ABSTRACT
Mutualistic nutrient cycling in the coral-algae symbiosis depends on limited nitrogen (N) availability for algal symbionts. Denitrifying prokaryotes capable of reducing nitrate or nitrite to dinitrogen could thus support coral holobiont functioning by limiting N availability. Octocorals show some of the highest denitrification rates among reef organisms; however, little is known about the community structures of associated denitrifiers and their response to environmental fluctuations. Combining 16S rRNA gene amplicon sequencing with nirS in-silico PCR and quantitative PCR, we found differences in bacterial community dynamics between two octocorals exposed to excess dissolved organic carbon (DOC) and concomitant warming. Although bacterial communities of the gorgonian Pinnigorgia flava remained largely unaffected by DOC and warming, the soft coral Xenia umbellata exhibited a pronounced shift toward Alphaproteobacteria dominance under excess DOC. Likewise, the relative abundance of denitrifiers was not altered in P. flava but decreased by 1 order of magnitude in X. umbellata under excess DOC, likely due to decreased proportions of Ruegeria spp. Given that holobiont C:N ratios remained stable in P. flava but showed a pronounced increase with excess DOC in X. umbellata, our results suggest that microbial community dynamics may reflect the nutritional status of the holobiont. Hence, denitrifier abundance may be directly linked to N availability. This suggests a passive regulation of N cycling microbes based on N availability, which could help stabilize nutrient limitation in the coral-algal symbiosis and thereby support holobiont functioning in a changing environment.
IMPORTANCE Octocorals are important members of reef-associated benthic communities that can rapidly replace scleractinian corals as the dominant ecosystem engineers on degraded reefs. Considering the substantial change in the (a)biotic environment that is commonly driving reef degradation, maintaining a dynamic and metabolically diverse microbial community might contribute to octocoral acclimatization. Nitrogen (N) cycling microbes, in particular denitrifying prokaryotes, may support holobiont functioning by limiting internal N availability, but little is known about the identity and (a)biotic drivers of octocoral-associated denitrifiers. Here, we show contrasting dynamics of bacterial communities associated with two common octocoral species, the soft coral Xenia umbellata and the gorgonian Pinnigorgia flava after a 6-week exposure to excess dissolved organic carbon under concomitant warming conditions. The specific responses of denitrifier communities of the two octocoral species aligned with the nutritional status of holobiont members. This suggests a passive regulation based on N availability in the coral holobiont.
KEYWORDS: nitrogen limitation, denitrification, microbial plasticity, Symbiodiniaceae-bacteria interactions, phase shifts
INTRODUCTION
Coral reefs are hot spots of marine biodiversity and primary productivity in oligotrophic tropical oceans (1). Corals, the ecosystem engineers of these reefs, are key to supporting these ecosystems (2). The symbiosis with intracellular dinoflagellate algae of the family Symbiodiniaceae is central to this ecological success as it enables corals access to heterotrophic as well as autotrophic nutrient sources to supporting growth and productivity (3, 4). In particular, the translocation of organic carbon (C) in the form of photosynthates by symbiotic algae is a major energy source for the coral host, which provides inorganic nutrients and carbon dioxide from its catabolism to supporting algal photosynthesis (5). However, the efficient symbiotic trade of C in the coral-algae symbiosis depends heavily on limited nitrogen (N) availability for the algae. Constant N limitation of algal symbionts is required to limit their populations and ensure the accumulation of excess photosynthates available for translocation (5, 6). Given the pronounced environmental fluctuations and seasonality in coral reefs, the functioning of symbiosis thus depends on active and/or passive regulation of nutrient availability for algal symbionts, summarized in (7). Importantly, the nutrient availability in the symbiosis does not depend on interactions of the host and its symbiotic algae alone (8, 9). Corals also associate with a diverse prokaryotic microbiome with varying degrees of taxonomic flexibility depending on host species and environmental conditions (10). Many members of the prokaryotic microbiome are or may be actively involved in the provision and recycling of limiting nutrients such as N, phosphorus (P), thereby altering nutrient availability for the holobiont (11–14). As such, the ecological success of corals likely depends on an intricate functional interplay of all its microbial associates. This diverse multispecies assemblage termed the coral holobiont extends the metabolic properties of its members and may help their rapid adaptation to changing environmental conditions (15).
For millions of years, the functional interplay between holobiont members has formed the basis of the ecological success of corals and the reefs they support (16–18). Yet in recent decades, anthropogenic activities have led to widespread coral mortality and reef degradation (19, 20). Global and local stressors such as ocean warming and labile DOC loading are known to disrupt holobiont functioning, resulting in coral bleaching (i.e., the collapse of the coral-algal symbiosis) or coral diseases (21–23). The breakdown of this symbiosis is not exclusively restricted to interactions between the coral host and their algal symbionts but involves other members of the holobiont as well (e.g., prokaryotes) (10, 24, 25). Given that the stability of the coral-algal symbiosis is dependent on N limitation of the algal symbionts, microbial N cycling may stabilize or destabilize holobiont functioning depending on the environmental conditions (7, 14, 26).
In particular, denitrifiers, that is, prokaryotes that encompass the reduction of nitrate or nitrite to dinitrogen gas (27), might help alleviate excess N stress of holobionts (7). Denitrifiers appear to be widely associated with many reef organisms and their activity was recently confirmed in corals from the Red Sea (28). Specifically, it has been observed that the activity of microbial denitrification in coral holobionts increased with environmental N levels (29). As such, changes in community structure, abundance, and activity of denitrifiers might directly affect coral holobiont functioning by altering N availability for other holobiont members. Yet, it remains unexplored how environmental changes, such as ocean warming and excess DOC, influence coral-associated denitrifying bacterial communities.
To date, most studies have focused on the bacterial community dynamics of scleractinian corals given their importance as reef ecosystem engineers (25, 30). In contrast, even though octocorals constitute highly abundant members of benthic communities of coral reefs, their bacterial community dynamics under environmental stress remain largely unexplored (31). Octocorals were recently shown to exhibit some of the highest denitrification rates among Red Sea reef organisms and substrates (32), indicating a potential importance of this microbial functional trait to octocoral health and holobiont functioning. However, the (a)biotic drivers of denitrifier abundance and community composition in octocoral holobionts are unknown. Here, we set out to explore the effects of excess DOC and concomitant warming on bacterial community structure with a focus on diversity and abundance of putative denitrifiers associated with the pulsating soft coral Xenia umbellata (Lamarck, 1816) and the gorgonian Pinnigorgia flava (Nutting, 1910). The experiment consisted of two consecutive phases. In phase 1, excess DOC enrichment was performed over the course of 21 days at varying concentrations via administration of daily glucose dosing. In phase 2, the continued DOC enrichment was combined with stepwise warming for 24 days (Fig. 1). By doing so, we aimed to (i) provide a comprehensive overview of the bacterial symbionts associated with two common and taxonomically distinct octocoral species and their response to excess DOC and ocean warming; and (ii) provide a comparative assessment on the abundance, diversity, and community dynamics of associated putative denitrifiers in these octocoral holobionts.
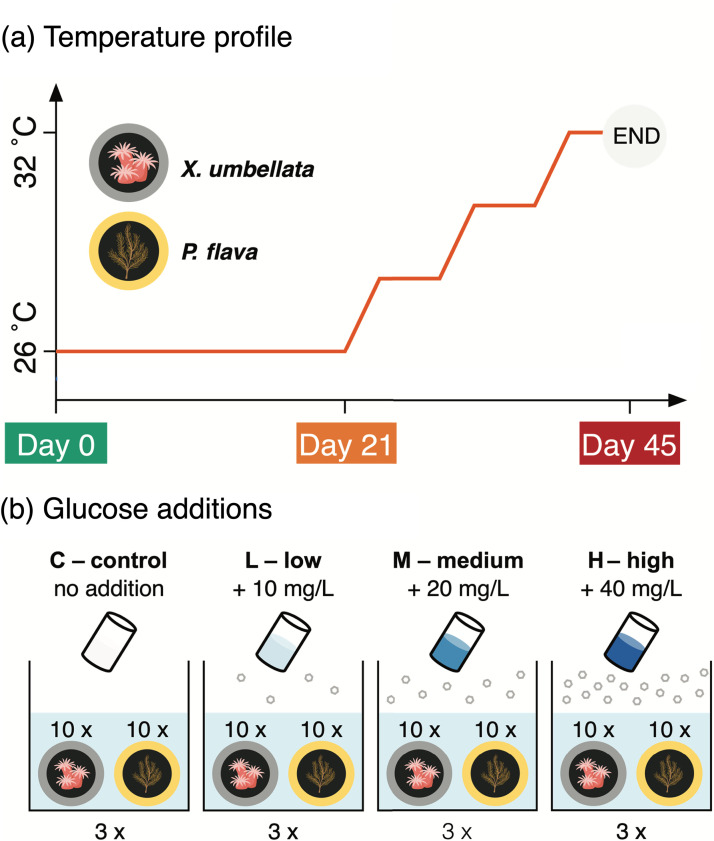
FIG 1.
Design of the conducted manipulative aquarium experiment. (a) Step-wise increases in temperature started from day 21 to day 45 with an increase in 2°C every 8 days. (b) Dissolved organic carbon (i.e., glucose) additions were applied daily throughout the experiment.
RESULTS
Overview of 16S rRNA gene sequencing data.
Over the duration of 45 days of the aquarium experiment with two octocoral species, using four levels of DOC concentrations and two different temperatures, a total of 78 coral fragments were collected for 16S rRNA gene amplicon sequencing on the Illumina MiSeq platform. Of these, 76 samples passed quality control, resulting in 8,145 amplicon sequence variants (ASVs) based on 1,150,262 sequences. Of total ASVs, 5,703 ASVs were present in both coral species. 1,847 ASVs were exclusively associated with the soft coral X. umbellata and 595 ASVs with the gorgonian P. flava only. Under undisturbed condition, the total number of bacterial ASVs associated with X. umbellata (6,344 ASVs at day 0) was higher than in P. flava (4,260 ASVs at day 0). However, over the course of our experiment, X. umbellata showed a decline in total ASV diversity regardless of DOC treatments (1,618 and 1,058 ASVs for day 21 and 45, respectively), as well as significantly reduced Inverse Simpson Index (two-way ANOVA, time-effect: F2, 24 = 14.97, P < 0.001, DOC-effect: F4, 24 = 5.76, P = 0.002). Specifically, compared to day 0, the Inverse Simpson Index experienced an 80% reduction at day 21 (Tukey's honestly significant difference [HSD], P = 0.003) and an 84% reduction at day 45 (Tukey's HSD, P < 0.001) (Fig. S1 and Fig. S2 in the supplemental material). In contrast, ASV numbers and Inverse Simpson Index remained stable in the P. flava bacterial microbiome (Two-way ANOVA, time-effect: F2, 26 = 1.57, P = 0.227, DOC-effect: F4, 26 = 0.20, P = 0.936; Fig. S1 and Fig. S2) throughout the experiment.
