Abstract
Bardet-Biedl syndrome (BBS) is a hereditary genetic disorder that results in numerous clinical manifestations including olfactory dysfunction. Of at least 21 BBS-related genes that can carry multiple mutations, a pathogenic mutation, BBS1M390R, is the single most common mutation of clinically diagnosed BBS outcomes. While the deletion of BBS related genes in mice can cause variable penetrance in different organ systems, the impact of the Bbs1M390R mutation in the olfactory system remains unclear. Using a clinically relevant knock-in mouse model homozygous for Bbs1M390R, we investigated the impact of the mutation on the olfactory system and tested the potential of viral-mediated, wildtype gene replacement therapy to rescue smell loss. The cilia of olfactory sensory neurons (OSNs) in Bbs1M390R/M390R mice were significantly shorter and fewer than those of wild-type mice. Also, both peripheral cellular odor detection and synaptic-dependent activity in the olfactory bulb were significantly decreased in the mutant mice. Furthermore, to gain insight into the degree to which perceptual features are impaired in the mutant mice, we used whole-body plethysmography to quantitatively measure odor-evoked sniffing. The Bbs1M390R/M390R mice showed significantly higher odor detection thresholds (reduced odor sensitivity) compared to wild-type mice, however, their odor discrimination acuity was still well maintained. Importantly, adenoviral expression of Bbs1 in OSNs restored cilia length and re-established both peripheral odorant detection and odor perception. Together, our findings further expand our understanding for the development of gene therapeutic treatment for congenital ciliopathies in the olfactory system.
Keywords: olfactory cilia, Bardet-Biedl syndrome, ciliopathies, gene therapy, whole body plethysmography, olfactory perception
Introduction
Olfaction is an important way for humans to communicate with the external environment (1) and defects in olfaction markedly decreases quality of life (2, 3). Population-based studies of olfactory dysfunction indicate that its prevalence varies between 2.7% and 24.5% and increases at older ages (4, 5). Still, olfactory dysfunction is often under-recognized, and the underlying mechanisms are not well understood. Recently, reports of the loss of sense of smell induced by COVID-19 have drawn more attention to the importance of olfaction (6–8) and added to the growing list of causes of olfactory loss. Another cause of olfactory dysfunction is a class of genetic disorders with defects in cilia, termed ciliopathies (9–11). Bardet-Biedl syndrome (BBS) is a ciliopathy characterized by numerous clinical manifestations in multiple ciliated organ systems including in the olfactory system (11, 12). Mutations in any of at least 21 BBSome related genes (13–22), encoding a protein complex consisting of eight BBS core proteins (BBS1, 2, 4, 5, 7, 8, 9, 18) (23, 24), can induce BBS. The BBSome is involved in the assembly and stabilization of the intraflagellar transport (IFT) complex and mainly functions as a cargo adaptor for IFT that regulates cilia biogenesis and ciliary protein trafficking (25, 26). The BBSome has varying functions in different types of cilia (27–30), explaining why genetic mutation or deletion of BBSome related genes in mice can cause variable penetrance in different organ systems (27, 28, 31, 32). Even within the olfactory system, the loss of specific BBSome core components shows differential impact on IFT trafficking (28), highlighting the need to understand the role of individual BBS proteins in the olfactory system, as well as in other ciliated organ systems.
As one of the most important BBSome subunits, BBS1 is directly involved in ciliary cargo protein recognition, binding, and trafficking (33–36). It has been discovered that a single point mutation, M390R, in BBS1 dissociates BBS4 from the BBSome core complex, thus impeding proper BBSome function (37). More importantly, BBS1 is the single most common gene responsible for clinically diagnosed BBS outcomes (31, 38, 39). The BBS1M390R mutation accounts for approximately 80% of BBS1 mutations (40). Individuals with the BBS1M390R mutation were reported to be anosmic (loss of sense of smell), as assessed with a smell identification test (12). A clinically relevant Bbs1M390R/M390R knock-in mouse model recapitulated the hallmark symptoms experienced by BBS patients, including retinal degeneration, male infertility, brain abnormalities, and obesity (31, 41). The mutant mice were suggested to have olfactory dysfunction utilizing the buried food-seeking test (31), however the potential for cognitive or movement impairment complicates interpretation of the results (42). It is therefore important to investigate the consequence of the Bbs1M390R mutation on the olfactory system at the cellular, tissue, and perceptual level.
Despite the large unmet medical need in olfaction, there are no curative therapeutic options for patients (43). Gene therapy offers a promising corrective treatment for olfactory dysfunction caused by ciliopathies. Our previous studies in ciliopathy mouse models provided proof of concept that olfactory impairment could be remedied by intranasal viral gene delivery of the wildtype gene (28, 43, 44). However, all these studies in BBS were conducted in genetic knockout (a complete and/or significant loss of protein) mouse models rather than a mouse model with a mutated gene, such as Bbs1M390R/M390R. Gene therapeutic restoration in a mouse model expressing a mutant protein could pose challenges, as the mutated protein may inhibit or compete with virally expressed wildtype protein (45). This is especially important because the genetic cause of BBS patients is predominantly through mutations in BBS related genes rather than gene deletions (40, 46, 47), therefore, adding cause to explore gene therapy in the mutant Bbs1M390R/M390R mice. Furthermore, previous investigation of olfactory dysfunction in ciliopathy mouse models did not assess the degree to which perceptual features, including odor detection threshold and odor discrimination ability, were impaired. For the development of curative treatments, it is critical to understand whether defects in perception could be restored by single gene replacement therapy. Importantly, in other ciliated systems, the potential of genetic restoration in the Bbs1M390R/M390R mouse model through genetic breeding demonstrated tissues specific effects (48). Specifically, the ectopic expression of human BBS1 was only able to rescue male infertility but not retinal degeneration (48). Therefore, in the context of the Bbs1M390R mutation it is unclear if all tissues are amenable to rescue with ectopic expression of wildtype Bbs1 gene. Together, this elucidates the need to assess if viral mediated delivery of gene therapy in Bbs1M390R/M390R mice can rescue olfactory dysfunction. In the present study, we characterized the olfactory phenotype from the cellular to perceptual level in the Bbs1M390R/M390R mouse model (31). Importantly, we demonstrated that wildtype gene replacement could restore olfactory sensory input to the mutant mice. This work highlights the potential for gene therapy in the treatment of congenital ciliopathies and provides a proof-of-concept for the restoration of cilia in other organ systems in a clinically relevant mouse model.
Materials and Methods
Mice.
Bbs1M390R/M390R mice (31) of both sexes (3–4 months of age) were bred and maintained in groups of five animals per cage under a 12-h light/dark cycle at the University of Florida. Bbs1M390R/M390R mice were of a 129/SvEv background. The breeding scheme involved crossing heterozygous mice (Bbs1M390R/+) and utilizing WT littermates as controls and homozygous mice as mutant mice for experimentation. All animal procedures were ethically reviewed and approved by the University Committee for the Use and Care of Animals at the University of Florida. The experiments were performed in accordance with the US National research Council’s Guide for the Care and Use of Laboratory Animals, the US Public Health Service’s Policy on Human Care and Use of Laboratory Animals, and Guide for the care and Use of Laboratory Animals. Genotyping was performed according to previously published work (31). The genotyping primers used were as following from 5’ to 3’, Wild Type Forward: GAA CCT TGA TTT GGG CTC TCC; Mutant Forward: GCT ACC CGT GAT ATT GCT GAA; Common Reverse: AGA TTT GAT CTC CCG ATC TGC.
cDNA constructs and adenovirus production.
Adenovirus MP-GFP, MP-mCherry, mCherry-M71, mCherry-Centrin-2, and BBS1-mCherry were described previously (25, 28, 43). Following the manufacturer’s protocols, all target protein expression cDNAs with fluorescent proteins expression cDNA were inserted into the pAd/CMV/V5-DEST expression vector (Gateway technology, Invitrogen, Waltham, MA, USA). Adenovirus was produced in HEK293 cells following the ViraPower protocol (Invitrogen, Waltham, MA, USA), isolated with the Virapur Adenovirus mini purification Virakit, dialyzed in 2.5% glycerol, 25 mM NaCl, and 20 mM Tris-HCl (pH 8.0) with a Slide-A-Lyzer dialysis cassette (Thermo Fisher Scientific, Waltham, MA, USA) at 4°C overnight and stored at −80 °C.
Intranasal viral administration.
Mice at 3–4 months of age were lightly anesthetized with isoflurane and received 20 μl of virus on 3 consecutive days. The viral infection was performed by a pulled 1-mL syringe (~0.5-mm tip) placed at the nostrils and administered during each inhalation. Then the mice were used for examination, 10 days after the third treatment to allow for adequate protein expression.
