Abstract
Tridax procumbens is a flowering plant of the Asteraceae family with a wide range of medicinal uses like anti-inflammatory, anti-diabetic, anti-microbial, immunomodulatory, etc. This study aimed to investigate the anti-cancerous activity of human lung cancer for targeting luteolin, a phytochemical of Tridax procumbens. The computational study has been done for studying the structural properties of luteolin. The drug-likeness of the molecule has been predicted by virtual screening of ADMET properties. The molecular docking technique of the in-silico method is performed to check the complex formation between protein and ligand. The reactivity and stability of the molecule are investigated with the help of molecular dynamics (MD) simulations. In the present work, we have tried to establish a strong candidature of any of the phytochemical of Tridax Procumbens as an inhibitor against human lung cancer.
Supplementary information
The online version contains supplementary material available at 10.1007/s11224-022-01882-7.
Keywords: DFT analysis, Molecular Docking, Molecular Dynamics Simulations, MCM7
Introduction
Cancer is one of the deadly diseases present in our world that majorly imparts in the global death ratio, yet it is incurable. It can be defined as the process of abnormal and uncontrolled cell division that occurs inside the human body and invade other parts, destroying body tissues [1]. It can affect any organ like the lung, kidney, intestine, uterus, brain, and even blood [2]. World Health Organization (WHO) has reported 9.6 million deaths, or one in six deaths, in 2018 and stated that cancer is the second leading cause of death worldwide [3]. The Globocan report 2020 issued by WHO reported that lung cancer is the second disease imparting 11.4% of the total cancer cases after breast cancer (11.7%) [4]. Lung cancer has the highest mortality rate of 2.21 million cases of the total deaths recorded from cancer in 2020 [4]. The dysregulation of deoxyribose nucleic acid (DNA) is the major reason for cancer initiation and progression in the human body [5]. Minichromosome maintenance (MCM) complex is the fifth most common type of cancer that leads to the evolution of the pre-replication process for DNA [6]. The regulation of MCM protein in the human body leads to the proliferation of various types of cancers [7]. MCM7 plays a vital role in cancer development and progression and acts as an initiator in eukaryotic DNA and G1/S cell cycle propagation [8]. It is one among the family of MCM DNA helicase comprising six conserved proteins called MCM2-7 [9]. It was first isolated from Saccharomyces cerevisiae (budding yeast) [10]. This process is called DNA replication licensing in which the MCM complex unwinds the bounded strands of the DNA [11]. This unwinding leads to genome duplication in proliferating cells and chromosomal defects. Such chromosomal defects result in tumorigenesis [12]. MCM proteins are found to be highly involved in human cancer evolution and malignant transformations. Thus, the MCM proteins are considered promising targets for cancer drug development.
On the other hand, in the last few decades, medicinal herbs have been shifted from fringe to mainstream use, and a more number of people seek remedies in herbal extracts [13, 14]. Plants have always been an important source of anti-cancerous supplements. Natural extracts share a large part in anticancer drugs available in the markets. Many studies have been reported for anti-cancer ailments from herbal extracts like gedunin for ovarian cancer [15], ginsenoside for breast cancer [16], cinnamon [17], and many more. These findings have motivated us to work specifically on herbal extracts. For the present work, we have considered a tropical plant Tridax procumbens. This plant has historical shreds of evidence for being used as a medicinal plant [18]. The plant is native to Asia, America, Africa and, Australia. In many countries, Tridax procumbens is widely used for the healing of open wounds [18]. This herb is also known for its wide range of pharmacological activities like anti-fungal [19], anti-inflammatory [20], anti-tubercular [21], hepatoprotective [22], anti-diabetic [23] etc. It is also known for curing asthma [24] and is a fulfilled herb with secondary metabolites like steroids, terpenoids, tannins, flavonoids, saponins, glycosides, and amino acids [25].
The virtual screening of pharmacological properties is done for Luteolin to study its drug-like resemblances. The energy optimization of the structure of Luteolin is done to investigate its stability. The binding of ligand to protein is accounted by performing molecular docking, and the stability of the complex formed by docking of protein and ligand is verified by performing the molecular dynamics simulations. In the present paper, an in silico study done with luteolin as an inhibitor will help in establishing the strong candidature of luteolin as a potential drug against human cancer.
Material and methods
Potential target protein structure for cancer MCM7 protease
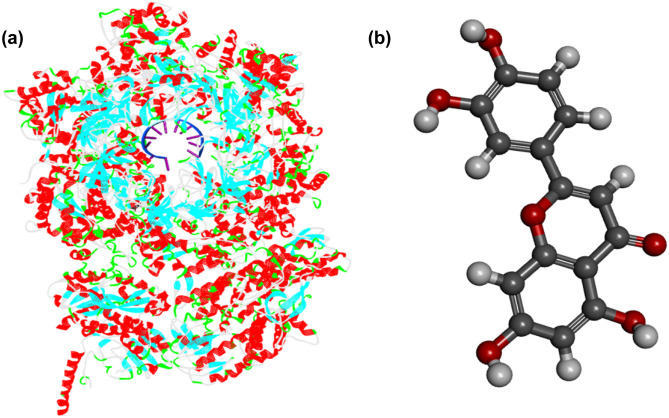
Human cancer encodes a large number of protein structures. The structure we have considered for this study is protease MCM7 (PDB ID: 6XTX, resolution: 3.29 Å). MCM7 can metastasize and destroy the living tissues in the body. The malignancies in which MCM7 is involved are hepatocellular carcinoma, head, and neck, esophagus, etc. [26]. Thus, it is suitable to design a study that will identify the compound with inhibitor activity for preventing replication of DNA of cancerous cells. The 3D structure of protein MCM7 is downloaded from “Protein Data Bank.” Removal of heteroatoms, water molecules, and addition of polar hydrogen is done with the help of the software “BIOVIA Discovery Studio Visualizer” (https://discover.3ds.com/discovery-studio-visualizer-download). The structure of receptor protease MCM7 with DNA backbone and basic pairs located at the center of the structure is shown in Fig. 1.
Fig. 1.
(a) Structure of receptor protease MCM7 (PDB ID: 6XTX) with DNA backbone and basic pairs are located at the center of the structure (backbone: blue, basic pairs: magenta), (b) structure of luteolin
Potential inhibitor: Tridax procumbens
Tridax procumbens is a phytochemical-rich plant constituting baicalin, tetrandrine, luteolin, apigenin, stigmasterol, catechin, epicatechin, quercetin, myricetin, gallocatechin, sitosterol, akuammidine, kaempferol, and many more [27]. Among all the phytochemicals, luteolin is a flavonoid having anti-cancerous properties [28]. Numerous works have been done so far reporting the anticancer properties of Luteolin [29]. Anticancer activity of luteolin is investigated against gastric cancers [30], breast cancer [31], prostate cancer [32], brain tumors [33], cervical cancer [34], skin cancer [35], etc. Thus, it makes sure that our vision to use it as an anti-cancer agent will not disappoint us. Luteolin is a flavonoid belonging to the vitamin B family. It is mainly present in many food supplements like parsley, broccoli, onion leaves, carrots, peppers, cabbages, apple skins, etc. [36]. It is highly added as a food supplement due to its anti-oxidative property. Various pre-clinical reports have proven that luteolin possesses a wide range of pharmacological activities like anti-hepatotoxic [37], hypotensive [38], anti-urolithiasis [39], hemostatic [40], antimicrobial or antibacterial activity [41], and many others. It is seen that the luteolin has preventive activity against several parasitic agents like Leishmania donovani [42] and plasmodium falciparum [43]. Many studies have been already done considering luteolin as a potential inhibitor against the Mpro protease of COVID-19 [44]. The structure of Luteolin is downloaded from the online database “PubChem” (https://pubchem.ncbi.nlm.nih.gov/) ID:5280445) (Fig. 1).
