Abstract
High-quality medical data is critical to the development and implementation of machine learning (ML) algorithms in healthcare; however, security, and privacy concerns continue to limit access. We sought to determine the utility of “synthetic data” in training ML algorithms for the detection of tuberculosis (TB) from inflammatory biomarker profiles. A retrospective dataset (A) comprised of 278 patients was used to generate synthetic datasets (B, C, and D) for training models prior to secondary validation on a generalization dataset. ML models trained and validated on the Dataset A (real) demonstrated an accuracy of 90%, a sensitivity of 89% (95% CI, 83–94%), and a specificity of 100% (95% CI, 81–100%). Models trained using the optimal synthetic dataset B showed an accuracy of 91%, a sensitivity of 93% (95% CI, 87–96%), and a specificity of 77% (95% CI, 50–93%). Synthetic datasets C and D displayed diminished performance measures (respective accuracies of 71% and 54%). This pilot study highlights the promise of synthetic data as an expedited means for ML algorithm development.
Keywords: Artificial intelligence, biomarkers, data accessibility, electronic medical record, privacy, simulation
1. Background
Access to high-quality medical data is often hard to acquire which can impede the development and implementation of artificial intelligence (AI)/machine learning (ML) algorithms in healthcare.1, 2, 3 Common sources of clinical data include electronic medical record (EMR) systems which are tightly regulated and often inaccessible to AI/ML developers due to patient privacy concerns.4 Additionally, extraction of clean data from EMR systems can be challenging due to platform limitations, accuracy of data, as well as prioritization of essential day-to-day operations by local institutional information technology (IT), teams over requests for datasets for developmental purposes.5, 6, 7 As an alternative, manual extraction of EMR data may be an option, but is at risk for transcription errors and is extremely time-consuming. Given these limitations, AI/ML developers often gravitate to other more easily accessible databases derived from clinical trials and/or unrelated research studies.8 Although clinical trial/research data could be more convenient to access due to availability, these datasets may not have been collected for the intended use, and also may not accurately represent “real world” practices.
The use of in silico (i.e., synthetic) data provides opportunities to accelerate the development of AI/ML models in healthcare.9,10 The synthetic data is produced based on using real-world observations to create a de-identified data set that emulates the “real data equivalent” appropriate for distribution to developers. This practice has been leveraged to great effect in the basic sciences and pharmaceutical industry for drug development,11,12 however, to date this paradigm has not been widely adopted in laboratory medicine for AI/ML. To this end, the goal of this paper is to determine the clinical utility of “synthetic data” trained ML algorithm and their performance measures.
Tuberculosis (TB) serves as a unique opportunity to evaluate the potential value of the synthetic-data trained ML algorithms to diagnose disease. Over 10 million people acquire TB annually despite advancements in therapeutics and diagnostic testing methods.13 Therefore, this infectious disease remains a persistent clinical concern which is responsible for significant morbidity and mortality, particularly in the developing world. Continued technological gaps in TB testing include the urgent need for robust biomarkers to enable identification of latent infection and increased sensitivity.14 The recent novel application of multiplex biomarker assays has demonstrated promise toward this goal and serves as our prototype for evaluating the utility of its synthetic data for training AI/ML algorithms.
The generation of effective TB-predictive ML algorithms is dependent on robust datasets for training and performance assessment. To that end, the first objective of this approach was to generate expanded synthetic datasets that are statistically similar to the original dataset, which contains recorded values from actual patients. Using the larger, deidentified synthetic data instead of the original, limited data will allow users to perform downstream analysis and train machine learning models on a larger dataset without exposing any confidential information about the patients.
2. Materials and methods
The anonymized retrospective dataset was derived from 278 patients who were initially recruited in Pakistan per World Health Organization (WHO) general guidelines15 for TB diagnostics from a recently published study that was conducted to evaluate a multiplex serologic panel for active tuberculosis patients.16 No patient identifiable information was available or shared (only raw multiplex serology data and the status of TB positivity and negativity).
