Significance
Hematopoietic cell transplantation is the gold standard treatment for many blood disorders, including blood cancers. Nonetheless, frequent post-transplant complications limit the long-term benefit of HCT. Here, we find that circulating cell-free DNA is a highly versatile analyte for monitoring of the most important complications of HCT: Graft-Versus-Host Disease, infection, graft failure and disease relapse. We show that these different therapeutic complications can be informed from a single cell-free DNA sequencing assay followed by disease-specific bioinformatic analyses. This test requires only low coverage DNA sequencing and is compatible with small volumes of blood. Cell-free DNA may improve the care of allogeneic HCT recipients by enabling earlier detection and better prediction of the complex array of complications that occur after HCT.
Keywords: hematopoietic cell transplant, cell-free DNA, graft-versus-host disease, infection, disease relapse
Abstract
Allogeneic hematopoietic cell transplantation (HCT) provides effective treatment for hematologic malignancies and immune disorders. Monitoring of posttransplant complications is critical, yet current diagnostic options are limited. Here, we show that cell-free DNA (cfDNA) in blood is a versatile analyte for monitoring of the most important complications that occur after HCT: graft-versus-host disease (GVHD), a frequent immune complication of HCT, infection, relapse of underlying disease, and graft failure. We demonstrate that these therapeutic complications are informed from a single assay, low-coverage bisulfite sequencing of cfDNA, followed by disease-specific bioinformatic analyses. To inform GVHD, we profile cfDNA methylation marks to trace the cfDNA tissues-of-origin and to quantify tissue-specific injury. To inform infection, we implement metagenomic cfDNA profiling. To inform cancer relapse, we implement analyses of tumor-specific genomic aberrations. Finally, to detect graft failure, we quantify the proportion of donor- and recipient-specific cfDNA. We applied this assay to 170 plasma samples collected from 27 HCT recipients at predetermined timepoints before and after allogeneic HCT. We found that the abundance of solid-organ–derived cfDNA in the blood at 1 mo after HCT is predictive of acute GVHD (area under the curve, 0.88). Metagenomic profiling of cfDNA revealed the frequent occurrence of viral reactivation in this patient population. The fraction of donor-specific cfDNA was indicative of relapse and remission, and the fraction of tumor-specific cfDNA was informative of cancer relapse. This proof-of-principle study shows that cfDNA has the potential to improve the care of allogeneic HCT recipients by enabling earlier detection and better prediction of the complex array of complications that occur after HCT.
More than 30,000 patients undergo allogeneic hematopoietic cell transplants (HCT) worldwide each year for treatment of malignant and nonmalignant hematologic diseases (1–3). Yet, a myriad of complications occur in this patient population. For example, up to 50% of patients experience graft-versus-host disease (GVHD), an immune response in which donor immune cells attack recipient tissues in the first year after transplantation (2, 4–6). Complications due to infection also occur frequently, mostly in the first year after transplantation, with bacterial and viral infection occurring in 52 and 57.9% of patients, respectively (7, 8). In addition, up to 50% of patients treated for malignant hematologic diseases suffer cancer relapse (9, 10). Last, graft failure is a major complication of HCT (11–14).
Patient monitoring for post-HCT complications relies on a complex combination of diagnostic assays. Early and accurate diagnosis of GVHD is critical to inform treatment decisions and to prevent serious long-term complications. Unfortunately, there are few noninvasive diagnostic options that reliably identify patients early after the onset of GVHD symptoms: In current practice, diagnosis of GVHD relies primarily on clinical symptoms and requires confirmation with invasive procedures, such as biopsies of the gastrointestinal tract, skin, or liver (15). Furthermore, there is a critical need for tools that can broadly and sensitively inform infection. A wide range of microorganisms can cause disease after HCT, and infection testing currently relies on a combination of bacterial culture that are slow and suffer from a high false-negative rate and viral PCRs which have limited multiplexity. To screen for cancer recurrence, the presence of cancer cells in the circulation is used as a prognostic marker for relapse and disease-free survival. Current monitoring options of minimal residual disease include flow-cytometry and qPCR. However, these technologies are insensitive to genetic and phenotypic changes (16). Donor chimerism is currently used to quantify engraftment, relapse, and graft loss but relies on the analysis of living cells and may not be sensitive to the high turnover rate of leukemic cells (17).
Here, we investigate the utility of circulating cell-free DNA (cfDNA) as a versatile analyte to monitor HCT recipients after transplantation. cfDNA in the blood of HCT recipients is a complex mixture of DNA from several sources: different tissues, microbes, donor cells, and tumor cells (18–20) (Fig. 1A). In this work, we demonstrate that a single assay, genome-wide methylation profiling of cfDNA, enables simultaneous monitoring of the major complications that arise after HCT. First, we show that methylation profiling by whole-genome bisulfite sequencing of cfDNA can be used to quantify the tissues-of-origin of cfDNA to thereby detect and quantify tissue injury due to GVHD after HCT. Second, we demonstrate the possibility to identify infectious agents via whole-genome bisulfite sequencing of cfDNA. Last, we show that the levels of donor- and tumor-derived cfDNA can inform engraftment, mixed chimerism, and cancer relapse. Together, this study provides a proof of principle that cfDNA profiling can be used to simultaneously monitor immune, cancer, and infectious complications and treatment failure after allogeneic HCT.
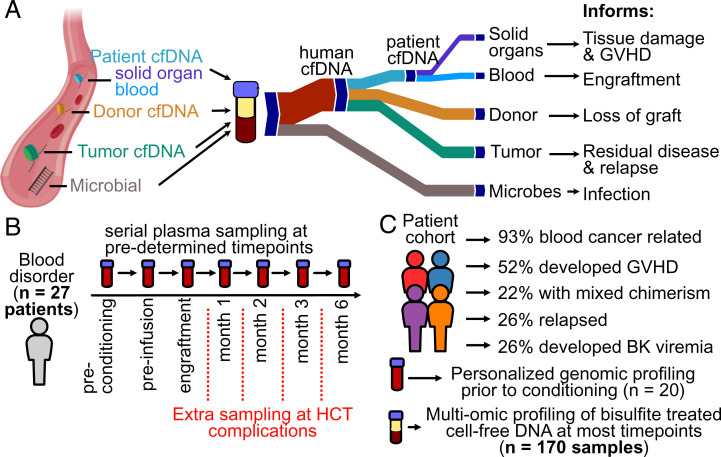
Fig. 1.
Study overview. (A) cfDNA origins inform diverse transplant events and complications. (B) Plasma from 27 HCT recipients was serially collected at seven or more predetermined timepoints. (C) Patient cohort characteristics.
Results
We performed a prospective cohort study to evaluate the utility of cfDNA to predict and monitor GVHD, infection, cancer relapse, and treatment failure after allogeneic HCT. We selected 27 adults that underwent allogeneic HCT and assayed a total of 170 serial plasma samples collected at seven predetermined timepoints, including before conditioning chemotherapy, on the day of but before hematopoietic cell infusion, after neutrophil engraftment (>500 neutrophils per microliter), and at 1, 2, 3, and 6 mo post-HCT (Fig. 1B). Additional samples were collected at the time of presentation of complications, such as symptoms of BK disease. The test cohort included patients with both malignant (n = 25) and nonmalignant blood disorders (n = 2) (Fig. 1C and SI Appendix, Table 1). Prior to conditioning, patient tumor cells were genotyped using a targeted deep sequencing panel (21) (n = 20 total, 6 patients with copy number alterations [CNAs]). In total, 14 patients developed acute GVHD (GVHD+) and 13 did not (GVHD-), four patients experienced graft failure, seven developed BK virus viremia, and five patients suffered cancer recurrence (Fig. 1C) (Materials and Methods and SI Appendix).
