Abstract
Proteins, as essential biomolecules, account for a large fraction of cell mass, and thus the synthesis of the complete set of proteins (i.e., the proteome) represents a substantial part of the cellular resource budget. Therefore, cells might be under selective pressures to optimize the resource costs for protein synthesis, particularly the biosynthesis of the 20 proteinogenic amino acids. Previous studies showed that less energetically costly amino acids are more abundant in the proteomes of bacteria that survive under energy-limited conditions, but the energy cost of synthesizing amino acids was reported to be weakly associated with the amino acid usage in Saccharomyces cerevisiae. Here we present a modeling framework to estimate the protein cost of synthesizing each amino acid (i.e., the protein mass required for supporting one unit of amino acid biosynthetic flux) and the glucose cost (i.e., the glucose consumed per amino acid synthesized). We show that the logarithms of the relative abundances of amino acids in S. cerevisiae’s proteome correlate well with the protein costs of synthesizing amino acids (Pearson’s r = −0.89), which is better than that with the glucose costs (Pearson’s r = −0.5). Therefore, we demonstrate that S. cerevisiae tends to minimize protein resource, rather than glucose or energy, for synthesizing amino acids.
Keywords: amino acid biosynthetic cost, constraint-based modeling, metabolic engineering, proteome constraint, Saccharomyces cerevisiae
Proteins perform diverse functions in living cells, and the total proteome accounts for a substantial fraction of cell mass. The biosynthesis of the building blocks (i.e., 20 proteinogenic amino acids) thus requires a large investment of resources, such as nutrients in the environments. Evidence showed that nutrient scarcity plays a key role in the evolution of protein sequences of marine microbes (1, 2), meaning that cellular amino acid composition could be optimized for resource conservation.
To quantitatively investigate the relationship between environmental nutrients and cellular amino acid compositions, the concept of amino acid biosynthetic cost was proposed, which estimates the energy or substrate consumed per amino acid synthesized based on metabolic network (3, 4). By assuming that energy is limiting to cell survival, studies showed that in diverse prokaryotic organisms, highly expressed proteins favor amino acids that have lower average energy costs (3, 5), meaning that the cellular proteome is biased toward amino acids whose biosynthesis require less amount of the limiting substrate, typically glucose.
However, a few studies showed that the traditional amino acid biosynthetic cost (i.e., energy cost of synthesizing amino acids) might be a weak descriptor of the amino acid composition of the yeast Saccharomyces cerevisiae (4, 6), a unicellular eukaryote that could derive from diverse ecological origins (7), among which substrates are mostly in excess. Thus, other resources might be optimized for synthesizing amino acids for organisms that have evolved in nutrient-rich environments.
Given that cells growing in nutrient-rich conditions could be constrained by a limited cellular proteome, which is an internal resource that should be optimized to balance various biological activities (8), we hypothesize that in yeast the amino acids that require more mass of protein resource for their biosynthesis are less frequently utilized and vice versa. To test this hypothesis, we estimated the protein cost (9) of synthesizing each amino acid, which represents the protein mass required for supporting one unit of amino acid biosynthetic flux. Subsequently, we investigated the relationship between the protein costs of synthesizing amino acids and their abundances in the proteome of S. cerevisiae.
