ABSTRACT
A nodule-inhabiting Paenibacillus sp. strain (UY79) isolated from wild peanut (Arachis villosa) was screened for its antagonistic activity against diverse fungi and oomycetes (Botrytis cinerea, Fusarium verticillioides, Fusarium oxysporum, Fusarium graminearum, Fusarium semitectum, Macrophomina phaseolina, Phomopsis longicolla, Pythium ultimum, Phytophthora sojae, Rhizoctonia solani, Sclerotium rolfsii, and Trichoderma atroviride). The results obtained show that Paenibacillus sp. UY79 was able to antagonize these fungi/oomycetes and that agar-diffusible compounds and volatile compounds (different from HCN) participate in the antagonism exerted. Acetoin, 2,3-butanediol, and 2-methyl-1-butanol were identified among the volatile compounds produced by strain UY79 with possible antagonistic activity against fungi/oomycetes. Paenibacillus sp. strain UY79 did not affect symbiotic association or growth promotion of alfalfa plants when coinoculated with rhizobia. By whole-genome sequence analysis, we determined that strain UY79 is a new species of Paenibacillus within the Paenibacillus polymyxa complex. Diverse genes putatively involved in biocontrol activity were identified in the UY79 genome. Furthermore, according to genome mining and antibiosis assays, strain UY79 would have the capability to modulate the growth of bacteria commonly found in soil/plant communities.
IMPORTANCE Phytopathogenic fungi and oomycetes are responsible for causing devastating losses in agricultural crops. Therefore, there is enormous interest in the development of effective and complementary strategies that allow the control of the phytopathogens, reducing the input of agrochemicals in croplands. The discovery of new strains with expanded antifungal activities and with a broad spectrum of action is challenging and of great future impact. Diverse strains belonging to the P. polymyxa complex have been reported to be effective biocontrol agents. Results presented here show that the novel discovered strain of Paenibacillus sp. presents diverse traits involved in antagonistic activity against a broad spectrum of pathogens and is a potential and valuable strain to be further assessed for the development of biofungicides.
KEYWORDS: biocontrol, nodule-inhabiting bacteria, Paenibacillus, volatile metabolites, fusaricidin, volatile compounds, metabolites
INTRODUCTION
Plant health depends on the existence of a proper balance between beneficial and pathogenic microorganisms with which plants coexist in natural environments. In unbalanced environments, pathogenic fungi and oomycetes may produce severe plant diseases. It has been estimated that crop losses due to fungal diseases are around 30% of world agricultural production (1). In healthy environments, various microorganisms able to suppress plant diseases, either by boosting the plant immune system or by direct inhibition of the pathogen, could be found (2). Microorganisms with the ability to control the development of phytopathogens belong to the group of biological control agents (BCAs) defined as “a natural enemy, antagonist, or other organism, used for pest control” (ISPM 05; International Standards for Phytosanitary Measures) (3).
Several mechanisms involved in the biocontrol of phytopathogens have been found in BCAs, ranging from hyperparasitism and predation to the production of lytic enzymes (4). Enzymes able to degrade the cell wall of fungi or oomycetes (chitinases, cellulases, β-1,3-glucanases) antagonize mycelial growth, while xylanases may act as elicitors of plant systemic resistance, enhancing plant resistance to pathogens (5, 6). Cellulases and β-glycosidases may be also implicated in facilitating plant tissue colonization, enabling bacterial endophytic lifestyle (7). Secretion of antibiotics and the generation of organic as well as inorganic volatile compounds (VCs) are other widely distributed biocontrol mechanisms (8–10). Successful BCAs generally express multiple biological traits that act additively and synergistically to efficiently suppress the pathogen (9). BCAs are ubiquitous constituents of soils and of plant microbiota. They can be found as endophytes of roots, stems, leaves, and flowers of diverse plant genera and in legume nodules (11–13). A remarkable feature of legumes is their ability to establish symbiotic associations with a group of bacteria known as rhizobia, belonging to the orders Rhizobiales and Burkholderiales. As part of this symbiosis, distinctive structures, called nodules, are elicited in the root (or occasionally in the stems) of the legume, where the biological nitrogen fixation process takes place (14). Legume nodules were considered for many years to be exclusively occupied by rhizobia, but strong evidence demonstrated that nodules also harbor a diverse community of microorganisms (13, 15–17). Major bacterial phyla that have been consistently found as nodule inhabitants include Actinobacteria, Firmicutes, and Proteobacteria. The presence of Agrobacterium, Arthrobacter, Acinetobacter, Bacillus, Bosea, Enterobacter, Micromonospora, Mycobacterium, Paenibacillus, Pseudomonas, and Stenotrophomonas genera has been reported, with strains of Bacillus spp. and Paenibacillus spp. frequently isolated from this niche (13, 15, 16). The role of nodule endophytes is unknown, but it has been found that some strains possess biocontrol activity against phytopathogens (17). This fact makes nodules an interesting and still poorly explored environment for the identification of strains with biocontrol activity against phytopathogenic fungi.
The goal of this work was to identify and characterize a nodule-inhabiting bacterium obtained from Arachis villosa (wild peanut) collected in a Uruguayan National Park. According to the genome analysis, strain UY79 is a new species of Paenibacillus that belongs to the Paenibacillus polymyxa group. Results obtained in vitro and in silico indicate that strain UY79 possesses a broad spectrum of antimicrobial activity, as well as a plethora of mechanisms putatively involved in antagonisms.
RESULTS AND DISCUSSION
Strain UY79 is a new species of Paenibacillus and belongs to the P. polymyxa group.
In this work, we isolated a strain from a surface-sterilized nodule of A. villosa with phenotypic traits different from those expected for rhizobia. According to 16S rRNA analysis, the isolate was identified as belonging to the Paenibacillus genus (see Fig. S1 in the supplemental material). Based on multilocus sequence analysis (MLSA) of concatenated sequences of 16S rRNA, gyrB, rpoB, recA, and recN genes retrieved from 22 sequenced genomes, Paenibacillus sp. strain UY79 grouped closely to Paenibacillus kribbensis, Paenibacillus brasilensis, and Paenibacillus peoriae type strains but in a different branch (Fig. 1). Moreover, phylogenetic affiliation of the 16S rRNA gene showed that P. kribbensis, P. peoriae, Paenibacillus jamilae (recently reclassified as P. polymyxa) (18), Paenibacillus ottowii, Paenibacillus terrae, P. brasilensis and P. polymyxa formed a monophyletic group (referred to as the Paenibacillus polymyxa complex [19]), and strain UY79 clearly grouped within the P. polymyxa complex (Fig. S1). Until very recently, no representatives of this group were isolated from root nodules. In 2021, Ali et al. (19) reported the isolation of P. peoriae from nodules of Robinia pseudoacacia and Dendrolobium triangulare and of P. kribbensis from Ormosia semicastrata nodules; therefore, as far as we know, Paenibacillus sp. UY79 is the third representative of the P. polymyxa complex to be isolated from root nodules. No correlation was found between the taxonomic identity and the origin of the strains (Fig. S1).
FIG 1.
Taxonomic affiliation of Paenibacillus sp. strain UY79. (A) Multilocus phylogenetic analysis based on 16S rRNA, gyrB, rpoB, recA, and recN gene concatenated sequences (8,535 positions) obtained from sequenced genomes. The tree was constructed using the MEGAX program, maximum likelihood method, and general time-reversible model. The numbers at each node are percentages of bootstrap replications calculated from 1,000 replicate trees. Strain UY79 is shown in red, and type strains are indicated with a superscript T. (B) Scores of average nucleotide identity (ANI). NCBI accession numbers of genomes are shown in parentheses.
By comparing the UY79 genome to the 22 sequenced genomes of the Paenibacillus species publicly available in GenBank, we found that values of average nucleotide identity (ANI) were below 88.9% in all cases (Fig. 1), indicating that the strain UY79 is a new Paenibacillus species.
The genome size of UY79 was calculated to be 4.9 Mb with 5,353 predicted coding sequences according to RAST 2.0, and the GC content was estimated to be 46.4% (Table S1). A great variation has been found in the genome sizes and GC content of Paenibacillus strains, with values ranging from 4 to 8.8 Mb for genome sizes and from 41% to 63% for GC content (20, 21). Therefore, both genome size and GC content of UY79 genome are within the lowest ranges found.
Strain UY79 exhibited a broad spectrum of antagonism against fungi and oomycetes mediated by the production of diffusible compounds.
Species of the genus Paenibacillus have been reported as members of the root-nodule microbiome (16, 17, 22), and while the function they exert in the nodule is not completely understood, many species have been reported to display antagonistic activities against different phytopathogenic fungi/oomycetes (7, 23). These observations prompted us to investigate the potential of Paenibacillus sp. strain UY79 as a biocontrol agent.
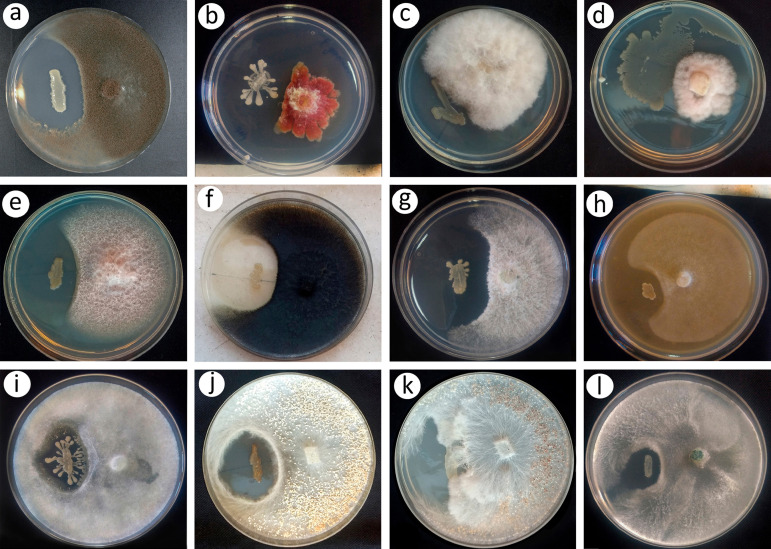
Results shown in Fig. 2 indicate that strain UY79 produced diffusible compounds in potato dextrose agar (PDA) (or V8 agar in the case of Phytophthora sojae) that were able to inhibit all 10 fungi and the two oomycetes tested. Although no quantitative evaluation was performed, qualitatively we can infer that strain UY79 produces an important mycelial growth inhibition of Botrytis cinerea A1 (Fig. 2a), Fusarium verticillioides A71 (Fig. 2e), Macrophomina phaseolina J431 (Fig. 2f), Phomopsis longicolla J429 (Fig. 2g), P. sojae Ps25 (Fig. 2h), and Rhizoctonia solani Rz01 (Fig. 2j), a moderate growth inhibition of Fusarium graminearum S127 (Fig. 2b), Pythium ultimum Py03 (Fig. 2i), and Trichoderma atroviride 1607 (Fig. 2l), and a slight growth inhibition of Fusarium oxysporum J38 (Fig. 2c), Fusarium semitectum J141 (Fig. 2d), and Sclerotium rolfsii 1948 (Fig. 2k). In addition, for F. verticillioides A71 facing UY79, a zone of mycelial lysis at the edge of the mycelial growth facing the bacterium was observed (Fig. 2e). In the case of R. solani Rz01 (Fig. 2j) and S. rolfsii 1948 (Fig. 2k), a lower density of resistance structures was detected in the proximity of the confrontation zone. It is interesting to note the dark brown pigmentation of strain UY79 when facing R. solani Rz01 (Fig. 2j).
