Significance
Sulfoquinovose, a sulfosugar derivative of glucose, is produced by most photosynthetic organisms and contains up to half of all sulfur in the biosphere. Several pathways for its breakdown are known, though they provide access to only half of the carbon in sulfoquinovose and none of its sulfur. Here, we describe a fundamentally different pathway within the plant pathogen Agrobacterium tumefaciens that features oxidative desulfurization of sulfoquinovose to access all carbon and sulfur within the molecule. Biochemical and structural analyses of the pathway’s key proteins provided insights how the sulfosugar is recognized and degraded. Genes encoding this sulfoquinovose monooxygenase pathway are present in many plant pathogens and symbionts, alluding to a possible role for sulfoquinovose in plant host–bacteria interactions.
Keywords: carbohydrate metabolism, sulfur cycle, oxidative desulfurization
Abstract
Catabolism of sulfoquinovose (SQ; 6-deoxy-6-sulfoglucose), the ubiquitous sulfosugar produced by photosynthetic organisms, is an important component of the biogeochemical carbon and sulfur cycles. Here, we describe a pathway for SQ degradation that involves oxidative desulfurization to release sulfite and enable utilization of the entire carbon skeleton of the sugar to support the growth of the plant pathogen Agrobacterium tumefaciens. SQ or its glycoside sulfoquinovosyl glycerol are imported into the cell by an ATP-binding cassette transporter system with an associated SQ binding protein. A sulfoquinovosidase hydrolyzes the SQ glycoside and the liberated SQ is acted on by a flavin mononucleotide-dependent sulfoquinovose monooxygenase, in concert with an NADH-dependent flavin reductase, to release sulfite and 6-oxo-glucose. An NAD(P)H-dependent oxidoreductase reduces the 6-oxo-glucose to glucose, enabling entry into primary metabolic pathways. Structural and biochemical studies provide detailed insights into the recognition of key metabolites by proteins in this pathway. Bioinformatic analyses reveal that the sulfoquinovose monooxygenase pathway is distributed across Alpha- and Betaproteobacteria and is especially prevalent within the Rhizobiales order. This strategy for SQ catabolism is distinct from previously described pathways because it enables the complete utilization of all carbons within SQ by a single organism with concomitant production of inorganic sulfite.
Sulfoquinovose (SQ; 6-deoxy-6-sulfoglucose) is an anionic sulfosugar found in plant and cyanobacterial sulfolipids and in S-layer proteins in archaea (1). It is estimated that SQ holds around half of all sulfur in the biosphere, with 10 billion tons produced each year in nature, and so its cycling is a significant component of the biogeochemical sulfur cycle (2). SQ is primarily found as the headgroup of the plant sulfolipid sulfoquinovosyl diacylglycerol, and its close association with photosynthetic membranes and proteins supports roles in their structure and function (3). Microbial communities play a dominant role in SQ cycling, and usually more than one tier of organism is required to completely assimilate this source of carbon and sulfur. Organisms with a tier 1 sulfoglycolysis pathway perform scission of the C3–C4 bond of SQ to give two three-carbon fragments; carbons 1 to 3 enter central metabolism, while carbons 4 to 6 bearing the sulfonate are excreted as dihydroxypropanesulfonate (DHPS) or sulfolactate (SL). Syntrophic organisms with a tier 2 pathway process DHPS and SL to access the remaining three-carbon fragment and release inorganic sulfur. To date, three tier 1 sulfoglycolysis pathways have been described: the sulfoglycolytic Embden-Meyerhof-Parnas (4), sulfoglycolytic Entner-Doudoroff (sulfo-ED) (5, 6), and sulfoglycolytic sulfofructose transaldolase pathways (7, 8). Tier 2 metabolism has been described for various specialized bacteria that "biomineralize" SL or DHPS to release inorganic sulfite, which under aerobic conditions is readily oxidized to sulfate (1). While many of the steps in the three sulfoglycolysis pathways differ, all three pathways share the presence of a specialized glycoside hydrolase, a sulfoquinovosidase (SQase), which catalyzes the hydrolysis of SQ glycosides, such as sulfoquinovosyl glycerol (SQGro), to release SQ (9, 10).
While the tier 1 and 2 pathways described to date require two or more organisms to complete the “biomineralization” of SQ, there is some evidence that this can also be accomplished by a single organism. Roy et al. have reported that an Agrobacterium strain from soil can completely consume SQ, with release of sulfate, although the genetic and biochemical details behind this process were not investigated (11). We previously reported that Agrobacterium tumefaciens C58 contains a functional SQase, with the ability to hydrolyze SQGro (9). However, analysis of its genome did not reveal any genes homologous to those expected for known sulfoglycolysis pathways.
Here, we investigate the “biomineralization” of SQ by A. tumefaciens (Agrobacterium fabrum) strain C58 and show that this organism effects the oxidoreductive desulfurization of SQ to release inorganic sulfite and glucose, which can feed into primary metabolism. We show that this pathway involves an SQ/SQGro solute-binding protein and associated ATP-binding cassette (ABC) transporter; an SQase to release SQ from its glycosides; a flavin-dependent SQ monooxygenase with paired flavin reductase to effect oxidative desulfurization of SQ to sulfite and 6-oxo-glucose (6-OG); and an NADPH-dependent oxidoreductase to reduce 6-OG to glucose. X-ray structures determined for each of these proteins in complex with relevant metabolites reveal the molecular basis of substrate binding and catalysis. We show through bioinformatics analyses that this pathway, which enables the complete assimilation of SQ, is distributed across Alpha- and Betaproteobacteria and is particularly well represented within the Rhizobiales order.
Results
Differential Expression of a Gene Cluster in the Presence SQ.
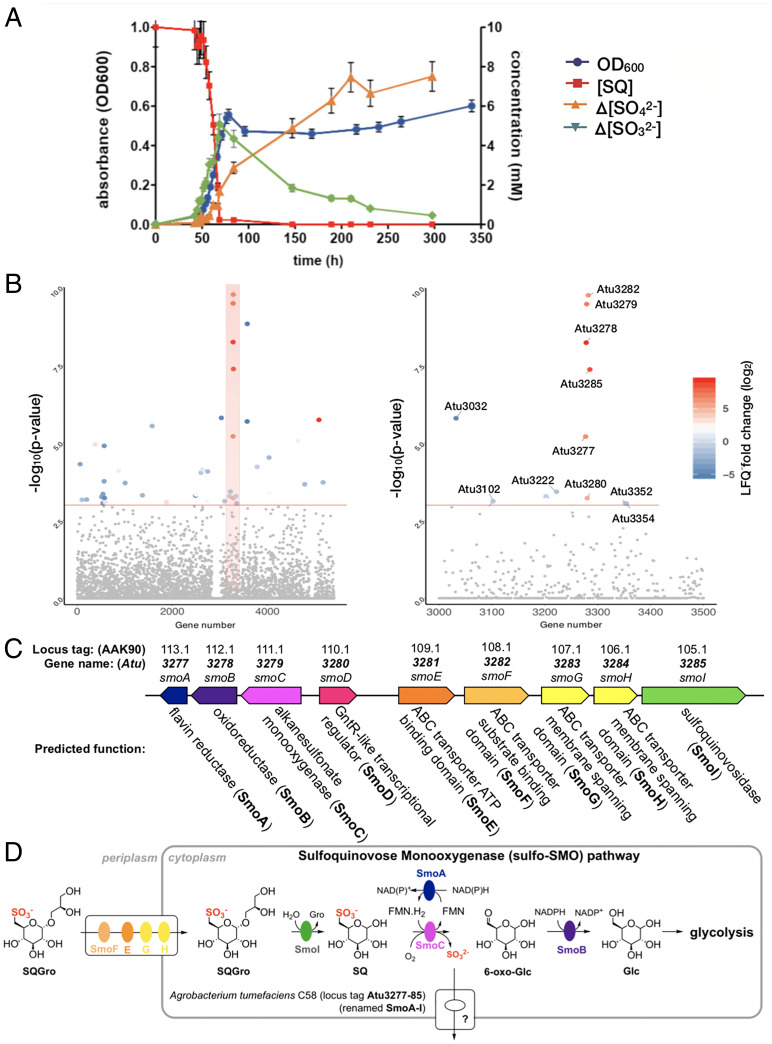
To determine if A. tumefaciens C58 can utilize SQ as a carbon source, we attempted to grow this organism in M9 minimal media containing SQ as the sole carbon source. A. tumefaciens C58 exhibited robust growth in this media, and analysis of spent culture supernatant failed to detect DHPS or SL. Instead, the culture supernatant accumulated sulfate, but with a lag between consumption of SQ and sulfate release (Fig. 1A and SI Appendix, Fig. S1), as was previously reported by Roy et al. for Agrobacterium sp. strain ABR2 (11). Noting that sulfite is generally released from organosulfonate degradation pathways (1, 12), we analyzed the supernatant for sulfite (SO32−) and observed that SQ consumption is coincident with production of sulfite, which slowly undergoes autooxidation to sulfate. To investigate the metabolism of the carbon skeleton of SQ, we cultured A. tumefaciens on 13C6-SQ (13) and analyzed the culture supernatant using 13C nuclear magnetic resonance spectroscopy (SI Appendix, Fig. S2). The only significant 13C-labeled product we could detect was 13C-bicarbonate, which formed transiently during exponential phase growth, and the 13C-labeled bicarbonate signal disappeared at stationary phase, presumably through exchange with atmospheric CO2. A. tumefaciens grew on other sulfoquinovosides, including SQGro and methyl α-sulfoquinovoside (MeSQ), but did not grow on other alkylsulfonates including DHPS, SL, sulfoacetic acid, taurine, pentanesulfonate, 2-(N-morpholino)ethanesulfonic acid (MES), 3-(N-morpholino)propanesulfonic acid (MOPS), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), cysteic acid, or methanesulfonic acid (SI Appendix, Fig. S3). Collectively, these data demonstrate that A. tumefaciens effects the complete metabolism of the carbon backbone of SQ with concomitant release of sulfite.
Fig. 1.
A. tumefaciens utilizes SQ and its glycosides as a carbon source. (A) Optical density of A. tumefaciens C58 culture (blue) and concentration of SQ ([SQ]) (red), change in concentration of sulfite (Δ[sulfite]) (green) and change in concentration of sulfate (Δ[sulfate]) (yellow), with respect to time. This data are representative of two independent experiments (SI Appendix, Fig. S1); error bars denote observational error (derived by propagation of estimated random errors). (B) Manhattan plot of comparative proteomics data for A. tumefaciens C58 grown on SQ versus glucose, demonstrating that the most heavily up-regulated proteins belong to a single gene cluster. (C) An illustration of the up-regulated cluster with automated annotations for each of the gene products. These would later be renamed smoABCDEFGHI to reflect the importance of the sulfoquinovose monooxygenase enzyme activity to this biochemical pathway. (D) An illustratation of the hypothetical roles played by the gene products of this pathway to complete the catabolism of SQGro.
