Abstract
The extent of tissue regeneration varies widely between species. Mammals have a limited regenerative capacity whilst lower vertebrates such as the zebrafish (Danio rerio), a freshwater teleost, can robustly regenerate a range of tissues, including the spinal cord, heart, and fin. The molecular and cellular basis of this altered response is one of intense investigation. In this review, we summarise the current understanding of the association between zebrafish regeneration and Hippo pathway function, a phosphorylation cascade that regulates cell proliferation, mechanotransduction, stem cell fate, and tumorigenesis, amongst others. We also compare this function to Hippo pathway activity in the regenerative response of other species. We find that the Hippo pathway effectors Yap/Taz facilitate zebrafish regeneration and that this appears to be latent in mammals, suggesting that therapeutically promoting precise and temporal YAP/TAZ signalling in humans may enhance regeneration and hence reduce morbidity.
Subject terms: Regeneration, Cell signalling, Stem-cell differentiation
Introduction
Many different organisms have the ability to regenerate, although the robustness, efficiency, and scope of this regeneration is varied. Invertebrates such as planarians and Hydra regenerate their entire body such that, when cut in half, each section forms an entire new organism1–4. At the other end of the scale, mammalian regeneration is limited, with adult animals often responding to injury with fibrotic scarring rather than regeneration5,6. Some mammalian tissues do regenerate, including the skin, intestine, liver, peripheral nervous system, and blood7–11, as well as foetal tissues12 but this capability is impaired in ageing systems13,14, which, along with a general lack of regenerative ability in most tissues, causes high morbidity in humans.
Midway on the scale from complete (invertebrate) to limited (mammalian) regeneration are lower vertebrates, including amphibians and fish. The zebrafish Danio rerio has the potential to completely regenerate multiple adult and embryonic organs, including the heart, fin, and many nervous system components15–20. First explored in the 1980s by Streisinger21,22, the zebrafish is regularly utilised in the study of adult and embryonic regeneration due to their rapid external development, relative low cost, ease of genetic manipulation, scalability, transparent juveniles, and high rate of regeneration, none of which are present in the mouse.
The cellular and molecular drivers of zebrafish regeneration have been the subject of intense research5,19. Effective replacement of lost or damaged cells requires a large pool of available healthy cells. Cell pools can be formed by multiple sources, including the activation of resident stem or progenitor cells (differentiation), the reversion of differentiated cells to a more immature pluripotent state (dedifferentiation), or the conversion of one differentiated cell type into another mature cell type (transdifferentiation)5,23. On a molecular level, these can be driven by epigenetic and gene expression changes, such as alterations in DNA methylation5,23–25, histone modifications5,26–29, regeneration-responsive enhancers28,30–33, and the activation of a range of key developmental signalling pathways, including Bmp, Fgf, Notch, RA, Shh, and Wnt/β-catenin (summarised in Table 1)34–77. In recent years, it has become evident that Hippo signalling (Fig. 1) plays a critical role in developmental and regenerative processes in both zebrafish and mammals. This is associated with the Hippo pathway’s role in regulating cell proliferation and migration, detecting and responding to changes in tissue tension, extracellular matrix, chemical cues, which consequently alter cell fates78–82.
Table 1.
A summary of major non-Hippo signalling pathways involved in zebrafish regeneration.
| Signalling pathway | Model | Role of pathway |
|---|---|---|
| BMP | Heart34 | Promotes CM proliferation and dedifferentiation |
| Tail Fin239–241,35,36 | Enhances proliferation and differentiation of osteoblasts in the blastema | |
| Calcineurin | Tail Fin37 | Regulates regeneration rate for positional information |
| Fgf | Spinal Cord188,38 | Increases glial bridge formation, neuronal proliferation, and neurite outgrowth |
| Tail Fin229,39–41 |
Promotes blastema formation and regenerative outgrowth Regulates regenerative growth rate |
|
| Lateral Line42 | Promotes support cell differentiation | |
| Igf | Heart43 | Enhances CM proliferation |
| Tail Fin44 | Promotes blastema cell proliferation and basal epithelium maintenance | |
| Jak/Stat3 | Heart45 | Promotes CM proliferation |
| Lateral Line46 | Increases progenitor cell proliferation and differentiation | |
| Liver47 |
Necessary for appropriate timing of progenitor cell-to-hepatocyte differentiation Establishes the correct number of biliary epithelial cells during regeneration |
|
| NF-κB | Heart48 | Promotes CM proliferation and dedifferentiation |
| Notch | Heart49 | Enhances CM proliferation |
| Spinal Cord50 | Inhibits motor neuron neurogenesis | |
| Tail Fin51,52 | Maintains blastema cells in a proliferative undifferentiated state | |
| Lateral Line274,53,54 | Reduces support cell proliferation | |
| Liver55–57 | Enhances biliary cell to hepatocyte conversion and differentiation of progenitor cells to biliary epithelial cells | |
| Nrg | Heart58 | Promotes CM proliferation |
| RA | Heart59,60 | Enhances CM proliferation and wound epithelium formation |
| Tail Fin59,61–63 |
Increases blastema and basal epidermis formation and patterning during regenerative outgrowth Restricts osteoprogenitor cells to boy ray regions |
|
| ROS | Heart64 | Recruits immune cells and primes heart for regeneration |
| Tail Fin65 | Promotes proliferation of stump epidermal cells | |
| Shh | Heart43 | Increases CM proliferation |
| Spinal Cord66,67 | Activates motor neuron neurogenesis | |
| Tail Fin35 | Promotes proliferation and differentiation of osteoblasts in the blastema | |
| Tgfβ | Tail Fin41,68 | Enhances cell migration and blastemal proliferation during outgrowth |
| Heart43,69,70 | Promotes CM proliferation and transient scar formation | |
| Wnt/β-catenin | Spinal Cord185,190 | Increases glial progenitor differentiation into neurons, axonal regrowth, and deposition of pro-regenerative collagen |
| Tail Fin212,239,71–73 | Enhances blastemal cell proliferation and osteoblast dedifferentiation | |
| Lateral Line279,53,74 | Promotes support cell dedifferentiation and proliferation, and hair cell formation | |
| Liver55,75–77 | Increases differentiation of biliary-derived progenitor cells into hepatocytes |
Fig. 1. Summary of the Hippo pathway signalling cascade and its stimuli.
The Hippo pathway is regulated by the integration of a range of upstream stimuli. This includes mechanotransductive elements (such as caveolae and Piezo signalling), metabolism, extracellular matrix and integrin signalling, transduction of extracellular stimuli via mitogenic growth factor signalling and GPCRs, cell polarity and cell–cell contacts. Activation of the Hippo pathway triggers a phosphorylation cascade that leads to the phosphorylation of the Hippo pathway effectors YAP/TAZ. Phosphorylation of YAP/TAZ redistributes YAP/TAZ to the cytoplasm, blocking TEAD-mediated gene expression. Hippo pathway inactivation prevents YAP/TAZ phosphorylation, allowing their nuclear translocation and hence TEAD-mediated gene expression. Note that MST1/2 (mammalian STE20-like kinase1/2) are encoded by STK4/3, and TAZ by WWTR1. Figure 1 is created in BioRender.com.
The core Hippo signalling pathway is comprised of a serine/threonine kinase phosphorylation cascade (Fig. 1), most of which were identified in genetic screens of Drosophila melanogaster for tumour suppressor genes83,84. Activity of this pathway is regulated by a range of stimuli, including mechanical signalling, cell shape, ECM stiffness, cell polarity, metabolism, and cell:cell contacts78,79,82,85–90, which are integrated to stimulate key kinases MST1/2 (the fly Hippo orthologs), STK25, and MAP4Ks when the Hippo pathway is active87,89,91–93. These kinases then phosphorylate, and so activate, LATS1/2, which phosphorylate the core Hippo effectors transcriptional co-activator YAP1 and its paralog TAZ on multiple conserved serine residues86,87,91,92,94,95. YAP1/TAZ phosphorylation triggers their retention in the cytoplasm via binding to protein 14-3-3, or ubiquitin-mediated degradation86,87,94–97. When the Hippo pathway is inactive these phosphorylations do not occur, resulting in YAP1/TAZ nuclear localisation, where they outcompete VGLL4 and bind to transcription factors TEAD1-487,98–101. Binding to TEADs stimulate the expression of a range of pro-proliferative, -oncogenic, -stemness, and -EMT genes, such as CTGF and CYR6178,87,90,98–100,102–104. Additional YAP1/TAZ transcription factors have also been identified87, but the most extensively studied are the TEADs. Zebrafish Hippo pathway genes have high genetic orthology to human genes, suggesting that this is an appropriate model in which to study Hippo pathway function (Fig. 2). Here we review the role of the Hippo pathway in the regeneration of a range of organs, including heart, spinal cord, tail fin, lateral line, and liver regeneration, with a focus on the zebrafish.
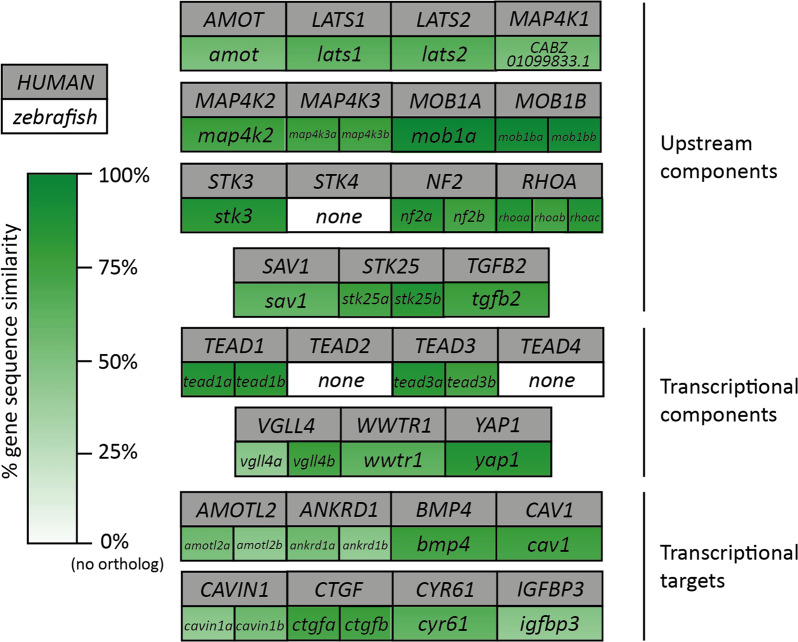
Fig. 2. Similarity between selected human and zebrafish Hippo pathway genes.
Direct gene sequence comparison between a sample of human and zebrafish Hippo pathway members and transcriptional targets shows a range of similarity scores, emphasizing a high degree of similarities between fish and human genes, while also highlighting that some Hippo pathway components appear to have no direct orthologs present in both species. WWTR1 encodes TAZ. STK4 encodes MST1 and STK3 encodes MST2 (in accordance with the consensus of the Hippo pathway field). CYR61 is also known as CCN1 and CTGF as CCN2. % gene sequence similarity identified using ensembl.org under orthology tab. ctgfb, nf2b, map4k2, and rhoaa-c could not be identified as orthologues in this manner, so manual BLAST comparison of genomic sequence (from GRCz11) was performed to give the values indicated.
Heart regeneration
Cardiovascular diseases are the primary cause of morbidity and mortality globally, with around half of these deaths caused by ischaemic heart disease leading to heart failure105. This is due to the limited regeneration capacity of the adult human heart, which responds to heart muscle damage with fibrosis and scarring rather than the reformation of contractile muscle106. A similar response is seen in other mammals (such as the mouse), which also show limited cardiac regeneration after experimental injury paradigms107. An exception is an enhanced heart regeneration potential in neonatal mice, but this is transient and is lost within the first week of life108, coinciding with a decrease in YAP1 transcriptional activity78, and the withdrawal of cardiomyocytes (CMs) from the cell cycle109. However, this regenerative ability in neonatal mice108 highlights that there may be therapeutic potential in reactivating the regenerative capacity in humans.
In contrast to restricted mammalian regeneration, both adult and embryonic zebrafish regenerate their heart fully following injury and even after multiple insults5,6,15,19,110–113 (Fig. 3). This extensive heart regeneration is the result of two key characteristics: a high level of existing CM proliferation (around 3% per week, compared to <1% per year in adult mice114 and humans115), and a permissive extracellular environment that stimulates it19,116. One major hurdle and pathological driver in mammalian heart regeneration is the formation of a fibrotic scar and non-permissive ECM at the injury area, replacing dead CMs with non-contractile elements such as collagen or fibroblasts rather than new CMs106,117 (Fig. 3B). However, in the zebrafish, although collagen and fibronectin does accumulate and a scar is formed, it is eliminated to allow effective regeneration113,118–120. This scarring is regulated by Hippo signalling, with cav-1, yap1, and ctgfa mutants having disrupted scar formation and hence regeneration121–123 (see Table 2 for a summary of these phenotypes). Heart injury promotes Ctgfa secretion into the ECM from endocardial cells, where it promotes the expression of pro-regenerative ECM genes (such as fibronectins and collagens)121. This expression allows for a transient scar, as shown by ctgfa mutants having a larger and more persistent scar, whilst ctgfa overexpression speeds scar resolution121. Similarly, yap1 mutants have an altered ECM composition at the injury site, resulting in increased scarring and impaired regeneration at early time points122. This alteration of the scar microenvironment by secretion of Hippo pathway transcriptional targets suggests that Hippo signalling may also indirectly regulate the infiltration and proliferation of CMs through a cell non-autonomous mechanism.
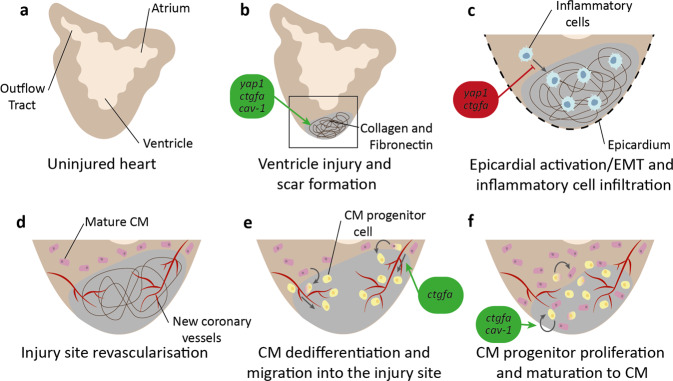
Fig. 3. Overview of zebrafish heart regeneration.
a Structure of the uninjured zebrafish adult heart. b Injury at the ventricle apex induces collagen and fibronectin deposition and scar formation. yap1, ctgfa, and cav-1 promote appropriate and transient scar formation. c Heart epicardium undergoes EMT and inflammatory cells (blue) infiltrate into the scar. yap1 and ctgfa inhibit inflammatory cell infiltration. d New coronary vessels form to revascularize the injury site. e Mature cardiomyocytes (CMs) (pink) dedifferentiate into progenitor cells (yellow) and migrate along the new coronary vessels into the injury site. ctgfa promotes CM migration. f CM progenitors proliferate to create a progenitor cell pool, which matures back to CMs to reform the heart muscle. ctgfa and cav-1 promote cell proliferation.
