Abstract
Objective:
Surveillance tools for early cancer detection are suboptimal, including hepatocellular carcinoma (HCC), and biomarkers are urgently needed. Extracellular vesicles (EVs) have gained increasing scientific interest due to their involvement in tumor initiation and metastasis, however, most extracellular RNA (exRNA) blood-based biomarker studies are limited to annotated genomic regions.
Design:
EVs were isolated with ultracentrifugation and nanoDLD and quality assessed by electron microscopy, immunoblotting, nanoparticle tracking, and deconvolution analysis. Genome-wide sequencing of the largely unexplored small exRNA landscape, including unannotated transcripts, identified and reproducibly quantified small RNA clusters (smRCs). Their key genomic features were delineated across biospecimens and EV isolation techniques in prostate cancer and HCC. Three independent exRNA cancer datasets with a total of 479 samples from 375 patients, including longitudinal samples, were utilized for this study.
Results:
ExRNA smRCs were dominated by uncharacterized, unannotated small RNA with a consensus sequence of 20bp. An unannotated 3-smRC signature was significantly overexpressed in plasma exRNA of HCC patients (p<0.01, n=157). An independent validation in a phase 2 biomarker case-control study revealed 86% sensitivity and 91% specificity for the detection of early HCC from controls at risk (n=209) (area under the ROC curve [AUC]: 0.87). The 3-smRC signature was independent of alpha-fetoprotein (p<0.0001) and a composite model yielded an increased AUC of 0.93.
Conclusion:
These findings directly lead to the prospect of a minimally-invasive, blood-only, operator-independent clinical tool for HCC surveillance, thus highlighting the potential of unannotated smRCs for biomarker research in cancer.
Keywords: next-generation sequencing, genomics, cancer surveillance, liver cancer
One sentence summary:
We employ a novel, data-driven approach to identify and characterize small RNA clusters from unannotated loci in extracellular vesicle-derived RNA across different cancer types, isolation techniques, and biofluids, facilitating discovery of a robust biomarker for detection of early stage liver cancer.
INTRODUCTION
Extracellular vesicles (EVs), including microvesicles and exosomes, are nanoparticles whose nucleic acid payload is capable of priming receptor cells to modify key cellular functions[1,2]. EVs are heterogeneous, both in terms of biogenesis and content[3]. While larger EVs such as apoptotic bodies mostly contain fragmented DNA, smaller EVs such as exosomes are enriched in non-coding, regulatory small RNAs[2,4]. In cancer, EVs are increasingly recognized as key players in tumor initiation and metastasis[5], mainly through miRNA trafficking, prompting their evaluation as early detection and treatment response biomarkers[6]. Importantly, most studies characterizing extracellular RNA (exRNA) and studying EV-related biomarkers apply conventional, reference-based, RNA sequencing approaches, and are thus limited to known annotated genomic regions (e.g., miRNA, snoRNA, lncRNA, etc.). However, small RNAs arise from thousands of endogenous genes and are part of the genomic ‘dark matter’ of highly abundant yet largely uncharacterized non-coding RNA, with emerging roles in regulating gene expression via post-transcriptional and translational mechanisms. In fact, relatively little attention has been paid to characterizing the general expression landscape of circulating EV small RNA and their precursors in this context regardless of biotype, especially for those expressed from unannotated genomic regions.
With a 5-year survival of 18%, liver cancer is the second most lethal malignancy after pancreatic cancer. Projections estimate more than 1 million deaths due to this cancer in 2030 worldwide[7]. Survival in patients enrolled in early detection programs of hepatocellular carcinoma (HCC), the most common form of primary liver cancer, doubles that of those not enrolled in surveillance[8]. However, implementation of surveillance among patients at high risk of HCC in the United States is very low (20%)[9] and the performance of recommended surveillance tools (i.e. ultrasound and serum alpha-fetoprotein (AFP)[10,11]) is suboptimal, with low sensitivity (63%) and moderate specificity (83%) for early stage HCC missing close to 40% of tumors[12]. Improvement in this area is urgently needed by developing better read-outs of oncogenesis and facilitating implementation of surveillance through minimally-invasive, operator-independent tools.
Our aim was to characterize the small exRNA expression landscape associated with circulating EVs, to identify novel small RNA biomarker candidates, and to test their clinical utility for the detection of early stage HCC among high-risk patients. Strongly departing from previous exRNA characterization studies, which are restricted to quantifying expression of known (i.e. annotated) transcripts[13,14], we adopt a different approach by de novo assembly and characterization of the small RNA expression landscape of exRNA, specifically including unannotated genomic regions across a HCC plasma EV dataset and a prostate cancer dataset with tumor and adjacent tissue, urine, and blood specimens. The latter dataset was used to define discrete loci called small RNA clusters (smRCs), delineate their key genomic properties, and quantify their reproducibility across biofluid and isolation techniques. In the second part of our study, two independant HCC cohorts were used to identify and validate potential candidates to discriminate early stage HCC and controls at high risk in the setting of a phase 2 biomarker case-control study. In summary, by using whole RNA sequencing of EV-associated exRNA, we describe novel clinically-relevant smRCs in circulating exRNA as an unrecognized source for biomarker discovery. In our study, a 3-smRC signature was able to discriminate between patients with early stage HCC and patients at high-risk for HCC with better performance compared to a meta-analysis on the current standard of surveillance by ultrasound +/− AFP[12].
MATERIALS AND METHODS
Patient enrollment and study cohorts
This study utilizes three independent cancer exRNA datasets (Fig. 1B).
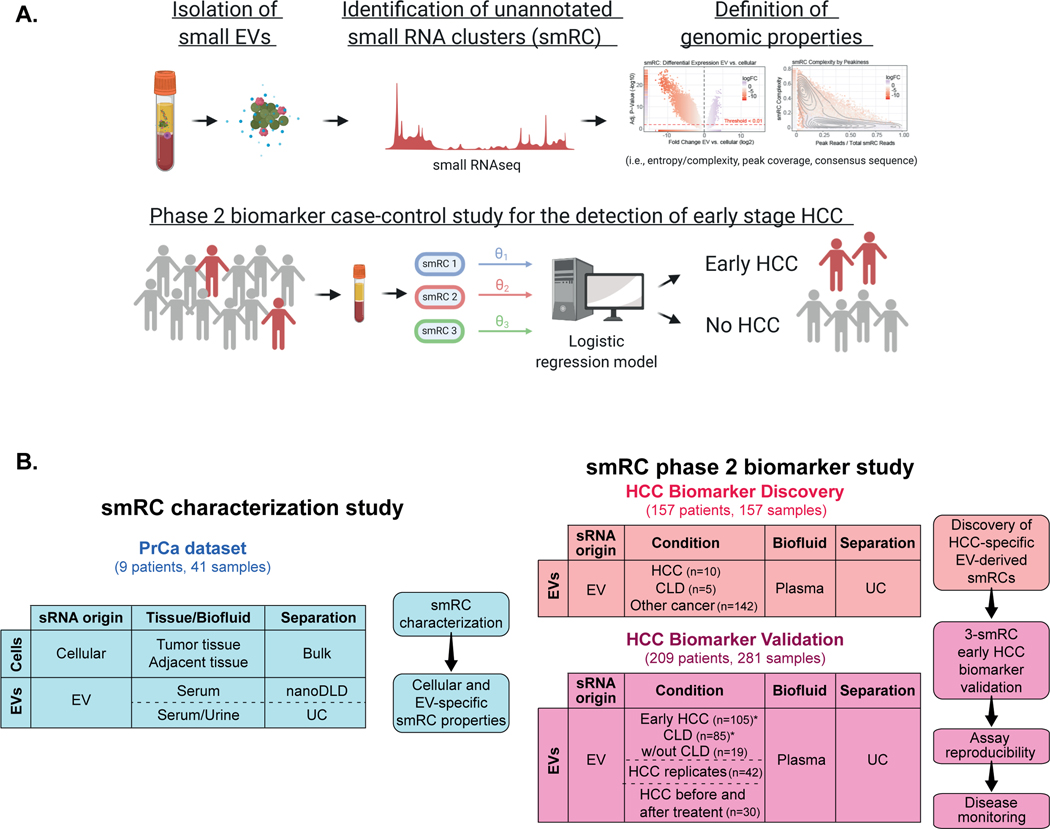
Figure 1. Study summary and flow chart for sample distribution.
(A) small RNA clusters (smRCs) from unannotated genomic regions were identified by unsupervised small RNA sequencing from circulating EVs and characterized. Their clinical utility was confirmed in a phase 2 biomarker case-control study for the detection of early stage HCC. (Created with BioRender.com.) (B) Schematic view of study flow diagram with different cohorts, and available specimen and separation method for each cohort. Three independent datasets with a total of 479 samples from 375 patients were included.
