Summary
Union of two gametes to form a zygote is a defining event in the life of sexual eukaryotes, yet the mechanisms that underlie cell-cell fusion during fertilization remain poorly characterized. Here, in studies of fertilization in the green alga, Chlamydomonas, we report identification of a membrane protein on minus gametes, Minus Adhesion Receptor 1 (MAR1), that is essential for the membrane attachment with plus gametes that immediately precedes lipid bilayer merger. We show that MAR1 forms a receptor pair with previously identified receptor FUS1 on plus gametes, whose ectodomain architecture we find is identical to a sperm adhesion protein conserved throughout plant lineages. Strikingly, before fusion, MAR1 is biochemically and functionally associated with the ancient, evolutionarily conserved eukaryotic class II fusion protein HAP2 on minus gametes. Thus, the integral membrane protein MAR1 provides a molecular link between membrane adhesion and bilayer merger during fertilization in Chlamydomonas.
Keywords: membrane fusion reaction, gamete adhesion, cell-cell fusion, MAR1, FUS1, GEX2, HAP2, Chlamydomonas, mating structure, fertilization, gamete fusion
eTOC blurb
Gamete fusion in organisms across kingdoms depends on fusogen HAP2. Pinello et al. demonstrate that receptor MAR1 is associated with HAP2 on Chlamydomonas minus gametes and binds to conserved adhesion protein FUS1/GEX2 on plus gametes, suggesting a mechanism that ensures bilayer merger is triggered only upon lineage-specific gamete membrane attachment.
Graphical Abstract
Introduction
Fusion of gametes to form a zygote during fertilization is a complex cell-cell interaction whose core cellular features have been conserved since the origin of eukaryotes. Initial cell-cell recognition, typically at cell surface locations entirely separate from the site of fusion, bring the two gametes together and induce signals within them to prepare for fusion (Dean, 2007; Dresselhaus et al., 2016; Ikawa et al., 2010; Snell and Goodenough, 2009). Upon the consequent gamete activation, the gametes undergo a second interaction by gamete-specific adhesion proteins at newly available sites on their plasma membranes specialized for fusion. Attachment of the plasma membranes is rapidly followed by merger of the lipid bilayers to complete the fusion reaction. The conservation of the cellular steps of fertilization suggests that at least some of the molecular underpinnings of the gamete membrane fusion reaction might also be conserved (Sankaranarayanan and Higashiyama, 2018; Tajima and Nishimura, 2018). Surprisingly, in spite of over a century of study of fertilization (Bianchi and Wright, 2020; Lillie, 1914), we still lack a comprehensive understanding in any single organism of the molecules or mechanisms that compose and regulate the gamete fusion reaction.
Studies in mouse have shown that the proteins IZUMO1 on sperm and JUNO on the egg form a complex that is essential for the sperm-egg membrane binding preceding fusion. The two proteins, which form the only receptor pair known to be essential for gamete membrane adhesion in any organism, are insufficient for fusion and a chordate gamete fusogen remains unidentified (Aydin et al., 2016; Bianchi et al., 2014; Inoue et al., 2005; Ohto et al., 2016). Several other mouse gamete proteins with mammalian orthologs have been reported as essential for fertilization, including CD9 on eggs (Le Naour et al., 2000; Miyado et al., 2000), and FIMP (Fujihara et al., 2020), SPACA6 (Barbaux et al., 2020; Lorenzetti et al., 2014), SOF1 (Noda et al., 2020), and TMEM95 (Lamas-Toranzo et al., 2020; Noda et al., 2020) on sperm, but they neither adhere nor fuse cells when heterologously expressed, and their molecular functions remain unclear.
Proteins with roles in gamete membrane interactions during the fusion reaction have been described in several other model organisms, including Prm1p in S. cerevisiae (Heiman and Walter, 2000); Spe-9, Spe-45, and EGG-1 & 2 in C. elegans (Kadandale et al., 2005; Nishimura et al., 2015; Singaravelu et al., 2015; Singson et al., 1998); P48/45, p47 and p230 in the malaria pathogens Plasmodium (van Dijk et al., 2010); the mating type proteins in ciliates (Cervantes et al., 2013); Bouncer in D. rerio (Herberg et al., 2018); GEX2 and DMP8 & 9 in the plant Arabidopsis thaliana (Cyprys et al., 2019; Engel et al., 2005; Mori et al., 2014; Takahashi et al., 2018); and FUS1 in the green alga Chlamydomonas reinhardtii (hereafter, Chlamydomonas) (Ferris et al., 1996; Misamore et al., 2003). The presence of immunoglobulin-like (Ig-like) domains in Spe-45, IZUMO1, GEX2, and FUS1 is consistent with the known roles of the domain in protein-protein interactions. This collection of adhesion proteins, however, has not been reported to possess a common overall domain architecture, and thus, a gamete adhesion protein family that spans unicellular and multicellular plants or animals has been absent.
In contrast, a single protein of conserved structure, HAP2, is essential for fertilization in unicellular and multicellular organisms across kingdoms, and was likely the primordial sexual fusogen (Camacho-Nuez et al., 2017; Cole et al., 2014; Ebchuqin et al., 2014; Hirai et al., 2008; Johnson et al., 2004; Liu et al., 2008; Mori et al., 2006; Okamoto et al., 2016; Ramakrishnan et al., 2019; Steele and Dana, 2009). Studies in Chlamydomonas, Plasmodium, and Tetrahymena showed that HAP2 was dispensable for gamete membrane adhesion but was required for bilayer merger (Cole et al., 2014; Liu et al., 2008). Remarkably, structural modeling and X-ray crystallography demonstrated an unambiguous structural homology between HAP2 and viral and developmental class II fusion proteins, including the envelope proteins from the Bunyavirales, Flaviviruses, and Alphaviruses and EFF-1 from C. elegans (Fedry et al., 2017; Perez-Vargas et al., 2014; Pinello et al., 2017; Valansi et al., 2017). More recent studies showed that Chlamydomonas HAP2, which is required only on minus gametes, indeed forms trimers in vivo and that trimer formation is essential for fusion (Zhang et al., 2021). Notably, HAP2 (also called GCS1) has not yet been detected in fungi or chordates, either because it was lost or because it evolved to become unrecognizable.
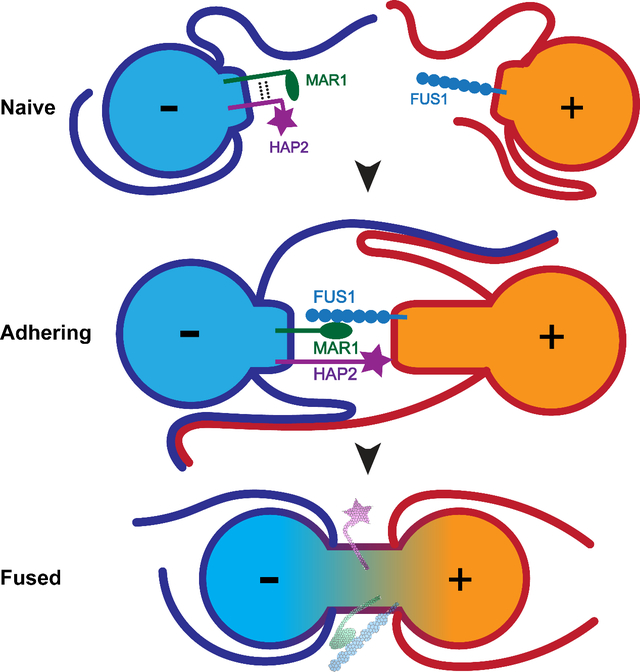
In Chlamydomonas, when plus and minus gametes are mixed together, they initially recognize and adhere to each other by their cilia through mating type-specific adhesion proteins that function only on the cilia, SAG1 in plus gametes and SAD1 in minus gametes (Snell and Goodenough, 2009) (Fig. 1A). Ciliary adhesion activates a protein kinase-dependent signaling pathway that leads to increases in intracellular cyclic AMP, thereby triggering gamete activation. During activation, gametes release their extracellular matrices (cell walls), recruit additional adhesion proteins to their ciliary membranes (Belzile et al., 2013; Ranjan et al., 2019), and assemble a membrane protuberance called a mating structure between their two cilia (Friedmann et al., 1968; Weiss et al., 1977). Ciliary adhesion and the accompanying vigorous motility of the cells bring the apical ends of pairs of gametes into alignment, leading to collisions between the two mating structures and adhesion of their tips. FUS1 is the plus gamete-specific membrane adhesion protein present on the mating structures of plus gametes, and plus gametes lacking FUS1 fail to adhere, fail to support formation of HAP2 trimers, and fail to fuse (Ferris et al., 1996; Misamore et al., 2003; Zhang et al., 2021). Within seconds of membrane adhesion, HAP2 present on the minus mating structure engages with the lipid bilayer of the plus mating structure to fuse the two membranes (Feng et al., 2018; Zhang et al., 2021), followed by rapid coalescence of the two gametes into a quadri-ciliated zygote (Fig. 1A). Nuclear fusion by the protein, GEX1, which is a member of the KAR5/GEX1 family of nuclear fusion proteins, follows soon thereafter (Beh et al., 1997; Engel et al., 2005; Ning et al., 2013).
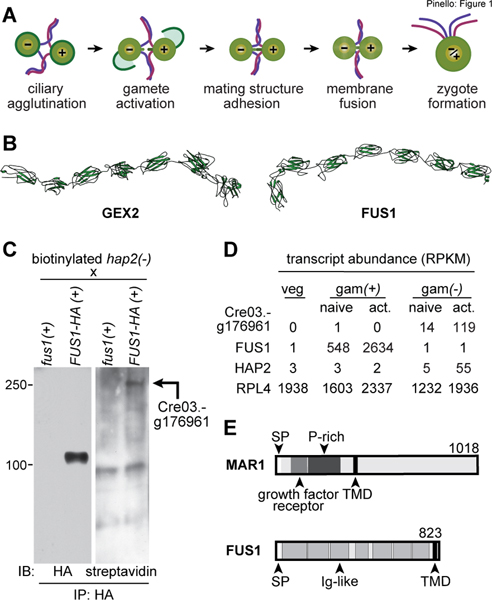
Figure 1. Relationship between FUS1 and plant sperm adhesion protein GEX2 and identification of a FUS1 binding protein in minus gametes.
(A) Illustration depicting steps in Chlamydomonas fertilization. (B) RaptorX structural models of the ectodomains of A. thaliana GEX2 (residues 28–1003) and C. reinhardtii FUS1 (residues 17–791). The N-terminus for each model is on the left. (C) Cre03.g176961 encodes a FUS1-HA-binding protein on minus gametes. Immunoblots (IB) probed with anti-HA antibodies (left) or streptavidin-HRP (right) showing anti-HA immunoprecipitates (IP) of lysate supernatants from live, biotinylated hap2(−) gametes that had been mixed with fus1(+) or FUS1-HA(+) gametes. Mass spectrometry identified the Cre03.g176961 biotinylated protein in the FUS1-HA(+) immunoprecipitates (Data S2). (D) Cre03.g176961 transcripts are specifically expressed in minus gametes. RPKM (Reads Per Kilobase per Million mapped reads) data from Ning et al. (2013) of the indicated transcripts (left) in vegetative cells (veg) and in naive and activated plus (gam(+)) and minus gametes (gam(−)). Results for constitutively expressed ribosomal protein RPL4 (Cre09.g397697) are also shown. (E) Domain architectures of MAR1 (top) and FUS1 (bottom) proteins, showing the signal peptide (SP) transmembrane domain (TMD) in each, the Ig-like folds (Ig-like) in FUS1, and the growth factor receptor cysteine-rich domain and proline-rich region (P-rich) in MAR1. (See also Table S1, Fig. S1, and Data S1, S2.)
Here, we report that Chlamydomonas FUS1 and previously described plant sperm adhesion protein, GEX2, are members of a broadly conserved protein family characterized by extracellular domains composed almost entirely of Ig-like domains. We identify a new, lineage-restricted protein on minus gametes, MAR1, that is a receptor for FUS1 on plus gametes. Formation of the FUS1-MAR1 pair is essential for the gamete membrane attachment that immediately precedes bilayer merger and is necessary for gamete fusion. Moreover, we find that MAR1 is biochemically associated with HAP2 and is required for proper HAP2 expression and localization at the minus mating structure. Thus, during the gamete membrane fusion reaction in Chlamydomonas, a lineage-specific protein, MAR1, functions at the nexus of conserved membrane adhesion protein, FUS1, and ancient membrane fusogen, HAP2.
Results
Chlamydomonas FUS1 and plant gamete adhesion protein, GEX2, share a common ectodomain architecture
We used the more powerful protein analysis methods available now to update the earlier report that the FUS1 ectodomain contained 5 Ig-like domains (Misamore et al., 2003). Our new analysis uncovered FUS1 orthologs in several other algal species (Yamamoto et al., 2021) and identified two more Ig-like domains in FUS1, for a total of 7 (Fig. 1B and Table S1). To our surprise, DELTA-BLAST, PSI-BLAST, and PROMALS3D analyses identified sequence and secondary structure similarities between the entire ectodomains of Chlamydomonas FUS1 and plant GEX2 sequences (including those of Arabidopsis thaliana, Oryza sativa, and Triticum dicoccoides; Table S1 and Data S1). Furthermore, in an HHPred query of the A. thaliana proteome with the FUS1 ectodomain, GEX2 was the top hit (E-value: 2.2e-13, Identities: 15%) and in an HHPred query of the Chlamydomonas proteome with the GEX2 ectodomain, FUS1 was the top hit (E-value: 8.7e-26, Identities: 16%).
Structural modeling using the coevolution-independent modeler, RaptorX contact prediction (Xu et al., 2021), showed similar tertiary structures for Chlamydomonas FUS1 and Arabidopsis GEX2 ectodomains (Fig. 1B and Table S1). Template-based homology modeling on all platforms tested (PHYRE2, SWISS-MODEL and HHPRED) found that Chlamydomonas FUS1 and Arabidopsis GEX2 have strong structural homologies to the same structures in the protein data bank, namely, the Dictyostelium gelation factor rod domain and bacterial invasin-intimin-like proteins, thereby demonstrating structural uniformity among their Ig-like domains (Fig S1). Taken together, our results indicate that FUS1 and GEX2 belong to a family of gamete cell surface adhesion proteins whose ectodomains are constituted by Ig-like domains.