Distinct bacterial communities and bacterial community responses of X. umbellata and P. flava.
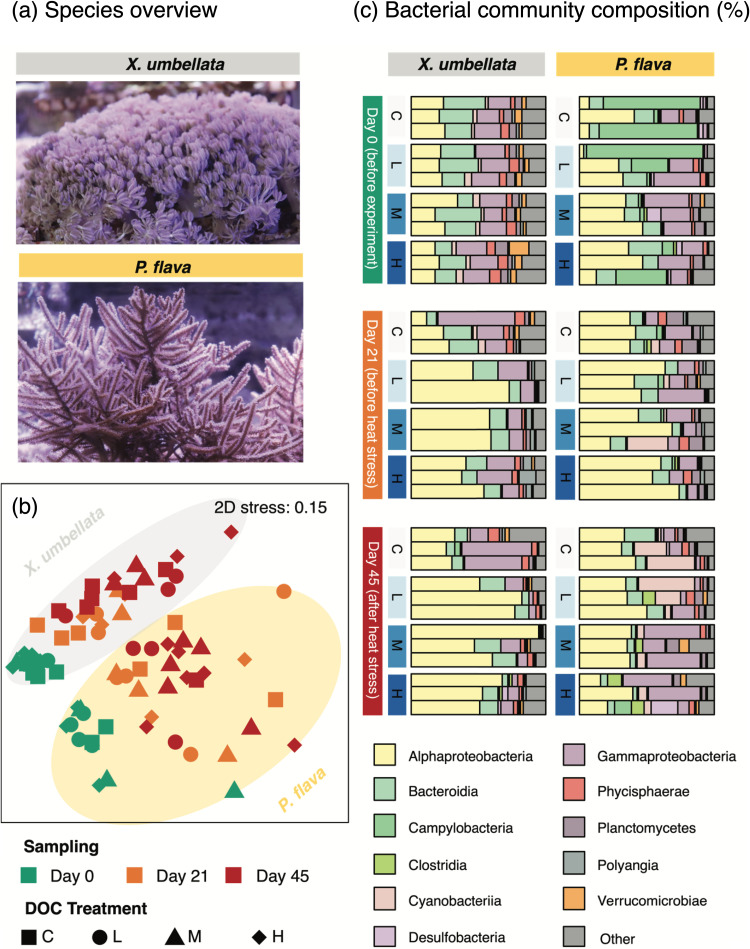
At day 0, that is, just prior to the start of the experiment, both octocoral species were associated with distinct dominant bacterial taxa (analysis of similarities [ANOSIM], R = 0.68, P = 0.001; Fig. 2b, c). In X. umbellata-associated bacterial communities, Alphaproteobacteria (25–30% proportion; ca. 70% of sequences affiliated to Rhodobacteraceae), Bacteroidia (20–25% proportion; ca. 75% of sequences affiliated to Kordia) and Gammaproteobacteria (15–25% proportion; ca. 70% of sequences affiliated to Alteromonadaceae) were identified as dominant classes. In contrast, P. flava was dominated by Alphaproteobacteria (20–40% proportion; ca. 65% of sequences affiliated to Rhodobacteraceae), Gammaproteobacteria (10–40% proportion; ca. 16% of sequences affiliated to Vibrionaceae) and Campylobacteria (15–80% proportion; ca. 70% of sequences affiliated to Arcobacteraceae) (Fig. 2c).
FIG 2.
Bacterial community compositions of corals X. umbellata and P. flava over the course of the experiment. (a) Representative photographs of corals. (b) Non-metric multidimensional scaling (NMDS) plot based on Bray-Curtis dissimilarity matrix of bacterial community compositions associated with coral samples at day 0, day 21, and day 45. (c) Stacked bar plots of bacterial community compositions of corals across different time points and DOC treatments. Stacked bar plots display the 10 most abundant bacterial classes (>1%, determined for each coral species separately).
X. umbellata bacterial communities significantly varied across time points (PERMANOVA, F2, 22 = 5.07, P = 0.001), and were significantly affected by excess DOC (PERMANOVA, F3, 22 = 1.92, P = 0.003) (Fig. 2c). At day 21, bacterial community structure of X. umbellata at control did not markedly change from day 0 (ANOSIM, R = 0.52, P = 0.132; Fig. 2c). Yet, bacterial communities under excess DOC separated well from the control (ANOSIM, R = 1.00, P = 0.132; Fig. 2c) by showing a 2-fold increase in the proportion of Alphaproteobacteria, primarily ASVs affiliated to Rhodobacteraceae (63%–75% proportion of Alphaproteobacteria) and Hyphomonadaceae (25–28%). Until day 45, following continuous excess DOC paired with a temperature increase to 32°C, X. umbellata bacterial communities were dominated by Alphaproteobacteria (mainly Rhodobacteraceae; 75–80% of all sequences in this class), a higher proportion than in corals with excess DOC at day 21 (ANOSIM, R = 1.00, P = 0.132; Fig. 2c). P. flava-associated bacterial communities differed significantly over time (PERMANOVA, F2, 24 = 2.60, P = 0.001). This was first and foremost driven by the 8-fold increased proportion of ASV5, which was affiliated with Paraspirulinaceae, in corals without excess DOC. In contrast, the bacterial community structure of P. flava did not significantly respond to excess DOC throughout the experiment (PERMANOVA, F3, 24 = 0.99, P = 0.500; Fig. 2c).
Pronounced effects of excess DOC on composition and abundance of putative denitrifiers associated with X. umbellata.
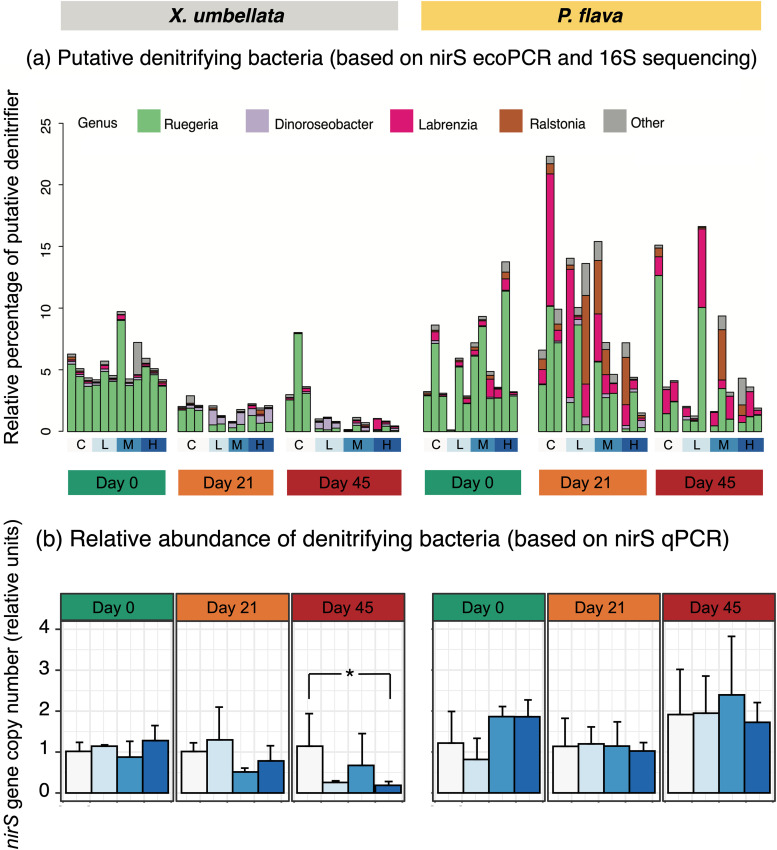
The nirS in-silico PCR and 16S rRNA gene sequencing retrieved a total of 97 ASVs distributed over 14 putative denitrifying genera from 76 octocoral samples. Putative denitrifying taxa were represented by 12 genera and 75 ASVs and accounted for approximately 10% of the X. umbellata bacterial community. The putative denitrifying community in P. flava varied more across samples than in X. umbellata, with denitrifiers being represented by 14 genera from 79 ASVs, accounting for up to 23% of the overall bacterial community. Ruegeria (ca. 90% of denitrifier-affiliated sequences, across 23 ASVs dominated by 60–65% ASV8 and 26–35% ASV11) showed its dominance in both octocoral denitrifying communities at day 0. Labrenzia (including ASV43, ASV126, and ASV179) was the second most dominant genus and occupied a higher proportion in P. flava (Fig. 3a).
FIG 3.
Community compositions and relative abundances of putative denitrifiers in corals X. umbellata and P. flava over the course of the experiment. (a) Relative proportions of denitrifier genera of corals X. umbellata and P. flava inferred by nirS in-silico PCR in relation to the total bacterial community from 16S rRNA gene sequencing. (b) Relative fold changes in copy numbers of nirS gene referenced to 16S rRNA gene and in relation to the day 0 control samples (n = 3) of corals X. umbellata and P. flava. Values are means ± SD, and the asterisk indicates statistically significant differences (*P < 0.05).
The denitrifying community of X. umbellata revealed a ca. 80% reduction in sequence proportion related to Ruegeria spp. and a ca. 30% increase in the proportion of sequences affiliated with Dinoroseobacter spp. after 21 days of excess DOC (Fig. 3a). However, this shift did not affect the cumulative relative abundance of putative denitrifiers, as estimated by nirS gene relative abundance (ANOVA, F3, 24 = 2.280, P = 0.105; Fig. 3b). Denitrifying community structure in P. flava showed no significant response to DOC, as well as a non-significant change in the nirS relative abundance based on qPCR (ANOVA, F3, 24 = 0.961, P = 0.427; Fig. 3b).
