Abstract
The nationwide COVID-19 epidemic ended in 2020, a few months after its outbreak in Wuhan, China at the end of 2019. Most COVID-19 cases occurred in Hubei Province, with a few local outbreaks in other provinces of China. A few studies have reported the early SARS-CoV-2 epidemics in several large cities or provinces of China. However, information regarding the early epidemics in small and medium-sized cities, where there are still traditionally large families and community culture is more strongly maintained and thus, transmission profiles may differ, is limited. In this study, we characterized 60 newly sequenced SARS-CoV-2 genomes from Anyang as a representative of small and medium-sized Chinese cities, compared them with more than 400 reference genomes from the early outbreak, and studied the SARS-CoV-2 transmission profiles. Genomic epidemiology revealed multiple SARS-CoV-2 introductions in Anyang and a large-scale expansion of the epidemic because of the large family size. Moreover, our study revealed two transmission patterns in a single outbreak, which were attributed to different social activities. We observed the complete dynamic process of single-nucleotide polymorphism development during community transmission and found that intrahost variant analysis was an effective approach to studying cluster infections. In summary, our study provided new SARS-CoV-2 transmission profiles representative of small and medium-sized Chinese cities as well as information on the evolution of SARS-CoV-2 strains during the early COVID-19 epidemic in China.
Keywords: SARS-CoV-2, Epidemiology, Community transmission, Single-nucleotide polymorphism, Intrahost variant
Highlights
-
•
The SARS-CoV-2 strains from multiple regions and multiple lineages together caused the outbreak of COVID-19 in Anyang.
-
•
The traditional family/community facilitates the widespread of SARS-CoV-2 in small and medium-sized Chinese cities.
-
•
The iSNV analysis is an effective approach to studying cluster infections and reconstructing the transmission chain.
1. Introduction
Since the first case reported in December 2019, coronavirus disease 2019 (COVID-19) rapidly developed into a global pandemic over several months and became an unprecedented public health disaster in human history (Zhou et al., 2020). As of December 2021, there have been more than 260 million confirmed COVID-19 cases and more than 5 million deaths worldwide (https://covid19.who.int/). A series of variants with increased infectivity and vaccine resistance have successively emerged in different parts of the world (Boehm et al., 2021; England, 2020; Faria et al., 2021; Tegally et al., 2021), casting a shadow over the expectation of ending the COVID-19 pandemic in a short time via vaccine herd immunity (Gupta, 2021). At present, the global COVID-19 pandemic is still far from over.
Molecular epidemiology is an important scientific approach to studying the epidemics of infectious diseases and has played an unprecedentedly significant role in combating the global COVID-19 pandemic. In the past year and a half, molecular epidemiological studies worldwide have identified numerous newly emerging and potentially threatening SARS-CoV-2 lineages, provided a thorough understanding of the COVID-19 epidemic dynamics in various countries and cities, and determined the SARS-CoV-2 sources in regional outbreaks. In China, some molecular epidemiological studies have focused on the early COVID-19 epidemic in large cities, including Beijing (Du et al., 2020), Shanghai (Zhang et al., 2020b), and Guangdong (Lu et al., 2020), and mainly analyzed the circulation characteristics of local SARS-CoV-2 strains based on viral genome sequences obtained by next-generation sequencing (NGS). After the nationwide COVID-19 epidemic ended in early 2020, research attention shifted to regional COVID-19 outbreaks that successively arose in multiple Chinese cities (Cao et al., 2020; Pang et al., 2020; Shiwei et al., 2021; Xiang et al., 2020) and were generally caused by different SARS-CoV-2 variants imported from abroad. These studies aimed to trace virus sources through viral phylogenetic and genome variant analyses; however, the reconstruction of transmission chains/networks still relied on epidemiological information, including travel history, onset time, and close contacts, which were generally obtained from personal statements of infected patients and lacked the support of objective evidence.
The current study was carried out in Anyang, a city located in Henan Province in central China, midway between Wuhan and Beijing, with a population of more than five million. Unlike the large metropolises studied in previous studies, Anyang is a representative of small and medium-sized inland cities with traditional Chinese family and community structures. In particular, this study focused on family and community SARS-CoV-2 transmission events. Through epidemiological investigation, high-throughput sequencing, and clinical testing, we comprehensively studied the local largest community outbreaks. We report the transmission characteristics of early SARS-CoV-2 strains from multiple levels, including the city, family/community, and infector-infectee pair levels. Our findings provide new insights into the transmission and evolution of SARS-CoV-2 in Chinese small and medium-sized cities during the early COVID-19 pandemic.
2. Materials and methods
2.1. Patients and sample collection
In total, 53 symptomatic and 12 asymptomatic patients confirmed as having SARS-CoV-2 infection between January 24 and February 23 in 2020 in Anyang were enrolled in this study. Oropharyngeal swabs and serum samples were collected from each patient at various time points between infection confirmation and discharge from the hospital. All specimens were aliquoted and stored at −80 °C. Epidemiological investigation was performed for all patients and their close contacts.
2.2. RNA extraction, RT-qPCR, genome sequencing, and enzyme-linked immunosorbent assay (ELISA)
The oropharyngeal swabs were collected, and viral RNA was extracted using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany). To detect the viral RNA, RT-qPCR targeting two regions of ORF1_ab and N was performed using the 2019-nCoV detection kit (Bio-Germ, Shanghai, China) per the manufacturer’s instructions. We preferentially used the RNA extracted from the oropharyngeal swabs collected at the time of infection confirmation for NGS. Five samples were sequenced on an Illumina NovaSeq system using a metagenomic sequencing strategy, the other samples were sequenced on an MGI MGISEQ-2000 instrument using a multiplex PCR amplicon sequencing strategy. The sequencing reads were assembled against the SARS-CoV-2 reference genome WIV-04 (GISAID accession ID, EPI_ISL_402124) using Geneious (v.10.2.6; Biomatters Ltd, Auckland, Zealand) and CLC Genomics Workbench (v.12.0.3; QIAGEN, Aarhus, Denmark). Nested PCR and Sanger sequencing were used to fill gaps in incomplete genome sequences. Finally, 60 genomic consensus sequences were obtained and deposited to CNCB-NGDC (Supplementary Table S4). IgM and IgG against the spike protein receptor-binding domain and nucleocapsid protein were measured using an ELISA kit generated in-house (Zhang et al., 2020a).
2.3. Phylogenetic and variant analyses
We downloaded all SARS-CoV-2 genome sequences collected in Chinese mainland before March 31, 2020 from GISAID (https://www.gisaid.org/) and ordered them according to sampling time. We randomly selected one of every three contiguous genome sequences to produce a mainland SARS-CoV-2 genome sequence subset. For the selection of foreign SARS-CoV-2 genomes, we referred to the global SARS-CoV-2 phylogeny built with Nextstrain (https://nextstrain.org/ncov/global). The Chinese mainland, foreign, and Anyang genome sequences were combined in a preliminary dataset. Genome sequences with low sequencing quality were removed. Root-to-tip analysis was performed using TempEST v.1.5.3 to assess the presence of a temporal signal in the preliminary dataset (Rambaut et al., 2016). A few genome sequences for which genetic divergence and sampling date were inconsistent were removed. The remaining genome sequences were aligned using MAFFT v.7.402 (Katoh et al., 2019). Finally, a formal dataset including 277 Chinese mainland, 155 foreign, and 60 Anyang genome sequences was established. SARS-CoV-2 phylogenies based on Bayesian inference were constructed under a general time reversal nucleotide substitution model (empirical base frequency and gamma-distributed rate variation), a strict molecular clock model, and a coalescent model with constant size, using BEAST v.1.10.4 with a chain length of 4 × 108, sampling every 4,000 steps (Suchard et al., 2018). TRACER v.1.7.1 (Rambaut et al., 2018) was employed to evaluate convergence for all parameters, and all values of effective sample size were above 200. A maximum clade credibility tree was constructed using TreeAnnotator v.1.8.4 and was visualized in iTOL v.6.1.1 (Letunic and Bork, 2021). To show clustering within families/communities, we constructed a TCS haplotype network based on the alignment of the 60 Anyang SARS-CoV-2 genomes using PopART v.1.7 (Leigh and Bryant, 2015).