A prostate cancer cohort, which we termed the ‘smRC characterization’ cohort (n=9 patients, total of 41 samples). This cohort served to define and study the properties of smRCs in exRNA. For this dataset, de-identified data and biospecimens from human subjects consented under ongoing institutional review board (IRB)-approved protocols at the Icahn School of Medicine at Mount Sinai (GCO# 14–0318, 15–1135 and 10–1180) were collected from prostate cancer patients undergoing prostatectomy. Specifically, biospecimens included prostate cancer and adjacent prostate non-tumoral tissue from biopsy or prostatectomy, urine, and serum, where applicable. Each of these protocols involves the prospective collection of clinical data (e.g., demographics, baseline characteristics, treatments, and outcomes).
A HCC ‘biomarker discovery’ cohort (n=157 patients, total of 157 samples) to identify differentially expressed smRCs between HCC patients and controls with chronic liver disease, and patients with non-HCC malignancies to test the HCC-specificity of our biomarkers.
An independent HCC ‘biomarker validation’ cohort (n=209 patients, total of 281 samples, including 42 patients with replicates to assess assay reproducibility and 30 patients with samples before and after HCC treatment) to confirm their clinical utility in a phase 2 biomarker case-control study for detection of early stage HCC.
Samples for the HCC ‘biomarker discovery’ and ‘biomarker validation’ cohorts were collected from consented patients enrolled in an IRB-approved protocol to derive new HCC biomarkers from blood (HS-15–00540) or provided by the Tisch Cancer Institute Biorepository (HSM#10–00135) at the Icahn School of Medicine at Mount Sinai. Thus, HCC cases and controls were collected from the same clinical setting for the biomarker discovery and validation cohorts. Importantly, for the HCC biomarker discovery cohort, cases and controls were matched for age, gender, presence of cirrhosis, and etiology (Supplementary Table S1). Small RNA sequencing data from patients with other (non-HCC) malignancies were downloaded from exRNA atlas (https://exrna-atlas.org/, RRID:SCR_017221, including n=100 colon cancer, n=6 pancreatic adenocarcinoma, and n=36 prostate cancer patients, respectively).
For the phase 2 biomarker case-control validation study, we included three patient populations: 1) HCC cases limited to very early or early stage patients according to the BCLC classification[7] (i.e., stages 0 or A). All HCC patients were treatment-naïve at the time of blood sampling, 2) Patients with liver cirrhosis or different forms of chronic liver disease (CLD) at high-risk for HCC as per clinical practice guidelines[10,11], but without radiological evidence of HCC at the time of blood collection, 3) patients with benign liver nodules (e.g., hemangioma) without chronic liver disease. However, the latter were not considered in the logistic regression model. HCC diagnosis was made according to the criteria of the European Association for the Study of the Liver (EASL)[11]. In a subset of patients (n=30) sequential blood samples were available before and after these patients had received HCC treatment. Response was assessed according to modified RECIST criteria[15]. Liver cirrhosis was diagnosed based on histology, or non-invasively through combined transient elastography, imaging or laboratory evidence of liver dysfunction and portal hypertension. Patients with concurrent malignancies were excluded.
Methods and supplementary data on Sample collection and enrichment of EVs from human plasma, serum, and urine, Characterization of EV-enriched isolates, RNA extraction, small library preparation and next-generation sequencing, trimming, alignement, deconvolution analysis, smRC definition and properties, HCC smRC biomarker selection, reverse transcriptase quantitative polymerase chain reaction (RT-qPCR), smRC overlap with known biotypes and prostate cancer motif sequences can be found in the supplementary material.
Data Analysis
Following the guidelines of the Early Detection Research Network by the National Cancer Institute and the white paper on biomarker development in cancer[16], we conducted a phase 2 biomarker case-control study for early detection of HCC. This set of guidelines is generally used by academic and industry research who conduct biomarker studies and has been adopted by the US Food and Drug Administration as the benchmark for approval of new diagnostic devices. According to this paper, a phase 2 biomarker study is a retrospective case-control study to estimate the true positive rates and false positive rates or area-under-the-receiver-operating-curve (AUC) for the clinical biomarker assay and to assess its ability to distinguish subjects with cancer from subjects without cancer. Although the primary aim of this study was to assess the performance of our novel early HCC detection biomarker test, we wanted to put our results into clinical context by comparing them to the current gold-standard for early HCC surveillance (i.e. abdominal ultrasound and AFP)[10,11]. Based on the largest meta-analysis on surveillance for HCC, sensitivity of ultrasound and AFP is 63% for early stage tumors with a specificity of 83%[12]. We powered this study to detect an increase in sensitivity from 63% to 80% and specificity from 83% to 95% when comparing ultrasound and AFP to our new test. Given an alpha of 0.05 and a power (1-ß) of 90%, the number of samples needed to detect this difference based on asymptotic normal distribution theory[17] was 89 cases (early HCC) and 83 controls (patients at high risk of HCC, CLD) (simulations using different exact or approximate confidence intervals for the difference of binomial proportions[18] resulted in 93 cases and 82 controls; online application: https://mwsill.shinyapps.io/sample_size_diagnostic_test/).
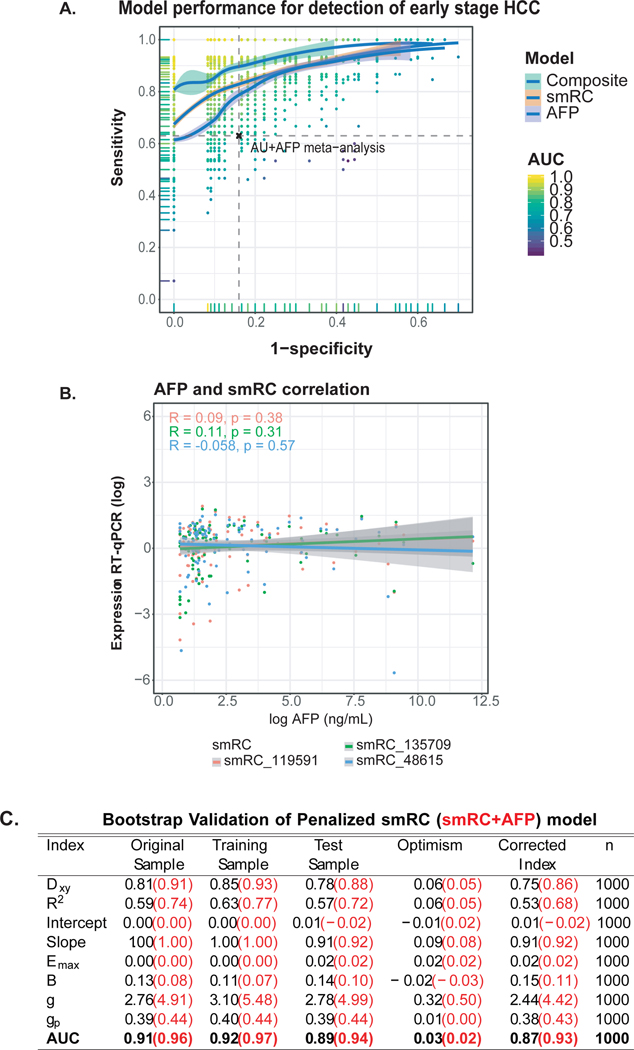
The analysis of the phase 2 biomarker case-control study to test the performance of the 3-smRC signature for early detection of HCC was limited to early stage HCC (n=105) and controls at risk for HCC (CLD, n=85) to represent the optimal population of interest.[11,16] We used penalized maximum likelihood techniques, bootstrap and cross-validation to estimate and control for model optimism, RT-qPCR batch plate effects, and over-fitting, and also rigorously computed the positive and negative predictive power estimates of our 3-smRC early detection signature. We computed a number of indices of model performance, discrimination measures, and calibration measures under bootstrap resampling (n = 1000), as summarized in Fig. 6C, extended in Supplementary Table S2, in order to demonstrate model performance and estimate generalization error by averaging performance across bootstrap resampling. In the first row, the key measure of discrimination Somers’ Dxy is the rank correlation between the observed and predicted response values, which in the case of logistic regression for a binary response reduces to simply Dxy = 2(c – ½), where c is Harrel’s c-statistic and equal to the AUC of the ROC for the early HCC vs. CLD prediction. In the case of the smRC model we immediately deduce that the bootstrap adjusted AUC is ½ + 3/8 = 7/8 = 0.875. Modest adjusted modified R2 ~ 0.52 is observed, combined with bootstrap-adjusted slope and intercept indicating modest and acceptably low over-fitting. Relatively bootstrap-adjusted low Emax (the maximum error in predicted probabilities), modest Brier score (B), very low unreliability index (U), high discrimination (D), high quality (Q = D – U), also indicate a reasonably robust model. Also, the bootstrap adjusted total Gini’s mean difference for based on the smRC model is a healthy 2.44, which robustly represents typical log-odds differences between early HCC and CLD patients predicted by the model. Converting this early HCC log-odds estimate to an early HCC probability prediction, we see that the typical predicted probability gap between early HCC and CLD patients is 38%. Finally, we compute the partial mean gini-scores of the smRC model predictors and find that the smRCs themselves have by far the largest termwise log-odds compared to any technical variance covariates (e.g., batch). We note in passing that repeated cross-validation gave similar results for Dxy and adjusted Slope (Fig. 6C, extended in Supplementary Table S2).
