Abstract
Toxoplasma gondii is a worldwide food-borne parasite that can infect almost all warm-blooded animals, including humans. To date, there are no effective drugs to prevent or eradicate T. gondii infection. Recent studies have shown that probiotics could influence the relationship between the microbiota and parasites in the host. Koumiss has been used to treat many diseases based on its probiotic diversity. Therefore, we explored the effect of koumiss on T. gondii infection via its effect on the host intestinal microbiota. BALB/c mice were infected with T. gondii and treated with PBS, koumiss and mares’ milk. Brain cysts were counted, and long-term changes in the microbiota and the effect of koumiss on gut microbiota were investigated with high-throughput sequencing technology. The results suggested that koumiss treatment significantly decreased the cyst counts in the brain (P < 0.05). Moreover, T. gondii infection changed the microbiota composition, and koumiss treatment increased the relative abundance of Lachnospiraceae and Akkermansia muciniphila, which were associated with preventing T. gondii infection. Moreover, koumiss could inhibit or ameliorate T. gondii infection by increasing the abundance of certain bacteria that control unique metabolic pathways. The study not only established a close interaction among the host, intracellular pathogens and intestinal microbiota but also provided a novel focus for drug development to prevent and eradicate T. gondii infection.
Subject terms: Infection, Drug development, Public health
Introduction
Toxoplasma gondii is a worldwide intracellular protozoan parasite that can infect almost all cell types, including humans, livestock, companion animals and wildlife1. It is estimated that nearly one-third of the human population has been exposed to T. gondii2. Infection may occur mainly by ingesting raw vegetables and water containing sporulated oocysts or uncooked meat containing tissular cysts or by vertical transmission3,4. Toxoplasmosis can cause health problems in humans, such as encephalitis, pneumonitis and myocarditis, and even result in death in immunodeficient patients2. Moreover, infection can cause foetus miscarriage, congenital malformations and other symptoms during pregnancy5. Once a human is infected with T. gondii, tachyzoites massively proliferate and subsequently persist for a long time as dormant cysts, preferentially in the brain and muscles6.
Chronic infection was previously considered benign7, but recent studies have shown that it is responsible for neurological disorder diseases, such as bipolar disease, schizophrenia and epilepsy, although the causal relationships are unclear8–10. In addition, dormant cysts can be reactivated and lead to severe consequences, including encephalitis, when host immunity is defective11. Nevertheless, effective drugs that prevent or eradicate T. gondii tissue cyst formation have not been identified, although 50 compounds affecting over 20 different pathways or mechanisms were tested over the last three decades7. Therefore, the identification of a significant method for treating chronic T. gondii infection holds great promise.
Recently, the gut microbiota, which can affect the gastrointestinal system and other distal organs in the body, including the brain, has attracted more attention. Many studies have made great progress by establishing correlations between intestinal microbiota and diseases, such as hepatic encephalopathy, Alzheimer’s disease and schizophrenia, and treatment effects, such as antidepressant effects12–15. Many studies have shown that there is a close interaction among parasites, microbial communities, and host immune responses16. Furthermore, probiotics can disrupt the relationship between the parasite and associated microbiota by increasing the gut microbiota diversity to change the microbiota composition17. However, to date, no study has linked gut microbiota with toxoplasmosis treatment.
Koumiss is a traditional fermented drink product of nomadic people in central Asia, which is made by fermenting mares’ milk with lactic acid bacteria and yeasts18. The rich microbial microbiota caused by spontaneous fermentation determines its nutritional and medical value. Guo et al. separated 6 bacterial phyla, represented by 126 genera and 49 species, and 3 fungal phyla, represented by 59 genera and 57 species, from 11 artisanal koumiss samples19. In China, koumiss was the earliest beverage used in food therapy with an obvious auxiliary treatment effect in digestive diseases20. In addition, koumiss has been widely used in treating hyperlipidemia and chronic atrophic gastritis and for promoting immunomodulation and anti-inflammatory effects because of its probiotic diversity21–23.
In this study, we explored the possibility that koumiss consumption treats or alleviates toxoplasmosis by modulating the intestinal microbiota of mice chronically infected by T. gondii. After mice infected by T. gondii were treated with Koumiss, the brain cysts were counted, and the 16S rRNA gene sequencing of the gut microbiota was determined with high-throughput sequencing technology. This study reveals the effects of koumiss consumption in treating toxoplasmosis by establishing the relationship between T. gondii and the gut microbiota in the host and provides a new idea for drug development.
Results
Infection of mice with T. gondii and cyst counting
The feces of the C, G, S and X groups were collected on 7 dpi, 14 dpi, 21 dpi, 28 dpi, 35 dpi, and 42 dpi, respectively. Moreover, the brain tissue of the C, G, S and X groups at 42 dpi was collected and homogenized to count cysts. As shown in Fig. 1, the cyst counts in the brains of the G, S and X groups were 14.5/units, 8.33/units, and 13/units, respectively. The cyst counts of the S group were significantly lower than those of the G group (P < 0.05), indicating that koumiss may have a regulating effect on T. gondii infection.
Figure 1.
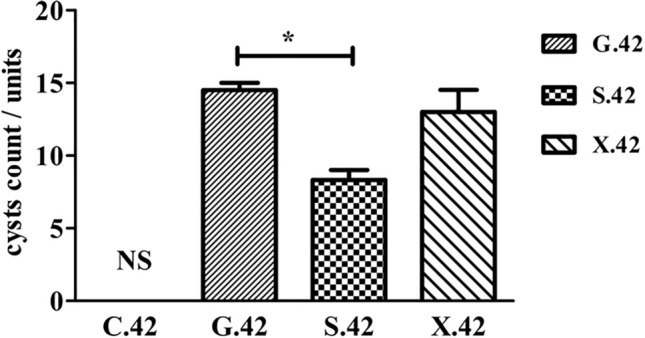
The cyst counts of the C, G, S and X groups at 42 dpi.