We performed comparative proteomic experiments to identify changes associated with the growth of A. tumefaciens on SQ compared to glucose at midlog phase (Fig. 1B). The largest and most significant change we observed was an increase in the abundance of proteins encoded by a single cluster of genes (Atu3277-Atu3285) for cells grown on SQ. Proteins encoded by Atu3283 and Atu3284 were not observed; however, they are predicted to be integral membrane proteins that can be difficult to detect using conventional proteomic workflows (14). Thus, the gene cluster Atu3277-Atu3285, which was subsequently renamed smoA-smoI, appeared to be important for growth on SQ (Fig. 1C). While the protein encoded by Atu3285 was previously identified as an SQase (9), the proteins encoded by other genes in the cluster were not annotated with functions that were consistent with any sulfoglycolysis pathway, suggesting that A. tumefaciens uses a different approach for the catabolism of SQ. The automated annotations ascribed to the respective gene products in the cluster, which included a putative ABC transporter system, sulfonate monooxygenase, short-chain dehydrogenase/reductase (SDR) oxidoreductase, flavin reductase, and exporters, enabled development of a hypothetical biochemical pathway that could explain the complete assimilation of SQ by A. tumefaciens (Fig. 1D). We proceeded to biochemically validate this hypothesis and gain structural insights into the proteins involved.
Atu3282 (smoF) Encodes an ABC Transporter Solute-binding Protein that Binds SQGro.
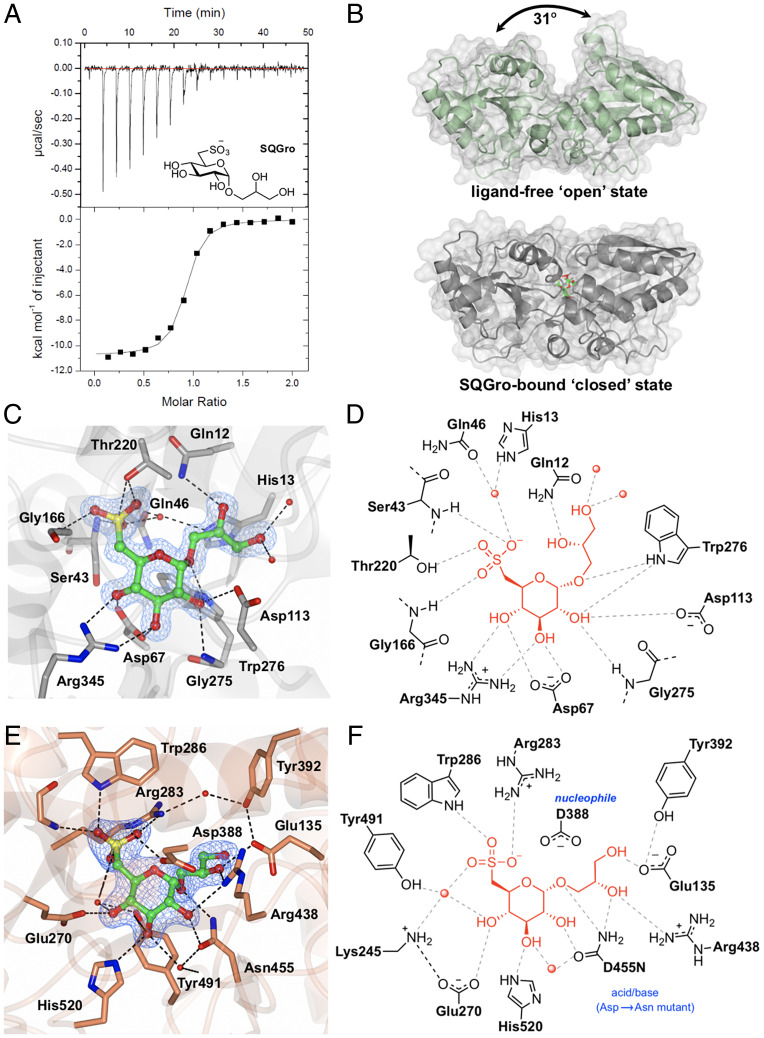
Within the gene cluster identified through proteomics, Atu3281 (smoE), Atu3283 (smoG), and Atu3284 (smoH) were annotated as an ABC transporter system, with Atu3282 (smoF) encoding an associated periplasmic solute-binding protein. The substrate preferences of solute-binding proteins are useful for assigning functions to their associated ABC transporters (15). Accordingly, we produced recombinant SmoF (SI Appendix, Fig. S4) and demonstrated that it binds SQGro with Kd = 0.29 ± 0.17 μM (ΔH = −11 ± 0.4 kcal mol−1, ΔS = −7 ± 2 cal mol−1 deg−1) (Fig. 2A and SI Appendix, Fig. S5 and Table S3). No binding was observed for the stereochemically related monosaccharides D-glucose and D-glucuronic acid.
Fig. 2.
Biochemical and structural analyses of the SQGro-binding protein SmoF (Atu3282) and SQase SmoI (Atu3285). (A) Isothermal titration calorimogram for SmoF titrated against its cognate ligand 2’R-SQGro. The data are representative of two independent experiments (SI Appendix, Fig. S5). (B) Ribbon diagrams (with transparent surface) for the open and closed (liganded) conformations of SmoF. The 2’R-SQGro is bound tightly in the interdomain cleft and is inaccessible to the bulk solvent in the closed conformation. (C) Interactions between protein and ligand within the SmoF•2’R-SQGro complex: SmoF is in gray, 2’R-SQGro is in green, and the 2Fo − Fc map at 1.5σ is in blue. (D) An illustration highlighting key interactions from C. (E) Interactions between protein and ligand within the complex pf SmoI-D455N SQase and 2’R-SQGro: SmoI is in gold, 2’R-SQGro is in green, and the 2Fo − Fc map at 1.5σ is in blue. (F) An illustration highlighting key interactions from E: red spheres represent ordered water molecules; dotted lines represent proposed hydrogen bonds.
To delineate how SmoF recognizes its ligand, we used X-ray diffraction methods to obtain a high-resolution three-dimensional (3D) structure of SmoF in its ligand-free apo state and in complex with SQGro (Fig. 2B and SI Appendix, Table S4). Like most ABC transporter solute-binding proteins, SmoF possesses two globular domains with a similar α/β fold forming a deep cleft lined with aromatic and polar residues to capture the ligand. Comparisons of the structures for ligand-free SmoF and the SQGro complex revealed a large conformational change in the protein resulting from interdomain rotation upon SQGro binding. The relative movement of domains was assessed using the DynDom server, which indicated a hinge rotation of 31° about four linker regions connecting the two domains (SI Appendix, Fig. S6). SQGro is buried deep within the interdomain cleft, and residues from both domains accommodate this ligand through a network of hydrogen-bonding interactions (Fig. 2 C and D). The sulfonate of SQGro, which is the defining feature of this sulfosugar, is accommodated by hydrogen bonds to the sidechain of Thr220 (2.6 Å), backbone amides of Gly166 (3 Å) and Ser43 (2.8 Å), and an ordered water molecule that in turn hydrogen bonds to the sidechain of His13 (3 Å) and Gln46 (3.2 Å) (Fig. 2 C and D). These and the other interactions in the SQGro-bound “closed” state stabilized SmoF substantially, as evidenced by a 15 °C increase in the protein melting temperature (SI Appendix, Fig. S7).
The Structural Basis of SQGro Recognition by the SQase Atu3285 (SmoI).
We previously reported that Atu3285 (smoI) encodes an SQase that preferentially hydrolyzes 2'R-SQGro, the natural stereoisomer of this glycoside (9). To understand the molecular basis of the preference SmoI has for this stereoisomer, we determined the 3D structure of a pseudo-Michaelis complex: the inactive acid/base mutant SmoI-D455N in complex with 2'R-SQGro (Fig. 2 E and F). SmoI-D455N•SQGro crystallized with four protomers in the asymmetric unit, each showing unambiguous density of the substrate bound at the active site. As described previously, the overall fold is an (ɑ/β)8 barrel appended with small β sheet domain and the sulfonate group is recognized by the Arg283/Trp286/Tyr491 triad8. Arg438 and Glu135 make hydrogen-bonding interactions with the glyceryl aglycone of 2'R-SQGro. Only Arg438 interacts with the C2-hydroxyl group of the glyceryl aglycone, and thus this residue appears to drive selectivity for the 2'R-SQGro stereoisomer.
Atu3277 (smoA) Encodes a Flavin Mononucleotide Reductase.
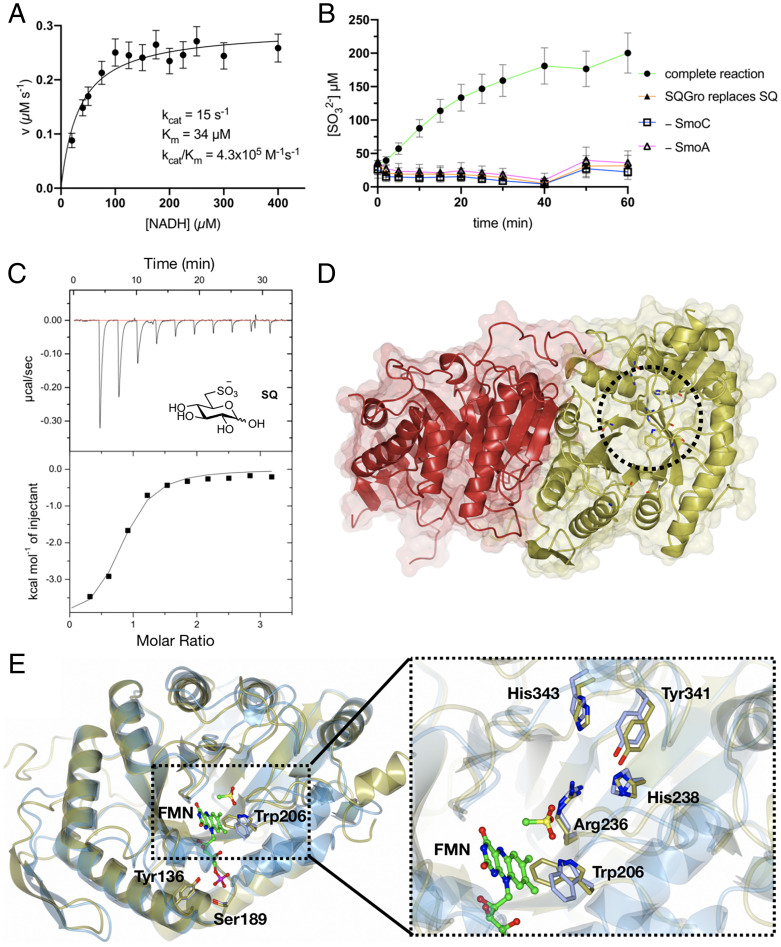
SmoA, annotated as a flavin reductase, was recombinantly expressed in Escherichia coli and maintained a yellow color throughout purification, suggesting that it had copurified with a flavin cofactor. A sample of this protein was heat-denatured to release the cofactor and the supernatant analyzed by liquid chromatography–mass spectromtery (LC-MS) to reveal that flavin mononucleotide (FMN) was the sole detectable flavin (SI Appendix, Fig. S8). Michaelis-Menten kinetics were conducted for SmoA with saturating FMN and nicotinamide adenine dinucleotide (NADH) or nicotinamide adenine dinucleotide phosphate (NADPH) to determine which of these reductants was preferred by the enzyme. With NADH, the kinetic parameters were KM = 35 ± 5 µM, kcat = 14.5 ± 0.5 s−1, and kcat/KM = 4.1 × 105 M−1 s−1, while for NADPH, saturation was not observed and kcat/KM = 6.8 × 102 M−1 s−1, indicating that NADH is the preferred cofactor for SmoA (Fig. 3A and SI Appendix, Figs. S9 and S10). Owing to difficulties in obtaining structural data for this enzyme, we also studied a close homolog from Rhizobium oryzae (RoSmoA, UniProt accession number: A0A1X7D6Q3), which possesses a syntenic gene cluster to Atu3277-Atu3285. Recombinant RoSmoA also copurified with FMN (SI Appendix, Fig. S8) and utilized the NADH cofactor with KM = 16 ± 5 µM, kcat = 33 ± 2 s−1, and kcat/KM = 2.1 × 106 M−1 s−1 (SI Appendix, Fig. S9).
