Abstract
Nanoparticles (NPs) that can activate macrophages infected with the tuberculosis causative pathogen Mycobacterium tuberculosis, could be an effective host directed therapy for the disease. In this study, curdlan was conjugated to poly(lactic-co-glycolic acid) (PLGA) to produce immunotherapeutic NPs. Various physicochemical characterizations were used to evaluate the curdlan-PLGA copolymer and the NPs. Molecular dynamics and simulation studies were used to characterize the interaction between curdlan, on the polymer and on NPs, with the Dectin-1 macrophage receptor. NPs with varying curdlan densities were evaluated for their effects on the production of pro- and anti-inflammatory cytokines in M. tuberculosis infected RAW264.7 macrophages. The killing efficacy of the NPs against intracellular M. tuberculosis was assessed. Physicochemical characterization of the curdlan-PLGA copolymer and NPs indicated successful formation of curdlan-PLGA copolymer and NPs of varying curdlan density (0–8% w/w) had sizes between 330 and 453 nm. Modelling studies showed curdlan to have a strong affinity for Dectin-1. Cytotoxicity assays showed the NPs to be non-toxic over 72 h. The proinflammatory cytokine TNF-α was found to be significantly upregulated by the NPs. The NPs reduced intracellular M. tuberculosis burden over 72 h. These NPs are a promising host directed therapy for intracellular eradication of M. tuberculosis.
Keywords: Immunotherapy and tuberculosis, PLGA immunotherapeutic nanoparticles, PLGA nanoparticles and tuberculosis, Host-directed therapy and nanoparticles
Introduction
Tuberculosis (TB) is one of the leading infectious disease killer globally. Although the incidence has been falling worldwide, nearly 10 million people fall ill each year, and an estimated 1.4 million deaths occurred in 2019 due to TB.1 This scenario combined with the fact that microbial drug resistance is on the rise and has also occurred soon after new antibiotic drugs have been introduced for clinical use,2,3 makes the need for new therapeutics, with novel mechanisms of action for the treatment of TB, critical.
The TB causative organism, Mycobacterium tuberculosis, spreads via inhalation and once in the lungs, is resident within macrophages. M. tuberculosis is able to dampen the anti-bacterial response of macrophages, which enables residence and multiplication4 within these cells. Therefore, there is an opportunity for the development of host-directed therapies for the treatment of TB.5–7
Nanoparticles (NPs) can be used to stimulate macrophages into a bactericidal state to eliminate intracellular M. tuberculosis.4 The surface of the NPs can be functionalized with ligands to interact with specific macrophage surface receptors involved in macrophage activation. In previous work, we have shown that 1,3-β-glucan adsorbed or conjugated to polymeric NPs could stimulate macrophages.8,9 The polysaccharide 1,3-β-glucan is well known to stimulate macrophages to produce pro-inflammatory signals (reactive oxygen and nitrogen species (ROS/RNS)). β-glucans bind to the Dectin-1 present on macrophage surfaces, subsequently activating various downstream signal transduction pathways which promote pro-inflammatory gene expression as well as intracellular ROS/RNS production. Pro-inflammatory cytokines produced through Dectin-1 activation include IL-12 and TNF-α,10,11 which are known to be crucial in the control of M. tuberculosis.12,13
The goal of this study is to determine the efficacy of a curdlan (1,3-β-glucan) functionalized poly(lactic-co-glycolic acid) PLGA NP against M. tuberculosis infected macrophages. These NPs would be expected to activate macrophages leading to death of intracellular M. tuberculosis in the absence of antibiotic drugs. Molecular dynamics and simulation studies have been used to predict the mechanics involved in the interaction between curdlan and the Dectin-1 receptor allowing a better understanding of how the receptor is triggered to produce the immunomodulatory response. The efficacy of the NP system has been investigated in M. tuberculosis infected macrophages.
Experimental
Materials
1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC.HCl), N-hydroxysuccinimide (NHS), N,N-diisopropylethylamine (DIEA), curdlan from Alcaligenes faecalis, poly vinyl alcohol (PVA: MW, 13–23 kDa, partially hydrolysed (98%)), Resomer RG503H poly (D,L-lactide-co-glycolide) (50/50) with the terminal carboxylic acid (24–38 kDa) and non-acid terminated PLGA (MW, 30–60 kDa) were purchased from Sigma-Aldrich (St Louis, MO). All reagents were of analytical grade or above and were used as purchased unless otherwise stated. Deionised water was obtained from a Barnstead Easy-Pure (II) UV-ultrapure water system (Thermo Fisher Scientific, USA) and was used throughout the study.
RAW264.7 murine macrophages (ATCC TIB-71) were cultured in D10 (Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated Fetal Bovine Serum (FBS) both of which were obtained from Thermo Fisher), with passage number maintained below 30. Liquid mycobacterial cultures, M. tuberculosis ΔleuDΔpanD, M. tuberculosis ΔleuDΔpanD::pMV306hsp+lux, Table 1, were grown in 7H9-OGT –7H9 (Becton Dickinson), supplemented with 10% oleic acid–albumin–dextrose– catalase (OADC; Becton Dickinson, NJ, United States), 0.2% (v/v) glycerol (Sigma-Aldrich), 0.05% (v/v), Tween-80 (Sigma-Aldrich). Solid cultures were grown on 7H10 agar (Becton Dickinson, NJ). Additional supplements included pantothenate (24 μg/mL), leucine (50 μg/mL), hygromycin (50 μg/mL) (Invitrogen) and kanamycin (25 μg/mL) (Sigma-Aldrich, MO), the latter for maintenance of the pMV306hsp+lux plasmid. Phosphate-Buffered Saline (PBS) was obtained from Thermo Fisher and the MTT (3- (4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) together with the DMSO solution was purchased from Sigma-Aldrich. Secreted cytokines were analysed using a customized mouse Multiplex R&D luminex kit (Thermo Fisher).
Table 1.
Strains and plasmids used in this study.
| Strain/plasmid | Description | Source |
|---|---|---|
| M. tuberculosis ΔleuDΔpanD | Double leucine and pantothenate auxotroph of M. tuberculosis HygR | 14 |
| M. tuberculosis ΔleuDΔpanD:: pMV306hsp+lux | M. tuberculosis ΔleuDΔpanD transformed with episomal, KanR plasmid pMV306hsp + LuxCDABE | pMV306hsp + LuxCDABE15 Addgene plasmid number 26155 |
HygR, hygromycin resistant; KanR, kanamycin resistant
Methods
C-PLGA Copolymer
C-PLGA copolymer synthesis.
To synthesize NPs of C-PLGA NPs, a copolymer of curdlan-PLGA was first synthesized, following the procedure by Tukulula et al. with some modifications.8 Briefly, 50 mg of curdlan was dissolved in 5 mL of DMSO under N2 atmosphere at 60° C. NHS (10.6 mg) was added to a stirring solution of PLGA-COOH (954.8 mg) in 5 mL anhydrous DMF until dissolution at 0°C. EDC (19.1 mg) and DIEA (15.9 mg) were subsequently added to the solution and left to react at room temperature for 2–3 h. The activated PLGA–COOH solution was added dropwise to the dissolved curdlan and left to react for 48 h under inert gas. The product was precipitated with cold deionised water and washed several times. The conjugate was then lyophilised over three days.
