Significance
Metagenomic data have recently transformed our view of the role played by sulfur metabolism in anoxic environments by showing that this trait is much more widespread than previously believed. A key enzyme in sulfur metabolism is the dissimilatory sulfite reductase DsrAB that is ubiquitous in organisms with a reductive, oxidative, or disproportionating activity. However, the function of some dsr genes, such as dsrD, has so far been unknown despite its use as a functional marker to genomically assign the type of sulfur energy metabolism, sometimes with unclear results. Here, we disclose the function of DsrD as an activator of DsrAB that significantly increases its activity, providing important insights into the mechanism of this enzyme in different types of sulfur metabolism.
Keywords: sulfur metabolism, dissimilatory sulfite reductase, sulfate-reducing bacteria, sulfur disproportionation, allosteric activation
Abstract
Dissimilatory sulfur metabolism was recently shown to be much more widespread among bacteria and archaea than previously believed. One of the key pathways involved is the dsr pathway that is responsible for sulfite reduction in sulfate-, sulfur-, thiosulfate-, and sulfite-reducing organisms, sulfur disproportionators and organosulfonate degraders, or for the production of sulfite in many photo- and chemotrophic sulfur-oxidizing prokaryotes. The key enzyme is DsrAB, the dissimilatory sulfite reductase, but a range of other Dsr proteins is involved, with different gene sets being present in organisms with a reductive or oxidative metabolism. The dsrD gene codes for a small protein of unknown function and has been widely used as a functional marker for reductive or disproportionating sulfur metabolism, although in some cases this has been disputed. Here, we present in vivo and in vitro studies showing that DsrD is a physiological partner of DsrAB and acts as an activator of its sulfite reduction activity. DsrD is expressed in respiratory but not in fermentative conditions and a ΔdsrD deletion strain could be obtained, indicating that its function is not essential. This strain grew less efficiently during sulfate and sulfite reduction. Organisms with the earliest forms of dsrAB lack the dsrD gene, revealing that its activating role arose later in evolution relative to dsrAB.
The reduction of sulfite is a key step in dissimilatory sulfate reduction (DSR), a microbial process performed by anaerobic bacteria or archaea, which derive energy from reducing sulfate to hydrogen sulfide (1, 2). DSR drives the biogeochemical sulfur cycle and has a strong influence on other element cycles and on the redox balance of the oceans and atmosphere. It has a particular impact in marine sediments where it accounts for up to 50% of carbon mineralization (3, 4) and prevents methane emissions by its involvement in anaerobic methane oxidation (5, 6). Remarkably, the existence of a cryptic sulfur cycle, involving several sulfur compounds, means that DSR and other sulfur metabolisms are also important in low-sulfate environments, where fermentation and methanogenesis were thought to dominate (2, 4, 7), and through their action prevent methane production by diverting carbon flow and energy resources away from methanogens. In addition, sulfate-reducing prokaryotes (SRPs) are members of the human gut flora, where their production of sulfide has been associated with inflammatory bowel diseases and cancer (8–10).
In DSR, sulfite reduction is performed by the DsrAB dissimilatory sulfite reductase, an α2β2 tetrameric enzyme that uses a [4Fe-4S]-coupled siroheme as a catalytic cofactor (1, 11, 12). Beyond its role in sulfate reduction, DsrAB is also a key enzyme in other types of microbial sulfur metabolism, being present in a wide range of other organisms including sulfur, thiosulfate, and sulfite reducers; sulfur disproportionators; organosulfonate degraders and in many photo- and chemotrophic sulfur-oxidizing prokaryotes (SOPs), where it operates in the reverse direction and is designated as reverse-acting DsrAB (rDsrAB) (2, 7, 13). DsrAB is an ancient enzyme whose two paralogous genes, dsrA and dsrB, evolved from the duplication of a simpler sulfite reductase gene (14, 15), and phylogenetically it is organized in three main families comprising the archaeal and bacterial reductive enzymes and the oxidative rDsrABs (16–18). The phylogenetic studies support a reductive origin for this enzyme and indicate that dissimilatory sulfite reduction likely preceded DSR (19, 20). The dsrAB genes are widely used as ecologic and phylogenetic markers of dissimilatory sulfur metabolism, and recent studies have uncovered that this metabolic trait is much more widespread in both bacteria and archaea than previously believed (18, 20–29).
Contrary to assimilatory and other dissimilatory sulfite reductases (13), DsrAB does not reduce sulfite directly to sulfide. Instead, DsrAB requires the action of its partner protein DsrC (12) to catalyze a four-electron reduction of sulfite to a S0 valence state, in the form of a trisulfide bound to DsrC (30). The DsrC-trisulfide is believed to be later reduced to HS− and DsrC by the DsrMKJOP transmembrane complex (12, 30, 31). All organisms that have dsrAB genes also have dsrC and minimally the dsrMK genes (12, 32). The presence of genes for the dsr pathway, which can include up to 15 genes, is characteristic of oxidative or reductive sulfite metabolism and has been used to tentatively discriminate between SRPs, SOPs, and organisms with other types of sulfur-based energy metabolism. For example, genes that have been considered characteristic of SOPs are rdsrAB and the dsrEFH genes that code for a sulfur donor protein to DsrC (7, 12, 33) and dsrL that is essential for sulfur oxidation and is a physiological partner for rDsrAB (34, 35), whereas the dsrD gene is considered to be characteristic of reductive sulfur metabolism (18, 21–23, 36). However, as highlighted in several studies, metabolic assignments based solely on genetic composition are far from clear in some cases, as it is the case for some organisms that have dsrEFH and/or dsrL along with reductive dsrAB (18, 21, 27, 35, 37). For a clear assignment of genes to metabolism types, it is obviously critical to have a good understanding of the function performed by the proteins encoded by those genes. An important advancement was the recent elucidation of the function of DsrL as an electron acceptor for rDsrAB coupled to the reduction of NAD+ (38) and the subsequent identification of different classes of DsrL proteins, with DsrL-2 being present in organisms with a reductive sulfur metabolism, where it acts as an electron donor for DsrAB, while oxidizing NADPH (35). These studies were critical to clarify that DsrL is also present in organisms with a reductive/disproportionating type metabolism. In contrast, the function of DsrD was so far completely unknown.
The dsrD gene, first identified in 1995 by Karkhoff-Schweizer and colleagues (39), is absent from SOPs and is present in most organisms encoding a reductive enzyme (i.e., most SRPs; sulfur disproportionators; and sulfite-, thiosulfate-, and organosulfonate-reducing organisms), being fused with dsrB in Bilophila wadsworthia (1, 12, 40, 41). Notably, it is absent in archaea containing early-diverging reductive DsrAB homologs (18, 20). The dsrD gene is usually found immediately downstream of dsrAB, and DsrD is among the highest expressed proteins (37, 42, 43), which together with its widespread distribution and genetic location all suggest an important role in the dissimilatory reduction of sulfite. In Desulfovibrio vulgaris Hildenborough (D. vulgaris H), a model SRP, dsrD was found to be downregulated under stationary phase conditions (44) and was the most downregulated gene when cells were exposed to high sulfide concentrations (10 mM vs. 1 mM) (45). In this organism, DsrD is composed of 78 amino acids, and its crystallographic structure displays a winged helix-turn-helix (wHTH) motif characteristic of DNA-binding proteins, which led to the suggestion that this protein could be a transcription regulator of the genes involved in DSR (46). DsrD also presents a high content of conserved lysine residues that could point to a role in anion binding, such as SO42−/SO32−/HS−, but this was not confirmed (39, 47). In addition, Desulfobacterota DsrD proteins contain a well-conserved motif (YWSSGSTT) with no function assigned, and the absence of cysteine residues or cofactors suggests that this protein is unlikely to be involved in electron transfer or in sulfur chemistry like DsrC (30, 39, 46). In this study, we report in vivo and in vitro studies, which provide a detailed functional and physiological characterization of DsrD. The in vivo studies were performed with the bacterium D. vulgaris H from Desulfobacterota (48), which is genetically tractable, and the in vitro biochemical assays were performed with proteins from the thermophilic euryarchaeon Archaeoglobus fulgidus, from which DsrAB can be purified in active form, in contrast to D. vulgaris H (30).