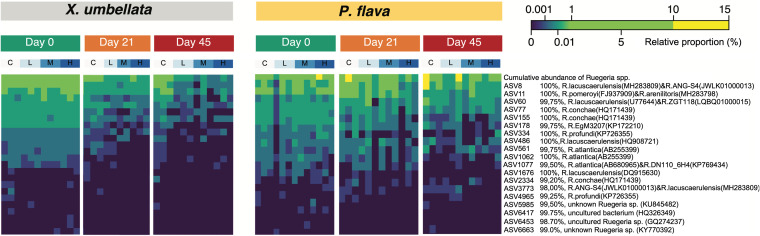
The denitrifiers of X. umbellata revealed significant changes in community structure across treatments at day 45. This was primarily due to the reduced proportion of Ruegeria spp. under excess DOC and concomitant warming (Fig. 3a, Fig. 4). This shift in the community was further reflected in a significant decrease in nirS relative abundance in corals under excess DOC (ANOVA, F6, 24 = 2.45, P = 0.05; Fig. 3b), especially when comparing the high DOC treatment (at a concentration of 40 mg L−1) to the control (Tukey's HSD, P = 0.03). Conversely, denitrifiers in P. flava showed no significant change in community structure or cumulative relative abundance (nirS qPCR, ANOVA, F6, 24 = 0.93, P = 0.49; Fig. 3b) in the presence of excess DOC and warming.
FIG 4.
Relative proportions of putative denitrifying Ruegeria spp. ASVs in total bacterial communities associated with corals X. umbellata and P. flava across different time points and DOC treatments.
Physiological changes in X. umbellata subjected to excess DOC and warming.
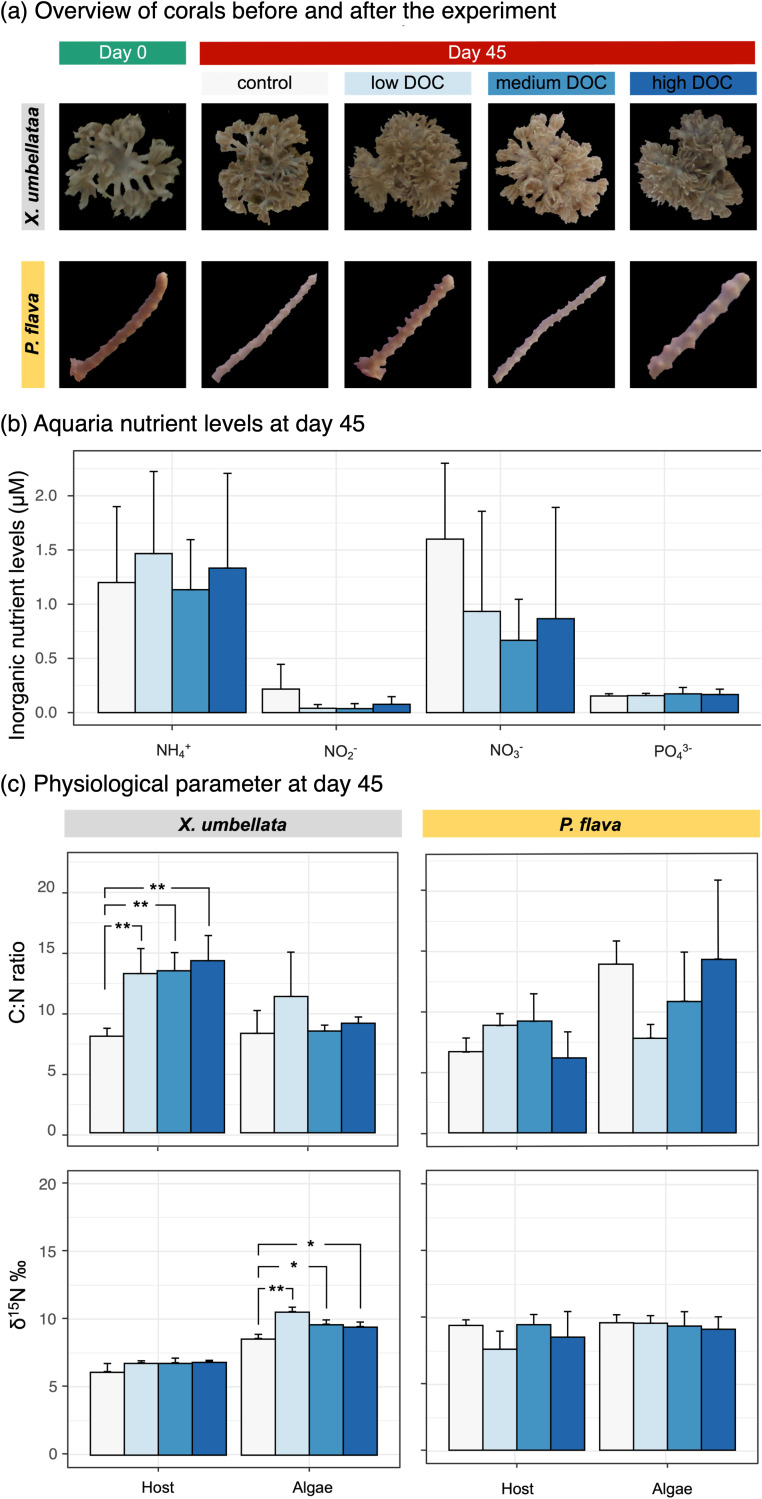
No change in phenotype was observed for the two octocoral species with excess DOC for 21 days. Even at the end of the experiment after combined excess DOC and warming conditions, X. umbellata still maintained a healthy appearance across excess DOC groups (Fig. 5a). In contrast, moderate bleaching was observed in P. flava under higher DOC concentrations (i.e., 20 mg L−1, 40 mg L−1; Fig. 5a). At day 45, X. umbellata with excess DOC showed 30% higher C:N ratios in host tissues compared to their control counterparts (ANOVA, F3, 8 = 12.59, P = 0.002; Fig. 5c), while no significant difference was observed in C:N ratios of Symbiodiniaceae (ANOVA, F3, 8 = 1.002, P = 0.44; Fig. 5c). This was accompanied by a moderate albeit not significant increase in host δ15N signatures (ANOVA, F3, 8 = 2.7, P = 0.116), as well as a 25% increase in δ15N signatures of Symbiodiniaceae under excess DOC (ANOVA, F3, 8 = 19.44, P < 0.001; Fig. 5c). There was no statistically significant difference across treatments in C:N ratios of P. flava host (ANOVA, F3, 8 = 2.435, P = 0.14) and Symbiodiniaceae (ANOVA, F3, 8 = 2.508, P = 0.133), nor in δ15N‰ signatures of host (ANOVA, F3, 8 = 1.396, P = 0.313) and Symbiodiniaceae (ANOVA, F3, 8 = 0.234, P = 0.87) (Fig. 5c).
FIG 5.
Physiological changes in corals X. umbellata and P. flava at the end of the experiment. (a) Phenotypes of corals X. umbellata and P. flava before and after the experiment. (b) Seawater inorganic nutrient levels in all aquaria at the end of the experiment. (c) Elemental (carbon and nitrogen) changes in both coral holobionts. Values are means ± SD, and the asterisk indicates statistically significant differences (*P < 0.05, **P < 0.01).
At the end of the experiment, seawater nutrient levels did not significantly differ among treatments with regard to NH4+ (ANOVA, F3, 8 = 0.069, P = 0.975) and PO43- (ANOVA: F3, 8 = 10.08, P = 0.969) levels (Fig. 5b). Nevertheless, NO2- (0.22 ± 0.19 μM) and NO3- (1.60 ± 0.57 μM) concentrations from the control were 2- to 4-fold higher (NO2-, ANOVA, F3, 8 = 1.502, P = 0.286; NO3-, ANOVA, F3, 8 = 0.741, P = 0.557; Fig. 5b) than those of aquaria with excess DOC (NO2-: 0.05 ± 0.04 μM; NO3-: 0.84 ± 0.66 μM).
DISCUSSION
N cycling microbes, including denitrifiers, are considered key players in the coral holobiont, as they might help in maintaining a N-limited state critical for the functioning of the coral-algae symbiosis (7, 33, 34). In the current study, we provide an assessment of the overall bacterial and denitrifying community structures associated with two octocoral species as well as their abundances and dynamics under excess DOC and concomitant warming in a 6-week aquarium experiment. We found pronounced octocoral species-specific responses to excess DOC in bacterial community structure in general, and in denitrifier community composition and abundance, which aligned with patterns in C:N ratios of host tissues. Taken together, our results suggest a link between octocoral-associated denitrifiers and nutritional status (i.e., N availability of the holobiont), as well as a passive regulation of denitrifier communities as discussed below.
Distinct bacterial community structures between octocoral species.
Bacterial community structure and diversity were distinct between two octocoral species X. umbellata and P. flava at the beginning of the experiment, even though these corals had been maintained in the same aquaria for years. At the bacterial class level, P. flava was initially dominated by Alpha- and Gammaproteobacteria as well as Campylobacteria; X. umbellata was dominated by Alphaproteobebacteria, Bacteroidia, and Gammaproteobacteria. Our results are in line with previous findings that octocoral-associated bacterial communities are species-specific (35–38). This also concurs with the abundantly reported host-specific microbial assemblages for scleractinian corals, as well as a structural similarity to scleractinian coral bacterial communities at the class level (15, 25, 30, 39, 40). The gorgonian P. flava showed a lower bacterial diversity, which is consistent with previous reports (36, 37). Due to the broad leaf-like morphology and comparatively small polyps, algal symbionts may maximize photosynthetic activity to sustain a high autotrophic capacity in gorgonians (41). The invariable C:N ratio in the P. flava holobiont in our study suggests a relatively poor heterotrophic capacity, lending support to previous observations by Baker et al. (41). The low heterotrophic capacity of P. flava may restrict bacterial diversity (42). Limited niche space for bacterial associates due to morphological and physiological features of P. flava may be responsible for the observed low bacterial diversity. Being an octocoral, P. flava does not build a porous calcium carbonate skeleton that may offer niche space for a number of specialized endolithic microbes, as known from scleractinian corals (43, 44), nor does it secrete a thick surface mucus layer like Xenia sp. (45) that may provide favorable habitat for prokaryotic growth, including that of anaerobes (46, 47).
