Abstract
The histone chaperone FACT (FAcilitates Chromatin Transcription) plays an essential role in transcription and DNA replication by its dual functions on nucleosome assembly to maintain chromatin integrity and nucleosome disassembly to destabilize nucleosome and facilitate its accessibility simultaneously. Mono-ubiquitination at Lysine 119 of H2A (ubH2A) has been suggested to repress transcription by preventing the recruitment of FACT at early elongation process. However, up to date, how ubH2A directly affects FACT on nucleosome assembly and disassembly remains elusive. In this study, we demonstrated that the dual functions of FACT are differently regulated by ubH2A. The H2A ubiquitination does not affect FACT’s chaperone function in nucleosome assembly and FACT can deposit ubH2A–H2B dimer on tetrasome to form intact nucleosome. However, ubH2A greatly restricts FACT binding on nucleosome and inhibits its activity of nucleosome disassembly. Interestingly, deubiquitination of ubH2A rescues the nucleosome disassembly function of FACT to activate gene transcription. Our findings provide mechanistic insights of how H2A ubiquitination affects FACT in breaking nucleosome and maintaining its integrity, which sheds light on the biological function of ubH2A and various FACT’s activity under different chromatin states.
Graphical Abstract
Graphical Abstract.
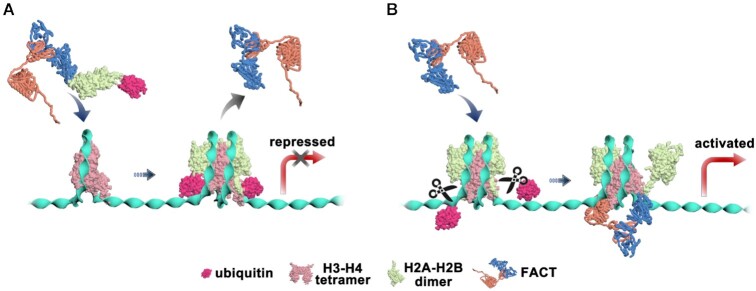
ubH2A restricts FACT binding on nucleosome, but maintains FACT's chaperone function.
INTRODUCTION
Genomic DNA in eukaryotes is packaged into chromatin by histones, which provides a structural platform for all DNA-related processes, including transcription and DNA replication. Nucleosome, the basic unit of chromatin, is composed of a histone octamer with two copies of each histone H2A, H2B, H3 and H4, which is wrapped with 147 base pairs (bp) of DNA about 1.7 turns in a left-handed manner (1,2). Nucleosomes generally act as repressors to limit the access of nuclear proteins to DNA. During the chromatin-based processes, the nucleosome barrier needs to be temporarily removed to expose DNA, and then rapidly restored afterwards to preserve the original epigenetic identity (3,4). The FACT (FAcilitates Chromatin Transcription) complex, initially identified as a chaperone of histone H2A–H2B dimer (5), is highly conserved among eukaryotes and essential for cell viability (6,7). Previous studies have found that FACT not only facilitates the progression of DNA and RNA polymerases on chromatin templates (5,8), but also functions to maintain the genome-wide integrity of chromatin (9–15). Recently, the in vitro structural and single-molecular investigations have revealed the functions of FACT by facilitating both the nucleosome disassembly and re-assembly (16–18). FACT has been found to tether both the (H3–ṇH4)2 tetramer and H2A–H2B dimer on DNA by its two subunits SPT16 (Suppressor of Ty 16) and SSRP1 (Structure-specific recognition protein-1), which function distinctly but coordinately to mediate nucleosome assembly/disassembly. However, how the dual functions of FACT in nucleosome assembly and disassembly are regulated by different epigenetic factors remains unclear.
H2A mono-ubiquitination is a prevalent post-translational modification of histone H2A, in which the 76 amino acid protein ubiquitin is covalently attached through its C-terminus to the ϵ-amino group of lysine 119 of H2A (ubH2A). About 5–15% of total H2A has been reported to be ubiquitinated in higher eukaryotes (19). H2A ubiquitination has been reported to tightly associate with transcription, replication and DNA damage repair (20,21). The H2A ubiquitination site Lysine 119 is located at the beginning of the C-terminal tails of H2A at DNA entry/exit point of nucleosome and also at the end of the H2A docking domain (residues 80–119) which accounts for the binding of H2A–H2B dimer to the (H3–ṇH4)2 tetramer in histone octamer (2). Single-molecular investigation has shown that ubH2A dramatically stabilizes the nucleosome by blocking DNA peeling from histone octamer (22). Genome-wide analysis in mammalian cells have revealed that ubH2A is broadly distributed on the genome, and mostly associated with repressed genes (23). H2A ubiquitination has been found to prevent H3K4 di- and tri-methylation by methyltransferase MLL3, which are reported to activate gene transcription (24). Furthermore, a strong correlation between ubH2A and the polycomb repressive complexes PRC1 (the major H2A ubiquitination ligase), as well as gene repression marker H3K27me3 (methylated by PRC2), has been observed in stem cells and other cell types (25–29). In addition, the chromatin immunoprecipitation (ChIP) analysis in RAW 264.7 cells have revealed that ubH2A might block SPT16 recruitment on chemokine genes to inhibit RNA polymerase II release at the early stage of elongation (30). But to date, how ubH2A directly regulates FACT on nucleosome dynamics is still poorly understood.
In this work, we directly investigated the effects of ubH2A on the functions of FACT by manipulating nucleosomes with magnetic tweezers at the single molecule level. We revealed that the functions of FACT on nucleosome assembly and disassembly are differently regulated by ubH2A. H2A ubiquitination inhibits FACT binding on nucleosome and blocks FACT disassembling nucleosome. Deubiquitination of ubH2A nucleosome rescues the role of FACT in nucleosome disassembly in vitro. Genome-wide and specific gene analysis also identified that ubH2A inhibits the binding of FACT to repress gene transcription and the deubiquitination of ubH2A induces the binding of FACT to activate gene. Strikingly, we revealed that FACT can deposit ubH2A–H2B dimer on tetrasome to form intact nucleosome. H2A ubiquitination does not affect FACT’s chaperone function on nucleosome assembly. Different regulation of ubH2A on the functions of FACT on nucleosome assembly and disassembly, provides a detailed molecular mechanism by which histone chaperone and histone modification coordinately remodel the nucleosome dynamics during gene transcription.
MATERIALS AND METHODS
Nucleosome reconstitution
Recombinant histones H2A, H2B, H3, H4 were cloned and purified as previously described (31). ubH2A protein was purchased from Hefei KS-V Peptide Biological Technology Co., Ltd, which is chemically synthetized by utilizing standard solid phase peptide synthesis (SPPS) and peptide fragments ligation reactions. 409 bp DNA template containing a single 601 sequence for magnetic tweezers’ analysis (the constructs of the DNA template was shown in Supplementary Figure S1C) was prepared by PCR using a biotin (bio)-labeled forward primer and a three-digoxigenin (3 dig)-labeled reverse primer. Respective histone octamers, tetramers and dimers were reconstituted as previously described (32). Equimolar amounts of individual histones in unfolding buffer (7 M guanidinium HCl, 20 mM Tris–HCl, pH 7.5, 10 mM DTT) were dialyzed into refolding buffer (2 M NaCl, 10 mM Tris–HCl, pH 7.5, 1 mM EDTA, 5 mM 2-mercaptoethanol), and purified using Superdex 200 column. Nucleosomes and tetrasomes were assembled using the salt-dialysis method as previously described (32). Histone octamers/tetrasomes and DNA templates were mixed in TEN buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA, 2 M NaCl) and dialyzed for 16–18 h at 4°C in TEN buffer, which was continuously diluted by slowly pumping in TE buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA) to lower the concentration of NaCl from 2 to 0.6 M. Samples were collected after final dialysis in HE buffer (10 mM HEPES, pH 8.0, 0.1 mM EDTA) for 4 hr. The stoichiometry of histone octamer to DNA template was determined by AFM and gel shift analysis.
Purification of FACT
For the purification of FACT, Sf9 cells were infected with baculovirus containing Flag-SPT16, or His6-SSRP1 and incubated at 27°C for 72 h. SPT16 and SSRP1 were co-expressed for FACT complex. Proteins were purified as previously described with minor modifications (16). The infected cells were collected by centrifugation and resuspended with lysis buffer (20 mM Tris–HCl, pH 8.0, 150 mM NaCl, 5% glycerol, 1 mM PMSF). The recombinant FACT complex was purified in two steps. First, the cell extracts were incubated with anti-Flag M2-agarose (Sigma) beads at 4°C for 8 hr, and then eluted in the presence of 0.5 mg/ml Flag peptide (Sigma). Second, the proteins were further purified through Heparin HP column. The purified proteins were dialyzed against BC-100 buffer (100 mM NaCl, 10 mM Tris–HCl, pH 8.0, 0.5 mM EDTA, 20% glycerol, 1 mM DTT, 1 mM PMSF) and stored at −80°C.
