Abstract
The rise of single-molecule localization microscopy (SMLM) and related super-resolution methods over the past 15 years has revolutionized how we study biological and materials systems. In this perspective, we reflect on the underlying philosophy of how diffraction-unlimited pictures containing rich spatial and functional information may gradually emerge through the local accumulation of single-molecule measurements. Starting with the basic concepts, we analyze the uniqueness and opportunities in building up the final picture one molecule at a time. Then, after brief introductions to the more established multicolor and three-dimensional measurements, we highlight emerging efforts to extend SMLM to new dimensions and functionalities as fluorescence polarization, emission spectra, and molecular motions, and discuss rising opportunities and future directions. With single molecules as our quanta, the bottom-up accumulation approach provides a powerful conduit for multidimensional microscopy at the nanoscale.
Graphical Abstract
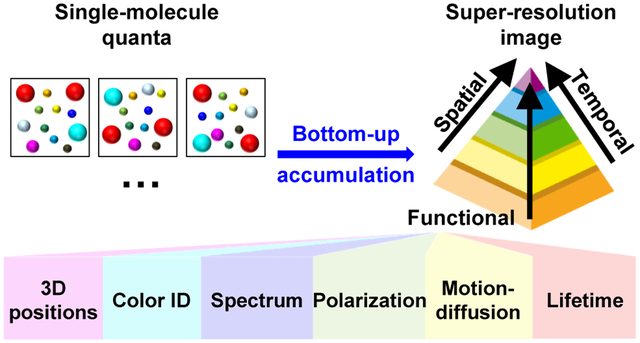
Building up diffraction-unlimited pictures one molecule at a time
The double-slit experiment offers a striking demonstration of the mystery of quantum mechanics:1,2 as single particles (quanta), e.g., photons or electrons, are allowed to pass through one at a time, even though each quantum offers minimal information, accumulation of the positions of many independently measured quanta gradually gives rise to the underlying picture from the bottom up, with continuously improving signal-to-noise ratio over time.
It is thus remarkable that over the past ~15 years, an analogous process overcomes the diffraction-limited resolution barrier of microscopy to reach ~10 nm spatial resolutions, but here the quanta are single molecules. In these approaches, which we collectively discuss as single-molecule localization microscopy (SMLM), single fluorescent molecules sparsely emitting in the wide field, e.g., as a small subset of the probes labeled to an intracellular structure, are recorded with a high-sensitivity camera (Figure 1a). As the single-molecule images are well isolated from each other, they can be separately evaluated for their super-resolved positions (at ~10 nm precision) and other properties (Figure 1b) over a minimal background. Although the resultant, few-molecule positions and additional parameters appear insignificant on their own, a fluorescence on-off switching mechanism is introduced to allow different, random subsets of the labeled probes to be detected over different camera frames (Figure 1c). Thus, by accumulating the results of a large amount (often >105) of molecules over many (often >104) frames, a super-resolved image, potentially with additional high-dimensional information, gradually emerges (Figure 1e) over the time scale of minutes. This SMLM scheme is first achieved through the photoswitching of fluorescent dyes (STORM3) and fluorescent proteins (FPs) [(F)PALM4,5]. The reversible binding of fluorescent probes to the target (PAINT6) provides another powerful strategy.
Figure 1.
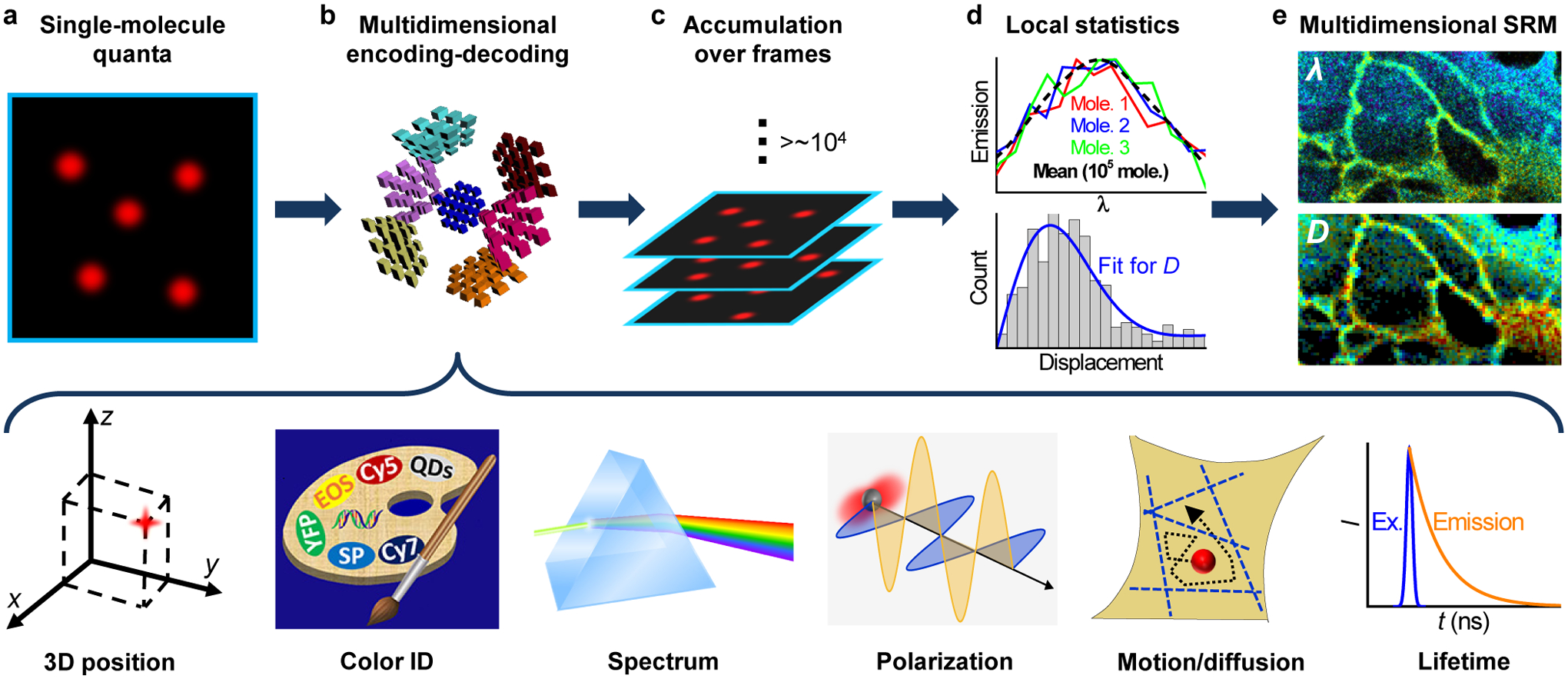
A bottom-up approach toward multidimensional super-resolution microscopy. (a) Emitting single molecules are kept at a low density in the wide field, so that they could be each independently evaluated for their nanoscale positions and high-dimensional properties. (b) Examples of multidimensional single-molecule observables that may be encoded-decoded, including localization in 3D, color identity, spectrum, fluorescence polarization, motion and diffusion, and fluorescence lifetime. (c) Stochastic sampling of single molecules over many camera frames, e.g., through photoswitching or diffusional exchange, to enable accumulation of the single-molecule quanta. (d) The accumulated single-molecule measurements enable local statistics to extract meaningful parameters at the nanoscale. (e) Resultant multidimensional super-resolution images, with the possibility to integrate measurements of different dimensions. The palette scheme in (b) is adapted from ref120.
Whereas many excellent review articles have discussed the glorious resolution and other technical aspects of SMLM,7–12 as well as the broader field of super-resolution microscopy (aka “nanoscopy”),13–15 in this perspective we reflect on the underlying philosophy of how high-resolution, potentially multidimensional pictures gradually emerge through the local accumulation of single-molecule measurements carried out at low densities. In particular, whereas earlier efforts on SMLM focus on achieving multicolor capabilities and resolving the three-dimensional (3D) positions, we highlight recent work that generalizes SMLM by locally accumulating new dimensionalities of the single-molecule signal, including fluorescence polarization, emission spectra, and transient displacements, which enables the construction of functional maps of varying new parameters at the super-resolution level.
The single-molecule quanta: uniqueness and opportunities
The above-described mechanism to generate high-resolution images through single-molecule accumulation challenges the mindset of traditional microscopy. With typical top-down approaches, including super-resolution methods as stimulated emission depletion (STED) microscopy,16 high resolution is achieved by shrinking the effective detection region to the smallest possible size; resolution is defined by this size, and the signal is the ensemble sum of all fluorophores in the detection region.
In contrast, SMLM works from the bottom up: it imposes no spatial confinement in illumination or detection, but utilizes the sparsity of single molecules in the wide field to enable their individual super-localization at the nanoscale (Figure 1a), and builds up the final image through accumulating the single-molecule quanta over time (Figure 1c). Resolution is now defined by how precisely one can determine the position of each molecule;17,18 whereas this trait does limit SMLM to systems in which single molecules can be detected with adequate signal-to-noise ratios, very high resolutions, down to a few nanometers, can be achieved using very bright fluorophores.19
Meanwhile, the sparsity of molecules in the wide-field implies that each molecule can be measured and analyzed independently. Excellent opportunities thus arise for the parallel detection of diverse single-molecule properties (Figure 1b), including the three-dimensional positions, fluorescence polarization, emission spectra, and molecular motions, for multiple molecules in the same camera frame. The fluorescence on-off switching mechanism, then, provides a convenient way to stochastically sample different molecules over many frames (Figure 1c), thus versatile measurements of single-molecule properties at ultrahigh throughputs.
Together, as the achieved single-molecule measurements offer the ultimate sensitivity and resolution for fluorescence detection, the rapid accumulation of massive amounts of molecules with super-localized positions enables local statistical analysis (Figure 1d) to overcome the potentially low signal-to-noise ratio (e.g., for spectra) and the stochastic nature of single-molecule behavior (e.g., for molecular diffusion). High-resolution images can thus be constructed with valuable insights into diverse local structural and functional properties (Figure 1e), as analyzed in detail below.
Technically, it is further helpful to comment on the unique data format of SMLM due to its single-molecule origin, a topic under-discussed in the literature. Traditional digital micrographs record pixelated images, i.e., bitmaps presenting intensities in a two-dimensional array. Although the pixel-based format is readily stored on a computer, it does not well accommodate image operations as subpixel shifting, non-integral scaling, non-90° rotations, and shearing. SMLM, in contrast, amasses a list containing the positions (coordinates) and other possible attributes like brightness, single-molecule image shape, and spectrum, of all the detected molecules. The length of this list is proportional to the number of molecules detected in the experiment, rather than defined by the size of the view. Moreover, although the raw single-molecule data, as recorded by the camera, are pixelated, the super-localized single-molecule positions take continuous values that depend little on pixelation.17 We thus enter an interesting regime in which each molecule reports a quantized description of the underlying structure in a continuous coordinate system.
The above “point-cloud” data format of SMLM affords exceptional versatilities for image processing and data mining. For example, subpixel image drifts, which often occur during SMLM data acquisition, are readily removed by shifting the molecular coordinates in different frames. Meanwhile, no distortion or loss of information is expected in SMLM for image operations like scaling, rotation, and projection. Consequently, it becomes straightforward to translate the SMLM data between different coordinates. This feature is particularly helpful for matching and comparing different datasets, both for multi-view referencing (below) and for correlation with other microscopy methods.20 For the final presentation of results, the continuous spatial coordinates and the near-continuous (1 over >104 frames) timestamps further offer great flexibility in how to spatiotemporally divide the detected molecules to best define structures and extract local functional parameters. The resultant tradeoffs between spatial resolution, temporal resolution, and sensitivity are discussed below.
