Cai et al. describe their finding of cellular diversity in tuberculous pleural effusion (TPE). They establish a link between clonal expansion and lineage plasticity of T cells and reveal the involvement of granzyme K–expressing CD8 T cells in TPE pathogenesis.
Abstract
Orchestration of an effective T lymphocyte response at infection sites is critical for protection against Mycobacterium tuberculosis (Mtb) infection. However, the local T cell immunity landscape in human tuberculosis is poorly defined. Tuberculous pleural effusion (TPE), caused by Mtb, is characterized by an influx of leukocytes to the pleural space, providing a platform suitable for delineating complex tissue responses to Mtb infection. Using single-cell transcriptomics and T cell receptor sequencing, we analyzed mononuclear cell populations in paired pleural fluid and peripheral blood of TPE patients. While all major cell clusters were present in both tissues, their relative proportions varied significantly by anatomic location. Lineage tracking analysis revealed subsets of CD8 and CD4 T cell populations with distinct effector functions specifically expanded at pleural sites. Granzyme K–expressing CD8 T cells were preferentially enriched and clonally expanded in pleural fluid from TPE, suggesting that they are involved in the pathogenesis of the disease. The findings collectively reveal the landscape of local T cell immunity in tuberculosis.
Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb) infection, remains a global health problem, with millions of deaths each year (Floyd et al., 2018). Decades of immunological studies on TB, performed in both humans and animal models, have revealed several cell-mediated immune mechanisms that are critical for protection against Mtb infection (Bold and Ernst, 2012; Canaday et al., 2001; Caruso et al., 1999; Lalvani et al., 1998; Lewinsohn et al., 1998; Scanga et al., 2000). However, most previous studies of human TB were based on the analysis of cells from peripheral blood (Berry et al., 2010), which might not accurately reflect Mtb-specific immune responses that occur at local sites of infection. Understanding the inflammatory mediators and immune cells present locally in Mtb-infected tissues in humans has lagged.
Pleural effusion is a common disease with multiple known etiologies. Tuberculous pleural effusion (TPE) is characterized by intense accumulation of exudative fluid and inflammatory cells in the pleural space of Mtb-infected individuals (Vorster et al., 2015; Wang et al., 2018). Parapneumonic effusion (PPE) and malignant pleural effusion (MPE) are the other two exudative non-TPE conditions. PPE is mainly associated with bacterial infections and is characterized by neutrophil-dominant inflammation (Light, 2006). MPE is accompanied by the presence of malignant cells or tissues. The majority of MPE cases are caused by metastatic disease: lung cancer is one of the most common malignancies causing MPE worldwide (Porcel et al., 2015). It is documented that TPE, MPE, and PPE show indistinguishable blood biochemistry and routine examination (Hooper et al., 2010; Vorster et al., 2015).
Lymphocytes have been shown to dominate the leukocyte infiltrates in TPE, although neutrophils and macrophages may also be presented (Mitra et al., 2005). Lymphocytic pleural fluid (PF) is observed in ∼90% of TPE cases (Lee et al., 2016). The inflammation can resolve spontaneously within a few weeks or months without chemotherapy (San José et al., 1999; Shaw et al., 2019). The mechanism underlying the successful self-elimination of PF in TPE is incompletely understood but believed to be mediated by the compartmentalized T helper 1 cells (Th1) immune response (Jalapathy et al., 2004). Indeed, various studies have demonstrated that the lymphocyte population in PF predominantly comprises Th1 cells capable of producing high levels of IFN-γ (da Cunha Lisboa et al., 2019; Sharma et al., 2002). Therefore, understanding the heterogeneity, clonotype, migration, and effector functions of T cells in PF from TPE will provide important insights into antimycobacterial responses at the tissue level. The compartmentalized immune response in the pleural space of patients with TPE provides an excellent clinical setting to dissect the local immune response to Mtb.
Single-cell RNA sequencing (scRNA-seq) can be used to reliably identify and analyze closely related cell populations, including cells in complex microenvironments (Zhang et al., 2018; Zheng et al., 2017). Single-cell TCR sequencing (scTCR-seq) is a powerful tool that can define the dynamics of clonal expansion within T cell populations, which is particularly useful in situations where high cellular turnover is expected. scTCR-seq can thus be used to measure clonal expansion and track clonal lineages across tissues.
By combining scRNA-seq and scTCR-seq data from individual cells, in this study we correlated transcriptional status with antigen specificity, modeled clonal expansion dynamics alongside T cell populations, and investigated T cell phenotypic plasticity in different contexts. We identified 25 unique T cell subsets with distinct tissue distribution patterns and determined the connectivity and potential developmental paths of these subsets. Importantly, these studies uncovered a previously unknown strong association between granzyme K (GZMK)–expressing CD8 T cells and TPE disease.
Results
scRNA-seq profiling reveals high T cell heterogeneity in PF from patients with TPE
Although the lymphocyte population in PF favors a Th1 response (da Cunha Lisboa et al., 2019; Sharma et al., 2002), the T cell populations in PF are complex (Tong and Shi, 2013). Single-cell technologies are powerful tools that can reveal the composition of T cell populations to further understand the T cell response to Mtb infection. We first aimed to explore the cellular diversity in PF from patients with TPE. We obtained fresh PF and paired blood samples from four patients with TPE (Fig. 1 A and Table S1) before anti-TB treatment. We then performed coupled scRNA-seq and scTCR-seq on these samples. One peripheral blood mononuclear cell (PBMC) sample did not pass quality control due to poor unique molecular identifiers (UMIs) and median gene counts (Table S2). Therefore, the analysis included three PBMC and four pleural fluid mononuclear cell (PFMC) samples. After filtering the scRNA-seq data (see Materials and methods), we retained a total of 64,517 cell transcriptomes for analysis across the four patients (Fig. S1 A and Table S3).
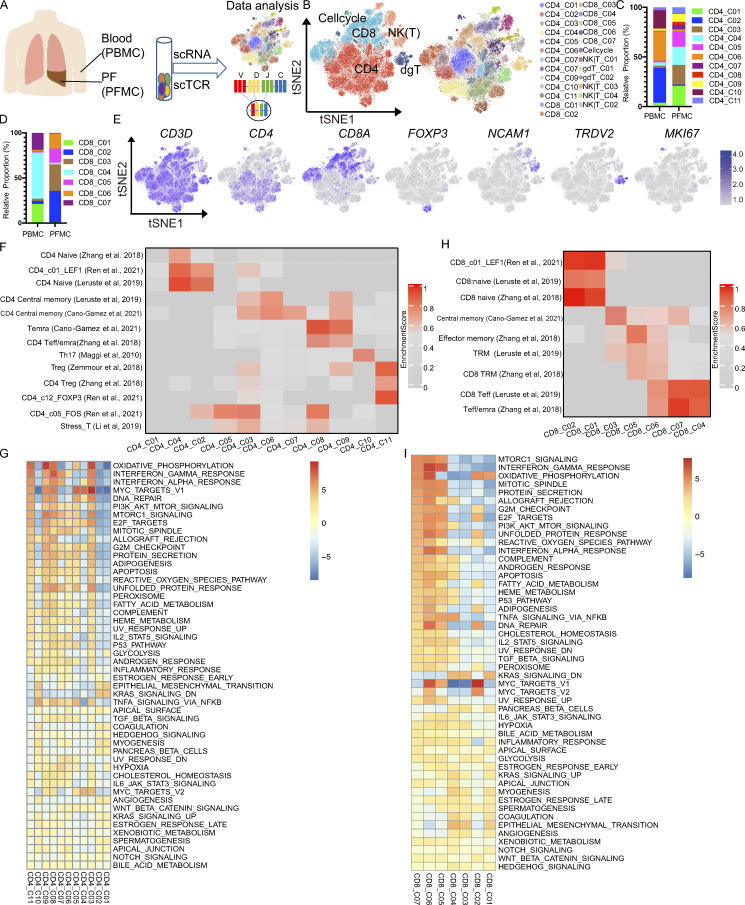
Figure 1.
Single-cell immune profiling of T cell heterogeneity in TPE. (A) The experimental workflow for defining and comparing T cells between the blood and PF in TPE. (B) tSNE of the T cell profile, with each cell color-coded for the main cell types and associated cell subsets from four TPE patients (PBMC, n = 3; PFMC n = 4). (C) The fraction of cells for CD4 T cell subsets in blood and PF from four TPE patients. (D) The fraction of cells for CD8 T cell subsets in blood and PF from four TPE patients. (E) tSNE plot of expression levels of selected genes in different clusters indicated by the colored oval. (F and G) Individual cell enrichment of indicated selected signatures or marker genes from published dataset; the enrichment scores are shown in the heatmap. (F) CD4 subsets. (G) CD8 subsets. Temra, effector memory or effector; Teff, effector; Trm, tissue resident memory. (H) Differences in pathway activities scored per cell by GSEA between the different CD4 T clusters from four TPE patients. (I) Differences in pathway activities scored per cell by GSVA between the different CD8 T clusters from four TPE patients.
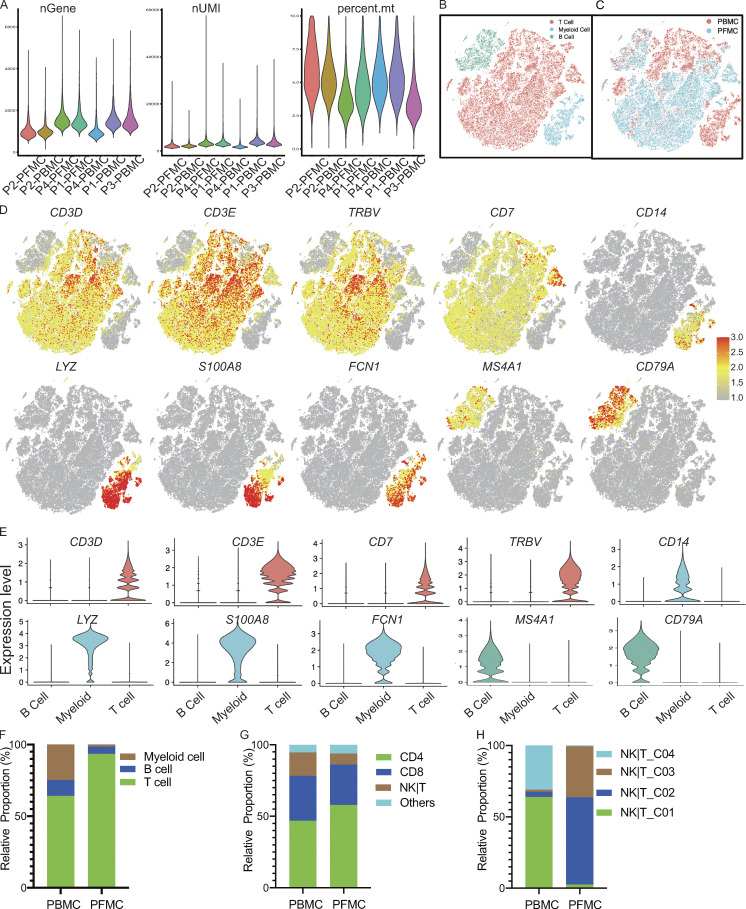
Figure S1.
scRNA-seq of cells from TPE. (A) Consistency of cell capture and identification in scRNA-seq. Number of UMIs (nUMIs) and genes (nGene) identified and fraction of reads mapped to mitochondrial (mt) genes across all samples (n = 4). (B) All samples merged single cells with the associated main cell types. (C) All samples merged single cells with the corresponding tissues (PBMCs and PFMCs). (D) tSNE plot of expression levels of selected genes in different cell types indicated by the colored ovals. (E) Violin plot of expression levels of selected genes in different cell types. (F) The relative proportion of cells for main cell types in blood and PF from TPE (n = 4). (G) The relative proportion of cells for T cell subsets in the T cell fraction in blood and PF from TPE (n = 4). (H) The relative proportion of cells for NK|T subsets in blood and PF from TPE (n = 4).
Consistent with our previous report (Cai et al., 2020), among PBMCs, 63.9, 24.7, and 11.4% were T, myeloid, and B cells, respectively. However, the PFMCs largely comprised T cells (93.5%), followed by B and myeloid cells (5.1 and 1.3%, respectively; Fig. S1, B–F; and Table S3). Therefore, we investigated this T cell fraction further. Unsupervised clustering using the Seurat package (Butler et al., 2018) and based on gene expression identified five distinct clusters: CD4, CD8, NK or other T (NK|T), γδT cells, and cell cycle from the T cell fraction from the four TPE patients (Fig. 1 B). T cells from PBMCs and PFMCs (n = 52,268) consisted largely of CD4 T cells (46.8% in PBMCs and 58.0% in PFMCs), followed by CD8 T cells (31.7% in PBMCs and 28.2% in PFMCs) and NK|T cells (16.4% in PBMCs and 7.8% in PFMCs; Fig. S1, G and H; and Table S2).
Further subclustering revealed a high level of heterogeneity within the T cell compartment, with 25 different subsets (Figs. 1 B and S2 A). Most subsets were present in PBMCs and PFMCs, although the relative proportions varied in a tissue-dependent manner (Fig. 1, C and D; and Table S2). Among the 25 different clusters, we identified 11 subsets of CD4 T cells, 7 subsets of CD8 T cells, 4 subsets of NK|T cells, and 3 additional clusters corresponding to γδT cells (expressing TRDV1 and TRDV2 and enriched in the γδT signature; Savas et al., 2018), and cells enriched in the cycling signature gene expression (MKI67; Fig. 1 E; Fig. S2, A–C; and Table S4).