Figure 6. Performance of 3-smRC signature in a phase 2 biomarker case-control study.
(A) ROC curve for maximized gain-of-certainty across repeated cross validation. Each point represents a pair of sensitivities and specificities that maximize gain-in-certainty (i.e. sensitivity + specificity) from a test validation ROC curve, whose AUC colors the point. The loess curves trace the best density fit of points across this space, with 95% confidence intervals shown in gray. (B) AFP and smRC correlation plot. (C) Bootstrap validation parameters for smRC and smRC+AFP model. Dxy: Somers’ rank correlation between the observed HCC status and predicted HCC probabilities; Emax: maximum absolute calibration error on probability scale; B: Brier score; g: Gini’s mean difference of log-odds between HCC and CLD; gp: Gini’s mean difference in probability scale; AUC: Area Under the Receiver Operating Curve (ROC).
We next repeated the penalized maximum likelihood estimation procedure using a model with both smRCs and AFP readings included, given that a log likelihood ratio test for an AFP term was highly significant (p < 1e-8). Computing the same indices of model performance across bootstrap resampling (n = 1000), we found dramatically better performance for a combined model including our 3-smRC signature and AFP compared to the 3-smRC signature alone as shown in Fig. 6C, with bootstrap adjusted AUC ~ 0.93, lower overall error and evidence for overfitting, a much smaller Brier score of 0.11, and a dramatic increase in the Gini indices such that a typical early-HCC – CLD predicted probability difference was 43% (Fig. 6C). Finally, even though balanced accuracy is not a proper scoring rule, we estimated the maximized balanced accuracy landscape by subjecting the smRC logistic regression model for HCC risk to a cross-validation repeated 1000 times (i.e., a random 85% training, 15% testing split repeated 1,000 times) and computing maximizing sensitivity and specificity on the test ROCs.
For descriptive statistics, continuous variables are reported as median and categorical variables as counts and percentages. We used the Fisher’s exact test and the Student’s t-test to compare differences between categorical and continuous variables, respectively. Pearson’s or Spearman’s correlation coefficients were computed for correlation of continuous variables as indicated. Boxplot center line shows median, box limits show upper and lower quartiles, whiskers show 1.5x interquartile range, and points represent outliers. Error bars represent the 95% confidence intervals. All statistical analyses were conducted on Rstudio (R version 3.5.0, RRID:SCR_000432).
RESULTS
Our study is summarized in Fig. 1A. It utilizes three independent cancer exRNA datasets (Fig. 1B): 1) A prostate cancer cohort, which we termed the ‘smRC characterization’ cohort, to define and study the properties of smRCs in exRNA (n=9 patients, total of 41 samples). 2) A HCC ‘biomarker discovery’ cohort (n=157 patients) to identify differentially expressed smRCs between HCC patients and controls with chronic liver disease, and patients with non-HCC malignancies to test the HCC-specificity of our biomarkers. 3) An independent HCC ‘biomarker validation’ cohort (n=209 patients, total of 281 samples, including 42 patients with replicates and 30 patients with longitudinal samples before and after HCC treatment) to confirm their clinical utility in a phase 2 biomarker case-control study for detection of early stage HCC. In total, our study included 479 samples from 375 patients.
Characterization studies confirm enrichment of small EV from blood and urine
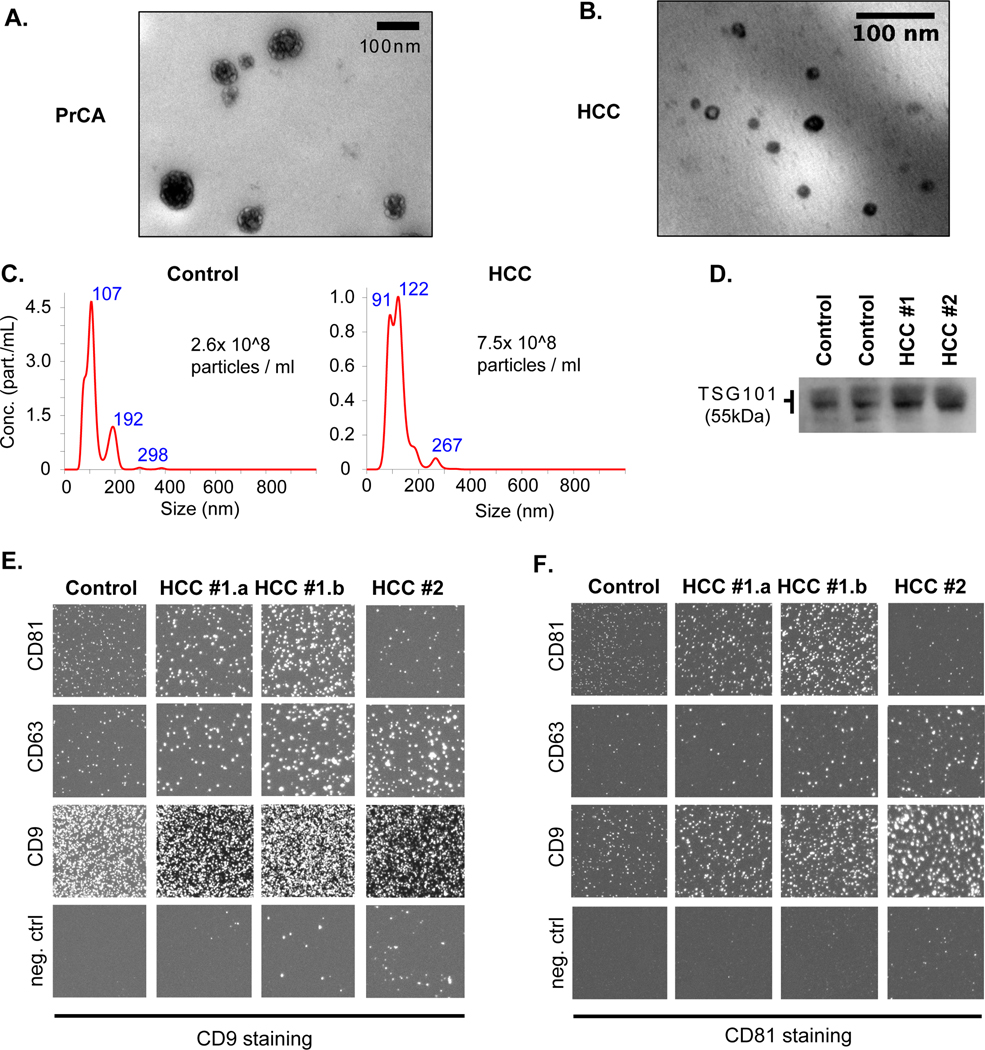
In all cohorts, differential ultracentrifugation (UC) was employed to enrich for EV isolates, and quality assessment of EV isolates was guided by recommendations of the International Society of Extracellular Vesicles[19] Specifically, we used transmission electron microscopy (TEM), nanoparticle tracking analysis (NTA), immuno-labeling with Western Blotting for intracellular (i.e., tumor susceptibility gene 101 protein, TSG101) and Exoview™ for transmembrane (i.e., tetraspanins CD9, CD63, CD81) vesicle proteins in a subset of samples (Fig. 2). This suggested an enrichment for small EVs (median size of 120 nm on NTA) with compatible morphology on NTA and TEM (Fig. 2A-C), and expression of typical markers for small EV populations with a dominance of CD9/CD81 and CD9/CD63 co-expression, and a paucity of CD63/CD81 coexpression (Fig. 2D-F). Additionally, for the ‘smRC characterization’ cohort in prostate cancer, we isolated EVs from a subset of patients (n=5) using the ‘lab-on-chip’ technology nanoDLD[20] (DLD) for serum samples. We also isolated purely cellular small RNA (<300 nt) from prostate cancer and adjacent non-cancerous tissue of the same patients to quantify exRNA-isolation technology, biofluid, and exRNA-specific variance in small RNA profiles, respectively. Part of our prostate cancer dataset has been included in an exRNA-atlas based deconvolution analysis published earlier[4]. In that study, an independent analysis found that our UC and nanoDLD isolation methods specifically isolate low- (cargo type 1) and variable (cargo type 4) density vesicles with minimum contamination from lipoproteins (cargo type 2) and argonaute proteins (cargo type 3B)[4]. For this study, we have now performed the same computational deconvolution analysis for our HCC ‘biomarker discovery’ dataset to determine carrier types and found that cargo type 4 was preferentially enriched (Supplementary Fig. S1A). In fact, cargo type 4 is associated with vesicles in the 60 – 150 nm size range, which were purified consistently with nanoDLD, and also the lowest-density OptiPrep fractions 1–3 from serum and plasma[4]. Cargo type enrichments associated with low density vesicles, lipoproteins, AGO2-positive ribonucleoproteins (RNPs), and AGO-2 negative RNPs were significantly depleted (Supplementary Fig. S1). Together, these results confirm a successful enrichment of small extracellular vesicles from a variety of biospecimens of prostate cancer and HCC patients with our methods.
