Abstract
SARS-CoV-2, a novel coronavirus and an etiologic agent for the current global health emergency, causes acute infection of the respiratory tract leading to severe disease and significant mortality. Ever since the start of SARS-CoV-2, also known as the COVID-19 pandemic, countless uncertainties have been revolving around the pathogenesis and epidemiology of the SARS-CoV-2 infection. While air pollution has been shown to be strongly correlated to increased SARS-CoV-2 morbidity and mortality, whether environmental pollutants such as ground-level ozone affects the susceptibility of individuals to SARS-CoV-2 is not yet established. To investigate the impact of ozone inhalation on the expression levels of signatures associated with host susceptibility to SARS-CoV-2, we analyzed lung tissues collected from mice that were sub-chronically exposed to air or 0.8 ppm ozone for three weeks (4 h/night, 5 nights/week), and analyzed the expression of signatures associated with host susceptibility to SARS-CoV-2. SARS-CoV-2 entry into the host cells is dependent on the binding of the virus to the host cellular receptor, angiotensin-converting enzyme (ACE2), and its subsequent proteolytic priming by the host-derived protease, transmembrane protease serine 2 (TMPRSS2). The Ace2 transcripts were significantly elevated in the parenchyma, but not in the extrapulmonary airways and alveolar macrophages, from ozone-exposed mice. The TMPRSS2 protein and Tmprss2 transcripts were significantly elevated in the extrapulmonary airways, parenchyma, and alveolar macrophages from ozone-exposed mice. A significant proportion of additional known SARS-CoV-2 host susceptibility genes were upregulated in alveolar macrophages and parenchyma from ozone-exposed mice. Our data indicate that the unhealthy levels of ozone in the environment may predispose individuals to severe SARS-CoV-2 infection. Given the severity of this pandemic and the challenges associated with direct testing of host-environment interactions in clinical settings, we believe that this ozone exposure-based study informs the scientific community of the potentially detrimental effects of the ambient ozone levels in determining the host susceptibility to SARS-CoV-2.
Subject terms: Environmental impact, Acute inflammation
Introduction
SARS-CoV-2, a novel coronavirus and an etiologic agent of the current global health emergency, causes acute infection of the respiratory tract leading to severe disease and significant mortality1. SARS-CoV-2 entry into the host cells is dependent upon the binding of the viral spike (S) protein to the host cellular receptor, angiotensin converting enzyme (ACE2), and its proteolytic priming by the host-derived protease, transmembrane protease serine 2 (TMPRSS2)2. Therefore, the host susceptibility to SARS-CoV-2 could vary based on the expression of the host susceptibility proteins including ACE2 and TMPRSS23–5. For example, increased expression of the host-derived protease, TMPRSS2, may promote the priming of SARS-CoV-2, thus resulting in increased infectivity and disease severity. The current literature indicates that individuals have varied susceptibility to SARS-CoV-2 that may be dependent on age6,7, gender8, underlying comorbidities9, and environmental pollution10,11. However, the list of factors determining varied susceptibilities of the human population to SARS-CoV-2 remains incomplete.
Nearly one-third of the United States population lives in areas with unhealthy levels of ozone12,13. While it is already known that the unhealthy levels of ozone increase the risk for developing cardiopulmonary health problems14–20, it is unclear whether the ambient ozone levels regulate the expression of host susceptibility proteins to SARS-CoV-2 and, in turn, account for the varied susceptibilities of the human population to SARS-CoV-2. Addressing this critical question is highly relevant in terms of increasing our mechanistic understanding of the host-air pollution (environment) interactions underlying the SARS-CoV-2 pathogenesis and epidemiology, and for developing future preventive and therapeutic strategies.
To begin to understand the impact of ozone inhalation on the host susceptibility to SARS-CoV-2, we analyzed lung tissues collected from mice that were sub-chronically exposed to filtered air or 0.8 ppm ozone for three weeks (4 h/night, 5 nights/week)21, and analyzed the expression of the gene and protein signatures associated with host susceptibility to SARS-CoV-2. We used western blotting and immunohistochemistry for assessing expression levels of TMPRSS2 protein in three different lung tissue compartments. To determine the RNA levels of Tmprss2 and Ace2 in a cell-specific manner, in situ gene expression was assessed for Tmprss2 and Ace2 genes using RNAscope approach. Finally, the mRNA expression levels of 33 known SARS-CoV-2 host susceptibility genes were assessed in three lung tissue compartments using RNASeq approach. Our findings indicate that host-environment interactions may modulate the expression of host susceptibility proteins to SARS-CoV-2 and prime the host to manifest severe respiratory illness following SARS-CoV-2 infection.