A minus gamete protein that binds to FUS1
We took advantage of the ease of preparing biochemical quantities of pure gametes in Chlamydomonas to identify proteins on minus [(−)] gametes that bound to FUS1 on plus [(+)] gametes. hap2(−) gametes were surface labeled with biotin, mixed with fus1::FUS1-HA(+) gametes, and immunoprecipitation, immunoblotting, and mass spectrometry were used to identify biotinylated minus gamete proteins that associated with FUS1-HA. Use of the fusion-defective hap2(−) gametes to block fertilization at the stage of mating structure adhesion maximized opportunities for adhesion protein interactions and prevented the degradation of FUS1 that is coincident with gamete fusion (Liu et al., 2010). As a control, we also mixed biotinylated hap2(−) gametes with fus1(+) gametes, which lack FUS1. FUS1-HA was pulled down efficiently by anti-HA antibody, and as expected, staining was absent in the fus1 control samples (Fig. 1C, left panel). Importantly, streptavidin blotting showed a biotinylated protein migrating with an apparent molecular mass of ~250 kDa that was co-immunoprecipitated only in the FUS1-HA-containing sample (Fig. 1C, right panel).
Specific expression of Cre03.g176961 in minus gametes
The protein encoded by gene Cre03.g176961 exhibited high coverage in mass spectrometry analysis of the 250 kDa region of samples from FUS1 immunoprecipitates and was absent in fus1 samples (Data S2). Importantly, previous gene expression studies in Chlamydomonas vegetative cells and naive and activated gametes (Ning et al., 2013) showed that Cre03.g176961 transcripts were uniquely present in minus gametes and upregulated upon activation (Fig. 1D). This expression signature is similar to that of other fertilization-essential transcripts in minus gametes (Ning et al., 2013), including HAP2, and distinct from those of plus-specific FUS1 and constitutively expressed ribosomal transcript RPL4 (Fig. 1D), making the Cre03.g176961 protein, hereafter called Minus Adhesion Receptor 1 (MAR1), a strong candidate for a FUS1 binding partner.
Based on the annotation in Phytozome (Goodstein et al., 2012), and confirmatory RT-PCR characterization of the transcript, MAR1 (GenBank: KT288268) encodes a 1018-residue, single-pass transmembrane protein with a predicted molecular mass of 99 kDa. Analysis of the amino acid sequence predicted a signal peptide, a 365-residue ectodomain, a single transmembrane domain, and a 608-residue cytoplasmic domain (Fig. 1E; scale-drawing of FUS1 shown for comparison). Two notable features of the MAR1 ectodomain are a growth factor receptor cysteine-rich domain near the N-terminus (residues 78–159) and a proline-rich segment (residues 142 to 365) that includes 5 repeats of a “PPSPX” motif. The latter motif was previously identified in other Chlamydomonas proteins, including a cell wall protein, GP1 (Ferris et al., 2001); and the ciliary adhesion proteins, SAG1 and SAD1 (Ferris et al., 2005). In the MAR1 cytoplasmic domain, structural homology searches found segments with likeness to the ABA receptor kinase from Arabidopsis (PDB: 5XD6, residues 512–620), and collagen (PDB: 1YGV, residues 619–1018). Detectable MAR1 sequence homologies in other organisms, however, were scant, with only a small number of algal relatives containing putative MAR1 orthologs (Data S2).
We introduced a FLAG-tagged MAR1 transgene driven by its endogenous promoter into hap2(−) cells, and hap2::HAP2-HA(−) cells. The MAR1-FLAG protein was expressed in minus gametes and absent in minus vegetative cells (Fig. 2A). Consistent with the biotinylation studies, and in spite of a predicted molecular mass of 107 kDa, MAR1-FLAG appeared as a ~250 kDa protein in SDS-PAGE. We are uncertain of the basis for this anomalous behavior, but the several proline-rich regions might contribute to its altered migration pattern (Ferris et al., 2001). Immunoblot analyses of lysates from equal numbers of transgene-expressing gametes indicated that the three gamete-specific proteins, MAR1, HAP2 and FUS1, were expressed at similar levels (Data S2).
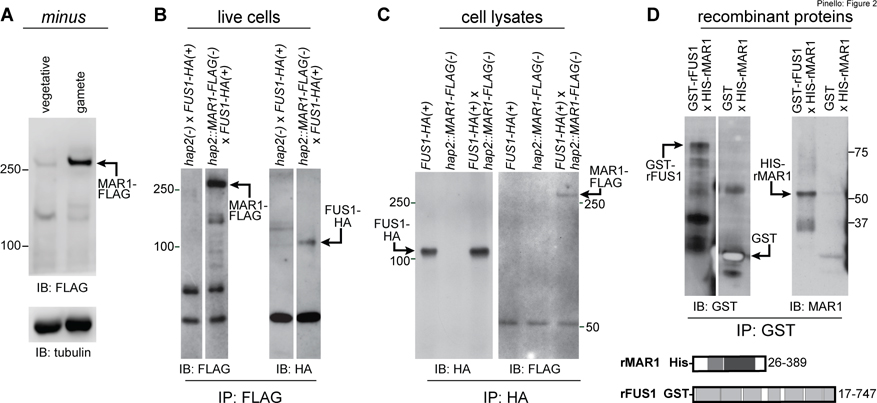
Figure 2. MAR1 on minus gametes directly interacts with FUS1 on plus gametes.
(A) MAR1-FLAG is enriched in minus gametes compared to minus vegetative cells. Immunoblot (IB) with anti-FLAG antibodies showing equal cell equivalents of hap2::MAR1-FLAG; HAP2-HA(−) vegetative cells and gametes (upper panel). Lower panel shows a tubulin loading control. (B) MAR1-FLAG interacts with FUS1-HA during mating structure adhesion. FUS1-HA(+) gametes were mixed for 30 min with hap2::MAR1-FLAG(−) or hap2(−) gametes, and lysate supernatants from the mixed gametes were subjected to immunoprecipitation (IP) with anti-FLAG antibodies followed by immunoblotting with anti-FLAG (left) or anti-HA antibodies (right). (C) FUS1 and MAR1 interact with each other when separately prepared cell lysates are mixed. Separately lysed mixtures of FUS1-HA(+) with hap2(−) gametes and hap2::MAR1-FLAG(−) with fus1(+) gametes were either kept separate (controls; left and middle lanes) or mixed together (right lane) and subjected to immunoprecipitation with anti-HA antibodies followed by immunoblotting with anti-HA (left) or anti-FLAG (right) antibodies (see also Fig. S2). (D) The recombinant GST-tagged ectodomain of FUS1 (GST-rFUS1) interacts with the recombinant His-tagged ectodomain of MAR1 (His-rMAR1). GST-rFUS1 and GST protein in bacterial lysates were bound to glutathione beads followed by incubation of those beads with lysates from bacteria expressing His-rMAR1. Proteins eluted with reduced glutathione were immunoblotted with anti-GST (upper left) or anti-MAR1 antibodies (upper right). Illustrations depicting the ectodomain residues in His-rMAR1 and GST-rFUS1 recombinant proteins (lower panel). White spaces indicate superfluous lanes digitally eliminated in blots in B and D. Protein interaction experiments were carried out at least 3 times.
MAR1 is a receptor for FUS1
In vivo and in vitro protein interaction assays were consistent with the biotinylation results and showed that MAR1 and FUS1 bound to each other. In in vivo assays with live cell mixtures, fus1::FUS1-HA(+) gametes were mixed with hap2::MAR1-FLAG(−) gametes for 30 minutes to allow gamete activation and mating structure adhesion without membrane fusion, followed by disruption of the samples in lysis buffer, immunoprecipitation with anti-FLAG antibody, and immunoblotting with anti-HA antibody. hap2(−) gametes (i. e., expressing only un-tagged endogenous MAR1) mixed with fus1::FUS1-HA(+) gametes were used as a control. As shown in Fig. 2B, FUS1-HA was present in the FLAG immunoprecipitates of the hap2::MAR1-FLAG(−) gametes that had been mixed with fus1::FUS1-HA(+) gametes, but not in the control immunoprecipitates. Related experiments, in which separately prepared detergent lysates of activated hap2::MAR1-FLAG(−) gametes and fus1::FUS1-HA(+) gametes were mixed together followed by immunoprecipitation and immunoblotting as above, also showed that the two proteins were associated with each other (Fig. 2C). Additional experiments with lysates of cells expressing IFT172-FLAG mixed with FUS-HA lysates and with lysates of cells expressing IMP3-HA mixed with MAR1-FLAG lysates demonstrated the specificity of the interaction between MAR1-FLAG and FUS1-HA (Fig. S2). Finally, we found that bacterially expressed, His-tagged MAR1 ectodomain (His-MAR1) was precipitated by the GST-tagged FUS1 ectodomain (rFUS1) but not by GST alone (Fig. 2D). Together, these results demonstrated that the ectodomains of MAR1 and FUS1 directly interacted with each other.
Mating structure adhesion is severely impaired in mar1 minus gametes and fusion is blocked
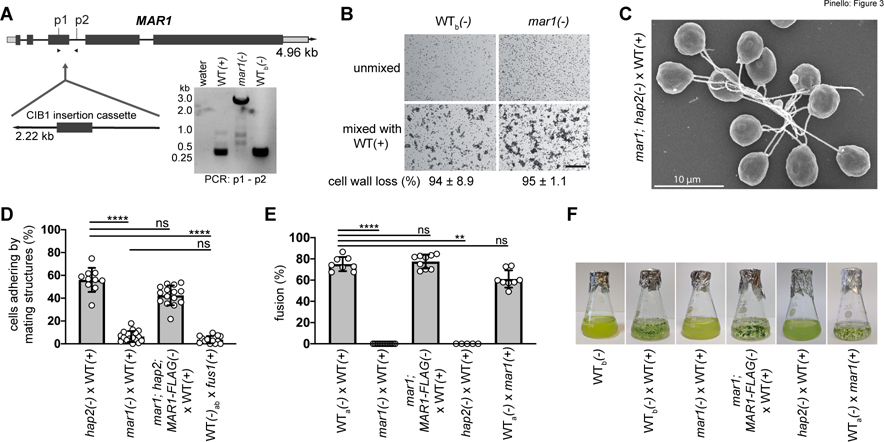
To test whether the MAR1 and FUS1 interaction detected biochemically was important functionally, we examined the fertilization phenotypes of a Chlamydomonas CLiP library mutant minus strain, LMJ.RY0402.185187 (hereafter, called mar1), which is annotated to have an insertion in the MAR1 gene (Li et al., 2019). PCR across the predicted insertion site showed that the CIB1 plasmid had indeed inserted into the 3rd exon of MAR1, thereby disrupting its coding sequence (Fig. 3A, Fig. S3). The mar1(−) mutant cells showed no detectable morphological or growth phenotype when cultured as vegetative cells. Furthermore, when placed into N-free medium overnight, the mar1(−) cells differentiated into minus gametes that were fully capable of ciliary adhesion with wild type (WT) plus gametes upon mixing, forming characteristic, large clusters of plus and minus gametes adhering to each other by their cilia (Fig. 3B low magnification DIC images; Fig. 3C SEM image). Gamete activation, as quantitatively assessed by cell wall loss, was also indistinguishable in mixtures of mar1(−) gametes or WT(−) control gametes with WT(+) gametes (Fig. 3B, bottom).
Figure 3. MAR1 is essential for mating structure adhesion and gamete fusion.
(A) Illustration of the MAR1 gene showing the location of the disrupting CIB1 insertion cassette in the mar1(−) mutant (CLiP: LMJ.RY0402.185187). Dark grey rectangles and lines represent exons and introns, respectively. Light grey rectangles represent 5’ and 3’ UTRs. DNA gel electrophoresis image (lower right) shows results of genotyping PCRs across the mar1 insertion site using primers p1 and p2 and DNA template from WT(+), mar1(−), and WT(−) cells, and a water template control. The 2.6 kB amplicons from mar1(−) cells were sequenced to confirm the CIB1 insertion location in the 3rd exon of the MAR1 coding sequence (Fig. S3). (B) Ciliary agglutination and gamete activation are unperturbed in the mar1(−) gametes. Differential interference contrast (DIC) micrographs of live, WTb(−) and mar1(−) gametes before (top) and 10 min after (bottom) mixing with WT(+) gametes; scale bar 0.2 mm. The large clusters of cells visible in the bottom images form when multiple cells interact with each other by their cilia. The percent of cells that lost their cell walls after mixing (4 biological replicates) is indicated below the images (± SD). (C) Scanning electron micrograph of mar1; hap2(−) gametes experiencing ciliary adhesion with WT(+) gametes. (D) mar1(−) gametes fail to undergo mating structure adhesion with WT(+) gametes. Results are from 10 to 16 biological replicates per group analyzed using one-way non-parametric ANOVA with Dunn’s post-test; ****P<0.0001; ns, not significant; bars are means; error bars, SD. (E-F) MAR1 is required for gamete fusion. Gamete fusion and zygote formation were assessed microscopically at 10 min (E) and macroscopically at 1–3 days (F) after mixing of the indicated gametes. For (E), results from 5 to 14 biological replicates per group were analyzed using one-way non-parametric ANOVA with Dunn’s post-test; ****P<0.0001 and **P=0.0027; bars are means; error bars, SD. Images in (F) are representative of one to three biological replicates. 21gr was the WT(+) gamete strain used as controls in these experiments. WT(−) gametes used as controls were hap2::HAP2-HA(−) (WTa) and parental CLiP mutant strain, CC-5325(−) (WTb).
mar1(−) gametes, however, were severely impaired in their ability to undergo mating structure adhesion with WT(+) gametes. Whereas a mean of 56% of cells adhered by their mating structures in the fusion-blocked mixtures of hap2(−) gametes with WT(+) gametes, only 6% adhered in the mixtures of mar1(−) gametes with WT(+) gametes. This adhesion-defective phenotype was indistinguishable from that observed in mixtures of WT(−) gametes mixed with fus1(+) gametes (Fig. 3D and Misamore et al., 2003). Introduction of the MAR1-FLAG transgene into mar1(−) gametes also harboring the hap2 gene-disruption, rescued mating structure adhesion with WT(+) gametes to levels analogous to that of mixtures of hap2(−) and WT(+) gametes. These results demonstrated a central role for MAR1 in mating structure adhesion.
