Significance
Pseudouridine is among the most-abundant RNA modifications. We present a framework for conceptualizing how eukaryotic pseudouridine synthases select their substrates. This work reveals the structure of yeast pseudouridine synthase 7 (Pus7) and presents cell-based and biochemical investigations of enzyme binding and activity. We demonstrate that Pus7 interacts promiscuously with RNAs containing UGUAR sequences. Our observations raise the question of why these enzymes only modify <5% of UGUAR sequences in the transcriptome, suggesting that factors beyond inherent enzyme properties—such as protein localization, local RNA structure, and RNA–protein interactions—principally shape Pus7 substrate selection. These findings support a role for Pus7 in providing cells with a mechanism to rapidly alter protein synthesis in response to cellular conditions.
Keywords: pseudouridine, RNA modification, Pus7, TruD, structure
Abstract
Pseudouridine (Ψ) is a ubiquitous RNA modification incorporated by pseudouridine synthase (Pus) enzymes into hundreds of noncoding and protein-coding RNA substrates. Here, we determined the contributions of substrate structure and protein sequence to binding and catalysis by pseudouridine synthase 7 (Pus7), one of the principal messenger RNA (mRNA) modifying enzymes. Pus7 is distinct among the eukaryotic Pus proteins because it modifies a wider variety of substrates and shares limited homology with other Pus family members. We solved the crystal structure of Saccharomyces cerevisiae Pus7, detailing the architecture of the eukaryotic-specific insertions thought to be responsible for the expanded substrate scope of Pus7. Additionally, we identified an insertion domain in the protein that fine-tunes Pus7 activity both in vitro and in cells. These data demonstrate that Pus7 preferentially binds substrates possessing the previously identified UGUAR (R = purine) consensus sequence and that RNA secondary structure is not a strong requirement for Pus7-binding. In contrast, the rate constants and extent of Ψ incorporation are more influenced by RNA structure, with Pus7 modifying UGUAR sequences in less-structured contexts more efficiently both in vitro and in cells. Although less-structured substrates were preferred, Pus7 fully modified every transfer RNA, mRNA, and nonnatural RNA containing the consensus recognition sequence that we tested. Our findings suggest that Pus7 is a promiscuous enzyme and lead us to propose that factors beyond inherent enzyme properties (e.g., enzyme localization, RNA structure, and competition with other RNA-binding proteins) largely dictate Pus7 substrate selection.
Posttranscriptional modifications to the four standard RNA nucleosides increase the structural and functional complexity of RNAs. The C5-glycosidic isomer of uridine, pseudouridine (Ψ), is incorporated into multiple RNA species including transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), and eukaryotic messenger RNAs (mRNAs) (Fig. 1A). While Ψ has been studied in the context of noncoding RNAs for decades, the significance of Ψ in mRNAs is less well understood. Like all mRNA modifications, Ψ has the potential to affect every step in the life cycle of an mRNA (1). In line with this idea, mRNA pseudouridylation has been implicated as a regulator of human alternative splicing, yeast protein synthesis, and toxoplasma mRNA metabolism (2–4). Nonetheless, despite its prevalence, how cells select mRNAs for Ψ modification and the impacts of individual mRNA Ψ-sites on biological processes remain to be established.
Fig. 1.
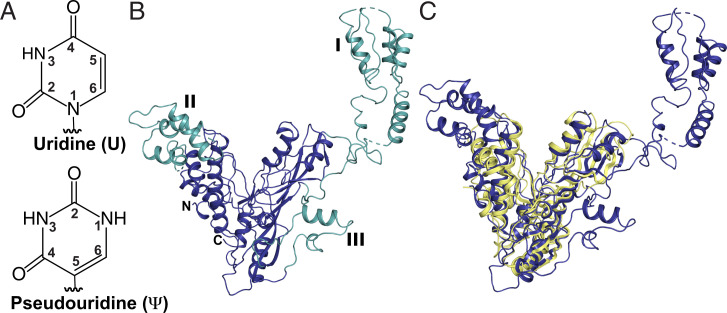
S. cerevisiae Pus7 structure. (A) Uridine and pseudouridine. (B) X-ray structure of Pus7 at 3.2-Å resolution (PDB: 7MZV). The structurally conserved, V-shaped enzyme core housing the PUS and TRUD domains (blue). The three eukaryotic-specific insertions (green) are numbered I through III. (C) Superimposition of the S. cerevisiae Pus7 (blue) and E. coli TruD (yellow, PDB: 1SB7) structures demonstrating the structural conservation of the enzyme’s catalytic core.
Ψ is installed into RNAs in all organisms by pseudouridine synthases (Pus). This large class of enzymes is categorized into six families: TruA, TruB, RluA, RsuA, TruD, and Pus10 (SI Appendix, Fig. S1). Despite the observation that many Pus enzymes are not required for cellular viability in nonstressed conditions, wild-type cells outcompete Pus-deficient cells, suggesting that these enzymes confer a fitness advantage (5–7). Consistent with this, Pus proteins markedly enhance cellular fitness under heat shock (8–11). Furthermore, mutations in the human TruA, TruB, and TruD family members (Pus1, Pus4, and Pus7, respectively) are linked to inherited diseases such as mitochondrial myopathy and sideroblastic anemia and intellectual disabilities (12–15). In addition to their enzymatic roles, there is emerging evidence that some Pus proteins can have alternative functions in the cell—acting as a tRNA folding chaperones or prions (16–18). One of the next horizons for the Pus field will be deconvoluting the contributions of each of the varied Pus activities (noncoding RNA– and mRNA-modifying or nonenzymatic) to gene expression.
Bacterial Pus family members have been studied for decades, and their structures, chemical mechanisms, and modes of RNA target selection are well characterized (19, 20). However, it is unclear why some eukaryotic Pus enzymes exhibit an increased substrate scope, modifying hundreds of mRNAs in addition to their traditional noncoding targets (21, 22). The enzymes responsible for catalyzing the bulk of Ψ incorporation into eukaryotic mRNAs are Pus1, Pus4, and Pus7 (23, 24). Recent transcriptome-wide Ψ-mapping and reporter studies indicate that mRNA secondary structure and sequence contribute to substrate selection by human Pus1 and Pus4 (25, 26). Pus7 is distinct from Pus1 and 4, as it shares ∼10% sequence identity with these enzymes and incorporates Ψ into a particularly diverse set of RNAs (tRNA, tRNA fragments, small nuclear RNA [snRNA], rRNA, and mRNA) (23, 24, 27–29). Pus7 has been shown to preferentially incorporate Ψ into UGUAR (U = U converted to Ψ, R = A or G) consensus sequences (23, 24, 28). However, this sequence motif does not explain how Pus7 selects its substrates, because only a limited set (<5%) of the UGUAR sequences present in RNAs are Pus7 targets.