Fig. 3.
Biochemical and structural analyses of the flavin reductase SmoA and SQ monooxygenase SmoC. (A) Michaelis-Menten kinetics for SmoA-catalyzed reduction of FMN by NADH. The data are representative of two independent replicates (SI Appendix, Fig. S10); error bars denote observational errors (derived by propagation of estimated random error). (B) SmoC activity assessed using sulfite release assay with Ellman’s reagent in the presence of FMN, flavin reductase, NADH, and SQ. The data are representative of two independent experiments (SI Appendix, Fig. S11); error bars denote observational error (derived by propagation of estimated random errors). (C) Isothermal titration calorimogram of interaction of SmoC with SQ as determined by ITC. The data are representative of two independent experiments (SI Appendix, Fig. S13). (D) Transparent molecular surface and ribbon diagram of RoSmoC homodimer showing cofactor binding pocket and active site (dotted circle). (E) Alternative orientation of RoSmoC monomer (in gold) overlaid with the MsuD·FMN·CH3SO3− complex (7K14.pdb in ice blue) showing FMN from the latter. Expansion shows view of proposed substrate-binding pocket and conserved residues lining the active site of RoSmoC.
Atu3279 (smoC) Encodes an SQ Monooxygenase that Desulfurizes SQ.
SmoC is annotated as an alkanesulfonate monooxygenase, though it possesses only 30% sequence identity with the well-characterized alkanesulfonate monooxygenase SsuD, from E. coli. SsuD catalyzes the reduced flavin mononucleotide (FMNH2)- and O2-dependent oxidation of alkanesulfonates to produce the corresponding aldehyde and sulfite, with a preference for pentanesulfonate (16). The mechanism of this and related enzymes has been intensively studied yet remains enigmatic. The transformation is thought to involve initial formation of a C4a-peroxy or N5-peroxy flavin species on-enzyme. One mechanism posits that the terminal peroxide oxygen attacks the sulfonate sulfur of the substrate before undergoing a rearrangement to effect C-S bond fissure and release of the aldehyde and sulfite products (SI Appendix, Fig. S12A) (17). An alternative mechanism suggests the peroxide deprotonates C6, which is then oxidized to an α-hydroxysulfonate that undergoes elimination to produce sulfite and the aldehyde (SI Appendix, Fig. S12B) (18). To demonstrate activity for recombinant SmoC (SI Appendix, Fig. S4), we adapted assays developed for SsuD that use Ellman’s reagent to detect sulfite released by the enzyme (19). Direct detection of the putative sugar product, 6-OG, is not trivial, as this molecule exists as a complex equilibrium of (hemi)acetals and hydrates that have poor stability. Thus, SmoC was incubated with SQ in the presence of SmoA, FMN, and NADH, which generate FMNH2 in situ, and the concentration of sulfite determined periodically using Ellman’s reagent (Fig. 3B). Maximal substrate conversion was ∼200 µM (SI Appendix, Fig. S12C), which is commensurate with the solubility of molecular oxygen in water under standard conditions, with peak activity observed at pH 8.5 (SI Appendix, Fig. S12D). No activity was observed when SQ was replaced with other sulfonates, including SQGro (the precursor to SQ) or HEPES (an unrelated sulfonate) demonstrating that, unlike the promiscuous SsuD, SmoC has high specificity for SQ (SI Appendix, Fig. S12C). As such, the hydrolysis of SQGro by SmoI necessarily precedes oxidative desulfurization by SmoC. This observation is further supported by isothermal titration calorimetry (ITC), where SQ was found to bind SmoC with Kd = 3 µM in the absence of any flavin-based cofactors, whereas no binding was detected for SQGro (Fig. 3C and SI Appendix, Fig. S13 and Table S3). The unique SQ monooxygenase activity of SmoC defines this pathway: it is the enzyme that effects fissure of the C–S bond in SQ, and so it was chosen as the namesake for this gene cluster, and Atu3277-Atu3285 was renamed the SQ MonoOxygenase cluster (smoA-I).
While we could readily crystallize SmoC, these crystals only diffracted to a maximum resolution of 3.4 Å. The corresponding low-resolution map suggested that SmoC exists as a dimer, which was confirmed in solution by size exclusion chromatography–multiangle light scattering (SEC-MALS) (SI Appendix, Fig. S14). To obtain structural information for an SQ monooxygenase, we turned to the homolog from R. oryzae (RoSmoC). Recombinant RoSmoC exhibited similar activity and substrate selectivity for SQ to SmoC (SI Appendix, Fig. S12E) and provided crystals that diffracted to 1.9 Å. Importantly, the low-resolution structure of A. tumefaciens SmoC superimposed with the high-resolution RoSmoC structures with a peptide backbone rmsd of 0.4 Å across the entire structure, providing confidence that both enzymes shared a common structure and function (SI Appendix, Fig. S15). Both SQ monooxygenases consist of a core (α/β)8 triose-phosphate isomerase (TIM) barrel with three additional insertion regions, analogous to monooxygenases from the bacterial luciferase family. The protomers exist as a homodimer that buries 4,697 Å2 of surface area, amounting to 18% of total accessible surface area for each protomer (Fig. 3D). Pairwise structural analysis using the distance matrix alignment (DALI) server identified close relationships to a putative luciferase-like monooxygenase (3RAO.pdb) with an rmsd of 2.4 over 314 residues and a Z score of 34.3, the FMNH2-dependent methanesulfonate monooxygenase MsuD (7K14.pdb, rmsd 2.0/322 residues, Z score of 41.0), and the FMNH2-dependent alkanesulfonate monooxygenase SsuD (1M41.pdb, rmsd 1.8/317 residues, Z score of 41.2).
Comparisons of the RoSmoC structure with MsuD (7K14.pdb) in complex with FMN enabled identification of the FMN binding site for RoSmoC: a deep hydrophobic pocket that accommodates the isoalloxazine ring system and extends out to the protein-solvent interface, which is gated by conserved phosphate-binding residues Tyr136 and Ser189 (Fig. 3E) (18). The close structural and functional relationship of RoSmoC to MsuD is evident from the conservation of a putative sulfonate binding site comprised of the side-chains Trp206, Arg236, His238, Tyr341, and His343 (18). Aside from conferring these enzymes with an ability to bind sulfonates, these conserved active site residues have been suggested to contribute to the stabilization of a peroxyflavin intermediate in MsuD and SsuD (18, 19). Efforts to obtain crystals of a RoSmoC–SQ complex were unsuccessful, limiting further insights into the origin of enzyme specificity toward SQ over other sulfonates.
Atu3278 (smoB) Encodes an Oxidoreductase that Converts 6-OG to Glucose.
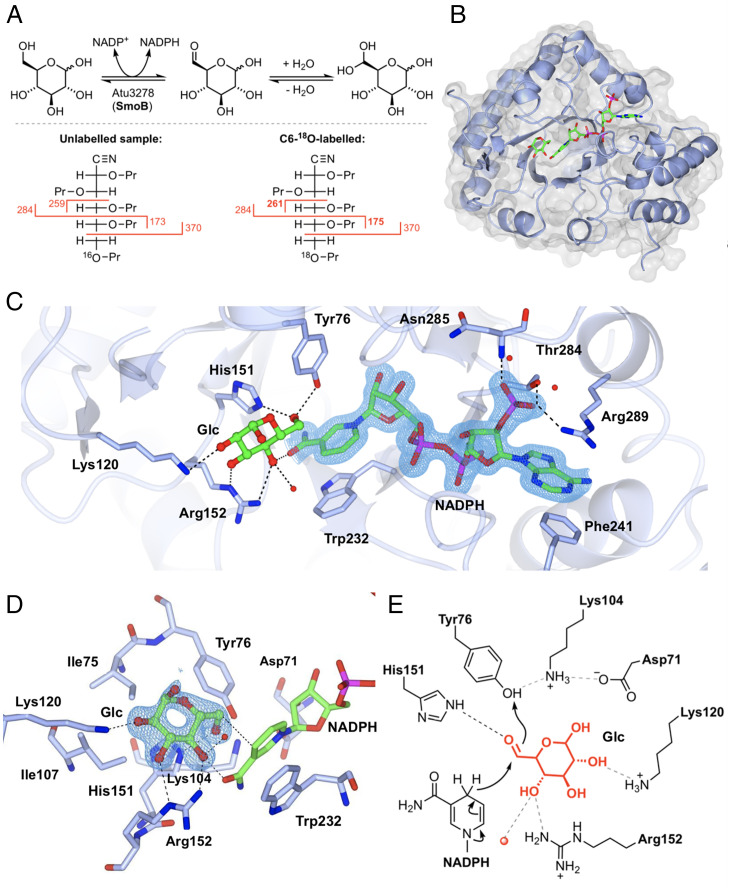
SmoB is annotated as an SDR, and we had hypothesized that it was responsible for reduction of 6-OG to glucose (Fig. 1D). Since 6-OG is difficult to study directly, we tested our hypothesis by looking for SmoB-mediated isotope incorporation into glucose at equilibrium (Fig. 4A). Assuming our hypothesis to be correct, and as a consequence of microscopic reversibility, incubation of SmoB with a nicotinamide cofactor and glucose in H218O should result in transient formation of 6-OG, rapid and reversible hydration/dehydration with H218O to compete-out 16O at C6 for 18O, and reduction to give 6-18O-glucose. In parallel to this process, 18O incorporation will occur at C1 of glucose through a similar series of hydration/dehydration reactions. Before proceeding with these experiments, we used ITC to establish which nicotamide cofactor was suitable for SmoB: NADPH bound to SmoB with Kd ∼2 µM, while no binding was observed for NADH (SI Appendix, Fig. S16 and Table S3). Thus, glucose pre-equilibrated in H218O was incubated with SmoB and NADP+ and then analyzed by mass spectrometry to reveal the formation of a product 4 Da greater in mass than glucose, presumably due to the incorporation of two 18O atoms into glucose. The crude reaction mixture was subjected to peracetylation (Ac2O/pyridine) and then LC-MS analysis to confirm that the +4 Da product coeluted with authentic D-glucose-pentaacetate (SI Appendix, Fig. S17). To determine that the 18O label was incorporated at C6 of glucose, we used electron-impact gas chromatography–mass spectrometry (GC-MS), which required conversion of the reaction product to the acyclic pentapropionate aldonitrile (SI Appendix, Fig. S18) (20). This approach provided diagnostic C1-C5 and C5-C6 fragment ions. The 18O-labeled product gave a C5-C6 fragment that was 2 mass units higher (m/z 173 versus 175), whereas the C1-C5 fragment was the same as unlabeled glucose reference (m/z 370), demonstrating that the 18O is incorporated at C6. Only enzymatic reactions conducted in the presence of NADP+ produced product labeled with 18O at C6: NAD+ failed to produce any product, supporting our observations by ITC and defining the cofactor specificity of SmoB.
