Abstract
Purpose of Review
Loss of chromosome 7 has long been associated with adverse-risk myeloid malignancy. In the last decade, CUX1 has been identified as a critical tumor suppressor gene located within a commonly deleted segment of chromosome arm 7q. Additional genes encoded on 7q have also been identified as bona fide myeloid tumor suppressors, further implicating chromosome 7 deletions in disease pathogenesis. This review will discuss the clinical implications of del(7q) and CUX1 mutations, both in disease and clonal hematopoiesis, and synthesize recent literature on CUX1 and other chromosome 7 tumor suppressor genes.
Recent Findings
Two major studies, including a new mouse model, have been published that support a role for CUX1 inactivation in the development of myeloid neoplasms. Additional recent studies describe the cellular and hematopoietic effects from loss of the 7q genes LUC7L2 and KMT2C/MLL3, and the implications of chromosome 7 deletions in clonal hematopoiesis.
Summary
Mounting evidence supports CUX1 as being a key chromosome 7 tumor suppressor gene. As 7q encodes additional myeloid regulators and tumor suppressors, improved models of chromosome loss are needed to interrogate combinatorial loss of these critical 7q genes.
Keywords: CUX1, 7q, monosomy 7, contiguous gene syndrome, myeloid neoplasia
Introduction
Loss of all or part of chromosome 7 [−7/del(7q)] is among the most common chromosomal abnormalities in high-risk myeloid disease [1]. The high frequency of −7/del(7q) suggests chromosome 7 harbors tumor suppressor genes (TSGs) important to disease pathogenesis, and −7/del(7q) has therefore been the subject of intense investigation. However, a major challenge in the identification of candidate tumor suppressors is the lack of recurrent second-hit mutations on the remaining allele [2,3]. These observations suggest that chromosome 7 TSGs likely act in a haploinsufficient manner, whereby single-copy loss of a gene produces a mutant phenotype, in contrast to Knudson’s classical two-hit hypothesis of tumor suppressors [4].
In an alternative attempt to map candidate TSGs, minimally-deleted regions (MDR) have been identified at the cytogenetic bands 7q22, 7q34, and 7q35-36 by aligning commonly deleted segments of 7q [2,3]. In 2013, CUX1 was identified as one of the most significantly differentially expressed genes within the 7q22 MDR in −7/del(7q) leukemias, with ~50% expression compared to cases with both copies of CUX1 [5]. CUX1 is a non-clustered homeobox transcription factor, and knockdown of the ortholog of CUX1 in Drosophila melanogaster leads to myeloid cell hyperplasia [5]. In addition to CUX1, 7q contains multiple additional TSGs and myeloid regulators (Table 1). In this review we focus on recent findings regarding CUX1 and other 7q-encoded genes, including the splicing factor LUC7L2 and the histone lysine methyltransferase KMT2C/MLL3. We discuss chromosome 7 deletions in clonal hematopoiesis of indeterminate potential (CHIP), briefly review approaches to model del(7q), and endorse the concept of 7q as a contiguous gene syndrome (CGS) region in which combined loss of multiple dose-sensitive TSGs contributes to disease.
Table 1: 7q genes implicated in myeloid disease based on clinical and experimental data.
The cellular function, deletion phenotypes in cells and animal models, and clinical associations are provided with supporting references. Cell deletion phenotype is in hematopoietic cells unless otherwise specified.
| Gene Name (location) |
Cellular Function | Cellular Deletion Phenotype |
Hematopoietic Murine Deletion Phenotype |
CHIP gene? | Clinical Associations |
|---|---|---|---|---|---|
| SAMD9/SAMD9L (7q21.1) | Endosomal fusion protein, terminating surface receptor signaling [6] | Persistent cytokine receptor signaling and cytokine hypersensitivity [6] | Enhanced HSC colony-forming potential and in vivo reconstitution [6] | No | Germline activating mutations cause MIRAGE syndrome [8] |
| Regulates protein synthesis rate [7*] | Gain of function mutations interfere with ribosome assembly in K562 cells [7*] | Late MDS in germline +/− and −/− mice [6] | Mutant allele lost through monosomy 7 via adaptation by aneuploidy [8-10] | ||
| ACHE (7q22.1) (CDR) | Hydrolyzes acetylcholine; associated with stress hematopoiesis [11] | Enhanced proliferation and decreased apoptosis in mouse bone marrow cultures[12] Impaired erythroid differentiation in human CD34+ cells [13*] |
Increased neutrophil cell number in +/− mice [14] | No | None reported |
| CUX1 (7q22.1)(CDR) | Homeobox transcription factor [15] | Enhanced proliferation, activation of PI3K-AKT signaling [17] | Mild monocytosis in shCux1mid mice, increasing monocytosis and lethal anemia in shCux1low mice [17] | Yes [19-22*] | Significantly decreased expression in −7/del(7q) leukemias [5] |
| Recruited to sites of DNA double strand breaks [16*] | Decreased apoptosis in hematopoietic progenitors [18**] | Increased mean RBC volume in hematopoietic-specific +/− mice, monocytosis and anemia in −/− mice [18**] | Inactivating mutations found in MDS, AML, and MDS/MPN overlap syndromes [23-25] | ||
| Impaired DNA damage response [16*] | shCux1low and −/− mice develop MDS/MPN-like disease [17, 18**] | Inactivating mutations associated with poor survival in MDS [24,25] | |||
| shCux1low mice treated with alkylating agents develop a rapid, fatal t-MN [16*] | |||||
| RASA4 (7q22.1)(CDR) | RAS GTPase-activating protein [26] | Elevated ERK phosphorylation in macrophages after FcyR stimulation [26] | No overt disease; germline −/− mice have impaired macrophage phagocytosis [26] | No | Promoter hypermethylation in JMML [27] |
| KMT2E (7q22.3) | Epigenetic regulator, capable of binding H3K4 methylation [28] | Cell cycle arrest in lung fibroblasts and HCT116 cells [29] | No overt disease; germline −/− mice have impaired neutrophil maturation, decreased RBC and hematocrit [31-33] | No | High expression associated with favorable outcome in cytogenetically normal AML [34] |
| Reported catalytically inactive [28] | Increased ROS, impaired DNA damage response in +/− and −/− hematopoietic progenitors [30] | +/− mice have mild reduction in thymocytes and splenocyte, decreased RBC and hematocrit, increased RBC distribution width [14, 32] | |||
| DOCK4 (7q31.1) | GTPase activator [35,36] | RBC cytoskeletal defects, decreased erythroid colony formation in human CD34+ cells [35,36] | No reported hematopoietic phenotype in germline −/− mice [14,37] | No | Significantly decreased expression in MDS, associated with overall decreased survival in MDS [35,36] |
| MKLN1 (7q32.3) | Organization of F-actin networks [38,39] | Decreased retrograde intracellular transport in neurons [39] | No reported hematopoietic phenotype in germline +/− or −/− mice [39] | No | Associated with inherited predisposition for MPN [40] |
| Mutations observed in relapsed pediatric AML [41] | |||||
| TRIM24 (7q33) | Nuclear receptor co-regulator; RING-domain E3 ubiquitin ligase [42,43] | Increased proliferation in human CD34+ cells [13*] | Germline −/− mice develop hepatocellular carcinoma but have no hematopoietic phenotype [46] | No | High expression in AML reported, associated with poor survival [45] |
| Targets p53 for degradation [44] | Decreased proliferation in AML-193 and Kasumi-1 cells [45] | **Conflicting evidence for TRIM24 as hematopoietic oncogene or TSG** | |||