Results and Discussion
DsrD Interacts Nonspecifically with DNA.
Since the 2003 determination of the DsrD crystal structure revealing a winged-helix motif similar to those present in Z-DNA and B-DNA binding proteins, DsrD was associated with a possible DNA-binding function, eventually regulating the dsr genes (46). However, no experimental data have been reported so far supporting this possible function. We tested whether DsrD is able to bind to DNA and namely to the dsrAB promoter region. For this, we performed a competitive electrophoretic mobility shift assay by incubating A. fulgidus DsrD with a 981-bp DNA fragment from the A. fulgidus dsrAB promoter region and with the kanamycin-resistance gene (803 bp) as a nonspecific DNA fragment and control. In both situations, we observed a DNA shift in the presence of DsrD that was dependent on the DsrD concentration (SI Appendix, Fig. S1), indicating that DsrD can bind to DNA, as might be expected from its high content of positively charged lysine residues, but that this binding is unspecific. This does not support a regulatory function for DsrD, which is also disfavored by its very high cellular levels, in the same order as DsrAB or even more (37, 42, 43), which point to a direct role in dissimilatory metabolism.
DsrD Is Produced in Respiratory Conditions.
The presence of DsrD was analyzed in several growth conditions. For this, D. vulgaris H wild-type (WT) cells were grown in Missouri (MO) media in fermentative conditions (Pyr) and in respiratory conditions with lactate/sulfate (LS4), ethanol/sulfate (ES4), lactate/thiosulfate (LT), lactate/sulfite (LS3), and H2/acetate/sulfate (HS4); and then cells were collected at late exponential phase. The crude extracts were analyzed by Western blot against DsrD and DsrB as an internal control (Fig. 1A). The Western blot results indicate that DsrD is more abundant under LS4, LT, and LS3 conditions; less abundant in ES4 and HS4 conditions; and apparently absent in fermentative conditions. In contrast, the production of DsrB is constant in all growth conditions tested, which is in line with the constitutive expression of dsrAB. Interestingly, it was also under LS4, LT, and LS3 growth conditions that D. vulgaris H WT reached higher cell densities. These findings suggest a role for DsrD under respiratory conditions and support a direct function in sulfite reduction.
Fig. 1.
DsrD expression and growth studies D. vulgaris H strains. (A) Western blot of DsrD (1) and DsrB (2) expression in different growth media. (B to D) Growth curves of WT (blue), ΔdsrD mutant (orange), and ΔdsrD + pMO-dsrD complemented strain (gray) are show in the following panels: (B) 60 mM pyruvate (Pyr 60), (C) 30 mM lactate/30 mM sulfate (LS4 30/30), (D) 15 mM lactate/10 mM sulfite (LS3 15/10), and (E) 30 mM lactate/20 mM sulfite (LS3 30/20). Data points are mean ± SD, n = 3 independent experiments.
Deletion of dsrD Affects Respiratory Growth.
The dsrD gene could be deleted in D. vulgaris H to create the D. vulgaris H ΔdsrD strain through replacement by a kanamycin cassette. A complemented strain, D. vulgaris H ΔdsrD + pMO-dsrD, was also generated by inserting a plasmid encoding dsrD (pMO-dsrD) in D. vulgaris H ΔdsrD. Strains D. vulgaris H WT, D. vulgaris H ΔdsrD, and D. vulgaris H ΔdsrD + pMO-dsrD were grown in fermentative conditions with pyruvate and in respiratory conditions with lactate and sulfate or sulfite (Fig. 1 B–E, Table 1).
Table 1. Growth parameters of D. vulgaris H strains, maximal optical density at 600 nm (OD600) and doubling time (Td).
| Strain | Pyr 60 | LS4 30/30 | LS3 15/10 | LS3 30/20 | ||||
| Td (h) | Max. OD600 | Td (h) | Max. OD600 | Td (h) | Max. OD600 | Td (h) | Max. OD600 | |
| WT | 31.1 ± 0.8 | 0.147 ± 0.005 | 5.0 ± 0.3 | 0.82 ± 0.02 | 2.8 ± 0.2 | 0.81 ± 0.02 | 2.9 ± 0.1 | 1.26 ± 0.02 |
| ΔdsrD | 27.6 ± 3.1 | 0.155 ± 0.017 | 10.0 ± 0.5 | 0.80 ± 0.02 | 30.3 ± 1.4 | 0.36 ± 0.01 | 24.6 ± 5.5 | 0.56 ± 0.02 |
| ΔdsrD + pMO-dsrD | 30.9 ± 3.5 | 0.133 ± 0.015 | 8.6 ± 0.4 | 0.76 ± 0.01 | 5.4 ± 0.7 | 0.58 ± 0.01 | 4.2 ± 0.3 | 0.99 ± 0.02 |
Data are mean ± SD (n = 3 independent experiments).
Under fermentative conditions, with pyruvate as the sole carbon and energy source, all three strains presented a similar growth behavior, with identical doubling times and maximum cell densities. However, the same was not observed under LS4 respiratory conditions (30 mM lactate/30 mM sulfate [LS4 30/30]), where D. vulgaris H ΔdsrD presented a twofold slower growth than D. vulgaris H WT (P < 0.01). This growth difference between WT and ΔdsrD strains was even more evident with sulfite as the electron acceptor (15 mM lactate/10 mM sulfite [LS3 15/10]), resulting in a higher difference in doubling time (10× slower) and a significant decrease in the maximum cell density attained (P < 0.01). In medium containing a higher sulfite concentration (30 mM lactate/20 mM sulfite [LS3 30/20]), the ΔdsrD strain took a long time to initiate exponential growth (∼75 h), while in LS3 15/10, there was virtually no lag phase. The effect on maximum cell density in the ΔdsrD strain vs. the D. vulgaris H WT was similar in LS3 15/10 and LS3 30/20, i.e., a decrease of ∼55% for the ΔdsrD strain. The D. vulgaris H WT phenotype in respiratory conditions was only partially recovered in the D. vulgaris H ΔdsrD + pMO-dsrD strain in LS4 and LS3 conditions, suggesting a reduced expression level of DsrD in the complemented strain relative to the WT. These results show that DsrD is not strictly essential, as we could obtain a ΔdsrD deletion strain in D. vulgaris H, in contrast to DsrC (30). However, the deleted strain could not grow as efficiently by sulfate or sulfite reduction, whereas DsrD did not seem to be involved in fermentative growth conditions.
Identification of DsrD Physiological Partners.