2.1. Synthetic data generation
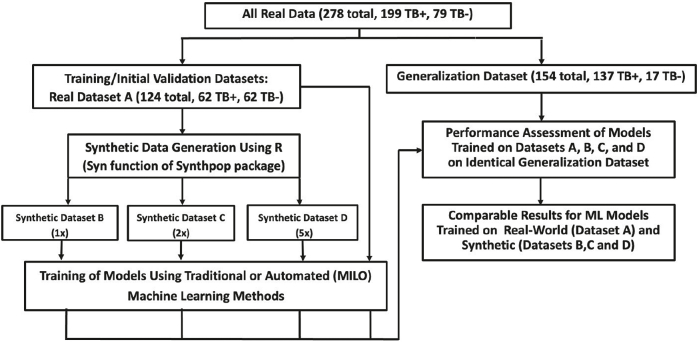
Synthetic datasets were trained and tested on the real-world dataset derived from the aforementioned 278 subjects with and without TB [Fig. 1]. Study subjects were tested on a multiplex serology platform for 31 TB antigen biomarkers. The data was divided into datasets for training and initial validation and a generalization dataset as depicted in the study design diagram. Dataset A (real data) was used for training and initial validation as well as synthetic data generation and was comprised of 124 cases (62 TB positive and 62 TB negative). Similar to Dataset A, the secondary generalization dataset is comprised of the remaining real-world data (154 total cases, 137 TB positive and 17 TB negative) which is used to validate the models trained on the real and synthetic datasets.
Fig. 1.
Study design.
Synthetic data were derived from Dataset A (the real dataset) which was used to produce three different synthetic datasets (B, C, and D). Dataset B represents a one-to-one ratio of the synthetic data to the real data acquired from dataset A, while Dataset C and D are the expanded synthetic datasets representing a one to two and a one to five ratios to Dataset A. Datasets B, C, and D were developed using R statistical software with the synthpop package. We created an R script that reads the original dataset containing the real data from Dataset A within its comma-separated values (.csv) file into a separate data frame. We then randomly shuffled the entire dataset and divided the data frame into two, one containing the target feature “TB-31” and the other containing the rest of the columns (features). Then we created the synthetic dataset using the features as a source. This was done by calling the syn function of the synthpop library in R software (R-project. org). The syn function accepts a parameter m that is defined as number of synthetic copies of the original (observed) data to be generated. We re-ran the function 3 times with m=1, m=2, and m=5 to generate 1 time, 2 times, and 5 times the rows in the original dataset features, respectively.
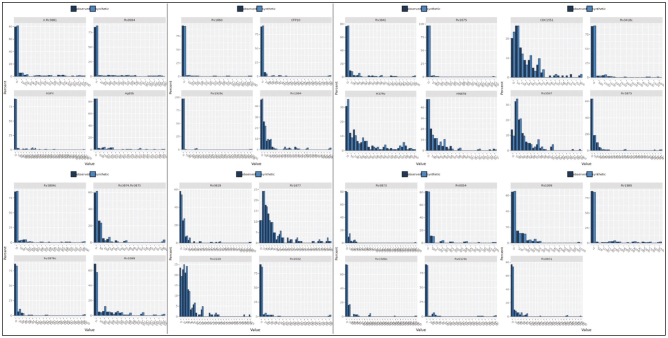
Once the synthetic features were generated, their respective datasets were then statistically compared with the original real dataset (Dataset A) as seen in Fig. 2. This includes calculating 1st quartile, median, and 3rd quartile values. Visual representation was also done to ensure that the distribution of data between the original and synthetic datasets is as close as possible. In addition to a similar distribution of each feature individually, it is also important to ensure that the relationship between features resembles the original dataset as closely as possible. A correlation matrix was therefore utilized to show differences in the relationship between variables.
Fig. 2.
Distribution of Dataset A vs. Dataset B.
Next, we combined the target feature with the synthetic features. For the synthetic dataset with m=1, we directly combined the target feature with the synthetic features one to one using the cbind function in R to generate our final 1-time synthetic dataset (Dataset B). For the ×2 synthetic dataset (expanded one to two to create Dataset C) and ×5 synthetic dataset (expanded one to five to create Dataset D), we took each set of the synthetic datasets generated from synthpop (×5 and ×2) and then used cbind function separately on each set to combine the target feature. This resulted in 2 data frames (for the ×2) and 5 data frames (for the ×5), respectively. We then merged these multiple data frames into a single, final combined dataset using the rbind function to generate our final ×2 (Dataset C) and ×5 (Dataset D) synthetic data frame.
