Summary
The thymus has a high capacity to support the differentiation of ILCs, especially when E protein transcription factors are ablated. Whether it contributes to the homeostasis of ILC pools in tissues is not clear. Single-cell RNA sequencing analysis shows a substantial amount of ILC precursors in wild type but not athymic nude blood. The precursors express CD3 intracellularly (ic) but not on the surface. The abundance of Lin−CD127+CD62L+icCD3ε+ precursors varies with age, peaking at 2–3 months. These cells can differentiate into various ILC subsets on OP9-DL1 stroma in vitro. In the lung, small intestine, and epidermis, icCD3ε+ cells differentiate into diverse ILC subsets in different tissue environments in steady state. Helminth infection promotes their differentiation toward functional ILC2s. Thus, the thymus appears to play a role in replenishing ILC pools in different peripheral tissues. Because thymic activity is age-dependent, this finding may help explain age-related differences in immune responses.
Subject areas: Immunology, Components of the immune system, Omics, Transcriptomics
Graphical abstract
Highlights
-
•
Single-cell RNA sequencing detects thymus-dependent (td) ILC precursors in the blood
-
•
Intracellular (ic) but not surface CD3ε marks td-ILCs in the blood and tissues
-
•
Blood td-ILCs differentiate into distinct ILC subsets in vitro
-
•
Helminth infection promotes the maturation of icCD3ε+ ILC2s
Immunology; Components of the immune system; Omics; Transcriptomics;
Introduction
Innate lymphoid cells (ILCs), grouped as ILC1, ILC2, and ILC3, represent an emergent class of immune cells that act as first responders in the immune response, integrating environmental cues from mucosal tissues and microbes (Artis and Spits, 2015; Eberl et al., 2015; Vivier et al., 2018). The differentiation of ILCs is believed to take place in the bone marrow or fetal liver. Multipotent lymphoid primed progenitors or common lymphoid progenitors have been shown to give rise to all subsets of ILCs in addition to B and T lymphocytes. A series of intermediate progenitors have been identified in the bone marrow; some can give rise to both NK and the three ILC subsets whereas others only generate ILCs (Vivier et al., 2018; Walker et al., 2019; Yang and Bhandoola, 2016). ILCs are also thought to be replenished from tissue-resident ILC progenitors though the identities of these progenitors in different tissues are not well established. In human blood, multipotent ILC progenitors have been characterized but the mouse counterparts have not been well documented (Lim et al., 2017; Nagasawa et al., 2019).
With the exception of NK cells of thymic origin (Li et al., 2010; Vargas et al., 2011; Veinotte et al., 2006), the thymus had not been considered a site for ILC development until we discovered mass production of ILC2s due to the thymus-specific ablation of the function of E protein transcription factors (Wang et al., 2017). In our Id1 transgenic mice (called Id1 hereafter) where the Id1 inhibitor of E proteins is expressed using the proximal promoter of the Lck gene (pLck) at the early T cell progenitor (ETP) stage, ILC2s were found to be 10- to 100-fold enriched in the thymus and throughout the body (Wang et al., 2017). Likewise, ablation of two E protein genes, Tcf3 and Tcf12, using plck-Cre resulted in similar accumulation of ILC2s in the double knockout (called dKO hereafter) mice (Qian et al., 2019; Wang et al., 2017). In support of the ILC2-producing potential in wild-type (WT) thymus, a fraction of ILC2s in the lung of WT but not athymic nude mice have been shown to carry rearranged T cell receptor (TCR) genes, which suggested their thymic history of differentiation (Qian et al., 2019; Shin et al., 2020). In the thymus-specific plck-Cre reporter mice, we detected reporter expression in blood ILCs and in peripheral tissues in a thymus-dependent manner (Qian et al., 2019). Using in vitro culture on OP9-DL1 stromal cells, the CD4 and CD8 double-negative thymocytes at DN1 (ETP) and DN3 stages exhibited robust potential to differentiate into ILC2s. Additionally, fetal thymic organ culture studies illustrated the potential of the thymus to support ILC2 differentiation after ablation of E protein genes (Miyazaki et al., 2017). Examination of pre- and postnatal thymi also detected small numbers of ILC2s (Ferreira et al., 2021; Jones et al., 2018; Kernfeld et al., 2018).
Despite evidence suggesting the capability of the thymus to support ILC differentiation and the potential of thymocytes to differentiate into ILCs, outstanding questions are how the thymus contributes to the pools of ILCs, the magnitude of thymic contribution under homeostasis, and the phenotype of thymus-dependent ILCs. In this report, we address these questions by using single-cell RNA sequencing (scRNAseq) and different mouse models. We show that several subsets of the lineage negative (Lin−) and Thy1+ population (enriched of ILCs) in the blood are dramatically reduced in athymic nude mice compared to WT controls, which suggest the thymus dependence of these subsets. These cells are further defined as CD127+CD62L+intracellular CD3ε+(icCD3ε+) and their frequencies peak at around 2–3 months of age. Lin−CD127+CD62L+ cells can differentiate into ILC subsets on OP9-DL1 stromal cells in vitro. In the periphery, icCD3ε+ ILCs were found in the lung, small intestine, and epidermis of WT but not athymic nude mice. These cells respond to helminth infection by producing ILC2 cytokines. Together, these data suggest that the thymus serves as a source of ILC precursors and replenishes tissue ILC pools.
Results
Single-cell RNA sequencing analyses identify ILC precursors in the blood whose levels correlate with the functionality of the thymus
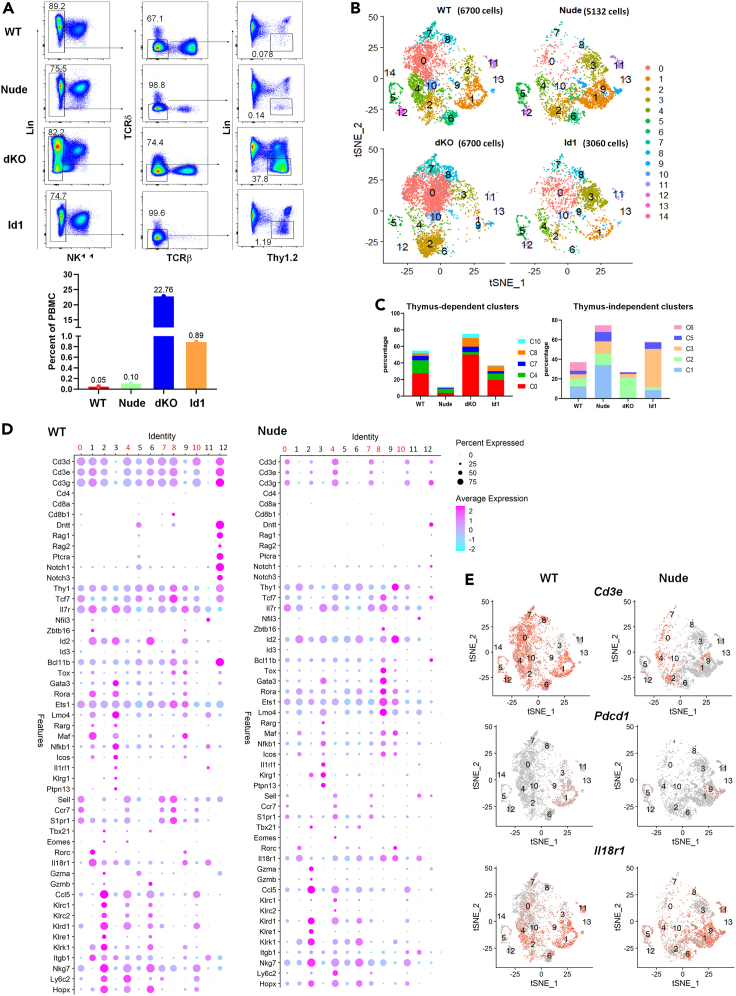
If the thymus were to continuously supply ILCs to peripheral tissues, it would first export them to the blood. We thus used scRNAseq to analyze blood Lin−Thy1+ cells from 4 different strains of 2-months-old mice and compared the levels of different subsets. Besides WT mice, we utilized athymic nude mice which are devoid of thymus-derived cells. Id1 and dKO mice have excessive amounts of ILC2s made in the thymus from DN1 and DN3 cells, respectively (Qian et al., 2019; Wang et al., 2017). Figure 1A shows the sorting profiles and the abundance of the Lin−Thy1+ subset in each strain. Remarkably, the frequencies of this population in Id1 and dKO mice are 19- and 484-fold higher than that in WT mice, respectively.
Figure 1.
Single-cell RNA sequencing analyses identifies ILC precursors in the blood whose levels correlate with the functionality of the thymus
(A) Sorting profiles for indicated strains of mice. Cells in the last gate were used for scRNAseq. The frequencies of these cells in PBMCs are shown in the bar graph.
(B) T-distributed stochastic neighbor embedding (t-SNE) map of ILCs in different strains (WT, Nude, dKO, and Id1). Cluster numbers were marked. Numbers in parenthesis show the number of cells sequenced.
(C) Percentage of cells in thymus-dependent (top) and independent (bottom) clusters.
(D) Dot plot showing the expression of genes of interest in each cluster (0–12) in WT and nude mice.