Figure 2. Quality assessment of EV enrichment process for exRNA extractions from human blood samples.
(A, B) Transmission electron microscopy image of prostate cancer serum isolate (A) and HCC plasma isolate (B). (C) Nanoparticle tracking analysis (Nanosight®) results in the plasma isolate of a control (left) and HCC patient (right) with corresponding size distribution and estimated particle concentration. (D) Western Blotting image of protein lysate from isolate against TSG101 (~55 kDa) in two control (left) and two HCC (right) patients. (E,F) Immunolabeling of the isolate with Exoview™. Isolates were captured by indicated antibodies (CD81, CD63, CD9, control IgG) on a chip and stained with CD9 (E) or CD81 (F) antibodies to visualize different EV subpopulations in one control and three HCC samples (#1.a and #1.b represent technical replicates from the same patient).
Identification and characterization of small RNA clusters from unannotated exRNA
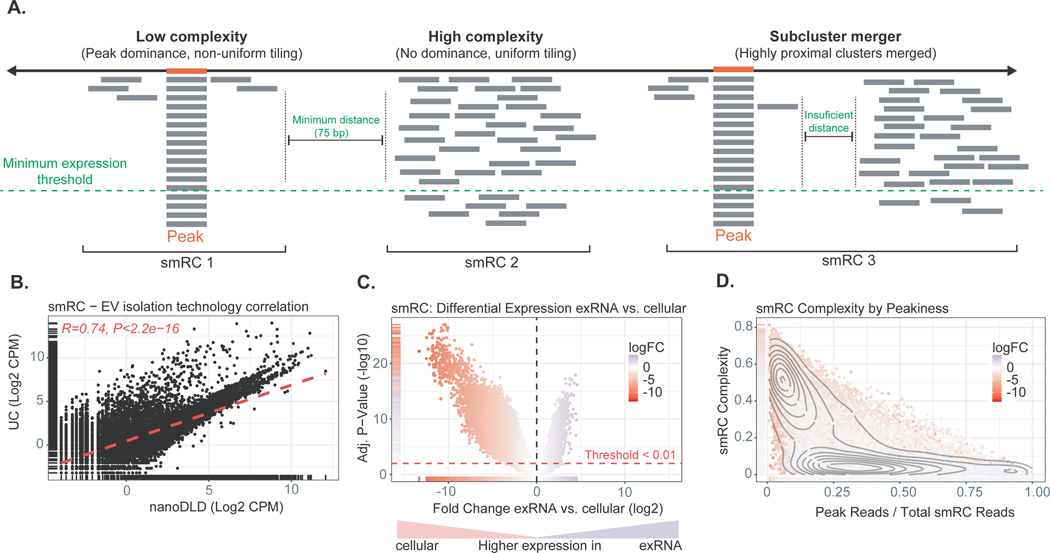
In the HCC ‘biomarker discovery’ cohort, we detected recurrent small clusters of contiguous genomic regions with sufficient alignment coverage in unannotated regions. Normally, we would disregard them as part of our standard RNA sequencing analytical pipeline. However, these small RNA clusters (termed ‘smRCs’) presented with a dominant peak sequence on many occasions, suggesting a non-random pattern, which prompted us to further investigate their genomic properties and potential role as cancer biomarkers. First, we captured the known heterogeneous genome-wide expression of clusters of small RNA precursors[21], each of which can give rise to multiple functional small RNA products, by defining clusters of small RNA reads (i.e., smRCs, Fig. 3A). Adjacent smRCs are merged if they overlap within a minimal padding threshold (75 bp), and we define the key properties of smRCs: a) entropy (i.e., read tiling efficiency or complexity), b) peak coverage, and c) consensus sequence of each smRC (see below). The set of all smRCs, computed once for all samples, is essentially the paired set of all accumulation loci of small RNA expression and their peak-coverage consensus sequences, and constitutes a smoothed, de-novo assembled small RNA expression landscape with a standard count matrix.
Figure 3. Key properties of small RNA clusters (smRCs).
(A) Minimum coverage and sub-read length minimal spacing define smRCs. Read tiling complexity captures heterogeneity of smRC read distribution. (B) Correlation of smRC expression across different EV enrichment methods (i.e., ultracentrifugation, UC, and nanoDLD). (C) Volcano plot for differential expression between smRC of cellular versus exRNA origin. (D) smRC complexity as a function of peak coverage colored by differential smRC expression between cellular and exRNA origin. smRCs enriched in exRNAs (purple) present with low complexity and higher peak coverage, whereas cellular smRCs (red) are more frequently of high complexity and lower peak coverage.
To determine whether smRCs were specific to blood or present in other compartments and tissue types, we expanded our analysis and delineated key genomic properties of smRCs in our ‘smRC characterization’ prostate cancer dataset, where we had access to different biological sample types (blood, urine, tumoral and non-tumoral adjacent tissue) and different EV isolation methods (ultracentrifugation and nanoDLD[22,23]). In order to profile the maximal coverage and overall distribution of expression within smRCs associated with exRNA, we defined two quantities. First, a ‘peak’ coverage which is simply the ratio of reads in the smRC peak to total smRC coverage, and second, a tiling complexity measure which is the ratio of unique read nucleotide sequences to total smRC coverage. Since almost all small RNAs arise from post-transcriptional processing of larger RNA precursors, the quantification of alignment patterns is largely an empirical task for which measures of maxima (peak coverage) and heterogeneity (tiling complexity) become crucial tools to classify these patterns. smRCs with low tiling complexity are those with a non-uniform tiling of transcripts and a relative dominance of equal reads forming a peak. Here, the term peak-coverage consensus sequence is referring to the sequence of the dominant transcript (left panel of Fig 3A). In contrast, smRCs with high tiling complexity are clusters of transcripts with a uniform tiling of reads and few peaks, which can result in a fairly long genomic region covered by this cluster (middle panel of Fig. 3A). The mean length of the consensus peak sequence was 20 nt (ranging from 15 to 100 nt in length). In contrast, the mean genomic length of smRCs was 674 nt (Supplementary Fig. S2A). Technical reproducibility of smRC quantification included comparing two different EV enrichment methods in serum (UC and nanoDLD), and different biofluid compartments (urine and serum) of the same patients. We found a high correlation between enrichment methods (spearman R ~ 0.74, p < 2.2e-16, Fig. 3B) with over 80% of smRCs detected by both methods above the 20th percentile of expression (Supplementary Fig. S3A). We found a modest correlation between different biofluid compartments (i.e. urine and serum) using UC (spearman R ~ 0.45, p < 1e-16, see Supplementary Fig. S3B+C for self-reproducibility).
Taken together, we robustly identified smRCs, including in unannotated regions, across biospecimens and enrichment methods by an unsupervised data-driven view of the entire small RNA landscape.
ExRNA smRCs are enriched for non-coding transcripts from unannotated regions
Well-expressed smRCs possessed a heteroscedastic count variance profile which facilitated usual differential expression analysis via linear modeling (Supplementary Fig. S2B). The total number and magnitude of overexpressed smRCs in cells was significantly higher than in exRNA (Fig. 3C). However, we observed a significant difference in the complexity of smRCs found in exRNA compared to cells, and found that the major contributor of smRC variable expression was RNA origin (with low complexity typical in exRNA- versus high complexity typical of cellular smRC origin, Supplementary Fig. S2C, Fig. 3D). Indeed, the bimodal pattern reveals a clear separation between cellular smRCs, which overwhelmingly have relatively high tiling complexity, and exRNA smRCs that have much stronger evidence for high relative peak coverages and low complexity. The mean size of the peak within smRCs was slightly higher than the minimal trimmed read length, and was significantly different between exRNA-derived and cell-derived (16.5 bp versus 22.6 bp, p < 1 e-16). exRNA-associated smRCs preferentially overlap unannotated small RNA species compared to cellular smRCs (Supplementary Fig. S4, Supplementary Table S3). Finally, we orthogonally validated the expression of three unannotated smRCs from the prostate cancer dataset by correlating RNA sequencing data with RT-qPCR (Supplementary Fig. S3D, Supplementary Table S4).