The multicolor and 3D challenges
As SMLM builds up images by accumulating single molecules, to achieve multicolor (multi-probe) SMLM necessitates the correct identification of each detected molecule. This goal is first realized in STORM through the controlled photoactivation of the same emitter dye paired with different activator dyes.21 As fluorescence emission is from a single emitter dye, this approach precludes chromatic aberration between color channels. On the flip side, non-specific activation leads to high color crosstalks. In contrast, sequential SMLM of spectrally well-separated probes22,23 achieves lower crosstalk. However, the typical filter-switching scheme is slow, and alignment between channels is difficult when convolved with sample drift. Exciting with different lasers using a multi-band filter set24,25 overcomes these limitations. Still, the use of fluorophores of substantially different spectra incurs chromatic aberrations, and different probes may not achieve the best performance in the same imaging buffer. These issues are partly addressed with the ratiometric and spectrally resolved approaches below. Separately, multicolor SMLM may also be achieved by reusing the same fluorophore via the sequential labeling-imaging of different targets,26,27 but at the expense of time.
For 3D imaging, the single-molecule origin of SMLM similarly demands the axial position (depth z) of every detected molecule. A common approach is to encode z into the shape of the single-molecule image, or effectively, the point spread function (PSF) of the microscope system. In 3D-STORM,28 a cylindrical lens is simply inserted into the optical path to induce astigmatism, i.e., a shift in the focal plane for two orthogonal directions, so that the PSF elongates in opposite directions for molecules below and above the focal plane, thus enabling depth encoding/decoding. Meanwhile, numerous efforts have created other depth-encoded PSFs (“PSF engineering”) to enable extended working z ranges and other functionalities.29
Multi-view referencing for 3D, multicolor, and polarization SMLM
In addition to encoding into the PSF shape, comparing the behavior of the same molecules in carefully designed, complementary views offers another powerful means to extract single-molecule properties, a strategy we discuss here collectively as multi-view referencing.
As SMLM, partly owing to its stochastic on-off switching mechanism, often yields a broad distribution of photon counts between molecules, it is difficult to directly extract useful information from the absolute single-molecule brightness. By referring between complementary views, the relative (ratiometric) brightness becomes relevant. Meanwhile, alterations in single-molecule position and image shape can be further engineered into the different views. Technically, as discussed above, the point-cloud data type of SMLM is ideal for mapping molecules between different coordinates. Mismatches in finding the same molecules across different views are further minimal, considering that with the typical sparsity condition of SMLM, less than one molecule is detected within the diffraction limit in each frame.
For 3D-SMLM, biplane FPALM30 thus employs a beamsplitter to create a pair of views with shifted focal planes (e.g., green box in Figure 2a). Depth information is thus decoded by comparing the images of the same single molecules recorded in the two views (Figure 2b)30,31. The use of a secondary objective lens at the backside of the sample offers another way to obtain two views. Besides simply combining these two views with the above cylindrical-lens scheme,32 more sophisticated 3D approaches contrive single-molecule interferometry (e.g., Figure 2e): comparison of the single-molecule intensities,33 plus PSF shapes,34,35 in the resultant multiple interferometry views (e.g., Figure 2f) helps determine z with outstanding precisions.
Figure 2.
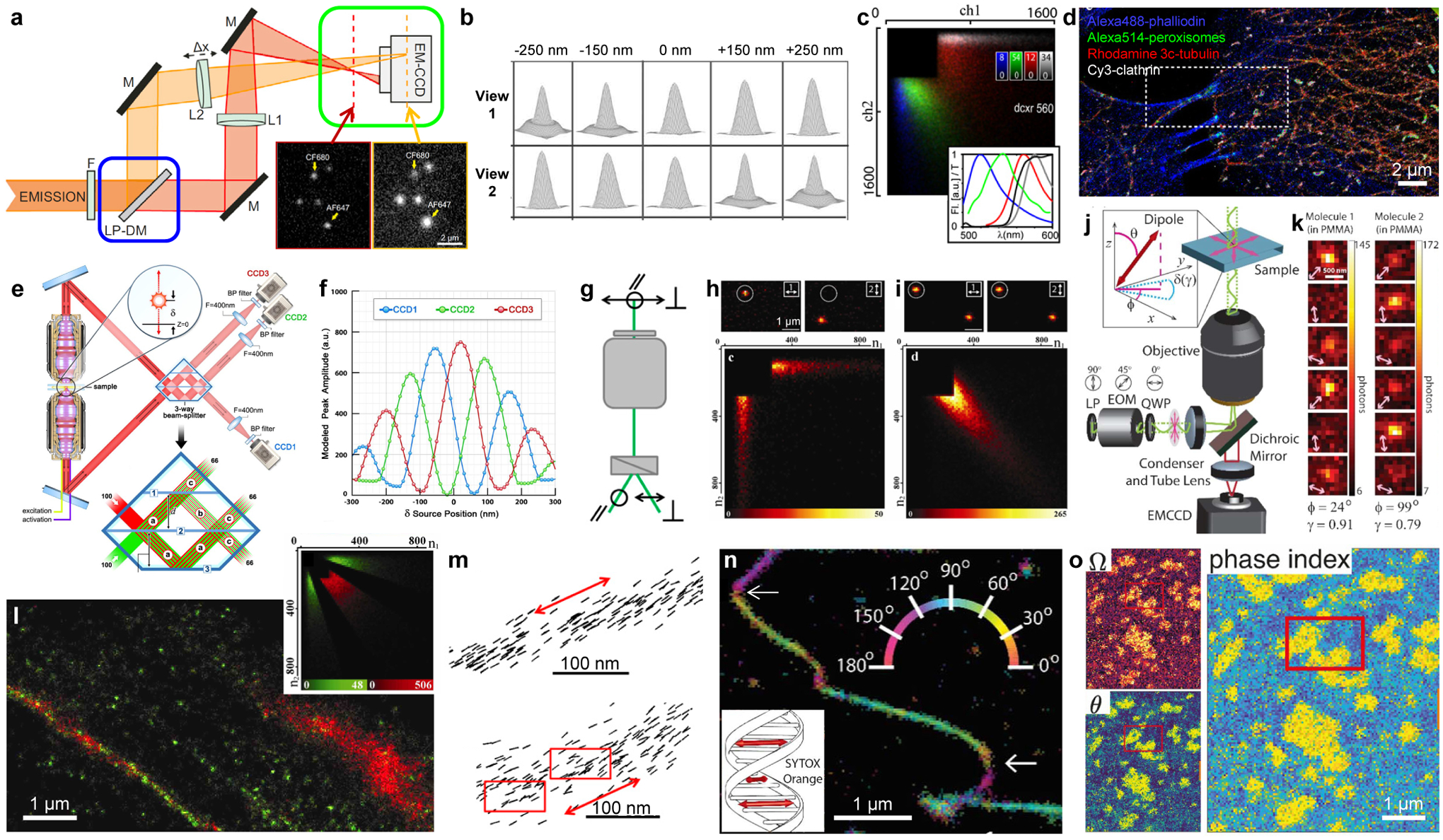
Multi-view referencing for 3D, multicolor, and polarization SMLM. (a) Integration of biplane 3D imaging (green box: shifted focal planes between two views) and ratiometric color detection (blue box: fluorescence split into long- and short-wavelength components). LP-DM: long-pass dichroic mirror. Inset: single-molecule images obtained in the two views. (b) Simulated images of a point source at different axial positions for two views with a 500-nm focal shift. (c) Density heatmaps of photon counts recorded in the long- and short-wavelength channels, for individual molecules of four different dyes with their emission spectra (colored curves) and the transmission of the dichroic mirror (black curve) shown in the inset. (d) Four-color SMLM of a fixed cell by separating the four probes based on (c). (e) 3D-SMLM based on multiphase interference between fluorescence collected from two opposing objective lenses. (f) Expected brightness detected by the three cameras in (e) for single molecules at different axial positions. (g) Splitting the fluorescence into two orthogonal polarizations. (h) (top) Fluorescence images of single pcRhB molecules in PMMA, recorded in two channels of orthogonal polarizations. (bottom) Density heatmaps of photon counts recorded in the two channels for different single molecules. (i) Same as (h), but for pcRhB in mowiol. (j) An electro-optic modulator (EOM) rotates the linear polarization direction of the excitation laser in consecutive frames. (k) Resultant images of two rhodamine 101 molecules in PMMA, showing dissimilar changes in brightness in consecutive frames. (l) Classification of single β-actin-tdEosFP molecules in the SMLM image into immobile (green) and mobile (red) fractions based on the brightness in two channels of orthogonal polarizations (inset). (m) Single-molecule orientations measured for Alexa Fluor 488-phalloidin labeled to two actin fibers in fixed cells. Red arrows: averaged fiber direction. Red boxes: regions of structural heterogeneity. (n) Color-coded orientation-resolved SMLM image for SYTOX Orange labeled to a DNA strand in vitro. Arrows: abrupt bends. Inset: absorption dipole moment of the dye is perpendicular to the DNA axis. (o) Orientation-resolved SMLM image of Nile Red in a phase-separated supported lipid bilayer, shown as maps of solid angle (Ω), polar angle (θ), and combined phase index. (a) is from ref42. (b) is from ref31. (c,d) are from ref37. (e,f) are from ref33. (g,m) are from ref48. (h,i,l) are from ref47. (j,k,n) are from ref51. (o) is from ref49.
For multicolor SMLM, splitting the single-molecule fluorescence into different views based on the wavelength, e.g., using a dichroic mirror (e.g., blue box in Figure 2a), enables the ratiometric color identification of spectrally overlapping probes.36–39 Here, a unique feature rises due to the single-molecule nature of SMLM: unmixing between different fluorophores, as often required in bulk spectral microscopy,40,41 becomes unnecessary. Consequently, by just comparing the single-molecule brightness in two views corresponding to the long- and short-wavelength components (e.g., Figure 2c), up to 4 probes can be concurrently imaged (Figure 2d), albeit with increased crosstalk as more color channels are added. The split long- and short-wavelength channels may be further focused to different planes to integrate biplane 3D localization (Figure 2a).42
Multi-view referencing has also greatly facilitated fluorescence polarization and anisotropy detections,43–45 which provide valuable insights into molecular orientations and rotational mobility. Whereas for SMLM, it remains difficult to extract polarization orientations from the PSF shape, polarizing beamsplitters provide an intuitive way to split the fluorescence (e.g., Figure 2g), so that polarization can be evaluated for single molecules by comparing their intensity ratios between the different split views (e.g., Figure 2hi),46–48 with the additional option of PSF engineering.44,49 In a different approach, the detected fluorescence is unsplit, but the linear polarization direction of the excitation laser is alternated in consecutive camera frames (Figure 2j).50,51 The modulated single-molecule intensities over different frames thus create a mechanism for self-referencing within the same imaging channel (Figure 2k). Together, these approaches have enabled the high-throughput accumulation of single-molecule fluorescence anisotropy, orientation, and wobble behaviors. With the concurrently obtained super-resolved position of each molecule, they thus resolved, at the nanoscale, protein rotational mobility in live (Figure 2l) and fixed cells,46,47,52 orientational order in biological filaments including actin fibers (Figure 2m),48 DNA strands (Figure 2n),48,51 and amyloid fibrils,53,54 and compositional heterogeneity in supported lipid bilayers (Figure 2o).49
The fluorescence spectrum
The integration of spectroscopy and microscopy offers powerful pathways to multiplexed and functional imaging.40,41 Although the fluorescence spectrum has been a key component of single-molecule experiments,55,56 typical approaches adopt the top-down strategies from bulk spectral microscopy,40,41 so that the fluorescence excitation and/or detection are confined to the smallest possible spot, e.g., in a confocal setting, to ensure the signal is from one molecule at a time. Such single-spot approaches afford low throughputs and usually cannot resolve multiple molecules within the diffraction limit.