Methods
Animal husbandry, experimental design, and ozone exposure
Seven-week-old C57BL/6 J mice were procured from Jackson Laboratory (Bar Harbor, ME). Mice were maintained in individually ventilated, hot-washed cages on a 12-h dark/light cycle. Mice were housed in polycarbonate cages and fed a regular diet and water ad libitum. All animal experimentation procedures were approved by LSU Institutional Animal Care and Use Committee (IACUC) and performed in accordance with the ethical guidelines and regulations. The authors complied with the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines. Ozone was generated by an ozone generator (TSE Systems, Chesterfield, MO), and the ozone levels were monitored by UV photometric ozone analyzer (Envia Altech Environment, Geneva, IL). Data acquisition was done through DACO monitoring software (TSE Systems, Chesterfield, MO). Control mice were kept in a chamber supplied by filtered air (Air). Animals were exposed to ozone (800 ppb; 4 h/night, 5 nights/week, for 3 weeks) or air. Further details have been published previously21.
Necropsy and tissue harvesting, micro-dissection of the extrapulmonary airways, the parenchyma, and purification of airspace macrophages
Animals were euthanized and tissues were collected for RNA isolation or histological analyses, as described previously21. Briefly, 12–16h after final exposure, the ozone- or air-exposed mice were anesthetized with an intraperitoneal injection of 2,2,2-tribromoethanol (250 mg/kg; Sigma-Aldrich, St Louis, MO). Midline laparotomy was performed to expose and severe inferior vena cava for exsanguination. Thereafter, thoracotomy was performed, and lungs were lavaged with a calculated volume (Body-weight in grams × 0.035 × 1000 = volume in µl) of ice-cold calcium- and magnesium-free Dulbecco’s phosphate-buffered saline (DPBS). Left lung lobes were formalin-fixed for immunohistochemical analyses. CD11b negative (CD11b−) BALF cells were harvested using CD11b-microbead kit, according to the manufacturer’s recommendations (Miltenyi Biotech, MA). Extrapulmonary airways and parenchymal regions of lungs were dissected, as described previously21.
Immunohistochemistry for TMPRSS2
The immunohistochemical staining for TMPRSS2 was performed using previously published procedure22–24. Briefly, formalin-fixed, paraffin-embedded 5 µm lung sections were used for immunohistochemical localization of TMPRSS2. Sections were deparaffinized with Xylene and rehydrated with graded ethanol. Heat-induced antigen-retrieval was performed using a Citrate buffer (pH 6.0). Endogenous peroxidases were quenched with 3% hydrogen peroxide (10 min at room temperature). After blocking with 3% goat serum for 30 min, sections were incubated for 2h at room temperature with rabbit polyclonal TMPRSS2 primary antibody (ab214462; Abcam Cambridge, MA). The sections were then processed using VECTASTAIN Elite ABC HRP Kit (Vector Laboratories, Burlingame, CA), followed by chromogenic substrate conversion to insoluble colored precipitate using ImmPACT NovaRED HRP substrate Kit (Vector Laboratories, Burlingame, CA). Sections were counterstained with Gill’s Hematoxylin-I, dehydrated, and coverslipped with mounting media (H-5000, Vector Laboratories, Burlingame, CA).
Western blotting for TMPRSS2
Whole lung homogenates aliquots were separated by SDS-PAGE (NuPAGE 4–12% Bis–Tris gradient gel; Life Technologies, CA) and transferred to the PVDF membrane. Rabbit polyclonal TMPRSS2 primary antibody (ab214462; Abcam Cambridge, MA) and mouse monoclonal alpha-tubulin (T5168, Sigma-Aldrich, MO) were used. Protein bands were visualized using secondary antibodies (Alexa fluor 680 Goat anti-rabbit IgG or Alexa fluor 800 Goat anti-mouse IgG) and acquired using Odyssey CLx, Imager (LI-COR, NE)25.
In situ localization of Tmprss2 mRNA
Formalin-fixed and paraffin-embedded 5 µm lung sections were used for in situ localization of Tmprss2 mRNA using RNAscope technologies, as reported previously21,23. Briefly, formalin-fixed, paraffin-embedded 5 µm lung sections were deparaffinized with Xylene and rehydrated with graded ethanol. In situ hybridization was performed using Advanced Cell Diagnostics (ACD) proprietary RNAScope Technology (ACD, Newark, CA). Predesigned transcript-specific probes were used to hybridize Tmprss2 and Ace2 transcripts in the airway sections, and an RNAScope 2.5 HD Duplex Assay Kit (ACD) was used to amplify the transcript signals.
RNA isolation and quality assessment, construction of sequencing library, RNA sequencing and gene expression analyses, and data availability
The detailed methodologies have been previously published21. The raw data have been submitted to the Gene Expression Omnibus (GEO) database. The data is available via https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE156799 . Heat maps for normalized gene expression values (Z-scores) were generated using GraphPad Prism 9.0 (GraphPad Software, La Jolla, CA).