To establish the determinants of Pus7 substrate selection, we first solved a crystal structure of Saccharomyces cerevisiae Pus7. Comparing the yeast and human Pus7 structures with their prokaryotic TruD counterparts, we noticed a large eukaryotic-specific insertion (insertion domain I, ID-I) sitting atop the active site in two distinct conformations. Both of the observed conformations for this domain are well positioned to potentially interact with incoming RNA substrates. Our biochemical and cell-based studies of Pus7 enzymes lacking ID-I (Pus7ΔIDI) support this supposition. We also evaluated the impact of RNA sequence and structure on Pus7 substrate binding and modification. Our results indicate that Pus7 preferentially associates with RNAs containing a UGUAR consensus sequence but only minimally discriminates between binding different RNA structures. The enzyme has a greater degree of selectivity during catalysis than binding, with the rate constant and extent of pseudouridylation increasing on RNAs with less predicted-structure both in vitro and in cells. Nonetheless, despite these preferences, Pus7 modifies all of the UGUAR containing substrates that we presented it in vitro. Our observations lead us to propose an opportunistic model for Pus7 substrate selection. In this model, Pus7 rapidly samples RNAs, binding and modifying any UGUAR sequence that it can access for long enough. This model is supported by observations that UGUAR sequences in structured contexts are modified more rapidly and efficiently at elevated temperatures, when RNA structures are destabilized. These findings extend the current framework for thinking about Pus enzymes by proposing that protein compartmentalization, local RNA structure, and RNA–protein interactions are among the most-substantive determinants for Pus7 substrate discrimination.
Results
S. cerevisiae Pus7 Structure Reveals Flexibility in the Architecture of the Eukaryotic Insertion Domains.
Pus7, like other TruD family members, shares little sequence identity with the four other Pus families (TruA, TruB, RluA, and RsuA) (30). Available structures of TruD from Escherichia coli (Protein Data Bank [PDB]: 1SI7, 1ZSW, and 1SB7), Methanosarcina mazei (PDB: 1Z2Z), and Pus7 from humans (PDB: 5KKP) show that the core of the enzyme has a V-shape formed by two conserved domains: the catalytic PUS domain and the TRUD domain (31–33). The structure of the PUS domain is highly conserved across all pseudouridine synthases (34), with the invariant catalytic aspartate falling on a β-strand that lies across the center of this domain. The TRUD domain is a mixed αβ fold conserved across all TruD homologs (30–33). This domain is oriented adjacent to the PUS domain, and together they form a catalytic cleft (V) lined with positively charged residues. The TRUD and PUS domains are connected by an extensive loop-rich region at the base of the cleft where the active site sits. Structural comparison of the five biological assemblies present in the three E. coli TruD structures demonstrates the ability of these core domains to flex toward each other by 18° (32). This motion could be facilitated by the hinge-like, loop-rich region connecting the domains and potentially help the enzyme to “clamp” down on RNA substrates.
Archaeal and eukaryotic TruD homologs contain large insertions at specific sites in the TRUD domain (33). Eukaryotic homologs possess two additional unique insertions decorating the catalytic domain (31–33). The largest of these eukaryotic-specific insertions, ID-I, is a small domain first visualized in the structure of human Pus7 (PDB: 5KKP). This domain is conserved in yeast Pus7 (SI Appendix, Fig. S2) (30). Eukaryotic members of the TruD family target a much-larger range of RNAs than their bacterial counterparts, which only modify U13 on tRNAGlu (30). Given their conservation and the expanded substrate selection observed, it stands to reason that structural features (insertions) unique to the eukaryotic enzymes might play a crucial role in the recognition and modification of their additional substrates.
To that end, we sought to develop a structural basis for understanding how eukaryotic insertions can contribute to Pus7 substrate selection and modification. We used a combination of molecular replacement (using human Pus7 as a model) and single-wavelength anomalous dispersion phasing to obtain a crystal structure of S. cerevisiae Pus7 to 3.2-Å resolution (Fig. 1B and SI Appendix, Table S1 and Fig. S2) (PDB: 7MZV). The core of the Pus7 structure strongly resembles that of TruD (RMSD = 3.74 Å based on 144 atoms), adopting the characteristic V-shape conformation of the PUS and TRUD domains (Fig. 1C and SI Appendix, Fig. S2). The active site sits near the bottom of the cleft adjacent to the hinge region where the two domains interface. The universally conserved catalytic aspartate (D256) resides on a loop between β3 and β4, is surrounded by residues conserved within the TruD family (K61, F67, E71, N305, and F307) (SI Appendix, Fig. S3) and sits ∼4 Å closer to the conserved active site residues in Pus7 than in TruD (SI Appendix, Fig. S2). Such positioning is consistent with the location of the catalytic Asp in the cocrystal structure of TruB with an RNA (PDB: 1K8W) (32). The most-notable structural difference between Pus7 and TruD is the presence of the three eukaryotic-specific insertions that decorate the enzyme’s V-shaped core (ID-I [aa 75 to 215], II [Ins-II, aa 365 to 443], and III [Ins-III, aa 594 to 634]) (Fig. 1B). These insertions are connected to the catalytic domain by flexible linkers.
A structural alignment of yeast and human Pus7 reveals that the three insertions adopt similar folds and are present at equivalent locations in both structures (SI Appendix, Fig. S2). ID-I folds into an extensive domain that is anchored to the top exterior (side opposite the active site) of the PUS domain by flexible linkers. ID-I is observed in two distinct orientations above the active site (SI Appendix, Fig. S2). Ins-III, the smallest insertion, is located on the exterior of the catalytic domain near the hinge region. Ins-II is a helical bundle perched atop the TRUD domain, and together, ID-I and Ins-II elongate the cleft between the PUS and TRUD domains. These insertions are ideally positioned to potentially interact with RNA substrates. Comparison of human and yeast Pus7 structures demonstrates that ID-I and Ins-II can adopt different orientations relative to the core of the protein. In yeast Pus7, ID-I extends laterally away from the protein, while in human Pus7, it is slightly rotated to be positioned directly above the active site. This suggests that ID-I can move as a rigid body, both swiveling laterally (akin to a flag on a pole) over the top of or away from the active site. Ins-II is also found in slightly different orientations in each structure, hinting to its rotational and translational freedom of motion. Ins-II in yeast Pus7 is pivoted away from the active site but angled in toward the catalytic cleft in human Pus7. Though the physiological relevance of these orientations is not immediately apparent, the freedom of motion and inherent flexibility could enable Ins-II or ID-I to play a part in recognizing regions of the substrate distal to the site of modification.
To determine whether ID-I shares homology with known RNA-binding motifs, we performed a protein structure comparison search with the DALI server (35). Using ID-I from both the yeast and human Pus7 structures, our results indicate that ID-I shares strong structural homology with the RNA-binding R3H domains. The strongest match was to a Poly(A)-specific ribonuclease domain (PDB: 2A1S, Z = 6.3, %ID =21) that utilizes an R3H motif to bind single-stranded nucleic acids. This motif contains an invariant arginine separated from a conserved histidine by three residues. Sequence alignments demonstrate that these residues are present and conserved in ID-I (SI Appendix, Fig. S3). Additionally, the structure of the R3H motif is characterized by two α-helices packed against a three-strand β-sheet, which aligns well with the structures of ID-I in yeast and human Pus7 (SI Appendix, Fig. S4). ID-I’s predicted ability to interact with RNAs and its stark positional variance relative to the active site suggest that ID-I may serve as an attenuator of RNA-binding.