Notably, and mirroring the phenotypes observed in both the adhesion-defective fus1(+) gametes (Ferris et al., 1996; Misamore et al., 2003) and fusion-defective hap2(−) gametes, loss of MAR1 completely abrogated gamete fusion as quantified by counts of quadri-ciliated zygotes at 10–30 min after mixing (Fig. 3E, 10 min results) and by visual examination after overnight incubation to detect appearance of immotile zygotes, which appear as heterogeneous, green flocculant material in liquid cultures (Fig. 3F). The fusion defect of mar1(−) gametes was rescued with the MAR1-FLAG transgene (Fig. 3E, F), and the extent of fusion 10 minutes after mixing mar1;MAR1-FLAG(−) gametes with WT(+) gametes (77% fusion) was indistinguishable from that in mixtures of WT gametes (75% fusion), consistent with the results of the macroscopic assay (Fig. 3F). As expected, and demonstrating that MAR1 functions only in minus gametes during fertilization, plus gametes bearing the mar1 mutation were fully competent to fuse with WT(−) gametes (61%). These results showed that MAR1 was essential in minus gametes for zygote formation.
Furthermore, analysis of F1 and F2 progeny from crosses of MAR1-FLAG-rescued mar1(−) gametes mixed with plus gametes (Fig. S3) confirmed that the adhesion- and fusion-defective phenotypes of mar1(−) gametes co-segregated with the mar1 mutant genotype. As expected, crosses of mar1(−) gametes with WT(+) gametes failed to produce any progeny. Taken together, our biochemical and genetic results demonstrated that the direct interaction of mating structure membrane proteins MAR1 on minus gametes and FUS1 on plus gametes mediated the membrane attachment between the two gametes that occurs after their mutual recognition and activation, and that formation of this receptor pair was essential for the gamete membrane fusion reaction.
MAR1-FLAG is localized at the minus mating structure and is lost rapidly after gamete fusion
Confirming that MAR1 functions at the cell surface, brief incubation of live hap2::MAR1-FLAG;HAP2-HA(−) gametes with pronase resulted in loss of nearly all of MAR1-FLAG (Fig. 4A, top), and as expected, only the surface-expressed upper isoform (S) of HAP2 (Fig. 4A, middle, Liu et al., 2008). Immunostaining of mar1;MAR1-FLAG;hap2;HAP2-HA(−) gametes with anti-FLAG antibodies further showed that MAR1-FLAG was present as a punctum between the two cilia at the apical ends of the minus gametes (Fig. 4B), the location of the minus mating structure. MAR1-FLAG remained localized at the minus mating structure after gamete activation (Fig. S4). Double immunostaining of mar1;MAR1-FLAG;hap2;HAP2-HA(−) gametes with anti-HA and anti-FLAG antibodies further confirmed their co-localization at the minus mating structure (Fig. 4C). Finally, similar to loss of HAP2 and FUS1 after fusion, and as part of a block to polygamy (Liu et al., 2010), 30 minutes after ~70% of mar1::MAR1-FLAG(−) gametes had fused with WT(+) gametes, MAR1-FLAG had become nearly undetectable (Fig. 4D).
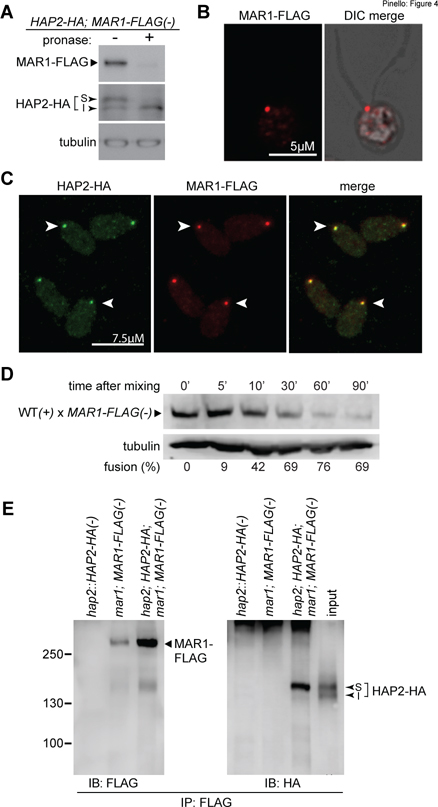
Figure 4. MAR1 co-localizes with HAP2 on the surface of the minus mating structure and is biochemically associated with the fusogen.
(A) MAR1-FLAG is expressed at the cell surface. Live hap2::HAP2-HA; MAR1-FLAG(−) gametes were incubated with or without pronase (0.05%) for 20 min followed by immunoblotting with anti-FLAG and anti-HA antibodies. The surface (S) and internal (I) isoforms of HAP2-HA are indicated. (B) MAR1-FLAG is localized at the minus mating structure at the apical end of minus gametes between the two cilia. Fluorescence alone and fluorescence-merged DIC single z-section images of a naive mar1;MAR1-FLAG;hap2;HAP2-HA(−) gamete immunostained with anti-FLAG antibodies (see also Fig. S4). (C) MAR1-FLAG staining is coincident with HAP2-HA at the minus gamete mating structure (arrowheads). mar1;MAR1-FLAG;hap2;HAP2-HA(−) gametes were immunostained with anti-FLAG and anti-HA antibodies. (D) MAR1-FLAG is rapidly lost after gamete fusion. mar1::MAR1-FLAG(−) gametes were mixed with WT(+) gametes and assayed for fusion and for MAR1-FLAG at the indicated times after mixing. The lower panel shows tubulin loading controls. Percent fusion is shown below the blots. (E) MAR1 is biochemically associated with HAP2. Minus gametes expressing both MAR1-FLAG and HAP2-HA were activated by mixing with adhesion-defective fus1(+) gametes for 60 min, followed by immunoprecipitation with anti-FLAG antibodies and immunoblotting with anti-FLAG (left panel) or anti-HA (right panel) antibodies. Strains lacking either MAR1-FLAG or HAP2-HA were controls; input (represents ~2.6% of the cell equivalents loaded in the eluate lane) for the mar1;MAR1-FLAG;hap2;HAP2-HA(−) sample is shown on the right panel. HAP2-MAR1 interaction experiments were carried out at least 4 times. Surface (S) and internal (I) isoforms of HAP2-HA are indicated.
Biochemical and functional assays demonstrate an association between MAR1 and fusogen HAP2
To test for a biochemical interaction between these temporally co-functioning, co-localized proteins, we immunoprecipitated MAR1-FLAG from the lysates of activated minus gametes expressing both MAR1-FLAG and HAP2-HA and used immunoblotting to assess whether HAP2-HA was also present in the immunoprecipitates. As shown in Fig. 4E, HAP2-HA was indeed present in the anti-FLAG immunoprecipitates, but absent in control immunoprecipitates from activated hap2::HAP2-HA(−) gametes (lacking MAR1-FLAG), and activated mar1; MAR1-FLAG(−) gametes (lacking HAP2-HA). Notably, we consistently found that MAR1 preferentially associated with the upper, membrane-surface-expressed isoform (S) of HAP2 (Fig. 4E). Despite the association, MAR1-FLAG expression (Fig. 5A) and localization (Fig. 5B) in hap2(−) gametes were indistinguishable from that of minus gametes expressing WT HAP2-HA. Furthermore, as reported here (Fig. 3D) and previously (Feng et al., 2018; Liu et al., 2008; Zhang et al., 2021), hap2(−) gametes were fully capable of undergoing mating structure adhesion with plus gametes. Thus, MAR1 expression, localization, and function were independent of HAP2.
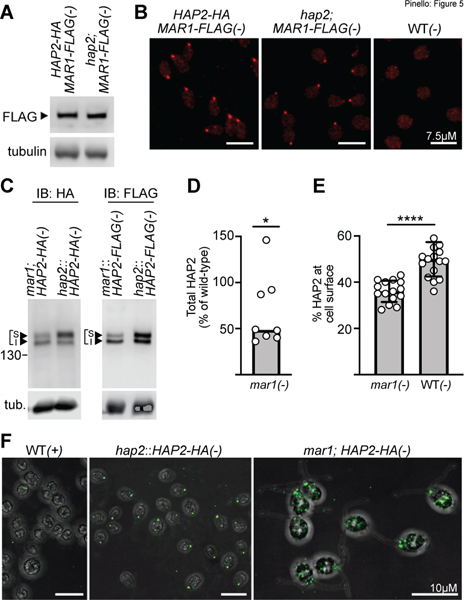
Figure 5. HAP2 expression and localization depend on MAR1, but MAR1 properties are independent of HAP2.
(A-B) MAR1 expression and localization are independent of HAP2. (A) Immunoblots of MAR1-FLAG (upper panel) in lysates of mar1;MAR1-FLAG;hap2;HAP2-HA(−) gametes (left side) and mar1;MAR1-FLAG;hap2(−) gametes (right side). Tubulin is shown as a loading control (lower panel). (B) MAR1-FLAG immunolocalization in mar1;MAR1-FLAG;hap2;HAP2-HA(−) gametes (left), and mar1;hap2;MAR1-FLAG(−) gametes (middle) by fluorescence microscopy. Negative control (right) was WT(−) gametes expressing neither HAP2-HA nor MAR1-FLAG. (C-F) HAP2 expression and mating structure localization are impaired in minus gametes lacking MAR1. (C) Representative immunoblots from lysates of HAP2-HA-expressing (left panel) or HAP2-FLAG-expressing (right panel) minus gametes in the absence (left lanes) or presence (right lanes) of WT MAR1. Controls (right lanes) were hap2::HAP2-HA(−) and hap2::HAP2-FLAG(−) gametes which contained the endogenous WT MAR1 gene; lower panel shows tubulin (tub) loading controls. (D) Quantification of total HAP2-HA and HAP2-FLAG in immunoblots that was normalized to tubulin for each sample. Median HAP2 expression (bar) in mar1(−) gametes was 49% of that in minus gametes expressing WT MAR1; Wilcoxon Signed Rank test, *P=0.04. Each open circle represents a biological replicate mar1(−) sample (n=8) and is expressed as a percentage of the median signal present in WT MAR1(−) samples (n=5). (E) Quantification of the percentage of total HAP2 present in the upper isoform of the HAP2 doublet that was determined from immunoblot signals of HAP2-HA or HAP2-FLAG expressed in mar1(−) or WT MAR1(−) gametes. The mean percent of cell surface HAP2 in mar1(−) gametes was 36% (n=15) compared to 50% in WT MAR1(−) gametes (n=17); two-tailed unpaired t test with Welch’s correction; ****P<0.0001; bars are mean; error bars, SD. (F) Confocal z-stack composite images of anti-HA immunostained mar1;HAP2-HA(−) gametes (lacking MAR1, right), hap2::HAP2-HA(−) gametes (containing WT MAR1, middle), and control WT(+) gametes (left). The HA immunofluorescence of the mar1;HAP2-HA(−) gametes was substantially dimmer than that of the hap2::HAP2-HA(−) gametes, and the relative brightness was increased here to allow visualization of the altered HAP2-HA distribution (see also Fig. S5).
On the other hand, the expression and localization of HAP2 in minus gametes lacking MAR1 were substantially altered. Total HAP2 protein expression in mar1;HAP2-HA(−) and mar1::HAP2-FLAG(−) gametes was reduced 2-fold compared to that of strains expressing HAP2 in a WT MAR1 background (representative blots, Fig. 5C and Fig. S5; quantification of multiple independent experiments, Fig. 5D). Moreover, quantitative densitometry measurements of immunoblots across multiple biological replicates also showed that the mean percentage of HAP2 present in the upper, surface-expressed isoform was reduced from 50% in WT(−) gametes to 36% in the mar1(−) gametes, representing a reduction of 28% (Fig. 5E; Fig. S5).
Absence of MAR1 also substantially altered HAP2 localization. Immunofluorescent staining and confocal microscopy of HAP2-HA in minus gametes containing WT MAR1 showed that the predominant localization of HAP2-HA was, as shown above (Fig. 4C) and previously (Fedry et al., 2017; Feng et al., 2018; Liu et al., 2010; Zhang et al., 2021), as a single spot at the site of the minus mating structure (Fig. 5F, middle panel, Fig. S5). Low intensity, non-specific anti-HA staining was observed in negative control WT(+) gamete samples (Fig. 5F, left panel), as well as in small amounts within cells and elsewhere on the slide in other samples (as shown above in Figs. 4C). On the other hand, HAP2-HA localization in minus gametes lacking MAR1 was strikingly different. Consistent with the immunoblotting results, the immunofluorescence signal in mar1(−) gametes was much lower (Fig. S5) and was only infrequently detected as a single apically-localized spot. Fig. 5F, right panel shows an image whose relative brightness was increased to allow visualization of the cellular localization of this reduced amount of HAP2-HA. Often, HAP2-HA in the mar1(−) gametes appeared as smaller puncta close to the site of the mating structure, but was predominately found throughout the cell body as multiple, discrete puncta of varying sizes (Fig. 5F, Fig. S5). Thus, proper expression and localization of HAP2-HA at the minus mating structure depended upon MAR1.
Discussion
We investigated the proteins and protein interactions that underlie membrane adhesion and bilayer merger during the gamete membrane fusion reaction in Chlamydomonas. We determined that the ectodomain of FUS1, the previously identified adhesion protein on the plus gamete mating structure, is predicted to be constituted entirely by a linear array of Ig-like domains, a domain architecture that BLAST searches and structural modeling show is similar to that of GEX2, a gamete adhesion protein present throughout land plants. We also identified a lineage-restricted, minus gamete-specific membrane protein, MAR1, that is localized at the surface of the minus mating structure and that binds directly to FUS1. Like FUS1, MAR1 is essential for mating structure adhesion and for gamete fusion. These interacting proteins are now the second gamete membrane adhesion receptor pair demonstrated to be essential for fertilization (Bianchi et al., 2014). In addition to interacting with FUS1, MAR1 is also associated with HAP2 on minus gametes and essential for proper HAP2 expression and localization during gametogenesis. Our results suggest that the membrane adhesion immediately preceding HAP2-dependent bilayer merger during fertilization in organisms across Viridiplantae is mediated by an interaction between a member of the conserved FUS1/GEX2 family of adhesion proteins and a lineage-restricted binding partner on the cognate gamete.
