Abstract
COVID-19 is a highly contagious disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The case-fatality rate is significantly higher in older patients and those with diabetes, cancer or cardiovascular disorders. The human proteins, angiotensin-converting enzyme 2 (ACE2), transmembrane protease serine 2 (TMPRSS2) and basigin (BSG), are involved in high-confidence host-pathogen interactions with SARS-CoV-2 proteins. We considered these three proteins as seed nodes and applied the random walk with restart method on the human interactome to construct a protein-protein interaction sub-network, which captures the effects of viral invasion. We found that ‘Insulin resistance’, ‘AGE-RAGE signaling in diabetic complications’ and ‘adipocytokine signaling’ were the common pathways associated with diabetes, cancer and cardiovascular disorders. The association of these critical pathways with aging and its related diseases explains the molecular basis of COVID-19 fatality. We further identified drugs that have effects on these proteins/pathways based on gene expression studies. We particularly focused on drugs that significantly downregulate ACE2 along with other critical proteins identified by the network-based approach. Among them, COL-3 had earlier shown activity against acute lung injury and acute respiratory distress, while entinostat and mocetinostat have been investigated for non-small-cell lung cancer. We propose that these drugs can be repurposed for COVID-19.
Keywords: SARS-CoV-2, coronavirus, disease comorbidity analysis, protein-protein interactions, biological networks, biological pathways, drug repurposing
1. Introduction
The COVID-19 outbreak started in the Wuhan city of China in December 2019, and rapidly spread to other parts of the world [1]. It was declared as a pandemic by the World Health Organization in March 2020, and by mid of May 2020, it had spread to more than 200 countries infecting more than 4 million people and causing more than 300000 deaths [2]. Coronaviruses have caused two major pandemics in the past - severe acute respiratory syndrome (SARS), which originated from China in 2003, and Middle East respiratory syndrome (MERS), which originated from Saudi Arabia in 2012 [3], [4]. Several research groups are working on the development of drugs and vaccines against these viruses. There are several ongoing clinical trials of drugs such as chloroquine, lopinavir, ritonavir, nafamostat, camostat and famotidine [5], [6]. However, currently there are no therapeutics which have been considered safe and effective for the treatment of COVID-19 [7].
The viral entry into human cells is carried out by the spike (S) protein of SARS-CoV-2. Three human proteins have been identified as host receptors for the viral invasion into human cells - angiotensin-converting enzyme 2 (ACE2) [8], transmembrane protease serine 2 (TMPRSS2) [9] and basigin (BSG/CD147) [10]. In addition, a number of human proteins involved in host-pathogen protein-protein interactions with the virus have been identified recently [11]. In another recent study the authors identified a number of host-pathogen interactions for SARS-CoV-2 by assembling CoV-associated host proteins from four known human coronaviruses (SARS-CoV, MERS-CoV, HCoV-229E, and HcoV-NL63) [12]. There are few studies based on gene co-expression analysis of ACE2, TMPRSS2 and other co-expressed genes in the lung cell [13], [14]. Some other studies have also analyzed the differential gene expression, but these studies are also restricted to a single cell type [15], [16], [17], [18].
Apart from causing pneumonia, COVID-19 may also cause damage to other organs such as the heart, liver, and kidneys, as well as to organ systems such as the circulatory and the immune system. Patients eventually die of multiple organ failure, shock, acute respiratory distress syndrome, heart failure and renal failure. Diseases like diabetes, cardiovascular disorders and cancer are risk factors for severe patients compared to non-severe patients [19]. It has also been observed that COVID-19 has more severe effects on older people and people with aforementioned lifestyle diseases than younger people [20]. Network-based approach has been effectively used to understand the disease mechanism and comorbidities of SARS-CoV and HIV infections [21]. In this work, we first re-constructed a host protein-protein interaction (PPI) network based on known information about initial host contacts of the virus and their neighborhood. The aim of this study is to carry out an integrated analysis of PPI, their association to diseases, genes, pathways and drug associations to explain the molecular basis of fatal comorbidities of COVID-19 with other diseases like diabetes, cardiovascular diseases and cancer. We have identified important proteins and pathways associated with these diseases and drugs that can be repurposed for COVID-19 using gene expression data of drug molecules and heterogeneous network information.
2. Methods
The workflow implemented in the study is shown in Fig. F1 of the supplementary file, which can be found on the Computer Society Digital Library at http://doi.ieeecomputersociety.org/10.1109/TCBB.2021.3075299. We start by identifying the local human PPI network around the three proteins which interact with SARS-CoV-2 proteins. We then identified the fatal comorbidities associated with the proteins in the PPI network, and carried out the pathway analysis to identify the critical pathways associated with the fatal comorbidities. Finally, we carried out drug analysis to show which drugs may have adverse effects in patients with comorbidities and identified those which may be effective.
2.1. COVID-19 Related Host PPI Network
The human PPI data was obtained from a recent study [22], which integrates experimentally validated interactions from 15 bioinformatics and systems biology databases. The interactome data was updated based on the recent versions of the constituent databases. The details about the construction of human interactome are provided in section S2 of the supplementary file, available online. The host PPI network associated with COVID-19 infection, called COVID-19 host PPI network, was constructed by considering the PPIs around the SARS-CoV-2 protein receptors in the human interactome. The three reported receptors, ACE2 [8], TMPRSS2 [9] and BSG [10], were considered as seed nodes for the execution of random walk with restart (RWR) algorithm on the human interactome. RWR is a ranking algorithm, which engages a walker to inspect the global network to determine the closeness between two proteins in the PPI network [23], [24], [25]. The walker starts the journey from the reported receptors (seed nodes) and travels randomly to all other proteins in the PPI network, but is forced to return to the seed proteins with a restart probability, r, of 0.7. The closeness of the proteins to the reported receptors is computed as a probability vector, Pi, which represents the probability of each protein in the PPI network at each step i as

|
where, P0 is the initial probability vector and AT is the transpose of the column-normalized adjacency matrix of the network. Pi+1, the final outcome of the RWR calculations, is considered to be converged when (Pi+1) – Pi < 1*10−6, indicating that the probability vector was stable. The significant proteins from RWR analysis were filtered based on RWR score cutoff of 0.0002.
The RWR algorithm is highly dependent on the network topology and may pick up several proteins unrelated to the seed proteins [23]. To screen these false positives, a permutation test was performed where RWR was employed 1000 times, each time using randomly generated seed proteins from the PPI network [23], [24], [25]. A p-value for each protein predicted by RWR was calculated, which captures the significance of the predicted proteins. The p-value was computed as

|
Here, Θ represented the number of RWRs on randomly generated seed proteins in which the probability of the RWR-predicted protein, prot, is higher than that of the seed nodes. Proteins with p-value lower than 0.05 were considered statistically significant for further analysis [23]. The sub-network of human interactome obtained from the RWR analysis can be analyzed to understand the mechanism of viral invasion.
2.2. Disease Comorbidity
The disease-gene association information was obtained by considering high confidence experimentally validated associations reported in DisGeNet [26], OMIM [27], ClinVar [28] and PheGenI [29] databases. The disease terms from these resources were mapped on the Disease Ontology database [30] to obtain uniform ontology. The disease classification in the Disease Ontology database was used to extend the disease-gene association information to the disease group. The diseases associated with the genes in COVID-19 host PPI network were identified as possible comorbidities, and were further analyzed. 79 disease terms belonging to the 6 disease groups (Fig. 2) were associated with one or more of the 30 genes from the COVID-19 host PPI network, as shown in Fig. F3 of the supplementary file, available online.
Fig. 2.