Pus7 Enhances S. cerevisiae Viability under Translational Stress.
Pus7 enhances cellular fitness under temperature stress and modifies a wider variety of mRNA targets under heat shock (24). Given the recently discovered links between Pus7 mutations and neurological defects, we hypothesized that Pus7 is important for cellular health under additional stress conditions (12). To test this supposition, we compared the growth of wild-type and pus7Δ S. cerevisiae cells under 15 different conditions: multiple temperatures (22 °C, 30 °C, and 37 °C), elevated salt concentrations (NaCl and MgCl2), varied pH (pH 4.5 and 8.5), carbon sources (glucose, sucrose, and galactose), proteosome stress (MG132), and translational stress (puromycin, paromomycin, cycloheximide, and hygromycin). Cell growth was assessed by spot-plating on solid media and growth curves in liquid media (SI Appendix, Figs. S5 and S6). pus7Δ cells do not exhibit a growth defect relative to wild-type cells in YPD (yeast extract peptone dextrose) media at 22 °C or 30 °C but are sensitive to increased temperature (37 °C) as previously reported (11). We did not observe any carbon source–dependent growth changes between the wild-type and the knockout cells. The pus7Δ strain has a slight sensitivity to high concentrations of NaCl but not MgCl2. This is consistent with high-throughput studies that identified pus7Δ as one of ∼300 yeast knockouts that are more sensitive than wild-type cells to hyperosmotic (1 M NaCl) stress (36).
Translation inhibitors had the largest impact on pus7Δ growth relative to wild-type cells. pus7Δ cells are more sensitive to puromycin, cycloheximide, and hygromycin and exhibit a decreased sensitivity to paromomycin (SI Appendix, Figs. S5 and S6). This makes sense as Pus7 modifies both tRNAs and mRNAs, which could impact translation. To test hypothesis, we analyzed available ribosome-profiling data sets for pus7Δ (37, 38). We observe that ribosome occupancy is increased on Pus7 targeted mRNA codons in pus7Δ cells (SI Appendix, Fig. S7), consistent with recent reports that mRNA pseudouridinyltion slows translation elongation (3, 39). Together, our results indicate that Pus7 is likely to be particularly important when cellular translation is under stress.
Conserved Pus7 Active Site Residues Enhance RNA Modification.
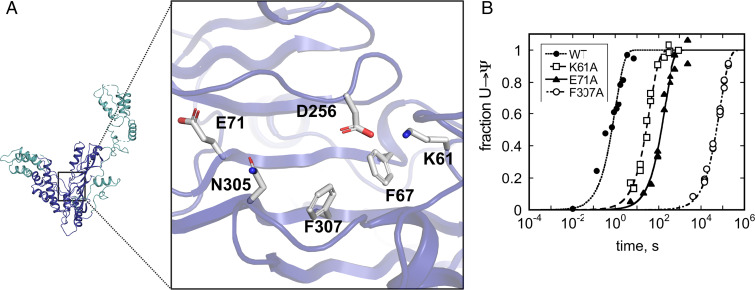
Our crystal structure revealed that the Pus7 active site is similar to that of TruD and suggests that ID-I might be positioned to contribute to enzyme function. We next wanted to test whether conserved Pus7 active site residues and ID-I enhance the ability of the enzyme to modify a reported mRNA substrate, CDC8 (24). To accomplish this, we measured the single-turnover rate constants (kobs) for Ψ incorporation into a 61 nt–long region of CDC8 by wild-type and mutant enzymes (D256A, K61A, E71A, F67A, H161A, N305A, F307A, F307Y, and Pus7ΔID-I) (Fig. 2 and Table 1). In these experiments, the Pus7 enzymes (2 to 10 μM) were each incubated with 3H-labeled CDC8 RNA (<100 nM) and 3H release upon conversion of U to Ψ was monitored at discrete time points (40). As expected, mutation of the catalytic D256 residue to alanine abolishes Pus7 activity, with no Ψ formation observed after 16 h (41). Alanine substitution of the nearby active site residues K61, F67, and E71 reduced the kobs for CDC8 modification by 40- to 200-fold, consistent with the proposed role of these residues in substrate positioning during catalysis in TruD. Mutations to the conserved active site NxF motif had larger impacts on Pus7 activity, with N305A and F307A mutants decreasing kobs by up to 50,000-fold relative to the wild-type enzyme. The F307A defect was partially rescued by a F307Y mutation (400-fold reduction in kobs), suggesting that the F307 base stacks with the target uridine to enhance CDC8 modification. In contrast to the active site mutants, kobs is unchanged by the ID-I point mutation H161A. However, removal of ID-I (Pus7ΔID-I) reduced kobs by twofold (SI Appendix, Table S2). These data reveal that conserved active site residues are important for Ψ modification by Pus7, while ID-I does not have a large influence on the rate determining step for Pus7 under saturating enzyme concentrations.
Fig. 2.
Pus7 active site residues enhance catalysis. (A) Conserved Pus7 active site residues investigated in this study. (B) Time courses for Ψ incorporation into a CDC8 mRNA by saturating concentrations of (●) wild-type Pus7 and Pus7 active site mutants ([□] K61A, [▲] E71A, and [○] F307A). The single-turnover rate constants (kobs) for alanine substitutions of all of the residues displayed in A are reported in Table 1.
Table 1.
Wild-type and mutant Pus7 single turnover rate constants and dissociation constants for CDC8
| PUS7 | kobs (s−1)* | KD,app1 (nM)† |
| WT | 9.9 ± 1.0 × 10−1 | 75 ± 15 |
| K61A | 2.6 ± 0.1 × 10−2 | 180 ± 40 |
| F67A | 4.6 ± 0.2 × 10−3 | 480 ± 50 |
| E71A | 5.2 ± 0.3 × 10−3 | 210 ± 50 |
| H161A | 6.9 ± 0.9 × 10−1‡ | 170 ± 40 |
| D256A | N.A.§ | 60 ± 15 |
| N305A | 4.0 ± 0.1 × 10−4 | 230 ± 60 |
| F307A | 1.3 ± 0.1 × 10−5 | 340 ± 170 |
| F307Y | 2.6 ± 0.1 × 10−3 | N.D.¶ |
| ΔID1 | 3.8 ± 0.6 × 10−1‡ | 160 ± 40 |
*kobs values were determined by tritium release assays using 2 μM PUS7 and <100 nM CDC8 substrate. At least three replicate curves were collected for each mutant.
†KD,app1 values were determined by EMSAs (n ≥ 2) using Pus7 containing the catalytically inactive D256A mutation in addition to the mutation indicated in the first column.
‡kobs value determined using 10 μM Pus7 and <100 nM CDC8 substrate.
§N.A., no activity.
¶N.D., not determined.
Multiple Pus7 Enzymes Bind to Unmodified and Modified CDC8.