Different treatment methods changed the abundance of the intestinal microbiota in mice
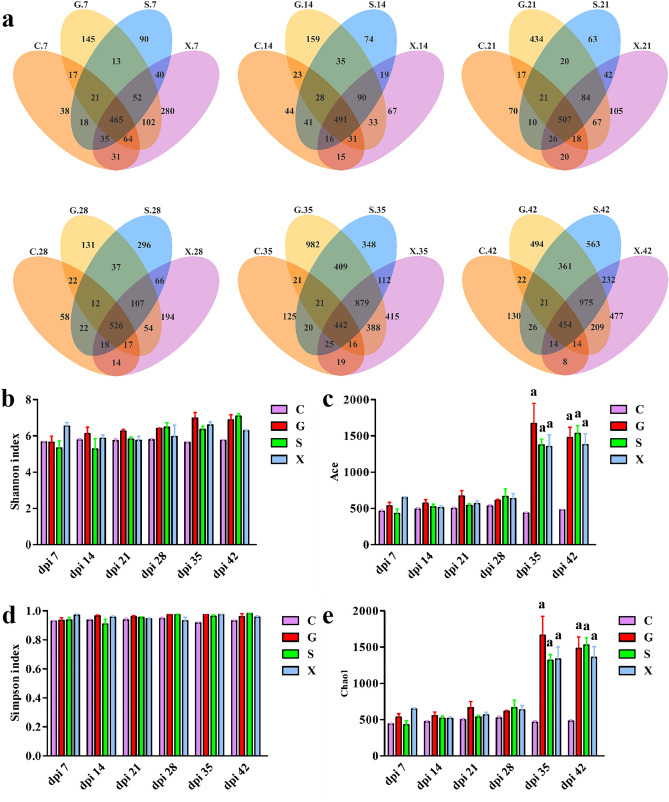
In the present study, we determined the 16S rRNA gene sequence of the gut microbiota in the C, G, S and X groups at 7 dpi, 14 dpi, 21 dpi, 28 dpi, 35 dpi and 42 dpi. All effective tags of samples were clustered with 97% identity into OTUs, which were used to better analyze species composition. The OTUs of overlapping parts peaked at 28 dpi, indicating the most similarity in different groups. However, the total number of OTUs at 35 dpi was significantly higher than before, and greater differences in species composition were observed among the groups (Fig. 2a). As shown in Fig. 2b, d, the Shannon and Simpson indices, which describe species diversity and uniformity, were not significantly different among the groups. In addition, the Ace and Chao1 indices of the G, S and X groups at 35 dpi and 42 dpi were dramatically higher than those of the C group (P < 0.001) (Fig. 2c, e). The results showed that the G, S and X groups had higher microbiota richness at 35 dpi and 42 dpi, which was consistent with the Venn diagram results.
Figure 2.
Different treatment methods change the intestinal microbiota diversity of mice. (a) Venn diagram showing OTUs of the C, G, S and X groups at 7 dpi, 14 dpi, 21 dpi, 28 dpi, 35 dpi and 42 dpi. Overlapping parts represent shared OTUs among groups. (b) Shannon, (c) Ace, (d) Simpson, and (e) Chao1 indices in α-diversity analyses. a indicates P < 0.001.
The community structure of microbiota at phylum level
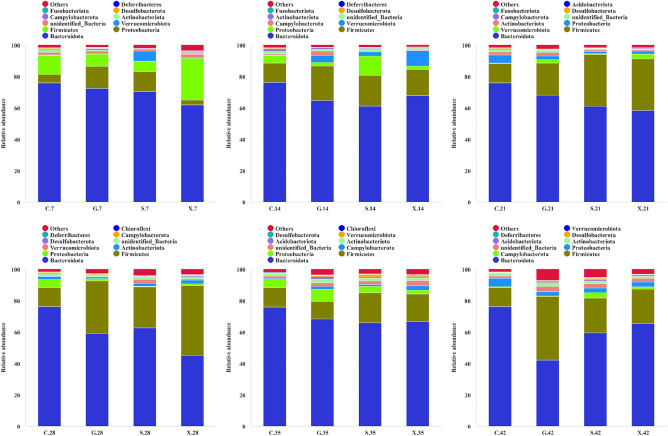
The community structure of samples at different taxonomic levels on the basis of OTUs, including phylum, class, order, family, genus and species, was determined. In this study, 59 phyla, represented by 745 genera and 357 species, were identified. The composition of the microbiota at the phylum level, whose abundances were ranked in the top 10, was mainly analyzed. The community structure of the C group on 7 dpi included Bacteroidota (76.1%), Firmicutes (12.4%), Proteobacteria (5.2%), Actinobacteriota (1.9%) and others (Fig. 3). After infection with T. gondii, Bacteroidota was less abundant in all treatment groups than in the C group, although it had the highest abundance. Nevertheless, the abundance of Firmicutes exhibited an obvious increase in the G, S and X groups until 28 dpi. The G group exhibited the maximum increase from 7.8 (7 dpi) to 40.5% (42 dpi). On 28 dpi, the abundance of Fusobacteriota decreased to nearly 0 in the G, S and X groups. Moreover, the abundance of Actinobacteriota maintained a stable increase in the S group and was close to that in the C group at 42 dpi.
Figure 3.
Community structure of microbiota at the phylum level.
