Abstract
During the SARS-CoV-2 pandemic, novel and traditional vaccine strategies have been deployed globally. We investigated whether antibodies stimulated by mRNA vaccination (BNT162b2), including third-dose boosting, differ from those generated by infection or adenoviral (ChAdOx1-S and Gam-COVID-Vac) or inactivated viral (BBIBP-CorV) vaccines. We analyzed human lymph nodes after infection or mRNA vaccination for correlates of serological differences. Antibody breadth against viral variants is lower after infection compared with all vaccines evaluated but improves over several months. Viral variant infection elicits variant-specific antibodies, but prior mRNA vaccination imprints serological responses toward Wuhan-Hu-1 rather than variant antigens. In contrast to disrupted germinal centers (GCs) in lymph nodes during infection, mRNA vaccination stimulates robust GCs containing vaccine mRNA and spike antigen up to 8 weeks postvaccination in some cases. SARS-CoV-2 antibody specificity, breadth, and maturation are affected by imprinting from exposure history and distinct histological and antigenic contexts in infection compared with vaccination.
Keywords: COVID-19, BioNTech-Pfizer, BNT162b2, Moderna, mRNA-1273, Astra Zeneca, ChAdOx1-S, Sputnik V, Gam-COVID-Vac, Sinopharm, BBIBP-CorV, vaccine, SARS-CoV-2, imprinting, SARS-CoV-2 variants of concern, Delta variant, endemic coronaviruses, antibodies, lymph node germinal center, autopsy
Graphical abstract
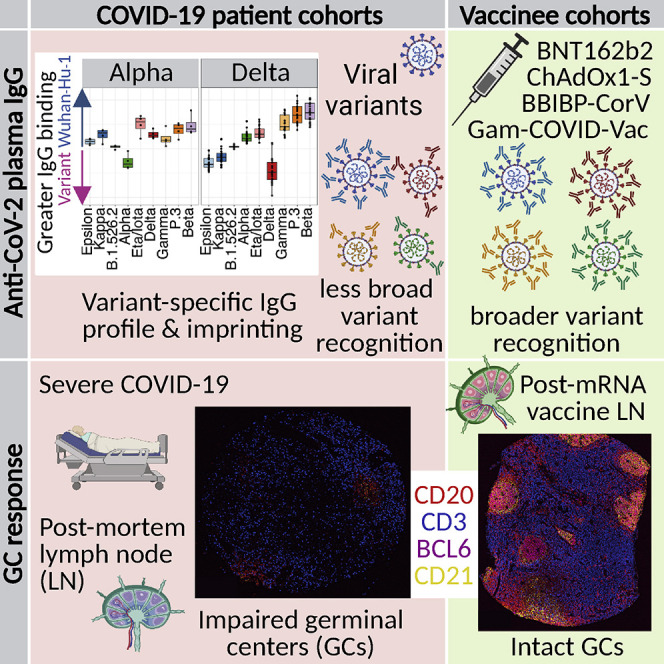
Human antibody responses to SARS-CoV-2 differ between vaccination and infection, with mRNA vaccination inducing more productive lymph node GC responses and several vaccine types stimulating IgG antibodies capable of recognizing a broader range of viral variants.
Introduction
The urgent need for countermeasures against the coronavirus disease 2019 (COVID-19) pandemic has spurred the rapid development of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccines of diverse formulations. mRNA vaccines BNT162b2 (BioNTech-Pfizer) and mRNA-1273 (Moderna/NIAID) have demonstrated high efficacy and safety in clinical trials for COVID-19 prevention (Baden et al., 2021; Polack et al., 2020; Walsh et al., 2020). Additional COVID-19 vaccines including adenoviral vectored vaccines ChAdOx1-S (Astra Zeneca) (Voysey et al., 2021), Ad26.COV2.S (Johnson & Johnson) (Sadoff et al., 2021), and Gam-COVID-Vac (Sputnik V) and inactivated viral vaccines such as BBIBP-CorV (Sinopharm) also have reported efficacy. Correlates of vaccine-elicited protection from COVID-19 are the titers of neutralizing antibodies to SARS-CoV-2, and the concentration of antibodies binding to spike or receptor-binding domain (RBD) (Earle et al., 2021; Gilbert et al., 2022; Khoury et al., 2021; Röltgen and Boyd, 2021). Most neutralizing antibodies target the RBD and prevent binding to angiotensin-converting enzyme 2 (ACE2) receptor (Greaney et al., 2021a; Yuan et al., 2021). Current SARS-CoV-2 vaccines all contain or induce the expression of antigens similar to those of the early Wuhan-Hu-1 viral isolate, but differ in elicited binding and neutralizing antibody responses, with higher responses from mRNA vaccines compared with adenovirus-vectored or inactivated virus vaccines (Dashdorj et al., 2021a, 2021b). It remains to be determined precisely how the immune system responds to mRNA and other vaccine platforms compared with SARS-CoV-2 infection. Data from RBD variant antigen yeast display and pseudotyped virus neutralization show that RBD epitope targeting by polyclonal serum antibodies is narrower in infected patients compared with mRNA-1273 vaccinees (Greaney et al., 2021b).
Several SARS-CoV-2 variants of concern with mutations in the spike gene have emerged and spread globally, with differing abilities to evade neutralizing antibody responses elicited by Wuhan-Hu-1 infection or vaccination. The most immune-evasive variants, including the recent Omicron variant, have alterations in epitopes containing amino acid E484 (Garcia-Beltran et al., 2021; Greaney et al., 2021a; Hoffmann et al., 2021). The appearance of virus variants, waning antibody levels after infection or vaccination (Falsey et al., 2021; Levin et al., 2021), and breakthrough infections in previously immunized individuals (Keehner et al., 2021) indicate that periodic vaccine boosting of immunity to SARS-CoV-2 is warranted. Third doses of mRNA-1273 (Chu et al., 2021) and BNT162b2 (Falsey et al., 2021) administered several months after the second dose prompt an increase in neutralizing antibodies greater than the peak following initial vaccination doses. mRNA-1273 vaccination followed by mRNA booster vaccines expressing Beta spike gives higher neutralizing titers to Wuhan-Hu-1-like SARS-CoV-2 compared with the Beta variant (Choi et al., 2021; Wu et al., 2021), suggesting that some degree of immune imprinting, or preferential responses to the viral variants initially encountered by the immune system, may affect the development of antibodies against new viral variants (Wheatley et al., 2021).
Germinal center (GC) responses in human lymphoid tissues enable antibody affinity maturation and durable serological and memory B cell responses although extrafollicular B cell responses are also reported (Elsner and Shlomchik, 2020; Lam et al., 2020; Woodruff et al., 2020). The degree to which SARS-CoV-2 infections or different vaccines stimulate GC responses and differ in factors such as the quantity, persistence, and localization of antigen in lymph nodes (LNs) and other lymphoid tissues are important open questions. Approaches such as fine-needle aspiration (FNA) are being increasingly used to study LN-derived cells from healthy human subjects (Havenar-Daughton et al., 2020; Lederer et al., 2022; Turner et al., 2021). Disrupted LN GCs have been reported in autopsies of deceased COVID-19 patients (Haslbauer et al., 2021; Kaneko et al., 2020), while elevated frequencies of GC B cells are seen after mRNA vaccination in healthy individuals (Turner et al., 2021) and lower GC B cell frequencies after mRNA vaccination of immunocompromised individuals (Lederer et al., 2022). To date, no direct comparison of LN GC histology and cellular composition, combined with measurement of viral or vaccine antigen quantity, persistence, and distribution in draining LN sites of COVID-19 patients and vaccinees has been reported.
Here, we compare antibody responses in BNT162b2 mRNA vaccine recipients following first, second, and third vaccine doses to antibody responses of COVID-19 patients. We find differences in the magnitude, isotype profiles, SARS-CoV-2 spike domain specificity, and breadth of binding antibody responses to a panel of nine viral variants in addition to Wuhan-Hu-1. Anti-RBD IgG binding to SARS-CoV-2 variants of concern and interest in recipients of four different vaccines (BNT162b2, ChAdOx1-S, Gam-COVID-Vac, and BBIBP-CorV) and in COVID-19 patients shows greater binding breadth for viral variant RBDs following all vaccines compared with Wuhan-Hu-1 infection. We quantify a strong imprinting effect of prior vaccination with Wuhan-Hu-1 spike antigen on antibody specificities following breakthrough infection with viral variants. Histological analysis of draining LN shows marked impairment of GCs in severe COVID-19 compared with mRNA vaccination, higher quantities, and persistence of spike antigen accumulated in the GCs of mRNA vaccinees and detectable vaccine RNA in GCs for up to 2 months post-second dose.
Results
Magnitude and waning of anti-SARS-CoV-2 IgG following BNT162b2 vaccination and response to third-dose boost
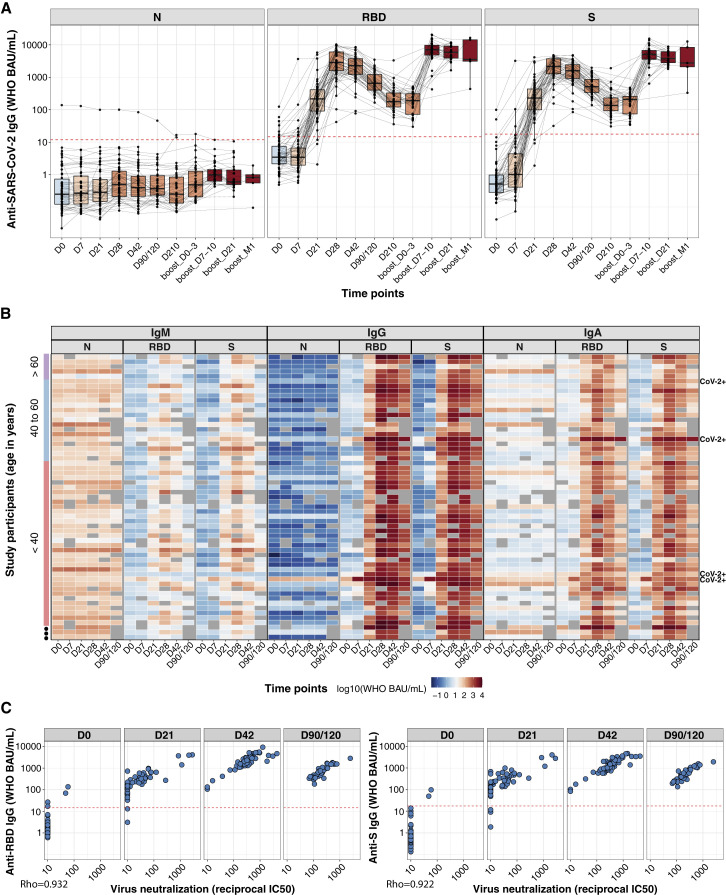
We measured anti-SARS-CoV-2 antibodies for nucleocapsid (N), full spike, and RBD in Stanford BNT162b2 study participant plasma samples using multiplexed electrochemiluminescence (ECL) assays (Meso Scale Discovery, MSD), in WHO binding antibody units (BAU). The first and second vaccine doses were at days 0 and 21, with the third-dose boosting at approximately 9 months. Plasma samples were collected in a time course up to 7 months after the first dose and up to 1 month after the third dose. Four of the 59 vaccine recipients had a history of SARS-CoV-2 reverse-transcription quantitative polymerase chain reaction (RT-qPCR)-confirmed infection (CoV-2+) prior to vaccination. IgG for spike protein and RBD in vaccinees reached their initial peak at day 28 after the first dose (Figures 1A and 1B). IgG binding to spike and RBD was highly correlated with SARS-CoV-2 neutralization titers (Arunachalam et al., 2021) (Figure 1C). By 9 months, spike-specific IgG had decreased approximately 20-fold from the maximum, but the third-dose boost raised IgG concentrations above the prior peak within 1 week. IgG specific for N protein, which is not encoded in the vaccine, was negative throughout the study in 54 of the previously uninfected vaccinees, but one participant seroconverted for anti-N IgG between days 90 and 210 after the prime, indicating a breakthrough infection (Figure 1A). CoV-2+ vaccinees had accelerated RBD and spike IgG responses after the first dose, and detectable anti-N IgG was unaffected by vaccination (Figure S1 A).
Figure 1.
Magnitude and duration of anti-SARS-CoV-2 IgG following BNT162b2 vaccination and third-dose boost
(A) Anti-SARS-CoV-2 N, RBD, and spike (S) antibody responses are shown for plasma samples from individuals who received BNT162b2 prime (D0, n = 59 individuals), second dose (D21, n = 58 individuals), and third dose (around month 9, n = 36 individuals) vaccination. Box-whisker plots of the anti-SARS-CoV-2 IgG concentrations in WHO BAU/mL show the interquartile range as the box and the whisker ends as the most extreme values within 1.5 times the interquartile range below the 25% quantile and above the 75% quantile. Red dashed lines indicate the cutoff values for the positivity of each assay (MSD, package insert).
(B) Heatmap showing the development of antibody responses in longitudinal samples collected at D0, D7, D21, D28, D42, and D90/120 time points post-prime vaccination (x axis). WHO BAU/mL Ig concentrations are displayed for study participants sorted by age (y axis, color coded). Rows are labeled on the right with “CoV-2+” for participants with a previous SARS-CoV-2 RT-qPCR positive test result.
(C) Correlations between anti-RBD and anti-spike IgG binding antibody concentrations in WHO BAU/mL and SARS-CoV-2 virus neutralization assays. Spearman rank correlation (coefficient = Rho, displayed in the plot for each assay comparison) was used to assess the strength of correlation between binding antibody concentrations and virus neutralization results. Red dashed lines indicate the cutoff values for the positivity of each assay (MSD, package insert).
Figure S1.