Computational method for structural analysis
The ground state energy optimization is done with the help of the software “Gaussian 09” [45] and method B3LYP with a standard 6-311G basis set [46, 47]. To visualize the Gaussian results, Gauss View 5 molecular visualization program has been used [48]. Optimized geometry is used to derive the Mulliken charge distribution and molecular electrostatic potential (MEP) surface map that helps in explaining the chemical stability of the molecule. The frontier molecular orbitals (FMO) (highest occupied molecular orbital and lowest unoccupied molecular orbital) energies are also derived with the help of optimized geometry. These HOMO–LUMO energies are used for computing ionization potential (IP), energy gap (ΔE), electron affinity (EA), chemical potential (CP), electronegativity (χ), softness (S), and hardness (η) [49]. These parameters are considered global reactivity parameters and help in determining the chemical reactivity of the molecule.
Drug-likeness and ADMET properties
We have done the virtual screening of drug-likeness rules, and ADMET properties of Luteolin. Lipinski’s rule, MDDR-like rule, Veber’s rule, Ghose filter, Egan rule, Muegge rule, lipophilicity, water-solubility, etc., are examples of drug-likeness rules [50]. Some of the drug-likeness rules are molecular weight < 500 g/mol, hydrogen bond donors < 5, hydrogen bond acceptor < 10, MLOGP (n-octanol–water partition coefficient) < 4.15, molar refractivity should be between 40 and 130, log P ranging between − 0.4 and + 5.6, solubility (log S) > − 5.7 [51]. Additionally, absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties are also important in drug designing as they signify whether the compound undergoes proper metabolic processes in the human body or is toxic [52]. In the present work, all the drug-likeness and ADMET features are listed with the help of the online database SwissADME.
Prediction of activity spectra for substances (PASS)
Cytotoxicity prediction is done with the help of the freely accessible online platform CLC-Pred (Cell Line Cytotoxicity Predictor) [53]. CLC-Pred is a cytotoxicity predictor used in in-silico studies that can predict the cytotoxic effect of chemical compounds by the virtue of PASS technology. Near about 4000 kinds of biological activities like toxic and adverse effects, mechanisms of action, interaction with metabolic enzymes and transporters, pharmacological effects influence on gene expression, etc. can be predicted by PASS [54]. The structure of luteolin is submitted in smiles format. Prediction of the activity spectrum of a compound is estimated in terms of probable activity (Pa) and probable inactivity (Pi). Pa and Pi values vary between 0.000 and 1.000 [55]. The activities following condition Pa > Pi are considered as possible for a particular compound. Experimental pharmacological action is considered high if the Pa > 0.7 and low if 0.5 < Pa < 0.7 [56]. Data can also be extracted in structured data file format. Analysis of data extracted from CLC-Pred is interpreted based on their IG50 (half-maximal inhibitory growth), IC50 (half maximal inhibitory concentration), and % inhibition (of activity) values [57]. Compounds having IG50 and IC50 values 10,000 nM and inhibition of more than 50% are considered as active [58].
Prediction of cardiac toxicity
Prediction of cardiac toxicity detects the cardio-related harms by the consumption of the drug. Pred-hERG 4.2 is a freely accessible validated web server that is used for the identification of cardiotoxic blockers of the compound [59]. The human ether-a-go-go-related gene (hERG) is a cardiac repolarizer that mainly encodes a protein and also activates the rectifier potassium channel (IKr) [60]. Heartbeat delay is the main caution of dysfunctioning of hERG which may often cause sudden death [61]. The structure of luteolin is submitted to Pred-hERG in smileys format. Potency, confidence, applicability domain, and probability map are recorded as the results [62]. To be non-cardio toxic, the confidence value should not exceed 0.26 for any compound [63]. The fragments representing hERG blockage are indicated in the probability map.
Molecular docking study
For molecular docking studies, Chain 4 and Chain 7 are chosen based on the AGS cocrystal found naturally in the CryoEM structure of human MCM7 structure (PDB ID: 6XTX) [64]. Missing residues in Chain 4 and 7 structures are completed using SWISS-MODEL (https://swissmodel.expasy.org/). Based on the cocrystal AGS in the 6LXT structure, the active site coordinates are determined as x: 211.170, y: 201.937, and z: 140.544, and the grid box volume is chosen as 20*20*20 Å3. Molecular docking is performed with both Autodock Vina and Glide to validate the results [65, 66]. The optimized luteolin structure for molecular docking studies is obtained from the DFT study. The protein and ligand input pdbqt files required for Autodock Vina docking are created with AutoDockTools-1.5.6. For Glide docking, the protein structure is prepared with the “Protein Preparation Wizard” in Schrödinger Maestro 12.8 and the ligand structure is prepared with the “LigPrep” module using OPLS4 force field. Protein–ligand interactions are visualized using BIOVIA Discovery Studio Visualizer v21 and UCSF Chimera v1.15 software. The docked structure by Glide of Luteolin with chain 7 and chain 4 of 6XTX is further used for performing MD simulations.
Molecular dynamics simulations
The software “Gromacs 2019.2 version” is used for performing the molecular dynamics simulation [67, 68]. The protein preparation topology is created with Gromos 43A1 force field and SCP water model. The ligand topology file is obtained from the GlycoBioChem PRODRG2 server (http://davapc1.bioch.dundee.ac.uk/cgi-bin/prodrg) [69]. The protein–ligand complex is simulated for 300 ps in canonical (amount of substance (N), pressure (P) and temperature (T) — NPT) and 300 ps isothermal-isobaric (amount of substance (N), volume (V), and equilibrium steps temperature (T) — NVT) ensembles. The molecular dynamics simulations run for 100 ns. The root mean square deviation (RMSD) and root mean square fluctuation (RMSF) is calculated to study the stability of the complex during the simulation. RMSD and RMSF help in the prediction of the atomic positions and the complex stability of the molecule undergoing simulation.
Results and discussions
Analysis of structural properties
Optimized structure and charge analysis
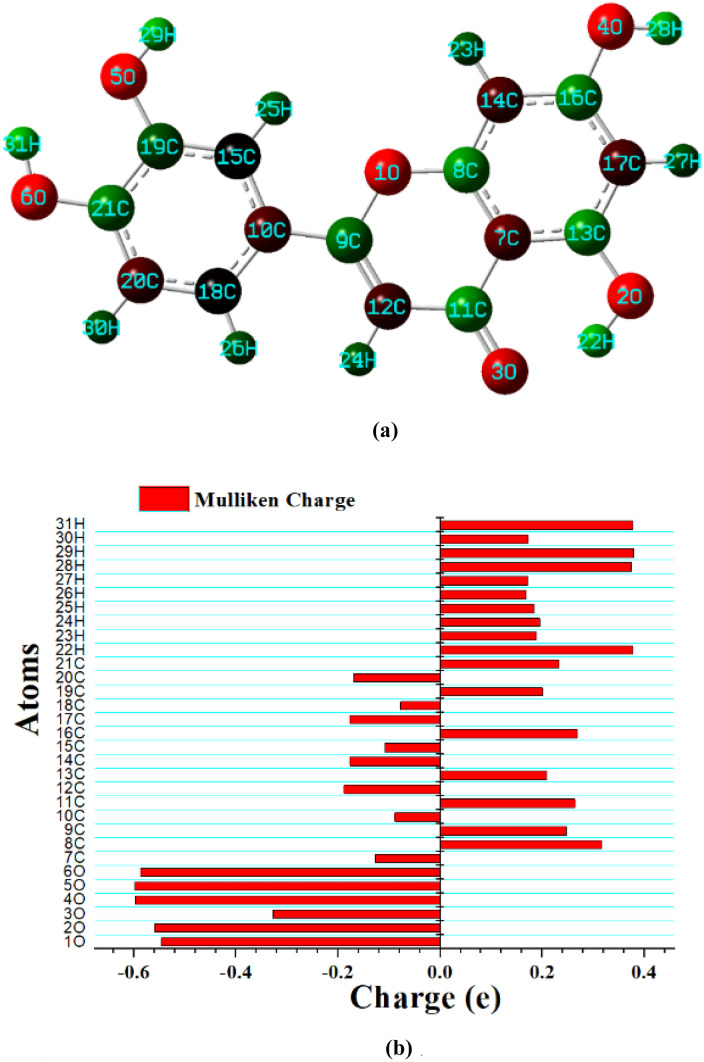
The optimized geometry of luteolin seems to be planar with the total electrostatic potential energy of − 1028.92 au, and dipole moment 4.85 Debye. The high dipole moment of the probe molecule suggests the bioactivity of the structure that strengthens the formation of the bond between the drug and targeted protein [70]. Bond length (Å) and bond angle (°) between different atoms of optimized geometry are mentioned in SD.1 and SD.2.