To identify potential physiological partners of DsrD, a pull-down assay was performed, taking advantage of the Strep-tagged DsrD expressed by the complemented strain. For that, cells of D. vulgaris H WT and D. vulgaris H ΔdsrD + pMO-dsrD were grown in MO LS3 30/20 conditions where a phenotype for ΔdsrD was more evident. The crude extracts were subjected to Strep-tag affinity chromatography to isolate DsrD and coelute potential physiological partners. D. vulgaris H WT was used as a control to discard proteins with an intrinsic ability to adhere to the resin. The eluted fractions from both strains were analyzed in a Tricine sodium dodecyl sulfate polyacrylamide gel electrophoresis (Tricine-SDS-PAGE) gel (Fig. 2A), and six bands were evident in the eluted fraction of D. vulgaris H ΔdsrD + pMO-dsrD, expressing Strep-tagged DsrD. Only two of the six bands were also faintly visible in the D. vulgaris H WT control assay. The identity of the bands was determined by mass spectrometry (SI Appendix, Table S1). Two bands of ∼140 kDa and ∼22 kDa were identified as pyruvate carboxylase, an enzyme that contains biotin as a prosthetic group, which explains its intrinsic binding to the Strep-tactin resin. However, only the ∼22-kDa band is present in the D. vulgaris H WT eluate, which is likely the result of protein degradation from the 140-kDa protein. GroL (a 60-kDa chaperonin) was also identified in both the D. vulgaris H ΔdsrD + pMO-dsrD and WT eluted fractions, suggesting an intrinsic affinity of this highly expressed protein to bind the resin and/or its involvement in folding of recombinant DsrD. Two unique bands of ∼49 kDa and ∼43 kDa were present in the D. vulgaris H ΔdsrD + pMO-dsrD eluted fraction and absent in the control eluate. These were identified as DsrA and DsrB, respectively, revealing that DsrD pulls down DsrAB, which indicates there is a direct interaction between these proteins. Curiously, a band corresponding to DsrC (∼12 kDa) was not detected in the D. vulgaris H ΔdsrD + pMO-dsrD eluate, and its absence was confirmed by Western blot analysis (SI Appendix, Fig. S2). This supports the idea that the previously observed covalent cross-link between D. vulgaris H DsrC and the DsrAB siroheme (11) is not physiological and occurs during the long DsrAB purification process.
Fig. 2.
Pull-down and interaction assays. (A) Proteins coeluting with DsrD-Strep in pull-down assay with the D. vulgaris H complemented strain (ΔdsrD + pMO-dsrD) and WT (negative control). (B) Sensorgrams of DsrD interaction with increasing concentrations of DsrAB (15.6 nM to 1,000 nM) using SPR. Sensorgrams were run in duplicate. (C) Binding curve of DsrD (32 nM) with DsrAB (8.5 μM to 0.259 nM) using MST. Data points are mean ± SD, n = 3 independent experiments.
Hittel and Voordouw did not detect DsrD in partially purified DsrAB preparations (47), whereas Shatsky and colleagues reported DsrAB being copurified with tagged DsrD, and DsrD being copurified with tagged DsrA or DsrB with lower yields (49), suggesting a transient binding of DsrD to DsrAB and not the formation of a stable complex. Both studies corroborate our findings since we also did not detect DsrD when purifying DsrAB/C from D. vulgaris H (11).
DsrD Interacts Strongly with DsrAB.
To further characterize the interaction between DsrD and DsrAB, two approaches were used, namely, surface plasmon resonance (SPR) and microscale thermophoresis (MST), using proteins from A. fulgidus. A. fulgidus DsrAB was isolated from cell extracts (30, 50), and A. fulgidus DsrC and DsrD were recombinantly produced in Escherichia coli. SPR is a gold standard method for measuring binding kinetics without the need for labeling. DsrD was immobilized on a nitrilotriacetic acid (NTA) sensor chip by the C-terminal His-tag, and its interaction was studied with increasing concentrations of DsrAB (Fig. 2B). The interaction observed could be detected even at low concentrations of DsrAB. Steady-state conditions could not be achieved (even by increasing the association time), which means that no kinetic model could fit the experimental data and so the association and dissociation rate constants could not be obtained. Nevertheless, an estimate for the equilibrium dissociation constant (KD) of 540 ± 20 nM was derived from duplicate sensorgrams using the maximum response reached. This value suggests a high affinity for the DsrD-DsrAB complex. To validate these results with a different technique, we chose MST, which is a highly sensitive technique for studying protein-protein interactions, and uses labeling via amine residues at the protein surface but does not require protein immobilization. The association curve presents a sigmoidal shape with a KD of 350 ± 60 nM, showing that DsrD binds strongly to DsrAB (Fig. 2C). The KDs obtained by the two techniques are in the same order of magnitude, confirming a strong interaction between DsrD and DsrAB.
DsrD Is an Activator of DsrAB Activity.
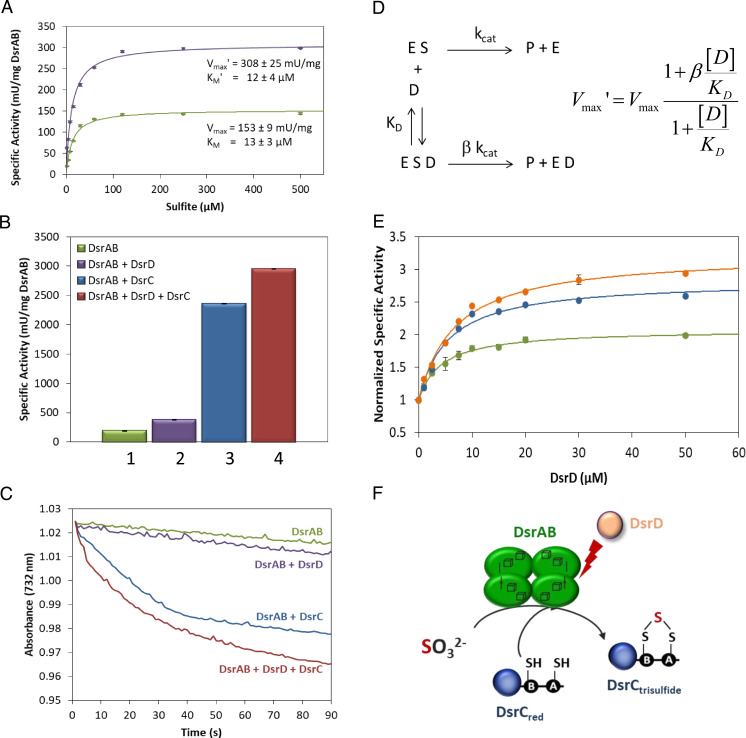
We showed previously that DsrC acts as a cosubstrate in sulfite reduction by DsrAB (30), so we next studied how DsrD affects the activity of DsrAB/DsrC, using the A. fulgidus proteins. First, we studied the effect of DsrD on sulfite reduction by DsrAB alone, which has a low level of activity. These experiments showed that in the presence of DsrD, the maximum activity (Vmax) of sulfite reduction by DsrAB doubled (308 ± 25 mU/mg) relative to its absence (153 ± 9 mU/mg) (Fig. 3A). However, the affinity for sulfite measured by the Michaelis constant (Km) was not affected (Km = 13 ± 3 μM without, and 12 ± 4 μM with DsrD). Mass spectrometry of DsrD at the end of the reaction showed an unaltered molecular mass relative to the purified DsrD, revealing that DsrD is not modified during the process, in contrast to what happens with DsrC (SI Appendix, Fig. S3).
Fig. 3.
Kinetic assays of the effect of DsrD on A. fulgidus DsrAB activity. (A) Sulfite reduction assays in the absence (green) and presence of excess DsrD (50 μM; purple). Data points are mean ± SD, n = 3 independent experiments. (B) Specific activity of A. fulgidus DsrAB, isolated (1), in the presence of 50 μM DsrD (2), in the presence of 10 μM DsrC (3), and in the presence of both DsrD and DsrC (4). (C) Kinetic traces of sulfite reduction activity by DsrAB as in (B) (representative traces). (D) Kinetic model for sulfite (S) reduction by DsrAB (E) with binding of an activator (D; in this case DsrD) to the enzyme saturated with substrate (ES), and its effect on the catalytic constant. kcat and βkcat are catalytic rate constants and KD is the equilibrium dissociation constant. The equation describes the dependence of Vmax′ on the concentration of DsrD (detailed description in SI Appendix). (E) Normalized specific activity of DsrAB (Vmax′/Vmax) as a function of DsrD at three different concentrations of DsrC. Sulfite is present at 500 μM (substrate saturation) and DsrC at 0, 10 μM, and 20 μM in green, blue, and orange, respectively. Parameters KD and β were obtained from the nonlinear fit of the data to the equation (solid lines). Data points are mean ± SD, n = 2 independent experiments. (F) Schematic representation of sulfite reduction involving DsrAB, DsrC, and DsrD.