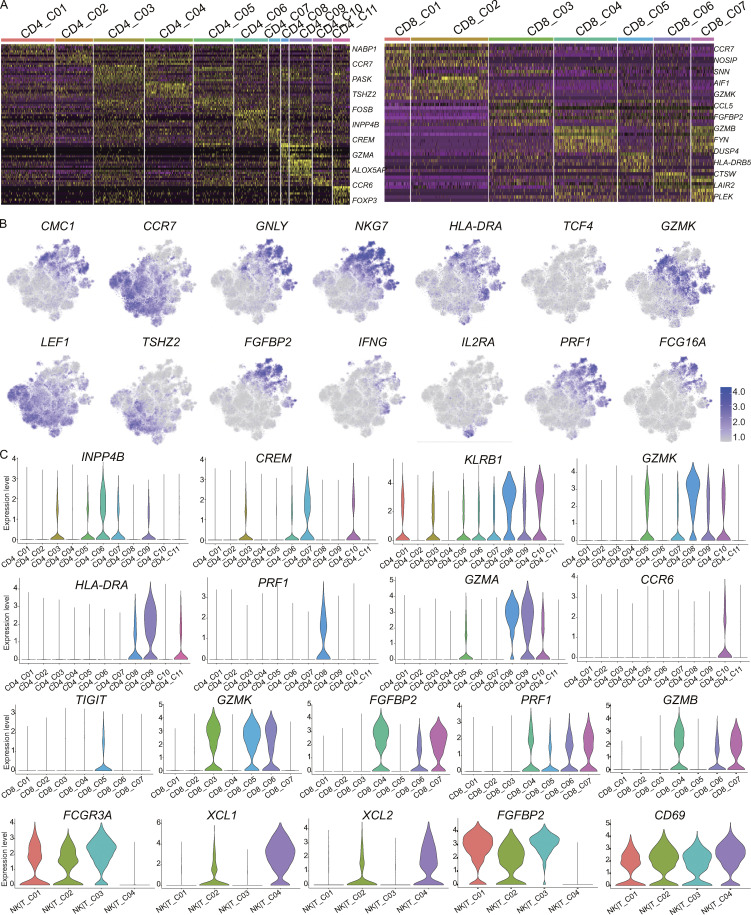
Figure S2.
Expression of marker genes for T cell subsets. (A) Differential expression analysis comparing CD4 subsets (left) and CD8 subsets (right) from the blood and PF. Heatmaps indicate the up- and down-regulated genes in CD4 T cell and CD8 T cell subsets, respectively. (B) tSNE plot of expression levels of selected genes in different cell subsets. (C) Violin plot of expression levels of selected genes in different cell subsets.
For detailed annotation of the CD4 T and CD8 T subsets, marker-gene signatures collected from published datasets (Table S5; Zemmour et al., 2018; Zhang et al., 2018; Leruste et al., 2019; Li et al., 2019; Cano-Gamez et al., 2020; Ren et al., 2021) were used for the cell identity or phenotyping determination. Based on these published cell marker references, we identified two naive CD4 T cell subsets, CD4_C02 and CD4_C04, that highly expressed CCR7 and LEF1 and were enriched in naive signatures (Fig. 1 F and Table S4). Two subsets, CD4_C03 and CD4_C05, exhibited a stress phenotype (Fig. 1 F; Li et al., 2019) and were predominant in PFMCs (Fig. 1 C). CD4_C06 and CD4_C07 were enriched in a central memory signature with high ANXA1 and INPP4B expression in CD4_C06 and high levels of CREM and KLF6 in CD4_C07 (Fig. S2, B and C; and Table S4). Two effector memory or effector subsets, CD4_C08 and CD4_C09, differed in marker gene expression. CD4_C08, a CD4 CTL subset characterized by high KLRB1, GZMK, GZMA, and PRF1 expression, was enriched in PF, while CD4_C09 displayed Th1 phenotype based on high IFNG expression with a Th1 signature (Fig. 1 F; Zhang et al., 2018); Th17 (CD4_C11, expressing KLRB1 and CCR6; Maggi et al., 2010); and CD4 regulatory T cell (CD4_C11, expressing FOXP3 and IL2RA/CD25; Fig. 1 F and Table S4; Zemmour et al., 2018). Gene signatures of CD4_C01, which specifically highly expressed NABP1, were not enriched in any of the referenced signatures (Fig. 1 F). We further explored the functions of these subsets by pathway analysis. Gene set enrichment analysis (GSEA) showed that CD4_C08 and CD4_C09 exhibited similar patterns, with up-regulation in Myc-targets, strong IFN-γ and IFN-α responses, and high rates of transcriptional activity, suggesting immune activity (Fig. 1 G). Interestingly, two clusters, CD4_C03 and CD4_C05, with stress phenotypes that differed in the IFN-γ and IFN-α response, IL2-Stat5 signaling, were suggestive of different responses to IFNs (Fig. 1 G).
We noted a similarly high level of diversity among the CD8 T cell subsets. Compared with the referenced gene signatures, we also identified naive CD8 T cells (CD8_C01 and CD8_C02), central memory CD8 T cells (CD8_C03), CD8 effector memory T cells (CD8_C05), and three terminal effector CD8 T cells (CD8_C04, CD8_C06, and CD8_C07; Fig. 1 H). Two blood-enriched terminal effector CD8 T cell subsets, CD8_C04 and CD8_C07, both expressed high levels of GZMH, ZEB2, GZMB, NKG7, and FGFBP2, but the latter subset exhibited a higher level of HLA-DRs and CX3CR1 (Table S4), while PF-enriched CD8 effector T cells (CD8_C06) exhibited high expression of GZMA, GZMK, GZMH, CCL5, and NKG7, with low expression of GZMB (Fig. 1, D and H; and Fig. S2 C). We noted that CD8_C05 and CD8_C06 shared a tissue-resistant phenotype (Fig. 1 H). We found that some pathways were differentially regulated among the CD8 T cell subsets. For example, CD8_C05 and CD8_C06 exhibited high mTORC1 signaling and strong IFN-γ and IFN-α responses, indicating cell activation (Fig. 1 I). Although CD8_C04 and CD8_C07 were both enriched in terminal effector signatures, the functions of these subsets differed based on their GSEA results. Similar to CD8_C06, CD8_C07 showed high mTORC1 signaling and strong IFN-γ and IFN-α responses, while CD8_C04 exhibited low Myc targets, reactive oxygen species pathways, and fatty acid metabolism, indicating low transcriptional and metabolic activity (Lambrechts et al., 2018).
NK|T_01, NK|T_03, and NK|T_04 cells highly expressed the circulating NK (cNK) cell markers FCGR3A, FGFBP2, and SPON2 (Cai et al., 2020) and the transcription factor T-bet (TBX21). NK|T_02 cells highly expressed the tissue-resident NK (rNK) cell marker CD69, as well as XCL1 and XCL2, but a low level of FCGR3A (Fig. S2, B and C; and Table S4). NK|T_C02 (cNK) and NK|T_C03 (rNK) were enriched in cells from the PF compared with cells from the blood, indicating the existence of both cNK cells and rNK cells in the PF.
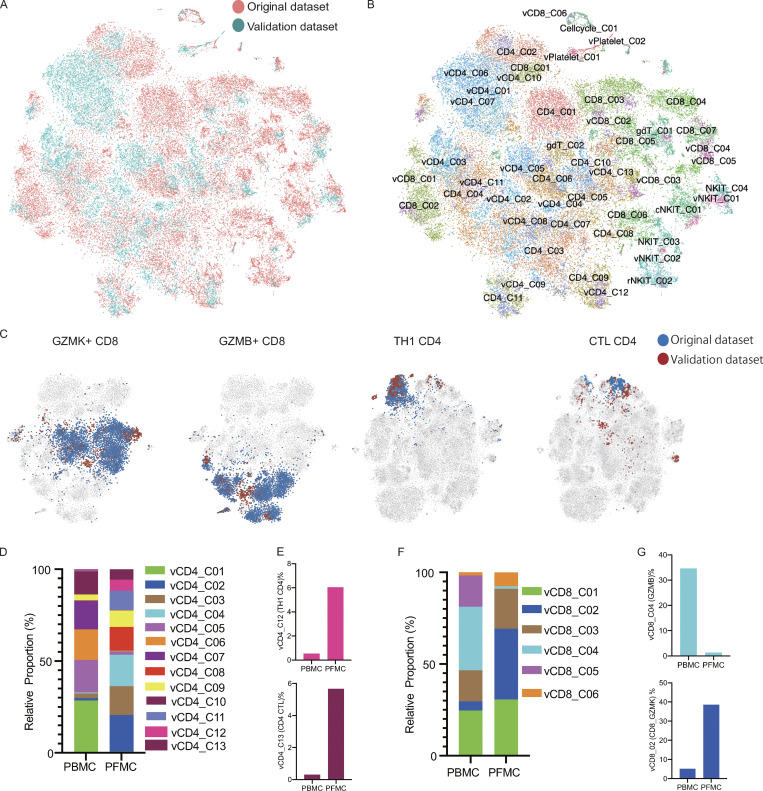
A validation cohort of a further 27,212 single cells from two additional patients with TPE (two CD3-enriched PBMCs and paired PFMCs) revealed that most of the CD4 and CD8 subsets detected in the original set of four patients could also be identified in these two additional patients (Fig. S3, A and B), indicating that the vast majority of cell subtypes originally identified could be readily recovered in TPE from the two additional patients (Table S6). Importantly, the enrichment of Th1–like CD4 and GZMK-expressing CD8 T cells in PF was also replicated in the additional two patients with TPE (Fig. S3, C–G).
Figure S3.
Analysis of the validation dataset. (A and B) tSNE of the combined original (n = 4) and validation (n = 2) data with each cell color-coded for the different datasets (A) or associated cell subsets (B). For integrating two datasets, the datasets were normalized to find variable features using the R package Seurat (v3.2.0) with default parameters, and 30 PCs were selected for the tSNE analysis, with default parameters. CD4/CD8 from the original dataset; vCD4/vCD8 from the validation dataset. (C) tSNE of the GZMK-expressing CD8, GZMB-expressing CD8, Th1 CD4, and CTL CD4 T cells from the original and validation datasets. (D) The relative proportion of cells of the CD4-cell subsets in the T cell fraction of the validation dataset (n = 2). (E) The fraction of cells for Th1 CD4 and CTL CD4 in CD4 T cells from blood and PF in the validation dataset (n = 2). (F) The relative proportion of cells of the CD8 T cells subsets in the T cell fraction of the validation dataset (n = 2). (G) The fraction of cells of the GZMB-expressing CD8 and GZMK-expressing CD8 T cells in CD8 T cell from blood and PF in the validation dataset (n = 2).
Together, these unbiased analyses delineated the extent of T cell heterogeneity at the site of Mtb infection in TPE and identified that Th1-like CD4 cells and GZMK-expressing CD8 cells predominated in the PF.
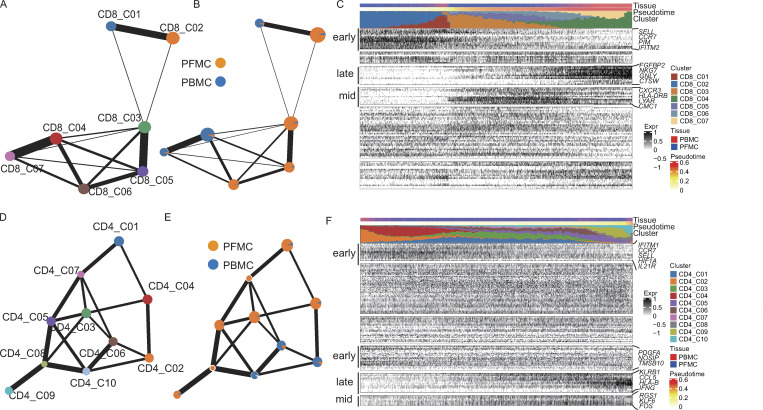
Pseudo-time analysis reveals distinct CD4 and CD8 T cell differentiation trajectories in PBMCs and PFMCs
The complete transcriptome data for many T cells allowed us to gain insights into the functional states and relationships among these cells. To analyze global connectivity and the potential trajectory topology in the T cell state transitions, we applied partition-based graph abstraction (PAGA; Wolf et al., 2019), which provides an interpretable, graph-like map of the data manifold, to order CD4 or CD8 T cells in pseudo-time and determine their differential trajectories. The PAGA map revealed several nodes with high connectivity between CD8 T cell subsets that represented potential trans-differentiation bridges. Specifically, CD8_C03 seemed to be an intermediate state, connecting to all other clusters with naive (CD8_C01 and CD8_C02) to activated CD8 T cell subsets. In particular, we observed high connectivity between CD8_C03 and CD8_C05 and between CD8_C04 and CD8_C07 (Fig. 2, A and B).
Figure 2.
Pseudo-time analysis reveals distinct CD4 and CD8 T cell differentiation trajectories in PBMCs and PFMCs. (A and B) PAGA analysis of CD8 T cell pseudo-time: the associated cell type (A) and the corresponding status (PBMC, n = 3; and PFMC, n = 4; B) are shown. (C) Reconstructed PAGA paths for the differentiation of the identified CD8 T cells from TPE. (D and E) PAGA analysis of CD8 T cell pseudo-time: the associated cell type (D) and the corresponding status (PBMC and PFMC; E) are shown. (F) Reconstructed PAGA paths for the differentiation of the identified CD4 T cells from TPE.