FIG 2.
In vitro antagonistic activity of Paenibacillus sp. strain UY79 against fungi and oomycetes analyzed by the dual plate assay. Antagonistic activity of strain UY79 (left streak) was visualized as a growth inhibition zone of the fungi/oomycetes. Fungi/oomycetes include Botrytis cinerea A1 (a), Fusarium graminearum S127 (b), Fusarium oxysporum J38 (c), Fusarium semitectum J41 (d), Fusarium verticillioides A71 (e), Macrophomina phaseolina J431 (f), Phomopsis longicolla J429 (g), Phytophthora sojae Ps25 (h), Pythium ultimum Py03 (i), Rhizoctonia solani Rz01 (j), Sclerotium rolfsii 1948 (k), and Trichoderma atroviride 1607 (l). Dual plate assays were performed on PDA medium, except for P. sojae Ps25, which was grown on V8 agar medium. All assays were performed in triplicate, and representative plates are shown.
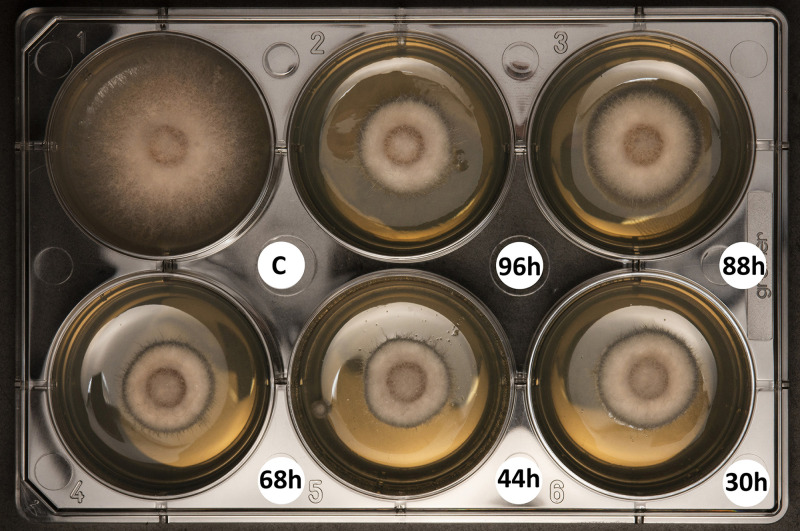
As strain UY79 proved to be capable of producing diffusible compounds with antimicrobial activity in the presence of the fungi/oomycetes, we wondered if it was also able to produce them when grown alone in liquid medium. F. verticilloides A71 was randomly selected as the target for this assay. As shown in Fig. 3, compounds that inhibited F. verticillioides A71 growth were present in cell-free supernatant of UY79 cultures after 30 h of incubation. The inhibition phenotype was maintained in 44-, 68-, 88-, and 96-h cultures, suggesting that either the antifungal compound production was maintained for up to 96 h or that the produced compounds were active for such a period.
FIG 3.
Cell-free supernatants of Paenibacillus sp. strain UY79 exhibit antifungal activity against Fusarium verticillioides A71. Cell-free supernatants of cultures grown for 30, 44, 68, 88, and 96 h were included in PDA medium (1:1). PDB medium instead of cell-free supernatant was used as a control (C). The experiment was performed in triplicate, and a representative plate is shown.
Strain UY79 produces volatile compounds with antagonistic activity against fungi and oomycetes.
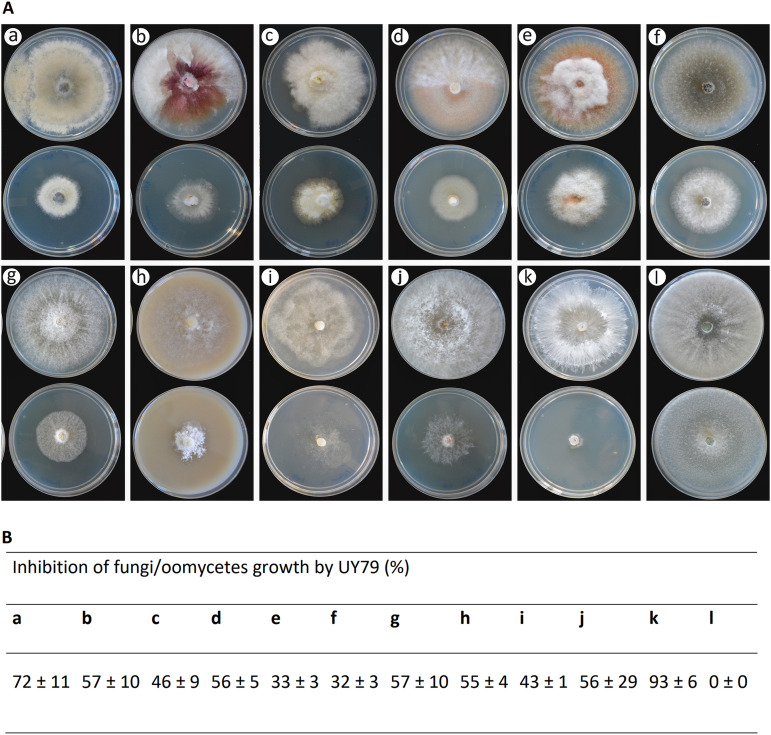
As shown in Fig. 4, Paenibacillus sp. strain UY79 produces VCs with antagonistic activity, being able to inhibit the growth of 9 out of 10 fungi analyzed and the two oomycetes analyzed (Fig. 4A). Growth of 7 phytopathogens, out of the 11 assessed, was inhibited more than 50% (Fig. 4B). This was the case for B. cinerea A1 (a), F. graminearum S127 (b), F. semitectum J41 (d), P. longicolla J429 (g), P. sojae Ps25 (h), R. solani Rz01 (j), and S. rolfsii 1948 (k) (Fig. 4A and B). Remarkably, growth of S. rolfsii 1948 (k) and B. cinerea A1 (a) was severely compromised (93% ± 6% and 72% ± 11%, respectively) when facing strain UY79. M. phaseolina J431 (f) and F. verticilliodes A71 (e) were the least affected, resulting in approximately 30% of mycelial inhibition by UY79 VCs (Fig. 4B). As shown in Fig. 4A, altered pigmentation was observed in F. graminearum S127 (b) and M. phaseolina J431 (f), and to a lesser extent in F. semitectum J41 (d) and F. verticillioides A71 (e), as a response to VCs produced by strain UY79. It is interesting to note that a lower density of hyphae was observed in R. solani Rz01 (j) while, in contrast, a higher density of hyphae was produced by P. sojae Ps25 (h) (Fig. 4A). Although T. atroviride 1607 (l) did not show radial growth inhibition, an alteration in the colony pattern was observed. Together, these results indicate that strain UY79 produces VCs with a broad spectrum of antimicrobial activity and that fungus/oomycete responses depend on each particular microorganism.
FIG 4.
Volatile compounds produced by Paenibacillus sp. strain UY79 exert antagonistic activity against different fungi and oomycetes. Antagonism was analyzed by the two-base plate method. (A) Production of volatile compounds with antimicrobial activity was evidenced as growth inhibition of the fungi/oomycetes facing strain UY79 (bottom plates) in comparison with the fungal growth of the fungi/oomycetes not facing the bacterium (upper plates). Fungi/oomycetes analyzed were Botrytis cinerea A1 (a), Fusarium graminearum S127 (b), Fusarium oxysporum J38 (c), Fusarium semitectum J41 (d), Fusarium verticillioides A71 (e), Macrophomina phaseolina J431 (f), Phomopsis longicolla J429 (g), Phytophthora sojae Ps25 (h), Pythium ultimum Py03 (i), Rhizoctonia solani Rz01 (j), Sclerotium rolfsii 1948 (k), and Trichoderma atroviride 1607 (l). Assays were performed on PDA medium except for P. sojae Ps25, which was grown on V8 agar medium. The experiment was done in triplicate. (A) Photographs of one representative plate per treatment. (B) Percentage of mycelial growth inhibition (mean values ± standard deviation).
Since hydrogen cyanide (HCN) is a VC with well-known antimicrobial activity (24) and its production has been reported in some species of Paenibacillus (25), we assessed the ability of strain UY79 to produce this compound. According to the picrate-filter paper method, no production of HCN was observed in either PDA or tryptic soy agar (TSA) medium, either with or without the addition of glycine (data not shown). These results suggest that antimicrobial VCs other than HCN are being produced by strain UY79.
Identification by gas chromatography-mass spectrometry (GC-MS) of VCs produced by Paenibacillus sp. strain UY79 grown for 36 h or 7 days on PDA medium showed that acetoin (3-hydroxy-2-butanone) was the most abundant VC produced (>65% relative abundance). Other compounds detected were 2-methyl-1-butanol (4.3 to 5.3%), 2-methyl-1-propanol (1.7 to 2.5%), 3-methyl-1-butanol (0.9 to 2.7%), and 2,3-butanediol (1.1 to 1.7%) (Table 1; Table S2). No relevant variations were observed in the composition or the main relative content of VCs produced by UY79, neither at different times of growth (36 h or 7 days) nor with different sorption times (10 min or 30 min) (Table S2; Fig. S2A).
TABLE 1.