| HIPK2 (7q34)(CDR) | Serine/threonine nuclear kinase [47-49] | Decreased p53 activation and apoptosis in MCF-7 cells [47] | No reported hematopoietic phenotype in germline −/− mice [14, 51] | No | Low frequency missense mutations in MDS and AML [49] |
| Phosphorylates p53 to activate apoptosis [47] | Decreased erythroid expansion and differentiation in human CD34+ cells [50] | ||||
| Increased cisplatin resistance in RKO colon cancer and H1299 lung cancer cell lines [48] | |||||
| LUC7L2 (7q34)(CDR) | Splicing factor, co-localizes with U1 snRNP [52,53*,54*] | Altered splicing in K562 and HeLa cells [53*,54*] | No overt disease; Increased platelet volume in germline +/− mice [14] | Yes [19,20,22*] | Heterozygous inactivating mutations observed in MDS and AML [52,56] |
| Decreased expression of glycolysis genes; metabolic shift to OXPHOS in K562 and HeLa cells [53*,54*] | No reported hematopoietic phenotype in other germline −/− mice [55] | Decreased expression associated with reduced survival in MDS [52,56] | |||
| ATP6V0E2 (7q36.1)(CDR) | Intracellular proton pump [50] | Decrease erythroid expansion and differentiation in human CD34+ cells [50] | None reported | No | None reported |
| CUL1 (7q36.1)(CDR) | E3 ubiquitin ligase; transcriptional repressor [57-59*] | Increased transcription of c-MYC target genes in HeLa cells [59*] | Germline −/− is embryonic lethal [57] | No | Rare mutations observed in myeloid neoplasms [52] |
| Deletion in T-cell lineage yields T-cell lymphomas [58] | |||||
| EZH2 (7q36.1)(CDR) | Catalytic component of Polycomb Repressive Complex 2, places H3K27 methylation [60] | Decreased H3K27 methylation, partial compensation by EZH1 [61, 62] | Conditional knockout mice observed up to 30 weeks do not develop myeloid disease [63] | Yes [19,20,22*,65*,66**] | Mono-allelic and bi-allelic inactivating mutations observed in myeloid disease [3,67,68] |
| Transplant recipients of Ezh2−/− cells develop mixed disease, including T-cell lymphoma and very late MDS [62,64] | Inactivating mutations associated with poor prognosis in MDS and drug resistance in AML [69,70] | ||||
| KMT2C (7q36.1)(CDR) | Core component of COMPASS complex, places H3K4 methylation [71] | Decreased H3K4me1 at some enhancer regions; compensation by KMT2D [73**] | No overt disease; germline and hematopoietic-specific +/− and −/− mice have increased HSC number and self-renewal and splenomegaly [14,73**] | No | Mutations observed in AML, possibly over-represented due to pseudogene [75-77] |
| Recruited to sites of DNA double strand breaks [72*] | Decreased expression of DNA damage response genes after knockdown in HTB9 bladder cancer cells [74] | Mono-allelic knockout accelerates shNf1, p53−/− leukemogenesis [75] | Mutations observed in relapsed pediatric AML [41] | ||
| Selective advantage in +/− and −/− HSCs after chemotherapy [73**] |
+/−: heterozygous deletion
+/−: homozygous deletion
HSC: hematopoietic stem cell
RBC: red blood cell
MDS: myelodysplastic syndrome
MPN: myeloproliferative neoplasm
t-MN: therapy-related myeloid neoplasm
AML: acute myeloid leukemia
JMML: juvenile myelomonocytic leukemia
OXPHOS: oxidative phosphorylation
Clinical Features and Implications of −7/del(7q)
Chromosome 7 alterations in hematologic malignancies are almost always deletions or copy-neutral loss of heterozygosity (LOH), in contrast to solid tumors where amplifications are observed [78]. −7/del(7q) occurs in a wide range of myeloid diseases, including 5-10% of acute myeloid leukemias (AML) and adult myelodysplastic syndromes (MDS), 40% of pediatric MDS, 40% of myeloid neoplasms arising from cancer predisposition syndromes, and 50% of therapy-related myeloid neoplasms (t-MN) [9,79-81]. −7/del(7q) is associated with higher-risk MDS, faster time to transformation to AML, and poor overall survival in AML, and is therefore considered an adverse prognostic event [3,79]. Within these diseases, chromosome 7 deletions often co-occur with 5q deletions and gains of chromosome 8, but also frequently occur as isolated cytogenetic events [9,79-81]. Together, these findings strongly suggest a role for chromosome 7 deletions in disease pathogenesis.
A major unanswered question is to what degree −7/del(7q) influences disease initiation and pathogenesis. Analyses of clonal hierarchies in AML, t-MN, and CHIP suggest chromosome 7 alterations are early events [82-87]. In CHIP, seemingly healthy individuals with no history of hematologic malignancy harbor low-frequency alterations in genes associated with leukemia in their blood; these individuals are at an increased risk of development of a hematologic malignancy, but it remains unclear why only some individuals progress to disease [84]. Two recent studies by Gao et al. and Saiki et al. examined the combined landscape of somatic variants and copy number alterations in CHIP in large cohorts from Memorial Sloan Kettering and BioBank Japan, respectively [65*,66**]. Both studies identified chromosome 7 deletions and LOH at similar levels as previous CHIP studies [84-87]. Saiki et al. further found that individuals with del(7q) and 7q LOH had significantly increased risk for development of hematologic malignancy, particularly myeloid disease [66**]. The risks associated with chromosome 7 abnormalities were similar to those of 17p deletions or LOH, the chromosome arm encoding TP53, indicating that del(7q) is a biomarker for risk of disease progression that warrants close monitoring [66**].
There is also evidence that −7/del(7q) functions as a driver event in disease. In a study of pediatric MDS, 30% of patients with −7/del(7q) had no other detectable cytogenetic or molecular abnormalities in the coding region of the genome [9]. Though it is possible that non-coding changes were present but not detected, it is compelling evidence that −7/del(7q) alone may be sufficient to promote MDS. Spontaneous remission of monosomy 7 has also been observed in children with MDS, albeit rarely, with subsequent resolution of disease, further suggesting −7 is critical for enabling the disease state [88]. In addition to occurring alone, −7/del(7q) can coexist with other somatic and karyotypic alterations, most commonly complex karyotypes or RAS pathway mutations [75,89]. While RAS pathway mutations function as oncogenic drivers in a number of other cancer types, RAS pathway mutations typically arise late in AML development [79,90,91]. Additionally, RAS mutations are not typically observed in CHIP, and, in contrast to -7/del(7q), do not have prognostic impact in MDS and AML [65*,66**,92,93]. Therefore, even in the context of additional mutations, multiple lines of evidence point to −7/del(7q) as a driver of disease.
Cell extrinsic factors likely also influence cells with −7/del(7q). −7/del(7q) is found in up to 50% of t-MNs, second cancers arising after treatment for a primary malignancy, and is particularly associated with prior alkylating agent therapy [81]. −7/del(7q) is also found in hematopoietic cells of benzene-exposed workers as well as AML in elderly patients, which often resembles t-MNs [94,95]. These data suggest −7 may be selected for in the context of environmental exposures and aging, similar to PPM1D and TP53 mutations promoting fitness during chemotherapy [96,97]. Identifying the mechanism by which loss of chromosome 7 genes increases fitness in response to different environmental pressures remains an outstanding question.