Results and Discussion
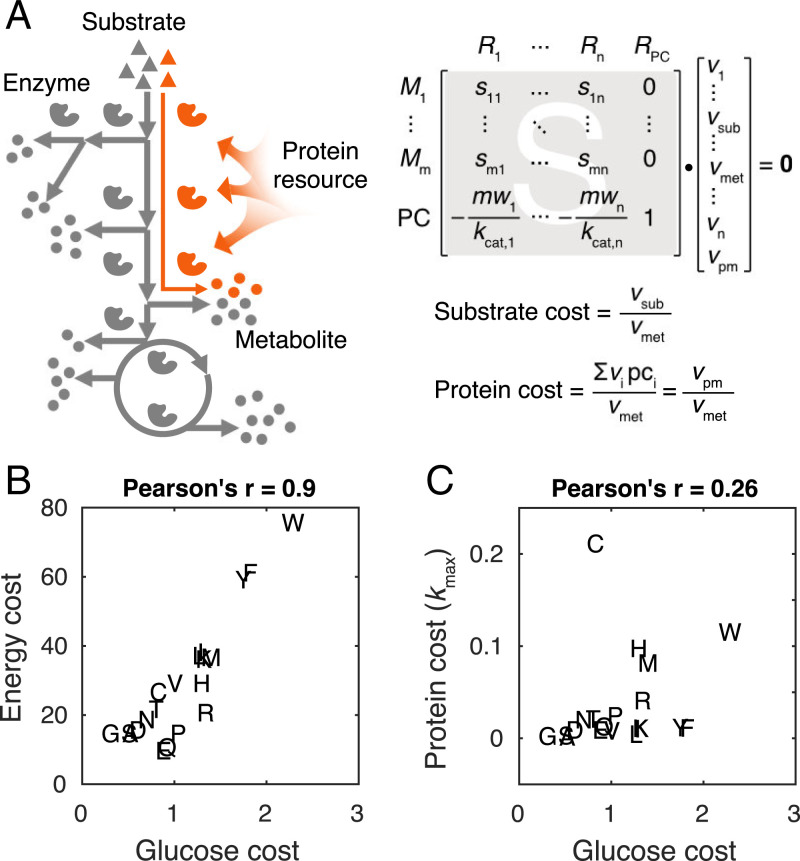
We present a general framework to estimate substrate and protein cost of synthesizing a metabolite using a genome-scale metabolic model (GEM) integrated with protein cost information of individual enzymatic reactions (Fig. 1A), which is calculated by molecular weight over turnover rate (kcat) of the corresponding enzyme (9). The substrate (or protein) cost of synthesizing the metabolite can be calculated by the substrate uptake rate (or the total protein mass that supports the needed rates of reactions involved in the metabolite synthesis pathway) over the metabolite synthesis rate (Fig. 1A). For estimating the substrate cost, there is ensured balancing of redox and energy, such that the cost represents true overall stoichiometric costs.
Fig. 1.
Estimation of glucose and protein costs of synthesizing amino acids. (A) The modeling framework to estimate substrate and protein costs of synthesizing a metabolite of interest. By directing the flux from the substrate to the metabolite—for example, maximizing the metabolite synthesis rate (vmet) while fixing the substrate uptake rate (vsub)—all the involved reactions and their rates (vi) can be obtained. In addition, the total protein mass (vpm) that supports the flux distribution can be obtained, which is the sum of the products of rates (vi) and protein costs (pci) of individual reactions. (B) The comparison between estimated glucose costs and previously reported energy costs of synthesizing amino acids (12). (C) The comparison between estimated glucose and protein costs of synthesizing amino acids. Amino acids are shown by one-letter abbreviations. Energy cost: high-energy phosphate bond consumed per amino acid synthesized. Glucose cost: glucose consumed per amino acid synthesized. Protein cost: protein mass (gram protein per gram cell dry weight) required per amino acid biosynthetic flux.
To estimate the substrate (i.e., glucose) and protein costs of synthesizing amino acids in S. cerevisiae, we used the GEM Yeast8 (10) integrated with the in vivo enzyme catalytic rate (kapp) dataset (11) (SI Appendix). Note that we adopted the maximum kapp (kmax) across conditions to calculate the generic protein costs of synthesizing amino acids, referred to as kmax-based protein costs, which could represent the potential. With model simulations (SI Appendix), we estimated glucose and protein costs of synthesizing 20 amino acids in S. cerevisiae (Dataset S1). We found that the estimated glucose costs correlate well with the reported energy costs in yeast (12) (Fig. 1B). Notably, there is no correlation between the estimated glucose and protein costs (Fig. 1C), which might be caused by some amino acids with high glucose costs but low protein costs, and vice versa. Furthermore, we performed amino acid substitution analysis to compare costs between pairs of amino acids (SI Appendix). The results (Dataset S2) are consistent with our findings: that is, tryptophan, phenylalanine, and tyrosine have higher glucose costs while cysteine, tryptophan, histidine, and methionine have higher protein costs than the others.
In addition to kmax, we also used condition-specific kapp to estimate the protein costs of synthesizing amino acids for yeast cells under diverse conditions (SI Appendix), in which glucose is the sole carbon source (Dataset S3). We found that the protein costs of synthesizing amino acids estimated for various conditions correlate well with each other, as well as the kmax-based protein costs, and none of these different estimated protein costs correlates with the glucose costs of synthesizing amino acids (Dataset S4).
