Summary
Neurodegenerative diseases are characterized by progressive cell loss leading to disruption of the structure and function of the central nervous system. Amyotrophic lateral sclerosis (ALS) was among the first of these disorders modeled in patient-specific iPSCs, and recent findings have translated into some of the earliest iPSC-inspired clinical trials. Focusing on ALS as an example, we evaluate the status of modeling neurodegenerative diseases using iPSCs, including methods for deriving and using disease-relevant neuronal and glial lineages. We further highlight the remaining challenges in exploiting the full potential of iPSC technology for understanding and potentially treating neurodegenerative diseases such as ALS.
Keywords: Pluripotent stem cells, clinical translation, disease modeling, neurodegenerative diseases, ALS, FTD, motor neurons: cortical neurons, astrocytes, microglia
Introduction
Neurodegenerative diseases such as Amyotrophic Lateral Sclerosis (ALS), Parkinson’s disease (PD), and Alzheimer’s disease (AD), are characterised by a high level of etiological heterogeneity, with diverse genetic causes, environmental factors and complex pathophysiologies played out in the multicellular environment of the aging nervous system. This complexity poses significant challenges for in vitro modeling. However, the advent of human induced pluripotent stem cell (hiPSC) technology (Takahashi and Yamanaka, 2006) has dramatically changed our ability to create physiologically relevant in vitro models for such diseases. As the genetic background of the cell donor is maintained during cell reprogramming, hiPSCs provide an excellent means to evaluate the effect of disease-causing mutations on the relevant, otherwise non-accessible cell types in neurodegenerative disorders, such as neurons and glial cells. In addition to monoculture differentiation protocols, more complex in vitro models, including multicellular and three-dimensional (3D) culture compositions, are now also becoming available to capture disease-relevant cellular interactions.
ALS has a significant monogenic contribution to causation, and the cell types (principally spinal motor neurons) affected by the disease can be produced to a high level of purity in vitro from stem cells. It therefore serves as an exemplar for the use of hiPSCs for discovery and translational neuroscience with implications more widely for the fields of neurodegenerative disease modeling and stem cell differentiation. In this review, we critically evaluate hiPSC differentiation protocols for the most ALS-relevant cell types (spinal motor neurons, cortical neurons, astrocytes and microglia) and summarize the impact of these models on our understanding of ALS pathogenesis. We also provide a perspective on the use of multicellular and 3D models, then review the remaining challenges and possible solutions in hiPSC research, including emerging technologies, and the role of hiPSC-based findings in improving prospects for successful clinical trials in ALS.
Amyotrophic lateral sclerosis: disease mechanisms and pathology
Amyotrophic lateral sclerosis (ALS), is a neurodegenerative disorder with a prevalence of ~5/100,000 people and a mean age of onset of ~64 years (life-time risk ~1:400) in populations of European genetic heritage (Chio et al., 2013; Kiernan et al., 2011). Characterized by the degeneration of MNs in the cortex, brainstem, and spinal cord, ALS results in progressive muscle weakness and paralysis, with death typically due to neuromuscular respiratory failure a median of 2.5 years from symptom onset, although there is considerable heterogeneity, with approximately 5% of cases surviving for more than 10 years (Chio et al., 2011).
Approximately 90% ALS cases are considered sporadic (sALS), in the absence of a family history of ALS or the related condition frontotemporal dementia (FTD). However, in ~10% of all ALS patients, including in a significant minority of apparently sALS cases, a disease-determining genetic variant can be identified, suggesting that the genetic contribution to ALS risk is substantially driven by rare variants (Talbot et al., 2018). The commonest mutation, a dynamic hexanucleotide repeat expansion (HRE) GGGGCC (G4C2) in the first intron of C9orf72 (DeJesus-Hernandez et al., 2011; Renton et al., 2011), accounts for 20-50% of fALS cases and 3-5% of sALS patients, respectively (Kiernan et al., 2021). In another 10-20% of fALS and 1-2% of sALS cases, mutations are identified in the SOD1 gene (Rosen et al., 1993). Mutations in TARDBP (encoding for the 43 kDa transactive response DNA binding protein TDP-43) and FUS are each found in 5% of fALS and less than 1% of sALS cases (Sreedharan et al., 2008; Vance et al., 2009), while a multitude of other, less commonly mutated genes, including MAT3, OPTN, UBQLN2, TBK1, and NEK1, are also known to cause fALS and sALS. Based on founder effects, the contribution of genetic mutations to ALS varies between populations of different geographic origin. For instance, mutations in C9orf72 are rare in Japan (Ogaki et al., 2012), while mutations in TARDBP are common in Sardinia (Borghero et al., 2014).
Neuropathologically, almost all ALS cases (~97%), except for the ~3% caused by mutations in the FUS and SOD1 genes, show nuclear clearing and cytoplasmic aggregation of TDP-43 in neuronal and non-neuronal cells (Neumann et al., 2006). This characteristic TDP-43 pathology is also found in 40-50% of patients with isolated frontotemporal degeneration (FTD), a neurodegenerative disease characterized clinically by deterioration of social cognition and language, with relative preservation of memory, and pathologically by frontotemporal lobar degeneration (FTLD). Clinical overlap exists between ALS and FTD, with 3-5% of ALS patients showing overt behavioral variant FTD, while more subtle forms of loss of executive functions are found in up to 50% of ALS patients (Beeldman et al., 2018; Talbot et al., 2018). This suggests that FTLD-TDP and ALS share substantial biological features and potential treatment strategies (Burrell et al., 2016).
Over the past decades of ALS research, a multitude of in vitro and in vivo models have been created in an effort to recapitulate and study the processes that lead to MN degeneration in ALS patients. In agreement with data from neuroimaging and postmortem studies, these models suggest that ALS arises through a combination of primary neuronal damage with contributions to pathogenesis from non-neuronal cells such as glia (Vahsen et al., 2021). Despite this increasing understanding of ALS pathophysiology, there are currently only two licensed therapeutic options for ALS patients, riluzole and edaravone, which only very moderately improve survival and rate of progression (Kiernan et al., 2021). A large number of drug candidates showing promising results in preclinical models have so far failed to translate into benefits for patients. With the notable exception of antisense oligonucleotides (ASOs), which are currently undergoing clinical trials in patients carrying mutations in the SOD1 or C9orf72 genes, there is no prospect yet of substantial therapeutic options for ALS patients.
The failed development of new drugs that alter natural history can, in part, be explained by a lack of accurate disease models. ALS research has heavily relied on animal models overexpressing mutant forms of SOD1, which only occur in a small fraction of ALS cases, and crucially do not show TDP-43 aggregation. Inherent species differences in gene expression and the functional organization of the motor system also contribute to the difficultly in translating findings from animal models to humans (Herculano-Houzel et al., 2015; Lin et al., 2014; Schieber, 2007). hiPSC technology has offered a new and powerful tool to circumvent such issues through the development of in vitro differentiation protocols for the most ALS-relevant cell types that are discussed below. Methods which directly re-program fibroblasts into relevant cell types, by bypassing the hiPSC stage, are complementary to hiPSC technology for the modeling of conditions such as ALS, since some age-dependent molecular profiles are preserved while they are lost upon hiPSC reprogramming (Mertens et al., 2015). This concept has been reviewed in detail elsewhere (Mertens et al., 2018).
Differentiation of hiPSCs into motor neurons and cortical neurons
From hiPSCs to spinal motor neurons
The direct differentiation of hiPSCs to spinal MNs, which we will refer here as MNs, is the first essential step toward their use in the study of neurodevelopment, neurodegeneration, and potential future use in the clinic. Mimicking embryonic neurodevelopment, the generation of hiPSC-derived MNs involves several now clearly defined developmental steps:
Neural induction starts with inhibition of bone morphogenetic protein BMP and transforming growth factor beta (TGFb) signalling (referred as dual-SMAD inhibition) (Chambers et al., 2009) as well as inhibition of WNT signalling (Wilson et al., 2001), following the neural default model (Munoz-Sanjuan and Brivanlou, 2002). A positive role for fibroblast growth factor (FGF) signaling has been described in the chick embryo (Wilson et al., 2000) but may not be required for anterior neuroecotderm induction in hiPSCs.
Rostral neural progenitors can be induced to adopt a more posterior positional identity, governed by a response to caudalizing signals (i.e. retinoic acid, RA) (Durston et al., 1998; Muhr et al., 1999). Alternatively, posterior neural precursors can be induced directly in the presence of FGF and WNT signalling (Peljto et al., 2010) via induction of a transient posterior precursor referred to as neuro-mesodermal precursor (Lippmann et al., 2015). While the RA-based caudalization yields mostly brachial level progenitors, the WNT/FGF based pattern provides access to trunk and lumbar progenitors.
The resulting spinal progenitor cells then acquire a MN progenitor identity, induced by the ventralizing action of sonic hedgehog (SHH) signaling (Briscoe and Ericson, 2001). The activation of the SHH pathway inhibits the expression of dorsal progenitors in a concentration-dependent manner that mimics the response of primary spinal progenitor cells (Wichterle et al., 2002).
It has become clear that the systematic variation in the identity, timing and concentration of the patterning factors that hiPSCs are exposed to in the early phases can determine the efficiency, identity, and functional maturity of the final cultures, and may explain the discordant results reported by different groups when modelling ALS in hiPSC-derived MNs. The first reported generation of hiPSCs and successful differentiation to MNs from an ALS patient came from the Eggan laboratory in 2008 (Dimos et al., 2008). The authors used a directed differentiation protocol previously developed for mouse and human embryonic stem cells (hESCs) to produce spinal MNs (Lee et al., 2007; Li et al., 2005; Wichterle et al., 2002). This was soon followed by a comprehensive MN differentiation protocol for hESCs and hiPSCs (Hu and Zhang, 2009), and subsequently by numerous publications describing variations in MN differentiation protocols used in ALS research (Table 1).
Table 1:
Spinal MN differentiation protocols used in ALS studies
| Reference | Induction compounds | Protocol length (days) |
Culture Purity (%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SMAD inhibition | Patterning | ||||||||||
| Chir | LDN | SB | Compound C |
RA | SHH | SAG | HB9 | Tuj1 | ChAT | ||
| (Dimos et al., 2008) | ✓ | ✓ | 45 | 20 | - | - | |||||
| (Hu and Zhang, 2009) | ✓ | ✓ | 56 | >40 | 30-40 | - | |||||
| (Karumbayaram et al., 2009) | ✓ | ✓ | 90-95 | - | - | - | |||||
| (Patani et al., 2011) | ✓ | 55 | - | - | - | ||||||
| (Amoroso et al., 2013) | ✓ | ✓ | ✓ | ✓ | 35 | - | - | - | |||
| (Sareen et al., 2013) | ✓ | 75 | - | - | - | ||||||
| (Maury et al., 2015) | ✓ | ✓ | ✓ | ✓ | ✓ | 35 | - | - | - | ||
| (Qu et al., 2014) | ✓ | ✓ | 30 | - | 70 | - | |||||
| (Kiskinis et al., 2014) | ✓ | ✓ | ✓ | ✓ | 55 | - | - | - | |||
| (Devlin et al., 2015) | ✓ | ✓ | ✓ | 130 | 44 | 81 | - | ||||
| (Du et al., 2015) | ✓ | ✓ | ✓ | 28 | - | - | - | ||||
| (Calder et al., 2015) | ✓ | ✓ | ✓ | 20 | 40 | - | - | ||||
| (Bursch et al., 2019) | ✓ | ✓ | ✓ | ✓ | ✓ | >42 | - | 89 | 27 | ||
| (Dafinca et al., 2020) | ✓ | ✓ | ✓ | ✓ | 30 | - | 90 | 90 | |||
| (Ababneh et al., 2020) | ✓ | ✓ | ✓ | ✓ | 30 | 35 | 80 | 80 | |||
| (Mehta et al., 2021) | ✓ | ✓ | ✓ | ✓ | ✓ | 35 | - | - | - | ||
Spinal MN differentiation protocols based on dual SMAD inhibition for the neural induction step rely on small molecule compounds, such as SB431542 (SB, an inhibitor of activin-nodal signaling) and LDN193189 (LDN, an inhibitor of BMP signaling). Several alternative BMP inhibitors have been used including dorsomorphin, recombinant noggin and DMH1 (Neely et al., 2012). In some instances, dual SMAD inhibition is combined with CHIR99021 (CHIR, an activator of the Wnt signaling pathway) that promote posterior identities and neuroepithelial proliferation (Li et al., 2011). Although now commonly used in MN differentiation protocols in ALS (Amoroso et al., 2013; Devlin et al., 2015; Kiskinis et al., 2014; Maury et al., 2015), it is noteworthy that the first studies on MN differentiations did not include a SMAD inhibition stage (Dimos et al., 2008; Hu and Zhang, 2009).
After initial neural induction, hiPSCs must be directed towards a caudal and ventral identity using RA and recombinant SHH or its agonist (SAG) respectively (Wichterle et al., 2002). In 2015, Maury and colleagues performed a comprehensive study of the effect of varying the concentration and timing of these two morphogens on differentiation efficiency as well as impact on MN subtype identity (Maury et al., 2015). This protocol has been used in several recent studies for C9orf72 hiPSC-MNs (Dafinca et al., 2020; Mehta et al., 2021; Selvaraj et al., 2018). Overall, the concentration of RA in different protocols ranges from 100 nM (Hu and Zhang, 2009) to 1 μM (Dafinca et al., 2016; Karumbayaram et al., 2009; Maury et al., 2015), while one study showed the generation of MNs in the absence of activators of retinoid signaling (Patani et al., 2011). Conversely, Calder et al showed in 2015 that efficient MN generation can be obtained by early exposure to RA in the absence of SHH pathway agonists via suppression of GLI3 signaling (Calder et al., 2015).
Before maturation, neurons are generally dissociated and plated at lower densities to stimulate neurite outgrowth. Depending on the protocol, this stage can occur as early as 9 days (Maury et al., 2015; Qu et al., 2014) or as late as 45 days post-induction (Devlin et al., 2015). When final neural maturation is induced, the growth medium is usually supplemented with brain-derived neurotrophic factor (BDNF) and glial-derived neurotrophic factor (GDNF) (Dafinca et al., 2020; Dafinca et al., 2016; Devlin et al., 2015; Dimos et al., 2008; Hu and Zhang, 2009; Kiskinis et al., 2014; Qu et al., 2014; Selvaraj et al., 2018), as well as ciliary neurotrophic factor (CNTF) (Devlin et al., 2015; Dimos et al., 2008; Karumbayaram et al., 2009; Kiskinis et al., 2014) or IGF-1 (Dafinca et al., 2020; Dafinca et al., 2016; Hu and Zhang, 2009; Qu et al., 2014).
Despite the well-defined phases of MN differentiation, the marked variability in differentiation protocols, as well as in the identity of the final cultures, even between different hiPSC lines differentiated in parallel, requires objective assessment of MN identity and purity if studies are to be meaningfully compared. Postmitotic MNs can be identified by specific MN transcription factors, such as ISL1 and HB9, while more mature MNs express choline acetyltransferase (ChAT) and the vesicular acetylcholine neurotransmitter transporter (vAChT) (Amoroso et al., 2013; Karumbayaram et al., 2009). While all MNs will initially follow this pattern of expression, it has become clear that during the differentiation process only a subset of these proteins may be expressed (Amoroso et al., 2013; Maury et al., 2015). Depending on the markers used to define MN identity, the purity of MN cultures reported in the ALS literature ranges from 20-30% (Amoroso et al., 2013; Dimos et al., 2008), 40-50% (Devlin et al., 2015; Hu and Zhang, 2009; Maury et al., 2015) to 95% (Du et al., 2015). One strategy to reduce heterogeneity of MN cultures is to enrich for MNs by fluorescence activated cell sorting (FACS) of neural progenitors, for example by adenoviral transduction with HB9:GFP (Amoroso et al., 2013; Wainger et al., 2014). However, restriction of HB9 expression to early development favors selection of immature neurons which may not be the most relevant disease models for neurodegenerative disorders.
Assessment of the functional maturity of hiPSC-MNs by electrophysiology involve the presence of action potentials and sustained trains of action potentials which occur only at later stages of MN differentiation (Hu and Zhang, 2009). In heterogeneous cultures MNs are generally identified and patch-clamped on the basis of size, not molecular identity. Studies focusing on electrophysiological properties in ALS hiPSC-MNs therefore typically use extended maturation periods (Devlin et al., 2015). However, it is important to note that different subtypes of MNs have different electrophysiological properties such as rate and amplitude of action potential and the presence of action potentials alone is not sufficient to categorize a MN as mature. Other parameters of functional maturity in MN cultures include evidence of synaptic connectivity such as the presence of miniature excitatory or inhibitory postsynaptic currents. Furthermore, whole cell patch clamping is a technically challenging, low throughout method that may be inefficient at determining the real variation present within a neuronal culture. Therefore, there is growing effort to implement optical readouts of neuronal activity that can be monitored across thousands of neurons in parallel such as Calcium imaging, use of fluorescent voltage indicators combined with optogenetic stimulation (“all optical electrophysiology”) (Kiskinis et al., 2018) or activity measurements using multielectrode arrays (MEA) (Wainger et al., 2014), methods that may provide a better representation of overall network activity and connectivity in heterogeneous cultures (Ronchi et al., 2021).
From hiPSCs to cortical neurons
In many neural conversion protocols, hiPSCs undergo the formation of radially organized neuroepithelia (neural rosettes) (Zhang et al., 2001). The neural rosettes assume by default a primitive anterior identity (Pankratz et al., 2007) and yield glutamatergic forebrain neurons with dorsal telencephalic identity in the absence of morphogens (Elkabetz et al., 2008; Li et al., 2009). A default anterior and dorsal identity is also observed when using dual-SMAD inhibition-based neural induction protocols (Chambers et al., 2009).