Live en face confocal imaging.
Virally transduced animals were euthanized with CO2, rapidly decapitated, and bisected along the cranial midline, and then the olfactory turbinates were exposed by removing the septal tissue. The tissue was placed with the turbinate surface facing down in a bath of freshly oxygenated artificial cerebrospinal fluid (ACSF) and used for confocal imaging.
Immunohistochemistry.
Mice were anesthetized with isoflurane and perfused with 4% paraformaldehyde (PFA) in 1x PBS. After decapitation, the mouse heads were post-fixed in 4% PFA overnight at 4 °C and then decalcified in 0.5 M EDTA/1x PBS for 3 days at 4 °C. Following, snouts were cryoprotected in 10%, 20% and 30% sucrose in 1x PBS for 2 h, 2 h and overnight, respectively, at 4 °C. Following, the tissue was frozen in optimal cutting temperature (OCT) compound and cut into sections (10–12 μm) on a Leica cryostat (Leica Biosystems, Buffalo Grove, IL, USA). The following primary antibodies and concentrations were used: goat anti-OMP (1:1000; 544–10001, Wako, Richmond, VA, USA); mouse anti-acetylated α-tubulin (1:1000; clone 6–11 B-1, T6793, Sigma-Aldrich, St. Louis, MO, USA), rabbit anti-tyrosine hydroxylase (1:500, AB152, Millipore, Burlington, MA, USA). Standard immunostaining was performed as previously described (28).
Electro-olfactogram.
EOG responses to odor delivery were recorded from multiple turbinates using a MultiClamp 700A amplifier controlled by pClamp software (Molecular Devices, San Jose, CA). After euthanizing with CO2, the olfactory turbinates of the mice were exposed for EOG recording. All odorants were diluted in DMSO and mixed to the final dilution in ultrapure water. Then odorants, amyl acetate (AA), hexanal, 1-octanol, and eugenol, were delivered in vapor-phase along with the humidified airflow to the surface of the tissue. Tissues were allowed 1 min between subsequent odorant deliveries to reduce the adaptation of the EOG response to the previous odorant. Electrodes (1–3 MΩ) were made from borosilicate glass capillaries and filled with 0.5% SeaPlaque agarose in PBS. The data were analyzed with Clampfit (Molecular Devices, San Jose, CA).
Whole-body plethysmography.
Odor-induced sniffing responses were recorded from a whole-body plethysmograph chamber and controlled by pClamp (Molecular Devices, San Jose, CA) software. All odorants were diluted in mineral oil in log series and delivered in the vapor phase with constant air delivery (1L/min) into the chamber. Animals were habituated to the experimental setting for three days prior the experiments so they could adapt to the background odor, plethysmograph chamber, and possible pressure changes which may be associated with odor delivery. The animals were placed in the plethysmograph for 20min/day with 10 times mineral oil (Sigma-Aldrich, St. Louis, MO, USA) vapor delivery. The recording was done in 4 subsequent days, each consisting of 10 trials of mineral oil followed by the presentation of an odorant at 10−8, 10−7, 10−6, 10−5, 10−4, 10−3, 10−2 and 10−1 Torr (1 Torr = 133.32 Pa). The odor detection threshold data was collected from the response of each mouse to 4 different odorants. Sniffing frequency ratios (sniffing rate 5 seconds pre vs 5 seconds after odor delivery) were calculated with Clampfit (Molecular Devices, San Jose, CA) and compared.
Odor discrimination.
The experiment was performed using a whole-body plethysmograph experimental setting. Mice were subjected to this test the day after the completion of three days mineral oil habituation (20min/day, 10times mineral oil vapor delivery). To ensure all mice could detect the odor, odorants were diluted in mineral oil to 10−3 Torr (1 Torr = 133.32 Pa) vapor pressure. Following 10 trials of mineral oil delivery, 10−3 Torr of odorant was delivered 10 times, then followed by an additional odorant in the same manner. Sniffing frequency ratios were calculated with Clampfit (Molecular Devices, San Jose, CA) and compared.
Statistical analysis.
Results are presented as mean ± SEM of at least three independent experiments. GraphPad Prism 6 software (GraphPad, San Diego, CA, USA) was used for statistical tests. Student’s t-test was used for comparison of the results between two groups and one-way ANOVA was used to calculate the statistical significance among multiple groups. *, **, *** and **** indicates p <0.05, p <0.01, p <0.001, and p <0.0001 statistical differences compared to control or untreated groups, respectively. Images were processed using Adobe Photoshop CS6 (Adobe, San Jose, CA, USA).
Results
Bbs1M390R mutation reduces olfactory cilia length and number.
In mammals, olfactory sensory neurons (OSNs) are bipolar neurons that extend their multiple non-motile sensory cilia to the apical surface of the olfactory epithelium (OE) forming a meshwork that allows for efficient odorant detection. In order to understand the penetrance and pathogenic mechanism of the Bbs1M390R mutation within the olfactory system, we characterized the olfactory phenotype of Bbs1M390R/M390R mice at several levels. First, coronal sections of the OE were immunostained for olfactory marker protein (OMP) to label mature OSNs and the olfactory cilia were immuno-labeled with anti-acetylated α-tubulin (Acet. Tubulin), a post-translational modification of tubulin that is highly concentrated in cilia (43). In the wildtype (WT) control group, acetylated α-tubulin displayed robust staining of the cilia layer, shown at the apical surface along the turbinates (Fig. 1A). However, in the Bbs1M390R/M390R mice there was a noticeable reduction in acetylated α-tubulin signal, suggesting an overall loss of olfactory ciliation (Fig. 1B). Interestingly, the Bbs1M390R/M390R mice did not show a reduction in acetylated α-tubulin on the apical surface of the adjacent respiratory epithelium (Fig. 1B), which like the OE possess cells with multiple cilia. This is consistent with other BBS deletion mouse models, where previous work showed lack of impact on respiratory ciliation (28, 43). Next, to better resolve cilia from individual OSNs and allow for quantification of cilia length and number, we used intranasal inoculation of adenovirus 5 encoding a fluorescently tagged cilia marker protein, myristoylated-palmitoylated (MP)-mCherry combined with live en face imaging of the surface of the OE (25) (Fig. 1C). The olfactory cilia in Bbs1M390R/M390R mice were significantly shorter and fewer than those of WT mice (Fig. 1D–F).
Figure 1. Olfactory cilia loss in Bbs1M390R/M390R mutant mice.

(A&B) Representative confocal images of coronal sections through nasal epithelium of (A) WT control and (B) Bbs1M390R/M390R mutant mice, immunostained for acetylated α-tubulin (Acet. Tubulin) to reveal ciliary microtubules and olfactory marker protein (OMP) to mark mature OSNs. The olfactory epithelium (OE) and respiratory epithelium (RE) line the turbinates of the nasal cavity (NC). Bbs1M390R/M390R mutants showed global acetylated α-tubulin signal reduction on the OE apical surface. (C) Representative live en face confocal images of ectopically expressed MP-mCherry in OSN cilia with Bbs1M390R/M390R OSNs possessing fewer and shorter cilia than control group. (D) Cumulative distribution of cilia lengths from en face confocal images of (black) WT and (red) Bbs1M390R/M390R OSNs. (E&F) Quantification of OSN cilia length (E) and cilia number (F) per OSN. N = 4 mice (3months old) for each group. (G) Representative images from (top) control and (bottom) Bbs1M390R/M390R OSNs infected with mCherry-Centrin2 depicting loss of basal bodies in mutant mice. (H) Quantification of basal body numbers demonstrated a decrease in Bbs1M390R/M390R OSNs (n = 53 OSNs) compared to control (n = 45 OSNs). Student’s unpaired t- test, ****p < 0.0001. Values represent means ± SEM. Scale bar, 500μm (A, B) and 40 μm (insets of A, B), 10 μm (C), 5 μm (G).
To further confirm the reduction of cilia number and understand the impact of the Bbs1M390R mutation on the basal body, which are modified centrioles located at the base of cilia that are necessary for cilia formation, we measured the number of basal bodies in the dendritic knobs of WT and Bbs1M390R/M390R mice by en face imaging. Basal bodies were identified by ectopically expressed mCherry-Centrin-2, and their analysis within the dendritic knob revealed a reduced number in Bbs1M390R/M390R OSNs compared with WT controls (Fig. 1G&H). Our results demonstrated that OSNs with Bbs1M390R mutation retain the capacity to assemble basal bodies but fail to maintain normal basal body number, which is consistent with the result that Bbs1M390R/M390R mice have fewer cilia than WT mice (Fig. 1F). Together, these observations show that Bbs1M390R mutation affects olfactory cilia maintenance.