Anti-SARS-CoV-2 Ig antibody responses in plasma and saliva following BNT162b2 vaccination, related to Figure 1
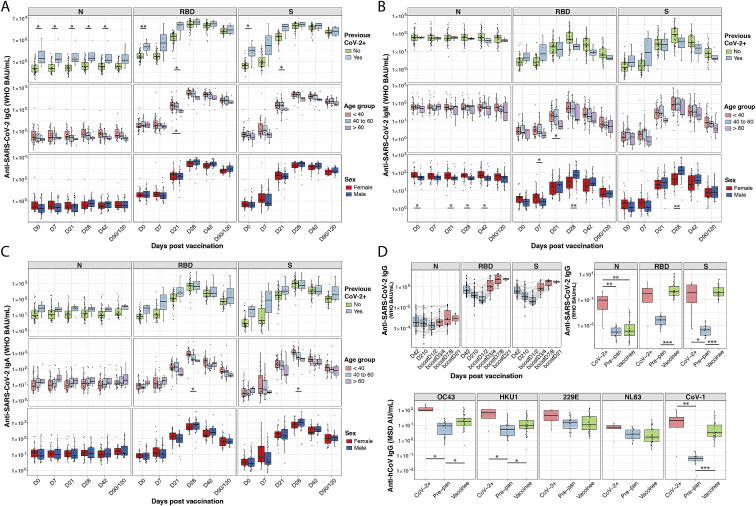
Anti-SARS-CoV-2 N, RBD, and spike (S) IgG (A), IgM (B), and IgA (C) responses are shown for plasma from individuals who received BNT162b2 prime (D0, n = 59) and second dose (D21, n = 58) vaccination. Box-whisker plots of the WHO binding arbitrary unit (BAU/mL) anti-SARS-CoV-2 concentrations show the interquartile range as the box and the whisker ends as the most extreme values within 1.5 times the interquartile range below the 25% quantile and above the 75% quantile. Comparisons between groups of previously SARS-CoV-2-infected (CoV-2+) versus noninfected individuals, and female versus male were by the two-sided Wilcoxon rank sum test; comparison between age groups (<40; 40–60; >60 years) was done using pairwise Wilcoxon rank sum test with Bonferroni correction. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(D) Anti-SARS-CoV-2 N, RBD, and S IgG concentrations in BAU/mL are shown for saliva from individuals who received BNT162b2 prime/boost and third dose vaccination (upper left panel). Anti-SARS-CoV-2 N, RBD, and S (upper right panel) concentrations in BAU/mL, as well as anti-SARS-CoV-1 and anti-HCoV-OC43, -HKU1, -NL63, and -229E S IgG (lower panel) concentrations in MSD arbitrary units (AU/mL), are shown for saliva collected on D42 after BNT162b2 prime vaccination (vaccinee), around D42 post-symptom onset for COVID-19 patients (CoV-2+), and before the onset of the COVID-19 pandemic for pre-pandemic healthy human controls (Pre-pan). Box-whisker plots of anti-SARS-CoV-2 IgG concentrations show the interquartile range as the box and the whisker ends as the most extreme values within 1.5 times the interquartile range below the 25% quantile and above the 75% quantile. Statistical test for significance between groups (CoV-2+; Pre-pan, vaccinee) was performed using pairwise Wilcoxon rank sum test with Bonferroni correction. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
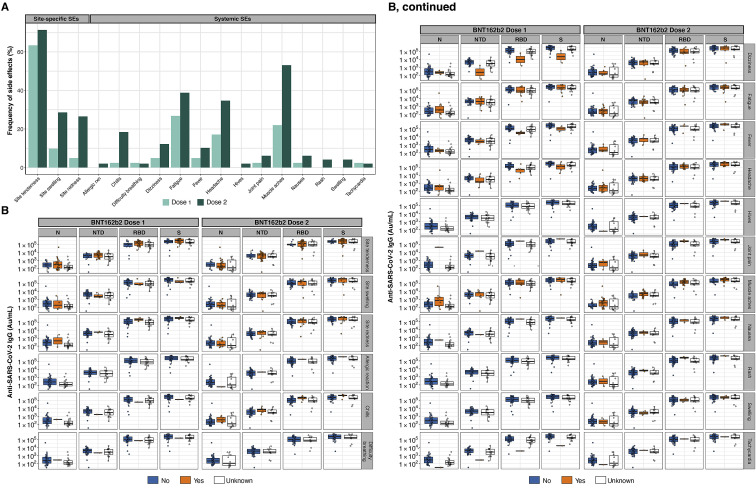
BNT162b2 recipients had weak IgM and IgA responses to spike and RBD compared with their IgG responses. Robust IgG responses were seen in all age groups (Figures 1B and S1A–S1C). Convalescent COVID-19 patients and BNT162b2 vaccinees had similar low saliva IgG concentrations for spike and RBD, several orders of magnitude lower than those detected in plasma (Figure S1D). As in plasma, saliva IgG peaked at 1 week after third-dose boosting, at higher values than the peak after the second dose (Figure S1D). Reported side effects after vaccination showed no relationship to plasma IgG responses (Figures S2 A and S2B).
Figure S2.
The magnitude of antibody responses is not correlated with reported vaccine-associated side effects (SEs), related to Figure 1
(A) Frequency of site-specific and systemic vaccine-associated SEs after prime (light green) and second dose (dark green) BNT162b2 vaccination.
(B) Box-whisker plots of the MSD AU/mL anti-SARS-CoV-2 IgG concentrations in BNT162b2 vaccinee plasma collected on D28 postvaccination show the interquartile range as the box and the whisker ends as the most extreme values within 1.5 times the interquartile range below the 25% quantile and above the 75% quantile. For a given SE (rows), vaccinees were grouped according to no SE reported (“No,” colored in blue) or SE reported (“Yes,” colored in orange). Vaccinees where SEs were unknown are shown as white boxplots.
BNT162b2 vaccination and Wuhan-Hu-1 SARS-CoV-2 infection stimulate distinct antibody isotypes and endemic coronavirus antibody responses
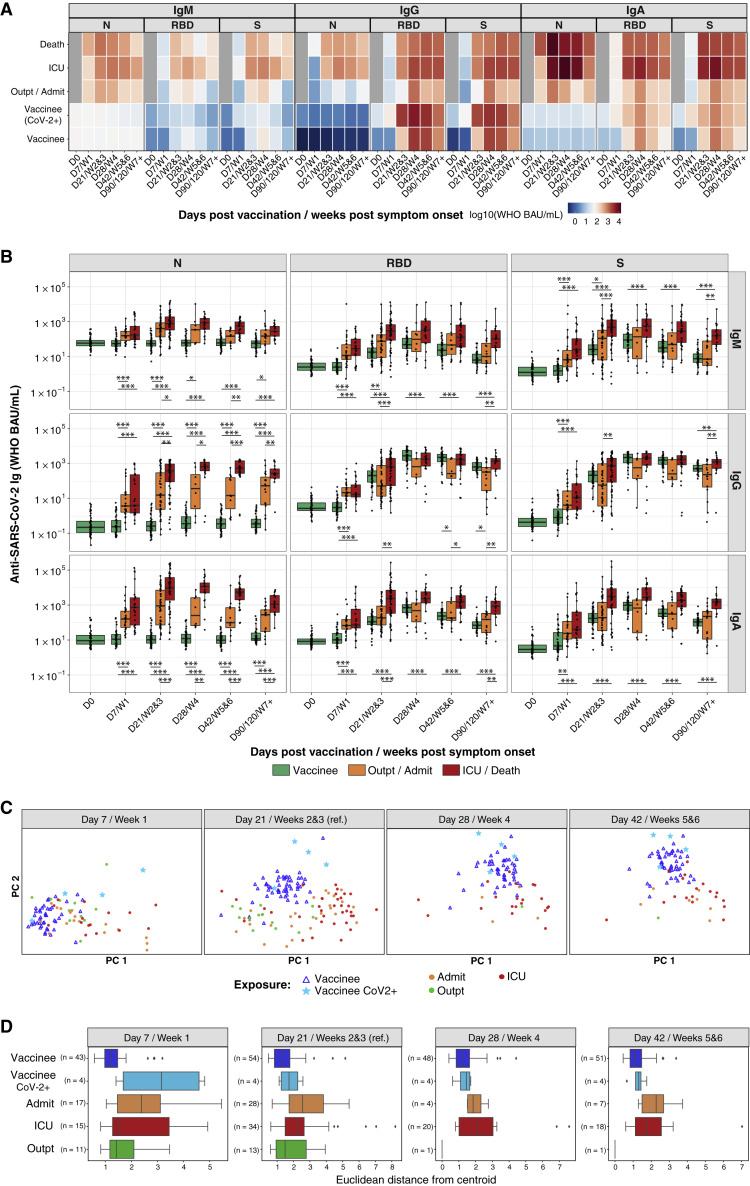
Severe COVID-19 stimulates higher SARS-CoV-2-specific antibody titers than asymptomatic infection or mild illness (Long et al., 2020; Röltgen et al., 2020). We compared antibody isotype concentrations specific for spike and RBD in COVID-19 patients (Stanford cohort 1 of this study) from the initial months of the pandemic (Röltgen et al., 2020) with the responses of the Stanford BNT162b2 vaccinees (Figures 2A and 2B). Patients were classified as outpatients, admitted patients not requiring care in the intensive care unit (ICU), ICU patients, and those who died from their illness. Stanford BNT162b2 vaccinee RBD and spike IgG concentrations were comparable to those of severely ill patients and higher than those of mildly or moderately ill patients for anti-RBD antibodies at day 42 (Figures 2A and 2B). The BNT162b2 vaccine induced a highly IgG-polarized serological response with minimal IgM- and IgA-binding spike and RBD (Figures 2A and 2B). Principal component analysis (PCA) showed clustered and homogeneous SARS-CoV-2 spike and spike-domain-specific serological responses in BNT162b2 vaccinees compared with infected patients, as quantified by smaller distances for vaccinated participants from the group centroid (Figures 2C and 2D).
Figure 2.
BNT162b2 vaccination and SARS-CoV-2 infection elicit distinct antibody profiles
(A and B) Anti-SARS-CoV-2 N, RBD, and spike (S) IgM, IgG, and IgA antibody responses are shown for individuals who received BNT162b2 prime (D0) and second (D21) vaccination doses and for COVID-19 patients. (A) The heatmap shows the development of antibody responses in longitudinal samples from vaccinees/patients collected at D0, D7/week 1, D21/weeks 2 and 3, D28/week 4, D42/weeks 5 and 6, and D90/120/≥week 7 after vaccination/COVID-19 symptom onset (x axis). The color scale encodes the median values of log10 WHO BAU/mL Ig concentrations. (B) Box-whisker plots show the development of antibody responses in longitudinal samples from vaccinees/patients collected at D0, D7/week 1, D21/weeks 2 and 3, D28/week 4, D42/weeks 5 and 6, and D90/120/≥week 7 after vaccination/COVID-19 symptom onset (x axis). Box-whisker plots show the interquartile range as the box and the whisker ends as the most extreme values within 1.5 times the interquartile range below the 25% quantile and above the 75% quantile. Statistical test: pairwise Wilcoxon rank sum test with Bonferroni correction. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Individuals were classified as outpatients (Outpt) and hospital-admitted patients (Admit); ICU patients and those who died from their illness (Death); and vaccinees who had (CoV-2+) or had not had a positive SARS-CoV-2 test in the past.
(C) PCA of anti-SARS-CoV-2 RBD, N-terminal domain, and S (but not N) IgM, IgG, and IgA concentrations across BNT162b2 vaccinees and Wuhan-Hu-1-infected Stanford COVID-19 patient cohort 1 at different time points after vaccination/COVID-19 symptom onset visualized on a consistent PCA reference created using D21/weeks 2 and 3 as a reference time point.
(D) Distribution of Euclidean distances between BNT162b2 vaccinee samples and their centroid, compared with Wuhan-Hu-1-infected Stanford COVID-19 patient cohort 1 samples and their centroid, at different time points after vaccination/COVID-19 symptom onset.
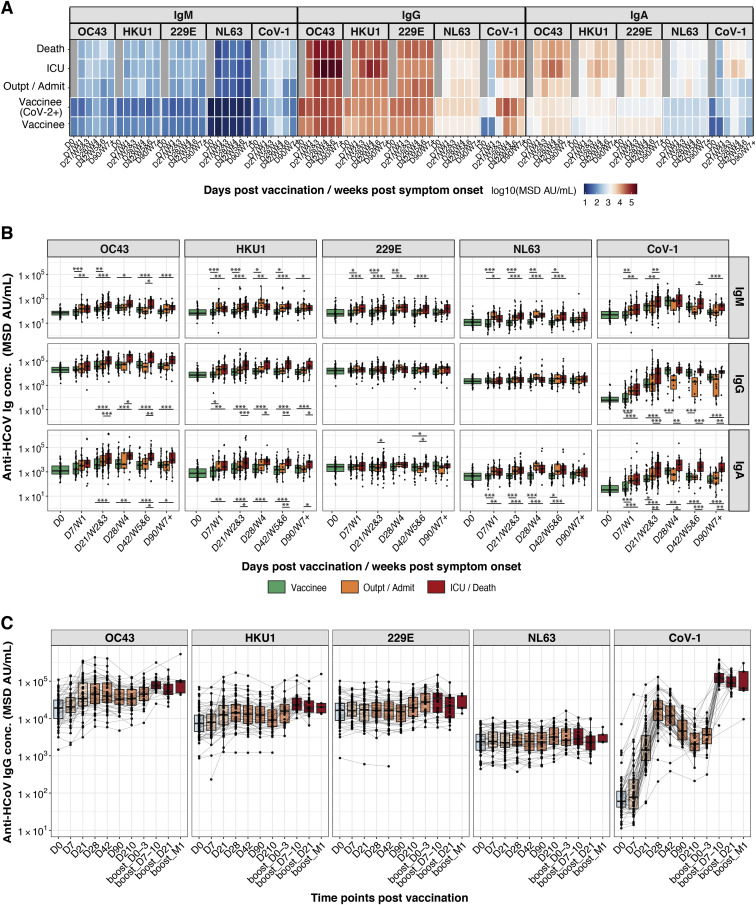
SARS-CoV-2 vaccinees and COVID-19 patients showed boosting of SARS-CoV-1 spike antibodies, but infected patients showed greater boosting of spike IgG and IgA for endemic human betacoronaviruses OC43 and HKU1 (Figures S3 A and S3B). The third BNT162b2 vaccine dose further increased vaccinee titers to SARS-CoV-1, OC43, and HKU1 (Figure S3C). Antibodies to the spike antigens of the endemic human alphacoronaviruses NL63 and 229E were not boosted (Figure S3).
Figure S3.
BNT162b2 vaccination produces less broad serological responses to endemic human coronaviruses (HCoVs) compared with SARS-CoV-2 infection, related to Figure 2
(A and B) Anti-SARS-CoV-1 spike and anti-HCoV-OC43, -HKU1, -NL63, and -229E spike IgM, IgG, and IgA antibody responses are shown for individuals who received BNT162b2 prime (D0) and boost (D21) vaccination doses and for COVID-19 patients. (A) The heatmap shows the development of antibody responses in longitudinal samples from vaccinees/patients collected at/during D0, D7/week 1, D21/weeks 2 and 3, D28/week 4, D42/weeks 5 and 6, and D90/≥week 7 after vaccination/COVID-19 symptom onset (x axis). The color scale encodes the median values of log10 MSD AU/mL concentrations. (B) Box-whisker plots show the development of antibody responses in longitudinal samples from vaccinees/patients collected at/during D0, D7/week 1, D21/weeks 2 and 3, D28/week 4, D42/weeks 5 and 6, and D90/≥week 7 after vaccination/COVID-19 symptom onset (x axis). Box-whisker plots show the interquartile range as the box and the whisker ends as the most extreme values within 1.5 times the interquartile range below the 25% quantile and above the 75% quantile. Statistical test: pairwise Wilcoxon rank sum test with Bonferroni correction. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Individuals were classified as vaccinees who have not been previously exposed to SARS-CoV-2 (vaccinees), outpatients (Outpt) and hospital-admitted patients (Admit), and ICU patients and those who died from their illness (Death).
(C) Box-whisker plots show anti-SARS-CoV-1 spike and anti-HCoV spike antibody responses in plasma samples from individuals who received BNT162b2 prime (D0, n = 59 individuals), second dose (D21, n = 58 individuals), and third dose (around month 9, n = 36 individuals) vaccination. Box-whisker plots show the interquartile range as the box and the whisker ends as the most extreme values within 1.5 times the interquartile range below the 25% quantile and above the 75% quantile.