Most current cortical neuron differentiation protocols do not selectively yield neurons from a particular cortical layer but lead to a mixture of deep and upper layer neurons. Their relevance to ALS, which is characterized by restricted degeneration of specific subsets of cortical neurons affecting corticofugal neurons in the motor cortex and their connections, is therefore less clear. In addition, there is considerable interest in modelling the related neurodegenerative disease FTD, which can occur as part of the ALS disease spectrum, but even less is understood of the specific cortical neuronal subtypes undergoing degeneration. We will briefly discuss here the specific protocols used in C9orf72-ALS/FTD research to generate hiPSC-derived cortical neurons in terms of length and how they differ at various stages (Table 2). However, further efforts are clearly needed in the field to yield cortical neuron differentiation protocols more directly relevant to ALS/FTD research.
Table 2:
Cortical neuron differentiation protocols used in C9orf72-ALS/FTD studies
| Reference | Induction compounds | Expansion phase |
Final maturation factors |
Protocol length (days) |
|---|---|---|---|---|
| (Shi et al., 2012) Used later in (Dafinca et al., 2016) | Dorsomorphin (10 uM) SB (10 uM) N2, B27 | Neurospheres | - | 80 |
| (Almeida et al., 2013) Used later in (Lopez-Gonzalez et al., 2016; Lopez-Gonzalez et al., 2019; Maor-Nof et al., 2021) | bFGF, N2, B27 | Neurospheres | BDNF, GDNF | 72 |
| (Bilican et al., 2012) (Livesey et al., 2014) Used later in (Selvaraj et al., 2018) | N2, B27, Forskolin (10 μM) | Monolayer | BDNF, GDNF | 56 |
Cortical neuron differentiation protocols currently used for ALS/FTD can be broadly defined by three main stages: hiPSCs convert to cortical neurepithelial stem cells, then to progenitor cells, followed by differentiation and maturation into cortical neurons able to fire action potentials and form synapses.
Several studies in which cortical neurons have been differentiated from ALS hiPSCs were based on a protocol by (Delaloy et al., 2010), and have used cells carrying the G4C2 repeat expansion in C9orf72 (e.g. (Almeida et al., 2013; Freibaum et al., 2015; Yuva-Aydemir et al., 2019). Similar to the cortical neuron differentiation protocol by Shi et al. (Shi et al., 2012), the first phase involves dissociation of hiPSC colonies and embryoid body (EB) formation. After a week, EBs are typically attached to laminin-coated dishes and allowed to form neural rosettes (e.g. (Almeida et al., 2013; Shi et al., 2012), which are then lifted and maintained as neurospheres in suspension for 1-2 weeks. Final differentiation is initiated by plating dissociated neurospheres on dishes coated with poly-D-lysine and laminin either without further addition of other growth factors (Shi et al., 2012) or with the addition of BDNF and GDNF to support neurite outgrowth (e.g. (Almeida et al., 2013; Dafinca et al., 2016; Selvaraj et al., 2018).
In addition to the extrinsic factor-based MN and cortical differentiation strategies listed in this section, several groups have developed transcription factor-driven differentiation strategies. Those include the forced expression of Neurogenin 2 (NGN2), ISL1 and LHX3 to convert control, sALS, and C9orf72 ALS/FTD patient-derived iPSCs into induced MNs (iMNs) (Shi et al., 2019; Shi et al., 2018) based on earlier work in mouse ESCs (Mazzoni et al., 2013). Transcription factor-based conversion of iPSCs into iMNs has been used for screening assays and for candidate evaluation studies in the context of SOD1, TARDBP, C9orf72 and sALS (Imamura et al., 2017). Forced expression of Neurogenin 1 (NGN1) and NGN2 (Busskamp et al., 2014; Lam et al., 2017), or NGN2 alone (Zhang et al., 2013), was also sufficient for the rapid induction of cortical-like neurons (iNs) from hiPSCs and can be combined with extrinsic patterning strategies (Nehme et al., 2018) to enhance cortical marker expression. NGN2-driven differentiation was adapted by Shlevkov et al. to perform a high-content screen which identified 6 small-molecule regulators of the axonal transport of mitochondria, some of which rescued transport deficits in iPSC MNs derived from a SOD1-ALS patient (Shlevkov et al., 2019).
Differentiation of hiPSCs into astrocytes and microglia
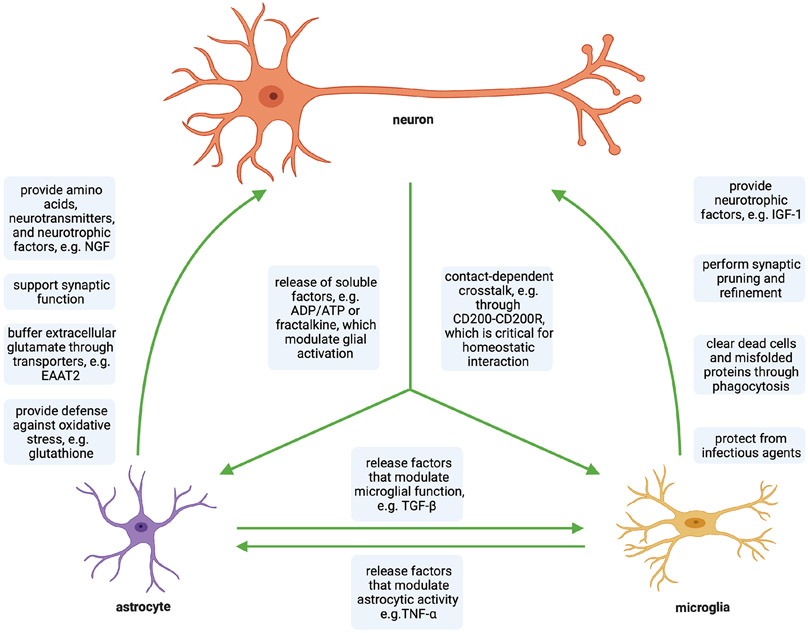
In neurodegenerative disorders such as ALS, astrocytes and microglia are thought to play a role in pathogenesis by promoting an inflammatory state in the brain (neuroinflammation). In the brain and spinal cord, astrocytes and microglia exert their primary functions on neurons, but also signal to one other (Matejuk and Ransohoff, 2020). These physiological, tricellular interactions are summarized in Figure 1. While changes in those interactions have been implicated in ALS (Filipi et al., 2020), it remains unclear what triggers neuroinflammation and how those changes contribute to neurodegeneration in ALS. Nevertheless, controlling neuroinflammation by modulating the communication between MNs and glia is under active study as a potential therapeutic strategy for ALS. The recent development of new differentiation protocols for hiPSC-derived astrocytes and microglia has opened up the opportunity to study the effect of ALS-causing mutations on glial cells as well as the interaction between human glial cells and MNs in vitro. Because the number of astrocyte and microglia differentiation protocols used in ALS studies is still rather limited, we will cover differentiation strategies that have been utilized in both ALS and non-ALS studies.
Figure 1. Physiological interactions between neurons, microglia and astrocytes in the brain and spinal cord:
Neuronal and glial cells interact through various contact-dependent and -independent mechanisms under homeostatic conditions.
From hiPSCs to astrocytes
Protocols to differentiate astrocytes from hiPSCs (Table 3) typically consist of four steps: (1) differentiation of hiPSCs into neural progenitor cells (NPCs); (2) neural patterning to specify astrocytes to defined regions of the CNS; (3) long-term culture or induction of gliogenic switch; and (4) astrocyte terminal differentiation and maturation (reviewed in (Krencik and Zhang, 2011; Tyzack et al., 2016).
Table 3. Existing astrocyte differentiation protocols from hiPSCs.
Protocols used in non-ALS studies are indicated in black, while protocols in ALS studies are indicated in blue.
| Study | Critical factors for terminal differentiation |
Length | Presence of serum in terminal differentiation media |
|---|---|---|---|
| (Hu et al., 2010) | cAMP (1 uM) | > 3 months | No |
| (Krencik and Zhang, 2011) | CNTF (10 ng/ml) | > 3 months | No |
| (Emdad et al., 2012) | CNTF (20 ng/ml) | ~ 5 weeks | No |
| (Juopperi et al., 2012) | Chemically not defined commercial media (ScienCell) | > 3 months | Yes |
| (Lafaille et al., 2012) | 5% FBS | 2–3 months | Yes |
| (Serio et al., 2013) (Birger et al., 2019) (Zhao et al., 2020) | CNTF (10 ug/ml) | ~ 2 months | No |
| (Shaltouki et al., 2013) | CNTF (5ng/ml) BMP (10ng/ml) FGF2 (8ng/ml) 1% FBS Activin A (10ng/ml) Heregulin1b (10ng/ml) IGFI analog (200ng/ml) |
~ 5 weeks from the NSC stage | Yes |
| (Roybon et al., 2013) | hrFGF1 (0.1 to 100ng/ml, then 50ng/ml) hrFGF2 (0.1 to 100ng/ml, then 50ng/ml) |
> 80 days | Yes |
| (Sareen et al., 2014) | EGF (100ng/ml) FGF2 (100ng/ml) CNTF (−) |
> 2 months | No |
| (Mormone et al., 2014) | CNTF (20 ng/ml) | 28-35 days | No |
| (Meyer et al., 2014) (Hautbergue et al., 2017) (Varcianna et al., 2019) | 10% FBS | > 7 days from induced NPCs (iNPCs) | Yes |
| (Holmqvist et al., 2015) | FGF2 (20 ng/ml) EGF (100 ng/ml) |
~ 45 days | No |
| (Pasca et al., 2015; Sloan et al., 2017) | - | From day 100 | No |
| (Du et al., 2015) (Madill et al., 2017) (NB: Based on (Krencik and Zhang, 2011)) | 10% FBS | > 3 months | Yes |
| (Zhou et al., 2016) | Ascorbic Acid (0.2mM) | > 28 days | No |
| (Yasui et al., 2017) | LIF (1/1.000, Wako) 1% and 5% FBS |
~ 28 days from hNPCs | Yes |
| (Tew et al., 2017) and (Soubannier et al., 2020) | Chemically not defined commercial media (ScienCell) | ~ 30 days from hNPCs | Yes |
| (Hall et al., 2017) (Smethurst et al.,2020) | BMP4 (10 ng/ml) LIF (10 ng/ml) |
~ 3 months | No |
| (Perriot et al., 2018) | CNTF (20 ng/ml) | 2–3 months | No |
| (Canals et al., 2018) | dbcAMP (500 mg/ml) heparin-binding EGF-like growth factor (5 ng/ml) BMP4 (10 ng/ml) CNTF (10 ng/ml) |
< 28 days | No |
| (Lundin et al., 2018) | FGF2 (8 ng/mL) heregulin 1β (10 ng/mL) IGF1 (200 ng/mL) ActivinA (10 ng/mL) |
> 28 days | No |
| (Li et al., 2018) | BMP4 (10 ng/mL) CNTF (10 ng/mL) |
4-7 weeks | No |
| (Rosati et al., 2018) | 2% FBS | > 5 weeks from neurospheres | Yes |
| (Tchieu et al., 2019) | HB-EGF (10ng/ml) CNTF (10 ng/mL) LIF: 10ng/ml |
> 4 weeks | No |
| (Bradley et al., 2019) | BMP4 (10 ng/ml) CNTF (10 ng/ml |
~ 3 months | No |
| (Raman et al., 2020) | BMP4 (10 ng/mL) heregulin-β (10 ng/mL) CNTF (10 ng/mL) |
> 50 days | No |
| (Barbar et al., 2020) | T3 (60ng/mL) cAMP (1 μM) Ascorbic Acid (20 μg/mL) |
> 2 months | No |
| (Leventoux et al., 2020) | BDNF (10 ng/ml) GDNF (10 ng/ml) |
From day 48 | No |
| (Peteri et al., 2021) | CNTF (20 ng/mL) | > 75 days | No |
Astrocyte differentiation protocols from hiPSCs are listed in table 3. During development, gliogenesis follows neurogenesis. In the late embryonic stage and early postnatal period, astrocyte progenitors are specified from NPCs via Notch signalling (Chambers et al., 2001). In vitro, NPCs can be derived from hiPSCs using 3D differentiation methods (Eiraku et al., 2008; Falk et al., 2012; Watanabe et al., 2005) or 2D monolayer strategies such as dual SMAD inhibition (Chambers et al., 2009. Regional specification of NPCs is obtained through the combination of growth factors and cytokines (i.e. FGF8, RA, SHH, BMP), which allows the generation of spinal cord-, midbrain-, and dorsal or ventral-forebrain specific astrocytes (Hjorth, 1993; Holmqvist et al., 2015; Krencik et al., 2011; Krencik and Zhang, 2011; Li et al., 2018; Roybon et al., 2013). Astrocyte lineage specification (gliogenic switch) via long-term NPC expansion and subsequent terminal astrocytic differentiation are then performed in media supplemented with various factors alone or in combination such as cAMP, CNTF, LIF, EGF, FGF2, BMP, or in presence of serum.
In the initial protocols for the derivation of astrocytes from hiPSCs, NPCs were expanded in the presence of RA, SHH and cAMP, cultured in suspension for two months in media consisting of DMEM/F12, N1 supplement and cAMP, after which they were cultured on plastic for an additional 7 days (Hu et al., 2010). Because gliogenesis follows neurogenesis during embryonic development, generation of astrocytes from hiPSCs can be protracted, requiring differentiation periods ranging from 3 to 6 months, making hiPSC studies laborious, costly and difficult to standardize (Studer et al., 2015). A recent method to efficiently generate astrocytes in 4 to 7 weeks from the hiPSC stage uses CRISPR/Cas9-mediated inducible expression of the transcription factor nuclear factor I A (NFIA), or NFIA in combination with SRY-box transcription factor 9 (SOX9) (Li et al., 2018). The rapid, NFIA and SOX9 mediated transcriptional fate conversion was also reported for directly generating mouse astrocytes from postnatal skin fibroblast (Caiazzo et al., 2015). An alternative strategy is the use of NFIA to rapidly trigger the gliogenic switch in early spinal or cortical patterned NPCs followed by the astrocytic differentiation of those glial competent precursors into spinal cord or cortex- related astrocytes from hiPSCs using LIF or serum-based differentiation protocols (Tchieu et al., 2019).
To selectively enrich for astrocytes, glial-committed NPCs were positively sorted for A2B5 by magnetic selection followed by re-plating in neurobasal medium containing CNTF for an additional 2 weeks (Mormone et al., 2014). Alternatively, a nearly pure astrocyte population can be isolated from 3D human cerebral cortical spheroids (hCS), which, after 590 days in culture, were dissociated and immunopanned using anti-Thy1 to harvest neurons and anti-HepaCAM to harvest astrocytes (Sloan et al., 2017), methods developed previously for the isolation of astrocytes from the adult brain. CD49f is another marker which has been proposed to purify human fetal astrocytes in the spinal cord and spinal astrocytes derived from hiPSCs (Barbar et al., 2020). After FACS purification, CD49f+ astrocytes showed trophic support of neurons, glutamate uptake and phagocytic capability. In response to inflammatory stimuli, CD49f+ astrocytes acquired an A1-like reactive state characterized by impaired phagocytosis and glutamate uptake as well as loss of capability to support neuronal maturation. Moreover, conditioned medium from CD49f+ A1 astrocytes was toxic to human and rodent neurons.
However, for precise modeling of region-specific neurodegenerative disorders, it is important to consider that astrocytes possess heterogeneous region-specific morphologies and functions. In rodents, astrocytes are morphologically classified into two major subtypes: protoplasmic and fibrous, which are widely distributed in the brain and spinal cord (reviewed in (Tabata, 2015)). In the human cortex, two additional astrocyte subtypes exist: interlaminar and varicose projection astrocytes (Oberheim et al., 2012). These four astrocyte subtypes are characterized by different GFAP expression and morphology. In addition, functional differences exist between spinal and cortical, ventral and dorsal regions (as reviewed in (Tyzack et al., 2016). For instance, cortical astrocytes express high levels of the glutamate transporter GLT-1, whereas spinal astrocytes express lower levels of GLT-1 but express glycine receptors. The hiPSC-derived astrocytes described to date still possess immature morphological and functional characteristics compared to adult astrocytes and are most likely mixed populations of different astrocyte subtypes, morphologies, and functions. Although some laboratories have attempted to generate cortical or spinal specific astrocytes, their region-specific functions have not been investigated in depth, and we are still lacking efficient ways to guide the astrocyte differentiation toward a well-defined dorsal vs ventral profile. Proper morphological and functional characterization will be key to understanding the pathogenesis of region-specific neurodegenerative disorders, and critical for precise hiPSC disease modeling.
From hPSCs to microglia
While protocols for the differentiation of hiPSCs into astrocytes have now been available for more than a decade, hiPSC-derived microglia have become available only recently (Muffat et al., 2016). These differentiation protocols principally aim to mimic microglial ontogeny, which is distinct from that of Myb-dependent blood macrophages, as microglia arise from Myb-independent yolk sac-derived progenitors (Ginhoux et al., 2010; Gomez Perdiguero et al., 2015; Kierdorf et al., 2013; Schulz et al., 2012). During neurodevelopment, these progenitors migrate into the brain to undergo final maturation to microglia in concert with neurons and then maintain their population by self-renewal (Bruttger et al., 2015). hiPSC-derived microglia are therefore more suitable for disease modelling compared to blood monocyte-derived macrophages, which are employed by some researchers as a proxy for brain macrophages. Several articles have recently reviewed the commonalities and differences between protocols for the differentiation of hiPSC-derived microglia (e.g. (Haenseler and Rajendran, 2019; Hasselmann and Blurton-Jones, 2020; Hedegaard et al., 2020; Speicher et al., 2019; Wurm et al., 2021)). Here, we briefly focus on key principles and limitations of current methods.