The Genome-to-Variants tool (https://bigd.big.ac.cn/ncov/online/tool/variation) in the RCoV19 of CNCB-NGDC was used to scan single nucleotide polymorphisms (SNPs) in the viral genomes. A SNP distribution map of the 60 Anyang SARS-CoV-2 genomes was drawn using the Gene Structure Display Server (GSDS 2.0) (Hu et al., 2015). Low-frequency variants were detected using CLC Genomics Workbench (v.12.0.3; QIAGEN, Aarhus, Denmark). Referring to previous studies (Lythgoe et al., 2021; Xiao et al., 2020), two rounds of intra-host single nucleotide variation (iSNV) calling were performed. In the first round, iSNVs were called using the following conservative criteria: (1) required significance >1.0; (2) minimum coverage ≥100 reads at iSNV site; (3) minor allele frequency (MAF) > 5%. After inter-individual iSNVs were identified, the second round of iSNV calling focused on the nucleotide sites of inter-individual iSNVs, using relatively relaxed criteria (MAF >1%). This iSNV calling strategy can filter false-positive sites caused by sequencing errors, while iSNVs with lower MAF at sites of inter-individual iSNVs are detected and withheld. In this study, SNP and iSNVs were defined in reference to a previous study (Zhang et al., 2021).
3. Results
3.1. Epidemiological characteristics of the COVID-19 epidemic in Anyang
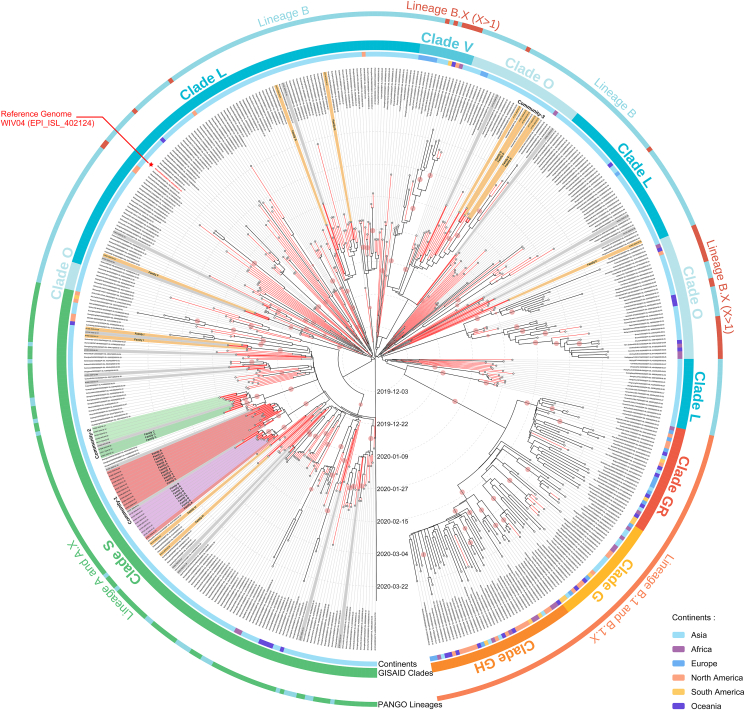
We obtained 60 nearly full-length genomes without 5′ or 3′ flanking regions, accounting for 92.3% of all local COVID-19 cases. Combining the Anyang SARS-CoV-2 genomes and global SARS-CoV-2 reference genomes, we constructed a comprehensive dataset including 492 virus genome sequences representing the SARS-CoV-2 strains circulating between December 26, 2019 and March 31, 2020. We first evaluated the presence of a temporal signal in this dataset using root-to-tip analysis. The correlation coefficient was 0.7641, indicating a good temporal signal (Supplementary Fig. S1). Subsequently, we performed Bayesian inference of phylogeny and the analysis results were explained according to two major SARS-CoV-2 nomenclature systems, namely Phylogenetic Assignment of Named Global Outbreak (Pango) (Rambaut et al., 2020) and Global Initiative of Sharing All Influenza Data (GISAID) (Tang et al., 2020). We found that three major lineages/clades of SARS-CoV-2 were circulating globally before March 31, 2020, including lineages A (clade S), B (clade L), and B.1-B.1.X (clades G, GR, and GH) (Fig. 1). Specifically, lineage A (clade S) and lineage B (clade L) were the dominant lineages before March 2020, and strains of these lineages were mainly circulating in Asian countries and regions. In contrast, lineage B.1 (clade G) and its descendant B.1.X (clades GR and GH) replaced lineages A and B and became the dominant lineages worldwide in March 2020. Nearly all SARS-CoV-2 genomes collected in Chinese mainland, including all Anyang genomes, belonged to lineages A and B (clades S and L). Very few SARS-CoV-2 Chinese genomes were assigned to lineage B.1-B.1.X (clade G), but the sample collection records showed that most of them were related to overseas imports. According to Bayesian inference of phylogeny, SARS-CoV-2 was probably first introduced into human society in November 2019 [geometric mean: 2019.873, 95% highest posterior density (HPD): 2019.818–2019.923], which has been also previously suggested (Gomez-Carballa et al., 2020; Nie et al., 2020) (Fig. 1).
Fig. 1.
Bayesian maximum clade credibility tree of the SARS-CoV-2 genome sequences. All reference genome sequences were obtained from samples collected between December 2019 and March 2020, as indicated in the Methods. Branch lines of the Chinese genome sequences are shown in red, and the Anyang genome sequences are shown in red and bold. Anyang clustered cases are highlighted with a colored background, whereas non-clustered cases are presented with a grey background. Family or community information is indicated on the branches. The inner, middle, and outer rings represent the sampling location, Pango lineage, and GISAID clade, respectively, of the genome sequences. Phylogenetic clusters with posterior probability values > 0.75 are marked with pale red circles. The reference genome WIV04 is indicated in red font and marked with a red asterisk.