These data demonstrate that EV-associated smRCs predominantly present with a small number of highly covered peaks (i.e low complexity and high peak coverage) compared to cellularderived smRCs. They preferentially capture non-coding small RNA compared to protein-coding RNA, but are also significantly enriched in unannotated genomic regions.
Identification of an HCC-specific 3-smRC signature in plasma exRNA
Given their high biological and technological independent reproducibility, tractable statistical properties, and unique ability to discriminate concentrations of exRNA-specific small RNA, we computed the smRC profile of our ‘HCC biomarker discovery’ cohort of 15 patients, including 10 patients with HCC and 5 controls at risk for HCC matched for age, sex, and etiology of the underlying liver disease (Supplementary Table S1). We found that exRNA-derived smRCs were differentially expressed between HCC and controls. In fact, 250 smRCs were enough to distinguish them (Supplementary Fig. S5A). This led us to hypothesize that smRCs could be useful tools for early HCC detection. We selected the three top differentially expressed and low-complexity smRCs for further biomarker analysis (see Supplementary Methods) and confirmed differential expression between HCC and controls at risk (Supplementary Fig. 5B). Additional analysis in a cohort of 142 patients with non-HCC malignancies (100 colon cancer, 6 pancreatic adenocarcinoma, and 36 prostate cancer patients) further confirmed their HCC specificity (Supplementary Fig. 5B). We orthogonally validated the differential expression of our 3-smRC signature in this ‘HCC biomarker discovery’ cohort using RT-qPCR. Pearson’s correlation coefficient was higher than 0.6 for all three smRCs when comparing data from small RNA sequencing and RT-qPCR (n=15, p<0.05, Supplementary Fig. S5C-E). The three smRCs were located in regions of chromosomes 3q, 8q (both unannotated intergenic regions), and 10q (intronic region of SGPL1) (Supplementary Table S4). Altogether, smRCs are able to discriminate HCC and controls including a 3-smRC signature that was orthogonally validated by RT-qPCR.
EV-associated small RNA clusters are overexpressed in patients with early stage HCC compared to high-risk controls
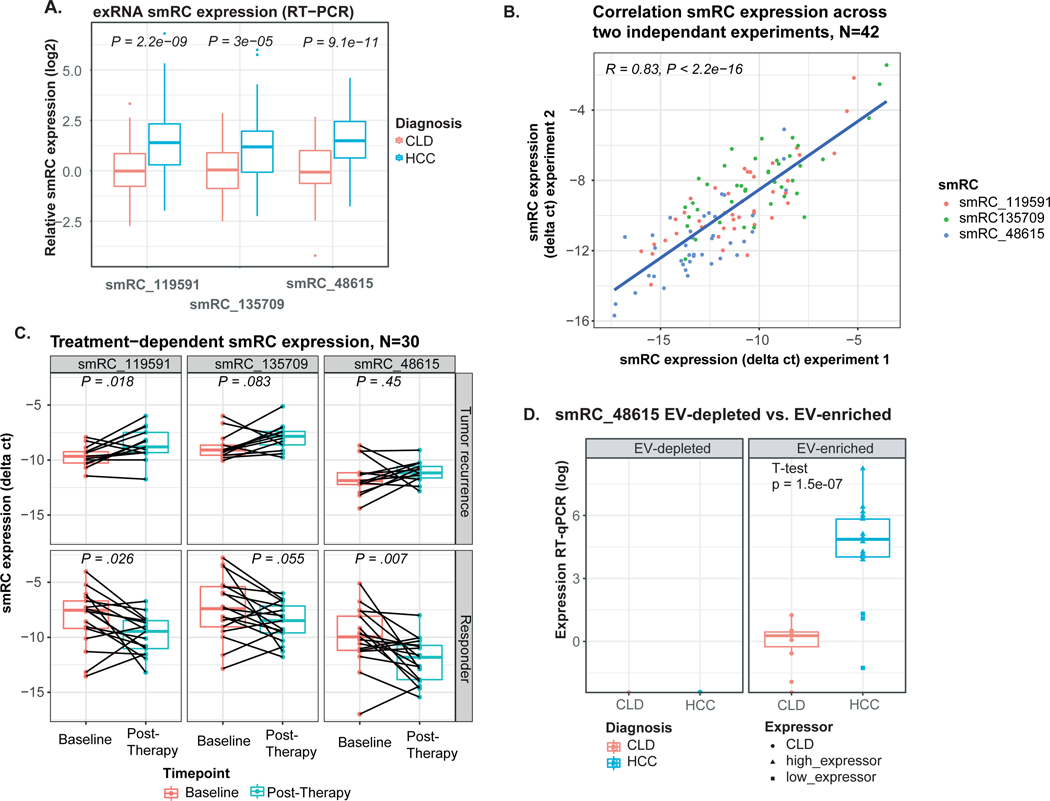
To determine the clinical utility of smRCs in exRNA, we designed a phase 2 biomarker case-control study following the recommendations from the Early Detection Research Network (EDRN) from the National Cancer Institute[16]. In detail, we aimed at assessing the performance of our 3-smRC signature as a novel early detection biomarker in HCC. Unlike many studies in this setting[24], we only enrolled patients with HCC at an early stage (Barcelona Clinic Liver Cancer classification (BCLC) stage 0 or A[7]), who can be cured with either surgery or ablation[7]. Crucially, our control cohort is the target population for HCC surveillance as defined in clinical practice guidelines[10,11] and a recent white paper on biomarker development for HCC by the International Liver Cancer Association[25]. We included 209 patients: n=105 treatment-naive, early stage HCC, n=85 control patients with cirrhosis and/or chronic liver disease (CLD) at high-risk for HCC enrolled in HCC surveillance (Table 1), and n=19 individuals without chronic liver disease (non-CLD). Our main matching criteria was prevalence of cirrhosis as we believe this was potentially the strongest variable that could affect the performance of our biomarker. By comparing HCC and CLD groups, we did not observe clinically significant differences in prevalence of cirrhosis, variables associated with liver function (bilirubin, albumin), or etiology. As expected, HCC cases were slightly older and predominantly male gender compared to CLD (Table 1). However, age and gender did not impact smRC expression (Supplementary Fig. 6A+B). We confirmed significant overexpression of our 3-smRC signature in plasma of early stage HCC patients compared to CLD controls with RT-qPCR (p<3e-5, Fig. 4A, see Supplementary Fig. S6C for comparison with non-CLD patients). To confirm the reproducibility of our biomarker analysis, we repeated the quantification of our 3-smRC early detection signature in 42 patients. This included EV enrichment from plasma, RNA extraction and RT-qPCR. These two independent experiments yielded a correlation coefficient of 0.83 (p<0.001, Fig. 4B). Longitudinal analysis in a subset of 30 patients with available sequential blood samples before and after HCC treatment revealed that smRC expression dynamics correlate with tumor response in these patients. In patients without early tumor recurrence after resection (n=13), smRC expression levels significantly decreased compared to baseline (paired t-test, Fig. 4C). Additional experiments showed significantly higher expression of smRC-48615 in EV-enriched isolates as opposed to EV-depleted plasma (n=30 patients, Fig. 4D), suggesting the smRC signal is in fact EV-associated.
Table 1.
Clinical characteristics
| CLD, N = 851 | HCC, N = 1051 | P-value2 | |
|---|---|---|---|
| Age | 0.15 | ||
| ≤60 years | 38 (45%) | 34 (33%) | |
| >60 years | 47 (55%) | 68 (67%) | |
| Gender | <0.001 | ||
| Female | 38 (45%) | 20 (20%) | |
| Male | 47 (55%) | 82 (80%) | |
| Cirrhosis (Yes) | 61 (72%) | 68 (67%) | 0.6 |
| Bilirubin | 0.5 | ||
| ≤1.2 mg/dL | 55 (67%) | 62 (73%) | |
| >1.2 mg/dL | 27 (33%) | 23 (27%) | |
| Albumin | 0.10 | ||
| ≥3.5 g/dL | 62 (77%) | 54 (64%) | |
| <3.5 g/dL | 19 (23%) | 31 (36%) | |
| Etiology | 0.4 | ||
| Non-viral | 17 (31%) | 40 (39%) | |
| Viral | 37 (69%) | 62 (61%) | |
| Tumor stage (BCLC) | |||
| Very Early (Stage 0) | n.a. | 22 (21%) | |
| Early (Stage A) | n.a. | 83 (79%) | |
| Single Nodule | n.a. | 92 (90%) | |
| Largest nodule (cm) | n.a. | 2.9 (2.0, 4.6) | |
| AFP (ng/mL) * | 4 (2, 5) | 8 (4, 92) | <0.001 |
Statistics presented: median (IQR); n (%);
Statistical tests performed: Wilcoxon rank-sum test; chi-square test of independence;
Upper limit of normal 9 ng/mL. AFP, alpha fetoprotein, BCLC, Barcelona Clinic for Liver Cancer, n.a., not applicable.