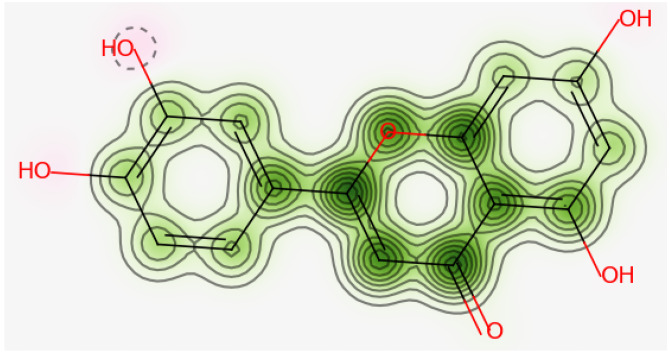
The graphical representation of Mulliken charges shows the positive impact of hydrogen atoms and the negative impact of carbon and oxygen atoms (Fig. 2b). Atom 5O bonded to benzene ring shows the maximum negative charge of 0.664e. 6O and 4O also show near about maximum negative charges of 0.609e and 0.614e respectively (SD.3). On the other hand, 22H, 28H, 29H, and 31H are the hydrogen atoms showing the highest positive charges of 0.396e, 0.381e 0.396e, and 0.400e, respectively (SD.3). A high variation of charge within the molecule is observed for C atoms. High positive and negative effect in atoms shows the possibility of intramolecular charge transfer property within the Luteolin.
Fig. 2.
(a) Optimized structure of Luteolin. Red atoms are oxygen, dark green is hydrogen, light green is carbon, and brown are the carbons bonded to hydrogen. (b) Plot showing Mulliken charge distribution of Luteolin. The plot shows the positive behavior of hydrogen and the negative behavior of oxygen
Frontier Molecular Orbital (FMO) analysis
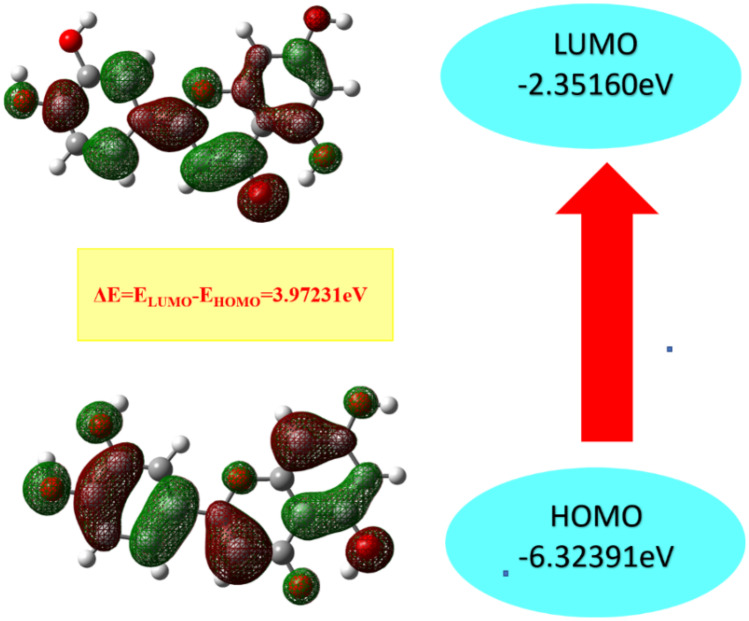
The chemical reactivity, stability, and optical properties of a molecule are derived by HOMO and LUMO energies. Koopman’s theorem is used for calculating different FMO related parameters (Table 1) [71].
| 1 |
| 2 |
| 3 |
| 4 |
| 5 |
Table 1.
Chemical parameters of luteolin (all values are in eV and S is in (eV)−1)
| S No | Molecular property | Values |
|---|---|---|
| 1 | HOMO | − 6.32 |
| 2 | LUMO | − 2.35 |
| 3 | Energy gap (ΔE) | 3.97 |
| 4 | Ionization potential (IP) | 6.32 |
| 5 | Electron affinity (EA) | 2.35 |
| 6 | Chemical potential (CP) | − 4.33 |
| 7 | Electronegativity (χ) | 4.33 |
| 8 | Hardness (η) | 1.98 |
| 9 | Softness (S) | 0.50 |
The energy gap between the HOMO and LUMO energies is found to be 3.97 eV (Fig. 3). This confirms the delocalization of electrons within the molecule. The value of IP (6.32 eV) shows the tendency of the molecule to easily loose the free electrons. The value of EA (2.35 eV) is reported for the molecule that shows the atoms attract the electrons more easily. The value of χ is calculated to be 4.33 eV. All the FMO parameters confirm the high chemical reactivity of the Luteolin molecule. The high value of η (1.986 eV) and low value of S (0.503 eV) shows the stability of the molecule. These parameters show the chemical reactivity of the system.
Fig. 3.
HOMO and LUMO with respective energies and energy difference
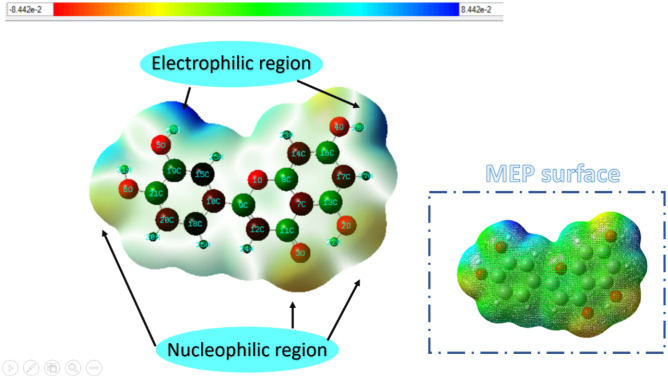
MEP analysis
The reactivity of any molecule is determined by examining the molecular electrostatic potentiality of the molecule. MEP helps in the determination of active sites of the molecule. Figure 4 shows the MEP surface indicating electrophilic regions from blue color and nucleophilic regions in yellow color. There is some portion of the molecule which is neutral or does not participate in the chemical reaction. In the MEP surface, the green-colored region shows the neutral part of the molecule. The electron-rich locations shaded in yellow color indicate that the molecule has plenty of electrons for undergoing chemical reactions, and the maximum negative region for electrophile attack shown in blue color validates the possibility of the molecule being highly chemically reactive [72].
Fig. 4.
MEP surface showing the electrophilic region in blue color and nucleophilic region in yellow portions
Analysis of drug likeness and ADMET properties
Luteolin follows Lipinski’s rule of five without any violation with a molecular mass of 286.24 g/mol, logP < 5, H-donor < 5, and H-acceptor < 10. Along with showing high gastrointestinal absorption and moderate solubility, Ghose’s filter, Egan’s rule, Veber’s rule, and Mugge’s rule. Table 2 lists the properties and their responses for luteolin, lipophilicity, or partition coefficient which is an important virtue of virtual screening which accounts for the affinity of the drug in the lipid environment. For a compound to be used as a drug, the lipophilicity value should be positive to be stable in lipid medium, and logP should be less than 5 to be a potential drug. For luteolin, all log P parameters are less than 5. Also, luteolin has shown positive behavior for being water-soluble. Luteolin also shows high gastronomical intestinal absorption which shows it can be a better drug for human consumption.
