility of exome sequencing for kidney disease. N Engl J Med 380: 142–151, 2019. 10.1056/NEJMoa1806891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Ven AT, Connaughton DM, Ityel H, Mann N, Nakayama M, Chen J, Vivante A, Hwang DY, Schulz J, Braun DA, Schmidt JM, Schapiro D, Schneider R, Warejko JK, Daga A, Majmundar AJ, Tan W, Jobst-Schwan T, Hermle T, Widmeier E, Ashraf S, Amar A, Hoogstraaten CA, Hugo H, Kitzler TM, Kause F, Kolvenbach CM, Dai R, Spaneas L, Amann K, Stein DR, Baum MA, Somers MJG, Rodig NM, Ferguson MA, Traum AZ, Daouk GH, Bogdanović R, Stajić N, Soliman NA, Kari JA, El Desoky S, Fathy HM, Milosevic D, Al-Saffar M, Awad HS, Eid LA, Selvin A, Senguttuvan P, Sanna-Cherchi S, Rehm HL, MacArthur DG, Lek M, Laricchia KM, Wilson MW, Mane SM, Lifton RP, Lee RS, Bauer SB, Lu W, Reutter HM, Tasic V, Shril S, Hildebrandt F: Whole-exome sequencing identifies causative mutations in families with congenital anomalies of the kidney and urinary tract. J Am Soc Nephrol 29: 2348–2361, 2018. 10.1681/ASN.2017121265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torres VE, Chapman AB, Devuyst O, Gansevoort RT, Grantham JJ, Higashihara E, Perrone RD, Krasa HB, Ouyang J, Czerwiec FS; TEMPO 3:4 Trial Investigators: Tolvaptan in patients with autosomal dominant polycystic kidney disease. N Engl J Med 367: 2407–2418, 2012. 10.1056/NEJMoa1205511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torres VE, Chapman AB, Devuyst O, Gansevoort RT, Perrone RD, Koch G, Ouyang J, McQuade RD, Blais JD, Czerwiec FS, Sergeyeva O; REPRISE Trial Investigators: Tolvaptan in later-stage autosomal dominant polycystic kidney disease. N Engl J Med 377: 1930–1942, 2017. 10.1056/NEJMoa1710030 [DOI] [PubMed] [Google Scholar]
- 22.Malone AF, Phelan PJ, Hall G, Cetincelik U, Homstad A, Alonso AS, Jiang R, Lindsey TB, Wu G, Sparks MA, Smith SR, Webb NJ, Kalra PA, Adeyemo AA, Shaw AS, Conlon PJ, Jennette JC, Howell DN, Winn MP, Gbadegesin RA: Rare hereditary COL4A3/COL4A4 variants may be mistaken for familial focal segmental glomerulosclerosis. Kidney Int 86: 1253–1259, 2014. 10.1038/ki.2014.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jayasinghe K, Stark Z, Kerr PG, Gaff C, Martyn M, Whitlam J, Creighton B, Donaldson E, Hunter M, Jarmolowicz A, Johnstone L, Krzesinski E, Lunke S, Lynch E, Nicholls K, Patel C, Prawer Y, Ryan J, See EJ, Talbot A, Trainer A, Tytherleigh R, Valente G, Wallis M, Wardrop L, West KH, White SM, Wilkins E, Mallett AJ, Quinlan C: Clinical impact of genomic testing in patients with suspected monogenic kidney disease [published online ahead of print September 17, 2020]. Genet Med 10.1038/s41436-020-00963-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Panels (utilized in this study) and the genes included in each of them. Download Supplemental Table 1, PDF file, 263 KB (263KB, pdf)
"Other" indications for referral. Download Supplemental Table 2, PDF file, 263 KB (263KB, pdf)
Detection rates of different tests among indications for referral. Download Supplemental Table 3, PDF file, 263 KB (263KB, pdf)
Impact on management in patients with partial diagnosis. Download Supplemental Table 4, PDF file, 263 KB (263KB, pdf)
Demographics, phenotype, genetic variants’ information, clinician’s comments and recommendations for patients without diagnostic result who were found to have variants of uncertain significance (VUS). Download Supplemental Table 5, PDF file, 263 KB (263KB, pdf)