Bbs1M390R/M390R OSNs have impaired odor detection.
To test if olfactory cilia loss results in functional impairment, electro-olfactogram (EOG) recordings were performed as a measurement of peripheral odor detection. EOG records the change in electrical potentials of a population of OSNs in response to vaporized odor delivery. Compared with WT controls, Bbs1M390R/M390R showed a significantly reduced EOG signal amplitude generated from presentation of increasing concentrations of amyl acetate (AA) as well as several other odorants (Fig. 2A&B). These data show that the Bbs1M390R mutation impairs odor detection at the level of the OE. BBS proteins have been implicated in the trafficking of ciliary membrane proteins, including G protein-coupled receptors (GPCRs) (46, 47). Therefore, we assessed if olfactory receptors (ORs), which are members of the GPCR superfamily and critically important for olfactory signal transduction, were localized to the olfactory cilia. Co-expression of ectopically expressed, MP-GFP and mCherry-M71 showed that ectopic odorant receptor M71 localized to the residual OSN cilia of Bbs1M390R/M390R animals (SFig.1). Even with impaired OSN cilia length and number, Bbs1M390R/M390R mutants retain the capacity for trafficking of odorant receptor M71 to the cilia of OSNs, which suggests that the reduced EOG response in Bbs1M390R/M390R mice is not due to the mislocalization of olfactory signaling components.
Figure 2. Bbs1M390R/M390R mice OSNs have impaired peripheral odor detection, reduced TH and smaller glomeruli.

(A) Representative electro-olfactogram (EOG) recordings from the surface of the olfactory epithelium of WT control and mutant mice in response to different concentration of amyl acetate (AA). Arrowheads indicate the time of odor delivery. (B) Quantified EOG data showing the Bbs1M390R/M390R mice had reduced amyl acetate (AA), hexanal, 1-octanol, and eugenol responses compared with WT control group. 10 WT and 11 Bbs1M390R/M390R mice (3months old) were used for the experiment. Two way ANOVA, **p <0.01, *** p <0.001, and ****p <0.0001. Values represent means ± SEM. (C) Immunolabeling coronal sections of the olfactory bulb with TH, a marker for neuronal activity. (D) Quantification of normalized TH intensity showing mutant mice (0.579 ± 0.0735) had a decrease in normalized TH intensity compared to WT (WT 1.0 ± 0.0735). (E) 4′,6-diamidino-2-phenylindole (DAPI) stained nuclei indicates the glomerular area (red circles). (F) Normalized glomerular area quantification. Bbs1M390R/M390R mice have a smaller glomerular area (0.778 ± 0.0617) compared to their WT littermate (1.0 ± 0.0617). N value of 4 WT and 4 Bbs1M390R/M390R. Student’s unpaired t-test, ***p <0.001, ****p <0.0001. Values represent means ± SEM. Scale bar, 50 μm.
After odorants bind to ORs present in cilia, the signal is then transduced within the OSN, which forms synaptic connections with second-order neurons in the olfactory bulb (OB), specifically in spherical structures called glomeruli (49). We investigated if mutating BBS1 would impact synaptic activity by immunostaining for tyrosine hydroxylase (TH), a marker for synaptic activity, and OMP, to identify glomeruli. Bbs1M390R/M390R mice showed a reduction in TH intensity per glomerulus compared to their WT littermate controls, shown in the representative image (Fig. 2C) and quantified data (Fig. 2D). Then we investigated if the reduction of afferent input would impact the size of glomeruli. As shown in Fig. 2E&F, Bbs1M390R/M390R mice had smaller glomerular area compared to that of WT. Together, the data show that Bbs1M390R mutation results in a reduction of odor-stimulated synaptic activity in the OB.
The Bbs1M390R/M390R mice show a higher odor detection threshold.
It is important to determine whether reduced olfactory cilia length causes olfactory function defects at the whole animal level. Standard testing of the olfactory function includes assessing the odor detection threshold and conducting an odor discrimination test. Traditional operant-based assays to measure odor detection thresholds and odor discrimination require odor association training and learning (50, 51). However, cilia deficient mice have severely impaired odor detection making it difficult to complete odor associated training. Further, global mutation of Bbs1 may affect neuronal cilia function (52) which could impact learning(53). Together, these factors would confound results from odor-association tasks. An alternative approach that is independent of associative learning is the odor habituation/dishabituation test, as it takes advantage of a rodent’s tendency to investigate novel smells (54, 55), to assay odor detection sensitivity and odor discrimination (56). The cotton tip-based odor habituation/dis-habituation test has been widely used to assess olfactory function of olfactory deficient animals, however, large inconsistency and variability can be easily introduced by both the observer and the experimental setting. Taking advantage of the innate property of mice in which they increase their sniffing rate upon detection of a novel odorant, we combined the cross-habituation assay and whole-body plethysmography (Fig. 3A&B) to directly measure odor-evoked sniffing. This strategy largely reduces potential systematic error and provides a robust and sensitive behavioral platform to quantify the influence of cilia loss on odor perception (57).
Figure 3. Bbs1M390R/M390R mice exhibit a higher odor detection threshold.

(A) Representative plethysmograph traces prior to and during delivery of amyl acetate 10−6 M (arrow). Applicant of odorant (arrow) failed to elicit bouts of high frequency sniffing in Bbs1M390R/M390R (bottom) which were readily apparent in the WT mouse (top). (B) Detection thresholds of 9 WT, 12 Bbs1M390R/M390R (average of 4 odors / mouse) from an ascending staircase paradigm indicating reduced odorant sensitivity (increased detection thresholds) in Bbs1M390R/M390R mice. Each mouse was delivered 10 trials of vaporized mineral oil followed by presentation of an odorant at 10−8, 10− 7, 10−6, 10−5, 10−4, 10−3, 10−2, and 10−1 Torr. Sniffing frequency ratios (sniffing Hz pre vs during odor) were compared between groups. Student’s unpaired t-test, ****p < 0.0001. ***p < 0.001. **p < 0.01.*p < 0.05. Values represent means ± SEM.
Animals were habituated to the experimental setting for two days prior to the testing. Mineral oil was delivered 10 times to control for variables and then odorants, diluted in mineral oil, were delivered at different vapor pressures in constant airflow. The WT control mice showed an increased sniffing rate following 10−8 Torr of odor delivery (1 Torr = 133.32 Pa), indicating the detection of odorant (Fig. 3B). However, the Bbs1M390R/M390R mice did not increase their sniffing rate until the delivery of odorants at 10−4 Torr (Fig. 3B). Compared with control mice, Bbs1M390R/M390R mice showed odor detection thresholds that required significantly higher vapor pressure of odorants. Importantly, Bbs1M390R/M390R mice showed a similar response as WT mice following 10−3 Torr of odor delivery, which suggests that Bbs1M390R/M390R mice can detect the odor with the vapor pressure equal or higher than 10−3 Torr. These data show that Bbs1M390R/M390R mice have reduced odor detection sensitivity.
The Bbs1M390R/M390R mice have normal odor discrimination acuity.
Humans and other animals can detect and discriminate thousands of structurally different odorants including differentiating between enantiomers and even molecules that differ by a single carbon (58). To examine the odor discrimination ability of the Bbs1M390R/M390R mice, whole-body plethysmography combined with an odor cross-habituation assay was performed. For the odor discrimination experiments, odorants were diluted to 10−3 Torr, a vapor pressure that Bbs1M390R/M390R mice can detect, as to not confound the discrimination assay (Fig. 3B). When an odorant was repeatedly presented to the WT mouse, the sniffing response to the odorant attenuated as the mouse habituated (Fig.4A). However, when a different odorant was presented, and if it could be discriminated from the previous habituated odor, the response then increased. Importantly, after the presentation of three structurally different odorants, results showed that Bbs1M390R/M390R mice retain normal gross odor discrimination acuity (Fig.4A). We further tested the fine odor discrimination ability of Bbs1M390R/M390R mice by using two pairs of odorants with similar structures. Specifically, we tested limonene (+) and limonene (−) which represent a pair of enantiomers (58) as well as butanol and pentanol, alcohols with only a single carbon difference (59). Surprisingly, Bbs1M390R/M390R mice still retained comparable fine odor discrimination acuity as WT mice (Fig. 4B). These data indicate that the Bbs1M390R/M390R mice have normal odor discrimination acuity despite significant impairment in peripheral odor detection.
Figure 4. Bbs1M390R/M390R mice have normal gross and fine odor discrimination acuity.