We isolated cfDNA from plasma (0.5 to 1.9 mL per sample) and implemented whole-genome bisulfite sequencing and bioinformatic analyses to profile epigenetic and genetic marks within cfDNA that may inform the diverse complications that arise after HCT. We implemented a single-stranded DNA (ssDNA) library preparation to obtain sequence information after bisulfite conversion (22, 23). This ssDNA library preparation avoids degradation of adapter-bound molecules, which is common for WGBS library preparations that rely on ligation of methylated adapters before bisulfite conversion and avoids amplification biases inherent to WGBS library preparations that implement random priming (24). We obtained 39 ± 14 million paired-end reads per sample, corresponding to 0.96 ± 0.4-fold per-base human genome coverage and achieved a high bisulfite conversion efficiency (99.4 ± 0.4%). We used paired-end read mapping to characterize the length of bisulfite-treated cfDNA at single-nucleotide resolution and to investigate potential degradation of cfDNA due to bisulfite treatment. This analysis revealed a fragmentation profile similar to the fragmentation profile previously reported for plasma cfDNA that was not subjected to bisulfite treatment (25). The mode of fragments longer than 100 base pairs (bp) was 165 ± 7 bp, and Fourier analysis revealed a 10.4-bp periodicity in the fragment length profile (SI Appendix, Fig. 1). A second peak at 60 to 90 bp in the fragment length profile is characteristic of single-stranded library preparation methods and was reported previously (22, 26). Overall, we do not find evidence of significant cfDNA fragmentation due to bisulfite treatment, in line with a recent report (27).
Temporal Dynamics of cfDNA Tissues-of-Origin in Response to Conditioning Therapy and HCT.
We first examined the utility of cfDNA tissues-of-origin by methylation profiling to identify organ injury due to GVHD after HCT in plasma samples obtained prior to the clinical diagnosis of GVHD. To quantify the relative proportion of cfDNA derived from different vascularized tissues and hematologic cell types, we analyzed cfDNA methylation profiles against a reference set of methylation profiles of pure cell and tissue types (28–32) (samples with sequencing depth greater than 0.1×, 138 reference tissues; Materials and Methods and Fig. 2 A and B and Dataset 1). We computed the absolute concentration of tissue-specific cfDNA by multiplying the proportion of tissue-specific cfDNA with the concentration of total host-derived cfDNA (Materials and Methods). The most striking features seen in the data include the following: 1) a decrease in blood-cell–specific cfDNA fraction in response to conditioning therapy performed to deplete the patient’s hematopoietic cells and its subsequent increase at engraftment (Fig. 2C); 2) an increase in total cfDNA concentration at engraftment (Fig. 2D); 3) a decrease in total cfDNA concentration after 180 d (SI Appendix, Fig. 2); and 4) an association between tissue-specific cfDNA and the incidence of GVHD (Statistical Analysis).
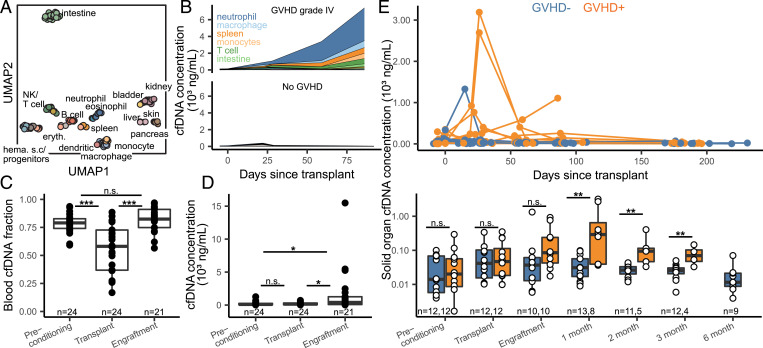
Fig. 2.
Host-derived cfDNA dynamics before and after HCT. (A) Uniform manifold approximation and projection (UMAP) dimensional reduction of cell and tissue methylation profiles. Individual tissues are colored by UMAP coordinates using a linear gradient where each of the four corners is either cyan, magenta, yellow, or black. (B) Examples of cfDNA dynamics in the case of severe GVHD (patient 003, Top) and no GVHD (patient 017, Bottom) in the first 3 mo posttransplant. (C and D) Effect of conditioning and HCT infusion on cfDNA composition (C) and absolute concentration (D). (E) Solid-organ–derived cfDNA concentration in plasma. Top: solid-organ cfDNA and days posttransplant for each patient timepoint. Bottom: solid-organ cfDNA by timepoint. Samples are removed from analysis if plasma was collected after GVHD diagnosis. *P < 0.05; **P < 0.01; ***P < 0.001.
cfDNA Tissues-of-Origin by Methylation Profiling to Monitor GVHD.
We next examined these features in more detail to explore the utility of these measurements to monitor immune related complications of HCT. Prior to conditioning, neutrophils, erythrocyte progenitors, and monocytes were the major contributors of cfDNA in plasma (22.9, 12.4, and 12.2%, respectively, average cfDNA concentration 208 ± 280 ng/mL plasma). A variety of HCT conditioning regimens have been developed with varying degrees of organ toxicity and myelosuppression. Most patients in our cohort received reduced intensity conditioning therapy (n = 25), whereas two patients received myeloablative conditioning therapy. Comparison of cfDNA tissues-of-origin in plasma before and after conditioning showed a significant drop in blood-derived cfDNA, as expected from the function of the conditioning therapy (mean proportion of hematopoietic cell cfDNA decreased from 78 ± 8 to 55 ± 22%, P value = 9.6 × 10−5; Fig. 2C). The proportion of blood-derived cfDNA increased to 82 ± 11% at engraftment (P value = 1.4 × 10−5; Fig. 2C). The most notable effect of stem cell infusion and engraftment was a significant increase in the absolute concentration of cfDNA (mean human-derived cfDNA concentration from 204 ng/mL on day of transplant to 1,763 ng/mL at engraftment [P = 0.021]; Fig. 2D).
We next evaluated the performance of a cfDNA tissue-of-origin measurement to predict GVHD (Fig. 2E). We defined GVHD here as the clinical manifestation of any stage of the disease within the first 6 mo post-HCT (GVHD+; Materials and Methods). We excluded samples collected after GVHD diagnosis, as these patients received additional GVHD treatment. We found that the concentration of solid-organ–specific cfDNA was significantly elevated for patients in the GVHD+ group at month 1, 2, and 3 (P = 0.0025, 0.0032, and 0.0044, respectively) but not at the two pretransplant timepoints (P = 0.71 prior to conditioning and P = 0.93 prior to hematopoietic cell infusion) (Fig. 2E). Receiver operating characteristic analysis (ROC) of the performance of cfDNA as a predictive marker of GVHD yielded an area under the curve (AUC) of 0.88, 0.95, and 0.96 at engraftment and months 1, 2, and 3, respectively. These results support the notion that cfDNA predicts GVHD occurrence as early as 1 mo after HCT (mean solid organ cfDNA of 873 and 39 ng/mL plasma for GVHD+ and GVHD−, respectively; ROC AUC = 0.88; P = 0.0025) (Fig. 2E). Importantly, we found that solid organ cfDNA concentration was more indicative of GVHD than total cfDNA concentration (SI Appendix, Fig. 3), highlighting the importance of methylation profiling for GVHD detection.