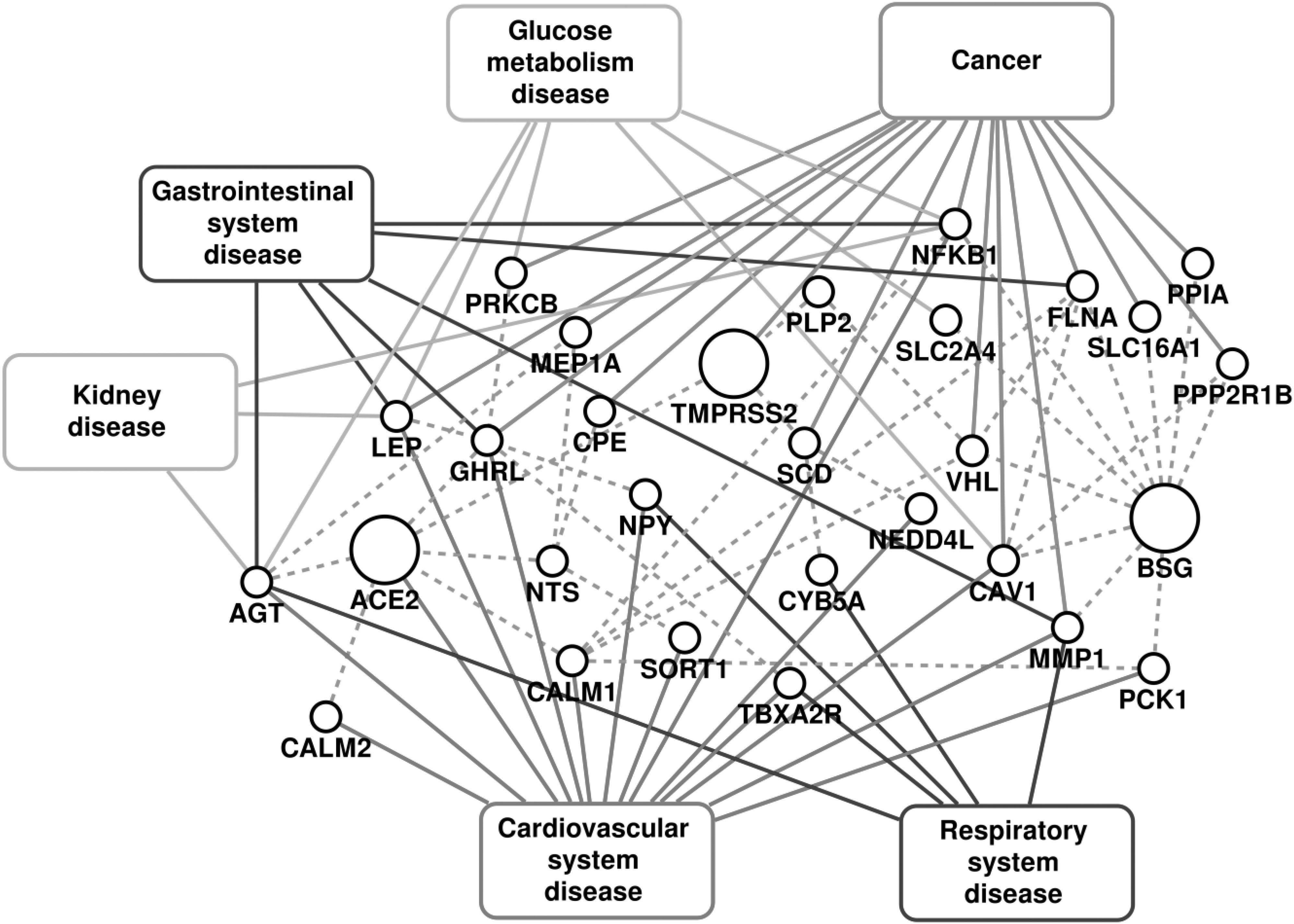
PPI associated with the COVID-19 infection and diseases associated with them. The dashed lines represent the protein-protein interactions.
2.3. Functional Analysis
The gene ontology (GO) analysis and pathway analysis were carried out using the gene set enrichment analysis web server, Enrichr [31]. The set of proteins associated with different disease groups were used as input in Enrichr to identify the statistically significant pathways among all the human pathways reported in KEGG database (2019 release) [32]. The pathways and GO terms with adjusted p-value < 0.05 computed using Fisher's exact test were considered as significant in the study. The significant GO terms for biological processes were obtained from the Gene Ontology resource (2018 release) [33] using Enrichr by considering all the proteins in the COVID-19 host PPI network. The relationship between the biological processes was analyzed using the REVIGO webserver [34].
2.4. Identification of Drug Molecules Using Gene Expression Data
Drug Gene Budger [35] was used to explore drugs and small molecules in L1000 data [36] that significantly regulated ACE2 expression (both upregulated and downregulated) along with other critical proteins of the COVID-19 host network. Small molecules that lead to differential gene expression of important proteins, captured by log2 fold change (LFC) higher than 1.5 for upregulation and less than −1.5 for downregulation, were considered for further analyses. Drugs which are responsible for significant over expression of ACE2 can increase the risk of COVID-19. On the other hand, drugs which significantly downregulate the ACE2 expression were further explored for their potential therapeutic application against COVID-19. Mechanism of action, clinical trial stages and structural details of drugs were analysed using DrugBank [37], PubChem [38], L1000FWD [39] and ClinicalTrials.gov (https://clinicaltrials.gov/). Drugbank is a knowledgebase consisting of pharmacological and clinical level information about drugs such as side effects and drug interactions. PubChem is a repository of chemical information where users can search chemicals and their chemical and physical properties, biological activities, safety and toxicity information among others. L1000 FireWorks Display (L1000FWD) shows effects of various perturbations on cells along with perturbation attributes such as mechanism of action. CrinicalTrials.gov is a repository of all clinical studies conducted across the world. These data sources were analysed to find mechanism of action, clinical trial stages and structural details of drugs. Drugs which were approved by FDA or undergoing clinical trials were considered for further analysis. An edge between an FDA-approved drug that induced a gene expression less than −1.5 log2 fold change was established with the respective gene to develop a drug-gene network.
All scripts were written in Python and Perl, while network analysis was performed in Cytoscape [40].
3. Results and Discussion
We have analyzed the biological processes, pathways and disease comorbidities of COVID-19 by considering the protein-protein interactions around the point of viral entry into the host cell through its receptors, ACE2, TMPRSS2 and BSG. The PPI neighborhood of ACE2, TMPRSS2 and BSG obtained from RWR (COVID-19 host PPI network) comprises of 65 proteins and 105 edges, as shown in Fig. 1. The progression of the viral infection affects human health through various biological processes carried out by the proteins in the neighborhood of the receptors in the human interactome. We identified the significant GO terms for biological processes associated with the proteins in the COVID-19 host PPI. The statistically significant biological processes and the relationship between them through the COVID-19 host PPI are shown in supplementary Fig. F2, available online. The processes are colored (Fig. F2) based on the dispensability score obtained from REVIGO, where higher scores are represented by darker shades of gray [34]. The COVID-19 host PPI comprises of proteins that regulate critical metabolic processes of the human body. These include the processes involved in immune response, glucose metabolism, vasoconstriction, protein metabolism and intracellular transport. Although the molecular basis of the effect of COVID-19 on these biological processes can be validated experimentally through protein assays and gene expression data analysis, the network-based approach provides a systems level understanding of the effect on the interconnected biological processes.
Fig. 1.