Toxoplasma gondii infection influenced the gut microbiota composition of the mice
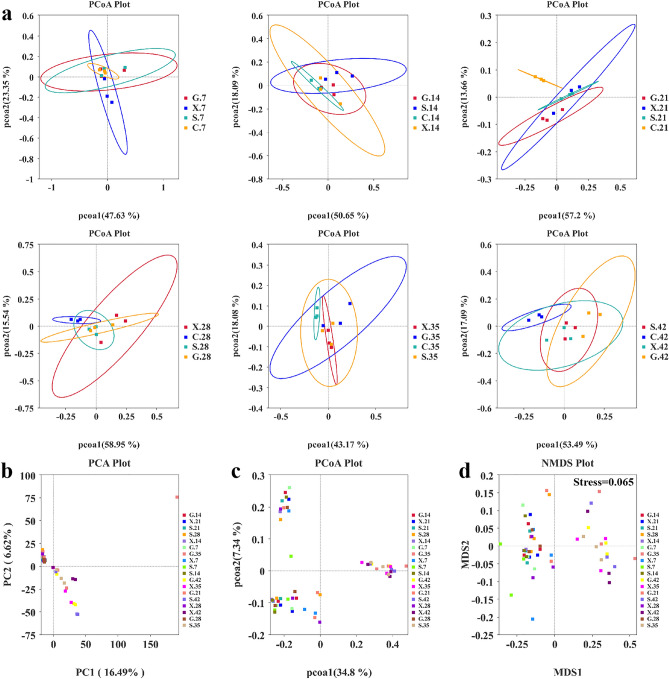
Multivariate statistical methods based on UniFrac distance were used to analyze the changes in the gut microbiota in the mice infected by T. gondii. The gut microbiota composition of the G, S and X groups was markedly distinct from that of the C group, which was clustered separately beginning on 21 dpi (Fig. 4a). The results suggested that T. gondii infection resulted in alterations in gut microbiota composition. Moreover, the PCA plot demonstrated that the microbiota community structure of the G, S and X groups at 35 dpi and 42 dpi was separated from the G, S and X groups at 7, 14, 21 and 28 dpi by PC1 (16.49% of the explained variance) (Fig. 4b). There was also a separation of 35 and 42 dpi from 7, 14, 21 and 28 dpi in the G, S and X groups by PCoA1 (34.8% of the explained variance) (Fig. 4c). NMDS was applied to overcome the shortcomings of PCA and PCoA and determine the species information of samples. The same result was observed on the two-dimensional plane in the form of points, indicating more significant variation in the intestinal microbiota composition caused by T. gondii infection over time (Fig. 4d).
Figure 4.
Alterations in gut microbiota caused by T. gondii infection. (a) PCoA of the weighted UniFrac values for the C, G, S and X group on 7 dpi, 14 dpi, 21 dpi, 28 dpi, 35 dpi and 42 dpi. (b) PCA and (c) PCoA of the unweighted UniFrac values for the G, S and X groups at 7 dpi, 14 dpi, 21 dpi, 28 dpi, 35 dpi and 42 dpi. (d) NMDS plot showing the differences in gut microbiota composition based on the unweighted UniFrac distance. A stress < 0.2 means that the NMDS plot can accurately reflect differences among samples.
The impact of Koumiss treatment on the intestinal microbiota of mice infected with T. gondii
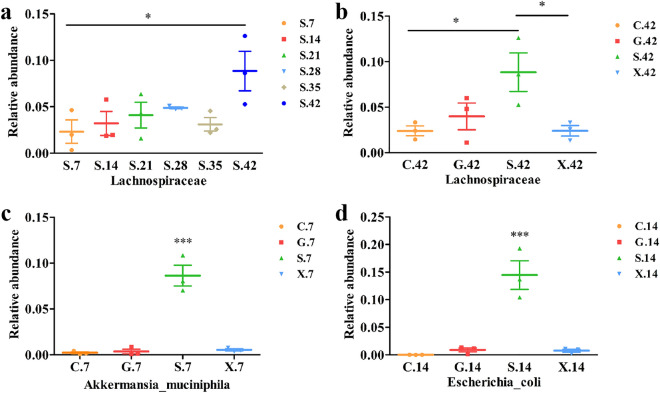
To study the impact of koumiss on the microbiota of mice infected with T. gondii, we compared the differences in the species among different groups using Tukey’s test. Koumiss treatment changed the intestinal microbiota composition in mice infected with T. gondii, such as the abundances of Actinobacteriota, Lactobacillaceae and Acetobacter. The most striking finding was that the relative abundance of Lachnospiraceae continuously increased after the mice were orally gavaged with koumiss (Fig. 5a). There was a significant difference between 7 and 42 dpi (P < 0.05). Furthermore, there were significant differences on 42 dpi between the S and C group (P < 0.05), S and X group (P < 0.05), respectively (Fig. 5b). More importantly, the relative abundance of Akkermansia muciniphila in the S group on 7 dpi was dramatically different from that in the C, G and X groups (P < 0.001) (Fig. 5c) Moreover, there was a significant increase in the relative abundance of Escherichia coli in the S group compared with that in the C, G and X groups on 14 dpi (P < 0.001) (Fig. 5d).
Figure 5.
The impact of Koumiss treatment on the intestinal microbiota of mice infected with T. gondii. (a) The relative changes in Lachnospiraceae abundance in the S group on 7 dpi, 14 dpi, 21 dpi, 28 dpi, 35 dpi and 42 dpi. (b) The relative abundance of Lachnospiraceae in the C, G, S and X groups at 42 dpi. (c) The relative abundance of Akkermansia muciniphila in the C, G, S and X groups at 7 dpi. (d) The relative abundance of Escherichia coli in the C, G, S and X groups at 14 dpi. *P < 0.05; ***P < 0.001.
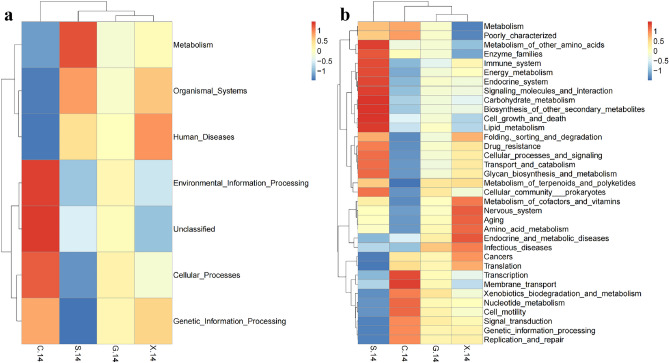
Koumiss treatment caused changes in the microbiota functional profile
Tax4Fun analysis was carried out to understand the differences in the function of the intestinal microbiota among the treatment groups. The Tax4Fun software package is well-known for its high accuracy in predicting the function of environmental samples, such as the intestine and soil. The key is to compare 16S rRNA gene sequencing data with the KEGG database to achieve functional annotation. The results showed that at 14 dpi, the mice in the S group had more bacteria than those in the C, G and X groups that functioned in metabolism, including glycan biosynthesis and metabolism, carbohydrate metabolism, energy metabolism, biosynthesis of other secondary metabolites and endocrine metabolism (Fig. 6a, b). There was also an obvious increase in cellular processes and signaling, drug resistance, immune system and signaling molecules and interactions in the S group at 14 dpi. However, membrane transport in the S group at 14 dpi showed a downward trend compared with that in the C group (Fig. 6b).