The 60 Anyang SARS-CoV-2 genomes were distributed in multiple small clusters in lineages A and B, and many of them were located next to virus genomes from other Chinese cities, indicating that there were multiple geographic and lineage sources of the SARS-CoV-2 strains that caused the epidemic in Anyang. This was in line with the epidemiological statistics of the infected cases, which showed that local cases infected in Anyang, imported cases from Wuhan, and imported cases from other cities accounted for 69.23% (45 cases), 18.46% (12 cases), and 13.31% (8 cases), respectively. Besides Wuhan, other cities related to imported cases in Anyang included Beijing, Hefei (the capital of Anhui Province), Jinan (the capital of Shandong Province), Yichang (a city of Hubei Province), and Zhuzhou and Yueyang (two cities of Hunan Province). Among these imported cases, 8 out of 21 had produced next-generation cases after they arrived in Anyang, and nearly all these infection events occurred in local families and communities (families A–I and communities 1–3). Notably, transmission events in communities 1 and 2 accounted for half of Anyang infected cases, and their SARS-CoV-2 genome sequences formed two prominent clusters, which were obvious divergent from other small clusters in lineage A (Supplementary Fig. S2). In view of the high proportion of family and community transmission cases as well as the large scale of single family/community transmission, the prevention and control of family/community transmissions are very important to curb the COVID-19 epidemic in small and medium-sized cities in China.
Next, we investigated the variant profiles of all 492 genomes. According to Bayesian inference of phylogeny, the average substitution rate was 1.123 × 10–3 (95% HPD interval: 9.735 × 10–4 and 1.274 × 10–3) substitution/site/year, which was similar to previous calculation results based on a genome dataset of approximately the same period (Koyama et al., 2020). Statistics of the China National Genome Database (https://bigd.big.ac.cn/ncov/variation/annotation) on early circulating SARS-CoV-2 strains in China showed that the highest number of nucleic acid mutations occurred in ORF1ab, followed by the S and N genes. These variant characteristics were also observed in the Anyang SARS-CoV-2 genomes (Supplementary Fig. S3). Referring to the reference genome WIV04, we identified a total of 93 SNPs in the 60 Anyang SARS-CoV-2 genomes, including 36 synonymous variants, 52 non-synonymous variants, 2 other variants (one in the 5′-untranslated region and one in an intergenetic region), and 3 deletions (Supplementary Fig. S4). As lineage A was the predominant lineage in Anyang, not surprisingly, two feature variants in lineage A (clade S), C8782T and T28144C, were the two most common SNPs in the Anyang SARS-CoV-2 genomes.
3.2. Epidemiological investigation of two family transmission events
As family/community transmission is a major driver of epidemics in cities, to better understand the SARS-CoV-2 transmission in families and communities, we carried out a comprehensive analysis of the family transmission events in families A and B (the relationships among patients and family members are listed in Supplementary Table S1) based on epidemiological investigation, genome analyses, and clinical testing.
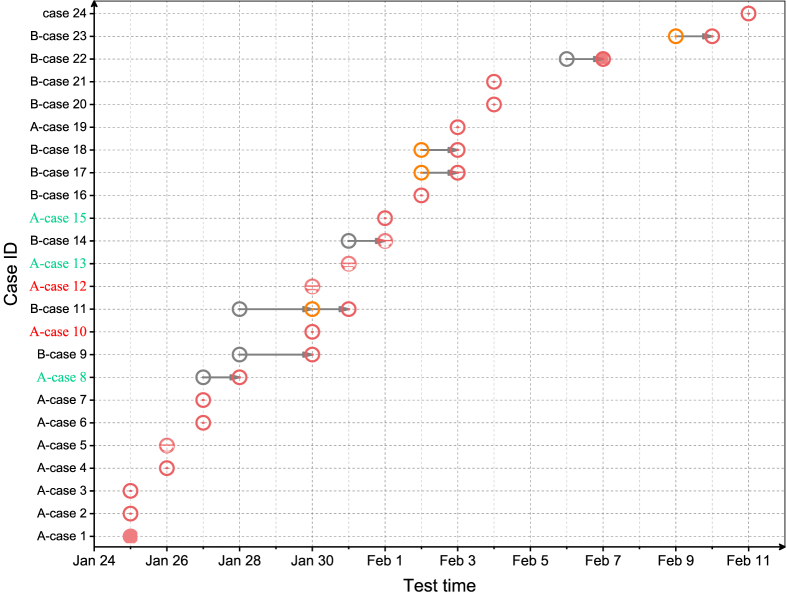
First, we outlined the basic transmission events in the two families using the epidemiological data. The transmission event in family A, related to a presumed asymptomatic superspreader, attracted nationwide attention in the initial period of the COVID-19 epidemic. According to a previous study on this transmission event, case 8, who traveled back from Wuhan to Anyang (January 20, 2020), transmitted the disease to many family members, but remained asymptomatic herself (Bai et al., 2020). However, the previous study only included the six earliest patients and was far from revealing the whole transmission event. The current epidemiological investigation showed that this family transmission event involved 13 infected cases with complex epidemiological links. Specifically, between January 4, 2020 and January 15, 2020, case 1 and several of her relatives (cases 2, 3, 5) and a family friend (case 4) were nursing an elderly family member (case 1’s father) in the hospital. During this period, several other relatives (cases 6, 8, 12, and 13) occasionally went to the hospital to visit the elderly family member. Subsequently, all of the above persons and another two persons (case 7, a friend of case 1, and case 15, a distant relative) attended the funeral of the elderly family member during January 16 to January 19. Case 10 (case 6’s father) and case 19 (another friend of family A) had not been to the hospital or the funeral, but case 10 lived with case 6 during this period, and case 19 had close contact with multiple members of family A after the funeral. As shown in Fig. 2, the family A members were successively confirmed as having SARS-CoV-2 infection between January 25, 2020 and February 3, 2020. Except for case 8, no members of family A had a history of traveling to Wuhan.
Fig. 2.
Chronological order of infection confirmation for community 1 (families A and B). Red and grey open circles represent positive and negative viral RT-qPCR test results, respectively. Orange open circles represent uncertain viral RT-qPCR test results. Half-filled red circles represent positive IgM test results. Filled red circles represent positive IgM and IgG test results. Cases 8, 13, and 15 were asymptomatic; cases 10, 12 were two elderly patients who received tests after they were overwhelmed by infection. Serum samples from the day of infection confirmation for IgM and IgG tests were not available for cases 4, 10, 15, 18, 23.
The transmission event in family B mainly involved three small families. The small families of cases 9 (including cases 9, 11, and 18) and 14 (including cases 14 and 16) lived in a large house together with their mother (case 17). Case 9 was the sister-in-law of case 14, and they had often helped each other with the household. Case 20 was the son of case 9. He, his wife (case 21), and their son lived in another house, but they frequently visited his parents (cases 9 and 11) and her father (case 23). Case 22 was a neighbor of case 9, and she and case 9 regularly visited each other. Unlike family A members, family B members did not participate in a large family gathering before their disease onset, but most of them lived very closely in a community, and the members of the small families had relatively frequent interpersonal contacts, especially during the Chinese Spring Festival (January 25). Case 9 was the first confirmed case of family B, followed by her daughter, case 11. Between January 30 and February 10, as many as 10 members of family B were confirmed as having SARS-CoV-2 infection (Fig. 2). None of them had a history of traveling of Wuhan.
The epidemiological investigation revealed that 15 members of family A, including cases 1, 2, 3, 5, and 6, had had lunch and supper in a restaurant where case 9 of family B worked, on January 16, 2020 (the first day of the funeral). Investigators had previously suspected an epidemiological link between families A and B, but except for the contact history, they did not find convincing evidence to support this speculation.
