Abstract
The SARS-CoV-2 Omicron variant with increased fitness is spreading rapidly worldwide. Analysis of cryo-EM structures of the spike (S) from Omicron reveals amino acid substitutions forging interactions that stably maintain an active conformation for receptor recognition. The relatively more compact domain organization confers improved stability and enhances attachment but compromises the efficiency of the viral fusion step. Alterations in local conformation, charge, and hydrophobic microenvironments underpin the modulation of the epitopes such that they are not recognized by most NTD- and RBD-antibodies, facilitating viral immune escape. Structure of the Omicron S bound with human ACE2, together with the analysis of sequence conservation in ACE2 binding region of 25 sarbecovirus members, as well as heatmaps of the immunogenic sites and their corresponding mutational frequencies, sheds light on conserved and structurally restrained regions that can be used for the development of broad-spectrum vaccines and therapeutics.
Keywords: COVID-19, SARS-CoV-2 variants, Omicron, infectivity, immune evasion, receptor recognition, cryo-EM structure, stability, fusogenicity
Graphical abstract
Mutations in the SARS-CoV-2 Omicron variant improve spike trimer stability that supports viral attachment but appears to compromise viral fusion. They also perturb the confirmation of antigenic sites for antibody recognition, which may contribute to immune evasion.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been continuously evolving through mutations in its viral genome, resulting in increased transmissibility, infectivity, and immune escape as observed among emerging variants of concerns (VOCs). Four widely circulated VOCs, Alpha, Beta, Gamma, and Delta, have been previously characterized, among which the Beta variant showed the greatest magnitude of immune evasion from neutralizing antibodies, whereas Delta exhibited dramatically enhanced transmission and infectivity (Davies et al., 2021; Mlcochova et al., 2021; Wang et al., 2021b). A newly identified VOC, named the Omicron variant (B.1.1.529), with an unprecedented number of mutations, is quickly spreading worldwide. Although the Delta variant remains the most prevalent strain currently, the Omicron variant is likely to become dominant by early 2022 as predicted by mathematical models (Kumar et al., 2021). The Omicron spike (S) harbors over 30 amino acid substitutions, 15 of which are in the receptor binding domain (RBD). These include three new clusters: (1) S371L, S373P and S375F, (2) N440K and G446S, and (3) Q493, G496, Q498, and Y505H, as well as other accumulated mutations, such as K417N, S477N, T478K, E484A, and N501Y (Figure 1 A), presumably conferring greater resistance to neutralizing antibodies and vaccine-induced humoral immunity (Cameroni et al., 2021; Cao et al., 2021; Liu et al., 2021b). Three small deletions (Δ69-70, Δ143-145, and Δ212), one 3-residue insertion (ins214EPE), and four substitutions (A67V, T95I, G142D, and N211I) in the N-terminal domain (NTD) probably alter its local conformation, presumably disrupting the original antigenic site existing in the wild type (WT) and also affecting viral infectivity (Figure 1A). In addition, mutations near by the furin cleavage site like H655Y, N679K, and P681H might be implicated in proteolytic activation since the P681R substitution identified in Delta could enhance viral fusogenicity and pathogenicity (Saito et al., 2021). Preliminary data suggest that the Omicron variant escapes almost all clinically approved antibody therapeutics, significantly impairs humoral immunity elicited by natural infection and vaccination, and possesses higher transmission rates among household contacts than those of the Delta variant, attributing to a higher risk of yet another resurgence of the pandemic (Lu et al., 2021; Wong et al., 2021). SARS-CoV-2 can enter into host cells through endosomes as well as the cellular surface, with inhibition of the activity of both the endosomal cathepsin L and the cell-surface membrane protein TMPRSS2 required to fully block its entry (Hoffmann et al., 2020). The entry for SARS-CoV-2 involves interactions of S with the ACE2 receptor and known or unidentified attachment cofactors, and subsequent priming of the protein by host cell proteases (Lan et al., 2020; Shang et al., 2020; Wang et al., 2020). These two key events advance the life cycle of the virus from the prefusion to the postfusion stage, leading to the fusion of the viral membrane with that of the host cell (Hoffmann et al., 2020; Zhu et al., 2021); details of which are not clearly understood. A set of substitutions, generally comprising of no more than three amino acids, in the RBD of four previous VOCs, have been shown to be associated with an increased affinity toward ACE2 (Zhou et al., 2021). Surprisingly, the Omicron RBD has nine substitutions (K417N, G446S, S477N, E484A, Q493R, G496S, Q498R, N501Y, and Y505H), that are located on the ACE2 binding interface, considerably affecting the receptor recognition, cell tropism, and entry pattern, particularly in combination with other mutations such as H655Y, N679K, and P681H (Figure 1A). Thus, understanding the underlying molecular basis for the enhanced transmissibility, immune resistance, and virological properties of the Omicron variant may facilitate development of intervention strategies to halt this looming crisis.
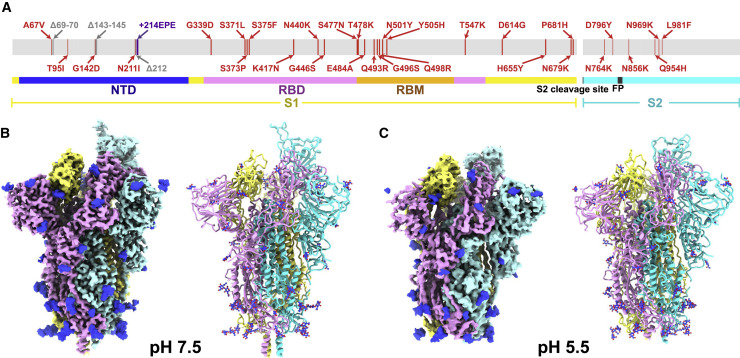
Figure 1.
Overall structures of the Omicron S-trimer
(A) Schematic diagram of the full-length SARS-CoV-2 spike (S) protein sequence with substitutions, deletions, and insertions illustrated in red, gray, and purple, respectively.
(B and C) Cryo-EM structures of the Omicron S-trimer at serological and endosomal pH. Structures are shown as surface (left) and ribbon (right). The three subunits of S protein are colored in yellow, cyan, and magenta, respectively.
See also Figures S1 and S2.
Results
Cryo-EM structures of the Omicron spike at serological and endosomal pH
Based on the first reported genome sequence of the Omicron variant (Figure 1A), we expressed prefusion-stabilized soluble trimeric ectodomain (S-trimer), which contains GSAS mutations at the furin cleavage site and 6P mutations along with the T4 fibritin trimerization domain (Lv et al., 2020; Wrapp et al., 2020), and purified the protein to homogeneity by affinity chromatography and size-exclusion chromatography (Figure S1 ). To explore the effect of endosomal pH on triggering conformational alterations, purified S-trimer was dialyzed against PBS buffer (pH 7.5) and acidic solution (0.1M Sodium citrate, pH 5.5), separately. We determined asymmetric cryo-EM reconstructions of the Omicron S-trimer at a resolution of 3.3 Å and 3.9 Å under neutral and acidic pH conditions (Figures 1B, 1C, and S1 and Table S1), respectively. Previous cryo-EM structures had revealed two prevalent prefusion conformations (but not limited to two) for SARS-CoV-2 S-trimer, including Alpha, Beta, and Kappa variants: a single-up conformation and all-down conformation, related to the positioning of the RBD (Cai et al., 2021). By contrast, only one conformational state with one “up” RBD and two “down” RBDs was observed in structures of the Omicron S-trimer, either at pH 7.5 or pH 5.5 (Figures 1B and 1C). Distinct from multiple orientations of RBD in the S-trimer at pH 5.5 (Zhou et al., 2020), structural heterogeneity in the Omicron S-trimer seems largely decreased, even when examined at endosomal pH. Overall structures of neutral and acidic Omicron S-trimer are indistinguishable, apart from some disorder on the NTD and RBM of the acidic S-trimer (Figures 1B, 1C, and S2 ).
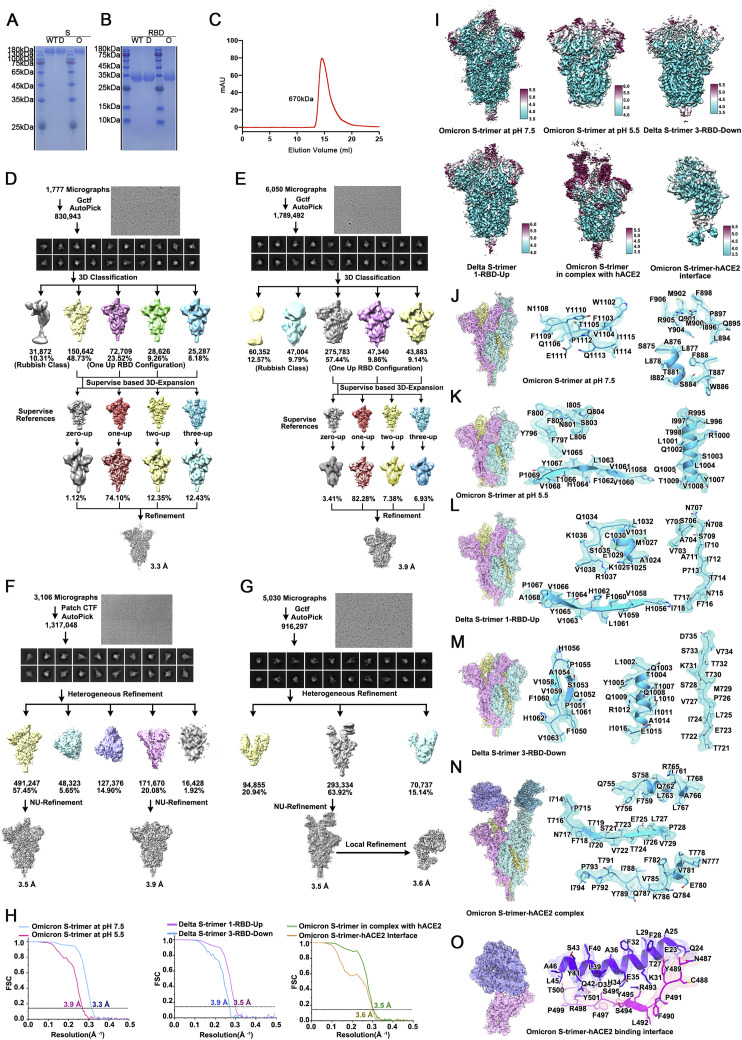
Figure S1.