Figure 6.
Heatmaps of KEGG functional classification of different groups at 14 dpi in Level 1 (a) and Level 2 (b).
Discussion
Toxoplasma gondii is a food-borne parasite found worldwide that can infect almost all warm-blooded animals, including humans24. Chronic T. gondii infection often results in few clinical symptoms in immunocompetent individuals. However, some studies have suggested that it can result in changes in immunity, brain transcriptomic signatures, microbiota, and even behavior and personality in the host6,25,26. Some studies have revealed that a healthy intestinal microbiota is a complex ecological community that can carry out critical physiological functions with the intestinal mucosa of the host27. Moreover, changes in the gut microbiota composition can prevent or alleviate intestinal protozoan infection and affect the ultimate outcomes of parasitic diseases. There is also a close connection among the host, microbiota and pathogens. An interesting example is that a certain gut microbiota composition in the host can impede gut colonization by helminths or reduce the colonization time28. To date, there is no study that associates the host microbiota with the treatment of T. gondii infection. Shao et al. only discussed the changes in the intestinal microbiota 13 days and 21 days after infection with T. gondii25. In this study, we investigated not only the effect of koumiss treatment on the intestinal microbiota of mice chronically infected with T. gondii but also the long-term changes in the gut microbiota caused by T. gondii infection, including the changes that occur in the acute infection stage, chronic infection stage and cyst formation stage.
In this study, we determined the 16S rRNA gene sequences of the gut microbiota of the C, G, S and X groups at 7 dpi, 14 dpi, 21 dpi, 28 dpi, 35 dpi and 42 dpi. The intestinal microbiota community structure at the phylum level was mainly analyzed. Bacteroidota was the most abundant phylum in the C, G, S and X groups, and the relative abundance of Bacteroidota decreased after infection with T. gondii. There was an obvious increase in the relative abundance of Firmicutes in all the experimental groups, which was different from a previous study25. In a previous study, mice were infected with 100 purified sporulated oocysts of the ME49 strain, which may have caused inflammation in the intestine and was associated with a low F/B (Firmicutes/Bacteroidota) index29. In this study, mice were inoculated with 3 cysts to establish chronic infection, and no intestinal inflammation was observed (data not shown), but this treatment resulted in a high F/B index. The gut-brain axis plays an important and complex bidirectional role in communication between the intestine and the central nervous system (CNS) and is involved in different pathways and systems, including the autonomic and enteric nervous system, the endocrine system, the hypothalamic–pituitary–adrenal axis (HPA), the immune system, and the regulation of the microbiota and its metabolites30–32. Therefore, we considered that one of the reasons that explains our results is that parasites might influence the intestinal microbiota through the gut-brain axis. Moreover, this parasite is distributed all around the host during chronic infection, which makes the mechanism more difficult to understand; this is an interesting phenomenon worthy of further study.
In addition, we found that T. gondii infection dramatically changed the community structure of the microbiota. The C group and other treatment groups showed an obvious separation in the PCoA plot at 21 dpi, indicating that T. gondii infection would alter the intestinal microbiota composition, which was consistent with previous studies25. Nevertheless, the C group was not separately clustered from the other treatment groups at 14 dpi. This result may have occurred because a large number of parasites had appeared in the brain. The strains of T. gondii and the inoculation dose and time may affect the distribution of T. gondii. Zenner et al. confirmed that T. gondii first reached the mesenteric lymph nodes (MLNs) through the intestinal wall, then reached spleen, and finally reached other organs33. Saeij et al. thought that a virulent strain of T. gondii first appeared in the head and neck, whereas an attenuated strain appeared preferentially in the abdomen34. The changes in the microbiota composition caused by T. gondii in the brain were also associated with the gut-brain axis. Interestingly, there was also an apparent separation of 35 and 42 dpi from 7, 14, 21 and 28 dpi in the G, S and X groups, which indicated that the community structure of the gut microbiota could change over time during T. gondii infection. The results are shown in the PCA plot, PCoA plot, NMDS plot and Venn diagrams. The Ace and Chao1 indices of the G, S and X groups were obviously increased at 35 and 42 dpi. The results demonstrated that there was an obvious increase in the gut microbiota diversity during T. gondii cyst formation and that this increased diversity persisted for a long time. Traditionally, pathogen infection is thought to decrease the microbiota richness. However, the results of this study were completely opposite of that previous theory. This discovery has important implications for understanding the connection between pathogens and the intestinal microbiota in the host.
The effect of koumiss treatment on intestinal microbiota was also discussed in this study. It should be noted that the infected mice treated with koumiss were not significantly different from the untreated mice in terms of OTUs, the Shannon index or the Simpson index. However, we still found that koumiss treatment altered the relative abundance of certain bacteria, such as Actinobacteriota, Lactobacillaceae and Acetobacter. The most important alteration was that the relative abundance of Lachnospiraceae continuously increased after the mice infected with T. gondii were orally gavaged with koumiss. There were significant differences in the relative abundance of Lachnospiraceae between the S and C group, the S and X group at 42 dpi. Lachnospiraceae are a family of obligate anaerobic bacteria that are abundant in healthy humans. Lachnospiraceae can facilitate colonization resistance against intestinal pathogens by producing the short-chain fatty acids (SCFAs) acetate and butyrate35, converting primary to secondary bile acids36 or producing lantibiotics37. Therefore, it was thought that the reduction in the numbers of cysts in the brain in the S group, a key metric indicating decreased parasitic burden during chronic T. gondii infection, may be associated with a high level of Lachnospiraceae. Moreover, the results published in Science showed that Lachnospiraceae can allow mice to survive after receiving a lethal dose of total body radiation by promoting hematopoiesis restoration and gastrointestinal repair38. More importantly, Lachnospiraceae abundances can be altered by changes in diet39. This will be further explored for the treatment of T. gondii infection. Furthermore, we expectedly found that the relative abundance of A. muciniphila in the S group at 7 dpi dramatically differed from that in the C, G and X groups (P < 0.001). A. muciniphila is an emerging beneficial bacterium that controls the essential regulatory system of glucose and energy metabolism40. In addition, A. muciniphila can also decrease blood lipid levels and affect metabolic modulation, immune response regulation, and gut health maintenance41–44. Interestingly, it was also found that A. muciniphila could facilitate the expulsion of intestinal Trichinella spiralis through the development of goblet cell hyperplasia and secretion of mucus45,46. Although A. muciniphila was isolated from human feces in 2004, the relationship between A. muciniphila and intracellular pathogens is still unknown. This study provides a novel focus for establishing connections between these species through the intestinal microbiota.