Mono-nucleosome Pull-down assay
Biotin-labeled DNA was prepared by PCR with biotin-labeled primer. 1 μg Biotin-labeled mono-nucleosomes and streptavidin-agarose-beads (10 μl) were mixed in BC300 buffer (20 mM Tris–HCl, pH 7.5, 10% glycerol, 300 mM NaCl, 0.1% NP-40, 500 μg/ml BSA), then incubated with the FACT complexes overnight at 4°C. Then the beads were washed with the wash buffer (20 mM Tris–HCl, pH 7.5, 10% glycerol, 300 mM NaCl, 0.5% NP-40) for 3 times, 10 min each time. The proteins were eluted with SDS-loading buffer (50 mM Tris, pH 6.8, 2% SDS, 0.1% Bromophenol blue, 10% glycerol, 1% beta-mercaptoethanol) and analyzed by western blot.
SILAC mass spectrometry
For the SILAC pull-down assays, mono-nucleosomes were immobilized on dynabeads Myone C1. Immobilized H2A mono-nucleosomes were incubated with light nuclear extract and immobilized ubH2A mono-nucleosomes were incubated with heavy nuclear extract in binding buffer (20 mM Tris–HCl, pH 7.6, 10% glycerol, 150 mM KCl, 0.1% NP-40, 1 mM DTT, 50 ng/μl BSA, protease inhibitors). After 2 hr incubation, pull-down materials were washed with binding buffer for 5 times, and then eluted with SDS-loading buffer. Equal amount of H2A (light) and ubH2A (heavy) pull-down samples were combined and subjected to in-gel digestion and LC–MS/MS analysis. In the reverse experiment, immobilized ubH2A mono-nucleosomes were incubated with light nuclear extract and immobilized H2A mono-nucleosomes were incubated with heavy nuclear extract. Samples were analyzed on a Q Exactive equipped with an Easy n-LC 1000 HPLC system (Thermo Scientific). The raw data were analyzed with Proteome Discovery version 2.2.0.388 using Sequest HT search engine for protein identification and Percolator for FDR (false discovery rate) analysis. The Uniprot human protein database (updated on October 2017) was used for searching the data, and then analyzed by R (http://www.r-project.org/).
Flag-IP assay
10 μl anti-Flag M2-agarose (Sigma) beads for each reaction was washed by PBS twice and pre-cleaned in BC300 buffer (20 mM Tris–HCl, pH 7.5, 10% glycerol, 300 mM NaCl, 0.1% NP-40, 500 μg/ml BSA) for 1 h at 4°C. Then FACT, H2A–H2B dimer and ubH2A–H2B dimer were added. After overnight incubation at 4°C, beads were washed 3 times with the wash buffer (20 mM Tris–HCl, pH 7.5, 10% glycerol, 300 mM NaCl, 0.5% NP-40), 10 min each time. Finally, proteins were eluted by SDS-loading buffer and analyzed by western blot.
In vitro histone deubiquitination assays
In vitro histone deubiquitination assays were performed as described previously with some modifications (33). 1 μg mono-nucleosomes (ubH2A) were incubated with recombinant USP21 in deubiquitination reaction buffer (50 mM Tris–HCl, pH 7.5, 50 mM NaCl, 5 mM DTT) at 30°C for 1 h. The reaction was terminated by the addition of SDS–loading buffer and proteins were resolved on SDS–PAGE and blotted with the anti-H2A antibody and anti-USP21 antibody.
Binding assay
Tetrasomes were reconstituted on 87 bp 601 DNA fragments using the salt-dialysis method as described above (32). FACT was preincubated with H2A–H2B dimer or ubH2A–H2B dimer for 30 min at 4°C, and then mixed with the reconstituted tetrasomes in the binding buffer (10 mM HEPES, pH 8.0, 1 mM EDTA, 60 mM NaCl) at 30°C for 1 hr. The samples were analyzed by 5% native PAGE electrophoresis in 0.25× TBE buffer (22.5 mM Tris, pH 8.0, 22.5 mM boric acid, 0.5 mM EDTA) for 1 hr at 80 V.
AFM analysis
Reconstituted nucleosome samples were prepared in HE buffer with DNA concentrations of 20 ng/μl. The samples were fixed with 0.1% glutaraldehyde (Fluka) in HE buffer on ice for 30 min. Rinse the mixture with 500 μl HE buffer for 2–3 times in a vivaspin500 column. Spin the column at 15 000 g for 2–5 min at 4°C to get rid of the liquids. Then dilute the sample solution in HE buffer with concentrations of 0.5–1 ng/μl. Drop 20 μl spermidine (1 mM) onto newly cleaved mica surface and incubate for 10 min. Then rinse the mica with 200 μl ddH2O for 4 times and blow dry mica surface briefly with nitrogen gas. Add 10 μl sample solution onto mica surface, incubate for 10–15 min. Finally, wash the mica with 200 μl ddH2O for 3 times and blow dry gently. The prepared AFM samples were examined using ScanAsyst Mode of AFM (MultiMode 8 SPM system, BRUKER).
Single-molecule magnetic tweezers analysis
The single-molecule stretching experiments were performed by magnetic tweezers. As shown in Figure 1C, the two ends of the 409 bp DNA construct were tethered via digoxigenin and anti-digoxigenin ligation to a glass coverslip and via biotin-streptavidin ligation to a 2.8 μm diameter Dynabeads (M280, Invitrogen Norway). Two small NdFeB magnets on the DNA constructs were controlled to pull on the Dynabeads and thus stretch the DNA molecule. The real time position of the bead was monitored by a CCD camera (MC1362, Mikrotron) at 200 Hz through an inverted microscope objective (UPLSAPO60XO, NA 1.35, Olympus). The extension (end to end distance) of the DNA construct was determined at nanometer resolution by analyzing the diffraction pattern of Dynabeads (34).
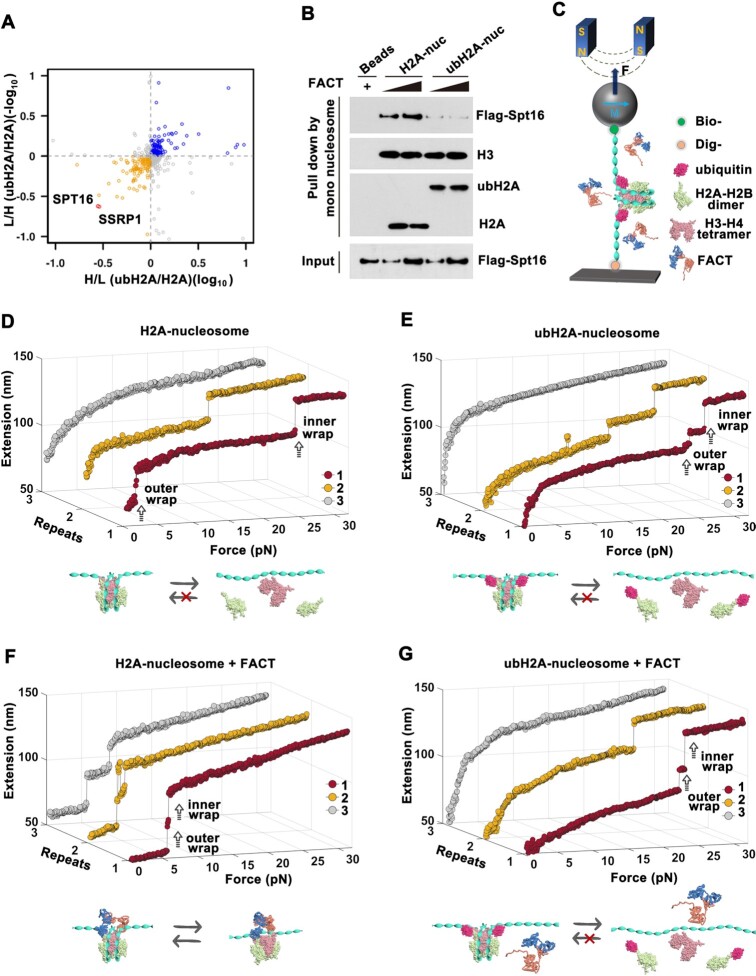
Figure 1.
ubH2A inhibits the binding of FACT on nucleosome and blocks the remodeling function of FACT. (A) Quantitative SILAC-based mass spectrometry analysis of mono-nucleosome pull-down assays. H2A and ubH2A nucleosomes were incubated with heavy or light nuclear extracts of HEK293T cells (n = 4 biologically independent experiments). (B) Mono-nucleosome pull-down assay of H2A and ubH2A mono-nucleosome incubated with the purified FACT. The immunoblots are representative of three independent experiments. (C) A schematic representation of single-molecule stretching experiment (not to scale): a single H2A or ubH2A mono-nucleosome is tethered between a coverslip and a superparamagnetic bead. (D–G) Repeated stretching measurements of H2A and ubH2A mono-nucleosome without (D, E) and with (F, G) FACT. The cartoons below represent the relative dynamic process of the nucleosome, respectively. In each stretching cycle, the force is increased up to 32 pN at a loading rate of 0.1 pN/s.
To anchor the nucleosome sample, the surface of coverslip needs to be functionalized. The coverslips were cleaned in Piranha Solution for 2 h, and then coated with 70 μl Sigmacote (Sigma-Aldrich) for 5 s. After that, the polystyrene beads (2 μm, QDSpher) were pipetted on the coverslip and put on a hot plate at 150°C for 5 min to melt on the coverslip. These firmly bound beads were served as reference beads for drift correction. To coat the coverslip with anti-digoxigenin, the flow cell was incubated with 100 μl anti-digoxigenin (0.1 mg/ml) at 37°C for 2 h. To avoid the non-specific bond between samples and the coverslip, the flow cell was incubated with passivation buffer (10 mg/ml BSA, 1 mM EDTA, 10 mM phosphate buffer, pH 7.4, 10 mg/ml PluronicF127 surfactant (Sigma-Aldrich), 3 mM NaN3) overnight at 37°C. The flow cell was ready for measurement.