DsrC also induces a strong increase in DsrAB activity, which is characterized by a fast phase that occurs while DsrC is being consumed (30). Next, we investigated the influence of DsrD on the DsrAB physiological reaction, i.e., sulfite reduction in the presence of DsrC. The activity-promoting effect of DsrD is also observed in the presence of DsrC, with a 25% increase in activity vs. that observed in the presence of only DsrAB and DsrC (Fig. 3B). This rate increase is reflected in a shorter fast phase of DsrC reduction, as expected, since the same amount of reduced DsrC is consumed in a shorter period (Fig. 3C). These data indicate that DsrD acts as an activator of DsrAB.
To further characterize the effect of DsrD, we evaluated the enzymatic activity of DsrAB with increasing concentrations of DsrD and saturating concentrations of sulfite. Under these conditions, specific activity is equal to Vmax, and we may study its dependence on the concentration of DsrD. The maximal activity in the presence of DsrD (Vmax′) can be expressed as a function of Vmax, a dimensionless parameter β, the KD, and the concentration of DsrD according to a simple kinetic model (Fig. 3D) that assumes fast equilibrium between DsrAB and DsrD. Normalized data were fit to the equation derived from the model to obtain the KD and the parameter β that describes the effect of DsrD on the catalytic rate constant. The value of β is 2.08 (green curve in Fig. 3E), which is consistent with the twofold effect of DsrD on DsrAB catalysis (Fig. 3A). The KD value for the same curve is 4.25 μM, which is 10× higher than the value determined by MST in the absence of sulfite. The difference is likely due to the very different experimental conditions used; the MST data were obtained at 25 °C, whereas the kinetic data were obtained at 60 °C. The higher temperature used in the kinetic assays should favor the dissociation of DsrD due to the increase in entropy of the products.
A similar kinetic characterization was performed with two concentrations of DsrC (blue and orange curves in Fig. 3E). The data show that the value of β increases in the presence of DsrC (2.83 and 3.24 at 10 μM and 20 μM DsrC, respectively) indicating a synergistic effect between DsrD and DsrC with respect to catalysis. On the other hand, the affinity of DsrD toward DsrAB seems to decrease slightly in the presence of DsrC (KD = 5.47 μM and 6.83 μM at 10 μM and 20 μM DsrC, respectively). These results are in line with an activator role for DsrD since it enhances the catalytic activity of DsrAB independently of the presence of DsrC (Fig. 3F), while not affecting the affinity for sulfite. They also agree with the observation that DsrD is not essential for growth in D. vulgaris H but contributes to its efficiency, as revealed by the behavior of the ΔdsrD strain that cannot grow as efficiently as the WT during sulfate/sulfite respiration. Since DsrD is not modified during DsrAB catalysis (SI Appendix, Fig. S3), it is likely that the activating role of DsrD is associated with a conformational alteration in DsrAB that leads to increased activity, meaning that DsrD acts as an allosteric activator of DsrAB. In the gut pathogen B. wadsworthia, the dissimilatory sulfite reductase was isolated as DsrABD, and the dsrD gene is fused to dsrB (41). The C terminus of DsrB is distant from the active site (11), which suggests that DsrD binds far away from the substrate pocket. Future modeling studies will be performed to try to elucidate the DsrD binding site and possible conformational alterations induced by DsrD on DsrAB.
Role of DsrAB and DsrD in Different Sulfur Metabolisms.
Elucidating the physiological function of DsrD is essential to fully understand the mechanism of DsrAB and is also highly relevant because of their widespread use as functional marker proteins to discriminate between different types of sulfur energy metabolism. In particular, it is important to address whether DsrD is only associated with a reductive DsrAB or if it could also function in oxidative metabolism. Some organisms from newly discovered phyla encode DsrAB and DsrD, which are considered characteristic of reductive metabolism, but also DsrL and/or DsrEFH, which were thought to be exclusive of SOPs (18, 21, 22, 27), making it unclear if in these organisms sulfur metabolism occurs in a reductive, disproportionating or oxidative mode. The recent description of DsrL-2 in several organisms with a reductive DsrAB, where it acts as electron donor for this enzyme while oxidizing NADPH, shows that DsrL proteins can be involved in reductive metabolism (35). On the other hand, it has been suggested that reductive DsrAB may operate reversibly in an oxidative mode and that its presence together with DsrD may not be indicative of reductive metabolism.
In some of the cases where DsrAB is present together with DsrD and DsrL, the organisms are capable of disproportionation, where reductive and oxidative sulfur metabolism occur concurrently. One example is Desulfurivibrio alkaliphilus, which couples sulfide oxidation to nitrate reduction with production of elemental sulfur, which is then disproportionated (37), as shown before for other Desulfobulbaceae (51–53), including recently for cable bacteria (54). The fate of elemental sulfur during this process has not been clarified and may involve its oxidation to sulfite, which has been shown to be an intermediate during disproportionation of sulfur or thiosulfate (52, 53). Disproportionation of sulfite then involves its oxidation to sulfate by adenosine triphosphate (ATP) sulfurylase and adenosine 5′-phosphosulfate (APS) reductase, both of which are readily reversible enzymes, as well as its reduction to sulfide, performed by DsrABCD. Cable bacteria are another important example of organisms performing sulfide oxidation coupled with oxygen or nitrate reduction and containing reductive DsrABCD (55). They are closely related to the genus Desulfobulbus that contain several disproportionators, also pointing to the involvement of elemental sulfur disproportionation in their energy metabolism (55), which was recently confirmed (54). Thus, it is reasonable to assume that disproportionation of sulfur, thiosulfate, and sulfite involves DsrAB (and DsrD) operating in a reductive direction. The reversibility of the reductive DsrAB, although theoretically possible, has never been shown and may imply protein adaptation, as suggested by the existence of separate phylogenetic groups of rDsrAB and DsrC for the reverse reaction, operating with different proteins (e.g., DsrEFH). The presence of DsrEFH indicates the need for dedicated proteins to transfer sulfur onto DsrC, which are not present in simple sulfate reducers and most disproportionators. Nevertheless, it should be pointed out that disproportionation of elemental sulfur can apparently also occur without the involvement of DsrAB, as suggested by proteomic studies of Desulfurella amilsii, an organism that can grow by elemental sulfur or thiosulfate reduction and also by elemental sulfur disproportionation (56). In D. amilsii, a high abundance of DsrAB, DsrC, and DsrMKJOP was only observed during thiosulfate reduction and not during sulfur disproportionation. This organism also does not have ATP sulfurylase and APS reductase, and sulfite was not detected during disproportionation, suggesting a different mechanism for this process (56).
Importantly, no dsrD is present in the genomes of SOPs with oxidative-type rDsrAB (18, 35). Thus, the reduction of intracellularly formed sulfite from sulfur compound disproportionation and/or thiosulfate or organosulfonate reduction can explain the presence of reductive DsrAB/DsrD/DsrC and DsrL-2 in organisms that do not perform DSR. In the case of disproportionation, the intermediate production of elemental sulfur and/or sulfite means that the pathway for sulfide oxidation is not a simple reversal of the canonical sulfate reduction pathway.
In conclusion, our results clarify the functional role of DsrD as an allosteric activator of DsrAB catalysis in sulfite reduction, providing important insights into our molecular understanding of dissimilatory sulfur metabolism. The absence of DsrD in organisms encoding the putative earliest forms of DsrAB, such as Crenarchaeota, Candidatus Rokubacteria, and Candidatus Hydrothermarchaeota (18, 20), indicates that DsrD appeared later in the evolutionary history of the dsr genes, which is in line with its nonessential function, as observed in the present studies. In terms of its use as a functional marker gene, we postulate that dsrD when found together with dsrAB, dsrC, and dsrMK genes is likely evidence of reductive sulfur metabolism, which may be part of a larger disproportionating scheme.
