Abstract
Pancreatic ductal adenocarcinomas (PDAC) represent one of the deadliest cancers worldwide. Survival is still low due to diagnosis at an advanced stage and resistance to treatment. Herein, we review the main types of liquid biopsy able to help in both prognosis and adaptation of treatments.
Keywords: pancreatic ductal adenocarcinoma, liquid biopsy, exosomes, prognosis, personalized treatment, circulating free DNA (cfDNA), circulating free microRNAs
Introduction
Biopsies represent a fundamental tool for clinical assessment, aiding with the diagnosis and management of the disease. In the oncological setting, tissue biopsy with fragments of primary/metastatic tumors has been traditionally used for the histological classification of disease and, more recently, for the genetic mutational profiling of cancer. However, evolving techniques for the analysis of histological samples have highlighted the limitations associated with such assessment (1). In addition, concerns over single-biopsy bias have been raised, with Gerlinger et al. demonstrating intertumoral and intratumoral heterogeneity between different sites on the primary tumor and its metastasis (2). Therefore, single-biopsy risks underestimating the complexity of the histological and genomic landscape of the tumor and serial monitoring with repeated tissue biopsies does not prove easily feasible. These limitations have highlighted the need for a minimally invasive approach for the systematic and real-time monitoring of cancer. In the recent years, the minimally-invasive liquid biopsy technique has emerged, utilizing body effluents (mainly blood) to identify cancer biomarkers, and assess tumor biochemistry and genetic status on a systemic level (3–5). Types and categories of liquid biopsy can be classified by the type of material analyzed and the type of analysis to be performed (6). A significant positive with respect to using liquid biopsies rather than tissue biopsies is that we can capture all the genetic material released from all the tumor sites, thereby enabling us to portray whole tumor heterogeneity (7).
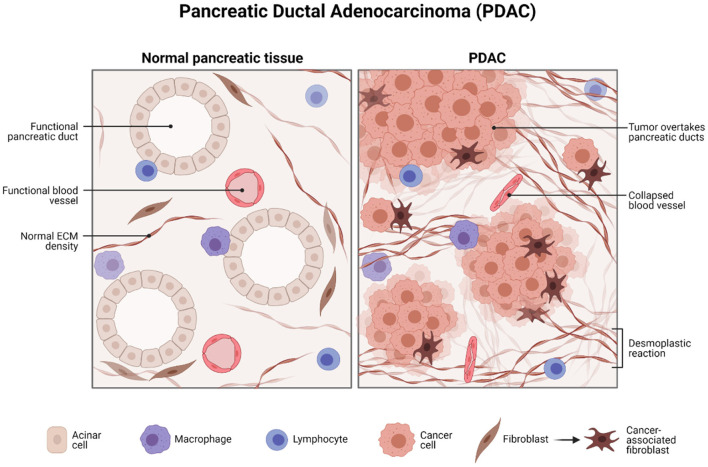
Pancreatic ductal adenocarcinoma (PDAC) represents one of the deadliest cancers worldwide. Survival is still low due to diagnosis at an advanced stage and resistance to treatment. There is a drive to identify biomarker/s that facilitates in the diagnosis, prognosis, and development of personalized treatment strategies. This study will focus on the potential role of liquid biopsy as a prognostic and theranostic tool for managing PDAC (Figure 1).
Figure 1.
Histology of pancreatic ductal adenocarcinoma. Created with BioRender.com.
Pancreatic Ductal Adenocarcinoma: Current Methods of Diagnosis and Their Challenges
Pancreatic ductal adenocarcinoma represents the most common type of pancreatic cancer and one of the most aggressive solid tumor malignancies. PDAC poses a significant challenge. Given its typical silent course and late presentation by which the tumor is mainly unresectable, 1-year overall survival (OS) and 5-year OS are 24 and 5% (or less), respectively (8, 9). Epidemiological data suggest that PDAC is the 7th leading cause of global cancer-related deaths and 3rd leading cause in the United States (US) (10, 11). Furthermore, by 2030, it is predicted that PDAC will be the 2nd most common cause of cancer deaths, preceded by lung cancer (12).
Aside from asymptomatic disease, the clinical presentation of PDAC may include weight loss, abdominal pain, jaundice, steatorrhea (10, 13), and less commonly type 2 diabetes mellitus (14) and thromboembolic disease (15). In addition, patients may develop pancreatic exocrine insufficiency (PEI), secondary to tumor-driven obstructive damage to the secreting component of the pancreas (16).
Guidelines dictate that the preferential investigation to aid with diagnosis in patients with suspected pancreatic cancer is a pancreatic protocol CT, with secondary fluorodeoxyglucose (FDG)-PET/CT and endoscopic ultrasound (EUS)-guided tissue sampling should the diagnosis remain unclear (17). Surveillance for pancreatic cancer should be considered in patients with an inherited high risk: family history, hereditary pancreatitis, and genetic mutations [Serine Protease 1 (PRSS1), breast cancer gene 1/2 (BRCA1/2), partner and localizer of BRCA2 (PALB2), cyclin dependent kinase inhibitor 2A (CDKN2A), mutl homolog 1 (MLH1), muts homolog 2/6 (MSH2/6), PMS1 Homolog 2, Mismatch Repair System Component (PMS2)], by way of magnetic resonance imaging (MRI) magnetic resonance cholangiopancreatography (MRCP) or EUS (17). Upon confirmation of the diagnosis of pancreatic cancer, staging should be performed, including FDG-PET/CT in patients with the localized disease on CT when planning for surgery, radiotherapy, or systemic therapy. MRI, EUS, and laparoscopy with laparoscopic US may also be performed to support decisions for management (17).
Endoscopic US-guided tissue sampling is an invasive surgical approach to cytological and histological analysis of tumors and does not overcome concerns over a single biopsy bias. However, given an appropriate efficacy study, liquid biopsy provides the scope for minimally invasive sampling as a systemic level diagnostic tool for tumor analysis.
Management of PDAC and Methods of Surveillance
Pancreatic ductal adenocarcinoma exhibits rapid tumor progression, with limited efficacy of current therapeutic drug regimes in locoregional and metastatic disease (18). In resectable and borderline resectable cases of PDAC, mainstay treatment remains surgical resection in combination with chemoradiotherapy; however, by the time of clinical presentation, the advanced tumor stage renders over 80% of cases as unresectable (19, 20). Metal stenting to relieve biliary and duodenal obstructions may be performed in patients not suitable for resection (unresectable or not fit for surgery). Gemcitabine-based chemotherapy and combination chemotherapy, including folfirinox (irinotecan, oxaliplatin, 5-fluorouracil, and leucovorin), demonstrate a moderate benefit for OS (21). Novel immunotherapeutics for PDAC prove promising for targeted therapy, with pembrolizumab as an the United States Food and Drug Administration (FDA)-approved programmed cell death protein 1 (PD-1) checkpoint inhibitor for unresectable and metastatic mismatch repair-deficient/microsatellite instability-high (dMMR/MSI-H) solid tumors (22) and other agents under investigation in clinical trials (23–28). Nutritional support with replacement therapy, pain management, and psychological support also proves essential for holistic patient care.