Purification, characterization, and Cryo-EM single-particle analysis of Delta and Omicron S trimer, related to Figures 1, 2, and 6
(A) SDS-PAGE analysis of the Wild-type, Delta (marked as D) and Omicron (marked as O) S trimer. (B) SDS-PAGE analysis of the WT, Delta and Omicron RBD. (C) Gel filtration profile of the affinity-purified Omicron S-trimer. Flowcharts for structure determinations of (D) Omicron S-trimer at pH 7.5, (E) Omicron S-trimer at pH 5.5, (F) Delta S-trimer at pH 7.5, (G) Omicron S-trimer in complex with hACE2 at pH 7.5. (H) The gold-standard FSC curves of overall maps of Omicron S-trimer at pH 7.5, Omicron S-trimer at pH 5.5, Delta S-trimer at pH 7.5, Omicron S-trimer in complex with hACE2 at pH 7.5 and local maps of the binding interface. (I) Local resolution assessments of cryo-EM maps using ResMap are shown. Cryo-EM maps of (J) Omicron S-trimer at pH 7.5, (K) Omicron S-trimer at pH 5.5, (L) Delta S-trimer with 1-RBD-up, (M) Delta S-trimer with 3-RBD-down, (N) Omicron S-trimer in complex with hACE2 at pH 7.5 and (O) local maps of the binding interface. Residues are shown as sticks with oxygen colored in red, nitrogen colored in blue and sulfurs colored in yellow. Three different protomers of S trimer are colored in cyan, pink and yellow, respectively.
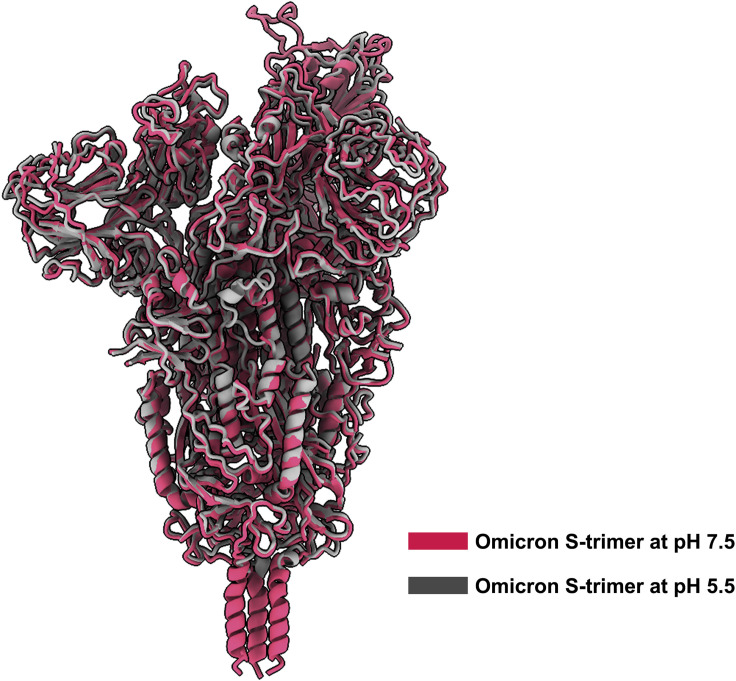
Figure S2.
Superimposition of overall structures of Omicron S-trimers solved at pH 7.5 and pH 5.5, related to Figure 1
The structure of Omicron S-trimer (gray) at pH 5.5 is superimposed to Omicron S-trimer (red) at pH 5.5. No notable structural differences were observed except for some disorder on NTD and RBM in structure solved at pH 5.5.
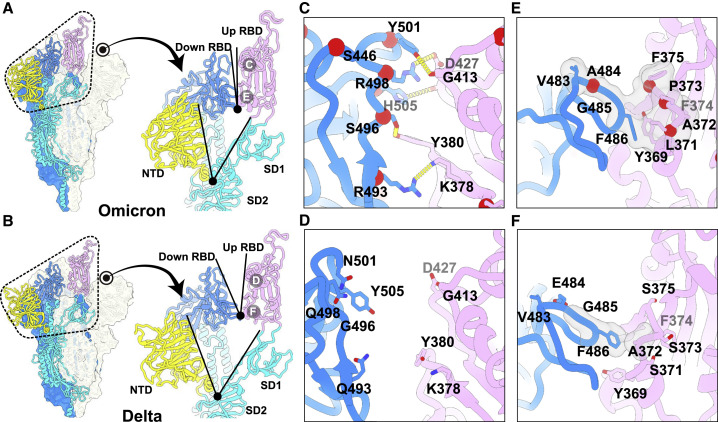
Structural features underpinning the up configuration
To further provide insights into the structural heterogeneity, we also determined near-atomic structures of the Delta S-trimer using the same strategy (Figure S1 and Table S1). In line with WT and most variants, Delta S-trimer exhibits two distinct conformational states corresponding to a closed form with all three RBDs down and an open form with one RBD up (Figure S3 ). To investigate structural basis for the stabilized up conformation, we further scrutinized these structures and used S-trimer structures from Alpha, Beta, Gamma, and Delta for in-depth comparison (Figures 2 and S3). Strikingly, a highly compact domain organization at the interface between the up S-monomer (defined as mol A) and its adjacent down RBD (defined as mol B) was observed in Omicron (Figures 2A and 2B). Compared to the other S-trimers, the domain shifts include a >12° and >6° clockwise rotation of the NTD (mol A, colored in yellow) and down RBD (mol B, colored in blue), respectively, together with a counterclockwise rotation of the SD1 (mol A, colored in cyan) and up RBD (mol A, colored in violet), underpinning the up configuration with buried areas of 450 Å2 between two contacted RBDs (Figures 2A, 2B, and S3). These extensive contacts were primarily due to hydrophilic and hydrophobic interactions, which were largely mediated by the nine mutations within two patches (Figures 2C and 2E). Substitutions of Q493R, G496S, Q498R, N501Y, and Y505H in the Omicron RBD (mol B) and K378, Y380, D427, and G413 from the up RBD (mol A) established five hydrogen bonds not observed in Delta (Figures 2C and 2D). Furthermore, four-residue mutations with completely opposite characteristics from hydrophilic to hydrophobic, including S371L, S373P, S375F, and E484A, together with Y369, A373, F374, V483, G485, and F486 have resulted in massive hydrophobic interactions (Figures 2E and 2F). Previous studies observed that distal regions of the S, such as furin cleavage loop and D614G, can lead to allosteric effects on RBD open/down disposition (Yurkovetskiy et al., 2020). Apart from these, the FPPR segment (fusion peptide proximal region, residues 828–853) and 630 loop (residues 620–640) are also proposed to modulate the S stability and structural rearrangements (Cai et al., 2021). For the Omicron, the FPPR and 630 loop were well resolved in the RBD-down conformation, whereas they were partially ordered in the RBD-up conformation (Figure S3), which is also consistent with a recent report that the FPPR segment and 630 loop facilitate clamping down of the RBDs in the closed form (Cai et al., 2021). These structural observations explain the decreased structural heterogeneity for Omicron S.
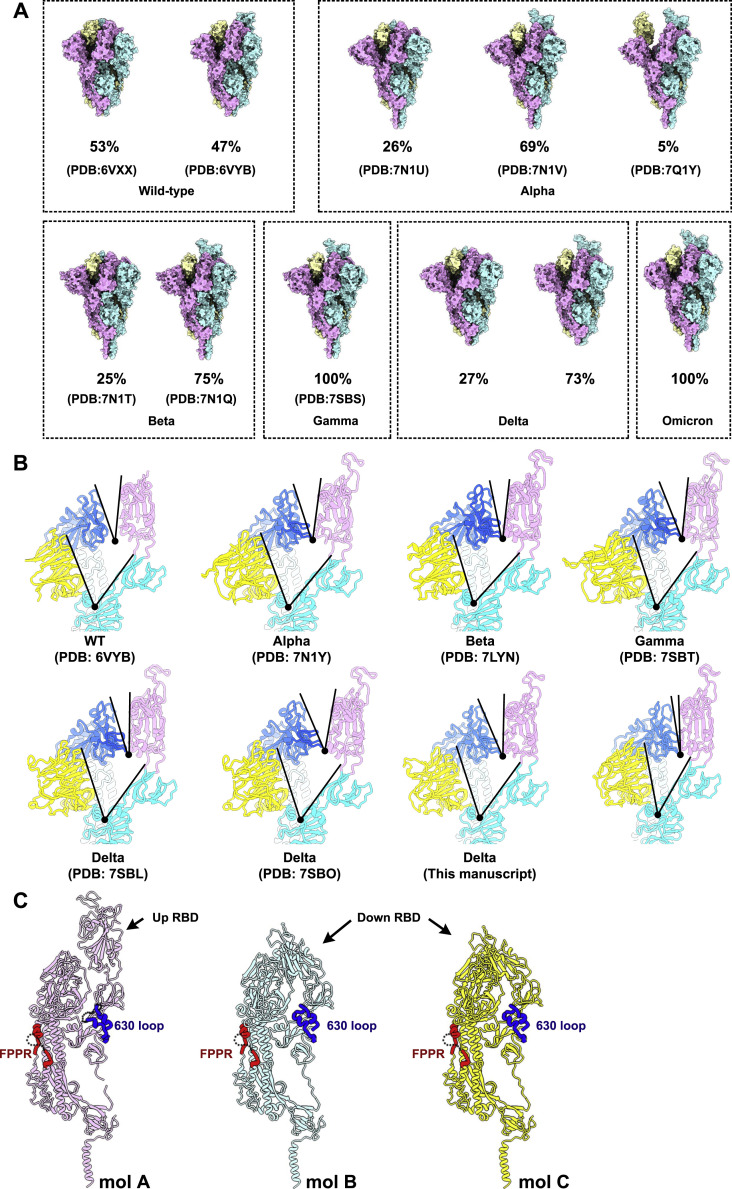
Figure S3.