Measuring discrimination acuity of WT and Bbs1M390R/M390R mice by odor cross-habituation assay using a whole-body plethysmograph chamber. (A) For the normal gross odor discrimination assay, the mouse was delivered 10 trials of vaporized mineral oil followed by a presentation of amyl acetate (10−3 Torr), 10 times in a row, followed by cineole in the same manner, and hexanal. 5 WT and 6 Bbs1M390R/M390R mice. (B) To assess fine odor discrimination acuity, the mouse was delivered 10 trials of vaporized mineral oil followed by a presentation of limonene (–) (10−3 Torr), 10 times in a row, followed by limonene (+) in the same manner, and butanol and pentanol. 6 mice per group. Sniffing frequency ratios (sniffing Hz pre vs during odor) were compared between groups. Values represent means ± SEM.
Ectopically expressed BBS1 rescues the Bbs1M390R/M390R olfactory cilia length, peripheral odor detection and odor detection threshold.
To examine the potential of gene replacement therapy to correct BBS olfactory deficits, we used intra-nasal adenoviral delivery of wildtype Bbs1-mCherry in Bbs1M390R/M390R OSNs. Using live en face confocal imaging, we examined whether ectopic expression of the WT Bbs1 gene could restore olfactory cilia of mutant animals. Compared with non-treated Bbs1M390R/M390R OSNs, BBS1-mCherry positive olfactory cilia length was significantly increased (Fig. 5A&B). Following, EOG was performed to test if the partial restoration of cilia length was sufficient to rescue the impaired peripheral odor detection. Compared with untreated Bbs1M390R/M390R mice, the mice receiving Bbs1-mCherry showed significantly higher odor-evoked responses to high concentrations of AA delivery (Fig.5C). Finally, to further test the effect of gene therapy as a potential treatment in the odor perception in BBS1 M390R-associated hyposmia, we examined odor detection thresholds in Bbs1-mCherry treated mice. Interestingly, 10 days post virus infection, Bbs1M390R/M390R mice with the expression of BBS1 showed an increased sniffing rate following 10−8 Torr odor delivery (Fig. 5D). Taken together, these data show that wildtype gene replacement in a subset of OSNs rescues Bbs1M390R/M390R olfactory cilia length, and in turn restores both peripheral odor detection and odor detection threshold.
Figure 5. WT BBS1 gene replacement rescues olfactory cilia length, peripheral odor detection and odor detection threshold in Bbs1M390R/M390R mice.
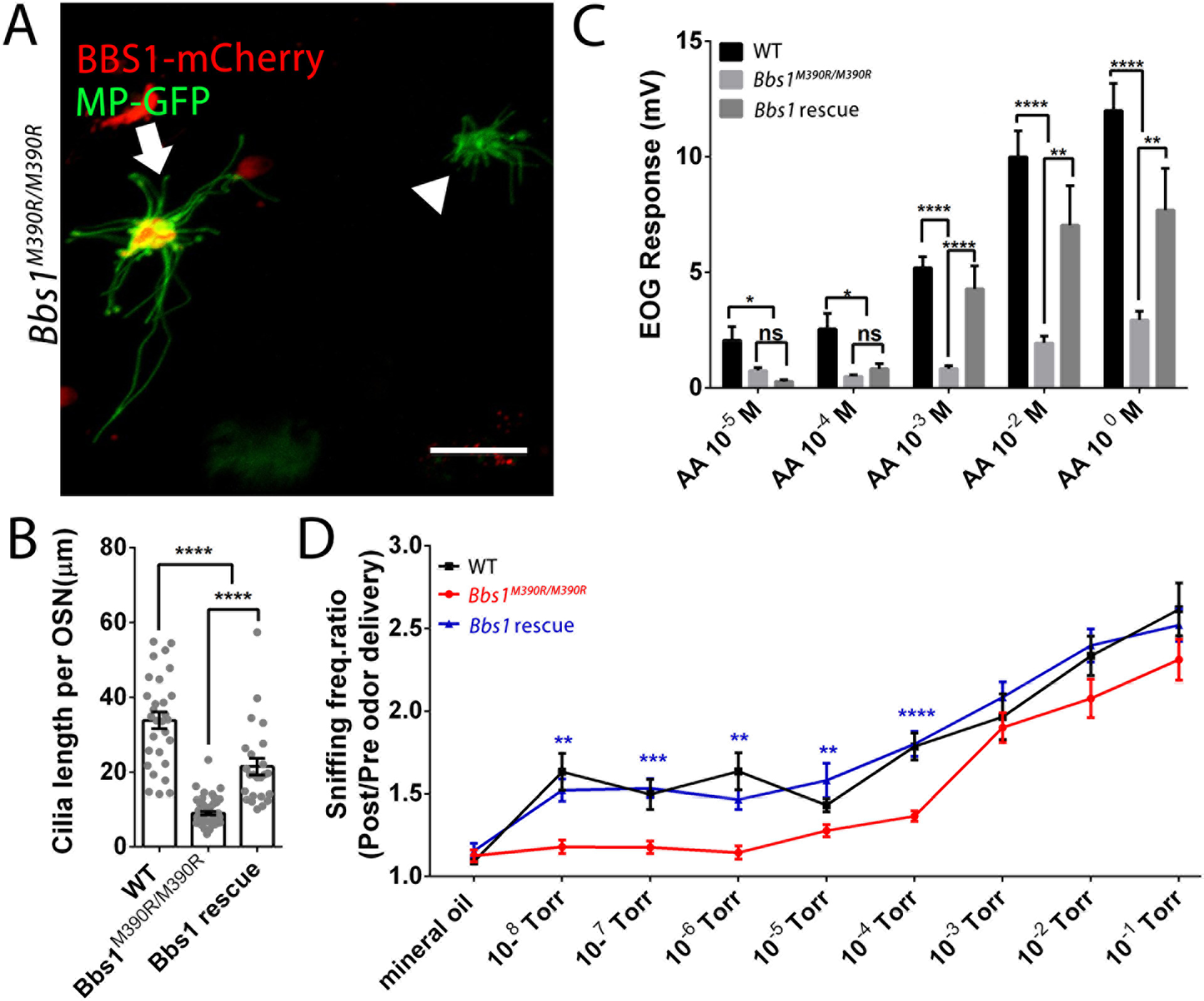
(A) Representative live en face confocal images of ectopically expressed BBS1-mCherry in Bbs1M390R/M390R OSNs restoring the olfactory cilia length. Scale bar, 10μm (B) Quantification of olfactory cilia length per OSN for WT, Bbs1M390R/M390R, and Bbs1M390R/M390R rescue. (C) Quantified EOG data showing the BBS1-mCherry treatment significantly rescue Bbs1M390R/M390R mice odor detection under high concentration of amyl acetate (AA) stimulation condition. (D) Odor detection threshold can be rescued by ectopic expression of BBS1-mCherry. The mouse was delivered 10 trials of vaporized mineral oil followed by the presentation of an odorant at 10−8, 10− 7, 10−6, 10−5, 10−4, 10−3, 10−2, and 10−1 Torr (4 different odors/mouse). Sniffing frequency ratios (sniffing Hz pre vs during odor) were compared between Bbs1M390R/M390R and Bbs1M390R/M390R rescue groups. 11 Bbs1M390R/M390R mice were used in the rescue group. One way ANOVA, ****p < 0.0001. ***p < 0.001. **p < 0.01.*p < 0.05. Values represent means ± SEM.
Discussion
Our work investigated the penetrance of the Bbs1M390R mutation on cellular odor detection in OSNs including the concomitant effects of sensory deprivation by cilia loss on olfactory perception and the extent of recovery following restoration of cilia to OSNs. Compared with WT mice, Bbs1M390R/M390R mice showed significantly shorter and fewer olfactory cilia, impaired odorant detection at the periphery, reduced synaptic-dependent activity within the OB, and had higher odor detection thresholds. However, the mutant mice retained normal odor discrimination acuity. Importantly, using adenoviral gene replacement of the WT Bbs1 gene, the olfactory cilia length was partially rescued in terminally differentiated neurons, which incredibly was sufficient to re-establish both peripheral odorant detection and odor detection thresholds. Notably, although this is not the first report of applying a whole-body plethysmograph system to measure olfactory perception in rodents, the current study is the first to use this setting to assess olfactory impairments in a clinically relevant ciliopathy model and to evaluate the therapeutic effect of gene replacement in odor detection sensitivity.