Surveillance during the management of PDAC and after curative surgery also plays a role in the risk stratification and identifying prognostic factors in patients. First, baseline performance status (PS) of patient has been recognized as an essential clinical biomarker and independent prognostic factor for treatment response. Poor baseline has been negatively correlated with chemotherapeutic response rates, including gemcitabine, and less favorable clinical outcomes (29, 30). Second, biological prognostic biomarkers in PDAC have also been suggested, including carbohydrate antigen 19-9 (CA 19-9) and activin A. Elevated baseline CA 19-9 (pancreatic tumor marker) and activin A (transforming growth factor beta (TGF-β) superfamily cytokine) have been correlated with worse prognosis, with a role in postoperative recurrence and metastatic spread, respectively (31, 32). Results from an early meta-analysis that looked at the specificity and sensitivity of the use of common serum biomarkers highlighted that for carcinoembryonic antigen (CEA) and CA 19-9. The mean sensitivity and specificity estimates for CEA were 44.2 and 84.8%, respectively, and for CA 19-9, the mean sensitivity and specificity were 78.2 and 82.8%, respectively. The analysis included numerous studies that include patients with benign pancreatic disease including chronic pancreatitis and cholelithiasis (33). Due to a range in sensitivities and specificities, the importance of having markers to monitor patient outcomes for treatment planning. An intensified chemotherapy regimen has been advised in patients with elevated CA 19-9 (31) and activin A neutralizing antibodies have been studied for decreasing tumor metastasis (32).
Use of Liquid Biopsies in the Detection and Surveillance in PDAC
First, with respect to the type of material analyzed, tumor-released markers that may be detected in liquid biopsy include circulating tumor cells (CTCs), circulating free nucleic acids [circulating free DNA (cfDNA), circulating free RNA (cfRNA), circulating free microRNA (cfmiRNA)], exosomes, and tumor-educated platelets (TEPs) (6, 34, 35).
Circulating tumor cells are among the most extensively studied markers and represent cancer cells that have gained plasticity and motility by epithelial–mesenchymal transition (EMT), extravasating from the tumor primary and into systemic circulation (35, 36). Principally, CTCs can lead to cancer metastasis through intravasation into distant tissue and, therefore, prove critical in cancer diagnostics and prognostics. In the context of cancer, cfDNA, cfRNA, and cfmiRNA are nucleic acid fragments discharged into circulation from apoptotic cells and necrotic tumor cells (35). Important to recognize is that most cfDNA is non-malignant in origin. However, when cfDNA derives from tumor cells, it is called circulating tumor DNA (ctDNA), carrying tumor-specific mutations (37); there is evidence suggesting elevated levels of cfDNA in the later stages of cancer (38). Circulating free nucleic acids are unstable in blood and other sources for genetic analysis include circulating microvesicles (exosomes) and TEPs (35, 39, 40).
Liquid biopsy can also be categorized utilizing the type of analysis including small- and large-scales mutation analysis, with targeted deep sequencing and next-generation sequencing, and analysis of structural change and copy number alterations (6). Mutational profiling recognizes point mutations, insertions, deletions, and genomic strands, which may aid with the metric classification of prognosis, resistance, disease burden, and therapeutic efficacy of medicines (41–43). Structural analysis using the chain termination method (Sanger method) involves the identification of single nucleotide polymorphisms (SNPs), structural and copy number variations, and focal mutational profiling as above (6, 44). cfDNA can also be studied to identify copy number alterations (45). Thus, liquid biopsy appears promising and demonstrates suitability for application into conventional cancer care (46–49).
Circulating Tumor Cells in PDAC
The concept of the liquid biopsy in PDAC is to ultimately paint a molecular genetic landscape of PDAC, which will improve the characterization of PDAC heterogeneity, aid in the early detection and treatment surveillance, with minimal circulating tumor material (50). CTCs and circulating tumor-derived proteins have been used clinically including CA 19-9 and prostate-specific antigen (PSA) used for detecting and surveillance pancreatic cancer and prostate cancer, respectively. There is hope that by utilizing CTCs or circulating tumor-derived proteins, we would be able to develop a marker test that is highly sensitive and specific in detecting PDAC (51, 52).
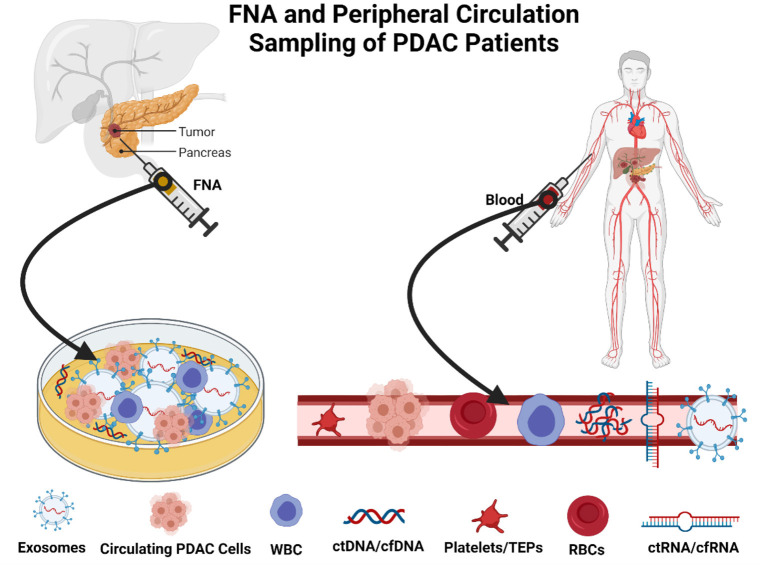
The methodology relies on the use of low volume micromaterials released from the PDAC, which releases components, including cell-free DNA (cfDNA), cell-free RNA (cfRNA), extracellular vesicles (EVs), and CTCs, into bodily fluids including blood, fine needle aspirates at biopsy, and saliva (53, 54). CTCs comprise a low percentage of circulating material released from the PDAC tumors. However, despite their small collective proportion of circulating tumor biomaterial, it is also found in clusters and can be found as single cells (55). Singular cells are often detected in the peripheral circulation and can be seen in blood biopsies (Figure 2). There are relative obstacles in using CTCs as a biomarker for early detection and surveillance of PDAC (54, 56). First, there is a fact that the presence of CTCs does not always correlate to the presence of malignant invasive PDAC and, as aforementioned, the low yield of CTCs in peripheral circulation; the theoretical reasoning for the low yield of CTCs is primarily due to their relative size mainly among other factors (57, 58). CTCs are often 3–5 times the size of capillary openings before entering central circulation via the portal vein. Therefore, the larger CTCs remain trapped either within the capillaries and the smaller CTCs enter peripheral circulation (59–61). The anatomical location and blood drainage that supplies the pancreas will directly drain into the portal vein via the splenic vein. This further implicates the choice of attaining peripheral CTCs and whether the use of peripheral blood and presents portal vein sampling as a preferential option for point of sampling to allow for early diagnosis (62, 63), disease stratification, and longitudinal monitoring of therapeutic response in patients with pancreatic. CTCs are exceedingly heterogeneous concerning PDAC tumor biology (62, 64, 65), which is often a benefit compared to a singular tumor biopsy. Several techniques are described to isolate CTCs and their derived proteins including mass spectrometry and proteomics. Antibody-affinity binding-based technology has been documented as highly specific and sensitive in detecting novel biomarkers from liquid biopsies (66, 67). For PDAC, in the context of utilizing CTCs and other components of a liquid biopsy are compared with the diagnostic marker, CA19-9 (68). Proteins of interest from liquid biopsies are glypican 1 (GPC1), epithelial cellular adhesion molecule (EpCAM), CD45, CD69, tissue inhibitor of metalloproteinase 1 (TIMP1), thrombospondin-2 (THBS2), leucine-rich alpha-2-glycoprotein 1 (LRG1) and also, more importantly, kirsten rat sarcoma viral oncogene homolog (KRAS) mutations (69, 70). Blood metabolomics has also been indicated as a method of detecting early stages of PDAC and surveillance of the recurrence of PDAC. Recent investigations highlight that the plasma metabolites such as acetylspermidine, diacetylspermine, indole derivatives, and lysophosphatidylcholines could distinguish PDAC from healthy subjects benign pancreatic disorders (68, 71).