Conformational comparation of WT and VOC S trimers, related to Figure 2
(A) Surface presentations of WT S-trimer (Walls et al., 2020), Alpha S-trimer (Cai et al., 2021), Beta S-trimer (Cai et al., 2021), Gamma S-trimer (Zhang et al., 2021a), Delta S-trimer (Zhang et al., 2021a) and Omicron S-trimer were shown with each protomer in different colors (cyan, pink and yellow). The ratio of S-trimer with different conformations is shown in the figure. (B) Shown here are the focused views of interprotomer RBD-to-RBD contacts of WT (Walls et al., 2020), Alpha (Cai et al., 2021), Beta (Gobeil et al., 2021), Gamma (Zhang et al., 2021a), Delta (Zhang et al., 2021a) and Omicron S-trimers. All structures are displayed in the same orientation. Angles formed by up RBD and its adjacent down RBD (top), as well as NTD and its inner-protomeric SD1-SD2 axis (bottom) are marked out. (C) Three protomers (mol A, magenta; mol B, cyan and mol C, yellow) of Omicron S trimer are shown as ribbon in the same orientation. Structures of FPPR and 630 loop on each protomer are accentuated in bold red and bold blue, respectively.
Figure 2.
Structural features underpinning the up configuration
(A and B) Left: representation of Omicron (A) and Delta (B) S-trimer in a prefusion conformation with one protomer in “open” state. The “up-state” protomer is shown as ribbon with NTD, RBD, and S2 colored in yellow, magenta, and cyan, respectively; the neighboring two protomers in “down” state are shown as surface in blue and gray, respectively. Right: zoomed-in view of the interprotomer RBD-to-RBD contact outlined in black dotted line in (A and B) with angles formed by up RBD and an adjacent down RBD (top), as well as NTD and its inner-protomeric SD1-SD2 axis (bottom) marked out.
(C and E) Zoomed-in view of interaction details of two independent interfaces for Omicron (A).
(D and F) Zoomed-in view of interaction details of two independent interfaces for Delta (B).
The mutated residues are shown as sphere in red with symbols, and the residues involved in the interactions are shown as sticks. The hydron bonds are shown as yellow dashed lines (C and D) and hydrophobic network is highlighted in gray (E and F). See also Figures S1 and S3.
Improved stability and decreased viral fusion ability
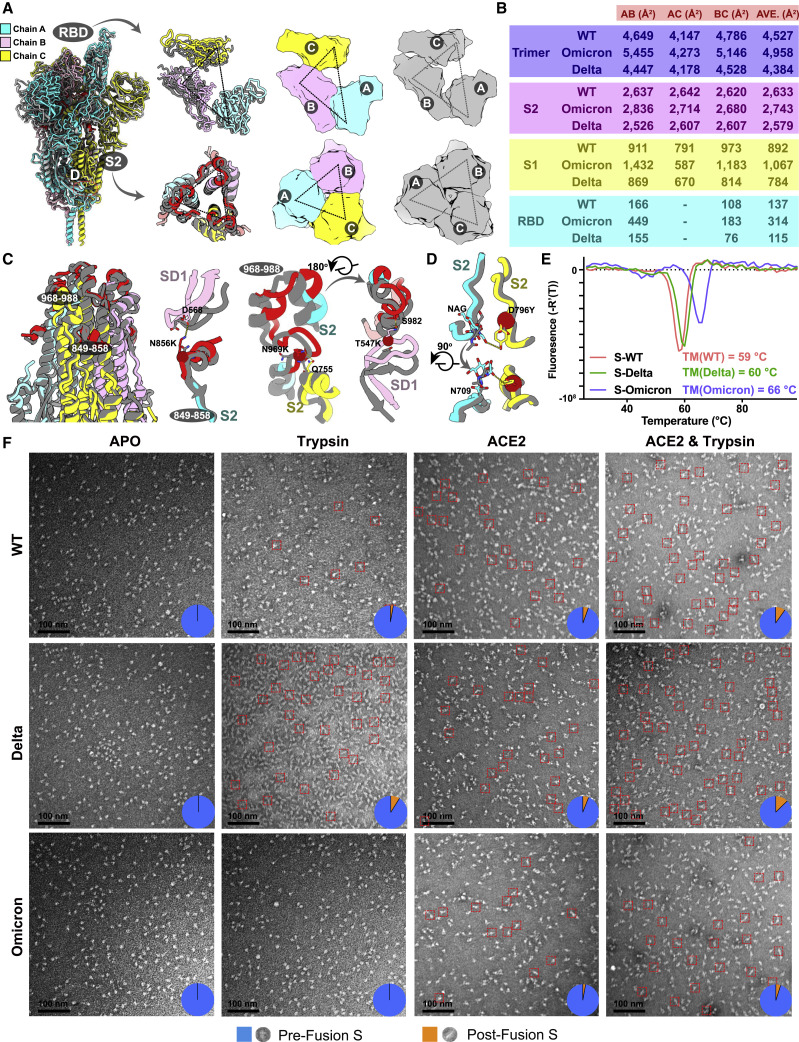
Viral stability, particularly that of surface proteins like S-trimer, closely correlates with its entry efficiency, viral transmission, adaptability, and immunogenicity (Laha et al., 2020). In spite of adopting an up configuration, the Omicron S-trimer exhibits a much more compact architecture in the regions formed by three copies of S2 and RBD, representing a highly stabilized conformation (Figure 3 A). Consequently, the Omicron S-trimer possesses substantially increased intersubunit (mol A–mol B) interactions of up to 5,455 Å2, when compared to its counterparts from Delta with buried areas of 4,447 Å2 (Figure 3B). Similarly, improved intersubunit contacts from either S2-S2 or S1-S1 are also clearly observed, thereby conferring enhanced stability for the Omicron S-trimer in prefusion conformation (Figure 3B). To further investigate the molecular basis for observed tight domain organization in Omicron, superimposition of S2 from Delta and Omicron revealed two local conformational shifts at intersubunit interfaces (residues 849–858 and 968–988) (Figure 3C). In the broader context of the S-trimer, the N856K, N969K, and T547K (in SD1) changes from a short side chain to a long one building up three hydrogen bonds with D658 (in SD1), Q755, and S982 from neighboring subunits, pulling the three subunits closer (Figure 3C). Additionally, the substitution of D796Y can stabilize the sugar at residue N709 from its adjacent subunit through forming a hydrogen bond (Figure 3D). In line with structural observations, thermal stability assays verified that the Omicron S-trimer was more stable than those from WT and Delta (Figure 3E).
Figure 3.
Improved stability and decreased fusogenicity
(A) Superimposition of the structure of the Omicron S-trimer (color scheme is same as in Figure 1B) onto the structure of the Delta S-trimer state (gray). Right: top views of the RBD (top) and S2 (bottom) show the intersubunit contacts of the Omicron and Delta S-trimers.
(B) Buried surface areas between two neighboring protomers or S2- and S1-subunits or RBD domains. A, protomer A; B, protomer B; C, protomer C; as labeled in (A).
(C) Snapshot of the S2 subunit from (A). Two regions with substantially conformational alterations in Omicron are highlighted in red and shown separately. Residues involved in the formation of hydrogen bonds are shown as sticks. The key mutated residues are shown as sphere in red.
(D) The substitution of D796Y in Omicron can stabilize a sugar at N709 from its neighboring protomer.
(E) Thermal stability of WT (red), Delta (green), and Omicron (blue) S-trimer.
(F) Fusogenicity evaluation of WT, Delta, and Omicron S-trimer triggered by treatment of trpsin or trpsin and ACE2. Negative staining images of WT (top), Delta (middle), and Omicron (bottom) S-trimer after incubation with trypsin, ACE2, and both trypsin and ACE2 at conditions described in the method.
In general, improved stability, to some extent, might increase the persistence of the Omicron variant in the exposed environments, posing a higher risk of transmission among household contacts when compared to the Delta variant. Theoretically, improved stability can facilitate viral attachment to the host cells via increasing receptor recognition efficiency; however, viral membrane fusion may be compromised. To test this, analysis of trypsin- or trypsin- and ACE2-mediated S fusogenic conformational rearrangements (Walls et al., 2019) evaluated by negatively stained EM was performed (Figure 3F). Analysis of the negatively stained sample showed that the Omicron S-trimer largely remained in the prefusion conformation and was relatively stable in presence of trypsin and ACE2 at room temperature (Figure 3F). While the same treatment led to formation of more postfusion rosettes for the Delta S-trimer, suggesting that the viral fusion efficiency for the Omicron S-trimer is possibly declined (Figure 3F). These results also largely match the experimental observations of a significant reduction in syncitia formation during the Omicron infection (Meng et al., 2021), despite the presence of the P681H substitution, a similar change of P681R, which is known to favor S1/S2 cleavage and enhance viral fusogenicity in Delta (Saito et al., 2021).