Ciliopathies are highly pleiotropic and typically display a large spectrum of abnormalities within different organ systems (28, 60, 61). This variability in penetrance and phenotype is true within BBS. For instance, Bbs4−/− mice had cilia with normal distribution and abundance in the brain (27) and respiratory system (28), but abnormally long cilia in kidney cells (32), and dramatically reduced cilia length and number in olfactory neurons (28). These discrepancies appear to persist in the Bbs1M390R/M390R mice that showed relatively normal respiratory cilia (12, 62) that was confirmed by our immunostaining (Fig. 1A&B) but elongated cilia in ependymal cells that reside in the brain (31). As we report here and similar to other BBS models (28, 43, 63), the Bbs1M390R mutation reduced OSN ciliation (Fig. 1). Interestingly, the differences in phenotype between tissues is not limited to cilia morphology but also may include the cellular function of the BBSome itself. Previous reports in other ciliated systems showed that the BBSome is integral for the ciliary localization of receptors (27, 64), however in the olfactory system ORs localized properly in olfactory cilia in both the Bbs1M390R/M390R (SFig. 1) and Bbs4−/− mouse models (28). Together, these highlight unique aspects of olfactory cilia but also the necessity of studying the penetrance of BBS in individual ciliated organ systems.
Defects in glomeruli, including structure and synaptic activity, have been reported in multiple ciliopathy mouse models (28, 43, 44). Consistent with other BBS mouse models, impaired afferent activity indicated by decreased tyrosine hydroxylase (TH) staining and a reduction in glomerular size were also observed in Bbs1M390R/M390R mice (Fig. 2C–F). Interestingly, it appears that a larger decrease in glomerular size resulted from the deletion of BBS1 in OSNs (Bbs1osnKO) (43), compared to Bbs1M390R/M390R mice. While this would require a comparative study to confirm, multiple factors may contribute to any potential differences, including genetic backgrounds, age, and the gene mutation rather than deletion. The glomerular size discrepancy could be induced by differences in the genetic backgrounds of the mouse models(65). The Bbs1M390R/M390R mouse model was of pure 129/SvEv genetic background, and Bbs1osnKO was of mixed genetic background with 129/SvEv and 129P2/OlaHsd. Additionally, there is a difference in the age of mice investigated in these two studies whereby juvenile (P21) Bbs1osnKO mice and adult (3m) Bbs1M390R/M390R mice were used. It is unknown if the timing of BBS1 loss of function impacts the glomerular size. Importantly, Bbs1M390R/M390R mice represent a global mutation model while Bbs1osnKO mice a conditional knock-out of BBS1 in mature OSNs. Regardless of any potential anatomical difference between BBS1 models, it is clear that loss or mutation in BBS1 impacts glomeruli in the OB.
Odor detection sensitivity and odor discrimination ability are major perceptual features of olfaction. Olfactory function in BBS patients has been tested clinically utilizing common smell identification tests such as UPSIT and B-SIT (12, 66), which neither measure odor detection threshold nor odor discrimination directly. It is now apparent from patients and mouse models that ciliopathies entail loss of cellular odor detection and disruption of gross olfactory-mediated behaviors (12, 28, 31, 43, 44, 63, 67–69). Several globally mutated ciliopathy mouse models (Bbs2−/−, Bbs4−/−, and Bbs1M390R/M390R) displayed olfactory deficits in the buried food seeking behavior assay (31, 68). However the interpretation of changes to odor-guided behaviors in these models can be confounded by the disruption of ciliation or ciliary function in other cell types. For example, cognitive defects or disruptions to mobility (i.e. obesity or motor control) that may occur with ciliopathies can affect movement(42) and hence can skew results in an assay dependent on timed movement for discovery. In addition, the buried food seeking behavior assay has limitations in examining olfactory function because of the variability in motivation for food seeking behavior (70, 71). Importantly, ciliopathy studies to date have examined gross odor-guided behaviors but not olfactory perceptual measures (31, 44). Here, we employed two different olfactory assays, both in the context of a whole-body plethysmograph, to quantify odor detection thresholds (an ascending staircase paradigm) (Fig. 3) and odor discrimination acuity (odor cross-habituation paradigm) (Fig. 4) in Bbs1M390R/M390R mice. These assays take advantage of the innate behavior that mice have of spontaneously increasing their sniffing rate to investigate novel stimuli. We were able to precisely quantify Bbs1M390R/M390R mice odor detection threshold and dissociate odor discrimination from odor detection sensitivity. Our data revealed that Bbs1M390R/M390R is a hyposmic model rather than an anosmic model, which is consistent with the result that Bbs1M390R OSNs, although shortened still maintain residual olfactory cilia. Importantly, our results are also consistent with BBS patients falling within the range of hyposmic to anosmic depending on the mutation. This shows that our preclinical mouse model recapitulates the symptoms of olfactory dysfunction experienced by BBS patients and justifies the use of this mouse model to further understand the pathogenesis of BBS. Knowing the odor detection threshold of mice allowed us to measure their ability to discriminate odors. By applying odor at a vapor pressure that can be detected by Bbs1M390R/M390R mice, we showed that these animals have normal odor discrimination acuity. It is still unclear if BBS patients have defects in odor discrimination. Also, odor detection threshold has not been quantitatively evaluated in BBS patients. Therefore, even if discrimination assays had been completed on patients, results may be confounded by testing odorants at concentrations below their detection threshold. The assessment in mice, however, provides novel insights into the mechanism of dysfunction in olfactory perception in BBS, which may aid in the rational design of a more accurate diagnostic test. Our results that Bbs1M390R/M390R mice had significant loss of olfactory input but retained both gross and fine odor discrimination, highlight the tremendous spare capacity of the olfactory system which likely helps to maintain the integrity of the neural circuitry necessary for odor perception. These observations are encouraging because, unlike other systems that are degenerative such as the retina, they highlight the therapeutic advantage of the olfactory system.
Previous work has shown that in ciliopathy mouse models, intranasal viral gene replacement can rescue ciliation in OSNs and restore their odor detection capacity (28, 43, 44). A limitation of these studies is that they only utilized genetic knockout BBS mouse models. Here, we used a clinically relevant BBS1 mutant mouse model, where the hypomorphic allele still expresses BBS1 protein (31, 72). However, a concern with overexpression of the WT Bbs1 gene in Bbs1M390R/M390R OSNs was whether the WT protein can compete with the endogenously expressed mutant BBS1 to efficiently incorporate into the BBSome and thus allow for proper BBSome assembly and function (37). Fortunately, we were able to rescue the olfactory deficits in the Bbs1M390R/M390R mice, suggesting that proper function of the BBSome can be restored by ectopic WT Bbs1 expression. Interestingly, analysis of the dynamic behavior of the BBSome throughout the primary cilium demonstrated a rapid recovery (halftime of 2–6 seconds) in a photobleaching assay, indicating that the BBSome complex has a relatively fast turnover rate (73). Said data suggests that the WT BBS1 protein is assembled de novo into the BBSome rather than replacing the mutant BBS1 protein in the pre-existing BBSome. However, further mechanistic studies are needed to better understand the dynamics of BBSome in OSNs.
While the restoration of odor-evoked peripheral OSN responses is meaningful, true curative therapies should also include the return of odor perception. In the context of sensory deprivation due to loss of cilia, it is still largely unknown what degree of restoration is necessary to recover perceptual features. Here we showed that WT Bbs1 gene replacement could rescue sensory input and restore the odor detection threshold to Bbs1M390R/M390R mice. Our previous data showed that our non-invasive method induced ectopic gene expression in less than 15% of mature OSNs (44). Together, even with a low infection rate, the promising restoration of olfactory sensitivity suggests that olfactory dysfunction in ciliopathy patients may be rescued by a similar approach. These findings are especially intriguing since the ectopic expression of the human BBS1 transgene in Bbs1M390R/M390R mice showed a tissue dependent restoration of select BBS phenotypes (48). Specifically, the breeding of Bbs1M390R/M390R with a transgenic mouse overexpressing human BBS1 (under the CAG promotor) remedied infertility but not retinal degeneration. The authors acknowledge that differential expression of the BBS1 transgene could account for the tissue dependent rescue. Nevertheless, this highlights the need to investigate curative therapies in targeted tissues. Further, these findings underscore the attractiveness of the olfactory system as a target for gene therapy, as restoration of cilia to only a subset OSNs is sufficient for rescue of olfactory perception.
We acknowledge the limitations inherent in the use of transgenic inbred animals, that possess identical genetic backgrounds. The heterogeneity and symptomatic variability in BBS patients have been at least partly explained by secondary-variants, which underscores the need for consideration of mutational load in BBS (74–76). However, the benefit of utilizing mouse models is that it allows for the elucidation of the precise function of the target protein and the investigation of how a single genetic defect leads to pathology. Work in this preclinical mouse model expands our understanding of the penetrance of olfactory dysfunction induced by Bbs1M390R mutation and provides proof of concept that olfactory dysfunction could be remedied by intranasal viral gene delivery of the wildtype gene. This represents just the beginning of studying the pathogenesis of BBS and developing therapies for BBS. Further efforts are needed to explore how this mutation affects olfactory function in human patients and therapeutic potential.
