Abstract
A critical barrier to the development of a human immunodeficiency virus (HIV) cure is the lack of a scalable animal model that enables robust evaluation of eradication approaches prior to testing in humans. We established a humanized mouse model of latent HIV infection by transplanting “J-Lat” cells, Jurkat cells harboring a latent HIV provirus encoding an enhanced green fluorescent protein (GFP) reporter, into irradiated adult NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice. J-Lat cells exhibited successful engraftment in several tissues including spleen, bone barrow, peripheral blood, and lung, in line with the diverse natural tissue tropism of HIV. Administration of tumor necrosis factor (TNF)-α, an established HIV latency reversal agent, significantly induced GFP expression in engrafted cells across tissues, reflecting viral reactivation. These data suggest that our murine latency (“μ-Lat”) model enables efficient determination of how effectively viral eradication agents, including latency reversal agents, penetrate and function in diverse anatomical sites harboring HIV in vivo.
Keywords: Humanized mouse, HIV, HIV latency, latency reversal, shock and kill, antiviral gene therapy
Introduction
The advent of antiretroviral therapy (ART) has dramatically reduced morbidity and mortality for human immunodeficiency virus (HIV)-infected individuals with access to healthcare in resource-rich countries. However, despite years of potent therapy, eradication of infection is not achieved due to the persistence of HIV latently-infected cells during treatment(1). Accumulating evidence suggest that “non-AIDS” cardiovascular, renal and hepatic diseases are amplified by HIV infection, and the immune system may exhibit premature senescence even among patients with complete viral suppression(2). Moreover, although enormous progress has been made to provide ART in resource-limited settings, there are huge economic, political and operational challenges to reach the goal of universal access to lifelong treatment. These realities have created a pronounced interest in developing HIV cure strategies.
A number of approaches to achieve a sterilizing or functional cure for HIV infection are currently under investigation, including therapeutic vaccination, immunomodulatory approaches, therapeutic HIV latency reversal (the “shock and kill” strategy), as well as a number of gene therapy approaches(3,4). In all scenarios, it will be critical to have proper diagnostic tools and models in place to comprehensively evaluate performance and safety prior to deployment in a clinical setting. A critical barrier to the development of an HIV cure is the lack of an accessible and scalable preclinical animal model that enables robust evaluation of candidate eradication approaches prior to testing in humans(5). As a result, many promising curative approaches never graduate past the petri dish stage. Infection of nonhuman primates (NHP) with simian immunodeficiency virus (SIV) is an option and has been utilized extensively to study HIV/AIDS pathogenesis(6,7). Recent advancements have been made in optimizing ART regimens to achieve durable virus suppression and thus enable evaluation of HIV cure strategies in the SIV-NHP model(8–10). However, biological limitations remain since this model uses SIV and might not recapitulate human host-HIV interactions and HIV latency mechanisms(11–15). In addition, NHP experiments involve complex ethical considerations, and the high costs and labor requirements only allow small numbers of animals to be utilized in any given trial, limiting statistical power and generalizability(11).
Mouse models represent another alternative with lower costs, more convenient husbandry requirements, as well as greater scalability. In the context of HIV studies, a wide range of small animal models have been developed comprising knockout mouse (16–18), transgenic mouse(19–23), and humanized mouse models(6,24–29). Humanized mice are established by xenotransplantation of human cells or tissues in immunodeficient mouse strains. Most strains used in HIV research are derivatives of severe combined immunodeficient (SCID) mice, which harbor mutations in the gene coding for a DNA-dependent protein kinase catalytic subunit (Prkdc). These mice are “humanized” using two approaches: 1) human cells are injected with or without prior irradiation of mice or 2) portions of tissue are surgically implanted. Different cells have been used for injection, including human peripheral blood mononuclear cells (PBMCs), as in the hu-PBL-SCID mouse model(30), obtained from healthy(31) or HIV-infected ART suppressed individuals(32), or human hematopoietic stem cells (HSCs), as in the hu-HSC mouse model(33), or the more recently developed T-cell-only(34) and myeloid-only(35) mouse models (ToM and MoM). Implantation of fetal thymus and liver tissue fragments are used for the SCID-hu thy/liv(36,37) and bone marrow/liver/thymus (BLT)(38,39) mouse models. Although all these model systems have contributed to our understanding of HIV pathogenesis and persistence, key limitations remain that need to be addressed in order to fully exploit the potential of these small animal models in HIV cure research. Hu-PBL-SCID mice struggle with the development of graft versus host disease (GVHD) which renders this model inapplicable for long-term studies involving HIV persistence. The generation of SCID-hu thy/liv and BLT mice is limited due to the need for surgical implantation and a limited supply of tissue. Moreover, these models as well as hu-HSC, ToM and MoM mice rely on the engraftment of cells or tissues typically derived from human fetal specimens, which in light of recent changes in U.S. federal policies face significant challenges, as ethical, legal, and political considerations surrounding the use of fetal tissue in scientific research have made it increasingly difficult to obtain such material(40). A major limitation shared by all these models remains the low frequency of HIV latently-infected cells, which impacts the applicability of these models as robust in vivo test bases for HIV cure strategies.
In the present study, we therefore pursued a new approach and transplanted J-Lat 11.1 cells (J-Lat cells) into irradiated adult NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice. J-Lat cell clones have been derived from Jurkat cells, an immortalized human T lymphocyte cell line. Each cell harbors a single latent HIV provirus encoding an enhanced green fluorescent protein (GFP) reporter in place of the nef gene(41). The proviruses within the J-Lat clones are typically latent due to epigenetic repression(42) and were selected to be responsive to TNF-α stimulation, resulting in viral LTR-driven GFP expression. J-Lat 11.1 cells were chosen for this model as they displayed the greatest viral reactivation upon TNF-α treatment in cell culture settings with respect to other clones(43). Our data show robust engraftment of J-Lat cells in several tissues three weeks after injection as well as significant reactivation of these latently-infected cells in vivo upon intravenous administration of the established latency reversal agent (LRA) tumor necrosis factor (TNF)-α. By applying an established and widely-utilized HIV latency reporter cell line(41,43,44), we circumvent the need for fetal or any other donor-derived tissue, and achieve high frequencies of HIV latently-infected cells in vivo on a short experimental time scale. In addition, the presence of the GFP reporter cassette in the integrated viral genome provides for quick and convenient assessment of viral reactivation using flow cytometry or microscopy. Moreover, by taking advantage of an HIV reporter cell line, our murine latency (μ-Lat) model represents a highly reproducible platform that allows for a simple transition from in vitro screening approaches to an in vivo system enabling the evaluation of bioavailability and pharmacokinetics of candidate regimens. Although not intended to serve as a pathophysiology model, we present this approach as a scalable, accessible, and cost-effective preclinical testbed to evaluate the safety, tolerability, and performance of HIV cure strategies in distinct anatomical niches.
Materials and Methods
Cell culture and treatment:
J-Lat 11.1 cells (kindly provided by Dr. Eric Verdin) contain an integrated latent full-length HIV genome harboring a mutation in the env gene and GFP in place of the nef gene as a reporter for transcriptional activity of the provirus(45). J-Lat 11.1 cells were grown in media composed of RPMI 1640 (Gibco) supplemented with 10% fetal bovine serum (FBS) (Corning) and 1% Penicillin-Streptomycin (Gibco). Cells were cultured at 37°C in a humidified incubator containing 5% CO2. To test the potency of latency reversal agents (LRAs), 1 x 106 cells in 1 ml RPMI were left untreated (negative control), incubated in 0.5% DMSO (vehicle control), 20 nM PMA/1 μM Ionomycin (positive control) or 20 ng/μl TNF-α for 24h.
Mice:
The work was approved by the Institutional Animal Care and Use Committee guidelines at Covance Laboratories, Inc. (San Carlos, CA) under Animal Welfare Assurance A3367-01 and protocol number IAC 2185 / ANS 2469. Animal husbandry was carried out according to the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Mice were sacrificed in accordance with the guidelines from the American Veterinary Medical Association.
Adult female and male mice (≥8 weeks old) were included in this study and were maintained at Vitalant Research Institute (VRI). Breeding pairs of NSG mice were obtained from Jackson Laboratories (Bar Harbor, ME), and were bred and maintained at VRI in a vivarium free from >40 murine pathogens as determined through biannual nucleic acid testing (Mouse Surveillance Plus PRIA; Charles River) of sentinel mice exposed to mixed bedding. Mice were maintained in sterile, disposable microisolator cages (Innovive, Inc.), which were changed every 14 days. Environmental enrichment was provided by autoclaved cotton Nestlets (Ancare Corp.) and GLP-certified Bio-Huts (Bio-Serv). Feed consisted of sterile, irradiated diet of Teklad Global 19% protein diet (Envigo) with free access to sterile-filtered, acidified water (Innovive, Inc.). Several days prior to radiation and 3 weeks following radiation, mice were fed with irradiated Global 2018 rodent diet with 4100 ppm Uniprim® (Envigo).
J-Lat cell surface marker staining:
For each staining, 1 x 106 J-Lat cells were washed once with PBS (Gibco), resuspended in 100 μl PBS, and stained with Zombie dye (Cat. #423105, BioLegend) according to manufacturer’s protocol (1:100 dilution) to enable subsequent discrimination between live and dead cells. 10 min after incubation with the Zombie dye, human CD45-PE (clone HI30, Cat. #304039, BioLegend), CD4-PE (clone OKT4, Cat. #317410, BioLegend), TCR α/β-PE (clone IP26, Cat. #306708, BioLegend), CD27-PE (clone M-T271, Cat. #356406, BioLegend), CD147-APC (clone HIM6, Cat. #306214, BioLegend), CD29-APC (clone TS2/16, Cat. #303008, BioLegend), and HLA-ABC-APC/Cy7 (clone W6/32, Cat. #311426, BioLegend) antibodies were added respectively, and were incubated for another 20-30 min at room temperature (RT) in the dark. Cells were then washed with 2 ml of cell staining buffer (Cat. #420201, BioLegend), resuspended in 300 μl PBS and measured using an LSR II flow cytometer (BD Biosciences). Respective Isotype stained samples were used as control. At least 10,000 events were recorded for each sample.