To anchor the nucleosome to the coverslip in the flow cell, we first injected the nucleosome into the flow cell and incubated for 15 min. The digoxin-labeled end of the nucleosome was bound to the anti-digoxin coated coverslip. Then, we rinsed away the unbound nucleosome with HE and injected the beads (M280 Invitrogen Norway) into the flow cell and incubated for 15 min. The bio-labeled end of nucleosome was anchored to the streptavidin-coated beads (35).
The applied tensions on the super paramagnetic beads arise from the strong magnetic field gradient of the two magnets. The tensions are tuned by adjusting the magnets position in z-direction. Based on the geometry of a pendulum, the stretching force on the nucleosome in z-direction was calculated as 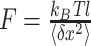 , according to the equipartition theorem, where
, according to the equipartition theorem, where 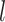 is the extension of DNA and
is the extension of DNA and 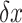 is the fluctuation of the bead in x-direction (36). However, the camera acquisition frequency is finite. The measurement of
is the fluctuation of the bead in x-direction (36). However, the camera acquisition frequency is finite. The measurement of 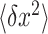 in real space appears systematic error at forces greater than ∼10 pN. In our LabView software, the improved force calibration based on the power-spectral-density (PSD) analysis by considering both the translational and the rotational motions of the beads is achieved (37). To calibrate the force in the force-extension measurement, the force calculation for a 10 000 bp DNA tethered between the bead (M280) and the coverslip was carried out. At each magnet position, the x-position of the bead was recorded for 5 min and the corresponding force was calibrated by improved power-spectral-density (PSD) analysis. We repeated force measurements for 10 independent DNA tethers. The relationship between force and magnet position can be fitted well with a single exponential function (38).
in real space appears systematic error at forces greater than ∼10 pN. In our LabView software, the improved force calibration based on the power-spectral-density (PSD) analysis by considering both the translational and the rotational motions of the beads is achieved (37). To calibrate the force in the force-extension measurement, the force calculation for a 10 000 bp DNA tethered between the bead (M280) and the coverslip was carried out. At each magnet position, the x-position of the bead was recorded for 5 min and the corresponding force was calibrated by improved power-spectral-density (PSD) analysis. We repeated force measurements for 10 independent DNA tethers. The relationship between force and magnet position can be fitted well with a single exponential function (38).
The real-time position (x, y, z) of the bead at various forces was recorded by comparing the diffraction pattern of the bead with calibration images at various distances from the focal point of the objective. In our LabView software, the quadrant-interpolation (QI) algorithm was applied to trace the three-dimensional position of the beads in the flow cell (39). The QI algorithm enables highly parallel single molecule experiments and reduces the pixel bias. In our measurements, we can trace about 50 samples simultaneously. These parallel measurements help us to trace the dynamics of folding and unfolding at high-throughput level. All measurements were carried out at 25°C.
To trace the conformation transition of nucleosome, the force-extension curve was measured (Figure 1D and E). To reveal the detail of the structural transition, we tuned the magnets position in z-direction by moving the magnets at 0.1 pN/s continuously and the corresponding force on the nucleosome changed from 0.1 to 32.0 pN, during which the extension of the nucleosome was recorded. After that, force was reduced to 0.1 pN rapidly and waited for 3 min, and then repeated the stretching cycle on the same nucleosome. More than 100 measurements were carried out for H2A and ubH2A nucleosomes. To investigate how FACT affects the nucleosome dynamics, we anchored the nucleosome on magnetic tweezers, injected FACT and incubated for 30 min, and then rinsed away the unbound FACT with HE buffer. To investigate how FACT mediates nucleosome assembly, we anchored the tetrasome on magnetic tweezers, injected FACT with or without (ub)H2A–H2B dimer and incubated for 30 min, and then rinsed away the unbound proteins with HE buffer.The real-time trajectories of the individual nucleosomes or tetrasomes were then traced by magnetic tweezers. The statistics for the stretching experiments, including the number of stretching repeats and the fraction of nucleosome shown the behavior as indicated in the relative figure, are listed in the Supplementary Table S1. The commercial magnetic beads (M280, Invitrogen) contain anisotropic and nonuniformly distributed magnetic nanoparticles (40). Under the external magnetic field, the overall anisotropy axis of the beads aligns with the external field resulting in an off-axis movement. In our experiments, the diverse mechanical response of the nucleosome samples under small tensions is due to the bead's rotation in the external field, which has little effects on the mechanical disruptions of nucleosomes under large tensions.
Real-time measurement of deubiquitylating process for ubH2A nucleosomes
To investigate the real-time deubiquitinated process, a combination of force-clamp measurement force-ramp measurement was performed. We firstly changed the force continuously from 0 to 7 pN and held at 7 pN for 3 min. Then, we injected 50 μl mixture of USP21 (40 nM) and FACT (50 nM) into the flow cell and continued to hold at 7 pN for longer time to capture the deubiquitinating process. After observing the real-time deubiquitinating process, we carried out the force-ramp measurement three times on the same sample from 0 to 32 pN at 0.1 pN/s to confirm whether FACT can bind deubiquitinated ubH2A nucleosome.
mESC cell line
For the HA-AID-Ring1B mESCs, we first established Tir1 (E3 ligase for AID) stable expression cell line. Then we knocked a HA-AID coding sequence into the endogenous Ring1b locus by CRISPR-Cas9-mediated genome editing. The guide sequence was 5′-GCACAGCCTGAGACATTTCT-3′. To more efficiently decrease ubH2A, we further knocked out Ring1a using the guide sequence: 5′-CACAGAATGCCAGCAAAACG-3′. IAA (25 ug/ml) was used to degrade HA-AID-Ring1B.
ChIP
ubH2A ChIP and SSRP1 ChIP were performed as previously described (41). mESCs were fixed with 1% formaldehyde for 10 min at room temperature. Fixation was stopped by adding glycine to 0.125 M. Fixed mESCs were resuspended in nuclei lysis buffer (50 mM Tris–HCl, pH 8.0, 10 mM EDTA, 1% SDS, protease inhibitors) and sonicated to 300–500-bp-sized fragments. Protein A Dynabeads and protein G Dynabeads (Life Technology) were pre-incubated with ubH2A antibody (CST, #8240) and SSRP1 antibody (BioLegend, 609710) for 6 h at 4°C. Then sonicated chromatin was incubated with beads coated with ubH2A antibody and SSRP1 antibody overninght at 4°C. The beads were subjected to extensive washes with RIPA150 (50 mM Tris–HCl, pH 8.0,150 mM NaCl, 1 mM EDTA, pH 8.0, 0.1% SDS, 1% Triton X-100, 0.1% sodium deoxycholate) once, RIPA500 (50 mM Tris–HCl, pH 8.0, 0.5 M NaCl, 1 mM EDTA, pH 8.0, 0.1% SDS, 1% Triton X-100, 0.1% sodium deoxycholate) twice, RIPA-LiCl (50 mM Tris–HCl, pH 8.0, 1 mM EDTA pH 8.0, 1% NP-40, 0.7% sodium deoxycholate, 0.5 M LiCl) twice and 1× TE twice. The beads were resuspended with freshly made direct elution buffer (10 mM Tris–HCl, pH 8.0, 300 mM NaCl, 5 mM EDTA, pH 8.0, 0.5% SDS). The mix was vortexed at 65°C for 30 min, then placed on a magnetic stand for 2 min; the supernatant was then collected and de-crosslinked by adding SDS to a final concentration of 1% and incubated at 65°C for 6 hrs, followed by treatment with RNase A and proteinase K. DNA was extracted with phenol:chloroform:isoamyl (25:24:1) and precipitated with 75% ethanol and 300 mM NaAc followed by qPCR analysis. 50 and 100 μg chromatin were used for ubH2A and SSRP1 ChIP respectively. All the qPCR experiments were performed on the real-time PCR (ABI 7300, USA) using FastStart Universal SYBR Green Master (ROX). The primer pairs used for the qPCR experiments were the following: Hsf5: sense 5′-GTTCTGAAACCTCCGCTGGC-3′, antisense 5′-CGACACAAGGCTACCTTCGC-3′; Foxn4: sense 5′-GTCGCAGCTCGATCCGTTT-3′, antisense 5′-CCGCCCGCTGAAATCTCCTT-3′; Lrrfip1: sense 5′-CTATCGGATCGCGGCTGTCA-3′, antisense 5′-TTAGCTGGAACCACACCGCA-3′; Rasd2: sense 5′-GTAACGTAAGCTGCCAGGCG-3′, antisense 5′-AGCCAGGTGCTCGAAGGTTT-3′. ChIP enrichment are normalized to the input. The relative % of input: ubH2A samples are normalized to the last time point, SSRP1 samples are normalized to the first time point. The results were calculated by three independent replications.
RNA-seq
The total RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer's instruction. For poly(A) RNA-seq, libraries were prepared according to the Illumina TruSeq protocol and sequenced using a HiSeq2000 system at Berry Genomics.