Table 2.
Physiochemical, drug-likeness, pharmacokinetics, lipophilicity, and medicinal chemistry of luteolin
| Physicochemical properties | |
|---|---|
| Formula | C15H10O6 |
| Molecular weight | 286.24 g/mol |
| Num. H-bond acceptors | 6 |
| Num. H-bond donors | 4 |
| Molar Refractivity | 76.01 |
| TPSA | 111.13 Å2 |
| Lipophilicity | |
|---|---|
| Log Po/w (iLOGP) | 1.86 |
| Log Po/w (XLOGP3) | 2.53 |
| Log Po/w (WLOGP) | 2.28 |
| Log Po/w (MLOGP) | − 0.03 |
| Log Po/w (SILICOS-IT) | 2.03 |
| Consensus Log Po/w | 1.73 |
| Water solubility | |
|---|---|
| Log S (ESOL) | − 3.71 |
| Solubility | 5.63e-02 mg/ml; 1.97 × 10−4 mol/l |
| Class | Soluble |
| Log S (Ali) | − 4.51 |
| Solubility | 8.84e-03 mg/ml; 3.09 × 10−5 mol/l |
| Class | Moderately soluble |
| Log S (SILICOS-IT) | − 3.82 |
| Solubility | 4.29e − 02 mg/ml; 1.50 × 10−4 mol/l |
| Class | Soluble |
| Pharmacokinetics | |
|---|---|
| GI absorption | High |
| BBB permeant | No |
| P-gp substrate | No |
| Log Kp (skin permeation) | − 6.25 cm/s |
| Drug-likeness | |
|---|---|
| Lipinski | Yes; 0 violation |
| Ghose | Yes |
| Veber | Yes |
| Egan | Yes |
| Muegge | Yes |
| Bioavailability Score | 0.55 |
| Medicinal chemistry | |
|---|---|
| PAINS | 1 alert: catechol_A |
| Brenk | 1 alert: catechol |
| Lead-likeness | Yes |
| Synthetic accessibility | 3.02 |
PASS analysis
The considered molecule luteolin is widely preferred for the treatment of hypertension, inflammatory disorders, and cancer. Data given in SD.4 displays one of the breast carcinomas for Pa > 0.3 which matches with one of its known therapeutic applications. Cytotoxicity against many cell lines is also predicted including oligodendroglioma, colon adenocarcinoma, hepatoblastoma, and many more (SD.4). For Pa > 0.5, cancer line Hs 683 (oligodendroglioma) is predicted.
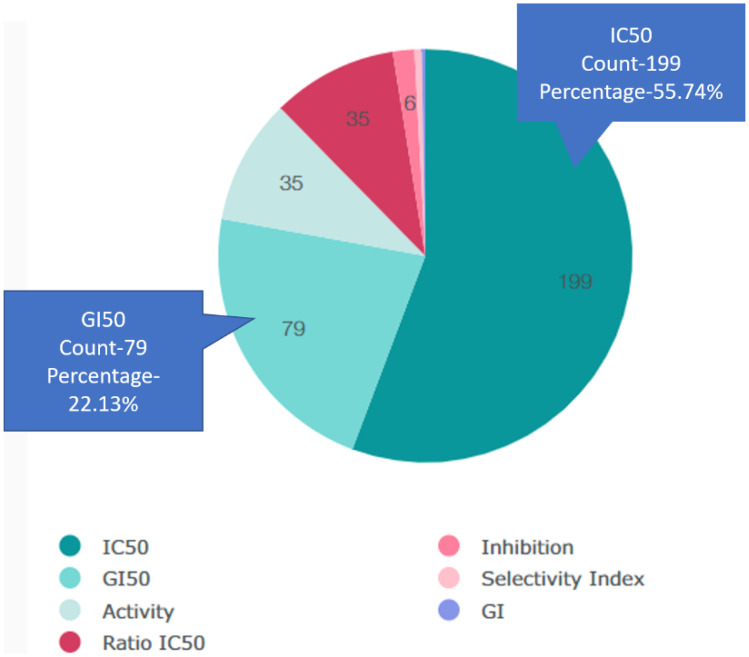
Predicted activity (Pa) with value 0.523 and probable inactivity (Pi) with value 0.049 is obtained for luteolin. Results verify that the probable activity (Pa) score is very close to 1 and the probable inactivity (Pi) score is very close to 0. Half maximal inhibitory concentration (IC50) which shows the extent of inhibiting of any compound against a specific biological function shows more than 50% probability in luteolin which is considered as an active compound. Figure 5 shows the percent share of possible biological activities possessed by luteolin. Cancer cell growth anti-proliferation is explained as GI50 activity.
Fig. 5.
Pie chart showing types of possible biological activities in Luteolin. Share of IC50 and GI50 is also shown with probability percentage
Cardiac toxicity analysis
The value of the applicability domain for luteolin is observed to be 0.19 which lies in the acceptable range (less than 0.26). The probability map shown in Fig. 6 indicates the presence of both positive and negative contributions of atoms or fragments to the hERG blockage. From Fig. 6, it is noticeable that the pink-colored region near the OH groups in the probability map indicates the decreased hERG blockage region. The set of atoms constituting oxygen promotes the hERG cardiac potentiality of the molecule. The low value of the applicability domain predicted luteolin as non-cardiotoxic with a 50% confidence value.
Fig. 6.
Probability map of luteolin showing hERG blockage promoting regions in pink shade
Molecular docking analysis
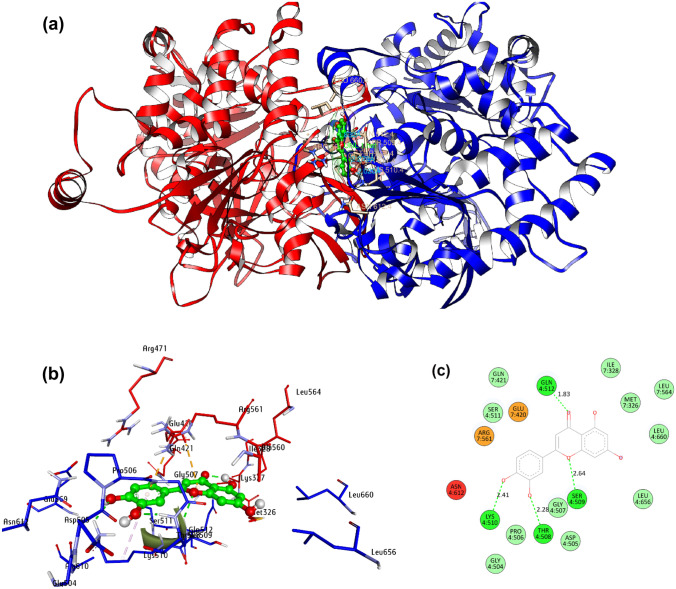
After performing multiple times, docking of the ligand with protein the first binding pose is considered as the best binding pose with binding affinity − 8.4 kcal/mol and four hydrogen bonds (Fig. 7). All the conventional hydrogen bond interaction details are mentioned in Table 3 with common active site amino acid residues Gln512, Ser509, Thr508, and Lys510. All these results show that the first docked pose is the best pose of luteolin in the binding site of MCM7. It also validates the potentiality of luteolin to be used as a drug against MCM7 cancer. The 2D structure of donor–acceptor interactions is also verified to locate the hydrogen bonds associated with the complex (Fig. 7).
Fig. 7.
(a) 3D view of the whole protein and ligand obtained from Autodock Vina. (b) Molecular docking binding pose of chain 4 (blue), chain 7 (red), and luteolin (green) at MCM7 protein active site. (c) Schematic protein–ligand molecular interactions diagram including H bond lengths
Table 3.