To evaluate the ability of this assay to pinpoint the site of GVHD, we quantified the burden of skin-derived cfDNA in the blood of GVHD-negative individuals (n = 13) and individuals who developed cutaneous GVHD (n = 12). We found that plasma samples from individuals with GVHD had a higher burden of skin-derived cfDNA prior to clinical diagnosis of skin GVHD when compared to samples from individuals who did not develop cutaneous GVHD (mean skin cfDNA of 7.2 and 2.1 ng/mL plasma, respectively; P = 0.045 for samples collected after engraftment and before clinical diagnosis; SI Appendix, Fig. 4). Skin-derived cfDNA concentration was the highest among patients who developed severe (grade 3 or 4) skin GVHD (mean skin cfDNA of 13.1 and 2.1 ng/mL plasma between samples from patients with severe skin GVHD and patients who were GVHD negative; P = 0.017, SI Appendix, Fig. 4). The number of samples from patients diagnosed with hepatic and gastrointestinal GVHD was insufficient to test the performance of the assay to identify GVHD-related injury to the liver or gut (n = 3 and 5, respectively). These results highlight the promise of whole-genome bisulfite sequencing to identify the site of incidence of GVHD, although larger follow-on studies are needed to confirm these results.
Plasma Virome Screening after HCT.
cfDNA from microbes can be detected in the circulation, providing a means to screen for infection via metagenomic cfDNA sequencing (22, 33–36). This may be a particularly powerful approach in the context of HCT, given the high incidence of infectious complications and the broad range of microorganisms that can cause disease in HCT. To test this concept, we mined all cfDNA data for microbial-derived sequences. In a previous study, we found close agreement between the abundance of organisms measured by shotgun sequencing of untreated and bisulfite-treated cfDNA, confirming the possibility to perform metagenomic cfDNA sequencing by WGBS (37). To identify microbial-derived cfDNA after WGBS, we first identified and removed host-related sequences, and we aligned the remaining unmapped reads to a set of microbial reference genomes (0.2 ± 0.4% of total reads, Materials and Methods). We implemented a background correction algorithm to remove contributions due to alignment noise and environmental contamination and compared species abundances by the relative abundance of species reads to human reads (35, 38) (relative genomic equivalents [RGE]).
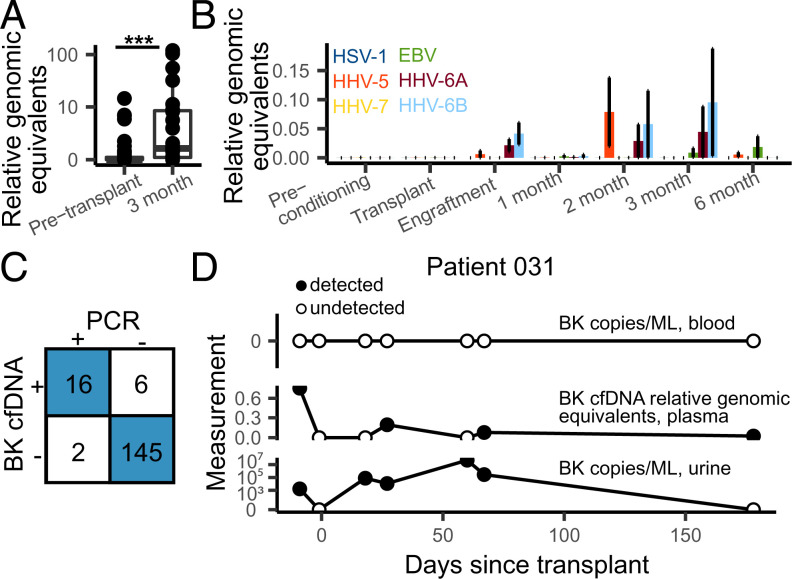
Using this procedure, we found a significant increase after HCT in the burden of cfDNA derived from DNA viruses (DNA sequencing is not sensitive to RNA molecules, average RGE of 1.34 and 26.1, preconditioning and month 3, respectively; P = 0.0090), most of which are not part of systematic clinical testing. Viruses from the Anelloviridae family were the most abundant (463 occurrences of an Anelloviridae species). We and others have reported a link between the abundance in plasma of Anelloviridae and the degree of immunosuppression in transplantation (39, 40). In line with these observations, the increase in cfDNA derived from DNA viruses was largely due to an increase in the burden of Anelloviridae in the first months after HCT (Fig. 3A). Herpesviridae and Polyomaviridae frequently establish latent infection in adults and may reactivate after allogeneic HCT (41). We identified cfDNA from human Herpesviridae and Polyomaviridae in 100 of 170 samples from 26 of 27 patients (Fig. 3 B and C).
Fig. 3.
Infectome screening in HCT patients. (A) Relative genomic equivalents of Anelloviruses detected before transplant (preconditioning and transplant timepoints) and the 3 mo timepoint. (B) Relative genomic equivalents of human herpesviruses by timepoint. Error bars represent SEM. (C) Concordance between clinically validated BK PCR test (in blood) and BK cfDNA identification. (D) BK abundances in blood (PCR test, Top), plasma (cfDNA, Middle), and urine (PCR test, Bottom) in patient 031.
BK Polyomavirus PCR tests are routinely performed in this patient population due to the frequent complications related to BK Polyomavirus. We tested the sensitivity of the cfDNA assay against a clinically validated BK Polyomavirus PCR screening test and found strong concordance (sensitivity = 0.89, specificity = 0.98; SI Appendix, Fig. 5 and Materials and Methods). For 4/6 discordant readouts, where the cfDNA test detected BK polyomavirus and the PCR test did not (in blood), three were from a patient with clinically confirmed reactivation of the virus in the urine (Fig. 3D). These findings demonstrate the possibility to sensitively screen for infectious complications after HCT via cfDNA.
Tumor- and Donor-Specific cfDNA Inform Cancer Relapse and Loss of Engraftment.