J-Lat cell transplantation into mice:
If not otherwise stated, J-Lat cell transplantation was performed as follows: mice were irradiated with 175 cGy radiation dose using a RS2000 X-Ray Biological Irradiator (RAD Source Technologies, Inc.) 3 hours prior to cell transplantation. Mice were transplanted with 10 x 106 J-Lat cells in a volume of 200 μl by tail vein injection. J-Lat cell transplanted mice as well as control mice (left untransplanted) were sacrificed and analyzed within 25 days of cell transplantation.
Engraftment analysis of J-Lat cells in mice by flow cytometry:
Transplanted mice were sacrificed by cervical dislocation 3 weeks post injection (or as indicated in figure legends). In one experiment, BM, brain, gut, intraepithelial lymphocytes (IEL) from gut tissue, heart, spleen, lung, lymph nodes, and PB were harvested. In subsequent experiments, selected tissues were harvested including BM, spleen, lung, and PB. Single cell suspensions from the BM, spleen, and PB were prepared as described previously by Beyer et al.(46). Brain, gut, heart, lymph node, and lung specimens were processed as follows: after harvest, all samples were stored in PBS on ice. Brain and lymph node specimens were washed twice in PBS, brain specimens additionally cut into small fragments (3-4 mm2), followed by mashing the tissue fragments with a pestle and passaging the cell suspensions through 70 μm cell strainers. Gut, heart, and lung specimens were washed twice with PBS and then cut into 3-4 mm2 pieces in a petri dish in 1 ml digestion solution consisting of 1 mg/ml DE Collagenase (Cat. #011-1040, VitaCyte) and 100 U/ml DNase I (Cat. #D5025-15KU, Sigma-Aldrich) final concentration in HBSS (Gibco). Fragments were transferred to 50 ml falcon tubes, digestion solution was added to a final volume of up to 5 ml, and samples were incubated at 37°C for 30 min (gut, heart) or 50 min (lung). IEL from gut tissue were isolated as supernatant after digestion incubation. Afterwards, up to 5 ml of stop solution, consisting of 0.5% BSA (Cat. #A2153, MilliPoreSigma) and 2 mM EDTA (Cat. #E0306, Teknova) final concentration in PBS, was added to each sample to end the enzymatic digestion reaction. Single cell suspensions were prepared by passage through a 70 μm cell strainer and washed with PBS. For the analysis of J-Lat cell engraftment, single cells from harvested mouse tissues were stained with human CD147-APC or human CD147-APC combined with human CD29-APC, and additional Pacific Blue conjugate, mouse-specific lineage markers including CD45-Pacific Blue (clone 30-F11, Cat. #103126, BioLegend), TER-119-Pacific Blue (clone TER-119, Cat. #116232, BioLegend), and H-2KD-Pacific Blue antibodies (clone SF1-1.1, Cat. #116616, BioLegend). Cells were incubated for 30 min at RT in the dark. Zombie dye staining detected in the APC-Cy7 channel was used for discrimination of live and dead cells. Following staining, cells were washed and run on an LSR II flow cytometer, where at least 10,000 events were recorded per sample. Flow cytometry data were analyzed using FlowJo software (FlowJo, Inc.).
Treatment of J-Lat engrafted mice with latency reversal agent TNF-α:
Mice engrafted with J-Lat cells were treated with recombinant human TNF-α (Cat. #PHC3011, Gibco), a potent and well-known LRA. Briefly, NSG mice were transplanted with J-Lat cells and 3 weeks post-transplantation, mice received TNF-α (diluted in 200 μl PBS) intravenously at a dose of 20 μg/mouse(47). NSG mice injected with J-Lat cells and treated with PBS were used as vehicle control group.
After 24h of TNF-α treatment, mice were sacrificed to determine viral reactivation in PB, BM, lung and spleen tissues. Latency reversal of the HIV provirus was analyzed by comparing GFP expression of J-Lat cells in tissues of TNF-α treated mice vs the vehicle control group. GFP expression of engrafted J-Lat cells was determined by flow cytometry as described above.
Data analysis:
All data were analyzed using GraphPad Prism version 8.2 and are presented as mean ± the standard error of the mean (SEM). Applied statistical tests and analyses are described in figure legends. A P value of ≤0.05 was considered as statistically significant. Individual P values are indicated in figures.
Results
The human cell surface proteins CD147 and CD29 enable discrimination of J-Lat cells from mouse cells.
Our model involves transplantation of J-Lat cells (immortalized human T cells harboring a latent HIV provirus with a GFP reporter reflecting viral transcriptional activity) into irradiated adult NSG mice(41). We therefore searched for cell surface markers that identify engrafted J-Lat cells in a background of mouse cells, exhibiting three key features: 1) universal expression across J-Lat cells, 2) high per-cell expression on J-Lat cells, and 3) absence of expression on the surface of mouse cells.
Our candidate panel included seven cell surface proteins commonly known to be expressed in human CD4+ T cells: CD45, CD4, TCR α/β, CD27, CD147, CD29, and HLA-ABC. Four of these cell surface proteins were found to be expressed universally among the J-Lat population: CD45, CD147, CD29 and HLA-ABC (Figure 1A). The mean fluorescence intensity (MFI) of the four proteins was measured, reflecting the relative abundance of each protein on the cell surface. CD147 exhibited the highest MFI, followed by CD29 (Figure 1B). We next tested if the CD29 and CD147 antibodies (both APC conjugated to achieve maximum signal to noise ratio in the APC-channel) showed detectable binding to mouse cells obtained from different tissues. We harvested bone marrow (BM), lung, brain, gut, intraepithelial lymphocytes (IEL) from gut tissue, lymph nodes, heart and spleen from an untransplanted control mouse. In addition, based on Hoggatt et al.(48), who reported that blood parameters such as cell composition significantly varied depending on sampling manner and site, we performed peripheral blood (PB) harvest in three different ways: by retro-orbital bleed (r.-o.), tail vein bleed, or heart bleed. Human CD29 and CD147 staining was combined with antibodies targeting the mouse-specific lineage markers CD45, TER-119, and H-2Kd for a positive identification of mouse cells. While 100% of cultured J-Lat cells expressed CD29 and CD147 and were negative for mouse-specific markers (Figure 1C), the frequency of CD147+/CD29+ cells was negligible or absent across mouse tissues (Figure 1D and E). Most tissues harbored no CD147+/CD29+ cells, while 0.06% CD147+/CD29+ cells were detected in the lung and 1.37% CD147+/CD29+ cells were detected in PB upon heart bleed (Figure 1E). For both samples, lung and PB heart bleed, these percentages represent a single event detected to be positive for CD147/CD29 (Figure 1D and E). Thus, we concluded that among the tested cell surface proteins, human CD147 combined with CD29 function as a specific marker pair for the identification of J-Lat cells engrafted in mouse tissues.
Figure 1: The cell surface proteins CD147 and CD29 are abundantly and specifically expressed in J-Lat 11.1 cells.
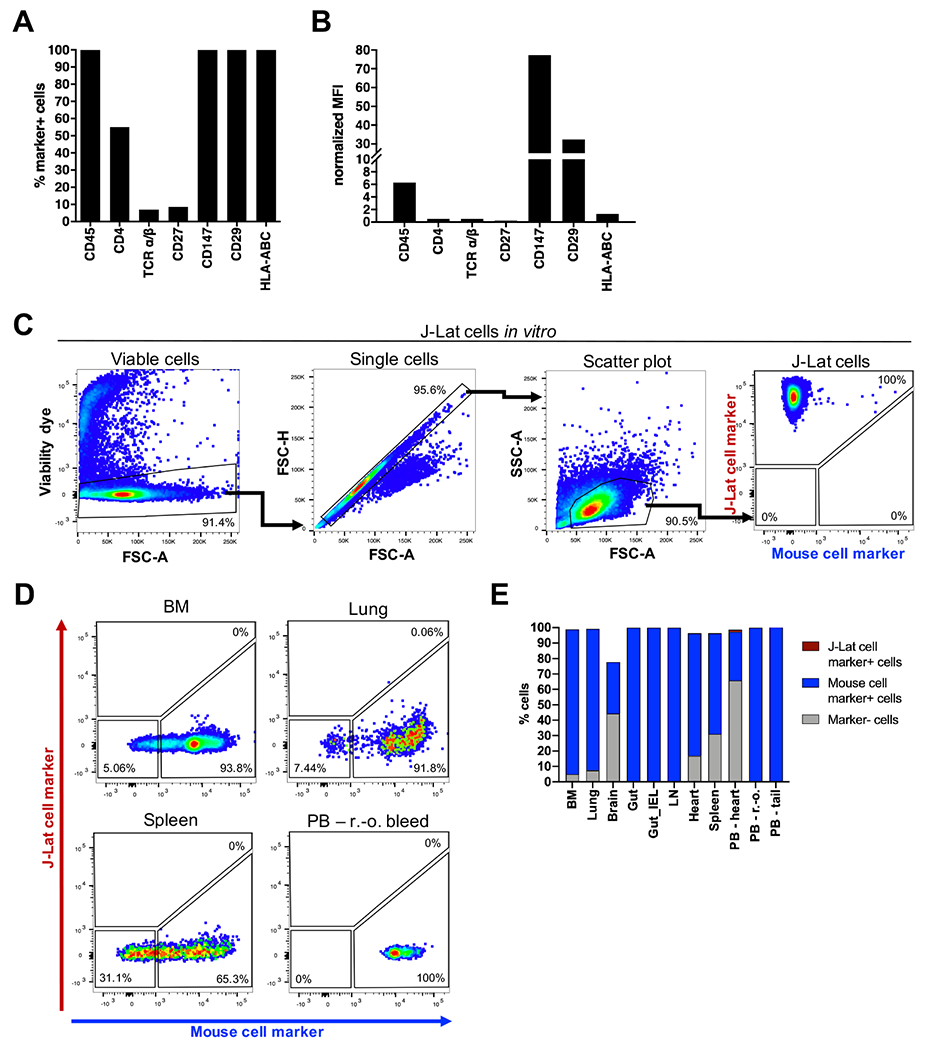
1 x 106 J-Lat cells were stained with antibodies targeting select cell surface proteins to (A) measure the frequency of marker-positive cells and (B) mean fluorescence intensity (MFI) of proteins on the cell surface using flow cytometry. The MFI of marker signal positive cells was normalized to signal positive cells of respective isotype control stained samples. A representative experiment is summarized in the bar graphs. (C) Gating strategy to identify human J-Lat cells upon multicolor staining with CD147/CD29-APC (J-Lat cell marker) and CD45/TER-119/H-2Kd-Pacific Blue (mouse cell marker) in vitro. (D) Representative flow cytometry plots show multicolor staining of single cell suspensions prepared from different mouse tissues obtained from an untransplanted control animal. Mouse cells were stained, measured, and analyzed in the same way as in vitro J-Lat cells to evaluate specificity and background signal of human CD29 and CD147 antibodies. (E) Single cell suspensions were prepared from 8 mouse tissues and three different harvest methods for peripheral blood (PB) to determine the applicability of the multicolor staining for subsequent engraftment studies. BM = bone marrow, IEL = intraepithelial lymphocytes, LN = lymph node, PB = peripheral blood, r.-o. = retro-orbital.
