Abstract
Background:
Multiple pharmacogenomic studies have identified the synonymous genomic variant rs7853758 (G>A, L461L) and the intronic variant rs885004 in SLC28A3 as statistically associated with a lower incidence of anthracycline–induced cardiotoxicity (AIC). However, the true causal variant(s), the cardioprotective mechanism of this locus, the role of SLC28A3 and other solute carrier (SLC) transporters in AIC, and the suitability of SLC transporters as targets for cardioprotective drugs has not been investigated.
Methods:
Six well–phenotyped, doxorubicin–treated pediatric patients from the original association study cohort were re–recruited and human induced pluripotent stem cell–derived cardiomyocytes were generated. Patient-specific doxorubicin–induced cardiotoxicity (DIC) was then characterized using assays of cell viability, activated caspase 3/7, and doxorubicin uptake. The role of SLC28A3 in DIC was then queried using overexpression and knockout of SLC28A3 in isogenic hiPSCs using a CRISPR/Cas9. Fine–mapping of the SLC28A3 locus was then completed after SLC28A3 resequencing and an extended in silico haplotype and functional analysis. Genome editing of potential causal variant was done using cytosine base editor. SLC28A3-AS1 overexpression was done using a lentiviral plasmid-based transduction and was validated using stranded RNA-Seq after ribosomal RNA depletion. Drug screening was done using the Prestwick drug library (n = 1200) followed by in vivo validation in mice. The effect of desipramine on DOX cytotoxicity was also investigated in eight cancer cell lines.
Results:
Here, using the most commonly used anthracycline, doxorubicin, we demonstrate that patient–derived cardiomyocytes recapitulate the cardioprotective effect of the SLC28A3 locus and that SLC28A3 expression influences the severity of DIC. Using Nanopore–based fine–mapping and base editing we identify a novel cardioprotective SNP rs11140490 in the SLC28A3 locus which exerts its effect by regulating an antisense long noncoding–RNA (SLC28A3-AS1) that overlaps with SLC28A3. Using high–throughput drug screening in patient–derived cardiomyocytes and whole organism validation in mice, we identify the SLC competitive inhibitor desipramine as protective against DIC.
Conclusions:
This work demonstrates the power of the human induced pluripotent stem cell model to take a SNP from a statistical association through to drug discovery, providing human cell–tested data for clinical trials to attenuate DIC.
Keywords: Doxorubicin–induced cardiotoxicity, GWAS, solute carriers, human induced pluripotent stem cells, cardiomyocytes, cardioprotectant
Introduction
Doxorubicin (DOX), a cytotoxic anthracycline antibiotic, is a common anti–cancer agent used to treat a wide variety of adult and childhood cancers. The cardiotoxicity of anthracyclines has been documented to be occur in 9% of treated adult patients1, and on average occurs in just 3.5 months after the last chemotherapy dose and 98% of patients experience cardiotoxicity within the first year1. Early cardiotoxicity leads to dose limitation or treatment discontinuation to the detriment of therapy. The cardiotoxicity of doxorubicin is also well–understood to be dose–dependent, with 65% and 85% of cancer patients experiencing a decline in left ventricular ejection fraction (LVEF) when treated with DOX dose of 550 and 700 mg/m2, respectively2. However, even despite attempts to severely limit cumulative dose, cardiotoxicity occurs in 14.5% of breast cancer patients receiving the most common 240 mg/m2 cumulative dose3.
Pharmacogenomic research has attempted to discover predictive DNA biomarkers for anthracycline–induced cardiotoxicity (AIC) and has so far identified about 75 AIC–associated loci4, 5. However, the true connection between these loci and cardiotoxicity is far from proven, as the vast majority of AIC pharmacogenomic studies lack functional validation of the identified associations. As a result, there are currently no FDA–approved genetic biomarkers being used in routine clinical practice to predict AIC6, and only a single on–market drug, dexrazoxane is approved to decrease the incidence of AIC.
SLC28A3 encodes solute carrier transporter family 28 member 3 and is the most robustly replicated AIC–associated cardioprotective loci. This locus was initially discovered in a large multi–center pediatric candidate gene association study (CGAS), identifying two single nucleotide polymorphisms (SNPs), rs7853758 (G>A, L461L), which is in a coding region but synonymous, and rs885004 which is located in intron 8. Both SNPs are in high linkage disequilibrium. rs7853758 is highly associated with a lower risk of developing DIC in both discovery (n = 188, Padj = 0.0071, OR = 0.29) and replication (n = 156, Padj = 0.0072, OR = 0.33) cohorts7. Importantly, this genetic association was replicated in a third additional multicenter independent cohort of 218 patients8. The sensitivity and specificity (95% CI) of rs7853758 is 17.4 (7.8–31.4) and 64.6 (58.8–70.1), respectively 9. Despite identification of this SNP through two replication cohorts in CGAS, the mechanisms by which a synonymous variant can influence AIC is unclear. Thus, the validity of this this locus in relation to AIC, the true causal variant and the cardioprotective mechanism of this locus, the role of SLC28A3 and other solute carrier (SLC) transporters in AIC, and the suitability of SLC transporters as targets for cardioprotective drugs are critical unanswered questions.
Here, we show that patient–specific hiPSC–CMs recapitulate the cardioprotective effect of SNP rs7853758. SLC28A3 knockdown and overexpression using CRISPR/Cas9 reduces and increases DOX uptake into cardiomyocytes respectively, altering their sensitivity to DOX, thus confirming the role of this locus in DIC. Fine–mapping of the SLC28A3 locus uncovered that rather than the original CGAS–identified synonymous SNP (rs7853758), it is actually the linked SNP rs11140490 that is the causal cardioprotective variant within that locus. Further mechanistic studies showed that rs11140490 exerts its action by regulating a SLC28A3–overlapping antisense long non–coding RNA SLC28A3-AS1. Screening for other potential cardiac–specific SLC transporters in relation to DIC revealed that SLC22A4 and SLC22A17 are also implicated in DIC. Finally screening a drug library using hiPSC–CMs followed by in vivo validation in a mouse model of DIC discovered that the SLC competitive inhibitor, desipramine protects against DIC by without hindering DOX chemotherapy efficacy. Together these findings provide a novel genetic test for rs11140490 that can identify patients who are protected from DIC and two potential therapeutic options, either using the lncRNA SLC28A3-AS1 or developing a derivative of desipramine to attenuate DIC.
Methods
The data, analytic methods, and study materials are available other researchers upon reasonable request for purposes of reproducing the results. Detailed methods are provided in the Supplementary Information online. RNA–seq data have been deposited in Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) with accession code GSE165731.
Human induced pluripotent cell derivation and cardiac differentiation.
Protocols and consents were approved by the Northwestern University and University of British Columbia Institutional Review Boards. Six well–phenotyped, doxorubicin–treated pediatric patients from the original association study cohort were re–recruited with informed consent. Peripheral blood mononuclear cells were isolated from blood and reprogramed to hiPSCs using CytoTune–iPS 2.0 Sendai Reprogramming Kit (Invitrogen)10. SNP karyotyping was performed using a whole–genome Infinium HumanCytoSNP–12 BeadChip Array (Illumina). Differentiation into cardiomyocytes was performed according to previously described protocol with some modifications11, 12 including the use of a TNNT2-driven antibiotic selection cassette for cardiomyocyte purification.
CRISPR/Cas9–mediated gene knockout and overexpression.
To generate SLC28A3 knockout gRNA expression vectors, gRNA targeting the start codon designed with minimal predicted off–target effect13. Each gRNA was annealed and inserted into pSpCas9(BB)-2A-Puro (PX459) V2.0 (48138, Addgene) plasmid that expresses puromycin resistance gene for downstream antibiotic selection, in addition to Cas9. 106 cells were electroporated with 5 μg PX459 plasmid and positive clones were selected 24 h post transfection using puromycin treatment for 48 h. To generate SLC28A3 overexpressing cells, Human SLC28A3 Sequence-Verified cDNA was first amplified and cloned under the CAG promoter of a pAAVS1–Nst–CAG–DEST gateway cloning vector (80489, Addgene). AAVS1 gRNA expression vector14 (pXAT2, Addgene 80494), which expresses gRNA and Cas9, was used to target AAVS1 locus in the first intron of the PPP1R12C gene14. Cells were then electroporated with AAVS1 targeting plasmid and SLC28A3 overexpression donor plasmid. Positive clones were selected using neomycin treatment for 14 days.
Patient-specific doxorubicin–induced cardiotoxicity (DIC) characterization.
Patient-specific DIC was characterized by assays of cell viability using CellTiter–Glo 2.0 (Promega) and activated caspase 3/7 using Caspase 3/7–Glo (Promega) that were used per manufacturer’s instructions. Doxorubicin uptake was quantified using flow cytometry by measuring DOX intrinsic fluorescence-PE 1 and 3 h post doxorubicin treatment and normalized to baseline fluorescence. All cells were stained with NucRed Live ReadyProbes Reagent (Invitrogen) to monitor cell viability.
SLC28A3 locus genetic fine mapping.
Fine–mapping of the SLC28A3 locus was then completed after SLC28A3 resequencing using MinION Nanopore sequencer and an extended in silico haplotype and functional analysis.
Genome editing of potential causal variant rs11140490.
Locus-specific base-editor protein complex and the gRNA were designed using Beditor 15, and the designed gRNA was cloned in the gRNA expressing plasmid (73797, Addgene). Then 1 × 106 cells were electroporated with 4 μg of the base editor expressing plasmid (pSI-Target-AID-NG, 119861, Addgene) and 4 μg of the gRNA expressing plasmid (lenti sgRNA (MS2) _puro, 73797, Addgene). Cells were then selected with puromycin 24 h post transfection for 48 h, clones were hand-picked, the target locus was PCR-amplified and sanger-sequenced to confirm the SNP editing in all clones.
SLC28A3-AS1 overexpression in isogenic hiPSCs.
The SLC28A3-AS1 cDNA was cloned into pLenti-C-Myc-DDK-IRES-Puro lentiviral vector (Origene) which was then co–transfected with packaging plasmids psPAX2 (Addgene 12260) and pMD2.G (Addgene 12259) into Lenti-X 293T cells (Takara) to generate lentivirus. Virus–containing supernatant was collected at 48– and 72–hours post–transfection. Lentivirus was concentrated 1:100 from cleared supernatant using PEG-iT (SBI). Isogeneic hiPSCs were then transduced and positive clones were selected with puromycin for seven days to generate ISOSLC28A3-AS1. SLC28A3-AS1 overexpression was confirmed using stranded RNA–Seq after ribosomal RNA depletion.