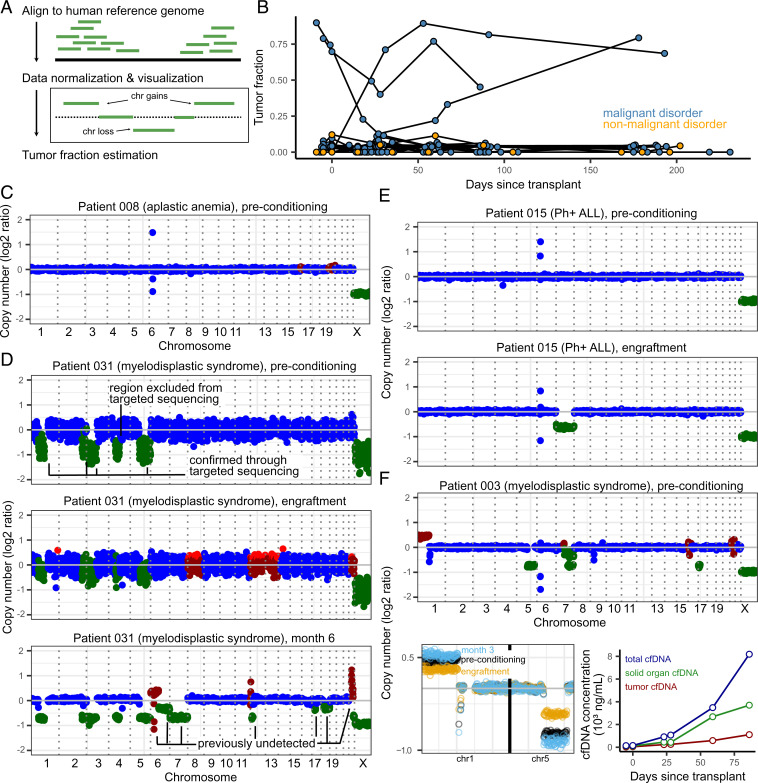
Many studies have established the utility of circulating tumor-specific cfDNA for early cancer detection and monitoring of minimal residual disease. Here, we assessed the utility of cfDNA profiling of cancer-associated CNAs as an approach to detect the presence of leukemia-derived DNA in plasma (Fig. 4A) (42). At the Dana-Farber Cancer Institute, chromosomal aberrations related to malignant blood disorders are examined using a clinically validated, targeted, ultra-deep sequencing assay [pre transplant peripheral blood mononuclear cells (PBMC); n = 20 patients, Rapid Heme Panel (RHP) (21)]. Using RHP data, we identified six patients with CNAs. We next analyzed all cfDNA WGBS sequence data and found the cfDNA assay was able to detect leukemia-specific CNAs before transplant in two of these patients (Fig. 4 B, D, and F, patients 003 [mortality] and 031 [no mortality]). Relative copy number changes were used to estimate the fraction of cell free originating from tumor cells (Fig. 4 A and B and Materials and Methods). Last, we found that the genome-wide cfDNA assay enabled detection of CNAs in regions not included in the RHP (Fig. 4D), underlining the importance of a genome-wide approach (43). Copy number changes were detected in a single sample from a patient that did not have alterations detected in a RHP taken over 100 d prior to their preconditioning timepoint.
Fig. 4.
(A) Overview of tumor fraction estimation using CNAs. (B) Tumor fractions as measured through ichorCNA at all collected timepoints. Patients without malignant disease and without CNAs (as identified through targeted sequencing) were used to gauge the error in tumor fraction measured by ichorCNA (up to 12%). (C) Example of a CAN profile in a patient with a nonmalignant blood disorder (with no alterations expected). The few outliers in the coverage plot for patient 008 are likely due to errors in sequence mapping. Genome-wide plots in (C–F) (Top only in F) are colored by ichorCNA’s identification of a given region as neutral (blue), gained (red), or lost (green). (D–F) CAN profiles of three patients with measurable CNA-based tumor fractions. (D) Patient 015 was found to have loss of chromosome 7 at the time of engraftment and in all subsequent samples. (E) Patient 031, over the course of their treatment, developed additional, clinically undetected structural variants. (F) Patient 003 (deceased on day 91) had detectable tumor fraction and clinical evidence of GVHD. Solid-organ–derived cfDNA was higher than the tumor load (line plot, Right-hand side). Top: genome-wide coverage plot. Bottom Left: copy number profiles on chromosomes 1 and 5 show a decrease in copy number changes at engraftment (yellow) and subsequent increase at month 3 (blue), when compared to the preconditioning timepoint (black). Bottom Right: Tumor- and solid-organ–derived cfDNA concentration at all available timepoints for patient 003. Patients 003, 008, 015, and 031 were all male–male donor–recipient pairs.
We highlight three cases that exemplify the utility of continuous patient monitoring. First, cfDNA monitoring for patient 031 detected new CNAs after HCT, suggesting the expansion of a subclonal tumor population over the course of treatment (Fig. 4D and SI Appendix, Fig. 6). In this patient, we estimated tumor fractions of 90% at preconditioning, 23% at engraftment, and 79% at month 6. Bone marrow biopsies performed 8.5 mo after transplant (outside the timeframe of the current study) revealed hypocellular marrow consistent with acute myeloid leukemia. Second, profiling of patient 015 diagnosed with Philadelphia chromosome positive ALL (Ph+ALL) revealed the presence of monosomy 7 at engraftment and all subsequent timepoints (Fig. 4E and SI Appendix, Fig. 7). Clinical chimerism testing based on short-tandem repeat PCR amplification for this same patient showed full engraftment of donor cells, and bone marrow examination showed no evidence of leukemia relapse. Cytogenic analysis performed after transplant confirmed monosomy 7 in donor cells for this patient, highlighting the utility of an untargeted sequencing assay to identify rare transplantation events. Last, for patient 003 (Fig. 4F and SI Appendix, Fig. 8), who was diagnosed with severe GVHD (cutaneous stage 4, overall grade IV; unresolved; mortality day 91), cfDNA tissue-of-origin profiling revealed an increase in solid-organ–derived cfDNA in addition to increasing tumor-derived cfDNA load (Fig. 4F), potentially pointing toward a joint GVHD and relapse.
Plasma Donor–Derived cfDNA as a Marker of Mixed Chimerism.
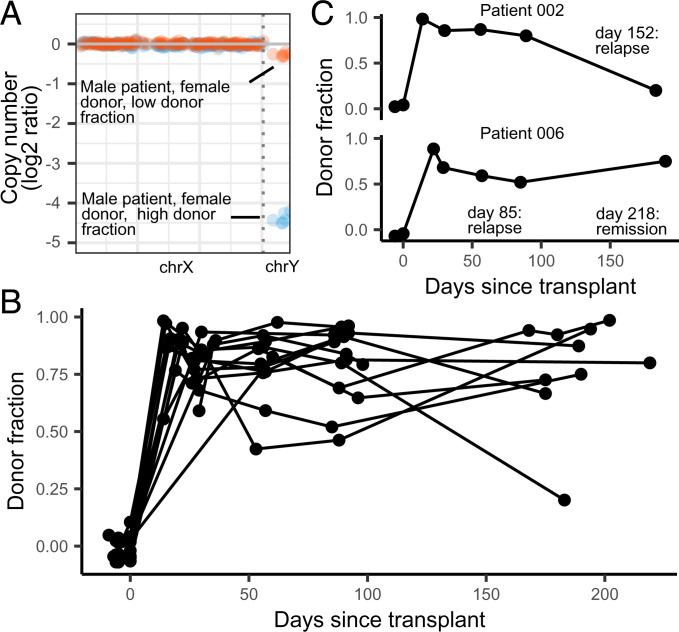
Measurements of donor–recipient chimerism are a routine part of clinical monitoring and can inform cancer relapse and loss the donor stem cell graft (44, 45). These measurements are performed on isolated hematopoietic cell populations and do not account for the turnover rate of cells, which are often higher in leukemic cells than in normal cells (46). Therefore, it has been proposed that chimerism analysis of cfDNA may offer complimentary information to traditional cell-based chimerism analysis (17, 47). Here, we show the feasibility of measuring donor-derived cfDNA (dd-cfDNA) by leveraging the relative abundance of X and Y chromosomes in sex-mismatched recipient pairs (samples with depth of sequencing > 0.1×; Fig. 5A). We analyzed samples collected prior to transplantation to assess the error rate of this measurement (mean donor fraction 0.0 ± 4.6%). We found that the donor fraction is highest at engraftment (86 ± 13%) and remains constant in the absence of complications (Fig. 5B and SI Appendix, Figs. 9 and 10). We highlight two examples from patients who experienced HCT complications (Fig. 5C). In Patient 002, we observed a gradual decrease in dd-cfDNA after engraftment, with a sharp drop on day 183. This patient developed disease relapse on day 152, and the gradual decrease in dd-cfDNA preceded relapse. Similarly, in patient 006, we observed a steady drop in dd-cfDNA after engraftment, prior to disease relapse on day 85. The fraction of dd-cfDNA for this patient subsequently increased at month six, before the patient entered remission (day 218). While these data show decreases in donor fraction in patients who experience relapse, changes in donor fraction in HCT patients may also be due to other clinical issues, such as loss of engraftment. Taken together, these data suggest that dd-cfDNA is an informative biomarker for HCT monitoring and can be used in conjunction with other cfDNA features to inform levels of donor cell engraftment and quantification of residual recipient cells and relapse.