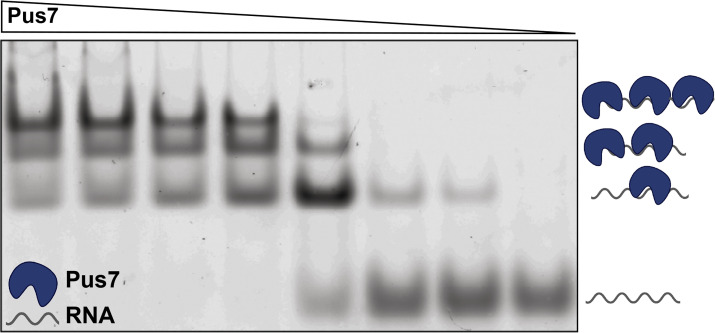
To assess the contributions of active site residues and ID-I to RNA substrate-binding, we performed electrophoretic mobility shift assays (EMSAs) with a 5′-fluorescein labeled CDC8 and a series of catalytically inactive Pus7 mutants (D256A mutant background). We observed a single band shift at low enzyme concentrations (<50 nM), indicating the formation of a Pus7-CDC8 (ES) complex (Fig. 3). For all of the mutants evaluated the singly bound species is supershifted when enzyme concentrations are increased (Fig. 3 and SI Appendix, Fig. S8). These additional bands indicate the formation of complexes that have multiple Pus7 proteins associated with CDC8 (ESEn complexes). We speculate that these lower-affinity binding events reflect nonspecific interactions between Pus7 and the RNA phosphodiester backbone. This may be a general property of Pus enzymes, as we also observe a similar binding behavior for the Pus family member Pus1 interacting with CDC8 (SI Appendix, Fig. S9). For both enzymes, the transition from the 1:1 complex to the n+1:1 complex occurs over a very narrow concentration range, reflecting the large number of nonspecific binding sites available on each RNA. Based on these observations, we considered several binding models (SI Appendix, Fig. S10) and ultimately fit our data to a simplified mechanism in which Pus7 independently binds a unique site tightly (KD,app1: low nM) and multiple additional sites with reduced affinities (KD,app2: high nM–μM) (SI Appendix, Extended Methods). Such a model is supported by stopped-flow studies with 5′-fluorescein–labeled CDC8 and unlabeled D256A Pus7. At low enzyme concentrations, we observed a single exponential phase, whose rate constant (kobs1) is linearly dependent on enzyme concentration (SI Appendix, Fig. S11). As we increase enzyme concentration, a second phase emerged (kobs2) consistent with our EMSAs indicating that multiple proteins bind to Pus7 RNA targets.
Fig. 3.
Multiple Pus7 proteins bind to CDC8 RNA. The association of increasing concentrations of catalytically inactive D256A Pus7 with limiting amounts of 5′-fluorescein-labeled CDC8 visualized on a nondenaturing agarose gel. Increased concentrations of D256A resulted in multiple binding events.
We applied our binding model to obtain KD,app1 values for D256A, D256A/K61A, D256A/F67A, D256A/E71A, D256A/H161A, D256A/N305A, D256A/F307A, and D256A/Pus7ΔID-I binding to CDC8 (Table 1). D256A binds CDC8 tightly with KD,app1 = 60 ± 15 nM, consistent with the KD estimated for D256A from our stopped-flow assays (kon,app = 4.3 × 108 M−1s−1, koff,app = 35 s−1, and koff,app/kon,app = KD ∼85 nM) (SI Appendix, Fig. S10). Additional active site point mutations increased the KD,app1 for Pus7 binding CDC8 by two- to eightfold relative to D256A (Table 1 and SI Appendix, Fig. S8). Similarly, removal of ID-I (D256A/Pus7ΔID-I) increased the KDapp,1 for CDC8 by twofold. Many enzymes bind their products with different affinities than their substrates. To test whether Pus7 discriminates between unmodified and Ψ-modified transcripts, we measured the KDapp,1 values for Pus7 and Pus7ΔID-I binding to modified CDC8. We found that wild-type Pus7 bound Ψ-modified CDC8 sequences with an affinity similar to its catalytically inactive counterpart (D256A) for an unmodified CDC8 substrate (Table 1). In contrast, Pus7ΔID-I has an eightfold weaker affinity for a Ψ-modified CDC8 than D256A/Pus7ΔID-I. This indicates that Pus7 does not distinguish between substrate or product during binding and that ID-I promotes this lack of discrimination.
ID-I Influences the Extent of Ψ Incorporation in Full-Length mRNAs In Vitro and in Cells.
Our binding studies suggest that ID-I has the potential to impact substrate selection. We tested this hypothesis by evaluating Ψ incorporation under conditions in which Pus7 must distinguish between all cellular RNAs. Pus7 and Pus7ΔID-I proteins (50 μM) were incubated for 10 min at 30 °C with 150 μg of total RNA purified from pus7Δ cells. The extent of Ψ incorporation on previously reported Pus7 modified substrates (ARG5,6, BET2, TEF5, RTC3(U77), RTC3(U288), TEF2(U555), and TEF2 (U1104)) was measured using N-cyclohexyl-N′-β-(4-methylmorpholinium)ethylcarbodiimide–reverse transcription and ligation-assisted PCR analysis of Ψ modification (CLAP) (42). The RNAs treated with Pus7 and Pus7ΔID-I exhibited a similar range of stoichiometries (6 to 40% versus 7 to 60%) (SI Appendix, Fig. S12). While the range of Ψ incorporation levels is similar, the presence of ID-I impacted the extent of Ψ addition on four of the seven substrates that we examined—with Pus7ΔID-I incorporating Ψ at lower levels than Pus7 in ARG5,6, BET2, and TEF2(U555) and adding more Ψ in RTC3(U288) (SI Appendix, Fig. S13). We verified that ID-I influences how frequently Pus7 incorporates Ψ in cells by using CLAP to measure Ψ-levels on BET2, RTC3(U77), RTC3(U288), TEF2(U555), and TEF2 (U1104) mRNAs purified from cells containing or lacking ID-I (SI Appendix, Fig. S13). Both in vitro and in cells, ID-I does not have a uniform effect on the substrates that we examined. Together, these data suggest that ID-I may act as a rheostat to fine-tune how Pus7 interacts with individual sequences. The idea that ID-I makes subtle contributions to Pus7 function is supported by our observation that cells expressing Pus7ΔID-I do not have a growth defect (SI Appendix, Figs. S14 and S15).
Pus7 Tightly Binds RNAs with an Array of Sequences and Secondary Structures.
The sequences targeted by Pus7 in cells are common in RNAs, yet only a small subset of potential target uridines are converted to Ψ. Pus1 and Pus4 are reported to use RNA secondary structure to recognize their substrates, and we wondered whether RNA structure similarly dictates which uridines Pus7 modifies (25, 26). To begin asking this question, we predicted the secondary structure context of S. cerevisae mRNA sequences modified by Pus7 (23). For each sequence, we modeled a 100-nucleotide region surrounding the site of Ψ incorporation. Secondary structure models were obtained using two different folding algorithms in the RNAstructure software package (free energy minimization and maximum expected accuracy) (23, 43). These predictions indicate that in cells Pus7 modifies Us in a wide variety of structural contexts including unstructured regions, loops, bulges, and helices (SI Appendix, Fig. S16). We noted that multiple Pus7 consensus sequences are often present within a single targeted mRNA. Therefore, we also examined the structural context of nonmodified sequences within Pus7 targeted mRNAs. Comparison of our structural models suggest that targeted uridines more commonly exist in less-structured contexts than nontargeted uridines present on the same mRNAs (SI Appendix, Fig. S16).