Materials and Methods
Construction of D. vulgaris H Strains.
A D. vulgaris H strain lacking the dsrD gene (ΔdsrD) was constructed by double homologous recombination. The vector for insertion of a ΩKm cassette in dsrD was constructed via sequence ligation independent cloning according to Li and Elledge (57), using D. vulgaris H chromosomal DNA as the template and primers #1, #2, #3, and #4 to flank the upstream and downstream regions of dsrD; using the pSC27 vector and primers #5 and #6 for the kanamycin cassette; and using the pMO719 vector and primers #7 and #8 for the pUC-ori and spectinomycin resistance gene. The assembled plasmid (pMOIP17) was transformed into E. coli α-select Silver Efficiency (Bioline) and plated on Luria Bertani (LB) agar plates containing kanamycin and spectinomycin. After sequence confirmation, the plasmid was electroporated into D. vulgaris H cells, and double recombinants were selected in Missouri yeast (MOY) solid medium with 30 mM of sodium pyruvate by secondary antibiotic screening, as described in Keller et al. (58). Colonies resistant to G418 but sensitive to spectinomycin were selected and grown in medium containing G418, an analog of kanamycin that is more effective in D. vulgaris H (59). The absence of the dsrD gene in the D. vulgaris ΔdsrD mutant strain was verified by PCR using primers #9 and #10 and chromosomal DNA extracted from the screened cells.
A D. vulgaris H ΔdsrD + pMO-dsrD complemented strain was created by inserting the pMO-dsrD vector into D. vulgaris H ΔdsrD mutant cells by electroporation. To create pMO-dsrD, dsrD was amplified from D. vulgaris H genomic DNA using primers #9 and #10 and cloned in pET22b(+) using NdeI and XhoI restriction sites, originating in the pET-dsrD-D. vulgaris H vector. The XhoI restriction site was mutated to Eco47III using primer #11, and then dsrD was amplified using primers #12 and #13 and cloned in pMOIP12P using NdeI and Eco47III, inserting a Strep-tag at the C terminus of dsrD, creating the pMO-dsrD vector.
A descriptive list of plasmids and strains (SI Appendix, Table S2) and primers (SI Appendix, Table S3) used throughout this study is presented in the SI Appendix.
Growth Studies.
D. vulgaris H WT and the two modified strains were grown anaerobically at 37 °C in MO medium (59). Under respiratory conditions, 15 or 30 mM sodium lactate was added as the electron donor, and sodium sulfate (30 mM), sodium sulfite (10 or 20 mM) or sodium thiosulfate (10 or 20 mM) was added as the terminal electron acceptor. For fermentative conditions, 60 mM sodium pyruvate was used, and lactate and sulfate were omitted from the culture media. No yeast extract was used in the growth experiments, but all media were inoculated with 2% (vol/vol) fresh precultured cells grown in MOY medium (0.5 g/L yeast extract) containing 60 mM sodium pyruvate and 3 mM sodium sulfate. The optical density at 600 nm (OD600) of the cultures was monitored at various time points. Antibiotics G418 at 400 μg/mL and spectinomycin at 100 μg/mL were added to the media according to the strain. However, in the final growth curves, only spectinomycin was used. Statistically significant relationships were determined using ANOVA, with a P < 0.05 considered statistically significant.
Electrophoretic Mobility Shift Assay.
A 981-bp fragment from the dsrAB promoter region was obtained by PCR using the A. fulgidus VC-16 genome as the template and primers #16 and #17. As a negative control, the 803-bp gene for kanamycin resistance was amplified from the pSC27 plasmid, using primers #18 and #19. Based on Grimm et al. (60), 200 fmol of both DNA fragments were incubated with A. fulgidus DsrD (10 to 100 pmol) in incubation buffer (5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer [pH 7.8], 20 mM KCl, 0.02% [vol/vol] Tween 20, and 1 mM Tris(2-carboxyethyl)phosphine (TCEP)) for 15 min at 70 °C. The samples were loaded in a 1.5% agarose gel stained with GreenSafe Premium (NZYtech) in 0.5% Tris-Borate-EDTA (TBE) buffer. The electrophoresis was run at 90 V and the gel analyzed under ultroviolet light.
Western Blot Analysis.
D. vulgaris H WT and modified strains were grown in MO medium with the designated electron donor/acceptor, collected at the end of exponential phase, and centrifuged at 3,000 × g for 10 min. The cell pellets were mechanically disrupted with glass beads using a Minilys homogenizer (Bertin Technologies) and centrifuged at 17,000 × g for 20 min at 4 °C. The protein concentration in the crude cell extracts was determined by the Bradford method (BioRad) with bovine serum albumin as the standard (NZYTech). Crude cell extracts (40 μg) were analyzed in Tricine-SDS-PAGE gels (10% acrylamide [wt/vol]) and proteins transferred to 0.45-µm PVDF (polyvinylidene difluoride) membranes (Roche) (transfer buffer: 48 mM Tris-HCl [pH 9.2] and 39 mM glycine) using a Mini Trans-Blot electrophoretic transfer cell (BioRad) at 100 V for 7 min (DsrD), 8 min (DsrC), or 30 min (DsrB) at 4 °C. Membranes for DsrD analysis were treated with blocking buffer (20 mM Tris-HCl [pH 7.6], 150 mM NaCl, and 5% [wt/vol] nonfat milk) for 1 h at room temperature (RT), incubated with TBST (20 mM Tris-HCl [pH 7.6], 150 mM NaCl, and 0.05% [vol/vol] Tween 20) plus anti-DsrD antibody at a 1:5,000 dilution for 1 h at RT, and then incubated with TBST plus anti-rabbit Immunoglobulin G (IgG)-alkaline phosphatase antibody (Sigma-Aldrich) at a 1:15,000 dilution for 45 min at RT. Membranes for DsrB and DsrC analysis were incubated with TBST, 1.2% (wt/vol) nonfat milk, and primary antibody (anti-DsrB at a 1:8,300 dilution and anti-DsrC at a 1:3,300 dilution) for 1 h at RT and then incubated with anti-rabbit IgG-alkaline phosphatase antibody at a 1:15,000 dilution in TBST for 45 min at RT. Between every incubation step, the membranes were washed three times with TBS (20 mM Tris-HCl [pH 7.6] and 150 mM NaCl). All protein detections were performed by incubating the membranes in alkaline phosphatase buffer (100 mM Tris-HCl [pH 9.5], 100 mM NaCl, and 250 µM MgCl2) and nitro-blue tetrazolium chloride/5-bromo-4-chloro-3-indolylphosphate toluidine (Sigma-Aldrich). The anti-DsrB antibody was produced in rabbits with synthesized peptides based on the D. vulgaris H DsrB amino acid sequence, and anti-DsrC and anti-DsrD antibodies were produced from purified proteins at Davids Biotechnologie GmbH.
Pull-Down Assay.
D. vulgaris H WT and ΔdsrD + pMO-dsrD cells were grown in MO medium supplemented with 30 mM sodium lactate and 20 mM sodium sulfite. Cells were collected at the end of exponential phase, centrifuged, and disrupted as described previously for the Western blot analysis. Crude extracts were loaded in gravity flow columns containing Strep-Tactin resin (IBA Lifesciences) equilibrated with buffer W (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, and 10% [vol/vol] glycerol). After five column volumes of washing with buffer W, the Strep-tagged DsrD was eluted with buffer W plus 2.5 mM D-desthiobiotin. The pull-down assay of the two strains was normalized by loading in the Strep-Tactin column the same amount of total protein of each crude extract and performing elution with identical volumes throughout the process. The desthiobiotin-eluted fractions (14 µg for ΔdsrD + pMO-dsrD and the equivalent volume for D. vulgaris H WT) were analyzed in a 10% Tricine-SDS-PAGE gel, and proteins were identified by mass spectrometry.
Production of Recombinant A. fulgidus DsrD.