FUS1/GEX2 joins HAP2 and KAR5/GEX1 to make a trio of protein families central to sexual reproduction in organisms from unicellular green algae to flowering plants
The Immunoglobulin Superfamily (IgSF) is large and diverse, and many of its members function in intercellular adhesion in prokaryotes and eukaryotes (Chatterjee et al., 2020; Honig and Shapiro, 2020). Several proteins important for sperm-egg interactions in metazoans possess Ig-like domains (Nishimura and L’Hernault, 2016), the best characterized being the sperm adhesion protein IZUMO1, whose single Ig-like domain forms a portion of the interface between IZUMO1 and its egg binding partner, JUNO (Aydin et al., 2016; Ohto et al., 2016). The previously reported presence of Ig-like domains in Chlamydomonas FUS1 (Misamore et al., 2003) and plant GEX2 (Mori et al., 2014) called further attention to the widespread use these domains in sexual reproduction. Our findings here indicate, however, that the Ig-like domains constitute the entire functional portion of FUS1 and nearly the entire ectodomain of GEX2. The expansive binding repertoire of vertebrate antibody proteins, which are composed entirely of immunoglobulin domains, is provided by structural variations in a subset of their domains. Future structural studies should provide insights into whether similar variations in particular Ig-like domains of FUS1/GEX2 family members underlie their ability to interact with gamete receptor proteins across taxa.
Remarkably, uncovering the breadth of conservation within this family of gamete adhesion proteins now indicates that at least three of the core functions unique to eukaryotic sexual reproduction are carried out by three protein families conserved across all plant lineages: (1) gamete membrane adhesion mediated by FUS1/GEX2 family members, (2) membrane merger mediated by HAP2 family members, and (3) pronuclear fusion mediated by the KAR5/GEX1 family of nuclear envelope proteins (Mori et al., 2014; Ning et al., 2013; Wong and Johnson, 2010). The HAP2 and KAR5/GEX1 families are not restricted to plant lineages and are essential for sexual reproduction in organisms from green algae and protists to multicellular plants and animals. The conservation of domain architectures within members of each of these three families suggests that while the evolutionary pressures on reproductive proteins often lead to rapid changes in their primary amino acid sequences, this diversification may take place on a relatively unchanging structural backbone (Ferris et al., 1997; Mori et al., 2014; Swanson and Vacquier, 2002).
MAR1 is bifunctional and interacts with FUS1 on plus gametes and HAP2 on minus gametes during the membrane fusion reaction
Our biochemical results with endogenous proteins in gamete lysates and with bacterially expressed proteins indicated that interaction between FUS1 and MAR1 ectodomains was independent of Chlamydomonas-specific post-translational modifications and that it was direct. One potential binding site in the MAR1 ectodomain could be within its proline-rich region, which contains several PPSPX repeats akin to the poly-proline repeats of algal and higher plant hydroxyproline-rich glycoproteins (HPRGs, Ferris et al., 2005; Ferris et al., 2001) that support other types of protein-protein interactions. The MAR1 growth factor receptor domain, which is similar to growth factor motifs found in other gamete interaction proteins such as a mating type protein in the ciliate, T. thermophila (Cervantes et al., 2013); and Spe-9 in C. elegans (Singson et al., 1998), could also be a potential protein interaction site.
Furthermore, results from several independent methods were all consistent with a biochemical and functional association between MAR1 and the class II fusogen HAP2 on the minus mating structure. Total HAP2 expression as well as the portion of HAP2 at the cell-surface were reduced in minus gametes lacking MAR1, HAP2 was mis-localized in gametes lacking MAR1, and MAR1 was biochemically associated with the cell-surface form of HAP2. One interpretation of these findings is that, like the partner protein interactors of viral class II fusion proteins, MAR1 participates in biosynthesis and trafficking of HAP2 to the minus mating structure. For example, in the absence of their partner E2 proteins, the E1 fusogens of Sindbis virus and Semliki Forest Virus are inefficiently transported to the plasma membranes of their host cells (Andersson et al., 1997; Carleton et al., 1997). Independently of a role in trafficking, MAR1 might also stabilize HAP2 at the mating structure.
It will be important to determine whether the MAR1-HAP2 association is direct or indirect and to identify the regions of MAR1 and HAP2 that participate in this association. If the interaction is indirect, a handful of Chlamydomonas membrane proteins whose transcripts show minus gamete-specific expression similar to MAR1 (Ning et al., 2013) would be good candidates for a linking protein; including Cre03.g175926, which has some sequence similarity to MAR1. If the interaction is direct, possible cytoplasmic domain interaction sites within HAP2 and MAR1 are a 237-residue segment of HAP2 essential for its mating structure localization (Liu et al., 2015) and a 400-residue segment within MAR1 with a predicted structural likeness to collagen.
Before a structure was available, Wong et al. (2010) demonstrated that the fusion capacity of Arabidopsis HAP2 was retained when its ectodomain was exchanged with the HAP2 ectodomain from a closely related species, but was not when the swap was with the ectodomain of a more distantly related species, indicating that the ectodomain of HAP2 was important in lineage-specific functions. Recent protein structures of the Chlamydomonas trimeric HAP2 ectodomain have brought to light regions within loops of domain I that may participate in such lineage-specific protein-protein interactions (Baquero et al., 2019; Feng et al., 2018). Furthermore, comparisons between the HAP2 structures of Chlamydomonas, Arabidopsis, and Trypanosoma cruzi showed substantial, taxa-specific differences in their fusion loops, supporting a possible lineage-specific role for protein interactions of this region (Fedry et al., 2018).
Our discovery that the lineage-specific protein MAR1 functions at the nexus of two broadly conserved, fusion-essential protein families leads to the speculation that the cognate gamete adhesion receptor pairs in other organisms might also be composed of one conserved protein and another that is lineage specific. The IZUMO1-JUNO receptor pair would support such a model, as JUNO is specific to mammals, but IZUMO1 is present throughout vertebrates (Bianchi et al., 2014; Grayson, 2015). Even this model is overly simplistic, however, since the FUS1/GEX2 family member in Chlamydomonas is present in the gamete lacking HAP2, whereas the FUS1/GEX2 family members in land plants are present in the gamete expressing HAP2.
Nevertheless, our findings raise the exciting possibility that MAR1 binding to Chlamydomonas FUS1 triggers conformational changes within HAP2 required for its conversion to fusion-driving trimers (Zhang et al., 2021). In class I fusogen-dependent Paramyxoviruses, the interaction of a fusogen-associated adhesion protein with its membrane receptor activates the fusion protein (Navaratnarajah et al., 2020). Future studies on the FUS1-MAR1-HAP2 axis have the potential to illuminate new, conserved regions in HAP2 that act before its function in bilayer merger in organisms across plant taxa as well as in the many unicellular organisms and metazoans that also depend on HAP2 for gamete fusion.
Limitations of the study
Given that FUS1 and HAP2 are each members of conserved protein families that function in the gamete membrane fusion reaction, it is possible that they also interact directly with each other. Such an interaction could be essential for the fusion reaction but would have gone undetected in these studies. Also, our studies were focused on the role of MAR1 in HAP2 cellular properties and functions before plus and minus gametes bind to each other at their mating structures, but our results are silent on whether FUS1 binding to MAR1 alters the MAR1-HAP2 interaction. In addition, MAR1 or the MAR1-HAP2 association might contribute to structural properties of the minus mating structure that are important for fusion but that can be determined by ultrastructural studies of cells or by structural studies with proteins expressed in vitro.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. William J. Snell (wsnell1@umd.edu)
Materials availability
Materials generated in this study including plasmids and new Chlamydomonas strains are available upon request from the lead contact or direct purchase from the Chlamydomonas Resource center (https://www.chlamycollection.org/).
Data and code availability
All other data reported in this paper are available from the lead contact upon request.
No large-scale datasets or new code were generated in this study.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Chlamydomonas cell culture
Cells were grown vegetatively in liquid TAP medium under a 13:11 h light-dark cycle at 22°C. Gametogenesis was induced by transferring vegetative cells into N-free medium followed by overnight agitation on a shaker in continuous light.
Chlamydomonas stocks
Existing Chlamydomonas strains used for experiments were 21gr(+), fus1(+), fus1::FUS1-HA(+), hap2(−), hap2::HAP2-HA(−), CC-5313(+), mar1(−) also called LMJ.RY0402.185187(−), and CC-5325(−). Transgenic strains generated in this study and used for experiments were made by stable transformation and crosses of the above strains. The following transgenic Chlamydomonas strains were generated and are designated as follows by numeric or alphanumeric codes in brackets [with coded independently-isolated strains separated by commas] within the given genotypes: mar1;MAR1-FLAG(+)[3d-1]; mar1::MAR1-FLAG(−) [MF-c7]; mar1;MAR1-FLAG(−)[111,115,147]; mar1(−)[106, 133, 144]; mar1;hap2(−)[73; 98]; mar1;hap2(+)[148,156]; mar1;hap2; MAR1-FLAG(−)[78,146]; mar1;HAP2-HA(−)[122]; mar1;hap2;HAP2-HA(−)[132]; mar1::HAP2-FLAG(−)[HF-6, HF-10]; hap2::HAP2-HA;MAR1-FLAG(−)[CC-5284]; and mar1;MAR1-FLAG;hap2;HAP2-HA(−)[102,114,131]. Names denote strain origins and genotype; lowercase italics indicates the presence of a mutant gene, uppercase italics indicates the presence of the wild-type gene, double colons (::) indicate strains where transgene introduction was done through direct transformation by electroporation, and semi-colons (;) indicate the presence of a transgene introduced in this case, through genetic crosses of a previously-transformed parental strain and isolated in a progeny strain. Further details regarding the authentication and descriptions of the transgenic strains used in this study are provided in Figure S3 and Data S4.
Chlamydomonas transformation
Chlamydomonas cells were transformed by electroporation of plasmid DNA encoding the tagged HAP2 or MAR1 transgenes using a BioRad GenePulser Xcell (Shimogawara et al., 1998). SpeI-linearized MAR1-FLAG or SbfI-linearized HAP2-FLAG plasmid DNA were transformed into hap2(−) and mar1(−) cells. The sequences of the plasmid DNAs used for transformation and of PCR products across the insertion site of the mar1(−) CLiP mutant, LMJ.RY0402.185187, were confirmed by analysis at the MGH CCIB DNA core and Eurofins, respectively. Positive transformants were selected for further analysis based on their growth on zeocin (10 μg/mL, Invitrogen) TAP-agar plates, presence of the ble gene product assessed by PCR of Chelex-100-extracted template DNA (Nouemssi et al., 2020) using primers Zeo_F1 and Zeo_R1, and immunoblotting of gamete lysates for expression of the FLAG-tagged proteins. Data S3 and S4 provide further details on plasmid construction, identification of transformants based on growth on selective media, PCR screening, and expression of tagged proteins.
Chlamydomonas crosses and progeny selection
For crosses (Figure S3), equal numbers of the indicated plus and minus gametes were mixed together and plated onto paromomycin (15 μg/mL, Sigma) TAP-agar plates at 0.5, 1, 1.5, and 2 h after mixing. Agar plates from crosses were exposed to continuous light for 16–24 h, then placed in the dark to allow zygote maturation. After at least 7 days in the dark, any remaining unfused cells were scraped towards the edges of the plate with a sterile razor blade and their further growth prevented by exposure of the plate to chloroform vapors for 30–60 sec. The thick cell walls of the zygospores remained attached to the agar and were resistant to the cytolytic effects of the chloroform vapors. After incubation under a 13:11 light-dark cycle at 21°C for 2 weeks to allow germination and visible colony growth, 20 – 30 colonies (each representing the vegetative haploid descendants of 4 initial meiotic progeny) were picked and resuspended in N-free medium with shaking for 1 h to obtain a single-cell suspension, then re-plated onto TAP-agar plates to allow for growth of single colonies composed of individual meiotic progeny. Resulting colonies were screened for mating type, genotype and phenotype by PCR, western blotting, and behavior in mixing assays (Data S4) to isolate the desired genotypes.
METHOD DETAILS
Cell surface biotinylation
To prepare hap2(−) gametes for surface biotinylation, they were activated in db-cAMP buffer for 30 m (Sigma; Misamore et al., 2003) before a 30 m incubation in 1 mg/mL Sulfo-NHS-LC-Biotin (ThermoFisher). Biotinylation was stopped by addition of 20 mM Tris (pH 7.3) followed by 3 washes in N-free medium. Activated, biotinylated hap2(−) gametes were mixed with an equal number of FUS1-HA(+) gametes, or, as a control, with fus1(+) gametes, for 30 m followed by lysis in 4 mL RIPA buffer (20 mM Tris, 150 mM NaCI, 1% NP-40, 0.5% DOC, 0.1% SDS and protease inhibitor cocktail, Wilson et al., 1999) and sonication 3 times (10 s each) on ice. The lysate was cleared by centrifugation for 30 m at 15,000 × g and incubated for 4 h at 4°C with anti-HA monoclonal antibody (Santa Cruz) and protein A sepharose (Sigma). The protein A sepharose was washed 4 × with 1/3 RIPA buffer (20 mM Tris, 150 mM NaCI, 0.33% NP-40, 0.17% DOC, 0.03% SDS with protease inhibitor cocktail) and eluted with 1 mg/mL HA peptide as described previously (Ning et al., 2013), or by boiling in sample buffer. Biotinylated proteins in the eluate were detected on immunoblots using streptavidin-HRP (Jackson ImmunoResearch). The 250 kDa regions of similarly prepared samples were analyzed by mass spectrometry at the Proteomics Core at UT Southwestern Medical Center (Dallas, TX), identifying Cre03.g176961 peptides with 21.8% coverage of the MAR1 sequence (Data S2).