Polymer Characterization.
Polymers (curdlan, PLGA and C-PLGA) were characterized using 1H NMR on a Bruker Avance spectrometer at 400 MHz in deuterated dimethyl sulfoxide (DMSO-d6). Chemical shifts are referenced to undeuterated solvent signals at δH 2.5 and reported in ppm.
Thermal analysis was performed using a TGA 4000 (Pyris 6 TGA) in ramp mode from 30°C to 600°C at a rate of 10°C/min under nitrogen. Hot stage microscopy was performed on an Olympus SZX7 hot stage microscope in ramp mode from 30°C to 600°C at a rate of 10 °C/min in silicon oil. Samples were placed on a glass slide with a drop of silicon oil and protected with a cover slip.
The interaction of curdlan and the C-PLGA copolymer with Dectin-1 receptor was explored through a molecular docking simulation using Schrodinger Meastro GLIDE docking SP (standard precise mode) program as previously reported.14 Briefly, a short curdlan polymer (consisting of four β-glucan monomers) was generated from the polymer module of Meastro. The protein and ligand (curdlan polymer) were prepared and docked using the default parameters. The binding energy was estimated and the interactions between the curdlan and the amino acids in the Dectin-1 binding site were elucidated using Pymol (Schrödinger LL. The PyMOL Molecular Graphics System 2016 version 2.2. pymol. org/2/support. html. Accessed March. 2019;5) and Molecular Operating Environment (MOE) (Molecular Operating Environment, The Chemical Computing Group, v. 2016.08, http://www.chemcomp.com software). To explore the orientation of curdlan in the curdlan-PLGA copolymer, a short molecular dynamics simulation was conducted with UCSF Chimera using AMBER ff14SB force field and charges were assigned with AM1-BCC.
Molecular Operating Environment MOE 2019.01 was used to prepare three-dimensional protein model of Dectin-1 (RSCB PDB ID: 2CL8) for docking. The binding site was the position of the co-crystalized ligand in the protein model. Molecular docking simulation was carried out with the MOE Dock module using the induce-fit model.
C-PLGA NP
NP Synthesis.
An emulsion-solvent evaporation technique was developed to synthesize the NPs having varying densities of curdlan (2, 5 and 8% w/w).15–18 Briefly, 2 mL of a 1% w/v polymer solution (mixture of C-PLGA and PLGA) in a DMSO: DCM mixture (1:9) was added drop-wise to 10 mL of a PVA solution in de-ionised water (0.5% w/v) under probe sonication (Bandelin Sonoplus UW2070 + HD 2070 Berlin, Germany) for 7 min, 55% power, 70 Watt). A mixture of C-PLGA and PLGA was used to produce the desired concentration of curdlan functionalized NPs. For example, to synthesize 2% w/w curdlan loaded PLGA NPs, a ratio of 4 parts PLGA: 1-part C-PLGA was used. The same procedure was used to synthesize NPs containing 5% and 8% w/w curdlan loaded PLGA NPs. PLGA NPs were synthesized using only a solution of PLGA polymer. The solvent was subsequently removed in vacuo at 35°C and the NPs collected through centrifugation at 10 000 × g for 5 min, followed by washing once with de-ionized water and lyophilization over 72 h in the presence of sucrose (1% w/v) as a cryoprotectant.
NP Characterization.
NP characteristics of hydrodynamic diameter, polydispersity index (PDI) and zeta potential, were measured using a Malvern Zetasizer Nano ZS (Malvern Instruments Ltd, Worcestershire, UK). NPs were suspended in de-ionized water (1 mg/mL) and measurements were conducted at 25°C in triplicate.
The stability of the NPs was also investigated. The NPs were incubated at 37°C in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10 % Fetal Bovine Serum (FBS) at a concentration of 1 mg/mL. Particle size, PDI and zeta potential were monitored over 72 h.
To determine the presence of curdlan on the surface of the NPs, XPS analysis was performed. Data were collected using a Kratos Axis Ultra DLD, using an Al (monochromatic) anode, equipped with charge neutralizer. The operating pressure was kept below 5 × 10−9 Torr. For wide/survey XPS scans, the following parameters were used: emission current was kept at 10 mA and the anode (HT) voltage at 15 kV. The resolution used to acquire wide/survey scans was at 160 eV pass energy using a hybrid lens in the slot mode. The spectra were deconvoluted using OriginPro 8 MA, USA. For the quantitative studies, the area under the peaks was used for the atomic percentages obtained from elements present on the survey spectra and the deconvoluted spectra. The temperature of the sample was kept at 19°C and no effect of the beam damage was observed.
Molecular docking was used to gain insight into the probable interaction of a hypothesized model of the C-PLGA NPs with Dectin-1. The structure of the NPs was modelled using Schrödinger Material Science 4.1 suite (Schrödinger Materials Science Suite 2021–2, Schrödinger, LLC, New York, NY, 2021). Briefly, a spherical hydrocarbon cage was used to represent a NP (Volume = 264.4 Å and Area = 225.5 Å) onto which four monomers of curdlan were attached at different points. The model was prepared for molecular docking by protonation at pH 7, addition of explicit hydrogen and energy minimized with the Amber10: EHT force field. Five poses were generated for the modelled C-PLGA copolymers nanoparticle and the poses were evaluated with the GBVI/WSA ΔG scoring function. UCSF Chimera was used to visualize the poses.
Biological Studies
Cytotoxicity of NPs to Macrophages.
RAW264.7 macrophage viability after NP exposure was measured using the MTT assay. Absorbance values were translated into cell survival measurements and expressed as percentage cell viability relative to untreated controls (Eq. 1).
| (1) |
The assay was conducted to compare the means of the administered NP concentrations at 72 h for the NP formulations (PLGA, 2%, 5%, and 8% w/w C-PLGA). Each formulation was tested in quadruplicate and the results are representative of triplicate biological repeats.
Stimulation of infected Macrophages and killing efficacy of intracellular M. tuberculosis. Bacterial strains.
Mycobacterial cells were cultured in 7H9 supplemented with 10% OADC, 0.2% (v/v) glycerol, 0.05% (v/v) Tween-80 (7H9-OGT), and the necessary antibiotics for plasmid maintenance (kanamycin (25 μg/mL), hygromycin (50 μg/mL)). Additionally, M. tuberculosis ΔleuDΔpanD liquid cultures were supplemented with leucine (50 μg/mL) and pantothenate (24 μg/mL) and incubated at 37°C with shaking at 180 rpm (Labotec Orbital shaking incubator (Labotec). Reporter strains expressing the bacterial luciferase operon of plasmid pMV306hsp + LuxCDABE19 were previously obtained through electroporation as described by Snapper et al. (1990).20 Bioluminescence from luxCDABE gene expression was used to measure the changes in bacterial numbers for the M. tuberculosis ΔleuDΔpanD::pMV306hsp+lux strain. Solid cultures of M. tuberculosis ΔleuDΔpanD::pMV306hsp+lux were grown on 7H10 agar containing leucine (50 μg/mL) and pantothenate (24 μg/mL). Plasmid pMV306hsp + LuxCDABE was obtained from Addgene; reference number 2615519.