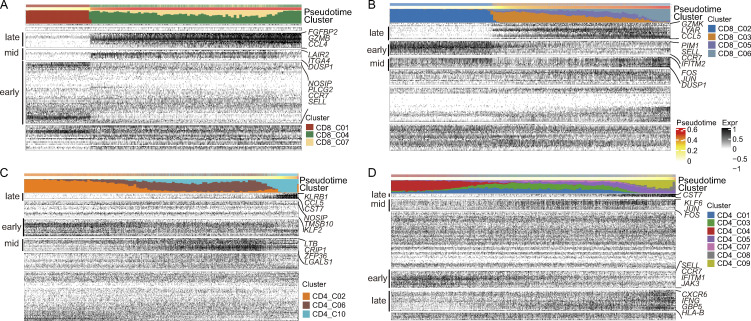
To understand the biological processes driving pseudo-time components, we investigated which genes covaried in expression with pseudo-time. We clustered the top 20 genes identified as significantly covarying with PAGA pseudo-time and identified groups of genes that were expressed early, mid/mid to late, and late (Fig. 2 C). In keeping with the cluster distribution, naive markers, such as CCR7 and SELL, were expressed early, while effector markers (FGFBP2, NKG7, GNLY, and CTSW) were expressed late (Fig. 2 C). Interestingly, we observed distinct differentiation pathways of blood-enriched CD8 subsets (CD8_C04 and CD8_C07) and PF-enriched CD8 subsets (CD8_C03, CD8_C05, and CD8_C06). We then analyzed blood-enriched and PF-enriched CD8 subsets separately. We found distinct differentiation patterns between blood and PF. For example, the genes expressed late in blood CD8 T cells were FGFBP2, GZMB, and CCL4, while those expressed late in PF CD8 T cells were GZMK, LYAR, and CCL5 (Fig. 2 B; and Fig. S4, A and B).
Figure S4.
Pseudo-time analysis of CD4 and CD8 differentiation trajectories in PBMCs and PFMCs. (A) Reconstructed PAGA paths for differentiation of the identified CD8 T cells from blood (n = 3). (B) Reconstructed PAGA paths for differentiation of the identified CD8 T cells from PF (n = 4). (C) Reconstructed PAGA paths for differentiation of the identified CD4 T cells from blood (n = 3). (D) Reconstructed PAGA paths for differentiation of the identified CD8 T cells from PF (n = 4).
We next traced the lineage relationships of CD4 T cells using the same approach. The PAGA analysis revealed a gradient of T cell differentiation from the naive clusters (CD4_C02 and CD4_C04) to activated clusters and a potential conversion among CD4 subsets (Fig. 2, D and E). The CD4_C03 cells (mainly from PFMCs) and CD4_C06 cells (mainly from PBMCs) were in different directions in the pseudo-time trajectory node and were connected with other CD4 subsets. CD4_C08 and CD4_C09 were positioned at the opposite end of naive CD4 clusters (Fig. 2 D). These identified differentiation trajectories were associated with gradually increased CCL5, KLRB1, and CST7 expression or decreased CCR7 and SELL expression, both of which are associated with CD4 T cell activation (Fig. 2 F). An intriguing observation was the identification of CD4_C08 representing a potential intermediate T cell state on a Th1-like cell differential pathway and gradually progressing toward the CD4_C09 cluster (Fig. 2 D). We confirmed a gradual differentiation of the Th1-like lineage, leading to the temporal acquisition of cytotoxic cells expressing KLRB1, GZMK, GZMA, and PFR1 in CD4_C08 and the terminal differentiation of a subset with a Th1-like profile (CD4_C09). In addition, we observed a specific signal for T cell activation markers (CXCR6, IFNG, GBP5, and HLA-B) in the PF CD4 subset, as suggested by the pseudo-time cluster distribution (Fig. S4, C and D).
Based on the trajectory analyses, we revealed a specific differential feature of CD4 and CD8 T cells in PF, which suggests a differential impact of the PF microenvironment on the differentiation of T cells, conceivably reflecting differential availability of antigen or differential exposure to Mtb between blood and PF. In addition, we inferred that CD8_C03 and CD4_C08 cells might serve as intermediate populations in the PF, which could be useful for therapeutic strategies targeting these intermediate populations.
TCR analysis provides evidence of clonal expansion in TPE
Interactions between TCRs and antigens presented by the MHC are critical for adaptive immunity. When T cells recognize cognate antigen, they undergo clonal expansion (Mayer et al., 2019). Therefore, the relative abundance of TCR sequences may indicate T cell clonal expansion patterns and T cell differentiation history (Han et al., 2015). The scTCR-seq of PBMCs and PFMCs from the same TPE samples enabled us to understand the extent of T cell clonal expansion and lineage plasticity.
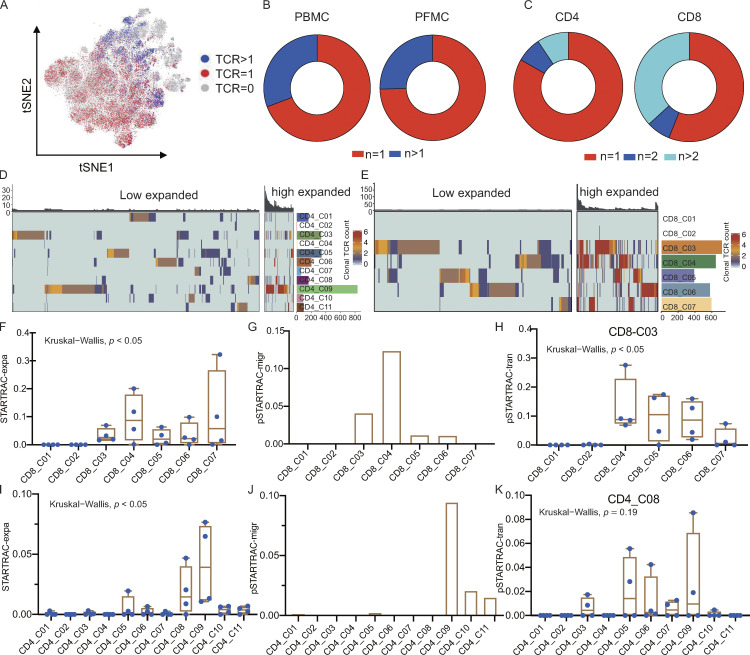
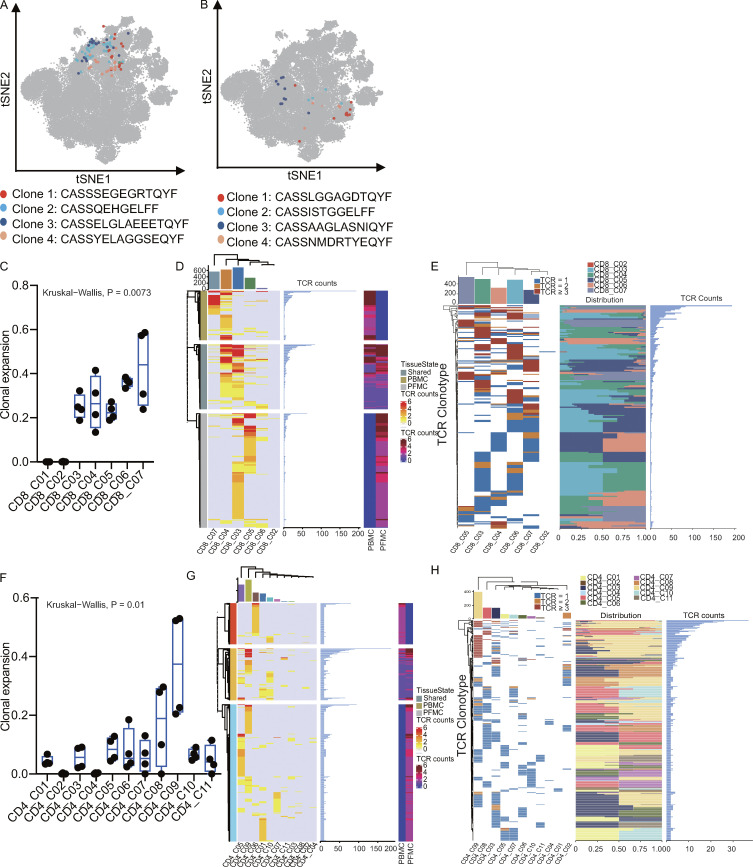
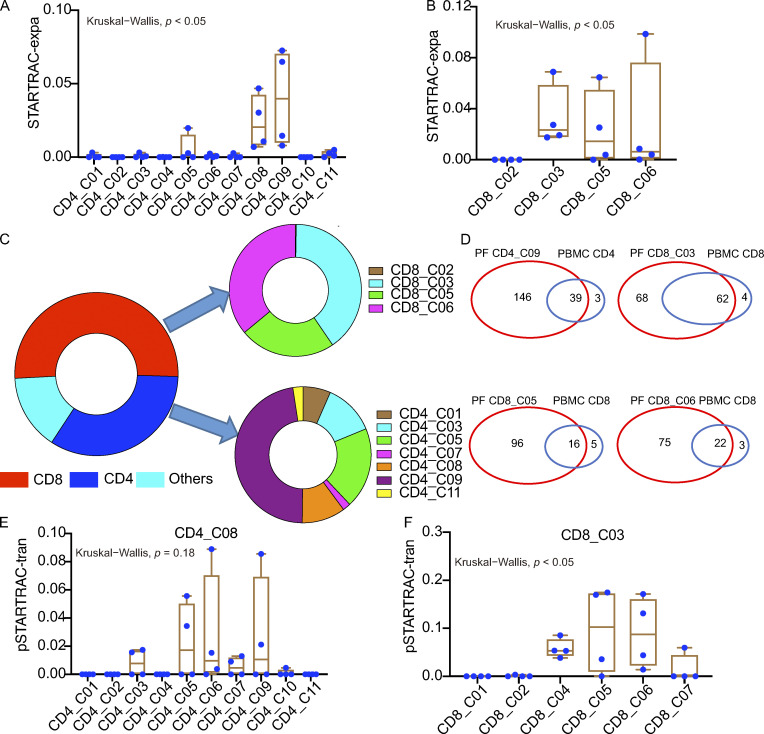
We recovered 5,900 and 18,007 coupled, full-length TCR sequences from PBMCs and PFMCs, respectively (Fig. 3 A and Table S7). While most TCRs were unique, 30.7 and 25.4% of the TCRs in PBMCs and PFMCs, respectively, were present in at least two cells (Fig. 3 B and Table S7). The clonally expanded cells were mostly CD8 T cells (65%), although we also detected CD4 T cell clones in nearly all patients (Fig. 3 C and Table S7). Interestingly, we found several identical TCRs across different CD8 or CD4 T cell clusters, suggesting that these cells might have had common ancestors but developed into different cell types (Fig. S5, A and B; and Table S8). We observed highly expanded clones (n ≥ 5) distributed in CD4 and CD8 subsets (Fig. 3, D and E). These highly expanded CD8 clones were in both PBMCs (30.2%) and PFMCs (69.8%). Conversely, most of the highly expanded CD4 clones were observed in the PF rather than in the blood. Together, these data suggest different clonal expansion patterns between the blood and PF in TPE.
Figure 3.
TCR distribution and clonality analysis. (A) The TCR distribution of T cells. Gray (n = 0), red (n = 1), and blue (n > 1). (B and C) TCR distribution in different tissue types (B) and cell types (C). (D and E) Phenotypic analysis of low expanded (TCR <5) and high expanded (TCR ≥5) clonotypes in CD4 and CD8 T cell subsets. Individual T cells are grouped by TCR sequence; each bar above the heatmaps represents a distinct TCR sequence. (F) Clonal expansion levels of CD8 T cell clusters quantified by STARTRAC-expa for each patient (n = 4); Kruskal–Wallis H tests were performed to analyze differences in the STARTRAC results. (G) Migration potentials of CD8 subsets by pSTARTRAC-migr indices between blood and PF. The shared clonotypes in each CD8 subset (all patients combined) between blood and PF was analyzed by pSTARTRAC-migr. (H) Developmental transition of CD8_C03 cells with other CD8 cells, quantified by pSTARTRAC-tran indices for each patient (n = 4); Kruskal–Wallis H tests were performed to analyze differences. (I) Clonal expansion levels of CD4 T cell clusters quantified by STARTRAC-expa for each patient (n = 4); Kruskal–Wallis H tests were performed to analyze differences. (J) Migration potentials of CD4 subsets by pSTARTRAC-migr indices between blood and PF. The shared clonotypes in each CD4 subset (all patients combined) between blood and PF was analyzed by pSTARTRAC-migr. (K) Developmental transition of CD4_C08 cells with other CD4 cells quantified by pairwise STARTRAC-tran indices for each patient (n = 4). Kruskal–Wallis H tests were performed to analyze differences.
Figure S5.