Fig. 5.
Donor fractions and days posttransplant in sex-mismatched patients. (A) The donor fraction is measured from the relative coverage of sex chromosomes (Materials and Methods). (B) Donor fraction in all sex-mismatched patients. (C) Donor fraction in two patients who experienced disease relapse.
Discussion
In this work, we have introduced a cfDNA assay with the potential to simultaneously screen for the most important complications that arise after allogeneic HCT. This work was inspired by recent studies that have shown that cfDNA is an analyte with utility in 1) monitoring rejection after solid-organ transplantation (48–51); 2) screening for infection and viral reactivation (33, 36, 37); and 3) early detection of cancer or relapse of disease (43, 52, 53).
Numerous studies have demonstrated that dd-cfDNA in the blood of solid-organ transplant recipients is a quantitative marker of solid-organ transplant injury (49, 50), and a variety of commercial cfDNA assays are already in use (48, 54–57). We reasoned that cfDNA may also inform tissue injury due to GVHD after HCT. To quantify cfDNA derived from any tissue, we implemented bisulfite sequencing of cfDNA to profile cytosine methylation marks which are comprised within cfDNA and are cell-, tissue-, and organ-type specific. We found that the burden of cfDNA from solid organs is predictive of the onset of GVHD as early as 1 mo after HCT. Protein biomarkers have previously been investigated for diagnosis and prediction of GVHD (58, 59). ST2 and REG3-α, which both derive from the gastrointestinal tract, are two such biomarkers with the strongest predictive power. The cfDNA assay presented here has inherent advantages over these protein biomarker technologies. First, cfDNA may provide a generalizable approach to measure injury to any tissue, whereas protein injury markers may not be available for all cell and tissue types. Second, because the concentration of tissue-specific DNA can be directly related to the degree of cellular injury (37, 60–62), cfDNA may offer a measure of injury that can be trended over time.
Whole-genome sequencing is not only responsive to human host-derived cfDNA but also to microbial cfDNA in the blood circulation. Several recent studies have demonstrated the value of metagenomic cfDNA sequencing to screen for infection in a variety of clinical settings, including urinary tract infection (33, 37), sepsis (36), and invasive fungal disease (63). In HCT, metagenomic cfDNA sequencing has been used to identify pathogens in blood before clinical onset of bloodstream infections (64). Here, we explored the potential to identify viral-derived cfDNA in plasma of HCT recipients using whole-genome bisulfite sequencing. This approach revealed the frequent presence of cfDNA from anelloviruses, cytomegalovirus, herpesvirus 6, Epstein–Barr virus, and polyomavirus in the blood of HCT recipients. Anelloviruses were common in this cohort and, while rarely pathogenic, can be used as a surrogate for the degree of immunosuppression in transplant patients (39, 40, 65). We demonstrate sensitive detection of BK virus cfDNA for patients that were BK virus positive in blood and for a patient that tested negative for BK virus in the blood but tested positive for BK virus in the urine, which may indicate that the cfDNA assay reported here has a higher sensitivity than clinical PCR assays. The assay reported here has the potential to simultaneously inform about GVHD, from the tissues-of-origin of host cfDNA and infection, from metagenomic analysis of microbial cfDNA. Compared to conventional metagenomic sequencing, this assay requires one additional experimental step to bisulfite convert cfDNA, which can be completed within ∼2 h and is compatible with multiple existing next-generation sequencing workflows. However, it remains important to note that this metagenomic sequencing assay is insensitive to RNA viruses and microbes for which a reference genome is not available.
Circulating tumor DNA has been shown to be a highly sensitive molecule for the detection of minimal residual disease (43, 66). The identification of solid-tumor–derived circulating nucleic acids relies on the identification of single-nucleotide polymorphisms or CNAs (42, 52) or detection of changes in DNA fragmentation patterns (53, 67, 68). In this work, we focused on structural variants of malignant disease to detect tumor-specific cfDNA and found evidence of subclonal expansion, newly acquired mutations, and simultaneous occurrence of GVHD and cancer relapse. Future studies where whole-genome sequencing is performed on the primary tumor cells may uncover tumor-associated single nucleotide polymorphisms (SNPs) and be used in conjunction with CNA analysis to improve detection of circulating tumor DNA in malignant blood disease (43).
Donor-derived cells and, recently, dd-cfDNA have been explored as markers for GVHD, loss of graft, and recurrence of disease (19, 47). We observed increased amounts of dd-cfDNA at engraftment, and these levels remained elevated in the absence of HCT complications. For patients that suffered relapse of disease, we observed a decrease in the burden of dd-cfDNA, potentially due to suppression of normal marrow cells by leukemic cells or increases in recipient tissue damage (19, 69). Studies by Duque-Afonso et al. and Sharon et al. reported elevated amounts of transplant recipient cfDNA in cases of GVHD, suggesting patient tissue contributions to the cfDNA mixture (19, 47). Interestingly, Duque-Afonso et al. also observed increased recipient cfDNA at the time of relapse and progressive disease, suggesting that donor-derived (or recipient-derived) cfDNA alone may not be sufficient in distinguishing different important complications of HCT, supporting the need for an assay that is informative of the tissues-of-origin of cfDNA. Follow-on studies characterizing the relationship between dd-cfDNA dynamics and HCT complications such as graft loss and relapse are necessary to characterize the sensitivity and specificity of our assay.
This is a proof-of-principle study with several limitations that can be addressed in future work. First, the scope of the current study with 27 patients was not powered to detect any association of cfDNA with acute GVHD involving organs other than skin (liver and gut). Our results suggest that cfDNA tissue-of-origin profiling is predictive of acute GVHD, but larger studies will be needed to extend the current observations to other sites of organ damage and to assess its utility in detecting and diagnosing chronic GVHD. In addition, larger studies, including patient populations with diverse HCT complications, are necessary to resolve the origins of cfDNA in cases of relapse of disease. Despite these potential limitations, we have shown here that cfDNA is a versatile analyte to monitor HCT patients, and our data highlights the importance of comprehensive monitoring all origins of cfDNA to assess the most severe complications of HCT.
Materials and Methods
Study Cohort.
We performed a nested case-control study within a prospective cohort of adult patients undergoing allogeneic HCT at Dana-Farber Cancer Institute. Patients were followed for 6 mo after HCT. Patients were selected for this study on a rolling basis and were placed in the GVHD case or control groups based on clinical manifestation of the disease within the first 6 mo after HCT. The study was approved by the Dana-Farber/Harvard Cancer Center’s Office of Human Research Studies. All patients provided written informed consent.
For this study, we used 170 blood samples collected from 27 allogeneic HCT recipients from August 2018 to August 2019. Baseline patient characteristics were recorded. Covariates of interest included human leukocyte antigen (HLA) matching, donor relatedness, and donor–recipient sex mismatch (SI Appendix, Table 1). Date of onset of GVHD as well as GVHD prophylaxis and treatment regimens were documented. GVHD was diagnosed clinically and pathologically. GVHD severity was graded according to the Glucksberg criteria (43). Other clinical events of interest included the development of bloodstream infections, BK polyomavirus disease, and clinical disease from other DNA viruses.