Luteolin molecular interaction details at the active site of MCM7 protein obtained from Glide XP and Autodock Vina tools
| Protein/docking tools | Interacting residues | Distance (Å) | Category | Type |
|---|---|---|---|---|
|
DNA replication licensing factor MCM7 (PDB ID: 6XTX) Glide XP (− 8.5 kcal/mol) |
4:Tyr465 | 2.23237 | Hydrogen bond | Conventional hydrogen bond |
| 4:Gln512 | 2.12171 | Conventional hydrogen bond | ||
| 4:Tyr652 | 2.0905 | Conventional hydrogen bond | ||
| 7:Glu420 | 1.70975 | Conventional hydrogen bond | ||
| 4:Leu656 | 2.93413 | Hydrophobic | Pi-Sigma | |
| 4:Thr508 | 5.03603 | Amide-Pi stacked | ||
| 4:Ser509 | 4.2699 | Amide-Pi stacked | ||
| 4:Leu660 | 5.06686 | Pi-Alkyl | ||
| 7:Ala560 | 4.91514 | Pi-Alkyl | ||
| 7:Ala560 | 4.90091 | Pi-Alkyl | ||
| 7:Arg561 | 5.05345 | Pi-Alkyl | ||
| 7:Leu564 | 5.16814 | Pi-Alkyl | ||
|
DNA replication licensing factor MCM7 (PDB ID: 6XTX) Autodock Vina (− 8.4 kcal/mol) |
4:Thr508 | 2.27927 | Hydrogen bond | Conventional hydrogen bond |
| 4:Ser509 | 2.64023 | Conventional hydrogen bond | ||
| 4:Lys510 | 2.40961 | Conventional hydrogen bond | ||
| 4:Gln512 | 1.83235 | Conventional hydrogen bond | ||
| 7:Arg561 | 3.88199 | Electrostatic | Pi-Cation | |
| 7:Glu420 | 4.24757 | Pi-Anion | ||
| 4:Lys510 | 5.14426 | Hydrophobic | Pi-Alkyl |
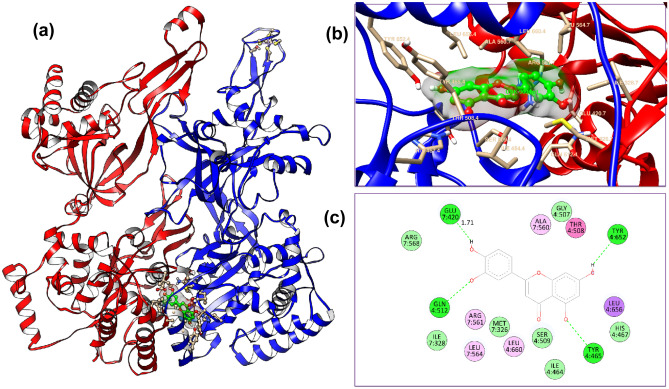
The docking results obtained by Schrodinger software are shown in Fig. 8. The Glide XP score of − 10 or less is considered as the good binding score. In case the hydrophobic interactions are associated with the binding site, the Glide XP score of − 8 or − 9 is also considered as the good binding score. In our case, the Glide XP score obtained by the docking of luteolin with MCM7 has the value of − 8.516 which is very close. The amino acid residues Glu420, Gln512, Tyr652, and Tyr465 are associated with conventional bond formation. The pi-sigma interaction is represented by Leu565 residue. Leu564, Leu660, Ala560, and Alg561 residues are associated with the pi-alkyl bond. The pose with glide emodel value -66.445 is selected as the best pose. Molecular interaction details obtained from Glide XP and Autodock Vina tools are given in Table 3.
Fig. 8.
(a) Docking mode of protein and ligand by Schrodinger-Glide XP docking of protein active site with Chain 7 (red), and chain 4 (blue) with MCM7, (b) 3D interaction of donor–acceptor interaction obtained by docking of Luteolin with MCM7, (c) 2D view of protein–ligand interaction showing conventional, carbon and pi-donor H-bonds
Molecular dynamics simulations
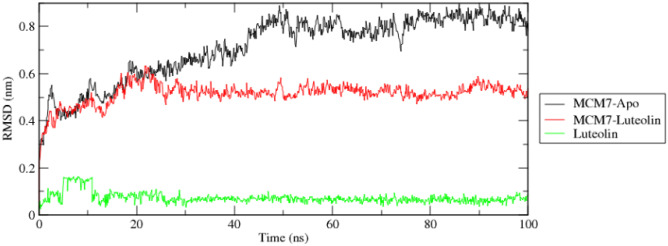
Root mean square deviation (RMSD)
The RMSD is calculated to determine the protein–ligand stability and functionality of the complex. This provides an idea about the structural deviation of the protein–ligand complex during the time trajectory of 100 ns (Fig. 9). The RMSD deviation of luteolin gave a rise up to 0.2 nm in the initial 10 s and remained constant for the whole simulation time. Unlike luteolin, the RMSD of the apoprotein rise for initial 45 s of the simulation time. This rise in RMSD value increases up to 1 nm and fluctuates from 0.6 to 1 nm for the next 55 s. RMSD graph of MCM7-luteolin complex shows intermediate values of apoprotein graph and luteolin graph. The RMSD value of the complex fluctuates from 0.2 to 0.6 nm consistently. The consistent value of RMSD for complex shows that the complex remains stable during the whole simulation time.
Fig. 9.
Root mean square deviation (RMSD) of MCM7 apoprotein and its complex with luteolin for time trajectory from 0 to 100 ns
Root mean square fluctuations (RMSF)
The RMSF give insights into the protein residue fluctuations for the complex. The RMSF plot for chain 4 and chain 7 of the 6XTX protein complexed with luteolin is analyzed for 100 ns of the simulation time. The RMSF plot of the complex is shown by red color, and apoprotein is shown by black color (Fig. 10). The plot for both the chains shows frequent fluctuations that validate the high flexibility of the complex. The plot indicates that the binding of the complex did not get affected throughout the simulation time. Thus, the RMSF analysis validates that the process of simulation did not alter the complex resulting in the stable protein–ligand complex.
Fig. 10.
