Abstract
Helicobacter pylori isolates vary between geographic regions. Certain H. pylori genotypes may be associated with disease outcome. Thirty-eight children underwent diagnostic upper endoscopy at four medical centers and were retrospectively analyzed to determine if H. pylori virulence genes were associated with endoscopic disease severity, histologic parameters, and host demographics. The H. pylori virulence genotype was analyzed by a reverse hybridization line probe assay and type-specific PCR. Endoscopic ulcers or erosions were found in 17 (45%) patients, with 13 (34%) of these patients having antral nodularity. Histological gastritis, of varying severity, was present in all children. Four patients harbored more than one H. pylori strain: one subject had both cagA+ and cagA-negative strains, while three patients harbored either two different cagA-negative strains (two children) or two cagA+ strains (one child). There were 28 (74%) cagA+ isolates; 19 were associated with the vacA s1b genotype, 7 were associated with the vacA s1a genotype, 1 was associated with the vacA s1c genotype, and 1 was associated with the s2 genotype. Of 14 cagA-negative isolates, 6 were vacA s2 genotype, 4 were vacA s1b, 3 were vacA s1a, and 1 was vacA s1c. Nine of ten (90%) Hispanics had similar H. pylori strains (vacA s1b,m1), and all Asian-Canadian children were infected by strains with vacA s1c genotype. No correlation between H. pylori strain and endoscopic or histopathologic abnormalities was found. This study provides a baseline framework of North American children and their H. pylori strains, serving as a powerful epidemiological tool for prospective investigations to better understand the transmission and evolution of diverse disease outcomes.
Using DNA fingerprinting, restriction fragment length polymorphism, and multilocus enzyme analysis, Helicobacter pylori strains isolated from adults have demonstrated considerable heterogeneity in selected genes (1, 3, 17, 34). The different H. pylori genes have shown distinct geographic distribution and correlation with severity of disease. van Doorn et al. (35, 38) demonstrated that vacA alleles have specific distributions across different ethnic groups and geographic regions; for example, the vacA s1c H. pylori strains are found exclusively in persons of Asian descent. Also, specific H. pylori genotypes (in particular, cagA, vacA, cagE-picB, and iceA) are considered more virulent strains since they are associated with more severe gastroduodenal disease in adults (18, 25, 27, 37, 38, 41, 43). For example, vacA type s1a strains have been isolated more frequently in adults with peptic ulcer disease and are associated with increased gastric epithelial damage (2, 41). An additional H. pylori virulence gene, iceA (induced by contact with epithelium), has been more commonly observed in H. pylori strains isolated from adults with peptic ulcer disease compared to those with gastritis alone (6, 7, 38).
The cagA gene is closely associated with the vacA s1 genotype and is considered a marker for severe host disease (6, 7, 21, 24, 26, 28, 30, 38). Using serology, Elitsur et al. (13) estimated that the prevalences of anti-CagA antibodies among asymptomatic children were 54 and 69% among symptomatic children (P < 0.05). More recently, Yahav et al. (42) has shown that anti-CagA seropositive H. pylori-infected children have more severe gastroduodenal disease and worse outcomes (i.e., more difficult to eradicate and longer time for disease resolution) than H. pylori-infected children who are CagA seronegative.
Studies of genetic variability of H. pylori in children have been restricted to single-center, serological analysis of the cagA pathogenicity island (8, 9, 22, 26). Moreover, there have been no studies that have evaluated pediatric H. pylori isolates in correlation with quantitative histopathologic data in infected children. Accordingly, a multicenter pediatric study was undertaken to determine if different H. pylori genotypes are associated with disease severity, are seen in specific ethnic groups, or have a restricted geographic distribution. In this retrospective study, we investigated the role of the virulence genes cagA, vacA, and iceA in gastroduodenal disease in children by analyzing H. pylori strains obtained from four different sites in North America using PCR and a reverse hybridization assay. The H. pylori genotype was also correlated with demographic, endoscopic, and histologic data.
MATERIALS AND METHODS
Patient population, endoscopy, and pathology data.
This study included a random sample of patients from four centers (Miami Children's Hospital, Miami, Fla.; Rainbow Babies and Children's Hospital, Cleveland, Ohio; Hospital for Sick Children, University of Toronto, Toronto, Ontario, Canada; and Children's Healthcare of Atlanta at Egleston, Atlanta, Ga.) in which an H. pylori culture was available for genotyping. The H. pylori specimen was obtained during a diagnostic fiberoptic upper endoscopy, which was performed at the discretion of the pediatric gastroenterologist because of the subject's persistent gastrointestinal symptoms and signs. The study cohort was accrued over a 3-year study period, and patients were selected for analysis using a random numbering scheme. All patients were treated at each center with eradication H. pylori therapy as described previously (11).