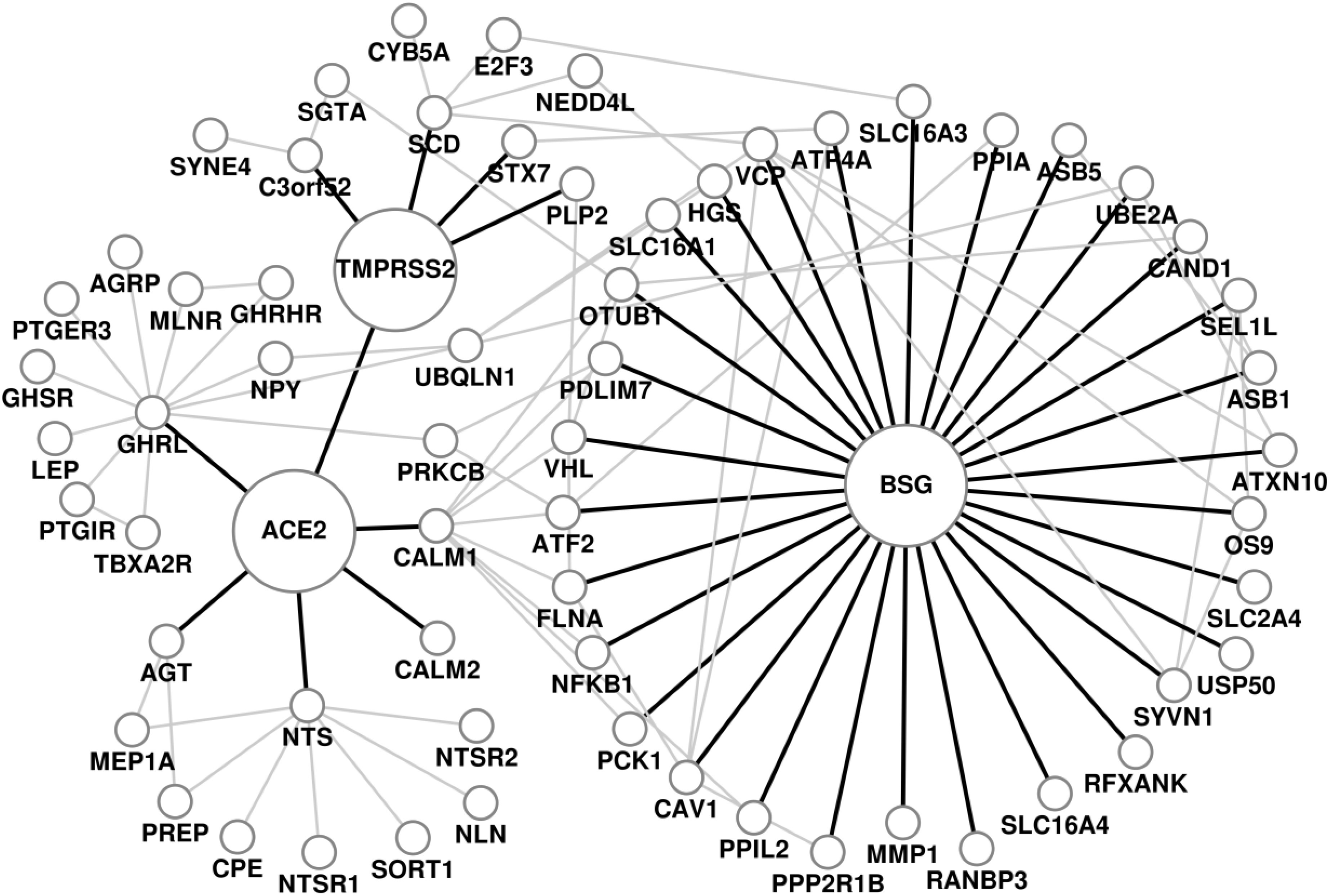
COVID-19 host protein-protein interaction network. The receptors of SARS-CoV2, ACE2, TMPRSS2 and BSG, and the edges connecting them to their immediate neighbors are highlighted.
3.1. Disease Comorbidity
The impact of COVID-19 has been observed to be severe in patients with cancer, cardiovascular diseases, diabetes and gastrointestinal disorders [1], [41], [42], [43]. The fatalities in these critical care patients have been mostly due to the original comorbidity leading to multiple organ failure [42], [44]. These complex conditions alter many biological processes and pathways in the human body. Here, we investigate the effect of COVID-19 through the host PPI and analyze the important pathways affected in the severe comorbidities due to the infection. Fig. 2 shows the disease association of the PPI network for respiratory diseases, cardiovascular diseases, cancers, glucose metabolism disorders, kidney diseases and gastrointestinal diseases, and the detailed association with each disease within the disease group is given in supplementary Fig. F3 and table T1, available online. The high comorbidity of COVID-19 with several diseases of different disease groups can be attributed to the critical genes that are associated with multiple disease groups. These include Angiotensinogen (AGT), Nuclear factor kappa B subunit 1 (NFKB1), Caveolin 1 (CAV1), Leptin (LEP), Ghrelin (GHRL) and Von Hippel-Lindau (VHL). For all the disease groups with fatal comorbidity, we identified the pathways associated with genes in the COVID-19 host PPI network. The significant pathways identified are shown in Fig. 3 and the detailed list of pathways is provided in supplementary table T2, available online.
Fig. 3.
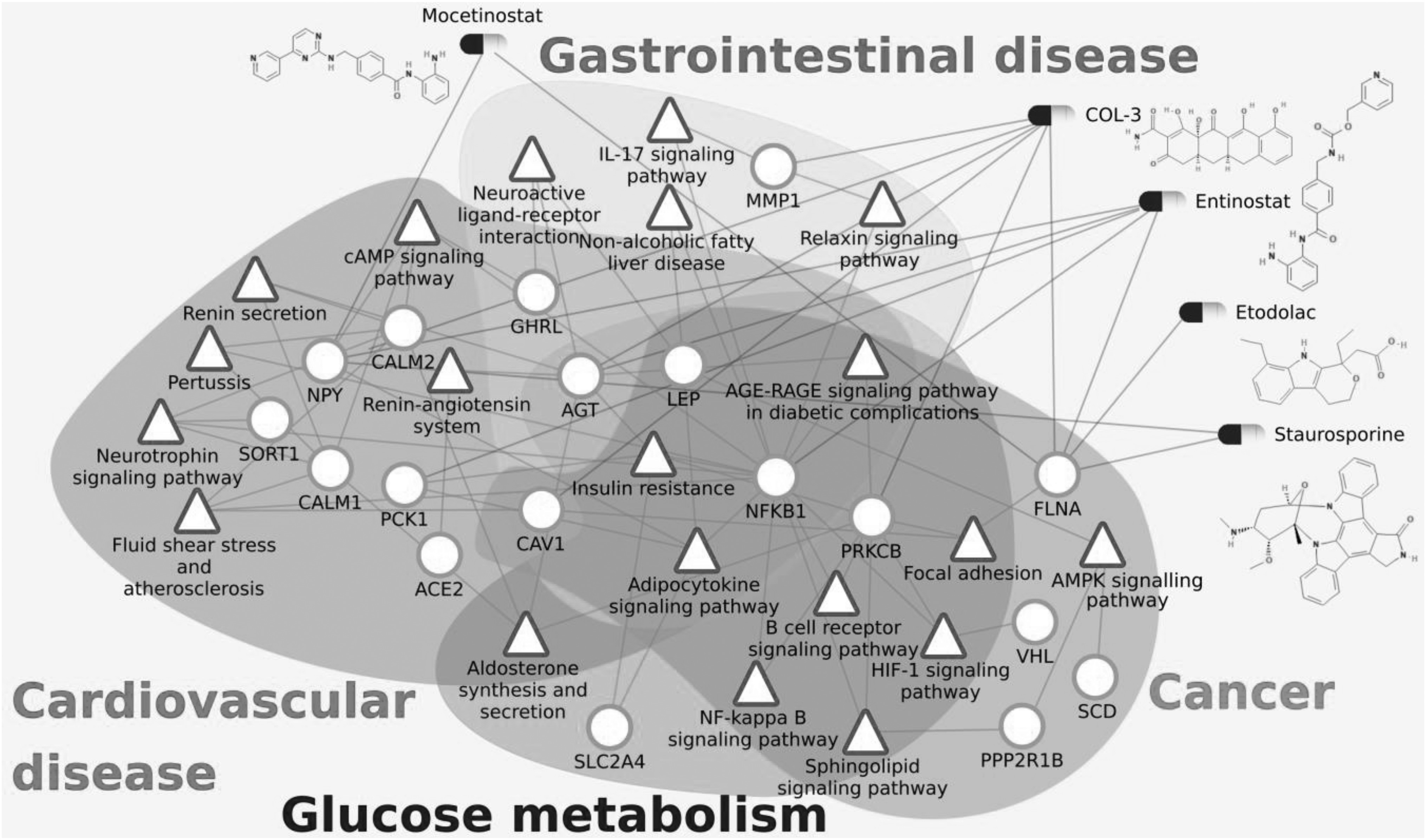
Heterogeneous network comprising of the pathways (green triangles) responsible for fatal comorbidities related to COVID-19, genes (red circles) and drugs (as capsules) which affect them. Important proteins and corresponding significant pathways are shown. All drugs shown here are also connected with ACE2 as they downregulate ACE2 gene expression but the edges between ACE2 and the drugs are not shown here for simplicity.