RNA-seq and gene expression analysis
The pair-end RNA-seq reads were mapped to the Mus musculus mm9 gene annotation model using Subread (42), with the default parameters, and the only uniquely mapped reads were used for further analysis. The normalization was performed with R package DEseq2 (43).
ChIP-seq analysis
The SSRP1 and ubH2A ChIP-seq data were downloaded from Gene Expression Omnibus (GEO: GSE90906, PRJNA604675) (41,44). All the ChIP-seq data were mapped to the mouse genome (mm9) using bowtie2 (45) with the default parameters. Low quality reads were removed by samtools (46) and only uniquely mapped reads which mapping to a unique genomic location and strand were kept. Enriched peaks were called using MACS (47). The p-value cutoff for peak detection was 1e-5 with MACS14.
RESULTS
H2A ubiquitination inhibits the binding of FACT on nucleosome
As previously investigated by coimmunoprecipitation (Co-IP) studies of endogenous SPT16 with unmodified Flag-H2A or Flag-ubH2A stably expressed in HEK293 cells, SPT16 has been found to specifically interact with H2A, but not with ubH2A (30). To reveal how ubH2A affects FACT binding to nucleosome, the mono-nucleosomes containing unmodified H2A or ubH2A were assembled on biotin-labelled 601 DNA template, and incubated with SILAC (Stable Isotope Labelling with Amino acids in Cell culture) -treated nuclear extracts of HEK293T. The SILAC-based mass spectrometry with mono-nucleosomes pull-down assays were conducted to identify the unmodified H2A or ubH2A specific binding protein factors (please find details in Methods). As shown in Figure 1A, the two subunits of FACT, SPT16 and SSRP1 are both enriched on unmodified H2A mono-nucleosome, as compared to ubH2A mono-nucleosome. To exclude the indirect effect in nuclear extracts, the recombinant FACT with SPT16 and SSRP1 were purified and incubated with biotin-labeled H2A or ubH2A mono-nucleosomes (Supplementary Figure S1A and B). The mono-nucleosomes pull-down assays verified that the purified FACT has a higher affinity for the H2A nucleosome (Figure 1B). The results indicated that H2A ubiquitination directly inhibits the binding of FACT at the nucleosome level.
H2A ubiquitination blocks FACT disassembling nucleosome
To further investigate how ubH2A affects FACT’s functions on nucleosome dynamics, single-molecule magnetic tweezers were employed to trace the structure transition of unmodified H2A and ubH2A mono-nucleosome. The H2A and ubH2A mono-nucleosomes were reconstituted onto a 409 bp DNA fragment with one Widom 601 nucleosome positioning sequence, as characterized by AFM images (Supplementary Figure S1C). Two ends of DNA fragment were modified with digoxin and biotin to specifically tether with coverslip and paramagnetic bead respectively (Figure 1C). The real-time trajectories of the individual nucleosomes were traced while the tension was continuously increased up to 32 pN. For each nucleosome, three repeated stretching cycles were performed to quantitatively assess the stability and reversibility of H2A and ubH2A nucleosomes.
In the absence of FACT, two extension jumps corresponding to disruptions of the outer and inner DNA wrap were observed in the force-extension curves for both H2A and ubH2A nucleosome (Figure 1D and E, Repeat 1), consistent with previous results of ours (16,22,48) and others (49,50). As defined by previous work, the nucleosome core particle, composed of a histone octamer wrapped by DNA about 1.7 turns, typically displays a well-characterized two-step unfolding dynamics. The first step at lower force with step size of about 21 nm, corresponds to the unraveling of outer nucleosomal DNA wrap, which mainly stabilized by the interacion between H2A/H2B dimer with DNA; and the second one at higher force with step size of about 25 nm, is associated with the unraveling of inner DNA wrap, stabilized by the interacion between H3/H4 with DNA (16,22,48). For H2A nucleosome, the rupture forces, indicating the mechanical stability of nucleosome, were observed at 3.1 ± 1.2 pN (mean ± standard deviation) and 22.2 ± 4.1 pN for the outer and inner DNA wrap respectively (Figure 1D, Repeat 1). Concurrently, in line with our previous work (22), the two-step disruption of the ubH2A nucleosome occurring at much higher forces of 14.6 ± 5.6 pN and 24.0 ± 5.0 pN (Figure 1E, Repeat 1), indicating the much higher mechanical stability of ubH2A-nucleosome. The rupture forces for the H2A and ubH2A nucleosome were statistically analyzed as shown in Supplementary Figure S1D. As for the repeated stretching experiments, the reassembly properties of the H2A and ubH2A nucleosome were examined (Figure 1D and E). For both H2A and ubH2A nucleosome, only the first stretching cycle showed the typical two-step disruption of intact nucleosome (Supplementary Figure S1E), which indicated that the nucleosome structure cannot be reassembled correctly due to the displacement of core histones from DNA (51).
To investigate how FACT affects the nucleosome dynamics, we anchored the nucleosome on magnetic tweezers, injected FACT and incubated for 30 mins, and then rinsed away the unbound FACT with HE buffer. The real-time trajectories of the individual nucleosomes were then traced by magnetic tweezers. After incubation with FACT, the H2A nucleosome was disrupted at much lower forces (<10 pN), indicating that FACT greatly impairs the stability of H2A nucleosome (Figure 1F). More importantly, in the repeated stretching measurements with magnetic tweezers, H2A nucleosome maintains the unfolding pathway with similar force response in each stretching cycle, suggesting that FACT tethers the histones to the DNA and allows the reassembly of H2A nucleosome when the nucleosome is fully disrupted, which is consistent with our previous results (16). Intriguingly, FACT does not affect the unfolding dynamics of ubH2A nucleosomes. No matter with or without the incubation with FACT, the ubH2A nucleosomes present a similar unrepeated unfolding pathway: irreversible two-step extension jumps at 14.2 ± 8.1 pN for disruption of the outer wrap and 24.4 ± 3.5 pN for disruption of the inner wrap. The statistics for the stretching experiments are shown in Supplementary Figure S1D, E and Supplementary Table S1. The results strongly implied that ubH2A inhibits the binding of FACT onto nucleosome and blocks the function of FACT on nucleosome (Figure 1G).
Deubiquitination of ubH2A rescues the remodeling function of FACT on nucleosome
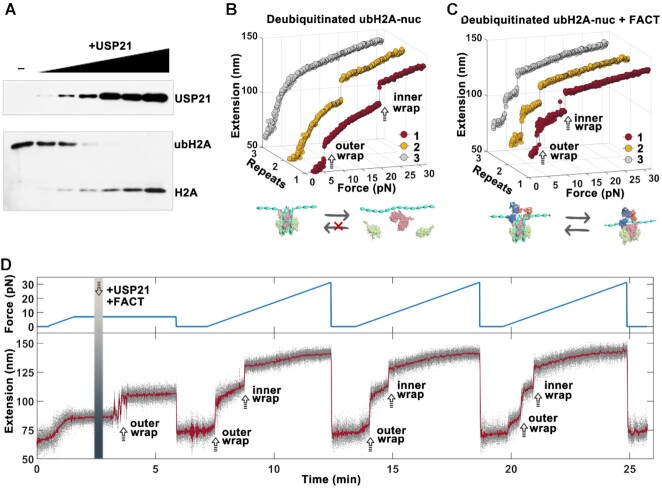
We have shown that FACT cannot bind to ubH2A nucleosome to regulate the nucleosome dynamics. To further confirm whether the inhibition is caused by H2A ubiquitination, we repeated the experiments after deubiquitination of the ubH2A nucleosome. The deubiquitination of ubH2A nucleosome was performed using USP21, a well-defined deubiquitinase for ubH2A (52). The ubH2A nucleosomes were successfully deubiquitinated by USP21 in vitro with high efficiency, as examined by western blot experiments (Figure 2A). Force-extension measurements showed that the disruption mode for deubiquitinated ubH2A-nucleosomes restores to that for H2A-nucleosome, with the rupture force at 3.6 ± 1.6 pN for the outer wrap and 23.2 ± 2.4 pN for the inner wrap (Figure 2B). Likewise, FACT can remodel the deubiquitinated ubH2A nucleosomes effectively as H2A-nucleosomes, by breaking the nucleosome stability (<10 pN) and maintaining its integrity in repeated stretching cycles as shown in Figure 2C. The rupture forces and reassembly property for the deubiquitinated ubH2A nucleosome were statistically analyzed (Supplementary Figure S2A and B).
Figure 2.
Deubiquitination of ubH2A rescues the remodeling function of FACT on nucleosome. (A) In vitro deubiquitination assay for ubH2A nucleosome incubated with increasing amounts of USP21. Western blot experiments show that ubH2A-nucleosomes are successfully deubiquitinated by USP21 with high efficiency. (B, C) Repeated stretching measurements of deubiquitinated ubH2A-nucleosome without (B) and with (C) FACT. (D) The real-time trace of the deubiquitinating processes of USP21 on one ubH2A nucleosome and its effect on FACT’s function. The tension first increases to 7.0 pN continuously at 0.1 pN/s, and then holds at 7.0 pN for 3 min before USP21 and FACT introduced. The deubiquitination process of USP21 on ubH2A nucleosome was observed by the 20 nm extension jump corresponding the disruption of outer DNA wrap of deubiquitinated ubH2A nucleosome at 7 pN. Then the tension was released to 0 pN for 3 min and stretched to 32 pN at 0.1 pN/s for three times to observe the remodeling function of FACT on the same nucleosome.