Endoscopic diagnoses in the stomach were defined, for the purpose of this study, as follows: normal gross appearance, erosions (anatomic location in stomach was designated), ulcers (anatomic location), and nodularity (anatomic location). Endoscopic pictures from each endoscopy were independently reviewed by one of the authors (B.D.G.) in order to standardize endoscopic reporting for each patient. From each child, a minimum of four endoscopic gastric biopsies were obtained: one for on-site rapid urease testing, one for culture, and at least two for histopathology (one from the antrum and another from the corpus).
The two biopsies used for histopathologic evaluation were formalin fixed, paraffin embedded, and stained with hematoxylin and eosin. In order to obtain uniform grading of the inflammatory infiltrate, one pathologist retrospectively reviewed the biopsies using the visual analog scale from the Updated Sydney Classification of Gastritis (10). The Updated Sydney Classification of Gastritis grades the following histopathological features on a scale that goes from absent to marked, namely, (i) the amounts of bacteria, (ii) neutrophils, and (iii) mononuclear inflammatory cells, as well as (iv) the degree of atrophy and intestinal metaplasia present in a biopsy. When necessary, silver impregnation stains were used to confirm identification of bacteria in the gastric biopsies. For the final pathology diagnosis, the gastritis was classified as either chronic active, if both neutrophils and mononuclear inflammatory cells were present, or chronic if only mononuclear cells were seen.
Bacterial cultures.
Three centers (Cleveland, Toronto, and Atlanta) had the microbiological laboratory capability to permit on-site primary cultures. In these centers, biopsies (ca. 0.1 to 0.2 mg/biopsy) were homogenized under aseptic conditions in either 1.5 ml of sterile saline or transport medium (vial containing 1.5 ml of brucella broth and 20% sterile glycerol) and a loopful of homogenated biopsy tissue streaked onto both nonselective (brain heart infusion [BHI] agar with 5% sheep blood) and selective (Skirrows) media using previously described techniques (31–33). Briefly, agar plates were placed under microaerobic conditions (5% O2, 10% CO2, 85% N2) at 37°C and incubated for 5 to 10 days until small, gray, translucent colonies consistent with the morphology of H. pylori were obtained. Biochemical analyses were performed for catalase, oxidase, and urease activity, dark-field microscopy was used for morphology and viability confirmation, and flagellum stain was used to confirm H. pylori at each site before freezing and storage of primary cultures. Gastric biopsies at Miami were placed immediately into cryovials containing 1 ml of Trypticase soy medium with 20% glycerol and then shipped on dry ice to the laboratory of B.D.G. at Emory University, where primary isolation was performed as described above. Once isolated, H. pylori cultures from each participating center were shipped, without accompanying clinical or epidemiological information, to the Centers for Disease Control and Prevention (CDC) for further analysis.
Molecular analysis.
Once received at the CDC, H. pylori isolates were cultured on BHI agar plates containing 5% sheep blood (Becton Dickinson) for 3 to 5 days at 37°C under microaerobic conditions. H. pylori was harvested from plates by suspension in 2 ml of sterile 0.9% NaCl solution and sedimented by centrifugation at 10,000 rpm for 2 min. For DNA extraction, bacteria were resuspended in 400 ml of 10 mM Tris-HCl (pH 8.0), 5 mM EDTA, 0.1% sodium dodecyl sulfate, and 0.1 mg of proteinase K per ml and incubated for 2 to 4 h at 55°C. Proteinase K was inactivated by incubation at 95°C for 10 min. The lysate was clarified by centrifugation at 14,000 rpm for 2 min. The supernatant was diluted 1/100 in sterile water and directly employed for PCR. vacA and cagA genotypes were determined by multiplex PCR and reverse hybridization on a line probe assay (LiPA), as described earlier (38, 39). Briefly, parts of the vacA s and m allele regions, as well as the cagA gene, were amplified. PCR products were reverse hybridized to a LiPA strip, comprising specific probes for vacA, s1A, s1b, s1c, s2, vacA m1, m2a, m2b, and cagA. iceA genotypes were determined by type-specific PCR, as described earlier (15, 36, 38). The PCR primers for cagA, vacA, and iceA used in this study have been published previously (26, 38).