Establishment and optimization of J-Lat cell engraftment in humanized mice.
In the context of establishing a humanized mouse model, we examined and optimized several parameters simultaneously to achieve a highly reproducible and robust in vivo platform (Figure 2).
Figure 2: Schematic representation of parameters that were examined and optimized to establish the μ-Lat model.
J-Lat cell engraftment kinetics, J-Lat cell dosage, irradiation of recipient mice, and testing of J-Lat cell engraftment in different mouse strains were investigated.
In the following pilot experiments, we focused on J-Lat cell engraftment in the BM (Figure 3). To measure the kinetics of J-Lat cell engraftment, we harvested the BM at different time points post J-Lat cell injection (Figure 3A). We examined the effect of varying cell doses (Figure 3B) and irradiation of recipient mice (Figure 3C) on J-Lat cell engraftment levels, and compared engraftment levels in NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CMV-IL3,CSF2,KITLG)1Eav/MloySzJ (NSG-3GS) and NSG mice (Figure 3D). We detected J-Lat cells in the BM 18 days post cell injection (pci), and detection levels reached a significant increase 25 days pci compared to 1 and 3 days pci with an average of 16.3±8.6% engrafted J-Lat cells (Figure 3A). Using a J-Lat cell injection dose of 10 x 106 J-Lat cells compared to 5 x 106 J-Lat cells per mouse led to a higher but not significant increase in engraftment levels in the BM of nonirradiated NSG-3GS mice (Figure 3B). Nonirradiated NSG-3GS recipient mice harbored lower frequencies of engrafted J-Lat cells in the BM compared to irradiated NSG-3GS recipient mice (Figure 3C). Again, this difference was not significant but showed a slight increase in J-Lat cell engraftment levels when recipient mice were irradiated. Finally, when testing engraftment levels in different mouse strains, we found significantly higher numbers of J-Lat cells in NSG mice (Figure 3D). J-Lat cell engraftment levels in the BM 3 weeks pci of 10 x 106 J-Lat cells were 2.4 fold higher in NSG mice compared to NSG-3GS mice (Figure 3D). Based on our results, we concluded that irradiation of recipient NSG mice and a cell dose of 10 x 106 J-Lat cells per mouse administered through intravenous injection resulted in efficient and reproducible cell engraftment levels which peaked 3 weeks following cell transplantation.
Figure 3: Optimal engraftment levels of J-Lat cells are achieved 3 weeks post injection in irradiated NSG mice using 10 x 106 J-Lat cells for transplantation.
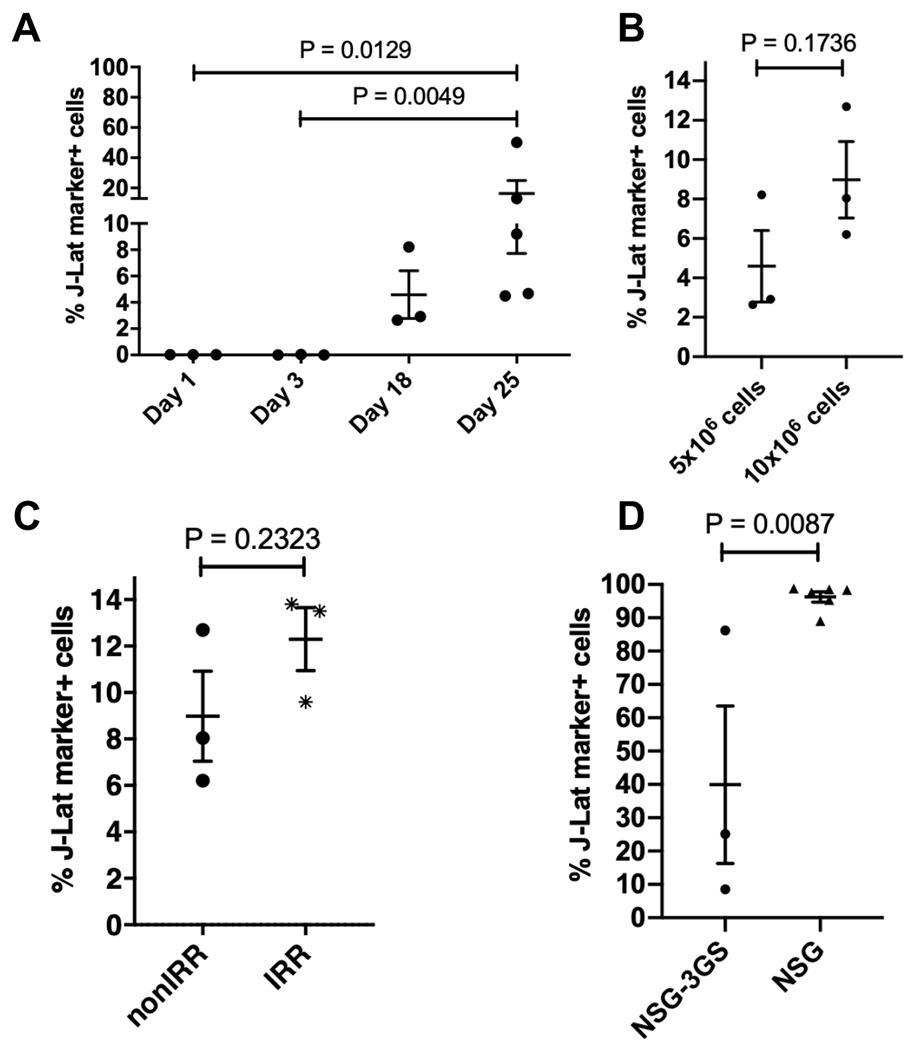
The parameters described in Figure 2 were examined and optimized concomitantly by focusing on engraftment levels in the BM. (A) J-Lat cell engraftment kinetics: Nonirradiated NSG-3GS mice were transplanted with 5 x 106 J-Lat cells and the BM harvested at indicated time points post cell injection. (B) J-Lat cell injection dosage: J-Lat cell engraftment levels in the BM of nonirradiated NSG-3GS were compared using different initial injection cell doses of 5 x 106 J-Lat cells (BM harvested 18 days post cell injection) or 10 x 106 J-Lat cells (BM harvested 14 days post cell injection). (C) Irradiation of recipient mice: NSG-3GS recipient mice were nonirradiated (nonIRR) or irradiated (IRR) before transplantation of 10 x 106 J-Lat cells, and engraftment levels were determined two weeks post cell transplantation. (D) Mouse strain selection: 10 x 106 J-Lat cells were injected either into irradiated NSG-3GS or irradiated NSG mice and engraftment levels in the BM were measured 3 weeks post cell injection and 24h post TNF-α treatment. Each dot represents an individual animal for the respective condition. Each plot represents an independent experiment with n ≥ 3. P values in (A) were determined using the nonparametric Kruskal-Wallis test with the uncorrected Dunn’s test for multiple comparisons. P values in (B-D) were determined using an unpaired, two-tailed t-test. A standard P<0.05 significance threshold was used.
J-Lat cells engraft successfully in several tissues in transplanted NSG mice.
Using the determined experimental conditions, we investigated J-Lat cell engraftment in several tissues, as listed above, that we harvested from 5 irradiated NSG mice 3 weeks pci (Figure 4). To analyze the expression of J-Lat and mouse markers on cells obtained from different mouse tissues, we followed the gating strategy as described before (see Figure 1C and Figure 4A and B). J-Lat cell engraftment levels were evaluated based on the frequency, MFI, and the absolute number of CD147+/CD29+ cells in the respective processed tissues (Figure 4C–E and Supplementary Figure 1). We found engrafted J-Lat cells in several tissues including the BM, lung, spleen, and PB (Figure 4A and B). The mean frequency of engrafted J-Lat cells varied across tissues from approximately 30% in the BM to 0.7% in the spleen (Figure 4C). While the MFI of the engrafted J-Lat cells was comparable within and across tissues (Figure 4D), the absolute number of engrafted J-Lat cells varied drastically. We detected the highest J-Lat cell amount in the BM, followed by the amount in the lung and then the spleen (Figure 4E). We did not detect engrafted J-Lat cells in the gut, IEL from the gut or heart tissues (Figure 4C–E). Processing brain and lymph node tissues yielded only low overall cell numbers that were evaluable (Supplementary Figure 1A and B). While J-Lat cells were identified in processed brain tissues at high frequency (Supplementary Figure 1C), the actual number of engrafted J-Lat cells was on average 5 cells (Supplementary Figure 1E). We thus decided to exclude the brain in subsequent experiments due to the low overall cell numbers. We did not detect engrafted J-Lat cells in the lymph node (Supplementary Figure 1B–E). When focusing on engraftment in the PB, we found that heart and r.-o. bleed yielded a substantial higher frequency of engrafted J-Lat cells compared to tail vein bleed (Figure 4C). However, PB harvest by r.-o. bleeding showed the highest MFI, and more importantly, the highest absolute number of engrafted J-Lat cells (Figure 4D and E). Taken together, we found different engraftment levels based on the sample collection method as observed by Hoggatt et al.(48). Thus, due to considerable engraftment levels and practicality of collection, we decided to harvest PB in subsequent experiments via r.-o. bleeding.