Case 24 was a female resident of Anyang and was confirmed as having SARS-CoV-2 infection on February 11, 2020. She was the only infected case in her family, and neither she nor her family had a history of traveling outside of Anyang. By the end of the local COVID-19 epidemic, the infection source of case 24 had not been identified.
3.3. Molecular epidemiological analysis of the two transmission events in families A and B
To find experimental evidence to reconstruct the two transmission events in families A and B, we took multiple molecular epidemiological approaches. First, the phylogenetic tree in Fig. 1 showed that the 13 virus genomes of family A, the 8 virus genomes of family B (no genome sequence was available for case 23, and partial regions of the case 22 genome were available, but could not be used in the phylogenetic analysis) and the virus genome of case 24 were clustered into a distinct cluster (community 1) with a cluster-specific SNP, T11418C, and this SNP was absent in all other early SARS-CoV-2 genomes of China (Supplementary Fig. S2). The close phylogenetic relationships and the shared cluster-specific SNP suggested that family A, family B, and case 24 probably had epidemiological links and that the virus that had caused their infections could be traced to a common source.
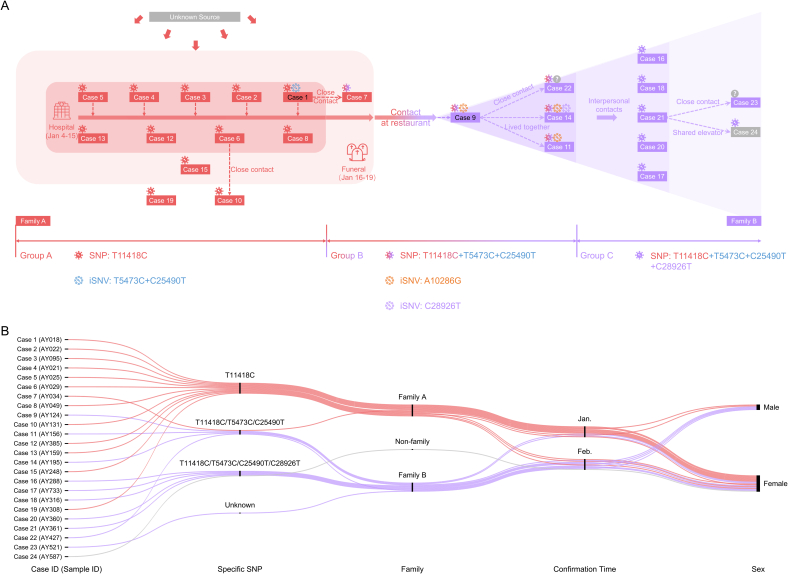
Next, using the genome consensuses, we performed a SNP analysis and identified four inter-individual SNPs, including T11418C, T5473C, C25490T, and C28926T (Fig. 3A, Supplementary Table S2). These accumulated SNPs in the virus genome were like “scale marks” on the virus transmission chain. All genome consensuses of the infected cases could be classified into three SNP groups based on these “scale marks” (Fig. 3A). SNP-group A, including nearly all cases of family A except case 7 (cases 1–6, 8, 10, 12–13, 15, 19), was characterized by only one variant, T11418C; SNP-group B, including case 7 of family A and four cases of family B (cases 9, 11, 14, 22), had three SNPs, namely T11418C, T5473C, and C25490C; SNP-group C, including the other cases of family B (cases 16–18, 20–21, 23) and case 24, carried four SNPs, namely T114418, T5473C, C25490T, and C28926T. The increase in SNPs over the three SNP groups presented a clear family- or time-dependent pattern (Fig. 3B), which outlined an axis of virus transmission in the two families and case 24 and demonstrated the virus transmission links among them. In conclusion, the virus was transmitted from family A to family B, then to case 24.
Fig. 3.
Epidemiological information of and significant virus variants in infected cases in community 1 (families A and B). A Schematic representation of transmission events in community 1 reconstructed on the basis of epidemiological investigation and SNP and iSNV analyses. B Alluvial diagram of significant SNPs, confirmation times, and sex of the infected cases in families A and B.
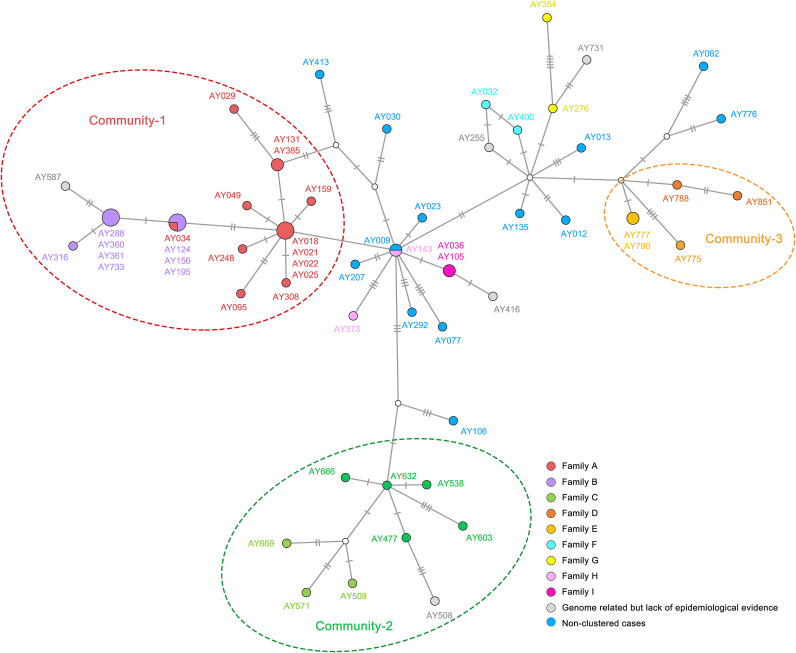
To more closely examine the transmission links among family A, family B and case 24 in the Anyang epidemic, based on the 60 Anyang SARS-CoV-2 genome consensuses, we constructed a genome haplotype network (Fig. 4). The network showed that virus genomes within the same family/community had a short distance to each other, and all families/communities could be easily distinguished. The virus genomes of family A, family B, and Case 24 (AY587) were distributed on the same long branch, which diverged from the other SARS-CoV-2 genomes of Anyang. On the long branch, the genomes of SNP-group A were distributed closer to the branch root, whereas those of SNP-group B were distributed in the middle of the branch, and the those of SNP-group C were distributed closer to the branch tip. The genome distribution pattern in the network also suggested virus transmission from family A to family B and then to case 24, corroborating the SNP analysis results.
Fig. 4.
Haplotype network of Anyang SARS-CoV-2 genomes. Family cases are indicated in different colors, and three communities are circled. Each short line crossing the linking lines represents a SNP.