Analysis of TCR expansion and sharing status between subsets or tissues. (A) The spatial distribution of cells, showing only those CD8 T cells sharing the same TCRs in colors. (B) The spatial distribution of cells, showing only those CD4 T cells sharing the same TCRs in colors. Four different clonotypes are shown, with different colors. (C) The percentage of clonal expansion in CD8 T cell clusters. Clonal expansion rate = total of cells in expansion clonotypes (TCR > 1) cells ÷ total of cells with TCR. (D) Analysis of shared clonotypes in CD8 T cells between PBMCs and PFMCs. Left: PBMC specific TCRs (top), shared PBMC and PFMC TCRs (middle), and PFMC-specific TCRs (bottom) in CD8 T cell subsets. Middle: The number of cells with identical TCRs for each clonotype from the left panel. Right: The distribution of different clonotypes in PBMCs and PFMCs (PBMC, n = 3, PFMC, n = 4). (E) Analysis of shared clonotypes in CD8 subsets. Left: TCR sequences are grouped by CD8 subsets; each bar in the heatmaps represents a distinct TCR sequence. Middle: The distribution of cells with identical TCRs in CD8 T cell subsets. Right: The number of cells with identical TCRs for each clonotype from left panel. (F) The percentage of clonal expansion in CD4 T cell clusters. (G) Analysis of shared clonotypes in CD4 T cell between PBMCs and PFMCs. Left: PBMC-specific TCRs (top), shared PBMC and PFMC TCRs (middle), and PFMC-specific TCRs (bottom) in CD4 T cell subsets. Middle: The number of cells with identical TCRs for each clonotype from the left panel. Right: The distribution of different clonotypes in PBMCs and PFMCs. (H) Analysis of shared clonotypes in CD4 subsets. Left: TCR sequences are grouped by CD4 subsets; each bar in the heatmaps represents a distinct TCR sequence. Middle: The distribution of cells with identical TCRs in CD4 T cell subsets. Right: The number of cells with identical TCRs for each clonotype from the left panel.
Single T cell analysis by RNA sequencing and TCR tracking (STARTRAC) has recently been used in other studies to analyze scTCR-seq data (Bhatt et al., 2021; Zhang et al., 2018), so we next performed STARTRAC analysis of the TCRs to study the relationships among different T cell subsets. STARTRAC incorporates several unique indices to quantitatively describe the degree of clonal expansion (expa), migration (migr), and developmental transition (tran) of T cell clusters upon TCR tracking (Zhang et al., 2018). Focusing on CD8 T cells, the STARTRAC-expa index revealed that naive T cell clusters, CD8_C01 and CD8_C02, displayed negligible clonal expansion, as expected. In contrast, other clusters had undergone clonal expansion. Among them, CD8_C07 (effector cells) exhibited the highest degree of clonal expansion (Fig. 3 F). We also compared the clonal expansion rate by directly calculating the percentage of T cells with clonal TCRs in each cluster. Similar results were observed, with CD8_C01 and CD8_C02 showing low clonal expansion and CD8_C07 showing high clonal expansion (Fig. S5 C). Pairwise (p)STARTRAC-migr analysis revealed a high degree of TCR sharing between CD8_C04 cells found in the blood and PF, whereas the cluster with the highest level of clonal expansion (CD8_C07) exhibited tissue exclusivity (Figs. 3 G and S5 D). Accordingly, CD8_C04 cells expressed different sets of genes associated with migration, including those encoding chemokines and other trafficking-related molecules (CCL5, S1PR5, F2R, and CCL4; Table S4), which supports their capability of circulating in the periphery and homing to the site of infection in TPE. Notably, pSTARTRAC-tran analysis of CD8 subsets indicated that CD8_C03 was highly associated with the other four CD8 clusters (CD8_C04, CD8_C05, CD8_C06, and CD8_C07; Fig. 3 H), which was also observed by directly calculating the shared clonotypes among CD8 subsets (Fig. S5 E). These results suggest that CD8_C03 represents a transitional state shared by multiple effector-cell populations. This result further supported the pseudo-time analysis that CD8_C03 is connected with these CD8 subsets.
In contrast to CD8 cells, CD4 cells exhibited lower levels of clonal expansion overall (Fig. 3, C and D). Among all the CD4 clusters, CD4_C09 exhibited the highest level of clonal expansion, followed by CD4_C08 (Fig. 3, D and I; and Fig. S5 F). The cells comprising these two clusters were mainly from the PF (Fig. 1 C). pSTARTRAC-migr analysis of CD4 clusters revealed that CD4_C09 cells were associated with the highest level of mobility between blood and PF, followed by CD4_C10 cells (Fig. 3 J). Consistently, CD4_C09 highly expressed CXCR6 and CXCR3, while CD4_C10 highly expressed CCR6 (Table S4), suggesting that the clusters may be enriched with distinct Th cell populations, such as Th1 and Th17. Finally, pSTARTRAC-tran analysis suggested that TCR pairs were shared between CD4_C08 and CD4_C09 and between CD4_C08 and CD4_C05 (Fig. 3 K), which supported the trajectories analysis that CD4_C08 cells were developmentally linked to CD4_C05 and CD4_C09 cells (Fig. 2 D).
We also calculated shared clonotypes in each cluster and in different clusters between blood and PF to evaluate the extent of tissue migration and state transition, respectively. The results showed that CD4_C09 and CD8_C04 shared more clonotypes between blood and PF (Fig. S5, D and G) and that CD4_C08 and CD8_C03 shared more clonotypes with other CD4 and CD8 subsets, respectively (Fig. S5, E and H). These results were consistent with our STARTRAC results. Altogether, these data enabled us to link TCR information to various T cell activation and differentiation states and to reveal a previously unknown clonal expansion landscape in CD4 and CD8 T cell subsets in TPE.
Distinct highly expanded T cell clonotypes in PF-enriched T cell subsets
The above results suggested that several subsets, such as CD4_C08, CD4_C09, CD8_C03, CD8_C05, and CD8_C06, dominated the PF with clonal expansion (Fig. 1 C and Fig. 3, F and I). We therefore further analyzed the TCR characteristics of these subsets from the PF, which potentially represented locally expanded clonotypes and likely Mtb-specific T cells. We thus performed STARTRAC analysis on these cells and observed that CD4_C08 and CD4_C09 were enriched with highly clonally expanded clones among PF CD4 clusters (Fig. 4 A). Similarly, among the CD8 cells in the PF, CD8_C03, CD8_C05, and CD8_C06 exhibited signs of clonal expansion among the PF-infiltrating CD8 clusters (Fig. 4 B). Notably, clones with high levels of expansion (n ≥ 5) in the PF were CD8_C03, CD8_C05, and CD8_C06 clusters and Th1-like CD4 T cell clusters (CD4_C09; Fig. 4 C). All other clusters in the PF showed low levels of clonal outgrowth (Fig. 4 C). Notably, only PF CD8_C03 shared some TCR clonotypes from blood; most of the expanded TCR clonotypes in the Th1-like CD4 T cells, PF CD8_CD5, and PF CD8_C06 differed from those seen in the blood (Fig. 4 D and Fig. S5, B and E). This unexpected finding suggested that T cells at the site of infection do not simply originate from passive trafficking through the peripheral lymph organs. Our data thus suggest that the clonal accumulation of both CD4_C09 and PF-infiltrating CD8 T cells is likely a result of local T cell proliferation and activation in the PF. However, we could not formally rule out the possibility that these clonal T cells were amplified in the lung and then percolated into the pleura.
Figure 4.
STARTRAC analysis of PF-enriched T cell clonal expansion and developmental transition. (A) Clonal expansion levels of CD4 T cells from PF quantified by STARTRAC-expa for each patient (n = 4); Kruskal–Wallis H tests were performed to analyze differences in the STARTRAC results. (B) Clonal expansion levels of CD8 T cells from PF quantified by STARTRAC-expa for each patient (n = 4). (C) Highly expanded clonotype (TCR ≥5) distribution in cell types in PF. (D) Shared expanded clonotypes in PF-enrich CD4 and CD8 subsets between PF and blood. (E) Developmental transition of PF CD4_C08 cells with other PF-infiltrating CD4 cells quantified by pSTARTRAC-tran indices for each patient (n = 4). (F) Developmental transition of PF CD8_C03 cells with other PF-infiltrating CD8 cells quantified by pSTARTRAC-tran indices for each patient (n = 4).
We next used STARTRAC to analyze the transition potential between the PF-infiltrating clusters. pSTARTRAC-tran indicated that PF CD4_C08 shared TCRs with other CD4 subsets, while PF CD8_C03 shared TCRs with CD8_C05 and CD8_C06, indicating their transitional status in the PF (Fig. 4, E and F). Overall, our TCR repertoire analysis of PF T cells identified distinct, highly expanded T cell clonotypes in PF-enriched T cell subsets. In addition, most of the highly expanded clonotypes were exclusive to the PF, indicating their potential for recognizing Mtb-associated antigens and undergoing local expansion.
Gene expression profiling of PF-enriched T cells reveals a distinct transcriptional profile
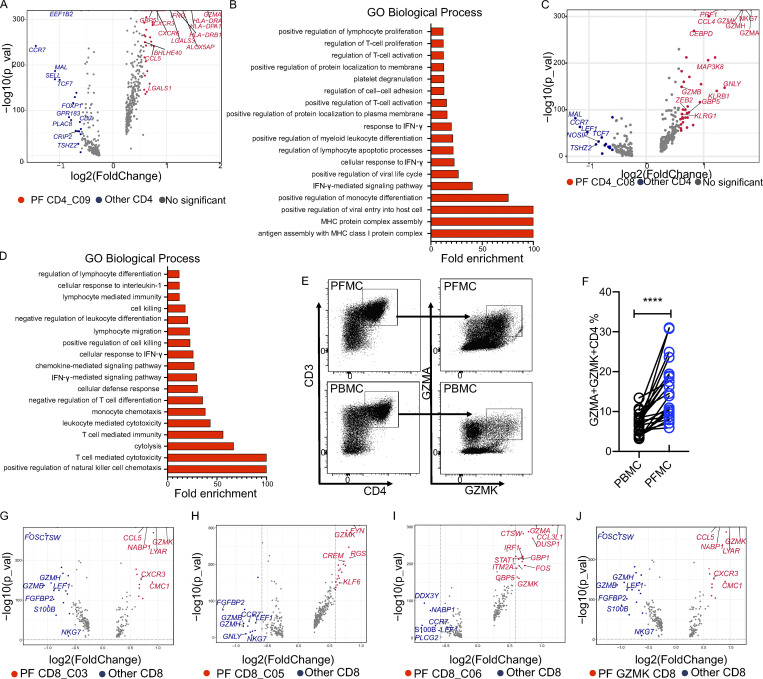
Having identified high levels of T cell heterogeneity and clonal expansion at the site of Mtb infection in TPE by scRNA-seq and scTCR-seq, we next wanted to understand the gene expression signature of these subsets from PF. We therefore analyzed the differential gene expression profile of these subsets. Among the CD4 clusters enriched in the PF, we found that the CD4_C09 subset expressed high levels of IFNG (Table S4). scRNA-seq allowed us to analyze the gene expression signature of this cluster isolated from the PF. We found 49 differentially expressed genes (DEGs; adjusted P value <0.01 and fold-change >1.5) specifically expressed in PF CD4_C09 compared with other CD4 cells (Fig. 5 A and Table S9). Besides known IFN-γ–associated genes (HLA-DRA, IFNG, GBP5, and CCL5), we also identified novel genes up-regulated in this cluster, including ALOX5AP, LGASL3, and GZMA, indicating that these cells might also have cytotoxic functions (Table S9; Maehara et al., 2020). Gene ontology (GO) enrichment analysis showed that the up-regulated genes in PF CD4_C09 were associated with antigen processing, positive regulation of lymphocyte proliferation, and positive regulation of myeloid leukocyte differentiation (Fig. 5 B). In addition, we found higher IFNG and CXCR6 expression levels in cells from PF CD4_C09 than in cells from blood CD4_C09, indicative of a high terminal-effector Th1 cell response in the PF (Table S10). Besides CD4_C09, we identified another cluster as a specific PF-enriched CD4 CTL cluster (Fig. 1 C): CD4_C08, which highly expressed 39 genes associated with CTL functions, such as GZMA, NKG7, GZMK, PRF1, KLRB1, and GNLY (Fig. 5 C and Table S9). GO results confirmed that these cells were highly activated compared with other CD4 cells and were involved in mediating cytolysis (Fig. 5 D). This finding was further confirmed by flow cytometry analysis of PFMCs from patients with TPE. In line with the RNA-seq data, we detected a subset of GZMA- and GZMK-expressing CD4 T cells in PF, supporting the presence of this CTL CD4 subset in PF (n = 22, cohort II in Table S1; and Fig. 5, E and F).
Figure 5.
Gene expression characterization associated with PF-infiltrating T cells. (A) Volcano plot showing DEGs in PF CD4_C09 versus other CD4 T cells (n = 4). Each red dot denotes an individual gene passing P value (val) and fold-difference (P < 0.01; average fold-change >1.5) thresholds. (B) GO analysis of genes positively expressed in PF CD4_C09. (C) Volcano plot showing DEGs in PF CD4_C08 versus other CD4 T cells (n = 4). Each red dot denotes an individual gene passing P value and fold-difference thresholds. (D) GO analysis of genes positively expressed in PF CD4_C08. (E) Gating strategy for GZMK+GZMA+ CD4 T cells by flow cytometry. (F) The frequency of GZMK+GZMA+ CD4 T cells in paired PBMCs and PFMCs from TPE patients (n = 22). A paired t test was used to analyze differences in paired samples; ****, P < 0.0001. (G–J) Volcano plot showing DEGs in CD8_C03 (G), CD8_C05 (H), CD8_C06 (I), and GZMK-expressing CD8 (J). Each red or blue dot denotes an individual gene passing P value and fold-difference thresholds (P < 0.01; average fold-change >1.5).