Considering all the potential therapeutic advantages of the olfactory system, smell loss in BBS represents a strong candidate for gene therapy. For example, in addition to the spare capacity for olfactory function described above, the OE is accessible in a noninvasive manner for viral vector delivery. Further, OSNs, unlike retinal cells, tolerate overexpression of the BBS1 protein (77). Although we used adenovirus in this study as proof of concept, our previous work shows that the OE is conducive to infection by adeno-associated viruses (AAV) (43). The use of AAV is clinically advantageous as it allows for stable incorporation of genetic material, induces only a mild immune response, and is overall considered safe (78). Importantly, the first in vivo gene therapy approved by the FDA utilized AAV for the treatment of retinal degeneration in Leber congenital amarosis, a ciliopathy (79). Given the regenerative capacity of the OE, it is unclear if stable incorporation is necessary. What is advantageous is that rather than progressive degeneration that occurs with ciliopathies in the retina and other sensory systems, the OE maintains a large therapeutic window due to lifelong neurogenesis. Therefore, it is not unrealistic to consider that olfactory function in patients could be fully restored by rescuing olfactory input using gene therapy. Our work provides a viable path to develop therapeutic options for treating olfactory loss in patients and produces a more complete understanding of the mechanisms involved in the restoration of function in ciliopathies.
Supplementary Material
Acknowledgments
We thank D. Wesson (University of Florida) for providing technical assistance and guidance in setting up the whole-body plethysmograph. This work was supported by the National Institute of Deafness and Other Communication Disorders R01DC019345 (J.R.M) and T32DC015994 (J.C.H).
Nonstandard Abbreviations
- BBS
Bardet-Biedl syndrome
- OSNs
olfactory sensory neurons
- OE
olfactory epithelium
- IFT
intraflagellar transport
- OB
olfactory bulb
- ACSF
artificial cerebrospinal fluid
- OMP
olfactory marker protein
- Acet. Tubulin
acetylated α-tubulin
- TH
tyrosine hydroxylase
- AA
amyl acetate
- GPCR
G Protein-Coupled Receptor
- EOG
electro-olfactogram
- MP
myristoylated-palmitoylated
Footnotes
Conflict of Interest Statement
The authors declare no competing or financial interest in connection with this article.
References
- 1.Sarafoleanu C, Mella C, Georgescu M, and Perederco C (2009) The importance of the olfactory sense in the human behavior and evolution. J Med Life 2, 196–198 [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffman HJ, Ishii EK, and MacTurk RH (1998) Age-related changes in the prevalence of smell/taste problems among the United States adult population. Results of the 1994 disability supplement to the National Health Interview Survey (NHIS). Ann N Y Acad Sci 855, 716–722 [DOI] [PubMed] [Google Scholar]
- 3.Hummel T, and Nordin S (2005) Olfactory disorders and their consequences for quality of life. Acta Otolaryngol 125, 116–121 [DOI] [PubMed] [Google Scholar]
- 4.Current Otorhinolaryngology Reports. Springer, 8210; Aarhus, V-DK, Denmark [Google Scholar]
- 5.Hüttenbrink KB, Hummel T, Berg D, Gasser T, and Hähner A (2013) Olfactory dysfunction: common in later life and early warning of neurodegenerative disease. Dtsch Arztebl Int 110, 1–7, e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meng X, Deng Y, Dai Z, and Meng Z (2020) COVID-19 and anosmia: A review based on up-to-date knowledge. Am J Otolaryngol 41, 102581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menni C, Valdes AM, Freidin MB, Sudre CH, Nguyen LH, Drew DA, Ganesh S, Varsavsky T, Cardoso MJ, El-Sayed Moustafa JS, Visconti A, Hysi P, Bowyer RCE, Mangino M, Falchi M, Wolf J, Ourselin S, Chan AT, Steves CJ, and Spector TD (2020) Real-time tracking of self-reported symptoms to predict potential COVID-19. Nat Med 26, 1037–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parma V, Ohla K, Veldhuizen MG, Niv MY, Kelly CE, Bakke AJ, Cooper KW, Bouysset C, Pirastu N, Dibattista M, Kaur R, Liuzza MT, Pepino MY, Schopf V, Pereda-Loth V, Olsson SB, Gerkin RC, Rohlfs Dominguez P, Albayay J, Farruggia MC, Bhutani S, Fjaeldstad AW, Kumar R, Menini A, Bensafi M, Sandell M, Konstantinidis I, Di Pizio A, Genovese F, Ozturk L, Thomas-Danguin T, Frasnelli J, Boesveldt S, Saatci O, Saraiva LR, Lin C, Golebiowski J, Hwang LD, Ozdener MH, Guardia MD, Laudamiel C, Ritchie M, Havlicek J, Pierron D, Roura E, Navarro M, Nolden AA, Lim J, Whitcroft KL, Colquitt LR, Ferdenzi C, Brindha EV, Altundag A, Macchi A, Nunez-Parra A, Patel ZM, Fiorucci S, Philpott CM, Smith BC, Lundstrom JN, Mucignat C, Parker JK, van den Brink M, Schmuker M, Fischmeister FPS, Heinbockel T, Shields VDC, Faraji F, Santamaria E, Fredborg WEA, Morini G, Olofsson JK, Jalessi M, Karni N, D’Errico A, Alizadeh R, Pellegrino R, Meyer P, Huart C, Chen B, Soler GM, Alwashahi MK, Welge-Lussen A, Freiherr J, de Groot JHB, Klein H, Okamoto M, Singh PB, Hsieh JW, Author GG, Reed DR, Hummel T, Munger SD, and Hayes JE (2020) More Than Smell-COVID-19 Is Associated With Severe Impairment of Smell, Taste, and Chemesthesis. Chem Senses 45, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zimmerman K, and Yoder BK (2015) SnapShot: Sensing and Signaling by Cilia. Cell 161, 692–692.e691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Badano JL, Mitsuma N, Beales PL, and Katsanis N (2006) The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet 7, 125–148 [DOI] [PubMed] [Google Scholar]
- 11.Oh EC, and Katsanis N (2012) Cilia in vertebrate development and disease. Development 139, 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulaga HM, Leitch CC, Eichers ER, Badano JL, Lesemann A, Hoskins BE, Lupski JR, Beales PL, Reed RR, and Katsanis N (2004) Loss of BBS proteins causes anosmia in humans and defects in olfactory cilia structure and function in the mouse. Nat Genet 36, 994–998 [DOI] [PubMed] [Google Scholar]
- 13.Katsanis N, Beales PL, Woods MO, Lewis RA, Green JS, Parfrey PS, Ansley SJ, Davidson WS, and Lupski JR (2000) Mutations in MKKS cause obesity, retinal dystrophy and renal malformations associated with Bardet-Biedl syndrome. Nat Genet 26, 67–70 [DOI] [PubMed] [Google Scholar]
- 14.Slavotinek AM, and Biesecker LG (2000) Phenotypic overlap of McKusick-Kaufman syndrome with bardet-biedl syndrome: a literature review. Am J Med Genet 95, 208–215 [PubMed] [Google Scholar]
- 15.Mykytyn K, Braun T, Carmi R, Haider NB, Searby CC, Shastri M, Beck G, Wright AF, Iannaccone A, Elbedour K, Riise R, Baldi A, Raas-Rothschild A, Gorman SW, Duhl DM, Jacobson SG, Casavant T, Stone EM, and Sheffield VC (2001) Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nat Genet 28, 188–191 [DOI] [PubMed] [Google Scholar]
- 16.Nishimura DY, Searby CC, Carmi R, Elbedour K, Van Maldergem L, Fulton AB, Lam BL, Powell BR, Swiderski RE, Bugge KE, Haider NB, Kwitek-Black AE, Ying L, Duhl DM, Gorman SW, Heon E, Iannaccone A, Bonneau D, Biesecker LG, Jacobson SG, Stone EM, and Sheffield VC (2001) Positional cloning of a novel gene on chromosome 16q causing Bardet-Biedl syndrome (BBS2). Hum Mol Genet 10, 865–874 [DOI] [PubMed] [Google Scholar]
- 17.Nishimura DY, Swiderski RE, Searby CC, Berg EM, Ferguson AL, Hennekam R, Merin S, Weleber RG, Biesecker LG, Stone EM, and Sheffield VC (2005) Comparative genomics and gene expression analysis identifies BBS9, a new Bardet-Biedl syndrome gene. Am J Hum Genet 77, 1021–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ansley SJ, Badano JL, Blacque OE, Hill J, Hoskins BE, Leitch CC, Kim JC, Ross AJ, Eichers ER, Teslovich TM, Mah AK, Johnsen RC, Cavender JC, Lewis RA, Leroux MR, Beales PL, and Katsanis N (2003) Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature 425, 628–633 [DOI] [PubMed] [Google Scholar]