Whether the effects of −7 and del(7q) are equivalent remains an open question. Monosomy 7 and del(7q) are often grouped together clinically despite differing mechanisms of occurrence: monosomy 7 results from a chromosome segregation failure, whereas del(7q) results from chromosomal breakage [98]. Some studies have assessed −7 separately from del(7q) and report better prognosis for del(7q) in AML and MDS [99, 100], though others have found no difference [101]. The concept that −7 is prognostically worse than del(7q) is perplexing as the majority of implicated chromosome 7 TSGs are located on 7q. Additionally, there is heterogeneity in the breakpoints for 7q deletions; whether different deletions spanning distinct genes carry unique prognostic implications remains unclear.
CUX1 Mutations in Myeloid Disease
CUX1, previously known as CUTL1 and CCAAT displacement protein (CDP), is a ubiquitously expressed, non-clustered homeobox transcription factor that is both evolutionarily and functionally conserved from Drosophila melanogaster to humans. This review will focus on the role of CUX1 mutations in myeloid disease; please see ref. [102] for the role of CUX1 in other models [102].
CUX1 is one of the few chromosome 7 genes that is recurrently mutated in cancer, with mutations identified in 2-4% of myeloid diseases including AML, MDS, and MDS/myeloproliferative neoplasms (MPN) [24,25]. CUX1 is also mutated in 1-5% of various solid tumors [25]. CUX1 mutation patterns fit a signature representative of TSGs, characterized by frameshift or nonsense alterations distributed throughout the coding frame [25,78,103] (Figure 1). Further, bi-allelic CUX1 mutations are rare, suggesting haploinsufficiency [24]. MDS and AML patients with inactivating CUX1 mutations have decreased survival compared to those with wild type CUX1, with overall survival mirroring patients with −7/del(7q) [25]. Our lab has shown that CUX1 knockdown in human CD34+ hematopoietic stem and progenitor cells results in a gene signature similar to patients with −7/del(7q) [17]. CUX1 mutations have also been identified in CHIP, indicating CUX1 inactivation can be an early event similar to −7/del(7q) [20,22]. Collectively, the clinical data strongly implicate CUX1 inactivation in myeloid disease development and support CUX1 being a critical 7q TSG.
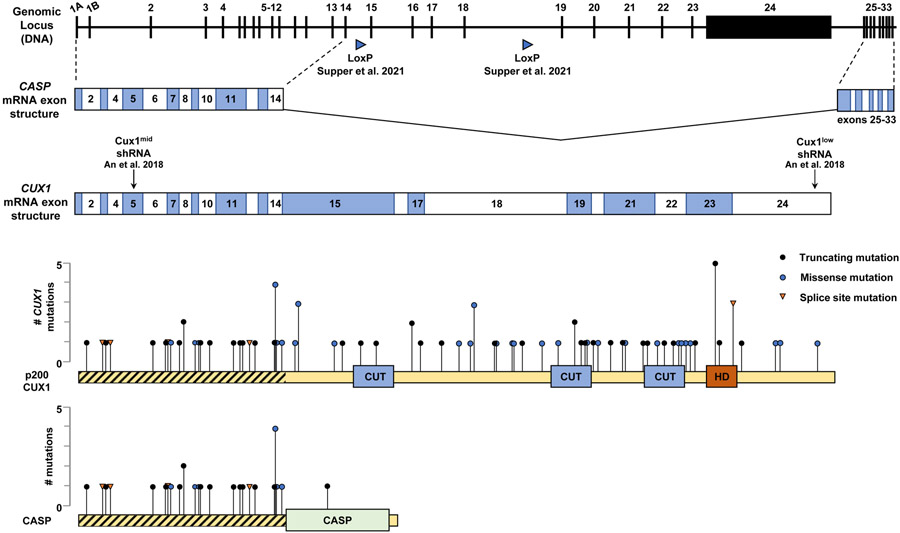
Figure 1. Structures of CASP and CUX1.
The genomic locus of CUX1 has two alternative start sites (exons 1A and 1B) and contains 33 exons which encode two gene products, CUX1 and CASP. The locus organization is conserved between humans and mice. CUX1 contains 24 exons; CASP is spliced from exons 1-14 and 25-33. The CUX1 NM_181552 mRNA exon structure is shown with Cux1mid and Cux1low shRNA targeting locations from ref. [17]; the LoxP Cre recombination sites from ref. [18**] are shown below the genomic locus. The p200 CUX1 protein is depicted below the exon structure with the 4 DNA binding domains depicted; exon length is drawn to scale to match the protein. Overlaid is a plot of CUX1 mutations from AACR Project GENIE disease classes ”Leukemia”, “Myelodysplastic Syndromes”, “Myeloproliferative Neoplasms”, and “Myelodysplastic/Myeloproliferative Neoplasms” [ref. 103]. The distribution of mutations fits a pattern representative of tumor suppressor genes [ref. 78]. A plot of CASP is shown below CUX1; there is only a single mutation within the CASP exons not shared with CUX1. Regions shared by CUX1 and CASP are hatched.
The cellular function of CUX1 and the role of CUX1 loss in myeloid malignancies is still under active exploration. Investigation of CUX1 is complicated by the complexity of the locus. The CUX1 gene is large, spanning 340 kilobases and 33 exons, with multiple RNA and protein isoforms [104]. Hematopoietic cells, however, only express the full length p200 CUX1 protein [105*]. The p200 isoform contains four DNA-binding domains, consisting of three CUT-repeat domains and one homeodomain (Figure 1) [106]. CUX1 is further complicated by being one of the few mammalian genes that shares exons with a second, independent gene, namely CASP (Cux1 Alternative Splice Product) [107]. Exons 15 to 24 are unique to CUX1 and contain the four DNA-binding domains (Figure 1). CASP does not have DNA-binding domains, nor is it located in the nucleus. Instead, CASP is a highly-expressed Golgi-associated protein, thought to be involved in vesicle transport [108]. Unfortunately, CASP and CUX1 isoforms are routinely aggregated in genomics datasets, such as RNA sequencing, making it a challenge to parse out independent roles of CUX1 and CASP. Likewise, unless antibodies are carefully vetted for reactivity to either CUX1, CASP, or both, investigators can be misled by subsequent results [105*].
Due in part to this complexity, and the requirement for Cux1 during development, establishment of traditional Cux1 knockout mice has been challenging [109]. To circumvent these issues, our lab developed inducible shRNA-based murine models for Cux1 knockdown, reducing CUX1 protein levels to 54% (Cux1mid) or 12% (Cux1low) in thymocytes [17]. The Cux1mid shRNA targets an exon shared by all Cux1 and Casp transcripts and approximates CUX1 haploinsufficiency, whereas the Cux1low model affects CUX1-encoding transcripts only (Figure 1). Ubiquitous shRNA expression in Cux1mid mice leads to a normocytic anemia and splenomegaly, while Cux1low mice develop an MDS/MPN-like disease with fatal anemia, supporting the notion that Cux1 is a dose-sensitive TSG [17]. These models further suggest the effects of mutations in shared exons can likely be attributed to CUX1 disruption and not CASP, as the disease caused by the Cux1low shRNA (which does not target Casp mRNA) is more severe than that in the Cux1mid mice (which does target Casp mRNA). Additionally, there are few reported mutations in exons unique to CASP, and there is currently no evidence CASP plays a role in human disease (Figure 1) [103,108] . Recently, Supper et al. reported a Cux1 knockout model in which exons 15-18 were excised in the hematopoietic compartment driven by Vav1-iCre [18**]. This approach, which avoids Casp isoforms, removes the first two DNA-binding domains and ablates protein expression in an allele-dependent manner in splenocytes. Similar to Cux1mid mice, Cux1+/− mice develop mild anemia and bone marrow dysplasia [17,18**]. This phenotype is exacerbated upon full Cux1 loss, with Cux1−/− mice developing an MDS/MPN-like disease, akin to Cux1low mice [17,18**]. The authors further show Cux1 loss cooperates with a Flt3ITD/+ mutation to accelerate disease, though it is worth noting that FLT3 mutations are not enriched in −7/del(7q) leukemias [18**,89]. Still, this second model provides compelling evidence for the pathogenesis of Cux1 loss in myeloid disease.