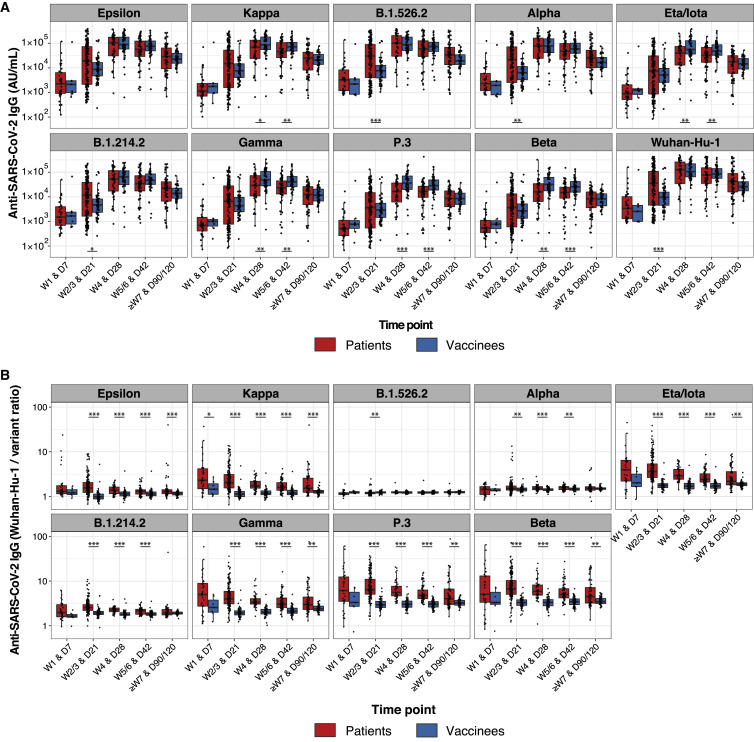
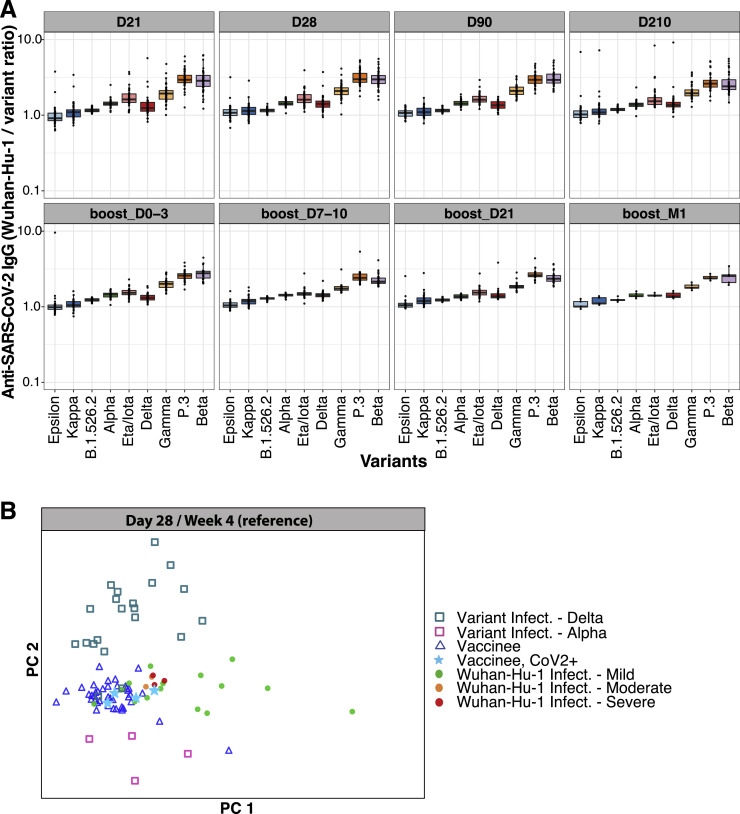
Greater breadth of IgG binding to viral variants following BNT162b2 vaccination compared with infection with Wuhan-Hu-1 SARS-CoV-2
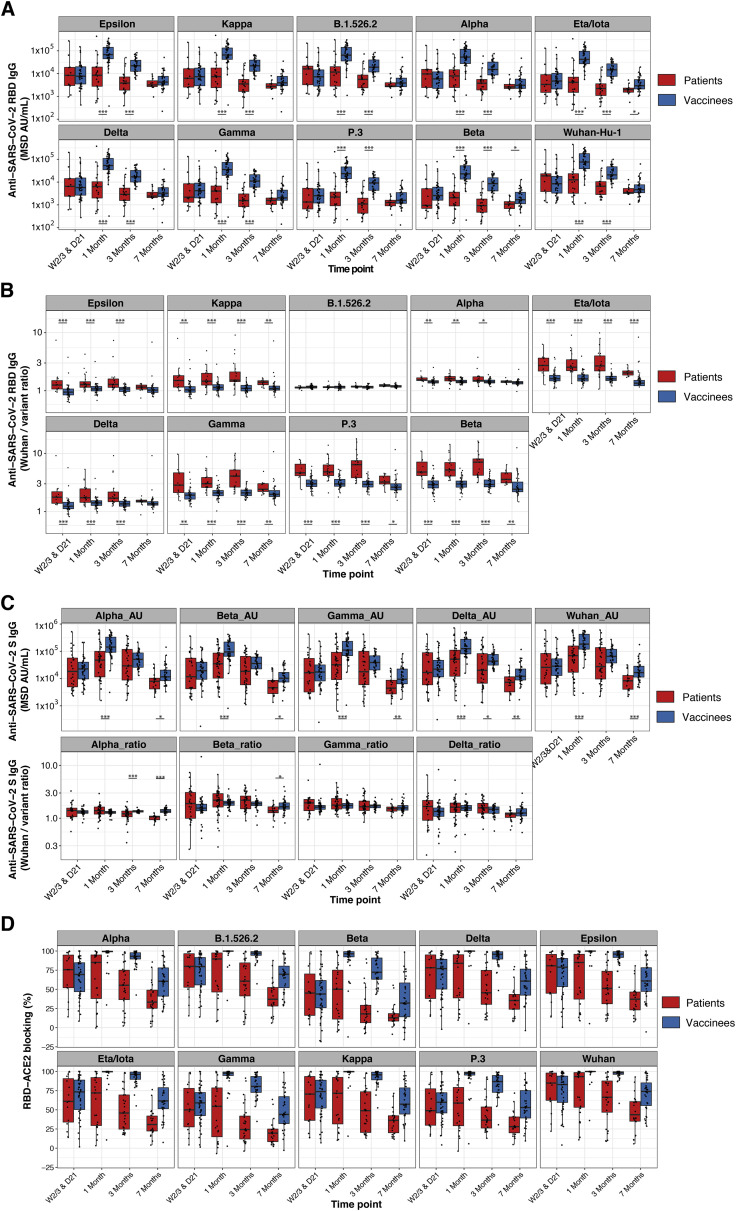
Immune-evasive SARS-CoV-2 variants have spread globally (Harvey et al., 2021; Plante et al., 2021; Röltgen and Boyd, 2021). We compared plasma IgG responses to the RBDs of nine different SARS-CoV-2 variants of concern and interest in BNT162b2 vaccinees and COVID-19 patients, using multiplexed MSD ECL assays. For RBD antigens from Epsilon, Kappa, B.1.526.2, B.1.214.2, Alpha, Eta/Iota, Gamma, P.3, and Beta variants, both vaccinee and infected patient IgG showed the greatest decrease in binding to Beta, Gamma, and P.3 variants relative to Wuhan-Hu-1 (Figure 3 A). To quantify the differences in variant RBD binding by vaccinee and patient plasma IgG, we calculated the ratios of anti-RBD IgG concentrations for Wuhan-Hu-1 compared with viral variants, with higher ratios indicating greater binding of Wuhan-Hu-1 RBD compared with variant RBD (Figure 3B). COVID-19 patients showed a greater IgG binding bias for Wuhan-Hu-1 RBD compared with variant RBDs in the initial weeks post-onset of symptoms; in contrast, BNT162b2 vaccinee IgG had relatively greater breadth of binding to variant RBDs and less preference for Wuhan-Hu-1 RBD. Over time, infected patient plasma samples showed improvement in variant RBD binding relative to Wuhan-Hu-1 RBD, suggesting evolution of the antibody response through at least 7 weeks post-onset of symptoms (Figure 3). BNT162b2 vaccinee IgG Wuhan-Hu-1 to variant RBD binding ratios did not change from day 21 onward. The greater breadth of variant RBD binding (including the Delta variant) by vaccinee IgG compared with COVID-19 patient IgG was seen in a second, independent validation cohort (Stanford cohort 2) of predominantly mildly ill COVID-19 patients. Greater Wuhan-Hu-1 to variant RBD IgG binding ratios were found in weeks 2–3, month 1, month 3, and month 7 in infected patients compared with vaccinees (Figures S4 A and S4B), with improvement in variant recognition over time in the infected patients. Notably, the increased breadth of vaccinee IgG compared with COVID-19 patient IgG binding to viral variant antigens was greatest for RBD, the main target of neutralizing antibodies, and was decreased or not detected when whole spike antigens were tested (Figure S4C). Functional blocking of ACE2 binding to RBD was concordant with the RBD-specific IgG concentrations measured in these populations (Figures S4A and S4D).
Figure 3.
Greater breadth of IgG binding to SARS-CoV-2 RBD variants following BNT162b2 vaccination compared with infection with Wuhan-Hu-1 SARS-CoV-2
Anti-SARS-CoV-2 Wuhan-Hu-1 and viral variant RBD IgG responses are shown for Stanford individuals who received BNT162b2 vaccination and for Wuhan-Hu-1-infected COVID-19 Stanford patient cohort 1 at different time points after vaccination/COVID-19 symptom onset. Box-whisker plots show the interquartile range as the box and the whisker ends as the most extreme values within 1.5 times the interquartile range below the 25% quantile and above the 75% quantile. Significance between patient and vaccinee groups were tested with two-sided Wilcoxon rank sum test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001
(A) Anti-RBD IgG concentrations.
(B) Ratios of anti-Wuhan-Hu-1 to variant RBD IgG concentration.
Figure S4.
Greater breadth of IgG binding to SARS-CoV-2 variant RBDs following BNT162b2 vaccination compared with infection with Wuhan-Hu-1 SARS-CoV-2 (validation cohort), related to Figure 3
(A and B) Anti-SARS-CoV-2 Wuhan-Hu-1 and viral variant RBD IgG responses are shown for Stanford individuals who received BNT162b2 vaccination and for Wuhan-Hu-1-infected COVID-19 Stanford patient cohort 2 at different time points after vaccination/COVID-19 symptom onset. Box-whisker plots show the interquartile range as the box and the whisker ends as the most extreme values within 1.5 times the interquartile range below the 25% quantile and above the 75% quantile. Significance between groups was tested with two-sided Wilcoxon rank sum test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(A) Anti-RBD IgG concentrations.
(B) Ratios of anti-Wuhan-Hu-1 to variant RBD IgG concentration.
(C) Anti-SARS-CoV-2 Wuhan-Hu-1 and viral variant spike IgG responses as anti-spike IgG concentrations (upper panels) and as ratios of anti-Wuhan-Hu-1 to variant spike IgG concentration (lower panels) are shown for Stanford individuals who received BNT162b2 vaccination and for Wuhan-Hu-1-infected COVID-19 Stanford patient cohorts 1 and 2 samples. Box-whisker plots show the interquartile range as the box and the whisker ends as the most extreme values within 1.5 times the interquartile range below the 25% quantile and above the 75% quantile. Significance between groups was tested with two-sided Wilcoxon rank sum test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(D) Percentage blocking of ACE2 binding to RBD of specified viral variants by plasma antibodies of BNT162b2 vaccinees and Stanford patient cohort 2 samples.
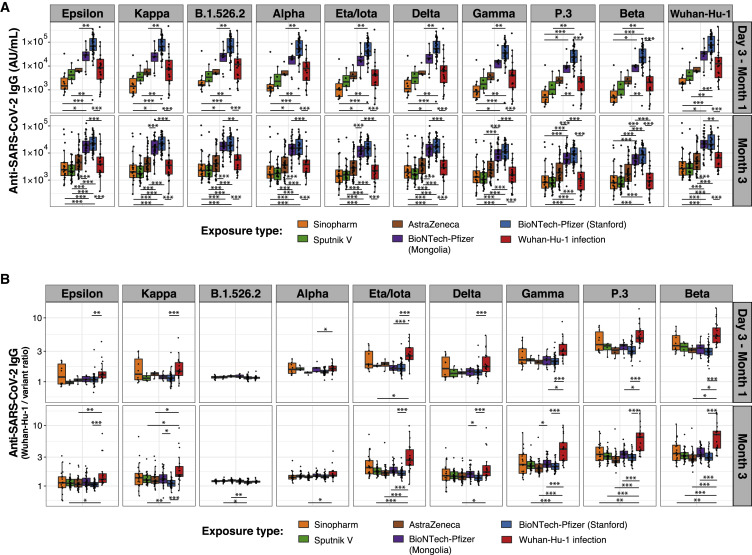
Improved IgG binding to viral variants is consistent across four COVID-19 vaccines (BTN162b2, ChAdOx1-S, Gam-COVID-Vac, and BBIBP-CorV) compared with infection
Several COVID-19 vaccines, including mRNA, viral vector-based, and inactivated virus vaccines, have been approved for use internationally. Varying efficacy and antibody responses from the vaccines have been reported (Baden et al., 2021; Dashdorj et al., 2021a, 2021b; Polack et al., 2020; Sadoff et al., 2021; Voysey et al., 2021). We compared IgG responses in Stanford COVID-19 cohort 2 patients and BNT162b2 vaccinees with those of participants in a Mongolian observational study deploying four different COVID-19 vaccines: the mRNA vaccine BNT162b2, adenoviral vectored vaccines ChAdOx1-S (Astra Zeneca) and Gam-COVID-Vac (Sputnik V), and an alum-adjuvanted, inactivated viral vaccine BBIBP-CorV (Sinopharm). RBD-specific IgG concentrations for Wuhan-Hu-1 and all viral variants measured (Epsilon, Kappa, B.1.526.2, Delta, Alpha, Eta/Iota, Gamma, P.3, and Beta) differed greatly between vaccine groups, with BNT162b2 eliciting the highest antibody levels, followed by Astra Zeneca, Sputnik V, and Sinopharm vaccination (Figure 4 A). IgG concentration differences between vaccines were significant for most viral variant RBDs. Stanford BNT162b2 vaccinees compared with Mongolian BNT162b2 vaccinees had higher IgG concentrations at early time points, likely due to differences in the timing of sample collection (Stanford days 28 and 90; Mongolian participants variable time points before 1 and 3 months) (Figure 4A). Despite the different vaccine compositions and magnitudes of antibody responses, all four vaccines elicited IgG with relatively greater breadth of viral variant RBD binding compared with that of infected patients (Figure 4B).
Figure 4.
Greater breadth of IgG binding to SARS-CoV-2 variant RBDs following vaccination with four different vaccines compared with infection with Wuhan-Hu-1 SARS-CoV-2
Anti-SARS-CoV-2 Wuhan-Hu-1 and viral variant RBD IgG responses are shown for individuals who received BNT162b2 (BioNTech-Pfizer), ChAdOx1-S (Astra Zeneca), Gam-COVID-Vac (Sputnik V), and BBIBP-CorV (Sinopharm) vaccination and for Wuhan-Hu-1-infected COVID-19 Stanford patient cohort 2 within 1 month and around 3 months after vaccination/COVID-19 symptom onset. Box-whisker plots show the interquartile range as the box and the whisker ends as the most extreme values within 1.5 times the interquartile range below the 25% quantile and above the 75% quantile. Significance between groups was tested with pairwise Wilcoxon rank sum test with Bonferroni correction. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001
(A) Anti-RBD IgG concentrations.
(B) Ratios of anti-Wuhan-Hu-1 to variant RBD IgG concentration.