Table 2.
Overview of zebrafish phenotypes seen when Hippo pathway components are disrupted.
| Gene | Activity level | Disruption method | Allele created | Model | Phenotype |
|---|---|---|---|---|---|
| amotl2a | − | MO/TALEN | N/A/fu45, fu46 | LL development261 |
Overproliferation in trailing edge of pLLP Increased pLLP size and cell number Reduced pLLP migration speed Increased number of neuromasts |
| cav-1α | − | TALEN | pd1094, pd1104 | Heart regeneration123 | Impaired recovery after injury, injury-induced CM proliferation, and scar resolution |
| MO | N/A | LL development136 | Reduced number and maturation of hair cells and neuromasts | ||
| ctgfa | − | TALEN | bns50 | Heart regeneration121 |
Reduced CM proliferation, expression of pro-regenerative ECM genes, and CM migration along the coronary vasculature to repopulate the wound Increased collagenous scarring |
| SC regeneration197 | Reduced functional recovery after injury, glial cell proliferation and bridging, and axon regeneration | ||||
| + | hsp70:ctgfa OE plasmid | pd97 | Heart regeneration121 | Increased recovery after injury, CM proliferation, resolution of collagen deposition, and expression of pro-regenerative ECM genes | |
| SC regeneration197 | Enhanced functional recovery after injury, glial bridging, and axon growth | ||||
| CRISPR | zf3090 | Tail fin regeneration234 | Increased tissue stiffness, contractility, and ECM deposition | ||
| lats2 | − | CRISPR | mw87 | Cancer311 |
Increased lethality Formation of peripheral nerve sheath tumours by 3mpf |
| nf2a | − | MO | N/A | Liver development299 | Hepatomegaly, dilated bile duct, and extrahepatic choledochal cysts |
| sav1 | − | CRISPR | mw95 | Liver development301 | Biliary dysgenesis, altered hepatocyte morphology and polarity, and biliary cell dysplastic morphology and increased expansion |
| stk3 | − | TALEN | mw96 | Liver development301 | Biliary dysgenesis, altered hepatocyte morphology and polarity, and biliary cell dysplastic morphology and increased expansion |
| wwtr1 | − | MO | N/A | Tail fin regeneration149 | Lack of skeletal ossification |
| yap1 | − | TALEN | mw48 | Heart regeneration122 |
Improper scar formation Reduced ability to secrete collagen at the injury site Increased macrophage infiltration in the scar, monocyte chemotactic gene expression, space between the epicardium and myocardium, and CM proliferation |
| LL regeneration277 | Reduced progenitor cell maturation and proliferation | ||||
| Liver development147 | Reduced liver size | ||||
| hsp70:DN-Yap plasmid | zf621 | SC regeneration193 | Impaired functional recovery after injury, axon growth, and glial bridging | ||
| Tail fin regeneration232,233 |
Reduced recovery after injury, cell proliferation, and osteoprogenitor differentiation into osteoblasts Defects in bone formation |
||||
| 5 µM verteporfin | N/A | LL regeneration277 | Defective supporting cell, hair cell and mantle cell proliferation and hair cell maturation | ||
| LL development259–262 | Reduced number of neuromasts and hair cells, pLLP size, number of cells in the pLLP, mechanoreceptor differentiation, and Wnt signalling component expression | ||||
| + | Tol2 (myl7:3SA-myc yap1) | N/A | Heart regeneration122 | Increased CM proliferation | |
| hsp70:CA-Yap plasmid | zf622 | Tail fin regeneration232 | Impaired recovery after injury, increased cell proliferation | ||
| CA-Yap1 mRNA injection (in lpar2b MO) | N/A | LL development262 | Increased pLLP size, and number of neuromasts and proliferating cells in the pLLP | ||
| I-SceI (lf:Yap1) | N/A | Liver development300 | Hepatomegaly | ||
| yap1;wwtr1 | − | CRISPR | N/A | SC regeneration193 | Impaired functional recovery after injury |
This table is non-exhaustive and primarily covers developmental and regenerative phenotypes described in this review. Many other Hippo pathway mutants and morphants exist (e.g. wwtr1 alleles bns35, swu46, swu47, va4, mw49, ncv114, and fu55). See individual gene pages on zfin.org for a complete list.
The immune system creates a permissive microenvironment for regeneration (Fig. 3C). This is clearly demonstrated in the medaka, a teleost species closely related to the zebrafish. The medaka displays limited heart regeneration, a finding that is surprising considering their evolutionary similarity to the zebrafish124,125. This limited regeneration is, at least in part, due to the medaka’s delay in macrophage recruitment to the injury site124. When recapitulated in the zebrafish by clodronate liposome-mediated macrophage depletion, these macrophage defects cause compromised neovascularisation and CM proliferation and consequently severe defects in heart regeneration124. This is due to the role of macrophages and other immune cell components such as Treg cells in many areas of cardiac regeneration, including enhancing neovascularisation, CM proliferation, and scar resolution via the production of pro-regenerative factors, with inhibition of inflammation and timely immune cell recruitment inhibiting regeneration124,126–129.
However, this pro-regenerative effect of the immune system is not simple. Yap1-Ctgfa signalling, shown to enhance cardiac regeneration, also negatively regulates the migration and infiltration of macrophages into the injury site121,122, suggesting that inhibiting macrophage infiltration promotes cardiac regeneration. Similarly, yap1 KO fish have increased macrophage infiltration in the scar and increased monocyte chemotactic gene expression122, and ctgfa KO promotes the chemokine receptor gene cxcr3.1 in the heart to increase M1 macrophage polarisation and so enhance inflammatory signalling121, and both KO lines have defective regeneration. This apparent contradiction may be due to differences between experimental paradigms in investigating immune cell function in regeneration—it has been shown that the type of immune cells recruited, and the different regenerative stages alter the functional role of the immune system in regeneration128. An alternative explanation for this apparent discrepancy could be due to the requirement for tight spatio-temporal control of the immune system function during regeneration. This is shown by disruption of reparative regeneration after both immune system hyperactivation121,122 and excessive inhibition124,126,127,129. Another potential reason for the inconsistency is that various immune cell types likely react differently to the injury, and so the Hippo pathway may respond in a range of ways to the same trigger. Therefore, the extent of activation or inhibition in these studies will greatly impact the results. Further in-depth studies are needed in order to fully elucidate the detailed spatiotemporal inflammatory response including revealing the exact immune cell types involved in regeneration and thereby the role of the Hippo pathway in the immune system’s contribution to cardiac regeneration.
Hippo pathway signalling has also been linked to the epicardium, which is activated after heart injury in the zebrafish (Fig. 3C)15,130. The epicardium promotes regeneration, potentially by functioning as a cellular scaffold that generates epicardial-derived cells which differentiate into myofibroblasts and perivascular fibroblasts in the injured myocardium131. This may then act in a paracrine manner to induce CM proliferation and neoangiogenesis131. Epicardial activation has not yet been linked to the Hippo pathway in zebrafish heart regeneration. However, in the developing mouse, Hippo components are expressed in both the proepicardium and epicardium, and deletion of either Yap or Taz in the mouse gives coronary defects and impacts on epicardial cell proliferation, EMT, and specification of cell fate132. Similar developmental cardiac defects can be seen in a range of Hippo pathway component mutants in the zebrafish112,133–152, suggesting that this role of the Hippo pathway may be conserved between mammals and teleosts.
After injury, existing differentiated CMs undergo limited dedifferentiation, upregulate the embryonic cardiogenesis gene gata4, and proliferate113,153–156. These CMs migrate to the injury site along newly-formed coronary vasculature157–162 (Fig. 3E), where they proliferate further and differentiate to replace dead CMs and form new functional heart muscle163 (Fig. 3F). CM proliferation is promoted by a range of signalling pathways, including Nrg, Tgfβ, Igf, and the Hippo pathway (Table 1). Disruption of the Hippo pathway-regulated genes cav-1α and ctgfa inhibits CM proliferation and repopulation of the injury area121,123. Similarly, TGFβ-mediated activation of regulatory elements upstream of ctgfa promotes CM proliferation at the injury site32. cav-1α and ctgfa are induced after injury in epicardial and endocardial cells respectively, suggesting a role for these cells in cell non-autonomous regulation of CM function, such as in ECM secretion in response to extracellular stimuli121,123. Disruption of cav-1α and ctgfa results in defective heart regeneration121,123, whilst overexpression of ctgfa and yap1 has the opposite effect121,122. Disrupting Hippo signalling in mammals gives comparable results. In pigs, CM-specific knockdown of Sav (which results in increased YAP activity164,165) increases CM proliferation and improves heart function after myocardial infarction165. Similar outcomes are observed when Yap1 is disrupted in mice, causing heart regeneration defects through decreased CM proliferation166–171, whilst heart regeneration (and CM proliferation) is stimulated after Yap1 activation167–169, potentially due to the Hippo pathway’s link to cytoskeletal and ECM regulation170. However, the opposite effect is observed when Hippo signalling is disrupted in murine cardiac fibroblasts172,173. Deletion of Yap1/Taz in these fibroblasts results in improved cardiac function after myocardial infarction through modulation of the fibrotic and fibroinflammatory response172. Enhanced Yap1/Taz signalling (through either Yap1 overexpression or Lats1/2 deletion) has the opposing effect, with mice displaying elevated fibrotic responses173,172. This apparent contradiction between the role of the Hippo pathway in CMs and cardiac fibroblasts supports a model where the Hippo pathway functions differently in different cell types.
CMs in zebrafish ctgfa mutants also fail to migrate along the coronary vasculature to infiltrate the wound, despite no changes in revascularisation, potentially as a result of alterations in cytoskeletal gene expression in a cell autonomous regulation of CM infiltration121,123. Supporting this, data using in vitro primary rat cultures of cardiac fibroblasts show that Yap1 siRNA-mediated knockdown reduces expression of factors associated with cytoskeletal motility and ECM adhesion, although these results have not been recapitulated in zebrafish, CMs, or in vivo122.
In summary, the Hippo signalling pathway enhances cardiac regeneration by temporal activation of Yap1/Taz and promotes normal cardiovascular development. Yap1/Taz promote appropriate scar formation and potentially prevent overactivation of the immune response, which, when combined, increase scar resolution, spatiotemporal CM proliferation, and thereby cardiac regeneration. Taking advantage of this regenerative capacity may hold therapeutic potential in the treatment of human MI. For example, pharmacological regulation of the Hippo pathway could modulate CM proliferation and fate plasticity156,174, promoting scarless healing in the adult heart and reducing disease burden. Recent work disrupting Hippo signalling in pigs after myocardial infarction165 suggests, in a clinically relevant model system, that this could be possible. However, precise cell type-specific modulation of the Hippo pathway will be vital to realise its full potential, as the Hippo pathway has been shown to have different functions in the cell types involved. For example, heart function is improved after injury in mammals when YAP activity is increased in CMs165 but also when Yap1/Taz is deleted in cardiac fibroblasts172.
Spinal cord regeneration
The Hippo pathway is also associated with regeneration after spinal cord injury (SCI) in the zebrafish. After SCI in humans and other mammals, the affected axons and neurons are destroyed and a non-permissive scar is formed in the place of new cells, commonly resulting in lifelong disability175–177. However, both adult and larval zebrafish robustly and effectively regenerate their spinal cords after injury, with viable axon regrowth over the lesion site and return of full swimming function within weeks after injury18,19,178–182 (Fig. 4).
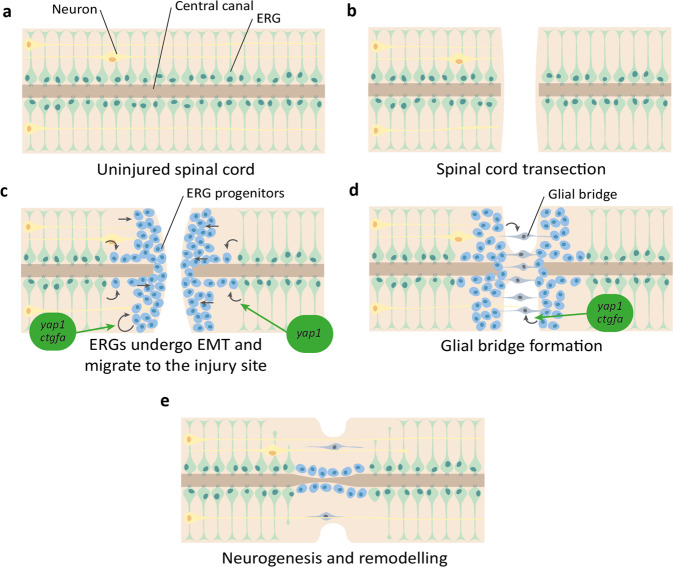
Fig. 4. Overview of zebrafish spinal cord regeneration.
a Structure of the uninjured spinal cord, with ependymal radial glia (ERG) (green) lining the central canal and motor neurons (yellow). b Spinal cord transection disrupts neuronal processes. c ERGs undergo EMT to form ERG progenitors (blue) and migrate to the site of injury. yap1 promotes EMT of ERGs, and yap1 and ctgfa promote progenitor proliferation. d ERG progenitors extend processes across the injury site to form a glial bridge (grey). yap1 and ctgfa promote the formation of the glial bridge. e Neuronal processes extend across the injury site, guided by the glial bridge to promote remodelling and reformation of the spinal cord.
For functional recovery in the spinal cord, new and existing cells must proliferate, migrate to the injury site, bridge the lesion, and differentiate to reintegrate with existing distal neuronal circuitry183. Neurogenesis from tissue-resident progenitors is a vital step for this to occur in zebrafish, which is promoted by multiple signalling pathways, including Wnt/β-catenin, Fgf, Shh, and is inhibited by Notch signalling (Table 1). The tissue-resident progenitors responsible for cell proliferation and bridging are thought to be the ventral ependymal radial glia (ERG)181,184–186. These cells have general functions during development and adulthood in maintaining spinal cord homoeostasis such as sealing the blood-brain barrier and maintaining ion balance, but also proliferate and differentiate into a range of neuronal cell types after injury181,183,187.
To allow new cell processes to traverse the lesion site, a glial bridge is formed. After injury, ERGs migrate to the lesion and elongate to form an astroglial bridge over the lesion, along which axons can grow to innervate distal targets (Fig. 4C–E). This is driven by pro-regenerative gene expression (e.g. col12a1a/b and tenascin-c), interactions with other cell types such as Schwann cells, and additional environmental cues180,182,184,188–190. Zebrafish glial bridging shares clear morphological and functional similarities with the bridging observed during mammalian peripheral nerve regeneration (which occurs to a much greater extent than mammalian CNS regeneration)183,191–193, indicating that this common process may be manipulated in the human for therapeutic benefit.