Statistical analyses
Student’s t-test was used to determine significant differences among groups. Statistical analyses were performed using GraphPad Prism 9.0 (GraphPad Software, La Jolla, CA). All data were expressed as mean ± standard error of the mean (SEM). A p-value < 0.05 was considered statistically significant.
Results and discussion
TMPRSS2 is essential for the proteolytic priming of viral spike (S) protein of the SARS-CoV-2 following its binding to host receptor, ACE2. In fact, a recent study elegantly demonstrated that host cell entry of SARS-CoV-2 can be blocked by a clinically proven inhibitor of TMPRSS2 indicating the critical importance of TMPRSS2 in determining SARS-CoV-2 infectivity2. Therefore, the host susceptibility to SARS-CoV-2 could vary based on the expression of the host susceptibility proteins including ACE2 and TMPRSS23–5. Individuals have varied susceptibility to SARS-CoV-2 that may be dependent on various factors including air pollution10,11. Air pollution levels correlate strongly with increased morbidity and mortality due to SARS-CoV-226–28. While it is already known that the unhealthy levels of ozone, one of the 6 criteria air pollutants, increase the risk for developing cardiopulmonary health problems14–20, it is unclear whether the ambient ozone regulates the levels of expression of host susceptibility proteins to SARS-CoV-2 and in turn accounts, in part, for the varied susceptibilities of the human population to SARS-CoV-2. Therefore, we sought to test the hypothesis that ozone induces the expression of TMPRSS2 in lung tissue.
As compared to the filtered air-exposed mice, the ozone-exposed mice had significantly elevated levels of TMPRSS2 protein in the whole lung lysate (Fig. 1A and B; Supplemental Fig. 1). These results demonstrate that ozone exposure increases the expression of TMPRSS2 protein in the lungs of mice. Next, we performed immunohistochemical staining on lung sections to visualize cell-specific localization of TMPRSS2. Interestingly, while the TMPRSS2 staining was evident in the airway epithelium and alveolar macrophages from filtered air-exposed mice, the staining intensity was remarkably increased in the airway epithelial cells, alveolar epithelial cells, and alveolar macrophages from ozone-exposed mice (Fig. 1C). These results clearly demonstrate that ozone increases the expression of TMPRSS2 in the lung tissue in a cell-specific manner.
Figure 1.
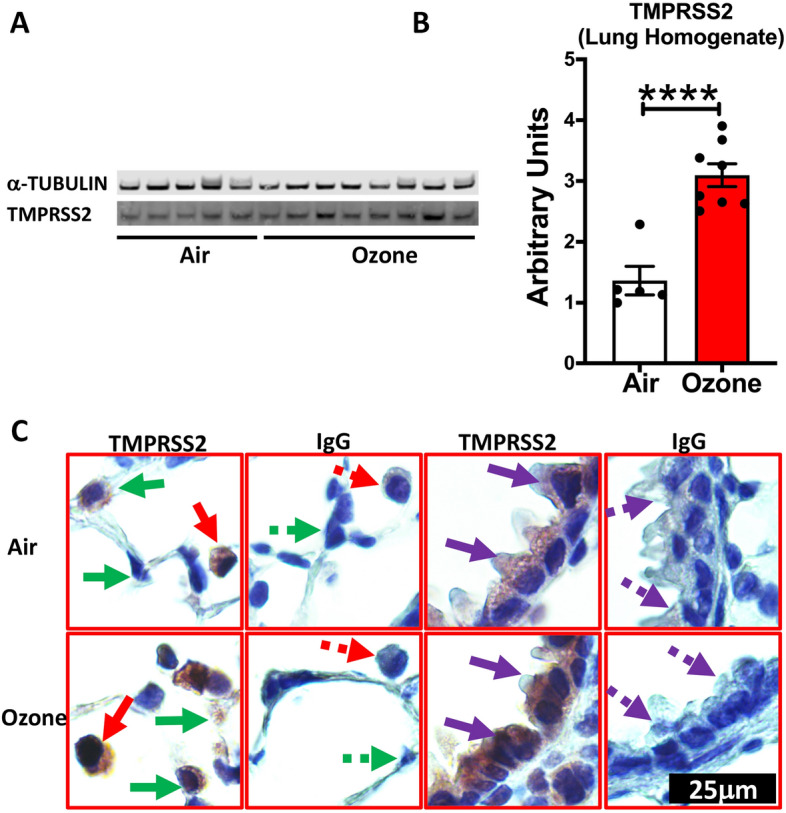
TMPRSS2 protein expression is upregulated in the lungs of ozone -exposed mice. Western blot representative gel image (A) showing bands for TMPRSS2 protein and alpha-tubulin loading control, and band intensity analyses (B, Bar Graph) on the whole lung homogenate from air- and ozone-exposed mice (n = 6–8). (C) Immunohistochemical staining for TMPRSS2 in macrophages (solid red arrow), alveolar epithelial cells (solid green arrow), and bronchiolar epithelial cells (solid purple arrow). Negatively stained cells are indicated by dotted arrows in lung sections that were incubated with antibody (IgG) control. All images were captured at the same magnification.