The dsrD gene (locus tag: AF0425) was amplified by PCR from the A. fulgidus VC-16 genome, using primers #14 and #15. The digested PCR product was cloned into a pET-22b(+) expression vector (Novagen), which allows the insertion of a 6×-His tag sequence at the C terminus of DsrD. The cloned vector was verified by sequencing. The recombinant plasmid was transformed into E. coli BL21-Gold(DE3) competent cells (Stratagene) containing the pRARE2 expression vector, for the expression of the rare codons present in A. fulgidus. Transformed cells were grown at 37 °C in LB medium supplemented with ampicillin (100 μg/mL) and chloramphenicol (30 μg/mL) until an OD600 of 0.5 was reached. At this point, isopropyl-β-D-thiogalactopyranoside was added to a final concentration of 500 μM, and growth was continued for another 5 h. Then, E. coli cells were collected by centrifugation at 7,930 × g for 12 min at 4 °C and stored at −20 °C.
The cell pellet was resuspended in buffer K (25 mM potassium phosphate buffer [pH 7.4], 300 mM NaCl, and 20 mM imidazole) in the presence of DNase (Sigma-Aldrich) and a protease inhibitor mixture (Roche) and disrupted in a French press at 6.2 MPa. Cell lysates were centrifuged at 17,000 × g for 20 min at 4 °C. After filtration with a 0.2-μm polyethersulfone (PES) membrane filter, the supernatant was loaded a HiTrap immobilized metal-ion affinity chromatography (IMAC) high performance column (GE Healthcare) equilibrated with buffer K. An additional step of washing with 30 mM imidazole was performed in order to remove DNA contamination. DsrD was eluted with 25 mM potassium phosphate buffer (pH 7.4), 300 mM NaCl, and 150 mM imidazole and then concentrated and dialyzed to 25 mM potassium phosphate buffer (pH 7.4) by ultrafiltration (3-kDa cutoff; Amicon Millipore). Protein concentration was determined using the bicinchoninic acid assay (BCA) method (Merck Millipore), and purity was analyzed in a 10% Tricine-SDS-PAGE gel (SI Appendix, Fig. S4). The purified DsrD was kept under anaerobic conditions.
Purification of A. fulgidus DsrAB.
A. fulgidus VC-16 (DSM 4304) was grown in sulfate-thiosulfate-lactate medium in 300 L fermenter at 83 °C, as previously described (61), without addition of dithionite. Frozen cells were resuspended in 25 mM potassium phosphate buffer pH 7.0 (buffer A), homogenized, and disrupted in a French press at 6.9 MPa in the presence of DNase (Sigma-Aldrich) and a protease inhibitor mixture (Roche). The lysate was centrifuged at 7,930 × g for 20 min, followed by ultracentrifugation at 138,000 × g for 2 h. The purification protocol followed two consecutive ionic exchange chromatography steps, as described in detail in Santos et al. (30). The purity of DsrAB was analyzed by a 10% Tricine-SDS-PAGE gel (SI Appendix, Fig. S4), and the protein concentration was determined using the absorption coefficient (ε) of 60 mM−1 cm−1 at 593 nm (62). The purified DsrAB was kept under anoxic conditions to prevent prolonged contact with O2, which leads to gradual activity loss.
Production of A. fulgidus DsrC.
The A. fulgidus VC16 dsrC gene (AF2228) was amplified by PCR using genomic DNA and primers (#20/#21) with NdeI and EcoRI restriction sites, respectively. The amplified product was ligated into a pET-28a(+) vector (Novagen) resulting in pET-His-dsrC-AF and inserting a His-tag at the N terminus of DsrC. Then, as described in Santos et al. (30) and using the same primers (#22/#23 primers), the two DsrC structural Cys (C77 and C85) were replaced to Ala by site-directed mutagenesis using the NZYMutagenesis kit (NZYTech), generating pET-His-dsrCC77A/C85A-AF. This facilitates an analysis of the DsrC redox state by MalPEG/SDS-PAGE without affecting its functional role. The cloned vector was verified by sequencing and transformed into E. coli BL21-Gold(DE3) competent cells (Stratagene) containing the pRARE2 expression vector. The DsrC expression procedure was identical to that described for DsrD.
In contrast to the purification of A. fulgidus DsrD, the cell pellet was resuspended in buffer X (25 mM potassium phosphate buffer [pH 7.4], 300 mM NaCl, and 30 mM imidazole) in the presence of DNase (Sigma-Aldrich) and disrupted in a French press at 6.2 MPa. The lysate was centrifuged at 17,000 × g for 20 min at 4 °C, and the supernatant was subjected to heat shock at 70 °C for 10 min followed by centrifugation at 17,000 × g for 15 min at 4 °C in order to remove E. coli–precipitated proteins. After filtration with a 0.2-μm PES membrane filter, the supernatant was loaded in a HiTrap IMAC HP column (GE Healthcare) equilibrated with buffer X. DsrC was eluted with 25 mM potassium phosphate buffer (pH 7.4), 300 mM NaCl, and 150 mM imidazole and was then concentrated and dialyzed to 25 mM potassium phosphate buffer (pH 7.4) by ultrafiltration (10-kDa cutoff; Amicon Millipore). The purity of DsrC was analyzed in a 10% Tricine-SDS-PAGE gel (SI Appendix, Fig. S4), and the protein concentration was determined using an ε of 24 mM−1 cm−1 at 280 nm. Before its use in activity assays, DsrC was reduced with 5 mM dithiothreitol during 30 min at 37 °C, and excess reductant was removed with a HiTrap desalting column (GE Healthcare). Reduced DsrC was concentrated, if necessary, by ultrafiltration and kept under anaerobic conditions. DsrC in different redox states was analyzed by a gel-shift assay using methoxy-polyethylene glycol maleimide (MalPEG) (Fluka), as previously described (63).
DsrAB Activity Assay.
The enzymatic reduction of sulfite was performed inside a Coy anaerobic chamber (98% N2 and 2% H2) at 60 °C using Zn-reduced methyl viologen (Sigma-Aldrich) as the electron donor. The activity assays were run in 50 mM KPi (pH 7.0); 1 mM Zn-reduced methyl viologen (MV+); 50 nM of DsrAB in the absence or presence of DsrD (range from 1 to 50 µM); and 0, 10, or 20 μM of DsrC. The reaction was started by addition of sodium sulfite (range from 1 to 500 μM) after a 5-min incubation of DsrAB in the reaction buffer, followed by a 1-min incubation with DsrD and 1-min incubation with DsrC. The oxidation of MV+ was monitored at 732 nm (ε = 3.15 mM−1 cm−1) in a Shimadzu UV-1800 spectrophotometer.
Mass Spectrometry.
For protein identification, protein bands were excised from a 10% Tricine-SDS-PAGE gel and subjected to in-gel tryptic digestion followed by peptide identification in a MALDI-TOF/TOF (matrix-assisted laser desorption/ionization-time of flight) analyzer (Applied Biosystems 4800plus). The data were analyzed in a combined mode using Mascot search engine and National Center for Biotechnology Information (NCBI) database.
For intact mass determination, DsrD was analyzed before and after addition to the sulfite reductase assay. The protein buffer was exchanged to 20 mM ammonium acetate (pH 7.2) before analysis, using a Micro Bio-Spin 6 size exclusion spin column (Bio-Rad). The protein solution was mixed with 10 mg/mL Sinapinic acid (Sigma) in 50% (vol/vol) acetonitrile and 5% (vol/vol) formic acid (liquid chromatography–mass spectrometry (LC/MS) grade; Fisher) in a 1:1 ratio, and then this solution was applied directly onto the MALDI plate and allowed to air dry. The data were acquired in Linear Mid Mass Positive mode using a 5800 MALDI-TOF/TOF (AB Sciex) mass spectrometer and TOF/TOF Series Explorer Software v.4.1.0 (AB Sciex). External calibration was performed using a Protein MALDI-MS Calibration Kit (MSCAL3, ProteoMass). MS data were obtained by the ITQB/iBET UniMS Mass Spectrometry Unit, Oeiras, Portugal.