Typically, hiPSC microglial differentiation protocols consist of four steps (Table 4): (1) initial patterning into cells corresponding to the primitive streak ; (2) differentiation into hemangioblasts and primitive hematopoietic stem cells; (3) differentiation into myeloid progenitor cells; (4) terminal microglial differentiation and maturation. These milestones are achieved by protocols of diverging complexity and duration, either by simply exposing hiPSCs to different growth factors and cytokines, or additionally employing sophisticated sorting methods (FACS or MACS), or by inducing hypoxia. The initial patterning step can be performed via 3D EB formation (Haenseler et al., 2017; Muffat et al., 2016), or in 2D monolayers (Abud et al., 2017; Douvaras et al., 2017; Guttikonda et al., 2021; McQuade et al., 2018; Pandya et al., 2017; Takata et al., 2017). The differentiation of hemogenic endothelial cells into primitive haematopoetic stem cells is achieved by the addition of cytokines including IL-3 and M-CSF. This leads to the formation of yolk sac-like structures, progressively releasing myeloid progenitor cells into the culture medium, which can be simply harvested and re-plated for terminal differentiation or further enriched by FACS or MACS. Terminal differentiation into hiPSC-derived microglia is typically achieved by continuous CSF1R engagement, typically using IL-34. The resulting hiPSC-derived microglia generated by different protocols show similar properties: they are motile and functionally active in phagocytosis and secretion, and their transcriptome is similar to human fetal microglia. However, the regional identity of these cells, for instance, whether they represent spinal or cortical microglia, is usually ill-defined, emphasizing that there remains a need for better characterization of cells generated using currently available protocols.
Table 4. Existing microglia differentiation protocols from hiPSCs.
Protocols used in non-ALS studies are indicated in black. One protocol used in ALS studies is indicated in blue.
| Study | Critical factors for terminal differentiation |
Length | Compatibility with other cell types demonstrated? |
Presence of serum in terminal media |
|---|---|---|---|---|
| (Muffat et al., 2016) | M-CSF (5 ng/mL) IL-34 (100 ng/mL) |
~74 d | hiPSC-derived neuroglial cultures (2D and 3D) | no |
| (Haenseler et al., 2017) | IL-34 (100 ng/mL) optional GM-CSF (10 ng/mL) | ~45 d | hiPSC-derived cortical neurons | no |
| (Abud et al., 2017) | M-CSF (25 ng/mL) IL-34 (100 ng/mL) TGFβ-1 (50 ng/mL) CD200 (100 ng/mL) CX3CL1 (100 ng/mL) |
~38 d | rat hippocampal neurons, hiPSC-derived brain organoids (3D); transplantation into the cortex of MITRG mice and the hippocampi of AD mice | no |
| (Douvaras et al., 2017) | IL-34 (100 ng/mL) GM-CSF (10 ng/mL) |
~45-60 d | no | no |
| (Pandya et al., 2017) | dependent on co-culture with human astrocytes IL-3 (20 ng/mL) GM-CSF (20 ng/mL) M-CSF (20 ng/mL) FBS (10%) |
~29 d | only for murine iPSC-derived microglia differentiated using this protocol | yes |
| (Takata et al., 2017) | M-CSF (50 ng/mL) | ~46 d | co-culture with hiPSC-derived neurons | no |
| (Amos et al., 2017) | ScienCell Research Microglia Medium (undefined) mixed 1:1 with media containing M-CSF (10 ng/mL) GM-CSF (10 ng/mL) IL-34 (10 ng/mL) TGFβ-1 (2 ng/mL) FBS (1%) |
~40 d | no | yes |
| (McQuade et al., 2018) (NB: simplified protocol based on (Abud et al., 2017); used in (Lorenzini et al., 2020)) | M-CSF (25 ng/mL) IL-34 (100 ng/mL) TGFβ-1 (50 ng/mL) CD200 (100 ng/mL) CX3CL1 (100 ng/mL) |
~38 d | transplantation into the cortex and hippocampi of MITRG mice | no |
| (Brownjohn et al., 2018) (NB: based on (Haenseler et al., 2017)) | IL-34 (100 ng/mL) GM-CSF (10 ng/mL) FBS (10%) |
~24-39 d | hiPSC-derived brain organoids (3D) | yes |
| (Konttinen et al., 2019) | M-CSF (10 or 5 ng/mL) IL-34 (10 or 100 ng/mL) FBS (10%) |
~24-42 d | hiPSC-derived neurons (3D) and cerebral brain organoids | yes |
| (Xu et al., 2019) (NB: based on (Abud et al., 2017)) | ScienCell Research Microglia Medium (undefined) M-CSF (25 ng/mL) GM-CSF (25 ng/mL) IL-34 (50 ng/mL) TGF~-1 (50 ng/mL) IGF-1 (25 ng/mL) |
~37 d | no | yes |
| (Guttikonda et al., 2021) | IL-34 (100 ng/mL) M-CSF (20 ng/mL) FBS (10%) |
~24 d | tri-culture with hiPSC-derived astrocytes and cortical neurons | yes |
| (Reich et al., 2020) (NB: based on (Haenseler et al., 2017)) | IL-34 (100 ng/mL) M-CSF (25 ng/mL) TGFβ-1 (50 ng/mL) |
~28-32 d | hiPSC-derived neurons | no |
Insights into ALS pathophysiology from hiPSC models
A wide range of cellular phenotypes have been described in models of ALS. A major challenge is how to relate these to the pathophysiology of the human disease in a way that improves the prospect of identifying disease-modifying drugs. It is fundamental to appreciate that ALS is a clinical syndrome in which neurodegeneration occurs in the complex network which produces voluntary movement (Talbot et al., 2018), with multiple genetic and non-genetic contributions. Disease modelling in ALS is likely to uncover common cellular phenotypes across different mutations (eg; alterations in excitability, defects in axonal transport), but these may be driven by distinct differences in upstream biological pathways.
Animal models, particularly rodents, continue to have a place in disease modelling in ALS and have provided important clues into core pathogenic pathways and the relative contribution of neuronal and non-neuronal cells in the context of intact tissue architecture, especially the neuromuscular synapse, which shows common features in its organization across species. However, animal models based on genetic manipulation, such as overexpression of human genes in mice from heterologous promoters designed to force a ‘strong’ phenotype, may not faithfully recapitulate the key pathways in human ALS, and may amplify interspecies differences in biology and response to therapy. Complementing vertebrate models, the fruit fly Drosophila Melanogaster and yeast models have been employed by multiple labs in ALS research to gain additional insights into ALS pathogenesis and for drug discovery due to the recapitulation of specific aspects of ALS pathology and the availability of tools to rapidly manipulate gene and protein expression (reviewed in (Di Gregorio and Duennwald, 2018; Liguori et al., 2021; Zhang et al., 2018). In contrast, post-mortem tissue from ALS patients enables access to the full spectrum of disease heterogeneity but only captures a static snapshot at the end-stage of disease. Not all MNs are equally susceptible to degeneration in ALS, and those cells remaining at autopsy may reflect populations relatively resistant to the disease process, making the differentiation between disease drivers and compensatory mechanisms in post-mortem tissue samples difficult.
hiPSCs present a complementary platform to vertebrate, fly and yeast models and post-mortem tissue, facilitating investigation of relevant disease processes in the context of the specific human cell-types targeted in ALS, across all genotypes, and potentially a way to address the difficult problem of modelling sporadic disease. With increased knowledge of neurodevelopment, hiPSCs will provide access to the full repertoire of ALS-relevant human cell types at scale, enabling high-throughput assays such as genetic and chemical screens (Fujimori et al., 2018; Imamura et al., 2017). An up-to-date list of ALS models using hiPSC technology as well as their main findings is provided in Table 5.
Table 5.
ALS models for C9orf72, SOD1, TARDBP and FUS mutations using hiPSC technology
| Mutation | Cell types | Purpose and application |
Key findings/take home messages |
Reference |
|---|---|---|---|---|
| SOD1 | MNs | Defining phenotypes directly linked to SOD1A4V mutation |
|
(Kiskinis et al., 2014) |
| SOD1 | MNs | Studying role of protein inclusions |
|
(Chen et al., 2014) |
| SOD1; C9orf72; FUS | MNs | Electrophysiological properties of MNs |
|
(Wainger et al., 2014) |
| SOD1; FUS | MNs | Determining whether divergent causal mutations exhibit common dysfunctional pathways |
|
(Bhinge et al., 2017) |
| SOD1; TARDBP; C9orf72 | MNs | Screen to repurpose existing drugs for ALS |
|
(Imamura et al., 2017) |
| C9orf72 | MNs | Studying mechanisms of C9 HRE pathology |
|
(Sareen et al., 2013) |
| C9orf72 (FTD) | Neurons | Understanding C9orf72 pathology in FTD |
|
(Almeida et al., 2013) |
| C9orf72 | MNs | Studying the toxic gain-of-function role of C9orf72 in ALS |
|
(Donnelly et al., 2013) |
| C9orf72 | Neurons | Investigating HRE RNA-induced toxicity |
|
(Zhang et al., 2015a) |
| C9orf72 | Neurons | Studying RAN mediated toxicity |
|
(Freibaum et al., 2015) |
| C9orf72; TARDBP | MNs | Characterization of electrophysiological properties of ALS MNs |
|
(Devlin et al., 2015) |
| C9orf72 | Cortical neurons, MNs | Phenotypic characterization of cortical and spinal MNs |
|
(Dafinca et al., 2016) |
| C9orf72 | MNs; Neurons | Understanding C9ORF72 HRE-mediated toxicity |
|
(Lopez-Gonzalez et al., 2016) |
| C9orf72; sALS | Astrocytes, MNs | Investigating mechanisms of impaired proteostasis in MNs |
|
(Madill et al., 2017) |
| C9orf72 | Astrocytes, MNs | Role of astrocytes in humanized system |
|
(Zhao et al., 2020) |
| C9orf72 | Astrocytes, MNs | Studying astrocyte-mediated toxicity |
|
(Birger et al., 2019) |
| C9orf72 | MNs | Investigating proteins with altered nucleocytoplasmic distribution in C9ORF72 HRE-ALS |
|
(Ortega et al., 2020) |
| C9orf72; SOD1; sALS | MNs | Cross comparison of multiOMIC datasets in hiPSC-MNs and patient tissues |
|
(Wong and Venkatachalam, 2019) |
| TARDBP | MNs, neurons | Investigating cell-autonomous mechanisms of TARDBP pathology |
|
(Bilican et al., 2012) |
| TARDBP | MNs | Developing platform for disease modeling and screening |
|
(Egawa et al., 2012) |
| TARDBP | astrocytes, MNs | Investigating non-cell autonomous contribution to TARDBP ALS |
|
(Serio et al., 2013) |
| TARDBP | MNs | Investigate composition and role of TDP-43 aggregates in ALS |
|
(Chou et al., 2018) |
| TARDBP | MNs | Studying mechanisms of TARDBP pathology |
|
(Kreiter et al., 2018) |
| TARDBP | MNs | Comparing calcium dysregulation in C9orf72 and TDP-43 iPS-derived MNs |
|
(Dafinca et al., 2020) |
| TARDBP | MNs | Gaining insights into the role of SGs in pathophysiology |
|
(Fang et al., 2019) |
| FUS | MNs | Studying FUS-induced pathology |
|
(Higelin et al., 2016) |
| FUS | MNs | Mechanisms of FUS pathology |
|
(Ichiyanagi et al., 2016) |
| FUS | MNs | Mechanisms of FUS pathology |
|
(Guo et al., 2017b) |
| FUS | MNs; Neurons | Investigating the role of stress granules in ALS |
|
(Marrone et al., 2018) |
| FUS | hiPSCs; MNs | Understanding nuclear-cytoplasmic transport defects due to FUS mutations |
|
(Naumann et al., 2018) |
| FUS | MNs | Understanding the effect of FUS mutations on neuromuscular junction |
|
(Picchiarelli et al., 2019) |
| sALS | MNs | Modeling sALS |
|
(Burkhardt et al., 2013) |
| sALS | astrocytes, MNs | Investigating astrocyte-mediated toxicity to MNs in human system |
|
(Re et al., 2014) |
| sALS | MNs | Studying non-genetic forms of ALS |
|
(Alves et al., 2015) |
| sALS | MN; SkM | Developing platform for NMJ studies |
|
(Osaki et al., 2018) |
| sALS; FUS; TARDBP; SOD1 | MNs | Develop multi-phenotypic screen to cluster heterogeneous sALS lines |
|
(Fujimori et al., 2018) |
Motor neuron models
i). C9orf72
HRE mutations in the first intron of C9orf72 can be associated with pure ALS, pure FTD or ALS/FTD within the same pedigree, suggesting that the mutation acts on cortical neurons or spinal MNs in concert with complex genetic interactors and age-related cellular events. Given the challenges in generating rodent models of C9orf72-related neurodegeneration (Mordes et al., 2020), hiPSCs represent a unique tool to study the differential effect of the mutation on neuron subtypes and the identification of potential modifiers.
MNs generated from C9orf72 hiPSCs consistently exhibit the characteristic RNA foci and dipeptide repeat-associated non-AUG (RAN) pathology of C9orf72-ALS/FTD (Almeida et al., 2019; Donnelly et al., 2013; Gendron et al., 2017; Sareen et al., 2013), and variable degrees of reduced C9orf72 protein expression (haploinsufficiency) (Donnelly et al., 2013) (Sareen et al., 2013). Although TDP-43 mislocalisation and aggregation is a core feature of C9orf72-ALS/FTD at post-mortem, few studies to date have convincingly demonstrated this in hiPSC-MNs. Whether this means that TDP-43 dysregulation in C9orf72-ALS can be dissociated from other aspects of pathophysiology, or whether further development in cell maturation is required for full pathological processing of TDP-43 is unknown. However, a range of phenotypes reported in C9orf72 lines suggest that the mutation is associated with reduced firing capacity, sensitivity to glutamate toxicity, impaired nucleocytoplasmic transport and altered vesicular and synaptic trafficking (Dafinca et al., 2016; Donnelly et al., 2013; Freibaum et al., 2015; Sareen et al., 2013; Zhang et al., 2015a). The use of hiPSC-MNs has been an important pre-clinical tool in demonstrating the effectiveness of ASO therapy currently in clinical trials (Sareen et al., 2013). C9orf72 hiPSC models therefore remain an important tool for investigating pathophysiology and for validation of drugs targeting key aspects of C9orf72-ALS pathogenesis.
ii). SOD1
SOD1 was the first gene in which mutations were found in ALS, and it is therefore not surprising that SOD1 models have dominated translational research for the last 20 years. However, with the exception of ASO therapy now in Phase 2 clinical trials, the results have been disappointing (Miller et al., 2020). Rodent models tended to emphasize the effects of high levels of misfolded mutant SOD1 on neurons and the contribution of glia and neuroinflammation, but despite a vast literature on work from transgenic mouse models, the mechanism whereby SOD1 mutations cause ALS is still unclear. hiPSC models showed that MNs in monoculture display phenotypes which can be disease relevant and independent of other cells types such as astrocytes. For instance, increased apoptosis, fewer neurites, reduced soma size, and neurite length specifically in MNs derived from a patient carrying the severe SOD1A4V mutant were rescued with genetic correction using a two-step, zinc-finger nuclease (ZFN)-mediated gene targeting strategy (Kiskinis et al., 2014). Other studies also confirmed the effect of mutant SOD1 on neurofilament dynamics, axonal growth and function (Chen et al., 2014; Kim et al., 2020) and that misfolded mutant SOD1 is present in basal culture conditions. However, direct non-cell autonomous toxic effects of mutant SOD1-expressing astrocytes on MN survival have also been demonstrated and are discussed in the section on “non-neuronal cell types” below. Overall, SOD1 mutations have been less extensively studied in hiPSCs compared to C9orf72 or TARDBP.
iii). TARDBP
Given that the majority of ALS cases show TDP-43 pathology at autopsy, hiPSC-MNs carrying mutations in its gene TARDBP are of particular interest in potentially revealing a mechanism for loss of TDP-43 regulation in ALS. Studies have consistently shown that detergent insoluble TDP-43 levels are increased in the cytoplasm, though not generally in the form of clear aggregates. MNs showed reduced growth and survival (e.g. (Serio et al., 2013)), alterations in neurofilament organization and axonal vesicular trafficking (e.g. (Kreiter et al., 2018)), and a range of alterations in mitochondrial and other sub-cellular organelle dysfunction (e.g. (Dafinca et al., 2020)). Without genetic manipulation to specifically label TDP-43, which could promote artefacts, it is not possible to study its dynamic movement between nucleus and cytoplasm and in phase transition to form membraneless organelles which may be critical to the initiation of the pathological cascades driving observed phenotypes.
Of particular interest for its clinical potential, a study from Fang and colleagues performed a high-content screen of compounds to alter stress granule properties and TDP-43 aggregate-like structures in hiPSC-MNs carrying mutations in TARDBP or FUS (Fang et al., 2019). Interestingly, compounds containing extended planar aromatic moieties decreased the recruitment of RNA-binding proteins such as TDP-43 into stress granules leading to reduced formation of TDP-43 aggregate-like structures, suggesting that such compounds could be used to treat ALS patients with TDP-43 pathology.
iv). FUS
Mutations in FUS account for a small minority of familial cases (<5%) and are associated with characteristic TDP-43 negative neuropathological changes at autopsy. Therefore, although an RNA binding protein, FUS may exert its pathological effects through distinct mechanisms from most ALS cases. In accordance with postmortem tissue, FUS cytoplasmic mislocalization and accumulation has been recapitulated across multiple studies of (FUS) mutant hiPSC-MNs(Guo et al., 2017b; Higelin et al., 2016; Ichiyanagi et al., 2016; Marrone et al., 2018; Naumann et al., 2018). In addition to alterations in splicing (Ichiyanagi et al., 2016), FUS has been implicated in altered DNA damage repair (Higelin et al., 2016) and, of particular interest given the lower MN predominant pattern typical of FUS-ALS, in axonal RNA transport (Guo et al., 2017b) and NMJ stability (Picchiarelli et al., 2019).
v). Studies directly comparing different ALS-causing genes and sALS
One caveat to hiPSC studies comparing phenotypes across genotypes is that often such studies are done independently, thus, variability from cell purity, maturation, and the type of MNs generated from the differentiation protocol, as noted later, all act as confounding variables. A study that directly compared electrophysiological properties across hiPSC-MNs derived from SOD1A4V, FUSM511FS, FUSH517Q, and C9orf72 demonstrated that despite differences in firing patterns, all genotypes exhibited hyperexcitability that correlated with reduced cell survival and could be reversed with ezogabine (retigabine), a Kv7 or KCNQ voltage-gated potassium channel activator (Wainger et al., 2014). These data contrasted with studies demonstrating hypoexcitability in ALS hiPSC-MN cultures (Dafinca et al., 2020; Devlin et al., 2015). Longitudinal measurements of excitability in both TDP43M337V and C9orf72 hiPSC-MNs have shown that MNs initially exhibit hyperexcitability which converts to hypoexcitability upon prolonged culture (Dafinca et al., 2020).