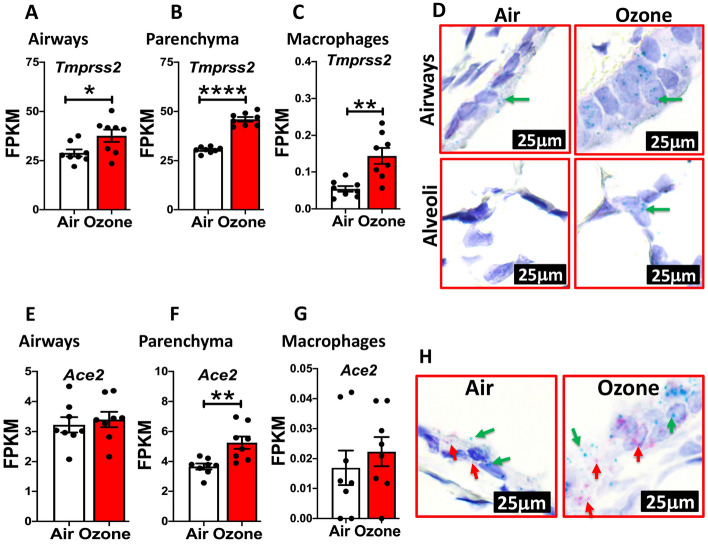
In order to test whether TMPRSS2 protein expression correlates with mRNA expression in a cell-specific manner, we analyzed RNASeq dataset from extrapulmonary airways (Fig. 2A), parenchyma (Fig. 2B), and alveolar macrophages (Fig. 2C) from filtered air- and ozone-exposed mice for Tmprss2 transcript levels. As expected, the fragment per kilobase per million mapped (FPKM) reads for Tmprss2 were significantly upregulated in all the three tissue compartments in ozone-exposed as compared to filtered air-exposed mice. We further confirmed these findings for cell-specificity in ozone-exposed airways and alveoli using RNAscope-based in situ hybridization. This assay also showed significantly increased signals for Tmprss2 transcripts in both the airway epithelial cells and the alveolar epithelial cells of ozone-exposed compared to filtered air-exposed mice (Fig. 2D). These data suggest that the changes in the protein levels of TMPRSS2 are a result of changes at the level of gene expression indicating that ozone directly or indirectly differentially regulates the gene expression of Tmprss2.
Figure 2.
Tmprss2 mRNA expression is upregulated in the lungs of ozone -exposed mice. FPKM values obtained from RNAseq data set obtained from airways (A), parenchyma (B), and alveolar macrophages (C) were used to quantify relative expression levels of Tmprss2 mRNA in air- versus ozone-exposed mice (n = 8). The data are expressed as mean (± SEM). Student’s t-Test; *P < 0.05, **P < 0.01, ****P < 0.0001. (D) RNAscope-based in situ hybridization for Tmprss2 transcripts (green dots representing punctate staining for Tmprss2 mRNA in airway epithelial cells (Top) and alveolar epithelial cells (bottom) in air- (left) and ozone-exposed (right) mice. All images were captured at the same magnification. FPKM values obtained from RNAseq data set obtained from airways (E), parenchyma (F), and alveolar macrophages (G) were used to quantify relative expression levels of Ace2 mRNA in air- versus ozone-exposed mice (n = 8). The data are expressed as means (± SEM). Student’s t-Test ** P < 0.01. (H) Duplex RNAscope-based in situ hybridization for Tmprss2 (green puncta, green arrow) and Ace2 (red puncta, red arrow) in the airways of air- (left) and ozone-exposed (right) mice. All images were captured at the same magnification.
Next, to test whether ozone exposure also alters the expression of Ace2 transcripts, we analyzed RNASeq dataset from extrapulmonary airways (Fig. 2E), parenchyma (Fig. 2F), and alveolar macrophages (Fig. 2G) from filtered air- and ozone-exposed mice for Ace2 transcript levels. While the FPKM reads for Ace2 were significantly upregulated in the parenchyma compartment (Fig. 2F) of ozone-exposed mice, the expression levels of Ace2 in ozone-exposed extrapulmonary airways (Fig. 2E) and macrophage compartments (Fig. 2G) were not significantly different from filtered air-exposed mice. In situ hybridization revealed increased staining for Ace2 transcripts in the airways of ozone-exposed mice (Fig. 2H). These data suggest that the ozone exposure results in a parallel increase in the expression of the two most critical SARS-CoV-2 host susceptibility genes.