Relevant VCs produced by Paenibacillus sp. strain UY79 and by S. rolfsii 1948 grown alone or facing each othera
| Compoundb |
Paenibacillus sp. UY79 |
S. rolfsii 1948 |
S. rolfsii 1948 vs Paenibacillus sp. UY79 |
|||
|---|---|---|---|---|---|---|
| RT (min)c | Area (%) | RT (min) | Area (%) | RT (min) | Area (%) | |
| 2-Methyl-1-propanol | 2.274 | 1.70 | 2.299 | 3.36 | 2.285 | 2.27 |
| 2-Ethylfuran | NDd | ND | 3.382 | 0.39 | ND | ND |
| 2,5-Dimethylfuran | ND | ND | 3.512 | 1.13 | 3.510 | 0.14 |
| 3-Hydroxy-2-butanone | 4.043 | >65 | ND | ND | 4.094 | 50.08 |
| 3-Methyl-1-butanol | 4.391 | 2.66 | 4.409 | 5.66 | 4.399 | 2.75 |
| 2-Methyl-1-butanol | 4.495 | 5.31 | ND | ND | 4.501 | 5.00 |
| 2,3-Butanediol | 6.301 | 1.14 | ND | ND | ND | ND |
VCs were collected from bacterial cultures grown for 7 days and extracted for 30 min with SPME fibers.
Compound was identified by comparison of its mass spectrum against reference libraries (NIST 08, Wiley 139).
RT, retention time.
ND, not detected.
Taking into account the work published by Ebadzadsahrai et al. (26) showing that the VCs produced by bacterial-fungal cocultures were not the sum of those of monocultures, we also analyzed the composition of the volatilome produced when strain UY79 was grown facing S. rolfsii 1948. This fungus was selected for the assay, as its growth was severely compromised when facing strain UY79 (Fig. 4A and B). Results obtained indicate that the compositions of VCs produced by cocultures correspond to those produced by both strains when grown alone (Table 1; Fig. S2B). It is worth noting that among the compounds produced by the UY79 strain, 2-methyl-1-propanol and 3-methyl-1-butanol were also produced by S. rolfsii 1948, whereas acetoin, 2,3-butanediol and 2-methyl-1-butanol were produced only by strain UY79 (Table 1; Fig. S2B). It is well known that acetoin and 2,3-butanediol are two compounds able to promote plant growth and induce plant systemic resistance (27). Khalaf and Raizada (28) reported that out of 37 isolates of Paenibacillus sp. analyzed, 18 of them were able to produce acetoin and 2,3-butanediol and that the ability to produce these compounds correlates with antagonism (28). Recently, Wu et al. (29) reported that acetoin was the main volatile organic compound produced by a Bacillus amyloloquefaciens strain and that this compound was able to reduce mycelial growth of B. cinerea although with low antifungal activity. As far as we know, the mechanism involved in fungal antagonism exerted by acetoin has not yet been unraveled. In the case of 2-methyl-1-butanol, Raza et al. (30) reported that P. polymyxa strain WR-2 was able to produce 2-methyl-1-butanol, which completely inhibited the growth of F. oxysporum. Suppression of mycelial growth, conidial germination, and appressorium formation is among the mechanisms described to be involved in the antagonistic effect of 2-methyl-1-butanol against fungi (31). In summary, previous works show that these three compounds may participate to different degrees in biological control against fungi (28, 30, 31).
Investigations of the volatilome (including organic and inorganic VCs) produced by bacteria are a topic that has aroused great interest. Using VCs for biocontrol of plant pathogens presents some advantages, as they may induce systemic resistance in plants, can be used in conditions where physical contact between the pathogen and the BCA is not possible, and leave less residuals in the environment once applied (23). Furthermore, VCs may participate in a plethora of interactions intra- and interkingdom (32).
Strain UY79 produced cell wall-degrading enzymes, siderophores, and proteolytic enzymes.
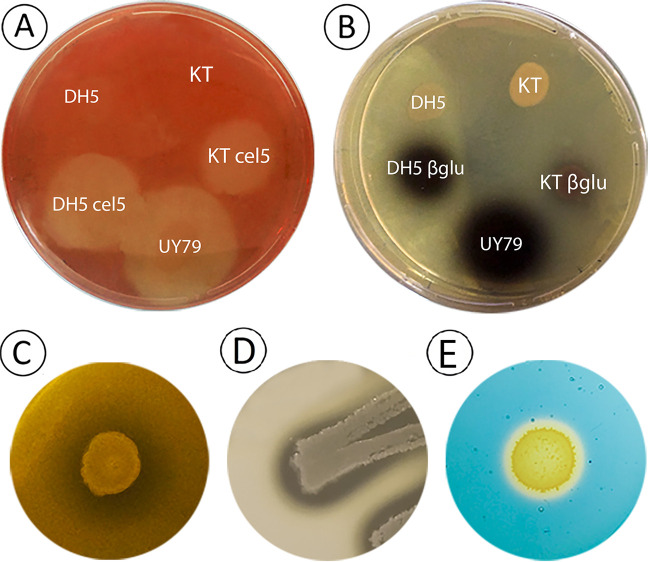
Other putative antagonistic traits, such as the production of cell wall-degrading enzymes, siderophore production, and proteolytic activity, were assessed. As shown in Fig. 5, strain UY79 exhibited cellulase (Fig. 5A), β-glucosidase (β-glu) (Fig. 5B), xylanase (Fig. 5C), and protease (Fig. 5D) activity. Remarkably, strain UY79 displayed better cellulase and β-glucosidase activity than the positive controls, which are laboratory strains harboring a plasmid containing a constitutively expressed cellulase or β-glucosidase, respectively. Xylanase activity was evidenced even without the need of Congo red staining, suggesting high enzymatic activity. These results show that strain UY79 harbors a suite of cell wall-degrading enzymes which might be responsible for the antagonism mediated by diffusible compounds. However, it should be noted that the presence of these hydrolytic activities or encoding genes does not necessarily indicate that they are involved in biological control. For instance, Ali et al. (19) recently found that β-1,3-glucanase and chitinase activity exhibited by some Paenibacillus sp. strains were not involved in their antifungal activity.
FIG 5.
Paenibacillus sp. strain UY79 exhibits cellulase, β-glucosidase, xylanase, and protease activity, as well as siderophore production. (A) Cellulase activity was detected as a clear halo around colonies grown on carboxymethyl cellulose (CMC)-containing medium after staining with Congo red. E. coli DH5α cel5 (DH5 cel5) and P. putida KT2440 cel5 (KT cel5) were used as positive controls, while E. coli DH5α (DH5) and P. putida KT2440 (KT) were used as negative controls. (B) Beta-glucosidase activity was detected as a dark halo around colonies grown on esculin-containing medium. E. coli DH5α β-glu (DH5 βglu) and P. putida KT2440 β-glu (KT βglu) were used as positive controls, while E. coli DH5α (DH5) and P. putida KT2440 (KT) were used as negative controls. (C) Xylanase activity was detected as a clear halo around Paenibacillus sp. UY79 colonies on xylan-containing medium. (D) Proteolytic activity was detected as a clear halo around Paenibacillus sp. UY79 colonies on skim milk-containing medium. (E) Siderophore production was detected in CAS medium as an orange halo around bacterial colonies.
Regarding siderophore production, the ability to produce iron-chelating compounds was detected in CAS medium (Fig. 5D). Different roles have been assigned to siderophores produced by plant-associated bacteria. They may be responsible for pathogenicity, such as the siderophore produced by Erwinia chrysanthemi, or may promote plant growth through iron solubilization or by controlling phytopathogens (33–35). We currently do not have enough information to ascertain the putative role of siderophore(s) produced by strain UY79 in biocontrol; nonetheless, considering that the antagonistic assays performed in this work were done in iron-sufficient PDA medium, we can speculate that siderophore(s) was not being produced and therefore did not contribute to the observed antifungal activity.
Main plant growth promotion (PGP) traits were not present in Paenibacillus sp. strain UY79.
No nifH gene was detected by PCR (data not shown), and indole-3-acetic acid (IAA)-like compounds and phosphate solubilization compounds could not be detected by in vivo analysis (Fig. S3). In agreement with these results, we did not find nifH homologous genes nor genes putatively involved in auxin production or phosphate solubilization (Table S3).
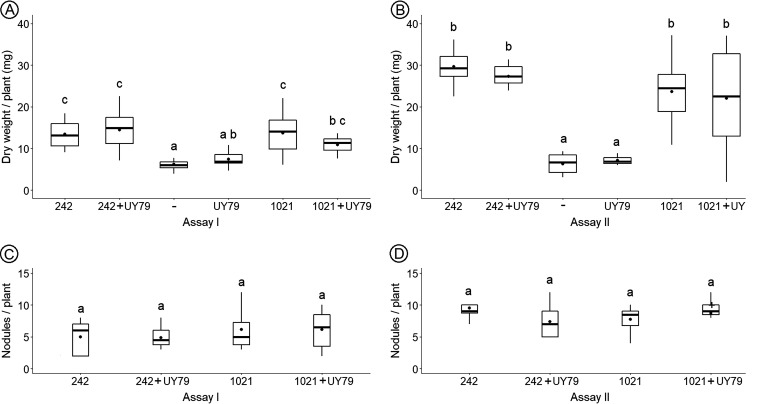
These results together with the observation that inoculation of alfalfa plants with strain UY79 did not enhance plant dry weight (Fig. 6A and B) indicate that strain UY79 did not display direct PGP activity.
FIG 6.
Paenibacillus sp. UY79 does not affect plant growth promotion exerted by rhizobia. Evaluations of plant growth (A and B) and nodule number per plant (C and D) in response to inoculation are shown. Results of assay I are depicted in panels A and C, and results of assay II are depicted in panels B and D. Medicago sativa cv. crioula plants were grown in N-free medium. E. meliloti 242, E. meliloti 1021, and Paenibacillus sp. UY79 strains were used to inoculate alfalfa seedlings separately or as a mixture (1:1) of the rhizobium strain with Paenibacillus sp. UY79 (242+UY79 and 1021+UY79, respectively). A negative control (–) without bacteria was also included. Data from each graph were independently analyzed as described in Materials and Methods, and different letters in the same graph indicate significant differences.
Paenibacillus sp. strain UY79 did not affect alfalfa (Medicago sativa L.) plant growth promotion by rhizobia.
Keeping in mind the potential use of strain UY79 as a biopesticide for diverse crops, and considering that alfalfa is the most important legume crop in cultivated areas after soybean (36), we assessed the effect of UY79 on alfalfa plant growth promotion exerted by rhizobia. The results obtained indicate that promotion of plant shoot dry weight (Fig. 6A and B) and number of nodules per plant (Fig. 6C and D) were not significantly affected by strain UY79.