Species-specific responses of octocoral-associated bacterial communities to excess DOC.
Little information on drivers of octocoral microbial dynamics is available. Here we showed that octocorals exhibit species-specific responses to excess DOC and concomitant warming with regard to their bacterial communities. Over time, excess DOC led to conspicuous changes in the bacterial community structure of X. umbellata, as characterized by a marked continuous increase in Rhodobacteraceae, making up to 80% of the overall bacterial community at the end of the experiment. We cannot disentangle the direct and indirect effects of DOC on associated microbiomes. While based on qPCR data of 16S rRNA gene, we found an overall increase of bacterial abundance in X. umbellata under excess DOC. In contrast, no such change in response to excess DOC was observed in the bacterial community structure or abundance of P. flava. The absence of change in bacterial community structure in P. flava aligns with prior reports that gorgonian corals host a group of extremely robust bacterial communities over large geographic scales (48), seasonal variations (37) and even in the presence of environmental stressors such as increased temperatures and UV radiation (35). These observations suggest a differential “flexibility” or plasticity in bacterial community responses, as recently reported for a number of scleractinian corals (49).
This apparent interspecific plasticity of octocoral microbiomes may be explained by the initial differences in bacterial diversity and community structure between these two octocoral species. In general, coral bacterial microbiomes with greater diversity likely show a higher degree of functional redundancy and may exhibit more flexible associations in response to environmental change, which align with the ecological resilience of the holobiont (10, 25, 50–52). In contrast, the lower diversity of the bacterial community associated with P. flava might suggest bacteria are highly selected and likely inhabit host-constructed niches, as previously reported (53). These may be dominated by a few specialized species of high abundance (49) and vary less under environmental perturbation (54).
Observations of such a species-specificity may pertain to functional traits of the animal host and/or algal symbionts, such as heterotrophic capacity, nutritional status, or ecological resilience, including but not limited to heat tolerance as well as (a)biotic environmental drivers (25, 54–56). Differences in trophic strategies between these two octocoral species in particular may be of potential importance for their contrasting responses of the overall bacterial community under excess DOC. Xenia sp. is thought to be entirely autotrophic (57). However, Fabricius and colleague argued that Xenia sp. may exhibit a mixotrophic lifestyle, that is, relying on autotrophic and heterotrophic food sources (58). Our findings of increased C:N ratios in X. umbellata under excess DOC lend support to the latter. Indeed, the capacity to feed heterotrophically may allow X. umbellata to benefit from the surrounding DOC, thereby also creating a favorable environment for heterotrophic bacterial propagation, which may subsequently alter the overall bacterial community structure.
X. umbellata is known for its distinctive pulsation behavior, which may contribute to its ecological resilience by mixing of the surface boundary layer (59, 60). Of note, excess DOC remarkably enhanced their pulsation rates (61) and net primary productions (62), but whether this increase in pulsation may have an effect on the microbial plasticity of X. umbellata remains to be determined. Some endosymbiotic Symbiodiniaceae are known to feed heterotrophically (63), suggesting that excess DOC in X. umbellata may also promote the heterotrophic growth of Symbiodiniaceae. Given Symbiodiniaceae-associated bacteria often form a major component of the coral microbiome (64, 65), changes in Symbiodiniaceae physiology or even community composition likely contribute to microbial community dynamics observed in the present study.
Distinct patterns of microbial plasticity between two octocoral species appeared to be primarily driven by the increase of Rhodobacteraceae (ASV1: Roseobacter clade CHAB-I-5; ASV9 and ASV13: unclassified Rhodobacteraceae) exclusively in X. umbellata. This observation contrasts the findings of (49) reporting on the structural inflexibility in the bacterial microbiome of the scleractinian coral Pocillopora verrucosa under excess DOC. The pronounced increase in sequences proportions affiliated to multiple Rhodobacteraceae ASVs however align well with genomic evidence and ecological observations (67). Rhodobacteraceae are known for their considerable versatility with regard to C utilization, which allows them to thrive and rapidly proliferate in high-DOC environments (67). Due to the intragenomic heterogeneity, 16S rRNA gene sequencing using hypervariable region V3-V4 could overestimate the prokaryotic diversity of our investigated octocoral species (77). However, this overestimated diversity caused negligible effects on our main conclusion. As we compared samples across the same species under different conditions, the relative error remained the same and the differences are still a reflection of differences in treatments.
A passive regulation of denitrifiers in octocoral holobiont functioning.
Coral-associated diazotrophs (N-fixing prokaryotes) have previously been suggested to play a central role in supporting holobiont fitness and functioning when the surrounding environmental N availability is low (7, 26). In contrast, denitrifiers have been proposed to be important in maintaining the coral-algae symbiosis in a N-limited state (7), yet our understanding of whether their abundance in the coral holobiont is of physiological or ecological relevance remains poor. On average, the proportion of putative denitrifiers in the microbiome was higher in P. flava than in X. umbellata. This difference between host species may be attributed to their distinct trophic strategies. Microbial community compositions in coral holobionts are highly selective due to different host functional or life history traits pertaining to e.g., development, physiology, and metabolism (17, 40, 68). Due to a relatively low capacity to obtain (in)organic N sources from the surrounding environment, P. flava is likely to rely more on symbiotic N cycling microbes to acquire or remove N to fulfill its metabolic requirements (13, 28, 69).
In the presence of N, bioavailable C sources including glucose usually favor denitrification and hence the growth of denitrifiers (70). However, excess glucose caused no stimulating effects on the relative abundance of denitrifiers in our study. This apparent contradiction might be explained by the following two considerations: first, denitrifier populations might be regulated by N availability; consequently, denitrifier abundance may not necessarily increase in the presence of high DOC loads in a N-limited environment. Second, if denitrifiers were not limited by environmental N availability, different denitrifying taxa may exhibit differential preferences for C sources. For instance, some Rhodobacteraceae taxa dominate microbial glucose uptake in coastal North Sea waters (71), while others may be suppressed by allochthonous glucose input, but follow fluctuations in population dynamics of primary producers (72).
Importantly, a reduction in the cumulative relative abundance of denitrifiers (as reflected in nirS relative gene copies quantified by qPCR) during DOC enrichment was exclusively observed in X. umbellata. In contrast to P. flava, this soft coral species showed an increase in C:N ratios in the animal host under excess DOC at the end of the experiment. As increased C:N ratios imply a relative decrease of N availability, associated changes in the nutritional status of the host may directly impact its interaction with associated denitrifiers. The notion of reduced N availability for holobiont members is further corroborated by the increase in δ15N signatures, potentially indicating a reduced uptake of inorganic N from the seawater and an increased retention of N within the holobiont.
A passive regulation of N cyclers could thereby directly support overall holobiont functioning under fluctuating environmental conditions. In periods of low N availability, reduced denitrifier abundance (and therefore overall denitrification activity) might reduce the competition for N source in the coral holobiont and favor N uptake by the algae to support their growth. Likewise, rapid growth of denitrifiers during periods of excess N availability could increase the competition for N source between holobiont members, thereby alleviating excess N stress, and ultimately stabilizing the coral-algal symbiosis.
Can denitrifiers provide new insights into Symbiodiniaceae-bacteria interactions?
By inference, all four dominant denitrifiers identified in our study appear to form close associations with Symbiodiniaceae instead of coral hosts. The predominant putative denitrifier Ruegeria spp., formerly named Silicibacter spp., are known as dinoflagellate-associated bacteria. They are attracted to and capable of catabolizing degrading DMSP produced by the dinoflagellate host (73). Labrenzia spp. were previously reported to be core microbiome members of Symbiodiniaceae in the coral holobiont (64, 65). Given their ability to produce DMSP (an osmolyte and powerful scavenger of reactive oxygen species), the consistent association of Labrenzia spp. potentially assist in reducing oxidative stress of Symbiodiniaceae (65). Likewise, Dinoroseobacter spp. were previously shown to supply their dinoflagellate host with essential nutrients, specifically the essential vitamins B1 and B12, to support their growth in particular under the nutrient-limitation state (74). Ralstonia spp. were identified as intracellular, i.e., occurring within the endodermal coral host cells in close proximity to the Symbiodiniaceae, and were proposed to be implicated in the functioning of the coral-algae symbiosis (53).
Notably, we observed differential responses of different putative denitrifying taxa to experimental treatments. Specifically, an increasing proportion of Dinoroseobacter spp. concurs with a decreasing proportion of Ruegeria spp. in X. umbellata with excess DOC. At this point, the causes and consequences of altered denitrifier community structure and abundance for octocoral holobiont functioning remain to be determined. Functional studies will be required to link these microbiome dynamics with related denitrifying activities and nutritional states of holobionts to disentangle the potential role of denitrifiers in octocoral holobiont fitness and functioning. Further localization of denitrifiers in the intact symbiosis could help us better understand the interactions between octocoral-associated N cycling microbes with other members of the holobiont: the animal host, Symbiodiniaceae, and other microbes in a changing environment.
Conclusion.
Phase shifts on coral reef ecosystems have been linked to several environmental stressors. Among those, the detrimental effects of excess DOC (typically associated with sewage and reduced water quality) on scleractinian corals have received considerable attentions in the past (21, 22, 75). Changes in the activity, abundance and community structure of N cycling microbes have been discussed as critical components to corals’ response to environmental DOC loading. While an imbalance in N cycling significantly affected the structure and functions of bacterial microbiomes associated with scleractinian corals, very little information is available for octocorals, which are globally abundant particularly on reefs undergoing phase shifts (66, 76). Here, we provide insights into the dynamics of octocoral-associated bacterial communities with an emphasis on putative denitrifying communities under excess DOC and warming. The dynamics of denitrifiers aligned with the nutritional status of the octocoral host, which implies their critical role in regulating internal nutrient availability of the holobiont in a changing environment. To obtain a better understanding on the interactions between octocoral holobiont members, future studies should expand to a comparative taxonomic framework of octocoral host species and link their functional and life history traits to their microbial communities, in particular potentially critical functional groups such as N cyclers. Integrated holistic approaches combining “-omics” approaches and cultivation-dependent methods may aid such a challenging endeavor.