Figure 4. smRC expression in ‘HCC biomarker validation’ cohort.
(A) Expression for each smRC between HCC patients and chronic liver disease controls (CLD) (center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers). (B) Correlation of biomarker analysis for all three smRCs in 42 patients across two independent experiments, including EV enrichment from plasma, exRNA extraction and RT-qPCR. (C) Longitudinal analysis of smRC expression in 30 patients with available sequential blood samples before and after HCC treatment (responders n=13, tumor recurrence n=17, paired t-test). Displayed is the smRC expression as delta between ct values of the spike-in control and respective smRC; smaller delta equals higher expression of the smRC. (D) Expression of smRC-48615 in EV-enriched isolates and EV-depleted plasma. Displayed are samples from HCC and CLD controls. Triangles indicate HCC samples with relatively high expression, rectangles indicate samples with lower expression.
In summary, we report on three smRCs with differential expression between early stage HCC and controls at high-risk, who represent the target population for surveillance programs, including replicates and longitudinal samples before and after HCC treatment.
A 3-smRC signature predicts early stage hepatocellular carcinoma
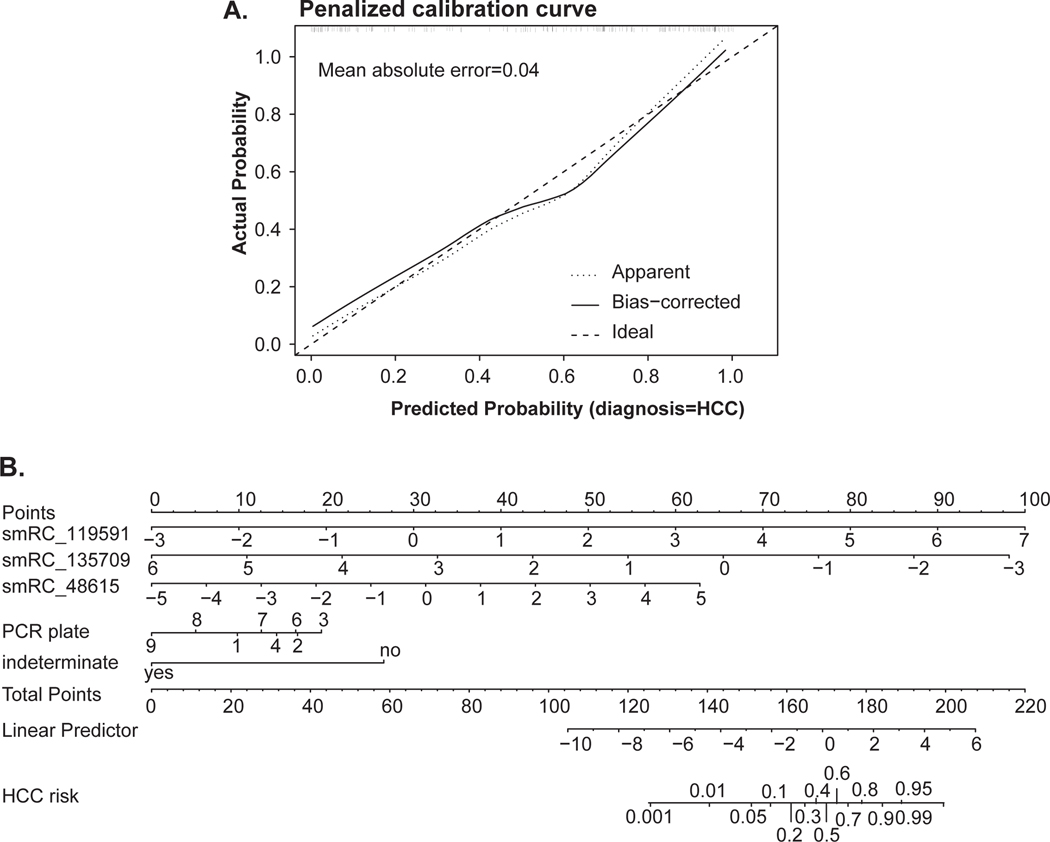
To leverage the collective power of all three smRCs to predict early HCC risk, we built a parsimonious logistic regression model to discriminate between early HCC patients and CLD controls using smRC expression and adjusting for the RT-qPCR sequencing batch effect. Importantly, this model excluded patients without chronic liver disease (non-CLD), because these patients are not recommended to be part of surveillance for HCC. This analysis allowed us to test if there is a well calibrated and predictive association between smRC expression and early HCC detection using an appropriate number of effective degrees of freedom in our model. We used penalized maximum likelihood techniques, bootstrap and cross-validation to estimate and control for model optimism[26], RT-qPCR batch plate effects, and over-fitting of our 3-smRC early detection signature. The logistic regression model was well calibrated with a low mean absolute probability error (0.04) to predict early HCC (Fig. 5A), low Brier score (B = 0.15), high AUC (0.87), and high Gini mean difference in predicted log-odds between HCC and CLD patients (2.44) adjusted under bootstrap (n = 1,000) resampling (Fig. 6C, Supplementary Table S2). Predicted HCC risk via smRC expression can be visualized via a patient nomogram to provide an individual estimate of HCC risk (Fig. 5B). In order to estimate sensitivity and specificity measures at plausible decision points, we applied the logistic regression model to a 85/15 split of the biomarker validation set for training and testing respectively. Averaging over 1,000 iterations, we recovered 86% sensitivity and 91% specificity with a positive predictive value (i.e. true positive rate) of 89% on average by maximizing the balanced accuracy of the test ROC curves (Supplementary Fig. S7 and Fig. 6A+C). The area under the ROC curve (AUC) for our 3-smRC model was 0.87. Finally, a likelihood ratio test between an AFP-only early HCC detection model and one incorporating both AFP and our 3-smRC early detection signature showed that our smRCs add significant predictive power to AFP alone (p<0.0001). As expected, AFP levels and expression of our 3-smRC signatures were not correlated (Fig. 6B), which suggest that both capture complementary signals for early HCC detection. Indeed, a blood-based composite model of our 3-smRC signature and AFP yielded an increased AUC of 0.93, lower Brier score of 0.11, and better test performance (85% sensitivity, 94% specificity, positive predictive value of 95%, Fig. 6A+C, Supplementary Table S2). These data confirm that our plasma 3-smRC signature robustly yields high accuracy in predicting early stage HCC among patients at high-risk, independent of AFP. A composite model including our 3-smRC signature and AFP further enhances its performance.
Figure 5.
(A) Calibration curve for penalized smRC logistic regression model to predict early HCC, with mean error 0.04. (B) Nomogram for 3-smRC signature to predict early stage HCC.
DISCUSSION
Our study provides a conceptually novel solution to a key barrier in the field of exRNA-derived cancer biomarkers. We strongly depart from previous exRNA characterization studies, which are restricted to quantifying expression of known (i.e. annotated) transcripts[13,14]. Thus, we do not discard the substantial component of unannotated exRNA, or simply focus on a particular RNA biotype (e.g. miRNA)[27,28]. Instead, we provide a novel, scalable, and data-driven view of the entire small exRNA landscape unfettered by incomplete and emerging prior knowledge. This approach allowed us to identify and validate novel circulating biomarkers for the detection of curable, early stage HCC.
By de novo characterizing the unknown non-coding small exRNA landscape across EV isolation technologies, biofluid, and cancer type, we have defined the key properties of exRNA-associated smRCs, including their clinical application in early cancer detection. We have used a comprehensive dataset from prostate cancer patients to establish an exRNA-specific smRC feature set from which we mine their key statistical properties and develop selection criteria. These properties indicate that the tractable smRC-based quantification of novel, unannotated, small RNA expression signatures is feasible across different EV isolation techniques applied to different biofluids, potentially offering a completely novel, data-driven strategy for increasing the sensitivity of EV-associated biomarker discovery. It is important to emphasize that multiple small functional noncoding RNA can arise from transcriptional post-processing of a single larger RNA precursor gene (e.g. endogenous siRNAs of plants[29] and animals [30], miRNA hairpins yielding miRNA*[31], and piRNAs[32]), so smRCs estimate the overlooked underlying expression profile of small RNA precursor genes and thereby facilitate accurate quantification, differential expression, and motif discovery of unknown, heterogeneous, small RNA dominated exRNA payloads. In this sense, smRCs might more accurately measure the information content of exRNA.