Structural basis for altered antigenic characteristics
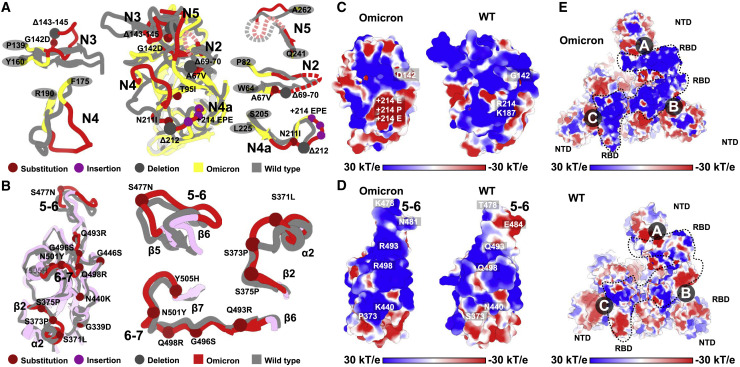
The overall architectures of the Omicron S-trimer resemble those of WT and other VOCs in the corresponding conformation (Figures 2A and S3). However, the Omicron variant can escape the majority of existing SARS-CoV-2 neutralizing antibodies (NAbs) and humoral immune responses elicited by natural infection or vaccinations, indicative of a considerably altered antigenic structure. The RBD and NTD are two main targets of neutralization in SARS-CoV-2 and other coronaviruses (Cerutti et al., 2021; Dejnirattisai et al., 2021; McCallum et al., 2021; Zhang et al., 2021b). Superimpositions of RBD and NTD of the WT and Omicron S clearly revealed local conformational alterations in antigenic loops (Figures 4A and 4B). Among these, the most striking differences are in the NTD, which contains three small deletions, one 3-residue insertion and four substitutions (Figure 1A). The mutation A67V together with Δ69-70 triggers the conformational changes in N2 loop. Similarly, N211I and Δ212, as well as ins214EPE, alter the configuration of the loop (defined as N4a) encompassing residues 209–216 located adjacent to N4 loop (Figures 4A and S4 ). Remarkably, the substitution of G142D as well as Δ143-145 leads to a reconfiguration of N3 loop from a hair-pin fold to a loose loop. This reorganization, together with the N4a loop, further mediates conformational changes in its neighboring loops N4 and N5 (Figures 4A and S4). Of note, an antigenic supersite comprising N3 and N5 is completely altered in Omicron (Cerutti et al., 2021; McCallum et al., 2021; Zhang et al., 2021b), structurally explaining the observation that nearly all NTD-targeting NAbs have lost their activities against Omicron. Two major antigenic sites consisting of the 5-6 loop (residues 475–485) and 6-7 loop (residues 492–505) are widely targeted by very potent NAbs via blocking ACE2 binding (Figures 4B and S4) but can be substantially impaired by T478K, E484K, and N501Y substitutions observed in several VOCs (Dejnirattisai et al., 2021; Wang et al., 2021b). Concernedly, newly occurred mutations, S477N, Q493R, G496S, Q498R, and Y505H, together with three already existing mutations, further alter the antigenic characteristics, leading to a striking evasion of the antibody. Substitution like S371L, S373P, and S375F also cause a clear conformational change in the loop connecting α2 and β2, another antigenic site that is generally conserved across sarbecoviruses (Lv et al., 2020; Martinez et al., 2021) (Figures 4B and S4). In addition to local conformational alterations, the Omicron RBD and NTD exhibit an increased positively charged and negatively charged surface, respectively, when compared to those of WT (Figures 4C–4E).
Figure 4.
Structural basis for altered antigenic characteristics
(A and B) Superimpositions of the structures of the Omicron NTD (A) and RBD (B) onto WT NTD and RBD. Structures here are shown as ribbon with substitutions, insertions, and deletions depicted as spheres in brown, purple, and dark, respectively. Color scheme for Omicron is same as in Figure 1B; the NTD and RBD from WT are colored in gray. Loops of NTD (A) and RBD (B) in Omicron with significantly conformational changes relative to WT were colored in red and zoomed in.
(C and D) Electrostatic surface of Omicron (left) and WT (right) NTD (C) and RBD (D). Mutations resulting in dramatic electrostatic changes are labeled.
(E) Electrostatic surface of the Omicron (top) and WT (bottom) S-trimer viewed from a top-down perspective. The distinct differences in charge distribution are marked out in dotted lines.
See also Figure S4.
Figure S4.
Secondary structure of the Omicron RBD and NTD, related to Figure 4
Primary sequence and secondary structural elements of the Omicron RBD and NTD, the five-pointed star represents the inserted residues, the blue short line represents the deleted residues and the red box represents the mutated residues.
Structural dissection of the evasion of neutralization of five classes of antibodies
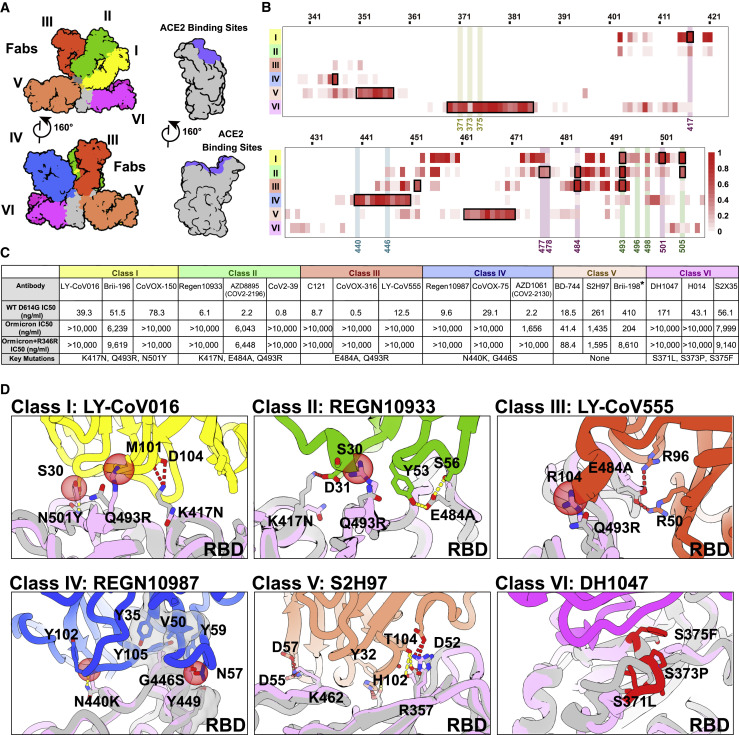
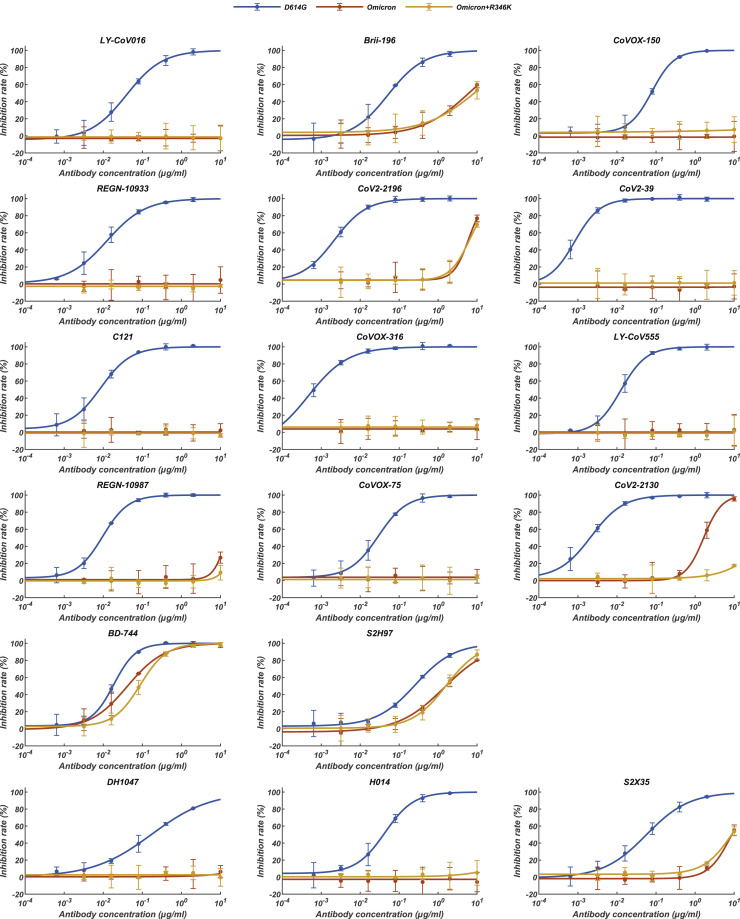
RBD-targeting NAbs can generally be categorized into six classes (from I to VI) based on cluster analysis on epitope from 280 available RBD-NAb complex structures (Figure 5 A) (Wang et al., 2021a) that are also related to the four/five groups on the basis of competition with the hACE2 for binding to S (Barnes et al., 2020; Hastie et al., 2021; Yuan et al., 2021). We constructed an antigenic heatmap for RBD using the 280 NAb complex structures to estimate in vivo antibody recognition frequencies on the RBD (Figure 5B). The first three classes of antibodies targeting the RBM with partially overlapped epitopes are highly potent by way of blocking the interactions between SARS-CoV-2 and ACE2. Class I antibodies, primarily derived from IGHV3-53/IGHV3-66 with short HCDR3s, recognize only the up RBD and make significant contacts with K417, Q493, N501, and Y505 (Figure 5B). Class III antibodies bound to RBD either in up or down configuration extensively associate with E484, Q493, and partially with L452 (Figure 5B). Class II antibodies bind the patch between sites I and III, frequently interacting with S477, T478, E484, Q493, and Y505 (Figure 5B). Class IV antibodies attach to the right shoulder of RBD with relatively condensed epitopes comprising residues 440–450 (Figure 5B). Class V and VI antibodies, generally cross reactive to sarbecoviruses, target two cryptic epitopes, consisting of residues 351–357, 462–471, and 368–385, respectively, which are only accessible when at least one RBD is in the open state (Figure 5B).
Figure 5.
Structural dissection of the evasion of neutralization of antibodies
(A) Surface representation of RBD in complex with six types of NAbs. RBD is colored in gray and the six representative Fab fragments belonging to six classes are colored as follows: class I, yellow; class II, green; class III, red; class IV, blue; class V, brown; class VI, magenta.
(B) Heatmap represents the frequency of RBD residues recognized by NAbs from six classes. Mutations present in Omicron RBD are marked out and highlighted.
(C) Summary of representative NAbs from each of six classes. Neutralizing titer (IC50) of each NAb against WT and Omicron is enumerated. The key residues involved in immune evasion for each class are also listed below. The IC50 data of Brii-198 was marked with “∗,” which represents the data referred from the available publication (Liu et al., 2021b). All neutralization assays were performed in biological triplicates.
(D) Binding interface between RBD and representative NAbs. All structures are shown as ribbon with the key residues shown with sticks. The clashes between RBD and NAb are shown as red sphere; salt bridges and hydrogen bonds are presented as red dashed lines and yellow dashed lines, respectively. Fab fragments of LY-CoV016, REGN10933, LY-CoV555, REGN10987, S2H97, and DH1047, representatives of classes I, II, III, IV, V, and VI, respectively, are colored according to the class they belong to; WT RBD is colored in gray; Omicron RBD is colored in light purple.
See also Figure S5.