Fig. 4.
Biochemical and structural analyses of 6-OG reductase SmoB. (A) Top: Equilibrium oxygen exchange at C-6 of Glc via 6-OG facilitated by SmoB when incubated with NADP+ in H218O. Bottom: Derivatization and MS fragmentation allows localization of 18O to C6 of Glc. (B) Transparent molecular surface and ribbon diagram of SmoB in complex with NADPH and Glc. (C) Closeup view of SmoB•NADPH•Glc ternary complex. Backbone and carbon atoms of SmoB are shown in ice blue, and NADPH and glucose are shown in cylinder format. Electron density for NADPH corresponds to the 2Fo − Fc map in blue at levels of 1σ. (D) Substrate-binding pocket of SmoB depicting hydrogen-bonding interactions of glucose with the active site residues, including the conserved catalytic residues Asp71, Lys-104, His151, and Tyr76. Electron density corresponds to the 2Fo − Fc map (in blue) at levels of 1σ. The geometry of the SmoB-Glc complex indicates the likely trajectory of hydride addition to 6-OG. (E) Proposed mechanism of SmoB catalyzed reduction of 6-OG by NADPH showing hydride transfer from C4 of nicotinamide ring of NADPH to C6 carbonyl and Y76 (within the catalytic tetrad) as the proton donor. The red sphere is a bound water molecule; dotted lines are proposed hydrogen bonds.
We determined the 3D structure of SmoB using X-ray diffraction methods. This initial structure revealed that SmoB exists as a compact trimer; however, the C-terminal His6-tag in this construct occupied the putative active site of adjoining subunits, making cocrystallization with cofactors difficult (SI Appendix, Fig. S19). To overcome this issue, SmoB was subcloned into a different vector and expressed with a cleavable N-terminal purification tag. This protein maintained the same catalytic activity, and SEC-MALS confirmed it remained a trimer in solution (SI Appendix, Fig. S20). This SmoB construct was cocrystallized with NADPH and a ternary product complex obtained by soaking crystals with D-glucose (Fig. 4B). These crystals diffracted to a resolution of 1.5 Å, and the resulting model revealed that SmoB is an (ɑ/β)8 TIM barrel fold with a C-terminal cofactor binding site. The overall fold has high structural conservation with members of the aldo-keto reductase (AKR) superfamily. SmoB binds NADPH with the 2'-phosphate oxygens hydrogen-bonded to Thr284, Arg289, and backbone amide of Asn285 and the adenine ring stacked between Arg289 and Phe241 at the C terminus (Fig. 4C). NADPH binds in an extended anti-conformation, and the nicotinamide ring is located at the base of the substrate-binding pocket. Trp232 makes a π-π stacking interaction with the nicotinamide ring that positions the reactive center (C4) at a distance of 3 Å from C-6 of glucose, appropriate for hydride transfer (Fig. 4D). Within the SmoB•NADP+•glucose complex, glucose interacts with Arg152 (2.9 Å) and Lys120 (3 Å) as well as His151 (2.8 Å) and Tyr76 (2.7 Å) within the conserved catalytic tetrad His/Tyr/Lys/Asp that is common to the AKR superfamily (Fig. 4E) (21).
SMO Pathways Occur in the Alphaproteobacteria and Betaproteobacteria.
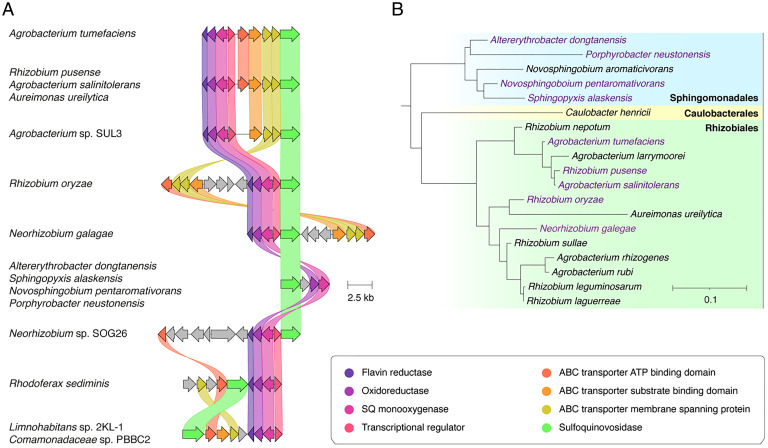
To ascertain how widespread this pathway for SQ utilization might be, a Multigene BLAST search was conducted of the nonredundant protein set of the National Center for Biotechnology Information (NCBI) for gene clusters that contain homologous SQases and SQ monoxygenases. This identified many putative smo gene clusters across the Agrobacterium and Rhizobium genus within the Rhizobiales order and evidence of some broader expansion into the Alphaproteobacteria and Betaproteobacteria classes (Fig. 5). Among these putative smo gene clusters, some were syntenic while others were substantially rearranged (nonsyntenic) or modified to make use of other (non-ABC) transporter systems. The use of diverse transport systems is not surprising: a similar phenomenon has been observed for the sulfo-ED pathway (5, 6). Indeed, sulfo-ED gene clusters have been identified in several Rhizobiales (5, 6), suggesting that there has been ample opportunity for genetic exchanges between these pathways during their evolution.
Fig. 5.
Prevalence of the SMO pathway. (A) Architecture of the SMO gene cluster in A. tumefaciens and homologous gene clusters in other organisms. Colored links indicate ≥30% protein sequence similarity. Only those clusters encoding putative SQ monoxygenases and SQases were annotated as putative SMO gene clusters. (B) A phylogenetic tree demonstrating the diversity of organisms possessing putative SMO gene clusters. The tree was constructed by pruning of the All-Species Living Tree Project’s 16s rRNA-based LTP release 132 (https://www.arb-silva.de/projects/living-tree/).
Discussion
While existing pathways for the breakdown of SQ require two different organisms and involve scission of the carbon chain into two three-carbon fragments, we describe here a fundamentally different approach that features complete utilization of the SQ carbon skeleton. The SMO pathway includes several proteins with hitherto undescribed activities, including an SQGro-binding protein; an FMNH2- and O2-dependent SQ monooxygenase that defines this “SMO” pathway by catalyzing scission of the C–S bond in SQ; and an oxidoreductase dedicated to the NADPH-dependent reduction of 6-OG to glucose. Like all other sulfoglycolytic pathways studied to date, the SMO pathway also possesses a conserved SQase, which is essential for liberating SQ from its precursor glycoside SQGro (9, 10).
The SMO pathway is reminiscent of other sugar-metabolizing pathways in bacteria. For example, the SmoI (SQase), SmoF (SQGro-binding protein), and SmoE/G/H (ABC transporter) proteins encoded by the smo cluster are analogous to MalP (maltodextrin phosphorylase), MalE (maltose binding protein), and MalF/G/K (ABC transporter) encoded by the mal operon of E. coli that imports and degrades maltose (22). Additionally, the SmoC (SQ monooxygenase) and SmoA (flavin reductase) proteins of the SMO pathway are reminiscent of the SsuD (FMNH2-dependent alkylsulfonate monooxygenase) and SsuE (NADPH-dependent FMN reductase) pair encoded by the ssu operon of E. coli that degrades alkanesulfonates (16). Indeed, it is likely that the SMO pathway arose through the recombination and neofunctionalization of analogous sugar- and sulfonate-metabolizing pathways.
Through structural analysis, we identified key residues involved in sulfosugar recognition and processing in order to provide greater confidence to bioinformatic analyses of putative smo gene clusters, an approach that has proven valuable for the identification of tier 1 sulfoglycolytic pathways (9, 23, 24). This includes the Thr220-Gly166-Ser43-H2O(His13-Gln46) cluster of SmoF for the recognition of SQGro; the Arg283-Tryp286-H2O(Tyr491) triad of SmoI for the recognition of SQGro; and the Trp206-Arg236-His238-Tyr341-His343 constellation of SmoC for the recognition of SQ. Given the importance of the SQ monooxygenase SmoC to the SMO pathway, further empirical and computational work is warranted to understand what interactions drive its selectivity for SQ, which lies in contrast with the promiscuity exhibited by alkanesulfonate monoxygenases like SsuD.
The prevalence of the SMO pathway in Alphaproteobacteria of the Rhizobiales order is intriguing, since many bacteria of this order are plant symbionts or pathogens. Indeed, those bacteria that do not possess an SMO pathway often possess a complementary tier 1 sulfo-ED pathway (5). Accordingly, it appears that plant sulfolipid catabolism is important for Rhizobiales, whether they be plant pathogens/symbionts or free-living organisms adopting an oligotrophic saprophytic lifestyle in substrate replete with decaying plant tissues. Symbiotic bacteria of the Rhizobiales order reside within the root nodules of their plant host, where they harness four-carbon substrates from the host for energy and central metabolism (25). Speck et al. showed that sulfonate utilization gene clusters were expressed by the plant symbiont Bradyrhizobium diazoefficiens USDA 110 within these nodules and that this may be important for utilizing diverse sulfur sources to support symbiotic and possibly free-living lifestyles (26). With sulfolipid representing a large and accessible pool of sulfur in plants, one possible purpose of the SMO pathway may be to salvage sulfur for these bacteria. This is an important distinction between the SMO pathway and the tier 1 sulfoglycolytic pathways: the latter supports two-member microbial communities containing a second member with a tier 2 pathway to provide access to the sulfur of SQ (27). In this sense, use of the SMO pathway, which enables the complete utilization of the carbon skeleton and access to the sulfur of the monosaccharide, can be considered a “selfish” metabolic strategy and could provide an advantage in the highly competitive soil environment or in the absence of other bacterial species within colonized plant tissues. Combined with the pathway’s requirement for molecular oxygen to effect C–S bond fissure, this may explain why the SMO pathway occurs within those bacteria that are commonly associated with plants. Understanding how the SMO and tier 1 pathways impact fitness within different environmental niches remains an important question, with answers that have significant implications for understanding plant diseases and symbioses as well as soil chemistry.
Materials and Methods
Specialist Reagents.
SQ and methyl α-sulfoquinovoside were purchased from MCAT GmbH, and (13C6)SQ, glycer-1-yl α-sulfoquinovoside (SQGro), and dicyclohexylammonium sulfolactate and cyclohexylammonium dihydroxypropanesulfonate were synthesized as described (13, 28). All other sulfonates were purchased from Sigma-Aldrich.