Glucose Metabolism Disorder. The genes AGT, LEP, PRKCB, NFKB1, CAV1 and SLC2A4 of the COVID-19 host PPI are associated with glucose metabolism diseases (Fig. 2). They regulate ‘insulin resistance’, ‘AGE-RAGE signaling pathway in diabetic complications’, ‘adipocytokine signaling pathway’, ‘B cell receptor signaling pathway’, ‘NF-kappa B signaling pathway’, ‘aldosterone synthesis and secretion’, ‘HIF-1 signaling pathway’ and ‘sphingolipid signaling pathway’.
Cardiovascular Diseases. The genes that can lead to cardiovascular diseases (Fig. 2) are involved in pathways that are responsible for ‘renin secretion’, ‘renin-angiotensin system’, ‘fluid shear stress and atherosclerosis’, ‘aldosterone synthesis and secretion’, ‘insulin resistance’, ‘AGE-RAGE signaling pathway in diabetic complications’, ‘adipocytokine signaling pathway’, ‘neurotrophin signaling pathway’ and ‘cAMP signaling pathway’. There are 14 genes of the COVID-19 host PPI network that are associated with cardiovascular diseases. Among them, AGT, ACE2, CALM1 and CALM2 are the critical genes associated with most of the cardiovascular diseases (supplementary Fig. F3, available online) and significant pathways (supplementary Table T2, available online), which are all connected in the immediate neighborhood of ACE2.
Cancer. The cancer pathways associated with the COVID-19 host PPIs are ‘HIF-1 signaling pathway’, ‘sphingolipid signaling pathway’, ‘AMPK signaling pathway’, ‘B cell receptor signaling pathway’, ‘NF-kappa B signaling pathway’, ‘insulin resistance’, ‘AGE-RAGE signaling pathway in diabetic complications’, ‘adipocytokine signaling pathway’ and ‘focal adhesion’. The genes PRKCB, NFKB1, VHL, PPP2RB1 and CAV1 are associated with different cancer types (supplementary Fig. F3, available online) and significant pathways (supplementary Table T2, available online).
Gastrointestinal Diseases. The genes AGT, LEP, GHRL, NFKB1, MMP1 and FLNA of the COVID-19 host PPI are known to be associated with gastrointestinal diseases. The pathways of gastrointestinal diseases are ‘relaxin signaling pathway’, ‘IL-17 signaling pathway’, ‘insulin resistance’, ‘AGE-RAGE signaling pathway in diabetic complications’, ‘adipocytokine signaling pathway’, ‘non-alcoholic fatty liver disease (NAFLD)’ and ‘neuroactive ligand-receptor interaction’. The genes that can lead to gastrointestinal diseases are shown in Fig. 2.
AGT is associated with the most number of diseases, which belong to the categories of cardiovascular, respiratory, glucose metabolism, kidney and gastrointestinal diseases. It is the only precursor of all angiotensin peptides and regulates blood pressure and homeostasis of water and sodium through the renin–angiotensin system (RAS) [45]. The RAS pathway is known to be associated with cardiovascular diseases, respiratory diseases, glucose metabolism, kidney disease and gastrointestinal diseases [46], [47], [48], [49]. NFKB1 is a transcription factor of proinflammatory molecules and is an important regulator of innate and adaptive immunity, cell proliferation, stress responses and apoptosis. It is therefore associated with pathogenic infections, diabetes, kidney and liver diseases, and cancer [50], [51]. CAV1 is a membrane protein associated with endocytosis, extracellular matrix organization, cholesterol distribution, cell migration and signaling [52]. It is associated with diabetes, cancer and cardiovascular diseases. LEP and GHRL regulate the energy homeostasis in the body by storage of fat and appetite regulation respectively [53], [54]. These are associated with glucose metabolism disorders, gastrointestinal disorders and some forms of cancer. VHL is a tumor suppressor gene associated with many forms of cancer (supplementary Fig. F3, available online) [55].
There are three critical pathways that are affected in all severe disease groups – ‘insulin resistance’, ‘AGE-RAGE signaling pathway in diabetic complications’ and ‘adipocytokine signaling pathway’. The progression of COVID-19 infection leading to severe conditions and fatality can be explained based on these critical pathways.
Insulin Resistance. Insulin resistance is a characteristic feature of the most prevalent metabolic disorder, type 2 diabetes, and is associated with cardiovascular diseases and cancer [56]. Hyperinsulinemia can lead to hypertension by the activation of the sympathetic nervous system, renal sodium retention, altered transmembrane cation transport and growth-promoting effects of vascular smooth muscle cells [57]. Hypertension along with dyslipidemia caused by insulin resistance can lead to cardiovascular conditions, especially coronary artery disease [58]. The increase in level of insulin can also stimulate the synthesis of sex steroids that can promote cellular proliferation and inhibit apoptosis, leading to cancer [59], [60]. Insulin resistance can also be associated with cancer through the overproduction of reactive oxygen species that can damage DNA, contributing to mutagenesis and carcinogenesis. The genes AGT, NFKB1, PRKCB and SLC2A4 of the COVID-19 host PPI are associated with insulin resistance.
AGE-RAGE Signaling Pathway in Diabetic Complications. Advanced glycation end products (AGEs) are the products of non-enzymatic glycation and oxidation of proteins and lipids that accumulate in diabetes [61], [62]. Increased levels of AGEs, especially carboxymethyllysine (CML), have been observed to be associated with atherosclerosis, coronary artery disease and heart failure [63]. The indirect vascular effects of elevated AGEs such as coronary dysfunction, atherosclerosis and thrombosis, and their direct effects on myocardium, lead to heart failure. The increased levels of these highly reactive AGEs induce persistent inflammation and oxidative stress, which lead to cancer [64], [65].
Adipocytokine Signaling Pathway. Adipocytokines are secreted by the adipose tissues, which signal key metabolic organs such as liver, muscle and pancreas, to maintain metabolic homeostasis through the adipocytokine signaling pathway [66]. Leptin (LEP) is the major adipocytokine which controls appetite and is associated with obesity. However, it also stimulates oxidative stress, inflammation, thrombosis, arterial stiffness, angiogenesis and atherogenesis. It is associated with diabetes, cardiovascular conditions, chronic kidney diseases and cancer [67], [68].
It can be observed that all the three pathways common to the fatal comorbidities are also related to aging [69], [70], [71], [72]. This is in congruence with the age-wise distribution of case fatality rate of COVID-19 observed in China and Italy [73], [74]. The case-fatality rate for patients older than 80 years is as high as 20 percent whereas it reduces to less than 1 percent for patients younger than 60 years of age. Aging leads to altered levels of insulin, AGEs and LEP in the body, which are further affected in COVID-19 patients through these pathways. It can be concluded from these observations that the three critical pathways play a significant role in regulating the severity of COVID-19 and risk of mortality, through the comorbid conditions such as diabetes, cardiovascular diseases, cancer, gastrointestinal disorders and higher age that are all associated with these three pathways.
3.2. Potential Drugs for COVID-19
The spike protein of SARS-CoV-2 mainly binds to the target cells through ACE2 [43]. The ACE2 is expressed in epithelial cells of the lungs, intestine, kidneys, and blood vessels [75]. Studies based on SARS-CoV infected mice model suggest that over-expression of human ACE2 enhanced disease severity [76], [77], [78]. Drug Gene Budger (DGB) [35] was used to explore existing drugs and small molecules that regulate ACE2 expression from L1000 data [36]. The analysis of expression data helps in understanding the importance of proteins and pathways identified in this study and their relationship with SARS-CoV-2 receptors. The objective is to identify drugs that can increase the chances of COVID-19 infection in patients with comorbidites and explore the existing drugs which can be repurposed for COVID-19.