SDS-PAGE and Immunoblotting
For gamete lysates, 2.5 × 107 gametes were suspended in 225 μL N-free medium, and lysed by adding 75 μL of 4x SDS-PAGE sample buffer (1× concentration in sample = 40 mM Tris, pH 6.8, 1% SDS, 5% glycerol, 0.0003% Bromophenol blue, 1 mM EDTA and either 100 mM DTT or 5 mM TCEP) at 95°C for 5 −10 m. 2.5 × 106 cell equivalents were subjected to electrophoresis on SDS polyacrylamide tris-glycine gels (7 or 8%), transferred to PVDF membranes, blocked in 5% milk + TBSt buffer, and probed with either rat anti-HA (1/1000; Sigma, 3F10) or mouse anti-FLAG (1/5000, Sigma, M2) primary antibodies, followed by goat-anti-rat HRP or goat-anti-mouse HRP (1/5000; Millipore) secondary antibodies diluted in 0.5% milk + TBSt buffer. Tubulin was used as a loading control and was detected by probing with mouse anti-α-tubulin primary antibodies (1/5000; Sigma, B512). Immunoblots were developed with SuperSignal West Femto (Thermo Scientific) and exposed to film or scanned with a C-digit blot scanner (LI-COR). Images were processed using Image Studio software (LI-COR) and Adobe Illustrator and Photoshop.
Protease treatments
Protease treatments were performed as described previously (Liu et al., 2010) with minor modifications. To assess surface localization of HAP2-HA and HAP2-FLAG, live minus gametes were treated with 0.05% trypsin for 20 m followed by 3 washes in N-free media with 0.1 mg/mL Trypsin inhibitor from chicken egg white (Sigma) before lysis in SDS-PAGE sample buffer. Surface localization of MAR1-FLAG and HAP2-HA was also assessed by treating live gametes with 1% pronase (1.0 mg/mL, Sigma) for 20 m followed by 3 washes in N-free media with 1 mM PMSF before lysis in SDS-PAGE sample buffer.
Immunoprecipitations
For protein association assays, lysates were prepared from the indicated strains after gamete activation by mixing gametes for 30 – 60 m with fusion-defective fus1(+) or hap2(−) gametes. The mixed gametes were disrupted by lysis in cold detergent buffers supplemented with protease inhibitor cocktail (Roche); RIPA or 1% Triton X-100 (20 mM Tris, 150 mM NaCI, 1% Triton X-100) as indicated, then lysates were cleared by centrifugation for 30 m at 15,000 × g. Detergent supernatants were incubated rotating for 4 h at 4°C with protein A agarose or sepharose and a 1/100 dilution of anti-FLAG (Sigma, mouse M2 monoclonal or rabbit polyclonal) or anti-HA (Santa Cruz, mouse F-7 monoclonal or AbClonal, rabbit polyclonal) antibodies. The immunoprecipitates were either boiled directly in sample buffer or eluted with FLAG peptide (Sigma, 200 μg/mL) or HA peptide (Roche, 1 mg/mL). The presence of the tagged forms of FUS1, HAP2 and MAR1 proteins in minus gamete lysate input samples and immunoprecipitation eluates was assessed by SDS-PAGE followed by immunoblotting.
Recombinant protein production and interaction assays
For recombinant protein production, the expression vectors GST-rFUS1 and HIS-rMAR1 plasmids were transformed into BL21(DE3) (New England Biolabs) competent E. coli cells. Bacterial cells expressing the recombinant proteins were grown with shaking overnight at 37°C in liquid LB media containing antibiotics and then 0.5 mL was inoculated into 50 mL LB cultures. The cultures were shaken for 1 hour at 37°C and induced with IPTG (1 mM) for 3 h at 30°C. Bacteria were lysed in 0.9 mL lysis buffer (20 mM Tris pH 8.0, 150 mM NaCI, 5 mM DTT, 1 mg/mL lysozyme, 5 μg/mL DNase I, 10 μg/mL RNase A, and protease inhibitor cocktail) on ice for 30 m. Triton X-100 was added to 1% (final concentration), then samples were sonicated on ice for 1 m, cleared by centrifugation at 4°C 15,000 × g for 30 m and used in GST pull-down assays. BL21/pGST-FUS1 supernatants were incubated with glutathione beads for 1 hour and washed 3× with washing buffer (20 mM Tris, 150 mM NaCI, 0.3% Triton X-100, protease inhibitor cocktail). The glutathione beads with bound GST-FUS1 were incubated with BL21/pHis-MAR1 lysate overnight at 4°C, washed 3× with washing buffer, and incubated with elution buffer (50 mM Tris pH 8.0, 20 mM reduced glutathione, 0.3% Triton X-100, protease inhibitor cocktail). Recombinant GST protein was used as a control for the recombinant protein interaction assays.
MAR1 polyclonal antibody generation
Antibodies against MAR1 peptides (1 = TQPPRPPWPPRPPPAPPPS, residues 164–182; and 2 = QIPQAPRWPYPQLPSWPPAS, residues 214–233) were generated in rabbits by YenZym Antibodies (Brisbane, CA). Only polyclonal antibodies generated against peptide 1 were used. Antibodies were purified on peptide-conjugated affinity columns and specificity verified by immunoblotting with recombinant protein samples with and without MAR1 (Data S3).
Indirect immunofluorescence microscopy
Immunofluorescent staining of gametes was performed as described previously (Belzile et al., 2013; Feng et al., 2018; Ranjan et al., 2019) with some modifications. Immunofluorescent staining of gametes was performed on samples of approximately 1 × 107 live gametes in N-free medium that were allowed to settle and adhere for 10–30 m on 0.1% poly-L-lysine coated slides or coverslips. Excess media and non-adhered cells were gently removed by pipetting before samples were plunged into ice-cold methanol for 20 m. In some cases, cells were pre-fixed with 4% paraformaldehyde for 10 m prior to methanol fixation to better preserve cilia (Ranjan et al., 2019). For single staining with anti-HA antibodies (1/100, Sigma, 3F10) or anti-FLAG antibodies (1/100, Sigma, M2), gametes were blocked for 30 – 60 m in diluted goat serum (5% goat serum, 0.1% BSA, 0.5% Triton X-100, 1% cold-water fish gelatin in PBS). For double staining with anti-HA (1/100, Roche, 3F10) and anti-FLAG antibodies (1/100, Sigma, M2), gametes were blocked in diluted goat and donkey serum (5% donkey serum, 10% goat serum, 0.1% BSA, 0.5% Triton X-100, 1% cold-water fish gelatin in PBS). Primary antibodies were diluted in blocking sera and incubated with samples for 2 h at 20°C or overnight at 4°C, washed 3× with PBSt (PBS + 0.5% Triton X-100), and incubated with blocking-sera-diluted secondary antibodies for 2 h at 20°C. For anti-HA staining, the secondary antibody used was Alexa Fluor 488-conjugated goat anti-rat IgG (1/400, Invitrogen). For anti-FLAG staining, the secondary antibody used was Alexa Fluor 594-conjugated goat anti-mouse IgG (1/400, Invitrogen). Samples were washed 3× with PBSt and coverslips were mounted using either Fluoromount-G (Southern Biotech) or Prolong Gold antifade reagent (Invitrogen) and sealed with nail polish. Immunofluorescence images of gametes were captured on a Leica SP5 X Laser Scanning confocal microscope with a Leica 63x/1.4 NA oil objective lens. Images are individual z-sections or z-stack composites as indicated in the figure legends and were processed with Adobe photoshop and Leica LASAF software.
Differential interference contrast microscopy
Ciliary adhesion between minus and plus gametes was assessed by differential interference contrast (DIC) microscopy using an inverted Axiovert 135 TV microscope fitted with an ORCA-ER (Hamamatsu) digital camera and a 5x/0.15 NA DIC Epiplan NEOFLUAR objective lens controlled by StreamPix software (NorPix). Scale bars were drawn based on similarly acquired image taken of the known grid lengths of an Improved Neubauer hemocytometer slide.
Scanning electron microscopy
For examination of ciliary adhesion by scanning electron microscopy, WT(+) gametes were mixed with mar1;hap2(−) gametes for 30 m, washed 10x in sterile-filtered N-free media, fixed for 20 m in Parducz fixative (1 part saturated HgCl2, 6 parts 2%OsO4)(Parducz, 1967), then aliquots of the cells suspended in fixative were transferred onto 0.1% poly-L-lysine coated coverslips for an additional 45 m. Coverslips were washed 10× in ddH2O, then dehydrated by sequential washes (3× each, 2 m/wash) in 70%, 95%, and 100% ethanol solutions. Coverslips were critical point dried from CO2 and mounted on stubs over double-sided tape before being sputter-coated with gold–palladium (60%:40%) (Balzers Med 010) and observed using a scanning electron microscope (Hitachi S-4700 FESEM) at an accelerating voltage of 5 kV in high vacuum.
Chlamydomonas bioassays
Assays used to quantify gamete activation, mating structure adhesion and cell-cell fusion were performed as previously described (Feng et al., 2018) with minor modifications. Gamete activation was assayed by cell wall loss, as quantified by the amount of OD435-detectable chlorophyll released into the supernatant upon treatment of mixed gametes with a detergent buffer (Feng et al., 2018; Snell, 1982; Wang et al., 2006). Equal numbers of plus and minus gametes of the indicated genotypes were mixed for 10 m at a concentration of 5 × 107 cells/mL before being added to 1.6 volumes of 4°C N-free medium containing 0.075% Triton-X 100 and 5mM EDTA (pH8), vortexed, centrifuged at 8700 × g for 30 s, and OD435exp of supernatant determined by a Nanodrop 2000 spectrophotometer. The maximum OD435max (representing 100% cell wall loss) for individual samples was then obtained after resuspending and sonicating samples for 12 s with a Microson XL sonicator. Percent cell wall loss was calculated as: [(OD435max − OD435exp)/ OD435max] × 100. Mating structure adhesion was assayed in plus and minus gametes mixed in equal numbers at a concentration of 5.0×107cells/mL for 10 m, followed by fixation in an equal volume of 5% glutaraldehyde (Feng et al., 2018). Ciliary adhesions were disrupted by pipetting 20× with a 200 μL pipette tip. Glutaraldehyde fixation coupled with the light agitation from pipetting disrupts flagellar adhesions leaving pairs of plus and minus gametes attached only by their mating structures (Feng et al., 2018; Forest, 1983). The percent of gametes attached by their mating structures was calculated as: [(2 × number of pairs) / (2 × number of pairs + number of single gametes)] × 100. Gamete fusion was assayed by the microscopic enumeration of quadri-ciliated cells from equal numbers of plus and minus gametes mixed for 10 m and fixed in an equal volume of 5% glutaraldehyde as previously described (Feng et al., 2018). The percent of gamete fusion was calculated as: [(2 × number of quadri-ciliated cells) / (2 × number of quadri-ciliated cells + number of single gametes)] × 100. For assessment of gamete activation, 4 independent cell wall loss experiments were performed for each cross tested. For adhesion and fusion experiments, at least 200 cells (as either singles or pairs) were counted in each of at least 5 biological replicates per cross. Gamete strains used for quantification of gamete activation, mating structure adhesion, and gamete fusion were the F1 and F2 progeny strains derived from crosses of the original mar1(−) CLiP mutant strain LMJ.RY0402.185187, which had been transformed with the MAR1-FLAG encoding transgene (Figure S3 and Data S3 and S4). Preliminary results gathered using the LMJ.RY0402.185187(−) gametes agreed with the phenotypes observed in the mar1(−) progeny strains.
Sequence homology detection and protein structure modeling
The similarity of the ectodomain sequence of Chlamydomonas reinhardtii FUS1 (GenBank#:AAC49416, residues 18–791) to plant GEX2 species was detected by DELTA-BLAST (Boratyn et al., 2012) and PSI-BLAST (Altschul et al., 1997; Aravind and Koonin, 1999), with significant hits to GEX2 family members detected upon iterative searches (Table S1). The online tool PROMALS3D (Pei and Grishin, 2007) was used to construct the FUS1 and GEX2 ectodomain sequence alignment shown in Data S1. The RaptorX Structure Prediction web server hosting the CASP12 and CASP13 top-ranked contact prediction method for modeling protein structure from a given amino acid sequence irrespective of coevolution information (Källberg et al., 2012; Xu, 2019; Xu et al., 2021) was used to construct structural models of the C. reinhardtii FUS1 and A. thaliana GEX2 ectodomains and to identify previously undetected Ig-like domains. Images of the FUS1 and GEX2 structural models (Fig. 1B) were generated using PyMol software (Schrodinger, LLC). Structural homologies of GEX2, FUS1 and MAR1 (GenBank: KT288268) with existing structures in the Protein Data Bank were detected using the RaptorX template-based structural modeling server (Källberg et al., 2012), PHYRE2 (Kelley et al., 2015), SWISS-MODEL (Waterhouse et al., 2018) and HHPRED (Zimmermann et al., 2018).
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of HAP2 signals
Surface and total tagged HAP2 protein signals on digital images of immunoblots were quantified using Image Studio Digits software (LI-COR) and tubulin was used for normalization. For quantification of relative protein abundance median signal intensities were determined within rectangles drawn around the upper, surface band of HAP2, the lower, internal band of HAP2, and α-tubulin on digital images of immunoblots. Background correction was performed by ImageStudioDigits software (LI-COR) using a right/left border setting of 2 on each rectangle. Total HAP2 signal was calculated as the sum of the signals of the upper and lower HAP2 bands. To compare total HAP2 proportions respective to tubulin loading controls across multiple blots, lane normalization factors were calculated for both total HAP2 and α-tubulin as described by the vendor (LI-COR). The percentage of surface-expressed HAP2 for each sample was calculated as [(signal of HAP2 upper band)/(Total HAP2 signal)] ×100. For quantification of HAP2-HA immunofluorescence signals, system-optimized z-sections of immunostained mar1;HAP2-HA(−), hap2;HAP2-HA(−), and hap2(−) naive gametes were acquired by the Leica SP5 X Laser Scanning confocal microscope using equivalent gain, laser, and zoom factor settings. Max projection images were created of the z-sections using Leica LAS X core module software and the lasso tool was used to draw a region of interest (ROI) around each cell in a given field of view and record its mean value intensity within the Ax488 channel. HAP2-HA signals of individual cells from nine fields of view were measured for the mar1;HAP2-HA(−), hap2;HAP2-HA(−) samples, and six fields of view were measured for hap2(−) samples.