Statistical analyses.
Data were entered into Epi Info 6.1 (CDC, U.S. Department of Health and Human Services, 1995) and analyzed by using software SAS (version 6.12). Pearson rank or a chi-squared (χ2) test was used to assess the relationship between individual H. pylori genotypes and the endoscopic diagnoses and between the H. pylori genotypes and the subject's geographic origin. Unconditional logistic regression analysis was employed to assess differences in the histopathologic and endoscopic parameters between groups (age, gender, and geographic origin), while controlling for H. pylori genotype.
RESULTS
Patient demographics.
A total of 38 subjects were selected: 13 from Cleveland, 12 from Atlanta, 8 from Miami, and 5 from Toronto, Canada. The median age was 9.4 years (range, 3 to 16 years). Of the study cohort, 61% was male and 39% was female. All children studied resided in North America and included a variety of ethnic groups. A total of 16 (42%) of the children studied were Caucasian 11 (29%) were black (of these 3 were of mixed Caucasian-black heritage, and 1 was black-Hispanic), and 2 were Asian (second-generation Chinese-Canadian and third-generation Vietnamese-Canadian).
Endoscopic findings.
Ulcers or erosions were found in 17 patients: 1 had esophageal ulcers, 4 of 17 (24%) patients had gastric ulcers, and 12 of 17 (70%) had duodenal or pyloric channel ulcers. No history of nonsteroidal anti-inflammatory agents taken during the month prior to endoscopy was elicited. Of the 38 children, 13 (34%) had antral nodularity; 2 of these 13 had diffuse gastric nodularity. The stomachs of 25 of 38 (66%) children did not have nodularity, erosions, or ulcers on endoscopy and were considered to have a normal macroscopic appearance.
Histopathology and grading of inflammation severity.
Adequate histopathological analysis was performed in 33 cases. In four patients the biopsies were too superficial, and in one case slides or paraffin blocks were not available for review. In each of the 33 cases, H. pylori was demonstrated with densities in the mucosa ranging from mild to marked. All of the patients studied had histologic evidence of chronic inflammation. With the exception of five patients, all had active inflammatory infiltrate component (i.e., the presence of polymorphonuclear leukocytes). Of the patients, 21 had a marked degree of inflammation, 4 had moderate amounts, and 8 had mild inflammatory infiltrates. Eosinophils were observed in 87% of the patients. The number of lymphoid follicles in gastric biopsies in H. pylori-infected children ranged from 0 to 4 (mean, 3.2). Three patients had atrophy in the antrum; in two of them it was classified as moderate, and in the other one it was classified as mild. Two of the children with atrophy also had focal, mild intestinal metaplasia.
Molecular analysis of H. pylori strains.
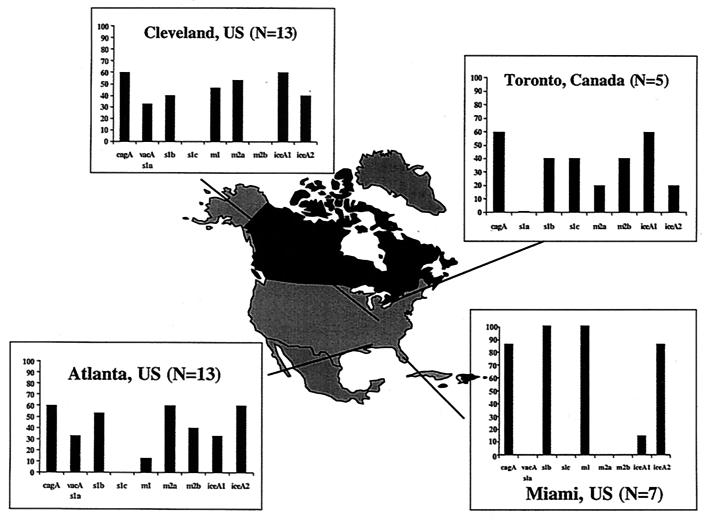
The distribution of pediatric H. pylori strain genotypes (cagA, vacA s and m alleles, and iceA alleles) corresponding to each center are depicted in Fig. 1. Evidence of infection by more than one H. pylori strain was found in four patients: two from Cleveland and two from Atlanta. One case had cagA+ and cagA-negative strains, while the other three patients had either two cagA-negative strains (two cases) or two cagA+ strains (one case). The latter three cases showed two H. pylori strains in the same isolate since there was evidence of different vacA s and m genes. Of the 34 cases where only one H. pylori strain was identified, 25 (74%) were cagA+ and 9 (26%) were cagA negative. The vacA and ice genotypes varied through the H. pylori strains. Of the 28 cagA+ isolates, 19 (68%) were associated with the vacA s1b genotype, 7 (25%) were associated with the vacA s1a genotype, 1 (4%) was associated with the vacA s1c genotype, and 1 (4%) was associated with the s2 genotype. Among the 14 cagA-negative isolates, 6 (43%) were vacA s2 4 (29%) were vacA s1b, 3 (21%) were vacA s1a, and 1 (7%) was vacA s1c. The vacA s2 genotype was always associated with the vacA m2 genotype.