To gain insights into the function of PF-enriched CD8 T cell subsets, we identified genes specifically expressed in PF-enriched CD8 clusters. We obtained 14, 17, and 20 DEGs in PF-enriched CD8_C03, CD8_C05, and CD8_C06, respectively (Fig. 5, G–I; and Table S9). Interestingly, we found that all three clusters expressed low levels of FGFBP2, GZMH, and GZMB, but high levels of LYAR and GZMK, which have not been well characterized in the context of TB (Fig. 5, G–J). In addition, we observed a gradual increase in PRF1 expression in CD8_C03, CD8_C05, and CD8_C06, indicating different levels of cytotoxic activity in these clusters (Fig. S2 C).
In summary, these data not only confirmed that the IFN-γ response is increased in the PF from TPE compared with the response in blood but also uncovered PF-specific gene signatures of CD4 CTLs and GZMK-expressing CD8 T cells, which could be further explored as additional markers to define PF-enriched CD4 and CD8 T cell clusters.
GZMK is produced in PF of TPE patients and inducible in CD8 T cells upon activation in vitro
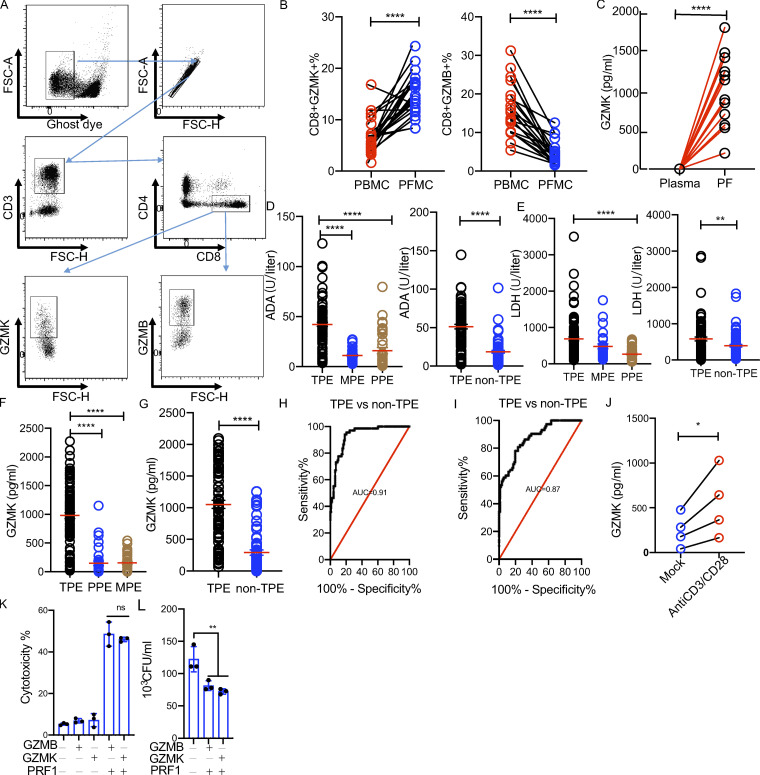
Our data thus far showed that GZMK is specifically and highly expressed by PF-enriched CD8 T cells, while GZMB is almost absent in these cells (Fig. S2 C). We validated the scRNA-seq results using flow cytometry. Consistent with the scRNA-seq data, we found that the frequency of GZMK-expressing CD8 T cells was significantly increased in PF compared with that in blood, whereas the frequency of GZMB CD8 T cells was decreased in PF (Fig. 6, A and B). It has previously been reported that GZMK can be secreted by CD8 T cells (Mogilenko et al., 2021), we asked whether GZMK was specifically increased in PF. By determining the levels of GZMK in paired plasma and PF samples from TPE patients (n = 12), we found that the GZMK level was significantly increased in PF (Fig. 6 C), suggesting that GZMK is specifically expressed at the site of Mtb infection (cohort II in Table S1). We next explored whether GZMK was specifically increased in PF from TPE. To do this, the GZMK levels in PF from TPE were compared with those from non-TPE in two independent cohorts (cohort III, TPE, n = 98; PPE, n = 36; MPE, n = 31; and cohort IV, TPE, n = 88; non-TPE, n = 73; Table S1). In line with previous studies that showed that levels of adenosine deaminase (ADA) and lactate dehydrogenase (LDH) are increased in TPE (Daniil et al., 2007; Valdés et al., 2015; Wang et al., 2018), we found that the level of ADA and LDH were significantly increased in TPE in both cohorts (Fig. 6, D and E). Of note, we found that GZMK levels in PF from TPE were significantly higher than those from non-TPE in both of the two independent cohorts (Fig. 6, F and G). The area under the curve for GZMK was 0.91 and 0.87 in cohorts III and IV, respectively (Fig. 6, H and I), indicating that GZMK is useful for distinguishing TPE from non-TPE.
Figure 6.
GZMK is produced in PF of TPE patients and is inducible in CD8 T cells upon activation in vitro. (A) Gating strategy of GZMK+ or GZMB+ CD8 T cells by flow cytometry. FSC, forward scatter. (B) The frequency of GZMK+ CD8 or GZMB+ CD8 T cells in paired PBMCs and PFMCs from TPE patients (n = 22). A paired t test was used to analyze differences in paired samples; ****, P < 0.0001. (C) The level of GZMK protein expression in paired plasma and PF samples (n = 12); A paired t test was used to analyze differences in paired samples; ****, P < 0.0001. (D) The level of ADA in the PF from TPE, PPE, MPE, and non-TPE patients. Left: Cohort III (TPE, n = 98; PPE n = 36; and MPE n = 31). Right: Cohort IV (TPE, n = 88; and non-TPE, n = 73); One-way ANOVA Newman–Keuls multiple comparison test was used to compare differences among multiple groups; ****, P < 0.0001; unpaired t test was used to analyze differences between two groups; ****, P < 0.0001. (E) The level of ADA in the PF from TPE, PPE, MPE, and non-TPE patients. Left: Cohort III. Right: Cohort IV. One-way ANOVA Newman–Keuls multiple comparison test was used to compare differences among multiple groups; ****, P < 0.0001; unpaired t test was used to analyze differences between two groups; **, P < 0.01. (F) The level of GZMK protein in the PF from TPE, PPE, and MPE patients in cohort III. (G) The level of GZMK protein in the PF from TPE and non-MPE patients in cohort IV. One-way ANOVA Newman–Keuls multiple comparison test was used to compare differences among multiple groups; ****, P < 0.0001; unpaired t test was used to analyze differences between two groups; ****, P < 0.0001. (H) Receiver operater characteristic curve for GZMK to separate TPE from non-TPE in cohort III. (I) Receiver operater characteristic curve for GZMK to separate TPE from non-TPE in cohort IV. (J) The level of GZMK protein in CD8 from PFMCs (n = 4) with or without anti-CD3/CD28 stimulation; unpaired t test was used to analyze differences between two groups; *, P < 0.05. (K) The cytotoxicity of GZMK and GZMB with or without perforin in THP-1–derived macrophages. Purified GZMK (10 µg/ml) and GZMB (10 µg/ml) with or without Perforin 1 were added to THP-1–derived macrophages. Cytotoxicity was determined by WST-1 assays; one-way ANOVA Newman–Keuls multiple comparison test was used to compare differences among multiple groups; ns, P > 0.05. (L) The bactericidal activity of GZMK and GZMB in Mtb-infected THP-1–derived macrophages. Purified GZMK (10 µg/ml) and GZMB (10 µg/ml) with or without perforin 1 were added to Mtb-infected macrophages. CFUs were determined in 24 h. The experiments were replicated three times. One-way ANOVA Newman–Keuls multiple comparison test was used to compare differences among multiple groups; **, P < 0.01. The data represent means ± SEM.
The impressive enrichment of GZMK-expressing CD8 T cells in PF prompted us to further characterize the potential function of GZMK-expressing CD8 T cells in TPE. However, due to limitations of the technique, we could not isolate GZMK-expressing CD8 T cells for the purpose of defining their functions. We therefore first investigated whether GZMK-expressing CD8 T cells contribute to the increasing level of GZMK in PF. We isolated CD8 T cells from PFMCs and stimulated them with anti-CD3/CD28. As expected, we found that GZMK was induced in CD8 T cells derived from PFMCs following anti-CD3/CD28 stimulation (Fig. 6 J), indicating that the increased level of GZMK in TPE is, at least partially, secreted by activated GZMK-expressing CD8 T cells. In addition, we determined the cytotoxic and bactericidal activity of purified GZMK and GZMB using an in vitro macrophage infection model. We found that GZMK and GZMB have cytotoxic and bactericidal activity in Mtb-infected macrophages (Fig. 6, K and L).
In summary, our experimental data corroborated our transcriptomics findings suggest that GZMK protein could be induced in CD8 T cells by Mtb. These findings provide new insights into the potential functions of these cells and suggest that the accumulation of GZMK-expressing CD8 T cells is an intrinsic and possibly key feature at sites of Mtb infection.
Discussion
Our understanding of the mechanisms orchestrating local immune responses to pathogens in humans is limited due to a lack of information about the immune landscape in tissues. Taking advantage of the nature of the localized immune response in TPE, we mapped the T cell landscape, identified novel T cell populations, and defined the dynamics of clonal expansion within T cell populations at the site of Mtb infection. By analyzing >52,900 cells from the PF and blood, we identified 25 subsets, including different types of PF-associated CD4 and CD8 cells. Our data not only recapitulated many of the T cell subsets in PF identified in previous studies (Cai et al., 2020; Chen et al., 2010), but also revealed highly heterogeneous T cell subsets with diverse functions at the site of infection. We further identified the gene expression signature in cell subsets contributing to the IFN-γ response and genes associated with CD8 subset-enriched PF. Combined with TCR data, we revealed the connectivity and potential developmental path of T cell subsets in TPE. Going forward, our transcriptomics data, together with the paired TCR information for individual T cells, ultimately constitutes a comprehensive resource for multidimensional T cell characterization, especially local T cell immunity in the context of Mtb infection.
Previous studies have shown that the lymphocyte population in the PF from TPE favors a Th1 response, with high levels of IFN-γ and other related cytokines (Hooper et al., 2009; Mitra et al., 2005; Seiscento et al., 2010; Sharma et al., 2002). The PF has a markedly higher proportion of Th1 cells than the peripheral blood, a phenomenon termed “compartmentalization,” which is thought to be responsible for the paucibacillary nature of these effusions and low yield on Mtb culture (Mitra et al., 2005; Rossi et al., 1987; Sharma et al., 2002). Consistent with these studies, we found a Th1-like CD4 cluster (CD4_C09) that showed high IFNG, TBX21, and CXCR3 expression (Di Cesare et al., 2009) and was predominant in the PF compared with its presence in blood. Although the single-cell sequencing approach may potentially underestimate the extent of anti-TB T cell responses, especially in peripheral blood, our coupled scRNA-seq and scTCR-seq data allowed us to perform in-depth analyses of this cluster, including its gene expression signature and clonality. Interestingly, we found that this cluster also highly expressed HLA-DRs and GZMA, indicating that these cells are likely multifunctional, as CD4 cells expressing HLA-DRs contribute to disease-associated inflammation by compromising regulatory T cell–mediated suppression in human TB (Viallard et al., 2001). We also found that PF CD4_C09 cells were clonally expanded, with most clonotypes in the PF differing from those found in the blood, suggesting that these cells do not simply originate from passive trafficking through the peripheral lymph organs, namely compartmentalization. Those PF-specific clones might recognize Mtb antigens at the site of infection. The amplification of these clones most likely augments their immune response functions, further magnifying the Th1 cell response in PF.
We also identified a CTL CD4 cluster (CD4_C08) enriched in the PF that shared similar gene expression characteristics with cytotoxic CD8 T cells (GZMK, KLRB1, GZMA, and PRF1; Li et al., 2019; Maehara et al., 2020). This CD4 cluster has not previously been reported in TPE. It has been hypothesized previously that CTL CD4 T cells, possibly generated through repeated antigen stimulation (Appay et al., 2002; Brown et al., 2006; Casazza et al., 2006), play a protective role against infectious diseases and cancer (Brown et al., 2009; Haabeth et al., 2014; Zheng et al., 2017). CTL CD4 T cells have been proposed to use various mechanisms to kill the target cells, with most underlying pathways not being mutually exclusive (Brown et al., 2009). The exact molecular mechanism shared by CD4_C08 and CD4_C09 cells remains unclear. Our data showed that CD4_C08 and CD4_C09 are closely linked to each other in that they both express GZMA and CXCR6, the latter being a chemokine receptor that is expressed by T cells upon activation (Kim et al., 2001), suggesting similarities in the function and trafficking patterns between the two populations. Interestingly, another study revealed that CTL CD4 T cells can produce Th1 cytokines, IFN-γ, and TNF-α upon ex vivo PMA/ionomycin stimulation (Hashimoto et al., 2019). It would be interesting to examine whether therapeutic strategies targeting these intermediate populations could promote the killing and/or activation of Mtb-infected macrophages. Regardless, our results clearly highlight an ongoing adaptive immune response in the PF from TPE and invite further investigations into the role played by CTL CD4 T cells in TB.