Growth Studies.
Cultures of A. tumefaciens C58 were grown in a phosphate-buffered mineral salts media (M9, pH 7.2), with glucose or SQ (10 mM) as the sole carbon source. Cultures were incubated at 30 °C (250 rpm), with adaptation and robust growth observed within 2 to 3 d. These were subcultured (1% inoculum) into the same media (10 mL) and grown at 30 °C (250 rpm). Bacterial growth was quantitated using a Varian Cary50 ultraviolet (UV)/visible spectrophotometer to measure OD600. Growth experiments were replicated twice.
Reducing Sugar Assay for Culture Supernatant.
The reducing sugar assay was performed according to the procedure of Blakeney and Mutton (29). This assay uses preprepared alkaline diluent and 4-hydroxybenzoic acid hydrazide (PAHBAH) working solution. Alkaline diluent was prepared by the addition of sodium hydroxide (20 g, 0.50 mol) to a solution of 0.10 M trisodium citrate (50 mmol, 500 mL) and 0.02 M calcium chloride (13 mmol, 500 mL). PAHBAH working solution was prepared by dissolving 4-hydroxybenzhydrazide (PAHBAH) (0.25 g, 1.6 mmol) in alkaline diluent (50 mL). The PAHBAH working solution should be made fresh shortly before use. To determine reducing sugar concentration, 0.90 mL of PAHBAH working solution was added to 0.10 mL of sample. The mixture was heated at 98 °C for 4 min, and then 0.5 mL of the mixture was diluted into 1.0 mL of deionized water and the absorbance read at 415 nm using a Varian Cary50 UV/visible spectrophotometer. Concentrations of SQ were determined with reference to a standard curve constructed using SQ.
Turbidometric Sulfate Assay for Culture Supernatant.
The sulfate assay was performed according to the procedure of Sörbo (30). This assay uses a barium sulfate–polyethylene glycol (Ba-PEG) reagent, which contains polyethylene glycol (PEG) to stabilize BaSO4 crystals and a small amount of preformed BaSO4 seed crystals to improve the reproducibility and linearity of the assay. The Ba-PEG reagent should be prepared fresh before use. Ba-PEG reagent was prepared by dissolving BaCl2 (42 mg, 0.20 mmol) and polyethylene glycol 6000 (0.75 g) in deionized water (5.0 mL). A small amount of Na2SO4 (10 µL, 50 mM) was added to this solution, with efficient magnetic stirring to generate preformed BaSO4 seed crystals. Individual sulfate assays were conducted as follows. An aliquot of culture supernatant obtained after pelleting of cells for 5 min at 5,000 g (typically 100 μL, containing a maximum of 2.5 µmol of Na2SO4) was diluted to 0.1 mL with deionized water before the addition of 0.5 M HCl (0.1 mL) followed by Ba-PEG reagent (0.1 mL). The mixture was mixed vigorously and the absorbance of the sample at 400 nm determined using a Varian Cary50 UV/visible spectrophotometer. Concentrations of sulfate were determined by reference to a standard curve constructed using Na2SO4. This curve was linear up to 2.5 µmol of Na2SO4.
Colorimetric Fuchsin Sulfite Assay for Culture Supernatant
The fuchsin sulfite assay was performed according to the procedures of Brychkova et al. (31) and Kurmanbayeva et al. (32). This procedure requires three preprepared solutions: Reagents A, B, and C. Reagent A was prepared by dissolution of basic fuchsin (4.0 mg, 12 µmol) in deionized water (8.25 mL) at 0 °C, prior to the addition of 98% H2SO4 (1.25 mL). Reagent B was prepared by diluting formaldehyde (36% in H2O, 0.32 mL) in deionized water (9.68 mL) at 0 °C. Reagent C was prepared by dilution of Reagent A (1 mL) in deionized water (7 mL), prior to the addition of solution reagent B (1 mL). Individual sulfite assays were performed by addition of Reagent C (516 µL) to a mixture of sample (72 µL) and 0.5 mM Na2SO3 (12 µL), with the latter providing a stable background signal for reference. The sample was incubated at 20 to 22 °C for 10 min and the absorbance of the sample at 570 nm determined using a Varian Cary50 UV/visible spectrophotometer. Concentrations of sulfite were determined by reference to a standard curve constructed using Na2SO3.
NMR Analysis of Metabolites Produced from (13C6)SQ.
M9 minimal media (5 mL) containing 10 mM glucose was inoculated with A. tumefaciens C58 and grown to stationary phase at 30 °C (250 rpm). A 50 μL aliquot of this culture was used to inoculate 2 mL of M9 minimal media containing 10 mM (13C6)SQ and the culture incubated at 30 °C (250 rpm). At OD600 0.27 and OD600 0.49, 950 μL samples of culture supernatant were diluted with 100 μL of D2O and 13C-NMR spectra acquired using a 400 MHz spectrophotomer (100 MHz for 13C).
Growth of A. tumefaciens C58 on Diverse Alkanesulfonates.
M9 minimal media (5 mL) containing 10 mM glucose was inoculated with A. tumefaciens C58 and grown to stationary phase at 30 °C (250 rpm). A 50 μL aliquot of this starter culture was used to inoculate 2 mL of M9 minimal media containing 10 mM of the alternative alkanesulfonate substrate: SQ (positive control), MeSQ, glycer-1-yl α-sulfoquinovoside (SQGro), dicyclohexylammonium sulfolactate, cyclohexylammonium dihydroxypropanesulfonate, sulfoacetic acid, taurine, sodium pentanesulfonate, cysteic acid, MOPS, HEPES, PIPES, MES, and methanesulfonic acid. Cultures were incubated for 30 d at 30 °C (250 rpm) with daily observations of optical density at 600 nm. Each experiment was performed in duplicate. Growth was observed on SQ (positive control), MeSQ, and SQGro but not on any other sulfonate. Control experiments established that A. tumefaciens grows on glucose in the presence and absence of cyclohexylamine or dicyclohexylamine and does not grow on cyclohexylamine or dicyclohexylamine alone.
Digestion of Samples for Quantitative Proteomics.
Freeze-dried A. tumefaciens whole-cell pellets were resuspended in 500 μL lysis buffer (4% sodium dodecyl sulfate (SDS), 50 mM Tris pH 8.5, 10 mM dithiothreitol (DTT)) and boiled at 95 °C for 10 min with shaking at 2,000 rpm to shear DNA and inactivate protease activity. Lysates were cooled to room temperature and protein concentration determined using a bichinchoninic acid (BCA) assay. Each sample (200 μg of protein) was acetone precipitated by mixing four volumes of ice-cold acetone with one volume of sample. Samples were precipitated overnight at −20 °C and then centrifuged at 4,000 × g for 10 min at 4 °C. The precipitated protein pellets were resuspended with 80% ice-cold acetone and precipitated for an additional 4 h at −20 °C. Samples were centrifuged at 17,000 × g for 10 min at 4 °C to collect precipitated protein, and the supernatant was discarded and excess acetone driven off at 65 °C for 5 min. Dried protein pellets were resuspended in 6 M urea, 2 M thiourea, and 40 mM NH4HCO3 and reduced/alkylated prior to digestion with Lys-C (1/200 wt/wt) and then trypsin (1/50 wt/wt) overnight as previously described (33). Digested samples were acidified to a final concentration of 0.5% formic acid and desalted using C18 stage tips (34) before analysis by LC-MS.
Quantitative Proteomics Using Reversed Phase LC-MS.
Purified peptides were resuspended in Buffer A* (2% acetonitrile [MeCN], 0.1% trifluoroacetic acid [TFA]) and separated using a Proflow-equipped Dionex Ultimate 3000 Ultra-Performance Liquid Chromatography system (Thermo Fisher Scientific) with a two-column chromatography setup composed of a PepMap100 C18 20 mm × 75 μm trap and a PepMap C18 500 mm × 75 μm analytical column (Thermo Fisher Scientific). Samples were concentrated onto the trap column at 5 μL min−1 with Buffer A (2% MeCN, 0.1% formic acid [FA]) for 6 min and then infused into an Orbitrap Q-Exactive HF Mass Spectrometer (Thermo Fisher Scientific) at 250 nL min−1. Peptides were separated using 124-min gradients altering the buffer composition from 2% Buffer B (80% MeCN, 0.1% FA) to 8% B over 14 min, then from 8% B to 30% B over 80 min, 30% B to 45% B over 10 min, and 45% B to 95% B over 2 min, holding at 95% B for 10, then dropped to 2% B over 1 min and holding at 2% B for the remaining 7 min. The Q-Exactive HF Mass Spectrometer was operated in a data-dependent mode automatically switching between the acquisition of a single Orbitrap MS scan (120,000 resolution) and a maximum of 20 tandem mass spectrometry scans (higher energy collisional dissociation normalized collision energy 28, maximum fill time 40 ms, automatic gain control 2 × 105 with a resolution of 15,000).
Mass Spectrometry Data Analysis.
Proteomics datasets were searched using MaxQuant (v1.5.3.3) (35) against the A. tumefaciens C58 proteome (Uniprot proteome id UP000000813, downloaded 27 January 2018, 5,344 entries). Searches were performed with carbamidomethylation of cysteine set as a fixed modification and oxidation of methionine as well as acetylation of protein N-termini allowed as variable modifications. The protease specificity was set to trypsin, allowing two miscleavage events with a maximum false discovery rate of 1.0% set for protein and peptide identifications. To enhance the identification of peptides between samples, the Match Between Runs option was enabled with a precursor match window set to 2 min and an alignment window of 10 min. For label-free quantitation (LFQ), the MaxLFQ option within MaxQuant (36) was enabled in addition to the requantification module. The resulting protein group output was processed within the Perseus (v1.4.0.6) (37) analysis environment to remove reverse matches and common protein contaminates prior. For LFQ comparisons, missing values were imputed using Perseus and Pearson correlations visualized using R. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (38) partner repository with the dataset identifier PXD014115.
Cloning.
Oligonucleotides encoding Atu3277 (SmoA), Atu3278 (SmoB), Atu3279 (SmoC), and Atu3282 (SmoF) were amplified by PCR using Phusion polymerase HF master mix (NEB), the appropriate primers listed in SI Appendix, Table S1 and A. tumefaciens C58 gDNA as template. Oligonucleotides encoding RoSmoA and RoSmoC were synthesized (IDT) to provide the sequences listed in SI Appendix, Table S1. These were cloned into the pET29b(+) vector at the NdeI and XhoI sites and sequence-verified by Sanger sequencing to give expression vectors for SmoA, SmoB, SmoC, SmoF, RoSmoA, and RoSmoC. Due to interference from the SmoB C-terminal His6-tag during structural studies, the smoB (Atu3278) gene was subcloned into the pET-YSBLIC3C vector (39) by PCR amplification with the relevant primers in SI Appendix, Table S1 and In-Fusion cloning (Clontech Laboratories, Inc.) into linearized YSBLIC3C vector according to the manufacturer’s protocol. The expression plasmid was sequence-verified by Sanger sequencing.
Protein Expression and Purification.