FIG. 1.
Map of North America depicting the location of the four participating centers (Miami, Fla.; Atlanta, Ga.; Cleveland, Ohio; and Toronto, Ontario, Canada) and the distribution of the vacA s and m regions, cagA, and the iceA1 and iceA2 genotypes of H. pylori strains obtained from the pediatric subjects. For each center, the prevalence of each type (s1a, s1b, s1c, s2, m1, m2a, and m2b; cagA+; and iceA1 and iceA2 genotypes) is given as a percentage of the total number of strains.
Correlation of H. pylori genotype and demographic data.
Two of the Caucasian children had more than one H. pylori isolate; both had two cagA-negative strains. The majority of Caucasian children (10 of 16) were cagA+ (5 from Cleveland, 2 from Atlanta, and 3 from Toronto). Their vacA and iceA genotypes were varied. Six Cuban-American children from Miami had cagA+ strains and vacA s1b, m1 alleles. Two of the black children had more than one H. pylori strain; one was from Cleveland, and the other was from Atlanta. Seven of the isolates from six of the black children were cagA+, and two were cagA-negative strains. The three children from Georgia of mixed black-white race showed a variety of cagA, vacA, and iceA genotypes. The two children from Asian decent were from Toronto and showed different cagA, vacA m, and iceA genotypes; however, they shared the same vacA s1c gene.
Correlation of H. pylori genotype with endoscopic and histopathologic diagnoses.
Endoscopy demonstrated ulcers or erosions in 12 (32%) of the children with cagA+ H. pylori and in 5 (13%) of the children with cagA-negative strains (P > 0.05; not significant). Gastric nodularity was present in 9 (24%) of the children with cagA+ H. pylori and in 4 (10%) of the children with cagA-negative strains (P > 0.05; not significant). Endoscopies classified as having normal gastric mucosa were seen in five (13%) children with cagA+ isolates and in three (8%) children with cagA-negative H. pylori strains (P > 0.05; not significant).
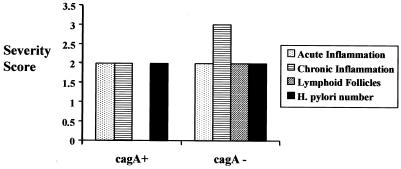
Among the 33 patients for whom pathology was available, marked gastritis was present in 13 (39%) children with cagA+ H. pylori and in 9 (27%) children with cagA-negative strains (Fig. 2). Moderate gastritis was seen only in 4 (12%) of children with cagA+ H. pylori. Mild inflammation was demonstrated in six (18%) children with cagA+ H. pylori and in two (6%) children with cagA-negative strains. As mentioned previously, atrophy was seen in two patients of Hispanic decent, both of whom harbored cagA+ isolates. Atrophy was also demonstrated in one of the children of Asian decent; this patient had a cagA-negative isolate.
FIG. 2.
Lack of correlation between pediatric H. pylori cagA status and the severity of antral inflammation as assessed by the four parameters used by the Updated Sydney Classification of Gastritis: acute inflammation, chronic inflammation, presence of lymphoid follicles, and H. pylori number. Although not shown, a similar lack of correlation was seen between the vacA or iceA genotype and the inflammation severity scores.
DISCUSSION
This study is the first multicenter genotypic analysis of H. pylori strains obtained from pediatric populations. We found a variety of H. pylori genotypes but could not demonstrate an association between the strain genotype and either the endoscopic features or the histopathologic findings of infected children. This lack of correlation between the H. pylori genotype and the pediatric gastroduodenal disease may be due, in part, to a highly selected symptomatic population evaluated upon referral to the pediatric gastroenterologists at tertiary-care, academic centers. Conversely, the bias of our patient population should have exaggerated the potential relationships between genotype and virulence. Specifically, since we only studied individuals who presumably had highly virulent disease resulting in clinical symptoms which then resulted in the child's referral to the subspecialist and endoscopic evaluation, the fact that we saw no relationship between genotype and H. pylori virulence genes is even more significant.