Increasing evidence supports the notion that CD8 T cells play pivotal roles in controlling Mtb infection through TCR-mediated cytotoxicity via granzymes (Kamath et al., 2004; Lewinsohn et al., 2003; Woodworth et al., 2008). A previous study demonstrated that GZMB can kill Mtb directly in the presence of granulysin, via multiple mechanisms (Dotiwala et al., 2017). In our study, we observed that heterogeneous CD8 T cell subsets in the PF displayed different phenotypes from those observed in the blood. Of note, clonally expanded CD8 subsets (CD8_C03, CD8_C05, and CD8_C06) in the PF expressed high levels of GZMK but low levels of GZMB. The preferential accumulation of GZMK-expressing CD8 T cells in the PF but not peripheral blood samples from patients with TPE, suggesting a locally induced differentiation process (at the very least, GZMK is up-regulated at the site of infection) possibly depending on TCR triggering, as we demonstrated that GZMK was induced in CD8 T cells upon anti-CD3/CD28 stimulation in vitro. Indirect support for this model comes from our observation that most clonotypes in these cells (based on scTCR-seq data) were exclusively observed in the PF, which is likely a result of local T cell proliferation and activation in PF rather than migration from the blood. This finding suggests an impact of the PF environment on the development of GZMK-expressing CD8 T cells, conceivably reflecting the differential availability of antigens or tissue factors or bacteria in the blood and PF.
Several recent studies have also observed a GZMK-expressing CD8 T subset in human tumors and aging individuals by using scRNA sequencing (Zheng et al., 2017; Guo et al., 2018; Zhang et al., 2018; Li et al., 2019; Mogilenko et al., 2021). Those GZMK-expressing CD8 T cells also express low levels of GZMB. GZMK-expressing CD8 T cells were shown to be enriched in tumor lesion sites and adjacent nontumorous mucosa (Zhang et al., 2018; Zheng et al., 2017). GZMK-expressing CD8 T cells have been reported to accumulate in tissues due to aging and contribute to the inflammatory response through the secretion of GZMK (Mogilenko et al., 2021). Based on our trajectory and TCR lineage results, GZMK-expressing CD8 T cells in TPE represent a late developmental stage with cytotoxic capability (based on the expression of PRF1). Our data demonstrated that purified GZMK has cytotoxic and bactericidal activity in Mtb-infected macrophages. Thus, the enrichment of GZMK-expressing CD8 T cells in PF may represent an active tissue-specific effector mechanism mediating tissue inflammation and provide a backup for the loss of GZMB-expressing cells at the site of Mtb infection. However, we cannot rule out the possibility that GZMK-expressing CD8 T cells promote immunopathology, as GZMK has the ability to induce macrophage cell death and increase the inflammatory response (Mogilenko et al., 2021; Turner et al., 2019), both of which could result in tissue damage in TB. Therefore, defining the exact role that GZMK-expressing CD8 T cells play during the course of TB disease warrants further investigation.
In summary, the results of this study provide crucial insights into the spectrum of T cell heterogeneity at the site of Mtb infection and yield several important findings that advance our understanding of T cell diversity in TPE. Our scRNA-seq and scTCR lineage tracing identified: (1) differential T cells in the blood and PF from TPE; (2) heterogeneous T cell subsets with diverse functions in the PF from TPE; and (3) distinct, highly expanded T cell clonotypes in T cell subsets enriched in the PF and a strong association between GZMK and TPE disease. This characterization provides a useful framework to now examine the role of T cell subsets in TB disease progression and to guide further investigations into the underlying characteristics that modulate T cell heterogeneity at the site of infection. This new knowledge concerning T cell composition and function at the site of infection might help us to identify new antigens for vaccine development.
Materials and methods
Ethics statement
This study was approved by the Institutional Review Board of the Shenzhen University School of Medicine, China, and written informed consent was obtained from each participant. All experiments and samplings were performed in accordance with ethical and biosafety protocols approved by our institution.
Participants, clinical samples, and diagnostic criteria
The patients recruited to this study were those with known causes that contributed to their PFs. The four study cohorts were recruited from Shenzhen Third People’s Hospital (China), Shenzhen Baoan People’s Hospital (China), and Yuebei Second People’s Hospital (China) between August 2017 and December 2020. Cohort I included six patients with TPE whose PBMCs and PFMCs were collected for scRNA-seq and scTCR-seq. Cohort II included 22 patients with TPE from whom paired blood and PF samples were obtained for flow cytometry analysis. Cohort III included 98 cases of TPE, 36 cases of PPE with pneumonia, and 31 cases of MPE with PF available for evaluating GZMK levels. Cohort IV included 88 cases of TPE and 73 cases of non-TPE (23 cases of PPE and 50 cases of MPE). The diagnosis of TPE was confirmed if a patient had exudative effusion and was culture-positive for Mtb (using PF, a pleural biopsy specimen, or sputum) and/or had evidence of TB based on pleural biopsy specimens positive for granulomatous inflammation with acid-fast bacilli. PPE was diagnosed as any effusion associated with bacterial pneumonia, based on the following: (a) lavage fluid or sputum cultures were Mtb negative during clinical follow-up, (b) new infiltration and clinical signs on chest radiography were evident and completely resolved following treatment with appropriate antibiotics, and (c) viral pathogens were not detected. A diagnosis of MPE was established by demonstration of malignant cells in PF and/or pleural biopsy specimens; histologically, there were 25 cases of adenocarcinoma and 6 of squamous cell carcinoma. Patients were excluded if they had received any invasive procedures directed into the pleural cavity or if they had suffered chest trauma within 3 mo before hospitalization. At the time of sample collection, none of the patients had received any anti-TB therapy, anticancer treatment, antibiotic therapy, corticosteroids, or nonsteroidal anti-inflammatory drugs. The characteristics of all cohorts are summarized in Table S1.
PBMC and PFMC isolation
Heparinized whole-blood samples were collected by venipuncture from all participants, and PF samples were collected from patients with TPE, pneumonia, or lung cancer. PBMCs were obtained by gradient separation of whole-blood samples over Ficoll-Hypaque density gradient (Ficoll-Paque Plus; Amersham Biosciences). PFMCs and supernatants were separated by centrifugation of ≤50 ml PF at 300 g for 5 min at 4°C. Isolated PBMCs and PFMCs were immediately used for flow cytometry or stored at −150°C. The plasma and supernatants from PF were stored at −80°C until further use.
10x Genomics single-cell sample processing and cDNA library preparation
Four paired PBMCs and PFMCs were subjected to 10x Genomics single-cell sequencing. For the validation cohort, cell suspensions from two paired PBMCs and PFMCs were enriched for CD3 using human CD3-microbeads, according to the manufacturer’s instructions (Miltenyi Biotec). CD3+ enriched single-cell suspensions were subjected to 10x Genomics single-cell sample processing and cDNA library preparation. Briefly, cell viability was assessed by trypan blue staining, and samples with viability >90% were prepared using Chromium Single Cell 5′ Library and Chromium Single Cell V(D)J Enrichment Kits, Human (10x Genomics), according to the manufacturer’s instructions. The PBMCs and PFMCs were washed once with PBS containing 0.05% BSA and resuspended in PBS containing 0.05% BSA to a final concentration of 700–1,200 cells/µl, as determined using a hemocytometer. Single cells were captured in droplets and subjected to reverse transcription (53°C for 45 min, 85°C for 5 min). The emulsions were broken, and the cDNA was purified before PCR amplification (98°C for 45 s; 13 cycles of 98°C for 20 s, 67°C for 30 s, and 72°C for 1 min; 72°C for 1 min). Amplified cDNA was then purified by SPRIselect beads and used for both 5′ gene expression library construction and TCR enrichment. For gene expression library construction, 50 ng purified cDNA was fragmented and end-repaired, double-sided size-selected with SPRIselect beads, PCR-amplified with sample indexing primers (98°C for 45 s; 14 cycles of 98°C for 20 s, 54°C for 30 s, and 72°C for 20 s; 72°C for 1 min), and double-sided size-selected again with SPRIselect beads. For TCR library construction, TCR transcripts were enriched from 2 µl amplified cDNA by PCR (98°C for 45s; 10 cycles of 98°C for 20s, 67°C for 30 s, and 72°C for 1 min; 72°C for 1 min). The scRNA-seq and scTCR-seq libraries were sequenced on an Illumina HiSeq X Ten system (Illumina).
Single-cell data preprocessing, gene expression quantification, and cell-type determination
The raw data from each sample were demultiplexed and aligned to the GRCh38 reference genome, and the UMI counts were quantified using the 10x Genomics Cell Ranger pipeline (v3.1.0).
Data analysis continued with the filtered barcode matrix files using the Seurat package (v3.2.0) after cells were recognized from droplets by DropletUtils (v1.6.1; Lun et al., 2019). One PBMC sample was removed due to the poor quality of median gene and UMI counts (Table S2). On average, 1,365 genes and 4,793 UMIs were obtained per cell, which is comparable with previously published droplet-based scRNA-seq studies of human cells (Zhang et al., 2019). Cells with >200 detected genes and <10% mitochondrial reads were considered valid and used for downstream analysis. To prevent clusters from being biased by mitochondrial transcript content, the gene expression values were scaled based on the cell mitochondrial transcript content.
Next, SCTransform was applied to normalize and find variable features within the single-cell gene expression data (Hafemeister and Satija, 2019). Clustering and differential expression analyses were performed using the R package Seurat, with default parameters (Wolock et al., 2019). Based on the PCElbowPlot, a certain number of principal components (PCs) were selected for the clustering analysis when that number reached the baseline of the SD of the PCs. Cell clusters were visualized using t-distributed stochastic neighbor embedding (tSNE; Satija et al., 2015). Cells were represented in a 2D tSNE plane, and clusters were identified and annotated according to known biological cell types using canonical marker genes or published reference gene signatures (Table S5). For differential gene expression, a model-based analysis of single-cell transcriptomics (MAST) test (Finak et al., 2015; log fold-change ≥0.25, minimum percentage 0.1, and minimum differential percentage >0.15) was used to select only those genes with an adjusted P value <0.05. For pathway enrichment analysis, the enrichGO function within clusterProfiler (v3.14.3) was used, with gene sets from the GO knowledge base. To identify T cell subtypes, T-like cells were extracted from the global data for downstream analyses. To prevent clusters from being biased by mitochondrial, ribosomal, metallothionein, or transcript content, gene expression values were scaled after deleting the relevant gene family. Then, LogNormalize and mean.var.plot were applied to normalize and find variable features within the single-cell gene expression data. Next, the downstream analysis was run in the same way as described above, including dimensional reduction, clustering, and finding cluster biomarkers. Signatures used for subset identity determination or phenotyping already published are referenced in each figure. See Table S5 for details.
Trajectory analysis
Trajectory analysis was calculated using PAGA in Scanpy (v1.5.1) with default parameters (Wolf et al., 2019), using the 20 most significant DEGs compared with the others for each selected cluster. The lineage trajectory analysis revealed a continuous cell type transition from the assigned discrete cell types.
GSEA
Pathway analyses were performed on the 50 hallmark pathways annotated in the molecular signature database (Subramanian et al., 2005). The gene sets we used were from the database MSigDB48 (v7.4). In the single-cluster enrichment analysis, the normalized and centered expression data were transformed to z-scores. For each cluster, the z-scores across cells were averaged per gene. The Wilcoxon rank sum test in the presto package (v1.0.0) was performed to obtain the rank of all genes. Then, we used the fgsea package (v1.17.1) to calculate GSEA enrichment scores and P values for each collection of gene sets. Briefly, the activity scores for each cell were compared using a generalized linear model. The outputs of these generalized linear models were visualized in heat maps.
TCR data analysis
The raw TCR data from each sample were demultiplexed and aligned to the GRCh38 reference genome, and clonotype counts were quantified using the 10x Genomics Cell Ranger pipeline (v3.1.0). For clonal expansion levels of CD4 and CD8 subsets, the percentage of cells with clonal TCR for each patient was calculated. Then, STARTRAC (v0.1.0) was applied to analyze different aspects of T cells based on paired single-cell transcriptomes and TCR sequences (Zhang et al., 2018). The STARTRAC indices, STARTRAC-expa, STARTRAC-migr, and STARTRAC-tran, measure the degree of clonal expansion, tissue migration, and state transition of T cell clusters upon TCR tracking, respectively. The details of the formulae for STARTRAC-expa, STARTRAC-migr, and STARTRAC-tran were described previously (Zhang et al., 2018). STARTRAC-expa ranges from 0 to 1, with 0 indicating no clonal expansion and 1 indicating that the cluster contains only one clonally expanded clonotype. If a cluster contains multiple clonotypes with distinct degrees of clonal expansion, STARTRAC-expa will be between 0 and 1, with a high STARTRAC-expa indicating high clonality. Both STARTRAC-migr and STARTRAC-tran are defined at two different levels (clonotypes and clusters), with clonotype-level definitions describing the extent of migration and state transition of a given clonotype and cluster-level definitions depicting the summary of such properties of all clonotypes within a cluster. For two T cell clusters with similar clonal expansion and clonal size, the one with clonal cells broadly distributed throughout various tissues would probably be more mobile (Zhang et al., 2018). Pairwise STARTRAC-migr (pSTARTRAC-migr) and pSTARTRAC-tran indices were also used for precise quantification between tissues or cell clusters. pSTARTRAC-migr uses the same formula as STARTRAC-migr and is used to measure the migration potential between two tissues. pSTARTRAC-tran uses the same formula as STARTRAC-migr and is used to measure the transition between two clusters (Zhang et al., 2018). All analyses were performed in the R v3.6.1 environment and Python v3.7.4.