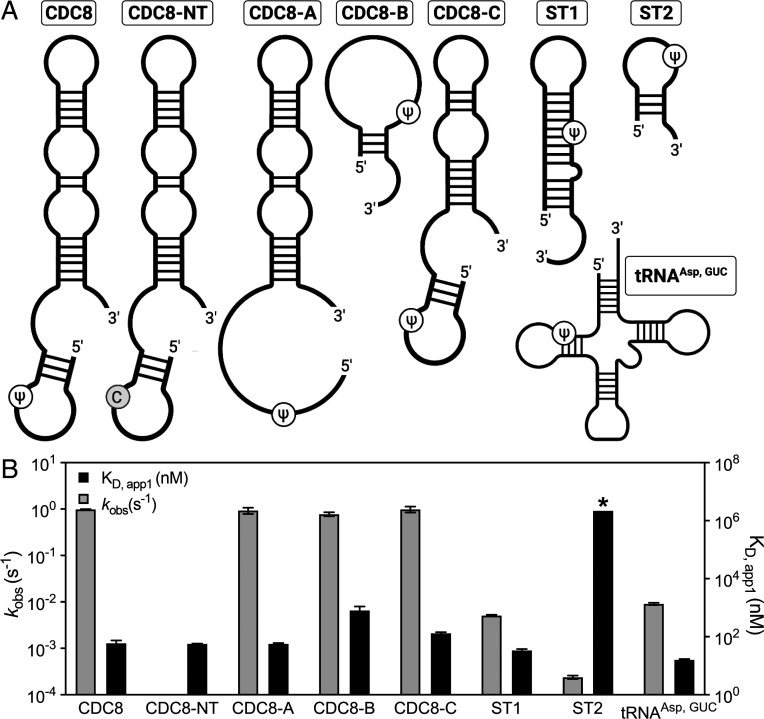
To experimentally evaluate the ability of Pus7 to interact with RNAs in a variety of structural contexts, we measured the binding of a catalytically inactive D256A Pus7 mutant to a series of 5′-fluorescein-labeled RNAs (Fig. 4A). These RNAs differ in both their sequence and secondary structures and include a natural tRNA target (tRNAAsp), three truncated CDC8 mRNAs (CDC8-A, CDC8-B, and CDC8-C), and two nonnatural substrates that place the target U in different structural contexts (ST1 and ST2). D256A bound to nearly all of the RNA substrates that we tested with similar affinities (16 to 130 nM) (SI Appendix, Table S3). Only the short (19 to 25 nt) CDC8-B and ST2 substrates significantly increased the KD,app1 for D256A, though the enzyme still bound the CDC8-B with a sub-μM dissociation constant (KD,app1 = 800 ± 320 nM) (Fig. 4 and SI Appendix, Table S3). These findings indicate that Pus7 has a substantial affinity for RNAs in general and, together with our stopped-flow binding data, lead us to propose that the enzyme rapidly searches for consensus sequences amid many nonspecific binding sites (SI Appendix, Fig. S17).
Fig. 4.
Pus7 can bind and modify a variety of RNA substrates. (A) Secondary structures of the RNAs investigated in this study. The substrate sequences are available in SI Appendix, Table S5. (B) KD,app1 (right y-axis, black bars) and kobs (left y-axis, gray bars) values for Pus7 binding and modifying the substrates displayed in A. The KD,app1 displayed for ST2 (*) is a lower limit for this value (SI Appendix, Fig. S9).
Pus7 Can Rapidly Incorporate Ψ on a Diverse Set of RNAs.
Our results indicate that PUS7 substrate-binding is largely independent of the RNA secondary structures and sequences that we investigated. We wondered whether RNA structural properties play a bigger role in determining the ability of Pus7 to modify substrates. To test this, we compared the single-turnover rate constants for Ψ incorporation into tRNAAsp, CDC8, CDC8-A/B/C, ST1, and ST2 (Figs. 4 and 5). We find that wild-type Pus7 modified the CDC8 mRNA substrates ∼10-fold faster than tRNAAsp (kobs,tRNA = 0.009 s−1 ± 0.0005 and kobs,cdc8 = 0.99 s−1 ± 0.1). The kobs,tRNA value that we measured is slower than previous studies of E. coli TruA, TruB, and RluA Pus enzymes, which have rate constants between 0.1 and 0.7 s−1 on their noncoding targets (44–46). As expected, no Ψ is incorporated into substrates when the target uridine is mutated to a cytidine (tRNAAspNT and CDC8NT). While the truncation of CDC8 (CDC8-A/B/C) does not alter the rate constant for pseudouridylation, we find that Pus7 incorporates Ψ 200- to 4,000-fold more slowly into the shorter nonnatural UGUAG-containing RNA sequences (ST1 and ST2). We anticipate that the reduced kobs for ST2 reflects its weak binding of Pus7 (Fig. 4 and SI Appendix, Fig. S8). Notably, the removal of ID-I (Pus7ΔID-I) partially recovered enzyme activity on ST1 (increasing kobs,ST1 by two-fold), suggesting that this domain may help to serve as a gatekeeper for substrate selection (Fig. 5). Together, our data demonstrate that Pus7 is capable of fully modifying any substrate containing a UGUAG target sequence, regardless of context. However, it does convert uridines to Ψ more quickly when they are present in regions of RNAs >25 nt in length predicted to be flexible single stranded.
Fig. 5.
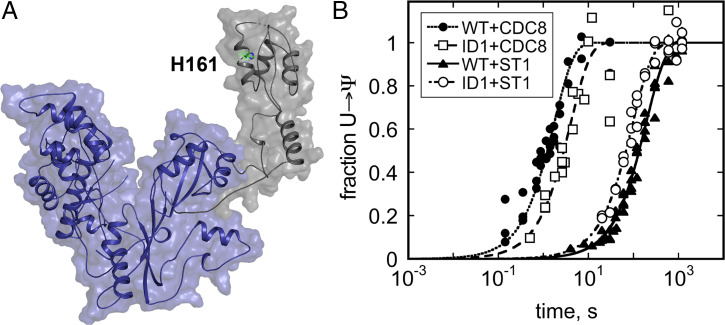
ID-I enhances Pus7 selectivity for CDC8 over ST1. (A) Crystal structure of Pus7 with ID-I shown in gray. The Pus7ΔID1 protein (blue) lacks ID-I (gray). (B) Time courses of Ψ incorporation into CDC8 and ST1 by wild-type Pus7 (●: CDC8; ▲: ST1) and Pus7ΔID1 (□: CDC8; ○: ST1). The single-turnover kobs values for these reactions are reported in SI Appendix, Table S4. ID-I enhances the ability of Pus to discriminate between CDC8 and ST1; wild-type kobs,CDC8/kobs,ST1 = 178, and Pus7ΔID1 kobs,CDC8/kobs,ST1 = 40.
Pus7 Activity toward Us Predicted to Be in Structured Regions Is Enhanced at Increased Temperatures.