In our study cohort, only the ethnic or racial origin of the infected host seemed to be a factor, which correlated with the H. pylori strain genotype. All children resided in North America and, despite the relative small sample size, the diversity of ethnic backgrounds is relatively reflective of the demographics found in both the United States and Canada, adding validity to the correlation between ethnicity or race and the H. pylori strain genotype. This retrospective study facilitated the creation of a network of medical centers with specific capabilities to determine a baseline of patients and H. pylori genotypes that will enable us to plan a prospective multicenter study. Prospective investigations employing this multicenter cohort will yield the numbers needed to ascertain the overall impact of H. pylori genotype on the spectrum of pediatric gastroduodenal disease. Moreover, the inclusion of additional centers, prospective enrollment, and better representation of the diverse ethnic makeup of North America and, thereby, the ability to sample multiple generations of children from different ethnic backgrounds may provide additional insight into the evolution of H. pylori genotypes in different populations worldwide.
Recently, investigators have demonstrated that distinct H. pylori genotypes have specific geographic distributions (35, 37, 38). In Europe, for example, a distribution gradient of the vacA s1 subtypes has been observed (i.e., vacA s1a genotype in individuals from northern Europe, England, Ireland, and Scotland), whereas in Central and South America, virtually all H. pylori strains contained the vacA s1b genotype and in East Asia the subtype s1c is observed most frequently (35, 38). In the present study, the two Asian children had a vacA s1c allele, and most of the H. pylori isolates from Hispanic children had a vacA s1b,m1 genotype. Although our study cohort was relatively small in size, these genotypes follow a geographic and ethnic distribution pattern similar to the one seen in adult populations.
This study highlights a number of important observations, such as providing evidence that multiple H. pylori genotypes can occur in infected children. It is possible that children, when first acquiring the infection, are colonized by multiple H. pylori strains. It has been postulated that over time, through natural selection influenced by host and bacterial factors, one specific H. pylori genotype predominates in infected adults (4, 20, 23, 31). This may be the reason why, in our previous studies, H. pylori-infected children have been found to have greater numbers of organisms compared to infected adults (40). Additionally, data from a nonhuman primate model of Helicobacter infection provide support for this hypothesis. Rhesus monkeys challenged with a mixture of seven genetically distinct H. pylori strains resulted in variable susceptibility to different genotypes during the acute phase of the infection compared to latter stages when one H. pylori strain predominated (12).
Onset of H. pylori infection is during childhood in most human populations (29). Although it is believed that both host factors and bacterial factors dictate eventual disease outcome, the natural history of H. pylori infection after childhood acquisition remains poorly characterized (5, 19, 20). In this study, only symptomatic pediatric patients were endoscoped, i.e., children with persistent upper gastrointestinal signs and symptoms warranting diagnostic upper endoscopy. Endoscopic abnormalities were evident in more than half of the children with H. pylori isolates which were cagA+, yet there were no significant differences in endoscopic or histologic diagnoses in those children harboring cagA compared to cagA-deficient strains. In this cohort of children, the prevalence of peptic ulcer disease was high for this age group. This is likely due to the retrospective selection of cases for study from tertiary care referral centers (i.e., selection bias). Future studies that incorporate a greater number of pediatric gastrointestinal centers from different geographic regions are necessary to eliminate some of this bias to the study sample and thus to better evaluate childhood gastroduodenal disease in correlation with bacterial genotype.
Finally, the observation of H. pylori-infected pediatric patients with atrophy and intestinal metaplasia is exceedingly uncommon. Pathologic diagnosis of atrophy has been controversial in adult populations because of the lack of strict diagnostic criteria, difficulties in performing the diagnosis in one biopsy, and poor reproducibility when assessing severity (14, 16, 32, 33). Clearly, prospective studies with larger numbers of children from multiple centers and geographic regions are needed to better define the spectrum of illness and natural history of disease following pediatric H. pylori infection, as well as to better understand the epidemiology and pathobiology of this infection; such studies are essential for developing more effective methods of eradication and prevention.
ACKNOWLEDGMENTS
Benjamin D. Gold is supported by a Public Health Service grant from the National Institute of Health (NIDDK 53708-01). Steven J. Czinn is supported by Public Health Service grant NIDDK 46461-01.