In order to induce glial cells to undergo bridging, ventral ERGs undergo an epithelial-to-mesenchymal transition (EMT)193 (Fig. 4C). EMT is a common feature of many cells activated by injury, and is linked to stem cell activation, increased cellular plasticity, and tissue remodelling194–196. Glial EMT is both necessary and sufficient to induce glial bridging, and is linked to Yap1-Ctgfa signalling193,197. yap1, wwtr1 (gene encoding Taz), and ctgfa are upregulated following SCI, with yap1 and ctgfa expression localised to bridging glia and ventral ERGs193,197. As well as inducing ctgfa expression in ventral ERGs, Yap1 promotes twist1a expression193. twist1a is an established EMT marker, activation of which directs a mesenchymal transition in Ctgfa+ ERGs, promoting glial bridging and functional spinal cord repair193.
Similar to heart regeneration, one major difference between the zebrafish and mammalian response to SCI is the formation of a glial scar. SCI causes vascular damage, oedema, and inflammation, resulting in widespread gliosis, necrosis, and apoptosis that eventually forms a glial/fibrotic scar in mammals, stretching beyond the site of the initial trauma and acting to prevent secondary damage but also preventing axon regrowth176,198,199. There is no significant scarring in the zebrafish, so there is no experimental work linking the Hippo pathway in zebrafish to scar resolution, however siRNA-mediated knockdown of the YAP1/TAZ-TEAD target gene Ctgf in rats reduces the glial scar and hence improves regeneration after SCI200, suggesting that YAP1/TAZ signalling may promote scar formation or impair scar resolution and outlining a potential therapeutic target for SCI treatment in mammals.
Loss of function mutations of yap1, wwtr1, and ctgfa all result in impaired functional recovery after SCI, with ctgfa and yap1 disruption causing a glia-specific cell proliferation reduction, resulting in impaired bridging and axon regeneration across the lesion site193,197. Exogenous administration of human CTGF to these ctgfa mutants reversed this defect197. This finding, and the similar finding that heart scar formation is larger and more persistent in ctgfa, yap1, and cav-1 mutants121–123 appears in contrast to that seen in the rat glial scar200, which found that knockdown of CTGF increased recovery through the clearance of scarring, and the current clinical trials which are targeting CTGF to reduce fibrosis and scarring201. This may be due to species differences in the function of the Hippo pathway, but this is not supported by the relatively high translatability of other studies between rodent and zebrafish. An alternative explanation might be that Yap/Taz-Ctgfa signalling has opposing effects at different stages of spinal cord regeneration, or that strictly regulated temporal activation/repression of signalling is key, although studies of this in mammals must be performed after the scar has been resolved, which currently presents an experimental challenge.
Yap1 signalling is also associated with the regenerative role of glial cells in other parts of the CNS, such as the retina. In the zebrafish, retinal damage induces reprogramming events where Müller glia are converted to a highly proliferative progenitor-like state, dividing asymmetrically to replace lost photoreceptors202–207. Yap1 knockdown blocks Müller glial cell proliferation and neurogenesis after light damage of the zebrafish retina208, suggesting a common role for yap1 in the regenerative functions of glial cells. Mammalian retinas usually do not have a proliferative, pro-regenerative, Müller glia response to injury. However, in the mouse, YAP promotes glial reprogramming, with YAP activation inducing Müller glia reprogramming to a highly proliferative, progenitor-like cell202,204. This suggests that promoting Yap1 signalling therapeutically may also promote CNS regeneration in humans.
These findings propose a model in which Yap1 senses the mechanical stress caused by SCI, enhancing ctgfa and twist1a expression to activate a pro-EMT and pro-proliferative transcriptional programme in ventral ERGs, promoting glial bridging, axon regeneration, and, consequently, functional recovery193. This model suggests that enhancing scar resolution, promoting EMT, enhancing CTGF signalling at later stages of regeneration, and identifying CTGF-responsive spinal cord cells may allow for the identification of a therapeutic target to promote mammalian spinal cord regeneration197. Targeting CTGF has been investigated in a variety of preclinical and clinical trials for multiple conditions, including muscular dystrophy and pancreatic cancer. For example, the monoclonal antibody Pamrevlumab has shown promise in trials for idiopathic pulmonary fibrosis209,210. However, these trials involve the inhibition of CTGF activity, rather than the enhancement that may be required to promote recovery201,211. Consequently, further insights must be obtained before translating these findings into an effective treatment option in humans.
Tail fin regeneration
Zebrafish and other teleosts regenerate their fins completely after multiple consecutive amputations212, a phenomenon that was studied as early as the 18th century213, and by the regeneration pioneer T. H. Morgan at the turn of the 20th century214–216. Fin regeneration occurs through epimorphic regeneration, a process characterised by the presence of a blastema early in regeneration (Fig. 5). This mass of undifferentiated proliferating progenitor cells at the site of injury is formed by mature cell dedifferentiation, which can then differentiate back into mature cells to generate an actively growing tissue that replaces the lost appendage217.
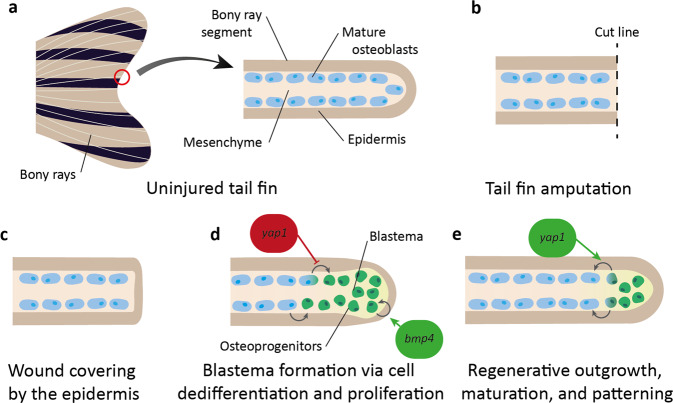
Fig. 5. Overview of zebrafish tail fin regeneration (adult), focussing on osteoblast regeneration of bony rays.
a The uninjured tail fin of the adult zebrafish is formed of many bony rays, which each consist of epidermis surrounding mature osteoblasts (purple) in the mesenchyme. b Amputation of the tail fin disrupts the bony ray segment. c In the initial stages of tail fin regeneration the epidermis covers the wound. d Osteoblasts and other mature cells dedifferentiate and proliferate at the wound tip to form a blastema with osteoprogenitors (green). yap1 inhibits osteoblast dedifferentiation and bmp4 enhances blastema cell proliferation. e The bony ray segment extends through maturation of the progenitor cells back to their original cell type. yap1 promotes osteoprogenitor maturation.
There is not yet direct evidence for a role for the Hippo pathway in dedifferentiation in the zebrafish caudal fin blastema, but in other in vivo models, both mammalian and invertebrate, the Hippo pathway maintain stemness, promote proliferation, and revert differentiated cells to a progenitor cell state81,218–224. In the zebrafish, dedifferentiated cells proliferate to form a large pool of progenitor cells in the blastema (Fig. 5D).
Blastema formation is enhanced and maintained by a range of developmental signalling pathways, including Hippo, Wnt/β-catenin, Igf, Notch, Fgf, Shh, Tgfβ, (Table 1) as well as inflammatory signals such as Il1β and Hsp90α225–227. The concentration gradient of these signalling pathways gives positional information along the proximodistal axis of the injured tissue, ensuring that structures are reformed at the correct location and that the tissue grows at an appropriate rate, halting when the previous size and shape is reached16,228–231. Hippo signalling is one such signalling pathway with activity changes in proximodistal expression. In the high cell density distal blastema, Yap1 is mainly cytoplasmic (and so inactive), whilst in the low density proximal blastema, it becomes mainly nuclear (active)232. Yap1 is also localised to α-catenin and F-actin when in the cytoplasm232. This suggests that the heterogeneous cell densities within the blastema could be transduced through cell junctions and the cytoskeleton232. These mechanical properties then impact Yap1 localisation, which alters the regenerative capacity of the fin232. For example, yap1 disruption impairs cell proliferation and alters key signalling pathways, including promoting Wnt and reducing Bmp signalling after fin injury232,233. This results in an accumulation of osteoprogenitors and prevention of osteoblast differentiation, and so defective regeneration233. Ctgfa levels are also increased following fin injury, and disruption of its regulatory sequences induces increased tissue stiffness and ECM deposition234.
Tail fin progenitor cells are not multipotent. Instead, cells remain lineage restricted235,236. The osteoblast is one such cell type. After injury, these cells dedifferentiate, proliferate, and mature to only give rise to osteoblasts in the regenerate (Fig. 5)236–238. More specifically, injury induces differentiated mature osteoblasts close to the injury site, which usually form the bony rays of the fin, to lose expression of late and intermediate osteoblast differentiation markers (such as osteocalcin and osterix) and undergo a Wnt/β-catenin-mediated EMT to gain progenitor markers and generate osteoprogenitor cells, which migrate to the blastema and proliferate in a Fgf-dependent manner237,239. These progenitors then undergo Bmp-mediated maturation into osteoblasts239 (Fig. 5D), a process that is associated with the Hippo pathway233,239. This link to osteoblast formation and function is most dramatically illustrated by wwtr1 disruption in embryonic zebrafish, which results in a complete lack of skeletal ossification149. Similarly, disruption of yap1 results in major bone defects and impaired fin regeneration, caused by an inhibition of osteoprogenitor cell maturation, giving an increased osteoprogenitor pool with a downregulation of intermediate and mature gene markers233. These defects are mediated by a reduction in Bmp signalling (which usually promotes maturation into osteoblasts239). In wild-type fish, Yap1 promotes Bmp signalling in a cell non-autonomous manner, restricting osteoprogenitors to the distal blastema (where Yap1 is inactive), and promotes osteoblast formation in the proximal blastema (where Yap1 is active)233. bmp4 is also associated with tail fin regeneration. Bmp4 is expressed in the distal blastema, and its inhibition reduces fin outgrowth after injury due to reduced proliferation of blastema cells240,241. This data suggests that Yap1 functions in the blastema to mechanotransduce tension changes and control the fate and migration of specific cell types in the amputated fin, regulating the precise control of tissue growth, potentially through the expression of ECM factors such as Ctgfa232,234.
The Hippo pathway is also associated with the differentiation of osteoblasts from mesenchymal stem cells (MSCs) during development, which generate neurons, adipocytes, skeletal muscle, and osteoblasts242. In in vitro studies, TAZ promotes osteoblast differentiation from MSCs via activation of Runx2-dependent gene transcription whilst inhibiting adipocyte differentiation via repression of PPARγ signalling149. CTGF also promotes osteoblast differentiation from MSCs in vitro243. Similar data are observed in mice, where YAP1 and TAZ promote bone formation and repair through their regulation of the osteoblast lineage244,245. Osteoblast lineage-specific Yap1 KO mice have reduced osteoblast differentiation and increased adipocyte formation, an effect that is diminished following increased β-catenin expression, demonstrating the importance of Wnt/β-catenin signalling in this process245. However, the role of the Hippo pathway in osteoblast differentiation is contested, with some in vitro studies suggesting that YAP1/TAZ suppress osteoblast differentiation and bone formation, and increase adipogenesis246,247, so more work is required to elucidate this complexity.
Zebrafish tail fin regeneration is most closely associated with limb regeneration, which does not occur in mammals or other higher vertebrates, although the mouse has been found in some instances to regenerate the digit tip in both newborns and adults248. Appendage regeneration does occur in certain amphibians such as salamanders as well as some invertebrates, and the Drosophila yap1 ortholog yki has been shown to promote wing disc regeneration249. Regeneration of an entire limb in mammals appears unlikely, but work in the zebrafish tail fin and other systems suggests that Hippo signalling may play an important role and promoting it could enhance regenerative capacity of specific aspects of limb regeneration, such as enhanced bone regeneration after breaks.
Hair cell regeneration in the lateral line
The lateral line is a mechanosensitive organ in fish and other aquatic amphibians that detects motion of the external liquid, aiding feeding and social behaviour as well as orientation in currents. In zebrafish, this rapidly developing organ is formed of sixty small clusters of cells (termed neuromasts) in adulthood (expanded from an initial eight in larvae)250, located along with the head (anterior lateral line) and trunk (posterior lateral line, pLL) in stereotyped positions17. Neuromasts consist of a group of hair cells with stereocilia projecting out of the skin and into the surrounding water, mechanical movement of which triggers sensation, and surrounding interdigitating supporting cells and mantle cells (Fig. 6A). Hair cells are innervated by ribbon synapses with afferent sensory neurons17 that project to the hindbrain, where they exhibit a somatotopy similar to the tonotopy seen in mammalian cochlear afferent projections251.
Fig. 6. Overview of neuromast regeneration.
a Uninjured neuromasts consist of hair cells (green) with cilia projecting into the external liquid, support cells (blue), mantle cells (orange), and afferent sensory neurons (red) that project to the brain. b Administration of aminoglycosides or Cu2+ causes specific hair cell death. c Support cell proliferation increases and cells transdifferentiate into hair cells. yap1 promotes support cell transdifferentiation. d Hair cell cilia regrowth restores neuromast function.
During early zebrafish development, a pLL primordium (pLLP) is generated behind the otic vesicle, forming a mass of cells that migrates along the flank beneath the skin, depositing protoneuromasts at periodic intervals252–255. The deposition of protoneuromasts and their development into mature neuromasts is mediated by Wnt/β-catenin, Notch, and Fgf signalling pathways, and is reviewed elsewhere17,256. These migrating cells must maintain a cohesive structure through high levels of expression of E-cadherin and tight junctions. In mammalian epithelial cells, E-Cadherin is a key upstream regulator of YAP1/TAZ257,258, indicating a potential role for Hippo signalling in this process. In fact, the Hippo pathway is linked to lateral line development in the zebrafish, as indicated by the induced expression of Yap1, Amotl2a, and Cav-1α in the developing lateral line136,259–261, and how disrupting these proteins functions impact lateral line formation. Downregulation of Cav-1α reduces the number of neuromasts formed136 and disruption of yap1 triggers a range of phenotypes, including a reduction in primordium size, reduced number of neuromasts, and a decrease in hair cell number259–262. Amotl2a negatively regulates Yap1 in the developing lateral line, limiting proliferation and so restricting the size of the pLLP, coupling with Notch signalling (which upregulates Yap1 to promote proliferation) to ensure correct pLLP size is reached259,261.
Analysis of the transcriptome of yap1-deficient embryos shows multiple gene expression changes, including those involved in the Wnt/β-catenin signalling pathway260, and lysophosphatidic acid (LPA)262. One of these factors is Prox1a, a target of β-catenin that aids hair cell differentiation in the lateral line260,263. Analysis of yap1- and prox1a-deficient embryos shows that yap1 deficiency recapitulates the prox1a deficiency phenotype of reduced hair cell number and impaired mechanoreceptor differentiation. These yap1 phenotypes are rescued by the administration of prox1a mRNA, suggesting that Yap1 functions by promoting Prox1a activity, so regulating hair cell maturation260. In the lateral line, the LPA receptor Lpar2b is expressed in the pLL and neuromasts, and its loss-of-function phenocopies yap1 KD262. LPA inhibits the Hippo kinase module, consequently activating YAP1/TAZ264,265 and stimulating cell proliferation, migration, and differentiation262,266. In the zebrafish specifically, LPA affects early development, promoting vascular and midline development, left-right patterning, and cell migration during gastrulation, amongst others267–270. In the pLL Lpar2b regulates Yap1 phosphorylation, suggesting that LPA signalling controls both primordium size and neuromast number by regulating Yap1 activity262. These results suggest that the Hippo pathway promotes appropriate size and cell function in lateral line development through a range of signalling pathways, many of which are also associated with other developmental processes.