Mouse model of doxorubicin–induced cardiomyopathy and drug administration.
Procedures followed were in accordance with Stanford University’s institutional guidelines. In vivo validation was done using C57BL/6J 10 week old male mice were co-treated with doxorubicin (NovaPlus) and water as a control vehicle (n = 10), or with desipramine (Sigma) as experimental groups (n = 8). At day 0, mice were treated with doxorubicin (3 mg kg−1) intraperitoneally twice a week alone or with desipramine by Alzet pump infusion (20 mg kg−1 day−1) for 3 weeks (day 0–day 21). For the control group, we treated mice with corn oil in the same schedule as desipramine administration. We recorded an echocardiogram once a week (day 0, day 7, day 14, and day 21) and terminated the experiment at day 21.
Statistical Analysis.
Data were analyzed in R version 4.0.3 and graphed in GraphPad Prism 6. Detailed statistical information is included in the corresponding figure legends. Data were presented as mean ± SEM. Comparisons were conducted via one way–ANOVA test, or an unpaired two–tailed Student’s t–test with significant differences defined as P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), and P < 0.0001 (****). Our sample size (3 patients in each category) was based on the feasibility of handling this number of hiPSC lines. For dose response curves, log-logistic non-linear regression model was used to estimate the value of the four parameters, and t–statistic was used to test for significant difference in LD50 between different groups using “drc” package16 in R.
Results
Investigation of the protective role of variant rs7853758
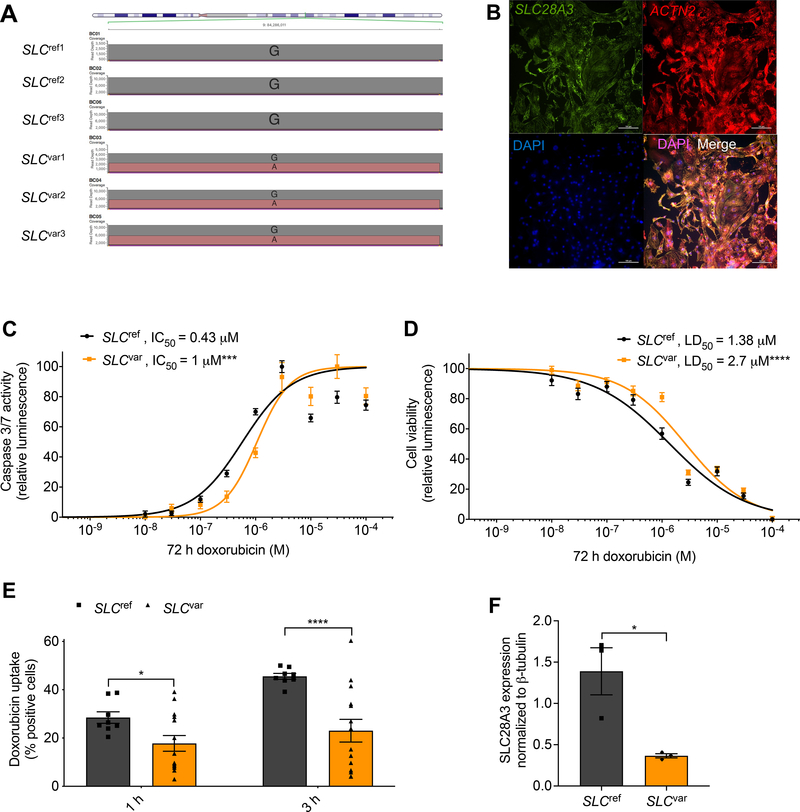
Six well–phenotyped, doxorubicin–treated pediatric patients from the original CGAS cohort were specifically re–recruited according to the original inclusion criteria17 (Supplementary Table 1). These included three patients who were heterozygous for the rs7853758 variant and were protected from DIC (SLCvar1, SLCvar2, SLCvar3; collectively SLCvar), and three control patients who did not carry this protective variant (they carry the reference allele) and developed DIC after the DOX therapy (SLCref1, SLCref2, SLCref3; collectively SLCref). Detailed patient data including age, sex, ethnicity, type of cancer, treatment regimen, and presence or absence or DIC are provided in Supplementary Table 2. hiPSC lines were established from patients’ peripheral blood using non–integrating (Sendai virus–based) reprogramming and our well–established protocols18, 19. These lines showed normal hiPSC morphology (Supplementary Fig. 1a), expressed high levels of undifferentiated cell markers (Supplementary Figs. 1b–c), and were karyotypically normal (Supplementary Fig. 2). The genotypes of the rs7853758 SNP were validated using Nanopore–based sequencing (Fig. 1a). Cardiomyocytes were generated using our established chemically defined, small molecule–based monolayer differentiation system11, 20 (Supplementary Fig. 3a) along with a TNNT2-based antibiotic selection cassette which consistently produces cardiomyocytes which are 80–98% TNNT2+ (Supplementary Figs. 3b–d). hiPSC-CMs express SLC28A3 throughout the cell (Fig. 1b).
Figure 1. Patient-specific hiPSC–CMs recapitulate the cardioprotective effect of SLC28A3 variant rs7853758.
Comparison of hiPSC–CMs derived from three patients harboring the heterozygous rs7853758 variant and were protected from DIC after DOX treatment (SLCvar1, SLCvar2, SLCvar3; collectively SLCvar), to hiPSC-CMs from three control patients who did not carry this protective SNP and developed DIC upon same DOX treatment (SLCref1, SLCref2, SLCref3 SLCref3SLCref3; collectively SLCref). A, Nanopore sequencing reads at SNP rs7853758 locus confirming its genotypes in all patient–derived hiPSC lines. B, Immunofluorescent staining showing the expression and localization of SLC28A3 throughout the cell in patient–derived hiPSC–CMs. C, Effect of DOX (72 h) on cell viability in SLCvar (n = 126) and SLCref (n = 81) hiPSC–CMs measured by a CellTiter-Glo 2.0 assay. D, Effect of DOX (72 h) on apoptosis measured by activated caspase 3/7 in SLCvar (n = 20) and SLCref (n = 20) hiPSC–CMs. E, Assessment of DOX uptake via measurement of percentage of cells with DOX intrinsic fluorescence using a flow cytometry-based assay in patient–derived hiPSC–CMs (n = 8–13). F, SLC28A3 expression in SLCref (n = 3), and SLCvar (n = 3) hiPSC-CMs using western blot. n = full independent experimental replicates, Error bars, s.e.m, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 by unpaired two-tailed Student’s t-test (E and F). For (C and D) log-logistic non-linear regression model was used to estimate the value of the four parameters, and t-statistic was used to test for significant difference in LD50 between different groups.
Patient–specific hiPSC–CMs recapitulate the cardioprotective effect of rs7853758 against DIC.
We first sought to model the cardioprotective effect of rs7853758 variant in patient–specific hiPSC–CMs. A cell viability assay completed at 72 h post DOX treatment demonstrated that the three SLCvar lines recapitulated the protective effect of SNP (rs7853758 G>A, L461L) (LD50 = 2.7 μM, P <0.0001, n = 126) as compared to the SLCref lines (LD50 = 1.38 μM, n = 81) (Fig. 1c and Supplementary Fig. 12a). Consistently, apoptosis as quantified by a caspase–3 and –7 activity assay completed 72 h post DOX treatment was significantly lower in SLCvar lines (IC50 = 1 μM, P = 0.001, n = 20) compared to the SLCref lines (IC50 = 0.43 μM, n = 20) (Fig. 1d). Since SLC28A3 is an uptake transporter, we hypothesized that the lower level of DIC in the SLCvar lines might be due to reduced DOX uptake as a result of impaired SLC28A3 function in these patients. To test this, we used a flow cytometry–based assay and found that intracellular DOX uptake was ~50% lower in SLCvar as compared to SLCref (Fig. 1e), which is consistent with the magnitude LD50 (1.38 μM vs. 2.7 μM) and IC50 (0.43 μM and 1 μM) changes on our in vitro DIC assays (Fig. 1c). To investigate whether this cardioprotective effect seen in SLCvar is due to altered SLC28A3 protein expression, we then quantified SLC28A3 in our patient–specific hiPSC–CMs using western blot. SLC28A3 expression in SLCvar was significantly lower than SLCref (P = 0.03) (Fig. 1f, and Supplementary Fig. 12b).
SLC28A3 altered expression affects doxorubicin–induced cardiotoxicity in hiPSC–CMs.