The sparsity of single molecules in SMLM, however, creates a situation in which the fluorescence from many molecules in the wide field, as spatially “self-confined” individual point sources, can be parallelly dispersed spectrally without interfering with each other (Figure 3ab). After adding the on-off switching mechanism native to SMLM, millions of single-molecule spectra can thus be collected in minutes, a concept dubbed as “ultrahigh-throughput single-molecule spectroscopy”.57,58 This wide-field spectral imaging scheme, however, is complicated by the issue that the spatial and spectral information of a randomly located molecule are convolved. Referencing with an undispersed image of the same molecules readily decouples the two: this secondary view may be obtained using a secondary objective lens from the backside of the sample,57 a beamsplitter (e.g., Figure 3a),59 or the zeroth order of a grating.60 Separately, the spectral/color information may be encoded into the PSF shape, which eliminates the need for a reference image but does not record the actual spectrum.61,62
Figure 3.
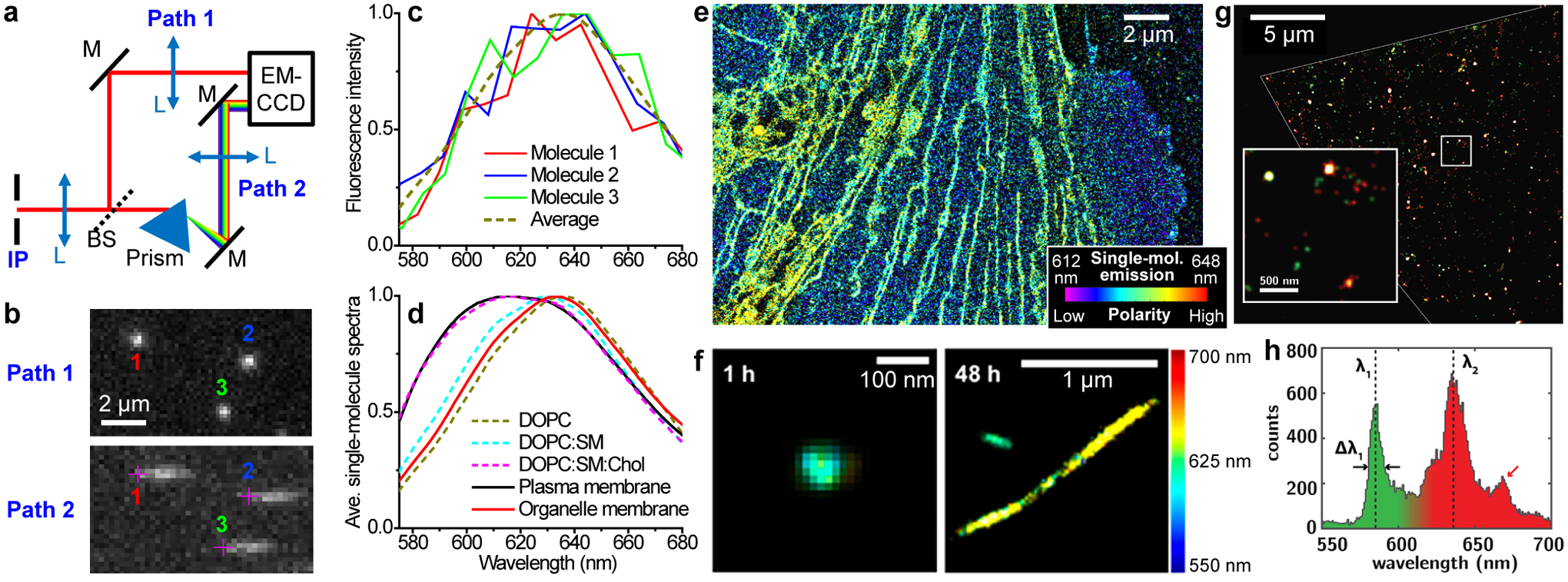
Spectrally resolved SMLM unveils nanoscale heterogeneities in biological and materials systems. (a) Schematic of a beamsplitter-based system. IP, intermediate image plane of the microscope; BS, beamsplitter. (b) A small region of single-molecule images (top) and spectra (bottom) concurrently acquired in a 6 ms snapshot from Paths 1 and 2 in (a), respectively, for Nile Red molecules in a supported lipid bilayer (SLB). Crosses in the spectral channel denote the spectral position of 590 nm for each molecule, as obtained by referring to the positions of the same molecules in the image channel. (c) Spectra of the three molecules in (b), compared to that averaged from 280,898 single molecules from the same sample. (d) Averaged spectra of single Nile Red molecules at the live-cell plasma membrane and at the nanoscale organelle membranes, versus that at SLBs of different compositions. (e) SR-SMLM image of a Nile Red-labeled live PtK2 cell. Color presents single-molecule spectral mean. (f) SR-SMLM image of Nile Red-labeled α-synuclein aggregates after 1 h (left) and 48 h (right) incubation. (g) SR-SMLM image of fluorescent defects in a flake of hexagonal boron nitride, colored by emission wavelength. (h) Distribution of center emission wavelengths for individual defects in (g). (a–e) are from ref70. (f) is from ref72. (g,h) is from ref66.
With the concurrently obtained spatial and spectral information of millions of single molecules, the resultant spectrally resolved SMLM (SR-SMLM) data are first employed for multicolor imaging.57,59,60 By comparing the measured spectrum of each detected molecule with standards, four spectrally overlapped fluorophores ~10 nm apart in emission wavelength are thus separated in immunolabeled cells with ≲ 1% misidentification.57 The capability to identify different fluorophore species is also utilized to characterize diverse chemical and materials systems, including single-molecule reaction pathways,63,64 heterogeneities in polymers,65 and defects in two-dimensional materials (Figure 3gh).66,67
The integration of SR-SMLM with environmentally sensitive fluorophores further enabled functional super-resolution microscopy.68 In particular, with the solvatochromic dye Nile Red and derivatives, SR-SMLM has mapped out, at the nanoscale, local variations in chemical polarity for cellular and artificial lipid membranes (Figure 3de),69–71 in vitro protein aggregates (Figure 3f),69,72 and surface adlayers.73 In these experiments, the accumulation and averaging of single-molecule spectra, through the gradual buildup over many camera frames, help improve the otherwise low signal-to-noise ratios of individual molecules (Figure 3c). Yet at the same time, the high spatial resolution achieved in super-localizing each molecule allows local spectra to be obtained for arbitrary regions of interest at the nanoscale. These capabilities, for example, helped unveil spectral differences for Nile Red molecules labeled to the plasma membrane vs. the membranes of nanoscale intracellular organelles with excellent contrast (Figure 3de).70 Separately, an analogous wide-field spectral imaging strategy has been applied to detect voltage-induced spectral shifts in the emission of single nanoparticles.74
Molecular motion and diffusion
Molecular motion and diffusion underlie vital (bio)physical and chemical processes that govern the complex, dynamic behaviors of systems far away from the thermodynamic equilibrium, e.g., the living cell. While single-particle tracking (SPT) has been an established tool for studying single-molecule motion and diffusion at the nanoscale,75–77 historically the requirement to resolve different trajectories with diffraction-limited optics seriously limits the number of particles (molecules) that can be tracked in a sample.
The advent of SMLM has inspired new approaches to achieve SPT at drastically increased densities. Thus, by employing photoactivation analogous to PALM/STORM78–80 and probe exchange analogous to PAINT,81–84 thousands of molecules can now be tracked in the same view (Figure 4a–c), yet with excellent spatial sparsity, by spreading out the measurement in the temporal dimension over many camera frames. We refer in-depth discussion to Reference85.
Figure 4.
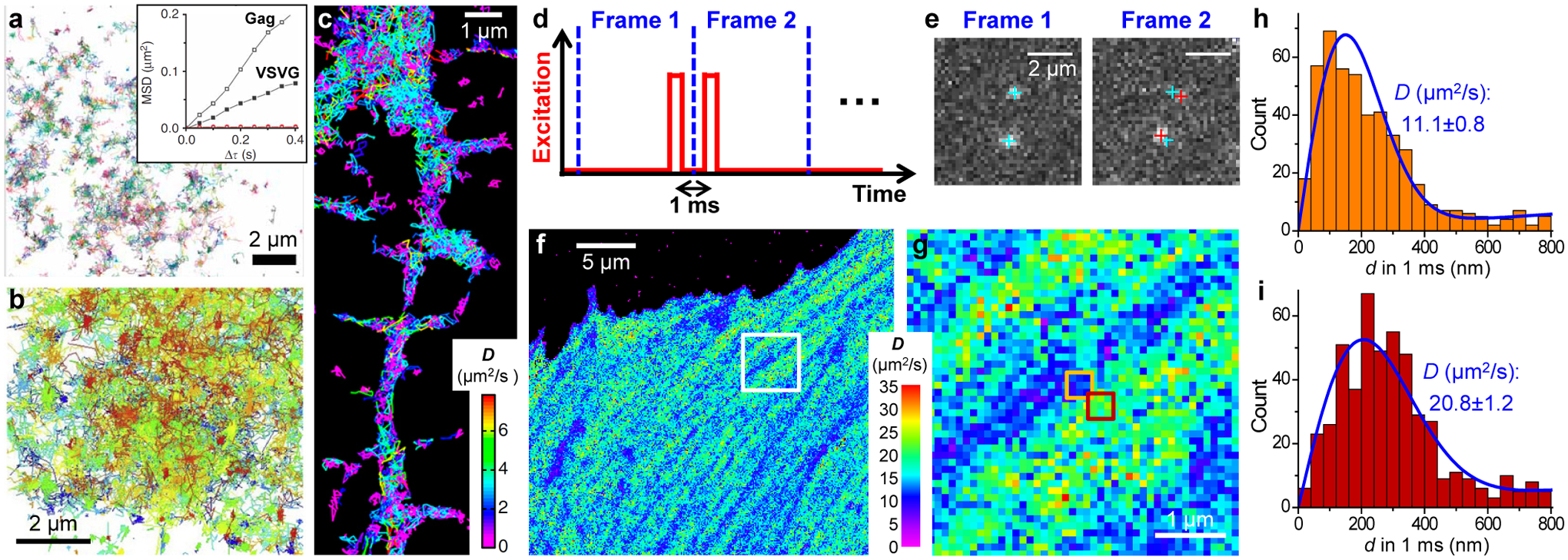
From single-molecule motion to nanoscale diffusivity mapping. (a) High-density single-molecule tracking in a live COS-7 cell via photoactivation of the Eos FP tagged to the membrane protein VSVG. Each color indicates a different single-molecule trajectory. Inset: mean-squared displacement as a function of time lag for two trajectories of Gag and VSVG. (b) High-density single-molecule tracking of GPI-GFP in the plasma membrane of a live COS-7 cell through the in situ binding of anti-GFP-AT647Ns. (c) High-density single-molecule tracking of the lipophilic dye DiI in the plasma membrane of a live neuron. Color presents the fitted diffusion coefficients D for each molecule. (d) Detecting the transient displacements of single molecules by applying a pair of closely timed stroboscopic excitation pulses across two tandem camera frames. (e) Resultant images recorded in the two frames for two mEos3.2 FP molecules freely diffusing inside a living cell. Cyan and red crosses: positions of the two molecules in Frame 1 and Frame 2, respectively. (f) SMdM D map obtained from ~104 pairs of the above excitation pulses by binning the resultant single-molecule displacements onto 100×100 nm2 grids for local fitting to a diffusion model. (g) Zoom-in of the white box in (f). (h,i) Distribution of 1-ms single-molecule displacements for two adjacent 300 × 300-nm2 areas [orange and red boxes in (g)]. Blue curves: fits with resultant D values and uncertainties labeled. (a) is from ref78. (b) is from ref81. (c) is from ref79. (d–i) are from ref87.