REFERENCES
- 1.Achtman M, Azuma T, Berg D E, Ito Y, Morelli G, Pan Z J, Suerbaum S, Thompson S A, van der Ende A, van Doorn L J. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol. 1999;32:459–470. doi: 10.1046/j.1365-2958.1999.01382.x. [DOI] [PubMed] [Google Scholar]
- 2.Atherton J C. The clinical relevance of strain types of Helicobacter pylori. Gut. 1997;40:701–703. doi: 10.1136/gut.40.6.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaser M J. Heterogeneity of Helicobacter pylori. Eur J Gastroenterol Hepatol. 1997;9(Suppl. 1):S3–S7. doi: 10.1007/978-94-009-1792-7_3. [DOI] [PubMed] [Google Scholar]
- 4.Blaser M J. Hypotheses on the pathogenesis and natural history of Helicobacter pylori-induced inflammation. Gastroenterology. 1992;102:720–727. doi: 10.1016/0016-5085(92)90126-j. [DOI] [PubMed] [Google Scholar]
- 5.Blaser M J. Hypothesis: the changing relationships of Helicobacter pylori and humans: implications for health and disease. J Infect Dis. 1999;179:1523–1530. doi: 10.1086/314785. [DOI] [PubMed] [Google Scholar]
- 6.Censini S, Lange C, Xiang Z, Crabtree J E, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Covacci A, Telford J L, Del Giudice G, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 8.Day A S, Jones N L, Lynett J T, Jennings H A, Fallone C A, Beech R, Sherman P M. cagE is a virulence factor associated with Helicobacter pylori-induced duodenal ulceration in children. J Infect Dis. 2000;181:1370–1375. doi: 10.1086/315394. [DOI] [PubMed] [Google Scholar]
- 9.Day A S, Sherman P M. Helicobacter pylori infection, host genes, and disease outcome. Pediatr Res. 2000;47:703. doi: 10.1203/00006450-200006000-00001. [DOI] [PubMed] [Google Scholar]
- 10.Dixon M F, Genta R M, Yardley J H, Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 1996;20:1161–1181. doi: 10.1097/00000478-199610000-00001. [DOI] [PubMed] [Google Scholar]
- 11.Drumm B, Koletzko S, Oderda G. Helicobacter pylori infection in children: a consensus statement. Medical position paper: report of the European Paediatric Task Force on Helicobacter pylori on a Consensus Conference, Budapest, Hungary, September 1998. J Pediatr Gastroenterol Nutr. 2000;30:207–213. doi: 10.1097/00005176-200002000-00020. [DOI] [PubMed] [Google Scholar]
- 12.Dubois A, Berg D E, Incecik E T, Fiala N, Heman-Ackah L M, Del Valle J, Yang M, Wirth H P, Perez-Perez G I, Blaser M J. Host specificity of Helicobacter pylori strains and host responses in experimentally challenged nonhuman primates. Gastroenterology. 1999;116:90–96. doi: 10.1016/s0016-5085(99)70232-5. [DOI] [PubMed] [Google Scholar]
- 13.Elitsur Y, Neace C, Werthammer M C, Triest W E. Prevalence of CagA, VacA antibodies in symptomatic and asymptomatic children with Helicobacter pylori infection. Helicobacter. 1999;4:100–105. doi: 10.1046/j.1523-5378.1999.98530.x. [DOI] [PubMed] [Google Scholar]
- 14.el-Zimaity H M, Graham D Y, al-Assi M T, Malaty H, Karttunen T J, Graham D P, Huberman R M, Genta R M. Interobserver variation in the histopathological assessment of Helicobacter pylori gastritis. Hum Pathol. 1996;27:35–41. doi: 10.1016/s0046-8177(96)90135-5. [DOI] [PubMed] [Google Scholar]
- 15.Figueiredo C, Quint W G, Sanna R, Sablon E, Donahue J P, Xu Q, Miller G G, Peek R M, Jr, Blaser M J, van Doorn L J. Genetic organization and heterogeneity of the iceA locus of Helicobacter pylori. Gene. 2000;246:59–68. doi: 10.1016/s0378-1119(00)00054-8. [DOI] [PubMed] [Google Scholar]