Next, we calculated the relative abundances of amino acids in the proteomes of S. cerevisiae cells under diverse conditions (Dataset S5) based on absolute proteomics data (SI Appendix). We found that the relative abundances of amino acids correlate strongly across conditions with the lowest Pearson’s r being above 0.96 (Dataset S6), meaning that the yeast amino acid composition is conserved across environmental conditions. The conserved amino acid composition could be explained by the fact that amino acid frequencies of more than 90% individual proteins in S. cerevisiae’s proteome significantly correlate with the average relative abundances of amino acids (Dataset S7). Accordingly, we can use the average relative abundances of amino acids among conditions as a proxy (Dataset S5).
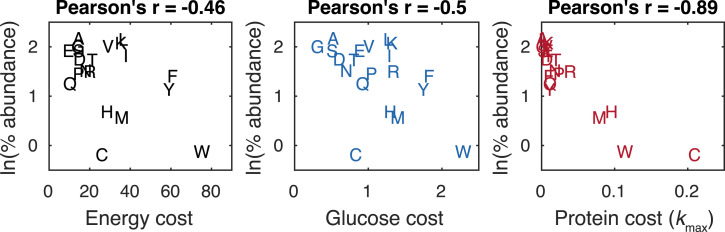
To investigate the relationship between costs and yeast amino acid composition, we adopted a published theoretical model, which deduced a negative linear relationship between amino acid biosynthetic costs and the logarithms of the relative abundances of amino acids by minimizing biosynthetic cost and simultaneously maximizing sequence diversity (13). Note that the sequence diversity accounts for diverse characteristics of various amino acids that are required for protein structure and function. Therefore, we transformed the average relative abundances of amino acids to a logarithmic scale and then correlated them with costs. We found that the energy and glucose costs of synthesizing amino acids correlate weakly with the logarithms of the average relative abundances of amino acids (Fig. 2), in line with previous findings (4, 6). Interestingly, the kmax-based protein costs of synthesizing amino acids considerably improve the correlation (Fig. 2). Therefore, S. cerevisiae tends to minimize protein resource, rather than glucose or energy, for synthesizing amino acids.
Fig. 2.
Correlation of the logarithms of the average amino acid relative abundances with amino acid energy (12), glucose, and protein costs of synthesizing amino acids. Amino acids are shown by one-letter abbreviations.
Furthermore, we found that the condition-specific protein costs of synthesizing amino acids correlate with the logarithms of the relative abundances of amino acids under the corresponding conditions better than glucose costs (Dataset S8), in line with the results using kmax-based protein costs (Fig. 2). While the Pearson’s r of the correlation using condition-specific protein costs differ from various conditions, we found that the correlation is better at rapid growth (growth rate ≥ 0.2/h) conditions than in slow ones (Dataset S8). This suggests that yeast cells at faster growth optimize better the proteome allocation for synthesizing amino acids than at slower growth, which is consistent with the findings that proteome resource allocated to metabolism is more limited in fast-growing cells (14).
Due to the limited coverage of the S. cerevisiae enzyme kinetic data, many reactions in the model were assigned with nonyeast kcat (15), which might affect the estimated protein costs. Therefore, we examined the contributions of three types of enzyme kinetic data (yeast kmax, yeast in vitro kcat, and nonyeast kcat) to the estimated protein costs of synthesizing amino acids (SI Appendix). We found that yeast kmax and in vitro kcat can account for a large fraction of the estimated protein cost for most amino acids, with the notable exception of tryptophan, whose protein cost is mostly contributed by nonyeast kcat (Dataset S9). By removing tryptophan, we still found that the correlation between the logarithms of the average relative abundances of amino acids and protein costs of synthesizing amino acids (Pearson’s r = −0.89) is better than glucose costs (Pearson’s r = −0.28), suggesting that our hypothesis is valid. Moreover, given that yeast in vitro kcat might also differ from in vivo kmax (11), we performed random sampling for all the yeast in vitro kcat and nonyeast kcat simultaneously to reestimate protein costs of synthesizing 20 amino acids and then correlated them with the logarithms of the average relative abundances of amino acids (SI Appendix). By sampling 1,000 times we obtained 1,000 Pearson’s r values and found that less than 5% of them were greater than the Pearson’s r value of using glucose costs (−0.5). Therefore, our conclusion is robust to the uncertainties of yeast in vitro kcat and nonyeast kcat.