The high regenerative capacity of the amphibian lateral line was first observed in the salamander271,272 but has since been observed in multiple organisms, including the zebrafish273 (Fig. 6). This is in contrast to the limited regeneration of mammalian hair cells, e.g. of the inner ear256. The majority of regenerated lateral line hair cells are formed by symmetric asynchronous division of support cells in the first 20 hours post injury254,274,275, where mitotic division of one support cell gives rise to two hair cells276 (Fig. 6C). The molecular and cellular triggers of this regeneration include pathways involved in lateral line development—Wnt/β-catenin, Notch, and Fgf signalling—as well as novel factors such as the Jak/Stat3 pathway (Table 1), which balance self-renewal, hair cell differentiation, and the risk of overgrowth.
The Hippo pathway links to lateral line regeneration277. The expression pattern of supporting cells during regeneration is reminiscent of expression in the migrating primordium during lateral line development, which is silenced when leading progenitors differentiate into mature supporting cells and hair cells259,278,279. This includes the expression of Hippo components cav-1 and ctgfa, which are upregulated in the support cells of both the zebrafish lateral line and the mouse inner ear280. In addition, after severe hair cell injury, Yap1 is activated in hair cell precursors, and regeneration is impaired in yap1 mutants277. Yap1 activation may occur through cell junction damage and resulting loss of junction-associated proteins such as Amotl2a, which usually restricts Yap1 activity in the lateral line261,277. Activated Yap1 upregulates lin28a transcription, an RNA-binding protein that regulates the translation of mRNAs involved in developmental timing, pluripotency and metabolism281. This promotes a Yap1-lin28a-let7-Wnt signalling axis that is both necessary and sufficient to promote progenitor cell activation and hence neuromast regeneration. The Yap1-lin28a-let7-Wnt signalling axis has other roles in dedifferentiation, including zebrafish retinal regeneration, mammalian embryonic inner ear development, and in vitro reprogramming of stem cell cultures282–285.
In summary, Yap1/Taz signalling in progenitor support cells is triggered after hair cell injury, promoting their differentiation towards hair cells via a Wnt signalling pathway, and enhancing recovery. Promoting Yap1/Taz signalling may also have therapeutic benefits in humans. The hair cells of the inner ear do not regenerate256 but have high similarity to zebrafish lateral line hair cells. This includes similar expression patterns of mechanosensitive ion channel and tip link genes and responses to key signalling pathways and ototoxic insults256,286–290, and so targeting the Hippo pathway to promote the regeneration of inner ear hair cells to combat age-related hearing decline may be a viable approach.
Liver regeneration
Despite limited mammalian regeneration of many organs, both mammals and zebrafish can regenerate their livers efficiently through the proliferation of differentiated hepatocytes, regaining liver function through epimorphic regrowth and compensatory enlargement of liver lobes291,292 (Fig. 7). However, this capacity of hepatocytes to repopulate the liver in humans can be overwhelmed by chronic or severe injury, resulting in liver failure that is only treatable by liver transplantation. There are many functional, cellular, and structural similarities between mammalian and zebrafish liver, both of which can regenerate their liver after more chronic insults,292 making the zebrafish a useful model to study the development and regeneration of the liver. However, limited research has been performed investigating the role of the Hippo pathway in zebrafish liver regeneration, although much work on this topic has been performed in the mouse. After experimental murine liver injury, YAP1 protein levels increase, with increased nuclear localisation in the liver and enhanced expression of downstream YAP1/TAZ target genes220,293,294. In a mouse model with Yap deletion in hepatocytes, bile duct ligation results in hepatic necrosis, reduced hepatocyte proliferation, and increased mortality220,295 compared to wild-type mice, suggesting a key role for Hippo signalling in mammalian liver regeneration.
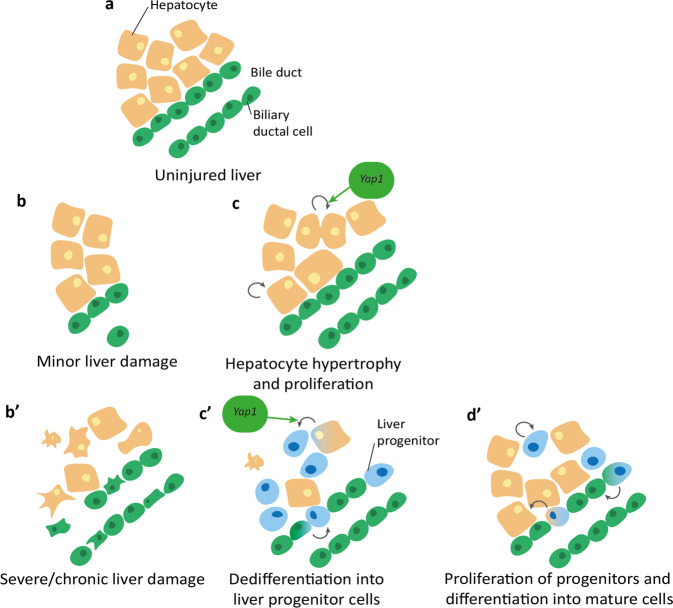
Fig. 7. Overview of liver regeneration after minor (b, c) and severe (b’, c’, d’) injury.
a Healthy (uninjured) zebrafish liver consists of multiple cell types hepatocytes (orange) and bile ducts comprising of biliary ductal cells (green). b Minor liver injury such as partial hepatectomy removes portions of the liver and the associated cells. c Liver recovery after minor liver damage involves hypertrophy and increased proliferation of remaining cells. Yap1 promotes hepatocyte proliferation. b’ Chronic or severe liver damage causes widespread cell death and necrosis. c’ Remaining cells dedifferentiate into liver progenitor cells, promoted by Yap1. d’ Progenitor cells proliferate then differentiate into mature hepatocytes and biliary ductal cells.
One method posited to promote liver regeneration is the recapitulation of developmental processes to generate progenitor-like cells that repopulate the liver after hepatocyte loss. Supporting this, after severe liver injury biliary cells have been shown to transdifferentiate into hepatocytes via a dedifferentiated progenitor-like state to repopulate the liver291,292. Hippo signalling is implicated in multiple cell fate transitions during liver regeneration in the mouse296,297. This includes YAP signalling activation by the alteration of cholangiocytes’ epigenome and transcriptome to aid their restoration of normal hepatocyte and cholangiocyte number296. YAP also associates with factors such as Arid1a to promote the induction of liver progenitor-like cell-enriched genes297.
The Hippo pathway is linked to liver development in both mammalian and zebrafish livers, and likely regulates cell fate plasticity in this process. In mice, YAP1 overexpression causes hepatomegaly that is reversible upon cessation of YAP1 signalling, suggesting a function for YAP1 in regulation of cell proliferation and hepatocyte function221,298. Hepatomegaly is also observed in the zebrafish after Yap1 overexpression or nf2a disruption299,300, whilst conversely yap1-/- fish have reduced liver size147. Other structural defects observed when disrupting upstream Hippo pathway components in the zebrafish include dilated bile ducts299, biliary dysgenesis301, and extrahepatic choledochal cysts299. Yap1 has also been linked to metabolism in the zebrafish liver, where it stimulates nucleotide biosynthesis to promote tissue growth through increasing glutamine synthetase and glucose transporter glut1 expression147,300,302.
The Hippo pathway’s role in hepatocyte development is thought to be vital in its role in the liver as hepatocytes are the predominant cell type in the liver and are key to liver function292. Appropriate Hippo pathway function is essential in the maintenance of mature hepatocytes, with hepatocyte-specific Nf2 loss in mice leading to hepatocyte dedifferentiation into highly renewable progenitors303, and overexpression causing a dysplastic hepatocyte morphology221. YAP1 is also associated with the formation of bile ducts in the developing mouse304, and with the function of the bile ducts (which promote immune cell recruitment and function) in the regenerating adult mouse liver305. Similarly, stk3 and sav1 zebrafish mutants (which both result in increased Yap1 activity) display altered hepatocyte morphology and polarity alongside biliary cell disruption301. Overall, these data suggest a conserved role for the Hippo pathway in structural liver, hepatocyte and biliary cell function between mammals and zebrafish. This implies that the Hippo pathway may also have a role in zebrafish liver regeneration, although this research is still in its infancy and will need further detailed investigation before conclusions can be drawn.
Conclusion
The zebrafish is a powerful model system for the study of regeneration due to their rapid external development, relative low cost, transparent juvenile stages and robust reparative regeneration as well as the availability of a range of established genetic tools and other experimental procedures to study these. In this review, the role of the Hippo pathway in zebrafish regeneration is summarised, with the finding that Yap1/Taz signalling often enhances regeneration through the promotion of cell proliferation, progenitor cell dedifferentiation and maturation, EMT, and scar resolution, as well as linking to key developmental pathways. The phenotypes resulting from the disruption of Hippo pathway components is summarised in Table 2.
The positive effect of Yap1/Taz signalling on regeneration in the zebrafish, which appears to be latent in mammals, suggests some therapeutic potential in promoting YAP/TAZ signalling to enhance mammalian regeneration. However, this must be carefully investigated, as many of the processes associated with enhanced regeneration are linked to an increased risk of cancer, such as an elevated cell proliferation rate, cellular heterogeneity, and increased stemness306. In fact, dysregulation of the Hippo pathway and thereby pathological hyperactivation of YAP1/TAZ promotes carcinogenesis in most, if not all, types of solid tumours102,307,308. The zebrafish may therefore be vital in the elucidation of this association between cancer and regeneration, which could allow us to manipulate regenerative potential without impacting carcinogenesis or vice versa. One way to do this could be through the utilisation of zebrafish Hippo pathway-induced cancer models, which recapitulate human findings in that manipulation of Hippo signalling can trigger tumour formation309–311. However, the field of Hippo signalling in the zebrafish is still relatively new, and so much work must be performed to bridge the gaps that are currently preventing its translation to the clinic, particularly the study of the molecular and cellular drivers of the Hippo pathway’s effects on both regeneration and development (Box 1).
Box 1: Outstanding questions.
What are the interactions and feedback between Hippo signalling and the immune system in regeneration that result in the current contradictory observations described? Does the Hippo pathway function differently in distinct immune processes, or are these apparent contradictions simply due to differences in experimental design?
Enhanced Yap1/Taz signalling is linked to increased risk of tumorigenesis. Is there a way to activate Yap1/Taz signalling spatiotemporally and precisely in specific cell types to promote regeneration without increasing cancer risk?
Can these findings be translated to humans? If so, can they be manipulated for therapeutic purposes, such as triggering regeneration in non-regenerating organs (e.g. heart and CNS) or promoting regeneration after chronic injury (e.g. liver)?
Zebrafish are an attractive model to study Hippo pathway dynamics in vivo, as it is amenable to live imaging with genetically encoded biosensors and tagged proteins. What are the dynamics of the Hippo pathway components in different cell types in vivo during development and regeneration in vertebrae?
Acknowledgements
Work ongoing in the Gram Hansen lab is supported by a University of Edinburgh Chancellor’s Fellowship as well as by Worldwide Cancer Research (19-0238) and LifeArc-CSO. Additional funding has been obtained from the Bone Cancer Research Trust (BCRT), Sarcoma UK (SUK202.2016), the Wellcome Trust-University of Edinburgh Institutional Strategic Support Fund (ISSF3). Y.F. received a Wellcome Trust Sir Henry Dale Fellowship (100104/Z/12/Z); Cancer Research UK early detection project award (C38363/A26931) and a Cancer Research UK pioneering award (C38363/A25107). S.E.R. is funded by a Wellcome Trust PhD studentship [108906/Z/15/Z]. Figures, besides Fig. 1, created in Adobe Illustrator.