On a molecular level, CUX1 preferentially binds enhancer elements and acts as a transcriptional activator or repressor in a context-dependent manner [15,17,110]. Recently, our lab reported that CUX1 loss also impacts the epigenetic landscape of cells, both basally and in the context of irradiation-induced DNA damage [16*]. After irradiation, CUX1−/− cells show an impaired DNA damage response with decreased H3K27me2/3 and H3K9me2/3 at double-strand breaks, marks normally associated with DNA repair [16*,111,112]. These changes indicate a novel epigenetic, non-transcriptional role for CUX1. Further, Cux1-deficient cells continue to proliferate after alkylating agent exposure, ultimately leading to alkylator-induced t-MN in Cux1-deficient mice [16*]. Given the epidemiologic connection between −7/del(7q) t-MNs and alkylating agent chemotherapy, this study provides a missing mechanistic link between del(7q) and t-MN – ie. CUX1 is required for normal recognition and repair of chemotherapy-induced DNA damage [16*,113*]. Importantly, restoration of CUX1 levels post-genotoxic stress prevented transformation in this model, indicating that i) sustained CUX1-deficiency is required for t-MN maintenance, and ii) targeting putative negative regulators of CUX1 may be a therapeutic avenue for myeloid disease with CUX1 mutations or deletions [16*]. The cellular functions of CUX1 and consequences of CUX1 loss are summarized in Figure 2.
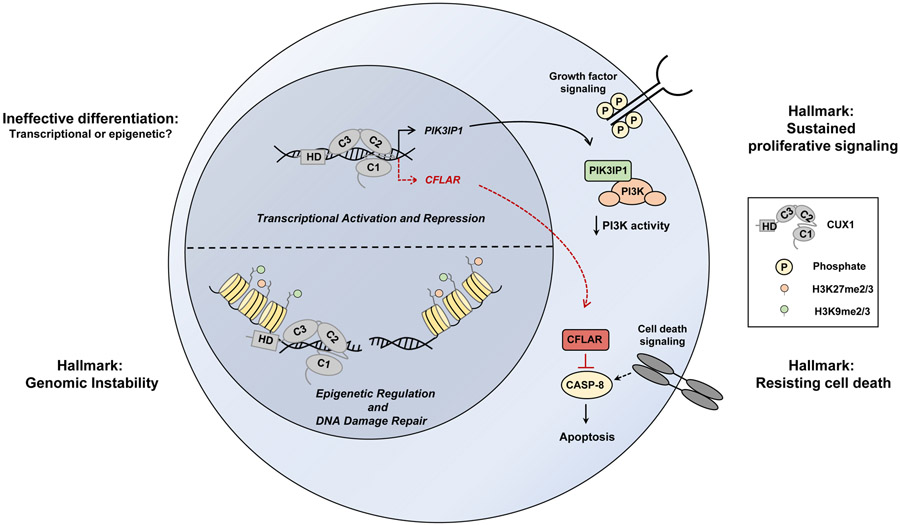
Figure 2. Cellular functions of CUX1 and consequences of CUX1 loss in hematopoietic cells.
CUX1 is involved in transcription, DNA damage repair, proliferation, and differentiation. One target gene of CUX1 is PIK3IP1, which inhibits PI3K activity [ref. 17, ref. 18**]. Loss of CUX1 results in decreased PIK3IP1 expression and increased PI3K-AKT signaling, promoting proliferation and resembling the “Sustained proliferative signaling” Hallmark of Cancer [ref. 114]. CUX1 also downregulates expression of CFLAR, an anti-apoptotic protein that inhibits caspase-8 [ref. 18**]. Loss of CUX1 results in alleviation of CFLAR repression and apoptosis resistance, promoting the hallmark “Resisting cell death” [ref. 114]. CUX1 also regulates epigenetic histone marks and functions in epigenetic-driven DNA repair; CUX1 loss results in sustained DNA damage, resembling the hallmark “Genomic instability” [ref. 114]. CUX1 loss also results in ineffective erythropoiesis and impaired differentiation, though the mechanism remains unknown [ref. 17, ref. 18**]. CUX1 is depicted alone on the DNA strand for simplicity.
7q as a Contiguous Gene Syndrome Region
A contiguous gene syndrome is a genetic disorder caused by large-scale chromosomal alterations affecting copy number, leading to dosage imbalance of multiple neighboring genes [115]. In addition to CUX1, multiple bona fide TSGs and myeloid regulators have been identified on 7q, many of which are also mutated in myeloid and solid tumors and yield hematopoietic phenotypes when deleted in mice (Table 1). We propose reframing chromosome 7 MDRs as CGS regions in cancer, similar to those observed on 5q and 8p [116,117]. Here we highlight the recent literature on the 7q-encoded genes EZH2, LUC7L2, and KMT2C/MLL3, and discuss potential interactions with combined CUX1 deficiency.
Similar to CUX1, the 7q genes EZH2 and, less commonly, LUC7L2, are also mutated in CHIP and are located in 7q MDRs [3,22]. Of note, EZH2 is among the only 7q genes observed to have recurrent bi-allelic inactivation in myeloid disease, suggesting a canonical tumor suppressive role for EZH2 in these diseases [3,4]. In the recent CHIP study from Gao et al., every event of chromosome 7 copy-neutral loss of heterozygosity co-localized with an EZH2 mutation, implicating this alteration was selected to eliminate the remaining wild-type EZH2 allele [65*]. EZH2 encodes the catalytic component of the Polycomb Repressive Complex 2, a major H3K27 methyltransferase complex, and loss of Ezh2 in murine hematopoietic stem cells results in myelodysplasia with late development of myelodysplastic disorders [62]. As inactivating mutations in EZH2 also carry a poor prognosis in MDS, there may be a compounding interaction upon combined loss of EZH2 and CUX1 in the context of del(7q), particularly as both proteins converge on the regulation of H3K27 methylation [16*,70].