In conclusion, we estimated glucose and protein costs of synthesizing proteinogenic amino acids in S. cerevisiae using a GEM integrated with enzyme kinetic data, and demonstrated that the protein costs of synthesizing amino acids outperform the traditional glucose or energy costs in quantitatively describing amino acid composition of S. cerevisiae. Our findings open the possibility for expanding the cost-minimization principle by resource allocation under proteome constraints (16). Additionally, our modeling framework can be readily applied to calculate protein costs of producing chemicals, which could provide valuable angles besides substrate costs in the field of metabolic engineering.
Materials and Methods
To implement protein cost information, the enzymatic reactions in Yeast8 (10) were integrated with either in vivo kmax or condition-specific kapp (11) based on the enzyme-constrained modeling frameworks (17, 18). For the reactions without available in vivo data, in vitro kcat or nonyeast data were assigned instead (15). Details of all the materials and methods are provided in SI Appendix. The data and codes are available at https://github.com/SysBioChalmers/Amino_acid.
Supplementary Material
Acknowledgments
We acknowledge funding from the European Union’s Horizon 2020 research and innovation program under Grant Agreement 686070 and from the Knut and Alice Wallenberg Foundation.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114622119/-/DCSupplemental.
Data Availability
Data have been deposited in GitHub (https://github.com/SysBioChalmers/Amino_acid). All other study data are included in the article and/or supporting information.
References
- 1.Grzymski J. J., Dussaq A. M., The significance of nitrogen cost minimization in proteomes of marine microorganisms. ISME J. 6, 71–80 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mende D. R., et al. , Environmental drivers of a microbial genomic transition zone in the ocean’s interior. Nat. Microbiol. 2, 1367–1373 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Akashi H., Gojobori T., Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 99, 3695–3700 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton M. D., Delneri D., Oliver S. G., Rattray M., Bergman C. M., Evolutionary systems biology of amino acid biosynthetic cost in yeast. PLoS One 5, e11935 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heizer E. M. Jr., et al. , Amino acid cost and codon-usage biases in 6 prokaryotic genomes: A whole-genome analysis. Mol. Biol. Evol. 23, 1670–1680 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Raiford D. W., et al. , Do amino acid biosynthetic costs constrain protein evolution in Saccharomyces cerevisiae? J. Mol. Evol. 67, 621–630 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Peter J., et al. , Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556, 339–344 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y., Nielsen J., Mathematical modelling of proteome constraints within metabolism. Curr. Opin. Syst. Biol. 25, 50–56 (2021). [Google Scholar]
- 9.Chen Y., Nielsen J., Energy metabolism controls phenotypes by protein efficiency and allocation. Proc. Natl. Acad. Sci. U.S.A. 116, 17592–17597 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu H., et al. , A consensus S. cerevisiae metabolic model Yeast8 and its ecosystem for comprehensively probing cellular metabolism. Nat. Commun. 10, 3586 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y., Nielsen J., In vitro turnover numbers do not reflect in vivo activities of yeast enzymes. Proc. Natl. Acad. Sci. U.S.A. 118, e2108391118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner A., Energy constraints on the evolution of gene expression. Mol. Biol. Evol. 22, 1365–1374 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Krick T., et al. , Amino acid metabolism conflicts with protein diversity. Mol. Biol. Evol. 31, 2905–2912 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metzl-Raz E., et al. , Principles of cellular resource allocation revealed by condition-dependent proteome profiling. eLife 6, e28034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y., Li F., Mao J., Chen Y., Nielsen J., Yeast optimizes metal utilization based on metabolic network and enzyme kinetics. Proc. Natl. Acad. Sci. U.S.A. 118, e2020154118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scott M., Gunderson C. W., Mateescu E. M., Zhang Z., Hwa T., Interdependence of cell growth and gene expression: Origins and consequences. Science 330, 1099–1102 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Sánchez B. J., et al. , Improving the phenotype predictions of a yeast genome-scale metabolic model by incorporating enzymatic constraints. Mol. Syst. Biol. 13, 935 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bekiaris P. S., Klamt S., Automatic construction of metabolic models with enzyme constraints. BMC Bioinformatics 21, 19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data have been deposited in GitHub (https://github.com/SysBioChalmers/Amino_acid). All other study data are included in the article and/or supporting information.