Assessing the macrophage and intracellular bacterial responses to stimulation by the NPs.
RAW264.7 cells were seeded at a concentration of 5 × 104 cells per well in a white solid bottom, 96-well tissue culture plate and incubated overnight at 37°C, 5% CO2 (Thermo Steri-Cycle CO2 incubator, Marshall Scientific). Macrophage cells were infected with M. tuberculosis ΔleuDΔpanD::pMV306hsp+lux and M. tuberculosis ΔleuDΔpanD at an MOI of 10:1. Sampson et al. (2004) and Mouton et al. (2019) have previously characterized and compared M. tuberculosis ΔleuDΔpanD with non-attenuated M. tuberculosis H37Rv and provided data supporting the use of this attenuated strain as a model organism in TB research.21,22 In preparation for infection, the mycobacterial cells were sonicated at 37 kHz for 12 min to disperse clumps and filtered through a 40 μm cell strainer. Cells were washed in D10 and the absorbance (OD600) adjusted to 1. This was achieved by measuring the OD of the bacterial cultures prior to washing and then resuspending the cell pellet in a volume of D10 that would result in an adjusted OD600 = 1. The D10 media was continuously supplemented with leucine (50 μg/mL) and pantothenate (24 μg/mL) to support the growth of M. tuberculosis ΔleuDΔpanD cells. Once absorbance was adjusted, 100 μL of cell suspension was added to each well and incubated at 37°C, 5% CO2 for 3 h. Following the 3 h uptake period, cells were washed once with PBS, and 200 μL D10 containing (1:100) penicillin/streptomycin was added to the wells and incubated at 37°C, 5% CO2 for 1 h to kill any remaining extracellular, un-phagocytosed bacteria. Cells were then washed three times with PBS. To the NP treated wells, 100 mL fresh D10 and 100 mL of a particular NPs suspended in D10 (supplemented with leucine (50 μg/mL) and pantothenate (24 μg/mL)) was added. NP suspensions were UV-sterilized for 30 minutes prior to before addition to the macrophages. Rifampicin treated wells (2 μg/mL) (positive control), untreated wells with M. tuberculosis ΔleuDΔpanD::pMV306hsp+lux (negative control) and untreated wells with M. tuberculosis ΔleuDΔpanD (background luminescence control) received 200 mL fresh D10 only (supplemented with leucine (50 μg/mL) and pantothenate (24 μg/mL)).
In addition, in vitro cultured M. tuberculosis ΔleuDΔpanD:: pMV306hsp+lux (i.e. in the absence of macrophages) was also treated with each of the NPs. in vitro growth was assessed to be able to draw comparisons between intracellular and in vitro growth effects and provide a proof of concept on the importance of macrophage interaction with the NPs towards bacterial killing. Based on the MTT cytotoxicity results, four NP formulations at a concentration of 1 mg/mL were added to the bacteria. Each treatment was evaluated in quadruplicate (technical repeats) and the experiment itself done in triplicate (biological repeats). Bioluminescence of the bacterial luciferase operon on the pMV306hsp+LuxCDABE provides a real time proxy of bacterial cell numbers and was used to measure the intracellular bacterial load at 24h, 48h and 72h. A FLUOstar microplate reader (FLUOstar Omega plate reader, BMG LABTECH) was used and the luminescence gain setting automatically adjusted to 3600. The mean luminescence of the WT M. tuberculosis ΔleuDΔpanD cells was subtracted from each of the respective wells to obtain the plotted relative light units (RLU’S). Confirmatory colony forming unit (CFU) plating was performed after 72h on 7H10 agar supplemented with 10% OADC, 0.2% (v/v) glycerol, 1eucine (50 μg/mL) and pantothenate (24 μg/mL). Briefly, after removal of the supernatant, macrophages were lysed with dH2O and serial dilutions of the lysates plated and incubated at 37 °C for 21 days.
Cell culture supernatants of the macrophage only controls, untreated macrophages containing M. tuberculosis ΔleuDΔpanD:: pMV306hsp-lux, and M. tuberculosis ΔleuDΔpanD::pMV306hsp-lux treated with each of the four NP formulations (PLGA, 2%, 5% and 8% C-PLGA) were collected. The cell supernatants were collected in Spin- X® centrifuge tubes and possible extracellular bacilli were removed by centrifugation (1 min, 10,000 rpm) and the flow-through was stored at −20 °C. A customized mouse Multiplex Immunoassay (R&D Systems, WhiteSci) was used to simultaneously quantify Interferongamma (IFN-γ), tumor necrosis factor-alpha (TNF-α), interleukin (IL)-1β, IL-4, IL-10, and IL-12. Cytokine concentrations were measured on a Bio-Plex platform (Bio Plex ™, Bio-Rad laboratories). The lower limit of quantification (LLOQ) was 11.933 pg/ml, 2.414 pg/ml, 224.536 pg/ml, 60.243 pg/ml, 13.734 pg/ml and 44.492 pg/ml for IFN-γ, TNF-α, IL-1β, IL-4, IL-10 and IL-12, respectively.
Statistical Analysis
Data analyses were performed for the DLS data using the one-way ANOVA (GraphPad Prism 6.0), time studies for the DLS data were analyzed using two-way ANOVA (GraphPad Prism 6.0). Differences between data sets were considered statistically significant when p < 0.05. Biological data were treated with a one-way ANOVA (Graph- Pad Prism v7.05), conducted between the different treatments followed by a Tukey’s multiple comparisons test and a p < 0.05 value was considered to be statistically significant.
Results and Discussion
C-PLGA Copolymer Characterization
NMR Spectroscopy
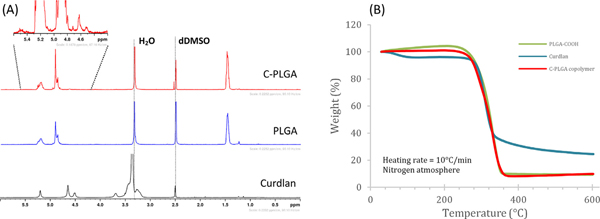
Proton NMR was performed to confirm the formation of curdlan-PLGA copolymer. The presence of curdlan peaks between 4.4. and 4.7 ppm of the C-PLGA copolymer spectrum suggests the success of the synthesis, Fig. 1(A). The ratio of curdlan to PLGA peaks in the curdlan-PLGA copolymer suggests minimal incorporation of curdlan as was expected.8
Fig. 1.