LUC7L2 encodes a splicing factor, and inactivating LUC7L2 mutations have been identified in both MDS and AML [52,56]. Splicing factor mutations occur in over 50% of MDS cases but are challenging to characterize due to poor overlap of alternative splicing events [118]. Two new studies independently report an unexpected downregulation of glycolysis genes following LUC7L2 loss, with subsequent shifting of metabolism toward oxidative phosphorylation [53*,54*]. Both studies identify exon skipping as a mechanism of decreased gene expression, and link alternative splicing events to glucose metabolism, a novel mechanism not previously ascribed to splicing factor mutations [53*,54*,118]. Recent studies have also shed new light on the H3K4 methyltransferase KMT2C/MLL3. KMT2C mutations are not frequently detected in CHIP, though mutations are found in AML and Kmt2c haploinsufficiency enhances leukemogenesis [73*,75]. Chen et al. characterized two novel knockout models of Kmt2c and report increased self-renewal in hematopoietic stem cells and a selective advantage of Kmt2c mutant cells in the presence of chemotherapy, though the mice do not develop any overt malignancies [73**]. Chang et al. reported that, similar to CUX1, KMT2C is recruited to sites of DNA damage, and loss of KMT2C results in decreased expression of DNA damage response genes [72,74*]. Given CUX1 involvement in the DNA damage response, combined loss of CUX1 and KMT2C may synergize and further promote development of a t-MN [16*,73**]. Collectively, these findings indicate -7/del(7q) likely deregulates multiple cellular pathways involved in myeloid disease including cell signaling, energy metabolism, RNA splicing, DNA repair, and epigenetic regulation. Whether combinatorial loss of 7q genes acts in an additive or epistatic fashion remains an important, unanswered question.
Modeling del(7q)
Given the existence of multiple 7q TSGs, it is essential to innovate new models to interrogate combined gene deficiency. The lack of chromosomal synteny between humans and mice is a barrier to generating mouse models with large-scale deletions, and the variations in 7q deletion locations and length make determining boundaries challenging [119,120]. Alternative models include the use of induced pluripotent stem cells derived from del(7q) MDS patients, however these cells are difficult to culture and can undergo spontaneous dosage correction, restoring the missing chromosome 7 segment to the diploid state [50]. Recently, CRISPR-Cas9 has been used to simultaneously target multiple loci on different chromosomes to model CHIP [121,122]; multiplex CRISPR-Cas9-based gene deletion may therefore be a novel means to model del(7q) that circumvents the challenges of other approaches.
Conclusion
As efforts to define the role of −7/del(7q) continue, clinical evidence is mounting that chromosome 7 deletions and CUX1 mutations can be early, driving events. Emerging data indicate that certain pressures, such as genotoxic therapy, can select for CUX1-deficient clones, and this fitness advantage likely corresponds with the inherent drug resistance of malignancies arising from these clones. To understand the spectrum of environmental exposures that select for CUX1-deficient clones and to identify at-risk individuals, it is imperative that clinical and research CHIP sequencing panels probe for both CUX1 and del(7q) going forward. Finally, several 7q TSGs have functions that converge on similar pathways. While mechanistic studies of 7q genes have traditionally focused on individual genes, studies investigating combined gene deletions are warranted to refine our understanding of how −7/del(7q) drives malignancy.
Key Points.
Chromosome 7 alterations are early, driving events in myeloid disease pathogenesis.
CUX1 mutations also occur early in myeloid disease, and sustained CUX1 loss is necessary for disease maintenance.
Chromosome arm 7q encodes multiple myeloid TSGs and regulators, suggesting the existence of contiguous gene syndrome region(s).
Acknowledgements
The authors thank Angela Stoddart for helpful comments and careful reading of the manuscript. The authors acknowledge the American Association for Cancer Research (AACR) and its financial and material support in the development of the AACR Project Genomics Evidence Neoplasia Information Exchange (GENIE) registry, as well as members of the consortium for their commitment to data sharing.
Financial support and sponsorship
Megan E. McNerney is supported in part by NIH R01 HL142782, NIH R01 CA231880, American Cancer Society Research Scholar Award 132457-RSG-18-171-01-LIB, and NIH/NCI P30 CA014599. Matthew RM Jotte is supported by NIH T32 GM007281.
Abbreviations:
- TSG
tumor suppressor gene
- MDR
minimally-deleted region
- CHIP
clonal hematopoiesis of indeterminate potential
- CGS
contiguous gene syndrome
- LOH
loss of heterozygosity
- AML
acute myeloid leukemia
- MDS
myelodysplastic syndrome
- T-MN
therapy-related myeloid neoplasm
- MPN
myeloproliferative neoplasm
Footnotes
Conflicts of Interest: None declared.
References
- 1.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016; 127:2391–2405. [DOI] [PubMed] [Google Scholar]
- 2.Le Beau MM, Espinosa R, Davis EM, et al. Cytogenetic and molecular delineation of a region of chromosome 7 commonly deleted in malignant myeloid diseases. Blood 1996; 88:1930–1935. [PubMed] [Google Scholar]
- 3.Jerez A, Sugimoto Y, Makishima H, et al. Loss of heterozygosity in 7q myeloid disorders: clinical associations and genomic pathogenesis. Blood 2012; 119:6109–6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knudson AG. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A 1971; 68:820–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McNerney ME, Brown CD, Wang X, et al. CUX1 is a haploinsufficient tumor suppressor gene on chromosome 7 frequently inactivated in acute myeloid leukemia. Blood 2013; 121:975–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagamachi A, Matsui H, Asou H, et al. Haploinsufficiency of SAMD9L, an endosome fusion facilitator, causes myeloid malignancies in mice mimicking human diseases with monosomy 7. Cancer Cell 2013; 24:305–317. [DOI] [PubMed] [Google Scholar]
- 7. Thomas ME, Abdelhamed S, Hiltenbrand R, et al. Pediatric MDS and bone marrow failure-associated germline mutations in SAMD9 and SAMD9L impair multiple pathways in primary hematopoietic cells. Leukemia 2021; 35:3232–3244.33731850 * This study links alterations in ribosome formation and protein synthesis rates to gain-of-function SAMD9/SAMD9L mutations.
- 8.Narumi S, Amano N, Ishii T, et al. SAMD9 mutations cause a novel multisystem disorder, MIRAGE syndrome, and are associated with loss of chromosome 7. Nat Genet 2016; 48:792–797. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz JR, Ma J, Lamprecht T, et al. The genomic landscape of pediatric myelodysplastic syndromes. Nat Commun. 2017; 8:1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong JC, Bryant V, Lamprecht T, et al. Germline SAMD9 and SAMD9L mutations are associated with extensive genetic evolution and diverse hematologic outcomes. JCI Insight 2018; 3:e121086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pick M, Perry C, Lapidot T, et al. Stress-induced cholinergic signaling promotes inflammation-associated thrombopoiesis. Blood 2006; 107:3397–3406. [DOI] [PubMed] [Google Scholar]
- 12.Soreq H, Patinkin D, Lev-Lehman E, et al. Antisense oligonucleotide inhibition of acetylcholinesterase gene expression induces progenitor cell expansion and suppresses hematopoietic apoptosis ex vivo. Proc Natl Acad Sci U S A 1994; 91:7907–7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baeten JT, Liu W, Preddy IC, et al. CRISPR screening in human hematopoietic stem and progenitor cells reveals an enrichment for tumor suppressor genes within chromosome 7 commonly deleted regions. Leukemia 2021, under revision. * This study screens chromosome 7 genes for regulators of proliferation and erythroid differentiation.
- 14.Meehan TF, Conte N, West DB, et al. Disease model discovery from 3,328 gene knockouts by The International Mouse Phenotyping Consortium. Nat Genet 2017; 49:1231–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arthur RK, An N, Khan S, McNerney ME. The haploinsufficient tumor suppressor, CUX1, acts as an analog transcriptional regulator that controls target genes through distal enhancers that loop to target promoters. Nucleic Acids Research 2017; 45:6350–6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Imgruet MK, Lutze J, An N, et al. Loss of a 7q gene, CUX1, disrupts epigenetic-driven DNA repair and drives therapy-related myeloid neoplasms. Blood 2021; 138:790–805.34473231 * The authors report CUX1 involvement in regulation of epigenetic histone marks, and provide a mechanistic link between 7q gene loss and development of alkylating agent-induced t-MN.