- 16.Genta R M, Rugge M. Gastric precancerous lesions: heading for an international consensus. Gut. 1999;45(Suppl. 1):I5–I8. doi: 10.1136/gut.45.2008.i5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibson J R, Slater E, Xerry J, Tompkins D S, Owen R J. Use of an amplified-fragment length polymorphism technique to fingerprint and differentiate isolates of Helicobacter pylori. J Clin Microbiol. 1998;36:2580–2585. doi: 10.1128/jcm.36.9.2580-2585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Go M F, Kapur V, Graham D Y, Musser J M. Population genetic analysis of Helicobacter pylori by multilocus enzyme electrophoresis: extensive allelic diversity and recombinational population structure. J Bacteriol. 1996;178:3934–3938. doi: 10.1128/jb.178.13.3934-3938.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gold B D. Pediatric Helicobacter pylori infection: clinical manifestations, diagnosis, and therapy. Curr Top Microbiol Immunol. 1999;241:71–102. doi: 10.1007/978-3-642-60013-5_6. [DOI] [PubMed] [Google Scholar]
- 20.Graham D Y. Helicobacter pylori infection in the pathogenesis of duodenal ulcer and gastric cancer: a model. Gastroenterology. 1997;113:1983–1991. doi: 10.1016/s0016-5085(97)70019-2. [DOI] [PubMed] [Google Scholar]
- 21.Karaolis D K, Somara S, Maneval D R, Jr, Johnson J A, Kaper J B. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature. 1999;399:375–379. doi: 10.1038/20715. [DOI] [PubMed] [Google Scholar]
- 22.Kolho K L, Karttunen R, Heikkila P, Lindahl H, Rautelin H. Gastric inflammation is enhanced in children with CagA-positive Helicobacter pylori infection. Pediatr Infect Dis J. 1999;18:337–341. doi: 10.1097/00006454-199904000-00005. [DOI] [PubMed] [Google Scholar]
- 23.Kuipers E J, Perez-Perez G I, Meuwissen S G, Blaser M J. Helicobacter pylori and atrophic gastritis: importance of the cagA status. J Natl Cancer Inst. 1995;87:1777–1780. doi: 10.1093/jnci/87.23.1777. [DOI] [PubMed] [Google Scholar]
- 24.Li L, Kelly L K, Ayub K, Graham D Y, Go M F. Genotypes of Helicobacter pylori obtained from gastric ulcer patients taking or not taking NSAIDs. Am J Gastroenterol. 1999;94:1502–1507. doi: 10.1111/j.1572-0241.1999.01133.x. [DOI] [PubMed] [Google Scholar]
- 25.Marshall D G, Coleman D C, Sullivan D J, Xia H, O'Morain C A, Smyth C J. Genomic DNA fingerprinting of clinical isolates of Helicobacter pylori using short oligonucleotide probes containing repetitive sequences. J Appl Bacteriol. 1996;81:509–517. doi: 10.1111/j.1365-2672.1996.tb03540.x. [DOI] [PubMed] [Google Scholar]
- 26.Miehlke S, Genta R M, Graham D Y, Go M F. Molecular relationships of Helicobacter pylori strains in a family with gastroduodenal disease. Am J Gastroenterol. 1999;94:364–368. doi: 10.1111/j.1572-0241.1999.859_u.x. [DOI] [PubMed] [Google Scholar]
- 27.Miehlke S, Kibler K, Kim J G, Figura N, Small S M, Graham D Y, Go M F. Allelic variation in the cagA gene of Helicobacter pylori obtained from Korea compared to the United States. Am J Gastroenterol. 1996;91:1322–1325. [PubMed] [Google Scholar]
- 28.Mobley H L. Helicobacter pylori factors associated with disease development. Gastroenterology. 1997;113:S21–S28. doi: 10.1016/s0016-5085(97)80006-6. [DOI] [PubMed] [Google Scholar]
- 29.Parsonnet J. The incidence of Helicobacter pylori infection. Aliment Pharmacol Ther. 1995;9:45–51. [PubMed] [Google Scholar]
- 30.Peek R M, Jr, Vaezi M F, Falk G W, Goldblum J R, Perez-Perez G I, Richter J E, Blaser M J. Role of Helicobacter pylori cagA(+) strains and specific host immune responses on the development of premalignant and malignant lesions in the gastric cardia. Int J Cancer. 1999;82:520–524. doi: 10.1002/(sici)1097-0215(19990812)82:4<520::aid-ijc9>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 31.Rollan A, Giancaspero R, Fuster F, Acevedo C, Figueroa C, Hola K, Schulz M, Duarte I. The long-term reinfection rate and the course of duodenal ulcer disease after eradication of Helicobacter pylori in a developing country. Am J Gastroenterol. 2000;95:50–56. doi: 10.1111/j.1572-0241.2000.01700.x. [DOI] [PubMed] [Google Scholar]