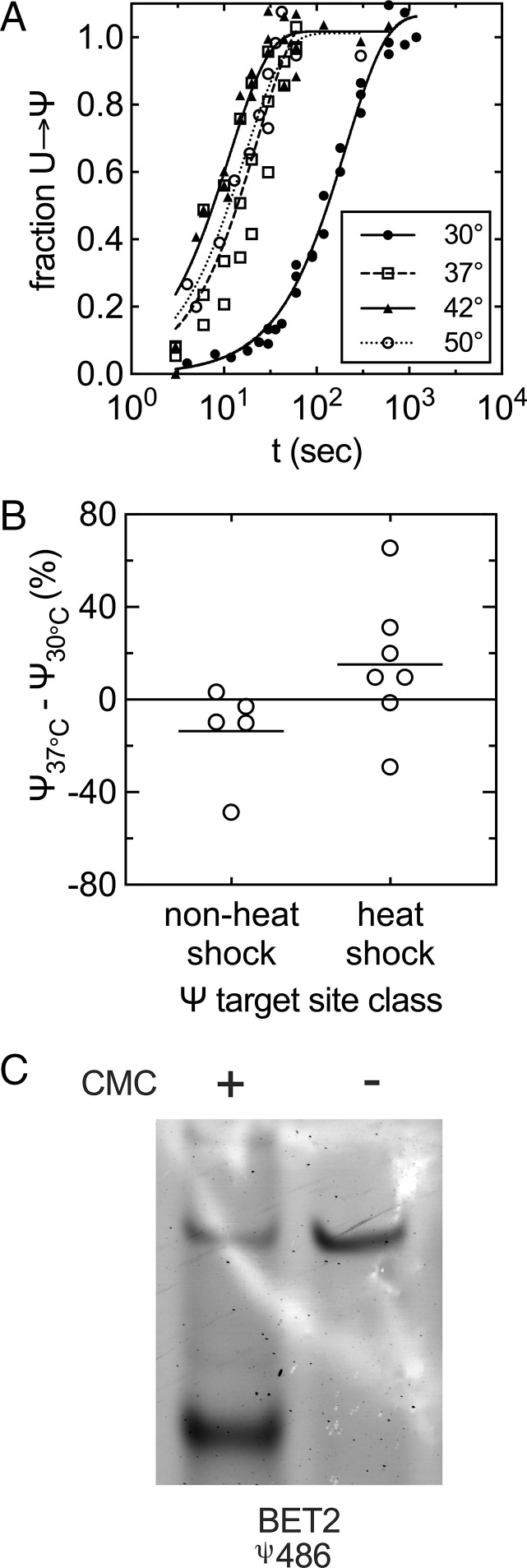
We further explored our observation that Pus7 appears to be more active on less structured UGUAR sequences by measuring the rate constants for Ψ incorporation into the ST1 substrate at elevated temperatures (37 °C, 42 °C, and 50 °C) where the structural stability of the RNA is reduced and the molecule should be more dynamic. If, as we hypothesize, base-pairing limits Pus7 activity toward substrates, then we anticipated that heating samples should increase pseudouridylation. Indeed, we find that kobs,ST1 is increased by 18-fold between 30 °C and 42 °C (Fig. 6A and SI Appendix, Fig. S19). There is still an enhancement in kobs,ST1 at 50 °C, albeit less than that at 42 °C, despite being closer to the Tm of ST1. We modeled the thermostability of Pus7, and our model indicates that Pus7 is not stable at 50 °C (SI Appendix, Fig. S20), likely accounting for the decrease in activity observed at this temperature.
Fig. 6.
Pus7 is more active at elevated temperatures on substrates with UGUAR sequences predicted to be in secondary structures. (A) Time courses of Ψ incorporation into ST1 by wild-type Pus7 at varying temperatures (●: 30 °C; □: 37 °C; ▲: 42 °C; and ○: 50 °C). The single-turnover kobs values for these reactions are reported in SI Appendix, Fig. S19. (B) Differences in the stoichiometry of Ψ incorporation at 30 °C and 37 °C in full-length RNA substrates measured by CLAP. The level of Ψ addition is generally enhanced at sites that were only detectable under heat shock in Schwartz et al. (24). (C) Representative CLAP gel of BET2 pseudouridylated target site from total RNA purified from BY4741 yeast pus7Δ::kanMX. Black arrow denotes the truncated, pseudouridylated product. The upper band is the unmodified, full-length product.
In addition to our in vitro observations, we modeled the structures of 20 randomly selected mRNAs reported to be modified by Pus7 under heat shock at 30 °C and 45 °C (SI Appendix, Fig. S20). Comparison of these models reveals that most of the targeted Us that we modeled are in different, often less-structured contexts at 45 °C than at 30 °C (SI Appendix, Fig. S13). We tested if the predicted decrease in substrate structure correlates with increased modificaiton levels by measuring the degree of Ψ incorporation on nine full-length Pus7 substrates (ARG5,6, BET2, TEF5, RTC3(U77), RTC3(U288), TEF2(U555), TEF2 (U1104), U2snRNA(U35), and U2snRNA(U56)) at different temperatures by CLAP (42). In these assays, we reacted 150 μg of total RNA purified from pus7Δ cells with 50 μM of purified Pus7 at 30 °C and 37 °C for 10 min. The extent of pseudouridylation increased at 37 °C on all but one of the “heat shock” targets with more predicted secondary structure (TEF5, RTC3(U77), RTC3(U288), TEF2(U555), and TEF2 (U1104)) (Fig. 6B). In contrast, the level of pseudourdinylation on targets previously observed in Ψ-mapping studies at 30 °C (ARG5,6, BET2, and U2snRNA(U35)) or under nutrient starvation (U2snRNA(U56)) were either unaffected or decreased (Fig. 6B).
Discussion
Our fundamental understanding of the structure and mechanism of pseudouridine synthases is largely built on foundational studies of bacterial enzymes that exclusively modify noncoding RNAs. The discovery of pseudouridine in eukaryotic mRNAs, coupled with the identification of heritable diseases caused by mutations to human Pus enzymes, have ignited a renewed interest in eukaryotic pseudouridine synthases. While all Pus enzymes share a structurally conserved catalytic core, the eukaryotic enzymes possess additional insertions with unidentified functions. Pus7, a homolog of the bacterial TruD enzyme, is among the Pus enzymes that modify the largest number of mRNAs in eukaryotes (23, 24). We solved the crystal structure of S. cerevisiae Pus7, revealing the inherent flexibility in the form and position of Pus7 eukaryotic-specific insertions (Fig. 1B). The largest insertion (ID-I) is a positively charged domain connected to the Pus7 core by long flexible linkers. Normal mode analyses of our structure with elNémo and DynOmics Portal 1.0, along with the differing positions of ID-I observed in the two Pus7 structures, suggest that this domain is very flexible. ID-I appears capable of swinging away the core of the molecule, which may allow it to form contacts with RNA substrates distal from their modification sites. Our findings that the removal of ID-I (Pus7ΔIDI) reduces RNA-binding and enhances substrate discrimination are consistent with such a model (Fig. 5 and Table 1). Nonetheless, in contrast to the large effect of active site mutations on substrate catalysis (38- to 74,000-fold), the impacts of ID-I deletion both in vivo and in vitro are modest (two- to fourfold reduction in Ψ incorporation) and indicate that the role of ID-I is more likely to fine-tune RNA substrate selection (Fig. 5 and SI Appendix, Fig. S13).