SPR Analysis.
The SPR experiments were performed at 25 °C on a BIAcore 2000 instrument (GE Healthcare). DsrD was immobilized in a NTA sensor chip (GE Healthcare) by the His-tag tail present at the C terminus. All the assays were performed with 10 mM HEPES (pH 7.4), 150 mM NaCl, 50 μM ethylenediamine tetraacetic acid (EDTA), and 0.005% (vol/vol) Tween 20 running buffer. DsrD (11 nM) was bound to a previously activated flow cell at a flow rate of 5 μL/min in the presence of Ni (500 μM NiCl2 in running buffer) in order to have an immobilization of around 50 resonance units (RUs). Another activated flow cell was similarly treated with running buffer in the absence of DsrD (control cell). Interaction experiments with DsrAB were performed with a 3-min injection of increasing concentrations of DsrAB (range from 15.6 nM to 1,000 nM) at a flow rate of 30 μL/min. At the end of sample injection, the NTA sensor chip was regenerated with a regeneration solution (350 mM EDTA in running buffer) for 1 min at a flow rate of 10 μL/min before a new cycle of surface activation and immobilization. Sensorgrams were obtained by subtracting the unspecific binding to the control flow cell in order to remove buffer artifacts and normalizing to the baseline injection. Equilibrium KD was determined from duplicate experiments and according to RUsteady-state = (RUmax × [analyte])/(KD + [analyte]).
MST Analysis.
The MST experiments were performed at 25 °C on a Monolith NT.115 instrument (NanoTemper). For the DsrD labeling, the procedure of the Monolith NT Protein Labeling Kit RED–NHS second generation was followed, which contains a dye that reacts efficiently with amines to form highly stable dye-protein-conjugates. A range of nonlabeled DsrAB concentrations (from 8.5 μM to 0.259 nM) was incubated with 32 nM of labeled DsrD in MST buffer (25 mM potassium phosphate [pH 7.0], 150 mM NaCl, and 0.05% [vol/vol] Tween 20). The samples were loaded into glass capillaries, and the MST analysis was carried out using 95% light-emitting diode (LED) power and medium MST power. Data were analyzed using the MO.Affinity Analysis v2.3 software (NanoTemper) from triplicate experiments.
Supplementary Material
Acknowledgments
We thank Christiane Dahl from the University of Bonn for critically reading the manuscript. This work was funded by the Fundação para a Ciência e Tecnologia (Portugal) through Fellowships PD/BD/135488/2018 (A.C.C.B.), PD/BD/128204/2016 (D.F.), and SFRH/BPD/79823/2011 (S.S.V.); Grants PTDC/BIA-MIC/6512/2014 and PTDC/BIA-BQM/29118/2017; and Research unit Molecular and Structural Microbiology (MOSTMICRO-ITQB) (Grants UIDB/04612/2020 and UIDP/04612/2020). The European Union’s Horizon 2020 Research and Innovation Program (Grant Agreement No. 810856) is also acknowledged.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2118880119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Rabus R., et al. , A post-genomic view of the ecophysiology, catabolism and biotechnological relevance of sulphate-reducing prokaryotes. Adv. Microb. Physiol. 66, 55–321 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Wasmund K., Mußmann M., Loy A., The life sulfuric: Microbial ecology of sulfur cycling in marine sediments. Environ. Microbiol. Rep. 9, 323–344 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowles M. W., Mogollón J. M., Kasten S., Zabel M., Hinrichs K.-U., Global rates of marine sulfate reduction and implications for sub-sea-floor metabolic activities. Science 344, 889–891 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Jørgensen B. B., Findlay A. J., Pellerin A., The biogeochemical sulfur cycle of marine sediments. Front. Microbiol. 10, 849 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGlynn S. E., Chadwick G. L., Kempes C. P., Orphan V. J., Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526, 531–535 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Wegener G., Krukenberg V., Riedel D., Tegetmeyer H. E., Boetius A., Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 526, 587–590 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Dahl C., “A biochemical view on the biological sulfur cycle” in Environmental Technologies to Treat Sulphur Pollution (Principles and Engineering, 2020), pp. 55–96. [Google Scholar]
- 8.Carbonero F., Benefiel A. C., Alizadeh-Ghamsari A. H., Gaskins H. R., Microbial pathways in colonic sulfur metabolism and links with health and disease. Front. Physiol. 3, 448 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devkota S., Chang E. B., Interactions between diet, bile acid metabolism, gut microbiota, and inflammatory bowel diseases. Dig. Dis. 33, 351–356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ridlon J. M., Wolf P. G., Gaskins H. R., Taurocholic acid metabolism by gut microbes and colon cancer. Gut Microbes 7, 201–215 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliveira T. F., et al. , The crystal structure of Desulfovibrio vulgaris dissimilatory sulfite reductase bound to DsrC provides novel insights into the mechanism of sulfate respiration. J. Biol. Chem. 283, 34141–34149 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venceslau S. S., Stockdreher Y., Dahl C., Pereira I. A. C., The “bacterial heterodisulfide” DsrC is a key protein in dissimilatory sulfur metabolism. Biochim. Biophys. Acta 1837, 1148–1164 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Simon J., Kroneck P. M. H., Microbial sulfite respiration. Adv. Microb. Physiol. 62, 45–117 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Wagner M., Roger A. J., Flax J. L., Brusseau G. A., Stahl D. A., Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180, 2975–2982 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhillon A., Goswami S., Riley M., Teske A., Sogin M., Domain evolution and functional diversification of sulfite reductases. Astrobiology 5, 18–29 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Loy A., et al. , Reverse dissimilatory sulfite reductase as phylogenetic marker for a subgroup of sulfur-oxidizing prokaryotes. Environ. Microbiol. 11, 289–299 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Müller A. L., Kjeldsen K. U., Rattei T., Pester M., Loy A., Phylogenetic and environmental diversity of DsrAB-type dissimilatory (bi)sulfite reductases. ISME J. 9, 1152–1165 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anantharaman K., et al. , Expanded diversity of microbial groups that shape the dissimilatory sulfur cycle. ISME J. 12, 1715–1728 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chernyh N. A., et al. , Dissimilatory sulfate reduction in the archaeon ‘Candidatus Vulcanisaeta moutnovskia’ sheds light on the evolution of sulfur metabolism. Nat. Microbiol. 5, 1428–1438 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Colman D. R., et al. , Phylogenomic analysis of novel Diaforarchaea is consistent with sulfite but not sulfate reduction in volcanic environments on early Earth. ISME J. 14, 1316–1331 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan S., et al. , Insights into ecological role of a new deltaproteobacterial order Candidatus Acidulodesulfobacterales by metagenomics and metatranscriptomics. ISME J. 13, 2044–2057 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hausmann B., et al. , Peatland Acidobacteria with a dissimilatory sulfur metabolism. ISME J. 12, 1729–1742 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dalcin Martins P., et al. , Viral and metabolic controls on high rates of microbial sulfur and carbon cycling in wetland ecosystems. Microbiome 6, 138 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker B. J., Lazar C. S., Teske A. P., Dick G. J., Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria. Microbiome 3, 14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKay L. J., et al. , Co-occurring genomic capacity for anaerobic methane and dissimilatory sulfur metabolisms discovered in the Korarchaeota. Nat. Microbiol. 4, 614–622 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Zecchin S., et al. , Rice paddy Nitrospirae carry and express genes related to sulfate respiration: Proposal of the new genus “Candidatus Sulfobium.” Appl. Environ. Microbiol. 84, e02224-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thiel V., et al. , “Candidatus Thermonerobacter thiotrophicus,” a non-phototrophic member of the Bacteroidetes/Chlorobi with dissimilatory sulfur metabolism in hot spring mat communities. Front. Microbiol. 