(A) 1H NMR of the starting materials curdlan, PLGA and final product curdlan-PLGA copolymer (run in DMSO-d6) and (B) TGA thermogram curves of PLGA-COOH, curdlan and curdlan-PLGA copolymerC-PLGA).
TGA Analysis
Thermogravimetric analysis was performed over a temperature range of 30–600°C for curdlan, PLGA-COOH and C-PLGA, Fig. 1(B). As expected, curdlan shows two stages of mass loss,8,23 the first weight loss of 4.6% w/w between 30 and 150°C is attributed to the desorption of adsorbed and bound water. The second weight loss of 70.3% w/w between 250°C and 400°C can be attributed to curdlan decomposition. Both the carboxylated PLGA and curdlan-PLGA copolymer show a single weight loss stage due to thermal decomposition from ± 220°C, which follows that observed by Tukulula et al.8 The data in Fig. 1 (B) also shows the decomposition of C-PLGA and PLGA-COOH with the copolymer C-PLGA showing a slightly faster decomposition rate than PLGA-COOH, indicating possible reduced thermal stability due to the addition of the curdlan polymer on the carboxylated PLGA which introduces weaker links.24,25
Hot stage Microscopy
Hot stage microscopy (HSM) enabled visual analysis of sample morphology, melting point and degradation over a set temperature range. Supplementary Fig. 1 shows the morphology of curdlan, PLGA-COOH and the copolymer product curdlan-PLGA at different stages during heating. Visible transformations were seen at the degradation temperatures, 261°C for curdlan, 262°C for PLGA-COOH and 256°C for curdlan-PLGA copolymer. Microscopy indicates that PLGA-COOH is the only compound that melts (123.4°C, not shown), whilst both curdlan and curdlan-PLGA copolymer tend to lose shape/structure and change colour in the case of curdlan and PLGA-COOH. At 600°C the compound morphologies are distinctly unique from each other confirming the formation of the curdlan-PLGA copolymer.
Molecular Docking and Molecular Dynamics
Molecular interaction of Curdlan with Dectin-1.
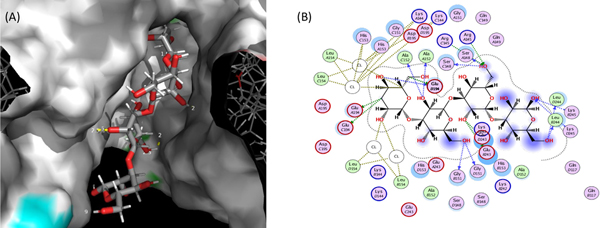
Fig. 2(A) shows the first binding pose of curdlan (4 monomers) within the Dectin-1 binding site. The hydrogen bond was visualized with Pymol at this setting: distance = or < 2.0 Angstrom and 20° angle. It was observed that 3 monomers of the polymer may be sufficient for interaction with Dectin-1. The binding energy of the curdlan tetramer to Dectin-1 was −7.54 kcal/mol, suggesting a strong affinity of curdlan for Dectin-1. When visualized on MOE (Fig. 2(B)) at distance = or > 2.0 Angstrom and 20° angle, the number of hydrogen bonds displayed via hydrogen bond donors on curdlan were 11 bonds depicted by the blue arrows (ALA152 (2 bonds), GLU194, GLY151 (2 bonds), LEU244 (2 bonds), LYS245 (2 bonds), SER148 (2 bonds)). Whilst the green arrows which represent hydrogen bonds formed by hydrogen bond acceptors on curdlan, a total of 7 hydrogen bonds (GLU194 (3 bonds), ARG145 (2 bonds), GLU243 (2 bonds)). The majority of hydroxyl groups on the monomers of curdlan are amenable to hydrogen bond formation and may account for the number of hydrogen bonds observed.
Fig. 2.
(A) 3D rendering of the Dectin-1 active site interacting with 4 monomers of the curdlan substrate (stick model), (B) 2D rendering showing the spatial interaction of curdlan, 4-monomers (skeletal formula) with amino acids from Dectin-1. Both figures are represented within physiological fluid.
Molecular Dynamics Simulation of the C-PLGA Copolymer Chain.
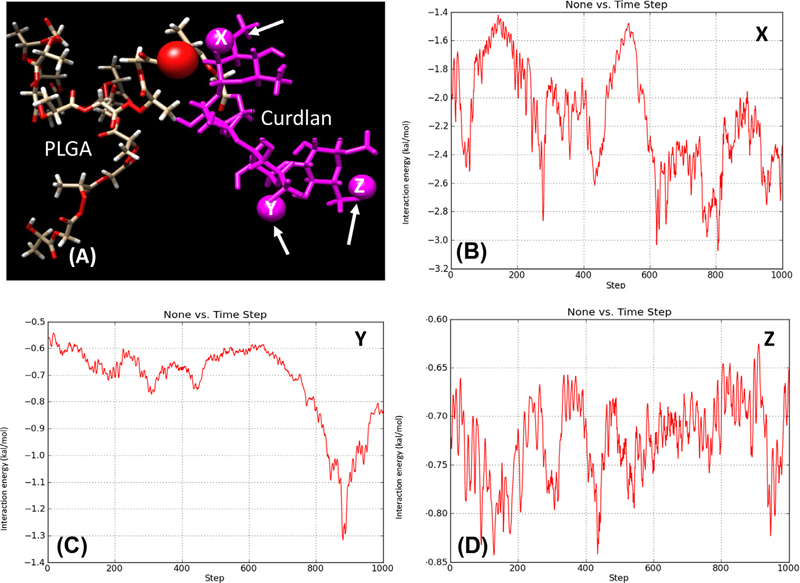
Molecular dynamics simulation of a 5-monomer curdlan polymer linked to the PLGA polymer was performed over 1000-time steps (Supplementary Video 1). This enabled a better understanding of how curdlan on the copolymer orientates to allow interaction with Dectin-1.
Analysis of the simulation showed the root mean squared deviation (RMSD) and kinetic energy of the entire copolymer structure changed significantly towards the end of the simulation (Supplementary Fig. 2). This indicates significant movement and change of positions by the atoms in the copolymer i.e. the copolymer was highly flexible. This might be advantageous during synthesis due to the readiness of the copolymer system to move and fold over into a stable conformation (as observed by the lower RSMD at the towards the end of the simulation).
Due to the flexibility of the copolymer and knowing the affinity of the curdlan hydroxyl groups for Dectin-1, an investigation into the extent of curdlan hydroxyl group interaction with PLGA was investigated. Fig. 3(A) shows the diagrammatic representation of the interaction between PLGA and curdlan. Fig. 3(C–E) show plots of interaction energy (hydrogen bonding) between three selected hydroxyl groups on curdlan and a hydroxyl group on PLGA. The results showed that three hydroxyl groups, selected from different parts of the curdlan polymer interacted to different degrees with the PLGA polymer over the 1000-step simulation. The selected hydroxyl group on the curdlan labelled X (in Fig. 3(B)) had the greatest tendency to interact with the selected portion of the PLGA polymer with interaction energy between − 1.4 kcal/mol and − 3.2 kcal/mol over the 1000-step simulation. This was followed by the hydroxyl group labelled Y on the curdlan polymer, Fig. 3(C) which showed interaction energy between − 0.55 kcal/mol and − 1.32 kcal/mol, with the greatest interaction occurring towards the end of the simulation as the curdlan polymer folded over towards the PLGA polymer. The least interaction (between − 0.63 kcal/mol and − 0.83 kcal/mol) was observed between the hydroxyl group labelled Z on the curdlan polymer, Fig. 3 (D) and the selected portion of PLGA. Therefore, the further away the hydroxyl group is on the curdlan polymer the less the chance of interacting with the PLGA polymer and the higher the chance of such monomers being available for the desired interaction with Dectin-1.