Figure 4: J-Lat cells engraft successfully in several tissues in transplanted NSG mice.
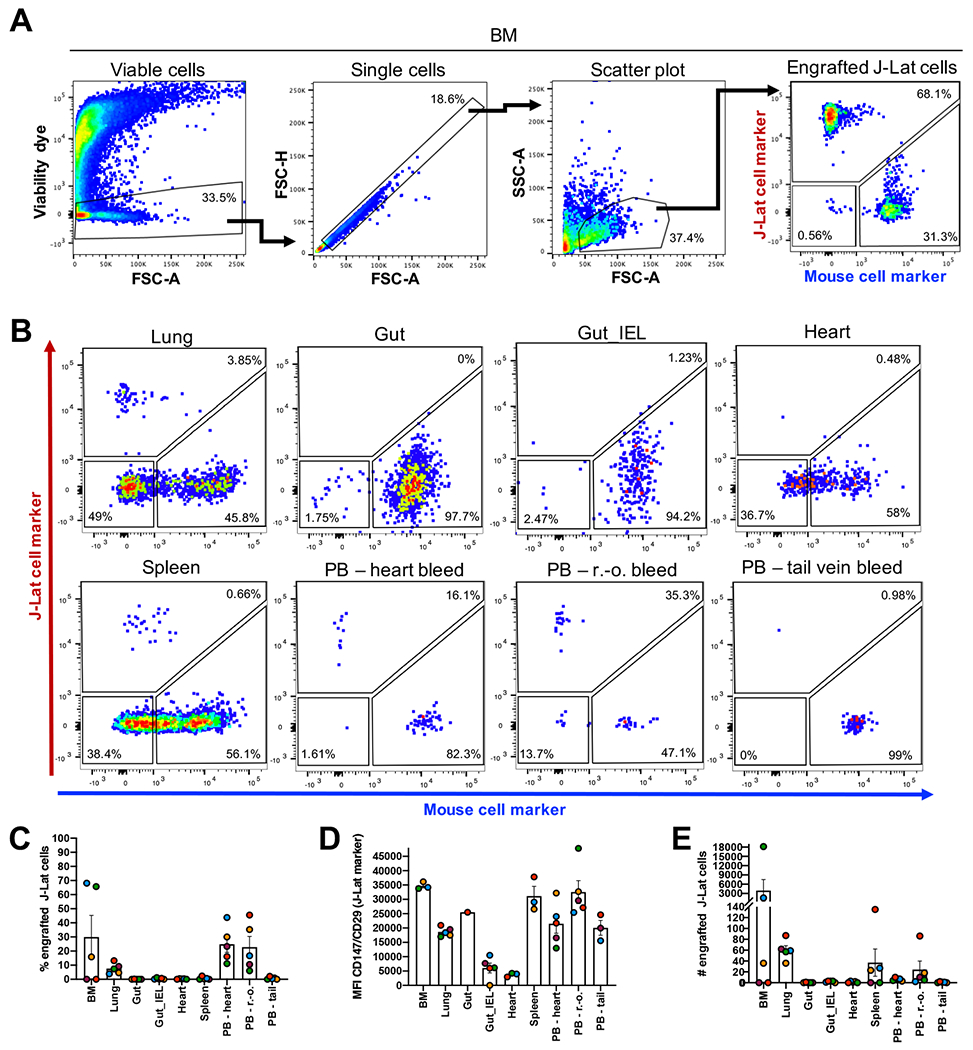
(A) Gating strategy to identify human J-Lat cells in harvested mouse tissues exemplified here with BM. (B) Representative flow cytometry plots are shown for engraftment levels across tissue sites observed at necropsy and engraftment levels in PB (n = 5) using three different harvest approaches: r.-o. bleeding, tail vein bleeding, and heart bleeding. Bar graphs summarize (C) frequency, (D) MFI, and (E) number of engrafted J-Lat cells in the respective tissue. Each data point represents an individual animal. Colors indicate tissues harvested from the same animal. Error bars show the standard error of the mean (SEM).
Based on these results and parameter thresholds applied in concert (frequency > 0.005; MFI > 15,000; absolute number > 10), we focused in subsequent experiments on J-Lat engraftment in the BM, lung, spleen and PB via r.-o. bleed and established a suitable engraftment and sample collection protocol to drive the model.
J-Lat cells maintain low GFP expression upon engraftment in selected tissues.
Once we confirmed reproducible and robust engraftment in several tissues, we wanted to investigate GFP expression levels of engrafted J-Lat cells. As described before, the GFP reporter integrated into the latent provirus in J-Lat cells is under the control of the proviral LTR and thus reflects viral transcriptional activity. High GFP expression levels would thus indicate spontaneous viral reactivation in vivo and render the model inapplicable for testing LRAs. We measured J-Lat cell engraftment levels and GFP background signal of engrafted J-Lat cells in the selected tissues (BM, lung, spleen, PB r.-o.) of 10 NSG mice 3 weeks pci (Figure 5). We applied our previously described gating strategy to identify engrafted J-Lat cells based on CD147/CD29 expression and measured GFP signal within this compartment (Figure 5A–C). Engraftment of J-Lat cells was detected in the BM, lung, spleen, and PB r.-o. as observed before (Figure 5D). Across these tissues, GFP expression levels of engrafted J-Lat cells remained low and frequencies did not exceed an average of 4% GFP-positive cells (Figure 5E). The results showed that while these mouse tissues harbored substantial levels of HIV latently-infected cells in vivo, engraftment of J-Lat cells in the tested tissues did not lead to spontaneous reactivation of viral transcription.
Figure 5: Engrafted J-Lat cells exhibit low basal levels of GFP expression 3 weeks post cell transplantation.
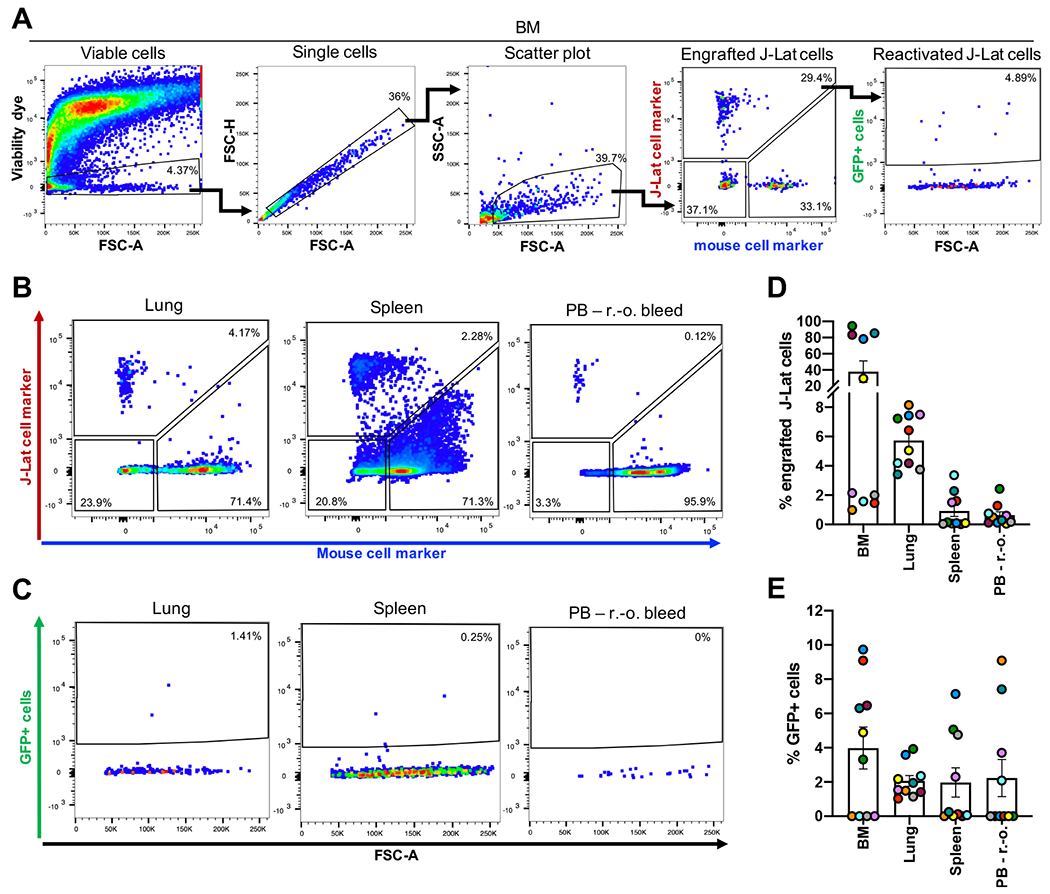
(A) Gating strategy to identify human J-Lat cells and GFP background signal of engrafted J-Lat cells in harvested mouse tissues illustrated here with a representative BM sample. Representative flow cytometry plots are shown for (B) engraftment levels and (C) GFP background signal across selected tissues observed at necropsy (n = 10). Bar graphs summarize (D) cell frequency and (E) GFP expression levels of engrafted J-lat cells. Each data point represents an individual animal. Colors indicate tissues harvested from the same animal. Error bars show SEM.
TNF-α treatment induces GFP expression in tissue engrafted J-Lat cells reflecting reactivation of latent provirus in vivo.