However, as the starting point of SPT is usually to obtain long single-particle trajectories to enable their individual diffusivity evaluation (e.g., Figure 4a inset and Figure 4c), the application is often limited to slow diffusions in spatially confined systems, e.g., lipid membranes.77 In contrast, for an unbound FP that rapidly diffuses inside the mammalian cytoplasm, severe motion-blur occurs at the typical imaging framerates. Although stroboscopic illumination overcomes motion-blur,86 molecules readily diffuse out of focus between consecutive frames, thus forbidding tracking.
To overcome these challenges, a recent method, single-molecule displacement/diffusivity mapping (SMdM),87 forgoes trajectories, but instead focuses on obtaining the transient displacements of single molecules. This is achieved by applying a pair of closely timed (~1 ms center-to-center) stroboscopic excitation pulses across two tandem camera frames (Figure 4d). By cross-referencing the single-molecule positions super-localized in the two paired frames (Figure 4e), the nanoscale displacements of single molecules (d) are thus evaluated for the ~1 ms transient window, during which period molecules remain in focus. The obtained d values, however, are not individually meaningful. Owing to the stochastic nature of diffusion, even for a fixed diffusion coefficient D, the displacement of a molecule in any given time window obeys a distribution rather than settles on a single value. The philosophy of bottom-up buildup now holds the key to extracting useful local parameters: after repeating the above paired excitation scheme for ~104 cycles, the accumulated d values, as single-molecule quanta, are spatially binned, e.g., onto 100×100 nm2 grids, so that the distribution of d in each bin is fit to a diffusion model (Figure 4hi). Color-plotting the resultant D values for each spatial bin thus yields a super-resolution map of local diffusivity (Figure 4fg). Additional parameters, including the principal direction of diffusion, can be further extracted by accumulating 2D displacement vectors.88
Integration and combination
The above approaches to measure the different properties of single molecules do not necessarily interfere with each other. It is thus often possible to integrate different measurements into the same experiment, so that multidimensional information can be extracted from the same molecules. The integration of 3D-SMLM and multicolor-SMLM is achieved early on.89 SR-SMLM and 3D-SMLM is also readily integrated by inserting a cylindrical lens into the spectrally undispersed single-molecule reference images.57 Single-molecule emission from four fluorophores is thus concurrently recorded and well distinguished in the resultant four-dimensional (3D spatial + 1D spectral) super-resolution data (Figure 5a–c). Polarization detection has been concurrently integrated with both cylindrical lens-based 3D-localization and SR-SMLM-like wide-field single-particle spectroscopy for tracking quantum rods (Figure 5de).90 A recent study on live-cell membranes integrates SMdM with 3D-SMLM and SR-SMLM (Figure 5fg), and thus resolved and differentiated between nanoscale diffusional heterogeneities of different origins.88 Meanwhile, a machine-learning approach has simultaneously extracted both 3D and color information from unmodified PSFs to achieve two-color 3D-SMLM.91 It remains interesting to see how related approaches could help unleash multidimensional information in single-molecule signals and guide future experiments.92,93
Figure 5.
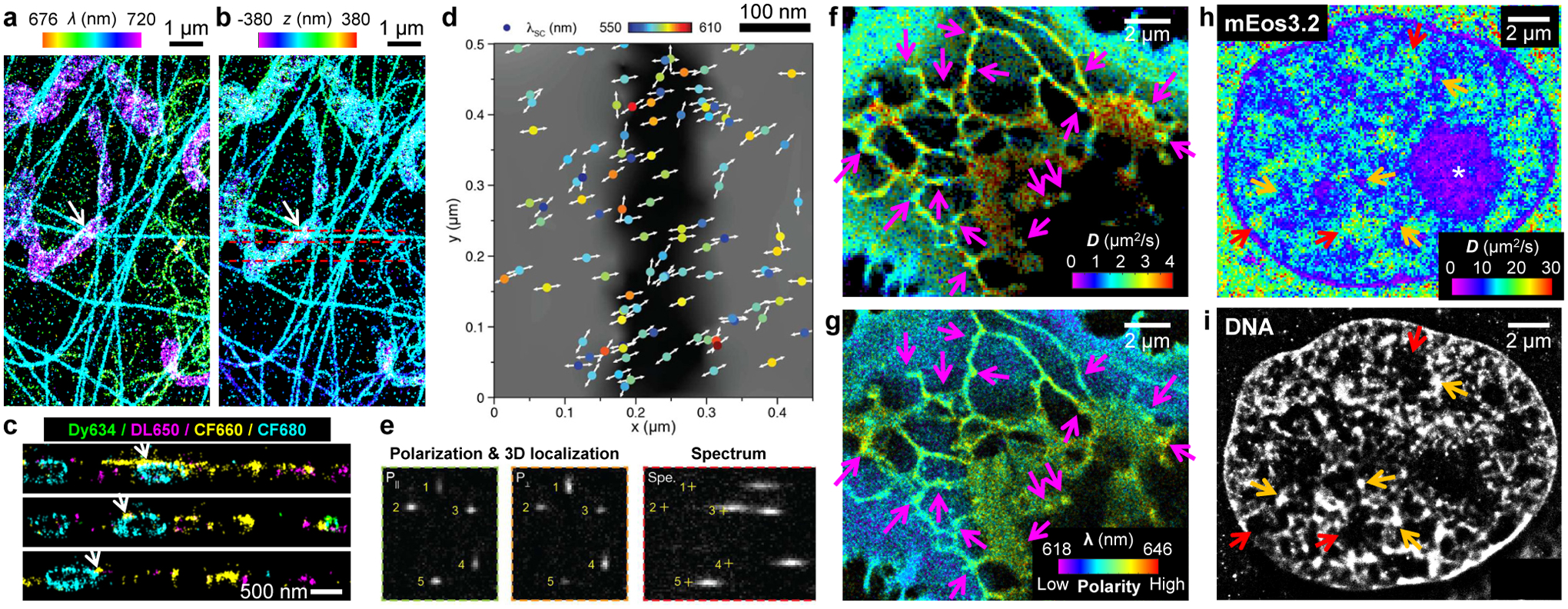
Integration and combination of multidimensional single-molecule signals. (a,b) Integration of SR-SMLM (a; color for emission wavelength) and 3D-SMLM (b; color for axial depth z) for a fixed cell labeled by four spectrally overlapped dyes. (c) Vertical sections along the three dashed lines in (b). Here each molecule is categorized as one of the four dyes and accordingly recolored. (d) Concurrent detections of orientation (arrows) and emission wavelength (color) for single quantum rods in the wide field. (e) Three complementary views are obtained for single quantum rods: two for polarization directions plus 3D localizations (left two panels) and one for spectra in the wide field (right). (f,g) Concurrent SMdM (f; color for diffusivity D) and SR-SMLM (g; color for emission wavelength) for Nile Red in cellular membranes, showing reduced D but unchanged emission wavelength at endoplasmic reticulum-plasma membrane contact sites (arrows). (h,i) Correlated SMdM (colored for D) of the mEos3.2 FP in the nucleus of a mammalian cell (h) and SMLM of the same cell with a DNA stain (i). Asterisk: reduced D in the nucleolus. Red and orange arrows: highest and lowest D in the SMdM image, coinciding with regions devoid of DNA and of high local DNA density in the SMLM image, respectively. (a–c) are from ref57. (d,e) are from ref90. (f,g) are from ref88. (h,i) are from ref87.
Combination and correlation of data from different experiments present another important direction. Whereas correlative approaches for super-resolution microscopy have been previously reviewed,20 recent SMdM experiments provide additional, cogent cases:87 by correlating SMdM diffusivity maps of unbound FPs with SMLM spatial images of probes labeling actin and DNA (Figure 5hi), it is thus shown that the nanoscale diffusivity variations in the cytoplasm and the nucleus correlate with the ultrastructure of the actin cytoskeleton and the genome, respectively.
Tradeoffs between spatial resolution, temporal resolution, and functionality
Whereas the spatial resolution of SMLM is ultimately limited by how precisely each molecule is localized (“optical resolution”),17,18 additional tradeoffs exist between the final effective spatial resolution, temporal resolution, and accuracies of the intended functional parameters.
For the construction of SMLM images, previous studies and reviews have well examined the tradeoff between spatial and temporal resolutions.9,11,94 As SMLM images are formed from individual molecules separately reporting their own positions, one can reasonably assume that features smaller than the typical spacing between the detected molecules are not captured. Formally, the Nyquist criterion argues that structures finer than 2/ρ1/dim, where ρ is the density of detected molecules and dim is the dimensionality of the image, cannot be resolved. Thus, for typical images to be rendered in two dimensions, structural details improve as the square root of the count of detected molecules. For stationary structures like fixed cells, this consideration is helpful in deciding when enough frames of single-molecule raw data have been collected. For dynamic structures, e.g., live cells, the recorded dataset may be divided in time to construct multiple SMLM images as a series. Finer division yields higher temporal resolutions, but at the expense of structural details due to insufficient localizations. Conversely, dividing into fewer time brackets increases structural details in each SMLM image, but loses fast dynamics and is vulnerable to motion blur.
Faster accumulation of single-molecule data eases the above contention between spatial and temporal resolutions. One approach is to record at increased framerates while concurrently accelerating single-molecule on-off switching. This strategy, however, is limited by the camera readout rate, probe properties, as well as cell health under the high laser powers needed to squeeze out the fluorescence in the reduced time. Another strategy is to increase the count of emitting molecules in each frame. However, as the molecules crowd up, their signals start to overlap, and one deviates from the key assumption in SMLM that each molecule is independently measured and analyzed. Multiple-emitter fitting,95,96 compressed sensing,97 and deep learning98,99 are among the various approaches to handle dense SMLM data with overlapping PSFs.