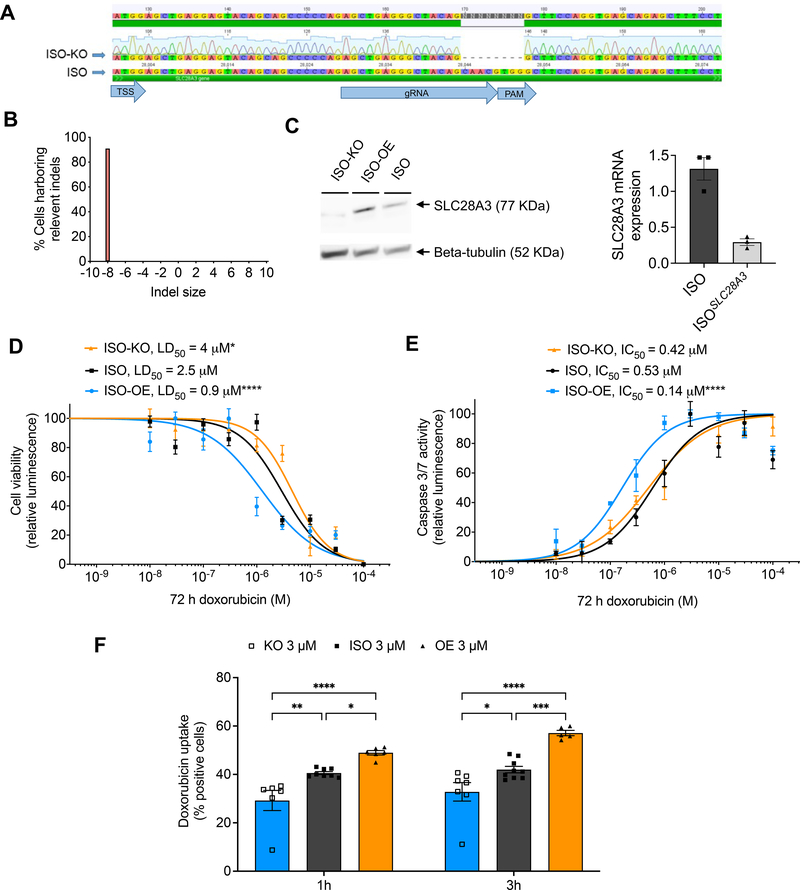
To further confirm the implication of SLC28A3 in DIC regardless of patient-specific genetic, transcriptomic, and environmental background and to isolate the effect of SLC28A3 in relation to DIC, we next examined whether a gain or loss of function of SLC28A3 altered DIC in an isogenic hiPSC line (ISO). The ISO line was derived from a healthy individual and its whole genome sequencing confirmed that it doesn’t harbor any other DIC–associated loci identified by the original study. SLC28A3 overexpression (ISO–OE) and knockout (ISO–KO) lines were generated via a CRISPR/Cas9–mediated approach. Disturbance at start codon region was confirmed by Sanger sequencing (Fig. 2a–b) and altered SLC28A3 expression in these lines was confirmed by RT-PCR and western blot showing 90% downregulation of SLC28A3 in ISO-KO (Fig. 2c). The effect of SLC28A3 OE and KO on in vitro DIC was investigated using the above cell viability and caspase assays at 72 h post DOX treatment. The cell viability assay showed that the ISO–OE hiPSC–CMs (LD50 = 0.9 μM) were ~3.3–fold and 1.4–fold more sensitive to DOX as compared to ISO–KO (LD50 = 4 μM, P = 0.03) and ISO (LD50 = 2.5 μM, P < 0.0001), respectively (Fig. 2d). Likewise, caspase activity was ~2.6–fold higher in ISO–OE (LD50 = 0.14 μM) as compared to ISO (LD50 = 0.53 μM, P <0.0001) (Fig. 2e). We next sought to investigate the effect of SLC28A3 KO and OE on DOX intracellular uptake. DOX uptake was significantly higher in ISO–OE as compared to ISO both at 1 h (P = 0.035) and 3 h (P < 0.0017) post DOX treatment (Fig. 2f). Similarly, DOX uptake was significantly lower in ISO–KO as compared to ISO both at 1 h (P = 0.0009) and 3 h (P = 0.0006) post DOX treatment (Fig. 2f). These findings show that SLC28A3 is implicated in DIC regulation through affecting DOX uptake into cardiomyocytes.
Figure 2. SLC28A3 expression affects the severity of DIC by regulating DOX uptake into cardiomyocytes.
A, Validation of CRISPR/Cas9–mediated SLC28A3 knockout (KO) in an isogenic hiPSC line detected by Sanger sequencing, showing 8 bp deletion downstream of the transcription start site (TSS). PAM, protospacer adjacent motif. B, Demonstration that 91% of the cell population acquire the introduced deletion. C, Validation of KO and AAVS1-based SLC28A3 overexpression (OE) by western blot and RT-PCR. D, Effect of DOX (72 h) on viability in ISO (n = 45), ISO–OE (n = 14), and ISO–KO (n = 6) hiPSC–CMs. E, Effect of doxorubicin (72 h) on apoptosis measured by activated caspase 3/7 in ISO (n = 8), ISO–OE (n = 10), and ISO–KO (n = 6) hiPSC–CMs. F, Assessment of DOX uptake via measurement of DOX intrinsic fluorescence using flow cytometry-based assay (n = 6–9). n = full independent experimental replicates, Error bars, s.e.m, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 by unpaired two-tailed Student’s t-test (f). For (d and e) log-logistic non–linear regression model was used to estimate the value of the four parameters, and t-statistic was used to test for significant difference in LD50 between different groups.
Fine mapping at the SLC28A3 locus prioritizes potential causal variant.
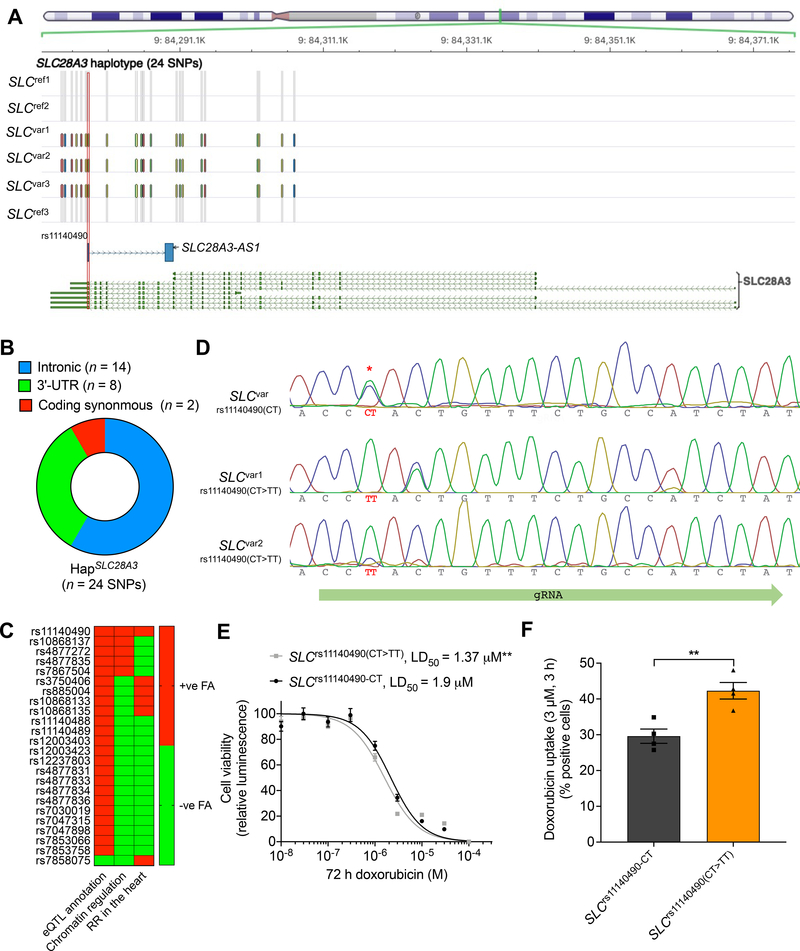
rs7853758 is a coding synonymous SNP located in exon 14 and thus does not affect the amino acid sequence. Because of the tag–SNP and linkage disequilibrium (LD) issues associated with GWAS21, we expected that rs7853758 is linked (coinherited) to additional SNPs including the causal one(s). To elucidate this, we next fine–mapped the SLC28A3 locus to identify the potential causal variant. We sequenced the SLC28A3 gene in all six SLCvar and SLCref patients using a Nanopore MinION sequencer and SNPs were called using Nanopolish22 (see online methods). For all patients, Nanopore genotypes of the original association study hit, rs7853758 were in concordance with the GWAS–chip genotypes (Fig. 1a). In total 133 SNPs were identified all of which have at least one variant allele in at least one patient (Supplementary Fig. 4e). The vast majority of identified SNPs were intronic (n = 93), 25 SNPs were located in 5’–UTR, 12 SNPs were located in 3’–UTR, in addition to three coding SNPs of which two were synonymous and one non–synonymous (Supplementary Table 6). We then examined which SNPs were exclusively co–inherited in cardioprotected patients and identified a cardioprotective haplotype, HapSLC28A3 comprising 24 SNPs that is co–inherited only in cardioprotected patients. These SNPs are distributed as follows, eight SNPs are located in the 3'–UTR, 14 SNPs are intronic, and two are coding synonymous SNPs (Figs. 3a–b and Supplementary Table 6).
Figure 3. Fine–mapping at the SLC28A3–SLC28A3-AS1 locus identifies rs11140490 as the potential causal cardioprotective variant.
A, Location of the HapSLC28A3 comprising 24 SNPs that are co–inherited only in SLCvar protected patients. SNP rs11140490 marked by red rectangle is located at the splice site of the first exon of an overlapping long non–coding RNA, SLC28A3-AS1 (adapted from Magdy et al.29). B, Consequence of co–inherited HapSLC28A3 SNPs (n = 24). C, Overall prioritization of candidate causal SNPs based on functional annotation analyses including, eQTL annotation, chromatin regulatory analyses, and overlapping with regulatory regions (RR) in cardiac tissues. FA, functional annotation (adopted from Magdy et al.29). D, Editing of rs11140490 (CT>TT) in two patient-specific hiPSC lines using cytosine base editor (Target-AID-NG), rs11140490 is marked by red asterisk. E, Effect of DOX (72 h) on viability in SLCvar-rs11140490 (CT) (2 lines, n = 31) and SLCvar-rs11140490(CT>TT edited) (2 lines, n = 75) hiPSC–CMs. F, Assessment of DOX uptake via measurement percentage of cells with DOX intrinsic fluorescence using flow cytometry-based assay in SLCvar-rs11140490 (CT) and SLCvar-rs11140490(CT>TT edited) (n = 4). n = full independent experimental replicates, Error bars, s.e.m, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 by unpaired two-tailed Student’s t-test (F). For (E) log-logistic non-linear regression model was used to estimate the value of the four parameters, and t-statistic was used to test for significant difference in LD50 between different groups.
Interestingly, seven SNPs within HapSLC28A3 are located within a long non–coding RNA, ensemble gene id: ENSG00000233262 that we called “SLC28A3-AS1” (submitted to HGNC registry) that overlaps with SLC28A3 forming another haplotype, HapSLC28A3-AS1 (Fig. 3a, Supplementary Figs. 5a–b). HapSLC28A3-AS1 consists of SNPs, rs11140490 (A>G), rs10868135 (T>C), rs4877831 (C>G), rs4877833 (T>C), rs7853066 (A>G), rs7853758 (G>A), and rs7030019 (A>G).
In order to validate and confirm the linkage disequilibrium pattern of the Nanopore–identified cardioprotective haplotype, HapSLC28A3, we investigated the haplotype structure and allelic frequency of this cardioprotective haplotype on a wider population level in 99 individuals of the CEU (Utah Residents (CEPH) with Northern and Western European Ancestry) population, the same ethnic population of the study cohort. This analysis showed that the 24 SNPs constituting the HapSLC28A3 were in high LD with an average D’ and R2 of 0.99 and 0.84, respectively (Supplementary Figs. 5a–b and Supplementary Table 11). Regarding HapSLC28A3-AS1, seven structures were identified, Hap–ISLC28A3-AS1 to Hap–VIISLC28A3-AS1 (Supplementary Fig. 5c). In that, Hap–ISLC28A3-AS1 consists of the reference alleles for all seven SNPs (ATCTAGA) and is inherited in 71.7% of the examined population, whereas Hap–IISLC28A3-AS1 comprises the variant alleles for all seven SNPs (GCGCGAG) and is inherited in 17.7% of the examined population (Supplementary Fig. 5c). This finding confirms the linkage disequilibrium pattern identified by the Nanopore pipeline in all of the study patients.