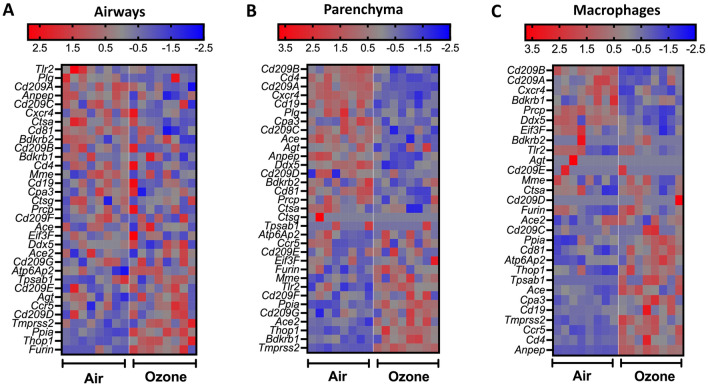
Finally, to determine the effect of ozone exposure on the expression levels of genes known to be involved in the host response to SARS-CoV-2, we compared the normalized z-scores of 33 known genes associated with host susceptibility to SARS-CoV-229–37. Of the three tissues analyzed, while only four of the total 33 genes, i.e., Furin, Thop1, Ppia, and Tmprss2, were significantly increased in the extrapulmonary airways of ozone-exposed versus filtered air-exposed mice (Fig. 3A); in the lung parenchyma, nearly half of the host susceptibility genes were upregulated while the rest half were downregulated in ozone-exposed versus filtered air-exposed mice (Fig. 3B). In contrast to the extrapulmonary airways and the lung parenchyma, a major proportion of the host susceptibility genes including those of Tmprss2, Ace, Anpep, Cd4, and Ccr5 were significantly upregulated in the CD11b- lung macrophages from ozone-exposed versus filtered air-exposed mice (Fig. 3C) indicating a large effect of ozone exposure on SARS-CoV-2 host susceptibility genes in CD11b− lung macrophages.
Figure 3.
Heat maps depicting normalized gene expression values (Z-scores) of genes associated with SARS-CoV-2 host susceptibility in airways (A), parenchyma (B), and alveolar macrophages (C) from air- and ozone-exposed mice.
In terms of cell-specific responses, our data clearly indicate the upregulated expression of Tmprss2 in alveolar macrophages. In addition to the alveolar macrophages, the interstitial macrophages may also exhibit similar expression trends for Tmprss2 and other host susceptibility genes. While the interstitial macrophages are anatomically restricted to the interstitial spaces, the recently recruited inflammatory macrophages in SARS-CoV-2-infected rhesus macaques have been shown to exhibit interstitial cell-specific surface marker expression38. In addition, ozone exposure also induces the recruitment of CD11b + exudative macrophages into the lungs of mice39. In our study, since we selectively depleted CD11b + immune cells, the data presented in the current study reflect upregulated expression of Tmprss2 and other host susceptibility genes specifically in CD11b− macrophages. It is quite possible that these inflammatory CD11b + macrophages may have even more exaggerated upregulation of Tmprss2 and other host susceptibility genes. Within the parenchyma, in addition to the retained alveolar macrophages, a variety of immune cell types including myeloid cells such as neutrophils, eosinophils, classical dendritic cells and basophils, and adaptive immune cells such as B and T lymphocytes, and innate lymphoid cells such as ILC2 might have also contributed to the overall increase in the expression of Tmprss2 and other host susceptibility genes including Ace2. For instance, CD4 + T cells, but not CD8 + T cells are also known to express Tmprss2 and Ace2 and these two host susceptibility proteins act in concert with CD4 to mediate SARS-CoV-2 infection40. Whether ozone exposure upregulates expression of Tmprss2 and other host susceptibility genes in immune cells other than alveolar macrophages remains unexplored and requires further cell-specific transcriptomic and proteomic studies.
This study also has some limitations. First, mice are not susceptible to SARS-CoV-2 infection because SARS-CoV-2 is incapable of using murine ortholog of ACE2 for entering the host cells41,42. Therefore, we could not directly test the effect of ozone exposure on SARS-CoV-2 infectivity in mice. Future studies focused on the effect of ozone exposure on humanized ACE2 (hACE2) mice are essential. Second, the effect of ozone exposure on the expression of host susceptibility proteins in aged mice remains unexplored. Since elderly and older adults are high-risk patients for developing severe SARS-CoV-2 infection43,44, further investigations on three-dimensional relationship between ozone pollution, old age, and SARS-CoV-2 infectivity are warranted.