- 19.Badano JL, Ansley SJ, Leitch CC, Lewis RA, Lupski JR, and Katsanis N (2003) Identification of a novel Bardet-Biedl syndrome protein, BBS7, that shares structural features with BBS1 and BBS2. Am J Hum Genet 72, 650–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang AP, Beck JS, Yen HJ, Tayeh MK, Scheetz TE, Swiderski RE, Nishimura DY, Braun TA, Kim KY, Huang J, Elbedour K, Carmi R, Slusarski DC, Casavant TL, Stone EM, and Sheffield VC (2006) Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11). Proc Natl Acad Sci U S A 103, 6287–6292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiang AP, Nishimura D, Searby C, Elbedour K, Carmi R, Ferguson AL, Secrist J, Braun T, Casavant T, Stone EM, and Sheffield VC (2004) Comparative genomic analysis identifies an ADP-ribosylation factor-like gene as the cause of Bardet-Biedl syndrome (BBS3). Am J Hum Genet 75, 475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, Lewis RA, Green JS, Parfrey PS, Leroux MR, Davidson WS, Beales PL, Guay-Woodford LM, Yoder BK, Stormo GD, Katsanis N, and Dutcher SK (2004) Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117, 541–552 [DOI] [PubMed] [Google Scholar]
- 23.Loktev AV, Zhang Q, Beck JS, Searby CC, Scheetz TE, Bazan JF, Slusarski DC, Sheffield VC, Jackson PK, and Nachury MV (2008) A BBSome subunit links ciliogenesis, microtubule stability, and acetylation. Dev Cell 15, 854–865 [DOI] [PubMed] [Google Scholar]
- 24.Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peränen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, and Jackson PK (2007) A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129, 1201–1213 [DOI] [PubMed] [Google Scholar]
- 25.Williams CL, McIntyre JC, Norris SR, Jenkins PM, Zhang L, Pei Q, Verhey K, and Martens JR (2014) Direct evidence for BBSome-associated intraflagellar transport reveals distinct properties of native mammalian cilia. Nat Commun 5, 5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wingfield JL, Lechtreck KF, and Lorentzen E (2018) Trafficking of ciliary membrane proteins by the intraflagellar transport/BBSome machinery. Essays Biochem [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berbari NF, Lewis JS, Bishop GA, Askwith CC, and Mykytyn K (2008) Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc Natl Acad Sci U S A 105, 4242–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uytingco CR, Williams CL, Xie C, Shively DT, Green WW, Ukhanov K, Zhang L, Nishimura DY, Sheffield VC, and Martens JR (2019) BBS4 is required for intraflagellar transport coordination and basal body number in mammalian olfactory cilia. J Cell Sci 132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McClintock TS, Khan N, Xie C, and Martens JR (2020) Maturation of the Olfactory Sensory Neuron and Its Cilia. Chem Senses 45, 805–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu Y, Garrison JE, Kim G, Schmitz AR, Searby CC, Zhang Q, Datta P, Nishimura DY, Seo S, and Sheffield VC (2017) BBSome function is required for both the morphogenesis and maintenance of the photoreceptor outer segment. PLoS Genet 13, e1007057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis RE, Swiderski RE, Rahmouni K, Nishimura DY, Mullins RF, Agassandian K, Philp AR, Searby CC, Andrews MP, Thompson S, Berry CJ, Thedens DR, Yang B, Weiss RM, Cassell MD, Stone EM, and Sheffield VC (2007) A knockin mouse model of the Bardet-Biedl syndrome 1 M390R mutation has cilia defects, ventriculomegaly, retinopathy, and obesity. Proc Natl Acad Sci U S A 104, 19422–19427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mokrzan EM, Lewis JS, and Mykytyn K (2007) Differences in renal tubule primary cilia length in a mouse model of Bardet-Biedl syndrome. Nephron Exp Nephrol 106, e88–96 [DOI] [PubMed] [Google Scholar]
- 33.Su X, Driscoll K, Yao G, Raed A, Wu M, Beales PL, and Zhou J (2014) Bardet-Biedl syndrome proteins 1 and 3 regulate the ciliary trafficking of polycystic kidney disease 1 protein. Hum Mol Genet 23, 5441–5451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei Q, Zhang Y, Li Y, Zhang Q, Ling K, and Hu J (2012) The BBSome controls IFT assembly and turnaround in cilia. Nat Cell Biol 14, 950–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klink BU, Gatsogiannis C, Hofnagel O, Wittinghofer A, and Raunser S (2020) Structure of the human BBSome core complex. Elife 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nozaki S, Katoh Y, Kobayashi T, and Nakayama K (2018) BBS1 is involved in retrograde trafficking of ciliary GPCRs in the context of the BBSome complex. PLoS One 13, e0195005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Q, Yu D, Seo S, Stone EM, and Sheffield VC (2012) Intrinsic protein-protein interaction-mediated and chaperonin-assisted sequential assembly of stable bardet-biedl syndrome protein complex, the BBSome. J Biol Chem 287, 20625–20635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Novas R, Cardenas-Rodriguez M, Irigoín F, and Badano JL (2015) Bardet-Biedl syndrome: Is it only cilia dysfunction? FEBS Lett 589, 3479–3491 [DOI] [PubMed] [Google Scholar]
- 39.Khan SA, Muhammad N, Khan MA, Kamal A, Rehman ZU, and Khan S (2016) Genetics of human Bardet-Biedl syndrome, an updates. Clin Genet 90, 3–15 [DOI] [PubMed] [Google Scholar]
- 40.Mykytyn K, Nishimura DY, Searby CC, Shastri M, Yen HJ, Beck JS, Braun T, Streb LM, Cornier AS, Cox GF, Fulton AB, Carmi R, Luleci G, Chandrasekharappa SC, Collins FS, Jacobson SG, Heckenlively JR, Weleber RG, Stone EM, and Sheffield VC (2002) Identification of the gene (BBS1) most commonly involved in Bardet-Biedl syndrome, a complex human obesity syndrome. Nat Genet 31, 435–438 [DOI] [PubMed] [Google Scholar]
- 41.Drack AV, Dumitrescu AV, Bhattarai S, Gratie D, Stone EM, Mullins R, and Sheffield VC (2012) TUDCA slows retinal degeneration in two different mouse models of retinitis pigmentosa and prevents obesity in Bardet-Biedl syndrome type 1 mice. Invest Ophthalmol Vis Sci 53, 100–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forsyth RL, and Gunay-Aygun M (1993) Bardet-Biedl Syndrome Overview. In GeneReviews((R)) (Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Mirzaa G, and Amemiya A, eds), Seattle (WA) [Google Scholar]
- 43.Williams CL, Uytingco CR, Green WW, McIntyre JC, Ukhanov K, Zimmerman AD, Shively DT, Zhang L, Nishimura DY, Sheffield VC, and Martens JR (2017) Gene Therapeutic Reversal of Peripheral Olfactory Impairment in Bardet-Biedl Syndrome. Mol Ther 25, 904–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Green WW, Uytingco CR, Ukhanov K, Kolb Z, Moretta J, McIntyre JC, and Martens JR (2018) Peripheral Gene Therapeutic Rescue of an Olfactory Ciliopathy Restores Sensory Input, Axonal Pathfinding, and Odor-Guided Behavior. J Neurosci 38, 7462–7475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Veitia RA (2007) Exploring the molecular etiology of dominant-negative mutations. Plant Cell 19, 3843–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fattahi Z, Rostami P, Najmabadi A, Mohseni M, Kahrizi K, Akbari MR, Kariminejad A, and Najmabadi H (2014) Mutation profile of BBS genes in Iranian patients with Bardet-Biedl syndrome: genetic characterization and report of nine novel mutations in five BBS genes. J Hum Genet 59, 368–375 [DOI] [PubMed] [Google Scholar]
- 47.Manara E, Paolacci S, D’Esposito F, Abeshi A, Ziccardi L, Falsini B, Colombo L, Iarossi G, Pilotta A, Boccone L, Guerri G, Monica M, Marta B, Maltese PE, Buzzonetti L, Rossetti L, and Bertelli M (2019) Mutation profile of BBS genes in patients with Bardet-Biedl syndrome: an Italian study. Ital J Pediatr 45, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cring MR, Meyer KJ, Searby CC, Hedberg-Buenz A, Cave M, Anderson MG, Wang K, and Sheffield VC (2021) Ectopic expression of BBS1 rescues male infertility, but not retinal degeneration, in a BBS1 mouse model. Gene Ther [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagayama S, Homma R, and Imamura F (2014) Neuronal organization of olfactory bulb circuits. Front Neural Circuits 8, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Linster C, Johnson BA, Morse A, Yue E, and Leon M (2002) Spontaneous versus reinforced olfactory discriminations. J Neurosci 22, 6842–6845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sundberg H, Doving K, Novikov S, and Ursin H (1982) A method for studying responses and habituation to odors in rats. Behav Neural Biol 34, 113–119 [DOI] [PubMed] [Google Scholar]