To eliminate the probability that the causal variant might be located in one of the adjacent genes to the SLC28A3 / SLC28A3-AS1 target locus, we did an extended LD analysis to include all variants that are located with 1 MB up and down–stream of the target locus in both European (the same ethnic population of the original genotype–phenotype association study) and all ethnicity populations. These analyses did not identify any other SNPs that are linked to rs7853758 and are not included in our original haplotype analysis. This eliminates the probability that the causal cardioprotective variant is located in adjacent genes (Supplementary Fig. 6).
In order to prioritize cardioprotective haplotype SNPs, we investigated the regulatory properties of all candidate SNPs (n = 24). Using the Encyclopedia of DNA Elements (ENCODE) and Roadmap Epigenomics23 data and DeepSEA24 algorithm, we examined the functional effect of each SNP on altering chromatin features (transcription factors, DNase hypersensitive site, and histone marks) binding sites. Among all SNPs, rs11140490 and rs4877835 had the top chromatin regulatory effect as both SNPs predicted to alter the binding site of 206 and 204 chromatin features, respectively (Supplementary Fig. 4f, Supplementary Tables 7 and 8). Moreover, SNP rs11140490 has the most substantial regulatory effect as it is predicted to alter the binding sites of 43 features with log2 fold change of ≥1, whereas rs4877835 is predicted to alter the binding sites of only 4 features with log2 fold change of ≥1. Unsurprisingly, the primary study significant association, rs7853758 does not show any significant chromatin regulatory effect (Supplementary Fig. 4g).
Since DIC affects cardiomyocytes, we performed an additional regulatory analysis exclusively focusing on human cardiac tissue, and for that we used ensemble regulatory build that includes transcription factors, histone mark, and DNase hypersensitive regions. Six SNPs, rs11140490, rs4877835, rs4877831, rs7047898, rs885004, and rs10868137 were found to be located in at least one regulatory region in human cardiac tissue (Supplementary Table 9). Finally, to investigate further regulatory consequences of these candidate SNPs, we used the Genotype–Tissue Expression (GTEx) project database (https://www.gtexportal.org/home/) to investigate which of the identified candidate SNPs have been shown to be an expression quantitative trait loci (eQTL). All candidate SNPs except rs7858075 have been previously identified as eQTL in cultured fibroblasts, thyroid, and brain tissues (Supplementary Table 10). Although the eQTLs identified by the GTEx are located in non-cardiac tissues, these associations emphasize the regulatory function of these SNPs. These findings when taken together suggest that SNP rs11140490, interestingly located at the splice site of the first exon of SLC28A3-AS1, is the SNP with the highest likelihood to be the causal cardioprotective SNP (Fig. 3c).
Editing rs11140490 in hiPSC–CMs confirms its causality in relation to protection against DIC.
After we prioritized rs11140490 to be the top candidate causal variant within the DIC–associated SLC28A3–SLC28A3-AS1 locus, we went on to confirm the causality of this variant. Using a base–editor mediated approach, we edited the SNP rs11140490 in hiPSCs from two SLCvar patients that harbor the heterozygous genotype CT back to the reference genotype TT. The cytosine base editor that we have used is composed of a catalytically inactive “dead” Cas9 (dCas9) fused to (CBE) cytidine deaminase that converts C•G base pair to a T•A base pair. Importantly, the CBE does not induce a DNA cut and thus helped us avoid the mono-allelic genomic deletions and loss-of-heterozygosity problem associated with the homology directed repair (HDR)-based genomic editing approaches25. Positive hiPSC clones were differentiated into cardiomyocytes and finally DIC and DOX uptake were quantified using the above assays. SLCrs11140490(CT>TT) cardiomyocytes were more sensitive to DOX (LD50 = 1.37 μM, P = 0.005) as compared to SLCrs11140490(CT) LD50 = 1.9 μM) (Fig. 3e). Moreover, DOX uptake was significantly higher in SLCrs11140490(CT>TT) when compared to SLC rs11140490(CT) at 3 h post DOX treatment (P = 0.006) (Fig. 3f). This finding confirms that the SNP rs11140490 is the causal cardioprotective SNP affecting DIC.
Variant rs11140490 exert its cardioprotective effect by regulating the long non–coding RNA, SLC28A3-AS1.
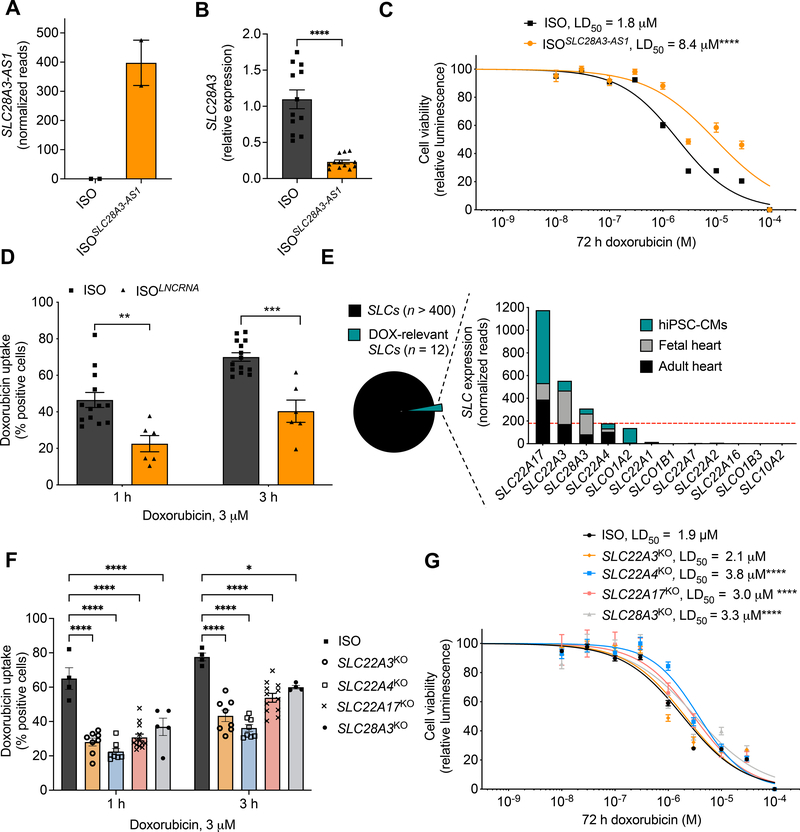
Next, we investigated the cardioprotective mechanism of rs11140490. This variant is located at the splice site of the first exon of the anti–sense SLC28A3-AS1 that overlaps with SLC28A3. Thus, we hypothesized that SNP rs11140490 might exert its cardioprotective action via regulating the transcription of SLC28A3-AS1, which in turn alters the expression of doxorubicin–related genes including SLC28A3, and eventually alters patients’ susceptibility to DIC. To test this hypothesis, we investigated the effect of altered expression of SLC28A3-AS1 on DIC phenotype. We overexpressed SLC28A3-AS1 in an isogenic cell line (ISOSLC28A3-AS1) by transducing isogenic hiPSCs with the SLC28A3-AS1 cDNA cloned into pLenti-MYC-DDK-IRES-Puro lentivirus expression vector (ORIGENE, PS100069). SLC28A3-AS1 overexpression was confirmed by stranded RNA-Seq after ribosomal RNA depletion (Fig. 4a). We then assessed the effect SLC28A3-AS1 overexpression on SLC28A3 expression in cardiomyocytes and showed that SLC28A3 is significantly downregulated in ISOSLC28A3-AS1 cardiomyocytes after DOX treatment (Fig. 4b). We then investigated the effect of SLC28A3-AS1 overexpression on DIC and DOX uptake as before. ISOSLC28A3-AS1 hiPSC–CMs were significantly more resistant to DOX (LD50 = 8.4 μM, P < 0.0001) as compared to ISO (LD50 = 1.8 μM) (Fig. 4c). Moreover, DOX uptake was significantly impaired in ISOSLC28A3-AS1 cardiomyocytes when compare to ISO both at 1 h (P = 0.001) and 3 h (P < 0.001) post DOX treatment (Fig. 4d). These results show that the regulation of SLC28A3-AS1 is a potential mechanism by which SNP rs11140490 exerts its cardioprotective effect.
Figure 4. The cardioprotective role of SLC28A3-AS1.
A, Relative Expression of SLC28A3-AS1 in ISO hiPSC compared to ISO transduced by viral plasmid encoding SLC28A3-AS1 cDNA to overexpress SLC28A3-AS1 (ISOSLC28A3-AS1) assessed by RNA–Seq (n = 2). B, SLC28A3 relative expression in ISO and ISOSLC28A3-AS1 hiPSC-CMs (n = 5–11) assessed by real-time PCR. C, Effect of SLC28A3-AS1 overexpression on cell viability after DOX (72 h) treatment, ISO (n = 17), ISOSLC28A3-AS1 (n = 12). D, Effect of SLC28A3-AS1 overexpression on DOX uptake 1 h and 3 h post DOX treatment (n = 6–14). E, Relative human cardiomyocyte expression of SLC transporters (n = 12) previously identified as transporting DOX or a DOX metabolite and/or by genetic associations with DOX clinical outcomes. Red dashed line denoted for the expression cutoff for SLC transporter selection. F, Effect of knocking out DOX–relevant SLC transporters on DOX uptake into patient–derived cardiomyocytes [SLC28A3KO, SLC22A4KO, SLC22A3KO, and SLC22A17KO (n = 5–13)]. G, Effect of knocking out potential cardiac-specific SLC transporters on cell viability after DOX treatment [SLC28A3KO (n = 14), SLC22A4KO (n = 58), SLC22A3KO (n = 17), and SLC22A17KO, (n = 10), ISO (n = 128)]. n = full independent experimental replicates, Error bars, s.e.m, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 by unpaired two-tailed Student’s t-test (B, D, and F). For (C and G) log-logistic non-linear regression model was used to estimate the value of the four parameters, and t-statistic was used to test for significant difference in LD50 between different groups.
Implication of other SLC transporters in DIC regulation.