MATERIALS AND METHODS
Coral preparation and maintenance.
The soft coral X. umbellata and the gorgonian P. flava were cultivated (temperature: 26 ± 0.5°C; pH 7.8 ± 0.2; salinity: 35 ± 3‰) for more than 2 years at the Marine Ecology Department of the University of Bremen. Coral species were identified by barcoding gene (COI, mutS and 28S rRNA gene) amplification and sanger sequencing. Small X. umbellata fragments (1-2 cm in side length) were cut from 5 mother colonies (5 × 7×12 cm) and fixed on cubical-shaped calcium carbonate coral holders (1 × 1 cm) with rubber bands. After fragments recovered from fragmentation for 7 days and attached to the holders, rubber bands were removed. Simultaneously, branches of 3–4 cm in height were cut from 4 mother colonies (18 × 1×24 cm) of P. flava, and subsequently attached to coral holders using aquarium moss coral fix glue. Thereafter, 120 fragments of X. umbellata and 120 fragments of P. flava were distributed among 12 experimental aquaria tanks (water volume 50 L) and acclimated for 10 days prior to the experiment. Each aquarium was equipped with a thermostat (3613 aquarium heater, 75 W 220–240 V), a pump (EHEIM Compact On 300 pump) and a protein skimmer that were all purchased from EHEIM GmbH and Co. KG in Germany to maintain stable aquarium conditions. Additionally, LED lights (Royal Blue-matrix module and Ultra Blue White 1:1-matrix module, WALTRAt day time LED light, Germany) were used to simulate day - night rhythm of 12 h-12 h at the intensity of 120.8 ± 10.2 μmol quanta m−2 s−1. No additional feeding was provided and 10% of the artificial seawater was exchanged daily to maintain stable water parameters. To verify stable environmental conditions, salinity (C: 35.39 ± 0.05; L: 35.38 ± 0.08; M: 35.50 ± 0.08; H: 35.11 ± 0.09) and pH (C: 8.15 ± 0.01; L: 8.05 ± 0.02; M: 8.05 ± 0.02; H: 8.03 ± 0.02) were monitored throughout the experiment (Fig. S4 in the supplemental material).
Experimental design, sampling and DNA extraction.
In the first phase of experiment, temperature was kept constant at 26°C for 21 days (Fig. 1). Daily additions of glucose to the aquaria based on a stock solution (d-Glucose, 40 mg ml−1) were used to simulate four different glucose enrichment levels: control (no addition), low (equivalent to 10 mg L−1), medium (equivalent to 20 mg L−1) and high (equivalent to 40 mg L−1). Daily measurements of total organic carbon (TOC) using a TOC-L analyzer (Shimadzu, Japan) were used to approximate glucose levels in the aquaria and the daily dosing of glucose was adjusted accordingly to achieve the desired enrichment levels. For the second phase, temperature was increased gradually in all aquaria, ultimately adding 2°C every 8 days to a final temperature of 32°C. The range of 26–32°C represents a latitudinal gradient of mean maximum temperature from north to south across the Red Sea, where X. umbellata is widely abundant (78). The temperature maximum of 32°C is also close to the thermal physiological limit of this species (79). During this gradual ramping phase, all DOC treatments were continued as described above (Fig. 1). The experiment was terminated at day 45.
Corals for molecular analysis were collected at day 0 (before DOC additions, as baseline data), after the first phase of the experiment (day 21, DOC treatments at 26°C), and after the second phase (day 45, DOC treatments at 32°C). For this, fresh coral samples were collected and immediately frozen in liquid nitrogen and stored at −80°C until further processing. Additionally, samples for physiological measurements and aquarium seawater inorganic nutrients were collected at day 45 as outlined below. Frozen coral samples were ground into powder over liquid nitrogen using mortar and pestle. Genomic DNA was extracted according to the instruction of Quick-DNA Universal Kit Quick Protocol for Solid Tissue (ZYMO RESEARCH, USA). Afterwards, DNA was quantified by spectrophotometry at 260 nm and 280 nm (Tecan Infinite 200 PRO, Austria) and quality-checked by 1% (wt/vol) agarose gel electrophoresis (Biometra Horizon 58, Germany).
Illumina MiSeq 16S rRNA gene sequencing and sequence analysis.
The 16S rRNA gene amplicon sequencing was conducted at LGC genomics (Berlin, Germany). The hypervariable regions V3-V4 of the bacterial 16S rRNA gene were amplified and sequenced using the primer pair S-d-Bact-0341-b-S-17 (5′-CCTACGGGNGGCWGCAG-3′) and S-d-Bact-0785-a-A-21 (5′-GACTACHVGGGTATCTAATCC-3′) (80). The PCRs included 1–10 ng of DNA extract (total volume 1 μl), 15 pmol of each forward primer and reverse primer in 20 μl volume of 1 x MyTaq buffer containing 1.5 units MyTaq DNA polymerase (Bioline GmbH, Germany) and 2 μl of BioStabII PCR Enhancer (Sigma-Aldrich Co., Germany). For each sample, the forward and reverse primers had the same 10-nt barcode sequence for multiplexing. PCRs were carried out for 35 cycles using the following parameters: 1 min 96°C predenaturation; 96°C denaturation for 15s, 55°C annealing for 30s, 70°C extension for 90s, hold at 8°C. The Illumina library was pooled, and size selected by preparative gel electrophoresis, and the sequencing was conducted on the Illumina MiSeq platform in a 2 × 300 bp paired-end run using V3 Chemistry.
After demultiplexing and removal of primer sequences from the raw paired-end reads by LGC genomics, further sequence processing was performed according to the DADA2 (1.14.1) pipeline for the generation of exact amplicon sequence variants (ASVs) (81). Specifically, sequences were filtered and quality trimmed to 225 bp (forward) and 235 bp (reverse) at a maximum expected error rate of 5. Trimmed sequences were pooled and used for error learning and denoising. In total, 77 samples were pooled with 3,858,215 reads in 1,264,025 unique sequences. Denoised sequences were merged, followed by chimeras’ removal according to default parameters. A total of 26,902 chimeras were identified out of 40,839 ASVs, singletons generated during the merging step were removed. ASVs between 400 to 430 bp were retained and taxonomically classified by “assignTaxonomy” based on the SILVA database release 138 (82). ASVs that matched chloroplast and mitochondrial sequences were removed prior to further analysis.
An in silico PCR for the nitrite reductase nirS primer pair, nirS-1F (5′-CCTAYTGGCCGCCRCART-3′) and nirS-qR (5′-TCCMAGCCRCCRTCRTGCAG-3′) (83) was used to characterize the putative denitrifier community. The program ecoPCR (84) from OBI tools 1.01.22 was launched against the Ensembl Bacteria release 42 with a maximum three mismatches and a zero mismatch zone of 2 bp at the 3′ end of each primer, retaining fragments between 50 bp and 500 bp. Resultant sequences from ecoPCR with the fragment size between 224 and 227 bp were blasted against the GenBank Nucleotide database (NCBI nucleotide BLAST, date accessed 2020/04/05), and sequences that were not identified as originating from denitrifying nirS were removed. The genus affiliation of the remaining nirS fragments was used as potential denitrifying taxa to recover denitrifier communities based on our 16S rRNA gene sequencing results. In addition, ASV sequences affiliated to the predominant genus were aligned to SILVA 138, and the accession numbers of nearest relatives from SILVA Incremental Aligner (SINA) output were used to obtain a higher resolved taxonomic path.
Quantification real-time PCR (qPCR) of denitrifying nirS gene.
We assessed the denitrification potential in the coral holobiont via the relative quantification of nirS gene, which catalyzes the conversion of nitrite to nitric oxide in the denitrification cascade (85) and has been previously used to determine denitrifier abundance and diversity (27, 83, 86). The nirS gene was amplified using the same primer pair nirS-1F, qR (83) previously used for in silico PCR as outlined above, and validated with Sanger sequencing (StarSEQ, Mainz, Germany). The relative quantification of nirS gene abundance was done by qPCR using the CFX96TM Touch Real-Time PCR Detection System (BIO-RAD, USA) by SensiFAST™ SYBR No-ROX Kit (Bioline, USA). CT values of nirS gene amplicons (as a proxy of denitrifier abundance) were referenced against CT values of 16S rRNA gene amplicons (as a reference for total bacterial abundance) using the primer-pair Bact-16S_784F: 5′-AGGATTAGATACCCTGGTA-3′ and Bact-16S_1061R: 5′-CRRCACGAGCTGACGAC-3′ (87), according to the delta-delta Ct method (2-ΔΔCt) (88). 10-20 ng of DNA extract was used for 16S rRNA gene and nirS qPCRs. Final cycling conditions consisted of a hot-start activation at 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 10 s, annealing at 65°C (nirS gene in X. umbellata) or 60°C (nirS gene in P. flava and 16S rRNA gene for both species) for 20 s, and extension at 72°C for 30 s. Final extension was carried out at 72°C for 10 s followed by a melting curve from 65 to 95°C with increase of 0.5°C steps every 5 s. The qPCR efficiency was validated by calibration curves of genomic DNA from E. coli (ATCC 25922) targeting 16S rRNA gene and DSM 428 Alcaligenes eutropus H16 targeting nirS gene separately.
Seawater inorganic nutrient and coral elemental analysis.
Nutrient samples were collected in triplicates at the end of the experiment (i.e., day 45). 50 ml of aquaria seawater was collected through 0.45 μm filters in 50 ml sterilized centrifuge tubes, and immediately frozen at −20°C until further analysis. Nutrient levels were measured spectrophotometrically using the Infinite 200 PRO (Tecan Infinite 200 PRO, Austria) according to reference 89. Coral samples for elemental and isotope analysis were collected at day 45. Fresh coral fragments were immediately frozen in liquid nitrogen and stored at −80°C until further processing. Corals were thawed at room temperature and homogenized for 30 s at 3,500 rpm with an Ultra Turrax (IKA, Germany). The resulting homogenized coral slurry was separated into coral tissue and algal symbiont fractions by centrifugation at 3,000 × g for 5 min (Eppendorf, Germany). The host fraction, that is, the resulting supernatant, was carefully removed by pipetting without disturbing the algal symbiont pellet. Algal symbiont pellets were resuspended in 0.22 μm filtered seawater (FSW) in sterilize 2 ml Eppendorf tubes, and host and algal samples were dried at 40°C for 1 week. After the dried matter was pulverized using clean mini pestles; 1.0 mg of coral tissue or algal symbiont sample was used for measuring total nitrogen content (TN). Further, 1.0 mg of coral tissue or algal symbiont sample mixed with 200 μl 1 mol L−1 HCl was used for analyzing total organic carbon content (TCorg). TN and TCorg were analyzed in an elemental analyzer (Euro EA, Germany), Isotopic δ15N and δ13C signatures were measured using an Isotope Ratio MS (Thermo Fisher, USA), following reference 90.