(E) Feature plots showing the expression of indicated genes in WT and nude mice.
Analyses using the Seurat program (Butler et al., 2018; Stuart et al., 2019) generated 15 clusters labeled in the order of their frequencies, among which clusters 0, 4, 7, 8, and 10 are markedly reduced in nude mice compared to WT mice, suggesting that the existence of these subsets requires the thymus (Figures 1B and 1C). Although these clusters may represent heterogeneous subsets, we designate them collectively as thymus-dependent ILCs (td-ILCs). td-ILCs constituted 55% of circulating Lin−Thy1+ cells in WT mice, highlighting their relative abundance among circulating ILCs. Interestingly, they are highly enriched in dKO mice, accounting for 75% of the Lin−Thy1+ population and 17% of the total peripheral blood mononuclear cells (PBMCs).
In contrast, cluster 3 constitutes 39% of Lin−Thy1+ cells in Id1 mice but only 4% and 12% in WT and nude mice, respectively (Figures 1B and 1C). Considering the frequency of Lin−Thy1+ cells, cluster 3 cells are more abundant in dKO and Id1 mice by over 100-fold compared to WT mice. Because dKO and Id1 mice are highly enriched of ILC2s, cluster 3 may encompass ILC2s. Despite their thymic origin, cluster 3 cells in Id1 and dKO mice possess similar transcriptional profiles as those in WT and athymic mice such that they are all assigned to the same cluster. Interestingly, athymic mice possess more of these cells than WT mice, perhaps due to lack of suppression by T regulatory cells or homeostatic competition from T cells (Rigas et al., 2017).
We next analyzed levels of the genes known to be expressed in T cells and ILCs in each cluster in WT and athymic mice (Figure 1D). None of the clusters expressed CD4 or CD8, thus excluding the possibility of contamination with conventional αβ T cells. Cluster 12 in WT mice expresses genes found in immature T cells but the same cluster found in athymic mice did not express these T cell-specific genes (Figure 1D). Interestingly, the td-ILC clusters (cluster0, 4, 7, 8, and 10) as well as other clusters such as clusters 1 and 2 in WT mice possess transcripts coding for different CD3 chains. However, compared to other Cd3 expressing clusters, td-ILCs distinctively express Sell and S1pr1, which encode a ligand or receptor necessary for cells to exit the thymus (Matloubian et al., 2004; Weinreich et al., 2011) (Figure 1D). In addition, td-ILCs express Il7ra and Tcf7 general markers for all ILCs (De Obaldia and Bhandoola, 2015), but transcripts for Tbx21, Eomes, Gata3, and Rorc, which specify ILC subsets were not detected.
Besides td-ILCs, cluster 1 expresses a higher level of Il7r, Id2, Rora, and Maf, as well as Zbtb16 and Rorc, which hinted the inclusion of ILC3s (Figure 1D). Curiously, these cells also expressed Cd3 genes in WT mice but not in nude mice, suggesting cluster 1 cells in WT and nude mice may be of different origins (Figures 1D and 1E). Cluster 1 in both WT and nude mice also express Pdcd1 and Il18r1, which mark ILC progenitors (Figure 1E). It thus appears that cluster 1 contains both ILC progenitors (ILCPs) and ILC3s but further fractionation could not separate these cell types into ILCP and ILC3 sub-clusters (data not shown). Cluster 2 harbors a list of genes related to NK cells along with Tbx21 and Eomes, thus suggesting that this cluster contains NK or ILC1s. Cluster 3 best fits the description of ILC2s based on the expression of ILC2 signature genes such as Gata3, Lmo4, Rora, and Il1rl1 (Figure 1D).
Examination of individual genes in feature plots revealed interesting patterns: while most of the cells have Il7r and Tcf7, only the thymus-dependent clusters express Sell (Figure S1). Interestingly, Id2 levels are high in thymus-independent clusters but the thymus-dependent ones expressed Id3 instead. Tbx21 and Rorc marked ILC1s and ILC3s in clusters 2 and 1, respectively. Gata3 is more widely expressed but its level is higher in cluster 3.
Age-dependent output of blood td-ILCs and their differentiation
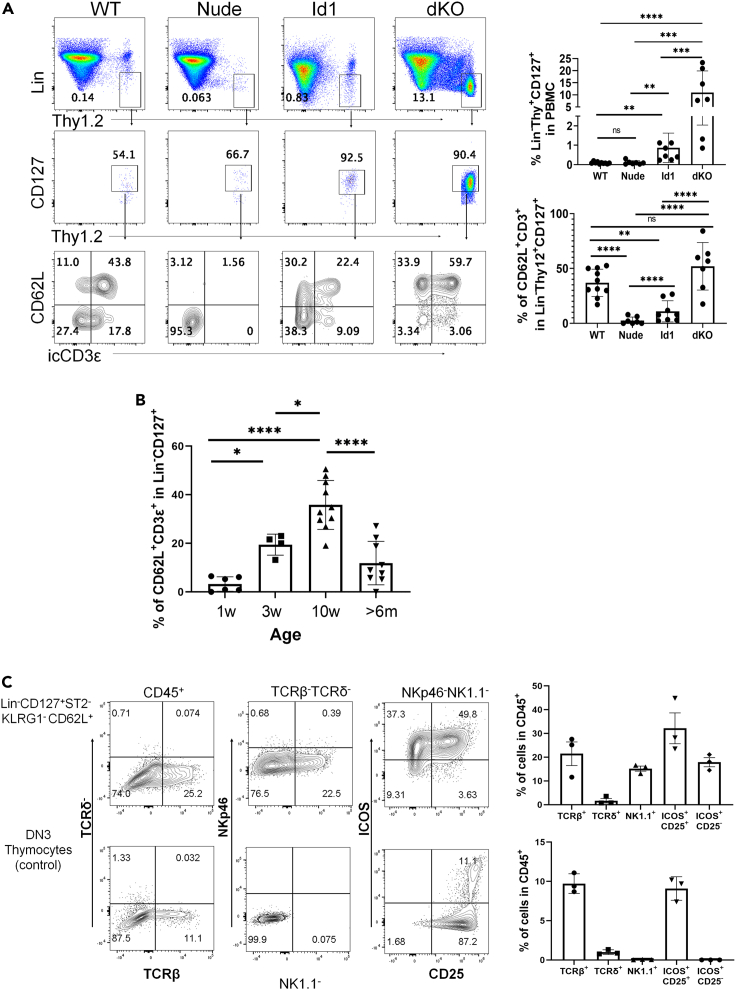
To verify the scRNAseq data using flow cytometry, we chose CD127 (encoded by Il7r), CD62L (encoded by Sell) and icCD3ε to label thymus-dependent ILCs (Figure 2A). We first evaluated the levels of Lin−Thy1+IL7R+icCD3ε+CD62L+ cells in the blood of WT, nude, dKO, and Id1 mice at 2 months of age. Consistent with the scRNAseq data, a high proportion of WT Lin−Thy1+CD127+ cells are icCD3ε+, most of which are CD62L+, suggesting that these cells are enriched of td-ILCs. As expected, dKO mice harbor a large number of such cells but nude mice had few of them, thus confirming that the frequencies of these cells correlate with the presence of a thymus. Because ILC2 differentiation in Id1 mice occurs in DN1 cells, which express low levels of CD3, it is not surprising that lower percentages of icCD3ε+ cells were detected.
Figure 2.
Age-dependent output of blood td-ILCs and their differentiation
(A) Peripheral blood of indicated strains of mice were analyzed. The frequencies of Lin−Thy1+CD127+ and subsequent CD62L+icCD3ε+ subsets are quantified.
(B) The average frequency of blood CD62L+icCD3ε+ cells of the Lin−Thy1+CD127+ compartment in WT mice of indicated ages. Data are represented as mean ± SD. One-way ANOVA was used for determining the statistical significance of data shown in (A) and (B). ∗p<0.05, ∗∗<0.01, ∗∗∗p<0.001, ∗∗∗∗p<0.0001, ns, not significant.
(C) In vitro differentiation on OP9-DL stroma of the indicated cell populations (800 cells per well) in 30 ng/mL of each IL-2, IL-7, and stem cell factor. Cells were analyzed on day 9 of the culture and the frequencies of indicated subsets are shown in the graphs. Data shown are representative of three independent experiments.
Next, we examined the frequencies of td-ILCs in correlation with age, ranging from 7 days to over 6 months old (Figure 2B). The percentage of icCD3ε+CD62L+ cells in the Lin−Thy1+CD127+ fraction in the blood was quantified. There was a gradual increase in the frequencies of td-ILCs until 2–3 months of age and that decreased significantly in mice over 6 months old. These results suggest that td-ILCs are primarily generated postnatally and their production depends on a functional thymus. Schneider et al. showed that a major wave of ILC2 expansion in different tissues occurs at the neonatal stage (Schneider et al., 2019). It is thus unlikely that td-ILCs, if at all, contribute significantly at this stage of expansion but may replenish the ILC pools in adulthood.