Figure 2.
Method of sampling in patients with pancreatic ductal adenocarcinoma (PDAC). Created with BioRender.com.
Often, the metabolites mentioned are combined with tumor associated proteins of PDAC, including CA 19-9, to monitor early detection of PDAC and tumor progression. The highly diagnostic and prognostic value of using CTCs from liquid biopsies, particularly from solid gastrointestinal tumors, is primarily due to CTCs from direct natural shedding of CTCs and detachment from a single or multiple loci of the primary PDAC tumor as well as metastases into the peripheral circulation and often are the primary sources of metastases (72, 73). The immunobiology of CTCs captures the whole-body burden of tumor than single point tissue biopsies; they highlight the tumor heterogeneity. The favoring of CTCs rather than exosomes, ctDNA, cfDNA, ctRNA, or cfRNA or any other components of liquid biopsies due to its stored biogenetic information encodes for various expressive influential genes and proteins, particular in the development of a complex, comprehensive landscape of tumor heterogeneity and evolution (74, 75). Thus, CTCs play an integral role in testing current and new therapeutics; the additional benefit of using CTCs includes the tumor heterogeneity, which can be developed into an ex-vivo culture that genetically mirrors the whole PDAC tumor (76, 77). This allows clinicians to assess personalized therapeutic regimes and identify and predict the success of therapeutic regimes tested and potential new immunotherapies developed (78). Processes including reverse transcription-PCR (RT-PCR) and ultra/standard density centrifugation have been used to isolate CTCs of PDAC tumor cells from whole blood based on CEA, cytokeratin-20 (CK-20), and EpCAM (79–81) (Table 1).
Table 1.
Sampling materials and methods of clinical application with PDAC samples.
| Type of materials | Diagnostic detection | Method of detection | References |
|---|---|---|---|
| ctDNA/ctRNA | Amplification, deletion and translocation Gene driver mutations, Chromosomal abnormalities |
PCR Whole genome sequencing Next-generation sequencing |
Fabbri et al. (82) |
| CTCs | Gene expression Cell Proliferation/growth Cell differentiation Cell mobility Apoptosis |
Next-generation sequencing Microarray testing qRT-PCR CTC Chip sequencing EPISOT Phenotypic analysis: immunostaining, FACS and LC-MS |
Pantel et al. (37) |
| miRNA | Gene fusion transcripts RNA expression |
Next-generation sequencing Microarray testing qRT-PCR |
Tesfaye et al. (83) |
| TEPs | Gene expression | Next-generation sequencing RNA sequencing |
Best et al. (84) Joosse et al. (40) |
PDAC, pancreatic ductal adenocarcinoma; LC-MS, liquid chromatography-mass spectrometry; FACS, fluorescence-activated single cell sorting; qRT-PCR, real time quantitative reverse transcription polymerase chain reaction.
Circulating Tumor DNA and RNA
Circulating Tumor DNA
Circulating tumor DNA of PDACs is representative of a short genomic fragment coding for the PDAC tumor spanning all the chromosomes. ctDNA is found in peripheral circulation, which often originates from the necrosis or apoptosis of the primary PDAC tumor or from metastatic PDAC (85). ctDNA also encompasses a proportion of circulating cfDNA from PDAC tumor cells. The general base pair size of circulating ctDNA is usually about 150–160 base pairs; often, these pairs are associated with nucleosomes, contributing to ctDNA detection (86). In the evolving epigenetic landscapes of pancreatic cancer, the ability to access the DNA of PDAC tumors is imperative to orchestrate a personalized precision therapeutic regime (87). Nucleosomes facilitate access to DNA due to several metabolic reactions. These remodeled nucleosomes move along the DNA and expose or cover the base regions in particular transcription factor binding sites. In a typical physiological environment, these nucleosomes interact with and unify many nuclear components and processes required for gene expression (88, 89). Remarkably, there are mutations present in multiple of the components that facilitate the remodeling of nucleosomes that have also been explicitly found in PDAC, including members of the switch/sucrose non-fermentable (SWI/SNF) family: brahma-related gene-1 (BRG1), SWI/SNF Related, Matrix Associated, Actin Dependent Regulator Of Chromatin, Subfamily A, Member 4 (SMARCA4), BRG-associated factor 250a (BAF250a (ARID1A)) or BRG-associated factor 250b (BAF250b (ARID1B)) and brahma homolog (BRM (SMARCA2)) among others, suggesting a crucial role for nucleosome positioning in the establishing PDAC cancer phenotype and heterogeneity (90–92). The exposed non-coding regions of RNA or DNA, controlled by the nucleosomes, have a direct impact on the epigenetics of PDAC. Due to their prevalence as circulating exosomes encapsulated in body fluids, these exosomes and nucleosomes offer potential therapeutic targets as well as biomarkers for diagnosis and monitoring disease progression (93, 94).
Circulating tumor DNA presents as precious markers particularly for being: (1) a biomarker of PDAC for early detection of the onset of PDAC and surveillance of PDAC recurrence and (2) a biomarker, which facilitates decision-making to permit the most appropriate therapeutic regime (95, 96). Aforementioned, including the critical roles in PDAC detection and treatment, ctDNA has been highlighted in several studies to correlate to PDAC tumor burden, primarily more sensitive in detecting advanced PDAC cancer detection than early-stage PDAC malignancy (97). Furthermore, studies have highlighted that plasma KRAS and epidermal growth factor receptor (EGFR) mutations act as the most predictive and reliable biomarker for detecting and surveillance PDAC, particularly KRAS G12D mutation (98, 99). Reportedly, there is a significant association between the KRAS G12D mutation and the appearance with “micrometastasis” within patients with PDAC, thereby allowing clinicians to stratify high-risk and low-risk patients preoperatively and guide appropriate initial treatment regimes (100).
With respect to ctDNA, there are a number of widely used applications in the identification and quantification technique of ctDNA. For example, identifying EGFR-tyrosine kinase (PTK) mutations from ctDNA for diagnostic purposes in patients with non-small cell lung cancer (NSCLC) (101, 102). The identification of mutations in the EGFR-PTK pathways and testing potential therapeutics involving EGFR-PTK inhibitors and highlighting patients who are specifically sensitized or resistant to this treatment method. ctDNA has also been used in assisting in the detection and recurrence surveillance for colorectal and breast cancers. Utilizing ctDNA monitoring in these cancer types has been extremely promising in highlighting appropriate patients for particular treatment pathways and identify specific therapeutic regimes that will have the most clinical benefit (103, 104).