After we provided proof of principle for the importance of SLC transporters in DIC by showing that SLC28A3 expression and genomic variants affect the severity of DIC, we examined other potential SLC transporters that might affect DIC. To date, there are more than 450 identified SLC transporters, 12 of which have been shown to either transport DOX or a DOX metabolite and/or their genes harbor SNPs that are significantly associated with DOX clinical outcomes26. We examined which of these 12 SLC transporters are expressed in adult heart tissue, fetal heart tissue, and hiPSC–CMs. Only three additional SLC transporters met these criteria, SLC22A4, SLC22A3, SLC22A17 were selected for further investigation (Fig. 4e). For each of these transporters, we generated CRISPR/Cas9–mediated KO hiPSC (SLC22A4KO, SLC22A3KO, and SLC22A17KO) and differentiated them to cardiomyocytes (Supplementary Fig. 7). We examined the effect of altered expression of each candidate transporter on DOX intracellular uptake and cell viability after DOX treatment. Expectedly, SLC22A3, SLC22A4, SLC28A3, and SLC22A17 knockout resulted in a significant decrease in DOX uptake in isogenic hiPSC–CMs (Fig. 4f). DIC quantification revealed that SLC22A4KO cardiomyocytes (LD50 = 3.8 μM, P < 0.0001, n = 58), SLC28A3KO cardiomyocytes (LD50 = 3.3 μM, P < 0.0001, n = 17), and SLC22A17KO cardiomyocytes (LD50 = 3 μM, P = 0.005, n = 10) were less sensitive to DOX and more protected against DIC as compared to ISO (LD50 = 1.9 μM, P < 0.0001, n = 128). Whereas, knocking out SLC22A3 had a detectable yet not significant effect on DIC (LD50 = 2.1 μM, P = 0.5, n = 17) (Fig. 4g).
High–throughput drug screening reveals the SLC inhibitor, desipramine as a novel cardioprotective drug against DIC in hiPSC–CMs and murine.
Since SLC transporters affect DIC and represent well–founded druggable targets, we went on to examine whether SLC substrates can alter DOX uptake and subsequently regulate DIC in hiPSC–CMs. To test this, we first screened 17 drugs with well-established roles in affecting SLC transporter efficacy in relation to DOX uptake (Supplementary Table 12). In order eliminate potential toxicity from high doses of the SLC substrate drugs, LD50 was determined in our ISO cardiomyocytes for the 17 drugs to determine relevant maximum tolerable dose (Supplementary Fig. 8). This cherry–picked screening revealed that desipramine was the only substrate that altered DOX uptake at both 1 h and 3 h post DOX treatment. DOX uptake was significantly lower in cells co–treated with desipramine (3 μM) plus doxorubicin (3 μM) as compared to cells treated with doxorubicin (3 μM) only, after 1 h (P = 0.008) and 3 h (P = 0.04) posttreatment (Supplementary Figs. 9a–b). Since desipramine significantly decreased doxorubicin transport into cardiomyocytes, we next inspected whether lower doxorubicin intracellular concentration in desipramine pre–treated cells affect the magnitude of DIC.
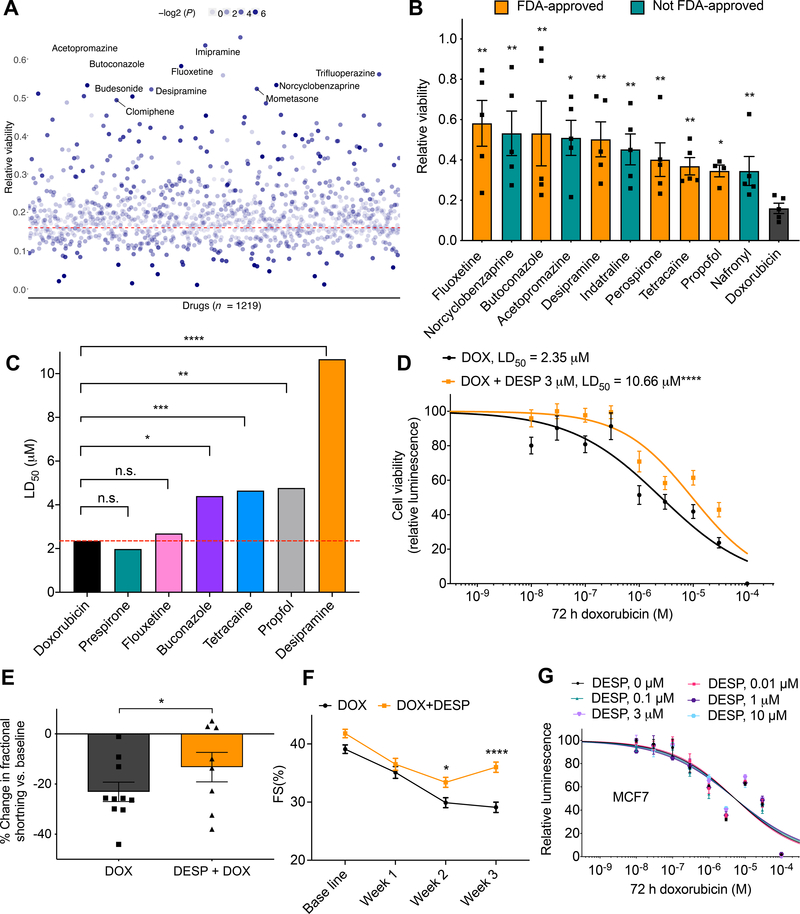
Next, we sought to expand our screening for novel cardioprotectants by testing the cardioprotective role of the Prestwick drug library that includes 1219 drugs. Cells were pre–treated with 3 μM of relevant drugs for 24 h, then co–treated with relevant drug (3 μM) and doxorubicin (10 μM) for 72 h after which DIC was quantified. For each plate, untreated and doxorubicin (10 μM) only treated cells were included to serve as negative and positive controls, respectively. Drug library screening reveled 34 cardioprotective molecules that attenuated cell death after DOX treatment (Fig. 5a). Drug repurposing is significantly faster and more cost–effective than de novo drug discovery approaches, and thus we focused on the top FDA–approved drugs identified from our screening. Interestingly, the top ten significant cardio–protectants included six FDA approved drugs; fluoxetine, butoconcazole, perospirone, tetracaine, propofol, and desipramine (Fig. 5b), of these drugs, desipramine again showed the most significant protection against DOX–induced cardiomyocyte death (P = 0.007, 2.5–fold) (Fig. 5a–b). To further validate these results, we then investigated the cardioprotective effect of these FDA–approved cardioprotective drugs against 10 log–doses of doxorubicin. This analysis showed that desipramine has the strongest cardioprotective effect when compared to cells treated only with DOX (LD50 = 10.66 μM, P < 0.0001), followed by propofol (LD50 = 4.77 μM, P = 0.003), then tetracaine (LD50 = 4.65 μM, P < 0.001), then butoconazole (LD50 = 4.4 μM, P = 0.01) (Fig. 5c–d, Supplementary Fig. 9c).
Figure 5. Multi-modality drug screening identifies desipramine as a novel effective cardioprotectant against DIC.
A, Prestwick drug library screening (n = 1219) in relation to DIC (n = 5). All drugs were used at 3 μM. Red dashed line represents cell viability 72 h post DOX (10 μM) treatment; the top ten significant cardioprotective drugs based on cell viability are labeled. B, Bar plot showing top ten significant cardioprotective (based on P value) compared to DOX alone (72 h, 10 μM) treated cells. Non–FDA–approved drugs are represented by teal bars. C, Further validation of top FDA-approved drugs (identified from the Prestwick library screening) against 10 log–doses of doxorubicin. LD50, median lethal dose. D, Effect of co-treatment of desipramine (3 μM) and doxorubicin (72 h) on hiPSC-CM viability [DOX (n = 42), DOX + DESP (n = 35)]. E, Percent change in ventricular fraction shortening (FS) normalized to baseline, after 3 weeks of doxorubicin treatment (3 mg/kg, ip, n = 10) compared co-treatment (n = 8) of desipramine (20 mg/kg/day, Alzet pump) and doxorubicin (3 mg/kg, ip) in mice. F, Ventricular fractional shortening at baseline, 1-, 2-, 3-weeks post treatment. G, Assessment of cell viability of MCF breast cancer cell line after 72 h of DOX and desipramine (DESP) cotreatment (n = 12–20). f = full independent experimental replicates, Error bars, s.e.m, *P < 0.05, **P ≤ 0.01, ***P < 0.001, ****P < 0.0001 by unpaired two-tailed Student’s t-test (A-C and E) and by ANOVA with post-hoc testing (F). For (D and G) log-logistic non-linear regression model was used to estimate the value of the four parameters, and t-statistic was used to test for significant difference in LD50 between different groups.
Based on these findings, we selected desipramine as our lead cardioprotective drug, and thus we then investigated whether or not desipramine could attenuate DIC in vivo. For that, we treated mice with doxorubicin (3 mg kg−1 intraperitoneal twice weekly for 3 weeks) plus desipramine (20 mg kg−1 day−1 infusion for 3 weeks), or water as a vehicle control. Doxorubicin treatment results in a steady decline in cardiac function, as assessed by fractional shortening. Critically, cardiac function was significantly less attenuated by doxorubicin at three weeks with desipramine (P < 0.05), compared with vehicle treatment (Fig. 5e–f, Supplementary Fig. 12b–c, and Supplementary Table 13). To rule out the possibility that desipramine could diminish DOX chemotherapeutic effect, we studied eight cancer cell lines, representing breast, liver, colorectal, prostate, uterus, cervix, and bone cancers and found that co–treatment with desipramine did not impede the anticancer efficacy of DOX in any of these lines (Fig. 5f, and Supplementary Fig. 10).
Discussion
The identification of reliable predictive genomic biomarkers for DIC and the discovery of efficient cardioprotectants are indispensable to enhance the clinical utility of doxorubicin in cancer treatment. The identification of gene variants predictive of altering DIC through GWAS has provided impetus for developing platforms to confirm these GWAS hits, moving them from ‘association’ to ‘confirmed mechanism’. In this work we demonstrated that the patient–specific hiPSC–CM model is ideal for studying the implication of transporter inhibitors and genetic variants in DIC. We show that patient–specific cardiomyocytes do indeed recapitulate the cardioprotective effect of the CGAS-identified SLC28A3 locus; confirm for the first time the role of SLC28A3 in DIC independent of patient–specific genetic background; and critically reveal that another SNP within this cardioprotective locus, rs11140490 has the highest likelihood to be causal. Furthermore, this platform allowed us to discover that the SLC competitive inhibitor, desipramine protects against DIC without hampering DOX chemotherapy efficacy.