We also explored the differentiation potential of blood td-ILCs using OP9-DL1 culture (Figure 2C). Because icCD3ε cannot be used for live cell sorting, we isolated Lin−CD127+KLRG1−ST2−CD62L+ cells, which is a heterogeneous population, and plated them on OP9-DL1 stroma. Our pilot experiment showed that the differentiation of these cells depended on IL-2; the culture was thus carried out in the presence of IL-2, IL-7, and stem cell factor. DN3 thymocytes were used as a positive control for ILC2 differentiation. The cells were analyzed on day 9 for the expression of surface markers including NK1.1, NKp46, ICOS, and CD25 as well as TCRβ and TCRδ. A small fraction of the cells carried TCRβ, which may be due to de novo differentiation from some immature T precursors included in the population or contaminating T cells that expanded in the presence of IL-2 and IL-7. Of the TCR− cells, about 20% expressed NK1.1 but not NKp46, suggesting that the cells are likely of NK/ILC1 lineage. The majority of the NK1.1− fraction displayed ICOS, of which more than half also expressed CD25, characteristic of ILC2s. ILC3s may also be present in the ICOS+ fraction. These results suggest that the Lin−CD127+KLRG1−ST2−CD62L+ population includes progenitors capable of generating T cells and/or ILC subsets. It would also be interesting to test if the same population of cells from nude mice has reduced differentiation potentials but the demand for a large number of mice for isolating these cells makes the experiment unfeasible. It should be noted that due to the heterogeneity of the population and the arbitrary in vitro condition used, the true potentials of these cells need to be investigated at the single-cell level and in different tissue environments.
Intracellular CD3 marks thymus-dependent ILC precursors in peripheral tissues
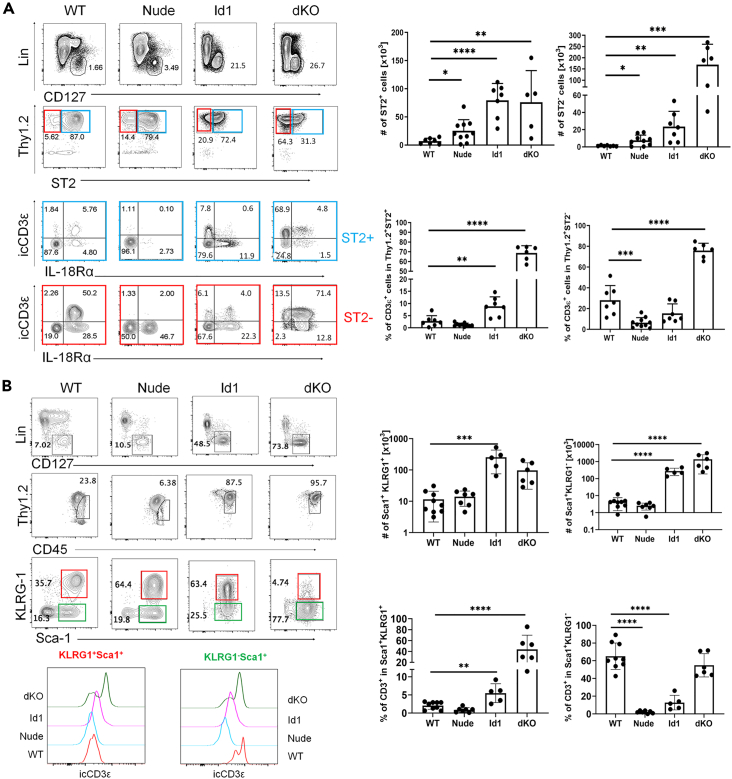
To identify thymus-dependent ILC or ILC precursors in peripheral tissues, we stained lung, small intestine, and skin cells using flow cytometry. In the lung, conventional ILC2s were identified as Lin−CD127+Thy1+ST2+ whereas immature ILC2s and other ILCs may be in the ST2− counterpart (Figure 3A). We then interrogated these two subsets for the levels of icCD3ε and IL18R1. IL18R1 is recently shown to mark ILC precursors in the lung (Ghaedi et al., 2020; Zeis et al., 2020). In WT mice, a small fraction of ST2+ cells expressed CD3ε and/or IL18R1 but a high proportion of ST2− cells were icCD3ε+ and/or IL18R1+. In contrast, nude mice had few icCD3ε+ cells in both ST2+ and ST2− subsets, suggesting that icCD3ε marks thymus-dependent cells. In nude mice, despite the marked reduction of icCD3ε+ cells, the total numbers of ST2+ and ST2− cells did not decrease, likely due to the expansion of other cells. In contrast, dKO mice, in which massive amounts of ILC2 and ILC precursors are produced from icCD3ε+ committed T cells in the thymus, had over 100 times more ST2+ or ST2− cells in the lung, most of which are icCD3ε+. Because ILC2s in Id1 mice originate from CD3ε− multipotent early T cell progenitors, the ST2+ and ST2− cells in the lung were largely icCD3ε− even though large amount of such cells were present (Figure 3A).
Figure 3.
Intracellular CD3 marks thymus-dependent ILC precursors in peripheral tissues
(A) Analyses of lung ILCs in indicated strains. The lineage cocktail and special efforts to exclude T and NK cells are described in the STAR Methods. ST2+ and ST2− subsets are color coded in blue and red. Bar graphs show cumulative data from at least three experiments. Unpaired Student's test was used for determining the statistical significance. ∗p<0.05, ∗∗<0.01, ∗∗∗p<0.001, ∗∗∗∗p<0.0001.
(B) Analyses of small intestine ILCs in indicated strains. KLRG1+ and KLRG1− subsets are color coded in red and green. Bar graphs show cumulative data from at least three experiments. One-way ANOVA was used for determining the statistical significance. ∗∗<0.01, ∗∗∗p<0.001, ∗∗∗∗p<0.0001. Data are represented as mean ± SD.
In the small intestine, Lin−CD127+CD45hiSca-1+ cells were divided into KLRG1+ and KLRG1−. The former is considered conventional mature ILC2s while the latter may include immature ILC2s or other ILCs (Figure 3B). icCD3ε was mostly detected in the KLRG1− subset in wild-type mice but not in nude mice. When the icCD3ε+ cells were absent in nude mice, other cells made up the total numbers due to homeostatic expansion. dKO mice had icCD3ε+ cells in both KLRG1+ and KLRG1− fractions but Id1 mice did not express CD3ε in either population (Figure 3B).
We have also examined the epidermis and found high frequencies of icCD3ε+ cells in the Lin−Thy1+ICOS− cells in WT but not nude mice, suggesting that these cells are associated with the thymus (Figures S2A and S2B). Although the Lin−Thy1+ICOS+ population is known to contain ILC2s (Schneider et al., 2019), the ICOS− subset might contain immature ILC2s or other ILCs (Figure S2C).
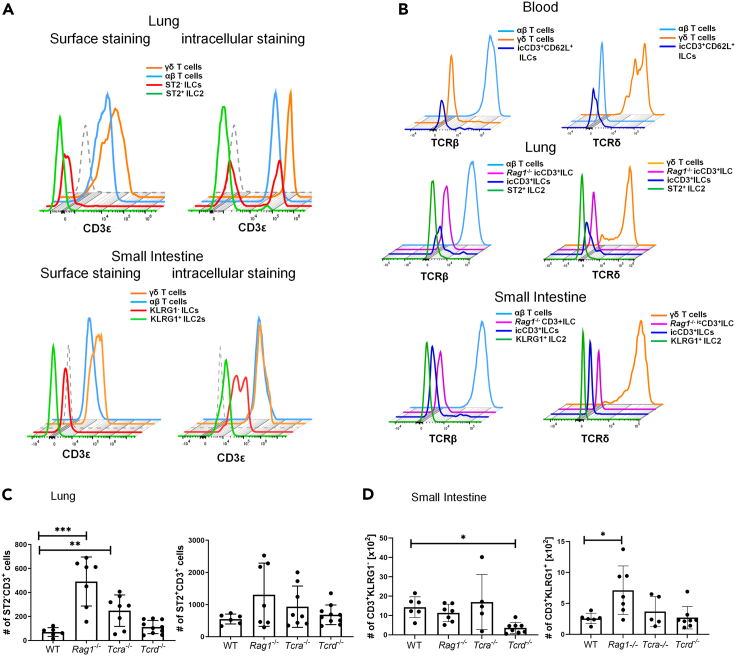
Lin−CD127+icCD3ε+ cells are distinct from conventional T cells
To rule out that icCD3ε+ cells are of T cell lineage even though they are negative for either TCRβ or TCRδ as well as CD4 and CD8 surface markers. We first demonstrated that CD3ε expression in ILCs in both lung and small intestine could only be detected using intracellular but not surface staining (Figure 4A). In contrast, either αβ or γδ T cells were labeled without permeabilizing the cells. Furthermore, we showed that these icCD3ε+ cells have no intracellular TCRβ or TCRδ (Figure 4B). Finally, we examined levels of icCD3ε+ in the lung and small intestine of Rag1−/−, Tcra−/-, and Tcrd−/- mice, which lack αβ and/or γδ T cells. The levels of icCD3ε+ cells in the lung of these mutants were similar or higher compared to WT mice (Figure 4C). The increase in mice lacking αβ T cells is probably due to diminished homeostatic competition between T cells and ILCs or the absence of Treg-mediated suppression. In the small intestine, icCD3ε+ cells were also readily detectable even though the amount in the KLRG1− fraction were reduced in Tcrd−/− mice (Figure 4D). This may be explained by a partial contamination of non-conventional γδ T cells which downregulated their TCRs or ILC differentiation in the thymus depends on TCRγδ signaling.