Variant-specific serological responses following Alpha and Delta SARS-CoV-2 infection and immune imprinting after vaccination
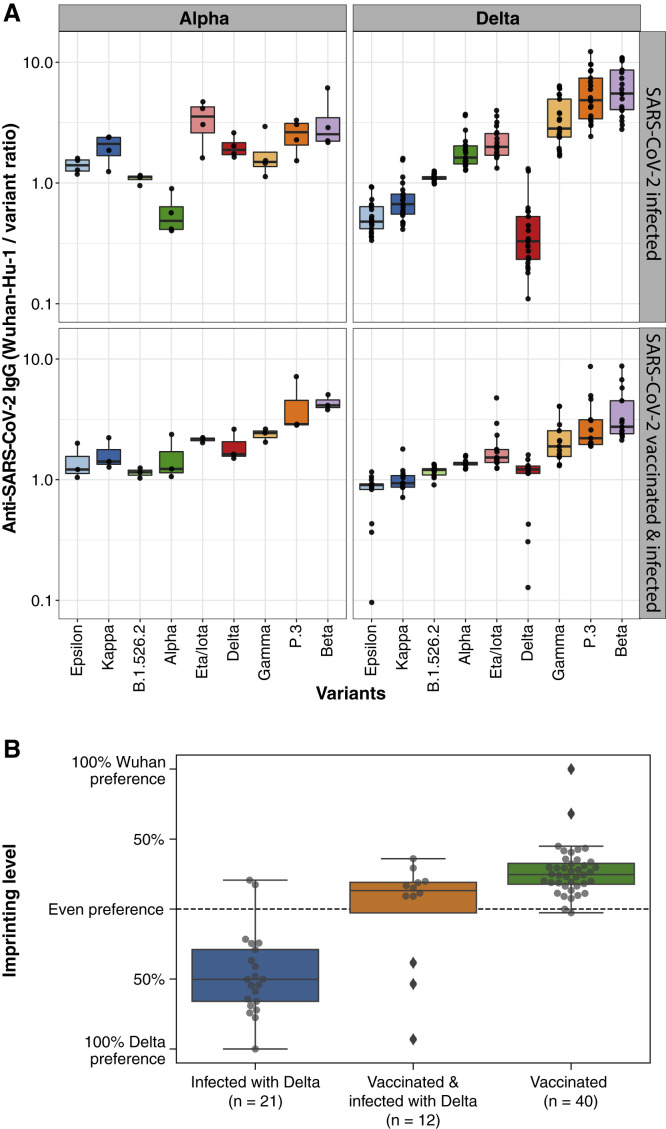
Immune imprinting, a phenomenon in which primary exposure to an antigen forms epitope-specific B cell memory and affects future B cell and antibody responses against variant epitopes, has been studied in influenza infection and vaccination. COVID-19 patients and BNT162b2 vaccinees who were only exposed to Wuhan-Hu-1 antigens in this study exhibit a consistent hierarchy in IgG binding concentrations to the different SARS-CoV-2 variant RBDs relative to the Wuhan-Hu-1 RBD, decreasing from Epsilon, Kappa, B.1.526.2, Delta, Eta/Iota, Gamma, P.3 to Beta (Figure S5 A). To test for imprinting of the serological response to variant RBDs, we first analyzed the ratios of Wuhan-Hu-1 to variant RBD IgG concentrations in COVID-19 patients who were infected with Alpha or Delta variants, confirmed by allele-specific RT-qPCR testing or viral sequencing. IgG from Alpha- or Delta-variant-infected patients with no history of COVID-19 vaccination or prior SARS-CoV-2 infection preferentially bound Alpha and Delta variant RBDs, respectively, compared with Wuhan-Hu-1 RBD (Figure 5 A, upper panels). Delta infection also elicited higher IgG concentrations to other variant RBDs containing L452R such as Epsilon and Kappa, compared with Wuhan-Hu-1 (Figure 5A, upper right). PCA of variant RBD-specific IgG responses in vaccinees and variant-infected patients (Figure S5B) highlights the distinct serological responses elicited by infection with the variant viruses. To test whether prior exposure to one SARS-CoV-2 RBD variant causes imprinting of humoral immunity, we analyzed plasma from individuals vaccinated with Wuhan-Hu-1-like antigens and subsequently infected with Alpha or Delta variants (Figure 5A, lower panels). Despite breakthrough infection with Alpha or Delta viral variants, the vaccinated individuals showed patterns of IgG binding to viral variant RBDs similar to those of individuals exposed to only Wuhan-Hu-1. We quantified the degree of imprinting of IgG specificity by log transforming the ratios of IgG binding to pairs of antigens (e.g., Wuhan-Hu-1 RBD compared with Delta RBD) for individual samples, then rescaling to range from −100% to +100% corresponding to the maximal preference for each antigen observed in other plasma specimens, including those from individuals exposed only to a single antigen variant (Figure 5B).
Figure S5.
Anti-SARS-CoV-2 RBD IgG signatures following BNT162b2 vaccination and SARS-CoV-2 infection, related to Figure 5
(A) Ratios of anti-Wuhan-Hu-1 to variant RBD IgG concentration are shown for Stanford individuals who received BNT162b2 vaccination at different time points after the second dose (D21, n = 58 individuals) and third dose (around month 9, n = 36 individuals) vaccination. Box-whisker plots show the interquartile range as the box and the whisker ends as the most extreme values within 1.5 times the interquartile range below the 25% quantile and above the 75% quantile.
(B) Principal component analysis (PCA) of anti-SARS-CoV-2 Wuhan-Hu-1 and viral variant RBD IgG concentrations across Stanford BNT162b2 vaccinees, Stanford COVID-19 patient cohort 2, and SARS-CoV-2-variant-infected patients.
Figure 5.
Variant-specific serological signature following Alpha and Delta SARS-CoV-2 infection
(A) Anti-Wuhan-Hu-1 to variant RBD IgG concentration ratios are shown for individuals with primary SARS-CoV-2 Alpha or Delta variant infection (upper panels) or secondary variant infection after vaccination (lower panels). Box-whisker plots show the interquartile range as the box and the whisker ends as the most extreme values within 1.5 times the interquartile range below the 25% quantile and above the 75% quantile.
(B) Anti-SARS-CoV-2 variant IgG binding preference levels of BNT162b2 vaccinees on day 28 postvaccination and of previously vaccinated or nonvaccinated individuals infected with the SARS-CoV-2 Delta variant.
LN GC impairment in severe SARS-CoV-2 infection but robust development following SARS-CoV-2 mRNA vaccination
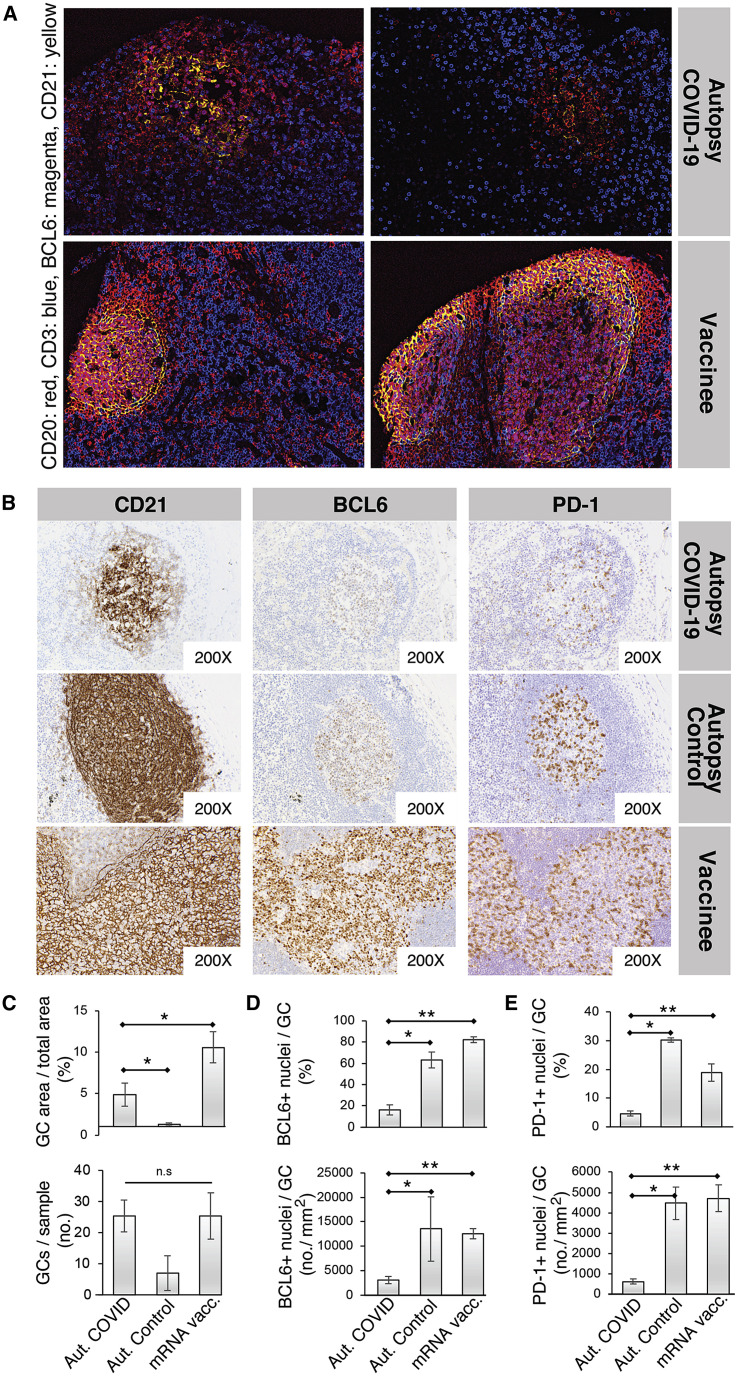
The differences in viral variant RBD IgG binding between SARS-CoV-2-infected patients and recipients of the four COVID-19 vaccines suggest that the organization of the humoral immune responses in secondary lymphoid tissues may differ between infection and vaccination, potentially due to direct effects of the viral infection, differences in innate immune stimuli between vaccination and infection, or the quantity or localization of viral antigens, among other possibilities. Previous studies have revealed a loss of GCs and a reduction in BCL6+ GC B cells in severe acute SARS-CoV-2 infection, raising the possibility that humoral responses may be altered or subverted by the virus (Kaneko et al., 2020). It is unclear whether draining LN immune responses to SARS-CoV-2 infection in the lungs differ from those elicited in axillary LNs following deltoid intramuscular mRNA vaccination. To compare GC architecture in response to SARS-CoV-2 infection and vaccination, we obtained peribronchial LN tissues from seven COVID-19 patients and three control autopsy cases as well as axillary LN core needle biopsies of seven individuals vaccinated with mRNA-1273 or BNT162b2. Importantly, core needle biopsy sample tissue volumes were suitable for the assessment of LN histoarchitecture. Vaccinee axillary LN biopsies were from the ipsilateral (same-side) arm vaccinated. Controls were thoracic LNs from individuals who succumbed to prepandemic non-COVID-19 pneumonias and contain GCs likely due to ongoing adaptive immune responses elicited by other antigens. LN histology for COVID-19 patients and vaccinees was evaluated with four-color codetection by indexing (CODEX) immunofluorescence analysis for CD20, CD3, BCL6, and CD21, which are markers of B cells, T cells, GC B cells (or T follicular helper [Tfh] cells), and follicular dendritic cells, respectively (Figures 6A and S6A), as well as by single-color immunohistochemical stains for these markers and the Tfh cell marker PD-1 (Figure 6 B). GCs were poorly formed in the severely ill COVID-19 patient peribronchial LNs compared with the axillary LNs of vaccinees, with disrupted CD21+ follicular dendritic cell networks and decreased BCL6+ cells (including GC B cells and Tfh cells) and PD-1+ cells (consistent with Tfh cells) within GCs (Figures 6A–6E). Disruption of CD21+ follicular dendritic cell networks was seen in both primary and secondary follicles in LNs from COVID-19 patients (Figure S6 B). mRNA vaccination was associated with follicular hyperplasia with fully developed GC architecture, including robust induction of GC B cells, Tfh cells, and extensive follicular dendritic cell networks (Figures 6A and 6B).
Figure 6.
Disrupted LN GCs in COVID-19 patients versus mRNA vaccinees
(A) Representative LN GC histology of COVID-19 patients and vaccinees evaluated with four-color Codex immunofluorescence analysis for CD20 (B cells), CD3 (T cells), BCL6 (GC B cells [major subset] and follicular helper T cells [minor subset]), and CD21 (follicular dendritic cells).
(B) Representative immunohistochemistry of GCs with CD21 (left), BCL6 (middle), and PD-1 (right) in peribronchial LNs of an autopsy patient who died of COVID-19, a control autopsy patient who died from a non-COVID-19 pneumonia (prepandemic), and in an axillary LN of a patient vaccinated with a SARS-CoV-2 mRNA vaccine.
(C–E) Relative proportion (upper) and absolute number (lower) of GCs in LNs (C), of BCL6+ cells within GCs (D), and of PD-1+ cells within GCs (E) from COVID-19 autopsy patients (n = 6), control autopsy patients (n = 3), and mRNA-vaccinated patients (n = 7). Quantification performed in QuPath digital pathology analysis software. Wilcoxon rank sum test was used to calculate p values. Error bars represent mean ± SEM. ∗p < 0.03; ∗∗p < 0.003.
Figure S6.
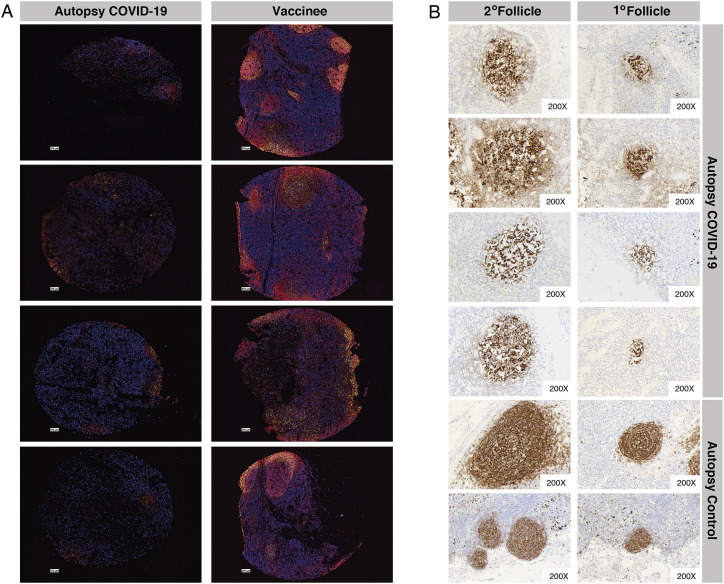
Disrupted LN GCs in COVID-19 patients versus mRNA vaccinees, related to Figure 6
(A) LN GC histology for COVID-19 patients (left) and mRNA vaccinees (right) evaluated with four-color codetection by indexing (Codex) immunofluorescence analysis for CD20 (red), CD3 (blue), BCL6 (magenta), and CD21 (yellow) markers of B cells, T cells, GC B cells (or T follicular helper cells), and follicular dendritic cells, respectively.
(B) Representative CD21 immunohistochemistry of secondary (left) and primary (right) follicles of four autopsy patients who died of COVID-19 and two control autopsy patients.
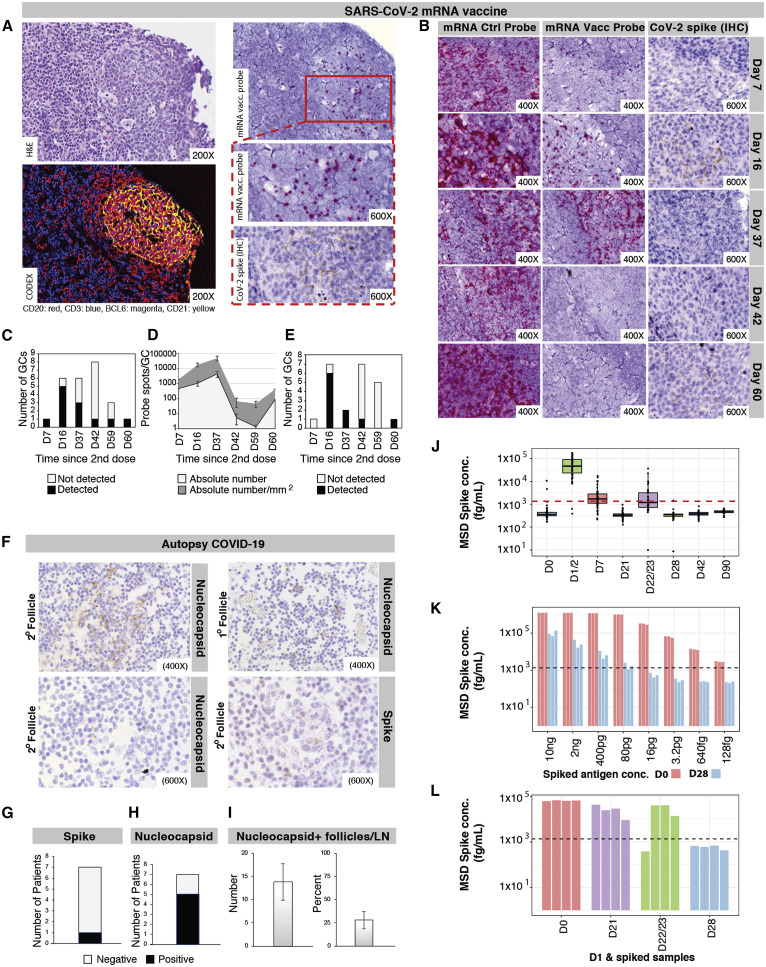
Prolonged detection of vaccine mRNA in LN GCs and spike antigen in LN GCs and blood following SARS-CoV-2 mRNA vaccination
The biodistribution, quantity, and persistence of vaccine mRNA and spike antigen after vaccination and viral antigens after SARS-CoV-2 infection are incompletely understood but are likely to be major determinants of immune responses. We performed in situ hybridization with control and SARS-CoV-2 vaccine mRNA-specific RNAScope probes in the core needle biopsies of the ipsilateral axillary LNs that were collected 7–60 days after the second dose of mRNA-1273 or BNT162b2 vaccination and detected vaccine mRNA collected in the GCs of LNs on days 7, 16, and 37 postvaccination, with lower but still appreciable specific signal at day 60 (Figures 7 A–7E). Only rare foci of vaccine mRNA were seen outside of GCs. Axillary LN core needle biopsies of nonvaccinees (n = 3) and COVID-19 patient specimens were negative for vaccine probe hybridization. Immunohistochemical staining for spike antigen in mRNA-vaccinated patient LNs varied between individuals but showed abundant spike protein in GCs 16 days post-second dose, with spike antigen still present as late as 60 days post-second dose. Spike antigen localized in a reticular pattern around the GC cells, similar to staining for follicular dendritic cell processes (Figure 7B). COVID-19 patient LNs showed lower quantities of spike antigen but a rare GC had positive staining (Figure 7F). Immunohistochemical staining for N antigen in peribronchial LN secondary and primary follicles of COVID-19 patients (Figures 7F–7I) was positive in five of the seven patients, with a mean percentage of nucleocapsid-positive follicles of more than 25%.