Fig. 3.
(A) A plot of interaction energy (hydrogen bonding) between three selected hydroxyl groups (X, Y and Z indicated by arrows) on curdlan (magenta) and a hydroxyl group on PLGA. Figures (B) to (D) represent the interaction energy of the individual hydroxyl groups X, Y and Z with the red atom of PLGA respectively.
Overall, the simulation (Supplementary Video 1) showed there is a good probability that the curdlan polymer may interact with portions of the PLGA polymer within this copolymer system, thus limiting the exposure of the correct curdlan polymer orientation to allow interaction with Dectin-1. From the video and the molecular docking simulations mentioned above, it can be deduced that a curdlan chain length of more than 3 monomers is desirable in the copolymer to make allowance for the dynamics displayed while exposing a sufficient portion of the curdlan polymer for the desired interaction with Dectin-1.
Modeling C-PLGA NP and its interactions with Dectin-1.
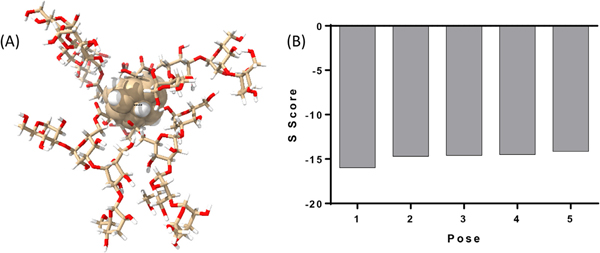
To predict the interaction of the modelled C-PLGA NP with the Dectin-1 receptor, the NP model was docked against the binding site of Dectin-1 using the induce-fit model in MOE 2019.01. Fig. 4(A) depicts the model of the C-PLGA NP. The NP core is represented by the central sphere and attached to the surface are 4 chains of curdlan tetramers, represented as sticks. The generalized Born volume integral (GBVI) model was used to generate the S score for five poses. A higher negative S score is a predictor of better interaction with the protein and more stable NP-protein complex. The estimated free energy of binding (S score) of the five poses of C-PLGA NP are presented in Fig. 4(B). Each pose represents a possible energy minimized spatial orientation of the modelled C-PLGA NP relative to Dectin-1. The high S score for all the poses suggests that the C-PLGA NP has a strong interaction with Dectin-1.
Fig. 4.
(A) A diagrammatic representation of the spherical NP core with 4 monomers of the curdlan-PLGA copolymer attached to the surface. (B) The estimated free energy of binding (S score) of the five poses of C-PLGA nanoparticle.
Supplementary Fig. 3 shows both the hydrogen bonds (A) and Van der Waals (B) interactions of the C-PLGA NP with the amino acid residues in Dectin-1. Supplementary Fig. 3(A) showed a total of 193 hydrogen bonds with hydrogen bond detection setting at a distance tolerance of 2.5 Å and angle tolerance of 20°. The molecular docking studies thus predicted the modelled C-PLGA NP with the curdlan moieties on its surface can potentially bind strongly to Dectin-1 to illicit the immunomodulatory response.
C-PLGA NP Characterization
Size, PDI, Zeta Potential
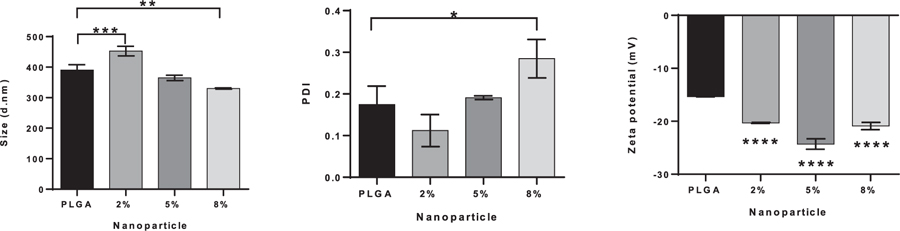
Fig. 5 shows the mean sizes, size distributions and zeta potentials of the NPs. The values obtained fall within previously observed ranges8 and represent mean ± standard deviation (SD) where n = 3. The NP sizes were 390 ± 18 nm, 453 ± 16 nm, 365 ± 9 nm and 330 ± 2 nm for the PLGA NPs, 2%, 5% and 8% C-PLGA NPs respectively. Statistical analysis shows a significant difference between PGLA NPs and the 2% as well as the 8% C-PLGA NPs, which are substantially larger and smaller than the PLGA NPs respectively. The observed decrease in size from 2% to 8% corresponds with the increase in curdlan and this behaviour can be attributed to the capping effect of the curdlan at higher concentration.26 The size distributions are similar and show a low size distribution for the various NPs with a slight significant difference between the PLGA NPs and 8% C-PLGA NPs. These values range from 0.17 ± 0.04, 0.11 ± 0.04, 0.19 ± 0.004 and 0.28 ± 0.05 for the PLGA to 8% C-PLGA NPs. The NP charge represented by the zeta potential shows the NPs had a net negative charge. The zeta potentials point to good stability of the particles with PLGA NPs showing a value of −15.4 ± 0.06 mV, 2% C-PLGA NPs showing −20.3 ± 0.1 mV and the 5% C-PLGA NPs with −24.3 ± 0.98 mV. Statistical analysis showed there is a significant difference between PLGA NPs and the rest of the C-PLGA NPs, with p values < 0.05 and may suggest that curdlan aids in NP stability.
Fig. 5.
Size (A), polydispersity index (PDI) (B) and zeta potential (C) of PLGA and % C-PLGA NPs. NPs suspended in de-ionized water (1 mg/mL) were prepared and measurements were conducted at 25°C. Results shown are the mean and standard deviation of n = 3 replicates. Statistical differences are indicated as follows: * is p ≤ 0.05, ** is p ≤ 0.01, *** is p ≤ 0.001 and **** is p ≤ 0.0001. Statistical differences in (C) are between the PLGA NPs and 2%, 5% and 8% C-PLGA NPs respectively.