Statistical analysis.
All statistical analyses were performed in R studio 3.6.1, specifically vegan for multivariate statistics (91) and ggplot2 for visualization (92). Alpha diversity indices were calculated based on repeated (n = 100) random subsampling of ASVs to the minimum library size at sequencing depth 3,000. Data normality was determined by the Shapiro–Wilk test, and statistical differences between different time points and DOC treatments for each species was tested using two-way analysis of variance (ANOVA) with Tukey’s HSD as a post hoc comparison. Beta dispersion of samples was conducted for each coral species using function “betadisper”. Beta diversity was evaluated between species, time points and DOC treatments by non-metric multidimensional scaling (NMDS) plot based on Bray-Curtis dissimilarities of relative ASV proportions. Analysis of similarities (ANOSIM) was used to illustrate the dissimilarity of bacterial community structures between two coral species at day 0. To illustrate the significant difference in bacterial variations at ASV level from different time points and DOC treatments, permutational multivariate analysis of variance (PERMANOVA, ‘adonis’ function, 999 permutations) based on Bray-Curtis dissimilarity was applied along with ANOSIM as post hoc comparisons. An ANOSIM R value close to 1 suggests a strong separation between groups, while values close to 0 indicates an overlap between groups (93). After log transformation to meet data normality, homogeneity of variance and independence, the qPCR data was analyzed by two-way ANOVA and Tukey’s HSD. Seawater inorganic nutrient levels in aquaria and elemental changes in animal host and algal symbionts respectively were analyzed by one-way ANOVA and Tukey’s HSD.
Data availability.
The Sanger sequencing data derived from COI, mutS, and 28S rRNA gene PCR amplicons for octocoral identification have been deposited in the European Nucleotide Archive (ENA) under the project accession number PRJEB43824. Primer-clipped DNA sequences generated by Miseq 16S rRNA amplicon sequencing were deposited on NCBI under BioProject accession number PRJNA718022.
ACKNOWLEDGMENTS
We thank Svea Vollstedt, Nadim Katzer, Edoardo Zelli, Meghan Kennedy, Rassil Nafeh and Claudia Dessi for their support with animal husbandry and during aquarium experiments. Many thanks to Achim Meyer and Sonja Peters for their assistance with qPCR. We also thank Dorothee Dasbach and Matthias Birkicht for helping with elemental, isotope and nutrient measurements. This work was supported by the grants Wi 2677/9-1 and Wi 2677/16-1 from the German Research Foundation (DFG) to C.W. and the China Scholarship Council (CSC) 201807565016. N.X. acknowledges support by the Open Access Publication Funds of Alfred-Wegener-Institut Helmholtz-Zentrum für Polar- und Meeresforschung.
We declare no conflict of interest.
C.W., A.G., N.X. designed and conceived the research; N.X., S.M.S.-G. performed aquarium project; C.W., A.G. contributed coral samples and reagents; N.X., C.H. analyzed sequencing data and conducted statistical analysis; N.X., C.P., N.R. conducted data visualization, interpretation, and wrote the paper with improvement suggestions from all co-authors. All authors gave final approval for publication.
Footnotes
Supplemental material is available online only.
Contributor Information
Nan Xiang, Email: nanxiang@uni-bremen.de.
Ning-Yi Zhou, Shanghai Jiao Tong University.
REFERENCES
- 1.Muscatine L, Kaplan I. 1994. Resource partitioning by reef corals as determined from stable isotope composition. II. 15N of zooxanthellae and animal tissue versus depth. Pac Sci 48:304–312. [Google Scholar]
- 2.Wild C, Huettel M, Klueter A, Kremb SG, Rasheed MY, Jørgensen BB. 2004. Coral mucus functions as an energy carrier and particle trap in the reef ecosystem. Nature 428:66–70. 10.1038/nature02344. [DOI] [PubMed] [Google Scholar]
- 3.Muscatine L, Porter JW. 1977. Reef corals: mutualistic symbioses adapted to nutrient-poor environments. Bioscience 27:454–460. 10.2307/1297526. [DOI] [Google Scholar]
- 4.Muscatine L. 1990. The role of symbiotic algae in carbon and energy flux in reef corals. Coral Reefs 25:75–87. [Google Scholar]
- 5.Falkowski PG, Dubinsky Z, Muscatine L, McCloskey L. 1993. Population control in symbiotic corals. Bioscience 43:606–611. 10.2307/1312147. [DOI] [Google Scholar]
- 6.Dubinsky Z, Jokiel PL. 1994. Ratio of energy and nutrient fluxes regulates symbiosis between zooxanthellae and corals. Pac Sci 48:313–324. [Google Scholar]
- 7.Rädecker N, Pogoreutz C, Voolstra CR, Wiedenmann J, Wild C. 2015. Nitrogen cycling in corals: the key to understanding holobiont functioning? Trends Microbiol 23:490–497. 10.1016/j.tim.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Benavides M, Houlbrèque F, Camps M, Lorrain A, Grosso O, Bonnet S. 2016. Diazotrophs: a non-negligible source of nitrogen for the tropical coral Stylophora pistillata. J Exp Biol 219:2608–2612. 10.1242/jeb.139451. [DOI] [PubMed] [Google Scholar]
- 9.Bednarz VN, Grover R, Maguer J-F, Fine M, Ferrier-Pagès C. 2017. The assimilation of diazotroph-derived nitrogen by scleractinian corals depends on their metabolic status. mBio 8:e02058-16. 10.1128/mBio.02058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voolstra CR, Ziegler M. 2020. Adapting with microbial help: microbiome flexibility facilitates rapid responses to environmental change. BioEssays 42:2000004. 10.1002/bies.202000004. [DOI] [PubMed] [Google Scholar]
- 11.Lesser MP, Mazel CH, Gorbunov MY, Falkowski PG. 2004. Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science 305:997–1000. 10.1126/science.1099128. [DOI] [PubMed] [Google Scholar]
- 12.Raina J-B, Tapiolas D, Willis BL, Bourne DG. 2009. Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl Environ Microbiol 75:3492–3501. 10.1128/AEM.02567-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pogoreutz C, Rädecker N, Cárdenas A, Gärdes A, Wild C, Voolstra CR. 2017. Nitrogen fixation aligns with nifH abundance and expression in two coral trophic functional groups. Front Microbiol 8:1187. 10.3389/fmicb.2017.01187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pogoreutz C, Rädecker N, Cárdenas A, Gärdes A, Voolstra CR, Wild C. 2017. Sugar enrichment provides evidence for a role of nitrogen fixation in coral bleaching. Glob Chang Biol 23:3838–3848. 10.1111/gcb.13695. [DOI] [PubMed] [Google Scholar]
- 15.Rohwer F, Seguritan V, Azam F, Knowlton N. 2002. Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser 243:1–10. 10.3354/meps243001. [DOI] [Google Scholar]
- 16.Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. 2007. The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol 5:355–362. 10.1038/nrmicro1635. [DOI] [PubMed] [Google Scholar]
- 17.Bourne DG, Webster NS. 2013. Coral Reef Bacterial Communities. Springer, Berlin, Germany. [Google Scholar]
- 18.Pogoreutz C, Voolstra CR, Rädecker N, Weis V, Cardenas A, Raina J-B. 2020. Chapter 7. The coral holobiont highlights the dependence of cnidarian animal hosts on their associated microbes. In Bosch TCG, Hadfield MG (ed), Cellular Dialogues in the Holobiont. Taylor Francis, CRC Press, Boca Raton, FL. 10.1201/9780429277375. [DOI] [Google Scholar]
- 19.Hughes TP, Kerry JT, Álvarez-Noriega M, Álvarez-Romero JG, Anderson KD, Baird AH, Babcock RC, Beger M, Bellwood DR, Berkelmans R, Bridge TC, Butler IR, Byrne M, Cantin NE, Comeau S, Connolly SR, Cumming GS, Dalton SJ, Diaz-Pulido G, Eakin CM, Figueira WF, Gilmour JP, Harrison HB, Heron SF, Hoey AS, Hobbs J-PA, Hoogenboom MO, Kennedy EV, Kuo C-Y, Lough JM, Lowe RJ, Liu G, McCulloch MT, Malcolm HA, McWilliam MJ, Pandolfi JM, Pears RJ, Pratchett MS, Schoepf V, Simpson T, Skirving WJ, Sommer B, Torda G, Wachenfeld DR, Willis BL, Wilson SK. 2017. Global warming and recurrent mass bleaching of corals. Nature 543:373–377. 10.1038/nature21707. [DOI] [PubMed] [Google Scholar]
- 20.Hoegh-Guldberg O. 1999. Climate change, coral bleaching and the future of the world's coral reefs. Mar Freshw Res 50:839–866. 10.1071/MF99078. [DOI] [Google Scholar]
- 21.Kuntz N, Kline D, Sandin S, Rohwer F. 2005. Pathologies and mortality rates caused by organic carbon and nutrient stressors in three Caribbean coral species. Mar Ecol Prog Ser 294:173–180. 10.3354/meps294173. [DOI] [Google Scholar]
- 22.Kline D, Kuntz N, Breitbart M, Knowlton N, Rohwer F. 2006. Role of elevated organic carbon levels and microbial activity in coral mortality. Mar Ecol Prog Ser 314:119–125. 10.3354/meps314119. [DOI] [Google Scholar]
- 23.Bourne D, Iida Y, Uthicke S, Smith-Keune C. 2008. Changes in coral-associated microbial communities during a bleaching event. ISME J 2:350–363. 10.1038/ismej.2007.112. [DOI] [PubMed] [Google Scholar]