Koumiss treatment also influenced the function of the microbiota in the intestine. The results showed that at 14 dpi, the mice in the S group had more bacteria than those in the C, G and X groups that functioned in metabolism, such as carbohydrate metabolism, lipid metabolism and amino acid metabolism. Host metabolites can not only promote the growth of T. gondii but also directly govern its development and differentiation47. The glycolysis pathway is very important for the T. gondii tachyzoite stage, and this pathway not only provides a carbon source for fatty acid synthesis but also drives T. gondii invasion of host cells48. After hosts are infected by T. gondii, dendritic cells and macrophages drive an immune response but also improve the dependence of metabolism on glycolysis, which supports T. gondii growth49. A. muciniphila may inhibit T. gondii growth by regulating glucose and energy metabolism. Furthermore, research has suggested that T. gondii infection increases the relative abundance of Escherichia coli in the intestine50. This may be the reason why the relative abundance of Escherichia coli was higher in the S group than in the C, G and X groups at 14 dpi. Lipids also participate in innate immunity and may play an important role in controlling T. gondii infection51. Lachnospiraceae may facilitate colonization resistance against intestinal pathogens through the lipid metabolism pathway. T. gondii infection also interferes with arginine uptake, synthesis and metabolization in the host cell. A study demonstrated that T. gondii induced the integrated stress response (ISR) by depleting arginine from host cells, which caused increased expression of the host cationic amino acid transporter CAT1 to ensure a constant supply of arginine52. Compared with that in uninfected cells, the expression of arginine succinate synthase 1 (ASS) in T. gondii-infected cells was upregulated by fivefold53. A study also indicated that T. gondii differentiated toward bradyzoites when essential nutrient supplies, such as arginine and cysteine, were limited54. Moreover, we strikingly found that the S group had fewer bacteria that function in membrane transport at 14 dpi. T. gondii secretes rhoptries (ROPs) and dense granules (GRAs) that interfere with biological processes in host cells, resulting in slight evasion of host cell autonomous defense mechanisms55. Secreted proteins are also regarded as nonspecific small molecule channels that facilitate the flux of metabolites between the parasitophorous vacuole (PV) space and the host cytoplasm56. Therefore, koumiss may inhibit T. gondii energy supplementation by reducing metabolite transport. In addition, it was found that at 14 dpi, the S group had more bacteria in the organismal system, including the immune system, which were beneficial for preventing T. gondii infection.
In conclusion, we investigated brain cyst counts and explored long-term changes in the microbiota and the effect of koumiss treatment on the microbiota in mice infected with T. gondii. Strikingly, we found that koumiss treatment significantly reduced brain cyst counts in mice infected with T. gondii. Moreover, T. gondii infection altered the intestinal microbiota community structure, whereas koumiss treatment could enhance the relative abundance of Lachnospiraceae and A. muciniphila; this effect of ameliorating T. gondii infection occurred via special metabolic pathways. The study established a close interaction among the host, intracellular pathogens and intestinal microbiota and provided a novel direction for preventing and eradicating T. gondii infection.
Materials and methods
Animals and parasites
One hundred and forty-four 6-week-old female BALB/c mice, weighing 15–18 g, were purchased from SPF (Beijing) Biotechnology Co., Ltd. The mice (three per cage) were given ad libitum access to food and water and housed under a 12 h light/dark cycle. The animal protocols were reviewed and approved by the Inner Mongolia Agricultural University Laboratory Animal Welfare and Animal Experimental Ethical Inspection Committee (approval No.: 2020-037). All experiments were performed in accordance with relevant guidelines and regulations. And the study was conducted in compliance with the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines57. The T. gondii Prugniaud strain was obtained from the National Animal Protozoa Laboratory of China Agricultural University. After 1 week of acclimation, the mice were randomly divided into Control group (C, n = 36), Infection group (G, n = 36), Koumiss group (S, n = 36) and Mare milk group (X, n = 36). Then, the G, S and X groups were infected orally with 200 µL sterile phosphate buffered solution (PBS) containing 3 tissular cysts, while the C group was infected orally with the same volume of sterile PBS alone. The G, S and X groups were orally gavaged with 100 µL PBS, koumiss and mares’ milk at two-day intervals. Koumiss samples and fresh mare milk samples obtained from the Abaga banner of Xilin Gol League were used in their original form.
Samples collection and cyst counting
Six mice were selected randomly from each group and individually placed in clean and sterile cages. Then, the feces were collected aseptically within two hours, and the mice were euthanized by cervical dislocation. Mice of the C, G, S and X group were euthanized at 7 days post infection (dpi), 14 dpi, 21 dpi, 28 dpi, 35 dpi, and 42 dpi. The feces were treated with liquid nitrogen and stored at − 80 °C. The mice were sacrificed at 42 dpi, the brains were homogenized, and the cysts were counted three times as described previously58.
DNA extraction and PCR amplification
Genomic DNA was extracted from feces using the cetyltrimethylammonium ammonium bromide (CATB) method. The purity and concentration of the nucleic acids were determined by 1% agarose gel electrophoresis. The 16S rRNA genes (V3–V4 region) were amplified with specific primers with barcodes. Amplification was performed with primers with the following sequences: forward primer 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′). Thermal cycling consisted of initial denaturation at 98 °C for 1 min, 30 cycles of denaturation at 98 °C for 10 s, annealing at 50 °C for 30 s, and elongation at 72 °C for 30 s, and a final elongation step at 72 °C for 5 min. The PCR products were detected by 2% agarose gel electrophoresis and purified with a Qiagen Gel Extraction Kit (Qiagen, Germany).