The assessment of NP properties in cell culture media is crucial for understanding their effects on cells in culture. In this work, NP stability was examined in DMEM with 10% FBS, Supplementary Fig. 4. The average NP size decreased to 288 ± 12 nm, 413 ± 15 nm, 291 ± 4 nm and 308 ± 8 nm for the PLGA NPs, 2%, 5% and 8% C-PLGA NPs respectively. This is due to the presence of proteins in the media resulting in the formation of a protein corona which affects the NP size, PDI and zeta potential. As the NPs are not protected with polyethylene glycol, they experience a higher extent of protein binding.27 Both NP size and size distribution remain relatively constant over a 72-h period, whilst the zeta potential of the NPs increased over time. Statistical analysis confirmed there was a significant difference between the PDI observed at the start and after 72 h for the various NPs (p < 0.05). This observation is possibly an effect of the protein corona.
Quantitative Surface Analysis of the NPs
Quantitative analysis of the NP surface was performed using XPS. The chemical bonds of elements present on the surface of the NPs have specific binding energies which are dependent on their chemical environment enabling a determination of the functional groups present, and by deconvoluting the spectra, a calculation of the relative percentage chemical composition can be made, Fig. 6.
Fig. 6.
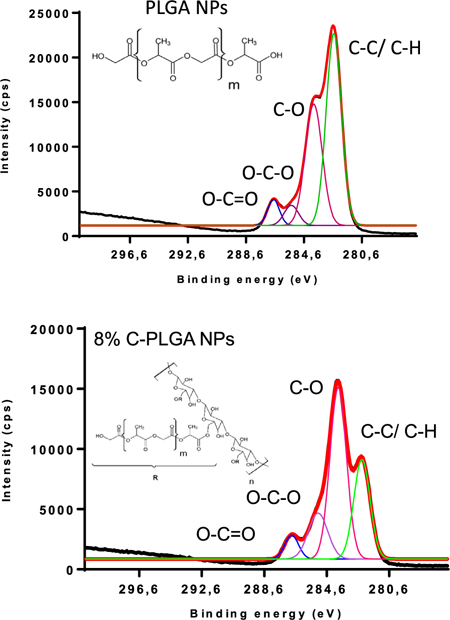
XPS spectra (black) showing the calculated deconvoluted spectra (red) summing the individual peaks under the curve. These individual peaks represent the intensities of the C 1s binding energies on the surface of the PLGA and 8 % C-PLGA NPs.
For the purposes of determining the presence of curdlan on the surface of the NPs, a comparison between the carbon species of the PLGA NPs and most concentrated curdlan containing, 8% w/w C-PLGA NPs was performed. The PLGA and 8% w/w C-PLGA NP C1s spectra shows the presence of four distinct carbonyl functional groups, namely C-C/C-H, C-O, O-C-O and O-C=O (Table 2). There is a clear decrease in the C-C/C-H composition from 52.5% in the PLGA NPs to 28.5% in the 8% C-PLGA NPs, which can be attributed to the presence of curdlan, containing high amounts of oxygen atoms. As expected, there is an increase in the percent composition of C-O and O-C-O from 36.9 and 5.2% to 50.0 and 15.7% respectively due to the presence of the curdlan. Finally, the O-C=O composition between the two NPs remains fairly constant. This data suggests the presence of curdlan on the surface of the 8% curdlan containing PLGA NPs.
Table 2.
Binding energies and composition for C 1s in PLGA and 8% C-PLGA NPs.
| NP |
PLGA NPs |
8% C-PLGA NPs |
||
|---|---|---|---|---|
| Species | Binding energy (eV) | Composition (%) | Binding energy (eV) | Composition (%) |
| C-C/C-H | 285.0 | 52.5 | 284.9 | 28.5 |
| C-O | 286.4 | 36.9 | 286.4 | 50.0 |
| O-C-O | 288.0 | 5.2 | 287.7 | 15.7 |
| O-C=O | 289.2 | 5.5 | 289.4 | 5.8 |
Biological Studies
Cytotoxicity of Nanoparticles to RAW264.7Macrophages
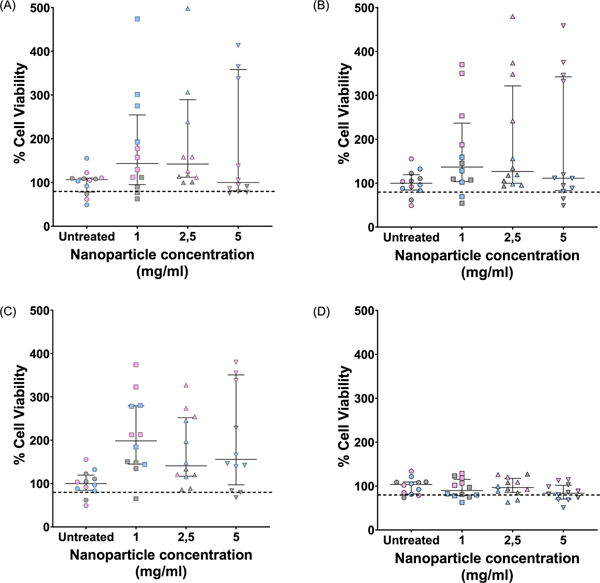
The toxicity of the 4 different NP formulations was assessed at 3 different concentrations over a 72 h period. All NPs showed no cytotoxicity towards RAW264.7 macrophages with the average values of three biological repeats remaining above 80% cell viability (Fig. 7), which was in agreement with previous reports.8,28–30 No clear dose dependent trend was observed and variability in cell viability (as evidenced by the interquartile range) was present (Fig. 7). Interestingly, for the PLGA, 2% and 5% C-PLGA NPs, it was observed that the addition of the NPs resulted in viability values higher than 100% possibly suggesting stimulation of macrophage proliferation.
Fig. 7.
MTT cytotoxicity assessment of the C-PLGA NP formulations administered at 3 different concentrations on murine RAW264.7 macrophages after 72h. The macrophages were exposed to PLGA NPs (A), and C-PLGA NPs with 2% (B), 5% (C) and 8% (D) (w/w) C-PLGA NPs. Results shown are the median and interquartile range of n = 3 biological replicates (quadruplicate technical repeats, the grey, pink and blue represents data from a single biological repeat). The horizontal dotted line represents 80% viability cut-off as determined through ISO standards.
Stimulation of TB Infected Macrophages and Death of Intracellular M. Tuberculosis
Cytokine Concentrations.