All vectors were transformed into “T7 Express” E. coli (NEB), except for the vector encoding SmoF (Atu3282), which was transformed into “Shuffle T7” E. coli (NEB), and all were plated onto Lysogeny Broth (LB)-agar (50 μg mL−1 kanamycin) and incubated at 37 °C for 16 h. A single colony was used to inoculate 10 mL of LB media containing 50 µg mL−1 kanamycin, and the cultures were incubated at 37 °C for 16 h. These starter cultures were used to inoculate 1,000 mL of S-broth (35 g tryptone, 20 g yeast extract, 5 g NaCl, pH 7.4) containing 50 µg mL−1 kanamycin, which was incubated with shaking (250 rpm) at 37 °C until it reached an OD600 of 0.8. Each culture was cooled to room temperature, isopropyl thiogalactoside added to a final concentration of 400 µM, and incubation with shaking (200 rpm) continued at 18 °C for 19 h. Cells were harvested by centrifugation at 8,000 × g for 20 min at 4 °C and then resuspended in 40 mL binding buffer (50 mM NaPi, 300 mM NaCl, 5 mM imidazole, pH 7.5) containing protease inhibitor (Roche cOmplete EDTA-free protease inhibitor mixture) and lysozyme (0.1 mg mL−1) by nutating at 4 °C for 30 min. Benzonase (1 µL, 250 U) was added to the mixture then lysis was effected by sonication [10× (15 s on/45 s off) at 45% amplitude]. The lysate was centrifuged at 18,000 × g for 20 min at 4 °C and the supernatant collected. The supernatants were filtered (0.45 µm) and loaded onto a 1 mL HiTrap TALON IMAC column (GE). The column was washed with 3 × 10 mL of binding buffer, and then the protein was eluted using elution buffer (50 mM NaPi, 300 mM NaCl, 400 mM imidazole, pH 7.5). Fractions containing product, as judged by SDS-PAGE, were further purified by size exclusion chromatography on a HiPrep 16/60 Sephacryl S-200 HR column (GE) using 50 mM NaPi, 150 mM NaCl, pH 7.5 (Atu3277 SmoA; Atu3278, SmoB; Atu3279, SmoC) or 50 mM sodium citrate, 150 mM NaCl, pH 5.5 (Atu3282, SmoF) as buffer (SI Appendix, Fig. S2). SmoI (Atu3285 or SmoI) was prepared as previously described (9).
SEC-MALS Analyses.
Experiments were conducted on a system comprising a Wyatt HELEOS-II multiangle light scattering detector and a Wyatt rEX refractive index detector linked to a Shimadzu liquid chromatography (LC) system (SPD-20A UV detector, LC20-AD isocratic pump system, DGU-20A3 degasser, and SIL-20A autosampler). Experiments were conducted at room temperature (20 ± 2 °C). Solvents were filtered through a 0.2 µm filter prior to use, and a 0.1 µm filter was present in the flow path. The column was equilibrated with >2 CV of buffer (50 mM NaPi, 300 mM NaCl pH 7.4) before use and buffer was infused at the working flow rate until baselines for UV, light scattering, and refractive index detectors were all stable. The sample injection volume was 100 µL of protein at 6 mg mL−1 in 50 mM NaPi buffer, 300 mM NaCl pH 7.4. Shimadzu LC Solutions software was used to control the LC and Astra V software for the HELEOS-II and rEX detectors. The Astra data collection was 1 min shorter than the LC solutions run to maintain synchronization. Blank buffer injections were used as appropriate to check for carryover between sample runs. Data were analyzed using the Astra V software. Molecular weights were estimated using the Zimm fit method with degree 1. A value of 0.158 was used for protein refractive index increment (dn/dc).
ITC.
ITC experiments were performed using a MicroCal iTC200 (GE Healthcare) at 25 °C, with a 750 rpm stirring speed and a reference power of 10 µCal⋅s−1. Proteins and substrates were equilibrated into degassed and filter-sterilized buffer (50 mM NaPi, 200 mM NaCl, pH 7.4 for SmoC/F and 25 mM NaPi, pH 7.5 for SmoB). Protein concentration was determined by BCA assay (Thermo Fisher) before initiating experiments. For SmoC–SQ binding, 600 µM SQ was titrated into the ITC cell containing 40 µM SmoC as a series of 10 × 3.94 µL injections with a preinjection of 1 × 0.4 µL. For SmoF–SQGro binding, 200 µM SQGro was titrated into the ITC cell containing 20 µM SmoF as a series of 15 × 2.94 µL injections with a preinjection of 1 × 0.4 µL. The delay between injections was set at 120 s, with an initial injection delay of 60 s. For SmoB-NAD(P)H binding, 1 mM NADH was titrated into the ITC cell containing 40 μM SmoB as a series of 19 × 3 μL injections with a preinjection of 1 × 4 μL. The delay between injections was set at 150 s, with an initial injection delay of 180 s. All data analysis was performed in MicroCal ITC Origin Analysis software (Malvern).
Nanodifferential Scanning Fluorescence Analysis of SmoF.
Thermal stability analysis for SmoF in the presence and absence of SQGro ligand was performed on a Prometheus NT.48 (NanoTemper) at 15% excitation, scanning from 20 °C to 65 °C at 0.5 °C min−1. All protein samples were at a concentration of 1 mg mL−1 in 50 mM citrate, 150 mM NaCl at pH 5.5, with a 10 μL capillary load per sample. Data acquisition and analysis was performed with PR.ThermControl (NanoTemper) software.
Identification of the Flavin Cofactor that Copurified with SmoA.
An amount of 100 µL of recombinant flavin reductase (SmoA or RoSmoA) at a concentration of 20 mg mL−1 in 50 mM Tris, 150 mM NaCl, pH 8.5 was heated at 90 °C for 10 min. The sample was clarified by centrifugation (16,000 × g, 10 min, 4 °C) and the supernatant filtered (0.2 µm). Samples were analyzed by LC-MS on an Agilent system (G6125B mass detector, 1290 Infinity G7120A high speed pump, 1290 Infinity G7129B autosampler, and 1290 Infinity G7117B diode array detector). Conditions for LC were as follows: column: Phenomenex 00B-4752-AN Luna Omega 1.6 µm PS C18 100Å (50 × 2.1 mm); injection volume: 1 μL; gradient: 3 to 100% B over 20 min (solvent A: water + 0.1% FA; solvent B: MeCN + 0.1% FA); flow rate: 0.6 mL min−1; DAD: 254 and 214 nm.
Michaelis-Menten Kinetic Analyses of SmoA and RoSmoA.
Reactions were conducted at 25 °C in 96-well plate format and involved the addition of SmoA or RoSmoA (final concentration of 20 nM for NADH and 500 nM for NADPH) to 20 to 800 µM NAD(P)H in 50 mM NaPi, 150 mM NaCl, 30 µM FMN, 0.01% BSA, pH 7.4 at a total volume of 100 µL. The progress of the enzyme-catalyzed conversion of NAD(P)H to NAD(P)+ was monitored by measuring loss of absorbance at 340 nM over time using an Envision Multimodal Plate Reader (Perkin-Elmer). Initial rates for each reaction were calculated after first subtracting the rate of spontaneous NAD(P)H oxidation (determined using an enzyme-free control) and an empirically determined extinction coefficient for NAD(P)H under these conditions. Each initial rate was determined in triplicate and fit to a Michaelis-Menten equation using Prism 8 (GraphPad).
Sulfoquinovose Monooxygenase Assay.
This SQ monooxygenase activity assay is based on a previously described alkanesulfonate monooxygenase activity assays (19) and uses Ellman’s reagent to quantify sulfite released by these enzymes. A 2 mL reaction containing 1 mM SQ, 1 mM NADH, 3 µM FMN, 0.01% (wt/vol) BSA, 100 nM SmoA or RoSmoA, and 300 nM SQ monooxygenase (SmoC or RoSmoC) in buffer (25 mM Tris pH 9.1, 25 mM NaCl) was incubated at 30 °C, along with controls lacking reaction components or using alternate sulfonate substrates. Reactions were initiated by the addition of SmoA or RoSmoA to the mixture. Sulfite concentration in the samples was determined at discrete time points by quenching 40 µL of the reaction in 160 µL of Ellman’s reagent (0.125 mg mL−1 in 25 mM NaPi pH 7.0, prepared fresh) within a 96-well plate. After 60 s, the absorbance of the sample at 405 nm was determined using an Envision Multimodal Plate Reader (Perkin-Elmer). The sulfite concentration was interpolated using a calibration curve generated under these conditions: a linear relationship between sulfite concentration and absorbance at 405 nm was observed for 5 to 1,000 µM Na2SO3. The activity of SQ monooxygenases at different pH was determined by modifying the buffer in the aforementioned reactions (MES: pH 6.0, 6.5 and Tris: pH 7.0, 7.5, 8.0, 8.5, 9.1) using an endpoint of t = 30 min.
Equilibrium Isotope Labeling Using SmoB
In order to prelabel the anomeric position, glucose was incubated in 98% H218O with heating at 80 °C for 2 d, then evaporated to dryness to give C1-18O-labeled glucose. Labeling was determined to be 95% by mass spectrometry based on intensities of the M and M+2 peaks. Using H218O buffer (100 mM potassium phosphate, pH 7.0), NAD+ and NADP+ were each added at 0.05 molar equivalent to C1-18O-glucose and SmoB. Four control experiments were conducted: one without enzyme, one without NAD+ and NADP+, one in H216O, and one in H216O with unlabeled glucose. Reactions were monitored by mass spectrometry. Only in the experimental sample containing enzyme, H218O, and NAD+/NADP+ was an M+4 signal observed, and this reached a maximum intensity after 72 h. Two additional reactions were performed using SmoB, glucose, and either NADP+ or NAD+ in H218O, and only the reaction containing NADP+ generated the M+4 species. To confirm that the M+4 species was glucose with two 18O labels, we studied the product by HPLC. However, under aqueous HPLC conditions, the 18O-label at C1 is lost through chemical exchange with solvent. Therefore, we acetylated the product to form the pentaacetate to ensure no exchange at the anomeric position during HPLC analysis. The aforementioned reaction mixture was evaporated under reduced pressure. The crude residue was treated with acetic anhydride in pyridine (1:2, 1 mL) overnight. The product was extracted with ethyl acetate and washed with saturated CuSO4 to remove pyridine. The organic solution containing peracetylated glucose was analyzed by LC-MS on an Agilent system (G6125B mass detector, 1290 Infinity G7120A high speed pump, 1290 Infinity G7129B autosampler, and 1290 Infinity G7117B diode array detector). Conditions for LC were as follows: column: Phenomenex 00B-4752-AN Luna Omega 1.6 µm PS C18 100Å (50 × 2.1 mm); injection volume: 1 μL; gradient: 0 to 65% B over 20 min (solvent A: water + 0.1% FA; solvent B: MeCN + 0.1% FA); flow rate: 0.6 mL min−1. Peaks with m/z 413 [M+Na]+, m/z 415 [M+2+Na]+, and m/z 417 [M+4+Na]+ had the same retention time as an authentic glucose pentaacetate standard.
GC-MS Analysis of Isotopically Labeled Carbohydrates.