Drugs that Increase the Possibility of COVID-19. Drugs that are responsible for overexpression of ACE2 can increase the probability of COVID-19 infection. Age and gender are other important determinants of ACE2 expression as it is observed that the expression is significantly higher in older people and males [79], [80]. The ACE2 expression is substantially increased in patients with type 1 and type 2 diabetes, who are treated with ACE inhibitors like moexipril (Drug Bank id: DB00691) [81] or angiotensin II type-I receptor blockers (ARBs). Hypertension is also treated with ACE inhibitors and ARBs, which results in an upregulation of ACE2. ACE2 can also be increased by thiazolidinediones and ibuprofen [43]. Based on gene-expression data from L1000, we identified few additional drugs which are responsible for significant over expression of ACE2. We hypothesise that these drugs can increase the chance of COVID-19. The ACE2 expression is significantly upregulated by Danusertib (LFC 1.8), which is studied for hormone refractory prostate cancer [82]. Trichostatin A is an anti-cancer drug, which also possesses antifungal and antibiotic properties. It inhibits the activation of the PI3K/Akt and ERK1/2 pathways [83] and significantly upregulates ACE2 expression (LFC 2).
Drugs That Downregulate ACE2 Expression. The drugs which significantly downregulate ACE2 activity, can be considered as probable therapeutics against COVID-19. Drugs that significantly downregulate ACE2 gene expression and at the same time inhibit other important proteins and pathways of the host sub-network (shown in Fig. 3) were identified from L1000 dataset [36]. Some recent articles indicate that hydroxychloroquine and chloroquine inhibit terminal glycosylation of ACE2 [84], [85]. As a result, ACE2 becomes less efficient in interacting with the SARS-CoV-2 spike protein, thus inhibiting viral entry. It will be interesting to identify other drugs that affect ACE2.
The complete list of drugs that downregulate ACE2 and at least one important protein of the COVID-19 host PPI network is shown in Table 1. COL-3 is chemically modified tetracycline-3 [6-dimethyl-6-deoxy-4-de(dimethylamino) tetracycline] which is also known as incyclinide or CMT-3. It has been extensively studied as a potential new therapeutic agent for allergic conditions, inflammatory conditions (i.e., arthritis, acute respiratory distress syndrome, septic shock syndrome, acne and rosacea), neoplastic diseases (i.e., colon carcinoma, prostate cancer) and infectious (fungal) diseases. COL-3 has been used in trials for HIV infection and brain and central nervous system tumors [86]. Most importantly, COL-3 has shown promising results against acute lung injury and acute respiratory distress in animal models [87]. It significantly downregulates ACE2 (LFC -1.8) and other important proteins in the COVID-19 host PPI network, including AGT, CAV1, FLNA, MMP1, NPY and PRKCB (see Fig. 3).
TABLE 1. Drugs From L1000 Dataset Which Significantly Downregulate ACE2 Along With Other Critical Proteins of the COVID-19 Related Human PPI Sub-Network (Fig. 3).
| S. No. | Drug name | Host genes downregulated | LFC for ACE2 | Status of drug |
|---|---|---|---|---|
| 1. | COL-3 | ACE2, AGT, CAV1, FLNA, MMP1, NPY, PRKCB | −1.8 | Phase 2 completed |
| 2 | Entinostat | ACE2, AGT, FLNA, NFKB, NPY, PCK1 | −1.8 | Phase 3 recruiting |
| 3 | Mocetinostat | ACE2, FLNA, NPY | −1.7 | Phase 2 completed |
| 4 | Staurosporine | ACE2, FLNA, NPY | −1.5 | Phase 2 completed |
| 5 | Etodolac | ACE2, FLNA | −1.6 | FDA approved |
The log2 Fold Change (LFC) Data is Shown Only for ACE2.
Interestingly, the list of drugs include investigational anti-cancer drugs entinostat, mocetinostat and alvespimycin. Entinostat and mocetinostat are benzamide-containing histone deacetylase (HDAC) inhibitors that downregulate ACE2 expression (LFC −1.8 and −1.7 respectively as shown in Table 1). Entinostat [88] is under investigation for the treatment of non-small-cell lung cancer and epigenetic therapy. It is also found to downregulate other important proteins in the COVID-19 host PPI network, including AGT, FLNA, NFKB, NPY and PCK1. Mocetinostat [89] is currently in phase 2 clinical trials for the treatment of various lymphoid and myeloid malignancies [90]. Alvespimycin is a derivative of geldanamycin, which is known to reduce acute respiratory distress syndrome [91], [92]. Alvespimycin inhibits HSP90 and its regulation of cell signaling pathways. It downregulates ACE2 expression (LFC −2.11) and at the same time significantly downregulates CAV1, FLNA, MMP1, NPY which are part of the COVID-19 host PPI network. In fact, geldanamycin itself downregulates ACE2 (LFC −1.2). However, Alvespimycin has been terminated in phase 2 clinical trials. Staurosporine, a natural product isolated from Streptomyces staurosporeus, is a potent PKC inhibitor which downregulates ACE2 expression (LFC −1.5) [93], [94].
Etodolac [95] is a non-steroidal anti-inflammatory moderate painkiller drug used in rheumatoid arthritis and osteoarthritis. It significantly downregulates ACE2 expression (LFC −1.5). The anti-inflammatory effects of etodolac result from inhibition of the cyclooxygenase enzymes (COX), specially COX-2. COX-2 is part of the NFKB pathway, an important pathway of the COVID-19 host PPI sub-network. It was also found to downregulate FLNA of the host sub-network, which is also part of the NFKB pathway.
4. Conclusion
Diseases are regulated by a complex network of protein-protein interactions. The impact of a pathogenic infection on human health is governed by the host-pathogen interactions, and it propagates through the PPI network to affect biological processes. We have used the human PPI network to explain the molecular basis of the relationships between COVID-19 and other diseases that have high case-fatality rate. We started with three high confidence host contacts of SARS-CoV-2, viz., ACE2, TMPRSS2 and BSG, and then identified the local network around them in the human interactome using the RWR method. We could identify the proteins and pathways that are implicated in cancer, cardiovascular disease, diabetes and gastrointestinal disorders from this local network. The pathways ‘Insulin resistance’, ‘AGE-RAGE signaling in diabetic complications’ and ‘adipocytokine signaling’ were found to be associated with all the fatal comorbidities and are also known to be associated with ageing, which has been reported in several studies as one of the major risk factors in COVID-19 patients. These observations suggest that the severity of disease progression in SARS-CoV-2 infection and the risk of mortality is higher for patients with comorbidities associated with the three critical pathways.
The present study is an attempt to integrate publicly available multi-omics data including protein-protein interactions, disease-gene associations, pathway information, data of FDA-approved drugs and drugs undergoing clinical trials, and the differential gene-expression of drugs to provide a systems level understanding of the propagation of COVID-19 in patients with fatal comorbidities, and also comment on drug indications. We identified 5 drugs that can significantly downregulate the primary receptor of SARS-CoV-2, ACE2, along with other important proteins of the host PPI sub-network. Among them, COL-3 has previously shown activity against acute lung injury and acute respiratory distress, while entinostat and mocetinostat are in clinical trials for non-small-cell lung cancer. We opine that these drugs can be investigated further for their therapeutic value and repurposed against COVID-19.
The inferences presented in this work are based on a holistic approach to understand the critical comorbidities of COVID-19 based on protein-protein interactions. Although we have only considered experimentally validated data, the reliability of the analysis presented in the work is dependent on the authenticity of the data available across the various databases. The molecular basis of comorbidities and potential drugs proposed here are preliminary indications requiring experimental validation.
Acknowledgments
The authors would like to thank Dr. Rajgopal Srinivasan, Dr. Siladitya Padhi, Dr. Navneet Bung and Sowmya Ramaswamy Krishnan for helpful discussions. The authors declare that they are employees of Tata Consultancy Services Limited—a commercial company. The company has provided support for this study in the form of salaries to authors, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. Broto Chakrabarty and Dibyajyoti Das contributed equally to this work.