Root mean square fluctuations (RMSF) graph of MCM7 in apoprotein (black) and its complex (red) with luteolin for chain 4 and chain 7 of the protein for time trajectory of 100 ns
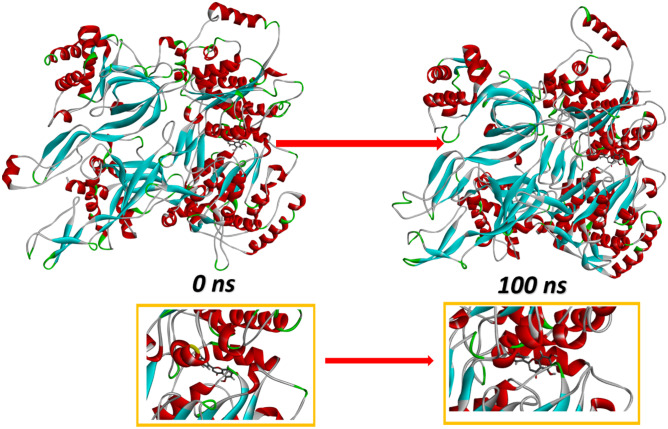
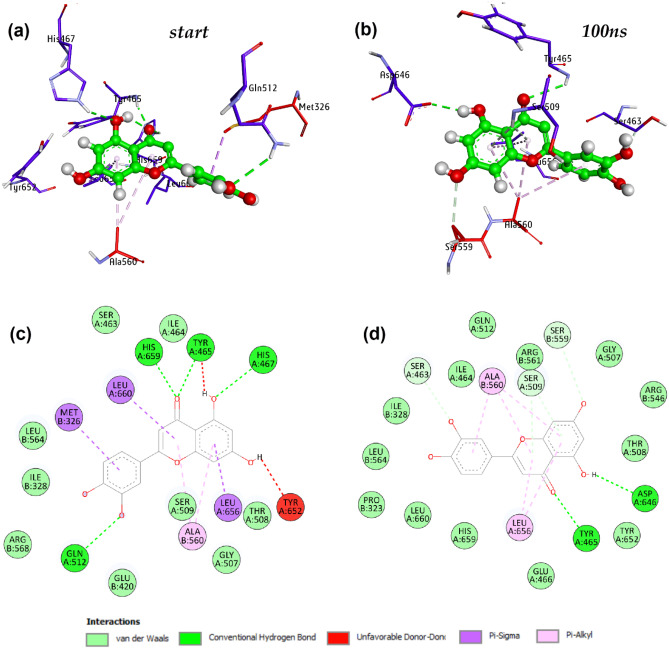
Protein–ligand interactions from MD simulations
The position of the ligand was observed on the protein before and after simulation and is illustrated in Fig. 11. The interacting residues associated with the protein–ligand complex after docking were His457, Tyr465, Gln512, Met326, Tyr465, His659, Tyr652, and Ala560. This complex was used for the simulation. Thus, the amino acid residues associated to the intramolecular hydrogen bonding of the ligand with protein are His457, Tyr465, Gln512, Met326, Tyr465, His659, Tyr652, and Ala560. The residues associated in the binding after simulation differ from the residues associated to initial complex. After the 100 ns of simulation trajectory, the amino acid residues associated with the protein–ligand complex were obtained as Asp646, Tyr465, Ser509, Ser463, Ala560, Ser559, and Leu656. The association of the amino acid residues to the protein–ligand complex before and after 100 ns of simulation time is illustrated in Fig. 12a, b, respectively. Before simulation, the residues His659, Tyr465, His467, and Gln512 impart in the formation of conventional hydrogen bond that are highly polar in nature due to the presence of hydrogen bonds. But after the simulation of the system for 100 ns, only Tyr465 and Asp465 residues impart in the conventional hydrogen bond formation. The residues Ser463, Ile464, Leu564, Ile328, Arg568, Glu420, Gly507, Ser509, and Thr508 are attached to the complex before simulation forming the Van der Waal forces (Fig. 12c), but after 100 ns, the residues Gln512, Ile464, Ser463, Ile328, Leu564, Pro323, Leu660, His659, Glu466, Tyr652, Thr508, Arg546, Gly507, and Arg561 are associated to the protein–ligand complex (Fig. 12d). These residues represent the Van der Waal forces that make covalent bonds, and these covalent bonds are non-polar in nature. Thus, ligand interaction with protein at 100 ns varies from the initial complex obtained from docking. The interacting residues also change over time leading to polar conventional hydrogen bonds and some non-polar Van der Waal interactions.
Fig. 11.
Diagram showing position of the ligand bonded to protein at 0 ns and 100 ns of molecular dynamics simulations of MCM7-Luteolin complex
Fig. 12.
Binding poses and schematic protein–ligand interactions diagram at 0 ns and 100 ns of molecular dynamics simulations of MCM7-Luteolin complex
Conclusion
This study has utilized comprehensive in-silico techniques for determining the anti-cancerous activity of luteolin. The high value of the dipole moment of the luteolin implies the bioactivity of the molecule. Therefore, it can better support the bond formation between the drug and targeted protein. Results of charge analysis show the property of charge transfer of the compound. FMO parameters like ΔE, IP, EA, CP, and χ show values favorable enough to ideally justify its chemical stability. The high value of η and the low value of S show the stiffness and the stability of the molecule. The presence of electrophilic and nucleophilic regions justifies the displacement of the electron cloud and hence intermolecular charge transfer within the luteolin molecule. Throughout the virtual screening of pharmacokinetic properties like Lipinski rules, Ghose’s filter, Egan’s rule, Veber’s rule, and Mugge’s rule, etc., luteolin proved itself a beneficial compound that shows drug-like behavior. Along with showing positive results in ADMET properties, it responded to PASS and cardio-toxic analysis positively. The value of active probability for biological activities is much higher than the inactive probability that validates the probability of biological activities within the molecule. 0.19 is the value for the applicability domain which is much less than 0.26 which is considered the cut-off value for cardio-toxicity. Thus, luteolin does not have any cardio-toxic behavior and can be preferred as a potential drug against MCM7 cancer. Furthermore, the results of molecular docking revealed the binding score of the best binding site is found to be − 8.4 kcal/mol which is quite good. The RMSD and RMSF values obtained by MD simulations have proved that the selection of docking pose seems to be correct. The amino acid residues associated to the binding pose of the ligand with the protein changes over the simulation time showing the point mutation in protein. It also conveys the availability of polar and nonpolar interactions. The protein ligand complex maintains stability throughout the simulation time. Summarizing all the in-silico study results directs toward the strong candidature of Luteolin as a potential inhibitor. We believe that our present study would provide a lead in drug development from luteolin for preventing DNA replication.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Ankara University-Scientific Research Unit for supplying the Schöridenger software program purchased under grant project number BAP-21B0237004. Molecular dynamics simulations reported were performed utilizing TÜBİTAK (The Scientific and Technological Research Council of Turkey), ULAKBİM (The Turkish Academic Network and Information Center), and High Performance and Grid Computing Center (TRUBA resources).
Author contribution
Shradha Lakhera: data curation, writing- original draft preparation, visualization, investigation, software, validation. Meenakshi Rana: conceptualization, methodology, writing — reviewing and editing, supervision. Kamal Devlal: conceptualization, writing — reviewing and editing. Ismail Celik: software, validation, reviewing and editing. Rohitash Yadav: data curation, software, validation
Availability of data and material
Protein structure: https://www.rcsb.org/. Ligand structure: https://pubchem.ncbi.nlm.nih.gov/. ADMET properties: http://www.swissadme.ch/. Molecular docking: https://swissmodel.expasy.org/. Docking analysis: https://disCover.3ds.com/discovery-studio-visualizer-download. Ligand topology: http://davapc1.bioch.dundee.ac.uk/cgi-bin/prodrg.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gupta SP. Quantitative structure- activity relationship studies on anticancer drugs. Chem Rev. 1994;94:1507–1551. doi: 10.1021/cr00030a003. [DOI] [Google Scholar]
- 2.Fu J, Zhou B, Zhang L, Balaji KS, Wei C, Liu X, Chen H, Peng J, Fu J. Expressions and significances of the angiotensin-converting enzyme 2 gene, the receptor of SARS-CoV-2 for COVID-19. Mol Biol Rep. 2020;47:4383–4392. doi: 10.1007/s11033-020-05478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organisation health report on Cancer (2018) https://www.who.int/health-topics/cancer#:~:text=Cancer%20is%20the%20second%20leading,in%20six%20deaths%2C%20in%202018
- 4.IARC- GLOBOCAN (2020) New Global Cancer Data. https://www.uicc.org/news/globocan-2020-new-global-cancer
- 5.Nugent M. MicroRNA function and dysregulation in bone tumors: the evidence to date. Cancer Manag Res. 2014;6:15–25. doi: 10.2147/CMAR.S53928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Richards TA, Aves SJ (2009) Ancient diversification of eukaryotic MCM DNA replication proteins. BMC Evol Biol 9. 10.1186/1471-2148-9-60 [DOI] [PMC free article] [PubMed]