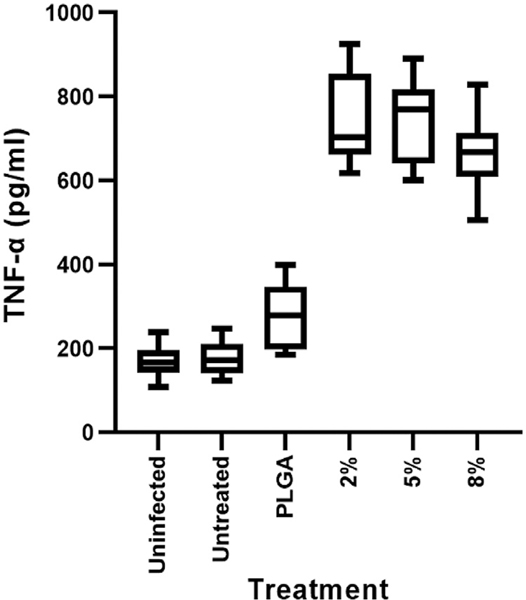
Intracellular cultured M. tuberculosis ΔleuDΔpanD::pMV306hsp-lux were treated with 5 mg/mL of the PLGA, 2%, 5% and 8% w/w C-PLGA NPs and the expression of bioluminescence was tracked for 72 h. A rifampicin (2 μg/mL) treated positive control was included to provide an indication of what is expected if bacterial cell death is occurring. Additionally, untreated, negative controls were included. TNF-α secretion levels were significantly upregulated (p < 0.0001) in the supernatant of the M. tuberculosis infected macrophages treated with 2%, 5% and 8% C-PLGA NPs compared to PLGA only NP treated cells, the untreated controls and untreated-uninfected macrophage (Fig. 8). The other cytokines, IFN-γ, lL-1β, 1L-4,1L-10, and 1L-12, were found below the detection limit (< OOR).
Fig. 8.
TNF-α cytokine secretion by RAW264.7 murine macrophages due to NP stimulation after 72h. Macrophages infected with M. tuberculosis ΔleuDΔpanD:: pMV306hsp-lux were treated with either PLGA, 2%, 5% and 8% C-PLGA NPs. Controls included uninfected and untreated macrophages (Uninfected) as well as M. tuberculosis ΔleuDΔpanD::pMV306hsp-lux infected and untreated macrophages (Untreated). Results represent 6 data points expressed as the median and IQR (3 technical repeats and 2 biological repeats).
The importance of the pro-inflammatory cytokine TNF- α in activating macrophages and reducing bacterial burden is well documented and various studies have shown the aggravation of TB pathological outcomes with the addition of TNF-α neutralizing agents.31,32 In one of these studies, Lin et al. (2010) showed with cynomolgus macaques, that treatment with TNF-α neutralizing agents led to disease dissemination by 8 weeks post-infection and that anti-TNF-α agents caused disease reactivation in latently infected subjects.33 However, it is important to find an inflammatory balance in host directed therapies. A notable issue frequently arising is the possible induction of a cytokine storm; an excessive inflammatory response frequently associated with increased levels of TNF-α.34 This balance could easily be tipped in favour of the pathogen and alter the pathological outcome of TB treatment thus the aim should be to strike a balance between toxicity and efficacy.
Impact of NPs on intracellular M. tuberculosis bacterial burden.
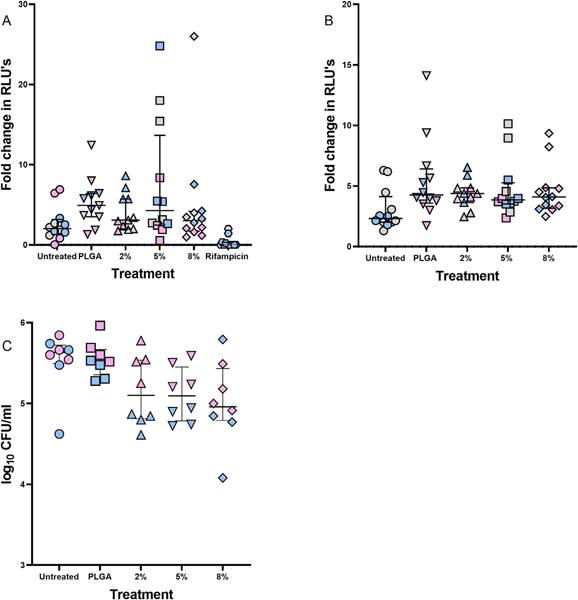
We next explored whether the NP effect on macrophage phenotype (as evidenced by elevated TNF- α production) impacted on bacterial growth and survival. To do this, we assessed intracellular bacterial burden by measuring luminescence as well as by serial dilution plating following NP treatment over 72 h. Luminescence readings and CFU counts (Fig. 9) demonstrated reduced intracellular bacterial burdens following treatment with C-PLGA NPs. Although the fold change in RLUs or CFU counts was not statistically significant, the data suggests that these NPs can impact the growth and survival of intracellular M. tuberculosis.
Fig. 9.
Effect of NPs on M. tuberculosis cell numbers as a fold-change between 72 h and 24 h. Intracellular M. tuberculosis ΔleuDΔpanD::pMV306hsp-lux within RAW264.7 macrophages (A) were compared with in vitro M. tuberculosis ΔleuDΔpanD::pMV306hsp-lux cell numbers (B), by tracking luminescence over 72 h. (C) Determination of intracellular M. tuberculosis ΔleuDΔpanD::pMV306hsp-lux burden by CFU plating after 72 h. The grey, pink and blue represents data from a single biological repeat. Data is expressed as the median and interquartile range of n = 2 biological repeats and n = 4 technical repeats.
Curdlan on the surface of the NPs is anticipated to interact with the Dectin-1 receptor on macrophages stimulating the Syk-dependent activation of the NF-κB pathway.35 This pathway is known to upregulate the production of oxidative species, Ca2+ levels and proinflammatory cytokines such as TNF-α, as well as enhancing the phagocytic tendency of macrophages.8,29,35 This would be expected to culminate in a macrophage-specific antibacterial effect.8,36
Conclusion
Successful synthesis of the immunomodulatory curdlan-PLGA copolymer and C-PLGA NPs was verified using various physicochemical techniques. Bioinformatics approaches predicted a strong interaction between the curdlan on the NP surface and Dectin-1 receptor. Pro-inflammatory cytokine production was observed following macrophage stimulation with curdlan functionalized PLGA NPs, along with a trend towards a reduction in intracellular bacterial burden. These NPs show promise as a host-directed immunotherapy for TB, and warrant further investigation.
Supplementary Material
Acknowledgements
Research reported in this publication was supported by the Fogarty International Center of the National Institutes of Health under Award Number K43TW010371 granted to AD and 5R01AI152109 from the National Institute of Allergy and Infectious Diseases (NIAID) awarded to SLS and AD. SLS is funded by the South African Research Chairs Initiative of the Department of Science and Technology and National Research Foundation (NRF) of South Africa, award number UID 86539. The authors acknowledge the SA MRC Centre for TB Research and DST/NRF Centre of Excellence for Biomedical Tuberculosis Research for financial support for this work. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the SA MRC or the NRF.
Footnotes
Supplementary Materials
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.xphs.2021.09.012.
References
- 1.World Health Organization. Global Tuberculosis Report 2020. 2020. p. 1–232.