Conflict of Interest
Broto Chakrabarty, Dibyajyoti Das, Gopalakrishnan Bulusu and Arijit Roy were employed by the company Tata Consultancy Services Limited.
Biographies

Broto Chakrabarty received the bachelor of technology degree in bioinformatics from the Jaypee University of Information Technology, the MS degree in research, and the PhD degree in bioinformatics from the International Institute of Information Technology, Hyderabad, India. He is currently a scientist with TCS Innovation Labs, Hyderabad. He is currently working on the application of network theory for the analyses of protein structures, protein dynamics, and systems level analysis of protein–protein interactions to understand the complex disease mechanisms. His research focuses on biological network analysis.

Dibyajyoti Das received the bachelors of technology degree in biotechnology from the Heritage Institute of Technology, Kolkata, the masters of technology degree in bioinformatics from the University of Hyderabad, Hyderabad. He is currently a scientist with TCS Innovation Labs, Hyderabad, with more than 10 years of experience. His research interests include applications of deep learning, machine learning, and network theory to understand biological systems.

Gopalakrishnan Bulusu studied chemistry from the University of Madras in 1981 and from the University of Hyderabad in 1983, and received the PhD degree in molecular biophysics from the Indian Institute of Science, Bengaluru, in 1989. From 1990 to 1992, he was a research associate in the area of structural biology with the Universities of Paris and from 1993 to 1996, with Cambridge. From 1997 to 2001, before he joined Dr. Reddy's Laboratories, Hyderabad, he taught medicinal chemistry for five years with the National Institute of Pharmaceutical Education and Research, where he lead the Molecular Modelling and Drug Design Department in 2001. In 2003, he joined R&D Division of Tata Consultancy Services, where he led the Computational Structural Biology group until he retired in 2020. He is currently an adjunct professor with the Centre for Computational Natural Sciences and Bioinformatics, International Institute of Information Technology, Hyderabad, and with the Dr. Reddy's Institute of Life Sciences, University of Hyderabad Campus. He has authored more than 50 research papers and two book chapters. His research interests include the areas of bioinformatics, structural biology, and drug design. He is a life member of the Indian Biophysical Society, wherein he is currently a vice president. He was the member of the ACS and ISCB.

Arijit Roy received the PhD degree in chemistry from the Indian Institute of Technology, Kharagpur. He is currently a senior scientist with TCS Innovation Labs, Hyderabad. He was a postdoc with the Institut de Biologie Structurale, Grenoble, France, and the Stony Brook University, NY, USA. He also develops rare event simulation methods to study biological systems. His research interests include drug design using deep neural network based methods, network biology, and drug repurposing.
Contributor Information
Broto Chakrabarty, Email: broto.c@tcs.com.
Dibyajyoti Das, Email: dibyajyoti.das@tcs.com.
Gopalakrishnan Bulusu, Email: g.bulusu@tcs.com.
Arijit Roy, Email: roy.arijit3@tcs.com.
References
- [1].Huang C., et al. , “Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China,” Lancet, vol. 395, no. 10223, pp. 497–506, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bedford J., et al. , “COVID-19: Towards controlling of a pandemic,” Lancet, vol. 395, no. 10229, pp. 1015–1018, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Drosten C., et al. , “Identification of a novel coronavirus in patients with severe acute respiratory syndrome,” New Engl. J. Med., vol. 348, no. 20, pp. 1967–1976, May 2003. [DOI] [PubMed] [Google Scholar]
- [4].Zaki A. M., van Boheemen S., Bestebroer T. M., Osterhaus A. D. M. E., and Fouchier R. A. M., “Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia,” New Engl. J. Med., vol. 367, no. 19, pp. 1814–1820, Nov. 2012. [DOI] [PubMed] [Google Scholar]
- [5].Dong L., Hu S., and Gao J., “Discovering drugs to treat coronavirus disease 2019 (COVID-19),” Drug Discov. Ther., vol. 14, no. 1, pp. 58–60, 2020. [DOI] [PubMed] [Google Scholar]
- [6].Shaffer L., “15 drugs being tested to treat COVID-19 and how they would work,” Nat. Med., May 15, 2020. [Online]. Available: https://www.nature.com/articles/d41591-020-00019-9 doi: 10.1038/d41591-020-00019-9 [DOI] [PubMed]
- [7].Cao B., et al. , “A trial of Lopinavir-Ritonavir in adults hospitalized with severe COVID-19,” New Engl. J. Med., vol. 382, no. 19, pp. 1787—1799, Mar. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhou P., et al. , “A pneumonia outbreak associated with a new coronavirus of probable bat origin,” Nature, vol. 579, no. 7798, pp. 270–273, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hoffmann M., et al. , “SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor,” Cell, vol. 181, no. 2, pp. 271–280, Apr. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wang K., et al. , “CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells,” Signal Transduct. Target. Ther., vol. 5, no. 1, Dec. 2020, Art. no. 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gordon D. E., et al. , “A SARS-CoV-2 protein interaction map reveals targets for drug repurposing,” Nature, vol. 583, pp. 459–468, Apr. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhou Y., Hou Y., Shen J., Huang Y., Martin W., and Cheng F., “Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2,” Cell Discov., vol. 6, 2020, Art. no. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cava C., Bertoli G., and Castiglioni I., “In silico discovery of candidate drugs against COVID-19,” Viruses, vol. 12, no. 4, Apr. 2020, Art. no. 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cordero A. I. H., et al. , “Gene expression network analysis provides potential targets against SARS-CoV-2,” Sci. Rep., vol. 10, no. 1, Dec. 2020, Art. no. 21863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ouyang Y., et al. , “Downregulated gene expression spectrum and immune responses changed during the disease progression in patients with COVID-19,” Clin. Infect. Dis., vol. 71, no. 16, pp. 2052–2060, Nov. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yuan J., Fan D., Xue Z., Qu J., and Su J., “Co-expression of mitochondrial genes and ACE2 in cornea involved in COVID-19,” Invest. Ophthalmol. Vis. Sci., vol. 61, no. 12, Oct. 2020, Art. no. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kusmartseva I., et al. , “Expression of SARS-CoV-2 entry factors in the pancreas of normal organ donors and individuals with COVID-19,” Cell Metab., vol. 32, no. 6, pp. 1041–1051.e6, Dec. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ramesh P., Veerappapillai S., and Karuppasamy R., “Gene expression profiling of corona virus microarray datasets to identify crucial targets in COVID-19 patients,” Gene Rep., vol. 22, Mar. 2021, Art. no. 100980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yang J., et al. , “Prevalence of comorbidities in the novel Wuhan coronavirus (COVID-19) infection: A systematic review and meta-analysis,” Int. J. Infect. Dis., vol. 94, pp. 91–95, May 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yang X., et al. , “Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study,” Lancet Respir. Med., vol. 8, no. 5, pp. 475–481, May 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Moni M. A. and Liò P., “Network-based analysis of comorbidities risk during an infection: SARS and HIV case studies,” BMC Bioinf., vol. 15, Oct. 2014, Art. no. 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Cheng F., Kovács I. A., and Barabási A.-L., “Network-based prediction of drug combinations,” Nat. Commun., vol. 10, no. 1, 2019, Art. no. 1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li L., Wang Y., An L., Kong X., and Huang T., “A network-based method using a random walk with restart algorithm and screening tests to identify novel genes associated with Menière's disease,” PLoS One, vol. 12, no. 8, 2017, Art. no. e0182592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chen Y. and Xu R., “Network-based gene prediction for Plasmodium falciparum malaria towards genetics-based drug discovery,” BMC Genomic , vol. 16, no. Suppl 7, 2015, Art. no. S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Das D., Krishnan S. R., Roy A., and Bulusu G., “A network-based approach reveals novel invasion and Maurer's clefts-related proteins in Plasmodium falciparum,” Mol. Omics, vol. 15, no. 6, pp. 431–441, 2019. [DOI] [PubMed] [Google Scholar]