Timepoints.
Standard timepoints for plasma collection were determined prior to patient recruitment and included preconditioning (the day of their first conditioning dose, prior to receiving treatment), transplantation (the day of transplantation, prior to transfusion), engraftment (detailed in Engraftment) and months 1, 2, 3, and 6 after transplant. In the event of BK-related symptoms, disease, or reactivation, additional timepoints were collected. In the case of two timepoints overlapping, the sample was preferentially labeled as engraftment, month 1, month 3, or month 6 (in that order).
Engraftment.
Neutrophil engraftment was considered when blood samples contained an absolute neutrophil count greater or equal than 500 cell per microliter blood on two separate measurements.
Relapse.
Disease relapse was defined through standard criteria for each underlying disease.
Mixed Chimerism.
Mixed chimerism is broadly defined as 5 to 95% T cells of donor origin (44). Here, we used a criteria of <75% T cells of donor origin to characterize mixed chimerism. Only timepoints obtained after engraftment were considered.
Clinical BK Polyomavirus Disease Identification.
Patients were identified as BK virus disease positive when they presented BK-related urinary symptoms that correlated with positive BK qPCR test in either urine or blood (>105 copies/mL in urine, >0 copies/mL in blood; Viracor BK qPCR test, reference No. 2500) and did not have evidence of any other cause of genitourinary pathology at the time of symptom onset.
Blood Sample Collection and Plasma Extraction.
Blood samples were collected through standard venipuncture in ethylenediaminetetraacetic acid tubes (Becton Dickinson, reference No. 366643) on admission, before the beginning of the conditioning chemotherapy, on the day of HCT after the completion of the conditioning chemotherapy, at engraftment (usually 14 to 21 d after HCT), and at months 1, 2, 3 and 6 post-HCT. Plasma was extracted through blood centrifugation (2,000 rpm for 10 min using a Beckman Coulter Allegra 6R centrifuge) and stored in 0.5- to 2-mL aliquots at −80 °C. Plasma samples were shipped from the Dana-Farber Cancer Institute to Cornell University on dry ice.
Nucleic Acid Control Preparation.
Synthetic oligos were prepared (IDT, SI Appendix, Table 2), mixed in equal proportions, and diluted at ∼150 ng/uL. At the time of cfDNA extraction, 8 uL control was added to 1,992 µL 1× phosphate-buffered saline (PBS) and processed as a sample in all downstream experiments.
cfDNA Extraction.
cfDNA was extracted according to manufacturer recommendations (Qiagen Circulating Nucleic Acid Kit, reference No. 55114; elution volume, 45 µL). Eluted DNA was quantified using a Qubit 3.0 Fluorometer (using 2 µL eluted DNA). Measured cfDNA concentration was obtained using the following formula:
Whole-Genome Bisulfite Sequencing.
cfDNA and nucleic acid controls were bisulfite treated according to manufacturer recommendations (Zymo Methylation Lightning Kit, reference No. D5030). Sequencing libraries were prepared using a previously described single-stranded library preparation protocol. Libraries were quality controlled through DNA fragment analysis (Agilent fragment analyzer) and sequenced on an Illumina NextSeq550 machine using 2 ± 75-bp reads. Nucleic acid controls were sequenced at ∼1% of the total sequencing lane.
Human Genome Alignment.
Adapter sequences were trimmed using BBTools (70). The Bismark alignment tool (71) was used to align reads to the human genome (version hg19), remove PCR duplicates, and calculate methylation densities.
Reference Tissue Methylation Profiles and Tissue-of-Origin Measurement.
Reference tissue methylomes were obtained from publicly available databases (28–32) (Dataset 1). Genomic coordinates from different sources were normalized and converted to a standard four-column bed file (columns: chromosome, start, end, and methylation fraction) using hg19 assembly coordinates. Methylation profiles were grouped by tissue type, and differentially methylated regions were found using Metilene (72). Tissues and cell types of origin were determined using quadratic programming as previously described (37).
Donor Fraction.
Donor fractions were calculated by measuring the relative coverage of X and Y chromosomes in sex-mismatched donor–recipient pairs. Coverage was summed across binned, 500–base pair windows and adjusted for mappability and guanine-cytosine content using HMMcopy (33, 73).
Tumor Fraction.
ichorCNA (42) (version 2.0) was used to detect CNAs and estimate tumor fraction in patients with cancer. A window size of 1 MB along with a ploidy of(2,3) and a wide range of nontumor restart fractions were used to calculate coverage on autosomal chromosomes. Coverage was normalized using a panel of normals generated from the plasma of five healthy donors (IRB 1910009101). The plasma used for the panel of normals was processed using the same workflow as described in the whole-genome bisulfite sequencing methods to account for experimental and sequencing artifacts. The normalized coverage profile for each sample was then used to detect CNAs and estimate tumor fraction.
Metagenomic Alignment and Quantification of Microbial cfDNA.
After WGBS, reads were adapter trimmed using BBTools (70), and short reads are merged with FLASH (74). Sequences were aligned to a C-to-T converted genome using Bismark (71). Unmapped reads were BLASTed (75) using hs-blastn (76) to a list of C-to-T–converted microbial reference genomes. A relative abundance of all detected organisms was determined using GRAMMy (77), and relative genomic abundances are measured as previously described (35). Microbial cfDNA fraction was calculated by dividing the unique number of reads mapping to microbial species (after adjusting for the length of each microbial genome in the reference set) to the total number of adapter-trimmed reads. Human fraction is estimated as 1 − microbial fraction. Microbial species were then filtered for environmental contamination and alignment noise using previously described methods (38).
cfDNA Concentration.
cfDNA concentration of a specific tissue or microbe is calculated as follows:
cfDNA BK Polyomavirus Identification.
In our unbiased metagenomic cfDNA assay, we use RGE, the relative coverage of a given viral genome compared to the host (hg19) coverage, to compare the microbial abundance of samples with different sequencing depths. Therefore, a BK polyomavirus RGE > 0 implies that at least one cfDNA molecule mapped to the BK genome after filtering for batch effects and the presence of contamination in negative controls (see nucleic acid control section). Here, we defined an RGE > 0 as positive for BK virus. To determine this RGE threshold, we used samples from patients with no detected BK through blood and urine PCR as true negatives (n =18) and samples from patients with detectable BK virus through blood PCR as true positives (n = 121). Comparing the RGE from these groups revealed significant differences in the amount of BK-derived cfDNA (mean 1.527 ± 3.292 versus 0.001 ± 0.007 RGE in clinically positive and negative samples, respectively; P < 2.2 × 10−16). ROC analysis on this data also revealed an AUC of 0.942 (SI Appendix, Fig. 5). Comparing the RGE of the false-positives (RGE = 0.024, 0.032, and 0.07) to the lower-end RGEs of true positives (RGE = 0.00, 0.00, 0.0299, 0.038, and 0.09), we opted to minimize false-negatives with a cutoff of RGE > 0.
Depth of Coverage.
The depth of sequencing was measured by summing the depth of coverage for each mapped base pair on the human genome after duplicate removal and dividing by the total length of the human genome (hg19, without unknown bases).
Bisulfite Conversion Efficiency.
We estimated bisulfite conversion efficiency by quantifying the rate of C[A/T/C] methylation in human-aligned reads [using MethPipe (78)], which are rarely methylated in mammalian genomes.