Pathological phenotypes and sensitivity to stressors highlight both the overlap and heterogeneity captured by hiPSC models of ALS. Mitochondrial dysfunction and oxidative stress in hiPSC-MNs seem to be shared across multiple genotypes including SOD1A4V, C9orf72, TDP-43and sALS (Alves et al., 2015; Dafinca et al., 2016; Egawa et al., 2012; Kiskinis et al., 2014). However, transcriptomic and proteomic comparisons of C9orf72 and SOD1 hiPSC-MNs revealed differentially altered pathways, also reflected in expression profiles from MNs isolated from post-mortem spinal cord (Kiskinis et al., 2014; Wong and Venkatachalam, 2019). Multiple SOD1 mutant hiPSC-MNs have been linked with ER stress; however, TDP-43 MNs exhibit sensitivity to phophoinositide 3-kinase but not ER stress (Bhinge et al., 2017; Bilican et al., 2012). Conversely, nucleocytoplasmic transport defects have been detected in C9orf72 and TDP43 patient hiPSC-MNs but not with SOD1 mutants, although mislocalization of nuclear pore complex proteins has been reported in SOD1 patients (Chou et al., 2018; Freibaum et al., 2015; Ortega et al., 2020; Wong and Venkatachalam, 2019; Zhang et al., 2015a). Together these data suggest that different genotypes may achieve MN degeneration via divergent mechanisms.
Studies from sALS hiPSCs particularly highlight the ability of hiPSCs to recapitulate the heterogeneous nature of ALS without known genetic origin. Burkhardt et al. ((Burkhardt et al., 2013)) showed that forebrain cortical neurons and spinal MNs derived from three of 16 sALS patient hiPSCs exhibited TDP-43 aggregate-like structures. These aggregate-like structures seemed to recapitulate the pathology in post-mortem tissue samples from one of the same patients from which the hiPSC were derived. This was important as it led to a high-content chemical screen in both forebrain cortical neurons and spinal MNs which identified FDA-approved drugs (i.e. the cardiac glycoside Digoxin) as potential inhibitors of TDP-43 aggregate-like structures, demonstrating the feasibility of patient-derived hiPSC-based disease for drug screening. However, it is noteworthy that this is the only study that has shown TDP-43 aggregate-like structures in iPSC models and that the prototypical ALS neuropathology of TDP-43 aggregation has not been convincingly recapitulated in any other hiPSC-based study to-date. An extensive comparison of hiPSC-MNs derived from fALS patients with SOD1, FUS, and TDP43 mutations as well as 32 sALS patients across multiple cellular phenotypes, including neurite length, cell death, and abnormal protein aggregation, demonstrated variability across these in vitro phenotypes that correlated with patient clinical heterogeneity (Fujimori et al., 2018).
In a study analyzing the transcriptome of hiPSC-MNs from patients carrying mutations in the valosin containing protein (VCP) gene increased intron retention (IR) was identified as a dominant feature of the splicing program during early MN differentiation, which was also observed in independent RNAseq data sets from SOD1 and FUS MNs, with the most significant increase of IR in the Splicing Factor Proline and Glutamine rich (SFPQ) transcript (Luisier et al., 2018). As a result, the SFPQ protein binds extensively to its retained intron and is lost from the nuclei of VCP, FUS and SOD1 MNs. This was also observed in mouse transgenic ALS models and human postmortem tissue from sALS cases, identifying nuclear loss of SFPQ as a unifying hallmark across fALS and sALS.
In a study from 2018, Shi and colleagues found that C9orf72 interacted with endosomes and was required for normal vesicle trafficking and lysosomal biogenesis in hiPSC-MNs. HRE-mediated C9orf72 haploinsufficiency caused neurodegeneration through i) accumulation of glutamate receptors and excitotoxicity and ii) impaired clearance of neurotoxic DPRs (Shi et al., 2018). In 2019, the same group found that iMNs from C9orf72 and several sALS patients showed impaired autophagosome formation and aberrant accumulation of glutamate receptors (Shi et al., 2019). Moreover, treatment with the anticoagulation-deficient form of activated protein C (3K3A-APC) was able to rescue these defects in both C9orf72 and sALS iMNs, decreased DPRs and restored TDP-43 localization. Of note, 3K3A-APC also decreased glutamate receptor expression and proteostasis in vivo in C9orf72 gain- and loss-of-function mouse models. Particularly important for its clinical potential, this study also identified that the ability of 3K3A-APC to rescue ALS iMN survival was dependent on its ability to activate protease-activated receptor 1 (PAR1), which identifies PAR1 as a therapeutic target for both C9orf72 and sALS (Shi et al., 2019).
Non-neuronal cell types
Numerous studies reported activation of astrocytes and microglia in post-mortem tissue samples from ALS patients and in murine models of ALS, implicating neuroinflammation as an important feature of ALS (Vahsen et al., 2021). Murine and human astrocytes differ in their size and complexity (Oberheim et al., 2009) and microglia exhibit important species-specific differences in receptor expression and response to stimuli (Smith and Dragunow, 2014). Thus, hiPSC models have a potentially important role to play in dissecting the interaction of these cells with MNs in ALS pathophysiology, and to identify effective therapies that target astrocytes and microglia and therefore neuroinflammation.
Astrocytes expressing mutant SOD1 killed spinal primary and embryonic mouse stemcell derived MNs through soluble factors (Nagai et al., 2007). Similarly, MNs derived from mouse ESCs carrying the mutant SOD1G93A allele showed neurodegenerative properties when co-cultured with SOD1G93A glial cells (Di Giorgio et al., 2007). Astrocytes isolated from mutant SOD1-overexpressing mice or from postmortem brain tissue of sALS patients displayed toxicity to mouse- and human PSC-derived MNs in vitro (Brites and Vaz, 2014; Haidet-Phillips et al., 2011; Marchetto et al., 2008). Interestingly, this toxicity is specific for MNs as other neuronal subtypes in the same dish remained unaffected (Di Giorgio et al., 2008). Studies on C9orf72 hiPSC astrocytes showed that the C9orf72 mutation leads to both cell- autonomous astrocyte pathology and non- cell autonomous effects on MN pathophysiology. C9orf72 astrocytes co- cultured with MNs caused MNs to undergo a progressive loss of action potential output due to decreases in the magnitude of voltage- activated Na+and K+ currents. Notably, this phenotype was reversed by CRISPR/Cas- 9 mediated excision of the C9orf72 repeat expansion (Zhao et al., 2020). Media conditioned by C9orf72 hiPSC astrocytes increased oxidative stress in wild type MNs, suggesting this contributes to MN degeneration in C9orf72 ALS, although it is noteworthy that this study was conducted in C9orf72 hiPSC astrocytes and hiPSC astrocytes from non-affected donors that were not genetically matched (Birger et al., 2019).
Studies on TARDBP hiPSC astrocytes are scarce in comparison, but TARDBP hiPSC astrocytes have increased TARDBP expression, higher levels of cytoplasmic TDP-43 aggregation, and decreased cell survival compared to wild type controls, but co-culture with MNs did not induce MN cell death (Serio et al., 2013). In addition, a recent study showed that hiPSC MNs are more vulnerable to TDP-43 aggregation and toxicity compared with their astrocyte counterparts, and that astrocytes provide protection from seeded aggregation within MNs by reducing (mislocalized) cytoplasmic TDP-43, TDP-43 aggregation and cell toxicity (Smethurst et al., 2020).
To date, only one study has investigated hiPSC microglia derived from an ALS patient. C9orf72 hiPSC-derived microglia displayed a heightened immune response as well as altered expression of endosomal marker early endosome antigen 1 and lysosomal associated membrane protein 1, which was also confirmed in patient post-mortem tissue samples (Lorenzini et al., 2020). Investigating hiPSC-derived microglia in the context of ALS is crucial, considering the growing evidence for the involvement of microglia in ALS pathophysiology. Importantly, hiPSC-derived microglia express ALS-relevant genes, including C9orf72, SOD1, and TDP-43 (Haenseler et al., 2017), increasing the likelihood of identifying phenotypes in hiPSC-derived microglia from ALS patients carrying mutations in these genes. It will be of particular interest to investigate hiPSC-derived microglia in co-culture paradigms in order to further elucidate their putative toxic properties to MNs. Interestingly, co-culture studies demonstrated that astrocytes derived from C9orf72 but not TDP43 M337 hiPSCs reduced hiPSC-derived MN functional output and survival (Birger et al., 2019; Madill et al., 2017; Serio et al., 2013; Zhao et al., 2020). Systematic investigation of glia across different genotypes in parallel will be essential in future studies to exclude the confounding effects of technical variation.
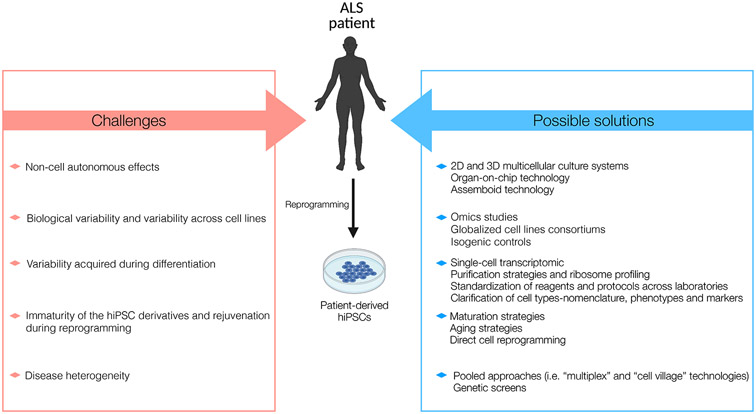
5. Challenges, possible solutions, and emerging technologies in hiPSC research
Despite the potential of hiPSCs to model ALS in vitro, several challenges remain. These, together with possible solutions and emerging technologies in hiPSC research, are summarized in Figure 2.
Figure 2. Challenges and possible solutions in modeling ALS using hiPSC technology.
Several challenges, such as weak technical reproducibility and lack of modelling inter-cellular crosstalk, remain to be resolved to improve disease modelling with hiPSC models. Amongst others, possible solutions include cell purification strategies and the development of more complex, multicellular and three-dimensional culture systems.
Multicellular and 3D culture models to study non-cell autonomous effects in ALS
While simplified hiPSC-derived neuronal, astrocytic, and microglial monoculture paradigms allow the study of cell-autonomous effects of disease-causing mutations, modelling crosstalk between neurons and glia is crucial to understanding their non-cell-autonomous interaction in disease. Co-culture with hiPSC-derived neuronal cells facilitates glial maturation in vitro, as this supports, for example, the ramified homeostatic state of microglia (Haenseler et al., 2017) and the highly branched homeostatic state of astrocytes (Enright et al., 2020). Astrocyte/MN co-cultures from hiPSCs have already been harnessed to study ALS patient-derived cells (Smethurst et al., 2020), and protocols for hiPSC-derived MN/microglia co-culture are in development. However, even more complex in vitro paradigms that incorporate and model the physiological interactions between all three cell types would be ideal. A triculture model of hiPSC-derived cortical neurons, astrocytes, and microglia has been recently described in the context of Alzheimer Disease (AD) (Guttikonda et al., 2021). A similar approach to study ALS will require adaptations to ensure hiPSC-derived MNs, microglia, and astrocytes reflect the spinal subtypes of these cells. Interestingly, recent evidence suggests the existence of multiple different and diverse subtypes of MNs in the mouse spinal cord (Alkaslasi et al., 2021; Blum et al., 2021). It would be intriguing to analyze how the transcriptome of iPSC-derived MNs in monoculture and co-culture corresponds to individual MN subtypes seen in vivo, and whether modifications to existing protocols could lead to an enrichment of different subtypes.
In the CNS, signaling from oligodendrocytes and infiltrating peripheral immune cells, such as macrophages, T-cells and NK cells, also contribute to ALS (Vahsen et al., 2021). In the peripheral nervous system (PNS), skeletal muscle (SkMs) and Schwann cell dysfunction might also play a role in ALS disease onset and progression (Arbour et al., 2017). A ‘motor unit-on-a-chip’ system (Osaki et al., 2020) has been used to test the effect of ALS candidate compounds on MN survival in a microenvironment that recapitulates the crosstalk between MNs and SkMs through the neuromuscular junction (NMJ).
An even more ambitious approach to the investigation of a large range of cell types in 3D is to combine multiple organoids in so-called ‘assembloids’ to model aspects of cellular crosstalk that occur between different tissues or organs (Andersen et al., 2020). Although variability between multiple organoids/assembloids remains a challenge, this technology may be important in understanding more complex, connectivity-dependent mechanisms across neuropsychiatric and neurodegenerative disorders including ALS (Marton and Pasca, 2020).
Overcoming sources of variability in hiPSC production
The emergence of hiPSC-derived models of ALS has coincided with widespread adoption of high-throughput technologies for the unbiased appraisal of the genome, transcriptome, proteome, and epigenome, allowing the in-depth characterization of hiPSC-derived neurons and glia. However, such studies have also demonstrated considerable variability even for cells from the same individual, across independent iPSC inductions and subsequent differentiations.
An extensive study of undifferentiated hiPSCs by the “Human-Induced Pluripotent Stem Cells Initiative” has shown that the primary cause of variability between cell lines is due to genetic variation between individuals, which explains 5.2–26.3% of methylation, gene expression and RNA sequencing differences as well as 21.4–45.8% of differences found in protein immunostaining (Kilpinen et al., 2017). The same study also found significant effects of cell culture conditions and copy number alterations acquired during reprogramming but only minor effects of passage number and biological sex. Other studies have focused on differences between hiPSC lines and established hESC lines, and have identified differences in regional methylation patterns and regulatory gene expression, suggesting an epigenetic memory of the somatic cell of origin (Ohi et al., 2011). However, the degree of variability from such bias appears significantly smaller than the genetic background of the donor, and likely abates after passaging (Bock et al., 2011; Burrows et al., 2016; Nishino et al., 2011). Concerns about reproducibility have therefore accompanied hiPSC-derived disease models including ALS.
One approach to address donor variability has been to greatly increase the number of control and patient replicates, as has been done in the “NeuroLINCS” consortium in ALS (Keenan et al., 2018). The availability of well-characterized cell lines, through international consortia like “Answer ALS” (https://www.answerals.org/) represents an opportunity to overcome the limitations of hiPSC donor variability by analyzing data from MNs differentiated from more than 1,000 patients and control hiPSC lines. In addition to the increased cost associated with scaling up hiPSC production, the challenge of such large, often multicenter, efforts is the introduction of additional sources of variation, related to technical differences in experimental methods, operator-driven differences and sequencing batch effects, which together can obscure the biology of interest (Volpato et al., 2018). On the other hand, the design of such studies enables the identification of these biases, which may otherwise pass by unnoticed in smaller scale experimental designs, and can allow unwanted variation to be regressed out of datasets using appropriate bioinformatic and statistical methods. However, there is a risk that disease-relevant variation may be lost in this process leading to the generation of an ‘idealized dataset’ (Risso et al., 2014).
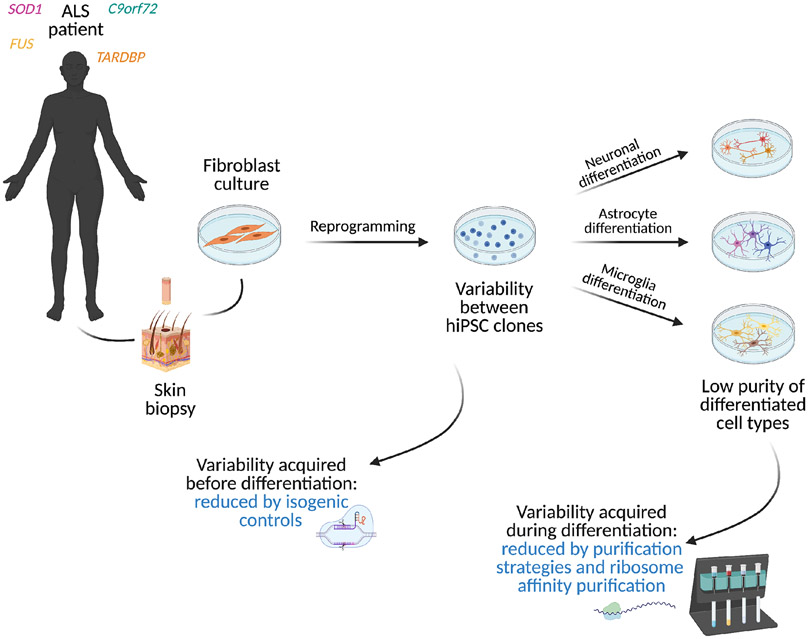
Advances in genomic engineering have significantly improved the ability to control for genetic variability between individuals by allowing the correction of disease-causing mutations in hiPSCs to generate isogenic controls. (Figure 3). Isogenic hiPSCs lines have been developed using CRISPR/Cas9 technology for several genetic mutations in ALS: SOD1 (Bhinge et al., 2017; Bursch et al., 2019; Imamura et al., 2017; Kiskinis et al., 2014; Wang et al., 2017) C9orf72 hexanucleotide repeat expansions (Abo-Rady et al., 2020; Bursch et al., 2019; Dafinca et al., 2020; Lopez-Gonzalez et al., 2019; Selvaraj et al., 2018),TARDBP (Tann et al., 2019),and FUS (R521C) (Bursch et al., 2019; Guo et al., 2017a; Wang et al., 2017).