Author contributions
S.E.R. and C.G.H. drafted the manuscript. S.E.R., Y.F., and C.G.H. revised the manuscript. C.G.H. and Y.F. supervised the project. S.E.R. prepared the figures.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wittlieb J, Khalturin K, Lohmann JU, Anton-Erxleben F, Bosch TCG. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc. Natl. Acad. Sci. USA. 2006;103:6208–6211. doi: 10.1073/pnas.0510163103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Wolfswinkel JC, Wagner DE, Reddien PW. Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell. 2014;15:326–339. doi: 10.1016/j.stem.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ivankovic M, et al. Model systems for regeneration: planarians. Development. 2019;146:dev167684. doi: 10.1242/dev.167684. [DOI] [PubMed] [Google Scholar]
- 4.Vogg MC, Galliot B, Tsiairis CD. Model systems for regeneration: Hydra. Development. 2019;146:dev177212. doi: 10.1242/dev.177212. [DOI] [PubMed] [Google Scholar]
- 5.Zhao A, Qin H, Fu X. What determines the regenerative capacity in animals? Bioscience. 2016;66:735–746. [Google Scholar]
- 6.Poss KD, Keating MT, Nechiporuk A. Tales of regeneration in zebrafish. Dev. Dyn. 2003;226:202–210. doi: 10.1002/dvdy.10220. [DOI] [PubMed] [Google Scholar]
- 7.Mao SA, Glorioso JM, Nyberg SL. Liver regeneration. Transl. Res. 2014;163:352–362. doi: 10.1016/j.trsl.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plikus MV, et al. Epithelial stem cells and implications for wound repair. Semin. Cell Dev. Biol. 2012;23:946–953. doi: 10.1016/j.semcdb.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong AW, Meng Z, Guan K-L. The Hippo pathway in intestinal regeneration and disease. Nat. Rev. Gastroenterol. Hepatol. 2016;13:324–337. doi: 10.1038/nrgastro.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheib J, Höke A. Advances in peripheral nerve regeneration. Nat. Rev. Neurol. 2013;9:668–676. doi: 10.1038/nrneurol.2013.227. [DOI] [PubMed] [Google Scholar]
- 11.Flach J, Milyavsky M. Replication stress in hematopoietic stem cells in mouse and man. Mutat. Res. - Fundam. Mol. Mech. Mutagen. 2018;808:74–82. doi: 10.1016/j.mrfmmm.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Larson BJ, Longaker MT, Lorenz HP. Scarless fetal wound healing: a basic science review. Plast. Reconstr. Surg. 2010;126:1172–1180. doi: 10.1097/PRS.0b013e3181eae781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sousounis, K., Baddour, J. A. & Tsonis, P. A. Aging and regeneration in vertebrates. Curr. Top. Dev. Biol.108, 217–246 (2014). [DOI] [PubMed]
- 14.Yun MH. Changes in regenerative capacity through lifespan. Int. J. Mol. Sci. 2015;16:25392–25432. doi: 10.3390/ijms161025392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith KA, Mommersteeg MTM. Talkin’ ‘bout regeneration: new advances in cardiac regeneration using the zebrafish. Curr. Opin. Physiol. 2020;14:48–55. [Google Scholar]
- 16.Sehring, I. M. & Weidinger, G. Recent advancements in understanding fin regeneration in zebrafish. Wiley Interdiscip. Rev. Dev. Biol. 9, e367 (2020). [DOI] [PubMed]
- 17.Thomas, E. D., Cruz, I. A., Hailey, D. W. & Raible, D. W. There and back again: development and regeneration of the zebrafish lateral line system. Wiley Interdiscip. Rev. Dev. Biol. 4, 1–16 (2015). [DOI] [PMC free article] [PubMed]
- 18.Cigliola V, Becker CJ, Poss KD. Building bridges, not walls: spinal cord regeneration in zebrafish. Dis. Model. Mech. 2020;13:dmm044131. doi: 10.1242/dmm.044131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gemberling M, Bailey TJ, Hyde DR, Poss KD. The zebrafish as a model for complex tissue regeneration. Trends Genet. 2013;29:611. doi: 10.1016/j.tig.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marques IJ, Lupi E, Mercader N. Model systems for regeneration: zebrafish. Development. 2019;146:dev167692. doi: 10.1242/dev.167692. [DOI] [PubMed] [Google Scholar]
- 21.Streisinger G, Singer F, Walker C, Knauber D, Dower N. Segregation analyses and gene-centromere distances in zebrafish. Genetics. 1986;112:311–319. doi: 10.1093/genetics/112.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Streisinger G, Walker C, Dower N, Knauber D, Singer F. Production of clones of homozygous diploid zebra fish (Brachydanio rerio) Nature. 1981;291:293–296. doi: 10.1038/291293a0. [DOI] [PubMed] [Google Scholar]
- 23.Jopling C, Boue S, Belmonte JCI. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nat. Rev. Mol. Cell Biol. 2011;12:79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- 24.Powell C, Grant AR, Cornblath E, Goldman D. Analysis of DNA methylation reveals a partial reprogramming of the Müller glia genome during retina regeneration. Proc. Natl. Acad. Sci. USA. 2013;110:19814–19819. doi: 10.1073/pnas.1312009110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirose K, Shimoda N, Kikuchi Y. Transient reduction of 5-methylcytosine and 5-hydroxymethylcytosine is associated with active DNA demethylation during regeneration of zebrafish fin. Epigenetics. 2013;8:899–906. doi: 10.4161/epi.25653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart S, Tsun Z-Y, Belmonte JCI. A histone demethylase is necessary for regeneration in zebrafish. Proc. Natl. Acad. Sci. USA. 2009;106:19889–19894. doi: 10.1073/pnas.0904132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfefferli, C., Müller, F., Jazwinska, A. & Wicky, C. Specific NuRD components are required for fin regeneration in zebrafish. BMC Biol. 12, 30 (2014). [DOI] [PMC free article] [PubMed]
- 28.Goldman JA, et al. Resolving heart regeneration by replacement histone profiling. Dev. Cell. 2017;40:392–404. doi: 10.1016/j.devcel.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Golenberg N, et al. Citrullination regulates wound responses and tissue regeneration in zebrafish. J. Cell Biol. 2020;219:e201908164. doi: 10.1083/jcb.201908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, W. et al. Changes in regeneration-responsive enhancers shape regenerative capacities in vertebrates. Science309, eaaz3090 (2020). [DOI] [PMC free article] [PubMed]
- 31.Kang J, et al. Modulation of tissue repair by regeneration enhancer elements. Nature. 2016;532:201–206. doi: 10.1038/nature17644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfefferli, C. & Jazwinska, A. The careg element reveals a common regulation of regeneration in the zebrafish myocardium and fin. Nat. Commun. 8, 15151 (2017). [DOI] [PMC free article] [PubMed]
- 33.Thompson JD, et al. Identification and requirements of enhancers that direct gene expression during zebrafish fin regeneration. Development. 2020;147:dev191262. doi: 10.1242/dev.191262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu C-C, et al. Spatially Resolved Genome-wide Transcriptional Profiling Identifies BMP Signaling as Essential Regulator of Zebrafish Cardiomyocyte Regeneration. Dev. Cell. 2016;36:36–49. doi: 10.1016/j.devcel.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 35.Quint E, et al. Bone patterning is altered in the regenerating zebrafish caudal fin after ectopic expression of sonic hedgehog and bmp2b or exposure to cyclopamine. Proc. Natl Acad. Sci. 2002;99:8713–8718. doi: 10.1073/pnas.122571799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schebesta M, Lien C-L, Engel FB, Keating MT. Transcriptional Profiling of Caudal Fin Regeneration in Zebrafish. ScientificWorldJournal. 2006;6:38–54. doi: 10.1100/tsw.2006.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kujawski S, et al. Calcineurin Regulates Coordinated Outgrowth of Zebrafish Regenerating Fins. Dev. Cell. 2014;28:573–587. doi: 10.1016/j.devcel.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 38.Goldshmit, Y. et al. Different Fgfs have distinct roles in regulating neurogenesis after spinal cord injury in zebrafish. Neural Dev. 13, (2018). [DOI] [PMC free article] [PubMed]
- 39.Poss KD, et al. Roles for Fgf Signaling during Zebrafish Fin Regeneration. Dev. Biol. 2000;222:347–358. doi: 10.1006/dbio.2000.9722. [DOI] [PubMed] [Google Scholar]
- 40.Whitehead GG, Makino S, Lien C-L, Keating M. T. fgf20 is essential for initiating zebrafish fin regeneration. Sci. (80-.). 2005;310:1957–1960. doi: 10.1126/science.1117637. [DOI] [PubMed] [Google Scholar]
- 41.König D, Page L, Chassot B, Jazwinska A. Dynamics of actinotrichia regeneration in the adult zebrafish fin. Dev. Biol. 2018;433:416–432. doi: 10.1016/j.ydbio.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 42.Lee SG, et al. Myc and Fgf Are Required for Zebrafish Neuromast Hair Cell Regeneration. PLoS One. 2016;11:e0157768. doi: 10.1371/journal.pone.0157768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi W-Y, et al. In vivo monitoring of cardiomyocyte proliferation to identify chemical modifiers of heart regeneration. Development. 2013;140:660–666. doi: 10.1242/dev.088526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chablais F, Jazwinska A. IGF signaling between blastema and wound epidermis is required for fin regeneration. Development. 2010;137:871–879. doi: 10.1242/dev.043885. [DOI] [PubMed] [Google Scholar]
- 45.Fang Y, et al. Translational profiling of cardiomyocytes identifies an early Jak1/Stat3 injury response required for zebrafish heart regeneration. Proc. Natl Acad. Sci. 2013;110:13416–13421. doi: 10.1073/pnas.1309810110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang J, et al. The stat3/socs3a Pathway Is a Key Regulator of Hair Cell Regeneration in Zebrafish stat3/socs3a Pathway: Regulator of Hair Cell Regeneration. J. Neurosci. 2012;32:10662–10673. doi: 10.1523/JNEUROSCI.5785-10.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khaliq M, et al. Stat3 Regulates Liver Progenitor Cell-Driven Liver Regeneration in Zebrafish. Gene Expr. 2018;18:157–170. doi: 10.3727/105221618X15242506133273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karra R, Knecht AK, Kikuchi K, Poss KD. Myocardial NF-κB activation is essential for zebrafish heart regeneration. Proc. Natl Acad. Sci. 2015;112:13255–13260. doi: 10.1073/pnas.1511209112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao L, et al. Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc. Natl Acad. Sci. 2014;111:1403–1408. doi: 10.1073/pnas.1311705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dias TB, Yang Y-J, Ogai K, Becker T, Becker CG. Notch Signaling Controls Generation of Motor Neurons in the Lesioned Spinal Cord of Adult Zebrafish. J. Neurosci. 2012;32:3245–3252. doi: 10.1523/JNEUROSCI.6398-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grotek B, Wehner D, Weidinger G. Notch signaling coordinates cellular proliferation with differentiation during zebrafish fin regeneration. Development. 2013;140:1412–1423. doi: 10.1242/dev.087452. [DOI] [PubMed] [Google Scholar]
- 52.Münch J, González-Rajal A, de la Pompa JL. Notch regulates blastema proliferation and prevents differentiation during adult zebrafish fin regeneration. Development. 2013;140:1402–1411. doi: 10.1242/dev.087346. [DOI] [PubMed] [Google Scholar]
- 53.Romero-Carvajal A, et al. Regeneration of Sensory Hair Cells Requires Localized Interactions between the Notch and Wnt Pathways. Dev. Cell. 2015;34:267–282. doi: 10.1016/j.devcel.2015.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pinto-Teixeira F, et al. Inexhaustible hair-cell regeneration in young and aged zebrafish. Biol. Open. 2015;4:903–909. doi: 10.1242/bio.012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang M, et al. Antagonistic Interaction Between Wnt and Notch Activity Modulates the Regenerative Capacity of a Zebrafish Fibrotic Liver Model. Hepatology. 2014;60:1753–1766. doi: 10.1002/hep.27285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He J, Lu H, Zou Q, Luo L. Regeneration of Liver After Extreme Hepatocyte Loss Occurs Mainly via Biliary Transdifferentiation in Zebrafish. Gastroenterology. 2014;146:789–800. doi: 10.1053/j.gastro.2013.11.045. [DOI] [PubMed] [Google Scholar]
- 57.Ko S, et al. Hdac1 Regulates Differentiation of Bipotent Liver Progenitor Cells During Regeneration via Sox9b and Cdk8. Gastroenterology. 2019;156:187–202. doi: 10.1053/j.gastro.2018.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gemberling M, Karra R, Dickson AL, Poss KD. Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. Elife. 2015;4:e05871. doi: 10.7554/eLife.05871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mathew LK, et al. Comparative expression profiling reveals an essential role for Raldh2 in epimorphic regeneration. J. Biol. Chem. 2009;284:33642–33653. doi: 10.1074/jbc.M109.011668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kikuchi K, et al. Retinoic Acid Production by Endocardium and Epicardium Is an Injury Response Essential for Zebrafish Heart Regeneration. Dev. Cell. 2011;20:397–404. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blum N, Begemann G. Retinoic acid signaling controls the formation, proliferation and survival of the blastema during adult zebrafish fin regeneration. Development. 2012;139:107–116. doi: 10.1242/dev.065391. [DOI] [PubMed] [Google Scholar]
- 62.Blum N, Begemann G. Osteoblast de- and redifferentiation are controlled by a dynamic response to retinoic acid during zebrafish fin regeneration. Development. 2015;142:2894–2918. doi: 10.1242/dev.120204. [DOI] [PubMed] [Google Scholar]
- 63.Blum N, Begemann G. Retinoic acid signaling spatially restricts osteoblasts and controls ray-interray organization during zebrafish fin regeneration. Development. 2015;142:2888–2893. doi: 10.1242/dev.120212. [DOI] [PubMed] [Google Scholar]
- 64.Han P, et al. Hydrogen peroxide primes heart regeneration with a derepression mechanism. Cell Res. 2014;24:1091–1107. doi: 10.1038/cr.2014.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gauron, C. et al. Sustained production of ROS triggers compensatory proliferation and is required for regeneration to proceed. Sci. Rep. 3, (2013). [DOI] [PMC free article] [PubMed]
- 66.Reimer MM, et al. Sonic hedgehog is a polarized signal for motor neuron regeneration in adult zebrafish. J. Neurosci. 2009;29:15073–15082. doi: 10.1523/JNEUROSCI.4748-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reimer MM, et al. Dopamine from the Brain Promotes Spinal Motor Neuron Generation during Development and Adult Regeneration. Dev. Cell. 2013;25:478–491. doi: 10.1016/j.devcel.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 68.Jazwinska A, Badakov R, Keating MT. Activin-βA Signaling Is Required for Zebrafish Fin Regeneration. Curr. Biol. 2007;17:1390–1395. doi: 10.1016/j.cub.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 69.Chablais F, Jazwinska A. The regenerative capacity of the zebrafish heart is dependent on TGFβ signaling. Development. 2012;139:1921–1930. doi: 10.1242/dev.078543. [DOI] [PubMed] [Google Scholar]
- 70.Dogra, D. et al. Opposite effects of Activin type 2 receptor ligands on cardiomyocyte proliferation during development and repair. Nat. Commun. 8, (2017). [DOI] [PMC free article] [PubMed]
- 71.Kawakami Y, et al. Wnt/β-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 2006;20:3232–3237. doi: 10.1101/gad.1475106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stoick-Cooper CL, et al. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134:479–489. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- 73.Wehner D, et al. Wnt/β-catenin Signaling Defines Organizing Centers that Orchestrate Growth and Differentiation of the Regenerating Zebrafish Caudal Fin. Cell Rep. 2014;6:467–481. doi: 10.1016/j.celrep.2013.12.036. [DOI] [PubMed] [Google Scholar]
- 74.Head JR, Gacioch L, Pennisi M, Meyers JR. Activation of Canonical Wnt/Beta-Catenin Signaling Stimulates Proliferation in Neuromasts in the Zebrafish Posterior Lateral Line. Dev. Dyn. 2013;242:832–846. doi: 10.1002/dvdy.23973. [DOI] [PubMed] [Google Scholar]