Figure 4.
Lin-CD127+icCD3ε+ cells are distinct from conventional T cells
(A) Detection of CD3ε by intracellular and surface staining. Histograms show CD3ε levels in the indicated subsets of cells in the indicated tissues.
(B) Intracellular levels of TCRβ or TCRδ in icCD3ε+ ILCs in comparison to T cells or Rag1−/− ILCs.
(C) Quantification of the frequencies of icCD3ε+ cells in Lin−CD127+ ST2+ and ST2− subsets in the lung of the indicated strain of mice.
(D) Quantification of the frequencies of icCD3ε+ cells in Lin−CD127+CD45hi KLRG1+ and KLRG1− subsets in the small intestine of the indicated strain of mice. ∗p<0.05, ∗∗<0.01, ∗∗∗p<0.001. Data are represented as mean ± SD.
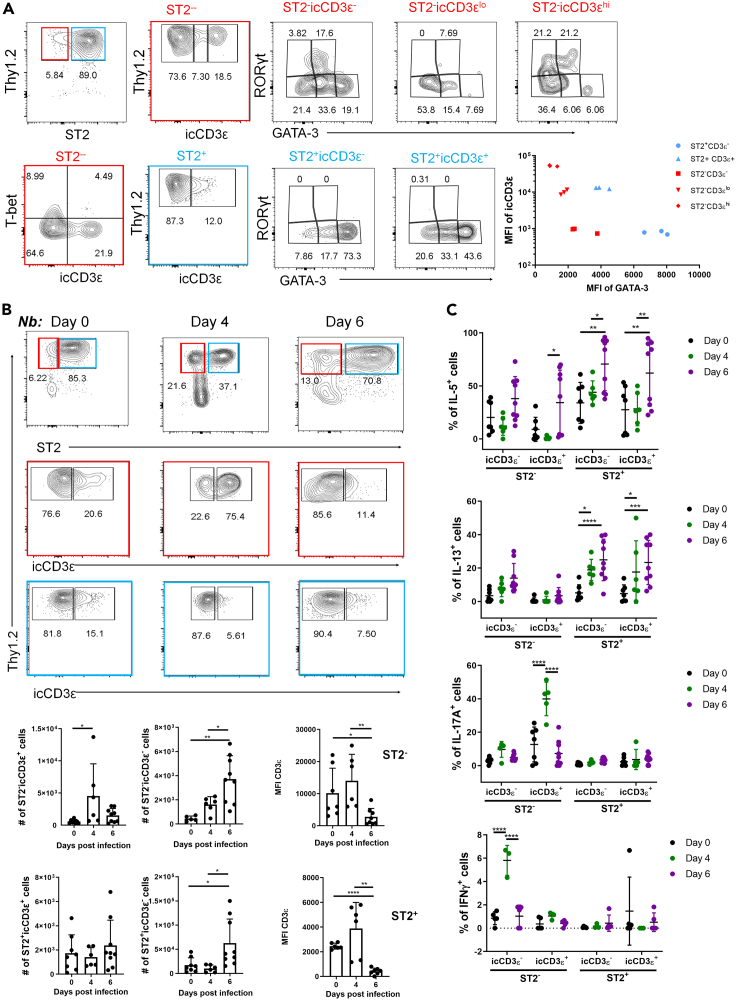
icCD3ε+ cells mature into ILC2 and ILC3 in the lung
To investigate the differentiation potentials of the icCD3ε+ cells in WT lung, we analyzed Lin−Thy1+CD127+ST2+ or ST2− fractions for the expression of signature transcription factors, T-bet, GATA3, and RORγt (Figure 5A). Very few cells expressed T-bet in the ST2− fraction while the ST2+ subset expressed mostly GATA3. Interestingly, we observed an inverse correlation between icCD3ε versus GATA3 and/or ST2 (Figure 5A). Specifically, when the ST2− fraction was divided into icCD3ε high, low, and negative subsets, the mean fluorescence intensities of GATA3 in each subset trended oppositely. In the ST2+ compartment, icCD3ε+ cells harbor lower levels of GATA3 compared to the icCD3ε− ones but their GATA3 levels were higher than those in ST2− cells. CD3 expression is possibly downregulated as the newly arrived ILC precursors differentiate into ILC2s. In addition, icCD3ε+ST2−cells also expressed RORγt together with low levels of GATA3, which has been found in ILC3s (Zhong et al., 2020). We further analyzed the ILC3 potential by assessing CCR6 and NKp46 expression in the ST2− compartment (Figure S3). While NKp46 was detected on few cells, CCR6 was found in a substantial fraction of icCD3ε+ cells and some of them are CCR6+RORγt+. In contrast, icCD3ε− cells displayed lower levels of CCR6. These results suggest a potential of icCD3ε+ST2− cells to differentiate along the ILC3 path (Figures 5A and S3).
Figure 5.
icCD3ε+ cells mature into ILC2 and ILC3 in the lung
(A) Lung Lin−CD127+ cells of 2-month-old WT mice were analyzed for the expression of indicated transcription factors in triplicates. Subsequent gates from ST2− and ST2+ cells are color coded in red and blue. Mean fluorescence intensities of icCD3ε and GATA3 in indicated subsets are plotted on the right.
(B) Analyses of Lin−CD127+ cells of WT lung at indicated time points post subcutaneous infection with 500 stage3 Nb. Cell numbers or MFI of icCD3ε of indicated subsets are shown in the bar graphs.
(C) Cytokine expression in the indicated subsets from experiments described in (B) was measured after stimulation with PMA and Ionomycin for 3.5 h. One-way ANOVA was used for determining the statistical significance. ∗p<0.05, ∗∗<0.01, ∗∗∗p<0.001, ∗∗∗∗p<0.0001. Data are represented as mean ± SD.
Next, we tested the type 2 immune responses of icCD3ε+ cells after 4 and 6 days of infection with helminths, Nippostrongylus brasiliensis (Nb) (Figure 5B). On day 4 post infection, the worms mainly reside in the lung, causing massive tissue damage whereas by day 6, they move to the small intestine before being expulsed. There was a marked increase in ST2− cells on day 4, which could be due to an influx of icCD3ε+ ILC precursors from the blood and/or due to local expansion of such cells. The majority of the ST2− cells are icCD3ε+ on day 4 but become icCD3ε− on day 6, possibly owing to the maturation of the precursors toward ST2+ ILC2s. Consistently, ST2+ cells, mostly icCD3ε−, did not increase until day 6.
Production of IL-5 and IL-13 were boosted on day 6 in the icCD3ε+ and icCD3ε−ST2+ cells and to a lesser extent in the ST2− cells (Figure 5C). Additionally, on day 4, IL-17 was detected in icCD3ε+ST2−cells, which correlates with presence of CCR6 and/or RORγt+ ILC3s in this population. The IL-17-producing cells are unlikely inflammatory ILC2s because they do not express KLRG1 (Huang et al., 2015). At this time point, a small number of IFNγ-producing cells were found in the icCD3ε−ST2− subset, which agrees with the existence of some T-bet+ cells which may be ILC1s. Collectively, these data suggest that the ST2−icCD3ε+ cells in the lung can differentiate into ILC2s and ILC3s, and helminth infection accelerates their differentiation toward ILC2s.