To gain information about the types of NAbs that could either completely lose or show a dramatic reduction in neutralizing activities against Omicron, we performed pseudovirus assays. The neutralization activity profile of 18 well-characterized antibodies from all six classes (Figure 5C), except for Brii-198, whose neutralizing datasets were published recently, was evaluated (Liu et al., 2021b). Among these, neutralization of five of the six classes of NAbs was strikingly escaped by Omicron (Figures 5C and S5 ). For Class I NAbs represented by LY-CoV016, substitutions of Q493R and N501Y with longer side chains induced steric clashes with Y102, M101 from HCDR3, and S30 from LCDR1, respectively; mutation K417N further broke the salt bridge with D104 from HCDR3, leading to inactivity in binding to Omicrom S (Figure 5D). Regarding Class II antibodies, e.g., REGN10933, changes of K417N and E484A disrupted hydrogen bonds established by D31 from LCDR1 and Y563, S56 from HCDR2, respectively; mutation Q493R also directly clashed with S30 from LCDR1, resulting in the antibody losing its capacity to bind Omicron S (Figure 5D). Similarly, the mutation Q493R caused severe clashes with R104 from HCDR3, and E484A abolished charge interactions with R50 from HCDR2 and R96 from LCDR3, resulting in the inability of Class III antibodies exemplified by LY-CoV555 to bind to the Omicron S (Figure 5D). In addition to existing mutations like K417N, E484A, and N501Y, the Q493R mutation, or a similar mutation, Q493K, have previously been detected through in vitro resistance mapping efforts or in immunocompromised hosts (Baum et al., 2020; Choi et al., 2020; Clark et al., 2021; Focosi et al., 2021; Nabel et al., 2021; Weisblum et al., 2020). The Q493R mutation acts as one extra key immune escape site in Omicron, which in conjunction with local conformational changes caused by other nearby mutations, leads to greater resistance to Class I–III antibodies via steric clashes. Disruption of the hydrophobic microenvironment constructed by interactions between V445, G447, and P499 from RBD and Y35; V50, Y59, and Y105 from HCDRs of REGN10987, a representative antibody from Class IV antibodies by the G446S substitution; and the moderate clash between mutation N440K with Y102 from HCDR3 reduces binding affinities of the antibodies to Omicron S dramatically (Cao et al., 2021) (Figure 5D). As mentioned above, substitutions of S371L, S373P, and S375F drove a distinct conformational shift, stabilizing two adjacent RBDs (one up and one down) (Figures 2E and 3B), which consequently altered the relatively conserved antigenic site, mainly targeted by Class VI antibodies (Figure 5D). Fortunately, epitopes of Class V antibodies are mostly beyond the mutated patch, thereby less affected by Omicron, although most NAbs from this class are less potent. Given that an additional R346K mutation has been frequently detected in Omicron, accounting for ∼26% of the cases, we also constructed another pseudovirus (Omicron+R346K) containing this mutation for assessing neutralization by these representative NAbs (Figure 5C). The overall results resembled those observed for Omicron, with the exception that the neutralizing activities of AZD1061 (CoV2-2130) were further diminished (Figures 5C and S5). In summary, we dissected the evasion of neutralization of the five classes of NAbs by Omicron and identified two unidentified immune escape sites: G446S and S371L/S373P/S375F, which together with existing substitutions confer greater resistance to five of the six classes of RBD-antibodies.
Figure S5.
Pseudovirus neutralization of representative NAb drugs against SARS-CoV-2 variants, related to Figure 5
Pseudovirus (VSV-based) assays were performed using Huh-7 cells. Data are collected from three biological replicates and represented as mean ± s.d.
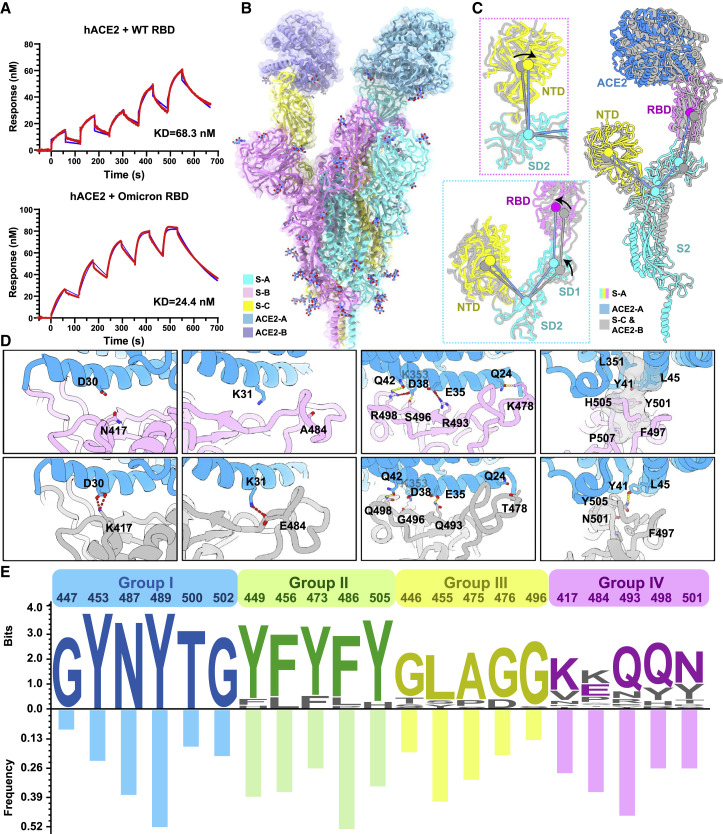
Molecular determinants for enhanced binding affinity to human ACE2
Unexpectedly, nine out of fifteen substitutions in the Omicron RBD are located on the human ACE2 binding interface, which considerably affects receptor recognition. We measured the binding affinities of WT and Omicron RBD to ACE2 by surface plasmon resonance (SPR), in which the CM5 biosensor was labeled with amine-coupling ACE2 and flooded with the Omicron or WT RBD in the flow-through. (Figure 6 A). The Omicron RBD showed a 2.8-fold increased binding affinity to human ACE2 compared to the WT RBD (Figure 6A). To further unveil molecular details of the binding, we determined cryo-EM structures of the Omicron S-trimer in complex with human ACE2 at 3.5 Å (Figures 6B and S1 and Tables S1 and S2). The structure showed two copies of ACE2 bound to two RBDs (mol A and mol C) in the up conformation. The stabilized up S-monomer showed no notable conformational alterations upon ACE2 binding, but the other up S-monomer had a wider angle between NTD, SD2, SD1, and RBD that was triggered by ACE2 (Figure 6C), suggesting that the stabilized up configuration is tolerant to the interference by ACE2 binding, probably antibody binding as well. Local refinement of the RBD-ACE2 region resulted in a reliable density map for analysis of the mode of interaction between Omicron S and ACE2 (Figure S1). Structural comparisons of the key interactions at the interface between the Omicron/WT and ACE2 revealed that new substitutions of T478K, Q493R, G496S, and Q498R strengthened the binding of Omicron to ACE2 by establishing hydrogen bonds or salt bridges with Q24, E35, K353, and D38, respectively (Figure 6D). Together with the N501Y, a mutation known to improve the binding affinity by 6-fold (Zhou et al., 2021), these mutations facilitate the binding of Omicron to ACE2. Meanwhile, mutations K417N and E484A decreased binding affinity to ACE2 via breaking two hydrogen bonds in Omicron. This reduction in affinity is offset by the interactions forged by other mutations. Overall, the Omicron variant possesses improved binding to ACE2.
Figure 6.
Molecular determinants for enhanced binding affinity to human ACE2
(A) Binding affinity of hACE2 with WT RBD (top) or Omicron RBD (bottom) measured by SPR.
(B) Overall structure of the Omicron S-trimer in complex with hACE2. Three copies of S monomer were colored in yellow, cyan, and magenta, respectively; two hACE2 molecules bound to RBD were colored in purple and blue, respectively.
(C) Superimposition of two S monomer-hACE2 molecules with a focused alignment on S2 subunit. Color scheme for the S monomer in a stabilized up conformation is same as in Figure 1B, and the other S monomer is colored in gray. Insets represent the structural shifts of NTD and SD2 (left-top) and altered angles (left-bottom) formed by NTD, SD2, SD1, and RBD that were triggered by ACE2 binding.
(D) Changes at the interfaces between WT RBD (PDB: 6M0J) (Lan et al., 2020) and Omicron RBD with hACE2. hACE2, WT RBD, and Omicron RBD are colored in blue, gray, and magenta, respectively; key mutated residues were shown as sticks with the abolished (left four) and newly established (right four) bonds denoted in dashed lines. Salt bridges, red; hydron bonds, yellow. Hydrophobic network is highlighted in gray.
(E) Analysis of sequence conservation and antigenicity frequency on residues involved in ACE2 binding. The logo plot represents the conservation of these residues from 25 sarbecoviruses, and the histogram shows the antigenic frequency of the same residues targeted by NAbs. Logos and bars of four types of residues are colored with blue (identical residues), green (homologous residues), yellow (conditionally altered residues), and pink (highly diverse residues), respectively. Amino acid sequences of these residues involved in hACE2 binding are from WT SARS-CoV-2, 17 SARS-CoV-2 variants (Alpha, Beta, Gamma, Delta, Lambda, Mu, Delta plus, Omicron, Eta, Lota, Kappa, Theta, Iota, B.1.1.318, B.1.620, and C.1.2, C.363), SARS-CoV-1, pangolin coronavirus (GX/P2V/2017 and GD/1/2019), bat coronavirus (WIV, RaTG13 and LYRa11), and civet coronavirus of 007/2004.
See also Figures S1 and S6.