Pus7 is distinguished from other pseudouridine synthases by the apparent variety of substrates that it has been reported to modify in cells. Our in vitro biochemical assays support this idea, as we find that Pus7 can bind and pseudouridylate UGUAR motifs in diverse sequence and structural contexts (Fig. 4). Our kinetic and CLAP data indicate that although Pus7 can modify uridines predicted to be in strong secondary structures, it is most active on Us in regions with less-predicted structure (Figs. 4 and 6 and SI Appendix, Fig. S19). The relatively slow kobs value that we measured for tRNAAsp, in which the targeted uridine is base paired, exemplifies this trend. These biochemical observations are in line with the predicted structural contexts of uridines targeted by Pus7 in yeast cells. The models we obtained using RNAstructure reveal a trend for Pus7 substrate selection in which modified UGUAR sequences are in less-structured regions than unmodified UGUAR sequences (SI Appendix, Fig. S16). These observations can help to partially rationalize which UGUAR sites Pus7 modifies in cells.
Under heat shock, Pus7 modifies 15-fold more mRNAs (24). Consistent with this, we found that the ST1 RNA, predicted to contain a base-paired UGUAG sequence, is modified more efficiently at elevated temperatures when the stability of the base-paired region of the molecule is significantly reduced (Fig. 6A and SI Appendix, Fig. S19). Additionally, we measured the stoichiometry of Ψ incorporation in full-length RNAs previously reported to be modified under either unstressed or heat-shock conditions (23, 24). We saw that heat shock–induced sites are more efficiently modified at higher temperatures than Ψ sites detected in unstressed cells (Fig. 6B). This is notable because Pus7 should be less stable (SI Appendix, Fig. S20) and presumably less active at elevated temperatures. Our data suggest that increased RNA dynamics are more important than having optimal enzyme activity on these substrates.
Although our data indicate that Pus7 more quickly modifies structurally unconstrained Us, we find that if left for long enough (2 to 10 min) Pus7 converts 100% of Us to Ψs in all of the model UGUAG-containing sequences that we present it (Fig. 4). Similarly, we also observed that Pus1 is able to modify sequences not predicted to contain its preferred secondary structure if allowed to react for 30 min (SI Appendix, Fig. S9). Since a significant portion of Us identified as Pus7 targets in sequencing studies are predicted to be in structured regions, a simplistic model in which Pus7 only interacts with single-stranded uridines does not satisfactorily explain either our in vitro studies or the breadth of targets identified by Ψ-mapping in cells. Our findings suggest that instead of identifying motifs that Pus7 can modify, we need to address why this promiscuous enzyme does not modify every UGUAR sequence in cells. We propose that Pus7 rapidly samples RNA sequences and opportunistically selects substrates that contain an accessible (even if only transiently) UGUAR sequence motif (SI Appendix, Fig. S15). Such a mechanism is reminiscent of DNA glycosylases that use facilitated diffusion to quickly scan nonspecific sites in their search for damaged bases (47, 48).
The work that we present here suggests that Pus7 is a promiscuous enzyme and that factors beyond inherent enzyme properties (e.g., enzyme localization, RNA structure, and competition with other RNA-binding proteins) significantly contribute to shaping Pus7 substrate selection (49, 50). This idea is exemplified by the observed relocalization of Pus7 to the cytoplasm and subsequent increased substrate scope under heat shock (24). The importance of protein localization and cellular conditions to Pus target selection is unlikely to be unique to Pus7. Pus4 was recently reported relocalize to the cytoplasm and have increased activity toward its mRNA substrates when it is in a prion conformation (18). Collectively, these findings indicate that the environment of RNA substrates, which remodels in response to changing cellular conditions, plays a previously unrecognized role in determining the Ψ modification landscape.
Methods
Native and SeMe-labeled S. cerevisiae Pus7 and Pus 1 proteins were expressed in E. coli BL21(DE3) cells and purified on sequential Ni(NTA) and Resource Q columns. Pus7 crystals were obtained by the sitting-drop vapor diffusion. Diffraction data were collected at Advanced Photon Source and processed using XDS. Phaser was used to obtain a molecular replacement solution and REFAC5 was used for model refinement. The quality of the final Pus7 model was assessed with MolProbity. Wild-type and pus7Δ growth were assessed in YPD supplemented with either NaCl, MgSO4, puromycin, cycloheximide, hygromycin B, MG132 and 1.5 to 3 mg/mL paromomycin, or yeast extract peptone supplemented with either glucose, galactose, or sucrose. All RNAs (SI Appendix, Table S5) were prepared by run-off T7 transcription of DNA oligonucleotides and labeled (5′ fluorescein or 3H) for binding, stopped-flow, and single-turnover experiments. Pus7 substrate-binding was evaluated by EMSA, and stopped-flow and modification were monitored by 3H-release. The stoichiometries of Ψ incorporation at specific sites in full-length mRNAs modifications were measured using CLAP. Detailed procedures and reaction conditions for all experiments are provided in SI Appendix, Extended Methods.
Supplementary Material
Acknowledgments
This work was funded by the University of Michigan start-up funds (M.K. and K.S.K.), Rackham Merit Fellowship (M.K.F.), NIH awards R35 GM128836 (K.S.K.), R01 GM117141 (M.K.), and T32 GM008597 (M.K.F.), and Research Corporation for Science Advancement Cottrell Scholar Award (K.S.K.). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the Life Sciences Collaborative Access Team (LS-CAT) Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). Biorender was used to create Figs. 5A and 6.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2109708119/-/DCSupplemental.
Data Availability
Crystal structure data have been deposited in Protein Data Bank (7MZV). All other study data are included in the article.