Some chemical pesticides affect the rhizobium-legume symbiosis (37, 38); therefore, the fact that strain UY79 did not interfere with symbiosis or biological nitrogen fixation exerted by symbionts, at least in alfalfa, is an additional advantage when considering its use as a biological control agent.
Assessment of antibacterial activity.
Considering the potential use of strain UY79 as a biological control agent, and therefore its possible release into the environment, we investigated its capability to coexist with or to inhibit diverse soil- and plant-associated bacteria. As shown in Fig. S4, the antibiosis phenotype was diverse and depended on the target strain, as well as on the medium in which the assay was performed. According to the observed phenotype, we classified the inhibition as follows: type I, moderate growth of UY79 and an inhibition halo in the drop; type II, poor growth of UY79 and an inhibition halo around the drop; type III, good growth of UY79 and an inhibition halo in the drop; and type IV, good growth of UY79 and inhibition halo around the drop. Strain UY79 exerted no growth inhibition and coexisted with rhizobial strains Ensifer meliloti 1021, Rhizobium tropici CIAT 899, and Paraburkholderia sp. strain UYPR4.13, while for Bradyrhizobium elkanii U-1301 and U-1302, type I inhibition was observed. Interestingly, in the case of Cupriavidus necator UYPR2.512, a medium-dependent phenotype was observed, as UY79 showed no inhibition in tryptone-yeast extract (TY) medium and type II inhibition on TSA. This medium-dependent phenotype was also observed for Gram-positive Bacillus subtilis ATCC 6633, which was highly inhibited in TY (type IV inhibition) but was not affected in TSA, where both strains coexisted. When other PGP bacteria were assessed, strain UY79 exerted type III inhibition in Azospirillum brasilense Sp7 and Streptomyces sp. strain UYFA156, while in the case of Pseudomonas, strain UY79 was not capable of growing. Finally, in the case of the plant pathogen Erwinia carotovora SCC3193, no inhibition by strain UY79 was observed under the condition assayed. In order to investigate whether the antibacterial compounds produced by strain UY79 were present in the supernatant of UY79 cultures, we performed a similar assay but with a drop of a 45-μm-filtered supernatant of UY79 over the confluent lawn of the target strain. No inhibition was detected by this approach (data not shown). This result suggests that other factors are required to generate the inhibitory phenotype, such as a physical contact between strain UY79 and target strains or a bacterial biofilm formation. Another plausible explanation is that antibacterial compounds present in the supernatant did not reach a concentration sufficient to exert inhibition.
Genome mining of genes putatively involved in antimicrobial response.
Further, we manually scored the RAST 2.0 annotation of the UY79 draft genome, searching for functions involved in the biological control of phytopathogenic fungi and oomycetes. Several genes involved in the synthesis of hydrolytic enzymes were identified (Table S3), among them seven cellulases, three β-glucosidases, 16 xylanases, two chitinases, and one protease. Also, genes putatively coding for nonribosomal peptide synthetase or polyketide clusters (NRPS/PKS) were identified. Among the six detected clusters identified, three showed more than 40% similarity with known NRPS reported to be involved in the synthesis of tridecaptin (100% cluster similarity), fusaricidin B (100% cluster similarity), and tridecaptin (40% cluster similarity), respectively. The remaining three NRPS did not show similarity with known clusters. We also found one ribosomally synthesized and posttranslationally modified peptide (RiPP) cluster putatively involved in the synthesis of a lasso peptide identified as a paeninodin (40% cluster similarity). Fusaricidins are nonribosomal peptides (NRPs), consisting of a cyclic lipopeptide with six amino acids and an unusual fatty acid chain of 15-guanidino-3-hydroxypentadecanoic acid (39). Fusaricidins exhibit antagonistic activity against diverse fungi as well as against Gram-negative bacteria (40). In the case of tridecaptins, these molecules consist of linear acylated tridecapeptides that exhibit a strong selective antibacterial activity against Gram-negative bacteria and moderate activity against Gram-positive bacteria (41, 42) and have been reported as produced by Bacillus and Paenibacillus species, including by strains of the P. polymyxa complex (43, 44). Paeninodin is a RiPP lasso peptide with a unique structure comprising 16 to 21 residues, produced by clusters of a unique and well conserved organization (45). Interestingly, it has been found that some lasso peptides possess antimicrobial and antiviral activities.
Considering the results of GC-MS analysis, we decided to mine the UY79 genome for genes putatively involved in the biosynthesis of acetoin, 2,3-butanediol, and 2-methyl-1-butanol. As shown in Table S3, genes putatively involved in acetoin, 2,3-butanediol, and 2-methyl-1-butanol were identified. Acetoin can be synthesized through the condensation of two pyruvate molecules to form α-acetolactate by an acetolactate synthase. Then, α-acetolactate can be converted to (3R)-acetoin in a reaction catalyzed by α-acetolactate decarboxylase (ald), or alternatively, α-acetolactate can spontaneously decompose into diacetyl through oxidative decarboxylation. Diacetyl can subsequently be reduced into (3S)-acetoin in a reaction catalyzed by a diacetyl reductase (46, 47). No homolog to the ald gene was found in the genome of UY79, but a presumptive diacetyl reductase-encoding gene was (Table S3). Therefore, acetoin might be synthesized in UY79 by diacetyl reduction. Acetoin may be further reduced to 2,3-butanediol by a butanediol-dehydrogenase (47), which was also identified in the UY79 genome (Table S3). Concerning genes putatively involved in 2-methyl-1-butanol, we identified by genome mining an amino acid aminotransferase (EC 2.6.1.42), a 2-ketoacid decarboxylase, and an alcohol dehydrogenase (EC 1.1.1.1), which may be responsible for the conversion of ketoisovalerate to 2-methylisobutyaldehyde and then to 2-methyl-1-butanol (48).
Concerning other traits presumably relevant to rhizosphere colonization, endophytic fitness, or biocontrol performance, the genome was mined for genes involved in bacterial motility and in siderophore biosynthesis. As shown in Table S3, some genes probably involved in motility were identified. Genes involved in the biosynthesis of the achromobactin-like siderophore class and genes putatively involved in the internalization of bacillibactin, ferrichrome, and other siderophores were also detected (Table S3).
It is interesting to note that by genome mining, we found the presence of different systems putatively involved in antibiosis and bacteriophage production, as well as several regions involved in the CRISPR/Cas adaptative antiviral immune response (Table S3). These results together with the ability to produce diverse diffusible and volatile compounds against fungi/oomycetes suggest that strain UY79 could either coexist or interfere with various soil- and plant-associated microorganisms, perhaps modulating the microbiota associated with plants, as has been described for other nodule-inhabiting Paenibacillus strains (17, 49).
In conclusion, we identify a new nodule-inhabiting Paenibacillus species that belongs to the P. polymyxa group. The wide spectrum of its antagonistic effect, together with the diversity of mechanisms putatively involved (diffusible and volatile compounds, hydrolytic enzymes, iron scavenging), makes Paenibacillus sp. strain UY79 a promising biocontrol agent.
MATERIALS AND METHODS
Microbes, media, and growth conditions used in this work.
The microorganisms used in this work are listed in Table 2. The bacterial strain UY79 was isolated from a root nodule of Arachis villosa, a native Uruguayan legume, collected in Nuevo Berlin, Río Negro, Uruguay (32°59′04.00′′S 58°03′48.20′′W). The nodule was surface sterilized for 2 min with a solution of 10 mM HgCl2 in 0.1 N HCl, followed by seven washes with sterile distilled water. The surface-sterilized nodule was crushed with a sterile glass rod and streaked on yeast-mannitol agar (50) supplemented with 1 g/L glutamate (YMAG). Plates were incubated at 30°C and checked every day for bacterial growth. Colonies phenotypically different from rhizobia were selected, cultured in YMG broth, and stored at −80°C with 25% (vol/vol) glycerol.
TABLE 2.
Microbial strains used in this work and relevant characteristics
| Strain | Characteristics relevant to this work | Source or referencea |
|---|---|---|
| Bacteria | ||
| Azospirillum brasilense SP7 | PGPR soil bacterium; used in bacterial antibiosis assays | 80 |
| Bacillus subtilis ATCC 6633 | Reference strain; used in bacterial antibiosis assays | ATCC |
| Bradyrhizobium elkanii U-1301 | Commercial rhizobium inoculant for soybeans; used in bacterial antibiosis assays | MGAP |
| Bradyrhizobium elkanii U-1302 | Commercial rhizobium inoculant for soybeans; used in bacterial antibiosis assays | MGAP |
| Paraburkholderia sp. UYPR1.413 | Rhizobium; used in bacterial antibiosis assays | 53 |
| Cupriavidus necator UYPR2.512 | Rhizobium; used in bacterial antibiosis assays | 53 |
| Escherichia coli DH5α | Laboratory strain; used as a negative control in some hydrolytic-enzyme assays | 81 |
| Escherichia coli DH5α Cel5 | E. coli DH5α harboring a plasmid containing endoglucanase Cel5A from Bacillus subtilis, which confers cellulase activity; used as positive control in cellulolytic assays | 82 |
| Escherichia coli DH5α β-glu | E. coli DH5α harboring a plasmid with a metagenomic DNA fragment containing a glycosyl hydrolase which confers beta-glucosidase activity; used as positive control in beta-glucosidase assays | 82 |
| Ensifer meliloti 1021 | Rhizobium; used in bacterial antibiosis assays | 83 |
| Erwinia caratovora SCC3193 | Soil bacterium, phytopathogen; used in bacterial antibiosis assays | 84 |
| Microbacterium sp. UYFA68 | AIA producing strain; used as positive control in AIA assays | 85 |
| Pseudomonas putida KT2440 Cel5 | P. putida KT2440 harboring a plasmid containing endoglucanase Cel5A from B. subtilis, which confer cellulase activity; used as a positive control in cellulolytic assays | BIOGEM |
| Pseudomonas putida KT2440 β-glu | P. putida KT2440 harboring a plasmid with a metagenomic DNA fragment containing a glycosyl hydrolase which confers beta-glucosidase activity; used as a positive control in beta-glucosidase assays | BIOGEM |
| Pantoea sp. UYSB45 | Phosphate-solubilizing strain; used as a positive control in phosphate solubilization assays | 86 |
| Paenibacillus sp. UY79 | Isolated from an Arachis villosa nodule | This work |
| Pseudomonas fluorescens UP148 | Soil bacterium, PGPR; used in bacterial antibiosis assays | 87 |
| Pseudomonas fluorescens UP61 | Soil bacterium, HCN-producing strain; used in bacterial antibiosis assays | 88 |
| Pseudomonas protegenes Pf-5 | Soil bacterium, PGPR; used in bacterial antibiosis assays | 89 |
| Pseudomonas putida KT2440 | Laboratory strain; used as a negative control in some hydrolytic-enzyme assays | 90 |
| Rhizobium tropici CIAT 899 | Rhizobium; used in bacterial antibiosis assays | 91 |
| Streptomyces sp. UYFA 156 | Soil bacterium, PGPR; used in bacterial antibiosis assays | 85 |
| Fungi | ||
| Botrytis cinerea Pers. A1 | Phytopathogenic fungus isolated from lemon plant | 92 |
| Fusarium graminearum Schwabe S127 | Phytopathogenic fungus isolated from sorghum grain | Mycology lab |
| Fusarium oxysporum Schltdl J38 | Phytopathogenic fungus isolated from soybean | Mycology lab |
| Fusarium semitectum Berk. & Ravenel J41 | Phytopathogenic fungus isolated from soybean | Mycology lab |
| Fusarium verticillioides (Sacc.) Nirenberg A71 | Phytopathogenic fungus isolated from corn grain | Mycology lab |
| Macrophomina phaseolina (Tassi) Goid J431 | Phytopathogenic fungus isolated from soybean | Mycology lab |
| Phomopsis longicolla Hobbs J429 | Phytopathogenic fungus isolated from soybean | Mycology lab |
| Rhizoctonia solani J.G. Kühn Rz01 | Phytopathogenic fungus isolated from soybean | INIA |
| Sclerotium rolfsii Sacc. 1948 | Phytopathogenic fungus isolated from sweet beet | Mycology lab |
| Trichoderma atroviridae P.Karst 1607 | Biocontrol agent against fungal diseases of plants isolated from peat | Mycology lab |
| Oomycetes | ||
| Phythophtora sojae Kaufm. & Gerd Ps25 | Phytopathogenic oomycete | INIA |
| Pythium ultimum Trow Py03 | Phytopathogenic oomycete | INIA |
MGAP, Ministerio de Agricultura y Pesca, Uruguay (U-1301 [or SEMIA 587], U-1302 [or SEMIA 5019]); BIOGEM, Instituto de Investigaciones Biológicas (IIBCE)—laboratory strain collection; Mycology lab, Sección Micología, Facultad de Ciencias-Universidad de la República—laboratory strain collection; INIA, Instituto Nacional de Investigación Agropecuaria, Programa Cultivos de Secano, Estación Experimental La Estanzuela—laboratory strain collection.