Statistical Analyses
Data were analyzed using Prism 9.0 software (GraphPad) and are presented as median or mean ± SD with or without raw data points for individual biological replicates as indicated in the figure legends. Significance was defined as a p-value <0.05. In part to determine whether data met assumptions of the statistical approach (for parametric vs. non-parametric tests), data from experimental groups were first subjected to Shapiro-Wilk, D’Agostino & Pearson and Kolmogorov-Smirnov normality tests. The statistical tests used to analyze data from each experiment, what n represents, and the exact value of n are indicated in the figure legends.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat monoclonal anti-HA (clone 3F10) | Roche | Cat#11867423001 (Lot#42155800) |
| Mouse monoclonal anti-FLAG M2 | Sigma | Cat#F1804 (Lot#SLBJ4607V) |
| Rabbit polyclonal anti-HA | AbClonal | Cat#AE036 (Lot#356155811) |
| Rabbit polyclonal anti-FLAG | Sigma | Cat#F7425 |
| Mouse anti-HA | Santa Cruz | Cat#SC-7392 |
| Mouse anti-FLAG (clone M2) | Sigma | Cat#F-1804 |
| Mouse monoclonal anti-α-tubulin (B512) | Sigma | Cat#T6074 (Lot#037M4804V) |
| Goat anti-rat IgG HRP | EMD Millipore | Cat#AP136 (Lot#3270737) |
| Goat anti-mouse IgG HRP | EMD Millipore | Cat#12-349 (Lot#2985412) |
| Alexa Fluor 488 Goat anti-rat IgG (H+L) | Invitrogen | Cat#A11006 (Lot#2299157) |
| Alexa Fluor 594 Goat anti-mouse IgG (H+L) | Invitrogen | Cat#A11005 (Lot#1830459) |
| Alexa Fluor 488 Donkey Anti-Rat | Invitrogen | Cat#A48269 |
| Alexa Fluor 594 Donkey anti-mouse | Invitrogen | Cat#A32744 |
| Mouse monoclonal anti-GST | Santa Cruz | Cat#SC-138 |
| Rabbit polyclonal anti-MAR1 (Custom-generated against MAR1 peptide: TQPPRPPWPPRPPPAPPPS, residues 164–182) | YenZym™ | Rabbit#YZ5485-6 (Lot#50317) |
| Bacterial and virus strains | ||
| B21 (DE3) cells | New England Biolabs | Cat#C2527I |
| Chemicals, peptides, and recombinant proteins | ||
| Protease Inhibitor cocktail for Plant cell and tissue extracts | Roche | Cat#11836153001 (Lot#25765800) |
| Poly-l-lysine | Sigma | Cat#P8920 |
| Fluoromount-G | Southern Biotech | Cat#0100-01 (Lot#D1810-Q490) |
| Prolong Gold antifade reagent | Invitrogen | Cat#P36934 (Lot#2322656) |
| SuperSignal West Femto Max Sensitivity substrate | ThermoFisher Scientific | Cat#34095 (Lot#WE325003) |
| Protein A Sepharose 4B Fast Flow from S. aureus | Sigma | Cat#P9424 (Lot#MKBZ075IV) |
| lysozyme | ThermoFisher Scientific | Cat#89833 |
| IPTG | ThermoFisher Scientific | Cat#34060 |
| HRP-conjugated streptavidin | Jackson ImmunoResearch | Cat#016030084 |
| Paraformaldehyde | Aldrich | Cat#441244 (Lot#MKCB6246) |
| Phire Hot Start II DNA Polymerase | ThermoFisher Scientific | Cat#F-122 (Lot#00760841) |
| Chelex 100 Resin | BioRad | Cat#142-1253 (Lot #: 64404479) |
| Agar, microbiology tested suitable for plant cell culture | Sigma | Cat#A1296 (Lot#BCCD6437) |
| 30% Acrylamide/Bis Solution 37.5:1 | BioRad | Cat#1610158 (Lot#64316706) |
| Ammonium persulfate | Sigma | Cat#A3678 (Lot#MKBW1233V) |
| N,N,N’,N’- Tetramethyl ethylenediamine (TEMED) | Sigma | Cat#T9281 (Lot#BCCF2825) |
| TCEP-HCl | GoldBio | Cat#TCEP10 (Lot#9915.052616A) |
| DTT | Sigma | Cat#D0632 (Lot#SLBW1508) |
| HgCl2 | ThermoFisher Scientific | Cat#M155I |
| OsO4 | Electron Microscopy Sciences | Cat#19140 |
| Glutaraldehyde solution, grade II, 25% in H2O | Sigma | Cat#G-6257 (Lot#55H0619) |
| Dibutyryl-cAMP | Sigma | Cat#D0627 |
| EZ-Link™ Sulfo-NHS-LC-Biotin | ThermoFisher Scientific | Cat#21335 (Lot#QJ222159) |
| Pronase | Roche | Cat#10165921001 (Lot#70506922) |
| Trypsin from bovine pancreas | Sigma | Cat#T8003 (Lot#SLBM232IV) |
| Trypsin inhibitor from chicken egg white | Sigma | Cat#T2011 (Lot#SLBP7284V) |
| Methanol | Pharmco | Cat#339000000 (Lot#C19C12006) |
| Sodium orthovanadate (Na3O4V) | Aldrich | Cat#450243 (Lot#MKBZ693OV) |
| NaF | Sigma | Cat#201154 |
| Phenylmethylsulfonylfluoride (PMSF) | Roche | Cat#15460420 (Lot#10837091001) |
| Gelatin from cold water fish skin | Sigma | Cat#G7765 (Lot#SLBS7921) |
| Goat Serum | Sigma | Cat#G9023 (Lot#SLCB4567) |
| Donkey Serum | Sigma | Cat#D9663 (Lot#SLCD2393) |
| Sodium deoxycholate | Sigma | Cat#D6750 (Lot#126K0061) |
| Triton X-100 | Sigma | Cat#X-100 (Lot#SLBM7930V) |
| Transcriptor First Strand cDNA Synthesis kit | Roche | Cat#04379012001 (Lot#10769023) |
| DNase I | Ambion | Cat#AM2222 |
| RNase A | Thermo Scientific | Cat#EN0531 |
| Glutathione beads | Sigma | Cat#G4510 |
| Glutathione, reduced | Sigma | Cat#G6013 |
| Infusion Dry-down PCR Cloning Kit | Takara (Clontech) | Cat#639602 |
| HA peptide | Roche | Cat#11666975001 |
| FLAG peptide | Sigma | Cat#F4799 |
| Experimental models: Organisms/strains | ||
| C. reinhardtii: LMJ.RY0402.185187(−) or mar1(−): Cre03.g176961::pCIB(−) | CLiP Library (Li et al., 2019) https://www.chlamylibrary.org/index | LMJ.RY0402.185187 |
| C. reinhardtii: progeny 106: mar1(−) | This paper | N/A |
| C. reinhardtii: progeny 133: mar1(−) | This paper | N/A |
| C. reinhardtii: progeny 144: mar1(−) | This paper | N/A |
| C. reinhardtii: WT(−): cw15(−) | Chlamydomonas Resource Center https://www.chlamycollection.org/ | CC-5325 |
| C. reinhardtii: CC-5313: Cre12g541400::pSL72(+) | Chlamydomonas Resource Center | CC-5313 |
| C. reinhardtii: progeny 3d-1: mar1;MAR1-FLAG(+) | This paper | N/A |
| C. reinhardtii: MF-c7: mar1::MAR1-FLAG(−) | This paper | N/A |
| C. reinhardtii: progeny 111: mar1;MAR1-FLAG(−) | This paper | N/A |
| C. reinhardtii: progeny 115: mar1;MAR1-FLAG(−) | This paper | N/A |
| C. reinhardtii: progeny 147: mar1;MAR1-FLAG(−) | This paper | N/A |
| C. reinhardtii: progeny 73: mar1;hap2(−) | This paper | N/A |
| C. reinhardtii: progeny 98: mar1;hap2(−) | This paper | N/A |
| C. reinhardtii: progeny 148: mar1;hap2(+) | This paper | N/A |
| C. reinhardtii: progeny 156: mar1;hap2(+) | This paper | N/A |
| C. reinhardtii: FUS1-HA(+): fus1::FUS1-HA(+) | (Liu et al., 2010) | N/A |
| C. reinhardtii: fus1(+): imp1-15 (fus1) | Chlamydomonas Resource Center (Ferris et al., 1996) | CC-1158 |
| C. reinhardtii: HAP2-FLAG(−): hap2[40d4]::HAP2-FLAG-ble(−) | Chlamydomonas Resource Center (Liu et al., 2010) | CC-5309 |
| C. reinhardtii: HAP2-HA(−) or WT(−): hap2[40d4]::HAP2-HA(−) | Chlamydomonas Resource Center (Feng et al., 2018; Liu et al., 2015) | CC-5295 |
| C. reinhardtii: hap2(−): [40d4] B215.Cre16.g674852::pMN56(−) | Chlamydomonas Resource Center (Liu et al., 2015) | CC-5281 |
| C. reinhardtii: WT(+): 21gr | Chlamydomonas Resource Center (Proschold et al., 2005; Sager, 1955) | CC-1690 |
| C. reinhardtii: IFT172-FLAG: fla11-2; FLAG-IFT172 (1G3) | Chlamydomonas Resource Center (Lin et al., 2018) | CC-5466 |
| C. reinhardtii: IMP3-HA: imp3::IMP3-HA | Chlamydomonas Resource Center (Lin et al., 2013) | CC-5149 |
| C. reinhardtii: progeny 78: mar1;hap2;MAR1-FLAG(−) | This paper | N/A |
| C. reinhardtii: progeny 146: mar1;hap2; MAR1-FLAG(−) | This paper | N/A |
| C. reinhardtii: progeny 122: mar1;HAP2-HA (−) | This paper | N/A |
| C. reinhardtii: progeny 132: mar1;hap2;HAP2-HA(−) | This paper | N/A |
| C. reinhardtii: HF-6: mar1::HAP2-FLAG(−) | This paper | N/A |
| C. reinhardtii: progeny HF-10: mar1::HAP2-FLAG(−) | This paper | N/A |
| C. reinhardtii: hap2::HAP2-HA;MAR1-FLAG(−) | Chlamydomonas Resource Center | CC-5284 |
| C. reinhardtii: progeny 102: mar1;MAR1-FLAG;hap2;HAP2-HA(−) | This paper | N/A |
| C. reinhardtii: progeny 114: mar1;MAR1-FLAG;hap2;HAP2-HA(−) | This paper | N/A |
| C. reinhardtii: progeny 131: mar1;MAR1-FLAG;hap2;HAP2-HA(−) | This paper | N/A |
| Oligonucleotides | ||
| HSV-TEV-FLAG: CAGCCAGAACTCGCCCCGGAAGACCCCGAGGATGATCGA TCCGGACCGGAGAACCTGTATTTCCAGGGAGCCATCCCC ACGACGGAGAACCTGTACTTCCAGGTGGACGCCAACTGC CGGCCCGGCTCCTCCATGGACTACAAGGACCACGATGGC GATTACAAGGATCACGACATCGACTACAAGGACGACGAC GACAAG |
This paper | N/A |
| p1 Mar1_185187_F2 genotyping: CACCGCCTGGAACTACTCCTC |
This paper | N/A |
| p2 Mar1_185187_R2 genotyping: TTGGGTCTGGTTTAGATTTGGGCTGG |
This paper | N/A |
| RB1 CC-5313 genotyping: ATGGGGCGGTATCGGAGGAAAAG |
This paper | N/A |
| C12g541400.R2 CC-5313 genotyping: TGTAGCAAGGCTCCCGCTTCTAGTTG |
This paper | N/A |
| Zeo_F1 zeocin genotyping: GCTGCATGTGCACAGTCACGCTGTCTC |
This paper | N/A |
| Zeo_R1 zeocin genotyping: GGATCCCACACACCTGCCCGTCT |
This paper | N/A |
| Fus1_P3_Fw plus gamete genotyping: GACCATCGTAGAGCGCTCTCACCAATTG |
This paper | N/A |
| Fus1_P3_Rev plus gamete genotyping: CCGCTACGCTTCTGCGTCTTGATAGTC |
This paper | N/A |
| Nit_p1_F hap2-2 genotyping: GGTCGAGGTGCCGTAAAGC |
This paper | N/A |
| Nit_p2_R hap2-2 genotyping: CCTGATGCCCCTTCAACCG |
This paper | N/A |
| L8 MAR1 gene amplification: CGGCGGCTTTGTTCCGTTTGTTTG |
This paper | N/A |
| R5B MAR1 gene amplification: CAGTCTGCGTTTCCCTTCTCGTGG |
This paper | N/A |
| G176961.R20 ble insertion into MAR1-FLAG: CCGCGAATTCactagtGCTGCATGTGCACAGTCACGC |
This paper | N/A |
| L20 ble insertion into MAR1-FLAG: GCCCCGTGCTcctaggGGATCCCACACACCTGCCC |
This paper | N/A |
| PGenD.s ble insertion into HAP2-FLAG: actagtGGATCCCACACACCTGCCCGTCTG |
This paper | N/A |
| PGenD.as ble insertion into HAP2-FLAG: tctagaGCTGCATGTGCACAGTCACGCTGTCT |
This paper | N/A |
| g176961-L3 HIS-MAR1: TCCGGACTTGCCTTTTTAGC |
This paper | N/A |
| g176961-R10 HIS-MAR1: GGCTTCCTACAGGCCCACCA |
This paper | N/A |
| Recombinant DNA | ||
| Plasmid. pGenD-Ble | Chlamydomonas Resource Center (Fischer and Rochaix, 2001) | https://www.chlamycollection.org/ |
| pGEM-T-easy vector, MAR1 | Promega | Cat#A1360 (Lot#219942) |
| PET28a(+) vector, HIS-MAR1 | GenScript assisted in cloning, see Supplemental Data S3 | N/A |
| Plasmid. HAP2-HA: pYJ36 | (Liu et al., 2008) | N/A |
| Plasmid. HAP2- FLAG: C2925/pYJHSVTEVFL-zeo | This paper | N/A |
| Plasmid. pGEX-2T empty vector | Sigma | Cat#GE28-9546-53 |
| Plasmid. GST-rFUS1: pGEX-2T FUS1 | (Misamore et al., 2003) | N/A |
| Plasmid. MAR1-FLAG: DH5 α/pYJFUS1BHSVFL+Z | This paper | N/A |
| Software and algorithms | ||
| Leica LAS X core module | Leica Microsystem Inc. | https://www.leica-microsystems.com |
| StreamPix 5.8.1.0 | NorPix | https://www.norpix.com/products/streampix/streampix.php |
| GraphPad Prism Version 9.0.0 | GraphPad | https://www.graphpad.com/ |
| Image Studio™ | LI-COR | https://www.licor.com/bio/image-studio/ |
| PyMol Software | Schrodinger, LLC | https://www.schrodinger.com/products/pymol |
| Adobe Illustrator and Photoshop | Adobe | https://www.adobe.com/products/catalog.html |
| RaptorX | (Källberg et al., 2012; Xu, 2019; Xu et al., 2021) | http://raptorx.uchicago.edu/ |
| PROMALS3D | (Pei and Grishin, 2007) | http://prodata.swmed.edu/promals3d/promals3d.php |
| DELTA-BLAST | (Boratyn et al., 2012) | https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&PROGRAM=blastp&BLAST_PROGRAMS=deltaBlast |
| PSI-BLAST | (Altschul et al., 1997; Aravind and Koonin, 1999) | https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&PROGRAM=blastp&BLAST_PROGRAMS=psiBlast |
| PHYRE2 | (Kelley et al., 2015) | http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index |
| SWISS-MODEL | (Waterhouse et al., 2018) | https://swissmodel.expasy.org/ |
| HHPRED | (Zimmermann et al., 2018) | https://toolkit.tuebingen.mpg.de/tools/hhpred |
Highlights.