- 17.An N, Khan S, Imgruet MK, et al. Gene dosage effect of CUX1 in a murine model disrupts HSC homeostasis and controls the severity and mortality of MDS. Blood 2018; 131:2682–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Supper E, Rudat S, Iyer V, et al. Cut-like homeobox 1 (CUX1) tumor suppressor gene haploinsufficiency induces apoptosis evasion to sustain myeloid leukemia. Nat Commun 2021; 12:2482.33931647 ** The authors generate a novel hematopoietic-specific knockout of Cux1 with similar phenotypes as ref. [17], supporting a role for CUX1 in activation in myeloid disease. They also report cooperation between Cux1 loss and Flt3ITD mutations.
- 19.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014; 371:2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshizato T, Dumitriu B, Hosokawa K, et al. Somatic mutations and clonal hematopoiesis in aplastic anemia. N Engl J Med 2015; 373:35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zink F, Stacey SN, Norddahl GL, et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood 2017; 130:742–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Robertson NA, Latorre-Crespo E, Terradas-Terradas M, et al. Longitudinal dynamics of clonal hematopoiesis identifies gene-specific fitness effects. BioRvix 2021. 10.1101/2021.05.27.446006. * This BioRxiv preprint reports that CHIP mutations in CUX1, LUC7L2, and EZH2 confer a selective fitness advantage.
- 23.Lindsley RC, Saber W, Mar BG, et al. Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N Engl J Med 2017; 37:536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aly M, Ramdzan ZM, Nagata Y, et al. Distinct clinical and biological implications of CUX1 in myeloid neoplasms. Blood Adv 2019; 3:2164–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong CC, Martincorena I, Rust AG, et al. Inactivating CUX1 mutations promote tumorigenesis. Nat Genet 2014; 46:33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Guo J, Dzhagalov I, He Y-W. An essential function for the calcium-promoted Ras inactivator in Fcgamma receptor-mediated phagocytosis. Nat Immunol 2005; 6:911–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poetsch AR, Lipka DB, Witte T, et al. RASA4 undergoes DNA hypermethylation in resistant juvenile myelomonocytic leukemia. Epigenetics 2014; 9:1252–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mas-y-Mas S, Barbon M, Teyssier C, et al. The human mixed lineage leukemia 5 (MLL5), a sequentially and structurally divergent SET domain-containing protein with no intrinsic catalytic activity. PLoS ONE 2016; 11:e0165139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng F, Liu J, Zhou SH, et al. RNA interference against mixed lineage leukemia 5 resulted in cell cycle arrest. Int J Biochem Cell Biol 2008; 40:2472–2481. [DOI] [PubMed] [Google Scholar]
- 30.Tasdogan A, Kumar S, Allies G, et al. DNA damage-induced HSPC malfunction depends on ROS accumulation downstream of IFN-1 signaling and Bid mobilization. Cell Stem Cell 2016; 19:752–767. [DOI] [PubMed] [Google Scholar]
- 31.Heuser M, Yap DB, Leung M, et al. Loss of Mll5 results in pleiotropic hematopoietic defects, reduced neutrophil immune function, and extreme sensitivity to DNA demethylation. Blood 2009; 113:1432–1443. [DOI] [PubMed] [Google Scholar]
- 32.Madan V, Madan B, Brykczynska U, et al. Impaired function of primitive hematopoietic cells in mice lacking the Mixed-Lineage-Leukemia homolog Mll5. Blood 2009; 113:1444–1454. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Wong J, Klinger M, Tran MT, et al. MLL5 Contributes to hematopoietic stem cell fitness and homeostasis. Blood 2009; 113:1455–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Damm F, Oberacker T, Thol F, et al. Prognostic importance of histone methyltransferase MLL5 expression in acute myeloid leukemia. J Clin Oncol 2011; 29:682–689. [DOI] [PubMed] [Google Scholar]
- 35.Zhou L, Opalinska J, Sohal D, et al. Aberrant epigenetic and genetic marks are seen in myelodysplastic leukocytes and reveal Dock4 as a candidate pathogenic gene on chromosome 7q. J Biol Chem 2011; 286:25211–25223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sundaravel S, Duggan R, Bhagat T, et al. Reduced DOCK4 expression leads to erythroid dysplasia in myelodysplastic syndromes. Proc Natl Acad Sci U S A 2015; 112:E6359–E6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo D, Peng Y, Wang L, et al. Autism-like social deficit generated by Dock4 deficiency is rescued by restoration of Rac1 activity and NMDA receptor function. Mol Psychiatry 2021; 26:1505–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valiyaveettil M, Bentley AA, Gursahaney P, et al. Novel role of the muskelin-RanBP9 complex as a nucleocytoplasmic mediator of cell morphology regulation. J Cell Biol 2008; 182:727–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heisler FF, Loebrich S, Pechmann Y, et al. Muskelin regulates actin filament- and microtubule-based GABA(A) receptor transport in neurons. Neuron 2011;70:66–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bao EL, Nandakumar SK, Liao X, et al. Inherited myeloproliferative neoplasm risk affects haematopoietic stem cells. Nature 2020; 586:769–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McNeer NA, Philip J, Geiger H, et al. Genetic mechanisms of primary chemotherapy resistance in pediatric acute myeloid leukemia. Leukemia 2019; 33:1934–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teyssier C, Ou CY, Khetchoumian K, et al. Transcriptional intermediary factor 1α mediates physical interaction and functional synergy between the coactivator-associated arginine methyltransferase 1 and glucocorticoid receptor-interacting protein 1 nuclear receptor coactivators. Molecular Endocrinology 2006; 20:1276–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tisserand J, Khetchoumian K, Thibault C, et al. Tripartite motif 24 (Trim24/Tif1α) tumor suppressor protein is a novel negative regulator of interferon (IFN)/signal transducers and activators of transcription (STAT) signaling pathway acting through retinoic acid receptor α (Rarα) inhibition. J Biol Chem 2011; 286:33369–33379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allton K, Jain AK, Herz HM, et al. Trim24 targets endogenous p53 for degradation. Proc Natl Acad Sci U S A 2009; 106:11612–11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li C, Xin H, Shi Y, Mu J. Knockdown of TRIM24 suppresses growth and induces apoptosis in acute myeloid leukemia through downregulation of Wnt/GSK-3β/β-catenin signaling. Hum Exp Toxicol 2020; 39:1725–1736. [DOI] [PubMed] [Google Scholar]
- 46.Khetchoumian K, Teletin M, Tisserand J, et al. Loss of Trim24 (Tif1alpha) gene function confers oncogenic activity to retinoic acid receptor alpha. Nat Genet 2007; 39:1500–1506. [DOI] [PubMed] [Google Scholar]
- 47.D’Orazi G, Cecchinelli B, Bruno T, et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol 2002; 4:11–19. [DOI] [PubMed] [Google Scholar]
- 48.Di Stefano V, Rinaldo C, Sacchi A, et al. Homeodomain-interacting protein kinase-2 activity and p53 phosphorylation are critical events for cisplatin-mediated apoptosis. Exp Cell Res 2004; 293:311–320. [DOI] [PubMed] [Google Scholar]
- 49.Li X-L, Arai Y, Harada H, et al. Mutations of the HIPK2 gene in acute myeloid leukemia and myelodysplastic syndrome impair AML1- and p53-mediated transcription. Oncogene 2007; 26:7231–7239. [DOI] [PubMed] [Google Scholar]
- 50.Kotini AG, Chang C-J, Boussaad I, et al. Functional analysis of a chromosomal deletion associated with myelodysplastic syndromes using isogenic human induced pluripotent stem cells. Nat Biotechnol 2015; 33:646–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sjölund J, Pelorosso FG, Quigley DA, DelRosario R, Balmain A. Identification of Hipk2 as an essential regulator of white fat development. Proc Natl Acad Sci U S A 2014; 111:7373–7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hosono N, Makishima H, Jerez A, et al. Recurrent genetic defects on chromosome 7q in myeloid neoplasms. Leukemia 2014; 28:1348–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Daniels NJ, Hershberger CE, Gu X, et al. Functional analyses of human LUC7-like proteins involved in splicing regulation and myeloid neoplasms. Cell Rep 2021; 35:108989.33852859 * This study identifies a decrease in expression of glycolysis genes after knockdown of the 7q gene LUC7L2, due to altered splicing events.