Library construction and sequencing
Following library construction by the TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, USA), the library was sequenced on the Illumina NovaSeq sequencing platform, and 250 bp paired-end reads were generated. The measurement was repeated three times for each sample. The reads were merged with FLASH (VI.2.7, http://ccb.jhu.edu/software/FLASH/) and filtered rigorously according to the QIIME (V1.9.1, http://qiime.org/scripts/split libraries fastq.html/) quality control process59. After the tags were compared with the reference database (Silva database, https://www.arb-silva.de/), chimera sequences were removed, and the effective tags were finally obtained.
Biological information analyses
All effective tags of samples were clustered with 97% identity into operational taxonomic units (OTUs) by the Uparse algorithm (V7.0.1001, http://www.drive5.com/uparse/). The taxonomic information was obtained by species annotation analysis, and the community composition of samples was counted individually at each classification level, including the Kingdom, Phylum, Class, Order, Family, Genus and Species levels. The least amount of data in the sample was used as the standard for homogenization, which was the basis for subsequent alpha diversity analysis and beta diversity analysis. Principal component analysis (PCA), a method of variance decomposition based on Euclidean distances, was used to reduce the dimension of multidimensional data and extract the most important elements in the data60. In addition, to overcome the shortcomings of linear models, such as PCA and principal coordinates analysis (PCoA), nonmetric multidimensional scaling (NMDS) was also applied to determine the species information of the samples, and the results are expressed in the form of points on the two-dimensional plane61.
Statistical analysis
Alpha diversity index analyses of the samples were performed with QIIME by calculating the Chao1, Shannon, Simpson and Ace indices. Shannon and Simpson indexes were used to describe the species diversity and equitability. The higher the community diversity was, the greater the Shannon and Simpson indexes were. Chao1 and Ace indexes reflected on the number of species and OTUs respectively. Among all the indices at the level of Alpha-diversity, Shannon index presents remarkable characteristics. It was worth noting that Shannon and Simpson indexes would decrease when the relative abundance of species in a community exceeded a certain limitation. The limit was about 0.67 for the Simpson index and about 0.7 for the Shannon index. QIIME also served to calculate UniFrac distance. R software was used to draw PCA, PCoA and NMDS diagrams. GraphPad Prism software was also used for data analyses and plots. Kyoto Encyclopedia for Genes and Genomics (KEGG) classifications were performed with the Tax4Fun software package. Statistically significant differences in species composition between groups were determined using Tukey’s test. P < 0.05 indicated a significant difference.
Acknowledgements
This research was mainly funded by National Natural Science Foundation of China (32160548, 31760489) and Science and Technology Plan Project of Food Science and Engineering College Inner Mongolia Agricultural University, China (SPKJ202011).
Author contributions
Study concept and design: X.Y. and W.H. Samples collection and experiment: W.H., X.J., Y.S., J.G. and X.Y. Analysis and interpretation of data: X.Y. and W.H. Manuscript writing: W.H. Manuscript review and editing: X.Y. and J.G. Supervision: X.Y. and J.G. Funding acquisition and project administration: X.Y.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xinlei Yan and Wenying Han.
Contributor Information
Xinlei Yan, Email: yanxinlei1987620@foxmail.com.
Jun Guo, Email: guojunge@imau.edu.cn.
References
- 1.Joanne PW, Dubey JP. Toxoplasmosis of Animals and Humans. 3. CRC Press; 2010. p. 4. [Google Scholar]
- 2.Weiss LM, Dubey JP. Toxoplasmosis: A history of clinical observations. Int. J. Parasitol. 2009;39(8):895–901. doi: 10.1016/j.ijpara.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tenter AM, Heckeroth AR, Weiss LM. Toxoplasma gondii: From animals to human. Int. J. Parasitol. 2000;30(12–13):1217–1258. doi: 10.1016/S0020-7519(00)00124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heddergott M, Frantz AC, Stubbe M, Stubbe A, Ansorge H, Osten-Sacken N. Seroprevalence and risk factors of Toxoplasma gondii infection in invasive raccoons (Procyon lotor) in Central Europe. Parasitol. Res. 2017;116(8):2335–2340. doi: 10.1007/s00436-017-5518-7. [DOI] [PubMed] [Google Scholar]
- 5.Chessa G, Chisu V, Porcu R, Masala G. Molecular characterization of Toxoplasma gondii type II in sheep abortion in Sardinia, Italy. Parasite. 2014 doi: 10.1051/parasite/2014007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu RS, et al. Transcriptomic profiling of mouse brain during acute and chronic infections by Toxoplasma gondii oocysts. Front. Microbiol. 2020 doi: 10.3389/fmicb.2020.570903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paula JL, Ruben RB. Perspective on current and emerging drugs in the treatment of acute and chronic toxoplasmosis. Postgrad. Med. 2019;131(8):589–596. doi: 10.1080/00325481.2019.1655258. [DOI] [PubMed] [Google Scholar]
- 8.Sutterland AL, et al. Beyond the association Toxoplasma gondii in schizophrenia, bipolar disorder, and addiction: Systematic review and meta-analysis. Acta Psychiatr. Scand. 2015;132(3):161–179. doi: 10.1111/acps.12423. [DOI] [PubMed] [Google Scholar]
- 9.Elsheikha HM, Zhu XQ. Toxoplasma gondii infection and schizophrenia: An inter-kingdom communication perspective. Curr. Opin. Infect. Dis. 2016;29(3):311–318. doi: 10.1097/QCO.0000000000000265. [DOI] [PubMed] [Google Scholar]