FUS1/GEX2 proteins mediate gamete adhesion in unicellular and multicellular plants
FUS1 on Chlamydomonas plus gametes binds directly to MAR1 on minus gametes
MAR1-FUS1 receptor pair formation is essential for gamete adhesion and fusion
MAR1 is biochemically and functionally associated with conserved fusogen HAP2
Acknowledgments
We thank UMD colleagues Jocelyn Chen and Drs. Jun Zhang, Mayanka Awasthi, and Peeyush Ranjan; UT Southwestern colleagues Drs. Muqing Cao, Wenhao Li, Jue Ning, and Saikat Mukhopadhyay; Institut Pasteur colleagues Drs. Felix Rey, Eduard Salazar, and Ignacio Fernandez; and colleague, Ursula Goodenough, Washington University, St. Louis, for helpful discussions; Dr. Tim Maugel of the UMD Laboratory for Biological Ultrastructure for assistance with SEM; and Drs. Kate Luby-Phelps and Abhijit Bugde (UT Southwestern Medical Center, Live Cell Imaging Core) and Amy Beaven (UMD, Imaging Core) for guidance with light microscopy. Funding was provided by NIH GM56778 and GM122565 to W.J.S. and F32-GM126735 to J.F.P.
Footnotes
Declaration of Interests: The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, and Lipman DJ (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson H, Barth BU, Ekström M, and Garoff H (1997). Oligomerization-dependent folding of the membrane fusion protein of Semliki Forest virus. J. Virol. 71, 9654–9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, and Koonin EV (1999). Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol. 287, 1023–1040. 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]
- Aydin H, Sultana A, Li S, Thavalingam A, and Lee JE (2016). Molecular architecture of the human sperm IZUMO1 and egg JUNO fertilization complex. Nature 534, 562–565. 10.1038/nature18595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquero E, Fedry J, Legrand P, Krey T, and Rey FA (2019). Species-Specific Functional Regions of the Green Alga Gamete Fusion Protein HAP2 Revealed by Structural Studies. Structure 27, 113–124 e114. 10.1016/j.str.2018.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaux S, Ialy-Radio C, Chalbi M, Dybal E, Homps-Legrand M, Do Cruzeiro M, Vaiman D, Wolf J-P, and Ziyyat A (2020). Sperm SPACA6 protein is required for mammalian Sperm-Egg Adhesion/Fusion. Sci. Rep 10, 1–15. 10.1038/s41598-020-62091-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beh CT, Brizzio V, and Rose MD (1997). KAR5 encodes a novel pheromone-inducible protein required for homotypic nuclear fusion. J. Cell Biol. 139, 1063–1076. 10.1083/jcb.139.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzile O, Hernandez-Lara CI, Wang Q, and Snell WJ (2013). Regulated membrane protein entry into flagella is facilitated by cytoplasmic microtubules and does not require IFT. Curr. Biol. 23, 1460–1465. 10.1016/j.cub.2013.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi E, Doe B, Goulding D, and Wright GJ (2014). Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature 508, 483–487. 10.1038/nature13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi E, and Wright GJ (2020). Find and fuse: Unsolved mysteries in sperm–egg recognition. PLoS Biol. 18, e3000953. 10.1371/journal.pbio.3000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, and Brunak S (2004). Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4, 1633–1649. 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Boratyn GM, Schäffer AA, Agarwala R, Altschul SF, Lipman DJ, and Madden TL (2012). Domain enhanced lookup time accelerated BLAST. Biol Direct 7, 12. 10.1186/1745-6150-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho-Nuez M, Hernandez-Silva DJ, Castaneda-Ortiz EJ, Paredes-Martinez ME, Rocha-Martinez MK, Alvarez-Sanchez ME, Mercado-Curiel RF, Aguilar-Tipacamu G, and Mosqueda J (2017). Hap2, a novel gene in Babesia bigemina is expressed in tick stages, and specific antibodies block zygote formation. Parasit Vectors 10, 568. 10.1186/s13071-017-2510-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton M, Lee H, Mulvey M, and Brown DT (1997). Role of glycoprotein PE2 in formation and maturation of the Sindbis virus spike. J. Virol. 71, 1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes MD, Hamilton EP, Xiong J, Lawson MJ, Yuan D, Hadjithomas M, Miao W, and Orias E (2013). Selecting one of several mating types through gene segment joining and deletion in Tetrahymena thermophila. PLoS Biol. 11, e1001518. 10.1371/journal.pbio.1001518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Basak AJ, Nair AV, Duraivelan K, and Samanta D (2020). Immunoglobulin-fold containing bacterial adhesins: molecular and structural perspectives in host tissue colonization and infection. FEMS Microbiol. Lett. 368, fnaa220. 10.1093/femsle/fnaa220. [DOI] [PubMed] [Google Scholar]
- Cole ES, Cassidy-Hanley D, Fricke Pinello J, Zeng H, Hsueh M, Kolbin D, Ozzello C, Giddings T Jr., Winey M, and Clark TG (2014). Function of the male-gamete-specific fusion protein HAP2 in a seven-sexed ciliate. Curr. Biol. 24, 2168–2173. 10.1016/j.cub.2014.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyprys P, Lindemeier M, and Sprunck S (2019). Gamete fusion is facilitated by two sperm cell-expressed DUF679 membrane proteins. Nature Plants 5, 253–257. [DOI] [PubMed] [Google Scholar]
- Dean J (2007). The enigma of sperm-egg recognition in mice. Soc Reprod Fertil Suppl 63, 359–365. [PubMed] [Google Scholar]
- Dresselhaus T, Sprunck S, and Wessel GM (2016). Fertilization Mechanisms in Flowering Plants. Curr. Biol. 26, R125–139. 10.1016/j.cub.2015.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebchuqin E, Yokota N, Yamada L, Yasuoka Y, Akasaka M, Arakawa M, Deguchi R, Mori T, and Sawada H (2014). Evidence for participation of GCS1 in fertilization of the starlet sea anemone Nematostella vectensis: implication of a common mechanism of sperm-egg fusion in plants and animals. Biochem. Biophys. Res. Commun. 451, 522–528. 10.1016/j.bbrc.2014.08.006. [DOI] [PubMed] [Google Scholar]
- Engel ML, Holmes-Davis R, and McCormick S (2005). Green sperm. Identification of male gamete promoters in Arabidopsis. Plant Physiol. 138, 2124–2133. 10.1104/pp.104.054213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedry J, Forcina J, Legrand P, Pehau-Arnaudet G, Haouz A, Johnson M, Rey FA, and Krey T (2018). Evolutionary diversification of the HAP2 membrane insertion motifs to drive gamete fusion across eukaryotes. PLoS Biol. 16, e2006357. 10.1371/journal.pbio.2006357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedry J, Liu Y, Pehau-Arnaudet G, Pei J, Li W, Tortorici MA, Traincard F, Meola A, Bricogne G, Grishin NV, et al. (2017). The Ancient Gamete Fusogen HAP2 Is a Eukaryotic Class II Fusion Protein. Cell 168, 904–915 e910. 10.1016/j.cell.2017.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Dong X, Pinello J, Zhang J, Lu C, Iacob RE, Engen JR, Snell WJ, and Springer TA (2018). Fusion surface structure, function, and dynamics of gamete fusogen HAP2. Elife 7, e39772. 10.7554/eLife.39772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Pavlovic C, Fabry S, and Goodenough UW (1997). Rapid evolution of sex-related genes in Chlamydomonas. Proc. Natl. Acad. Sci. USA 94, 8634–8639. 10.1073/pnas.94.16.8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Waffenschmidt S, Umen JG, Lin H, Lee JH, Ishida K, Kubo T, Lau J, and Goodenough UW (2005). Plus and minus sexual agglutinins from Chlamydomonas reinhardtii. Plant Cell 17, 597–615. 10.1105/tpc.104.028035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Woessner JP, and Goodenough UW (1996). A sex recognition glycoprotein is encoded by the plus mating-type gene fus1 of Chlamydomonas reinhardtii. Mol. Biol. Cell 7, 1235–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Woessner JP, Waffenschmidt S, Kilz S, Drees J, and Goodenough UW (2001). Glycosylated polyproline II rods with kinks as a structural motif in plant hydroxyproline-rich glycoproteins. Biochemistry 40, 2978–2987. 10.1021/bi0023605. [DOI] [PubMed] [Google Scholar]
- Fischer N, and Rochaix JD (2001). The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol. Genet. Genomics 265, 888–894. 10.1007/s004380100485. [DOI] [PubMed] [Google Scholar]
- Forest CL (1983). Specific contact between mating structure membranes observed in conditional fusion-defective Chlamydomonas mutants. Exp. Cell Res. 148, 143–154. 10.1016/0014-4827(83)90194-5. [DOI] [PubMed] [Google Scholar]
- Friedmann I, Colwin AL, and Colwin LH (1968). Fine-structural aspects of fertilization in Chlamydomonas reinhardi. J. Cell Sci. 3, 115–128. [DOI] [PubMed] [Google Scholar]
- Fujihara Y, Lu YG, Noda T, Oji A, Larasati T, Kojima-Kita K, Yu ZF, Matzuk RM, Matzuk MM, and Ikawa M (2020). Spermatozoa lacking Fertilization Influencing Membrane Protein (FIMP) fail to fuse with oocytes in mice. Proc. Natl. Acad. Sci. USA 117, 9393–9400. 10.1073/pnas.1917060117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, and Rokhsar DS (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–1186. 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson P (2015). Izumo1 and Juno: the evolutionary origins and coevolution of essential sperm-egg binding partners. R. Soc. Open Sci. 2, 150296. ARTN 150296 10.1098/rsos.150296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger ZA, Brown MS, Isberg RR, and Bjorkman PJ (1999). Crystal structure of invasin: A bacterial integrin-binding protein. Science 286, 291–295. DOI 10.1126/science.286.5438.291. [DOI] [PubMed] [Google Scholar]
- Heiman MG, and Walter P (2000). Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J. Cell Biol. 151, 719–730. 10.1083/jcb.151.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herberg S, Gert KR, Schleiffer A, and Pauli A (2018). The Ly6/uPAR protein Bouncer is necessary and sufficient for species-specific fertilization. Science 361, 1029–1033. 10.1126/science.aat7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M, Arai M, Mori T, Miyagishima SY, Kawai S, Kita K, Kuroiwa T, Terenius O, and Matsuoka H (2008). Male fertility of malaria parasites is determined by GCS1, a plant-type reproduction factor. Curr. Biol. 18, 607–613. 10.1016/j.cub.2008.03.045. [DOI] [PubMed] [Google Scholar]
- Honig B, and Shapiro L (2020). Adhesion Protein Structure, Molecular Affinities, and Principles of Cell-Cell Recognition. Cell 181, 520–535. 10.1016/j.cell.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa M, Inoue N, Benham AM, and Okabe M (2010). Fertilization: a sperm’s journey to and interaction with the oocyte. J. Clin. Invest. 120, 984–994. 10.1172/JCI41585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue N, Ikawa M, Isotani A, and Okabe M (2005). The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature 434, 234–238. 10.1038/nature03362. [DOI] [PubMed] [Google Scholar]
- Johnson MA, von Besser K, Zhou Q, Smith E, Aux G, Patton D, Levin JZ, and Preuss D (2004). Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168, 971–982. 10.1534/genetics.104.029447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT (1999). Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202. 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Kadandale P, Stewart-Michaelis A, Gordon S, Rubin J, Klancer R, Schweinsberg P, Grant BD, and Singson A (2005). The egg surface LDL receptor repeat-containing proteins EGG-1 and EGG-2 are required for fertilization in Caenorhabditis elegans. Curr. Biol. 15, 2222–2229. 10.1016/j.cub.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Käll L, Krogh A, and Sonnhammer ELL (2007). Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 35, W429–432. 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Källberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, and Xu J (2012). Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 7, 1511–1522. 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, and Sternberg MJE (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamas-Toranzo I, Hamze JG, Bianchi E, Fernandez-Fuertes B, Perez-Cerezales S, Laguna-Barraza R, Fernandez-Gonzalez R, Lonergan P, Gutierrez-Adan A, Wright GJ, et al. (2020). TMEM95 is a sperm membrane protein essential for mammalian fertilization. Elife 9, e53913. ARTN e53913 10.7554/eLife.53913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Naour F, Rubinstein E, Jasmin C, Prenant M, and Boucheix C (2000). Severely reduced female fertility in CD9-deficient mice. Science 287, 319–321. 10.1126/science.287.5451.319. [DOI] [PubMed] [Google Scholar]
- Li X, Patena W, Fauser F, Jinkerson RE, Saroussi S, Meyer MT, Ivanova N, Robertson JM, Yue R, Zhang R, et al. (2019). A genome-wide algal mutant library and functional screen identifies genes required for eukaryotic photosynthesis. Nat. Genet. 51, 627–635. 10.1038/s41588-019-0370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie FR (1914). Studies of fertilization. VI. The mechanism of fertilization in Arbacia. J. Exp. Zool. 16, 523–590. 10.1002/jez.1400160404. [DOI] [Google Scholar]