Flow cytometry and intracellular cytokine staining
PBMCs and PFMCs from TPE were obtained for flow cytometry as previously described (Cai et al., 2020). The samples were then stained with antibodies against CD3 (SK7), CD4 (RPA-T4), and CD8 (SK1; BD Biosciences) plus ghost dye (Tonbo Biosciences) for 30 min at 4°C. Following fixation and permeabilization for 30 min at room temperature, the cells were stained with mAbs against GZMB (GM26E7), GZMK (GB11), and GZMA (CB9; BD Biosciences) for 30 min at room temperature. All antibodies were validated by the manufacturer for flow cytometry application. The cells were resuspended in 200 µl of 2% paraformaldehyde, acquired using FACSDiva software (BD Biosciences), and analyzed using FlowJo software v10.
ELISA
The levels of GZMK (Multi Sciences) and GZMB (Neobioscience) in plasma and PF samples were determined by ELISA, according to the manufacturer’s instructions.
ADA and LDH assay
The activity of ADA and LDH in PF was measured on a fully automated chemistry analyzer (AU5800; Beckman Coulter). The operation was performed in strict accordance with the standard SOP. All experiments were performed in accordance with the manufacturers’ protocols.
Primary human CD8 T cell isolation and in vitro activation
PFMCs from patients with TPE were used to isolate CD8 T cells. CD8 T cells were isolated from four TPE PFMCs using magnetic beads (Miltenyi Biotec), according to the manufacturer’s protocol. Isolated CD8 T cells were cultured in RPMI 1640 supplemented with 10% heat-inactivated FBS. Then, CD8 T cells were activated with anti-CD3/CD28 (Gibco) for at 37°C for 12 h. GZMK levels were measured in cell culture supernatants by ELISA, as described above.
GZMK and GZMB activity assay
Human monocytic THP-1 cells (TIB-202; ATCC) were grown in RPMI 1640 supplemented with l-glutamine (2 mM) and 10% heat-inactivated FBS (all from Gibco Life Technologies). The THP-1 cells were treated with 40 ng/ml PMA (Sigma-Aldrich) in 12-well plates at 37°C for 24 h and allowed to differentiate into macrophages. The cells were then incubated with fresh prewarmed RPMI 1640 (10% FBS and 2 mM L-glutamine), infected with virulent Mtb H37Rv at a multiplicity of infection of 5 for 4 h and then washed three times with PBS. Purified GZMK or GZMB (gifts from Dr. Feng Shao of the National Institute of Biological Sciences, Beijing, China) were then added with or without Perforin1 (RPB317Hu01; Cloud-Clone Corp). Cell cytotoxicity was measured by an LDH assay using a Cytotoxicity Assay kit (Promega) in 24 h. For the CFU assay, the cells were lysed in 0.1% SDS, plated at various dilutions on 7H10 plates, and incubated at 37°C for 2 to 4 wk.
Statistical analyses
A one-way ANOVA Newman–Keuls multiple comparison test was used to compare differences among multiple groups. An unpaired t test was used to analyze differences between two groups. A paired t test was used to analyze differences in paired samples. All statistical analyses were performed in GraphPad Prism (v7.0). Two-tailed statistical tests were conducted, and a P value <0.05 was considered statistically significant.
Online supplemental material
Fig. S1 shows the consistency of cell capture and the cell markers for cell subsets. Fig. S2 shows the marker genes for CD4 and CD8 T cell subsets. Fig. S3 shows the comparison between the original dataset and the validation dataset. Fig. S4 shows the pseudo-time of CD4 and CD8 differentiation trajectories in PBMCs and PFMCs. Fig. S5 shows the TCR expansion and sharing status between subsets or tissues. Table S1 shows the demographic characteristics of the study populations. Table S2 shows the characteristics of scRNA-seq of the eight samples. Table S3 shows the cell numbers and frequencies of all subsets in PBMCs or PFMCs from TPE. Table S4 shows the marker genes of all subsets identified by scRNA-seq. Table S5 shows the reference gene signature list. Table S6 shows the comparison of CD4 and CD8 subsets between the original and validation datasets. Table S7 shows the TCR distribution in PBMCs or PFMCs. Table S8 shows the identical TCRs shared between the CD8 and CD4 subsets. Table S9 shows the gene expression characterization associated with PF-infiltrating T subsets. Table S10 shows the DEGs of PBMC CD4_C09 versus PFMC CD4_C09.
Supplementary Material
shows the demographic characteristics of the study populations.
shows the characteristics of scRNA-seq of the eight samples.
shows the cell numbers and frequencies of all subsets in PBMCs or PFMCs from TPE.
shows the marker genes of all subsets identified by scRNA-seq.
shows the reference gene signature list.
shows the comparison of CD4 and CD8 subsets between the original and validation datasets.
shows the TCR distribution in PBMCs or PFMCs.
shows the identical TCRs shared between the CD8 and CD4 subsets.
shows the gene expression characterization associated with PF-infiltrating T subsets.
shows the DEGs of PBMC CD4_C09 versus PFMC CD4_C09.
Acknowledgments
The authors gratefully acknowledge all the study participants and study staff for their help and cooperation during the study and Dr. J. Tamanini (Shenzhen University and ETediting) for editing the manuscript before submission. The authors also thank Dr. Z. Cheng from St. Jude Children’s Research Hospital for informative discussions.
Y. Cai, Y. Dai, C.G. Feng, and X. Chen are supported by the National Natural Science Foundation of China (81871255, 82100015, 82072252, and 91942315) and the Guangdong Provincial Key Laboratory of Regional Immunity and Diseases (2019B030301009).
Author contributions: X. Chen, Y. Cai, and C.G. Feng conceived and designed the experiments. Y. Cai, Y. Wang, C.G. Feng, and X. Chen drafted the manuscript. Y. Cai, C. Shi, Y. Dai, F. Li, Y. Xu, and P. Zhang performed the experiments. F. Kong, Z. Wen, and G. Deng collected the samples. Y. Cai, Y. Wang, C. Shi, B. Kang, and Q. Zhou analyzed the data. A. Singhal and Q. Yang provided advice on the project design and data interpretation. All authors critically reviewed and approved the manuscript.
Data availability
The sequencing raw data and processed data used in this paper are available in GSA (Genome Sequence Archive in the BIG Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences) under accession numbers HRA000910 and HRA00036.
References
- Appay, V., Zaunders J.J., Papagno L., Sutton J., Jaramillo A., Waters A., Easterbrook P., Grey P., Smith D., McMichael A.J., et al. 2002. Characterization of CD4(+) CTLs ex vivo. J. Immunol. 168:5954–5958. 10.4049/jimmunol.168.11.5954 [DOI] [PubMed] [Google Scholar]
- Berry, M.P., Graham C.M., McNab F.W., Xu Z., Bloch S.A., Oni T., Wilkinson K.A., Banchereau R., Skinner J., Wilkinson R.J., et al. 2010. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 466:973–977. 10.1038/nature09247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt, D., Kang B., Sawant D., Zheng L., Perez K., Huang Z., Sekirov L., Wolak D., Huang J.Y., Liu X., et al. 2021. STARTRAC analyses of scRNAseq data from tumor models reveal T cell dynamics and therapeutic targets. J. Exp. Med. 218:e20201329. 10.1084/jem.20201329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bold, T.D., and Ernst J.D.. 2012. CD4+ T cell-dependent IFN-γ production by CD8+ effector T cells in Mycobacterium tuberculosis infection. J. Immunol. 189:2530–2536. 10.4049/jimmunol.1200994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D.M., Dilzer A.M., Meents D.L., and Swain S.L.. 2006. CD4 T cell-mediated protection from lethal influenza: perforin and antibody-mediated mechanisms give a one-two punch. J. Immunol. 177:2888–2898. 10.4049/jimmunol.177.5.2888 [DOI] [PubMed] [Google Scholar]
- Brown, D.M., Kamperschroer C., Dilzer A.M., Roberts D.M., and Swain S.L.. 2009. IL-2 and antigen dose differentially regulate perforin- and FasL-mediated cytolytic activity in antigen specific CD4+ T cells. Cell. Immunol. 257:69–79. 10.1016/j.cellimm.2009.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, A., Hoffman P., Smibert P., Papalexi E., and Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Y., Dai Y., Wang Y., Yang Q., Guo J., Wei C., Chen W., Huang H., Zhu J., Zhang C., et al. 2020. Single-cell transcriptomics of blood reveals a natural killer cell subset depletion in tuberculosis. EBioMedicine. 53:102686. 10.1016/j.ebiom.2020.102686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaday, D.H., Wilkinson R.J., Li Q., Harding C.V., Silver R.F., and Boom W.H.. 2001. CD4(+) and CD8(+) T cells kill intracellular Mycobacterium tuberculosis by a perforin and Fas/Fas ligand-independent mechanism. J. Immunol. 167:2734–2742. 10.4049/jimmunol.167.5.2734 [DOI] [PubMed] [Google Scholar]
- Cano-Gamez, E., Soskic B., Roumeliotis T.I., So E., Smyth D.J., Baldrighi M., Willé D., Nakic N., Esparza-Gordillo J., Larminie C.G.C., et al. 2020. Single-cell transcriptomics identifies an effectorness gradient shaping the response of CD4+ T cells to cytokines. Nat. Commun. 11:1801. 10.1038/s41467-020-15543-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso, A.M., Serbina N., Klein E., Triebold K., Bloom B.R., and Flynn J.L.. 1999. Mice deficient in CD4 T cells have only transiently diminished levels of IFN-gamma, yet succumb to tuberculosis. J. Immunol. 162:5407–5416. [PubMed] [Google Scholar]
- Casazza, J.P., Betts M.R., Price D.A., Precopio M.L., Ruff L.E., Brenchley J.M., Hill B.J., Roederer M., Douek D.C., and Koup R.A.. 2006. Acquisition of direct antiviral effector functions by CMV-specific CD4+ T lymphocytes with cellular maturation. J. Exp. Med. 203:2865–2877. 10.1084/jem.20052246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Zhang M., Liao M., Graner M.W., Wu C., Yang Q., Liu H., and Zhou B.. 2010. Reduced Th17 response in patients with tuberculosis correlates with IL-6R expression on CD4+ T Cells. Am. J. Respir. Crit. Care Med. 181:734–742. 10.1164/rccm.200909-1463OC [DOI] [PubMed] [Google Scholar]
- da Cunha Lisboa, V., Ribeiro-Alves M., da Silva Corrêa R., Ramos Lopes I., Mafort T.T., Santos A.P., Porto Amadeu T., Rufino R., and Silva Rodrigues L.. 2019. Predominance of Th1 Immune Response in Pleural Effusion of Patients with Tuberculosis among Other Exudative Etiologies. J. Clin. Microbiol. 58:e00927-19. 10.1128/JCM.00927-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniil, Z.D., Zintzaras E., Kiropoulos T., Papaioannou A.I., Koutsokera A., Kastanis A., and Gourgoulianis K.I.. 2007. Discrimination of exudative pleural effusions based on multiple biological parameters. Eur. Respir. J. 30:957–964. 10.1183/09031936.00126306 [DOI] [PubMed] [Google Scholar]
- Di Cesare, A., Di Meglio P., and Nestle F.O.. 2009. The IL-23/Th17 axis in the immunopathogenesis of psoriasis. J. Invest. Dermatol. 129:1339–1350. 10.1038/jid.2009.59 [DOI] [PubMed] [Google Scholar]
- Dotiwala, F., Sen Santara S., Binker-Cosen A.A., Li B., Chandrasekaran S., and Lieberman J.. 2017. Granzyme B Disrupts Central Metabolism and Protein Synthesis in Bacteria to Promote an Immune Cell Death Program. Cell. 171:1125–1137.e11. 10.1016/j.cell.2017.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak, G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A.K., Slichter C.K., Miller H.W., McElrath M.J., Prlic M., et al. 2015. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16:278. 10.1186/s13059-015-0844-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd, K., Glaziou P., Zumla A., and Raviglione M.. 2018. The global tuberculosis epidemic and progress in care, prevention, and research: an overview in year 3 of the End TB era. Lancet Respir. Med. 6:299–314. 10.1016/S2213-2600(18)30057-2 [DOI] [PubMed] [Google Scholar]
- Guo, X., Zhang Y., Zheng L., Zheng C., Song J., Zhang Q., Kang B., Liu Z., Jin L., Xing R., et al. 2018. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 24:978–985. 10.1038/s41591-018-0045-3 [DOI] [PubMed] [Google Scholar]
- Haabeth, O.A., Tveita A.A., Fauskanger M., Schjesvold F., Lorvik K.B., Hofgaard P.O., Omholt H., Munthe L.A., Dembic Z., Corthay A., and Bogen B.. 2014. How Do CD4(+) T Cells Detect and Eliminate Tumor Cells That Either Lack or Express MHC Class II Molecules? Front. Immunol. 5:174. 10.3389/fimmu.2014.00174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister, C., and Satija R.. 2019. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20:296. 10.1186/s13059-019-1874-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, A., Glanville J., Hansmann L., and Davis M.M.. 2015. Corrigendum: Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat. Biotechnol. 33:210. 10.1038/nbt0215-210c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, K., Kouno T., Ikawa T., Hayatsu N., Miyajima Y., Yabukami H., Terooatea T., Sasaki T., Suzuki T., Valentine M., et al. 2019. Single-cell transcriptomics reveals expansion of cytotoxic CD4 T cells in supercentenarians. Proc. Natl. Acad. Sci. USA. 116:24242–24251. 10.1073/pnas.1907883116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper, C.E., Lee Y.C., and Maskell N.A.. 2009. Interferon-gamma release assays for the diagnosis of TB pleural effusions: hype or real hope? Curr. Opin. Pulm. Med. 15:358–365. 10.1097/MCP.0b013e32832bcc4e [DOI] [PubMed] [Google Scholar]