Figure 3. Overcoming biological variability through isogenic controls and cell purification strategies.
hiPSCs are typically generated by reprogramming skin biopsy-derived fibroblasts into pluripotency. Reprogramming-induced variability between different hiPSC clones can be reduced by generating isogenic control lines, where the mutant allele is corrected through gene-editing. Variability acquired during differentiation can be reduced by cell purification and ribosome affinity purification strategies.
Of note, the correction of HRE in C9orf72 resulted in three non-overlapping datasets between three laboratories (Dafinca et al., 2020). The lack of reproducibility may stem from differences in the CRISPR/Cas9 strategy design between the three experiments but may also reflect the variability of MN differentiations or even be a product of CRISPR/Cas9 off-target effects, which remain poorly characterized and difficult to detect (Zhang et al., 2015b). Furthermore, there is increasing appreciation of deleterious on-target effects after CRISPR/Cas9 editing, for instance mono-allelic deletions or loss of heterozygosity (Weisheit et al., 2020) . Improvements are continuously taking place in the development of this technology and better tools are already available (i.e. CRISPR/Cpf1, Hi-Fi Cas9) to circumvent some of the previous problems in terms of targeting accuracy and efficiency of homology-directed repair.
In the context of isogenic lines, the NIH Intramural Center for Alzheimer’s and Related Dementias recently initiated the largest iPSC genome engineering project to date, termed “iPSC Neurodegenerative Disease Initiative” (iNDI) (Ramos et al., 2021). This project will generate a large series of iPSC lines by engineering a number of disease-causing mutations into deeply characterized parental lines derived from control individuals to model AD, dementia with Lewy bodies/Parkinson’s disease dementia (DLB/PDD), ALS and FTD, and other adult-onset neurodegenerative disorders.
Overcoming sources of variability in hiPSC differentiation
In addition to intrinsic differences between parent lines due to genetics, the differentiation process gives rise to heterogeneous cell populations which differ in both number and composition, with significant amounts of interneurons and microglia reported in culture in some studies (Ho et al., 2021; Thiry et al., 2020). Although the identity of MNs in culture typically corresponds to hindbrain and rostral cord, the relative composition of bulk cultures may also obscure important differences in the relative maturity and subtype of cell populations. Single cell characterization (using transcriptomics and cell sorting) will become an essential technique to dissect MN biology in heterogenous monocultures, and, importantly, will enable appraisal of co-cultures and multicellular model systems, both in 2D and 3D. Sorting of hiPSC-derived cells based on their expressed markers (Haenseler et al., 2017) or exogenously introduced transgenic markers (Kiskinis et al., 2014; Shi et al., 2018), while allowing an assessment of overall subtype composition in bulk cultures, introduces stress through a necessary dissociation step, with cells exhibiting significant rate of cell death, which may bias analysis and subsequent culture.
Another purification technique, which may complement the search for cell specific outputs from hiPSC-derived monocultures and co-cultures, is translating ribosome affinity purification (TRAP), which uses tagged ribosomal proteins under cell-type-specific promoters and which has been successfully applied to capture translated RNA from cell populations of interest in mouse brains. The feasibility of this technique in hiPSC-derived cortical neurons has recently been established, although this experiment was performed using a ubiquitous promoter requiring enrichment using MACS (Rodrigues et al., 2020).
The significant variation in protocols for the differentiation of hiPSC-derived cells with critical roles in ALS argues for harmonization of the criteria used to obtain and characterize the differentiated cell types. An example of such an approach, which might be applied to other cell types relevant to ALS, has recently helped to clarify reactive astrocyte nomenclature, phenotype and markers with the goal to reduce astrocyte heterogeneity and promote the development of universal astrocyte-based biomarkers and therapies (Escartin et al., 2021).
Overcoming immaturity of hiPSC derivatives
ALS is an age-related neurodegenerative disorder. Because hiPSC derivatives more closely resemble a fetal than an adult stage, the biological landscape of these cells may reflect developmental pathways or the pre-symptomatic phase of ALS and may fail to capture ALS disease phenotypes (reviewed in (Guo et al., 2017a)). A detailed overview on promoting aging in hiPSC models of ALS can be found elsewhere (Ziff and Patani, 2019); therefore, we focus on key principles here. Electrical stimulation, including using optogenetics and chemogenetics, metabolic stimulation and multicellular and 3D culture approaches, may all promote maturation. Promoting aging through overexpression of progerin, a truncated form of lamin A associated with premature aging, has been shown to augment disease phenotypes in a model of PD (Miller et al., 2013). However, whether this approach is physiologically relevant to mimic ‘normal’ aging in an iPSC-derived model is uncertain.
Because cellular rejuvenation occurs during hiPSC reprogramming, potentially repairing the macromolecular damage that could contribute to neurodegenerative phenotypes (Studer et al., 2015), another strategy is to bypass the PSC stage by direct conversion of fibroblasts into neurons or glia (Hautbergue et al., 2017; Mertens et al., 2015; Meyer et al., 2014; Varcianna et al., 2019). Directly reprogrammed i-astrocytes (iNPC- As), for example, retain the age-related features of the donor fibroblasts and thus have the potential to capture more relevant disease phenotypes (Gatto et al., 2021). Epigenetic changes are reset during reprogramming, which may reduce the utility of hiPSC-derived cells to model responses to environmental stimuli relevant to ALS, while direct lineage reprogramming approaches more faithfully preserve epigenetic memory (Meyer et al., 2014). However, the cell fate stability of directly reprogrammed cells is unclear, and because fibroblasts have limited self-renewal, reprogrammed cells are often non expandable (Chambers and Studer, 2011).
Overcoming ALS heterogeneity with multiplexed, pooled approaches and genetic screens
The variable penetrance of some ALS-determining mutations and the complex genetic contribution to sporadic ALS, based on a combination of common variants of small effect and rarer variants of stronger effect, suggests that there remain many genetic factors to be uncovered. Multiplexed or novel pooled approaches have the potential to identify ALS-relevant mechanisms that are masked in conventional smaller scale experimental setups. One possible approach is the usage of high-content image-based assays, which have been successfully used to distinguish fibroblasts derived from 12 spinal muscular atrophy (SMA) patients and 12 healthy control individuals based on high-throughput imaging and machine learning analysis of morphological criteria (Yang et al., 2019). A complimentary technique, PRobe-based Imaging for Sequential Multiplexing (PRISM) (Tomov et al., 2021), which permits the simultaneous imaging of 10 or more molecular targets using fluorescent DNA-conjugated antibody probes, has been applied to evaluate cell identity and population composition of iPSC-derived cortical neuron and MN cultures. The combined application of these or similar multiplexed techniques to ALS-relevant cell types has the potential to characterize disease signals and multiple contributing gene and protein targets at large-scale.
A different option is pooled technology, such as a ‘village-in-a-dish’ in which cells from up to 100 donors are mixed together, fed, passaged, stimulated in a shared environment and scored for phenotypes (Mitchell et al., 2020). After sorting or selecting for the phenotype of interest, the genomic DNA is sequenced in the resulting village, and computational analysis reveals the proportion of cells from each donor contributing to phenotypic variation which can then be mapped back to genomic variation. This permits hypothesis-driven testing of multiple disease phenotypes across many disease-associated mutations, allowing for the simultaneous increase in throughput in both the number of genotypes and phenotypes that can be studied and decreasing inter-line assay variability. Pooled approaches could be employed to uncover phenotypes across large sALS cohorts or to compare pathomechanisms in cells differentiated from individuals carrying ALS-causing mutations in different genes.
A novel approach to studying the role of genetic factors in ALS is to use CRISPR-interference (CRISPR-i)-based platforms for genetic screens combined with single-cell RNA-sequencing technologies, as shown by Tian and colleagues (Tian et al., 2019). CRISPR-i uses a catalytic version of Cas9 (dCas9) which has no DNA cutting activity but is fused to a transcriptional repressor domain that suppresses the expression of human genes. Tian et al. implemented this technique in hiPSCs and hiPSC-derived neurons to reveal neuron-specific genes that are essential for neurons but not for hiPSCs by single-cell RNA-sequencing. The identification of genes which selectively promote neuronal survival in hiPSC-derived neurons may give important insights into neurodegenerative diseases such as ALS.
Translation of promising drug candidates from hiPSC models into human ALS in current clinical trials
Despite the challenges that remain to be overcome, hiPSC modeling approaches have the potential to accelerate translation of drugs into clinical trials by demonstrating target engagement, notably with ASO treatment, and by providing a screening platform for ALS-relevant phenotypes. In vitro disease modeling using hiPSC technology has led to at least three clinical trials of drug candidates for ALS.
In SOD1 and C9orf72 hiPSC models, ezogabine, a potassium Kv7 channel activator approved by the US Food and Drug Administration for epilepsy, reduced neuronal excitability (Wainger et al., 2014). Based on the in vitro data, the investigators moved directly from hiPSC modeling into a multicenter phase 2 randomized clinical trial of 2 doses of ezogabine in patients with ALS and found that ezogabine decreased cortical and spinal MN excitability, though the study was not powered to detect an effect on disease progression (Wainger et al., 2021). Of note, ezogabine has been previously used in humans and has an established safety profile, which made it substantially more straightforward to move directly into phase 2 studies based on the hiPSC-related work from (Wainger et al., 2014).
Imamura et al. identified src/c-Abl inhibitors as therapeutic targets in a high throughput screen assessing survival of SOD1 hiPSC-MNs. The most promising src/c-Abl inhibitor bosutinib further rescued viability of hiPSC-MNs derived from patients with TDP43 and C9orf72 mutations and two of three people with sALS, suggesting a common target across fALS genotypes and between fALS and sALS. Furthermore, a 3D neuromuscular junction (NMJ) organ-on-a-chip model using hiPSC-MNs derived from an sALS patient co-cultured with muscle skeletal bundles verified that bosutinib co-treated with rapamycin rescued the reduced muscle contractions in sALS cultures (Osaki et al., 2018). Bosutinib is currently in clinical trials to test efficacy in ALS patients.
Ropinirole, a non-ergoline dopamine agonist used to treat the symptoms of PD, showed positive effects on suppression of neurite retraction and cell death in hiPSC-MNs from patients with TARDBP or FUS mutations, and notably also on cells from patients with sALS. Putative mechanisms of action of ropinirole are 1) suppression of oxidative stress; 2) inhibition of TDP-43 and FUS aggregation; 3) improvement of mitochondrial function, (Okano et al., 2020). A randomized, double-blind, placebo-controlled, single-center, and open-label continuation phase I/IIa clinical trial, was designed to explore the safety, tolerability and efficacy of ropinirole hydrochloride as an ALS treatment (Morimoto et al., 2019).
In further developing iPSCs as validation tool for disease modelling and drug discovery it will be crucial to demonstrate that drugs which successfully rescue phenotypes in iPSC-derived models predict therapeutic efficacy in patients.
Conclusion
hiPSC technology permits the generation of ALS-relevant cell types, both in the CNS and PNS, allowing for the creation of highly relevant disease models that capture the specific genetic signature of a given patient. Using hiPSC -MNs, a variety of cell-autonomous mechanisms have been elucidated, which are thought to be shared between fALS and sALS, such as the dysfunction of nucleocytoplasmic transport, stress granule processing, and autophagy. hiPSC -derived astrocytes from ALS patients have been used to investigate the role of these cells in ALS pathophysiology through their effect on MNs. Differentiation protocols for hiPSC-derived microglia have also recently become available, but research on their role in ALS is still emerging. More complex, multicellular and 3D cultures systems can be used to investigate cell-cell interactions in vitro that were not previously feasible in monotypic cell configurations. Protocols to differentiate hiPSCs into other cell types potentially contributing to the disease pathogenesis such as skeletal muscle, or cells that are relatively resistant to degeneration in ALS, such as oculomotor neurons, will additionally increase the battery of tools to investigate ALS.
Several technical caveats of hiPSC-derived models remain to be resolved. The variety of differentiation protocols in use for the different ALS relevant cell types, with resultant inconsistency between the identity of differentiated cells as well as the results obtained by different laboratories, may be overcome by the use of isogenic controls and enrichment of pure cell fractions. Additional important aspects of hiPSC biology require investigation to clarify their utility for the study of an age-related neurodegenerative disease like ALS. The relative immaturity of hiPSC-derived cells may require more sophisticated approaches to in vitro maturation to reveal how the cellular tolerance to genetic mutations present from conception breaks down with aging.
While many important questions remain, clinical trials based on hiPSC-disease modeling studies are now emerging. The ability of hiPSC models to reflect unique aspects of disease can also be expected to contribute to more informed patient stratification in trials, and to the identification of which cell type is most likely to be the target of treatments.
Acknowledgements
Figures were created with BioRender.com.
EG is supported by Rubicon (2020/30766/ZONMW). LS is supported by the National Institutes of Health (NIH, USA) R01NS093872, R21NS116545, R01AG054720 and core grant P30CA008748, the Starr Foundation, the Department of Defense AL200169 - W81XWH2110140 and by Project ALS. BFV is supported by the University of Oxford Clarendon Fund, St John’s College Oxford, the Oxford-Medical Research Council Doctoral Training Partnership, and the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre. The views expressed are those of the authors and not necessarily those of the National Health Service, the NIHR, or the Department of Health and Social Care. RD is supported by a Celgene/BMS Fellowship. Work in KT laboratory is supported by funding from the MND Association.