- 75.Poulain M, Ober EA. Interplay between Wnt2 and Wnt2bb controls multiple steps of early foregutderived organ development. Development. 2011;138:3557–3568. doi: 10.1242/dev.055921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goessling W, et al. APC mutant zebrafish uncover a changing temporal requirement for wnt signaling in liver development. Dev. Comp. Immunol. 2008;320:161–174. doi: 10.1016/j.ydbio.2008.05.526. [DOI] [PubMed] [Google Scholar]
- 77.Choi T-Y, Ninov N, Stainier DYR, Shin D. Extensive Conversion of Hepatic Biliary Epithelial Cells to Hepatocytes After Near Total Loss of Hepatocytes in Zebrafish. Gastroenterology. 2014;146:776–788. doi: 10.1053/j.gastro.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moya IM, Halder G. Hippo–YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol. 2019;20:211–226. doi: 10.1038/s41580-018-0086-y. [DOI] [PubMed] [Google Scholar]
- 79.Panciera T, Azzolin L, Cordenonsi M, Piccolo S. Mechanobiology of YAP and TAZ in physiology and disease. Nat. Publ. Gr. 2017;18:758–770. doi: 10.1038/nrm.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fu V, Plouffe SW, Guan K-L. The Hippo pathway in organ development, homeostasis, and regeneration. Curr. Opin. Cell Biol. 2017;49:99–107. doi: 10.1016/j.ceb.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao B, Tumaneng K, Guan K-L. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat. Cell Biol. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ardestani A, Lupse B, Maedler K. Hippo signaling: key emerging pathway in cellular and whole-body metabolism. Trends Endocrinol. Metab. 2018;29:492–509. doi: 10.1016/j.tem.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 83.Kim W, Jho E. The history and regulatory mechanism of the Hippo pathway. BMB Rep. 2018;51:106–118. doi: 10.5483/BMBRep.2018.51.3.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Davis JR, Tapon N. Hippo signalling during development. Development. 2019;146:dev167106. doi: 10.1242/dev.167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–185. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 86.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gram Hansen C, Moroishi T, Guan K-L. YAP and TAZ: a nexus for Hippo signaling and beyond. Trends Cell Biol. 2015;25:499–513. doi: 10.1016/j.tcb.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meng, Z., Moroishi, T. & Guan, K.-L. Mechanisms of Hippo pathway regulation. Genes Dev. 30, 1–17 (2016). [DOI] [PMC free article] [PubMed]
- 89.Fulford A, Tapon N, Ribeiro PS. Upstairs, downstairs: spatial regulation of Hippo signalling. Curr. Opin. Cell Biol. 2017;51:22–32. doi: 10.1016/j.ceb.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 90.Rausch V, Gram Hansen C. The Hippo pathway, YAP/TAZ, and the plasma membrane. Trends Cell Biol. 2020;30:32–48. doi: 10.1016/j.tcb.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 91.Meng, Z. et al. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat. Commun. 6, 8357 (2015). [DOI] [PMC free article] [PubMed]
- 92.Zheng Y, et al. Identification of happyhour/MAP4K as alternative Hpo/Mst-like kinases in the Hippo kinase cascade. Dev. Cell. 2015;34:642–655. doi: 10.1016/j.devcel.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lim S, et al. Identification of the kinase STK25 as an upstream activator of LATS signaling. Nat. Commun. 2019;10:1547. doi: 10.1038/s41467-019-09597-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao B, Li L, Tumaneng K, Wang C-Y, Guan K-L. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF^β-TRCP. Genes Dev. 2010;22:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lei Q-Y, et al. TAZ Promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the Hippo pathway. Mol. Cell. Biol. 2008;28:2426–2436. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu C-Y, et al. The Hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCF^β-TRCP E3 ligase. J. Biol. Chem. 2010;285:37159–37169. doi: 10.1074/jbc.M110.152942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang S, et al. YAP antagonizes innate antiviral immunity and is targeted for lysosomal degradation through IKKε-mediated phosphorylation. Nat. Immunol. 2017;18:733–743. doi: 10.1038/ni.3744. [DOI] [PubMed] [Google Scholar]
- 98.Zhang H, et al. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J. Biol. Chem. 2009;284:13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–4069. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- 100.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hillmer, R. E. & Link, B. A. The roles of Hippo signaling transducers Yap and Taz in chromatin remodeling. Cells8, 502 (2019). [DOI] [PMC free article] [PubMed]
- 102.Salem, O. & Gram Hansen, C. The Hippo pathway in prostate cancer. Cells8, 370 (2019). [DOI] [PMC free article] [PubMed]
- 103.Yu F-X, Zhao B, Guan K-L. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rausch V, et al. The Hippo pathway regulates caveolae expression and mediates flow response via caveolae. Curr. Biol. 2019;29:1–14. doi: 10.1016/j.cub.2018.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Riching AS, Song K. Cardiac regeneration: new insights into the frontier of ischemic heart failure therapy. Front. Bioeng. Biotechnol. 2021;8:637538. doi: 10.3389/fbioe.2020.637538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.St. John Sutton MG, Sharpe N. Left ventricular remodeling after myocardial infarction: pathophysiology and therapy. Circulation. 2000;101:2981–2988. doi: 10.1161/01.cir.101.25.2981. [DOI] [PubMed] [Google Scholar]
- 107.Engel FB, Hsieh PCH, Lee RT, Keating MT. FGF1/p38 MAP kinase inhibitor therapy induces cardiomyocyte mitosis, reduces scarring, and rescues function after myocardial infarction. Proc. Natl. Acad. Sci. USA. 2006;103:15546–15551. doi: 10.1073/pnas.0607382103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Porrello ER, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J. Mol. Cell. Cardiol. 1996;28:1737–1746. doi: 10.1006/jmcc.1996.0163. [DOI] [PubMed] [Google Scholar]
- 110.Bise, T., Sallin, P., Pfefferli, C. & Jazwinska, A. Multiple cryoinjuries modulate the efficiency of zebrafish heart regeneration. Sci. Rep. 10, 11551 (2020). [DOI] [PMC free article] [PubMed]
- 111.Sehring IM, Jahn C, Weidinger G. Zebrafish fin and heart: what’s special about regeneration? Curr. Opin. Genet. Dev. 2016;40:48–56. doi: 10.1016/j.gde.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 112.Wang J, et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138:3421–3430. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 114.Senyo SE, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bergmann O, et al. Dynamics of cell generation and turnover in the human heart. Cell. 2015;161:1566–1575. doi: 10.1016/j.cell.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 116.Foglia MJ, Poss KD. Building and re-building the heart by cardiomyocyte proliferation. Development. 2016;143:729–740. doi: 10.1242/dev.132910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bassat E, et al. The extracellular matrix protein agrin promotes heart regeneration in mice. Nature. 2017;547:179–184. doi: 10.1038/nature22978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.González-Rosa JM, Martín V, Peralta M, Torres M, Mercader N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development. 2011;138:1663–1674. doi: 10.1242/dev.060897. [DOI] [PubMed] [Google Scholar]
- 119.Wang J, Karra R, Dickson AL, Poss KD. Fibronectin is deposited by injury-activated epicardial cells and is necessary for zebrafish heart regeneration. Dev. Biol. 2013;382:427–435. doi: 10.1016/j.ydbio.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lowe V, et al. Neuropilin 1 mediates epicardial activation and revascularization in the regenerating zebrafish heart. Development. 2019;146:dev174482. doi: 10.1242/dev.174482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mukherjee D, et al. Ccn2a is an injury-induced matricellular factor that promotes cardiac regeneration in zebrafish. Development. 2021;148:dev193219. doi: 10.1242/dev.193219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Flinn MA, Jeffery BE, O’Meara CC, Link BA. Yap is required for scar formation but not myocyte proliferation during heart regeneration in zebrafish. Cardiovasc. Res. 2019;115:570–577. doi: 10.1093/cvr/cvy243. [DOI] [PubMed] [Google Scholar]
- 123.Cao J, et al. Single epicardial cell transcriptome sequencing identifies caveolin 1 as an essential factor in zebrafish heart regeneration. Development. 2016;143:232–243. doi: 10.1242/dev.130534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lai S-L, et al. Reciprocal analyses in zebrafish and medaka reveal that harnessing the immune response promotes cardiac regeneration. Elife. 2017;6:e25605. doi: 10.7554/eLife.25605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Furutani-Seiki M, Wittbrodt J. Medaka and zebrafish, an evolutionary twin study. Mech. Dev. 2004;121:629–637. doi: 10.1016/j.mod.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 126.Huang W-C, et al. Treatment of Glucocorticoids inhibited early immune responses and impaired cardiac repair in adult zebrafish. PLoS One. 2013;8:e66613. doi: 10.1371/journal.pone.0066613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.de Preux Charles, A.-S., Bise, T., Baier, F., Marro, J. & Jazwinska, A. Distinct effects of inflammation on preconditioning and regeneration of the adult zebrafish heart. Open Biol.6, 160102 (2016). [DOI] [PMC free article] [PubMed]
- 128.Godwin JW, Pinto AR, Rosenthal NA. Chasing the recipe for a pro-regenerative immune system. Semin. Cell Dev. Biol. 2017;61:71–79. doi: 10.1016/j.semcdb.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hui SP, et al. Zebrafish regulatory T cells mediate organ-specific regenerative programs. Dev. Cell. 2017;43:659–672. doi: 10.1016/j.devcel.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 130.Stewart, K. M. R., Walker, S. L., Baker, A. H., Riley, P. R. & Brittan, M. Hooked on heart regeneration: the zebrafish guide to recovery. Cardiovasc. Res.10.1093/cvr/cvab214 (2021). [DOI] [PMC free article] [PubMed]
- 131.González-Rosa JM, Peralta M, Mercader N. Pan-epicardial lineage tracing reveals that epicardium derived cells give rise to myofibroblasts and perivascular cells during zebrafish heart regeneration. Dev. Biol. 2012;370:173–186. doi: 10.1016/j.ydbio.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 132.Singh A, et al. Hippo signaling mediators Yap and Taz are required in the epicardium for coronary vasculature development. Cell Rep. 2016;15:1384–1393. doi: 10.1016/j.celrep.2016.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Miesfeld JB, et al. Yap and Taz regulate retinal pigment epithelial cell fate. Development. 2015;142:3021–3032. doi: 10.1242/dev.119008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hildebrand, S. et al. The E-cadherin/AmotL2 complex organizes actin filaments required for epithelial hexagonal packing and blastocyst hatching. Sci. Rep.7, 9540 (2017). [DOI] [PMC free article] [PubMed]
- 135.Hultin, S. et al. Amotl2 links VE-cadherin to contractile actin fibres necessary for aortic lumen expansion. Nat. Commun.5, 3743 (2014). [DOI] [PubMed]
- 136.Nixon SJ, et al. Caveolin-1 is required for lateral line neuromast and notochord development. J. Cell Sci. 2007;120:2151–2161. doi: 10.1242/jcs.003830. [DOI] [PubMed] [Google Scholar]
- 137.Flinn, M. A. et al. Llgl1 regulates zebrafish cardiac development by mediating Yap stability in cardiomyocytes. Development147, dev193581 (2020). [DOI] [PMC free article] [PubMed]
- 138.Fang P-K, et al. Caveolin-1α and -1β perform nonredundant roles in early vertebrate development. Am. J. Pathol. 2006;169:2209–2222. doi: 10.2353/ajpath.2006.060562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dai X, et al. Phosphorylation of angiomotin by Lats1/2 kinases inhibits F-actin binding, cell migration, and angiogenesis. J. Biol. Chem. 2013;288:34041–34051. doi: 10.1074/jbc.M113.518019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nagasawa-Masuda A, Terai K. Yap/Taz transcriptional activity is essential for vascular regression via Ctgf expression and actin polymerization. PLoS One. 2017;12:e0174633. doi: 10.1371/journal.pone.0174633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nakajima H, et al. Flow-dependent endothelial YAP regulation contributes to vessel maintenance. Dev. Cell. 2017;40:523–536. doi: 10.1016/j.devcel.2017.02.019. [DOI] [PubMed] [Google Scholar]
- 142.Astone M, et al. Zebrafish mutants and TEAD reporters reveal essential functions for Yap and Taz in posterior cardinal vein development. Sci. Rep. 2018;8:1–15. doi: 10.1038/s41598-018-27657-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Grimm L, et al. Yap1 promotes sprouting and proliferation of lymphatic progenitors downstream of Vegfc in the zebrafish trunk. Elife. 2019;8:e42881. doi: 10.7554/eLife.42881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Xu D, et al. Scribble influences cyst formation in autosomal-dominant polycystic kidney disease by regulating Hippo signaling pathway. FASEB J. 2018;32:4394–4407. doi: 10.1096/fj.201701376RR. [DOI] [PubMed] [Google Scholar]
- 145.Park M-H, et al. CCN1 interlinks integrin and hippo pathway to autoregulate tip cell activity. Elife. 2019;8:e46012. doi: 10.7554/eLife.46012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dooley CM, et al. The gene regulatory basis of genetic compensation during neural crest induction. PLOS Genet. 2019;15:e1008213. doi: 10.1371/journal.pgen.1008213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cox AG, et al. Yap regulates glucose utilization and sustains nucleotide synthesis to enable organ growth. EMBO J. 2018;37:e100294. doi: 10.15252/embj.2018100294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pappalardo A, et al. Thyroid development in zebrafish lacking Taz. Mech. Dev. 2015;138:268–278. doi: 10.1016/j.mod.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 149.Hong J-H, et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Sci. (80-.) 2005;309:1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 150.Yi X, et al. The effector of Hippo signaling, Taz, is required for formation of the micropyle and fertilization in zebrafish. PLoS Genet. 2019;15:e1007408. doi: 10.1371/journal.pgen.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lo HP, et al. The caveolin–cavin system plays a conserved and critical role in mechanoprotection of skeletal muscle. J. Cell Biol. 2015;210:833–849. doi: 10.1083/jcb.201501046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kim M, Kim M, Lee M-S, Kim C-H, Lim D-S. The MST1/2-SAV1 complex of the Hippo pathway promotes ciliogenesis. Nat. Commun. 2014;5:1–14. doi: 10.1038/ncomms6370. [DOI] [PubMed] [Google Scholar]