To further confirm that deubiquitination of ubH2A nucleosome rescues the remodeling function of FACT on the nucleosome, the real-time deubiquitinating process of USP21 on ubH2A nucleosomes and its effect on FACT’s function on nucleosome dynamics were traced (Figure 2D). The real-time deubiquitinating process of ubH2A nucleosome was monitored at a constant force of 7 pN. Before introducing USP21, no extension jump was observed because of the higher stability of ubH2A nucleosome. After additions of USP21 and FACT, a ∼20 nm extension jump was observed, corresponding to the disruption of the outer wrap due to the deubiquitination of ubH2A. After that, the repeated force-extension measurements were performed on the same nucleosome from 0 to 32 pN. Two extension jumps were identically observed at lower force <10 pN in each stretching cycle, indicating FACT could remodel the deubiquitinated ubH2A nucleosome (Figure 2B). As a control, if USP21 was introduced alone, the real-time deubiquitination process can be observed, but the structure of deubiquitinated ubH2A nucleosome was irreversible in the subsequent stretching cycles (Supplementary Figure S2C). On the other hand, if FACT was added alone, no deubiquitination process can be observed, and a typical unfolding process of the ubH2A nucleosome was observed in the subsequent stretching experiments, which indicated that FACT cannot remodel ubH2A nucleosome (Supplementary Figure S2D). Taken together, these results showed that the deubiquitination of ubH2A nucleosome rescues the remodeling functions of FACT on the nucleosome, which further revealed that ubH2A acts as an inhibitor to prevent FACT’s binding and remodeling at nucleosome level.
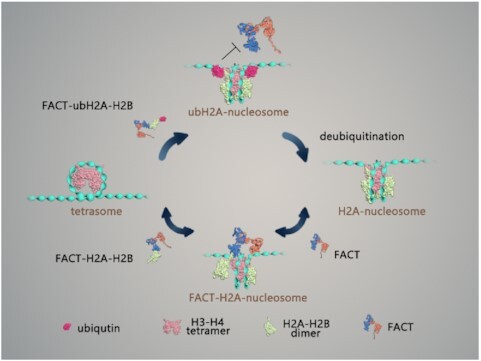
FACT can deposit ubH2A–H2B dimer to form intact nucleosome
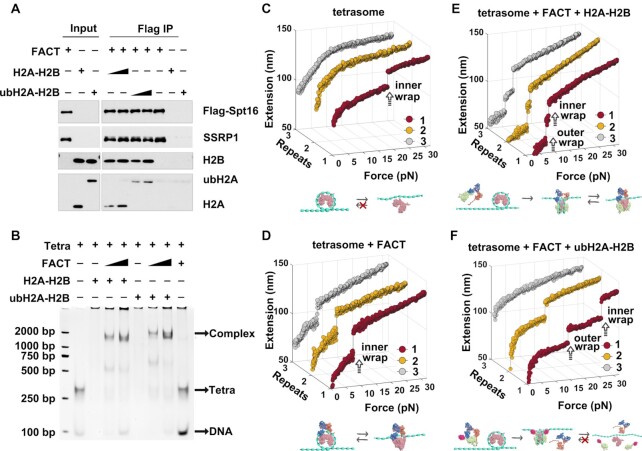
We have shown that ubH2A inhibits FACT’s function on the nucleosome disassembly. How does ubH2A regulate the nucleosome assembly function of FACT? To address this question, we investigated the FACT’s deposition ability of ubH2A–H2B dimers onto the (H3–H4)2 tetrasome (the particles made of an (H3–ṇH4)2 tetramer wrapped with DNA). We first checked the binding property of FACT to H2A–H2B dimer or ubH2A–H2B dimer. The recombinant FACT with Flag-SPT16 and SSRP1 were purified and incubated with recombinant H2A–H2B dimer or ubH2A–H2B dimer. The immunoprecipitation assays showed that ubH2A does not affect the binding property of FACT to ubH2A–H2B dimer. FACT binds both H2A–H2B dimer and ubH2A–H2B dimer with the similar efficiency (Figure 3A). We then examined whether ubH2A affects the deposit of the FACT-ubH2A–H2B on the (H3–ṇH4)2 tetrasome. The gel electrophoresis analysis showed that FACT can promote the loading of H2A–H2B as well as ubH2A–H2B to the tetrasomes to form a ternary complex with the FACT-H2A–H2B and the (H3–ṇH4)2 tetrasome, indicating the ubH2A does not affect the loading of the FACT-ubH2A–H2B complex on the (H3–ṇH4)2 tetrasome (Figure 3B). The results indicated that FACT promotes the deposit of ubH2A–H2B and H2A–H2B dimer onto the (H3–ṇH4)2 tetrasome with the similar efficiency.
Figure 3.
FACT can deposit ubH2A–H2B dimer onto tetrasome to form intact nucleosome. (A) Immunoprecipitation assays of the recombinant FACT with Flag-SPT16 and SSRP1 incubated with recombinant H2A–H2B or ubH2A–H2B dimer. The results showed that ubH2A does not affect the binding property of FACT on H2A–H2B dimer. (B) The gel electrophoresis analysis of the binding of FACT-H2A–H2B or FACT-ubH2A–H2B with (H3–ṇH4)2 tetrasome, which showed that FACT can promote the binding of H2A–H2B to tetrasomes to form a ternary complex with FACT-H2A–H2B and (H3–ṇH4)2 tetrasome, and the ubH2A does not affect the binding of FACT-H2A–H2B on (H3–ṇH4)2 tetrasome. (C–F) The repeated stretching measurements of the mono-tetrasome alone (C), with FACT alone (D), with FACT and H2A–H2B dimer (E), and with FACT and ubH2A–H2B dimer (F), respectively. The cartoons below represent the relative dynamic process of the tetrasome or nucleosome, respectively.
We further confirmed the effect of ubH2A on the FACT-mediated nucleosome assembly by magnetic tweezers. Mono-(H3–ṇH4)2 tetrasomes were reconstituted in vitro onto the 409 bp DNA fragment with one Widom 601 nucleosome positioning sequence. The reconstituted mono-tetrasomes were well positioned in the middle of DNA, as characterized by AFM images (Supplementary Figure S3A). Repeated force-extension measurements of the mono-tetrasomes showed the irreversible one-step extension jump corresponding to the unwrapping of tetrasome, with the rupture force at 22.3 ± 5.2 pN, similar to the unfolding of the inner wrap of nucleosome (Figure 3C). To investigate how ubH2A affects the FACT-mediated nucleosome assembly, we anchored the tetrasome on magnetic tweezers, injected FACT with or without H2A–H2B dimer and incubated for 30 min, and then rinsed away the unbound proteins with HE buffer. After incubation with FACT only, one-step extension jump was observed repeatedly with the lower rupture force 7.2 ± 2.8 pN (Figure 3D), indicating that FACT can interact with the tetrasome to impair its stability and maintain its integrity (Supplementary Figure S3B). After incubation with both FACT and H2A–H2B dimer, the repeated two-step extension jumps were observed at 5.6 ± 2.1 and 7.7 ± 3.4 pN respectively, indicating that FACT can deposit H2A–H2B dimers onto the tetrasome to form an intact nucleosome, and keep modulating the stability and reversibility of the reassembled nucleosome (Figure 3E), which are consistent with our previous results (16). Interestingly, after incubation with both FACT and ubH2A–H2B dimer, we can observe an irreversible two-step extension jumps with the rupture forces at 15.0 ± 7.1 and 22.9 ± 5.8 pN, similar to the unfolding process of the ubH2A nucleosome (Figure 3F). The rupture forces and reassembly property for the FACT-deposited H2A and ubH2A nucleosome were statistically analyzed as shown in Supplementary Figure S3C and D. The results indicated that FACT can deposit the ubH2A–H2B dimers onto the tetrasome to form intact ubH2A nucleosome. Once the intact ubH2A nucleosome is formed, FACT is excluded from the nucleosome, as revealed in the stretching cycles. In conclusion, ubH2A does not affect the nucleosome assembly property of FACT to mediate the deposition of H2A–H2B dimers onto the tetrasome to form intact nucleosome.