Finally, we sought to determine if HIV LRA administration would result in viral reactivation in vivo. Viral reactivation was measured based on the frequency of GFP expressing cells within engrafted J-Lat cells following LRA treatment (Figure 6A).
Figure 6: TNF-α treatment reactivates latent HIV in vivo in the μ-Lat model.
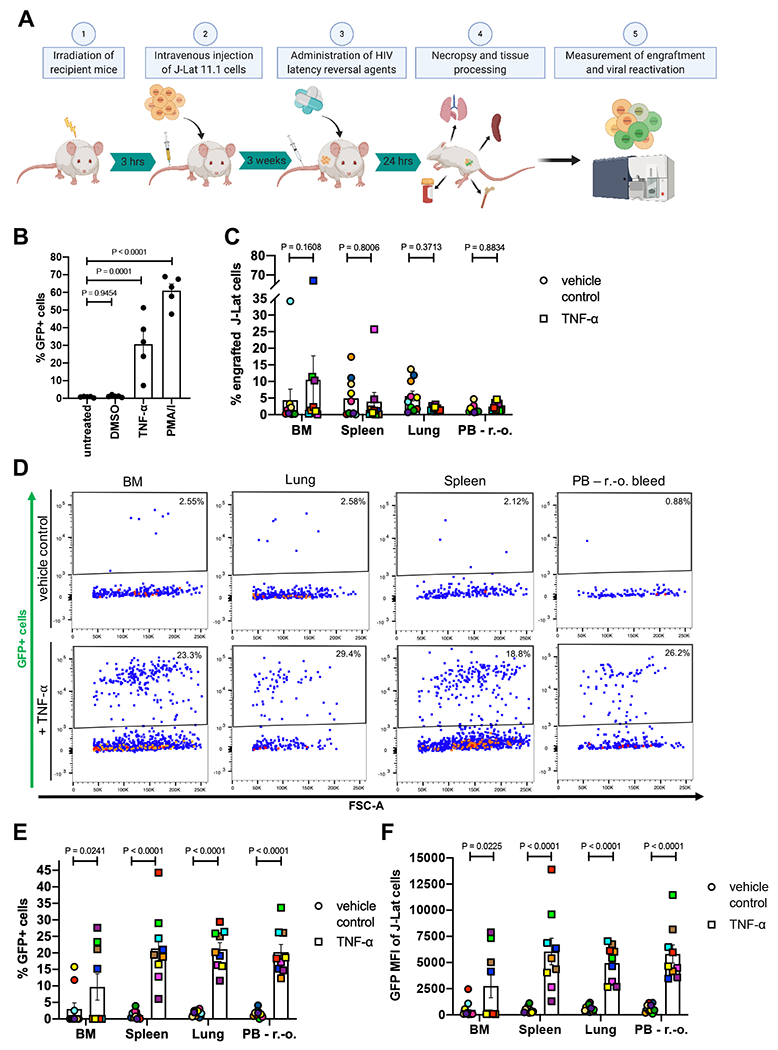
(A) Schematic representation of the procedure to test LRAs in vivo using the μ-Lat model: (1) Mice receiving cells of interest for transplantation are irradiated 3 hours prior to cell injection. (2) Each mouse receives 10 x 106 J-Lat cells via intravenous injection. (3) Three weeks post injection (21 days) mice are treated for 24h with LRAs of interest, followed by (4) necropsy, tissue harvest and processing, and preparation of single-cell suspensions. (5) Single-cell suspensions are stained for J-Lat and mouse cell markers to assess engraftment levels via flow cytometry. Reactivation of latent provirus following LRA treatment is assessed by measuring GFP expression via flow cytometry (the J-Lat provirus contains an LTR-driven GFP reporter). (B) J-Lat cells were treated in vitro for 24h with 0.5% DMSO (vehicle control), 20 ng/μl TNF-α, 20 nM PMA/1 μM Ionomycin (positive control) or left untreated (negative control) to assess viral reactivation based on GFP expression (n = 5). (C) J-Lat engraftment levels were measured 24h post tail vein injection of 20 μg TNF-α (n = 9) or vehicle control (PBS, n = 10) in BM, spleen, lung and PB of mice. (D) Representative flow cytometry plots demonstrating the effect of vehicle control (top panel) or TNF-α treatment (bottom panel) on viral reactivation (GFP expression) in engrafted J-Lat cells. The induction of GFP expression among engrafted J-Lat cells across tissues is summarized as (E) frequency of GFP+ cells and (F) GFP MFI. One-way ANOVA and uncorrected Fisher’s LSD for multiple comparisons was used to analyze (B) in vitro reactivation data, and a mixed effects model and uncorrected Fisher’s LSD for multiple comparisons was used to analyze (C) in vivo engraftment and (E-F) viral reactivation data. Colors indicate specific animals and tissues obtained from the same animal. Error bars show SEM.
The proviruses within the J-Lat family of clones were selected to be responsive to TNF-α stimulation, resulting in viral LTR-driven GFP expression(41). Previous studies have shown, that among the established clones, J-Lat 11.1 cells displayed greatest viral reactivation upon TNF-α treatment in cell culture settings(43). 24h in vitro treatment of J-Lat cells with 20 ng/μl TNF-α resulted in a significant increase of GFP-positive cells compared to mock-treated (0.5% DMSO) and untreated cells, which were largely GFP-negative (Figure 6B). Therefore, we performed in vivo reactivation experiments using 20 μg of TNF-α as an LRA (with PBS as a vehicle control)(47). 24h following TNF-α tail vein injection, effects on tissue engraftment and reactivation of latent provirus were estimated in a cross-sectional manner at necropsy, comparing 9 animals treated with TNF-α versus 10 vehicle control animals. J-Lat cell engraftment and GFP expression of J-Lat cells were analyzed as described in Figure 5A. Analyses of all tissues demonstrated no significant effect of TNF-α treatment on J-Lat cell engraftment levels within the respective compartment (Figure 6C). However, TNF-α treatment did lead to significant increases in the frequency of GFP-positive cells as well as GFP MFI among engrafted J-Lat cells (Figure 6D–F). Comparing the two populations (TNF-α vs vehicle control) demonstrated a GFP expression fold-change of 3.2 in the BM, 17.8 in the spleen, 11.7 in the lung, and 12.7 in the PB, illustrating potent and significant viral reactivation.
Discussion
The development of an HIV cure will be accelerated by the deployment of a convenient and cost-effective animal model that enables the determination of an agent’s therapeutic efficacy as it permeates through tissue-specific barriers and the circulatory, respiratory and excretory systems. We present a novel humanized mouse model of HIV latency that aims to address this gap. In this study, we established a reliable method to discriminate between J-Lat and mouse cells (Figure 1 and Figure 4) and demonstrated robust engraftment of these cells in multiple tissues sites (Figure 4 and Figure 5). Significant viral reactivation was observed in TNF-α-treated animals with respect to vehicle control-treated animals (Figure 6D–F). Importantly, although in vivo treatment with TNF-α induced GFP expression and thus viral reactivation by 3.2 (BM), 17.8 (spleen), 11.7 (lung), and 12.7 (PB) -fold compared to vehicle control treated animals (Figure 6E), TNF-α treatment in vitro induced a 25.3-fold increase in GFP expressing cells (Figure 6B). Moreover, the observed variability in TNF-α-mediated viral reactivation across tissue sites demonstrates that the efficacy of latency reversal approaches can differ dramatically between distinct anatomical niches due to pharmacokinetic factors and drug penetration efficiencies. These data emphasize the importance of testing the efficacies of promising and potent regimens in an in vivo system, and reinforce the relevance of the μ-Lat model.
Although not intended to serve as a pathophysiology model, the μ-Lat platform offers key advantages over existing preclinical models focused on HIV eradication. Firstly, the model is highly efficient; the timeframe to generate mouse colonies that are ready for administration of experimental therapies is approximately three weeks. In comparison, even the simplified humanized mouse model introduced by Kim et al.(31) requires four weeks for robust engraftment of intraperitoneally injected human PBMCs in NSG mice and an additional 5 weeks of incubation upon HIV infection. Secondly, the presence of the GFP reporter cassette in the integrated J-Lat viral genome allows for rapid and convenient assessment of HIV latency reversal in vivo, eliminating the requirement for PCR or culture-based diagnostics to determine LRA potency. Thirdly, the model is characterized by high frequencies of HIV latently-infected cells (e.g. in some experiments exceeding 30% engraftment in BM), and these cells are distributed in a number of relevant anatomical sites that are central to HIV persistence. This stands in contrast to organoid-based models (e.g. the SCID-hu thy/liv mouse model) that exclusively involves viral colonization within a xenografted tissue(49). Lastly, the model is highly scalable due to the low costs and limited labor requirements of the method involving the easily and widely available J-Lat cell line (provided free of charge via the NIH AIDS Reagent Program). Importantly, the clonal nature of cell lines promotes consistency and high reproducibility across laboratories, while allowing for an easy transition from the petri-dish stage to an in vivo evaluation system.
Similar to other animal models, there are caveats associated with the μ-Lat model. Although the GFP reporter present within the J-Lat cell conveniently broadcasts transactivation of the viral LTR and induction of viral transcription (informative precursor steps that may lead to the production of viral antigen and release of virus), other assays will need to be applied to the μ-Lat model to specifically quantify viral production or clearance of infected cells mediated by a candidate HIV eradication agent. In addition, the efficiency of the model is largely driven by the administration of an HIV latently-infected cell line, rather than differentiated primary cells. As presented here, a single latent clone with a single proviral integration site was injected into mice. Proviral integration site is known to affect viral latency and responsiveness to LRA administration(4,50–52). However, this issue could be ameliorated by mixing different HIV latently-infected cell clones at specific ratios, thereby achieving higher integration site heterogeneity to more accurately represent in vivo variability. For instance, there are 11 J-Lat clones currently available via the NIH AIDS Reagent Program, each of which is characterized by a distinct proviral integration site. These clones often behave differently from each other as well when exposed to LRAs in vitro, likely reflecting a diversity of molecular mechanisms reinforcing viral latency across clones(43). This mechanistic diversity could further enhance the predictive potential of the μ-Lat model.