A 0.1 µL aliquot of SmoB-glucose reaction mixture (containing ∼2.5 nmol glucose) was transferred to a gas chromatography (GC) vial insert (deactivated) together with 1 nmol scyllo-inositol as an internal standard. Samples were derivatized as described in Antoniewicz et al. (20), with minor modifications. Briefly, samples were dried (in vacuo, 35 °C with a 40 µL methanol wash), followed by addition of hydroxylamine hydrochloride (20 mg mL−1 in 25 µL pyridine) and incubation at 90 °C for 1 h. Vials were cooled briefly at 20 to 22 °C before the addition of propionic anhydride (50 µL) and incubation at 60 °C for 30 min. Samples were evaporated to dryness under a stream of nitrogen at 60 °C and resuspended in ethyl acetate (40 µL). Control samples of U-12C-glucose, U-13C-glucose, 1,2-13C2-glucose, and 6,6-2H2-glucose were also prepared at a 2.5 nmol scale in the assay buffer mixture. Samples were blinded for analysis. The derivatized labeled glucose samples (SI Appendix, Fig. S13 and Table S7) were analyzed by GC-MS using a DB5 capillary column (J&W Scientific, 30 m, 250 μm inner diameter, 0.25 μm film thickness) with a 10 m inert duraguard. The injector insert and GC-MS transfer line temperatures were 270 °C and 250 °C, respectively. The oven temperature gradient was programmed as follows: 70 °C (1 min); 70 °C to 295 °C at 12.5 °C min−1; 295 °C to 320 °C at 25 °C min−1; 320 °C for 2 min. Glucose and scyllo-inositol were identified by reference to authentic standards. A calibration curve was generated using glucose standard in assay buffer (starting concentration 50 nmol, 2-fold dilution series). SI Appendix, Fig. S12 shows the fraction of labeled fragments, corrected for isotope natural abundance by DExSI analysis (40).
Protein Crystallization.
Initial crystallization screening was performed using commercially available INDEX (Hampton Research), PACT premier, and CSSI/II (Molecular Dimensions) screens in 96-well sitting drop trays. Further optimization was carried out in a 48-well sitting drop or 24-well hanging-drop format to obtain optimal crystals for X-ray diffraction. Unless otherwise stated, all crystals were grown at 20 °C.
Crystals of apo-SmoF were obtained by mixing 0.15 µL of protein stock (50 mg mL−1 protein in 50 mM citrate, 150 mM NaCl, pH 5.5) with 0.15 µL mother liquor (0.3 M ammonium acetate, 0.1 M Bis-Tris, 25% wt/vol PEG 3350, pH 5.5) housed in a Rigaku Xtaltrak plate hotel to enable consistent growth and monitoring at 6 °C. Crystals were harvested with nylon CryoLoops (Hampton Research) and cryopreserved in liquid nitrogen without additional cryoprotectants.
Crystals of SmoF were initially obtained by mixing 0.15 µL of protein stock (3.5 mg mL−1 protein with 2'R-SQGro at a 1:10 molar ratio in 50 mM citrate, 150 mM NaCl, pH 5.5) with 0.15 µL mother liquor (30% [wt/vol] polyethylene glycol 4000, 0.2 M sodium acetate, 0.1 M Tris chloride, pH 8.5). The resulting crystals were used to prepare a seed stock by mixing the crystallization drop with 100 µL mother liquor and vortexing for 60 s with one Teflon bead. An optimization plate was set up with drops comprised of 0.1 µL of various mother liquors (28–36% [wt/vol] polyethylene glycol 4000, 0.2 M sodium acetate, 0.1 M Tris chloride, pH 7.1 to 9.1), 50 nL seed stock solution, and 0.15 µL protein stock (4 mg mL−1 protein with 2'R-SQGro at a 1:10 molar ratio in 50 mM citrate, 150 mM NaCl, pH 5.5). A single crystal grown at 31.8% (wt/vol) polyethylene glycol 4000, 0.2 M sodium acetate, 0.1 M Tris chloride, pH 8.95, was harvested with a nylon CryoLoop (Hampton Research) and cryopreserved in liquid nitrogen with 25% (vol/vol) ethylene glycol as cryoprotectant.
Crystals of SmoI-D455N-E370A-E371A were obtained by mixing 0.4 µL of protein stock (35 mg mL−1 protein in 50 mM NaPi, 300 mM NaCl, pH 7.4) with 0.5 μL mother liquor (26% PEG 3350 wt/vol, 0.2 M KSCN, 0.1 M Bis-Tris propane, pH 6.5). Crystals were soaked with solid SQGro in mother liquor for 2 min prior to harvesting with nylon CryoLoops (Hampton Research) and cryopreserved without additional cryoprotectants.
Crystals of apo-SmoC were obtained by mixing 0.6 µL of protein stock (60 mg mL−1 protein in 50 mM Tris, 300 mM NaCl, pH 7.5) with 0.5 μL mother liquor (0.2 M NaCl, 0.1 M MES pH 6, 26% PEG 6000 wt/vol, and 10 mM SQ-glucitol). Crystals of apo-RoSmoC were obtained by mixing 0.1 µL of protein stock (11.7 mg mL−1 protein in 50 mM Tris, 300 mM NaCl, pH 7.5) with 0.2 μL mother liquor (0.2M NaNO3, 20% PEG 3350 wt/vol, and 10 mM SQ). Crystals were harvested with nylon CryoLoops (Hampton Research) and cryopreserved in liquid nitrogen without additional cryoprotectants.
Crystals of SmoB-apo (YSBLIC3C construct) were obtained by mixing 0.15 µL of protein stock (20 mg mL−1 protein in 50 mM NaPi, 150 mM NaCl, pH 7.4) with 0.15 µL mother liquor (0.2 M sodium malonate dibasic monohydrate, 0.1 M Bis-Tris propane pH 8.5, 20% wt/vol PEG 3350). For the SmoB•NADPH complex, crystals were obtained by mixing 0.15 µL of protein stock (20 mg mL−1 protein in 50 mM NaPi, 150 mM NaCl, 2 mM NADPH, pH 7.4) with 0.15 µL mother liquor (0.1 M succinic acid, sodium dihydrogen phosphate, glycine buffer [SPG buffer, Qiagen], 25% wt/vol PEG 1500 at pH 6.0). For the SmoB•NADPH•Glc complex, crystals were obtained in a hanging drop by mixing 1 µL of protein stock (13 mg mL−1 protein in 50 mM NaPi 150 mM NaCl, pH 7.4) with 1 μL of mother liquor (2 mM NADPH, 0.1 M SPG [Qiagen], 25% wt/vol PEG 1500 at pH 6). Crystals were soaked with solid glucose in mother liquor for 1 min prior to harvesting with nylon CryoLoops (Hampton Research) and cryopreserved without additional cryoprotectants.
X-ray Data Collection, Processing, and Refinement.
The data were processed and integrated using XDS (41) and scaled using SCALA (42) included in the Xia2 processing system (43). Data reduction was performed with AIMLESS, and resolution was cut until CC1/2 = 0.5. The structure of the SmoI•SQGro complex was determined using molecular replacement using 5OHS (9) as the initial model. For SmoF, the structure was solved by molecular replacement using PHASER (44) with a search model created from Protein Data Bank (PDB) ID: 6DTQ (45). The structure of RoSmoC was solved by molecular replacement using the ensemble based on PDB ID: 1M41 (19) as an initial search model. The structure of SmoB was determined using molecular replacement with the monomer of an AKR from S. enterica (PDB ID: 4R9O) as the initial model. The apo-SmoF structure was solved using a dissected C-terminal domain of the SmoF•SQGro structure. Structures were built and refined by iterative cycles using Coot (46) and REFMAC (47) or Phenix (48), the latter employing local noncrystallographic symmetry restraints. Following building and refinement of the protein and water molecules, clear residual density was observed in the omit maps for cocomplex structures, and respective ligands were modeled into these. The coordinate and refinement library files were prepared using ACEDRG (49). The final structures gave Rcryst and Rfree values along with data and refinement statistics that are presented in SI Appendix, Tables S4–S6. Data were collected at Diamond Light Source, Didcot, Oxfordshire, UK, on beamlines I24 (SmoI-D455N•SQGro, to 2.15 Å; SmoF-apo, to 1.88 Å), I04 (RoSmoC to 1.75 Å), and I04-1 (SmoC-apo, to 3.2 Å; SmoB-apo_YSBLIC3C, to 1.5 Å; SmoB-apo; pET29a; SmoB•NADPH and SmoB•NADPH•Glc) and at the Australian Synchrotron using the MX2 beamline (At3282•SQGro complex, to 1.7 Å). The coordinate files and structure factors have been deposited in the PDB with the coordinate accession numbers 7OFX (SmoI-D455N•SQGro), 7NBZ (SmoF-apo), 7OFY (SmoF•SQGro), 7OH2 (RoSmoC), 7OLF (SmoC-apo), 7BBY (SmoB-apo; pET29a), 7BBZ (SmoB-apo; YSBLIC3C), 7BC0 (SmoB•NADPH), and 7BC1 (SmoB•NADPH•Glc).
Structure-Based Analyses.
Crystal packing interactions were analyzed using the protein interactions, surfaces, and assemblies (PISA) server (50). Structural comparisons and structure-based sequence alignments were conducted using PDB25 search on DALI server against a representative subset of the PDB (51). All structure figures were generated using ccp4mg (52).
Bioinformatic Analysis SMO Pathway Prevalence.
Each gene within the A. tumefaciens C58 SMO gene cluster (Atu3277-Atu3285) was submitted as a query to the NCBI BLASTp algorithm to search a database comprised of nonredundant protein sequences with A. tumefaciens (taxid: 358) sequences excluded. Standard algorithm parameters were used, except the maximum target sequences was set to 10,000. Results were filtered to only retain protein sequences with E value ≤1.19 × 10−51. The corresponding nucleotide accession numbers for each protein from all nine searches were extracted and combined and duplicates removed to provide a list of candidate genome sequences. This was converted into a reference library for MultiGeneBLAST (53) and queried using the A. tumefaciens C58 SMO gene cluster. Clusters identified by this workflow with both an SQ monoxygenase and SQase homolog were regarded as putative SMO gene clusters. Clusters representative of the observed diversity were visualized using Clinker (54). A phylogenetic tree of species possessing a putative SMO gene cluster was generated by pruning the All-Species Living Tree Project’s 16s rRNA release 132 (55) using iTOL (56).