The SLC28A3 genetic variant rs7853758 is the most robustly replicated AIC strongly associated cardioprotective loci. We show that rs7853758 is in perfect LD with 23 other SNPs of which 22 are non–coding variants, forming the cardioprotective HapSLC28A3. Pinpointing causal SNP within this locus is crucial for clinical translation because testing for the causal variant guarantees the detection of the best possible clinical correlation with AIC. Almost 93% of phenotype–associated genetic variants are non–coding27. Using the ENCODE project dataset, it has been shown that ~80% of GWAS–identified non–coding SNPs are not the causal SNP28. Using our recently developed cost–effective fine mapping pipeline29, we found that rs11140490, but not the CGAS–identified hit rs7853758, is the variant with the highest likelihood to be causal in DIC.
Editing only the rs11140490 cardioprotective genotype (CT) back to the reference genotype (TT) in patient–specific hiPSC–CMs renders these cardiomyocytes more susceptible to DIC confirming the causality of this novel variants. Interestingly, editing rs11140490 increase the sensitivity of hiPSC-CMs by ~28% which is modest and thus denotes that there might be more SNPs within the HapSLC28A3 that interact in an additive manner with rs11140490 to predispose to protection against DIC
Going forward, we propose that a simple clinical test to detect the presence of rs11140490 can be used to predict that a patient will be less likely to experience DIC and that, with future clinical trials, it may be possible for these patients to be treated with a longer duration (higher cumulative dose) of doxorubicin to enhance the efficacy of their chemotherapy. Similarly, the rs11140490 genetic testing could be employed as a part of a polygenic cardiotoxicity risk stratification score for doxorubicin-containing chemotherapy regimens such that the protective effect of rs11140490 could balance out for a risk factor that would have otherwise prevented the administration of a relevant doxorubicin-containing chemotherapy regimen to a particular patient.
SLC28A3 encodes a cardiac–specific uptake transporter that has no/marginal expression in several cancer cells and hence, represents a highly druggable target to screen for cardioprotective agents. A large number of drugs have been identified as being trafficked by SLC transporters which explains the substantial role of these transporters in both drug pharmacokinetics and pharmacodynamics and emphasizes the importance of this class of transporters in drug response disposition30. Our high–throughput drug screening discovered that treating patient–specific cardiomyocytes with the SLC inhibitor desipramine protects against DIC through decreasing the intracellular uptake of DOX into human heart. Our results suggest that a single dose of 3 μM desipramine 24 h before the administration of DOX in addition to another dose of 3 μM desipramine co–administered with DOX is sufficient for protecting against DIC.
Desipramine is a tricyclic antidepressant sold under that brand name Norpramin, that was first patented in 196231. The typical adult dose of desipramine 100 mg to 200 mg/day. In more severely ill patients, dosage may be further increased gradually to 300 mg/day if necessary. The typical therapeutic concentration is 100–300 ng/ml with daily dosing. Our 3 μM in vitro dose would be 798 ng/ml, but importantly, our preliminary data suggest that just two doses or potentially one dose per cycle of doxorubicin would be required to attenuate DIC. We would therefore suggest that these desipramine doses are potentially within the acceptable clinical range, although further animal model work to confirming dosing strategy is required prior to clinical trial.
Desipramine is far from an ideal drug, being the most potent sodium channel blocker among its group and causing cardiotoxicity when used chronically. Desipramine-treated patients have been shown to have significantly lower rates of sinus pauses and junctional rhythm, but significantly higher rates of single or paired premature atrial contractions and runs of supraventricular tachycardia as well as lengthening of the QT interval. The solution to this is to develop a desipramine derivative without the sodium channel blockade effect, although approach eliminates the primary advantage of repurposing a drug like desipramine to attenuate DIC. Still, the advantage of using our hiPSC-CM platform for subsequent testing will be a major advantage in such an effort
The majority (56%) of candidate drugs have failed in clinical trial due to the lack of efficacy, most likely because the pre-clinical models used to test the drug does not recapitulate what happens in humans32 or in the specific target cell type. Additionally, studies staring with genetic correlations in genes encoding targets increases the success rate in clinical development by 2–fold33. Patient-derived hiPSC–CMs provides a unique platform that firstly permits a thorough validation of GWAS–identified AIC–associated loci, and secondly recapitulate alteration in DIC phenotype in a human–relevant manner. Hence the utilization of hiPSC–CMs in the development of cardioprotectants substantially improves the potential of developing novel derivatives of desipramine that have the same SLC28A3 competitive inhibition effect as desipramine without the well–known side effects common to the tricyclic antidepressant drug family.
Clearly hiPSC–CMs do not fully mimic the human whole–body model and do not recapitulate all the steps of drug pharmaco–kinetics and –dynamics. However, current hiPSC–CM generation methodologies have enhanced the robustness, purity, maturation, and scalability to a point where these cells are suitable for a wide-range of disease modelling and drug response assays34–42. Here we demonstrated that patient–specific cardiomyocytes recapitulate intra–individual variability in genomic–dependent DIC susceptibility. We show that hiPSC–CMs are appropriate to study drug response–associated loci especially for genes with a known mechanism of action such as transporter–encoding genes. The integration of CRISPR/Cas9–based genetic editing to our patient–specific hiPSC model proves to be a powerful tool in identify causal genetic variations in relation to a specific drug–response. The expansion of the utility of patient–specific hiPSC–CMs to study additional anthracycline–relevant loci as well as those of other anti-cancer agents will help to identify patients/population–specific chemotherapeutic–induced cardiotoxicity genetic biomarkers and new cardioprotective agents. Ultimately, the information derived from this platform may allow physicians to tailor chemotherapeutics doses based on patient genotype, bringing the promise of personalized medicine to the field of cardio–oncology.
Supplementary Material
Clinical Perspective.
What Is New?
This study shows that patient–specific cardiomyocytes recapitulate the cardioprotective effect of the CGAS-identified SLC28A3 locus and we functionally confirm for the first time the role of SLC28A3 in doxorubicin–induced cardiotoxicity (DIC).
A novel genetic variant, rs11140490 is the potential causal variant in the SLC28A3 cardioprotective locus.
The solute carrier transporter inhibitor desipramine protects against DIC through decreasing the intracellular uptake of doxorubicin into the heart.
What Are the Clinical Implications?
We provide two potential therapeutic options to attenuate DIC, either repurposing FDA-approved desipramine or therapy with long non-coding RNA SLC28A3-AS1.
We propose that a simple clinical test to detect the presence of rs11140490 can be used to predict that a patient will be less likely to experience DIC and that, with future clinical trials, it may be possible for these patients to be treated with a longer duration (higher cumulative dose) of doxorubicin to enhance the efficacy of their chemotherapy.
Acknowledgments
P.W.B. and T.M. designed the research. T.M., M.J., H.–H.K., C.W., D.L–L., H.F., M.R.–T., M.G., H.J., and P.W.B. performed the experiments. G.J. and D.B. designed and the performed mouse experiments. C.J.D.R. and B.C.C determined the AIC phenotype to be used patient recruitment and provided patient samples. P.W.B. supervised the project. T.M. and P.W.B. wrote the manuscript with input from all other authors.
Funding Sources
This work was supported by NIH grants K99/R00 HL121177, R01 CA220002, R01 CA261898, American Heart Association Transformational Project Award 18TPA34230105, and the Fondation Leducq (P.W.B.), Canadian Cancer Society (C.J.R., B.C.C., P.W.B.), Michael Smith Foundation for Health Research Scholar Award (C.J.R.), and NIH R33 HL123655 (D.B., B.C.C.). Patients from which hiPSC were derived were recruited by the Canadian Pharmacogenomics Network for Drug Safety (B.C.C. and C.J.R.) which has received grant funding from the Canadian Institutes for Health Research (CIHR), the CIHR Drug Safety and Effectiveness Network, Genome Canada, Genome British Columbia, and the British Columbia Provincial Health Services Authority.