2.2. ML training and generalization
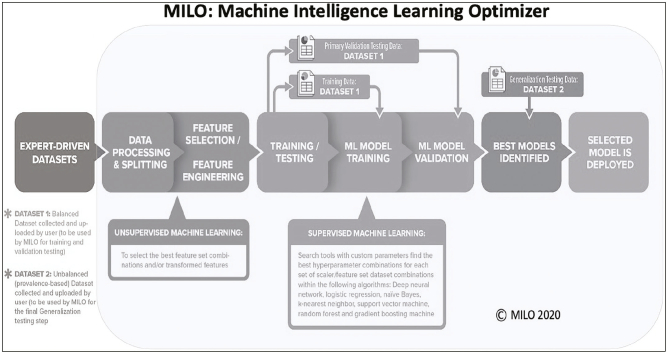
Both the real dataset (A) and the respective synthetic datasets (B, C, and D) were used to train the ML algorithms produced using: (a) traditional non-automated manual coding techniques of an optimized random forest (RF) algorithm followed by (b) our automated machine learning (auto-ML) Machine Learning Intelligence (MILO) platform (MILO ML, LLC, Sacramento, CA). For the non-automated traditional ML approach, the RF algorithm in the R software package was used to train the models. Four different models were subsequently trained: on the original real dataset (non-synthetic Dataset A), on the ×1 synthetic dataset (Dataset B), on the ×2 synthetic dataset (Dataset C), and on the ×5 synthetic dataset (Dataset D). All models generated above were secondarily validated on a separate “real” (nonsynthetic) generalization dataset to test and measure performance for all the aforementioned models that were constructed from the original real (non-synthetic) and synthetic datasets. The same training and validation steps described above (in the non-automated RF approach) were also repeated through our automated machine learning approach through the Auto-ML platform MILO. As described previously,17, 18, 19 the MILO platform incorporates an automated data processor, a data feature selector and data transformer, followed by multiple supervised ML model building approaches that make use of its custom hyperparameter search tools that help identify the optimal hyperparameter combinations for each of the seven algorithms utilized within MILO (neural network/multi-layer perceptron, logistic regression (LR), naïve Bayes (NB), k-nearest neighbor (k-NN), support vector machine (SVM), random forest (RF), and XGBoost gradient boosting machine (GBM) techniques) [Fig. 3].
Fig. 3.
Overview of MILO workflow.
2.3. Traditional statistical analysis
Traditional statistics was also performed on each dataset via JMP Software (SAS Institute, Cary, NC). Data was also assessed for normality using the Ryan-Joiner Test. Continuous parametric variables were analyzed using the 2-sample t-test. A P value <0.05 was considered statistically significant with ROC analysis also performed to compare TB biomarker performance.
3. Results
3.1. Demographics
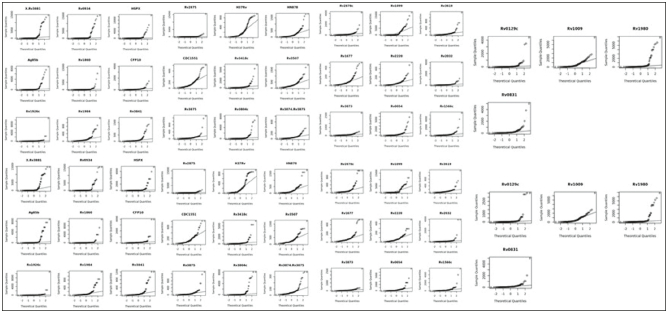
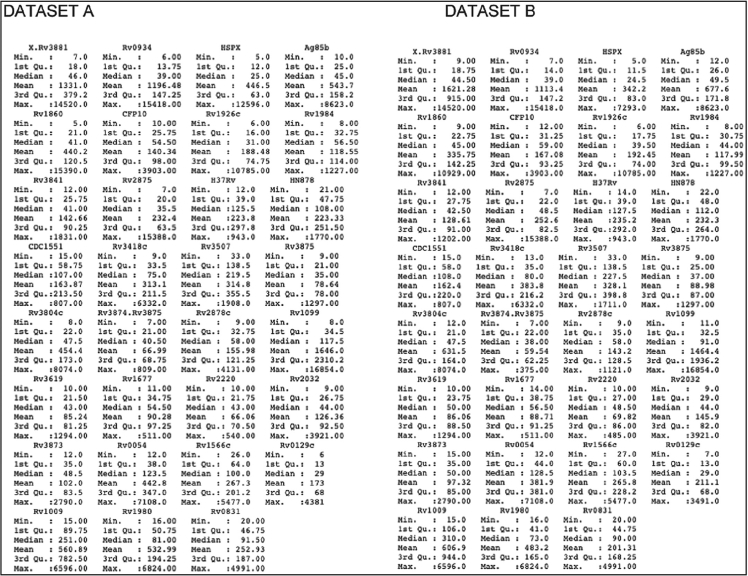
Fig. 2 illustrates histogram distributions for synthetic versus real data observed and Fig. 4 illustrates QQ plot for synthetic versus real data. Table 1 provides descriptive statistics for biomarkers used as features in ML training for datasets A and B.