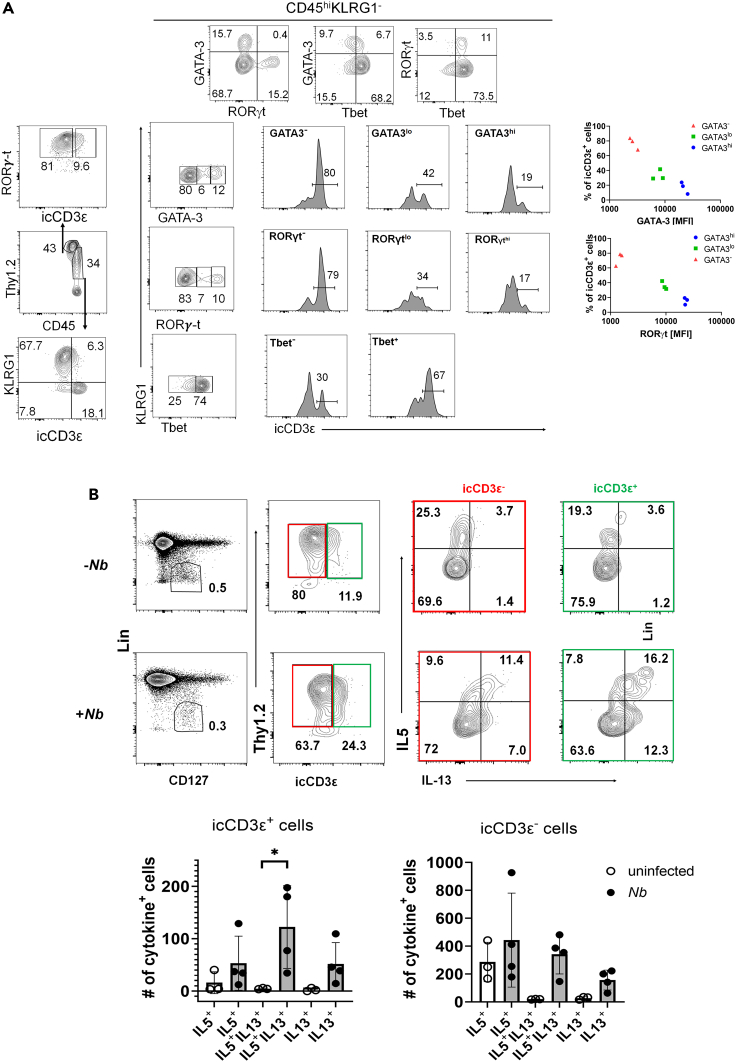
Differentiation of icCD3ε+ cells in the small intestine
The microenvironment of the small intestine likely impacts ILC precursors differently from the lung. Conventional ILC3s are found in the Lin−CD127+CD45lo fraction but ILC2s are included in the CD45hi fraction (Figure 6A). Though small fractions of ILC3s and KLRG1+ ILC2s are icCD3ε+, the majority of icCD3ε+ cells are included in the CD45hiKLRG1− subset. Unlike the findings in the lung, a large proportion of these cells expressed T-bet, which suggest an ILC1 and/or NK potential (Figure 6A). Some of these T-bet+ cells also expressed RORγt, which might be due to the plasticity between ILC1s and ILC3s (Figure 6A). However, there were also cells expressing GATA3 or RORγt at varying levels. When the KLRG1− cells were divided into negative, low, and high fractions based on the fluorescence intensity of GATA3 or RORγt, the percentages of icCD3ε+ cells were found to be inversely correlated with the intensities (Figure 6A).
Figure 6.
Differentiation of icCD3ε+ cells in the small intestine
(A) Lin−CD127+ cells from WT small intestine were first divided into CD45lo and CD45hi. icCD3ε levels were determined for each subset. Transcription factor levels in CD45hi KLRG1− cells were further analyzed and the correlation between the percentages of icCD3ε+ cells and the levels of GATA3 or RORγt are plotted on the right.
(B) MLN cells from mice with or without Nb infection for 6 days were analyzed. Lin−CD127+ cells were separated into Thy1+icCD3ε− and Thy1+icCD3ε+ subsets, which were then scored for IL-5 and IL-13 expression. Numbers of cytokine+ cells were calculated. One-way ANOVA was used for determining the statistical significance. ∗p<0.05. Data are represented as mean ± SD.
We next evaluated the responses of icCD3ε+ cells in mesenteric lymph nodes (MLN) to helminth infection. Six days after Nb infection, Lin−CD127+ cells were first divided into icCD3ε+ and icCD3ε−, and cytokine expression in these cells were analyzed (Figure 6B). While little expression of IL-17 or IFNγ was detected (data not shown), Nb infection markedly increased IL-5- and IL-13-expressing cells, particularly IL-13+ cells. This occurs in both icCD3ε+ and icCD3ε− subsets, suggesting that icCD3ε+ ILC precursors can differentiate into functional ILC2s.
Discussion
Our scRNAseq analyses revealed the compositions of ILCs in mouse blood. These findings help clarify the issue whether ILCs circulate in the blood in steady state. Previous studies using parabionts of two mouse strains suggested that ILCs, at least ILC2s, do not interchange via circulation at steady state (Gasteiger et al., 2015). Our data showed a detectable fraction of the Lin−Thy1+ cells represent ILC2s (cluster 3). Subsets resembling ILC1s and ILC3s (clusters 2 and 1, respectively) are also found. Perhaps, the exchange rate between circulating and tissue ILC2s is very low at steady state and differs in different tissues (Huang et al., 2018). More importantly, we have found that td-ILCs (included in clusters 0 as well as clusters 4, 7, 8, and 10) represented the largest fraction of Lin−Thy1+ cells and appeared to be subsets of precursors of ILCs. Because the frequencies of these precursors are greatly diminished in the athymic nude mice and markedly augmented in two mouse strains, dKO and Id1, which undergo accelerated ILC differentiation in the thymus, it is tempting to postulate that these cells are derived from the thymus. This is also supported by our previous finding that the generation of blood tdTomato+ ILCs from thymus-specific plck-Cre/ROSA-26-stop-tdTomato donor bone marrow required the thymus in the hosts (Qian et al., 2019). However, additional direct evidence would strengthen such a notion. Nonetheless, these findings provide evidence for the presence of mouse ILC precursors in the circulation and are consistent with findings in human blood. In human blood, the ILC precursors also constitute a sizable fraction of the Lin−CD127+ subset and they are heterogeneous with regard to their ILC differentiation potentials (Lim et al., 2017; Nagasawa et al., 2019). In addition to bone marrow-derived precursors, thymic progenitors were also suggested to generate CD5+ ILCs (Alisjahbana et al., 2021; Nagasawa et al., 2017).
The characteristics of thymus-dependent ILC precursors include the expression of genes encoding different CD3 subunits and genes for cell surface molecules required for exiting the thymus, CD62L, and S1PR1. While CD62L and S1PR1 enable the ILC precursors to leave the thymus (Matloubian et al., 2004; Teng et al., 2011), CD3 expression suggests that these cells diverge from the T cell path after T cell commitment. Indeed, dKO mice, in which E protein genes are ablated starting at the DN3 stage (Wang et al., 2017), harbors an extremely large amount of cluster 0 cells. This notion is also consistent with the observation of rearranged TCRβ and TCRγ genes in lung ILC2s of WT but not nude mice, and the ability of purified DN3 thymocytes to differentiate into ILC2s in vitro (Qian et al., 2019; Shin et al., 2020). A remaining question is what triggers the ILC divergence from T cell development. It has recently been reported that RORα plays a key role in driving ILC fate away from the T cell lineage in fetal thymus (Ferreira et al., 2021). However, because E protein transcription factors are powerful suppressors of ILC2 differentiation, one might imagine that upregulation of their inhibitors, namely Id proteins, could promote ILC differentiation. Indeed, Id3 is known to be transiently activated following pre-TCR signaling (Bain et al., 2001). Id3 is also highly expressed in γδ T cells, thus creating opportunities for ILC differentiation from T cell precursors (Lauritsen et al., 2009). Consistently, our scRNAseq data showed that Id3 was specifically expressed in thymus-dependent clusters. Inducible ablation of E proteins in thymocytes in an ILC2 differentiation culture for 24 or 72 h led to an increase in Rora expression (Peng et al., 2020; Qian et al., 2019). However, overexpression of E proteins in ILC2s did not repress Rora transcription, thus suggesting an indirect mechanism of the crosstalk between RORα and E proteins (Peng et al., 2020). Further studies are needed to gain insight into the crossroads leading to T and ILC fates in the thymus and all of the intrinsic factors involved in driving each direction.
We have shown that intracellular but not surface CD3ε marks thymus-dependent ILCs in peripheral tissues such as the lung, small intestine, and epidermis because nude mice have very few of icCD3ε+ cells. Interestingly, most of the icCD3ε+ cells reside in the compartments containing immature ILC2s and other ILC subsets, e.g., ST2− in the lung, KLRG1−- in the small intestine, and ICOS− in the epidermis. This suggests that the icCD3ε+ cells likely arrive from the blood as precursors and differentiate under the influence of their respective tissue environments. In fact, we have observed ongoing ILC2 as well as ILC3 differentiation from icCD3ε+ cells in the lung or small intestine. Upregulation of signature transcription factors, GATA3 and RORγt, correlated inversely with CD3ε expression. It is possible that these icCD3ε+ ST2− or KLRG1− cells eventually become ST2+ or KLRG1+ cells. This is particularly apparent in mice infected with helminths, which stimulate the production of type 2 cytokines in both icCD3ε+ and icCD3ε− cells in the lung and MLN, respectively. Our data also agree with the study where ST2−IL18R+ cells in the lung were shown to be ILC precursors, which are from the blood (Ghaedi et al., 2020; Zeis et al., 2020). Their scRNAseq data also showed Cd3e expression in the cluster containing these precursors but not in differentiated cells. In athymic mice, the frequencies of icCD3ε+IL-18R+ cells were greatly reduced.
In addition, different tissue environments may foster the maturation of the precursors into different subsets. In the ST2− compartment of the lung, icCD3ε+ cells express GATA3 and RORγt, which are signature transcription factors of ILC2s and ILC3s. However, the KLRG1− fraction of the small intestine possess a high percentage of T-bet+ icCD3ε+ cells, which are indicative of the NK/ILC1 fate. Our result from in vitro differentiation cultures showed the potentials of the Lin-CD127+CD62L+ precursors, which encompass heterogeneous populations, to differentiate into various NK/ILC subsets. It is thus unclear if the different ILC subsets in the lung and small intestine are the progenies of distinct precursors with different bias or the results of different induction signals in different tissues.