- 153.Jopling C, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kikuchi K, et al. Primary contribution to zebrafish heart regeneration by gata4+ cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gupta V, et al. An injury-responsive gata4 program shapes the zebrafish cardiac ventricle. Curr. Biol. 2013;23:1221–1227. doi: 10.1016/j.cub.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang R, et al. In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature. 2013;498:497–502. doi: 10.1038/nature12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Harrison MRM, et al. Chemokine-guided angiogenesis directs coronary vasculature formation in zebrafish. Dev. Cell. 2015;33:442–454. doi: 10.1016/j.devcel.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kikuchi K, et al. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development. 2011;138:2895–2902. doi: 10.1242/dev.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kim J, et al. PDGF signaling is required for epicardial function and blood vessel formation in regenerating zebrafish hearts. Proc. Natl Acad. Sci. USA. 2010;107:17206–17210. doi: 10.1073/pnas.0915016107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Marín-Juez R, et al. Coronary revascularization during heart regeneration is regulated by epicardial and endocardial cues and forms a scaffold for cardiomyocyte repopulation. Dev. Cell. 2019;51:503–515. doi: 10.1016/j.devcel.2019.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Marín-Juez R, et al. Fast revascularization of the injured area is essential to support zebrafish heart regeneration. Proc. Natl Acad. Sci. USA. 2016;113:11237–11242. doi: 10.1073/pnas.1605431113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lepilina A, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 163.Itou J, et al. Migration of cardiomyocytes is essential for heart regeneration in zebrafish. Development. 2012;139:4133–4142. doi: 10.1242/dev.079756. [DOI] [PubMed] [Google Scholar]
- 164.Tapon N, et al. Salvador promotes both cell cycle exit and apoptosis in drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 165.Liu S, et al. Gene therapy knockdown of Hippo signaling induces cardiomyocyte renewal in pigs after myocardial infarction. Sci. Transl. Med. 2021;13:eabd6892. doi: 10.1126/scitranslmed.abd6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Del Re DP, et al. Yes-associated protein isoform 1 (Yap1) promotes cardiomyocyte survival and growth to protect against myocardial ischemic injury. J. Biol. Chem. 2013;288:3977–3988. doi: 10.1074/jbc.M112.436311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.von Gise A, et al. YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc. Natl Acad. Sci. USA. 2012;109:2394–2399. doi: 10.1073/pnas.1116136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Xin M, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc. Natl. Acad. Sci. USA. 2013;110:13839–13844. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lin Z, et al. Cardiac-specific YAP activation improves cardiac function and survival in an experimental murine MI model. Circ. Res. 2014;115:354–363. doi: 10.1161/CIRCRESAHA.115.303632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Morikawa Y, Heallen T, Leach J, Xiao Y, Martin JF. Dystrophin-glycoprotein complex sequesters Yap to inhibit cardiomyocyte proliferation. Nature. 2017;547:227–231. doi: 10.1038/nature22979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mia, M. M. & Singh, M. K. The Hippo signaling pathway in cardiac development and diseases. Front. Cell Dev. Biol.7, 211 (2019). [DOI] [PMC free article] [PubMed]
- 172.Mia, M. M. et al. Loss of Yap/Taz in cardiac fibroblasts attenuates adverse remodelling and improves cardiac function. Cardiovasc. Res. 10.1093/cvr/cvab205 (2021). [DOI] [PubMed]
- 173.Xiao Y, et al. Hippo pathway deletion in adult resting cardiac fibroblasts initiates a cell state transition with spontaneous and self-sustaining fibrosis. Genes Dev. 2019;33:1491–1505. doi: 10.1101/gad.329763.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tian Y, et al. A microRNA-Hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice. Sci. Transl. Med. 2015;7:279ra38. doi: 10.1126/scitranslmed.3010841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Courtine G, Sofroniew MV. Spinal cord repair: advances in biology and technology. Nat. Med. 2019;25:898–908. doi: 10.1038/s41591-019-0475-6. [DOI] [PubMed] [Google Scholar]
- 176.Tran AP, Warren PM, Silver J. The biology of regeneration failure and success after spinal cord injury. Physiol. Rev. 2018;98:881–917. doi: 10.1152/physrev.00017.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hilton BJ, Moulson AJ, Tetzlaff W. Neuroprotection and secondary damage following spinal cord injury: concepts and methods. Neurosci. Lett. 2017;652:3–10. doi: 10.1016/j.neulet.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 178.Becker T, Wullimann MF, Becker CG, Bernhardt RR, Schachner M. Axonal regrowth after spinal cord transection in adult zebrafish. J. Comp. Neurol. 1997;377:577–595. doi: 10.1002/(sici)1096-9861(19970127)377:4<577::aid-cne8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 179.Becker CG, et al. L1.1 is involved in spinal cord regeneration in adult zebrafish. J. Neurosci. 2004;24:7837–7842. doi: 10.1523/JNEUROSCI.2420-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Becker T, Becker CG. Dynamic cell interactions allow spinal cord regeneration in zebrafish. Curr. Opin. Physiol. 2020;14:64–69. [Google Scholar]
- 181.Becker CG, Becker T. Neuronal regeneration from ependymo-radial glial cells: cook, little pot, cook! Dev. Cell. 2015;32:516–527. doi: 10.1016/j.devcel.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 182.Rasmussen JP, Sagasti A. Learning to swim, again: axon regeneration in fish. Exp. Neurol. 2017;287:318–330. doi: 10.1016/j.expneurol.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 183.Ghosh, S. & Hui, S. P. Regeneration of zebrafish CNS: adult neurogenesis. Neural Plast.2016, 5815439 (2016). [DOI] [PMC free article] [PubMed]
- 184.Reimer MM, et al. Motor neuron regeneration in adult zebrafish. J. Neurosci. 2008;28:8510–8516. doi: 10.1523/JNEUROSCI.1189-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Briona LK, Poulain FE, Mosimann C, Dorsky RI. Wnt/ß-catenin signaling is required for radial glial neurogenesis following spinal cord injury. Dev. Biol. 2015;403:15–21. doi: 10.1016/j.ydbio.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ohnmacht J, et al. Spinal motor neurons are regenerated after mechanical lesion and genetic ablation in larval zebrafish. Development. 2016;143:1464–1474. doi: 10.1242/dev.129155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kuscha V, et al. Lesion-induced generation of interneuron cell types in specific dorsoventral domains in the spinal cord of adult zebrafish. J. Comp. Neurol. 2012;520:3604–3616. doi: 10.1002/cne.23115. [DOI] [PubMed] [Google Scholar]
- 188.Goldshmit Y, et al. Fgf-dependent glial cell bridges facilitate spinal cord regeneration in Zebrafish. J. Neurosci. 2012;32:7477–7492. doi: 10.1523/JNEUROSCI.0758-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yu Y-M, et al. The extracellular matrix glycoprotein tenascin-C promotes locomotor recovery after spinal cord injury in adult zebrafish. Neuroscience. 2011;183:238–250. doi: 10.1016/j.neuroscience.2011.03.043. [DOI] [PubMed] [Google Scholar]
- 190.Wehner, D. et al. Wnt signaling controls pro-regenerative Collagen XII in functional spinal cord regeneration in zebrafish. Nat. Commun. 8, 126 (2017). [DOI] [PMC free article] [PubMed]
- 191.Gomez-Sanchez JA, et al. After nerve injury, lineage tracing shows that myelin and Remak Schwann cells elongate extensively and branch to form repair Schwann cells, which shorten radically on remyelination. J. Neurosci. 2017;37:9086–9099. doi: 10.1523/JNEUROSCI.1453-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Parrinello S, et al. EphB signaling directs peripheral nerve regeneration through Sox2-dependent Schwann cell sorting. Cell. 2010;143:145–155. doi: 10.1016/j.cell.2010.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Klatt Shaw, D. et al. Localized EMT reprograms glial progenitors to promote spinal cord repair. Dev. Cell56, 613.e7–626.e7(2021). [DOI] [PMC free article] [PubMed]
- 194.Wilson MM, Weinberg RA, Lees JA, Guen VJ. Emerging mechanisms by which EMT programs control stemness. Trends Cancer. 2020;6:775–780. doi: 10.1016/j.trecan.2020.03.011. [DOI] [PubMed] [Google Scholar]
- 195.Ye X, Weinberg RA. Epithelial-mesenchymal plasticity: a central regulator of cancer progression. Trends Cell Biol. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Jessen KR, Arthur-Farraj P. Repair Schwann cell update: adaptive reprogramming, EMT, and stemness in regenerating nerves. Glia. 2019;67:421–437. doi: 10.1002/glia.23532. [DOI] [PubMed] [Google Scholar]
- 197.Mokalled MH, et al. Injury-induced ctgfa directs glial bridging and spinal cord regeneration in zebrafish. Science. 2016;354:630–634. doi: 10.1126/science.aaf2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bradbury, E. J. & Burnside, E. R. Moving beyond the glial scar for spinal cord repair. Nat. Commun. 10, 3879 (2019). [DOI] [PMC free article] [PubMed]
- 199.Hu, R. et al. Glial scar and neuroregeneration: histological, functional, and magnetic resonance imaging analysis in chronic spinal cord injury. J. Neurosurg.13, 169–180 (2010). [DOI] [PubMed]
- 200.Wang Y, et al. Lentivirus-mediated silencing of the CTGF gene suppresses the formation of glial scar tissue in a rat model of spinal cord injury. Spine J. 2018;18:164–172. doi: 10.1016/j.spinee.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 201.NIH, U. S. National Library of Medicine. ClinicalTrials.govhttps://clinicaltrials.gov/ct2/results?cond=&term=ctgf&cntry=&state=&city=&dist (2021).
- 202.Hamon A, et al. Linking YAP to Müller glia quiescence exit in the degenerative retina. Cell Rep. 2019;27:1712–1725. doi: 10.1016/j.celrep.2019.04.045. [DOI] [PubMed] [Google Scholar]
- 203.Hamon A, Roger JE, Yang X-J, Perron M. Müller glial cell-dependent regeneration of the neural retina: an overview across vertebrate model systems. Dev. Dyn. 2016;245:727–738. doi: 10.1002/dvdy.24375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Rueda EM, et al. The Hippo pathway blocks mammalian retinal Müller glial cell reprogramming. Cell Rep. 2019;27:1637–1649. doi: 10.1016/j.celrep.2019.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wan J, Goldman D. Retina regeneration in zebrafish. Curr. Opin. Genet. Dev. 2016;40:41–47. doi: 10.1016/j.gde.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Bernardos RL, Barthel LK, Meyers JR, Raymond PA. Late-stage neuronal progenitors in the retina are radial Müller glia that function as retinal stem cells. J. Neurosci. 2007;27:7028–7040. doi: 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Thummel R, Kassen SC, Montgomery JE, Enright JM, Hyde DR. Inhibition of Müller glial cell division blocks regeneration of the light-damaged zebrafish retina. Dev. Neurobiol. 2008;68:492–408. doi: 10.1002/dneu.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hoang T, et al. Gene regulatory networks controlling vertebrate retinal regeneration. Science. 2020;370:eabb8598. doi: 10.1126/science.abb8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Richeldi L, et al. Pamrevlumab, an anti-connective tissue growth factor therapy, for idiopathic pulmonary fibrosis (PRAISE): a phase 2, randomised, double-blind, placebo-controlled trial. Lancet Respir. Med. 2020;8:25–33. doi: 10.1016/S2213-2600(19)30262-0. [DOI] [PubMed] [Google Scholar]
- 210.Raghu G, et al. FG-3019 anti-connective tissue growth factor monoclonal antibody: results of an open-label clinical trial in idiopathic pulmonary fibrosis. Eur. Respir. J. 2016;47:1481–1491. doi: 10.1183/13993003.01030-2015. [DOI] [PubMed] [Google Scholar]
- 211.Ramazani Y, et al. Connective tissue growth factor (CTGF) from basics to clinics. Matrx Biol. 2018;68–69:44–66. doi: 10.1016/j.matbio.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 212.Azevedo AS, Grotek B, Jacinto A, Weidinger G, Saúde L. The regenerative capacity of the zebrafish caudal fin is not affected by repeated amputations. PLoS One. 2011;6:e22820. doi: 10.1371/journal.pone.0022820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Broussonet, M. Observations sur la regeneration de quelques parties du corps des poissons. Hist. d. l’Acad. R. des Sci. 105, 625–641 (1786).
- 214.Morgan, T. H. Regeneration (Macmillan, 1901).
- 215.Morgan TH. Regeneration in teleosts. Arch. f.ür. Entwickslungsmech Org. 1900;10:120–131. [Google Scholar]
- 216.Sunderland ME. Regeneration: Thomas hunt Morgan’s window into development. J. Hist. Biol. 2010;43:325–361. doi: 10.1007/s10739-009-9203-2. [DOI] [PubMed] [Google Scholar]
- 217.Pfefferli, C. & Jazwinska, A. The art of fin regeneration in zebrafish. Regeneration2, 72–83 (2015). [DOI] [PMC free article] [PubMed]
- 218.Cao X, Pfaff SL, Gage FH. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 2008;22:3320–3334. doi: 10.1101/gad.1726608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Panciera T, et al. Induction of expandable tissue-specific stem/progenitor cells through transient expression of YAP/TAZ. Cell Stem Cell. 2016;19:725–737. doi: 10.1016/j.stem.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Patel SH, Camargo FD, Yimlamai D. Hippo signaling in the liver regulates organ size, cell fate, and carcinogenesis. Gastroenterology. 2017;152:533–545. doi: 10.1053/j.gastro.2016.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Camargo FD, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 222.Karpowicz P, Perez J, Perrimon N. The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration. Development. 2010;138:4135–4145. doi: 10.1242/dev.060483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Mo J-S, Park HW, Guan K-L. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014;15:642–656. doi: 10.15252/embr.201438638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Lian I, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hasegawa T, et al. Transient inflammatory response mediated by interleukin-1β is required for proper regeneration in zebrafish fin fold. Elife. 2017;6:e22716. doi: 10.7554/eLife.22716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Li J, et al. Expression analysis of Hsp90α and cytokines in zebrafish caudal fin regeneration. Dev. Comp. Immunol. 2021;116:103922. doi: 10.1016/j.dci.2020.103922. [DOI] [PubMed] [Google Scholar]
- 227.Nguyen-Chi M, et al. TNF signaling and macrophages govern fin regeneration in zebrafish larvae. Cell Death Dis. 2017;8:e2979. doi: 10.1038/cddis.2017.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Tornini VA, et al. Live monitoring of Blastemal cell contributions during appendage regeneration. Curr. Biol. 2016;26:2981–2991. doi: 10.1016/j.cub.2016.08.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132:5173–5183. doi: 10.1242/dev.02101. [DOI] [PubMed] [Google Scholar]
- 230.Shibata E, Liu Z, Kawasaki T, Sakai N, Kawakami A. Robust and local positional information within a fin ray directs fin length during zebrafish regeneration. Dev. Growth, Differ. 2018;60:354–364. doi: 10.1111/dgd.12558. [DOI] [PubMed] [Google Scholar]
- 231.Nechiporuk A, Keating MT. A proliferation gradient between proximal and msxb-expressing distal blastema directs zebrafish fin regeneration. Development. 2002;129:2607–2617. doi: 10.1242/dev.129.11.2607. [DOI] [PubMed] [Google Scholar]
- 232.Mateus R, et al. Control of tissue growth by Yap relies on cell density and F-actin in zebrafish fin regeneration. J. Cell Sci. 2015;128:e1.2–e1.2. doi: 10.1242/dev.119701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Brandão AS, et al. Yap induces osteoblast differentiation by modulating Bmp signalling during zebrafish caudal fin regeneration. J. Cell Sci. 2019;132:jcs231993. doi: 10.1242/jcs.231993. [DOI] [PubMed] [Google Scholar]
- 234.Moro A, et al. MicroRNA-dependent regulation of biomechanical genes establishes tissue stiffness homeostasis. Nat. Cell Biol. 2019;21:348–358. doi: 10.1038/s41556-019-0272-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Stewart S, Stankunas K. Limited dedifferentiation provides replacement tissue during zebrafish fin regeneration. Dev. Biol. 2012;365:339–349. doi: 10.1016/j.ydbio.2012.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Tu S, Johnson SL. Fate restriction in the growing and regenerating zebrafish fin. Dev. Cell. 2011;20:725–732. doi: 10.1016/j.devcel.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Knopf F, et al. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev. Cell. 2011;20:713–724. doi: 10.1016/j.devcel.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 238.Sousa S, et al. Differentiated skeletal cells contribute to blastema formation during zebrafish fin regeneration. Development. 2011;138:3897–3905. doi: 10.1242/dev.064717. [DOI] [PubMed] [Google Scholar]