Applying our approach to a separate HCC plasma-based exRNA dataset (n=157), we derived a 3-smRC (unannotated), HCC-specific signature, which was then validated in an independent HCC cohort (‘HCC biomarker validation cohort’, n=209) to discriminate patients with incipient HCC from controls at high-risk of cancer. Despite the significant Wilcoxon p values demonstrating non-random separation of groups, the variability of smRC expression across HCC, and in fact CLD, was our primary motivation to turn to the more appropriate analytic setting of a full logistic regression model, which can not only leverage the combined predictive power of even partially correlated smRC biomarker candidates, but can effectively regress out technical bias (e.g. sequencing batch plate effects from RT-qPCR). Within this context, extra-sample error can be robustly estimated using standard bootstrap resampling techniques, leading directly to robust estimates of calibration and prediction error and power (Fig. 4B) that accounts for over-fitting. Generalizability of our results will be improved by external cohorts, ideally in a prospective setting, which are limitations of our study. Nevertheless, bootstrap resampling with replacement and cross-validation with a 85/15 test/training split both gave similar estimates of model optimism and extra-sample generalizability when averaged over 1000 iterations, which have previously been shown to be robust predictors of extra-sample performance[33]. Guided by recommendations of the International Society of Extracellular Vesicles[19], a thorough characterization of our isolates suggested a predominant enrichment for small EVs as a likely origin of our smRC signal. However, we cannot rule out contamination of other nanoparticles and we acknowledge technology-based differences for morphology assessment of EV isolates between nanoparticle tracking analysis (NTA) and transmission electron microscopy (TEM), particularly for size estimation, as reported previously[34].
Importantly, our exRNA-derived smRC signature was developed as a method for early HCC detection in the context of cancer surveillance and not as a HCC diagnostic tool. There is a subtle but very crucial difference between these two clinical scenarios which directly determined the patient population we deliberately selected for this study, as extensively outlined in clinical guidelines[10,11] and a recent white paper on HCC biomarker development[25]. Briefly, they explicitly underscore the urgent clinical need for new tools to detect patients with early stage HCC, as they can be cured if diagnosed at this stage. Other malignant liver tumors (e.g. cholangiocarcinoma) and associated metastases rarely occur in patients with cirrhosis and are not the target of liver cancer surveillance programs[7]. Nevertheless, we have confirmed the HCC specificity of our 3-smRC signature in a dataset of 142 patients with other malignancies. We purposely chose to test our early detection biomarker candidates in the context of the hardest possible scenario of distinguishing between chronic liver disease and very early, curable, HCC. Our signature is independently validated in more than 200 patients, where we demonstrate its ability to accurately detect patients with early stage HCC, including technical replicates and longitudinal samples before and after HCC treatment. We demonstrate that our 3-smRC signature (86% sensitivity, 91% specificity) not only outperforms the recommended surveillance tools (serum alpha-fetoprotein (AFP) combined with abdominal ultrasound: 63% sensitivity, 83% specificity)[12] for early stage HCC detection, but is complementary to AFP and in combination further maximizes HCC detection rates. There are other approaches currently under evaluation for early HCC detection using other liquid biopsy analytes[35], mostly involving circulating DNA. Blood-based DNA mutation[36], methylation[24,37], and DNA end-motif profiling[38] studies have shown comparable performance to our 3-smRC signature. The main difference with our study is that most of them included HCC patients at more advanced stages[39] as opposed to our exclusively early-stage cohort, and/or healthy controls, which might bias the performance of the tests.
Our study is a phase 2 biomarker case-control study according to the white paper by Pepe et al. on phases of biomarker development[16] and the Early Detection Research Network guidelines by the National Cancer Institute. This design includes the use of unannotated small RNA sequences with the aim of assessing the performance of our 3 markers to discriminate early stage HCC and controls at high-risk by inferring sensitivity and specificity of our test. Additionally, we wanted to put our results into clinical context. Therefore, we powered our study to compare against the performance of ultrasound and AFP for the detection of early stage HCC according to the largest meta-analysis available. Subsequent studies directly comparing our 3-smRC signature against ultrasound and AFP in a prospective setting will follow (i.e. phase 3/4 biomarker studies).
Despite not yet having a clear functional role in oncogenesis apart from suggestive enrichments in key RNA binding protein motifs (Supplementary Fig. S8), our findings strongly suggest that unannotated smRCs enable a robust, blood-based, minimally invasive, operator independent surveillance test for HCC, which is a major unmet clinical need in at-risk patients. In our study, a 3-smRC signature was able to detect HCC at an early tumor stage allowing patients to receive curative therapies, offering the potential to generalize this strategy to other cancer types as well. While further validation in phase 3/4 biomarker studies will pave the way for its clinical implementation, this study highlights complex, heterogeneous, non-coding and unannotated small RNA payloads of exRNA and their emergence as a powerful modality for biomarker discovery in cancer.
Supplementary Material
SUMMARY BOX.
What is already known about this subject?
Current surveillance tools for the detection of early stage HCC are suboptimal. EV-based early detection biomarkers have historically been plagued by poor reproducibility and are biased towards studying well-characterized miRNA across multiple indications.
What are the new findings?
Using a novel approach to reproducibly quantify and characterize the largely unexplored landscape of unannotated small RNA expression signatures from payloads of circulating EVs, we identified unannotated biomarkers capable of detecting early stage HCC with high accuracy.
How might it impact on clinical practice in the foreseeable future?
These findings directly lead to the prospect of a minimally-invasive, blood-only, operator-independent hepatocellular carcinoma surveillance biomarker.
ACKNOWLEDGEMENTS
The authors thank the office of Scientific Computing and the Genomics Core Facility at the Icahn School of Medicine at Mount Sinai (ISMMS) for providing computational resources and staff expertise, as well as the ISMMS Tissue Biorepository for providing some of the samples. The authors further thank Dr. Veronica Sanchez-Gonzalez (NanoView Biosciences, Boston, MA) for helping with the Exoview™ analysis.
Patient and Public Involvement (PPI) statement:
Patients or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Grant support: JvF is supported by the German Research Foundation (FE1746/1–1, FE1746/3–1) and the Clinician Scientist Program at University Medical Center Hamburg. TGL is supported by the Grant for Studies Broadening from the Spanish Association for the Study of the Liver (Asociación Española para el Estudio del Hígado, AEEH). AJC is supported by the National Cancer Institute Ruth L. Kirschstein NRSA Institutional Research Training Grant (CA078207). MEA, JELD, XC and BL were supported by the Icahn Institute of Genomics and Multiscale Biology. IL is supported by a grant from the Swiss National Science Foundation, from Foundation Roberto & Gianna Gonella and Foundation SICPA. PKH is supported by the German Research Foundation (HA8754/1–1). DD is supported by the Grant for Studies Broadening from the Spanish Association for the Study of the Liver (Asociación Española para el Estudio del Hígado, AEEH) and the Cancer Research Grant from Nuovo Soldati Foundation. JML is supported by grants from the U.S. Department of Defense (CA150272P3), European Commission Framework Program 7 (HEPTROMIC, proposal number 259744) and Horizon 2020 Program (HEPCAR, proposal number 667273–2), the Asociación Española Contra el Cáncer (AECC), Samuel Waxman Cancer Research Foundation, Spanish National Health Institute (SAF2013–41027) and Grup de Recerca Consolidat – Recerca Translacional en Oncologia Hepàtica. AGAUR (Generalitat de Catalunya), SGR 1162. CCC is supported by NCI grant P01-CA087497 and NIH grant U54-OD020353. AV is supported by the U.S. Department of Defense (CA150272P3).
Declaration of interests: JvF, BL, and AV are inventors in a provisional patent application for the 3-smRC signature. DDA received consulting fees from Almylam and Novartis. AV has received consulting fees from Boehringer-Ingelheim, Guidepoint and Fujifilm; advisory board fees from Gilead, Exact Sciences, Nucleix and NGM Pharmaceuticals; and research support from Eisai Pharmaceuticals. The remaining authors have nothing to declare in relation to this manuscript.