Beyond concerns regarding integration site heterogeneity, the immortalized nature of the J-Lat cell line may impact molecular and regulatory pathways that affect HIV latency. However, the widespread usage of the J-Lat model and its derivative clones to examine viral latency and to evaluate the efficacy of HIV cure strategies in vitro speak to the model’s utility(53–62). Ample data from primary cell-based models of latency and experiments involving ex vivo administration of LRAs to cells obtained from HIV-infected individuals on ART suggest that differences between applied models and between individuals can have dramatic effects on the establishment, maintenance, and reversal of HIV latency(43,63,64). Moreover, profiling of HIV latency in multiple tissue sites has demonstrated that the nature of viral latency may even vary extensively within a single infected individual(65–67). Therefore, it needs to be considered that any applied model system or cells from any given individual may introduce biases that impact generalizability, just as a cell line-based system(26).
Our work described here represents a proof of concept, demonstrating that the engraftment of a cell line-based model of HIV latency may constitute a useful testbed for HIV cure strategies. This general approach is highly versatile and should allow for a broad range of infected cell types to be examined in vivo. For example, HIV latency in the myeloid compartment is likely critical to viral persistence(35,68–71), and multiple reports suggest this compartment may respond quite differently to curative approaches, as compared to lymphoid reservoirs(72,73). It may be fruitful to inject the U1 HIV chronically-infected promonocytic cell line(74) into NSG mice to examine LRA responses in myeloid cells. Building further on the myeloid theme, the central nervous system (CNS) compartment is an important viral sanctuary site in the setting of ART(73,75–78), and HIV eradication approaches will almost certainly face unique challenges in this niche(78,79). As direct injection of human cells into the murine CNS has been used successfully as an engraftment approach(80–84), site-specific injection of the recently developed HC69.5 HIV latently-infected microglial cell line(85,86) into the brains of NSG mice may provide a convenient platform to gauge CNS-focused cure strategies.
Beyond preclinical investigation of LRAs, the μ-Lat framework may provide a convenient model system to evaluate gene therapy-based HIV eradication approaches. Gene therapy approaches targeting HIV infection generally fall into two categories: 1) Gene editing can be used to target or “excise” the HIV provirus directly in infected cells as an eradication approach(87,88) or 2) Editing can be used to modulate host cells to render them refractory to HIV infection and/or potentiate antiviral immune responses(89). In the former case, in vivo delivery of the gene therapy modality will likely be necessary to pervasively attack the HIV reservoir within a broad range of tissue sites in infected individuals. The μ-Lat model is well-suited to investigate gene delivery in this context, as it is characterized by robust engraftment of HIV latently-infected cells into diverse anatomical niches. This will allow for efficient assessment of gene therapy vector dissemination and antiviral function across anatomic sites. The LTR-driven GFP cassette within the J-Lat integrated provirus may facilitate this assessment; direct targeting of the GFP sequence may be used to examine vector trafficking and delivery, while specific targeting of the HIV LTR as a cure approach would be associated with a convenient readout (relative loss of GFP expression upon induced latency reversal).
In summary, the μ-Lat model is optimized for efficient and scalable evaluation of select HIV eradication approaches in vivo, allowing determination of therapeutic efficacy in addition to essential safety, tolerability, and pharmacokinetic parameters. Further development and diversification of the μ-Lat model system may enable convenient testing of HIV eradication approaches, including antiviral gene therapy strategies, in a range of cell types and tissue sites in vivo.
Supplementary Material
Acknowledgments
This study was supported by the National Institutes of Health grants R01 AI150449 (SKP) and R01 MH112457 (SKP), and was additionally supported by a grant from the National Institutes of Health, University of California San Francisco-Gladstone Institute of Virology & Immunology Center for AIDS Research (P30 AI027763). We thank Dr. Roland Schwarzer for valuable input and careful review of the manuscript.
Nonstandard abbreviations
- ART
antiretroviral therapy
- BM
bone marrow
- GFP
green fluorescence reporter
- GVHD
graft versus host disease
- HIV
human Immunodeficiency virus
- HSC
hematopoietic stem cell
- IEL
intraepithelial lymphocytes
- LN
lymph node
- LRA
latency reversal agent
- LTR
long terminal repeats
- MFI
mean fluorescence intensity
- NHP
nonhuman primates
- NSG
NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ
- NSG-3GS
NOD.Cg-Prkdcscid Il2rgtm1Wj/ Tg(CMV-IL3,CSF2,KITLG)1Eav/MloySzJ
- PBMC
peripheral blood mononuclear cell
- PMA
phorbol-myristate-acetate
- r.-o.
retro-orbital
- SCID
Severe combined immunodeficiency
- SIV
simian immunodeficiency virus
- TNF
tumor necrosis factor
Footnotes
Conflict of interest statement
The authors have declared that no conflict of interest exists.
References
- 1.Wong JK, Hezareh M, Günthard HF, Havlir DV, Ignacio CC, Spina CA, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science (80- ) [Internet]. 1997. Nov 14 [cited 2019 Nov 15];278(5341):1291–5. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9360926 [DOI] [PubMed] [Google Scholar]
- 2.Piot P, Bartos M, Ghys PD, Walker N, Schwartländer B. The global impact of HIV/AIDS. Nature [Internet]. 2001. Apr 19 [cited 2019 Nov 15];410(6831):968–73. Available from: http://www.nature.com/articles/35073639 [DOI] [PubMed] [Google Scholar]
- 3.Pitman MC, Lau JSY, McMahon JH, Lewin SR. Barriers and strategies to achieve a cure for HIV. Lancet HIV [Internet]. 2018. Jun [cited 2019 Nov 15];5(6):e317–28. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0031938416312148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwarzer Gramatica, Greene. Reduce and Control: A Combinatorial Strategy for Achieving Sustained HIV Remissions in the Absence of Antiretroviral Therapy. Viruses [Internet]. 2020. Feb 8;12(2):188. Available from: https://www.mdpi.com/1999-4915/12/2/188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Policicchio BB, Pandrea I, Apetrei C. Animal Models for HIV Cure Research. Front Immunol [Internet]. 2016. Jan 28 [cited 2019 Nov 15];7(JAN):12. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26858716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nixon CC, Mavigner M, Silvestri G, Garcia JV. In vivo models of human immunodeficiency virus persistence and cure strategies. J Infect Dis. 2017;215(Suppl 3):S142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denton PW, Søgaard OS, Tolstrup M. Using animal models to overcome temporal, spatial and combinatorial challenges in HIV persistence research. J Transl Med [Internet]. 2016. Dec 9;14(1):44. Available from: http://www.translational-medicine.com/content/14/1/44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shytaj IL, Norelli S, Chirullo B, Della Corte A, Collins M, Yalley-Ogunro J, et al. A highly intensified ART regimen induces long-term viral suppression and restriction of the viral reservoir in a simian AIDS model. PLoS Pathog. 2012;8(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinoso JB, Rabi SA, Blankson JN, Gama L, Mankowski JL, Siliciano RF, et al. A Simian Immunodeficiency Virus-Infected Macaque Model To Study Viral Reservoirs That Persist during Highly Active Antiretroviral Therapy. J Virol. 2009;83(18):9247–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.North TW, Higgins J, Deere JD, Hayes TL, Villalobos A, Adamson L, et al. Viral Sanctuaries during Highly Active Antiretroviral Therapy in a Nonhuman Primate Model for AIDS. J Virol. 2010;84(6):2913–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar N, Chahroudi A, Silvestri G. Animal models to achieve an HIV cure. Curr Opin HIV AIDS [Internet]. 2016. Jul;11(4):432–41. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0031938416312148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature [Internet]. 2004. Jul 7;430(6999):569–73. Available from: http://www.nature.com/articles/nature02777 [DOI] [PubMed] [Google Scholar]
- 13.Mariani R, Chen D, Schröfelbauer B, Navarro F, König R, Bollman B, et al. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114(1):21–31. [DOI] [PubMed] [Google Scholar]