Limitations of the study
It remains to be determined the extent to which thymus-dependent ILC precursors contribute to the overall pools of ILC subsets. Although icCD3ε is a good marker for such ILC precursors, its downregulation after ILC maturation renders them indistinguishable from ILCs from other sources. Fate-mapping studies using plck-Cre had also been exploited but the reporter was not ideal (Qian et al., 2019). Additional Cre transgenes, which is specifically expressed at the early stage of T cell development, are necessary for evaluating the contribution of thymus-derived ILCs in steady-state and pathophysiological conditions. Nevertheless, the relatively high frequencies of circulating ILC precursors associated the thymus in adult mice and their high propensity to differentiate in peripheral tissues highlight the under-appreciated role of the thymus to ILC homeostasis.
STAR★Methods
Key resources table
| REAGENT OR RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Syrian hamster anti-Mouse KLRG1 PerCP-eFluor710 | Thermo Fisher Scientific | Cat# 46-5893-82, clone 2F1, RRID:AB_10670282 |
| Syrian hamster anti-mouse-human KLRG1 (MAFA) PE-dazzle 594 | Biolegend | Cat# 138424, clone 2F1/KLRG1, RRID:AB_2564051 |
| Syrian hamster anti-mouse/human KLRG1 (MAFA) Brilliant Violet 605 | Biolegend | Cat# 138419, clone 2F1/KLRG1 RRID:AB_2563357 |
| Rat anti-mouse CD127 (IL-7Ralpha) FITC | Biolegend | Cat# 135007, clone A7434, RRID:AB_1937231 |
| Rat anti-mouse CD127 (IL-7Ralpha) PE | Biolegend | Cat# 135010, clone A7434, RRID:AB_1937251 |
| Rat anti-mouse CD127 (IL-7Ralpha) APC | Biolegend | Cat# 135012, clone A7434, RRID:AB_1937216 |
| Rat anti-mouse CD127 (IL-7Ralpha) PE/Dazzle 594 | Biolegend | Cat# 135032, clone A7434, RRID:AB_2564217 |
| Armenian hamster anti-mouse CD3ε BUV496 | BD Biosciences | Cat# 612955, clone 145-2C11, RRID:AB_2870231 |
| Armenian hamster anti-mouse CD3ε APC | BioLegend | Cat# 100312, clone 145-2C11, RRID:AB_312677 |
| Rat anti-mouse CD90.2 Brilliant Violet 785 | Biolegend | Cat# 105331, clone 30-H12, RRID:AB_2562900 |
| Rat anti-mouse CD90.2 (Thy1.2) APC/Fire 750 | Biolegend | Cat# 140326, clone 53-2.1, RRID:AB_2650963 |
| Rat anti-mouse CD90.2 antibody FITC | Biolegend | Cat# 105306, clone 30-H12, RRID:AB_313177 |
| Mouse anti-mouse NK-1.1 Brilliant Violet 605 | Biolegend | Cat# 108753, clone PK136, RRID:AB_2686977 |
| Mouse anti-mouse NK-1.1 Alexa Fluor 647 | Biolegend | Cat# 108720, clone PK136, RRID:AB_2132713 |
| Rat anti-mouse CD62L antibody PerCP/Cyanine5.5 | Biolegend | Cat# 104432, clone MEL-14, RRID:AB_2285839 |
| Armenian hamster anti-mouse TCR gamma/delta APC | Biolegend | Cat# 118116, clone GL3, RRID:AB_1731813 |
| Armenian hamster anti-mouse TCR gamma/delta FITC | Biolegend | Cat# 118106, clone GL3, RRID:AB_313830 |
| Armenian hamster anti-mouse TCR gamma/delta APC/Fire 750 | Biolegend | Cat# 118136, clone GL3, RRID:AB_2650828 |
| Armenian hamster anti-mouse TCR gamma/delta Brilliant Violet 421 | Biolegend | Cat# 118120, clone GL3, RRID:AB_2562566 |
| Armenian hamster anti-mouse TCR gamma/delta PerCP/Cyanine5.5 | Biolegend | Cat# 118118, clone GL3, RRID:AB_10612756 |
| Armenian hamster anti-mouse TCR gamma/delta Brilliant Violet 605 | Biolegend | Cat# 118129, clone GL3, RRID:AB_2563356 |
| Armenian hamster anti-mouse TCR beta chain APC | Biolegend | Cat# 109212, clone H57-597, RRID:AB_313435 |
| Armenian hamster anti-mouse TCR beta chain APC/Fire750 | Biolegend | Cat# 109246, clone H57-597, RRID:AB_2629697 |
| Armenian hamster anti-mouse TCR beta chain Brilliant Violet BV421 | Biolegend | Cat# 109230, clone H57-597, RRID:AB_2562562 |
| Armenian hamster anti-mouse TCR beta chain PerCP/Cyanine5.5 | Biolegend | Cat# 109228, clone H57-597, RRID:AB_1575173 |
| Armenian hamster anti-mouse TCR beta chain Brilliant Violet 605 | Biolegend | Cat# 109241, clone H57-597, RRID:AB_2629563 |
| Streptavidin Brilliant Violet 711 | BD Biosciences | Cat# 563262, RRID:AB_2869478 |
| Rat anti-mouse CD45 Brilliant Violet BV785 | Biolegend | Cat# 103149, clone 30-F11, RRID:AB_2564590 |
| Mouse anti-mouse CD45.2 PE/Dazzle 594 | Biolegend | Cat# 109846, clone 104, RRID:AB_2564177 |
| Rat anti-mouse Ly-6A/E (Sca-1) PerCP/Cyanine5.5 | Biolegend | Cat# 108123, clone D7, RRID:AB_893619 |
| Rat anti-mouse Ly-6A/E (Sca-1) FITC | Biolegend | Cat# 108106, clone D7, RRID:AB_313343 |
| Rat anti-Hu/Mo IL-5 eFluor 450 | Thermo Fisher Scientific | Cat# 48-7052-82, clone TRFK5, RRID:AB_2802295 |
| Rat anti-mouse/human IL-5 APC | Biolegend | Cat# 504306, clone TRFK5, RRID:AB_315330 |
| Armeninan hamster anti-human/mouse/rat CD278 (ICOS) Brillian Violet BV785 | Biolegend | Cat# 313533, clone C398.4A, RRID:AB_2629728 |
| Rat anti-mouse IFN gamma eFluor 450 | Thermo Fisher Scientific | Cat# 48-7311-82, clone XMG1.2, RRID:AB_1834366 |
| Rat anti-mouse IL-33Ralpha (ST2) PE | Biolegend | Cat# 146608, clone DIH4, RRID:AB_2728177 |
| Rat anti-mouse IL-33Ralpha (IL1RL1, ST2) Brilliant Violet BV421 | Biolegend | Cat# 145309, clone DIH9, RRID:AB_2565634 |
| Rat anti-mouse CD218a (IL-18Ra) | Thermo Fisher Scientific | Cat# 12-5183-80, clone P3TUNYA , RRID:AB_2572616 |
| Rat anti-mouse IL-13 PE | Thermo Fisher Scientific | Cat# 12-7133-82, clone eBio13A, RRID:AB_763559 |
| Rat anti-mouse IL-17A PerCP/Cyanine5.5 | Biolegend | Cat# 506920, clone TC11-18H10.1, RRID:AB_961384 |
| Rat anti-mouse IFN-gamma Alexa Fluor 488 | Biolegend | Cat# 505813, clone XMG1.2, RRID:AB_493312 |
| Rat anti-mouse IL-13 Alexa Fluor 488 | Thermo Fisher Scientific | Cat# 53-7133-82, clone eBio13A, RRID:AB_2016708 |
| Rat biotin anti-mouse CD5 | Biolegend | Cat# 100604, clone 53-7.3, RRID:AB_312733 |
| Hamster biotin anti-mouse γδ T-cell receptor | BD Biosciences | Cat# 553176, clone GL3, RRID:AB_394687 |
| Armenian hamster biotin anti-mouse TCR beta chain | Biolegend | Cat# 109204, clone H57-597, RRID:AB_313427 |
| Rat biotin anti-mouse Ly-6G/Ly-6C (Gr-1) | Biolegend | Cat# 108404, clone RB6-8C5, RRID:AB_313369 |
| Rat biotin anti-mouse CD8a | Biolegend | Cat# 100704, clone 53-6.7, RRID:AB_312743 |
| Rat biotin anti-mouse CD4 | Biolegend | Cat# 100404, clone GK1.5, RRID:AB_312689 |
| Rat biotin anti-mouse/human CD45R/B220 | Biolegend | Cat# 103204, clone RA3-6B2, RRID:AB_312989 |
| Rat biotin anti-mouse/human CD11b antibody | Biolegend | Cat# 101204, clone M1/70, RRID:AB_312787 |
| Armenian hamster biotin anti-mouse CD11c | Biolegend | Cat# 117304, clone N418, RRID:AB_313773 |
| Mouse biotin anti-mouse NK-1.1 | Biolegend | Cat# 108704, clone PK136, RRID:AB_313391 |
| Armenian hamster biotin FcεRIα | Biolegend | Cat# 134304, clone MAR-1, RRID:AB_1626106 |
| Rat biotin anti-mouse TER-119/Erythroid Cells | Biolegend | Cat# 116204, clone Ter-119, RRID:AB_313705 |
| Mouse biotin anti-mouse CD19 | Biolegend | Cat# 101504, clone MB19-1, RRID:AB_312823 |
| Rat biotinylated anti-mouse CD49b/Pan- NK cells | BD Biosciences | Cat# 553856,clone DX5, RRID:AB_395092 |
| Rat TruStain FcX (anti-mouse CD16/32) | Biolegend | Cat# 101320, clone 93, RRID:AB_1574975 |
| Rat anti human/mouse GATA-3 | Thermo Fisher Scientific | Cat# 46-9966-42, clone TWAJ, RRID:AB_10804487 |
| Mouse anti-mouse RORγt Brillian Violet BV421 | BD Biosciences | Cat# 562894, clone Q31-378, RRID:AB_2687545 |
| Mouse anti-T-bet PE/Dazzle 594 | Biolegend | Cat# 644828, clone 4B10, RRID:AB_2565677 |
| Critical commercial assays | ||
| BD Cytofix/Cytoperm Fixation/PermeablizationKit | BD Biosciences | Cat# 554714, RRID:AB_2869008 |
| True-Nuclear Transcription Factor Buffer Set | Biolegend | Cat# 424401 |
| Deposited data | ||
| Single-cell RNAseq | Dr Sun Lab | GEO: GSE176357 |
| Experimental models: Organisms/strains | ||
| Id1tg/tg mice | The Sun Lab | N/A |
| plck-Cre, HEBF/FE2AF/F (dKO) mice | From Yuan Zhuang at Duke University, bred in-house | N/A |
| B6(SJL)-Foxn1nu-2J;GrsrJ mice | Jackson Laboratory | Stock No: 016195 |
| C57BL/6J mice | Jackson Laboratory | Stock No: 000664 |
| Nippostrongylus Brasiliensis | From Joseph F. Urban, Jr at USDA, maintained in house | N/A |
| Software and algorithms | ||
| FlowJo 10.6.1 | FlowJo | https://www.flowjo.com/solutions/flowjo/downloads/previous-versions |
| GraphPad PRISM 7.0 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Seurat 3.0 | Butler et al., 2018 | https://satijalab.org/seurat/ |
Resource availability
Lead contact
Xiao-Hong Sun, Oklahoma Medical Research Foundation, 825 NE 13th Street, Oklahoma City, OK, Tel: 405-271-7103, Fax: 405-271-7128, E-mail: sunx@omrf.org.