To reveal more hidden evidence, based on the high-throughput sequencing data, we conducted iSNV analysis. We found four iSNVs with low allele frequency, which were either shared by multiple individuals or overlapped with the inter-individual SNPs described above (Fig. 3A, Supplementary Fig. S2). Specifically, T5473C (2.79%) and C25490T (1.60%) were two iSNVs in case 1 of SNP-group A. After inter-individual transmission, they became signature SNPs of SNP-group B cases (Fig. 3A). Similarly, C28926T initially was an iSNV (22.41%) in case 14 of SNP-group B. After inter-individual transmission, it became a newly added signature SNP of SNP-group C cases (Fig. 3A). A10286G was an inter-individual iSNV only found in SNP-group B cases (no reads covered this site in the NGS data of case 22), but it was lost during transmission (Fig. 3A). This iSNV analysis revealed more detailed links between the two families. Combined with the contact history, these data allowed us to determine several infector-infectee pairs, including case 1-case 7, case 1-case 9, case 9-case 11, case 9-case 14, and case 9-case 22. Of note, we excluded two other inter-individual iSNVs, C15157A and C241T. C15157A was only found in the amplicon-based sequencing data and the quality of the NGS reads covering this site was quite poor, suggesting that it might have been a false-positive result probably caused by sequencing bias or error. C241T was identified as a SNP or iSNV in multiple family/community transmissions and multiple non-clustered cases. According to statistics of the China National Center for Bioinformation-National Genomics Data Center (CNCB-NGDC), C241T rapidly replaced the corresponding wild-type allele in the virus population and became a feature SNP of clade G since February 2020, and some studies have shown that C241 may confer an advantage to SARS-CoV-2 transmission (Chaudhari et al., 2021; Luo et al., 2021). Considering that C241T was relatively prevalent in the Anyang virus genomes and it could not be ruled out that it arose spontaneously, we excluded it from the family/community-specific variants.
The results of the multiple molecular epidemiological approaches, including lineage phylogeny, SNP, genome haplotype network, and iSNV analyses, were highly consistent, providing not only crucial experimental evidence to support the previous speculation that the two families had an epidemiological link, but also an outline of virus transmission. Unexpectedly, we also found the infection source of case 24, which was related to the family B transmission event, especially the cases within SNP-group C. Based on all these analyses, we reason that the two family transmission events and case 24 can be included in a large community transmission, namely community 1, which was also the largest community transmission event in Anyang, accounting for 35% of the infected cases in the COVID-19 epidemic.
3.4. Different introduction and transmission patterns of SARS-CoV-2 in families A and B
Based on the new evidence obtained in the molecular epidemiological analyses, we conducted a complementary investigation to reconstruct an elaborate virus transmission process and compared the different characteristics of the two family transmissions.
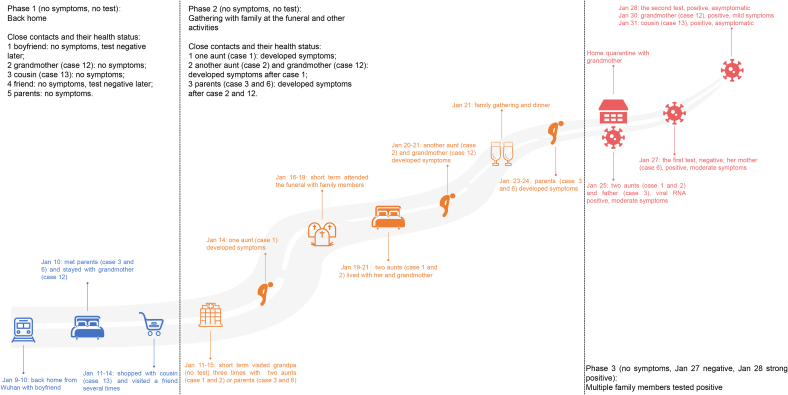
We re-evaluated the possibility that case 8 was the infection source of her family. First, the updated epidemiological information did not support that she arrived at home carrying the virus (Fig. 5), although she had a history of traveling to Wuhan. Case 8 had long-hours or high-frequency contact with at least four persons (her boyfriend, a close friend, her grandmother, and a younger female cousin) during January 9 to 14. Her boyfriend traveled back from Wuhan with her and they sat together on the train for several hours. Her close friend companied her to go shopping and dining after she arrived. However, both persons tested RT-qPCR negative throughout the outbreak. In contrast, all infected cases were her relatives. Second, as shown in Fig. 5, the infection confirmation times of case 8 and her two closely contacted relatives (grandmother/case 12 and younger female cousin/case 13) were later than those of some other members of family A, which implied that case 8 and her two relatives were probably not among the earliest infected cases in the family. In contrast with case 8, the five persons who had been undertaking a lot of nursing work in the hospital and organizational work at the funeral (cases 1–5), were the earliest confirmed cases and were confirmed around the same time (January 25 and 26), followed by other members of family A. Particularly, case 1 tested positive for IgM and IgG on the day of confirmation, although cases 2, 3, and 5 did not (serum of case 4 was not obtained on the day of confirmation), which implied that case 1 was infected earlier than the other cases, which was confirmed by the disease onset record of the epidemiological investigation. Third, the clinical testing results suggested that the case 8 infection was probably transient and mild. The RT-qPCR test result of case 8 was negative even on January 27, the day before her infection was confirmed. In subsequent multiple viral RNA tests, all results were negative. Moreover, case 8 tested negative for IgM and IgG antibodies throughout the epidemic. Collectively, the epidemiological and clinical data did not support that case 8 was the first member to be infected in her family or that she had been persistently shedding virus in her family. Thus, case 8 was likely a recipient of infection rather than a superspreader.
Fig. 5.
Epidemiological timeline of case 8 before she was confirmed as having SARS-CoV-2 infection. Three phases can be distinguished in the contact history from arriving home to infection confirmation.
Although the source of infection was difficult to determine, the process and pattern of SARS-CoV-2 transmission in family A were relatively clear. First, SARS-CoV-2 entered family A before the funeral as the virus was transmitted from family A at the beginning of the funeral. Case 7, who was a close friend of case 1, attended the funeral only on the first two days (January 16 and 17) and had been consoling case 1 during this period, and later got infected with SARS-CoV-2 (Fig. 3A). Case 9 only had contact with family A members in her restaurant on the first day of the funeral (January 16), and later also got infected (Fig. 3A). We speculated that the first round of family transmission occurred in the hospital rather than at the funeral. Second, the first round of family transmission was probably due to multiple introductions from the same source. Except for cases 7, 15, and 19, who were infected either at the funeral or after the funeral, the infection confirmation dates of the other family A members were concentrated within one week (January 25 to 31), which suggested that most family A members were probably exposed to the same infection source within a short time. Besides the infection timeline, the virus genome variant pattern also suggested this transmission character (Supplementary Table S3). The genome consensuses of the presumed earliest infected cases (cases 1, 2, 4, and 5) carried only three SNPs, including a family-specific SNP (T11418C) and two feature SNPs of lineage A (C8782T and T28144C), and were completely the same. Compared with these four cases, the other infected cases of family A carried one to three additional individual-specific SNPs, and none of the new individual-specific SNPs later developed into a family-dominant mutation, which implied that in family A, no long virus transmission chain was formed.
Epidemiological and variant analyses showed that SARS-CoV-2 entered family B via case 9, who was infected by a member of family A at her restaurant. Family B members were confirmed as having SARS-CoV-2 infection between January 30 (case 9) and February 11 (case 23), which was a longer period than that in family A, probably because this family had not held large family gatherings. The virus genome variant patterns of family B also supported this transmission character (Supplementary Table S3). The two iSNVs with low allele frequency (T5473C and C25490T) in case 1 of family A were transmitted to case 9 and became two SNPs, which were later transmitted to other cases in family B. Likewise, the iSNV C28926T that arose in case 14 was transmitted to the subsequent cases in family B and became a SNP. At the end of virus transmission in family B, a significant molecular signature comprising four SNPs, T11418C, T5473C, C25490T, and C28926T, was formed. The explicit first infected case and the process of SNP accumulation suggested that SARS-CoV-2 entered family B through a single introduction, followed by cascade transmission, which was very different from the virus transmission pattern in family A.