Circulating Tumor RNA
Circulating tumor RNA is also included in the group of important circulating biomarkers extracted from liquid biopsies. Among the essential circulating biomarkers in peripheral blood include ctRNAs mentioned above, but subclassification of ctRNAs also includes: miRNAs, TEPs, and metabolites (105–107). The importance of ctRNA is that they express particular non-coding regions of genetic transcripts of PDAC tumors. The significance of ctRNA is implicated when investigating the RNA expression profiles of extracted components of the liquid biopsy (108). Studies have shown that the RNA profiles of TEPs have been shown to discriminate between being in a patient with PDAC tumors compared to TEPs in circulation in the presence of healthy tissues. This is primarily due to platelets RNA biomarker signatures altering by the presence of cancer (109, 110). Microarray studies investigating platelets from the control group of inflammatory diseases identified 22 differentially expressed genes. The expression levels of these genes are increased in patients with cancer (84). Platelet markers that alter significantly in the presence of cancer have been identified as beta-2-microglobulin (B2M), pro-platelet basic protein (PPBP), thymosin beta 4 X-Linked (TMSB4X), and platelet factor 4 (PF4) (111). Due to their molecular uniqueness in the presence of cancers, TEPs can also subclassify multiple types of gastrointestinal solid cancers (107). MicroRNA (miR) identification has a prominent role in PDAC detection, monitoring, and surveillance.
Several significant circulating miR molecular signatures have been important in the early detection and surveillance of PDAC (112). Several studies highlight the miRs: miR-17-5p, miR-21, miR-155, miR-196a, miR-17-5p, and miR-21. All of which have been shown to have a considerably high diagnostic sensitivity and specificity (113, 114). When investigating and treating patients at early stages of PDAC and treating patients with premalignant pancreatic lesions, including mucinous cystic neoplasms (MCNs) and intraductal papillary mucosal neoplasms (IPMNs), miRs play a pivotal role in indicating a malignant transformation and initiating the appropriate treatment at the correct timepoint (115). When analyzing the expression of miR-191, miR-451a, and miR-21 in pancreatic cancer and IPMNs, we see that the expression levels dramatically increased in all the three miRs when comparing the IPMN cohort vs. the PDAC cohort (114, 116). These miRs have be shown to have a greater diagnostic process when compared to the conventional clinical markers including CA-19.9 when monitoring the transformation of premalignant lesion of IPMN to cancerous early-/late-stage PDAC (117).
Circulating EVs
Extracellular vesicles are essentially circulating nano-sized proportions containing nucleoprotein material products of cell types they have originated from, more importantly in this instance from tumor cells including from PDAC (118, 119). These vesicles are formed by distinct loading mechanisms that encapsulate RNA and protein profiles from origin parent cells, thus highlighting a sophisticated and selective loading mechanism employed. Hence, EV has gained significant interest and is highly sought after as potential diagnostic biomarkers. The genetically significant proteins that EV hold often facilitate the mapping and interpretation and the genetic and metabolic landscape of the tumor stem from (120). EVs have a considerably longer half-life when compared to ctDNA, ctRNA, or circulating miR. They are highly abundant in peripheral circulation due to their consistent production from PDAC tumors, irrespective of the rates of tumor cell necrosis, apoptosis, or invasion (121). These factors impact their use as markers for early detection and increase the ability to be yielded from a number of sources including ascites, blood, and saliva (122, 123). Particularly in regards to PDAC samples and attaining a traditional fine needle aspiration (FNA) biopsy can be problematic often yields low volume and low-quality tissue for conventional tissue diagnosis protocol (124). However, using EVs from these FNA biopsies samples, clinicians can explore protein and genetic analysis for necessary diagnostics for specifically targeted therapy (125).
MicroRNA has been highlighted as a potential key identifier for PDAC. The level of expression of their genetic signature theoretically correlates to improved sensitivity and specificity of microarrays for detecting PDAC (126, 127). In reference to future directions of the use of exosomes, EV and CTCs, for clinical activity, are that we need to develop a means in which we can increase the rate of sensitivity and specificity in regard to accuracy of testing and selection of genetic materials including nucleic acids and proteins to unwind the heterogeneous genomic picture of the PDAC tumors (128).
Conclusion
The availability of liquid biopsy has allowed clinicians to study peripheral blood and FNA biopsy samples of patients with PDAC for clinical applications including early detection and surveillance, to optimize overall survival outcomes in affected patients. Exosomes, EVs CTC, ctRNA, and ctDNA have been investigated to provide a matching analysis of PDAC tumor genomic profiles confirming heterogeneity; this finding is essential in highlighting potential therapeutic targets against PDAC variants personalizing precision medicine for each patient afflicted by PDAC. The clinical relevance of liquid biopsies and genomic profiling of PDAC has not only been used for the monitoring of early-to-late PDAC and surveillance of recurrence, but the sampling of the genetic material of exosomes, EVs CTC, ctRNA, and ctDNA assists with the treatment monitoring and for prognostic purposes in patients. Large-scale retrospective and prospective studies are required to rigorously assess the validity of the use of liquid biopsies safely in the clinical environment to manage PDAC in patients.