- Hooper, C., Lee Y.C., and Maskell N.. BTS Pleural Guideline Group . 2010. Investigation of a unilateral pleural effusion in adults: British Thoracic Society Pleural Disease Guideline 2010. Thorax. 65(Suppl 2):ii4–ii17. 10.1136/thx.2010.136978 [DOI] [PubMed] [Google Scholar]
- Jalapathy, K.V., Prabha C., and Das S.D.. 2004. Correlates of protective immune response in tuberculous pleuritis. FEMS Immunol. Med. Microbiol. 40:139–145. 10.1016/S0928-8244(03)00303-1 [DOI] [PubMed] [Google Scholar]
- Kamath, A.B., Woodworth J., Xiong X., Taylor C., Weng Y., and Behar S.M.. 2004. Cytolytic CD8+ T cells recognizing CFP10 are recruited to the lung after Mycobacterium tuberculosis infection. J. Exp. Med. 200:1479–1489. 10.1084/jem.20041690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C.H., Rott L., Kunkel E.J., Genovese M.C., Andrew D.P., Wu L., and Butcher E.C.. 2001. Rules of chemokine receptor association with T cell polarization in vivo. J. Clin. Invest. 108:1331–1339. 10.1172/JCI13543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalvani, A., Brookes R., Wilkinson R.J., Malin A.S., Pathan A.A., Andersen P., Dockrell H., Pasvol G., and Hill A.V.. 1998. Human cytolytic and interferon gamma-secreting CD8+ T lymphocytes specific for Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA. 95:270–275. 10.1073/pnas.95.1.270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts, D., Wauters E., Boeckx B., Aibar S., Nittner D., Burton O., Bassez A., Decaluwé H., Pircher A., Van den Eynde K., et al. 2018. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 24:1277–1289. 10.1038/s41591-018-0096-5 [DOI] [PubMed] [Google Scholar]
- Lee, J., Lim J.K., Yoo S.S., Lee S.Y., Cha S.I., Park J.Y., and Kim C.H.. 2016. Different characteristics of tuberculous pleural effusion according to pleural fluid cellular predominance and loculation. J. Thorac. Dis. 8:1935–1942. 10.21037/jtd.2016.06.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leruste, A., Tosello J., Ramos R.N., Tauziède-Espariat A., Brohard S., Han Z.Y., Beccaria K., Andrianteranagna M., Caudana P., Nikolic J., et al. 2019. Clonally Expanded T Cells Reveal Immunogenicity of Rhabdoid Tumors. Cancer Cell. 36:597–612.e8. 10.1016/j.ccell.2019.10.008 [DOI] [PubMed] [Google Scholar]
- Lewinsohn, D.M., Alderson M.R., Briden A.L., Riddell S.R., Reed S.G., and Grabstein K.H.. 1998. Characterization of human CD8+ T cells reactive with Mycobacterium tuberculosis-infected antigen-presenting cells. J. Exp. Med. 187:1633–1640. 10.1084/jem.187.10.1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinsohn, D.A., Heinzel A.S., Gardner J.M., Zhu L., Alderson M.R., and Lewinsohn D.M.. 2003. Mycobacterium tuberculosis-specific CD8+ T cells preferentially recognize heavily infected cells. Am. J. Respir. Crit. Care Med. 168:1346–1352. 10.1164/rccm.200306-837OC [DOI] [PubMed] [Google Scholar]
- Li, H., van der Leun A.M., Yofe I., Lubling Y., Gelbard-Solodkin D., van Akkooi A.C.J., van den Braber M., Rozeman E.A., Haanen J.B.A.G., Blank C.U., et al. 2019. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell. 176:775–789.e18. 10.1016/j.cell.2018.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light, R.W. 2006. Parapneumonic effusions and empyema. Proc. Am. Thorac. Soc. 3:75–80. 10.1513/pats.200510-113JH [DOI] [PubMed] [Google Scholar]
- Lun, A.T.L., Riesenfeld S., Andrews T., Dao T.P., Gomes T., and Marioni J.C.. participants in the 1st Human Cell Atlas Jamboree . 2019. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20:63. 10.1186/s13059-019-1662-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehara, T., Kaneko N., Perugino C.A., Mattoo H., Kers J., Allard-Chamard H., Mahajan V.S., Liu H., Murphy S.J., Ghebremichael M., et al. 2020. Cytotoxic CD4+ T lymphocytes may induce endothelial cell apoptosis in systemic sclerosis. J. Clin. Invest. 130:2451–2464. 10.1172/JCI131700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi, L., Santarlasci V., Capone M., Peired A., Frosali F., Crome S.Q., Querci V., Fambrini M., Liotta F., Levings M.K., et al. 2010. CD161 is a marker of all human IL-17-producing T-cell subsets and is induced by RORC. Eur. J. Immunol. 40:2174–2181. 10.1002/eji.200940257 [DOI] [PubMed] [Google Scholar]
- Mayer, A., Zhang Y., Perelson A.S., and Wingreen N.S.. 2019. Regulation of T cell expansion by antigen presentation dynamics. Proc. Natl. Acad. Sci. USA. 116:5914–5919. 10.1073/pnas.1812800116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, D.K., Sharma S.K., Dinda A.K., Bindra M.S., Madan B., and Ghosh B.. 2005. Polarized helper T cells in tubercular pleural effusion: phenotypic identity and selective recruitment. Eur. J. Immunol. 35:2367–2375. 10.1002/eji.200525977 [DOI] [PubMed] [Google Scholar]
- Mogilenko, D.A., Shpynov O., Andhey P.S., Arthur L., Swain A., Esaulova E., Brioschi S., Shchukina I., Kerndl M., Bambouskova M., et al. 2021. Comprehensive Profiling of an Aging Immune System Reveals Clonal GZMK+ CD8+ T Cells as Conserved Hallmark of Inflammaging. Immunity. 54:99–115.e12. 10.1016/j.immuni.2020.11.005 [DOI] [PubMed] [Google Scholar]
- Porcel, J.M., Gasol A., Bielsa S., Civit C., Light R.W., and Salud A.. 2015. Clinical features and survival of lung cancer patients with pleural effusions. Respirology. 20:654–659. 10.1111/resp.12496 [DOI] [PubMed] [Google Scholar]
- Ren, X., Wen W., Fan X., Hou W., Su B., Cai P., Li J., Liu Y., Tang F., Zhang F., et al. 2021. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 184:1895–1913.e19. 10.1016/j.cell.2021.01.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, G.A., Balbi B., and Manca F.. 1987. Tuberculous pleural effusions. Evidence for selective presence of PPD-specific T-lymphocytes at site of inflammation in the early phase of the infection. Am. Rev. Respir. Dis. 136:575–579. 10.1164/ajrccm/136.3.575 [DOI] [PubMed] [Google Scholar]
- San José, M.E., Valdés L., Saavedra M.J., De Vega J.M., Alvarez D., Viñuela J., Penela P., Valle J.M., and Seoane R.. 1999. Lymphocyte populations in tuberculous pleural effusions. Ann. Clin. Biochem. 36:492–500. 10.1177/000456329903600413 [DOI] [PubMed] [Google Scholar]
- Satija, R., Farrell J.A., Gennert D., Schier A.F., and Regev A.. 2015. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33:495–502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas, P., Virassamy B., Ye C., Salim A., Mintoff C.P., Caramia F., Salgado R., Byrne D.J., Teo Z.L., Dushyanthen S., et al. Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer (kConFab) . 2018. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat. Med. 24:986–993. 10.1038/s41591-018-0078-7 [DOI] [PubMed] [Google Scholar]
- Scanga, C.A., Mohan V.P., Yu K., Joseph H., Tanaka K., Chan J., and Flynn J.L.. 2000. Depletion of CD4(+) T cells causes reactivation of murine persistent tuberculosis despite continued expression of interferon gamma and nitric oxide synthase 2. J. Exp. Med. 192:347–358. 10.1084/jem.192.3.347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiscento, M., Vargas F.S., Acencio M.M., Teixeira L.R., Capelozzi V.L., Sales R.K., and Antonangelo L.. 2010. Pleural fluid cytokines correlate with tissue inflammatory expression in tuberculosis. Int. J. Tuberc. Lung Dis. 14:1153–1158. [PubMed] [Google Scholar]
- Sharma, S.K., Mitra D.K., Balamurugan A., Pandey R.M., and Mehra N.K.. 2002. Cytokine polarization in miliary and pleural tuberculosis. J. Clin. Immunol. 22:345–352. 10.1023/A:1020604331886 [DOI] [PubMed] [Google Scholar]
- Shaw, J.A., Diacon A.H., and Koegelenberg C.F.N.. 2019. Tuberculous pleural effusion. Respirology. 24:962–971. 10.1111/resp.13673 [DOI] [PubMed] [Google Scholar]
- Subramanian, A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, Z.H., and Shi H.Z.. 2013. Subpopulations of helper T lymphocytes in tuberculous pleurisy. Tuberculosis (Edinb.). 93:279–284. 10.1016/j.tube.2013.02.014 [DOI] [PubMed] [Google Scholar]
- Turner, C.T., Zeglinski M.R., Richardson K.C., Zhao H., Shen Y., Papp A., Bird P.I., and Granville D.J.. 2019. Granzyme K Expressed by Classically Activated Macrophages Contributes to Inflammation and Impaired Remodeling. J. Invest. Dermatol. 139:930–939. 10.1016/j.jid.2018.09.031 [DOI] [PubMed] [Google Scholar]
- Valdés, L., San-José E., Ferreiro L., Golpe A., González-Barcala F.J., Toubes M.E., Rodríguez-Álvarez M.X., Álvarez-Dobaño J.M., Rodríguez-Núñez N., Rábade C., and Gude F.. 2015. Predicting malignant and tuberculous pleural effusions through demographics and pleural fluid analysis of patients. Clin. Respir. J. 9:203–213. 10.1111/crj.12125 [DOI] [PubMed] [Google Scholar]
- Viallard, J.F., Bloch-Michel C., Neau-Cransac M., Taupin J.L., Garrigue S., Miossec V., Mercie P., Pellegrin J.L., and Moreau J.F.. 2001. HLA-DR expression on lymphocyte subsets as a marker of disease activity in patients with systemic lupus erythematosus. Clin. Exp. Immunol. 125:485–491. 10.1046/j.1365-2249.2001.01623.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorster, M.J., Allwood B.W., Diacon A.H., and Koegelenberg C.F.. 2015. Tuberculous pleural effusions: advances and controversies. J. Thorac. Dis. 7:981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Zhou Q., Zhai K., Wang Y., Liu J.Y., Wang X.J., Wang Z., Zhang J.C., Tong Z.H., and Shi H.Z.. 2018. Diagnostic accuracy of interleukin 27 for tuberculous pleural effusion: two prospective studies and one meta-analysis. Thorax. 73:240–247. 10.1136/thoraxjnl-2016-209718 [DOI] [PubMed] [Google Scholar]
- Wolf, F.A., Hamey F.K., Plass M., Solana J., Dahlin J.S., Göttgens B., Rajewsky N., Simon L., and Theis F.J.. 2019. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 20:59. 10.1186/s13059-019-1663-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolock, S.L., Lopez R., and Klein A.M.. 2019. Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst. 8:281–291.e9. 10.1016/j.cels.2018.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodworth, J.S., Wu Y., and Behar S.M.. 2008. Mycobacterium tuberculosis-specific CD8+ T cells require perforin to kill target cells and provide protection in vivo. J. Immunol. 181:8595–8603. 10.4049/jimmunol.181.12.8595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemmour, D., Zilionis R., Kiner E., Klein A.M., Mathis D., and Benoist C.. 2018. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat. Immunol. 19:291–301. 10.1038/s41590-018-0051-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Yu X., Zheng L., Zhang Y., Li Y., Fang Q., Gao R., Kang B., Zhang Q., Huang J.Y., et al. 2018. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 564:268–272. 10.1038/s41586-018-0694-x [DOI] [PubMed] [Google Scholar]
- Zhang, Q., He Y., Luo N., Patel S.J., Han Y., Gao R., Modak M., Carotta S., Haslinger C., Kind D., et al. 2019. Landscape and Dynamics of Single Immune Cells in Hepatocellular Carcinoma. Cell. 179:829–845.e20. 10.1016/j.cell.2019.10.003 [DOI] [PubMed] [Google Scholar]
- Zheng, C., Zheng L., Yoo J.K., Guo H., Zhang Y., Guo X., Kang B., Hu R., Huang J.Y., Zhang Q., et al. 2017. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell. 169:1342–1356.e16. 10.1016/j.cell.2017.05.035 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the demographic characteristics of the study populations.
shows the characteristics of scRNA-seq of the eight samples.
shows the cell numbers and frequencies of all subsets in PBMCs or PFMCs from TPE.
shows the marker genes of all subsets identified by scRNA-seq.
shows the reference gene signature list.
shows the comparison of CD4 and CD8 subsets between the original and validation datasets.
shows the TCR distribution in PBMCs or PFMCs.
shows the identical TCRs shared between the CD8 and CD4 subsets.
shows the gene expression characterization associated with PF-infiltrating T subsets.
shows the DEGs of PBMC CD4_C09 versus PFMC CD4_C09.
Data Availability Statement
The sequencing raw data and processed data used in this paper are available in GSA (Genome Sequence Archive in the BIG Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences) under accession numbers HRA000910 and HRA00036.