9, 3159 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carr S. A., et al. , Carboxydotrophy potential of uncultivated Hydrothermarchaeota from the subseafloor crustal biosphere. ISME J. 13, 1457–1468 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hua Z. S., et al. , Genomic inference of the metabolism and evolution of the archaeal phylum Aigarchaeota. Nat. Commun. 9, 2832 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santos A. A., et al. , A protein trisulfide couples dissimilatory sulfate reduction to energy conservation. Science 350, 1541–1545 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Pires R. H., et al. , Characterization of the Desulfovibrio desulfuricans ATCC 27774 DsrMKJOP complex—a membrane-bound redox complex involved in the sulfate respiratory pathway. Biochemistry 45, 249–262 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Grein F., Ramos A. R., Venceslau S. S., Pereira I. A. C., Unifying concepts in anaerobic respiration: Insights from dissimilatory sulfur metabolism. Biochim. Biophys. Acta 1827, 145–160 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Stockdreher Y., et al. , Cytoplasmic sulfurtransferases in the purple sulfur bacterium Allochromatium vinosum: Evidence for sulfur transfer from DsrEFH to DsrC. PLoS One 7, e40785 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lübbe Y. J., Youn H. S., Timkovich R., Dahl C., Siro(haem)amide in Allochromatium vinosum and relevance of DsrL and DsrN, a homolog of cobyrinic acid a,c-diamide synthase, for sulphur oxidation. FEMS Microbiol. Lett. 261, 194–202 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Löffler M., Wallerang K. B., Venceslau S. S., Pereira I. A. C., Dahl C., The iron-sulfur favoprotein DsrL as NAD(P)H:acceptor oxidoreductase in oxidative and reductive dissimilatory sulfur metabolism. Front. Microbiol. 11, 578209 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flieder M., et al. , Novel taxa of Acidobacteriota implicated in seafloor sulfur cycling. ISME J. 15, 3159–3180 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thorup C., Schramm A., Findlay A. J., Finster K. W., Schreiber L., Disguised as a sulfate reducer: Growth of the deltaproteobacterium Desulfurivibrio alkaliphilus by sulfide oxidation with nitrate. mBio 8, e00671-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Löffler M., et al. , DsrL mediates electron transfer between NADH and rDsrAB in Allochromatium vinosum. Environ. Microbiol. 22, 783–795 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Karkhoff-Schweizer R. R., Huber D. P. W., Voordouw G., Conservation of the genes for dissimilatory sulfite reductase from Desulfovibrio vulgaris and Archaeoglobus fulgidus allows their detection by PCR. Appl. Environ. Microbiol. 61, 290–296 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ø. Larsen, Lien T., Birkeland N. K., A novel organization of the dissimilatory sulfite reductase operon of Thermodesulforhabdus norvegica verified by RT-PCR. FEMS Microbiol. Lett. 203, 81–85 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Laue H., Friedrich M., Ruff J., Cook A. M., Dissimilatory sulfite reductase (desulfoviridin) of the taurine-degrading, non-sulfate-reducing bacterium Bilophila wadsworthia RZATAU contains a fused DsrB-DsrD subunit. J. Bacteriol. 183, 1727–1733 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keller K. L., Wall J. D., Genetics and molecular biology of the electron flow for sulfate respiration in desulfovibrio. Front. Microbiol. 2, 135 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otwell A. E., Callister S. J., Zink E. M., Smith R. D., Richardson R. E., Comparative proteomic analysis of Desulfotomaculum reducens MI-1: Insights into the metabolic versatility of a Gram-positive sulfate- and metal-reducing bacterium. Front. Microbiol. 7, 191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W., et al. , Global transcriptomic analysis of Desulfovibrio vulgaris on different electron donors. Antonie van Leeuwenhoek 89, 221–237 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Caffrey S. M., Voordouw G., Effect of sulfide on growth physiology and gene expression of Desulfovibrio vulgaris Hildenborough. Antonie van Leeuwenhoek 97, 11–20 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Mizuno N., Voordouw G., Miki K., Sarai A., Higuchi Y., Crystal structure of dissimilatory sulfite reductase D (DsrD) protein—possible interaction with B- and Z-DNA by its winged-helix motif. Structure 11, 1133–1140 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Hittel D. S., Voordouw G., Overexpression, purification and immunodetection of DsrD from Desulfovibrio vulgaris Hildenborough. Antonie van Leeuwenhoek 77, 271–280 (2000). [DOI] [PubMed] [Google Scholar]
- 48.Waite D. W., et al. , Proposal to reclassify the proteobacterial classes Deltaproteobacteria and Oligoflexia, and the phylum Thermodesulfobacteria into four phyla reflecting major functional capabilities. Int. J. Syst. Evol. Microbiol. 70, 5972–6016 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Shatsky M., et al. , Bacterial interactomes: Interacting protein partners share similar function and are validated in independent assays more frequently than previously reported. Mol. Cell. Proteomics 15, 1539–1555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schiffer A., et al. , Structure of the dissimilatory sulfite reductase from the hyperthermophilic archaeon Archaeoglobus fulgidus. J. Mol. Biol. 379, 1063–1074 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Fuseler K., Krekeler D., Sydow U., Cypionka H., A common pathway of sulfide oxidation by sulfate-reducing bacteria. FEMS Microbiol. Lett. 144, 129–134 (1996). [Google Scholar]
- 52.Cypionka H., Smock A. M., Böttcher M. E., A combined pathway of sulfur compound disproportionation in Desulfovibrio desulfuricans. FEMS Microbiol. Lett. 166, 181–186 (1998). [Google Scholar]
- 53.Finster K., Microbiological disproportionation of inorganic sulfur compounds. J. Sulfur Chem. 29, 281–292 (2008). [Google Scholar]
- 54.Müller H., Marozava S., Probst A. J., Meckenstock R. U., Groundwater cable bacteria conserve energy by sulfur disproportionation. ISME J. 14, 623–634 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kjeldsen K. U., et al. , On the evolution and physiology of cable bacteria. Proc. Natl. Acad. Sci. U.S.A. 116, 19116–19125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Florentino A. P., et al. , Insight into the sulfur metabolism of Desulfurella amilsii by differential proteomics. Environ. Microbiol. 21, 209–225 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li M. Z., Elledge S. J., Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4, 251–256 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Keller K. L., Wall J. D., Chhabra S., Methods for engineering sulfate reducing bacteria of the genus Desulfovibrio. Methods Enzymol. 497, 503–517 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Zane G. M., Yen H. C., Wall J. D., Effect of the deletion of qmoABC and the promoter-distal gene encoding a hypothetical protein on sulfate reduction in Desulfovibrio vulgaris Hildenborough. Appl. Environ. Microbiol. 76, 5500–5509 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grimm F., Dobler N., Dahl C., Regulation of dsr genes encoding proteins responsible for the oxidation of stored sulfur in Allochromatium vinosum. Microbiology (Reading) 156, 764–773 (2010). [DOI] [PubMed] [Google Scholar]
- 61.Stetter K. O., Lauerer G., Thomm M., Neuner A., Isolation of extremely thermophilic sulfate reducers: Evidence for a novel branch of archaebacteria. Science 236, 822–824 (1987). [DOI] [PubMed] [Google Scholar]
- 62.Dahl C., Kredich N. M., Deutzmann R., Trüper H. G., Dissimilatory sulphite reductase from Archaeoglobus fulgidus: Physico-chemical properties of the enzyme and cloning, sequencing and analysis of the reductase genes. J. Gen. Microbiol. 139, 1817–1828 (1993). [DOI] [PubMed] [Google Scholar]
- 63.Venceslau S. S., et al. , Redox states of Desulfovibrio vulgaris DsrC, a key protein in dissimilatory sulfite reduction. Biochem. Biophys. Res. Commun. 441, 732–736 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.