A complementary epidemiological investigation showed that case 24 once visited the building where case 21 worked, and they took the same elevator several times. This well explains the close phylogenetic relationship and the same molecular SARS-CoV-2 genome signature between Case 24 and family B.
4. Discussion
Our study provided a scenario of the SARS-CoV-2 epidemic in Anyang, a city representative of small and medium-sized Chinese cities, during the early COVID-19 pandemic from a molecular epidemiological perspective. Further, it revealed that even in the early COVID-19 epidemic, both geographic and lineage sources of the SARS-CoV-2 strains were very complex in Anyang. In contrast to the epidemics in large cities and well-developed regions such as Beijing, Shanghai, and Guangdong, the entire Anyang epidemic was caused by domestic strains of lineages A and B as the city is located in central China and does not have international transport (Du et al., 2020; Lu et al., 2020; Zhang et al., 2020b). Although most imported cases were successfully intervened, SARS-CoV-2 spread from a few imported cases, expanding into a large-scale local epidemic. Thus, restricting population movement between and within cities is equally important to curb the development of the COVID-19 epidemic in small and medium-sized cities as in large cities. Moreover, unlike several regional outbreaks after the nationwide epidemic, which were generally related to large public facilities, such as markets, airports, theaters, and hospitals, the Anyang epidemics were mainly driven by family/community transmissions. Given that nearly all infected cases in Anyang were acquaintances of each other, including relatives, friends, and neighbors, more intervention measures should be implemented within families and communities during COVID-19 epidemics in small-medium-sized cities.
To understand how family/community transmission could expand the epidemic in the city and characterize family/community transmissions, we used multiple approaches, including epidemiological investigation, genome analyses, and clinical testing, to comprehensively study the largest local community transmission events. Our study provided a perspective on SARS-CoV-2 transmission in Chinese traditional families and communities. We identified two patterns of SARS-CoV-2 transmission in community transmission events involving two large families. SARS-Cov-2 was introduced in family A by multiple introductions over a short period, followed by a rapid expansion. In contrast, in family B, the virus was introduced once, followed by a cascade of transmission events. Our findings indicate that the transmission pattern largely depends on the mode of interpersonal activity, which in small and medium-sized Chinese cities is greatly associated with the family and community structures. In China, small families generally live together, especially in the wide suburbs and rural areas, and family members have frequent social interaction. Like in community 1, the transmission events in communities 2 and 3 in Anyang also occurred in traditional communities (Fig. 1, Supplementary Fig. S5). Obviously, clustered infections or outbreaks are more likely in traditional communities, for which it is more difficult to trace the infection source and clarify the transmission chain because of the very complex contact networks. Therefore, in families and communities, more prevention measures should be implemented, such as maintaining a moderate physical distance among family members who live together and minimizing family gatherings during traditional festivals. In short, the prevention of family and community transmission is key to the prevention of SARS-CoV-2 outbreaks in small and medium-sized Chinese cities.
Based on genome consensus sequence alignment, SNPs were identified and used for the molecular epidemiological analysis. Since the onset of the COVID-19 pandemic, researchers have often used SNPs and molecular signatures composed of SNPs to trace outbreaks at different scales. In the Boston epidemic, researchers found that the SARS-CoV-2 genomes related to two superspreading events harbored different SNPs, and on the basis of these molecular signatures, the link between individual clusters and wider community spread was clarified (Lemieux et al., 2020). In China, a significant example is the Beijing Xinfadi market outbreak. Seventy-two virus genomes from this outbreak were assigned to lineage B.1.1 and shared the same molecular signature, which comprised seven SNPs and was mainly carried by European strains (Pang et al., 2020). Based on this evidence, the strain that caused the market outbreak was considered to have been imported from Europe through food cold-chain logistics. Likewise, three molecular signature patterns related to three community transmission events in Anyang were detected in our study. Like community 1, communities 2 and 3 carried specific SNPs, which constituted their molecular signatures in this study (Supplementary Figs. S4 and S5). Although molecular signatures or specific SNP have been widely used to validate the source strain in successive regional COVID-19 outbreaks, iSNVs have rarely been used to this end. A critical issue in the study of early SARS-CoV-2 strains is that the number of SNPs is very low, which in turn limits obtaining sufficient useful information for analyses. To address this problem, we used combined SNP data and iSNV data. Four significant iSNVs (T5473C, C25490T, C28926T, and A10286G) with minor allele frequencies not only provided evidence to clarify the modes of SARS-Cov-2 transmission, but also corroborated the SNP data and epidemiological findings. Therefore, our study showed that iSNV analysis was an effective approach to studying family/community transmission and early SARS-CoV-2 strains.
Surprisingly, we observed that dominant variants, including variants that arose in individuals, were fixed in the virus genome through inter-individual transmission and subsequently spread to the whole population. This phenomenon has been termed “evolution in action” and has been previously observed in SARS-CoV-2 strains (Lythgoe et al., 2021). Moreover, we observed different transmission consequences of inter-individual iSNVs. Among the four significant inter-individual iSNVs identified in this study, three (T5473C, C25490T, and C28926T) finally developed into dominant variants in the community 1 population. The fourth, A10286G, maintained a low allele frequency after the initial inter-individual transmission (from case 9 to case 14), but was completely lost in subsequent inter-individual transmissions (from case 14 to other members of family B). Two studies in the UK (Lythgoe et al., 2021) and Austria (Popa et al., 2020) have reported similar scenarios of mutation formation and discussed the transmission bottleneck of SARS-CoV-2, which we did not due to the limited numbers of infector-infectee pairs. Moreover, a few studies have reported a high SARS-CoV-2 genetic diversity within the same host between samples collected at different times and between samples collected from different body parts (Ruan et al., 2021; Wang et al. 2021a, 2021b). Therefore, we suggest that SNP and iSNV data should be combined with epidemiological data rather than be used alone in the study of transmission chains. In particular, infector-infectee pairs should be cautiously determined and their relationships should be validated by a definite contact history and reliable variant evidence.
5. Conclusions
In conclusion, we described a COVID-19 epidemic in Anyang, a city representative of small to medium-sized Chinese cities, as well as virus transmission in traditional families and communities. Our findings provide new insights into the early Chinese COVID-19 epidemic and into the transmission and evolution of early SARS-CoV-2 strains.
Data availability
All the data generated during the current study are included in the manuscript.
Ethics statement
This study was approved by the ethics review committee of Anyang Municipal Center for Disease Control and Prevention. Written informed consent for the use of clinical samples and clinical data was obtained from all patients involved in this study.
Author contributions
Li Yang: methodology, investigation, data curation, formal analysis, visualization, validation, writing-original draft. Si Hao-Rui: software, data curation, formal analysis. Zhu Yan: resources, investigation. Xie Nan: investigation. Li Bei: resources, investigation. Zhang Xiang-Ping: investigation. Han Jun-Feng: investigation. Bao Hong-Hong: investigation. Yang Yong: investigation. Zhao Kai: investigation. Hou Zi-Yuan: investigation. Cheng Si-Jia: investigation. Zhang Shuan-Hu: project administration, supervision. Shi Zheng-Li: project administration, conceptualization, supervision, writing-review & edit, funding acquisition. Zhou Peng: project administration, conceptualization, supervision, validation, writing-review &edit, funding acquisition.