Materials availability
This study did not generate new unique reagents.
Data and code availability
•Single cell RNAseq data has been deposited in NCBI. The accession number for the scRNA-seq data reported in this paper is GEO: GSE176357 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE176357).
•This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
•Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Mice
C57BL/6J, plck-Cre and B6(SJL)-Foxn1nu−2J;GrsrJ (B6 nude) mice were purchased from the Jackson Laboratory. plck-Id1 transgenic mice (Id1) and plck-Cre;E2Af/f;HEBf/f (dKO) mice were as described (Kim et al., 1999; Wojciechowski et al., 2007). Both male and female mice were used in the experiments and no significant differences between the genders were observed. Unless indicated otherwise, mice of 2-3 months of age were used. All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee policies at the Oklahoma Medical Research Foundation.
Method details
Isolation and analyses of hematopoietic cells from non-lymphoid tissues
Single cells from lung and lamina propria of the small intestine were isolated essentially as described (Qian et al., 2019). To isolate cells from the skin on the back of the animals, the skin of WT but not nude mice was shaved, and subcutaneous tissues were removed. Skin was then cut into small pieces and incubated in a solution containing 0.5% trypsin and 2 mM EDTA in pre-warmed HBSS buffer supplemented with 10mM HEPES and 2% inactivated newborn calf serum (INCS) for 45 min at 37°C to separate epidermis from dermis. The epidermis layer was transferred to Mg/Ca-containing HBSS supplemented with 10mM HEPES; 10% INCS, and 5 μg/mL DNase, and dispersed using a syringe. Single cells suspensions were passed through 70μm cell strainers. Cells were collected by centrifugation and the pellets were washed with cold RPMI1400 supplemented with 10mM HEPES and 2% INCS before staining for flow cytometry.
The lineage (Lin) cocktail included biotinylated anti-FcεR, anti-B220, anti-CD19, anti-Mac-1, anti-Gr-1, anti-CD11c, anti-Ter-119, anti-CD5, anti-CD8α, and anti-CD4 followed by staining with conjugated streptavidin. To ensure complete elimination of NK and T cells in the Lin− gate, cells were typically stained with anti-NK1.1, anti-TCRβ, and anti-TCRδ, each conjugated to a different fluorophore. NK and T cells were gated out prior to further analyses. If NK and/or T cells were not stained separately, biotinylated antibodies were included in the lineage cocktail. Mean fluorescence intensity was determined using geometric mean in the FlowJo program. Flow cytometric analysis was performed on an LSR-II (BD Biosciences) or Aurora (Cytek Biosciences).
Single cell RNA sequencing
Peripheral blood treated with ACD anti-coagulant were pooled from 3 to 12 mice depending on the strains. PBMCs were recovered using equal volume of 2% Dextran in PBS (MW 55,000). Blood Lin−Thy1+ cells enriched of ILCs were sorted and used in cDNA library preparation per manufacturer's protocol (10x Genomics). The generated libraries were sequenced using an Illumina NovaSeq 6000.
scRNAseq data processing
The Cell Ranger of pipeline version 3.1.0 was used to perform sample demultiplexing, barcode processing, and single-cell 3′ counting. The Linux command cellranger mkfastq was applied to demultiplex raw base call (BCL) files from the illumine NovaSeq6000 sequencer, into sample-specific fastq files. Then, fastq files for each sample were processed with cellranger count, which was used to align samples to mm10 genome, filter and quantify.
Seurat R package (version 3.2.2) was used to process datasets from different samples and canonical correlation analysis was adopted for data integration and potential batch effect correction (Butler et al., 2018; Stuart et al., 2019). Principal component analysis (PCA) was performed on the scaled matrix with 2000 most variable genes using the first 20 principal components. Both tSNE and UMAP dimensional reductions were carried out using the first 20 PCs to obtain two-dimensional representations of the cell states. For clustering, we used the function FindClusters that implements a shared nearest neighbor modularity optimization-based clustering algorithm on 20 PCs with resolution ranging from 0.5 to 1.0 explored for analysis.
Quantification and statistical analysis
Statistical analysis was performed with GraphPad Prism. Unless indicated otherwise, data were expressed as means ± SD. Student's t-test was used for two-group comparison. One-way analysis of variance (ANOVA) was used for multiple group comparisons. ∗p < 0.05, ∗∗<0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Acknowledgements
We thank Dr. Yuan Zhuang for providing plck-Cre; E2Af/+; HEBf/+ mice and Vincent Peng for critical reading of the manuscript. We thank Monica Davis for maintenance of the mouse colonies. We are grateful to the Clinical Genomics Center and the Flow cytometry facility at the Oklahoma Medical Research Foundation for outstanding technical assistance. This work was supported by grants from the NIH (1R01AI126851) and the Presbyterian Health Foundation to X.-H.S., as well as the Oklahoma Center for the Advancement of Science and Technology (HR16-085) to W.R.C. X.-H.S. holds the Lew and Mira Ward Chair in Biomedical Research at the Oklahoma Medical Research Foundation. The Aurora flow cytometer was purchased with support from NIH grant 1S10OD028479-01.
Author contributions
S.B., A.P., M.M., and X.-H. S. performed the experiments; K.L. carried out bioinformatics analyses; S.B., A.P., and X.-H. Sun designed the study. S.B., A.P., K.L., W.R.C., and X.-H. S. analyzed the data and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science.
Published: February 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103732.