For high-dimensional SMLM beyond structure, the achieved accuracies for the intended functional parameters also depend on the count of detected molecules. For measurements that work with intensities, including various forms of multi-view referencing and SR-SMLM, the obtained signal-to-noise ratios, based on simple fluorescence summing-averaging, should improve to the square root of the count of molecules N. For SMdM, as discussed above, diffusion coefficients are obtained by fitting the distribution of single-molecule transient displacements. For simple random-walk models, the uncertainty of this fit scales inversely with the square root of the degree of freedom, N–1. These square-root dependencies warrant strategic spatiotemporal binning of the data, so that improved confidences in local functional parameters are achieved in reasonable manners by either increasing the size of the spatial bin, at the expense of spatial resolution, or allowing longer accumulation times, at the expense of temporal resolution. It is further worth mentioning that to meet the criterion that the signals from different molecules do not interfere with each other, the allowed single-molecule density in high-dimensional SMLM may be yet lower than that of the image-only SMLM, e.g., the increased footprint taken by the dispersed spectra in SR-SMLM57 and the need to avoid multiple emitters within the search radius between tandem frames in SMdM.87 Recent successes with deep learning in analyzing overlapped, complex PSFs (Figure 6j)93,99 point to potential ways to alleviate this issue.
Figure 6.
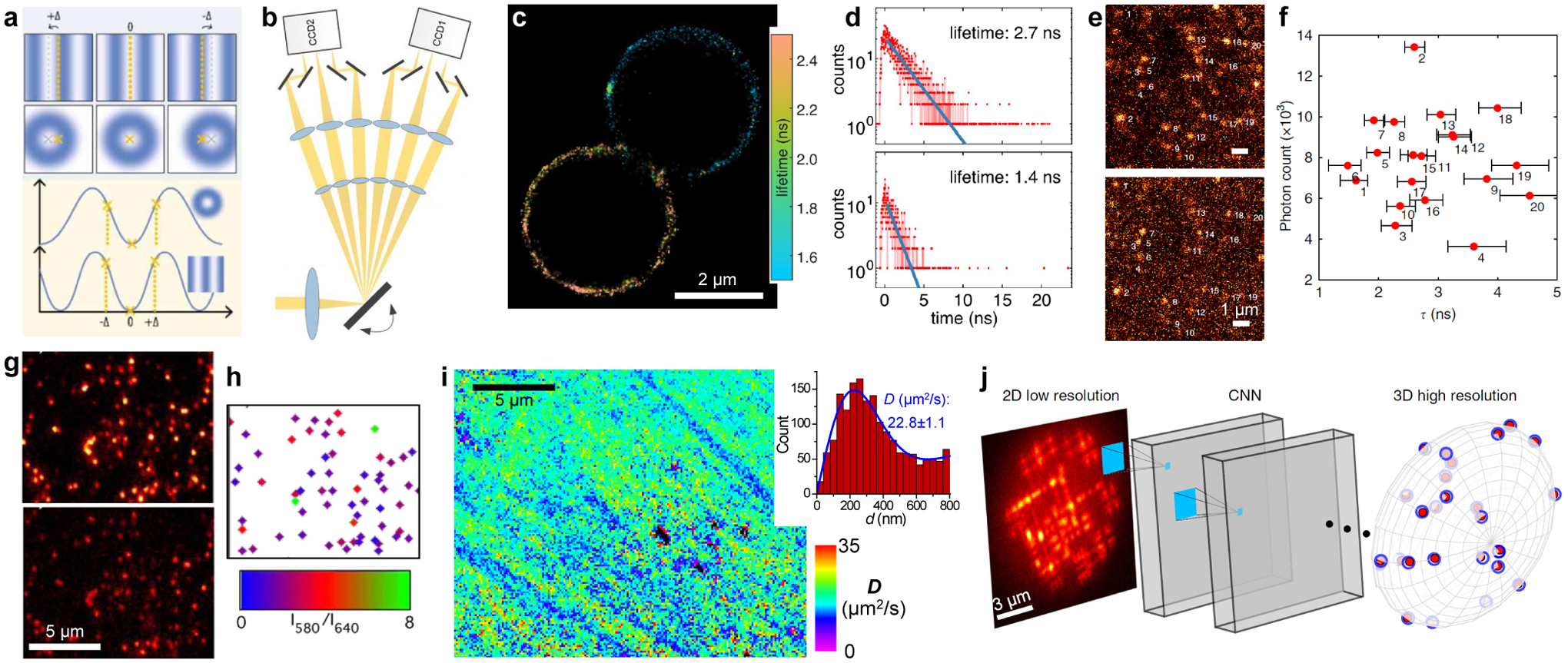
Rising opportunities. (a) Improving single-molecule localization via modulating the illumination pattern. (b) In one implementation, the wide-field image is fast scanned between six recording positions, so that six images under different synchronized illumination patterns are concurrently recorded in every frame. (c) Lifetime-resolved SMLM image of two beads labeled by two different dyes, obtained using a confocal setup. (d) Histograms of photon arrival time for two single molecules in (c). Blue lines: exponential fits. (e) Wide-field fluorescence images of single Alexa Fluor 532 molecules, with (top) and without (bottom) gating at 1.6 ns with a Pockels cell. (f) Brightness vs. the estimated lifetime for the molecules numbered in (e), based on the relative intensities in the two views. (g) Wide-field fluorescence images of single C-SNARF-1 molecules in an aluminosilicate film in 580 (top) and 640 (bottom) nm emission channels. (h) Emission ratio image for identified single molecules. (i) SMdM diffusivity map obtained using the non-switchable FP mEmerald. Inset: Distribution of 1-ms single-molecule displacements for a typical region. (j) A convolutional neural network (CNN) receives a raw image of overlapping complex PSFs and outputs a 3D high-resolution volume. (a) is from ref104. (b) is from ref105. (c,d) are from ref112. (e,f) are from ref113. (g,h) are from ref135. (i) is from ref87. (j) is from ref99.
Rising opportunities and future directions
Since its demonstration ~15 years ago, SMLM and related methods have progressed rapidly, hence enormous opportunities for wide-ranging fields.
Optical designs.
Continued innovations in both illumination and detection designs have kept pushing the achievable spatiotemporal resolution, sample depth,100 and throughput101 of SMLM. In particular, new methods have emerged that substantially enhance the localization precision of single molecules via the modulation of the illumination pattern (Figure 6a),102 remarkably with both patterns analogous to that used in STED microscopy103,104 and that used in structured illumination microscopy.105–108 Interestingly, the multi-view referencing concept discussed above is utilized here to improve localization precision, whereby the brightness of the same single molecules is referred between altered illumination patterns in different views (Figure 6b)105,108 or over time.103,104,106,107 The latter strategy may be compared with the above-discussed polarization SMLM approach in which the illumination polarization direction is modulated in consecutive camera frames (Figure 2jk).50,51 Given the recent demonstration that frame-synchronized modulation of the illumination wavelength enables fast excitation spectral microscopy in the wide-field,109 it is further interesting to speculate whether similar tactics may extend SMLM to the new dimension of the fluorescence excitation spectrum.
Another potentially important future dimension for SMLM is the fluorescence lifetime, an often-examined parameter in functional microscopy.110,111 A recent study achieves lifetime-resolved SMLM through confocal scanning, thus resolving two spectrally similar fluorescent probes in labeled beads (Figure 6cd) and fixed cells.112 Although the confocal scheme employed in this study benefits from an established technique, the single-spot scanning approach notably limits the imaging speed. Recently demonstrated wide-field single-molecule fluorescence lifetime approaches, e.g., via comparing the intensity between gated and ungated channels (Figure 6ef)113 and the use of special detectors,114 if applicable to SMLM, could help bring the throughput to be comparable to the typical wide-field SMLM experiments. The potential to thus map out local environments at the nanoscale represents an exciting possibility.
Beyond the nanosecond dynamics of fluorescence lifetime, it is further tempting to speculate whether SMLM could be pushed to the ultrafast regime. Although ultrafast spectroscopy has been achieved for isolated single molecules,115,116 it remains unclear whether such experiments may be carried out with high throughput and/or achieve the signal on-off switching needed for imaging multiple molecules within the diffraction limit. The extension of SMLM to non-fluorescence single-molecule detection schemes117 presents another challenge. In particular, recent advances in interferometric scattering microscopy have enabled the label-free detection and super-localization of single proteins,118 thus pointing to novel avenues to achieving SMLM and encoding functions.
Molecular designs.
Molecular designs have been at the heart of SMLM development. Recent reviews have well summarized the desired characteristics of, as well as the multifaceted efforts toward, new SMLM probes.119–122 An ideal probe would emit strongly as single molecules and possess either good photoswitching [for photoactivation-based approaches like STORM and (F)PALM] or reversible-binding (for PAINT) properties. For live-cell experiments, labeling specificity, cell toxicity, and membrane permeability are additional concerns; for the last issue, new approaches have enabled the high-throughput intracellular delivery of membrane non-permeable probes for SMLM applications.123,124
Beyond the usual efforts to attain brighter probes, recent work has further utilized novel interactions between biomolecules for encoding functional information into SMLM.125,126 In particular, utilizing the highly controllable interactions of DNA strands, new probes have enabled the SMLM imaging of cellular traction forces127,128 and single-molecule-based super-resolution labeling.129,130
The rise of high-dimensional SMLM methods beyond the traditional multicolor and 3D efforts affords new challenges and opportunities. For polarization-resolved SMLM, one recurring finding is that the detected single-molecule polarization orientation, as well as its dynamics, depends strongly on both the molecular structure of the fluorescent probe and how it is tagged to the target. For example, when actin fibers in fixed cells are labeled by dye-linked phalloidin, three different dyes respectively exhibit parallel, perpendicular, and random orientations with respect to the fiber direction.48 Similarly, contrasting orientational behaviors are found for dyes that insert differently into DNA strands51 and lipid bilayers.49 Thus, the future design and utilization of probes for polarization-resolved SMLM would benefit greatly from well-defined probe-target interactions, in addition to the common SMLM quest for bright, switchable probes.
Meanwhile, while SR-SMLM has been highly successful in resolving local chemical polarity, experiments have largely relied on a single, classic solvatochromic dye, Nile Red. A recent study reports the rational design for a Nile Red derivative tailored for the SR-SMLM of live-cell plasma membranes.71 Still, beyond chemical polarity, many other functional parameters131–133 could potentially be encoded into the single-molecule spectra, hence tremendous opportunities for probe development. For example, pH-dependent spectral variations have been measured for sparse single pH-indicator molecules in the wide field through ratiometric fluorescence detection (Figure 6gh),134,135 yet it remains to be shown how such probes may achieve on-off switching for SMLM, as well as be introduced into dynamic systems like live cells.
In connection with the above discussions, it is worth mentioning that SMdM opens up possibilities to work with fluorescently non-switching probes. In SMdM, the paired stroboscopic pulses across tandem frames (Figure 4d) leave ample time between the unpaired pulses for probes outside the illuminated area to diffuse in, hence a mechanism to replenish photobleached molecules without photoactivation [as required in STORM/(F)PALM]. Meanwhile, the stroboscopic excitation eliminates the need for target-binding to register clear single-molecule images (as required in PAINT). Thus, SMdM has been achieved with conventional, non-photoactivatable and non-binding fluorophores, e.g., the common FP mEmerald (Figure 6i).87 The prospect of thus being able to utilize any bright probes, together with the recently demonstrated integration of SMdM and SR-SMLM (Figure 5fg),88 bodes well for the future incorporation of diverse sensor probes. In particular, whereas single-molecule Förster resonance energy transfer (smFRET)136 has been a great tool for studying the dynamics of biomolecules in vitro, recent work has demonstrated its application in live cells.137 The possible integration of smFRET with SMdM and related SMLM approaches could thus unlock great potentials for probing intracellular dynamics at the super-resolution level.