Fig. 4.
QQ plot of Dataset A vs. Dataset B: The figure shows the Q-Q (quantile-quantile) plot for each attribute in the original dataset and the synthetic dataset. It shows that the distribution of each attribute is similar across the two datasets.
Table 1.
Descriptive statistics for biomarkers in Dataset A (real data) vs. Dataset B (synthetic ×1).
3.2. Machine learning performance
The nonautomated traditional (manual programming) ML approaches trained on Dataset A (real data) identified an RF model that produced an accuracy of 77.3%, with clinical sensitivity and specificity of 74.5% (95% CI, 66.3–81.5%) and 100% (95% CI, 80.5–100%) respectively. Using the same technique, RF models built on Dataset B showed an accuracy of 61.6%, with clinical sensitivity of 60.6% (95% CI, 51.9–68.8%) and specificity of 70.6% (95% CI, 44.0–89.7%). Random forest models built on Datasets C and D respectively yielded accuracies of 64.2% and 38.9%. Clinical sensitivity, were respectively, 63.5% (95% CI, 54.9–71.6%) and 40.2% (95% CI, 31.9–48.9%), and specificity was 70.6% (95% CI, 44.0–89.7%) and 29.4% (95% CI, 10.3–56.0%). ROC_AUC for the manually programmed RF models trained from Datasets A, B, C, and D were 0.97, 0.73, 0.83, and 0.68, respectively [Table 2].
Table 2.
Performance comparison of the models trained on real data versus synthetic data.
|
Model performances based on the “real” secondary dataset |
Trained on dataset A real data (95% CI) |
Trained on dataset B (synthetic data ×1) (95% CI) |
Trained on dataset C (synthetic data ×2) (95% CI) |
Trained on dataset D (synthetic data ×5) (95% CI) |
|---|---|---|---|---|
| MILO’s best models | MILO GBM | MILO SVM | MILO DNN | MILO DNN |
| ROC-AUC | 0.95 (0.87–1) | 0.83 (0.63–1) | 0.91 (0.8–1) | 0.55 (0.48–0.62) |
| Accuracy | 90 (84–95) | 91 (85–95) | 71 (63–78) | 54 (46–62) |
| Sensitivity | 89 (83–94) | 93 (87–96) | 67 (59–75) | 49 (40–58) |
| Specificity | 100 (81–100) | 77 (50–93) | 100 (81–100) | 94 (71–99) |
| MILO’s best RF models | MILO RF | MILO RF | MILO RF | MILO RF |
| ROC-AUC | 0.96 (0.82–1) | 0.77 (0.67–0.87) | 0.87 (0.77–0.97) | 0.66 (0.52–0.8) |
| Accuracy | 89 (83–93) | 71 (63–78) | 74 (66–81) | 56 (48–64) |
| Sensitivity | 88 (81–93) | 69 (60–76) | 72 (64–80) | 53 (44–61) |
| Specificity | 100 (81–100) | 88 (64–99) | 88 (64–99) | 82 (57–96) |
| Non-MILO RF models | Non-MILO RF | Non-MILO RF | Non-MILO RF | Non-MILO RF |
| ROC-AUC | 0.97 (0.94–1) | 0.73 (0.60–0.88) | 0.83 (0.71–0.92) | 0.68 (0.57–0.82) |
| Accuracy | 77 (70–84) | 62 (54–69) | 64 (56–72) | 39 (31–47) |
| Sensitivity | 75 (66–82) | 61 (52–69) | 64 (55–72) | 40 (32–49) |
| Specificity | 100 (81–100) | 71 (44–90) | 71 (44–90) | 29 (10–56) |
DNN = deep neural network, GBM = gradient boosting machine, RF = random forest, SVM = support vector machine.