Author Contributions
DO-B, GS, and NC: conceptualization, methodology, software, validation, formal analysis, investigation, resources, data curation, writing—original draft preparation, writing—review and editing, and visualization. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1.Postel M, Roosen A, Laurent-Puig P, Taly V, Wang-Renault SF. Droplet-based digital PCR and next generation sequencing for monitoring circulating tumor DNA: a cancer diagnostic perspective. Expert Rev Mol Diagn. (2018) 18:7–17. 10.1080/14737159.2018.1400384 [DOI] [PubMed] [Google Scholar]
- 2.Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. (2012) 366:883–92. 10.1056/NEJMoa1113205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu D, Park BH. Liquid biopsy: unlocking the potentials of cell-free DNA. Virchows Arch. (2017) 471:147–54. 10.1007/s00428-017-2137-8 [DOI] [PubMed] [Google Scholar]
- 4.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. (2014) 6:224ra24. 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. (2013) 10:472–84. 10.1038/nrclinonc.2013.110 [DOI] [PubMed] [Google Scholar]
- 6.Arneth B. Update on the types and usage of liquid biopsies in the clinical setting: a systematic review. BMC Cancer. (2018) 18:527. 10.1186/s12885-018-4433-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamyabi N, Bernard V, Maitra A. Liquid biopsies in pancreatic cancer. Expert Rev Anticancer Ther. (2019) 19:869–78. 10.1080/14737140.2019.1670063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 9.Reyes-Gibby CC, Chan W, Abbruzzese JL, Xiong HQ, Ho L, Evans DB, et al. Patterns of self-reported symptoms in pancreatic cancer patients receiving chemoradiation. J Pain Symptom Manage. (2007) 34:244–52. 10.1016/j.jpainsymman.2006.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. (2019) 69:7–34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 11.Ilic M, Ilic I. Epidemiology of pancreatic cancer. World J Gastroenterol. (2016) 22:9694. 10.3748/wjg.v22.i44.9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. (2014) 74:2913–21. 10.1158/0008-5472.CAN-14-0155 [DOI] [PubMed] [Google Scholar]
- 13.Porta M, Fabregat X, Malats N, Guarner L, Carrato A, de Miguel A, et al. Exocrine pancreatic cancer: symptoms at presentation and their relation to tumour site and stage. Clin Transl Oncol. (2005) 7:189–97. 10.1007/BF02712816 [DOI] [PubMed] [Google Scholar]
- 14.De Souza A, Khawaja KI, Masud F, Saif MW. Metformin and pancreatic cancer: is there a role? Cancer Chemother Pharmacol. (2016) 77:235–42. 10.1007/s00280-015-2948-8 [DOI] [PubMed] [Google Scholar]
- 15.Khorana AA. Cancer and coagulation. Am J Hematol. (2012) 87 (Suppl. 1):S82–7. 10.1002/ajh.23143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vujasinovic M, Valente R, Del Chiaro M, Permert J, Löhr JM. Pancreatic exocrine insufficiency in pancreatic cancer. Nutrients. (2017) 9:183. 10.3390/nu9030183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Guideline Alliance (UK) . Pancreatic cancer in adults: diagnosis and management. London: National Institute for Health and Care Excellence; (2018). 156:591–600. [PubMed] [Google Scholar]
- 18.Löhr M. Is it possible to survive pancreatic cancer? Nat Clin Pract Gastroenterol Hepatol. (2006) 3:236–7. 10.1038/ncpgasthep0469 [DOI] [PubMed] [Google Scholar]
- 19.Buanes TA. Role of surgery in pancreatic cancer. World J Gastroenterol. (2017) 23:3765. 10.3748/wjg.v23.i21.3765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White RR, Reddy S, Tyler DS. The role of chemoradiation therapy in locally advanced pancreatic cancer. HPB. (2005) 7:109–13. 10.1080/13651820510028963a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adamska A, Domenichini A, Falasca M. Pancreatic ductal adenocarcinoma: current and evolving therapies. Int J Mol Sci. (2017) 18:1338. 10.3390/ijms18071338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Administration U. S. F. and D. U.S. Food and Drug Administration. KEYTRUDA® (pembrolizumab) injection, highlights of prescribing information . FDA. (2021) 1:2. [Google Scholar]
- 23.Hardacre JM, Mulcahy M, Small W, Talamonti M, Obel J, Krishnamurthi S, et al. Addition of algenpantucel-L immunotherapy to standard adjuvant therapy for pancreatic cancer: a phase 2 study. J Gastrointest Surg. (2013) 17:94–100. 10.1007/s11605-012-2064-6 [DOI] [PubMed] [Google Scholar]
- 24.Hewitt DB, Nissen N, Hatoum H, Musher B, Seng J, Coveler AL, et al. A phase 3 randomized clinical trial of chemotherapy with or without algenpantucel-L (HyperAcute-Pancreas) immunotherapy in subjects with borderline resectable or locally advanced unresectable pancreatic cancer. Ann Surg. (2022) 275:45–53. 10.1097/SLA.0000000000004669 [DOI] [PubMed] [Google Scholar]
- 25.Nagai K, Adachi T, Harada H, Eguchi S, Sugiyama H, Miyazaki Y. Dendritic cell-based immunotherapy pulsed with wilms tumor 1 peptide and mucin 1 as an adjuvant therapy for pancreatic ductal adenocarcinoma after curative resection: a phase I/IIa clinical trial. Anticancer Res. (2020) 40:5765–76. 10.21873/anticanres.14593 [DOI] [PubMed] [Google Scholar]
- 26.O'Hara MH, O'Reilly EM, Varadhachary G, Wolff RA, Wainberg ZA, Ko AH, et al. CD40 agonistic monoclonal antibody APX005M (sotigalimab) and chemotherapy, with or without nivolumab, for the treatment of metastatic pancreatic adenocarcinoma: an open-label, multicentre, phase 1b study. Lancet Oncol. (2021) 22:118–31. 10.1016/S1470-2045(20)30532-5 [DOI] [PubMed] [Google Scholar]
- 27.Byrne KT, Betts CB, Mick R, Sivagnanam S, Bajor DL, Laheru DA, et al. Neoadjuvant selicrelumab, an agonist CD40 antibody, induces changes in the tumor microenvironment in patients with resectable pancreatic cancer. Clin Cancer Res. (2021) 27:4574–86. 10.1158/1078-0432.CCR-21-1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng L, Ding D, Edil BH, Judkins C, Durham JN, Thomas DL, 2nd, et al. Vaccine-induced intratumoral lymphoid aggregates correlate with survival following treatment with a neoadjuvant and adjuvant vaccine in patients with resectable pancreatic adenocarcinoma. Clin Cancer Res. (2021) 27:1278–86. 10.1158/1078-0432.CCR-20-2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Louvet C, Labianca R, Hammel P, Lledo G, Zampino MG, André T, et al. Gemcitabine in combination with oxaliplatin compared with gemcitabine alone in locally advanced or metastatic pancreatic cancer: results of a GERCOR and GISCAD phase III trial. J Clin Oncol. (2005) 23:3509–16. 10.1200/JCO.2005.06.023 [DOI] [PubMed] [Google Scholar]
- 30.Van Cutsem E, van de Velde H, Karasek P, Oettle H, Vervenne WL, Szawlowski A, et al. Phase III trial of gemcitabine plus tipifarnib compared with gemcitabine plus placebo in advanced pancreatic cancer. J Clin Oncol. (2004) 22:1430–8. 10.1200/JCO.2004.10.112 [DOI] [PubMed] [Google Scholar]
- 31.Berardi R, Mandolesi A, Pellei C, Maccaroni E, Onofri Onofri A. (2013) Prognostic Factors in Pancreatic Cancer: The Role of Perineural Vascular Vascular and Lymphatic Invasion and of Ca19-9. J Gastroint Dig Syst. 3:134. 10.4172/2161-069X.100013430348713 [DOI] [Google Scholar]
- 32.Mancinelli G, Torres C, Krett N, Bauer J, Castellanos K, McKinney R, et al. Role of stromal activin A in human pancreatic cancer and metastasis in mice. Sci Rep. (2021) 11:7986. 10.1038/s41598-021-87213-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poruk KE, Gay DZ, Brown K, Mulvihill JD, Boucher KM, Scaife CL, et al. The clinical utility of CA 19-9 in pancreatic adenocarcinoma: diagnostic and prognostic updates. Curr Mol Med. (2013) 13:340–51. 10.2174/1566524011313030003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poulet G, Massias J, Taly V. Liquid biopsy: general concepts. Acta Cytol. (2019) 63:449–55. 10.1159/000499337 [DOI] [PubMed] [Google Scholar]