Non–Standard Abbreviations and Acronyms
- AIC
Anthracycline–induced cardiotoxicity
- CGAS
Candidate gene association study
- DES
Desipramine
- DIC
Doxorubicin–induced cardiotoxicity
- DOX
Doxorubicin
- eQTL
Expression quantitative trait loci
- GWAS
Genome–wide association study
- ISO
Isotype
- SLC
Solute carrier
Footnotes
Conflict of Interest Disclosures. None declared
Supplemental Materials
References
- 1.Cardinale D, Colombo A, Bacchiani G, Tedeschi I, Meroni CA, Veglia F, Civelli M, Lamantia G, Colombo N, Curigliano G, et al. Early detection of anthracycline cardiotoxicity and improvement with heart failure therapy. Circulation. 2015;131:1981–8. [DOI] [PubMed] [Google Scholar]
- 2.Swain SM, Whaley FS and Ewer MS. Congestive heart failure in patients treated with doxorubicin: A retrospective analysis of three trials. Cancer. 2003;97:2869–2879. [DOI] [PubMed] [Google Scholar]
- 3.Avila MS, Ayub-Ferreira SM, de Barros Wanderley MR Jr., das Dores Cruz F, Goncalves Brandao SM, Rigaud VOC, Higuchi-Dos-Santos MH, Hajjar LA, Kalil Filho R, Hoff PM, et al. Carvedilol for Prevention of Chemotherapy-Related Cardiotoxicity: The CECCY Trial. J Am Coll Cardiol. 2018;71:2281–2290. [DOI] [PubMed] [Google Scholar]
- 4.Magdy T, Burmeister BT and Burridge PW. Validating the pharmacogenomics of chemotherapy-induced cardiotoxicity: What is missing? Pharmacol Ther. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aminkeng F, Ross CJD, Rassekh SR, Rieder MJ, Bhavsar AP, Sanatani S, Bernstein D, Hayden MR, Amstutz U and Carleton BC. Pharmacogenomic screening for anthracycline-induced cardiotoxicity in childhood cancer. Br J Clin Pharmacol. 2017;83:1143–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan LL and Lyon AR. Role of Biomarkers in Prediction of Cardiotoxicity During Cancer Treatment. Curr Treat Options Cardiovasc Med. 2018;20:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visscher H, Ross CJD, Rassekh SR, Barhdadi A, Dubé M-P, Al-Saloos H, Sandor GS, Caron HN, van Dalen EC, Kremer LC, et al. Pharmacogenomic prediction of anthracycline-induced cardiotoxicity in children. J Clin Oncol. 2012;30:1422–1428. [DOI] [PubMed] [Google Scholar]
- 8.Visscher H, Ross CJD, Rassekh SR, Sandor GSS, Caron HN, van Dalen EC, Kremer LC, van der Pal HJ, Rogers PC, Rieder MJ, et al. Validation of variants in SLC28A3 and UGT1A6 as genetic markers predictive of anthracycline-induced cardiotoxicity in children. Pediatr Blood Cancer. 2013;60:1375–1381. [DOI] [PubMed] [Google Scholar]
- 9.Aminkeng F, Ross CJD, Rassekh SR, Hwang S, Rieder MJ, Bhavsar AP, Smith A, Sanatani S, Gelmon KA, Bernstein D, et al. Recommendations for genetic testing to reduce the incidence of anthracycline-induced cardiotoxicity. Br J Clin Pharmacol. 2016;82:683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fusaki N, Ban H, Nishiyama A, Saeki K and Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, et al. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11:855–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burridge PW, Holmstrom A and Wu JC. Chemically Defined Culture and Cardiomyocyte Differentiation of Human Pluripotent Stem Cells. Curr Protoc Hum Genet. 2015;87:21 3 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA and Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oceguera-Yanez F, Kim SI, Matsumoto T, Tan GW, Xiang L, Hatani T, Kondo T, Ikeya M, Yoshida Y, Inoue H, et al. Engineering the AAVS1 locus for consistent and scalable transgene expression in human iPSCs and their differentiated derivatives. Methods. 2016;101:43–55. [DOI] [PubMed] [Google Scholar]
- 15.Dandage R, Despres PC, Yachie N and Landry CR. beditor: A Computational Workflow for Designing Libraries of Guide RNAs for CRISPR-Mediated Base Editing. Genetics. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritz C, Baty F, Streibig JC and Gerhard D. Dose-Response Analysis Using R. PLoS ONE. 2015;10:e0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aminkeng F, Bhavsar AP, Visscher H, Rassekh SR, Li Y, Lee JW, Brunham LR, Caron HN, van Dalen EC, Kremer LC, et al. A coding variant in RARG confers susceptibility to anthracycline-induced cardiotoxicity in childhood cancer. Nat Genet. 2015;47:1079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Churko JM, Burridge PW and Wu JC. Generation of human iPSCs from human peripheral blood mononuclear cells using non-integrative Sendai virus in chemically defined conditions. Methods Mol Biol. 2013;1036:81–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diecke S, Lu J, Lee J, Termglinchan V, Kooreman NG, Burridge PW, Ebert AD, Churko JM, Sharma A, Kay MA, et al. Novel codon-optimized mini-intronic plasmid for efficient, inexpensive, and xeno-free induction of pluripotency. Sci Rep. 2015;5:8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burridge PW, Holmstrom A and Wu JC. Chemically Defined Culture and Cardiomyocyte Differentiation of Human Pluripotent Stem Cells. Curr Protoc Hum Genet. 2015;87:21 3 1–21 3 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magdy T and Burridge PW. The future role of pharmacogenomics in anticancer agent-induced cardiovascular toxicity. Pharmacogenomics. 2018;19:79–82. [DOI] [PubMed] [Google Scholar]
- 22.Loman NJ, Quick J and Simpson JT. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods. 2015;12:733–5. [DOI] [PubMed] [Google Scholar]
- 23.Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou J and Troyanskaya OG. Predicting effects of noncoding variants with deep learning-based sequence model. Nat Methods. 2015;12:931–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weisheit I, Kroeger JA, Malik R, Klimmt J, Crusius D, Dannert A, Dichgans M and Paquet D. Detection of Deleterious On-Target Effects after HDR-Mediated CRISPR Editing. Cell Rep. 2020;31:107689. [DOI] [PubMed] [Google Scholar]
- 26.Magdy T, Burmeister BT and Burridge PW. Validating the pharmacogenomics of chemotherapy-induced cardiotoxicity: What is missing? Pharmacol Ther. 2016;168:113–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaub MA, Boyle AP, Kundaje A, Batzoglou S and Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magdy T, Kuo HH and Burridge PW. Precise and Cost-Effective Nanopore Sequencing for Post-GWAS Fine-Mapping and Causal Variant Identification. iScience. 2020;23:100971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giacomini KM, Huang SM, Tweedie DJ, Benet LZ, Brouwer KL, Chu X, Dahlin A, Evers R, Fischer V, Hillgren KM, et al. Membrane transporters in drug development. Nat Rev Drug Discov. 2010;9:215–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersen J, Kristensen AS, Bang-Andersen B and Stromgaard K. Recent advances in the understanding of the interaction of antidepressant drugs with serotonin and norepinephrine transporters. Chem Commun (Camb). 2009:3677–92. [DOI] [PubMed] [Google Scholar]
- 32.Arrowsmith J and Miller P. Trial watch: phase II and phase III attrition rates 2011–2012. Nat Rev Drug Discov. 2013;12:569. [DOI] [PubMed] [Google Scholar]
- 33.Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, Floratos A, Sham PC, Li MJ, Wang J, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47:856–60. [DOI] [PubMed] [Google Scholar]
- 34.Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, Feldman O, Gepstein A, Arbel G, Hammerman H, et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. [DOI] [PubMed] [Google Scholar]
- 35.Malan D, Zhang M, Stallmeyer B, Müller J, Fleischmann BK, Schulze-Bahr E, Sasse P and Greber B. Human iPS cell model of type 3 long QT syndrome recapitulates drug-based phenotype correction. Basic Res Cardiol. 2016;111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carvajal-Vergara X, Sevilla A, D’Souza SL, Ang Y-S, Schaniel C, Lee D-F, Yang L, Kaplan AD, Adler ED, Rozov R, et al. Patient-specific induced pluripotent stem cell derived models of LEOPARD syndrome. Nature. 2010;465:808–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J and Dolmetsch RE. Using iPS cells to investigate cardiac phenotypes in patients with Timothy Syndrome. Nature. 2011;471:230–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim C, Wong J, Wen J, Wang S, Wang C, Spiering S, Kan NG, Forcales S, Puri PL, Leone TC, et al. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature. 2013;494:105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, Navarrete EG, Hu S, Wang L, Lee A, et al. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med. 2012;4:130ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, Yuan H, Jiang D, Zhang D, Zangi L, et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med. 2014;20:616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Magdy T and Burridge PW. Unraveling Difficult Answers: From Genotype to Phenotype in Coronary Artery Disease. Cell Stem Cell. 2019;24:203–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drawnel FM, Boccardo S, Prummer M, Delobel F, Graff A, Weber M, Gérard R, Badi L, Kam-Thong T, Bu L, et al. Disease modeling and phenotypic drug screening for diabetic cardiomyopathy using human induced pluripotent stem cells. Cell Rep. 2014;9:810–821. [DOI] [PubMed] [Google Scholar]
- 43.Chou BK, Gu H, Gao Y, Dowey SN, Wang Y, Shi J, Li Y, Ye Z, Cheng T and Cheng L. A facile method to establish human induced pluripotent stem cells from adult blood cells under feeder-free and xeno-free culture conditions: a clinically compliant approach. Stem Cells Transl Med. 2015;4:320–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuo HH, Gao X, DeKeyser JM, Fetterman KA, Pinheiro EA, Weddle CJ, Fonoudi H, Orman MV, Romero-Tejeda M, Jouni M, et al. Negligible-Cost and Weekend-Free Chemically Defined Human iPSC Culture. Stem Cell Reports. 2020;14:256–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim D, Paggi JM, Park C, Bennett C and Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao Y, Smyth GK and Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Love MI, Huber W and Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmittgen TD and Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. [DOI] [PubMed] [Google Scholar]
- 49.Wick RR, Judd LM, Gorrie CL and Holt KE. Completing bacterial genome assemblies with multiplex MinION sequencing. Microbial genomics. 2017;3:e000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Coster W, D'Hert S, Schultz DT, Cruts M and Van Broeckhoven C. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics. 2018;34:2666–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G and Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F and Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cingolani P, Patel VM, Coon M, Nguyen T, Land SJ, Ruden DM and Lu X. Using Drosophila melanogaster as a Model for Genotoxic Chemical Mutational Studies with a New Program, SnpSift. Front Genet. 2012;3:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Narasimhan V, Danecek P, Scally A, Xue Y, Tyler-Smith C and Durbin R. BCFtools/RoH: a hidden Markov model approach for detecting autozygosity from next-generation sequencing data. Bioinformatics. 2016;32:1749–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Durinck S, Spellman PT, Birney E and Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Braun D, Kim TD, le Coutre P, Köhrle J, Hershman JM and Schweizer U. Tyrosine kinase inhibitors noncompetitively inhibit MCT8-mediated iodothyronine transport. The Journal of clinical endocrinology and metabolism. 2012;97:E100–5. [DOI] [PubMed] [Google Scholar]