Algorithm and theory.
The development of SMLM and related methods goes hand-in-hand with algorithms, from how the single-molecule signals are generated and collected to the analysis, presentation, and interpretation of the collected data.11,138–140 Whereas we are thus guaranteed to continue witnessing algorithms coevolve with optical and molecular designs at all levels, emerging efforts to incorporate machine learning afford fresh insights.93 While we have already discussed several specific examples above (e.g., Figure 6j), generally, machine learning offers facile ways to achieve near-optimal solutions without presumptions of underlying models or formulas. Instead, optimizations at different levels, from the design and analysis of PSFs to the formation and presentation of final images, are driven by the big training data. Here, SMLM readily provides millions of experimental PSFs; yet, to construct the perfect training sets, it often remains a challenge to associate each PSF with the “ground truth”.
At a more fundamental level, the new paradigm of constructing functional super-resolution images through the single-molecule quanta also calls for new theoretical discussions. Whereas the traditional top-down approach of microscopy reports the assembly behavior of many molecules, SMLM arrives at the final picture from the bottom up, and the exceptionally large (~106) statistics of single-molecule measurements may afford new insights on their own. For example, when SR-SMLM is applied with Nile Red to probe the cell membrane, statistics of the single-molecule spectra unveil the existence of a low-polarity phase in cholesterol-treated cells independent of spatial information.70 When compared to the classic single-molecule experiments, the local accumulation philosophy of SMLM and related methods is further unique. For example, in SPT, single particles are tracked over consecutive frames to enable the individual fitting of their trajectories. This particle-centered approach implies each particle is unique and/or “remembers” its history, but overlooks the issue that as a particle diffuses, it samples different locations that may be characterized by dissimilar local properties. SMdM, in contrast, assumes all probe molecules equivalent, but accumulates, for each fixed location, how fast these identical molecules travel. This location-centered strategy is powerful in unveiling spatial heterogeneity, and together with the other high-dimensional SMLM approaches discussed above, raises interesting questions on how to best define physicochemical parameters in heterogeneous systems at the nanoscale.
Conclusion.
Together, with single molecules as our quanta, the bottom-up accumulation philosophy of SMLM provides a powerful conduit for multidimensional microscopy beyond the diffraction limit. We look forward to the future development and application of related methods: when we look back in another 15 years, it would be a real surprise if new innovations had not advanced far beyond the realm discussed in this perspective.
ACKNOWLEDGMENTS
We thank all past and current lab members for contributing to related work. We acknowledge support from the National Science Foundation (CHE-1554717), the National Institute of General Medical Sciences of the National Institutes of Health (DP2GM132681), the Beckman Young Investigator Program, the Packard Fellowships for Science and Engineering, and the Bakar Fellows Award. K.X. is a Chan Zuckerberg Biohub investigator.
REFERENCES
- (1).Dempster AJ; Batho HF Light quanta and interference. Phys. Rev 1927, 30, 644–648. [Google Scholar]
- (2).Aspden RS; Padgett MJ; Spalding GC Video recording true single-photon double-slit interference. Am. J. Phys 2016, 84, 671–677. [Google Scholar]
- (3).Rust MJ; Bates M; Zhuang X Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 2006, 3, 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Betzig E; Patterson GH; Sougrat R; Lindwasser OW; Olenych S; Bonifacino JS; Davidson MW; Lippincott-Schwartz J; Hess HF Imaging intracellular fluorescent proteins at nanometer resolution. Science 2006, 313, 1642–1645. [DOI] [PubMed] [Google Scholar]
- (5).Hess ST; Girirajan TPK; Mason MD Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J 2006, 91, 4258–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Sharonov A; Hochstrasser RM Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc. Natl. Acad. Sci. U. S. A 2006, 103, 18911–18916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Patterson G; Davidson M; Manley S; Lippincott-Schwartz J Superresolution imaging using single-molecule localization. Annu. Rev. Phys. Chem 2010, 61, 345–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Xu K; Shim S-H; Zhuang X Super-resolution imaging through stochastic switching and localization of single molecules: an overview. In Far-Field Optical Nanoscopy; Tinnefeld P, Eggeling C, Hell SW, Eds.; Springer: Berlin, 2015; pp 27–64. [Google Scholar]
- (9).Liu Z; Lavis LD; Betzig E Imaging live-cell dynamics and structure at the single-molecule level. Mol. Cell 2015, 58, 644–659. [DOI] [PubMed] [Google Scholar]
- (10).Sauer M; Heilemann M Single-molecule localization microscopy in eukaryotes. Chem. Rev 2017, 117, 7478–7509. [DOI] [PubMed] [Google Scholar]
- (11).Baddeley D; Bewersdorf J Biological insight from super-resolution microscopy: what we can learn from localization-based images. Annu. Rev. Biochem 2018, 87, 965–989. [DOI] [PubMed] [Google Scholar]
- (12).Möckl L; Moerner WE Super-resolution microscopy with single molecules in biology and beyond-essentials, current trends, and future challenges. J. Am. Chem. Soc 2020, 142, 17828–17844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Sahl SJ; Hell SW; Jakobs S Fluorescence nanoscopy in cell biology. Nat. Rev. Mol. Cell Biol 2017, 18, 685–701. [DOI] [PubMed] [Google Scholar]
- (14).Sigal YM; Zhou R; Zhuang X Visualizing and discovering cellular structures with super-resolution microscopy. Science 2018, 361, 880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Schermelleh L; Ferrand A; Huser T; Eggeling C; Sauer M; Biehlmaier O; Drummen GPC Super-resolution microscopy demystified. Nat. Cell Biol 2019, 21, 72–84. [DOI] [PubMed] [Google Scholar]
- (16).Blom H; Widengren J Stimulated emission depletion microscopy. Chem. Rev 2017, 117, 7377–7427. [DOI] [PubMed] [Google Scholar]
- (17).Thompson RE; Larson DR; Webb WW Precise nanometer localization analysis for individual fluorescent probes. Biophys. J 2002, 82, 2775–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Deschout H; Cella Zanacchi F; Mlodzianoski M; Diaspro A; Bewersdorf J; Hess ST; Braeckmans K Precisely and accurately localizing single emitters in fluorescence microscopy. Nat. Methods 2014, 11, 253–66. [DOI] [PubMed] [Google Scholar]
- (19).Vaughan JC; Jia S; Zhuang XW Ultrabright photoactivatable fluorophores created by reductive caging. Nat. Methods 2012, 9, 1181–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Hauser M; Wojcik M; Kim D; Mahmoudi M; Li W; Xu K Correlative super-resolution microscopy: new dimensions and new opportunities. Chem. Rev 2017, 117, 7428–7456. [DOI] [PubMed] [Google Scholar]
- (21).Bates M; Huang B; Dempsey GT; Zhuang XW Multicolor super-resolution imaging with photoswitchable fluorescent probes. Science 2007, 317, 1749–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Bock H; Geisler C; Wurm CA; Von Middendorff C; Jakobs S; Schonle A; Egner A; Hell SW; Eggeling C Two-color far-field fluorescence nanoscopy based on photoswitchable emitters. Appl. Phys. B-Lasers Opt 2007, 88, 161–165. [Google Scholar]
- (23).Shroff H; Galbraith CG; Galbraith JA; White H; Gillette J; Olenych S; Davidson MW; Betzig E Dual-color superresolution imaging of genetically expressed probes within individual adhesion complexes. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 20308–20313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Jones SA; Shim SH; He J; Zhuang XW Fast, three-dimensional super-resolution imaging of live cells. Nat. Methods 2011, 8, 499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Wilmes S; Staufenbiel M; Lisse D; Richter CP; Beutel O; Busch KB; Hess ST; Piehler J Triple-color super-resolution imaging of live cells: resolving submicroscopic receptor organization in the plasma membrane. Angew. Chem.-Int. Edit 2012, 51, 4868–4871. [DOI] [PubMed] [Google Scholar]
- (26).Jungmann R; Avendano MS; Woehrstein JB; Dai MJ; Shih WM; Yin P Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods 2014, 11, 313–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Tam J; Cordier GA; Borbely JS; Alvarez AS; Lakadamyali M Cross-talk-free multi-color STORM imaging using a single fluorophore. PLoS One 2014, 9, e101772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Huang B; Wang W; Bates M; Zhuang X Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 2008, 319, 810–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).von Diezmann A; Shechtman Y; Moerner WE Three-dimensional localization of single molecules for super-resolution imaging and single-particle tracking. Chem. Rev 2017, 117, 7244–7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Juette MF; Gould TJ; Lessard MD; Mlodzianoski MJ; Nagpure BS; Bennett BT; Hess ST; Bewersdorf J Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat. Methods 2008, 5, 527–529. [DOI] [PubMed] [Google Scholar]
- (31).Ram S; Prabhat P; Chao J; Ward ES; Ober RJ High accuracy 3D quantum dot tracking with multifocal plane microscopy for the study of fast intracellular dynamics in live cells. Biophys. J 2008, 95, 6025–6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Xu K; Babcock HP; Zhuang X Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton. Nat. Methods 2012, 9, 185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Shtengel G; Galbraith JA; Galbraith CG; Lippincott-Schwartz J; Gillette JM; Manley S; Sougrat R; Waterman CM; Kanchanawong P; Davidson MW et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc. Natl. Acad. Sci. U. S. A 2009, 106, 3125–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Aquino D; Schonle A; Geisler C; von Middendorff C; Wurm CA; Okamura Y; Lang T; Hell SW; Egner A Two-color nanoscopy of three-dimensional volumes by 4Pi detection of stochastically switched fluorophores. Nat. Methods 2011, 8, 353–359. [DOI] [PubMed] [Google Scholar]
- (35).Huang F; Sirinakis G; Allgeyer ES; Schroeder LK; Duim WC; Kromann EB; Phan T; Rivera-Molina FE; Myers JR; Irnov I et al. Ultra-high resolution 3D imaging of whole cells. Cell 2016, 166, 1028–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Bossi M; Folling J; Belov VN; Boyarskiy VP; Medda R; Egner A; Eggeling C; Schonle A; Hell SW Multicolor far-field fluorescence nanoscopy through isolated detection of distinct molecular species. Nano Lett 2008, 8, 2463–2468. [DOI] [PubMed] [Google Scholar]