Discussion
Viruses latch on to receptors using conserved residues. Many members of the β-Coronavirus lineage B (termed sarbecoviruses), including SARS-CoV and SARS-CoV-2 variants and civet-, bat-, and pangolin-derived sarbecoviruses, can utilize human ACE2 to enter host cells (Liu et al., 2021a; Wrobel et al., 2021). Interestingly, SARS-CoV-2 variants and sarbecoviruses from pangolin have higher affinities for ACE2 than SARS-CoV, as do sarbecoviruses originating from civets and bats, which might be explained by the affinity-enhancing mutations present in these viruses (Liu et al., 2021a; Starr et al., 2020). An analysis of the conservation of protein sequence around the receptor binding sites of 25 reported sarbecovirus members relying on human ACE2 for cellular entry reveals that 11 residues of the virus out of a total of 21 residues that engage with the receptor are highly conserved (Figure 6E). The amino acids of the virus involved in binding ACE2 can be categorized into four groups based on their conservation and essential roles in binding: (1) group I includes six identical residues such as G447, Y453, N487, Y489, T500, and G502; (2) group II consists of five homologous residues like Y449/F/H, F456/L, Y473/F, F486/L, and Y505H; (3) group III has five conditionally altered residues like G446/S/T, L455/S/Y, A475/P/S, G476/D, and G496/S; and lastly, (4) group IV includes five highly diverse residues, K417/V/N/R/T, E484/K/P/Q/V/A, Q493/N/E/R/Y, Q498/Y/H/R, and N501/Y/T/D/S (Figure 6E). Mutations in the first two groups of amino acids are strictly constrained, presumably because they act as molecular determinants in either retaining basic affinity for ACE2 or ensuring proper protein folding, while substitutions in the last two groups of amino acids are, to some extent, tolerated or even enhance ACE2 binding and are frequently observed in various VOCs (Figure 6E). Of note, amino acids belonging to the group IV that dramatically affect affinity include residues at positions 417 (e.g., K417), 484 (e.g., K484), 493 (e.g., R493), 498 (e.g., R498), and 501 (e.g., Y501). These amino acids are also known to vary across sarbecoviruses. Furthermore, consensus mutations, including G446T/S, F456L, Y473F, and G476D are observed in low-affinity group (SARS-CoV and civet and bat sarbecoviruses), indicative of a concerted role for these mutations in regulating affinity for ACE2 (Figure S6 ). In addition, the sequence and structural analysis revealed substantial plasticity in a complicated network formed by multiple substitutions, as some mutations probably increase polar contacts while others may impair hydrophobic interactions. Our results are largely supported by deep mutational scanning assays, unveiling the details of molecular interactions involved in ACE2 binding (Figure S6) (Starr et al., 2020).
Figure S6.
Analysis of sequence conservation and antigenicity frequency on residues of RBD involved in ACE2 binding, related to Figure 6
(A) Multiple sequence alignment of residues involved in ACE2 binding from 25 representative sarbecovirus members. (B) The height of an amino acid code indicates its preference on a given site of RBM with respect to ACE2 binding (top) and RBD expression (bottom), as is shown previously. Residues are classified into four groups: the identical residues, homologous residues, conditionally altered residues and diverse residues, which are colored in blue, green, orange and red, respectively.
In addition to the pursuit of greater transmissibility and infectivity, viral evolution is primarily driven by immune escape as well. Our immunogenic and mutational heatmaps for RBD using the 280 NAb complex structures to estimate in vivo NAb-targeting frequencies and viral mutation frequencies revealed an overall positive correlation between hot immunogenic sites and areas with high mutation frequencies, but with an exception for some sites involved in ACE2 binding (Figure 6E). Only three of the top ten hottest immunogenic residues contained substitutions (Q493R, E484K/A, and Y505H) in circulating SARS-CoV-2 variants, among which Q493R and Y505H are newly acquired in Omicron (Figure 6E). Particularly, hot immunogenic residues, such as F486, Y489, Y449, N487, and F456, are mostly from the groups I and II type of residues within ACE2 binding sites, and mutations at these positions have not yet been described among circulating variants (Figure 6E). In other words, substitutions of these hot immunogenic sites are best avoided because they probably sacrifice binding activity to ACE2, which could prove to be fatal to the virus. These correlates clearly explain why mutations at these positions would not be selected. In line with our findings, several ACE2-mimic antibodies have already been shown to broadly cross-neutralize sarbecoviruses (Cameroni et al., 2021; Park et al., 2021). The conserved and structurally constrained region for ACE2 recognition revealed in this study would rationalize the development of broad-spectrum vaccine and antibody therapeutics.
Limitations of the study
Cryo-EM structures of the Omicron S-trimer solved at serological and endosomal pH in this study reveal amino acid substitutions forging interactions that stably maintain an active conformation for receptor recognition. Future effort in determining the in situ structure of the Omicron virions could provide more insights into functional significance into these structural features. Moreover, the increased thermal stability of the Omicron S-trimer correlates with decreased fusogenicity, consistent with structural observations of more compact domain organization and newly established intersubunit interactions by amino acid substitutions. It will be important to further test the improved viral attachment and impaired viral fusion ability based on biochemical analysis with virological investigation approaches.
STAR★Methods
Key resources table
| REAGENT or RESOURCES | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| DH5α Chemically Competent Cell | Invitrogen | Cat#12034013 |
| G∗ΔG-VSV | Kerafast | Cat#EH1020-PM |
| Cell lines | ||
| HEK293F cells | Thermo Fisher | Cat # 11625019 |
| Huh7 cells | NIFDC | N/A |
| Chemicals, peptides, and recombinantproteins | ||
| RPMI 1640 Medium | Thermo Fisher | Cat # 12633012 |
| Trypsin | Thermo Fisher | Lot# TG269188 |
| DMEM | Thermo Fisher | Cat#11965092 |
| 1x PBS, pH 7.4 | Thermo Fisher | Cat # 10010031 |
| Fetal bovine serum (FBS) | Thermo Fisher | Cat # 10099 |
| D-luciferin | Thermo Fisher | Cat # L2916 |
| Uranyl Formate | Electron Microscopy China | Cat # GZ02625 |
| Critical commercial reagents | ||
| Superose 6 Increase 10/300 GL | GE healthcare | Cat # 29091596 |
| SYPRO protein gel stain | Thermo Fisher | Cat # S6650 |
| NiNTA Biosensors | Octet | Cat # 18-5102 |
| AR2G Biosensors | Octet | Cat # 18-5095 |
| CM5 Biosensor | Cytiva | Lot # 10310113 |
| Recombinant DNA | ||
| SARS-CoV-2 S gene, residues 1-1208, pcDNA | GenScript ProBio | GenBank: MN908947 |
| SARS-CoV-2 Delta S gene, residues 1-1208, 6P and T19R, G142D, EF156-157del, R158G, L452R, T478K, D614G, P681R, D950N, T4 fibritin trimerization motif,6xHis, pcDNA | This manuscript | N/A |
| SARS-CoV-2 Omicron S gene, residues 1-1208, 6P and A67V, Δ69-70, T95I, G142D, Δ143-145, Δ211, L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493K, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F, T4 fibritin trimerization motif,6xHis, pcDNA | This manuscript | N/A |
| Deposited data | ||
| Local map of Omicron S-trimer in complex with ACE2 | This manuscript | PDB ID 7WGC, EMD-32483 |
| Omicron S-trimer in complex with ACE2 | This manuscript | PDB ID 7WGB, EMD-32482 |
| Delta S-trimer (1 RBD Up) | This manuscript | PDB ID 7WG9, EMD-32481 |
| Delta S-trimer (3 RBD Down) | This manuscript | PDB ID 7WG8, EMD-32480 |
| Omicron S-trimer at pH 5.5 | This manuscript | PDB ID 7WG7, EMD-32479 |
| Omicron S-trimer at pH 7.5 | This manuscript | PDB ID 7WG6, EMD-32478 |
| Software | ||
| igraph (1.2.5) | N/A | https://cran.r-project.org/web/packages/igraph/index.html |
| Gctf program (v1.06) | N/A | https://www2.mrc-lmb.cam.ac.uk/download/gctf/ |
| RELION (v3.07) | Zivanov et al., 2018 | https://www2.mrc-lmb.cam.ac.uk/relion |
| UCSF Chimera | N/A | https://www.cgl.ucsf.edu/chimera |
| UCSF ChimeraX | N/A | https://www.rbvi.ucsf.edu/chimerax/ |
| PHENIX | N/A | https://www.phenix-online.org |
| Coot | N/A | https://www2.mrc-lmb.cam.ac.uk/Personal/pemsley/coot |
| cryoSPARC 3.2.4 | N/A | https://cryosparc.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Xiangxi Wang (xiangxi@ibp.ac.cn).
Materials availability
All reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Cell lines
HEK293T and Huh-7 cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS). The cultures were maintained at 37°C in an incubator supplied with 8% CO2.
SARS-CoV-2 pseudovirus
The SARS-CoV-2 pseudovirus was constructed as previously described using VSV pseudotyped virus (G∗ΔG-VSV) (Nie et al., 2020). SARS-CoV-2 pseudovirus carrying D614G, Omicron, and Omicron (R346K) mutations was constructed and used, as described previously (Cao et al., 2021). Omicron pseudovirus contains the following mutations: A67V, H69del, V70del, T95I, G142D, V143del, Y144del, Y145del, N211del, L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F.
Method details
Protein expression and purification
The plasmids encoding the full-length spike (S) protein (residues 1-1028) of wild-type SARS-CoV-2 (GenBank: MN908947) was used as the template for the construction of the S gene of Delta (T19R, G142D, EF156-157del, R158G, L452R, T478K, D614G, P681R, D950N) and Omicron (A67V, Δ69-70, T95I, G142D, Δ143-145, Δ211, L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493K, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F) by overlapping PCR. All the S gene constructs have six proline substitutions at residues 817, 892, 899, 942, 986, and 987 and two alanine substitutions at residues 683 and 685 and a C-terminal T4 fibritin foldon domain to facilitate the protein expression and stabilization of the trimer conformation (ACROBiosystems, Cat No. SPN-C52Hz). All the constructs described above were attached with a C-terminal six-His for protein purification. To obtain these proteins, the plasmids constructed above were transiently transfected into HEK293 F cells grown in suspension at 37°C in a rotating, humidified incubator supplied with 8% CO2 and maintained at 130 rpm. After incubation for 72 h, the supernatant was harvested, concentrated, and exchanged into the binding buffer by tangential flow filtration cassette. The protein of interest was separated by affinity chromatography using resin attached with Ni-NTA and subjected to additional purification by size exclusion chromatography using a Superose 6 10/300 column (GE Healthcare) in 20 mM Tris pH 7.5 and 200 mM NaCl.