Footnotes
Declaration of interests
L.S. is a scientific co-founder and paid consultant of BlueRock Therapeutics and he is listed as an inventor of several patents owned by MSKCC related to hiPSC-differentiation technologies.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ababneh NA, Scaber J, Flynn R, Douglas A, Barbagallo P, Candalija A, Turner MR, Sims D, Dafinca R, Cowley SA, et al. (2020). Correction of amyotrophic lateral sclerosis related phenotypes in induced pluripotent stem cell-derived motor neurons carrying a hexanucleotide expansion mutation in C9orf72 by CRISPR/Cas9 genome editing using homology-directed repair. Hum Mol Genet 29, 2200–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abo-Rady M, Kalmbach N, Pal A, Schludi C, Janosch A, Richter T, Freitag P, Bickle M, Kahlert AK, Petri S, et al. (2020). Knocking out C9ORF72 Exacerbates Axonal Trafficking Defects Associated with Hexanucleotide Repeat Expansion and Reduces Levels of Heat Shock Proteins. Stem Cell Reports 14, 390–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, Yeromin AV, Scarfone VM, Marsh SE, Fimbres C, et al. (2017). iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron 94, 278–293 e279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkaslasi MR, Piccus ZE, Hareendran S, Silberberg H, Chen L, Zhang Y, Petros TJ, and Le Pichon CE (2021). Single nucleus RNA-sequencing defines unexpected diversity of cholinergic neuron types in the adult mouse spinal cord. Nat Commun 12, 2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida S, Gascon E, Tran H, Chou HJ, Gendron TF, Degroot S, Tapper AR, Sellier C, Charlet-Berguerand N, Karydas A, et al. (2013). Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol 126, 385–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida S, Krishnan G, Rushe M, Gu Y, Kankel MW, and Gao FB (2019). Production of poly(GA) in C9ORF72 patient motor neurons derived from induced pluripotent stem cells. Acta Neuropathol 138, 1099–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves CJ, Dariolli R, Jorge FM, Monteiro MR, Maximino JR, Martins RS, Strauss BE, Krieger JE, Callegaro D, and Chadi G (2015). Gene expression profiling for human iPS-derived motor neurons from sporadic ALS patients reveals a strong association between mitochondrial functions and neurodegeneration. In Front Cell Neurosci, pp. 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoroso MW, Croft GF, Williams DJ, O'Keeffe S, Carrasco MA, Davis AR, Roybon L, Oakley DH, Maniatis T, Henderson CE, et al. (2013). Accelerated high-yield generation of limb-innervating motor neurons from human stem cells. J Neurosci 33, 574–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos PJ, Fung S, Case A, Kifelew J, Osnis L, Smith CL, Green K, Naydenov A, Aloi M, Hubbard JJ, et al. (2017). Modulation of Hematopoietic Lineage Specification Impacts TREM2 Expression in Microglia-Like Cells Derived From Human Stem Cells. ASN Neuro 9, 1759091417716610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J, Revah O, Miura Y, Thom N, Amin ND, Kelley KW, Singh M, Chen X, Thete MV, Walczak EM, et al. (2020). Generation of Functional Human 3D Cortico-Motor Assembloids. Cell 183, 1913–1929 e1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbour D, Vande Velde C, and Robitaille R (2017). New perspectives on amyotrophic lateral sclerosis: the role of glial cells at the neuromuscular junction. J Physiol 595, 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbar L, Jain T, Zimmer M, Kruglikov I, Sadick JS, Wang M, Kalpana K, Rose IVL, Burstein SR, Rusielewicz T, et al. (2020). CD49f Is a Novel Marker of Functional and Reactive Human iPSC-Derived Astrocytes. Neuron 107, 436–453 e412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeldman E, Raaphorst J, Klein Twennaar M, Govaarts R, Pijnenburg YAL, de Haan RJ, de Visser M, and Schmand BA (2018). The cognitive profile of behavioural variant FTD and its similarities with ALS: a systematic review and meta-analysis. J Neurol Neurosurg Psychiatry 89, 995–1002. [DOI] [PubMed] [Google Scholar]
- Bhinge A, Namboori SC, Zhang X, VanDongen AMJ, and Stanton LW (2017). Genetic Correction of SOD1 Mutant iPSCs Reveals ERK and JNK Activated AP1 as a Driver of Neurodegeneration in Amyotrophic Lateral Sclerosis. In Stem Cell Reports, pp. 856–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilican B, Serio A, Barmada SJ, Nishimura AL, Sullivan GJ, Carrasco M, Phatnani HP, Puddifoot CA, Story D, Fletcher J, et al. (2012). Mutant induced pluripotent stem cell lines recapitulate aspects of TDP-43 proteinopathies and reveal cell-specific vulnerability. In Proc Natl Acad Sci U S A, pp. 5803–5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birger A, Ben-Dor I, Ottolenghi M, Turetsky T, Gil Y, Sweetat S, Perez L, Belzer V, Casden N, Steiner D, et al. (2019). Human iPSC-derived astrocytes from ALS patients with mutated C9ORF72 show increased oxidative stress and neurotoxicity. EBioMedicine 50, 274–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum JA, Klemm S, Shadrach JL, Guttenplan KA, Nakayama L, Kathiria A, Hoang PT, Gautier O, Kaltschmidt JA, Greenleaf WJ, et al. (2021). Single-cell transcriptomic analysis of the adult mouse spinal cord reveals molecular diversity of autonomic and skeletal motor neurons. Nat Neurosci 24, 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, et al. (2011). Reference Maps of Human ES and iPS Cell Variation Enable High-Throughput Characterization of Pluripotent Cell Lines. Cell 144, 439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghero G, Pugliatti M, Marrosu F, Marrosu MG, Murru MR, Floris G, Cannas A, Parish LD, Occhineri P, Cau TB, et al. (2014). Genetic architecture of ALS in Sardinia. Neurobiol Aging 35, 2882.e2887–2882.e2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley RA, Shireman J, McFalls C, Choi J, Canfield SG, Dong Y, Liu K, Lisota B, Jones JR, Petersen A, et al. (2019). Regionally specified human pluripotent stem cell-derived astrocytes exhibit different molecular signatures and functional properties. Development 146, dev170910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, and Ericson J (2001). Specification of neuronal fates in the ventral neural tube. Curr Opin Neurobiol 11, 43–49. [DOI] [PubMed] [Google Scholar]
- Brites D, and Vaz AR (2014). Microglia centered pathogenesis in ALS: insights in cell interconnectivity. Front Cell Neurosci 8, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownjohn PW, Smith J, Solanki R, Lohmann E, Houlden H, Hardy J, Dietmann S, and Livesey FJ (2018). Functional Studies of Missense TREM2 Mutations in Human Stem Cell-Derived Microglia. Stem Cell Reports 10, 1294–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruttger J, Karram K, Wortge S, Regen T, Marini F, Hoppmann N, Klein M, Blank T, Yona S, Wolf Y, et al. (2015). Genetic Cell Ablation Reveals Clusters of Local Self-Renewing Microglia in the Mammalian Central Nervous System. Immunity 43, 92–106. [DOI] [PubMed] [Google Scholar]
- Burkhardt MF, Martinez FJ, Wright S, Ramos C, Volfson D, Mason M, Garnes J, Dang V, Lievers J, Shoukat-Mumtaz U, et al. (2013). A cellular model for sporadic ALS using patient-derived induced pluripotent stem cells. In Mol Cell Neurosci, pp. 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell JR, Halliday GM, Kril JJ, Ittner LM, Gotz J, Kiernan MC, and Hodges JR (2016). The frontotemporal dementia-motor neuron disease continuum. Lancet 388, 919–931. [DOI] [PubMed] [Google Scholar]
- Burrows CK, Banovich NE, Pavlovic BJ, Patterson K, Gallego Romero I, Pritchard JK, and Gilad Y (2016). Genetic Variation, Not Cell Type of Origin, Underlies the Majority of Identifiable Regulatory Differences in iPSCs. PLOS Genetics 12, e1005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursch F, Kalmbach N, Naujock M, Staege S, Eggenschwiler R, Abo-Rady M, Japtok J, Guo W, Hensel N, Reinhardt P, et al. (2019). Altered calcium dynamics and glutamate receptor properties in iPSC-derived motor neurons from ALS patients with C9orf72, FUS, SOD1 or TDP43 mutations. Hum Mol Genet 28, 2835–2850. [DOI] [PubMed] [Google Scholar]
- Busskamp V, Lewis NE, Guye P, Ng AH, Shipman SL, Byrne SM, Sanjana NE, Murn J, Li Y, Li S, et al. (2014). Rapid neurogenesis through transcriptional activation in human stem cells. Mol Syst Biol 10, 760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazzo M, Giannelli S, Valente P, Lignani G, Carissimo A, Sessa A, Colasante G, Bartolomeo R, Massimino L, Ferroni S, et al. (2015). Direct conversion of fibroblasts into functional astrocytes by defined transcription factors. Stem Cell Reports 4, 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calder EL, Tchieu J, Steinbeck JA, Tu E, Keros S, Ying SW, Jaiswal MK, Cornacchia D, Goldstein PA, Tabar V, et al. (2015). Retinoic Acid-Mediated Regulation of GLI3 Enables Efficient Motoneuron Derivation from Human ESCs in the Absence of Extrinsic SHH Activation. J Neurosci 35, 11462–11481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canals I, Ginisty A, Quist E, Timmerman R, Fritze J, Miskinyte G, Monni E, Hansen MG, Hidalgo I, Bryder D, et al. (2018). Rapid and efficient induction of functional astrocytes from human pluripotent stem cells. Nat Methods 15, 693–696. [DOI] [PubMed] [Google Scholar]
- Chambers CB, Peng Y, Nguyen H, Gaiano N, Fishell G, and Nye JS (2001). Spatiotemporal selectivity of response to Notch1 signals in mammalian forebrain precursors. Development 128, 689–702. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, and Studer L (2009). Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 27, 275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, and Studer L (2011). Cell fate plug and play: direct reprogramming and induced pluripotency. Cell 145, 827–830. [DOI] [PubMed] [Google Scholar]
- Chen H, Qian K, Du Z, Cao J, Petersen A, Liu H, Blackbourn LW, Huang C-L, Errigo A, Yin Y, et al. (2014). Modeling ALS with iPSCs reveals that mutant SOD1 misregulates neurofilament balance in motor neurons. In Cell Stem Cell, pp. 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chio A, Calvo A, Moglia C, Mazzini L, Mora G, and group P.s. (2011). Phenotypic heterogeneity of amyotrophic lateral sclerosis: a population based study. J Neurol Neurosurg Psychiatry 82, 740–746. [DOI] [PubMed] [Google Scholar]
- Chio A, Logroscino G, Traynor BJ, Collins J, Simeone JC, Goldstein LA, and White LA (2013). Global epidemiology of amyotrophic lateral sclerosis: a systematic review of the published literature. Neuroepidemiology 41, 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C-C, Zhang Y, Umoh ME, Vaughan SW, Lorenzini I, Liu F, Sayegh M, Donlin-Asp PG, Chen YH, Duong DM., et al. (2018). TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. In Nat Neurosci, pp. 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafinca R, Barbagallo P, Farrimond L, Candalija A, Scaber J, Ababneh NA, Sathyaprakash C, Vowles J, Cowley SA, and Talbot K (2020). Impairment of Mitochondrial Calcium Buffering Links Mutations in C9ORF72 and TARDBP in iPS-Derived Motor Neurons from Patients with ALS/FTD. Stem Cell Reports 14, 892–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafinca R, Scaber J, Ababneh N, Lalic T, Weir G, Christian H, Vowles J, Douglas AG, Fletcher-Jones A, Browne C, et al. (2016). C9orf72 Hexanucleotide Expansions Are Associated with Altered Endoplasmic Reticulum Calcium Homeostasis and Stress Granule Formation in Induced Pluripotent Stem Cell-Derived Neurons from Patients with Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. Stem Cells 34, 2063–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. (2011). Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, Young WL, Ivey KN, and Gao FB (2010). MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell 6, 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin AC, Burr K, Borooah S, Foster JD, Cleary EM, Geti I, Vallier L, Shaw CE, Chandran S, and Miles GB (2015). Human iPSC-derived motoneurons harbouring TARDBP or C9ORF72 ALS mutations are dysfunctional despite maintaining viability. Nat Commun 6, 5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio FP, Boulting GL, Bobrowicz S, and Eggan KC (2008). Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. In Cell Stem Cell, pp. 637–648. [DOI] [PubMed] [Google Scholar]
- Di Giorgio FP, Carrasco MA, Siao MC, Maniatis T, and Eggan K (2007). Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat Neurosci 10, 608–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Gregorio SE, and Duennwald ML (2018). ALS Yeast Models—Past Success Stories and New Opportunities. Frontiers in Molecular Neuroscience 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, et al. (2008). Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science 321, 1218–1221. [DOI] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang P-W, Pham JT, Haeusler AR, Heusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, et al. (2013). RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. In Neuron, pp. 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douvaras P, Sun B, Wang M, Kruglikov I, Lallos G, Zimmer M, Terrenoire C, Zhang B, Gandy S, Schadt E, et al. (2017). Directed Differentiation of Human Pluripotent Stem Cells to Microglia. Stem Cell Reports 8, 1516–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du ZW, Chen H, Liu H, Lu J, Qian K, Huang CL, Zhong X, Fan F, and Zhang SC (2015). Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nat Commun 6, 6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durston AJ, van der Wees J, Pijnappel WW, and Godsave SF (1998). Retinoids and related signals in early development of the vertebrate central nervous system. Curr Top Dev Biol 40, 111–175. [DOI] [PubMed] [Google Scholar]
- Egawa N, Kitaoka S, Tsukita K, Naitoh M, Takahashi K, Yamamoto T, Adachi F, Kondo T, Okita K, Asaka I, et al. (2012). Drug screening for ALS using patient-specific induced pluripotent stem cells. In Sci Transl Med, pp. 145ra104. [DOI] [PubMed] [Google Scholar]
- Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, and Sasai Y (2008). Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3, 519–532. [DOI] [PubMed] [Google Scholar]
- Elkabetz Y, Panagiotakos G, Al Shamy G, Socci ND, Tabar V, and Studer L (2008). Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev 22, 152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emdad L, D'Souza SL, Kothari HP, Qadeer ZA, and Germano IM (2012). Efficient differentiation of human embryonic and induced pluripotent stem cells into functional astrocytes. Stem Cells Dev 21, 404–410. [DOI] [PubMed] [Google Scholar]
- Enright HA, Lam D, Sebastian A, Sales AP, Cadena J, Hum NR, Osburn JJ, Peters SKG, Petkus B, Soscia DA, et al. (2020). Functional and transcriptional characterization of complex neuronal co-cultures. Sci Rep 10, 11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escartin C, Galea E, Lakatos A, O'Callaghan JP, Petzold GC, Serrano-Pozo A, Steinhauser C, Volterra A, Carmignoto G, Agarwal A, et al. (2021). Reactive astrocyte nomenclature, definitions, and future directions. Nat Neurosci 24, 312–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk A, Koch P, Kesavan J, Takashima Y, Ladewig J, Alexander M, Wiskow O, Tailor J, Trotter M, Pollard S, et al. (2012). Capture of neuroepithelial-like stem cells from pluripotent stem cells provides a versatile system for in vitro production of human neurons. PLoS One 7, e29597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang MY, Markmiller S, Vu AQ, Javaherian A, Dowdle WE, Jolivet P, Bushway PJ, Castello NA, Baral A, Chan MY, et al. (2019). Small-Molecule Modulation of TDP-43 Recruitment to Stress Granules Prevents Persistent TDP-43 Accumulation in ALS/FTD. Neuron 103, 802–819.e811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipi T, Hermanova Z, Tureckova J, Vanatko O, and Anderova AM (2020). Glial Cells-The Strategic Targets in Amyotrophic Lateral Sclerosis Treatment. J Clin Med 9, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, Badders N, Valentine M, Miller BL, Wong PC, et al. (2015). GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525, 129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori K, Ishikawa M, Otomo A, Atsuta N, Nakamura R, Akiyama T, Hadano S, Aoki M, Saya H, Sobue G, et al. (2018). Modeling sporadic ALS in iPSC-derived motor neurons identifies a potential therapeutic agent. Nat Med 24, 1579–1589. [DOI] [PubMed] [Google Scholar]
- Gatto N, Dos Santos Souza C, Shaw AC, Bell SM, Myszczynska MA, Powers S, Meyer K, Castelli LM, Karyka E, Mortiboys H, et al. (2021). Directly converted astrocytes retain the ageing features of the donor fibroblasts and elucidate the astrocytic contribution to human CNS health and disease. Aging Cell 20, e13281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, Chew J, Stankowski JN, Hayes LR, Zhang YJ, Prudencio M, Carlomagno Y, Daughrity LM, Jansen-West K, Perkerson EA, et al. (2017). Poly(GP) proteins are a useful pharmacodynamic marker for C9ORF72-associated amyotrophic lateral sclerosis. Sci Transl Med 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, et al. (2010). Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F, et al. (2015). Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Fumagalli L, Prior R, and Van Den Bosch L (2017a). Current Advances and Limitations in Modeling ALS/FTD in a Dish Using Induced Pluripotent Stem Cells. Front Neurosci 11, 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Naujock M, Fumagalli L, Vandoorne T, Baatsen P, Boon R, Ordovás L, Patel A, Welters M, Vanwelden T, et al. (2017b). HDAC6 inhibition reverses axonal transport defects in motor neurons derived from FUS-ALS patients. Nat Commun 8, 861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttikonda SR, Sikkema L, Tchieu J, Saurat N, Walsh RM, Harschnitz O, Ciceri G, Sneeboer M, Mazutis L, Setty M, et al. (2021). Fully defined human pluripotent stem cell-derived microglia and tri-culture system model C3 production in Alzheimer's disease. Nat Neurosci 24, 343–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenseler W, and Rajendran L (2019). Concise Review: Modeling Neurodegenerative Diseases with Human Pluripotent Stem Cell-Derived Microglia. Stem Cells 37, 724–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenseler W, Sansom SN, Buchrieser J, Newey SE, Moore CS, Nicholls FJ, Chintawar S, Schnell C, Antel JP, Allen ND, et al. (2017). A Highly Efficient Human Pluripotent Stem Cell Microglia Model Displays a Neuronal-Co-culture-Specific Expression Profile and Inflammatory Response. Stem Cell Reports 8, 1727–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haidet-Phillips AM, Hester ME, Miranda CJ, Meyer K, Braun L, Frakes A, Song S, Likhite S, Murtha MJ, Foust KD, et al. (2011). Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. Nat Biotechnol 29, 824–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall CE, Yao Z, Choi M, Tyzack GE, Serio A, Luisier R, Harley J, Preza E, Arber C, Crisp SJ, et al. (2017). Progressive Motor Neuron Pathology and the Role of Astrocytes in a Human Stem Cell Model of VCP-Related ALS. Cell Rep 19, 1739–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselmann J, and Blurton-Jones M (2020). Human iPSC-derived microglia: A growing toolset to study the brain's innate immune cells. Glia 68, 721–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautbergue GM, Castelli LM, Ferraiuolo L, Sanchez-Martinez A, Cooper-Knock J, Higginbottom A, Lin YH, Bauer CS, Dodd JE, Myszczynska MA, et al. (2017). SRSF1-dependent nuclear export inhibition of C9ORF72 repeat transcripts prevents neurodegeneration and associated motor deficits. Nat Commun 8, 16063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedegaard A, Stodolak S, James WS, and Cowley SA (2020). Honing the Double-Edged Sword: Improving Human iPSC-Microglia Models. Front Immunol 11, 614972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S, Catania K, Manger PR, and Kaas JH (2015). Mammalian Brains Are Made of These: A Dataset of the Numbers and Densities of Neuronal and Nonneuronal Cells in the Brain of Glires, Primates, Scandentia, Eulipotyphlans, Afrotherians and Artiodactyls, and Their Relationship with Body Mass. Brain, Behavior and Evolution 86, 145–163. [DOI] [PubMed] [Google Scholar]