- 59.Damaraju VL, Weber D, Kuzma M, Cass CE and Sawyer MB. Selective Inhibition of Human Equilibrative and Concentrative Nucleoside Transporters by BCR-ABL Kinase Inhibitors: IDENTIFICATION OF KEY hENT1 AMINO ACID RESIDUES FOR INTERACTION WITH BCR-ABL KINASE INHIBITORS. J Biol Chem. 2016;291:18809–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Braun D and Schweizer U. Authentic bosutinib inhibits triiodothyronine transport by monocarboxylate transporter 8. Thyroid. 2014;24:926–7. [DOI] [PubMed] [Google Scholar]
- 61.Yin J, Duan H and Wang J. Impact of Substrate-Dependent Inhibition on Renal Organic Cation Transporters hOCT2 and hMATE1/2-K-Mediated Drug Transport and Intracellular Accumulation. J Pharmacol Exp Ther. 2016;359:401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsuda M, Terada T, Ueba M, Sato T, Masuda S, Katsura T and Inui K. Involvement of human multidrug and toxin extrusion 1 in the drug interaction between cimetidine and metformin in renal epithelial cells. J Pharmacol Exp Ther. 2009;329:185–91. [DOI] [PubMed] [Google Scholar]
- 63.Shitara Y, Itoh T, Sato H, Li AP and Sugiyama Y. Inhibition of transporter-mediated hepatic uptake as a mechanism for drug-drug interaction between cerivastatin and cyclosporin A. J Pharmacol Exp Ther. 2003;304:610–6. [DOI] [PubMed] [Google Scholar]
- 64.Shitara Y, Takeuchi K, Nagamatsu Y, Wada S, Sugiyama Y and Horie T. Long-lasting inhibitory effects of cyclosporin A, but not tacrolimus, on OATP1B1- and OATP1B3-mediated uptake. Drug Metab Pharmacokinet. 2012;27:368–78. [DOI] [PubMed] [Google Scholar]
- 65.Craddock AL, Love MW, Daniel RW, Kirby LC, Walters HC, Wong MH and Dawson PA. Expression and transport properties of the human ileal and renal sodium-dependent bile acid transporter. The American journal of physiology. 1998;274:G157–69. [DOI] [PubMed] [Google Scholar]
- 66.Schroeder A, Eckhardt U, Stieger B, Tynes R, Schteingart CD, Hofmann AF, Meier PJ and Hagenbuch B. Substrate specificity of the rat liver Na(+)-bile salt cotransporter in Xenopus laevis oocytes and in CHO cells. The American journal of physiology. 1998;274:G370–5. [DOI] [PubMed] [Google Scholar]
- 67.Taguchi T, Masuo Y, Kogi T, Nakamichi N and Kato Y. Characterization of Long-Lasting Oatp Inhibition by Typical Inhibitor Cyclosporine A and In Vitro-In Vivo Discrepancy in Its Drug Interaction Potential in Rats. J Pharm Sci. 2016;105:2231–9. [DOI] [PubMed] [Google Scholar]
- 68.Pahwa S, Alam K, Crowe A, Farasyn T, Neuhoff S, Hatley O, Ding K and Yue W. Pretreatment With Rifampicin and Tyrosine Kinase Inhibitor Dasatinib Potentiates the Inhibitory Effects Toward OATP1B1- and OATP1B3-Mediated Transport. J Pharm Sci. 2017;106:2123–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu Q, Wang C, Meng Q, Liu Q, Sun H, Peng J, Ma X, Kaku T and Liu K. OAT1 and OAT3: targets of drug-drug interaction between entecavir and JBP485. European journal of pharmaceutical sciences : official journal of the European Federation for Pharmaceutical Sciences. 2013;48:650–7. [DOI] [PubMed] [Google Scholar]
- 70.Kouzuki H, Suzuki H and Sugiyama Y. Pharmacokinetic study of the hepatobiliary transport of indomethacin. Pharmaceutical research. 2000;17:432–8. [DOI] [PubMed] [Google Scholar]
- 71.Takeda M, Khamdang S, Narikawa S, Kimura H, Hosoyamada M, Cha SH, Sekine T and Endou H. Characterization of methotrexate transport and its drug interactions with human organic anion transporters. J Pharmacol Exp Ther. 2002;302:666–71. [DOI] [PubMed] [Google Scholar]
- 72.Hu S, Mathijssen RH, de Bruijn P, Baker SD and Sparreboom A. Inhibition of OATP1B1 by tyrosine kinase inhibitors: in vitro-in vivo correlations. Br J Cancer. 2014;110:894–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taguchi T, Masuo Y, Sakai Y and Kato Y. Short-lasting inhibition of hepatic uptake transporter OATP1B1 by tyrosine kinase inhibitor pazopanib. Drug Metab Pharmacokinet. 2019;34:372–379. [DOI] [PubMed] [Google Scholar]
- 74.Oulianova N, Falk S and Berteloot A. Two-step mechanism of phlorizin binding to the SGLT1 protein in the kidney. The Journal of membrane biology. 2001;179:223–42. [DOI] [PubMed] [Google Scholar]
- 75.David-Silva A, Esteves JV, Morais M, Freitas HS, Zorn TM, Correa-Giannella ML and Machado UF. Dual SGLT1/SGLT2 Inhibitor Phlorizin Ameliorates Non-Alcoholic Fatty Liver Disease and Hepatic Glucose Production in Type 2 Diabetic Mice. Diabetes, metabolic syndrome and obesity : targets and therapy. 2020;13:739–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Urakami Y, Akazawa M, Saito H, Okuda M and Inui K. cDNA cloning, functional characterization, and tissue distribution of an alternatively spliced variant of organic cation transporter hOCT2 predominantly expressed in the human kidney. Journal of the American Society of Nephrology : JASN. 2002;13:1703–10. [DOI] [PubMed] [Google Scholar]
- 77.Bednarczyk D, Ekins S, Wikel JH and Wright SH. Influence of molecular structure on substrate binding to the human organic cation transporter, hOCT1. Mol Pharmacol. 2003;63:489–98. [DOI] [PubMed] [Google Scholar]
- 78.Ohashi R, Tamai I, Yabuuchi H, Nezu JI, Oku A, Sai Y, Shimane M and Tsuji A. Na(+)-dependent carnitine transport by organic cation transporter (OCTN2): its pharmacological and toxicological relevance. J Pharmacol Exp Ther. 1999;291:778–84. [PubMed] [Google Scholar]
- 79.van Montfoort JE, Müller M, Groothuis GM, Meijer DK, Koepsell H and Meier PJ. Comparison of "type I" and "type II" organic cation transport by organic cation transporters and organic anion-transporting polypeptides. J Pharmacol Exp Ther. 2001;298:110–5. [PubMed] [Google Scholar]
- 80.Cha SH, Sekine T, Fukushima JI, Kanai Y, Kobayashi Y, Goya T and Endou H. Identification and characterization of human organic anion transporter 3 expressing predominantly in the kidney. Mol Pharmacol. 2001;59:1277–86. [DOI] [PubMed] [Google Scholar]
- 81.Yabuuchi H, Tamai I, Nezu J, Sakamoto K, Oku A, Shimane M, Sai Y and Tsuji A. Novel membrane transporter OCTN1 mediates multispecific, bidirectional, and pH-dependent transport of organic cations. J Pharmacol Exp Ther. 1999;289:768–73. [PubMed] [Google Scholar]
- 82.Nozawa T, Tamai I, Sai Y, Nezu J and Tsuji A. Contribution of organic anion transporting polypeptide OATP-C to hepatic elimination of the opioid pentapeptide analogue [D-Ala2, D-Leu5]-enkephalin. J Pharm Pharmacol. 2003;55:1013–20. [DOI] [PubMed] [Google Scholar]
- 83.Vavricka SR, Van Montfoort J, Ha HR, Meier PJ and Fattinger K. Interactions of rifamycin SV and rifampicin with organic anion uptake systems of human liver. Hepatology (Baltimore, Md). 2002;36:164–72. [DOI] [PubMed] [Google Scholar]
- 84.Fattinger K, Cattori V, Hagenbuch B, Meier PJ and Stieger B. Rifamycin SV and rifampicin exhibit differential inhibition of the hepatic rat organic anion transporting polypeptides, Oatp1 and Oatp2. Hepatology (Baltimore, Md). 2000;32:82–6. [DOI] [PubMed] [Google Scholar]
- 85.Cui Y, König J, Leier I, Buchholz U and Keppler D. Hepatic uptake of bilirubin and its conjugates by the human organic anion transporter SLC21A6. J Biol Chem. 2001;276:9626–30. [DOI] [PubMed] [Google Scholar]
- 86.Sekine T, Cha SH, Tsuda M, Apiwattanakul N, Nakajima N, Kanai Y and Endou H. Identification of multispecific organic anion transporter 2 expressed predominantly in the liver. FEBS letters. 1998;429:179–82. [DOI] [PubMed] [Google Scholar]
- 87.Hirano M, Maeda K, Shitara Y and Sugiyama Y. Drug-drug interaction between pitavastatin and various drugs via OATP1B1. Drug Metab Dispos. 2006;34:1229–36. [DOI] [PubMed] [Google Scholar]
- 88.Minematsu T and Giacomini KM. Interactions of tyrosine kinase inhibitors with organic cation transporters and multidrug and toxic compound extrusion proteins. Mol Cancer Ther. 2011;10:531–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shen H, Yang Z, Zhao W, Zhang Y and Rodrigues AD. Assessment of vandetanib as an inhibitor of various human renal transporters: inhibition of multidrug and toxin extrusion as a possible mechanism leading to decreased cisplatin and creatinine clearance. Drug Metab Dispos. 2013;41:2095–103. [DOI] [PubMed] [Google Scholar]
- 90.Cho SK, Kim CO, Park ES and Chung JY. Verapamil decreases the glucose-lowering effect of metformin in healthy volunteers. Br J Clin Pharmacol. 2014;78:1426–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oostendorp RL, van de Steeg E, van der Kruijssen CM, Beijnen JH, Kenworthy KE, Schinkel AH and Schellens JH. Organic anion-transporting polypeptide 1B1 mediates transport of Gimatecan and BNP1350 and can be inhibited by several classic ATP-binding cassette (ABC) B1 and/or ABCG2 inhibitors. Drug Metab Dispos. 2009;37:917–23. [DOI] [PubMed] [Google Scholar]
- 92.Cvetkovic M, Leake B, Fromm MF, Wilkinson GR and Kim RB. OATP and P-glycoprotein transporters mediate the cellular uptake and excretion of fexofenadine. Drug Metab Dispos. 1999;27:866–71. [PubMed] [Google Scholar]
- 93.Radchenko M, Symersky J, Nie R and Lu M. Structural basis for the blockade of MATE multidrug efflux pumps. Nat Commun. 2015;6:7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang L, Schaner ME and Giacomini KM. Functional characterization of an organic cation transporter (hOCT1) in a transiently transfected human cell line (HeLa). J Pharmacol Exp Ther. 1998;286:354–61. [PubMed] [Google Scholar]
- 95.Gorboulev V, Ulzheimer JC, Akhoundova A, Ulzheimer-Teuber I, Karbach U, Quester S, Baumann C, Lang F, Busch AE and Koepsell H. Cloning and characterization of two human polyspecific organic cation transporters. DNA and cell biology. 1997;16:871–81. [DOI] [PubMed] [Google Scholar]
- 96.Wu X, Huang W, Ganapathy ME, Wang H, Kekuda R, Conway SJ, Leibach FH and Ganapathy V. Structure, function, and regional distribution of the organic cation transporter OCT3 in the kidney. American journal of physiology Renal physiology. 2000;279:F449–58. [DOI] [PubMed] [Google Scholar]
- 97.Wu X, George RL, Huang W, Wang H, Conway SJ, Leibach FH and Ganapathy V. Structural and functional characteristics and tissue distribution pattern of rat OCTN1, an organic cation transporter, cloned from placenta. Biochim Biophys Acta. 2000;1466:315–27. [DOI] [PubMed] [Google Scholar]
- 98.Wu X, Huang W, Prasad PD, Seth P, Rajan DP, Leibach FH, Chen J, Conway SJ and Ganapathy V. Functional characteristics and tissue distribution pattern of organic cation transporter 2 (OCTN2), an organic cation/carnitine transporter. J Pharmacol Exp Ther. 1999;290:1482–92. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.