Figure 7.
Localization of SARS-CoV-2 proteins and vaccine mRNA in LNs
(A) Representative LN GC after mRNA vaccination showing hematoxylin and eosin staining (upper left), four-color Codex staining (lower left), in situ hybridization of a SARS-CoV-2 mRNA vaccine-specific probe (upper right [lower magnification] and middle right [greater magnification]), and immunohistochemical (IHC) staining for spike antigen (lower right). Vaccine mRNA probe hybridization was visualized by colorimetric development with Fast Red chromogen, and positive IHC staining for spike antigen was visualized as granular brown color from 3,3′-diaminobenzidine (DAB) reagent.
(B) Representative in situ hybridization of an RNAScope control probe (left panels) and SARS-CoV-2 mRNA vaccine-specific probe (middle panels) within ipsilateral axillary core needle LN biopsies of female patients 7–60 days after second mRNA-1273 or BNT162b2 dose. Probe hybridization is indicated by red chromogen spots. IHC signal for spike antigen (right panels) is detected as granular brown staining.
(C) Quantification of SARS-CoV-2 mRNA vaccine-specific probe-staining GCs in vaccinated LN biopsies.
(D) Quantification of positive SARS-CoV-2 mRNA vaccine-specific probe spots per GC in vaccinee LNs. Error bars represent mean ± SEM.
(E) Spike-protein-positive GC quantification from IHC staining of vaccinee LNs.
(F) IHC staining for spike (lower right panel) and nucleocapsid (upper panels and lower left panel) antigens in representative sections of COVID-19 patient peribronchial LNs. Nucleocapsid detection in primary (upper right panel) and secondary (upper left panel) LN follicles.
(G) Due to the low frequency of detection of spike antigen in COVID-19 patient LNs, quantification is presented as the number of patients with positive staining in their LN specimens.
(H) Quantification of the number of COVID-19 patients with LN follicles positive for nucleocapsid IHC staining.
(I) Number and percentage of nucleocapsid-positive follicles by IHC in COVID-19 patient LNs. Error bars represent mean ± SEM.
(J) Spike concentration measured in plasma samples collected before and at several time points after BNT162b2 vaccination, with the red dotted line indicating the cutoff for positive.
(K) Spike concentrations were measured in plasma samples collected from BNT162b2 vaccinees on D0 (spike negative) or D28 (spike positive) spiked with different concentrations of recombinant spike protein. Black dotted line = cutoff for positive.
(L) Spike concentration measured in plasma samples collected from BNT162b2 vaccinees on D0, D21, D22/23, and D28 mixed with the same plasma sample collected from one BNT162b2 vaccinee on D1 (spike positive). Black dotted line = cutoff for positive.
Spike protein was detected in the plasma of 96% of the vaccinees at days 1–2 (median spike concentration of 47 pg/mL) and in 63% at day 7 (median spike concentration of 1.7 pg/mL) after the prime vaccine dose. In contrast, spike antigen detection after the vaccine boost on day 21 was reduced, with half of the study participants being positive on days 1–2 (median spike concentration of 1.2 pg/mL) and only one individual on day 7 post-boost (Figure 7J). We suspected that high concentrations of spike-specific antibodies developed by vaccinees within the first 2–3 weeks after the prime vaccine dose could impede detection of spike antigen by competing for spike-binding sites with the anti-spike reagent antibodies in the antigen assay. To test this hypothesis, we added different concentrations of recombinant spike protein to spike-negative vaccinee plasma samples collected on day 0 (n = 3) and day 28 (n = 3) after the prime vaccination. While the recombinant spike protein could readily be detected in day 0 plasma samples, only high concentrations of the antigen led to a positive signal when mixed with the day 28 samples (Figure 7K). We then mixed a spike-positive plasma sample collected one day after vaccination with spike-negative plasma samples collected on days 0, 21, 22–23, and 28 (n = 4 each). Spike antigen detection levels were high in the mix of day 1 and day 0 samples, decreased in the mix of day 1 and day 21, and day 1 and day 22–23 samples, and below the cutoff for positive in the mix of day 1 and day 28 samples (Figure 7L). Together, our results are consistent with spike-specific antibodies blocking the detection of the antigen in antigen capture-based assays.
Discussion
One of the positive developments amid the global calamity of the SARS-CoV-2 pandemic has been the rapid design, production, and deployment of a variety of vaccines, including remarkably effective mRNA vaccines encoding the viral spike (Baden et al., 2021; Polack et al., 2020). We find that BNT162b2 vaccination produces IgG responses to spike and RBD at concentrations as high as those of severely ill COVID-19 patients and follows a similar time course. Unlike infection, which stimulates robust but short-lived IgM and IgA responses, vaccination shows a pronounced bias for IgG production even at early time points. These responses were similar across the adult age range in our study. The relative absence of IgM and IgA responses suggests a potent effect of the vaccine formulation in driving early and extensive IgG class-switching, potentially as a result of the reported T helper type 1-polarized CD4+ T cell responses stimulated by vaccine components (Lederer et al., 2020; Lindgren et al., 2017; Pardi et al., 2018). Our data demonstrate that vaccinee plasma and saliva spike and RBD-specific IgG concentrations decrease from their peak values by approximately 20-fold by 9 months after primary vaccination but quickly exceed prior peak concentrations in 7–8 days after boosting with a third vaccine dose.
Correlates of immunological protection from SARS-CoV-2 infection following vaccination or prior infection are still under investigation. Analysis of Moderna mRNA-1273 and AstraZeneca ChAdOx1-S responses highlights the overall similarity of correlate of protection results for spike-binding antibody and neutralizing antibody assays (Feng et al., 2021; Gilbert et al., 2022). We compared spike or RBD-binding antibody responses with Wuhan-Hu-1 SARS-CoV-2 neutralization data in BNT162b2 vaccinees and confirmed the high correlation of these assay results, supporting the interpretation that sensitive, precise, and validated commercial multiplexed antigen-binding assays with a wide dynamic range, such as the MSD ECL assays in this study, will be valuable in providing standardized correlates of protection data for vaccines as the pandemic continues. Particularly in the context of viral variants, it will be important to determine whether predictions of vulnerability to infection or severe disease can be improved by adding data from other immunological assays, including T cell measurements.
Differences in B cell responses to SARS-CoV-2 infection and vaccination may be reflected in the binding breadth of antibodies to different SARS-CoV-2 variants. We find that plasma of individuals who received prime/boost BNT162b2 vaccination, as well as individuals who received adenoviral vectored (ChAdOx1-S or Gam-COVID-Vac) or inactivated virus (BBIBP-CorV) COVID-19 vaccines show consistent patterns of binding to variant RBDs with modest decreases compared with Wuhan-Hu-1 RBD binding. In contrast, COVID-19 patients produce antibody responses with significantly greater Wuhan-Hu-1 RBD binding preference and lower breadth of variant RBD binding. These differences between vaccinee and COVID-19 patient IgG variant antigen binding were greatest for the RBD, the target of most neutralizing antibodies and were diminished when full spike antigen with its greater number of non-neutralizing epitopes was tested. These results, covering many clinically relevant viral variant antigens and several vaccine modalities, are consistent with findings for RBD-binding IgG in mRNA-1273 vaccinees compared with infected patients (Greaney et al., 2021b). Notably, COVID-19 patients with Alpha or Delta variant infections display characteristic serological profiles specific to the RBD of the infecting variant, indicating that SARS-CoV-2 variant serotyping may be useful for epidemiological studies of populations to determine exposure to circulating SARS-CoV-2 variants. Both vaccinees and COVID-19 patients exposed to Wuhan-Hu-1 antigens show the greatest decreases in antibody binding to RBD variants harboring E484 alterations, including Beta and Gamma. Although susceptibility to infection by viral variants is common to both vaccinated and convalescent populations, particularly as antibody titers decrease over time (Israel et al., 2022; Levin et al., 2021), our findings lead to the prediction that antibodies derived from infection may provide somewhat decreased protection against virus variants compared with comparable concentrations of antibodies stimulated by vaccination.
As additional variants of SARS-CoV-2 appear over time, individuals will acquire distinct immunological histories depending on which vaccines they received and which viral variants infected them. The idea that “imprinting” by a prior antigen exposure can shape, either positively or negatively, the response to a subsequent variant is well established in studies of influenza viruses and has been implicated in birth-year differences in susceptibility to particular avian influenza viruses (Gostic et al., 2016). We find that prior vaccination with Wuhan-Hu-1-like antigens followed by infection with Alpha or Delta variants gives rise to plasma antibody responses with apparent Wuhan-Hu-1-specific imprinting manifesting as relatively decreased responses to the variant virus epitopes, compared with unvaccinated patients infected with those variant viruses. While current booster vaccinations are still based on the Wuhan-Hu-1-like antigens, vaccine manufacturers are in the process of evaluating updated vaccine-encoding sequences from one or more circulating variants. Initial results from the third-dose boosting with Beta-spike-encoding mRNA vaccines after prior second-dose mRNA-1273 vaccination are consistent with our findings of significant imprinting of serological responses by the first antigen encountered (Choi et al., 2021; Chu et al., 2021), indicating that vaccine-derived imprinting affects subsequent antibody responses stimulated by vaccination as well as infection. The extent to which vaccine boosting or infection with different variants will effectively elicit antibody responses to new epitopes or rather increase responses to the epitopes of antigens encountered previously, as in the “original antigenic sin” phenomenon described for influenza virus infection and vaccination (Arevalo et al., 2020; Zhang et al., 2019), will be an important topic of ongoing study. The degree of imprinting may depend on the particular variants and the order in which they are introduced to the individual’s immune system and the number of exposures, such as the number of vaccine doses received. Additional data for evaluating the magnitude of these effects and their consequences for protection from infection are likely to become available in coming months, as individuals with different histories of SARS-CoV-2 vaccination or viral variant infection become infected with the more highly mutated Omicron variant (https://covdb.stanford.edu/page/mutation-viewer/#omicron). As a practical consideration, the very-high spike-specific IgG concentrations generated by mRNA vaccination and periodic additional booster doses may be able to compensate for relatively decreased binding to new viral variant antigens, potentially decreasing the public health impact of antibody response imprinting if vaccine boosting is widely adopted.
We hypothesized that differences in the serological responses observed in SARS-CoV-2 infection compared with vaccination, particularly those related to variant-antigen-binding breadth, could be related to the anatomical sites where the viral antigens are encountered, the quantity of viral antigens in those anatomical sites, differences in the cell populations stimulated in secondary lymphoid tissues, and potential damage to immunological tissues during infection. With CODEX multiplexed immunofluorescence microscopy and immunohistochemical microscopy, we identified follicular hyperplasia with robust axillary LN GCs after mRNA (BNT162b2 or mRNA-1273) vaccination, containing CD21+ follicular dendritic cell networks, BCL6+ B cells, and PD-1+ cells at significantly higher frequencies compared with those in peribronchial LNs of deceased COVID-19 patients. These findings demonstrate greater stimulation of GC B cells and Tfh cells in vaccination and normal functional organization of GC follicular dendritic cells. Loss or impairment of GCs in patients with severe COVID-19 suggests that SARS-CoV-2 viral infection subverts the humoral immune response, by directly damaging immune cells or as a secondary effect of inflammatory responses to infection (Feng et al., 2020; Kaneko et al., 2020). The observed extended presence of vaccine mRNA and spike protein in vaccinee LN GCs for up to 2 months after vaccination was in contrast to rare foci of viral spike protein in COVID-19 patient LNs. We hypothesize that the abundant spike antigen in the GCs of mRNA vaccine recipient LNs may contribute to the increased breadth of viral variant RBD binding by IgG seen after vaccination, potentially due to high antigen concentrations stimulating B cells with lower affinity for Wuhan-Hu-1 spike epitopes and better binding to variant epitopes. Persistent vaccine RNA and spike antigen at elevated concentrations in vaccinee LNs could result in less strict selection for higher-affinity B cells in the immune response compared with situations where antigen is more limiting (Cirelli et al., 2019). However, our observation that all vaccine modalities (mRNA, adenoviral, and inactivated virus) stimulated greater viral variant breadth of IgG binding than infection could indicate that some other aspects of SARS-CoV-2 infection underlie these differences, such as alteration of GC function.
Prepandemic analysis of a model RNA vaccine for yellow fever virus in a rhesus macaque at 16 h postvaccination showed that vaccine RNA in LN cell suspensions was detected predominantly in professional antigen-presenting cells including monocytes, classical dendritic cells, and B cells at this early time point (Lindsay et al., 2019). Data from follicular dendritic cells were not reported. Our histological data from SARS-CoV-2 mRNA-vaccinated humans at considerably later time points (7–60 days post-second dose) show vaccine RNA almost entirely in GCs, distributed primarily between the nuclei of GC cells, similar to the pattern seen by immunostaining for follicular dendritic cell processes or B cell cytoplasm. Additional co-localization studies with higher resolution may be required to determine more exactly which specific cell types harbor mRNA vaccine and spike antigen in humans following COVID-19 mRNA vaccination and infection and may provide further mechanistic insights into the basis for the differences in serological responses after vaccination compared with infection.
At least some portion of spike antigen generated after administration of BNT162b2 becomes distributed into the blood. We detected spike antigen in 96% of vaccinees in plasma collected 1–2 days after the prime injection, with antigen levels reaching as high as 174 pg/mL. The range of spike antigen concentrations in the blood of vaccinees at this early time point largely overlaps with the range of spike antigen concentrations reported in plasma in a study of acute infection (Ogata et al., 2020) although a small number of infected individuals had higher concentrations in the ng/mL range. At later time points after vaccination, the concentrations of spike antigen in blood quickly decrease although spike is still detectable in plasma in 63% of vaccinees 1 week after the first dose. A practical finding in our study is that the detection of spike antigen in plasma samples is impeded after second dose BNT162b2 vaccination, likely due to the formation of circulating immune complexes of anti-spike antibodies and spike protein, masking the antigen epitopes of the capture and detection antibodies that form the basis of antigen detection assays, similar to assay interference that has been reported for other diseases (Bollinger et al., 1992; Lima et al., 2014; Miles et al., 1993).