As a comparison, the best MILO RF models (automated ML approach) showed slightly better performance within the various datasets evaluated. The RF MILO model built on the real dataset A showed an accuracy of 89% with a clinical sensitivity and specificity of 89% (95% CI, 83-93%) and 100% (95% CI, 81-100%), respectively. The best performing MILO RF model based on the synthetic dataset B (1:1 ratio with the real data) showed an accuracy of 71% and clinical sensitivity and specificity of 69% (95% CI, 60–76%) and 88% (95% CI, 64–99%), respectively. The best MILO RF model based on the expanded synthetic dataset C (×2) showed an accuracy of 74% with a clinical sensitivity and specificity of 72% (95% CI, 64–80%) and 88% (95% CI, 64–99%), respectively, while the best performing MILO RF model based on the expanded synthetic dataset D (×5) showed an accuracy of 56% with a clinical sensitivity and specificity of 53% (95% CI, 44–61%) and 82% (95% CI, 57–96%), respectively. ROC AUC for the MILO RF models trained from Datasets A, B, C, and D were 0.96, 0.77, 0.87, and 0.66, respectively [Table 2].
The overall best performing models (on the real and the synthetic datasets) were shown to be the MILO non-RF models. The best overall MILO model (a GBM model) built on the real dataset A showed an accuracy of 90% with a clinical sensitivity and specificity of 89% (95% CI, 83–94%) and 100% (95% CI, 81–100%), respectively. The best performing MILO model (an SVM model) based on the synthetic dataset B (1:1 ratio with the real data) showed an accuracy of 91% and clinical sensitivity and specificity of 93% (95% CI, 87–96%) and 77% (95% CI, 50–93%), respectively. The best overall MILO model used a neural network technique and based on the expanded synthetic dataset C (×2) showed an accuracy of 71% with a clinical sensitivity and specificity of 67% (95% CI, 59–75%) and 100% (95% CI, 81–100%), respectively, whereas the best performing overall MILO model based on the expanded synthetic (×5) dataset D (also a neural network model) showed an accuracy of 54% with a clinical sensitivity and specificity of 49% (95% CI, 40–58%) and 94% (95% CI, 71–99%) respectively. ROC AUC for the best (non-RF) MILO models trained from Datasets A, B, C, and D were 0.95, 0.83, 0.91, and 0.55, respectively [Table 2].
Overall, as shown above, compared to the random forest models evaluated through the non-automated (non-MILO) approach, the best MILO models (including the best MILO RF models) were shown to perform slightly better in both real and the synthetic dataset-trained models evaluated [Table 2]. Also, the overall ROC-AUC comparison measures, within the various models and datasets (in both the MILO and non-MILO approaches) showed the real dataset to be the best performing data followed by the expanded synthetic dataset ×2 when compared to the unexpanded (×1) synthetic dataset B and the expanded ×5 synthetic dataset D.
4. Conclusions
This work provides proof-of-concept for the utility of converting real-world patient datasets to synthetic datasets to aid in the development of ML models for differentiating TB positive and negative patients from complex serologic datasets. Importantly, the synthetic datasets allowed development of models with good performance characteristics upon validation in a secondary, real-world generalization dataset. This was true of models which were developed from both traditional (non-automated derived RF models) as well as the models derived from our automated ML approach. However, the overall MILO approach was able to find the better performing models within the synthetic datasets evaluated which supports its use within such settings to emulate real-world data modeling. The MILO approach also displayed that such approach is not model-specific, with its best performing models employing an array of algorithms (i.e., neural network, gradient boosting machine, and support vector machine), depending on the synthetic dataset utilized [Table 2]. Although models trained on datasets with the artificially increased sized (×5) synthetic data (Dataset D) showed decreased performance, the trend in this study showed that the unexpanded (×1) dataset (Dataset B) had the best overall accuracy while the slightly expanded (×2) synthetic dataset (Dataset C) yielding the overall best models, based on the ROC-AUC, within these synthetic datasets regardless of the modeling approach employed, MILO auto-ML or non-MILO RF.