Statistical Analysis.
Statistical analysis was performed in R (version 3.5). All tests were performed using a two-sided Wilcoxon test.
Supplementary Material
Acknowledgments
We thank the Cornell Genomics Center for help with sequencing assays, the Cornell Bioinformatics facility for computational assistance, the Pasquarello Tissue Bank at the Dana-Farber Cancer Institute for sample processing and cryopreservation, and members of the I.D.V. laboratory for helpful discussions. We thank Francoise Vermeulen of the Cornell Statistical Consulting Unit for helpful discussion. This work was supported by R01AI146165 (to M.P.C., F.M.M., J.R., and I.D.V.), R21AI133331 (to I.D.V.), R21AI124237 (to I.D.V.), DP2AI138242 (to I.D.V.), and a National Sciences and Engineering Research Council of Canada fellowship PGS-D3 (to A.P.C.).
Footnotes
Competing interest statement: A.P.C., M.P.C., P.B., J.R., and I.D.V. have submitted patents related to the presented work. M.P.C. reports grants from McGill Interdisciplinary Initiative in Infection and Immunity, grants from Canadian Institutes of Health Research, during the conduct of the study; personal fees from GEn1E Lifesciences (as a member of the scientific advisory board), personal fees from nplex biosciences (as a member of the scientific advisory board), outside the submitted work. M.P.C. and I.D.V. are co-founders of Kanvas Biosciences and own equity in the company. P.B. is currently an employ at Kanvas Biosciences and owns equity in the company. I.D.V. is a member of the Scientific Advisory Board of Karius Inc. J.R. receives research funding from Amgen, Equillium, Kite/Gilead, and Novartis and serves on Data Safety Monitoring Committees for AvroBio and Scientific Advisory Boards for Akron Biotech, Clade Therapeutics, Garuda Therapeutics, Immunitas Therapeutics, LifeVault Bio, Novartis, Rheos Medicines, Talaris Therapeutics, and TScan Therapeutics.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2113476118/-/DCSupplemental.
Data Availability
Sequence data is being deposited at the database for phenotypes and genotypes (dbGaP, https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001564.v3.p1). All code used to generate figures and analyze primary data are available at GitHub (https://github.com/alexpcheng/gvhd_submission) (79).
References
- 1.Gratwohl A., et al. , Worldwide Network of Blood and Marrow Transplantation, Hematopoietic stem cell transplantation: A global perspective. JAMA 303, 1617–1624 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McDonald-Hyman C., Turka L. A., Blazar B. R., Advances and challenges in immunotherapy for solid organ and hematopoietic stem cell transplantation. Sci. Transl. Med. 7, 280rv2–280rv2 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niederwieser D., et al. , Hematopoietic stem cell transplantation activity worldwide in 2012 and a SWOT analysis of the Worldwide Network for Blood and Marrow Transplantation Group including the global survey. Bone Marrow Transplant. 51, 778–785 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paczesny S., Discovery and validation of graft-versus-host disease biomarkers. Blood 121, 585–594 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrara J. L. M., Deeg H. J., Graft-versus-host disease. N. Engl. J. Med. 324, 667–674 (1991). [DOI] [PubMed] [Google Scholar]
- 6.Ferrara J. L. M., Chaudhry M. S., GVHD: Biology matters. Blood Adv. 2, 3411–3417 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuster M. G., et al. , Infections in hematopoietic cell transplant recipients: Results from the organ transplant infection project, a multicenter, prospective, cohort study. Open Forum Infect. Dis. 4, ofx050 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Czyzewski K., et al. , Epidemiology, outcome and risk factors analysis of viral infections in children and adolescents undergoing hematopoietic cell transplantation: Antiviral drugs do notprevent epstein–barr virus reactivation. Infect. Drug Resist. 12, 3893–3902 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J.-H., et al. , Allogeneic hematopoietic cell transplantation for acute leukemia in first relapse or second remission. Korean J. Hematol. 45, 95–101 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mawad R., Lionberger J. M., Pagel J. M., Strategies to reduce relapse after allogeneic hematopoietic cell transplantation in acute myeloid leukemia. Curr. Hematol. Malig. Rep. 8, 132–140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Passweg J. R., et al. , Donor characteristics affecting graft failure, graft-versus-host disease, and survival after unrelated donor transplantation with reduced-intensity conditioning for hematologic malignancies. Biol. Blood Marrow Transplant. 17, 1869–1873 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Satwani P., et al. , Transplantation-related mortality, graft failure, and survival after reduced-toxicity conditioning and allogeneic hematopoietic stem cell transplantation in 100 consecutive pediatric recipients. Biol. Blood Marrow Transplant. 19, 552–561 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Davies S. M., et al. , Engraftment and survival after unrelated-donor bone marrow transplantation: A report from the national marrow donor program. Blood 96, 4096–4102 (2000). [PubMed] [Google Scholar]
- 14.Gooley T. A., et al. , Reduced mortality after allogeneic hematopoietic-cell transplantation. N. Engl. J. Med. 363, 2091–2101 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogelsang G. B., Lee L., Bensen-Kennedy D. M., Pathogenesis and treatment of graft-versus-host disease after bone marrow transplant. Annu. Rev. Med. 54, 29–52 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Jentzsch M., et al. , Clinical challenges and consequences of measurable residual disease in non-APL acute myeloid leukemia. Cancers (Basel) 11, 1625 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aljurf M., et al. , Chimerism analysis of cell-free DNA in patients treated with hematopoietic stem cell transplantation may predict early relapse in patients with hematologic malignancies. Biotechnol. Res. Int. 2016, 8589270 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buedts L., Vandenberghe P., Circulating cell-free DNA in hematological malignancies. Haematologica 101, 997–999 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharon E., et al. , Quantification of transplant-derived circulating cell-free DNA in absence of a donor genotype. PLoS Comput. Biol. 13, e1005629 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camargo J. F., et al. , Next-generation sequencing of microbial cell-free DNA for rapid noninvasive diagnosis of infectious diseases in immunocompromised hosts. F1000 Res. 8, 1194 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kluk M. J., et al. , Validation and implementation of a custom next-generation sequencing clinical assay for hematologic malignancies. J. Mol. Diagn. 18, 507–515 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burnham P., et al. , Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma. Sci. Rep. 6, 27859 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gansauge M.-T., Meyer M., Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat. Protoc. 8, 737–748 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Miura F., Enomoto Y., Dairiki R., Ito T., Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 40, e136–e136 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang P., Lo Y. M. D., The long and short of circulating cell-free DNA and the ins and outs of molecular diagnostics. Trends Genet. 32, 360–371 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Snyder M. W., Kircher M., Hill A. J., Daza R. M., Shendure J., Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 164, 57–68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Werner B., et al. , Circulating cell-free DNA from plasma undergoes less fragmentation during bisulfite treatment than genomic DNA due to low molecular weight. PLoS One 14, e0224338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.ENCODE Project Consortium, The ENCODE (ENCyclopedia Of DNA elements) project. Science 306, 636–640 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Bujold D., et al. , The international human epigenome consortium data portal. Cell Syst. 3, 496–499.e2 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Albrecht F., List M., Bock C., Lengauer T., DeepBlue epigenomic data server: Programmatic data retrieval and analysis of epigenome region sets. Nucleic Acids Res. 44 (W1), W581–W586 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernstein B. E., et al. , The NIH Roadmap Epigenomics Mapping Consortium, The NIH roadmap epigenomics mapping consortium. Nat. Biotechnol. 28, 1045–1048 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernández J. M., et al. , BLUEPRINT Consortium, The BLUEPRINT data analysis portal. Cell Syst. 3, 491–495.e5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burnham P., et al. , Urinary cell-free DNA is a versatile analyte for monitoring infections of the urinary tract. Nat. Commun. 