References
- 1.Jones J. D., Monroe J., Koutmou K. S., A molecular-level perspective on the frequency, distribution, and consequences of messenger RNA modifications. Wiley Interdiscip. Rev. RNA 11, e1586 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez N. M., et al. , Pseudouridine synthases modify human pre-mRNA co-transcriptionally and affect splicing. bioRxiv [Preprint]. 10.1101/2020.08.29.273565 (Accessed 31 August 2020). [DOI]
- 3.Levi O., Arava Y. S., Pseudouridine-mediated translation control of mRNA by methionine aminoacyl tRNA synthetase. Nucleic Acids Res. 49, 432–443 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakamoto M. A., Lovejoy A. F., Cygan A. M., Boothroyd J. C., mRNA pseudouridylation affects RNA metabolism in the parasite Toxoplasma gondii. RNA 23, 1834–1849 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gutgsell N., et al. , Deletion of the Escherichia coli pseudouridine synthase gene truB blocks formation of pseudouridine 55 in tRNA in vivo, does not affect exponential growth, but confers a strong selective disadvantage in competition with wild-type cells. RNA 6, 1870–1881 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Del Campo M., Kaya Y., Ofengand J., Identification and site of action of the remaining four putative pseudouridine synthases in Escherichia coli. RNA 7, 1603–1615 (2001). [PMC free article] [PubMed] [Google Scholar]
- 7.Breslow D. K., et al. , A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods 5, 711–718 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urbonavicius J., Durand J. M., Björk G. R., Three modifications in the D and T arms of tRNA influence translation in Escherichia coli and expression of virulence genes in Shigella flexneri. J. Bacteriol. 184, 5348–5357 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishida K., et al. , Pseudouridine at position 55 in tRNA controls the contents of other modified nucleotides for low-temperature adaptation in the extreme-thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res. 39, 2304–2318 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinghorn S. M., O’Byrne C. P., Booth I. R., Stansfield I., Physiological analysis of the role of truB in Escherichia coli: A role for tRNA modification in extreme temperature resistance. Microbiology (Reading) 148, 3511–3520 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Sinha H., et al. , Sequential elimination of major-effect contributors identifies additional quantitative trait loci conditioning high-temperature growth in yeast. Genetics 180, 1661–1670 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Brouwer A. P. M., et al. , Variants in PUS7 cause intellectual disability with speech delay, microcephaly, short stature, and aggressive behavior. Am. J. Hum. Genet. 103, 1045–1052 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeharia A., et al. , Mitochondrial myopathy, sideroblastic anemia, and lactic acidosis: An autosomal recessive syndrome in Persian Jews caused by a mutation in the PUS1 gene. J. Child Neurol. 20, 449–452 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Bykhovskaya Y., Casas K., Mengesha E., Inbal A., Fischel-Ghodsian N., Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA). Am. J. Hum. Genet. 74, 1303–1308 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaheen R., et al. , A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum. Genet. 135, 707–713 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keffer-Wilkes L. C., Veerareddygari G. R., Kothe U., RNA modification enzyme TruB is a tRNA chaperone. Proc. Natl. Acad. Sci. U.S.A. 113, 14306–14311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keffer-Wilkes L. C., Soon E. F., Kothe U., The methyltransferase TrmA facilitates tRNA folding through interaction with its RNA-binding domain. Nucleic Acids Res. 48, 7981–7990 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia D. M., et al. , A prion accelerates proliferation at the expense of lifespan. eLife 10, 10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Penzo M., Guerrieri A. N., Zacchini F., Treré D., Montanaro L., RNA pseudouridylation in physiology and medicine: For better and for worse. Genes (Basel) 8, 301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamma T., Ferré-D’Amaré A. R., Pseudouridine synthases. Chem. Biol. 13, 1125–1135 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Rintala-Dempsey A. C., Kothe U., Eukaryotic stand-alone pseudouridine synthases – RNA modifying enzymes and emerging regulators of gene expression? RNA Biol. 14, 1185–1196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boccaletto P., et al. , MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlile T. M., et al. , Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwartz S., et al. , Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Safra M., Nir R., Farouq D., Vainberg Slutskin I., Schwartz S., TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code. Genome Res. 27, 393–406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlile T. M., et al. , mRNA structure determines modification by pseudouridine synthase 1. Nat. Chem. Biol. 15, 966–974 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guzzi N., et al. , Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 173, 1204–1216.e26 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Behm-Ansmant I., et al. , The Saccharomyces cerevisiae U2 snRNA:pseudouridine-synthase Pus7p is a novel multisite-multisubstrate RNA:Psi-synthase also acting on tRNAs. RNA 9, 1371–1382 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma X., Zhao X., Yu Y. T., Pseudouridylation (Psi) of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J. 22, 1889–1897 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaya Y., Ofengand J., A novel unanticipated type of pseudouridine synthase with homologs in bacteria, archaea, and eukarya. RNA 9, 711–721 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ericsson U. B., Nordlund P., Hallberg B. M., X-ray structure of tRNA pseudouridine synthase TruD reveals an inserted domain with a novel fold. FEBS Lett. 565, 59–64 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Hoang C., Ferre-D’Amare A. R., Crystal structure of the highly divergent pseudouridine synthase TruD reveals a circular permutation of a conserved fold. RNA 10, 1026–1033 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaya Y., Del Campo M., Ofengand J., Malhotra A., Crystal structure of TruD, a novel pseudouridine synthase with a new protein fold. J. Biol. Chem. 279, 18107–18110 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Mueller E. G., Chips off the old block. Nat. Struct. Biol. 9, 320–322 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Holm L., Using Dali for protein structure comparison. Methods Mol. Biol. 2112, 29–42 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Yoshikawa K., et al. , Comprehensive phenotypic analysis for identification of genes affecting growth under ethanol stress in Saccharomyces cerevisiae. FEMS Yeast Res. 9, 32–44 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Blanchet S., et al. , Deciphering the reading of the genetic code by near-cognate tRNA. Proc. Natl. Acad. Sci. U.S.A. 115, 3018–3023 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chou H. J., Donnard E., Gustafsson H. T., Garber M., Rando O. J., Transcriptome-wide analysis of roles for tRNA modifications in translational regulation. Mol. Cell 68, 978–992.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eyler D. E., et al. , Pseudouridinylation of mRNA coding sequences alters translation. Proc. Natl. Acad. Sci. U.S.A. 116, 23068–23074 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Czekay D. P., Schultz S. K., Kothe U., Assaying the molecular determinants and kinetics of RNA pseudouridylation by H/ACA snoRNPs and stand-alone pseudouridine synthases. Methods Mol. Biol. 2298, 357–378 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Veerareddygari G. R., Singh S. K., Mueller E. G., The pseudouridine synthases proceed through a glycal intermediate. J. Am. Chem. Soc. 138, 7852–7855 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang W., Eckwahl M. J., Zhou K. I., Pan T., Sensitive and quantitative probing of pseudouridine modification in mRNA and long noncoding RNA. RNA 25, 1218–1225 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bellaousov S., Reuter J. S., Seetin M. G., Mathews D. H., RNAstructure: Web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 41, W471–W474 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wright J. R., Keffer-Wilkes L. C., Dobing S. R., Kothe U., Pre-steady-state kinetic analysis of the three Escherichia coli pseudouridine synthases TruB, TruA, and RluA reveals uniformly slow catalysis. RNA 17, 2074–2084 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang L., Pookanjanatavip M., Gu X., Santi D. V., A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. Biochemistry 37, 344–351 (1998). [DOI] [PubMed] [Google Scholar]
- 46.Ramamurthy V., Swann S. L., Spedaliere C. J., Mueller E. G., Role of cysteine residues in pseudouridine synthases of different families. Biochemistry 38, 13106–13111 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Taylor E. L., Kesavan P. M., Wolfe A. E., O’Brien P. J., Distinguishing specific and nonspecific complexes of alkyladenine DNA glycosylase. Biochemistry 57, 4440–4454 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Esadze A., Stivers J. T., Facilitated diffusion mechanisms in DNA base excision repair and transcriptional activation. Chem. Rev. 118, 11298–11323 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaidyanathan P. P., AlSadhan I., Merriman D. K., Al-Hashimi H. M., Herschlag D., Pseudouridine and N6-methyladenosine modifications weaken PUF protein/RNA interactions. RNA 23, 611–618 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerber A. P., Herschlag D., Brown P. O., Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2, E79 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystal structure data have been deposited in Protein Data Bank (7MZV). All other study data are included in the article.