Strain UY79 was then routinely grown in tryptic soy broth/agar (TSB/TSA; BD) or potato dextrose broth/agar (PDB/PDA; Oxoid Ltd.) at either 30°C or 25°C, as indicated throughout the article. Fungi and oomycetes were routinely grown on PDA at 25°C, except for Phytophthora sojae Ps25, which was grown on V8 agar medium (51). Strains of fungi and oomycetes used in this work were obtained from two culture collections: Laboratorio de Micología, Facultad de Ciencias, and Instituto Nacional de Investigación Agropecuaria, La Estanzuela.
Phylogenetic affiliation of strain UY79 using 16S rRNA.
Genomic DNA was purified using the Zymo Quick-DNA fungal/bacterial miniprep kit as described by the manufacturer.
An almost complete sequence (ca. 1,400 bp) of the 16S rRNA gene was obtained by PCR amplification using universal primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) (52) as previously described (53). Amplicons were sequenced at Macrogen, Inc. (Seoul, Korea). Forward and reverse sequences were assembled and curated using the DNA Baser V3 sequence assembler. The sequence obtained was deposited in the NCBI GenBank database under accession number MT973969. Identification of bacterial genus was accomplished using the “Identify” tool at the EzBioCloud server (54) (https://www.ezbiocloud.net/identify).
To evaluate if strain UY79 was phylogenetically related to other Paenibacillus isolated from similar environments, a phylogenetic tree was constructed including 16S rRNA gene sequences from strain UY79, 33 Paenibacillus species type strains isolated from soil, compost, rhizosphere, or different plant compartments and tissues (root nodule, root, leaf, seed), and two pathogenic strains (Paenibacillus larvae ATCC 9545 and Paenibacillus lautus NBRC 15380). Sequences were retrieved from EzBioCloud. Sequences were aligned with MAFFT v7.453, and Gblock v0.91b was used to remove poorly aligned positions. A maximum likelihood tree was assembled in MEGA X based on the Kimura two-parameter model (+G+I) (55, 56). The robustness of the tree branches was estimated with 1,000 bootstrap pseudoreplicates.
Sequencing, assembly, annotation, and mining of Paenibacillus sp. strain UY79 genome.
The genome of strain UY79 was sequenced by the paired-end-sequencing method using the Illumina TrueSeq platform (Macrogen, Seoul, Korea). Low-quality sequences were removed using Trim Galore 0.4.4 with the following line command: trim_galore –paired –three_prime_clip_R1 10 –three_prime_clip_R2 10 –length 50 UY79_1.fastq UY79_2.fastq. Filtered sequences were used for de novo assembly using Velvet Assembler 2.2.5 with the following line command: VelvetOptimiser.pl -s 19 -e 81 -d AssemVelvetOptimizer -f '-shortPaired -fastq -separate UY79_1_val_1.fq UY79_2_val_2.fq' -t 8 –. The genome was annotated by using the NCBI Prokaryotic Genome Annotation Pipeline (57) and the Rapid Annotation using Subsystem Technology (RAST 2.0) (58). The strain UY79 genome sequence was deposited in the NCBI GenBank database under accession number JAFFQR010000000. To identify gene clusters putatively encoding secondary metabolites with antimicrobial activity, we used the web server ANTISMASH 5.2.0 based on profile hidden Markov models of specific genes (59). The PROPHAGE HUNTER and PHASTER web servers were used to predict and annotate bacteriophage genes in the bacterial genome (60, 61). The BLAST tool (62) on the NCBI web server was used for manual annotation of some genes.
Multilocus sequence analysis and average nucleotide identity test.
A phylogenetic analysis based on multilocus sequences (MLSA) was performed using 16S rRNA, gyrB, rpoB, recA, and recN concatenated gene sequences (8,535 positions) retrieved from the sequenced Paenibacillus species genomes publicly available in the EzBioCloud database (http://www.ezbiocloud.net/eztaxon) (54). Sequences were aligned with MAFFT v7.453, and Gblock v0.91b was used to remove poorly aligned positions. A maximum likelihood tree was assembled in MEGA X, based on the maximum likelihood method and general time-reversible model (+G+I) (63). The robustness of the tree branches was estimated with 1,000 bootstrap pseudoreplicates.
The average nucleotide identity (ANI) score between the species included in the MLSA was calculated using the ANI calculator tool from EzBioCloud (54) (https://www.ezbiocloud.net/tools/ani).
In vitro antagonistic activity of strain UY79 against fungi and oomycetes.
For antagonism assays, strain UY79 was grown in 5 mL of TSB for 16 h at 30°C and 200 rpm. Macrophomina phaseolina J431, Rhizoctonia solani Rz01, Pythium ultimum Py03, Fusarium graminearum S127, Fusarium oxysporum J38, Phomopsis longicolla J429, Sclerotium rolfsii 1948, Fusarium verticillioides A71, Fusarium semitectum J41, Botrytis cinerea A1, and Trichoderma atroviride 1607 were cultured on PDA, and Phytophthora sojae Ps25 was cultured on V8 agar medium, at 25°C for 5 days.
To evaluate the antifungal activity of agar-diffusible compounds produced by strain UY79, a dual plate assay was performed as described by Geels and Schippers (64). Briefly, a 0.9-cm mycelial agar plug from the leading edge of the fungus/oomycete culture, previously grown for 5 days at 25°C, was placed on fresh PDA medium. Strain UY79 was streaked as a small line approximately 3 cm away from the mycelial plug. Plates were incubated at 25°C for 2 to 8 days, until radial growth of the mycelia reached the edge of the plate opposite of the side with the bacterial inoculum. The mycelial growth diameter and formation of inhibition zones around bacterial growth were recorded. Three independent assays were performed.
To detect the presence of antifungal compounds in bacterial cultures grown in liquid medium, strain UY79 was grown in 100 mL of PDB medium at 25°C and 200 rpm. After 30, 44, 68, 88, and 96 h, 10 mL was collected and centrifuged at 10,000 × g for 10 min, and supernatants were filtered through a 0.45-μm filter. Cell-free supernatants were mixed at an equal ratio with melted (45°C) PDA made with 40 g/L agar. For the negative control, PDB medium was used instead of cell-free supernatant. It is important to highlight that no significant differences were observed between fungal growth in PDA or in PDA diluted at an equal ratio with distilled water (data not shown). Two milliliters of the mixture was poured into a 6-well plate. A 0.5-cm mycelial plug from a fresh culture of F. verticillioides was placed in the center of the well and incubated for 3 days at 25°C, and mycelial growth was measured. Fungal growth on PDA medium without the addition of supernatant was used as a control. Three independent assays were performed.
To evaluate the production of volatile compounds (VCs) with antifungal activity, a dual plate assay (65) was conducted in PDA or V8 agar as indicated. Briefly, strain UY79 was grown in 5 mL of TSB for 16 h at 30°C and 200 rpm, and 100 μL of the culture was spread as a lawn in a TSA plate. A 0.9-cm mycelial plug of the fungi/oomycetes was placed in the center of a PDA plate. Both uncovered plates were placed facing each other and were sealed with Parafilm to prevent VC leakage. As controls, plates containing the fungi/oomycetes were positioned facing plates with PDA or V8 agar medium as indicated. The plates were incubated at 25°C until the mycelia of controls reached the edge of the petri dish. Antagonism was determined by measuring the percentage of growth inhibition (GI%) as follows: GI% = [(R − r) × 100]/R, where R is the radius of the mycelia of the control fungus/oomycete not facing the bacterium and r is the radius of the fungus/oomycete facing the bacterium. Three independent biological replicates were performed.
Production of the volatile compound hydrogen cyanide was assessed qualitatively by the picrate-filter paper method (66). A dual plate assay was performed, where picrate-embedded filter paper was placed in the cover of a petri dish, face to face with a lawn of bacteria. The plate was sealed with Parafilm to prevent VC leakage. Both PDA and TSA media, either with the addition of 4.4 g/L of glycine or in its absence, were assessed. Cyanogenic activity was visualized as a change in color of the filter paper from yellow to orange. Pseudomonas fluorescens UP61 was used as a positive control, and medium without bacteria was used as a negative control.