Limitations of the study
Data from SARS-CoV-2-infected clinical cohorts and vaccinated individuals in this study are observational. Longitudinal data for COVID-19 vaccine responses are derived predominantly from BNT162b2 mRNA vaccine recipients at Stanford, with data for the other four COVID-19 vaccines at a single postvaccination time point per individual. To make precise, internally controlled comparisons of polyclonal antibody responses to different viral variant antigens, we used multiplexed ECL assays of antibody binding to RBD, rather than virus neutralization assays, therefore our data do not reflect potentially functional antibodies binding to the spike N-terminal domain, or antibodies that may have other activities in vivo. In the analysis of imprinting of serological responses, plasma specimens were not available from the period after vaccination but before variant virus infection, precluding direct comparison of antibody specificities pre- and post-infection. Additional epidemiological studies will be needed to evaluate the clinical impact of antibody response imprinting on susceptibility to infection by new viral variants and the severity of disease in infected patients. LN histological comparisons between COVID-19 patients and vaccinees have the limitations that the infected patient specimens were limited to those with severe disease, the number of individuals analyzed was relatively low (seven COVID-19 patients and seven vaccinees), and the LN sampling was not done prospectively at predetermined time points after vaccination or infection. The serological analysis in this study is of polyclonal antibody responses; evaluation of the clonal B cell and plasma-cell populations producing these antibody mixtures in comparable numbers of subjects in infection and vaccination will likely be required for further mechanistic understanding.
Taken together, these results underscore important differences between SARS-CoV-2 antibody responses produced by vaccination versus infection. Key questions for the months and years ahead include the duration of effective vaccine-stimulated serological responses after third-dose boosting or other repeated exposures, particularly for the recent Omicron variant and other variants that will emerge in the future, and the safety and efficacy of variant-targeting vaccine boosters in previously vaccinated or infected individuals. Further mechanistic investigations into the differences in antibody breadth elicited by vaccination and infection are needed to define the roles of T cell help, antibody affinity maturation, GC function, and innate immune responses to vaccine components, as well as the cellular and subcellular distribution of vaccine RNA and expressed antigen in lymphoid tissues. Lessons from the antibody responses to the initial SARS-CoV-2 variants are likely to be important both for preparing for future additional variants of this virus, as well as improving vaccination strategies for other pathogens such as influenza virus.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Sulfo-tag conjugated anti-human IgG | Meso Scale Discovery | Cat#D21ADF-3 |
| Sulfo- conjugated anti-human IgM | Meso Scale Discovery | Cat#D21ADD-3 |
| Sulfo-tag conjugated anti-human IgA | Meso Scale Discovery | Cat#D21ADE-3 |
| ACE2 calibration reagent | Meso Scale Discovery | Cat#C01ADG-2 |
| Reference Standard 1 | Meso Scale Discovery | Cat#C00ADK-2 |
| Anti-CD20 (clone SP32), for CODEX | Abcam | Cat#ab236434 |
| Anti-CD3 (clone EP449E), for CODEX | Akoya Biosciences | Cat#4450027 |
| Anti-CD21 (clone EP3093), for CODEX | Akoya Biosciences | Cat#4450027 |
| Anti-BCL6 (clone IG191E/A8), for CODEX | Biolegend | Cat#648301; RRID: AB_2274637 |
| Anti-CD20 (clone L26), for IHC | Dako | Cat#M0755; RRID: AB_2282030 |
| Anti-CD3 (clone 2GV6), for IHC | Ventana | Cat#790-4341; RRID: AB_2335978 |
| Anti-CD21 (clone EP3093), for IHC | Ventana | Cat#760-4438 |
| Anti-BCL6 (clone GL191E/A8), for IHC | Ventana | Cat#760-4241; RRID: AB_2335965 |
| Anti-PD-1 (clone NAT105), for IHC | Cell-Marque | Cat#315M-96; RRID: AB_1160829 |
| Anti-CovNP T62 (polyclonal) | Sino Biological | Cat#40143-T62; RRID: AB_2892769 |
| Anti-SARS-CoV-2 spike (clone 1A9) | GeneTex | Cat#GTX632604; RRID: AB_2864418 |
| Biological samples | ||
| Plasma and saliva samples from 59 individuals vaccinated with BNT162b2 vaccine | This paper | N/A |
| Plasma samples from 196 individuals vaccinated with BNT162b2, ChAdOx1-S, Gam-COVID-Vac, BBIBP-CorV vaccines | ND Dashdorj, Ulaanbaatar, Mongolia | http://www.onomfoundation.org |
| Plasma samples from 188 patients infected with SARS-CoV-2 | This paper | N/A |
| Plasma samples from 50 individuals infected with SARS-CoV-2 variants | This paper | N/A |
| 37 plasma and 20 saliva samples from healthy human control individuals | Sean N. Parker Center for Allergy & Asthma Research | https://med.stanford.edu/allergyandasthma.html |
| Seven post-mortem peribronchial lymph nodes from patients who died of COVID-19 | Banner Health | https://www.bannerhealth.com/ |
| Three post-mortem peribronchial lymph nodes from pre-pandemic control patients | Banner Health | https://www.bannerhealth.com/ |
| Seven axillary lymph node core needle biopsies from individuals vaccinated with BNT162b2 or mRNA-1273 | This paper | N/A |
| Three axillary lymph node core needle biopsies from unvaccinated individuals | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Sulfo-tag conjugated human ACE2 protein | Meso Scale Discovery | Cat#D21ADG-3 |
| MSD GOLD Read Buffer B | Meso Scale Discovery | Cat#R60AM-2 |
| RNAscope Probe-V-nCoV2019-S | Advanced cell diagnostics | Cat#848561 |
| RNAscope Probe S-encoding-mRNA-1273-C1 (targeting 101-1488 of Spike-encoding_contig_assembled_from_Moderna_mRNA-1273_vaccine) | Advanced cell diagnostics | Cat#1104251-C1 |
| RNAscope Probe S-encoding-BNT-162b2-C1 (targeting 101-1143 of Spike-encoding_contig_assembled_from_BioNTech/Pfizer_BNT-162b2_vaccine) | Advanced cell diagnostics | Cat#1104241-C1 |
| RNA ISH Positive Control Probe PPIB | Advanced cell diagnostics | Cat#RS7755 |
| SARS-CoV-2 Spike protein | ATUM, custom | N/A |
| Bond Aspirating Probe Cleaning Solution | Leica Microsystems | Cat#CS9100 |
| Bond Dewax Solution | Leica Microsystems | Cat#AR9222 |
| Bond Epitope Retrieval 2 | Leica Microsystems | Cat#AR9640 |
| Bond Epitope Retrieval 1 | Leica Microsystems | Cat#AR9961 |
| Bond Enzyme Pre-treatment Kit | Leica Microsystems | Cat#AR9551 |
| Bond Mixing Stations | Leica Microsystems | Cat#S21.1971 |
| Bond Open Containers, 30mL | Leica Microsystems | Cat#OP309700 |
| Bond Open Containers, 7mL | Leica Microsystems | Cat#OP79193 |
| Bond Polymer Refine Kit | Leica Microsystems | Cat#DS9800 |
| Bond Primary Antibody Diluent | Leica Microsystems | Cat#AR9352 |
| Bond Slide Labs and Ribbon | Leica Microsystems | Cat#S21.4564 |
| Bond Titration Kit | Leica Microsystems | Cat#OPT9049 |
| Bond Titration Container | Leica Microsystems | Cat#OTP9719 |
| Bond Universal Covertiles | Leica Microsystems | Cat#S21.4611 |
| Bond Wash Solution | Leica Microsystems | Cat#AR9590 |
| Hematoxylin II | Ventana | Cat#790-2208 |
| Bluing, 760-2037EZ Prep Solution (10X) | Ventana | Cat#950-102 |
| OptiView Detection Kit | Ventana | Cat#760-700 |
| Protease 1 | Ventana | Cat#760-2018 |
| Protease 2 | Ventana | Cat#760-2019 |
| Reaction Buffer (10X) | Ventana | Cat#950-300 |
| SSC Solution | Ventana | Cat#950-110 |
| Ultra CC1 Solution | Ventana | Cat#950-224 |
| Ultra LCS Solution | Ventana | Cat#650-210 |
| UltraView Universal DAB Detection Kit | Ventana | Cat#760-500 |
| Vantage Clear Overlay | Ventana | Cat#1749400 |
| Dako Target Retrieval at pH9 | Aligent | Cat#S2368 |
| Hydrogen Peroxide 30% (diluted to 3%) | Thermo-Fisher | Cat#H325500 |
| Normal Horse Serum 2.5% | Victor Labs | Cat#S-2012 |
| ImmPress HRP Universal Secondary Antibody | Victor Labs | Cat#MP-7500 |
| Dako Liquid DAB+ Substrate Chromogen System | Aligent | Cat#K3468 |
| Critical commercial assays | ||
| V-PLEX Coronavirus Panel 2 (IgG) Kit | Meso Scale Discovery | Cat#K15369U |
| V-PLEX Coronavirus Panel 2 (IgM) Kit | Meso Scale Discovery | Cat#K15370U |
| V-PLEX Coronavirus Panel 2 (IgA) Kit | Meso Scale Discovery | Cat#K15371U |
| V-PLEX SARS-CoV-2 Panel 9 (IgG) Kit | Meso Scale Discovery | Cat#K15448U |
| V-PLEX SARS-CoV-2 Panel 11 (IgG) Kit | Meso Scale Discovery | Cat#K15455U |
| V-PLEX SARS-CoV-2 Panel 11 (ACE2) Kit | Meso Scale Discovery | Cat#K15458U |
| V-PLEX SARS-CoV-2 Panel 20 (IgG) Kit | Meso Scale Discovery | Cat#K15551U |
| S-PLEX SARS-CoV-2 Spike Kit | Meso Scale Discovery | Cat#K150ADJS |
| RNAScope 2.5 HD Assay-RED kit | Advanced cell diagnostics | Cat#322350 |
| Deposited data | ||
| Electrochemiluminescence data | This paper; Mendeley Data | https://doi.org/10.17632/j8r94pfrj6.1 |
| Original code | This paper; Github | https://github.com/boyd-lab/covid-infection-vs-vaccination |
| Original code | This paper; Zenodo | https://doi.org/10.5281/zenodo.5854880 |
| Virus neutralization antibody data from individuals vaccinated with BNT162b2 | Arunachalam et al. (2021) | DOI:10.1038/s41586-021-03791-x |
| Software and algorithms | ||
| R version 4.0.5 base packages | The R Foundation | https://www.rstudio.com/products/rstudio/download/ |
| R version 4.0.5 ggplot2 package | The R Foundation | https://cran.r-project.org/web/packages/ggplot2/index.html |
| QuPath version 0.2.3 | Bankhead et al. (2017) | PMID: 29203879 https://qupath.github.io/ |
| Python version 3.7.10 | Python Software Foundation | https://www.python.org |
| CODEX® MAV | Akoya Biosciences | https://help.codex.bio |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Dr. Scott D. Boyd (publications_scott_boyd@stanford.edu).
Materials availability
This study did not generate new unique reagents.
Experimental models and subject details
Plasma and saliva samples from Stanford BNT162b2 vaccinees
To study antibody responses after first, second, and third dose vaccination with BNT162b2, we collected longitudinal blood and saliva samples from 59 vaccinees (29 were women, 27 were men, 3 were unknown, and all donors were adults between the ages of 19 to 79 years). Baseline blood samples were collected on day 0 before or immediately after the first vaccine dose. Individuals received their second dose on day 21 and a third dose about 9 months after the prime. Blood sample collection after prime vaccination was scheduled for days 1, 7, 21, 22, 28, and 42 with blood draws ± one day from the assigned time point, or days 90, 120, or 210 ± 1 week from the assigned time point. In addition, blood samples were collected on days 0 to 3, 7 to 10, 21, and 28 after the third vaccine dose. Saliva samples were collected on day 42 after the first dose, as well as before, and 1 or 2, 3 or 4, 7, and 21 days after the third vaccine dose. Peripheral blood was collected in vacutainer cell preparation tubes (CPTs) containing sodium citrate. Plasma was isolated and stored at -80°C. Saliva was collected from study participants, centrifuged, and supernatants were stored at −80°C. All BNT162b2 vaccine study participants provided informed consent under Stanford University Institutional Review Board approved protocol IRB-55689.
Plasma and saliva samples from Stanford COVID-19 patients
Blood and saliva samples were collected between March and December 2020 from COVID-19 patients who reported to Stanford Healthcare-associated clinical sites. SARS-CoV-2 infection was confirmed for all patients by RT-qPCR of nasopharyngeal swabs as described (Corman et al., 2020; Hogan et al., 2020). Blood samples were collected in heparin- or EDTA-coated vacutainers. After centrifugation for collection of plasma or saliva, samples were stored at −80°C. The use of these samples for antibody testing was approved by the Stanford University Institutional Review Board (Protocols IRB-48973 and IRB-55689).
Stanford COVID-19 patient cohort 1 included 530 plasma samples collected from 100 moderately to severely ill COVID-19 patients (52 were women and 48 were men; ages ranged from 1 to 95 years; 23 were outpatients, 33 were admitted to hospital without needing ICU care, 20 were treated in the ICU and 24 died of COVID-19) between March 2020 and August 2020. ELISA serology data for Wuhan-Hu-1 SARS-CoV-2-specific antibodies in these specimens have been reported previously (Röltgen et al., 2020).
Stanford COVID-19 patient cohort 2 was included as a validation cohort of 87 samples from 74 mostly mildly ill patients who had blood sample draws between March and December 2020, at approximately 21 days (n = 15 samples), 1 month (n = 23 samples), 3 months (n = 27 samples) and 7 months (n = 22 samples) after positive RT-qPCR testing for SARS-CoV-2 infection. Of those patients, 37 were women, 34 were men and 3 were unknown. Donors were 19 to 72 years of age and in terms of disease severity, 59 were mildly ill, 6 were moderate ill and 9 had a severe/critical disease course. Specimen time points were selected to match those of Stanford BNT162b2 vaccinee sample collections. Saliva samples were collected from five COVID-19 patients.
Stanford SARS-CoV-2 variant infection cohort blood samples were collected from COVID-19 patients during acute infection with SARS-CoV-2 Alpha (n = 7) or Delta (n = 34) variants. Samples were from 20 women and 21 men, all between 2 and 92 years of age. SARS-CoV-2 genotyping data were obtained using a multiplex, mutation-specific RT-qPCR targeting N501Y, E484K, and L452R, as previously described (Wang et al., 2021). Samples from the first multiplexed reaction suspected to contain the Alpha variant were analyzed with a second confirmatory genotyping RT-qPCR assay to detect mutations encoding the N501Y amino acid change, as described (Dashdorj et al., 2021a).
Plasma samples from Mongolian vaccinees
To study SARS-CoV-2 variant-specific IgG responses elicited by different COVID-19 vaccines, we tested plasma samples collected in July 2021 from 196 Mongolian vaccine study participants (109 were women, 87 were men, all were adults between 20 and 85 years of age) who had been fully vaccinated with one of four COVID-19 vaccines: BioNTech-Pfizer BNT162b2 (n = 47), AstraZeneca ChAdOx1-S (n = 50), Sputnik V Gam-COVID-Vac (n = 45) and Sinopharm BBIBP-CorV (n = 54). Participants were recruited by public announcement and volunteers were enrolled after signing the consent form approved by the Ethics Review Board at the Ministry of Health of Mongolia. SARS-CoV-2 pseudotyped virus neutralization and RBD-ACE2 blocking data on the same samples have been reported previously (Dashdorj et al., 2021a). Peripheral blood was collected in CPT, centrifuged for collection of plasma, and stored at −80°C.
Healthy human control (HHC) plasma and saliva
37 plasma and 20 saliva samples from HHCs collected before the onset of the COVID-19 pandemic for studies at the Sean N. Parker Center for Allergy & Asthma Research were used to verify pre-pandemic antibody binding concentrations to the different coronavirus antigens, and manufacturer-provided cutoffs for positive serology assay results. Use of these samples was approved by the Stanford University Institutional Review Board (Protocols IRB-8629 and IRB-60171). No demographic information was available for these samples.