H2A ubiquitination inhibits the binding of FACT to repress gene transcription
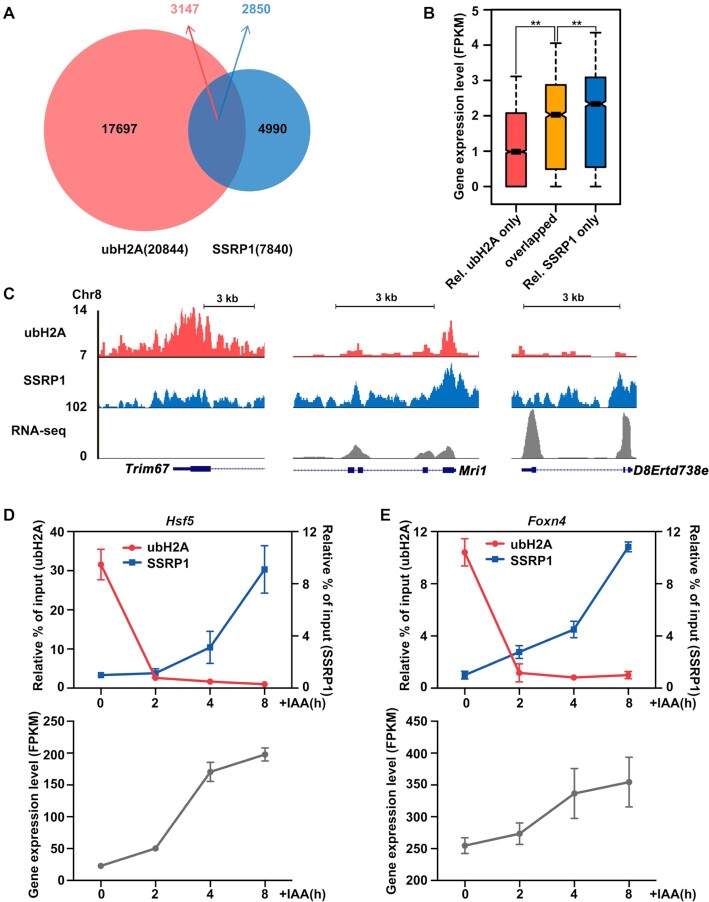
To further explore how ubH2A cooperates with FACT to regulate gene expression, we analyzed the genome-wide correlation between the levels of ubH2A and SSPR1 (one subunit of FACT complex). We performed ChIP-seq analysis for ubH2A and SSRP1 in mESCs (41,44). A total of 20 844 ubH2A and 7840 SSRP1 enrichment sites were identified by model-based analysis of ChIP-seq peak calling (Figure 4A). Among them, 3174 ubH2A peaks overlapped with 2850 SSRP1 peaks (overlapped region in Figure 4A). We analyzed the gene expression level between the genes relatively enriched with ubH2A only, SSRP1 only, and both ubH2A/SSRP1 overlapped. Our results showed that FACT facilitates gene transcription, with SSRP1 only enriched genes significantly more active than the relative ubH2A only enriched and ubH2A/SSRP1 overlapped genes; while ubH2A inhibits gene transcription, with ubH2A only enriched genes significantly more repressed than the SSRP1 only and ubH2A/SSRP1 overlapped genes (Figure 4B for the whole genome and Figure 4C for specific gene regions), consistent with previous studies.
Figure 4.
Genome-wide and specific gene analysis on the correlation of ubH2A and FACT on gene transcription. (A) Venn diagram showing the overlap of ubH2A (red) and SSRP1 (blue) peaks in mESCs. A total of 3147 ubH2A peaks overlapped with 2,850 SSRP1 peaks; the majority of ubH2A peaks (17 697) and SSRP1 peaks (4990) did not overlap. (B) Genome-wide analysis on the expression level of genes relatively enriched with ubH2A only, relatively enriched with SSRP1 only, and ubH2A/SSRP1 overlapped.**Significant difference (P < 0.01) according to Wilcoxon signed-rank test. (C) The distribution of ubH2A and FACT at indicated genes as well as their corresponding expression levels, for genes relatively enriched for ubH2A only (left column), ubH2A/SSRP1 overlapped (middle column), and relatively enriched for SSRP1 only (right column). The chromatin regions enriched for FACT are correlated with low level of ubH2A and associated with highly transcribed genes. (D, E) ChIP-qPCR analysis of the level of ubH2A and SSRP1 on the promoter regions of Hsf5 and Foxn4 (up) and the RNA-seq analysis of gene expression level with FPKM (fragments per kilobase of transcript per million fragments sequenced) related RNA-seq analysis (bottom) after IAA addition. The data represent means ± S.D. ChIP enrichments are normalized to the input. Relative % of input: ubH2A samples are normalized to the last time point, SSRP1 samples are normalized to the first time point.
The genes in the ubH2A/SSRP1 overlapped region in Figure 4A are the genes regulated by both ubH2A and FACT. But the static enrichment data from ChIP-seq analysis of cell polulation cannot give imformation of the dynamic interplay of ubH2A and FACT on transcription. To further explore the dynamic regulation of ubH2A and SSRP1 on the gene transcription, several genes from the overlapped region were selected to investigate by time-course ChIP-qPCR analysis. We dynamically decreased the level of ubH2A by degrading the endogenous Ring1B (the major H2A ubiquitination ligase) in Ring1a–/– mESC using an auxin-inducible degradation system (AID), and then monitored the related effect on FACT recruitment and gene expression level. Our results showed that ubH2A decreased significantly within 2 h after the addition of IAA (indole-3-acetic acid) (Supplementary Figure S4A). We then examined the dynamic regulation of ubH2A and SSRP1 on several genes selected in the overlapped regions which can be regulated by both ubH2A and FACT, with the typical examples of Hsf5, Foxn4, Lrrfip1 and Rasd2 shown in Figure 4D–E and S4B–C. Time-course ChIP-qPCR analysis revealed that the rapidly decrease of ubH2A at the promoter region was accompanied by gradually increased SSRP1 after IAA addition, which was also accompanied by the activation of gene transcription as demonstrated by RNA-seq analysis. The results confirmed that ubH2A inhibits the binding of FACT to repress gene transcription and the deubiquitination of ubH2A induces the binding of FACT to activate gene.
DISCUSSION
In eukaryotes, organisms and cells inherit genetic information encoded in DNA sequence, as well as epigenetic information from parental generation, which regulates the activity of gene expression. The histone chaperone FACT has been found to play essential roles in nucleosome disassembly and assembly during many chromatin-related processes, including gene transcription, DNA replication, and repair. In these processes, the nucleosome barrier needs to be temporarily disassembled to expose DNA for processing, and then be rapidly reassembled to protect the DNA and preserve the original epigenetic identity (3,4). Previous studies have found that FACT appears to function not only in destabilizing nucleosome to facilitate the progression of DNA and RNA polymerases on chromatins, but also in establishing and maintaining the genome-wide integrity of chromatin structure (9–15). In addition, recent in vitro structural and single-molecular investigations have revealed that FACT can tether both the (H3–ṇH4)2 tetramer and H2A–H2B dimer on DNA by its two subunits SPT16 and SSRP1, which functions distinctly but coordinately on the nucleosome disassembly and assembly (16–18). FACT has been shown to functionally correlate with different histone modifications or variants in distinct processes, including cooperating with ubH2A and ubH2B in gene transcription, associating with CENP-C or CENP-T/-W to function at centromeres, or promoting H2A.X deposition in DNA repair (30,53–56).
Mono-ubiquitination at Lysine 119 of H2A, as the first identified histone ubiquitination, is a prevalent post-translational modification in higher eukaryotic organisms with about 5–15% of total H2A ubiquitinated (19,57). The ubH2A are primarily associated with gene repression in the context of chromatin, with a strong correlation to the polycomb repressive complexes PRC1, the major H2A ubiquitination ligase, and H3K27me3 methylated by PRC2 in many cell types (25–29). Previous study has revealed that ubH2A might block the recruitment of Spt16 at protein level, in which the effect of ubH2A on the binding of FACT was investigated by coimmunoprecipitation (Co-IP) studies of endogenous SPT16 with unmodified Flag-H2A or Flag-ubH2A stably expressed in HEK293 cells (30). However, how ubH2A directly regulates the functions of FACT on nucleosome dynamics is poorly understood.
Here, we revealed the different regulation of H2A ubiquitination on the functions of FACT in nucleosome disassembly and assembly. On one hand, we found that ubH2A directly inhibits the binding of FACT on nucleosome and blocks the FACT’s function in nucleosome disassembly. Our SILAC-based mass spectrometry with mono-nucleosomes pull-down assays showed that both the two subunits of FACT, SPT16 and SSRP1 are preferentially enriched on unmodified H2A mono-nucleosomes compared to ubH2A mono-nucleosomes, indicating ubH2A may inhibit the binding of FACT on nucleosomes. The results were verified by the mono-nucleosomes pull-down assays using purified FACT complex, which also showed that ubH2A directly inhibits the binding of FACT on nucleosomes, which completely blocks the nucleosome disassembly function of FACT. Using single-molecule magnetic tweezers, we found that no matter with or without FACT, the ubH2A nucleosomes display a similar unfolding pathway: irreversible two-step transitions at higher forces of about 15 pN for disruption of the outer wrap and about 24 pN for disruption of the inner wrap. The results indicated that the H2A ubiquitination completely blocks the nucleosome disassembly function of FACT. As a comparison, for the unmodified H2A nucleosomes, FACT greatly impairs the stability of H2A nucleosome and helps to maintain the integrity of H2A nucleosome when the nucleosome is fully disrupted. In line with this, real-time tracking of the deubiquitination process and its effect on FACT functions on the same ubH2A nucleosome in vitro showed that the deubiquitination of ubH2A nucleosome can rescue the nucleosome disassembly function of FACT. Genome-wide and specific gene analysis also showed that ubH2A inhibits the binding of FACT to repress gene transcription and the deubiquitination of ubH2A induces the binding of FACT to activate gene. On the other hand, we found that ubH2A does not affect the FACT’s chaperone function in nucleosome assembly. Binding assays showed that the ubH2A does not affect both the binding between FACT and H2A–H2B dimer, as well as the loading of the FACT-H2A–H2B complex onto the (H3–ṇH4)2 tetrasome. In addition, magnetic tweezers experiments showed that ubH2A does not affect the chaperone property of FACT to mediate the deposition of H2A–H2B dimers onto the tetrasome to form intact nucleosome.