- (37).Testa I; Wurm CA; Medda R; Rothermel E; von Middendorf C; Folling J; Jakobs S; Schonle A; Hell SW; Eggeling C Multicolor fluorescence nanoscopy in fixed and living cells by exciting conventional fluorophores with a single wavelength. Biophys. J 2010, 99, 2686–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Gunewardene MS; Subach FV; Gould TJ; Penoncello GP; Gudheti MV; Verkhusha VV; Hess ST Superresolution imaging of multiple fluorescent proteins with highly overlapping emission spectra in living cells. Biophys. J 2011, 101, 1522–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Zhang YD; Schroeder LK; Lessard MD; Kidd P; Chung J; Song YB; Benedetti L; Li YM; Ries J; Grimm JB et al. Nanoscale subcellular architecture revealed by multicolor three-dimensional salvaged fluorescence imaging. Nat. Methods 2020, 17, 225–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Zimmermann T; Rietdorf J; Pepperkok R Spectral imaging and its applications in live cell microscopy. FEBS Lett 2003, 546, 87–92. [DOI] [PubMed] [Google Scholar]
- (41).Gao L; Smith RT Optical hyperspectral imaging in microscopy and spectroscopy - a review of data acquisition. J. Biophotonics 2015, 8, 441–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Winterflood CM; Platonova E; Albrecht D; Ewers H Dual-color 3D superresolution microscopy by combined spectral-demixing and biplane imaging. Biophys. J 2015, 109, 3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Jameson DM; Ross JA Fluorescence polarization/anisotropy in diagnostics and imaging. Chem. Rev 2010, 110, 2685–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Backlund MP; Lew MD; Backer AS; Sahl SJ; Moerner WE The role of molecular dipole orientation in single-molecule fluorescence microscopy and implications for super-resolution imaging. ChemPhysChem 2014, 15, 587–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Shroder DY; Lippert LG; Goldman YE Single molecule optical measurements of orientation and rotations of biological macromolecules. Methods Appl. Fluoresc 2016, 4, 042004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Gould TJ; Gunewardene MS; Gudheti MV; Verkhusha VV; Yin SR; Gosse JA; Hess ST Nanoscale imaging of molecular positions and anisotropies. Nat. Methods 2008, 5, 1027–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Testa I; Schonle A; Middendorff CV; Geisler C; Medda R; Wurm CA; Stiel AC; Jakobs S; Bossi M; Eggeling C et al. Nanoscale separation of molecular species based on their rotational mobility. Opt. Express 2008, 16, 21093–21104. [DOI] [PubMed] [Google Scholar]
- (48).Cruz CAV; Shaban HA; Kress A; Bertaux N; Monneret S; Mavrakis M; Savatier J; Brasselet S Quantitative nanoscale imaging of orientational order in biological filaments by polarized superresolution microscopy. Proc. Natl. Acad. Sci. U. S. A 2016, 113, E820–E828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Lu J; Mazidi H; Ding T; Zhang O; Lew MD Single-molecule 3D orientation imaging reveals nanoscale compositional heterogeneity in lipid membranes. Angew. Chem. Int. Ed 2020, 59, 17572–17579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Ha T; Enderle T; Chemla DS; Selvin PR; Weiss S Single molecule dynamics studied by polarization modulation. Phys. Rev. Lett 1996, 77, 3979–3982. [DOI] [PubMed] [Google Scholar]
- (51).Backer AS; Lee MY; Moerner WE Enhanced DNA imaging using super-resolution microscopy and simultaneous single-molecule orientation measurements. Optica 2016, 3, 659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Fu G; Tu LC; Zilman A; Musser SM Investigating molecular crowding within nuclear pores using polarization-PALM. eLife 2017, 6, e28716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Shaban HA; Valades-Cruz CA; Savatier J; Brasselet S Polarized super-resolution structural imaging inside amyloid fibrils using Thioflavine T. Sci. Rep 2017, 7, 12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Ding T; Wu T; Mazidi H; Zhang O; Lew MD Single-molecule orientation localization microscopy for resolving structural heterogeneities between amyloid fibrils. Optica 2020, 7, 602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Xie XS; Trautman JK Optical studies of single molecules at room temperature. Annu. Rev. Phys. Chem 1998, 49, 441–480. [DOI] [PubMed] [Google Scholar]
- (56).Moerner WE; Fromm DP Methods of single-molecule fluorescence spectroscopy and microscopy. Rev. Sci. Instrum 2003, 74, 3597–3619. [Google Scholar]
- (57).Zhang Z; Kenny SJ; Hauser M; Li W; Xu K Ultrahigh-throughput single-molecule spectroscopy and spectrally resolved super-resolution microscopy. Nat. Methods 2015, 12, 935–938. [DOI] [PubMed] [Google Scholar]
- (58).Yan R; Moon S; Kenny SJ; Xu K Spectrally resolved and functional super-resolution microscopy via ultrahigh-throughput single-molecule spectroscopy. Acc. Chem. Res 2018, 51, 697–705. [DOI] [PubMed] [Google Scholar]
- (59).Mlodzianoski MJ; Curthoys NM; Gunewardene MS; Carter S; Hess ST Super-resolution imaging of molecular emission spectra and single molecule spectral fluctuations. PLoS One 2016, 11, e0147506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Dong B; Almassalha L; Urban BE; Nguyen TQ; Khuon S; Chew TL; Backman V; Sun C; Zhang HF Super-resolution spectroscopic microscopy via photon localization. Nat. Commun 2016, 7, 12290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Shechtman Y; Weiss LE; Backer AS; Lee MY; Moerner WE Multicolour localization microscopy by point-spread-function engineering. Nat. Photonics 2016, 10, 590–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Smith C; Huisman M; Siemons M; Grunwald D; Stallinga S Simultaneous measurement of emission color and 3D position of single molecules. Opt. Express 2016, 24, 4996–5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Kim D; Zhang Z; Xu K Spectrally resolved super-resolution microscopy unveils multipath reaction pathways of single spiropyran molecules. J. Am. Chem. Soc 2017, 139, 9447–9450. [DOI] [PubMed] [Google Scholar]
- (64).Sansalone L; Zhang Y; Mazza MMA; Davis JL; Song KH; Captain B; Zhang HF; Raymo FM High-throughput single-molecule spectroscopy resolves the conformational isomers of BODIPY chromophores. J. Phys. Chem. Lett 2019, 10, 6807–6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Urban BE; Dong B; Nguyen TQ; Backman V; Sun C; Zhang HF Subsurface super-resolution imaging of unstained polymer nanostructures. Sci. Rep 2016, 6, 28156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Comtet J; Glushkov E; Navikas V; Feng J; Babenko V; Hofmann S; Watanabe K; Taniguchi T; Radenovic A Wide-field spectral super-resolution mapping of optically active defects in hexagonal boron nitride. Nano Lett 2019, 19, 2516–2523. [DOI] [PubMed] [Google Scholar]
- (67).Stern HL; Wang R; Fan Y; Mizuta R; Stewart JC; Needham LM; Roberts TD; Wai R; Ginsberg NS; Klenerman D et al. Spectrally resolved photodynamics of individual emitters in large-area monolayers of hexagonal boron nitride. ACS Nano 2019, 13, 4538–4547. [DOI] [PubMed] [Google Scholar]
- (68).Yan R; Wang B; Xu K Functional super-resolution microscopy of the cell. Curr. Opin. Chem. Biol 2019, 51, 92–97. [DOI] [PubMed] [Google Scholar]
- (69).Bongiovanni MN; Godet J; Horrocks MH; Tosatto L; Carr AR; Wirthensohn DC; Ranasinghe RT; Lee JE; Ponjavic A; Fritz JV et al. Multi-dimensional super-resolution imaging enables surface hydrophobicity mapping. Nat. Commun 2016, 7, 13544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Moon S; Yan R; Kenny SJ; Shyu Y; Xiang L; Li W; Xu K Spectrally resolved, functional super-resolution microscopy reveals nanoscale compositional heterogeneity in live-cell membranes. J. Am. Chem. Soc 2017, 139, 10944–10947. [DOI] [PubMed] [Google Scholar]
- (71).Danylchuk DI; Moon S; Xu K; Klymchenko AS Tailor-made switchable solvatochromic probes for live-cell super-resolution imaging of plasma membrane organization. Angew. Chem.-Int. Edit 2019, 58, 14920–14924. [DOI] [PubMed] [Google Scholar]
- (72).Lee JE; Sang JC; Rodrigues M; Carr AR; Horrocks MH; De S; Bongiovanni MN; Flagmeier P; Dobson CM; Wales DJ et al. Mapping surface hydrophobicity of α-synuclein oligomers at the nanoscale. Nano Lett 2018, 18, 7494–7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Xiang L; Wojcik M; Kenny SJ; Yan R; Moon S; Li W; Xu K Optical characterization of surface adlayers and their compositional demixing at the nanoscale. Nat. Commun 2018, 9, 1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Kuo Y; Li J; Michalet X; Chizhik A; Meir N; Bar-Elli O; Chan E; Oron D; Enderlein J; Weiss S Characterizing the quantum-confined stark effect in semiconductor quantum dots and nanorods for single-molecule electrophysiology. ACS Photonics 2018, 5, 4788–4800. [Google Scholar]
- (75).Manzo C; Garcia-Parajo MF A review of progress in single particle tracking: from methods to biophysical insights. Rep. Prog. Phys 2015, 78, 124601. [DOI] [PubMed] [Google Scholar]
- (76).Shen H; Tauzin LJ; Baiyasi R; Wang WX; Moringo N; Shuang B; Landes CF Single particle tracking: from theory to biophysical applications. Chem. Rev 2017, 117, 7331–7376. [DOI] [PubMed] [Google Scholar]
- (77).Kusumi A; Tsunoyama TA; Hirosawa KM; Kasai RS; Fujiwara TK Tracking single molecules at work in living cells. Nat. Chem. Biol 2014, 10, 524–532. [DOI] [PubMed] [Google Scholar]
- (78).Manley S; Gillette JM; Patterson GH; Shroff H; Hess HF; Betzig E; Lippincott-Schwartz J High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods 2008, 5, 155–157. [DOI] [PubMed] [Google Scholar]
- (79).Shim SH; Xia C; Zhong G; Babcock HP; Vaughan JC; Huang B; Wang X; Xu C; Bi GQ; Zhuang X Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 13978–13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Hoze N; Nair D; Hosy E; Sieben C; Manley S; Herrmann A; Sibarita JB; Choquet D; Holcman D Heterogeneity of AMPA receptor trafficking and molecular interactions revealed by superresolution analysis of live cell imaging. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 17052–17057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Giannone G; Hosy E; Levet F; Constals A; Schulze K; Sobolevsky AI; Rosconi MP; Gouaux E; Tampe R; Choquet D et al. Dynamic superresolution imaging of endogenous proteins on living cells at ultra-high density. Biophys. J 2010, 99, 1303–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Winckler P; Lartigue L; Giannone G; De Giorgi F; Ichas F; Sibarita JB; Lounis B; Cognet L Identification and super-resolution imaging of ligand-activated receptor dimers in live cells. Sci. Rep 2013, 3, 2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Masson JB; Dionne P; Salvatico C; Renner M; Specht CG; Triller A; Dahan M Mapping the energy and diffusion landscapes of membrane proteins at the cell surface using high-density single-molecule imaging and Bayesian inference: application to the multiscale dynamics of glycine receptors in the neuronal membrane. Biophys. J 2014, 106, 74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Tas RP; Chazeau A; Cloin BMC; Lambers MLA; Hoogenraad CC; Kapitein LC Differentiation between oppositely oriented microtubules controls polarized neuronal transport. Neuron 2017, 96, 1264–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Cognet L; Leduc C; Lounis B Advances in live-cell single-particle tracking and dynamic super-resolution imaging. Curr. Opin. Chem. Biol 2014, 20, 78–85. [DOI] [PubMed] [Google Scholar]