- [26].Piñero J., et al. , “DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants,” Nucleic Acids Res. , vol. 45, no. D1, pp. D833–D839, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Amberger J. S., Bocchini C. A., Schiettecatte F., Scott A. F., and Hamosh A., “OMIM.org: Online mendelian inheritance in man (OMIM), an online catalog of human genes and genetic disorders,” Nucleic Acids Res. , vol. 43, pp. D789–D798, Jan. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Landrum M. J., et al. , “ClinVar: Public archive of interpretations of clinically relevant variants,” Nucleic Acids Res. , vol. 44, no. D1, pp. D862–D868, Jan. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ramos E. M., et al. , “Phenotype-genotype integrator (PheGenI): Synthesizing genome-wide association study (GWAS) data with existing genomic resources,” Eur. J. Hum. Genet., vol. 22, no. 1, pp. 144–147, Jan. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kibbe W. A., et al. , “Disease ontology 2015 update: An expanded and updated database of human diseases for linking biomedical knowledge through disease data,” Nucleic Acids Res., vol. 43, pp. D1071–D1078, Jan. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kuleshov M. V., et al. , “Enrichr: A comprehensive gene set enrichment analysis web server 2016 update,” Nucleic Acids Res. , vol. 44, no. W1, pp. W90–W97, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kanehisa M., Sato Y., Kawashima M., Furumichi M., and Tanabe M., “KEGG as a reference resource for gene and protein annotation,” Nucleic Acids Res. , vol. 44, pp. D457–D462, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].The Gene Ontology Consortium, “The gene ontology resource: 20 years and still going strong,” Nucleic Acids Res., vol. 47, pp. D330–D338, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Supek F., Bošnjak M., Škunca N., and Šmuc T., “REVIGO summarizes and visualizes long lists of gene ontology terms,” PLoS One, vol. 6, no. 7, 2011, Art. no. e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wang Z., He E., Sani K., Jagodnik K. M., Silverstein M. C., and Ma'ayan A., “Drug gene budger (DGB): An application for ranking drugs to modulate a specific gene based on transcriptomic signatures,” Bioinformatics, vol. 35, no. 7, pp. 1247–1248, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Subramanian A., et al. , “A next generation connectivity map: L1000 platform and the first 1,000,000 profiles,” Cell, vol. 171, no. 6, pp. 1437–1452, Nov. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wishart D. S., et al. , “DrugBank 5.0: A major update to the DrugBank database for 2018,” Nucleic Acids Res. , vol. 46, no. D1, pp. D1074–D1082, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kim S., et al. , “PubChem 2019 update: Improved access to chemical data,” Nucleic Acids Res. , vol. 47, no. D1, pp. D1102–D1109, Jan. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang Z., Lachmann A., Keenan A. B., and Ma'ayan A., “L1000FWD: Fireworks visualization of drug-induced transcriptomic signatures,” Bioinformatics, vol. 34, no. 12, pp. 2150–2152, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Shannon P., et al. , “Cytoscape: A software environment for integrated models of biomolecular interaction networks,” Genome Res. , vol. 13, no. 11, pp. 2498–2504, Nov. 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wang D., et al. , “Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China,” JAMA, vol. 323, no. 11, pp. 1061–1069, Feb. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wang T., et al. , “Comorbidities and multi-organ injuries in the treatment of COVID-19,” Lancet, vol. 395, no. 10228, 2020, Art. no. e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Fang L., Karakiulakis G., and Roth M., “Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection?,” Lancet Respir Med., vol. 8, no. 4, Mar. 2020, Art. no. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Liang W., et al. , “Cancer patients in SARS-CoV-2 infection: A nationwide analysis in China,” Lancet Oncol. , vol. 21, no. 3, pp. 335–337, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lu H., Cassis L. A., Kooi C. W. V., and Daugherty A., “Structure and functions of angiotensinogen,” Hypertens. Res., vol. 39, no. 7, pp. 492–500, Jul. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wu C.-H., Mohammadmoradi S., Chen J. Z., Sawada H., Daugherty A., and Lu H. S., “Renin-angiotensin system and cardiovascular functions,” Arterioscler Thromb. Vasc. Biol., vol. 38, no. 7, pp. e108–e116, Jul. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Marshall R. P., “The pulmonary renin-angiotensin system,” Curr. Pharmaceut. Des., vol. 9, no. 9, pp. 715–722, 2003. [DOI] [PubMed] [Google Scholar]
- [48].Joseph J. J., Tcheugui J. B. E., Effoe V. S., Hsueh W. A., Allison M. A., and Golden S. H., “Renin-angiotensin-aldosterone system, glucose metabolism and incident type 2 diabetes mellitus: MESA,” J. Amer. Heart Assoc., vol. 7, no. 17, 2018, Art. no. e009890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Remuzzi G., Perico N., Macia M., and Ruggenenti P., “The role of renin-angiotensin-aldosterone system in the progression of chronic kidney disease,” Kidney Int. Suppl., no. 99, pp. 57–65, Dec. 2005. [DOI] [PubMed]
- [50].Patel S. and Santani D., “Role of NF-kappa B in the pathogenesis of diabetes and its associated complications,” Pharmacol. Rep., vol. 61, no. 4, pp. 595–603, Aug. 2009. [DOI] [PubMed] [Google Scholar]
- [51].Cartwright T., Perkins N. D., and Wilson C. L., “NFKB1: A suppressor of inflammation, ageing and cancer,” FEBS J. , vol. 283, no. 10, pp. 1812–1822, 2016. [DOI] [PubMed] [Google Scholar]
- [52].Nwosu Z. C., Ebert M. P., Dooley S., and Meyer C., “Caveolin-1 in the regulation of cell metabolism: A cancer perspective,” Mol. Cancer, vol. 15, Nov. 2016, Art. no. 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Margetic S., Gazzola C., Pegg G. G., and Hill R. A., “Leptin: A review of its peripheral actions and interactions,” Int. J. Obesity Rel. Metab. Disord., vol. 26, no. 11, pp. 1407–1433, Nov. 2002. [DOI] [PubMed] [Google Scholar]
- [54].Sato T., Nakamura Y., Shiimura Y., Ohgusu H., Kangawa K., and Kojima M., “Structure, regulation and function of ghrelin,” J. Biochem., vol. 151, no. 2, pp. 119–128, Feb. 2012. [DOI] [PubMed] [Google Scholar]
- [55].Kim W. Y. and Kaelin W. G., “Role of VHL gene mutation in human cancer,” J. Clin. Oncol., vol. 22, no. 24, pp. 4991–5004, Dec. 2004. [DOI] [PubMed] [Google Scholar]
- [56].Yaribeygi H., Farrokhi F. R., Butler A. E., and Sahebkar A., “Insulin resistance: Review of the underlying molecular mechanisms,” J. Cell. Physiol., vol. 234, no. 6, pp. 8152–8161, Jun. 2019. [DOI] [PubMed] [Google Scholar]
- [57].McFarlane S. I., Banerji M., and Sowers J. R., “Insulin resistance and cardiovascular disease,” J. Clin. Endocrinol. Metab., vol. 86, no. 2, pp. 713–718, Feb. 2001. [DOI] [PubMed] [Google Scholar]
- [58].Ginsberg H. N., “Insulin resistance and cardiovascular disease,” J. Clin. Investig., vol. 106, no. 4, pp. 453–458, Aug. 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Arcidiacono B., et al. , “Insulin resistance and cancer risk: An overview of the pathogenetic mechanisms,” Exp. Diabetes Res., vol. 2012, vol. 2012, Art. no. 789174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Orgel E. and Mittelman S. D., “The links between insulin resistance, diabetes, and cancer,” Curr. Diabetes Rep., vol. 13, no. 2, pp. 213–222, Apr. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Singh R., Barden A., Mori T., and Beilin L., “Advanced glycation end-products: A review,” Diabetologia, vol. 44, no. 2, pp. 129–146, Feb. 2001. [DOI] [PubMed] [Google Scholar]