- 14.Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, et al. SAMHD1 restricts HIV-1 infection in resting CD4 + T cells. Nat Med. 2012;18(11):1682–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manel N, Littman DR. Hiding in plain sight: How HIV evades innate immune responses. Cell [Internet]. 2011;147(2):271–4. Available from: 10.1016/j.cell.2011.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rehwinkel J, Maelfait J, Bridgeman A, Rigby R, Hayward B, Liberatore RA, et al. SAMHD1-dependent retroviral control and escape in mice. EMBO J [Internet]. 2013;32(18):2454–62. Available from: 10.1038/emboj.2013.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behrendt R, Schumann T, Gerbaulet A, Nguyen LA, Schubert N, Alexopoulou D, et al. Mouse SAMHD1 Has Antiretroviral Activity and Suppresses a Spontaneous Cell-Intrinsic Antiviral Response. Cell Rep [Internet]. 2013. Aug;4(4):689–96. Available from: https://linkinghub.elsevier.com/retrieve/pii/S2211124713003999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wittmann S, Behrendt R, Eissmann K, Volkmann B, Thomas D, Ebert T, et al. Phosphorylation of murine SAMHD1 regulates its antiretroviral activity. Retrovirology. 2015;12(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seay K, Qi X, Zheng JH, Zhang C, Chen K, Dutta M, et al. Mice Transgenic for CD4-Specific Human CD4, CCR5 and Cyclin T1 Expression: A New Model for Investigating HIV-1 Transmission and Treatment Efficacy. PLoS One. 2013;8(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rahim MMA, Chrobak P, Hu C, Hanna Z, Jolicoeur P. Adult AIDS-Like Disease in a Novel Inducible Human Immunodeficiency Virus Type 1 Nef Transgenic Mouse Model: CD4+ T-Cell Activation Is Nef Dependent and Can Occur in the Absence of Lymphophenia. Vol. 83, Journal of Virology. 2009. p. 11830–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanna Z, Weng X, Kay DG, Poudrier J, Lowell C, Jolicoeur P. The Pathogenicity of Human Immunodeficiency Virus (HIV) Type 1 Nef in CD4C/HIV Transgenic Mice Is Abolished by Mutation of Its SH3-Binding Domain, and Disease Development Is Delayed in the Absence of Hck. Vol. 75, Journal of Virology. 2001. p. 9378–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dickie P, Felser J, Eckhaus M, Bryant J, Silver J, Marinos N, et al. HIV-associated nephropathy in transgenic mice expressing HIV-1 genes. Virology. 1991;185(1):109–19. [DOI] [PubMed] [Google Scholar]
- 23.Maung R, Hoefer MM, Sanchez AB, Sejbuk NE, Medders KE, Desai MK, et al. CCR5 Knockout Prevents Neuronal Injury and Behavioral Impairment Induced in a Transgenic Mouse Model by a CXCR4-Using HIV-1 Glycoprotein 120. J Immunol [Internet]. 2014. Aug 15;193(4):1895–910. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0031938416312148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marsden MD, Zack JA. Studies of retroviral infection in humanized mice. Virology [Internet]. 2015. May;479–480(1):297–309. Available from: https://linkinghub.elsevier.com/retrieve/pii/S004268221500029X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Victor Garcia J Humanized mice for HIV and AIDS research. Curr Opin Virol. 2016;19:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitney JB, Brad Jones R. In Vitro and In Vivo Models of HIV Latency. In: Zhang L, Lewin SR, editors. HIV Vaccines and Cure : The Path Towards Finding an Effective Cure and Vaccine [Internet]. Singapore: Springer Singapore; 2018. p. 241–63. Available from: 10.1007/978-981-13-0484-2_10 [DOI] [Google Scholar]
- 27.Krishnakumar V, Durairajan SSK, Alagarasu K, Li M, Dash AP. Recent updates on mouse models for human immunodeficiency, influenza, and dengue viral infections. Viruses. 2019;11(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou J, Takahashi M, Burnett JC, Li S, Rossi JJ, Li H, et al. HIV Replication and Latency in a Humanized NSG Mouse Model during Suppressive Oral Combinational Antiretroviral Therapy. J Virol. 2018;92(7):e02118–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai F, Chen Q. Humanized mouse models for the study of infection and pathogenesis of human viruses. Vol. 10, Viruses. 2018. p. 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mosier DE, Gulizia RJ, Baird SM, Wilson DB. Transfer of a functional human immune system to mice with severe combined immunodeficiency. Nature. 1988;335(6187):256–9. [DOI] [PubMed] [Google Scholar]
- 31.Kim KCKCKCKC, Choi BS, Kim KCKCKCKC, Park KH, Lee HJ, Cho YK, et al. A Simple Mouse Model for the Study of Human Immunodeficiency Virus. AIDS Res Hum Retroviruses. 2016;32(2):194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flerin NC, Bardhi A, Zheng JH, Korom M, Folkvord J, Kovacs C, et al. Establishment of a Novel Humanized Mouse Model To Investigate In Vivo Activation and Depletion of Patient-Derived HIV Latent Reservoirs. Silvestri G, editor. J Virol [Internet]. 2019. Jan 9;93(6):1–18. Available from: http://jvi.asm.org/lookup/doi/10.1128/JVI.02051-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lapidot T, Pflumio F, Doedens M, Murdoch B, Williams DE, Dick JE. Cytokine stimulation of multilineage hematopoiesis from immature human cells engrafted in SCID mice. Science (80- ) [Internet]. 1992;255(5048):1137–41. Available from: https://science.sciencemag.org/content/255/5048/1137 [DOI] [PubMed] [Google Scholar]
- 34.Honeycutt JB, Wahl A, Archin N, Choudhary S, Margolis D, Garcia JV. HIV-1 infection, response to treatment and establishment of viral latency in a novel humanized T cell-only mouse (TOM) model. Retrovirology [Internet]. 2013;10(1):1. Available from: Retrovirology [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honeycutt JB, Wahl A, Baker C, Spagnuolo RA, Foster J, Zakharova O, et al. Macrophages sustain HIV replication in vivo independently of T cells Find the latest version : Macrophages sustain HIV replication in vivo independently of T cells. J Clin Invest. 2016;126(4):1353–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCune J, Namikawa R, Kaneshima H, Shultz L, Lieberman M, Weissman I. The SCID-hu mouse: murine model for the analysis of human hematolymphoid differentiation and function. Science (80- ) [Internet]. 1988. Sep 23;241(4873):1632–9. Available from: https://science.sciencemag.org/content/241/4873/1632 [DOI] [PubMed] [Google Scholar]
- 37.Namikawa R, Weilbaecher KN, Kaneshima H, Yee EJ, McCune JM. Longterm human hematopoiesis in the SCID-hu mouse. J Exp Med. 1990;172(4):1055–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lan P, Tonomura N, Shimizu A, Wang S, Yang YG. Reconstitution of a functional human immune system in immunodeficient mice through combined human fetal thymus/liver and CD34+ cell transplantation. Blood. 2006;108(2):487–92. [DOI] [PubMed] [Google Scholar]
- 39.Melkus MW, Estes JD, Padgett-Thomas A, Gatlin J, Denton PW, Othieno FA, et al. Humanized mice mount specific adaptive and innate immune responses to EBV and TSST-1. Nat Med. 2006;12(11):1316–22. [DOI] [PubMed] [Google Scholar]
- 40.McCune JM, Weissman IL. The Ban on US Government Funding Research Using Human Fetal Tissues: How Does This Fit with the NIH Mission to Advance Medical Science for the Benefit of the Citizenry? [Internet]. Vol. 13, Stem Cell Reports. 2019. [cited 2019 Nov 19]. p. 777–86. Available from: http://www.ncbi.nlm.nih.gov/pubmed/31722191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jordan A, Bisgrove D, Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. 2003. [cited 2017 Jun 26];22(8). Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC154479/pdf/cdg188.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoszko M, De Crignis E, Rokx C, Khalid MM, Lungu C, Palstra RJ, et al. Small Molecule Inhibitors of BAF; A Promising Family of Compounds in HIV-1 Latency Reversal. EBioMedicine [Internet]. 2016;3:108–21. Available from: 10.1016/j.ebiom.2015.11.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spina CA, Anderson J, Archin NM, Bosque A, Chan J, Famiglietti M, et al. An In-Depth Comparison of Latent HIV-1 Reactivation in Multiple Cell Model Systems and Resting CD4+ T Cells from Aviremic Patients. Emerman M, editor. PLoS Pathog [Internet]. 2013. Dec 26;9(12):e1003834. Available from: 10.1371/journal.ppat.1003834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spivak AM, Planelles V. Novel Latency Reversal Agents for HIV-1 Cure. Annu Rev Med [Internet]. 2018. Jan 29 [cited 2018 Aug 15];69(1):421–36. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5892446/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jordan A, Defechereux P, Verdin E. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 2001;20(7):1726–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beyer AI, Muench MO. Comparison of human hematopoietic reconstitution in different strains of immunodeficient mice. Stem Cells Dev. 2017;26(2):102–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slørdal L, Muench MO, Warren DJ, Moore MAS. Radioprotection by murine and human tumor-necrosis factor: Dose-dependent effects on hematopoiesis in the mouse. Eur J Haematol. 1989;43(5):428–34. [DOI] [PubMed] [Google Scholar]
- 48.Hoggatt J, Hoggatt AF, Tate TA, Fortman J, Pelus LM. Bleeding the laboratory mouse: Not all methods are equal. Exp Hematol [Internet]. 2016;44(2):132–137.e1. Available from: 10.1016/j.exphem.2015.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Namikawa R, Kaneshima H, Lieberman M, Weissman IL, McCune JM. Infection of the SCID-hu mouse by HIV-1. Science (80- ). 1988;242(4886):1684–6. [DOI] [PubMed] [Google Scholar]
- 50.Symons J, Cameron PU, Lewin SR. HIV integration sites and implications for maintenance of the reservoir. Curr Opin HIV AIDS [Internet]. 2018. Mar;13(2):152–9. Available from: http://insights.ovid.com/crossref?an=01222929-201803000-00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson EM, Maldarelli F. The role of integration and clonal expansion in HIV infection: Live long and prosper 11 Medical and Health Sciences 1108 Medical Microbiology Ben Berkhout, Alexander Pasternak. Retrovirology [Internet]. 2018;15(1):1–22. Available from: 10.1186/s12977-018-0448-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, et al. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science (80- ) [Internet]. 2014. Jul 11;345(6193):179–83. Available from: https://www.sciencemag.org/lookup/doi/10.1126/science.1254194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abdel-Mohsen M, Chavez L, Tandon R, Chew GM, Deng X, Danesh A, et al. Human Galectin-9 Is a Potent Mediator of HIV Transcription and Reactivation. PLoS Pathog [Internet]. 2016;12(6):1–28. Available from: 10.1371/journal.ppat.1005677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang G, Mendes EA, Kaiser P, Wong DP, Tang Y, Cai I, et al. Synergistic Reactivation of Latent HIV Expression by Ingenol-3-Angelate, PEP005, Targeted NF-kB Signaling in Combination with JQ1 Induced p-TEFb Activation. PLoS Pathog. 