In conclusion, this study suggests a possible role of the host-environment (ozone pollution) interactions in modulating the susceptibility of the human population to SARS-CoV-2. Since TMPRSS2 is essential for the proteolytic processing of several coronaviruses including SARS-CoV-1, MERS, as well as influenza A virus45,46, our findings have implications beyond SARS-CoV-2 infections. Taken together, this study presents a novel finding that will have a significant and immediate impact on our understanding of the pathogenesis and epidemiology of the SARS-CoV-2 pandemic.
Supplementary Information
Acknowledgements
We thank Sherry Ring for assistance with histological tissue processing. We acknowledge the GeneLab for providing core services and access to Bioinformatics software. The GeneLab is supported by Institutional Development Awards (IDeA) from the National Institute of General Medical Sciences (Grants P20 GM103424 and P20 GM130555).
Author contributions
Y.S. and S.P. conceived and designed the research; T.V. performed the RNA in situ hybridization assays; K.P. performed the immunohistochemical staining; I.C. performed immunoblotting assays. Y. S., T.V., K.P., and I.C., maintained the animal colony, performed ozone exposures, and conducted animal necropsies. S.P. and Y.S. analyzed the histopathological, immunohistochemical, and RNA in situ hybridization data and wrote and reviewed the manuscript for intellectual contents.
Funding
This work was supported by the National Institute of Environmental Health Sciences Grant R01ES030125 (to Y.S.).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Thao Vo, Kshitiz Paudel and Ishita Choudhary.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-04906-8.
References
- 1.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–80 e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ragia G, Manolopoulos VG, Munoz MNM. Inhibition of SARS-CoV-2 entry through the ACE2/TMPRSS2 pathway: A promising approach for uncovering early COVID-19 drug therapies. Eur. J. Clin. Pharmacol. 2020;76(12):1623–1630. doi: 10.1007/s00228-020-02963-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterjee SK, Saha S, Munoz MNM. Molecular pathogenesis, immunopathogenesis and novel therapeutic strategy against COVID-19. Front. Mol. Biosci. 2020;7:196. doi: 10.3389/fmolb.2020.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradding P, Richardson M, Hinks TSC, Howarth PH, Choy DF, Arron JR, Wenzel SE, Siddiqui S. ACE2, TMPRSS2, and furin gene expression in the airways of people with asthma-implications for COVID-19. J. Allergy Clin. Immunol. 2020;146:208–211. doi: 10.1016/j.jaci.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lingappan K, Karmouty-Quintana H, Davies J, Akkanti B, Harting MT. Understanding the age divide in COVID-19: Why are children overwhelmingly spared? Am. J. Physiol. Lung Cell. Mol. Physiol. 2020;319:L39–L44. doi: 10.1152/ajplung.00183.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inde Z. et al Age-dependent regulation of SARS-CoV-2 cell entry genes and cell death programs correlates with COVID-19 disease severity. bioRxiv (2020). [DOI] [PMC free article] [PubMed]
- 8.Di Stadio A, Della Volpe A, Ralli M, Ricci G. Gender differences in COVID-19 infection. The estrogen effect on upper and lower airways can it help to figure out a treatment? Eur. Rev. Med. Pharmacol. Sci. 2020;24:5195–6. doi: 10.26355/eurrev_202005_21298. [DOI] [PubMed] [Google Scholar]
- 9.Di Sanyaolu A, et al. Comorbidity and its impact on patients with COVID-19. SN. Compr. Clin. Med. 2020 doi: 10.1007/s42399-020-00363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu X. et al. Exposure to air pollution and COVID-19 mortality in the United States: A nationwide cross-sectional study. medRxiv (2020).