Supplemental information
References
- Alisjahbana A., Gao Y., Sleiers N., Evren E., Brownlie D., von Kries A., Jorns C., Marquardt N., Michaelsson J., Willinger T. CD5 surface expression marks intravascular human innate lymphoid cells that have a distinct ontogeny and migrate to the lung. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.752104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artis D., Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- Bain G., Cravatt C.B., Loomans C., Alberola-Ila J., Hedrick S.M., Murre C. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras- ERK MAPK cascade. Nat. Immunol. 2001;2:165–171. doi: 10.1038/84273. [DOI] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Obaldia M.E., Bhandoola A. Transcriptional regulation of innate and adaptive lymphocyte lineages. Annu. Rev. Immunol. 2015;33:607–642. doi: 10.1146/annurev-immunol-032414-112032. [DOI] [PubMed] [Google Scholar]
- Eberl G., Colonna M., Di Santo J.P., McKenzie A.N. Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science. 2015;348 doi: 10.1126/science.aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira A.C.F., Szeto A.C.H., Heycock M.W.D., Clark P.A., Walker J.A., Crisp A., Barlow J.L., Kitching S., Lim A., Gogoi M., et al. RORalpha is a critical checkpoint for T cell and ILC2 commitment in the embryonic thymus. Nat. Immunol. 2021;22:166–178. doi: 10.1038/s41590-020-00833-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger G., Fan X., Dikiy S., Lee S.Y., Rudensky A.Y. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science. 2015;350:981–985. doi: 10.1126/science.aac9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaedi M., Shen Z.Y., Orangi M., Martinez-Gonzalez I., Wei L., Lu X., Das A., Heravi-Moussavi A., Marra M.A., Bhandoola A., et al. Single-cell analysis of RORalpha tracer mouse lung reveals ILC progenitors and effector ILC2 subsets. J. Exp. Med. 2020;217 doi: 10.1084/jem.20182293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Guo L., Qiu J., Chen X., Hu-Li J., Siebenlist U., Williamson P.R., Urban J.F., Jr., Paul W.E. IL-25-responsive, lineage-negative KLRG1(hi) cells are multipotential 'inflammatory' type 2 innate lymphoid cells. Nat. Immunol. 2015;16:161–169. doi: 10.1038/ni.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Mao K., Chen X., Sun M.A., Kawabe T., Li W., Usher N., Zhu J., Urban J.F., Jr., Paul W.E., et al. S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science. 2018;359:114–119. doi: 10.1126/science.aam5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R., Cosway E.J., Willis C., White A.J., Jenkinson W.E., Fehling H.J., Anderson G., Withers D.R. Dynamic changes in intrathymic ILC populations during murine neonatal development. Eur. J. Immunol. 2018;48:1481–1491. doi: 10.1002/eji.201847511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernfeld E.M., Genga R.M.J., Neherin K., Magaletta M.E., Xu P., Maehr R. Asingle-cell transcriptomic atlas of thymus organogenesis resolves cell types and developmental maturation. Immunity. 2018;48:1258–1270. doi: 10.1016/j.immuni.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Peng X.C., Sun X.H. Massive apoptosis of thymocytes in T-cell-deficient Id1 transgenic mice. Mol.Cell Biol. 1999;19:8240–8253. doi: 10.1128/mcb.19.12.8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritsen J.P., Wong G.W., Lee S.Y., Lefebvre J.M., Ciofani M., Rhodes M., Kappes D.J., Zuniga-Pflucker J.C., Wiest D.L. Marked induction of the helix-loop-helix protein Id3 promotes the gammadelta T cell fate and renders their functional maturation Notch independent. Immunity. 2009;31:565–575. doi: 10.1016/j.immuni.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Burke S., Wang J., Chen X., Ortiz M., Lee S.C., Lu D., Campos L., Goulding D., Ng B.L., et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329:85–89. doi: 10.1126/science.1188063. 1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A.I., Li Y., Lopez-Lastra S., Stadhouders R., Paul F., Casrouge A., Serafini N., Puel A., Bustamante J., Surace L., et al. Systemic human ILC precursors provide asubstrate for tissue ILC differentiation. Cell. 2017;168:1086–1100. doi: 10.1016/j.cell.2017.02.021. [DOI] [PubMed] [Google Scholar]
- Matloubian M., Lo C.G., Cinamon G., Lesneski M.J., Xu Y., Brinkmann V., Allende M.L., Proia R.L., Cyster J.G. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- Miyazaki M., Miyazaki K., Chen K., Jin Y., Turner J., Moore A.J., Saito R., Yoshida K., Ogawa S., Rodewald H.R., et al. The E-id protein Axis specifies adaptive lymphoid cell identity and suppresses thymic innate lymphoid cell development. Immunity. 2017;46:818–834. doi: 10.1016/j.immuni.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa M., Germar K., Blom B., Spits H. Human CD5(+) innate lymphoid cells are functionally immature and their development from CD34(+) progenitor cells is regulated by Id2. Front Immunol. 2017;8:1047. doi: 10.3389/fimmu.2017.01047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa M., Heesters B.A., Kradolfer C.M.A., Krabbendam L., Martinez-Gonzalez I., de Bruijn M.J.W., Golebski K., Hendriks R.W., Stadhouders R., Spits H., et al. KLRG1 and NKp46 discriminate subpopulations of human CD117(+)CRTH2(-) ILCs biased toward ILC2 or ILC3. J. Exp. Med.Jem. 2019;216:1762–1776. doi: 10.1084/jem.20190490. 20190490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng V., Georgescu C., Bakowska A., Pankow A., Qian L., Wren J.D., Sun X.H. E proteins orchestrate dynamic transcriptional cascades implicated in the suppression of the differentiation of group 2 innate lymphoid cells. J. Biol. Chem. 2020;295:14866–14877. doi: 10.1074/jbc.RA120.013806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L., Bajana S., Georgescu C., Peng V., Wang H.C., Adrianto I., Colonna M., Alberola-Ila J., Wren J.D., Sun X.H. Suppression of ILC2 differentiation from committed T cell precursors by E protein transcription factors. J. Exp. Med. 2019;216:884–899. doi: 10.1084/jem.20182100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigas D., Lewis G., Aron J.L., Wang B., Banie H., Sankaranarayanan I., Galle-Treger L., Maazi H., Lo R., Freeman G.J., et al. Type 2 innate lymphoid cell suppression by regulatory T cells attenuates airway hyperreactivity and requires inducible T-cell costimulator-inducible T-cell costimulator ligand interaction. J. Allergy Clin.Immunol. 2017;139:1468–1477.e2. doi: 10.1016/j.jaci.2016.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C., Lee J., Koga S., Ricardo-Gonzalez R.R., Nussbaum J.C., Smith L.K., Villeda S.A., Liang H.E., Locksley R.M. Tissue-resident group 2 innate lymphoid cells differentiate by layered ontogeny and in situ perinatal priming. Immunity. 2019;50:1425–1438. doi: 10.1016/j.immuni.2019.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S.B., Lo B.C., Ghaedi M., Scott R.W., Li Y., Messing M., Hernaez D.C., Cait J., Murakami T., Hughes M.R., et al. Abortive gammadeltaTCR rearrangements suggest ILC2s are derived from T-cell precursors. Blood Adv. 2020;4:5362–5372. doi: 10.1182/bloodadvances.2020002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e1. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F., Zhou Y., Jin R., Chen Y., Pei X., Liu Y., Dong J., Wang W., Pang X., Qian X., et al. The molecular signature underlying the thymic migration and maturation of TCRalphabeta+ CD4+ CD8 thymocytes. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas C.L., Poursine-Laurent J., Yang L., Yokoyama W.M. Development of thymic NK cells from double negative 1 thymocyte precursors. Blood. 2011;118:3570–3578. doi: 10.1182/blood-2011-06-359679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veinotte L.L., Greenwood C.P., Mohammadi N., Parachoniak C.A., Takei F. Expression of rearranged TCRgamma genes in natural killer cells suggests a minor thymus-dependent pathway of lineage commitment. Blood. 2006;107:2673–2679. doi: 10.1182/blood-2005-07-2797. [DOI] [PubMed] [Google Scholar]
- Vivier E., Artis D., Colonna M., Diefenbach A., Di Santo J.P., Eberl G., Koyasu S., Locksley R.M., McKenzie A.N.J., Mebius R.E., et al. Innate lymphoid cells: 10 Years on. Cell. 2018;174:1054–1066. doi: 10.1016/j.cell.2018.07.017. [DOI] [PubMed] [Google Scholar]
- Walker J.A., Clark P.A., Crisp A., Barlow J.L., Szeto A., Ferreira A.C.F., Rana B.M.J., Jolin H.E., Rodriguez-Rodriguez N., Sivasubramaniam M., et al. Polychromicreporter mice reveal unappreciated innate lymphoid cell progenitor heterogeneity and elusive ILC3 progenitors in bone marrow. Immunity. 2019;51:104–118 e107. doi: 10.1016/j.immuni.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.C., Qian L., Zhao Y., Mengarelli J., Adrianto I., Montgomery C.G., Urban J.F., Jr., Fung K.M., Sun X.H. Downregulation of E Protein activity augments an ILC2 differentiation program in the thymus. J. Immunol. 2017;198:3149–3156. doi: 10.4049/jimmunol.1602009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M.A., Jameson S.C., Hogquist K.A. Postselection thymocyte maturation and emigration are independent of IL-7 and ERK5. J. Immunol. 2011;186:1343–1347. doi: 10.4049/jimmunol.1002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski J., Lai A., Kondo M., Zhuang Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J. Immunol. 2007;178:5717–5726. doi: 10.4049/jimmunol.178.9.5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Bhandoola A. The development of adult innate lymphoid cells. Curr.Opin.Immunol. 2016;39:114–120. doi: 10.1016/j.coi.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeis P., Lian M., Fan X., Herman J.S., Hernandez D.C., Gentek R., Elias S., Symowski C., Knopper K., Peltokangas N., et al. In Situ maturation and tissue adaptation of type 2 innate lymphoid cell progenitors. Immunity. 2020;53:775–792. doi: 10.1016/j.immuni.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C., Zheng M., Cui K., Martins A.J., Hu G., Li D., Tessarollo L., Kozlov S., Keller J.R., Tsang J.S., et al. Differential expression of the transcription factor GATA3 specifies lineage and functions of innate lymphoid cells. Immunity. 2020;52:83–95 e84. doi: 10.1016/j.immuni.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
•Single cell RNAseq data has been deposited in NCBI. The accession number for the scRNA-seq data reported in this paper is GEO: GSE176357 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE176357).
•This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
•Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.