Axillary LN core biopsies and post-mortem peribronchial LN tissues
To analyze and compare GC architecture in response to COVID-19 vaccination and SARS-CoV-2 infection, we collected axillary LN core needle biopsies from BNT162b2 or mRNA-1273 vaccinees, and excised post-mortem peribronchial LNs from patients who died of COVID-19. For the selection of vaccinee tissues, we performed a retrospective search of our pathology archives and medical records between January 2021 and June 2021 for female patients who received either mRNA-1273 or BNT162b2 vaccination and subsequently underwent an ipsilateral axillary LN core needle biopsy due to mammographic findings and routine clinical care. Seven patients underwent biopsy one to eight weeks after vaccination with their second dose of mRNA vaccine. Three unvaccinated females undergoing axillary LN core biopsy for routine clinical care and mammographic findings served as controls. We included seven peribronchial LNs from two female and five male patients who died of COVID-19 before August 2020, one to three weeks after symptom onset. Control post-mortem peribronchial LN biopsies were from pre-pandemic patients who died of non-COVID-19 causes. Autopsies were done by the Arizona Study of Aging and Neurodegenerative Disorders Brain and Body Donation Program (Beach et al., 2015). Analysis of these tissues was approved by Stanford University Institutional Review Board Protocol IRB-48973.
Method details
MSD ECL binding assays
Plasma samples from vaccinees and COVID-19 patients were heat-inactivated at 56°C for 30 minutes and tested using multiplexed ECL detection in a 96-well plate format with MSD® V-PLEX® serology panels and instrumentation according to the manufacturer’s instructions. V-PLEX COVID-19 Coronavirus Panel 2 kits were used to detect IgM, IgG, and IgA antibodies to SARS-CoV-2 N, S1 NTD, RBD, and spike antigens and to spike proteins of SARS-CoV-1 and other HCoVs including HCoV-OC43, HCoV-HKU1, HCoV-NL63, and HCoV-229E. V-PLEX SARS-CoV-2 Panel 9 and 11 kits were used to determine IgG antibody concentrations and RBD-ACE2 blocking antibody percentages to different SARS-CoV-2 variant RBDs, with Alpha, Beta, Gamma, Epsilon, Kappa, Eta/Iota, B.1.526.2, P.3 and Wuhan-Hu-1 present in both panel 9 and 11, and with B.1.214.2 in panel 9 and Delta in panel 11. V-PLEX SARS-CoV-2 Panel 20 kits were used to determine IgG antibody concentrations to Alpha, Beta, Gamma, Delta, and Wuhan-Hu-1 SARS-CoV-2 variant spike proteins. Plasma samples were analyzed in duplicate at a 1:5,000 (for IgG binding assays) or a 1:10 (for RBD-ACE2 blocking assays) dilution in MSD diluent. Coronavirus-specific antibodies were detected with anti-human IgM, IgG, or IgA antibodies, or indirectly with human ACE2 protein (for RBD-ACE2 blocking assays) conjugated to SULFO-TAG™ ECL labels and read with a MESO® QuickPlex® SQ 120 instrument. Cutoff values for positive antibody test results for Wuhan-Hu-1 antigens were determined by the manufacturer based on sera from 200 pre-pandemic healthy adults and 214 PCR-confirmed COVID-19 patients. We tested an additional 37 healthy adult pre-pandemic plasma specimens to evaluate the manufacturer’s cutoff values, and to determine cutoffs for positive binding to variant virus antigens, defined as the mean plus three standard deviations of the results from the pre-pandemic specimens. Antibody binding ratios for Wuhan-Hu-1 and viral variant antigens were only calculated for specimens that were above the cutoff values for positive results. Saliva samples were analyzed in duplicate at a 1:5 dilution in MSD diluent 2. Each plate contained duplicates of a 7-point calibration curve with serial dilution of a reference standard, a blank well and three positive control samples. Calibration curves were used to calculate antibody unit concentrations by backfitting ECL signals measured for each sample to the curve.
MSD ECL spike antigen detection
SARS-CoV-2 spike antigen was quantified in plasma samples using an antigen capture ECL immunoassay platform (Meso Scale Discovery). S-PLEX® SARS-CoV-2 spike Kit assays were performed according to manufacturer instructions. A 7-point calibration curve and negative control consisting of assay diluent were run in duplicate on each plate. Plates were read using a MESO QuickPlex SQ 120 instrument. Raw signals were converted to a concentration based on linear regression to the 7-point calibration curve. Recombinant SARS-CoV-2 spike protein used for plasma spiking experiments was made by ATUM (https://www.atum.bio/).
Histology, immunohistochemistry, and in situ hybridization
LN core needle and autopsy tissue samples were fixed in formalin and embedded in paraffin (FFPE), and sectioned. Once unstained slides were generated and initial Hematoxylin and Eosin (H&E) stained sections were analyzed, two distinct 0.6 mm areas from each LN sample were cored out of each tissue block and re-embedded to construct a tissue microarray (TMA). Immunohistochemistry was performed on four-micron sections using standard automated or manual methods including deparaffinization, peroxidase blocking, antigen retrieval, primary and secondary antibody incubation, detection with 3,3′-Diaminobenzidine (DAB) development, and counterstaining. Assays were performed on Roche Ventana (Tucson, AZ) Ultra instruments using Ventana Optiview detection, or Leica (Buffalo Grove, IL) Bond III instruments using Leica Polymer Refine detection or manually using Dako (Carpenteria, CA) Target Retrieval (TR) and Liquid DAB+ Substrate Chromogen System with ImmPress (Vector, Burlingame, CA) secondary antibodies.
For in situ hybridization, manual methods were used as previously described (Cloutier et al., 2021), using manufacturer-recommended protocols with the RNAScope 2.5 HD Assay-RED kit and probes from Advanced Cell Diagnostics (Newark, CA). Two SARS-CoV-2 vaccine probes were developed to target bases 101–1143 of the spike encoding sequence of the BNT162b2 vaccine or bases 101–1488 of the spike encoding sequence of the mRNA-1273 vaccine. Both probes recognized SARS-CoV-2 mRNA vaccine, thus only the SARS-CoV-2 vaccine probe recognizing bases 101–1488 of mRNA-1273 vaccine are presented. To assess the specificity of SARS-CoV-2 RNAScope vaccine probes, they were tested against SARS-CoV-2 infected placental tissue, in addition to staining for SARS-CoV-2 viral probe which targets bases 21631–23303 of the S-gene. SARS-CoV-2 RNAScope vaccine probes did not recognize SARS-CoV-2 virus. In addition, for each tissue tested there were internal negative control areas which did not react with SARS-CoV-2 vaccine probe.
Whole slide imaging (WSI) and quantitative image analysis
Whole slide images of immunohistochemistry-stained slides for BCL6, PD-1 and CD21 were scanned at 40X magnification (0.25 μM per pixel) on an Aperio AT2 scanner (Leica Biosystems, Nussloch, Germany) in ScanScope Virtual Slide (SVS) format. Total lymphoid tissue and individual GCs were annotated as regions of interests (ROIs) using the QuPath open-source WSI software (Bankhead et al., 2017). GCs were defined as B cell areas with CD21+ follicular dendritic cell networks and BCL6+ nuclei. Primary follicles were defined as B cell areas with CD21+ follicular dendritic cell networks without BCL6+ nuclei. For each GC ROI, the standard positive cell detection function was used to identify positive and negative cells with a single threshold of 0.2 when scoring the cell DAB OD mean. For autopsy BCL6 analysis (COVID-19 and control) the threshold was adjusted to 0.05 given the dim expression of BCL6. For each ROI, the area in mm2, number positive cells per mm2, and percent positive cells were calculated by QuPath.
A TMA slide containing two distinct 0.6 mm cores of each of the mRNA vaccine (n = 7) and vaccine control (n = 3) biopsies hybridized with SARS-CoV-2 mRNA RNAScope vaccine probe was scanned at 40× magnification (0.25 μM per pixel) on an Aperio AT2 scanner in SVS format. Total lymphoid tissue and individual GCs were annotated as ROIs using QuPath. For each GC ROI, the number of spots/clusters of RNAScope probe was detected using the QuPath subcellular detection option per manufacture instructions. Our detection parameters were as follows: detection threshold = 0.6; split by intensity. Our split and cluster parameters were as follows: expected spot size = 2 μm2, min spot size = 2 μm2, and max spot size = 3 μm2.
Co-detection by indexing (Codex)
All antibodies used for Codex were first screened, titrated, and validated by individual staining on FFPE human tonsil tissue samples. Standard manual immunohistochemistry was used to crossvalidate antibodies with the same, nonconjugated antibody clones listed in the key resource table. Tissue preparation for Codex was undertaken by obtaining 8-μm-thick sections from FFPE tissue blocks that were immobilized on charged square glass coverslips coated with polylysine (Electron Microscopy Sciences Hatfield, PA) prepared according to the manufacturer’s instructions. The coated glass coverslips were stained with a cocktail containing nucleotide-barcoded primary antibodies. The coverslips underwent nuclear staining (DAPI) and were loaded on the stage of an automated inverted fluorescence microscope connected to the robotic fluidic system known as CODEX (Akoya Biosciences, Marlborough, MA). In addition to the nuclear stain, fluorophore-tagged complementary nucleotide sequences (reporters) were used to iteratively reveal three antibodies at a time per cycle. Two additional blank cycles at the beginning and at the end of the antibody reveal cycles were added for purposes of subtraction of autofluorescence background. Automated image acquisition, processing, segmentation, and fluidics exchange were conducted using an Akoya Codex instrument and Codex driver software (Akoya Biosciences) (Black et al., 2021; Goltsev et al., 2018; Schürch et al., 2020). Composite images providing multiplex fluorescent signals of stained cell types, cellular niches, and tissue architecture were captured using CODEX® MAV and FIJI software (Akoya Biosciences).
Quantification and statistical analysis
Statistical tests were performed in R using base packages for statistical analysis and the ggplot2 package for graphics. Box-whisker plots show median (horizontal line), interquartile range (box), and the end of the lower whisker representing the smallest observation greater than or equal to the 25% quantile minus 1.5 times the interquartile range, and the end of the upper whisker representing the largest observation less than or equal to the 75% quantile plus 1.5 times the interquartile range. In serological analyses where statistical significance was tested, significance was defined as: ∗∗∗p value < 0.001; ∗∗p value < 0.01; ∗p value ≤ 0.05.
For the principal component analysis (PCA) we log-transformed, calculated z-scores, and ran PCA on MSD antibody concentration measurements or Wuhan-Hu-1/variant RBD IgG concentration ratios from a reference time point after COVID-19 vaccination or SARS-CoV-2 infection using Python v3.7.10 and packages numpy v1.19.1, pandas v1.2.5, and scikit-learn v1.0. We then applied these transformations to matching data from all other time points, enabling us to visualize the change over time in these serology measurements on a consistent PCA reference. Plots were created with Python packages matplotlib version 3.3.2 and seaborn version 0.11.2.
For the analysis of the homogeneity or dispersion of serology measurements in groups differing by vaccination or infection status (Figures 2C and 2D) at a particular time point, we plotted each group’s distribution of Euclidean distances to its centroid (calculated with Python package scipy version 1.6.2). These distance distributions were consistent when calculated in the raw measurement data space of arbitrary units (AU) for the MSD ECL assay or in the transformed PC1 and PC2 space after embedding the raw measurements into the PCA space created from one reference time point.
Ratios of concentrations of IgG binding to Wuhan-Hu-1 RBD compared to variant virus RBDs were plotted for specimens with IgG binding above the cutoff for positive binding to Wuhan-Hu-1 RBD (Figures 3, 4 and S4) to avoid distortion of ratios by samples without specific binding. Ratios of IgG binding to spike antigens were calculated in a similar manner (Figure S4C, lower panel). Corresponding IgG concentrations for samples used to calculate ratios were plotted for reference (Figures 3A, 4A, S4A and S4C upper panel). To quantify serological imprinting from prior Wuhan-Hu-1 antigen exposure from vaccination on subsequent responses to breakthrough infection with the Delta variant, we first computed the ratio of Wuhan-Hu-1 RBD-binding level to Delta RBD-binding level. Here, a ratio of one indicates even preference, while ratios greater than one indicate preferential binding of Wuhan-Hu-1 over Delta. Each ratio is symmetric with its inverse; for example, a Wuhan-Hu-1/Delta binding ratio of 4/5 indicates the same degree of preference for Delta binding as the ratio 5/4 indicates for Wuhan-Hu-1 binding preference. We then log-transformed the ratios, which sets the even preference level at zero, with positive values corresponding to Wuhan-Hu-1 preference, and makes these values symmetric around zero (e.g., a value of -0.2 indicates the same level of preference for Delta as +0.2 does for Wuhan-Hu-1 binding). Finally, we rescaled the negative and positive values separately to the ranges -100% to 0 and 0 to +100%. The resulting magnitudes are binding preferences relative to the maximum binding preference observed for a particular variant, including data from individuals only exposed to Wuhan-Hu-1 or individual variants. In particular, -100% refers to the maximum observed binding preference towards Delta. We plotted the distributions of these binding preference levels for BNT162b2 vaccinees without known prior SARS-CoV-2 infection, for Delta infection cases with no recorded prior infection or vaccine exposure, and for Delta breakthrough infections following Wuhan-Hu-1 wild-type vaccination. An example binding preference level of +30% for a Delta breakthrough infection case suggests this individual is at 30% of the most imprinted state for Wuhan-Hu-1 preference. A binding preference of -20% would instead suggest that, following infection, this individual lost imprinting to Wuhan-Hu-1 and gained preference for binding the Delta variant.
Acknowledgments
We acknowledge Lilit Grigoryan, Yupeng Feng, Meera Trisal, Florian Wimmers, and the Stanford Clinical and Translational Research Unit (CTRU) for their help in sample processing. We thank the research participants as well as the clinical staff who contributed to these studies. We thank ATUM (https://www.atum.bio/) for providing recombinant SARS-CoV-2 spike protein. The graphical abstract was created with BioRender.com.
This work was supported by NIH/NIAID R01AI127877, R01AI130398, and U19AI057229 (S.D.B.), a pilot project award from the Bill & Melinda Gates Foundation (S.D.B. and S.C.A.N.), NIH/NCI 1U54CA260517 (S.D.B.), NIH/NIAID HHSN272201700013C (G.B.S.), NIH/NIA P30AG019610-20S1 (T.G.B.), an endowment to S.D.B. from the Crown Family Foundation, an anonymous philanthropic donation to the Boyd lab, an American Heart Association Postdoctoral Fellowship and Arnold O. Beckman Independence Award to M.J.S., an Early Postdoc Mobility Fellowship Stipend to O.F.W. from the Swiss National Science Foundation (SNSF), and an NSF Graduate Research Fellowship to M.Z.
Author contributions
S.D.B., K.C.N., B.P., and M.M.D. conceptualized and designed the trial; K.R., S.C.A.N., O.S., M.L.T., Y.N., and S.D.B. conceptualized and designed the study; A.S.L., M.M.S., J.L., and S.B.S. coordinated and performed blood collections under the supervision of K.C.N. and R.S.C.; C.C., F.Y., O.F.W., R.A.H., P.S.A., E.H., A.S.L., M.M., I.C., F.G., V.M., C.L., and M.J.S. collected and processed samples; K.R., S.C.A.N., O.S., S.F.Y., O.F.W., D.S., A.W., D.C., and S.Z. performed the experiments; E.P., N.J.D., N.D.D., R.M., G.E.S., T.G.B., G.W.C., J.L.W., J.N.W., Y.N., M.L.T., G.B.S., and B.A.P. provided reagents and/or samples and/or protocols; K.R., S.C.A.N., O.S., S.F.Y., F.Y., M.Z., G.B.S., and S.D.B. analyzed the data and/or performed statistical analyses; K.R., S.C.A.N., O.S., and S.D.B. wrote the manuscript; all authors provided intellectual contributions and edited and approved the manuscript.
Declaration of interests
S.D.B. has consulted for Regeneron, Sanofi, Novartis, and Janssen on topics unrelated to this study and owns stock in AbCellera Biologics. K.C.N. reports grants from the National Institute of Allergy and Infectious Diseases (NIAID), Food Allergy Research & Education (FARE), End Allergies Together (EAT), National Heart Lung and Blood Institute (NHLBI), and National Institute of Environmental Health Sciences (NIEHS). K.C.N. is Director of FARE and World Allergy Organization (WAO) Center of Excellence at Stanford; Adviser at Cour Pharmaceuticals; Cofounder of Before Brands, Alladapt, Latitude, and IgGenix; National Scientific Committee member for the Immune Tolerance Network (ITN) of NIAID; recipient of a Research Sponsorship from Nestle; Consultant and Advisory Board Member at Before Brands, Alladapt, IgGenix, NHLBI, and ProBio; and Data and Safety Monitoring Board member at NHLBI. J.L.W., J.N.W., and G.B.S. are employees of Meso Scale Diagnostics (MSD).