- 11.Liang D, Shi L, Zhao J, Liu P, Sarnat JA, Gao S, Schwartz J, Liu Y, Ebelt ST, Scovronick N, Chang HH. Urban air pollution may enhance COVID-19 case-fatality and mortality rates in the United States. Innovation (N Y) 2020;1(3):100047. doi: 10.1016/j.xinn.2020.100047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lange SS, Mulholland SE, Honeycutt ME. What are the net benefits of reducing the ozone standard to 65 ppb? An alternative analysis. Int. J. Environ. Res. Publ. Health. 2018;15(8):1586. doi: 10.3390/ijerph15081586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dionisio KL, Nolte CG, Spero TL, Graham S, Caraway N, Foley KM, Isaacs KK. Characterizing the impact of projected changes in climate and air quality on human exposures to ozone. J. Expo. Sci. Environ. Epidemiol. 2017;27:260–270. doi: 10.1038/jes.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raza A, Dahlquist M, Jonsson M, Hollenberg J, Svensson L, Lind T, Ljungman PLS. Ozone and cardiac arrest: The role of previous hospitalizations. Environ. Pollut. 2019;245:1–8. doi: 10.1016/j.envpol.2018.10.042. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Wu S, Pan L, Xu J, Shan J, Yang X, Dong W, Deng F, Chen Y, Shima M, Guo X. Short-term effects of various ozone metrics on cardiopulmonary function in chronic obstructive pulmonary disease patients: Results from a panel study in Beijing China. Environ. Pollut. 2018;232:358–366. doi: 10.1016/j.envpol.2017.09.030. [DOI] [PubMed] [Google Scholar]
- 16.Koman PD, Mancuso P. Ozone exposure, cardiopulmonary health, and obesity: A substantive review. Chem. Res. Toxicol. 2017;30:1384–1395. doi: 10.1021/acs.chemrestox.7b00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dye JA, Ledbetter AD, Schladweiler MC, Costa DL, Kodavanti UP. Whole body plethysmography reveals differential ventilatory responses to ozone in rat models of cardiovascular disease. Inhal. Toxicol. 2015;27(Suppl 1):14–25. doi: 10.3109/08958378.2014.954167. [DOI] [PubMed] [Google Scholar]
- 18.Dye JA, Costa DL, Kodavanti UP. Executive summary: Variation in susceptibility to ozone-induced health effects in rodent models of cardiometabolic disease. Inhal. Toxicol. 2015;27(Suppl 1):105–115. doi: 10.3109/08958378.2014.995388. [DOI] [PubMed] [Google Scholar]
- 19.Day DB, Xiang J, Mo J, Li F, Chung M, Gong J, Weschler CJ, Ohman-Strickland PA, Sundell J, Weng W, Zhang Y, Zhang JJ. Association of ozone exposure with cardiorespiratory pathophysiologic mechanisms in healthy adults. JAMA Intern. Med. 2017;177:1344–1353. doi: 10.1001/jamainternmed.2017.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balmes JR. Long-term exposure to ozone and cardiopulmonary mortality: Epidemiology strikes again. Am. J. Respir. Crit. Care Med. 2019;200:958–959. doi: 10.1164/rccm.201906-1105ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choudhary I, Vo T, Paudel K, Patial S, Saini Y. Compartment-specific transcriptomics of ozone-exposed murine lungs reveals sex and cell type-associated perturbations relevant to mucoinflammatory lung diseases. Am. J. Physiol. Lung Cell Mol. Physiol. 2020;320(1):L99–L125. doi: 10.1152/ajplung.00381.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis BW, Choudhary I, Paudel K, Mao Y, Sharma R, Wang Y, Deshane JS, Boucher RC, Patial S, Saini Y. The innate lymphoid system is a critical player in the manifestation of mucoinflammatory airway disease in mice. J. Immunol. 2020;205:1695–1708. doi: 10.4049/jimmunol.2000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis BW, Vo T, Choudhary I, Kidder A, Bathula C, Ehre C, Wakamatsu N, Patial S, Saini Y. Ablation of IL-33 suppresses Th2 responses but is accompanied by sustained mucus obstruction in the scnn1b transgenic mouse model. J. Immunol. 2020;204:1650–1660. doi: 10.4049/jimmunol.1900234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis BW, Sultana R, Sharma R, Noel A, Langohr I, Patial S, Penn AL, Saini Y. Early postnatal secondhand smoke exposure disrupts bacterial clearance and abolishes immune responses in muco-obstructive lung disease. J. Immunol. 2017;199:1170–1183. doi: 10.4049/jimmunol.1700144. [DOI] [PubMed] [Google Scholar]
- 25.Saini Y, Dang H, Livraghi-Butrico A, Kelly EJ, Jones LC, O'Neal WK, Boucher RC. Gene expression in whole lung and pulmonary macrophages reflects the dynamic pathology associated with airway surface dehydration. BMC Gen. 2014;15:726. doi: 10.1186/1471-2164-15-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woodby B, Arnold MM, Valacchi G. SARS-CoV-2 infection, COVID-19 pathogenesis, and exposure to air pollution: What is the connection? Ann. N. Y. Acad. Sci. 2020;1468(1):15–38. doi: 10.1111/nyas.14512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Villeneuve PJ, Goldberg MS. Methodological considerations for epidemiological studies of air pollution and the SARS and COVID-19 coronavirus outbreaks. Environ. Health Perspect. 