S trimer thermal stability assay
PaSTRy was performed with SYPRO Orange (Invitrogen, Carlsbad, USA) as fluorescent probes to detect the exposed hydrophobic residues by an MX3005 qPCR instrument (Agilent, Santa Clara, USA). Here, we set up pH = 7.4 25 μL reaction system which contained 10 μg of target protein, i.e., S trimer of WT, Delta and Omicron, 1000x SYPRO Orange, and ramped up the temperature from 25°C to 99°C. Fluorescence was recorded in triplicate at an interval of 1°C.
Bio-layer interferometry
Bio-layer interferometry (BLI) experiments were run on an Octet Red 96e machine (Fortebio). To measure the binding affinities of RBD from different variants with ACE2, His-tagged WT RBD, Delta RBD or Omicron RBD were immobilized onto NTA biosensors (Fortebio) and three-fold serial dilutions of ACE2 were used as analytes. Also, the ACE2 was immobilized onto AR2G biosensors (Fortebio), as RBD of WT, Delta and Omicron as the analytes. Data were then recorded using software Data Acquisition 11.1 (Fortebio) and analyzed using software Data Analysis HT 11.1 (Fortebio) with a 1: 1 fitting model. After the Omicron RBD was loaded onto the NTA biosensor, the human ACE2 (or orthologs of other species) containing buffer solution was passed over the bound RBD. Repeated experiments yielded substantially lower affinities, some even returning negative results, whereas the WT RBD seemed to yield expected binding affinities with ACE2, indicative of the unsuitability of the use of Omicron RBD as immobilization phase. In addition, nonspecific binding of the Omicron RBD to AR2G biosensor was clearly detected when the amine-coupling ACE2 was immobilized onto AR2G biosensor in BLI assay, which also led to unreliable results. Thus, we chose SPR assay for binding affinity measurement.
Surface plasmon resonance
ACE2 was immobilized onto CM5 sensor chips using a Biacore T100 (GE Healthcare). Serial dilutions of purified WT and Omicron RBD were injected, ranging in concentrations from 250 to 15.6 nM. The resulting data were fitted to a 1:1 binding model using Biacore Evaluation Software (GE Healthcare).
Pseudovirus neutralization assay
The pseudovirus neutralization assays were performed using Huh-7 cell lines, as described previously (Cao et al., 2020). Briefly, various concentrations of antibodies (5-fold serial dilution using DMEM) were mixed with the same volume of SARS-CoV-2 pseudovirus in a 96 well-plate. The mixture was incubated for 1 h at 37°C and supplied with 5% CO2. Next, pre-mixed Huh-7 cells were added to all wells, incubated for 24 h at 37°C supplied with 5% CO2. After incubation, the supernatants were removed, and D-luciferin reagents (Invitrogen) were added to each well. Luciferase activity was measured by using a microplate spectrophotometer (PerkinElmer EnSight). The inhibition rate is calculated by comparing the OD value to the negative and positive control wells. IC50 was determined by a four-parameter logistic regression using GraphPad Prism 9.0.1 (GraphPad Software Inc.).
Conformational change analysis using negative staining EM
The WT, Delta, and Omicron S-trimer at 1 mg/mL were incubated with hACE2 at a ratio of 9 hACE2 molecules per S trimer overnight on ice. Samples were diluted to a concentration of 0.03 mg/mL before being imaged and negatively stained for EM. For trypsin treatment, samples were added with 1.6 μg/mL trypsin at room temperature for 15 min and used for imaging by negative staining EM.
Cryo-EM sample preparation, data collection
The Delta S-trimer, Omicron S-trimer (at neutral or acidic pH), or Omicron S trimer (at neutral pH) mixed with hACE2 was dropped onto the pre-glow-discharged holey carbon-coated gold grid (C-flat, 300-mesh, 1.2/1.3, Protochips In.), blotted for 6 s with no force in 100% relative humidity and immediately plunged into the liquid ethane using Vitrobot (FEI). Cryo-EM datasets of these complexes were collected at 300 kV with an FEI Titan Krios microscope (FEI). Movies (32 frames, each 0.2 s, total dose of 60 e− Å-2) were recorded using a K3 Summit direct detector with a defocus range between 1.5- 2.5 μm. Automated single particle data acquisition was carried out by SerialEM, with a calibrated magnification of 22,500, yielding a final pixel size of 1.07 Å.
Cryo-EM data processing
A total of 3,106, 1,777, 6,050, and 5,030 micrographs of Delta S-trimer, Omicron S-trimer under neutral and acidic pH conditions, and Omicron S-trimer mixed with hACE2 at pH 7.5 were recorded. All the micrographs were processed with MotionCor2 in Relion3.0. The CTF value of each micrograph was estimated by Gctf. Then 1,317,048, 830,943, 1,789,492 and 916,297 particles of Delta S-trimer, Omicron S-trimer under neutral and acidic pH conditions and neutral Omicron S-trimer mixed with hACE2 were picked and extracted by Relion3.0. Apo S and S-trimer-hACE2 complex were extracted with a 3202-pixel box and a 3602-pixel box, respectively. Reference-free 2D alignment by cryoSPARC (Punjani et al., 2017) was applied for all the particles. Based on the results of 2D alignment, 926,504 and 458,926 particles of Delta S-trimer and neutral Omicron S-trimer complexed with hACE2 were selected and applied for Heterogeneous Refinement by cryoSPARC. Then 309,137 and 479,164 particles of Omicron S-trimer under neutral and acidic pH were selected and applied for template-guided 3D-classification by Relion3.0 (Zivanov et al., 2018). The 3D classification, using SARS-CoV-2 structure (zero-up) (EMD-21452) as initial model was performed. The results yielded only one configuration with one RBD up, apart from the rubbish class (∼10% particles). To excluded the existence of other conformations of Omicron S-trimer, a second round of 3D expansion using structures with zero-up (EMD-21452), one-up (EMD-21457), two-up (EMD-24127) or three up (EMD-30333) as references was carried out and only one up configuration was observed. For all the classifications, no symmetry was imposed. When the potential conformation for each structure was produced, particles from each candidate model were selected and processed by non-uniform auto-refinement and postprocessing in cryoSPARC to generate the final cryo-EM maps for Delta S-trimer, Omicron S-trimer under neutral and acidic pH conditions and Omicron S-trimer-hACE2 complex at pH 7.5. To improve the resolution of the interface between RBD and hACE2, the local refinement (Wang et al., 2019; Yang et al., 2020) was used to obtain the final resolution of the focused interfaces, which contained the interfaces of RBD and hACE2 investigated here as described previously. The resolution was determined based on the gold-standard Fourier shell correlation (threshold = 0.143) and evaluated by ResMap (Kucukelbir et al., 2014). All dataset processing is shown in Figure. S2 and also summarized in Table S1.
Model fitting and refinement
The atom models of the complexes were generated by first fitting the chains of native apo SARS-CoV-2 S trimer (PDB number of 6VYB) (Walls et al., 2020) and ACE2 (PDB number of 6M0J) (Lan et al., 2020) into the obtained cryo-EM densities by Chimera. Then the structure was manually adjusted and corrected according to the protein sequences and cryo-EM densities in Coot, and finally, real-space refinement was performed by Phenix. Details of the refinement statistics of the complexes are summarized in Table S1.
Quantification and statistical analysis
Bio-layer interferometry assays, as well as surface plasmon resonance assays, were performed in triplicates. Neutralization assays were performed in biological triplicates.
Acknowledgments
We thank Dr. Xiaojun Huang, Dr. Xujing Li, and Dr. Lihong Chen for cryo-EM data collection at the Center for Biological Imaging (CBI) in the Institution of Biophysics, CAS. We also thank Dr. Yuanyuan Chen, Zhenwei Yang, and Bingxue Zhou for technical support on Bio-layer interferometry (BLI) and surface plasmon resonance (SPR). Work was supported by the Strategic Priority Research Program (XDB29010000 and XDB37030000), CAS (YSBR-010), National Key Research and Development Program (2020YFA0707500 and 2018YFA0900801), Beijing Municipal Science and Technology Project (Z201100005420017), and Ministry of Science and Technology of China (EKPG21-09 and CPL-1233). X.W. was supported by Ten Thousand Talent Program and the NSFS Innovative Research Group (no. 81921005). K.W. and N.W. were supported by the Special Research Assistant Project of the Chinese Academy of Medical Sciences.
Author contributions
X.W., Y.C., and L.C. designed the study; Z.C., P.L., N.W., L.W., K.F., Q.Z., K.W., R.C., R.F., Z.J., M.Y., G.X., B.Z., W.F., T.C., L.F., Y.W., X.R., and P.Y performed experiments; P.L., N.W., L.W., and L.C. prepared the cryo-EM samples and solved the structures; all authors analyzed data; X.W., Y.C., and L.C. wrote the manuscript with input from all co-authors.
Declaration of interests
X.S.X. and Y.C. are inventors on the patent application of DXP-604 and BD series antibodies. X.S.X. and Y.C. are founders of Singlomics Biopharmaceuticals Inc. Other authors declare no competing interests.
Published: January 25, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2022.01.019.
Supplemental information
Data and code availability
-
•
The cryo-EM maps have been deposited at the Electron Microscopy Data Bank (www.ebi.ac.uk/emdb) and are available under accession numbers: EMDB: 32478 (Omicron S-trimer at pH 7.5), EMDB: 32479 (Omicron S-trimer at pH 5.5), EMDB: 32480 (Delta S-trimer with 3 RBD close), EMDB: 32481 (Delta S-trimer with 1 RBD open), EMDB: 32482 (Omicron S-trimer in complex with hACE2), EMDB: 32483 (Local optimized reconstruction of Omicron S-trimer in complex with hACE2). Atomic models corresponding to EMDB: 32478, EMDB: 32479, EMDB: 32480, EMDB: 32481, EMDB: 32482, EMDB: 32483 have been deposited in the Protein Data Bank (www.rcsb.org) and are available under accession numbers, PDB: 7WG6, PDB: 7WG7, PDB: 7WG8, PDB: 7WG9, PDB: 7WGB, PDB: 7WGC, respectively.