- 24.Kushmaro A, Banin E, Loya Y, Stackebrandt E, Rosenberg E. 2001. Vibrio shiloi sp. nov., the causative agent of bleaching of the coral Oculina patagonica. Int J Syst Evol Microbiol 51:1383–1388. 10.1099/00207713-51-4-1383. [DOI] [PubMed] [Google Scholar]
- 25.Ziegler M, Seneca FO, Yum LK, Palumbi SR, Voolstra CR. 2017. Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat Commun 8:14213–14218. 10.1038/ncomms14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cardini U, Bednarz VN, Naumann MS, van Hoytema N, Rix L, Foster RA, Al-Rshaidat MM, Wild C. 2015. Functional significance of dinitrogen fixation in sustaining coral productivity under oligotrophic conditions. Proc Biol Sci 282:20152257. 10.1098/rspb.2015.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braker G, Fesefeldt A, Witzel K-P. 1998. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl Environ Microbiol 64:3769–3775. 10.1128/AEM.64.10.3769-3775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tilstra A, Roth F, El-Khaled YC, Pogoreutz C, Rädecker N, Voolstra CR, Wild C. 2021. Relative abundance of nitrogen cycling microbes in coral holobionts reflects environmental nitrate availability. R Soc Open Sci 8:201835. 10.1098/rsos.201835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El-Khaled YC, Roth F, Tilstra A, Rädecker N, Karcher DB, Kürten B, Jones BH, Voolstra CR, Wild C. 2020. In situ eutrophication stimulates dinitrogen fixation, denitrification, and productivity in Red Sea coral reefs. Mar Ecol Prog Ser 645:55–66. 10.3354/meps13352. [DOI] [Google Scholar]
- 30.Ziegler M, Roik A, Porter A, Zubier K, Mudarris MS, Ormond R, Voolstra CR. 2016. Coral microbial community dynamics in response to anthropogenic impacts near a major city in the central Red Sea. Mar Pollut Bull 105:629–640. 10.1016/j.marpolbul.2015.12.045. [DOI] [PubMed] [Google Scholar]
- 31.Van De Water JAJM, Allemand D, Ferrier-Pagès C. 2018. Host-microbe interactions in octocoral holobionts - recent advances and perspectives. Microbiome 6. 10.1186/s40168-018-0431-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El-Khaled YC, Roth F, Rädecker N, Tilstra A, Karcher DB, Kürten B, Jones BH, Voolstra CR, Wild C. 2021. Nitrogen fixation and denitrification activity differ between coral-and algae-dominated Red Sea reefs. Sci Rep 11:1–15. 10.1038/s41598-021-90204-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ngugi DK, Ziegler M, Duarte CM, Voolstra CR. 2020. Genomic blueprint of glycine betaine metabolism in coral metaorganisms and their contribution to reef nitrogen budgets. Iscience 23:101120. 10.1016/j.isci.2020.101120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rädecker N, Pogoreutz C, Gegner HM, Cárdenas A, Roth F, Bougoure J, Guagliardo P, Wild C, Pernice M, Raina J-B, Meibom A, Voolstra CR. 2021. Heat stress destabilizes symbiotic nutrient cycling in corals. Proc Natl Acad Sci USA 118:e2022653118. 10.1073/pnas.2022653118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCauley M, Jackson CR, Goulet TL. 2020. Microbiomes of Caribbean octocorals vary over time but are resistant to environmental change. Front Microbiol 11:1272. 10.3389/fmicb.2020.01272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van de Water JA, Melkonian R, Voolstra CR, Junca H, Beraud E, Allemand D, Ferrier-Pagès C. 2017. Comparative assessment of Mediterranean gorgonian-associated microbial communities reveals conserved core and locally variant bacteria. Microb Ecol 73:466–478. 10.1007/s00248-016-0858-x. [DOI] [PubMed] [Google Scholar]
- 37.Van De Water JAJM, Voolstra CR, Rottier C, Cocito S, Peirano A, Allemand D, Ferrier-Pagès C. 2018. Seasonal Stability in the Microbiomes of Temperate Gorgonians and the Red Coral Corallium rubrum Across the Mediterranean Sea. Microb Ecol 75:274–288. 10.1007/s00248-017-1006-y. [DOI] [PubMed] [Google Scholar]
- 38.Osman EO, Suggett DJ, Voolstra CR, Pettay DT, Clark DR, Pogoreutz C, Sampayo EM, Warner ME, Smith DJ. 2020. Coral microbiome composition along the northern Red Sea suggests high plasticity of bacterial and specificity of endosymbiotic dinoflagellate communities. Microbiome 8:8–16. 10.1186/s40168-019-0776-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Littman RA, Willis BL, Pfeffer C, Bourne DG. 2009. Diversities of coral-associated bacteria differ with location, but not species, for three acroporid corals on the Great Barrier Reef. FEMS Microbiol Ecol 68:152–163. 10.1111/j.1574-6941.2009.00666.x. [DOI] [PubMed] [Google Scholar]
- 40.Ziegler M, Grupstra CGB, Barreto MM, Eaton M, Baomar J, Zubier K, Al-Sofyani A, Turki AJ, Ormond R, Voolstra CR. 2019. Coral bacterial community structure responds to environmental change in a host-specific manner. Nat Commun 10. 10.1038/s41467-019-10969-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baker DM, Freeman CJ, Knowlton N, Thacker RW, Kim K, Fogel ML. 2015. Productivity links morphology, symbiont specificity and bleaching in the evolution of Caribbean octocoral symbioses. ISME J 9:2620–2629. 10.1038/ismej.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meistertzheim A-L, Lartaud F, Arnaud-Haond S, Kalenitchenko D, Bessalam M, Le Bris N, Galand PE. 2016. Patterns of bacteria-host associations suggest different ecological strategies between two reef building cold-water coral species. Deep Sea Res Part I: Oceanographic Res Papers 114:12–22. 10.1016/j.dsr.2016.04.013. [DOI] [Google Scholar]
- 43.Yang S-H, Lee STM, Huang C-R, Tseng C-H, Chiang P-W, Chen C-P, Chen H-J, Tang S-L. 2016. Prevalence of potential nitrogen-fixing, green sulfur bacteria in the skeleton of reef-building coral Isopora palifera. Limnol Oceanogr 61:1078–1086. 10.1002/lno.10277. [DOI] [Google Scholar]
- 44.Pernice M, Raina J-B, Rädecker N, Cárdenas A, Pogoreutz C, Voolstra CR. 2020. Down to the bone: the role of overlooked endolithic microbiomes in reef coral health. ISME J 14:325–334. 10.1038/s41396-019-0548-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bednarz VN, Naumann MS, Niggl W, Wild C. 2012. Inorganic nutrient availability affects organic matter fluxes and metabolic activity in the soft coral genus Xenia. J Exp Biol 215:3672–3679. 10.1242/jeb.072884. [DOI] [PubMed] [Google Scholar]
- 46.Kellogg C. 2004. Tropical Archaea: diversity associated with the surface microlayer of corals. Mar Ecol Prog Ser 273:81–88. 10.3354/meps273081. [DOI] [Google Scholar]
- 47.Sharon G, Rosenberg E. 2008. Bacterial growth on coral mucus. Curr Microbiol 56:481–488. 10.1007/s00284-008-9100-5. [DOI] [PubMed] [Google Scholar]
- 48.Correa H, Haltli B, Duque C, Kerr R. 2013. Bacterial communities of the gorgonian octocoral Pseudopterogorgia elisabethae. Microb Ecol 66:972–985. 10.1007/s00248-013-0267-3. [DOI] [PubMed] [Google Scholar]
- 49.Pogoreutz C, Rädecker N, Cárdenas A, Gärdes A, Wild C, Voolstra CR. 2018. Dominance of Endozoicomonas bacteria throughout coral bleaching and mortality suggests structural inflexibility of the Pocillopora verrucosa microbiome. Ecol Evol 8:2240–2252. 10.1002/ece3.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reshef L, Koren O, Loya Y, Zilber-Rosenberg I, Rosenberg E. 2006. The coral probiotic hypothesis. Environ Microbiol 8:2068–2073. 10.1111/j.1462-2920.2006.01148.x. [DOI] [PubMed] [Google Scholar]
- 51.Torda G, Donelson JM, Aranda M, Barshis DJ, Bay L, Berumen ML, Bourne DG, Cantin N, Foret S, Matz M, Miller DJ, Moya A, Putnam HM, Ravasi T, Van Oppen MJH, Thurber RV, Vidal-Dupiol J, Voolstra CR, Watson S-A, Whitelaw E, Willis BL, Munday PL. 2017. Rapid adaptive responses to climate change in corals. Nat Clim Chang 7:627–636. 10.1038/nclimate3374. [DOI] [Google Scholar]
- 52.Louca S, Polz MF, Mazel F, Albright MBN, Huber JA, O'Connor MI, Ackermann M, Hahn AS, Srivastava DS, Crowe SA, Doebeli M, Parfrey LW. 2018. Function and functional redundancy in microbial systems. Nat Ecol Evol 2:936–943. 10.1038/s41559-018-0519-1. [DOI] [PubMed] [Google Scholar]
- 53.D Ainsworth T, Krause L, Bridge T, Torda G, Raina J-B, Zakrzewski M, Gates RD, Padilla-Gamiño JL, Spalding HL, Smith C, Woolsey ES, Bourne DG, Bongaerts P, Hoegh-Guldberg O, Leggat W. 2015. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J 9:2261–2274. 10.1038/ismej.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gardner SG, Camp EF, Smith DJ, Kahlke T, Osman EO, Gendron G, Hume BC, Pogoreutz C, Voolstra CR, Suggett DJ. 2019. Coral microbiome diversity reflects mass coral bleaching susceptibility during the 2016 El Niño heat wave. Ecol Evol 9:938–956. 10.1002/ece3.4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bednarz VN, van de Water JA, Rabouille S, Maguer JF, Grover R, Ferrier‐Pagès C. 2019. Diazotrophic community and associated dinitrogen fixation within the temperate coral Oculina patagonica. Environ Microbiol 21:480–495. 10.1111/1462-2920.14480. [DOI] [PubMed] [Google Scholar]