Published: January 25, 2022
Data and code availability
Raw data from all serology Figures have been deposited on Mendeley at https://doi.org/10.17632/j8r94pfrj6.1 and are publicly available as of the date of publication. All original code has been deposited on the Zenodo platform at https://doi.org/10.5281/zenodo.5854880 and is publicly available at the Github repository https://github.com/boyd-lab/covid-infection-vs-vaccination as of the date of publication. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Arevalo C.P., Sage V.L., Bolton M.J., Eilola T., Jones J.E., Kormuth K.A., Nturibi E., Balmaseda A., Gordon A., Lakdawala S.S., et al. Original antigenic sin priming of influenza virus hemagglutinin stalk antibodies. PNAS. 2020;117:17221–17227. doi: 10.1073/pnas.1920321117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam P.S., Scott M.K.D., Hagan T., Li C., Feng Y., Wimmers F., Grigoryan L., Trisal M., Edara V.V., Lai L., et al. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature. 2021;596:410–416. doi: 10.1038/s41586-021-03791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankhead P., Loughrey M.B., Fernández J.A., Dombrowski Y., McArt D.G., Dunne P.D., McQuaid S., Gray R.T., Murray L.J., Coleman H.G., et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 2017;7:16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach T.G., Adler C.H., Sue L.I., Serrano G., Shill H.A., Walker D.G., Lue L., Roher A.E., Dugger B.N., Maarouf C., et al. Arizona study of aging and neurodegenerative disorders and brain and body donation program. Neuropathology. 2015;35:354–389. doi: 10.1111/neup.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black S., Phillips D., Hickey J.W., Kennedy-Darling J., Venkataraaman V.G., Samusik N., Goltsev Y., Schürch C.M., Nolan G.P. CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat. Protoc. 2021;16:3802–3835. doi: 10.1038/s41596-021-00556-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger R.C., Kline R.L., Francis H.L., Moss M.W., Bartlett J.G., Quinn T.C. Acid dissociation increases the sensitivity of p24 antigen detection for the evaluation of antiviral therapy and disease progression in asymptomatic human immunodeficiency virus-infected persons. J. Infect. Dis. 1992;165:913–916. doi: 10.1093/infdis/165.5.913. [DOI] [PubMed] [Google Scholar]
- Choi A., Koch M., Wu K., Chu L., Ma L., Hill A., Nunna N., Huang W., Oestreicher J., Colpitts T., et al. Safety and immunogenicity of SARS-CoV-2 variant mRNA vaccine boosters in healthy adults: an interim analysis. Nat. Med. 2021;27:2025–2031. doi: 10.1038/s41591-021-01527-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu L., Montefiori D., Huang W., Nestorova B., Chang Y., Carfi A., Edwards D.K., Oestreicher J., Legault H., Girard B., et al. Immune memory response after a booster injection of mRNA-1273 for severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) MedRxiv. 2021 09.29.21264089. [Google Scholar]
- Cirelli K.M., Carnathan D.G., Nogal B., Martin J.T., Rodriguez O.L., Upadhyay A.A., Enemuo C.A., Gebru E.H., Choe Y., Viviano F., et al. Slow delivery immunization enhances HIV neutralizing antibody and germinal center responses via modulation of immunodominance. Cell. 2019;177:1153–1171.e28. doi: 10.1016/j.cell.2019.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier J.M., Allard G., Bean G.R., Hornick J.L., Charville G.W. PDGFB RNA in situ hybridization for the diagnosis of dermatofibrosarcoma protuberans. Mod. Pathol. 2021;34:1521–1529. doi: 10.1038/s41379-021-00800-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashdorj N.J., Dashdorj N.D., Mishra M., Danzig L., Briese T., Lipkin W.I., Mishra N. Molecular and serological investigation of the 2021 COVID-19 case surge in Mongolian vaccinees. MedRxiv. 2021 doi: 10.1001/jamanetworkopen.2021.48415. 08.11.21261915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashdorj N.J., Wirz O.F., Röltgen K., Haraguchi E., Buzzanco A.S., Sibai M., Wang H., Miller J.A., Solis D., Sahoo M.K., et al. Direct comparison of antibody responses to four SARS-CoV-2 vaccines in Mongolia. Cell Host Microbe. 2021;29:1738–1743.e4. doi: 10.1016/j.chom.2021.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earle K.A., Ambrosino D.M., Fiore-Gartland A., Goldblatt D., Gilbert P.B., Siber G.R., Dull P., Plotkin S.A. Evidence for antibody as a protective correlate for COVID-19 vaccines. Vaccine. 2021;39:4423–4428. doi: 10.1016/j.vaccine.2021.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsner R.A., Shlomchik M.J. Germinal center and extrafollicular B cell responses in vaccination, immunity, and autoimmunity. Immunity. 2020;53:1136–1150. doi: 10.1016/j.immuni.2020.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsey A.R., Frenck R.W., Walsh E.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Bailey R., Swanson K.A., Xu X., et al. SARS-CoV-2 neutralization with BNT162b2 vaccine dose 3. N. Engl. J. Med. 2021;385:1627–1629. doi: 10.1056/NEJMc2113468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Phillips D.J., White T., Sayal H., Aley P.K., Bibi S., Dold C., Fuskova M., Gilbert S.C., Hirsch I., et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:2032–2040. doi: 10.1038/s41591-021-01540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Diao B., Wang R., Wang G., Wang C., Tan Y., Liu L., Wang C., Liu Y., Liu Y., et al. The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directly decimates human spleens and lymph nodes. MedRxiv. 2020 03.27.20045427. [Google Scholar]
- Garcia-Beltran W.F., Denis K.J.S., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2021;185:457–466. doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert P.B., Montefiori D.C., McDermott A.B., Fong Y., Benkeser D., Deng W., Zhou H., Houchens C.R., Martins K., Jayashankar L., et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2022;375:43–50. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goltsev Y., Samusik N., Kennedy-Darling J., Bhate S., Hale M., Vazquez G., Black S., Nolan G.P. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell. 2018;174:968–981.e15. doi: 10.1016/j.cell.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostic K.M., Ambrose M., Worobey M., Lloyd-Smith J.O. Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science. 2016;354:722–726. doi: 10.1126/science.aag1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Loes A.N., Gentles L.E., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Antibodies elicited by mRNA-1273 vaccination bind more broadly to the receptor binding domain than do those from SARS-CoV-2 infection. Sci. Transl. Med. 2021;13:eabi9915. doi: 10.1126/scitranslmed.abi9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., et al. COVID-19 Genomics UK (COG-UK) Consortium SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbauer J.D., Matter M.S., Stalder A.K., Tzankov A. Histomorphological patterns of regional lymph nodes in COVID-19 lungs. Pathologe. 2021;42:89–97. doi: 10.1007/s00292-021-00945-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C., Newton I.G., Zare S.Y., Reiss S.M., Schwan B., Suh M.J., Hasteh F., Levi G., Crotty S. Normal human lymph node T follicular helper cells and germinal center B cells accessed via fine needle aspirations. J. Immunol. Methods. 2020;479 doi: 10.1016/j.jim.2020.112746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Krüger N., Schulz S., Cossmann A., Rocha C., Kempf A., Nehlmeier I., Graichen L., Moldenhauer A.S., Winkler M.S., et al. The Omicron variant is highly resistant against antibody-mediated neutralization – implications for control of the COVID-19 pandemic. Cell. 2021;185:447–456. doi: 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan C.A., Sahoo M.K., Huang C., Garamani N., Stevens B., Zehnder J., Pinsky B.A. Comparison of the Panther Fusion and a laboratory-developed test targeting the envelope gene for detection of SARS-CoV-2. J. Clin. Virol. 2020;127:104383. doi: 10.1016/j.jcv.2020.104383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel A., Shenhar Y., Green I., Merzon E., Golan-Cohen A., Schäffer A.A., Ruppin E., Vinker S., Magen E. Large-scale study of antibody titer decay following BNT162b2 mRNA vaccine or SARS-CoV-2 infection. Vaccines. 2022;10, 64 doi: 10.3390/vaccines10010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko N., Kuo H.H., Boucau J., Farmer J.R., Allard-Chamard H., Mahajan V.S., Piechocka-Trocha A., Lefteri K., Osborn M., Bals J., et al. Loss of Bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell. 2020;183:143–157.e13. doi: 10.1016/j.cell.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keehner J., Horton L.E., Binkin N.J., Laurent L.C., SEARCH Alliance, Pride D., Longhurst C.A., Abeles S.R., Torriani F.J. Resurgence of SARS-CoV-2 infection in a highly vaccinated health system workforce. N. Engl. J. Med. 2021;385:1330–1332. doi: 10.1056/NEJMc2112981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- Lam J.H., Smith F.L., Baumgarth N. B cell activation and response regulation during viral infections. Viral Immunol. 2020;33:294–306. doi: 10.1089/vim.2019.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer K., Bettini E., Parvathaneni K., Painter M.M., Agarwal D., Lundgreen K.A., Weirick M., Muralidharan K., Castano D., Goel R.R., et al. Germinal center responses to SARS-CoV-2 mRNA vaccines in healthy and immunocompromised individuals. Cell. 2022 doi: 10.1016/j.cell.2022.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer K., Castaño D., Gómez Atria D., Oguin T.H., Wang S., Manzoni T.B., Muramatsu H., Hogan M.J., Amanat F., Cherubin P., et al. SARS-CoV-2 mRNA vaccines foster potent antigen-specific germinal center responses associated with neutralizing antibody generation. Immunity. 2020;53:1281–1295.e5. doi: 10.1016/j.immuni.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin E.G., Lustig Y., Cohen C., Fluss R., Indenbaum V., Amit S., Doolman R., Asraf K., Mendelson E., Ziv A., et al. Waning immune humoral response to BNT162b2 Covid-19 vaccine over 6 months. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2114583. NEJMoa2114583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima Mda R., Nogueira R.M., Filippis A.M., Nunes P.C., Sousa C.S., Silva M.H., Santos F.B. A simple heat dissociation method increases significantly the ELISA detection sensitivity of the nonstructural-1 glycoprotein in patients infected with DENV type-4. J. Virol. Methods. 2014;204:105–108. doi: 10.1016/j.jviromet.2014.02.031. [DOI] [PubMed] [Google Scholar]
- Lindgren G., Ols S., Liang F., Thompson E.A., Lin A., Hellgren F., Bahl K., John S., Yuzhakov O., Hassett K.J., et al. Induction of robust B cell responses after influenza mRNA vaccination is accompanied by circulating hemagglutinin-specific ICOS+ PD-1+ CXCR3+ T follicular helper cells. Front. Immunol. 2017;8:1539. doi: 10.3389/fimmu.2017.01539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay K.E., Bhosle S.M., Zurla C., Beyersdorf J., Rogers K.A., Vanover D., Xiao P., Araínga M., Shirreff L.M., Pitard B., et al. Visualization of early events in mRNA vaccine delivery in non-human primates via PET-CT and near-infrared imaging. Nat. Biomed. Eng. 2019;3:371–380. doi: 10.1038/s41551-019-0378-3. [DOI] [PubMed] [Google Scholar]
- Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., Liao P., Qiu J.F., Lin Y., Cai X.F., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Miles S.A., Balden E., Magpantay L., Wei L., Leiblein A., Hofheinz D., Toedter G., Stiehm E.R., Bryson Y. Rapid serologic testing with immune-complex-dissociated HIV p24 antigen for early detection of HIV infection in neonates. Southern California Pediatric AIDS Consortium. N. Engl. J. Med. 1993;328:297–302. doi: 10.1056/NEJM199302043280501. [DOI] [PubMed] [Google Scholar]
- Ogata A.F., Maley A.M., Wu C., Gilboa T., Norman M., Lazarovits R., Mao C.P., Newton G., Chang M., Nguyen K., et al. Ultra-sensitive serial profiling of SARS-CoV-2 antigens and antibodies in plasma to understand disease progression in COVID-19 patients with severe disease. Clin. Chem. 2020;66:1562–1572. doi: 10.1093/clinchem/hvaa213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Naradikian M.S., Parkhouse K., Cain D.W., Jones L., Moody M.A., Verkerke H.P., Myles A., Willis E., et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 2018;215:1571–1588. doi: 10.1084/jem.20171450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J.A., Mitchell B.M., Plante K.S., Debbink K., Weaver S.C., Menachery V.D. The variant gambit: COVID-19’s next move. Cell Host Microbe. 2021;29:508–515. doi: 10.1016/j.chom.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röltgen K., Boyd S.D. Antibody and B cell responses to SARS-CoV-2 infection and vaccination. Cell Host Microbe. 2021;29:1063–1075. doi: 10.1016/j.chom.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röltgen K., Powell A.E., Wirz O.F., Stevens B.A., Hogan C.A., Najeeb J., Hunter M., Wang H., Sahoo M.K., Huang C., et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci. Immunol. 2020;5:eabe0240. doi: 10.1126/sciimmunol.abe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoff J., Gray G., Vandebosch A., Cárdenas V., Shukarev G., Grinsztejn B., Goepfert P.A., Truyers C., Fennema H., Spiessens B., et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against Covid-19. N. Engl. J. Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürch C.M., Bhate S.S., Barlow G.L., Phillips D.J., Noti L., Zlobec I., Chu P., Black S., Demeter J., McIlwain D.R., et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell. 2020;182:1341–1359.e19. doi: 10.1016/j.cell.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J.S., O’Halloran J.A., Kalaidina E., Kim W., Schmitz A.J., Zhou J.Q., Lei T., Thapa M., Chen R.E., Case J.B., et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature. 2021;596:109–113. doi: 10.1038/s41586-021-03738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E.E., Frenck R.W., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Miller J.A., Verghese M., Sibai M., Solis D., Mfuh K.O., Jiang B., Iwai N., Mar M., Huang C., et al. Multiplex SARS-CoV-2 genotyping reverse transcriptase PCR for population-level variant screening and epidemiologic surveillance. J. Clin. Microbiol. 2021;59:e0085921. doi: 10.1128/JCM.00859-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley A.K., Fox A., Tan H.X., Juno J.A., Davenport M.P., Subbarao K., Kent S.J. Immune imprinting and SARS-CoV-2 vaccine design. Trends Immunol. 2021;42:956–959. doi: 10.1016/j.it.2021.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff M.C., Ramonell R.P., Nguyen D.C., Cashman K.S., Saini A.S., Haddad N.S., Ley A.M., Kyu S., Howell J.C., Ozturk T., et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat. Immunol. 2020;21:1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Choi A., Koch M., Ma L., Hill A., Nunna N., Huang W., Oestreicher J., Colpitts T., Bennett H., et al. Preliminary analysis of safety and immunogenicity of a SARS-CoV-2 variant vaccine booster. MedRxiv. 2021 doi: 10.1038/s41591-021-01527-y. 05.05.21256716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Wilson I.A. Recognition of the SARS-CoV-2 receptor binding domain by neutralizing antibodies. Biochem. Biophys. Res. Commun. 2021;538:192–203. doi: 10.1016/j.bbrc.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Stacey H.D., Mullarkey C.E., Miller M.S. Original antigenic sin: how first exposure shapes lifelong anti–influenza virus immune responses. J. Immunol. 2019;202:335–340. doi: 10.4049/jimmunol.1801149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data from all serology Figures have been deposited on Mendeley at https://doi.org/10.17632/j8r94pfrj6.1 and are publicly available as of the date of publication. All original code has been deposited on the Zenodo platform at https://doi.org/10.5281/zenodo.5854880 and is publicly available at the Github repository https://github.com/boyd-lab/covid-infection-vs-vaccination as of the date of publication. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.