-
•
This study did not generate custom computer code.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
References
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Lavine C.L., Rawson S., Peng H., Zhu H., Anand K., Tong P., Gautam A., et al. Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants. Science. 2021;373:642–648. doi: 10.1126/science.abi9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameroni E., Saliba C., Bowen J.E., Rosen L.E., Culap K., Pinto D., De Marco A., Zepeda S.K., di Iulio J., Zatta F. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2021 doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y.R., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R. B. 1.1. 529 escapes the majority of SARS-CoV-2 neutralizing antibodies of diverse epitopes. bioRxiv. 2021 doi: 10.1101/2021.12.07.470392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti G., Guo Y., Zhou T., Gorman J., Lee M., Rapp M., Reddem E.R., Yu J., Bahna F., Bimela J. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host & Microbe. 2021;29:819–833. e817. doi: 10.1016/j.chom.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., Solomon I.H., Kuo H.-H., Boucau J., Bowman K., et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. J. Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.A., Clark L.E., Pan J., Coscia A., McKay L.G., Shankar S., Johnson R.I., Brusic V., Choudhary M.C., Regan J. SARS-CoV-2 evolution in an immunocompromised host reveals shared neutralization escape mechanisms. Cell. 2021;184:2605–2617.e2618. doi: 10.1016/j.cell.2021.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., et al. CMMID COVID-19 Working Group. COVID-19 Genomics UK (COG-UK) Consortium Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372 doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Zhou D., Ginn H.M., Duyvesteyn H.M., Supasa P., Case J.B., Zhao Y., Walter T.S., Mentzer A.J., Liu C. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell. 2021;184:2183–2200.e2122. doi: 10.1016/j.cell.2021.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focosi D., Novazzi F., Genoni A., Dentali F., Gasperina D.D., Baj A., Maggi F. Emergence of SARS-COV-2 Spike Protein Escape Mutation Q493R after Treatment for COVID-19. Emerg. Infect. Dis. 2021;27:2728–2731. doi: 10.3201/eid2710.211538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobeil S.M., Janowska K., McDowell S., Mansouri K., Parks R., Stalls V., Kopp M.F., Manne K., Saunders K., Edwards R.J., et al. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation and antigenicity. Science. 2021 doi: 10.1126/science.abi6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie K.M., Li H., Bedinger D., Schendel S.L., Dennison S.M., Li K., Rayaprolu V., Yu X., Mann C., Zandonatti M., et al. CoVIC-DB team1 Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: A global consortium study. Science. 2021;374:472–478. doi: 10.1126/science.abh2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A., Sigworth F.J., Tagare H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Thambiraja T.S., Karuppanan K., Subramaniam G. Omicron and Delta Variant of SARS-CoV-2: A Comparative Computational Study of Spike Protein. Journal of medical virology. 2021 doi: 10.1002/jmv.27526. [DOI] [PubMed] [Google Scholar]
- Laha S., Chakraborty J., Das S., Manna S.K., Biswas S., Chatterjee R. Characterizations of SARS-CoV-2 mutational profile, spike protein stability and viral transmission. Infect. Genet. Evol. 2020;85:104445. doi: 10.1016/j.meegid.2020.104445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Liu K., Pan X., Li L., Yu F., Zheng A., Du P., Han P., Meng Y., Zhang Y., Wu L. Binding and molecular basis of the bat coronavirus RaTG13 virus to ACE2 in humans and other species. Cell. 2021;184:3438–3451.e3410. doi: 10.1016/j.cell.2021.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Iketani S., Guo Y., Chan J.F.-W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S. Striking Antibody Evasion Manifested by the Omicron Variant of SARS-CoV-2. Nature. 2021 doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- Lu L., Mok B.W.-Y., Chen L., Chan J.M.-C., Tsang O.T.-Y., Lam B.H.-S., Chuang V.W.-M., Chu A.W.-H., Chan W.-M., Ip J.D. Neutralization of SARS-CoV-2 Omicron variant by sera from BNT162b2 or Coronavac vaccine recipients. Clinical Infectious Diseases. 2021 doi: 10.1093/cid/ciab1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Z., Deng Y.-Q., Ye Q., Cao L., Sun C.-Y., Fan C., Huang W., Sun S., Sun Y., Zhu L., et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020;369:1505–1509. doi: 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D.R., Schäfer A., Gobeil S., Li D., De la Cruz G., Parks R., Lu X., Barr M., Stalls V., Janowska K. A broadly cross-reactive antibody neutralizes and protects against sarbecovirus challenge in mice. Science translational medicine. 2021:eabj7125. doi: 10.1126/scitranslmed.abj7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347.e2316. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng B., Ferreira I., Abdullahi A., Kemp S.A., Goonawardane N., Papa G., Fatihi S., Charles O., Collier D., Choi J. SARS-CoV-2 Omicron spike mediated immune escape, infectivity and cell-cell fusion. bioRxiv. 2021 doi: 10.1101/2021.12.17.473248. [DOI] [Google Scholar]
- Mlcochova P., Kemp S.A., Dhar M.S., Papa G., Meng B., Ferreira I.A.T.M., Datir R., Collier D.A., Albecka A., Singh S., et al. Indian SARS-CoV-2 Genomics Consortium (INSACOG) Genotype to Phenotype Japan (G2P-Japan) Consortium. CITIID-NIHR BioResource COVID-19 Collaboration SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599:114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel K.G., Clark S.A., Shankar S., Pan J., Clark L.E., Yang P., Coscia A., McKay L.G., Varnum H.H., Brusic V. Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain. Science. 2021:eabl6251. doi: 10.1126/science.abl6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M., et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020;9:680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.-J., De Marco A., Starr T.N., Liu Z., Pinto D., Walls A.C., Zatta F., Zepeda S.K., Bowen J., Sprouse K.S., et al. Antibody-mediated broad sarbecovirus neutralization through ACE2 molecular mimicry. Science. 2021 doi: 10.1126/science.abm8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Saito A., Irie T., Suzuki R., Maemura T., Nasser H., Uriu K., Kosugi Y., Shirakawa K., Sadamasu K., Kimura I., et al. Genotype to Phenotype Japan (G2P-Japan) Consortium Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature. 2021 doi: 10.1038/s41586-021-04266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310.e1220. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Xiong X., Park Y.-J., Tortorici M.A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026–1039.e1015. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e286. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Zhao D., Wang J., Zhang Y., Wang M., Gao Y., Li F., Wang J., Bu Z., Rao Z., Wang X. Architecture of African swine fever virus and implications for viral assembly. Science. 2019;366:640–644. doi: 10.1126/science.aaz1439. [DOI] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e899. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Cao Y., Zhou Y., Wu J., Jia Z., Hu Y., Yisimayi A., Fu W., Wang L., Liu P. A third dose of inactivated vaccine augments the potency, breadth, and duration of anamnestic responses against SARS-CoV-2. medRxiv. 2021 doi: 10.1101/2021.09.02.21261735. [DOI] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C., Muecksch F., Rutkowska M., Hoffmann H.-H., Michailidis E., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.-C., Au A.K.-W., Chen H., Yuen L.L.-H., Li X., Lung D.C., Chu A.W.-H., Ip J.D., Chan W.-M., Tsoi H.-W. The Lancet Regional Health–Western Pacific; 2021. Transmission of Omicron (B. 1.1. 529)-SARS-CoV-2 Variant of Concern in a designated quarantine hotel for travelers: a challenge of elimination strategy of COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel A.G., Benton D.J., Xu P., Calder L.J., Borg A., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. Structure and binding properties of Pangolin-CoV spike glycoprotein inform the evolution of SARS-CoV-2. Nat. Commun. 2021;12:837. doi: 10.1038/s41467-021-21006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Yang P., Wang N., Chen Z., Su D., Zhou Z.H., Rao Z., Wang X. Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation. Protein Cell. 2020;11:339–351. doi: 10.1007/s13238-020-00710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Huang D., Lee C.D., Wu N.C., Jackson A.M., Zhu X., Liu H., Peng L., van Gils M.J., Sanders R.W., et al. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science. 2021;373:818–823. doi: 10.1126/science.abh1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183:739–751.e738. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Xiao T., Cai Y., Lavine C.L., Peng H., Zhu H., Anand K., Tong P., Gautam A., Mayer M.L., et al. Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant. Science. 2021;374:1353–1360. doi: 10.1126/science.abl9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Cao L., Gao X.-S., Zheng B.-Y., Deng Y.-Q., Li J.-X., Feng R., Bian Q., Guo X.-L., Wang N. A proof of concept for neutralizing antibody-guided vaccine design against SARS-CoV-2. National Science Review. 2021;8:nwab053. doi: 10.1093/nsr/nwab053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Tsybovsky Y., Gorman J., Rapp M., Cerutti G., Chuang G.-Y., Katsamba P.S., Sampson J.M., Schön A., Bimela J. Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host & Microbe. 2020;28:867–879.e865. doi: 10.1016/j.chom.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M., Tuekprakhon A., Nutalai R. Evidence of escape of SARS-CoV-2 variant B. 1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361.e2346. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Deng Y.-Q., Zhang R.-R., Cui Z., Sun C.-Y., Fan C.-F., Xing X., Huang W., Chen Q., Zhang N.-N. Double lock of a potent human therapeutic monoclonal antibody against SARS-CoV-2. National Science Review. 2021;8:nwaa297. doi: 10.1093/nsr/nwaa297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. elife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The cryo-EM maps have been deposited at the Electron Microscopy Data Bank (www.ebi.ac.uk/emdb) and are available under accession numbers: EMDB: 32478 (Omicron S-trimer at pH 7.5), EMDB: 32479 (Omicron S-trimer at pH 5.5), EMDB: 32480 (Delta S-trimer with 3 RBD close), EMDB: 32481 (Delta S-trimer with 1 RBD open), EMDB: 32482 (Omicron S-trimer in complex with hACE2), EMDB: 32483 (Local optimized reconstruction of Omicron S-trimer in complex with hACE2). Atomic models corresponding to EMDB: 32478, EMDB: 32479, EMDB: 32480, EMDB: 32481, EMDB: 32482, EMDB: 32483 have been deposited in the Protein Data Bank (www.rcsb.org) and are available under accession numbers, PDB: 7WG6, PDB: 7WG7, PDB: 7WG8, PDB: 7WG9, PDB: 7WGB, PDB: 7WGC, respectively.
-
•
This study did not generate custom computer code.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.