- 56.Camp EF, Kahlke T, Nitschke MR, Varkey D, Fisher NL, Fujise L, Goyen S, Hughes DJ, Lawson CA, Ros M, Woodcock S, Xiao K, Leggat W, Suggett DJ. 2020. Revealing changes in the microbiome of Symbiodiniaceae under thermal stress. Environ Microbiol 22:1294–1309. 10.1111/1462-2920.14935. [DOI] [PubMed] [Google Scholar]
- 57.Schlichter D, Svoboda A, Kremer BP. 1983. Functional autotrophy of Heteroxenia fuscescens (Anthozoa: Alcyonaria): carbon assimilation and translocation of photosynthates from symbionts to host. Mar Biol 78:29–38. 10.1007/BF00392968. [DOI] [Google Scholar]
- 58.Fabricius K, Klumpp D. 1995. Widespread mixotrophy in reef-inhabiting soft corals:the influence of depth, and colony expansion and contraction on photosynthesis. Mar Ecol Prog Ser 125:195–204. 10.3354/meps125195. [DOI] [Google Scholar]
- 59.Kremien M, Shavit U, Mass T, Genin A. 2013. Benefit of pulsation in soft corals. Proc Natl Acad Sci USA 110:8978–8983. 10.1073/pnas.1301826110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wild C, Naumann MS. 2013. Effect of active water movement on energy and nutrient acquisition in coral reef-associated benthic organisms. Proc Natl Acad Sci USA 110:8767–8768. 10.1073/pnas.1306839110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vollstedt S, Xiang N, Simancas-Giraldo SM, Wild C. 2020. Organic eutrophication increases resistance of the pulsating soft coral Xenia umbellata to warming. PeerJ 8:e9182. 10.7717/peerj.9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simancas-Giraldo SM, Xiang N, Kennedy MM, Nafeh R, Zelli E, Wild C. 2021. Photosynthesis and respiration of the soft coral Xenia umbellata respond to warming but not to organic carbon eutrophication. PeerJ 9:e11663. 10.7717/peerj.11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jeong HJ, Yoo YD, Kang NS, Lim AS, Seong KA, Lee SY, Lee MJ, Lee KH, Kim HS, Shin W, Nam SW, Yih W, Lee K. 2012. Heterotrophic feeding as a newly identified survival strategy of the dinoflagellate Symbiodinium. Proc Natl Acad Sci USA 109:12604–12609. 10.1073/pnas.1204302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matthews JL, Raina JB, Kahlke T, Seymour JR, van Oppen MJ, Suggett DJ. 2020. Symbiodiniaceae‐bacteria interactions: rethinking metabolite exchange in reef‐building corals as multi‐partner metabolic networks. Environ Microbiol 22:1675–1687. 10.1111/1462-2920.14918. [DOI] [PubMed] [Google Scholar]
- 65.Lawson CA, Raina JB, Kahlke T, Seymour JR, Suggett DJ. 2018. Defining the core microbiome of the symbiotic dinoflagellate, Symbiodinium. Environ Microbiol Rep 10:7–11. 10.1111/1758-2229.12599. [DOI] [PubMed] [Google Scholar]
- 66.Loya Y, Sakai K, Yamazato K, Nakano Y, Sambali H, van Woesik R. 2001. Coral bleaching: the winners and the losers. Ecol Lett 4:122–131. 10.1046/j.1461-0248.2001.00203.x. [DOI] [Google Scholar]
- 67.Cárdenas A, Neave MJ, Haroon MF, Pogoreutz C, Rädecker N, Wild C, Gärdes A, Voolstra CR. 2018. Excess labile carbon promotes the expression of virulence factors in coral reef bacterioplankton. ISME J 12:59–76. 10.1038/ismej.2017.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Webster NS, Reusch TBH. 2017. Microbial contributions to the persistence of coral reefs. ISME J 11:2167–2174. 10.1038/ismej.2017.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tilstra A, El-Khaled YC, Roth F, Radecker N, Pogoreutz C, Voolstra CR, Wild C. 2019. Denitrification aligns with N2 fixation in red sea corals. Sci Rep 9:19460. 10.1038/s41598-019-55408-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beauchamp EG, Trevors JT, Paul JW. 1989. Carbon sources for bacterial denitrification, p 113–142. In Advances in soil science. Springer, New York, NY. 10.1007/978-1-4613-8847-0_3. [DOI] [Google Scholar]
- 71.Alonso C, Pernthaler J. 2006. Roseobacter and SAR11 dominate microbial glucose uptake in coastal North Sea waters. Environ Microbiol 8:2022–2030. 10.1111/j.1462-2920.2006.01082.x. [DOI] [PubMed] [Google Scholar]
- 72.Allers E, Gómez-Consarnau L, Pinhassi J, Gasol JM, Šimek K, Pernthaler J. 2007. Response of Alteromonadaceae and Rhodobacteriaceae to glucose and phosphorus manipulation in marine mesocosms. Environ Microbiol 9:2417–2429. 10.1111/j.1462-2920.2007.01360.x. [DOI] [PubMed] [Google Scholar]
- 73.Miller TR, Hnilicka K, Dziedzic A, Desplats P, Belas R. 2004. Chemotaxis of Silicibacter sp. strain TM1040 toward dinoflagellate products. Appl Environ Microbiol 70:4692–4701. 10.1128/AEM.70.8.4692-4701.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wagner-Döbler I, Ballhausen B, Berger M, Brinkhoff T, Buchholz I, Bunk B, Cypionka H, Daniel R, Drepper T, Gerdts G, Hahnke S, Han C, Jahn D, Kalhoefer D, Kiss H, Klenk H-P, Kyrpides N, Liebl W, Liesegang H, Meincke L, Pati A, Petersen J, Piekarski T, Pommerenke C, Pradella S, Pukall R, Rabus R, Stackebrandt E, Thole S, Thompson L, Tielen P, Tomasch J, von Jan M, Wanphrut N, Wichels A, Zech H, Simon M. 2010. The complete genome sequence of the algal symbiont Dinoroseobacter shibae: a hitchhiker's guide to life in the sea. ISME J 4:61–77. 10.1038/ismej.2009.94. [DOI] [PubMed] [Google Scholar]
- 75.Haas AF, Fairoz MF, Kelly LW, Nelson CE, Dinsdale EA, Edwards RA, Giles S, Hatay M, Hisakawa N, Knowles B, Lim YW, Maughan H, Pantos O, Roach TN, Sanchez SE, Silveira CB, Sandin S, Smith JE, Rohwer F. 2016. Global microbialization of coral reefs. Nat Microbiol 1:16042. 10.1038/nmicrobiol.2016.42. [DOI] [PubMed] [Google Scholar]
- 76.Norström A, Nyström M, Lokrantz J, Folke C. 2009. Alternative states on coral reefs: beyond coral–macroalgal phase shifts. Mar Ecol Prog Ser 376:295–306. 10.3354/meps07815. [DOI] [Google Scholar]
- 77.Sun D-L, Jiang X, Wu QL, Zhou N-Y. 2013. Intragenomic heterogeneity of 16S rRNA genes causes overestimation of prokaryotic diversity. Appl Environ Microbiol 79:5962–5969. 10.1128/AEM.01282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Benayahu Y, Loya Y. 1984. Life history studies on the Red Sea soft coral Xenia macrospiculata Gohar, 1940. I. Annual dynamics of gonadal development. Biol Bull 166:32–43. 10.2307/1541428. [DOI] [Google Scholar]
- 79.Chaidez V, Dreano D, Agusti S, Duarte CM, Hoteit I. 2017. Decadal trends in Red Sea maximum surface temperature. Sci Rep 7. 10.1038/s41598-017-08146-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glockner FO. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. 2016. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mosier AC, Francis CA. 2010. Denitrifier abundance and activity across the San Francisco Bay estuary. Environ Microbiol Rep 2:667–676. 10.1111/j.1758-2229.2010.00156.x. [DOI] [PubMed] [Google Scholar]
- 84.Ficetola G, Coissac E, Zundel S, Riaz T, Shehzad W, Bessière J, Taberlet P, Pompanon F. 2010. An in silico approach for the evaluation of DNA barcodes. BMC Genomics 11:434. 10.1186/1471-2164-11-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zumft WG. 1997. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61:533–616. 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee JA, Francis CA. 2017. Deep nirS amplicon sequencing of San Francisco Bay sediments enables prediction of geography and environmental conditions from denitrifying community composition. Environ Microbiol 19:4897–4912. 10.1111/1462-2920.13920. [DOI] [PubMed] [Google Scholar]
- 87.Andersson AF, Lindberg M, Jakobsson H, Bäckhed F, Nyrén P, Engstrand L. 2008. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3:e2836. 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 89.Grasshoff K, Kremling K, Ehrhardt M. 2009. Methods of seawater analysis. John Wiley & Sons. 10.1002/9783527613984. [DOI] [Google Scholar]
- 90.Moncada C, Hassenrück C, Gärdes A, Conaco C. 2019. Microbial community composition of sediments influenced by intensive mariculture activity. FEMS Microbiol Ecol 95:fiz006. 10.1093/femsec/fiz006. [DOI] [PubMed] [Google Scholar]
- 91.Oksanen J. 2015. Vegan: an introduction to ordination. https://cran.r-project.org/web/packages/vegan/vignettes/intro-vegan.pdf.
- 92.Warnes MGR, Bolker B, Bonebakker L, Gentleman R, Huber W. 2016. Package ‘gplots’. Various R programming tools for plotting data. https://github.com/talgalili/gplots.
- 93.Ramette A. 2007. Multivariate analyses in microbial ecology. FEMS Microbiol Ecol 62:142–160. 10.1111/j.1574-6941.2007.00375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4. Download AEM.01886-21-s0001.pdf, PDF file, 2.1 MB (2.1MB, pdf)
Data Availability Statement
The Sanger sequencing data derived from COI, mutS, and 28S rRNA gene PCR amplicons for octocoral identification have been deposited in the European Nucleotide Archive (ENA) under the project accession number PRJEB43824. Primer-clipped DNA sequences generated by Miseq 16S rRNA amplicon sequencing were deposited on NCBI under BioProject accession number PRJNA718022.