2015;11(7):1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Darcis G, Kula A, Bouchat S, Fujinaga K, Corazza F, Ait-Ammar A, et al. An In-Depth Comparison of Latency-Reversing Agent Combinations in Various In Vitro and Ex Vivo HIV-1 Latency Models Identified Bryostatin-1+JQ1 and Ingenol-B+JQ1 to Potently Reactivate Viral Gene Expression. PLoS Pathog. 2015;11(7):1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reuse S, Calao M, Kabeya K, Guiguen A, Gatot JS, Quivy V, et al. Synergistic activation of HIV-1 expression by deacetylase inhibitors and prostratin: Implications for treatment of latent infection. PLoS One. 2009;4(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brogdon J, Ziani W, Wang X, Veazey RS, Xu H. In vitro effects of the small-molecule protein kinase C agonists on HIV latency reactivation. Sci Rep [Internet]. 2016;6(November):1–8. Available from: 10.1038/srep39032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen D, Wang H, Aweya JJ, Chen Y, Chen M, Wu X, et al. HMBA Enhances Prostratin-Induced Activation of Latent HIV-1 via Suppressing the Expression of Negative Feedback Regulator A20/TNFAIP3 in NF- B Signaling. Biomed Res Int. 2016;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rochat MA, Schlaepfer E, Speck RF. Promising Role of Toll-Like Receptor 8 Agonist in Concert with Prostratin for Activation of Silent HIV. J Virol. 2017;91(4):1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klase Z, Yedavalli VSRKRK, Houzet L, Perkins M, Maldarelli F, Brenchley J, et al. Activation of HIV-1 from Latent Infection via Synergy of RUNX1 Inhibitor Ro5-3335 and SAHA. Luban J, editor. PLoS Pathog [Internet]. 2014. Mar 20;10(3):e1003997. Available from: 10.1371/journal.ppat.1003997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bouchat S, Delacourt N, Kula A, Darcis G, Van Driessche B, Corazza F, et al. Sequential treatment with 5-aza-2′-deoxycytidine and deacetylase inhibitors reactivates HIV -1 . EMBO Mol Med. 2016;8(2):117–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kutsch O, Wolschendorf F, Planelles V. Facts and Fiction: Cellular Models for High Throughput Screening for HIV-1 Reactivating Drugs. Curr HIV Res [Internet]. 2011. Dec 1;9(8):568–78. Available from: http://www.eurekaselect.com/openurl/content.php?genre=article&issn=1570-162X&volume=9&issue=8&spage=568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bradley T, Ferrari G, Haynes BF, Margolis DM, Browne EP. Single-Cell Analysis of Quiescent HIV Infection Reveals Host Transcriptional Profiles that Regulate Proviral Latency. Cell Rep [Internet]. 2018. Oct;25(1):107–117.e3. Available from: https://linkinghub.elsevier.com/retrieve/pii/S2211124718314505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Telwatte S, Morón-López S, Aran D, Kim P, Hsieh C, Joshi S, et al. Heterogeneity in HIV and cellular transcription profiles in cell line models of latent and productive infection: implications for HIV latency. Retrovirology [Internet]. 2019;16(1):32. Available from: 10.1186/s12977-019-0494-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dahl V, Gisslen M, Hagberg L, Peterson J, Shao W, Spudich S, et al. An example of genetically distinct HIV type 1 variants in cerebrospinal fluid and plasma during suppressive therapy. J Infect Dis. 2014;209(10):1618–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blackard J T. HIV Compartmentalization: A Review on a Clinically Important Phenomenon. Curr HIV Res. 2012;10(2):133–42. [DOI] [PubMed] [Google Scholar]
- 67.Telwatte S, Lee S, Somsouk M, Hatano H, Baker C, Kaiser P, et al. Gut and blood differ in constitutive blocks to HIV transcription, suggesting tissue-specific differences in the mechanisms that govern HIV latency. PLoS Pathog. 2018;14(11):1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thompson KA, Cherry CL, Bell JE, McLean CA. Brain cell reservoirs of latent virus in presymptomatic HIV-infected individuals. Am J Pathol [Internet]. 2011;179(4):1623–9. Available from: 10.1016/j.ajpath.2011.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Castellano P, Prevedel L, Valdebenito S, Eugenin EA. HIV infection and latency induce a unique metabolic signature in human macrophages. Sci Rep [Internet]. 2019;9(1):1–14. Available from: 10.1038/s41598-019-39898-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wong ME, Jaworowski A, Hearps AC. The HIV Reservoir in Monocytes and Macrophages. Front Immunol [Internet]. 2019. Jun 26;10(June). Available from: 10.3389/fimmu.2019.01435/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gama L CJACVRACGSPDFEQSSEBBLMMPKBSMLMJMGFOS. Myeloid and CD4 T Cells Comprise the Latent Reservoir in Antiretroviral Therapy-Suppressed SIVmac251-Infected Macaques. Silvestri G, Bennink JR, editors. MBio [Internet]. 2019. Aug 20;10(4):1–19. Available from: 10.1128/mBio.01659-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gray LR, On H, Roberts E, Lu HK, Moso MA, Raison JA, et al. Toxicity and in vitro activity of HIV-1 latency-reversing agents in primary CNS cells. J Neurovirol [Internet]. 2016. Aug 4;22(4):455–63. Available from: 10.1007/s13365-015-0413-4 [DOI] [PubMed] [Google Scholar]
- 73.Hellmuth J, Valcour V, Spudich S. CNS reservoirs for HIV: implications for eradication. J virus Erad [Internet]. 2015;1(2):67–71. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26430703%0Ahttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4586130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Folks TM, Justement J, Kinter A, Dinarello CA, Fauci AS, Folks TM, et al. Cytokine-Induced Expression of HIV-1 in a Chronically Infected Promonocyte Cell Line. Science (80- ) [Internet]. 1987;238(4828):800–2. Available from: https://www.jstor.org/stable/1700363 [DOI] [PubMed] [Google Scholar]
- 75.Joseph SB, Arrildt KT, Sturdevant CB, Swanstrom R. HIV-1 target cells in the CNS. J Neurovirol [Internet]. 2015. Jun 19;21(3):276–89. Available from: http://link.springer.com/10.1007/s13365-014-0287-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barat C, Proust A, Deshiere A, Leboeuf M, Drouin J, Tremblay MJ. Astrocytes sustain long-term productive HIV-1 infection without establishment of reactivable viral latency. Glia [Internet]. 2018. Jul [cited 2018 Aug 15];66(7):1363–81. Available from: 10.1002/glia.23310 [DOI] [PubMed] [Google Scholar]
- 77.Edén A, Fuchs D, Hagberg L, Nilsson S, Spudich S, Svennerholm B, et al. HIV-1 Viral Escape in Cerebrospinal Fluid of Subjects on Suppressive Antiretroviral Treatment. J Infect Dis. 2010;202(12):1819–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Churchill M, Nath A. Where does HIV hide? A focus on the central nervous system. Curr Opin HIV AIDS [Internet]. 2013. May;8(3):165–9. Available from: http://content.wkhealth.com/linkback/openurl?sid=WKPTLP:landingpage&an=01222929-201305000-00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nath A, Clements JE. Eradication of HIV from the brain: Reasons for pause. Aids. 2011;25(5):577–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tyor WR, Power C, Gendelman HE, Markham RB. A model of human immunodeficiency virus encephalitis in scid mice. Proc Natl Acad Sci U S A. 1993;90(18):8658–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Potula R, Poluektova L, Knipe B, Chrastil J, Heilman D, Dou H, et al. Inhibition of indoleamine 2,3-dioxygenase (IDO) enhances elimination of virus-infected macrophages in an animal model of HIV-1 encephalitis. Blood. 2005;106(7):2382–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Poluektova LY, Munn DH, Persidsky Y, Gendelman HE. Generation of Cytotoxic T Cells Against Virus-Infected Human Brain Macrophages in a Murine Model of HIV-1 Encephalitis. J Immunol [Internet]. 2002. Apr 15;168(8):3941–9. Available from: 10.4049/jimmunol.168.8.3941 [DOI] [PubMed] [Google Scholar]
- 83.Poluektova L, Gorantla S, Faraci J, Birusingh K, Dou H, Gendelman HE. Neuroregulatory Events Follow Adaptive Immune-Mediated Elimination of HIV-1-Infected Macrophages: Studies in a Murine Model of Viral Encephalitis. J Immunol [Internet]. 2004. Jun 15;172(12):7610–7. Available from: 10.4049/jimmunol.172.12.7610 [DOI] [PubMed] [Google Scholar]
- 84.Persidsky Y, Limoges J, McComb R, Bock P, Baldwin T, Tyor W, et al. Human immunodeficiency virus encephalitis in SCID mice. Am J Pathol. 1996;149(3):1027–53. [PMC free article] [PubMed] [Google Scholar]
- 85.Garcia-Mesa Y, Jay TR, Checkley MA, Luttge B, Dobrowolski C, Valadkhan S, et al. Immortalization of primary microglia: a new platform to study HIV regulation in the central nervous system. J Neurovirol [Internet]. 2017. [cited 2018 Sep 5];23(1):47–66. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5329090/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alvarez-Carbonell D, Garcia-Mesa Y, Milne S, Das B, Dobrowolski C, Rojas R, et al. Toll-like receptor 3 activation selectively reverses HIV latency in microglial cells. Retrovirology [Internet]. 2017;14(1):9. Available from: http://retrovirology.biomedcentral.com/articles/10.1186/s12977-017-0335-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yin C, Zhang T, Qu X, Zhang Y, Putatunda R, Xiao X, et al. In Vivo Excision of HIV-1 Provirus by saCas9 and Multiplex Single-Guide RNAs in Animal Models. Mol Ther [Internet]. 2017. May 3 [cited 2018 Aug 16];25(5):1168–86. Available from: http://www.sciencedirect.com/science/article/pii/S1525001617301107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dash PK, Kaminski R, Bella R, Su H, Mathews S, Ahooyi TM, et al. Sequential LASER ART and CRISPR Treatments Eliminate HIV-1 in a Subset of Infected Humanized Mice. Nat Commun [Internet]. 2019. Dec 2;10(1):1–20. Available from: 10.1038/s41467-019-10366-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, et al. Gene Editing of CCR5 in Autologous CD4 T Cells of Persons Infected with HIV. N Engl J Med [Internet]. 2014. Mar 6 [cited 2019 Nov 15];370(10):901–10. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24597865 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