- 2.Richter E, Rüsch-Gerdes S, Hillemann D. First linezolid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2007;51(4):1534–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wasserman S, Louw G, Ramangoaela L, et al. Linezolid resistance in patients with drug-resistant TB and treatment failure in South Africa. J Antimicrob Chemother. 2019;74(8):2377–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bekale RB, Du Plessis S-M, Hsu N-J, et al. Mycobacterium tuberculosis and interactions with the host immune system: opportunities for nanoparticle based immunotherapeutics and vaccines. Pharm Res. 2019;36(1):1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young C, Walzl G. Du Plessis N. Therapeutic host-directed strategies to improve outcome in tuberculosis. Mucosal Immunol. 2020;13(2):190–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kilinç G, Saris A, Ottenhoff THM, Haks MC. Host-directed therapy to combat mycobacterial infections*. Immunol Rev. 2021:(December 2020):1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maphasa RE, Meyer M, Dube A. The macrophage response to mycobacterium tuberculosis and opportunities for autophagy inducing nanomedicines for tuberculosis therapy. Front Cell Infect Microbiol. 2021;10(February):1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tukulula M, Hayeshi R, Fonteh P, et al. Curdlan-conjugated PLGA nanoparticles possess macrophage stimulant activity and drug delivery capabilities. Pharm Res. 2015;32(8):2713–2726. [DOI] [PubMed] [Google Scholar]
- 9.Dube A, Reynolds JL, Law WC, Maponga CC, Prasad PN, Morse GD. Multimodal nanoparticles that provide immunomodulation and intracellular drug delivery for infectious diseases. Nanomed Nanotechnol Biol Med. 2014;10(4):831–838. [DOI] [PubMed] [Google Scholar]
- 10.Goodridge HS, Wolf AJ, Underhill DM. B-Glucan recognition by the innate immune system. Immunol Rev. 2009;230(1):38–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan GCF, Chan WK, Sze DMY. The effects of beta-glucan on human immune and cancer cells. J Hematol Oncol. 2009;2(25). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs M, Togbe D, Fremond C, et al. Tumor necrosis factor is critical to control tuberculosis infection. Microbes Infect. 2007;9(5):623–628. [DOI] [PubMed] [Google Scholar]
- 13.Quesniaux VFJ, Jacobs M, Allie N, et al. TNF in host resistance to tuberculosis infection. Curr Dir Autoimmun. 2010:11157–11179. [DOI] [PubMed] [Google Scholar]
- 14.Li D, Meng X, Wang Q, et al. Consensus scoring model for the molecular docking study of Mtor kinase inhibitor. J Mol Graph Model. 2017:7981–7987. [DOI] [PubMed] [Google Scholar]
- 15.Zhou J, Patel TR, Sirianni RW, et al. Highly penetrative, drug-loaded nanocarriers improve treatment of glioblastoma. Proc Natl Acad Sci USA. 2013;110(29):11751–11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCall RL, Sirianni RW. PLGA nanoparticles formed by single- or double-emulsion with vitamin E-TPGS. J Vis Exp. 2013:82e51015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong YI, Shim YH, Kim C, Lim GT, Choi KC, Yoon C. Effect of cryoprotectants on the reconstitution of surfactant-free nanoparticles of poly(DL-lactide-co-glycolide). J Microencapsul. 2005;22(6):593–601. [DOI] [PubMed] [Google Scholar]
- 18.Yadav K, Yadav D, Yadav M, Kumar S. Noscapine loaded PLGA nanoparticles prepared using oil-in-water emulsion solvent evaporation method. J Nanopharm Drug Deliv. 2016;3(1):97–105. [Google Scholar]
- 19.Andreu N, Zelmer A, Fletcher T, et al. Optimisation of bioluminescent reporters for use with mycobacteria. PLoS One. 2010;5(5):e10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snapper SB, Melton RE, Mustafa S, Kieser T, Jr WRJ. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4(11):1911–1919. [DOI] [PubMed] [Google Scholar]
- 21.Mouton JM, Heunis T, Dippenaar A, Gallant JL, Kleynhans L, Sampson SL. Comprehensive characterization of the attenuated double auxotroph mycobacterium tuberculosisΔleudΔpanCD as an alternative to h37Rv. Front Microbiol. 2019;10 (AUG):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sampson SL, Dascher CC, Sambandamurthy VK, et al. Protection elicited by a double leucine and pantothenate auxotroph of mycobacterium tuberculosis in guinea pigs. Infect Immun. 2004;72(5):3031–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bothara SB, Singh S. Thermal studies on natural polysaccharide. Asian Pac J Trop Biomed. 2012;2(2):S1031–S1035. [Google Scholar]
- 24.Shi L Characterization of the flame retardancy of EVA copolymer by plasma grafting of acrylic acid. Eur Polym J. 2000:362611–362615. [Google Scholar]
- 25.Khayet M, Nasef MM, Mengual JI. Radiation grafted poly (ethylene terephthalate) - graft -polystyrene pervaporation membranes for organic /organic separation. J Membr Sci. 2005:26377–26395. [Google Scholar]
- 26.Liu Q, Duan B, Zhang L. Progress in rigid polysaccharide-based nanocomposites with therapeutic functions. J. Mater. Chem. B, 2017;5690–5713. [DOI] [PubMed] [Google Scholar]
- 27.Samkange T, D’Souza S, Obikeze K, Dube A. Influence of PEGylation on PLGA nanoparticle properties, hydrophobic drug release and interactions with human serum albumin. J Pharm Pharmacol. 2019;71(10):1497–1507. [DOI] [PubMed] [Google Scholar]
- 28.Makadia HK, Siegel SJ. Poly lactic-co-glycolic acid (PLGA) as biodegradable controlled drug delivery carrier. Polymers. 2011;3(3):1377–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dube A, Lemmer Y, Hayeshi R, et al. State of the art and future directions in nanomedicine for tuberculosis. Expert Opin Drug Deliv. 2013;10(12):1725–1734. [DOI] [PubMed] [Google Scholar]
- 30.Chen X, Gao C. Influences of surface coating of PLGA nanoparticles on immune activation of macrophages. J Mater Chem B. 2018;6(14):2065–2077. [DOI] [PubMed] [Google Scholar]
- 31.Churchyard GJ, Kaplan G, Fallows D, Wallis RS, Onyebujoh P, Rook GA. Advances in immunotherapy for tuberculosis treatment. Clin Chest Med. 2009;30(4):769–782. [DOI] [PubMed] [Google Scholar]
- 32.Cavalcanti YVN, Brelaz MCA, Neves JKDAL, Ferraz JC, Pereira VRA. Role of TNF-alpha, IFN-gamma, and IL-10 in the development of pulmonary tuberculosis. Pulm Med. 2012. 20121–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin PL, Flynn JL. Understanding latent tuberculosis: a moving target. J Immunol. 2010:18515–18522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gustafson HH, Holt-Casper D, Grainger DW, Ghandehari H. Nanoparticle uptake: the phagocyte problem. Nano Today. 2015;10(4):487–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagener M, Hoving JC, Ndlovu H, Marakalala MJ. Dectin-1-Syk-CARD9 signaling pathway in TB immunity. Front Immunol. 2018;9:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tukulula M, Gouveia L, Paixao P, Hayeshi R, Naicker B, Dube A. Functionalization of PLGA Nanoparticles with 1,3-β-glucan Enhances the Intracellular Pharmacokinetics ofRifampicin in Macrophages. Pharm Res. 2018;35(6):111–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.