2020;128:95001. doi: 10.1289/EHP7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Domingo JL, Marques M, Rovira J. Influence of airborne transmission of SARS-CoV-2 on COVID-19 pandemic. Rev. Environ. Res. 2020;188:109861. doi: 10.1016/j.envres.2020.109861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kucher AN, Babushkina NP, Sleptcov AA, Nazarenko MS. Genetic control of human infection with SARS-CoV-2. Russ. J. Genet. 2021;57:627–641. doi: 10.1134/S1022795421050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng M, Karki R, Williams EP, Yang D, Fitzpatrick E, Vogel P, Jonsson CB, Kanneganti TD. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 2021;22:829–838. doi: 10.1038/s41590-021-00937-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ni W, Yang X, Yang D, Bao J, Li R, Xiao Y, Hou C, Wang H, Liu J, Yang D, Xu Y, Cao Z, Gao Z. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care. 2020;24:422. doi: 10.1186/s13054-020-03120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson BA, Xie X, Bailey AL, Kalveram B, Lokugamage KG, Muruato A, Zou J, Zhang X, Juelich T, Smith JK, Zhang L, Bopp N, Schindewolf C, Vu M, Vanderheiden A, Winkler ES, Swetnam D, Plante JA, Aguilar P, Plante KS, Popov V, Lee B, Weaver SC, Suthar MS, Routh AL, Ren P, Ku Z, An Z, Debbink K, Diamond MS, Shi PY, Freiberg AN, Menachery VD. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature. 2021;591:293–299. doi: 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao H, Xu LH, Yamada Y, Liu DX. Coronavirus spike protein inhibits host cell translation by interaction with eIF3f. PLoS One. 2008;3:e1494. doi: 10.1371/journal.pone.0001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silva-Aguiar RP, Peruchetti DB, Rocco PRM, Schmaier AH, PMR ES, Martins MA, Carvalho VF, Pinheiro AAS, Caruso-Neves C. Role of the renin-angiotensin system in the development of severe COVID-19 in hypertensive patients. Am. J. Physiol. Lung Cell Mol. Physiol. 2020;319:L596–L602. doi: 10.1152/ajplung.00286.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neidleman J, Luo X, George AF, McGregor M, Yang J, Yun C, Murray V, Gill G, Greene WC, Vasquez J, Lee SA, Ghosn E, Lynch KL, Roan NR. Distinctive features of SARS-CoV-2-specific T cells predict recovery from severe COVID-19. Cell Rep. 2021;36:109414. doi: 10.1016/j.celrep.2021.109414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LoPresti M, Beck DB, Duggal P, Cummings DAT, Solomon BD. The role of host genetic factors in coronavirus susceptibility: Review of animal and systematic review of human literature. Am. J. Hum. Genet. 2020;107:381–402. doi: 10.1016/j.ajhg.2020.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amraei R, Yin W, Napoleon MA, Suder EL, Berrigan J, Zhao Q, Olejnik J, Chandler KB, Xia C, Feldman J, Hauser BM, Caradonna TM, Schmidt AG, Gummuluru S, Muhlberger E, Chitalia V, Costello CE, Rahimi N. CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2. ACS Cent. Sci. 2021;7:1156–1165. doi: 10.1021/acscentsci.0c01537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Upadhyay, A.A. et al. TREM2+ and interstitial macrophages orchestrate airway inflammation in SARS-CoV-2 infection in rhesus macaques. bioRxiv (2021). [DOI] [PMC free article] [PubMed]
- 39.Tighe RM, Li Z, Potts EN, Frush S, Liu N, Gunn MD, Foster WM, Noble PW, Hollingsworth JW. Ozone inhalation promotes CX3CR1-dependent maturation of resident lung macrophages that limit oxidative stress and inflammation. J. Immunol. 2011;187:4800–4808. doi: 10.4049/jimmunol.1101312. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 40.Gustavo, G. et al. View ORCID ProfileAlessandro S. Farias: SARS-CoV-2 uses CD4 to infect t helper lymphocytes. medRxiv 10.1101/2020.09.25.20200329 (2020).
- 41.Zhao Y, Zhao Z, Wang Y, Zhou Y, Ma Y, Zuo W. Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020;202:756–759. doi: 10.1164/rccm.202001-0179LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nanda A, Vura N, Gravenstein S. COVID-19 in older adults. Aging Clin. Exp. Res. 2020;32:1199–1202. doi: 10.1007/s40520-020-01581-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perrotta F, Corbi G, Mazzeo G, Boccia M, Aronne L, D'Agnano V, Komici K, Mazzarella G, Parrella R, Bianco A. COVID-19 and the elderly: Insights into pathogenesis and clinical decision-making. Aging Clin. Exp. Res. 2020;32:1599–1608. doi: 10.1007/s40520-020-01631-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hatesuer B, Bertram S, Mehnert N, Bahgat MM, Nelson PS, Pohlmann S, Schughart K. Tmprss2 is essential for influenza H1N1 virus pathogenesis in mice. PLoS Pathog. 2013;9:e1003774. doi: 10.1371/journal.ppat.1003774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iwata-Yoshikawa N, Okamura T, Shimizu Y, Hasegawa H, Takeda M, Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J. Virol. 2019;93:e01815–18. doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.