- 32.Schenk B E, Kuipers E J, Klinkenberg-Knol E C, Bloemena E C, Sandell M, Nelis G F, Snel P, Festen H P, Meuwissen S G. Atrophic gastritis during long-term omeprazole therapy affects serum vitamin B12 levels. Aliment Pharmacol Ther. 1999;13:1343–1346. doi: 10.1046/j.1365-2036.1999.00616.x. [DOI] [PubMed] [Google Scholar]
- 33.Schultze V, Hackelsberger A, Gunther T, Miehlke S, Roessner A, Malfertheiner P. Differing patterns of Helicobacter pylori gastritis in patients with duodenal, prepyloric, and gastric ulcer disease. Scand J Gastroenterol. 1998;33:137–142. doi: 10.1080/00365529850166851. [DOI] [PubMed] [Google Scholar]
- 34.Shortridge V D, Stone G G, Flamm R K, Beyer J, Versalovic J, Graham D W, Tanaka S K. Molecular typing of Helicobacter pylori isolates from a multicenter U. S. clinical trial by ureC restriction fragment length polymorphism. J Clin Microbiol. 1997;35:471–473. doi: 10.1128/jcm.35.2.471-473.1997. . (Erratum, 36:1468, 1998.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Doorn L J, Figueiredo C, Megraud F, Pena S, Midolo P, Queiroz D M, Carneiro F, Vanderborght B, Pegado M D, Sanna R, De Boer W, Schneeberger P M, Correa P, Ng E K, Atherton J, Blaser M J, Quint W G. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology. 1999;116:823–830. doi: 10.1016/s0016-5085(99)70065-x. [DOI] [PubMed] [Google Scholar]
- 36.van Doorn L J, Figueiredo C, Rossau R, Jannes G, van Asbroek M, Sousa J C, Carneiro F, Quint W G. Typing of Helicobacter pylori vacA gene and detection of cagA gene by PCR and reverse hybridization. J Clin Microbiol. 1998;36:1271–1276. doi: 10.1128/jcm.36.5.1271-1276.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Doorn L J, Figueiredo C, Sanna R, Pena S, Midolo P, Ng E K, Atherton J C, Blaser M J, Quint W G. Expanding allelic diversity of Helicobacter pylori vacA. J Clin Microbiol. 1998;36:2597–2603. doi: 10.1128/jcm.36.9.2597-2603.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Doorn L J, Figueiredo C, Sanna R, Plaisier A, Schneeberger P, de Boer W, Quint W. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology. 1998;115:58–66. doi: 10.1016/s0016-5085(98)70365-8. [DOI] [PubMed] [Google Scholar]
- 39.van Doorn L J, Henskens Y, Nouhan N, Verschuuren A, Vreede R, Herbink P, Ponjee G, van Krimpen K, Blankenburg R, Scherpenisse J, Quint W. The efficacy of laboratory diagnosis of Helicobacter pylori infections in gastric biopsy specimens is related to bacterial density and vacA, cagA, and iceA genotypes. J Clin Microbiol. 2000;38:13–17. doi: 10.1128/jcm.38.1.13-17.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitney A E, Guarner J, Hutwagner L, Gold B D. Histopathological differences between Helicobacter pylori gastritis in children and adults. Ann Diagn Pathol. 2000;4:1–7. doi: 10.1053/adpa.2000.17871. [DOI] [PubMed] [Google Scholar]
- 41.Xiang Z, Censini S, Bayeli P F, Telford J L, Figura N, Rappuoli R, Covacci A. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect Immun. 1995;63:94–98. doi: 10.1128/iai.63.1.94-98.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yahav J, Fradkin A, Weisselberg B, Diver-Haver A, Shmuely H, Jonas A. Relevance of CagA positivity to clinical course of Helicobacter pylori infection in children. J Clin Microbiol. 2000;38:3534–3537. doi: 10.1128/jcm.38.10.3534-3537.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaoka Y, Kodama T, Kita M, Imanishi J, Kashima K, Graham D Y. Relation between clinical presentation, Helicobacter pylori density, interleukin 1β and 8 production, and cagA status. Gut. 1999;45:804–811. doi: 10.1136/gut.45.6.804. [DOI] [PMC free article] [PubMed] [Google Scholar]