- 239.Stewart S, Gomez AW, Armstrong BE, Henner A, Stankunas K. Sequential and opposing activities of Wnt and BMP coordinate zebrafish bone regeneration. Cell Rep. 2014;6:482–498. doi: 10.1016/j.celrep.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Smith A, Avaron F, Guay D, Padhi BK, Akimenko MA. Inhibition of BMP signaling during zebrafish fin regeneration disrupts fin growth and scleroblast differentiation and function. Dev. Biol. 2006;299:438–454. doi: 10.1016/j.ydbio.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 241.Murciano C, et al. Ray–interray interactions during fin regeneration of Danio rerio. Dev. Biol. 2002;252:214–224. doi: 10.1006/dbio.2002.0848. [DOI] [PubMed] [Google Scholar]
- 242.Halder G, Dupont S, Piccolo S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat. Rev. Mol. Cell Biol. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- 243.Luo Q, et al. Connective tissue growth factor (CTGF) is regulated by Wnt and bone morphogenetic proteins signaling in osteoblast differentiation of mesenchymal stem cells. J. Biol. Chem. 2004;279:55958–55968. doi: 10.1074/jbc.M407810200. [DOI] [PubMed] [Google Scholar]
- 244.Kegelman CD, et al. Skeletal cell YAP and TAZ combinatorially promote bone development. FASEB J. 2018;32:2706–2721. doi: 10.1096/fj.201700872R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Pan, J.-X. et al. YAP promotes osteogenesis and suppresses adipogenic differentiation by regulating β-catenin signaling. Bone Res.6, 18 (2018). [DOI] [PMC free article] [PubMed]
- 246.Seo E, et al. SOX2 regulates YAP1 to maintain stemness and determine cell fate in the osteo-adipo lineage. Cell Rep. 2013;3:2075–2087. doi: 10.1016/j.celrep.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Xiong J, Almeida M, O’Brien CA. The YAP/TAZ transcriptional co-activators have opposing effects at different stages of osteoblast differentiation. Bone. 2018;112:1–9. doi: 10.1016/j.bone.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Rinkevich Y, Lindau P, Ueno H, Longaker MT, Weissman IL. Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature. 2011;476:409–414. doi: 10.1038/nature10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Grusche FA, Degoutin JL, Richardson HE, Harvey KF. The Salvador/Warts/Hippo pathway controls regenerative tissue growth in Drosophila melanogaster. Dev. Biol. 2011;350:255–266. doi: 10.1016/j.ydbio.2010.11.020. [DOI] [PubMed] [Google Scholar]
- 250.Nuñez VA, et al. Postembryonic development of the posterior lateral line in the zebrafish. Evol. Dev. 2009;11:391–404. doi: 10.1111/j.1525-142X.2009.00346.x. [DOI] [PubMed] [Google Scholar]
- 251.Alexandre D, Ghysen A. Somatotopy of the lateral line projection in larval zebrafish. Proc. Natl. Acad. Sci. USA. 1999;96:7558–7562. doi: 10.1073/pnas.96.13.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ledent V. Postembryonic development of the posterior lateral line in zebrafish. Development. 2002;129:597–604. doi: 10.1242/dev.129.3.597. [DOI] [PubMed] [Google Scholar]
- 253.Sarrazin AF, Nuñez VA, Sapède D, Tassin V, Dambly-Chaudière C. Origin and early development of the posterior lateral line system of zebrafish. J. Neurosci. 2010;30:8234–8244. doi: 10.1523/JNEUROSCI.5137-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Lush ME, Piotrowski T. Sensory hair cell regeneration in the zebrafish lateral line. Dev. Dyn. 2014;243:1187–1202. doi: 10.1002/dvdy.24167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Lecaudey V, Cakan-Akdogan G, Norton WHJ, Gilmour D. Dynamic Fgf signaling couples morphogenesis and migration in the zebrafish lateral line primordium. Development. 2008;135:2695–2705. doi: 10.1242/dev.025981. [DOI] [PubMed] [Google Scholar]
- 256.Atkinson PJ, Huarcaya Najarro E, Sayyid ZN, Cheng AG. Sensory hair cell development and regeneration: similarities and differences. Development. 2015;142:1561–1571. doi: 10.1242/dev.114926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Kim N-G, Koh E, Chen X, Gumbiner BM. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc. Natl. Acad. Sci. USA. 2011;108:11930–11935. doi: 10.1073/pnas.1103345108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Benham-Pyle BW, Pruitt BL, Nelson WJ. Mechanical strain induces E-cadherin-dependent Yap1 and β-catenin activation to drive cell cycle entry. Science. 2015;348:1024–1027. doi: 10.1126/science.aaa4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Kozlovskaja-Gumbriene A, et al. Proliferation-independent regulation of organ size by Fgf/Notch signaling. Elife. 2017;6:e21049. doi: 10.7554/eLife.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Loh SL, et al. Zebrafish yap1 plays a role in differentiation of hair cells in posterior lateral line. Sci. Rep. 2014;4:1–9. doi: 10.1038/srep04289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Agarwala S, et al. Amotl2a interacts with the Hippo effector Yap1 and the Wnt/β-catenin effector Lef1 to control tissue size in zebrafish. Elife. 2015;4:e08201. doi: 10.7554/eLife.08201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Wang, X. et al. Lpar2b controls lateral line tissue size by regulating Yap1 activity in zebrafish. Front. Mol. Neurosci.11, 34 (2018). [DOI] [PMC free article] [PubMed]
- 263.Pistocchi, A. et al. The zebrafish prospero homolog prox1 is required for mechanosensory hair cell differentiation and functionality in the lateral line. BMC Dev. Biol. 9, 58 (2009). [DOI] [PMC free article] [PubMed]
- 264.Yung YC, Stoddard NC, Chun J. LPA receptor signaling: pharmacology, physiology, and pathophysiology. J. Lipid Res. 2014;55:1192–1214. doi: 10.1194/jlr.R046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Yu F-X, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Okudaira S, Yukiura H, Aoki J. Biological roles of lysophosphatidic acid signaling through its production by autotaxin. Biochimie. 2010;92:698–706. doi: 10.1016/j.biochi.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 267.Lai S-L, et al. Autotaxin/Lpar3 signaling regulates Kupffer’s vesicle formation and left-right asymmetry in zebrafish. Development. 2012;139:4439–4448. doi: 10.1242/dev.081745. [DOI] [PubMed] [Google Scholar]
- 268.Frisca, F. et al. Role of ectonucleotide pyrophosphatase/phosphodiesterase 2 in the midline axis formation of zebrafish. Nature6, 37678 (2016). [DOI] [PMC free article] [PubMed]
- 269.Yukiura H, et al. Autotaxin regulates vascular development via multiple Lysophosphatidic Acid (LPA) receptors in zebrafish. J. Biol. Chem. 2011;286:43972–43983. doi: 10.1074/jbc.M111.301093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Lee S-J, et al. LPA1 is essential for lymphatic vessel development in zebrafish. FASEB J. 2008;22:3706–3715. doi: 10.1096/fj.08-106088. [DOI] [PubMed] [Google Scholar]
- 271.Stone LS. The development of lateral-line sense organs in amphibians observed in living and vital-stained preparations. J. Comp. Neurol. 1933;57:507–540. [Google Scholar]
- 272.Stone LS. Further experimental studies of the development of lateral-line sense organs in amphibians observed in living preparations. J. Comp. Neurol. 1937;68:83–115. [Google Scholar]
- 273.Dijkgraaf S. The functional and significance of the lateral-line organs. Biol. Rev. 1962;38:51–105. doi: 10.1111/j.1469-185x.1963.tb00654.x. [DOI] [PubMed] [Google Scholar]
- 274.Ma EY, Rubel EW, Raible DW. Notch signaling regulates the extent of hair cell regeneration in the zebrafish lateral line. J. Neurosci. 2008;28:2261–2273. doi: 10.1523/JNEUROSCI.4372-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Namdaran P, Reinhart KE, Owens KN, Raible DW, Rubel EW. Identification of modulators of hair cell regeneration in the zebrafish lateral line. J. Neurosci. 2012;32:3516–3528. doi: 10.1523/JNEUROSCI.3905-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.López-Schier H, Hudspeth AJ. A two-step mechanism underlies the planar polarization of regenerating sensory hair cells. Proc. Natl. Acad. Sci. USA. 2006;103:18615–18620. doi: 10.1073/pnas.0608536103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Ye Z, et al. Yap-lin28a axis targets let7-Wnt pathway to restore progenitors for initiating regeneration. Elife. 2020;9:e55771. doi: 10.7554/eLife.55771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Aman A, Piotrowski T. Wnt/β-catenin and Fgf signaling control collective cell migration by restricting chemokine receptor expression. Dev. Cell. 2008;15:749–761. doi: 10.1016/j.devcel.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 279.Jiang, L., Romero-Carvajal, A., Haug, J. S., Seidel, C. W. & Piotrowski, T. Gene-expression analysis of hair cell regeneration in the zebrafish lateral line. Proc. Natl. Acad. Sci. USA111, E1383–E1392 (2014). 10.1073/pnas.1402898111 [DOI] [PMC free article] [PubMed]
- 280.Giffen, K. P., Liu, H., Kramer, K. L. & He, D. Z. Expression of protein-coding gene orthologs in zebrafish and mouse inner ear non-sensory supporting cells. Front. Neurosci. 13, 1117 (2019). [DOI] [PMC free article] [PubMed]
- 281.Jin J, et al. Evidence that Lin28 stimulates translation by recruiting RNA helicase A to polysomes. Nucleic Acids Res. 2011;39:3724–3734. doi: 10.1093/nar/gkq1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Sci. (80-.) 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 283.Zhang J, et al. LIN28 regulates stem cell metabolism and conversion to primed pluripotency. Cell Stem Cell. 2016;19:66–80. doi: 10.1016/j.stem.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 284.Golden EJ, Benito-Gonzalez A, Doetzlhofer A. The RNA-binding protein LIN28B regulates developmental timing in the mammalian cochlea. Proc. Natl. Acad. Sci. USA. 2015;112:E3864–E3873. doi: 10.1073/pnas.1501077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Ramachandran R, Fausett BV, Goldman D. Ascl1a regulates Müller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat. Cell Biol. 2010;12:1101–1107. doi: 10.1038/ncb2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Corey DP, et al. TRPA1 is a candidate for the mechanosensitive transduction channel of vertebrate hair cells. Nature. 2004;432:723–730. doi: 10.1038/nature03066. [DOI] [PubMed] [Google Scholar]
- 287.Siemens J, et al. Cadherin 23 is a component of the tip link in hair-cell stereocilia. Nature. 2004;428:950–955. doi: 10.1038/nature02483. [DOI] [PubMed] [Google Scholar]
- 288.Söllner C, et al. Mutations in cadherin 23 affect tip links in zebrafish sensory hair cells. Nature. 2004;428:955–959. doi: 10.1038/nature02484. [DOI] [PubMed] [Google Scholar]
- 289.Santos F, Macdonald G, Rubel EW, Raible DW. Lateral line hair cell maturation is a determinant of aminoglycoside susceptibility in zebrafish (Danio rerio) Hear. Res. 2006;213:25–33. doi: 10.1016/j.heares.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 290.Buck LMJ, Winter MJ, Redfern WS, Whit TT. Ototoxin-induced cellular damage in neuromasts disrupts lateral line function in larval zebra fish. Hear. Res. 2012;284:67–81. doi: 10.1016/j.heares.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 291.Forbes SJ, Newsome PN. Liver regeneration—mechanisms and models to clinical application. Nat. Rev. Gastroenterol. Hepatol. 2016;13:473–485. doi: 10.1038/nrgastro.2016.97. [DOI] [PubMed] [Google Scholar]
- 292.Wang, S., Miller, S. R., Ober, E. A. & Sadler, K. C. Making it new again: insight into liver development, regeneration, and disease from zebrafish. Curr. Top. Dev. Biol. 124, 161–195 (2017). [DOI] [PMC free article] [PubMed]
- 293.Wang C, et al. Differences in Yes-associated protein and mRNA levels in regenerating liver and hepatocellular carcinoma. Mol. Med. Rep. 2012;5:410–414. doi: 10.3892/mmr.2011.640. [DOI] [PubMed] [Google Scholar]
- 294.Grijalva JL, et al. Dynamic alterations in Hippo signaling pathway and YAP activation during liver regeneration. Am. J. Physiol. Gastrointest. Liver Physiol. 2014;307:G196–G204. doi: 10.1152/ajpgi.00077.2014. [DOI] [PubMed] [Google Scholar]
- 295.Bai H, et al. Yes-associated protein regulates the hepatic response after bile duct ligation. Hepatology. 2012;56:1097–1107. doi: 10.1002/hep.25769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Aloia L, et al. Epigenetic remodelling licences adult cholangiocytes for organoid formation and liver regeneration. Nat. Cell Biol. 2019;21:1321–1333. doi: 10.1038/s41556-019-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Li W, et al. A homeostatic arid1a-dependent permissive chromatin state licenses hepatocyte responsiveness to liver-injury-associated YAP signaling. Cell Stem Cell. 2019;25:54–68. doi: 10.1016/j.stem.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 298.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Sadler KC, Amsterdam A, Soroka C, Boyer J, Hopkins N. A genetic screen in zebrafish identifies the mutants vps18, nf2 and foie gras as models of liver disease. Development. 2005;132:3561–3572. doi: 10.1242/dev.01918. [DOI] [PubMed] [Google Scholar]
- 300.Cox AG, et al. Yap reprograms glutamine metabolism to increase nucleotide biosynthesis and enable liver growth. Nat. Cell Biol. 2016;18:886–896. doi: 10.1038/ncb3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Brandt ZJ, Echert AE, Bostrom JR, North PN, Link BA. Core Hippo pathway components act as a brake on Yap/Taz in the development and maintenance of the biliary network. Development. 2020;4:dev.184242. doi: 10.1242/dev.184242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Driskill JH, Pan D. The Hippo pathway in liver homeostasis and pathophysiology. Annu. Rev. Pathol. Mech. Dis. 2021;16:299–322. doi: 10.1146/annurev-pathol-030420-105050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Yimlamai D, et al. Hippo pathway activity influences liver cell fate. Cell. 2014;157:1324–1338. doi: 10.1016/j.cell.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Molina, L. M. et al. Compensatory hepatic adaptation accompanies permanent absence of intrahepatic biliary network due to YAP1 loss in liver progenitors. Cell Rep. 36, 109310 (2021). [DOI] [PMC free article] [PubMed]
- 305.Verboven E, et al. Regeneration defects in Yap and Taz mutant mouse livers are caused by bile duct disruption and cholestasis. Gastroenterology. 2021;160:847–862. doi: 10.1053/j.gastro.2020.10.035. [DOI] [PubMed] [Google Scholar]
- 306.MacCarthy-Morrogh L, Martin P. The hallmarks of cancer are also the hallmarks of wound healing. Sci. Signal. 2020;13:eaay8690. doi: 10.1126/scisignal.aay8690. [DOI] [PubMed] [Google Scholar]
- 307.Moroishi T, Gram Hansen C, Guan K-L. The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer. 2015;15:73–79. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat. Rev. Drug Discov. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Merkes C, et al. Ewing sarcoma Ewsa protein regulates chondrogenesis of Meckel’s Cartilage through modulation of Sox9 in zebrafish. PLoS One. 2015;10:e0116627. doi: 10.1371/journal.pone.0116627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Mayrhofer M, et al. A novel brain tumour model in zebrafish reveals the role of YAP activation in MAPK- and PI3K-induced malignant growth. Dis. Model. Mech. 2017;10:15–28. doi: 10.1242/dmm.026500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Brandt ZJ, North PN, Link BA. Somatic mutations of lats2 cause peripheral nerve sheath tumors in zebrafish. Cells. 2019;8:972. doi: 10.3390/cells8090972. [DOI] [PMC free article] [PubMed] [Google Scholar]