- [62].Ramasamy R., Yan S. F., and Schmidt A. M., “Receptor for AGE (RAGE): Signaling mechanisms in the pathogenesis of diabetes and its complications,” Ann. New York Acad. Sci., vol. 1243, pp. 88–102, Dec. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hegab Z., Gibbons S., Neyses L., and Mamas M. A., “Role of advanced glycation end products in cardiovascular disease,” World J. Cardiol., vol. 4, no. 4, pp. 90–102, Apr. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Turner D. P., “Advanced glycation end-products: A biological consequence of lifestyle contributing to cancer disparity,” Cancer Res., vol. 75, no. 10, pp. 1925–1929, May 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Schröter D., and Höhn A., “Role of advanced glycation end products in carcinogenesis and their therapeutic implications,” Curr. Pharmaceut. Des., vol. 24, no. 44, pp. 5245–5251, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Cao H., “Adipocytokines in obesity and metabolic disease,” J. Endocrinol., vol. 220, no. 2, pp. 47–59, Feb. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Dutta D., Ghosh S., Pandit K., Mukhopadhyay P., and Chowdhury S., “Leptin and cancer: Pathogenesis and modulation,” Indian J. Endocrinol. Metab., vol. 16, no. Suppl 3, pp. S596–S600, Dec. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Katsiki N., Mikhailidis D. P., and Banach M., “Leptin, cardiovascular diseases and type 2 diabetes mellitus,” Acta Pharmacologica Sinica, vol. 39, no. 7, pp. 1176–1188, Jul. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Fink R. I., Kolterman O. G., Griffin J., and Olefsky J. M., “Mechanisms of insulin resistance in aging,” J. Clin. Invest., vol. 71, no. 6, pp. 1523–1535, Jun. 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Ryan A. S., “Insulin resistance with aging: Effects of diet and exercise,” Sports Med., vol. 30, no. 5, pp. 327–346, Nov. 2000. [DOI] [PubMed] [Google Scholar]
- [71].Gulcelik N. E., Halil M., Ariogul S., and Usman A., “Adipocytokines and aging: Adiponectin and leptin,” Minerva Endocrinol., vol. 38, no. 2, pp. 203–210, Jun. 2013. [PubMed] [Google Scholar]
- [72].Chaudhuri J., et al. , “The role of advanced glycation end products in aging and metabolic diseases: Bridging association and causality,” Cell Metab. , vol. 28, no. 3, pp. 337–352, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Epidemiology Group of Emergency Response Mechanism of Novel Coronavirus Pneumonia, China Center for Disease Control Prevention, “The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China,” Chinese J. Epidemiol., vol. 41, no. 2, pp. 145–151, Feb. 2020. [Google Scholar]
- [74].Onder G., Rezza G., and Brusaferro S., “Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy,” JAMA, vol. 323, no. 18, pp. 1775–1776, Mar. 2020. [DOI] [PubMed] [Google Scholar]
- [75].Wan Y., Shang J., Graham R., Baric R. S., and Li F., “Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus,” J. Virol., vol. 94, no. 7, 2020, Art. no. e00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Yang X.-H., et al. , “Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection,” Comput. Med., vol. 57, no. 5, pp. 450–459, Oct. 2007. [PubMed] [Google Scholar]
- [77].Sommerstein R., Kochen M. M., Messerli F. H., and Gräni C., “Coronavirus disease 2019 (COVID-19): Do Angiotensin-converting enzyme inhibitors/angiotensin receptor blockers have a biphasic effect?,” J. Amer. Heart Assoc., vol. 9, no. 7, 2020, Art. no. e016509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Monteil V., et al. , “Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2,” Cell, vol. 181, no. 4, pp. 905–913, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Fernández-Atucha A., et al. , “Sex differences in the aging pattern of renin-angiotensin system serum peptidases,” Biol. Sex Differences, vol. 8, 2017, Art. no. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Walters T. E., Kalman J. M., Patel S. K., Mearns M., Velkoska E., and Burrell L. M., “Angiotensin converting enzyme 2 activity and human atrial fibrillation: Increased plasma angiotensin converting enzyme 2 activity is associated with atrial fibrillation and more advanced left atrial structural remodelling,” Europace, vol. 19, no. 8, pp. 1280–1287, Aug. 2017. [DOI] [PubMed] [Google Scholar]
- [81].Chrysant G. S. and Nguyen P. K., “Moexipril and left ventricular hypertrophy,” Vasc. Health Risk Manage. , vol. 3, no. 1, pp. 23–30, 2007. [PMC free article] [PubMed] [Google Scholar]
- [82].Meulenbeld H. J., et al. , “Randomized phase II study of danusertib in patients with metastatic castration-resistant prostate cancer after docetaxel failure,” BJU Int , vol. 111, no. 1, pp. 44–52, Jan. 2013. [DOI] [PubMed] [Google Scholar]
- [83].Ma J., et al. , “Trichostatin A, a histone deacetylase inhibitor, suppresses proliferation and promotes apoptosis of esophageal squamous cell lines,” Mol. Med. Rep., vol. 11, no. 6, pp. 4525–4531, Jun. 2015. [DOI] [PubMed] [Google Scholar]
- [84].Wang M., et al. , “Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro,” Cell Res , vol. 30, no. 3, pp. 269–271, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Vincent M. J., et al. , “Chloroquine is a potent inhibitor of SARS coronavirus infection and spread,” Virol. J. , vol. 2, Aug. 2005, Art. no. 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Viera M. H., Perez O. A., and Berman B., “Incyclinide,” Drugs Future, vol. 32, no. 3, pp. 209–214, 2007. [Google Scholar]
- [87].Bosma K. J., Taneja R., and Lewis J. F., “Pharmacotherapy for prevention and treatment of acute respiratory distress syndrome: Current and experimental approaches,” Drugs, vol. 70, no. 10, pp. 1255–1282, Jul. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Connolly R. M., Rudek M. A., and Piekarz R., “Entinostat: A promising treatment option for patients with advanced breast cancer,” Future Oncol., vol. 13, no. 13, pp. 1137–1148, Jun. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Gerson S. L., Caimi P. F., William B. M., and Creger R. J., “Pharmacology and molecular mechanisms of antineoplastic agents for hematologic malignancies,” in Hematology, Hoffman R., et al., Eds., 7th ed. Philadelphia, U.K.: Elsevier, 2018, pp. 849–912. [Google Scholar]
- [90].Sheikh S., Bekheet M., Olzscha H., and La Thangue N. B., “Predicting and monitoring responses to epigenetic drugs,” in Drug Discovery in Cancer Epigenetics, Egger G. and Arimondo P., Eds. Waltham, MA, USA: Academic, 2016, pp. 373–406. [Google Scholar]
- [91].Lancet J. E., et al. , “Phase I study of the heat shock protein 90 inhibitor alvespimycin (KOS-1022, 17-DMAG) administered intravenously twice weekly to patients with acute myeloid leukemia,” Leukemia, vol. 24, no. 4, pp. 699–705, Apr. 2010. [DOI] [PubMed] [Google Scholar]
- [92].Wang C., et al. , “Geldanamycin reduces acute respiratory distress syndrome and promotes the survival of mice infected with the highly virulent H5N1 influenza virus,” Front. Cell Infect. Microbiol., vol. 7, 2017, Art. no. 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Omura S., et al. , “A new alkaloid AM-2282 of streptomyces origin. Taxonomy, fermentation, isolation and preliminary characterization,” J. Antibiotics, vol. 30, no. 4, pp. 275–282, Apr. 1977. [DOI] [PubMed] [Google Scholar]
- [94].Tamaoki T., Nomoto H., Takahashi I., Kato Y., Morimoto M., and Tomita F., “Staurosporine, a potent inhibitor of phospholipid/Ca++dependent protein kinase,” Biochem. Biophys. Res. Commun., vol. 135, no. 2, pp. 397–402, Mar. 1986. [DOI] [PubMed] [Google Scholar]
- [95].Humber L. G., “Etodolac: The chemistry, pharmacology, metabolic disposition, and clinical profile of a novel anti-inflammatory pyranocarboxylic acid,” Med. Res. Rev., vol. 7, no. 1, pp. 1–28, Mar. 1987. [DOI] [PubMed] [Google Scholar]