- 7.Hua C, Zhao G, Li Y, Bie L. Minichromosome maintenance (MCM) family as potential diagnostic and prognostic tumor markers for human gliomas. BMC Cancer. 2014;14:526. doi: 10.1186/1471-2407-14-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hyrien O (2016) How MCM loading and spreading specify eukaryotic DNA replication initiation sites. F1000Research 5 2063. 10.12688/f1000research.9008.1 [DOI] [PMC free article] [PubMed]
- 9.Boos D, Frigola J, Diffley JFX. Activation of the replicative DNA helicase: breaking up is hard to do. Curr Opin Cell Biol. 2012;24:423–430. doi: 10.1016/j.ceb.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 10.Donovan S, Harwood J, Drury LS, Diffley JFX. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. PNAS. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leman AR, Noguchi E. The replication fork: understanding the eukaryotic replication machinery and the challenges to genome duplication. Genes. 2013;4:1–32. doi: 10.3390/genes4010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petropoulou C, Kotantaki P, Karamitros D, Taraviras S. Cdt1 and Geminin in cancer: markers or triggers of malignant transformation. Front Biosci. 2008;13:4485–4494. doi: 10.2741/3018. [DOI] [PubMed] [Google Scholar]
- 13.Sen S, Chakraborty R, De B. Challenges and opportunities in the advancement of herbal medicine: India’s position and role in a global context. J Herb Med. 2011;1:67–75. doi: 10.1016/j.hermed.2011.11.001. [DOI] [Google Scholar]
- 14.Lakhera S, Devlal K, Ghosh A, Rana M (2021) In silico investigation of phytoconstituents of medicinal herb piper longum against SARS-CoV-2 by molecular docking and molecular dynamics analysis. Results Chem 100199. 10.1016/j.rechem.2021.100199 [DOI] [PMC free article] [PubMed]
- 15.Matulonis U, Sood A, Fallowfield L. Ovarian cancer. Nat Rev Dis Primers. 2016;2:16061. doi: 10.1038/nrdp.2016.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan Z, Wei B, Deng J, Mi Y, Dong Y, Zhu C, Fu R, Qu L, Fan D. The anti-tumor effect of ginsenoside Rh4 in MCF-7 breast cancer cells in vitro and in vivo. Biochem Biophys Res Commun. 2018;499:482–487. doi: 10.1016/j.bbrc.2018.03.174. [DOI] [PubMed] [Google Scholar]
- 17.Sadeghi S, Davoodvandi A, Pourhanifeh MH, Sharifi N, Nezhad RA, Sahebnasagh R, Moghadam SA, Sahebkar A, Mirzaei H. Anti-cancer effects of cinnamon: insights into its apoptosis effects. Eur J Med Chem. 2019;178:131–140. doi: 10.1016/j.ejmech.2019.05.067. [DOI] [PubMed] [Google Scholar]
- 18.Ambulkar S, Ambulkar P, Deshmukh MP, Budhrani AB (2020) Experimental evaluation of wound healing activity of various dosage forms of Tridax procumbens. Indian J Forensic Med Toxicol 14
- 19.Andriana Y, Xuan TD, Quy TN, Minh TN, Van TM, Viet TD. Antihyperuricemia, antioxidant, and antibacterial activities of Tridax procumbens L.". Foods. 2019;8:1–21. doi: 10.3390/foods8010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berlin Grace VM, Viswanathan S, David Wilson D et al (2020) Significant action of Tridax procumbens L. leaf extract on reducing the TNF-α and COX-2 gene expressions in induced inflammation site in Swiss albino mice. Inflammopharmacol 28:929–938. 10.1007/s10787-019-00634-0 [DOI] [PubMed]
- 21.Bhagat V, Kondawar M (2019) Antitubercular potential of dendrophthoe falcate (l.) And Tridax procumbens (l.) Plants extracts against h37rv stain of mycobacteria tuberculosis. Int J Pharm Sci Res 10:251–259. 10.13040/IJPSR.0975-8232.10(1).251-59
- 22.Ravikumar V, Shivashangari KS, Devaki T (2005) Hepatoprotective activity of Tridax procumbens against d-galactosamine/lipopolysaccharide-induced hepatitis in rats. J Ethnopharmacol 101:55–60. 10.1016/j.jep.2005.03.019 [DOI] [PubMed]
- 23.Bhagat VC, Kondawar MS. A comprehensive review on phytochemistry and pharmacological use of Tridax procumbens Linn. J pharmacogn phytochem. 2019;8:01–10. [Google Scholar]
- 24.Ikewuchi J. Effect of aqueous extract of Tridax procumbens Linn on plasma electrolytes of salt loaded rats, Pakistan. J Nutr. 2010;9:103–105. [Google Scholar]
- 25.Sujitha R, Sharmila R (2018) Phytochemical analysis and in vitro anticancer activity of Tridax procumbens linn. World J Pharm Res 7:867–878. 10.20959/wjpr201810-12319
- 26.Qiu Y, Wang W, Zhang B, Mei L, Shi Z. MCM7 amplification and overexpression promote cell proliferation, colony formation and migration in esophageal squamous cell carcinoma by activating the AKT1/mTOR signaling pathway. Oncol Rep. 2017;37:3590–3596. doi: 10.3892/or.2017.5614. [DOI] [PubMed] [Google Scholar]
- 27.Barik P, Talukdar P (2018) Established phytoligands from Tridax procumbens linn. Against bacterial dna-gyrase b receptor: molecular docking approach. World J Pharm Res 7. 10.20959/wjpr201812-12576
- 28.Imran M, Rauf A, Izneid TA, Nadeem M, Shariati MA, Khan IA, Imran A, Orhan IE, Rizwan M, Atif M, Gondal TA, Mubarak MS. Luteolin, a flavonoid, as an anticancer agent: a review. Biomed Pharmacother. 2019;112:108612. doi: 10.1016/j.biopha.2019.108612. [DOI] [PubMed] [Google Scholar]
- 29.Yu Q, Zhang M, Ying Q. Decrease of AIM2 mediated by luteolin contributes to non-small cell lung cancer treatment. Cell Death Dis. 2019;10:218. doi: 10.1038/s41419-019-1447-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Closas RG, Gonzalez CA, Agudo A, Riboli E. Intake of specific carotenoids and flavonoids and the risk of gastric cancer in Spain. Cancer Causes Control. 1999;10:71–75. doi: 10.1023/A:1008867108960. [DOI] [PubMed] [Google Scholar]
- 31.Rana M, Jain A, Rani V, Chowdhury P. Glutathione capped core/shell CdSeS/ZnS quantum dots as a medical imaging tool for cancer cells. Inorg Chem Commun. 2020;112:107723. doi: 10.1016/j.inoche.2019.107723. [DOI] [Google Scholar]
- 32.Fang J, Zhou Q, Shi XI, Jiang BH (2007) Luteolin inhibits insulin-like growth factor 1 receptor signaling in prostate cancer cells. Carcinogenesis 28:713–723. 10.1093/carcin/bgl189 [DOI] [PubMed]
- 33.Ganai SA, Sheikh FA, Baba ZA, Mir MA, Mantoo MA, Yatoo MA. Anticancer activity of the plant flavonoid luteolin against preclinical models of various cancers and insights on different signalling mechanisms modulated. Phytother Res. 2021;35:3509–3532. doi: 10.1002/ptr.7044. [DOI] [PubMed] [Google Scholar]
- 34.Horinaka M, Yoshida T, Shiraishi T, Nakata S, Wakada M, Nakanishi R, Nishino H, Sakai T. The combination of TRAIL and luteolin enhances apoptosis in human cervical cancer HeLa cells. Biochem Biophys Res Commun. 2005;333:833–838. doi: 10.1016/j.bbrc.2005.05.179. [DOI] [PubMed] [Google Scholar]
- 35.Byun S, Lee KW, Jung SK, Lee EJ, Hwang MK, Lim SH, Bode AM, Lee HJ, Dong Z (2010) Luteolin inhibits protein kinase Cε and c-Src activities and UVB-induced skin cancer. Cancer Res 70. 10.1158/0008-5472.CAN-09-4093 [DOI] [PubMed]
- 36.Khajuria R, Singh S, Bahl A (2019) General introduction and sources of flavonoids. In: Singh Tuli H. (eds) Current Aspects of Flavonoids: Their Role in Cancer Treatment, Springer, Singapore. 10.1007/978-981-13-5874-6_1
- 37.Wagh S, Shinde G. Protective effect of Tridax procumbens Linn against isoniazid induced hepatic damage. J Pharm Res. 2011;4:3612–3614. [Google Scholar]