- 35.Mader S, Pantel K. Liquid biopsy: current status and future perspectives. Oncol Res Treat. (2017) 40:404–8. 10.1159/000478018 [DOI] [PubMed] [Google Scholar]
- 36.Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem. (2013) 59:110–8. 10.1373/clinchem.2012.194258 [DOI] [PubMed] [Google Scholar]
- 37.Pantel K, Alix-Panabières C. Real-time liquid biopsy in cancer patients: fact or fiction? Cancer Res. (2013) 73:6384–8. 10.1158/0008-5472.CAN-13-2030 [DOI] [PubMed] [Google Scholar]
- 38.Rouvinov K, Mermershtain W, Dresler H, Ariad S, Riff R, Shani-Shrem N, et al. Circulating cell-free DNA levels in patients with metastatic renal cell carcinoma. Oncol Res Treat. (2017) 40:707–10. 10.1159/000479523 [DOI] [PubMed] [Google Scholar]
- 39.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. (2016) 126:1208–15. 10.1172/JCI81135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joosse SA, Pantel K. Tumor-educated platelets as liquid biopsy in cancer patients. Cancer Cell. (2015) 28:552–4. 10.1016/j.ccell.2015.10.007 [DOI] [PubMed] [Google Scholar]
- 41.Messadi DV. Diagnostic aids for detection of oral precancerous conditions. Int J Oral Sci. (2013) 5:59–65. 10.1038/ijos.2013.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shyr D, Liu Q. Next generation sequencing in cancer research and clinical application. Biol Proced. (2013) 15:4. 10.1186/1480-9222-15-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malapelle U, Pisapia P, Rocco D, Smeraglio R, di Spirito M, Bellevicine C, et al. Next generation sequencing techniques in liquid biopsy: focus on non-small cell lung cancer patients. Transl Lung Cancer Res. (2016) 5:505–510. 10.21037/tlcr.2016.10.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mardis ER, Wilson RK. Cancer genome sequencing: a review. Hum Mol Genet. (2009) 18:R163–8. 10.1093/hmg/ddp396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Østrup O, Ahlborn LB, Lassen U, Mau-Sørensen M, Nielsen FC. Detection of copy number alterations in cell-free tumor DNA from plasma. BBA Clin. (2017) 7:120–26. 10.1016/j.bbacli.2017.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molina-Vila MA, Mayo-de-Las-Casas C, Giménez-Capitán A, Jordana-Ariza N, Garzón M, Balada A, et al. Liquid biopsy in non-small cell lung cancer. Front Med. (2016) 3:69. 10.3389/fmed.2016.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin CQ, Yuan CH, Qu Z, Guan Q, Chen H, Wang FB. Liquid biopsy of hepatocellular carcinoma: circulating tumor-derived biomarkers. Dis Markers. (2016) 2016:1427849. 10.1155/2016/1427849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shankar GM, Balaj L, Stott SL, Nahed B, Carter BS. Liquid biopsy for brain tumors. Expert Rev Mol Diagn. (2017) 17:943–47. 10.1080/14737159.2017.1374854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lousada-Fernandez F, Rapado-Gonzalez O, Lopez-Cedrun JL, Lopez-Lopez R, Muinelo-Romay L, Suarez-Cunqueiro MM. Liquid biopsy in oral cancer. Int J Mol Sci. (2018) 19:1704. 10.3390/ijms19061704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qi ZH, Xu HX, Zhang SR, Xu JZ, Li S, Gao HL, et al. The significance of liquid biopsy in pancreatic cancer. J Cancer. (2018) 9:3417–26. 10.7150/jca.24591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X, Shi S, Zhang B, Ni Q, Yu X, Xu J. Circulating biomarkers for early diagnosis of pancreatic cancer: facts and hopes. Am J Cancer Res. (2018) 8:332–53. [PMC free article] [PubMed] [Google Scholar]
- 52.Hayes B, Murphy C, Crawley A, O'Kennedy R. Developments in point-of-care diagnostic technology for cancer detection. Diagnostics. (2018) 8:39. 10.3390/diagnostics8020039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.García-Silva S, Gallardo M, Peinado H. DNA-loaded extracellular vesicles in liquid biopsy: tiny players with big potential? Front cell Dev Biol. (2020) 8:622579. 10.3389/fcell.2020.622579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu W, Hurley J, Roberts D, Chakrabortty SK, Enderle D, Noerholm M, et al. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol. (2021) 32:466–77. 10.1016/j.annonc.2021.01.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brock G, Castellanos-Rizaldos E, Hu L, Coticchia C, Skog J. Liquid biopsy for cancer screening, patient stratification and monitoring. Transl. Cancer Res. (2015) 4:280–90. 10.3978/j.issn.2218-676X.2015.06.05 [DOI] [Google Scholar]
- 56.Lee J.-S., Park SS, Lee YK, Norton JA, Jeffrey SS. Liquid biopsy in pancreatic ductal adenocarcinoma: current status of circulating tumor cells and circulating tumor DNA. Mol Oncol. (2019) 13:1623–50. 10.1002/1878-0261.12537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pimienta M, Edderkaoui M, Wang R, Pandol S. The potential for circulating tumor cells in pancreatic cancer management. Front Physiol. (2017) 8:381. 10.3389/fphys.2017.00381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martini V, Timme-Bronsert S, Fichtner-Feigl S, Hoeppner J, Kulemann B. Circulating tumor cells in pancreatic cancer: current perspectives. Cancers. (2019) 11:1659. 10.3390/cancers11111659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krog BL, Henry MD. Biomechanics of the circulating tumor cell microenvironment. Adv Exp Med Biol. (2018) 1092:209–33. 10.1007/978-3-319-95294-9_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kowalik A, Kowalewska M, Gózdz S. Current approaches for avoiding the limitations of circulating tumor cells detection methods-implications for diagnosis and treatment of patients with solid tumors. Transl Res. (2017) 185:58–84.e15. 10.1016/j.trsl.2017.04.002 [DOI] [PubMed] [Google Scholar]
- 61.Micalizzi DS, Maheswaran S, Haber DA. A conduit to metastasis: circulating tumor cell biology. Genes Dev. (2017) 31:1827–40. 10.1101/gad.305805.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buscail E, Alix-Panabières C, Quincy P, Cauvin T, Chauvet A, Degrandi O, et al. High clinical value of liquid biopsy to detect circulating tumor cells and tumor exosomes in pancreatic ductal adenocarcinoma patients eligible for up-front surgery. Cancers. (2019) 11:1656. 10.3390/cancers11111656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Catenacci DV, Chapman CG, Xu P, Koons A, Konda VJ, Siddiqui UD, et al. Acquisition of portal venous circulating tumor cells from patients with pancreaticobiliary cancers by endoscopic ultrasound. Gastroenterology. (2015) 149:1794–803.e4. 10.1053/j.gastro.2015.08.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stephenson D, Nahm C, Chua T, Gill A, Mittal A, de Reuver P, et al. Circulating and disseminated tumor cells in pancreatic cancer and their role in patient prognosis: a systematic review and meta-analysis. Oncotarget. (2017) 8:107223–36. 10.18632/oncotarget.19928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu X, Li C, Li J, Yu T, Zhou G, Cheng J, et al. Detection of CTCs in portal vein was associated with intrahepatic metastases and prognosis in patients with advanced pancreatic cancer. J Cancer. (2018) 9:2038–45. 10.7150/jca.23989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soda N, Rehm BHA, Sonar P, Nguyen NT, Shiddiky MJA. Advanced liquid biopsy technologies for circulating biomarker detection. J Mater Chem B. (2019) 7:6670–704 10.1039/C9TB01490J [DOI] [PubMed] [Google Scholar]