Identification of VCs produced by UY79 and by S. rolfsii 1948 when grown alone or when facing each other.
An assay similar to that conducted for the evaluation of antifungal activity of VCs was set up. Paenibacillus sp. strain UY79 was grown on PDA for 36 h or 7 days as indicated, and S. rolfsii 1948 and cultures of both microbes facing each other were grown on PDA for 7 days. Uninoculated PDA was used as a control. VCs produced were collected by using the solid-phase microextraction (SPME) technique (67) with a StableFlex fiber of divinylbenzene/carboxen/polydimethylsiloxane (50 μm divinylbenzene/30 μm carboxen/polydimethylsiloxane) (Supelco, USA). The SPME fiber was inserted between the plates and exposed for either 10 or 30 min to the VCs, depending on the sample assayed, as indicated in Table 1. Then, the fiber was desorbed into the GC port at 250°C for 1 min and separated in a GC-MS (Shimadzu QP2010 Ultra) equipped with a TR-5MS column (30 m, 0.25-mm inside diameter, 0.25 μm). The initial oven temperature was 33°C, which was held for 3 min and then raised to 80°C at a rate of 3°C/min, from 80°C to 180°C at 10°C/min, and from 180°C to 240°C at 40°C/min, and then held at 240°C for 2 min. The mass spectrometer was operated in the electron ionization mode at 70 eV and 225°C and scanned from 30 to 500 m/z. For VC identification, mass spectra were compared with those of the Wiley 139 and the NIST08/NIST08s mass spectrometry library. The GC peak area was used to estimate the relative abundance for each VC.
Determination of siderophore production and xylanase, β-glucosidase, cellulase, and protease activities.
Strain UY79 was grown in TSB for 48 h at 30°C and 200 rpm, and 10-μL drops were spotted in each of the media to be assayed. Three independent assays were performed in all the experiments. Siderophore production was assessed as the formation of an orange halo around the colony by using the chrome azurol S (CAS) agar plate method (68). To evaluate xylanase activity, the bacterium was grown on TSA containing 0.5% (wt/vol) xylan beechwood as a substrate. Xylanase activity was visualized as a clear halo around the colony (69). β-Glucosidase activity was assessed on TSA containing 0.2% (wt/vol) esculin and 0.03% (wt/vol) FeCl3. The esculetin released from esculin by β-glucosidase action was detected as a dark halo around the colonies (70). Escherichia coli DH5α β-glu and Pseudomonas putida KT2440 β-glu were used as positive controls, while E. coli DH5α and P. putida KT2440 were used as negative controls.
Cellulolytic activity was assayed on bacterial cultures grown on TSA containing 0.5% (wt/vol) carboxymethyl cellulose. Cellulase activity was detected using Congo red staining (71). Briefly, colonies were overlaid with 0.05% (wt/vol) Congo red, incubated for 10 min at room temperature, washed with distilled water, and incubated for 10 min with 1 M NaCl. An orange halo around colonies in the red background indicates cellulose-hydrolyzing activity. E. coli DH5α and P. putida KT2440 harboring the endoglucanase Cel5A from Bacillus subtilis, which confers cellulase activity, were used as positive controls, while E. coli DH5α and P. putida KT2440 were used as negative controls.
Proteolytic activity was assessed on bacterial cultures grown on TSA diluted medium (1:100) containing 5% skim milk. Protease activity was detected as a clear halo around the colony (72).
In vitro evaluation of direct plant growth-promoting activities.
In order to determine if strain UY79 has the potential to fix nitrogen, the presence of the nifH gene (a gene that encodes the nitrogenase iron protein) was assessed by PCR using the PolF (5′-TGCGAYCCSAARGCBGACTC-3′) and PolR (5′-ATSGCCATCATYTCRCCGGA-3′) primers, which amplify a 350-bp intragenic region (73). Ensifer meliloti 1021 was used as a positive control.
Indole-3-acetic acid (IAA) production by strain UY79 was determined by a colorimetric assay using the Salkowski reagent according to the method described by Gordon and Weber (74). Briefly, an IAA-producing strain (Microbacterium sp. strain UYFA 68) and strain UY79 were grown in 6 mL of TSB with or without 100 μg/mL tryptophan. The IAA concentration in cultures was determined from a calibration curve determined with pure IAA. Each treatment was performed three times.
To evaluate the ability to solubilize phosphate, bacteria were grown in NBRIP (75) or PVK (76) medium containing rock phosphate or (PO4)2Ca3 as the sole phosphate source. Solubilization of phosphate was visualized as a clear halo around the colony. Pantoea sp. strain UYSB45 was used as a positive control. Three independent assays were performed.
Effect of strain UY79 in alfalfa plant growth promotion exerted by two rhizobial strains.
Seeds of Medicago sativa var. crioula were surface sterilized with 10 mM HgCl2 in 0.1 N HCl as previously described (77). Surface-sterilized seeds were germinated at 30°C on petri dishes containing water with 0.8% (wt/vol) agar. After germination, seedlings were transferred into glass plant tubes containing 15 mL of Jensen’s N-free medium (50) with 0.8% (wt/vol) agar. Seedlings were inoculated simultaneously with Paenibacillus sp. strain UY79 and either E. meliloti 242 or E. meliloti 1021. Inoculation was done using ca. 1 × 106 CFU of each strain. The experiment included the following controls: plants solely inoculated with E. meliloti 242, E. meliloti 1021, or strain UY79. A control without bacteria was included. Plants were grown at 24°C with a photoperiod of 16 h of light and 8 h of darkness. Two independent assays were performed, and plants were harvested 6 or 10 weeks postinoculation (assays I and II, respectively). The dry weight of the aerial portion and the nodule number per plant were recorded. Eight plants (one plant per tube) were used per condition. Experiments were independently analyzed using the nonparametric Kruskal-Wallis test, and pairwise comparisons were done using the Wilcoxon test for dry weight and an analysis of variance (ANOVA) with post hoc Tukey’s honestly significant difference (HSD) test for nodule count. An alpha of 0.05 was used as the significance cutoff value for all statistical analyses.
Assessment of antibiosis activity against different soil- and plant-associated bacteria.
To investigate if strain UY79 displayed antibacterial activity, 11 Gram-negative strains (belonging to the Bradyrhizobium, Ensifer, Rhizobium, Cupriavidus, Paraburkholderia, Azospirillum, and Erwinia genera) and two Gram-positive strains (B. subtilis ATCC 6633 and Streptomyces sp. UYFA156) were used as target strains (Table 2). The soft-agar overlay assay was carried out according to James et al. (78) and Rao et al. (79) with slight modifications. Briefly, target strains were grown until late exponential phase in either TSB or TY medium, and 50 μL of the bacterial culture was used to inoculate 25 mL of soft agar (45°C) and poured into either TSA or TY plates, respectively. A 10-μL drop of a UY79 culture grown in TSB for 24 h at 30°C was spotted onto the inoculated solidified soft agar, and plates were incubated at 30°C. Antibiosis was considered positive when a zone of inhibition, around or in the spots containing the UY79 inoculum, was observed. The assay was performed twice.
Data availability.
UY79 gene sequence data were deposited in the NCBI GenBank database under accession numbers MT973969 and JAFFQR010000000.
ACKNOWLEDGMENTS
We acknowledge Karen Malán from the Analytical Platform at IIBCE for her great assistance in the GC-MS utilization.
This work was partially supported by PEDECIBA Química/Biología.
Footnotes
Supplemental material is available online only.
Contributor Information
Andrés Costa, Email: acosta@iibce.edu.uy.
Elena Fabiano, Email: efabiano@iibce.edu.uy.
Hideaki Nojiri, University of Tokyo.