- 54. Jourdain AA, Begg BE, Mick E, et al. Loss of LUC7L2 and U1 snRNP subunits shifts energy metabolism from glycolysis to OXPHOS. Mol Cell 2021; 81:1905–1919.e12.33852893 * This study also identifies a decrease in expression of glycolysis genes following LUC7L2 loss, with a subsequent increase in oxidative phosphorylation.
- 55.Li C, Feng L, Luo W-W, et al. The RNA-binding protein LUC7L2 mediates MITA/STING intron retention to negatively regulate innate antiviral response. Cell Discov 2021; 7(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh H, Lane AA, Correll M, et al. Putative RNA-splicing gene LUC7L2 on 7q34 represents a candidate gene in pathogenesis of myeloid malignancies. Blood Cancer J 2013; 3:e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dealy MJ, Nguyen KV, Lo J, et al. Loss of Cul1 results in early embryonic lethality and dysregulation of cyclin E. Nat Genet 1999; 23:245–248. [DOI] [PubMed] [Google Scholar]
- 58.Piva R, Liu J, Chiarle R, Podda A, Pagano M, Inghirami G. In vivo interference with Skp1 function leads to genetic instability and neoplastic transformation. Mol Cell Biol 2002; 22:8375–8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sweeney MA, Iakova P, Maneix L, et al. The ubiquitin ligase Cullin-1 associates with chromatin and regulates transcription of specific c-MYC target genes. Sci Rep 2020; 10:13942.32811853 * This report describes a novel function for the 7q gene CUL1, an E3 ubiquitin ligase, in regulation of c-MYC target genes.
- 60.Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002; 298:1039–1043. [DOI] [PubMed] [Google Scholar]
- 61.Neff T, Sinha AU, Kluk MJ, et al. Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proc Natl Acad Sci U S A 2012; 109:5028–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mochizuki-Kashio M, Aoyama K, Sashida G, et al. Ezh2 loss in hematopoietic stem cells predisposes mice to develop heterogeneous malignancies in an Ezh1-dependent manner. Blood 2015; 126:1172–1183. [DOI] [PubMed] [Google Scholar]
- 63.Shimizu T, Kubovcakova L, Nienhold R, et al. Loss of Ezh2 synergizes with JAK2-V617F in initiating myeloproliferative neoplasms and promoting myelofibrosis. J Exp Med 2016; 213:1479–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sashida G, Harada H, Matsui H, et al. Ezh2 loss promotes development of myelodysplastic syndrome but attenuates its predisposition to leukaemic transformation. Nat Commun 2014; 5:4177. [DOI] [PubMed] [Google Scholar]
- 65. Gao T, Ptashkin R, Bolton KL, et al. Interplay between chromosomal alterations and gene mutations shapes the evolutionary trajectory of clonal hematopoiesis. Nat Commun 2021; 12:338.33436578 * This study examines both somatic mutations and copy number alterations in CHIP.
- 66. Saiki R, Momozawa Y, Nannya Y, et al. Combined landscape of single-nucleotide variants and copy number alterations in clonal hematopoiesis. Nat Med 2021; 27:1239–1249.34239136 ** This is the first study to report increased risk of developing myeloid disease for individuals with del(7q) CHIP.
- 67.Ernst T, Chase AJ, Score J, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet 2010; 42:722–726. [DOI] [PubMed] [Google Scholar]
- 68.Nikoloski G, Langemeijer SMC, Kuiper RP, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet 2010; 42:665–667. [DOI] [PubMed] [Google Scholar]
- 69.Göllner S, Oellerich T, Agrawal-Singh S, et al. Loss of the histone methyltransferase EZH2 induces resistance to multiple drugs in acute myeloid leukemia. Nat Med 2017; 23:69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sakhdari A, Class C, Montalban-Bravo G, et al. Loss of EZH2 protein expression in myelodysplastic syndrome correlates with EZH2 mutation and portends a worse outcome. Blood 2019; 134:3016–3016. [Google Scholar]
- 71.Miller T, Krogan NJ, Dover J, et al. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci U S A 2001; 98:12902–12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chang A, Liu L, Ashby JM, et al. Recruitment of KMT2C/MLL3 to DNA damage sites mediates DNA damage responses and regulates PARP inhibitor sensitivity in cancer. Cancer Res 2021; 81:3358–3373.33853832 * The authors demonstrate a role for the 7q gene KMT2C/MLL3 in the DNA damage response.
- 73. Chen R, Okeyo-Owuor T, Patel RM, et al. Kmt2c mutations enhance HSC self-renewal capacity and convey a selective advantage after chemotherapy. Cell Rep 2021; 34:108751.33596429 ** This study reports that heterozygous mutations in the 7q gene Kmt2c confer a selective advantage to hematopoietic stem cells after chemotherapy.