- 10.Shiojiri D, Kinai E, Teruya K, Kikuchi Y, Oka S. Combination of Clindamycin and Azithromycin as alternative treatment for Toxoplasma gondii encephalitis. Emerg. Infect. Dis. 2019;25(4):841–843. doi: 10.3201/eid2504.181689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wohlfert EA, Blader IJ, Wilson EH. Brains and Brawn: Toxoplasma infections of the central nervous system and skeletal muscle. Trends Parasitol. 2017;33(7):519–531. doi: 10.1016/j.pt.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen ZF, et al. The role of intestinal bacteria and gut-brain axis in Hepatic encephalopathy. Front. Cell. Infect. Microbiol. 2020 doi: 10.3389/fcimb.2020.595759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang LW, et al. Antidepressant effects and mechanisms of the total Iridoids of Valeriana jatamansi on the brain-gut axis. Planta Med. 2019;86(3):172–179. doi: 10.1055/a-1068-9686. [DOI] [PubMed] [Google Scholar]
- 14.Marta S, Katarzyna DL, Satler DB, Donata K, Ewa B, Jerzy L. The gut microbiome alterations and inflammation-driven pathogenesis of Alzheimer's Disease—A critical review. Mol. Neurobiol. 2019;56(3):1841–1851. doi: 10.1007/s12035-018-1188-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan XX, Kang YL, Zhuo CJ, Huang XF, Song XQ. The gut microbiota promotes the pathogenesis of schizophrenia via multiple pathways. Biochem. Biophys. Res. Commun. 2019;512(2):373–380. doi: 10.1016/j.bbrc.2019.02.152. [DOI] [PubMed] [Google Scholar]
- 16.Candela M, Biagi E, Turroni S, Vitali B, Brigidi P. Mechanisms involved in the intestinal interaction between host and bifidobacteria. Microb. Ecol. Health Dis. 2008;20(4):189–192. doi: 10.1080/08910600802333483. [DOI] [Google Scholar]
- 17.Partida-Rodriguez O, et al. Human intestinal microbiota: Interaction between parasites and the host immune response. Arch. Med. Res. 2017;48(8):690–700. doi: 10.1016/j.arcmed.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 18.Askar K, Gerard L, Nawel A, Christian M, Gaukhar K. Fermented mare milk product (Qymyz, Koumiss) Int. Dairy J. 2021 doi: 10.1016/J.IDAIRYJ.2021.105065. [DOI] [Google Scholar]
- 19.Guo L, et al. Study of bacterial and fungal community structures in traditional koumiss from Inner Mongolia. J. Dairy Sci. 2019;102(3):1972–1984. doi: 10.3168/jds.2018-15155. [DOI] [PubMed] [Google Scholar]
- 20.Xu SZ. Dietotherapy and medical value of mare's milk and sour mare's milk. Chin. J. Ethnic Med. 2012;18(02):21–22. doi: 10.16041/j.cnki.cn15-1175.2012.02.002. [DOI] [Google Scholar]
- 21.Hou QC, et al. Koumiss consumption modulates gut microbiota, increases plasma high density cholesterol, decreases immunoglobulin G and albumin. J. Funct. Foods. 2019;52:469–478. doi: 10.1016/j.jff.2018.11.023. [DOI] [Google Scholar]
- 22.Li CK, et al. Koumiss consumption alleviates symptoms of patients with chronic atrophic gastritis: A possible link to modulation of gut microbiota. J. Nutr. Oncol. 2017;2(01):36–51. [Google Scholar]
- 23.Rong JJ, et al. Probiotic and anti-inflammatory attributes of an isolate Lactobacillus helveticus NS8 from Mongolian fermented koumiss. BMC Microbiol. 2015;15(1):196–206. doi: 10.1186/s12866-015-0525-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melo RPB, et al. Atypical Toxoplasma gondii genotype from a sheep and a pig on Fernando de Noronha Island, Brazil, showed different mouse virulence profiles. Parasitol. Res. 2019;119(1):351–356. doi: 10.1007/s00436-019-06522-4. [DOI] [PubMed] [Google Scholar]
- 25.Shao DY, et al. Changes to the gut microbiota in mice induced by infection with Toxoplasma gondii. Acta. Trop. 2020 doi: 10.1016/j.actatropica.2019.105301. [DOI] [PubMed] [Google Scholar]
- 26.Holliman RE. Toxoplasmosis, behaviour and personality. J. Infect. 1997;35(2):105–110. doi: 10.1016/S0163-4453(97)91380-3. [DOI] [PubMed] [Google Scholar]
- 27.Matsuki, T. & Tanaka, R. Function of the human gut microbiota. In The Human Microbiota and Microbiome 90–106 (2014).
- 28.Reynolds LA, Brett FB, Maizels RM. Cohabitation in the intestine: Interactions among Helminth parasites, bacterial microbiota, and host immunity. J. Immunol. 2015;195(9):4059–4066. doi: 10.4049/jimmunol.1501432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins SM. A role for the gut microbiota in IBS. Nat. Rev. Gastroenterol. Hepatol. 2014;11(8):497–505. doi: 10.1038/nrgastro.2014.40. [DOI] [PubMed] [Google Scholar]
- 30.Duvallet C, Gibbons SM, Gurry T, Irizarry RA, Alm EJ. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 2017;8(1):1784. doi: 10.1038/s41467-017-01973-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burberry A, et al. C9orf72 suppresses systemic and neural inflammation induced by gut microbes. Nature. 2020;582(7810):89–94. doi: 10.1038/s41586-020-2288-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blacher E, et al. Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature. 2019;572(7770):474–480. doi: 10.1038/s41586-019-1443-5. [DOI] [PubMed] [Google Scholar]
- 33.Zenner L, Darcy F, Capron AC, Esbron-Delauw MF. Toxoplasma gondii: Kinetics of the dissemination in the host tissues during the acute phase of infection of mice and rats. Exp. Parasitol. 1998;90(1):86–94. doi: 10.1006/expr.1998.4301. [DOI] [PubMed] [Google Scholar]