Although this study shows that synthetic datasets can be employed for diagnostic modeling studies, the fact remains that the models trained on the real dataset outperformed the models that were trained on the synthetic data, regardless of the size of the synthetic data employed. Therefore, there remains a need for continually improving such synthetic datasets to help build models that can ultimately closely mimic the performance of the models that were based on real datasets. Continued improved methods in dataset processing may in the future allow manufacture of larger sample sizes with more realistic variations which may closely reflect the original real-world dataset. More important than boosting the size of the clinical datasets at this time is the capability of making them available with fewer patient privacy concerns. In that regard, we have shown that these synthetic datasets retain similar distribution of features, relationships among features, and most importantly the ability to train models which subsequently exhibit good performance in the desired task as measured against the secondary generalization dataset (real-world data not altered by the synthetic data generation process).
The development of deployable AI/ML algorithms with real-world utility is reliant upon the availability of robust datasets of sufficient size for model training, prior to validation and performance assessment on secondary generalization datasets.1, 2, 3 With the now widespread availability of computational storage for large datasets and processing speed to facilitate high-throughput algorithm training, AI/ML models are widely used in a range of applications from image recognition to control of autonomous vehicles.20 However, there currently appears to be an underutilization of these methods to solve challenges in healthcare given the widespread penetrance and successful implementation of AI/ML elsewhere in modern times.21 Clinical medicine at first glance appears to be an ideal application for these methods, given that high-impact diagnostic, prognostic, and treatment decisions are often made based on interpretation and synthesis of multiple quantitative and complex data elements. In addition, the advent of the EMR means that vast quantities of clinical data have been accumulated over the past several decades, and this only continues to accelerate with the advent of new diagnostic modalities with even larger data outputs (e.g., genomics and proteomics).
The successful application of AI/ML methods in other fields outside of healthcare has been less challenging, since available data may be widely disseminated and used for development in an open-source fashion. In contrast, healthcare data is heavily restricted due to patient privacy regulations.4 Access to such data must proceed through a very time-consuming and highly regulated process requiring researchers to submit a specific protocol defining the dataset required, how it will be developed, and outcome measures.1,2,5 This necessarily onerous process means that development on clinical data is highly limited, and there is a disconnect between data related to critical healthcare challenges, and the developers with the expertise to create models which may solve them. Often the clinical personnel who are most acutely aware of these needs do not have the specific data science expertise required for robust development, validation, and deployment of useful AI/ML algorithms. On the other hand, data scientists often lack the clinical background needed to define the scope of the critical tasks that AI/ML can be brought to bear in the healthcare domain.22,23 More importantly, although public datasets exist in other fields for developers, data scientists often lack access to clinical datasets, crippling development in this critical, high-impact field.
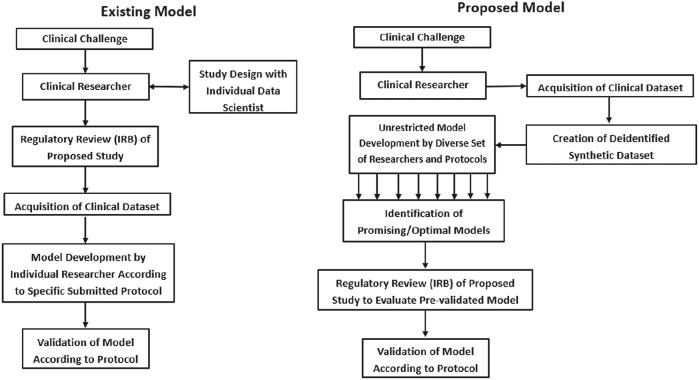
We propose a relatively new paradigm [Fig. 5] to address this shortcoming in the field, in which deidentified synthetic datasets may be made more accessible for development purposes. Developers may more freely explore these datasets, increasing the probability of discovery of optimal algorithms and diagnostic models. Models with great promise may subsequently be tested on additional real-world datasets, which may at that point require appropriate compliance with traditional institutional review board protocol. However, this step would only need to be taken after identification of suitable models, shifting the burden of regulatory compliance toward the generalization and validation phase, rather than prior to development. This would remove a now rate-limiting step which greatly impinges on AI/ML development in healthcare.
Fig. 5.
Paradigm for AI/ML development in healthcare. Synthetic data may help to improve access to clinical data if it is shown to reduce regulatory hurdles.