- Lin H, Guo S, and Dutcher SK (2018). RPGRIP1L helps to establish the ciliary gate for entry of proteins. J. Cell Sci. 131. 10.1242/jcs.220905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HW, Miller ML, Granas DM, and Dutcher SK (2013). Whole Genome Sequencing Identifies a Deletion in Protein Phosphatase 2A That Affects Its Stability and Localization in Chlamydomonas reinhardtii. PLoS Genet 9. ARTN e1003841 10.1371/journal.pgen.1003841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Misamore MJ, and Snell WJ (2010). Membrane fusion triggers rapid degradation of two gamete-specific, fusion-essential proteins in a membrane block to polygamy in Chlamydomonas. Development 137, 1473–1481. 10.1242/dev.044743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Pei J, Grishin N, and Snell WJ (2015). The cytoplasmic domain of the gamete membrane fusion protein HAP2 targets the protein to the fusion site in Chlamydomonas and regulates the fusion reaction. Development 142, 962–971. 10.1242/dev.118844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Tewari R, Ning J, Blagborough AM, Garbom S, Pei J, Grishin NV, Steele RE, Sinden RE, Snell WJ, and Billker O (2008). The conserved plant sterility gene HAP2 functions after attachment of fusogenic membranes in Chlamydomonas and Plasmodium gametes. Genes Dev. 22, 1051–1068. 10.1101/gad.1656508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzetti D, Poirier C, Zhao M, Overbeek PA, Harrison W, and Bishop CE (2014). A transgenic insertion on mouse chromosome 17 inactivates a novel immunoglobulin superfamily gene potentially involved in sperm-egg fusion. Mamm. Genome 25, 141–148. 10.1007/s00335-013-9491-x. [DOI] [PubMed] [Google Scholar]
- Misamore MJ, Gupta S, and Snell WJ (2003). The Chlamydomonas Fus1 protein is present on the mating type plus fusion organelle and required for a critical membrane adhesion event during fusion with minus gametes. Mol. Biol. Cell 14, 2530–2542. 10.1091/mbc.e02-12-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyado K, Yamada G, Yamada S, Hasuwa H, Nakamura Y, Ryu F, Suzuki K, Kosai K, Inoue K, Ogura A, et al. (2000). Requirement of CD9 on the egg plasma membrane for fertilization. Science 287, 321–324. 10.1126/science.287.5451.321. [DOI] [PubMed] [Google Scholar]
- Mori T, Igawa T, Tamiya G, Miyagishima S-Y, and Berger F (2014). Gamete attachment requires GEX2 for successful fertilization in Arabidopsis. Curr. Biol. 24, 170–175. 10.1016/j.cub.2013.11.030. [DOI] [PubMed] [Google Scholar]
- Mori T, Kuroiwa H, Higashiyama T, and Kuroiwa T (2006). GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat. Cell Biol. 8, 64–71. 10.1038/ncb1345. [DOI] [PubMed] [Google Scholar]
- Navaratnarajah CK, Generous AR, Yousaf I, and Cattaneo R (2020). Receptor-mediated cell entry of paramyxoviruses: Mechanisms, and consequences for tropism and pathogenesis. J. Biol. Chem. 295, 2771–2786. 10.1074/jbc.REV119.009961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necci M, Piovesan D, Dosztányi Z, and Tosatto SCE (2017). MobiDB-lite: fast and highly specific consensus prediction of intrinsic disorder in proteins. Bioinformatics 33, 1402–1404. 10.1093/bioinformatics/btx015. [DOI] [PubMed] [Google Scholar]
- Nielsen H (2017). Predicting Secretory Proteins with SignalP. Methods in Molecular Biology (Clifton, N.J.) 1611, 59–73. 10.1007/978-1-4939-7015-5_6. [DOI] [PubMed] [Google Scholar]
- Ning J, Otto TD, Pfander C, Schwach F, Brochet M, Bushell E, Goulding D, Sanders M, Lefebvre PA, Pei J, et al. (2013). Comparative genomics in Chlamydomonas and Plasmodium identifies an ancient nuclear envelope protein family essential for sexual reproduction in protists, fungi, plants, and vertebrates. Genes Dev. 27, 1198–1215. 10.1101/gad.212746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura H, and L’Hernault SW (2016). Gamete interactions require transmembranous immunoglobulin-like proteins with conserved roles during evolution. Worm 5, e1197485. 10.1080/21624054.2016.1197485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura H, Tajima T, Comstra HS, Gleason EJ, and L’Hernault SW (2015). The Immunoglobulin-like Gene spe-45 Acts during Fertilization in Caenorhabditis elegans like the Mouse Izumo1 Gene. Curr. Biol. 25, 3225–3231. 10.1016/j.cub.2015.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Lu YG, Fujihara Y, Oura S, Koyano T, Kobayashi S, Matzuk MM, and Ikawa M (2020). Sperm proteins SOF1, TMEM95, and SPACA6 are required for sperm-oocyte fusion in mice. Proc. Natl. Acad. Sci. USA 117, 11493–11502. 10.1073/pnas.1922650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouemssi SB, Ghribi M, Beauchemin R, Meddeb-Mouelhi F, Germain H, and Desgagne-Penix I (2020). Rapid and Efficient Colony-PCR for High Throughput Screening of Genetically Transformed Chlamydomonas reinhardtii. Life 10, 186. 10.3390/life10090186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto U, Ishida H, Krayukhina E, Uchiyama S, Inoue N, and Shimizu T (2016). Structure of IZUMO1-JUNO reveals sperm-oocyte recognition during mammalian fertilization. Nature 534, 566–569. 10.1038/nature18596. [DOI] [PubMed] [Google Scholar]
- Okamoto M, Yamada L, Fujisaki Y, Bloomfield G, Yoshida K, Kuwayama H, Sawada H, Mori T, and Urushihara H (2016). Two HAP2-GCS1 homologs responsible for gamete interactions in the cellular slime mold with multiple mating types: Implication for common mechanisms of sexual reproduction shared by plants and protozoa and for male-female differentiation. Dev. Biol. 415, 6–13. 10.1016/j.ydbio.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagni M, Ioannidis V, Cerutti L, Zahn-Zabal M, Jongeneel CV, Hau J, Martin O, Kuznetsov D, and Falquet L (2007). MyHits: improvements to an interactive resource for analyzing protein sequences. Nucleic Acids Res. 35, W433–437. 10.1093/nar/gkm352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parducz B (1967). Ciliary movement and coordination in ciliates. Int. Rev. Cytol. 21, 91–128. 10.1016/s0074-7696(08)60812-8. [DOI] [PubMed] [Google Scholar]
- Pei J, and Grishin NV (2007). PROMALS: towards accurate multiple sequence alignments of distantly related proteins. Bioinformatics 23, 802–808. 10.1093/bioinformatics/btm017. [DOI] [PubMed] [Google Scholar]
- Perez-Vargas J, Krey T, Valansi C, Avinoam O, Haouz A, Jamin M, Raveh-Barak H, Podbilewicz B, and Rey FA (2014). Structural basis of eukaryotic cell-cell fusion. Cell 157, 407–419. 10.1016/j.cell.2014.02.020. [DOI] [PubMed] [Google Scholar]
- Pinello JF, Lai AL, Millet JK, Cassidy-Hanley D, Freed JH, and Clark TG (2017). Structure-Function Studies Link Class II Viral Fusogens with the Ancestral Gamete Fusion Protein HAP2. Curr. Biol. 27, 651–660. 10.1016/j.cub.2017.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proschold T, Harris EH, and Coleman AW (2005). Portrait of a species: Chlamydomonas reinhardtii. Genetics 170, 1601–1610. 10.1534/genetics.105.044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan C, Maier S, Walker RA, Rehrauer H, Joekel DE, Winiger RR, Basso WU, Grigg ME, Hehl AB, Deplazes P, and Smith NC (2019). An experimental genetically attenuated live vaccine to prevent transmission of Toxoplasma gondii by cats. Sci. Rep. 9, 1474. 10.1038/s41598-018-37671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan P, Awasthi M, and Snell WJ (2019). Transient Internalization and Microtubule-Dependent Trafficking of a Ciliary Signaling Receptor from the Plasma Membrane to the Cilium. Curr. Biol. 29, 2942–2947 e2942. 10.1016/j.cub.2019.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R (1955). Inheritance in the Green Alga Chlamydomonas Reinhardi. Genetics 40, 476–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaranarayanan S, and Higashiyama T (2018). Capacitation in Plant and Animal Fertilization. Trends Plant Sci. 23, 129–139. 10.1016/j.tplants.2017.10.006. [DOI] [PubMed] [Google Scholar]
- Shimogawara K, Fujiwara S, Grossman A, and Usuda H (1998). High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148, 1821–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singaravelu G, Rahimi S, Krauchunas A, Rizvi A, Dharia S, Shakes D, Smith H, Golden A, and Singson A (2015). Forward Genetics Identifies a Requirement for the Izumo-like Immunoglobulin Superfamily spe-45 Gene in Caenorhabditis elegans Fertilization. Curr. Biol. 25, 3220–3224. 10.1016/j.cub.2015.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singson A, Mercer KB, and L’Hernault SW (1998). The C. elegans spe-9 gene encodes a sperm transmembrane protein that contains EGF-like repeats and is required for fertilization. Cell 93, 71–79. 10.1016/s0092-8674(00)81147-2. [DOI] [PubMed] [Google Scholar]
- Snell WJ (1982). Study of the release of cell wall degrading enzymes during adhesion of Chlamydomonas gametes. Exp. Cell Res. 138, 109–119. 10.1016/0014-4827(82)90096-9. [DOI] [PubMed] [Google Scholar]
- Snell WJ, and Goodenough UW (2009). Flagellar adhesion, flagellar-generated signaling, and gamete fusion during mating. In The Chlamydomonas Sourcebook, Witman GB, ed. (Elsevier; ), pp. 369–394. [Google Scholar]
- Steele RE, and Dana CE (2009). Evolutionary history of the HAP2/GCS1 gene and sexual reproduction in metazoans. PLoS One 4, e7680. 10.1371/journal.pone.0007680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, and Vacquier VD (2002). The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3, 137–144. 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Tajima T, and Nishimura H (2018). Immunoglobulin-Like Domains Have an Evolutionarily Conserved Role During Gamete Fusion in C. elegans and Mouse. In Origin and Evolution of Biodiversity, Pontarotti P, ed. (Springer International Publishing; ), pp. 163–179. [Google Scholar]
- Takahashi T, Mori T, Ueda K, Yamada L, Nagahara S, Higashiyama T, Sawada H, and Igawa T (2018). The male gamete membrane protein DMP9/DAU2 is required for double fertilization in flowering plants. Development 145, dev170076. 10.1242/dev.170076. [DOI] [PubMed] [Google Scholar]
- Valansi C, Moi D, Leikina E, Matveev E, Graña M, Chernomordik LV, Romero H, Aguilar PS, and Podbilewicz B (2017). Arabidopsis HAP2/GCS1 is a gamete fusion protein homologous to somatic and viral fusogens. J. Cell Biol. 216, 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk MR, van Schaijk BC, Khan SM, van Dooren MW, Ramesar J, Kaczanowski S, van Gemert GJ, Kroeze H, Stunnenberg HG, Eling WM, et al. (2010). Three members of the 6-cys protein family of Plasmodium play a role in gamete fertility. PLoS Pathog 6, e1000853. 10.1371/journal.ppat.1000853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Pan J, and Snell WJ (2006). Intraflagellar Transport Particles Participate Directly in Cilium-Generated Signaling in Chlamydomonas. Cell 125, 549–562. 10.1016/j.cell.2006.02.044. [DOI] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RL, Goodenough DA, and Goodenough UW (1977). Membrane differentiations at sites specialized for cell fusion. J. Cell Biol. 72, 144–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NF, O’Connell JS, Lu M, and Snell WJ (1999). Flagellar adhesion between mt(+) and mt(−) Chlamydomonas gametes regulates phosphorylation of the mt(+)-specific homeodomain protein GSP1. J. Biol. Chem. 274, 34383–34388. 10.1074/jbc.274.48.34383. [DOI] [PubMed] [Google Scholar]
- Wong JL, and Johnson MA (2010). Is HAP2-GCS1 an ancestral gamete fusogen? Trends Cell Biol. 20, 134–141. 10.1016/j.tcb.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Wong JL, Leydon AR, and Johnson MA (2010). HAP2(GCS1)-dependent gamete fusion requires a positively charged carboxy-terminal domain. PLoS Genet. 6, e1000882. 10.1371/journal.pgen.1000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J (2019). Distance-based protein folding powered by deep learning. Proc. Natl. Acad. Sci. USA 116, 16856–16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JB, McPartlon M, and Li J (2021). Improved protein structure prediction by deep learning irrespective of co-evolution information. Nat Mach Intell 3, 601–+. 10.1038/s42256-021-00348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Hamaji T, Kawai-Toyooka H, Matsuzaki R, Takahashi F, Nishimura Y, Kawachi M, Noguchi H, Minakuchi Y, Umen JG, et al. (2021). Three genomes in the algal genus Volvox reveal the fate of a haploid sex-determining region after a transition to homothallism. P Natl Acad Sci USA 118. ARTN e2100712118 10.1073/pnas.2100712118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Pinello JF, Fernandez I, Baquero E, Fedry J, Rey FA, and Snell WJ (2021). Species-specific gamete recognition initiates fusion-driving trimer formation by conserved fusogen HAP2. Nat Commun 12, 4380. 10.1038/s41467-021-24613-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann L, Stephens A, Nam SZ, Rau D, Kubler J, Lozajic M, Gabler F, Soding J, Lupas AN, and Alva V (2018). A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 430, 2237–2243. 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All other data reported in this paper are available from the lead contact upon request.
No large-scale datasets or new code were generated in this study.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.