- 34.Saeij JPJ, Boyle JP, Grigg ME, Arrizabalaga G, Boothroyd JC. Bioluminescence imaging of Toxoplasma gondii infection in living mice reveals dramatic differences between strains. Infect. Immun. 2005;73(2):695–702. doi: 10.1128/IAI.73.2.695-702.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Byndloss MX, et al. Microbiota-activated PPAR-g signaling inhibits dysbiotic Enterobacteriaceae expansion. Science. 2017;357(6351):570–575. doi: 10.1126/science.aam9949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Studer N, et al. Functional intestinal Bile Acid 7 alpha-dehydroxylation by Clostridium scindens associated with protection from Clostridium difficile infection in a gnotobiotic mouse model. Front. Cell. Infect. Microbiol. 2016;6:191. doi: 10.3389/fcimb.2016.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim SG, et al. Microbiotaderived lantibiotic restores resistance against vancomycin-resistant Enterococcus. Nature. 2019;572(7771):665–666. doi: 10.1038/s41586-019-1501-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo H, et al. Multi-omics analyses of radiation survivors identify radioprotective microbes and metabolites. Science. 2020;370(6516):549–560. doi: 10.1126/science.aay9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hasani A, Ebrahimzadeh S, Hemmati F, Khabbaz A, Hasani A, Gholizadeh P. The role of Akkermansia muciniphila in obesity, diabetes and atherosclerosis. J. Med. Microbiol. 2021 doi: 10.1099/JMM.0.001435. [DOI] [PubMed] [Google Scholar]
- 41.Dao MC, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: Relationship with gut microbiome richness and ecology. Gut. 2016;65(3):426–436. doi: 10.1136/gutjnl-2014-308778. [DOI] [PubMed] [Google Scholar]
- 42.Zhai QX, Feng SS, Arjan N, Chen W. A next generation probiotic, Akkermansia muciniphila. Crit. Rev. Food Sci. Nutr. 2019;59(19):3227–3236. doi: 10.1080/10408398.2018.1517725. [DOI] [PubMed] [Google Scholar]
- 43.Plovier H, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 2017;23(1):107–113. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 44.Ashrafian F, et al. Akkermansia muciniphila-derived extracellular vesicles as a mucosal delivery vector for amelioration of obesity in mice. Front. Microbiol. 2019;10:2155. doi: 10.3389/fmicb.2019.02155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin XM, et al. β-Glucan-triggered Akkermansia muciniphila expansion facilitates the expulsion of intestinal helminth via TLR2 in mice. Carbohydr. Polym. 2022;275:118719. doi: 10.1016/J.CARBPOL.2021.118719. [DOI] [PubMed] [Google Scholar]
- 46.Ishikawa N, Wakelin D, Mahida YR. Role of T helper 2 cells in intestinal goblet cell hyperplasia in mice infected with Trichinella spiralis. Gastroenterology. 1997;113(2):542–549. doi: 10.1053/gast.1997.v113.pm9247474. [DOI] [PubMed] [Google Scholar]
- 47.Kloehn J, Hammoudi PM, Soldati-Favre D. Metabolite salvage and restriction during infection—A tug of war between Toxoplasma gondii and its host. Curr. Opin. Biotechnol. 2021;68:104–114. doi: 10.1016/j.copbio.2020.09.015. [DOI] [PubMed] [Google Scholar]
- 48.Nitzsche R, Zagoriy V, Lucius R, Gupta N. Metabolic cooperation of glucose and glutamine is essential for the lytic cycle of obligate intracellular parasite Toxoplasma gondii. J. Biol. Chem. 2016;291(1):126–141. doi: 10.1074/jbc.M114.624619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hargrave KE, Woods S, Millington O, Chalmers S, Westrop GD, Roberts CW. Multi-omics studies demonstrate Toxoplasma gondii-induced metabolic reprogramming of murine dendritic cells. Front. Cell. Infect. Microbiol. 2019 doi: 10.3389/fcimb.2019.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raetz M, et al. Parasite-induced TH1 cells and intestinal dysbiosis cooperate in IFN-γ-dependent elimination of Paneth cells. Nat. Immunol. 2013;14(2):136–142. doi: 10.1038/ni.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vallochi AL, Teixeira L, Oliveira KDS, Maya-Monteiro CM, Bozza PT. Lipid droplet, a key player in host–parasite interactions. Front. Immunol. 2018;9:1022. doi: 10.3389/fimmu.2018.01022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Augusto L, Amin PH, Wek RC, Sullivan WJ. Regulation of arginine transport by GCN2 eIF2 kinase is important for replication of the intracellular parasite Toxoplasma gondii. PLoS Pathog. 2019 doi: 10.1371/journal.ppat.1007746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olson WJ, Di Genova BM, Gallego-Lopez G, Dawson AR, Stevenson D, Amador-Noguez D. Dual metabolomic profiling uncovers Toxoplasma manipulation of the host metabolome and the discovery of a novel parasite metabolic capability. PLoS Pathog. 2020 doi: 10.1371/journal.ppat.1008432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fox BA, Gigley JP, Bzik DJ. Toxoplasma gondii lacks the enzymes required for de novo arginine biosynthesis and arginine starvation triggers cyst formation. Int. J. Parasitol. 2004;34(3):323–331. doi: 10.1016/j.ijpara.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 55.Lima TS, Lodoen MB. Mechanisms of human innate immune evasion by Toxoplasma gondii. Front. Cell. Infect. Microbiol. 2019 doi: 10.3389/fcimb.2019.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gold DA, et al. The Toxoplasma dense granule proteins GRA17 and GRA23 mediate the movement of small molecules between the host and the parasitophorous vacuole. Cell Host Microbe. 2015;17(5):642–652. doi: 10.1016/j.chom.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Percie-du-Sert N, et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. PLoS Biol. 2020;18:e3000410. doi: 10.1371/journal.pbio.3000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Braga-Silva CF, et al. Homeopathic remedy made from Toxoplasma gondii reduces the number of bradyzoites in mice infected by the protozoan. Eur. J. Integr. Med. 2015;7(5):517–524. doi: 10.1016/j.eujim.2015.05.002. [DOI] [Google Scholar]
- 59.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Avershina E, Trine F, Knut RD. Novo semi-alignment of 16S rRNA gene sequences for deep phylogenetic characterization of next generation sequencing data. Microbes Environ. 2013;28(2):211–216. doi: 10.1264/jsme2.ME12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kruskal JB. Nonmetric multidimensional scaling: A numerical method. Psychometrika. 1964;29:115–129. doi: 10.1007/BF02289694. [DOI] [Google Scholar]