REFERENCES
- 1.Savary S, Willocquet L, Pethybridge SJ, Esker P, McRoberts N, Nelson A. 2019. The global burden of pathogens and pests on major food crops. Nat Ecol Evol 3:430–439. 10.1038/s41559-018-0793-y. [DOI] [PubMed] [Google Scholar]
- 2.Lugtenberg B, Kamilova F. 2009. Plant-growth-promoting Rhizobacteria. Annu Rev Microbiol 63:541–556. 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 3.Legein M, Smets W, Vandenheuvel D, Eilers T, Muyshondt B, Prinsen E, Samson R, Lebeer S. 2020. Modes of action of microbial biocontrol in the phyllosphere. Front Microbiol 11:1619. 10.3389/fmicb.2020.01619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narayanasamy P. 2013. Mechanisms of action of fungal biological control agents, p 99–200. In Biological management of diseases of crops. Progress in biological control, vol 15. Springer, Dordrecht, The Netherlands. [Google Scholar]
- 5.Veliz EA, Martínez-Hidalgo P, Hirsch AM. 2017. Chitinase-producing bacteria and their role in biocontrol. AIMS Microbiol 3:689–705. 10.3934/microbiol.2017.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Q, Sun R, Ni M, Yu J, Li Y, Yu C, Dou K, Ren J, Chen J. 2017. Identification of a novel fungus, Trichoderma asperellum GDFS1009, and comprehensive evaluation of its biocontrol efficacy. PLoS One 12:e0179957. 10.1371/journal.pone.0179957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rybakova D, Cernava T, Köberl M, Liebminger S, Etemadi M, Berg G. 2016. Endophytes-assisted biocontrol: novel insights in ecology and the mode of action of Paenibacillus. Plant Soil 405:125–140. 10.1007/s11104-015-2526-1. [DOI] [Google Scholar]
- 8.Morath SU, Hung R, Bennett JW. 2012. Fungal volatile organic compounds: a review with emphasis on their biotechnological potential. Fungal Biol Rev 26:73–83. 10.1016/j.fbr.2012.07.001. [DOI] [Google Scholar]
- 9.Tilocca B, Cao A, Migheli Q. 2020. Scent of a killer: microbial volatilome and its role in the biological control of plant pathogens. Front Microbiol 11:41. 10.3389/fmicb.2020.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma IP, Chandra S, Kumar N, Chandra D. 2017. PGPR: heart of soil and their role in soil fertility. In Meena VMP, Bisht J, Pattanayak A (ed), Agriculturally important microbes for sustainable agriculture. Springer, Singapore. [Google Scholar]
- 11.Eljounaidi K, Lee SK, Bae H. 2016. Bacterial endophytes as potential biocontrol agents of vascular wilt diseases—review and future prospects. Biol Control 103:62–68. 10.1016/j.biocontrol.2016.07.013. [DOI] [Google Scholar]
- 12.Hong CE, Park JM. 2016. Endophytic bacteria as biocontrol agents against plant pathogens: current state-of-the-art. Plant Biotechnol Rep 10:353–357. 10.1007/s11816-016-0423-6. [DOI] [Google Scholar]
- 13.Martínez-Hidalgo P, Hirsch AM. 2017. The nodule microbiome: N2-fixing rhizobia do not live alone. Phytobiomes J 1:70–82. 10.1094/PBIOMES-12-16-0019-RVW. [DOI] [Google Scholar]
- 14.Andrews M, Andrews ME. 2017. Specificity in legume-rhizobia symbioses. Int J Mol Sci 18:705. 10.3390/ijms18040705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Meyer SE, De Beuf K, Vekeman B, Willems A. 2015. A large diversity of non-rhizobial endophytes found in legume root nodules in Flanders (Belgium). Soil Biol Biochem 83:1–11. 10.1016/j.soilbio.2015.01.002. [DOI] [Google Scholar]
- 16.Leite J, Fischer D, Rouws LF, Fernandes-Junior PI, Hofmann A, Kublik S, Schloter M, Xavier GR, Radl V. 2016. Cowpea nodules harbor non-rhizobial bacterial communities that are shaped by soil type rather than plant genotype. Front Plant Sci 7:2064. 10.3389/fpls.2016.02064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen BL, Pessotti RC, Fischer MS, Collins A, El-Hifnawi L, Liu MD, Traxler MF. 2020. Cooperation, competition, and specialized metabolism in a simplified root nodule microbiome. mBio 11:e01917-20. 10.1128/mBio.01917-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwak M-J, Choi S-B, Ha S-m, Kim EH, Kim B-Y, Chun J. 2020. Genome-based reclassification of Paenibacillus jamilae Aguilera et al. 2001 as a later heterotypic synonym of Paenibacillus polymyxa (Prazmowski 1880) Ash et al. 1994. Int J Syst Evol Microbiol 70:3134–3138. 10.1099/ijsem.0.004140. [DOI] [PubMed] [Google Scholar]
- 19.Ali MA, Lou Y, Hafeez R, Li X, Hossain A, Xie T, Lin L, Li B, Yin Y, Yan J, An Q. 2021. Functional analysis and genome mining reveal high potential of biocontrol and plant growth promotion in nodule-inhabiting bacteria within Paenibacillus polymyxa complex. Front Microbiol 11:618601. 10.3389/fmicb.2020.618601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie J, Shi H, Du Z, Wang T, Liu X, Chen S. 2016. Comparative genomic and functional analysis reveal conservation of plant growth promoting traits in Paenibacillus polymyxa and its closely related species. Sci Rep 6:21329. 10.1038/srep21329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu H, Qin S, Lan Y, Liu M, Cao X, Qiao D, Cao Y, Cao Y. 2017. Comparative genomic analysis of Paenibacillus sp. SSG-1 and its closely related strains reveals the effect of glycometabolism on environmental adaptation. Sci Rep 7:5720. 10.1038/s41598-017-06160-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharaf H, Rodrigues RR, Moon J, Zhang B, Mills K, Williams MA. 2019. Unprecedented bacterial community richness in soybean nodules vary with cultivar and water status. Microbiome 7:63. 10.1186/s40168-019-0676-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rybakova D, Rack-Wetzlinger U, Cernava T, Schaefer A, Schmuck M, Berg G. 2017. Aerial warfare: a volatile dialogue between the plant pathogen Verticillium longisporum and its antagonist Paenibacillus polymyxa. Front Plant Sci 8:1294. 10.3389/fpls.2017.01294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramette A, Frapolli M, Défago G, Moënne-Loccoz Y. 2003. Phylogeny of HCN synthase-encoding hcnBC genes in biocontrol fluorescent pseudomonads and its relationship with host plant species and HCN synthesis ability. Mol Plant Microbe Interact 16:525–535. 10.1094/MPMI.2003.16.6.525. [DOI] [PubMed] [Google Scholar]
- 25.Beims H, Bunk B, Erler S, Mohr KI, Spröer C, Pradella S, Günther G, Rohde M, von der Ohe W, Steinert M. 2020. Discovery of Paenibacillus larvae ERIC V: phenotypic and genomic comparison to genotypes ERIC I-IV reveal different inventories of virulence factors which correlate with epidemiological prevalences of American Foulbrood. Int J Med Microbiol 310:151394. 10.1016/j.ijmm.2020.151394. [DOI] [PubMed] [Google Scholar]
- 26.Ebadzadsahrai G, Higgins Keppler EA, Soby SD, Bean HD. 2020. Inhibition of fungal growth and induction of a novel volatilome in response to Chromobacterium vaccinii volatile organic compounds. Front Microbiol 11:1035. 10.3389/fmicb.2020.01035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryu CM, Farag MA, Hu CH, Reddy MS, Wei HX, Paré PW, Kloepper JW. 2003. Bacterial volatiles promote growth in Arabidopsis. Proc Natl Acad Sci USA 100:4927–4932. 10.1073/pnas.0730845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khalaf EM, Raizada MN. 2018. Bacterial seed endophytes of domesticated cucurbits antagonize fungal and oomycete pathogens including powdery mildew. Front Microbiol 9:42. 10.3389/fmicb.2018.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Y, Zhou J, Li C, Ma Y. 2019. Antifungal and plant growth promotion activity of volatile organic compounds produced by Bacillus amyloliquefaciens. Microbiologyopen 8:e00813. 10.1002/mbo3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raza W, Yuan J, Ling N, Huang Q, Shen Q. 2015. Production of volatile organic compounds by an antagonistic strain Paenibacillus polymyxa WR-2 in the presence of root exudates and organic fertilizer and their antifungal activity against Fusarium oxysporum f. sp. niveum. Biol Control 80:89–95. 10.1016/j.biocontrol.2014.09.004. [DOI] [Google Scholar]
- 31.Toffano L, Fialho MB, Pascholati SF. 2017. Potential of fumigation of orange fruits with volatile organic compounds produced by Saccharomyces cerevisiae to control citrus black spot disease at postharvest. Biol Control 108:77–82. 10.1016/j.biocontrol.2017.02.009. [DOI] [Google Scholar]
- 32.Weisskopf L, Schulz S, Garbeva P. 2021. Microbial volatile organic compounds in intra-kingdom and inter-kingdom interactions. Nat Rev Microbiol 19:391–404. 10.1038/s41579-020-00508-1. [DOI] [PubMed] [Google Scholar]
- 33.Powell PE, Szaniszlo PJ, Cline GR, Reid CPP. 1982. Hydroxamate siderophores in the iron nutrition of plants. J Plant Nutr 5:653–673. 10.1080/01904168209362994. [DOI] [Google Scholar]
- 34.Gu S, Yang T, Shao Z, Wang T, Cao K, Jousset A, Friman VP, Mallon C, Mei X, Wei Z, Xu Y, Shen Q, Pommier T. 2020. Siderophore-mediated interactions determine the disease suppressiveness of microbial consortia. mSystems 5:e00811-19. 10.1128/mSystems.00811-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franza T, Mahé B, Expert D. 2005. Erwinia chrysanthemi requires a second iron transport route dependent of the siderophore achromobactin for extracellular growth and plant infection. Mol Microbiol 55:261–275. 10.1111/j.1365-2958.2004.04383.x. [DOI] [PubMed] [Google Scholar]
- 36.Brambilla S, Frare R, Soto G, Jozefkowicz C, Ayub N. 2018. Absence of the nitrous oxide reductase gene cluster in commercial alfalfa inoculants is probably due to the extensive loss of genes during rhizobial domestication. Microb Ecol 76:299–302. 10.1007/s00248-018-1145-9. [DOI] [PubMed] [Google Scholar]
- 37.Mallik MAB, Tesfai K. 1985. Pesticidal effect on soybean-rhizobia symbiosis. Plant Soil 85:33–41. 10.1007/BF02197798. [DOI] [Google Scholar]
- 38.Moorman TB. 1989. A review of pesticide effects on microorganisms and microbial processes related to soil fertility. J Prod Agric 2:14–23. 10.2134/jpa1989.0014. [DOI] [Google Scholar]
- 39.Li Y, Chen S. 2019. Fusaricidin produced by Paenibacillus polymyxa WLY78 induces systemic resistance against Fusarium wilt of cucumber. Int J Mol Sci 20:5240. 10.3390/ijms20205240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kajimura Y, Kaneda M. 1996. Fusaricidin A, a new depsipeptide antibiotic produced by Bacillus polymyxa KT-8. Taxonomy, fermentation, isolation, structure elucidation and biological activity. J Antibiot 49:129–135. 10.7164/antibiotics.49.129. [DOI] [PubMed] [Google Scholar]
- 41.Cochrane SA, Lohans CT, Brandelli JR, Mulvey G, Armstrong GD, Vederas JC. 2014. Synthesis and structure-activity relationship studies of N-terminal analogues of the antimicrobial peptide Tridecaptin A1. J Med Chem 57:1127–1131. 10.1021/jm401779d. [DOI] [PubMed] [Google Scholar]