Increasing access to challenging problems in science and healthcare has previously resulted in solutions from unexpected sources. A user interface (Foldit) for the protein-folding software Rosetta allowed widespread access to non-scientists, who subsequently have provided solutions to difficult problems in protein structure prediction and design which previously challenged domain experts and existing algorithms.11,24 Increasing access to critical problems increases the likelihood of discovery of solutions by increasing throughput and diversification of possible solutions, both of which increase sampling depth and breadth. Indeed, a priori it may not be known which approach or algorithm may be optimal for a given task. Therefore, the best approach is to allow less restrictive research and development on each problem, rather than limiting model development on a particular task to the expertise and biases of the researcher with access to the dataset at hand. Increasing the connectivity of datasets and developers will inherently lead to improvements in model development and ultimately, potentially clinical outcomes. Computational researchers aim to avoid the trap of local energy minima (the best solution arising from one algorithm type or approach) and instead discover the true global minima (the true optimal model). In order to facilitate this, the social dynamics of the clinical healthcare and computational (AI/ML) researcher must allow widespread sampling of datasets by a diversity of approaches and personnel expertise. The most dramatic example of this is public distribution or crowd sourcing of such problems, but even expanding access to such problems beyond the confines of specifically approved protocols would represent a major enhancement to development of such protocols. To this end, we believe that adoption of an iterative model which makes clinical data available in a synthetic form prior to validation in a clinical setting may be a key step to realizing the full potential of AI/ML in healthcare.
A limitation to our study is that it is based on one dataset with relatively modest size and focused on TB. Further studies are needed to evaluate the impact of synthetic data for this and other medical disciplines.
Access to high-quality and accurate health record data remains an ongoing challenge for both routine patient care as well as for AI/ML development. Production of synthetic data derived from “real world” parameters provides opportunities to accelerate the development of AI/ML methods when data access remains limited. The use of synthetic data for training ML approaches to predict TB is feasible and supports further investigations in other disease states.
Financial support and sponsorship
Nil.
Author contributions
Hooman Rashidi served as PI and co-corresponding author for the paper. He developed and validated the ML algorithms used in the manuscript. Significantly contributed to the writing, review, and editing of the manuscript. Imran Khan served as co-investigator for this study and provided the TB dataset. He contributed to the writing and review of the manuscript. Luke T. Dang served as co-investigator for the study and supported the synthetic data analytics and non-automated machine learning development. He also produced Fig. 1, Fig. 5 for this paper. Samer Albahra developed and validated the ML algorithms used in the manuscript and helped with the ML studies validating the synthetic datasets. Ujjwal Ratan served as co-investigator for the study and provided the synthetic data analytics and non-automated machine learning development from Amazon Web Services. Wilson To served as co-investigator for the study and supported the synthetic data analytics and non-automated machine learning development from Amazon Web Services. Prathima Srinivas served as co-investigator for the study and supported the synthetic data analytics and nonautomated machine learning development from Amazon Web Services. Jeffery Wajda served as co-investigator for the study and contributed to the manuscript from the UC Davis Health perspective. Nam Tran serves as co-investigator for the paper. He contributed to the statistical review and writing of the manuscript. Performed basic statistical analysis of the study data as well as supporting the ML algorithm development with H.R. significantly contributed to the review and editing of the manuscript with co-authors.
Declaration of Competing Interest
Dr. Rashidi is a co-inventor of MILO and owns shares in MILO-ML, LLC. Dr. Albahra is a co-inventor of the MILO software and owns shares in MILO-ML, LLC. Dr. Tran is a co-inventor of the MILO software and owns shares in MILO-ML, LLC. He is also a consultant for Roche Diagnostics and Roche Molecular Systems.