9, 2412 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kowarsky M., et al. , Numerous uncharacterized and highly divergent microbes which colonize humans are revealed by circulating cell-free DNA. Proc. Natl. Acad. Sci. U.S.A. 114, 9623–9628 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Vlaminck I., et al. , Temporal response of the human virome to immunosuppression and antiviral therapy. Cell 155, 1178–1187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blauwkamp T. A., et al. , Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat. Microbiol. 4, 663–674 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Cheng, A. P., et al. , A cell-free DNA metagenomic sequencing assay that integrates the host injury response to infection. Proc. Natl. Acad. Sci. U.S.A. 116, 18738–18744 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burnham P., et al. , Separating the signal from the noise in metagenomic cell-free DNA sequencing. Microbiome 8, 18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomblyn M., et al. , Center for International Blood and Marrow Research; National Marrow Donor program; European Blood and MarrowTransplant Group; American Society of Blood and Marrow Transplantation; Canadian Blood and Marrow Transplant Group; Infectious Diseases Society of America; Society for Healthcare Epidemiology of America; Association of Medical Microbiology and Infectious Disease Canada; Centers for Disease Control and Prevention, Guidelines for preventing infectious complications among hematopoietic cell transplantation recipients: A global perspective. Biol. Blood Marrow Transplant. 15, 1143–1238 (2009). Correction in: Biol. Blood Marrow Transplant. 16, 294 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitz J., et al. , The value of torque teno virus (TTV) as a marker for the degree of immunosuppression in adult patients after hematopoietic stem cell transplantation (HSCT). Biol. Blood Marrow Transplant. 26, 643–650 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Marr K. A., Delayed opportunistic infections in hematopoietic stem cell transplantation patients: A surmountable challenge. Hematology Am. Soc. Hematol. Educ. Program 2012, 265–270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adalsteinsson V. A., et al. , Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 8, 1324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zviran A., et al. , Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nat. Med. 26, 1114–1124 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baron F., Sandmaier B. M., Chimerism and outcomes after allogeneic hematopoietic cell transplantation following nonmyeloablative conditioning. Leukemia 20, 1690–1700 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Olsson R., et al. , Graft failure in the modern era of allogeneic hematopoietic SCT. Bone Marrow Transplant. 48, 537–543 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Holyoake T. L., Jiang X., Drummond M. W., Eaves A. C., Eaves C. J., Elucidating critical mechanisms of deregulated stem cell turnover in the chronic phase of chronic myeloid leukemia. Leukemia 16, 549–558 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Duque-Afonso J., et al. , Cell-free DNA characteristics and chimerism analysis in patients after allogeneic cell transplantation. Clin. Biochem. 52, 137–141 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Sigdel T. K., et al. , A rapid noninvasive assay for the detection of renal transplant injury. Transplantation 96, 97–101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Vlaminck I., et al. , Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci. Transl. Med. 6, 241ra77 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Vlaminck I., et al. , Noninvasive monitoring of infection and rejection after lung transplantation. Proc. Natl. Acad. Sci. U.S.A. 112, 13336–13341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snyder T. M., Khush K. K., Valantine H. A., Quake S. R., Universal noninvasive detection of solid organ transplant rejection. Proc. Natl. Acad. Sci. U.S.A. 108, 6229–6234 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mouliere F., et al. , Detection of cell-free DNA fragmentation and copy number alterations in cerebrospinal fluid from glioma patients. EMBO Mol. Med. 10, e9323 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mouliere F., et al. , High fragmentation characterizes tumour-derived circulating DNA. PLoS One 6, e23418 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grskovic M., et al. , Validation of a clinical-grade assay to measure donor-derived cell-free DNA in solid organ transplant recipients. J. Mol. Diagn. 18, 890–902 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Beck J., et al. , Digital droplet PCR for rapid quantification of donor DNA in the circulation of transplant recipients as a potential universal biomarker of graft injury. Clin. Chem. 59, 1732–1741 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Gielis E. M., et al. , Cell-free DNA: An upcoming biomarker in transplantation. Am. J. Transplant. 15, 2541–2551 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Knight S. R., Thorne A., Lo Faro M. L., Donor-specific cell-free DNA as a biomarker in solid organ transplantation. A systematic review. Transplantation 103, 273–283 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Chen Y.-B., Cutler C. S., Biomarkers for acute GVHD: Can we predict the unpredictable? Bone Marrow Transplant. 48, 755–760 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Paczesny S., et al. , Three biomarker panel at day 7 and 14 can predict development of grade II-IV acute graft-versus-host disease. Blood 116, 675 (2010).20688963 [Google Scholar]
- 60.Sun K., et al. , Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl. Acad. Sci. U.S.A. 112, E5503–E5512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng A. P., et al. , Cell-free DNA tissues of origin by methylation profiling reveals significant cell, tissue, and organ-specific injury related to COVID-19 severity. Med (N Y) 2, 411–422.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moss J., et al. , Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 9, 5068 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hong D. K., et al. , Liquid biopsy for infectious diseases: Sequencing of cell-free plasma to detect pathogen DNA in patients with invasive fungal disease. Diagn. Microbiol. Infect. Dis. 92, 210–213 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Goggin K. P., et al. , Evaluation of plasma microbial cell-free DNA sequencing to predict bloodstream infection in pediatric patients with relapsed or refractory cancer. JAMA Oncol. 6, 552–556 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lecuit M., Eloit M., The human virome: New tools and concepts. Trends Microbiol. 21, 510–515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Diehl F., et al. , Circulating mutant DNA to assess tumor dynamics. Nat. Med. 14, 985–990 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mouliere F., et al. , Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 10, 4921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cristiano S., et al. , Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 570, 385–389 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miraki-Moud F., et al. , Acute myeloid leukemia does not deplete normal hematopoietic stem cells but induces cytopenias by impeding their differentiation. Proc. Natl. Acad. Sci. U.S.A. 110, 13576–13581 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bushnell B., Rood J., Singer E., BBMerge – Accurate paired shotgun read merging via overlap. PLoS One 12, e0185056 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krueger F., Andrews S. R., Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jühling F., et al. , metilene: Fast and sensitive calling of differentially methylated regions from bisulfite sequencing data. Genome Res. 26, 256–262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ha G., et al. , Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome Res. 22, 1995–2007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Magoč T., Salzberg S. L., FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 76.Chen Y., Ye W., Zhang Y., Xu Y., High speed BLASTN: An accelerated MegaBLAST search tool. Nucleic Acids Res. 43, 7762–7768 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xia L. C., Cram J. A., Chen T., Fuhrman J. A., Sun F., Accurate genome relative abundance estimation based on shotgun metagenomic reads. PLoS One 6, e27992 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song Q., et al. , A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. PLoS One 8, e81148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.A. P. Cheng, gvhd submission. GitHub. https://github.com/alexpcheng/gvhd_submission. Accessed 20 November 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data is being deposited at the database for phenotypes and genotypes (dbGaP, https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001564.v3.p1). All code used to generate figures and analyze primary data are available at GitHub (https://github.com/alexpcheng/gvhd_submission) (79).