- 38.Salahdeen HM, Yemitan OK, Alada ARA (2004) Effect of aqueous leaf extract of Tridax procumbens on blood pressure and heart rate in rats. Afr J Biomed Res 7. 10.4314/ajbr.v7i1.54062
- 39.Haytowitz DB, Bhagwat S, Holden JM. Sources of variability in the flavonoid content of foods. Procedia Food Sci. 2013;2:46–51. doi: 10.1016/j.profoo.2013.04.008. [DOI] [Google Scholar]
- 40.Lin Z, Fang Y, Huang A, Chen L, Guo S, Chen J (2014) Chemical constituents from Sedum aizoon and their hemostatic activity. Pharm Biol 52. 10.3109/13880209.2014.895019 [DOI] [PubMed]
- 41.Preveena J, Bhore SJ. Identification of bacterial endophytes associated with traditional medicinal plant Tridax procumbens Linn. Anc Sci Life. 2013;32:173–177. doi: 10.4103/0257-7941.123002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rebolledo GAG, Jonas SD, Arellanes MAJ. Natural compounds and extracts from Mexican medicinal plants with anti-leishmaniasis activity: an update, Asian Pacific. J Trop Med. 2017;10:1105–1110. doi: 10.1016/j.apjtm.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 43.Ghosh P, Biswas S, Biswas M, Dutta A, Sil A, Chatterjee S (2019) Morphological, ethno biological and phytopharmacological attributes of Tridax procumbens Linn. (Asteraceae): a review. Int J Sci Res Biol Sci 6:182–191. 10.26438/ijsrbs/v6i2.182191
- 44.Theoharides TC (2020) COVID-19, pulmonary mast cells, cytokine storms, and beneficial actions of luteolin. Biofactors 46:306–308. 10.1002/biof.1633 [DOI] [PMC free article] [PubMed]
- 45.Frisch MJ, Trucks GW, Schlegel HB et al (2009) Gaussian 09, Revision B. 01, Gaussian Inc., Wallingford CT 121:150–166
- 46.Becke AD (1993) Density‐functional thermochemistry. III. The role of exact exchange. J Chem Phys 98:5648. 10.1063/1.464913
- 47.Becke AD (1997) Density-functional thermochemistry. V. Systematic optimization of exchange-correlation functionals. J Chem Phys 107:8554–8560. 10.1063/1.475007
- 48.Dennington T, Keith J, Millam (2007) GaussView, Version 4.1.2, Semichem, Inc., Shawnee Mission KS
- 49.Yadav P, Rana M, Chowdhury P. DFT and MD simulation investigation of favipiravir as an emerging antiviral option against viral protease (3CLpro) of SARS-CoV-2. J Mol Struct. 2021;1246:131253. doi: 10.1016/j.molstruc.2021.131253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deb S, Reeves AA, Hopefl R, Bejusca R. ADME and pharmacokinetic properties of remdesivir: its drug interaction potential. Pharmaceuticals. 2021;14:655. doi: 10.3390/ph14070655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, Lee PW, Tang Y. A comprehensive source and free tool for assessment of chemical ADMET properties. ACS Publication Chem Inf Model. 2012;52:3099–3105. doi: 10.1016/j.molstruc.2017.09.108. [DOI] [PubMed] [Google Scholar]
- 52.Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 53.Schüttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 54.Lagunin A, Stepanchikova A, Filimonov D, Poroikov V. PASS: prediction of activity spectra for biologically active substances. Bioinformatics. 2000;16:747–748. doi: 10.1093/bioinformatics/16.8.747. [DOI] [PubMed] [Google Scholar]
- 55.Anzali S, Barnickel G, Cezanne B, Krug M, Filimonov D, Poroikov V. J Med Chem. 2001;44:2432–2437. doi: 10.1021/jm0010670. [DOI] [PubMed] [Google Scholar]
- 56.Chinnasamy P, Arumugam R. In silico prediction of anticarcinogenic bioactivities of traditional anti-inflammatory plants used by tribal healers in Sathyamangalam wildlife Sanctuary. India, EJBAS. 2018;5:265–279. doi: 10.1016/j.ejbas.2018.10.002. [DOI] [Google Scholar]
- 57.Thomas A, Peter P, Chandramohanakumar N. A profiling of anti-tumour potential of sterols in the mangrove fern Acrostichum aureum. Int J Pharmacogn Phytochem Res. 2016;8:1828–1832. [Google Scholar]
- 58.Nand M, Maiti P, Pant R, Kumari M, Chandra S, Pande V. Virtual screening of natural compounds as inhibitors of EGFR 696–1022 T790M associated with non-small cell lung cancer. Bioinformation. 2016;12:311–317. doi: 10.6026/97320630012311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Braga RC, Alves VM, Silva MF, Muratov E, Fourches D, Lião LM, Tropsha A, Andrade CH. Pred-hERG: A novel web- accessible computational tool for predicting cardiac toxicity. Mol Inform. 2015;34:698–701. doi: 10.1002/minf.201500040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee HM, Yu MS, Kazmi SR. Computational determination of hERG-related cardiotoxicity of drug candidates. BMC Bioinformatics. 2019;20:250. doi: 10.1186/s12859-019-2814-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hong JF, Baghdad O, Hwang JS, Uwe DH (2021) In silico assessment of human health risks caused by cyanotoxins from cyanobacteria. Biocell, Mendoza 45:65–77. 10.32604/biocell.2021.014154
- 62.Lagunin AA, Dubovskaja VI, Rudik AV, Pogodin PV, Druzhilovskiy DS, Gloriozova TA. CLC-Pred: A freely available web-service for in silico prediction of human cell line cytotoxicity for drug-like compounds. PLoS ONE. 2018;13:0191838. doi: 10.1371/journal.pone.0191838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Braga RC, Alves VM, Silva MFB, Muratov E, Fourches D, Liao LM, Tropsha A, Andrade CH. Pred-hERG: A novel web-accessible computational tool for predicting cardiac toxicity. Mol Inform. 2015;34:698–701. doi: 10.1002/minf.201500040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perez EA. Cardiac toxicity of ErbB2-targeted therapies: what do we know? Clin Breast Cancer. 2008;8:S114–S120. doi: 10.3816/CBC.2008.s.007. [DOI] [PubMed] [Google Scholar]
- 65.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Mainz DT. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein− ligand complexes. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 66.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bekker H, Berendsen HJC, Dijkstra EJ, Achterop S, van Drunen R, van der Spoel D, Sijbers A, Keegstra H (1993) Gromacs: a parallel computer for molecular dynamics simulations. Physics computing: Edited by R.A. de Groot and J. Nadrchal. World Sci Res 92:252–256
- 68.Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 69.Rzechorzek NJ, Hardwick SW, Jatikusumo VA, Chirgadze DY, Pellegrini L (2020) CryoEM structures of human CMG–ATPγS–DNA and CMG–AND-1 complexes. Nucleic Acids Res 48:6980–6995. 10.1093/nar/gkaa429 [DOI] [PMC free article] [PubMed]
- 70.Rana M, Chowdhury P. Nonlinear optical responses of organic based indole derivative: an experimental and computational study. Mater Today: Proc. 2020;28:241–245. doi: 10.1016/j.matpr.2020.01.598. [DOI] [Google Scholar]
- 71.Koopmans T. About the assignment of wave functions and eigenvalues for the individual electrons in an atom. Physica. 1934;1:104–113. doi: 10.1016/S0031-8914(34)90011-2. [DOI] [Google Scholar]
- 72.Rana M, Pooja P (2019) Chowdhury, Investigation on nonlinear optical responses of different pyrrole derivatives: a computational study. AIP Conf Proc 2136:040005. 10.1063/1.5120919
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Protein structure: https://www.rcsb.org/. Ligand structure: https://pubchem.ncbi.nlm.nih.gov/. ADMET properties: http://www.swissadme.ch/. Molecular docking: https://swissmodel.expasy.org/. Docking analysis: https://disCover.3ds.com/discovery-studio-visualizer-download. Ligand topology: http://davapc1.bioch.dundee.ac.uk/cgi-bin/prodrg.