References
- 1.Mayer-Schonberger V., Ingelsson E. Big data and medicine: A big deal? J Intern Med. 2017;289:418–429. doi: 10.1111/joim.12721. [DOI] [PubMed] [Google Scholar]
- 2.Singh R.P., Hom G.L., Abramoff M.D., et al. Current challenges and barriers to real-world artificial intelligence adoption for the health care system, provider, and the patient. Transl Vis Sci Technol. 2020;9:45. doi: 10.1167/tvst.9.2.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rashidi H.H., Tran N.K., Betts E.V., Howell L.P., Green R. Artificial intelligence and machine learning in pathology: The present landscape of supervised methods. Acad Pathol. 2019;6 doi: 10.1177/2374289519873088. 2374289519873088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agrawal R., Prabakaran S. Big data in digital healthcare: Lessons learnt and recommendations for general practice. Heredity (Edinb) 2020;124:525–534. doi: 10.1038/s41437-020-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller D.D. The medical AI insurgency: What physicians must know about data to practice with intelligent machines. NPJ Digit Med. 2019;2:62. doi: 10.1038/s41746-019-0138-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muthee V., Bochner A.F., Osterman A., et al. The impact of routine data quality assessments on electronic medical record data quality in Kenya. PLoS One. 2018;13 doi: 10.1371/journal.pone.0195362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner M.M., Hogan W.R. The accuracy of medication data in an outpatient electronic medical record. J Am Med Inform Assoc. 1996;3:234–244. doi: 10.1136/jamia.1996.96310637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah P., Kendall F., Khozin S., et al. Artificial intelligence and machine learning in clinical development: A translational perspective. NPJ Digit Med. 2019;2:69. doi: 10.1038/s41746-019-0148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobchev D.A., Pillai G.G., Karelson M. In silico machine learning methods in drug development. Curr Top Med Chem. 2014;14:1913–1922. doi: 10.2174/1568026614666140929124203. [DOI] [PubMed] [Google Scholar]
- 10.Nowok B., Raab G.M., Dibben C. Synthpop: Bespoke creation of synthetic data in R. J Stat Softw. 2016;74:1–26. [Google Scholar]
- 11.Koepnick B., Flatten J., Husain T., et al. De novo protein design by citizen scientists. Nature. 2019;570:390–394. doi: 10.1038/s41586-019-1274-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrero E., Dunham I., Sanseau P. In silico prediction of novel therapeutic targets using gene-disease associated data. J Transl Med. 2017;15:182. doi: 10.1186/s12967-017-1285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.CDC website https://www.cdc.gov/globalhealth/newsroom/topics/tb/index.html#:~:text=In%202018%2C%201.7%20billion%20people,1.5%20million%20lives%20each%20year Accessed on February26, 2021.
- 14.Walzl G., McNerney R., du Plessis N., et al. Tuberculosis: Advances and challenges in development of new diagnostics and biomarkers. Lancet Infect Dis. 2018;18:e199–e210. doi: 10.1016/S1473-3099(18)30111-7. [DOI] [PubMed] [Google Scholar]
- 15.World Health Organization Guidelines February 26, 2021. https://www.who.int/publications/guidelines/tuberculosis/en/
- 16.Khaliq A., Ravindran R., Hussainy S.F., et al. Field evaluation of a blood based test for active tuberculosis in endemic settings. Plos One. 2017;12 doi: 10.1371/journal.pone.0173359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tran N.K., Albahra S., Pham T.N., et al. Novel application of an automated-machine learning development tool for predicting burn sepsis: Proof of concept. Sci Rep. 2020;10:12354. doi: 10.1038/s41598-020-69433-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jen K.Y., Albahra S., Yen F., et al. Automated en masse machine learning model generation shows comparable performance as classic regression models for predicting delayed graft function in renal allografts. Transplantation. 2021;105:2646–2654. doi: 10.1097/TP.0000000000003640. [DOI] [PubMed] [Google Scholar]
- 19.Rashidi H.H., Makley A., Palmieri T.L., et al. Enhancing military burn- and trauma-related acute kidney injury prediction through an automated machine learning platform and point-of-care testing. Arch Pathol Lab Med. 2021;145:320–326. doi: 10.5858/arpa.2020-0110-OA. [DOI] [PubMed] [Google Scholar]
- 20.Forbes website https://www.forbes.com/sites/bernardmarr/2018/04/30/27-incredible-examples-of-ai-and-machine-learning-in-practice/?sh=19456af67502 Accessed on February 26, 2021.
- 21.Deist T.M., Patti A., Wang Z., Krane D., Sorenson T., Craft D. Simulation-assisted machine learning. Bioinformatics. 2019;35:4072–4080. doi: 10.1093/bioinformatics/btz199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garmire L.X., Gliske S., Nguyen Q.C., et al. The training of next generation data scientists in biomedicine. Pac Symp Biocomput. 2017;22:640–645. doi: 10.1142/9789813207813_0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunn M.C., Bourne P.E. Building the biomedical data science workforce. PLoS Biol. 2017;15 doi: 10.1371/journal.pbio.2003082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper S., Khatib F., Treuille A., et al. Predicting protein structures with a multiplayer online game. Nature. 2010;466:756–760. doi: 10.1038/nature09304. [DOI] [PMC free article] [PubMed] [Google Scholar]