- 52.Agassandian K, Patel M, Agassandian M, Steren KE, Rahmouni K, Sheffield VC, and Card JP (2014) Ciliopathy is differentially distributed in the brain of a Bardet-Biedl syndrome mouse model. PLoS One 9, e93484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pak TK, Carter CS, Zhang Q, Huang SC, Searby C, Hsu Y, Taugher R, Vogel T, Cychosz CC, Genova R, Moreira N, Stevens H, Wemmie J, Pieper AA, Wang K, and Sheffield VC (2020) A mouse model of Bardet-Biedl Syndrome has impaired fear memory, which is rescued by lithium treatment. bioRxiv, 2020.2010.2006.322883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wesson DW, Varga-Wesson AG, Borkowski AH, and Wilson DA (2011) Respiratory and sniffing behaviors throughout adulthood and aging in mice. Behav Brain Res 223, 99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang M, and Crawley JN (2009) Simple behavioral assessment of mouse olfaction. Curr Protoc Neurosci Chapter 8, Unit 8 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson DA, and Linster C (2008) Neurobiology of a simple memory. J Neurophysiol 100, 2–7 [DOI] [PubMed] [Google Scholar]
- 57.Johnson ME, Bergkvist L, Mercado G, Stetzik L, Meyerdirk L, Wolfrum E, Madaj Z, Brundin P, and Wesson DW (2020) Deficits in olfactory sensitivity in a mouse model of Parkinson’s disease revealed by plethysmography of odor-evoked sniffing. Sci Rep 10, 9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zou J, Wang W, Pan YW, Lu S, and Xia Z (2015) Methods to measure olfactory behavior in mice. Curr Protoc Toxicol 63, 11.18.11–11.18.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zou J, Pan YW, Wang Z, Chang SY, Wang W, Wang X, Tournier C, Storm DR, and Xia Z (2012) Targeted deletion of ERK5 MAP kinase in the developing nervous system impairs development of GABAergic interneurons in the main olfactory bulb and behavioral discrimination between structurally similar odorants. J Neurosci 32, 4118–4132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McIntyre JC, Williams CL, and Martens JR (2013) Smelling the roses and seeing the light: gene therapy for ciliopathies. Trends Biotechnol 31, 355–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reiter JF, and Leroux MR (2017) Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol 18, 533–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shah AS, Farmen SL, Moninger TO, Businga TR, Andrews MP, Bugge K, Searby CC, Nishimura D, Brogden KA, Kline JN, Sheffield VC, and Welsh MJ (2008) Loss of Bardet-Biedl syndrome proteins alters the morphology and function of motile cilia in airway epithelia. Proc Natl Acad Sci U S A 105, 3380–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tadenev AL, Kulaga HM, May-Simera HL, Kelley MW, Katsanis N, and Reed RR (2011) Loss of Bardet-Biedl syndrome protein-8 (BBS8) perturbs olfactory function, protein localization, and axon targeting. Proc Natl Acad Sci U S A 108, 10320–10325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin H, White SR, Shida T, Schulz S, Aguiar M, Gygi SP, Bazan JF, and Nachury MV (2010) The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 141, 1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mirich JM, Williams NC, Berlau DJ, and Brunjes PC (2002) Comparative study of aging in the mouse olfactory bulb. J Comp Neurol 454, 361–372 [DOI] [PubMed] [Google Scholar]
- 66.Iannaccone A, Mykytyn K, Persico AM, Searby CC, Baldi A, Jablonski MM, and Sheffield VC (2005) Clinical evidence of decreased olfaction in Bardet-Biedl syndrome caused by a deletion in the BBS4 gene. Am J Med Genet A 132A, 343–346 [DOI] [PubMed] [Google Scholar]
- 67.McEwen DP, Koenekoop RK, Khanna H, Jenkins PM, Lopez I, Swaroop A, and Martens JR (2007) Hypomorphic CEP290/NPHP6 mutations result in anosmia caused by the selective loss of G proteins in cilia of olfactory sensory neurons. Proc Natl Acad Sci U S A 104, 15917–15922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nishimura DY, Fath M, Mullins RF, Searby C, Andrews M, Davis R, Andorf JL, Mykytyn K, Swiderski RE, Yang B, Carmi R, Stone EM, and Sheffield VC (2004) Bbs2-null mice have neurosensory deficits, a defect in social dominance, and retinopathy associated with mislocalization of rhodopsin. Proc Natl Acad Sci U S A 101, 16588–16593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Uytingco CR, Green WW, and Martens JR (2019) Olfactory Loss and Dysfunction in Ciliopathies: Molecular Mechanisms and Potential Therapies. Curr Med Chem 26, 3103–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lehmkuhl AM, Dirr ER, and Fleming SM (2014) Olfactory assays for mouse models of neurodegenerative disease. J Vis Exp, e51804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peng M, Zhang C, Dong Y, Zhang Y, Nakazawa H, Kaneki M, Zheng H, Shen Y, Marcantonio ER, and Xie Z (2016) Battery of behavioral tests in mice to study postoperative delirium. Sci Rep 6, 29874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zaghloul NA, Liu Y, Gerdes JM, Gascue C, Oh EC, Leitch CC, Bromberg Y, Binkley J, Leibel RL, Sidow A, Badano JL, and Katsanis N (2010) Functional analyses of variants reveal a significant role for dominant negative and common alleles in oligogenic Bardet-Biedl syndrome. Proc Natl Acad Sci U S A 107, 10602–10607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Prasai A, Schmidt Cernohorska M, Ruppova K, Niederlova V, Andelova M, Draber P, Stepanek O, and Huranova M (2020) The BBSome assembly is spatially controlled by BBS1 and BBS4 in human cells. J Biol Chem 295, 14279–14290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kousi M, Soylemez O, Ozanturk A, Mourtzi N, Akle S, Jungreis I, Muller J, Cassa CA, Brand H, Mokry JA, Wolf MY, Sadeghpour A, McFadden K, Lewis RA, Talkowski ME, Dollfus H, Kellis M, Davis EE, Sunyaev SR, and Katsanis N (2020) Evidence for secondary-variant genetic burden and non-random distribution across biological modules in a recessive ciliopathy. Nat Genet 52, 1145–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lindstrand A, Frangakis S, Carvalho CM, Richardson EB, McFadden KA, Willer JR, Pehlivan D, Liu P, Pediaditakis IL, Sabo A, Lewis RA, Banin E, Lupski JR, Davis EE, and Katsanis N (2016) Copy-Number Variation Contributes to the Mutational Load of Bardet-Biedl Syndrome. Am J Hum Genet 99, 318–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Imhoff O, Marion V, Stoetzel C, Durand M, Holder M, Sigaudy S, Sarda P, Hamel CP, Brandt C, Dollfus H, and Moulin B (2011) Bardet-Biedl syndrome: a study of the renal and cardiovascular phenotypes in a French cohort. Clin J Am Soc Nephrol 6, 22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seo S, Mullins RF, Dumitrescu AV, Bhattarai S, Gratie D, Wang K, Stone EM, Sheffield V, and Drack AV (2013) Subretinal gene therapy of mice with Bardet-Biedl syndrome type 1. Invest Ophthalmol Vis Sci 54, 6118–6132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Daya S, and Berns KI (2008) Gene therapy using adeno-associated virus vectors. Clin Microbiol Rev 21, 583–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jacobson SG, Cideciyan AV, Ratnakaram R, Heon E, Schwartz SB, Roman AJ, Peden MC, Aleman TS, Boye SL, Sumaroka A, Conlon TJ, Calcedo R, Pang JJ, Erger KE, Olivares MB, Mullins CL, Swider M, Kaushal S, Feuer WJ, Iannaccone A, Fishman GA, Stone EM, Byrne BJ, and Hauswirth WW (2012) Gene therapy for leber congenital amaurosis caused by RPE65 mutations: safety and efficacy in 15 children and adults followed up to 3 years. Arch Ophthalmol 130, 9–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