How does the ubH2A affect the binding of FACT on nucleosome? The modified ubiquitin on the site of Lysine 119 at the beginning of H2A C-terminal tails and at the end of the H2A docking domain, is located at DNA entry/exit point of nucleosome (2), which dramatically stabilizes the nucleosome by blocking DNA peeling from histone octamer as we previously showed (22). Recent structural analysis on the FACT with nucleosome has revealed that the SPT16 middle domain may interact with one H2A/H2B dimer and SSRP1 middle domain holds the other H2A/H2B dimer at this point (17). These analysis suggested that the ubiquitin modified at this site may clash with the middle domain of SPT16 and SSRP1, and exclude the binding of FACT with nucleosome.
The distinct regulation of ubH2A on the FACT’s functions in nucleosome disassembly and assembly, as we revealed here, may help to safeguard the formation of ubH2A nucleosome and maintain the gene suppression property of ubH2A. The ubH2A on nucleosome inhibits the binding of FACT, which completely blocks the nucleosome disassembly function of FACT (Figure 5A). When the ubH2A–H2B dimer is eventually displaced from the nucleosome, the unstable sub-nucleosome may recruit FACT, which may function as a ‘rescue squad’, to deposit the ubH2A–H2B dimer to form an intact ubH2A nucleosome and maintain the repressed state of genes. In addition, the ubH2A has been found to be stably inherited during cell division for maintaining cell identity (41). The functional activity of FACT to chaperone ubH2A–H2B dimers may help to recycle and maintain the modified histones during DNA replication. To activate gene transcription, the ubH2A nucleosome needs to be deubiquitinated, which destabilizes the nucleosome and further recruits FACT to facilitate gene transcription by destabilizing the nucleosome (Figure 5B).
Figure 5.
Mechanism model proposed for the regulation of H2A ubiquitination on the function of FACT on nucleosomes. (A) H2A ubiquitination stabilizes the nucleosome and inhibits the binding of FACT to completely block the remodeling effect of FACT to repress gene transcription. (B) UbH2A does not affect FACT to chaperone H2A–H2B onto (H3–ṇH4)2 tetrasome to form intact nucleosome. For gene activation, ubH2A nucleosome needs to be deubiquitinated, which will destabilize the nucleosome and further activate the binding of FACT to remodel the nucleosome and facilitate gene transcription.
Taken together, we revealed a different regulation of ubH2A on FACT’s functions in nucleosome dynamics, which provides a new insight to understand how the FACT’s functions in nucleosome dynamics are regulated by different epigenetic factors. It will be of great interest to investigate the regulation of FACT’s functions in nucleosome dynamics by other related epigenetic factors, such as histone variants or chromatin remodelers, which may help to propose an unified model for FACT’s activity at the different chromatin states with different epigenetic information.
DATA AVAILABILITY
The RNA-seq data generated and analyzed in this study have been submitted to the NCBI Sequence Read Archive (SRA) database, under accession code PRJNA760283. Mouse ES cell ubH2A and SSRP1 ChIP-seq data were downloaded from PRJNA604675, GSE90906 respectively (41,44) and re-analysed. SILAC mass spectrometry data have been submitted to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD017105.
Supplementary Material
ACKNOWLEDGEMENTS
We thank T. Yao (National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences) for ordering reagents, and M. Zhang and J. Wang (Laboratory of Proteomics, Institute of Biophysics, Chinese Academy of Sciences) for SILAC mass spectrometry. We are also indebted to the colleagues whose work could not be cited due to the limitation of space.
Contributor Information
Yi-Zhou Wang, Shenzhen Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Shenzhen Key Laboratory of Agricultural Synthetic Biology, Genome Analysis Laboratory of the Ministry of Agriculture, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518124, China.
Cuifang Liu, National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Jicheng Zhao, National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Juan Yu, National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Anfeng Luo, Department of Immunology, School of Basic Medical Sciences, Advanced Innovation Center for Human Brain Protection, Capital Medical University, Beijing 100069, China.
Xue Xiao, National Laboratory for Condensed Matter Physics and Key Laboratory of Soft Matter Physics, Institute of Physics, Chinese Academy of Sciences, Beijing 100190, China.
Shuo-Xing Dou, National Laboratory for Condensed Matter Physics and Key Laboratory of Soft Matter Physics, Institute of Physics, Chinese Academy of Sciences, Beijing 100190, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Lu Ma, National Laboratory for Condensed Matter Physics and Key Laboratory of Soft Matter Physics, Institute of Physics, Chinese Academy of Sciences, Beijing 100190, China.
Peng-Ye Wang, National Laboratory for Condensed Matter Physics and Key Laboratory of Soft Matter Physics, Institute of Physics, Chinese Academy of Sciences, Beijing 100190, China; University of Chinese Academy of Sciences, Beijing 100049, China; Songshan Lake Materials Laboratory, Dongguan, Guangdong 523808, China.
Ming Li, National Laboratory for Condensed Matter Physics and Key Laboratory of Soft Matter Physics, Institute of Physics, Chinese Academy of Sciences, Beijing 100190, China.
Guohong Li, National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Jianbin Yan, Shenzhen Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Shenzhen Key Laboratory of Agricultural Synthetic Biology, Genome Analysis Laboratory of the Ministry of Agriculture, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518124, China.
Ping Chen, Department of Immunology, School of Basic Medical Sciences, Advanced Innovation Center for Human Brain Protection, Capital Medical University, Beijing 100069, China; National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Wei Li, National Laboratory for Condensed Matter Physics and Key Laboratory of Soft Matter Physics, Institute of Physics, Chinese Academy of Sciences, Beijing 100190, China; Songshan Lake Materials Laboratory, Dongguan, Guangdong 523808, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology of China [2019YFA0508903 to J.Z., 2018YFE0203302 to P.C., 2017YFA0504202 to G.L.]; National Natural Science Foundation of China [31991161, 31630041 to G.L., 32022014, 31871290 to P.C., 11874414 to W.L., 11774407 to S.-X.D., 21991133, 11874415 to P.-Y.W., 31770812 to L.M., 32070604 to J.Z., 32100470 to C.L.]; Beijing Municipal Science and Technology Committee [Z201100005320013 to G.L.]; Chinese Academy of Sciences (CAS) Key Research Program on Frontier Science [QYZDY-SSW-SMC020 to G.L., QYZDB-SSW-SLH045 to W.L.]; CAS Strategic Priority Research Program of Chinese Academy of Sciences [XDB37000000 to M.L.]; National Laboratory of Biomacromolecules [2020kf02 to W.L.]; National Key Research and Development Program [2016YFA0301500 to P.-Y.W.]; China Postdoctoral Science Foundation [2020M680711 to C.L.]; Guangdong Basic and Applied Basic Research Foundation [2019A1515110282 to Y. W.]; Elite Young Scientists Program of CAAS, Agricultural Science and Technology Innovation Program; HHMI International Research Scholar grant [55008737 to G.L.]. Funding for open access charge: National Key Research and Development Program.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kornberg R.D. Structure of chromatin. Annu. Rev. Biochem. 1977; 46:931–954. [DOI] [PubMed] [Google Scholar]
- 2. Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J.. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997; 389:251–260. [DOI] [PubMed] [Google Scholar]
- 3. Kaufman P.D., Rando O.J.. Chromatin as a potential carrier of heritable information. Curr. Opin. Cell Biol. 2010; 22:284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alabert C., Groth A.. Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 2012; 13:153–167. [DOI] [PubMed] [Google Scholar]
- 5. Belotserkovskaya R., Oh S., Bondarenko V.A., Orphanides G., Studitsky V.M., Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003; 301:1090–1093. [DOI] [PubMed] [Google Scholar]
- 6. Orphanides G., Wu W.-H., Lane W.S., Hampsey M., Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999; 400:284–288. [DOI] [PubMed] [Google Scholar]
- 7. Reinberg D., Sims R.J.. de FACTo nucleosome dynamics*. J. Biol. Chem. 2006; 281:23297–23301. [DOI] [PubMed] [Google Scholar]
- 8. Orphanides G., LeRoy G., Chang C.-H., Luse D.S., Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998; 92:105–116. [DOI] [PubMed] [Google Scholar]
- 9. Formosa T., Ruone S., Adams M.D., Olsen A.E., Eriksson P., Yu Y., Rhoades A.R., Kaufman P.D., Stillman D.J.. Defects in SPT16 or POB3 (yFACT) in saccharomyces cerevisiae cause dependence on the hir/hpc pathway: polymerase passage may degrade chromatin structure. Genetics. 2002; 162:1557–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mason P.B., Struhl K.. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 2003; 23:8323–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wittmeyer J., Joss L., Formosa T.. Spt16 and pob3 of saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase α. Biochemistry. 1999; 38:8961–8971. [DOI] [PubMed] [Google Scholar]
- 12. Lejeune E., Bortfeld M., White S.A., Pidoux A.L., Ekwall K., Allshire R.C., Ladurner A.G.. The chromatin-remodeling factor FACT contributes to centromeric heterochromatin independently of RNAi. Curr. Biol. 2007; 17:1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jamai A., Puglisi A., Strubin M.. Histone chaperone spt16 promotes redeposition of the original h3-h4 histones evicted by elongating RNA polymerase. Mol. Cell. 2009; 35:377–383. [DOI] [PubMed] [Google Scholar]