License statement:
The Submitting Author has the right to grant and does grant on behalf of all authors of the Work (as defined in the below author licence), an exclusive licence and/or a non-exclusive licence for contributions from authors who are: i) UK Crown employees; ii) where BMJ has agreed a CC-BY licence shall apply, and/or iii) in accordance with the terms applicable for US Federal Government officers or employees acting as part of their official duties; on a worldwide, perpetual, irrevocable, royalty-free basis to BMJ Publishing Group Ltd (“BMJ”) its licensees and where the relevant Journal is co-owned by BMJ to the co-owners of the Journal, to publish the Work in Gut and any other BMJ products and to exploit all rights, as set out in our licence.
The Submitting Author accepts and understands that any supply made under these terms is made by BMJ to the Submitting Author unless you are acting as an employee on behalf of your employer or a postgraduate student of an affiliated institution which is paying any applicable article publishing charge (“APC”) for Open Access articles. Where the Submitting Author wishes to make the Work available on an Open Access basis (and intends to pay the relevant APC), the terms of reuse of such Open Access shall be governed by a Creative Commons licence – details of these licences and which Creative Commons licence will apply to this Work are set out in our licence referred to above.
Footnotes
Data availability statement:
RNA-seq data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession numbers E-MTAB-8528 and E-MTAB-8529. Protocols for EV separation and characterization will be made available on EV TRACK (http://evtrack.org/) upon publication of the manuscript.
Code availability statement:
Upon publication, all code will be made publicly available at Dr. Losic GitHub site.
REFERENCES
- 1.Mathieu M, Martin-Jaular L, Lavieu G, et al. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol 2019;21:9–17. [DOI] [PubMed] [Google Scholar]
- 2.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science 2020;367. doi: 10.1126/science.aau6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 2018;19:213–28. [DOI] [PubMed] [Google Scholar]
- 4.Murillo OD, Thistlethwaite W, Rozowsky J, et al. exRNA Atlas Analysis Reveals Distinct Extracellular RNA Cargo Types and Their Carriers Present across Human Biofluids. Cell 2019;177:463–77.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kosaka N, Yoshioka Y, Fujita Y, et al. Versatile roles of extracellular vesicles in cancer. J Clin Invest 2016;126:1163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang KS, Im H, Hong S, et al. Multiparametric plasma EV profiling facilitates diagnosis of pancreatic malignancy. Sci Transl Med 2017;9. doi: 10.1126/scitranslmed.aal3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villanueva A. Hepatocellular Carcinoma. N Engl J Med 2019;380:1450–62. [DOI] [PubMed] [Google Scholar]
- 8.Choi DT, Kum H-C, Park S, et al. Hepatocellular Carcinoma Screening is Associated with Increased Survival of Patients with Cirrhosis. Clin Gastroenterol Hepatol Published Online First: 25 October 2018. doi: 10.1016/j.cgh.2018.10.031 [DOI] [PMC free article] [PubMed]
- 9.Singal AG, Yopp A, S Skinner C, et al. Utilization of hepatocellular carcinoma surveillance among American patients: a systematic review. J Gen Intern Med 2012;27:861–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marrero JA, Kulik LM, Sirlin CB, et al. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018;68:723–50. [DOI] [PubMed] [Google Scholar]
- 11.European Association for the Study of the Liver. Electronic address: easloffice@easloffice.eu, European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol Published Online First: 5 April 2018. doi: 10.1016/j.jhep.2018.03.019 [DOI]
- 12.Tzartzeva K, Obi J, Rich NE, et al. Surveillance Imaging and Alpha Fetoprotein for Early Detection of Hepatocellular Carcinoma in Patients With Cirrhosis: A Meta-analysis. Gastroenterology 2018;154:1706–18.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mjelle R, Dima SO, Bacalbasa N, et al. Comprehensive transcriptomic analyses of tissue, serum, and serum exosomes from hepatocellular carcinoma patients. BMC Cancer 2019;19:1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun N, Lee Y-T, Zhang RY, et al. Purification of HCC-specific extracellular vesicles on nanosubstrates for early HCC detection by digital scoring. Nat Commun 2020;11:4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lencioni R, Llovet JM. Modified RECIST (mRECIST) assessment for hepatocellular carcinoma. Semin Liver Dis 2010;30:52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pepe MS, Etzioni R, Feng Z, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst 2001;93:1054–61. [DOI] [PubMed] [Google Scholar]
- 17.Pepe MS. The Statistical Evaluation of Medical Tests for Classification and Prediction. Oxford University Press; 2003. [Google Scholar]
- 18.Agresti A, Coull BA. Approximate Is Better than ‘Exact’ for Interval Estimation of Binomial Proportions. The American Statistician. 1998;52:119. doi: 10.2307/2685469 [DOI] [Google Scholar]
- 19.Théry C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 2018;7:1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith JT, Wunsch BH, Dogra N, et al. Integrated nanoscale deterministic lateral displacement arrays for separation of extracellular vesicles from clinically-relevant volumes of biological samples. Lab Chip 2018;18:3913–25. [DOI] [PubMed] [Google Scholar]
- 21.Zhang W, Gao S, Zhou X, et al. Multiple distinct small RNAs originate from the same microRNA precursors. Genome Biol 2010;11:R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S-C, Wunsch BH, Hu H, et al. Broken flow symmetry explains the dynamics of small particles in deterministic lateral displacement arrays. Proc Natl Acad Sci U S A 2017;114:E5034–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wunsch BH, Smith JT, Gifford SM, et al. Nanoscale lateral displacement arrays for the separation of exosomes and colloids down to 20 nm. Nat Nanotechnol 2016;11:936–40. [DOI] [PubMed] [Google Scholar]
- 24.Xu R-H, Wei W, Krawczyk M, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater 2017;16:1155–61. [DOI] [PubMed] [Google Scholar]
- 25.Singal AG, Hoshida Y, Pinato DJ, et al. International Liver Cancer Association (ILCA) White Paper on Biomarker Development for Hepatocellular Carcinoma. Gastroenterology Published Online First: 8 March 2021. doi: 10.1053/j.gastro.2021.01.233 [DOI] [PMC free article] [PubMed]
- 26.Smith GCS, Seaman SR, Wood AM, et al. Correcting for optimistic prediction in small data sets. Am J Epidemiol 2014;180:318–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee YR, Kim G, Tak WY, et al. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int J Cancer 2019;144:1444–52. [DOI] [PubMed] [Google Scholar]
- 28.Jin X, Chen Y, Chen H, et al. Evaluation of Tumor-Derived Exosomal miRNA as Potential Diagnostic Biomarkers for Early-Stage Non-Small Cell Lung Cancer Using Next-Generation Sequencing. Clin Cancer Res 2017;23:5311–9. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y-X, Wang M, Wang X-J. Endogenous small RNA clusters in plants. Genomics Proteomics Bioinformatics 2014;12:64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piatek MJ, Werner A. Endogenous siRNAs: regulators of internal affairs. Biochem Soc Trans 2014;42:1174–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meijer HA, Smith EM, Bushell M. Regulation of miRNA strand selection: follow the leader? Biochem Soc Trans 2014;42:1135–40. [DOI] [PubMed] [Google Scholar]
- 32.Ozata DM, Gainetdinov I, Zoch A, et al. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet 2019;20:89–108. [DOI] [PubMed] [Google Scholar]
- 33.Harrell FE Jr. Regression Modeling Strategies: With Applications to Linear Models, Logistic and Ordinal Regression, and Survival Analysis. Springer, Cham: 2015. [Google Scholar]
- 34.Noble JM, Roberts LM, Vidavsky N, et al. Direct comparison of optical and electron microscopy methods for structural characterization of extracellular vesicles. J Struct Biol 2020;210:107474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Felden J, Garcia-Lezana T, Schulze K, et al. Liquid biopsy in the clinical management of hepatocellular carcinoma. Gut Published Online First: 3 September 2020. doi: 10.1136/gutjnl2019-320282 [DOI] [PubMed]
- 36.Qu C, Wang Y, Wang P, et al. Detection of early-stage hepatocellular carcinoma in asymptomatic HBsAg-seropositive individuals by liquid biopsy. Proc Natl Acad Sci U S A 2019;116:6308–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kisiel JB, Dukek BA, Kanipakam RVSR, et al. Hepatocellular Carcinoma Detection by Plasma Methylated DNA: Discovery, Phase I Pilot, and Phase II Clinical Validation. Hepatology Published Online First: 31 August 2018. doi: 10.1002/hep.30244 [DOI] [PMC free article] [PubMed]
- 38.Jiang P, Sun K, Peng W, et al. Plasma DNA End-Motif Profiling as a Fragmentomic Marker in Cancer, Pregnancy, and Transplantation. Cancer Discov 2020;10:664–73. [DOI] [PubMed] [Google Scholar]
- 39.Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.