- 67.Zhou B, Xu K, Zheng X, Chen T, Wang J, Song Y, et al. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct Target Ther. (2020) 5:144. 10.1038/s41392-020-00258-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fahrmann JF, Bantis LE, Capello M, Scelo G, Dennison JB, Patel N, et al. A plasma-derived protein-metabolite multiplexed panel for early-stage pancreatic cancer. J Natl Cancer Inst. (2019) 111:372–9. 10.1093/jnci/djy126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Elazezy M, Joosse SA. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput Struct Biotechnol J. (2018) 16:370–8. 10.1016/j.csbj.2018.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neumann MHD, Bender S, Krahn T, Schlange T. ctDNA and CTCs in liquid biopsy - current status and where we need to progress. Comput Struct Biotechnol J. (2018) 16:190–5. 10.1016/j.csbj.2018.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shaw VE, Lane B, Jenkinson C, Cox T, Greenhalf W, Halloran CM, et al. Serum cytokine biomarker panels for discriminating pancreatic cancer from benign pancreatic disease. Mol Cancer. (2014) 13:114. 10.1186/1476-4598-13-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Amantini C, Morelli MB, Nabissi M, Piva F, Marinelli O, Maggi F, et al. Expression profiling of circulating tumor cells in pancreatic ductal adenocarcinoma patients: biomarkers predicting overall survival. Front Oncol. (2019) 9:874. 10.3389/fonc.2019.00874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Konczalla L, Wöstemeier A, Kemper M, Karstens KF, Izbicki J, Reeh M. Clinical significance of circulating tumor cells in gastrointestinal carcinomas. Diagnostics. (2020) 10:192. 10.3390/diagnostics10040192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Woo D, Yu M. Circulating tumor cells as ‘liquid biopsies' to understand cancer metastasis. Transl Res. (2018) 201:128–35. 10.1016/j.trsl.2018.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen L, Bode AM, Dong Z. Circulating tumor cells: moving biological insights into detection. Theranostics. (2017) 7:2606–19. 10.7150/thno.18588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krempley BD, Yu KH. Preclinical models of pancreatic ductal adenocarcinoma. Chinese Clin Oncol. (2017) 6:25. 10.21037/cco.2017.06.15 [DOI] [PubMed] [Google Scholar]
- 77.Nelson SR, Walsh N. Genetic alterations featuring biological models to tailor clinical management of pancreatic cancer patients. Cancers. (2020) 12:1233. 10.3390/cancers12051233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Frappart PO, Hofmann TG. Pancreatic ductal adenocarcinoma (PDAC) organoids: the shining light at the end of the tunnel for drug response prediction and personalized medicine. Cancers. (2020) 12:2750. 10.3390/cancers12102750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brychta N, Drosch M, Driemel C, Fischer JC, Neves RP, Esposito I, et al. Isolation of circulating tumor cells from pancreatic cancer by automated filtration. Oncotarget. (2017) 8:86143–56. 10.18632/oncotarget.21026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sergeant G, Roskams T, van Pelt J, Houtmeyers F, Aerts R, Topal B. Perioperative cancer cell dissemination detected with a real-time RT-PCR assay for EpCAM is not associated with worse prognosis in pancreatic ductal adenocarcinoma. BMC Cancer. (2011) 11:47. 10.1186/1471-2407-11-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ferreira MM, Ramani VC, Jeffrey SS. Circulating tumor cell technologies. Mol. Oncol. (2016) 10:374–94. 10.1016/j.molonc.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fabbri A, Cossa M, Sonzogni A, Papotti M, Righi L, Gatti G, et al. Ki-67 labeling index of neuroendocrine tumors of the lung has a high level of correspondence between biopsy samples and surgical specimens when strict counting guidelines are applied. Virchows Arch. (2017) 470:153–164. 10.1007/s00428-016-2062-2 [DOI] [PubMed] [Google Scholar]
- 83.Tesfaye M, Savoldo B. Adoptive Cell Therapy in Treating Pediatric Solid Tumors. Curr Oncol Rep. (2018) 20:73. 10.1007/s11912-018-0715-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Best MG, Sol N, Kooi I, Tannous J, Westerman BA, Rustenburg F, et al. RNA-seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell. (2015) 28:666–76. 10.1016/j.ccell.2015.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rice A, del Rio Hernandez A. The mutational landscape of pancreatic and liver cancers, as represented by circulating tumor DNA. Front Oncol. (2019) 9:952. 10.3389/fonc.2019.00952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Keller L, Belloum Y, Wikman H, Pantel K. Clinical relevance of blood-based ctDNA analysis: mutation detection and beyond. Br J Cancer. (2021) 124:345–58. 10.1038/s41416-020-01047-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lomberk G, Dusetti N, Iovanna J, Urrutia R. Emerging epigenomic landscapes of pancreatic cancer in the era of precision medicine. Nat Commun. (2019) 10:3875. 10.1038/s41467-019-11812-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang SS, Xu J, Ji KY, Hwang C. Il. Epigenetic alterations in pancreatic cancer metastasis. Biomolecules. (2021) 11:1–14. 10.3390/biom11081082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hasan N, Ahuja N. The emerging roles of ATP-dependent chromatin remodeling complexes in pancreatic cancer. Cancers. (2019) 11:1859. 10.3390/cancers11121859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tsuda M, Fukuda A, Kawai M, Araki O, Seno H. The role of the SWI/SNF chromatin remodeling complex in pancreatic ductal adenocarcinoma. Cancer Sci. (2021) 112:490–7. 10.1111/cas.14768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pagliaroli L, Trizzino M. The evolutionary conserved SWI/SNF subunits ARID1A and ARID1B are key modulators of pluripotency and cell-fate determination. Front Cell Dev Biol. (2021) 9:449. 10.3389/fcell.2021.643361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hung YH, Hsu MC, Chen LT, Hung WC, Pan MR. Alteration of epigenetic modifiers in pancreatic cancer and its clinical implication. J Clin Med. (2019) 8:903. 10.3390/jcm8060903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lan B, Zeng S, Grützmann R, Pilarsky C. The role of exosomes in pancreatic cancer. Int J Mol Sci. (2019) 20:4332. 10.3390/ijms20184332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heredia-Soto V, Rodríguez-Salas N, Feliu J. Liquid biopsy in pancreatic cancer: are we ready to apply it in the clinical practice? Cancers. (2021) 13:1986. 10.3390/cancers13081986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nagai M, Sho M, Akahori T, Nakagawa K, Nakamura K. Application of liquid biopsy for surgical management of pancreatic cancer. Ann Gastroenterol Surg. (2020) 4:216–23. 10.1002/ags3.12317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jiang J, Ye S, Xu Y, Chang L, Hu X, Ru G, et al. Circulating tumor DNA as a Potential marker to detect minimal residual disease and predict recurrence in pancreatic cancer. Front Oncol. (2020) 10:1220. 10.3389/fonc.2020.01220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Singhi AD, Koay EJ, Chari ST, Maitra A. Early detection of pancreatic cancer: opportunities and challenges. Gastroenterology. (2019) 156:2024–40. 10.1053/j.gastro.2019.01.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Waters AM, Der CJ. KRAS: the critical driver and therapeutic target for pancreatic cancer. Cold Spring Harb Perspect Med. (2018) 8:a031435. 10.1101/cshperspect.a031435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Buscail L, Bournet B, Cordelier P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat Rev Gastroenterol Hepatol. (2020) 17:153–68. 10.1038/s41575-019-0245-4 [DOI] [PubMed] [Google Scholar]