Supplementary Material
Acknowledgments
Dr. Monica Doblin is thanked for the provision of A. tumefaciens strain C58. This work was supported in part by National Health and Medical Research Council of Australia (NHMRC) project grants GNT1100164 (N.E.S), GNT1174405 (D.B.A.), and GNT1139546 and GNT1139549 (E.D.G.-B.); the Leverhulme Trust grant RPG-2017-190 (G.J.D.); Australian Research Council grants DP180101957 and DP210100233 (S.J.W.) and DP210100362 (N.E.S.); and support from The Walter and Eliza Hall Institute of Medical Research, the Australian Cancer Research Fund, and a Victorian State Government Operational Infrastructure support grant (E.D.G.-B.). G.J.D. is supported by the Royal Society Ken Murray Research Professorship, E.D.G.-B. is supported by the Brian M. Davis Charitable Foundation Centenary Fellowship, M.J.M. is an NHMRC Principal Research Fellow, N.E.S. is supported by an Australian Research Council Future Fellowship (FT200100270), B.M.d.S. was supported by the Melbourne Research Scholarship, J.W.Y.M. was supported by a Sir John and Lady Higgins Scholarship, and M.P. was supported by an Australian Postgraduate Award. We acknowledge Dr. Johan P. Turkenburg and Sam Hart for assistance with X-ray data collection; the Diamond Light Source for access to beamlines I04, I24, and I04-1 under proposal number mx-18598; and the Australian Synchrotron, part of Australia's Nuclear Science and Technology Organisation, for access to the MX-2 beamline, which made use of the Australian Cancer Research Foundation detector. We thank the Melbourne Mass Spectrometry and Proteomics Facility of the Bio21 Molecular Science and Biotechnology Institute at The University of Melbourne for the support of mass spectrometry analysis and the Bioscience Technology Facility (University of York) for assistance with SEC-MALS analyses.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2116022119/-/DCSupplemental.
Data Availability
Structure coordinates have been deposited in the PDB (https://www.rcsb.org/) under accession codes 7OFX, 7OFY, 7NBZ, 7OH2, 7OLF, 7BBZ, 7BC0, 7BC1, and 7BBY. Proteomics data are available via ProteomeXchange (57, 58) (http://www.proteomexchange.org/) with the identifier PXD014115. Scripts used to screen for the related gene clusters listed in Fig. 5 are available on GitHub (https://github.com/jmui-unimelb/Gene-Cluster-Search-Pipeline) (59).
References
- 1.Goddard-Borger E. D., Williams S. J., Sulfoquinovose in the biosphere: Occurrence, metabolism and functions. Biochem. J. 474, 827–849 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Harwood J. L., Nicholls R. G., The plant sulpholipid—A major component of the sulphur cycle. Biochem. Soc. Trans. 7, 440–447 (1979). [DOI] [PubMed] [Google Scholar]
- 3.Mizusawa N., Wada H., The role of lipids in photosystem II. Biochim. Biophys. Acta 1817, 194–208 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Denger K., et al. , Sulphoglycolysis in Escherichia coli K-12 closes a gap in the biogeochemical sulphur cycle. Nature 507, 114–117 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Felux A. K., Spiteller D., Klebensberger J., Schleheck D., Entner-Doudoroff pathway for sulfoquinovose degradation in Pseudomonas putida SQ1. Proc. Natl. Acad. Sci. U.S.A. 112, E4298–E4305 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J., et al. , A sulfoglycolytic Entner-Doudoroff pathway in Rhizobium leguminosarum bv. trifolii SRDI565. Appl. Environ. Microbiol. 86, e00750-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frommeyer B., et al. , Environmental and intestinal phylum firmicutes bacteria metabolize the plant sugar sulfoquinovose via a 6-deoxy-6-sulfofructose transaldolase pathway. iScience 23, 101510 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y., et al. , A transaldolase-dependent sulfoglycolysis pathway in Bacillus megaterium DSM 1804. Biochem. Biophys. Res. Commun. 533, 1109–1114 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Abayakoon P., et al. , Structural and biochemical insights into the function and evolution of sulfoquinovosidases. ACS Cent. Sci. 4, 1266–1273 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Speciale G., Jin Y., Davies G. J., Williams S. J., Goddard-Borger E. D., YihQ is a sulfoquinovosidase that cleaves sulfoquinovosyl diacylglyceride sulfolipids. Nat. Chem. Biol. 12, 215–217 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Roy A. B., Hewlins M. J., Ellis A. J., Harwood J. L., White G. F., Glycolytic breakdown of sulfoquinovose in bacteria: A missing link in the sulfur cycle. Appl. Environ. Microbiol. 69, 6434–6441 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kertesz M. A., Riding the sulfur cycle—Metabolism of sulfonates and sulfate esters in gram-negative bacteria. FEMS Microbiol. Rev. 24, 135–175 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Abayakoon P., et al. , Comprehensive synthesis of substrates, intermediates and products of the sulfoglycolytic Embden-Meyerhoff-Parnas pathway. J. Org. Chem. 84, 2901–2910 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Speers A. E., Wu C. C., Proteomics of integral membrane proteins—Theory and application. Chem. Rev. 107, 3687–3714 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Davidson A. L., Dassa E., Orelle C., Chen J., Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72, 317–364 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Der Ploeg J. R., Iwanicka-Nowicka R., Bykowski T., Hryniewicz M. M., Leisinger T., The Escherichia coli ssuEADCB gene cluster is required for the utilization of sulfur from aliphatic sulfonates and is regulated by the transcriptional activator Cbl. J. Biol. Chem. 274, 29358–29365 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Thakur A., et al. , Substrate-dependent mobile loop conformational changes in alkanesulfonate monooxygenase from accelerated molecular dynamics. Biochemistry 59, 3582–3593 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Liew J. J. M., El Saudi I. M., Nguyen S. V., Wicht D. K., Dowling D. P., Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer. J. Biol. Chem. 297, 100823 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eichhorn E., Davey C. A., Sargent D. F., Leisinger T., Richmond T. J., Crystal structure of Escherichia coli alkanesulfonate monooxygenase SsuD. J. Mol. Biol. 324, 457–468 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Antoniewicz M. R., Kelleher J. K., Stephanopoulos G., Measuring deuterium enrichment of glucose hydrogen atoms by gas chromatography/mass spectrometry. Anal. Chem. 83, 3211–3216 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penning T. M., The aldo-keto reductases (AKRs): Overview. Chem. Biol. Interact. 234, 236–246 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dippel R., Boos W., The maltodextrin system of Escherichia coli: Metabolism and transport. J. Bacteriol. 187, 8322–8331 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma M., et al. , Dynamic structural changes accompany the production of dihydroxypropanesulfonate by sulfolactaldehyde reductase. ACS Catal. 10, 2826–2836 (2020). [Google Scholar]
- 24.Sharma M., et al. , Molecular basis of sulfosugar selectivity in sulfoglycolysis. ACS Cent. Sci. 7, 476–487 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Udvardi M., Poole P. S., Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol. 64, 781–805 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Speck J. J., James E. K., Sugawara M., Sadowsky M. J., Gyaneshwar P., An alkane sulfonate monooxygenase is required for symbiotic nitrogen fixation by Bradyrhizobium diazoefficiens (syn. Bradyrhizobium japonicum) USDA110T. Appl. Environ. Microbiol. 85, e01552-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denger K., Huhn T., Hollemeyer K., Schleheck D., Cook A. M., Sulfoquinovose degraded by pure cultures of bacteria with release of C3-organosulfonates: Complete degradation in two-member communities. FEMS Microbiol. Lett. 328, 39–45 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y., et al. , Concise synthesis of sulfoquinovose and sulfoquinovosyl diacylglycerides, and development of a fluorogenic substrate for sulfoquinovosidases. Org. Biomol. Chem. 18, 675–686 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Blakeney A. B., Mutton L. L., A simple colorimetric method for the determination of sugars in fruit and vegetables. J. Sci. Food Agric. 31, 889–897 (1980). [Google Scholar]
- 30.Sörbo B., “Sulfate: Turbidimetric and nephelometric methods” in Methods in Enzymology (Academic Press, 1987), Vol. 143, pp. 3–6. [DOI] [PubMed] [Google Scholar]
- 31.Brychkova G., Yarmolinsky D., Fluhr R., Sagi M., The determination of sulfite levels and its oxidation in plant leaves. Plant Sci. 190, 123–130 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Kurmanbayeva A., et al. , “Determination of total sulfur, sulfate, sulfite, thiosulfate, and sulfolipids in plants” in Plant Stress Tolerance: Methods and Protocols, Sunkar R. Ed. (Springer New York, 2017), pp 253–271. [DOI] [PubMed] [Google Scholar]
- 33.Scott N. E., et al. , Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell. Proteomics 10, M000031–MCP201 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rappsilber J., Mann M., Ishihama Y., Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Cox J., Mann M., MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Cox J., et al. , Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tyanova S., et al. , Visualization of LC-MS/MS proteomics data in MaxQuant. Proteomics 15, 1453–1456 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez-Riverol Y., et al. , The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47 (D1), D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fogg M. J., Wilkinson A. J., Higher-throughput approaches to crystallization and crystal structure determination. Biochem. Soc. Trans. 36, 771–775 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Dagley M. J., McConville M. J., DExSI: A new tool for the rapid quantitation of 13C-labelled metabolites detected by GC-MS. Bioinformatics 34, 1957–1958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evans P., Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Winter G., xia2: An expert system for macromolecular crystallography data reduction. J. Appl. Cryst. 43, 186–190 (2010). [Google Scholar]
- 44.Storoni L. C., McCoy A. J., Read R. J., Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Shukla S., et al. , Differential substrate recognition by maltose binding proteins influenced by structure and dynamics. Biochemistry 57, 5864–5876 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Murshudov G. N., Vagin A. A., Dodson E. J., Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Long F., et al. , AceDRG: A stereochemical description generator for ligands. Acta Crystallogr. D Struct. Biol. 73, 112–122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krissinel E., Henrick K., Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Holm L., Rosenström P., Dali server: Conservation mapping in 3D. Nucleic Acids Res. 38, W545-9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McNicholas S., Potterton E., Wilson K. S., Noble M. E., Presenting your structures: The CCP4mg molecular-graphics software. Acta Crystallogr. D Biol. Crystallogr. 67, 386–394 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Medema M. H., Takano E., Breitling R., Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol. Biol. Evol. 30, 1218–1223 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gilchrist C. L. M., Chooi Y. H., Clinker & clustermap.js: Automatic generation of gene cluster comparison figures. Bioinformatics (Oxford, England) 37, 2473–2475 (2021). [DOI] [PubMed] [Google Scholar]
- 55.Yilmaz P., et al. , The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–D648 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Letunic I., Bork P., Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47 (W1), W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deutsch E. W., et al. , The ProteomeXchange consortium in 2017: Supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 45 (D1), D1100–D1106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.E.D. Goddard-Borger, N. Scott, Agrobacterium sulfoquinovose metabolism. ProteomeXchange. http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD014115. Deposited 4 June 2019. [Google Scholar]
- 59.J. W.-Y. Mui, Gene cluster search pipeline. GitHub. https://github.com/jmui-unimelb/Gene-Cluster-Search-Pipeline. Deposited 10 November 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structure coordinates have been deposited in the PDB (https://www.rcsb.org/) under accession codes 7OFX, 7OFY, 7NBZ, 7OH2, 7OLF, 7BBZ, 7BC0, 7BC1, and 7BBY. Proteomics data are available via ProteomeXchange (57, 58) (http://www.proteomexchange.org/) with the identifier PXD014115. Scripts used to screen for the related gene clusters listed in Fig. 5 are available on GitHub (https://github.com/jmui-unimelb/Gene-Cluster-Search-Pipeline) (59).