- Higelin J, Demestre M, Putz S, Delling JP, Jacob C, Lutz A-K, Bausinger J, Huber A-K, Klingenstein M, Barbi G, et al. (2016). FUS Mislocalization and Vulnerability to DNA Damage in ALS Patients Derived hiPSCs and Aging Motoneurons. In Front Cell Neurosci, pp. 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjorth PS (1993). [Nursing diagnosis. European cooperation]. Sygeplejersken 93, 4–6, 30. [PubMed] [Google Scholar]
- Ho R, Workman MJ, Mathkar P, Wu K, Kim KJ, O’Rourke JG, Kellogg M, Montel V, Banuelos MG, Arogundade OA, et al. (2021). Cross-Comparison of Human iPSC Motor Neuron Models of Familial and Sporadic ALS Reveals Early and Convergent Transcriptomic Disease Signatures. Cell Systems 12, 159–175.e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist S, Brouwer M, Djelloul M, Diaz AG, Devine MJ, Hammarberg A, Fog K, Kunath T, and Roybon L (2015). Generation of human pluripotent stem cell reporter lines for the isolation of and reporting on astrocytes generated from ventral midbrain and ventral spinal cord neural progenitors. Stem Cell Res 15, 203–220. [DOI] [PubMed] [Google Scholar]
- Hu BY, Weick JP, Yu J, Ma LX, Zhang XQ, Thomson JA, and Zhang SC (2010). Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci U S A 107, 4335–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu BY, and Zhang SC (2009). Differentiation of spinal motor neurons from pluripotent human stem cells. Nat Protoc 4, 1295–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiyanagi N, Fujimori K, Yano M, Ishihara-Fujisaki C, Sone T, Akiyama T, Okada Y, Akamatsu W, Matsumoto T, Ishikawa M, et al. (2016). Establishment of In Vitro FUS-Associated Familial Amyotrophic Lateral Sclerosis Model Using Human Induced Pluripotent Stem Cells. In Stem Cell Reports, pp. 496–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura K, Izumi Y, Watanabe A, Tsukita K, Woltjen K, Yamamoto T, Hotta A, Kondo T, Kitaoka S, Ohta A, et al. (2017). The Src/c-Abl pathway is a potential therapeutic target in amyotrophic lateral sclerosis. Sci Transl Med 9, eaaf3962. [DOI] [PubMed] [Google Scholar]
- Juopperi TA, Kim WR, Chiang CH, Yu H, Margolis RL, Ross CA, Ming GL, and Song H (2012). Astrocytes generated from patient induced pluripotent stem cells recapitulate features of Huntington's disease patient cells. Mol Brain 5, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karumbayaram S, Novitch BG, Patterson M, Umbach JA, Richter L, Lindgren A, Conway AE, Clark AT, Goldman SA, Plath K, et al. (2009). Directed differentiation of human-induced pluripotent stem cells generates active motor neurons. Stem Cells 27, 806–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan AB, Jenkins SL, Jagodnik KM, Koplev S, He E, Torre D, Wang Z, Dohlman AB, Silverstein MC, Lachmann A, et al. (2018). The Library of Integrated Network-Based Cellular Signatures NIH Program: System-Level Cataloging of Human Cells Response to Perturbations. Cell Systems 6, 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierdorf K, Erny D, Goldmann T, Sander V, Schulz C, Perdiguero EG, Wieghofer P, Heinrich A, Riemke P, Holscher C, et al. (2013). Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat Neurosci 16, 273–280. [DOI] [PubMed] [Google Scholar]
- Kiernan MC, Vucic S, Cheah BC, Turner MR, Eisen A, Hardiman O, Burrell JR, and Zoing MC (2011). Amyotrophic lateral sclerosis. Lancet 377, 942–955. [DOI] [PubMed] [Google Scholar]
- Kiernan MC, Vucic S, Talbot K, McDermott CJ, Hardiman O, Shefner JM, Al-Chalabi A, Huynh W, Cudkowicz M, Talman P, et al. (2021). Improving clinical trial outcomes in amyotrophic lateral sclerosis. Nat Rev Neurol 17, 104–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpinen H, Goncalves A, Leha A, Afzal V, Alasoo K, Ashford S, Bala S, Bensaddek D, Casale FP, Culley OJ, et al. (2017). Common genetic variation drives molecular heterogeneity in human iPSCs. Nature 546, 370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BW, Ryu J, Jeong YE, Kim J, and Martin LJ (2020). Human Motor Neurons With SOD1-G93A Mutation Generated From CRISPR/Cas9 Gene-Edited iPSCs Develop Pathological Features of Amyotrophic Lateral Sclerosis. Front Cell Neurosci 14, 604171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiskinis E, Kralj JM, Zou P, Weinstein EN, Zhang H, Tsioras K, Wiskow O, Ortega JA, Eggan K, and Cohen AE (2018). All-Optical Electrophysiology for High-Throughput Functional Characterization of a Human iPSC-Derived Motor Neuron Model of ALS. Stem Cell Reports 10, 1991–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiskinis E, Sandoe J, Williams LA, Boulting GL, Moccia R, Wainger BJ, Han S, Peng T, Thams S, Mikkilineni S, et al. (2014). Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell 14, 781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konttinen H, Cabral-da-Silva MEC, Ohtonen S, Wojciechowski S, Shakirzyanova A, Caligola S, Giugno R, Ishchenko Y, Hernandez D, Fazaludeen MF, et al. (2019). PSEN1DeltaE9, APPswe, and APOE4 Confer Disparate Phenotypes in Human iPSC-Derived Microglia. Stem Cell Reports 13, 669–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiter N, Pal A, Lojewski X, Corcia P, Naujock M, Reinhardt P, Sterneckert J, Petri S, Wegner F, Storch A, et al. (2018). Age-dependent neurodegeneration and organelle transport deficiencies in mutant TDP43 patient-derived neurons are independent of TDP43 aggregation. In Neurobiol Dis, pp. 167–181. [DOI] [PubMed] [Google Scholar]
- Krencik R, Weick JP, Liu Y, Zhang ZJ, and Zhang SC (2011). Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat Biotechnol 29, 528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krencik R, and Zhang SC (2011). Directed differentiation of functional astroglial subtypes from human pluripotent stem cells. Nat Protoc 6, 1710–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafaille FG, Pessach IM, Zhang SY, Ciancanelli MJ, Herman M, Abhyankar A, Ying SW, Keros S, Goldstein PA, Mostoslavsky G, et al. (2012). Impaired intrinsic immunity to HSV-1 in human iPSC-derived TLR3-deficient CNS cells. Nature 491, 769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam RS, Töpfer FM, Wood PG, Busskamp V, and Bamberg E (2017). Functional Maturation of Human Stem Cell-Derived Neurons in Long-Term Cultures. PLoS One 12, e0169506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Shamy GA, Elkabetz Y, Schofield CM, Harrsion NL, Panagiotakos G, Socci ND, Tabar V, and Studer L (2007). Directed differentiation and transplantation of human embryonic stem cell-derived motoneurons. Stem Cells 25, 1931–1939. [DOI] [PubMed] [Google Scholar]
- Leventoux N, Morimoto S, Imaizumi K, Sato Y, Takahashi S, Mashima K, Ishikawa M, Sonn I, Kondo T, Watanabe H, et al. (2020). Human Astrocytes Model Derived from Induced Pluripotent Stem Cells. Cells 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Sun W, Zhang Y, Wei W, Ambasudhan R, Xia P, Talantova M, Lin T, Kim J, Wang X, et al. (2011). Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc Natl Acad Sci U S A 108, 8299–8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Tao Y, Bradley R, Du Z, Tao Y, Kong L, Dong Y, Jones J, Yan Y, Harder CRK, et al. (2018). Fast Generation of Functional Subtype Astrocytes from Human Pluripotent Stem Cells. Stem Cell Reports 11, 998–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XJ, Du ZW, Zarnowska ED, Pankratz M, Hansen LO, Pearce RA, and Zhang SC (2005). Specification of motoneurons from human embryonic stem cells. Nat Biotechnol 23, 215–221. [DOI] [PubMed] [Google Scholar]
- Li XJ, Zhang X, Johnson MA, Wang ZB, Lavaute T, and Zhang SC (2009). Coordination of sonic hedgehog and Wnt signaling determines ventral and dorsal telencephalic neuron types from human embryonic stem cells. Development 136, 4055–4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liguori F, Amadio S, and Volonté C (2021). Fly for ALS: Drosophila modeling on the route to amyotrophic lateral sclerosis modifiers. Cellular and Molecular Life Sciences 78, 6143–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Lin Y, Nery JR, Urich MA, Breschi A, Davis CA, Dobin A, Zaleski C, Beer MA, Chapman WC, et al. (2014). Comparison of the transcriptional landscapes between human and mouse tissues. Proc Natl Acad Sci U S A 111, 17224–17229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippmann ES, Williams CE, Ruhl DA, Estevez-Silva MC, Chapman ER, Coon JJ, and Ashton RS (2015). Deterministic HOX patterning in human pluripotent stem cell-derived neuroectoderm. Stem Cell Reports 4, 632–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey MR, Bilican B, Qiu J, Rzechorzek NM, Haghi G, Burr K, Hardingham GE, Chandran S, and Wyllie DJ (2014). Maturation of AMPAR composition and the GABAAR reversal potential in hPSC-derived cortical neurons. J Neurosci 34, 4070–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gonzalez R, Lu Y, Gendron TF, Karydas A, Tran H, Yang D, Petrucelli L, Miller BL, Almeida S, and Gao F-B (2016). Poly(GR) in C9ORF72-Related ALS/FTD Compromises Mitochondrial Function and Increases Oxidative Stress and DNA Damage in iPSC-Derived Motor Neurons. In Neuron, pp. 383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gonzalez R, Yang D, Pribadi M, Kim TS, Krishnan G, Choi SY, Lee S, Coppola G, and Gao FB (2019). Partial inhibition of the overactivated Ku80-dependent DNA repair pathway rescues neurodegeneration in C9ORF72-ALS/FTD. Proc Natl Acad Sci U S A 116, 9628–9633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzini I, Alsop E, Levy J, Gittings LM, Rabichow BE, Lall D, Moore S, Bustos L, Pevey R, Burciu C, et al. (2020). Activated iPSC-microglia from C9orf72 ALS/FTD patients exhibit endosomal-lysosomal dysfunction. bioRxiv, 2020.2009.2003.277459. [Google Scholar]
- Luisier R, Tyzack GE, Hall CE, Mitchell JS, Devine H, Taha DM, Malik B, Meyer I, Greensmith L, Newcombe J, et al. (2018). Intron retention and nuclear loss of SFPQ are molecular hallmarks of ALS. Nat Commun 9, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin A, Delsing L, Clausen M, Ricchiuto P, Sanchez J, Sabirsh A, Ding M, Synnergren J, Zetterberg H, Brolen G, et al. (2018). Human iPS-Derived Astroglia from a Stable Neural Precursor State Show Improved Functionality Compared with Conventional Astrocytic Models. Stem Cell Reports 10, 1030–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madill M, McDonagh K, Ma J, Vajda A, McLoughlin P, O'Brien T, Hardiman O, and Shen S (2017). Amyotrophic lateral sclerosis patient iPSC-derived astrocytes impair autophagy via non-cell autonomous mechanisms. In Mol Brain, pp. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maor-Nof M, Shipony Z, Lopez-Gonzalez R, Nakayama L, Zhang YJ, Couthouis J, Blum JA, Castruita PA, Linares GR, Ruan K, et al. (2021). p53 is a central regulator driving neurodegeneration caused by C9orf72 poly(PR). Cell 184, 689–708 e620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Muotri AR, Mu Y, Smith AM, Cezar GG, and Gage FH (2008). Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell 3, 649–657. [DOI] [PubMed] [Google Scholar]
- Marrone L, Poser I, Casci I, Japtok J, Reinhardt P, Janosch A, Andree C, Lee HO, Moebius C, Koerner E, et al. (2018). Isogenic FUS-eGFP iPSC Reporter Lines Enable Quantification of FUS Stress Granule Pathology that Is Rescued by Drugs Inducing Autophagy. In Stem Cell Reports, pp. 375–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton RM, and Pasca SP (2020). Organoid and Assembloid Technologies for Investigating Cellular Crosstalk in Human Brain Development and Disease. Trends Cell Biol 30, 133–143. [DOI] [PubMed] [Google Scholar]
- Matejuk A, and Ransohoff RM (2020). Crosstalk Between Astrocytes and Microglia: An Overview. Front Immunol 11, 1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maury Y, Come J, Piskorowski RA, Salah-Mohellibi N, Chevaleyre V, Peschanski M, Martinat C, and Nedelec S (2015). Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat Biotechnol 33, 89–96. [DOI] [PubMed] [Google Scholar]
- Mazzoni EO, Mahony S, Closser M, Morrison CA, Nedelec S, Williams DJ, An D, Gifford DK, and Wichterle H (2013). Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat Neurosci 16, 1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuade A, Coburn M, Tu CH, Hasselmann J, Davtyan H, and Blurton-Jones M (2018). Development and validation of a simplified method to generate human microglia from pluripotent stem cells. Mol Neurodegener 13, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta AR, Gregory JM, Dando O, Carter RN, Burr K, Nanda J, Story D, McDade K, Smith C, Morton NM, et al. (2021). Mitochondrial bioenergetic deficits in C9orf72 amyotrophic lateral sclerosis motor neurons cause dysfunctional axonal homeostasis. Acta Neuropathol 141, 257–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J, Paquola ACM, Ku M, Hatch E, Böhnke L, Ladjevardi S, McGrath S, Campbell B, Lee H, Herdy JR, et al. (2015). Directly Reprogrammed Human Neurons Retain Aging-Associated Transcriptomic Signatures and Reveal Age-Related Nucleocytoplasmic Defects. Cell Stem Cell 17, 705–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J, Reid D, Lau S, Kim Y, and Gage FH (2018). Aging in a Dish: iPSC-Derived and Directly Induced Neurons for Studying Brain Aging and Age-Related Neurodegenerative Diseases. Annu Rev Genet 52, 271–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Ferraiuolo L, Miranda CJ, Likhite S, McElroy S, Renusch S, Ditsworth D, Lagier-Tourenne C, Smith RA, Ravits J, et al. (2014). Direct conversion of patient fibroblasts demonstrates non-cell autonomous toxicity of astrocytes to motor neurons in familial and sporadic ALS. Proc Natl Acad Sci U S A 111, 829–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JD, Ganat YM, Kishinevsky S, Bowman RL, Liu B, Tu EY, Mandal PK, Vera E, Shim JW, Kriks S, et al. (2013). Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell 13, 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Cudkowicz M, Shaw PJ, Andersen PM, Atassi N, Bucelli RC, Genge A, Glass J, Ladha S, Ludolph AL, et al. (2020). Phase 1-2 Trial of Antisense Oligonucleotide Tofersen for SOD1 ALS. N Engl J Med 383, 109–119. [DOI] [PubMed] [Google Scholar]
- Mitchell JM, Nemesh J, Ghosh S, Handsaker RE, Mello CJ, Meyer D, Raghunathan K, de Rivera H, Tegtmeyer M, Hawes D, et al. (2020). Mapping genetic effects on cellular phenotypes with “cell villages”. bioRxiv, 2020.2006.2029.174383. [Google Scholar]
- Mordes DA, Morrison BM, Ament XH, Cantrell C, Mok J, Eggan P, Xue C, Wang JY, Eggan K, and Rothstein JD (2020). Absence of Survival and Motor Deficits in 500 Repeat C9ORF72 BAC Mice. Neuron 108, 775–783 e774. [DOI] [PubMed] [Google Scholar]
- Morimoto S, Takahashi S, Fukushima K, Saya H, Suzuki N, Aoki M, Okano H, and Nakahara J (2019). Ropinirole hydrochloride remedy for amyotrophic lateral sclerosis - Protocol for a randomized, double-blind, placebo-controlled, single-center, and open-label continuation phase I/IIa clinical trial (ROPALS trial). Regen Ther 11, 143–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mormone E, D'Sousa S, Alexeeva V, Bederson MM, and Germano IM (2014). "Footprint-free" human induced pluripotent stem cell-derived astrocytes for in vivo cell-based therapy. Stem Cells Dev 23, 2626–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffat J, Li Y, Yuan B, Mitalipova M, Omer A, Corcoran S, Bakiasi G, Tsai LH, Aubourg P, Ransohoff RM, et al. (2016). Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med 22, 1358–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhr J, Graziano E, Wilson S, Jessell TM, and Edlund T (1999). Convergent inductive signals specify midbrain, hindbrain, and spinal cord identity in gastrula stage chick embryos. Neuron 23, 689–702. [DOI] [PubMed] [Google Scholar]
- Munoz-Sanjuan I, and Brivanlou AH (2002). Neural induction, the default model and embryonic stem cells. Nat Rev Neurosci 3, 271–280. [DOI] [PubMed] [Google Scholar]
- Nagai M, Re DB, Nagata T, Chalazonitis A, Jessell TM, Wichterle H, and Przedborski S (2007). Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci 10, 615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann M, Pal A, Goswami A, Lojewski X, Japtok J, Vehlow A, Naujock M, Günther R, Jin M, Stanslowsky N, et al. (2018). Impaired DNA damage response signaling by FUS-NLS mutations leads to neurodegeneration and FUS aggregate formation. In Nature Communications, pp. 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely MD, Litt MJ, Tidball AM, Li GG, Aboud AA, Hopkins CR, Chamberlin R, Hong CC, Ess KC, and Bowman AB (2012). DMH1, a highly selective small molecule BMP inhibitor promotes neurogenesis of hiPSCs: comparison of PAX6 and SOX1 expression during neural induction. ACS Chem Neurosci 3, 482–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehme R, Zuccaro E, Ghosh SD, Li C, Sherwood JL, Pietilainen O, Barrett LE, Limone F, Worringer KA, Kommineni S, et al. (2018). Combining NGN2 Programming with Developmental Patterning Generates Human Excitatory Neurons with NMDAR-Mediated Synaptic Transmission. Cell Rep 23, 2509–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. (2006). Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133. [DOI] [PubMed] [Google Scholar]
- Nishino K, Toyoda M, Yamazaki-Inoue M, Fukawatase Y, Chikazawa E, Sakaguchi H, Akutsu H, and Umezawa A (2011). DNA Methylation Dynamics in Human Induced Pluripotent Stem Cells over Time. PLoS Genetics 7, e1002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberheim NA, Goldman SA, and Nedergaard M (2012). Heterogeneity of Astrocytic Form and Function. In Astrocytes: Methods and Protocols, Milner R, ed. (Totowa, NJ: Humana Press; ), pp. 23–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberheim NA, Takano T, Han X, He W, Lin JHC, Wang F, Xu Q, Wyatt JD, Pilcher W, Ojemann JG, et al. (2009). Uniquely hominid features of adult human astrocytes. In Journal of Neuroscience, pp. 3276–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogaki K, Li Y, Atsuta N, Tomiyama H, Funayama M, Watanabe H, Nakamura R, Yoshino H, Yato S, Tamura A, et al. (2012). Analysis of C9orf72 repeat expansion in 563 Japanese patients with amyotrophic lateral sclerosis. Neurobiol Aging 33, 2527.e2511–2526. [DOI] [PubMed] [Google Scholar]
- Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T, Qi Z, Downey SL, Manos PD, Rossi DJ, et al. (2011). Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nature Cell Biology 13, 541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano H, Yasuda D, Fujimori K, Morimoto S, and Takahashi S (2020). Ropinirole, a New ALS Drug Candidate Developed Using iPSCs. Trends Pharmacol Sci 41, 99–109. [DOI] [PubMed] [Google Scholar]
- Ortega JA, Daley EL, Kour S, Samani M, Tellez L, Smith HS, Hall EA, Esengul YT, Tsai Y-H, Gendron TF, et al. (2020). Nucleocytoplasmic Proteomic Analysis Uncovers eRF1 and Nonsense-Mediated Decay as Modifiers of ALS/FTD C9orf72 Toxicity. Neuron 106, 90–107.e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaki T, Uzel SGM, and Kamm RD (2018). Microphysiological 3D model of amyotrophic lateral sclerosis (ALS) from human iPS-derived muscle cells and optogenetic motor neurons. In Sci Adv, pp. eaat5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaki T, Uzel SGM, and Kamm RD (2020). On-chip 3D neuromuscular model for drug screening and precision medicine in neuromuscular disease. Nat Protoc 15, 421–449. [DOI] [PubMed] [Google Scholar]
- Pandya H, Shen MJ, Ichikawa DM, Sedlock AB, Choi Y, Johnson KR, Kim G, Brown MA, Elkahloun AG, Maric D, et al. (2017). Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat Neurosci 20, 753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz MT, Li XJ, Lavaute TM, Lyons EA, Chen X, and Zhang SC (2007). Directed neural differentiation of human embryonic stem cells via an obligated primitive anterior stage. Stem Cells 25, 1511–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, Kim CH, Park JY, O'Rourke NA, Nguyen KD, et al. (2015). Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods 12, 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patani R, Hollins AJ, Wishart TM, Puddifoot CA, Alvarez S, de Lera AR, Wyllie DJ, Compston DA, Pedersen RA, Gillingwater TH, et al. (2011). Retinoid-independent motor neurogenesis from human embryonic stem cells reveals a medial columnar ground state. Nat Commun 2, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peljto M, Dasen JS, Mazzoni EO, Jessell TM, and Wichterle H (2010). Functional diversity of ESC-derived motor neuron subtypes revealed through intraspinal transplantation. Cell Stem Cell 7, 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriot S, Mathias A, Perriard G, Canales M, Jonkmans N, Merienne N, Meunier C, El Kassar L, Perrier AL, Laplaud DA, et al. (2018). Human Induced Pluripotent Stem Cell-Derived Astrocytes Are Differentially Activated by Multiple Sclerosis-Associated Cytokines. Stem Cell Reports 11, 1199–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peteri UK, Pitkonen J, Utami KH, Paavola J, Roybon L, Pouladi MA, and Castren ML (2021). Generation of the Human Pluripotent Stem-Cell-Derived Astrocyte Model with Forebrain Identity. Brain Sci 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picchiarelli G, Demestre M, Zuko A, Been M, Higelin J, Dieterle S, Goy MA, Mallik M, Sellier C, Scekic-Zahirovic J, et al. (2019). FUS-mediated regulation of acetylcholine receptor transcription at neuromuscular junctions is compromised in amyotrophic lateral sclerosis. Nat Neurosci 22, 1793–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Q, Li D, Louis KR, Li X, Yang H, Sun Q, Crandall SR, Tsang S, Zhou J, Cox CL, et al. (2014). High-efficiency motor neuron differentiation from human pluripotent stem cells and the function of Islet-1. Nat Commun 5, 3449. [DOI] [PubMed] [Google Scholar]
- Raman S, Srinivasan G, Brookhouser N, Nguyen T, Henson T, Morgan D, Cutts J, and Brafman DA (2020). A Defined and Scalable Peptide-Based Platform for the Generation of Human Pluripotent Stem Cell-Derived Astrocytes. ACS Biomater Sci Eng 6, 3477–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos DM, Skarnes WC, Singleton AB, Cookson MR, and Ward ME (2021). Tackling neurodegenerative diseases with genomic engineering: A new stem cell initiative from the NIH. Neuron 109, 1080–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Re DB, Le Verche V, Yu C, Amoroso MW, Politi KA, Phani S, Ikiz B, Hoffmann L, Koolen M, Nagata T, et al. (2014). Necroptosis drives motor neuron death in models of both sporadic and familial ALS. In Neuron, pp. 1001–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M, Paris I, Ebeling M, Dahm N, Schweitzer C, Reinhardt D, Schmucki R, Prasad M, Kochl F, Leist M, et al. (2020). Alzheimer's Risk Gene TREM2 Determines Functional Properties of New Type of Human iPSC-Derived Microglia. Front Immunol 11, 617860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. (2011). A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D, Ngai J, Speed TP, and Dudoit S (2014). Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol 32, 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues DC, Mufteev M, Weatheritt RJ, Djuric U, Ha KCH, Ross PJ, Wei W, Piekna A, Sartori MA, Byres L, et al. (2020). Shifts in Ribosome Engagement Impact Key Gene Sets in Neurodevelopment and Ubiquitination in Rett Syndrome. Cell Rep 30, 4179–4196 e4111. [DOI] [PubMed] [Google Scholar]
- Ronchi S, Buccino AP, Prack G, Kumar SS, Schroter M, Fiscella M, and Hierlemann A (2021). Electrophysiological Phenotype Characterization of Human iPSC-Derived Neuronal Cell Lines by Means of High-Density Microelectrode Arrays. Adv Biol (Weinh) 5, e2000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati J, Ferrari D, Altieri F, Tardivo S, Ricciolini C, Fusilli C, Zalfa C, Profico DC, Pinos F, Bernardini L, et al. (2018). Establishment of stable iPS-derived human neural stem cell lines suitable for cell therapies. Cell Death Dis 9, 937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan JP, Deng HX, et al. (1993). Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362, 59–62. [DOI] [PubMed] [Google Scholar]
- Roybon L, Lamas NJ, Garcia AD, Yang EJ, Sattler R, Lewis VJ, Kim YA, Kachel CA, Rothstein JD, Przedborski S, et al. (2013). Human stem cell-derived spinal cord astrocytes with defined mature or reactive phenotypes. Cell Rep 4, 1035–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareen D, Gowing G, Sahabian A, Staggenborg K, Paradis R, Avalos P, Latter J, Ornelas L, Garcia L, and Svendsen CN (2014). Human induced pluripotent stem cells are a novel source of neural progenitor cells (iNPCs) that migrate and integrate in the rodent spinal cord. J Comp Neurol 522, 2707–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareen D, O'Rourke JG, Meera P, Muhammad AKMG, Grant S, Simpkinson M, Bell S, Carmona S, Ornelas L, Sahabian A, et al. (2013). Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. In Sci Transl Med, pp. 208ra149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieber MH (2007). Chapter 2 Comparative anatomy and physiology of the corticospinal system. In Handbook of Clinical Neurology, Eisen AA, and Shaw PJ, eds. (Elsevier; ), pp. 15–37. [DOI] [PubMed] [Google Scholar]
- Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SE, Pollard JW, et al. (2012). A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90. [DOI] [PubMed] [Google Scholar]
- Selvaraj BT, Livesey MR, Zhao C, Gregory JM, James OT, Cleary EM, Chouhan AK, Gane AB, Perkins EM, Dando O, et al. (2018). C9ORF72 repeat expansion causes vulnerability of motor neurons to Ca(2+)-permeable AMPA receptor-mediated excitotoxicity. Nat Commun 9, 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio A, Bilican B, Barmada SJ, Ando DM, Zhao C, Siller R, Burr K, Haghi G, Story D, Nishimura AL, et al. (2013). Astrocyte pathology and the absence of non-cell autonomy in an induced pluripotent stem cell model of TDP-43 proteinopathy. Proc Natl Acad Sci U S A 110, 4697–4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaltouki A, Peng J, Liu Q, Rao MS, and Zeng X (2013). Efficient generation of astrocytes from human pluripotent stem cells in defined conditions. Stem Cells 31, 941–952. [DOI] [PubMed] [Google Scholar]
- Shi Y, Hung ST, Rocha G, Lin S, Linares GR, Staats KA, Seah C, Wang Y, Chickering M, Lai J, et al. (2019). Identification and therapeutic rescue of autophagosome and glutamate receptor defects in C9ORF72 and sporadic ALS neurons. JCI Insight 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Kirwan P, and Livesey FJ (2012). Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc 7, 1836–1846. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lin S, Staats KA, Li Y, Chang WH, Hung ST, Hendricks E, Linares GR, Wang Y, Son EY, et al. (2018). Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat Med 24, 313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlevkov E, Basu H, Bray MA, Sun Z, Wei W, Apaydin K, Karhohs K, Chen PF, Smith JLM, Wiskow O, et al. (2019). A High-Content Screen Identifies TPP1 and Aurora B as Regulators of Axonal Mitochondrial Transport. Cell Rep 28, 3224–3237 e3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan SA, Darmanis S, Huber N, Khan TA, Birey F, Caneda C, Reimer R, Quake SR, Barres BA, and Pasca SP (2017). Human Astrocyte Maturation Captured in 3D Cerebral Cortical Spheroids Derived from Pluripotent Stem Cells. Neuron 95, 779–790 e776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smethurst P, Risse E, Tyzack GE, Mitchell JS, Taha DM, Chen YR, Newcombe J, Collinge J, Sidle K, and Patani R (2020). Distinct responses of neurons and astrocytes to TDP-43 proteinopathy in amyotrophic lateral sclerosis. Brain 143, 430–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, and Dragunow M (2014). The human side of microglia. In Trends Neurosci, pp. 125–135. [DOI] [PubMed] [Google Scholar]
- Soubannier V, Maussion G, Chaineau M, Sigutova V, Rouleau G, Durcan TM, and Stifani S (2020). Characterization of human iPSC-derived astrocytes with potential for disease modeling and drug discovery. Neurosci Lett 731, 135028. [DOI] [PubMed] [Google Scholar]
- Speicher AM, Wiendl H, Meuth SG, and Pawlowski M (2019). Generating microglia from human pluripotent stem cells: novel in vitro models for the study of neurodegeneration. Mol Neurodegener 14, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, et al. (2008). TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer L, Vera E, and Cornacchia D (2015). Programming and Reprogramming Cellular Age in the Era of Induced Pluripotency. Cell Stem Cell 16, 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata H (2015). Diverse subtypes of astrocytes and their development during corticogenesis. Front Neurosci 9, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, and Yamanaka S (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- Takata K, Kozaki T, Lee CZW, Thion MS, Otsuka M, Lim S, Utami KH, Fidan K, Park DS, Malleret B, et al. (2017). Induced-Pluripotent-Stem-Cell-Derived Primitive Macrophages Provide a Platform for Modeling Tissue-Resident Macrophage Differentiation and Function. Immunity 47, 183–198 e186. [DOI] [PubMed] [Google Scholar]
- Talbot K, Feneberg E, Scaber J, Thompson AG, and Turner MR (2018). Amyotrophic lateral sclerosis: the complex path to precision medicine. J Neurol 265, 2454–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tann JY, Wong LW, Sajikumar S, and Ibanez CF (2019). Abnormal TDP-43 function impairs activity-dependent BDNF secretion, synaptic plasticity, and cognitive behavior through altered Sortilin splicing. EMBO J 38, e100989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchieu J, Calder EL, Guttikonda SR, Gutzwiller EM, Aromolaran KA, Steinbeck JA, Goldstein PA, and Studer L (2019). NFIA is a gliogenic switch enabling rapid derivation of functional human astrocytes from pluripotent stem cells. Nat Biotechnol 37, 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcw J, Wang M, Pimenova AA, Bowles KR, Hartley BJ, Lacin E, Machlovi SI, Abdelaal R, Karch CM, Phatnani H, et al. (2017). An Efficient Platform for Astrocyte Differentiation from Human Induced Pluripotent Stem Cells. Stem Cell Reports 9, 600–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry L, Hamel R, Pluchino S, Durcan T, and Stifani S (2020). Characterization of Human iPSC-derived Spinal Motor Neurons by Single-cell RNA Sequencing. Neuroscience 450, 57–70. [DOI] [PubMed] [Google Scholar]
- Tian R, Gachechiladze MA, Ludwig CH, Laurie MT, Hong JY, Nathaniel D, Prabhu AV, Fernandopulle MS, Patel R, Abshari M, et al. (2019). CRISPR Interference-Based Platform for Multimodal Genetic Screens in Human iPSC-Derived Neurons. Neuron 104, 239–255.e212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomov ML, O'Neil A, Abbasi HS, Cimini BA, Carpenter AE, Rubin LL, and Bathe M (2021). Resolving cell state in iPSC-derived human neural samples with multiplexed fluorescence imaging. Commun Biol 4, 786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyzack G, Lakatos A, and Patani R (2016). Human Stem Cell-Derived Astrocytes: Specification and Relevance for Neurological Disorders. Curr Stem Cell Rep 2, 236–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahsen BF, Gray E, Thompson AG, Ansorge O, Anthony DC, Cowley SA, Talbot K, and Turner MR (2021). Non-neuronal cells in amyotrophic lateral sclerosis - from pathogenesis to biomarkers. Nat Rev Neurol 17, 333–348. [DOI] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. (2009). Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323, 1208–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varcianna A, Myszczynska MA, Castelli LM, O'Neill B, Kim Y, Talbot J, Nyberg S, Nyamali I, Heath PR, Stopford MJ, et al. (2019). Micro-RNAs secreted through astrocyte-derived extracellular vesicles cause neuronal network degeneration in C9orf72 ALS. EBioMedicine 40, 626–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpato V, Smith J, Sandor C, Ried JS, Baud A, Handel A, Newey SE, Wessely F, Attar M, Whiteley E, et al. (2018). Reproducibility of Molecular Phenotypes after Long-Term Differentiation to Human iPSC-Derived Neurons: A Multi-Site Omics Study. Stem Cell Reports 11, 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainger BJ, Kiskinis E, Mellin C, Wiskow O, Han SSW, Sandoe J, Perez NP, Williams LA, Lee S, Boulting G, et al. (2014). Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. In CellReports, pp. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainger BJ, Macklin EA, Vucic S, McIlduff CE, Paganoni S, Maragakis NJ, Bedlack R, Goyal NA, Rutkove SB, Lange DJ, et al. (2021). Effect of Ezogabine on Cortical and Spinal Motor Neuron Excitability in Amyotrophic Lateral Sclerosis: A Randomized Clinical Trial. In JAMA Neurol, pp. 186–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yi F, Fu L, Yang J, Wang S, Wang Z, Suzuki K, Sun L, Xu X, Yu Y, et al. (2017). CRISPR/Cas9-mediated targeted gene correction in amyotrophic lateral sclerosis patient iPSCs. Protein Cell 8, 365–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Kamiya D, Nishiyama A, Katayama T, Nozaki S, Kawasaki H, Watanabe Y, Mizuseki K, and Sasai Y (2005). Directed differentiation of telencephalic precursors from embryonic stem cells. Nat Neurosci 8, 288–296. [DOI] [PubMed] [Google Scholar]
- Weisheit I, Kroeger JA, Malik R, Klimmt J, Crusius D, Dannert A, Dichgans M, and Paquet D (2020). Detection of Deleterious On-Target Effects after HDR-Mediated CRISPR Editing. Cell Rep 31, 107689. [DOI] [PubMed] [Google Scholar]
- Wichterle H, Lieberam I, Porter JA, and Jessell TM (2002). Directed Differentiation of Embryonic Stem Cells into Motor Neurons. Cell 110, 385–397. [DOI] [PubMed] [Google Scholar]
- Wilson SI, Graziano E, Harland R, Jessell TM, and Edlund T (2000). An early requirement for FGF signalling in the acquisition of neural cell fate in the chick embryo. Curr Biol 10, 421–429. [DOI] [PubMed] [Google Scholar]
- Wilson SI, Rydstrom A, Trimborn T, Willert K, Nusse R, Jessell TM, and Edlund T (2001). The status of Wnt signalling regulates neural and epidermal fates in the chick embryo. Nature 411, 325–330. [DOI] [PubMed] [Google Scholar]
- Wong C-O, and Venkatachalam K (2019). Motor neurons from ALS patients with mutations in C9ORF72 and SOD1 exhibit distinct transcriptional landscapes. In Human Molecular Genetics, pp. 2799–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurm J, Konttinen H, Andressen C, Malm T, and Spittau B (2021). Microglia Development and Maturation and Its Implications for Induction of Microglia-Like Cells from Human iPSCs. Int J Mol Sci 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Zhang L, Liu G, Jiang N, Zhou W, and Zhang Y (2019). Pathological Changes in Alzheimer's Disease Analyzed Using Induced Pluripotent Stem Cell-Derived Human Microglia-Like Cells. J Alzheimers Dis 67, 357–368. [DOI] [PubMed] [Google Scholar]
- Yang SJ, Lipnick SL, Makhortova NR, Venugopalan S, Fan M, Armstrong Z, Schlaeger TM, Deng L, Chung WK, O'Callaghan L, et al. (2019). Applying Deep Neural Network Analysis to High-Content Image-Based Assays. SLAS Discov 24, 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui T, Uezono N, Nakashima H, Noguchi H, Matsuda T, Noda-Andoh T, Okano H, and Nakashima K (2017). Hypoxia Epigenetically Confers Astrocytic Differentiation Potential on Human Pluripotent Cell-Derived Neural Precursor Cells. Stem Cell Reports 8, 1743–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuva-Aydemir Y, Almeida S, Krishnan G, Gendron TF, and Gao FB (2019). Transcription elongation factor AFF2/FMR2 regulates expression of expanded GGGGCC repeat-containing C9ORF72 allele in ALS/FTD. Nat Commun 10, 5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Coyne AN, and Lloyd TE (2018). Drosophila models of amyotrophic lateral sclerosis with defects in RNA metabolism. Brain Res 1693, 109–120. [DOI] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, et al. (2015a). The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. In Nature, pp. 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SC, Wernig M, Duncan ID, Brustle O, and Thomson JA (2001). In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol 19, 1129–1133. [DOI] [PubMed] [Google Scholar]
- Zhang X-H, Tee LY, Wang X-G, Huang Q-S, and Yang S-H (2015b). Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Molecular Therapy - Nucleic Acids 4,e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, et al. (2013). Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78, 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Devlin AC, Chouhan AK, Selvaraj BT, Stavrou M, Burr K, Brivio V, He X, Mehta AR, Story D, et al. (2020). Mutant C9orf72 human iPSC-derived astrocytes cause non-cell autonomous motor neuron pathophysiology. Glia 68, 1046–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziff OJ, and Patani R (2019). Harnessing cellular aging in human stem cell models of amyotrophic lateral sclerosis. Aging Cell 18, e12862. [DOI] [PMC free article] [PubMed] [Google Scholar]