- 14. Kaplan C.D., Laprade L., Winston F.. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003; 301:1096–1099. [DOI] [PubMed] [Google Scholar]
- 15. Hainer S.J., Pruneski J.A., Mitchell R.D., Monteverde R.M., Martens J.A.. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev. 2011; 25:29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen P., Dong L., Hu M., Wang Y.-Z., Xiao X., Zhao Z., Yan J., Wang P.-Y., Reinberg D., Li M.et al.. Functions of FACT in breaking the nucleosome and maintaining its integrity at the single-nucleosome level. Mol. Cell. 2018; 71:284–293. [DOI] [PubMed] [Google Scholar]
- 17. Liu Y., Zhou K., Zhang N., Wei H., Luger K.. FACT caught in the act of manipulating the nucleosome. Nature. 2020; 577:426–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsunaka Y., Fujiwara Y., Oyama T., Hirose S., Morikawa K.. Integrated molecular mechanism directing nucleosome reorganization by human FACT. Genes Dev. 2016; 30:673–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goldknopf I.L., Taylor C.W., Baum R.M., Yeoman L.C., Olson M.O., Prestayko A.W., Busch H.. Isolation and characterization of protein A24, a ‘histone-like’ non-histone chromosomal protein. J. Biol. Chem. 1975; 250:7182–7187. [PubMed] [Google Scholar]
- 20. Frappier L., Verrijzer C.P.. Gene expression control by protein deubiquitinases. Curr. Opin. Genet. Dev. 2011; 21:207–213. [DOI] [PubMed] [Google Scholar]
- 21. Panier S., Durocher D. Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair. 2009; 8:436–443. [DOI] [PubMed] [Google Scholar]
- 22. Xiao X., Liu C., Pei Y., Wang Y.-Z., Kong J., Lu K., Ma L., Dou S.-X., Wang P.-Y., Li G.et al.. Histone H2A ubiquitination reinforces mechanical stability and asymmetry at the single-nucleosome level. J. Am. Chem. Soc. 2020; 142:3340–3345. [DOI] [PubMed] [Google Scholar]
- 23. Lee H.-G., Kahn T.G., Simcox A., Schwartz Y.B., Pirrotta V.. Genome-wide activities of polycomb complexes control pervasive transcription. Genome Res. 2015; 25:1170–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nakagawa T., Kajitani T., Togo S., Masuko N., Ohdan H., Hishikawa Y., Koji T., Matsuyama T., Ikura T., Muramatsu M.et al.. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 2008; 22:37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Endoh M., Endo T.A., Endoh T., Isono K.-i., Sharif J., Ohara O., Toyoda T., Ito T., Eskeland R., Bickmore W.A.et al.. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-Dependent repression of developmental genes to maintain ES cell identity. PLoS Genet. 2012; 8:e1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cohen I., Zhao D., Bar C., Valdes V.J., Dauber-Decker K.L., Nguyen M.B., Nakayama M., Rendl M., Bickmore W.A., Koseki H.et al.. PRC1 Fine-tunes gene repression and activation to safeguard skin development and stem cell specification. Cell Stem Cell. 2018; 22:726–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fursova N.A., Blackledge N.P., Nakayama M., Ito S., Koseki Y., Farcas A.M., King H.W., Koseki H., Klose R.J.. Synergy between variant PRC1 complexes defines polycomb-mediated gene repression. Mol. Cell. 2019; 74:1020–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scelfo A., Fernández-Pérez D., Tamburri S., Zanotti M., Lavarone E., Soldi M., Bonaldi T., Ferrari K.J., Pasini D. Functional landscape of PCGF proteins reveals both RING1A/B-Dependent-and RING1A/B-Independent-Specific activities. Mol. Cell. 2019; 74:1037–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blackledge N.P., Fursova N.A., Kelley J.R., Huseyin M.K., Feldmann A., Klose R.J.. PRC1 catalytic activity is central to polycomb system function. Mol. Cell. 2020; 77:857–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou W., Zhu P., Wang J., Pascual G., Ohgi K.A., Lozach J., Glass C.K., Rosenfeld M.G.. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol. Cell. 2008; 29:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li G., Margueron R., Hu G., Stokes D.L., Wang Y., Reinberg D. Highly compacted chromatin formed in vitro reflects the dynamics of transcription activation in vivo. Mol. Cell. 2010; 38:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen P., Zhao J., Wang Y., Wang M., Long H., Liang D., Huang L., Wen Z., Li W., Li X.et al.. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes Dev. 2013; 27:2109–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Joo H.Y., Zhai L., Yang C., Nie S., Erdjument-Bromage H., Tempst P., Chang C., Wang H.. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007; 449:1068–1072. [DOI] [PubMed] [Google Scholar]
- 34. Zhang Z., Menq C.-H.. Three-dimensional particle tracking with subnanometer resolution using off-focus images. Appl. Opt. 2008; 47:2361–2370. [DOI] [PubMed] [Google Scholar]
- 35. Smith S.B., Finzi L., Bustamante C.. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science. 1992; 258:1122–1126. [DOI] [PubMed] [Google Scholar]
- 36. Gosse C., Croquette V.. Magnetic tweezers: micromanipulation and force measurement at the molecular level. Biophys. J. 2002; 82:3314–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Daldrop P., Brutzer H., Huhle A., Kauert D.J., Seidel R.. Extending the range for force calibration in magnetic tweezers. Biophys. J. 2015; 108:2550–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen H., Fu H., Zhu X., Cong P., Nakamura F., Yan J.. Improved high-force magnetic tweezers for stretching and refolding of proteins and short DNA. Biophys. J. 2011; 100:517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Loenhout M.T.v., Kerssemakers J.W., Vlaminck I., Dekker C.. Non-bias-limited tracking of spherical particles, enabling nanometer resolution at low magnification. Biophys. J. 2012; 102:2362–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van Oene M.M., Dickinson L.E., Pedaci F., Kober M., Dulin D., Lipfert J., Dekker N.H.. Biological magnetometry: torque on superparamagnetic beads in magnetic fields. Phys Rev Lett. 2015; 114:218301. [DOI] [PubMed] [Google Scholar]
- 41. Zhao J., Wang M., Chang L., Yu J., Song A., Liu C., Huang W., Zhang T., Wu X., Shen X.et al.. RYBP/YAF2-PRC1 complexes and histone H1-dependent chromatin compaction mediate propagation of H2AK119ub1 during cell division. Nat. Cell Biol. 2020; 22:439–452. [DOI] [PubMed] [Google Scholar]
- 42. Liao Y., Smyth G.K., Shi W.. The subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013; 41:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L.. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010; 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mylonas C., Tessarz P.. Transcriptional repression by FACT is linked to regulation of chromatin accessibility at the promoter of ES cells. Life Sci. Alliance. 2018; 1:e201800085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Langmead B., Salzberg S.L.. Fast gapped-read alignment with bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.Genome Project Data Processing, S. . The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.et al.. Model-based analysis of chip-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li W., Chen P., Yu J., Dong L., Liang D., Feng J., Yan J., Wang P.-Y., Li Q., Zhang Z.et al.. FACT remodels the tetranucleosomal unit of chromatin fibers for gene transcription. Mol. Cell. 2016; 64:120–133. [DOI] [PubMed] [Google Scholar]
- 49. Mihardja S., Spakowitz A.J., Zhang Y., Bustamante C.. Effect of force on mononucleosomal dynamics. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:15871–15876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hall M.A., Shundrovsky A., Bai L., Fulbright R.M., Lis J.T., Wang M.D.. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat. Struct. Mol. Biol. 2009; 16:124–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Böhm V., Hieb A.R., Andrews A.J., Gansen A., Rocker A., Tóth K., Luger K., Langowski J.. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 2010; 39:3093–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gong L., Kamitani T., Millas S., Yeh E.T.H.. Identification of a novel isopeptidase with dual specificity for Ubiquitin- and NEDD8-conjugated proteins*. J. Biol. Chem. 2000; 275:14212–14216. [DOI] [PubMed] [Google Scholar]
- 53. Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase iI. Cell. 2006; 125:703–717. [DOI] [PubMed] [Google Scholar]
- 54. Fleming A.B., Kao C.-F., Hillyer C., Pikaart M., Osley M.. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell. 2008; 31:57–66. [DOI] [PubMed] [Google Scholar]
- 55. Prendergast L., Müller S., Liu Y., Huang H., Dingli F., Loew D., Vassias I., Patel D.J., Sullivan K.F., Almouzni G.. The CENP-T/-W complex is a binding partner of the histone chaperone FACT. Genes Dev. 2016; 30:1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Piquet S., Le Parc F., Bai S.K., Chevallier O., Adam S., Polo S,E.. The histone chaperone FACT coordinates H2A.X-Dependent signaling and repair of DNA damage. Mol. Cell. 2018; 72:888–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Goldknopf I.L., Busch H.. Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proc. Natl. Acad. Sci. U.S.A. 1977; 74:864–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data generated and analyzed in this study have been submitted to the NCBI Sequence Read Archive (SRA) database, under accession code PRJNA760283. Mouse ES cell ubH2A and SSRP1 ChIP-seq data were downloaded from PRJNA604675, GSE90906 respectively (41,44) and re-analysed. SILAC mass spectrometry data have been submitted to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD017105.