- 74.Rampias T, Karagiannis D, Avgeris M, et al. The lysine-specific methyltransferase KMT2C/MLL3 regulates DNA repair components in cancer. EMBO Rep 2019; 20:e46821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen C, Liu Y, Rappaport AR, et al. MLL3 Is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell 2014; 25:652–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arcipowski KM, Bulic M, Gurbuxani S, Licht JD. Loss of Mll3 catalytic function promotes aberrant myelopoiesis. PLoS One 2016; 11:e0162515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bowler TG, Pradhan K, Kong Y, et al. Misidentification of MLL3 and other mutations in cancer due to highly homologous genomic regions. Leukemia & Lymphoma 2019; 60:3132–3137. [DOI] [PubMed] [Google Scholar]
- 78.Davoli T, Xu AW, Mengwasser KE, et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 2013; 155:948–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 2016; 374:2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Haase D Cytogenetic features in myelodysplastic syndromes. Ann Hematol 2008; 87:515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smith SM, Le Beau MM, Huo D, et al. Clinical-cytogenetic associations in 306 patients with therapy-related myelodysplasia and myeloid leukemia: the University of Chicago series. Blood 2003; 102:43–52. [DOI] [PubMed] [Google Scholar]
- 82.Dimitriou M, Woll PS, Mortera-Blanco T, et al. Perturbed hematopoietic stem and progenitor cell hierarchy in myelodysplastic syndromes patients with monosomy 7 as the sole cytogenetic abnormality. Oncotarget 2016; 7:72685–72698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takahashi K, Wang F, Kantarjian H, et al. Copy number alterations detected as clonal hematopoiesis of indeterminate potential. Blood Adv 2017; 1:1031–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jacobs KB, Yeager M, Zhou W, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet 2012; 44:651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Laurie CC, Laurie CA, Rice K, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet 2012; 44:642–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Machiela MJ, Zhou W, Sampson JN, et al. Characterization of large structural genetic mosaicism in human autosomes. Am J Hum Genet 2015; 96:487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Loh P-R, Genovese G, Handsaker RE, et al. Insights into clonal haematopoiesis from 8,342 mosaic chromosomal alterations. Nature 2018; 559:350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mantadakis E, Shannon KM, Singer DA, et al. Transient monosomy 7: a case series in children and review of the literature. Cancer 1999; 85:2655–2661. [DOI] [PubMed] [Google Scholar]
- 89.McNerney ME, Brown CD, Peterson AL, et al. The spectrum of somatic mutations in high-risk acute myeloid leukaemia with −7/del(7q). Br J Haematol 2014; 166:550–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Almoguera C, Shibata D, Forrester K, et al. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell 1988; 53:549–554. [DOI] [PubMed] [Google Scholar]
- 91.Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med 1988; 319:525–532. [DOI] [PubMed] [Google Scholar]
- 92.Neubauer A, Dodge RK, George SL, et al. Prognostic importance of mutations in the ras proto-oncogenes in de novo acute myeloid leukemia. Blood 1994; 83:1603–1611. [PubMed] [Google Scholar]
- 93.Al-Kali A, Quintás-Cardama A, Luthra R, et al. Prognostic impact of RAS mutations in patients with myelodysplastic syndrome. Am J Hematol 2013; 88:365–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Appelbaum FR, Gundacker H, Head DR, et al. Age and acute myeloid leukemia. Blood 2006; 107:3481–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang L, Lan Q, Ji Z, et al. Leukemia-related chromosomal loss detected in hematopoietic progenitor cells of benzene-exposed workers. Leukemia 2012; 26:2494–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hsu JI, Dayaram T, Tovy A, et al. PPM1D mutations drive clonal hematopoiesis in response to cytotoxic chemotherapy. Cell Stem Cell 2018; 23:700–713.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wong TN, Miller CA, Jotte MRM, et al. Cellular stressors contribute to the expansion of hematopoietic clones of varying leukemic potential. Nat Commun 2018; 9:455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Griffiths AJF, Gelbart WM, Miller JH, Lewontin RC. Chapter 8: Chromosome Mutations. In: Modern Genetic Analysis. 1st edition. New York: W. H. Freeman; 1999. [Google Scholar]
- 99.Cordoba I, González-Porras JR, Nomdedeu B, et al. Better prognosis for patients with del(7q) than for patients with monosomy 7 in myelodysplastic syndrome. Cancer 2012; 118:127–133. [DOI] [PubMed] [Google Scholar]
- 100.Hasle H, Alonzo TA, Auvrignon A, et al. Monosomy 7 and deletion 7q in children and adolescents with acute myeloid leukemia: an international retrospective study. Blood 2007; 109:4641–4647. [DOI] [PubMed] [Google Scholar]
- 101.Hussain FTN, Nguyen EP, Raza S, et al. Sole abnormalities of chromosome 7 in myeloid malignancies: spectrum, histopathologic correlates, and prognostic implications. Am J Hematol 2012; 87:684–686. [DOI] [PubMed] [Google Scholar]
- 102.Sansregret L, Nepveu A. The multiple roles of CUX1: Insights from mouse models and cell-based assays. Gene 2008; 412:84–94. [DOI] [PubMed] [Google Scholar]
- 103.AACR Project GENIE Consortium. AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov 2017; 7:818–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rong Zeng W, Soucie E, Sung Moon N, et al. Exon/intron structure and alternative transcripts of the CUTL1 gene. Gene 2000; 241:75–85. [DOI] [PubMed] [Google Scholar]
- 105. Krishnan M, Senagolage MD, Baeten JT, et al. Genomic studies controvert the existence of the CUX1 p75 isoform. Sci Reports 2021; under revision. * This study identifies only the p200 CUX1 isoform in hematopoietic cells.
- 106.Aufiero B, Neufeld EJ, Orkin SH. Sequence-specific DNA binding of individual cut repeats of the human CCAAT displacement/cut homeodomain protein. Proc Natl Acad Sci U S A 1994; 91:7757–7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bürglin TR, Cassata G. Loss and gain of domains during evolution of cut superclass homeobox genes. Int J Dev Biol 2002; 46:115–123. [PubMed] [Google Scholar]
- 108.Gillingham AK, Pfeifer AC, Munro S. CASP, the alternatively spliced product of the gene encoding the CCAAT-displacement protein transcription factor, is a Golgi membrane protein related to giantin. Mol Biol Cell 2002; 13:3761–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tufarelli C, Fujiwara Y, Zappulla DC, Neufeld EJ. Hair defects and pup loss in mice with targeted deletion of the first cut repeat domain of the Cux/CDP homeoprotein gene. Dev Biol 1998; 200:69–81. [DOI] [PubMed] [Google Scholar]
- 110.Lievens PM, Donady JJ, Tufarelli C, Neufeld EJ. Repressor activity of CCAAT displacement protein in HL-60 myeloid leukemia cells. J Biol Chem 1995; 270:12745–12750. [DOI] [PubMed] [Google Scholar]
- 111.Zhang Y, Chang J-F, Sun J, et al. Histone H3K27 methylation modulates the dynamics of FANCD2 on chromatin to facilitate NHEJ and genome stability. J Cell Sci 2018; 131:jcs215525. [DOI] [PubMed] [Google Scholar]
- 112.Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, et al. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc Natl Acad Sci U S A 2014; 111:9169–9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Bolton KL, Ptashkin RN, Gao T, et al. Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat Genet 2020; 52:1219–1226.33106634 * This report adds to existing evidence that cancer therapies can select for CHIP clones that may develop into t-MNs.
- 114.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646–674. [DOI] [PubMed] [Google Scholar]
- 115.Schmickel RD. Contiguous gene syndromes: a component of recognizable syndromes. J Pediatr. 1986; 109:231–241. [DOI] [PubMed] [Google Scholar]
- 116.Stoddart A, Wang J, Fernald AA, et al. Cell intrinsic and extrinsic factors synergize in mice with haploinsufficiency for Tp53, and two human del(5q) genes, Egr1 and Apc. Blood 2014; 123:228–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xue W, Kitzing T, Roessler S, et al. A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc Natl Acad Sci U S A 2012; 109:8212–8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Qiu J, Zhou B, Thol F, et al. Distinct splicing signatures affect converged pathways in myelodysplastic syndrome patients carrying mutations in different splicing regulators. RNA 2016; 22:1535–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wong JCY, Zhang Y, Lieuw KH, et al. Use of chromosome engineering to model a segmental deletion of chromosome band 7q22 found in myeloid malignancies. Blood 2010; 115:4524–4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wong JC, Weinfurtner KM, Alzamora M del pilar, et al. Functional evidence implicating chromosome 7q22 haploinsufficiency in myelodysplastic syndrome pathogenesis. eLife 2015; 4:e07839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tothova Z, Krill-Burger JM, Popova KD, et al. Multiplex CRISPR/Cas9-based genome editing in human hematopoietic stem cells models clonal hematopoiesis and myeloid neoplasia. Cell Stem Cell 2017; 21(4):547–555.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Labuhn M, Perkins K, Matzk S, et al. Mechanisms of progression of myeloid preleukemia to transformed myeloid leukemia in children with down syndrome. Cancer Cell 2019; 36:123–138.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]