- (86).Xie XS; Choi PJ; Li GW; Lee NK; Lia G Single-molecule approach to molecular biology in living bacterial cells. Ann. Rev. Biophys 2008, 37, 417–444. [DOI] [PubMed] [Google Scholar]
- (87).Xiang L; Chen K; Yan R; Li W; Xu K Single-molecule displacement mapping unveils nanoscale heterogeneities in intracellular diffusivity. Nat. Methods 2020, 17, 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Yan R; Chen K; Xu K Probing nanoscale diffusional heterogeneities in cellular membranes through multidimensional single-molecule and super-resolution microscopy. J. Am. Chem. Soc 2020, 142, 18866–18873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Huang B; Jones SA; Brandenburg B; Zhuang XW Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat. Methods 2008, 5, 1047–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Dong B; Soetikno BT; Chen X; Backman V; Sun C; Zhang HF Parallel three-dimensional tracking of quantum rods using polarization-sensitive spectroscopic photon localization microscopy. ACS Photonics 2017, 4, 1747–1752. [Google Scholar]
- (91).Kim T; Moon S; Xu K Information-rich localization microscopy through machine learning. Nat. Commun 2019, 10, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Hershko E; Weiss LE; Michaeli T; Shechtman Y Multicolor localization microscopy and point-spread-function engineering by deep learning. Opt. Express 2019, 27, 6158–6183. [DOI] [PubMed] [Google Scholar]
- (93).Mockl L; Roy AR; Moerner WE Deep learning in single-molecule microscopy: fundamentals, caveats, and recent developments [Invited]. Biomed. Opt. Express 2020, 11, 1633–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Shroff H; Galbraith CG; Galbraith JA; Betzig E Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat. Methods 2008, 5, 417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Holden SJ; Uphoff S; Kapanidis AN DAOSTORM: an algorithm for high-density super-resolution microscopy. Nat. Methods 2011, 8, 279–280. [DOI] [PubMed] [Google Scholar]
- (96).Huang F; Schwartz SL; Byars JM; Lidke KA Simultaneous multiple-emitter fitting for single molecule super-resolution imaging. Biomed. Opt. Express 2011, 2, 1377–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Zhu L; Zhang W; Elnatan D; Huang B Faster STORM using compressed sensing. Nat. Methods 2012, 9, 721–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Nehme E; Weiss LE; Michaeli T; Shechtman Y Deep-STORM: super-resolution single-molecule microscopy by deep learning. Optica 2018, 5, 458–464. [Google Scholar]
- (99).Nehme E; Freedman D; Gordon R; Ferdman B; Weiss LE; Alalouf O; Naor T; Orange R; Michaeli T; Shechtman Y DeepSTORM3D: dense 3D localization microscopy and PSF design by deep learning. Nat. Methods 2020, 17, 734–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Liu S; Huh H; Lee SH; Huang F Three-dimensional single-molecule localization microscopy in whole-cell and tissue specimens. Annu. Rev. Biomed. Eng 2020, 22, 155–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Mahecic D; Testa I; Griffie J; Manley S Strategies for increasing the throughput of super-resolution microscopies. Curr. Opin. Chem. Biol 2019, 51, 84–91. [DOI] [PubMed] [Google Scholar]
- (102).Reymond L; Huser T; Ruprecht V; Wieser S Modulation-enhanced localization microscopy. J. Phys. Photon 2020, 2, 041001. [Google Scholar]
- (103).Balzarotti F; Eilers Y; Gwosch KC; Gynnå AH; Westphal V; Stefani FD; Elf J; Hell SW Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science 2017, 355, 606–612. [DOI] [PubMed] [Google Scholar]
- (104).Gwosch KC; Pape JK; Balzarotti F; Hoess P; Ellenberg J; Ries J; Hell SW MINFLUX nanoscopy delivers 3D multicolor nanometer resolution in cells. Nat. Methods 2020, 17, 217–224. [DOI] [PubMed] [Google Scholar]
- (105).Gu L; Li Y; Zhang S; Xue Y; Li W; Li D; Xu T; Ji W Molecular resolution imaging by repetitive optical selective exposure. Nat. Methods 2019, 16, 1114–1118. [DOI] [PubMed] [Google Scholar]
- (106).Reymond L; Ziegler J; Knapp C; Wang FC; Huser T; Ruprecht V; Wieser S SIMPLE: Structured illumination based point localization estimator with enhanced precision. Opt. Express 2019, 27, 24578–24590. [DOI] [PubMed] [Google Scholar]
- (107).Cnossen J; Hinsdale T; Thorsen RO; Siemons M; Schueder F; Jungmann R; Smith CS; Rieger B; Stallinga S Localization microscopy at doubled precision with patterned illumination. Nat. Methods 2020, 17, 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Gu L; Li Y; Zhang S; Zhou M; Xue Y; Li W; Xu T; Ji W Molecular-scale axial localization by repetitive optical selective exposure. Nat. Methods 2021, 18, 369–373. [DOI] [PubMed] [Google Scholar]
- (109).Chen K; Yan R; Xiang L; Xu K Excitation spectral microscopy for highly multiplexed fluorescence imaging and quantitative biosensing. Light Sci. Appl 2021, 10, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Berezin MY; Achilefu S Fluorescence lifetime measurements and biological imaging. Chem. Rev 2010, 110, 2641–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Rupsa D; Tiffany MH; Joe TS; Amani AG; Melissa CS Fluorescence lifetime imaging microscopy: fundamentals and advances in instrumentation, analysis, and applications. J. Biomed. Opt 2020, 25, 071203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Thiele JC; Helmerich DA; Oleksiievets N; Tsukanov R; Butkevich E; Sauer M; Nevskyi O; Enderlein J Confocal fluorescence-lifetime single-molecule localization microscopy. ACS Nano 2020, 14, 14190–14200. [DOI] [PubMed] [Google Scholar]
- (113).Bowman AJ; Klopfer BB; Juffmann T; Kasevich MA Electro-optic imaging enables efficient wide-field fluorescence lifetime microscopy. Nat Commun 2019, 10, 4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Oleksiievets N; Thiele JC; Weber A; Gregor I; Nevskyi O; Isbaner S; Tsukanov R; Enderlein J Wide-field fluorescence lifetime imaging of single molecules. J. Phys. Chem. A 2020, 124, 3494–3500. [DOI] [PubMed] [Google Scholar]
- (115).Brinks D; Hildner R; van Dijk EM; Stefani FD; Nieder JB; Hernando J; van Hulst NF Ultrafast dynamics of single molecules. Chem. Soc. Rev 2014, 43, 2476–91. [DOI] [PubMed] [Google Scholar]
- (116).Liebel M; Toninelli C; van Hulst NF Room-temperature ultrafast nonlinear spectroscopy of a single molecule. Nat. Photonics 2017, 12, 45–49. [Google Scholar]
- (117).Arroyo JO; Kukura P Non-fluorescent schemes for single-molecule detection, imaging and spectroscopy. Nat. Photonics 2015, 10, 11–17. [Google Scholar]
- (118).Young G; Kukura P Interferometric scattering microscopy. Annu Rev Phys Chem 2019, 70, 301–322. [DOI] [PubMed] [Google Scholar]
- (119).Ha T; Tinnefeld P Photophysics of fluorescent probes for single-molecule biophysics and super-resolution imaging. Annu. Rev. Phys. Chem 2012, 63, 595–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Li HL; Vaughan JC Switchable fluorophores for single-molecule localization microscopy. Chem. Rev 2018, 118, 9412–9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).Wang L; Frei MS; Salim A; Johnsson K Small-molecule fluorescent probes for live-cell super-resolution microscopy. J. Am. Chem. Soc 2019, 141, 2770–2781. [DOI] [PubMed] [Google Scholar]
- (122).Jradi FM; Lavis LD Chemistry of photosensitive fluorophores for single-molecule localization microscopy. ACS Chem. Biol 2019, 14, 1077–1090. [DOI] [PubMed] [Google Scholar]
- (123).Teng KW; Ishitsuka Y; Ren P; Youn Y; Deng X; Ge P; Lee SH; Belmont AS; Selvin PR Labeling proteins inside living cells using external fluorophores for microscopy. eLife 2016, 5, e20378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Moon S; Li W; Hauser M; Xu K Graphene-enabled, spatially controlled electroporation of adherent cells for live-cell super-resolution microscopy. ACS Nano 2020, 14, 5609–5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (125).Lu K; Vu CQ; Matsuda T; Nagai T Fluorescent protein-based indicators for functional super-resolution imaging of biomolecular activities in living cells. Int. J. Mol. Sci 2019, 20, 5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (126).Zhou X; Mehta S; Zhang J Genetically encodable fluorescent and bioluminescent biosensors light up signaling networks. Trends Biochem. Sci 2020, 45, 889–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Zhao Y; Pal K; Tu Y; Wang X Cellular force nanoscopy with 50 nm resolution based on integrin molecular tension imaging and localization. J. Am. Chem. Soc 2020, 142, 6930–6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (128).Brockman JM; Su H; Blanchard AT; Duan Y; Meyer T; Quach ME; Glazier R; Bazrafshan A; Bender RL; Kellner AV et al. Live-cell super-resolved PAINT imaging of piconewton cellular traction forces. Nat Methods 2020, 17, 1018–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (129).Kim SH; Liu Y; Hoelzel C; Zhang X; Lee T-H Super-resolution optical lithography with DNA. Nano Lett 2019, 19, 6035–6042. [DOI] [PubMed] [Google Scholar]
- (130).Liu N; Dai M; Saka SK; Yin P Super-resolution labelling with Action-PAINT. Nat. Chem 2019, 11, 1001–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (131).Yang ZG; Cao JF; He YX; Yang JH; Kim T; Peng XJ; Kim JS Macro-/micro-environment-sensitive chemosensing and biological imaging. Chem. Soc. Rev 2014, 43, 4563–4601. [DOI] [PubMed] [Google Scholar]
- (132).Wu D; Sedgwick AC; Gunnlaugsson T; Akkaya EU; Yoon J; James TD Fluorescent chemosensors: the past, present and future. Chem. Soc. Rev 2017, 46, 7105–7123. [DOI] [PubMed] [Google Scholar]
- (133).Hou JT; Ren WX; Li K; Seo J; Sharma A; Yu XQ; Kim JS Fluorescent bioimaging of pH: from design to applications. Chem. Soc. Rev 2017, 46, 2076–2090. [DOI] [PubMed] [Google Scholar]
- (134).Brasselet S; Moerner WE Fluorescence behavior of single-molecule pH-sensors. Single Molecules 2000, 1, 17–23. [Google Scholar]
- (135).Sun X; Xie J; Xu J; Higgins DA; Hohn KL Single-molecule studies of acidity distributions in mesoporous aluminosilicate thin films. Langmuir 2015, 31, 5667–5675. [DOI] [PubMed] [Google Scholar]
- (136).Lerner E; Cordes T; Ingargiola A; Alhadid Y; Chung S; Michalet X; Weiss S Toward dynamic structural biology: Two decades of single-molecule Förster resonance energy transfer. Science 2018, 359, eaan1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (137).Sustarsic M; Kapanidis AN Taking the ruler to the jungle: single-molecule FRET for understanding biomolecular structure and dynamics in live cells. Curr. Opin. Struct. Biol 2015, 34, 52–59. [DOI] [PubMed] [Google Scholar]
- (138).Nicovich PR; Owen DM; Gaus K Turning single-molecule localization microscopy into a quantitative bioanalytical tool. Nat. Protoc 2017, 12, 453–460. [DOI] [PubMed] [Google Scholar]
- (139).Khater IM; Nabi IR; Hamarneh G A review of super-resolution single-molecule localization microscopy cluster analysis and quantification methods. Patterns 2020, 1, 100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (140).Wu YL; Tschanz A; Krupnik L; Ries J Quantitative data analysis in single-molecule localization microscopy. Trends Cell Biol 2020, 30, 837–851. [DOI] [PubMed] [Google Scholar]