- 42.Upert G, Luther A, Obrecht D, Ermert P. 2021. Emerging peptide antibiotics with therapeutic potential. Med Drug Discov 9:100078. 10.1016/j.medidd.2020.100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aleti G, Sessitsch A, Brader G. 2015. Genome mining: prediction of lipopeptides and polyketides from Bacillus and related Firmicutes. Comput Struct Biotechnol J 13:192–203. 10.1016/j.csbj.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langendries S, Goormachtig S. 2021. Paenibacillus polymyxa, a Jack of all trades. Environ Microbiol 23:5659–5669. 10.1111/1462-2920.15450. [DOI] [PubMed] [Google Scholar]
- 45.Zhu S, Hegemann JD, Fage CD, Zimmermann M, Xie X, Linne U, Marahiel MA. 2016. Insights into the unique phosphorylation of the lasso peptide paeninodin. J Biol Chem 291:13662–13678. 10.1074/jbc.M116.722108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kandasamy V, Liu J, Dantoft SH, Solem C, Jensen PR. 2016. Synthesis of (3R)-acetoin and 2,3-butanediol isomers by metabolically engineered Lactococcus lactis. Sci Rep 6:36769–36769. 10.1038/srep36769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tinôco D, Pateraki C, Koutinas AA, Freire DMG. 2021. Bioprocess development for 2,3-butanediol production by Paenibacillus strains. Chem Bio Eng Rev 8:44–62. 10.1002/cben.202000022. [DOI] [Google Scholar]
- 48.Cann AF, Liao JC. 2008. Production of 2-methyl-1-butanol in engineered Escherichia coli. Appl Microbiol Biotechnol 81:89–98. 10.1007/s00253-008-1631-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han Q, Ma Q, Chen Y, Tian B, Xu L, Bai Y, Chen W, Li X. 2020. Variation in rhizosphere microbial communities and its association with the symbiotic efficiency of rhizobia in soybean. ISME J 14:1915–1928. 10.1038/s41396-020-0648-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vincent JM. 1970. A manual for the practical study of root-nodule bacteria. Blackwell Scientific Ltd., Oxford, United Kingdom. [Google Scholar]
- 51.Dhingra OB, Sinclair JB. 1995. Basic plant pathology methods, 2nd ed. CRC Press, Boca Raton, FL. [Google Scholar]
- 52.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taule C, Zabaleta M, Mareque C, Platero R, Sanjurjo L, Sicardi M, Frioni L, Battistoni F, Fabiano E. 2012. New betaproteobacterial Rhizobium strains able to efficiently nodulate Parapiptadenia rigida (Benth.) Brenan. Appl Environ Microbiol 78:1692–1700. 10.1128/AEM.06215-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J. 2017. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 56.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haft DH, DiCuccio M, Badretdin A, Brover V, Chetvernin V, O'Neill K, Li W, Chitsaz F, Derbyshire MK, Gonzales NR, Gwadz M, Lu F, Marchler GH, Song JS, Thanki N, Yamashita RA, Zheng C, Thibaud-Nissen F, Geer LY, Marchler-Bauer A, Pruitt KD. 2018. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res 46:D851–D860. 10.1093/nar/gkx1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T. 2019. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87. 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21. 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song W, Sun HX, Zhang C, Cheng L, Peng Y, Deng Z, Wang D, Wang Y, Hu M, Liu W, Yang H, Shen Y, Li J, You L, Xiao M. 2019. Prophage Hunter: an integrative hunting tool for active prophages. Nucleic Acids Res 47:W74–W80. 10.1093/nar/gkz380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 63.Nei M, Kumar N. 2000. Molecular evolution and phylogenetics. Oxford University Press, New York, NY. [Google Scholar]
- 64.Geels FP, Schippers B. 1983. Selection of antagonistic fluorescent Pseudomonas spp. and their root colonization and persistence following treatment of seed potatoes. J Phytopathol 108:193–206. 10.1111/j.1439-0434.1983.tb00579.x. [DOI] [Google Scholar]
- 65.Raza W, Ling N, Liu D, Wei Z, Huang Q, Shen Q. 2016. Volatile organic compounds produced by Pseudomonas fluorescens WR-1 restrict the growth and virulence traits of Ralstonia solanacearum. Microbiol Res 192:103–113. 10.1016/j.micres.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 66.Egan SV, Yeoh HH, Bradbury JH. 1998. Simple picrate paper kit for determination of the cyanogenic potential of cassava flour. J Sci Food Agric 76:39–48. . [DOI] [PubMed] [Google Scholar]
- 67.Di Francesco A, Ugolini L, Lazzeri L, Mari M. 2015. Production of volatile organic compounds by Aureobasidium pullulans as a potential mechanism of action against postharvest fruit pathogens. Biol Control 81:8–14. 10.1016/j.biocontrol.2014.10.004. [DOI] [Google Scholar]
- 68.Schwyn B, Neilands JB. 1987. Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56. 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 69.Kulkarni P, Gupta N. 2013. Screening and evaluation of soil fungal isolates for xylanase production. Res Sci Technol 5:33–36. [Google Scholar]
- 70.Eberhart B, Cross DF, Chase LR. 1964. Beta-glucosidase system of Neurospora crassa. I. Beta-glucosidase and cellulase activities of mutant and wild-type strains. J Bacteriol 87:761–770. 10.1128/jb.87.4.761-770.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim SJ, Lee CM, Han BR, Kim MY, Yeo YS, Yoon SH, Koo BS, Jun HK. 2008. Characterization of a gene encoding cellulase from uncultured soil bacteria. FEMS Microbiol Lett 282:44–51. 10.1111/j.1574-6968.2008.01097.x. [DOI] [PubMed] [Google Scholar]
- 72.Martínez-Rosales C, Castro-Sowinski S. 2011. Antarctic bacterial isolates that produce cold-active extracellular proteases at low temperature but are active and stable at high temperature. Polar Res 30:7123. 10.3402/polar.v30i0.7123. [DOI] [Google Scholar]
- 73.Poly F, Monrozier LJ, Bally R. 2001. Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res Microbiol 152:95–103. 10.1016/s0923-2508(00)01172-4. [DOI] [PubMed] [Google Scholar]
- 74.Gordon SA, Weber RP. 1951. Colorimetric estimation of indoleacetic acid. Plant Physiol 26:192–195. 10.1104/pp.26.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nautiyal CS. 1999. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol Lett 170:265–270. 10.1111/j.1574-6968.1999.tb13383.x. [DOI] [PubMed] [Google Scholar]
- 76.Pikovskaya RI. 1948. Mobilization of phosphorus in soil connection with the vital activity of some microbial species. Microbiology 17:362–370. [Google Scholar]
- 77.Pereira-Gómez M, Ríos C, Zabaleta M, Lagurara P, Galvalisi U, Iccardi P, Azziz G, Battistoni F, Platero R, Fabiano E. 2020. Native legumes of the Farrapos protected area in Uruguay establish selective associations with rhizobia in their natural habitat. Soil Biol Biochem 148:107854. 10.1016/j.soilbio.2020.107854. [DOI] [Google Scholar]
- 78.James SG, Holmström C, Kjelleberg S. 1996. Purification and characterization of a novel antibacterial protein from the marine bacterium D2. Appl Environ Microbiol 62:2783–2788. 10.1128/aem.62.8.2783-2788.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rao D, Webb JS, Kjelleberg S. 2005. Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudoalteromonas tunicata. Appl Environ Microbiol 71:1729–1736. 10.1128/AEM.71.4.1729-1736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tarrand JJ, Krieg NR, Döbereiner J. 1978. A taxonomic study of the Spirillum lipoferum group, with descriptions of a new genus, Azospirillum gen. nov. and two species, Azospirillum lipoferum (Beijerinck) comb. nov. and Azospirillum brasilense sp. nov. Can J Microbiol 24:967–980. 10.1139/m78-160. [DOI] [PubMed] [Google Scholar]
- 81.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 82.Alves LF, Meleiro LP, Silva RN, Westmann CA, Guazzaroni ME. 2018. Novel ethanol- and 5-hydroxymethyl furfural-stimulated β-glucosidase retrieved from a Brazilian secondary Atlantic forest soil metagenome. Front Microbiol 9:2556. 10.3389/fmicb.2018.02556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meade HM, Long SR, Ruvkun GB, Brown SE, Ausubel FM. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol 149:114–122. 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pirhonen M, Palva ET. 1988. Occurrence of bacteriophage T4 receptor in Erwinia carotovora. Mol Gen Genet 214:170–172. 10.1007/BF00340198. [DOI] [Google Scholar]
- 85.Vaz Jauri P, Taulé C, de los Santos MC, Fernandez B, Di Paolo A, Sotelo J, Battistoni F. 2020. Interactions between putatively endophytic bacteria and tall fescue (Festuca arundinacea): plant growth promotion and colonization in host and non-host cultivars. Plant Soil 451:207–220. 10.1007/s11104-019-04359-5. [DOI] [Google Scholar]
- 86.Mareque C, Taulé C, Beracochea M, Battistoni F. 2015. Isolation, characterization and plant growth promotion effects of putative bacterial endophytes associated with sweet sorghum (Sorghum bicolor (L) Moench). Ann Microbiol 65:1057–1067. 10.1007/s13213-014-0951-7. [DOI] [Google Scholar]
- 87.Bagnasco P, De La Fuente L, Gualtieri G, Noya F, Arias A. 1998. Fluorescent Pseudomonas spp. as biocontrol agents against forage legume root pathogenic fungi. Soil Biol Biochem 30:1317–1322. 10.1016/S0038-0717(98)00003-0. [DOI] [Google Scholar]
- 88.De La Fuente L, Thomashow L, Weller D, Bajsa N, Quagliotto L, Chernin L, Arias A. 2004. Pseudomonas fluorescens UP61 isolated from birdsfoot trefoil rhizosphere produces multiple antibiotics and exerts a broad spectrum of biocontrol activity. Eur J Plant Pathol 110:671–681. 10.1023/B:EJPP.0000041569.35143.22. [DOI] [Google Scholar]
- 89.Paulsen IT, Press CM, Ravel J, Kobayashi DY, Myers GSA, Mavrodi DV, DeBoy RT, Seshadri R, Ren Q, Madupu R, Dodson RJ, Durkin AS, Brinkac LM, Daugherty SC, Sullivan SA, Rosovitz MJ, Gwinn ML, Zhou L, Schneider DJ, Cartinhour SW, Nelson WC, Weidman J, Watkins K, Tran K, Khouri H, Pierson EA, Pierson LS, Thomashow LS, Loper JE. 2005. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat Biotechnol 23:873–878. 10.1038/nbt1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Regenhardt D, Heuer H, Heim S, Fernandez DU, Strömpl C, Moore ER, Timmis KN. 2002. Pedigree and taxonomic credentials of Pseudomonas putida strain KT2440. Environ Microbiol 4:912–915. 10.1046/j.1462-2920.2002.00368.x. [DOI] [PubMed] [Google Scholar]
- 91.Martínez-Romero E, Segovia L, Mercante FM, Franco AA, Graham P, Pardo MA. 1991. Rhizobium tropici, a novel species nodulating Phaseolus vulgaris L. beans and Leucaena sp. trees. Int J Syst Bacteriol 41:417–426. 10.1099/00207713-41-3-417. [DOI] [PubMed] [Google Scholar]
- 92.Ponce de León I, Oliver JP, Castro A, Gaggero C, Bentancor M, Vidal S. 2007. Erwinia carotovora elicitors and Botrytis cinerea activate defense responses in Physcomitrella patens. BMC Plant Biol 7:52. 10.1186/1471-2229-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4, Tables S1 to S3. Download AEM.01645-21-s0001.pdf, PDF file, 1.9 MB (1.9MB, pdf)
Data Availability Statement
UY79 gene sequence data were deposited in the NCBI GenBank database under accession numbers MT973969 and JAFFQR010000000.