Conflict of interest
The authors declare no competing interests.
Acknowledgements
This study was supported by the China National Science Foundation (Excellent Scholar Grants 81822028 and 82041013 to P.Z.), Ministry of Science and Technology of China (grant 2020YFC0840900 to P.Z.), and Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDB29010101 to Z.-L. S.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2022.01.030.
Contributor Information
Shuan-Hu Zhang, Email: ayzshlx@163.com.
Zheng-Li Shi, Email: zlshi@wh.iov.cn.
Peng Zhou, Email: peng.zhou@wh.iov.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Bai Y., Yao L., Wei T., Tian F., Jin D.Y., Chen L., Wang M. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020;323:1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm E., Kronig I., Neher R.A., Eckerle I., Vetter P., Kaiser L. Geneva centre for emerging viral D (2021) novel SARS-CoV-2 variants: the pandemics within the pandemic. Clin. Microbiol. Infect. 2021;27:1109–1117. doi: 10.1016/j.cmi.2021.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C., Hemuti M., Zhiyuan J., Xiang Z., Dayan W., Jun Z., Zhenguo G., Peipei L., Yang S., Zhixiao C., Yuchao W., Yao M., Guizhen W., Wenbo X., Xucheng F., Yong Z. Reemergent cases of COVID-19 — Xinjiang Uygur autonomous region, China, July 16, 2020. China CDC Weekly. 2020;2:761–763. doi: 10.46234/ccdcw2020.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhari A., Chaudhari M., Mahera S., Saiyed Z., Nathani N.M., Shukla S., Patel D., Patel C., Joshi M., Joshi C.G. In-Silico analysis reveals lower transcription efficiency of C241T variant of SARS-CoV-2 with host replication factors MADP1 and hnRNP-1. Inform Med Unlocked. 2021;25:100670. doi: 10.1016/j.imu.2021.100670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P., Ding N., Li J., Zhang F., Wang Q., Chen Z., Song C., Han K., Xie W., Liu J., Wang L., Wei L., Ma S., Hua M., Yu F., Wang L., Wang W., An K., Chen J., Liu H., Gao G., Wang S., Huang Y., Wu A.R., Wang J., Liu D., Zeng H., Chen C. Genomic surveillance of COVID-19 cases in Beijing. Nat. Commun. 2020;11:5503. doi: 10.1038/s41467-020-19345-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England P.H. 2020. Investigation of SARS-CoV-2 Variants of Concern: Technical Briefings.https://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201 [Google Scholar]
- Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S., Crispim M.A.E., Sales F.C., Hawryluk I., McCrone J.T., Hulswit R.J.G., Franco L.A.M., Ramundo M.S., de Jesus J.G., Andrade P.S., Coletti T.M., Ferreira G.M., Silva C.A.M., Manuli E.R., Pereira R.H.M., Peixoto P.S., Kraemer M.U., Gaburo N., Jr., Camilo C.D.C., Hoeltgebaum H., Souza W.M., Rocha E.C., de Souza L.M., de Pinho M.C., Araujo L.J.T., Malta F.S.V., de Lima A.B., Silva J.D.P., Zauli D.A.G., de S.F.A.C., Schnekenberg R.P., Laydon D.J., Walker P.G.T., Schluter H.M., Dos Santos A.L.P., Vidal M.S., Del Caro V.S., Filho R.M.F., Dos Santos H.M., Aguiar R.S., Modena J.L.P., Nelson B., Hay J.A., Monod M., Miscouridou X., Coupland H., Sonabend R., Vollmer M., Gandy A., Suchard M.A., Bowden T.A., Pond S.L.K., Wu C.H., Ratmann O., Ferguson N.M., Dye C., Loman N.J., Lemey P., Rambaut A., Fraiji N.A., Carvalho M., Pybus O.G., Flaxman S., Bhatt S., Sabino E.C. Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil. medRxiv. 2021 doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Carballa A., Bello X., Pardo-Seco J., Martinon-Torres F., Salas A. Mapping genome variation of SARS-CoV-2 worldwide highlights the impact of COVID-19 super-spreaders. Genome Res. 2020;30:1434–1448. doi: 10.1101/gr.266221.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.K. Will SARS-CoV-2 variants of concern affect the promise of vaccines? Nat. Rev. Immunol. 2021;21:340–341. doi: 10.1038/s41577-021-00556-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Jin J., Guo A.Y., Zhang H., Luo J., Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinf. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Platt D., Parida L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020;98:495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh J.W., Bryant D. popart: full-feature software for haplotype network construction. Methods in Ecology and Evolution. 2015;6:1110–1116. [Google Scholar]
- Lemieux J.E., Siddle K.J., Shaw B.M., Loreth C., Schaffner S.F., Gladden-Young A., Adams G., Fink T., Tomkins-Tinch C.H., Krasilnikova L.A., DeRuff K.C., Rudy M., Bauer M.R., Lagerborg K.A., Normandin E., Chapman S.B., Reilly S.K., Anahtar M.N., Lin A.E., Carter A., Myhrvold C., Kemball M.E., Chaluvadi S., Cusick C., Flowers K., Neumann A., Cerrato F., Farhat M., Slater D., Harris J.B., Branda J.A., Hooper D., Gaeta J.M., Baggett T.P., O’Connell J., Gnirke A., Lieberman T.D., Philippakis A., Burns M., Brown C.M., Luban J., Ryan E.T., Turbett S.E., LaRocque R.C., Hanage W.P., Gallagher G.R., Madoff L.C., Smole S., Pierce V.M., Rosenberg E., Sabeti P.C., Park D.J., MacInnis B.L. Phylogenetic analysis of SARS-CoV-2 in Boston highlights the impact of superspreading events. Science. 2020;371 doi: 10.1126/science.abe3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., du Plessis L., Liu Z., Hill V., Kang M., Lin H., Sun J., Francois S., Kraemer M.U.G., Faria N.R., McCrone J.T., Peng J., Xiong Q., Yuan R., Zeng L., Zhou P., Liang C., Yi L., Liu J., Xiao J., Hu J., Liu T., Ma W., Li W., Su J., Zheng H., Peng B., Fang S., Su W., Li K., Sun R., Bai R., Tang X., Liang M., Quick J., Song T., Rambaut A., Loman N., Raghwani J., Pybus O.G., Ke C. Genomic epidemiology of SARS-CoV-2 in Guangdong Province, China. Cell. 2020;181:997–1003. doi: 10.1016/j.cell.2020.04.023. e1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Yu F., Zhou M., Liu Y., Xia B., Zhang X., Liu J., Zhang J., Du Y., Li R., Wu L., Zhang X., Pan T., Guo D., Peng T., Zhang H. Engineering a reliable and convenient SARS-CoV-2 replicon system for analysis of viral RNA synthesis and screening of antiviral inhibitors. mBio. 2021;12:e02754–20. doi: 10.1128/mBio.02754-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lythgoe K.A., Hall M., Ferretti L., de Cesare M., MacIntyre-Cockett G., Trebes A., Andersson M., Otecko N., Wise E.L., Moore N., Lynch J., Kidd S., Cortes N., Mori M., Williams R., Vernet G., Justice A., Green A., Nicholls S.M., Ansari M.A., Abeler-Dorner L., Moore C.E., Peto T.E.A., Eyre D.W., Shaw R., Simmonds P., Buck D., Todd J.A., Connor T.R., Ashraf S., da Silva Filipe A., Shepherd J., Thomson E.C., Consortium C.