- 100.Guo S, Shi X, Shen J, Gao S, Wang H, Shen S, et al. Preoperative detection of KRAS G12D mutation in ctDNA is a powerful predictor for early recurrence of resectable PDAC patients. Br J Cancer. (2020) 122:857–67. 10.1038/s41416-019-0704-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Usui K, Yokoyama T, Naka G, Ishida H, Kishi K, Uemura K, et al. Plasma ctDNA monitoring during epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitor treatment in patients with EGFR-mutant non-small cell lung cancer (JP-CLEAR trial). Jpn J Clin Oncol. (2019) 49:554–8. 10.1093/jjco/hyz023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang J, Hui Y, Zhang Y, Zhang M, Ji B, Tian G, et al. Application of circulating tumor DNA as a biomarker for non-small cell lung cancer. Front Oncol. (2021) 11:725938. 10.3389/fonc.2021.725938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Adashek JJ, Janku F, Kurzrock R. Signed in blood: circulating tumor DNA in cancer diagnosis, treatment and screening. Cancers. (2021) 13:3600. 10.3390/cancers13143600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Calabuig-Fariñas S, Jantus-Lewintre E, Herreros-Pomares A, Camps C. Circulating tumor cells versus circulating tumor DNA in lung cancer-which one will win? Transl Lung Cancer Res. (2016) 5:466–82. 10.21037/tlcr.2016.10.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu L, Chen X, Petinrin OO, Zhang W, Rahaman S, Tang ZR, et al. Machine learning protocols in early cancer detection based on liquid biopsy: a survey. Life. (2021) 11:638. 10.3390/life11070638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Antunes-Ferreira M, Koppers-Lalic D, Würdinger T. Circulating platelets as liquid biopsy sources for cancer detection. Mol. Oncol. (2021) 15:1727–43. 10.1002/1878-0261.12859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hou J, Li X, Xie KP. Coupled liquid biopsy and bioinformatics for pancreatic cancer early detection and precision prognostication. Mol Cancer. (2021) 20:34. 10.1186/s12943-021-01309-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li Y, Al Hallak MN, Philip PA, Azmi AS, Mohammad RM. Non-coding rnas in pancreatic cancer diagnostics and therapy: focus on lncrnas, circrnas, and pirnas. Cancers. (2021) 13:4161. 10.3390/cancers13164161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Junqueira-Neto S, Batista IA, Costa JL, Melo SA. Liquid Biopsy beyond circulating tumor cells and cell-free DNA. Acta Cytol. (2019) 63:479–88. 10.1159/000493969 [DOI] [PubMed] [Google Scholar]
- 110.Guler GD, Ning Y, Ku CJ, Phillips T, McCarthy E, Ellison CK, et al. Detection of early stage pancreatic cancer using 5-hydroxymethylcytosine signatures in circulating cell free DNA. Nat Commun. (2020) 11:5270. 10.1038/s41467-020-18965-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Meng Y, Sun J, Zheng Y, Zhang G, Yu T, Piao H. Platelets: the emerging clinical diagnostics and therapy selection of cancer liquid biopsies. Onco Targets Ther. (2021) 14:3417–28. 10.2147/OTT.S311907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Khan IA, Rashid S, Singh N, Rashid S, Singh V, Gunjan D, et al. Panel of serum miRNAs as potential non-invasive biomarkers for pancreatic ductal adenocarcinoma. Sci Rep. (2021) 11:2824. 10.1038/s41598-021-82266-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang P, Zhuang L, Zhang J, Fan J, Luo J, Chen H, et al. The serum miR-21 level serves as a predictor for the chemosensitivity of advanced pancreatic cancer, and miR-21 expression confers chemoresistance by targeting FasL. Mol Oncol. (2013) 7:334–45. 10.1016/j.molonc.2012.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Carmicheal J, Patel A, Dalal V, Atri P, Dhaliwal AS, Wittel UA, et al. Elevating pancreatic cystic lesion stratification: current and future pancreatic cancer biomarker(s). Biochim Biophys Acta Rev Cancer. (2020) 1873:188318. 10.1016/j.bbcan.2019.188318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vicentini C, Calore F, Nigita G, Fadda P, Simbolo M, Sperandio N, et al. Exosomal miRNA signatures of pancreatic lesions. BMC Gastroenterol. (2020) 20:137. 10.1186/s12876-020-01287-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yang YF, Wang GY, He JL, Wu FP, Zhang YN. Overall survival of patients with KRAS wild-type tumor treated with FOLFOX/FORFIRI±cetuximab as the first-line treatment for metastatic colorectal cancer A meta-analysis. Medicine. (2017) 96:2–7. 10.1097/MD.0000000000006335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sethi V, Giri B, Saluja A, Dudeja V. Insights into the pathogenesis of pancreatic cystic neoplasms. Dig Dis Sci. (2017) 62:1778–86. 10.1007/s10620-017-4603-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zomer A, Vendrig T, Hopmans ES, van Eijndhoven M, Middeldorp JM, Pegtel DM. Exosomes: fit to deliver small RNA. Commun Integr Biol. (2010) 3:447–50. 10.4161/cib.3.5.12339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pegtel DM, van de Garde MDB, Middeldorp JM. Viral miRNAs exploiting the endosomal-exosomal pathway for intercellular cross-talk and immune evasion. Biochim Biophys Acta. (2011) 1809:715–21. 10.1016/j.bbagrm.2011.08.002 [DOI] [PubMed] [Google Scholar]
- 120.Han L, Lam EWF, Sun Y. Extracellular vesicles in the tumor microenvironment: old stories, but new tales. Mol Cancer. (2019) 18:59. 10.1186/s12943-019-0980-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dilsiz N. Role of exosomes and exosomal microRNAs in cancer. Futur Sci OA. (2020) 6:FSO465. 10.2144/fsoa-2019-0116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yáñez-Mó M, Siljander PR, Andreu Z, Zavec AB, Borràs FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. (2015) 4:27066. 10.3402/jev.v4.27066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Doyle LM, Wang MZ. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells. (2019) 8:727. 10.3390/cells8070727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Imaoka H, Sasaki M, Hashimoto Y, Watanabe K, Ikeda M. New era of endoscopic ultrasound-guided tissue acquisition: next-generation sequencing by endoscopic ultrasound-guided sampling for pancreatic cancer. J Clin Med. (2019) 8:1173. 10.3390/jcm8081173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Abhange K, Makler A, Wen Y, Ramnauth N, Mao W, Asghar W, et al. Small extracellular vesicles in cancer. Bioact Mater. (2021) 6:3705–43. 10.1016/j.bioactmat.2021.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang H, Ach RA, Curry B. Direct and sensitive miRNA profiling from low-input total RNA. RNA. (2007) 13:151–9. 10.1261/rna.234507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Giulietti M, Occhipinti G, Principato G, Piva F. Identification of candidate miRNA biomarkers for pancreatic ductal adenocarcinoma by weighted gene co-expression network analysis. Cell Oncol. (2017) 40:181–92. 10.1007/s13402-017-0315-y [DOI] [PubMed] [Google Scholar]
- 128.Ramamoorth M, Narvekar A. Non viral vectors in gene therapy- an overview. J Clin Diagn Res. (2015) 9:GE01–6. 10.7860/JCDR/2015/10443.5394 [DOI] [PMC free article] [PubMed] [Google Scholar]