-G.U., Bonsall D., Fraser C., Golubchik T. SARS-CoV-2 within-host diversity and transmission. Science. 2021;372 doi: 10.1126/science.abg0821. Oxford Virus Sequencing Analysis G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Q., Li X., Chen W., Liu D., Chen Y., Li H., Li D., Tian M., Tan W., Zai J. Phylogenetic and phylodynamic analyses of SARS-CoV-2. Virus Res. 2020;287:198098. doi: 10.1016/j.virusres.2020.198098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang X., Ren L., Wu S., Ma W., Yang J., Di L., Li J., Xiao Y., Kang L., Du S., Du J., Wang J., Li G., Zhai S., Chen L., Zhou W., Lai S., Gao L., Pan Y., Wang Q., Li M., Wang J., Huang Y., Wang J., Group C.-F.R., Group C.-L.T. Cold-chain food contamination as the possible origin of Covid-19 resurgence in Beijing. Natl. Sci. Rev. 2020;7:1861–1864. doi: 10.1093/nsr/nwaa264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa A., Genger J.W., Nicholson M.D., Penz T., Schmid D., Aberle S.W., Agerer B., Lercher A., Endler L., Colaco H., Smyth M., Schuster M., Grau M.L., Martinez-Jimenez F., Pich O., Borena W., Pawelka E., Keszei Z., Senekowitsch M., Laine J., Aberle J.H., Redlberger-Fritz M., Karolyi M., Zoufaly A., Maritschnik S., Borkovec M., Hufnagl P., Nairz M., Weiss G., Wolfinger M.T., von Laer D., Superti-Furga G., Lopez-Bigas N., Puchhammer-Stockl E., Allerberger F., Michor F., Bock C., Bergthaler A. Genomic epidemiology of superspreading events in Austria reveals mutational dynamics and transmission properties of SARS-CoV-2. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abe2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A. Posterior summarization in Bayesian Phylogenetics using tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O’Toole A., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Lam T.T., Max Carvalho L., Pybus O.G. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen) Virus Evol. 2016;2:vew007. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y., Hou M., Li J., Song Y., Wang H.-Y., He X., Zeng H., Lu J., Wen H., Chen C., Wu C.-I. One viral sequence for each host? – the neglected within-host diversity as the main stage of SARS-CoV-2 evolution. bioRxiv:2021. 2021:449205. 2006.2021. [Google Scholar]
- Shiwei L., Shuhua Y., Yinqi S., Baoguo Z., Huazhi W., Jinxing L., Wenjie T., Xiaoqiu L., Qi Z., Yunting X., Xifang L., Jianguo L., Yan G. A COVID-19 outbreak — Nangong city, Hebei Province, China, January 2021. China CDC Weekly. 2021;3:401–404. doi: 10.46234/ccdcw2021.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard M.A., Lemey P., Baele G., Ayres D.L., Drummond A.J., Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018;4:vey016. doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Wu C., Li X., Song Y., Yao X., Wu X., Duan Y., Zhang H., Wang Y., Qian Z., Cui J., Lu J. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020;7:1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., Mlisana K., von Gottberg A., Walaza S., Allam M., Ismail A., Mohale T., Glass A.J., Engelbrecht S., Van Zyl G., Preiser W., Petruccione F., Sigal A., Hardie D., Marais G., Hsiao N.Y., Korsman S., Davies M.A., Tyers L., Mudau I., York D., Maslo C., Goedhals D., Abrahams S., Laguda-Akingba O., Alisoltani-Dehkordi A., Godzik A., Wibmer C.K., Sewell B.T., Lourenco J., Alcantara L.C.J., Kosakovsky Pond S.L., Weaver S., Martin D., Lessells R.J., Bhiman J.N., Williamson C., de Oliveira T. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- Wang D., Wang Y., Sun W., Zhang L., Ji J., Zhang Z., Cheng X., Li Y., Xiao F., Zhu A., Zhong B., Ruan S., Li J., Ren P., Ou Z., Xiao M., Li M., Deng Z., Zhong H., Li F., Wang W.J., Zhang Y., Chen W., Zhu S., Xu X., Jin X., Zhao J., Zhong N., Zhang W., Zhao J., Li J., Xu Y. Population bottlenecks and intra-host evolution during human-to-human transmission of SARS-CoV-2. Front Med (Lausanne) 2021;8:585358. doi: 10.3389/fmed.2021.585358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang D., Zhang L., Sun W., Zhang Z., Chen W., Zhu A., Huang Y., Xiao F., Yao J., Gan M., Li F., Luo L., Huang X., Zhang Y., Wong S.S., Cheng X., Ji J., Ou Z., Xiao M., Li M., Li J., Ren P., Deng Z., Zhong H., Xu X., Song T., Mok C.K.P., Peiris M., Zhong N., Zhao J., Li Y., Li J., Zhao J. Intra-host variation and evolutionary dynamics of SARS-CoV-2 populations in COVID-19 patients. Genome Med. 2021;13:30. doi: 10.1186/s13073-021-00847-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z., Lingling M., Jianqun Z., Yong Z., Yang S., Zhijian B., Hong W., Ji W., Cao C., Jinbo X., Tianjiao J., Qian Y., Wenbo X., Dayan W., Wenqing Y. Reemergent cases of COVID-19 — Dalian city, Liaoning Province, China, July 22, 2020. China CDC Weekly. 2020;2:658–660. doi: 10.46234/ccdcw2020.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M., Liu X., Ji J., Li M., Li J., Yang L., Sun W., Ren P., Yang G., Zhao J., Liang T., Ren H., Chen T., Zhong H., Song W., Wang Y., Deng Z., Zhao Y., Ou Z., Wang D., Cai J., Cheng X., Feng T., Wu H., Gong Y., Yang H., Wang J., Xu X., Zhu S., Chen F., Zhang Y., Chen W., Li Y., Li J. Multiple approaches for massively parallel sequencing of SARS-CoV-2 genomes directly from clinical samples. Genome Med. 2020;12:57. doi: 10.1186/s13073-020-00751-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du R.H., Li B., Zheng X.S., Yang X.L., Hu B., Wang Y.Y., Xiao G.F., Yan B., Shi Z.L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microb. Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Tan Y., Ling Y., Lu G., Liu F., Yi Z., Jia X., Wu M., Shi B., Xu S., Chen J., Wang W., Chen B., Jiang L., Yu S., Lu J., Wang J., Xu M., Yuan Z., Zhang Q., Zhang X., Zhao G., Wang S., Chen S., Lu H. Viral and host factors related to the clinical outcome of COVID-19. Nature. 2020;583:437–440. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yin Q., Ni M., Liu T., Wang C., Song C., Liao L., Xing H., Jiang S., Shao Y., Chen C., Ma L. Dynamics of HIV-1 quasispecies diversity of participants on long-term antiretroviral therapy based on intrahost single-nucleotide variations. Int. J. Infect. Dis. 2021;104:306–314. doi: 10.1016/j.ijid.2021.01.015. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data generated during the current study are included in the manuscript.